{
"cells": [
{
"cell_type": "markdown",
"id": "31fb4635",
"metadata": {},
"source": [
"# How can I process tabular data files in Python?\n",
"\n",
"Words are useful, but what's more useful are the sentences and stories we build with them.\n",
"Similarly, while a lot of powerful, general tools are built into Python,\n",
"specialized tools built up from these basic units live in\n",
"libraries\n",
"that can be called upon when needed.\n",
"\n",
"Now that we have looked at some basic concepts like lists and loops, lets look at\n",
"digging into some of the data we have been provided!\n",
"\n",
"## Downloading our data\n",
"\n",
"In Colab, you can access the terminal of the remote machine by using `!` in front of Linux\n",
"bash commands. This means you can use the Linux command `wget` to download files from the internet.\n",
"\n",
"**Note: the file storage space on the remote machine you are using in Google Colab is not persistent:\n",
"the files and folders you upload/save will not still be there when you next log in. Please download\n",
"your work if you want to save it.**\n",
"\n",
"You will need to repeat these steps in any new notebook you use where you want to access the data.\n",
"\n",
"### Download files"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "900eb5ef",
"metadata": {},
"outputs": [],
"source": [
"# Download 2 files and store in the swc-python folder\n",
"!wget -P swc-python https://swcarpentry.github.io/python-novice-inflammation/data/python-novice-inflammation-data.zip \n",
"!wget -P swc-python https://swcarpentry.github.io/python-novice-inflammation/files/code/python-novice-inflammation-code.zip"
]
},
{
"cell_type": "markdown",
"id": "71aab84a",
"metadata": {},
"source": [
"### Unzip files"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "4a5a2bd9",
"metadata": {},
"outputs": [],
"source": [
"# Extract .zip files inside the folder swc-python/\n",
"!unzip /content/swc-python/python-novice-inflammation-code.zip -d /content/swc-python/\n",
"!unzip /content/swc-python/python-novice-inflammation-data.zip -d /content/swc-python/"
]
},
{
"cell_type": "markdown",
"id": "6e4ac452",
"metadata": {},
"source": [
"## Loading data into Python\n",
"\n",
"To begin processing the clinical trial inflammation data, we need to load it into Python.\n",
"We can do that using a library called\n",
"[NumPy](https://numpy.org/doc/stable \"NumPy Documentation\"), which stands for Numerical Python.\n",
"In general, you should use this library when you want to do fancy things with lots of numbers,\n",
"especially if you have matrices or arrays. To tell Python that we'd like to start using NumPy,\n",
"we need to [import](../learners/reference.md#import) it:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "d2f2f7df",
"metadata": {},
"outputs": [],
"source": [
"import numpy"
]
},
{
"cell_type": "markdown",
"id": "85205b58",
"metadata": {},
"source": [
"Importing a library is like getting a piece of lab equipment out of a storage locker and setting it\n",
"up on the bench. Libraries provide additional functionality to the basic Python package, much like\n",
"a new piece of equipment adds functionality to a lab space. Just like in the lab, importing too\n",
"many libraries can sometimes complicate and slow down your programs - so we only import what we\n",
"need for each program.\n",
"\n",
"Once we've imported the library, we can ask the library to read our data file for us:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "533850e4",
"metadata": {},
"outputs": [],
"source": [
"numpy.loadtxt(fname='inflammation-01.csv', delimiter=',')"
]
},
{
"cell_type": "markdown",
"id": "451c7a77",
"metadata": {},
"source": [
"The expression `numpy.loadtxt(...)` is a\n",
"[function call](../learners/reference.md#function-call)\n",
"that asks Python to run the [function](../learners/reference.md#function) `loadtxt` which\n",
"belongs to the `numpy` library.\n",
"The dot notation in Python is used most of all as an object attribute/property specifier or for invoking its method. `object.property` will give you the object.property value,\n",
"`object_name.method()` will invoke on object\\_name method.\n",
"\n",
"As an example, John Smith is the John that belongs to the Smith family.\n",
"We could use the dot notation to write his name `smith.john`,\n",
"just as `loadtxt` is a function that belongs to the `numpy` library.\n",
"\n",
"`numpy.loadtxt` has two [parameters](../learners/reference.md#parameter): the name of the file\n",
"we want to read and the [delimiter](../learners/reference.md#delimiter) that separates values\n",
"on a line. These both need to be character strings\n",
"(or [strings](../learners/reference.md#string) for short), so we put them in quotes.\n",
"\n",
"Since we haven't told it to do anything else with the function's output,\n",
"the [notebook](../learners/reference.md#notebook) displays it.\n",
"In this case,\n",
"that output is the data we just loaded.\n",
"By default,\n",
"only a few rows and columns are shown\n",
"(with `...` to omit elements when displaying big arrays).\n",
"Note that, to save space when displaying NumPy arrays, Python does not show us trailing zeros,\n",
"so `1.0` becomes `1.`.\n",
"\n",
"Our call to `numpy.loadtxt` read our file\n",
"but didn't save the data in memory.\n",
"To do that,\n",
"we need to assign the array to a variable. In a similar manner to how we assign a single\n",
"value to a variable, we can also assign an array of values to a variable using the same syntax.\n",
"Let's re-run `numpy.loadtxt` and save the returned data:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "762d1ef4",
"metadata": {},
"outputs": [],
"source": [
"data = numpy.loadtxt(fname='inflammation-01.csv', delimiter=',')"
]
},
{
"cell_type": "markdown",
"id": "4a95a1aa",
"metadata": {},
"source": [
"This statement doesn't produce any output because we've assigned the output to the variable `data`.\n",
"If we want to check that the data have been loaded,\n",
"we can print the variable's value:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "645d4236",
"metadata": {},
"outputs": [],
"source": [
"print(data)"
]
},
{
"cell_type": "markdown",
"id": "fb982871",
"metadata": {},
"source": [
"Now that the data are in memory,\n",
"we can manipulate them.\n",
"First,\n",
"let's ask what [type](../learners/reference.md#type) of thing `data` refers to:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "fda11a39",
"metadata": {},
"outputs": [],
"source": [
"print(type(data))"
]
},
{
"cell_type": "markdown",
"id": "500c9db2",
"metadata": {},
"source": [
"The output tells us that `data` currently refers to\n",
"an N-dimensional array, the functionality for which is provided by the NumPy library.\n",
"These data correspond to arthritis patients' inflammation.\n",
"The rows are the individual patients, and the columns\n",
"are their daily inflammation measurements."
]
},
{
"cell_type": "markdown",
"id": "f9d4908b",
"metadata": {},
"source": [
"## Data Type\n",
"\n",
"A Numpy array contains one or more elements\n",
"of the same type. The `type` function will only tell you that\n",
"a variable is a NumPy array but won't tell you the type of\n",
"thing inside the array.\n",
"We can find out the type\n",
"of the data contained in the NumPy array."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "a5015503",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"print(data.dtype)"
]
},
{
"cell_type": "markdown",
"id": "89db27f3",
"metadata": {},
"source": [
"This tells us that the NumPy array's elements are\n",
"[floating-point numbers](../learners/reference.md#floating-point-number)."
]
},
{
"cell_type": "markdown",
"id": "67618f66",
"metadata": {},
"source": [
"With the following command, we can see the array's [shape](../learners/reference.md#shape):"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "f25e4ecd",
"metadata": {},
"outputs": [],
"source": [
"print(data.shape)"
]
},
{
"cell_type": "markdown",
"id": "fe95757f",
"metadata": {},
"source": [
"The output tells us that the `data` array variable contains 60 rows and 40 columns. When we\n",
"created the variable `data` to store our arthritis data, we did not only create the array; we also\n",
"created information about the array, called [members](../learners/reference.md#member) or\n",
"attributes. This extra information describes `data` in the same way an adjective describes a noun.\n",
"`data.shape` is an attribute of `data` which describes the dimensions of `data`. We use the same\n",
"dotted notation for the attributes of variables that we use for the functions in libraries because\n",
"they have the same part-and-whole relationship.\n",
"\n",
"If we want to get a single number from the array, we must provide an\n",
"[index](../learners/reference.md#index) in square brackets after the variable name, just as we\n",
"do in math when referring to an element of a matrix. Our inflammation data has two dimensions, so\n",
"we will need to use two indices to refer to one specific value:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "78ae3117",
"metadata": {},
"outputs": [],
"source": [
"print('first value in data:', data[0, 0])"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "9538510b",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"print('middle value in data:', data[29, 19])"
]
},
{
"cell_type": "markdown",
"id": "1c559980",
"metadata": {},
"source": [
"The expression `data[29, 19]` accesses the element at row 30, column 20. While this expression may\n",
"not surprise you,\n",
"`data[0, 0]` might.\n",
"Programming languages like Fortran, MATLAB and R start counting at 1\n",
"because that's what human beings have done for thousands of years.\n",
"Languages in the C family (including C++, Java, Perl, and Python) count from 0\n",
"because it represents an offset from the first value in the array (the second\n",
"value is offset by one index from the first value). This is closer to the way\n",
"that computers represent arrays (if you are interested in the historical\n",
"reasons behind counting indices from zero, you can read\n",
"[Mike Hoye's blog post](https://exple.tive.org/blarg/2013/10/22/citation-needed/)).\n",
"As a result,\n",
"if we have an M×N array in Python,\n",
"its indices go from 0 to M-1 on the first axis\n",
"and 0 to N-1 on the second.\n",
"It takes a bit of getting used to,\n",
"but one way to remember the rule is that\n",
"the index is how many steps we have to take from the start to get the item we want.\n",
"\n",
"{alt=\"'data' is a 3 by 3 numpy array containing row 0: \\['A', 'B', 'C'\\], row 1: \\['D', 'E', 'F'\\], androw 2: \\['G', 'H', 'I'\\]. Starting in the upper left hand corner, data\\[0, 0\\] = 'A', data\\[0, 1\\] = 'B',data\\[0, 2\\] = 'C', data\\[1, 0\\] = 'D', data\\[1, 1\\] = 'E', data\\[1, 2\\] = 'F', data\\[2, 0\\] = 'G',data\\[2, 1\\] = 'H', and data\\[2, 2\\] = 'I', in the bottom right hand corner.\"}"
]
},
{
"cell_type": "markdown",
"id": "5079fef6",
"metadata": {},
"source": [
"## In the Corner\n",
"\n",
"What may also surprise you is that when Python displays an array,\n",
"it shows the element with index `[0, 0]` in the upper left corner\n",
"rather than the lower left.\n",
"This is consistent with the way mathematicians draw matrices\n",
"but different from the Cartesian coordinates.\n",
"The indices are (row, column) instead of (column, row) for the same reason,\n",
"which can be confusing when plotting data."
]
},
{
"cell_type": "markdown",
"id": "750e6310",
"metadata": {},
"source": [
"## Slicing data\n",
"\n",
"An index like `[30, 20]` selects a single element of an array,\n",
"but we can select whole sections as well.\n",
"For example,\n",
"we can select the first ten days (columns) of values\n",
"for the first four patients (rows) like this:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "a0af587b",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"print(data[0:4, 0:10])"
]
},
{
"cell_type": "markdown",
"id": "0d7b4872",
"metadata": {},
"source": [
"The [slice](../learners/reference.md#slice) `0:4` means, \"Start at index 0 and go up to,\n",
"but not including, index 4\". Again, the up-to-but-not-including takes a bit of getting used to,\n",
"but the rule is that the difference between the upper and lower bounds is the number of values in\n",
"the slice.\n",
"\n",
"We don't have to start slices at 0:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "924402cb",
"metadata": {},
"outputs": [],
"source": [
"print(data[5:10, 0:10])"
]
},
{
"cell_type": "markdown",
"id": "bf6e60f2",
"metadata": {},
"source": [
"We also don't have to include the upper and lower bound on the slice. If we don't include the lower\n",
"bound, Python uses 0 by default; if we don't include the upper, the slice runs to the end of the\n",
"axis, and if we don't include either (i.e., if we use ':' on its own), the slice includes\n",
"everything:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "cc128d1d",
"metadata": {},
"outputs": [],
"source": [
"small = data[:3, 36:]\n",
"print('small is:')\n",
"print(small)"
]
},
{
"cell_type": "markdown",
"id": "0b833f67",
"metadata": {},
"source": [
"The above example selects rows 0 through 2 and columns 36 through to the end of the array."
]
},
{
"cell_type": "markdown",
"id": "590f3ccb",
"metadata": {},
"source": [
"## Analyzing data\n",
"\n",
"NumPy has several useful functions that take an array as input to perform operations on its values.\n",
"If we want to find the average inflammation for all patients on\n",
"all days, for example, we can ask NumPy to compute `data`'s mean value:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "99212478",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"print(numpy.mean(data))"
]
},
{
"cell_type": "markdown",
"id": "1149a89b",
"metadata": {},
"source": [
"`mean` is a [function](../learners/reference.md#function) that takes\n",
"an array as an [argument](../learners/reference.md#argument)."
]
},
{
"cell_type": "markdown",
"id": "1ee00da9",
"metadata": {},
"source": [
"## Not All Functions Have Input\n",
"\n",
"Generally, a function uses inputs to produce outputs.\n",
"However, some functions produce outputs without\n",
"needing any input. For example, checking the current time\n",
"doesn't require any input."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "45eda2d9",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"import time\n",
"print(time.ctime())"
]
},
{
"cell_type": "markdown",
"id": "131b8668",
"metadata": {},
"source": [
"For functions that don't take in any arguments,\n",
"we still need parentheses (`()`)\n",
"to tell Python to go and do something for us.\n",
"\n",
"Let's use three other NumPy functions to get some descriptive values about the dataset.\n",
"We'll also use multiple assignment,\n",
"a convenient Python feature that will enable us to do this all in one line."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "104d2a29",
"metadata": {},
"outputs": [],
"source": [
"maxval, minval, stdval = numpy.amax(data), numpy.amin(data), numpy.std(data)\n",
"\n",
"print('maximum inflammation:', maxval)\n",
"print('minimum inflammation:', minval)\n",
"print('standard deviation:', stdval)"
]
},
{
"cell_type": "markdown",
"id": "3159d81a",
"metadata": {},
"source": [
"Here we've assigned the return value from `numpy.amax(data)` to the variable `maxval`, the value\n",
"from `numpy.amin(data)` to `minval`, and so on."
]
},
{
"cell_type": "markdown",
"id": "c5e35860",
"metadata": {},
"source": [
"## Mystery Functions in IPython\n",
"\n",
"How did we know what functions NumPy has and how to use them?\n",
"If you are working in IPython or in a Jupyter Notebook, there is an easy way to find out.\n",
"If you type the name of something followed by a dot, then you can use\n",
"[tab completion](../learners/reference.md#tab-completion)\n",
"(e.g. type `numpy.` and then press Tab)\n",
"to see a list of all functions and attributes that you can use. After selecting one, you\n",
"can also add a question mark (e.g. `numpy.cumprod?`), and IPython will return an\n",
"explanation of the method! This is the same as doing `help(numpy.cumprod)`.\n",
"Similarly, if you are using the \"plain vanilla\" Python interpreter, you can type `numpy.`\n",
"and press the Tab key twice for a listing of what is available. You can then use the\n",
"`help()` function to see an explanation of the function you're interested in,\n",
"for example: `help(numpy.cumprod)`."
]
},
{
"cell_type": "markdown",
"id": "c1aea732",
"metadata": {},
"source": [
"## Confusing Function Names\n",
"\n",
"One might wonder why the functions are called `amax` and `amin` and not `max` and `min` or why the other is called `mean` and not `amean`.\n",
"The package `numpy` does provide functions `max` and `min` that are fully equivalent to `amax` and `amin`, but they share a name with standard library functions `max` and `min` that come with Python itself.\n",
"Referring to the functions like we did above, that is `numpy.max` for example, does not cause problems, but there are other ways to refer to them that could.\n",
"In addition, text editors might highlight (color) these functions like standard library function, even though they belong to NumPy, which can be confusing and lead to errors.\n",
"Since there is no function called `mean` in the standard library, there is no function called `amean`."
]
},
{
"cell_type": "markdown",
"id": "9f74cb41",
"metadata": {},
"source": [
"When analyzing data, though,\n",
"we often want to look at variations in statistical values,\n",
"such as the maximum inflammation per patient\n",
"or the average inflammation per day.\n",
"One way to do this is to create a new temporary array of the data we want,\n",
"then ask it to do the calculation:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "d1e31d42",
"metadata": {},
"outputs": [],
"source": [
"patient_0 = data[0, :] # 0 on the first axis (rows), everything on the second (columns)\n",
"print('maximum inflammation for patient 0:', numpy.amax(patient_0))"
]
},
{
"cell_type": "markdown",
"id": "976c5f8f",
"metadata": {},
"source": [
"We don't actually need to store the row in a variable of its own.\n",
"Instead, we can combine the selection and the function call:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "fe49d3cd",
"metadata": {},
"outputs": [],
"source": [
"print('maximum inflammation for patient 2:', numpy.amax(data[2, :]))"
]
},
{
"cell_type": "markdown",
"id": "3893203e",
"metadata": {},
"source": [
"What if we need the maximum inflammation for each patient over all days (as in the\n",
"next diagram on the left) or the average for each day (as in the\n",
"diagram on the right)? As the diagram below shows, we want to perform the\n",
"operation across an axis:\n",
"\n",
"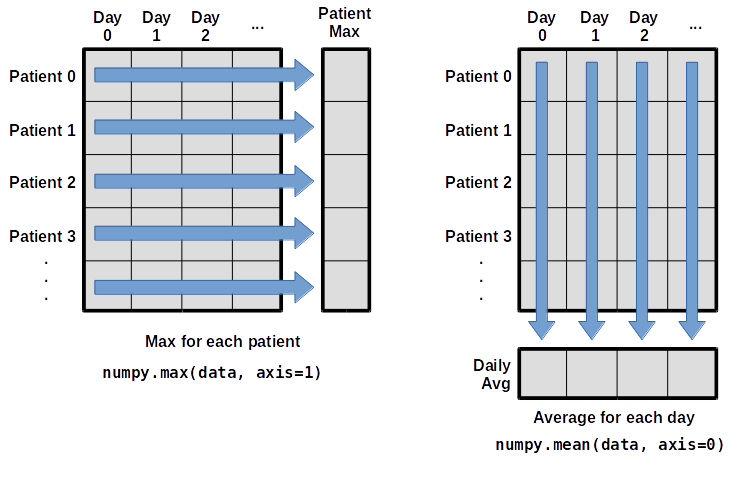{alt=\"Per-patient maximum inflammation is computed row-wise across all columns using numpy.amax(data, axis=1). Per-day average inflammation is computed column-wise across all rows using numpy.mean(data, axis=0).\"}\n",
"\n",
"To support this functionality,\n",
"most array functions allow us to specify the axis we want to work on.\n",
"If we ask for the average across axis 0 (rows in our 2D example),\n",
"we get:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "d31b8b9c",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"print(numpy.mean(data, axis=0))"
]
},
{
"cell_type": "markdown",
"id": "c5bab58f",
"metadata": {},
"source": [
"As a quick check,\n",
"we can ask this array what its shape is:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "f66f9657",
"metadata": {},
"outputs": [],
"source": [
"print(numpy.mean(data, axis=0).shape)"
]
},
{
"cell_type": "markdown",
"id": "b95aae65",
"metadata": {},
"source": [
"The expression `(40,)` tells us we have an N×1 vector,\n",
"so this is the average inflammation per day for all patients.\n",
"If we average across axis 1 (columns in our 2D example), we get:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "e5e11b76",
"metadata": {},
"outputs": [],
"source": [
"print(numpy.mean(data, axis=1))"
]
},
{
"cell_type": "markdown",
"id": "124c3e34",
"metadata": {},
"source": [
"which is the average inflammation per patient across all days."
]
},
{
"cell_type": "markdown",
"id": "2d08db33",
"metadata": {},
"source": [
"## Slicing Strings\n",
"\n",
"A section of an array is called a [slice](../learners/reference.md#slice).\n",
"We can take slices of character strings as well:"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "b35bbee7",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"element = 'oxygen'\n",
"print('first three characters:', element[0:3])\n",
"print('last three characters:', element[3:6])"
]
},
{
"cell_type": "markdown",
"id": "2d605f12",
"metadata": {},
"source": [
"- What is the value of `element[:4]`?\n",
"- What about `element[4:]`?\n",
"- Or `element[:]`?\n",
"- What is `element[-1]`?\n",
"- What is `element[-2]`?"
]
},
{
"cell_type": "markdown",
"id": "a0de7b1f",
"metadata": {},
"source": [
"Given those answers,\n",
"explain what `element[1:-1]` does."
]
},
{
"cell_type": "markdown",
"id": "47617ff3",
"metadata": {},
"source": [
"How can we rewrite the slice for getting the last three characters of `element`,\n",
"so that it works even if we assign a different string to `element`?\n",
"Test your solution with the following strings: `carpentry`, `clone`, `hi`.\n",
"\n",
"## Solution"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "6782473e",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"element = 'oxygen'\n",
"print('last three characters:', element[-3:])\n",
"element = 'carpentry'\n",
"print('last three characters:', element[-3:])\n",
"element = 'clone'\n",
"print('last three characters:', element[-3:])\n",
"element = 'hi'\n",
"print('last three characters:', element[-3:])"
]
},
{
"cell_type": "markdown",
"id": "0606e652",
"metadata": {},
"source": [
"## Thin Slices\n",
"\n",
"The expression `element[3:3]` produces an\n",
"[empty string](../learners/reference.md#empty-string),\n",
"i.e., a string that contains no characters.\n",
"If `data` holds our array of patient data,\n",
"what does `data[3:3, 4:4]` produce?\n",
"What about `data[3:3, :]`?"
]
},
{
"cell_type": "markdown",
"id": "a425026d",
"metadata": {},
"source": [
"## Stacking Arrays\n",
"\n",
"Arrays can be concatenated and stacked on top of one another,\n",
"using NumPy's `vstack` and `hstack` functions for vertical and horizontal stacking, respectively."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "1810d7cd",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"import numpy\n",
"\n",
"A = numpy.array([[1, 2, 3], [4, 5, 6], [7, 8, 9]])\n",
"print('A = ')\n",
"print(A)\n",
"\n",
"B = numpy.hstack([A, A])\n",
"print('B = ')\n",
"print(B)\n",
"\n",
"C = numpy.vstack([A, A])\n",
"print('C = ')\n",
"print(C)"
]
},
{
"cell_type": "markdown",
"id": "bf2b8b34",
"metadata": {},
"source": [
"Write some additional code that slices the first and last columns of `A`,\n",
"and stacks them into a 3x2 array.\n",
"Make sure to `print` the results to verify your solution.\n",
"\n",
"## Solution\n",
"\n",
"A 'gotcha' with array indexing is that singleton dimensions\n",
"are dropped by default. That means `A[:, 0]` is a one dimensional\n",
"array, which won't stack as desired. To preserve singleton dimensions,\n",
"the index itself can be a slice or array. For example, `A[:, :1]` returns\n",
"a two dimensional array with one singleton dimension (i.e. a column\n",
"vector)."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "01ead484",
"metadata": {},
"outputs": [],
"source": [
"D = numpy.hstack((A[:, :1], A[:, -1:]))\n",
"print('D = ')\n",
"print(D)"
]
},
{
"cell_type": "markdown",
"id": "c07556c8",
"metadata": {},
"source": [
"An alternative way to achieve the same result is to use Numpy's\n",
"delete function to remove the second column of A. If you're not\n",
"sure what the parameters of numpy.delete mean, use the help files."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "22adce3e",
"metadata": {},
"outputs": [],
"source": [
"D = numpy.delete(arr=A, obj=1, axis=1)\n",
"print('D = ')\n",
"print(D)"
]
},
{
"cell_type": "markdown",
"id": "790c09ea",
"metadata": {},
"source": [
"## Change In Inflammation\n",
"\n",
"The patient data is *longitudinal* in the sense that each row represents a\n",
"series of observations relating to one individual. This means that\n",
"the change in inflammation over time is a meaningful concept.\n",
"Let's find out how to calculate changes in the data contained in an array\n",
"with NumPy.\n",
"\n",
"The `numpy.diff()` function takes an array and returns the differences\n",
"between two successive values. Let's use it to examine the changes\n",
"each day across the first week of patient 3 from our inflammation dataset."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "e32fb577",
"metadata": {},
"outputs": [],
"source": [
"patient3_week1 = data[3, :7]\n",
"print(patient3_week1)"
]
},
{
"cell_type": "markdown",
"id": "ca361c21",
"metadata": {},
"source": [
"Calling `numpy.diff(patient3_week1)` would do the following calculations"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "d99b2e24",
"metadata": {},
"outputs": [],
"source": [
"[ 0 - 0, 2 - 0, 0 - 2, 4 - 0, 2 - 4, 2 - 2 ]"
]
},
{
"cell_type": "markdown",
"id": "f0afaf3a",
"metadata": {},
"source": [
"and return the 6 difference values in a new array."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "b9a4f05a",
"metadata": {},
"outputs": [],
"source": [
"numpy.diff(patient3_week1)"
]
},
{
"cell_type": "markdown",
"id": "bcae8197",
"metadata": {},
"source": [
"Note that the array of differences is shorter by one element (length 6).\n",
"\n",
"When calling `numpy.diff` with a multi-dimensional array, an `axis` argument may\n",
"be passed to the function to specify which axis to process. When applying\n",
"`numpy.diff` to our 2D inflammation array `data`, which axis would we specify?"
]
},
{
"cell_type": "markdown",
"id": "34da259d",
"metadata": {},
"source": [
"## Solution\n",
"\n",
"Since the row axis (0) is patients, it does not make sense to get the\n",
"difference between two arbitrary patients. The column axis (1) is in\n",
"days, so the difference is the change in inflammation -- a meaningful\n",
"concept."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "eb3df6d1",
"metadata": {
"lines_to_next_cell": 2
},
"outputs": [],
"source": [
"numpy.diff(data, axis=1)"
]
},
{
"cell_type": "markdown",
"id": "7301a29c",
"metadata": {},
"source": [
"If the shape of an individual data file is `(60, 40)` (60 rows and 40\n",
"columns), what would the shape of the array be after you run the `diff()`\n",
"function and why?"
]
},
{
"cell_type": "markdown",
"id": "816db011",
"metadata": {},
"source": [
"## Solution\n",
"\n",
"The shape will be `(60, 39)` because there is one fewer difference between\n",
"columns than there are columns in the data."
]
},
{
"cell_type": "markdown",
"id": "44cac4d0",
"metadata": {},
"source": [
"How would you find the largest change in inflammation for each patient? Does\n",
"it matter if the change in inflammation is an increase or a decrease?"
]
},
{
"cell_type": "markdown",
"id": "69c3a1df",
"metadata": {},
"source": [
"## Solution\n",
"\n",
"By using the `numpy.amax()` function after you apply the `numpy.diff()`\n",
"function, you will get the largest difference between days."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "aec7a55d",
"metadata": {},
"outputs": [],
"source": [
"numpy.amax(numpy.diff(data, axis=1), axis=1)"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "289d1b2c",
"metadata": {},
"outputs": [],
"source": [
"array([ 7., 12., 11., 10., 11., 13., 10., 8., 10., 10., 7.,\n",
" 7., 13., 7., 10., 10., 8., 10., 9., 10., 13., 7.,\n",
" 12., 9., 12., 11., 10., 10., 7., 10., 11., 10., 8.,\n",
" 11., 12., 10., 9., 10., 13., 10., 7., 7., 10., 13.,\n",
" 12., 8., 8., 10., 10., 9., 8., 13., 10., 7., 10.,\n",
" 8., 12., 10., 7., 12.])"
]
},
{
"cell_type": "markdown",
"id": "da19ff3b",
"metadata": {},
"source": [
"If inflammation values *decrease* along an axis, then the difference from\n",
"one element to the next will be negative. If\n",
"you are interested in the **magnitude** of the change and not the\n",
"direction, the `numpy.absolute()` function will provide that.\n",
"\n",
"Notice the difference if you get the largest *absolute* difference\n",
"between readings."
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "97bfc8ea",
"metadata": {},
"outputs": [],
"source": [
"numpy.amax(numpy.absolute(numpy.diff(data, axis=1)), axis=1)"
]
},
{
"cell_type": "code",
"execution_count": null,
"id": "fa01ef33",
"metadata": {},
"outputs": [],
"source": [
"array([ 12., 14., 11., 13., 11., 13., 10., 12., 10., 10., 10.,\n",
" 12., 13., 10., 11., 10., 12., 13., 9., 10., 13., 9.,\n",
" 12., 9., 12., 11., 10., 13., 9., 13., 11., 11., 8.,\n",
" 11., 12., 13., 9., 10., 13., 11., 11., 13., 11., 13.,\n",
" 13., 10., 9., 10., 10., 9., 9., 13., 10., 9., 10.,\n",
" 11., 13., 10., 10., 12.])"
]
},
{
"cell_type": "markdown",
"id": "d67919e5",
"metadata": {},
"source": [
"### Key points\n",
"\n",
"- Import a library into a program using `import libraryname`.\n",
"- Use the `numpy` library to work with arrays in Python.\n",
"- The expression `array.shape` gives the shape of an array.\n",
"- Use `array[x, y]` to select a single element from a 2D array.\n",
"- Array indices start at 0, not 1.\n",
"- Use `low:high` to specify a `slice` that includes the indices from `low` to `high-1`.\n",
"- Use `# some kind of explanation` to add comments to programs.\n",
"- Use `numpy.mean(array)`, `numpy.amax(array)`, and `numpy.amin(array)` to calculate simple statistics.\n",
"- Use `numpy.mean(array, axis=0)` or `numpy.mean(array, axis=1)` to calculate statistics across the specified axis.\n",
"\n"
]
}
],
"metadata": {
"jupytext": {
"cell_metadata_filter": "-all",
"main_language": "python",
"notebook_metadata_filter": "-all"
}
},
"nbformat": 4,
"nbformat_minor": 5
}