{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Use Case: Unmatched Instances Input"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Install Dependencies"
]
},
{
"cell_type": "code",
"execution_count": 10,
"metadata": {},
"outputs": [],
"source": [
"!pip install panoptica auxiliary rich numpy > /dev/null"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"If you installed the packages and requirements on your own machine, you can skip this section and start from the import section.\n",
"\n",
"### Setup Colab environment (optional) \n",
"Otherwise you can follow and execute the tutorial on your browser.\n",
"In order to start working on the notebook, click on the following button, this will open this page in the Colab environment and you will be able to execute the code on your own (*Google account required*).\n",
"\n",
"\n",
"  \n",
"\n",
"\n",
"Now that you are visualizing the notebook in Colab, run the next cell to install the packages we will use. There are few things you should follow in order to properly set the notebook up:\n",
"1. Warning: This notebook was not authored by Google. Click on 'Run anyway'.\n",
"1. When the installation commands are done, there might be \"Restart runtime\" button at the end of the output. Please, click it.\n",
"If you run the next cell in a Google Colab environment, it will **clone the 'tutorials' repository** in your google drive. This will create a **new folder** called \"tutorials\" in **your Google Drive**.\n",
"All generated file will be created/uploaded to your Google Drive respectively.\n",
"\n",
"After the first execution of the next cell, you might receive some warnings and notifications, please follow these instructions:\n",
" - 'Permit this notebook to access your Google Drive files?' Click on 'Yes', and select your account.\n",
" - Google Drive for desktop wants to access your Google Account. Click on 'Allow'.\n",
"\n",
"Afterwards the \"tutorials\" folder has been created. You can navigate it through the lefthand panel in Colab. You might also have received an email that informs you about the access on your Google Drive."
]
},
{
"cell_type": "code",
"execution_count": 11,
"metadata": {},
"outputs": [],
"source": [
"import sys\n",
"\n",
"# Check if we are in google colab currently\n",
"try:\n",
" import google.colab\n",
"\n",
" colabFlag = True\n",
"except ImportError as r:\n",
" colabFlag = False\n",
"\n",
"# Execute certain steps only if we are in a colab environment\n",
"if colabFlag:\n",
" # Create a folder in your Google Drive\n",
" from google.colab import drive\n",
"\n",
" drive.mount(\"/content/drive\")\n",
" # clone repository and set path\n",
" !git clone https://github.com/BrainLesion/tutorials.git /content/drive/MyDrive/tutorials\n",
" BASE_PATH = \"/content/drive/MyDrive/tutorials/panoptica\"\n",
" sys.path.insert(0, BASE_PATH)\n",
"\n",
"else: # normal jupyter notebook environment\n",
" BASE_PATH = \".\" # current working directory would be BraTs-Toolkit anyways if you are not in colab"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Setup Imports"
]
},
{
"cell_type": "code",
"execution_count": 12,
"metadata": {},
"outputs": [],
"source": [
"import numpy as np\n",
"from auxiliary.nifti.io import read_nifti\n",
"from rich import print as pprint\n",
"from panoptica import NaiveThresholdMatching, Panoptica_Evaluator, InputType\n",
"from panoptica.utils.segmentation_class import LabelGroup, SegmentationClassGroups"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Load Example Data"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
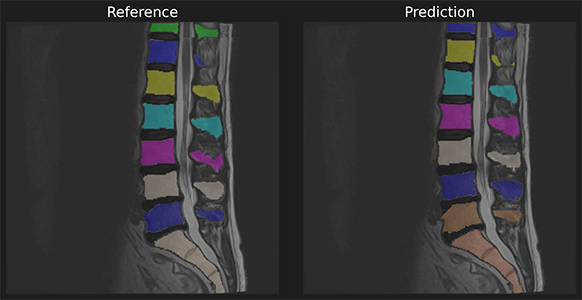
"To demonstrate we use a reference and predicition of spine a segmentation with unmatched instances.\n",
"\n",
"\n",
""
]
},
{
"cell_type": "code",
"execution_count": 13,
"metadata": {},
"outputs": [
{
"data": {
"text/plain": [
"(array([ 0, 2, 3, 4, 5, 6, 7, 8, 26, 102, 103, 104, 105,\n",
" 106, 107, 108, 202, 203, 204, 205, 206, 207, 208], dtype=uint8),\n",
" array([ 0, 3, 4, 5, 6, 7, 8, 9, 26, 103, 104, 105, 106,\n",
" 107, 108, 109, 203, 204, 205, 206, 207, 208, 209], dtype=uint8))"
]
},
"execution_count": 13,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"ref_masks = read_nifti(f\"{BASE_PATH}/spine_seg/unmatched_instance/ref.nii.gz\")\n",
"pred_masks = read_nifti(f\"{BASE_PATH}/spine_seg/unmatched_instance/pred.nii.gz\")\n",
"\n",
"# labels are unmatching\n",
"pred_masks[pred_masks == 27] = 26 # For later\n",
"np.unique(ref_masks), np.unique(pred_masks)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Run Evaluation"
]
},
{
"cell_type": "code",
"execution_count": 14,
"metadata": {},
"outputs": [],
"source": [
"# Define (optionally) semantic groups\n",
"# This means that only instance within one group can be matched to each other\n",
"segmentation_class_groups = SegmentationClassGroups(\n",
" {\n",
" \"vertebra\": LabelGroup(list(range(1, 11))),\n",
" \"ivd\": LabelGroup(list(range(101, 111))),\n",
" \"sacrum\": ([26], True),\n",
" \"endplate\": LabelGroup(list(range(201, 211))),\n",
" }\n",
")\n",
"# In this case, the label 26 can only be matched with label 26 (thats why have to ensure above that 26 exists in both masks, otherwise they wouldn't be matched)\n",
"\n",
"evaluator = Panoptica_Evaluator(\n",
" expected_input=InputType.UNMATCHED_INSTANCE,\n",
" instance_matcher=NaiveThresholdMatching(),\n",
" # If you want to use segmentation class groups, give it here as argument\n",
" segmentation_class_groups=segmentation_class_groups,\n",
")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Inspect Results\n",
"The results object allows access to individual metrics and provides helper methods for further processing"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"\n",
"### Group vertebra\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 7\n",
"Number of instances in prediction (num_pred_instances): 7\n",
"True Positives (tp): 7\n",
"False Positives (fp): 0\n",
"False Negatives (fn): 0\n",
"Recognition Quality / F1-Score (rq): 1.0\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.9631786034883428\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.9259373047661901 +- 0.009654749671578153\n",
"Panoptic Quality IoU (pq): 0.9259373047661901\n",
"Segmentation Quality Dsc (sq_dsc): 0.9615183012231253 +- 0.005245540988039026\n",
"Panoptic Quality Dsc (pq_dsc): 0.9615183012231253\n",
"Segmentation Quality ASSD (sq_assd): 0.16832296646947947 +- 0.01828381629759957\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): -0.005930868093584259 +- 0.010871203881221219\n",
"\n",
"\n",
"### Group ivd\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 7\n",
"Number of instances in prediction (num_pred_instances): 7\n",
"True Positives (tp): 7\n",
"False Positives (fp): 0\n",
"False Negatives (fn): 0\n",
"Recognition Quality / F1-Score (rq): 1.0\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.9423566613429801\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.8897861147389462 +- 0.029181150423413706\n",
"Panoptic Quality IoU (pq): 0.8897861147389462\n",
"Segmentation Quality Dsc (sq_dsc): 0.9414254100052913 +- 0.016436031942319355\n",
"Panoptic Quality Dsc (pq_dsc): 0.9414254100052913\n",
"Segmentation Quality ASSD (sq_assd): 0.29013503272997326 +- 0.05544330133482135\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): 0.020603174193257762 +- 0.03071580120223084\n",
"\n",
"\n",
"### Group sacrum\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 1\n",
"Number of instances in prediction (num_pred_instances): 1\n",
"True Positives (tp): 1\n",
"False Positives (fp): 0\n",
"False Negatives (fn): 0\n",
"Recognition Quality / F1-Score (rq): 1.0\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.9698239455931553\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.941415733208399 +- 0.0\n",
"Panoptic Quality IoU (pq): 0.941415733208399\n",
"Segmentation Quality Dsc (sq_dsc): 0.9698239455931553 +- 0.0\n",
"Panoptic Quality Dsc (pq_dsc): 0.9698239455931553\n",
"Segmentation Quality ASSD (sq_assd): 0.20907172118556794 +- 0.0\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): -0.011061174622567414 +- 0.0\n",
"\n",
"\n",
"### Group endplate\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 7\n",
"Number of instances in prediction (num_pred_instances): 7\n",
"True Positives (tp): 4\n",
"False Positives (fp): 3\n",
"False Negatives (fn): 3\n",
"Recognition Quality / F1-Score (rq): 0.5714285714285714\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.6793787581594264\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.54301762284604 +- 0.01014458743300687\n",
"Panoptic Quality IoU (pq): 0.31029578448345146\n",
"Segmentation Quality Dsc (sq_dsc): 0.7037824449992637 +- 0.008529812661560601\n",
"Panoptic Quality Dsc (pq_dsc): 0.40216139714243637\n",
"Segmentation Quality ASSD (sq_assd): 0.33450703853088465 +- 0.010995297631511717\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): -0.009548043713894769 +- 0.05397632450411714\n",
"\n"
]
}
],
"source": [
"# print all results\n",
"results = evaluator.evaluate(pred_masks, ref_masks, verbose=False)\n",
"# The groups will have the names specified above\n",
"for groupname, result in results.items():\n",
" print()\n",
" print(\"### Group\", groupname)\n",
" print(result)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
"
\n",
"\n",
"\n",
"Now that you are visualizing the notebook in Colab, run the next cell to install the packages we will use. There are few things you should follow in order to properly set the notebook up:\n",
"1. Warning: This notebook was not authored by Google. Click on 'Run anyway'.\n",
"1. When the installation commands are done, there might be \"Restart runtime\" button at the end of the output. Please, click it.\n",
"If you run the next cell in a Google Colab environment, it will **clone the 'tutorials' repository** in your google drive. This will create a **new folder** called \"tutorials\" in **your Google Drive**.\n",
"All generated file will be created/uploaded to your Google Drive respectively.\n",
"\n",
"After the first execution of the next cell, you might receive some warnings and notifications, please follow these instructions:\n",
" - 'Permit this notebook to access your Google Drive files?' Click on 'Yes', and select your account.\n",
" - Google Drive for desktop wants to access your Google Account. Click on 'Allow'.\n",
"\n",
"Afterwards the \"tutorials\" folder has been created. You can navigate it through the lefthand panel in Colab. You might also have received an email that informs you about the access on your Google Drive."
]
},
{
"cell_type": "code",
"execution_count": 11,
"metadata": {},
"outputs": [],
"source": [
"import sys\n",
"\n",
"# Check if we are in google colab currently\n",
"try:\n",
" import google.colab\n",
"\n",
" colabFlag = True\n",
"except ImportError as r:\n",
" colabFlag = False\n",
"\n",
"# Execute certain steps only if we are in a colab environment\n",
"if colabFlag:\n",
" # Create a folder in your Google Drive\n",
" from google.colab import drive\n",
"\n",
" drive.mount(\"/content/drive\")\n",
" # clone repository and set path\n",
" !git clone https://github.com/BrainLesion/tutorials.git /content/drive/MyDrive/tutorials\n",
" BASE_PATH = \"/content/drive/MyDrive/tutorials/panoptica\"\n",
" sys.path.insert(0, BASE_PATH)\n",
"\n",
"else: # normal jupyter notebook environment\n",
" BASE_PATH = \".\" # current working directory would be BraTs-Toolkit anyways if you are not in colab"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Setup Imports"
]
},
{
"cell_type": "code",
"execution_count": 12,
"metadata": {},
"outputs": [],
"source": [
"import numpy as np\n",
"from auxiliary.nifti.io import read_nifti\n",
"from rich import print as pprint\n",
"from panoptica import NaiveThresholdMatching, Panoptica_Evaluator, InputType\n",
"from panoptica.utils.segmentation_class import LabelGroup, SegmentationClassGroups"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Load Example Data"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"To demonstrate we use a reference and predicition of spine a segmentation with unmatched instances.\n",
"\n",
"\n",
""
]
},
{
"cell_type": "code",
"execution_count": 13,
"metadata": {},
"outputs": [
{
"data": {
"text/plain": [
"(array([ 0, 2, 3, 4, 5, 6, 7, 8, 26, 102, 103, 104, 105,\n",
" 106, 107, 108, 202, 203, 204, 205, 206, 207, 208], dtype=uint8),\n",
" array([ 0, 3, 4, 5, 6, 7, 8, 9, 26, 103, 104, 105, 106,\n",
" 107, 108, 109, 203, 204, 205, 206, 207, 208, 209], dtype=uint8))"
]
},
"execution_count": 13,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"ref_masks = read_nifti(f\"{BASE_PATH}/spine_seg/unmatched_instance/ref.nii.gz\")\n",
"pred_masks = read_nifti(f\"{BASE_PATH}/spine_seg/unmatched_instance/pred.nii.gz\")\n",
"\n",
"# labels are unmatching\n",
"pred_masks[pred_masks == 27] = 26 # For later\n",
"np.unique(ref_masks), np.unique(pred_masks)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Run Evaluation"
]
},
{
"cell_type": "code",
"execution_count": 14,
"metadata": {},
"outputs": [],
"source": [
"# Define (optionally) semantic groups\n",
"# This means that only instance within one group can be matched to each other\n",
"segmentation_class_groups = SegmentationClassGroups(\n",
" {\n",
" \"vertebra\": LabelGroup(list(range(1, 11))),\n",
" \"ivd\": LabelGroup(list(range(101, 111))),\n",
" \"sacrum\": ([26], True),\n",
" \"endplate\": LabelGroup(list(range(201, 211))),\n",
" }\n",
")\n",
"# In this case, the label 26 can only be matched with label 26 (thats why have to ensure above that 26 exists in both masks, otherwise they wouldn't be matched)\n",
"\n",
"evaluator = Panoptica_Evaluator(\n",
" expected_input=InputType.UNMATCHED_INSTANCE,\n",
" instance_matcher=NaiveThresholdMatching(),\n",
" # If you want to use segmentation class groups, give it here as argument\n",
" segmentation_class_groups=segmentation_class_groups,\n",
")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Inspect Results\n",
"The results object allows access to individual metrics and provides helper methods for further processing"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"\n",
"### Group vertebra\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 7\n",
"Number of instances in prediction (num_pred_instances): 7\n",
"True Positives (tp): 7\n",
"False Positives (fp): 0\n",
"False Negatives (fn): 0\n",
"Recognition Quality / F1-Score (rq): 1.0\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.9631786034883428\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.9259373047661901 +- 0.009654749671578153\n",
"Panoptic Quality IoU (pq): 0.9259373047661901\n",
"Segmentation Quality Dsc (sq_dsc): 0.9615183012231253 +- 0.005245540988039026\n",
"Panoptic Quality Dsc (pq_dsc): 0.9615183012231253\n",
"Segmentation Quality ASSD (sq_assd): 0.16832296646947947 +- 0.01828381629759957\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): -0.005930868093584259 +- 0.010871203881221219\n",
"\n",
"\n",
"### Group ivd\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 7\n",
"Number of instances in prediction (num_pred_instances): 7\n",
"True Positives (tp): 7\n",
"False Positives (fp): 0\n",
"False Negatives (fn): 0\n",
"Recognition Quality / F1-Score (rq): 1.0\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.9423566613429801\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.8897861147389462 +- 0.029181150423413706\n",
"Panoptic Quality IoU (pq): 0.8897861147389462\n",
"Segmentation Quality Dsc (sq_dsc): 0.9414254100052913 +- 0.016436031942319355\n",
"Panoptic Quality Dsc (pq_dsc): 0.9414254100052913\n",
"Segmentation Quality ASSD (sq_assd): 0.29013503272997326 +- 0.05544330133482135\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): 0.020603174193257762 +- 0.03071580120223084\n",
"\n",
"\n",
"### Group sacrum\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 1\n",
"Number of instances in prediction (num_pred_instances): 1\n",
"True Positives (tp): 1\n",
"False Positives (fp): 0\n",
"False Negatives (fn): 0\n",
"Recognition Quality / F1-Score (rq): 1.0\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.9698239455931553\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.941415733208399 +- 0.0\n",
"Panoptic Quality IoU (pq): 0.941415733208399\n",
"Segmentation Quality Dsc (sq_dsc): 0.9698239455931553 +- 0.0\n",
"Panoptic Quality Dsc (pq_dsc): 0.9698239455931553\n",
"Segmentation Quality ASSD (sq_assd): 0.20907172118556794 +- 0.0\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): -0.011061174622567414 +- 0.0\n",
"\n",
"\n",
"### Group endplate\n",
"\n",
"+++ MATCHING +++\n",
"Number of instances in reference (num_ref_instances): 7\n",
"Number of instances in prediction (num_pred_instances): 7\n",
"True Positives (tp): 4\n",
"False Positives (fp): 3\n",
"False Negatives (fn): 3\n",
"Recognition Quality / F1-Score (rq): 0.5714285714285714\n",
"\n",
"+++ GLOBAL +++\n",
"Global Binary Dice (global_bin_dsc): 0.6793787581594264\n",
"\n",
"+++ INSTANCE +++\n",
"Segmentation Quality IoU (sq): 0.54301762284604 +- 0.01014458743300687\n",
"Panoptic Quality IoU (pq): 0.31029578448345146\n",
"Segmentation Quality Dsc (sq_dsc): 0.7037824449992637 +- 0.008529812661560601\n",
"Panoptic Quality Dsc (pq_dsc): 0.40216139714243637\n",
"Segmentation Quality ASSD (sq_assd): 0.33450703853088465 +- 0.010995297631511717\n",
"Segmentation Quality Relative Volume Difference (sq_rvd): -0.009548043713894769 +- 0.05397632450411714\n",
"\n"
]
}
],
"source": [
"# print all results\n",
"results = evaluator.evaluate(pred_masks, ref_masks, verbose=False)\n",
"# The groups will have the names specified above\n",
"for groupname, result in results.items():\n",
" print()\n",
" print(\"### Group\", groupname)\n",
" print(result)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
"results['vertebra'][0].pq=0.9259373047661901\n",
"\n"
],
"text/plain": [
"results\u001b[1m[\u001b[0m\u001b[32m'vertebra'\u001b[0m\u001b[1m]\u001b[0m\u001b[1m[\u001b[0m\u001b[1;36m0\u001b[0m\u001b[1m]\u001b[0m.\u001b[33mpq\u001b[0m=\u001b[1;36m0\u001b[0m\u001b[1;36m.9259373047661901\u001b[0m\n"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"# get specific metric, e.g. pq\n",
"# Now we need to specify group first\n",
"pprint(f\"{results['vertebra'].pq=}\")"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
"results dict: \n",
"{\n",
" 'num_ref_instances': 7,\n",
" 'num_pred_instances': 7,\n",
" 'tp': 7,\n",
" 'fp': 0,\n",
" 'fn': 0,\n",
" 'prec': 1.0,\n",
" 'rec': 1.0,\n",
" 'rq': 1.0,\n",
" 'sq': 0.9259373047661901,\n",
" 'sq_std': 0.009654749671578153,\n",
" 'pq': 0.9259373047661901,\n",
" 'sq_dsc': 0.9615183012231253,\n",
" 'sq_dsc_std': 0.005245540988039026,\n",
" 'pq_dsc': 0.9615183012231253,\n",
" 'sq_assd': 0.16832296646947947,\n",
" 'sq_assd_std': 0.01828381629759957,\n",
" 'sq_rvd': -0.005930868093584259,\n",
" 'sq_rvd_std': 0.010871203881221219,\n",
" 'global_bin_dsc': 0.9631786034883428\n",
"}\n",
"\n"
],
"text/plain": [
"results dict: \n",
"\u001b[1m{\u001b[0m\n",
" \u001b[32m'num_ref_instances'\u001b[0m: \u001b[1;36m7\u001b[0m,\n",
" \u001b[32m'num_pred_instances'\u001b[0m: \u001b[1;36m7\u001b[0m,\n",
" \u001b[32m'tp'\u001b[0m: \u001b[1;36m7\u001b[0m,\n",
" \u001b[32m'fp'\u001b[0m: \u001b[1;36m0\u001b[0m,\n",
" \u001b[32m'fn'\u001b[0m: \u001b[1;36m0\u001b[0m,\n",
" \u001b[32m'prec'\u001b[0m: \u001b[1;36m1.0\u001b[0m,\n",
" \u001b[32m'rec'\u001b[0m: \u001b[1;36m1.0\u001b[0m,\n",
" \u001b[32m'rq'\u001b[0m: \u001b[1;36m1.0\u001b[0m,\n",
" \u001b[32m'sq'\u001b[0m: \u001b[1;36m0.9259373047661901\u001b[0m,\n",
" \u001b[32m'sq_std'\u001b[0m: \u001b[1;36m0.009654749671578153\u001b[0m,\n",
" \u001b[32m'pq'\u001b[0m: \u001b[1;36m0.9259373047661901\u001b[0m,\n",
" \u001b[32m'sq_dsc'\u001b[0m: \u001b[1;36m0.9615183012231253\u001b[0m,\n",
" \u001b[32m'sq_dsc_std'\u001b[0m: \u001b[1;36m0.005245540988039026\u001b[0m,\n",
" \u001b[32m'pq_dsc'\u001b[0m: \u001b[1;36m0.9615183012231253\u001b[0m,\n",
" \u001b[32m'sq_assd'\u001b[0m: \u001b[1;36m0.16832296646947947\u001b[0m,\n",
" \u001b[32m'sq_assd_std'\u001b[0m: \u001b[1;36m0.01828381629759957\u001b[0m,\n",
" \u001b[32m'sq_rvd'\u001b[0m: \u001b[1;36m-0.005930868093584259\u001b[0m,\n",
" \u001b[32m'sq_rvd_std'\u001b[0m: \u001b[1;36m0.010871203881221219\u001b[0m,\n",
" \u001b[32m'global_bin_dsc'\u001b[0m: \u001b[1;36m0.9631786034883428\u001b[0m\n",
"\u001b[1m}\u001b[0m\n"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"# get dict for further processing, e.g. for pandas\n",
"pprint(\"results dict: \", results[\"vertebra\"].to_dict())"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"\n",
"### Group vertebra\n",
"InputType.UNMATCHED_INSTANCE\n",
"Prediction array shape = (164, 399, 17) unique_values= [0 3 4 5 6 7 8 9]\n",
"Reference array shape = (164, 399, 17) unique_values= [0 2 3 4 5 6 7 8]\n",
"\n",
"InputType.MATCHED_INSTANCE\n",
"Prediction array shape = (164, 399, 17) unique_values= [0 2 3 4 5 6 7 8]\n",
"Reference array shape = (164, 399, 17) unique_values= [0 2 3 4 5 6 7 8]\n",
"\n",
"\n",
"### Group ivd\n",
"InputType.UNMATCHED_INSTANCE\n",
"Prediction array shape = (96, 406, 17) unique_values= [ 0 103 104 105 106 107 108 109]\n",
"Reference array shape = (96, 406, 17) unique_values= [ 0 102 103 104 105 106 107 108]\n",
"\n",
"InputType.MATCHED_INSTANCE\n",
"Prediction array shape = (96, 406, 17) unique_values= [ 0 102 103 104 105 106 107 108]\n",
"Reference array shape = (96, 406, 17) unique_values= [ 0 102 103 104 105 106 107 108]\n",
"\n",
"\n",
"### Group sacrum\n",
"InputType.UNMATCHED_INSTANCE\n",
"key UNMATCHED_INSTANCE not in intermediate steps, maybe the step was skipped?\n",
"InputType.MATCHED_INSTANCE\n",
"Prediction array shape = (140, 128, 17) unique_values= [ 0 26]\n",
"Reference array shape = (140, 128, 17) unique_values= [ 0 26]\n",
"\n",
"\n",
"### Group endplate\n",
"InputType.UNMATCHED_INSTANCE\n",
"Prediction array shape = (85, 385, 17) unique_values= [ 0 203 204 205 206 207 208 209]\n",
"Reference array shape = (85, 385, 17) unique_values= [ 0 202 203 204 205 206 207 208]\n",
"\n",
"InputType.MATCHED_INSTANCE\n",
"Prediction array shape = (85, 385, 17) unique_values= [ 0 202 204 206 207 209 210 211]\n",
"Reference array shape = (85, 385, 17) unique_values= [ 0 202 203 204 205 206 207 208]\n",
"\n"
]
}
],
"source": [
"# To inspect different phases, just use the returned intermediate_steps_data object\n",
"\n",
"import numpy as np\n",
"\n",
"for groupname, result in results.items():\n",
" print()\n",
" print(\"### Group\", groupname)\n",
" intermediate_steps_data = result.intermediate_steps_data\n",
" intermediate_steps_data.original_prediction_arr # yields input prediction array\n",
" intermediate_steps_data.original_reference_arr # yields input reference array\n",
"\n",
" # This works with all phases\n",
" for i in [InputType.UNMATCHED_INSTANCE, InputType.MATCHED_INSTANCE]:\n",
" try:\n",
" print(i)\n",
" pred = intermediate_steps_data.prediction_arr(i)\n",
" ref = intermediate_steps_data.reference_arr(i)\n",
" print(\n",
" \"Prediction array shape =\",\n",
" pred.shape,\n",
" \"unique_values=\",\n",
" np.unique(pred),\n",
" )\n",
" print(\n",
" \"Reference array shape =\", ref.shape, \"unique_values=\", np.unique(ref)\n",
" )\n",
" print()\n",
" except AssertionError as e:\n",
" print(e)\n",
" # This happens because Sacrum class group was set to single_instance, hence the Matching phase is skipped and there is no intermediate result for UNMATCHED_INSTANCE"
]
}
],
"metadata": {
"kernelspec": {
"display_name": "seg11panoptdev",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.11.6"
}
},
"nbformat": 4,
"nbformat_minor": 2
}