
Section 9 Antimicrobial Resistance
- William Fitzgerald
- Research Officer, Limerick Regional Veterinary Laboratory, Knockalisheen,Limerick, Ireland
Antimicrobial Resistance (AMR) is defined by the Office International des Epizooties (OIE) as the emergence of resistance amongst bacteria, viruses, fungi and parasites to products known as antimicrobials that are designed to treat them. The OIE has also stated that ‘the overuse and misuse of antimicrobials in human, animal and plant sectors has dramatically accelerated the emergence of AMR. Consequently, minimising the emergence and spread of AMR requires a co-ordinated, focused, multi-sectoral and multinational effort’.
The World Health Organisation (WHO) categorises antimicrobials used in human health as critically important, highly important and important to human health. Critically important antimicrobials (CIAs) should NOT be used as first line of treatment in animals –they should only be used when there is no effective alternative antimicrobials available for target species and indication–.
Additionally, some CIAs are further classified as Highest Priority Critically Important Antibiotics (HPCIAs). WHO Highest Priority CIA group contains antimicrobials licensed for use in veterinary medicine including 3rd and 4th generation cephalosporins, fluoroquinolones, macrolides, and polymyxins. Further information is available in the DAFM webpage (Department of Agriculture, Food and the Marine 2018).
Every year, the Veterinary Laboratory Service (VLS) handles between 1500–2000 bacterial isolates from multiple species; in 2018, there were 1,826 isolates. As part of DAFM plan to monitor AMR within the Irish animal health sector, VLS focuses on particular patterns of AMR, outlined in Figure 9.2.
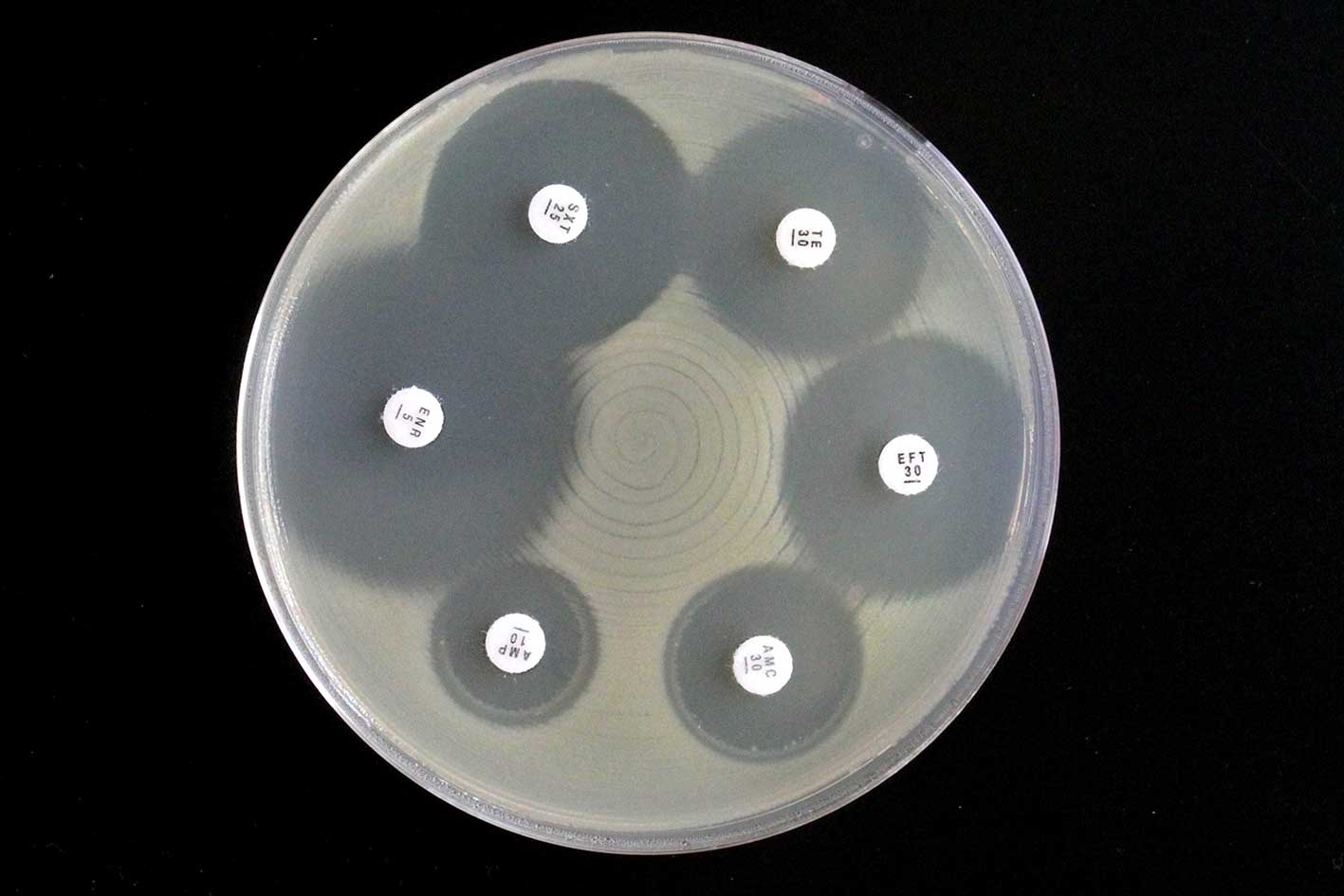
Figure 9.1: Antibiotic sensitivity test illustrating thin paper disks containing antibiotic placed on an agar plate with growing bacteria. Note the clear zone of inhibition, where bacteria growth around antibiotic disk has been inhibited. Photo: Pat Sheehan.
A number of antibiotic panels are used across VLS to initially assess levels of AMR nationally, the Respiratory, Enteric, Gram-positive and the Mastitis causing Gram-negative panels (Figure ). Statuary surveillance of AMR in zoonotic and commensal bacteria is carried out by CVRL as part of an EU- wide harmonised monitoring programme , details of which can be found here.
Disk Diffusion Test
The Kirby-Bauer test or disk diffusion test, is a standard tool for measuring the effectiveness of antimicrobials against pathogenic microorganisms (Figure 9.1). Antimicrobial-impregnated paper disks are placed on on a plate that is inoculated to allow growth of the bacteria and time for the agent to diffuse into the agar. As the drug moves through the agar, it establishes a concentration gradient. If the organism is susceptible to it, a clear zone will appear around the disk where growth has been inhibited.
The size of this zone of inhibition depends pon the sensitivity of the bacteria to the specific antimicrobial agent and the point at which the chemical’s minimum inhibitory concentration (MIC) is reached.
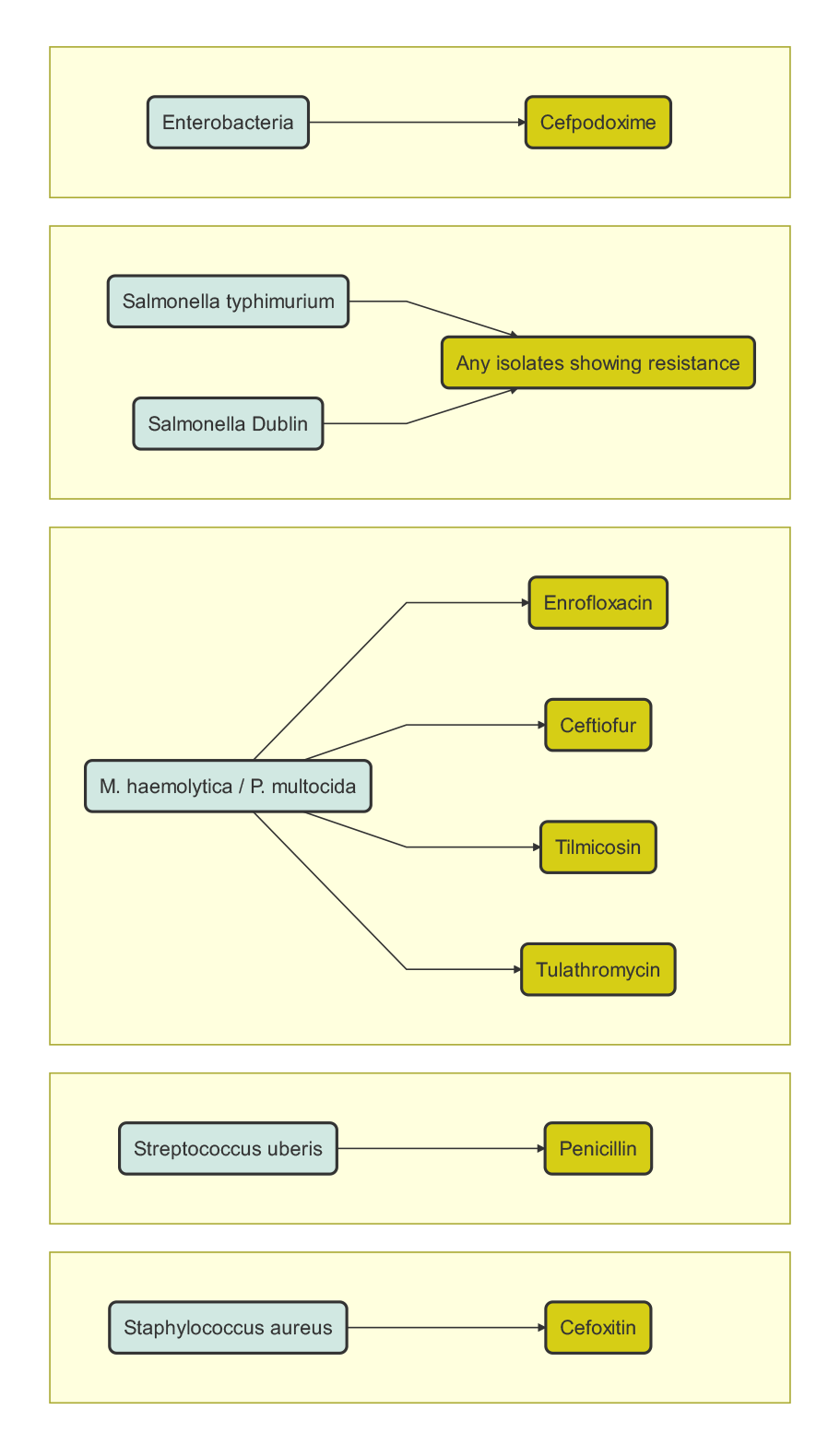
Figure 9.2: The significant AMR patterns that Veterinary Laboratory Service (VLS) monitors.
9.1 Staphylococcus aureus
In 2018, there were 407 isolates of Staphylococcus aureus in the VLS. Of these, 392 (96.3 per cent) were susceptible to Tetracycline, 407 (100 per cent) were susceptible to Trimethroprim-Sulphonamide, 403 (99 per cent) were susceptible to Amoxycillin-Clavulanate (Table 9.1).
9.2 Streptococcus uberis
In 2018, there were 291 Streptococcus uberis isolates in the VLS. Of these, 228 (78.3 per cent) were susceptible to Tetracyclines, 273 (93.8 per cent) were susceptible to Trimethoprim-Sulphonamide, 286 (98.2 per cent) was susceptible to Penicillin, 280 (96.2 per cent) susceptible to Ampicillin and 289 (99.3 per cent) were susceptible to Amoxycillin-Clavulanate (Table 9.1).
| S. aureus (n=407) | S. uberis (n=291) | |
|---|---|---|
| Ampicillin | 190 (46.6%) | 280 (96.2%) |
| Tetracycline | 392 (96.3%) | 228 (78.3%) |
| Trimethoprim-Sulphonamide | 407 (100%) | 273 (93.8%) |
| Florfenicol | Not Tested | Not tested |
| Amoxycillin-Clavulanate | 403 (99%) | 289 (99.3%) |
Case Study
Vancomycin resistant Enterococcus spp. (VRE) in a calf:
A single isolate of Vancomycin resistant Enterococcus faecium was confirmed in a calf using WGS. Enterococci are commensal Gram-positive bacteria that form part of the normal gut flora of animals and humans. Vancomycin is a member of the glycopeptide antimicrobial class and is of critical importance in the treatment of infections, caused by drug resistant organisms, such as Staphylococcus aureus. The use of the glycopeptide avoparicin, as a growth promoter in animals, was linked to the emergence of VRE in pigs and poultry and as a result, its use for this purpose was banned in 1996.
The level of VRE infection in Irish hospitals is one of the highest in Europe. Hospital outbreaks tend to be associated with a distinct, adapted subpopulation, which are often Ampicillin and Ciprofloxacin resistant. The latter is an unusual finding in animal derived isolates and the role of animals as a potential reservoir of resistance for human epidemic Enterococcus strains remains unclear.
9.3 Pasteurella multocida
In 2018, there were 181 isolates of Pasteurella multocida in the VLS. Of these, 173 (99.5 per cent) were susceptible to Tetracycline, 177 (97.7 per cent) was susceptible to Trimethoprim-Sulphonamide, 159 (87.8 per cent) were susceptible to Florfenicol, 179 (98.9 per cent) were susceptible to Amoxycillin-Clavulanate (Table 9.2).
Case Study
AMR in Pasteurella multocida
Pasteurella multocida frequently exists as a commensal bacterium of the respiratory tract in animals but it may act also as a primary or secondary pathogen. In 2018, almost 90 per cent of P. multocida* isolates were susceptible to all of the antimicrobials in the respiratory panel (Ampicillin, Amoxicillin-Clavulanate, Ceftiofur, Tetracycline, Sulfamethoxazole-Trimethoprim, Enrofloxacin, Florfenicol, Tulathromycin and Tilmicosin).
To date, 1 multi-drug resistant isolate of P. multocida from 2018 has undergone WGS. Resistance genes for Aminglycoside, Macrolide, Sulphonamide, and Tetracycline resistance were identified in the strain, which originated from the lung tissue of a weanling calf. The calf was one of at least seven that died during a large outbreak of respiratory disease and it had concurrent lungworm infestation. It is planned to continue with the genotypic characterisation of resistant P. multocida in 2019.
9.4 Mannheimia haemolytica
In 2018, there were 150 isolates of Mannheimia haemolytica. Of these, 135 (90 per cent) was susceptible to Tetracycline, 146 (97.3 per cent) were susceptible to Trimethoprim-Sulphonamide and 149 (99.3 per cent) were susceptible to Amoxycillin-Clavulanate and Florfenicol (Table 9.2).
| M. haemolytica (n=150) | P. multocida (n=181) | |
|---|---|---|
| Ampicillin | 143 (95.3%) | 169 (93.4%) |
| Tetracycline | 135 (90%) | 173 (95.6%) |
| Trimethoprim-Sulphonamide | 146 (97.3%) | 177 (97.8%) |
| Florfenicol | 149 (99.3%) | 159 (87.8%) |
| Amoxycillin-Clavulanate | 149 (99.3%) | 179 (98.9%) |
9.5 Escherichia coli
In 2018, there were 268 isolates of Escherichia coli in the VLS. Of these, 191 (71.2%) were susceptible to Tetracycline, 230 (85.8%) were susceptible to Trimethoprim-Sulphonamide and 229 (85.4%) were susceptible to Amoxycillin-Clavulanate (Table 9.3).
Case Study
Multiple Drug Resistant Escherichia coli on a suckler farm:
Escherichia coli was isolated from the organs and faeces of animals from a farm that was experiencing high mortality rates due to cases of enteritis and unresponsive post- operative infections. Evaluation of the bacterial genome confirmed that the strain was pathogenic to cattle and that its virulence genes were not targeted by commercially available vaccines.
Genes encoding resistance to beta lactam antimicrobials, included extended spectrum Beta-lactams, Fluoroquinolones, Phenicols, Aminoglycosides, Sulphonamides and Tetracyclines were identified.
| E. coli (n=268) | |
|---|---|
| Ampicillin | Not tested |
| Tetracycline | 191 (71.2%) |
| Trimethoprim-Sulphonamide | 230 (85.8%) |
| Florfenicol | Not Tested |
| Amoxycillin-Clavulanate | 229 (85.4%) |
Figure 3: Antibiotic panels used in the VLS and their constituent antibiotics
Respiratory Panel
- Ampicillin
- Amoxicillin Clavulanate
- Ceftiofur
- Tetracycline
- Sulfamethoxazole & Trimethoprim
- Enrofloxacin
- Florfenicol
- Tulathromycin
- Timlicosin
Enteric Panel
- Ampicillin
- Amoxicillin Clavulanate
- Ceftiofur
- Tetracycline
- Sulfamethoxazole & Trimethoprim
- Enrofloxacin
- Neomycin
- Cefpodoxime
- Streptomycin
Gram-Positive
- Ampicillin
- Amoxicillin Clavulanate
- Ceftiofur
- Tetracycline
- Sulfamethoxazole & Trimethoprim
- Cephalothin
- Cefoxitin
- Pirlimycin
- Kanamycin
- Erythromycin
- Cephalexin & Kanamycin
Mastitis Causing Gram-Negative Panel
- Ampicillin
- Amoxicillin Clavulanate
- Ceftiofur
- Tetracycline
- Sulfamethoxazole & Trimethoprim
- Enrofloxacin
- Neomycin
- Streptomycin
- Kanamycin
- Cefpodoxime
- Cephalexin & Kanamycin
9.5.1 Maldi-ToF
One of the more recent advancements in clinical veterinary bacteriology which has helped immensely in relation to identification of pathogens is the use of Maldi-ToF Mass Spectrometry. MALDI is the abbreviation for Matrix Assisted Laser Desorption/Ionization, TOF is Time of Flight. A portion of a colony of the microbe in question is placed onto the sample target and overlaid with matrix. The mass spectra generated are analyzed by dedicated software and compared with stored profiles. Species identification by this procedure is much faster, more accurate and cheaper than other procedures based on immunological or biochemical tests. Maldi ToF can also be used to predict the antibiotic susceptibility of a bacterial species. In 2018, this technique was used to confirm the identity of 588 isolates within DAFMs Veterinary Laboratory Service.

Figure 9.3: Sequencing platform for the generation of whole genome sequences. Photo: Rosemarie Slowey.
9.5.2 Whole Genome Sequencing
Sequencing of bacterial DNA sequences (whole genome sequencing, WGS) has been undertaken in the VLS since 2016. These sequences can be screened for antimicrobial resistance and virulence genes and sequences compared to evaluate the relatedness of the strains, which is particularly important in outbreak investigations. WGS has been used to confirm unusual resistance patterns initially detected in the RVLs.
References
Department of Agriculture, Food and the Marine. 2018. “Policy on Highest Priority Critically Important Antimicrobials.” Dept. of Agriculture, Food; the Marine. https://www.agriculture.gov.ie/media/migration/animalhealthwelfare/amr/PolicyHighestPriorityCriticallyImpAntimicrobials191118.pdf.
A cooperative effort between the VLS and the SAT Section of the DAFM