{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"(fcb-bridgedbnotebook)=\n",
"# Using BridgeDb web services\n",
"\n",
"\n",
"````{panels_fairplus}\n",
":identifier_text: FCB018\n",
":identifier_link: 'http://w3id.org/faircookbook/FCB018'\n",
":difficulty_level: 3\n",
":recipe_type: hands_on\n",
":reading_time_minutes: 30\n",
":intended_audience: data_manager, data_scientist \n",
":has_executable_code: yeah\n",
":recipe_name: Using BridgeDb web services\n",
"```` \n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"In this notebook I will present two use cases for BridgeDb with the purpose of identifier mapping: \n",
"* Mapping data from a recognized data source by BridgeDb to another recognized data source ([see here](https://github.com/bridgedb/BridgeDb/blob/2dba5780260421de311cb3064df79e16a396b887/org.bridgedb.bio/resources/org/bridgedb/bio/datasources.tsv)). For example mapping data identifiers from HGNC to Ensembl.\n",
"* Given a local identifier and a TSV mapping it to one of the BridgeDb data sources, how to map the local identifier to a different data source.\n",
"\n",
"[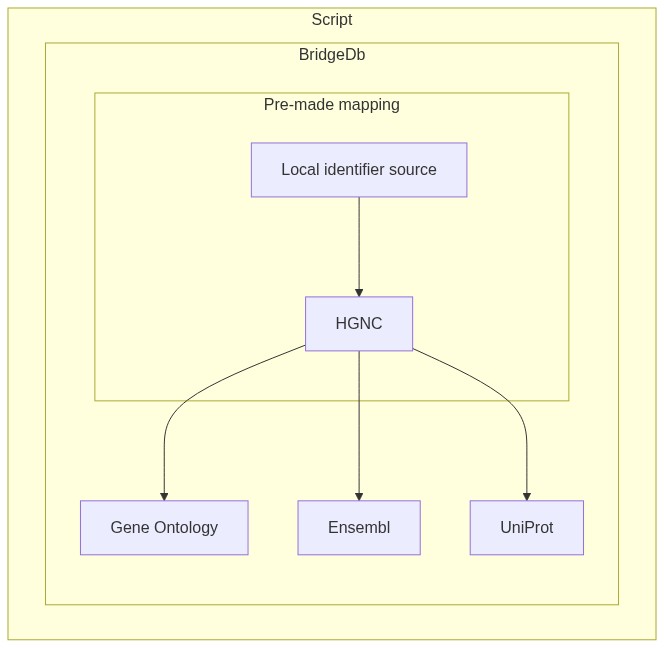](https://mermaid-js.github.io/mermaid-live-editor/#/edit/eyJjb2RlIjoiICAgIGdyYXBoIFRCIFxuICAgICAgc3ViZ3JhcGggU2NyaXB0XG5cbiAgICAgICAgICBzdWJncmFwaCBCcmlkZ2VEYlxuICAgICAgICAgICAgICBCW0FmZnldLS0-RVtHZW5lIE9udG9sb2d5XVxuICAgICAgICAgICAgICBCW0FmZnldLS0-RltFbnNlbWJsXVxuICAgICAgICAgICAgICBCW0FmZnldIC0tPiBIW1VuaVByb3RdXG4gICAgICAgICAgc3ViZ3JhcGggUHJlLW1hZGUgbWFwcGluZ1xuICAgICAgICAgICAgQVtMb2NhbCBpZGVudGlmaWVyIHNvdXJjZV0tLT5CW0hHTkNdXG4gICAgICAgICAgZW5kXG4gICAgICAgICAgZW5kXG4gICAgICAgIGVuZCIsIm1lcm1haWQiOnt9LCJ1cGRhdGVFZGl0b3IiOmZhbHNlfQ)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Querying the WS\n",
"\n",
"To query the Webservice we define below the url and the patterns for a single request and a batch request. You can find the docs [here](https://bridgedb.github.io/swagger/). We will use Python's requests library."
]
},
{
"cell_type": "code",
"execution_count": 7,
"metadata": {},
"outputs": [],
"source": [
"url = \"https://webservice.bridgedb.org/\""
]
},
{
"cell_type": "code",
"execution_count": 8,
"metadata": {},
"outputs": [],
"source": [
"single_request = url+\"{org}/xrefs/{source}/{identifier}\""
]
},
{
"cell_type": "code",
"execution_count": 9,
"metadata": {},
"outputs": [],
"source": [
"batch_request = url+\"{org}/xrefsBatch/{source}{}\""
]
},
{
"cell_type": "code",
"execution_count": 10,
"metadata": {},
"outputs": [],
"source": [
"import requests\n",
"import pandas as pd"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Here we define a method that will turn the web service response into a dataframe with columns corresponding to:\n",
"* The original identifier\n",
"* The data source that the identifier is part of\n",
"* The mapped identifier\n",
"* The data source for the mapped identifier"
]
},
{
"cell_type": "code",
"execution_count": 11,
"metadata": {},
"outputs": [],
"source": [
"def to_df(response, batch=False):\n",
" if batch:\n",
" records = []\n",
" for tup in to_df(response).itertuples():\n",
" if tup[3] != None:\n",
" for mappings in tup[3].split(','):\n",
" target = mappings.split(':', 1)\n",
" if len(target) > 1:\n",
" records.append((tup[1], tup[2], target[1], target[0]))\n",
" else:\n",
" records.append((tup[1], tup[2], target[0], target[0]))\n",
" return pd.DataFrame(records, columns = ['original', 'source', 'mapping', 'target'])\n",
" \n",
" return pd.DataFrame([line.split('\\t') for line in response.text.split('\\n')])"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Here we define the organism and the data source from which we want to map"
]
},
{
"cell_type": "code",
"execution_count": 12,
"metadata": {},
"outputs": [],
"source": [
"source = \"H\"\n",
"org = 'Homo sapiens'"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Case 1\n",
"\n",
"Here we first load the case 1 example data."
]
},
{
"cell_type": "code",
"execution_count": 21,
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
"\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" 0 | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" A1BG | \n",
"
\n",
" \n",
" | 1 | \n",
" A1CF | \n",
"
\n",
" \n",
" | 2 | \n",
" A2MP1 | \n",
"
\n",
" \n",
"
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" original | \n",
" source | \n",
" mapping | \n",
" target | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" A1BG | \n",
" HGNC | \n",
" uc002qsd.5 | \n",
" Uc | \n",
"
\n",
" \n",
" | 1 | \n",
" A1BG | \n",
" HGNC | \n",
" 8039748 | \n",
" X | \n",
"
\n",
" \n",
" | 2 | \n",
" A1BG | \n",
" HGNC | \n",
" GO:0072562 | \n",
" T | \n",
"
\n",
" \n",
" | 3 | \n",
" A1BG | \n",
" HGNC | \n",
" uc061drj.1 | \n",
" Uc | \n",
"
\n",
" \n",
" | 4 | \n",
" A1BG | \n",
" HGNC | \n",
" ILMN_2055271 | \n",
" Il | \n",
"
\n",
" \n",
" | ... | \n",
" ... | \n",
" ... | \n",
" ... | \n",
" ... | \n",
"
\n",
" \n",
" | 109 | \n",
" A2MP1 | \n",
" HGNC | \n",
" 16761106 | \n",
" X | \n",
"
\n",
" \n",
" | 110 | \n",
" A2MP1 | \n",
" HGNC | \n",
" 16761118 | \n",
" X | \n",
"
\n",
" \n",
" | 111 | \n",
" A2MP1 | \n",
" HGNC | \n",
" ENSG00000256069 | \n",
" En | \n",
"
\n",
" \n",
" | 112 | \n",
" A2MP1 | \n",
" HGNC | \n",
" A2MP1 | \n",
" H | \n",
"
\n",
" \n",
" | 113 | \n",
" A2MP1 | \n",
" HGNC | \n",
" NR_040112 | \n",
" Q | \n",
"
\n",
" \n",
"
\n",
"
114 rows × 4 columns
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" original | \n",
" source | \n",
" mapping | \n",
" target | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" A1BG | \n",
" HGNC | \n",
" uc002qsd.5 | \n",
" Uc | \n",
"
\n",
" \n",
" | 1 | \n",
" A1BG | \n",
" HGNC | \n",
" 8039748 | \n",
" X | \n",
"
\n",
" \n",
" | 2 | \n",
" A1BG | \n",
" HGNC | \n",
" GO:0072562 | \n",
" T | \n",
"
\n",
" \n",
" | 3 | \n",
" A1BG | \n",
" HGNC | \n",
" uc061drj.1 | \n",
" Uc | \n",
"
\n",
" \n",
" | 4 | \n",
" A1BG | \n",
" HGNC | \n",
" ILMN_2055271 | \n",
" Il | \n",
"
\n",
" \n",
" | ... | \n",
" ... | \n",
" ... | \n",
" ... | \n",
" ... | \n",
"
\n",
" \n",
" | 109 | \n",
" A2MP1 | \n",
" HGNC | \n",
" 16761106 | \n",
" X | \n",
"
\n",
" \n",
" | 110 | \n",
" A2MP1 | \n",
" HGNC | \n",
" 16761118 | \n",
" X | \n",
"
\n",
" \n",
" | 111 | \n",
" A2MP1 | \n",
" HGNC | \n",
" ENSG00000256069 | \n",
" En | \n",
"
\n",
" \n",
" | 112 | \n",
" A2MP1 | \n",
" HGNC | \n",
" A2MP1 | \n",
" H | \n",
"
\n",
" \n",
" | 113 | \n",
" A2MP1 | \n",
" HGNC | \n",
" NR_040112 | \n",
" Q | \n",
"
\n",
" \n",
"
\n",
"
114 rows × 4 columns
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" mapping | \n",
" local | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" ENSG00000121410 | \n",
" aa11 | \n",
"
\n",
" \n",
" | 1 | \n",
" ENSG00000148584 | \n",
" bb34 | \n",
"
\n",
" \n",
" | 2 | \n",
" ENSG00000256069 | \n",
" eg93 | \n",
"
\n",
" \n",
"
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" original | \n",
" source | \n",
" mapping | \n",
" target | \n",
" local | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" A1BG | \n",
" HGNC | \n",
" ENSG00000121410 | \n",
" En | \n",
" aa11 | \n",
"
\n",
" \n",
" | 1 | \n",
" A1CF | \n",
" HGNC | \n",
" ENSG00000148584 | \n",
" En | \n",
" bb34 | \n",
"
\n",
" \n",
" | 2 | \n",
" A2MP1 | \n",
" HGNC | \n",
" ENSG00000256069 | \n",
" En | \n",
" eg93 | \n",
"
\n",
" \n",
"
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" original | \n",
" source | \n",
" mapping | \n",
" target | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" A1BG | \n",
" HGNC | \n",
" uc002qsd.5 | \n",
" Uc | \n",
"
\n",
" \n",
" | 1 | \n",
" A1BG | \n",
" HGNC | \n",
" 8039748 | \n",
" X | \n",
"
\n",
" \n",
" | 2 | \n",
" A1BG | \n",
" HGNC | \n",
" GO:0072562 | \n",
" T | \n",
"
\n",
" \n",
" | 3 | \n",
" A1BG | \n",
" HGNC | \n",
" uc061drj.1 | \n",
" Uc | \n",
"
\n",
" \n",
" | 4 | \n",
" A1BG | \n",
" HGNC | \n",
" ILMN_2055271 | \n",
" Il | \n",
"
\n",
" \n",
" | ... | \n",
" ... | \n",
" ... | \n",
" ... | \n",
" ... | \n",
"
\n",
" \n",
" | 109 | \n",
" A2MP1 | \n",
" HGNC | \n",
" 16761106 | \n",
" X | \n",
"
\n",
" \n",
" | 110 | \n",
" A2MP1 | \n",
" HGNC | \n",
" 16761118 | \n",
" X | \n",
"
\n",
" \n",
" | 111 | \n",
" A2MP1 | \n",
" HGNC | \n",
" ENSG00000256069 | \n",
" En | \n",
"
\n",
" \n",
" | 112 | \n",
" A2MP1 | \n",
" HGNC | \n",
" A2MP1 | \n",
" H | \n",
"
\n",
" \n",
" | 113 | \n",
" A2MP1 | \n",
" HGNC | \n",
" NR_040112 | \n",
" Q | \n",
"
\n",
" \n",
"
\n",
"
114 rows × 4 columns
\n",
"