# Automatic Vestibular Schwannoma Segmentation using Deep Learning
This repository provides scripts for the segmentation of vestibular schwannoma on 3D T1 or T2 images,
based on the following papers:
* Shapey, J. et al. Segmentation of vestibular schwannoma from MRI, an open annotated dataset and baseline algorithm.
Scientific Data, 8(1), p.286. https://doi.org/10.1038/s41597-021-01064-w
* Wang, G. et al. Automatic Segmentation of Vestibular Schwannoma from T2-Weighted MRI by Deep
Spatial Attention with Hardness-Weighted Loss, MICCAI, pp 264-272, 2019.
* Shapey, J. et al. An artificial intelligence framework for automatic segmentation and volumetry of vestibular
schwannomas from contrast-enhanced T1-weighted and high-resolution T2-weighted MRI. Journal of Neurosurgery,
1(aop), pp.1-9.
## TCIA dataset
The dataset described in these publications is publicly available in The Cancer Imaging Archive (TCIA):
Shapey, J., Kujawa, A., Dorent, R., Wang, G., Bisdas, S., Dimitriadis, A., Grishchuck, D., Paddick, I., Kitchen,
N., Bradford, R., Saeed, S., Ourselin, S., & Vercauteren, T. (2021). Segmentation of Vestibular Schwannoma from
Magnetic Resonance Imaging: An Open Annotated Dataset and Baseline Algorithm [Data set]. The Cancer Imaging Archive.
https://doi.org/10.7937/TCIA.9YTJ-5Q73
Note, that one subject (VS-SEG-168) had to be removed from the data set because the data was incomplete.
TCIA offers to download the data set under a "Desciptive Folder Name" or a "Classic Folder Name". We recommend
choosing the Descriptive Folder Name and using the instruction under `prepocessing` to convert the
data set into a new data set with a more convenient folder structure and file names and to convert DICOM images and
segmentations into NIFTI files that are required as input to the automatic segmentation algorithm.
On TCIA, you can also download affine registration matrices (.tfm files) and the original segmentation contour lines
(JSON files) under the "Version 2" tab.
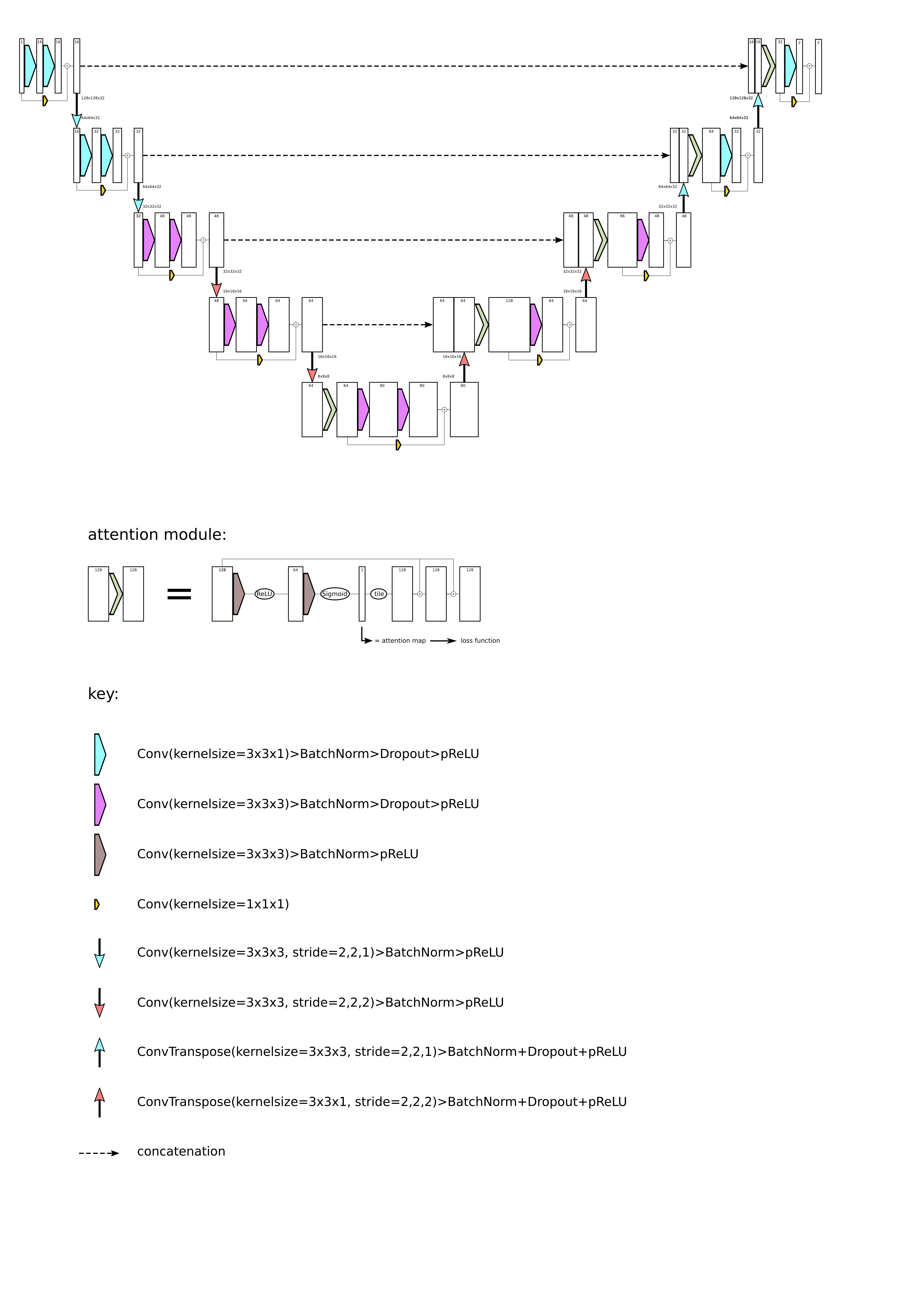
## Model
The implementation is based on [MONAI](https://monai.io/) and the network has a couple of differences
compared to the network described in the paper:
- the inputs and outputs of the two convolution+PRelu+BatchNorm modules that are found in each
layer are connected via a residual connection
- the downsampling is achieved by convolution with kernel_size=3 and stride=2, not by
maxpooling.
- each convolution (except from the convolutions of the attention module) has a dropout layer
following the PRelu activation
 ## Requirements
The following setup has been tested on Ubuntu 20.04.
* A CUDA compatible GPU with memory not less than 4GB is recommended.
* [CUDA Toolkit](https://developer.nvidia.com/cuda-toolkit). Suggested version 10.2 or above.
* [cuDNN](https://developer.nvidia.com/cudnn). Suggested version 7.6.5 or above.
* Python. Suggested version is 3.6.
The following Python libraries can be installed individually, or with a single command:
pip install -r requirements.txt
* [PyTorch](https://pytorch.org/get-started/locally/) (recommended 1.6.0)
pip install torch
* [MONAI](https://monai.io/) (recommended: 0.4.0)
pip install monai
* natsort (recommended: 7.0.1)
pip install natsort
* matplotlib (recommended: 3.3.1)
pip install matplotlib
* nibabel (recommended: 3.3.1)
pip install nibabel
* torchvision (recommended: 0.7.0)
pip install torchvision
* tensorboard (recommended: 2.3.0)
pip install tensorboard
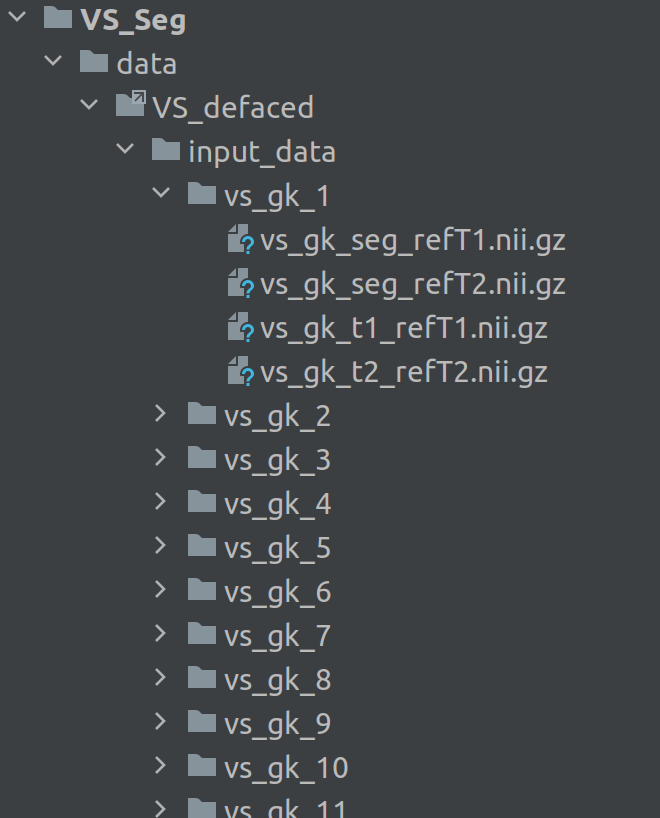
## How to store/access the data
After downloading the repository VS_Seg, create a folder VS_Seg/data and place your data in it,
e.g. the folder VS_defaced which contains the data as shown in the figure below.
## Requirements
The following setup has been tested on Ubuntu 20.04.
* A CUDA compatible GPU with memory not less than 4GB is recommended.
* [CUDA Toolkit](https://developer.nvidia.com/cuda-toolkit). Suggested version 10.2 or above.
* [cuDNN](https://developer.nvidia.com/cudnn). Suggested version 7.6.5 or above.
* Python. Suggested version is 3.6.
The following Python libraries can be installed individually, or with a single command:
pip install -r requirements.txt
* [PyTorch](https://pytorch.org/get-started/locally/) (recommended 1.6.0)
pip install torch
* [MONAI](https://monai.io/) (recommended: 0.4.0)
pip install monai
* natsort (recommended: 7.0.1)
pip install natsort
* matplotlib (recommended: 3.3.1)
pip install matplotlib
* nibabel (recommended: 3.3.1)
pip install nibabel
* torchvision (recommended: 0.7.0)
pip install torchvision
* tensorboard (recommended: 2.3.0)
pip install tensorboard
## How to store/access the data
After downloading the repository VS_Seg, create a folder VS_Seg/data and place your data in it,
e.g. the folder VS_defaced which contains the data as shown in the figure below.
 In case, the folder structure or file names are different from the suggested one, or to exclude cases follow
these instructions:
Open VS_Seg/Params/VSparams.py and make sure the function `load_T1_or_T2_data` returns `train_files, val_files,
test_files`where each of the returned variables is a list of dictionaries. Each dictionary represents an
image/label pair, where the key "image" corresponds to the path to the image file and the key "label"
corresponds to the path to the label file. `train_files` contains all dictionaries of paths to the training
images/labels, `val_files`to the validation images/labels and `test_files` to the testing images/labels.
# How to train
To start training, run the following command from a terminal in the VS_Seg repository:
python3 VS_train.py --results_folder_name $RESULTS_FOLDER_NAME 2> train_error_log.txt
optional parameters:
`--results_folder_name` followed by path to folder in which results are to be saved
`--split` followed by path to CSV file. See `params/split_TCIA.csv` as an example.
`--no_attention` removes attention module from neural network and attention maps from loss function
`--no_hardness` removes voxel hardness weighting from loss function
`--dataset T1` or `--dataset T2` to select the T1 or T2 dataset (default: T1)
`--debug` to run the algorithm on a small dataset, for only a couple of epochs
This script will create the folder VS_crop/model/results/ and the three sub-folders
* figures ⟶ will contain after training is completed:
- a png file of a central slice (center of mass slice of label) of the first
training set image after all pre-processing transformations have been applied to it, i.e. the way it is
passed into the neural network, and the corresponding label
- a png file showing training curves mean loss vs. epoch number and mean validation dice score vs.
epoch number
* logs ⟶ contains the full training log (training_log.txt). Can be used to track training process
during training.
* model ⟶ stores the model that performed best on the validation set (best_metric_model.pth)
* inferred_segmentations_nifti ⟶ contains the segmentations from inference (see next section)
Additionally, all output and error messages are logged in VS_Seg/train_error_log.txt
All model and optimizer parameters can be changed in the `__init__` function of class VSparams in
VS_Seg/Params/VSparams.py
# How to run inference
To start the training run the following command from a terminal in the VS_Seg repository:
python3 VS_inference.py --results_folder_name $RESULTS_FOLDER_NAME 2> inference_error_log.txt
and all other optional parameters have to match the values chosen in the training command.
The script will create the following sub-folders:
* figures ⟶ contains one png file per image in the test set showing the centre of mass slice (based
on ground truth label) with ground truth label and predicted label
* logs ⟶ test_log.txt contains individual and mean dice scores of test set
Additionally, all output and error messages are logged in VS_Seg/inference_error_log.txt
# Trained models
Results folders containing trained models based on T1 or T2 inputs and detailed inference results can be found here: https://doi.org/10.5281/zenodo.6323472
After downloading the folders and unzipping them, you can re-run inference with the following commands:
`python3 VS_inference.py --results_folder_name $RESUlTS_FOLDER_NAME_T1 --dataset T1`
`python3 VS_inference.py --results_folder_name $RESUlTS_FOLDER_NAME_T2 --dataset T2`
where $RESULTS_FOLDER_NAME_T1 and $RESULTS_FOLDER_NAME_T2 are the paths to the unzipped folders.
# Using Tensorboard to monitor training progress
The training algorithm will create a folder `runs` in the root folder and store the training progress data in it.
If tensorboard is installed the following command can be used to monitor the training progress:
`tensorboard --logdir ./runs`
In case, the folder structure or file names are different from the suggested one, or to exclude cases follow
these instructions:
Open VS_Seg/Params/VSparams.py and make sure the function `load_T1_or_T2_data` returns `train_files, val_files,
test_files`where each of the returned variables is a list of dictionaries. Each dictionary represents an
image/label pair, where the key "image" corresponds to the path to the image file and the key "label"
corresponds to the path to the label file. `train_files` contains all dictionaries of paths to the training
images/labels, `val_files`to the validation images/labels and `test_files` to the testing images/labels.
# How to train
To start training, run the following command from a terminal in the VS_Seg repository:
python3 VS_train.py --results_folder_name $RESULTS_FOLDER_NAME 2> train_error_log.txt
optional parameters:
`--results_folder_name` followed by path to folder in which results are to be saved
`--split` followed by path to CSV file. See `params/split_TCIA.csv` as an example.
`--no_attention` removes attention module from neural network and attention maps from loss function
`--no_hardness` removes voxel hardness weighting from loss function
`--dataset T1` or `--dataset T2` to select the T1 or T2 dataset (default: T1)
`--debug` to run the algorithm on a small dataset, for only a couple of epochs
This script will create the folder VS_crop/model/results/ and the three sub-folders
* figures ⟶ will contain after training is completed:
- a png file of a central slice (center of mass slice of label) of the first
training set image after all pre-processing transformations have been applied to it, i.e. the way it is
passed into the neural network, and the corresponding label
- a png file showing training curves mean loss vs. epoch number and mean validation dice score vs.
epoch number
* logs ⟶ contains the full training log (training_log.txt). Can be used to track training process
during training.
* model ⟶ stores the model that performed best on the validation set (best_metric_model.pth)
* inferred_segmentations_nifti ⟶ contains the segmentations from inference (see next section)
Additionally, all output and error messages are logged in VS_Seg/train_error_log.txt
All model and optimizer parameters can be changed in the `__init__` function of class VSparams in
VS_Seg/Params/VSparams.py
# How to run inference
To start the training run the following command from a terminal in the VS_Seg repository:
python3 VS_inference.py --results_folder_name $RESULTS_FOLDER_NAME 2> inference_error_log.txt
and all other optional parameters have to match the values chosen in the training command.
The script will create the following sub-folders:
* figures ⟶ contains one png file per image in the test set showing the centre of mass slice (based
on ground truth label) with ground truth label and predicted label
* logs ⟶ test_log.txt contains individual and mean dice scores of test set
Additionally, all output and error messages are logged in VS_Seg/inference_error_log.txt
# Trained models
Results folders containing trained models based on T1 or T2 inputs and detailed inference results can be found here: https://doi.org/10.5281/zenodo.6323472
After downloading the folders and unzipping them, you can re-run inference with the following commands:
`python3 VS_inference.py --results_folder_name $RESUlTS_FOLDER_NAME_T1 --dataset T1`
`python3 VS_inference.py --results_folder_name $RESUlTS_FOLDER_NAME_T2 --dataset T2`
where $RESULTS_FOLDER_NAME_T1 and $RESULTS_FOLDER_NAME_T2 are the paths to the unzipped folders.
# Using Tensorboard to monitor training progress
The training algorithm will create a folder `runs` in the root folder and store the training progress data in it.
If tensorboard is installed the following command can be used to monitor the training progress:
`tensorboard --logdir ./runs`
 ## Requirements
The following setup has been tested on Ubuntu 20.04.
* A CUDA compatible GPU with memory not less than 4GB is recommended.
* [CUDA Toolkit](https://developer.nvidia.com/cuda-toolkit). Suggested version 10.2 or above.
* [cuDNN](https://developer.nvidia.com/cudnn). Suggested version 7.6.5 or above.
* Python. Suggested version is 3.6.
The following Python libraries can be installed individually, or with a single command:
pip install -r requirements.txt
* [PyTorch](https://pytorch.org/get-started/locally/) (recommended 1.6.0)
pip install torch
* [MONAI](https://monai.io/) (recommended: 0.4.0)
pip install monai
* natsort (recommended: 7.0.1)
pip install natsort
* matplotlib (recommended: 3.3.1)
pip install matplotlib
* nibabel (recommended: 3.3.1)
pip install nibabel
* torchvision (recommended: 0.7.0)
pip install torchvision
* tensorboard (recommended: 2.3.0)
pip install tensorboard
## How to store/access the data
After downloading the repository VS_Seg, create a folder VS_Seg/data and place your data in it,
e.g. the folder VS_defaced which contains the data as shown in the figure below.
## Requirements
The following setup has been tested on Ubuntu 20.04.
* A CUDA compatible GPU with memory not less than 4GB is recommended.
* [CUDA Toolkit](https://developer.nvidia.com/cuda-toolkit). Suggested version 10.2 or above.
* [cuDNN](https://developer.nvidia.com/cudnn). Suggested version 7.6.5 or above.
* Python. Suggested version is 3.6.
The following Python libraries can be installed individually, or with a single command:
pip install -r requirements.txt
* [PyTorch](https://pytorch.org/get-started/locally/) (recommended 1.6.0)
pip install torch
* [MONAI](https://monai.io/) (recommended: 0.4.0)
pip install monai
* natsort (recommended: 7.0.1)
pip install natsort
* matplotlib (recommended: 3.3.1)
pip install matplotlib
* nibabel (recommended: 3.3.1)
pip install nibabel
* torchvision (recommended: 0.7.0)
pip install torchvision
* tensorboard (recommended: 2.3.0)
pip install tensorboard
## How to store/access the data
After downloading the repository VS_Seg, create a folder VS_Seg/data and place your data in it,
e.g. the folder VS_defaced which contains the data as shown in the figure below.
 In case, the folder structure or file names are different from the suggested one, or to exclude cases follow
these instructions:
Open VS_Seg/Params/VSparams.py and make sure the function `load_T1_or_T2_data` returns `train_files, val_files,
test_files`where each of the returned variables is a list of dictionaries. Each dictionary represents an
image/label pair, where the key "image" corresponds to the path to the image file and the key "label"
corresponds to the path to the label file. `train_files` contains all dictionaries of paths to the training
images/labels, `val_files`to the validation images/labels and `test_files` to the testing images/labels.
# How to train
To start training, run the following command from a terminal in the VS_Seg repository:
python3 VS_train.py --results_folder_name $RESULTS_FOLDER_NAME 2> train_error_log.txt
optional parameters:
`--results_folder_name` followed by path to folder in which results are to be saved
`--split` followed by path to CSV file. See `params/split_TCIA.csv` as an example.
`--no_attention` removes attention module from neural network and attention maps from loss function
`--no_hardness` removes voxel hardness weighting from loss function
`--dataset T1` or `--dataset T2` to select the T1 or T2 dataset (default: T1)
`--debug` to run the algorithm on a small dataset, for only a couple of epochs
This script will create the folder VS_crop/model/results/
In case, the folder structure or file names are different from the suggested one, or to exclude cases follow
these instructions:
Open VS_Seg/Params/VSparams.py and make sure the function `load_T1_or_T2_data` returns `train_files, val_files,
test_files`where each of the returned variables is a list of dictionaries. Each dictionary represents an
image/label pair, where the key "image" corresponds to the path to the image file and the key "label"
corresponds to the path to the label file. `train_files` contains all dictionaries of paths to the training
images/labels, `val_files`to the validation images/labels and `test_files` to the testing images/labels.
# How to train
To start training, run the following command from a terminal in the VS_Seg repository:
python3 VS_train.py --results_folder_name $RESULTS_FOLDER_NAME 2> train_error_log.txt
optional parameters:
`--results_folder_name` followed by path to folder in which results are to be saved
`--split` followed by path to CSV file. See `params/split_TCIA.csv` as an example.
`--no_attention` removes attention module from neural network and attention maps from loss function
`--no_hardness` removes voxel hardness weighting from loss function
`--dataset T1` or `--dataset T2` to select the T1 or T2 dataset (default: T1)
`--debug` to run the algorithm on a small dataset, for only a couple of epochs
This script will create the folder VS_crop/model/results/