



Enhancing Spatiotemporal Disease Progression Models via Latent Diffusion and Prior Knowledge
https://github.com/user-attachments/assets/28ad3693-5e3e-4f6e-9bbc-485424fbbee2
Installation •
Training •
CLI Application •
Cite
**NEWS**
* 🎉 BrLP has been awarded the runner-up position for the Media Best Paper Award at MICCAI 2025!
* 🎉 Our [extension paper](https://arxiv.org/pdf/2502.08560) has been accepted for publication in Medical Image Analysis (*IF=11.8*)
* 🎉 A [new paper](https://www.spiedigitallibrary.org/conference-proceedings-of-spie/13406/1340621/A-technical-assessment-of-latent-diffusion-for-Alzheimers-disease-progression/10.1117/12.3047135.short) from Vanderbilt University has replicated our results on the [BLSA](https://www.blsa.nih.gov/) dataset!
* 🆕 The [short guide on using the BrLP CLI](https://lemuelpuglisi.github.io/blog/2024/brlp/) is out!
* 🎉 BrLP has been nominated and shortlisted for the MICCAI Best Paper Award! (top <1%)
* 🎉 BrLP has been early-accepted and selected for **oral presentation** at [MICCAI 2024](https://conferences.miccai.org/2024/en/) (top 4%)!
## Table of Contents
- [Installation](#installation)
- [Data preparation](./REPR-DATA.md)
- [Training](#training)
- [Pretrained models](#pretrained-models)
- [Acknowledgements](#acknowledgements)
- [Citing our work](#citing)
## Installation
Download the repository, `cd` into the project folder and install the `brlp` package:
```console
pip install -e .
```
We recommend using a separate environment (see [Anaconda](https://www.anaconda.com/)). The code has been tested using python 3.9, however we expect it to work also with newer versions.
## Data preparation
Check out our document on [*Data preparation and study reproducibility*](./REPR-DATA.md). This file will guide you in organizing your data and creating the required CSV files to run the training pipelines.
## Training
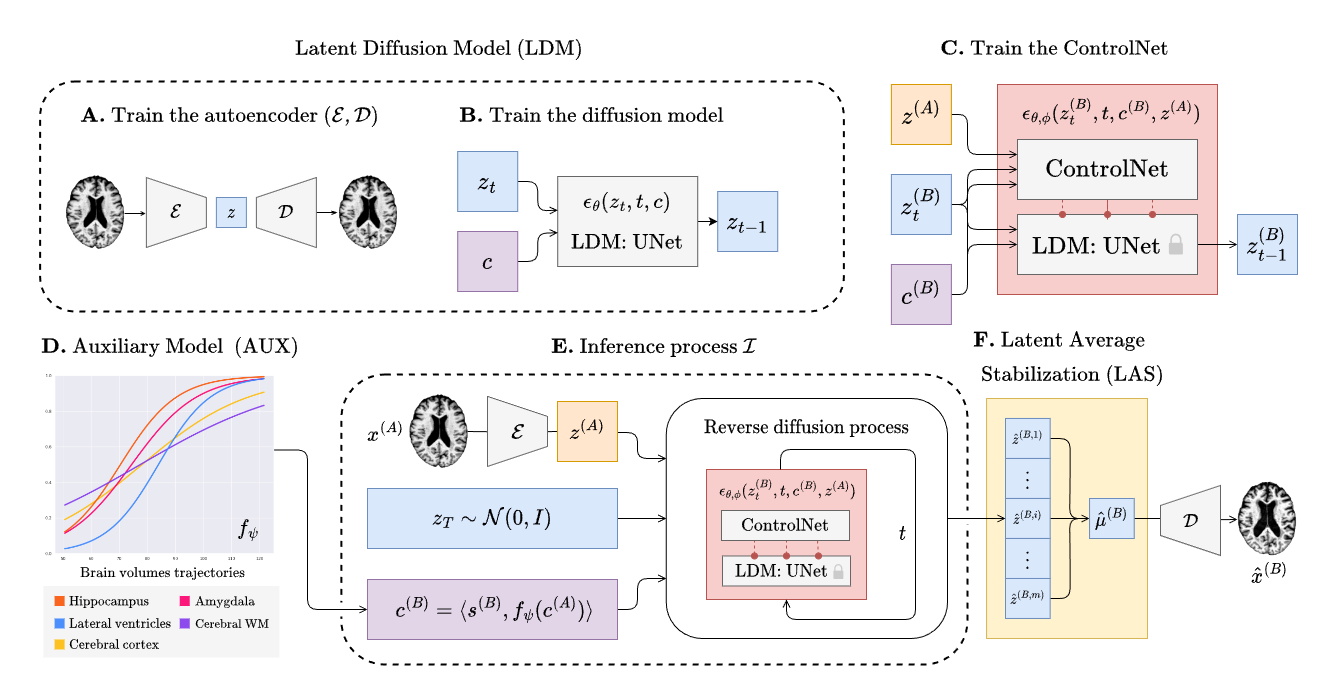
Training BrLP has 3 main phases that will be discribed in the subsequent sections. Every training (except for the auxiliary model) can be monitored using `tensorboard` as follows:
```console
tensorboard --logdir runs
```
### Train the autoencoder
Follow the commands below to train the autoencoder.
```console
# Create an output and a cache directory
mkdir ae_output ae_cache
# Run the training script
python scripts/training/train_autoencoder.py \
--dataset_csv /path/to/A.csv \
--cache_dir ./ae_cache \
--output_dir ./ae_output
```
Then extract the latents from your MRI data:
```console
python scripts/prepare/extract_latents.py \
--dataset_csv /path/to/A.csv \
--aekl_ckpt ae_output/autoencoder-ep-XXX.pth
```
Replace `XXX` to select the autoencoder checkpoints of your choice.
### Train the UNet
Follow the commands below to train the diffusion UNet. Replace `XXX` to select the autoencoder checkpoints of your choice.
```console
# Create an output and a cache directory:
mkdir unet_output unet_cache
# Run the training script
python scripts/training/train_diffusion_unet.py \
--dataset_csv /path/to/A.csv \
--cache_dir unet_cache \
--output_dir unet_output \
--aekl_ckpt ae_output/autoencoder-ep-XXX.pth
```
### Train the ControlNet
Follow the commands below to train the ControlNet. Replace `XXX` to select the autoencoder and UNet checkpoints of your choice.
```console
# Create an output and a cache directory:
mkdir cnet_output cnet_cache
# Run the training script
python scripts/training/train_controlnet.py \
--dataset_csv /path/to/B.csv \
--cache_dir unet_cache \
--output_dir unet_output \
--aekl_ckpt ae_output/autoencoder-ep-XXX.pth \
--diff_ckpt unet_output/unet-ep-XXX.pth
```
### Auxiliary models
Follow the commands below to train the DCM auxiliary model.
```console
# Create an output directory
mkdir aux_output
# Run the training script
python scripts/training/train_aux.py \
--dataset_csv /path/to/A.csv \
--output_path aux_output
```
We emphasize that any disease progression model capable of predicting volumetric changes over time is also viable as an auxiliary model for BrLP.
## Inference
Our package comes with a `brlp` command to use BrLP for inference. Check:
```console
brlp --help
```
The `--input` parameter requires a CSV file where you list all available data for your subjects. For an example, check to `examples/input.example.csv`. If you haven't segmented your input scans, `brlp` can perform this task for you using [SynthSeg](https://surfer.nmr.mgh.harvard.edu/fswiki/SynthSeg), but it requires that FreeSurfer >= 7.4 be installed. The `--confs` parameter specifies the paths to the models and other inference parameters, such as LAS $m$. For an example, check `examples/confs.example.yaml`.
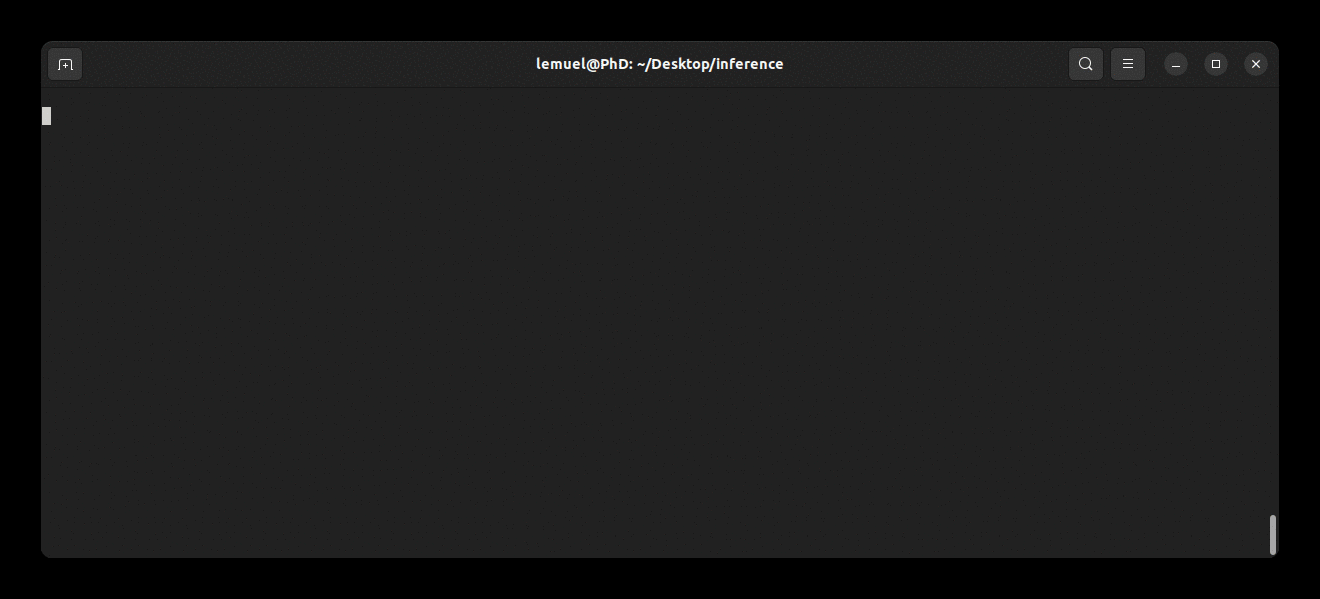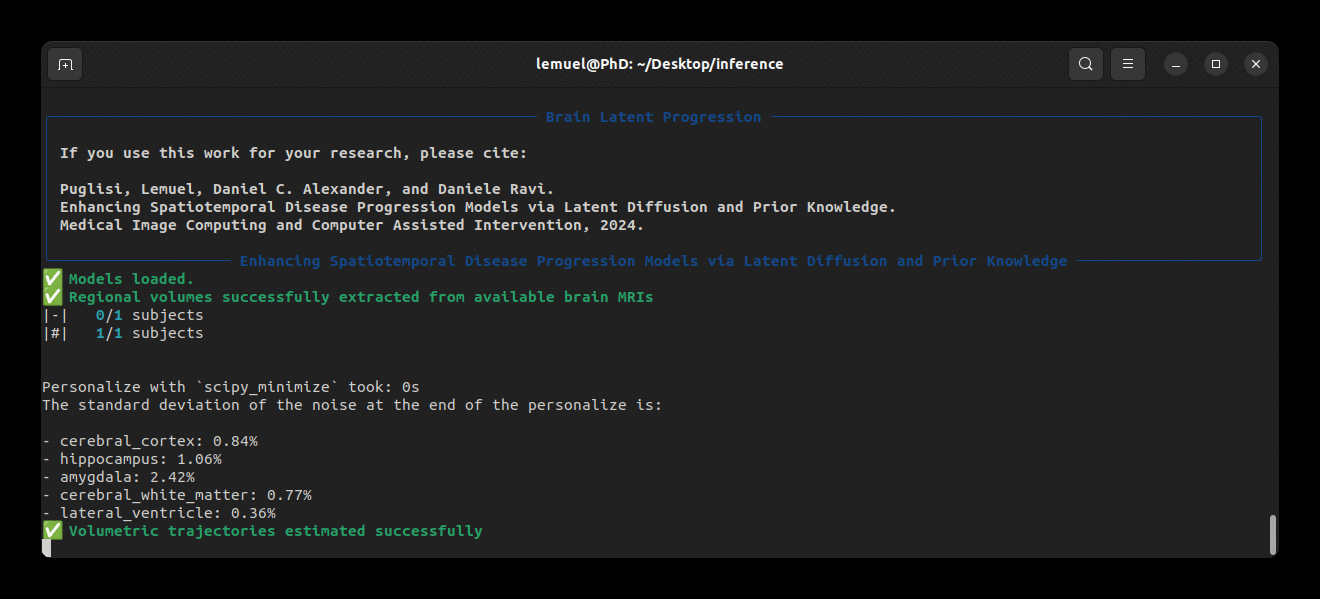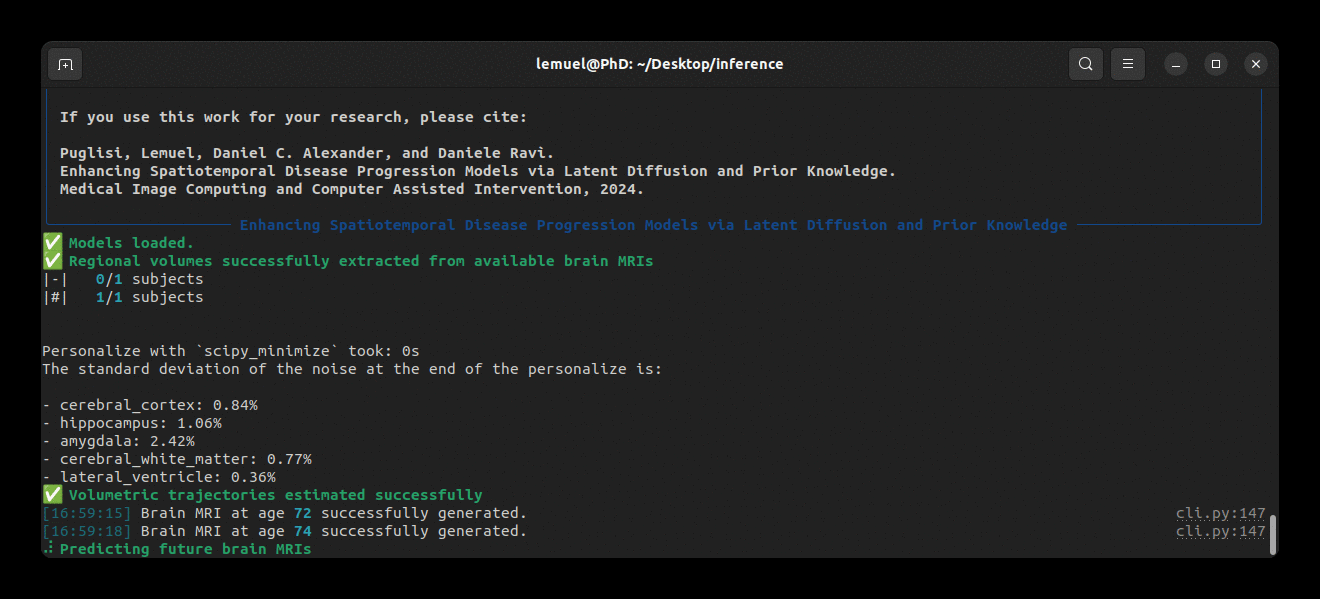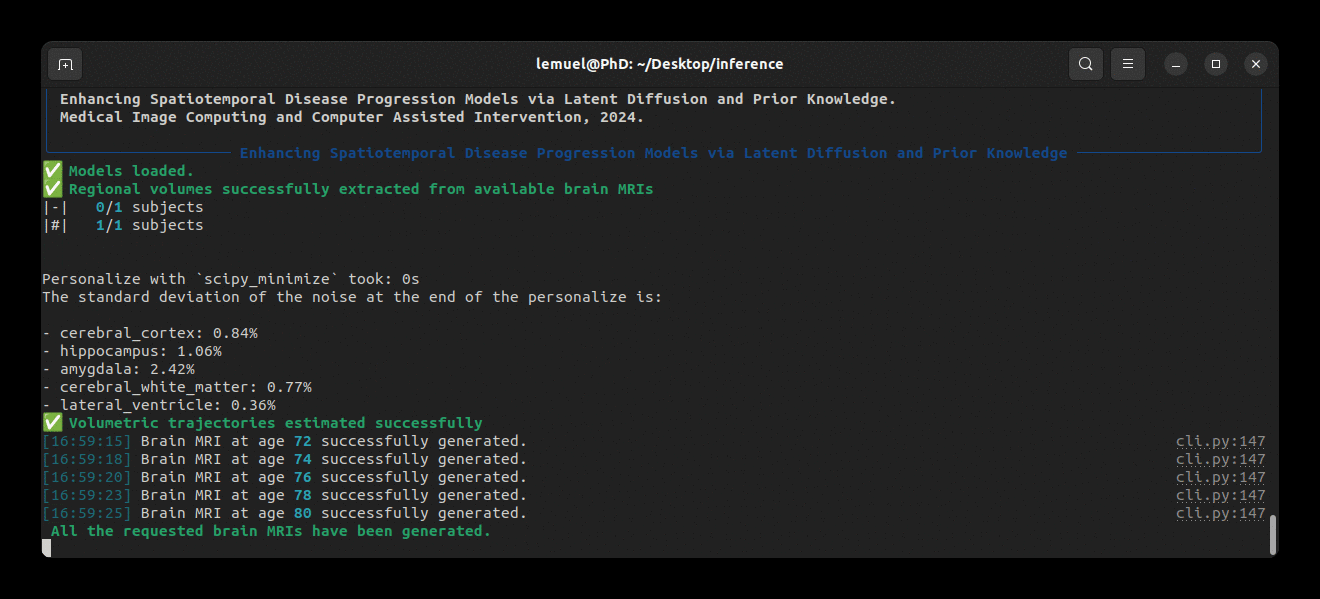
Running the program looks like this:
## Pretrained models
Download the pre-trained models for BrLP:
| Model | Weights URL |
| ---------------------- | ------------------------------------------------------------ |
| Autoencoder | [link](https://studentiunict-my.sharepoint.com/:u:/g/personal/pgllml99h18c351e_studium_unict_it/EUxUFIQtkQ1EvBqojYA5BAYByIyHbttca5Mx1cU4bC6q3A?e=sCXSUA) |
| Diffusion Model UNet | [link](https://studentiunict-my.sharepoint.com/:u:/g/personal/pgllml99h18c351e_studium_unict_it/EQT7KJTtfmRAguf8_utWeJIBUJPsRRgPZlt94s2vNbwVFw?e=IjHnx7) |
| ControlNet | [link](https://studentiunict-my.sharepoint.com/:u:/g/personal/pgllml99h18c351e_studium_unict_it/EYtVvH47dFJJnH8gtwSMA-MB8c3pm4_Z9g5F_IG1OKxW9Q?e=CzvGT4) |
| Auxiliary Models (DCM) | [link](https://studentiunict-my.sharepoint.com/:u:/g/personal/pgllml99h18c351e_studium_unict_it/EXJDQqLNCwBFkt2J6zg1kpwBS_1hAZoBfGy5AfcGOBZvHQ?e=Z05kOG) |
## Acknowledgements
We thank the maintainers of open-source libraries for their contributions to accelerating the research process, with a special mention of [MONAI](https://monai.io/) and its [GenerativeModels](https://github.com/Project-MONAI/GenerativeModels/tree/main) extension.
## Citing
Medical Image Analysis:
```bib
@article{puglisi2025brain,
title={Brain latent progression: Individual-based spatiotemporal disease progression on 3D brain MRIs via latent diffusion},
author={Puglisi, Lemuel and Alexander, Daniel C and Rav{\`\i}, Daniele},
journal={Medical Image Analysis},
year={2025}
}
```
MICCAI 2024 proceedings:
```bib
@inproceedings{puglisi2024enhancing,
title={Enhancing spatiotemporal disease progression models via latent diffusion and prior knowledge},
author={Puglisi, Lemuel and Alexander, Daniel C and Rav{\`\i}, Daniele},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={173--183},
year={2024},
organization={Springer}
}
```
SPIE Medical Imaging 2025 proceedings:
```bib
@inproceedings{mcmaster2025technical,
title={A technical assessment of latent diffusion for Alzheimer's disease progression},
author={McMaster, Elyssa and Puglisi, Lemuel and Gao, Chenyu and Krishnan, Aravind R and Saunders, Adam M and Ravi, Daniele and Beason-Held, Lori L and Resnick, Susan M and Zuo, Lianrui and Moyer, Daniel and others},
booktitle={Medical Imaging 2025: Image Processing},
volume={13406},
pages={505--513},
year={2025},
organization={SPIE}
}
```


