{
"cells": [
{
"cell_type": "markdown",
"id": "abd678aa-fb2a-47b7-bd31-2123bbea17e4",
"metadata": {},
"source": [
"# Replication: Agosta *et al*, 2013\n",
"\n",
"## Introduction\n",
"\n",
"This notebook attempts to replicate the following paper with the [PPMI](http://ppmi-info.org) dataset:\n",
"\n",
""
]
},
{
"cell_type": "markdown",
"id": "ab86edb8-d5ab-402a-b133-2a77530600a4",
"metadata": {},
"source": [
" "
]
},
{
"cell_type": "markdown",
"id": "9189c63b-94ad-469b-b9d0-328eb52cc8b8",
"metadata": {},
"source": [
"## Initial setup"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "52b4e9c2-4f75-43bc-b956-dbe9f1ba608d",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Installing notebook dependencies (see log in install.log)... \n",
"This notebook was run on 2022-07-21 15:38:37 UTC +0000\n"
]
}
],
"source": [
"import livingpark_utils\n",
"\n",
"utils = livingpark_utils.LivingParkUtils(\"agosta-etal\")\n",
"utils.notebook_init()\n",
"random_seed = 1"
]
},
{
"cell_type": "markdown",
"id": "ff5646ca-49a8-4072-b28c-cf8a82debb42",
"metadata": {
"tags": []
},
"source": [
"## PPMI cohort preparation"
]
},
{
"cell_type": "markdown",
"id": "003cc8c9-96aa-4772-af38-fb769c4fdff3",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"### Study data download"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "be13ea39-957e-440f-b27b-32113c81f6a0",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"required_files = [\n",
" \"iu_genetic_consensus_20220310.csv\",\n",
" \"Cognitive_Categorization.csv\",\n",
" \"Primary_Clinical_Diagnosis.csv\",\n",
" \"Demographics.csv\", # SEX\n",
" \"Socio-Economics.csv\", # EDUCYRS\n",
" \"PD_Diagnosis_History.csv\", # Disease duration\n",
" \"Montreal_Cognitive_Assessment__MoCA_.csv\",\n",
" \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\", # STROKE\n",
"]\n",
"\n",
"utils.download_ppmi_metadata(required_files)"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "a7719ba4-28c1-4b6e-a8a7-08e163c2dd94",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MDS_UPDRS_Part_III_clean.csv is now available\n",
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MRI_info.csv is now available\n"
]
}
],
"source": [
"# H&Y scores\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-treatment-and-on-off-status/main/PPMI medication and ON-OFF status.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"PPMI medication and ON-OFF status.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MRI_info.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-MRI-metadata/main/MRI metadata.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"MRI metadata.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")"
]
},
{
"cell_type": "markdown",
"id": "01671975-07d0-48a0-8774-4349abc3f090",
"metadata": {
"tags": []
},
"source": [
"### Inclusion criteria\n",
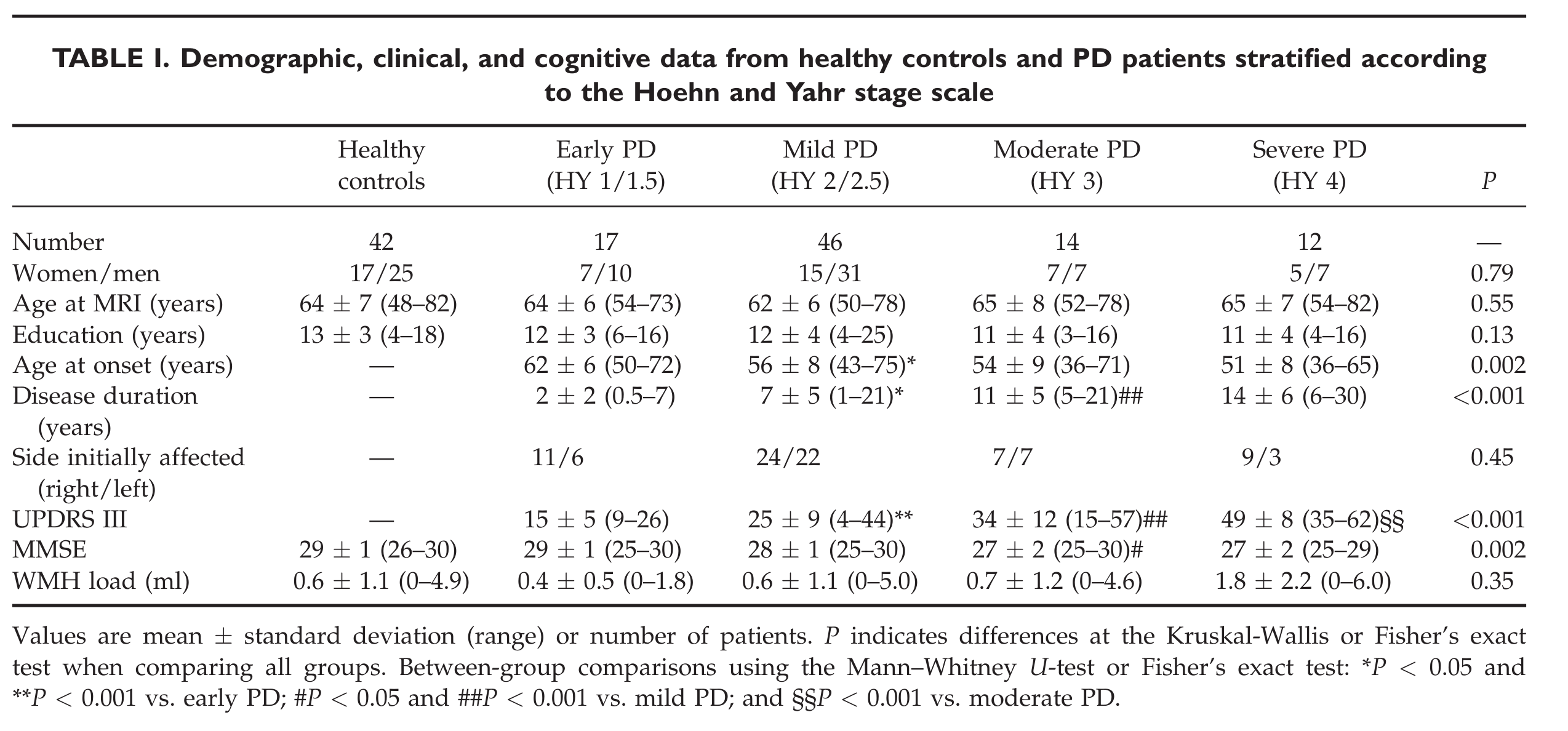
"\n",
"We obtain the following group sizes:\n",
"\n",
""
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "7984652c-62b1-465f-bfdb-dccbb9191dcb",
"metadata": {},
"outputs": [],
"source": [
"# H&Y during OFF time\n",
"# Exclusion criteria: Patients were excluded if they had:\n",
"# (a) parkin, leucine-rich repeat kinase 2 (LRRK2), and glu-\n",
"# cocerebrosidase (GBA) gene mutations; OK\n",
"# (b) cerebrovascular\n",
"# disorders, traumatic brain injury history, or intracranial\n",
"# mass; only stroke found in\n",
"#\n",
"# (c) other major neurological and medical diseases;\n",
"# (d) dementia: OK\n",
"\n",
"# DARTEL: 8-mm full width\n",
"# at half maximum (FWHM) Gaussian filter"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "b7589aaf-1a2f-411a-afbe-6aa8840fdddd",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"import pandas as pd\n",
"\n",
"# UPDRS3\n",
"updrs3 = pd.read_csv(op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PDSTATE\", \"NHY\", \"NP3TOT\"]\n",
"]\n",
"# Genetics\n",
"genetics = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"iu_genetic_consensus_20220310.csv\")\n",
")[[\"PATNO\", \"GBA_PATHVAR\", \"LRRK2_PATHVAR\", \"NOTES\"]]\n",
"# Cognitive Categorization\n",
"cog_cat = pd.read_csv(op.join(utils.study_files_dir, \"Cognitive_Categorization.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"COGSTATE\"]\n",
"]\n",
"# Diagnosis\n",
"diag = pd.read_csv(op.join(utils.study_files_dir, \"Primary_Clinical_Diagnosis.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PRIMDIAG\", \"OTHNEURO\"]\n",
"]\n",
"# MRI\n",
"mri = pd.read_csv(op.join(utils.study_files_dir, \"MRI_info.csv\"))[\n",
" [\"Subject ID\", \"Visit code\", \"Description\", \"Age\"]\n",
"]\n",
"mri.rename(columns={\"Subject ID\": \"PATNO\", \"Visit code\": \"EVENT_ID\"}, inplace=True)\n",
"# Demographics\n",
"demographics = pd.read_csv(op.join(utils.study_files_dir, \"Demographics.csv\"))[\n",
" [\"PATNO\", \"SEX\"]\n",
"]\n",
"# Soci-economics\n",
"socio_eco = pd.read_csv(op.join(utils.study_files_dir, \"Socio-Economics.csv\"))[\n",
" [\"PATNO\", \"EDUCYRS\"]\n",
"]\n",
"# Disease duration\n",
"disease_dur = utils.disease_duration()\n",
"# MoCA\n",
"moca = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"Montreal_Cognitive_Assessment__MoCA_.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"MCATOT\"]]\n",
"moca[\"MMSETOT\"] = moca[\"MCATOT\"].apply(utils.moca2mmse)\n",
"# Stroke\n",
"rem = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"STROKE\"]]"
]
},
{
"cell_type": "code",
"execution_count": 6,
"id": "7bdfd288-bf39-44c5-a194-a3baa873be64",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Number of controls: 46\n",
"Number of PD subjects: 125\n",
"Number of PD subject visits by H&Y score:\n"
]
},
{
"data": {
"text/html": [
"
"
]
},
{
"cell_type": "markdown",
"id": "9189c63b-94ad-469b-b9d0-328eb52cc8b8",
"metadata": {},
"source": [
"## Initial setup"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "52b4e9c2-4f75-43bc-b956-dbe9f1ba608d",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Installing notebook dependencies (see log in install.log)... \n",
"This notebook was run on 2022-07-21 15:38:37 UTC +0000\n"
]
}
],
"source": [
"import livingpark_utils\n",
"\n",
"utils = livingpark_utils.LivingParkUtils(\"agosta-etal\")\n",
"utils.notebook_init()\n",
"random_seed = 1"
]
},
{
"cell_type": "markdown",
"id": "ff5646ca-49a8-4072-b28c-cf8a82debb42",
"metadata": {
"tags": []
},
"source": [
"## PPMI cohort preparation"
]
},
{
"cell_type": "markdown",
"id": "003cc8c9-96aa-4772-af38-fb769c4fdff3",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"### Study data download"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "be13ea39-957e-440f-b27b-32113c81f6a0",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"required_files = [\n",
" \"iu_genetic_consensus_20220310.csv\",\n",
" \"Cognitive_Categorization.csv\",\n",
" \"Primary_Clinical_Diagnosis.csv\",\n",
" \"Demographics.csv\", # SEX\n",
" \"Socio-Economics.csv\", # EDUCYRS\n",
" \"PD_Diagnosis_History.csv\", # Disease duration\n",
" \"Montreal_Cognitive_Assessment__MoCA_.csv\",\n",
" \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\", # STROKE\n",
"]\n",
"\n",
"utils.download_ppmi_metadata(required_files)"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "a7719ba4-28c1-4b6e-a8a7-08e163c2dd94",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MDS_UPDRS_Part_III_clean.csv is now available\n",
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MRI_info.csv is now available\n"
]
}
],
"source": [
"# H&Y scores\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-treatment-and-on-off-status/main/PPMI medication and ON-OFF status.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"PPMI medication and ON-OFF status.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MRI_info.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-MRI-metadata/main/MRI metadata.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"MRI metadata.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")"
]
},
{
"cell_type": "markdown",
"id": "01671975-07d0-48a0-8774-4349abc3f090",
"metadata": {
"tags": []
},
"source": [
"### Inclusion criteria\n",
"\n",
"We obtain the following group sizes:\n",
"\n",
""
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "7984652c-62b1-465f-bfdb-dccbb9191dcb",
"metadata": {},
"outputs": [],
"source": [
"# H&Y during OFF time\n",
"# Exclusion criteria: Patients were excluded if they had:\n",
"# (a) parkin, leucine-rich repeat kinase 2 (LRRK2), and glu-\n",
"# cocerebrosidase (GBA) gene mutations; OK\n",
"# (b) cerebrovascular\n",
"# disorders, traumatic brain injury history, or intracranial\n",
"# mass; only stroke found in\n",
"#\n",
"# (c) other major neurological and medical diseases;\n",
"# (d) dementia: OK\n",
"\n",
"# DARTEL: 8-mm full width\n",
"# at half maximum (FWHM) Gaussian filter"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "b7589aaf-1a2f-411a-afbe-6aa8840fdddd",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"import pandas as pd\n",
"\n",
"# UPDRS3\n",
"updrs3 = pd.read_csv(op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PDSTATE\", \"NHY\", \"NP3TOT\"]\n",
"]\n",
"# Genetics\n",
"genetics = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"iu_genetic_consensus_20220310.csv\")\n",
")[[\"PATNO\", \"GBA_PATHVAR\", \"LRRK2_PATHVAR\", \"NOTES\"]]\n",
"# Cognitive Categorization\n",
"cog_cat = pd.read_csv(op.join(utils.study_files_dir, \"Cognitive_Categorization.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"COGSTATE\"]\n",
"]\n",
"# Diagnosis\n",
"diag = pd.read_csv(op.join(utils.study_files_dir, \"Primary_Clinical_Diagnosis.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PRIMDIAG\", \"OTHNEURO\"]\n",
"]\n",
"# MRI\n",
"mri = pd.read_csv(op.join(utils.study_files_dir, \"MRI_info.csv\"))[\n",
" [\"Subject ID\", \"Visit code\", \"Description\", \"Age\"]\n",
"]\n",
"mri.rename(columns={\"Subject ID\": \"PATNO\", \"Visit code\": \"EVENT_ID\"}, inplace=True)\n",
"# Demographics\n",
"demographics = pd.read_csv(op.join(utils.study_files_dir, \"Demographics.csv\"))[\n",
" [\"PATNO\", \"SEX\"]\n",
"]\n",
"# Soci-economics\n",
"socio_eco = pd.read_csv(op.join(utils.study_files_dir, \"Socio-Economics.csv\"))[\n",
" [\"PATNO\", \"EDUCYRS\"]\n",
"]\n",
"# Disease duration\n",
"disease_dur = utils.disease_duration()\n",
"# MoCA\n",
"moca = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"Montreal_Cognitive_Assessment__MoCA_.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"MCATOT\"]]\n",
"moca[\"MMSETOT\"] = moca[\"MCATOT\"].apply(utils.moca2mmse)\n",
"# Stroke\n",
"rem = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"STROKE\"]]"
]
},
{
"cell_type": "code",
"execution_count": 6,
"id": "7bdfd288-bf39-44c5-a194-a3baa873be64",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Number of controls: 46\n",
"Number of PD subjects: 125\n",
"Number of PD subject visits by H&Y score:\n"
]
},
{
"data": {
"text/html": [
"\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" PATNO | \n",
"
\n",
" \n",
" | NHY | \n",
" | \n",
"
\n",
" \n",
" \n",
" \n",
" | 1 | \n",
" 157 | \n",
"
\n",
" \n",
" | 2 | \n",
" 537 | \n",
"
\n",
" \n",
" | 3 | \n",
" 34 | \n",
"
\n",
" \n",
" | 4 | \n",
" 6 | \n",
"
\n",
" \n",
"
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" Healthy controls | \n",
" HY 1 | \n",
" HY 2 | \n",
" HY 3 | \n",
" HY 4 | \n",
"
\n",
" \n",
" \n",
" \n",
" | Number | \n",
" 42 | \n",
" 17 | \n",
" 46 | \n",
" 9 | \n",
" 2 | \n",
"
\n",
" \n",
" | Women / men | \n",
" 14 / 28 | \n",
" 7 / 10 | \n",
" 15 / 31 | \n",
" 4 / 5 | \n",
" 1 / 1 | \n",
"
\n",
" \n",
" | Age (years) | \n",
" 61 +/- 12 (33-83) | \n",
" 62 +/- 10 (40-79) | \n",
" 63 +/- 10 (42-78) | \n",
" 69 +/- 8 (55-80) | \n",
" 69 +/- 6 (64-73) | \n",
"
\n",
" \n",
" | Education (years) | \n",
" 16 +/- 3 (12-24) | \n",
" 14 +/- 3 (8-20) | \n",
" 15 +/- 3 (9-22) | \n",
" 14 +/- 3 (8-18) | \n",
" 20 +/- nan (20-20) | \n",
"
\n",
" \n",
" | Disease duration (years) | \n",
" — | \n",
" 4 +/- 3 (0-10) | \n",
" 4 +/- 3 (0-10) | \n",
" 4 +/- 2 (0-7) | \n",
" 2 +/- 2 (1-4) | \n",
"
\n",
" \n",
" | UPDRS III | \n",
" — | \n",
" 14 +/- 5 (6-22) | \n",
" 26 +/- 10 (9-51) | \n",
" 40 +/- 9 (29-56) | \n",
" nan +/- nan (nan-nan) | \n",
"
\n",
" \n",
" | MMSE | \n",
" 30 +/- 1 (28-30) | \n",
" 29 +/- 1 (26-30) | \n",
" 29 +/- 2 (22-30) | \n",
" 28 +/- 2 (26-30) | \n",
" 29 +/- 0 (29-29) | \n",
"
\n",
" \n",
"
\n",
"
 "
]
},
{
"cell_type": "markdown",
"id": "9189c63b-94ad-469b-b9d0-328eb52cc8b8",
"metadata": {},
"source": [
"## Initial setup"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "52b4e9c2-4f75-43bc-b956-dbe9f1ba608d",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Installing notebook dependencies (see log in install.log)... \n",
"This notebook was run on 2022-07-21 15:38:37 UTC +0000\n"
]
}
],
"source": [
"import livingpark_utils\n",
"\n",
"utils = livingpark_utils.LivingParkUtils(\"agosta-etal\")\n",
"utils.notebook_init()\n",
"random_seed = 1"
]
},
{
"cell_type": "markdown",
"id": "ff5646ca-49a8-4072-b28c-cf8a82debb42",
"metadata": {
"tags": []
},
"source": [
"## PPMI cohort preparation"
]
},
{
"cell_type": "markdown",
"id": "003cc8c9-96aa-4772-af38-fb769c4fdff3",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"### Study data download"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "be13ea39-957e-440f-b27b-32113c81f6a0",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"required_files = [\n",
" \"iu_genetic_consensus_20220310.csv\",\n",
" \"Cognitive_Categorization.csv\",\n",
" \"Primary_Clinical_Diagnosis.csv\",\n",
" \"Demographics.csv\", # SEX\n",
" \"Socio-Economics.csv\", # EDUCYRS\n",
" \"PD_Diagnosis_History.csv\", # Disease duration\n",
" \"Montreal_Cognitive_Assessment__MoCA_.csv\",\n",
" \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\", # STROKE\n",
"]\n",
"\n",
"utils.download_ppmi_metadata(required_files)"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "a7719ba4-28c1-4b6e-a8a7-08e163c2dd94",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MDS_UPDRS_Part_III_clean.csv is now available\n",
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MRI_info.csv is now available\n"
]
}
],
"source": [
"# H&Y scores\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-treatment-and-on-off-status/main/PPMI medication and ON-OFF status.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"PPMI medication and ON-OFF status.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MRI_info.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-MRI-metadata/main/MRI metadata.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"MRI metadata.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")"
]
},
{
"cell_type": "markdown",
"id": "01671975-07d0-48a0-8774-4349abc3f090",
"metadata": {
"tags": []
},
"source": [
"### Inclusion criteria\n",
"\n",
"We obtain the following group sizes:\n",
"\n",
""
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "7984652c-62b1-465f-bfdb-dccbb9191dcb",
"metadata": {},
"outputs": [],
"source": [
"# H&Y during OFF time\n",
"# Exclusion criteria: Patients were excluded if they had:\n",
"# (a) parkin, leucine-rich repeat kinase 2 (LRRK2), and glu-\n",
"# cocerebrosidase (GBA) gene mutations; OK\n",
"# (b) cerebrovascular\n",
"# disorders, traumatic brain injury history, or intracranial\n",
"# mass; only stroke found in\n",
"#\n",
"# (c) other major neurological and medical diseases;\n",
"# (d) dementia: OK\n",
"\n",
"# DARTEL: 8-mm full width\n",
"# at half maximum (FWHM) Gaussian filter"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "b7589aaf-1a2f-411a-afbe-6aa8840fdddd",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"import pandas as pd\n",
"\n",
"# UPDRS3\n",
"updrs3 = pd.read_csv(op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PDSTATE\", \"NHY\", \"NP3TOT\"]\n",
"]\n",
"# Genetics\n",
"genetics = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"iu_genetic_consensus_20220310.csv\")\n",
")[[\"PATNO\", \"GBA_PATHVAR\", \"LRRK2_PATHVAR\", \"NOTES\"]]\n",
"# Cognitive Categorization\n",
"cog_cat = pd.read_csv(op.join(utils.study_files_dir, \"Cognitive_Categorization.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"COGSTATE\"]\n",
"]\n",
"# Diagnosis\n",
"diag = pd.read_csv(op.join(utils.study_files_dir, \"Primary_Clinical_Diagnosis.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PRIMDIAG\", \"OTHNEURO\"]\n",
"]\n",
"# MRI\n",
"mri = pd.read_csv(op.join(utils.study_files_dir, \"MRI_info.csv\"))[\n",
" [\"Subject ID\", \"Visit code\", \"Description\", \"Age\"]\n",
"]\n",
"mri.rename(columns={\"Subject ID\": \"PATNO\", \"Visit code\": \"EVENT_ID\"}, inplace=True)\n",
"# Demographics\n",
"demographics = pd.read_csv(op.join(utils.study_files_dir, \"Demographics.csv\"))[\n",
" [\"PATNO\", \"SEX\"]\n",
"]\n",
"# Soci-economics\n",
"socio_eco = pd.read_csv(op.join(utils.study_files_dir, \"Socio-Economics.csv\"))[\n",
" [\"PATNO\", \"EDUCYRS\"]\n",
"]\n",
"# Disease duration\n",
"disease_dur = utils.disease_duration()\n",
"# MoCA\n",
"moca = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"Montreal_Cognitive_Assessment__MoCA_.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"MCATOT\"]]\n",
"moca[\"MMSETOT\"] = moca[\"MCATOT\"].apply(utils.moca2mmse)\n",
"# Stroke\n",
"rem = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"STROKE\"]]"
]
},
{
"cell_type": "code",
"execution_count": 6,
"id": "7bdfd288-bf39-44c5-a194-a3baa873be64",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Number of controls: 46\n",
"Number of PD subjects: 125\n",
"Number of PD subject visits by H&Y score:\n"
]
},
{
"data": {
"text/html": [
"
"
]
},
{
"cell_type": "markdown",
"id": "9189c63b-94ad-469b-b9d0-328eb52cc8b8",
"metadata": {},
"source": [
"## Initial setup"
]
},
{
"cell_type": "code",
"execution_count": 1,
"id": "52b4e9c2-4f75-43bc-b956-dbe9f1ba608d",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Installing notebook dependencies (see log in install.log)... \n",
"This notebook was run on 2022-07-21 15:38:37 UTC +0000\n"
]
}
],
"source": [
"import livingpark_utils\n",
"\n",
"utils = livingpark_utils.LivingParkUtils(\"agosta-etal\")\n",
"utils.notebook_init()\n",
"random_seed = 1"
]
},
{
"cell_type": "markdown",
"id": "ff5646ca-49a8-4072-b28c-cf8a82debb42",
"metadata": {
"tags": []
},
"source": [
"## PPMI cohort preparation"
]
},
{
"cell_type": "markdown",
"id": "003cc8c9-96aa-4772-af38-fb769c4fdff3",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"### Study data download"
]
},
{
"cell_type": "code",
"execution_count": 2,
"id": "be13ea39-957e-440f-b27b-32113c81f6a0",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"required_files = [\n",
" \"iu_genetic_consensus_20220310.csv\",\n",
" \"Cognitive_Categorization.csv\",\n",
" \"Primary_Clinical_Diagnosis.csv\",\n",
" \"Demographics.csv\", # SEX\n",
" \"Socio-Economics.csv\", # EDUCYRS\n",
" \"PD_Diagnosis_History.csv\", # Disease duration\n",
" \"Montreal_Cognitive_Assessment__MoCA_.csv\",\n",
" \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\", # STROKE\n",
"]\n",
"\n",
"utils.download_ppmi_metadata(required_files)"
]
},
{
"cell_type": "code",
"execution_count": 3,
"id": "a7719ba4-28c1-4b6e-a8a7-08e163c2dd94",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MDS_UPDRS_Part_III_clean.csv is now available\n",
"File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MRI_info.csv is now available\n"
]
}
],
"source": [
"# H&Y scores\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-treatment-and-on-off-status/main/PPMI medication and ON-OFF status.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"PPMI medication and ON-OFF status.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")\n",
"\n",
"# TODO: move this to livingpark_utils\n",
"import os.path as op\n",
"\n",
"file_path = op.join(utils.study_files_dir, \"MRI_info.csv\")\n",
"if not op.exists(file_path):\n",
" !(cd {utils.study_files_dir} && python -m wget \"https://raw.githubusercontent.com/LivingPark-MRI/ppmi-MRI-metadata/main/MRI metadata.ipynb\") # use requests to improve portability\n",
" npath = op.join(utils.study_files_dir, \"MRI metadata.ipynb\")\n",
" %run \"{npath}\"\n",
"print(f\"File {file_path} is now available\")"
]
},
{
"cell_type": "markdown",
"id": "01671975-07d0-48a0-8774-4349abc3f090",
"metadata": {
"tags": []
},
"source": [
"### Inclusion criteria\n",
"\n",
"We obtain the following group sizes:\n",
"\n",
""
]
},
{
"cell_type": "code",
"execution_count": 4,
"id": "7984652c-62b1-465f-bfdb-dccbb9191dcb",
"metadata": {},
"outputs": [],
"source": [
"# H&Y during OFF time\n",
"# Exclusion criteria: Patients were excluded if they had:\n",
"# (a) parkin, leucine-rich repeat kinase 2 (LRRK2), and glu-\n",
"# cocerebrosidase (GBA) gene mutations; OK\n",
"# (b) cerebrovascular\n",
"# disorders, traumatic brain injury history, or intracranial\n",
"# mass; only stroke found in\n",
"#\n",
"# (c) other major neurological and medical diseases;\n",
"# (d) dementia: OK\n",
"\n",
"# DARTEL: 8-mm full width\n",
"# at half maximum (FWHM) Gaussian filter"
]
},
{
"cell_type": "code",
"execution_count": 5,
"id": "b7589aaf-1a2f-411a-afbe-6aa8840fdddd",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Download skipped: No missing files!\n"
]
}
],
"source": [
"import pandas as pd\n",
"\n",
"# UPDRS3\n",
"updrs3 = pd.read_csv(op.join(utils.study_files_dir, \"MDS_UPDRS_Part_III_clean.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PDSTATE\", \"NHY\", \"NP3TOT\"]\n",
"]\n",
"# Genetics\n",
"genetics = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"iu_genetic_consensus_20220310.csv\")\n",
")[[\"PATNO\", \"GBA_PATHVAR\", \"LRRK2_PATHVAR\", \"NOTES\"]]\n",
"# Cognitive Categorization\n",
"cog_cat = pd.read_csv(op.join(utils.study_files_dir, \"Cognitive_Categorization.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"COGSTATE\"]\n",
"]\n",
"# Diagnosis\n",
"diag = pd.read_csv(op.join(utils.study_files_dir, \"Primary_Clinical_Diagnosis.csv\"))[\n",
" [\"PATNO\", \"EVENT_ID\", \"PRIMDIAG\", \"OTHNEURO\"]\n",
"]\n",
"# MRI\n",
"mri = pd.read_csv(op.join(utils.study_files_dir, \"MRI_info.csv\"))[\n",
" [\"Subject ID\", \"Visit code\", \"Description\", \"Age\"]\n",
"]\n",
"mri.rename(columns={\"Subject ID\": \"PATNO\", \"Visit code\": \"EVENT_ID\"}, inplace=True)\n",
"# Demographics\n",
"demographics = pd.read_csv(op.join(utils.study_files_dir, \"Demographics.csv\"))[\n",
" [\"PATNO\", \"SEX\"]\n",
"]\n",
"# Soci-economics\n",
"socio_eco = pd.read_csv(op.join(utils.study_files_dir, \"Socio-Economics.csv\"))[\n",
" [\"PATNO\", \"EDUCYRS\"]\n",
"]\n",
"# Disease duration\n",
"disease_dur = utils.disease_duration()\n",
"# MoCA\n",
"moca = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"Montreal_Cognitive_Assessment__MoCA_.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"MCATOT\"]]\n",
"moca[\"MMSETOT\"] = moca[\"MCATOT\"].apply(utils.moca2mmse)\n",
"# Stroke\n",
"rem = pd.read_csv(\n",
" op.join(utils.study_files_dir, \"REM_Sleep_Behavior_Disorder_Questionnaire.csv\")\n",
")[[\"PATNO\", \"EVENT_ID\", \"STROKE\"]]"
]
},
{
"cell_type": "code",
"execution_count": 6,
"id": "7bdfd288-bf39-44c5-a194-a3baa873be64",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Number of controls: 46\n",
"Number of PD subjects: 125\n",
"Number of PD subject visits by H&Y score:\n"
]
},
{
"data": {
"text/html": [
"