{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Tool to sample random 3D geological models from noddyverse 1M model suite\n",
" \n",
" \n",
"This notebook randomly samples and displays models from the 1 Million model suite, with the possibility to filter to specific deformation event sequences. \n",
" \n",
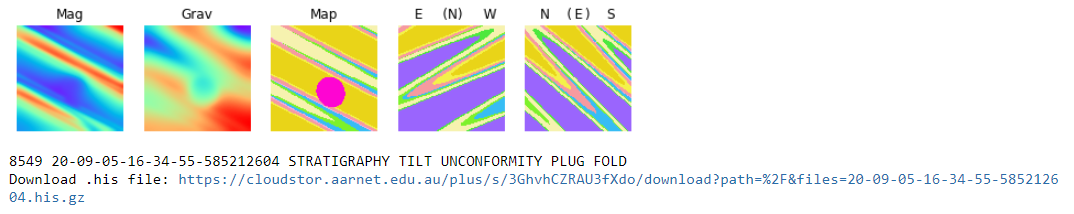
"For each model the magnetic response, the gravity response, the top surface, the north facing vertical section (looking from the south) and the west-facing vertical section (looking from the west) of the 4 x 4 x 4 km cube are displayed: \n",
" \n",
"\n",
" \n",
"The models consist of 20 m cubic voxels (200x200x200 voxels per model), and the history file used to define each model is provided as a link for each model. Windows software to read this history file is available at the Noddy Site, and the source code for a command line version of Noddy, as well as this notebook, is available from github The 1M models themselves are stored at https://cloudstor.aarnet.edu.au/plus/s/8ZT6tjOvoLWmLPx as individual files and as 343 tar files, one per history sequence from https://cloudstor.aarnet.edu.au/plus/s/UxnVSkHfnr7chW9. Each model displayed below has a link to the history file compressed with gzip, so you will need to ungzip it before loading it into Noddy. \n",
"\n",
"## Usage\n",
"\n",
"To use this notebook modify the parameters in the first cell and then both cells, with all outputs displayed at the bottom of the notebook. The code in noddyverse.py provides an example so users can see how to parse the individual files. \n",
" \n",
" \n",
"**his_filter** allows you to filter the models according to the event history, for example to only show histories whose last three events are a fold, then a fault, then a dyke use:\n",
" \n",
"his_filter=['FOLD','FAULT','DYKE']\n",
" \n",
"The full list of possible events (the first two, a base stratigraphy and a tilt are fixed) are:\n",
"\n",
"- 'FAULT' \n",
"- 'SHEAR-ZONE'\n",
"- 'FOLD' \n",
"- 'TILT' \n",
"- 'UNCONFORMITY' \n",
"- 'DYKE' \n",
"- 'PLUG'\n",
"- '' means all event types will be retained, so the command [ '' , '' , '' ] means all event types are available, and the default is [ 'DYKE' , 'FOLD' , 'DYKE' ]\n",
" \n",
"The same event labels can be used in all three positions\n",
" \n",
" \n",
"**display_number** defines how many randonly selected models to display"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"his_filter=['DYKE','FOLD','DYKE'] # FAULT SHEAR-ZONE FOLD TILT UNCONFORMITY DYKE PLUG default as loaded is to allow model sequences\n",
"\n",
"display_number=10 # number of randomly selected models to display"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"scrolled": false
},
"outputs": [],
"source": [
"from noddyverse import display_models\n",
"%matplotlib inline\n",
"display_models(his_filter,display_number)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"hide_input": false,
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.10"
}
},
"nbformat": 4,
"nbformat_minor": 2
}