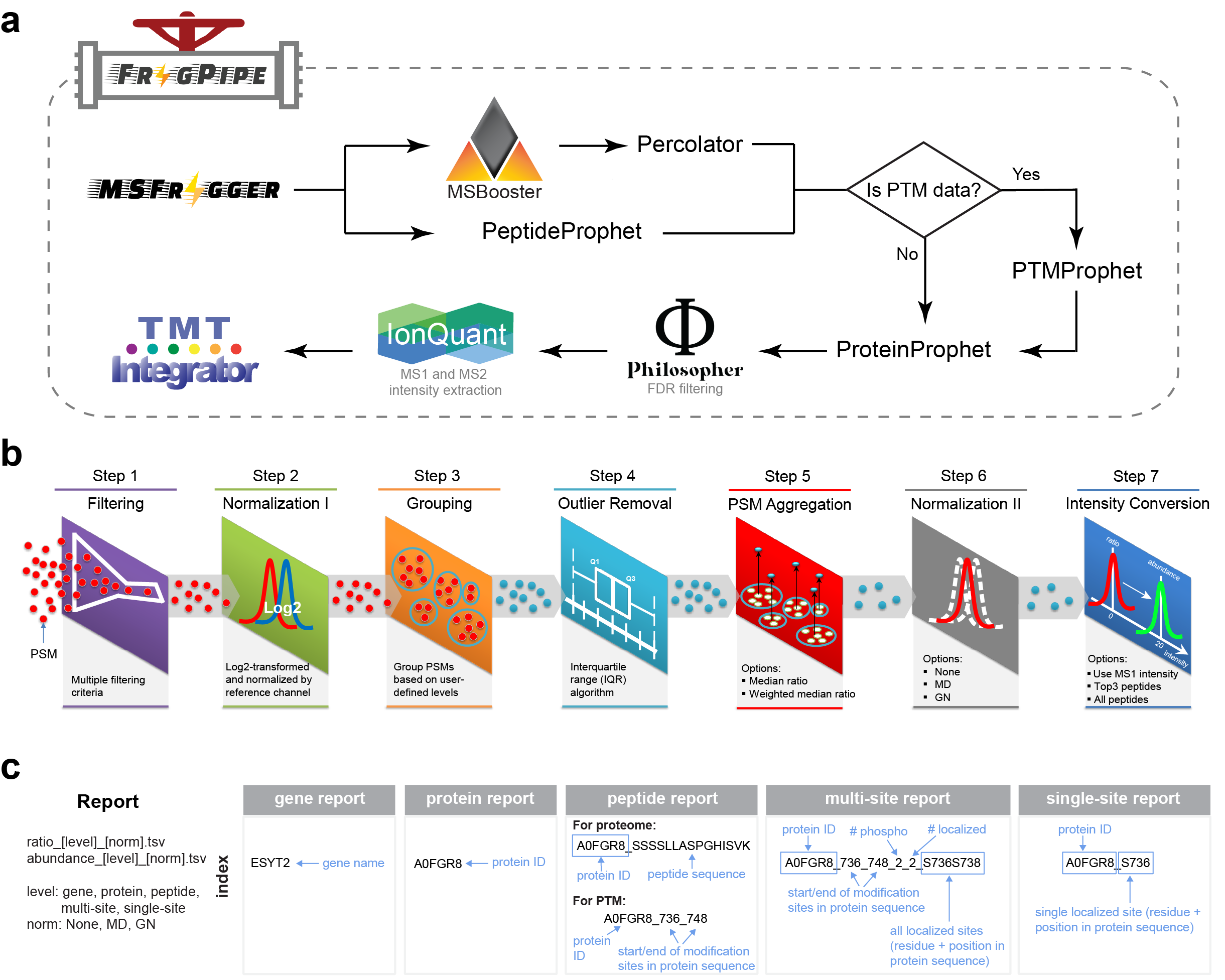
TMT-Integrator extracts and combines channel abundances from multiple isobaric labeling samples. It takes PSM tables as input, and generates quantification reports at the user-specified levels. TMT-Integrator currently provides four quantification options: gene, protein, peptide, and modified site levels. TMT-Integrator is included in [FragPipe](https://fragpipe.nesvilab.org/) with the Quant (Isobaric) options for complete analyses of isobaric labeling experiments.
There are seven steps in TMT-Integrator, (1) filtering, (2) normalization I, (3) grouping, (4) outlier removal, (5) PSM aggregation, (6) normalization II, (7) intensity conversion.
#### Best PSM selection
For improved quantification, we have the default option to include only PSMs that pass the following criteria:
* TMT/iTRAQ-labeled
* Reference channel intensity > 0
* Precursor ion purity >= 50%
* Summed (across all channels) MS2 intensity >= 5% (2.5% for phospho)
* PSM with the highest summed MS2 intensity among all PSMs identifying the same peptide ion in the sample/fraction (i.e. same LC-MS/MS run)
* Must not map (as unique or razor peptides) to contaminant proteins
* (Phospho: Must contain phosphorylation)
#### Normalization and log transformation
For each selected PSM, all channel intensities are divided by the reference channel, then log2 transformed, yielding normalized intensities.
#### PSM-level normalization (optional)
Within each multiplexed sample, selected PSMs are sorted and divided into groups by retention time. Within each retention time group, normalized intensities are subtracted from the median channel intensities.
#### Outlier removal (optional)
PSMs are grouped (by gene, protein, peptide, or modified site). Within these groups, the first quartile (Q1), third quartile (Q3) and interquartile range (IQR; Q3-Q1) are computed, and PSMs with intensities outside the boundaries of Q1-1.5xIQR and Q3+1.5xIQR are removed.
#### Protein-level normalization
There are two protein level normalization options: median centering or global normalization. With median centering, median normalized log2 intensity of the ith sample (m0) is subtracted from each channel so that the median ratio of each becomes 0. If global normalization is used, median centering is first performed, then the median deviation and median absolute deviation of centered values (m1 and m2) is calculated and used to scale all values: yij'=(yij/m2)xm1+m0, where yij is the log2 ratio in the ith group of the jth sample.
## Download
TMT-Integrator is included in [FragPipe](https://fragpipe.nesvilab.org/). The latest standalone version can be downloaded [here](https://github.com/Nesvilab/TMT-Integrator/releases/latest).
## Use
For most users, we recommend running TMT-Integrator through [FragPipe](https://fragpipe.nesvilab.org/) graphical user interface. See the TMT tutorials below, or other tutorials listed on the [FragPipe homepage](https://fragpipe.nesvilab.org/).
- [Basic tutorial for TMT data](https://fragpipe.nesvilab.org/docs/tutorial_tmt.html)
- [Tutorial for analyzing TMT data with multiple plexes](https://fragpipe.nesvilab.org/docs/tutorial_tmt-2plexes.html)
For command line uses, see the Philosopher tutorials for running TMT analyses [step-by-step](https://github.com/Nesvilab/philosopher/wiki/Step-by-step-TMT-analysis) or with the [pipeline](https://github.com/Nesvilab/philosopher/wiki/Pipeline-mode-for-TMT-analysis) command. These examples can also be used to run the tool separately (note that both label-free/MS1 and isobaric quantification must be run prior to TMT-Integrator):
`java -jar TMTIntegrator.jar TMTIntegrator.yaml PSM_Tables`
`java -jar -Xmx16g TMTIntegrator.jar TMTIntegrator.yaml /*_psm.tsv`
## TMT-Integrator settings in FragPipe GUI
The description for TMT-Integrator's options are listed as follows.
| Basic Options | Description | Default Value |
|--------------------------------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|-----------------------|
| Label type | Isobaric reagent product, supporting TMT-2, TMT-6, TMT-10, TMT-11, TMT-16, TMT-18, TMT-35 plex, and iTRAQ-4 and iTRAQ-8, et al. | Depends on experiment |
| Quant level | MS level for reporter ion intensity extraction. 2 for MS2 data; 3 for MS3 data. | 2 |
| Mass tolerance (ppm) | Reporter ion mass tolerance in PPM. | 20 |
| Define reference | Add an artificial reference channel if there is no reference channel in the sample ("Virtual") or use real reference channel if available ("Reference sample"). | Virtual |
| Ref sample tag | Unique tag to identify reference (Bridge) channels. | Bridge |
| Ref D sample tag (TMT-35) | Unique tag to identify reference (Pool) Deuterium channels. Only used for TMT 35-plex. | Pool |
| Group by | PSM aggregation to the specified level for data summarization (Gene level, Protein, Peptide sequence, Multiple PTM sites, single PTM site and All). | All |
| Normalization | Data normalization at sample level. None: no normalization; MD (median centering); GN (median centering + variance scaling); All: generate reports with all normalization options. | MD (median centering) |
| Log2 transform the intensity | Transform the intensity using log2 for report generation. | Enabled |
| Generate MSstats files (using philosopher) | Option to generate an MSstats-compatible report with Philosopher. Require selecting Philosopher as the "Intensity extraction tool". | Disabled |
| PTM Options | Description | Value |
|-------------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------|------------|
| Mod tag | PTM tag for generating PTM-specific reports, none: global data; S(79.9663),T(79.9663),Y(79.9663): phospho; K(114.0429): ubiquitin. | none |
| Use Glycan Compositions | For multi-mass report, index by glycan composition instead of mass to separate isomeric glycan compositions. Requires glycan assignment from PTM-Shepherd. | Disabled |
| Min site probability | Site localization confidence threshold. -1: global; 0: as determined by the search engine; above 0 (e.g., 0.75): min PTMProphet site probability. | -1 |
| Glycan FDR filter | (Optional) Remove PSMs not passing glycan FDR at specified level (q-value). Set to -1 to ignore. Requires glycan assignment from PTM-Shepherd. | -1 |
| Advanced Options | Description | Value |
|-------------------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|-----------------------------------------|
| Peptide-Gene uniqueness | Filter PSM based on their unique mapping to genes. Options include keeping all PSMs, no PSMs mapping to multiple genes in dataset and no PSMs mapping to multiple genes in FASTA. | Keep all PSMs |
| Peptide-Protein uniqueness | Filter PSM based on their unique mapping to proteins. Unique only: use peptides that are unique or confidently assigned to a single protein (or a single indistinguishable protein group). Unique+Razor: use 'unique plus razor' approach, wich each shared peptide assigned as razor to one protein (as classified by Philosopher and defined in psm.tsv file). | Unique+Razor |
| Min PSM probability | Minimum PSM probability threshold (on top of FDR filtering by Philosopher). | 0.9 |
| Min purity | Ion purity threshold. | 0.9 |
| Min Intensity (percent) | Remove low intensity PSMs with the summed reporter ion intensity in the lowest certain percentage of all PSMs. | 0.05 (regular data) or 0 (Astral data) |
| Min Resolution | Remove the PSM if there are any channels having resolutino less than the min resolution and SNR greater than or equal to 1 (only for RAW file formats). | 0 (regular data) or 45000 (Astral data) |
| Min best peptide probability | The minimum probability required for the highest probability PSM in each PSM group. | 0.9 |
| Min NTT | Minimum allowed number of enzymatic termini. | 0 |
| Aggregation method | Method to aggregate PSM abundances. Median: use median (default); Weighted: use precursor intensity-weighted abundances. | Weighted |
| Min SNR | Remove the PSM if all channels' summed SNR is less than the min SNR (only for RAW file formats). | 0 (regular data) or 1000 (Astral data) |
| Best PSM | Keep the best PSM only (highest summed TMT intensity) among all redundant PSMs within the same LC-MS run. | Enabled |
| PSM norm | Perform additional retention time-based normalization at the PSM level. | Disabled |
| Allow overlabel | Allow PSMs with TMT on S (when overlabeling on S) was allowed in the database search, limited to phospho TMT data. | Enabled |
| Allow unlabeled | Allow peptides with unlabled n-term (i.e. no TMT/iTRAQ label and no Acetyl at n-terminus). | Disabled |
| Outlier removal | Perform per-sample outlier removal within each PSM group during aggregation. | Enabled |
| Exclude proteins | Exclude proteins with the specified tag(s) at the beginning of the accession number (e.g. none: no exclusion; sp\|,tr\|: exclude proteins with sp\| or tr\|). | none |
| Ratio to Abundance conversion | Use MS1 precursor ion intensity or MS2 summed TMT reporter ion intensity in ratios to abundances conversion. | MS1 |
| Print reference intensity | Print individual reference sample intensities. | Disabled |