{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Calculation of the SVI "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"In this example the Standard Vegetation Index (SVI) is calculated from the Enhanced Vegetation Index (EVI) gained from satellite remote sensing. A MODIS vegetation product (MOD13Q1) with a temporal resolution of 16 days and a spatial resolution of 250m is used (https://lpdaac.usgs.gov/products/mod13q1v006/). "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Note: To execute a cell click on it and then press `Shift + Enter`. Alternatively run the entire notebook click on `Cell > Run All` (make sure all the configuration options are set correctly)."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Download and install Anaconda 3"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"To run this Jupyter Notebook you need python installed on your computer. We recommend downloading and installing Anaconda 3 (https://www.anaconda.com/download/) with the python 3.6 version as it includes a lot of useful packages."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Download MODIS data as Geotiffs from AppEEARS"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Access the [AppEEARS website](https://lpdaacsvc.cr.usgs.gov/appeears/) and create a user account (free) for USGS in case you do not already have one. Click on __Extract__ and __Area Sample__ and start a new request."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
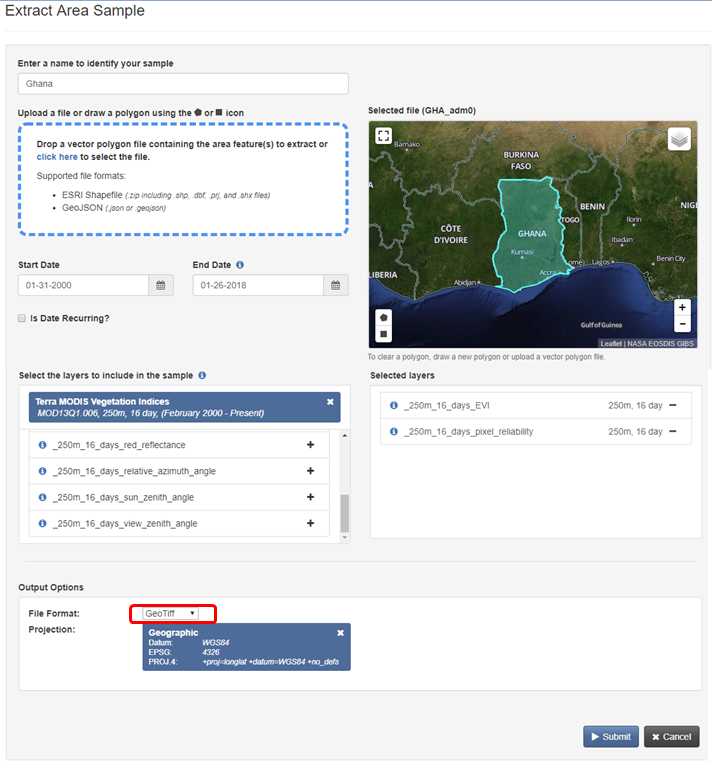
"1. Enter a request name. \n",
"2. Define your region of interest by specifying a __ESRI shapefile__ (zip) or drawing a __polygon__. Country shapefiles can be downloaded [here](http://www.gadm.org/country). \n",
"**Note:** The boundaries and names of the shapefiles do not imply official endorsement or acceptance by the United Nations.\n",
"3. Define the time period for your data: January __2000__ to __today__\n",
"4. Select the product: __MOD13Q1__ and the layers of interest: __EVI__ and __pixel_reliability__\n",
"5. Define the output as __Geotiff__ with __geographic Pojection__\n",
"6. Click on __Submit__"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
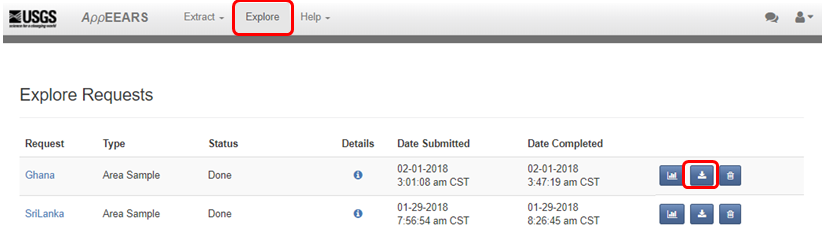
"Click on __Explore__ to check the status of the request. When the status says \"Done\" you can click on the __Download__ symbol. __Select all__ ordered datasets and start downloading the data."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
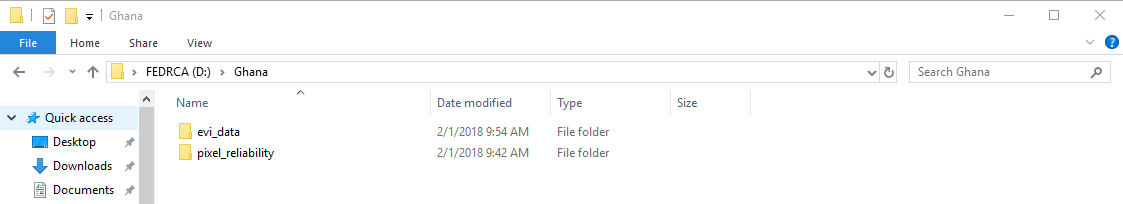
"After downloading store the __.tif__ files in two folders: one folder for the __evi_data__ and one for the __pixel_reliability__.\n",
"It is reccomended to use an external hard drive (or other mass storage media) to store your input and output data if possible. \n",
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Preparing the working environment"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"There are two possible approaches: \n",
"1. Install the required packages directly\n",
"2. Use the provided environment file to setup a new environment"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### 1. Installing the required packages directly"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The required package versions are: \n",
"* python 3.6 or 3.7\n",
"* gdal >= 3.0.1\n",
"* numpy >= 1.15.0\n",
"\n",
"The __gdal__ package needs to be installed and it must have Bigtiff support; and numpy may need to be updated. For these reasons it is recommended to use the __[conda-forge](https://conda-forge.org/)__ package repository as follows:\n",
"1. Open the __Anaconda Promt__ by searching for it in the windows start menu.\n",
"2. Type: `conda install -c conda-forge --no-pin gdal numpy` and hit `Enter`\n",
"\n",
"3. Type `y` and hit `Enter` if you are asked if you want to proceed.\n",
"\n",
"\n",
"__Note__: `--no-pin` is added to the install command in order to not restrict maximum package versions, and thus to avoid having certain packages be downgraded when installing gdal. If gdal and/or numpy do not meet the version requirements, then modify the install command to:\n",
"\n",
"`conda install -c conda-forge --no-pin gdal=3.0.1 numpy=1.15.0`"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### 2. Setup a new environment"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"All the relevant documentation can be found [here](https://conda.io/docs/user-guide/tasks/manage-environments.html).\n",
"\n",
"Carry out the following steps in order to setup the environment:\n",
"1. Download the environment file found [here](https://raw.githubusercontent.com/UN-SPIDER/drought-svi/master/environment.yml) (right click and choose `Save As`).\n",
"2. Open the __Anaconda Promt__ by searching for it in the windows start menu.\n",
"3. Navigate to where you have saved the environment file:\n",
"`cd C:\\my\\path`\n",
"4. Type: `conda env create -f environment.yml` and hit `Enter`\n",
"\n",
"After Step 1 verify that the file you has downloaded has the following structure:
\n",
"\n",
"name: unspider
\n",
"channels:\n",
" - conda-forge\n",
" - conda-forge/labels/broken\n",
" - defaults\n",
"dependencies:\n",
" - python=3.6\n",
" - gdal=3.0.1\n",
" - numpy=1.15.*\n",
" - matplotlib\n",
" - ipython\n",
" - jupyter\n",
" - ipywidgets\n",
" - tqdm
\n",
"\n",
"The line `- conda-forge/labels/broken` should only be inserted if an error saying a certain package can't be found occurs. \n",
"\n",
"The `unspider` environment should now be created. Make sure it is activated before running Jupyter or before running the script if you save it as a file (see the [general documentation](https://conda.io/docs/user-guide/tasks/manage-environments.html) for how to do this)."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Download this notebook and run it using Jupyter Notebook"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Download this Jupyter Notebook __[here](https://raw.githubusercontent.com/UN-SPIDER/drought-svi/master/UNSPIDER_SVI_recommended_practice.ipynb)__ (right click and choose `Save As`). Start Jupyter Notebook by searching for it in the Windows start menu or by typing `jupyter notebook` in the Anaconda Prompt and hitting `Enter`. After Jupyter Notebook opened in your web browser, search for the script on your computer and open it.\n",
"\n",
"[//]: #__[here](https://drive.google.com/drive/folders/1a3m5EbL2HGkkTLRA5U5V0oUH_J_V1DTT?usp=sharing)__\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Every cell has to be executed individually. A cell, which is still processing is marked by an `*` and a finished cell with a number. Cells are executed by clicking on the cell and hitting `Shift + Enter`. Alternatively run the entire notebook sequentially (i.e. one cell at a time) click on `Cell > Run All` (make sure all the configuration options are set correctly)."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"__Optional__: An alternative to running everything in Jupyter is to open this Jupyter Notebook in Jupyter and do:\n",
"\n",
"`File > Download as > Python (.py)`\n",
"\n",
"Then you run the script either in Spyder (an IDE that comes bundled with Anaconda) or in a terminal. This may yield better perfomance."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Nicer progress bars"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"During execution basic progress updates will be printed, however if you would like nicer and more informative progress bars (not just updates), then you will need to install the __tqdm__ package. To do so:\n",
"1. Open the __Anaconda Promt__ by searching for it in the windows start menu.\n",
"2. Type: `conda install tqdm` and hit `Enter`\n",
"3. Type `y` and hit `Enter` if you are asked if you want to proceed.\n",
"\n",
"When tqdm is used the bar on top always shows the overall progress, and the bar on the bottom shows the progress for a particular sub-process (such as generating the output images).\n",
"\n",
"__Note__: If you used the environment file to setup a new environment, then tqdm will already be installed in that environment. \n",
"\n",
"__Note2__: The latest version of tqdm has a bug which leads to the progress bar description field not getting printed fully when used in a Jupyter notebook. This issue has been reported and should be fixed quickly. The tqdm progress bars work as intended when the code is executed from a terminal.\n",
"\n",
"__Note3__: If you have tqdm installed but would prefer not to use it, modify the section dealing with tqdm in 'Import required packages' as follows:\n",
"```python\n",
"\"\"\"\n",
"# If tqdm is installed it will be used to display a nice progress bar\n",
"try:\n",
" from tqdm.autonotebook import tqdm\n",
" import sys\n",
"\n",
" # Overload built-in enumerate function\n",
" def enumerate(iterator, desc=None):\n",
" return builtins.enumerate(tqdm(iterator, desc=desc, file=sys.stdout,\n",
" leave=False))\n",
"except ImportError:\n",
"\"\"\"\n",
"tqdm = None\n",
"\n",
"# Overload built-in enumerate function\n",
"def enumerate(iterator, desc=None):\n",
" if desc is not None:\n",
" print(desc)\n",
" return builtins.enumerate(iterator)\n",
"```"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Import the required packages"
]
},
{
"cell_type": "code",
"execution_count": 1,
"metadata": {},
"outputs": [],
"source": [
"import os\n",
"import re\n",
"import gc\n",
"import builtins\n",
"import warnings\n",
"\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"from matplotlib import colors\n",
"\n",
"from osgeo import gdal\n",
"gdal.UseExceptions()\n",
"\n",
"\n",
"# If tqdm is installed it will be used to display a nice progress bar\n",
"try:\n",
" from tqdm.autonotebook import tqdm\n",
" import sys\n",
"\n",
" # Overload built-in enumerate function\n",
" def enumerate(iterator, desc=None):\n",
" return builtins.enumerate(tqdm(iterator, desc=desc, file=sys.stdout,\n",
" leave=False))\n",
"except ImportError:\n",
" tqdm = None\n",
"\n",
" # Overload built-in enumerate function\n",
" def enumerate(iterator, desc=None):\n",
" if desc is not None:\n",
" print(desc)\n",
" return builtins.enumerate(iterator)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Set data paths "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Input data paths:"
]
},
{
"cell_type": "code",
"execution_count": 2,
"metadata": {},
"outputs": [],
"source": [
"# Specify the name of your study area, this will be used for naming the output\n",
"# maps\n",
"study_area = \"Ghana\"\n",
"\n",
"# Specify the folder path where the EVI geotiffs are stored on your \n",
"# computer or external hard drive.\n",
"path_evi = \"D:\\Ghana\\evi_data\"\n",
"\n",
"# Specify the folder path where the pixel reliability geotiffs are stored on your \n",
"# computer or external hard drive.\n",
"path_pr = \"D:\\Ghana\\pixel_reliability\""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Output data paths:"
]
},
{
"cell_type": "code",
"execution_count": 4,
"metadata": {},
"outputs": [],
"source": [
"# Create and specify the folder where the output png-files should be stored\n",
"path_png = \"D:\\Ghana\\SVI_maps_Ghana_png\"\n",
"\n",
"# Create and specify the folder where the output tif-files should be stored\n",
"path_tif = \"D:\\Ghana\\SVI_maps_Ghana_tif\""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Function definitions"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"In the following cell, various functions are defined."
]
},
{
"cell_type": "code",
"execution_count": 5,
"metadata": {},
"outputs": [],
"source": [
"def get_doy(re_doy, string):\n",
" \"\"\"\n",
" Searches the input string for a DOY, and if one is found it returns a tuple\n",
" containing the DOY; else it returns None.\n",
"\n",
" :param re_doy: compiled re pattern used to match a DOY\n",
" :param string: input string that will be searched for a DOY\n",
" :return : if a DOY is found a tuple of the form\n",
" ( year, day of year), else None\n",
" \"\"\"\n",
" search_doy = re_doy.search(string)\n",
" if search_doy is None: # Case where no doy was found\n",
" return None\n",
" doy = search_doy.group(1)\n",
" year = int(doy[:4]) # Treat year values as integers\n",
" day = doy[4:] # Day values need to remain strings to be zero padded\n",
" return year, day\n",
"\n",
"\n",
"def check_prepare_files(evi_files, pr_files):\n",
" \"\"\"\n",
" Checks that for each evi file there is a corresponding pixel reliability\n",
" file. Then a dictionary is returned containing the sorted files lists for\n",
" each DOY.\n",
"\n",
" :param evi_files: list of evi files to be processed, containing a tuples of\n",
" the form ( file_path, year, day)\n",
" :param pr_files: list of pixel reliability files to be processed,\n",
" containing a tuples ofthe form\n",
" ( file_path, year, day)\n",
" :return: dictionary where keys are the available DOYs and values are\n",
" dictionaries for evi_files and pr_files countaining the sorted\n",
" evi_files and sorted pr_files.\n",
" \"\"\"\n",
" # Dictionary keys have properties similar enough to a mathematical set for\n",
" # our purposes\n",
" doy_dict = dict()\n",
"\n",
" for path, year, day in evi_files:\n",
" try:\n",
" doy_dict[day]['evi'].append((path, year, day))\n",
" except KeyError:\n",
" doy_dict[day] = dict()\n",
" doy_dict[day]['evi'] = [(path, year, day)]\n",
"\n",
" for path, year, day in pr_files:\n",
" try:\n",
" doy_dict[day]['pr'].append((path, year, day))\n",
" except KeyError:\n",
" try:\n",
" doy_dict[day]['pr'] = [(path, year, day)]\n",
" except KeyError:\n",
" doy_dict[day] = dict()\n",
" doy_dict[day]['pr'] = [(path, year, day)]\n",
"\n",
" if len(evi_files) != len(pr_files):\n",
" mismatch_doys = list()\n",
" for day in doy_dict.keys():\n",
" try:\n",
" if len(doy_dict[day]['evi']) != len(doy_dict[day]['pr']):\n",
" mismatch_doys.append(day)\n",
" except KeyError:\n",
" mismatch_doys.append(day)\n",
" mismatch_files = dict()\n",
" for day in mismatch_doys:\n",
" if any(i not in doy_dict[day]['pr'] for i in doy_dict[day]['evi']):\n",
" evi_years = [i[1] for i in doy_dict[day]['evi']]\n",
" pr_years = [i[1] for i in doy_dict[day]['pr']]\n",
" mismatch_evi = [i for i in evi_years if i not in pr_years]\n",
" mismatch_pr = [i for i in pr_years if i not in evi_years]\n",
"\n",
" # If the lists are empty future list comprehensions will fail\n",
" # to correctly exclude all files\n",
" if len(mismatch_evi) == 0:\n",
" mismatch_evi.append(-1)\n",
" if len(mismatch_pr) == 0:\n",
" mismatch_pr.append(-1)\n",
"\n",
" mismatch_files[day] = dict()\n",
" mismatch_files[day]['evi'] = [i[0] for i in doy_dict[day]['evi']\n",
" if i[1] in mismatch_evi]\n",
"\n",
" mismatch_files[day]['pr'] = [i[0] for i in doy_dict[day]['pr']\n",
" if i[1] in mismatch_pr]\n",
" war_msg = \"There is a mismatch in the number of evi data files and \"\n",
" war_msg += \"pixel reliability files.\\n\"\n",
" war_msg += \"The the mismatched files for each doy are:\\n\"\n",
" war_msg += str(mismatch_files)\n",
" war_msg += \"\\n\"\n",
" war_msg += \"These files will be ignored.\"\n",
" warnings.warn(war_msg)\n",
"\n",
" for day, v in mismatch_files.items():\n",
" for k_sub, v_sub in v.items():\n",
" doy_dict[day][k_sub] = [i for i in doy_dict[day][k_sub]\n",
" if i[0] not in v_sub]\n",
" else:\n",
" for day in doy_dict.keys():\n",
" doy_dict[day]['evi'] = sorted(doy_dict[day]['evi'],\n",
" key=lambda x: x[1])\n",
" doy_dict[day]['pr'] = sorted(doy_dict[day]['pr'],\n",
" key=lambda x: x[1])\n",
"\n",
" return doy_dict\n",
"\n",
"\n",
"def load_cloud_mask(files_list, cloud_mask):\n",
" \"\"\"\n",
" Loads all pixel reliability images into the cloud mask array.\n",
"\n",
" :param files_list: list of files to be processed, containing a tuples of\n",
" the form ( file_path, year, day)\n",
" :param cloud_mask: matrix containing the processed pixel reliability data\n",
" :return:\n",
" \"\"\"\n",
" for i, image in enumerate(files_list, \"Loading cloud_mask\"):\n",
" # Open the file from the pixel reliability file list\n",
" dataset = gdal.Open(image[0])\n",
" band = dataset.GetRasterBand(1)\n",
" # del dataset\n",
" data = band.ReadAsArray()\n",
"\n",
" # Write the data from each file to the array\n",
" cloud_mask[:, :, i] = data\n",
"\n",
" return cloud_mask\n",
"\n",
"\n",
"def prepare_cloud_mask(cloud_mask):\n",
" \"\"\"\n",
" Filters the pixel reliability array.\n",
"\n",
" :param cloud_mask: pixel reliability array\n",
" :return: filtered and rescaled pixel reliability matrix\n",
" \"\"\"\n",
" # Exchange value 0 with value 1 (Pixels with value 0 and 1 are used)\n",
" cloud_mask[cloud_mask == 0] = 1\n",
"\n",
" # Set values that are of no interest to us to NaN\n",
" # cloud_mask[cloud_mask != 1] = np.nan\n",
" cloud_mask[cloud_mask != 1] = np.nan\n",
"\n",
" # These are additional filters you may want to use\n",
" # Set value 2 to NA (Snow and Ice)\n",
" # cloud_mask[cloud_mask == 2] = np.nan\n",
"\n",
" # Set value 3 to NA (Cloud)\n",
" # cloud_mask[cloud_mask == 3] = np.nan\n",
"\n",
" # Set no data value (= -1) to NA\n",
" # cloud_mask[cloud_mask == -1] = np.nan\n",
"\n",
" # Set all values above 3 to NA\n",
" # cloud_mask[cloud_mask > 3] = np.nan\n",
"\n",
" return cloud_mask\n",
"\n",
"\n",
"def load_evi(files_list, cloud_mask):\n",
" \"\"\"\n",
" Loads a single geotiff into the evi matrix.\n",
"\n",
" :param files_list: list of files to be processed, containing a tuples of\n",
" the form ( file_path, year, day)\n",
" :param cloud_mask: matrix containing the processed pixel reliability data\n",
" :return: evi matrix with an additional geotiff of data\n",
" \"\"\"\n",
" for i, image in enumerate(files_list, \"Loading EVI\"):\n",
" # Open the file from the evi file list\n",
" dataset = gdal.Open(image[0])\n",
" band = dataset.GetRasterBand(1)\n",
" data = band.ReadAsArray()\n",
"\n",
" # Apply the cloud mask and write the data from each file to the array\n",
" # Note: the evi data is multiplied into the cloud_mask matrix in order\n",
" # to save RAM and speed up the computation\n",
" cloud_mask[:, :, i] *= data\n",
"\n",
" return cloud_mask\n",
"\n",
"\n",
"def prepare_evi(evi):\n",
" \"\"\"\n",
" Filters the evi to set all negative values to nan and rescales the data.\n",
"\n",
" :param evi: numpy array containing the results of multiplying the\n",
" cloud_mask data with the evi data\n",
" :return: filtered and rescaled input numpy array\n",
" \"\"\"\n",
" # Set negative values to nan\n",
" evi[evi < 0] = np.nan\n",
"\n",
" # Rescale the data\n",
" evi *= 0.0001\n",
"\n",
" return evi\n",
"\n",
"\n",
"def calculate_svi(evi):\n",
" \"\"\"\n",
" Calculates the SVI.\n",
"\n",
" The standard deviation is calculated using the following formula:\n",
" std = sqrt((E[X ** 2] - E[X] ** 2) * (N / N - 1))\n",
" std = sqrt((mean(x ** 2) - mean(x) ** 2) * (N / N - 1))\n",
"\n",
" Here N is the number of obervations, and the multiplier (N / N - 1) is the\n",
" adjustment factor necessary to convert the standard deviation to an\n",
" unbiased estimator of the variance of the infinite population.\n",
"\n",
" The SVI itself is calculated using the following formula:\n",
" SVI = (EVI - mean(EVI)) / std(EVI)\n",
"\n",
" A lot of the operations are done in place, such as squaring the evi matrix,\n",
" using it for something, and then taking the square root of it to get back\n",
" the original values. This may seem inefficient however it is much faster\n",
" than allocating large amount of memory and doing copy based operations.\n",
"\n",
" When it comes to numpy arrays, abbreviated code like this:\n",
" a += b\n",
" c /= d\n",
" Means that the operations are performed in place.\n",
"\n",
" Whereas operations like this:\n",
" a = a + b\n",
" c = c / d\n",
" First create a copy of the a and c arrays before performing the addition\n",
" and division. This leads to additional memory and computational overhead\n",
" due to creating a copy of the arrays.\n",
"\n",
" Since the input evi matrix can have only positive values squaring and\n",
" taking the square root in place causes no computational errors.\n",
"\n",
"\n",
" Note: The code for computing the mean and standard deviation is loosely\n",
" based on the built-in numpy methods for nanmean and nanstd.\n",
"\n",
" :param evi: numpy array containing the geotiff processing results\n",
" :return: evi array which has had its values replaced in-place with the SVI\n",
" values\n",
" \"\"\"\n",
" with warnings.catch_warnings():\n",
" warnings.simplefilter('ignore')\n",
" # Allocate memory to where intermittent data will be saved\n",
" shape = (evi.shape[0], evi.shape[1], 1)\n",
" num_non_nan = np.zeros(shape, dtype=np.float64)\n",
" sum_ = np.zeros(shape, dtype=np.float64)\n",
" sum_squares = np.zeros(shape, dtype=np.float64)\n",
"\n",
" # Create a mask that will say where nan values are, then negate it so\n",
" # it shows where the non nan values are\n",
" mask = ~np.isnan(evi)\n",
"\n",
" # Sum the non nan values\n",
" # Note: setting keepdims to True will allow to do efficient array wide\n",
" # broadcasting in future calculations (this greatly increases\n",
" # computation speed and reduces memory usage)\n",
" np.sum(mask, axis=2, out=num_non_nan, dtype=np.intp, keepdims=True)\n",
"\n",
" # Negate the mask so it shows where the nan values are\n",
" np.invert(mask, out=mask)\n",
"\n",
" # Substitute nan values with 0 in place\n",
" evi[mask] = 0\n",
"\n",
" # Calculate the sum\n",
" np.sum(evi, axis=2, out=sum_, dtype=np.float64, keepdims=True)\n",
"\n",
" # Square all values in place\n",
" np.square(evi, out=evi)\n",
"\n",
" # Calculate the sum of squares\n",
" np.sum(evi, axis=2, out=sum_squares, dtype=np.float64, keepdims=True)\n",
"\n",
" # Take the square root of evi to get back all the original values\n",
" np.sqrt(evi, out=evi)\n",
"\n",
" # Compute the mean by dividing the sum_ values in place\n",
" evi_mean = sum_ # Set easy to understand pointer\n",
" evi_mean /= num_non_nan # Divide in place\n",
"\n",
" # Square the mean in place\n",
" np.square(evi_mean, out=evi_mean)\n",
"\n",
" # Compute the standard deviation in place (saving it in sum_squares)\n",
" evi_std = sum_squares # Set easy to understand pointer\n",
" evi_std /= num_non_nan\n",
" evi_std -= evi_mean\n",
"\n",
" # Adjust the standard deviation to be an unbiased estimator of the\n",
" # variance of the infinite population\n",
" evi_std *= num_non_nan\n",
" num_non_nan -= 1\n",
" num_non_nan[num_non_nan <= 0] = np.nan # Avoid zero division errors\n",
"\n",
" # Finish calculating the standard deviation\n",
" evi_std /= num_non_nan\n",
" np.sqrt(evi_std, out=evi_std)\n",
"\n",
" # Take the square root in place to get back original mean\n",
" np.sqrt(evi_mean, out=evi_mean)\n",
"\n",
" # Replace 0 with nan in the evi array\n",
" evi[mask] = np.nan\n",
"\n",
" # Calculate the SVI\n",
" # Note: the svi is saved into the evi matrix in order to save RAM\n",
" svi = evi\n",
" svi -= evi_mean\n",
" svi /= evi_std\n",
"\n",
" return svi\n",
"\n",
"\n",
"def generate_png(file_name, data, breaks, extent):\n",
" \"\"\"\n",
" Generate an output png image representing the SVI for the input map area.\n",
"\n",
" :param file_name: file name to be used when saving the png image\n",
" :param data: numpy view onto the svi for a single year\n",
" :param breaks: standard deviation values that should be used as breaks in\n",
" the outputted png images\n",
" :param extent: tuple containing data for how to modify the output graphic\n",
" in order for it to scale correctly\n",
" :return:\n",
" \"\"\"\n",
" bounds = breaks # Set easy to understand pointer\n",
"\n",
" # Define the size of the figure (in inches)\n",
" fig, ax = plt.subplots(figsize=(5, 5))\n",
" plt.title(file_name)\n",
" cmap = colors.ListedColormap(['#4C0E0D', '#E72223', '#F19069', '#F9F6C6',\n",
" '#64B14B', '#04984A', '#00320E'])\n",
" norm = colors.BoundaryNorm(bounds, cmap.N)\n",
" cax = ax.imshow(data, cmap=cmap, norm=norm, extent=extent)\n",
" fig.colorbar(cax, cmap=cmap, norm=norm, boundaries=bounds, ticks=breaks)\n",
"\n",
" plt.savefig(os.path.join(path_png, file_name + \".png\"), dpi=100)\n",
" plt.close()\n",
"\n",
"\n",
"def generate_geotiff(file_name, data, geo_transform, projection):\n",
" \"\"\"\n",
" Generate an output geotiff image representing the SVI for the input map\n",
" area.\n",
"\n",
" :param file_name: file name to be used when saving the tif image\n",
" :param data: numpy view onto the svi for a single year\n",
" :param geo_transform: geo transform data to be used in saving the tif image\n",
" :param projection: projection data to be used in saving the tif image\n",
" :return:\n",
" \"\"\"\n",
" # Set geotiff output path\n",
" geotiff_path = os.path.join(path_tif, file_name + \".tif\")\n",
"\n",
" # Read columns from data array\n",
" cols = data.shape[1]\n",
" # Read rows from data array\n",
" rows = data.shape[0]\n",
"\n",
" # Set the driver to Geotiff\n",
" driver = gdal.GetDriverByName('GTiff')\n",
" # Create raster with shape of array as float64\n",
" out_raster = driver.Create(geotiff_path, cols, rows, 1, gdal.GDT_Float64)\n",
" # Read geo information from input file\n",
" out_raster.SetGeoTransform(geo_transform)\n",
" # Read band\n",
" out_band = out_raster.GetRasterBand(1)\n",
" # Set no data value to numpy's nan\n",
" out_band.SetNoDataValue(np.nan)\n",
" out_band.WriteArray(data)\n",
" # Set the projection according to the input file projection\n",
" out_raster.SetProjection(projection)\n",
" out_band.FlushCache()\n",
"\n",
"\n",
"def generate_images(files_list, svi, extent, geo_transform, projection):\n",
" \"\"\"\n",
" Generates all the output png and tif images.\n",
"\n",
" :param files_list: list of files to be processed, containing a tuples of\n",
" the form ( file_path, year, day)\n",
" :param svi: numpy array containing the svi\n",
" :param extent:\n",
" :param geo_transform: geo transform data to be used in saving the tif image\n",
" :param projection: projection data to be used in saving the tif image\n",
" :return:\n",
" \"\"\"\n",
" day = files_list[0][2]\n",
" years = [i[1] for i in files_list]\n",
"\n",
" # Calculate the standard deviation values that will be used to define the\n",
" # color scheme\n",
" std = np.nanstd(svi[:, :, 0])\n",
" std15 = 1.5 * std\n",
" std2 = 2 * std\n",
" breaks = [-4, -std2, -std15, -std, std, std15, std2, 4]\n",
"\n",
" for i, year in enumerate(years, \"Generating images\"):\n",
" file_name = \"SVI_{}_{}_{}\".format(study_area, day, year)\n",
" array = svi[:, :, i]\n",
" # Generating pngs for every time step\n",
" # Use pre-defined function to write pngs\n",
" generate_png(file_name, array, breaks, extent)\n",
"\n",
" # Generating geotiffs for every time step\n",
" # Use pre-defined function to write geotiffs\n",
" generate_geotiff(file_name, array, geo_transform, projection)\n",
" gc.collect() # Force garbage collection to save RAM"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Within the following cell the Modis files are loaded per DOY (Day of the year), the cloudmask from the pixel reliability dataset is applied, the EVI data is rescaled, the SVI is calculated and the SVI is saved as a smaller resolution PNG image and as a full scale Geotiff for each DOY and year of available data."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"__Note__: The script only works for files with file names which contain 'doy' followed by 7 digits indicating the year and day of the year (yyyyddd), since the DOY information is extracted from the file name. This corresponds to AppEEARS products, e.g. `MOD13Q1.006__250m_16_days_EVI_doy2001001_aid0001.tif` \n",
"\n",
"__Option__: If you want to use a different product (also geotiffs), you need to adapt `re_doy` to find the corresponding DOY sequences in the different product's filenames."
]
},
{
"cell_type": "code",
"execution_count": 6,
"metadata": {
"scrolled": true
},
"outputs": [
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "1ac74e97370e4b85b04eef48936fb86e",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"HBox(children=(IntProgress(value=0, description='Processing', max=2), HTML(value='')))"
]
},
"metadata": {},
"output_type": "display_data"
},
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"HBox(children=(IntProgress(value=0, description='Loading cloud_mask', max=19), HTML(value='')))"
]
},
"metadata": {},
"output_type": "display_data"
},
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"HBox(children=(IntProgress(value=0, description='Loading EVI', max=19), HTML(value='')))"
]
},
"metadata": {},
"output_type": "display_data"
},
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"HBox(children=(IntProgress(value=0, description='Generating images', max=19), HTML(value='')))"
]
},
"metadata": {},
"output_type": "display_data"
},
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"HBox(children=(IntProgress(value=0, description='Loading cloud_mask', max=19), HTML(value='')))"
]
},
"metadata": {},
"output_type": "display_data"
},
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"HBox(children=(IntProgress(value=0, description='Loading EVI', max=19), HTML(value='')))"
]
},
"metadata": {},
"output_type": "display_data"
},
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"HBox(children=(IntProgress(value=0, description='Generating images', max=19), HTML(value='')))"
]
},
"metadata": {},
"output_type": "display_data"
},
{
"name": "stdout",
"output_type": "stream",
"text": [
"\n"
]
}
],
"source": [
"# The following regular expression will match 'doy' followed by 7 digits,\n",
"# followed by 0 or more characters and ending in '.tif'\n",
"# It is used for building the initial file list of tif images that have a valid\n",
"# DOY structure\n",
"re_doy = re.compile(r\".*doy(\\d{7}).*\\.tif$\")\n",
"\n",
"# Build initial file list of tuples containing the filename, year and day for\n",
"# each file\n",
"# e.g. [( '/path/to/my_evi_doy2000193.tif', 2000, '193')]\n",
"# Note: the data types are stated in '<>'\n",
"evi_files = []\n",
"\n",
"# Create a list of files, which include the defined DOY in their filename\n",
"# for the EVI data\n",
"for _, _, files in os.walk(path_evi):\n",
" for file in files:\n",
" doy = get_doy(re_doy, file)\n",
" if doy is None:\n",
" continue\n",
" else:\n",
" evi_files.append((os.path.join(path_evi, file), doy[0], doy[1]))\n",
"\n",
"# Create a list of files, which include the defined DOY in their filename\n",
"# for the pixel reliability data\n",
"pr_files = []\n",
"\n",
"for _, _, files in os.walk(path_pr):\n",
" for file in files:\n",
" doy = get_doy(re_doy, file)\n",
" if doy is None:\n",
" continue\n",
" else:\n",
" pr_files.append((os.path.join(path_pr, file), doy[0], doy[1]))\n",
"\n",
"# Read an example file and define the shape of the data arrays\n",
"# Get the first file of the file list as example file\n",
"example_file = gdal.Open(evi_files[0][0])\n",
"\n",
"# Store necessary data from the example file\n",
"geo_transform = example_file.GetGeoTransform()\n",
"projection = example_file.GetProjection()\n",
"x_size = example_file.RasterXSize\n",
"y_size = example_file.RasterYSize\n",
"\n",
"del example_file # Save RAM\n",
"\n",
"# Preparing map reshaping\n",
"lon_start = geo_transform[0]\n",
"lon_stop = (x_size * geo_transform[1]) + geo_transform[0]\n",
"lon_step = geo_transform[1]\n",
"lat_start = geo_transform[3]\n",
"lat_stop = (y_size * geo_transform[5]) + geo_transform[3]\n",
"lat_step = geo_transform[5]\n",
"\n",
"extent = (lon_start, lon_stop, lat_stop, lat_start)\n",
"\n",
"# First make sure that image output directories exist, and if not then create\n",
"# them\n",
"# Check for png folder\n",
"if not os.path.exists(path_png):\n",
" os.makedirs(path_png)\n",
"\n",
"# Check for tif folder\n",
"if not os.path.exists(path_tif):\n",
" os.makedirs(path_tif)\n",
"\n",
"# Build a dictionary where keys are the available DOYs and values are sorted\n",
"# dictionaries for evi files and pr files\n",
"doy_dict = check_prepare_files(evi_files, pr_files)\n",
"\n",
"# The script creates some warnings due to the NA in the data.\n",
"# They are ignored by executing this cell.\n",
"warnings.filterwarnings('ignore')\n",
"\n",
"# If tqdm is available, use it.\n",
"# Note: both update_iter_desc and days_iterator are set in either case since it\n",
"# allows to avoid having many if clauses throughout the program and reduces\n",
"# code duplication.\n",
"if tqdm is not None:\n",
" def update_iter_desc(days_iterator, desc):\n",
" days_iterator.set_description(desc)\n",
" days_iterator = tqdm(sorted(doy_dict), desc=\"Processing\",\n",
" file=sys.stdout)\n",
"else:\n",
" def update_iter_desc(days_iterator, desc):\n",
" print(desc)\n",
" days_iterator = sorted(doy_dict)\n",
"\n",
"# Begin iterating through days of the year for which we have data.\n",
"for day in days_iterator:\n",
" # Provide a progress update\n",
" update_iter_desc(days_iterator, \"Processing DOY {}\".format(day))\n",
"\n",
" # Get the necessary files lists from doy_dict\n",
" evi_files_day = doy_dict[day]['evi']\n",
" pr_files_day = doy_dict[day]['pr']\n",
"\n",
" # Number of years for which we have data\n",
" num_years = len(evi_files_day)\n",
"\n",
" # Create a zero-filled 3D numpy array based on the example file\n",
" # dimensions\n",
" array_size = (y_size, x_size, num_years)\n",
" try:\n",
" # Adjust the size of the cloud mask array in place in order to save\n",
" # RAM and speed up processing\n",
" cloud_mask.resize(array_size)\n",
" except NameError:\n",
" # If this is the first iteration, cloud_mask will be undefined and\n",
" # trying to resize it will throw a NameError. This error is caught\n",
" # and cloud_mask is instantiated\n",
" cloud_mask = np.zeros(array_size, dtype=np.float64)\n",
"\n",
" # Reading the reliability data\n",
" cloud_mask = load_cloud_mask(pr_files_day, cloud_mask)\n",
"\n",
" # Preparing the reliability data\n",
" cloud_mask = prepare_cloud_mask(cloud_mask)\n",
"\n",
" # Reading the EVI data\n",
" evi = load_evi(evi_files_day, cloud_mask)\n",
"\n",
" # Preparing the EVI data\n",
" evi = prepare_evi(evi)\n",
"\n",
" update_iter_desc(days_iterator, \"Calculating SVI DOY {}\".format(day))\n",
" # Calculate SVI\n",
" svi = calculate_svi(evi)\n",
"\n",
" update_iter_desc(days_iterator, \"Generating images DOY {}\".format(day))\n",
" # Generating png images and geotiffs\n",
" generate_images(evi_files_day, svi, extent, geo_transform, projection)\n",
" \n",
" # Remove references to avoid array resize errors in future loops\n",
" del(evi, svi)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"***"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"__Note__: Using a large area of interest can lead to troubles due to a lack of memory (RAM). You can use the Task Manager to keep an eye on the memory usage while running this script. If you encounter problems, consider using a smaller area of interest (shapefile or polygon in AppEEARS) or a satellite product with a smaller resolution (500m (MOD13A1), 1km (MOD13A2))."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This is an example of the output PNG images. The geotiffs can be read by programs like QGIS, and used to produce maps.\n",
"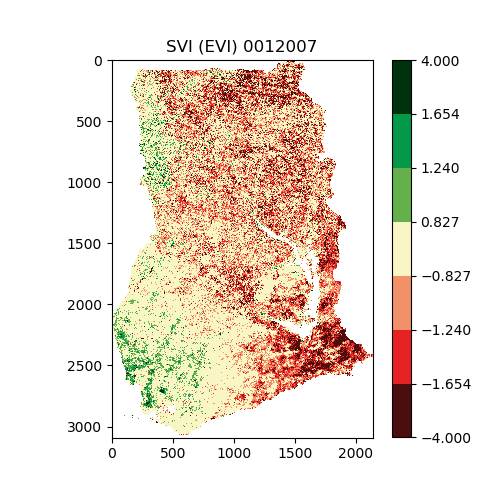"
]
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.3"
}
},
"nbformat": 4,
"nbformat_minor": 2
}