# Use-case 3: Classification, imjoy & python library usage
This use-case wraps the best performing approach of the [Kaggle Human Protein Atlas - Single Cell Classiication Challenge](https://www.kaggle.com/c/hpa-single-cell-image-classification).
It first segments the cells in the images using [a model for cell segmentation](https://bioimage.io/#/?id=10.5281%2Fzenodo.6200635) that predicts cell foreground and boundaries as input for a seeded watershed, with seeds from a [separate model for nucleus segmentation](https://bioimage.io/#/?id=10.5281%2Fzenodo.6200999).
The [HPA InceptionV3 model](https://bioimage.io/#/?id=10.5281%2Fzenodo.5910854) then classifies the individual cells.
We demonstrate how this approach can be wrapped for 3 different backends using the bioimageio models and consumer software.
## Usage in python library
The `hpa_app.py` scrips loads data from the hpa website, runs cell segmentation and then classifies each cell.
To this end it makes use of the python [bioimageio.core](https://github.com/bioimage-io/core-bioimage-io-python) library that allows to integrate bioimageio models into any python application.
It uses [napari](https://github.com/napari/napari) to visualize the segmentation and classification results. See the results or two images, one with nucleoplasm staining, the other with microtubulus staining below.
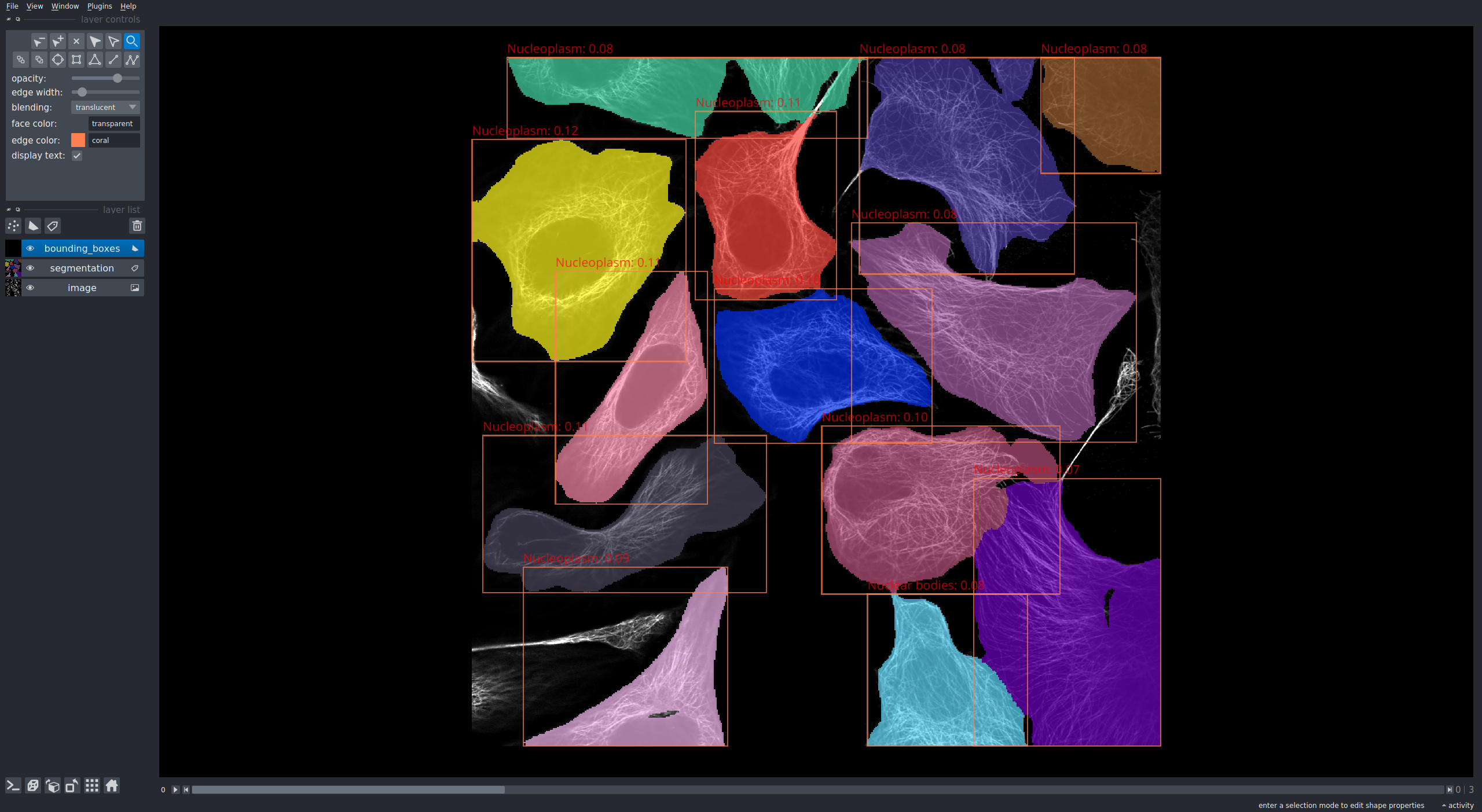
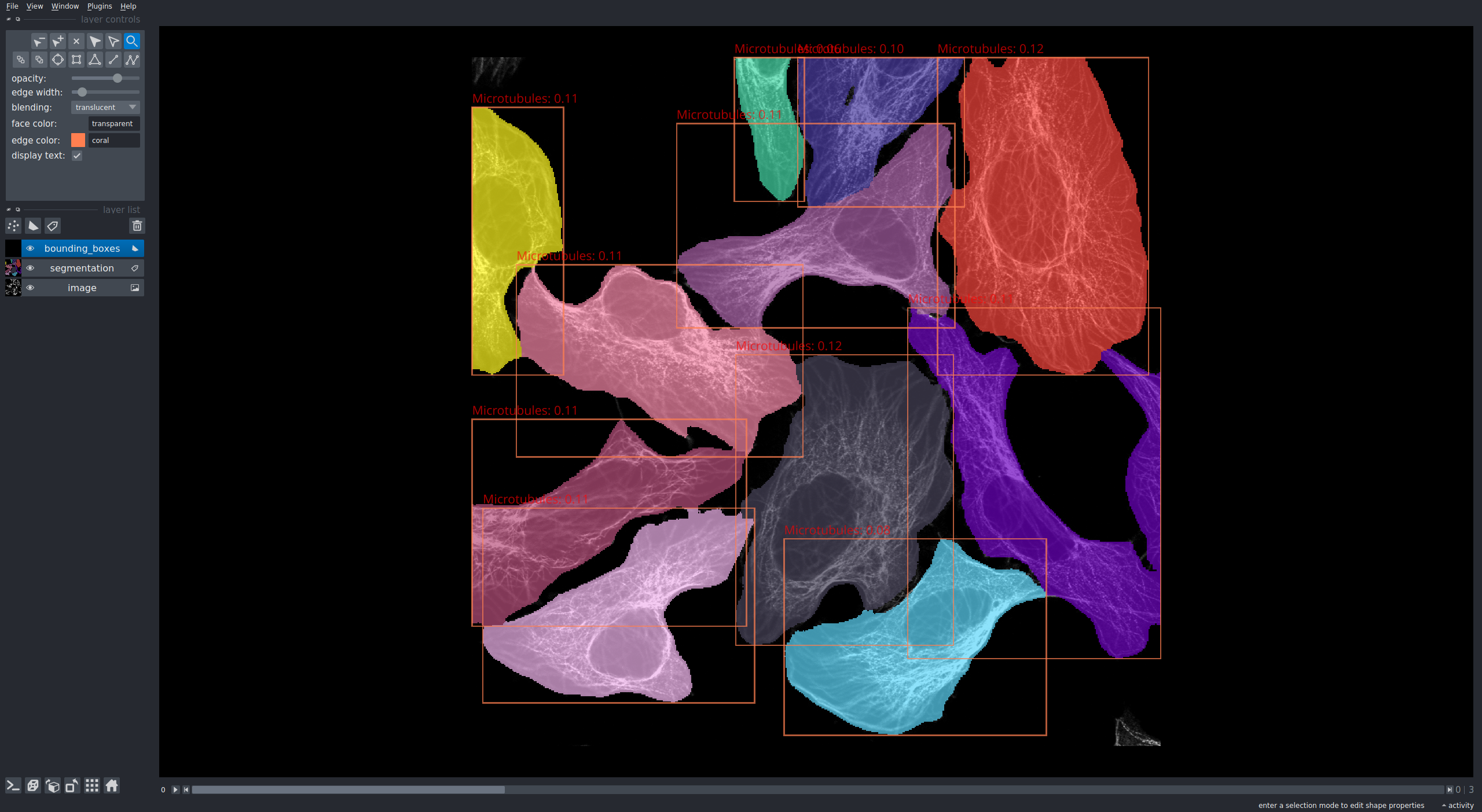 ### Dependencies
An conda environment that can run the app is defined in `environment.yaml`.
## Usage in imjoy / bioengine
The code for this use case is implemented in [this notebook](https://github.com/bioimage-io/use-cases/blob/main/case3-devtools/bioengine-model-runner-hpa-single-cell.ipynb).
### Dependencies
An conda environment that can run the app is defined in `environment.yaml`.
## Usage in imjoy / bioengine
The code for this use case is implemented in [this notebook](https://github.com/bioimage-io/use-cases/blob/main/case3-devtools/bioengine-model-runner-hpa-single-cell.ipynb).

 ### Dependencies
An conda environment that can run the app is defined in `environment.yaml`.
## Usage in imjoy / bioengine
The code for this use case is implemented in [this notebook](https://github.com/bioimage-io/use-cases/blob/main/case3-devtools/bioengine-model-runner-hpa-single-cell.ipynb).
### Dependencies
An conda environment that can run the app is defined in `environment.yaml`.
## Usage in imjoy / bioengine
The code for this use case is implemented in [this notebook](https://github.com/bioimage-io/use-cases/blob/main/case3-devtools/bioengine-model-runner-hpa-single-cell.ipynb).