Split-apply-combine and parallel computing
library(tidyverse)
library(gapminder)
library(stringr)
set.seed(1234)
theme_set(theme_minimal())Split-apply-combine
A common analytical pattern is to
- Split data into pieces,
- Apply some function to each piece,
- Combine the results back together again.
We have used this technique many times so far without explicitly identifying it as such.
dplyr::group_by()
gapminder %>%
group_by(continent) %>%
summarize(n = n())## # A tibble: 5 x 2
## continent n
## <fctr> <int>
## 1 Africa 624
## 2 Americas 300
## 3 Asia 396
## 4 Europe 360
## 5 Oceania 24gapminder %>%
group_by(continent) %>%
summarize(avg_lifeExp = mean(lifeExp))## # A tibble: 5 x 2
## continent avg_lifeExp
## <fctr> <dbl>
## 1 Africa 48.86533
## 2 Americas 64.65874
## 3 Asia 60.06490
## 4 Europe 71.90369
## 5 Oceania 74.32621for loops
countries <- unique(gapminder$country)
lifeExp_models <- vector("list", length(countries))
names(lifeExp_models) <- countries
for(i in seq_along(countries)){
lifeExp_models[[i]] <- lm(lifeExp ~ year,
data = filter(gapminder,
country == countries[[i]]))
}
head(lifeExp_models)## $Afghanistan
##
## Call:
## lm(formula = lifeExp ~ year, data = filter(gapminder, country ==
## countries[[i]]))
##
## Coefficients:
## (Intercept) year
## -507.5343 0.2753
##
##
## $Albania
##
## Call:
## lm(formula = lifeExp ~ year, data = filter(gapminder, country ==
## countries[[i]]))
##
## Coefficients:
## (Intercept) year
## -594.0725 0.3347
##
##
## $Algeria
##
## Call:
## lm(formula = lifeExp ~ year, data = filter(gapminder, country ==
## countries[[i]]))
##
## Coefficients:
## (Intercept) year
## -1067.8590 0.5693
##
##
## $Angola
##
## Call:
## lm(formula = lifeExp ~ year, data = filter(gapminder, country ==
## countries[[i]]))
##
## Coefficients:
## (Intercept) year
## -376.5048 0.2093
##
##
## $Argentina
##
## Call:
## lm(formula = lifeExp ~ year, data = filter(gapminder, country ==
## countries[[i]]))
##
## Coefficients:
## (Intercept) year
## -389.6063 0.2317
##
##
## $Australia
##
## Call:
## lm(formula = lifeExp ~ year, data = filter(gapminder, country ==
## countries[[i]]))
##
## Coefficients:
## (Intercept) year
## -376.1163 0.2277nest() and map()
# function to estimate linear model for gapminder subsets
le_vs_yr <- function(df) {
lm(lifeExp ~ year, data = df)
}
# split data into nests
(gap_nested <- gapminder %>%
group_by(continent, country) %>%
nest())## # A tibble: 142 x 3
## continent country data
## <fctr> <fctr> <list>
## 1 Asia Afghanistan <tibble [12 x 4]>
## 2 Europe Albania <tibble [12 x 4]>
## 3 Africa Algeria <tibble [12 x 4]>
## 4 Africa Angola <tibble [12 x 4]>
## 5 Americas Argentina <tibble [12 x 4]>
## 6 Oceania Australia <tibble [12 x 4]>
## 7 Europe Austria <tibble [12 x 4]>
## 8 Asia Bahrain <tibble [12 x 4]>
## 9 Asia Bangladesh <tibble [12 x 4]>
## 10 Europe Belgium <tibble [12 x 4]>
## # ... with 132 more rows# apply a linear model to each nested data frame
(gap_nested <- gap_nested %>%
mutate(fit = map(data, le_vs_yr)))## # A tibble: 142 x 4
## continent country data fit
## <fctr> <fctr> <list> <list>
## 1 Asia Afghanistan <tibble [12 x 4]> <S3: lm>
## 2 Europe Albania <tibble [12 x 4]> <S3: lm>
## 3 Africa Algeria <tibble [12 x 4]> <S3: lm>
## 4 Africa Angola <tibble [12 x 4]> <S3: lm>
## 5 Americas Argentina <tibble [12 x 4]> <S3: lm>
## 6 Oceania Australia <tibble [12 x 4]> <S3: lm>
## 7 Europe Austria <tibble [12 x 4]> <S3: lm>
## 8 Asia Bahrain <tibble [12 x 4]> <S3: lm>
## 9 Asia Bangladesh <tibble [12 x 4]> <S3: lm>
## 10 Europe Belgium <tibble [12 x 4]> <S3: lm>
## # ... with 132 more rows# combine the results back into a single data frame
library(broom)
(gap_nested <- gap_nested %>%
mutate(tidy = map(fit, tidy)))## # A tibble: 142 x 5
## continent country data fit tidy
## <fctr> <fctr> <list> <list> <list>
## 1 Asia Afghanistan <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 2 Europe Albania <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 3 Africa Algeria <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 4 Africa Angola <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 5 Americas Argentina <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 6 Oceania Australia <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 7 Europe Austria <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 8 Asia Bahrain <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 9 Asia Bangladesh <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## 10 Europe Belgium <tibble [12 x 4]> <S3: lm> <data.frame [2 x 5]>
## # ... with 132 more rows(gap_coefs <- gap_nested %>%
select(continent, country, tidy) %>%
unnest(tidy))## # A tibble: 284 x 7
## continent country term estimate std.error statistic
## <fctr> <fctr> <chr> <dbl> <dbl> <dbl>
## 1 Asia Afghanistan (Intercept) -507.5342716 40.484161954 -12.536613
## 2 Asia Afghanistan year 0.2753287 0.020450934 13.462890
## 3 Europe Albania (Intercept) -594.0725110 65.655359062 -9.048348
## 4 Europe Albania year 0.3346832 0.033166387 10.091036
## 5 Africa Algeria (Intercept) -1067.8590396 43.802200843 -24.379118
## 6 Africa Algeria year 0.5692797 0.022127070 25.727749
## 7 Africa Angola (Intercept) -376.5047531 46.583370599 -8.082385
## 8 Africa Angola year 0.2093399 0.023532003 8.895964
## 9 Americas Argentina (Intercept) -389.6063445 9.677729641 -40.258031
## 10 Americas Argentina year 0.2317084 0.004888791 47.395847
## # ... with 274 more rows, and 1 more variables: p.value <dbl>Parallel computing
Parallel computing (or processing) is a type of computation whereby many calculations or processes are carried out simultaneously.1 Rather than processing problems in serial (or sequential) order, the computer splits the task up into smaller parts that can be processed simultaneously using multiple processors. This is also called multithreading. By spliting the job up into simultaneous operations running in parallel, you complete your operation quicker, making the code more efficient. This approach works great with split-apply-combine because all the applied operations can be run independently. Why wait for the first chunk to complete if you can perform the operation on the second chunk at the same time?
Why use parallel computing
- Parallel computing imitates real life - in the real world, people use their brains to think in parallel - we multitask all the time without even thinking about it. Institutions are structured to process information in parallel, rather than in serial.
- It can be more efficient - by throwing more resources at a problem you can shorten the time to completion.
- You can tackle larger problems - more resources enables you to scale up the amount of data you process and potentially solve a larger problem.
- Distributed resources are cheaper than upgrading your own equipment. Why spend thousands of dollars beefing up your own laptop when you can instead rent computing resources from Google or Amazon for mere pennies?
Why not to use parallel computing
Limits to efficiency gains - Amdahl’s law defines theoretical limits to how much you can speed up computations via parallel computing. Because of this, you achieve diminishing returns over time.
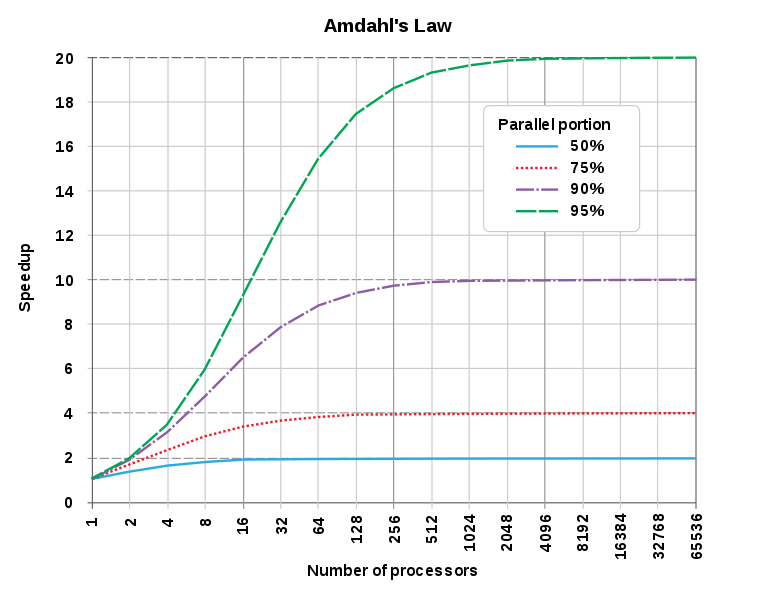
Amdahl’s Law from Wikipedia
- Complexity - writing parallel code can be more complicated than writing serial code, especially in R because it does not natively implement parallel computing - you have to explicitly build it into your script.
- Dependencies - your computation may rely on the output from the first set of tasks to perform the second tasks. If you compute the problem in parallel fashion, the individual chunks do not communicate with one another.
Parallel slowdown - parallel computing speeds up computations at a price. Once the problem is broken into separate threads, reading and writing data from the threads to memory or the hard drive takes time. Some tasks are not improved by spliting the process into parallel operations.
multidplyr
multidplyr is a work-in-progress package that implements parallel computing locally using dplyr. Rather than performing computations using a single core or processor, it spreads the computation across multiple cores. The basic sequence of steps is:
- Call
partition()to split the dataset across multiple cores. This makes a partitioned data frame, or aparty dffor short. - Each
dplyrverb applied to aparty dfperforms the operation independently on each core. It leaves each result on each core, and returns anotherparty df. - When you’re done with the expensive operations that need to be done on each core, you call
collect()to retrieve the data and bring it back to you local computer.
nycflights13::flights
Install multidplyr if you don’t have it already.
devtools::install_github("hadley/multidplyr")library(multidplyr)
library(nycflights13)Next, partition the flights data by flight number, compute the average delay per flight, and then collect the results:
flights1 <- partition(flights, flight)
flights2 <- summarize(flights1, dep_delay = mean(dep_delay, na.rm = TRUE))
flights3 <- collect(flights2)The dplyr code looks the same as usual, but behind the scenes things are very different. flights1 and flights2 are party dfs. These look like normal data frames, but have an additional attribute: the number of shards. In this example, it tells us that flights2 is spread across three nodes, and the size on each node varies from 1275 to 1286 rows. partition() always makes sure a group is kept together on one node.
flights2## Source: party_df [3,844 x 2]
## Shards: 3 [1,237--1,304 rows]
##
## # S3: party_df
## flight dep_delay
## <int> <dbl>
## 1 2 -0.5686275
## 2 3 3.6650794
## 3 4 7.5166240
## 4 6 8.5024155
## 5 8 6.9358974
## 6 10 24.3114754
## 7 11 6.8242991
## 8 12 28.2834646
## 9 15 10.2643080
## 10 19 10.0500000
## # ... with 3,834 more rowsPerformance
For this size of data, using a local cluster actually makes performance slower.
system.time({
flights %>%
partition() %>%
summarise(mean(dep_delay, na.rm = TRUE)) %>%
collect()
})## user system elapsed
## 0.474 0.066 0.968system.time({
flights %>%
group_by() %>%
summarise(mean(dep_delay, na.rm = TRUE))
})## user system elapsed
## 0.007 0.000 0.006That’s because there’s some overhead associated with sending the data to each node and retrieving the results at the end. For basic dplyr verbs, multidplyr is unlikely to give you significant speed ups unless you have 10s or 100s of millions of data points. It might however, if you’re doing more complex things.
gapminder
Let’s now return to gapminder and estimate separate linear regression models of life expectancy based on year for each country. We will use multidplyr to split the work across multiple cores. Note that we need to use cluster_library() to load the purrr package on every node.
# split data into nests
gap_nested <- gapminder %>%
group_by(continent, country) %>%
nest()
# partition gap_nested across the cores
gap_nested_part <- gap_nested %>%
partition(country)
# apply a linear model to each nested data frame
cluster_library(gap_nested_part, "purrr")
system.time({
gap_nested_part %>%
mutate(fit = map(data, function(df) lm(lifeExp ~ year, data = df)))
})## user system elapsed
## 0.002 0.000 0.113Compared to how long running it locally?
system.time({
gap_nested %>%
mutate(fit = map(data, function(df) lm(lifeExp ~ year, data = df)))
})## user system elapsed
## 0.109 0.002 0.112So it’s roughly 2 times faster to run in parallel. Admittedly you saved only a fraction of a second. In relative terms this is great, but in absolute terms it doesn’t mean much. This demonstrates it doesn’t always make sense to parallelize operations - only do so if you can make significant gains in computation speed. If each country had thousands of observations, the efficiency gains would have been more dramatic.
Acknowledgments
Session Info
devtools::session_info()## setting value
## version R version 3.4.3 (2017-11-30)
## system x86_64, darwin15.6.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## tz America/Chicago
## date 2018-04-24
##
## package * version date source
## assertthat 0.2.0 2017-04-11 CRAN (R 3.4.0)
## backports 1.1.2 2017-12-13 CRAN (R 3.4.3)
## base * 3.4.3 2017-12-07 local
## bindr 0.1.1 2018-03-13 CRAN (R 3.4.3)
## bindrcpp 0.2.2.9000 2018-04-08 Github (krlmlr/bindrcpp@bd5ae73)
## broom 0.4.4 2018-03-29 CRAN (R 3.4.3)
## cellranger 1.1.0 2016-07-27 CRAN (R 3.4.0)
## cli 1.0.0 2017-11-05 CRAN (R 3.4.2)
## colorspace 1.3-2 2016-12-14 CRAN (R 3.4.0)
## compiler 3.4.3 2017-12-07 local
## crayon 1.3.4 2017-10-03 Github (gaborcsardi/crayon@b5221ab)
## datasets * 3.4.3 2017-12-07 local
## devtools 1.13.5 2018-02-18 CRAN (R 3.4.3)
## digest 0.6.15 2018-01-28 CRAN (R 3.4.3)
## dplyr * 0.7.4.9003 2018-04-08 Github (tidyverse/dplyr@b7aaa95)
## evaluate 0.10.1 2017-06-24 CRAN (R 3.4.1)
## forcats * 0.3.0 2018-02-19 CRAN (R 3.4.3)
## foreign 0.8-69 2017-06-22 CRAN (R 3.4.3)
## gapminder * 0.3.0 2017-10-31 CRAN (R 3.4.2)
## ggplot2 * 2.2.1.9000 2018-04-24 Github (tidyverse/ggplot2@3c9c504)
## glue 1.2.0 2017-10-29 CRAN (R 3.4.2)
## graphics * 3.4.3 2017-12-07 local
## grDevices * 3.4.3 2017-12-07 local
## grid 3.4.3 2017-12-07 local
## gtable 0.2.0 2016-02-26 CRAN (R 3.4.0)
## haven 1.1.1 2018-01-18 CRAN (R 3.4.3)
## hms 0.4.2 2018-03-10 CRAN (R 3.4.3)
## htmltools 0.3.6 2017-04-28 CRAN (R 3.4.0)
## httr 1.3.1 2017-08-20 CRAN (R 3.4.1)
## jsonlite 1.5 2017-06-01 CRAN (R 3.4.0)
## knitr 1.20 2018-02-20 CRAN (R 3.4.3)
## lattice 0.20-35 2017-03-25 CRAN (R 3.4.3)
## lazyeval 0.2.1 2017-10-29 CRAN (R 3.4.2)
## lubridate 1.7.4 2018-04-11 CRAN (R 3.4.3)
## magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
## memoise 1.1.0 2017-04-21 CRAN (R 3.4.0)
## methods * 3.4.3 2017-12-07 local
## mnormt 1.5-5 2016-10-15 CRAN (R 3.4.0)
## modelr 0.1.1 2017-08-10 local
## munsell 0.4.3 2016-02-13 CRAN (R 3.4.0)
## nlme 3.1-137 2018-04-07 CRAN (R 3.4.4)
## parallel 3.4.3 2017-12-07 local
## pillar 1.2.1 2018-02-27 CRAN (R 3.4.3)
## pkgconfig 2.0.1 2017-03-21 CRAN (R 3.4.0)
## plyr 1.8.4 2016-06-08 CRAN (R 3.4.0)
## psych 1.8.3.3 2018-03-30 CRAN (R 3.4.4)
## purrr * 0.2.4 2017-10-18 CRAN (R 3.4.2)
## R6 2.2.2 2017-06-17 CRAN (R 3.4.0)
## Rcpp 0.12.16 2018-03-13 CRAN (R 3.4.4)
## readr * 1.1.1 2017-05-16 CRAN (R 3.4.0)
## readxl 1.0.0 2017-04-18 CRAN (R 3.4.0)
## reshape2 1.4.3 2017-12-11 CRAN (R 3.4.3)
## rlang 0.2.0.9001 2018-04-24 Github (r-lib/rlang@82b2727)
## rmarkdown 1.9 2018-03-01 CRAN (R 3.4.3)
## rprojroot 1.3-2 2018-01-03 CRAN (R 3.4.3)
## rstudioapi 0.7 2017-09-07 CRAN (R 3.4.1)
## rvest 0.3.2 2016-06-17 CRAN (R 3.4.0)
## scales 0.5.0.9000 2018-04-24 Github (hadley/scales@d767915)
## stats * 3.4.3 2017-12-07 local
## stringi 1.1.7 2018-03-12 CRAN (R 3.4.3)
## stringr * 1.3.0 2018-02-19 CRAN (R 3.4.3)
## tibble * 1.4.2 2018-01-22 CRAN (R 3.4.3)
## tidyr * 0.8.0 2018-01-29 CRAN (R 3.4.3)
## tidyselect 0.2.4 2018-02-26 CRAN (R 3.4.3)
## tidyverse * 1.2.1 2017-11-14 CRAN (R 3.4.2)
## tools 3.4.3 2017-12-07 local
## utils * 3.4.3 2017-12-07 local
## withr 2.1.2 2018-04-24 Github (jimhester/withr@79d7b0d)
## xml2 1.2.0 2018-01-24 CRAN (R 3.4.3)
## yaml 2.1.18 2018-03-08 CRAN (R 3.4.4)This work is licensed under the CC BY-NC 4.0 Creative Commons License.
