R basics
Overview
- Introduce the RStudio interface and panes
- Demonstrate some basic R/RStudio tips and tricks
- Distinguish scripts vs. R Markdown documents
- Define an RStudio project
- Clarify some assorted Git-related issues
RStudio interface
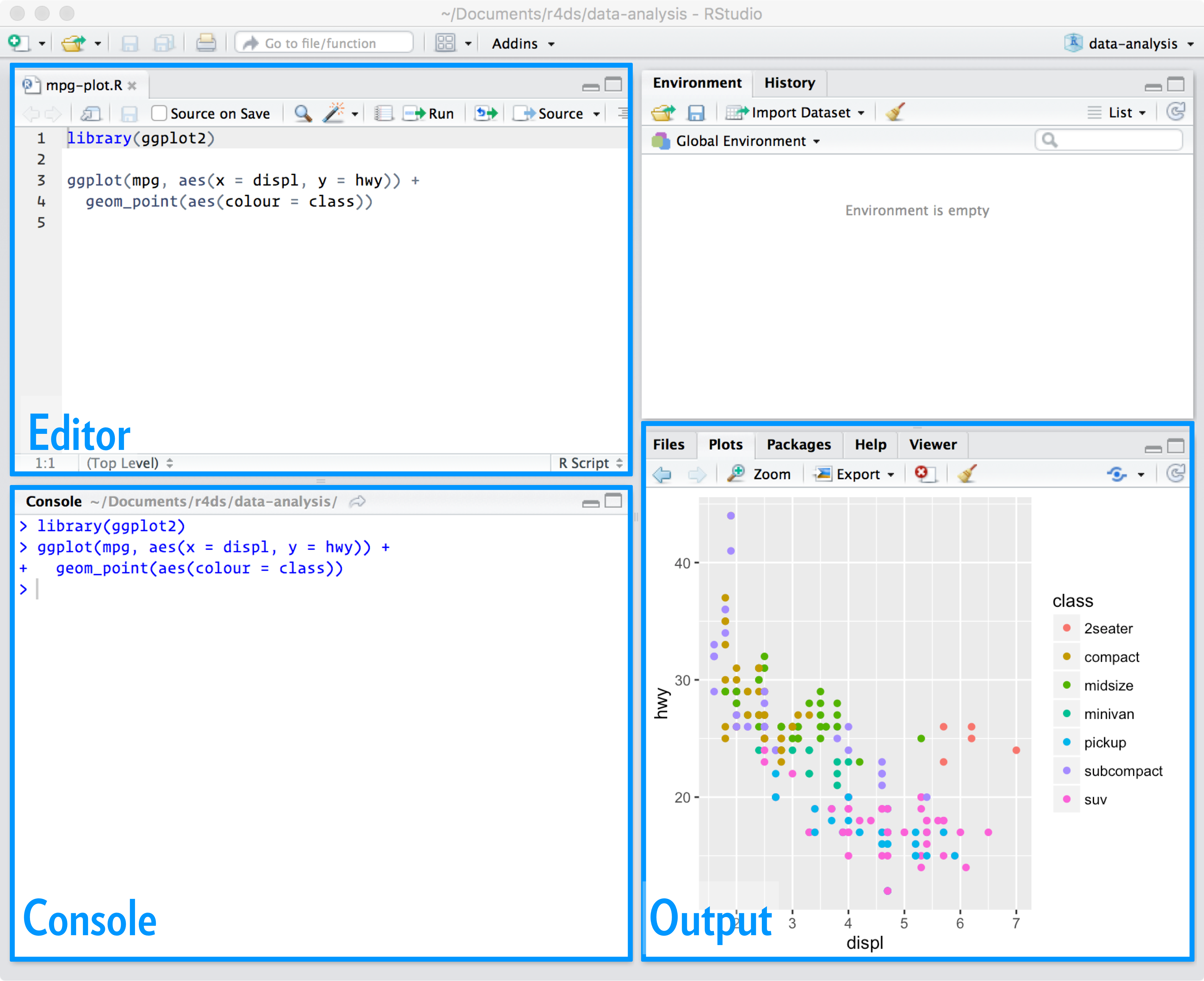
From R for Data Science
Launch RStudio.
Notice the default panes:
- Console (lower-left) - allows you to run code interactively in R.
- Editor (upper-left) - where you write scripts (more on that later).
- Environment/History (tabbed in upper right)
- Environment - shows a list of all objects currently in your workspace
- History - prints a list of all commands run in the R session
- Git - if you are using a Git-enabled repo, this will allow you to stage, commit, push, and pull to GitHub
- Files/Plots/Packages/Help (tabbed in lower right)
- Files - lists all files in the directory. Allows you to navigate through the file system.
- Plots - plots generated in the console and by R scripts will be displayed here.
- Packages - lists all the packages currently installed on your computer
- Help - interface to lookup help files for functions in R
Go into the Console, where we interact with the live R process.
Make an assignment and then inspect the object you just created.
x <- 3 * 4
x## [1] 12All R statements where you create objects (assignments) have this form:
objectName <- valueYou will make lots of assignments and the operator
<-is a pain to type. Don’t be lazy and use=, although it would work, because it will just sow confusion later. Instead, utilize RStudio’s keyboard shortcut: Alt/Option + - (the minus sign).
- Notice that RStudio automagically surrounds
<-with spaces, which demonstrates a useful code formatting practice. Code is miserable to read on a good day. Give your eyes a break and use spaces. - RStudio offers many handy keyboard shortcuts. Also, Alt+Shift+K brings up a keyboard shortcut reference card.
Assignment and object names
Object names cannot start with a digit and cannot contain certain other characters such as a comma or a space. You will be wise to adopt a convention for demarcating words in names.
i_use_snake_case
other.people.use.periods
evenOthersUseCamelCaseMake another assignment:
this_is_a_really_long_name <- 2.5To inspect this, try out RStudio’s completion facility: type the first few characters, press TAB, add characters until you disambiguate, then press return.
Make another assignment:
science_rules <- 2 ^ 3Let’s try to inspect:
sciencerules## Error in eval(expr, envir, enclos): object 'sciencerules' not foundsceince_rules## Error in eval(expr, envir, enclos): object 'sceince_rules' not foundImplicit contract with the computer / scripting language: Computer will do tedious computation for you. In return, you will be completely precise in your instructions. Typos matter. Case matters. Get better at typing.
Built-in functions
R has a mind-blowing collection of built-in functions that are accessed like so:
functionName(arg1 = val1, arg2 = val2, and so on)Let’s try using seq() which makes regular sequences of numbers and, while we’re at it, demo more helpful features of RStudio.
Type se in the console and hit TAB. A pop up shows you possible completions. Specify seq() by typing more to disambiguate or using the up/down arrows to select. Notice the floating tool-tip-type help that pops up, reminding you of a function’s arguments. If you want even more help, press F1 as directed to get the full documentation in the help tab of the lower right pane. Now open the parentheses and notice the automatic addition of the closing parenthesis and the placement of cursor in the middle. Type the arguments 1, 10 and hit return. RStudio also exits the parenthetical expression for you. IDEs are great.
seq(1, 10)## [1] 1 2 3 4 5 6 7 8 9 10The above also demonstrates something about how R resolves function arguments. You can always specify in name = value form. But if you do not, R attempts to resolve by position. So above, it is assumed that we want a sequence from = 1 that goes to = 10. Since we didn’t specify step size, the default value of by in the function definition is used, which ends up being 1 in this case. Be careful relying on this feature too often. If you use a function where you’re specifying more than 3 arguments, you might want to use name = value to avoid any later confusion.
Make this assignment and notice similar help with quotation marks.
yo <- "hello world"If you just make an assignment, you don’t get to see the value, so then you’re tempted to immediately inspect.
y <- seq(1, 10)
y## [1] 1 2 3 4 5 6 7 8 9 10This common action can be shortened by surrounding the assignment with parentheses, which causes assignment and “print to screen” to happen.
(y <- seq(1, 10))## [1] 1 2 3 4 5 6 7 8 9 10Not all functions have (or require) arguments:
date()## [1] "Tue Jan 3 12:56:45 2017"Now look at your workspace – in the upper right pane. The workspace is where user-defined objects accumulate. You can also get a listing of these objects with commands:
objects()## [1] "science_rules" "this_is_a_really_long_name"
## [3] "x" "y"
## [5] "yo"ls()## [1] "science_rules" "this_is_a_really_long_name"
## [3] "x" "y"
## [5] "yo"If you want to remove the object named y, you can do this
rm(y)To remove everything:
rm(list = ls())or click the broom in RStudio’s Environment pane.
Console editor vs. scripts vs. R Markdown
- Console editor
- Great for experimenting and interactive coding in R
- No record of past commands
- Must run one line at a time
- Script editor
- Build code in chunks, then run all at once
- Save as a
.Rfile (called an R script) - Can run:
- One line at a time (Cmd/Ctrl + Enter)
- Several lines at once (highlight the code with the cursor, then Cmd/Ctrl + Enter)
- Run the entire script at once (Cmd/Ctrl + Shift + S)
- Output is printed in the console
- Plots are displayed in the bottom-right pane
- You can split a complicated program/workflow into multiple and distinct R scripts (easier to organize large chunks of code)
- R Markdown
- Provides a unified authoring framework for data science
- Combines:
- Code
- Results
- Written commentary
- Displays output and plots within the document (can be changed)
- Good for a final report
- During class, usually better to work in an R script until you are comfortable using R Markdown for homework
For more information on R Markdown, read the R for Data Science chapter and read this short series of lessons on R Markdown.
Workflow: projects and working directories
Go to R for Data Science and read chapter 8 on workflows and R projects. Do not skip this chapter. It is brief, but contains lots of important information on:
- Your environment
- The working directory
- Navigating paths and directories
- Creating and using projects
If you don’t know what those terms refer to, go back and read the chapter. Right now.
Prove you understand projects by doing the following:
- Instruct RStudio not to preserve your workspace between sessions.
- Create a new, empty project called
my_project. Store it wherever you like on your computer, but you should quickly adopt some sort of directory structure for all your class-related projects. Create a new R script called
diamonds.R. Add the following code to the script, save it, and run the script:library(tidyverse) ggplot(diamonds, aes(carat, price)) + geom_hex() ggsave("diamonds.pdf") write_csv(diamonds, "diamonds.csv")- Quit RStudio. Go to your project folder using Windows Explorer or Finder (Mac). Verify you now have an
.Rprojfile as well as a PDF image and a csv file containing thediamondsdataset. Re-open RStudio by double-clicking on the
.Rprojfile. You should immediately see your previous working space containing the script and history. However your environment should be completely clean.
Assorted R stuff
- It is traditional to save R scripts with a
.Ror.rsuffix. Follow this convention unless you have some extraordinary reason not to. - Comments start with one or more
#symbols. Use them. RStudio helps you (de)comment selected lines with Ctrl+Shift+C (windows and linux) or Command+Shift+C (mac). - Clean out the workspace. That is, pretend like you’ve just revisited your project after a long absence. The broom icon or
rm(list = ls())will do this. When troubleshooting code, it is a good idea to do this, restart R (available from the Session menu), then re-run your analysis to truly check that the code you’re saving is complete and correct (or at least rule out obvious problems!). - This workflow will serve you well in the future:
- Create an RStudio project for an analytical project
- Keep inputs there (we’ll soon talk about importing)
- Keep scripts there; edit them, run them in bits or as a whole from there
- Keep outputs there (like the PDF written above)
- Avoid using the mouse for pieces of your analytical workflow, such as loading a dataset or saving a figure. It is terribly important for reproducility and for making it possible to retrospectively determine how a numerical table or PDF was actually produced (searching on local disk on filename, among
.Rfiles, will lead to the relevant script). You don’t know how to do this yet, but you will learn very soon.
Assorted things about Git and GitHub
.gitignore
By default, Git tracks all directories and files in your repository. Sometimes you may not want it to track everything. For instance, if you store a private API key or personally-identifiable data, you won’t want these files tracked by Git. If you did, when you push your repository to GitHub your private files will be shared with the world.
You could just store all of these files outside your repository, but that’s a pain and inconvenient. Instead, you can create a .gitignore file in your repository. This is a special file Git uses to determine what files it should ignore. Any file listed in .gitignore will not be tracked by Git.
When you create a new repository in GitHub (as opposed to forking an existing one), you have the option to add a template .gitignore file depending on what programming language you will use. For example, the default .gitignore file for R is
# History files
.Rhistory
.Rapp.history
# Session Data files
.RData
# Example code in package build process
*-Ex.R
# Output files from R CMD build
/*.tar.gz
# Output files from R CMD check
/*.Rcheck/
# RStudio files
.Rproj.user/
# produced vignettes
vignettes/*.html
vignettes/*.pdf
# OAuth2 token, see https://github.com/hadley/httr/releases/tag/v0.3
.httr-oauth
# knitr and R markdown default cache directories
/*_cache/
/cache/
# Temporary files created by R markdown
*.utf8.md
*.knit.md
.Rproj.userMost of these files are not sensitive, but are merely temporary work files that you don’t need to save and track using version control. You can specify files and directories by their full name, a partial name, or file extension. Starting with homework 2 I will always include a .gitignore in the repository, but for your own projects you will need to create these files as you find necessary.
Clone from the fork, not the master
Make sure whenever you clone a homework repository, use the url for the forked version, not the master repository. So for the first homework, I would use https://github.com/bensoltoff/hw01 when I clone the repo, not https://github.com/uc-cfss/hw01. If you use the master repo url, you will get an error when you try to push your changes to GitHub.
For an example, let’s say I wanted to make a contribution to ggplot2. I should fork the repo and clone the fork. Instead I goofed and cloned the original repo. When I try to push my change, I get an error message:
remote: Permission to hadley/ggplot2.git denied to bensoltoff.
fatal: unable to access 'https://github.com/hadley/ggplot2.git/': The requested URL returned error: 403I don’t have permission to edit the master repo on Hadley Wickham’s account.
How do I fix this? I could go back and clone the correct fork, but if I’ve already made several commits then I’ll lose all my work. Instead, I can change the upstream url: this changes the location Git tries to push my changes. To do this:
- Open up the shell
- Change the current working directory to your local project (should use the
cdcommand) List your existing remotes in order to get the name of the remote you want to change.
git remote -vorigin https://github.com/hadley/ggplot2.git (fetch) origin https://github.com/hadley/ggplot2.git (push)Change your remote’s URL to the fork with the
git remote set-urlcommand.git remote set-url originVerify that the remote URL has changed.
git remote -vorigin https://github.com/bensoltoff/ggplot2 (fetch) origin https://github.com/bensoltoff/ggplot2 (push)
Now I can push successfully to my fork, then submit a pull request.
Use the proper shell (GitBash for Windows)
Make sure to use the proper program when entering the shell. For Mac users, that is Terminal. For Windows users, that is GitBash: if you followed the setup instructions properly, you should have this program on your computer. Look for it under the Start Menu > Git > GitBash. If you try to use the Command Prompt, you will run into errors because it uses different commands than GitBash.
Acknowledgements
- This page is derived in part from “UBC STAT 545A and 547M”, licensed under the CC BY-NC 3.0 Creative Commons License.
Session Info
devtools::session_info()## Session info -------------------------------------------------------------## setting value
## version R version 3.4.3 (2017-11-30)
## system x86_64, darwin15.6.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## tz America/Chicago
## date 2018-03-29## Packages -----------------------------------------------------------------## package * version date source
## assertthat 0.2.0 2017-04-11 CRAN (R 3.4.0)
## backports 1.1.2 2017-12-13 CRAN (R 3.4.3)
## base * 3.4.3 2017-12-07 local
## bindr 0.1.1 2018-03-13 CRAN (R 3.4.3)
## bindrcpp 0.2 2017-06-17 CRAN (R 3.4.0)
## broom 0.4.3 2017-11-20 CRAN (R 3.4.1)
## cellranger 1.1.0 2016-07-27 CRAN (R 3.4.0)
## cli 1.0.0 2017-11-05 CRAN (R 3.4.2)
## colorspace 1.3-2 2016-12-14 CRAN (R 3.4.0)
## compiler 3.4.3 2017-12-07 local
## crayon 1.3.4 2017-10-03 Github (gaborcsardi/crayon@b5221ab)
## datasets * 3.4.3 2017-12-07 local
## devtools 1.13.5 2018-02-18 CRAN (R 3.4.3)
## digest 0.6.15 2018-01-28 CRAN (R 3.4.3)
## dplyr * 0.7.4.9000 2017-10-03 Github (tidyverse/dplyr@1a0730a)
## evaluate 0.10.1 2017-06-24 CRAN (R 3.4.1)
## forcats * 0.3.0 2018-02-19 CRAN (R 3.4.3)
## foreign 0.8-69 2017-06-22 CRAN (R 3.4.3)
## ggplot2 * 2.2.1 2016-12-30 CRAN (R 3.4.0)
## glue 1.2.0 2017-10-29 CRAN (R 3.4.2)
## graphics * 3.4.3 2017-12-07 local
## grDevices * 3.4.3 2017-12-07 local
## grid 3.4.3 2017-12-07 local
## gtable 0.2.0 2016-02-26 CRAN (R 3.4.0)
## haven 1.1.1 2018-01-18 CRAN (R 3.4.3)
## hms 0.4.2 2018-03-10 CRAN (R 3.4.3)
## htmltools 0.3.6 2017-04-28 CRAN (R 3.4.0)
## httr 1.3.1 2017-08-20 CRAN (R 3.4.1)
## jsonlite 1.5 2017-06-01 CRAN (R 3.4.0)
## knitr 1.20 2018-02-20 CRAN (R 3.4.3)
## lattice 0.20-35 2017-03-25 CRAN (R 3.4.3)
## lazyeval 0.2.1 2017-10-29 CRAN (R 3.4.2)
## lubridate 1.7.3 2018-02-27 CRAN (R 3.4.3)
## magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
## memoise 1.1.0 2017-04-21 CRAN (R 3.4.0)
## methods * 3.4.3 2017-12-07 local
## mnormt 1.5-5 2016-10-15 CRAN (R 3.4.0)
## modelr 0.1.1 2017-08-10 local
## munsell 0.4.3 2016-02-13 CRAN (R 3.4.0)
## nlme 3.1-131.1 2018-02-16 CRAN (R 3.4.3)
## parallel 3.4.3 2017-12-07 local
## pillar 1.2.1 2018-02-27 CRAN (R 3.4.3)
## pkgconfig 2.0.1 2017-03-21 CRAN (R 3.4.0)
## plyr 1.8.4 2016-06-08 CRAN (R 3.4.0)
## psych 1.7.8 2017-09-09 CRAN (R 3.4.1)
## purrr * 0.2.4 2017-10-18 CRAN (R 3.4.2)
## R6 2.2.2 2017-06-17 CRAN (R 3.4.0)
## Rcpp 0.12.15 2018-01-20 CRAN (R 3.4.3)
## readr * 1.1.1 2017-05-16 CRAN (R 3.4.0)
## readxl 1.0.0 2017-04-18 CRAN (R 3.4.0)
## reshape2 1.4.3 2017-12-11 CRAN (R 3.4.3)
## rlang 0.2.0 2018-02-20 cran (@0.2.0)
## rmarkdown 1.9 2018-03-01 CRAN (R 3.4.3)
## rprojroot 1.3-2 2018-01-03 CRAN (R 3.4.3)
## rstudioapi 0.7 2017-09-07 CRAN (R 3.4.1)
## rvest 0.3.2 2016-06-17 CRAN (R 3.4.0)
## scales 0.5.0 2017-08-24 cran (@0.5.0)
## stats * 3.4.3 2017-12-07 local
## stringi 1.1.7 2018-03-12 CRAN (R 3.4.3)
## stringr * 1.3.0 2018-02-19 CRAN (R 3.4.3)
## tibble * 1.4.2 2018-01-22 CRAN (R 3.4.3)
## tidyr * 0.8.0 2018-01-29 CRAN (R 3.4.3)
## tidyverse * 1.2.1 2017-11-14 CRAN (R 3.4.2)
## tools 3.4.3 2017-12-07 local
## utils * 3.4.3 2017-12-07 local
## withr 2.1.1 2017-12-19 CRAN (R 3.4.3)
## xml2 1.2.0 2018-01-24 CRAN (R 3.4.3)
## yaml 2.1.18 2018-03-08 CRAN (R 3.4.4)This work is licensed under the CC BY-NC 4.0 Creative Commons License.