{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# The Bias-Variance Tradeoff\n",
"> A Summary of lecture \"Machine Learning with Tree-Based Models in Python\n",
"\", via datacamp\n",
"\n",
"- toc: true \n",
"- badges: true\n",
"- comments: true\n",
"- author: Chanseok Kang\n",
"- categories: [Python, Datacamp, Machine_Learning]\n",
"- image: images/ensemble.png"
]
},
{
"cell_type": "code",
"execution_count": 2,
"metadata": {},
"outputs": [],
"source": [
"import pandas as pd\n",
"import numpy as np\n",
"import matplotlib.pyplot as plt\n",
"import seaborn as sns"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Generalization Error\n",
"- Supervised Learning - Under the Hood\n",
" - supervised learning: $y = f(x)$, $f$ is unknown.\n",
" - Goals\n",
" - Find a model $\\hat{f}$ that best approximates: $f: \\hat{f} \\approx f$\n",
" - $\\hat{f}$ can be Logistic Regression, Decision Tree, Neural Network,...\n",
" - Discard noise as much as possible\n",
" - **End goal**: $\\hat{f}$ should achieve a low predictive error on unseen datasets.\n",
"- Difficulties in Approximating $f$\n",
" - Overfitting: $\\hat{f}(x)$ fits the training set noise.\n",
" - Underfitting: $\\hat{f}$ is not flexible enough to approximate $f$.\n",
"- Generalization Error\n",
" - Generalization Error of $\\hat{f}$: Does $\\hat{f}$ generalize well on unseen data?\n",
" - It can be decomposed as follows:\n",
" $$ \\hat{f} = \\text{bias}^2 + \\text{variance} +\\text{irreducible error} $$\n",
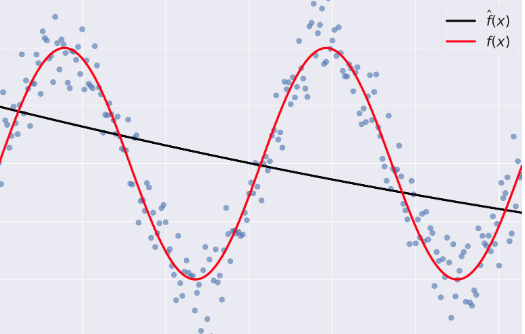
" - Bias: error term that tells you, on average, how much $\\hat{f} \\neq f$.\n",
"\n",
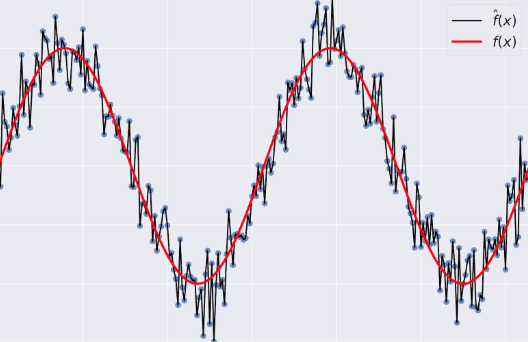
" - Variance: tells you how much $\\hat{f}$ is inconsistent over different training sets\n",
"\n",
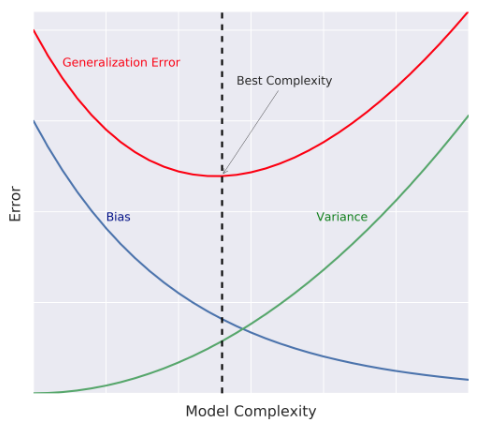
"- Model Complexity\n",
" - Model Complexity: sets the flexibility of $\\hat{f}$\n",
" - Example: Maximum tree depth, Minimum samples per leaf, ...\n",
"- Bias-Variance Tradeoff\n",
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Diagnose bias and variance problems\n",
"- Estimating the Generalization Error\n",
" - How do we estimate the generalization error of a model?\n",
" - Cannot be done directly because:\n",
" - $f$ is unknown\n",
" - usually you only have one dataset\n",
" - noise is unpredictable.\n",
" - Solution\n",
" - Split the data to training and test sets\n",
" - fit $\\hat{f}$ to the training set\n",
" - evaluate the error of $\\hat{f}$ on the unseen test set\n",
" - generalization error of $\\hat{f} \\approx$ test set error of $\\hat{f}$\n",
"- Better model Evaluation with Cross-Validation\n",
" - Test set should not be touched until we are confident about $\\hat{f}$'s performance\n",
" - Evaluating $\\hat{f}$ on training set: biased estimate, $\\hat{f}$ has already seen all training points\n",
" - Solution: Cross-Validation (CV)\n",
" - K-Fold CV\n",
" - Hold-Out CV\n",
"- K-Fold CV\n",
" $$\\text{CV error} = \\dfrac{E_1 + \\cdots + E_{10}}{10} $$\n",
"- Diagnose Variance Problems\n",
" - If $\\hat{f}$ suffers from **high variance**: CV error of $\\hat{f} >$ training set error of $\\hat{f}$\n",
" - $\\hat{f}$ is said to overfit the training set. To remedy overfitting:\n",
" - Decrease model complexity\n",
" - Gather more data, ...\n",
"- Diagnose Bias Problems\n",
" - if $\\hat{f}$ suffers from high bias: CV error of $\\hat{f} \\approx$ training set error of $\\hat{f} >>$ desired error.\n",
" - $\\hat{f}$ is said to underfit the training set. To remedy underfitting:\n",
" - Increase model complexity\n",
" - Gather more relevant features"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Instantiate the model\n",
"In the following set of exercises, you'll diagnose the bias and variance problems of a regression tree. The regression tree you'll define in this exercise will be used to predict the mpg consumption of cars from the auto dataset using all available features.\n",
"\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### Preprocess"
]
},
{
"cell_type": "code",
"execution_count": 4,
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
"\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" mpg | \n",
" displ | \n",
" hp | \n",
" weight | \n",
" accel | \n",
" origin | \n",
" size | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" 18.0 | \n",
" 250.0 | \n",
" 88 | \n",
" 3139 | \n",
" 14.5 | \n",
" US | \n",
" 15.0 | \n",
"
\n",
" \n",
" | 1 | \n",
" 9.0 | \n",
" 304.0 | \n",
" 193 | \n",
" 4732 | \n",
" 18.5 | \n",
" US | \n",
" 20.0 | \n",
"
\n",
" \n",
" | 2 | \n",
" 36.1 | \n",
" 91.0 | \n",
" 60 | \n",
" 1800 | \n",
" 16.4 | \n",
" Asia | \n",
" 10.0 | \n",
"
\n",
" \n",
" | 3 | \n",
" 18.5 | \n",
" 250.0 | \n",
" 98 | \n",
" 3525 | \n",
" 19.0 | \n",
" US | \n",
" 15.0 | \n",
"
\n",
" \n",
" | 4 | \n",
" 34.3 | \n",
" 97.0 | \n",
" 78 | \n",
" 2188 | \n",
" 15.8 | \n",
" Europe | \n",
" 10.0 | \n",
"
\n",
" \n",
"
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" mpg | \n",
" displ | \n",
" hp | \n",
" weight | \n",
" accel | \n",
" size | \n",
" origin_Asia | \n",
" origin_Europe | \n",
" origin_US | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" 18.0 | \n",
" 250.0 | \n",
" 88 | \n",
" 3139 | \n",
" 14.5 | \n",
" 15.0 | \n",
" 0 | \n",
" 0 | \n",
" 1 | \n",
"
\n",
" \n",
" | 1 | \n",
" 9.0 | \n",
" 304.0 | \n",
" 193 | \n",
" 4732 | \n",
" 18.5 | \n",
" 20.0 | \n",
" 0 | \n",
" 0 | \n",
" 1 | \n",
"
\n",
" \n",
" | 2 | \n",
" 36.1 | \n",
" 91.0 | \n",
" 60 | \n",
" 1800 | \n",
" 16.4 | \n",
" 10.0 | \n",
" 1 | \n",
" 0 | \n",
" 0 | \n",
"
\n",
" \n",
" | 3 | \n",
" 18.5 | \n",
" 250.0 | \n",
" 98 | \n",
" 3525 | \n",
" 19.0 | \n",
" 15.0 | \n",
" 0 | \n",
" 0 | \n",
" 1 | \n",
"
\n",
" \n",
" | 4 | \n",
" 34.3 | \n",
" 97.0 | \n",
" 78 | \n",
" 2188 | \n",
" 15.8 | \n",
" 10.0 | \n",
" 0 | \n",
" 1 | \n",
" 0 | \n",
"
\n",
" \n",
"
\n",
"
\n",
"\n",
"
\n",
" \n",
" \n",
" | \n",
" Age_std | \n",
" Total_Bilirubin_std | \n",
" Direct_Bilirubin_std | \n",
" Alkaline_Phosphotase_std | \n",
" Alamine_Aminotransferase_std | \n",
" Aspartate_Aminotransferase_std | \n",
" Total_Protiens_std | \n",
" Albumin_std | \n",
" Albumin_and_Globulin_Ratio_std | \n",
" Is_male_std | \n",
" Liver_disease | \n",
"
\n",
" \n",
" \n",
" \n",
" | 0 | \n",
" 1.247403 | \n",
" -0.420320 | \n",
" -0.495414 | \n",
" -0.428870 | \n",
" -0.355832 | \n",
" -0.319111 | \n",
" 0.293722 | \n",
" 0.203446 | \n",
" -0.147390 | \n",
" 0 | \n",
" 1 | \n",
"
\n",
" \n",
" | 1 | \n",
" 1.062306 | \n",
" 1.218936 | \n",
" 1.423518 | \n",
" 1.675083 | \n",
" -0.093573 | \n",
" -0.035962 | \n",
" 0.939655 | \n",
" 0.077462 | \n",
" -0.648461 | \n",
" 1 | \n",
" 1 | \n",
"
\n",
" \n",
" | 2 | \n",
" 1.062306 | \n",
" 0.640375 | \n",
" 0.926017 | \n",
" 0.816243 | \n",
" -0.115428 | \n",
" -0.146459 | \n",
" 0.478274 | \n",
" 0.203446 | \n",
" -0.178707 | \n",
" 1 | \n",
" 1 | \n",
"
\n",
" \n",
" | 3 | \n",
" 0.815511 | \n",
" -0.372106 | \n",
" -0.388807 | \n",
" -0.449416 | \n",
" -0.366760 | \n",
" -0.312205 | \n",
" 0.293722 | \n",
" 0.329431 | \n",
" 0.165780 | \n",
" 1 | \n",
" 1 | \n",
"
\n",
" \n",
" | 4 | \n",
" 1.679294 | \n",
" 0.093956 | \n",
" 0.179766 | \n",
" -0.395996 | \n",
" -0.295731 | \n",
" -0.177537 | \n",
" 0.755102 | \n",
" -0.930414 | \n",
" -1.713237 | \n",
" 1 | \n",
" 1 | \n",
"
\n",
" \n",
"
\n",
"