{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Plotly interactive Human Disease Network
Selecting the neighboring disorders on node click ##"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
" The nodes of the human disease network (HDN) are disorders,\n",
"and two disorders are connected if they share a genetic component. In order to identify the disorders that have common genetic components\n",
"with a given node, we are clicking that node and `on_click` is executed a callback that performs a kind of neighboring nodes selection."
]
},
{
"cell_type": "code",
"execution_count": 1,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Python version: 3.7.3\n"
]
}
],
"source": [
"import platform\n",
"print(f'Python version: {platform.python_version()}')"
]
},
{
"cell_type": "code",
"execution_count": 2,
"metadata": {},
"outputs": [
{
"data": {
"text/plain": [
"'4.8.1'"
]
},
"execution_count": 2,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"import plotly\n",
"plotly.__version__"
]
},
{
"cell_type": "code",
"execution_count": 3,
"metadata": {},
"outputs": [],
"source": [
"import igraph as ig\n",
"import numpy as np\n",
"import ast"
]
},
{
"cell_type": "code",
"execution_count": 4,
"metadata": {},
"outputs": [],
"source": [
"import plotly.graph_objs as go\n",
"import ipywidgets as ipw"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Read the `gml` file as an `igraph.Graph`:"
]
},
{
"cell_type": "code",
"execution_count": 5,
"metadata": {},
"outputs": [],
"source": [
"K = ig.Graph.Read_GML('human-disease.gml')"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The following function extracts from the graph K, the data needed to plot it via Plotly:"
]
},
{
"cell_type": "code",
"execution_count": 6,
"metadata": {},
"outputs": [],
"source": [
"def get_Plotly_netw(G, graph_layout, title='Plotly Interactive Human Disease Network',\n",
" source=None, flip='lr', width=950, height=790): #h=850\n",
" # G is an igraph.Graph instance\n",
" # graph_layout is an igraph.Layout instance\n",
" # title - the network title\n",
" # flip is one of the strings 'lr', 'ud' to get a pseudo-flip effect\n",
" # the igraph.Layout is referenced to the screen system of coords, and is upside-down flipped chonging the sign of y-coords\n",
" # the global HDN looks better with the left-right flipped layout, by changing the x-coords sign.\n",
" #width and height are the sizes of the plot area\n",
" \n",
" graph_layout = np.array(graph_layout)\n",
" \n",
" if flip == 'lr':\n",
" graph_layout[:,0] =- graph_layout[:,0]\n",
" elif flip == 'ud':\n",
" graph_layout[:,1] =- graph_layout[:,1] \n",
" else: \n",
" raise ValueError('There is no such a flip type')\n",
" \n",
" m = len(G.vs)\n",
" graph_edges = [e.tuple for e in G.es]# represent edges as tuples of end vertex indices\n",
" \n",
" Xn = [graph_layout[k][0] for k in range(m)]#x-coordinates of graph nodes(vertices)\n",
" Yn = [graph_layout[k][1] for k in range(m)]#y-coordinates -\"-\n",
" \n",
" Xe = []#list of edge ends x-coordinates\n",
" Ye = []#list of edge ends y-coordinates\n",
" for e in graph_edges:\n",
" Xe.extend([graph_layout[e[0]][0],graph_layout[e[1]][0], None])\n",
" Ye.extend([graph_layout[e[0]][1],graph_layout[e[1]][1], None]) \n",
"\n",
" size = [vertex['size'] for vertex in G.vs]\n",
" #define the Plotly graphical objects\n",
" \n",
" plotly_edges = go.Scatter(x=Xe,\n",
" y=Ye,\n",
" mode='lines',\n",
" line=dict(color='rgb(180,180,180)', width=0.75),\n",
" hoverinfo='none')\n",
" plotly_vertices = go.Scatter(x=Xn,\n",
" y=Yn,\n",
" mode='markers',\n",
" name='',\n",
" marker=dict(symbol='circle-dot',\n",
" size=size, \n",
" color=[vertex['color'] for vertex in G.vs], \n",
" line=dict(color='rgb(50,50,50)', width=0.5)\n",
" ),\n",
" text=[f\"{vertex['label']}
({vertex['disclass']})\" for vertex in G.vs],\n",
" hoverinfo='text')\n",
" \n",
" #Define the Plotly plot layout:\n",
" \n",
" plotly_plot_layout = dict(title=title, \n",
" width=width,\n",
" height=height,\n",
" showlegend=False,\n",
" xaxis_visible=False,\n",
" yaxis_visible=False, \n",
" margin=dict(t=100, b=5),\n",
" hovermode='closest',\n",
" paper_bgcolor='rgb(235,235,235)',\n",
" template='none'\n",
" )\n",
" \n",
" if source is not None:\n",
" annotations = [dict(showarrow=False, \n",
" text=source, \n",
" xref='paper', \n",
" yref='paper', \n",
" x=0, \n",
" y=-0.1, \n",
" xanchor='left', \n",
" yanchor='bottom', \n",
" font=dict(size=14) \n",
" )]\n",
" else:\n",
" annotations = []\n",
" \n",
" disorder_name = [vertex['label'] for vertex in G.vs]\n",
" for k, s in enumerate(size):\n",
" if s>= 20:\n",
" annotations.append(dict(text=disorder_name[k], \n",
" x=graph_layout[k][0], \n",
" y=graph_layout[k][1],\n",
" xref='x1', yref='y1',\n",
" font=dict(color='rgb(50,50,50)', size=10),\n",
" showarrow=False))\n",
" \n",
" plotly_plot_layout.update(annotations=annotations)\n",
" return go.FigureWidget(data=[plotly_edges, plotly_vertices], layout=plotly_plot_layout) "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Since the position of each node is recorded as a string, we convert it to a tuple of floats:"
]
},
{
"cell_type": "code",
"execution_count": 7,
"metadata": {},
"outputs": [],
"source": [
"pos = [ast.literal_eval(v['position']) for v in K.vs]\n",
"HDN_layout = np.array(pos)"
]
},
{
"cell_type": "code",
"execution_count": 8,
"metadata": {},
"outputs": [],
"source": [
"fig = get_Plotly_netw(K, HDN_layout)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
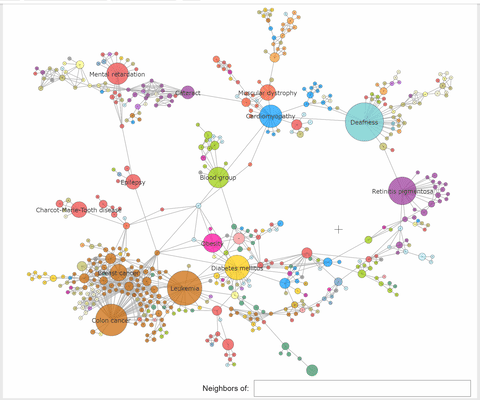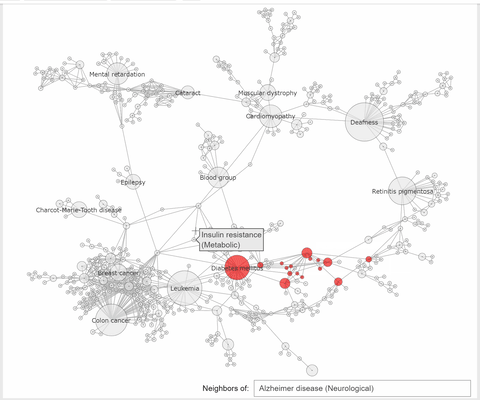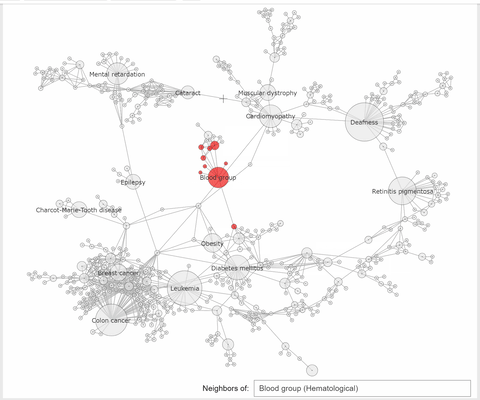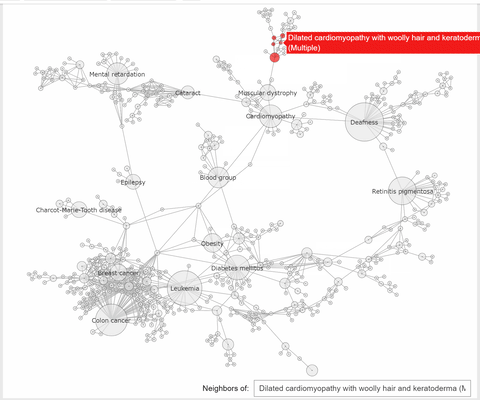
"The callback function associated to a node click event, extracts the neighbhoring nodes of the clicked one, color the subset\n",
"of these nodes with the same color, at choice, and the complementary nodes with gray. At the same time a `Text` widget displays the label and disorder class of the clicked disorder."
]
},
{
"cell_type": "code",
"execution_count": 9,
"metadata": {},
"outputs": [],
"source": [
"textbox = ipw.Text(value='',\n",
" description='Neighbors of:',\n",
" disabled=False,\n",
" continuous_update=True)\n",
"\n",
"textbox.layout = dict(margin='-35px 10px 10px 410px', width='450px') # place the textbox at the bottom right corner \n",
" # of the plotarea"
]
},
{
"cell_type": "code",
"execution_count": 10,
"metadata": {},
"outputs": [],
"source": [
"def select_neighbors(trace, points, state):\n",
" \n",
" if not points.point_inds:\n",
" return\n",
" ind = points.point_inds[0] \n",
" new_color = np.array(['rgba(230, 230, 230, 0.9)']*len(trace.x))\n",
" # extract the indices of the clicked vertex neighbors\n",
" I = np.append(np.unique(np.array(K.neighbors(ind, mode='out'))), [ind])\n",
" new_color[I] = K.vs[ind]['color'] #color all neighbors with the color of the clicked node or with 'rgba(240, 0, 0, 0.9)'\n",
" trace.marker.color = new_color \n",
" textbox.value = f\"{K.vs[ind]['label']} ({K.vs[ind]['disclass']})\""
]
},
{
"cell_type": "code",
"execution_count": 11,
"metadata": {},
"outputs": [
{
"data": {
"application/vnd.jupyter.widget-view+json": {
"model_id": "934c19fbd49c428683d72a03537e65d3",
"version_major": 2,
"version_minor": 0
},
"text/plain": [
"VBox(children=(FigureWidget({\n",
" 'data': [{'hoverinfo': 'none',\n",
" 'line': {'color': 'rgb(180,180,…"
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"fig.data[1].on_click(select_neighbors) \n",
"ipw.VBox([fig, textbox])"
]
},
{
"cell_type": "code",
"execution_count": 12,
"metadata": {},
"outputs": [
{
"data": {
"text/html": [
" \n"
],
"text/plain": [
""
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"%%html\n",
"
\n"
],
"text/plain": [
""
]
},
"metadata": {},
"output_type": "display_data"
}
],
"source": [
"%%html\n",
" "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.3"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.3"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
 "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.3"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.3"
}
},
"nbformat": 4,
"nbformat_minor": 4
}