{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Introduction\n",
"In this notebook, I'll show you a way how to you can connect JavaDoc comments with the Java nodes of jQAssistant's scan result. I'll elaborate the way to the solutions, because I hope that you can do a similar problem solving analysis (aka XML importing and wrangling) on your own."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Context\n",
"In a [blog post](https://www.feststelltaste.de/my-experiences-with-jqassistant-so-far/), [Yoann Buch got me to thinking about](https://www.feststelltaste.de/my-experiences-with-jqassistant-so-far/#comment-6) a how to add comments to the already existing class nodes in Neo4j scanned by jQAssistant). I meant it would be possible to do it with the Python library Pygments. I experimented with it a little bit, but it seems that it isn't going well. The main problem is the lack of structural information. "
]
},
{
"cell_type": "markdown",
"metadata": {
"collapsed": true
},
"source": [
"# Idea\n",
"I thought \"wait a minute, what about the JavaDoc that's generated in HTML\" and I thought one step further: \"If there are generators for HTML, is there a generator for XML, too?\". I just googled \"javadoc xml\" and found it: https://github.com/MarkusBernhardt/xml-doclet, \"A doclet to output javadoc as XML\" as Maven plugin. \n",
"\n",
"So the journey began..."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Implementation"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Preparing the code\n",
"In this prototype, I work with an old jQAssistant version 1.1.3 (because I haven't figured out how to read XML like described in [Dirk's answer at StackOverflow](http://www.stackoverflow.com/questions/31425610/error-when-scanning-xml-file-with-jqassistant) with the new version). \n",
"\n",
"I use an corresponding, old version of jQAssistant's [Spring Petclinic demo repo](https://github.com/buschmais/spring-petclinic/): \n",
" \n",
"git checkout f5811bf2ed9c5369a749cb90ef9e7a261de03760 .\n",
" "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Getting JavaDoc as XML\n",
"For getting all the JavaDoc from the source code, I just add the Maven plugin mentioned above additionally to the already existing ones:\n",
"```xml\n",
"\n",
" org.apache.maven.plugins\n",
" maven-javadoc-plugin\n",
" 2.10.4\n",
" \n",
" \n",
" xml-doclet\n",
" process-resources\n",
" \n",
" javadoc\n",
" \n",
" \n",
" com.github.markusbernhardt.xmldoclet.XmlDoclet\n",
" -d ${project.build.directory} -filename javadoc.xml\n",
" false\n",
" \n",
" com.github.markusbernhardt\n",
" xml-doclet\n",
" 1.0.5\n",
" \n",
" \n",
" \n",
" \n",
"\n",
"```"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Scanning the JavaDoc XML\n",
"In theory, I now have just to add this file to jQAssistant's scan configuration as <scanInclude>:\n",
"```xml\n",
"\n",
" ${project.build.directory}/javadoc.xml\n",
" xml:document\n",
"\n",
"```\n",
"I've added the <scope> to adivse jQAssistant to scan the whole XML content (according to this [StackOverflow answer](http://stackoverflow.com/questions/31425610/error-when-scanning-xml-file-with-jqassistant)).\n",
"\n",
"\n",
"Note: Unfortunately I haven't got this working as of today, i.e. the javadoc.xml isn't appearing in the database. So I've scanned the XML file in the target folder with an \n",
" \n",
"jqassistant.sh scan -f xml:document::javadoc.xml \n",
" \n",
"manually after scanning the project. This approach won't work with jQAssistant version 1.1.4+, so that's why I'm using an old version. I'll update this notebook when I've got this working with the newest version. "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## First build\n",
"I just built the complete project: \n",
" \n",
"mvn clean install \n",
"\n",
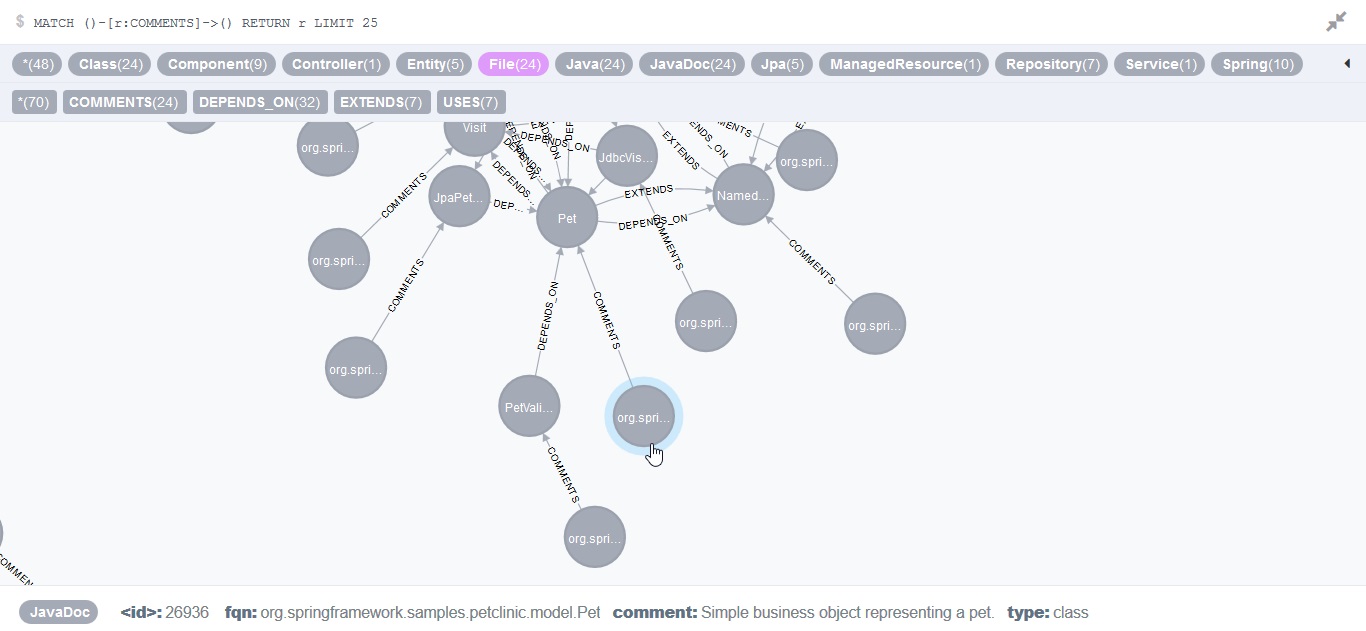
"jQAssistant places some nice graphs into the Neo4j database:"
]
},
{
"cell_type": "code",
"execution_count": 3,
"metadata": {
"collapsed": false
},
"outputs": [
{
"data": {
"application/javascript": [
"$(\"head\").append($(\"\").attr({\n",
" rel: \"stylesheet\",\n",
" type: \"text/css\",\n",
" href: \"https://cdnjs.cloudflare.com/ajax/libs/vis/4.8.2/vis.css\"\n",
"}));\n",
"require.config({ paths: { vis: 'https://cdnjs.cloudflare.com/ajax/libs/vis/4.8.2/vis.min' } }); require(['vis'], function(vis) { window.vis = vis; window.vis = vis; }); "
],
"text/plain": [
""
]
},
"metadata": {},
"output_type": "display_data"
},
{
"data": {
"text/html": [
"\n",
"\n",
"\n"
],
"text/plain": [
""
]
},
"execution_count": 3,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"import lib.neo4jupyter_mod as n4j\n",
"import py2neo\n",
"graph = py2neo.Graph()\n",
"\n",
"n4j.init_notebook_mode()\n",
"n4j.draw(graph, \n",
" n='n:Class { name: \"Pet\"}',\n",
" r=\"r:DEPENDS_ON\", \n",
" m=\"m:Class\", \n",
" options={\"Class\": \"name\"}, \n",
" limit=5)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Additionaly, the build outputs a nice XML file named javadoc.xml with all the existing comments into the target folder\n",
"```xml\n",
"\n",
"\n",
" \n",
" \n",
" In Servlet 3.0+ [..]\n",
" \n",
" \n",
" \n",
" \n",
" \n",
" \n",
" \n",
" \n",
" \n",
" \n",
" \n",
"...\n",
"```"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"I scanned this file already, i. e. it's also contained in the database. Let's have a look at it! \n",
" \n",
"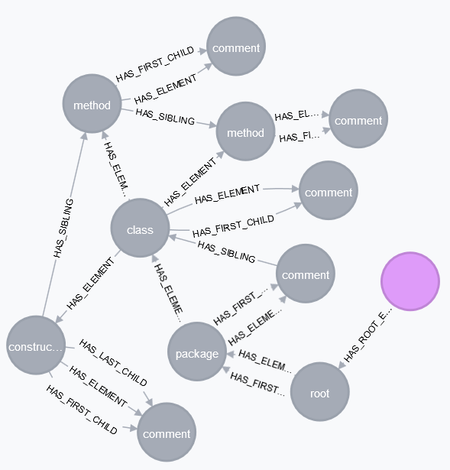\n",
"\n",
"The corresponding texts for the comments are in the children of the <comment> elements:"
]
},
{
"cell_type": "code",
"execution_count": 4,
"metadata": {
"collapsed": false,
"scrolled": true
},
"outputs": [
{
"data": {
"text/html": [
"\n",
"\n",
"\n"
],
"text/plain": [
""
]
},
"execution_count": 4,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"n4j.draw(graph, \n",
" n='n:Element { name: \"comment\"}',\n",
" m='m:Text',\n",
" options={\"Element\": \"name\", \"Text\": \"value\"}, \n",
" limit=10)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"But there is one problem left: The XML scanner scans **all** elements and places each element into a separate node. So if we have some HTML formatting in the JavaDoc like:\n",
"\n",
"```xml\n",
"\n",
" <code>Validator</code> for <code>Pet</code> forms.\n",
" <p>\n",
" We're not using Bean Validation annotations here because it is easier to define such validation rule in Java.\n",
" </p>\n",
"\n",
"```"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This leads to an \"interesting\" looking graph:\n",
"\n",
"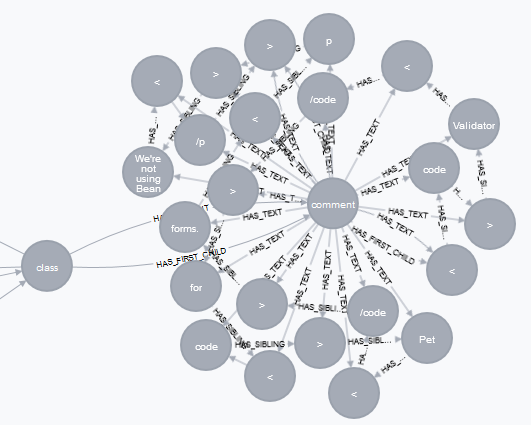"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"There's an easy solution for this, but let's first start with graph database action and solve that problem later."
]
},
{
"cell_type": "markdown",
"metadata": {
"collapsed": false
},
"source": [
"# Data Wrangling with Neo4j"
]
},
{
"cell_type": "code",
"execution_count": 5,
"metadata": {
"collapsed": false
},
"outputs": [
{
"data": {
"text/plain": [
"'\\nMATCH \\n(element:Element)-[:HAS_ELEMENT]->(Element { name : \"comment\"})-[:HAS_TEXT]->(doc_text:Text),\\n(element)-[:HAS_ATTRIBUTE]->(building_block:Attribute{name: \"qualified\"})\\nOPTIONAL MATCH\\n(element)-[:HAS_ATTRIBUTE]->(signature:Attribute{name: \"signature\"}),\\n(element)-[:HAS_ELEMENT]->(t:Element{name: \"return\"})-[:HAS_ATTRIBUTE]->(return_type:Attribute{name: \"qualified\"})\\nWHERE element.name =~ \"(method|class|interface|constructor)\"\\nWITH\\n id(element) as id,\\n // class and method\\n CASE element.name\\n WHEN \"method\"\\n THEN return_type.value + \" \" + SPLIT(building_block.value, \".\")[-1] + signature.value\\n WHEN \"constructor\"\\n THEN \"void ()\"\\n END as signature,\\n\\n CASE element.name\\n WHEN \"method\"\\n THEN SUBSTRING(building_block.value, 0, $subLength)SPLIT(building_block.value, \".\")\\n ELSE building_block.value\\n END as type_fqn,\\n\\n return_type.value as r,\\n reduce(s = \"\", x IN collect(doc_text) | s + x.value) as comment\\n\\nRETURN id, type_fqn, signature, comment\\n'"
]
},
"execution_count": 5,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"\"\"\"\n",
"MATCH \n",
"(element:Element)-[:HAS_ELEMENT]->(Element { name : \"comment\"})-[:HAS_TEXT]->(doc_text:Text),\n",
"(element)-[:HAS_ATTRIBUTE]->(building_block:Attribute{name: \"qualified\"})\n",
"OPTIONAL MATCH\n",
"(element)-[:HAS_ATTRIBUTE]->(signature:Attribute{name: \"signature\"}),\n",
"(element)-[:HAS_ELEMENT]->(t:Element{name: \"return\"})-[:HAS_ATTRIBUTE]->(return_type:Attribute{name: \"qualified\"})\n",
"WHERE element.name =~ \"(method|class|interface|constructor)\"\n",
"WITH\n",
" id(element) as id,\n",
" // class and method\n",
" CASE element.name\n",
" WHEN \"method\"\n",
" THEN return_type.value + \" \" + SPLIT(building_block.value, \".\")[-1] + signature.value\n",
" WHEN \"constructor\"\n",
" THEN \"void ()\"\n",
" END as signature,\n",
"\n",
" CASE element.name\n",
" WHEN \"method\"\n",
" THEN SUBSTRING(building_block.value, 0, $subLength)SPLIT(building_block.value, \".\")\n",
" ELSE building_block.value\n",
" END as type_fqn,\n",
"\n",
" return_type.value as r,\n",
" reduce(s = \"\", x IN collect(doc_text) | s + x.value) as comment\n",
"\n",
"RETURN id, type_fqn, signature, comment\n",
"\"\"\""
]
},
{
"cell_type": "code",
"execution_count": 6,
"metadata": {
"collapsed": false
},
"outputs": [
{
"data": {
"text/plain": [
"[]"
]
},
"execution_count": 6,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"create_comment_nodes_for_types=\"\"\"\n",
"MATCH \n",
"(element)-[:HAS_ELEMENT]->(class_comment:Element { name : \"comment\"})-[:HAS_TEXT]->(doc_text:Text),\n",
"(element:Element)-[:HAS_ATTRIBUTE]->(qualified:Attribute{name: \"qualified\"})\n",
"OPTIONAL MATCH\n",
"(element)-[:HAS_ATTRIBUTE]->(signature:Attribute{name: \"signature\"})\n",
"WHERE element.name =~ \"(method|class|interface|constructor)\"\n",
"WITH\n",
" id(element) as id,\n",
" element.name as type,\n",
" // class and method\n",
" CASE WHEN signature.value IS NULL THEN qualified.value ELSE qualified.value+signature.value END as key,\n",
" reduce(s = \"\", x IN collect(doc_text) | s + x.value) as text\n",
"\n",
"RETURN id, type, key, text\n",
"\"\"\"\n",
"graph.data(create_comment_nodes_for_types)"
]
},
{
"cell_type": "code",
"execution_count": 7,
"metadata": {
"collapsed": false
},
"outputs": [
{
"data": {
"text/plain": [
"[{'COUNT(javadoc)': 0}]"
]
},
"execution_count": 7,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"create_comment_nodes_for_classes=\"\"\"\n",
"MATCH \n",
"(package:Element { name : \"package\"})\n",
"-[:HAS_ELEMENT]->\n",
"(class:Element {name : \"class\"})\n",
"-[:HAS_ATTRIBUTE]->\n",
"(class_fqn:Attribute{name: \"qualified\"}),\n",
"(class_comment:Element { name : \"comment\"})\n",
"-[:HAS_TEXT]->(text:Text),\n",
"class-[:HAS_ELEMENT]->(class_comment)\n",
"\n",
"WITH DISTINCT \n",
" class.name as type_value, \n",
" class_fqn.value as fqn_value, \n",
" reduce(s = \"\", x IN collect(text) | s + x.value) as text_value\n",
" \n",
"CREATE (javadoc:JavaDoc { comment: text_value, type: type_value, fqn: fqn_value })\n",
"\n",
"RETURN COUNT(javadoc)\n",
"\"\"\"\n",
"graph.data(create_comment_nodes_for_classes)"
]
},
{
"cell_type": "code",
"execution_count": 8,
"metadata": {
"collapsed": false
},
"outputs": [
{
"data": {
"text/plain": [
"[{'rels': 0}]"
]
},
"execution_count": 8,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"create_relationship_query=\"\"\"\n",
"MATCH (type:Type), (javadoc:JavaDoc)\n",
"WHERE type.fqn = javadoc.fqn\n",
"MERGE (javadoc)-[r:COMMENTS]->(type)\n",
"RETURN COUNT(r) as rels\n",
"\"\"\"\n",
"graph.data(create_relationship_query)"
]
},
{
"cell_type": "code",
"execution_count": 9,
"metadata": {
"collapsed": false
},
"outputs": [
{
"data": {
"text/plain": [
"[{'COUNT(javadoc)': 0, 'COUNT(r)': 0}]"
]
},
"execution_count": 9,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"delete_comments_query=\"\"\"\n",
"MATCH (javadoc:JavaDoc)-[r:COMMENTS]->()\n",
"DELETE r, javadoc\n",
"RETURN COUNT(r), COUNT(javadoc)\n",
"\"\"\"\n",
"graph.data(delete_comments_query)"
]
}
],
"metadata": {
"anaconda-cloud": {},
"kernelspec": {
"display_name": "Python [Root]",
"language": "python",
"name": "Python [Root]"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.5.2"
}
},
"nbformat": 4,
"nbformat_minor": 0
}