---
output: github_document
---
```{r, include = FALSE}
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
options(tibble.print_min = 5, tibble.print_max = 5)
```
# dplyr  [](https://cran.r-project.org/package=dplyr)
[](https://github.com/tidyverse/dplyr/actions/workflows/R-CMD-check.yaml)
[](https://app.codecov.io/gh/tidyverse/dplyr)
## Overview
dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges:
* `mutate()` adds new variables that are functions of existing variables
* `select()` picks variables based on their names.
* `filter()` picks cases based on their values.
* `summarise()` reduces multiple values down to a single summary.
* `arrange()` changes the ordering of the rows.
These all combine naturally with `group_by()` which allows you to perform any operation "by group". You can learn more about them in `vignette("dplyr")`. As well as these single-table verbs, dplyr also provides a variety of two-table verbs, which you can learn about in `vignette("two-table")`.
If you are new to dplyr, the best place to start is the [data transformation chapter](https://r4ds.hadley.nz/data-transform) in R for Data Science.
## Backends
In addition to data frames/tibbles, dplyr makes working with other computational backends accessible and efficient. Below is a list of alternative backends:
- [arrow](https://arrow.apache.org/docs/r/) for larger-than-memory datasets, including on remote cloud storage like AWS S3, using the Apache Arrow C++ engine, [Acero](https://arrow.apache.org/docs/cpp/acero/overview.html).
- [dbplyr](https://dbplyr.tidyverse.org/) for data stored in a relational
database. Translates your dplyr code to SQL.
- [dtplyr](https://dtplyr.tidyverse.org/) for large, in-memory datasets.
Translates your dplyr code to high performance
[data.table](https://rdatatable.gitlab.io/data.table/) code.
- [duckplyr](https://duckplyr.tidyverse.org/) for large, in-memory
datasets. Translates your dplyr code to high performance
[duckdb](https://duckdb.org) queries with zero extra copies and
an automatic R fallback when translation isn’t possible.
- [sparklyr](https://spark.posit.co/) for very large datasets stored in
[Apache Spark](https://spark.apache.org).
## Installation
```{r, eval = FALSE}
# The easiest way to get dplyr is to install the whole tidyverse:
install.packages("tidyverse")
# Alternatively, install just dplyr:
install.packages("dplyr")
```
### Development version
To get a bug fix or to use a feature from the development version, you can install
the development version of dplyr from GitHub.
```{r, eval = FALSE}
# install.packages("pak")
pak::pak("tidyverse/dplyr")
```
## Cheat Sheet
[](https://cran.r-project.org/package=dplyr)
[](https://github.com/tidyverse/dplyr/actions/workflows/R-CMD-check.yaml)
[](https://app.codecov.io/gh/tidyverse/dplyr)
## Overview
dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges:
* `mutate()` adds new variables that are functions of existing variables
* `select()` picks variables based on their names.
* `filter()` picks cases based on their values.
* `summarise()` reduces multiple values down to a single summary.
* `arrange()` changes the ordering of the rows.
These all combine naturally with `group_by()` which allows you to perform any operation "by group". You can learn more about them in `vignette("dplyr")`. As well as these single-table verbs, dplyr also provides a variety of two-table verbs, which you can learn about in `vignette("two-table")`.
If you are new to dplyr, the best place to start is the [data transformation chapter](https://r4ds.hadley.nz/data-transform) in R for Data Science.
## Backends
In addition to data frames/tibbles, dplyr makes working with other computational backends accessible and efficient. Below is a list of alternative backends:
- [arrow](https://arrow.apache.org/docs/r/) for larger-than-memory datasets, including on remote cloud storage like AWS S3, using the Apache Arrow C++ engine, [Acero](https://arrow.apache.org/docs/cpp/acero/overview.html).
- [dbplyr](https://dbplyr.tidyverse.org/) for data stored in a relational
database. Translates your dplyr code to SQL.
- [dtplyr](https://dtplyr.tidyverse.org/) for large, in-memory datasets.
Translates your dplyr code to high performance
[data.table](https://rdatatable.gitlab.io/data.table/) code.
- [duckplyr](https://duckplyr.tidyverse.org/) for large, in-memory
datasets. Translates your dplyr code to high performance
[duckdb](https://duckdb.org) queries with zero extra copies and
an automatic R fallback when translation isn’t possible.
- [sparklyr](https://spark.posit.co/) for very large datasets stored in
[Apache Spark](https://spark.apache.org).
## Installation
```{r, eval = FALSE}
# The easiest way to get dplyr is to install the whole tidyverse:
install.packages("tidyverse")
# Alternatively, install just dplyr:
install.packages("dplyr")
```
### Development version
To get a bug fix or to use a feature from the development version, you can install
the development version of dplyr from GitHub.
```{r, eval = FALSE}
# install.packages("pak")
pak::pak("tidyverse/dplyr")
```
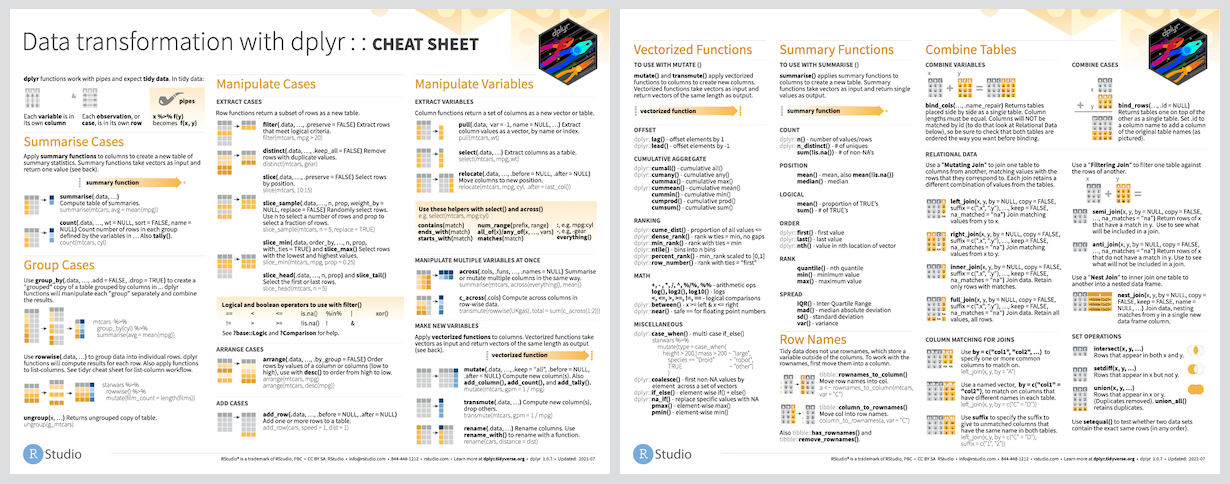
## Cheat Sheet
 ## Usage
```{r, message = FALSE}
library(dplyr)
starwars |>
filter(species == "Droid")
starwars |>
select(name, ends_with("color"))
starwars |>
mutate(name, bmi = mass / ((height / 100)^2)) |>
select(name:mass, bmi)
starwars |>
arrange(desc(mass))
starwars |>
group_by(species) |>
summarise(
n = n(),
mass = mean(mass, na.rm = TRUE)
) |>
filter(
n > 1,
mass > 50
)
```
## Getting help
If you encounter a clear bug, please file an issue with a minimal reproducible example on [GitHub](https://github.com/tidyverse/dplyr/issues). For questions and other discussion, please use [forum.posit.co](https://forum.posit.co/).
## Code of conduct
Please note that this project is released with a [Contributor Code of Conduct](https://dplyr.tidyverse.org/CODE_OF_CONDUCT).
By participating in this project you agree to abide by its terms.
## Usage
```{r, message = FALSE}
library(dplyr)
starwars |>
filter(species == "Droid")
starwars |>
select(name, ends_with("color"))
starwars |>
mutate(name, bmi = mass / ((height / 100)^2)) |>
select(name:mass, bmi)
starwars |>
arrange(desc(mass))
starwars |>
group_by(species) |>
summarise(
n = n(),
mass = mean(mass, na.rm = TRUE)
) |>
filter(
n > 1,
mass > 50
)
```
## Getting help
If you encounter a clear bug, please file an issue with a minimal reproducible example on [GitHub](https://github.com/tidyverse/dplyr/issues). For questions and other discussion, please use [forum.posit.co](https://forum.posit.co/).
## Code of conduct
Please note that this project is released with a [Contributor Code of Conduct](https://dplyr.tidyverse.org/CODE_OF_CONDUCT).
By participating in this project you agree to abide by its terms.
 [](https://cran.r-project.org/package=dplyr)
[](https://github.com/tidyverse/dplyr/actions/workflows/R-CMD-check.yaml)
[](https://app.codecov.io/gh/tidyverse/dplyr)
## Overview
dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges:
* `mutate()` adds new variables that are functions of existing variables
* `select()` picks variables based on their names.
* `filter()` picks cases based on their values.
* `summarise()` reduces multiple values down to a single summary.
* `arrange()` changes the ordering of the rows.
These all combine naturally with `group_by()` which allows you to perform any operation "by group". You can learn more about them in `vignette("dplyr")`. As well as these single-table verbs, dplyr also provides a variety of two-table verbs, which you can learn about in `vignette("two-table")`.
If you are new to dplyr, the best place to start is the [data transformation chapter](https://r4ds.hadley.nz/data-transform) in R for Data Science.
## Backends
In addition to data frames/tibbles, dplyr makes working with other computational backends accessible and efficient. Below is a list of alternative backends:
- [arrow](https://arrow.apache.org/docs/r/) for larger-than-memory datasets, including on remote cloud storage like AWS S3, using the Apache Arrow C++ engine, [Acero](https://arrow.apache.org/docs/cpp/acero/overview.html).
- [dbplyr](https://dbplyr.tidyverse.org/) for data stored in a relational
database. Translates your dplyr code to SQL.
- [dtplyr](https://dtplyr.tidyverse.org/) for large, in-memory datasets.
Translates your dplyr code to high performance
[data.table](https://rdatatable.gitlab.io/data.table/) code.
- [duckplyr](https://duckplyr.tidyverse.org/) for large, in-memory
datasets. Translates your dplyr code to high performance
[duckdb](https://duckdb.org) queries with zero extra copies and
an automatic R fallback when translation isn’t possible.
- [sparklyr](https://spark.posit.co/) for very large datasets stored in
[Apache Spark](https://spark.apache.org).
## Installation
```{r, eval = FALSE}
# The easiest way to get dplyr is to install the whole tidyverse:
install.packages("tidyverse")
# Alternatively, install just dplyr:
install.packages("dplyr")
```
### Development version
To get a bug fix or to use a feature from the development version, you can install
the development version of dplyr from GitHub.
```{r, eval = FALSE}
# install.packages("pak")
pak::pak("tidyverse/dplyr")
```
## Cheat Sheet
[](https://cran.r-project.org/package=dplyr)
[](https://github.com/tidyverse/dplyr/actions/workflows/R-CMD-check.yaml)
[](https://app.codecov.io/gh/tidyverse/dplyr)
## Overview
dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges:
* `mutate()` adds new variables that are functions of existing variables
* `select()` picks variables based on their names.
* `filter()` picks cases based on their values.
* `summarise()` reduces multiple values down to a single summary.
* `arrange()` changes the ordering of the rows.
These all combine naturally with `group_by()` which allows you to perform any operation "by group". You can learn more about them in `vignette("dplyr")`. As well as these single-table verbs, dplyr also provides a variety of two-table verbs, which you can learn about in `vignette("two-table")`.
If you are new to dplyr, the best place to start is the [data transformation chapter](https://r4ds.hadley.nz/data-transform) in R for Data Science.
## Backends
In addition to data frames/tibbles, dplyr makes working with other computational backends accessible and efficient. Below is a list of alternative backends:
- [arrow](https://arrow.apache.org/docs/r/) for larger-than-memory datasets, including on remote cloud storage like AWS S3, using the Apache Arrow C++ engine, [Acero](https://arrow.apache.org/docs/cpp/acero/overview.html).
- [dbplyr](https://dbplyr.tidyverse.org/) for data stored in a relational
database. Translates your dplyr code to SQL.
- [dtplyr](https://dtplyr.tidyverse.org/) for large, in-memory datasets.
Translates your dplyr code to high performance
[data.table](https://rdatatable.gitlab.io/data.table/) code.
- [duckplyr](https://duckplyr.tidyverse.org/) for large, in-memory
datasets. Translates your dplyr code to high performance
[duckdb](https://duckdb.org) queries with zero extra copies and
an automatic R fallback when translation isn’t possible.
- [sparklyr](https://spark.posit.co/) for very large datasets stored in
[Apache Spark](https://spark.apache.org).
## Installation
```{r, eval = FALSE}
# The easiest way to get dplyr is to install the whole tidyverse:
install.packages("tidyverse")
# Alternatively, install just dplyr:
install.packages("dplyr")
```
### Development version
To get a bug fix or to use a feature from the development version, you can install
the development version of dplyr from GitHub.
```{r, eval = FALSE}
# install.packages("pak")
pak::pak("tidyverse/dplyr")
```
## Cheat Sheet
 ## Usage
```{r, message = FALSE}
library(dplyr)
starwars |>
filter(species == "Droid")
starwars |>
select(name, ends_with("color"))
starwars |>
mutate(name, bmi = mass / ((height / 100)^2)) |>
select(name:mass, bmi)
starwars |>
arrange(desc(mass))
starwars |>
group_by(species) |>
summarise(
n = n(),
mass = mean(mass, na.rm = TRUE)
) |>
filter(
n > 1,
mass > 50
)
```
## Getting help
If you encounter a clear bug, please file an issue with a minimal reproducible example on [GitHub](https://github.com/tidyverse/dplyr/issues). For questions and other discussion, please use [forum.posit.co](https://forum.posit.co/).
## Code of conduct
Please note that this project is released with a [Contributor Code of Conduct](https://dplyr.tidyverse.org/CODE_OF_CONDUCT).
By participating in this project you agree to abide by its terms.
## Usage
```{r, message = FALSE}
library(dplyr)
starwars |>
filter(species == "Droid")
starwars |>
select(name, ends_with("color"))
starwars |>
mutate(name, bmi = mass / ((height / 100)^2)) |>
select(name:mass, bmi)
starwars |>
arrange(desc(mass))
starwars |>
group_by(species) |>
summarise(
n = n(),
mass = mean(mass, na.rm = TRUE)
) |>
filter(
n > 1,
mass > 50
)
```
## Getting help
If you encounter a clear bug, please file an issue with a minimal reproducible example on [GitHub](https://github.com/tidyverse/dplyr/issues). For questions and other discussion, please use [forum.posit.co](https://forum.posit.co/).
## Code of conduct
Please note that this project is released with a [Contributor Code of Conduct](https://dplyr.tidyverse.org/CODE_OF_CONDUCT).
By participating in this project you agree to abide by its terms.