


a. The above reaction system was incubated at 50 °C for 15 minutes (inserting 1-2 fragments), 30 minutes (inserting 3 fragments simultaneously), or 60 minutes (inserting 4-5 fragments simultaneously). If subsequent operations cannot be carried out immediately after the reaction is completed, the reaction sample can be stored at -20 °C.
b. Take 5uL of the reaction sample from the previous step and add it to receptive cells such as 50-100 uL of DH5a (note that the volume of the added DNA sample should not exceed 1/10 of the receptive volume). Gently mix and place the mixture on ice for 30 minutes.
c. Heat shock in a 42 °C water bath for 90 seconds, then quickly return to the ice bath and let stand for 3-5 minutes. c. Add 500 uL of antibiotic free SOC or LB culture medium, gently mix well, and shake at 37 °C for 1 hour.
d. Centrifuge the bacterial solution at 5000 g for 1 minute to precipitate the bacterial body. Suck off most of the culture medium, leaving about 50-100 uL of culture medium, and resuspend the bacterial body. Then apply all evenly onto LB plates containing appropriate antibiotics and incubate overnight in a 37 °C incubator.
e. The next day, select clones from the plate for colony PCR or extract plasmids for enzyme digestion identification, or directly select several clones for sequencing identification.
a. Cultivation of Escherichia coli: Use a needle to pick up WT and culture them in shake tubes with 150 μL amp-resistant LB liquid culture medium at 37 °C,900 rpm for 24 hours.
b. Add 2 μL precultured bacterial solution to 150 μL automatic induction culture medium, culture at 37 °C, 900 rpm for 16 hours.
Preparation of DES solvent:c. TEA buffer:triethanolamine (50mM, pH 7.4)
d. 95% choline chloride and ethylene glycol (1:2): choline chloride 139.62g + ethylene glycol 124.14 g, heat and stir at 80 °C, and diluted to 95% with TEA buffer
e. 30% choline chloride and acetamide (1:2): choline chloride 139.62 g + acetamide 118.14 g , heat and stir at 8 °C, and diluted to 30% with TEA buffer
f. 30% tetrabutylammonium bromide and ethylene glycol (1:2): tetrabutylammonium bromide 339.33 g + ethylene glycol 124.14 g , heat and stir at 80 °C, and diluted to 30% with TEA buffer
g. 75% choline chloride and ethylene glycol (1:2): choline chloride 139.62 g + ethylene glycol 124.14 g , heat and stir at 80 °C, and diluted to 75% with TEA buffer

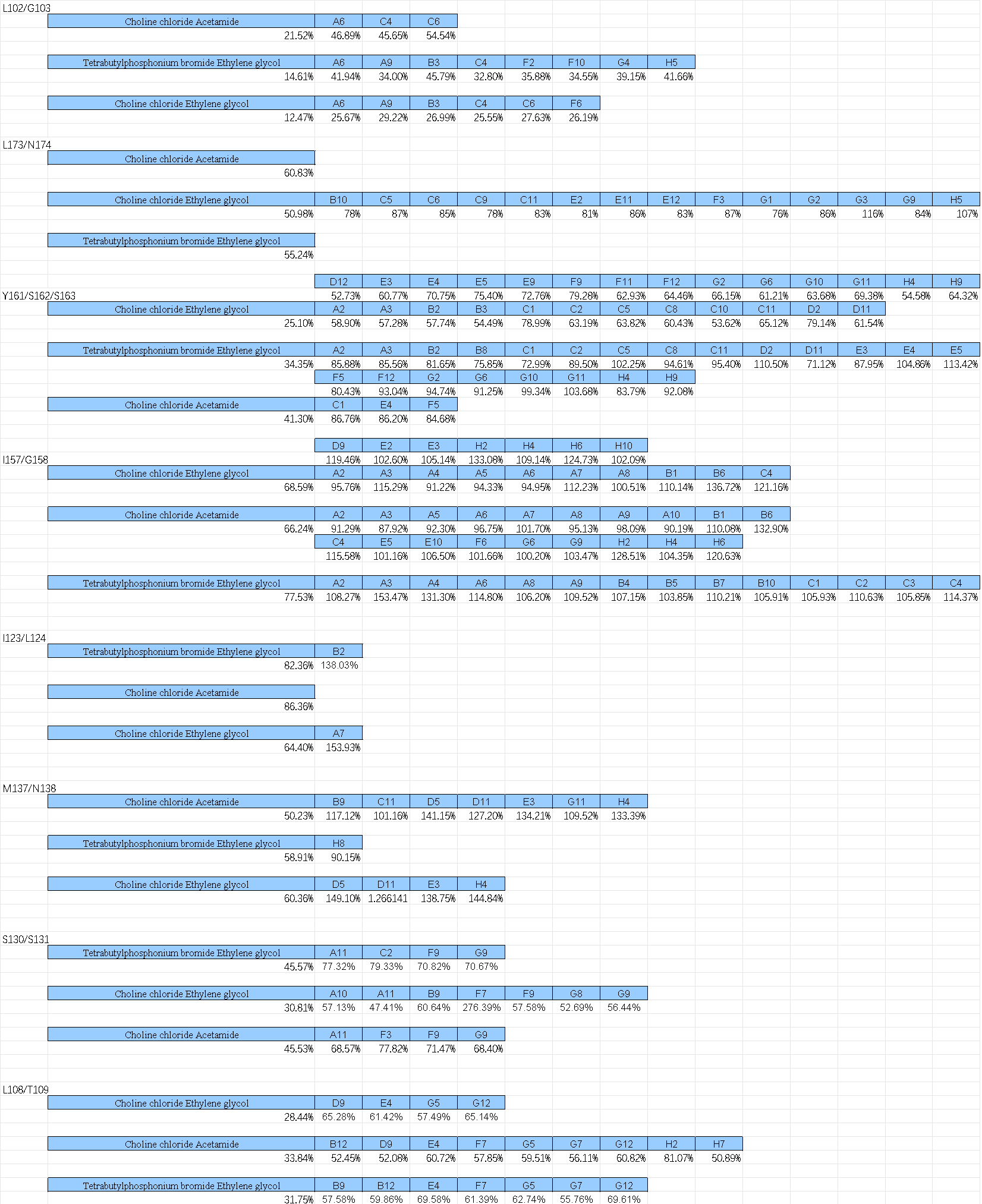
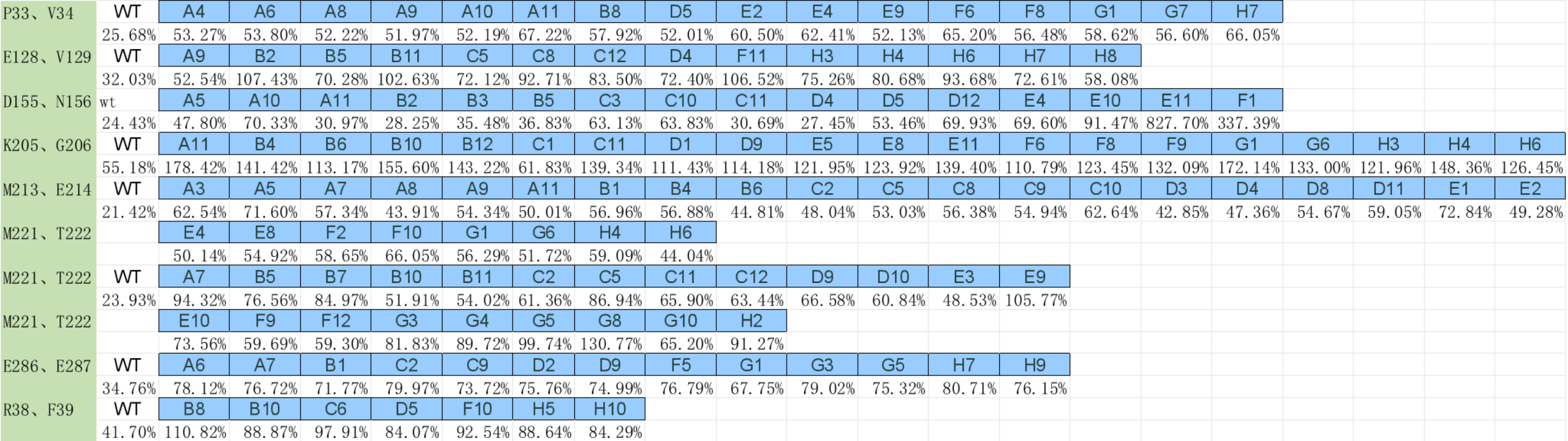
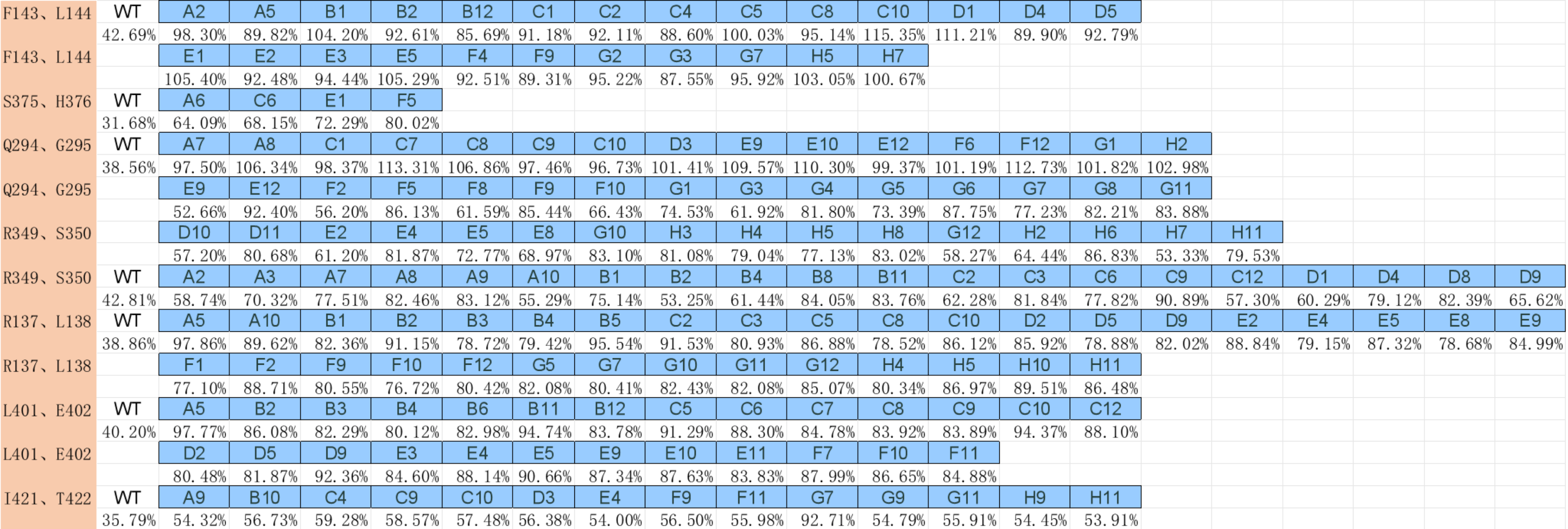
(a)-(c) Residual activity of BSLA WT at different DES concentrations. (b)The standard deviation to evaluate the applicability of the 96-well MTP-based screening system for directed BSLA WT evolution. From left to right, the compounds are 95% (v/v) ChCl:ethylene glycol, 30% (v/v) ChCl:acetamide, and 30% (v/v) TBPB:ethylene glycol, which used for the screening system to obtain residual activity of 30-40% of the BSLA WT.
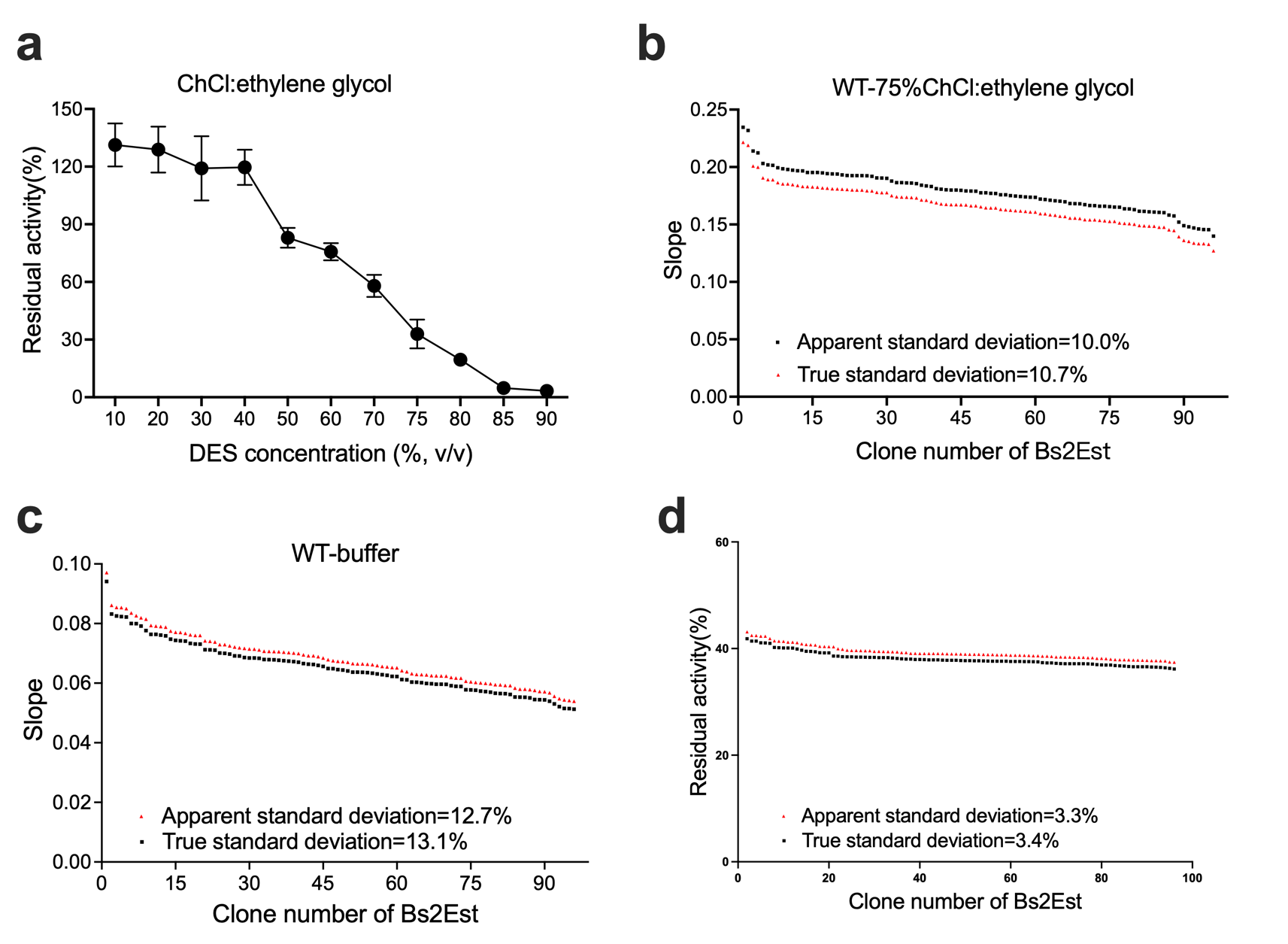 Figure 5.
Determine the solution for the high-throughput screening system.
Figure 5.
Determine the solution for the high-throughput screening system.
After excluding the DES types that tend to solidify at room temperature or are not easy to handle with high viscosity, we used choline chloride and ethylene glycol in a 1:2 ratio to configure concentrations of the solution.
a. 10X DNA Loading buffer
b. 15000 bp DNA Marker
c. PCR product
d. Nucleic acid gel dye(1 g agarose +100mL 1X TAE)
e. Buffer solution for nucleic acid gel electrophoresis()dilute 50X TAE to 1X with
f. RO
a. Add 100 mL 1X TAE to 1 g agar powder and add it into the conical flask specially made for agarose gel.
b. Boil in the microwave three times until completely dissolved, cool until not too hot to touch (45-55 °C), add 10 uL of nucleic acid dye, shake gently, and pour into the prepared mold, with a thickness of 60mm. Wait for it to solidify, then pull out the comb vertically to avoid damaging the sample hole.
c. Store the prepared nucleic acid gel in the lunch box.
e. Add 1X TAE buffer solution to the electrophoresis tank, and place the prepared gel run (large hole recovery gel and small hole verification gel, with the gel hole located at the negative electrode), with the buffer solution not passing through the sample hole.
f. Sampling: Select the appropriate size marker, mix the sample with the loading buffer, and then load the sample.
g. Set the voltage and press "run" to run the glue for about 30 minutes.
h. Take out the nucleic acid gel and observe it with a light microscope.
a. Pay attention to protection and wear gloves when operating!
b. To make glue, dilute TAE should be used, heated and cooled to 45 °C before adding nucleic acid dye. The prepared glue can be stored at 4 °C for one week.
c. The adhesive runs from the negative electrode to the positive electrode.
a. Take 100 µL of thawed receptive cells on ice, add the target plasmid, gently mix well, and let stand on ice for 30 minutes.
b. Heat shock the sample in a 42 °C water bath for 45-60 seconds, transfer it quickly to an ice bath, and let it stand for 2 minutes (do not shake the sample during the process of standing on ice, otherwise the conversion efficiency will be reduced).
c. Add 700 µL of sterile liquid medium (SOB or LB) without antibiotics to the centrifuge tube, mix well, and resuscitate at 37 °C and 200 rpm for 60 minutes.
d. According to the needs of the experiment, draw different volumes of resuscitation fluid and evenly spread it on the SOB or Lysogeny broth containing corresponding antibiotics, and put the plate upside down in the 37 °C incubator for overnight culture.
The bacterial liquid was transferred to the liquid LB supplemented with 1% 100 mg/mL Amp, at the same time. Pre cultivation system: 200 μL liquid LB + 1% 100 mg/mL Amp +bacterial broth
2 μL pre cultured bacterial solution, add self induced culture medium, culture them in 4 mL LB shake tubes with amp-resistant properties at 37 °C, 900 rpm for 16 hours.
a. Generally, the bacterial solution used for seed preservation needs to be thicker, so the cultivation time needs to be overnight;
b. Under the condition of Aseptic technique, add 500 uL bacterial solution and 500 uL 50% glycerol solution into the sterilized 2 mL cryopreservation tube respectively, mix gently and fully;
c. Glycerol cryopreserved bacteria can be directly stored in a -80 °C refrigerator.The frozen bacteria can be stored under this condition for several years. Subsequent freezing and thawing operations will reduce the shelf life.
(Note: Items that come into contact with the bacterial solution, such as gun heads, cryotubes, and 50% glycerol solution, must be in a sterile state and ensure that the operation is carried out under sterile conditions.)
a. Turn on the computer and enzyme marker power;
b. Place the enzyme label plate correctly. Do not put it in with a cover to prevent it from getting stuck inside the instrument; If the sample is accidentally spilled, it must be cleaned in a timely manner; Do not touch the card slot area when transferring the enzyme label board to prevent hand injuries;
c. Open the software and set parameters such as wavelength, temperature, and oscillation frequency;
d. Select the scanning hole position;
e. Conduct a scan, and the instrument reading is relatively accurate between 0.4 and 1.0, with an upper limit of 4.0, if the value is too large, dilute it before proceeding with the measurement; After scanning, save the data in a personal folder;
f. Remove the enzyme marker board and turn off the device.
a. Pre-culture experiment content: Toothpicks were dipped into glycerol bacteria in 96-well plates, in which four parallel experiments were done for each mutant, and they were precultured with LBamp. Take 150 μL from each well in the glycerol plate into the medium in microtiter plate, (37°C, 70% humidity, 900rpm, 24h) the obtained pre-culture solution can be used for the subsequent main culture experiments.
b. The main culture experiment content: 96-well plate as a container, take 2uL of pre-culture solution in the modified auto-induced Amp medium (components) to express protein, (37 °C, 70% humidity, 900 rpm, 16 hours).
c. Enzyme activity assay: The culture was centrifuged (4°C, 2000 rpm, 20 min) to obtain the crude enzyme, which was used for further enzyme activity assay. The esterase activity of BSLA and Bs2Est was determined using p-nitrophenyl butyrate (pNPB) as substrate. 95 μL DES/Buffer and 5 μL WT(Variants)/EV were incubated for 2 h at room temperature, and then the A410 nm was measured on the microtiter plate reader (Biotek Synergy HI, USA), and the release of pNP was recorded in 8 minutes.

LB-medium: Yeast extract 5 g/L, Tryptone 10 g/L, NaCl 10 g/L
Autoinduction medium: The media include: media components, buffer components and induction components
| Composition | Weight |
|---|---|
| Peptone (aus Casein, Roth) | 12 g |
| Yeast extract (Merck) | 24 g |
| Glycerol (Roth) | 5 g |
| Add dd. H2O to 800 mL | |
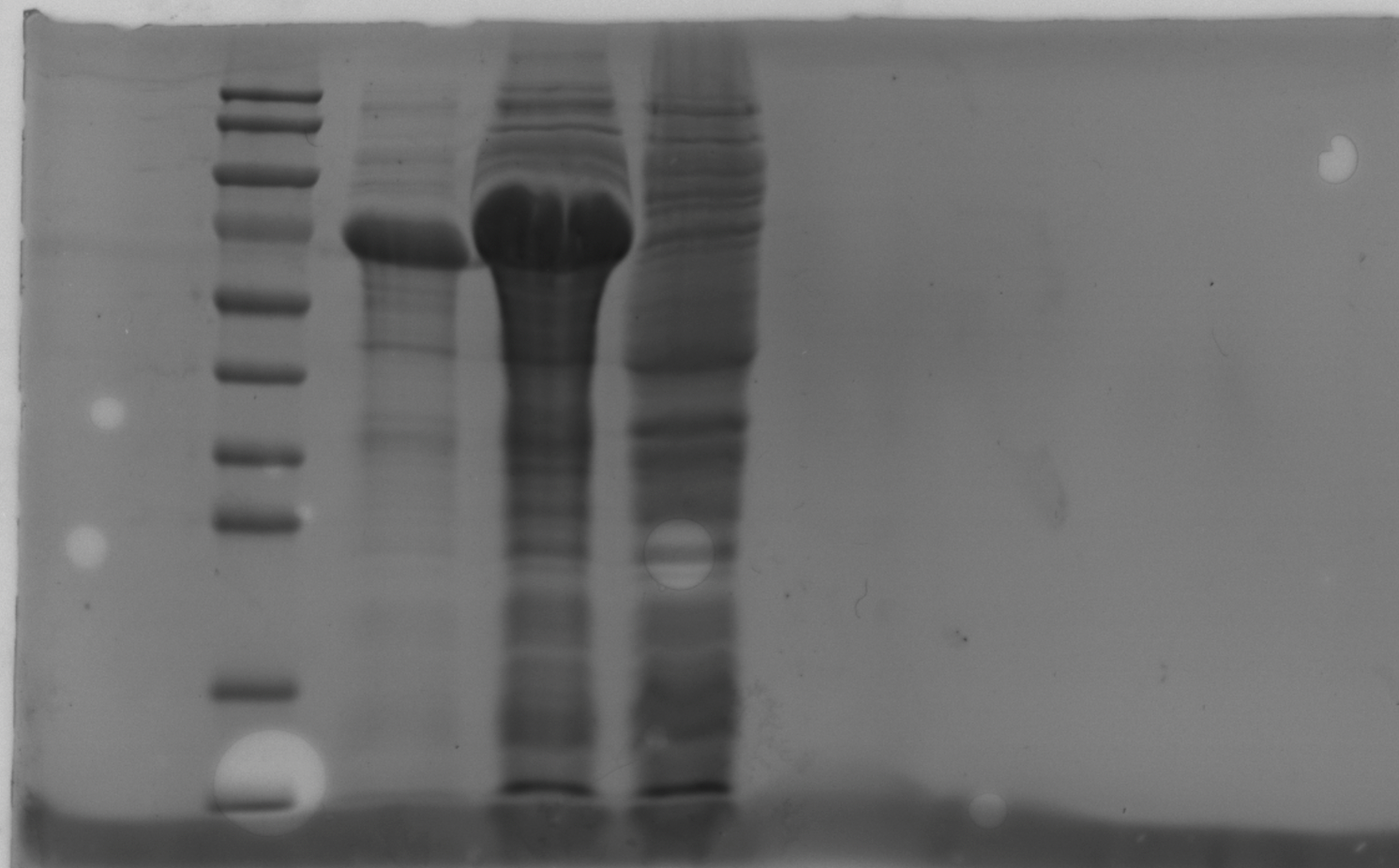
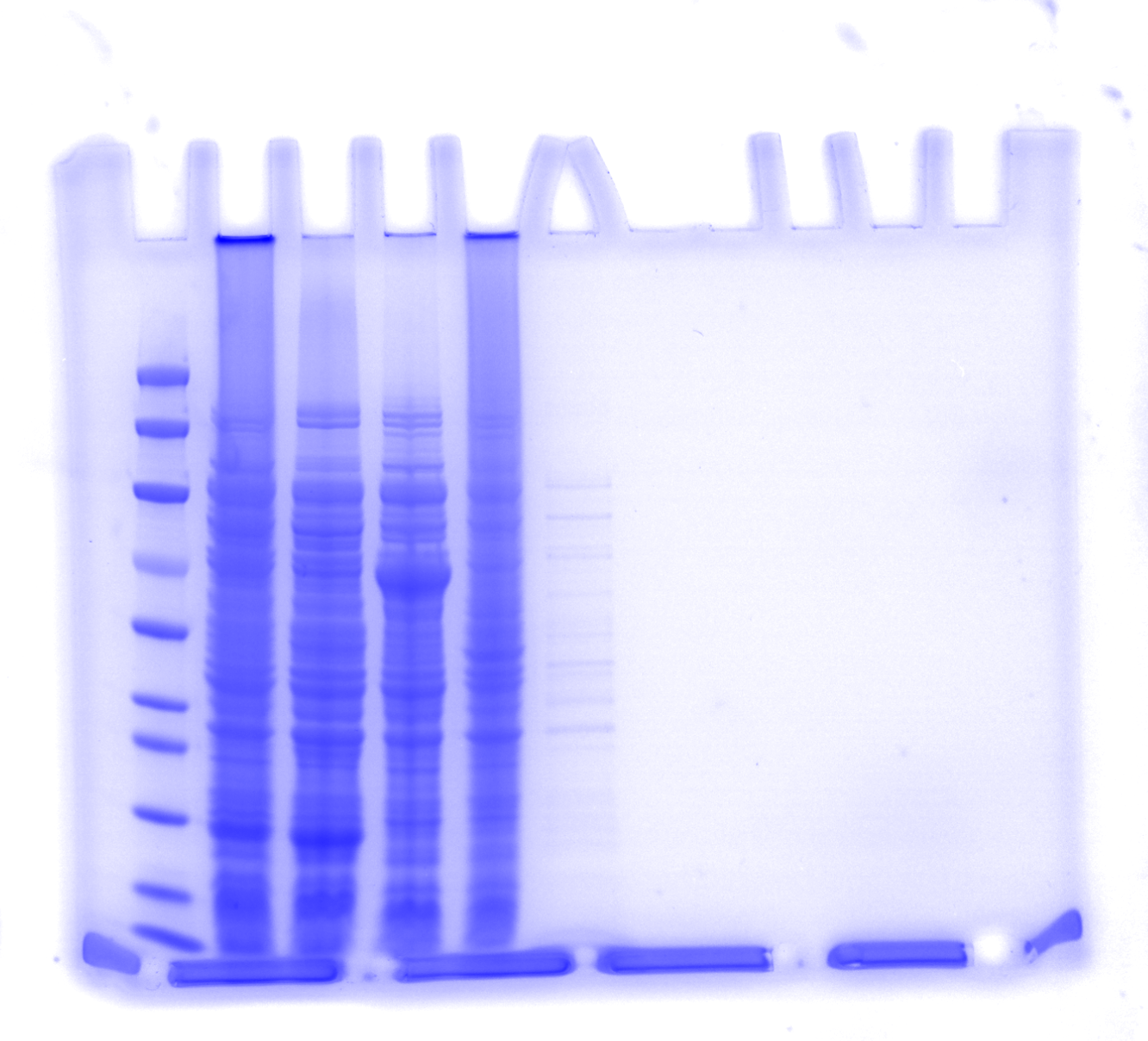
a. 50 mM potassium phosphate buffer (pH 8.0) 1000 mL
b. 50 mM sodium phosphate buffer, 300 mM sodium chloride, and 20 mM imidazole, pH 8.0
c. 50 mM sodium phosphate buffer, 300 mM sodium chloride, and 50 mM imidazole, pH 8.0
d. 50 mM sodium phosphate buffer, 300 mM sodium chloride, and 100 mM imidazole, pH 8.0
e. 50 mM sodium phosphate buffer, 300 mM sodium chloride, and 250 mM imidazole, pH 8.0
f. 50 mM sodium phosphate buffer, 300 mM sodium chloride, and 500 mM imidazole, pH 8.0
Toothpick dip the bacterial liquid into 4 ml LB: First, use a toothpick to transfer the bacterial liquid into a tube containing 4 mL of LB medium. LB medium is a nutrient-rich medium commonly used for culturing bacteria such as Escherichia coli. 37 °C, 220 rpm, 12 hours: Place the tube containing the bacterial liquid in a constant temperature shaker and culture it at 37 °C and 220 rpm for 12 hours. This step aims to allow the bacterial liquid to grow and proliferate under suitable temperature and agitation conditions. Inoculate 1% volume of the cultured liquid into 100 mL LB, 37 °C, 220 rpm, 2 hours: Take out 1% (by volume) of the liquid cultured in the previous step and add it to a large container containing 100 mL of auto-induction medium. Then, continue to culture it at 30°C for 16h.
First, use a toothpick to transfer the bacterial liquid into a tube containing 4 mL of LB medium. LB medium is a nutrient-rich medium commonly used for culturing bacteria such as Escherichia coli. 37 °C, 220 rpm, 12 hours: Place the tube containing the bacterial liquid in a constant temperature shaker and culture it at 37 °C and 220 rpm for 12 hours. This step aims to allow the bacterial liquid to grow and proliferate under suitable temperature and agitation conditions. Inoculate 1% volume of the cultured liquid into 100 mL LB, 37 °C, 220 rpm, 2 hours: Take out 1% (by volume) of the liquid cultured in the previous step and add it to a large container containing 100 mL of LB medium.
Then, continue to culture it at 37 °C and 220 rpm for 2 hours. This step aims to scale up the volume of the bacterial liquid to provide more cells for expression. 0.1 mM IPTG, 20 °C, 180 rpm, 24 hours: Add IPTG (Isopropyl β-D-1-thiogalactopyranoside) at a concentration of 0.1 millimolar to the culture medium to induce the expression of the target protein. Next, continue to culture it at 20 °C and 180 rpm for 24 hours. IPTG is a synthetic compound that can mimic signals bacteria receive in their natural environment, thus triggering the expression of the target protein. E. coli cells were harvested by centrifugation at 11600 g for 10 min at 4 °C, and then washed twice with proper buffer (pH 7.5). The washed cells were resuspended in 50 mM potassium phosphate buffer (pH 8.0) and disrupted by sonication (10-sec pulse on, 15-sec pulse off, AMP 20%).
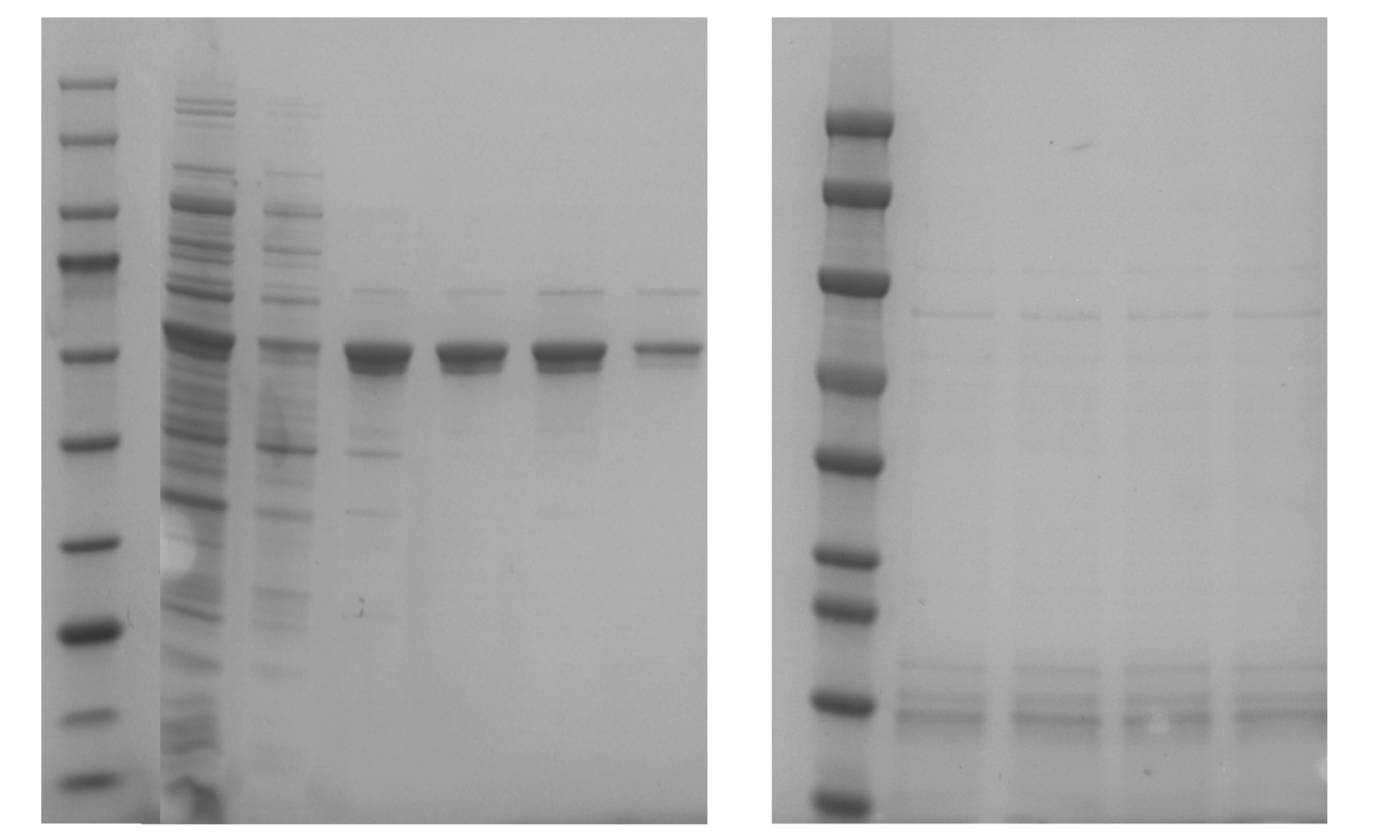
Pre-culture and master culture of all beneficial mutants of BSLA and Bs2Est. The expressed supernatant was taken for determination of different DES concentrations. Different concentrations of DES solution are diluted with buffer TAE. The method of measuring enzyme activity is consistent with the previous method of constructing a mutant library.
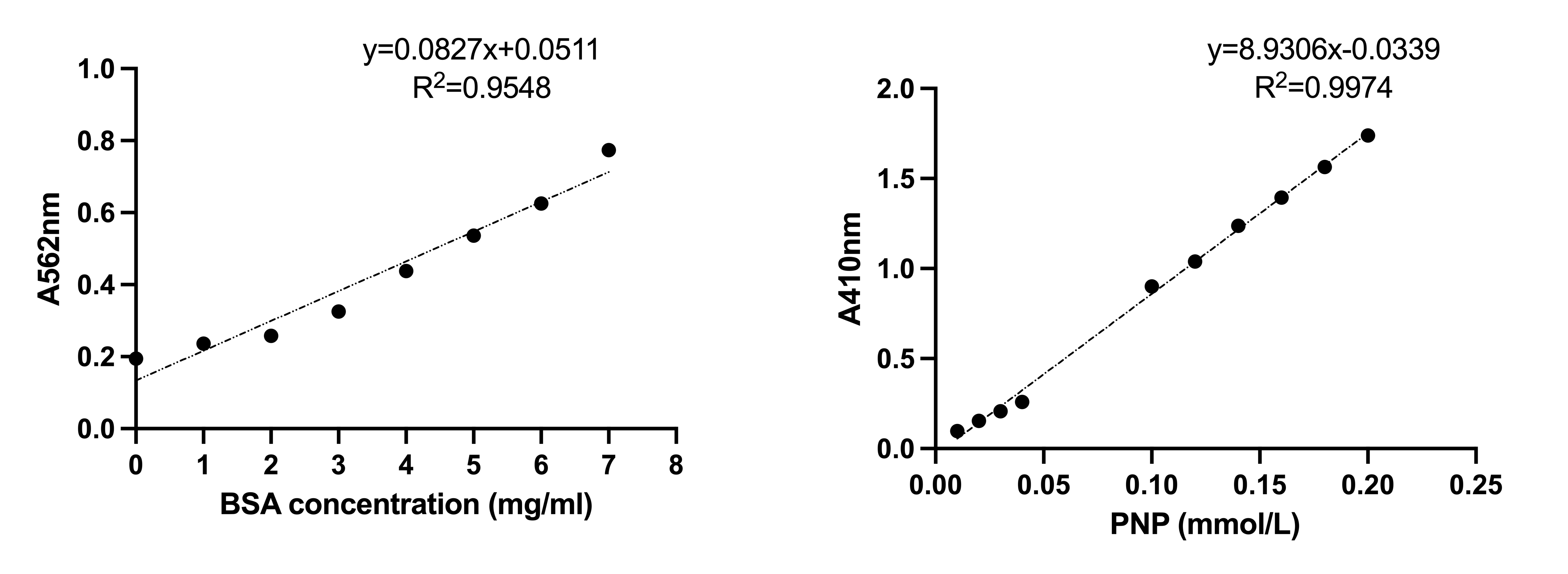
Kcat/KM assay with purified enzyme. Configure different concentrations of PNP to determine the 410 nm absorbance to plot the standard curve of PNP. All purified mutants are assayed for protein concentration and protein standard curves are plotted. Change the substrate pNPB concentration and monitor the absorbance value after 5 min of the reaction. Kcat/KM is obtained by Graphpad.
Pre-culture and master culture of all beneficial mutants of BSLA and Bs2Est. The expressed supernatant was taken for determination of different DES concentrations. Different concentrations of DES solution are diluted with buffer TAE. The method of measuring enzyme activity is consistent with the previous method of constructing a mutant library.
Kcat/KM assay with purified enzyme. Configure different concentrations of PNP to determine the 410 nm absorbance to plot the standard curve of PNP. All purified mutants are assayed for protein concentration and protein standard curves are plotted. Change the substrate pNPB concentration and monitor the absorbance value after 5 min of the reaction.Kcat/KM is obtained by Graphpad.
References
Sheldon, R. A.; Woodley, J. M. Role of Biocatalysis in Sustainable Chemistry. Chemical Reviews 2017. https://doi.org/10.1021/acs.chemrev.7b00203.
Cui, H.; Vedder, M.; Zhang, L.; Jaeger, K.-E.; Schwaneberg, U.; Davari, M. D. Polar Substitutions on the Surface of a Lipase Substantially Improve Tolerance in Organic Solvents. ChemSusChem 2022, 15 (9), e202102551. https://doi.org/10.1002/cssc.202102551.
Xu, P.; Liang, S.; Zong, M.-H.; Lou, W.-Y. Ionic Liquids for Regulating Biocatalytic Process: Achievements and Perspectives. Biotechnology Advances 2021. https://doi.org/10.1016/j.biotechadv.2021.107702.
Abbott, A. P.; Capper, G.; Davies, D. L.; Rasheed, R. K.; Tambyrajah, V. Novel Solvent Properties of Choline Chloride/Urea mixturesElectronic Supplementary Information (ESI) Available: Spectroscopic Data. See Http://Www.Rsc.Org/Suppdata/Cc/B2/B210714g/. Chem. Commun. 2003, No. 1, 70-71. https://doi.org/10.1039/b210714g.
Danait-Nabar, S.; Singhal, R. S. Investigation into the Chemical Modification of α-Amylase Using Octenyl Succinic Anhydride: Enzyme Characterisation and Stability Studies. Bioprocess Biosyst Eng 2023, 46 (5), 645–664. https://doi.org/10.1007/s00449-023-02850-z.
Acevedo-Rocha, C. G.; Hoebenreich, S.; Reetz, M. T. Iterative Saturation Mutagenesis: A Powerful Approach to Engineer Proteins by Systematically Simulating Darwinian Evolution. In Directed Evolution Library Creation; Gillam, E. M. J., Copp, J. N., Ackerley, D., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, 2014; Vol. 1179, pp 103–128. https://doi.org/10.1007/978-1-4939-1053-3_7.
Taklimi, S. M.; Divsalar, A.; Ghalandari, B.; Ding, X.; Di Gioia, M. L.; Omar, K. A.; Saboury, A. A. Effects of Deep Eutectic Solvents on the Activity and Stability of Enzymes. Journal of Molecular Liquids 2023, 377, 121562. https://doi.org/10.1016/j.molliq.2023.121562.
Wang, M.; Cui, H.; Gu, C.; Li, A.; Qiao, J.; Schwaneberg, U.; Zhang, L.; Wei, J.; Li, X.; Huang, H. Engineering All-Round Cellulase for Bioethanol Production. ACS Synth. Biol. 2023, 12 (7), 2187–2197. https://doi.org/10.1021/acssynbio.3c00289.
Qiao, J.; Sheng, Y.; Wang, M.; Li, A.; Li, X.; Huang, H. Evolving Robust and Interpretable Enzymes for the Bioethanol Industry. Angew Chem Int Ed 2023, 62 (12), e202300320. https://doi.org/10.1002/anie.202300320.
Cui, H.; Eltoukhy, L.; Zhang, L.; Markel, U.; Jaeger, K.; Davari, M. D.; Schwaneberg, U. Less Unfavorable Salt Bridges on the Enzyme Surface Result in More Organic Cosolvent Resistance. Angew. Chem. Int. Ed. 2021, 60 (20), 11448–11456. https://doi.org/10.1002/anie.202101642.