{
"cells": [
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"\n",
" \n",
" \n",
" \n",
"# Parallel Computing in Python with Dask\n",
"\n",
"## James Bourbeau\n",
"### Software Engineer, Quansight\n",
"#### Data-Driven Wisconsin 2019\n",
"\n",
"https://github.com/jrbourbeau/ddw-dask"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Outline\n",
"\n",
"- [Overview: What is/why Dask?](#Overview)\n",
"\n",
"- [High Level Collections](#High-Level-Collections)\n",
"\n",
" - [Dask Arrays](#Dask-Arrays)\n",
" \n",
" - [Dask DataFrames](#Dask-DataFrames)\n",
"\n",
"- [Dask Delayed](#Dask-Delayed)\n",
"\n",
"- [Schedulers](#Schedulers)\n",
"\n",
" - [Single Machine Schedulers](#Single-Machine-Schedulers)\n",
"\n",
" - [Distributed Scheduler](#Distributed-Scheduler)\n",
" \n",
"- [Scalable Machine Learning with Dask-ML](#Scalable-Machine-Learning-with-Dask-ML)\n",
"\n",
"- [Additional Resources](#Additional-Resources)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Overview\n",
"\n",
"[ [Back to top](#Outline) ]"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## What is Dask?\n",
"\n",
"- Dask is a flexible, open source library for parallel computing in Python\n",
"\n",
" - GitHub: https://github.com/dask/dask\n",
" \n",
" - Documentation: https://docs.dask.org\n",
"\n",
"- Scales the existing Python ecosystem"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Why Dask?\n",
"\n",
"- Enables parallel and larger-than-memory computations\n",
"\n",
"- Uses familiar APIs you're used to from projects like NumPy, Pandas, and scikit-learn\n",
"\n",
"- Allows you to scale existing workflows with minimal code changes\n",
"\n",
"- Dask works on your laptop, but also scales out to large clusters\n",
"\n",
"- Offers great built-in diagnosic tools"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Components of Dask\n",
"\n",
"
\n",
" \n",
"# Parallel Computing in Python with Dask\n",
"\n",
"## James Bourbeau\n",
"### Software Engineer, Quansight\n",
"#### Data-Driven Wisconsin 2019\n",
"\n",
"https://github.com/jrbourbeau/ddw-dask"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Outline\n",
"\n",
"- [Overview: What is/why Dask?](#Overview)\n",
"\n",
"- [High Level Collections](#High-Level-Collections)\n",
"\n",
" - [Dask Arrays](#Dask-Arrays)\n",
" \n",
" - [Dask DataFrames](#Dask-DataFrames)\n",
"\n",
"- [Dask Delayed](#Dask-Delayed)\n",
"\n",
"- [Schedulers](#Schedulers)\n",
"\n",
" - [Single Machine Schedulers](#Single-Machine-Schedulers)\n",
"\n",
" - [Distributed Scheduler](#Distributed-Scheduler)\n",
" \n",
"- [Scalable Machine Learning with Dask-ML](#Scalable-Machine-Learning-with-Dask-ML)\n",
"\n",
"- [Additional Resources](#Additional-Resources)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Overview\n",
"\n",
"[ [Back to top](#Outline) ]"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## What is Dask?\n",
"\n",
"- Dask is a flexible, open source library for parallel computing in Python\n",
"\n",
" - GitHub: https://github.com/dask/dask\n",
" \n",
" - Documentation: https://docs.dask.org\n",
"\n",
"- Scales the existing Python ecosystem"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Why Dask?\n",
"\n",
"- Enables parallel and larger-than-memory computations\n",
"\n",
"- Uses familiar APIs you're used to from projects like NumPy, Pandas, and scikit-learn\n",
"\n",
"- Allows you to scale existing workflows with minimal code changes\n",
"\n",
"- Dask works on your laptop, but also scales out to large clusters\n",
"\n",
"- Offers great built-in diagnosic tools"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Components of Dask\n",
"\n",
" \n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Task Graphs"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"def inc(i):\n",
" return i + 1\n",
"\n",
"def add(a, b):\n",
" return a + b\n",
"\n",
"a, b = 1, 12\n",
"c = inc(a)\n",
"d = inc(b)\n",
"output = add(c, d)\n",
"\n",
"print(f'output = {output}')"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This computation can be encoded in the following task graph:"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"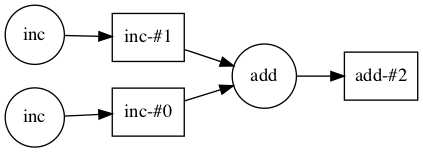"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
" \n",
"- Graph of inter-related tasks with dependencies between them\n",
"\n",
"- Circular nodes in the graph are Python function calls\n",
"\n",
"- Square nodes are Python objects that are created by one task as output and can be used as inputs in another task"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# High Level Collections\n",
"\n",
"[ [Back to top](#Outline) ]"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Dask Arrays\n",
"\n",
"- Dask arrays are chunked, n-dimensional arrays\n",
"\n",
"- Can think of a Dask array as a collection of NumPy `ndarray` arrays\n",
"\n",
"- Dask arrays implement a large subset of the NumPy API using blocked algorithms\n",
"\n",
"- For many purposes Dask arrays can serve as drop-in replacements for NumPy arrays"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"
\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Task Graphs"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"def inc(i):\n",
" return i + 1\n",
"\n",
"def add(a, b):\n",
" return a + b\n",
"\n",
"a, b = 1, 12\n",
"c = inc(a)\n",
"d = inc(b)\n",
"output = add(c, d)\n",
"\n",
"print(f'output = {output}')"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This computation can be encoded in the following task graph:"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
" \n",
"- Graph of inter-related tasks with dependencies between them\n",
"\n",
"- Circular nodes in the graph are Python function calls\n",
"\n",
"- Square nodes are Python objects that are created by one task as output and can be used as inputs in another task"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# High Level Collections\n",
"\n",
"[ [Back to top](#Outline) ]"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Dask Arrays\n",
"\n",
"- Dask arrays are chunked, n-dimensional arrays\n",
"\n",
"- Can think of a Dask array as a collection of NumPy `ndarray` arrays\n",
"\n",
"- Dask arrays implement a large subset of the NumPy API using blocked algorithms\n",
"\n",
"- For many purposes Dask arrays can serve as drop-in replacements for NumPy arrays"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
" "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"import numpy as np\n",
"import dask.array as da"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_np = np.arange(1, 50, 3)\n",
"arr_np"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can create a Dask array in a similar manner, but need to specify a `chunks` argument to tell Dask how to break up the underlying array into chunks."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_da = da.arange(1, 50, 3, chunks=5) # Each chunk is 5 items long\n",
"type(arr_da)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_da # Dask arrays have nice HTML output in Jupyter notebooks"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask arrays look and feel like NumPy arrays. For example, they have `dtype` and `shape` attributes"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(arr_da.dtype)\n",
"print(arr_da.shape)"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"Dask arrays are _lazily_ evaluated. The result from a computation isn't computed until you ask for it. Instead, a Dask task graph for the computation is produced. You can visualize the task graph using the `visualize()` method."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "-"
}
},
"outputs": [],
"source": [
"arr_da.visualize()"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"To compute a task graph call the `compute()` method"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_da.compute() # We'll go into more detail about .compute() later on"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The result of this computation is a fimilar NumPy `ndarray`"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"type(arr_da.compute())"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask arrays support a large portion of the NumPy interface:\n",
"\n",
"- Arithmetic and scalar mathematics: `+`, `*`, `exp`, `log`, ...\n",
"\n",
"- Reductions along axes: `sum()`, `mean()`, `std()`, `sum(axis=0)`, ...\n",
"\n",
"- Tensor contractions / dot products / matrix multiply: `tensordot`\n",
"\n",
"- Axis reordering / transpose: `transpose`\n",
"\n",
"- Slicing: `x[:100, 500:100:-2]`\n",
"\n",
"- Fancy indexing along single axes with lists or numpy arrays: `x[:, [10, 1, 5]]`\n",
"\n",
"- Array protocols like `__array__` and `__array_ufunc__`\n",
"\n",
"- Some linear algebra: `svd`, `qr`, `solve`, `solve_triangular`, `lstsq`, ...\n",
"\n",
"- ...\n",
"\n",
"See the [Dask array API docs](http://docs.dask.org/en/latest/array-api.html) for full details about what portion of the NumPy API is implemented for Dask arrays."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Blocked Algorithms\n",
"\n",
"Dask arrays are implemented using _blocked algorithms_. These algorithms break up a computation on a large array into many computations on smaller peices of the array. This minimizes the memory load (amount of RAM) of computations and allows for working with larger-than-memory datasets in parallel."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = da.random.random(20, chunks=5)\n",
"x"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"result = x.sum()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can build more complex computations using the familiar NumPy operations we're used to."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"x = da.random.random(size=(1_000, 1_000), chunks=(250, 500))\n",
"x"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"result = (x + x.T).sum(axis=0).mean()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can even perform computations on larger-than-memory arrays!"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = da.random.random(size=(40_000, 40_000), chunks=(2_000, 2_000))"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x.nbytes / 1e9 # Size of array in gigabytes"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result = (x + x.T).sum(axis=0).mean()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask.diagnostics import ProgressBar\n",
"\n",
"with ProgressBar():\n",
" result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Note**: Dask can be used to scale other array-like libraries that support the NumPy `ndarray` interface. For example, [pydata/sparse](https://sparse.pydata.org/en/latest/) for sparse arrays or [CuPy](https://cupy.chainer.org/) for GPU-accelerated arrays."
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Dask DataFrames\n",
"\n",
"[Pandas](http://pandas.pydata.org/pandas-docs/stable/) is great for dealing with tabular datasets that can fit in memory on a single machine. Dask becomes useful when dealing with larger-than-memory datasets. We saw that a Dask arrays are composed of many NumPy arrays, chunked along one or more dimensions. It's similar for Dask DataFrames: a Dask DataFrame is composed of many Pandas DataFrames and the partitioning happens only along the index.\n",
"\n",
"- Dask DataFrames are a collection of Pandas DataFrames\n",
"\n",
"- Dask DataFrames implement a large subset of the Pandas API\n",
"\n",
"- Backed by blocked algorithms that allow for parallel and out of core computation\n",
"\n",
"- For many purposes Dask DataFrames can serve as drop-in replacements for Pandas DataFrames"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"
"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"import numpy as np\n",
"import dask.array as da"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_np = np.arange(1, 50, 3)\n",
"arr_np"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can create a Dask array in a similar manner, but need to specify a `chunks` argument to tell Dask how to break up the underlying array into chunks."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_da = da.arange(1, 50, 3, chunks=5) # Each chunk is 5 items long\n",
"type(arr_da)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_da # Dask arrays have nice HTML output in Jupyter notebooks"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask arrays look and feel like NumPy arrays. For example, they have `dtype` and `shape` attributes"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"print(arr_da.dtype)\n",
"print(arr_da.shape)"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"Dask arrays are _lazily_ evaluated. The result from a computation isn't computed until you ask for it. Instead, a Dask task graph for the computation is produced. You can visualize the task graph using the `visualize()` method."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "-"
}
},
"outputs": [],
"source": [
"arr_da.visualize()"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"To compute a task graph call the `compute()` method"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"arr_da.compute() # We'll go into more detail about .compute() later on"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The result of this computation is a fimilar NumPy `ndarray`"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"type(arr_da.compute())"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask arrays support a large portion of the NumPy interface:\n",
"\n",
"- Arithmetic and scalar mathematics: `+`, `*`, `exp`, `log`, ...\n",
"\n",
"- Reductions along axes: `sum()`, `mean()`, `std()`, `sum(axis=0)`, ...\n",
"\n",
"- Tensor contractions / dot products / matrix multiply: `tensordot`\n",
"\n",
"- Axis reordering / transpose: `transpose`\n",
"\n",
"- Slicing: `x[:100, 500:100:-2]`\n",
"\n",
"- Fancy indexing along single axes with lists or numpy arrays: `x[:, [10, 1, 5]]`\n",
"\n",
"- Array protocols like `__array__` and `__array_ufunc__`\n",
"\n",
"- Some linear algebra: `svd`, `qr`, `solve`, `solve_triangular`, `lstsq`, ...\n",
"\n",
"- ...\n",
"\n",
"See the [Dask array API docs](http://docs.dask.org/en/latest/array-api.html) for full details about what portion of the NumPy API is implemented for Dask arrays."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Blocked Algorithms\n",
"\n",
"Dask arrays are implemented using _blocked algorithms_. These algorithms break up a computation on a large array into many computations on smaller peices of the array. This minimizes the memory load (amount of RAM) of computations and allows for working with larger-than-memory datasets in parallel."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = da.random.random(20, chunks=5)\n",
"x"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"result = x.sum()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can build more complex computations using the familiar NumPy operations we're used to."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"x = da.random.random(size=(1_000, 1_000), chunks=(250, 500))\n",
"x"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"outputs": [],
"source": [
"result = (x + x.T).sum(axis=0).mean()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can even perform computations on larger-than-memory arrays!"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = da.random.random(size=(40_000, 40_000), chunks=(2_000, 2_000))"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x.nbytes / 1e9 # Size of array in gigabytes"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result = (x + x.T).sum(axis=0).mean()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask.diagnostics import ProgressBar\n",
"\n",
"with ProgressBar():\n",
" result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Note**: Dask can be used to scale other array-like libraries that support the NumPy `ndarray` interface. For example, [pydata/sparse](https://sparse.pydata.org/en/latest/) for sparse arrays or [CuPy](https://cupy.chainer.org/) for GPU-accelerated arrays."
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Dask DataFrames\n",
"\n",
"[Pandas](http://pandas.pydata.org/pandas-docs/stable/) is great for dealing with tabular datasets that can fit in memory on a single machine. Dask becomes useful when dealing with larger-than-memory datasets. We saw that a Dask arrays are composed of many NumPy arrays, chunked along one or more dimensions. It's similar for Dask DataFrames: a Dask DataFrame is composed of many Pandas DataFrames and the partitioning happens only along the index.\n",
"\n",
"- Dask DataFrames are a collection of Pandas DataFrames\n",
"\n",
"- Dask DataFrames implement a large subset of the Pandas API\n",
"\n",
"- Backed by blocked algorithms that allow for parallel and out of core computation\n",
"\n",
"- For many purposes Dask DataFrames can serve as drop-in replacements for Pandas DataFrames"
]
},
{
"cell_type": "markdown",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
" "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import pandas as pd\n",
"import dask.dataframe as dd"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask DataFrames support many of the same data I/O methods as Pandas. For example, \n",
"\n",
"- `read_csv` \\ `to_csv`\n",
"- `read_hdf` \\ `to_hdf`\n",
"- `read_json` \\ `to_json`\n",
"- `read_parquet` \\ `to_parquet`"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Can specify a `chunksize` argument to set the number of rows per partition"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"ddf = dd.read_hdf('example_data.hdf', key='dataframe', chunksize=25)\n",
"ddf"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The `npartitions` attribute tells us how many Pandas DataFrames make up our Dask DataFrame"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"ddf.npartitions"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Like Dask arrays, Dask DataFrames are lazily evaluated. Here, the `dd.read_hdf` function wraps several calls to `pd.read_hdf`, once for each partition of the Dask DataFrame."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"ddf.visualize()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask DataFrames cover a well-used portion of the Pandas API:\n",
"\n",
"- Elementwise operations: `df.x` + `df.y`, `df * df`\n",
"\n",
"- Row-wise selections: `df[df.x > 0]`\n",
"\n",
"- Loc: `df.loc[4.0:10.5]`\n",
"\n",
"- Common aggregations: `df.x.max()`, `df.max()`\n",
"\n",
"- Is in: `df[df.x.isin([1, 2, 3])]`\n",
"\n",
"- Datetime/string accessors: `df.timestamp.month`\n",
"\n",
"- Froupby-aggregate (with common aggregations): `df.groupby(df.x).y.max()`, `df.groupby('x').max()`\n",
"\n",
"- ...\n",
"\n",
"See the [Dask DataFrame API docs](http://docs.dask.org/en/latest/dataframe-api.html) for full details about what portion of the Pandas API is implemented for Dask DataFrames."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"col_mean = ddf['col_1'].mean()\n",
"col_mean"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"col_mean.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"col_mean.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Dask Delayed\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"Sometimes problems don’t fit nicely into one of the high-level collections like Dask arrays or Dask DataFrames. In these cases, you can parallelize custom algorithms using the lower-level Dask `delayed` interface. This allows one to manually create task graphs with a light annotation of normal Python code."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import time\n",
"import random\n",
"\n",
"def inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1\n",
"\n",
"def double(x):\n",
" time.sleep(random.random())\n",
" return 2 * x\n",
" \n",
"def add(x, y):\n",
" time.sleep(random.random())\n",
" return x + y "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"data = [1, 2, 3, 4]\n",
"\n",
"output = []\n",
"for x in data:\n",
" a = inc(x)\n",
" b = double(x)\n",
" c = add(a, b)\n",
" output.append(c)\n",
"\n",
"total = sum(output)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask `delayed` wraps function calls and delays their execution. `delayed` functions record what we want to compute (a function and input parameters) as a task in a graph that we’ll run later on parallel hardware by calling `compute`."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask import delayed"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"@delayed\n",
"def lazy_inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"lazy_inc"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"inc_output = lazy_inc(3) # lazily evaluate inc(3)\n",
"inc_output"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"inc_output.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Using `delayed` functions, we can build up a task graph for the particular computation we want to perform"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"double_inc_output = lazy_inc(inc_output)\n",
"double_inc_output"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"double_inc_output.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"double_inc_output.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can use `delayed` to make our previous example computation lazy by wrapping all the function calls with delayed"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import time\n",
"import random\n",
"\n",
"@delayed\n",
"def inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1\n",
"\n",
"@delayed\n",
"def double(x):\n",
" time.sleep(random.random())\n",
" return 2 * x\n",
"\n",
"@delayed\n",
"def add(x, y):\n",
" time.sleep(random.random())\n",
" return x + y"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"data = [1, 2, 3, 4]\n",
"\n",
"output = []\n",
"for x in data:\n",
" a = inc(x)\n",
" b = double(x)\n",
" c = add(a, b)\n",
" output.append(c)\n",
"\n",
"total = delayed(sum)(output)\n",
"total"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"total.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"I highly recommend checking out the [Dask delayed best practices](http://docs.dask.org/en/latest/delayed-best-practices.html) page to avoid some common pitfalls when using `delayed`. "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Schedulers\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"High-level collections like Dask arrays and Dask DataFrames, as well as the low-level `dask.delayed` interface build up task graphs for a computation. After these graphs are generated, they need to be executed (potentially in parallel). This is the job of a task scheduler. Different task schedulers exist within Dask. Each will consume a task graph and compute the same result, but with different performance characteristics. "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask has two different classes of schedulers: single-machine schedulers and a distributed scheduler."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Single Machine Schedulers\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"Single machine schedulers provide basic features on a local process or thread pool and require no setup (only use the Python standard library). The different single machine schedulers Dask provides are:\n",
"\n",
"- `'threads'`: The threaded scheduler executes computations with a local `multiprocessing.pool.ThreadPool`. The threaded scheduler is the default choice for Dask arrays, Dask DataFrames, and Dask delayed. \n",
"\n",
"- `'processes'`: The multiprocessing scheduler executes computations with a local `multiprocessing.Pool`.\n",
"\n",
"- `'single-threaded'`: The single-threaded synchronous scheduler executes all computations in the local thread, with no parallelism at all. This is particularly valuable for debugging and profiling, which are more difficult when using threads or processes."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"You can configure which scheduler is used in a few different ways. You can set the scheduler globally by using the `dask.config.set(scheduler=)` command"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import dask\n",
"\n",
"dask.config.set(scheduler='threads')\n",
"total.compute(); # Will use the multi-threading scheduler"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"or use it as a context manager to set the scheduler for a block of code"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"with dask.config.set(scheduler='processes'):\n",
" total.compute() # Will use the multi-processing scheduler"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"or even within a single compute call"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.compute(scheduler='threads'); # Will use the multi-threading scheduler"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The `num_workers` argument is used to specify the number of threads or processes to use"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.compute(scheduler='threads', num_workers=4);"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Distributed Scheduler\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"Despite having \"distributed\" in it's name, the distributed scheduler works well on both single and multiple machines. Think of it as the \"advanced scheduler\".\n",
"\n",
"The Dask distributed cluster is composed of a single centralized scheduler and one or more worker processes. A `Client` object is used as the user-facing entry point to interact with the cluster.\n",
"\n",
"
"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import pandas as pd\n",
"import dask.dataframe as dd"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask DataFrames support many of the same data I/O methods as Pandas. For example, \n",
"\n",
"- `read_csv` \\ `to_csv`\n",
"- `read_hdf` \\ `to_hdf`\n",
"- `read_json` \\ `to_json`\n",
"- `read_parquet` \\ `to_parquet`"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Can specify a `chunksize` argument to set the number of rows per partition"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"ddf = dd.read_hdf('example_data.hdf', key='dataframe', chunksize=25)\n",
"ddf"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The `npartitions` attribute tells us how many Pandas DataFrames make up our Dask DataFrame"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"ddf.npartitions"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Like Dask arrays, Dask DataFrames are lazily evaluated. Here, the `dd.read_hdf` function wraps several calls to `pd.read_hdf`, once for each partition of the Dask DataFrame."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"ddf.visualize()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask DataFrames cover a well-used portion of the Pandas API:\n",
"\n",
"- Elementwise operations: `df.x` + `df.y`, `df * df`\n",
"\n",
"- Row-wise selections: `df[df.x > 0]`\n",
"\n",
"- Loc: `df.loc[4.0:10.5]`\n",
"\n",
"- Common aggregations: `df.x.max()`, `df.max()`\n",
"\n",
"- Is in: `df[df.x.isin([1, 2, 3])]`\n",
"\n",
"- Datetime/string accessors: `df.timestamp.month`\n",
"\n",
"- Froupby-aggregate (with common aggregations): `df.groupby(df.x).y.max()`, `df.groupby('x').max()`\n",
"\n",
"- ...\n",
"\n",
"See the [Dask DataFrame API docs](http://docs.dask.org/en/latest/dataframe-api.html) for full details about what portion of the Pandas API is implemented for Dask DataFrames."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"col_mean = ddf['col_1'].mean()\n",
"col_mean"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"col_mean.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"col_mean.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Dask Delayed\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"Sometimes problems don’t fit nicely into one of the high-level collections like Dask arrays or Dask DataFrames. In these cases, you can parallelize custom algorithms using the lower-level Dask `delayed` interface. This allows one to manually create task graphs with a light annotation of normal Python code."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import time\n",
"import random\n",
"\n",
"def inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1\n",
"\n",
"def double(x):\n",
" time.sleep(random.random())\n",
" return 2 * x\n",
" \n",
"def add(x, y):\n",
" time.sleep(random.random())\n",
" return x + y "
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"data = [1, 2, 3, 4]\n",
"\n",
"output = []\n",
"for x in data:\n",
" a = inc(x)\n",
" b = double(x)\n",
" c = add(a, b)\n",
" output.append(c)\n",
"\n",
"total = sum(output)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask `delayed` wraps function calls and delays their execution. `delayed` functions record what we want to compute (a function and input parameters) as a task in a graph that we’ll run later on parallel hardware by calling `compute`."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask import delayed"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"@delayed\n",
"def lazy_inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"lazy_inc"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"inc_output = lazy_inc(3) # lazily evaluate inc(3)\n",
"inc_output"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"inc_output.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Using `delayed` functions, we can build up a task graph for the particular computation we want to perform"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"double_inc_output = lazy_inc(inc_output)\n",
"double_inc_output"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"double_inc_output.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"double_inc_output.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can use `delayed` to make our previous example computation lazy by wrapping all the function calls with delayed"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import time\n",
"import random\n",
"\n",
"@delayed\n",
"def inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1\n",
"\n",
"@delayed\n",
"def double(x):\n",
" time.sleep(random.random())\n",
" return 2 * x\n",
"\n",
"@delayed\n",
"def add(x, y):\n",
" time.sleep(random.random())\n",
" return x + y"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"data = [1, 2, 3, 4]\n",
"\n",
"output = []\n",
"for x in data:\n",
" a = inc(x)\n",
" b = double(x)\n",
" c = add(a, b)\n",
" output.append(c)\n",
"\n",
"total = delayed(sum)(output)\n",
"total"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.visualize()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"total.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"I highly recommend checking out the [Dask delayed best practices](http://docs.dask.org/en/latest/delayed-best-practices.html) page to avoid some common pitfalls when using `delayed`. "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Schedulers\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"High-level collections like Dask arrays and Dask DataFrames, as well as the low-level `dask.delayed` interface build up task graphs for a computation. After these graphs are generated, they need to be executed (potentially in parallel). This is the job of a task scheduler. Different task schedulers exist within Dask. Each will consume a task graph and compute the same result, but with different performance characteristics. "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dask has two different classes of schedulers: single-machine schedulers and a distributed scheduler."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Single Machine Schedulers\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"Single machine schedulers provide basic features on a local process or thread pool and require no setup (only use the Python standard library). The different single machine schedulers Dask provides are:\n",
"\n",
"- `'threads'`: The threaded scheduler executes computations with a local `multiprocessing.pool.ThreadPool`. The threaded scheduler is the default choice for Dask arrays, Dask DataFrames, and Dask delayed. \n",
"\n",
"- `'processes'`: The multiprocessing scheduler executes computations with a local `multiprocessing.Pool`.\n",
"\n",
"- `'single-threaded'`: The single-threaded synchronous scheduler executes all computations in the local thread, with no parallelism at all. This is particularly valuable for debugging and profiling, which are more difficult when using threads or processes."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"You can configure which scheduler is used in a few different ways. You can set the scheduler globally by using the `dask.config.set(scheduler=)` command"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import dask\n",
"\n",
"dask.config.set(scheduler='threads')\n",
"total.compute(); # Will use the multi-threading scheduler"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"or use it as a context manager to set the scheduler for a block of code"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"with dask.config.set(scheduler='processes'):\n",
" total.compute() # Will use the multi-processing scheduler"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"or even within a single compute call"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.compute(scheduler='threads'); # Will use the multi-threading scheduler"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The `num_workers` argument is used to specify the number of threads or processes to use"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.compute(scheduler='threads', num_workers=4);"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Distributed Scheduler\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"Despite having \"distributed\" in it's name, the distributed scheduler works well on both single and multiple machines. Think of it as the \"advanced scheduler\".\n",
"\n",
"The Dask distributed cluster is composed of a single centralized scheduler and one or more worker processes. A `Client` object is used as the user-facing entry point to interact with the cluster.\n",
"\n",
" \n",
" \n",
"\n",
"Deploying a remote Dask cluster involves some [additional setup](https://distributed.dask.org/en/latest/setup.html). There are several projects for easily deploying a Dask cluster on commonly used computing resources:\n",
"\n",
"- [Dask-Kubernetes](https://kubernetes.dask.org/en/latest/) for deploying Dask using native Kubernetes APIs\n",
"- [Dask-Yarn](https://yarn.dask.org/en/latest/) for deploying Dask on YARN clusters\n",
"- [Dask-MPI](http://mpi.dask.org/en/latest/) for deploying Dask on existing MPI environments\n",
"- [Dask-Jobqueue](https://jobqueue.dask.org/en/latest/) for deploying Dask on job queuing systems (e.g. PBS, Slurm, etc.)\n",
"\n",
"\n",
"Setting up the distributed scheduler locally just involves creating a `Client` object, which lets you interact with the \"cluster\" (local threads or processes on your machine)."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask.distributed import Client"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"client = Client(threads_per_worker=4, n_workers=1)\n",
"client"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Note**: when we create a distributed scheduler `Client`, by default it registers itself as the default Dask scheduler. All `.compute()` calls will automatically start using the distributed scheduler unless otherwise told to use a different scheduler. \n",
"\n",
"The distributed scheduler has many features:\n",
"\n",
"- [Sophisticated memory management](https://distributed.dask.org/en/latest/memory.html)\n",
"\n",
"- [Data locality](https://distributed.dask.org/en/latest/locality.html)\n",
"\n",
"- [Adaptive deployments](https://distributed.dask.org/en/latest/adaptive.html)\n",
"\n",
"- [Cluster resilience](https://distributed.dask.org/en/latest/resilience.html)\n",
"\n",
"- ...\n",
"\n",
"See the [Dask distributed documentation](https://distributed.dask.org) for full details about all the distributed scheduler features.\n",
"\n",
"For this talk, I'd like to highlight two of the distributed scheduler features: real time diagnostics and the futures interface."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = da.ones((20_000, 20_000), chunks=(400, 400))\n",
"result = (x + x.T).sum(axis=0).mean()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Futures interface\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"The Dask distributed scheduler implements a superset of Python's [`concurrent.futures`](https://docs.python.org/3/library/concurrent.futures.html) interface that allows for finer control and asynchronous computation."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import time\n",
"import random\n",
"\n",
"def inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1\n",
"\n",
"def double(x):\n",
" time.sleep(random.random())\n",
" return 2 * x\n",
" \n",
"def add(x, y):\n",
" time.sleep(random.random())\n",
" return x + y "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can run these functions locally"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"inc(1)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Or we can submit them to run remotely on a Dask worker node"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"future = client.submit(inc, 1)\n",
"future"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The `submit` function sends a function and arguments to the distributed scheduler for processing. It returns a `Future` object that refer to remote data on the cluster. The `Future` returns immediately while the computations run remotely in the background. There is no blocking of the local Python session."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"If you wait a moment, and then check on the future again, you'll see that it has finished."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"future"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Once the computation for a `Future` is complete, you can retrieve the result using the `.result()` method"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"future.result()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### Specifying task dependencies\n",
"\n",
"Much like the `delayed` interface, we can submit tasks based on other futures. This will create a dependency between the inputs and outputs. Dask will track the execution of all tasks and ensure that downstream tasks are run at the proper time and place and with the proper data."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = client.submit(inc, 1)\n",
"y = client.submit(double, 2)\n",
"z = client.submit(add, x, y)\n",
"z"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"z.result()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"As an example, we can submit many tasks that depend on each other in a `for`-loop"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"zs = []\n",
"\n",
"for i in range(64):\n",
" x = client.submit(inc, i) # x = inc(i)\n",
" y = client.submit(double, x) # y = inc(x)\n",
" z = client.submit(add, x, y) # z = inc(y)\n",
" zs.append(z)\n",
"\n",
"total = client.submit(sum, zs)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.result()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### Custom computation: Tree summation\n",
"\n",
"As an example of a non-trivial algorithm, consider the classic tree reduction. We accomplish this with a nested `for`-loop and a bit of normal Python logic.\n",
"\n",
"```\n",
"finish total single output\n",
" ^ / \\\n",
" | c1 c2 neighbors merge\n",
" | / \\ / \\\n",
" | b1 b2 b3 b4 neighbors merge\n",
" ^ / \\ / \\ / \\ / \\\n",
"start a1 a2 a3 a4 a5 a6 a7 a8 many inputs\n",
"```"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can easily scale our distributed cluster up or down depending on our needs"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"client.cluster.scale(3) # ask for 3 4-thread workers"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"L = zs\n",
"while len(L) > 1:\n",
" new_L = []\n",
" for i in range(0, len(L), 2):\n",
" future = client.submit(add, L[i], L[i + 1]) # add neighbors\n",
" new_L.append(future)\n",
" L = new_L # swap old list for new"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"del future, L, new_L, total"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### Building a computation dynamically\n",
"\n",
"We can even dynamically submit tasks based on the output of other tasks. This gives more flexibility in situations where the computations may evolve over time.\n",
"\n",
"For this, we can use operations like [`as_completed()`](https://distributed.dask.org/en/latest/api.html#distributed.as_completed), which returns futures in the order in which they complete."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask.distributed import as_completed\n",
"\n",
"zs = client.map(inc, zs)\n",
"seq = as_completed(zs)\n",
"\n",
"while seq.count() > 2: # at least two futures left\n",
" a = next(seq)\n",
" b = next(seq)\n",
" new = client.submit(add, a, b) # add them together\n",
" seq.add(new) # add new future back into loop"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This was a brief demo of the distributed scheduler. It's has lots of other cool features not touched on here. For more information, check out the [Distributed documentation](https://distributed.dask.org). "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Scalable Machine Learning with Dask-ML\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"[Dask-ML](http://ml.dask.org/) is a Python library for scalable machine learning in Python. Build on top of Dask collections and supports the scikit-learn API. \n",
"\n",
"Three different approaches are taken to scaling modern machine learning algorithms:\n",
"\n",
"- Parallelize scikit-learn directly\n",
"\n",
"- Reimplement scalable algorithms with Dask arrays\n",
"\n",
"- Partner with other distributed libraries (like XGBoost and TensorFlow)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask_ml.datasets import make_classification\n",
"from dask_ml.model_selection import train_test_split\n",
"from dask_ml.metrics import accuracy_score\n",
"from dask_ml.linear_model import LogisticRegression"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X, y = make_classification(n_samples=1000,\n",
" n_features=2,\n",
" n_classes=2,\n",
" random_state=2,\n",
" chunks=10)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X_train, X_test, y_train, y_test = train_test_split(X, y,\n",
" test_size=0.3,\n",
" random_state=2)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"clf = LogisticRegression(max_iter=2)\n",
"clf.fit(X_train, y_train)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"y_pred = clf.predict(X_test)\n",
"accuracy_score(y_test, y_pred)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Additional Resources\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"- Dask links:\n",
"\n",
" - GitHub repository: https://github.com/dask/dask\n",
"\n",
" - Documentation: https://docs.dask.org\n",
"\n",
" - Dask examples repository: https://github.com/dask/dask-examples\n",
"\n",
"- There are lots of great Dask tutorial from various conference on YouTube. For example:\n",
"\n",
" - \"Parallelizing Scientific Python with Dask\" @ SciPy 2018: [YouTube](https://www.youtube.com/watch?v=mqdglv9GnM8)\n",
" \n",
" - \"Scalable Machine Learning with Dask\" @ SciPy 2018: [YouTube](https://www.youtube.com/watch?v=ccfsbuqsjgI)\n",
"\n",
"- If you have a Dask usage questions, please ask it on [Stack Overflow with the #dask tag](https://stackoverflow.com/questions/tagged/dask). Dask developers monitor this tag and will answer questions.\n",
"\n",
"- If you run into a bug, feel free to file a report on the [Dask GitHub issue tracker](https://github.com/dask/dask/issues)."
]
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.3"
}
},
"nbformat": 4,
"nbformat_minor": 4
}
\n",
" \n",
"\n",
"Deploying a remote Dask cluster involves some [additional setup](https://distributed.dask.org/en/latest/setup.html). There are several projects for easily deploying a Dask cluster on commonly used computing resources:\n",
"\n",
"- [Dask-Kubernetes](https://kubernetes.dask.org/en/latest/) for deploying Dask using native Kubernetes APIs\n",
"- [Dask-Yarn](https://yarn.dask.org/en/latest/) for deploying Dask on YARN clusters\n",
"- [Dask-MPI](http://mpi.dask.org/en/latest/) for deploying Dask on existing MPI environments\n",
"- [Dask-Jobqueue](https://jobqueue.dask.org/en/latest/) for deploying Dask on job queuing systems (e.g. PBS, Slurm, etc.)\n",
"\n",
"\n",
"Setting up the distributed scheduler locally just involves creating a `Client` object, which lets you interact with the \"cluster\" (local threads or processes on your machine)."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask.distributed import Client"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"client = Client(threads_per_worker=4, n_workers=1)\n",
"client"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Note**: when we create a distributed scheduler `Client`, by default it registers itself as the default Dask scheduler. All `.compute()` calls will automatically start using the distributed scheduler unless otherwise told to use a different scheduler. \n",
"\n",
"The distributed scheduler has many features:\n",
"\n",
"- [Sophisticated memory management](https://distributed.dask.org/en/latest/memory.html)\n",
"\n",
"- [Data locality](https://distributed.dask.org/en/latest/locality.html)\n",
"\n",
"- [Adaptive deployments](https://distributed.dask.org/en/latest/adaptive.html)\n",
"\n",
"- [Cluster resilience](https://distributed.dask.org/en/latest/resilience.html)\n",
"\n",
"- ...\n",
"\n",
"See the [Dask distributed documentation](https://distributed.dask.org) for full details about all the distributed scheduler features.\n",
"\n",
"For this talk, I'd like to highlight two of the distributed scheduler features: real time diagnostics and the futures interface."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = da.ones((20_000, 20_000), chunks=(400, 400))\n",
"result = (x + x.T).sum(axis=0).mean()"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"result.compute()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Futures interface\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"The Dask distributed scheduler implements a superset of Python's [`concurrent.futures`](https://docs.python.org/3/library/concurrent.futures.html) interface that allows for finer control and asynchronous computation."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"import time\n",
"import random\n",
"\n",
"def inc(x):\n",
" time.sleep(random.random())\n",
" return x + 1\n",
"\n",
"def double(x):\n",
" time.sleep(random.random())\n",
" return 2 * x\n",
" \n",
"def add(x, y):\n",
" time.sleep(random.random())\n",
" return x + y "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can run these functions locally"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"inc(1)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Or we can submit them to run remotely on a Dask worker node"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"future = client.submit(inc, 1)\n",
"future"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"The `submit` function sends a function and arguments to the distributed scheduler for processing. It returns a `Future` object that refer to remote data on the cluster. The `Future` returns immediately while the computations run remotely in the background. There is no blocking of the local Python session."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"If you wait a moment, and then check on the future again, you'll see that it has finished."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"future"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Once the computation for a `Future` is complete, you can retrieve the result using the `.result()` method"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"future.result()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### Specifying task dependencies\n",
"\n",
"Much like the `delayed` interface, we can submit tasks based on other futures. This will create a dependency between the inputs and outputs. Dask will track the execution of all tasks and ensure that downstream tasks are run at the proper time and place and with the proper data."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"x = client.submit(inc, 1)\n",
"y = client.submit(double, 2)\n",
"z = client.submit(add, x, y)\n",
"z"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"z.result()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"As an example, we can submit many tasks that depend on each other in a `for`-loop"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"%%time\n",
"\n",
"zs = []\n",
"\n",
"for i in range(64):\n",
" x = client.submit(inc, i) # x = inc(i)\n",
" y = client.submit(double, x) # y = inc(x)\n",
" z = client.submit(add, x, y) # z = inc(y)\n",
" zs.append(z)\n",
"\n",
"total = client.submit(sum, zs)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"total.result()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### Custom computation: Tree summation\n",
"\n",
"As an example of a non-trivial algorithm, consider the classic tree reduction. We accomplish this with a nested `for`-loop and a bit of normal Python logic.\n",
"\n",
"```\n",
"finish total single output\n",
" ^ / \\\n",
" | c1 c2 neighbors merge\n",
" | / \\ / \\\n",
" | b1 b2 b3 b4 neighbors merge\n",
" ^ / \\ / \\ / \\ / \\\n",
"start a1 a2 a3 a4 a5 a6 a7 a8 many inputs\n",
"```"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"We can easily scale our distributed cluster up or down depending on our needs"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"client.cluster.scale(3) # ask for 3 4-thread workers"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"L = zs\n",
"while len(L) > 1:\n",
" new_L = []\n",
" for i in range(0, len(L), 2):\n",
" future = client.submit(add, L[i], L[i + 1]) # add neighbors\n",
" new_L.append(future)\n",
" L = new_L # swap old list for new"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"del future, L, new_L, total"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"#### Building a computation dynamically\n",
"\n",
"We can even dynamically submit tasks based on the output of other tasks. This gives more flexibility in situations where the computations may evolve over time.\n",
"\n",
"For this, we can use operations like [`as_completed()`](https://distributed.dask.org/en/latest/api.html#distributed.as_completed), which returns futures in the order in which they complete."
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask.distributed import as_completed\n",
"\n",
"zs = client.map(inc, zs)\n",
"seq = as_completed(zs)\n",
"\n",
"while seq.count() > 2: # at least two futures left\n",
" a = next(seq)\n",
" b = next(seq)\n",
" new = client.submit(add, a, b) # add them together\n",
" seq.add(new) # add new future back into loop"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"This was a brief demo of the distributed scheduler. It's has lots of other cool features not touched on here. For more information, check out the [Distributed documentation](https://distributed.dask.org). "
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Scalable Machine Learning with Dask-ML\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"[Dask-ML](http://ml.dask.org/) is a Python library for scalable machine learning in Python. Build on top of Dask collections and supports the scikit-learn API. \n",
"\n",
"Three different approaches are taken to scaling modern machine learning algorithms:\n",
"\n",
"- Parallelize scikit-learn directly\n",
"\n",
"- Reimplement scalable algorithms with Dask arrays\n",
"\n",
"- Partner with other distributed libraries (like XGBoost and TensorFlow)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from dask_ml.datasets import make_classification\n",
"from dask_ml.model_selection import train_test_split\n",
"from dask_ml.metrics import accuracy_score\n",
"from dask_ml.linear_model import LogisticRegression"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X, y = make_classification(n_samples=1000,\n",
" n_features=2,\n",
" n_classes=2,\n",
" random_state=2,\n",
" chunks=10)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"X_train, X_test, y_train, y_test = train_test_split(X, y,\n",
" test_size=0.3,\n",
" random_state=2)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"clf = LogisticRegression(max_iter=2)\n",
"clf.fit(X_train, y_train)"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"y_pred = clf.predict(X_test)\n",
"accuracy_score(y_test, y_pred)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Additional Resources\n",
"\n",
"[ [Back to top](#Outline) ]\n",
"\n",
"- Dask links:\n",
"\n",
" - GitHub repository: https://github.com/dask/dask\n",
"\n",
" - Documentation: https://docs.dask.org\n",
"\n",
" - Dask examples repository: https://github.com/dask/dask-examples\n",
"\n",
"- There are lots of great Dask tutorial from various conference on YouTube. For example:\n",
"\n",
" - \"Parallelizing Scientific Python with Dask\" @ SciPy 2018: [YouTube](https://www.youtube.com/watch?v=mqdglv9GnM8)\n",
" \n",
" - \"Scalable Machine Learning with Dask\" @ SciPy 2018: [YouTube](https://www.youtube.com/watch?v=ccfsbuqsjgI)\n",
"\n",
"- If you have a Dask usage questions, please ask it on [Stack Overflow with the #dask tag](https://stackoverflow.com/questions/tagged/dask). Dask developers monitor this tag and will answer questions.\n",
"\n",
"- If you run into a bug, feel free to file a report on the [Dask GitHub issue tracker](https://github.com/dask/dask/issues)."
]
}
],
"metadata": {
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.7.3"
}
},
"nbformat": 4,
"nbformat_minor": 4
}