Adjust p-values produced by geom_pwc() on a ggplot.
This is mainly useful when using facet, where p-values are generally
computed and adjusted by panel without taking into account the other panels.
In this case, one might want to adjust after the p-values of all panels together.
Arguments
- p
a ggplot
- layer
An integer indicating the statistical layer rank in the ggplot (in the order added to the plot).
- p.adjust.method
method for adjusting p values (see
p.adjust). Has impact only in a situation, where multiple pairwise tests are performed; or when there are multiple grouping variables. Ignored when the specified method is"tukey_hsd"or"games_howell_test"because they come with internal p adjustment method. Allowed values include "holm", "hochberg", "hommel", "bonferroni", "BH", "BY", "fdr", "none". If you don't want to adjust the p value (not recommended), use p.adjust.method = "none".- label
character string specifying label. Can be:
the column containing the label (e.g.:
label = "p"orlabel = "p.adj"), wherepis the p-value. Other possible values are"p.signif", "p.adj.signif", "p.format", "p.adj.format".an expression that can be formatted by the
glue()package. For example, when specifyinglabel = "Wilcoxon, p = \{p\}", the expression {p} will be replaced by its value.a combination of plotmath expressions and glue expressions. You may want some of the statistical parameter in italic; for example:
label = "Wilcoxon, italic(p)= {p}"
.
- hide.ns
can be logical value (
TRUEorFALSE) or a character vector ("p.adj"or"p").- symnum.args
a list of arguments to pass to the function
symnumfor symbolic number coding of p-values. For example,symnum.args = list(cutpoints = c(0, 0.0001, 0.001, 0.01, 0.05, Inf), symbols = c("****", "***", "**", "*", "ns")).In other words, we use the following convention for symbols indicating statistical significance:
ns: p > 0.05*: p <= 0.05**: p <= 0.01***: p <= 0.001****: p <= 0.0001
- output
character. Possible values are one of
c("plot", "stat_test"). Default is "plot".
Examples
# Data preparation
#:::::::::::::::::::::::::::::::::::::::
df <- ToothGrowth
df$dose <- as.factor(df$dose)
# Add a random grouping variable
df$group <- factor(rep(c("grp1", "grp2"), 30))
head(df, 3)
#> len supp dose group
#> 1 4.2 VC 0.5 grp1
#> 2 11.5 VC 0.5 grp2
#> 3 7.3 VC 0.5 grp1
# Boxplot: Two groups by panel
#:::::::::::::::::::::::::::::::::::::::
# Create a box plot
bxp <- ggboxplot(
df, x = "supp", y = "len", fill = "#00AFBB",
facet.by = "dose"
)
# Make facet and add p-values
bxp <- bxp + geom_pwc(method = "t_test")
bxp
 # Adjust all p-values together after
ggadjust_pvalue(
bxp, p.adjust.method = "bonferroni",
label = "{p.adj.format}{p.adj.signif}", hide.ns = TRUE
)
# Adjust all p-values together after
ggadjust_pvalue(
bxp, p.adjust.method = "bonferroni",
label = "{p.adj.format}{p.adj.signif}", hide.ns = TRUE
)
 # Boxplot: Three groups by panel
#:::::::::::::::::::::::::::::::::::::::
# Create a box plot
bxp <- ggboxplot(
df, x = "dose", y = "len", fill = "#00AFBB",
facet.by = "supp"
)
# Make facet and add p-values
bxp <- bxp + geom_pwc(method = "t_test")
bxp
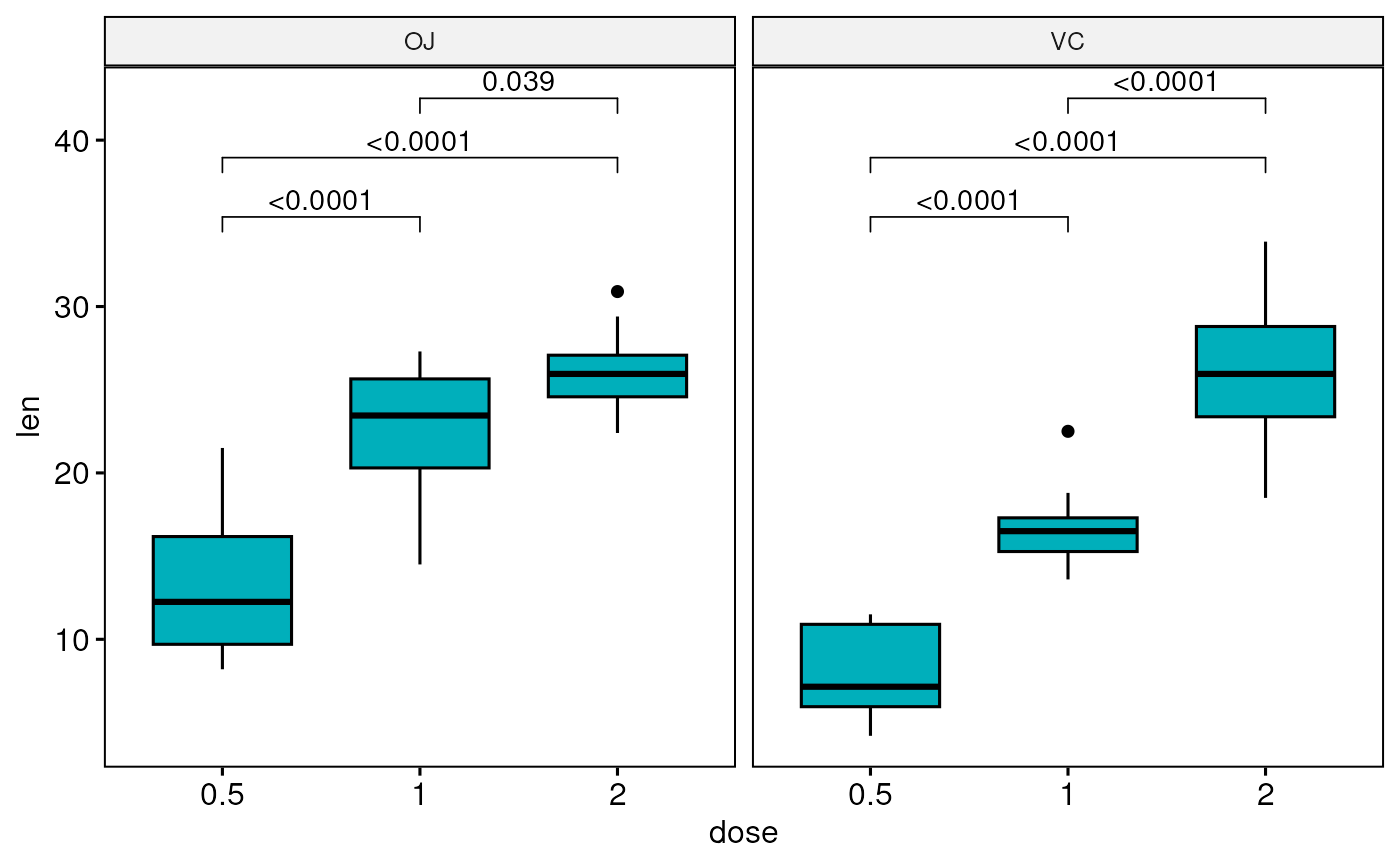
# Boxplot: Three groups by panel
#:::::::::::::::::::::::::::::::::::::::
# Create a box plot
bxp <- ggboxplot(
df, x = "dose", y = "len", fill = "#00AFBB",
facet.by = "supp"
)
# Make facet and add p-values
bxp <- bxp + geom_pwc(method = "t_test")
bxp
 # Adjust all p-values together after
ggadjust_pvalue(
bxp, p.adjust.method = "bonferroni",
label = "{p.adj.format}{p.adj.signif}"
)
# Adjust all p-values together after
ggadjust_pvalue(
bxp, p.adjust.method = "bonferroni",
label = "{p.adj.format}{p.adj.signif}"
)
