Comparison of polygenic scoring methods
1 Introduction
In this study we compare a range of polygenic scoring approaches within a reference standardised framework, whereby the polygenic scores are derived using a previously defined set of variants, and LD and minor allele frequency (MAF) are estimated using an ancestry matched reference genotype data sample. This ensures the performance of the methods will hold true when implementing them within a clinical context, which will require reference-standardisation of polygenic scoring.
2 Aims
- Compare the predictive utility of polygenic scores derived using a range of methods.
- Compare predictive utility of models including multiple PRSs based on difference parameters, to those using a single PRS selected using 10-fold cross-validation or pseudo-validation.
3 Methods
3.1 Samples
- UK Biobank
- TEDS
3.2 Outcomes
- UK Biobank
- Depression (binary)
- Intelligence (continuous)
- Body mass index (BMI - continuous)
- Height (continuous)
- Coronary Artery Disease (CAD - Binary)
- Type II Diabetes (T2D - Binary)
- Inflammatory Bowel Disorder (IBD - Binary)
- Rheumatoid arthritis (RheuArth - Binary)
- Multiple Slerosis (MultiScler - Binary)
- Prostate Cancer (Binary)
- Breast Cancer (Binary)
- TEDS
- ADHD traits (continuous)
- Height (continuous)
- Body mass index (BMI - continuous)
- GCSE scores (continuous)
3.3 Genotypic data
Both target sample underwent stringent quality control prior to imputation using the HRC-reference. After imputation, genotypes were converted to PLINK hard-calls, and only HapMap3 SNPs were retained. Eureopean individuals within the target samples were identified and retained if they were within the 3SD of the 1KG European mean of the first 100 principal components.
3.4 GWAS summary statistics
For each target phenotype, the largest independent phenotype-matched GWAS was selected for calculating polygenic scores. More information can be found in the table below.
Preparing GWAS sumstats table for UK Biobank phenotypes
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Phenotype_prep.config')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
library(data.table)
pheno<-fread('/users/k1806347/brc_scratch/Data/GWAS_sumstats/QC_sumstats_list_270121.csv')
ukb_pheno=c('Depression','Intelligence','BMI','Height','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
ukb_gwas=c('DEPR06','COLL01','BODY04','HEIG03','DIAB05','COAD01','CROH01','SCLE03','RHEU02','BRCA01','PRCA01')
ukb_prev=c(0.15,NA,NA,NA,0.05,0.03,0.013,0.00164,0.005,0.125,0.125)
ukb_dat<-data.frame(pheno=ukb_pheno,gwas=ukb_gwas,prev=ukb_prev)
pheno_ukb<-pheno[(pheno$code %in% ukb_gwas),]
pheno_ukb<-pheno_ukb[,c('code','phenotype_name','reference_year','pmid','n_cases','n_controls','n_total')]
names(pheno_ukb)<-c('code','trait','year','PMID','Ncases','Ncontrols','sample_size_discovery')
ldsc_res<-NULL
for(i in ukb_gwas){
tmp<-read.table(paste0(gwas_rep_munged,'/',i,'_herit.log'), sep='&')
}
pheno_ukb$ldsc_h2_observed<-NA
pheno_ukb$ldsc_h2_se<-NA
pheno_ukb$ldsc_lambda_GC<-NA
pheno_ukb$ldsc_intercept<-NA
pheno_ukb$ldsc_intercept_se<-NA
ldsc_logs<-list.files(path='/scratch/groups/ukbiobank/sumstats/munged', pattern='_herit.log')
ldsc_logs<-gsub('_herit.log','',ldsc_logs)
for(i in 1:dim(pheno_ukb)[1]){
if((pheno_ukb$code[i] %in% ldsc_logs)){
ldsc_logs_i<-read.fwf(paste0(gwas_rep_munged,'/',pheno_ukb$code[i],'_herit.log'),widths = 1000000)
pheno_ukb$ldsc_h2_observed[i]<-as.numeric(gsub(' .*','', gsub('Total Observed scale h2: ','',ldsc_logs_i[grepl('Total Observed scale h2: ', ldsc_logs_i$V1),])))
pheno_ukb$ldsc_h2_se[i]<-as.numeric(gsub("\\)",'', gsub(".*\\(",'', gsub('Total Observed scale h2: ','',ldsc_logs_i[grepl('Total Observed scale h2: ', ldsc_logs_i$V1),]))))
pheno_ukb$ldsc_lambda_GC[i]<-as.numeric(gsub('Lambda GC: ','',ldsc_logs_i[grepl('Lambda GC: ', ldsc_logs_i$V1),]))
pheno_ukb$ldsc_intercept[i]<-as.numeric(gsub(' .*','', gsub("Intercept: ",'',ldsc_logs_i[grepl("Intercept: ", ldsc_logs_i$V1),])))
pheno_ukb$ldsc_intercept_se[i]<-as.numeric(gsub("\\)",'', gsub(".*\\(",'', gsub("Intercept: ",'',ldsc_logs_i[grepl("Intercept: ", ldsc_logs_i$V1),]))))
}
}
pheno_ukb$pop_prev<-ukb_dat$prev[match(pheno_ukb$code, ukb_dat$gwas)]
pheno_ukb$Target_Phenotype<-ukb_dat$pheno[match(pheno_ukb$code, ukb_dat$gwas)]
pheno_ukb$Ncases<-as.numeric(gsub(',','',pheno_ukb$Ncases))
pheno_ukb$Ncontrols<-as.numeric(gsub(',','',pheno_ukb$Ncontrols))
pheno_ukb$samp_prev<-pheno_ukb$Ncases/(pheno_ukb$Ncases+pheno_ukb$Ncontrols)
h2l_R2 <- function(k, r2, p) {
# K baseline disease risk
# r2 from a linear regression model attributable to genomic profile risk score
# P proportion of sample that are cases
# calculates proportion of variance explained on the liability scale
#from ABC at http://www.complextraitgenomics.com/software/
#Lee SH, Goddard ME, Wray NR, Visscher PM. (2012) A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012 Apr;36(3):214-24.
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
h2l_R2 = C*r2 / (1 + C*theta*r2)
}
se_h2l_R2 <- function(k,h2,se, p) {
# K baseline disease risk
# r2 from a linear regression model attributable to genomic profile risk score
# P proportion of sample that are cases
# calculates proportion of variance explained on the liability scale
#from ABC at http://www.complextraitgenomics.com/software/
#Lee SH, Goddard ME, Wray NR, Visscher PM. (2012) A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012 Apr;36(3):214-24.
#SE on the liability (From a Taylor series expansion)
#var(h2l_r2) = [d(h2l_r2)/d(R2v)]^2*var(R2v) with d being calculus differentiation
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
se_h2l_R2 = C*(1-h2*theta)*se
}
pheno_ukb$ldsc_h2_observed<-as.numeric(pheno_ukb$ldsc_h2_observed)
pheno_ukb$ldsc_h2_se<-as.numeric(pheno_ukb$ldsc_h2_se)
pheno_ukb$h2_liab<-round(h2l_R2(k=pheno_ukb$pop_prev, r2=pheno_ukb$ldsc_h2_observed, p=pheno_ukb$samp_prev),3)
pheno_ukb$h2_liab_se<-round(se_h2l_R2(k=pheno_ukb$pop_prev, h2=pheno_ukb$ldsc_h2_observed, se=pheno_ukb$ldsc_h2_se, p=pheno_ukb$samp_prev),3)
pheno_ukb$ldsc_h2_observed<-paste0(pheno_ukb$ldsc_h2_observed," (", pheno_ukb$ldsc_h2_se,")")
pheno_ukb$ldsc_h2_se<-NULL
pheno_ukb$h2_liab[!is.na(pheno_ukb$Ncases)]<-paste0(pheno_ukb$h2_liab[!is.na(pheno_ukb$Ncases)]," (", pheno_ukb$h2_liab_se[!is.na(pheno_ukb$Ncases)],")")
pheno_ukb$h2_liab_se<-NULL
pheno_ukb$ldsc_intercept<-paste0(pheno_ukb$ldsc_intercept," (", pheno_ukb$ldsc_intercept_se,")")
pheno_ukb$ldsc_intercept_se<-NULL
pheno_ukb$Trait<-c('BMI','CAD','Intelligence',"Crohn's Disease",'Major Depression','T2D','Height','RheuArth','MultiScler','Prostate Cancer','Breast Cancer')
pheno_ukb<-pheno_ukb[match(ukb_dat$gwas,pheno_ukb$code),]
pheno_ukb<-pheno_ukb[,c('Trait','code','trait','year','PMID','Ncases','Ncontrols','sample_size_discovery','ldsc_h2_observed','h2_liab','ldsc_intercept','ldsc_lambda_GC')]
names(pheno_ukb)<-c('Target Phenotype','Code','GWAS Phenotype','Year','PMID','Ncase','Ncontrol','N','h2_obs','h2_liab','Intercept','Lambda')
write.csv(pheno_ukb, '/users/k1806347/brc_scratch/Data/GWAS_sumstats/UKBB_phenotype_GWAS_descrip.csv', row.names=F, quote=F)Show GWAS for UK Biobank phenotypes
| Target Phenotype | Code | GWAS Phenotype | Year | PMID | Ncase | Ncontrol | N | h2-obs (SE) | h2-liab (SE) | Intercept | Lambda |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Major Depression | DEPR06 | major depressive disorder | 2018 | 29700475 | 116404 | 314990 | 431394 | 0.0551 (0.0023) | 0.085 (0.004) | 1.0169 (0.0088) | 1.3894 |
| Intelligence | COLL01 | college completion | 2013 | 23722424 | NA | NA | 95427 | 0.1048 (0.0075) | NA | 1.0214 (0.0086) | 1.1940 |
| BMI | BODY04 | BMI | 2015 | 25673413 | NA | NA | 322154 | 0.1297 (0.0056) | NA | 0.6729 (0.0076) | 1.0772 |
| Height | HEIG03 | height | 2014 | 25282103 | NA | NA | 253288 | 0.312 (0.0141) | NA | 1.3254 (0.0185) | 2.0007 |
| T2D | DIAB05 | diabetes type 2 | 2017 | 28566273 | 26676 | 132532 | 159208 | 0.0781 (0.0053) | 0.124 (0.008) | 0.9984 (0.0083) | 1.1459 |
| CAD | COAD01 | coronary artery disease | 2015 | 26343387 | 60801 | 123504 | 184305 | 0.0661 (0.0045) | 0.057 (0.004) | 0.8875 (0.0074) | 1.0466 |
| Crohn’s Disease | CROH01 | crohn disease | 2015 | 26192919 | 5956 | 14927 | 20883 | 0.4751 (0.0538) | 0.542 (0.059) | 1.0316 (0.0092) | 1.1396 |
| MultiScler | SCLE03 | multiple sclerosis | 2011 | 21833088 | 9772 | 17376 | 27148 | 0.0493 (0.0262) | 0.021 (0.012) | 1.0599 (0.0085) | 1.0375 |
| RheuArth | RHEU02 | rheumatoid arthritis | 2014 | 24390342 | 14361 | 43923 | 58284 | 0.1406 (0.0176) | 0.102 (0.013) | 0.9511 (0.0076) | 1.0466 |
| Breast Cancer | BRCA01 | breast cancer | 2017 | 29059683 | 122977 | 105974 | 228951 | 0.1326 (0.0108) | 0.157 (0.013) | 1.1001 (0.0125) | 1.3581 |
| Prostate Cancer | PRCA01 | prostate cancer | 2017 | 29892016 | 79148 | 61106 | 140254 | 0.1635 (0.0238) | 0.198 (0.029) | 1.0804 (0.0153) | 1.2201 |
Preparing GWAS sumstats table for TEDS phenotypes
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Phenotype_prep.config')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
library(data.table)
pheno<-fread('/users/k1806347/brc_scratch/Data/GWAS_sumstats/QC_sumstats_list_270121.csv')
teds_pheno=c('Height21', 'BMI21', 'GCSE', 'ADHD')
teds_gwas=c('HEIG03', 'BODY11', 'EDUC03', 'ADHD04')
teds_prev=c(NA,NA,NA,0.05)
teds_dat<-data.frame(pheno=teds_pheno,gwas=teds_gwas,prev=teds_prev)
pheno_teds<-pheno[(pheno$code %in% teds_gwas),]
pheno_teds<-pheno_teds[,c('code','phenotype_name','reference_year','pmid','n_cases','n_controls','n_total')]
names(pheno_teds)<-c('code','trait','year','PMID','Ncases','Ncontrols','sample_size_discovery')
ldsc_res<-NULL
for(i in teds_gwas){
tmp<-read.table(paste0(gwas_rep_munged,'/',i,'_herit.log'), sep='&')
}
pheno_teds$ldsc_h2_observed<-NA
pheno_teds$ldsc_h2_se<-NA
pheno_teds$ldsc_lambda_GC<-NA
pheno_teds$ldsc_intercept<-NA
pheno_teds$ldsc_intercept_se<-NA
ldsc_logs<-list.files(path='/scratch/groups/ukbiobank/sumstats/munged', pattern='_herit.log')
ldsc_logs<-gsub('_herit.log','',ldsc_logs)
for(i in 1:dim(pheno_teds)[1]){
if((pheno_teds$code[i] %in% ldsc_logs)){
ldsc_logs_i<-read.fwf(paste0(gwas_rep_munged,'/',pheno_teds$code[i],'_herit.log'),widths = 1000000)
pheno_teds$ldsc_h2_observed[i]<-as.numeric(gsub(' .*','', gsub('Total Observed scale h2: ','',ldsc_logs_i[grepl('Total Observed scale h2: ', ldsc_logs_i$V1),])))
pheno_teds$ldsc_h2_se[i]<-as.numeric(gsub("\\)",'', gsub(".*\\(",'', gsub('Total Observed scale h2: ','',ldsc_logs_i[grepl('Total Observed scale h2: ', ldsc_logs_i$V1),]))))
pheno_teds$ldsc_lambda_GC[i]<-as.numeric(gsub('Lambda GC: ','',ldsc_logs_i[grepl('Lambda GC: ', ldsc_logs_i$V1),]))
pheno_teds$ldsc_intercept[i]<-as.numeric(gsub(' .*','', gsub("Intercept: ",'',ldsc_logs_i[grepl("Intercept: ", ldsc_logs_i$V1),])))
pheno_teds$ldsc_intercept_se[i]<-as.numeric(gsub("\\)",'', gsub(".*\\(",'', gsub("Intercept: ",'',ldsc_logs_i[grepl("Intercept: ", ldsc_logs_i$V1),]))))
}
}
pheno_teds$pop_prev<-teds_dat$prev[match(pheno_teds$code, teds_dat$gwas)]
pheno_teds$Target_Phenotype<-teds_dat$pheno[match(pheno_teds$code, teds_dat$gwas)]
pheno_teds$Ncases<-as.numeric(gsub(',','',pheno_teds$Ncases))
pheno_teds$Ncontrols<-as.numeric(gsub(',','',pheno_teds$Ncontrols))
pheno_teds$samp_prev<-pheno_teds$Ncases/(pheno_teds$Ncases+pheno_teds$Ncontrols)
h2l_R2 <- function(k, r2, p) {
# K baseline disease risk
# r2 from a linear regression model attributable to genomic profile risk score
# P proportion of sample that are cases
# calculates proportion of variance explained on the liability scale
#from ABC at http://www.complextraitgenomics.com/software/
#Lee SH, Goddard ME, Wray NR, Visscher PM. (2012) A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012 Apr;36(3):214-24.
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
h2l_R2 = C*r2 / (1 + C*theta*r2)
}
se_h2l_R2 <- function(k,h2,se, p) {
# K baseline disease risk
# r2 from a linear regression model attributable to genomic profile risk score
# P proportion of sample that are cases
# calculates proportion of variance explained on the liability scale
#from ABC at http://www.complextraitgenomics.com/software/
#Lee SH, Goddard ME, Wray NR, Visscher PM. (2012) A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012 Apr;36(3):214-24.
#SE on the liability (From a Taylor series expansion)
#var(h2l_r2) = [d(h2l_r2)/d(R2v)]^2*var(R2v) with d being calculus differentiation
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
se_h2l_R2 = C*(1-h2*theta)*se
}
pheno_teds$ldsc_h2_observed<-as.numeric(pheno_teds$ldsc_h2_observed)
pheno_teds$ldsc_h2_se<-as.numeric(pheno_teds$ldsc_h2_se)
pheno_teds$h2_liab<-round(h2l_R2(k=pheno_teds$pop_prev, r2=pheno_teds$ldsc_h2_observed, p=pheno_teds$samp_prev),3)
pheno_teds$h2_liab_se<-round(se_h2l_R2(k=pheno_teds$pop_prev, h2=pheno_teds$ldsc_h2_observed, se=pheno_teds$ldsc_h2_se, p=pheno_teds$samp_prev),3)
pheno_teds$ldsc_h2_observed<-paste0(pheno_teds$ldsc_h2_observed," (", pheno_teds$ldsc_h2_se,")")
pheno_teds$ldsc_h2_se<-NULL
pheno_teds$h2_liab[!is.na(pheno_teds$Ncases)]<-paste0(pheno_teds$h2_liab[!is.na(pheno_teds$Ncases)]," (", pheno_teds$h2_liab_se[!is.na(pheno_teds$Ncases)],")")
pheno_teds$h2_liab_se<-NULL
pheno_teds$ldsc_intercept<-paste0(pheno_teds$ldsc_intercept," (", pheno_teds$ldsc_intercept_se,")")
pheno_teds$ldsc_intercept_se<-NULL
pheno_teds$Trait<-c("ADHD symptoms",'BMI','GCSE','Height')
pheno_teds<-pheno_teds[match(teds_dat$gwas,pheno_teds$code),]
pheno_teds<-pheno_teds[,c('Trait','code','trait','year','PMID','Ncases','Ncontrols','sample_size_discovery','ldsc_h2_observed','h2_liab','ldsc_intercept','ldsc_lambda_GC')]
names(pheno_teds)<-c('Target Phenotype','Code','GWAS Phenotype','Year','PMID','Ncase','Ncontrol','N','h2_obs','h2_liab','Intercept','Lambda')
write.csv(pheno_teds, '/users/k1806347/brc_scratch/Data/GWAS_sumstats/TEDS_phenotype_GWAS_descrip.csv', row.names=F, quote=F)Show GWAS for TEDS phenotypes
| Target Phenotype | Code | GWAS Phenotype | Year | PMID | Ncase | Ncontrol | N | h2-obs (SE) | h2-liab (SE) | Intercept | Lambda |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Height | HEIG03 | height | 2014 | 25282103 | NA | NA | 253288 | 0.312 (0.0141) | NA | 1.3254 (0.0185) | 2.0007 |
| BMI | BODY11 | BMI | 2018 | 30124842 | NA | NA | 681275 | 0.1908 (0.0053) | NA | 1.1864 (0.0206) | 2.7872 |
| GCSE | EDUC03 | years educational attainment | 2018 | 30038396 | NA | NA | 766345 | 0.1066 (0.0026) | NA | 1.0301 (0.0137) | 2.0940 |
| ADHD symptoms | ADHD04 | ADHD | 2017 | 30478444 | 20183 | 35191 | 55374 | 0.3635 (0.0227) | 0.426 (0.026) | 1.0295 (0.0096) | 1.2431 |
3.5 Polygenic scoring
Polygenic scores were derived using the following methods:
- p-value thresholding and clumping (pT + clump).
- lassosum - Lasso-based shrinkage method.
- PRScs - Bayesian shrinkage method.
- SBLUP - Summary statistic-based BLUP estimates
- SBayesR - Bayesian shrinkage method
- LDpred - Bayesian shrinkage method
- DBSLMM - Bayesian shrinkage method
- LDpred2 - Bayesian shrinkage method
Polygenic scores were derived using a reference standardised pipeline. Two references were used for all methods, the European subset of the 1KG reference (described here), and an independant subset of 10K European individuals from UK Biobank (described here). In brief, all scores were derived using HapMap3 SNPs only, modelling LD based on these references Any HapMap3 missing in the target sample are imputed using the reference estimated allele frequency.
3.5.1 UK Biobank
pT + clump: Sparse
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J pt_clump
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/sbatch.sh
######
# UKBB ref
######
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir -p ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J pt_clump
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/pt_clump/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/pt_clump/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/pt_clump/sbatch.sh
pT + clump: Dense
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 25G
#SBATCH -t 72:00:00
#SBATCH -J pt_clump_dense
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_dense/Scaled_polygenic_scorer_dense.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.GWAS_sumstats_clumped.txt \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--prsice_path ${prsice_path} \
--rscript ${rscript} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/todo.txt | cut -d' ' -f1)%5 ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/sbatch.sh
# BODY04 and PRCA01 aren't finishing
# Run in batches
# Split phenotype files into batcheslibrary(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
pheno<-c('BMI', 'Prostate_Cancer')
for(i in pheno){
pheno_file<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',i,'.txt'))
pheno_file_split<-split(pheno_file, (as.numeric(rownames(pheno_file))-1) %/% 5000)
for(k in 1:length(pheno_file_split)){
write.table(pheno_file_split[[k]], paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',i,'.subset_',k,'.txt'), col.names=T, row.names=F, quote=F)
}
}. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
# BODY04
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/sbatch_BODY04.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -t 72:00:00
#SBATCH -J pt_clump_dense
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=BODY04
pheno=BMI
subset=${SLURM_ARRAY_TASK_ID}
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_dense/Scaled_polygenic_scorer_dense.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.subset_${subset}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.GWAS_sumstats_clumped.txt \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--prsice_path ${prsice_path} \
--rscript ${rscript} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/${gwas}.subset_${subset}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-10 ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/sbatch_BODY04.sh
# PRCA01
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/sbatch_PRCA01.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -t 72:00:00
#SBATCH -J pt_clump_dense
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=PRCA01
pheno=Prostate_Cancer
subset=${SLURM_ARRAY_TASK_ID}
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_dense/Scaled_polygenic_scorer_dense.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.subset_${subset}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.GWAS_sumstats_clumped.txt \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--prsice_path ${prsice_path} \
--rscript ${rscript} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/${gwas}.subset_${subset}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-10 ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/sbatch_PRCA01.sh
# Combine the score from the different batches
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
gwas<-c('BODY04', 'PRCA01')
for(i in gwas){
tmp<-NULL
for(k in 1:10){
print(k)
tmp<-rbind(tmp, fread(paste0(UKBB_output,'/PRS_for_comparison/1KG_ref/pt_clump_dense/',i,'.subset_',k,'/UKBB.subset.w_hm3.',i,'.profiles')))
}
dir.create(paste0(UKBB_output,'/PRS_for_comparison/1KG_ref/pt_clump_dense/',i))
fwrite(tmp, paste0(UKBB_output,'/PRS_for_comparison/1KG_ref/pt_clump_dense/',i,'/UKBB.subset.w_hm3.',i,'.profiles'), col.names=T, row.names=F, quote=F, sep=' ')
}pT + clump: Sparse (non-nested)
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J pt_clump_nonnested
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_nonnested/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_nonnested/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/sbatch.sh
lassosum
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.lassosum_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 15G
#SBATCH -n 5
#SBATCH -J lassosum
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_lassosum/Scaled_polygenic_scorer_lassosum.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/lassosum/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/lassosum/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--pheno_name ${gwas} \
--n_cores 5 \
--plink ${plink1_9} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/lassosum/sbatch.sh
######
# UKBB ref
######
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.lassosum_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 15G
#SBATCH -n 5
#SBATCH -J lassosum
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_lassosum/Scaled_polygenic_scorer_lassosum.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/lassosum/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/lassosum/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--pheno_name ${gwas} \
--n_cores 5 \
--plink ${plink1_9} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/lassosum/sbatch.sh
PRScs
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.PRScs_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J PRScs
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_PRScs/Scaled_polygenic_scorer_PRScs.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/PRScs/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/PRScs/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/PRScs/sbatch.sh
SBLUP
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.SBLUP_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBLUP
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBLUP/Scaled_polygenic_scorer_SBLUP.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBLUP/${gwas}/GWAS_sumstats_SBLUP.sblup.cojo \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBLUP/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBLUP/sbatch.sh
######
# UKBB ref
######
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.SBLUP_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBLUP
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBLUP/Scaled_polygenic_scorer_SBLUP.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBLUP/${gwas}/GWAS_sumstats_SBLUP.sblup.cojo \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBLUP/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBLUP/sbatch.sh
SBayesR
########
# 1KG ref
########
####
# Standard
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch.sh
####
# P<0.4
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_P4.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_P4/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_P4.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_P4.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_P4.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_P4.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_P4/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_P4/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}_P4/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_P4.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_P4.sh
####
# GCTB v2.03
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTB_203/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTB_203.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}_GCTB_203/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203.txt | cut -d' ' -f1)%5 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTB_203.sh
####
# GCTB v2.03 forced robust parameterisation
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203_robust.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTB_203_robust/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203_robust.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTB_203_robust.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203_robust.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203_robust.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}_GCTB_203_robust/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTB_203_robust.txt | cut -d' ' -f1)%5 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTB_203_robust.sh
########
# GCTB ref
########
####
# Standard
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}_GCTBref/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref.sh
####
# P<0.4
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_P4.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_P4/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_P4.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref_P4.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_P4.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_P4.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_P4/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_P4/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}_GCTBref_P4/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_P4.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref_P4.sh
####
# GCTB v2.03
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_GCTB_203/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref_GCTB_203.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}_GCTBref_GCTB_203/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref_GCTB_203.sh
####
# GCTB v2.03 with forced robust mode
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203_robust.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_GCTB_203_robust/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203_robust.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref_GCTB_203_robust.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203_robust.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203_robust.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203_robust/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203_robust/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas}_GCTBref_GCTB_203_robust/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/todo_GCTBref_GCTB_203_robust.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/sbatch_GCTBref_GCTB_203_robust.sh
########################
# Calculate polygenic scores using UKBB reference
########################
####
# Standard
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch.sh
####
# P<0.4
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_P4.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}_P4/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_P4.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch_P4.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_P4.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_P4.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_P4/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_P4/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas}_P4/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_P4.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch_P4.sh
####
# GCTB v2.03
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}_GCTB_203/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch_GCTB_203.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas}_GCTB_203/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch_GCTB_203.sh
####
# GCTB v2.03 robust parameterisation
####
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203_robust.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}_GCTB_203_robust/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203_robust.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch_GCTB_203_robust.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J SBayesR
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203_robust.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203_robust.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas}_GCTB_203_robust/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/todo_GCTB_203_robust.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/sbatch_GCTB_203_robust.sh
LDPred
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.LDPred_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J LDPred
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred/Scaled_polygenic_scorer_LDPred.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred/sbatch.sh
######
# UKBB ref
######
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.LDPred_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J LDPred
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred/Scaled_polygenic_scorer_LDPred.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred/sbatch.sh
LDPred2
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.LDPred_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -n 6
#SBATCH --nodes 1
#SBATCH -J LDPred2
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred2/Scaled_polygenic_scorer_LDPred2.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred2/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred2/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--n_cores 6 \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/todo.txt | cut -d' ' -f1)%5 ${UKBB_output}/PRS_for_comparison/1KG_ref/LDPred2/sbatch.sh
######
# UKBB ref
######
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.LDPred_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -n 6
#SBATCH -J LDPred2
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred2/Scaled_polygenic_scorer_LDPred2.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred2/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred2/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--n_cores 6 \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/LDPred2/sbatch.sh
DBSLMM
########
# 1KG ref
########
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.DBSLMM_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J DBSLMM
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_DBSLMM/Scaled_polygenic_scorer_DBSLMM.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/DBSLMM/${gwas}/1KGPhase3.w_hm3.${gwas}.dbslmm.GW.txt \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/DBSLMM/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/1KG_ref/DBSLMM/sbatch.sh
######
# UKBB ref
######
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
# Create directory
mkdir ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM
# Create file listing GWAS that haven't been processed.
> ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/todo.txt
for i in $(seq 1 11);do
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
if [ ! -f ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.DBSLMM_profiles ]; then
echo ${gwas_i} ${pheno_i} >> ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/todo.txt
fi
done
# Create shell script to run using sbatch
cat > ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 10G
#SBATCH -J DBSLMM
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/todo.txt)
pheno=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $2}' ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/todo.txt)
echo $gwas
echo $pheno
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_DBSLMM/Scaled_polygenic_scorer_DBSLMM.R \
--target_plink_chr ${UKBB_output}/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr \
--target_keep ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno}.txt \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/DBSLMM/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.dbslmm.GW.txt \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/DBSLMM/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/${gwas}/UKBB.subset.w_hm3.${gwas}
EOF
sbatch --array 1-$(wc -l ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/todo.txt | cut -d' ' -f1)%3 ${UKBB_output}/PRS_for_comparison/UKBB_ref/DBSLMM/sbatch.sh
3.5.2 TEDS
pT + clump: Sparse
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Calculate polygenic scores based on 1KG reference.
n=0
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 5G -p brc,shared -J pTclump /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/pt_clump/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
# Calculate polygenic scores based on UKBB reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J pTclump /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/pt_clump/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/pt_clump/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/pt_clump/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
pT + clump: Dense
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Calculate polygenic scores for for Europeans only for now.
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J PRS_dense /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_dense/Scaled_polygenic_scorer_dense.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.GWAS_sumstats_clumped.txt \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_dense/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--prsice_path ${prsice_path} \
--rscript ${rscript} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/pt_clump_dense/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
pT + clump: Sparse (non-nested)
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Calculate polygenic scores based on 1KG reference
n=0
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J pTclump /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_nonnested/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/pt_clump_nonnested/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/pt_clump_nonnested/${gwas}/TEDS.w_hm3.${gwas}.EUR
n=$(($n+1))
done
lassosum
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Calculate scores using the 1KG reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J lassosum /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_lassosum/Scaled_polygenic_scorer_lassosum.R \
--target_plink_chr /users/k1806347/brc_scratch/Data/TEDS/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/lassosum/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/lassosum/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--pheno_name ${gwas} \
--plink ${plink1_9} \
--n_cores 1 \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/lassosum/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
# Calculate scores using the UKBB reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J lassosum /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_lassosum/Scaled_polygenic_scorer_lassosum.R \
--target_plink_chr /users/k1806347/brc_scratch/Data/TEDS/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/lassosum/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/lassosum/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--pheno_name ${gwas} \
--plink ${plink1_9} \
--n_cores 1 \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/lassosum/${gwas}/TEDS.w_hm3.${gwas}.EUR
donePRScs
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Calculate scores using My 1KG reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J PRScs /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_PRScs/Scaled_polygenic_scorer_PRScs.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/PRScs/${gwas}_My1KGRef/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/PRScs/${gwas}_My1KGRef/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/PRScs/${gwas}_My1KGRef/TEDS.w_hm3.${gwas}.EUR
done
# Calculate scores using the PRScs 1KG reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J PRScs /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_PRScs/Scaled_polygenic_scorer_PRScs.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/PRScs/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/PRScs/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/PRScs/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
SBLUP
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBLUP /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBLUP/Scaled_polygenic_scorer_SBLUP.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBLUP/${gwas}/GWAS_sumstats_SBLUP.sblup.cojo \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBLUP/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBLUP/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBLUP /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBLUP/Scaled_polygenic_scorer_SBLUP.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBLUP/${gwas}/GWAS_sumstats_SBLUP.sblup.cojo \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBLUP/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/SBLUP/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
SBayesR
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
##########################
# Calculate polygenic scores using 1KG reference
##########################
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
# P<0.4
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_P4/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_P4/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}_P4/TEDS.w_hm3.${gwas}.EUR
done
# GCTB v2.03
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}_GCTB_203/TEDS.w_hm3.${gwas}.EUR
done
# GCTB v2.03 forced robust
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}_GCTB_203_robust/TEDS.w_hm3.${gwas}.EUR
done
##########################
# Calculate polygenic scores using UKBB reference
##########################
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 5G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/SBayesR/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
# P<0.4
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 5G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_P4/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_P4/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/SBayesR/${gwas}_P4/TEDS.w_hm3.${gwas}.EUR
done
# GCTB v2.03
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 5G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/SBayesR/${gwas}_GCTB_203/TEDS.w_hm3.${gwas}.EUR
done
# GCTB v2.03 forced robust
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 5G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/SBayesR/${gwas}_GCTB_203_robust/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/SBayesR/${gwas}_GCTB_203_robust/TEDS.w_hm3.${gwas}.EUR
done
##########################
# Calculate polygenic scores using GCTB reference
##########################
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}_GCTBref/TEDS.w_hm3.${gwas}.EUR
done
# P<0.4
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_P4/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_P4/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}_GCTBref_P4/TEDS.w_hm3.${gwas}.EUR
done
# GCTB V2.03
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}_GCTBref_GCTB_203/TEDS.w_hm3.${gwas}.EUR
done
# GCTB V2.03 forced robust
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J SBayesR /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_SBayesR/Scaled_polygenic_scorer_SBayesR.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203_robust/GWAS_sumstats_SBayesR.GW.snpRes \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/SBayesR/${gwas}_GCTBref_GCTB_203_robust/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/SBayesR/${gwas}_GCTBref_GCTB_203_robust/TEDS.w_hm3.${gwas}.EUR
done
LDPred
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Calculate scores using the 1KG reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J LDPred /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred/Scaled_polygenic_scorer_LDPred.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/LDPred/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
# Calculate scores using the UKBB reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J LDPred /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred/Scaled_polygenic_scorer_LDPred.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/LDPred/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
LDPred2
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Calculate scores using the 1KG reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J LDPred2 /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred2/Scaled_polygenic_scorer_LDPred2.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred2/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/LDPred2/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/LDPred2/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
# Calculate scores using the UKBB reference
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 10G -p brc,shared -J LDPred2 /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_LDPred2/Scaled_polygenic_scorer_LDPred2.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred2/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas} \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/LDPred2/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/LDPred2/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
DBSLMM
# Set required variables
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
###
# 1KG ref
###
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 5G -p brc,shared -J DBSLMM /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_DBSLMM/Scaled_polygenic_scorer_DBSLMM.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${Geno_1KG_dir}/Score_files_for_polygenic/DBSLMM/${gwas}/1KGPhase3.w_hm3.${gwas}.dbslmm.GW.txt \
--ref_scale ${Geno_1KG_dir}/Score_files_for_polygenic/DBSLMM/${gwas}/1KGPhase3.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/1KG_ref/DBSLMM/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
###
# UKB ref
###
for gwas in $(echo HEIG03 EDUC03 ADHD04 BODY11);do
sbatch -n 1 --mem 5G -p brc,shared -J DBSLMM /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer_DBSLMM/Scaled_polygenic_scorer_DBSLMM.R \
--target_plink_chr ${TEDS_output_dir}/Genotype/Harmonised/TEDS.w_hm3.QCd.AllSNP.chr \
--target_keep ${TEDS_output_dir}/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--ref_score ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/DBSLMM/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.dbslmm.GW.txt \
--ref_scale ${UKBB_output}/UKBB_ref/Score_files_for_polygenic/DBSLMM/${gwas}/UKBB.noPheno.EUR.10K.w_hm3.${gwas}.EUR.scale \
--ref_freq_chr ${UKBB_output}/UKBB_ref/genotype/UKBB.noPheno.EUR.10K.chr \
--plink ${plink1_9} \
--pheno_name ${gwas} \
--output ${TEDS_output_dir}/prs_comparison/UKBB_ref/DBSLMM/${gwas}/TEDS.w_hm3.${gwas}.EUR
done
3.6 Estimating predictive ability
Models containing a single predictor were derived using generalised linear model (GLM). Models containing multiple predictors were derived using elastic-net regularisation to reduce the likelihood of overfitting and account for multicollinearity when modelling highly correlated predictors. 10-fold cross validation was performed using 80% of individuals to identify optimal parameters, with subsequent test-set validation in the remaining 20% of individuals to estimate the predictive utility among individuals not included in the parameter selection process.
First compare the three pT+clump approaches using just using the 1KG reference. Then compare the pT+clump (sparse) and other PRS methods in a range of scenarios. I am seperating these analysese as the former is computationally intensive due to the many thresholds of PRSice.
Model building and evaluation was performed using an Rscript called Model_builder_V2.R (more information here).
3.6.1 UK Biobank
pT + clump comparison
##############################
# Evaluating predictive utility of pT + clump PRSs across multiple pTs individually and in combination
##############################
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Make required directories
for pheno_i in $(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer);do
mkdir -p /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs
done
# Create a file listing the predictors files
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs-dense.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_dense/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs_nonnested.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/pt_clump_nonnested/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.profiles
EOF
done
# Derive and evaluate models
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
prev=$(echo 0.15 NA NA NA 0.05 0.03 0.013 0.00164 0.005 0.125 0.125)
# pT + clump (sparse)
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs \
--n_core 2 \
--compare_predictors F \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs.predictor_groups
done
# pT + clump (dense)
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 50G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs-dense \
--n_core 2 \
--assoc F \
--compare_predictors T \
--eval_only T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs-dense.predictor_groups
done
# pT + clump (non-nested)
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs_nonnested \
--n_core 2 \
--compare_predictors F \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs_nonnested.predictor_groups
done
SBayesR approaches
SBayesR is performing poorly or some outcomes. I have contacted the authors of SBayesR for further advice. SBayesR seems sensitive to the assumption that each variant effect is estimated in the same sample. To fulfill this assumption authors suggest removing variants that deviate substantially from the sample size median. I have already done this but some GWAS did not have sample size information available and in two cases SBayesR did not perform well even after sample size truncation. Here, the authors suggest imputing sample size based on the SE and MAF of variants and resticting the GWAS to variants with a P<0.4. I also run the analysis using the SBayesR analysis using the GCTB-released LD reference to ensure this isn’t affecting the results.
################################
# Evaluate use of SBayesR PRS
################################
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Create a file listing the predictors files
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
# 1KG ref
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_P4/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTB_203/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTB_203_robust/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
# GCTB ref
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_P4.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_P4/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_GCTB_203.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_GCTB_203/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_GCTB_203_robust/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
# UKB ref
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}_P4/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}_GCTB_203/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
${UKBB_output}/PRS_for_comparison/UKBB_ref/SBayesR/${gwas_i}_GCTB_203_robust/UKBB.subset.w_hm3.${gwas_i}.SBayesR_profiles
EOF
done
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
prev=$(echo 0.15 NA NA NA 0.05 0.03 0.013 0.00164 0.005 0.125 0.125)
# Run with 1KG
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs_SBayesR \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups
done
# Run with 1KG and P4 threshold
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_P4.EUR-PRSs_SBayesR \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups
done
# Run with 1KG and GCTB v2.03
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups
done
# Run with 1KG and GCTB v2.03 with forced robust parameterisation
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups
done
# Run with GCTB ref
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref.EUR-PRSs_SBayesR \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref.EUR-PRSs_SBayesR.predictor_groups
done
# Run with GCTB ref with P4 threshold
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_P4.EUR-PRSs_SBayesR \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_P4.EUR-PRSs_SBayesR.predictor_groups
done
# Run with GCTB ref and GCTB v2.03
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_GCTB_203.EUR-PRSs_SBayesR \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_GCTB_203.EUR-PRSs_SBayesR.predictor_groups
done
# Run with GCTB ref and GCTB v2.03 and forced robust
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups
done
# Run with UKB reference
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups
done
# Run with UKB reference with P4 thresh
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_P4.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups
done
# Run with UKB reference with GCTB v2.03
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups
done
# Run with UKB reference with GCTB v2.03 with robust parameterisation
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
pheno_file_i=$(echo ${pheno_file} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups
done
Tabulate SBayesR heritability and convergence info for SBayesR
####
# Create a table comparing SBayesR analyses across LD references
####
library(data.table)
gwas<-c('DEPR06','COLL01','BODY04','HEIG03','DIAB05','COAD01','CROH01','SCLE03','RHEU02','EDUC03','ADHD04','BODY11','PRCA01','BRCA01')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
#Processing<-c('','_P4')
#Processing_name<-c('Original','P<0.4')
Processing<-c('','_P4','_GCTB_203','_GCTB_203_robust')
Processing_name<-c('Original','P<0.4','v2.03','v2.03 robust')
GCTB_hsq_all<-NULL
for(process in 1:length(Processing)){
for(i in 1:length(gwas)){
GCTB_log<-read.table(paste0(Geno_1KG_dir,'/Score_files_for_polygenic/SBayesR/', gwas[i],Processing[process],'/1KGPhase3.w_hm3.',gwas[i],'.log'), sep='&')
if(sum(grepl('Per variant N is not pres', GCTB_log$V1)) > 0 ){
perVar<-F
} else {
perVar<-T
}
if(sum(grepl('No results for chromosomes', GCTB_log$V1)) > 0 ){
Complete<-F
} else {
Complete<-T
}
hsq<-GCTB_log$V1[grepl('SNP-heritability estimate is ', GCTB_log$V1)]
hsq<-gsub('\\.$','',gsub('SNP-heritability estimate is ','',hsq))
hsq_est<-as.numeric(as.character(gsub(' .*','',hsq)))
hsq_se<-as.numeric(as.character(gsub("\\)",'',gsub(".*\\(SD=",'',hsq))))
tmp<-data.frame( Ref='1KG',
Process=Processing_name[process],
GWAS=gwas[i],
perVar=perVar,
Complete=Complete,
SBayesR_H2=paste0(round(hsq_est,3)," (SE=",round(hsq_se,3),")"))
GCTB_hsq_all<-rbind(GCTB_hsq_all, tmp)
}
GCTB_N_tab_UKB<-NULL
for(i in 1:length(gwas)){
GCTB_log<-read.table(paste0(UKBB_output,'/UKBB_ref/Score_files_for_polygenic/SBayesR/', gwas[i],Processing[process],'/UKBB.noPheno.EUR.10K.w_hm3.',gwas[i],'.log'), sep='&')
if(sum(grepl('Per variant N is not pres', GCTB_log$V1)) > 0 ){
perVar<-F
} else {
perVar<-T
}
if(sum(grepl('No results for chromosomes', GCTB_log$V1)) > 0 ){
Complete<-F
} else {
Complete<-T
}
hsq<-GCTB_log$V1[grepl('SNP-heritability estimate is ', GCTB_log$V1)]
hsq<-gsub('\\.$','',gsub('SNP-heritability estimate is ','',hsq))
hsq_est<-as.numeric(as.character(gsub(' .*','',hsq)))
hsq_se<-as.numeric(as.character(gsub("\\)",'',gsub(".*\\(SD=",'',hsq))))
tmp<-data.frame( Ref='UKB',
Process=Processing_name[process],
GWAS=gwas[i],
perVar=perVar,
Complete=Complete,
SBayesR_H2=paste0(round(hsq_est,3)," (SE=",round(hsq_se,3),")"))
GCTB_hsq_all<-rbind(GCTB_hsq_all, tmp)
}
GCTB_N_tab_GCTB<-NULL
for(i in 1:length(gwas)){
GCTB_log<-read.table(paste0(Geno_1KG_dir,'/Score_files_for_polygenic/SBayesR/', gwas[i],'_GCTBref',Processing[process],'/1KGPhase3.w_hm3.',gwas[i],'.log'), sep='&')
if(sum(grepl('Per variant N is not pres', GCTB_log$V1)) > 0 ){
perVar<-F
} else {
perVar<-T
}
if(sum(grepl('No results for chromosomes', GCTB_log$V1)) > 0 ){
Complete<-F
} else {
Complete<-T
}
hsq<-GCTB_log$V1[grepl('SNP-heritability estimate is ', GCTB_log$V1)]
hsq<-gsub('\\.$','',gsub('SNP-heritability estimate is ','',hsq))
hsq_est<-as.numeric(as.character(gsub(' .*','',hsq)))
hsq_se<-as.numeric(as.character(gsub("\\)",'',gsub(".*\\(SD=",'',hsq))))
tmp<-data.frame( Ref='GCTB',
Process=Processing_name[process],
GWAS=gwas[i],
perVar=perVar,
Complete=Complete,
SBayesR_H2=paste0(round(hsq_est,3)," (SE=",round(hsq_se,3),")"))
GCTB_hsq_all<-rbind(GCTB_hsq_all, tmp)
}
}
GCTB_hsq_all$SBayesR_H2_est<-as.numeric(gsub(' .*','',GCTB_hsq_all$SBayesR_H2))
GCTB_hsq_all$SBayesR_H2_se<-as.numeric(gsub("\\)",'',gsub(".*\\(SE=",'',GCTB_hsq_all$SBayesR_H2)))
GCTB_hsq_all$SBayesR_H2<-NULL
GCTB_hsq_all$Converge<-F
GCTB_hsq_all$Converge[GCTB_hsq_all$SBayesR_H2_est < 1.2 & GCTB_hsq_all$Complete == T]<-T
GCTB_hsq_all_use<-NULL
for(i in 1:length(gwas)){
for(k in c('1KG','UKB','GCTB')){
GCTB_hsq_all_i_k<-GCTB_hsq_all[GCTB_hsq_all$GWAS == gwas[i] & GCTB_hsq_all$Ref == k,]
GCTB_hsq_all_i_k$Use<-F
for(j in 1:length(Processing)){
if(GCTB_hsq_all_i_k$Converge[j] == T){
GCTB_hsq_all_i_k$Use[j]<-T
break()
}
}
GCTB_hsq_all_use<-rbind(GCTB_hsq_all_use, GCTB_hsq_all_i_k)
}
}
write.csv(GCTB_hsq_all_use, '/users/k1806347/brc_scratch/Analyses/PRS_comparison/SBayesR_H2_comp.csv', row.names=F, quote=F)
# Note. there are several GWAS that do not converge even with P<0.4.
# Estimates of h2 look more reasonable for the robust version of GCTB.Show SBayesR heritability and N info
| Ref | Process | GWAS | perVar | Complete | SBayesR_H2_est | SBayesR_H2_se | Converge | Use |
|---|---|---|---|---|---|---|---|---|
| 1KG | Original | DEPR06 | TRUE | TRUE | 0.035 | 0.002 | TRUE | TRUE |
| 1KG | P<0.4 | DEPR06 | TRUE | TRUE | 0.092 | 0.003 | TRUE | FALSE |
| 1KG | v2.03 | DEPR06 | TRUE | TRUE | 0.035 | 0.002 | TRUE | FALSE |
| 1KG | v2.03 robust | DEPR06 | TRUE | TRUE | 0.043 | 0.003 | TRUE | FALSE |
| UKB | Original | DEPR06 | TRUE | TRUE | 0.057 | 0.002 | TRUE | TRUE |
| UKB | P<0.4 | DEPR06 | TRUE | TRUE | 0.138 | 0.003 | TRUE | FALSE |
| UKB | v2.03 | DEPR06 | TRUE | TRUE | 0.057 | 0.002 | TRUE | FALSE |
| UKB | v2.03 robust | DEPR06 | TRUE | TRUE | 0.057 | 0.002 | TRUE | FALSE |
| GCTB | Original | DEPR06 | TRUE | TRUE | 0.058 | 0.002 | TRUE | TRUE |
| GCTB | P<0.4 | DEPR06 | TRUE | TRUE | 0.140 | 0.003 | TRUE | FALSE |
| GCTB | v2.03 | DEPR06 | TRUE | TRUE | 0.059 | 0.002 | TRUE | FALSE |
| GCTB | v2.03 robust | DEPR06 | TRUE | TRUE | 0.058 | 0.002 | TRUE | FALSE |
| 1KG | Original | COLL01 | FALSE | TRUE | 0.113 | 0.007 | TRUE | TRUE |
| 1KG | P<0.4 | COLL01 | FALSE | TRUE | 0.343 | 0.010 | TRUE | FALSE |
| 1KG | v2.03 | COLL01 | FALSE | TRUE | 0.115 | 0.007 | TRUE | FALSE |
| 1KG | v2.03 robust | COLL01 | FALSE | TRUE | 0.112 | 0.031 | TRUE | FALSE |
| UKB | Original | COLL01 | FALSE | TRUE | 0.162 | 0.007 | TRUE | TRUE |
| UKB | P<0.4 | COLL01 | FALSE | TRUE | 0.504 | 0.013 | TRUE | FALSE |
| UKB | v2.03 | COLL01 | FALSE | TRUE | 0.165 | 0.008 | TRUE | FALSE |
| UKB | v2.03 robust | COLL01 | FALSE | TRUE | 0.141 | 0.006 | TRUE | FALSE |
| GCTB | Original | COLL01 | FALSE | TRUE | 0.168 | 0.007 | TRUE | TRUE |
| GCTB | P<0.4 | COLL01 | FALSE | TRUE | 0.531 | 0.014 | TRUE | FALSE |
| GCTB | v2.03 | COLL01 | FALSE | TRUE | 0.171 | 0.007 | TRUE | FALSE |
| GCTB | v2.03 robust | COLL01 | FALSE | TRUE | 0.146 | 0.007 | TRUE | FALSE |
| 1KG | Original | BODY04 | TRUE | TRUE | 0.248 | 0.059 | TRUE | TRUE |
| 1KG | P<0.4 | BODY04 | TRUE | TRUE | 0.192 | 0.016 | TRUE | FALSE |
| 1KG | v2.03 | BODY04 | TRUE | TRUE | 0.255 | 0.064 | TRUE | FALSE |
| 1KG | v2.03 robust | BODY04 | TRUE | TRUE | 0.048 | 0.003 | TRUE | FALSE |
| UKB | Original | BODY04 | TRUE | TRUE | 1.397 | 0.680 | FALSE | FALSE |
| UKB | P<0.4 | BODY04 | TRUE | TRUE | 1.048 | 0.420 | TRUE | TRUE |
| UKB | v2.03 | BODY04 | TRUE | TRUE | 1.399 | 0.680 | FALSE | FALSE |
| UKB | v2.03 robust | BODY04 | TRUE | TRUE | 0.061 | 0.002 | TRUE | FALSE |
| GCTB | Original | BODY04 | TRUE | TRUE | 0.920 | 0.430 | TRUE | TRUE |
| GCTB | P<0.4 | BODY04 | TRUE | TRUE | 0.701 | 0.265 | TRUE | FALSE |
| GCTB | v2.03 | BODY04 | TRUE | TRUE | 0.957 | 0.461 | TRUE | FALSE |
| GCTB | v2.03 robust | BODY04 | TRUE | TRUE | 0.059 | 0.002 | TRUE | FALSE |
| 1KG | Original | HEIG03 | TRUE | TRUE | 0.718 | 0.109 | TRUE | TRUE |
| 1KG | P<0.4 | HEIG03 | TRUE | TRUE | 0.560 | 0.057 | TRUE | FALSE |
| 1KG | v2.03 | HEIG03 | TRUE | TRUE | 0.719 | 0.110 | TRUE | FALSE |
| 1KG | v2.03 robust | HEIG03 | TRUE | TRUE | 0.251 | 0.005 | TRUE | FALSE |
| UKB | Original | HEIG03 | TRUE | TRUE | 3.876 | 1.436 | FALSE | FALSE |
| UKB | P<0.4 | HEIG03 | TRUE | TRUE | 3.163 | 1.174 | FALSE | FALSE |
| UKB | v2.03 | HEIG03 | TRUE | TRUE | 3.877 | 1.437 | FALSE | FALSE |
| UKB | v2.03 robust | HEIG03 | TRUE | TRUE | 0.433 | 0.004 | TRUE | TRUE |
| GCTB | Original | HEIG03 | TRUE | FALSE | 2.806 | 1.142 | FALSE | FALSE |
| GCTB | P<0.4 | HEIG03 | TRUE | FALSE | 2.424 | 0.887 | FALSE | FALSE |
| GCTB | v2.03 | HEIG03 | TRUE | TRUE | 2.958 | 1.157 | FALSE | FALSE |
| GCTB | v2.03 robust | HEIG03 | TRUE | TRUE | 0.440 | 0.004 | TRUE | TRUE |
| 1KG | Original | DIAB05 | TRUE | TRUE | 0.065 | 0.005 | TRUE | TRUE |
| 1KG | P<0.4 | DIAB05 | TRUE | TRUE | 0.225 | 0.007 | TRUE | FALSE |
| 1KG | v2.03 | DIAB05 | TRUE | TRUE | 0.066 | 0.006 | TRUE | FALSE |
| 1KG | v2.03 robust | DIAB05 | TRUE | TRUE | 0.059 | 0.006 | TRUE | FALSE |
| UKB | Original | DIAB05 | TRUE | TRUE | 0.093 | 0.005 | TRUE | TRUE |
| UKB | P<0.4 | DIAB05 | TRUE | TRUE | 0.340 | 0.009 | TRUE | FALSE |
| UKB | v2.03 | DIAB05 | TRUE | TRUE | 0.094 | 0.005 | TRUE | FALSE |
| UKB | v2.03 robust | DIAB05 | TRUE | TRUE | 0.084 | 0.005 | TRUE | FALSE |
| GCTB | Original | DIAB05 | TRUE | TRUE | 0.092 | 0.005 | TRUE | TRUE |
| GCTB | P<0.4 | DIAB05 | TRUE | TRUE | 0.344 | 0.009 | TRUE | FALSE |
| GCTB | v2.03 | DIAB05 | TRUE | TRUE | 0.093 | 0.005 | TRUE | FALSE |
| GCTB | v2.03 robust | DIAB05 | TRUE | TRUE | 0.085 | 0.005 | TRUE | FALSE |
| 1KG | Original | COAD01 | FALSE | TRUE | 0.033 | 0.003 | TRUE | TRUE |
| 1KG | P<0.4 | COAD01 | FALSE | TRUE | 0.156 | 0.005 | TRUE | FALSE |
| 1KG | v2.03 | COAD01 | FALSE | TRUE | 0.036 | 0.003 | TRUE | FALSE |
| 1KG | v2.03 robust | COAD01 | FALSE | TRUE | 0.035 | 0.004 | TRUE | FALSE |
| UKB | Original | COAD01 | FALSE | TRUE | 0.077 | 0.019 | TRUE | TRUE |
| UKB | P<0.4 | COAD01 | FALSE | TRUE | 0.247 | 0.020 | TRUE | FALSE |
| UKB | v2.03 | COAD01 | FALSE | TRUE | 0.078 | 0.019 | TRUE | FALSE |
| UKB | v2.03 robust | COAD01 | FALSE | TRUE | 0.043 | 0.003 | TRUE | FALSE |
| GCTB | Original | COAD01 | FALSE | TRUE | 0.077 | 0.021 | TRUE | TRUE |
| GCTB | P<0.4 | COAD01 | FALSE | TRUE | 0.232 | 0.007 | TRUE | FALSE |
| GCTB | v2.03 | COAD01 | FALSE | TRUE | 0.072 | 0.016 | TRUE | FALSE |
| GCTB | v2.03 robust | COAD01 | FALSE | TRUE | 0.042 | 0.003 | TRUE | FALSE |
| 1KG | Original | CROH01 | FALSE | TRUE | 0.774 | 0.246 | TRUE | TRUE |
| 1KG | P<0.4 | CROH01 | FALSE | TRUE | 1.468 | 0.047 | FALSE | FALSE |
| 1KG | v2.03 | CROH01 | FALSE | TRUE | 0.905 | 0.301 | TRUE | FALSE |
| 1KG | v2.03 robust | CROH01 | FALSE | TRUE | 0.389 | 0.042 | TRUE | FALSE |
| UKB | Original | CROH01 | FALSE | FALSE | 0.640 | 0.141 | FALSE | FALSE |
| UKB | P<0.4 | CROH01 | FALSE | TRUE | 2.073 | 0.058 | FALSE | FALSE |
| UKB | v2.03 | CROH01 | FALSE | TRUE | 0.589 | 0.056 | TRUE | TRUE |
| UKB | v2.03 robust | CROH01 | FALSE | TRUE | 0.476 | 0.031 | TRUE | FALSE |
| GCTB | Original | CROH01 | FALSE | FALSE | 0.835 | 0.120 | FALSE | FALSE |
| GCTB | P<0.4 | CROH01 | FALSE | TRUE | 2.120 | 0.057 | FALSE | FALSE |
| GCTB | v2.03 | CROH01 | FALSE | TRUE | 0.933 | 0.164 | TRUE | TRUE |
| GCTB | v2.03 robust | CROH01 | FALSE | TRUE | 0.485 | 0.027 | TRUE | FALSE |
| 1KG | Original | SCLE03 | FALSE | TRUE | 0.689 | 0.077 | TRUE | TRUE |
| 1KG | P<0.4 | SCLE03 | FALSE | TRUE | 1.345 | 0.051 | FALSE | FALSE |
| 1KG | v2.03 | SCLE03 | FALSE | TRUE | 0.713 | 0.085 | TRUE | FALSE |
| 1KG | v2.03 robust | SCLE03 | FALSE | TRUE | 0.239 | 0.024 | TRUE | FALSE |
| UKB | Original | SCLE03 | FALSE | FALSE | 0.741 | 0.094 | FALSE | FALSE |
| UKB | P<0.4 | SCLE03 | FALSE | FALSE | 1.784 | 0.049 | FALSE | FALSE |
| UKB | v2.03 | SCLE03 | FALSE | TRUE | 0.742 | 0.092 | TRUE | TRUE |
| UKB | v2.03 robust | SCLE03 | FALSE | TRUE | 0.303 | 0.021 | TRUE | FALSE |
| GCTB | Original | SCLE03 | FALSE | FALSE | 0.767 | 0.126 | FALSE | FALSE |
| GCTB | P<0.4 | SCLE03 | FALSE | FALSE | 1.845 | 0.051 | FALSE | FALSE |
| GCTB | v2.03 | SCLE03 | FALSE | TRUE | 0.697 | 0.080 | TRUE | TRUE |
| GCTB | v2.03 robust | SCLE03 | FALSE | TRUE | 0.289 | 0.022 | TRUE | FALSE |
| 1KG | Original | RHEU02 | FALSE | TRUE | 0.244 | 0.011 | TRUE | TRUE |
| 1KG | P<0.4 | RHEU02 | FALSE | TRUE | 0.536 | 0.018 | TRUE | FALSE |
| 1KG | v2.03 | RHEU02 | FALSE | TRUE | 0.244 | 0.011 | TRUE | FALSE |
| 1KG | v2.03 robust | RHEU02 | FALSE | TRUE | 0.226 | 0.015 | TRUE | FALSE |
| UKB | Original | RHEU02 | FALSE | TRUE | 0.358 | 0.021 | TRUE | TRUE |
| UKB | P<0.4 | RHEU02 | FALSE | TRUE | 0.778 | 0.022 | TRUE | FALSE |
| UKB | v2.03 | RHEU02 | FALSE | TRUE | 0.365 | 0.033 | TRUE | FALSE |
| UKB | v2.03 robust | RHEU02 | FALSE | TRUE | 0.352 | 0.016 | TRUE | FALSE |
| GCTB | Original | RHEU02 | FALSE | TRUE | 0.358 | 0.012 | TRUE | TRUE |
| GCTB | P<0.4 | RHEU02 | FALSE | TRUE | 0.787 | 0.022 | TRUE | FALSE |
| GCTB | v2.03 | RHEU02 | FALSE | TRUE | 0.360 | 0.012 | TRUE | FALSE |
| GCTB | v2.03 robust | RHEU02 | FALSE | TRUE | 0.366 | 0.016 | TRUE | FALSE |
| 1KG | Original | EDUC03 | FALSE | TRUE | 0.063 | 0.002 | TRUE | TRUE |
| 1KG | P<0.4 | EDUC03 | FALSE | TRUE | 0.093 | 0.002 | TRUE | FALSE |
| 1KG | v2.03 | EDUC03 | FALSE | TRUE | 0.063 | 0.002 | TRUE | FALSE |
| 1KG | v2.03 robust | EDUC03 | FALSE | TRUE | 0.063 | 0.002 | TRUE | FALSE |
| UKB | Original | EDUC03 | FALSE | TRUE | 0.106 | 0.001 | TRUE | TRUE |
| UKB | P<0.4 | EDUC03 | FALSE | TRUE | 0.143 | 0.002 | TRUE | FALSE |
| UKB | v2.03 | EDUC03 | FALSE | TRUE | 0.106 | 0.001 | TRUE | FALSE |
| UKB | v2.03 robust | EDUC03 | FALSE | TRUE | 0.106 | 0.001 | TRUE | FALSE |
| GCTB | Original | EDUC03 | FALSE | TRUE | 0.108 | 0.001 | TRUE | TRUE |
| GCTB | P<0.4 | EDUC03 | FALSE | TRUE | 0.143 | 0.002 | TRUE | FALSE |
| GCTB | v2.03 | EDUC03 | FALSE | TRUE | 0.108 | 0.001 | TRUE | FALSE |
| GCTB | v2.03 robust | EDUC03 | FALSE | TRUE | 0.109 | 0.001 | TRUE | FALSE |
| 1KG | Original | ADHD04 | FALSE | TRUE | 0.254 | 0.013 | TRUE | TRUE |
| 1KG | P<0.4 | ADHD04 | FALSE | TRUE | 0.902 | 0.027 | TRUE | FALSE |
| 1KG | v2.03 | ADHD04 | FALSE | TRUE | 0.260 | 0.012 | TRUE | FALSE |
| 1KG | v2.03 robust | ADHD04 | FALSE | TRUE | 0.313 | 0.042 | TRUE | FALSE |
| UKB | Original | ADHD04 | FALSE | TRUE | 0.378 | 0.016 | TRUE | TRUE |
| UKB | P<0.4 | ADHD04 | FALSE | TRUE | 1.274 | 0.033 | FALSE | FALSE |
| UKB | v2.03 | ADHD04 | FALSE | TRUE | 0.385 | 0.015 | TRUE | FALSE |
| UKB | v2.03 robust | ADHD04 | FALSE | TRUE | 0.384 | 0.015 | TRUE | FALSE |
| GCTB | Original | ADHD04 | FALSE | TRUE | 0.390 | 0.016 | TRUE | TRUE |
| GCTB | P<0.4 | ADHD04 | FALSE | TRUE | 1.295 | 0.034 | FALSE | FALSE |
| GCTB | v2.03 | ADHD04 | FALSE | TRUE | 0.395 | 0.016 | TRUE | FALSE |
| GCTB | v2.03 robust | ADHD04 | FALSE | TRUE | 0.390 | 0.016 | TRUE | FALSE |
| 1KG | Original | BODY11 | TRUE | TRUE | 0.123 | 0.002 | TRUE | TRUE |
| 1KG | P<0.4 | BODY11 | TRUE | TRUE | 0.158 | 0.002 | TRUE | FALSE |
| 1KG | v2.03 | BODY11 | TRUE | TRUE | 0.124 | 0.002 | TRUE | FALSE |
| 1KG | v2.03 robust | BODY11 | TRUE | TRUE | 0.123 | 0.003 | TRUE | FALSE |
| UKB | Original | BODY11 | TRUE | TRUE | 0.213 | 0.003 | TRUE | TRUE |
| UKB | P<0.4 | BODY11 | TRUE | TRUE | 0.251 | 0.002 | TRUE | FALSE |
| UKB | v2.03 | BODY11 | TRUE | TRUE | 0.211 | 0.002 | TRUE | FALSE |
| UKB | v2.03 robust | BODY11 | TRUE | TRUE | 0.211 | 0.002 | TRUE | FALSE |
| GCTB | Original | BODY11 | TRUE | TRUE | 0.217 | 0.003 | TRUE | TRUE |
| GCTB | P<0.4 | BODY11 | TRUE | TRUE | 0.253 | 0.002 | TRUE | FALSE |
| GCTB | v2.03 | BODY11 | TRUE | TRUE | 0.217 | 0.003 | TRUE | FALSE |
| GCTB | v2.03 robust | BODY11 | TRUE | TRUE | 0.216 | 0.002 | TRUE | FALSE |
| 1KG | Original | PRCA01 | FALSE | TRUE | 0.124 | 0.004 | TRUE | TRUE |
| 1KG | P<0.4 | PRCA01 | FALSE | TRUE | 0.244 | 0.005 | TRUE | FALSE |
| 1KG | v2.03 | PRCA01 | FALSE | TRUE | 0.126 | 0.004 | TRUE | FALSE |
| 1KG | v2.03 robust | PRCA01 | FALSE | TRUE | 0.124 | 0.005 | TRUE | FALSE |
| UKB | Original | PRCA01 | FALSE | TRUE | 0.241 | 0.021 | TRUE | TRUE |
| UKB | P<0.4 | PRCA01 | FALSE | TRUE | 0.351 | 0.007 | TRUE | FALSE |
| UKB | v2.03 | PRCA01 | FALSE | TRUE | 0.242 | 0.022 | TRUE | FALSE |
| UKB | v2.03 robust | PRCA01 | FALSE | TRUE | 0.161 | 0.004 | TRUE | FALSE |
| GCTB | Original | PRCA01 | FALSE | TRUE | 0.195 | 0.020 | TRUE | TRUE |
| GCTB | P<0.4 | PRCA01 | FALSE | TRUE | 0.357 | 0.007 | TRUE | FALSE |
| GCTB | v2.03 | PRCA01 | FALSE | TRUE | 0.197 | 0.021 | TRUE | FALSE |
| GCTB | v2.03 robust | PRCA01 | FALSE | TRUE | 0.163 | 0.005 | TRUE | FALSE |
| 1KG | Original | BRCA01 | FALSE | TRUE | 0.107 | 0.003 | TRUE | TRUE |
| 1KG | P<0.4 | BRCA01 | FALSE | TRUE | 0.209 | 0.004 | TRUE | FALSE |
| 1KG | v2.03 | BRCA01 | FALSE | TRUE | 0.107 | 0.003 | TRUE | FALSE |
| 1KG | v2.03 robust | BRCA01 | FALSE | TRUE | 0.106 | 0.004 | TRUE | FALSE |
| UKB | Original | BRCA01 | FALSE | TRUE | 0.158 | 0.003 | TRUE | TRUE |
| UKB | P<0.4 | BRCA01 | FALSE | TRUE | 0.304 | 0.005 | TRUE | FALSE |
| UKB | v2.03 | BRCA01 | FALSE | TRUE | 0.158 | 0.003 | TRUE | FALSE |
| UKB | v2.03 robust | BRCA01 | FALSE | TRUE | 0.158 | 0.004 | TRUE | FALSE |
| GCTB | Original | BRCA01 | FALSE | TRUE | 0.160 | 0.003 | TRUE | TRUE |
| GCTB | P<0.4 | BRCA01 | FALSE | TRUE | 0.308 | 0.005 | TRUE | FALSE |
| GCTB | v2.03 | BRCA01 | FALSE | TRUE | 0.160 | 0.003 | TRUE | FALSE |
| GCTB | v2.03 robust | BRCA01 | FALSE | TRUE | 0.160 | 0.004 | TRUE | FALSE |
Compare all methods
# We need to read in polygenic scores based on a range of parameters, pseudovalidated scores as a seperate group.
# Create a file listing the predictors files
# For SBayesR, use P<0.4 score for those that do not converge.
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
GCTB_hsq_all<-read.csv('/users/k1806347/brc_scratch/Analyses/PRS_comparison/SBayesR_H2_comp.csv')
pheno<-c('Depression','Intelligence','BMI','Height','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas<-c('DEPR06','COLL01','BODY04','HEIG03','DIAB05','COAD01','CROH01','SCLE03','RHEU02','BRCA01','PRCA01')
for(i in 1:length(pheno)){
for(ref in c('1KG','UKBB')){
pred_file<-NULL
# pT+clump (sparse)
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/pt_clump/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.profiles'),
group='pT+clump'))
# lassosum
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/lassosum/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.lassosum_profiles'),
group='lassosum'))
if(ref == '1KG'){
# PRScs
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/PRScs/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.PRScs_profiles'),
group='PRScs'))
}
# SBLUP
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/SBLUP/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.SBLUP_profiles'),
group='SBLUP'))
# SBayesR
# Note use Robust implementation
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/SBayesR/',gwas[i],'_GCTB_203_robust/UKBB.subset.w_hm3.',gwas[i],'.SBayesR_profiles'),
group='SBayesR'))
# LDPred
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/LDPred/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.LDPred_profiles'),
group='LDPred'))
# DBSLMM
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/DBSLMM/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.DBSLMM_profiles'),
group='DBSLMM'))
# LDPred2
pred_file<-rbind(pred_file,data.frame( predictors=paste0(UKBB_output,'/PRS_for_comparison/',ref,'_ref/LDPred2/',gwas[i],'/UKBB.subset.w_hm3.',gwas[i],'.LDPred_profiles'),
group='LDPred2'))
# Write out list of predictors with groups
if(ref == '1KG'){
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'), row.names=F, col.names=T, quote=F)
} else {
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/UKBB.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'), row.names=F, col.names=T, quote=F)
}
}
}. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Run Model_builder_V2.R
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
prev=$(echo 0.15 NA NA NA 0.05 0.03 0.013 0.00164 0.005 0.125 0.125)
# 1KG reference
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 30G -n 3 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp \
--n_core 3 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp.predictor_groups
done
# UKBB reference
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 30G -n 3 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs.AllMethodComp \
--n_core 3 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs.AllMethodComp.predictor_groups
done
Compare all methods with PCs regressed
#############
# Regress project PCs from all polygenic scores
#############
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno<-c('Depression','Intelligence','BMI','Height','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas<-c('DEPR06','COLL01','BODY04','HEIG03','DIAB05','COAD01','CROH01','SCLE03','RHEU02','BRCA01','PRCA01')
# Read in PCs
pcs<-fread(paste0(UKBB_output,'/Projected_PCs/EUR/UKBB.w_hm3.EUR.eigenvec'))
# Extract the first 20PCs
pcs<-pcs[,c('FID','IID',paste0('PC',1:20))]
for(ref in c('1KG')){
for(i in 1:length(gwas)){
# Read in list of predictors
if(ref == '1KG'){
pred_file<-fread(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'))
} else {
pred_file<-fread(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/UKBB.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'))
}
for(file in 1:dim(pred_file)[1]){
# Read in predictors
file_i<-fread(pred_file$predictors[file])
# Merge with PCs
file_i_pcs<-merge(file_i, pcs, by=c('FID','IID'))
for(col_i in names(file_i_pcs)[grepl(gwas[i], names(file_i_pcs))]){
file_i_pcs_col_i<-file_i_pcs[,c(col_i,paste0('PC',1:20)), with=F]
names(file_i_pcs_col_i)[names(file_i_pcs_col_i) == col_i]<-'y'
# Regress covariates, scale residuals and round to three decimal places
file_i_pcs[[paste0(col_i,'_resid')]]<-round(as.numeric(scale(resid(lm(as.formula(paste0('y ~ ', paste(paste0('PC', 1:20), collapse=' + '))), data=file_i_pcs_col_i)))),3)
}
# Extract residual columns
file_i_resid<-file_i_pcs[,c('FID','IID',names(file_i_pcs)[grepl('_resid',names(file_i_pcs))]), with=F]
# Save the residuals
output<-gsub(paste0(ref,'_ref'),paste0(ref,'_ref_PCresid'),pred_file$predictors[file])
tmp<-sub('.*/','',output)
output_dir<-sub(paste0(tmp,'*.'),'',output)
system(paste0('mkdir -p ',output_dir))
write.table(file_i_resid, output, col.names=T, row.names=F, quote=F)
}
# Write out new list of predictors for residuals
pred_file_resid<-pred_file
pred_file_resid$predictors<-gsub(paste0(ref,'_ref'),paste0(ref,'_ref_PCresid'),pred_file$predictors)
if(ref == '1KG'){
write.table(pred_file_resid, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.PCresid.predictor_groups'), col.names=T, row.names=F, quote=F)
} else {
write.table(pred_file_resid, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/UKBB.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.PCresid.predictor_groups'), col.names=T, row.names=F, quote=F)
}
}
}. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config
# Run Model_builder_V2.R
pheno=$(echo Depression Intelligence BMI Height T2D CAD IBD MultiScler RheuArth Breast_Cancer Prostate_Cancer)
gwas=$(echo DEPR06 COLL01 BODY04 HEIG03 DIAB05 COAD01 CROH01 SCLE03 RHEU02 BRCA01 PRCA01)
prev=$(echo 0.15 NA NA NA 0.05 0.03 0.013 0.00164 0.005 0.125 0.125)
# 1KG reference
for i in $(seq 1 11);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 30G -n 3 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno ${UKBB_output}/Phenotype/PRS_comp_subset/UKBB.${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp.PCresid \
--n_core 3 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/${pheno_i}/Association_withPRSs/UKBB.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp.PCresid.predictor_groups
done
3.6.2 TEDS
pT + clump comparison
##############################
# Evaluating predictive utility of pT + clump PRSs across multiple pTs individually and in combination
##############################
# Make required directories
for pheno_i in $(echo Height21 BMI21 GCSE ADHD);do
mkdir -p /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs
done
# Create a file listing the predictors files
pheno=$(echo Height21 BMI21 GCSE ADHD)
gwas=$(echo HEIG03 BODY11 EDUC03 ADHD04)
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/pt_clump/${gwas_i}/TEDS.w_hm3.${gwas_i}.EUR.profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs-dense.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/pt_clump_dense/${gwas_i}/TEDS.w_hm3.${gwas_i}.EUR.profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs_nonnested.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/pt_clump_nonnested/${gwas_i}/TEDS.w_hm3.${gwas_i}.EUR.profiles
EOF
done
pheno=$(echo Height21 BMI21 GCSE ADHD)
gwas=$(echo HEIG03 BODY11 EDUC03 ADHD04)
prev=$(echo NA NA NA NA)
# pT+clump (sparse)
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs.predictor_groups
done
# pT+clump (dense)
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 20G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs-dense \
--n_core 2 \
--assoc T \
--compare_predictors T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs-dense.predictor_groups
done
# pT+clump (non-nested)
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs_nonnested \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs_nonnested.predictor_groups
done
SBayesR comparison
################################
# Evaluate use of multiple PRSs based on different shrinkage parameters, and use of pseudo-validated PRS (PRScs)
################################
# Create a file listing the predictors files
pheno=$(echo Height21 BMI21 GCSE ADHD)
gwas=$(echo HEIG03 BODY11 EDUC03 ADHD04)
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
# 1KG
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}_P4/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}_GCTB_203/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}_GCTB_203_robust/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
# GCTB
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_P4.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_P4/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_GCTB_203.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_GCTB_203/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/1KG_ref/SBayesR/${gwas_i}_GCTBref_GCTB_203_robust/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
# UKB ref
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/UKBB_ref/SBayesR/${gwas_i}/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/UKBB_ref/SBayesR/${gwas_i}_P4/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/UKBB_ref/SBayesR/${gwas_i}_GCTB_203/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
cat > /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups <<EOF
predictors
/users/k1806347/brc_scratch/Data/TEDS/prs_comparison/UKBB_ref/SBayesR/${gwas_i}_GCTB_203_robust/TEDS.w_hm3.${gwas_i}.EUR.SBayesR_profiles
EOF
done
# Derive and evaluate
pheno=$(echo Height21 BMI21 GCSE ADHD)
gwas=$(echo HEIG03 BODY11 EDUC03 ADHD04)
prev=$(echo NA NA NA NA)
####
# 1KG ref
####
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups
done
# P < 0.4
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_P4.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups
done
# GCTB v2.03
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups
done
# GCTB v2.03 robust
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups
done
####
# GCTB ref
####
# Original
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref.EUR-PRSs_SBayesR.predictor_groups
done
# P<0.4
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_P4.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_P4.EUR-PRSs_SBayesR.predictor_groups
done
# GCTB v2.03
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_GCTB_203.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_GCTB_203.EUR-PRSs_SBayesR.predictor_groups
done
# GCTB v2.03 robust
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups
done
###
# UKBB ref
###
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs_SBayesR.predictor_groups
done
# P<0.4
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_P4.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_P4.EUR-PRSs_SBayesR.predictor_groups
done
# GCTB v2.03
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_GCTB_203.EUR-PRSs_SBayesR.predictor_groups
done
# GCTB v2.03 robust
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 5G -n 1 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR \
--n_core 1 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}_GCTB_203_robust.EUR-PRSs_SBayesR.predictor_groups
done
Compare all methods
# We need to read in polygenic scores based on a range of parameters, pseudovalidated scores as a seperate group.
# Create a file listing the predictors files
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno<-c('Height21','BMI21','GCSE','ADHD')
gwas<-c('HEIG03','BODY11','EDUC03','ADHD04')
GCTB_hsq_all<-read.csv('/users/k1806347/brc_scratch/Analyses/PRS_comparison/SBayesR_H2_comp.csv')
Processing<-c('','_P4')
Processing_name<-c('Original','P<0.4')
for(i in 1:length(pheno)){
for(ref in c('1KG','UKBB')){
if(ref == '1KG'){
dat_path<-paste0(TEDS_output_dir,'/prs_comparison/1KG_ref/')
} else {
dat_path<-paste0(TEDS_output_dir,'/prs_comparison/UKBB_ref/')
}
pred_file<-NULL
# pT+clump (sparse)
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'pt_clump/',gwas[i],'/TEDS.w_hm3.',gwas[i],'.EUR.profiles'),
group='pT+clump'))
# lassosum
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'lassosum/',gwas[i],'/TEDS.w_hm3.',gwas[i],'.EUR.lassosum_profiles'),
group='lassosum'))
if(ref == '1KG'){
# PRScs
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'PRScs/',gwas[i],'/TEDS.w_hm3.',gwas[i],'.EUR.PRScs_profiles'),
group='PRScs'))
}
# SBLUP
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'SBLUP/',gwas[i],'/TEDS.w_hm3.',gwas[i],'.EUR.SBLUP_profiles'),
group='SBLUP'))
# SBayesR
# Use robust mode
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'SBayesR/',gwas[i],'_GCTB_203_robust/TEDS.w_hm3.',gwas[i],'.EUR.SBayesR_profiles'),
group='SBayesR'))
# LDPred
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'LDPred/',gwas[i],'/TEDS.w_hm3.',gwas[i],'.EUR.LDPred_profiles'),
group='LDPred'))
# DBSLMM
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'DBSLMM/',gwas[i],'/TEDS.w_hm3.',gwas[i],'.EUR.DBSLMM_profiles'),
group='DBSLMM'))
# LDPred2
pred_file<-rbind(pred_file,data.frame( predictors=paste0(dat_path,'LDPred2/',gwas[i],'/TEDS.w_hm3.',gwas[i],'.EUR.LDPred_profiles'),
group='LDPred2'))
# Write out list of predictors with groups
if(ref == '1KG'){
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/TEDS.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'), row.names=F, col.names=T, quote=F)
} else {
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/TEDS.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'), row.names=F, col.names=T, quote=F)
}
}
}# Run Model_builder_V2.R
pheno=$(echo Height21 BMI21 GCSE ADHD)
gwas=$(echo HEIG03 BODY11 EDUC03 ADHD04)
prev=$(echo NA NA NA NA)
# 1KG ref
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp.predictor_groups
done
# UKB ref
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs.AllMethodComp \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.UKBB_ref.${gwas_i}.EUR-PRSs.AllMethodComp.predictor_groups
done
Compare all methods with PCs regressed
#############
# Regress project PCs from all polygenic scores
#############
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
pheno<-c('Height21','BMI21','GCSE','ADHD')
gwas<-c('HEIG03','BODY11','EDUC03','ADHD04')
# Read in PCs
pcs<-fread(paste0('/users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.eigenvec'))
# Extract the first 20PCs
pcs<-pcs[,c('FID','IID',paste0('PC',1:20))]
for(ref in c('1KG')){
for(i in 1:length(gwas)){
# Read in list of predictors
if(ref == '1KG'){
pred_file<-fread(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/TEDS.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'))
} else {
pred_file<-fread(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/TEDS.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.predictor_groups'))
}
for(file in 1:dim(pred_file)[1]){
# Read in predictors
file_i<-fread(pred_file$predictors[file])
# Merge with PCs
file_i_pcs<-merge(file_i, pcs, by=c('FID','IID'))
for(col_i in names(file_i_pcs)[grepl(gwas[i], names(file_i_pcs))]){
file_i_pcs_col_i<-file_i_pcs[,c(col_i,paste0('PC',1:20)), with=F]
names(file_i_pcs_col_i)[names(file_i_pcs_col_i) == col_i]<-'y'
# Regress covariates, scale residuals and round to three decimal places
file_i_pcs[[paste0(col_i,'_resid')]]<-round(as.numeric(scale(resid(lm(as.formula(paste0('y ~ ', paste(paste0('PC', 1:20), collapse=' + '))), data=file_i_pcs_col_i)))),3)
}
# Extract residual columns
file_i_resid<-file_i_pcs[,c('FID','IID',names(file_i_pcs)[grepl('_resid',names(file_i_pcs))]), with=F]
# Save the residuals
output<-gsub(paste0(ref,'_ref'),paste0(ref,'_ref_PCresid'),pred_file$predictors[file])
tmp<-sub('.*/','',output)
output_dir<-sub(paste0(tmp,'*.'),'',output)
system(paste0('mkdir -p ',output_dir))
write.table(file_i_resid, output, col.names=T, row.names=F, quote=F)
}
# Write out new list of predictors for residuals
pred_file_resid<-pred_file
pred_file_resid$predictors<-gsub(paste0(ref,'_ref'),paste0(ref,'_ref_PCresid'),pred_file$predictors)
if(ref == '1KG'){
write.table(pred_file_resid, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/TEDS.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.PCresid.predictor_groups'), col.names=T, row.names=F, quote=F)
} else {
write.table(pred_file_resid, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/TEDS.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.PCresid.predictor_groups'), col.names=T, row.names=F, quote=F)
}
}
}
# Run Model_builder_V2.R
pheno=$(echo Height21 BMI21 GCSE ADHD)
gwas=$(echo HEIG03 BODY11 EDUC03 ADHD04)
prev=$(echo NA NA NA NA)
# 1KG ref
for i in $(seq 1 4);do
pheno_i=$(echo ${pheno} | cut -f ${i} -d ' ')
gwas_i=$(echo ${gwas} | cut -f ${i} -d ' ')
prev_i=$(echo ${prev} | cut -f ${i} -d ' ')
sbatch --mem 10G -n 2 -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2.R \
--pheno /users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_${pheno_i}.txt \
--keep /users/k1806347/brc_scratch/Data/TEDS/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep \
--out /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp.PCresid \
--n_core 2 \
--compare_predictors T \
--assoc T \
--outcome_pop_prev ${prev_i} \
--predictors /users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/${pheno_i}/Association_withPRSs/TEDS.w_hm3.${gwas_i}.EUR-PRSs.AllMethodComp.PCresid.predictor_groups
done
3.7 Estimating polygenicity of phenotypes
3.7.1 SBayesR
Show code using SBayesR results
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config')
gwas<-c('DEPR06','COLL01','HEIG03','BODY04','DIAB05','COAD01','CROH01','SCLE03','RHEU02','EDUC03','ADHD04','BODY11','PRCA01','BRCA01')
pop_prev<-c(0.15,NA,NA,NA,0.05,0.03,0.013,0.00164,0.005,NA,0.05,NA,0.125,0.125)
samp_prev<-c(0.28,NA,NA,NA,0.168,0.33,0.285,0.36,0.246,NA,0.364,NA,0.564,0.537)
h2l_R2 <- function(k, r2, p) {
# K baseline disease risk
# r2 from a linear regression model attributable to genomic profile risk score
# P proportion of sample that are cases
# calculates proportion of variance explained on the liability scale
#from ABC at http://www.complextraitgenomics.com/software/
#Lee SH, Goddard ME, Wray NR, Visscher PM. (2012) A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012 Apr;36(3):214-24.
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
h2l_R2 = C*r2 / (1 + C*theta*r2)
}
se_h2l_R2 <- function(k,h2,se, p) {
# K baseline disease risk
# r2 from a linear regression model attributable to genomic profile risk score
# P proportion of sample that are cases
# calculates proportion of variance explained on the liability scale
#from ABC at http://www.complextraitgenomics.com/software/
#Lee SH, Goddard ME, Wray NR, Visscher PM. (2012) A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012 Apr;36(3):214-24.
#SE on the liability (From a Taylor series expansion)
#var(h2l_r2) = [d(h2l_r2)/d(R2v)]^2*var(R2v) with d being calculus differentiation
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
se_h2l_R2 = C*(1-h2*theta)*se
}
res_all<-NULL
for(i in 1:length(gwas)){
# Read in SBayesR parRes
res<-fread(paste0(Geno_1KG_dir,'/Score_files_for_polygenic/SBayesR/',gwas[i],'_GCTBref_GCTB_203_robust/GWAS_sumstats_SBayesR.GW.parRes'))
if(is.na(pop_prev[i])){
res_all<-rbind(res_all, data.frame(GWAS=gwas[i],
hsq_mean=res$Mean[res$Par == 'hsq'],
hsq_sd=res$Mean[res$Par == 'hsq'],
NnzSnp_mean=res$Mean[res$Par == 'NnzSnp'],
NnzSnp_sd=res$SD[res$Par == 'NnzSnp']))
} else {
res_all<-rbind(res_all, data.frame(GWAS=gwas[i],
hsq_mean=h2l_R2(k=pop_prev[i], r2=res$Mean[res$Par == 'hsq'], p=samp_prev[i]),
hsq_sd=se_h2l_R2(k=pop_prev[i], h2=res$Mean[res$Par == 'hsq'], se=res$SD[res$Par == 'hsq'], p=samp_prev[i]),
NnzSnp_mean=res$Mean[res$Par == 'NnzSnp'],
NnzSnp_sd=res$SD[res$Par == 'NnzSnp']))
}
}
# Looks reasonable with robust paramterisation, except for BMI when using Locke et al sumstats.3.7.2 AVENGEME
3.7.2.1 UK Biobank
Show code using AVENGEME results
gwas<-c('DEPR06','COLL01','HEIG03','BODY04','DIAB05','COAD01','CROH01','SCLE03','RHEU02','PRCA01','BRCA01')
pheno<-c('Depression','Intelligence','Height','BMI','T2D','CAD','IBD', 'MultiScler','RheuArth','Prostate_Cancer','Breast_Cancer')
prev=c(0.15,NA,NA,NA,0.05,0.03,0.013,0.00164,0.005,0.125,0.125)
gwas_desc<-read.csv("/users/k1806347/brc_scratch/Data/GWAS_sumstats/UKBB_phenotype_GWAS_descrip.csv")
library(avengeme)
PRS_res<-list()
nsnp_logs<-list()
for(i in 1:length(gwas)){
res_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/UKBB.w_hm3.',gwas[i],'.EUR-PRSs.assoc.txt'), header=T, stringsAsFactors=F)
res_i<-res_i[1:dim(res_i)[1]-1,]
if(sum(grepl('Estimate',names(res_i))) == 1){
res_i$Z<-res_i$Estimate/res_i$SE
} else {
res_i$Z<-res_i$BETA/res_i$SE
}
PRS_res[[gwas[i]]]<-res_i
nsnp_log<-read.table(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/pt_clump/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.NSNP_per_pT'), header=T)
nsnp_logs[[gwas[i]]]<-nsnp_log
}
mod_res_all<-NULL
for(i in 1:length(gwas)){
if(is.na(prev[i])){
targ_N<-PRS_res[[gwas[i]]]$N[1]
mod_res<-estimatePolygenicModel(p=PRS_res[[gwas[i]]]$Z,
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
n=c(gwas_desc$N[gwas_desc$Code == gwas[i]], targ_N),
pupper = c(0,1e-8,1e-06,1e-04,1e-02,0.1,0.2,0.3,0.4,0.5),
binary = c(FALSE, FALSE),
prevalence = c(NA, NA),
sampling = c(NA, NA),
fixvg2pi02 = T,
alpha = 0.05)
mod_res_all<-rbind(mod_res_all,data.frame(Phenotype=pheno[i],
GWAS=gwas[i],
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
vg_est=mod_res$vg[1],
vg_lowCI=mod_res$vg[2],
vg_highCI=mod_res$vg[3],
pi0_est=mod_res$pi0[1],
pi0_lowCI=mod_res$pi0[2],
pi0_highCI=mod_res$pi0[3]))
} else {
targ_N<-PRS_res[[gwas[i]]]$N[1]
targ_N_Ca<-PRS_res[[gwas[i]]]$Ncas[1]
targ_N_Co<-PRS_res[[gwas[i]]]$Ncon[1]
mod_res<-estimatePolygenicModel(p=PRS_res[[gwas[i]]]$Z,
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
n=c(gwas_desc$N[gwas_desc$Code == gwas[i]], targ_N),
pupper = c(0,1e-8,1e-06,1e-04,1e-02,0.1,0.2,0.3,0.4,0.5),
binary = c(T, T),
prevalence = prev[i],
sampling = c(gwas_desc$Ncase[gwas_desc$Code == gwas[i]]/(gwas_desc$Ncase[gwas_desc$Code == gwas[i]]+gwas_desc$Ncontrol[gwas_desc$Code == gwas[i]]), targ_N_Ca/(targ_N_Ca+targ_N_Co)),
fixvg2pi02 = T,
alpha = 0.05)
mod_res_all<-rbind(mod_res_all,data.frame(Phenotype=pheno[i],
GWAS=gwas[i],
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
vg_est=mod_res$vg[1],
vg_lowCI=mod_res$vg[2],
vg_highCI=mod_res$vg[3],
pi0_est=mod_res$pi0[1],
pi0_lowCI=mod_res$pi0[2],
pi0_highCI=mod_res$pi0[3]))
}
}
# A pi0 of 0.98 divides the phenotype nicely into two groups
mod_res_all$HighPoly<-F
mod_res_all$HighPoly[mod_res_all$pi0_est < 0.96]<-T
# T2D, IBD, MultiScler, RheuArth, Prostate_Cancer, and Breast_Cancer are the low polygenicity outcomes.
write.csv(mod_res_all, '/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/PRS_AVENGME_res.csv', row.names=F)3.7.2.2 TEDS
Show code using AVENGEME results
prev=c(NA,NA,NA,NA)
pheno<-c('Height21', 'BMI21', 'GCSE', 'ADHD')
gwas<-c('HEIG03', 'BODY11', 'EDUC03', 'ADHD04')
gwas_desc<-read.csv("/users/k1806347/brc_scratch/Data/GWAS_sumstats/TEDS_phenotype_GWAS_descrip.csv")
library(avengeme)
PRS_res<-list()
nsnp_logs<-list()
for(i in 1:length(gwas)){
res_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/TEDS.w_hm3.',gwas[i],'.EUR-PRSs.assoc.txt'), header=T, stringsAsFactors=F)
res_i<-res_i[1:dim(res_i)[1]-1,]
if(sum(grepl('Estimate',names(res_i))) == 1){
res_i$Z<-res_i$Estimate/res_i$SE
} else {
res_i$Z<-res_i$BETA/res_i$SE
}
PRS_res[[gwas[i]]]<-res_i
nsnp_log<-read.table(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/pt_clump/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.NSNP_per_pT'), header=T)
nsnp_logs[[gwas[i]]]<-nsnp_log
}
mod_res_all<-NULL
for(i in 1:length(gwas)){
if(is.na(prev[i])){
targ_N<-PRS_res[[gwas[i]]]$N[1]
mod_res<-estimatePolygenicModel(p=PRS_res[[gwas[i]]]$Z,
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
n=c(gwas_desc$N[gwas_desc$Code == gwas[i]], targ_N),
pupper = c(0,1e-8,1e-06,1e-04,1e-02,0.1,0.2,0.3,0.4,0.5),
binary = c(FALSE, FALSE),
prevalence = c(NA, NA),
sampling = c(NA, NA),
fixvg2pi02 = T,
alpha = 0.05)
mod_res_all<-rbind(mod_res_all,data.frame(Phenotype=pheno[i],
GWAS=gwas[i],
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
vg_est=mod_res$vg[1],
vg_lowCI=mod_res$vg[2],
vg_highCI=mod_res$vg[3],
pi0_est=mod_res$pi0[1],
pi0_lowCI=mod_res$pi0[2],
pi0_highCI=mod_res$pi0[3]))
} else {
targ_N<-PRS_res[[gwas[i]]]$N[1]
targ_N_Ca<-PRS_res[[gwas[i]]]$Ncas[1]
targ_N_Co<-PRS_res[[gwas[i]]]$Ncon[1]
mod_res<-estimatePolygenicModel(p=PRS_res[[gwas[i]]]$Z,
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
n=c(gwas_desc$N[gwas_desc$Code == gwas[i]], targ_N),
pupper = c(0,1e-8,1e-06,1e-04,1e-02,0.1,0.2,0.3,0.4,0.5),
binary = c(T, T),
prevalence = prev[i],
sampling = c(gwas_desc$Ncase[gwas_desc$Code == gwas[i]]/(gwas_desc$Ncase[gwas_desc$Code == gwas[i]]+gwas_desc$Ncontrol[gwas_desc$Code == gwas[i]]), targ_N_Ca/(targ_N_Ca+targ_N_Co)),
fixvg2pi02 = T,
alpha = 0.05)
mod_res_all<-rbind(mod_res_all,data.frame(Phenotype=pheno[i],
GWAS=gwas[i],
nsnp=nsnp_logs[[gwas[i]]]$NSNP[length(nsnp_logs[[gwas[i]]]$NSNP)],
vg_est=mod_res$vg[1],
vg_lowCI=mod_res$vg[2],
vg_highCI=mod_res$vg[3],
pi0_est=mod_res$pi0[1],
pi0_lowCI=mod_res$pi0[2],
pi0_highCI=mod_res$pi0[3]))
}
}
# A pi0 of 0.96 identifies all phenotype in TEDS to high polygenicity, which is accurate.
mod_res_all$HighPoly<-F
mod_res_all$HighPoly[mod_res_all$pi0_est < 0.96]<-T
write.csv(mod_res_all, '/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/PRS_AVENGME_res.csv', row.names=F)4 Results
Create a sample size table
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
# Read in a log file for each phenotype and extracr the number of individuals
# UKB
gwas<-c('DEPR06','COLL01','HEIG03','BODY04','DIAB05','COAD01','CROH01','SCLE03','RHEU02','PRCA01','BRCA01')
pheno<-c('Depression','Intelligence','Height','BMI','T2D','CAD','IBD', 'MultiScler','RheuArth','Prostate_Cancer','Breast_Cancer')
keep<-fread('/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/ukb18177_glanville_post_qc_id_list.UpdateIDs.fam')
UKB_N_tab<-NULL
for(i in 1:length(pheno)){
pheno_i<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',pheno[i],'.txt'))
names(pheno_i)<-c('FID','IID','pheno')
pheno_i_keep<-merge(pheno_i, keep, by.x=c('FID','IID'), by.y=c('V1','V2'))
if(length(unique(pheno_i_keep$pheno)) > 2){
UKB_N_tab<-rbind(UKB_N_tab, data.frame(Phenotype=pheno[i],
N=dim(pheno_i_keep)[1],
Ncase=NA,
Ncont=NA))
} else {
UKB_N_tab<-rbind(UKB_N_tab, data.frame(Phenotype=pheno[i],
N=dim(pheno_i_keep)[1],
Ncase=sum(pheno_i_keep$pheno == 0),
Ncont=sum(pheno_i_keep$pheno == 1)))
}
}
write.csv(UKB_N_tab, '/users/k1806347/brc_scratch/Analyses/PRS_comparison/UKBB_outcomes_for_prediction/UKB_N_table.csv', row.names=F, quote=F)
# TEDS
pheno<-c('Height21', 'BMI21', 'GCSE', 'ADHD')
gwas<-c('HEIG03', 'BODY11', 'EDUC03', 'ADHD04')
keep<-fread(paste0(TEDS_output_dir,'/Projected_PCs/Ancestry_idenitfier/TEDS.w_hm3.AllAncestry.EUR.keep'))
TEDS_N_tab<-NULL
for(i in 1:length(pheno)){
pheno_i<-fread(paste0('/users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_',pheno[i],'.txt'))
names(pheno_i)<-c('FID','IID','pheno')
pheno_i_keep<-merge(pheno_i, keep, by.x=c('FID','IID'), by.y=c('V1','V2'))
TEDS_N_tab<-rbind(TEDS_N_tab, data.frame(Phenotype=pheno[i],
N=dim(pheno_i_keep)[1],
Ncase=NA,
Ncont=NA))
}
write.csv(TEDS_N_tab, '/users/k1806347/brc_scratch/Analyses/PRS_comparison/TEDS_outcomes_for_prediction/TEDS_N_table.csv', row.names=F, quote=F)Show sample size tables
| Phenotype | N | Ncase | Ncont |
|---|---|---|---|
| Depression | 50000 | 25000 | 25000 |
| Intelligence | 50000 | NA | NA |
| Height | 50000 | NA | NA |
| BMI | 50000 | NA | NA |
| T2D | 50000 | 35112 | 14888 |
| CAD | 50000 | 25000 | 25000 |
| IBD | 50000 | 46539 | 3461 |
| MultiScler | 50000 | 48863 | 1137 |
| RheuArth | 50000 | 46592 | 3408 |
| Prostate_Cancer | 50000 | 47073 | 2927 |
| Breast_Cancer | 50000 | 41488 | 8512 |
| Phenotype | N | Ncase | Ncont |
|---|---|---|---|
| Height21 | 5455 | NA | NA |
| BMI21 | 5220 | NA | NA |
| GCSE | 7296 | NA | NA |
| ADHD | 7880 | NA | NA |
Summarise and plot the results
Files<-c('UKBB','UKBB.UKBB_ref','TEDS','TEDS.UKBB_ref')
Samples<-c('UKBB','UKBB','TEDS','TEDS')
Files_descrip<-c('UKB target with 1KG ref','UKB target with UKB ref','TEDS target with 1KG ref', 'TEDS target with UKB ref')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
meta_res_eval<-NULL
meta_res_comp<-NULL
meta_res_eval_plots_cross<-list()
meta_res_eval_plots_indep<-list()
meta_res_eval_diff_plots_cross<-list()
meta_res_eval_diff_plots_indep<-list()
meta_res_eval_diff_matrix_cross<-list()
meta_res_eval_diff_matrix_indep<-list()
for(file in 1:length(Files)){
if(Samples[file] == 'UKBB'){
pheno<-c('Depression','Intelligence','Height','BMI','T2D','CAD','IBD', 'MultiScler','RheuArth','Prostate_Cancer','Breast_Cancer')
gwas<-c('DEPR06','COLL01','HEIG03','BODY04','DIAB05','COAD01','CROH01','SCLE03','RHEU02','PRCA01','BRCA01')
} else {
pheno<-c('Height21', 'BMI21', 'GCSE', 'ADHD')
gwas<-c('HEIG03', 'BODY11', 'EDUC03', 'ADHD04')
}
if(Files[file] == 'UKBB' | Files[file] == 'TEDS'){
##################
# Create a table showing different pT+clump methods
##################
# This is just a quick check that the dense and non-nested approaches improve prediction
res_eval<-NULL
for(i in 1:length(pheno)){
pT_clump<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs.pred_eval.txt'), header=T, stringsAsFactors=F)
pT_clump$Method<-"pT+clump (sparse)"
Param_tmp<-sort(as.numeric(gsub('e.','e-',gsub('_group','',gsub(paste0(gwas[i],'.'),'',pT_clump$Model)))))
pT_clump$Param<-as.character(as.numeric(gsub('e.','e-',gsub('_group','',gsub(paste0(gwas[i],'.'),'',pT_clump$Model)))))
pT_clump$Param[dim(pT_clump)[1]]<-'Multi-PRS'
pT_clump<-pT_clump[match(c(Param_tmp,'Multi-PRS'), pT_clump$Param),]
pT_clump$Model<-NULL
pT_clump$Group<-NA
pT_clump$Group[pT_clump$CrossVal_R == max(abs(pT_clump$CrossVal_R[pT_clump$Param != 'Multi-PRS']))]<-'10FCVal'
pT_clump$Group[pT_clump$Param == 'Multi-PRS']<-'Multi-PRS'
pT_clump<-pT_clump[!is.na(pT_clump$Group),]
pT_clump_nn<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs_nonnested.pred_eval.txt'), header=T, stringsAsFactors=F)
pT_clump_nn$Method<-"pT+clump (non-nested)"
pT_clump_nn$Param<-as.character(as.numeric(gsub('e.','e-',gsub('_group','',gsub(paste0(gwas[i],'.'),'',pT_clump_nn$Model)))))
for(j in (length(pT_clump_nn$Param)-1):1){
if(j==1){
pT_clump_nn$Param[j]<-paste0('0 - ',pT_clump_nn$Param[j])
} else {
pT_clump_nn$Param[j]<-paste0(pT_clump_nn$Param[j-1],' - ',pT_clump_nn$Param[j])
}
}
pT_clump_nn$Param[dim(pT_clump_nn)[1]]<-'Multi-PRS'
pT_clump_nn$Model<-NULL
pT_clump_nn$Group<-NA
pT_clump_nn$Group[pT_clump_nn$CrossVal_R == max(abs(pT_clump_nn$CrossVal_R[pT_clump_nn$Param != 'Multi-PRS']))]<-'10FCVal'
pT_clump_nn$Group[pT_clump_nn$Param == 'Multi-PRS']<-'Multi-PRS'
pT_clump_nn<-pT_clump_nn[!is.na(pT_clump_nn$Group),]
pT_clump_dense<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs-dense.pred_eval.txt'), header=T, stringsAsFactors=F)
pT_clump_dense$Method<-"pT+clump (dense)"
Param_tmp<-sort(as.numeric(gsub('e.','e-',gsub('_group','',gsub(paste0(gwas[i],'.'),'',pT_clump_dense$Model)))))
pT_clump_dense$Param<-as.character(as.numeric(gsub('e.','e-',gsub('_group','',gsub(paste0(gwas[i],'.'),'',pT_clump_dense$Model)))))
pT_clump_dense$Param[dim(pT_clump_dense)[1]]<-'Multi-PRS'
pT_clump_dense<-pT_clump_dense[match(c(Param_tmp,'Multi-PRS'), pT_clump_dense$Param),]
pT_clump_dense$Model<-NULL
pT_clump_dense$Group<-NA
pT_clump_dense$Group[pT_clump_dense$CrossVal_R == max(abs(pT_clump_dense$CrossVal_R[pT_clump_dense$Param != 'Multi-PRS']))][1]<-'10FCVal'
pT_clump_dense$Group[pT_clump_dense$Param == 'Multi-PRS']<-'Multi-PRS'
pT_clump_dense<-pT_clump_dense[!is.na(pT_clump_dense$Group),]
res_eval_i<-do.call(rbind, list(pT_clump, pT_clump_nn, pT_clump_dense))
res_eval_i$Phenotype<-pheno[i]
res_eval_i$CrossVal_OR<-NULL
res_eval_i$CrossVal_LowCI<-NULL
res_eval_i$CrossVal_HighCI<-NULL
res_eval_i$Cross_LiabR2<-NULL
res_eval_i$Cross_AUC<-NULL
res_eval_i$CrossVal_Ncas<-NULL
res_eval_i$CrossVal_Ncon<-NULL
res_eval_i$IndepVal_OR<-NULL
res_eval_i$IndepVal_LowCI<-NULL
res_eval_i$IndepVal_HighCI<-NULL
res_eval_i$Indep_LiabR2<-NULL
res_eval_i$Indep_AUC<-NULL
res_eval_i$IndepVal_Ncas<-NULL
res_eval_i$IndepVal_Ncon<-NULL
res_eval<-rbind(res_eval, res_eval_i)
}
res_eval<-res_eval[,c('Phenotype','Method','Group','Param','CrossVal_R','CrossVal_R_SE','CrossVal_pval','IndepVal_R','IndepVal_R_SE','IndepVal_pval')]
names(res_eval)[3]<-'Model'
write.csv(res_eval, paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',Samples[file],'_pT_clump_eval.csv'), row.names=F, quote=F)
}
##################
# Organise, make tables and figures for per phenotype results
##################
res_eval<-NULL
res_comp<-NULL
res_eval_plots_cross<-list()
res_eval_plots_indep<-list()
res_eval_diff_plots_cross<-list()
res_eval_diff_plots_indep<-list()
res_eval_diff_matrix_cross<-list()
res_eval_diff_matrix_indep<-list()
for(i in 1:length(pheno)){
if(grepl('UKBB_ref',Files[file])){
res_eval_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.pred_eval.txt'), header=T, stringsAsFactors=F)
res_comp_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs.AllMethodComp.pred_comp.txt'), header=T, stringsAsFactors=F)
} else {
res_eval_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.pred_eval.txt'), header=T, stringsAsFactors=F)
res_comp_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.pred_comp.txt'), header=T, stringsAsFactors=F)
}
# Idenitfy model names for 10FCVal, multi-PRS and pseudoval
selected_models<-NULL
##
# pT+clump (sparse)
##
# Identify best pT in 10FCVal
res_eval_i_per_pT<-res_eval_i[grepl('pT.clump.PredFile', res_eval_i$Model),]
selected_models<-rbind(selected_models,data.frame(Test='pT+clump.10FCVal', Label=
res_eval_i_per_pT[res_eval_i_per_pT$CrossVal_R == max(res_eval_i_per_pT$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='pT+clump.MultiPRS', Label='pT.clump'))
##
# lassosum
##
# Identify best pT in 10FCVal
res_eval_i_per_lassosum<-res_eval_i[grepl('lassosum.PredFile', res_eval_i$Model),]
selected_models<-rbind(selected_models,data.frame(Test='lassosum.10FCVal', Label=
res_eval_i_per_lassosum[res_eval_i_per_lassosum$CrossVal_R == max(res_eval_i_per_lassosum$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='lassosum.MultiPRS', Label='lassosum'))
# Identify pseudoVal
lassosum_val<-read.table(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/lassosum/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.log'), sep='&')
lassosum_val_s<-as.numeric(gsub('s = ','',lassosum_val$V1[grepl('s = ',lassosum_val$V1)]))
lassosum_val_lambda<-as.numeric(gsub('lambda =','',lassosum_val$V1[grepl('lambda =',lassosum_val$V1)]))
lassosum_val_param<-paste0('s',lassosum_val_s,'.lambda',lassosum_val_lambda)
lassosum_val_param<-substr(lassosum_val_param, 1, nchar(lassosum_val_param)-1)
selected_models<-rbind(selected_models,data.frame(Test='lassosum.PseudoVal', Label=res_eval_i[grepl(lassosum_val_param, res_eval_i$Model),]$Model[1]))
##
# PRScs
##
# Identify best pT in 10FCVal
res_eval_i_per_PRScs<-res_eval_i[grepl('PRScs.PredFile', res_eval_i$Model),]
res_eval_i_per_PRScs<-res_eval_i_per_PRScs[!grepl('phiauto', res_eval_i_per_PRScs$Model),]
selected_models<-rbind(selected_models,data.frame(Test='PRScs.10FCVal', Label=res_eval_i_per_PRScs[res_eval_i_per_PRScs$CrossVal_R == max(res_eval_i_per_PRScs$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='PRScs.MultiPRS', Label='PRScs'))
# Identify pseudoval
res_eval_i_per_PRScs<-res_eval_i[grepl('PRScs.PredFile', res_eval_i$Model),]
res_eval_i_per_PRScs<-res_eval_i_per_PRScs[grepl('phiauto', res_eval_i_per_PRScs$Model),]
selected_models<-rbind(selected_models,data.frame(Test='PRScs.PseudoVal', Label=res_eval_i_per_PRScs[res_eval_i_per_PRScs$CrossVal_R == max(res_eval_i_per_PRScs$CrossVal_R),]$Model[1]))
##
# SBLUP
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='SBLUP.Inf', Label='SBLUP'))
##
# SBayesR
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='SBayesR.PseudoVal', Label='SBayesR'))
##
# LDPred1
##
# Identify best pT in 10FCVal
res_eval_i_per_LDPred<-res_eval_i[grepl('LDPred.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred<-res_eval_i_per_LDPred[!grepl('.LDpred.inf', res_eval_i_per_LDPred$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.10FCVal', Label=res_eval_i_per_LDPred[res_eval_i_per_LDPred$CrossVal_R == max(res_eval_i_per_LDPred$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.MultiPRS', Label='LDPred'))
# Identify pseudoval
res_eval_i_per_LDPred<-res_eval_i[grepl('LDPred.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred<-res_eval_i_per_LDPred[grepl('.LDpred.inf', res_eval_i_per_LDPred$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.Inf', Label=res_eval_i_per_LDPred[res_eval_i_per_LDPred$CrossVal_R == max(res_eval_i_per_LDPred$CrossVal_R),]$Model[1]))
##
# LDPred2
##
# Identify best pT in 10FCVal
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[!grepl('beta.inf|beta.auto', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.10FCVal', Label=res_eval_i_per_LDPred2[res_eval_i_per_LDPred2$CrossVal_R == max(res_eval_i_per_LDPred2$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.MultiPRS', Label='LDPred2'))
# Identify pseudoval
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[grepl('beta.inf', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.Inf', Label=res_eval_i_per_LDPred2$Model))
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[grepl('beta.auto', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.PseudoVal', Label=res_eval_i_per_LDPred2$Model))
##
# DBSLMM
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='DBSLMM.PseudoVal', Label='DBSLMM'))
##
# All methods
##
selected_models<-rbind(selected_models,data.frame(Test='All.MultiPRS', Label='All'))
###
# Subset selected models and format
###
# Eval
selected_models$Label_2<-paste0(gsub('_group','',selected_models$Label),'_group')
res_eval_i_select<-res_eval_i[(res_eval_i$Model %in% selected_models$Label_2),]
res_eval_i_select<-merge(res_eval_i_select, selected_models[c('Test','Label_2')], by.x='Model',by.y='Label_2')
res_eval_i_select$Phenotype<-pheno[i]
if(sum(names(res_eval_i_select) == 'CrossVal_OR') == 0){
res_eval_i_select$CrossVal_OR<-NA
res_eval_i_select$CrossVal_LowCI<-NA
res_eval_i_select$CrossVal_HighCI<-NA
res_eval_i_select$Cross_LiabR2<-NA
res_eval_i_select$Cross_AUC<-NA
res_eval_i_select$CrossVal_Ncas<-NA
res_eval_i_select$CrossVal_Ncon<-NA
res_eval_i_select$IndepVal_OR<-NA
res_eval_i_select$IndepVal_LowCI<-NA
res_eval_i_select$IndepVal_HighCI<-NA
res_eval_i_select$Indep_LiabR2<-NA
res_eval_i_select$Indep_AUC<-NA
res_eval_i_select$IndepVal_Ncas<-NA
res_eval_i_select$IndepVal_Ncon<-NA
}
res_eval_i_select<-res_eval_i_select[c('Phenotype','Test',"CrossVal_R","CrossVal_R_SE",'CrossVal_OR','CrossVal_LowCI','CrossVal_HighCI','Cross_LiabR2','Cross_AUC','CrossVal_pval','CrossVal_N','CrossVal_Ncas','CrossVal_Ncon',"IndepVal_R","IndepVal_R_SE",'IndepVal_OR','IndepVal_LowCI','IndepVal_HighCI','Indep_LiabR2','Indep_AUC','IndepVal_pval','IndepVal_N','IndepVal_Ncas','IndepVal_Ncon')]
res_eval<-rbind(res_eval,res_eval_i_select)
# Comp
selected_models$Label_3<-gsub('_group','',selected_models$Label)
res_comp_i_select<-res_comp_i[(res_comp_i$Model_1 %in% selected_models$Label_3) & (res_comp_i$Model_2 %in% selected_models$Label_3),]
res_comp_i_select<-merge(res_comp_i_select, selected_models[c('Test','Label_3')], by.x='Model_1',by.y='Label_3')
names(res_comp_i_select)[names(res_comp_i_select) == 'Test']<-'Model_1_Test'
res_comp_i_select<-merge(res_comp_i_select, selected_models[c('Test','Label_3')], by.x='Model_2',by.y='Label_3')
names(res_comp_i_select)[names(res_comp_i_select) == 'Test']<-'Model_2_Test'
res_comp_i_select$Label<-NULL
res_comp_i_select$Phenotype<-pheno[i]
res_comp_i_select<-res_comp_i_select[c('Phenotype','Model_1_Test','Model_2_Test','Model_1_Cross_R','Model_2_Cross_R','Cross_R_diff','Cross_R_diff_pval','Model_1_Indep_R','Model_2_Indep_R','Indep_R_diff','Indep_R_diff_pval')]
res_comp<-rbind(res_comp,res_comp_i_select)
###
# Create plot showing performance of each method
###
library(ggplot2)
library(cowplot)
res_eval_i_select$Method<-gsub('\\..*','',res_eval_i_select$Test)
res_eval_i_select$Model<-gsub('.*\\.','',res_eval_i_select$Test)
res_eval_i_select$Method<-factor(res_eval_i_select$Method, levels=c('pT+clump','lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
res_eval_i_select$Model<-factor(res_eval_i_select$Model, levels=c('10FCVal','MultiPRS','PseudoVal','Inf'))
res_eval_plots_cross[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=CrossVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R-CrossVal_R_SE, ymax=CrossVal_R+CrossVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$CrossVal_R[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$CrossVal_R[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y') +
theme(legend.position="top", legend.title = element_blank(), legend.box="vertical", legend.margin=margin()) +
guides(fill=guide_legend(nrow=2))
res_eval_plots_indep[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=IndepVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R-IndepVal_R_SE, ymax=IndepVal_R+IndepVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$IndepVal_R[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$IndepVal_R[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y') +
theme(legend.position="top", legend.title = element_blank(), legend.box="vertical", legend.margin=margin()) +
guides(fill=guide_legend(nrow=2))
###
# Create plot showing performance of each method compared to pT+clump
###
res_eval_i_select$CrossVal_R_diff<-res_eval_i_select$CrossVal_R-res_eval_i_select$CrossVal_R[res_eval_i_select$Test == "pT+clump.10FCVal"]
res_eval_diff_plots_cross[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=CrossVal_R_diff, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R_diff-CrossVal_R_SE, ymax=CrossVal_R_diff+CrossVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$CrossVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$CrossVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y') +
theme(legend.position="top", legend.title = element_blank(), legend.box="vertical", legend.margin=margin()) +
guides(fill=guide_legend(nrow=2))
res_eval_i_select$IndepVal_R_diff<-res_eval_i_select$IndepVal_R-res_eval_i_select$IndepVal_R[res_eval_i_select$Test == "pT+clump.10FCVal"]
res_eval_diff_plots_indep[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=IndepVal_R_diff, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R_diff-IndepVal_R_SE, ymax=IndepVal_R_diff+IndepVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$IndepVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$IndepVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y') +
theme(legend.position="top", legend.title = element_blank(), legend.box="vertical", legend.margin=margin()) +
guides(fill=guide_legend(nrow=2))
###
# Create plot showing matrix of differences with significance
###
library(reshape2)
res_comp_i_select$Cross_R_diff[res_comp_i_select$Model_1_Test == res_comp_i_select$Model_2_Test]<-NA
res_comp_i_select$Indep_R_diff[res_comp_i_select$Model_1_Test == res_comp_i_select$Model_2_Test]<-NA
res_comp_i_select$Cross_R_diff_catagory<-'NA'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff < -0.002]<-'-0.025 - -0.002'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff < -0.025]<-'-0.08 - -0.025'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff < -0.08]<-'< -0.08'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > -0.002 & res_comp_i_select$Cross_R_diff < 0.002]<-'-0.002 - 0.002'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > 0.002]<-'0.002 - 0.025'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > 0.025]<-'0.025 - 0.08'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > 0.08]<-'> 0.08'
res_comp_i_select$Cross_R_diff_catagory<-factor(res_comp_i_select$Cross_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
res_comp_i_select$Indep_R_diff_catagory<-'NA'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff < -0.002]<-'-0.025 - -0.002'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff < -0.025]<-'-0.08 - -0.025'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff < -0.08]<-'< -0.08'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > -0.002 & res_comp_i_select$Indep_R_diff < 0.002]<-'-0.002 - 0.002'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > 0.002]<-'0.002 - 0.025'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > 0.025]<-'0.025 - 0.08'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > 0.08]<-'> 0.08'
res_comp_i_select$Indep_R_diff_catagory<-factor(res_comp_i_select$Indep_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
res_comp_i_select$cross_star<-' '
res_comp_i_select$cross_star[res_comp_i_select$Cross_R_diff_pval < 0.05]<-'*'
res_comp_i_select$cross_star[res_comp_i_select$Cross_R_diff_pval < 1e-3]<-'**'
res_comp_i_select$cross_star[res_comp_i_select$Cross_R_diff_pval < 1e-6]<-'***'
res_comp_i_select$indep_star<-' '
res_comp_i_select$indep_star[res_comp_i_select$Indep_R_diff_pval < 0.05]<-'*'
res_comp_i_select$indep_star[res_comp_i_select$Indep_R_diff_pval < 1e-3]<-'**'
res_comp_i_select$indep_star[res_comp_i_select$Indep_R_diff_pval < 1e-6]<-'***'
library(RColorBrewer)
res_eval_diff_matrix_cross[[pheno[i]]]<-ggplot(data = res_comp_i_select, aes(Model_2_Test, Model_1_Test, fill=Cross_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=pheno[i], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=res_comp_i_select, aes(Model_2_Test, Model_1_Test, label = cross_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(res_comp_i_select$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
res_eval_diff_matrix_indep[[pheno[i]]]<-ggplot(data = res_comp_i_select, aes(Model_2_Test, Model_1_Test, fill=Indep_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=pheno[i], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=res_comp_i_select, aes(Model_2_Test, Model_1_Test, label = indep_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(res_comp_i_select$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
}
write.csv(res_eval, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'.csv'), row.names=F, quote=F)
write.csv(res_comp, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'_diff.csv'), row.names=F, quote=F)
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_CrossVal_',Files[file],'.png'), units='px', res=300, width=2450, height=1000*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_plots_cross, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_IndepVal_',Files[file],'.png'), units='px', res=300, width=2450, height=1000*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_plots_indep, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_pTDiff_CrossVal_',Files[file],'.png'), units='px', res=300, width=2450, height=1000*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_diff_plots_cross, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_pTDiff_IndepVal_',Files[file],'.png'), units='px', res=300, width=2450, height=1000*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_diff_plots_indep, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'_diff_CrossVal.png'), units='px', res=300, width=2450, height=1750*length(res_eval_diff_matrix_cross))
print(plot_grid(plotlist=res_eval_diff_matrix_cross, ncol = 1))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'_diff_IndepVal.png'), units='px', res=300, width=2450, height=1750*length(res_eval_diff_matrix_cross))
print(plot_grid(plotlist=res_eval_diff_matrix_indep, ncol = 1))
dev.off()
###################
# Average results across phenotypes, creates tables and figures
###################
res_eval$Method<-gsub('\\..*','',res_eval$Test)
res_eval$Model<-gsub('.*\\.','',res_eval$Test)
res_eval$Phenotype<-factor(res_eval$Phenotype, level=unique(res_eval$Phenotype))
res_eval$Method<-factor(res_eval$Method, level=c("pT+clump",'lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
res_eval$Model<-factor(res_eval$Model, level=c('10FCVal','MultiPRS','PseudoVal','Inf'))
res_eval<-res_eval[order(res_eval$Phenotype, res_eval$Method, res_eval$Model),]
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
if(Samples[file] == 'UKBB'){
for(pheno_i in 1:length(pheno)){
if(pheno_i==1){
pheno_i_dat<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-pheno_i_dat
} else {
pheno_i_dat<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-merge(pheno_dat, pheno_i_dat, by=c('FID','IID'), all=T)
}
}
} else {
for(pheno_i in 1:length(pheno)){
if(pheno_i==1){
pheno_i_dat<-fread(paste0('/users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-pheno_i_dat
} else {
pheno_i_dat<-fread(paste0('/users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-merge(pheno_dat, pheno_i_dat, by=c('FID','IID'), all=T)
}
}
}
# Calculate corelation between all phenotypes
cors<-abs(cor(as.matrix(pheno_dat[,-1:-2]), use='p'))
cors<-cors[unique(res_eval$Phenotype,),unique(res_eval$Phenotype,)]
cors[is.na(cors)]<-0
# Run aggregate function
library(MAd)
meta_res_eval<-NULL
for(i in unique(res_eval$Test)){
res_eval_i<-res_eval[res_eval$Test == i,]
res_eval_i$Sample<-'Target'
cors_i<-cors[(dimnames(cors)[[1]] %in% res_eval_i$Phenotype),(dimnames(cors)[[1]] %in% res_eval_i$Phenotype)]
CrossVal_agg_res_eval_i<-agg(id=Sample, es=CrossVal_R, var=CrossVal_R_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_eval_i)
IndepVal_agg_res_eval_i<-agg(id=Sample, es=IndepVal_R, var=IndepVal_R_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_eval_i)
meta_res_eval<-rbind(meta_res_eval, data.frame( Test=i,
CrossVal_R=CrossVal_agg_res_eval_i$es[1],
CrossVal_R_SE=sqrt(CrossVal_agg_res_eval_i$var[1]),
IndepVal_R=IndepVal_agg_res_eval_i$es[1],
IndepVal_R_SE=sqrt(IndepVal_agg_res_eval_i$var[1])))
}
# Calculate difference between methods and p-value using aggregate
# Truncate difference p-values to stop 0 values
res_comp$Cross_R_diff_pval[res_comp$Cross_R_diff_pval == 0]<-1e-320
res_comp$Indep_R_diff_pval[res_comp$Indep_R_diff_pval == 0]<-1e-320
meta_res_diff<-NULL
for(i in unique(res_eval$Test)){
for(k in unique(res_eval$Test)){
meta_res_eval_i<-meta_res_eval[meta_res_eval$Test == i,]
meta_res_eval_k<-meta_res_eval[meta_res_eval$Test == k,]
res_comp_i_k<-res_comp[res_comp$Model_1_Test == i & res_comp$Model_2_Test == k,]
# Calculate diff SE based on p-value
for(j in 1:dim(res_comp_i_k)[1]){
if(res_comp_i_k$Cross_R_diff_pval[j] == 1){
res_comp_i_k$Cross_R_diff_pval[j]<-1-0.001
}
if(res_comp_i_k$Indep_R_diff_pval[j] == 1){
res_comp_i_k$Indep_R_diff_pval[j]<-1-0.001
}
}
res_comp_i_k$Cross_R_diff_z<-qnorm(res_comp_i_k$Cross_R_diff_pval/2)
res_comp_i_k$Cross_R_diff_SE<-res_comp_i_k$Cross_R_diff/res_comp_i_k$Cross_R_diff_z
res_comp_i_k$Indep_R_diff_z<-qnorm(res_comp_i_k$Indep_R_diff_pval/2)
res_comp_i_k$Indep_R_diff_SE<-res_comp_i_k$Indep_R_diff/res_comp_i_k$Indep_R_diff_z
res_comp_i_k$Sample<-'A'
cors_i<-cors[(dimnames(cors)[[1]] %in% res_comp_i_k$Phenotype),(dimnames(cors)[[1]] %in% res_comp_i_k$Phenotype)]
CrossVal_agg_res_comp_i_k<-agg(id=Sample, es=Cross_R_diff, var=Cross_R_diff_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_comp_i_k)
IndepVal_agg_res_eval_i<-agg(id=Sample, es=Indep_R_diff, var=Indep_R_diff_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_comp_i_k)
meta_res_diff<-rbind(meta_res_diff, data.frame(Test_ref=i,
Test_targ=k,
CrossVal_R_diff=CrossVal_agg_res_comp_i_k$es[1],
CrossVal_R_diff_SE=sqrt(CrossVal_agg_res_comp_i_k$var[1]),
IndepVal_R_diff=IndepVal_agg_res_eval_i$es[1],
IndepVal_R_diff_SE=sqrt(IndepVal_agg_res_eval_i$var[1])))
}
}
meta_res_diff$CrossVal_R_diff_Z<-meta_res_diff$CrossVal_R_diff/meta_res_diff$CrossVal_R_diff_SE
meta_res_diff$CrossVal_R_diff_P<-2*pnorm(-abs(meta_res_diff$CrossVal_R_diff_Z))
meta_res_diff$IndepVal_R_diff_Z<-meta_res_diff$IndepVal_R_diff/meta_res_diff$IndepVal_R_diff_SE
meta_res_diff$IndepVal_R_diff_P<-2*pnorm(-abs(meta_res_diff$IndepVal_R_diff_Z))
meta_res_diff<-meta_res_diff[,c("Test_ref","Test_targ","CrossVal_R_diff","CrossVal_R_diff_Z","CrossVal_R_diff_P","IndepVal_R_diff","IndepVal_R_diff_Z","IndepVal_R_diff_P")]
meta_res_diff$CrossVal_R_diff_Z[meta_res_diff$CrossVal_R_diff == 0]<-0
meta_res_diff$CrossVal_R_diff_P[meta_res_diff$CrossVal_R_diff == 0]<-1
meta_res_diff$IndepVal_R_diff_Z[meta_res_diff$IndepVal_R_diff == 0]<-0
meta_res_diff$IndepVal_R_diff_P[meta_res_diff$IndepVal_R_diff == 0]<-1
# Calculate percentage improvement
meta_res_diff$CrossVal_R_diff_perc<-NA
for(i in unique(meta_res_diff$Test_ref)){
meta_res_diff$CrossVal_R_diff_perc[meta_res_diff$Test_ref == i]<-meta_res_diff$CrossVal_R_diff[meta_res_diff$Test_ref == i]/meta_res_eval$CrossVal_R[meta_res_eval$Test == i]*100
}
meta_res_diff$IndepVal_R_diff_perc<-NA
for(i in unique(meta_res_diff$Test_ref)){
meta_res_diff$IndepVal_R_diff_perc[meta_res_diff$Test_ref == i]<-meta_res_diff$IndepVal_R_diff[meta_res_diff$Test_ref == i]/meta_res_eval$IndepVal_R[meta_res_eval$Test == i]*100
}
write.csv(meta_res_eval, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'_meta.csv'), row.names=F, quote=F)
meta_res_diff<-meta_res_diff[,c("Test_ref","Test_targ","CrossVal_R_diff",'CrossVal_R_diff_perc',"CrossVal_R_diff_Z","CrossVal_R_diff_P","IndepVal_R_diff",'IndepVal_R_diff_perc',"IndepVal_R_diff_Z","IndepVal_R_diff_P")]
write.csv(meta_res_diff, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'_diff_meta.csv'), row.names=F, quote=F)
###
# Create plot showing performance of each method
###
# To avoid unequal groups, insert NA for missing Model/Method pairs
meta_res_eval$Method<-gsub('\\..*','',meta_res_eval$Test)
meta_res_eval$Model<-gsub('.*\\.','',meta_res_eval$Test)
meta_res_eval$Method<-factor(meta_res_eval$Method, levels=c("pT+clump",'lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
meta_res_eval$Model<-factor(meta_res_eval$Model, levels=c('10FCVal','MultiPRS','PseudoVal','Inf'))
for(i in unique(meta_res_eval$Method)){
for(k in unique(meta_res_eval$Model)){
if(dim(meta_res_eval[meta_res_eval$Method == i & meta_res_eval$Model == k,])[1] > 0){
next
} else {
dud<-data.frame(Test=NA,
CrossVal_R=NA,
CrossVal_R_SE=NA,
IndepVal_R=NA,
IndepVal_R_SE=NA,
Method=i,
Model=k)
meta_res_eval<-rbind(meta_res_eval,dud)
}
}
}
res_eval<-res_eval[,c('Phenotype','Test','CrossVal_R','IndepVal_R','Method','Model')]
for(i in unique(res_eval$Method)){
for(k in unique(res_eval$Model)){
if(dim(res_eval[res_eval$Method == i & res_eval$Model == k,])[1] > 0){
next
} else {
dud<-data.frame(Phenotype=NA,
Test=paste0(i,'.',k),
CrossVal_R=NA,
IndepVal_R=NA,
Method=i,
Model=k)
res_eval<-rbind(res_eval,dud)
}
}
}
library(ggplot2)
library(cowplot)
meta_res_eval_plots_cross[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=CrossVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R-CrossVal_R_SE, ymax=CrossVal_R+CrossVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y')
meta_res_eval_plots_indep[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=IndepVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R-IndepVal_R_SE, ymax=IndepVal_R+IndepVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y')
if(Samples[file] == 'UKBB'){
meta_res_eval_plots_cross[[Files[file]]]<-meta_res_eval_plots_cross[[Files[file]]] + coord_cartesian(ylim=c(0,0.25), clip="on")
meta_res_eval_plots_indep[[Files[file]]]<-meta_res_eval_plots_indep[[Files[file]]] + coord_cartesian(ylim=c(0,0.25), clip="on")
} else {
meta_res_eval_plots_cross[[Files[file]]]<-meta_res_eval_plots_cross[[Files[file]]] + coord_cartesian(ylim=c(0,0.375), clip="on")
meta_res_eval_plots_indep[[Files[file]]]<-meta_res_eval_plots_indep[[Files[file]]] + coord_cartesian(ylim=c(0,0.375), clip="on")
}
###
# Create plot showing performance of each method compared to pT+clump
###
# Calculate differences at the average and phenotype specific level
meta_res_eval$CrossVal_R_diff<-meta_res_eval$CrossVal_R-meta_res_eval$CrossVal_R[which(meta_res_eval$Test == "pT+clump.10FCVal")]
meta_res_eval$IndepVal_R_diff<-meta_res_eval$IndepVal_R-meta_res_eval$IndepVal_R[which(meta_res_eval$Test == "pT+clump.10FCVal")]
res_eval$CrossVal_R_diff<-NA
res_eval$IndepVal_R_diff<-NA
for(i in pheno){
res_eval$CrossVal_R_diff[which(res_eval$Phenotype == i)]<-res_eval$CrossVal_R[which(res_eval$Phenotype == i)]-res_eval$CrossVal_R[which(res_eval$Test == "pT+clump.10FCVal" & res_eval$Phenotype == i)]
res_eval$IndepVal_R_diff[which(res_eval$Phenotype == i)]<-res_eval$IndepVal_R[which(res_eval$Phenotype == i)]-res_eval$IndepVal_R[which(res_eval$Test == "pT+clump.10FCVal" & res_eval$Phenotype == i)]
}
meta_res_eval_diff_plots_cross[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=CrossVal_R_diff, fill=Model)) +
geom_point(data=res_eval, mapping=aes(x=Method, y=CrossVal_R_diff, colour=Model), position=position_jitterdodge(jitter.width = 0.4, dodge.width = 0.7), alpha=0.3) +
geom_errorbar(aes(ymin=CrossVal_R_diff-CrossVal_R_SE, ymax=CrossVal_R_diff+CrossVal_R_SE), width=0.9, position=position_dodge(width = 0.7)) +
geom_point(stat="identity", position=position_dodge(0.7), size=2, shape=23, colour='black') +
labs(y="Difference in\nCorrelations (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y') +
ylim(c(-0.08,0.12))
meta_res_eval_diff_plots_indep[[Files[file]]] <- ggplot(meta_res_eval, aes(x=Method, y=IndepVal_R_diff, fill=Model)) +
geom_point(data=res_eval, mapping=aes(x=Method, y=IndepVal_R_diff, colour=Model), position=position_jitterdodge(jitter.width = 0.4, dodge.width = 1), alpha=0.3) +
geom_errorbar(aes(ymin=IndepVal_R_diff-IndepVal_R_SE, ymax=IndepVal_R_diff+IndepVal_R_SE), width=0.9, position=position_dodge(width = 1)) +
geom_point(stat="identity", position=position_dodge(1), size=2, shape=23, colour='black') +
labs(y="Difference in\nCorrelations (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y') +
ylim(c(-0.08,0.12))
###
# Create plot showing matrix of differences with significance
###
library(reshape2)
meta_res_diff$CrossVal_R_diff[meta_res_diff$Test_ref == meta_res_diff$Test_targ]<-NA
meta_res_diff$IndepVal_R_diff[meta_res_diff$Test_ref == meta_res_diff$Test_targ]<-NA
meta_res_diff$Cross_R_diff_catagory<-'NA'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.002]<-'-0.025 - -0.002'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.025]<-'-0.08 - -0.025'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.08]<-'< -0.08'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > -0.002 & meta_res_diff$CrossVal_R_diff < 0.002]<-'-0.002 - 0.002'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.002]<-'0.002 - 0.025'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.025]<-'0.025 - 0.08'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.08]<-'> 0.08'
meta_res_diff$Cross_R_diff_catagory<-factor(meta_res_diff$Cross_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
meta_res_diff$Indep_R_diff_catagory<-'NA'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.002]<-'-0.025 - -0.002'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.025]<-'-0.08 - -0.025'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.08]<-'< -0.08'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > -0.002 & meta_res_diff$IndepVal_R_diff < 0.002]<-'-0.002 - 0.002'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.002]<-'0.002 - 0.025'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.025]<-'0.025 - 0.08'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.08]<-'> 0.08'
meta_res_diff$Indep_R_diff_catagory<-factor(meta_res_diff$Indep_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
meta_res_diff$cross_star<-' '
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 0.05]<-'*'
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 1e-3]<-'**'
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 1e-6]<-'***'
meta_res_diff$indep_star<-' '
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 0.05]<-'*'
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 1e-3]<-'**'
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 1e-6]<-'***'
library(RColorBrewer)
meta_res_eval_diff_matrix_cross[[Files[file]]]<-ggplot(data = meta_res_diff, aes(Test_targ, Test_ref, fill=Cross_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=Files_descrip[file], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1)) +
coord_fixed() +
geom_text(data=meta_res_diff, aes(Test_ref, Test_targ, label = cross_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(meta_res_diff$Cross_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
meta_res_eval_diff_matrix_indep[[Files[file]]]<-ggplot(data = meta_res_diff, aes(Test_targ, Test_ref, fill=Indep_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=Files_descrip[file], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=meta_res_diff, aes(Test_targ, Test_ref, label = indep_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(meta_res_diff$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
}
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_CrossVal_meta.png', units='px', res=300, width=3400, height=2000)
plot_grid(plotlist=meta_res_eval_plots_cross, ncol = 2, nrow = 2)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_IndepVal_meta.png', units='px', res=300, width=3400, height=2000)
plot_grid(plotlist=meta_res_eval_plots_indep, ncol = 2, nrow = 2)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_pTDiff_CrossVal_meta.png', units='px', res=300, width=3400, height=2000)
plot_grid(plotlist=meta_res_eval_diff_plots_cross, ncol = 2, nrow = 2)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_pTDiff_IndepVal_meta.png', units='px', res=300, width=3400, height=2000)
plot_grid(plotlist=meta_res_eval_diff_plots_indep, ncol = 2, nrow = 2)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_diff_CrossVal_meta.png', units='px', res=300, width=4000, height=4000)
plot_grid(plotlist=meta_res_eval_diff_matrix_cross, ncol = 2, nrow = 2)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_diff_IndepVal_meta.png', units='px', res=300, width=4000, height=4000)
plot_grid(plotlist=meta_res_eval_diff_matrix_indep, ncol = 2, nrow = 2)
dev.off()
# UKB 1KG ref only
tiff('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_UKB_1KG_IndepVal_meta.tiff', units='px', res=300, width=1750, height=2000, compression = 'lzw')
plot_A<-meta_res_eval_plots_indep[[1]] +
labs(title=NULL)
plot_B<-meta_res_eval_diff_plots_indep[[1]] +
labs(title=NULL) +
ylim(c(-0.075,0.075))
plot_grid(plotlist=list(plot_A, plot_B), nrow = 2, labels=c('A','B'))
dev.off()
tiff('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_diff_UKB_1KG_IndepVal_meta.tiff', units='px', res=300, width=2000, height=2000, compression = 'lzw')
meta_res_eval_diff_matrix_indep[[1]] + labs(title=NULL)
dev.off()Summarise and plot results split by polygenicity
Files<-c('UKBB.HighPoly','UKBB.LowPoly')
Samples<-c('UKBB','UKBB')
Files_descrip<-c('UKB target with 1KG ref:\nHigh polygenicity','UKB target with 1KG ref:\nLow polygenicity')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
meta_res_eval<-NULL
meta_res_comp<-NULL
meta_res_eval_plots_cross<-list()
meta_res_eval_plots_indep<-list()
meta_res_eval_diff_plots_cross<-list()
meta_res_eval_diff_plots_indep<-list()
meta_res_eval_diff_matrix_cross<-list()
meta_res_eval_diff_matrix_indep<-list()
for(file in 1:length(Files)){
if(file == 1){
pheno<-c('Depression','Intelligence','BMI','Height','CAD')
gwas<-c('DEPR06','COLL01','BODY04','HEIG03','COAD01')
} else {
pheno<-c('T2D','IBD','MultiScler','RheuArth','Prostate_Cancer','Breast_Cancer')
gwas<-c('DIAB05','CROH01','SCLE03','RHEU02','PRCA01','BRCA01')
}
##################
# Organise, make tables and figures for per phenotype results
##################
res_eval<-NULL
res_comp<-NULL
res_eval_plots_cross<-list()
res_eval_plots_indep<-list()
res_eval_diff_plots_cross<-list()
res_eval_diff_plots_indep<-list()
res_eval_diff_matrix_cross<-list()
res_eval_diff_matrix_indep<-list()
for(i in 1:length(pheno)){
res_eval_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.pred_eval.txt'), header=T, stringsAsFactors=F)
res_comp_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.pred_comp.txt'), header=T, stringsAsFactors=F)
# Idenitfy model names for 10FCVal, multi-PRS and pseudoval
selected_models<-NULL
##
# pT+clump (sparse)
##
# Identify best pT in 10FCVal
res_eval_i_per_pT<-res_eval_i[grepl('pT.clump.PredFile', res_eval_i$Model),]
selected_models<-rbind(selected_models,data.frame(Test='pT+clump.10FCVal', Label=
res_eval_i_per_pT[res_eval_i_per_pT$CrossVal_R == max(res_eval_i_per_pT$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='pT+clump.MultiPRS', Label='pT.clump'))
##
# lassosum
##
# Identify best pT in 10FCVal
res_eval_i_per_lassosum<-res_eval_i[grepl('lassosum.PredFile', res_eval_i$Model),]
selected_models<-rbind(selected_models,data.frame(Test='lassosum.10FCVal', Label=
res_eval_i_per_lassosum[res_eval_i_per_lassosum$CrossVal_R == max(res_eval_i_per_lassosum$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='lassosum.MultiPRS', Label='lassosum'))
# Identify pseudoVal
lassosum_val<-read.table(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/lassosum/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.log'), sep='&')
lassosum_val_s<-as.numeric(gsub('s = ','',lassosum_val$V1[grepl('s = ',lassosum_val$V1)]))
lassosum_val_lambda<-as.numeric(gsub('lambda =','',lassosum_val$V1[grepl('lambda =',lassosum_val$V1)]))
lassosum_val_param<-paste0('s',lassosum_val_s,'.lambda',lassosum_val_lambda)
lassosum_val_param<-substr(lassosum_val_param, 1, nchar(lassosum_val_param)-1)
selected_models<-rbind(selected_models,data.frame(Test='lassosum.PseudoVal', Label=res_eval_i[grepl(lassosum_val_param, res_eval_i$Model),]$Model[1]))
##
# PRScs
##
# Identify best pT in 10FCVal
res_eval_i_per_PRScs<-res_eval_i[grepl('PRScs.PredFile', res_eval_i$Model),]
res_eval_i_per_PRScs<-res_eval_i_per_PRScs[!grepl('phiauto', res_eval_i_per_PRScs$Model),]
selected_models<-rbind(selected_models,data.frame(Test='PRScs.10FCVal', Label=res_eval_i_per_PRScs[res_eval_i_per_PRScs$CrossVal_R == max(res_eval_i_per_PRScs$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='PRScs.MultiPRS', Label='PRScs'))
# Identify pseudoval
res_eval_i_per_PRScs<-res_eval_i[grepl('PRScs.PredFile', res_eval_i$Model),]
res_eval_i_per_PRScs<-res_eval_i_per_PRScs[grepl('phiauto', res_eval_i_per_PRScs$Model),]
selected_models<-rbind(selected_models,data.frame(Test='PRScs.PseudoVal', Label=res_eval_i_per_PRScs[res_eval_i_per_PRScs$CrossVal_R == max(res_eval_i_per_PRScs$CrossVal_R),]$Model[1]))
##
# SBLUP
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='SBLUP.Inf', Label='SBLUP'))
##
# SBayesR
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='SBayesR.PseudoVal', Label='SBayesR'))
##
# LDPred1
##
# Identify best pT in 10FCVal
res_eval_i_per_LDPred<-res_eval_i[grepl('LDPred.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred<-res_eval_i_per_LDPred[!grepl('.LDpred.inf', res_eval_i_per_LDPred$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.10FCVal', Label=res_eval_i_per_LDPred[res_eval_i_per_LDPred$CrossVal_R == max(res_eval_i_per_LDPred$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.MultiPRS', Label='LDPred'))
# Identify pseudoval
res_eval_i_per_LDPred<-res_eval_i[grepl('LDPred.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred<-res_eval_i_per_LDPred[grepl('.LDpred.inf', res_eval_i_per_LDPred$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.Inf', Label=res_eval_i_per_LDPred[res_eval_i_per_LDPred$CrossVal_R == max(res_eval_i_per_LDPred$CrossVal_R),]$Model[1]))
##
# LDPred2
##
# Identify best pT in 10FCVal
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[!grepl('beta.inf|beta.auto', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.10FCVal', Label=res_eval_i_per_LDPred2[res_eval_i_per_LDPred2$CrossVal_R == max(res_eval_i_per_LDPred2$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.MultiPRS', Label='LDPred2'))
# Identify pseudoval
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[grepl('beta.inf', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.Inf', Label=res_eval_i_per_LDPred2$Model))
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[grepl('beta.auto', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.PseudoVal', Label=res_eval_i_per_LDPred2$Model))
##
# DBSLMM
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='DBSLMM.PseudoVal', Label='DBSLMM'))
##
# All methods
##
selected_models<-rbind(selected_models,data.frame(Test='All.MultiPRS', Label='All'))
###
# Subset selected models and format
###
# Eval
selected_models$Label_2<-paste0(gsub('_group','',selected_models$Label),'_group')
res_eval_i_select<-res_eval_i[(res_eval_i$Model %in% selected_models$Label_2),]
res_eval_i_select<-merge(res_eval_i_select, selected_models[c('Test','Label_2')], by.x='Model',by.y='Label_2')
res_eval_i_select$Phenotype<-pheno[i]
if(sum(names(res_eval_i_select) == 'CrossVal_OR') == 0){
res_eval_i_select$CrossVal_OR<-NA
res_eval_i_select$CrossVal_LowCI<-NA
res_eval_i_select$CrossVal_HighCI<-NA
res_eval_i_select$Cross_LiabR2<-NA
res_eval_i_select$Cross_AUC<-NA
res_eval_i_select$CrossVal_Ncas<-NA
res_eval_i_select$CrossVal_Ncon<-NA
res_eval_i_select$IndepVal_OR<-NA
res_eval_i_select$IndepVal_LowCI<-NA
res_eval_i_select$IndepVal_HighCI<-NA
res_eval_i_select$Indep_LiabR2<-NA
res_eval_i_select$Indep_AUC<-NA
res_eval_i_select$IndepVal_Ncas<-NA
res_eval_i_select$IndepVal_Ncon<-NA
}
res_eval_i_select<-res_eval_i_select[c('Phenotype','Test',"CrossVal_R","CrossVal_R_SE",'CrossVal_OR','CrossVal_LowCI','CrossVal_HighCI','Cross_LiabR2','Cross_AUC','CrossVal_pval','CrossVal_N','CrossVal_Ncas','CrossVal_Ncon',"IndepVal_R","IndepVal_R_SE",'IndepVal_OR','IndepVal_LowCI','IndepVal_HighCI','Indep_LiabR2','Indep_AUC','IndepVal_pval','IndepVal_N','IndepVal_Ncas','IndepVal_Ncon')]
res_eval<-rbind(res_eval,res_eval_i_select)
# Comp
selected_models$Label_3<-gsub('_group','',selected_models$Label)
res_comp_i_select<-res_comp_i[(res_comp_i$Model_1 %in% selected_models$Label_3) & (res_comp_i$Model_2 %in% selected_models$Label_3),]
res_comp_i_select<-merge(res_comp_i_select, selected_models[c('Test','Label_3')], by.x='Model_1',by.y='Label_3')
names(res_comp_i_select)[names(res_comp_i_select) == 'Test']<-'Model_1_Test'
res_comp_i_select<-merge(res_comp_i_select, selected_models[c('Test','Label_3')], by.x='Model_2',by.y='Label_3')
names(res_comp_i_select)[names(res_comp_i_select) == 'Test']<-'Model_2_Test'
res_comp_i_select$Label<-NULL
res_comp_i_select$Phenotype<-pheno[i]
res_comp_i_select<-res_comp_i_select[c('Phenotype','Model_1_Test','Model_2_Test','Model_1_Cross_R','Model_2_Cross_R','Cross_R_diff','Cross_R_diff_pval','Model_1_Indep_R','Model_2_Indep_R','Indep_R_diff','Indep_R_diff_pval')]
res_comp<-rbind(res_comp,res_comp_i_select)
}
###################
# Average results across phenotypes, creates tables and figures
###################
res_eval$Method<-gsub('\\..*','',res_eval$Test)
res_eval$Model<-gsub('.*\\.','',res_eval$Test)
res_eval$Phenotype<-factor(res_eval$Phenotype, level=unique(res_eval$Phenotype))
res_eval$Method<-factor(res_eval$Method, level=c("pT+clump",'lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
res_eval$Model<-factor(res_eval$Model, level=c('10FCVal','MultiPRS','PseudoVal','Inf'))
res_eval<-res_eval[order(res_eval$Phenotype, res_eval$Method, res_eval$Model),]
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
for(pheno_i in 1:length(pheno)){
if(pheno_i==1){
pheno_i_dat<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-pheno_i_dat
} else {
pheno_i_dat<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-merge(pheno_dat, pheno_i_dat, by=c('FID','IID'), all=T)
}
}
# Calculate corelation between all phenotypes
cors<-abs(cor(as.matrix(pheno_dat[,-1:-2]), use='p'))
cors<-cors[unique(res_eval$Phenotype,),unique(res_eval$Phenotype,)]
cors[is.na(cors)]<-0
# Run aggregate function
library(MAd)
meta_res_eval<-NULL
for(i in unique(res_eval$Test)){
res_eval_i<-res_eval[res_eval$Test == i,]
res_eval_i$Sample<-'Target'
cors_i<-cors[(dimnames(cors)[[1]] %in% res_eval_i$Phenotype),(dimnames(cors)[[1]] %in% res_eval_i$Phenotype)]
CrossVal_agg_res_eval_i<-agg(id=Sample, es=CrossVal_R, var=CrossVal_R_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_eval_i)
IndepVal_agg_res_eval_i<-agg(id=Sample, es=IndepVal_R, var=IndepVal_R_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_eval_i)
meta_res_eval<-rbind(meta_res_eval, data.frame( Test=i,
CrossVal_R=CrossVal_agg_res_eval_i$es[1],
CrossVal_R_SE=sqrt(CrossVal_agg_res_eval_i$var[1]),
IndepVal_R=IndepVal_agg_res_eval_i$es[1],
IndepVal_R_SE=sqrt(IndepVal_agg_res_eval_i$var[1])))
}
# Calculate difference between methods and p-value using aggregate
# Truncate difference p-values to stop 0 values
res_comp$Cross_R_diff_pval[res_comp$Cross_R_diff_pval == 0]<-1e-320
res_comp$Indep_R_diff_pval[res_comp$Indep_R_diff_pval == 0]<-1e-320
meta_res_diff<-NULL
for(i in unique(res_eval$Test)){
for(k in unique(res_eval$Test)){
meta_res_eval_i<-meta_res_eval[meta_res_eval$Test == i,]
meta_res_eval_k<-meta_res_eval[meta_res_eval$Test == k,]
res_comp_i_k<-res_comp[res_comp$Model_1_Test == i & res_comp$Model_2_Test == k,]
# Calculate diff SE based on p-value
for(j in 1:dim(res_comp_i_k)[1]){
if(res_comp_i_k$Cross_R_diff_pval[j] == 1){
res_comp_i_k$Cross_R_diff_pval[j]<-1-0.001
}
if(res_comp_i_k$Indep_R_diff_pval[j] == 1){
res_comp_i_k$Indep_R_diff_pval[j]<-1-0.001
}
}
res_comp_i_k$Cross_R_diff_z<-qnorm(res_comp_i_k$Cross_R_diff_pval/2)
res_comp_i_k$Cross_R_diff_SE<-res_comp_i_k$Cross_R_diff/res_comp_i_k$Cross_R_diff_z
res_comp_i_k$Indep_R_diff_z<-qnorm(res_comp_i_k$Indep_R_diff_pval/2)
res_comp_i_k$Indep_R_diff_SE<-res_comp_i_k$Indep_R_diff/res_comp_i_k$Indep_R_diff_z
res_comp_i_k$Sample<-'A'
cors_i<-cors[(dimnames(cors)[[1]] %in% res_comp_i_k$Phenotype),(dimnames(cors)[[1]] %in% res_comp_i_k$Phenotype)]
CrossVal_agg_res_comp_i_k<-agg(id=Sample, es=Cross_R_diff, var=Cross_R_diff_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_comp_i_k)
IndepVal_agg_res_eval_i<-agg(id=Sample, es=Indep_R_diff, var=Indep_R_diff_SE^2, cor=cors_i, method="BHHR", mod=NULL, data=res_comp_i_k)
meta_res_diff<-rbind(meta_res_diff, data.frame(Test_ref=i,
Test_targ=k,
CrossVal_R_diff=CrossVal_agg_res_comp_i_k$es[1],
CrossVal_R_diff_SE=sqrt(CrossVal_agg_res_comp_i_k$var[1]),
IndepVal_R_diff=IndepVal_agg_res_eval_i$es[1],
IndepVal_R_diff_SE=sqrt(IndepVal_agg_res_eval_i$var[1])))
}
}
meta_res_diff$CrossVal_R_diff_Z<-meta_res_diff$CrossVal_R_diff/meta_res_diff$CrossVal_R_diff_SE
meta_res_diff$CrossVal_R_diff_P<-2*pnorm(-abs(meta_res_diff$CrossVal_R_diff_Z))
meta_res_diff$IndepVal_R_diff_Z<-meta_res_diff$IndepVal_R_diff/meta_res_diff$IndepVal_R_diff_SE
meta_res_diff$IndepVal_R_diff_P<-2*pnorm(-abs(meta_res_diff$IndepVal_R_diff_Z))
meta_res_diff<-meta_res_diff[,c("Test_ref","Test_targ","CrossVal_R_diff","CrossVal_R_diff_Z","CrossVal_R_diff_P","IndepVal_R_diff","IndepVal_R_diff_Z","IndepVal_R_diff_P")]
meta_res_diff$CrossVal_R_diff_Z[meta_res_diff$CrossVal_R_diff == 0]<-0
meta_res_diff$CrossVal_R_diff_P[meta_res_diff$CrossVal_R_diff == 0]<-1
meta_res_diff$IndepVal_R_diff_Z[meta_res_diff$IndepVal_R_diff == 0]<-0
meta_res_diff$IndepVal_R_diff_P[meta_res_diff$IndepVal_R_diff == 0]<-1
# Calculate percentage improvement
meta_res_diff$CrossVal_R_diff_perc<-NA
for(i in unique(meta_res_diff$Test_ref)){
meta_res_diff$CrossVal_R_diff_perc[meta_res_diff$Test_ref == i]<-meta_res_diff$CrossVal_R_diff[meta_res_diff$Test_ref == i]/meta_res_eval$CrossVal_R[meta_res_eval$Test == i]*100
}
meta_res_diff$IndepVal_R_diff_perc<-NA
for(i in unique(meta_res_diff$Test_ref)){
meta_res_diff$IndepVal_R_diff_perc[meta_res_diff$Test_ref == i]<-meta_res_diff$IndepVal_R_diff[meta_res_diff$Test_ref == i]/meta_res_eval$IndepVal_R[meta_res_eval$Test == i]*100
}
write.csv(meta_res_eval, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'_meta_.csv'), row.names=F, quote=F)
meta_res_diff<-meta_res_diff[,c("Test_ref","Test_targ","CrossVal_R_diff",'CrossVal_R_diff_perc',"CrossVal_R_diff_Z","CrossVal_R_diff_P","IndepVal_R_diff",'IndepVal_R_diff_perc',"IndepVal_R_diff_Z","IndepVal_R_diff_P")]
write.csv(meta_res_diff, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_method_comp_',Files[file],'_diff_meta.csv'), row.names=F, quote=F)
###
# Create plot showing performance of each method
###
# To avoid unequal groups, insert NA for missing Model/Method pairs
meta_res_eval$Method<-gsub('\\..*','',meta_res_eval$Test)
meta_res_eval$Model<-gsub('.*\\.','',meta_res_eval$Test)
meta_res_eval$Method<-factor(meta_res_eval$Method, levels=c("pT+clump",'lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
meta_res_eval$Model<-factor(meta_res_eval$Model, levels=c('10FCVal','MultiPRS','PseudoVal','Inf'))
for(i in unique(meta_res_eval$Method)){
for(k in unique(meta_res_eval$Model)){
if(dim(meta_res_eval[meta_res_eval$Method == i & meta_res_eval$Model == k,])[1] > 0){
next
} else {
dud<-data.frame(Test=NA,
CrossVal_R=NA,
CrossVal_R_SE=NA,
IndepVal_R=NA,
IndepVal_R_SE=NA,
Method=i,
Model=k)
meta_res_eval<-rbind(meta_res_eval,dud)
}
}
}
res_eval<-res_eval[,c('Phenotype','Test','CrossVal_R','IndepVal_R','Method','Model')]
for(i in unique(res_eval$Method)){
for(k in unique(res_eval$Model)){
if(dim(res_eval[res_eval$Method == i & res_eval$Model == k,])[1] > 0){
next
} else {
dud<-data.frame(Phenotype=NA,
Test=paste0(i,'.',k),
CrossVal_R=NA,
IndepVal_R=NA,
Method=i,
Model=k)
res_eval<-rbind(res_eval,dud)
}
}
}
library(ggplot2)
library(cowplot)
meta_res_eval_plots_cross[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=CrossVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R-CrossVal_R_SE, ymax=CrossVal_R+CrossVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y')
meta_res_eval_plots_indep[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=IndepVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R-IndepVal_R_SE, ymax=IndepVal_R+IndepVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y')
if(Samples[file] == 'UKBB'){
meta_res_eval_plots_cross[[Files[file]]]<-meta_res_eval_plots_cross[[Files[file]]] + coord_cartesian(ylim=c(0,0.25), clip="on")
meta_res_eval_plots_indep[[Files[file]]]<-meta_res_eval_plots_indep[[Files[file]]] + coord_cartesian(ylim=c(0,0.25), clip="on")
} else {
meta_res_eval_plots_cross[[Files[file]]]<-meta_res_eval_plots_cross[[Files[file]]] + coord_cartesian(ylim=c(0,0.375), clip="on")
meta_res_eval_plots_indep[[Files[file]]]<-meta_res_eval_plots_indep[[Files[file]]] + coord_cartesian(ylim=c(0,0.375), clip="on")
}
###
# Create plot showing performance of each method compared to pT+clump
###
# Calculate differences at the average and phenotype specific level
meta_res_eval$CrossVal_R_diff<-meta_res_eval$CrossVal_R-meta_res_eval$CrossVal_R[which(meta_res_eval$Test == "pT+clump.10FCVal")]
meta_res_eval$IndepVal_R_diff<-meta_res_eval$IndepVal_R-meta_res_eval$IndepVal_R[which(meta_res_eval$Test == "pT+clump.10FCVal")]
res_eval$CrossVal_R_diff<-NA
res_eval$IndepVal_R_diff<-NA
for(i in pheno){
res_eval$CrossVal_R_diff[which(res_eval$Phenotype == i)]<-res_eval$CrossVal_R[which(res_eval$Phenotype == i)]-res_eval$CrossVal_R[which(res_eval$Test == "pT+clump.10FCVal" & res_eval$Phenotype == i)]
res_eval$IndepVal_R_diff[which(res_eval$Phenotype == i)]<-res_eval$IndepVal_R[which(res_eval$Phenotype == i)]-res_eval$IndepVal_R[which(res_eval$Test == "pT+clump.10FCVal" & res_eval$Phenotype == i)]
}
meta_res_eval_diff_plots_cross[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=CrossVal_R_diff, fill=Model)) +
geom_point(data=res_eval, mapping=aes(x=Method, y=CrossVal_R_diff, colour=Model), position=position_jitterdodge(jitter.width = 0.4, dodge.width = 0.7), alpha=0.3) +
geom_errorbar(aes(ymin=CrossVal_R_diff-CrossVal_R_SE, ymax=CrossVal_R_diff+CrossVal_R_SE), width=0.7, position=position_dodge(width = 0.7)) +
geom_point(stat="identity", position=position_dodge(0.7), size=2, shape=23, colour='black') +
labs(y="Difference in\nCorrelations (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y') +
ylim(c(-0.12,0.12))
meta_res_eval_diff_plots_indep[[Files[file]]] <- ggplot(meta_res_eval, aes(x=Method, y=IndepVal_R_diff, fill=Model)) +
geom_point(data=res_eval, mapping=aes(x=Method, y=IndepVal_R_diff, colour=Model), position=position_jitterdodge(jitter.width = 0.4, dodge.width = 1), alpha=0.3) +
geom_errorbar(aes(ymin=IndepVal_R_diff-IndepVal_R_SE, ymax=IndepVal_R_diff+IndepVal_R_SE), width=0.7, position=position_dodge(width = 1)) +
geom_point(stat="identity", position=position_dodge(1), size=2, shape=23, colour='black') +
labs(y="Difference in\nCorrelations (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y') +
ylim(c(-0.12,0.12))
###
# Create plot showing matrix of differences with significance
###
library(reshape2)
meta_res_diff$CrossVal_R_diff[meta_res_diff$Test_ref == meta_res_diff$Test_targ]<-NA
meta_res_diff$IndepVal_R_diff[meta_res_diff$Test_ref == meta_res_diff$Test_targ]<-NA
meta_res_diff$Cross_R_diff_catagory<-'NA'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.002]<-'-0.025 - -0.002'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.025]<-'-0.08 - -0.025'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.08]<-'< -0.08'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > -0.002 & meta_res_diff$CrossVal_R_diff < 0.002]<-'-0.002 - 0.002'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.002]<-'0.002 - 0.025'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.025]<-'0.025 - 0.08'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.08]<-'> 0.08'
meta_res_diff$Cross_R_diff_catagory<-factor(meta_res_diff$Cross_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
meta_res_diff$Indep_R_diff_catagory<-'NA'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.002]<-'-0.025 - -0.002'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.025]<-'-0.08 - -0.025'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.08]<-'< -0.08'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > -0.002 & meta_res_diff$IndepVal_R_diff < 0.002]<-'-0.002 - 0.002'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.002]<-'0.002 - 0.025'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.025]<-'0.025 - 0.08'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.08]<-'> 0.08'
meta_res_diff$Indep_R_diff_catagory<-factor(meta_res_diff$Indep_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
meta_res_diff$cross_star<-' '
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 0.05]<-'*'
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 1e-3]<-'**'
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 1e-6]<-'***'
meta_res_diff$indep_star<-' '
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 0.05]<-'*'
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 1e-3]<-'**'
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 1e-6]<-'***'
library(RColorBrewer)
meta_res_eval_diff_matrix_cross[[Files[file]]]<-ggplot(data = meta_res_diff, aes(Test_targ, Test_ref, fill=Cross_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=Files_descrip[file], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1)) +
coord_fixed() +
geom_text(data=meta_res_diff, aes(Test_ref, Test_targ, label = cross_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(meta_res_diff$Cross_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
meta_res_eval_diff_matrix_indep[[Files[file]]]<-ggplot(data = meta_res_diff, aes(Test_targ, Test_ref, fill=Indep_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=Files_descrip[file], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=meta_res_diff, aes(Test_targ, Test_ref, label = indep_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(meta_res_diff$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
}
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_CrossVal_meta_byPoly.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_plots_cross, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_IndepVal_meta_byPoly.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_plots_indep, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_pTDiff_CrossVal_meta_byPoly.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_diff_plots_cross, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_pTDiff_IndepVal_meta_byPoly.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_diff_plots_indep, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_diff_CrossVal_meta_byPoly.png', units='px', res=300, width=4000, height=2000)
plot_grid(plotlist=meta_res_eval_diff_matrix_cross, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PRS_method_comp_diff_IndepVal_meta_byPoly.png', units='px', res=300, width=4000, height=2000)
plot_grid(plotlist=meta_res_eval_diff_matrix_indep, ncol = 2, nrow = 1)
dev.off()Summarise and plot the results with PCs regressed
Files<-c('UKBB','TEDS')
Samples<-c('UKBB','TEDS')
Files_descrip<-c('UKB target with 1KG ref','TEDS target with 1KG ref')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
meta_res_eval<-NULL
meta_res_comp<-NULL
meta_res_eval_plots_cross<-list()
meta_res_eval_plots_indep<-list()
meta_res_eval_diff_plots_cross<-list()
meta_res_eval_diff_plots_indep<-list()
meta_res_eval_diff_matrix_cross<-list()
meta_res_eval_diff_matrix_indep<-list()
for(file in 1:length(Files)){
if(Samples[file] == 'UKBB'){
pheno<-c('Depression','Intelligence','Height','BMI','T2D','CAD','IBD', 'MultiScler','RheuArth','Prostate_Cancer','Breast_Cancer')
gwas<-c('DEPR06','COLL01','HEIG03','BODY04','DIAB05','COAD01','CROH01','SCLE03','RHEU02','PRCA01','BRCA01')
} else {
pheno<-c('Height21', 'BMI21', 'GCSE', 'ADHD')
gwas<-c('HEIG03', 'BODY11', 'EDUC03', 'ADHD04')
}
##################
# Organise, make tables and figures for per phenotype results
##################
res_eval<-NULL
res_comp<-NULL
res_eval_plots_cross<-list()
res_eval_plots_indep<-list()
res_eval_diff_plots_cross<-list()
res_eval_diff_plots_indep<-list()
res_eval_diff_matrix_cross<-list()
res_eval_diff_matrix_indep<-list()
for(i in 1:length(pheno)){
res_eval_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.PCresid.pred_eval.txt'), header=T, stringsAsFactors=F)
res_comp_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs.AllMethodComp.PCresid.pred_comp.txt'), header=T, stringsAsFactors=F)
# Idenitfy model names for 10FCVal, multi-PRS and pseudoval
selected_models<-NULL
##
# pT+clump (sparse)
##
# Identify best pT in 10FCVal
res_eval_i_per_pT<-res_eval_i[grepl('pT.clump.PredFile', res_eval_i$Model),]
selected_models<-rbind(selected_models,data.frame(Test='pT+clump.10FCVal', Label=
res_eval_i_per_pT[res_eval_i_per_pT$CrossVal_R == max(res_eval_i_per_pT$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='pT+clump.MultiPRS', Label='pT.clump'))
##
# lassosum
##
# Identify best pT in 10FCVal
res_eval_i_per_lassosum<-res_eval_i[grepl('lassosum.PredFile', res_eval_i$Model),]
selected_models<-rbind(selected_models,data.frame(Test='lassosum.10FCVal', Label=
res_eval_i_per_lassosum[res_eval_i_per_lassosum$CrossVal_R == max(res_eval_i_per_lassosum$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='lassosum.MultiPRS', Label='lassosum'))
# Identify pseudoVal
lassosum_val<-read.table(paste0('/users/k1806347/brc_scratch/Data/1KG/Phase3/Score_files_for_polygenic/lassosum/',gwas[i],'/1KGPhase3.w_hm3.',gwas[i],'.log'), sep='&')
lassosum_val_s<-as.numeric(gsub('s = ','',lassosum_val$V1[grepl('s = ',lassosum_val$V1)]))
lassosum_val_lambda<-as.numeric(gsub('lambda =','',lassosum_val$V1[grepl('lambda =',lassosum_val$V1)]))
lassosum_val_param<-paste0('s',lassosum_val_s,'.lambda',lassosum_val_lambda)
lassosum_val_param<-substr(lassosum_val_param, 1, nchar(lassosum_val_param)-1)
selected_models<-rbind(selected_models,data.frame(Test='lassosum.PseudoVal', Label=res_eval_i[grepl(lassosum_val_param, res_eval_i$Model),]$Model[1]))
##
# PRScs
##
# Identify best pT in 10FCVal
res_eval_i_per_PRScs<-res_eval_i[grepl('PRScs.PredFile', res_eval_i$Model),]
res_eval_i_per_PRScs<-res_eval_i_per_PRScs[!grepl('phiauto', res_eval_i_per_PRScs$Model),]
selected_models<-rbind(selected_models,data.frame(Test='PRScs.10FCVal', Label=res_eval_i_per_PRScs[res_eval_i_per_PRScs$CrossVal_R == max(res_eval_i_per_PRScs$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='PRScs.MultiPRS', Label='PRScs'))
# Identify pseudoval
res_eval_i_per_PRScs<-res_eval_i[grepl('PRScs.PredFile', res_eval_i$Model),]
res_eval_i_per_PRScs<-res_eval_i_per_PRScs[grepl('phiauto', res_eval_i_per_PRScs$Model),]
selected_models<-rbind(selected_models,data.frame(Test='PRScs.PseudoVal', Label=res_eval_i_per_PRScs[res_eval_i_per_PRScs$CrossVal_R == max(res_eval_i_per_PRScs$CrossVal_R),]$Model[1]))
##
# SBLUP
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='SBLUP.Inf', Label='SBLUP'))
##
# SBayesR
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='SBayesR.PseudoVal', Label='SBayesR'))
##
# LDPred1
##
# Identify best pT in 10FCVal
res_eval_i_per_LDPred<-res_eval_i[grepl('LDPred.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred<-res_eval_i_per_LDPred[!grepl('.LDpred.inf', res_eval_i_per_LDPred$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.10FCVal', Label=res_eval_i_per_LDPred[res_eval_i_per_LDPred$CrossVal_R == max(res_eval_i_per_LDPred$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.MultiPRS', Label='LDPred'))
# Identify pseudoval
res_eval_i_per_LDPred<-res_eval_i[grepl('LDPred.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred<-res_eval_i_per_LDPred[grepl('.LDpred.inf', res_eval_i_per_LDPred$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred1.Inf', Label=res_eval_i_per_LDPred[res_eval_i_per_LDPred$CrossVal_R == max(res_eval_i_per_LDPred$CrossVal_R),]$Model[1]))
##
# LDPred2
##
# Identify best pT in 10FCVal
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[!grepl('beta.inf|beta.auto', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.10FCVal', Label=res_eval_i_per_LDPred2[res_eval_i_per_LDPred2$CrossVal_R == max(res_eval_i_per_LDPred2$CrossVal_R),]$Model[1]))
# Identify multi pT
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.MultiPRS', Label='LDPred2'))
# Identify pseudoval
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[grepl('beta.inf', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.Inf', Label=res_eval_i_per_LDPred2$Model))
res_eval_i_per_LDPred2<-res_eval_i[grepl('LDPred2.PredFile', res_eval_i$Model),]
res_eval_i_per_LDPred2<-res_eval_i_per_LDPred2[grepl('beta.auto', res_eval_i_per_LDPred2$Model),]
selected_models<-rbind(selected_models,data.frame(Test='LDpred2.PseudoVal', Label=res_eval_i_per_LDPred2$Model))
##
# DBSLMM
##
# Identify PseudoVal
selected_models<-rbind(selected_models,data.frame(Test='DBSLMM.PseudoVal', Label='DBSLMM'))
##
# All methods
##
selected_models<-rbind(selected_models,data.frame(Test='All.MultiPRS', Label='All'))
###
# Subset selected models and format
###
# Eval
selected_models$Label_2<-paste0(gsub('_group','',selected_models$Label),'_group')
res_eval_i_select<-res_eval_i[(res_eval_i$Model %in% selected_models$Label_2),]
res_eval_i_select<-merge(res_eval_i_select, selected_models[c('Test','Label_2')], by.x='Model',by.y='Label_2')
res_eval_i_select$Phenotype<-pheno[i]
res_eval_i_select<-res_eval_i_select[c('Phenotype','Test',"CrossVal_R","CrossVal_R_SE","CrossVal_pval","IndepVal_R","IndepVal_R_SE","IndepVal_pval")]
res_eval<-rbind(res_eval,res_eval_i_select)
# Comp
selected_models$Label_3<-gsub('_group','',selected_models$Label)
res_comp_i_select<-res_comp_i[(res_comp_i$Model_1 %in% selected_models$Label_3) & (res_comp_i$Model_2 %in% selected_models$Label_3),]
res_comp_i_select<-merge(res_comp_i_select, selected_models[c('Test','Label_3')], by.x='Model_1',by.y='Label_3')
names(res_comp_i_select)[names(res_comp_i_select) == 'Test']<-'Model_1_Test'
res_comp_i_select<-merge(res_comp_i_select, selected_models[c('Test','Label_3')], by.x='Model_2',by.y='Label_3')
names(res_comp_i_select)[names(res_comp_i_select) == 'Test']<-'Model_2_Test'
res_comp_i_select$Label<-NULL
res_comp_i_select$Phenotype<-pheno[i]
res_comp_i_select<-res_comp_i_select[c('Phenotype','Model_1_Test','Model_2_Test','Model_1_Cross_R','Model_2_Cross_R','Cross_R_diff','Cross_R_diff_pval','Model_1_Indep_R','Model_2_Indep_R','Indep_R_diff','Indep_R_diff_pval')]
res_comp<-rbind(res_comp,res_comp_i_select)
###
# Create plot showing performance of each method
###
library(ggplot2)
library(cowplot)
res_eval_i_select$Method<-gsub('\\..*','',res_eval_i_select$Test)
res_eval_i_select$Model<-gsub('.*\\.','',res_eval_i_select$Test)
res_eval_i_select$Method<-factor(res_eval_i_select$Method, levels=c('pT+clump','lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
res_eval_i_select$Model<-factor(res_eval_i_select$Model, levels=c('10FCVal','MultiPRS','PseudoVal','Inf'))
res_eval_plots_cross[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=CrossVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R-CrossVal_R_SE, ymax=CrossVal_R+CrossVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$CrossVal_R[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$CrossVal_R[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
res_eval_plots_indep[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=IndepVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R-IndepVal_R_SE, ymax=IndepVal_R+IndepVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$IndepVal_R[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$IndepVal_R[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
###
# Create plot showing performance of each method compared to pT+clump
###
res_eval_i_select$CrossVal_R_diff<-res_eval_i_select$CrossVal_R-res_eval_i_select$CrossVal_R[res_eval_i_select$Test == "pT+clump.10FCVal"]
res_eval_diff_plots_cross[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=CrossVal_R_diff, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R_diff-CrossVal_R_SE, ymax=CrossVal_R_diff+CrossVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$CrossVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$CrossVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
res_eval_i_select$IndepVal_R_diff<-res_eval_i_select$IndepVal_R-res_eval_i_select$IndepVal_R[res_eval_i_select$Test == "pT+clump.10FCVal"]
res_eval_diff_plots_indep[[pheno[i]]] <- ggplot( res_eval_i_select, aes(x=Method, y=IndepVal_R_diff, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R_diff-IndepVal_R_SE, ymax=IndepVal_R_diff+IndepVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res_eval_i_select$IndepVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])-0.02, max(res_eval_i_select$IndepVal_R_diff[res_eval_i_select$Phenotype==pheno[i]])+0.025), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
###
# Create plot showing matrix of differences with significance
###
library(reshape2)
res_comp_i_select$Cross_R_diff[res_comp_i_select$Model_1_Test == res_comp_i_select$Model_2_Test]<-NA
res_comp_i_select$Indep_R_diff[res_comp_i_select$Model_1_Test == res_comp_i_select$Model_2_Test]<-NA
res_comp_i_select$Cross_R_diff_catagory<-'NA'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff < -0.002]<-'-0.025 - -0.002'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff < -0.025]<-'-0.08 - -0.025'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff < -0.08]<-'< -0.08'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > -0.002 & res_comp_i_select$Cross_R_diff < 0.002]<-'-0.002 - 0.002'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > 0.002]<-'0.002 - 0.025'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > 0.025]<-'0.025 - 0.08'
res_comp_i_select$Cross_R_diff_catagory[res_comp_i_select$Cross_R_diff > 0.08]<-'> 0.08'
res_comp_i_select$Cross_R_diff_catagory<-factor(res_comp_i_select$Cross_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
res_comp_i_select$Indep_R_diff_catagory<-'NA'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff < -0.002]<-'-0.025 - -0.002'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff < -0.025]<-'-0.08 - -0.025'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff < -0.08]<-'< -0.08'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > -0.002 & res_comp_i_select$Indep_R_diff < 0.002]<-'-0.002 - 0.002'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > 0.002]<-'0.002 - 0.025'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > 0.025]<-'0.025 - 0.08'
res_comp_i_select$Indep_R_diff_catagory[res_comp_i_select$Indep_R_diff > 0.08]<-'> 0.08'
res_comp_i_select$Indep_R_diff_catagory<-factor(res_comp_i_select$Indep_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
res_comp_i_select$cross_star<-' '
res_comp_i_select$cross_star[res_comp_i_select$Cross_R_diff_pval < 0.05]<-'*'
res_comp_i_select$cross_star[res_comp_i_select$Cross_R_diff_pval < 1e-3]<-'**'
res_comp_i_select$cross_star[res_comp_i_select$Cross_R_diff_pval < 1e-6]<-'***'
res_comp_i_select$indep_star<-' '
res_comp_i_select$indep_star[res_comp_i_select$Indep_R_diff_pval < 0.05]<-'*'
res_comp_i_select$indep_star[res_comp_i_select$Indep_R_diff_pval < 1e-3]<-'**'
res_comp_i_select$indep_star[res_comp_i_select$Indep_R_diff_pval < 1e-6]<-'***'
library(RColorBrewer)
res_eval_diff_matrix_cross[[pheno[i]]]<-ggplot(data = res_comp_i_select, aes(Model_2_Test, Model_1_Test, fill=Cross_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=pheno[i], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=res_comp_i_select, aes(Model_2_Test, Model_1_Test, label = cross_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(res_comp_i_select$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
res_eval_diff_matrix_indep[[pheno[i]]]<-ggplot(data = res_comp_i_select, aes(Model_2_Test, Model_1_Test, fill=Indep_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=pheno[i], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=res_comp_i_select, aes(Model_2_Test, Model_1_Test, label = indep_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(res_comp_i_select$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
}
dir.create(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/'))
write.csv(res_eval, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_',Files[file],'.csv'), row.names=F, quote=F)
write.csv(res_comp, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_',Files[file],'_diff.csv'), row.names=F, quote=F)
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_CrossVal_',Files[file],'.png'), units='px', res=300, width=2250, height=800*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_plots_cross, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_IndepVal_',Files[file],'.png'), units='px', res=300, width=2250, height=800*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_plots_indep, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_pTDiff_CrossVal_',Files[file],'.png'), units='px', res=300, width=2250, height=800*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_diff_plots_cross, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_pTDiff_IndepVal_',Files[file],'.png'), units='px', res=300, width=2250, height=800*ceiling(length(res_eval_plots_cross)/2))
print(plot_grid(plotlist=res_eval_diff_plots_indep, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_',Files[file],'_diff_CrossVal.png'), units='px', res=300, width=2250, height=1750*length(res_eval_diff_matrix_cross))
print(plot_grid(plotlist=res_eval_diff_matrix_cross, ncol = 1))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_',Files[file],'_diff_IndepVal.png'), units='px', res=300, width=2250, height=1750*length(res_eval_diff_matrix_cross))
print(plot_grid(plotlist=res_eval_diff_matrix_indep, ncol = 1))
dev.off()
###################
# Average results across phenotypes, creates tables and figures
###################
res_eval$Method<-gsub('\\..*','',res_eval$Test)
res_eval$Model<-gsub('.*\\.','',res_eval$Test)
res_eval$Phenotype<-factor(res_eval$Phenotype, level=unique(res_eval$Phenotype))
res_eval$Method<-factor(res_eval$Method, level=c("pT+clump",'lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
res_eval$Model<-factor(res_eval$Model, level=c('10FCVal','MultiPRS','PseudoVal','Inf'))
res_eval<-res_eval[order(res_eval$Phenotype, res_eval$Method, res_eval$Model),]
library(data.table)
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
if(Samples[file] == 'UKBB'){
for(pheno_i in 1:length(pheno)){
if(pheno_i==1){
pheno_i_dat<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-pheno_i_dat
} else {
pheno_i_dat<-fread(paste0(UKBB_output,'/Phenotype/PRS_comp_subset/UKBB.',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-merge(pheno_dat, pheno_i_dat, by=c('FID','IID'), all=T)
}
}
} else {
for(pheno_i in 1:length(pheno)){
if(pheno_i==1){
pheno_i_dat<-fread(paste0('/users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-pheno_i_dat
} else {
pheno_i_dat<-fread(paste0('/users/k1806347/brc_scratch/Data/TEDS/Phenotypic/Derived_outcomes/TEDS_',pheno[pheno_i],'.txt'))
names(pheno_i_dat)[3]<-pheno[pheno_i]
pheno_dat<-merge(pheno_dat, pheno_i_dat, by=c('FID','IID'), all=T)
}
}
}
# Calculate corelation between all phenotypes
cors<-abs(cor(as.matrix(pheno_dat[,-1:-2]), use='p'))
cors<-cors[unique(res_eval$Phenotype,),unique(res_eval$Phenotype,)]
cors[is.na(cors)]<-0
# Run aggregate function
library(MAd)
meta_res_eval<-NULL
for(i in unique(res_eval$Test)){
res_eval_i<-res_eval[res_eval$Test == i,]
res_eval_i$Sample<-'Target'
CrossVal_agg_res_eval_i<-agg(id=Sample, es=CrossVal_R, var=CrossVal_R_SE^2, cor=cors, method="BHHR", mod=NULL, data=res_eval_i)
IndepVal_agg_res_eval_i<-agg(id=Sample, es=IndepVal_R, var=IndepVal_R_SE^2, cor=cors, method="BHHR", mod=NULL, data=res_eval_i)
meta_res_eval<-rbind(meta_res_eval, data.frame( Test=i,
CrossVal_R=CrossVal_agg_res_eval_i$es[1],
CrossVal_R_SE=sqrt(CrossVal_agg_res_eval_i$var[1]),
IndepVal_R=IndepVal_agg_res_eval_i$es[1],
IndepVal_R_SE=sqrt(IndepVal_agg_res_eval_i$var[1])))
}
# Calculate difference between methods and p-value using aggregate
# Truncate difference p-values to stop 0 values
res_comp$Cross_R_diff_pval[res_comp$Cross_R_diff_pval == 0]<-1e-320
res_comp$Indep_R_diff_pval[res_comp$Indep_R_diff_pval == 0]<-1e-320
meta_res_diff<-NULL
for(i in unique(res_eval$Test)){
for(k in unique(res_eval$Test)){
meta_res_eval_i<-meta_res_eval[meta_res_eval$Test == i,]
meta_res_eval_k<-meta_res_eval[meta_res_eval$Test == k,]
res_comp_i_k<-res_comp[res_comp$Model_1_Test == i & res_comp$Model_2_Test == k,]
# Calculate diff SE based on p-value
for(j in 1:dim(res_comp_i_k)[1]){
if(res_comp_i_k$Cross_R_diff_pval[j] == 1){
res_comp_i_k$Cross_R_diff_pval[j]<-1-0.001
}
if(res_comp_i_k$Indep_R_diff_pval[j] == 1){
res_comp_i_k$Indep_R_diff_pval[j]<-1-0.001
}
}
res_comp_i_k$Cross_R_diff_z<-qnorm(res_comp_i_k$Cross_R_diff_pval/2)
res_comp_i_k$Cross_R_diff_SE<-res_comp_i_k$Cross_R_diff/res_comp_i_k$Cross_R_diff_z
res_comp_i_k$Indep_R_diff_z<-qnorm(res_comp_i_k$Indep_R_diff_pval/2)
res_comp_i_k$Indep_R_diff_SE<-res_comp_i_k$Indep_R_diff/res_comp_i_k$Indep_R_diff_z
res_comp_i_k$Sample<-'A'
CrossVal_agg_res_comp_i_k<-agg(id=Sample, es=Cross_R_diff, var=Cross_R_diff_SE^2, cor=cors, method="BHHR", mod=NULL, data=res_comp_i_k)
IndepVal_agg_res_eval_i<-agg(id=Sample, es=Indep_R_diff, var=Indep_R_diff_SE^2, cor=cors, method="BHHR", mod=NULL, data=res_comp_i_k)
meta_res_diff<-rbind(meta_res_diff, data.frame(Test_ref=i,
Test_targ=k,
CrossVal_R_diff=CrossVal_agg_res_comp_i_k$es[1],
CrossVal_R_diff_SE=sqrt(CrossVal_agg_res_comp_i_k$var[1]),
IndepVal_R_diff=IndepVal_agg_res_eval_i$es[1],
IndepVal_R_diff_SE=sqrt(IndepVal_agg_res_eval_i$var[1])))
}
}
meta_res_diff$CrossVal_R_diff_Z<-meta_res_diff$CrossVal_R_diff/meta_res_diff$CrossVal_R_diff_SE
meta_res_diff$CrossVal_R_diff_P<-2*pnorm(-abs(meta_res_diff$CrossVal_R_diff_Z))
meta_res_diff$IndepVal_R_diff_Z<-meta_res_diff$IndepVal_R_diff/meta_res_diff$IndepVal_R_diff_SE
meta_res_diff$IndepVal_R_diff_P<-2*pnorm(-abs(meta_res_diff$IndepVal_R_diff_Z))
meta_res_diff<-meta_res_diff[,c("Test_ref","Test_targ","CrossVal_R_diff","CrossVal_R_diff_Z","CrossVal_R_diff_P","IndepVal_R_diff","IndepVal_R_diff_Z","IndepVal_R_diff_P")]
meta_res_diff$CrossVal_R_diff_Z[meta_res_diff$CrossVal_R_diff == 0]<-0
meta_res_diff$CrossVal_R_diff_P[meta_res_diff$CrossVal_R_diff == 0]<-1
meta_res_diff$IndepVal_R_diff_Z[meta_res_diff$IndepVal_R_diff == 0]<-0
meta_res_diff$IndepVal_R_diff_P[meta_res_diff$IndepVal_R_diff == 0]<-1
# Calculate percentage improvement
meta_res_diff$CrossVal_R_diff_perc<-NA
for(i in unique(meta_res_diff$Test_ref)){
meta_res_diff$CrossVal_R_diff_perc[meta_res_diff$Test_ref == i]<-meta_res_diff$CrossVal_R_diff[meta_res_diff$Test_ref == i]/meta_res_eval$CrossVal_R[meta_res_eval$Test == i]*100
}
meta_res_diff$IndepVal_R_diff_perc<-NA
for(i in unique(meta_res_diff$Test_ref)){
meta_res_diff$IndepVal_R_diff_perc[meta_res_diff$Test_ref == i]<-meta_res_diff$IndepVal_R_diff[meta_res_diff$Test_ref == i]/meta_res_eval$IndepVal_R[meta_res_eval$Test == i]*100
}
write.csv(meta_res_eval, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_',Files[file],'_meta.csv'), row.names=F, quote=F)
meta_res_diff<-meta_res_diff[,c("Test_ref","Test_targ","CrossVal_R_diff",'CrossVal_R_diff_perc',"CrossVal_R_diff_Z","CrossVal_R_diff_P","IndepVal_R_diff",'IndepVal_R_diff_perc',"IndepVal_R_diff_Z","IndepVal_R_diff_P")]
write.csv(meta_res_diff, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PCresid/PRS_method_comp_',Files[file],'_diff_meta.csv'), row.names=F, quote=F)
###
# Create plot showing performance of each method
###
# To avoid unequal groups, insert NA for missing Model/Method pairs
meta_res_eval$Method<-gsub('\\..*','',meta_res_eval$Test)
meta_res_eval$Model<-gsub('.*\\.','',meta_res_eval$Test)
meta_res_eval$Method<-factor(meta_res_eval$Method, levels=c("pT+clump",'lassosum','PRScs','SBLUP','SBayesR','LDpred1','LDpred2','DBSLMM','All'))
meta_res_eval$Model<-factor(meta_res_eval$Model, levels=c('10FCVal','MultiPRS','PseudoVal','Inf'))
for(i in unique(meta_res_eval$Method)){
for(k in unique(meta_res_eval$Model)){
if(dim(meta_res_eval[meta_res_eval$Method == i & meta_res_eval$Model == k,])[1] > 0){
next
} else {
dud<-data.frame(Test=NA,
CrossVal_R=NA,
CrossVal_R_SE=NA,
IndepVal_R=NA,
IndepVal_R_SE=NA,
Method=i,
Model=k)
meta_res_eval<-rbind(meta_res_eval,dud)
}
}
}
res_eval<-res_eval[,c('Phenotype','Test','CrossVal_R','IndepVal_R','Method','Model')]
for(i in unique(res_eval$Method)){
for(k in unique(res_eval$Model)){
if(dim(res_eval[res_eval$Method == i & res_eval$Model == k,])[1] > 0){
next
} else {
dud<-data.frame(Phenotype=NA,
Test=paste0(i,'.',k),
CrossVal_R=NA,
IndepVal_R=NA,
Method=i,
Model=k)
res_eval<-rbind(res_eval,dud)
}
}
}
library(ggplot2)
library(cowplot)
meta_res_eval_plots_cross[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=CrossVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R-CrossVal_R_SE, ymax=CrossVal_R+CrossVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
meta_res_eval_plots_indep[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=IndepVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R-IndepVal_R_SE, ymax=IndepVal_R+IndepVal_R_SE), width=.2, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
if(Samples[file] == 'UKBB'){
meta_res_eval_plots_cross[[Files[file]]]<-meta_res_eval_plots_cross[[Files[file]]] + coord_cartesian(ylim=c(0,0.25), clip="on")
meta_res_eval_plots_indep[[Files[file]]]<-meta_res_eval_plots_indep[[Files[file]]] + coord_cartesian(ylim=c(0,0.25), clip="on")
} else {
meta_res_eval_plots_cross[[Files[file]]]<-meta_res_eval_plots_cross[[Files[file]]] + coord_cartesian(ylim=c(0,0.375), clip="on")
meta_res_eval_plots_indep[[Files[file]]]<-meta_res_eval_plots_indep[[Files[file]]] + coord_cartesian(ylim=c(0,0.375), clip="on")
}
###
# Create plot showing performance of each method compared to pT+clump
###
# Calculate differences at the average and phenotype specific level
meta_res_eval$CrossVal_R_diff<-meta_res_eval$CrossVal_R-meta_res_eval$CrossVal_R[which(meta_res_eval$Test == "pT+clump.10FCVal")]
meta_res_eval$IndepVal_R_diff<-meta_res_eval$IndepVal_R-meta_res_eval$IndepVal_R[which(meta_res_eval$Test == "pT+clump.10FCVal")]
res_eval$CrossVal_R_diff<-NA
res_eval$IndepVal_R_diff<-NA
for(i in pheno){
res_eval$CrossVal_R_diff[which(res_eval$Phenotype == i)]<-res_eval$CrossVal_R[which(res_eval$Phenotype == i)]-res_eval$CrossVal_R[which(res_eval$Test == "pT+clump.10FCVal" & res_eval$Phenotype == i)]
res_eval$IndepVal_R_diff[which(res_eval$Phenotype == i)]<-res_eval$IndepVal_R[which(res_eval$Phenotype == i)]-res_eval$IndepVal_R[which(res_eval$Test == "pT+clump.10FCVal" & res_eval$Phenotype == i)]
}
meta_res_eval_diff_plots_cross[[Files[file]]] <- ggplot( meta_res_eval, aes(x=Method, y=CrossVal_R_diff, fill=Model)) +
geom_point(data=res_eval, mapping=aes(x=Method, y=CrossVal_R_diff, colour=Model), position=position_jitterdodge(jitter.width = 0.4, dodge.width = 1), alpha=0.3) +
geom_errorbar(aes(ymin=CrossVal_R_diff-CrossVal_R_SE, ymax=CrossVal_R_diff+CrossVal_R_SE), width=0.7, position=position_dodge(width = 1)) +
geom_point(stat="identity", position=position_dodge(1), size=2, shape=23, colour='black') +
labs(y="Difference in\nCorrelations (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y') +
ylim(c(-0.12,0.12))
meta_res_eval_diff_plots_indep[[Files[file]]] <- ggplot(meta_res_eval, aes(x=Method, y=IndepVal_R_diff, fill=Model)) +
geom_point(data=res_eval, mapping=aes(x=Method, y=IndepVal_R_diff, colour=Model), position=position_jitterdodge(jitter.width = 0.4, dodge.width = 1), alpha=0.3) +
geom_errorbar(aes(ymin=IndepVal_R_diff-IndepVal_R_SE, ymax=IndepVal_R_diff+IndepVal_R_SE), width=0.7, position=position_dodge(width = 1)) +
geom_point(stat="identity", position=position_dodge(1), size=2, shape=23, colour='black') +
labs(y="Difference in\nCorrelations (SE)", x='', title=Files_descrip[file]) +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
geom_vline(xintercept = 1.5, linetype="dotted") +
geom_vline(xintercept = 2.5, linetype="dotted") +
geom_vline(xintercept = 3.5, linetype="dotted") +
geom_vline(xintercept = 4.5, linetype="dotted") +
geom_vline(xintercept = 5.5, linetype="dotted") +
geom_vline(xintercept = 6.5, linetype="dotted") +
geom_vline(xintercept = 7.5, linetype="dotted") +
geom_vline(xintercept = 8.5, linetype="dotted") +
background_grid(major = 'y', minor = 'y') +
ylim(c(-0.12,0.12))
###
# Create plot showing matrix of differences with significance
###
library(reshape2)
meta_res_diff$CrossVal_R_diff[meta_res_diff$Test_ref == meta_res_diff$Test_targ]<-NA
meta_res_diff$IndepVal_R_diff[meta_res_diff$Test_ref == meta_res_diff$Test_targ]<-NA
meta_res_diff$Cross_R_diff_catagory<-'NA'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.002]<-'-0.025 - -0.002'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.025]<-'-0.08 - -0.025'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff < -0.08]<-'< -0.08'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > -0.002 & meta_res_diff$CrossVal_R_diff < 0.002]<-'-0.002 - 0.002'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.002]<-'0.002 - 0.025'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.025]<-'0.025 - 0.08'
meta_res_diff$Cross_R_diff_catagory[meta_res_diff$CrossVal_R_diff > 0.08]<-'> 0.08'
meta_res_diff$Cross_R_diff_catagory<-factor(meta_res_diff$Cross_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
meta_res_diff$Indep_R_diff_catagory<-'NA'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.002]<-'-0.025 - -0.002'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.025]<-'-0.08 - -0.025'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff < -0.08]<-'< -0.08'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > -0.002 & meta_res_diff$IndepVal_R_diff < 0.002]<-'-0.002 - 0.002'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.002]<-'0.002 - 0.025'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.025]<-'0.025 - 0.08'
meta_res_diff$Indep_R_diff_catagory[meta_res_diff$IndepVal_R_diff > 0.08]<-'> 0.08'
meta_res_diff$Indep_R_diff_catagory<-factor(meta_res_diff$Indep_R_diff_catagory, level=rev(c('< -0.08','-0.08 - -0.025','-0.025 - -0.002','-0.002 - 0.002','0.002 - 0.025','0.025 - 0.08','> 0.08')))
meta_res_diff$cross_star<-' '
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 0.05]<-'*'
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 1e-3]<-'**'
meta_res_diff$cross_star[meta_res_diff$CrossVal_R_diff_P < 1e-6]<-'***'
meta_res_diff$indep_star<-' '
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 0.05]<-'*'
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 1e-3]<-'**'
meta_res_diff$indep_star[meta_res_diff$IndepVal_R_diff_P < 1e-6]<-'***'
res_eval_diff_matrix_indep[[pheno[i]]]<-ggplot(data = res_comp_i_select, aes(Model_2_Test, Model_1_Test, fill=Indep_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=pheno[i], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=res_comp_i_select, aes(Model_2_Test, Model_1_Test, label = indep_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(res_comp_i_select$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
library(RColorBrewer)
meta_res_eval_diff_matrix_cross[[Files[file]]]<-ggplot(data = meta_res_diff, aes(Test_targ, Test_ref, fill=Cross_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=Files_descrip[file], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1)) +
coord_fixed() +
geom_text(data=meta_res_diff, aes(Test_ref, Test_targ, label = cross_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(meta_res_diff$Cross_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
meta_res_eval_diff_matrix_indep[[Files[file]]]<-ggplot(data = meta_res_diff, aes(Test_targ, Test_ref, fill=Indep_R_diff_catagory))+
geom_tile(color = "white")+
labs(y='Test', x='Comparison', title=Files_descrip[file], fill='R difference') +
theme_minimal()+
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 8, hjust = 1))+
coord_fixed() +
geom_text(data=meta_res_diff, aes(Test_targ, Test_ref, label = indep_star), color = "black", size = 4, angle = 0, vjust=0.8) +
scale_fill_brewer(breaks = levels(meta_res_diff$Indep_R_diff_catagory), palette = "RdBu", drop=F, na.value='grey')
}
dir.create('/scratch/users/k1806347/Analyses/PRS_comparison/PCresid')
png('/scratch/users/k1806347/Analyses/PRS_comparison/PCresid/PRS_method_comp_CrossVal_meta.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_plots_cross, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PCresid/PRS_method_comp_IndepVal_meta.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_plots_indep, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PCresid/PRS_method_comp_pTDiff_CrossVal_meta.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_diff_plots_cross, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PCresid/PRS_method_comp_pTDiff_IndepVal_meta.png', units='px', res=300, width=3200, height=1000)
plot_grid(plotlist=meta_res_eval_diff_plots_indep, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PCresid/PRS_method_comp_diff_CrossVal_meta.png', units='px', res=300, width=4000, height=2000)
plot_grid(plotlist=meta_res_eval_diff_matrix_cross, ncol = 2, nrow = 1)
dev.off()
png('/scratch/users/k1806347/Analyses/PRS_comparison/PCresid/PRS_method_comp_diff_IndepVal_meta.png', units='px', res=300, width=4000, height=2000)
plot_grid(plotlist=meta_res_eval_diff_matrix_indep, ncol = 2, nrow = 1)
dev.off()Show AVENGEME polygenicity estimates table
| Phenotype | GWAS | HighPoly | pi0_est | pi0_lowCI | pi0_highCI |
|---|---|---|---|---|---|
| Depression | DEPR06 | TRUE | 0.879 | 0.864 | 0.892 |
| Intelligence | COLL01 | TRUE | 0.901 | 0.880 | 0.917 |
| Height | HEIG03 | TRUE | 0.953 | 0.951 | 0.956 |
| BMI | BODY04 | TRUE | 0.856 | 0.847 | 0.863 |
| T2D | DIAB05 | FALSE | 0.971 | 0.969 | 0.973 |
| CAD | COAD01 | TRUE | 0.953 | 0.948 | 0.957 |
| IBD | CROH01 | FALSE | 0.984 | 0.982 | 0.986 |
| MultiScler | SCLE03 | FALSE | 0.981 | 0.977 | 0.984 |
| RheuArth | RHEU02 | FALSE | 0.980 | 0.978 | 0.983 |
| Prostate_Cancer | PRCA01 | FALSE | 0.968 | 0.963 | 0.971 |
| Breast_Cancer | BRCA01 | FALSE | 0.970 | 0.967 | 0.973 |
| Height21 | HEIG03 | TRUE | 0.944 | 0.936 | 0.951 |
| BMI21 | BODY11 | TRUE | 0.843 | 0.817 | 0.866 |
| GCSE | EDUC03 | TRUE | 0.785 | 0.760 | 0.807 |
| ADHD | ADHD04 | TRUE | 0.910 | 0.862 | 0.937 |
4.1 1KG Reference
4.1.1 UKB
Show pT+clump method results
| Phenotype | Method | Model | CrossVal R (SE) | IndepVal R (SE) |
|---|---|---|---|---|
| Depression | pT+clump (sparse) | 10FCVal | 0.12 (0.005) | 0.118 (0.01) |
| Depression | pT+clump (sparse) | Multi-PRS | 0.127 (0.005) | 0.13 (0.01) |
| Depression | pT+clump (non-nested) | 10FCVal | 0.09 (0.005) | 0.086 (0.01) |
| Depression | pT+clump (non-nested) | Multi-PRS | 0.126 (0.005) | 0.13 (0.01) |
| Depression | pT+clump (dense) | 10FCVal | 0.12 (0.005) | 0.117 (0.01) |
| Depression | pT+clump (dense) | Multi-PRS | 0.126 (0.005) | 0.13 (0.01) |
| Intelligence | pT+clump (sparse) | 10FCVal | 0.084 (0.005) | 0.074 (0.01) |
| Intelligence | pT+clump (sparse) | Multi-PRS | 0.086 (0.005) | 0.08 (0.01) |
| Intelligence | pT+clump (non-nested) | 10FCVal | 0.063 (0.005) | 0.059 (0.01) |
| Intelligence | pT+clump (non-nested) | Multi-PRS | 0.086 (0.005) | 0.078 (0.01) |
| Intelligence | pT+clump (dense) | 10FCVal | 0.085 (0.005) | 0.076 (0.01) |
| Intelligence | pT+clump (dense) | Multi-PRS | 0.087 (0.005) | 0.079 (0.01) |
| Height | pT+clump (sparse) | 10FCVal | 0.304 (0.005) | 0.299 (0.01) |
| Height | pT+clump (sparse) | Multi-PRS | 0.32 (0.005) | 0.313 (0.009) |
| Height | pT+clump (non-nested) | 10FCVal | 0.26 (0.005) | 0.254 (0.01) |
| Height | pT+clump (non-nested) | Multi-PRS | 0.32 (0.005) | 0.312 (0.01) |
| Height | pT+clump (dense) | 10FCVal | 0.307 (0.005) | 0.299 (0.01) |
| Height | pT+clump (dense) | Multi-PRS | 0.32 (0.005) | 0.313 (0.01) |
| BMI | pT+clump (sparse) | 10FCVal | 0.249 (0.005) | 0.244 (0.01) |
| BMI | pT+clump (sparse) | Multi-PRS | 0.271 (0.005) | 0.274 (0.01) |
| BMI | pT+clump (non-nested) | 10FCVal | 0.184 (0.005) | 0.196 (0.01) |
| BMI | pT+clump (non-nested) | Multi-PRS | 0.271 (0.005) | 0.274 (0.01) |
| BMI | pT+clump (dense) | 10FCVal | 0.25 (0.005) | 0.242 (0.01) |
| BMI | pT+clump (dense) | Multi-PRS | 0.272 (0.005) | 0.275 (0.01) |
| T2D | pT+clump (sparse) | 10FCVal | 0.167 (0.005) | 0.173 (0.01) |
| T2D | pT+clump (sparse) | Multi-PRS | 0.212 (0.005) | 0.215 (0.01) |
| T2D | pT+clump (non-nested) | 10FCVal | 0.147 (0.005) | 0.153 (0.01) |
| T2D | pT+clump (non-nested) | Multi-PRS | 0.21 (0.005) | 0.213 (0.01) |
| T2D | pT+clump (dense) | 10FCVal | 0.168 (0.005) | 0.171 (0.01) |
| T2D | pT+clump (dense) | Multi-PRS | 0.211 (0.005) | 0.214 (0.01) |
| CAD | pT+clump (sparse) | 10FCVal | 0.135 (0.005) | 0.134 (0.01) |
| CAD | pT+clump (sparse) | Multi-PRS | 0.167 (0.005) | 0.166 (0.01) |
| CAD | pT+clump (non-nested) | 10FCVal | 0.098 (0.005) | 0.099 (0.01) |
| CAD | pT+clump (non-nested) | Multi-PRS | 0.167 (0.005) | 0.166 (0.01) |
| CAD | pT+clump (dense) | 10FCVal | 0.136 (0.005) | 0.137 (0.01) |
| CAD | pT+clump (dense) | Multi-PRS | 0.168 (0.005) | 0.167 (0.01) |
| IBD | pT+clump (sparse) | 10FCVal | 0.112 (0.005) | 0.112 (0.01) |
| IBD | pT+clump (sparse) | Multi-PRS | 0.128 (0.005) | 0.137 (0.01) |
| IBD | pT+clump (non-nested) | 10FCVal | 0.094 (0.005) | 0.107 (0.01) |
| IBD | pT+clump (non-nested) | Multi-PRS | 0.128 (0.005) | 0.138 (0.01) |
| IBD | pT+clump (dense) | 10FCVal | 0.113 (0.005) | 0.117 (0.01) |
| IBD | pT+clump (dense) | Multi-PRS | 0.127 (0.005) | 0.136 (0.01) |
| MultiScler | pT+clump (sparse) | 10FCVal | 0.09 (0.005) | 0.092 (0.01) |
| MultiScler | pT+clump (sparse) | Multi-PRS | 0.115 (0.005) | 0.116 (0.01) |
| MultiScler | pT+clump (non-nested) | 10FCVal | 0.079 (0.005) | 0.075 (0.01) |
| MultiScler | pT+clump (non-nested) | Multi-PRS | 0.114 (0.005) | 0.116 (0.01) |
| MultiScler | pT+clump (dense) | 10FCVal | 0.089 (0.005) | 0.084 (0.01) |
| MultiScler | pT+clump (dense) | Multi-PRS | 0.112 (0.005) | 0.115 (0.01) |
| RheuArth | pT+clump (sparse) | 10FCVal | 0.111 (0.005) | 0.103 (0.01) |
| RheuArth | pT+clump (sparse) | Multi-PRS | 0.131 (0.005) | 0.124 (0.01) |
| RheuArth | pT+clump (non-nested) | 10FCVal | 0.096 (0.005) | 0.078 (0.01) |
| RheuArth | pT+clump (non-nested) | Multi-PRS | 0.13 (0.005) | 0.124 (0.01) |
| RheuArth | pT+clump (dense) | 10FCVal | 0.111 (0.005) | 0.103 (0.01) |
| RheuArth | pT+clump (dense) | Multi-PRS | 0.132 (0.005) | 0.126 (0.01) |
| Prostate_Cancer | pT+clump (sparse) | 10FCVal | 0.152 (0.005) | 0.166 (0.01) |
| Prostate_Cancer | pT+clump (sparse) | Multi-PRS | 0.168 (0.005) | 0.175 (0.01) |
| Prostate_Cancer | pT+clump (non-nested) | 10FCVal | 0.138 (0.005) | 0.14 (0.01) |
| Prostate_Cancer | pT+clump (non-nested) | Multi-PRS | 0.169 (0.005) | 0.176 (0.01) |
| Prostate_Cancer | pT+clump (dense) | 10FCVal | 0.155 (0.005) | 0.17 (0.01) |
| Prostate_Cancer | pT+clump (dense) | Multi-PRS | 0.17 (0.005) | 0.18 (0.01) |
| Breast_Cancer | pT+clump (sparse) | 10FCVal | 0.179 (0.005) | 0.167 (0.01) |
| Breast_Cancer | pT+clump (sparse) | Multi-PRS | 0.2 (0.005) | 0.188 (0.01) |
| Breast_Cancer | pT+clump (non-nested) | 10FCVal | 0.168 (0.005) | 0.165 (0.01) |
| Breast_Cancer | pT+clump (non-nested) | Multi-PRS | 0.199 (0.005) | 0.187 (0.01) |
| Breast_Cancer | pT+clump (dense) | 10FCVal | 0.181 (0.005) | 0.163 (0.01) |
| Breast_Cancer | pT+clump (dense) | Multi-PRS | 0.198 (0.005) | 0.19 (0.01) |
Show cross-validation results
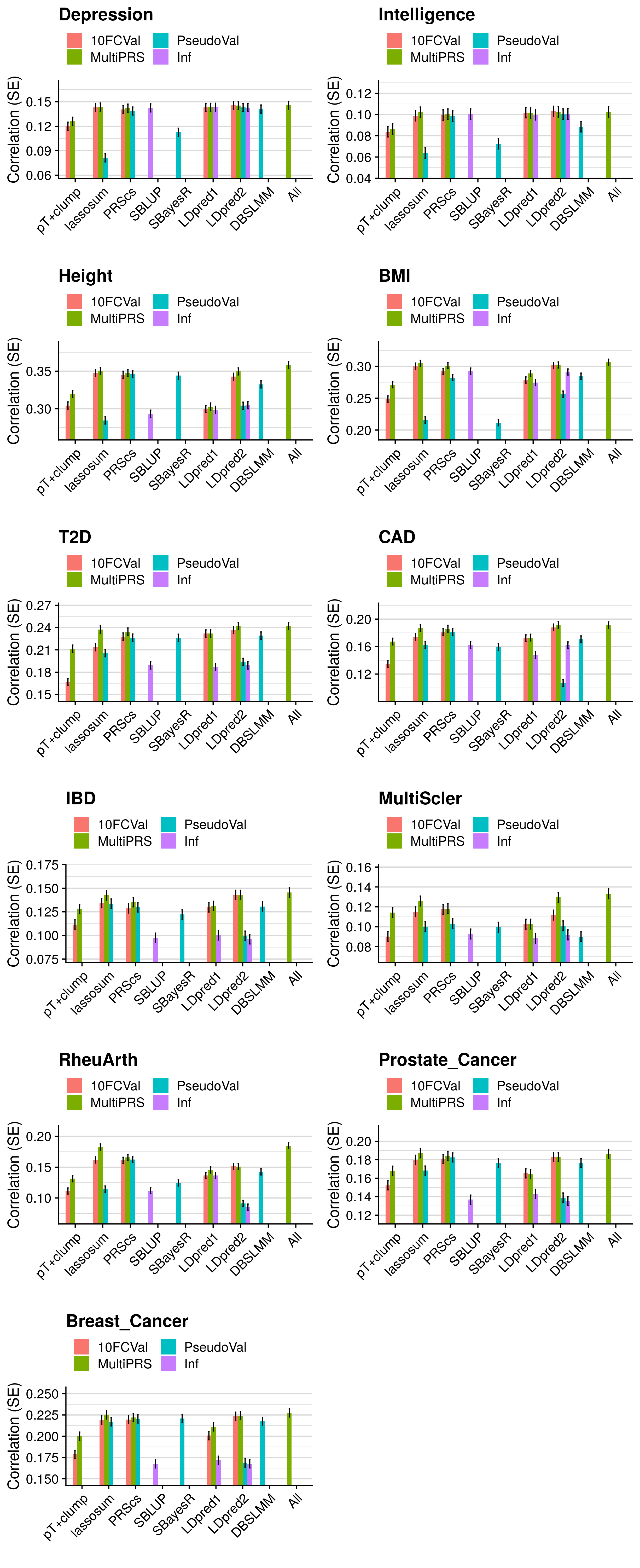
Predictive utility estimated using 10-fold cross-validation
Show test-validation results
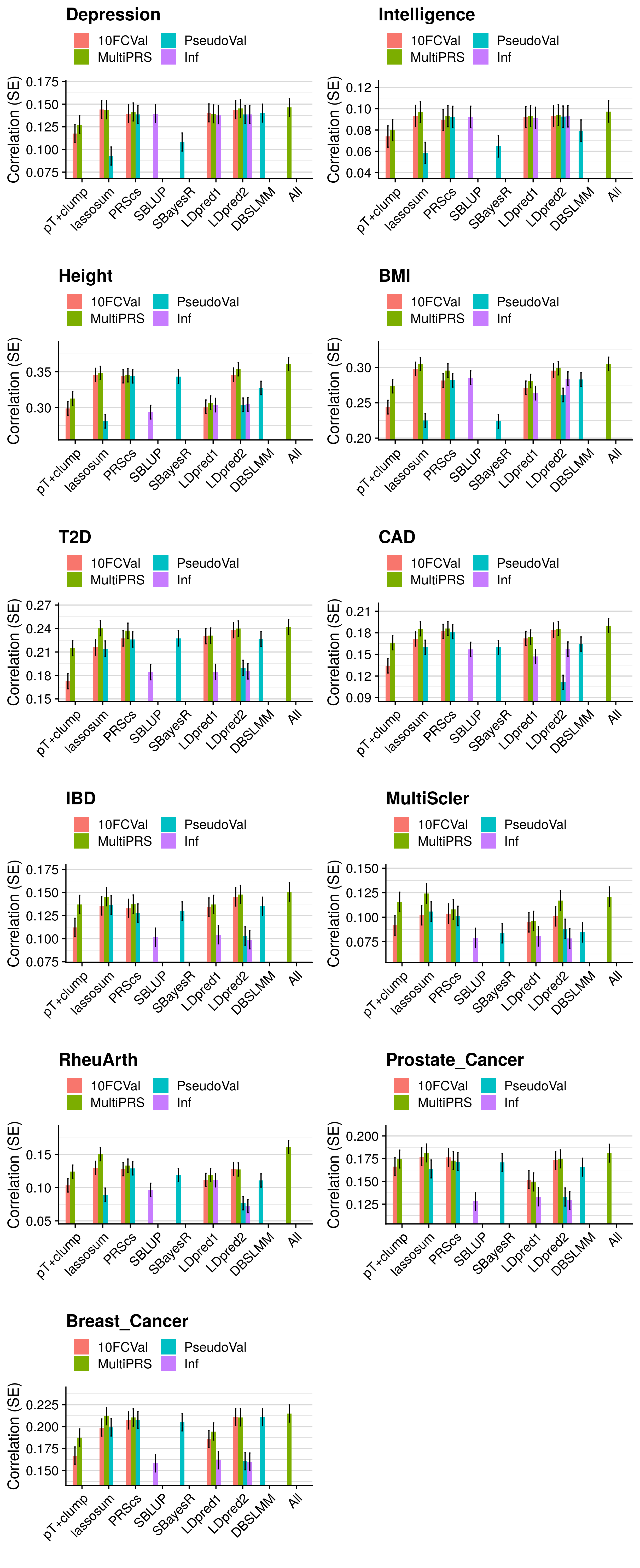
Predictive utility estimated using test set validation
Show UKBB results table
| Phenotype | Method | Model | CrossVal_R | CrossVal_R_SE | CrossVal_OR | CrossVal_LowCI | CrossVal_HighCI | Cross_LiabR2 | Cross_AUC | CrossVal_pval | IndepVal_R | IndepVal_R_SE | IndepVal_OR | IndepVal_LowCI | IndepVal_HighCI | Indep_LiabR2 | Indep_AUC | IndepVal_pval |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Depression | All | MultiPRS | 0.146 | 0.005 | 1.346 | 1.319 | 1.374 | 0.026 | 0.583 | 1.10e-188 | 0.146 | 0.010 | 1.348 | 1.295 | 1.404 | 0.026 | 0.582 | 5.97e-49 |
| Depression | DBSLMM | PseudoVal | 0.141 | 0.005 | 1.333 | 1.307 | 1.360 | 0.024 | 0.580 | 5.40e-177 | 0.140 | 0.010 | 1.331 | 1.278 | 1.385 | 0.024 | 0.578 | 4.73e-45 |
| Depression | lassosum | MultiPRS | 0.144 | 0.005 | 1.340 | 1.314 | 1.368 | 0.025 | 0.581 | 3.04e-183 | 0.144 | 0.010 | 1.341 | 1.288 | 1.396 | 0.025 | 0.580 | 2.86e-47 |
| Depression | lassosum | PseudoVal | 0.081 | 0.005 | 1.178 | 1.155 | 1.201 | 0.008 | 0.547 | 1.15e-59 | 0.093 | 0.010 | 1.206 | 1.159 | 1.255 | 0.010 | 0.552 | 1.47e-20 |
| Depression | lassosum | 10FCVal | 0.143 | 0.005 | 1.339 | 1.312 | 1.366 | 0.025 | 0.581 | 8.31e-182 | 0.144 | 0.010 | 1.342 | 1.288 | 1.397 | 0.025 | 0.580 | 1.84e-47 |
| Depression | LDpred1 | MultiPRS | 0.143 | 0.005 | 1.339 | 1.313 | 1.367 | 0.025 | 0.581 | 1.93e-182 | 0.139 | 0.010 | 1.328 | 1.276 | 1.383 | 0.023 | 0.578 | 1.63e-44 |
| Depression | LDpred1 | Inf | 0.143 | 0.005 | 1.340 | 1.313 | 1.367 | 0.025 | 0.581 | 1.22e-182 | 0.138 | 0.010 | 1.325 | 1.273 | 1.380 | 0.023 | 0.577 | 7.40e-44 |
| Depression | LDpred1 | 10FCVal | 0.143 | 0.005 | 1.339 | 1.312 | 1.366 | 0.025 | 0.581 | 6.49e-182 | 0.140 | 0.010 | 1.331 | 1.279 | 1.386 | 0.024 | 0.579 | 3.41e-45 |
| Depression | LDpred2 | MultiPRS | 0.145 | 0.005 | 1.345 | 1.318 | 1.373 | 0.025 | 0.582 | 8.96e-188 | 0.145 | 0.010 | 1.345 | 1.292 | 1.400 | 0.025 | 0.582 | 2.94e-48 |
| Depression | LDpred2 | 10FCVal | 0.146 | 0.005 | 1.346 | 1.319 | 1.374 | 0.026 | 0.582 | 1.56e-188 | 0.144 | 0.010 | 1.341 | 1.288 | 1.397 | 0.025 | 0.581 | 2.01e-47 |
| Depression | LDpred2 | PseudoVal | 0.143 | 0.005 | 1.339 | 1.312 | 1.366 | 0.025 | 0.581 | 4.58e-182 | 0.139 | 0.010 | 1.326 | 1.274 | 1.381 | 0.023 | 0.578 | 4.73e-44 |
| Depression | LDpred2 | Inf | 0.143 | 0.005 | 1.338 | 1.311 | 1.365 | 0.025 | 0.581 | 2.94e-181 | 0.139 | 0.010 | 1.326 | 1.274 | 1.381 | 0.023 | 0.577 | 4.98e-44 |
| Depression | PRScs | MultiPRS | 0.142 | 0.005 | 1.336 | 1.310 | 1.364 | 0.024 | 0.580 | 1.05e-179 | 0.141 | 0.010 | 1.334 | 1.282 | 1.389 | 0.024 | 0.579 | 8.04e-46 |
| Depression | PRScs | 10FCVal | 0.141 | 0.005 | 1.332 | 1.306 | 1.359 | 0.024 | 0.579 | 6.37e-176 | 0.139 | 0.010 | 1.329 | 1.276 | 1.383 | 0.023 | 0.578 | 1.27e-44 |
| Depression | PRScs | PseudoVal | 0.139 | 0.005 | 1.326 | 1.300 | 1.353 | 0.023 | 0.578 | 7.34e-171 | 0.139 | 0.010 | 1.326 | 1.274 | 1.381 | 0.023 | 0.579 | 4.89e-44 |
| Depression | pT+clump | MultiPRS | 0.126 | 0.005 | 1.292 | 1.266 | 1.318 | 0.019 | 0.571 | 1.41e-141 | 0.127 | 0.010 | 1.295 | 1.244 | 1.348 | 0.019 | 0.570 | 2.00e-37 |
| Depression | pT+clump | 10FCVal | 0.120 | 0.005 | 1.276 | 1.250 | 1.302 | 0.017 | 0.568 | 2.42e-128 | 0.118 | 0.010 | 1.269 | 1.220 | 1.321 | 0.017 | 0.564 | 3.61e-32 |
| Depression | SBayesR | PseudoVal | 0.113 | 0.005 | 1.256 | 1.231 | 1.282 | 0.015 | 0.564 | 4.36e-113 | 0.108 | 0.010 | 1.245 | 1.196 | 1.296 | 0.014 | 0.562 | 1.88e-27 |
| Depression | SBLUP | Inf | 0.143 | 0.005 | 1.337 | 1.310 | 1.364 | 0.024 | 0.581 | 1.78e-180 | 0.139 | 0.010 | 1.329 | 1.276 | 1.383 | 0.023 | 0.578 | 1.26e-44 |
| Intelligence | All | MultiPRS | 0.102 | 0.005 | NA | NA | NA | NA | NA | 8.80e-94 | 0.097 | 0.010 | NA | NA | NA | NA | NA | 1.69e-22 |
| Intelligence | DBSLMM | PseudoVal | 0.089 | 0.005 | NA | NA | NA | NA | NA | 1.58e-70 | 0.079 | 0.010 | NA | NA | NA | NA | NA | 1.71e-15 |
| Intelligence | lassosum | MultiPRS | 0.102 | 0.005 | NA | NA | NA | NA | NA | 3.25e-93 | 0.097 | 0.010 | NA | NA | NA | NA | NA | 2.78e-22 |
| Intelligence | lassosum | PseudoVal | 0.064 | 0.005 | NA | NA | NA | NA | NA | 2.07e-37 | 0.059 | 0.010 | NA | NA | NA | NA | NA | 4.55e-09 |
| Intelligence | lassosum | 10FCVal | 0.099 | 0.005 | NA | NA | NA | NA | NA | 1.37e-87 | 0.093 | 0.010 | NA | NA | NA | NA | NA | 9.43e-21 |
| Intelligence | LDpred1 | MultiPRS | 0.101 | 0.005 | NA | NA | NA | NA | NA | 1.10e-91 | 0.093 | 0.010 | NA | NA | NA | NA | NA | 1.05e-20 |
| Intelligence | LDpred1 | Inf | 0.100 | 0.005 | NA | NA | NA | NA | NA | 4.02e-89 | 0.092 | 0.010 | NA | NA | NA | NA | NA | 4.68e-20 |
| Intelligence | LDpred1 | 10FCVal | 0.102 | 0.005 | NA | NA | NA | NA | NA | 9.61e-93 | 0.092 | 0.010 | NA | NA | NA | NA | NA | 2.58e-20 |
| Intelligence | LDpred2 | MultiPRS | 0.103 | 0.005 | NA | NA | NA | NA | NA | 6.70e-94 | 0.094 | 0.010 | NA | NA | NA | NA | NA | 4.14e-21 |
| Intelligence | LDpred2 | 10FCVal | 0.103 | 0.005 | NA | NA | NA | NA | NA | 1.13e-94 | 0.093 | 0.010 | NA | NA | NA | NA | NA | 1.08e-20 |
| Intelligence | LDpred2 | PseudoVal | 0.100 | 0.005 | NA | NA | NA | NA | NA | 4.59e-90 | 0.093 | 0.010 | NA | NA | NA | NA | NA | 1.74e-20 |
| Intelligence | LDpred2 | Inf | 0.101 | 0.005 | NA | NA | NA | NA | NA | 1.99e-90 | 0.093 | 0.010 | NA | NA | NA | NA | NA | 1.34e-20 |
| Intelligence | PRScs | MultiPRS | 0.100 | 0.005 | NA | NA | NA | NA | NA | 4.92e-90 | 0.093 | 0.010 | NA | NA | NA | NA | NA | 1.15e-20 |
| Intelligence | PRScs | 10FCVal | 0.099 | 0.005 | NA | NA | NA | NA | NA | 1.64e-88 | 0.089 | 0.010 | NA | NA | NA | NA | NA | 3.13e-19 |
| Intelligence | PRScs | PseudoVal | 0.099 | 0.005 | NA | NA | NA | NA | NA | 7.97e-87 | 0.092 | 0.010 | NA | NA | NA | NA | NA | 2.13e-20 |
| Intelligence | pT+clump | MultiPRS | 0.086 | 0.005 | NA | NA | NA | NA | NA | 2.92e-67 | 0.080 | 0.010 | NA | NA | NA | NA | NA | 1.24e-15 |
| Intelligence | pT+clump | 10FCVal | 0.084 | 0.005 | NA | NA | NA | NA | NA | 2.38e-63 | 0.074 | 0.010 | NA | NA | NA | NA | NA | 1.32e-13 |
| Intelligence | SBayesR | PseudoVal | 0.072 | 0.005 | NA | NA | NA | NA | NA | 9.36e-48 | 0.065 | 0.010 | NA | NA | NA | NA | NA | 9.77e-11 |
| Intelligence | SBLUP | Inf | 0.100 | 0.005 | NA | NA | NA | NA | NA | 4.31e-90 | 0.092 | 0.010 | NA | NA | NA | NA | NA | 1.97e-20 |
| Height | All | MultiPRS | 0.358 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.361 | 0.009 | NA | NA | NA | NA | NA | 3.86e-305 |
| Height | DBSLMM | PseudoVal | 0.332 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.327 | 0.009 | NA | NA | NA | NA | NA | 1.77e-248 |
| Height | lassosum | MultiPRS | 0.350 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.348 | 0.009 | NA | NA | NA | NA | NA | 2.57e-283 |
| Height | lassosum | 10FCVal | 0.347 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.346 | 0.009 | NA | NA | NA | NA | NA | 1.43e-278 |
| Height | lassosum | PseudoVal | 0.285 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.281 | 0.010 | NA | NA | NA | NA | NA | 7.11e-181 |
| Height | LDpred1 | MultiPRS | 0.303 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.307 | 0.010 | NA | NA | NA | NA | NA | 6.68e-217 |
| Height | LDpred1 | Inf | 0.299 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.303 | 0.010 | NA | NA | NA | NA | NA | 4.27e-212 |
| Height | LDpred1 | 10FCVal | 0.300 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.301 | 0.010 | NA | NA | NA | NA | NA | 1.31e-208 |
| Height | LDpred2 | MultiPRS | 0.350 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.354 | 0.009 | NA | NA | NA | NA | NA | 1.57e-292 |
| Height | LDpred2 | 10FCVal | 0.343 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.346 | 0.009 | NA | NA | NA | NA | NA | 3.74e-279 |
| Height | LDpred2 | PseudoVal | 0.304 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.304 | 0.010 | NA | NA | NA | NA | NA | 1.33e-212 |
| Height | LDpred2 | Inf | 0.305 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.305 | 0.010 | NA | NA | NA | NA | NA | 1.41e-213 |
| Height | PRScs | MultiPRS | 0.347 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.345 | 0.009 | NA | NA | NA | NA | NA | 5.62e-278 |
| Height | PRScs | 10FCVal | 0.345 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.344 | 0.009 | NA | NA | NA | NA | NA | 1.44e-275 |
| Height | PRScs | PseudoVal | 0.346 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.344 | 0.009 | NA | NA | NA | NA | NA | 1.97e-275 |
| Height | pT+clump | MultiPRS | 0.320 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.313 | 0.009 | NA | NA | NA | NA | NA | 8.24e-226 |
| Height | pT+clump | 10FCVal | 0.304 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.299 | 0.010 | NA | NA | NA | NA | NA | 1.94e-205 |
| Height | SBayesR | PseudoVal | 0.344 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.343 | 0.009 | NA | NA | NA | NA | NA | 9.80e-275 |
| Height | SBLUP | Inf | 0.293 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.294 | 0.010 | NA | NA | NA | NA | NA | 5.63e-198 |
| BMI | All | MultiPRS | 0.307 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.305 | 0.010 | NA | NA | NA | NA | NA | 2.11e-214 |
| BMI | DBSLMM | PseudoVal | 0.285 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.283 | 0.010 | NA | NA | NA | NA | NA | 2.16e-183 |
| BMI | lassosum | MultiPRS | 0.305 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.305 | 0.010 | NA | NA | NA | NA | NA | 4.25e-214 |
| BMI | lassosum | PseudoVal | 0.216 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.225 | 0.010 | NA | NA | NA | NA | NA | 8.76e-115 |
| BMI | lassosum | 10FCVal | 0.300 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.298 | 0.010 | NA | NA | NA | NA | NA | 3.85e-204 |
| BMI | LDpred1 | MultiPRS | 0.289 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.281 | 0.010 | NA | NA | NA | NA | NA | 1.41e-180 |
| BMI | LDpred1 | Inf | 0.274 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.264 | 0.010 | NA | NA | NA | NA | NA | 9.50e-159 |
| BMI | LDpred1 | 10FCVal | 0.279 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.271 | 0.010 | NA | NA | NA | NA | NA | 4.25e-168 |
| BMI | LDpred2 | MultiPRS | 0.302 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.299 | 0.010 | NA | NA | NA | NA | NA | 1.20e-205 |
| BMI | LDpred2 | 10FCVal | 0.301 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.296 | 0.010 | NA | NA | NA | NA | NA | 6.63e-201 |
| BMI | LDpred2 | PseudoVal | 0.256 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.261 | 0.010 | NA | NA | NA | NA | NA | 1.56e-155 |
| BMI | LDpred2 | Inf | 0.291 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.284 | 0.010 | NA | NA | NA | NA | NA | 4.47e-185 |
| BMI | PRScs | MultiPRS | 0.301 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.296 | 0.010 | NA | NA | NA | NA | NA | 7.33e-201 |
| BMI | PRScs | 10FCVal | 0.292 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.282 | 0.010 | NA | NA | NA | NA | NA | 1.62e-181 |
| BMI | PRScs | PseudoVal | 0.282 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.282 | 0.010 | NA | NA | NA | NA | NA | 6.73e-182 |
| BMI | pT+clump | MultiPRS | 0.271 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.274 | 0.010 | NA | NA | NA | NA | NA | 2.95e-171 |
| BMI | pT+clump | 10FCVal | 0.249 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.244 | 0.010 | NA | NA | NA | NA | NA | 2.59e-135 |
| BMI | SBayesR | PseudoVal | 0.211 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.224 | 0.010 | NA | NA | NA | NA | NA | 1.09e-113 |
| BMI | SBLUP | Inf | 0.293 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.286 | 0.010 | NA | NA | NA | NA | NA | 5.09e-187 |
| T2D | All | MultiPRS | 0.242 | 0.005 | 1.703 | 1.665 | 1.741 | 0.062 | 0.650 | 0.00e+00 | 0.242 | 0.010 | 1.695 | 1.622 | 1.772 | 0.061 | 0.649 | 5.18e-133 |
| T2D | DBSLMM | PseudoVal | 0.229 | 0.005 | 1.656 | 1.619 | 1.693 | 0.055 | 0.642 | 0.00e+00 | 0.226 | 0.010 | 1.638 | 1.568 | 1.712 | 0.053 | 0.640 | 1.81e-116 |
| T2D | lassosum | MultiPRS | 0.237 | 0.005 | 1.687 | 1.650 | 1.725 | 0.059 | 0.647 | 0.00e+00 | 0.240 | 0.010 | 1.691 | 1.618 | 1.768 | 0.060 | 0.648 | 2.09e-131 |
| T2D | lassosum | PseudoVal | 0.206 | 0.005 | 1.567 | 1.533 | 1.602 | 0.044 | 0.627 | 0.00e+00 | 0.214 | 0.010 | 1.593 | 1.525 | 1.664 | 0.047 | 0.631 | 2.66e-104 |
| T2D | lassosum | 10FCVal | 0.214 | 0.005 | 1.599 | 1.564 | 1.634 | 0.048 | 0.632 | 0.00e+00 | 0.216 | 0.010 | 1.601 | 1.532 | 1.673 | 0.048 | 0.632 | 1.11e-105 |
| T2D | LDpred1 | MultiPRS | 0.232 | 0.005 | 1.666 | 1.629 | 1.703 | 0.057 | 0.644 | 0.00e+00 | 0.231 | 0.010 | 1.656 | 1.584 | 1.730 | 0.055 | 0.642 | 3.12e-121 |
| T2D | LDpred1 | Inf | 0.187 | 0.005 | 1.507 | 1.474 | 1.540 | 0.036 | 0.617 | 1.79e-311 | 0.184 | 0.010 | 1.493 | 1.430 | 1.559 | 0.035 | 0.615 | 3.26e-77 |
| T2D | LDpred1 | 10FCVal | 0.232 | 0.005 | 1.665 | 1.629 | 1.703 | 0.057 | 0.644 | 0.00e+00 | 0.230 | 0.010 | 1.653 | 1.582 | 1.727 | 0.055 | 0.642 | 2.09e-120 |
| T2D | LDpred2 | MultiPRS | 0.242 | 0.005 | 1.704 | 1.666 | 1.742 | 0.062 | 0.650 | 0.00e+00 | 0.240 | 0.010 | 1.689 | 1.616 | 1.765 | 0.060 | 0.647 | 5.19e-131 |
| T2D | LDpred2 | 10FCVal | 0.237 | 0.005 | 1.683 | 1.646 | 1.721 | 0.059 | 0.647 | 0.00e+00 | 0.238 | 0.010 | 1.680 | 1.608 | 1.756 | 0.059 | 0.646 | 1.30e-128 |
| T2D | LDpred2 | PseudoVal | 0.194 | 0.005 | 1.529 | 1.496 | 1.563 | 0.039 | 0.620 | 0.00e+00 | 0.190 | 0.010 | 1.510 | 1.446 | 1.577 | 0.037 | 0.618 | 1.13e-81 |
| T2D | LDpred2 | Inf | 0.189 | 0.005 | 1.514 | 1.481 | 1.547 | 0.037 | 0.618 | 1.51e-318 | 0.185 | 0.010 | 1.495 | 1.432 | 1.561 | 0.035 | 0.615 | 6.76e-78 |
| T2D | PRScs | MultiPRS | 0.235 | 0.005 | 1.676 | 1.639 | 1.713 | 0.058 | 0.645 | 0.00e+00 | 0.237 | 0.010 | 1.679 | 1.606 | 1.755 | 0.058 | 0.645 | 6.75e-128 |
| T2D | PRScs | 10FCVal | 0.228 | 0.005 | 1.650 | 1.614 | 1.687 | 0.055 | 0.642 | 0.00e+00 | 0.227 | 0.010 | 1.641 | 1.571 | 1.715 | 0.053 | 0.639 | 2.41e-117 |
| T2D | PRScs | PseudoVal | 0.226 | 0.005 | 1.645 | 1.609 | 1.682 | 0.054 | 0.640 | 0.00e+00 | 0.226 | 0.010 | 1.636 | 1.566 | 1.710 | 0.053 | 0.639 | 5.73e-116 |
| T2D | pT+clump | MultiPRS | 0.212 | 0.005 | 1.592 | 1.557 | 1.628 | 0.047 | 0.631 | 0.00e+00 | 0.215 | 0.010 | 1.598 | 1.529 | 1.670 | 0.048 | 0.633 | 6.05e-105 |
| T2D | pT+clump | 10FCVal | 0.167 | 0.005 | 1.440 | 1.409 | 1.472 | 0.029 | 0.604 | 1.44e-247 | 0.173 | 0.010 | 1.455 | 1.393 | 1.519 | 0.030 | 0.607 | 8.66e-68 |
| T2D | SBayesR | PseudoVal | 0.226 | 0.005 | 1.644 | 1.608 | 1.681 | 0.054 | 0.640 | 0.00e+00 | 0.227 | 0.010 | 1.641 | 1.570 | 1.714 | 0.053 | 0.640 | 1.97e-117 |
| T2D | SBLUP | Inf | 0.189 | 0.005 | 1.514 | 1.481 | 1.547 | 0.037 | 0.618 | 1.73e-318 | 0.184 | 0.010 | 1.492 | 1.429 | 1.558 | 0.035 | 0.614 | 4.04e-77 |
| CAD | All | MultiPRS | 0.191 | 0.005 | 1.485 | 1.454 | 1.516 | 0.027 | 0.609 | 0.00e+00 | 0.190 | 0.010 | 1.482 | 1.422 | 1.544 | 0.027 | 0.607 | 5.38e-82 |
| CAD | DBSLMM | PseudoVal | 0.171 | 0.005 | 1.420 | 1.391 | 1.449 | 0.022 | 0.598 | 1.03e-258 | 0.164 | 0.010 | 1.401 | 1.345 | 1.459 | 0.020 | 0.593 | 1.68e-61 |
| CAD | lassosum | MultiPRS | 0.187 | 0.005 | 1.473 | 1.443 | 1.503 | 0.026 | 0.607 | 1.50e-312 | 0.186 | 0.010 | 1.467 | 1.408 | 1.529 | 0.026 | 0.604 | 3.66e-78 |
| CAD | lassosum | PseudoVal | 0.162 | 0.005 | 1.394 | 1.366 | 1.423 | 0.020 | 0.592 | 6.96e-234 | 0.160 | 0.010 | 1.388 | 1.332 | 1.445 | 0.019 | 0.588 | 3.04e-58 |
| CAD | lassosum | 10FCVal | 0.174 | 0.005 | 1.430 | 1.401 | 1.460 | 0.023 | 0.599 | 5.99e-269 | 0.171 | 0.010 | 1.423 | 1.366 | 1.482 | 0.022 | 0.596 | 8.84e-67 |
| CAD | LDpred1 | MultiPRS | 0.173 | 0.005 | 1.428 | 1.399 | 1.457 | 0.022 | 0.599 | 1.24e-266 | 0.174 | 0.010 | 1.431 | 1.374 | 1.491 | 0.023 | 0.598 | 6.37e-69 |
| CAD | LDpred1 | Inf | 0.148 | 0.005 | 1.352 | 1.325 | 1.380 | 0.016 | 0.585 | 1.03e-193 | 0.147 | 0.010 | 1.351 | 1.297 | 1.406 | 0.016 | 0.584 | 1.57e-49 |
| CAD | LDpred1 | 10FCVal | 0.172 | 0.005 | 1.425 | 1.396 | 1.454 | 0.022 | 0.598 | 1.83e-263 | 0.172 | 0.010 | 1.425 | 1.368 | 1.484 | 0.022 | 0.597 | 2.22e-67 |
| CAD | LDpred2 | MultiPRS | 0.192 | 0.005 | 1.487 | 1.457 | 1.518 | 0.028 | 0.610 | 0.00e+00 | 0.186 | 0.010 | 1.468 | 1.409 | 1.530 | 0.026 | 0.604 | 2.44e-78 |
| CAD | LDpred2 | 10FCVal | 0.188 | 0.005 | 1.475 | 1.445 | 1.505 | 0.026 | 0.607 | 1.40e-314 | 0.184 | 0.010 | 1.462 | 1.403 | 1.523 | 0.025 | 0.604 | 8.81e-77 |
| CAD | LDpred2 | PseudoVal | 0.107 | 0.005 | 1.241 | 1.217 | 1.266 | 0.008 | 0.559 | 4.79e-102 | 0.111 | 0.010 | 1.252 | 1.203 | 1.303 | 0.009 | 0.561 | 7.84e-29 |
| CAD | LDpred2 | Inf | 0.162 | 0.005 | 1.394 | 1.366 | 1.422 | 0.019 | 0.593 | 8.28e-233 | 0.157 | 0.010 | 1.380 | 1.326 | 1.438 | 0.018 | 0.590 | 1.69e-56 |
| CAD | PRScs | MultiPRS | 0.186 | 0.005 | 1.469 | 1.439 | 1.499 | 0.026 | 0.607 | 4.20e-308 | 0.186 | 0.010 | 1.468 | 1.409 | 1.530 | 0.026 | 0.605 | 2.10e-78 |
| CAD | PRScs | 10FCVal | 0.181 | 0.005 | 1.454 | 1.424 | 1.484 | 0.025 | 0.604 | 3.86e-293 | 0.182 | 0.010 | 1.456 | 1.397 | 1.517 | 0.025 | 0.603 | 3.72e-75 |
| CAD | PRScs | PseudoVal | 0.181 | 0.005 | 1.453 | 1.423 | 1.483 | 0.024 | 0.603 | 6.93e-292 | 0.182 | 0.010 | 1.455 | 1.396 | 1.516 | 0.025 | 0.603 | 6.18e-75 |
| CAD | pT+clump | MultiPRS | 0.167 | 0.005 | 1.410 | 1.382 | 1.439 | 0.021 | 0.596 | 7.20e-249 | 0.166 | 0.010 | 1.407 | 1.351 | 1.466 | 0.021 | 0.593 | 6.59e-63 |
| CAD | pT+clump | 10FCVal | 0.135 | 0.005 | 1.315 | 1.289 | 1.342 | 0.013 | 0.577 | 5.47e-161 | 0.134 | 0.010 | 1.314 | 1.262 | 1.368 | 0.013 | 0.575 | 2.61e-41 |
| CAD | SBayesR | PseudoVal | 0.159 | 0.005 | 1.386 | 1.358 | 1.415 | 0.019 | 0.590 | 3.96e-226 | 0.160 | 0.010 | 1.387 | 1.332 | 1.444 | 0.019 | 0.589 | 4.38e-58 |
| CAD | SBLUP | Inf | 0.162 | 0.005 | 1.394 | 1.366 | 1.423 | 0.020 | 0.593 | 1.36e-233 | 0.157 | 0.010 | 1.379 | 1.325 | 1.437 | 0.018 | 0.590 | 2.87e-56 |
| IBD | All | MultiPRS | 0.146 | 0.005 | 1.558 | 1.510 | 1.608 | 0.049 | 0.644 | 3.88e-188 | 0.151 | 0.010 | 1.583 | 1.486 | 1.686 | 0.053 | 0.651 | 7.89e-52 |
| IBD | DBSLMM | PseudoVal | 0.131 | 0.005 | 1.503 | 1.457 | 1.552 | 0.039 | 0.632 | 7.13e-152 | 0.135 | 0.010 | 1.530 | 1.435 | 1.631 | 0.042 | 0.637 | 5.85e-42 |
| IBD | lassosum | MultiPRS | 0.142 | 0.005 | 1.546 | 1.498 | 1.595 | 0.047 | 0.641 | 3.20e-180 | 0.146 | 0.010 | 1.564 | 1.468 | 1.666 | 0.049 | 0.648 | 1.83e-48 |
| IBD | lassosum | PseudoVal | 0.134 | 0.005 | 1.518 | 1.471 | 1.567 | 0.041 | 0.634 | 5.40e-159 | 0.137 | 0.010 | 1.529 | 1.435 | 1.629 | 0.043 | 0.640 | 8.34e-43 |
| IBD | lassosum | 10FCVal | 0.134 | 0.005 | 1.519 | 1.471 | 1.567 | 0.042 | 0.632 | 2.71e-160 | 0.136 | 0.010 | 1.525 | 1.431 | 1.626 | 0.043 | 0.639 | 3.32e-42 |
| IBD | LDpred1 | MultiPRS | 0.131 | 0.005 | 1.522 | 1.474 | 1.572 | 0.040 | 0.631 | 2.75e-153 | 0.137 | 0.010 | 1.547 | 1.450 | 1.650 | 0.044 | 0.635 | 3.55e-43 |
| IBD | LDpred1 | Inf | 0.100 | 0.005 | 1.406 | 1.359 | 1.454 | 0.023 | 0.600 | 1.50e-89 | 0.104 | 0.010 | 1.425 | 1.332 | 1.525 | 0.025 | 0.603 | 1.30e-25 |
| IBD | LDpred1 | 10FCVal | 0.130 | 0.005 | 1.504 | 1.457 | 1.553 | 0.039 | 0.629 | 8.88e-150 | 0.134 | 0.010 | 1.522 | 1.428 | 1.622 | 0.042 | 0.631 | 1.93e-41 |
| IBD | LDpred2 | MultiPRS | 0.143 | 0.005 | 1.553 | 1.505 | 1.602 | 0.047 | 0.643 | 1.28e-181 | 0.148 | 0.010 | 1.571 | 1.474 | 1.674 | 0.051 | 0.648 | 4.75e-50 |
| IBD | LDpred2 | 10FCVal | 0.143 | 0.005 | 1.551 | 1.503 | 1.600 | 0.047 | 0.643 | 1.45e-181 | 0.145 | 0.010 | 1.557 | 1.461 | 1.659 | 0.049 | 0.647 | 2.49e-48 |
| IBD | LDpred2 | PseudoVal | 0.100 | 0.005 | 1.404 | 1.357 | 1.452 | 0.023 | 0.601 | 8.77e-89 | 0.103 | 0.010 | 1.422 | 1.328 | 1.522 | 0.024 | 0.605 | 5.74e-25 |
| IBD | LDpred2 | Inf | 0.096 | 0.005 | 1.390 | 1.343 | 1.438 | 0.021 | 0.597 | 1.99e-82 | 0.099 | 0.010 | 1.407 | 1.314 | 1.507 | 0.023 | 0.600 | 3.50e-23 |
| IBD | PRScs | MultiPRS | 0.135 | 0.005 | 1.516 | 1.469 | 1.565 | 0.042 | 0.635 | 1.52e-162 | 0.137 | 0.010 | 1.536 | 1.441 | 1.637 | 0.044 | 0.638 | 2.18e-43 |
| IBD | PRScs | 10FCVal | 0.129 | 0.005 | 1.494 | 1.448 | 1.543 | 0.038 | 0.628 | 2.34e-147 | 0.133 | 0.010 | 1.518 | 1.424 | 1.619 | 0.041 | 0.631 | 1.31e-40 |
| IBD | PRScs | PseudoVal | 0.130 | 0.005 | 1.499 | 1.452 | 1.547 | 0.039 | 0.629 | 1.62e-149 | 0.128 | 0.010 | 1.496 | 1.403 | 1.595 | 0.038 | 0.629 | 1.16e-37 |
| IBD | pT+clump | MultiPRS | 0.128 | 0.005 | 1.506 | 1.458 | 1.555 | 0.038 | 0.629 | 1.28e-145 | 0.137 | 0.010 | 1.548 | 1.451 | 1.651 | 0.044 | 0.635 | 3.80e-43 |
| IBD | pT+clump | 10FCVal | 0.112 | 0.005 | 1.449 | 1.401 | 1.498 | 0.029 | 0.616 | 5.73e-111 | 0.112 | 0.010 | 1.452 | 1.359 | 1.551 | 0.029 | 0.608 | 2.24e-29 |
| IBD | SBayesR | PseudoVal | 0.122 | 0.005 | 1.471 | 1.425 | 1.518 | 0.034 | 0.625 | 8.39e-133 | 0.130 | 0.010 | 1.499 | 1.407 | 1.597 | 0.039 | 0.637 | 6.55e-39 |
| IBD | SBLUP | Inf | 0.098 | 0.005 | 1.395 | 1.348 | 1.443 | 0.022 | 0.597 | 4.25e-85 | 0.102 | 0.010 | 1.416 | 1.323 | 1.517 | 0.024 | 0.600 | 2.44e-24 |
| MultiScler | All | MultiPRS | 0.133 | 0.005 | 1.550 | 1.493 | 1.609 | 0.079 | 0.718 | 1.31e-157 | 0.121 | 0.010 | 1.495 | 1.390 | 1.608 | 0.060 | 0.694 | 6.32e-34 |
| MultiScler | DBSLMM | PseudoVal | 0.090 | 0.005 | 1.424 | 1.367 | 1.483 | 0.036 | 0.663 | 1.19e-72 | 0.085 | 0.010 | 1.396 | 1.288 | 1.513 | 0.029 | 0.634 | 2.18e-17 |
| MultiScler | lassosum | MultiPRS | 0.126 | 0.005 | 1.542 | 1.484 | 1.603 | 0.070 | 0.708 | 5.35e-141 | 0.124 | 0.010 | 1.528 | 1.417 | 1.648 | 0.063 | 0.694 | 1.05e-35 |
| MultiScler | lassosum | PseudoVal | 0.100 | 0.005 | 1.449 | 1.393 | 1.508 | 0.044 | 0.686 | 1.32e-89 | 0.106 | 0.010 | 1.455 | 1.348 | 1.571 | 0.046 | 0.675 | 3.07e-26 |
| MultiScler | lassosum | 10FCVal | 0.115 | 0.005 | 1.530 | 1.468 | 1.594 | 0.059 | 0.673 | 4.31e-118 | 0.102 | 0.010 | 1.476 | 1.363 | 1.598 | 0.042 | 0.668 | 1.40e-24 |
| MultiScler | LDpred1 | MultiPRS | 0.103 | 0.005 | 1.510 | 1.448 | 1.575 | 0.047 | 0.674 | 2.92e-94 | 0.096 | 0.010 | 1.458 | 1.344 | 1.582 | 0.038 | 0.658 | 5.88e-22 |
| MultiScler | LDpred1 | Inf | 0.089 | 0.005 | 1.486 | 1.418 | 1.556 | 0.035 | 0.644 | 2.17e-70 | 0.081 | 0.010 | 1.440 | 1.315 | 1.578 | 0.026 | 0.630 | 6.95e-16 |
| MultiScler | LDpred1 | 10FCVal | 0.103 | 0.005 | 1.484 | 1.424 | 1.546 | 0.047 | 0.677 | 3.61e-94 | 0.095 | 0.010 | 1.426 | 1.318 | 1.544 | 0.037 | 0.659 | 1.98e-21 |
| MultiScler | LDpred2 | MultiPRS | 0.130 | 0.005 | 1.562 | 1.504 | 1.622 | 0.075 | 0.712 | 1.45e-149 | 0.117 | 0.010 | 1.503 | 1.394 | 1.621 | 0.056 | 0.686 | 8.03e-32 |
| MultiScler | LDpred2 | 10FCVal | 0.112 | 0.005 | 1.496 | 1.439 | 1.555 | 0.055 | 0.700 | 2.39e-111 | 0.101 | 0.010 | 1.453 | 1.343 | 1.571 | 0.042 | 0.670 | 4.30e-24 |
| MultiScler | LDpred2 | PseudoVal | 0.101 | 0.005 | 1.518 | 1.453 | 1.587 | 0.045 | 0.654 | 4.63e-91 | 0.088 | 0.010 | 1.456 | 1.335 | 1.588 | 0.032 | 0.645 | 1.09e-18 |
| MultiScler | LDpred2 | Inf | 0.092 | 0.005 | 1.501 | 1.434 | 1.571 | 0.037 | 0.641 | 9.98e-76 | 0.078 | 0.010 | 1.426 | 1.302 | 1.562 | 0.025 | 0.634 | 4.54e-15 |
| MultiScler | PRScs | MultiPRS | 0.118 | 0.005 | 1.514 | 1.456 | 1.574 | 0.062 | 0.692 | 2.61e-124 | 0.108 | 0.010 | 1.462 | 1.357 | 1.576 | 0.048 | 0.675 | 2.43e-27 |
| MultiScler | PRScs | 10FCVal | 0.118 | 0.005 | 1.536 | 1.474 | 1.600 | 0.061 | 0.675 | 3.97e-123 | 0.104 | 0.010 | 1.461 | 1.349 | 1.582 | 0.044 | 0.659 | 2.58e-25 |
| MultiScler | PRScs | PseudoVal | 0.103 | 0.005 | 1.447 | 1.393 | 1.504 | 0.047 | 0.686 | 7.63e-95 | 0.101 | 0.010 | 1.420 | 1.317 | 1.532 | 0.042 | 0.666 | 3.38e-24 |
| MultiScler | pT+clump | MultiPRS | 0.114 | 0.005 | 1.515 | 1.456 | 1.576 | 0.058 | 0.696 | 2.07e-116 | 0.116 | 0.010 | 1.513 | 1.400 | 1.635 | 0.055 | 0.677 | 4.31e-31 |
| MultiScler | pT+clump | 10FCVal | 0.090 | 0.005 | 1.444 | 1.385 | 1.507 | 0.036 | 0.665 | 5.26e-73 | 0.092 | 0.010 | 1.455 | 1.338 | 1.583 | 0.034 | 0.629 | 4.66e-20 |
| MultiScler | SBayesR | PseudoVal | 0.100 | 0.005 | 1.516 | 1.451 | 1.583 | 0.044 | 0.672 | 1.08e-88 | 0.084 | 0.010 | 1.440 | 1.317 | 1.575 | 0.028 | 0.643 | 4.95e-17 |
| MultiScler | SBLUP | Inf | 0.093 | 0.005 | 1.503 | 1.436 | 1.573 | 0.038 | 0.642 | 3.59e-77 | 0.079 | 0.010 | 1.427 | 1.304 | 1.562 | 0.025 | 0.635 | 2.90e-15 |
| RheuArth | All | MultiPRS | 0.185 | 0.005 | 1.644 | 1.596 | 1.692 | 0.065 | 0.670 | 2.11e-304 | 0.161 | 0.010 | 1.566 | 1.476 | 1.661 | 0.050 | 0.645 | 2.24e-59 |
| RheuArth | DBSLMM | PseudoVal | 0.142 | 0.005 | 1.524 | 1.478 | 1.571 | 0.038 | 0.639 | 6.14e-180 | 0.111 | 0.010 | 1.415 | 1.329 | 1.507 | 0.023 | 0.600 | 1.07e-28 |
| RheuArth | lassosum | MultiPRS | 0.183 | 0.005 | 1.627 | 1.581 | 1.675 | 0.064 | 0.669 | 2.97e-297 | 0.150 | 0.010 | 1.522 | 1.435 | 1.614 | 0.043 | 0.639 | 1.33e-51 |
| RheuArth | lassosum | PseudoVal | 0.114 | 0.005 | 1.454 | 1.407 | 1.502 | 0.025 | 0.610 | 1.14e-116 | 0.089 | 0.010 | 1.346 | 1.259 | 1.438 | 0.015 | 0.589 | 3.37e-19 |
| RheuArth | lassosum | 10FCVal | 0.162 | 0.005 | 1.560 | 1.516 | 1.607 | 0.050 | 0.654 | 3.05e-232 | 0.130 | 0.010 | 1.449 | 1.366 | 1.538 | 0.032 | 0.621 | 6.96e-39 |
| RheuArth | LDpred1 | MultiPRS | 0.145 | 0.005 | 1.544 | 1.497 | 1.592 | 0.040 | 0.640 | 4.70e-188 | 0.119 | 0.010 | 1.442 | 1.355 | 1.535 | 0.027 | 0.616 | 6.60e-33 |
| RheuArth | LDpred1 | Inf | 0.136 | 0.005 | 1.515 | 1.469 | 1.563 | 0.035 | 0.635 | 1.69e-165 | 0.111 | 0.010 | 1.415 | 1.328 | 1.507 | 0.023 | 0.611 | 7.94e-29 |
| RheuArth | LDpred1 | 10FCVal | 0.136 | 0.005 | 1.517 | 1.470 | 1.565 | 0.035 | 0.634 | 1.96e-165 | 0.112 | 0.010 | 1.419 | 1.332 | 1.511 | 0.024 | 0.607 | 4.10e-29 |
| RheuArth | LDpred2 | MultiPRS | 0.151 | 0.005 | 1.587 | 1.538 | 1.638 | 0.043 | 0.644 | 2.66e-202 | 0.127 | 0.010 | 1.486 | 1.394 | 1.583 | 0.031 | 0.619 | 1.85e-37 |
| RheuArth | LDpred2 | 10FCVal | 0.151 | 0.005 | 1.586 | 1.537 | 1.636 | 0.043 | 0.643 | 2.89e-203 | 0.128 | 0.010 | 1.490 | 1.399 | 1.588 | 0.031 | 0.618 | 5.07e-38 |
| RheuArth | LDpred2 | PseudoVal | 0.091 | 0.005 | 1.376 | 1.329 | 1.424 | 0.016 | 0.594 | 1.12e-74 | 0.077 | 0.010 | 1.309 | 1.221 | 1.403 | 0.011 | 0.581 | 1.66e-14 |
| RheuArth | LDpred2 | Inf | 0.085 | 0.005 | 1.353 | 1.307 | 1.401 | 0.014 | 0.588 | 2.55e-65 | 0.072 | 0.010 | 1.293 | 1.206 | 1.388 | 0.010 | 0.577 | 5.05e-13 |
| RheuArth | PRScs | MultiPRS | 0.166 | 0.005 | 1.576 | 1.530 | 1.622 | 0.052 | 0.660 | 5.65e-244 | 0.133 | 0.010 | 1.466 | 1.381 | 1.556 | 0.034 | 0.623 | 8.57e-41 |
| RheuArth | PRScs | 10FCVal | 0.161 | 0.005 | 1.547 | 1.503 | 1.593 | 0.049 | 0.657 | 5.26e-231 | 0.128 | 0.010 | 1.445 | 1.362 | 1.533 | 0.031 | 0.617 | 1.01e-37 |
| RheuArth | PRScs | PseudoVal | 0.162 | 0.005 | 1.551 | 1.507 | 1.597 | 0.050 | 0.658 | 1.57e-234 | 0.129 | 0.010 | 1.446 | 1.363 | 1.534 | 0.032 | 0.617 | 2.12e-38 |
| RheuArth | pT+clump | MultiPRS | 0.131 | 0.005 | 1.493 | 1.447 | 1.540 | 0.033 | 0.631 | 3.58e-153 | 0.124 | 0.010 | 1.466 | 1.377 | 1.562 | 0.029 | 0.623 | 1.28e-35 |
| RheuArth | pT+clump | 10FCVal | 0.111 | 0.005 | 1.425 | 1.380 | 1.471 | 0.023 | 0.614 | 1.72e-110 | 0.103 | 0.010 | 1.394 | 1.307 | 1.487 | 0.020 | 0.611 | 3.60e-25 |
| RheuArth | SBayesR | PseudoVal | 0.124 | 0.005 | 1.479 | 1.433 | 1.527 | 0.029 | 0.623 | 1.65e-137 | 0.119 | 0.010 | 1.461 | 1.370 | 1.558 | 0.027 | 0.619 | 6.12e-33 |
| RheuArth | SBLUP | Inf | 0.112 | 0.005 | 1.448 | 1.401 | 1.497 | 0.024 | 0.614 | 4.71e-112 | 0.097 | 0.010 | 1.380 | 1.291 | 1.475 | 0.018 | 0.597 | 3.43e-22 |
| Prostate_Cancer | All | MultiPRS | 0.186 | 0.005 | 1.655 | 1.606 | 1.705 | 0.172 | 0.701 | 2.12e-309 | 0.181 | 0.010 | 1.623 | 1.530 | 1.722 | 0.168 | 0.701 | 1.66e-74 |
| Prostate_Cancer | DBSLMM | PseudoVal | 0.176 | 0.005 | 1.636 | 1.588 | 1.687 | 0.155 | 0.690 | 5.72e-277 | 0.166 | 0.010 | 1.597 | 1.503 | 1.697 | 0.141 | 0.693 | 1.91e-62 |
| Prostate_Cancer | lassosum | MultiPRS | 0.187 | 0.005 | 1.683 | 1.633 | 1.735 | 0.173 | 0.699 | 1.66e-311 | 0.181 | 0.010 | 1.652 | 1.555 | 1.754 | 0.168 | 0.702 | 1.27e-74 |
| Prostate_Cancer | lassosum | PseudoVal | 0.168 | 0.005 | 1.630 | 1.581 | 1.681 | 0.141 | 0.683 | 6.67e-252 | 0.164 | 0.010 | 1.602 | 1.507 | 1.703 | 0.138 | 0.690 | 5.06e-61 |
| Prostate_Cancer | lassosum | 10FCVal | 0.180 | 0.005 | 1.654 | 1.605 | 1.705 | 0.161 | 0.693 | 1.67e-288 | 0.177 | 0.010 | 1.637 | 1.541 | 1.738 | 0.161 | 0.700 | 2.14e-71 |
| Prostate_Cancer | LDpred1 | MultiPRS | 0.164 | 0.005 | 1.620 | 1.570 | 1.671 | 0.135 | 0.679 | 1.64e-240 | 0.149 | 0.010 | 1.561 | 1.466 | 1.661 | 0.115 | 0.675 | 5.36e-51 |
| Prostate_Cancer | LDpred1 | Inf | 0.143 | 0.005 | 1.551 | 1.502 | 1.601 | 0.103 | 0.659 | 6.91e-182 | 0.133 | 0.010 | 1.525 | 1.429 | 1.627 | 0.092 | 0.655 | 1.04e-40 |
| Prostate_Cancer | LDpred1 | 10FCVal | 0.165 | 0.005 | 1.605 | 1.557 | 1.656 | 0.136 | 0.680 | 7.46e-243 | 0.152 | 0.010 | 1.555 | 1.462 | 1.653 | 0.119 | 0.675 | 9.66e-53 |
| Prostate_Cancer | LDpred2 | MultiPRS | 0.183 | 0.005 | 1.677 | 1.626 | 1.729 | 0.166 | 0.699 | 5.37e-298 | 0.175 | 0.010 | 1.653 | 1.555 | 1.757 | 0.156 | 0.703 | 2.23e-69 |
| Prostate_Cancer | LDpred2 | 10FCVal | 0.183 | 0.005 | 1.645 | 1.597 | 1.695 | 0.166 | 0.700 | 1.15e-298 | 0.173 | 0.010 | 1.616 | 1.523 | 1.715 | 0.154 | 0.703 | 3.31e-68 |
| Prostate_Cancer | LDpred2 | PseudoVal | 0.139 | 0.005 | 1.542 | 1.494 | 1.593 | 0.097 | 0.655 | 7.86e-172 | 0.133 | 0.010 | 1.528 | 1.432 | 1.631 | 0.091 | 0.657 | 1.24e-40 |
| Prostate_Cancer | LDpred2 | Inf | 0.135 | 0.005 | 1.532 | 1.483 | 1.583 | 0.092 | 0.651 | 8.62e-163 | 0.129 | 0.010 | 1.519 | 1.422 | 1.622 | 0.086 | 0.654 | 2.02e-38 |
| Prostate_Cancer | PRScs | MultiPRS | 0.184 | 0.005 | 1.659 | 1.609 | 1.709 | 0.167 | 0.699 | 5.42e-301 | 0.173 | 0.010 | 1.628 | 1.532 | 1.729 | 0.153 | 0.696 | 6.20e-68 |
| Prostate_Cancer | PRScs | 10FCVal | 0.181 | 0.005 | 1.637 | 1.589 | 1.687 | 0.162 | 0.696 | 4.74e-291 | 0.176 | 0.010 | 1.634 | 1.540 | 1.735 | 0.159 | 0.698 | 9.29e-71 |
| Prostate_Cancer | PRScs | PseudoVal | 0.183 | 0.005 | 1.643 | 1.594 | 1.693 | 0.165 | 0.698 | 9.65e-297 | 0.172 | 0.010 | 1.615 | 1.522 | 1.715 | 0.151 | 0.696 | 4.01e-67 |
| Prostate_Cancer | pT+clump | MultiPRS | 0.168 | 0.005 | 1.627 | 1.578 | 1.679 | 0.141 | 0.683 | 1.40e-251 | 0.175 | 0.010 | 1.640 | 1.542 | 1.744 | 0.156 | 0.691 | 2.85e-69 |
| Prostate_Cancer | pT+clump | 10FCVal | 0.152 | 0.005 | 1.587 | 1.538 | 1.638 | 0.116 | 0.669 | 3.61e-206 | 0.166 | 0.010 | 1.631 | 1.531 | 1.737 | 0.142 | 0.674 | 8.05e-63 |
| Prostate_Cancer | SBayesR | PseudoVal | 0.176 | 0.005 | 1.632 | 1.583 | 1.682 | 0.154 | 0.693 | 7.24e-277 | 0.171 | 0.010 | 1.599 | 1.507 | 1.697 | 0.150 | 0.697 | 1.83e-66 |
| Prostate_Cancer | SBLUP | Inf | 0.137 | 0.005 | 1.536 | 1.488 | 1.587 | 0.094 | 0.650 | 2.20e-166 | 0.128 | 0.010 | 1.514 | 1.418 | 1.617 | 0.085 | 0.653 | 7.05e-38 |
| Breast_Cancer | All | MultiPRS | 0.228 | 0.005 | 1.715 | 1.673 | 1.757 | 0.105 | 0.666 | 0.00e+00 | 0.215 | 0.010 | 1.655 | 1.577 | 1.736 | 0.090 | 0.654 | 6.31e-105 |
| Breast_Cancer | DBSLMM | PseudoVal | 0.217 | 0.005 | 1.681 | 1.640 | 1.722 | 0.096 | 0.660 | 0.00e+00 | 0.211 | 0.010 | 1.643 | 1.565 | 1.724 | 0.087 | 0.649 | 8.92e-101 |
| Breast_Cancer | lassosum | MultiPRS | 0.225 | 0.005 | 1.708 | 1.667 | 1.751 | 0.103 | 0.664 | 0.00e+00 | 0.212 | 0.010 | 1.646 | 1.568 | 1.727 | 0.088 | 0.653 | 6.68e-102 |
| Breast_Cancer | lassosum | PseudoVal | 0.217 | 0.005 | 1.682 | 1.641 | 1.723 | 0.096 | 0.657 | 0.00e+00 | 0.199 | 0.010 | 1.602 | 1.526 | 1.681 | 0.078 | 0.644 | 5.02e-90 |
| Breast_Cancer | lassosum | 10FCVal | 0.219 | 0.005 | 1.688 | 1.647 | 1.730 | 0.098 | 0.659 | 0.00e+00 | 0.199 | 0.010 | 1.601 | 1.525 | 1.680 | 0.077 | 0.644 | 7.52e-90 |
| Breast_Cancer | LDpred1 | MultiPRS | 0.211 | 0.005 | 1.657 | 1.617 | 1.698 | 0.090 | 0.655 | 0.00e+00 | 0.195 | 0.010 | 1.585 | 1.510 | 1.663 | 0.074 | 0.641 | 7.42e-86 |
| Breast_Cancer | LDpred1 | Inf | 0.172 | 0.005 | 1.523 | 1.486 | 1.562 | 0.060 | 0.627 | 2.25e-262 | 0.162 | 0.010 | 1.481 | 1.410 | 1.554 | 0.051 | 0.616 | 1.25e-59 |
| Breast_Cancer | LDpred1 | 10FCVal | 0.201 | 0.005 | 1.621 | 1.582 | 1.662 | 0.082 | 0.647 | 0.00e+00 | 0.186 | 0.010 | 1.559 | 1.485 | 1.636 | 0.068 | 0.634 | 1.68e-78 |
| Breast_Cancer | LDpred2 | MultiPRS | 0.224 | 0.005 | 1.703 | 1.662 | 1.745 | 0.102 | 0.664 | 0.00e+00 | 0.211 | 0.010 | 1.639 | 1.562 | 1.720 | 0.087 | 0.651 | 1.16e-100 |
| Breast_Cancer | LDpred2 | 10FCVal | 0.223 | 0.005 | 1.701 | 1.660 | 1.744 | 0.101 | 0.663 | 0.00e+00 | 0.211 | 0.010 | 1.642 | 1.565 | 1.724 | 0.087 | 0.650 | 4.71e-101 |
| Breast_Cancer | LDpred2 | PseudoVal | 0.169 | 0.005 | 1.514 | 1.477 | 1.552 | 0.058 | 0.625 | 3.86e-253 | 0.161 | 0.010 | 1.477 | 1.407 | 1.551 | 0.050 | 0.616 | 7.88e-59 |
| Breast_Cancer | LDpred2 | Inf | 0.168 | 0.005 | 1.510 | 1.474 | 1.548 | 0.057 | 0.625 | 3.29e-250 | 0.160 | 0.010 | 1.475 | 1.405 | 1.549 | 0.050 | 0.615 | 2.07e-58 |
| Breast_Cancer | PRScs | MultiPRS | 0.222 | 0.005 | 1.698 | 1.657 | 1.740 | 0.100 | 0.662 | 0.00e+00 | 0.210 | 0.010 | 1.641 | 1.564 | 1.723 | 0.087 | 0.651 | 1.59e-100 |
| Breast_Cancer | PRScs | 10FCVal | 0.220 | 0.005 | 1.688 | 1.647 | 1.730 | 0.098 | 0.661 | 0.00e+00 | 0.207 | 0.010 | 1.629 | 1.552 | 1.709 | 0.084 | 0.648 | 2.37e-97 |
| Breast_Cancer | PRScs | PseudoVal | 0.220 | 0.005 | 1.690 | 1.649 | 1.732 | 0.099 | 0.661 | 0.00e+00 | 0.208 | 0.010 | 1.630 | 1.553 | 1.710 | 0.084 | 0.649 | 7.26e-98 |
| Breast_Cancer | pT+clump | MultiPRS | 0.200 | 0.005 | 1.624 | 1.584 | 1.664 | 0.081 | 0.647 | 0.00e+00 | 0.188 | 0.010 | 1.564 | 1.490 | 1.641 | 0.069 | 0.636 | 6.73e-80 |
| Breast_Cancer | pT+clump | 10FCVal | 0.179 | 0.005 | 1.550 | 1.512 | 1.589 | 0.065 | 0.631 | 4.74e-284 | 0.167 | 0.010 | 1.498 | 1.427 | 1.573 | 0.054 | 0.624 | 1.69e-63 |
| Breast_Cancer | SBayesR | PseudoVal | 0.221 | 0.005 | 1.694 | 1.653 | 1.736 | 0.099 | 0.660 | 0.00e+00 | 0.205 | 0.010 | 1.619 | 1.543 | 1.699 | 0.082 | 0.648 | 2.51e-95 |
| Breast_Cancer | SBLUP | Inf | 0.168 | 0.005 | 1.510 | 1.473 | 1.548 | 0.057 | 0.624 | 5.33e-250 | 0.158 | 0.010 | 1.469 | 1.399 | 1.542 | 0.049 | 0.614 | 4.12e-57 |
Show cross-validation results compared to pT+clump
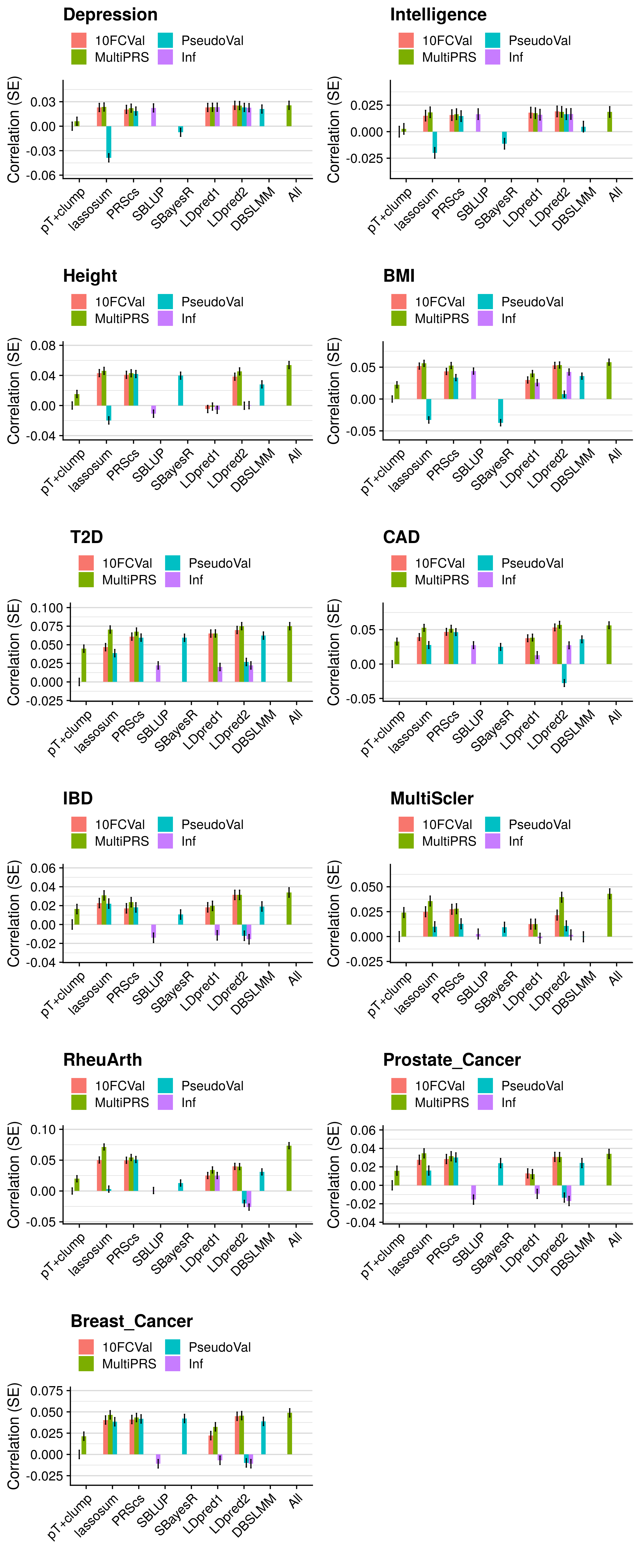
Predictive utility estimated using 10-fold cross-validation
Show test-validation results compared to pT+clump
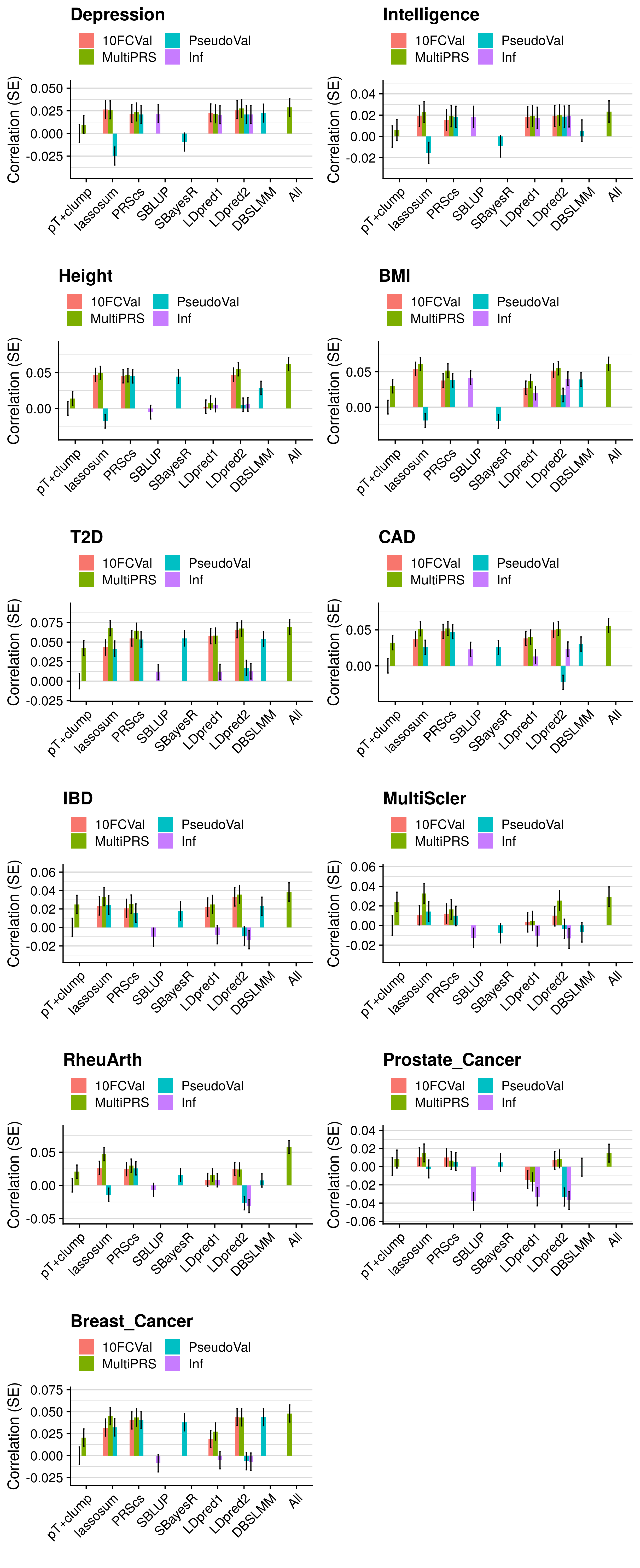
Predictive utility estimated using test set validation
Show difference results table
| Phenotype | Model 1 Test | Model 2 Test | Model 1 CrossVal R | Model 2 CrossVal R | CrossVal R Diff | CrossVal R Diff P | Model 1 IndepVal R | Model 2 IndepVal R | IndepVal R Diff | IndepVal R Diff P |
|---|---|---|---|---|---|---|---|---|---|---|
| Depression | LDpred2.PseudoVal | All.MultiPRS | 0.143 | 0.146 | -0.003 | 7.10e-01 | 0.139 | 0.146 | -0.008 | 8.95e-04 |
| Depression | pT+clump.10FCVal | All.MultiPRS | 0.120 | 0.146 | -0.026 | 2.26e-04 | 0.118 | 0.146 | -0.029 | 3.18e-06 |
| Depression | LDpred2.MultiPRS | All.MultiPRS | 0.145 | 0.146 | 0.000 | 9.60e-01 | 0.145 | 0.146 | -0.001 | 3.08e-01 |
| Depression | PRScs.PseudoVal | All.MultiPRS | 0.139 | 0.146 | -0.007 | 3.09e-01 | 0.139 | 0.146 | -0.008 | 3.26e-03 |
| Depression | LDpred1.Inf | All.MultiPRS | 0.143 | 0.146 | -0.002 | 7.34e-01 | 0.138 | 0.146 | -0.008 | 7.92e-04 |
| Depression | PRScs.MultiPRS | All.MultiPRS | 0.142 | 0.146 | -0.004 | 6.15e-01 | 0.141 | 0.146 | -0.005 | 1.65e-02 |
| Depression | LDpred2.Inf | All.MultiPRS | 0.143 | 0.146 | -0.003 | 6.76e-01 | 0.139 | 0.146 | -0.008 | 1.27e-03 |
| Depression | lassosum.10FCVal | All.MultiPRS | 0.143 | 0.146 | -0.003 | 6.99e-01 | 0.144 | 0.146 | -0.002 | 3.41e-01 |
| Depression | LDpred2.10FCVal | All.MultiPRS | 0.146 | 0.146 | 0.000 | 9.93e-01 | 0.144 | 0.146 | -0.002 | 8.12e-02 |
| Depression | lassosum.MultiPRS | All.MultiPRS | 0.144 | 0.146 | -0.002 | 7.62e-01 | 0.144 | 0.146 | -0.003 | 2.23e-01 |
| Depression | LDpred1.10FCVal | All.MultiPRS | 0.143 | 0.146 | -0.003 | 7.04e-01 | 0.140 | 0.146 | -0.006 | 2.85e-03 |
| Depression | All.MultiPRS | All.MultiPRS | 0.146 | 0.146 | 0.000 | 1.00e+00 | 0.146 | 0.146 | 0.000 | 1.00e+00 |
| Depression | SBayesR.PseudoVal | All.MultiPRS | 0.113 | 0.146 | -0.033 | 2.06e-06 | 0.108 | 0.146 | -0.038 | 2.01e-08 |
| Depression | LDpred1.MultiPRS | All.MultiPRS | 0.143 | 0.146 | -0.002 | 7.27e-01 | 0.139 | 0.146 | -0.007 | 8.24e-04 |
| Depression | pT+clump.MultiPRS | All.MultiPRS | 0.126 | 0.146 | -0.020 | 5.17e-03 | 0.127 | 0.146 | -0.019 | 2.17e-04 |
| Depression | lassosum.PseudoVal | All.MultiPRS | 0.081 | 0.146 | -0.064 | 3.26e-20 | 0.093 | 0.146 | -0.054 | 1.13e-08 |
| Depression | PRScs.10FCVal | All.MultiPRS | 0.141 | 0.146 | -0.005 | 4.70e-01 | 0.139 | 0.146 | -0.007 | 1.14e-02 |
| Depression | DBSLMM.PseudoVal | All.MultiPRS | 0.141 | 0.146 | -0.005 | 5.09e-01 | 0.140 | 0.146 | -0.006 | 2.37e-02 |
| Depression | SBLUP.Inf | All.MultiPRS | 0.143 | 0.146 | -0.003 | 6.44e-01 | 0.139 | 0.146 | -0.007 | 4.76e-03 |
| Depression | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.113 | 0.141 | -0.028 | 2.06e-15 | 0.108 | 0.140 | -0.032 | 1.06e-05 |
| Depression | LDpred2.Inf | DBSLMM.PseudoVal | 0.143 | 0.141 | 0.002 | 2.51e-01 | 0.139 | 0.140 | -0.002 | 5.81e-01 |
| Depression | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.120 | 0.141 | -0.021 | 1.69e-11 | 0.118 | 0.140 | -0.022 | 3.52e-04 |
| Depression | PRScs.10FCVal | DBSLMM.PseudoVal | 0.141 | 0.141 | 0.000 | 8.09e-01 | 0.139 | 0.140 | -0.001 | 8.48e-01 |
| Depression | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.139 | 0.141 | -0.002 | 1.47e-01 | 0.139 | 0.140 | -0.002 | 6.40e-01 |
| Depression | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.126 | 0.141 | -0.015 | 3.19e-02 | 0.127 | 0.140 | -0.013 | 2.22e-02 |
| Depression | LDpred1.Inf | DBSLMM.PseudoVal | 0.143 | 0.141 | 0.002 | 1.28e-01 | 0.138 | 0.140 | -0.002 | 5.17e-01 |
| Depression | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.144 | 0.141 | 0.002 | 7.22e-01 | 0.144 | 0.140 | 0.004 | 3.13e-01 |
| Depression | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.146 | 0.141 | 0.005 | 3.00e-03 | 0.144 | 0.140 | 0.004 | 2.32e-01 |
| Depression | lassosum.10FCVal | DBSLMM.PseudoVal | 0.143 | 0.141 | 0.002 | 2.83e-01 | 0.144 | 0.140 | 0.004 | 2.96e-01 |
| Depression | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.143 | 0.141 | 0.002 | 1.62e-01 | 0.140 | 0.140 | 0.000 | 9.35e-01 |
| Depression | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.143 | 0.141 | 0.002 | 7.56e-01 | 0.139 | 0.140 | -0.001 | 7.63e-01 |
| Depression | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.142 | 0.141 | 0.001 | 8.77e-01 | 0.141 | 0.140 | 0.001 | 6.67e-01 |
| Depression | All.MultiPRS | DBSLMM.PseudoVal | 0.146 | 0.141 | 0.005 | 5.09e-01 | 0.146 | 0.140 | 0.006 | 2.37e-02 |
| Depression | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.143 | 0.141 | 0.002 | 1.78e-01 | 0.139 | 0.140 | -0.002 | 5.93e-01 |
| Depression | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.141 | 0.141 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| Depression | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.145 | 0.141 | 0.004 | 5.43e-01 | 0.145 | 0.140 | 0.005 | 8.27e-02 |
| Depression | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.081 | 0.141 | -0.060 | 3.10e-35 | 0.093 | 0.140 | -0.047 | 7.68e-07 |
| Depression | SBLUP.Inf | DBSLMM.PseudoVal | 0.143 | 0.141 | 0.001 | 3.06e-01 | 0.139 | 0.140 | -0.001 | 8.02e-01 |
| Depression | SBayesR.PseudoVal | lassosum.MultiPRS | 0.113 | 0.144 | -0.031 | 8.66e-06 | 0.108 | 0.144 | -0.035 | 5.84e-07 |
| Depression | LDpred2.10FCVal | lassosum.MultiPRS | 0.146 | 0.144 | 0.002 | 7.67e-01 | 0.144 | 0.144 | 0.000 | 9.28e-01 |
| Depression | LDpred1.10FCVal | lassosum.MultiPRS | 0.143 | 0.144 | -0.001 | 9.40e-01 | 0.140 | 0.144 | -0.003 | 2.41e-01 |
| Depression | LDpred2.Inf | lassosum.MultiPRS | 0.143 | 0.144 | -0.001 | 9.11e-01 | 0.139 | 0.144 | -0.005 | 1.13e-01 |
| Depression | PRScs.10FCVal | lassosum.MultiPRS | 0.141 | 0.144 | -0.003 | 6.77e-01 | 0.139 | 0.144 | -0.004 | 2.01e-01 |
| Depression | PRScs.MultiPRS | lassosum.MultiPRS | 0.142 | 0.144 | -0.001 | 2.88e-01 | 0.141 | 0.144 | -0.002 | 3.95e-01 |
| Depression | pT+clump.MultiPRS | lassosum.MultiPRS | 0.126 | 0.144 | -0.017 | 1.21e-02 | 0.127 | 0.144 | -0.016 | 2.44e-03 |
| Depression | lassosum.MultiPRS | lassosum.MultiPRS | 0.144 | 0.144 | 0.000 | 1.00e+00 | 0.144 | 0.144 | 0.000 | 1.00e+00 |
| Depression | lassosum.PseudoVal | lassosum.MultiPRS | 0.081 | 0.144 | -0.062 | 6.03e-19 | 0.093 | 0.144 | -0.051 | 3.05e-07 |
| Depression | LDpred2.MultiPRS | lassosum.MultiPRS | 0.145 | 0.144 | 0.002 | 8.00e-01 | 0.145 | 0.144 | 0.002 | 5.81e-01 |
| Depression | LDpred1.MultiPRS | lassosum.MultiPRS | 0.143 | 0.144 | 0.000 | 9.64e-01 | 0.139 | 0.144 | -0.004 | 1.40e-01 |
| Depression | LDpred2.PseudoVal | lassosum.MultiPRS | 0.143 | 0.144 | 0.000 | 9.47e-01 | 0.139 | 0.144 | -0.005 | 1.05e-01 |
| Depression | LDpred1.Inf | lassosum.MultiPRS | 0.143 | 0.144 | 0.000 | 9.73e-01 | 0.138 | 0.144 | -0.005 | 8.90e-02 |
| Depression | pT+clump.10FCVal | lassosum.MultiPRS | 0.120 | 0.144 | -0.024 | 7.51e-04 | 0.118 | 0.144 | -0.026 | 3.61e-05 |
| Depression | lassosum.10FCVal | lassosum.MultiPRS | 0.143 | 0.144 | -0.001 | 9.35e-01 | 0.144 | 0.144 | 0.000 | 7.05e-01 |
| Depression | SBLUP.Inf | lassosum.MultiPRS | 0.143 | 0.144 | -0.001 | 8.76e-01 | 0.139 | 0.144 | -0.004 | 1.72e-01 |
| Depression | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.141 | 0.144 | -0.002 | 7.22e-01 | 0.140 | 0.144 | -0.004 | 3.13e-01 |
| Depression | All.MultiPRS | lassosum.MultiPRS | 0.146 | 0.144 | 0.002 | 7.62e-01 | 0.146 | 0.144 | 0.003 | 2.23e-01 |
| Depression | PRScs.PseudoVal | lassosum.MultiPRS | 0.139 | 0.144 | -0.005 | 4.78e-01 | 0.139 | 0.144 | -0.005 | 1.11e-01 |
| Depression | LDpred1.Inf | lassosum.PseudoVal | 0.143 | 0.081 | 0.062 | 3.29e-30 | 0.138 | 0.093 | 0.045 | 2.38e-05 |
| Depression | PRScs.10FCVal | lassosum.PseudoVal | 0.141 | 0.081 | 0.059 | 5.30e-38 | 0.139 | 0.093 | 0.047 | 3.11e-07 |
| Depression | LDpred2.MultiPRS | lassosum.PseudoVal | 0.145 | 0.081 | 0.064 | 5.00e-20 | 0.145 | 0.093 | 0.052 | 3.21e-08 |
| Depression | lassosum.PseudoVal | lassosum.PseudoVal | 0.081 | 0.081 | 0.000 | 1.00e+00 | 0.093 | 0.093 | 0.000 | 1.00e+00 |
| Depression | PRScs.PseudoVal | lassosum.PseudoVal | 0.139 | 0.081 | 0.057 | 7.05e-35 | 0.139 | 0.093 | 0.046 | 6.28e-07 |
| Depression | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.141 | 0.081 | 0.060 | 3.10e-35 | 0.140 | 0.093 | 0.047 | 7.68e-07 |
| Depression | SBLUP.Inf | lassosum.PseudoVal | 0.143 | 0.081 | 0.061 | 4.92e-29 | 0.139 | 0.093 | 0.047 | 1.63e-05 |
| Depression | LDpred2.Inf | lassosum.PseudoVal | 0.143 | 0.081 | 0.061 | 4.30e-29 | 0.139 | 0.093 | 0.046 | 2.66e-05 |
| Depression | pT+clump.10FCVal | lassosum.PseudoVal | 0.120 | 0.081 | 0.039 | 5.00e-11 | 0.118 | 0.093 | 0.025 | 3.46e-02 |
| Depression | LDpred2.10FCVal | lassosum.PseudoVal | 0.146 | 0.081 | 0.064 | 3.33e-39 | 0.144 | 0.093 | 0.051 | 1.38e-07 |
| Depression | lassosum.MultiPRS | lassosum.PseudoVal | 0.144 | 0.081 | 0.062 | 6.03e-19 | 0.144 | 0.093 | 0.051 | 3.05e-07 |
| Depression | LDpred1.10FCVal | lassosum.PseudoVal | 0.143 | 0.081 | 0.062 | 1.03e-31 | 0.140 | 0.093 | 0.048 | 4.83e-06 |
| Depression | All.MultiPRS | lassosum.PseudoVal | 0.146 | 0.081 | 0.064 | 3.26e-20 | 0.146 | 0.093 | 0.054 | 1.13e-08 |
| Depression | SBayesR.PseudoVal | lassosum.PseudoVal | 0.113 | 0.081 | 0.031 | 5.05e-10 | 0.108 | 0.093 | 0.015 | 1.27e-01 |
| Depression | lassosum.10FCVal | lassosum.PseudoVal | 0.143 | 0.081 | 0.062 | 8.99e-34 | 0.144 | 0.093 | 0.051 | 3.75e-07 |
| Depression | pT+clump.MultiPRS | lassosum.PseudoVal | 0.126 | 0.081 | 0.045 | 1.66e-10 | 0.127 | 0.093 | 0.035 | 9.54e-04 |
| Depression | LDpred2.PseudoVal | lassosum.PseudoVal | 0.143 | 0.081 | 0.062 | 3.09e-30 | 0.139 | 0.093 | 0.046 | 1.92e-05 |
| Depression | LDpred1.MultiPRS | lassosum.PseudoVal | 0.143 | 0.081 | 0.062 | 8.31e-19 | 0.139 | 0.093 | 0.047 | 1.06e-05 |
| Depression | PRScs.MultiPRS | lassosum.PseudoVal | 0.142 | 0.081 | 0.061 | 3.44e-18 | 0.141 | 0.093 | 0.049 | 3.36e-07 |
| Depression | lassosum.10FCVal | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.144 | 0.144 | 0.000 | 1.00e+00 |
| Depression | pT+clump.MultiPRS | lassosum.10FCVal | 0.126 | 0.143 | -0.017 | 1.54e-02 | 0.127 | 0.144 | -0.017 | 1.67e-03 |
| Depression | LDpred2.Inf | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 9.00e-01 | 0.139 | 0.144 | -0.005 | 1.19e-01 |
| Depression | SBayesR.PseudoVal | lassosum.10FCVal | 0.113 | 0.143 | -0.030 | 1.92e-17 | 0.108 | 0.144 | -0.036 | 6.11e-07 |
| Depression | PRScs.MultiPRS | lassosum.10FCVal | 0.142 | 0.143 | -0.001 | 9.05e-01 | 0.141 | 0.144 | -0.003 | 3.68e-01 |
| Depression | SBLUP.Inf | lassosum.10FCVal | 0.143 | 0.143 | -0.001 | 7.47e-01 | 0.139 | 0.144 | -0.004 | 1.76e-01 |
| Depression | lassosum.MultiPRS | lassosum.10FCVal | 0.144 | 0.143 | 0.001 | 9.35e-01 | 0.144 | 0.144 | 0.000 | 7.05e-01 |
| Depression | PRScs.10FCVal | lassosum.10FCVal | 0.141 | 0.143 | -0.002 | 1.84e-01 | 0.139 | 0.144 | -0.004 | 2.05e-01 |
| Depression | LDpred2.MultiPRS | lassosum.10FCVal | 0.145 | 0.143 | 0.002 | 7.38e-01 | 0.145 | 0.144 | 0.001 | 6.89e-01 |
| Depression | PRScs.PseudoVal | lassosum.10FCVal | 0.139 | 0.143 | -0.004 | 1.00e-02 | 0.139 | 0.144 | -0.005 | 1.16e-01 |
| Depression | LDpred2.PseudoVal | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 9.54e-01 | 0.139 | 0.144 | -0.005 | 1.27e-01 |
| Depression | LDpred1.MultiPRS | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 9.71e-01 | 0.139 | 0.144 | -0.005 | 1.43e-01 |
| Depression | pT+clump.10FCVal | lassosum.10FCVal | 0.120 | 0.143 | -0.023 | 5.41e-14 | 0.118 | 0.144 | -0.026 | 2.00e-05 |
| Depression | All.MultiPRS | lassosum.10FCVal | 0.146 | 0.143 | 0.003 | 6.99e-01 | 0.146 | 0.144 | 0.002 | 3.41e-01 |
| Depression | LDpred1.Inf | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 8.48e-01 | 0.138 | 0.144 | -0.006 | 9.80e-02 |
| Depression | DBSLMM.PseudoVal | lassosum.10FCVal | 0.141 | 0.143 | -0.002 | 2.83e-01 | 0.140 | 0.144 | -0.004 | 2.96e-01 |
| Depression | LDpred1.10FCVal | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 9.77e-01 | 0.140 | 0.144 | -0.004 | 2.29e-01 |
| Depression | lassosum.PseudoVal | lassosum.10FCVal | 0.081 | 0.143 | -0.062 | 8.99e-34 | 0.093 | 0.144 | -0.051 | 3.75e-07 |
| Depression | LDpred2.10FCVal | lassosum.10FCVal | 0.146 | 0.143 | 0.003 | 6.56e-02 | 0.144 | 0.144 | 0.000 | 9.83e-01 |
| Depression | LDpred2.10FCVal | LDpred1.MultiPRS | 0.146 | 0.143 | 0.002 | 7.33e-01 | 0.144 | 0.139 | 0.005 | 6.22e-02 |
| Depression | PRScs.PseudoVal | LDpred1.MultiPRS | 0.139 | 0.143 | -0.005 | 5.05e-01 | 0.139 | 0.139 | -0.001 | 8.22e-01 |
| Depression | LDpred2.Inf | LDpred1.MultiPRS | 0.143 | 0.143 | 0.000 | 9.47e-01 | 0.139 | 0.139 | -0.001 | 6.79e-01 |
| Depression | pT+clump.10FCVal | LDpred1.MultiPRS | 0.120 | 0.143 | -0.023 | 8.77e-04 | 0.118 | 0.139 | -0.022 | 3.17e-04 |
| Depression | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.113 | 0.143 | -0.031 | 1.13e-05 | 0.108 | 0.139 | -0.031 | 1.63e-05 |
| Depression | All.MultiPRS | LDpred1.MultiPRS | 0.146 | 0.143 | 0.002 | 7.27e-01 | 0.146 | 0.139 | 0.007 | 8.24e-04 |
| Depression | LDpred1.10FCVal | LDpred1.MultiPRS | 0.143 | 0.143 | 0.000 | 9.76e-01 | 0.140 | 0.139 | 0.001 | 2.93e-01 |
| Depression | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.139 | 0.139 | 0.000 | 1.00e+00 |
| Depression | PRScs.10FCVal | LDpred1.MultiPRS | 0.141 | 0.143 | -0.003 | 7.10e-01 | 0.139 | 0.139 | 0.000 | 9.60e-01 |
| Depression | lassosum.PseudoVal | LDpred1.MultiPRS | 0.081 | 0.143 | -0.062 | 8.31e-19 | 0.093 | 0.139 | -0.047 | 1.06e-05 |
| Depression | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.126 | 0.143 | -0.017 | 1.46e-02 | 0.127 | 0.139 | -0.012 | 2.81e-02 |
| Depression | LDpred1.Inf | LDpred1.MultiPRS | 0.143 | 0.143 | 0.000 | 9.91e-01 | 0.138 | 0.139 | -0.001 | 3.78e-01 |
| Depression | lassosum.MultiPRS | LDpred1.MultiPRS | 0.144 | 0.143 | 0.000 | 9.64e-01 | 0.144 | 0.139 | 0.004 | 1.40e-01 |
| Depression | PRScs.MultiPRS | LDpred1.MultiPRS | 0.142 | 0.143 | -0.001 | 8.77e-01 | 0.141 | 0.139 | 0.002 | 4.39e-01 |
| Depression | lassosum.10FCVal | LDpred1.MultiPRS | 0.143 | 0.143 | 0.000 | 9.71e-01 | 0.144 | 0.139 | 0.005 | 1.43e-01 |
| Depression | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.143 | 0.143 | 0.000 | 9.83e-01 | 0.139 | 0.139 | -0.001 | 6.94e-01 |
| Depression | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.141 | 0.143 | -0.002 | 7.56e-01 | 0.140 | 0.139 | 0.001 | 7.63e-01 |
| Depression | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.145 | 0.143 | 0.002 | 7.65e-01 | 0.145 | 0.139 | 0.006 | 1.41e-02 |
| Depression | SBLUP.Inf | LDpred1.MultiPRS | 0.143 | 0.143 | -0.001 | 9.11e-01 | 0.139 | 0.139 | 0.000 | 9.04e-01 |
| Depression | All.MultiPRS | LDpred1.Inf | 0.146 | 0.143 | 0.002 | 7.34e-01 | 0.146 | 0.138 | 0.008 | 7.92e-04 |
| Depression | LDpred2.PseudoVal | LDpred1.Inf | 0.143 | 0.143 | 0.000 | 7.84e-01 | 0.139 | 0.138 | 0.000 | 8.51e-01 |
| Depression | SBayesR.PseudoVal | LDpred1.Inf | 0.113 | 0.143 | -0.031 | 4.60e-17 | 0.108 | 0.138 | -0.030 | 3.90e-05 |
| Depression | LDpred2.MultiPRS | LDpred1.Inf | 0.145 | 0.143 | 0.002 | 7.74e-01 | 0.145 | 0.138 | 0.007 | 7.33e-03 |
| Depression | PRScs.PseudoVal | LDpred1.Inf | 0.139 | 0.143 | -0.005 | 9.05e-03 | 0.139 | 0.138 | 0.000 | 9.37e-01 |
| Depression | LDpred1.Inf | LDpred1.Inf | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.138 | 0.138 | 0.000 | 1.00e+00 |
| Depression | pT+clump.10FCVal | LDpred1.Inf | 0.120 | 0.143 | -0.023 | 8.46e-15 | 0.118 | 0.138 | -0.021 | 6.63e-04 |
| Depression | PRScs.MultiPRS | LDpred1.Inf | 0.142 | 0.143 | -0.001 | 8.68e-01 | 0.141 | 0.138 | 0.003 | 2.89e-01 |
| Depression | lassosum.PseudoVal | LDpred1.Inf | 0.081 | 0.143 | -0.062 | 3.29e-30 | 0.093 | 0.138 | -0.045 | 2.38e-05 |
| Depression | pT+clump.MultiPRS | LDpred1.Inf | 0.126 | 0.143 | -0.017 | 1.36e-02 | 0.127 | 0.138 | -0.011 | 5.12e-02 |
| Depression | LDpred1.MultiPRS | LDpred1.Inf | 0.143 | 0.143 | 0.000 | 9.91e-01 | 0.139 | 0.138 | 0.001 | 3.78e-01 |
| Depression | PRScs.10FCVal | LDpred1.Inf | 0.141 | 0.143 | -0.003 | 1.54e-01 | 0.139 | 0.138 | 0.001 | 7.41e-01 |
| Depression | LDpred2.Inf | LDpred1.Inf | 0.143 | 0.143 | -0.001 | 5.23e-01 | 0.139 | 0.138 | 0.000 | 8.72e-01 |
| Depression | SBLUP.Inf | LDpred1.Inf | 0.143 | 0.143 | -0.001 | 2.28e-01 | 0.139 | 0.138 | 0.001 | 3.88e-01 |
| Depression | DBSLMM.PseudoVal | LDpred1.Inf | 0.141 | 0.143 | -0.002 | 1.28e-01 | 0.140 | 0.138 | 0.002 | 5.17e-01 |
| Depression | lassosum.MultiPRS | LDpred1.Inf | 0.144 | 0.143 | 0.000 | 9.73e-01 | 0.144 | 0.138 | 0.005 | 8.90e-02 |
| Depression | LDpred1.10FCVal | LDpred1.Inf | 0.143 | 0.143 | 0.000 | 7.06e-01 | 0.140 | 0.138 | 0.002 | 1.61e-01 |
| Depression | lassosum.10FCVal | LDpred1.Inf | 0.143 | 0.143 | 0.000 | 8.48e-01 | 0.144 | 0.138 | 0.006 | 9.80e-02 |
| Depression | LDpred2.10FCVal | LDpred1.Inf | 0.146 | 0.143 | 0.002 | 8.69e-02 | 0.144 | 0.138 | 0.006 | 3.66e-02 |
| Depression | LDpred1.Inf | LDpred1.10FCVal | 0.143 | 0.143 | 0.000 | 7.06e-01 | 0.138 | 0.140 | -0.002 | 1.61e-01 |
| Depression | PRScs.MultiPRS | LDpred1.10FCVal | 0.142 | 0.143 | -0.001 | 9.00e-01 | 0.141 | 0.140 | 0.001 | 6.91e-01 |
| Depression | LDpred1.10FCVal | LDpred1.10FCVal | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| Depression | All.MultiPRS | LDpred1.10FCVal | 0.146 | 0.143 | 0.003 | 7.04e-01 | 0.146 | 0.140 | 0.006 | 2.85e-03 |
| Depression | SBayesR.PseudoVal | LDpred1.10FCVal | 0.113 | 0.143 | -0.030 | 1.31e-17 | 0.108 | 0.140 | -0.032 | 6.28e-06 |
| Depression | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.141 | 0.143 | -0.002 | 1.62e-01 | 0.140 | 0.140 | 0.000 | 9.35e-01 |
| Depression | pT+clump.MultiPRS | LDpred1.10FCVal | 0.126 | 0.143 | -0.017 | 1.53e-02 | 0.127 | 0.140 | -0.013 | 1.43e-02 |
| Depression | lassosum.MultiPRS | LDpred1.10FCVal | 0.144 | 0.143 | 0.001 | 9.40e-01 | 0.144 | 0.140 | 0.003 | 2.41e-01 |
| Depression | PRScs.10FCVal | LDpred1.10FCVal | 0.141 | 0.143 | -0.002 | 1.56e-01 | 0.139 | 0.140 | -0.001 | 7.84e-01 |
| Depression | lassosum.10FCVal | LDpred1.10FCVal | 0.143 | 0.143 | 0.000 | 9.77e-01 | 0.144 | 0.140 | 0.004 | 2.29e-01 |
| Depression | SBLUP.Inf | LDpred1.10FCVal | 0.143 | 0.143 | -0.001 | 4.78e-01 | 0.139 | 0.140 | -0.001 | 5.80e-01 |
| Depression | LDpred2.PseudoVal | LDpred1.10FCVal | 0.143 | 0.143 | 0.000 | 9.52e-01 | 0.139 | 0.140 | -0.002 | 3.64e-01 |
| Depression | pT+clump.10FCVal | LDpred1.10FCVal | 0.120 | 0.143 | -0.023 | 1.08e-14 | 0.118 | 0.140 | -0.023 | 1.47e-04 |
| Depression | LDpred2.MultiPRS | LDpred1.10FCVal | 0.145 | 0.143 | 0.002 | 7.42e-01 | 0.145 | 0.140 | 0.005 | 3.02e-02 |
| Depression | PRScs.PseudoVal | LDpred1.10FCVal | 0.139 | 0.143 | -0.004 | 5.82e-03 | 0.139 | 0.140 | -0.002 | 5.68e-01 |
| Depression | LDpred2.10FCVal | LDpred1.10FCVal | 0.146 | 0.143 | 0.003 | 1.97e-02 | 0.144 | 0.140 | 0.004 | 1.14e-01 |
| Depression | LDpred1.MultiPRS | LDpred1.10FCVal | 0.143 | 0.143 | 0.000 | 9.76e-01 | 0.139 | 0.140 | -0.001 | 2.93e-01 |
| Depression | LDpred2.Inf | LDpred1.10FCVal | 0.143 | 0.143 | 0.000 | 7.89e-01 | 0.139 | 0.140 | -0.002 | 3.44e-01 |
| Depression | lassosum.PseudoVal | LDpred1.10FCVal | 0.081 | 0.143 | -0.062 | 1.03e-31 | 0.093 | 0.140 | -0.048 | 4.83e-06 |
| Depression | LDpred1.Inf | LDpred2.MultiPRS | 0.143 | 0.145 | -0.002 | 7.74e-01 | 0.138 | 0.145 | -0.007 | 7.33e-03 |
| Depression | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.141 | 0.145 | -0.004 | 5.43e-01 | 0.140 | 0.145 | -0.005 | 8.27e-02 |
| Depression | SBLUP.Inf | LDpred2.MultiPRS | 0.143 | 0.145 | -0.003 | 6.82e-01 | 0.139 | 0.145 | -0.006 | 1.95e-02 |
| Depression | LDpred2.Inf | LDpred2.MultiPRS | 0.143 | 0.145 | -0.003 | 7.15e-01 | 0.139 | 0.145 | -0.007 | 1.84e-03 |
| Depression | pT+clump.10FCVal | LDpred2.MultiPRS | 0.120 | 0.145 | -0.025 | 3.00e-04 | 0.118 | 0.145 | -0.028 | 2.08e-05 |
| Depression | LDpred2.10FCVal | LDpred2.MultiPRS | 0.146 | 0.145 | 0.000 | 9.66e-01 | 0.144 | 0.145 | -0.001 | 1.66e-01 |
| Depression | PRScs.PseudoVal | LDpred2.MultiPRS | 0.139 | 0.145 | -0.007 | 3.35e-01 | 0.139 | 0.145 | -0.007 | 1.84e-02 |
| Depression | LDpred1.10FCVal | LDpred2.MultiPRS | 0.143 | 0.145 | -0.002 | 7.42e-01 | 0.140 | 0.145 | -0.005 | 3.02e-02 |
| Depression | All.MultiPRS | LDpred2.MultiPRS | 0.146 | 0.145 | 0.000 | 9.60e-01 | 0.146 | 0.145 | 0.001 | 3.08e-01 |
| Depression | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.113 | 0.145 | -0.033 | 2.64e-06 | 0.108 | 0.145 | -0.037 | 5.17e-08 |
| Depression | lassosum.10FCVal | LDpred2.MultiPRS | 0.143 | 0.145 | -0.002 | 7.38e-01 | 0.144 | 0.145 | -0.001 | 6.89e-01 |
| Depression | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.126 | 0.145 | -0.019 | 6.08e-03 | 0.127 | 0.145 | -0.018 | 1.15e-03 |
| Depression | lassosum.MultiPRS | LDpred2.MultiPRS | 0.144 | 0.145 | -0.002 | 8.00e-01 | 0.144 | 0.145 | -0.002 | 5.81e-01 |
| Depression | PRScs.10FCVal | LDpred2.MultiPRS | 0.141 | 0.145 | -0.005 | 5.03e-01 | 0.139 | 0.145 | -0.006 | 4.69e-02 |
| Depression | PRScs.MultiPRS | LDpred2.MultiPRS | 0.142 | 0.145 | -0.003 | 6.51e-01 | 0.141 | 0.145 | -0.004 | 1.09e-01 |
| Depression | lassosum.PseudoVal | LDpred2.MultiPRS | 0.081 | 0.145 | -0.064 | 5.00e-20 | 0.093 | 0.145 | -0.052 | 3.21e-08 |
| Depression | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.143 | 0.145 | -0.002 | 7.49e-01 | 0.139 | 0.145 | -0.007 | 1.12e-03 |
| Depression | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.143 | 0.145 | -0.002 | 7.65e-01 | 0.139 | 0.145 | -0.006 | 1.41e-02 |
| Depression | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.145 | 0.145 | 0.000 | 1.00e+00 | 0.145 | 0.145 | 0.000 | 1.00e+00 |
| Depression | LDpred1.Inf | LDpred2.10FCVal | 0.143 | 0.146 | -0.002 | 8.69e-02 | 0.138 | 0.144 | -0.006 | 3.66e-02 |
| Depression | PRScs.MultiPRS | LDpred2.10FCVal | 0.142 | 0.146 | -0.003 | 6.20e-01 | 0.141 | 0.144 | -0.003 | 2.94e-01 |
| Depression | LDpred2.MultiPRS | LDpred2.10FCVal | 0.145 | 0.146 | 0.000 | 9.66e-01 | 0.145 | 0.144 | 0.001 | 1.66e-01 |
| Depression | PRScs.PseudoVal | LDpred2.10FCVal | 0.139 | 0.146 | -0.007 | 7.06e-07 | 0.139 | 0.144 | -0.005 | 6.03e-02 |
| Depression | LDpred2.10FCVal | LDpred2.10FCVal | 0.146 | 0.146 | 0.000 | 1.00e+00 | 0.144 | 0.144 | 0.000 | 1.00e+00 |
| Depression | lassosum.MultiPRS | LDpred2.10FCVal | 0.144 | 0.146 | -0.002 | 7.67e-01 | 0.144 | 0.144 | 0.000 | 9.28e-01 |
| Depression | LDpred1.10FCVal | LDpred2.10FCVal | 0.143 | 0.146 | -0.003 | 1.97e-02 | 0.140 | 0.144 | -0.004 | 1.14e-01 |
| Depression | All.MultiPRS | LDpred2.10FCVal | 0.146 | 0.146 | 0.000 | 9.93e-01 | 0.146 | 0.144 | 0.002 | 8.12e-02 |
| Depression | SBayesR.PseudoVal | LDpred2.10FCVal | 0.113 | 0.146 | -0.033 | 1.84e-22 | 0.108 | 0.144 | -0.036 | 1.34e-07 |
| Depression | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.141 | 0.146 | -0.005 | 3.00e-03 | 0.140 | 0.144 | -0.004 | 2.32e-01 |
| Depression | pT+clump.MultiPRS | LDpred2.10FCVal | 0.126 | 0.146 | -0.019 | 5.16e-03 | 0.127 | 0.144 | -0.017 | 2.32e-03 |
| Depression | lassosum.PseudoVal | LDpred2.10FCVal | 0.081 | 0.146 | -0.064 | 3.33e-39 | 0.093 | 0.144 | -0.051 | 1.38e-07 |
| Depression | PRScs.10FCVal | LDpred2.10FCVal | 0.141 | 0.146 | -0.005 | 7.38e-04 | 0.139 | 0.144 | -0.004 | 1.30e-01 |
| Depression | lassosum.10FCVal | LDpred2.10FCVal | 0.143 | 0.146 | -0.003 | 6.56e-02 | 0.144 | 0.144 | 0.000 | 9.83e-01 |
| Depression | SBLUP.Inf | LDpred2.10FCVal | 0.143 | 0.146 | -0.003 | 1.22e-02 | 0.139 | 0.144 | -0.004 | 8.40e-02 |
| Depression | LDpred2.PseudoVal | LDpred2.10FCVal | 0.143 | 0.146 | -0.003 | 2.03e-02 | 0.139 | 0.144 | -0.005 | 1.56e-02 |
| Depression | pT+clump.10FCVal | LDpred2.10FCVal | 0.120 | 0.146 | -0.026 | 9.63e-16 | 0.118 | 0.144 | -0.026 | 4.47e-05 |
| Depression | LDpred2.Inf | LDpred2.10FCVal | 0.143 | 0.146 | -0.003 | 1.04e-02 | 0.139 | 0.144 | -0.005 | 1.72e-02 |
| Depression | LDpred1.MultiPRS | LDpred2.10FCVal | 0.143 | 0.146 | -0.002 | 7.33e-01 | 0.139 | 0.144 | -0.005 | 6.22e-02 |
| Depression | lassosum.10FCVal | LDpred2.PseudoVal | 0.143 | 0.143 | 0.000 | 9.54e-01 | 0.144 | 0.139 | 0.005 | 1.27e-01 |
| Depression | SBLUP.Inf | LDpred2.PseudoVal | 0.143 | 0.143 | -0.001 | 3.32e-01 | 0.139 | 0.139 | 0.001 | 4.84e-01 |
| Depression | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.113 | 0.143 | -0.030 | 3.37e-17 | 0.108 | 0.139 | -0.030 | 2.61e-05 |
| Depression | All.MultiPRS | LDpred2.PseudoVal | 0.146 | 0.143 | 0.003 | 7.10e-01 | 0.146 | 0.139 | 0.008 | 8.95e-04 |
| Depression | PRScs.PseudoVal | LDpred2.PseudoVal | 0.139 | 0.143 | -0.004 | 1.06e-02 | 0.139 | 0.139 | 0.000 | 9.95e-01 |
| Depression | lassosum.MultiPRS | LDpred2.PseudoVal | 0.144 | 0.143 | 0.000 | 9.47e-01 | 0.144 | 0.139 | 0.005 | 1.05e-01 |
| Depression | PRScs.MultiPRS | LDpred2.PseudoVal | 0.142 | 0.143 | -0.001 | 8.93e-01 | 0.141 | 0.139 | 0.003 | 3.45e-01 |
| Depression | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.145 | 0.143 | 0.002 | 7.49e-01 | 0.145 | 0.139 | 0.007 | 1.12e-03 |
| Depression | PRScs.10FCVal | LDpred2.PseudoVal | 0.141 | 0.143 | -0.002 | 1.80e-01 | 0.139 | 0.139 | 0.001 | 8.00e-01 |
| Depression | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.139 | 0.139 | 0.000 | 1.00e+00 |
| Depression | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.126 | 0.143 | -0.017 | 1.50e-02 | 0.127 | 0.139 | -0.011 | 5.36e-02 |
| Depression | pT+clump.10FCVal | LDpred2.PseudoVal | 0.120 | 0.143 | -0.023 | 1.43e-13 | 0.118 | 0.139 | -0.021 | 9.00e-04 |
| Depression | lassosum.PseudoVal | LDpred2.PseudoVal | 0.081 | 0.143 | -0.062 | 3.09e-30 | 0.093 | 0.139 | -0.046 | 1.92e-05 |
| Depression | LDpred1.Inf | LDpred2.PseudoVal | 0.143 | 0.143 | 0.000 | 7.84e-01 | 0.138 | 0.139 | 0.000 | 8.51e-01 |
| Depression | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.143 | 0.143 | 0.000 | 9.83e-01 | 0.139 | 0.139 | 0.001 | 6.94e-01 |
| Depression | LDpred1.10FCVal | LDpred2.PseudoVal | 0.143 | 0.143 | 0.000 | 9.52e-01 | 0.140 | 0.139 | 0.002 | 3.64e-01 |
| Depression | LDpred2.Inf | LDpred2.PseudoVal | 0.143 | 0.143 | 0.000 | 4.41e-01 | 0.139 | 0.139 | 0.000 | 9.66e-01 |
| Depression | LDpred2.10FCVal | LDpred2.PseudoVal | 0.146 | 0.143 | 0.003 | 2.03e-02 | 0.144 | 0.139 | 0.005 | 1.56e-02 |
| Depression | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.141 | 0.143 | -0.002 | 1.78e-01 | 0.140 | 0.139 | 0.002 | 5.93e-01 |
| Depression | pT+clump.MultiPRS | LDpred2.Inf | 0.126 | 0.143 | -0.017 | 1.69e-02 | 0.127 | 0.139 | -0.011 | 4.98e-02 |
| Depression | lassosum.MultiPRS | LDpred2.Inf | 0.144 | 0.143 | 0.001 | 9.11e-01 | 0.144 | 0.139 | 0.005 | 1.13e-01 |
| Depression | SBayesR.PseudoVal | LDpred2.Inf | 0.113 | 0.143 | -0.030 | 1.40e-16 | 0.108 | 0.139 | -0.030 | 3.32e-05 |
| Depression | All.MultiPRS | LDpred2.Inf | 0.146 | 0.143 | 0.003 | 6.76e-01 | 0.146 | 0.139 | 0.008 | 1.27e-03 |
| Depression | SBLUP.Inf | LDpred2.Inf | 0.143 | 0.143 | 0.000 | 6.23e-01 | 0.139 | 0.139 | 0.001 | 4.52e-01 |
| Depression | LDpred2.PseudoVal | LDpred2.Inf | 0.143 | 0.143 | 0.000 | 4.41e-01 | 0.139 | 0.139 | 0.000 | 9.66e-01 |
| Depression | PRScs.MultiPRS | LDpred2.Inf | 0.142 | 0.143 | -0.001 | 9.30e-01 | 0.141 | 0.139 | 0.003 | 3.50e-01 |
| Depression | LDpred2.MultiPRS | LDpred2.Inf | 0.145 | 0.143 | 0.003 | 7.15e-01 | 0.145 | 0.139 | 0.007 | 1.84e-03 |
| Depression | PRScs.10FCVal | LDpred2.Inf | 0.141 | 0.143 | -0.002 | 2.71e-01 | 0.139 | 0.139 | 0.001 | 8.04e-01 |
| Depression | LDpred1.Inf | LDpred2.Inf | 0.143 | 0.143 | 0.001 | 5.23e-01 | 0.138 | 0.139 | 0.000 | 8.72e-01 |
| Depression | LDpred1.MultiPRS | LDpred2.Inf | 0.143 | 0.143 | 0.000 | 9.47e-01 | 0.139 | 0.139 | 0.001 | 6.79e-01 |
| Depression | pT+clump.10FCVal | LDpred2.Inf | 0.120 | 0.143 | -0.023 | 5.47e-14 | 0.118 | 0.139 | -0.021 | 6.17e-04 |
| Depression | lassosum.PseudoVal | LDpred2.Inf | 0.081 | 0.143 | -0.061 | 4.30e-29 | 0.093 | 0.139 | -0.046 | 2.66e-05 |
| Depression | PRScs.PseudoVal | LDpred2.Inf | 0.139 | 0.143 | -0.004 | 2.44e-02 | 0.139 | 0.139 | 0.000 | 9.97e-01 |
| Depression | DBSLMM.PseudoVal | LDpred2.Inf | 0.141 | 0.143 | -0.002 | 2.51e-01 | 0.140 | 0.139 | 0.002 | 5.81e-01 |
| Depression | LDpred1.10FCVal | LDpred2.Inf | 0.143 | 0.143 | 0.000 | 7.89e-01 | 0.140 | 0.139 | 0.002 | 3.44e-01 |
| Depression | LDpred2.Inf | LDpred2.Inf | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.139 | 0.139 | 0.000 | 1.00e+00 |
| Depression | lassosum.10FCVal | LDpred2.Inf | 0.143 | 0.143 | 0.000 | 9.00e-01 | 0.144 | 0.139 | 0.005 | 1.19e-01 |
| Depression | LDpred2.10FCVal | LDpred2.Inf | 0.146 | 0.143 | 0.003 | 1.04e-02 | 0.144 | 0.139 | 0.005 | 1.72e-02 |
| Depression | PRScs.10FCVal | PRScs.MultiPRS | 0.141 | 0.142 | -0.002 | 8.29e-01 | 0.139 | 0.141 | -0.002 | 1.62e-01 |
| Depression | LDpred1.10FCVal | PRScs.MultiPRS | 0.143 | 0.142 | 0.001 | 9.00e-01 | 0.140 | 0.141 | -0.001 | 6.91e-01 |
| Depression | lassosum.MultiPRS | PRScs.MultiPRS | 0.144 | 0.142 | 0.001 | 2.88e-01 | 0.144 | 0.141 | 0.002 | 3.95e-01 |
| Depression | LDpred2.MultiPRS | PRScs.MultiPRS | 0.145 | 0.142 | 0.003 | 6.51e-01 | 0.145 | 0.141 | 0.004 | 1.09e-01 |
| Depression | All.MultiPRS | PRScs.MultiPRS | 0.146 | 0.142 | 0.004 | 6.15e-01 | 0.146 | 0.141 | 0.005 | 1.65e-02 |
| Depression | PRScs.PseudoVal | PRScs.MultiPRS | 0.139 | 0.142 | -0.004 | 6.10e-01 | 0.139 | 0.141 | -0.003 | 2.31e-01 |
| Depression | LDpred1.MultiPRS | PRScs.MultiPRS | 0.143 | 0.142 | 0.001 | 8.77e-01 | 0.139 | 0.141 | -0.002 | 4.39e-01 |
| Depression | pT+clump.MultiPRS | PRScs.MultiPRS | 0.126 | 0.142 | -0.016 | 2.10e-02 | 0.127 | 0.141 | -0.014 | 7.98e-03 |
| Depression | lassosum.PseudoVal | PRScs.MultiPRS | 0.081 | 0.142 | -0.061 | 3.44e-18 | 0.093 | 0.141 | -0.049 | 3.36e-07 |
| Depression | SBayesR.PseudoVal | PRScs.MultiPRS | 0.113 | 0.142 | -0.030 | 2.16e-05 | 0.108 | 0.141 | -0.033 | 1.95e-06 |
| Depression | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.141 | 0.142 | -0.001 | 8.77e-01 | 0.140 | 0.141 | -0.001 | 6.67e-01 |
| Depression | SBLUP.Inf | PRScs.MultiPRS | 0.143 | 0.142 | 0.000 | 9.65e-01 | 0.139 | 0.141 | -0.002 | 5.00e-01 |
| Depression | LDpred2.Inf | PRScs.MultiPRS | 0.143 | 0.142 | 0.001 | 9.30e-01 | 0.139 | 0.141 | -0.003 | 3.50e-01 |
| Depression | pT+clump.10FCVal | PRScs.MultiPRS | 0.120 | 0.142 | -0.022 | 1.52e-03 | 0.118 | 0.141 | -0.024 | 1.65e-04 |
| Depression | LDpred2.10FCVal | PRScs.MultiPRS | 0.146 | 0.142 | 0.003 | 6.20e-01 | 0.144 | 0.141 | 0.003 | 2.94e-01 |
| Depression | lassosum.10FCVal | PRScs.MultiPRS | 0.143 | 0.142 | 0.001 | 9.05e-01 | 0.144 | 0.141 | 0.003 | 3.68e-01 |
| Depression | LDpred2.PseudoVal | PRScs.MultiPRS | 0.143 | 0.142 | 0.001 | 8.93e-01 | 0.139 | 0.141 | -0.003 | 3.45e-01 |
| Depression | LDpred1.Inf | PRScs.MultiPRS | 0.143 | 0.142 | 0.001 | 8.68e-01 | 0.138 | 0.141 | -0.003 | 2.89e-01 |
| Depression | PRScs.MultiPRS | PRScs.MultiPRS | 0.142 | 0.142 | 0.000 | 1.00e+00 | 0.141 | 0.141 | 0.000 | 1.00e+00 |
| Depression | LDpred2.PseudoVal | PRScs.10FCVal | 0.143 | 0.141 | 0.002 | 1.80e-01 | 0.139 | 0.139 | -0.001 | 8.00e-01 |
| Depression | PRScs.10FCVal | PRScs.10FCVal | 0.141 | 0.141 | 0.000 | 1.00e+00 | 0.139 | 0.139 | 0.000 | 1.00e+00 |
| Depression | LDpred2.MultiPRS | PRScs.10FCVal | 0.145 | 0.141 | 0.005 | 5.03e-01 | 0.145 | 0.139 | 0.006 | 4.69e-02 |
| Depression | pT+clump.MultiPRS | PRScs.10FCVal | 0.126 | 0.141 | -0.015 | 3.71e-02 | 0.127 | 0.139 | -0.012 | 3.68e-02 |
| Depression | LDpred1.Inf | PRScs.10FCVal | 0.143 | 0.141 | 0.003 | 1.54e-01 | 0.138 | 0.139 | -0.001 | 7.41e-01 |
| Depression | LDpred1.MultiPRS | PRScs.10FCVal | 0.143 | 0.141 | 0.003 | 7.10e-01 | 0.139 | 0.139 | 0.000 | 9.60e-01 |
| Depression | pT+clump.10FCVal | PRScs.10FCVal | 0.120 | 0.141 | -0.021 | 2.06e-09 | 0.118 | 0.139 | -0.022 | 1.64e-03 |
| Depression | lassosum.PseudoVal | PRScs.10FCVal | 0.081 | 0.141 | -0.059 | 5.30e-38 | 0.093 | 0.139 | -0.047 | 3.11e-07 |
| Depression | PRScs.PseudoVal | PRScs.10FCVal | 0.139 | 0.141 | -0.002 | 1.70e-01 | 0.139 | 0.139 | -0.001 | 7.49e-01 |
| Depression | DBSLMM.PseudoVal | PRScs.10FCVal | 0.141 | 0.141 | 0.000 | 8.09e-01 | 0.140 | 0.139 | 0.001 | 8.48e-01 |
| Depression | SBLUP.Inf | PRScs.10FCVal | 0.143 | 0.141 | 0.002 | 3.23e-01 | 0.139 | 0.139 | 0.000 | 9.98e-01 |
| Depression | LDpred2.Inf | PRScs.10FCVal | 0.143 | 0.141 | 0.002 | 2.71e-01 | 0.139 | 0.139 | -0.001 | 8.04e-01 |
| Depression | lassosum.10FCVal | PRScs.10FCVal | 0.143 | 0.141 | 0.002 | 1.84e-01 | 0.144 | 0.139 | 0.004 | 2.05e-01 |
| Depression | LDpred2.10FCVal | PRScs.10FCVal | 0.146 | 0.141 | 0.005 | 7.38e-04 | 0.144 | 0.139 | 0.004 | 1.30e-01 |
| Depression | lassosum.MultiPRS | PRScs.10FCVal | 0.144 | 0.141 | 0.003 | 6.77e-01 | 0.144 | 0.139 | 0.004 | 2.01e-01 |
| Depression | LDpred1.10FCVal | PRScs.10FCVal | 0.143 | 0.141 | 0.002 | 1.56e-01 | 0.140 | 0.139 | 0.001 | 7.84e-01 |
| Depression | All.MultiPRS | PRScs.10FCVal | 0.146 | 0.141 | 0.005 | 4.70e-01 | 0.146 | 0.139 | 0.007 | 1.14e-02 |
| Depression | SBayesR.PseudoVal | PRScs.10FCVal | 0.113 | 0.141 | -0.028 | 9.79e-16 | 0.108 | 0.139 | -0.031 | 8.48e-06 |
| Depression | PRScs.MultiPRS | PRScs.10FCVal | 0.142 | 0.141 | 0.002 | 8.29e-01 | 0.141 | 0.139 | 0.002 | 1.62e-01 |
| Depression | All.MultiPRS | PRScs.PseudoVal | 0.146 | 0.139 | 0.007 | 3.09e-01 | 0.146 | 0.139 | 0.008 | 3.26e-03 |
| Depression | pT+clump.MultiPRS | PRScs.PseudoVal | 0.126 | 0.139 | -0.012 | 7.30e-02 | 0.127 | 0.139 | -0.011 | 5.35e-02 |
| Depression | LDpred2.PseudoVal | PRScs.PseudoVal | 0.143 | 0.139 | 0.004 | 1.06e-02 | 0.139 | 0.139 | 0.000 | 9.95e-01 |
| Depression | SBayesR.PseudoVal | PRScs.PseudoVal | 0.113 | 0.139 | -0.026 | 6.07e-14 | 0.108 | 0.139 | -0.030 | 1.56e-05 |
| Depression | LDpred2.MultiPRS | PRScs.PseudoVal | 0.145 | 0.139 | 0.007 | 3.35e-01 | 0.145 | 0.139 | 0.007 | 1.84e-02 |
| Depression | SBLUP.Inf | PRScs.PseudoVal | 0.143 | 0.139 | 0.004 | 2.82e-02 | 0.139 | 0.139 | 0.001 | 7.91e-01 |
| Depression | LDpred1.Inf | PRScs.PseudoVal | 0.143 | 0.139 | 0.005 | 9.05e-03 | 0.138 | 0.139 | 0.000 | 9.37e-01 |
| Depression | PRScs.10FCVal | PRScs.PseudoVal | 0.141 | 0.139 | 0.002 | 1.70e-01 | 0.139 | 0.139 | 0.001 | 7.49e-01 |
| Depression | LDpred2.Inf | PRScs.PseudoVal | 0.143 | 0.139 | 0.004 | 2.44e-02 | 0.139 | 0.139 | 0.000 | 9.97e-01 |
| Depression | PRScs.PseudoVal | PRScs.PseudoVal | 0.139 | 0.139 | 0.000 | 1.00e+00 | 0.139 | 0.139 | 0.000 | 1.00e+00 |
| Depression | LDpred2.10FCVal | PRScs.PseudoVal | 0.146 | 0.139 | 0.007 | 7.06e-07 | 0.144 | 0.139 | 0.005 | 6.03e-02 |
| Depression | pT+clump.10FCVal | PRScs.PseudoVal | 0.120 | 0.139 | -0.019 | 4.41e-08 | 0.118 | 0.139 | -0.021 | 2.43e-03 |
| Depression | PRScs.MultiPRS | PRScs.PseudoVal | 0.142 | 0.139 | 0.004 | 6.10e-01 | 0.141 | 0.139 | 0.003 | 2.31e-01 |
| Depression | lassosum.PseudoVal | PRScs.PseudoVal | 0.081 | 0.139 | -0.057 | 7.05e-35 | 0.093 | 0.139 | -0.046 | 6.28e-07 |
| Depression | lassosum.10FCVal | PRScs.PseudoVal | 0.143 | 0.139 | 0.004 | 1.00e-02 | 0.144 | 0.139 | 0.005 | 1.16e-01 |
| Depression | LDpred1.MultiPRS | PRScs.PseudoVal | 0.143 | 0.139 | 0.005 | 5.05e-01 | 0.139 | 0.139 | 0.001 | 8.22e-01 |
| Depression | lassosum.MultiPRS | PRScs.PseudoVal | 0.144 | 0.139 | 0.005 | 4.78e-01 | 0.144 | 0.139 | 0.005 | 1.11e-01 |
| Depression | LDpred1.10FCVal | PRScs.PseudoVal | 0.143 | 0.139 | 0.004 | 5.82e-03 | 0.140 | 0.139 | 0.002 | 5.68e-01 |
| Depression | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.141 | 0.139 | 0.002 | 1.47e-01 | 0.140 | 0.139 | 0.002 | 6.40e-01 |
| Depression | All.MultiPRS | pT+clump.MultiPRS | 0.146 | 0.126 | 0.020 | 5.17e-03 | 0.146 | 0.127 | 0.019 | 2.17e-04 |
| Depression | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.113 | 0.126 | -0.014 | 5.29e-02 | 0.108 | 0.127 | -0.019 | 1.76e-02 |
| Depression | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.143 | 0.126 | 0.017 | 1.46e-02 | 0.139 | 0.127 | 0.012 | 2.81e-02 |
| Depression | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.126 | 0.126 | 0.000 | 1.00e+00 | 0.127 | 0.127 | 0.000 | 1.00e+00 |
| Depression | lassosum.PseudoVal | pT+clump.MultiPRS | 0.081 | 0.126 | -0.045 | 1.66e-10 | 0.093 | 0.127 | -0.035 | 9.54e-04 |
| Depression | PRScs.10FCVal | pT+clump.MultiPRS | 0.141 | 0.126 | 0.015 | 3.71e-02 | 0.139 | 0.127 | 0.012 | 3.68e-02 |
| Depression | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.141 | 0.126 | 0.015 | 3.19e-02 | 0.140 | 0.127 | 0.013 | 2.22e-02 |
| Depression | SBLUP.Inf | pT+clump.MultiPRS | 0.143 | 0.126 | 0.016 | 1.91e-02 | 0.139 | 0.127 | 0.012 | 2.72e-02 |
| Depression | LDpred2.Inf | pT+clump.MultiPRS | 0.143 | 0.126 | 0.017 | 1.69e-02 | 0.139 | 0.127 | 0.011 | 4.98e-02 |
| Depression | pT+clump.10FCVal | pT+clump.MultiPRS | 0.120 | 0.126 | -0.006 | 3.84e-01 | 0.118 | 0.127 | -0.010 | 3.27e-04 |
| Depression | LDpred2.10FCVal | pT+clump.MultiPRS | 0.146 | 0.126 | 0.019 | 5.16e-03 | 0.144 | 0.127 | 0.017 | 2.32e-03 |
| Depression | PRScs.PseudoVal | pT+clump.MultiPRS | 0.139 | 0.126 | 0.012 | 7.30e-02 | 0.139 | 0.127 | 0.011 | 5.35e-02 |
| Depression | LDpred1.10FCVal | pT+clump.MultiPRS | 0.143 | 0.126 | 0.017 | 1.53e-02 | 0.140 | 0.127 | 0.013 | 1.43e-02 |
| Depression | LDpred1.Inf | pT+clump.MultiPRS | 0.143 | 0.126 | 0.017 | 1.36e-02 | 0.138 | 0.127 | 0.011 | 5.12e-02 |
| Depression | PRScs.MultiPRS | pT+clump.MultiPRS | 0.142 | 0.126 | 0.016 | 2.10e-02 | 0.141 | 0.127 | 0.014 | 7.98e-03 |
| Depression | lassosum.10FCVal | pT+clump.MultiPRS | 0.143 | 0.126 | 0.017 | 1.54e-02 | 0.144 | 0.127 | 0.017 | 1.67e-03 |
| Depression | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.143 | 0.126 | 0.017 | 1.50e-02 | 0.139 | 0.127 | 0.011 | 5.36e-02 |
| Depression | lassosum.MultiPRS | pT+clump.MultiPRS | 0.144 | 0.126 | 0.017 | 1.21e-02 | 0.144 | 0.127 | 0.016 | 2.44e-03 |
| Depression | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.145 | 0.126 | 0.019 | 6.08e-03 | 0.145 | 0.127 | 0.018 | 1.15e-03 |
| Depression | LDpred2.10FCVal | pT+clump.10FCVal | 0.146 | 0.120 | 0.026 | 9.63e-16 | 0.144 | 0.118 | 0.026 | 4.47e-05 |
| Depression | pT+clump.MultiPRS | pT+clump.10FCVal | 0.126 | 0.120 | 0.006 | 3.84e-01 | 0.127 | 0.118 | 0.010 | 3.27e-04 |
| Depression | LDpred2.Inf | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 5.47e-14 | 0.139 | 0.118 | 0.021 | 6.17e-04 |
| Depression | SBayesR.PseudoVal | pT+clump.10FCVal | 0.113 | 0.120 | -0.007 | 9.07e-02 | 0.108 | 0.118 | -0.009 | 2.85e-01 |
| Depression | PRScs.MultiPRS | pT+clump.10FCVal | 0.142 | 0.120 | 0.022 | 1.52e-03 | 0.141 | 0.118 | 0.024 | 1.65e-04 |
| Depression | SBLUP.Inf | pT+clump.10FCVal | 0.143 | 0.120 | 0.022 | 2.11e-14 | 0.139 | 0.118 | 0.022 | 2.20e-04 |
| Depression | lassosum.MultiPRS | pT+clump.10FCVal | 0.144 | 0.120 | 0.024 | 7.51e-04 | 0.144 | 0.118 | 0.026 | 3.61e-05 |
| Depression | PRScs.10FCVal | pT+clump.10FCVal | 0.141 | 0.120 | 0.021 | 2.06e-09 | 0.139 | 0.118 | 0.022 | 1.64e-03 |
| Depression | LDpred2.MultiPRS | pT+clump.10FCVal | 0.145 | 0.120 | 0.025 | 3.00e-04 | 0.145 | 0.118 | 0.028 | 2.08e-05 |
| Depression | lassosum.10FCVal | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 5.41e-14 | 0.144 | 0.118 | 0.026 | 2.00e-05 |
| Depression | LDpred2.PseudoVal | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 1.43e-13 | 0.139 | 0.118 | 0.021 | 9.00e-04 |
| Depression | LDpred1.MultiPRS | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 8.77e-04 | 0.139 | 0.118 | 0.022 | 3.17e-04 |
| Depression | pT+clump.10FCVal | pT+clump.10FCVal | 0.120 | 0.120 | 0.000 | 1.00e+00 | 0.118 | 0.118 | 0.000 | 1.00e+00 |
| Depression | All.MultiPRS | pT+clump.10FCVal | 0.146 | 0.120 | 0.026 | 2.26e-04 | 0.146 | 0.118 | 0.029 | 3.18e-06 |
| Depression | LDpred1.Inf | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 8.46e-15 | 0.138 | 0.118 | 0.021 | 6.63e-04 |
| Depression | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.141 | 0.120 | 0.021 | 1.69e-11 | 0.140 | 0.118 | 0.022 | 3.52e-04 |
| Depression | LDpred1.10FCVal | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 1.08e-14 | 0.140 | 0.118 | 0.023 | 1.47e-04 |
| Depression | lassosum.PseudoVal | pT+clump.10FCVal | 0.081 | 0.120 | -0.039 | 5.00e-11 | 0.093 | 0.118 | -0.025 | 3.46e-02 |
| Depression | PRScs.PseudoVal | pT+clump.10FCVal | 0.139 | 0.120 | 0.019 | 4.41e-08 | 0.139 | 0.118 | 0.021 | 2.43e-03 |
| Depression | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.126 | 0.113 | 0.014 | 5.29e-02 | 0.127 | 0.108 | 0.019 | 1.76e-02 |
| Depression | LDpred1.10FCVal | SBayesR.PseudoVal | 0.143 | 0.113 | 0.030 | 1.31e-17 | 0.140 | 0.108 | 0.032 | 6.28e-06 |
| Depression | PRScs.10FCVal | SBayesR.PseudoVal | 0.141 | 0.113 | 0.028 | 9.79e-16 | 0.139 | 0.108 | 0.031 | 8.48e-06 |
| Depression | LDpred2.10FCVal | SBayesR.PseudoVal | 0.146 | 0.113 | 0.033 | 1.84e-22 | 0.144 | 0.108 | 0.036 | 1.34e-07 |
| Depression | lassosum.10FCVal | SBayesR.PseudoVal | 0.143 | 0.113 | 0.030 | 1.92e-17 | 0.144 | 0.108 | 0.036 | 6.11e-07 |
| Depression | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.143 | 0.113 | 0.030 | 3.37e-17 | 0.139 | 0.108 | 0.030 | 2.61e-05 |
| Depression | lassosum.MultiPRS | SBayesR.PseudoVal | 0.144 | 0.113 | 0.031 | 8.66e-06 | 0.144 | 0.108 | 0.035 | 5.84e-07 |
| Depression | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.145 | 0.113 | 0.033 | 2.64e-06 | 0.145 | 0.108 | 0.037 | 5.17e-08 |
| Depression | All.MultiPRS | SBayesR.PseudoVal | 0.146 | 0.113 | 0.033 | 2.06e-06 | 0.146 | 0.108 | 0.038 | 2.01e-08 |
| Depression | PRScs.PseudoVal | SBayesR.PseudoVal | 0.139 | 0.113 | 0.026 | 6.07e-14 | 0.139 | 0.108 | 0.030 | 1.56e-05 |
| Depression | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.143 | 0.113 | 0.031 | 1.13e-05 | 0.139 | 0.108 | 0.031 | 1.63e-05 |
| Depression | pT+clump.10FCVal | SBayesR.PseudoVal | 0.120 | 0.113 | 0.007 | 9.07e-02 | 0.118 | 0.108 | 0.009 | 2.85e-01 |
| Depression | lassosum.PseudoVal | SBayesR.PseudoVal | 0.081 | 0.113 | -0.031 | 5.05e-10 | 0.093 | 0.108 | -0.015 | 1.27e-01 |
| Depression | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.113 | 0.113 | 0.000 | 1.00e+00 | 0.108 | 0.108 | 0.000 | 1.00e+00 |
| Depression | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.141 | 0.113 | 0.028 | 2.06e-15 | 0.140 | 0.108 | 0.032 | 1.06e-05 |
| Depression | SBLUP.Inf | SBayesR.PseudoVal | 0.143 | 0.113 | 0.030 | 2.11e-16 | 0.139 | 0.108 | 0.031 | 1.70e-05 |
| Depression | LDpred2.Inf | SBayesR.PseudoVal | 0.143 | 0.113 | 0.030 | 1.40e-16 | 0.139 | 0.108 | 0.030 | 3.32e-05 |
| Depression | LDpred1.Inf | SBayesR.PseudoVal | 0.143 | 0.113 | 0.031 | 4.60e-17 | 0.138 | 0.108 | 0.030 | 3.90e-05 |
| Depression | PRScs.MultiPRS | SBayesR.PseudoVal | 0.142 | 0.113 | 0.030 | 2.16e-05 | 0.141 | 0.108 | 0.033 | 1.95e-06 |
| Depression | LDpred2.10FCVal | SBLUP.Inf | 0.146 | 0.143 | 0.003 | 1.22e-02 | 0.144 | 0.139 | 0.004 | 8.40e-02 |
| Depression | PRScs.PseudoVal | SBLUP.Inf | 0.139 | 0.143 | -0.004 | 2.82e-02 | 0.139 | 0.139 | -0.001 | 7.91e-01 |
| Depression | LDpred1.10FCVal | SBLUP.Inf | 0.143 | 0.143 | 0.001 | 4.78e-01 | 0.140 | 0.139 | 0.001 | 5.80e-01 |
| Depression | LDpred2.Inf | SBLUP.Inf | 0.143 | 0.143 | 0.000 | 6.23e-01 | 0.139 | 0.139 | -0.001 | 4.52e-01 |
| Depression | SBayesR.PseudoVal | SBLUP.Inf | 0.113 | 0.143 | -0.030 | 2.11e-16 | 0.108 | 0.139 | -0.031 | 1.70e-05 |
| Depression | lassosum.10FCVal | SBLUP.Inf | 0.143 | 0.143 | 0.001 | 7.47e-01 | 0.144 | 0.139 | 0.004 | 1.76e-01 |
| Depression | pT+clump.MultiPRS | SBLUP.Inf | 0.126 | 0.143 | -0.016 | 1.91e-02 | 0.127 | 0.139 | -0.012 | 2.72e-02 |
| Depression | lassosum.MultiPRS | SBLUP.Inf | 0.144 | 0.143 | 0.001 | 8.76e-01 | 0.144 | 0.139 | 0.004 | 1.72e-01 |
| Depression | PRScs.10FCVal | SBLUP.Inf | 0.141 | 0.143 | -0.002 | 3.23e-01 | 0.139 | 0.139 | 0.000 | 9.98e-01 |
| Depression | PRScs.MultiPRS | SBLUP.Inf | 0.142 | 0.143 | 0.000 | 9.65e-01 | 0.141 | 0.139 | 0.002 | 5.00e-01 |
| Depression | LDpred1.MultiPRS | SBLUP.Inf | 0.143 | 0.143 | 0.001 | 9.11e-01 | 0.139 | 0.139 | 0.000 | 9.04e-01 |
| Depression | LDpred2.PseudoVal | SBLUP.Inf | 0.143 | 0.143 | 0.001 | 3.32e-01 | 0.139 | 0.139 | -0.001 | 4.84e-01 |
| Depression | LDpred1.Inf | SBLUP.Inf | 0.143 | 0.143 | 0.001 | 2.28e-01 | 0.138 | 0.139 | -0.001 | 3.88e-01 |
| Depression | LDpred2.MultiPRS | SBLUP.Inf | 0.145 | 0.143 | 0.003 | 6.82e-01 | 0.145 | 0.139 | 0.006 | 1.95e-02 |
| Depression | All.MultiPRS | SBLUP.Inf | 0.146 | 0.143 | 0.003 | 6.44e-01 | 0.146 | 0.139 | 0.007 | 4.76e-03 |
| Depression | SBLUP.Inf | SBLUP.Inf | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.139 | 0.139 | 0.000 | 1.00e+00 |
| Depression | DBSLMM.PseudoVal | SBLUP.Inf | 0.141 | 0.143 | -0.001 | 3.06e-01 | 0.140 | 0.139 | 0.001 | 8.02e-01 |
| Depression | pT+clump.10FCVal | SBLUP.Inf | 0.120 | 0.143 | -0.022 | 2.11e-14 | 0.118 | 0.139 | -0.022 | 2.20e-04 |
| Depression | lassosum.PseudoVal | SBLUP.Inf | 0.081 | 0.143 | -0.061 | 4.92e-29 | 0.093 | 0.139 | -0.047 | 1.63e-05 |
| Intelligence | LDpred2.PseudoVal | All.MultiPRS | 0.100 | 0.102 | -0.002 | 7.68e-01 | 0.093 | 0.097 | -0.005 | 1.16e-01 |
| Intelligence | pT+clump.10FCVal | All.MultiPRS | 0.084 | 0.102 | -0.019 | 8.46e-03 | 0.074 | 0.097 | -0.023 | 2.37e-04 |
| Intelligence | LDpred2.MultiPRS | All.MultiPRS | 0.103 | 0.102 | 0.000 | 9.48e-01 | 0.094 | 0.097 | -0.003 | 8.12e-02 |
| Intelligence | PRScs.PseudoVal | All.MultiPRS | 0.099 | 0.102 | -0.004 | 5.78e-01 | 0.092 | 0.097 | -0.005 | 1.50e-01 |
| Intelligence | LDpred1.Inf | All.MultiPRS | 0.100 | 0.102 | -0.003 | 7.11e-01 | 0.092 | 0.097 | -0.006 | 7.11e-02 |
| Intelligence | PRScs.MultiPRS | All.MultiPRS | 0.100 | 0.102 | -0.002 | 1.57e-01 | 0.093 | 0.097 | -0.004 | 1.30e-01 |
| Intelligence | LDpred2.Inf | All.MultiPRS | 0.101 | 0.102 | -0.002 | 7.90e-01 | 0.093 | 0.097 | -0.005 | 1.38e-01 |
| Intelligence | lassosum.10FCVal | All.MultiPRS | 0.099 | 0.102 | -0.003 | 6.21e-01 | 0.093 | 0.097 | -0.004 | 2.43e-01 |
| Intelligence | LDpred2.10FCVal | All.MultiPRS | 0.103 | 0.102 | 0.000 | 9.44e-01 | 0.093 | 0.097 | -0.004 | 6.03e-02 |
| Intelligence | lassosum.MultiPRS | All.MultiPRS | 0.102 | 0.102 | 0.000 | 7.70e-01 | 0.097 | 0.097 | -0.001 | 8.06e-01 |
| Intelligence | LDpred1.10FCVal | All.MultiPRS | 0.102 | 0.102 | -0.001 | 9.35e-01 | 0.092 | 0.097 | -0.005 | 5.55e-02 |
| Intelligence | All.MultiPRS | All.MultiPRS | 0.102 | 0.102 | 0.000 | 1.00e+00 | 0.097 | 0.097 | 0.000 | 1.00e+00 |
| Intelligence | SBayesR.PseudoVal | All.MultiPRS | 0.072 | 0.102 | -0.030 | 2.15e-05 | 0.065 | 0.097 | -0.033 | 4.97e-05 |
| Intelligence | LDpred1.MultiPRS | All.MultiPRS | 0.101 | 0.102 | -0.001 | 8.67e-01 | 0.093 | 0.097 | -0.004 | 1.06e-01 |
| Intelligence | pT+clump.MultiPRS | All.MultiPRS | 0.086 | 0.102 | -0.016 | 2.29e-02 | 0.080 | 0.097 | -0.017 | 2.02e-03 |
| Intelligence | lassosum.PseudoVal | All.MultiPRS | 0.064 | 0.102 | -0.039 | 4.97e-08 | 0.059 | 0.097 | -0.039 | 4.72e-06 |
| Intelligence | PRScs.10FCVal | All.MultiPRS | 0.099 | 0.102 | -0.003 | 6.74e-01 | 0.089 | 0.097 | -0.008 | 2.28e-02 |
| Intelligence | DBSLMM.PseudoVal | All.MultiPRS | 0.089 | 0.102 | -0.014 | 4.94e-02 | 0.079 | 0.097 | -0.018 | 3.11e-04 |
| Intelligence | SBLUP.Inf | All.MultiPRS | 0.100 | 0.102 | -0.002 | 7.70e-01 | 0.092 | 0.097 | -0.005 | 1.09e-01 |
| Intelligence | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.072 | 0.089 | -0.016 | 2.04e-04 | 0.065 | 0.079 | -0.015 | 9.17e-02 |
| Intelligence | LDpred2.Inf | DBSLMM.PseudoVal | 0.101 | 0.089 | 0.012 | 1.80e-06 | 0.093 | 0.079 | 0.013 | 7.09e-03 |
| Intelligence | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.084 | 0.089 | -0.005 | 2.18e-01 | 0.074 | 0.079 | -0.006 | 4.62e-01 |
| Intelligence | PRScs.10FCVal | DBSLMM.PseudoVal | 0.099 | 0.089 | 0.011 | 3.01e-05 | 0.089 | 0.079 | 0.010 | 5.47e-02 |
| Intelligence | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.099 | 0.089 | 0.010 | 1.36e-04 | 0.092 | 0.079 | 0.013 | 1.24e-02 |
| Intelligence | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.086 | 0.089 | -0.002 | 7.63e-01 | 0.080 | 0.079 | 0.000 | 9.54e-01 |
| Intelligence | LDpred1.Inf | DBSLMM.PseudoVal | 0.100 | 0.089 | 0.011 | 1.74e-05 | 0.092 | 0.079 | 0.012 | 1.96e-02 |
| Intelligence | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.102 | 0.089 | 0.014 | 5.47e-02 | 0.097 | 0.079 | 0.017 | 4.05e-04 |
| Intelligence | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.103 | 0.089 | 0.014 | 4.46e-10 | 0.093 | 0.079 | 0.014 | 3.02e-03 |
| Intelligence | lassosum.10FCVal | DBSLMM.PseudoVal | 0.099 | 0.089 | 0.010 | 3.00e-04 | 0.093 | 0.079 | 0.014 | 1.49e-02 |
| Intelligence | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.102 | 0.089 | 0.013 | 1.23e-07 | 0.092 | 0.079 | 0.013 | 1.10e-02 |
| Intelligence | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.101 | 0.089 | 0.013 | 7.18e-02 | 0.093 | 0.079 | 0.014 | 6.95e-03 |
| Intelligence | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.100 | 0.089 | 0.012 | 9.48e-02 | 0.093 | 0.079 | 0.014 | 4.48e-03 |
| Intelligence | All.MultiPRS | DBSLMM.PseudoVal | 0.102 | 0.089 | 0.014 | 4.94e-02 | 0.097 | 0.079 | 0.018 | 3.11e-04 |
| Intelligence | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.100 | 0.089 | 0.012 | 2.25e-06 | 0.093 | 0.079 | 0.013 | 7.90e-03 |
| Intelligence | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.089 | 0.089 | 0.000 | 1.00e+00 | 0.079 | 0.079 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.103 | 0.089 | 0.014 | 4.81e-02 | 0.094 | 0.079 | 0.015 | 1.08e-03 |
| Intelligence | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.064 | 0.089 | -0.025 | 5.98e-08 | 0.059 | 0.079 | -0.021 | 2.21e-02 |
| Intelligence | SBLUP.Inf | DBSLMM.PseudoVal | 0.100 | 0.089 | 0.012 | 2.24e-06 | 0.092 | 0.079 | 0.013 | 8.58e-03 |
| Intelligence | SBayesR.PseudoVal | lassosum.MultiPRS | 0.072 | 0.102 | -0.030 | 2.60e-05 | 0.065 | 0.097 | -0.032 | 1.03e-04 |
| Intelligence | LDpred2.10FCVal | lassosum.MultiPRS | 0.103 | 0.102 | 0.001 | 9.09e-01 | 0.093 | 0.097 | -0.004 | 1.08e-01 |
| Intelligence | LDpred1.10FCVal | lassosum.MultiPRS | 0.102 | 0.102 | 0.000 | 9.70e-01 | 0.092 | 0.097 | -0.005 | 1.22e-01 |
| Intelligence | LDpred2.Inf | lassosum.MultiPRS | 0.101 | 0.102 | -0.002 | 8.25e-01 | 0.093 | 0.097 | -0.004 | 2.08e-01 |
| Intelligence | PRScs.10FCVal | lassosum.MultiPRS | 0.099 | 0.102 | -0.003 | 7.07e-01 | 0.089 | 0.097 | -0.007 | 3.31e-02 |
| Intelligence | PRScs.MultiPRS | lassosum.MultiPRS | 0.100 | 0.102 | -0.002 | 2.11e-01 | 0.093 | 0.097 | -0.004 | 1.71e-01 |
| Intelligence | pT+clump.MultiPRS | lassosum.MultiPRS | 0.086 | 0.102 | -0.016 | 2.57e-02 | 0.080 | 0.097 | -0.017 | 4.08e-03 |
| Intelligence | lassosum.MultiPRS | lassosum.MultiPRS | 0.102 | 0.102 | 0.000 | 1.00e+00 | 0.097 | 0.097 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.PseudoVal | lassosum.MultiPRS | 0.064 | 0.102 | -0.038 | 6.16e-08 | 0.059 | 0.097 | -0.038 | 4.70e-06 |
| Intelligence | LDpred2.MultiPRS | lassosum.MultiPRS | 0.103 | 0.102 | 0.000 | 7.65e-01 | 0.094 | 0.097 | -0.003 | 2.60e-01 |
| Intelligence | LDpred1.MultiPRS | lassosum.MultiPRS | 0.101 | 0.102 | -0.001 | 9.03e-01 | 0.093 | 0.097 | -0.004 | 2.38e-01 |
| Intelligence | LDpred2.PseudoVal | lassosum.MultiPRS | 0.100 | 0.102 | -0.002 | 8.02e-01 | 0.093 | 0.097 | -0.004 | 1.72e-01 |
| Intelligence | LDpred1.Inf | lassosum.MultiPRS | 0.100 | 0.102 | -0.002 | 7.44e-01 | 0.092 | 0.097 | -0.005 | 1.32e-01 |
| Intelligence | pT+clump.10FCVal | lassosum.MultiPRS | 0.084 | 0.102 | -0.018 | 9.58e-03 | 0.074 | 0.097 | -0.023 | 5.90e-04 |
| Intelligence | lassosum.10FCVal | lassosum.MultiPRS | 0.099 | 0.102 | -0.003 | 6.52e-01 | 0.093 | 0.097 | -0.004 | 2.04e-01 |
| Intelligence | SBLUP.Inf | lassosum.MultiPRS | 0.100 | 0.102 | -0.002 | 8.04e-01 | 0.092 | 0.097 | -0.004 | 1.66e-01 |
| Intelligence | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.089 | 0.102 | -0.014 | 5.47e-02 | 0.079 | 0.097 | -0.017 | 4.05e-04 |
| Intelligence | All.MultiPRS | lassosum.MultiPRS | 0.102 | 0.102 | 0.000 | 7.70e-01 | 0.097 | 0.097 | 0.001 | 8.06e-01 |
| Intelligence | PRScs.PseudoVal | lassosum.MultiPRS | 0.099 | 0.102 | -0.004 | 6.08e-01 | 0.092 | 0.097 | -0.004 | 1.92e-01 |
| Intelligence | LDpred1.Inf | lassosum.PseudoVal | 0.100 | 0.064 | 0.036 | 3.63e-13 | 0.092 | 0.059 | 0.033 | 7.86e-04 |
| Intelligence | PRScs.10FCVal | lassosum.PseudoVal | 0.099 | 0.064 | 0.036 | 3.55e-17 | 0.089 | 0.059 | 0.031 | 2.19e-04 |
| Intelligence | LDpred2.MultiPRS | lassosum.PseudoVal | 0.103 | 0.064 | 0.039 | 4.56e-08 | 0.094 | 0.059 | 0.036 | 2.02e-05 |
| Intelligence | lassosum.PseudoVal | lassosum.PseudoVal | 0.064 | 0.064 | 0.000 | 1.00e+00 | 0.059 | 0.059 | 0.000 | 1.00e+00 |
| Intelligence | PRScs.PseudoVal | lassosum.PseudoVal | 0.099 | 0.064 | 0.035 | 2.40e-16 | 0.092 | 0.059 | 0.034 | 5.12e-05 |
| Intelligence | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.089 | 0.064 | 0.025 | 5.98e-08 | 0.079 | 0.059 | 0.021 | 2.21e-02 |
| Intelligence | SBLUP.Inf | lassosum.PseudoVal | 0.100 | 0.064 | 0.037 | 1.05e-13 | 0.092 | 0.059 | 0.034 | 5.20e-04 |
| Intelligence | LDpred2.Inf | lassosum.PseudoVal | 0.101 | 0.064 | 0.037 | 8.04e-14 | 0.093 | 0.059 | 0.034 | 4.46e-04 |
| Intelligence | pT+clump.10FCVal | lassosum.PseudoVal | 0.084 | 0.064 | 0.020 | 1.39e-04 | 0.074 | 0.059 | 0.015 | 1.38e-01 |
| Intelligence | LDpred2.10FCVal | lassosum.PseudoVal | 0.103 | 0.064 | 0.039 | 5.06e-21 | 0.093 | 0.059 | 0.034 | 2.88e-05 |
| Intelligence | lassosum.MultiPRS | lassosum.PseudoVal | 0.102 | 0.064 | 0.038 | 6.16e-08 | 0.097 | 0.059 | 0.038 | 4.70e-06 |
| Intelligence | LDpred1.10FCVal | lassosum.PseudoVal | 0.102 | 0.064 | 0.038 | 2.66e-18 | 0.092 | 0.059 | 0.034 | 9.94e-05 |
| Intelligence | All.MultiPRS | lassosum.PseudoVal | 0.102 | 0.064 | 0.039 | 4.97e-08 | 0.097 | 0.059 | 0.039 | 4.72e-06 |
| Intelligence | SBayesR.PseudoVal | lassosum.PseudoVal | 0.072 | 0.064 | 0.009 | 7.99e-02 | 0.065 | 0.059 | 0.006 | 5.42e-01 |
| Intelligence | lassosum.10FCVal | lassosum.PseudoVal | 0.099 | 0.064 | 0.035 | 1.97e-15 | 0.093 | 0.059 | 0.035 | 7.70e-05 |
| Intelligence | pT+clump.MultiPRS | lassosum.PseudoVal | 0.086 | 0.064 | 0.023 | 1.32e-03 | 0.080 | 0.059 | 0.021 | 1.36e-02 |
| Intelligence | LDpred2.PseudoVal | lassosum.PseudoVal | 0.100 | 0.064 | 0.037 | 7.66e-14 | 0.093 | 0.059 | 0.034 | 4.57e-04 |
| Intelligence | LDpred1.MultiPRS | lassosum.PseudoVal | 0.101 | 0.064 | 0.037 | 1.11e-07 | 0.093 | 0.059 | 0.035 | 1.04e-04 |
| Intelligence | PRScs.MultiPRS | lassosum.PseudoVal | 0.100 | 0.064 | 0.037 | 2.41e-07 | 0.093 | 0.059 | 0.034 | 2.23e-05 |
| Intelligence | lassosum.10FCVal | lassosum.10FCVal | 0.099 | 0.099 | 0.000 | 1.00e+00 | 0.093 | 0.093 | 0.000 | 1.00e+00 |
| Intelligence | pT+clump.MultiPRS | lassosum.10FCVal | 0.086 | 0.099 | -0.012 | 7.55e-02 | 0.080 | 0.093 | -0.013 | 1.31e-02 |
| Intelligence | LDpred2.Inf | lassosum.10FCVal | 0.101 | 0.099 | 0.002 | 3.76e-01 | 0.093 | 0.093 | 0.000 | 9.17e-01 |
| Intelligence | SBayesR.PseudoVal | lassosum.10FCVal | 0.072 | 0.099 | -0.026 | 1.78e-10 | 0.065 | 0.093 | -0.029 | 6.09e-04 |
| Intelligence | PRScs.MultiPRS | lassosum.10FCVal | 0.100 | 0.099 | 0.001 | 8.43e-01 | 0.093 | 0.093 | 0.000 | 9.49e-01 |
| Intelligence | SBLUP.Inf | lassosum.10FCVal | 0.100 | 0.099 | 0.001 | 4.26e-01 | 0.092 | 0.093 | -0.001 | 8.23e-01 |
| Intelligence | lassosum.MultiPRS | lassosum.10FCVal | 0.102 | 0.099 | 0.003 | 6.52e-01 | 0.097 | 0.093 | 0.004 | 2.04e-01 |
| Intelligence | PRScs.10FCVal | lassosum.10FCVal | 0.099 | 0.099 | 0.001 | 7.79e-01 | 0.089 | 0.093 | -0.004 | 3.10e-01 |
| Intelligence | LDpred2.MultiPRS | lassosum.10FCVal | 0.103 | 0.099 | 0.004 | 6.14e-01 | 0.094 | 0.093 | 0.001 | 8.00e-01 |
| Intelligence | PRScs.PseudoVal | lassosum.10FCVal | 0.099 | 0.099 | 0.000 | 8.12e-01 | 0.092 | 0.093 | -0.001 | 8.14e-01 |
| Intelligence | LDpred2.PseudoVal | lassosum.10FCVal | 0.100 | 0.099 | 0.001 | 4.70e-01 | 0.093 | 0.093 | -0.001 | 8.65e-01 |
| Intelligence | LDpred1.MultiPRS | lassosum.10FCVal | 0.101 | 0.099 | 0.002 | 7.41e-01 | 0.093 | 0.093 | 0.000 | 9.73e-01 |
| Intelligence | pT+clump.10FCVal | lassosum.10FCVal | 0.084 | 0.099 | -0.015 | 3.72e-07 | 0.074 | 0.093 | -0.019 | 9.65e-04 |
| Intelligence | All.MultiPRS | lassosum.10FCVal | 0.102 | 0.099 | 0.003 | 6.21e-01 | 0.097 | 0.093 | 0.004 | 2.43e-01 |
| Intelligence | LDpred1.Inf | lassosum.10FCVal | 0.100 | 0.099 | 0.001 | 6.51e-01 | 0.092 | 0.093 | -0.002 | 6.54e-01 |
| Intelligence | DBSLMM.PseudoVal | lassosum.10FCVal | 0.089 | 0.099 | -0.010 | 3.00e-04 | 0.079 | 0.093 | -0.014 | 1.49e-02 |
| Intelligence | LDpred1.10FCVal | lassosum.10FCVal | 0.102 | 0.099 | 0.003 | 7.14e-02 | 0.092 | 0.093 | -0.001 | 7.39e-01 |
| Intelligence | lassosum.PseudoVal | lassosum.10FCVal | 0.064 | 0.099 | -0.035 | 1.97e-15 | 0.059 | 0.093 | -0.035 | 7.70e-05 |
| Intelligence | LDpred2.10FCVal | lassosum.10FCVal | 0.103 | 0.099 | 0.004 | 1.72e-02 | 0.093 | 0.093 | 0.000 | 9.64e-01 |
| Intelligence | LDpred2.10FCVal | LDpred1.MultiPRS | 0.103 | 0.101 | 0.002 | 8.13e-01 | 0.093 | 0.093 | 0.000 | 9.88e-01 |
| Intelligence | PRScs.PseudoVal | LDpred1.MultiPRS | 0.099 | 0.101 | -0.003 | 6.95e-01 | 0.092 | 0.093 | -0.001 | 8.15e-01 |
| Intelligence | LDpred2.Inf | LDpred1.MultiPRS | 0.101 | 0.101 | -0.001 | 9.20e-01 | 0.093 | 0.093 | 0.000 | 9.15e-01 |
| Intelligence | pT+clump.10FCVal | LDpred1.MultiPRS | 0.084 | 0.101 | -0.017 | 1.33e-02 | 0.074 | 0.093 | -0.019 | 2.30e-03 |
| Intelligence | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.072 | 0.101 | -0.029 | 4.38e-05 | 0.065 | 0.093 | -0.028 | 6.61e-04 |
| Intelligence | All.MultiPRS | LDpred1.MultiPRS | 0.102 | 0.101 | 0.001 | 8.67e-01 | 0.097 | 0.093 | 0.004 | 1.06e-01 |
| Intelligence | LDpred1.10FCVal | LDpred1.MultiPRS | 0.102 | 0.101 | 0.001 | 9.33e-01 | 0.092 | 0.093 | -0.001 | 3.50e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.101 | 0.101 | 0.000 | 1.00e+00 | 0.093 | 0.093 | 0.000 | 1.00e+00 |
| Intelligence | PRScs.10FCVal | LDpred1.MultiPRS | 0.099 | 0.101 | -0.002 | 7.98e-01 | 0.089 | 0.093 | -0.004 | 2.64e-01 |
| Intelligence | lassosum.PseudoVal | LDpred1.MultiPRS | 0.064 | 0.101 | -0.037 | 1.11e-07 | 0.059 | 0.093 | -0.035 | 1.04e-04 |
| Intelligence | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.086 | 0.101 | -0.015 | 3.55e-02 | 0.080 | 0.093 | -0.013 | 2.26e-02 |
| Intelligence | LDpred1.Inf | LDpred1.MultiPRS | 0.100 | 0.101 | -0.001 | 8.37e-01 | 0.092 | 0.093 | -0.002 | 3.78e-01 |
| Intelligence | lassosum.MultiPRS | LDpred1.MultiPRS | 0.102 | 0.101 | 0.001 | 9.03e-01 | 0.097 | 0.093 | 0.004 | 2.38e-01 |
| Intelligence | PRScs.MultiPRS | LDpred1.MultiPRS | 0.100 | 0.101 | -0.001 | 8.94e-01 | 0.093 | 0.093 | 0.000 | 9.73e-01 |
| Intelligence | lassosum.10FCVal | LDpred1.MultiPRS | 0.099 | 0.101 | -0.002 | 7.41e-01 | 0.093 | 0.093 | 0.000 | 9.73e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.100 | 0.101 | -0.001 | 8.97e-01 | 0.093 | 0.093 | -0.001 | 8.30e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.089 | 0.101 | -0.013 | 7.18e-02 | 0.079 | 0.093 | -0.014 | 6.95e-03 |
| Intelligence | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.103 | 0.101 | 0.001 | 8.60e-01 | 0.094 | 0.093 | 0.001 | 6.99e-01 |
| Intelligence | SBLUP.Inf | LDpred1.MultiPRS | 0.100 | 0.101 | -0.001 | 8.99e-01 | 0.092 | 0.093 | -0.001 | 7.78e-01 |
| Intelligence | All.MultiPRS | LDpred1.Inf | 0.102 | 0.100 | 0.003 | 7.11e-01 | 0.097 | 0.092 | 0.006 | 7.11e-02 |
| Intelligence | LDpred2.PseudoVal | LDpred1.Inf | 0.100 | 0.100 | 0.001 | 5.80e-01 | 0.093 | 0.092 | 0.001 | 5.73e-01 |
| Intelligence | SBayesR.PseudoVal | LDpred1.Inf | 0.072 | 0.100 | -0.027 | 1.56e-10 | 0.065 | 0.092 | -0.027 | 1.78e-03 |
| Intelligence | LDpred2.MultiPRS | LDpred1.Inf | 0.103 | 0.100 | 0.003 | 7.04e-01 | 0.094 | 0.092 | 0.003 | 3.89e-01 |
| Intelligence | PRScs.PseudoVal | LDpred1.Inf | 0.099 | 0.100 | -0.001 | 4.73e-01 | 0.092 | 0.092 | 0.001 | 8.18e-01 |
| Intelligence | LDpred1.Inf | LDpred1.Inf | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.092 | 0.092 | 0.000 | 1.00e+00 |
| Intelligence | pT+clump.10FCVal | LDpred1.Inf | 0.084 | 0.100 | -0.016 | 4.03e-07 | 0.074 | 0.092 | -0.018 | 4.50e-03 |
| Intelligence | PRScs.MultiPRS | LDpred1.Inf | 0.100 | 0.100 | 0.001 | 9.41e-01 | 0.093 | 0.092 | 0.001 | 6.49e-01 |
| Intelligence | lassosum.PseudoVal | LDpred1.Inf | 0.064 | 0.100 | -0.036 | 3.63e-13 | 0.059 | 0.092 | -0.033 | 7.86e-04 |
| Intelligence | pT+clump.MultiPRS | LDpred1.Inf | 0.086 | 0.100 | -0.013 | 5.75e-02 | 0.080 | 0.092 | -0.012 | 5.65e-02 |
| Intelligence | LDpred1.MultiPRS | LDpred1.Inf | 0.101 | 0.100 | 0.001 | 8.37e-01 | 0.093 | 0.092 | 0.002 | 3.78e-01 |
| Intelligence | PRScs.10FCVal | LDpred1.Inf | 0.099 | 0.100 | 0.000 | 8.53e-01 | 0.089 | 0.092 | -0.002 | 5.80e-01 |
| Intelligence | LDpred2.Inf | LDpred1.Inf | 0.101 | 0.100 | 0.001 | 4.16e-01 | 0.093 | 0.092 | 0.001 | 4.56e-01 |
| Intelligence | SBLUP.Inf | LDpred1.Inf | 0.100 | 0.100 | 0.001 | 5.31e-01 | 0.092 | 0.092 | 0.001 | 5.93e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred1.Inf | 0.089 | 0.100 | -0.011 | 1.74e-05 | 0.079 | 0.092 | -0.012 | 1.96e-02 |
| Intelligence | lassosum.MultiPRS | LDpred1.Inf | 0.102 | 0.100 | 0.002 | 7.44e-01 | 0.097 | 0.092 | 0.005 | 1.32e-01 |
| Intelligence | LDpred1.10FCVal | LDpred1.Inf | 0.102 | 0.100 | 0.002 | 4.39e-02 | 0.092 | 0.092 | 0.001 | 7.49e-01 |
| Intelligence | lassosum.10FCVal | LDpred1.Inf | 0.099 | 0.100 | -0.001 | 6.51e-01 | 0.093 | 0.092 | 0.002 | 6.54e-01 |
| Intelligence | LDpred2.10FCVal | LDpred1.Inf | 0.103 | 0.100 | 0.003 | 3.29e-02 | 0.093 | 0.092 | 0.002 | 5.88e-01 |
| Intelligence | LDpred1.Inf | LDpred1.10FCVal | 0.100 | 0.102 | -0.002 | 4.39e-02 | 0.092 | 0.092 | -0.001 | 7.49e-01 |
| Intelligence | PRScs.MultiPRS | LDpred1.10FCVal | 0.100 | 0.102 | -0.002 | 8.29e-01 | 0.093 | 0.092 | 0.001 | 7.35e-01 |
| Intelligence | LDpred1.10FCVal | LDpred1.10FCVal | 0.102 | 0.102 | 0.000 | 1.00e+00 | 0.092 | 0.092 | 0.000 | 1.00e+00 |
| Intelligence | All.MultiPRS | LDpred1.10FCVal | 0.102 | 0.102 | 0.001 | 9.35e-01 | 0.097 | 0.092 | 0.005 | 5.55e-02 |
| Intelligence | SBayesR.PseudoVal | LDpred1.10FCVal | 0.072 | 0.102 | -0.029 | 6.95e-13 | 0.065 | 0.092 | -0.028 | 8.80e-04 |
| Intelligence | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.089 | 0.102 | -0.013 | 1.23e-07 | 0.079 | 0.092 | -0.013 | 1.10e-02 |
| Intelligence | pT+clump.MultiPRS | LDpred1.10FCVal | 0.086 | 0.102 | -0.015 | 2.86e-02 | 0.080 | 0.092 | -0.012 | 3.14e-02 |
| Intelligence | lassosum.MultiPRS | LDpred1.10FCVal | 0.102 | 0.102 | 0.000 | 9.70e-01 | 0.097 | 0.092 | 0.005 | 1.22e-01 |
| Intelligence | PRScs.10FCVal | LDpred1.10FCVal | 0.099 | 0.102 | -0.002 | 1.28e-01 | 0.089 | 0.092 | -0.003 | 3.87e-01 |
| Intelligence | lassosum.10FCVal | LDpred1.10FCVal | 0.099 | 0.102 | -0.003 | 7.14e-02 | 0.093 | 0.092 | 0.001 | 7.39e-01 |
| Intelligence | SBLUP.Inf | LDpred1.10FCVal | 0.100 | 0.102 | -0.001 | 2.40e-01 | 0.092 | 0.092 | 0.000 | 9.10e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred1.10FCVal | 0.100 | 0.102 | -0.002 | 2.60e-01 | 0.093 | 0.092 | 0.000 | 8.74e-01 |
| Intelligence | pT+clump.10FCVal | LDpred1.10FCVal | 0.084 | 0.102 | -0.018 | 2.02e-08 | 0.074 | 0.092 | -0.018 | 3.88e-03 |
| Intelligence | LDpred2.MultiPRS | LDpred1.10FCVal | 0.103 | 0.102 | 0.001 | 9.27e-01 | 0.094 | 0.092 | 0.002 | 4.14e-01 |
| Intelligence | PRScs.PseudoVal | LDpred1.10FCVal | 0.099 | 0.102 | -0.003 | 2.86e-02 | 0.092 | 0.092 | 0.000 | 9.47e-01 |
| Intelligence | LDpred2.10FCVal | LDpred1.10FCVal | 0.103 | 0.102 | 0.001 | 3.13e-01 | 0.093 | 0.092 | 0.001 | 6.60e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred1.10FCVal | 0.101 | 0.102 | -0.001 | 9.33e-01 | 0.093 | 0.092 | 0.001 | 3.50e-01 |
| Intelligence | LDpred2.Inf | LDpred1.10FCVal | 0.101 | 0.102 | -0.001 | 3.17e-01 | 0.093 | 0.092 | 0.001 | 7.87e-01 |
| Intelligence | lassosum.PseudoVal | LDpred1.10FCVal | 0.064 | 0.102 | -0.038 | 2.66e-18 | 0.059 | 0.092 | -0.034 | 9.94e-05 |
| Intelligence | LDpred1.Inf | LDpred2.MultiPRS | 0.100 | 0.103 | -0.003 | 7.04e-01 | 0.092 | 0.094 | -0.003 | 3.89e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.089 | 0.103 | -0.014 | 4.81e-02 | 0.079 | 0.094 | -0.015 | 1.08e-03 |
| Intelligence | SBLUP.Inf | LDpred2.MultiPRS | 0.100 | 0.103 | -0.002 | 7.62e-01 | 0.092 | 0.094 | -0.002 | 5.15e-01 |
| Intelligence | LDpred2.Inf | LDpred2.MultiPRS | 0.101 | 0.103 | -0.002 | 7.83e-01 | 0.093 | 0.094 | -0.001 | 6.09e-01 |
| Intelligence | pT+clump.10FCVal | LDpred2.MultiPRS | 0.084 | 0.103 | -0.019 | 8.16e-03 | 0.074 | 0.094 | -0.020 | 2.14e-03 |
| Intelligence | LDpred2.10FCVal | LDpred2.MultiPRS | 0.103 | 0.103 | 0.000 | 9.52e-01 | 0.093 | 0.094 | -0.001 | 4.31e-01 |
| Intelligence | PRScs.PseudoVal | LDpred2.MultiPRS | 0.099 | 0.103 | -0.004 | 5.71e-01 | 0.092 | 0.094 | -0.002 | 5.69e-01 |
| Intelligence | LDpred1.10FCVal | LDpred2.MultiPRS | 0.102 | 0.103 | -0.001 | 9.27e-01 | 0.092 | 0.094 | -0.002 | 4.14e-01 |
| Intelligence | All.MultiPRS | LDpred2.MultiPRS | 0.102 | 0.103 | 0.000 | 9.48e-01 | 0.097 | 0.094 | 0.003 | 8.12e-02 |
| Intelligence | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.072 | 0.103 | -0.030 | 2.02e-05 | 0.065 | 0.094 | -0.029 | 2.94e-04 |
| Intelligence | lassosum.10FCVal | LDpred2.MultiPRS | 0.099 | 0.103 | -0.004 | 6.14e-01 | 0.093 | 0.094 | -0.001 | 8.00e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.086 | 0.103 | -0.016 | 2.23e-02 | 0.080 | 0.094 | -0.014 | 1.34e-02 |
| Intelligence | lassosum.MultiPRS | LDpred2.MultiPRS | 0.102 | 0.103 | 0.000 | 7.65e-01 | 0.097 | 0.094 | 0.003 | 2.60e-01 |
| Intelligence | PRScs.10FCVal | LDpred2.MultiPRS | 0.099 | 0.103 | -0.003 | 6.67e-01 | 0.089 | 0.094 | -0.005 | 1.26e-01 |
| Intelligence | PRScs.MultiPRS | LDpred2.MultiPRS | 0.100 | 0.103 | -0.002 | 6.99e-02 | 0.093 | 0.094 | -0.001 | 6.47e-01 |
| Intelligence | lassosum.PseudoVal | LDpred2.MultiPRS | 0.064 | 0.103 | -0.039 | 4.56e-08 | 0.059 | 0.094 | -0.036 | 2.02e-05 |
| Intelligence | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.100 | 0.103 | -0.002 | 7.61e-01 | 0.093 | 0.094 | -0.002 | 5.30e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.101 | 0.103 | -0.001 | 8.60e-01 | 0.093 | 0.094 | -0.001 | 6.99e-01 |
| Intelligence | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.103 | 0.103 | 0.000 | 1.00e+00 | 0.094 | 0.094 | 0.000 | 1.00e+00 |
| Intelligence | LDpred1.Inf | LDpred2.10FCVal | 0.100 | 0.103 | -0.003 | 3.29e-02 | 0.092 | 0.093 | -0.002 | 5.88e-01 |
| Intelligence | PRScs.MultiPRS | LDpred2.10FCVal | 0.100 | 0.103 | -0.003 | 7.13e-01 | 0.093 | 0.093 | 0.000 | 9.78e-01 |
| Intelligence | LDpred2.MultiPRS | LDpred2.10FCVal | 0.103 | 0.103 | 0.000 | 9.52e-01 | 0.094 | 0.093 | 0.001 | 4.31e-01 |
| Intelligence | PRScs.PseudoVal | LDpred2.10FCVal | 0.099 | 0.103 | -0.004 | 1.94e-03 | 0.092 | 0.093 | -0.001 | 8.02e-01 |
| Intelligence | LDpred2.10FCVal | LDpred2.10FCVal | 0.103 | 0.103 | 0.000 | 1.00e+00 | 0.093 | 0.093 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.MultiPRS | LDpred2.10FCVal | 0.102 | 0.103 | -0.001 | 9.09e-01 | 0.097 | 0.093 | 0.004 | 1.08e-01 |
| Intelligence | LDpred1.10FCVal | LDpred2.10FCVal | 0.102 | 0.103 | -0.001 | 3.13e-01 | 0.092 | 0.093 | -0.001 | 6.60e-01 |
| Intelligence | All.MultiPRS | LDpred2.10FCVal | 0.102 | 0.103 | 0.000 | 9.44e-01 | 0.097 | 0.093 | 0.004 | 6.03e-02 |
| Intelligence | SBayesR.PseudoVal | LDpred2.10FCVal | 0.072 | 0.103 | -0.030 | 2.33e-14 | 0.065 | 0.093 | -0.028 | 4.22e-04 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.089 | 0.103 | -0.014 | 4.46e-10 | 0.079 | 0.093 | -0.014 | 3.02e-03 |
| Intelligence | pT+clump.MultiPRS | LDpred2.10FCVal | 0.086 | 0.103 | -0.016 | 1.91e-02 | 0.080 | 0.093 | -0.013 | 2.37e-02 |
| Intelligence | lassosum.PseudoVal | LDpred2.10FCVal | 0.064 | 0.103 | -0.039 | 5.06e-21 | 0.059 | 0.093 | -0.034 | 2.88e-05 |
| Intelligence | PRScs.10FCVal | LDpred2.10FCVal | 0.099 | 0.103 | -0.003 | 1.54e-02 | 0.089 | 0.093 | -0.004 | 2.02e-01 |
| Intelligence | lassosum.10FCVal | LDpred2.10FCVal | 0.099 | 0.103 | -0.004 | 1.72e-02 | 0.093 | 0.093 | 0.000 | 9.64e-01 |
| Intelligence | SBLUP.Inf | LDpred2.10FCVal | 0.100 | 0.103 | -0.003 | 3.38e-02 | 0.092 | 0.093 | -0.001 | 7.92e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred2.10FCVal | 0.100 | 0.103 | -0.003 | 1.99e-02 | 0.093 | 0.093 | -0.001 | 8.20e-01 |
| Intelligence | pT+clump.10FCVal | LDpred2.10FCVal | 0.084 | 0.103 | -0.019 | 1.51e-08 | 0.074 | 0.093 | -0.019 | 3.90e-03 |
| Intelligence | LDpred2.Inf | LDpred2.10FCVal | 0.101 | 0.103 | -0.002 | 3.96e-02 | 0.093 | 0.093 | 0.000 | 9.22e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.10FCVal | 0.101 | 0.103 | -0.002 | 8.13e-01 | 0.093 | 0.093 | 0.000 | 9.88e-01 |
| Intelligence | lassosum.10FCVal | LDpred2.PseudoVal | 0.099 | 0.100 | -0.001 | 4.70e-01 | 0.093 | 0.093 | 0.001 | 8.65e-01 |
| Intelligence | SBLUP.Inf | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 9.74e-01 | 0.092 | 0.093 | 0.000 | 8.88e-01 |
| Intelligence | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.072 | 0.100 | -0.028 | 3.97e-11 | 0.065 | 0.093 | -0.028 | 1.01e-03 |
| Intelligence | All.MultiPRS | LDpred2.PseudoVal | 0.102 | 0.100 | 0.002 | 7.68e-01 | 0.097 | 0.093 | 0.005 | 1.16e-01 |
| Intelligence | PRScs.PseudoVal | LDpred2.PseudoVal | 0.099 | 0.100 | -0.002 | 2.87e-01 | 0.092 | 0.093 | 0.000 | 9.51e-01 |
| Intelligence | lassosum.MultiPRS | LDpred2.PseudoVal | 0.102 | 0.100 | 0.002 | 8.02e-01 | 0.097 | 0.093 | 0.004 | 1.72e-01 |
| Intelligence | PRScs.MultiPRS | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 9.98e-01 | 0.093 | 0.093 | 0.000 | 8.84e-01 |
| Intelligence | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.103 | 0.100 | 0.002 | 7.61e-01 | 0.094 | 0.093 | 0.002 | 5.30e-01 |
| Intelligence | PRScs.10FCVal | LDpred2.PseudoVal | 0.099 | 0.100 | -0.001 | 6.14e-01 | 0.089 | 0.093 | -0.003 | 3.70e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.093 | 0.093 | 0.000 | 1.00e+00 |
| Intelligence | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.086 | 0.100 | -0.014 | 4.82e-02 | 0.080 | 0.093 | -0.013 | 4.22e-02 |
| Intelligence | pT+clump.10FCVal | LDpred2.PseudoVal | 0.084 | 0.100 | -0.017 | 4.12e-07 | 0.074 | 0.093 | -0.019 | 3.63e-03 |
| Intelligence | lassosum.PseudoVal | LDpred2.PseudoVal | 0.064 | 0.100 | -0.037 | 7.66e-14 | 0.059 | 0.093 | -0.034 | 4.57e-04 |
| Intelligence | LDpred1.Inf | LDpred2.PseudoVal | 0.100 | 0.100 | -0.001 | 5.80e-01 | 0.092 | 0.093 | -0.001 | 5.73e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.101 | 0.100 | 0.001 | 8.97e-01 | 0.093 | 0.093 | 0.001 | 8.30e-01 |
| Intelligence | LDpred1.10FCVal | LDpred2.PseudoVal | 0.102 | 0.100 | 0.002 | 2.60e-01 | 0.092 | 0.093 | 0.000 | 8.74e-01 |
| Intelligence | LDpred2.Inf | LDpred2.PseudoVal | 0.101 | 0.100 | 0.000 | 5.75e-01 | 0.093 | 0.093 | 0.000 | 7.00e-01 |
| Intelligence | LDpred2.10FCVal | LDpred2.PseudoVal | 0.103 | 0.100 | 0.003 | 1.99e-02 | 0.093 | 0.093 | 0.001 | 8.20e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.089 | 0.100 | -0.012 | 2.25e-06 | 0.079 | 0.093 | -0.013 | 7.90e-03 |
| Intelligence | pT+clump.MultiPRS | LDpred2.Inf | 0.086 | 0.101 | -0.014 | 4.49e-02 | 0.080 | 0.093 | -0.013 | 3.31e-02 |
| Intelligence | lassosum.MultiPRS | LDpred2.Inf | 0.102 | 0.101 | 0.002 | 8.25e-01 | 0.097 | 0.093 | 0.004 | 2.08e-01 |
| Intelligence | SBayesR.PseudoVal | LDpred2.Inf | 0.072 | 0.101 | -0.028 | 2.65e-11 | 0.065 | 0.093 | -0.028 | 8.83e-04 |
| Intelligence | All.MultiPRS | LDpred2.Inf | 0.102 | 0.101 | 0.002 | 7.90e-01 | 0.097 | 0.093 | 0.005 | 1.38e-01 |
| Intelligence | SBLUP.Inf | LDpred2.Inf | 0.100 | 0.101 | 0.000 | 6.15e-01 | 0.092 | 0.093 | 0.000 | 5.80e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred2.Inf | 0.100 | 0.101 | 0.000 | 5.75e-01 | 0.093 | 0.093 | 0.000 | 7.00e-01 |
| Intelligence | PRScs.MultiPRS | LDpred2.Inf | 0.100 | 0.101 | 0.000 | 9.75e-01 | 0.093 | 0.093 | 0.000 | 9.56e-01 |
| Intelligence | LDpred2.MultiPRS | LDpred2.Inf | 0.103 | 0.101 | 0.002 | 7.83e-01 | 0.094 | 0.093 | 0.001 | 6.09e-01 |
| Intelligence | PRScs.10FCVal | LDpred2.Inf | 0.099 | 0.101 | -0.001 | 5.31e-01 | 0.089 | 0.093 | -0.003 | 3.27e-01 |
| Intelligence | LDpred1.Inf | LDpred2.Inf | 0.100 | 0.101 | -0.001 | 4.16e-01 | 0.092 | 0.093 | -0.001 | 4.56e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.Inf | 0.101 | 0.101 | 0.001 | 9.20e-01 | 0.093 | 0.093 | 0.000 | 9.15e-01 |
| Intelligence | pT+clump.10FCVal | LDpred2.Inf | 0.084 | 0.101 | -0.017 | 1.09e-07 | 0.074 | 0.093 | -0.019 | 2.20e-03 |
| Intelligence | lassosum.PseudoVal | LDpred2.Inf | 0.064 | 0.101 | -0.037 | 8.04e-14 | 0.059 | 0.093 | -0.034 | 4.46e-04 |
| Intelligence | PRScs.PseudoVal | LDpred2.Inf | 0.099 | 0.101 | -0.002 | 2.33e-01 | 0.092 | 0.093 | 0.000 | 8.87e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.Inf | 0.089 | 0.101 | -0.012 | 1.80e-06 | 0.079 | 0.093 | -0.013 | 7.09e-03 |
| Intelligence | LDpred1.10FCVal | LDpred2.Inf | 0.102 | 0.101 | 0.001 | 3.17e-01 | 0.092 | 0.093 | -0.001 | 7.87e-01 |
| Intelligence | LDpred2.Inf | LDpred2.Inf | 0.101 | 0.101 | 0.000 | 1.00e+00 | 0.093 | 0.093 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.10FCVal | LDpred2.Inf | 0.099 | 0.101 | -0.002 | 3.76e-01 | 0.093 | 0.093 | 0.000 | 9.17e-01 |
| Intelligence | LDpred2.10FCVal | LDpred2.Inf | 0.103 | 0.101 | 0.002 | 3.96e-02 | 0.093 | 0.093 | 0.000 | 9.22e-01 |
| Intelligence | PRScs.10FCVal | PRScs.MultiPRS | 0.099 | 0.100 | -0.001 | 9.02e-01 | 0.089 | 0.093 | -0.004 | 3.44e-02 |
| Intelligence | LDpred1.10FCVal | PRScs.MultiPRS | 0.102 | 0.100 | 0.002 | 8.29e-01 | 0.092 | 0.093 | -0.001 | 7.35e-01 |
| Intelligence | lassosum.MultiPRS | PRScs.MultiPRS | 0.102 | 0.100 | 0.002 | 2.11e-01 | 0.097 | 0.093 | 0.004 | 1.71e-01 |
| Intelligence | LDpred2.MultiPRS | PRScs.MultiPRS | 0.103 | 0.100 | 0.002 | 6.99e-02 | 0.094 | 0.093 | 0.001 | 6.47e-01 |
| Intelligence | All.MultiPRS | PRScs.MultiPRS | 0.102 | 0.100 | 0.002 | 1.57e-01 | 0.097 | 0.093 | 0.004 | 1.30e-01 |
| Intelligence | PRScs.PseudoVal | PRScs.MultiPRS | 0.099 | 0.100 | -0.002 | 7.95e-01 | 0.092 | 0.093 | -0.001 | 7.53e-01 |
| Intelligence | LDpred1.MultiPRS | PRScs.MultiPRS | 0.101 | 0.100 | 0.001 | 8.94e-01 | 0.093 | 0.093 | 0.000 | 9.73e-01 |
| Intelligence | pT+clump.MultiPRS | PRScs.MultiPRS | 0.086 | 0.100 | -0.014 | 4.82e-02 | 0.080 | 0.093 | -0.013 | 1.80e-02 |
| Intelligence | lassosum.PseudoVal | PRScs.MultiPRS | 0.064 | 0.100 | -0.037 | 2.41e-07 | 0.059 | 0.093 | -0.034 | 2.23e-05 |
| Intelligence | SBayesR.PseudoVal | PRScs.MultiPRS | 0.072 | 0.100 | -0.028 | 7.59e-05 | 0.065 | 0.093 | -0.028 | 4.86e-04 |
| Intelligence | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.089 | 0.100 | -0.012 | 9.48e-02 | 0.079 | 0.093 | -0.014 | 4.48e-03 |
| Intelligence | SBLUP.Inf | PRScs.MultiPRS | 0.100 | 0.100 | 0.000 | 9.96e-01 | 0.092 | 0.093 | -0.001 | 8.46e-01 |
| Intelligence | LDpred2.Inf | PRScs.MultiPRS | 0.101 | 0.100 | 0.000 | 9.75e-01 | 0.093 | 0.093 | 0.000 | 9.56e-01 |
| Intelligence | pT+clump.10FCVal | PRScs.MultiPRS | 0.084 | 0.100 | -0.016 | 1.94e-02 | 0.074 | 0.093 | -0.019 | 2.92e-03 |
| Intelligence | LDpred2.10FCVal | PRScs.MultiPRS | 0.103 | 0.100 | 0.003 | 7.13e-01 | 0.093 | 0.093 | 0.000 | 9.78e-01 |
| Intelligence | lassosum.10FCVal | PRScs.MultiPRS | 0.099 | 0.100 | -0.001 | 8.43e-01 | 0.093 | 0.093 | 0.000 | 9.49e-01 |
| Intelligence | LDpred2.PseudoVal | PRScs.MultiPRS | 0.100 | 0.100 | 0.000 | 9.98e-01 | 0.093 | 0.093 | 0.000 | 8.84e-01 |
| Intelligence | LDpred1.Inf | PRScs.MultiPRS | 0.100 | 0.100 | -0.001 | 9.41e-01 | 0.092 | 0.093 | -0.001 | 6.49e-01 |
| Intelligence | PRScs.MultiPRS | PRScs.MultiPRS | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.093 | 0.093 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.PseudoVal | PRScs.10FCVal | 0.100 | 0.099 | 0.001 | 6.14e-01 | 0.093 | 0.089 | 0.003 | 3.70e-01 |
| Intelligence | PRScs.10FCVal | PRScs.10FCVal | 0.099 | 0.099 | 0.000 | 1.00e+00 | 0.089 | 0.089 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.MultiPRS | PRScs.10FCVal | 0.103 | 0.099 | 0.003 | 6.67e-01 | 0.094 | 0.089 | 0.005 | 1.26e-01 |
| Intelligence | pT+clump.MultiPRS | PRScs.10FCVal | 0.086 | 0.099 | -0.013 | 6.41e-02 | 0.080 | 0.089 | -0.010 | 1.03e-01 |
| Intelligence | LDpred1.Inf | PRScs.10FCVal | 0.100 | 0.099 | 0.000 | 8.53e-01 | 0.092 | 0.089 | 0.002 | 5.80e-01 |
| Intelligence | LDpred1.MultiPRS | PRScs.10FCVal | 0.101 | 0.099 | 0.002 | 7.98e-01 | 0.093 | 0.089 | 0.004 | 2.64e-01 |
| Intelligence | pT+clump.10FCVal | PRScs.10FCVal | 0.084 | 0.099 | -0.016 | 3.75e-06 | 0.074 | 0.089 | -0.016 | 1.93e-02 |
| Intelligence | lassosum.PseudoVal | PRScs.10FCVal | 0.064 | 0.099 | -0.036 | 3.55e-17 | 0.059 | 0.089 | -0.031 | 2.19e-04 |
| Intelligence | PRScs.PseudoVal | PRScs.10FCVal | 0.099 | 0.099 | -0.001 | 5.81e-01 | 0.092 | 0.089 | 0.003 | 4.06e-01 |
| Intelligence | DBSLMM.PseudoVal | PRScs.10FCVal | 0.089 | 0.099 | -0.011 | 3.01e-05 | 0.079 | 0.089 | -0.010 | 5.47e-02 |
| Intelligence | SBLUP.Inf | PRScs.10FCVal | 0.100 | 0.099 | 0.001 | 6.00e-01 | 0.092 | 0.089 | 0.003 | 3.81e-01 |
| Intelligence | LDpred2.Inf | PRScs.10FCVal | 0.101 | 0.099 | 0.001 | 5.31e-01 | 0.093 | 0.089 | 0.003 | 3.27e-01 |
| Intelligence | lassosum.10FCVal | PRScs.10FCVal | 0.099 | 0.099 | -0.001 | 7.79e-01 | 0.093 | 0.089 | 0.004 | 3.10e-01 |
| Intelligence | LDpred2.10FCVal | PRScs.10FCVal | 0.103 | 0.099 | 0.003 | 1.54e-02 | 0.093 | 0.089 | 0.004 | 2.02e-01 |
| Intelligence | lassosum.MultiPRS | PRScs.10FCVal | 0.102 | 0.099 | 0.003 | 7.07e-01 | 0.097 | 0.089 | 0.007 | 3.31e-02 |
| Intelligence | LDpred1.10FCVal | PRScs.10FCVal | 0.102 | 0.099 | 0.002 | 1.28e-01 | 0.092 | 0.089 | 0.003 | 3.87e-01 |
| Intelligence | All.MultiPRS | PRScs.10FCVal | 0.102 | 0.099 | 0.003 | 6.74e-01 | 0.097 | 0.089 | 0.008 | 2.28e-02 |
| Intelligence | SBayesR.PseudoVal | PRScs.10FCVal | 0.072 | 0.099 | -0.027 | 5.13e-11 | 0.065 | 0.089 | -0.025 | 2.80e-03 |
| Intelligence | PRScs.MultiPRS | PRScs.10FCVal | 0.100 | 0.099 | 0.001 | 9.02e-01 | 0.093 | 0.089 | 0.004 | 3.44e-02 |
| Intelligence | All.MultiPRS | PRScs.PseudoVal | 0.102 | 0.099 | 0.004 | 5.78e-01 | 0.097 | 0.092 | 0.005 | 1.50e-01 |
| Intelligence | pT+clump.MultiPRS | PRScs.PseudoVal | 0.086 | 0.099 | -0.012 | 8.64e-02 | 0.080 | 0.092 | -0.012 | 3.40e-02 |
| Intelligence | LDpred2.PseudoVal | PRScs.PseudoVal | 0.100 | 0.099 | 0.002 | 2.87e-01 | 0.093 | 0.092 | 0.000 | 9.51e-01 |
| Intelligence | SBayesR.PseudoVal | PRScs.PseudoVal | 0.072 | 0.099 | -0.026 | 2.18e-10 | 0.065 | 0.092 | -0.028 | 8.15e-04 |
| Intelligence | LDpred2.MultiPRS | PRScs.PseudoVal | 0.103 | 0.099 | 0.004 | 5.71e-01 | 0.094 | 0.092 | 0.002 | 5.69e-01 |
| Intelligence | SBLUP.Inf | PRScs.PseudoVal | 0.100 | 0.099 | 0.002 | 2.73e-01 | 0.092 | 0.092 | 0.000 | 9.81e-01 |
| Intelligence | LDpred1.Inf | PRScs.PseudoVal | 0.100 | 0.099 | 0.001 | 4.73e-01 | 0.092 | 0.092 | -0.001 | 8.18e-01 |
| Intelligence | PRScs.10FCVal | PRScs.PseudoVal | 0.099 | 0.099 | 0.001 | 5.81e-01 | 0.089 | 0.092 | -0.003 | 4.06e-01 |
| Intelligence | LDpred2.Inf | PRScs.PseudoVal | 0.101 | 0.099 | 0.002 | 2.33e-01 | 0.093 | 0.092 | 0.000 | 8.87e-01 |
| Intelligence | PRScs.PseudoVal | PRScs.PseudoVal | 0.099 | 0.099 | 0.000 | 1.00e+00 | 0.092 | 0.092 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.10FCVal | PRScs.PseudoVal | 0.103 | 0.099 | 0.004 | 1.94e-03 | 0.093 | 0.092 | 0.001 | 8.02e-01 |
| Intelligence | pT+clump.10FCVal | PRScs.PseudoVal | 0.084 | 0.099 | -0.015 | 1.37e-05 | 0.074 | 0.092 | -0.018 | 5.46e-03 |
| Intelligence | PRScs.MultiPRS | PRScs.PseudoVal | 0.100 | 0.099 | 0.002 | 7.95e-01 | 0.093 | 0.092 | 0.001 | 7.53e-01 |
| Intelligence | lassosum.PseudoVal | PRScs.PseudoVal | 0.064 | 0.099 | -0.035 | 2.40e-16 | 0.059 | 0.092 | -0.034 | 5.12e-05 |
| Intelligence | lassosum.10FCVal | PRScs.PseudoVal | 0.099 | 0.099 | 0.000 | 8.12e-01 | 0.093 | 0.092 | 0.001 | 8.14e-01 |
| Intelligence | LDpred1.MultiPRS | PRScs.PseudoVal | 0.101 | 0.099 | 0.003 | 6.95e-01 | 0.093 | 0.092 | 0.001 | 8.15e-01 |
| Intelligence | lassosum.MultiPRS | PRScs.PseudoVal | 0.102 | 0.099 | 0.004 | 6.08e-01 | 0.097 | 0.092 | 0.004 | 1.92e-01 |
| Intelligence | LDpred1.10FCVal | PRScs.PseudoVal | 0.102 | 0.099 | 0.003 | 2.86e-02 | 0.092 | 0.092 | 0.000 | 9.47e-01 |
| Intelligence | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.089 | 0.099 | -0.010 | 1.36e-04 | 0.079 | 0.092 | -0.013 | 1.24e-02 |
| Intelligence | All.MultiPRS | pT+clump.MultiPRS | 0.102 | 0.086 | 0.016 | 2.29e-02 | 0.097 | 0.080 | 0.017 | 2.02e-03 |
| Intelligence | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.072 | 0.086 | -0.014 | 4.71e-02 | 0.065 | 0.080 | -0.015 | 9.33e-02 |
| Intelligence | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.101 | 0.086 | 0.015 | 3.55e-02 | 0.093 | 0.080 | 0.013 | 2.26e-02 |
| Intelligence | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.086 | 0.086 | 0.000 | 1.00e+00 | 0.080 | 0.080 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.PseudoVal | pT+clump.MultiPRS | 0.064 | 0.086 | -0.023 | 1.32e-03 | 0.059 | 0.080 | -0.021 | 1.36e-02 |
| Intelligence | PRScs.10FCVal | pT+clump.MultiPRS | 0.099 | 0.086 | 0.013 | 6.41e-02 | 0.089 | 0.080 | 0.010 | 1.03e-01 |
| Intelligence | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.089 | 0.086 | 0.002 | 7.63e-01 | 0.079 | 0.080 | 0.000 | 9.54e-01 |
| Intelligence | SBLUP.Inf | pT+clump.MultiPRS | 0.100 | 0.086 | 0.014 | 4.79e-02 | 0.092 | 0.080 | 0.013 | 3.51e-02 |
| Intelligence | LDpred2.Inf | pT+clump.MultiPRS | 0.101 | 0.086 | 0.014 | 4.49e-02 | 0.093 | 0.080 | 0.013 | 3.31e-02 |
| Intelligence | pT+clump.10FCVal | pT+clump.MultiPRS | 0.084 | 0.086 | -0.003 | 7.09e-01 | 0.074 | 0.080 | -0.006 | 3.41e-02 |
| Intelligence | LDpred2.10FCVal | pT+clump.MultiPRS | 0.103 | 0.086 | 0.016 | 1.91e-02 | 0.093 | 0.080 | 0.013 | 2.37e-02 |
| Intelligence | PRScs.PseudoVal | pT+clump.MultiPRS | 0.099 | 0.086 | 0.012 | 8.64e-02 | 0.092 | 0.080 | 0.012 | 3.40e-02 |
| Intelligence | LDpred1.10FCVal | pT+clump.MultiPRS | 0.102 | 0.086 | 0.015 | 2.86e-02 | 0.092 | 0.080 | 0.012 | 3.14e-02 |
| Intelligence | LDpred1.Inf | pT+clump.MultiPRS | 0.100 | 0.086 | 0.013 | 5.75e-02 | 0.092 | 0.080 | 0.012 | 5.65e-02 |
| Intelligence | PRScs.MultiPRS | pT+clump.MultiPRS | 0.100 | 0.086 | 0.014 | 4.82e-02 | 0.093 | 0.080 | 0.013 | 1.80e-02 |
| Intelligence | lassosum.10FCVal | pT+clump.MultiPRS | 0.099 | 0.086 | 0.012 | 7.55e-02 | 0.093 | 0.080 | 0.013 | 1.31e-02 |
| Intelligence | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.100 | 0.086 | 0.014 | 4.82e-02 | 0.093 | 0.080 | 0.013 | 4.22e-02 |
| Intelligence | lassosum.MultiPRS | pT+clump.MultiPRS | 0.102 | 0.086 | 0.016 | 2.57e-02 | 0.097 | 0.080 | 0.017 | 4.08e-03 |
| Intelligence | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.103 | 0.086 | 0.016 | 2.23e-02 | 0.094 | 0.080 | 0.014 | 1.34e-02 |
| Intelligence | LDpred2.10FCVal | pT+clump.10FCVal | 0.103 | 0.084 | 0.019 | 1.51e-08 | 0.093 | 0.074 | 0.019 | 3.90e-03 |
| Intelligence | pT+clump.MultiPRS | pT+clump.10FCVal | 0.086 | 0.084 | 0.003 | 7.09e-01 | 0.080 | 0.074 | 0.006 | 3.41e-02 |
| Intelligence | LDpred2.Inf | pT+clump.10FCVal | 0.101 | 0.084 | 0.017 | 1.09e-07 | 0.093 | 0.074 | 0.019 | 2.20e-03 |
| Intelligence | SBayesR.PseudoVal | pT+clump.10FCVal | 0.072 | 0.084 | -0.011 | 1.92e-02 | 0.065 | 0.074 | -0.009 | 3.36e-01 |
| Intelligence | PRScs.MultiPRS | pT+clump.10FCVal | 0.100 | 0.084 | 0.016 | 1.94e-02 | 0.093 | 0.074 | 0.019 | 2.92e-03 |
| Intelligence | SBLUP.Inf | pT+clump.10FCVal | 0.100 | 0.084 | 0.017 | 7.89e-08 | 0.092 | 0.074 | 0.018 | 2.16e-03 |
| Intelligence | lassosum.MultiPRS | pT+clump.10FCVal | 0.102 | 0.084 | 0.018 | 9.58e-03 | 0.097 | 0.074 | 0.023 | 5.90e-04 |
| Intelligence | PRScs.10FCVal | pT+clump.10FCVal | 0.099 | 0.084 | 0.016 | 3.75e-06 | 0.089 | 0.074 | 0.016 | 1.93e-02 |
| Intelligence | LDpred2.MultiPRS | pT+clump.10FCVal | 0.103 | 0.084 | 0.019 | 8.16e-03 | 0.094 | 0.074 | 0.020 | 2.14e-03 |
| Intelligence | lassosum.10FCVal | pT+clump.10FCVal | 0.099 | 0.084 | 0.015 | 3.72e-07 | 0.093 | 0.074 | 0.019 | 9.65e-04 |
| Intelligence | LDpred2.PseudoVal | pT+clump.10FCVal | 0.100 | 0.084 | 0.017 | 4.12e-07 | 0.093 | 0.074 | 0.019 | 3.63e-03 |
| Intelligence | LDpred1.MultiPRS | pT+clump.10FCVal | 0.101 | 0.084 | 0.017 | 1.33e-02 | 0.093 | 0.074 | 0.019 | 2.30e-03 |
| Intelligence | pT+clump.10FCVal | pT+clump.10FCVal | 0.084 | 0.084 | 0.000 | 1.00e+00 | 0.074 | 0.074 | 0.000 | 1.00e+00 |
| Intelligence | All.MultiPRS | pT+clump.10FCVal | 0.102 | 0.084 | 0.019 | 8.46e-03 | 0.097 | 0.074 | 0.023 | 2.37e-04 |
| Intelligence | LDpred1.Inf | pT+clump.10FCVal | 0.100 | 0.084 | 0.016 | 4.03e-07 | 0.092 | 0.074 | 0.018 | 4.50e-03 |
| Intelligence | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.089 | 0.084 | 0.005 | 2.18e-01 | 0.079 | 0.074 | 0.006 | 4.62e-01 |
| Intelligence | LDpred1.10FCVal | pT+clump.10FCVal | 0.102 | 0.084 | 0.018 | 2.02e-08 | 0.092 | 0.074 | 0.018 | 3.88e-03 |
| Intelligence | lassosum.PseudoVal | pT+clump.10FCVal | 0.064 | 0.084 | -0.020 | 1.39e-04 | 0.059 | 0.074 | -0.015 | 1.38e-01 |
| Intelligence | PRScs.PseudoVal | pT+clump.10FCVal | 0.099 | 0.084 | 0.015 | 1.37e-05 | 0.092 | 0.074 | 0.018 | 5.46e-03 |
| Intelligence | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.086 | 0.072 | 0.014 | 4.71e-02 | 0.080 | 0.065 | 0.015 | 9.33e-02 |
| Intelligence | LDpred1.10FCVal | SBayesR.PseudoVal | 0.102 | 0.072 | 0.029 | 6.95e-13 | 0.092 | 0.065 | 0.028 | 8.80e-04 |
| Intelligence | PRScs.10FCVal | SBayesR.PseudoVal | 0.099 | 0.072 | 0.027 | 5.13e-11 | 0.089 | 0.065 | 0.025 | 2.80e-03 |
| Intelligence | LDpred2.10FCVal | SBayesR.PseudoVal | 0.103 | 0.072 | 0.030 | 2.33e-14 | 0.093 | 0.065 | 0.028 | 4.22e-04 |
| Intelligence | lassosum.10FCVal | SBayesR.PseudoVal | 0.099 | 0.072 | 0.026 | 1.78e-10 | 0.093 | 0.065 | 0.029 | 6.09e-04 |
| Intelligence | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.100 | 0.072 | 0.028 | 3.97e-11 | 0.093 | 0.065 | 0.028 | 1.01e-03 |
| Intelligence | lassosum.MultiPRS | SBayesR.PseudoVal | 0.102 | 0.072 | 0.030 | 2.60e-05 | 0.097 | 0.065 | 0.032 | 1.03e-04 |
| Intelligence | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.103 | 0.072 | 0.030 | 2.02e-05 | 0.094 | 0.065 | 0.029 | 2.94e-04 |
| Intelligence | All.MultiPRS | SBayesR.PseudoVal | 0.102 | 0.072 | 0.030 | 2.15e-05 | 0.097 | 0.065 | 0.033 | 4.97e-05 |
| Intelligence | PRScs.PseudoVal | SBayesR.PseudoVal | 0.099 | 0.072 | 0.026 | 2.18e-10 | 0.092 | 0.065 | 0.028 | 8.15e-04 |
| Intelligence | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.101 | 0.072 | 0.029 | 4.38e-05 | 0.093 | 0.065 | 0.028 | 6.61e-04 |
| Intelligence | pT+clump.10FCVal | SBayesR.PseudoVal | 0.084 | 0.072 | 0.011 | 1.92e-02 | 0.074 | 0.065 | 0.009 | 3.36e-01 |
| Intelligence | lassosum.PseudoVal | SBayesR.PseudoVal | 0.064 | 0.072 | -0.009 | 7.99e-02 | 0.059 | 0.065 | -0.006 | 5.42e-01 |
| Intelligence | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.072 | 0.072 | 0.000 | 1.00e+00 | 0.065 | 0.065 | 0.000 | 1.00e+00 |
| Intelligence | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.089 | 0.072 | 0.016 | 2.04e-04 | 0.079 | 0.065 | 0.015 | 9.17e-02 |
| Intelligence | SBLUP.Inf | SBayesR.PseudoVal | 0.100 | 0.072 | 0.028 | 3.63e-11 | 0.092 | 0.065 | 0.028 | 1.05e-03 |
| Intelligence | LDpred2.Inf | SBayesR.PseudoVal | 0.101 | 0.072 | 0.028 | 2.65e-11 | 0.093 | 0.065 | 0.028 | 8.83e-04 |
| Intelligence | LDpred1.Inf | SBayesR.PseudoVal | 0.100 | 0.072 | 0.027 | 1.56e-10 | 0.092 | 0.065 | 0.027 | 1.78e-03 |
| Intelligence | PRScs.MultiPRS | SBayesR.PseudoVal | 0.100 | 0.072 | 0.028 | 7.59e-05 | 0.093 | 0.065 | 0.028 | 4.86e-04 |
| Intelligence | LDpred2.10FCVal | SBLUP.Inf | 0.103 | 0.100 | 0.003 | 3.38e-02 | 0.093 | 0.092 | 0.001 | 7.92e-01 |
| Intelligence | PRScs.PseudoVal | SBLUP.Inf | 0.099 | 0.100 | -0.002 | 2.73e-01 | 0.092 | 0.092 | 0.000 | 9.81e-01 |
| Intelligence | LDpred1.10FCVal | SBLUP.Inf | 0.102 | 0.100 | 0.001 | 2.40e-01 | 0.092 | 0.092 | 0.000 | 9.10e-01 |
| Intelligence | LDpred2.Inf | SBLUP.Inf | 0.101 | 0.100 | 0.000 | 6.15e-01 | 0.093 | 0.092 | 0.000 | 5.80e-01 |
| Intelligence | SBayesR.PseudoVal | SBLUP.Inf | 0.072 | 0.100 | -0.028 | 3.63e-11 | 0.065 | 0.092 | -0.028 | 1.05e-03 |
| Intelligence | lassosum.10FCVal | SBLUP.Inf | 0.099 | 0.100 | -0.001 | 4.26e-01 | 0.093 | 0.092 | 0.001 | 8.23e-01 |
| Intelligence | pT+clump.MultiPRS | SBLUP.Inf | 0.086 | 0.100 | -0.014 | 4.79e-02 | 0.080 | 0.092 | -0.013 | 3.51e-02 |
| Intelligence | lassosum.MultiPRS | SBLUP.Inf | 0.102 | 0.100 | 0.002 | 8.04e-01 | 0.097 | 0.092 | 0.004 | 1.66e-01 |
| Intelligence | PRScs.10FCVal | SBLUP.Inf | 0.099 | 0.100 | -0.001 | 6.00e-01 | 0.089 | 0.092 | -0.003 | 3.81e-01 |
| Intelligence | PRScs.MultiPRS | SBLUP.Inf | 0.100 | 0.100 | 0.000 | 9.96e-01 | 0.093 | 0.092 | 0.001 | 8.46e-01 |
| Intelligence | LDpred1.MultiPRS | SBLUP.Inf | 0.101 | 0.100 | 0.001 | 8.99e-01 | 0.093 | 0.092 | 0.001 | 7.78e-01 |
| Intelligence | LDpred2.PseudoVal | SBLUP.Inf | 0.100 | 0.100 | 0.000 | 9.74e-01 | 0.093 | 0.092 | 0.000 | 8.88e-01 |
| Intelligence | LDpred1.Inf | SBLUP.Inf | 0.100 | 0.100 | -0.001 | 5.31e-01 | 0.092 | 0.092 | -0.001 | 5.93e-01 |
| Intelligence | LDpred2.MultiPRS | SBLUP.Inf | 0.103 | 0.100 | 0.002 | 7.62e-01 | 0.094 | 0.092 | 0.002 | 5.15e-01 |
| Intelligence | All.MultiPRS | SBLUP.Inf | 0.102 | 0.100 | 0.002 | 7.70e-01 | 0.097 | 0.092 | 0.005 | 1.09e-01 |
| Intelligence | SBLUP.Inf | SBLUP.Inf | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.092 | 0.092 | 0.000 | 1.00e+00 |
| Intelligence | DBSLMM.PseudoVal | SBLUP.Inf | 0.089 | 0.100 | -0.012 | 2.24e-06 | 0.079 | 0.092 | -0.013 | 8.58e-03 |
| Intelligence | pT+clump.10FCVal | SBLUP.Inf | 0.084 | 0.100 | -0.017 | 7.89e-08 | 0.074 | 0.092 | -0.018 | 2.16e-03 |
| Intelligence | lassosum.PseudoVal | SBLUP.Inf | 0.064 | 0.100 | -0.037 | 1.05e-13 | 0.059 | 0.092 | -0.034 | 5.20e-04 |
| Height | LDpred2.PseudoVal | All.MultiPRS | 0.304 | 0.358 | -0.054 | 3.80e-18 | 0.304 | 0.361 | -0.057 | 2.45e-27 |
| Height | pT+clump.10FCVal | All.MultiPRS | 0.304 | 0.358 | -0.054 | 5.46e-18 | 0.299 | 0.361 | -0.062 | 2.85e-32 |
| Height | LDpred2.MultiPRS | All.MultiPRS | 0.350 | 0.358 | -0.008 | 1.85e-01 | 0.354 | 0.361 | -0.007 | 1.45e-03 |
| Height | PRScs.PseudoVal | All.MultiPRS | 0.346 | 0.358 | -0.012 | 5.07e-02 | 0.344 | 0.361 | -0.017 | 3.90e-11 |
| Height | LDpred1.Inf | All.MultiPRS | 0.299 | 0.358 | -0.059 | 1.50e-21 | 0.303 | 0.361 | -0.057 | 9.27e-26 |
| Height | PRScs.MultiPRS | All.MultiPRS | 0.347 | 0.358 | -0.011 | 1.64e-19 | 0.345 | 0.361 | -0.015 | 1.33e-10 |
| Height | LDpred2.Inf | All.MultiPRS | 0.305 | 0.358 | -0.053 | 1.00e-17 | 0.305 | 0.361 | -0.056 | 4.58e-27 |
| Height | lassosum.PseudoVal | All.MultiPRS | 0.285 | 0.358 | -0.074 | 1.21e-31 | 0.281 | 0.361 | -0.080 | 7.75e-40 |
| Height | LDpred2.10FCVal | All.MultiPRS | 0.343 | 0.358 | -0.015 | 1.15e-02 | 0.346 | 0.361 | -0.015 | 2.57e-07 |
| Height | lassosum.MultiPRS | All.MultiPRS | 0.350 | 0.358 | -0.008 | 2.22e-01 | 0.348 | 0.361 | -0.012 | 2.46e-09 |
| Height | LDpred1.10FCVal | All.MultiPRS | 0.300 | 0.358 | -0.058 | 8.28e-21 | 0.301 | 0.361 | -0.060 | 5.55e-28 |
| Height | All.MultiPRS | All.MultiPRS | 0.358 | 0.358 | 0.000 | 1.00e+00 | 0.361 | 0.361 | 0.000 | 1.00e+00 |
| Height | SBayesR.PseudoVal | All.MultiPRS | 0.344 | 0.358 | -0.014 | 2.11e-02 | 0.343 | 0.361 | -0.017 | 2.98e-10 |
| Height | LDpred1.MultiPRS | All.MultiPRS | 0.303 | 0.358 | -0.055 | 1.04e-17 | 0.307 | 0.361 | -0.054 | 9.75e-25 |
| Height | pT+clump.MultiPRS | All.MultiPRS | 0.320 | 0.358 | -0.038 | 2.44e-09 | 0.313 | 0.361 | -0.048 | 2.13e-27 |
| Height | lassosum.10FCVal | All.MultiPRS | 0.347 | 0.358 | -0.011 | 7.97e-02 | 0.346 | 0.361 | -0.015 | 3.22e-10 |
| Height | PRScs.10FCVal | All.MultiPRS | 0.345 | 0.358 | -0.013 | 3.43e-02 | 0.344 | 0.361 | -0.017 | 4.24e-11 |
| Height | DBSLMM.PseudoVal | All.MultiPRS | 0.332 | 0.358 | -0.026 | 3.00e-05 | 0.327 | 0.361 | -0.033 | 5.35e-20 |
| Height | SBLUP.Inf | All.MultiPRS | 0.293 | 0.358 | -0.065 | 4.37e-25 | 0.294 | 0.361 | -0.067 | 1.41e-32 |
| Height | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.344 | 0.332 | 0.012 | 2.45e-06 | 0.343 | 0.327 | 0.016 | 1.18e-03 |
| Height | LDpred2.Inf | DBSLMM.PseudoVal | 0.305 | 0.332 | -0.028 | 9.47e-46 | 0.305 | 0.327 | -0.023 | 5.07e-09 |
| Height | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.304 | 0.332 | -0.028 | 2.05e-18 | 0.299 | 0.327 | -0.029 | 9.51e-06 |
| Height | PRScs.10FCVal | DBSLMM.PseudoVal | 0.345 | 0.332 | 0.013 | 3.97e-13 | 0.344 | 0.327 | 0.016 | 2.79e-06 |
| Height | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.346 | 0.332 | 0.014 | 1.74e-20 | 0.344 | 0.327 | 0.016 | 3.34e-08 |
| Height | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.320 | 0.332 | -0.013 | 4.90e-02 | 0.313 | 0.327 | -0.015 | 2.75e-03 |
| Height | LDpred1.Inf | DBSLMM.PseudoVal | 0.299 | 0.332 | -0.034 | 3.49e-70 | 0.303 | 0.327 | -0.024 | 4.78e-10 |
| Height | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.350 | 0.332 | 0.018 | 5.18e-03 | 0.348 | 0.327 | 0.021 | 4.97e-09 |
| Height | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.343 | 0.332 | 0.010 | 8.04e-10 | 0.346 | 0.327 | 0.019 | 2.03e-08 |
| Height | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.285 | 0.332 | -0.048 | 1.40e-36 | 0.281 | 0.327 | -0.046 | 1.21e-09 |
| Height | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.300 | 0.332 | -0.033 | 8.02e-57 | 0.301 | 0.327 | -0.026 | 1.81e-10 |
| Height | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.303 | 0.332 | -0.030 | 5.90e-06 | 0.307 | 0.327 | -0.021 | 2.55e-08 |
| Height | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.347 | 0.332 | 0.015 | 1.64e-02 | 0.345 | 0.327 | 0.018 | 1.40e-08 |
| Height | All.MultiPRS | DBSLMM.PseudoVal | 0.358 | 0.332 | 0.026 | 3.00e-05 | 0.361 | 0.327 | 0.033 | 5.35e-20 |
| Height | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.304 | 0.332 | -0.028 | 2.85e-47 | 0.304 | 0.327 | -0.024 | 2.29e-09 |
| Height | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.332 | 0.332 | 0.000 | 1.00e+00 | 0.327 | 0.327 | 0.000 | 1.00e+00 |
| Height | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.350 | 0.332 | 0.017 | 7.34e-03 | 0.354 | 0.327 | 0.026 | 2.99e-12 |
| Height | lassosum.10FCVal | DBSLMM.PseudoVal | 0.347 | 0.332 | 0.015 | 4.76e-14 | 0.346 | 0.327 | 0.018 | 4.86e-06 |
| Height | SBLUP.Inf | DBSLMM.PseudoVal | 0.293 | 0.332 | -0.039 | 9.41e-91 | 0.294 | 0.327 | -0.034 | 1.65e-18 |
| Height | SBayesR.PseudoVal | lassosum.MultiPRS | 0.344 | 0.350 | -0.006 | 3.17e-01 | 0.343 | 0.348 | -0.005 | 1.60e-01 |
| Height | LDpred2.10FCVal | lassosum.MultiPRS | 0.343 | 0.350 | -0.008 | 2.27e-01 | 0.346 | 0.348 | -0.002 | 4.76e-01 |
| Height | LDpred1.10FCVal | lassosum.MultiPRS | 0.300 | 0.350 | -0.051 | 5.85e-15 | 0.301 | 0.348 | -0.047 | 1.35e-18 |
| Height | LDpred2.Inf | lassosum.MultiPRS | 0.305 | 0.350 | -0.046 | 2.00e-12 | 0.305 | 0.348 | -0.044 | 2.03e-16 |
| Height | PRScs.10FCVal | lassosum.MultiPRS | 0.345 | 0.350 | -0.005 | 4.13e-01 | 0.344 | 0.348 | -0.005 | 5.36e-02 |
| Height | PRScs.MultiPRS | lassosum.MultiPRS | 0.347 | 0.350 | -0.003 | 6.20e-01 | 0.345 | 0.348 | -0.003 | 1.57e-01 |
| Height | pT+clump.MultiPRS | lassosum.MultiPRS | 0.320 | 0.350 | -0.031 | 1.90e-06 | 0.313 | 0.348 | -0.036 | 6.06e-16 |
| Height | lassosum.MultiPRS | lassosum.MultiPRS | 0.350 | 0.350 | 0.000 | 1.00e+00 | 0.348 | 0.348 | 0.000 | 1.00e+00 |
| Height | lassosum.10FCVal | lassosum.MultiPRS | 0.347 | 0.350 | -0.003 | 6.40e-01 | 0.346 | 0.348 | -0.003 | 5.59e-02 |
| Height | LDpred2.MultiPRS | lassosum.MultiPRS | 0.350 | 0.350 | -0.001 | 9.09e-01 | 0.354 | 0.348 | 0.005 | 6.88e-02 |
| Height | LDpred1.MultiPRS | lassosum.MultiPRS | 0.303 | 0.350 | -0.047 | 2.64e-13 | 0.307 | 0.348 | -0.042 | 7.22e-16 |
| Height | LDpred2.PseudoVal | lassosum.MultiPRS | 0.304 | 0.350 | -0.046 | 9.16e-13 | 0.304 | 0.348 | -0.045 | 1.49e-16 |
| Height | LDpred1.Inf | lassosum.MultiPRS | 0.299 | 0.350 | -0.052 | 1.69e-15 | 0.303 | 0.348 | -0.045 | 5.14e-17 |
| Height | pT+clump.10FCVal | lassosum.MultiPRS | 0.304 | 0.350 | -0.046 | 9.87e-13 | 0.299 | 0.348 | -0.050 | 8.33e-20 |
| Height | lassosum.PseudoVal | lassosum.MultiPRS | 0.285 | 0.350 | -0.066 | 2.70e-24 | 0.281 | 0.348 | -0.067 | 4.70e-31 |
| Height | SBLUP.Inf | lassosum.MultiPRS | 0.293 | 0.350 | -0.057 | 2.04e-18 | 0.294 | 0.348 | -0.055 | 2.87e-23 |
| Height | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.332 | 0.350 | -0.018 | 5.18e-03 | 0.327 | 0.348 | -0.021 | 4.97e-09 |
| Height | All.MultiPRS | lassosum.MultiPRS | 0.358 | 0.350 | 0.008 | 2.22e-01 | 0.361 | 0.348 | 0.012 | 2.46e-09 |
| Height | PRScs.PseudoVal | lassosum.MultiPRS | 0.346 | 0.350 | -0.004 | 5.09e-01 | 0.344 | 0.348 | -0.005 | 5.46e-02 |
| Height | LDpred1.Inf | lassosum.10FCVal | 0.299 | 0.347 | -0.049 | 4.71e-70 | 0.303 | 0.346 | -0.042 | 4.21e-14 |
| Height | PRScs.10FCVal | lassosum.10FCVal | 0.345 | 0.347 | -0.002 | 1.04e-01 | 0.344 | 0.346 | -0.002 | 5.20e-01 |
| Height | LDpred2.MultiPRS | lassosum.10FCVal | 0.350 | 0.347 | 0.002 | 7.25e-01 | 0.354 | 0.346 | 0.008 | 8.36e-03 |
| Height | lassosum.10FCVal | lassosum.10FCVal | 0.347 | 0.347 | 0.000 | 1.00e+00 | 0.346 | 0.346 | 0.000 | 1.00e+00 |
| Height | PRScs.PseudoVal | lassosum.10FCVal | 0.346 | 0.347 | -0.001 | 3.69e-01 | 0.344 | 0.346 | -0.002 | 5.02e-01 |
| Height | DBSLMM.PseudoVal | lassosum.10FCVal | 0.332 | 0.347 | -0.015 | 4.76e-14 | 0.327 | 0.346 | -0.018 | 4.86e-06 |
| Height | SBLUP.Inf | lassosum.10FCVal | 0.293 | 0.347 | -0.054 | 1.09e-80 | 0.294 | 0.346 | -0.052 | 1.12e-19 |
| Height | LDpred2.Inf | lassosum.10FCVal | 0.305 | 0.347 | -0.043 | 3.03e-54 | 0.305 | 0.346 | -0.041 | 1.71e-13 |
| Height | pT+clump.10FCVal | lassosum.10FCVal | 0.304 | 0.347 | -0.043 | 7.92e-53 | 0.299 | 0.346 | -0.047 | 1.39e-16 |
| Height | LDpred2.10FCVal | lassosum.10FCVal | 0.343 | 0.347 | -0.005 | 6.91e-03 | 0.346 | 0.346 | 0.000 | 9.23e-01 |
| Height | lassosum.MultiPRS | lassosum.10FCVal | 0.350 | 0.347 | 0.003 | 6.40e-01 | 0.348 | 0.346 | 0.003 | 5.59e-02 |
| Height | LDpred1.10FCVal | lassosum.10FCVal | 0.300 | 0.347 | -0.048 | 3.10e-67 | 0.301 | 0.346 | -0.045 | 1.12e-15 |
| Height | All.MultiPRS | lassosum.10FCVal | 0.358 | 0.347 | 0.011 | 7.97e-02 | 0.361 | 0.346 | 0.015 | 3.22e-10 |
| Height | SBayesR.PseudoVal | lassosum.10FCVal | 0.344 | 0.347 | -0.003 | 8.62e-02 | 0.343 | 0.346 | -0.002 | 5.64e-01 |
| Height | lassosum.PseudoVal | lassosum.10FCVal | 0.285 | 0.347 | -0.063 | 2.48e-99 | 0.281 | 0.346 | -0.065 | 1.96e-27 |
| Height | pT+clump.MultiPRS | lassosum.10FCVal | 0.320 | 0.347 | -0.028 | 1.79e-05 | 0.313 | 0.346 | -0.033 | 2.93e-13 |
| Height | LDpred2.PseudoVal | lassosum.10FCVal | 0.304 | 0.347 | -0.043 | 6.26e-55 | 0.304 | 0.346 | -0.042 | 1.17e-13 |
| Height | LDpred1.MultiPRS | lassosum.10FCVal | 0.303 | 0.347 | -0.044 | 7.58e-12 | 0.307 | 0.346 | -0.039 | 4.45e-13 |
| Height | PRScs.MultiPRS | lassosum.10FCVal | 0.347 | 0.347 | 0.000 | 9.78e-01 | 0.345 | 0.346 | 0.000 | 8.93e-01 |
| Height | lassosum.PseudoVal | lassosum.PseudoVal | 0.285 | 0.285 | 0.000 | 1.00e+00 | 0.281 | 0.281 | 0.000 | 1.00e+00 |
| Height | pT+clump.MultiPRS | lassosum.PseudoVal | 0.320 | 0.285 | 0.035 | 8.62e-08 | 0.313 | 0.281 | 0.032 | 6.86e-06 |
| Height | LDpred2.Inf | lassosum.PseudoVal | 0.305 | 0.285 | 0.020 | 8.20e-06 | 0.305 | 0.281 | 0.023 | 1.02e-02 |
| Height | SBayesR.PseudoVal | lassosum.PseudoVal | 0.344 | 0.285 | 0.059 | 1.17e-107 | 0.343 | 0.281 | 0.062 | 7.00e-31 |
| Height | PRScs.MultiPRS | lassosum.PseudoVal | 0.347 | 0.285 | 0.063 | 2.38e-23 | 0.345 | 0.281 | 0.064 | 1.13e-28 |
| Height | SBLUP.Inf | lassosum.PseudoVal | 0.293 | 0.285 | 0.009 | 5.90e-02 | 0.294 | 0.281 | 0.013 | 1.85e-01 |
| Height | lassosum.MultiPRS | lassosum.PseudoVal | 0.350 | 0.285 | 0.066 | 2.70e-24 | 0.348 | 0.281 | 0.067 | 4.70e-31 |
| Height | PRScs.10FCVal | lassosum.PseudoVal | 0.345 | 0.285 | 0.061 | 1.45e-106 | 0.344 | 0.281 | 0.063 | 1.20e-29 |
| Height | LDpred2.MultiPRS | lassosum.PseudoVal | 0.350 | 0.285 | 0.065 | 2.11e-23 | 0.354 | 0.281 | 0.073 | 8.00e-31 |
| Height | PRScs.PseudoVal | lassosum.PseudoVal | 0.346 | 0.285 | 0.062 | 6.18e-88 | 0.344 | 0.281 | 0.063 | 6.85e-24 |
| Height | LDpred2.PseudoVal | lassosum.PseudoVal | 0.304 | 0.285 | 0.020 | 1.89e-05 | 0.304 | 0.281 | 0.023 | 1.32e-02 |
| Height | LDpred1.MultiPRS | lassosum.PseudoVal | 0.303 | 0.285 | 0.018 | 5.30e-03 | 0.307 | 0.281 | 0.026 | 4.43e-03 |
| Height | pT+clump.10FCVal | lassosum.PseudoVal | 0.304 | 0.285 | 0.020 | 1.89e-08 | 0.299 | 0.281 | 0.018 | 1.14e-02 |
| Height | All.MultiPRS | lassosum.PseudoVal | 0.358 | 0.285 | 0.074 | 1.21e-31 | 0.361 | 0.281 | 0.080 | 7.75e-40 |
| Height | LDpred1.Inf | lassosum.PseudoVal | 0.299 | 0.285 | 0.014 | 2.14e-03 | 0.303 | 0.281 | 0.022 | 1.49e-02 |
| Height | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.332 | 0.285 | 0.048 | 1.40e-36 | 0.327 | 0.281 | 0.046 | 1.21e-09 |
| Height | LDpred1.10FCVal | lassosum.PseudoVal | 0.300 | 0.285 | 0.015 | 7.59e-04 | 0.301 | 0.281 | 0.020 | 2.68e-02 |
| Height | lassosum.10FCVal | lassosum.PseudoVal | 0.347 | 0.285 | 0.063 | 2.48e-99 | 0.346 | 0.281 | 0.065 | 1.96e-27 |
| Height | LDpred2.10FCVal | lassosum.PseudoVal | 0.343 | 0.285 | 0.058 | 1.10e-57 | 0.346 | 0.281 | 0.065 | 4.47e-19 |
| Height | LDpred2.10FCVal | LDpred1.MultiPRS | 0.343 | 0.303 | 0.040 | 9.80e-10 | 0.346 | 0.307 | 0.039 | 1.00e-25 |
| Height | PRScs.PseudoVal | LDpred1.MultiPRS | 0.346 | 0.303 | 0.043 | 2.77e-11 | 0.344 | 0.307 | 0.037 | 1.52e-17 |
| Height | LDpred2.Inf | LDpred1.MultiPRS | 0.305 | 0.303 | 0.002 | 7.79e-01 | 0.305 | 0.307 | -0.002 | 3.84e-01 |
| Height | pT+clump.10FCVal | LDpred1.MultiPRS | 0.304 | 0.303 | 0.001 | 8.33e-01 | 0.299 | 0.307 | -0.008 | 3.07e-01 |
| Height | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.344 | 0.303 | 0.041 | 2.48e-10 | 0.343 | 0.307 | 0.037 | 3.49e-08 |
| Height | All.MultiPRS | LDpred1.MultiPRS | 0.358 | 0.303 | 0.055 | 1.04e-17 | 0.361 | 0.307 | 0.054 | 9.75e-25 |
| Height | LDpred1.10FCVal | LDpred1.MultiPRS | 0.300 | 0.303 | -0.003 | 6.24e-01 | 0.301 | 0.307 | -0.006 | 1.77e-04 |
| Height | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.303 | 0.303 | 0.000 | 1.00e+00 | 0.307 | 0.307 | 0.000 | 1.00e+00 |
| Height | PRScs.10FCVal | LDpred1.MultiPRS | 0.345 | 0.303 | 0.042 | 7.93e-11 | 0.344 | 0.307 | 0.037 | 2.87e-13 |
| Height | lassosum.10FCVal | LDpred1.MultiPRS | 0.347 | 0.303 | 0.044 | 7.58e-12 | 0.346 | 0.307 | 0.039 | 4.45e-13 |
| Height | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.320 | 0.303 | 0.017 | 1.05e-02 | 0.313 | 0.307 | 0.006 | 3.22e-01 |
| Height | LDpred1.Inf | LDpred1.MultiPRS | 0.299 | 0.303 | -0.004 | 5.14e-01 | 0.303 | 0.307 | -0.003 | 5.54e-02 |
| Height | lassosum.MultiPRS | LDpred1.MultiPRS | 0.350 | 0.303 | 0.047 | 2.64e-13 | 0.348 | 0.307 | 0.042 | 7.22e-16 |
| Height | PRScs.MultiPRS | LDpred1.MultiPRS | 0.347 | 0.303 | 0.044 | 7.02e-12 | 0.345 | 0.307 | 0.039 | 1.96e-16 |
| Height | lassosum.PseudoVal | LDpred1.MultiPRS | 0.285 | 0.303 | -0.018 | 5.30e-03 | 0.281 | 0.307 | -0.026 | 4.43e-03 |
| Height | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.304 | 0.303 | 0.001 | 8.63e-01 | 0.304 | 0.307 | -0.003 | 2.62e-01 |
| Height | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.332 | 0.303 | 0.030 | 5.90e-06 | 0.327 | 0.307 | 0.021 | 2.55e-08 |
| Height | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.350 | 0.303 | 0.047 | 5.77e-13 | 0.354 | 0.307 | 0.047 | 3.26e-22 |
| Height | SBLUP.Inf | LDpred1.MultiPRS | 0.293 | 0.303 | -0.009 | 1.49e-01 | 0.294 | 0.307 | -0.013 | 1.97e-09 |
| Height | All.MultiPRS | LDpred1.Inf | 0.358 | 0.299 | 0.059 | 1.50e-21 | 0.361 | 0.303 | 0.057 | 9.27e-26 |
| Height | LDpred2.PseudoVal | LDpred1.Inf | 0.304 | 0.299 | 0.005 | 2.72e-06 | 0.304 | 0.303 | 0.000 | 8.80e-01 |
| Height | SBayesR.PseudoVal | LDpred1.Inf | 0.344 | 0.299 | 0.045 | 1.85e-40 | 0.343 | 0.303 | 0.040 | 5.63e-09 |
| Height | LDpred2.MultiPRS | LDpred1.Inf | 0.350 | 0.299 | 0.051 | 3.07e-15 | 0.354 | 0.303 | 0.050 | 2.82e-23 |
| Height | PRScs.PseudoVal | LDpred1.Inf | 0.346 | 0.299 | 0.048 | 1.85e-100 | 0.344 | 0.303 | 0.040 | 3.43e-19 |
| Height | LDpred1.Inf | LDpred1.Inf | 0.299 | 0.299 | 0.000 | 1.00e+00 | 0.303 | 0.303 | 0.000 | 1.00e+00 |
| Height | pT+clump.10FCVal | LDpred1.Inf | 0.304 | 0.299 | 0.006 | 1.48e-01 | 0.299 | 0.303 | -0.005 | 5.56e-01 |
| Height | PRScs.MultiPRS | LDpred1.Inf | 0.347 | 0.299 | 0.049 | 7.23e-15 | 0.345 | 0.303 | 0.042 | 7.00e-18 |
| Height | lassosum.10FCVal | LDpred1.Inf | 0.347 | 0.299 | 0.049 | 4.71e-70 | 0.346 | 0.303 | 0.042 | 4.21e-14 |
| Height | pT+clump.MultiPRS | LDpred1.Inf | 0.320 | 0.299 | 0.021 | 1.30e-03 | 0.313 | 0.303 | 0.009 | 1.30e-01 |
| Height | LDpred1.MultiPRS | LDpred1.Inf | 0.303 | 0.299 | 0.004 | 5.14e-01 | 0.307 | 0.303 | 0.003 | 5.54e-02 |
| Height | PRScs.10FCVal | LDpred1.Inf | 0.345 | 0.299 | 0.047 | 6.23e-71 | 0.344 | 0.303 | 0.040 | 1.44e-14 |
| Height | LDpred2.Inf | LDpred1.Inf | 0.305 | 0.299 | 0.006 | 6.28e-08 | 0.305 | 0.303 | 0.001 | 6.51e-01 |
| Height | SBLUP.Inf | LDpred1.Inf | 0.293 | 0.299 | -0.005 | 3.72e-07 | 0.294 | 0.303 | -0.010 | 9.68e-07 |
| Height | DBSLMM.PseudoVal | LDpred1.Inf | 0.332 | 0.299 | 0.034 | 3.49e-70 | 0.327 | 0.303 | 0.024 | 4.78e-10 |
| Height | lassosum.MultiPRS | LDpred1.Inf | 0.350 | 0.299 | 0.052 | 1.69e-15 | 0.348 | 0.303 | 0.045 | 5.14e-17 |
| Height | LDpred1.10FCVal | LDpred1.Inf | 0.300 | 0.299 | 0.001 | 4.80e-01 | 0.301 | 0.303 | -0.002 | 4.29e-01 |
| Height | lassosum.PseudoVal | LDpred1.Inf | 0.285 | 0.299 | -0.014 | 2.14e-03 | 0.281 | 0.303 | -0.022 | 1.49e-02 |
| Height | LDpred2.10FCVal | LDpred1.Inf | 0.343 | 0.299 | 0.044 | 3.56e-114 | 0.346 | 0.303 | 0.042 | 7.90e-28 |
| Height | LDpred1.Inf | LDpred1.10FCVal | 0.299 | 0.300 | -0.001 | 4.80e-01 | 0.303 | 0.301 | 0.002 | 4.29e-01 |
| Height | PRScs.MultiPRS | LDpred1.10FCVal | 0.347 | 0.300 | 0.048 | 2.94e-14 | 0.345 | 0.301 | 0.044 | 4.74e-19 |
| Height | LDpred1.10FCVal | LDpred1.10FCVal | 0.300 | 0.300 | 0.000 | 1.00e+00 | 0.301 | 0.301 | 0.000 | 1.00e+00 |
| Height | All.MultiPRS | LDpred1.10FCVal | 0.358 | 0.300 | 0.058 | 8.28e-21 | 0.361 | 0.301 | 0.060 | 5.55e-28 |
| Height | SBayesR.PseudoVal | LDpred1.10FCVal | 0.344 | 0.300 | 0.044 | 1.25e-39 | 0.343 | 0.301 | 0.042 | 3.56e-10 |
| Height | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.332 | 0.300 | 0.033 | 8.02e-57 | 0.327 | 0.301 | 0.026 | 1.81e-10 |
| Height | pT+clump.MultiPRS | LDpred1.10FCVal | 0.320 | 0.300 | 0.020 | 2.27e-03 | 0.313 | 0.301 | 0.012 | 6.28e-02 |
| Height | lassosum.MultiPRS | LDpred1.10FCVal | 0.350 | 0.300 | 0.051 | 5.85e-15 | 0.348 | 0.301 | 0.047 | 1.35e-18 |
| Height | PRScs.10FCVal | LDpred1.10FCVal | 0.345 | 0.300 | 0.045 | 5.11e-66 | 0.344 | 0.301 | 0.043 | 8.49e-16 |
| Height | lassosum.PseudoVal | LDpred1.10FCVal | 0.285 | 0.300 | -0.015 | 7.59e-04 | 0.281 | 0.301 | -0.020 | 2.68e-02 |
| Height | SBLUP.Inf | LDpred1.10FCVal | 0.293 | 0.300 | -0.006 | 4.58e-05 | 0.294 | 0.301 | -0.007 | 1.51e-02 |
| Height | LDpred2.PseudoVal | LDpred1.10FCVal | 0.304 | 0.300 | 0.004 | 1.21e-02 | 0.304 | 0.301 | 0.003 | 4.30e-01 |
| Height | pT+clump.10FCVal | LDpred1.10FCVal | 0.304 | 0.300 | 0.005 | 2.42e-01 | 0.299 | 0.301 | -0.002 | 7.79e-01 |
| Height | LDpred2.MultiPRS | LDpred1.10FCVal | 0.350 | 0.300 | 0.050 | 1.12e-14 | 0.354 | 0.301 | 0.053 | 3.39e-25 |
| Height | PRScs.PseudoVal | LDpred1.10FCVal | 0.346 | 0.300 | 0.046 | 1.46e-89 | 0.344 | 0.301 | 0.043 | 5.03e-20 |
| Height | LDpred2.10FCVal | LDpred1.10FCVal | 0.343 | 0.300 | 0.043 | 7.38e-96 | 0.346 | 0.301 | 0.045 | 3.02e-27 |
| Height | LDpred1.MultiPRS | LDpred1.10FCVal | 0.303 | 0.300 | 0.003 | 6.24e-01 | 0.307 | 0.301 | 0.006 | 1.77e-04 |
| Height | LDpred2.Inf | LDpred1.10FCVal | 0.305 | 0.300 | 0.005 | 3.51e-03 | 0.305 | 0.301 | 0.003 | 3.26e-01 |
| Height | lassosum.10FCVal | LDpred1.10FCVal | 0.347 | 0.300 | 0.048 | 3.10e-67 | 0.346 | 0.301 | 0.045 | 1.12e-15 |
| Height | LDpred1.Inf | LDpred2.MultiPRS | 0.299 | 0.350 | -0.051 | 3.07e-15 | 0.303 | 0.354 | -0.050 | 2.82e-23 |
| Height | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.332 | 0.350 | -0.017 | 7.34e-03 | 0.327 | 0.354 | -0.026 | 2.99e-12 |
| Height | SBLUP.Inf | LDpred2.MultiPRS | 0.293 | 0.350 | -0.056 | 4.21e-18 | 0.294 | 0.354 | -0.060 | 1.54e-30 |
| Height | LDpred2.Inf | LDpred2.MultiPRS | 0.305 | 0.350 | -0.045 | 3.82e-12 | 0.305 | 0.354 | -0.049 | 3.11e-25 |
| Height | pT+clump.10FCVal | LDpred2.MultiPRS | 0.304 | 0.350 | -0.045 | 2.80e-12 | 0.299 | 0.354 | -0.055 | 5.22e-20 |
| Height | LDpred2.10FCVal | LDpred2.MultiPRS | 0.343 | 0.350 | -0.007 | 2.74e-01 | 0.346 | 0.354 | -0.008 | 1.99e-05 |
| Height | PRScs.PseudoVal | LDpred2.MultiPRS | 0.346 | 0.350 | -0.003 | 5.86e-01 | 0.344 | 0.354 | -0.010 | 1.95e-04 |
| Height | LDpred1.10FCVal | LDpred2.MultiPRS | 0.300 | 0.350 | -0.050 | 1.12e-14 | 0.301 | 0.354 | -0.053 | 3.39e-25 |
| Height | All.MultiPRS | LDpred2.MultiPRS | 0.358 | 0.350 | 0.008 | 1.85e-01 | 0.361 | 0.354 | 0.007 | 1.45e-03 |
| Height | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.344 | 0.350 | -0.006 | 3.79e-01 | 0.343 | 0.354 | -0.010 | 5.78e-03 |
| Height | lassosum.PseudoVal | LDpred2.MultiPRS | 0.285 | 0.350 | -0.065 | 2.11e-23 | 0.281 | 0.354 | -0.073 | 8.00e-31 |
| Height | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.320 | 0.350 | -0.030 | 3.28e-06 | 0.313 | 0.354 | -0.041 | 2.79e-17 |
| Height | lassosum.MultiPRS | LDpred2.MultiPRS | 0.350 | 0.350 | 0.001 | 9.09e-01 | 0.348 | 0.354 | -0.005 | 6.88e-02 |
| Height | PRScs.10FCVal | LDpred2.MultiPRS | 0.345 | 0.350 | -0.004 | 4.83e-01 | 0.344 | 0.354 | -0.010 | 4.16e-04 |
| Height | PRScs.MultiPRS | LDpred2.MultiPRS | 0.347 | 0.350 | -0.002 | 7.06e-01 | 0.345 | 0.354 | -0.008 | 1.11e-03 |
| Height | lassosum.10FCVal | LDpred2.MultiPRS | 0.347 | 0.350 | -0.002 | 7.25e-01 | 0.346 | 0.354 | -0.008 | 8.36e-03 |
| Height | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.304 | 0.350 | -0.046 | 1.77e-12 | 0.304 | 0.354 | -0.050 | 1.98e-25 |
| Height | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.303 | 0.350 | -0.047 | 5.77e-13 | 0.307 | 0.354 | -0.047 | 3.26e-22 |
| Height | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.350 | 0.350 | 0.000 | 1.00e+00 | 0.354 | 0.354 | 0.000 | 1.00e+00 |
| Height | LDpred1.Inf | LDpred2.10FCVal | 0.299 | 0.343 | -0.044 | 3.56e-114 | 0.303 | 0.346 | -0.042 | 7.90e-28 |
| Height | PRScs.MultiPRS | LDpred2.10FCVal | 0.347 | 0.343 | 0.005 | 4.57e-01 | 0.345 | 0.346 | -0.001 | 8.19e-01 |
| Height | LDpred2.MultiPRS | LDpred2.10FCVal | 0.350 | 0.343 | 0.007 | 2.74e-01 | 0.354 | 0.346 | 0.008 | 1.99e-05 |
| Height | PRScs.PseudoVal | LDpred2.10FCVal | 0.346 | 0.343 | 0.004 | 1.41e-02 | 0.344 | 0.346 | -0.002 | 4.43e-01 |
| Height | LDpred2.10FCVal | LDpred2.10FCVal | 0.343 | 0.343 | 0.000 | 1.00e+00 | 0.346 | 0.346 | 0.000 | 1.00e+00 |
| Height | lassosum.MultiPRS | LDpred2.10FCVal | 0.350 | 0.343 | 0.008 | 2.27e-01 | 0.348 | 0.346 | 0.002 | 4.76e-01 |
| Height | LDpred1.10FCVal | LDpred2.10FCVal | 0.300 | 0.343 | -0.043 | 7.38e-96 | 0.301 | 0.346 | -0.045 | 3.02e-27 |
| Height | All.MultiPRS | LDpred2.10FCVal | 0.358 | 0.343 | 0.015 | 1.15e-02 | 0.361 | 0.346 | 0.015 | 2.57e-07 |
| Height | SBayesR.PseudoVal | LDpred2.10FCVal | 0.344 | 0.343 | 0.001 | 5.64e-01 | 0.343 | 0.346 | -0.003 | 5.79e-01 |
| Height | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.332 | 0.343 | -0.010 | 8.04e-10 | 0.327 | 0.346 | -0.019 | 2.03e-08 |
| Height | pT+clump.MultiPRS | LDpred2.10FCVal | 0.320 | 0.343 | -0.023 | 3.75e-04 | 0.313 | 0.346 | -0.033 | 2.63e-11 |
| Height | lassosum.10FCVal | LDpred2.10FCVal | 0.347 | 0.343 | 0.005 | 6.91e-03 | 0.346 | 0.346 | 0.000 | 9.23e-01 |
| Height | PRScs.10FCVal | LDpred2.10FCVal | 0.345 | 0.343 | 0.003 | 1.34e-01 | 0.344 | 0.346 | -0.002 | 5.29e-01 |
| Height | lassosum.PseudoVal | LDpred2.10FCVal | 0.285 | 0.343 | -0.058 | 1.10e-57 | 0.281 | 0.346 | -0.065 | 4.47e-19 |
| Height | SBLUP.Inf | LDpred2.10FCVal | 0.293 | 0.343 | -0.049 | 1.04e-131 | 0.294 | 0.346 | -0.052 | 1.95e-38 |
| Height | LDpred2.PseudoVal | LDpred2.10FCVal | 0.304 | 0.343 | -0.039 | 5.12e-120 | 0.304 | 0.346 | -0.042 | 3.55e-36 |
| Height | pT+clump.10FCVal | LDpred2.10FCVal | 0.304 | 0.343 | -0.038 | 1.51e-32 | 0.299 | 0.346 | -0.047 | 2.55e-13 |
| Height | LDpred2.Inf | LDpred2.10FCVal | 0.305 | 0.343 | -0.038 | 1.87e-116 | 0.305 | 0.346 | -0.041 | 2.73e-35 |
| Height | LDpred1.MultiPRS | LDpred2.10FCVal | 0.303 | 0.343 | -0.040 | 9.80e-10 | 0.307 | 0.346 | -0.039 | 1.00e-25 |
| Height | lassosum.PseudoVal | LDpred2.PseudoVal | 0.285 | 0.304 | -0.020 | 1.89e-05 | 0.281 | 0.304 | -0.023 | 1.32e-02 |
| Height | SBLUP.Inf | LDpred2.PseudoVal | 0.293 | 0.304 | -0.011 | 1.54e-21 | 0.294 | 0.304 | -0.010 | 4.40e-06 |
| Height | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.344 | 0.304 | 0.040 | 5.03e-32 | 0.343 | 0.304 | 0.040 | 7.25e-09 |
| Height | All.MultiPRS | LDpred2.PseudoVal | 0.358 | 0.304 | 0.054 | 3.80e-18 | 0.361 | 0.304 | 0.057 | 2.45e-27 |
| Height | PRScs.PseudoVal | LDpred2.PseudoVal | 0.346 | 0.304 | 0.042 | 1.81e-77 | 0.344 | 0.304 | 0.040 | 1.93e-18 |
| Height | lassosum.MultiPRS | LDpred2.PseudoVal | 0.350 | 0.304 | 0.046 | 9.16e-13 | 0.348 | 0.304 | 0.045 | 1.49e-16 |
| Height | PRScs.MultiPRS | LDpred2.PseudoVal | 0.347 | 0.304 | 0.043 | 4.39e-12 | 0.345 | 0.304 | 0.041 | 2.32e-17 |
| Height | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.350 | 0.304 | 0.046 | 1.77e-12 | 0.354 | 0.304 | 0.050 | 1.98e-25 |
| Height | PRScs.10FCVal | LDpred2.PseudoVal | 0.345 | 0.304 | 0.041 | 7.38e-56 | 0.344 | 0.304 | 0.040 | 2.85e-14 |
| Height | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.304 | 0.304 | 0.000 | 1.00e+00 | 0.304 | 0.304 | 0.000 | 1.00e+00 |
| Height | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.320 | 0.304 | 0.016 | 1.68e-02 | 0.313 | 0.304 | 0.009 | 1.55e-01 |
| Height | pT+clump.10FCVal | LDpred2.PseudoVal | 0.304 | 0.304 | 0.000 | 9.50e-01 | 0.299 | 0.304 | -0.005 | 5.30e-01 |
| Height | lassosum.10FCVal | LDpred2.PseudoVal | 0.347 | 0.304 | 0.043 | 6.26e-55 | 0.346 | 0.304 | 0.042 | 1.17e-13 |
| Height | LDpred1.Inf | LDpred2.PseudoVal | 0.299 | 0.304 | -0.005 | 2.72e-06 | 0.303 | 0.304 | 0.000 | 8.80e-01 |
| Height | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.303 | 0.304 | -0.001 | 8.63e-01 | 0.307 | 0.304 | 0.003 | 2.62e-01 |
| Height | LDpred1.10FCVal | LDpred2.PseudoVal | 0.300 | 0.304 | -0.004 | 1.21e-02 | 0.301 | 0.304 | -0.003 | 4.30e-01 |
| Height | LDpred2.Inf | LDpred2.PseudoVal | 0.305 | 0.304 | 0.001 | 7.33e-07 | 0.305 | 0.304 | 0.001 | 1.95e-02 |
| Height | LDpred2.10FCVal | LDpred2.PseudoVal | 0.343 | 0.304 | 0.039 | 5.12e-120 | 0.346 | 0.304 | 0.042 | 3.55e-36 |
| Height | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.332 | 0.304 | 0.028 | 2.85e-47 | 0.327 | 0.304 | 0.024 | 2.29e-09 |
| Height | pT+clump.MultiPRS | LDpred2.Inf | 0.320 | 0.305 | 0.015 | 2.24e-02 | 0.313 | 0.305 | 0.008 | 1.83e-01 |
| Height | lassosum.MultiPRS | LDpred2.Inf | 0.350 | 0.305 | 0.046 | 2.00e-12 | 0.348 | 0.305 | 0.044 | 2.03e-16 |
| Height | SBayesR.PseudoVal | LDpred2.Inf | 0.344 | 0.305 | 0.039 | 1.71e-31 | 0.343 | 0.305 | 0.039 | 9.77e-09 |
| Height | All.MultiPRS | LDpred2.Inf | 0.358 | 0.305 | 0.053 | 1.00e-17 | 0.361 | 0.305 | 0.056 | 4.58e-27 |
| Height | SBLUP.Inf | LDpred2.Inf | 0.293 | 0.305 | -0.011 | 2.10e-24 | 0.294 | 0.305 | -0.011 | 9.43e-07 |
| Height | LDpred2.PseudoVal | LDpred2.Inf | 0.304 | 0.305 | -0.001 | 7.33e-07 | 0.304 | 0.305 | -0.001 | 1.95e-02 |
| Height | PRScs.MultiPRS | LDpred2.Inf | 0.347 | 0.305 | 0.042 | 9.55e-12 | 0.345 | 0.305 | 0.041 | 2.21e-17 |
| Height | LDpred2.MultiPRS | LDpred2.Inf | 0.350 | 0.305 | 0.045 | 3.82e-12 | 0.354 | 0.305 | 0.049 | 3.11e-25 |
| Height | PRScs.10FCVal | LDpred2.Inf | 0.345 | 0.305 | 0.040 | 8.18e-56 | 0.344 | 0.305 | 0.039 | 3.04e-14 |
| Height | LDpred1.Inf | LDpred2.Inf | 0.299 | 0.305 | -0.006 | 6.28e-08 | 0.303 | 0.305 | -0.001 | 6.51e-01 |
| Height | LDpred1.MultiPRS | LDpred2.Inf | 0.303 | 0.305 | -0.002 | 7.79e-01 | 0.307 | 0.305 | 0.002 | 3.84e-01 |
| Height | pT+clump.10FCVal | LDpred2.Inf | 0.304 | 0.305 | 0.000 | 9.06e-01 | 0.299 | 0.305 | -0.006 | 4.72e-01 |
| Height | lassosum.10FCVal | LDpred2.Inf | 0.347 | 0.305 | 0.043 | 3.03e-54 | 0.346 | 0.305 | 0.041 | 1.71e-13 |
| Height | PRScs.PseudoVal | LDpred2.Inf | 0.346 | 0.305 | 0.041 | 7.07e-78 | 0.344 | 0.305 | 0.039 | 1.77e-18 |
| Height | DBSLMM.PseudoVal | LDpred2.Inf | 0.332 | 0.305 | 0.028 | 9.47e-46 | 0.327 | 0.305 | 0.023 | 5.07e-09 |
| Height | LDpred1.10FCVal | LDpred2.Inf | 0.300 | 0.305 | -0.005 | 3.51e-03 | 0.301 | 0.305 | -0.003 | 3.26e-01 |
| Height | LDpred2.Inf | LDpred2.Inf | 0.305 | 0.305 | 0.000 | 1.00e+00 | 0.305 | 0.305 | 0.000 | 1.00e+00 |
| Height | lassosum.PseudoVal | LDpred2.Inf | 0.285 | 0.305 | -0.020 | 8.20e-06 | 0.281 | 0.305 | -0.023 | 1.02e-02 |
| Height | LDpred2.10FCVal | LDpred2.Inf | 0.343 | 0.305 | 0.038 | 1.87e-116 | 0.346 | 0.305 | 0.041 | 2.73e-35 |
| Height | PRScs.10FCVal | PRScs.MultiPRS | 0.345 | 0.347 | -0.002 | 7.36e-01 | 0.344 | 0.345 | -0.001 | 1.70e-01 |
| Height | LDpred1.10FCVal | PRScs.MultiPRS | 0.300 | 0.347 | -0.048 | 2.94e-14 | 0.301 | 0.345 | -0.044 | 4.74e-19 |
| Height | lassosum.MultiPRS | PRScs.MultiPRS | 0.350 | 0.347 | 0.003 | 6.20e-01 | 0.348 | 0.345 | 0.003 | 1.57e-01 |
| Height | LDpred2.MultiPRS | PRScs.MultiPRS | 0.350 | 0.347 | 0.002 | 7.06e-01 | 0.354 | 0.345 | 0.008 | 1.11e-03 |
| Height | All.MultiPRS | PRScs.MultiPRS | 0.358 | 0.347 | 0.011 | 1.64e-19 | 0.361 | 0.345 | 0.015 | 1.33e-10 |
| Height | PRScs.PseudoVal | PRScs.MultiPRS | 0.346 | 0.347 | -0.001 | 8.63e-01 | 0.344 | 0.345 | -0.001 | 5.64e-02 |
| Height | LDpred1.MultiPRS | PRScs.MultiPRS | 0.303 | 0.347 | -0.044 | 7.02e-12 | 0.307 | 0.345 | -0.039 | 1.96e-16 |
| Height | pT+clump.MultiPRS | PRScs.MultiPRS | 0.320 | 0.347 | -0.028 | 2.02e-05 | 0.313 | 0.345 | -0.033 | 1.39e-13 |
| Height | lassosum.10FCVal | PRScs.MultiPRS | 0.347 | 0.347 | 0.000 | 9.78e-01 | 0.346 | 0.345 | 0.000 | 8.93e-01 |
| Height | SBayesR.PseudoVal | PRScs.MultiPRS | 0.344 | 0.347 | -0.003 | 6.00e-01 | 0.343 | 0.345 | -0.002 | 5.97e-01 |
| Height | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.332 | 0.347 | -0.015 | 1.64e-02 | 0.327 | 0.345 | -0.018 | 1.40e-08 |
| Height | SBLUP.Inf | PRScs.MultiPRS | 0.293 | 0.347 | -0.054 | 8.25e-18 | 0.294 | 0.345 | -0.052 | 2.04e-24 |
| Height | LDpred2.Inf | PRScs.MultiPRS | 0.305 | 0.347 | -0.042 | 9.55e-12 | 0.305 | 0.345 | -0.041 | 2.21e-17 |
| Height | pT+clump.10FCVal | PRScs.MultiPRS | 0.304 | 0.347 | -0.043 | 6.41e-12 | 0.299 | 0.345 | -0.046 | 1.53e-16 |
| Height | LDpred2.10FCVal | PRScs.MultiPRS | 0.343 | 0.347 | -0.005 | 4.57e-01 | 0.346 | 0.345 | 0.001 | 8.19e-01 |
| Height | lassosum.PseudoVal | PRScs.MultiPRS | 0.285 | 0.347 | -0.063 | 2.38e-23 | 0.281 | 0.345 | -0.064 | 1.13e-28 |
| Height | LDpred2.PseudoVal | PRScs.MultiPRS | 0.304 | 0.347 | -0.043 | 4.39e-12 | 0.304 | 0.345 | -0.041 | 2.32e-17 |
| Height | LDpred1.Inf | PRScs.MultiPRS | 0.299 | 0.347 | -0.049 | 7.23e-15 | 0.303 | 0.345 | -0.042 | 7.00e-18 |
| Height | PRScs.MultiPRS | PRScs.MultiPRS | 0.347 | 0.347 | 0.000 | 1.00e+00 | 0.345 | 0.345 | 0.000 | 1.00e+00 |
| Height | LDpred2.PseudoVal | PRScs.10FCVal | 0.304 | 0.345 | -0.041 | 7.38e-56 | 0.304 | 0.344 | -0.040 | 2.85e-14 |
| Height | PRScs.10FCVal | PRScs.10FCVal | 0.345 | 0.345 | 0.000 | 1.00e+00 | 0.344 | 0.344 | 0.000 | 1.00e+00 |
| Height | LDpred2.MultiPRS | PRScs.10FCVal | 0.350 | 0.345 | 0.004 | 4.83e-01 | 0.354 | 0.344 | 0.010 | 4.16e-04 |
| Height | pT+clump.MultiPRS | PRScs.10FCVal | 0.320 | 0.345 | -0.025 | 7.89e-05 | 0.313 | 0.344 | -0.031 | 3.23e-12 |
| Height | LDpred1.Inf | PRScs.10FCVal | 0.299 | 0.345 | -0.047 | 6.23e-71 | 0.303 | 0.344 | -0.040 | 1.44e-14 |
| Height | LDpred1.MultiPRS | PRScs.10FCVal | 0.303 | 0.345 | -0.042 | 7.93e-11 | 0.307 | 0.344 | -0.037 | 2.87e-13 |
| Height | pT+clump.10FCVal | PRScs.10FCVal | 0.304 | 0.345 | -0.041 | 1.14e-49 | 0.299 | 0.344 | -0.045 | 5.99e-16 |
| Height | lassosum.10FCVal | PRScs.10FCVal | 0.347 | 0.345 | 0.002 | 1.04e-01 | 0.346 | 0.344 | 0.002 | 5.20e-01 |
| Height | PRScs.PseudoVal | PRScs.10FCVal | 0.346 | 0.345 | 0.001 | 2.57e-01 | 0.344 | 0.344 | 0.000 | 9.64e-01 |
| Height | DBSLMM.PseudoVal | PRScs.10FCVal | 0.332 | 0.345 | -0.013 | 3.97e-13 | 0.327 | 0.344 | -0.016 | 2.79e-06 |
| Height | SBLUP.Inf | PRScs.10FCVal | 0.293 | 0.345 | -0.052 | 2.56e-80 | 0.294 | 0.344 | -0.050 | 3.49e-20 |
| Height | LDpred2.Inf | PRScs.10FCVal | 0.305 | 0.345 | -0.040 | 8.18e-56 | 0.305 | 0.344 | -0.039 | 3.04e-14 |
| Height | lassosum.PseudoVal | PRScs.10FCVal | 0.285 | 0.345 | -0.061 | 1.45e-106 | 0.281 | 0.344 | -0.063 | 1.20e-29 |
| Height | LDpred2.10FCVal | PRScs.10FCVal | 0.343 | 0.345 | -0.003 | 1.34e-01 | 0.346 | 0.344 | 0.002 | 5.29e-01 |
| Height | lassosum.MultiPRS | PRScs.10FCVal | 0.350 | 0.345 | 0.005 | 4.13e-01 | 0.348 | 0.344 | 0.005 | 5.36e-02 |
| Height | LDpred1.10FCVal | PRScs.10FCVal | 0.300 | 0.345 | -0.045 | 5.11e-66 | 0.301 | 0.344 | -0.043 | 8.49e-16 |
| Height | All.MultiPRS | PRScs.10FCVal | 0.358 | 0.345 | 0.013 | 3.43e-02 | 0.361 | 0.344 | 0.017 | 4.24e-11 |
| Height | SBayesR.PseudoVal | PRScs.10FCVal | 0.344 | 0.345 | -0.001 | 5.11e-01 | 0.343 | 0.344 | 0.000 | 8.89e-01 |
| Height | PRScs.MultiPRS | PRScs.10FCVal | 0.347 | 0.345 | 0.002 | 7.36e-01 | 0.345 | 0.344 | 0.001 | 1.70e-01 |
| Height | All.MultiPRS | PRScs.PseudoVal | 0.358 | 0.346 | 0.012 | 5.07e-02 | 0.361 | 0.344 | 0.017 | 3.90e-11 |
| Height | pT+clump.MultiPRS | PRScs.PseudoVal | 0.320 | 0.346 | -0.026 | 4.07e-05 | 0.313 | 0.344 | -0.031 | 4.76e-12 |
| Height | LDpred2.PseudoVal | PRScs.PseudoVal | 0.304 | 0.346 | -0.042 | 1.81e-77 | 0.304 | 0.344 | -0.040 | 1.93e-18 |
| Height | SBayesR.PseudoVal | PRScs.PseudoVal | 0.344 | 0.346 | -0.002 | 2.71e-01 | 0.343 | 0.344 | 0.000 | 9.17e-01 |
| Height | LDpred2.MultiPRS | PRScs.PseudoVal | 0.350 | 0.346 | 0.003 | 5.86e-01 | 0.354 | 0.344 | 0.010 | 1.95e-04 |
| Height | SBLUP.Inf | PRScs.PseudoVal | 0.293 | 0.346 | -0.053 | 1.45e-111 | 0.294 | 0.344 | -0.050 | 1.69e-26 |
| Height | LDpred1.Inf | PRScs.PseudoVal | 0.299 | 0.346 | -0.048 | 1.85e-100 | 0.303 | 0.344 | -0.040 | 3.43e-19 |
| Height | PRScs.10FCVal | PRScs.PseudoVal | 0.345 | 0.346 | -0.001 | 2.57e-01 | 0.344 | 0.344 | 0.000 | 9.64e-01 |
| Height | LDpred2.Inf | PRScs.PseudoVal | 0.305 | 0.346 | -0.041 | 7.07e-78 | 0.305 | 0.344 | -0.039 | 1.77e-18 |
| Height | PRScs.PseudoVal | PRScs.PseudoVal | 0.346 | 0.346 | 0.000 | 1.00e+00 | 0.344 | 0.344 | 0.000 | 1.00e+00 |
| Height | LDpred2.10FCVal | PRScs.PseudoVal | 0.343 | 0.346 | -0.004 | 1.41e-02 | 0.346 | 0.344 | 0.002 | 4.43e-01 |
| Height | pT+clump.10FCVal | PRScs.PseudoVal | 0.304 | 0.346 | -0.042 | 1.05e-47 | 0.299 | 0.344 | -0.045 | 1.11e-14 |
| Height | PRScs.MultiPRS | PRScs.PseudoVal | 0.347 | 0.346 | 0.001 | 8.63e-01 | 0.345 | 0.344 | 0.001 | 5.64e-02 |
| Height | lassosum.10FCVal | PRScs.PseudoVal | 0.347 | 0.346 | 0.001 | 3.69e-01 | 0.346 | 0.344 | 0.002 | 5.02e-01 |
| Height | lassosum.PseudoVal | PRScs.PseudoVal | 0.285 | 0.346 | -0.062 | 6.18e-88 | 0.281 | 0.344 | -0.063 | 6.85e-24 |
| Height | LDpred1.MultiPRS | PRScs.PseudoVal | 0.303 | 0.346 | -0.043 | 2.77e-11 | 0.307 | 0.344 | -0.037 | 1.52e-17 |
| Height | lassosum.MultiPRS | PRScs.PseudoVal | 0.350 | 0.346 | 0.004 | 5.09e-01 | 0.348 | 0.344 | 0.005 | 5.46e-02 |
| Height | LDpred1.10FCVal | PRScs.PseudoVal | 0.300 | 0.346 | -0.046 | 1.46e-89 | 0.301 | 0.344 | -0.043 | 5.03e-20 |
| Height | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.332 | 0.346 | -0.014 | 1.74e-20 | 0.327 | 0.344 | -0.016 | 3.34e-08 |
| Height | All.MultiPRS | pT+clump.MultiPRS | 0.358 | 0.320 | 0.038 | 2.44e-09 | 0.361 | 0.313 | 0.048 | 2.13e-27 |
| Height | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.344 | 0.320 | 0.024 | 1.67e-04 | 0.343 | 0.313 | 0.031 | 7.75e-10 |
| Height | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.303 | 0.320 | -0.017 | 1.05e-02 | 0.307 | 0.313 | -0.006 | 3.22e-01 |
| Height | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.320 | 0.320 | 0.000 | 1.00e+00 | 0.313 | 0.313 | 0.000 | 1.00e+00 |
| Height | lassosum.10FCVal | pT+clump.MultiPRS | 0.347 | 0.320 | 0.028 | 1.79e-05 | 0.346 | 0.313 | 0.033 | 2.93e-13 |
| Height | PRScs.10FCVal | pT+clump.MultiPRS | 0.345 | 0.320 | 0.025 | 7.89e-05 | 0.344 | 0.313 | 0.031 | 3.23e-12 |
| Height | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.332 | 0.320 | 0.013 | 4.90e-02 | 0.327 | 0.313 | 0.015 | 2.75e-03 |
| Height | SBLUP.Inf | pT+clump.MultiPRS | 0.293 | 0.320 | -0.026 | 6.12e-05 | 0.294 | 0.313 | -0.019 | 2.39e-03 |
| Height | LDpred2.Inf | pT+clump.MultiPRS | 0.305 | 0.320 | -0.015 | 2.24e-02 | 0.305 | 0.313 | -0.008 | 1.83e-01 |
| Height | pT+clump.10FCVal | pT+clump.MultiPRS | 0.304 | 0.320 | -0.015 | 1.90e-02 | 0.299 | 0.313 | -0.014 | 2.20e-06 |
| Height | LDpred2.10FCVal | pT+clump.MultiPRS | 0.343 | 0.320 | 0.023 | 3.75e-04 | 0.346 | 0.313 | 0.033 | 2.63e-11 |
| Height | PRScs.PseudoVal | pT+clump.MultiPRS | 0.346 | 0.320 | 0.026 | 4.07e-05 | 0.344 | 0.313 | 0.031 | 4.76e-12 |
| Height | LDpred1.10FCVal | pT+clump.MultiPRS | 0.300 | 0.320 | -0.020 | 2.27e-03 | 0.301 | 0.313 | -0.012 | 6.28e-02 |
| Height | LDpred1.Inf | pT+clump.MultiPRS | 0.299 | 0.320 | -0.021 | 1.30e-03 | 0.303 | 0.313 | -0.009 | 1.30e-01 |
| Height | PRScs.MultiPRS | pT+clump.MultiPRS | 0.347 | 0.320 | 0.028 | 2.02e-05 | 0.345 | 0.313 | 0.033 | 1.39e-13 |
| Height | lassosum.PseudoVal | pT+clump.MultiPRS | 0.285 | 0.320 | -0.035 | 8.62e-08 | 0.281 | 0.313 | -0.032 | 6.86e-06 |
| Height | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.304 | 0.320 | -0.016 | 1.68e-02 | 0.304 | 0.313 | -0.009 | 1.55e-01 |
| Height | lassosum.MultiPRS | pT+clump.MultiPRS | 0.350 | 0.320 | 0.031 | 1.90e-06 | 0.348 | 0.313 | 0.036 | 6.06e-16 |
| Height | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.350 | 0.320 | 0.030 | 3.28e-06 | 0.354 | 0.313 | 0.041 | 2.79e-17 |
| Height | LDpred2.10FCVal | pT+clump.10FCVal | 0.343 | 0.304 | 0.038 | 1.51e-32 | 0.346 | 0.299 | 0.047 | 2.55e-13 |
| Height | pT+clump.MultiPRS | pT+clump.10FCVal | 0.320 | 0.304 | 0.015 | 1.90e-02 | 0.313 | 0.299 | 0.014 | 2.20e-06 |
| Height | LDpred2.Inf | pT+clump.10FCVal | 0.305 | 0.304 | 0.000 | 9.06e-01 | 0.305 | 0.299 | 0.006 | 4.72e-01 |
| Height | SBayesR.PseudoVal | pT+clump.10FCVal | 0.344 | 0.304 | 0.040 | 8.80e-48 | 0.343 | 0.299 | 0.045 | 6.57e-16 |
| Height | PRScs.MultiPRS | pT+clump.10FCVal | 0.347 | 0.304 | 0.043 | 6.41e-12 | 0.345 | 0.299 | 0.046 | 1.53e-16 |
| Height | SBLUP.Inf | pT+clump.10FCVal | 0.293 | 0.304 | -0.011 | 7.04e-03 | 0.294 | 0.299 | -0.005 | 5.12e-01 |
| Height | lassosum.MultiPRS | pT+clump.10FCVal | 0.350 | 0.304 | 0.046 | 9.87e-13 | 0.348 | 0.299 | 0.050 | 8.33e-20 |
| Height | PRScs.10FCVal | pT+clump.10FCVal | 0.345 | 0.304 | 0.041 | 1.14e-49 | 0.344 | 0.299 | 0.045 | 5.99e-16 |
| Height | LDpred2.MultiPRS | pT+clump.10FCVal | 0.350 | 0.304 | 0.045 | 2.80e-12 | 0.354 | 0.299 | 0.055 | 5.22e-20 |
| Height | lassosum.PseudoVal | pT+clump.10FCVal | 0.285 | 0.304 | -0.020 | 1.89e-08 | 0.281 | 0.299 | -0.018 | 1.14e-02 |
| Height | LDpred2.PseudoVal | pT+clump.10FCVal | 0.304 | 0.304 | 0.000 | 9.50e-01 | 0.304 | 0.299 | 0.005 | 5.30e-01 |
| Height | LDpred1.MultiPRS | pT+clump.10FCVal | 0.303 | 0.304 | -0.001 | 8.33e-01 | 0.307 | 0.299 | 0.008 | 3.07e-01 |
| Height | pT+clump.10FCVal | pT+clump.10FCVal | 0.304 | 0.304 | 0.000 | 1.00e+00 | 0.299 | 0.299 | 0.000 | 1.00e+00 |
| Height | All.MultiPRS | pT+clump.10FCVal | 0.358 | 0.304 | 0.054 | 5.46e-18 | 0.361 | 0.299 | 0.062 | 2.85e-32 |
| Height | LDpred1.Inf | pT+clump.10FCVal | 0.299 | 0.304 | -0.006 | 1.48e-01 | 0.303 | 0.299 | 0.005 | 5.56e-01 |
| Height | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.332 | 0.304 | 0.028 | 2.05e-18 | 0.327 | 0.299 | 0.029 | 9.51e-06 |
| Height | LDpred1.10FCVal | pT+clump.10FCVal | 0.300 | 0.304 | -0.005 | 2.42e-01 | 0.301 | 0.299 | 0.002 | 7.79e-01 |
| Height | lassosum.10FCVal | pT+clump.10FCVal | 0.347 | 0.304 | 0.043 | 7.92e-53 | 0.346 | 0.299 | 0.047 | 1.39e-16 |
| Height | PRScs.PseudoVal | pT+clump.10FCVal | 0.346 | 0.304 | 0.042 | 1.05e-47 | 0.344 | 0.299 | 0.045 | 1.11e-14 |
| Height | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.320 | 0.344 | -0.024 | 1.67e-04 | 0.313 | 0.343 | -0.031 | 7.75e-10 |
| Height | LDpred1.10FCVal | SBayesR.PseudoVal | 0.300 | 0.344 | -0.044 | 1.25e-39 | 0.301 | 0.343 | -0.042 | 3.56e-10 |
| Height | PRScs.10FCVal | SBayesR.PseudoVal | 0.345 | 0.344 | 0.001 | 5.11e-01 | 0.344 | 0.343 | 0.000 | 8.89e-01 |
| Height | LDpred2.10FCVal | SBayesR.PseudoVal | 0.343 | 0.344 | -0.001 | 5.64e-01 | 0.346 | 0.343 | 0.003 | 5.79e-01 |
| Height | lassosum.PseudoVal | SBayesR.PseudoVal | 0.285 | 0.344 | -0.059 | 1.17e-107 | 0.281 | 0.343 | -0.062 | 7.00e-31 |
| Height | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.304 | 0.344 | -0.040 | 5.03e-32 | 0.304 | 0.343 | -0.040 | 7.25e-09 |
| Height | lassosum.MultiPRS | SBayesR.PseudoVal | 0.350 | 0.344 | 0.006 | 3.17e-01 | 0.348 | 0.343 | 0.005 | 1.60e-01 |
| Height | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.350 | 0.344 | 0.006 | 3.79e-01 | 0.354 | 0.343 | 0.010 | 5.78e-03 |
| Height | All.MultiPRS | SBayesR.PseudoVal | 0.358 | 0.344 | 0.014 | 2.11e-02 | 0.361 | 0.343 | 0.017 | 2.98e-10 |
| Height | PRScs.PseudoVal | SBayesR.PseudoVal | 0.346 | 0.344 | 0.002 | 2.71e-01 | 0.344 | 0.343 | 0.000 | 9.17e-01 |
| Height | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.303 | 0.344 | -0.041 | 2.48e-10 | 0.307 | 0.343 | -0.037 | 3.49e-08 |
| Height | pT+clump.10FCVal | SBayesR.PseudoVal | 0.304 | 0.344 | -0.040 | 8.80e-48 | 0.299 | 0.343 | -0.045 | 6.57e-16 |
| Height | lassosum.10FCVal | SBayesR.PseudoVal | 0.347 | 0.344 | 0.003 | 8.62e-02 | 0.346 | 0.343 | 0.002 | 5.64e-01 |
| Height | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.344 | 0.344 | 0.000 | 1.00e+00 | 0.343 | 0.343 | 0.000 | 1.00e+00 |
| Height | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.332 | 0.344 | -0.012 | 2.45e-06 | 0.327 | 0.343 | -0.016 | 1.18e-03 |
| Height | SBLUP.Inf | SBayesR.PseudoVal | 0.293 | 0.344 | -0.051 | 2.83e-47 | 0.294 | 0.343 | -0.050 | 1.32e-12 |
| Height | LDpred2.Inf | SBayesR.PseudoVal | 0.305 | 0.344 | -0.039 | 1.71e-31 | 0.305 | 0.343 | -0.039 | 9.77e-09 |
| Height | LDpred1.Inf | SBayesR.PseudoVal | 0.299 | 0.344 | -0.045 | 1.85e-40 | 0.303 | 0.343 | -0.040 | 5.63e-09 |
| Height | PRScs.MultiPRS | SBayesR.PseudoVal | 0.347 | 0.344 | 0.003 | 6.00e-01 | 0.345 | 0.343 | 0.002 | 5.97e-01 |
| Height | LDpred2.10FCVal | SBLUP.Inf | 0.343 | 0.293 | 0.049 | 1.04e-131 | 0.346 | 0.294 | 0.052 | 1.95e-38 |
| Height | PRScs.PseudoVal | SBLUP.Inf | 0.346 | 0.293 | 0.053 | 1.45e-111 | 0.344 | 0.294 | 0.050 | 1.69e-26 |
| Height | LDpred1.10FCVal | SBLUP.Inf | 0.300 | 0.293 | 0.006 | 4.58e-05 | 0.301 | 0.294 | 0.007 | 1.51e-02 |
| Height | LDpred2.Inf | SBLUP.Inf | 0.305 | 0.293 | 0.011 | 2.10e-24 | 0.305 | 0.294 | 0.011 | 9.43e-07 |
| Height | SBayesR.PseudoVal | SBLUP.Inf | 0.344 | 0.293 | 0.051 | 2.83e-47 | 0.343 | 0.294 | 0.050 | 1.32e-12 |
| Height | lassosum.PseudoVal | SBLUP.Inf | 0.285 | 0.293 | -0.009 | 5.90e-02 | 0.281 | 0.294 | -0.013 | 1.85e-01 |
| Height | pT+clump.MultiPRS | SBLUP.Inf | 0.320 | 0.293 | 0.026 | 6.12e-05 | 0.313 | 0.294 | 0.019 | 2.39e-03 |
| Height | lassosum.MultiPRS | SBLUP.Inf | 0.350 | 0.293 | 0.057 | 2.04e-18 | 0.348 | 0.294 | 0.055 | 2.87e-23 |
| Height | PRScs.10FCVal | SBLUP.Inf | 0.345 | 0.293 | 0.052 | 2.56e-80 | 0.344 | 0.294 | 0.050 | 3.49e-20 |
| Height | PRScs.MultiPRS | SBLUP.Inf | 0.347 | 0.293 | 0.054 | 8.25e-18 | 0.345 | 0.294 | 0.052 | 2.04e-24 |
| Height | LDpred1.MultiPRS | SBLUP.Inf | 0.303 | 0.293 | 0.009 | 1.49e-01 | 0.307 | 0.294 | 0.013 | 1.97e-09 |
| Height | LDpred2.PseudoVal | SBLUP.Inf | 0.304 | 0.293 | 0.011 | 1.54e-21 | 0.304 | 0.294 | 0.010 | 4.40e-06 |
| Height | LDpred1.Inf | SBLUP.Inf | 0.299 | 0.293 | 0.005 | 3.72e-07 | 0.303 | 0.294 | 0.010 | 9.68e-07 |
| Height | LDpred2.MultiPRS | SBLUP.Inf | 0.350 | 0.293 | 0.056 | 4.21e-18 | 0.354 | 0.294 | 0.060 | 1.54e-30 |
| Height | All.MultiPRS | SBLUP.Inf | 0.358 | 0.293 | 0.065 | 4.37e-25 | 0.361 | 0.294 | 0.067 | 1.41e-32 |
| Height | SBLUP.Inf | SBLUP.Inf | 0.293 | 0.293 | 0.000 | 1.00e+00 | 0.294 | 0.294 | 0.000 | 1.00e+00 |
| Height | DBSLMM.PseudoVal | SBLUP.Inf | 0.332 | 0.293 | 0.039 | 9.41e-91 | 0.327 | 0.294 | 0.034 | 1.65e-18 |
| Height | pT+clump.10FCVal | SBLUP.Inf | 0.304 | 0.293 | 0.011 | 7.04e-03 | 0.299 | 0.294 | 0.005 | 5.12e-01 |
| Height | lassosum.10FCVal | SBLUP.Inf | 0.347 | 0.293 | 0.054 | 1.09e-80 | 0.346 | 0.294 | 0.052 | 1.12e-19 |
| BMI | LDpred2.PseudoVal | All.MultiPRS | 0.256 | 0.307 | -0.050 | 3.99e-14 | 0.261 | 0.305 | -0.044 | 2.93e-15 |
| BMI | pT+clump.10FCVal | All.MultiPRS | 0.249 | 0.307 | -0.058 | 2.54e-18 | 0.244 | 0.305 | -0.061 | 2.24e-26 |
| BMI | LDpred2.MultiPRS | All.MultiPRS | 0.302 | 0.307 | -0.004 | 4.96e-01 | 0.299 | 0.305 | -0.006 | 3.94e-04 |
| BMI | PRScs.PseudoVal | All.MultiPRS | 0.282 | 0.307 | -0.024 | 2.41e-04 | 0.282 | 0.305 | -0.023 | 2.68e-09 |
| BMI | LDpred1.Inf | All.MultiPRS | 0.274 | 0.307 | -0.032 | 1.15e-06 | 0.264 | 0.305 | -0.041 | 7.32e-21 |
| BMI | PRScs.MultiPRS | All.MultiPRS | 0.301 | 0.307 | -0.005 | 4.21e-01 | 0.296 | 0.305 | -0.009 | 2.83e-07 |
| BMI | LDpred2.Inf | All.MultiPRS | 0.291 | 0.307 | -0.015 | 1.90e-02 | 0.284 | 0.305 | -0.021 | 6.88e-12 |
| BMI | lassosum.10FCVal | All.MultiPRS | 0.300 | 0.307 | -0.006 | 3.47e-01 | 0.298 | 0.305 | -0.007 | 3.42e-04 |
| BMI | LDpred2.10FCVal | All.MultiPRS | 0.301 | 0.307 | -0.005 | 4.39e-01 | 0.296 | 0.305 | -0.009 | 2.20e-07 |
| BMI | lassosum.MultiPRS | All.MultiPRS | 0.305 | 0.307 | -0.002 | 7.73e-01 | 0.305 | 0.305 | 0.000 | 8.84e-01 |
| BMI | LDpred1.10FCVal | All.MultiPRS | 0.279 | 0.307 | -0.028 | 2.31e-05 | 0.271 | 0.305 | -0.034 | 1.93e-16 |
| BMI | All.MultiPRS | All.MultiPRS | 0.307 | 0.307 | 0.000 | 1.00e+00 | 0.305 | 0.305 | 0.000 | 1.00e+00 |
| BMI | SBayesR.PseudoVal | All.MultiPRS | 0.211 | 0.307 | -0.095 | 1.34e-45 | 0.224 | 0.305 | -0.081 | 7.00e-27 |
| BMI | LDpred1.MultiPRS | All.MultiPRS | 0.289 | 0.307 | -0.018 | 6.81e-03 | 0.281 | 0.305 | -0.024 | 1.16e-12 |
| BMI | pT+clump.MultiPRS | All.MultiPRS | 0.271 | 0.307 | -0.036 | 7.60e-08 | 0.274 | 0.305 | -0.031 | 2.37e-12 |
| BMI | lassosum.PseudoVal | All.MultiPRS | 0.216 | 0.307 | -0.091 | 8.15e-42 | 0.225 | 0.305 | -0.080 | 5.47e-27 |
| BMI | PRScs.10FCVal | All.MultiPRS | 0.292 | 0.307 | -0.015 | 2.62e-02 | 0.282 | 0.305 | -0.024 | 2.66e-15 |
| BMI | DBSLMM.PseudoVal | All.MultiPRS | 0.285 | 0.307 | -0.022 | 8.66e-04 | 0.283 | 0.305 | -0.022 | 2.31e-09 |
| BMI | SBLUP.Inf | All.MultiPRS | 0.293 | 0.307 | -0.014 | 3.30e-02 | 0.286 | 0.305 | -0.019 | 4.50e-11 |
| BMI | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.211 | 0.285 | -0.073 | 6.58e-118 | 0.224 | 0.283 | -0.059 | 7.93e-21 |
| BMI | LDpred2.Inf | DBSLMM.PseudoVal | 0.291 | 0.285 | 0.007 | 9.40e-03 | 0.284 | 0.283 | 0.001 | 8.05e-01 |
| BMI | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.249 | 0.285 | -0.036 | 9.11e-24 | 0.244 | 0.283 | -0.039 | 1.02e-07 |
| BMI | PRScs.10FCVal | DBSLMM.PseudoVal | 0.292 | 0.285 | 0.007 | 2.41e-03 | 0.282 | 0.283 | -0.001 | 7.74e-01 |
| BMI | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.282 | 0.285 | -0.002 | 1.91e-01 | 0.282 | 0.283 | -0.001 | 7.51e-01 |
| BMI | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.271 | 0.285 | -0.014 | 4.17e-02 | 0.274 | 0.283 | -0.009 | 9.36e-02 |
| BMI | LDpred1.Inf | DBSLMM.PseudoVal | 0.274 | 0.285 | -0.010 | 2.10e-04 | 0.264 | 0.283 | -0.019 | 7.09e-04 |
| BMI | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.305 | 0.285 | 0.020 | 2.38e-03 | 0.305 | 0.283 | 0.022 | 9.20e-08 |
| BMI | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.301 | 0.285 | 0.017 | 1.43e-17 | 0.296 | 0.283 | 0.013 | 1.31e-03 |
| BMI | lassosum.10FCVal | DBSLMM.PseudoVal | 0.300 | 0.285 | 0.016 | 2.77e-12 | 0.298 | 0.283 | 0.015 | 1.04e-03 |
| BMI | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.279 | 0.285 | -0.006 | 4.46e-03 | 0.271 | 0.283 | -0.012 | 6.86e-03 |
| BMI | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.289 | 0.285 | 0.004 | 5.38e-01 | 0.281 | 0.283 | -0.002 | 6.32e-01 |
| BMI | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.301 | 0.285 | 0.017 | 1.16e-02 | 0.296 | 0.283 | 0.013 | 5.21e-04 |
| BMI | All.MultiPRS | DBSLMM.PseudoVal | 0.307 | 0.285 | 0.022 | 8.66e-04 | 0.305 | 0.283 | 0.022 | 2.31e-09 |
| BMI | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.256 | 0.285 | -0.028 | 3.12e-26 | 0.261 | 0.283 | -0.022 | 5.11e-05 |
| BMI | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.285 | 0.285 | 0.000 | 1.00e+00 | 0.283 | 0.283 | 0.000 | 1.00e+00 |
| BMI | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.302 | 0.285 | 0.017 | 8.02e-03 | 0.299 | 0.283 | 0.016 | 4.64e-05 |
| BMI | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.216 | 0.285 | -0.069 | 3.28e-99 | 0.225 | 0.283 | -0.058 | 7.97e-19 |
| BMI | SBLUP.Inf | DBSLMM.PseudoVal | 0.293 | 0.285 | 0.008 | 1.44e-03 | 0.286 | 0.283 | 0.003 | 5.94e-01 |
| BMI | SBayesR.PseudoVal | lassosum.MultiPRS | 0.211 | 0.305 | -0.093 | 5.75e-44 | 0.224 | 0.305 | -0.081 | 1.44e-25 |
| BMI | LDpred2.10FCVal | lassosum.MultiPRS | 0.301 | 0.305 | -0.003 | 6.27e-01 | 0.296 | 0.305 | -0.009 | 1.01e-03 |
| BMI | LDpred1.10FCVal | lassosum.MultiPRS | 0.279 | 0.305 | -0.026 | 7.75e-05 | 0.271 | 0.305 | -0.034 | 4.14e-13 |
| BMI | LDpred2.Inf | lassosum.MultiPRS | 0.291 | 0.305 | -0.014 | 3.98e-02 | 0.284 | 0.305 | -0.021 | 1.34e-08 |
| BMI | PRScs.10FCVal | lassosum.MultiPRS | 0.292 | 0.305 | -0.013 | 5.32e-02 | 0.282 | 0.305 | -0.023 | 8.25e-12 |
| BMI | PRScs.MultiPRS | lassosum.MultiPRS | 0.301 | 0.305 | -0.003 | 6.04e-01 | 0.296 | 0.305 | -0.009 | 3.02e-04 |
| BMI | pT+clump.MultiPRS | lassosum.MultiPRS | 0.271 | 0.305 | -0.034 | 3.77e-07 | 0.274 | 0.305 | -0.031 | 4.02e-11 |
| BMI | lassosum.MultiPRS | lassosum.MultiPRS | 0.305 | 0.305 | 0.000 | 1.00e+00 | 0.305 | 0.305 | 0.000 | 1.00e+00 |
| BMI | lassosum.PseudoVal | lassosum.MultiPRS | 0.216 | 0.305 | -0.089 | 2.62e-40 | 0.225 | 0.305 | -0.080 | 5.19e-27 |
| BMI | LDpred2.MultiPRS | lassosum.MultiPRS | 0.302 | 0.305 | -0.003 | 6.94e-01 | 0.299 | 0.305 | -0.006 | 2.85e-02 |
| BMI | LDpred1.MultiPRS | lassosum.MultiPRS | 0.289 | 0.305 | -0.016 | 1.56e-02 | 0.281 | 0.305 | -0.024 | 1.98e-09 |
| BMI | LDpred2.PseudoVal | lassosum.MultiPRS | 0.256 | 0.305 | -0.048 | 3.28e-13 | 0.261 | 0.305 | -0.044 | 1.18e-13 |
| BMI | LDpred1.Inf | lassosum.MultiPRS | 0.274 | 0.305 | -0.030 | 4.73e-06 | 0.264 | 0.305 | -0.041 | 3.69e-17 |
| BMI | pT+clump.10FCVal | lassosum.MultiPRS | 0.249 | 0.305 | -0.056 | 4.19e-17 | 0.244 | 0.305 | -0.061 | 6.01e-25 |
| BMI | lassosum.10FCVal | lassosum.MultiPRS | 0.300 | 0.305 | -0.004 | 5.14e-01 | 0.298 | 0.305 | -0.007 | 8.18e-05 |
| BMI | SBLUP.Inf | lassosum.MultiPRS | 0.293 | 0.305 | -0.012 | 6.56e-02 | 0.286 | 0.305 | -0.019 | 2.91e-08 |
| BMI | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.285 | 0.305 | -0.020 | 2.38e-03 | 0.283 | 0.305 | -0.022 | 9.20e-08 |
| BMI | All.MultiPRS | lassosum.MultiPRS | 0.307 | 0.305 | 0.002 | 7.73e-01 | 0.305 | 0.305 | 0.000 | 8.84e-01 |
| BMI | PRScs.PseudoVal | lassosum.MultiPRS | 0.282 | 0.305 | -0.022 | 7.11e-04 | 0.282 | 0.305 | -0.023 | 1.08e-07 |
| BMI | LDpred1.Inf | lassosum.PseudoVal | 0.274 | 0.216 | 0.059 | 3.28e-40 | 0.264 | 0.225 | 0.039 | 1.48e-05 |
| BMI | PRScs.10FCVal | lassosum.PseudoVal | 0.292 | 0.216 | 0.076 | 2.00e-64 | 0.282 | 0.225 | 0.057 | 3.90e-10 |
| BMI | LDpred2.MultiPRS | lassosum.PseudoVal | 0.302 | 0.216 | 0.086 | 8.31e-38 | 0.299 | 0.225 | 0.074 | 2.02e-22 |
| BMI | lassosum.PseudoVal | lassosum.PseudoVal | 0.216 | 0.216 | 0.000 | 1.00e+00 | 0.225 | 0.225 | 0.000 | 1.00e+00 |
| BMI | PRScs.PseudoVal | lassosum.PseudoVal | 0.282 | 0.216 | 0.067 | 9.48e-133 | 0.282 | 0.225 | 0.057 | 3.33e-25 |
| BMI | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.285 | 0.216 | 0.069 | 3.28e-99 | 0.283 | 0.225 | 0.058 | 7.97e-19 |
| BMI | SBLUP.Inf | lassosum.PseudoVal | 0.293 | 0.216 | 0.077 | 1.34e-60 | 0.286 | 0.225 | 0.061 | 1.28e-10 |
| BMI | LDpred2.Inf | lassosum.PseudoVal | 0.291 | 0.216 | 0.075 | 4.85e-59 | 0.284 | 0.225 | 0.059 | 3.09e-10 |
| BMI | pT+clump.10FCVal | lassosum.PseudoVal | 0.249 | 0.216 | 0.033 | 1.27e-11 | 0.244 | 0.225 | 0.019 | 5.37e-02 |
| BMI | LDpred2.10FCVal | lassosum.PseudoVal | 0.301 | 0.216 | 0.086 | 9.23e-109 | 0.296 | 0.225 | 0.071 | 1.69e-19 |
| BMI | lassosum.MultiPRS | lassosum.PseudoVal | 0.305 | 0.216 | 0.089 | 2.62e-40 | 0.305 | 0.225 | 0.080 | 5.19e-27 |
| BMI | LDpred1.10FCVal | lassosum.PseudoVal | 0.279 | 0.216 | 0.063 | 5.17e-94 | 0.271 | 0.225 | 0.046 | 9.26e-14 |
| BMI | All.MultiPRS | lassosum.PseudoVal | 0.307 | 0.216 | 0.091 | 8.15e-42 | 0.305 | 0.225 | 0.080 | 5.47e-27 |
| BMI | SBayesR.PseudoVal | lassosum.PseudoVal | 0.211 | 0.216 | -0.004 | 1.10e-01 | 0.224 | 0.225 | -0.001 | 8.44e-01 |
| BMI | lassosum.10FCVal | lassosum.PseudoVal | 0.300 | 0.216 | 0.085 | 1.13e-99 | 0.298 | 0.225 | 0.073 | 1.77e-19 |
| BMI | pT+clump.MultiPRS | lassosum.PseudoVal | 0.271 | 0.216 | 0.055 | 3.91e-16 | 0.274 | 0.225 | 0.049 | 3.37e-10 |
| BMI | LDpred2.PseudoVal | lassosum.PseudoVal | 0.256 | 0.216 | 0.041 | 8.43e-81 | 0.261 | 0.225 | 0.036 | 4.15e-17 |
| BMI | LDpred1.MultiPRS | lassosum.PseudoVal | 0.289 | 0.216 | 0.073 | 1.48e-27 | 0.281 | 0.225 | 0.056 | 3.22e-15 |
| BMI | PRScs.MultiPRS | lassosum.PseudoVal | 0.301 | 0.216 | 0.085 | 1.97e-37 | 0.296 | 0.225 | 0.071 | 1.32e-20 |
| BMI | lassosum.10FCVal | lassosum.10FCVal | 0.300 | 0.300 | 0.000 | 1.00e+00 | 0.298 | 0.298 | 0.000 | 1.00e+00 |
| BMI | pT+clump.MultiPRS | lassosum.10FCVal | 0.271 | 0.300 | -0.029 | 9.78e-06 | 0.274 | 0.298 | -0.024 | 1.37e-06 |
| BMI | LDpred2.Inf | lassosum.10FCVal | 0.291 | 0.300 | -0.009 | 2.07e-09 | 0.284 | 0.298 | -0.014 | 1.01e-05 |
| BMI | SBayesR.PseudoVal | lassosum.10FCVal | 0.211 | 0.300 | -0.089 | 9.78e-100 | 0.224 | 0.298 | -0.074 | 1.52e-18 |
| BMI | PRScs.MultiPRS | lassosum.10FCVal | 0.301 | 0.300 | 0.001 | 8.93e-01 | 0.296 | 0.298 | -0.002 | 3.71e-01 |
| BMI | SBLUP.Inf | lassosum.10FCVal | 0.293 | 0.300 | -0.008 | 1.15e-07 | 0.286 | 0.298 | -0.012 | 3.01e-05 |
| BMI | lassosum.MultiPRS | lassosum.10FCVal | 0.305 | 0.300 | 0.004 | 5.14e-01 | 0.305 | 0.298 | 0.007 | 8.18e-05 |
| BMI | PRScs.10FCVal | lassosum.10FCVal | 0.292 | 0.300 | -0.008 | 1.71e-07 | 0.282 | 0.298 | -0.016 | 3.45e-07 |
| BMI | LDpred2.MultiPRS | lassosum.10FCVal | 0.302 | 0.300 | 0.002 | 7.95e-01 | 0.299 | 0.298 | 0.001 | 7.13e-01 |
| BMI | PRScs.PseudoVal | lassosum.10FCVal | 0.282 | 0.300 | -0.018 | 2.39e-15 | 0.282 | 0.298 | -0.016 | 4.95e-04 |
| BMI | LDpred2.PseudoVal | lassosum.10FCVal | 0.256 | 0.300 | -0.044 | 4.85e-42 | 0.261 | 0.298 | -0.037 | 2.53e-08 |
| BMI | LDpred1.MultiPRS | lassosum.10FCVal | 0.289 | 0.300 | -0.012 | 7.66e-02 | 0.281 | 0.298 | -0.017 | 3.04e-05 |
| BMI | pT+clump.10FCVal | lassosum.10FCVal | 0.249 | 0.300 | -0.052 | 6.29e-69 | 0.244 | 0.298 | -0.054 | 6.65e-20 |
| BMI | All.MultiPRS | lassosum.10FCVal | 0.307 | 0.300 | 0.006 | 3.47e-01 | 0.305 | 0.298 | 0.007 | 3.42e-04 |
| BMI | LDpred1.Inf | lassosum.10FCVal | 0.274 | 0.300 | -0.026 | 1.56e-33 | 0.264 | 0.298 | -0.034 | 7.84e-15 |
| BMI | DBSLMM.PseudoVal | lassosum.10FCVal | 0.285 | 0.300 | -0.016 | 2.77e-12 | 0.283 | 0.298 | -0.015 | 1.04e-03 |
| BMI | LDpred1.10FCVal | lassosum.10FCVal | 0.279 | 0.300 | -0.022 | 1.97e-21 | 0.271 | 0.298 | -0.027 | 1.10e-08 |
| BMI | lassosum.PseudoVal | lassosum.10FCVal | 0.216 | 0.300 | -0.085 | 1.13e-99 | 0.225 | 0.298 | -0.073 | 1.77e-19 |
| BMI | LDpred2.10FCVal | lassosum.10FCVal | 0.301 | 0.300 | 0.001 | 4.36e-01 | 0.296 | 0.298 | -0.002 | 4.24e-01 |
| BMI | LDpred2.10FCVal | LDpred1.MultiPRS | 0.301 | 0.289 | 0.013 | 5.23e-02 | 0.296 | 0.281 | 0.015 | 1.03e-04 |
| BMI | PRScs.PseudoVal | LDpred1.MultiPRS | 0.282 | 0.289 | -0.006 | 3.36e-01 | 0.282 | 0.281 | 0.001 | 8.11e-01 |
| BMI | LDpred2.Inf | LDpred1.MultiPRS | 0.291 | 0.289 | 0.002 | 7.13e-01 | 0.284 | 0.281 | 0.003 | 4.59e-01 |
| BMI | pT+clump.10FCVal | LDpred1.MultiPRS | 0.249 | 0.289 | -0.040 | 2.05e-09 | 0.244 | 0.281 | -0.037 | 1.49e-07 |
| BMI | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.211 | 0.289 | -0.077 | 1.36e-30 | 0.224 | 0.281 | -0.057 | 8.42e-15 |
| BMI | All.MultiPRS | LDpred1.MultiPRS | 0.307 | 0.289 | 0.018 | 6.81e-03 | 0.305 | 0.281 | 0.024 | 1.16e-12 |
| BMI | LDpred1.10FCVal | LDpred1.MultiPRS | 0.279 | 0.289 | -0.010 | 1.27e-01 | 0.271 | 0.281 | -0.010 | 9.11e-05 |
| BMI | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.289 | 0.289 | 0.000 | 1.00e+00 | 0.281 | 0.281 | 0.000 | 1.00e+00 |
| BMI | PRScs.10FCVal | LDpred1.MultiPRS | 0.292 | 0.289 | 0.003 | 6.23e-01 | 0.282 | 0.281 | 0.001 | 8.88e-01 |
| BMI | lassosum.PseudoVal | LDpred1.MultiPRS | 0.216 | 0.289 | -0.073 | 1.48e-27 | 0.225 | 0.281 | -0.056 | 3.22e-15 |
| BMI | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.271 | 0.289 | -0.018 | 8.03e-03 | 0.274 | 0.281 | -0.007 | 2.00e-01 |
| BMI | LDpred1.Inf | LDpred1.MultiPRS | 0.274 | 0.289 | -0.014 | 3.12e-02 | 0.264 | 0.281 | -0.017 | 1.39e-08 |
| BMI | lassosum.MultiPRS | LDpred1.MultiPRS | 0.305 | 0.289 | 0.016 | 1.56e-02 | 0.305 | 0.281 | 0.024 | 1.98e-09 |
| BMI | PRScs.MultiPRS | LDpred1.MultiPRS | 0.301 | 0.289 | 0.013 | 1.68e-11 | 0.296 | 0.281 | 0.015 | 8.41e-05 |
| BMI | lassosum.10FCVal | LDpred1.MultiPRS | 0.300 | 0.289 | 0.012 | 7.66e-02 | 0.298 | 0.281 | 0.017 | 3.04e-05 |
| BMI | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.256 | 0.289 | -0.032 | 1.18e-06 | 0.261 | 0.281 | -0.020 | 3.79e-04 |
| BMI | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.285 | 0.289 | -0.004 | 5.38e-01 | 0.283 | 0.281 | 0.002 | 6.32e-01 |
| BMI | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.302 | 0.289 | 0.013 | 4.22e-02 | 0.299 | 0.281 | 0.018 | 6.82e-07 |
| BMI | SBLUP.Inf | LDpred1.MultiPRS | 0.293 | 0.289 | 0.004 | 5.61e-01 | 0.286 | 0.281 | 0.005 | 2.89e-01 |
| BMI | All.MultiPRS | LDpred1.Inf | 0.307 | 0.274 | 0.032 | 1.15e-06 | 0.305 | 0.264 | 0.041 | 7.32e-21 |
| BMI | LDpred2.PseudoVal | LDpred1.Inf | 0.256 | 0.274 | -0.018 | 1.50e-06 | 0.261 | 0.264 | -0.003 | 7.32e-01 |
| BMI | SBayesR.PseudoVal | LDpred1.Inf | 0.211 | 0.274 | -0.063 | 3.89e-43 | 0.224 | 0.264 | -0.040 | 1.51e-05 |
| BMI | LDpred2.MultiPRS | LDpred1.Inf | 0.302 | 0.274 | 0.028 | 2.73e-05 | 0.299 | 0.264 | 0.035 | 6.60e-15 |
| BMI | PRScs.PseudoVal | LDpred1.Inf | 0.282 | 0.274 | 0.008 | 3.92e-03 | 0.282 | 0.264 | 0.018 | 1.33e-03 |
| BMI | LDpred1.Inf | LDpred1.Inf | 0.274 | 0.274 | 0.000 | 1.00e+00 | 0.264 | 0.264 | 0.000 | 1.00e+00 |
| BMI | pT+clump.10FCVal | LDpred1.Inf | 0.249 | 0.274 | -0.026 | 2.12e-13 | 0.244 | 0.264 | -0.020 | 5.30e-03 |
| BMI | PRScs.MultiPRS | LDpred1.Inf | 0.301 | 0.274 | 0.027 | 4.87e-05 | 0.296 | 0.264 | 0.032 | 3.29e-13 |
| BMI | lassosum.PseudoVal | LDpred1.Inf | 0.216 | 0.274 | -0.059 | 3.28e-40 | 0.225 | 0.264 | -0.039 | 1.48e-05 |
| BMI | pT+clump.MultiPRS | LDpred1.Inf | 0.271 | 0.274 | -0.003 | 6.17e-01 | 0.274 | 0.264 | 0.010 | 1.15e-01 |
| BMI | LDpred1.MultiPRS | LDpred1.Inf | 0.289 | 0.274 | 0.014 | 3.12e-02 | 0.281 | 0.264 | 0.017 | 1.39e-08 |
| BMI | PRScs.10FCVal | LDpred1.Inf | 0.292 | 0.274 | 0.018 | 8.76e-13 | 0.282 | 0.264 | 0.018 | 3.37e-04 |
| BMI | LDpred2.Inf | LDpred1.Inf | 0.291 | 0.274 | 0.017 | 8.77e-18 | 0.284 | 0.264 | 0.020 | 2.85e-07 |
| BMI | SBLUP.Inf | LDpred1.Inf | 0.293 | 0.274 | 0.018 | 6.95e-21 | 0.286 | 0.264 | 0.022 | 3.52e-08 |
| BMI | DBSLMM.PseudoVal | LDpred1.Inf | 0.285 | 0.274 | 0.010 | 2.10e-04 | 0.283 | 0.264 | 0.019 | 7.09e-04 |
| BMI | lassosum.MultiPRS | LDpred1.Inf | 0.305 | 0.274 | 0.030 | 4.73e-06 | 0.305 | 0.264 | 0.041 | 3.69e-17 |
| BMI | LDpred1.10FCVal | LDpred1.Inf | 0.279 | 0.274 | 0.004 | 3.00e-02 | 0.271 | 0.264 | 0.007 | 5.85e-02 |
| BMI | lassosum.10FCVal | LDpred1.Inf | 0.300 | 0.274 | 0.026 | 1.56e-33 | 0.298 | 0.264 | 0.034 | 7.84e-15 |
| BMI | LDpred2.10FCVal | LDpred1.Inf | 0.301 | 0.274 | 0.027 | 3.02e-35 | 0.296 | 0.264 | 0.032 | 8.61e-13 |
| BMI | LDpred1.Inf | LDpred1.10FCVal | 0.274 | 0.279 | -0.004 | 3.00e-02 | 0.264 | 0.271 | -0.007 | 5.85e-02 |
| BMI | PRScs.MultiPRS | LDpred1.10FCVal | 0.301 | 0.279 | 0.023 | 5.93e-04 | 0.296 | 0.271 | 0.024 | 3.94e-09 |
| BMI | LDpred1.10FCVal | LDpred1.10FCVal | 0.279 | 0.279 | 0.000 | 1.00e+00 | 0.271 | 0.271 | 0.000 | 1.00e+00 |
| BMI | All.MultiPRS | LDpred1.10FCVal | 0.307 | 0.279 | 0.028 | 2.31e-05 | 0.305 | 0.271 | 0.034 | 1.93e-16 |
| BMI | SBayesR.PseudoVal | LDpred1.10FCVal | 0.211 | 0.279 | -0.067 | 4.00e-81 | 0.224 | 0.271 | -0.047 | 2.08e-11 |
| BMI | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.285 | 0.279 | 0.006 | 4.46e-03 | 0.283 | 0.271 | 0.012 | 6.86e-03 |
| BMI | pT+clump.MultiPRS | LDpred1.10FCVal | 0.271 | 0.279 | -0.008 | 2.60e-01 | 0.274 | 0.271 | 0.002 | 6.78e-01 |
| BMI | lassosum.MultiPRS | LDpred1.10FCVal | 0.305 | 0.279 | 0.026 | 7.75e-05 | 0.305 | 0.271 | 0.034 | 4.14e-13 |
| BMI | PRScs.10FCVal | LDpred1.10FCVal | 0.292 | 0.279 | 0.013 | 1.50e-06 | 0.282 | 0.271 | 0.010 | 6.51e-02 |
| BMI | lassosum.10FCVal | LDpred1.10FCVal | 0.300 | 0.279 | 0.022 | 1.97e-21 | 0.298 | 0.271 | 0.027 | 1.10e-08 |
| BMI | SBLUP.Inf | LDpred1.10FCVal | 0.293 | 0.279 | 0.014 | 1.93e-07 | 0.286 | 0.271 | 0.014 | 8.23e-03 |
| BMI | LDpred2.PseudoVal | LDpred1.10FCVal | 0.256 | 0.279 | -0.022 | 9.05e-18 | 0.261 | 0.271 | -0.010 | 5.67e-02 |
| BMI | pT+clump.10FCVal | LDpred1.10FCVal | 0.249 | 0.279 | -0.030 | 5.32e-16 | 0.244 | 0.271 | -0.027 | 2.79e-04 |
| BMI | LDpred2.MultiPRS | LDpred1.10FCVal | 0.302 | 0.279 | 0.024 | 3.64e-04 | 0.299 | 0.271 | 0.028 | 7.85e-11 |
| BMI | PRScs.PseudoVal | LDpred1.10FCVal | 0.282 | 0.279 | 0.004 | 3.05e-02 | 0.282 | 0.271 | 0.011 | 2.85e-03 |
| BMI | LDpred2.10FCVal | LDpred1.10FCVal | 0.301 | 0.279 | 0.023 | 4.56e-27 | 0.296 | 0.271 | 0.024 | 1.52e-08 |
| BMI | LDpred1.MultiPRS | LDpred1.10FCVal | 0.289 | 0.279 | 0.010 | 1.27e-01 | 0.281 | 0.271 | 0.010 | 9.11e-05 |
| BMI | LDpred2.Inf | LDpred1.10FCVal | 0.291 | 0.279 | 0.013 | 2.67e-06 | 0.284 | 0.271 | 0.013 | 1.73e-02 |
| BMI | lassosum.PseudoVal | LDpred1.10FCVal | 0.216 | 0.279 | -0.063 | 5.17e-94 | 0.225 | 0.271 | -0.046 | 9.26e-14 |
| BMI | LDpred1.Inf | LDpred2.MultiPRS | 0.274 | 0.302 | -0.028 | 2.73e-05 | 0.264 | 0.299 | -0.035 | 6.60e-15 |
| BMI | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.285 | 0.302 | -0.017 | 8.02e-03 | 0.283 | 0.299 | -0.016 | 4.64e-05 |
| BMI | SBLUP.Inf | LDpred2.MultiPRS | 0.293 | 0.302 | -0.010 | 1.47e-01 | 0.286 | 0.299 | -0.013 | 1.63e-05 |
| BMI | LDpred2.Inf | LDpred2.MultiPRS | 0.291 | 0.302 | -0.011 | 9.56e-02 | 0.284 | 0.299 | -0.015 | 7.18e-08 |
| BMI | pT+clump.10FCVal | LDpred2.MultiPRS | 0.249 | 0.302 | -0.053 | 9.71e-16 | 0.244 | 0.299 | -0.055 | 9.79e-19 |
| BMI | LDpred2.10FCVal | LDpred2.MultiPRS | 0.301 | 0.302 | -0.001 | 9.26e-01 | 0.296 | 0.299 | -0.003 | 3.91e-03 |
| BMI | PRScs.PseudoVal | LDpred2.MultiPRS | 0.282 | 0.302 | -0.020 | 2.74e-03 | 0.282 | 0.299 | -0.017 | 3.98e-05 |
| BMI | LDpred1.10FCVal | LDpred2.MultiPRS | 0.279 | 0.302 | -0.024 | 3.64e-04 | 0.271 | 0.299 | -0.028 | 7.85e-11 |
| BMI | All.MultiPRS | LDpred2.MultiPRS | 0.307 | 0.302 | 0.004 | 4.96e-01 | 0.305 | 0.299 | 0.006 | 3.94e-04 |
| BMI | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.211 | 0.302 | -0.091 | 9.68e-42 | 0.224 | 0.299 | -0.075 | 6.96e-23 |
| BMI | lassosum.10FCVal | LDpred2.MultiPRS | 0.300 | 0.302 | -0.002 | 7.95e-01 | 0.298 | 0.299 | -0.001 | 7.13e-01 |
| BMI | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.271 | 0.302 | -0.031 | 2.48e-06 | 0.274 | 0.299 | -0.025 | 3.60e-07 |
| BMI | lassosum.MultiPRS | LDpred2.MultiPRS | 0.305 | 0.302 | 0.003 | 6.94e-01 | 0.305 | 0.299 | 0.006 | 2.85e-02 |
| BMI | PRScs.10FCVal | LDpred2.MultiPRS | 0.292 | 0.302 | -0.010 | 1.23e-01 | 0.282 | 0.299 | -0.018 | 1.17e-06 |
| BMI | PRScs.MultiPRS | LDpred2.MultiPRS | 0.301 | 0.302 | -0.001 | 9.00e-01 | 0.296 | 0.299 | -0.003 | 1.91e-01 |
| BMI | lassosum.PseudoVal | LDpred2.MultiPRS | 0.216 | 0.302 | -0.086 | 8.31e-38 | 0.225 | 0.299 | -0.074 | 2.02e-22 |
| BMI | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.256 | 0.302 | -0.046 | 5.51e-12 | 0.261 | 0.299 | -0.038 | 2.79e-12 |
| BMI | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.289 | 0.302 | -0.013 | 4.22e-02 | 0.281 | 0.299 | -0.018 | 6.82e-07 |
| BMI | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.302 | 0.302 | 0.000 | 1.00e+00 | 0.299 | 0.299 | 0.000 | 1.00e+00 |
| BMI | LDpred1.Inf | LDpred2.10FCVal | 0.274 | 0.301 | -0.027 | 3.02e-35 | 0.264 | 0.296 | -0.032 | 8.61e-13 |
| BMI | PRScs.MultiPRS | LDpred2.10FCVal | 0.301 | 0.301 | 0.000 | 9.74e-01 | 0.296 | 0.296 | 0.000 | 9.90e-01 |
| BMI | LDpred2.MultiPRS | LDpred2.10FCVal | 0.302 | 0.301 | 0.001 | 9.26e-01 | 0.299 | 0.296 | 0.003 | 3.91e-03 |
| BMI | PRScs.PseudoVal | LDpred2.10FCVal | 0.282 | 0.301 | -0.019 | 1.72e-19 | 0.282 | 0.296 | -0.014 | 1.24e-03 |
| BMI | LDpred2.10FCVal | LDpred2.10FCVal | 0.301 | 0.301 | 0.000 | 1.00e+00 | 0.296 | 0.296 | 0.000 | 1.00e+00 |
| BMI | lassosum.MultiPRS | LDpred2.10FCVal | 0.305 | 0.301 | 0.003 | 6.27e-01 | 0.305 | 0.296 | 0.009 | 1.01e-03 |
| BMI | LDpred1.10FCVal | LDpred2.10FCVal | 0.279 | 0.301 | -0.023 | 4.56e-27 | 0.271 | 0.296 | -0.024 | 1.52e-08 |
| BMI | All.MultiPRS | LDpred2.10FCVal | 0.307 | 0.301 | 0.005 | 4.39e-01 | 0.305 | 0.296 | 0.009 | 2.20e-07 |
| BMI | SBayesR.PseudoVal | LDpred2.10FCVal | 0.211 | 0.301 | -0.090 | 3.56e-115 | 0.224 | 0.296 | -0.072 | 9.73e-20 |
| BMI | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.285 | 0.301 | -0.017 | 1.43e-17 | 0.283 | 0.296 | -0.013 | 1.31e-03 |
| BMI | pT+clump.MultiPRS | LDpred2.10FCVal | 0.271 | 0.301 | -0.030 | 4.44e-06 | 0.274 | 0.296 | -0.022 | 1.30e-05 |
| BMI | lassosum.PseudoVal | LDpred2.10FCVal | 0.216 | 0.301 | -0.086 | 9.23e-109 | 0.225 | 0.296 | -0.071 | 1.69e-19 |
| BMI | PRScs.10FCVal | LDpred2.10FCVal | 0.292 | 0.301 | -0.010 | 6.71e-09 | 0.282 | 0.296 | -0.014 | 1.57e-05 |
| BMI | lassosum.10FCVal | LDpred2.10FCVal | 0.300 | 0.301 | -0.001 | 4.36e-01 | 0.298 | 0.296 | 0.002 | 4.24e-01 |
| BMI | SBLUP.Inf | LDpred2.10FCVal | 0.293 | 0.301 | -0.009 | 4.62e-10 | 0.286 | 0.296 | -0.010 | 4.91e-04 |
| BMI | LDpred2.PseudoVal | LDpred2.10FCVal | 0.256 | 0.301 | -0.045 | 1.55e-56 | 0.261 | 0.296 | -0.035 | 2.65e-09 |
| BMI | pT+clump.10FCVal | LDpred2.10FCVal | 0.249 | 0.301 | -0.053 | 5.90e-66 | 0.244 | 0.296 | -0.052 | 8.97e-17 |
| BMI | LDpred2.Inf | LDpred2.10FCVal | 0.291 | 0.301 | -0.010 | 1.96e-17 | 0.284 | 0.296 | -0.012 | 3.34e-06 |
| BMI | LDpred1.MultiPRS | LDpred2.10FCVal | 0.289 | 0.301 | -0.013 | 5.23e-02 | 0.281 | 0.296 | -0.015 | 1.03e-04 |
| BMI | lassosum.10FCVal | LDpred2.PseudoVal | 0.300 | 0.256 | 0.044 | 4.85e-42 | 0.298 | 0.261 | 0.037 | 2.53e-08 |
| BMI | SBLUP.Inf | LDpred2.PseudoVal | 0.293 | 0.256 | 0.036 | 2.25e-21 | 0.286 | 0.261 | 0.025 | 1.56e-03 |
| BMI | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.211 | 0.256 | -0.045 | 3.19e-75 | 0.224 | 0.261 | -0.037 | 1.36e-14 |
| BMI | All.MultiPRS | LDpred2.PseudoVal | 0.307 | 0.256 | 0.050 | 3.99e-14 | 0.305 | 0.261 | 0.044 | 2.93e-15 |
| BMI | PRScs.PseudoVal | LDpred2.PseudoVal | 0.282 | 0.256 | 0.026 | 8.05e-29 | 0.282 | 0.261 | 0.021 | 1.23e-05 |
| BMI | lassosum.MultiPRS | LDpred2.PseudoVal | 0.305 | 0.256 | 0.048 | 3.28e-13 | 0.305 | 0.261 | 0.044 | 1.18e-13 |
| BMI | PRScs.MultiPRS | LDpred2.PseudoVal | 0.301 | 0.256 | 0.045 | 1.33e-11 | 0.296 | 0.261 | 0.035 | 1.57e-08 |
| BMI | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.302 | 0.256 | 0.046 | 5.51e-12 | 0.299 | 0.261 | 0.038 | 2.79e-12 |
| BMI | PRScs.10FCVal | LDpred2.PseudoVal | 0.292 | 0.256 | 0.036 | 1.82e-21 | 0.282 | 0.261 | 0.020 | 7.08e-03 |
| BMI | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.256 | 0.256 | 0.000 | 1.00e+00 | 0.261 | 0.261 | 0.000 | 1.00e+00 |
| BMI | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.271 | 0.256 | 0.015 | 2.85e-02 | 0.274 | 0.261 | 0.013 | 6.08e-02 |
| BMI | pT+clump.10FCVal | LDpred2.PseudoVal | 0.249 | 0.256 | -0.008 | 7.13e-02 | 0.244 | 0.261 | -0.017 | 4.56e-02 |
| BMI | lassosum.PseudoVal | LDpred2.PseudoVal | 0.216 | 0.256 | -0.041 | 8.43e-81 | 0.225 | 0.261 | -0.036 | 4.15e-17 |
| BMI | LDpred1.Inf | LDpred2.PseudoVal | 0.274 | 0.256 | 0.018 | 1.50e-06 | 0.264 | 0.261 | 0.003 | 7.32e-01 |
| BMI | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.289 | 0.256 | 0.032 | 1.18e-06 | 0.281 | 0.261 | 0.020 | 3.79e-04 |
| BMI | LDpred1.10FCVal | LDpred2.PseudoVal | 0.279 | 0.256 | 0.022 | 9.05e-18 | 0.271 | 0.261 | 0.010 | 5.67e-02 |
| BMI | LDpred2.Inf | LDpred2.PseudoVal | 0.291 | 0.256 | 0.035 | 1.42e-20 | 0.284 | 0.261 | 0.023 | 2.47e-03 |
| BMI | LDpred2.10FCVal | LDpred2.PseudoVal | 0.301 | 0.256 | 0.045 | 1.55e-56 | 0.296 | 0.261 | 0.035 | 2.65e-09 |
| BMI | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.285 | 0.256 | 0.028 | 3.12e-26 | 0.283 | 0.261 | 0.022 | 5.11e-05 |
| BMI | pT+clump.MultiPRS | LDpred2.Inf | 0.271 | 0.291 | -0.020 | 2.50e-03 | 0.274 | 0.284 | -0.011 | 6.64e-02 |
| BMI | lassosum.MultiPRS | LDpred2.Inf | 0.305 | 0.291 | 0.014 | 3.98e-02 | 0.305 | 0.284 | 0.021 | 1.34e-08 |
| BMI | SBayesR.PseudoVal | LDpred2.Inf | 0.211 | 0.291 | -0.080 | 2.75e-66 | 0.224 | 0.284 | -0.060 | 9.04e-11 |
| BMI | All.MultiPRS | LDpred2.Inf | 0.307 | 0.291 | 0.015 | 1.90e-02 | 0.305 | 0.284 | 0.021 | 6.88e-12 |
| BMI | SBLUP.Inf | LDpred2.Inf | 0.293 | 0.291 | 0.001 | 3.68e-02 | 0.286 | 0.284 | 0.001 | 2.93e-01 |
| BMI | LDpred2.PseudoVal | LDpred2.Inf | 0.256 | 0.291 | -0.035 | 1.42e-20 | 0.261 | 0.284 | -0.023 | 2.47e-03 |
| BMI | PRScs.MultiPRS | LDpred2.Inf | 0.301 | 0.291 | 0.010 | 1.24e-01 | 0.296 | 0.284 | 0.011 | 3.14e-04 |
| BMI | LDpred2.MultiPRS | LDpred2.Inf | 0.302 | 0.291 | 0.011 | 9.56e-02 | 0.299 | 0.284 | 0.015 | 7.18e-08 |
| BMI | PRScs.10FCVal | LDpred2.Inf | 0.292 | 0.291 | 0.001 | 6.06e-01 | 0.282 | 0.284 | -0.003 | 4.05e-01 |
| BMI | LDpred1.Inf | LDpred2.Inf | 0.274 | 0.291 | -0.017 | 8.77e-18 | 0.264 | 0.284 | -0.020 | 2.85e-07 |
| BMI | LDpred1.MultiPRS | LDpred2.Inf | 0.289 | 0.291 | -0.002 | 7.13e-01 | 0.281 | 0.284 | -0.003 | 4.59e-01 |
| BMI | pT+clump.10FCVal | LDpred2.Inf | 0.249 | 0.291 | -0.042 | 2.32e-41 | 0.244 | 0.284 | -0.040 | 2.47e-10 |
| BMI | lassosum.PseudoVal | LDpred2.Inf | 0.216 | 0.291 | -0.075 | 4.85e-59 | 0.225 | 0.284 | -0.059 | 3.09e-10 |
| BMI | PRScs.PseudoVal | LDpred2.Inf | 0.282 | 0.291 | -0.009 | 1.40e-03 | 0.282 | 0.284 | -0.002 | 6.72e-01 |
| BMI | DBSLMM.PseudoVal | LDpred2.Inf | 0.285 | 0.291 | -0.007 | 9.40e-03 | 0.283 | 0.284 | -0.001 | 8.05e-01 |
| BMI | LDpred1.10FCVal | LDpred2.Inf | 0.279 | 0.291 | -0.013 | 2.67e-06 | 0.271 | 0.284 | -0.013 | 1.73e-02 |
| BMI | LDpred2.Inf | LDpred2.Inf | 0.291 | 0.291 | 0.000 | 1.00e+00 | 0.284 | 0.284 | 0.000 | 1.00e+00 |
| BMI | lassosum.10FCVal | LDpred2.Inf | 0.300 | 0.291 | 0.009 | 2.07e-09 | 0.298 | 0.284 | 0.014 | 1.01e-05 |
| BMI | LDpred2.10FCVal | LDpred2.Inf | 0.301 | 0.291 | 0.010 | 1.96e-17 | 0.296 | 0.284 | 0.012 | 3.34e-06 |
| BMI | PRScs.10FCVal | PRScs.MultiPRS | 0.292 | 0.301 | -0.009 | 1.57e-01 | 0.282 | 0.296 | -0.014 | 3.55e-09 |
| BMI | LDpred1.10FCVal | PRScs.MultiPRS | 0.279 | 0.301 | -0.023 | 5.93e-04 | 0.271 | 0.296 | -0.024 | 3.94e-09 |
| BMI | lassosum.MultiPRS | PRScs.MultiPRS | 0.305 | 0.301 | 0.003 | 6.04e-01 | 0.305 | 0.296 | 0.009 | 3.02e-04 |
| BMI | LDpred2.MultiPRS | PRScs.MultiPRS | 0.302 | 0.301 | 0.001 | 9.00e-01 | 0.299 | 0.296 | 0.003 | 1.91e-01 |
| BMI | All.MultiPRS | PRScs.MultiPRS | 0.307 | 0.301 | 0.005 | 4.21e-01 | 0.305 | 0.296 | 0.009 | 2.83e-07 |
| BMI | PRScs.PseudoVal | PRScs.MultiPRS | 0.282 | 0.301 | -0.019 | 4.09e-03 | 0.282 | 0.296 | -0.014 | 5.30e-05 |
| BMI | LDpred1.MultiPRS | PRScs.MultiPRS | 0.289 | 0.301 | -0.013 | 1.68e-11 | 0.281 | 0.296 | -0.015 | 8.41e-05 |
| BMI | pT+clump.MultiPRS | PRScs.MultiPRS | 0.271 | 0.301 | -0.030 | 5.21e-06 | 0.274 | 0.296 | -0.022 | 6.32e-06 |
| BMI | lassosum.PseudoVal | PRScs.MultiPRS | 0.216 | 0.301 | -0.085 | 1.97e-37 | 0.225 | 0.296 | -0.071 | 1.32e-20 |
| BMI | SBayesR.PseudoVal | PRScs.MultiPRS | 0.211 | 0.301 | -0.090 | 6.17e-41 | 0.224 | 0.296 | -0.072 | 6.39e-20 |
| BMI | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.285 | 0.301 | -0.017 | 1.16e-02 | 0.283 | 0.296 | -0.013 | 5.21e-04 |
| BMI | SBLUP.Inf | PRScs.MultiPRS | 0.293 | 0.301 | -0.009 | 1.85e-01 | 0.286 | 0.296 | -0.010 | 7.74e-04 |
| BMI | LDpred2.Inf | PRScs.MultiPRS | 0.291 | 0.301 | -0.010 | 1.24e-01 | 0.284 | 0.296 | -0.011 | 3.14e-04 |
| BMI | pT+clump.10FCVal | PRScs.MultiPRS | 0.249 | 0.301 | -0.053 | 2.78e-15 | 0.244 | 0.296 | -0.052 | 6.57e-17 |
| BMI | LDpred2.10FCVal | PRScs.MultiPRS | 0.301 | 0.301 | 0.000 | 9.74e-01 | 0.296 | 0.296 | 0.000 | 9.90e-01 |
| BMI | lassosum.10FCVal | PRScs.MultiPRS | 0.300 | 0.301 | -0.001 | 8.93e-01 | 0.298 | 0.296 | 0.002 | 3.71e-01 |
| BMI | LDpred2.PseudoVal | PRScs.MultiPRS | 0.256 | 0.301 | -0.045 | 1.33e-11 | 0.261 | 0.296 | -0.035 | 1.57e-08 |
| BMI | LDpred1.Inf | PRScs.MultiPRS | 0.274 | 0.301 | -0.027 | 4.87e-05 | 0.264 | 0.296 | -0.032 | 3.29e-13 |
| BMI | PRScs.MultiPRS | PRScs.MultiPRS | 0.301 | 0.301 | 0.000 | 1.00e+00 | 0.296 | 0.296 | 0.000 | 1.00e+00 |
| BMI | LDpred2.PseudoVal | PRScs.10FCVal | 0.256 | 0.292 | -0.036 | 1.82e-21 | 0.261 | 0.282 | -0.020 | 7.08e-03 |
| BMI | PRScs.10FCVal | PRScs.10FCVal | 0.292 | 0.292 | 0.000 | 1.00e+00 | 0.282 | 0.282 | 0.000 | 1.00e+00 |
| BMI | LDpred2.MultiPRS | PRScs.10FCVal | 0.302 | 0.292 | 0.010 | 1.23e-01 | 0.299 | 0.282 | 0.018 | 1.17e-06 |
| BMI | pT+clump.MultiPRS | PRScs.10FCVal | 0.271 | 0.292 | -0.021 | 1.66e-03 | 0.274 | 0.282 | -0.008 | 1.42e-01 |
| BMI | LDpred1.Inf | PRScs.10FCVal | 0.274 | 0.292 | -0.018 | 8.76e-13 | 0.264 | 0.282 | -0.018 | 3.37e-04 |
| BMI | LDpred1.MultiPRS | PRScs.10FCVal | 0.289 | 0.292 | -0.003 | 6.23e-01 | 0.281 | 0.282 | -0.001 | 8.88e-01 |
| BMI | pT+clump.10FCVal | PRScs.10FCVal | 0.249 | 0.292 | -0.043 | 1.42e-45 | 0.244 | 0.282 | -0.038 | 8.95e-10 |
| BMI | lassosum.PseudoVal | PRScs.10FCVal | 0.216 | 0.292 | -0.076 | 2.00e-64 | 0.225 | 0.282 | -0.057 | 3.90e-10 |
| BMI | PRScs.PseudoVal | PRScs.10FCVal | 0.282 | 0.292 | -0.010 | 4.62e-04 | 0.282 | 0.282 | 0.000 | 9.59e-01 |
| BMI | DBSLMM.PseudoVal | PRScs.10FCVal | 0.285 | 0.292 | -0.007 | 2.41e-03 | 0.283 | 0.282 | 0.001 | 7.74e-01 |
| BMI | SBLUP.Inf | PRScs.10FCVal | 0.293 | 0.292 | 0.001 | 6.72e-01 | 0.286 | 0.282 | 0.004 | 1.48e-01 |
| BMI | LDpred2.Inf | PRScs.10FCVal | 0.291 | 0.292 | -0.001 | 6.06e-01 | 0.284 | 0.282 | 0.003 | 4.05e-01 |
| BMI | lassosum.10FCVal | PRScs.10FCVal | 0.300 | 0.292 | 0.008 | 1.71e-07 | 0.298 | 0.282 | 0.016 | 3.45e-07 |
| BMI | LDpred2.10FCVal | PRScs.10FCVal | 0.301 | 0.292 | 0.010 | 6.71e-09 | 0.296 | 0.282 | 0.014 | 1.57e-05 |
| BMI | lassosum.MultiPRS | PRScs.10FCVal | 0.305 | 0.292 | 0.013 | 5.32e-02 | 0.305 | 0.282 | 0.023 | 8.25e-12 |
| BMI | LDpred1.10FCVal | PRScs.10FCVal | 0.279 | 0.292 | -0.013 | 1.50e-06 | 0.271 | 0.282 | -0.010 | 6.51e-02 |
| BMI | All.MultiPRS | PRScs.10FCVal | 0.307 | 0.292 | 0.015 | 2.62e-02 | 0.305 | 0.282 | 0.024 | 2.66e-15 |
| BMI | SBayesR.PseudoVal | PRScs.10FCVal | 0.211 | 0.292 | -0.080 | 5.46e-70 | 0.224 | 0.282 | -0.058 | 2.74e-10 |
| BMI | PRScs.MultiPRS | PRScs.10FCVal | 0.301 | 0.292 | 0.009 | 1.57e-01 | 0.296 | 0.282 | 0.014 | 3.55e-09 |
| BMI | All.MultiPRS | PRScs.PseudoVal | 0.307 | 0.282 | 0.024 | 2.41e-04 | 0.305 | 0.282 | 0.023 | 2.68e-09 |
| BMI | pT+clump.MultiPRS | PRScs.PseudoVal | 0.271 | 0.282 | -0.011 | 9.08e-02 | 0.274 | 0.282 | -0.008 | 1.56e-01 |
| BMI | LDpred2.PseudoVal | PRScs.PseudoVal | 0.256 | 0.282 | -0.026 | 8.05e-29 | 0.261 | 0.282 | -0.021 | 1.23e-05 |
| BMI | SBayesR.PseudoVal | PRScs.PseudoVal | 0.211 | 0.282 | -0.071 | 2.10e-112 | 0.224 | 0.282 | -0.058 | 2.44e-20 |
| BMI | LDpred2.MultiPRS | PRScs.PseudoVal | 0.302 | 0.282 | 0.020 | 2.74e-03 | 0.299 | 0.282 | 0.017 | 3.98e-05 |
| BMI | SBLUP.Inf | PRScs.PseudoVal | 0.293 | 0.282 | 0.010 | 2.20e-04 | 0.286 | 0.282 | 0.004 | 4.96e-01 |
| BMI | LDpred1.Inf | PRScs.PseudoVal | 0.274 | 0.282 | -0.008 | 3.92e-03 | 0.264 | 0.282 | -0.018 | 1.33e-03 |
| BMI | PRScs.10FCVal | PRScs.PseudoVal | 0.292 | 0.282 | 0.010 | 4.62e-04 | 0.282 | 0.282 | 0.000 | 9.59e-01 |
| BMI | LDpred2.Inf | PRScs.PseudoVal | 0.291 | 0.282 | 0.009 | 1.40e-03 | 0.284 | 0.282 | 0.002 | 6.72e-01 |
| BMI | PRScs.PseudoVal | PRScs.PseudoVal | 0.282 | 0.282 | 0.000 | 1.00e+00 | 0.282 | 0.282 | 0.000 | 1.00e+00 |
| BMI | LDpred2.10FCVal | PRScs.PseudoVal | 0.301 | 0.282 | 0.019 | 1.72e-19 | 0.296 | 0.282 | 0.014 | 1.24e-03 |
| BMI | pT+clump.10FCVal | PRScs.PseudoVal | 0.249 | 0.282 | -0.034 | 9.68e-20 | 0.244 | 0.282 | -0.038 | 4.85e-07 |
| BMI | PRScs.MultiPRS | PRScs.PseudoVal | 0.301 | 0.282 | 0.019 | 4.09e-03 | 0.296 | 0.282 | 0.014 | 5.30e-05 |
| BMI | lassosum.PseudoVal | PRScs.PseudoVal | 0.216 | 0.282 | -0.067 | 9.48e-133 | 0.225 | 0.282 | -0.057 | 3.33e-25 |
| BMI | lassosum.10FCVal | PRScs.PseudoVal | 0.300 | 0.282 | 0.018 | 2.39e-15 | 0.298 | 0.282 | 0.016 | 4.95e-04 |
| BMI | LDpred1.MultiPRS | PRScs.PseudoVal | 0.289 | 0.282 | 0.006 | 3.36e-01 | 0.281 | 0.282 | -0.001 | 8.11e-01 |
| BMI | lassosum.MultiPRS | PRScs.PseudoVal | 0.305 | 0.282 | 0.022 | 7.11e-04 | 0.305 | 0.282 | 0.023 | 1.08e-07 |
| BMI | LDpred1.10FCVal | PRScs.PseudoVal | 0.279 | 0.282 | -0.004 | 3.05e-02 | 0.271 | 0.282 | -0.011 | 2.85e-03 |
| BMI | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.285 | 0.282 | 0.002 | 1.91e-01 | 0.283 | 0.282 | 0.001 | 7.51e-01 |
| BMI | All.MultiPRS | pT+clump.MultiPRS | 0.307 | 0.271 | 0.036 | 7.60e-08 | 0.305 | 0.274 | 0.031 | 2.37e-12 |
| BMI | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.211 | 0.271 | -0.060 | 1.50e-18 | 0.224 | 0.274 | -0.050 | 9.43e-10 |
| BMI | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.289 | 0.271 | 0.018 | 8.03e-03 | 0.281 | 0.274 | 0.007 | 2.00e-01 |
| BMI | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.271 | 0.271 | 0.000 | 1.00e+00 | 0.274 | 0.274 | 0.000 | 1.00e+00 |
| BMI | lassosum.PseudoVal | pT+clump.MultiPRS | 0.216 | 0.271 | -0.055 | 3.91e-16 | 0.225 | 0.274 | -0.049 | 3.37e-10 |
| BMI | PRScs.10FCVal | pT+clump.MultiPRS | 0.292 | 0.271 | 0.021 | 1.66e-03 | 0.282 | 0.274 | 0.008 | 1.42e-01 |
| BMI | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.285 | 0.271 | 0.014 | 4.17e-02 | 0.283 | 0.274 | 0.009 | 9.36e-02 |
| BMI | SBLUP.Inf | pT+clump.MultiPRS | 0.293 | 0.271 | 0.021 | 1.20e-03 | 0.286 | 0.274 | 0.012 | 3.00e-02 |
| BMI | LDpred2.Inf | pT+clump.MultiPRS | 0.291 | 0.271 | 0.020 | 2.50e-03 | 0.284 | 0.274 | 0.011 | 6.64e-02 |
| BMI | pT+clump.10FCVal | pT+clump.MultiPRS | 0.249 | 0.271 | -0.022 | 8.45e-04 | 0.244 | 0.274 | -0.030 | 8.69e-14 |
| BMI | LDpred2.10FCVal | pT+clump.MultiPRS | 0.301 | 0.271 | 0.030 | 4.44e-06 | 0.296 | 0.274 | 0.022 | 1.30e-05 |
| BMI | PRScs.PseudoVal | pT+clump.MultiPRS | 0.282 | 0.271 | 0.011 | 9.08e-02 | 0.282 | 0.274 | 0.008 | 1.56e-01 |
| BMI | LDpred1.10FCVal | pT+clump.MultiPRS | 0.279 | 0.271 | 0.008 | 2.60e-01 | 0.271 | 0.274 | -0.002 | 6.78e-01 |
| BMI | LDpred1.Inf | pT+clump.MultiPRS | 0.274 | 0.271 | 0.003 | 6.17e-01 | 0.264 | 0.274 | -0.010 | 1.15e-01 |
| BMI | PRScs.MultiPRS | pT+clump.MultiPRS | 0.301 | 0.271 | 0.030 | 5.21e-06 | 0.296 | 0.274 | 0.022 | 6.32e-06 |
| BMI | lassosum.10FCVal | pT+clump.MultiPRS | 0.300 | 0.271 | 0.029 | 9.78e-06 | 0.298 | 0.274 | 0.024 | 1.37e-06 |
| BMI | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.256 | 0.271 | -0.015 | 2.85e-02 | 0.261 | 0.274 | -0.013 | 6.08e-02 |
| BMI | lassosum.MultiPRS | pT+clump.MultiPRS | 0.305 | 0.271 | 0.034 | 3.77e-07 | 0.305 | 0.274 | 0.031 | 4.02e-11 |
| BMI | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.302 | 0.271 | 0.031 | 2.48e-06 | 0.299 | 0.274 | 0.025 | 3.60e-07 |
| BMI | LDpred2.10FCVal | pT+clump.10FCVal | 0.301 | 0.249 | 0.053 | 5.90e-66 | 0.296 | 0.244 | 0.052 | 8.97e-17 |
| BMI | pT+clump.MultiPRS | pT+clump.10FCVal | 0.271 | 0.249 | 0.022 | 8.45e-04 | 0.274 | 0.244 | 0.030 | 8.69e-14 |
| BMI | LDpred2.Inf | pT+clump.10FCVal | 0.291 | 0.249 | 0.042 | 2.32e-41 | 0.284 | 0.244 | 0.040 | 2.47e-10 |
| BMI | SBayesR.PseudoVal | pT+clump.10FCVal | 0.211 | 0.249 | -0.037 | 4.18e-14 | 0.224 | 0.244 | -0.020 | 4.33e-02 |
| BMI | PRScs.MultiPRS | pT+clump.10FCVal | 0.301 | 0.249 | 0.053 | 2.78e-15 | 0.296 | 0.244 | 0.052 | 6.57e-17 |
| BMI | SBLUP.Inf | pT+clump.10FCVal | 0.293 | 0.249 | 0.044 | 5.05e-47 | 0.286 | 0.244 | 0.042 | 1.09e-11 |
| BMI | lassosum.MultiPRS | pT+clump.10FCVal | 0.305 | 0.249 | 0.056 | 4.19e-17 | 0.305 | 0.244 | 0.061 | 6.01e-25 |
| BMI | PRScs.10FCVal | pT+clump.10FCVal | 0.292 | 0.249 | 0.043 | 1.42e-45 | 0.282 | 0.244 | 0.038 | 8.95e-10 |
| BMI | LDpred2.MultiPRS | pT+clump.10FCVal | 0.302 | 0.249 | 0.053 | 9.71e-16 | 0.299 | 0.244 | 0.055 | 9.79e-19 |
| BMI | lassosum.10FCVal | pT+clump.10FCVal | 0.300 | 0.249 | 0.052 | 6.29e-69 | 0.298 | 0.244 | 0.054 | 6.65e-20 |
| BMI | LDpred2.PseudoVal | pT+clump.10FCVal | 0.256 | 0.249 | 0.008 | 7.13e-02 | 0.261 | 0.244 | 0.017 | 4.56e-02 |
| BMI | LDpred1.MultiPRS | pT+clump.10FCVal | 0.289 | 0.249 | 0.040 | 2.05e-09 | 0.281 | 0.244 | 0.037 | 1.49e-07 |
| BMI | pT+clump.10FCVal | pT+clump.10FCVal | 0.249 | 0.249 | 0.000 | 1.00e+00 | 0.244 | 0.244 | 0.000 | 1.00e+00 |
| BMI | All.MultiPRS | pT+clump.10FCVal | 0.307 | 0.249 | 0.058 | 2.54e-18 | 0.305 | 0.244 | 0.061 | 2.24e-26 |
| BMI | LDpred1.Inf | pT+clump.10FCVal | 0.274 | 0.249 | 0.026 | 2.12e-13 | 0.264 | 0.244 | 0.020 | 5.30e-03 |
| BMI | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.285 | 0.249 | 0.036 | 9.11e-24 | 0.283 | 0.244 | 0.039 | 1.02e-07 |
| BMI | LDpred1.10FCVal | pT+clump.10FCVal | 0.279 | 0.249 | 0.030 | 5.32e-16 | 0.271 | 0.244 | 0.027 | 2.79e-04 |
| BMI | lassosum.PseudoVal | pT+clump.10FCVal | 0.216 | 0.249 | -0.033 | 1.27e-11 | 0.225 | 0.244 | -0.019 | 5.37e-02 |
| BMI | PRScs.PseudoVal | pT+clump.10FCVal | 0.282 | 0.249 | 0.034 | 9.68e-20 | 0.282 | 0.244 | 0.038 | 4.85e-07 |
| BMI | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.271 | 0.211 | 0.060 | 1.50e-18 | 0.274 | 0.224 | 0.050 | 9.43e-10 |
| BMI | LDpred1.10FCVal | SBayesR.PseudoVal | 0.279 | 0.211 | 0.067 | 4.00e-81 | 0.271 | 0.224 | 0.047 | 2.08e-11 |
| BMI | PRScs.10FCVal | SBayesR.PseudoVal | 0.292 | 0.211 | 0.080 | 5.46e-70 | 0.282 | 0.224 | 0.058 | 2.74e-10 |
| BMI | LDpred2.10FCVal | SBayesR.PseudoVal | 0.301 | 0.211 | 0.090 | 3.56e-115 | 0.296 | 0.224 | 0.072 | 9.73e-20 |
| BMI | lassosum.10FCVal | SBayesR.PseudoVal | 0.300 | 0.211 | 0.089 | 9.78e-100 | 0.298 | 0.224 | 0.074 | 1.52e-18 |
| BMI | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.256 | 0.211 | 0.045 | 3.19e-75 | 0.261 | 0.224 | 0.037 | 1.36e-14 |
| BMI | lassosum.MultiPRS | SBayesR.PseudoVal | 0.305 | 0.211 | 0.093 | 5.75e-44 | 0.305 | 0.224 | 0.081 | 1.44e-25 |
| BMI | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.302 | 0.211 | 0.091 | 9.68e-42 | 0.299 | 0.224 | 0.075 | 6.96e-23 |
| BMI | All.MultiPRS | SBayesR.PseudoVal | 0.307 | 0.211 | 0.095 | 1.34e-45 | 0.305 | 0.224 | 0.081 | 7.00e-27 |
| BMI | PRScs.PseudoVal | SBayesR.PseudoVal | 0.282 | 0.211 | 0.071 | 2.10e-112 | 0.282 | 0.224 | 0.058 | 2.44e-20 |
| BMI | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.289 | 0.211 | 0.077 | 1.36e-30 | 0.281 | 0.224 | 0.057 | 8.42e-15 |
| BMI | pT+clump.10FCVal | SBayesR.PseudoVal | 0.249 | 0.211 | 0.037 | 4.18e-14 | 0.244 | 0.224 | 0.020 | 4.33e-02 |
| BMI | lassosum.PseudoVal | SBayesR.PseudoVal | 0.216 | 0.211 | 0.004 | 1.10e-01 | 0.225 | 0.224 | 0.001 | 8.44e-01 |
| BMI | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.211 | 0.211 | 0.000 | 1.00e+00 | 0.224 | 0.224 | 0.000 | 1.00e+00 |
| BMI | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.285 | 0.211 | 0.073 | 6.58e-118 | 0.283 | 0.224 | 0.059 | 7.93e-21 |
| BMI | SBLUP.Inf | SBayesR.PseudoVal | 0.293 | 0.211 | 0.081 | 7.27e-68 | 0.286 | 0.224 | 0.062 | 3.83e-11 |
| BMI | LDpred2.Inf | SBayesR.PseudoVal | 0.291 | 0.211 | 0.080 | 2.75e-66 | 0.284 | 0.224 | 0.060 | 9.04e-11 |
| BMI | LDpred1.Inf | SBayesR.PseudoVal | 0.274 | 0.211 | 0.063 | 3.89e-43 | 0.264 | 0.224 | 0.040 | 1.51e-05 |
| BMI | PRScs.MultiPRS | SBayesR.PseudoVal | 0.301 | 0.211 | 0.090 | 6.17e-41 | 0.296 | 0.224 | 0.072 | 6.39e-20 |
| BMI | LDpred2.10FCVal | SBLUP.Inf | 0.301 | 0.293 | 0.009 | 4.62e-10 | 0.296 | 0.286 | 0.010 | 4.91e-04 |
| BMI | PRScs.PseudoVal | SBLUP.Inf | 0.282 | 0.293 | -0.010 | 2.20e-04 | 0.282 | 0.286 | -0.004 | 4.96e-01 |
| BMI | LDpred1.10FCVal | SBLUP.Inf | 0.279 | 0.293 | -0.014 | 1.93e-07 | 0.271 | 0.286 | -0.014 | 8.23e-03 |
| BMI | LDpred2.Inf | SBLUP.Inf | 0.291 | 0.293 | -0.001 | 3.68e-02 | 0.284 | 0.286 | -0.001 | 2.93e-01 |
| BMI | SBayesR.PseudoVal | SBLUP.Inf | 0.211 | 0.293 | -0.081 | 7.27e-68 | 0.224 | 0.286 | -0.062 | 3.83e-11 |
| BMI | lassosum.10FCVal | SBLUP.Inf | 0.300 | 0.293 | 0.008 | 1.15e-07 | 0.298 | 0.286 | 0.012 | 3.01e-05 |
| BMI | pT+clump.MultiPRS | SBLUP.Inf | 0.271 | 0.293 | -0.021 | 1.20e-03 | 0.274 | 0.286 | -0.012 | 3.00e-02 |
| BMI | lassosum.MultiPRS | SBLUP.Inf | 0.305 | 0.293 | 0.012 | 6.56e-02 | 0.305 | 0.286 | 0.019 | 2.91e-08 |
| BMI | PRScs.10FCVal | SBLUP.Inf | 0.292 | 0.293 | -0.001 | 6.72e-01 | 0.282 | 0.286 | -0.004 | 1.48e-01 |
| BMI | PRScs.MultiPRS | SBLUP.Inf | 0.301 | 0.293 | 0.009 | 1.85e-01 | 0.296 | 0.286 | 0.010 | 7.74e-04 |
| BMI | LDpred1.MultiPRS | SBLUP.Inf | 0.289 | 0.293 | -0.004 | 5.61e-01 | 0.281 | 0.286 | -0.005 | 2.89e-01 |
| BMI | LDpred2.PseudoVal | SBLUP.Inf | 0.256 | 0.293 | -0.036 | 2.25e-21 | 0.261 | 0.286 | -0.025 | 1.56e-03 |
| BMI | LDpred1.Inf | SBLUP.Inf | 0.274 | 0.293 | -0.018 | 6.95e-21 | 0.264 | 0.286 | -0.022 | 3.52e-08 |
| BMI | LDpred2.MultiPRS | SBLUP.Inf | 0.302 | 0.293 | 0.010 | 1.47e-01 | 0.299 | 0.286 | 0.013 | 1.63e-05 |
| BMI | All.MultiPRS | SBLUP.Inf | 0.307 | 0.293 | 0.014 | 3.30e-02 | 0.305 | 0.286 | 0.019 | 4.50e-11 |
| BMI | SBLUP.Inf | SBLUP.Inf | 0.293 | 0.293 | 0.000 | 1.00e+00 | 0.286 | 0.286 | 0.000 | 1.00e+00 |
| BMI | DBSLMM.PseudoVal | SBLUP.Inf | 0.285 | 0.293 | -0.008 | 1.44e-03 | 0.283 | 0.286 | -0.003 | 5.94e-01 |
| BMI | pT+clump.10FCVal | SBLUP.Inf | 0.249 | 0.293 | -0.044 | 5.05e-47 | 0.244 | 0.286 | -0.042 | 1.09e-11 |
| BMI | lassosum.PseudoVal | SBLUP.Inf | 0.216 | 0.293 | -0.077 | 1.34e-60 | 0.225 | 0.286 | -0.061 | 1.28e-10 |
| T2D | LDpred2.PseudoVal | All.MultiPRS | 0.194 | 0.242 | -0.048 | 1.79e-12 | 0.190 | 0.242 | -0.052 | 6.78e-17 |
| T2D | pT+clump.10FCVal | All.MultiPRS | 0.167 | 0.242 | -0.075 | 7.70e-28 | 0.173 | 0.242 | -0.069 | 5.83e-19 |
| T2D | LDpred2.MultiPRS | All.MultiPRS | 0.242 | 0.242 | 0.000 | 9.96e-01 | 0.240 | 0.242 | -0.002 | 2.13e-01 |
| T2D | PRScs.PseudoVal | All.MultiPRS | 0.226 | 0.242 | -0.015 | 2.34e-02 | 0.226 | 0.242 | -0.016 | 4.31e-05 |
| T2D | LDpred1.Inf | All.MultiPRS | 0.187 | 0.242 | -0.055 | 9.62e-16 | 0.184 | 0.242 | -0.057 | 4.48e-18 |
| T2D | PRScs.MultiPRS | All.MultiPRS | 0.235 | 0.242 | -0.007 | 2.83e-01 | 0.237 | 0.242 | -0.005 | 1.15e-01 |
| T2D | LDpred2.Inf | All.MultiPRS | 0.189 | 0.242 | -0.053 | 1.10e-14 | 0.185 | 0.242 | -0.057 | 3.30e-18 |
| T2D | lassosum.10FCVal | All.MultiPRS | 0.214 | 0.242 | -0.028 | 3.25e-05 | 0.216 | 0.242 | -0.026 | 1.79e-07 |
| T2D | LDpred2.10FCVal | All.MultiPRS | 0.237 | 0.242 | -0.005 | 4.46e-01 | 0.238 | 0.242 | -0.004 | 1.45e-01 |
| T2D | lassosum.MultiPRS | All.MultiPRS | 0.237 | 0.242 | -0.004 | 5.05e-01 | 0.240 | 0.242 | -0.001 | 5.62e-01 |
| T2D | LDpred1.10FCVal | All.MultiPRS | 0.232 | 0.242 | -0.010 | 1.43e-01 | 0.230 | 0.242 | -0.012 | 5.10e-04 |
| T2D | All.MultiPRS | All.MultiPRS | 0.242 | 0.242 | 0.000 | 1.00e+00 | 0.242 | 0.242 | 0.000 | 1.00e+00 |
| T2D | SBayesR.PseudoVal | All.MultiPRS | 0.226 | 0.242 | -0.016 | 2.17e-02 | 0.227 | 0.242 | -0.014 | 2.02e-04 |
| T2D | LDpred1.MultiPRS | All.MultiPRS | 0.232 | 0.242 | -0.010 | 5.11e-09 | 0.231 | 0.242 | -0.011 | 8.86e-04 |
| T2D | pT+clump.MultiPRS | All.MultiPRS | 0.212 | 0.242 | -0.030 | 8.65e-06 | 0.215 | 0.242 | -0.027 | 1.25e-07 |
| T2D | lassosum.PseudoVal | All.MultiPRS | 0.206 | 0.242 | -0.036 | 9.54e-08 | 0.214 | 0.242 | -0.027 | 8.48e-07 |
| T2D | PRScs.10FCVal | All.MultiPRS | 0.228 | 0.242 | -0.014 | 3.88e-02 | 0.227 | 0.242 | -0.015 | 1.01e-04 |
| T2D | DBSLMM.PseudoVal | All.MultiPRS | 0.229 | 0.242 | -0.013 | 6.19e-02 | 0.226 | 0.242 | -0.015 | 1.77e-05 |
| T2D | SBLUP.Inf | All.MultiPRS | 0.189 | 0.242 | -0.053 | 1.08e-14 | 0.184 | 0.242 | -0.057 | 1.24e-18 |
| T2D | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.226 | 0.229 | -0.003 | 2.83e-01 | 0.227 | 0.226 | 0.001 | 8.61e-01 |
| T2D | LDpred2.Inf | DBSLMM.PseudoVal | 0.189 | 0.229 | -0.040 | 1.66e-45 | 0.185 | 0.226 | -0.041 | 2.09e-13 |
| T2D | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.167 | 0.229 | -0.062 | 1.53e-47 | 0.173 | 0.226 | -0.054 | 2.59e-10 |
| T2D | PRScs.10FCVal | DBSLMM.PseudoVal | 0.228 | 0.229 | -0.001 | 5.03e-01 | 0.227 | 0.226 | 0.001 | 8.31e-01 |
| T2D | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.226 | 0.229 | -0.003 | 1.75e-01 | 0.226 | 0.226 | 0.000 | 9.03e-01 |
| T2D | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.212 | 0.229 | -0.018 | 1.01e-02 | 0.215 | 0.226 | -0.011 | 2.49e-02 |
| T2D | LDpred1.Inf | DBSLMM.PseudoVal | 0.187 | 0.229 | -0.042 | 4.35e-48 | 0.184 | 0.226 | -0.042 | 2.33e-13 |
| T2D | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.237 | 0.229 | 0.008 | 2.28e-01 | 0.240 | 0.226 | 0.014 | 1.14e-04 |
| T2D | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.237 | 0.229 | 0.008 | 8.86e-05 | 0.238 | 0.226 | 0.011 | 2.47e-03 |
| T2D | lassosum.10FCVal | DBSLMM.PseudoVal | 0.214 | 0.229 | -0.016 | 1.77e-07 | 0.216 | 0.226 | -0.011 | 6.96e-02 |
| T2D | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.232 | 0.229 | 0.003 | 1.96e-01 | 0.230 | 0.226 | 0.004 | 3.65e-01 |
| T2D | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.232 | 0.229 | 0.003 | 6.74e-01 | 0.231 | 0.226 | 0.005 | 2.77e-01 |
| T2D | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.235 | 0.229 | 0.005 | 4.27e-01 | 0.237 | 0.226 | 0.011 | 2.46e-03 |
| T2D | All.MultiPRS | DBSLMM.PseudoVal | 0.242 | 0.229 | 0.013 | 6.19e-02 | 0.242 | 0.226 | 0.015 | 1.77e-05 |
| T2D | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.194 | 0.229 | -0.035 | 2.16e-38 | 0.190 | 0.226 | -0.037 | 1.28e-11 |
| T2D | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.229 | 0.229 | 0.000 | 1.00e+00 | 0.226 | 0.226 | 0.000 | 1.00e+00 |
| T2D | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.242 | 0.229 | 0.013 | 6.08e-02 | 0.240 | 0.226 | 0.014 | 2.71e-04 |
| T2D | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.206 | 0.229 | -0.024 | 1.17e-10 | 0.214 | 0.226 | -0.012 | 9.36e-02 |
| T2D | SBLUP.Inf | DBSLMM.PseudoVal | 0.189 | 0.229 | -0.040 | 4.70e-46 | 0.184 | 0.226 | -0.042 | 4.78e-14 |
| T2D | SBayesR.PseudoVal | lassosum.MultiPRS | 0.226 | 0.237 | -0.011 | 1.02e-01 | 0.227 | 0.240 | -0.013 | 1.37e-03 |
| T2D | LDpred2.10FCVal | lassosum.MultiPRS | 0.237 | 0.237 | -0.001 | 9.22e-01 | 0.238 | 0.240 | -0.003 | 2.78e-01 |
| T2D | LDpred1.10FCVal | lassosum.MultiPRS | 0.232 | 0.237 | -0.005 | 4.23e-01 | 0.230 | 0.240 | -0.010 | 5.87e-04 |
| T2D | LDpred2.Inf | lassosum.MultiPRS | 0.189 | 0.237 | -0.048 | 1.60e-12 | 0.185 | 0.240 | -0.055 | 6.80e-18 |
| T2D | PRScs.10FCVal | lassosum.MultiPRS | 0.228 | 0.237 | -0.010 | 1.61e-01 | 0.227 | 0.240 | -0.013 | 2.89e-04 |
| T2D | PRScs.MultiPRS | lassosum.MultiPRS | 0.235 | 0.237 | -0.003 | 6.81e-01 | 0.237 | 0.240 | -0.003 | 2.05e-01 |
| T2D | pT+clump.MultiPRS | lassosum.MultiPRS | 0.212 | 0.237 | -0.026 | 1.46e-04 | 0.215 | 0.240 | -0.025 | 5.80e-08 |
| T2D | lassosum.MultiPRS | lassosum.MultiPRS | 0.237 | 0.237 | 0.000 | 1.00e+00 | 0.240 | 0.240 | 0.000 | 1.00e+00 |
| T2D | lassosum.PseudoVal | lassosum.MultiPRS | 0.206 | 0.237 | -0.032 | 3.12e-06 | 0.214 | 0.240 | -0.026 | 1.36e-07 |
| T2D | LDpred2.MultiPRS | lassosum.MultiPRS | 0.242 | 0.237 | 0.005 | 5.03e-01 | 0.240 | 0.240 | 0.000 | 9.04e-01 |
| T2D | LDpred1.MultiPRS | lassosum.MultiPRS | 0.232 | 0.237 | -0.005 | 4.32e-01 | 0.231 | 0.240 | -0.009 | 1.01e-03 |
| T2D | LDpred2.PseudoVal | lassosum.MultiPRS | 0.194 | 0.237 | -0.044 | 1.67e-10 | 0.190 | 0.240 | -0.051 | 7.52e-17 |
| T2D | LDpred1.Inf | lassosum.MultiPRS | 0.187 | 0.237 | -0.050 | 1.74e-13 | 0.184 | 0.240 | -0.056 | 2.15e-18 |
| T2D | pT+clump.10FCVal | lassosum.MultiPRS | 0.167 | 0.237 | -0.071 | 5.84e-25 | 0.173 | 0.240 | -0.068 | 6.94e-20 |
| T2D | lassosum.10FCVal | lassosum.MultiPRS | 0.214 | 0.237 | -0.024 | 4.82e-04 | 0.216 | 0.240 | -0.025 | 4.04e-09 |
| T2D | SBLUP.Inf | lassosum.MultiPRS | 0.189 | 0.237 | -0.048 | 1.59e-12 | 0.184 | 0.240 | -0.056 | 1.38e-18 |
| T2D | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.229 | 0.237 | -0.008 | 2.28e-01 | 0.226 | 0.240 | -0.014 | 1.14e-04 |
| T2D | All.MultiPRS | lassosum.MultiPRS | 0.242 | 0.237 | 0.004 | 5.05e-01 | 0.242 | 0.240 | 0.001 | 5.62e-01 |
| T2D | PRScs.PseudoVal | lassosum.MultiPRS | 0.226 | 0.237 | -0.011 | 1.09e-01 | 0.226 | 0.240 | -0.014 | 7.36e-05 |
| T2D | LDpred1.Inf | lassosum.PseudoVal | 0.187 | 0.206 | -0.019 | 2.56e-04 | 0.184 | 0.214 | -0.030 | 2.87e-03 |
| T2D | PRScs.10FCVal | lassosum.PseudoVal | 0.228 | 0.206 | 0.022 | 2.96e-10 | 0.227 | 0.214 | 0.013 | 6.75e-02 |
| T2D | LDpred2.MultiPRS | lassosum.PseudoVal | 0.242 | 0.206 | 0.036 | 9.62e-08 | 0.240 | 0.214 | 0.026 | 7.12e-06 |
| T2D | lassosum.PseudoVal | lassosum.PseudoVal | 0.206 | 0.206 | 0.000 | 1.00e+00 | 0.214 | 0.214 | 0.000 | 1.00e+00 |
| T2D | PRScs.PseudoVal | lassosum.PseudoVal | 0.226 | 0.206 | 0.021 | 5.80e-09 | 0.226 | 0.214 | 0.012 | 1.03e-01 |
| T2D | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.229 | 0.206 | 0.024 | 1.17e-10 | 0.226 | 0.214 | 0.012 | 9.36e-02 |
| T2D | SBLUP.Inf | lassosum.PseudoVal | 0.189 | 0.206 | -0.017 | 1.30e-03 | 0.184 | 0.214 | -0.030 | 3.09e-03 |
| T2D | LDpred2.Inf | lassosum.PseudoVal | 0.189 | 0.206 | -0.017 | 1.33e-03 | 0.185 | 0.214 | -0.029 | 4.15e-03 |
| T2D | pT+clump.10FCVal | lassosum.PseudoVal | 0.167 | 0.206 | -0.039 | 3.94e-34 | 0.173 | 0.214 | -0.042 | 4.23e-11 |
| T2D | LDpred2.10FCVal | lassosum.PseudoVal | 0.237 | 0.206 | 0.031 | 1.68e-30 | 0.238 | 0.214 | 0.023 | 1.38e-05 |
| T2D | lassosum.MultiPRS | lassosum.PseudoVal | 0.237 | 0.206 | 0.032 | 3.12e-06 | 0.240 | 0.214 | 0.026 | 1.36e-07 |
| T2D | LDpred1.10FCVal | lassosum.PseudoVal | 0.232 | 0.206 | 0.026 | 1.05e-19 | 0.230 | 0.214 | 0.016 | 5.85e-03 |
| T2D | All.MultiPRS | lassosum.PseudoVal | 0.242 | 0.206 | 0.036 | 9.54e-08 | 0.242 | 0.214 | 0.027 | 8.48e-07 |
| T2D | SBayesR.PseudoVal | lassosum.PseudoVal | 0.226 | 0.206 | 0.021 | 1.83e-18 | 0.227 | 0.214 | 0.013 | 5.67e-03 |
| T2D | lassosum.10FCVal | lassosum.PseudoVal | 0.214 | 0.206 | 0.008 | 6.43e-03 | 0.216 | 0.214 | 0.001 | 8.10e-01 |
| T2D | pT+clump.MultiPRS | lassosum.PseudoVal | 0.212 | 0.206 | 0.006 | 3.79e-01 | 0.215 | 0.214 | 0.001 | 9.15e-01 |
| T2D | LDpred2.PseudoVal | lassosum.PseudoVal | 0.194 | 0.206 | -0.012 | 1.71e-02 | 0.190 | 0.214 | -0.025 | 1.22e-02 |
| T2D | LDpred1.MultiPRS | lassosum.PseudoVal | 0.232 | 0.206 | 0.026 | 1.04e-04 | 0.231 | 0.214 | 0.017 | 2.81e-03 |
| T2D | PRScs.MultiPRS | lassosum.PseudoVal | 0.235 | 0.206 | 0.029 | 2.30e-05 | 0.237 | 0.214 | 0.023 | 9.22e-05 |
| T2D | lassosum.10FCVal | lassosum.10FCVal | 0.214 | 0.214 | 0.000 | 1.00e+00 | 0.216 | 0.216 | 0.000 | 1.00e+00 |
| T2D | pT+clump.MultiPRS | lassosum.10FCVal | 0.212 | 0.214 | -0.002 | 7.70e-01 | 0.215 | 0.216 | -0.001 | 9.07e-01 |
| T2D | LDpred2.Inf | lassosum.10FCVal | 0.189 | 0.214 | -0.025 | 4.13e-13 | 0.185 | 0.216 | -0.031 | 4.35e-06 |
| T2D | SBayesR.PseudoVal | lassosum.10FCVal | 0.226 | 0.214 | 0.013 | 1.35e-05 | 0.227 | 0.216 | 0.012 | 4.36e-02 |
| T2D | PRScs.MultiPRS | lassosum.10FCVal | 0.235 | 0.214 | 0.021 | 2.13e-03 | 0.237 | 0.216 | 0.021 | 1.55e-06 |
| T2D | SBLUP.Inf | lassosum.10FCVal | 0.189 | 0.214 | -0.025 | 3.02e-13 | 0.184 | 0.216 | -0.031 | 1.96e-06 |
| T2D | lassosum.MultiPRS | lassosum.10FCVal | 0.237 | 0.214 | 0.024 | 4.82e-04 | 0.240 | 0.216 | 0.025 | 4.04e-09 |
| T2D | PRScs.10FCVal | lassosum.10FCVal | 0.228 | 0.214 | 0.014 | 7.41e-10 | 0.227 | 0.216 | 0.012 | 1.22e-02 |
| T2D | LDpred2.MultiPRS | lassosum.10FCVal | 0.242 | 0.214 | 0.028 | 3.19e-05 | 0.240 | 0.216 | 0.024 | 1.59e-06 |
| T2D | PRScs.PseudoVal | lassosum.10FCVal | 0.226 | 0.214 | 0.013 | 4.14e-08 | 0.226 | 0.216 | 0.010 | 2.86e-02 |
| T2D | LDpred2.PseudoVal | lassosum.10FCVal | 0.194 | 0.214 | -0.020 | 8.22e-10 | 0.190 | 0.216 | -0.026 | 4.30e-05 |
| T2D | LDpred1.MultiPRS | lassosum.10FCVal | 0.232 | 0.214 | 0.018 | 6.86e-03 | 0.231 | 0.216 | 0.015 | 5.02e-05 |
| T2D | pT+clump.10FCVal | lassosum.10FCVal | 0.167 | 0.214 | -0.047 | 2.14e-26 | 0.173 | 0.216 | -0.043 | 8.50e-07 |
| T2D | All.MultiPRS | lassosum.10FCVal | 0.242 | 0.214 | 0.028 | 3.25e-05 | 0.242 | 0.216 | 0.026 | 1.79e-07 |
| T2D | LDpred1.Inf | lassosum.10FCVal | 0.187 | 0.214 | -0.027 | 3.27e-15 | 0.184 | 0.216 | -0.031 | 2.25e-06 |
| T2D | DBSLMM.PseudoVal | lassosum.10FCVal | 0.229 | 0.214 | 0.016 | 1.77e-07 | 0.226 | 0.216 | 0.011 | 6.96e-02 |
| T2D | LDpred1.10FCVal | lassosum.10FCVal | 0.232 | 0.214 | 0.018 | 3.68e-22 | 0.230 | 0.216 | 0.014 | 1.07e-04 |
| T2D | lassosum.PseudoVal | lassosum.10FCVal | 0.206 | 0.214 | -0.008 | 6.43e-03 | 0.214 | 0.216 | -0.001 | 8.10e-01 |
| T2D | LDpred2.10FCVal | lassosum.10FCVal | 0.237 | 0.214 | 0.023 | 1.36e-34 | 0.238 | 0.216 | 0.022 | 4.27e-09 |
| T2D | LDpred2.10FCVal | LDpred1.MultiPRS | 0.237 | 0.232 | 0.005 | 4.93e-01 | 0.238 | 0.231 | 0.007 | 8.48e-04 |
| T2D | PRScs.PseudoVal | LDpred1.MultiPRS | 0.226 | 0.232 | -0.006 | 4.13e-01 | 0.226 | 0.231 | -0.005 | 1.29e-01 |
| T2D | LDpred2.Inf | LDpred1.MultiPRS | 0.189 | 0.232 | -0.043 | 3.59e-10 | 0.185 | 0.231 | -0.046 | 2.78e-14 |
| T2D | pT+clump.10FCVal | LDpred1.MultiPRS | 0.167 | 0.232 | -0.065 | 2.36e-21 | 0.173 | 0.231 | -0.058 | 3.83e-13 |
| T2D | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.226 | 0.232 | -0.006 | 3.96e-01 | 0.227 | 0.231 | -0.004 | 4.27e-01 |
| T2D | All.MultiPRS | LDpred1.MultiPRS | 0.242 | 0.232 | 0.010 | 5.11e-09 | 0.242 | 0.231 | 0.011 | 8.86e-04 |
| T2D | LDpred1.10FCVal | LDpred1.MultiPRS | 0.232 | 0.232 | 0.000 | 9.86e-01 | 0.230 | 0.231 | -0.001 | 8.06e-02 |
| T2D | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.232 | 0.232 | 0.000 | 1.00e+00 | 0.231 | 0.231 | 0.000 | 1.00e+00 |
| T2D | PRScs.10FCVal | LDpred1.MultiPRS | 0.228 | 0.232 | -0.004 | 5.37e-01 | 0.227 | 0.231 | -0.004 | 2.60e-01 |
| T2D | lassosum.PseudoVal | LDpred1.MultiPRS | 0.206 | 0.232 | -0.026 | 1.04e-04 | 0.214 | 0.231 | -0.017 | 2.81e-03 |
| T2D | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.212 | 0.232 | -0.020 | 2.67e-03 | 0.215 | 0.231 | -0.016 | 1.96e-03 |
| T2D | LDpred1.Inf | LDpred1.MultiPRS | 0.187 | 0.232 | -0.045 | 4.85e-11 | 0.184 | 0.231 | -0.047 | 3.90e-16 |
| T2D | lassosum.MultiPRS | LDpred1.MultiPRS | 0.237 | 0.232 | 0.005 | 4.32e-01 | 0.240 | 0.231 | 0.009 | 1.01e-03 |
| T2D | PRScs.MultiPRS | LDpred1.MultiPRS | 0.235 | 0.232 | 0.003 | 7.10e-01 | 0.237 | 0.231 | 0.006 | 2.13e-02 |
| T2D | lassosum.10FCVal | LDpred1.MultiPRS | 0.214 | 0.232 | -0.018 | 6.86e-03 | 0.216 | 0.231 | -0.015 | 5.02e-05 |
| T2D | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.194 | 0.232 | -0.038 | 2.20e-08 | 0.190 | 0.231 | -0.041 | 5.68e-13 |
| T2D | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.229 | 0.232 | -0.003 | 6.74e-01 | 0.226 | 0.231 | -0.005 | 2.77e-01 |
| T2D | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.242 | 0.232 | 0.010 | 1.47e-01 | 0.240 | 0.231 | 0.009 | 7.28e-03 |
| T2D | SBLUP.Inf | LDpred1.MultiPRS | 0.189 | 0.232 | -0.043 | 3.54e-10 | 0.184 | 0.231 | -0.047 | 6.09e-15 |
| T2D | All.MultiPRS | LDpred1.Inf | 0.242 | 0.187 | 0.055 | 9.62e-16 | 0.242 | 0.184 | 0.057 | 4.48e-18 |
| T2D | LDpred2.PseudoVal | LDpred1.Inf | 0.194 | 0.187 | 0.007 | 1.43e-09 | 0.190 | 0.184 | 0.005 | 1.43e-02 |
| T2D | SBayesR.PseudoVal | LDpred1.Inf | 0.226 | 0.187 | 0.039 | 9.83e-20 | 0.227 | 0.184 | 0.043 | 4.40e-07 |
| T2D | LDpred2.MultiPRS | LDpred1.Inf | 0.242 | 0.187 | 0.055 | 7.95e-16 | 0.240 | 0.184 | 0.056 | 3.13e-17 |
| T2D | PRScs.PseudoVal | LDpred1.Inf | 0.226 | 0.187 | 0.039 | 3.13e-59 | 0.226 | 0.184 | 0.042 | 3.43e-18 |
| T2D | LDpred1.Inf | LDpred1.Inf | 0.187 | 0.187 | 0.000 | 1.00e+00 | 0.184 | 0.184 | 0.000 | 1.00e+00 |
| T2D | pT+clump.10FCVal | LDpred1.Inf | 0.167 | 0.187 | -0.020 | 4.17e-04 | 0.173 | 0.184 | -0.012 | 3.00e-01 |
| T2D | PRScs.MultiPRS | LDpred1.Inf | 0.235 | 0.187 | 0.048 | 2.98e-12 | 0.237 | 0.184 | 0.053 | 8.26e-20 |
| T2D | lassosum.PseudoVal | LDpred1.Inf | 0.206 | 0.187 | 0.019 | 2.56e-04 | 0.214 | 0.184 | 0.030 | 2.87e-03 |
| T2D | pT+clump.MultiPRS | LDpred1.Inf | 0.212 | 0.187 | 0.025 | 3.52e-04 | 0.215 | 0.184 | 0.031 | 1.76e-04 |
| T2D | LDpred1.MultiPRS | LDpred1.Inf | 0.232 | 0.187 | 0.045 | 4.85e-11 | 0.231 | 0.184 | 0.047 | 3.90e-16 |
| T2D | PRScs.10FCVal | LDpred1.Inf | 0.228 | 0.187 | 0.041 | 1.10e-60 | 0.227 | 0.184 | 0.043 | 5.70e-19 |
| T2D | LDpred2.Inf | LDpred1.Inf | 0.189 | 0.187 | 0.002 | 5.99e-02 | 0.185 | 0.184 | 0.001 | 7.02e-01 |
| T2D | SBLUP.Inf | LDpred1.Inf | 0.189 | 0.187 | 0.002 | 5.11e-02 | 0.184 | 0.184 | 0.000 | 9.57e-01 |
| T2D | DBSLMM.PseudoVal | LDpred1.Inf | 0.229 | 0.187 | 0.042 | 4.35e-48 | 0.226 | 0.184 | 0.042 | 2.33e-13 |
| T2D | lassosum.MultiPRS | LDpred1.Inf | 0.237 | 0.187 | 0.050 | 1.74e-13 | 0.240 | 0.184 | 0.056 | 2.15e-18 |
| T2D | LDpred1.10FCVal | LDpred1.Inf | 0.232 | 0.187 | 0.045 | 2.88e-57 | 0.230 | 0.184 | 0.046 | 1.11e-16 |
| T2D | lassosum.10FCVal | LDpred1.Inf | 0.214 | 0.187 | 0.027 | 3.27e-15 | 0.216 | 0.184 | 0.031 | 2.25e-06 |
| T2D | LDpred2.10FCVal | LDpred1.Inf | 0.237 | 0.187 | 0.050 | 1.11e-61 | 0.238 | 0.184 | 0.053 | 9.48e-20 |
| T2D | LDpred1.Inf | LDpred1.10FCVal | 0.187 | 0.232 | -0.045 | 2.88e-57 | 0.184 | 0.230 | -0.046 | 1.11e-16 |
| T2D | PRScs.MultiPRS | LDpred1.10FCVal | 0.235 | 0.232 | 0.003 | 6.97e-01 | 0.237 | 0.230 | 0.007 | 1.19e-02 |
| T2D | LDpred1.10FCVal | LDpred1.10FCVal | 0.232 | 0.232 | 0.000 | 1.00e+00 | 0.230 | 0.230 | 0.000 | 1.00e+00 |
| T2D | All.MultiPRS | LDpred1.10FCVal | 0.242 | 0.232 | 0.010 | 1.43e-01 | 0.242 | 0.230 | 0.012 | 5.10e-04 |
| T2D | SBayesR.PseudoVal | LDpred1.10FCVal | 0.226 | 0.232 | -0.006 | 1.62e-02 | 0.227 | 0.230 | -0.003 | 5.42e-01 |
| T2D | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.229 | 0.232 | -0.003 | 1.96e-01 | 0.226 | 0.230 | -0.004 | 3.65e-01 |
| T2D | pT+clump.MultiPRS | LDpred1.10FCVal | 0.212 | 0.232 | -0.020 | 2.87e-03 | 0.215 | 0.230 | -0.015 | 3.77e-03 |
| T2D | lassosum.MultiPRS | LDpred1.10FCVal | 0.237 | 0.232 | 0.005 | 4.23e-01 | 0.240 | 0.230 | 0.010 | 5.87e-04 |
| T2D | PRScs.10FCVal | LDpred1.10FCVal | 0.228 | 0.232 | -0.004 | 1.19e-02 | 0.227 | 0.230 | -0.003 | 3.63e-01 |
| T2D | lassosum.10FCVal | LDpred1.10FCVal | 0.214 | 0.232 | -0.018 | 3.68e-22 | 0.216 | 0.230 | -0.014 | 1.07e-04 |
| T2D | SBLUP.Inf | LDpred1.10FCVal | 0.189 | 0.232 | -0.043 | 2.35e-47 | 0.184 | 0.230 | -0.046 | 2.36e-15 |
| T2D | LDpred2.PseudoVal | LDpred1.10FCVal | 0.194 | 0.232 | -0.038 | 3.51e-41 | 0.190 | 0.230 | -0.041 | 2.89e-13 |
| T2D | pT+clump.10FCVal | LDpred1.10FCVal | 0.167 | 0.232 | -0.065 | 2.93e-57 | 0.173 | 0.230 | -0.058 | 1.33e-12 |
| T2D | LDpred2.MultiPRS | LDpred1.10FCVal | 0.242 | 0.232 | 0.010 | 1.41e-01 | 0.240 | 0.230 | 0.010 | 4.09e-03 |
| T2D | PRScs.PseudoVal | LDpred1.10FCVal | 0.226 | 0.232 | -0.005 | 8.16e-04 | 0.226 | 0.230 | -0.004 | 1.90e-01 |
| T2D | LDpred2.10FCVal | LDpred1.10FCVal | 0.237 | 0.232 | 0.005 | 1.18e-05 | 0.238 | 0.230 | 0.008 | 3.40e-04 |
| T2D | LDpred1.MultiPRS | LDpred1.10FCVal | 0.232 | 0.232 | 0.000 | 9.86e-01 | 0.231 | 0.230 | 0.001 | 8.06e-02 |
| T2D | LDpred2.Inf | LDpred1.10FCVal | 0.189 | 0.232 | -0.043 | 1.12e-46 | 0.185 | 0.230 | -0.045 | 1.15e-14 |
| T2D | lassosum.PseudoVal | LDpred1.10FCVal | 0.206 | 0.232 | -0.026 | 1.05e-19 | 0.214 | 0.230 | -0.016 | 5.85e-03 |
| T2D | LDpred1.Inf | LDpred2.MultiPRS | 0.187 | 0.242 | -0.055 | 7.95e-16 | 0.184 | 0.240 | -0.056 | 3.13e-17 |
| T2D | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.229 | 0.242 | -0.013 | 6.08e-02 | 0.226 | 0.240 | -0.014 | 2.71e-04 |
| T2D | SBLUP.Inf | LDpred2.MultiPRS | 0.189 | 0.242 | -0.053 | 9.26e-15 | 0.184 | 0.240 | -0.056 | 1.55e-17 |
| T2D | LDpred2.Inf | LDpred2.MultiPRS | 0.189 | 0.242 | -0.053 | 9.61e-15 | 0.185 | 0.240 | -0.055 | 3.12e-17 |
| T2D | pT+clump.10FCVal | LDpred2.MultiPRS | 0.167 | 0.242 | -0.075 | 7.08e-28 | 0.173 | 0.240 | -0.067 | 1.49e-18 |
| T2D | LDpred2.10FCVal | LDpred2.MultiPRS | 0.237 | 0.242 | -0.005 | 4.43e-01 | 0.238 | 0.240 | -0.002 | 4.18e-01 |
| T2D | PRScs.PseudoVal | LDpred2.MultiPRS | 0.226 | 0.242 | -0.015 | 2.30e-02 | 0.226 | 0.240 | -0.014 | 4.27e-04 |
| T2D | LDpred1.10FCVal | LDpred2.MultiPRS | 0.232 | 0.242 | -0.010 | 1.41e-01 | 0.230 | 0.240 | -0.010 | 4.09e-03 |
| T2D | All.MultiPRS | LDpred2.MultiPRS | 0.242 | 0.242 | 0.000 | 9.96e-01 | 0.242 | 0.240 | 0.002 | 2.13e-01 |
| T2D | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.226 | 0.242 | -0.016 | 2.13e-02 | 0.227 | 0.240 | -0.013 | 2.16e-03 |
| T2D | lassosum.10FCVal | LDpred2.MultiPRS | 0.214 | 0.242 | -0.028 | 3.19e-05 | 0.216 | 0.240 | -0.024 | 1.59e-06 |
| T2D | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.212 | 0.242 | -0.030 | 8.13e-06 | 0.215 | 0.240 | -0.025 | 7.69e-07 |
| T2D | lassosum.MultiPRS | LDpred2.MultiPRS | 0.237 | 0.242 | -0.005 | 5.03e-01 | 0.240 | 0.240 | 0.000 | 9.04e-01 |
| T2D | PRScs.10FCVal | LDpred2.MultiPRS | 0.228 | 0.242 | -0.014 | 3.84e-02 | 0.227 | 0.240 | -0.013 | 1.28e-03 |
| T2D | PRScs.MultiPRS | LDpred2.MultiPRS | 0.235 | 0.242 | -0.007 | 2.78e-01 | 0.237 | 0.240 | -0.003 | 3.51e-01 |
| T2D | lassosum.PseudoVal | LDpred2.MultiPRS | 0.206 | 0.242 | -0.036 | 9.62e-08 | 0.214 | 0.240 | -0.026 | 7.12e-06 |
| T2D | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.194 | 0.242 | -0.048 | 1.55e-12 | 0.190 | 0.240 | -0.050 | 6.01e-16 |
| T2D | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.232 | 0.242 | -0.010 | 1.47e-01 | 0.231 | 0.240 | -0.009 | 7.28e-03 |
| T2D | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.242 | 0.242 | 0.000 | 1.00e+00 | 0.240 | 0.240 | 0.000 | 1.00e+00 |
| T2D | LDpred1.Inf | LDpred2.10FCVal | 0.187 | 0.237 | -0.050 | 1.11e-61 | 0.184 | 0.238 | -0.053 | 9.48e-20 |
| T2D | PRScs.MultiPRS | LDpred2.10FCVal | 0.235 | 0.237 | -0.002 | 7.55e-01 | 0.237 | 0.238 | -0.001 | 7.47e-01 |
| T2D | LDpred2.MultiPRS | LDpred2.10FCVal | 0.242 | 0.237 | 0.005 | 4.43e-01 | 0.240 | 0.238 | 0.002 | 4.18e-01 |
| T2D | PRScs.PseudoVal | LDpred2.10FCVal | 0.226 | 0.237 | -0.010 | 3.37e-11 | 0.226 | 0.238 | -0.012 | 1.00e-04 |
| T2D | LDpred2.10FCVal | LDpred2.10FCVal | 0.237 | 0.237 | 0.000 | 1.00e+00 | 0.238 | 0.238 | 0.000 | 1.00e+00 |
| T2D | lassosum.MultiPRS | LDpred2.10FCVal | 0.237 | 0.237 | 0.001 | 9.22e-01 | 0.240 | 0.238 | 0.003 | 2.78e-01 |
| T2D | LDpred1.10FCVal | LDpred2.10FCVal | 0.232 | 0.237 | -0.005 | 1.18e-05 | 0.230 | 0.238 | -0.008 | 3.40e-04 |
| T2D | All.MultiPRS | LDpred2.10FCVal | 0.242 | 0.237 | 0.005 | 4.46e-01 | 0.242 | 0.238 | 0.004 | 1.45e-01 |
| T2D | SBayesR.PseudoVal | LDpred2.10FCVal | 0.226 | 0.237 | -0.010 | 1.45e-06 | 0.227 | 0.238 | -0.010 | 1.44e-02 |
| T2D | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.229 | 0.237 | -0.008 | 8.86e-05 | 0.226 | 0.238 | -0.011 | 2.47e-03 |
| T2D | pT+clump.MultiPRS | LDpred2.10FCVal | 0.212 | 0.237 | -0.025 | 2.32e-04 | 0.215 | 0.238 | -0.023 | 6.26e-06 |
| T2D | lassosum.PseudoVal | LDpred2.10FCVal | 0.206 | 0.237 | -0.031 | 1.68e-30 | 0.214 | 0.238 | -0.023 | 1.38e-05 |
| T2D | PRScs.10FCVal | LDpred2.10FCVal | 0.228 | 0.237 | -0.009 | 3.41e-09 | 0.227 | 0.238 | -0.011 | 4.46e-04 |
| T2D | lassosum.10FCVal | LDpred2.10FCVal | 0.214 | 0.237 | -0.023 | 1.36e-34 | 0.216 | 0.238 | -0.022 | 4.27e-09 |
| T2D | SBLUP.Inf | LDpred2.10FCVal | 0.189 | 0.237 | -0.048 | 5.94e-58 | 0.184 | 0.238 | -0.054 | 3.32e-20 |
| T2D | LDpred2.PseudoVal | LDpred2.10FCVal | 0.194 | 0.237 | -0.043 | 2.84e-54 | 0.190 | 0.238 | -0.048 | 6.64e-19 |
| T2D | pT+clump.10FCVal | LDpred2.10FCVal | 0.167 | 0.237 | -0.070 | 4.42e-69 | 0.173 | 0.238 | -0.065 | 2.26e-16 |
| T2D | LDpred2.Inf | LDpred2.10FCVal | 0.189 | 0.237 | -0.048 | 1.05e-58 | 0.185 | 0.238 | -0.053 | 6.63e-20 |
| T2D | LDpred1.MultiPRS | LDpred2.10FCVal | 0.232 | 0.237 | -0.005 | 4.93e-01 | 0.231 | 0.238 | -0.007 | 8.48e-04 |
| T2D | lassosum.10FCVal | LDpred2.PseudoVal | 0.214 | 0.194 | 0.020 | 8.22e-10 | 0.216 | 0.190 | 0.026 | 4.30e-05 |
| T2D | SBLUP.Inf | LDpred2.PseudoVal | 0.189 | 0.194 | -0.005 | 6.30e-16 | 0.184 | 0.190 | -0.005 | 1.98e-06 |
| T2D | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.226 | 0.194 | 0.033 | 7.95e-15 | 0.227 | 0.190 | 0.038 | 4.75e-06 |
| T2D | All.MultiPRS | LDpred2.PseudoVal | 0.242 | 0.194 | 0.048 | 1.79e-12 | 0.242 | 0.190 | 0.052 | 6.78e-17 |
| T2D | PRScs.PseudoVal | LDpred2.PseudoVal | 0.226 | 0.194 | 0.033 | 1.52e-47 | 0.226 | 0.190 | 0.036 | 2.87e-16 |
| T2D | lassosum.MultiPRS | LDpred2.PseudoVal | 0.237 | 0.194 | 0.044 | 1.67e-10 | 0.240 | 0.190 | 0.051 | 7.52e-17 |
| T2D | PRScs.MultiPRS | LDpred2.PseudoVal | 0.235 | 0.194 | 0.041 | 2.05e-09 | 0.237 | 0.190 | 0.047 | 2.24e-18 |
| T2D | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.242 | 0.194 | 0.048 | 1.55e-12 | 0.240 | 0.190 | 0.050 | 6.01e-16 |
| T2D | PRScs.10FCVal | LDpred2.PseudoVal | 0.228 | 0.194 | 0.034 | 3.45e-50 | 0.227 | 0.190 | 0.038 | 2.84e-17 |
| T2D | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.194 | 0.194 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| T2D | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.212 | 0.194 | 0.018 | 9.36e-03 | 0.215 | 0.190 | 0.025 | 1.59e-03 |
| T2D | pT+clump.10FCVal | LDpred2.PseudoVal | 0.167 | 0.194 | -0.027 | 1.72e-06 | 0.173 | 0.190 | -0.017 | 1.27e-01 |
| T2D | lassosum.PseudoVal | LDpred2.PseudoVal | 0.206 | 0.194 | 0.012 | 1.71e-02 | 0.214 | 0.190 | 0.025 | 1.22e-02 |
| T2D | LDpred1.Inf | LDpred2.PseudoVal | 0.187 | 0.194 | -0.007 | 1.43e-09 | 0.184 | 0.190 | -0.005 | 1.43e-02 |
| T2D | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.232 | 0.194 | 0.038 | 2.20e-08 | 0.231 | 0.190 | 0.041 | 5.68e-13 |
| T2D | LDpred1.10FCVal | LDpred2.PseudoVal | 0.232 | 0.194 | 0.038 | 3.51e-41 | 0.230 | 0.190 | 0.041 | 2.89e-13 |
| T2D | LDpred2.Inf | LDpred2.PseudoVal | 0.189 | 0.194 | -0.005 | 1.74e-18 | 0.185 | 0.190 | -0.004 | 1.65e-05 |
| T2D | LDpred2.10FCVal | LDpred2.PseudoVal | 0.237 | 0.194 | 0.043 | 2.84e-54 | 0.238 | 0.190 | 0.048 | 6.64e-19 |
| T2D | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.229 | 0.194 | 0.035 | 2.16e-38 | 0.226 | 0.190 | 0.037 | 1.28e-11 |
| T2D | pT+clump.MultiPRS | LDpred2.Inf | 0.212 | 0.189 | 0.023 | 1.07e-03 | 0.215 | 0.185 | 0.030 | 2.73e-04 |
| T2D | lassosum.MultiPRS | LDpred2.Inf | 0.237 | 0.189 | 0.048 | 1.60e-12 | 0.240 | 0.185 | 0.055 | 6.80e-18 |
| T2D | SBayesR.PseudoVal | LDpred2.Inf | 0.226 | 0.189 | 0.037 | 1.03e-17 | 0.227 | 0.185 | 0.042 | 7.51e-07 |
| T2D | All.MultiPRS | LDpred2.Inf | 0.242 | 0.189 | 0.053 | 1.10e-14 | 0.242 | 0.185 | 0.057 | 3.30e-18 |
| T2D | SBLUP.Inf | LDpred2.Inf | 0.189 | 0.189 | 0.000 | 9.69e-01 | 0.184 | 0.185 | -0.001 | 2.78e-01 |
| T2D | LDpred2.PseudoVal | LDpred2.Inf | 0.194 | 0.189 | 0.005 | 1.74e-18 | 0.190 | 0.185 | 0.004 | 1.65e-05 |
| T2D | PRScs.MultiPRS | LDpred2.Inf | 0.235 | 0.189 | 0.046 | 2.51e-11 | 0.237 | 0.185 | 0.052 | 2.32e-19 |
| T2D | LDpred2.MultiPRS | LDpred2.Inf | 0.242 | 0.189 | 0.053 | 9.61e-15 | 0.240 | 0.185 | 0.055 | 3.12e-17 |
| T2D | PRScs.10FCVal | LDpred2.Inf | 0.228 | 0.189 | 0.039 | 2.81e-57 | 0.227 | 0.185 | 0.042 | 4.31e-19 |
| T2D | LDpred1.Inf | LDpred2.Inf | 0.187 | 0.189 | -0.002 | 5.99e-02 | 0.184 | 0.185 | -0.001 | 7.02e-01 |
| T2D | LDpred1.MultiPRS | LDpred2.Inf | 0.232 | 0.189 | 0.043 | 3.59e-10 | 0.231 | 0.185 | 0.046 | 2.78e-14 |
| T2D | pT+clump.10FCVal | LDpred2.Inf | 0.167 | 0.189 | -0.022 | 1.07e-04 | 0.173 | 0.185 | -0.013 | 2.70e-01 |
| T2D | lassosum.PseudoVal | LDpred2.Inf | 0.206 | 0.189 | 0.017 | 1.33e-03 | 0.214 | 0.185 | 0.029 | 4.15e-03 |
| T2D | PRScs.PseudoVal | LDpred2.Inf | 0.226 | 0.189 | 0.037 | 3.50e-55 | 0.226 | 0.185 | 0.041 | 2.65e-18 |
| T2D | DBSLMM.PseudoVal | LDpred2.Inf | 0.229 | 0.189 | 0.040 | 1.66e-45 | 0.226 | 0.185 | 0.041 | 2.09e-13 |
| T2D | LDpred1.10FCVal | LDpred2.Inf | 0.232 | 0.189 | 0.043 | 1.12e-46 | 0.230 | 0.185 | 0.045 | 1.15e-14 |
| T2D | LDpred2.Inf | LDpred2.Inf | 0.189 | 0.189 | 0.000 | 1.00e+00 | 0.185 | 0.185 | 0.000 | 1.00e+00 |
| T2D | lassosum.10FCVal | LDpred2.Inf | 0.214 | 0.189 | 0.025 | 4.13e-13 | 0.216 | 0.185 | 0.031 | 4.35e-06 |
| T2D | LDpred2.10FCVal | LDpred2.Inf | 0.237 | 0.189 | 0.048 | 1.05e-58 | 0.238 | 0.185 | 0.053 | 6.63e-20 |
| T2D | PRScs.10FCVal | PRScs.MultiPRS | 0.228 | 0.235 | -0.007 | 3.22e-01 | 0.227 | 0.237 | -0.010 | 2.97e-05 |
| T2D | LDpred1.10FCVal | PRScs.MultiPRS | 0.232 | 0.235 | -0.003 | 6.97e-01 | 0.230 | 0.237 | -0.007 | 1.19e-02 |
| T2D | lassosum.MultiPRS | PRScs.MultiPRS | 0.237 | 0.235 | 0.003 | 6.81e-01 | 0.240 | 0.237 | 0.003 | 2.05e-01 |
| T2D | LDpred2.MultiPRS | PRScs.MultiPRS | 0.242 | 0.235 | 0.007 | 2.78e-01 | 0.240 | 0.237 | 0.003 | 3.51e-01 |
| T2D | All.MultiPRS | PRScs.MultiPRS | 0.242 | 0.235 | 0.007 | 2.83e-01 | 0.242 | 0.237 | 0.005 | 1.15e-01 |
| T2D | PRScs.PseudoVal | PRScs.MultiPRS | 0.226 | 0.235 | -0.008 | 2.32e-01 | 0.226 | 0.237 | -0.011 | 6.99e-06 |
| T2D | LDpred1.MultiPRS | PRScs.MultiPRS | 0.232 | 0.235 | -0.003 | 7.10e-01 | 0.231 | 0.237 | -0.006 | 2.13e-02 |
| T2D | pT+clump.MultiPRS | PRScs.MultiPRS | 0.212 | 0.235 | -0.023 | 7.51e-04 | 0.215 | 0.237 | -0.022 | 1.34e-05 |
| T2D | lassosum.PseudoVal | PRScs.MultiPRS | 0.206 | 0.235 | -0.029 | 2.30e-05 | 0.214 | 0.237 | -0.023 | 9.22e-05 |
| T2D | SBayesR.PseudoVal | PRScs.MultiPRS | 0.226 | 0.235 | -0.008 | 2.23e-01 | 0.227 | 0.237 | -0.010 | 3.18e-02 |
| T2D | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.229 | 0.235 | -0.005 | 4.27e-01 | 0.226 | 0.237 | -0.011 | 2.46e-03 |
| T2D | SBLUP.Inf | PRScs.MultiPRS | 0.189 | 0.235 | -0.046 | 2.48e-11 | 0.184 | 0.237 | -0.053 | 3.88e-20 |
| T2D | LDpred2.Inf | PRScs.MultiPRS | 0.189 | 0.235 | -0.046 | 2.51e-11 | 0.185 | 0.237 | -0.052 | 2.32e-19 |
| T2D | pT+clump.10FCVal | PRScs.MultiPRS | 0.167 | 0.235 | -0.068 | 6.35e-23 | 0.173 | 0.237 | -0.064 | 5.15e-15 |
| T2D | LDpred2.10FCVal | PRScs.MultiPRS | 0.237 | 0.235 | 0.002 | 7.55e-01 | 0.238 | 0.237 | 0.001 | 7.47e-01 |
| T2D | lassosum.10FCVal | PRScs.MultiPRS | 0.214 | 0.235 | -0.021 | 2.13e-03 | 0.216 | 0.237 | -0.021 | 1.55e-06 |
| T2D | LDpred2.PseudoVal | PRScs.MultiPRS | 0.194 | 0.235 | -0.041 | 2.05e-09 | 0.190 | 0.237 | -0.047 | 2.24e-18 |
| T2D | LDpred1.Inf | PRScs.MultiPRS | 0.187 | 0.235 | -0.048 | 2.98e-12 | 0.184 | 0.237 | -0.053 | 8.26e-20 |
| T2D | PRScs.MultiPRS | PRScs.MultiPRS | 0.235 | 0.235 | 0.000 | 1.00e+00 | 0.237 | 0.237 | 0.000 | 1.00e+00 |
| T2D | LDpred2.PseudoVal | PRScs.10FCVal | 0.194 | 0.228 | -0.034 | 3.45e-50 | 0.190 | 0.227 | -0.038 | 2.84e-17 |
| T2D | PRScs.10FCVal | PRScs.10FCVal | 0.228 | 0.228 | 0.000 | 1.00e+00 | 0.227 | 0.227 | 0.000 | 1.00e+00 |
| T2D | LDpred2.MultiPRS | PRScs.10FCVal | 0.242 | 0.228 | 0.014 | 3.84e-02 | 0.240 | 0.227 | 0.013 | 1.28e-03 |
| T2D | pT+clump.MultiPRS | PRScs.10FCVal | 0.212 | 0.228 | -0.016 | 1.74e-02 | 0.215 | 0.227 | -0.012 | 3.72e-02 |
| T2D | LDpred1.Inf | PRScs.10FCVal | 0.187 | 0.228 | -0.041 | 1.10e-60 | 0.184 | 0.227 | -0.043 | 5.70e-19 |
| T2D | LDpred1.MultiPRS | PRScs.10FCVal | 0.232 | 0.228 | 0.004 | 5.37e-01 | 0.231 | 0.227 | 0.004 | 2.60e-01 |
| T2D | pT+clump.10FCVal | PRScs.10FCVal | 0.167 | 0.228 | -0.061 | 3.89e-41 | 0.173 | 0.227 | -0.055 | 1.71e-09 |
| T2D | lassosum.PseudoVal | PRScs.10FCVal | 0.206 | 0.228 | -0.022 | 2.96e-10 | 0.214 | 0.227 | -0.013 | 6.75e-02 |
| T2D | PRScs.PseudoVal | PRScs.10FCVal | 0.226 | 0.228 | -0.001 | 3.63e-01 | 0.226 | 0.227 | -0.001 | 6.57e-01 |
| T2D | DBSLMM.PseudoVal | PRScs.10FCVal | 0.229 | 0.228 | 0.001 | 5.03e-01 | 0.226 | 0.227 | -0.001 | 8.31e-01 |
| T2D | SBLUP.Inf | PRScs.10FCVal | 0.189 | 0.228 | -0.039 | 1.65e-58 | 0.184 | 0.227 | -0.043 | 3.06e-20 |
| T2D | LDpred2.Inf | PRScs.10FCVal | 0.189 | 0.228 | -0.039 | 2.81e-57 | 0.185 | 0.227 | -0.042 | 4.31e-19 |
| T2D | lassosum.10FCVal | PRScs.10FCVal | 0.214 | 0.228 | -0.014 | 7.41e-10 | 0.216 | 0.227 | -0.012 | 1.22e-02 |
| T2D | LDpred2.10FCVal | PRScs.10FCVal | 0.237 | 0.228 | 0.009 | 3.41e-09 | 0.238 | 0.227 | 0.011 | 4.46e-04 |
| T2D | lassosum.MultiPRS | PRScs.10FCVal | 0.237 | 0.228 | 0.010 | 1.61e-01 | 0.240 | 0.227 | 0.013 | 2.89e-04 |
| T2D | LDpred1.10FCVal | PRScs.10FCVal | 0.232 | 0.228 | 0.004 | 1.19e-02 | 0.230 | 0.227 | 0.003 | 3.63e-01 |
| T2D | All.MultiPRS | PRScs.10FCVal | 0.242 | 0.228 | 0.014 | 3.88e-02 | 0.242 | 0.227 | 0.015 | 1.01e-04 |
| T2D | SBayesR.PseudoVal | PRScs.10FCVal | 0.226 | 0.228 | -0.002 | 5.85e-01 | 0.227 | 0.227 | 0.000 | 9.88e-01 |
| T2D | PRScs.MultiPRS | PRScs.10FCVal | 0.235 | 0.228 | 0.007 | 3.22e-01 | 0.237 | 0.227 | 0.010 | 2.97e-05 |
| T2D | All.MultiPRS | PRScs.PseudoVal | 0.242 | 0.226 | 0.015 | 2.34e-02 | 0.242 | 0.226 | 0.016 | 4.31e-05 |
| T2D | pT+clump.MultiPRS | PRScs.PseudoVal | 0.212 | 0.226 | -0.015 | 2.97e-02 | 0.215 | 0.226 | -0.011 | 6.43e-02 |
| T2D | LDpred2.PseudoVal | PRScs.PseudoVal | 0.194 | 0.226 | -0.033 | 1.52e-47 | 0.190 | 0.226 | -0.036 | 2.87e-16 |
| T2D | SBayesR.PseudoVal | PRScs.PseudoVal | 0.226 | 0.226 | 0.000 | 9.47e-01 | 0.227 | 0.226 | 0.001 | 8.05e-01 |
| T2D | LDpred2.MultiPRS | PRScs.PseudoVal | 0.242 | 0.226 | 0.015 | 2.30e-02 | 0.240 | 0.226 | 0.014 | 4.27e-04 |
| T2D | SBLUP.Inf | PRScs.PseudoVal | 0.189 | 0.226 | -0.037 | 2.05e-56 | 0.184 | 0.226 | -0.042 | 2.31e-19 |
| T2D | LDpred1.Inf | PRScs.PseudoVal | 0.187 | 0.226 | -0.039 | 3.13e-59 | 0.184 | 0.226 | -0.042 | 3.43e-18 |
| T2D | PRScs.10FCVal | PRScs.PseudoVal | 0.228 | 0.226 | 0.001 | 3.63e-01 | 0.227 | 0.226 | 0.001 | 6.57e-01 |
| T2D | LDpred2.Inf | PRScs.PseudoVal | 0.189 | 0.226 | -0.037 | 3.50e-55 | 0.185 | 0.226 | -0.041 | 2.65e-18 |
| T2D | PRScs.PseudoVal | PRScs.PseudoVal | 0.226 | 0.226 | 0.000 | 1.00e+00 | 0.226 | 0.226 | 0.000 | 1.00e+00 |
| T2D | LDpred2.10FCVal | PRScs.PseudoVal | 0.237 | 0.226 | 0.010 | 3.37e-11 | 0.238 | 0.226 | 0.012 | 1.00e-04 |
| T2D | pT+clump.10FCVal | PRScs.PseudoVal | 0.167 | 0.226 | -0.060 | 7.34e-39 | 0.173 | 0.226 | -0.053 | 4.90e-09 |
| T2D | PRScs.MultiPRS | PRScs.PseudoVal | 0.235 | 0.226 | 0.008 | 2.32e-01 | 0.237 | 0.226 | 0.011 | 6.99e-06 |
| T2D | lassosum.PseudoVal | PRScs.PseudoVal | 0.206 | 0.226 | -0.021 | 5.80e-09 | 0.214 | 0.226 | -0.012 | 1.03e-01 |
| T2D | lassosum.10FCVal | PRScs.PseudoVal | 0.214 | 0.226 | -0.013 | 4.14e-08 | 0.216 | 0.226 | -0.010 | 2.86e-02 |
| T2D | LDpred1.MultiPRS | PRScs.PseudoVal | 0.232 | 0.226 | 0.006 | 4.13e-01 | 0.231 | 0.226 | 0.005 | 1.29e-01 |
| T2D | lassosum.MultiPRS | PRScs.PseudoVal | 0.237 | 0.226 | 0.011 | 1.09e-01 | 0.240 | 0.226 | 0.014 | 7.36e-05 |
| T2D | LDpred1.10FCVal | PRScs.PseudoVal | 0.232 | 0.226 | 0.005 | 8.16e-04 | 0.230 | 0.226 | 0.004 | 1.90e-01 |
| T2D | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.229 | 0.226 | 0.003 | 1.75e-01 | 0.226 | 0.226 | 0.000 | 9.03e-01 |
| T2D | All.MultiPRS | pT+clump.MultiPRS | 0.242 | 0.212 | 0.030 | 8.65e-06 | 0.242 | 0.215 | 0.027 | 1.25e-07 |
| T2D | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.226 | 0.212 | 0.015 | 3.14e-02 | 0.227 | 0.215 | 0.012 | 2.29e-02 |
| T2D | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.232 | 0.212 | 0.020 | 2.67e-03 | 0.231 | 0.215 | 0.016 | 1.96e-03 |
| T2D | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.212 | 0.212 | 0.000 | 1.00e+00 | 0.215 | 0.215 | 0.000 | 1.00e+00 |
| T2D | lassosum.PseudoVal | pT+clump.MultiPRS | 0.206 | 0.212 | -0.006 | 3.79e-01 | 0.214 | 0.215 | -0.001 | 9.15e-01 |
| T2D | PRScs.10FCVal | pT+clump.MultiPRS | 0.228 | 0.212 | 0.016 | 1.74e-02 | 0.227 | 0.215 | 0.012 | 3.72e-02 |
| T2D | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.229 | 0.212 | 0.018 | 1.01e-02 | 0.226 | 0.215 | 0.011 | 2.49e-02 |
| T2D | SBLUP.Inf | pT+clump.MultiPRS | 0.189 | 0.212 | -0.023 | 1.06e-03 | 0.184 | 0.215 | -0.031 | 1.59e-04 |
| T2D | LDpred2.Inf | pT+clump.MultiPRS | 0.189 | 0.212 | -0.023 | 1.07e-03 | 0.185 | 0.215 | -0.030 | 2.73e-04 |
| T2D | pT+clump.10FCVal | pT+clump.MultiPRS | 0.167 | 0.212 | -0.045 | 8.52e-11 | 0.173 | 0.215 | -0.042 | 2.98e-11 |
| T2D | LDpred2.10FCVal | pT+clump.MultiPRS | 0.237 | 0.212 | 0.025 | 2.32e-04 | 0.238 | 0.215 | 0.023 | 6.26e-06 |
| T2D | PRScs.PseudoVal | pT+clump.MultiPRS | 0.226 | 0.212 | 0.015 | 2.97e-02 | 0.226 | 0.215 | 0.011 | 6.43e-02 |
| T2D | LDpred1.10FCVal | pT+clump.MultiPRS | 0.232 | 0.212 | 0.020 | 2.87e-03 | 0.230 | 0.215 | 0.015 | 3.77e-03 |
| T2D | LDpred1.Inf | pT+clump.MultiPRS | 0.187 | 0.212 | -0.025 | 3.52e-04 | 0.184 | 0.215 | -0.031 | 1.76e-04 |
| T2D | PRScs.MultiPRS | pT+clump.MultiPRS | 0.235 | 0.212 | 0.023 | 7.51e-04 | 0.237 | 0.215 | 0.022 | 1.34e-05 |
| T2D | lassosum.10FCVal | pT+clump.MultiPRS | 0.214 | 0.212 | 0.002 | 7.70e-01 | 0.216 | 0.215 | 0.001 | 9.07e-01 |
| T2D | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.194 | 0.212 | -0.018 | 9.36e-03 | 0.190 | 0.215 | -0.025 | 1.59e-03 |
| T2D | lassosum.MultiPRS | pT+clump.MultiPRS | 0.237 | 0.212 | 0.026 | 1.46e-04 | 0.240 | 0.215 | 0.025 | 5.80e-08 |
| T2D | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.242 | 0.212 | 0.030 | 8.13e-06 | 0.240 | 0.215 | 0.025 | 7.69e-07 |
| T2D | LDpred2.10FCVal | pT+clump.10FCVal | 0.237 | 0.167 | 0.070 | 4.42e-69 | 0.238 | 0.173 | 0.065 | 2.26e-16 |
| T2D | pT+clump.MultiPRS | pT+clump.10FCVal | 0.212 | 0.167 | 0.045 | 8.52e-11 | 0.215 | 0.173 | 0.042 | 2.98e-11 |
| T2D | LDpred2.Inf | pT+clump.10FCVal | 0.189 | 0.167 | 0.022 | 1.07e-04 | 0.185 | 0.173 | 0.013 | 2.70e-01 |
| T2D | SBayesR.PseudoVal | pT+clump.10FCVal | 0.226 | 0.167 | 0.059 | 4.88e-69 | 0.227 | 0.173 | 0.055 | 9.71e-16 |
| T2D | PRScs.MultiPRS | pT+clump.10FCVal | 0.235 | 0.167 | 0.068 | 6.35e-23 | 0.237 | 0.173 | 0.064 | 5.15e-15 |
| T2D | SBLUP.Inf | pT+clump.10FCVal | 0.189 | 0.167 | 0.022 | 1.08e-04 | 0.184 | 0.173 | 0.012 | 3.07e-01 |
| T2D | lassosum.MultiPRS | pT+clump.10FCVal | 0.237 | 0.167 | 0.071 | 5.84e-25 | 0.240 | 0.173 | 0.068 | 6.94e-20 |
| T2D | PRScs.10FCVal | pT+clump.10FCVal | 0.228 | 0.167 | 0.061 | 3.89e-41 | 0.227 | 0.173 | 0.055 | 1.71e-09 |
| T2D | LDpred2.MultiPRS | pT+clump.10FCVal | 0.242 | 0.167 | 0.075 | 7.08e-28 | 0.240 | 0.173 | 0.067 | 1.49e-18 |
| T2D | lassosum.10FCVal | pT+clump.10FCVal | 0.214 | 0.167 | 0.047 | 2.14e-26 | 0.216 | 0.173 | 0.043 | 8.50e-07 |
| T2D | LDpred2.PseudoVal | pT+clump.10FCVal | 0.194 | 0.167 | 0.027 | 1.72e-06 | 0.190 | 0.173 | 0.017 | 1.27e-01 |
| T2D | LDpred1.MultiPRS | pT+clump.10FCVal | 0.232 | 0.167 | 0.065 | 2.36e-21 | 0.231 | 0.173 | 0.058 | 3.83e-13 |
| T2D | pT+clump.10FCVal | pT+clump.10FCVal | 0.167 | 0.167 | 0.000 | 1.00e+00 | 0.173 | 0.173 | 0.000 | 1.00e+00 |
| T2D | All.MultiPRS | pT+clump.10FCVal | 0.242 | 0.167 | 0.075 | 7.70e-28 | 0.242 | 0.173 | 0.069 | 5.83e-19 |
| T2D | LDpred1.Inf | pT+clump.10FCVal | 0.187 | 0.167 | 0.020 | 4.17e-04 | 0.184 | 0.173 | 0.012 | 3.00e-01 |
| T2D | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.229 | 0.167 | 0.062 | 1.53e-47 | 0.226 | 0.173 | 0.054 | 2.59e-10 |
| T2D | LDpred1.10FCVal | pT+clump.10FCVal | 0.232 | 0.167 | 0.065 | 2.93e-57 | 0.230 | 0.173 | 0.058 | 1.33e-12 |
| T2D | lassosum.PseudoVal | pT+clump.10FCVal | 0.206 | 0.167 | 0.039 | 3.94e-34 | 0.214 | 0.173 | 0.042 | 4.23e-11 |
| T2D | PRScs.PseudoVal | pT+clump.10FCVal | 0.226 | 0.167 | 0.060 | 7.34e-39 | 0.226 | 0.173 | 0.053 | 4.90e-09 |
| T2D | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.212 | 0.226 | -0.015 | 3.14e-02 | 0.215 | 0.227 | -0.012 | 2.29e-02 |
| T2D | LDpred1.10FCVal | SBayesR.PseudoVal | 0.232 | 0.226 | 0.006 | 1.62e-02 | 0.230 | 0.227 | 0.003 | 5.42e-01 |
| T2D | PRScs.10FCVal | SBayesR.PseudoVal | 0.228 | 0.226 | 0.002 | 5.85e-01 | 0.227 | 0.227 | 0.000 | 9.88e-01 |
| T2D | LDpred2.10FCVal | SBayesR.PseudoVal | 0.237 | 0.226 | 0.010 | 1.45e-06 | 0.238 | 0.227 | 0.010 | 1.44e-02 |
| T2D | lassosum.10FCVal | SBayesR.PseudoVal | 0.214 | 0.226 | -0.013 | 1.35e-05 | 0.216 | 0.227 | -0.012 | 4.36e-02 |
| T2D | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.194 | 0.226 | -0.033 | 7.95e-15 | 0.190 | 0.227 | -0.038 | 4.75e-06 |
| T2D | lassosum.MultiPRS | SBayesR.PseudoVal | 0.237 | 0.226 | 0.011 | 1.02e-01 | 0.240 | 0.227 | 0.013 | 1.37e-03 |
| T2D | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.242 | 0.226 | 0.016 | 2.13e-02 | 0.240 | 0.227 | 0.013 | 2.16e-03 |
| T2D | All.MultiPRS | SBayesR.PseudoVal | 0.242 | 0.226 | 0.016 | 2.17e-02 | 0.242 | 0.227 | 0.014 | 2.02e-04 |
| T2D | PRScs.PseudoVal | SBayesR.PseudoVal | 0.226 | 0.226 | 0.000 | 9.47e-01 | 0.226 | 0.227 | -0.001 | 8.05e-01 |
| T2D | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.232 | 0.226 | 0.006 | 3.96e-01 | 0.231 | 0.227 | 0.004 | 4.27e-01 |
| T2D | pT+clump.10FCVal | SBayesR.PseudoVal | 0.167 | 0.226 | -0.059 | 4.88e-69 | 0.173 | 0.227 | -0.055 | 9.71e-16 |
| T2D | lassosum.PseudoVal | SBayesR.PseudoVal | 0.206 | 0.226 | -0.021 | 1.83e-18 | 0.214 | 0.227 | -0.013 | 5.67e-03 |
| T2D | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.226 | 0.226 | 0.000 | 1.00e+00 | 0.227 | 0.227 | 0.000 | 1.00e+00 |
| T2D | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.229 | 0.226 | 0.003 | 2.83e-01 | 0.226 | 0.227 | -0.001 | 8.61e-01 |
| T2D | SBLUP.Inf | SBayesR.PseudoVal | 0.189 | 0.226 | -0.037 | 9.69e-18 | 0.184 | 0.227 | -0.043 | 4.42e-07 |
| T2D | LDpred2.Inf | SBayesR.PseudoVal | 0.189 | 0.226 | -0.037 | 1.03e-17 | 0.185 | 0.227 | -0.042 | 7.51e-07 |
| T2D | LDpred1.Inf | SBayesR.PseudoVal | 0.187 | 0.226 | -0.039 | 9.83e-20 | 0.184 | 0.227 | -0.043 | 4.40e-07 |
| T2D | PRScs.MultiPRS | SBayesR.PseudoVal | 0.235 | 0.226 | 0.008 | 2.23e-01 | 0.237 | 0.227 | 0.010 | 3.18e-02 |
| T2D | LDpred2.10FCVal | SBLUP.Inf | 0.237 | 0.189 | 0.048 | 5.94e-58 | 0.238 | 0.184 | 0.054 | 3.32e-20 |
| T2D | PRScs.PseudoVal | SBLUP.Inf | 0.226 | 0.189 | 0.037 | 2.05e-56 | 0.226 | 0.184 | 0.042 | 2.31e-19 |
| T2D | LDpred1.10FCVal | SBLUP.Inf | 0.232 | 0.189 | 0.043 | 2.35e-47 | 0.230 | 0.184 | 0.046 | 2.36e-15 |
| T2D | LDpred2.Inf | SBLUP.Inf | 0.189 | 0.189 | 0.000 | 9.69e-01 | 0.185 | 0.184 | 0.001 | 2.78e-01 |
| T2D | SBayesR.PseudoVal | SBLUP.Inf | 0.226 | 0.189 | 0.037 | 9.69e-18 | 0.227 | 0.184 | 0.043 | 4.42e-07 |
| T2D | lassosum.10FCVal | SBLUP.Inf | 0.214 | 0.189 | 0.025 | 3.02e-13 | 0.216 | 0.184 | 0.031 | 1.96e-06 |
| T2D | pT+clump.MultiPRS | SBLUP.Inf | 0.212 | 0.189 | 0.023 | 1.06e-03 | 0.215 | 0.184 | 0.031 | 1.59e-04 |
| T2D | lassosum.MultiPRS | SBLUP.Inf | 0.237 | 0.189 | 0.048 | 1.59e-12 | 0.240 | 0.184 | 0.056 | 1.38e-18 |
| T2D | PRScs.10FCVal | SBLUP.Inf | 0.228 | 0.189 | 0.039 | 1.65e-58 | 0.227 | 0.184 | 0.043 | 3.06e-20 |
| T2D | PRScs.MultiPRS | SBLUP.Inf | 0.235 | 0.189 | 0.046 | 2.48e-11 | 0.237 | 0.184 | 0.053 | 3.88e-20 |
| T2D | LDpred1.MultiPRS | SBLUP.Inf | 0.232 | 0.189 | 0.043 | 3.54e-10 | 0.231 | 0.184 | 0.047 | 6.09e-15 |
| T2D | LDpred2.PseudoVal | SBLUP.Inf | 0.194 | 0.189 | 0.005 | 6.30e-16 | 0.190 | 0.184 | 0.005 | 1.98e-06 |
| T2D | LDpred1.Inf | SBLUP.Inf | 0.187 | 0.189 | -0.002 | 5.11e-02 | 0.184 | 0.184 | 0.000 | 9.57e-01 |
| T2D | LDpred2.MultiPRS | SBLUP.Inf | 0.242 | 0.189 | 0.053 | 9.26e-15 | 0.240 | 0.184 | 0.056 | 1.55e-17 |
| T2D | All.MultiPRS | SBLUP.Inf | 0.242 | 0.189 | 0.053 | 1.08e-14 | 0.242 | 0.184 | 0.057 | 1.24e-18 |
| T2D | SBLUP.Inf | SBLUP.Inf | 0.189 | 0.189 | 0.000 | 1.00e+00 | 0.184 | 0.184 | 0.000 | 1.00e+00 |
| T2D | DBSLMM.PseudoVal | SBLUP.Inf | 0.229 | 0.189 | 0.040 | 4.70e-46 | 0.226 | 0.184 | 0.042 | 4.78e-14 |
| T2D | pT+clump.10FCVal | SBLUP.Inf | 0.167 | 0.189 | -0.022 | 1.08e-04 | 0.173 | 0.184 | -0.012 | 3.07e-01 |
| T2D | lassosum.PseudoVal | SBLUP.Inf | 0.206 | 0.189 | 0.017 | 1.30e-03 | 0.214 | 0.184 | 0.030 | 3.09e-03 |
| CAD | LDpred2.PseudoVal | All.MultiPRS | 0.107 | 0.191 | -0.084 | 8.32e-36 | 0.111 | 0.190 | -0.079 | 5.04e-17 |
| CAD | pT+clump.10FCVal | All.MultiPRS | 0.135 | 0.191 | -0.056 | 2.96e-17 | 0.134 | 0.190 | -0.056 | 7.05e-13 |
| CAD | LDpred2.MultiPRS | All.MultiPRS | 0.192 | 0.191 | 0.001 | 9.15e-01 | 0.186 | 0.190 | -0.004 | 3.49e-02 |
| CAD | PRScs.PseudoVal | All.MultiPRS | 0.181 | 0.191 | -0.010 | 1.28e-01 | 0.182 | 0.190 | -0.008 | 2.50e-02 |
| CAD | LDpred1.Inf | All.MultiPRS | 0.148 | 0.191 | -0.043 | 6.91e-11 | 0.147 | 0.190 | -0.043 | 4.07e-10 |
| CAD | PRScs.MultiPRS | All.MultiPRS | 0.186 | 0.191 | -0.005 | 4.64e-01 | 0.186 | 0.190 | -0.004 | 1.57e-01 |
| CAD | LDpred2.Inf | All.MultiPRS | 0.162 | 0.191 | -0.029 | 9.67e-06 | 0.157 | 0.190 | -0.033 | 1.75e-08 |
| CAD | lassosum.10FCVal | All.MultiPRS | 0.174 | 0.191 | -0.017 | 8.96e-03 | 0.171 | 0.190 | -0.019 | 5.53e-05 |
| CAD | LDpred2.10FCVal | All.MultiPRS | 0.188 | 0.191 | -0.003 | 6.32e-01 | 0.184 | 0.190 | -0.006 | 2.74e-02 |
| CAD | lassosum.MultiPRS | All.MultiPRS | 0.187 | 0.191 | -0.004 | 5.88e-01 | 0.186 | 0.190 | -0.005 | 9.25e-02 |
| CAD | LDpred1.10FCVal | All.MultiPRS | 0.172 | 0.191 | -0.019 | 3.97e-03 | 0.172 | 0.190 | -0.018 | 2.08e-04 |
| CAD | All.MultiPRS | All.MultiPRS | 0.191 | 0.191 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| CAD | SBayesR.PseudoVal | All.MultiPRS | 0.159 | 0.191 | -0.032 | 1.82e-06 | 0.160 | 0.190 | -0.030 | 4.73e-07 |
| CAD | LDpred1.MultiPRS | All.MultiPRS | 0.173 | 0.191 | -0.018 | 9.43e-03 | 0.174 | 0.190 | -0.016 | 6.80e-04 |
| CAD | pT+clump.MultiPRS | All.MultiPRS | 0.167 | 0.191 | -0.024 | 5.79e-04 | 0.166 | 0.190 | -0.024 | 4.72e-06 |
| CAD | lassosum.PseudoVal | All.MultiPRS | 0.162 | 0.191 | -0.029 | 1.21e-05 | 0.160 | 0.190 | -0.030 | 1.47e-07 |
| CAD | PRScs.10FCVal | All.MultiPRS | 0.181 | 0.191 | -0.010 | 1.43e-01 | 0.182 | 0.190 | -0.008 | 2.77e-02 |
| CAD | DBSLMM.PseudoVal | All.MultiPRS | 0.171 | 0.191 | -0.020 | 1.85e-03 | 0.164 | 0.190 | -0.026 | 3.17e-07 |
| CAD | SBLUP.Inf | All.MultiPRS | 0.162 | 0.191 | -0.029 | 1.17e-05 | 0.157 | 0.190 | -0.033 | 1.03e-08 |
| CAD | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.159 | 0.171 | -0.011 | 2.87e-05 | 0.160 | 0.164 | -0.005 | 3.77e-01 |
| CAD | LDpred2.Inf | DBSLMM.PseudoVal | 0.162 | 0.171 | -0.009 | 2.55e-02 | 0.157 | 0.164 | -0.007 | 3.76e-01 |
| CAD | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.135 | 0.171 | -0.036 | 1.19e-14 | 0.134 | 0.164 | -0.030 | 1.35e-03 |
| CAD | PRScs.10FCVal | DBSLMM.PseudoVal | 0.181 | 0.171 | 0.011 | 5.71e-05 | 0.182 | 0.164 | 0.018 | 1.26e-03 |
| CAD | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.181 | 0.171 | 0.011 | 8.13e-05 | 0.182 | 0.164 | 0.017 | 1.35e-03 |
| CAD | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.167 | 0.171 | -0.003 | 6.39e-01 | 0.166 | 0.164 | 0.002 | 7.54e-01 |
| CAD | LDpred1.Inf | DBSLMM.PseudoVal | 0.148 | 0.171 | -0.023 | 5.12e-08 | 0.147 | 0.164 | -0.017 | 4.02e-02 |
| CAD | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.187 | 0.171 | 0.017 | 1.53e-02 | 0.186 | 0.164 | 0.021 | 3.14e-05 |
| CAD | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.188 | 0.171 | 0.017 | 2.79e-10 | 0.184 | 0.164 | 0.020 | 4.31e-04 |
| CAD | lassosum.10FCVal | DBSLMM.PseudoVal | 0.174 | 0.171 | 0.003 | 3.54e-01 | 0.171 | 0.164 | 0.007 | 3.30e-01 |
| CAD | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.172 | 0.171 | 0.002 | 5.90e-01 | 0.172 | 0.164 | 0.008 | 1.74e-01 |
| CAD | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.173 | 0.171 | 0.003 | 7.09e-01 | 0.174 | 0.164 | 0.010 | 9.42e-02 |
| CAD | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.186 | 0.171 | 0.015 | 2.54e-02 | 0.186 | 0.164 | 0.021 | 8.59e-06 |
| CAD | All.MultiPRS | DBSLMM.PseudoVal | 0.191 | 0.171 | 0.020 | 1.85e-03 | 0.190 | 0.164 | 0.026 | 3.17e-07 |
| CAD | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.107 | 0.171 | -0.064 | 1.52e-36 | 0.111 | 0.164 | -0.053 | 1.74e-07 |
| CAD | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.171 | 0.171 | 0.000 | 1.00e+00 | 0.164 | 0.164 | 0.000 | 1.00e+00 |
| CAD | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.192 | 0.171 | 0.021 | 2.07e-03 | 0.186 | 0.164 | 0.021 | 2.95e-05 |
| CAD | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.162 | 0.171 | -0.008 | 1.18e-02 | 0.160 | 0.164 | -0.004 | 4.99e-01 |
| CAD | SBLUP.Inf | DBSLMM.PseudoVal | 0.162 | 0.171 | -0.008 | 2.86e-02 | 0.157 | 0.164 | -0.007 | 3.48e-01 |
| CAD | SBayesR.PseudoVal | lassosum.MultiPRS | 0.159 | 0.187 | -0.028 | 5.61e-05 | 0.160 | 0.186 | -0.026 | 2.64e-05 |
| CAD | LDpred2.10FCVal | lassosum.MultiPRS | 0.188 | 0.187 | 0.001 | 9.30e-01 | 0.184 | 0.186 | -0.002 | 5.74e-01 |
| CAD | LDpred1.10FCVal | lassosum.MultiPRS | 0.172 | 0.187 | -0.015 | 2.75e-02 | 0.172 | 0.186 | -0.013 | 5.21e-03 |
| CAD | LDpred2.Inf | lassosum.MultiPRS | 0.162 | 0.187 | -0.025 | 2.32e-04 | 0.157 | 0.186 | -0.028 | 7.52e-07 |
| CAD | PRScs.10FCVal | lassosum.MultiPRS | 0.181 | 0.187 | -0.006 | 3.95e-01 | 0.182 | 0.186 | -0.004 | 2.86e-01 |
| CAD | PRScs.MultiPRS | lassosum.MultiPRS | 0.186 | 0.187 | -0.001 | 8.48e-01 | 0.186 | 0.186 | 0.000 | 9.14e-01 |
| CAD | pT+clump.MultiPRS | lassosum.MultiPRS | 0.167 | 0.187 | -0.020 | 3.66e-03 | 0.166 | 0.186 | -0.019 | 1.03e-04 |
| CAD | lassosum.MultiPRS | lassosum.MultiPRS | 0.187 | 0.187 | 0.000 | 1.00e+00 | 0.186 | 0.186 | 0.000 | 1.00e+00 |
| CAD | lassosum.PseudoVal | lassosum.MultiPRS | 0.162 | 0.187 | -0.025 | 2.77e-04 | 0.160 | 0.186 | -0.026 | 4.11e-07 |
| CAD | LDpred2.MultiPRS | lassosum.MultiPRS | 0.192 | 0.187 | 0.004 | 5.18e-01 | 0.186 | 0.186 | 0.000 | 9.41e-01 |
| CAD | LDpred1.MultiPRS | lassosum.MultiPRS | 0.173 | 0.187 | -0.014 | 4.03e-02 | 0.174 | 0.186 | -0.011 | 1.33e-02 |
| CAD | LDpred2.PseudoVal | lassosum.MultiPRS | 0.107 | 0.187 | -0.080 | 4.78e-31 | 0.111 | 0.186 | -0.074 | 1.23e-14 |
| CAD | LDpred1.Inf | lassosum.MultiPRS | 0.148 | 0.187 | -0.040 | 1.04e-08 | 0.147 | 0.186 | -0.038 | 1.90e-08 |
| CAD | pT+clump.10FCVal | lassosum.MultiPRS | 0.135 | 0.187 | -0.053 | 3.25e-14 | 0.134 | 0.186 | -0.052 | 6.58e-12 |
| CAD | lassosum.10FCVal | lassosum.MultiPRS | 0.174 | 0.187 | -0.013 | 5.13e-02 | 0.171 | 0.186 | -0.014 | 1.15e-04 |
| CAD | SBLUP.Inf | lassosum.MultiPRS | 0.162 | 0.187 | -0.025 | 2.69e-04 | 0.157 | 0.186 | -0.028 | 3.19e-07 |
| CAD | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.171 | 0.187 | -0.017 | 1.53e-02 | 0.164 | 0.186 | -0.021 | 3.14e-05 |
| CAD | All.MultiPRS | lassosum.MultiPRS | 0.191 | 0.187 | 0.004 | 5.88e-01 | 0.190 | 0.186 | 0.005 | 9.25e-02 |
| CAD | PRScs.PseudoVal | lassosum.MultiPRS | 0.181 | 0.187 | -0.006 | 3.65e-01 | 0.182 | 0.186 | -0.004 | 2.61e-01 |
| CAD | LDpred1.Inf | lassosum.PseudoVal | 0.148 | 0.162 | -0.015 | 2.07e-03 | 0.147 | 0.160 | -0.013 | 1.79e-01 |
| CAD | PRScs.10FCVal | lassosum.PseudoVal | 0.181 | 0.162 | 0.019 | 1.52e-09 | 0.182 | 0.160 | 0.022 | 5.08e-04 |
| CAD | LDpred2.MultiPRS | lassosum.PseudoVal | 0.192 | 0.162 | 0.030 | 1.85e-05 | 0.186 | 0.160 | 0.026 | 1.26e-06 |
| CAD | lassosum.PseudoVal | lassosum.PseudoVal | 0.162 | 0.162 | 0.000 | 1.00e+00 | 0.160 | 0.160 | 0.000 | 1.00e+00 |
| CAD | PRScs.PseudoVal | lassosum.PseudoVal | 0.181 | 0.162 | 0.019 | 3.52e-09 | 0.182 | 0.160 | 0.022 | 6.59e-04 |
| CAD | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.171 | 0.162 | 0.008 | 1.18e-02 | 0.164 | 0.160 | 0.004 | 4.99e-01 |
| CAD | SBLUP.Inf | lassosum.PseudoVal | 0.162 | 0.162 | 0.000 | 9.82e-01 | 0.157 | 0.160 | -0.003 | 7.61e-01 |
| CAD | LDpred2.Inf | lassosum.PseudoVal | 0.162 | 0.162 | 0.000 | 9.36e-01 | 0.157 | 0.160 | -0.002 | 7.91e-01 |
| CAD | pT+clump.10FCVal | lassosum.PseudoVal | 0.135 | 0.162 | -0.028 | 3.12e-08 | 0.134 | 0.160 | -0.026 | 1.05e-02 |
| CAD | LDpred2.10FCVal | lassosum.PseudoVal | 0.188 | 0.162 | 0.026 | 8.52e-18 | 0.184 | 0.160 | 0.024 | 6.63e-05 |
| CAD | lassosum.MultiPRS | lassosum.PseudoVal | 0.187 | 0.162 | 0.025 | 2.77e-04 | 0.186 | 0.160 | 0.026 | 4.11e-07 |
| CAD | LDpred1.10FCVal | lassosum.PseudoVal | 0.172 | 0.162 | 0.010 | 5.79e-04 | 0.172 | 0.160 | 0.012 | 3.11e-02 |
| CAD | All.MultiPRS | lassosum.PseudoVal | 0.191 | 0.162 | 0.029 | 1.21e-05 | 0.190 | 0.160 | 0.030 | 1.47e-07 |
| CAD | SBayesR.PseudoVal | lassosum.PseudoVal | 0.159 | 0.162 | -0.003 | 3.22e-01 | 0.160 | 0.160 | 0.000 | 9.68e-01 |
| CAD | lassosum.10FCVal | lassosum.PseudoVal | 0.174 | 0.162 | 0.012 | 3.63e-04 | 0.171 | 0.160 | 0.012 | 8.26e-02 |
| CAD | pT+clump.MultiPRS | lassosum.PseudoVal | 0.167 | 0.162 | 0.005 | 4.64e-01 | 0.166 | 0.160 | 0.006 | 3.08e-01 |
| CAD | LDpred2.PseudoVal | lassosum.PseudoVal | 0.107 | 0.162 | -0.055 | 3.35e-29 | 0.111 | 0.160 | -0.049 | 8.73e-07 |
| CAD | LDpred1.MultiPRS | lassosum.PseudoVal | 0.173 | 0.162 | 0.011 | 1.15e-01 | 0.174 | 0.160 | 0.014 | 1.74e-02 |
| CAD | PRScs.MultiPRS | lassosum.PseudoVal | 0.186 | 0.162 | 0.024 | 5.93e-04 | 0.186 | 0.160 | 0.026 | 2.09e-05 |
| CAD | lassosum.10FCVal | lassosum.10FCVal | 0.174 | 0.174 | 0.000 | 1.00e+00 | 0.171 | 0.171 | 0.000 | 1.00e+00 |
| CAD | pT+clump.MultiPRS | lassosum.10FCVal | 0.167 | 0.174 | -0.007 | 3.42e-01 | 0.166 | 0.171 | -0.005 | 4.12e-01 |
| CAD | LDpred2.Inf | lassosum.10FCVal | 0.162 | 0.174 | -0.012 | 2.75e-07 | 0.157 | 0.171 | -0.014 | 2.97e-03 |
| CAD | SBayesR.PseudoVal | lassosum.10FCVal | 0.159 | 0.174 | -0.014 | 4.68e-04 | 0.160 | 0.171 | -0.012 | 1.57e-01 |
| CAD | PRScs.MultiPRS | lassosum.10FCVal | 0.186 | 0.174 | 0.012 | 8.02e-02 | 0.186 | 0.171 | 0.014 | 6.67e-04 |
| CAD | SBLUP.Inf | lassosum.10FCVal | 0.162 | 0.174 | -0.012 | 3.21e-07 | 0.157 | 0.171 | -0.014 | 1.92e-03 |
| CAD | lassosum.MultiPRS | lassosum.10FCVal | 0.187 | 0.174 | 0.013 | 5.13e-02 | 0.186 | 0.171 | 0.014 | 1.15e-04 |
| CAD | PRScs.10FCVal | lassosum.10FCVal | 0.181 | 0.174 | 0.008 | 3.95e-04 | 0.182 | 0.171 | 0.011 | 1.35e-02 |
| CAD | LDpred2.MultiPRS | lassosum.10FCVal | 0.192 | 0.174 | 0.018 | 9.35e-03 | 0.186 | 0.171 | 0.014 | 5.13e-04 |
| CAD | PRScs.PseudoVal | lassosum.10FCVal | 0.181 | 0.174 | 0.007 | 8.71e-04 | 0.182 | 0.171 | 0.010 | 1.80e-02 |
| CAD | LDpred2.PseudoVal | lassosum.10FCVal | 0.107 | 0.174 | -0.067 | 5.51e-42 | 0.111 | 0.171 | -0.060 | 1.19e-09 |
| CAD | LDpred1.MultiPRS | lassosum.10FCVal | 0.173 | 0.174 | -0.001 | 9.14e-01 | 0.174 | 0.171 | 0.003 | 6.31e-01 |
| CAD | pT+clump.10FCVal | lassosum.10FCVal | 0.135 | 0.174 | -0.039 | 1.13e-30 | 0.134 | 0.171 | -0.037 | 5.16e-08 |
| CAD | All.MultiPRS | lassosum.10FCVal | 0.191 | 0.174 | 0.017 | 8.96e-03 | 0.190 | 0.171 | 0.019 | 5.53e-05 |
| CAD | LDpred1.Inf | lassosum.10FCVal | 0.148 | 0.174 | -0.026 | 3.08e-17 | 0.147 | 0.171 | -0.024 | 9.11e-05 |
| CAD | DBSLMM.PseudoVal | lassosum.10FCVal | 0.171 | 0.174 | -0.003 | 3.54e-01 | 0.164 | 0.171 | -0.007 | 3.30e-01 |
| CAD | LDpred1.10FCVal | lassosum.10FCVal | 0.172 | 0.174 | -0.002 | 5.61e-01 | 0.172 | 0.171 | 0.001 | 8.98e-01 |
| CAD | lassosum.PseudoVal | lassosum.10FCVal | 0.162 | 0.174 | -0.012 | 3.63e-04 | 0.160 | 0.171 | -0.012 | 8.26e-02 |
| CAD | LDpred2.10FCVal | lassosum.10FCVal | 0.188 | 0.174 | 0.014 | 2.30e-15 | 0.184 | 0.171 | 0.013 | 3.99e-04 |
| CAD | LDpred2.10FCVal | LDpred1.MultiPRS | 0.188 | 0.173 | 0.015 | 3.17e-02 | 0.184 | 0.174 | 0.010 | 2.96e-02 |
| CAD | PRScs.PseudoVal | LDpred1.MultiPRS | 0.181 | 0.173 | 0.008 | 2.49e-01 | 0.182 | 0.174 | 0.008 | 1.06e-01 |
| CAD | LDpred2.Inf | LDpred1.MultiPRS | 0.162 | 0.173 | -0.011 | 1.01e-01 | 0.157 | 0.174 | -0.017 | 1.26e-02 |
| CAD | pT+clump.10FCVal | LDpred1.MultiPRS | 0.135 | 0.173 | -0.039 | 2.70e-08 | 0.134 | 0.174 | -0.040 | 5.74e-06 |
| CAD | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.159 | 0.173 | -0.014 | 4.90e-02 | 0.160 | 0.174 | -0.014 | 1.98e-02 |
| CAD | All.MultiPRS | LDpred1.MultiPRS | 0.191 | 0.173 | 0.018 | 9.43e-03 | 0.190 | 0.174 | 0.016 | 6.80e-04 |
| CAD | LDpred1.10FCVal | LDpred1.MultiPRS | 0.172 | 0.173 | -0.001 | 8.82e-01 | 0.172 | 0.174 | -0.002 | 1.11e-01 |
| CAD | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.173 | 0.173 | 0.000 | 1.00e+00 | 0.174 | 0.174 | 0.000 | 1.00e+00 |
| CAD | PRScs.10FCVal | LDpred1.MultiPRS | 0.181 | 0.173 | 0.008 | 2.27e-01 | 0.182 | 0.174 | 0.008 | 9.95e-02 |
| CAD | lassosum.PseudoVal | LDpred1.MultiPRS | 0.162 | 0.173 | -0.011 | 1.15e-01 | 0.160 | 0.174 | -0.014 | 1.74e-02 |
| CAD | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.167 | 0.173 | -0.006 | 4.00e-01 | 0.166 | 0.174 | -0.008 | 2.06e-01 |
| CAD | LDpred1.Inf | LDpred1.MultiPRS | 0.148 | 0.173 | -0.025 | 2.21e-04 | 0.147 | 0.174 | -0.027 | 2.80e-07 |
| CAD | lassosum.MultiPRS | LDpred1.MultiPRS | 0.187 | 0.173 | 0.014 | 4.03e-02 | 0.186 | 0.174 | 0.011 | 1.33e-02 |
| CAD | PRScs.MultiPRS | LDpred1.MultiPRS | 0.186 | 0.173 | 0.013 | 6.28e-02 | 0.186 | 0.174 | 0.012 | 7.62e-03 |
| CAD | lassosum.10FCVal | LDpred1.MultiPRS | 0.174 | 0.173 | 0.001 | 9.14e-01 | 0.171 | 0.174 | -0.003 | 6.31e-01 |
| CAD | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.107 | 0.173 | -0.066 | 1.82e-21 | 0.111 | 0.174 | -0.063 | 1.29e-10 |
| CAD | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.171 | 0.173 | -0.003 | 7.09e-01 | 0.164 | 0.174 | -0.010 | 9.42e-02 |
| CAD | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.192 | 0.173 | 0.019 | 6.90e-03 | 0.186 | 0.174 | 0.012 | 7.09e-03 |
| CAD | SBLUP.Inf | LDpred1.MultiPRS | 0.162 | 0.173 | -0.011 | 1.10e-01 | 0.157 | 0.174 | -0.017 | 9.91e-03 |
| CAD | All.MultiPRS | LDpred1.Inf | 0.191 | 0.148 | 0.043 | 6.91e-11 | 0.190 | 0.147 | 0.043 | 4.07e-10 |
| CAD | LDpred2.PseudoVal | LDpred1.Inf | 0.107 | 0.148 | -0.041 | 4.08e-14 | 0.111 | 0.147 | -0.036 | 8.38e-04 |
| CAD | SBayesR.PseudoVal | LDpred1.Inf | 0.159 | 0.148 | 0.012 | 1.57e-02 | 0.160 | 0.147 | 0.012 | 2.03e-01 |
| CAD | LDpred2.MultiPRS | LDpred1.Inf | 0.192 | 0.148 | 0.044 | 1.66e-10 | 0.186 | 0.147 | 0.039 | 5.35e-09 |
| CAD | PRScs.PseudoVal | LDpred1.Inf | 0.181 | 0.148 | 0.033 | 1.80e-27 | 0.182 | 0.147 | 0.034 | 1.92e-08 |
| CAD | LDpred1.Inf | LDpred1.Inf | 0.148 | 0.148 | 0.000 | 1.00e+00 | 0.147 | 0.147 | 0.000 | 1.00e+00 |
| CAD | pT+clump.10FCVal | LDpred1.Inf | 0.135 | 0.148 | -0.013 | 9.34e-04 | 0.134 | 0.147 | -0.013 | 1.00e-01 |
| CAD | PRScs.MultiPRS | LDpred1.Inf | 0.186 | 0.148 | 0.038 | 3.22e-08 | 0.186 | 0.147 | 0.039 | 6.28e-10 |
| CAD | lassosum.PseudoVal | LDpred1.Inf | 0.162 | 0.148 | 0.015 | 2.07e-03 | 0.160 | 0.147 | 0.013 | 1.79e-01 |
| CAD | pT+clump.MultiPRS | LDpred1.Inf | 0.167 | 0.148 | 0.020 | 4.66e-03 | 0.166 | 0.147 | 0.019 | 1.87e-02 |
| CAD | LDpred1.MultiPRS | LDpred1.Inf | 0.173 | 0.148 | 0.025 | 2.21e-04 | 0.174 | 0.147 | 0.027 | 2.80e-07 |
| CAD | PRScs.10FCVal | LDpred1.Inf | 0.181 | 0.148 | 0.034 | 7.54e-28 | 0.182 | 0.147 | 0.035 | 1.93e-08 |
| CAD | LDpred2.Inf | LDpred1.Inf | 0.162 | 0.148 | 0.014 | 2.15e-09 | 0.157 | 0.147 | 0.010 | 3.06e-02 |
| CAD | SBLUP.Inf | LDpred1.Inf | 0.162 | 0.148 | 0.014 | 7.47e-10 | 0.157 | 0.147 | 0.010 | 3.51e-02 |
| CAD | DBSLMM.PseudoVal | LDpred1.Inf | 0.171 | 0.148 | 0.023 | 5.12e-08 | 0.164 | 0.147 | 0.017 | 4.02e-02 |
| CAD | lassosum.MultiPRS | LDpred1.Inf | 0.187 | 0.148 | 0.040 | 1.04e-08 | 0.186 | 0.147 | 0.038 | 1.90e-08 |
| CAD | LDpred1.10FCVal | LDpred1.Inf | 0.172 | 0.148 | 0.024 | 8.10e-16 | 0.172 | 0.147 | 0.025 | 4.36e-05 |
| CAD | lassosum.10FCVal | LDpred1.Inf | 0.174 | 0.148 | 0.026 | 3.08e-17 | 0.171 | 0.147 | 0.024 | 9.11e-05 |
| CAD | LDpred2.10FCVal | LDpred1.Inf | 0.188 | 0.148 | 0.040 | 2.11e-40 | 0.184 | 0.147 | 0.037 | 1.03e-09 |
| CAD | LDpred1.Inf | LDpred1.10FCVal | 0.148 | 0.172 | -0.024 | 8.10e-16 | 0.147 | 0.172 | -0.025 | 4.36e-05 |
| CAD | PRScs.MultiPRS | LDpred1.10FCVal | 0.186 | 0.172 | 0.014 | 4.44e-02 | 0.186 | 0.172 | 0.014 | 3.32e-03 |
| CAD | LDpred1.10FCVal | LDpred1.10FCVal | 0.172 | 0.172 | 0.000 | 1.00e+00 | 0.172 | 0.172 | 0.000 | 1.00e+00 |
| CAD | All.MultiPRS | LDpred1.10FCVal | 0.191 | 0.172 | 0.019 | 3.97e-03 | 0.190 | 0.172 | 0.018 | 2.08e-04 |
| CAD | SBayesR.PseudoVal | LDpred1.10FCVal | 0.159 | 0.172 | -0.013 | 1.80e-05 | 0.160 | 0.172 | -0.013 | 3.25e-02 |
| CAD | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.171 | 0.172 | -0.002 | 5.90e-01 | 0.164 | 0.172 | -0.008 | 1.74e-01 |
| CAD | pT+clump.MultiPRS | LDpred1.10FCVal | 0.167 | 0.172 | -0.005 | 4.88e-01 | 0.166 | 0.172 | -0.006 | 3.47e-01 |
| CAD | lassosum.MultiPRS | LDpred1.10FCVal | 0.187 | 0.172 | 0.015 | 2.75e-02 | 0.186 | 0.172 | 0.013 | 5.21e-03 |
| CAD | PRScs.10FCVal | LDpred1.10FCVal | 0.181 | 0.172 | 0.009 | 2.25e-04 | 0.182 | 0.172 | 0.010 | 5.38e-02 |
| CAD | lassosum.10FCVal | LDpred1.10FCVal | 0.174 | 0.172 | 0.002 | 5.61e-01 | 0.171 | 0.172 | -0.001 | 8.98e-01 |
| CAD | SBLUP.Inf | LDpred1.10FCVal | 0.162 | 0.172 | -0.010 | 5.47e-03 | 0.157 | 0.172 | -0.015 | 3.80e-02 |
| CAD | LDpred2.PseudoVal | LDpred1.10FCVal | 0.107 | 0.172 | -0.065 | 8.88e-39 | 0.111 | 0.172 | -0.061 | 1.00e-09 |
| CAD | pT+clump.10FCVal | LDpred1.10FCVal | 0.135 | 0.172 | -0.038 | 9.68e-17 | 0.134 | 0.172 | -0.038 | 3.33e-05 |
| CAD | LDpred2.MultiPRS | LDpred1.10FCVal | 0.192 | 0.172 | 0.020 | 4.30e-03 | 0.186 | 0.172 | 0.014 | 2.40e-03 |
| CAD | PRScs.PseudoVal | LDpred1.10FCVal | 0.181 | 0.172 | 0.009 | 3.27e-04 | 0.182 | 0.172 | 0.009 | 5.67e-02 |
| CAD | LDpred2.10FCVal | LDpred1.10FCVal | 0.188 | 0.172 | 0.016 | 6.63e-11 | 0.184 | 0.172 | 0.012 | 1.51e-02 |
| CAD | LDpred1.MultiPRS | LDpred1.10FCVal | 0.173 | 0.172 | 0.001 | 8.82e-01 | 0.174 | 0.172 | 0.002 | 1.11e-01 |
| CAD | LDpred2.Inf | LDpred1.10FCVal | 0.162 | 0.172 | -0.010 | 4.84e-03 | 0.157 | 0.172 | -0.015 | 4.51e-02 |
| CAD | lassosum.PseudoVal | LDpred1.10FCVal | 0.162 | 0.172 | -0.010 | 5.79e-04 | 0.160 | 0.172 | -0.012 | 3.11e-02 |
| CAD | LDpred1.Inf | LDpred2.MultiPRS | 0.148 | 0.192 | -0.044 | 1.66e-10 | 0.147 | 0.186 | -0.039 | 5.35e-09 |
| CAD | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.171 | 0.192 | -0.021 | 2.07e-03 | 0.164 | 0.186 | -0.021 | 2.95e-05 |
| CAD | SBLUP.Inf | LDpred2.MultiPRS | 0.162 | 0.192 | -0.030 | 1.72e-05 | 0.157 | 0.186 | -0.029 | 7.35e-08 |
| CAD | LDpred2.Inf | LDpred2.MultiPRS | 0.162 | 0.192 | -0.030 | 1.43e-05 | 0.157 | 0.186 | -0.028 | 1.04e-07 |
| CAD | pT+clump.10FCVal | LDpred2.MultiPRS | 0.135 | 0.192 | -0.057 | 1.59e-16 | 0.134 | 0.186 | -0.052 | 4.26e-11 |
| CAD | LDpred2.10FCVal | LDpred2.MultiPRS | 0.188 | 0.192 | -0.004 | 5.75e-01 | 0.184 | 0.186 | -0.002 | 2.79e-01 |
| CAD | PRScs.PseudoVal | LDpred2.MultiPRS | 0.181 | 0.192 | -0.011 | 1.20e-01 | 0.182 | 0.186 | -0.004 | 2.07e-01 |
| CAD | LDpred1.10FCVal | LDpred2.MultiPRS | 0.172 | 0.192 | -0.020 | 4.30e-03 | 0.172 | 0.186 | -0.014 | 2.40e-03 |
| CAD | All.MultiPRS | LDpred2.MultiPRS | 0.191 | 0.192 | -0.001 | 9.15e-01 | 0.190 | 0.186 | 0.004 | 3.49e-02 |
| CAD | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.159 | 0.192 | -0.032 | 2.97e-06 | 0.160 | 0.186 | -0.026 | 2.31e-05 |
| CAD | lassosum.10FCVal | LDpred2.MultiPRS | 0.174 | 0.192 | -0.018 | 9.35e-03 | 0.171 | 0.186 | -0.014 | 5.13e-04 |
| CAD | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.167 | 0.192 | -0.024 | 3.88e-04 | 0.166 | 0.186 | -0.020 | 2.15e-04 |
| CAD | lassosum.MultiPRS | LDpred2.MultiPRS | 0.187 | 0.192 | -0.004 | 5.18e-01 | 0.186 | 0.186 | 0.000 | 9.41e-01 |
| CAD | PRScs.10FCVal | LDpred2.MultiPRS | 0.181 | 0.192 | -0.010 | 1.34e-01 | 0.182 | 0.186 | -0.004 | 2.37e-01 |
| CAD | PRScs.MultiPRS | LDpred2.MultiPRS | 0.186 | 0.192 | -0.006 | 4.02e-01 | 0.186 | 0.186 | 0.000 | 9.78e-01 |
| CAD | lassosum.PseudoVal | LDpred2.MultiPRS | 0.162 | 0.192 | -0.030 | 1.85e-05 | 0.160 | 0.186 | -0.026 | 1.26e-06 |
| CAD | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.107 | 0.192 | -0.085 | 2.86e-34 | 0.111 | 0.186 | -0.075 | 6.43e-16 |
| CAD | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.173 | 0.192 | -0.019 | 6.90e-03 | 0.174 | 0.186 | -0.012 | 7.09e-03 |
| CAD | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.192 | 0.192 | 0.000 | 1.00e+00 | 0.186 | 0.186 | 0.000 | 1.00e+00 |
| CAD | LDpred1.Inf | LDpred2.10FCVal | 0.148 | 0.188 | -0.040 | 2.11e-40 | 0.147 | 0.184 | -0.037 | 1.03e-09 |
| CAD | PRScs.MultiPRS | LDpred2.10FCVal | 0.186 | 0.188 | -0.002 | 7.80e-01 | 0.186 | 0.184 | 0.002 | 4.80e-01 |
| CAD | LDpred2.MultiPRS | LDpred2.10FCVal | 0.192 | 0.188 | 0.004 | 5.75e-01 | 0.186 | 0.184 | 0.002 | 2.79e-01 |
| CAD | PRScs.PseudoVal | LDpred2.10FCVal | 0.181 | 0.188 | -0.007 | 5.17e-06 | 0.182 | 0.184 | -0.002 | 4.55e-01 |
| CAD | LDpred2.10FCVal | LDpred2.10FCVal | 0.188 | 0.188 | 0.000 | 1.00e+00 | 0.184 | 0.184 | 0.000 | 1.00e+00 |
| CAD | lassosum.MultiPRS | LDpred2.10FCVal | 0.187 | 0.188 | -0.001 | 9.30e-01 | 0.186 | 0.184 | 0.002 | 5.74e-01 |
| CAD | LDpred1.10FCVal | LDpred2.10FCVal | 0.172 | 0.188 | -0.016 | 6.63e-11 | 0.172 | 0.184 | -0.012 | 1.51e-02 |
| CAD | All.MultiPRS | LDpred2.10FCVal | 0.191 | 0.188 | 0.003 | 6.32e-01 | 0.190 | 0.184 | 0.006 | 2.74e-02 |
| CAD | SBayesR.PseudoVal | LDpred2.10FCVal | 0.159 | 0.188 | -0.028 | 7.04e-17 | 0.160 | 0.184 | -0.024 | 4.16e-04 |
| CAD | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.171 | 0.188 | -0.017 | 2.79e-10 | 0.164 | 0.184 | -0.020 | 4.31e-04 |
| CAD | pT+clump.MultiPRS | LDpred2.10FCVal | 0.167 | 0.188 | -0.021 | 2.88e-03 | 0.166 | 0.184 | -0.018 | 1.04e-03 |
| CAD | lassosum.PseudoVal | LDpred2.10FCVal | 0.162 | 0.188 | -0.026 | 8.52e-18 | 0.160 | 0.184 | -0.024 | 6.63e-05 |
| CAD | PRScs.10FCVal | LDpred2.10FCVal | 0.181 | 0.188 | -0.006 | 2.02e-05 | 0.182 | 0.184 | -0.002 | 5.10e-01 |
| CAD | lassosum.10FCVal | LDpred2.10FCVal | 0.174 | 0.188 | -0.014 | 2.30e-15 | 0.171 | 0.184 | -0.013 | 3.99e-04 |
| CAD | SBLUP.Inf | LDpred2.10FCVal | 0.162 | 0.188 | -0.026 | 2.86e-31 | 0.157 | 0.184 | -0.027 | 1.60e-09 |
| CAD | LDpred2.PseudoVal | LDpred2.10FCVal | 0.107 | 0.188 | -0.081 | 1.17e-77 | 0.111 | 0.184 | -0.073 | 1.06e-16 |
| CAD | pT+clump.10FCVal | LDpred2.10FCVal | 0.135 | 0.188 | -0.053 | 2.35e-49 | 0.134 | 0.184 | -0.050 | 9.99e-12 |
| CAD | LDpred2.Inf | LDpred2.10FCVal | 0.162 | 0.188 | -0.026 | 8.86e-33 | 0.157 | 0.184 | -0.026 | 1.70e-09 |
| CAD | LDpred1.MultiPRS | LDpred2.10FCVal | 0.173 | 0.188 | -0.015 | 3.17e-02 | 0.174 | 0.184 | -0.010 | 2.96e-02 |
| CAD | lassosum.10FCVal | LDpred2.PseudoVal | 0.174 | 0.107 | 0.067 | 5.51e-42 | 0.171 | 0.111 | 0.060 | 1.19e-09 |
| CAD | SBLUP.Inf | LDpred2.PseudoVal | 0.162 | 0.107 | 0.055 | 1.36e-25 | 0.157 | 0.111 | 0.046 | 1.53e-05 |
| CAD | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.159 | 0.107 | 0.053 | 1.47e-25 | 0.160 | 0.111 | 0.049 | 1.54e-06 |
| CAD | All.MultiPRS | LDpred2.PseudoVal | 0.191 | 0.107 | 0.084 | 8.32e-36 | 0.190 | 0.111 | 0.079 | 5.04e-17 |
| CAD | PRScs.PseudoVal | LDpred2.PseudoVal | 0.181 | 0.107 | 0.074 | 5.44e-54 | 0.182 | 0.111 | 0.071 | 2.35e-13 |
| CAD | lassosum.MultiPRS | LDpred2.PseudoVal | 0.187 | 0.107 | 0.080 | 4.78e-31 | 0.186 | 0.111 | 0.074 | 1.23e-14 |
| CAD | PRScs.MultiPRS | LDpred2.PseudoVal | 0.186 | 0.107 | 0.079 | 1.42e-29 | 0.186 | 0.111 | 0.075 | 2.50e-14 |
| CAD | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.192 | 0.107 | 0.085 | 2.86e-34 | 0.186 | 0.111 | 0.075 | 6.43e-16 |
| CAD | PRScs.10FCVal | LDpred2.PseudoVal | 0.181 | 0.107 | 0.075 | 2.12e-54 | 0.182 | 0.111 | 0.071 | 2.02e-13 |
| CAD | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.107 | 0.107 | 0.000 | 1.00e+00 | 0.111 | 0.111 | 0.000 | 1.00e+00 |
| CAD | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.167 | 0.107 | 0.060 | 5.92e-18 | 0.166 | 0.111 | 0.055 | 1.63e-08 |
| CAD | pT+clump.10FCVal | LDpred2.PseudoVal | 0.135 | 0.107 | 0.028 | 5.25e-07 | 0.134 | 0.111 | 0.023 | 3.98e-02 |
| CAD | lassosum.PseudoVal | LDpred2.PseudoVal | 0.162 | 0.107 | 0.055 | 3.35e-29 | 0.160 | 0.111 | 0.049 | 8.73e-07 |
| CAD | LDpred1.Inf | LDpred2.PseudoVal | 0.148 | 0.107 | 0.041 | 4.08e-14 | 0.147 | 0.111 | 0.036 | 8.38e-04 |
| CAD | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.173 | 0.107 | 0.066 | 1.82e-21 | 0.174 | 0.111 | 0.063 | 1.29e-10 |
| CAD | LDpred1.10FCVal | LDpred2.PseudoVal | 0.172 | 0.107 | 0.065 | 8.88e-39 | 0.172 | 0.111 | 0.061 | 1.00e-09 |
| CAD | LDpred2.Inf | LDpred2.PseudoVal | 0.162 | 0.107 | 0.055 | 6.72e-26 | 0.157 | 0.111 | 0.046 | 1.08e-05 |
| CAD | LDpred2.10FCVal | LDpred2.PseudoVal | 0.188 | 0.107 | 0.081 | 1.17e-77 | 0.184 | 0.111 | 0.073 | 1.06e-16 |
| CAD | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.171 | 0.107 | 0.064 | 1.52e-36 | 0.164 | 0.111 | 0.053 | 1.74e-07 |
| CAD | pT+clump.MultiPRS | LDpred2.Inf | 0.167 | 0.162 | 0.005 | 4.31e-01 | 0.166 | 0.157 | 0.009 | 2.36e-01 |
| CAD | lassosum.MultiPRS | LDpred2.Inf | 0.187 | 0.162 | 0.025 | 2.32e-04 | 0.186 | 0.157 | 0.028 | 7.52e-07 |
| CAD | SBayesR.PseudoVal | LDpred2.Inf | 0.159 | 0.162 | -0.002 | 6.33e-01 | 0.160 | 0.157 | 0.002 | 8.20e-01 |
| CAD | All.MultiPRS | LDpred2.Inf | 0.191 | 0.162 | 0.029 | 9.67e-06 | 0.190 | 0.157 | 0.033 | 1.75e-08 |
| CAD | SBLUP.Inf | LDpred2.Inf | 0.162 | 0.162 | 0.000 | 5.79e-01 | 0.157 | 0.157 | 0.000 | 7.37e-01 |
| CAD | LDpred2.PseudoVal | LDpred2.Inf | 0.107 | 0.162 | -0.055 | 6.72e-26 | 0.111 | 0.157 | -0.046 | 1.08e-05 |
| CAD | PRScs.MultiPRS | LDpred2.Inf | 0.186 | 0.162 | 0.024 | 4.86e-04 | 0.186 | 0.157 | 0.028 | 1.18e-08 |
| CAD | LDpred2.MultiPRS | LDpred2.Inf | 0.192 | 0.162 | 0.030 | 1.43e-05 | 0.186 | 0.157 | 0.028 | 1.04e-07 |
| CAD | PRScs.10FCVal | LDpred2.Inf | 0.181 | 0.162 | 0.020 | 3.52e-16 | 0.182 | 0.157 | 0.025 | 5.18e-07 |
| CAD | LDpred1.Inf | LDpred2.Inf | 0.148 | 0.162 | -0.014 | 2.15e-09 | 0.147 | 0.157 | -0.010 | 3.06e-02 |
| CAD | LDpred1.MultiPRS | LDpred2.Inf | 0.173 | 0.162 | 0.011 | 1.01e-01 | 0.174 | 0.157 | 0.017 | 1.26e-02 |
| CAD | pT+clump.10FCVal | LDpred2.Inf | 0.135 | 0.162 | -0.027 | 2.87e-16 | 0.134 | 0.157 | -0.023 | 5.01e-04 |
| CAD | lassosum.PseudoVal | LDpred2.Inf | 0.162 | 0.162 | 0.000 | 9.36e-01 | 0.160 | 0.157 | 0.002 | 7.91e-01 |
| CAD | PRScs.PseudoVal | LDpred2.Inf | 0.181 | 0.162 | 0.019 | 2.46e-15 | 0.182 | 0.157 | 0.024 | 7.55e-07 |
| CAD | DBSLMM.PseudoVal | LDpred2.Inf | 0.171 | 0.162 | 0.009 | 2.55e-02 | 0.164 | 0.157 | 0.007 | 3.76e-01 |
| CAD | LDpred1.10FCVal | LDpred2.Inf | 0.172 | 0.162 | 0.010 | 4.84e-03 | 0.172 | 0.157 | 0.015 | 4.51e-02 |
| CAD | LDpred2.Inf | LDpred2.Inf | 0.162 | 0.162 | 0.000 | 1.00e+00 | 0.157 | 0.157 | 0.000 | 1.00e+00 |
| CAD | lassosum.10FCVal | LDpred2.Inf | 0.174 | 0.162 | 0.012 | 2.75e-07 | 0.171 | 0.157 | 0.014 | 2.97e-03 |
| CAD | LDpred2.10FCVal | LDpred2.Inf | 0.188 | 0.162 | 0.026 | 8.86e-33 | 0.184 | 0.157 | 0.026 | 1.70e-09 |
| CAD | PRScs.10FCVal | PRScs.MultiPRS | 0.181 | 0.186 | -0.005 | 5.12e-01 | 0.182 | 0.186 | -0.004 | 6.57e-02 |
| CAD | LDpred1.10FCVal | PRScs.MultiPRS | 0.172 | 0.186 | -0.014 | 4.44e-02 | 0.172 | 0.186 | -0.014 | 3.32e-03 |
| CAD | lassosum.MultiPRS | PRScs.MultiPRS | 0.187 | 0.186 | 0.001 | 8.48e-01 | 0.186 | 0.186 | 0.000 | 9.14e-01 |
| CAD | LDpred2.MultiPRS | PRScs.MultiPRS | 0.192 | 0.186 | 0.006 | 4.02e-01 | 0.186 | 0.186 | 0.000 | 9.78e-01 |
| CAD | All.MultiPRS | PRScs.MultiPRS | 0.191 | 0.186 | 0.005 | 4.64e-01 | 0.190 | 0.186 | 0.004 | 1.57e-01 |
| CAD | PRScs.PseudoVal | PRScs.MultiPRS | 0.181 | 0.186 | -0.005 | 4.76e-01 | 0.182 | 0.186 | -0.004 | 6.60e-02 |
| CAD | LDpred1.MultiPRS | PRScs.MultiPRS | 0.173 | 0.186 | -0.013 | 6.28e-02 | 0.174 | 0.186 | -0.012 | 7.62e-03 |
| CAD | pT+clump.MultiPRS | PRScs.MultiPRS | 0.167 | 0.186 | -0.019 | 6.89e-03 | 0.166 | 0.186 | -0.020 | 1.85e-04 |
| CAD | lassosum.PseudoVal | PRScs.MultiPRS | 0.162 | 0.186 | -0.024 | 5.93e-04 | 0.160 | 0.186 | -0.026 | 2.09e-05 |
| CAD | SBayesR.PseudoVal | PRScs.MultiPRS | 0.159 | 0.186 | -0.026 | 1.28e-04 | 0.160 | 0.186 | -0.026 | 6.63e-05 |
| CAD | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.171 | 0.186 | -0.015 | 2.54e-02 | 0.164 | 0.186 | -0.021 | 8.59e-06 |
| CAD | SBLUP.Inf | PRScs.MultiPRS | 0.162 | 0.186 | -0.024 | 5.64e-04 | 0.157 | 0.186 | -0.029 | 3.18e-09 |
| CAD | LDpred2.Inf | PRScs.MultiPRS | 0.162 | 0.186 | -0.024 | 4.86e-04 | 0.157 | 0.186 | -0.028 | 1.18e-08 |
| CAD | pT+clump.10FCVal | PRScs.MultiPRS | 0.135 | 0.186 | -0.051 | 1.35e-13 | 0.134 | 0.186 | -0.052 | 3.08e-12 |
| CAD | LDpred2.10FCVal | PRScs.MultiPRS | 0.188 | 0.186 | 0.002 | 7.80e-01 | 0.184 | 0.186 | -0.002 | 4.80e-01 |
| CAD | lassosum.10FCVal | PRScs.MultiPRS | 0.174 | 0.186 | -0.012 | 8.02e-02 | 0.171 | 0.186 | -0.014 | 6.67e-04 |
| CAD | LDpred2.PseudoVal | PRScs.MultiPRS | 0.107 | 0.186 | -0.079 | 1.42e-29 | 0.111 | 0.186 | -0.075 | 2.50e-14 |
| CAD | LDpred1.Inf | PRScs.MultiPRS | 0.148 | 0.186 | -0.038 | 3.22e-08 | 0.147 | 0.186 | -0.039 | 6.28e-10 |
| CAD | PRScs.MultiPRS | PRScs.MultiPRS | 0.186 | 0.186 | 0.000 | 1.00e+00 | 0.186 | 0.186 | 0.000 | 1.00e+00 |
| CAD | LDpred2.PseudoVal | PRScs.10FCVal | 0.107 | 0.181 | -0.075 | 2.12e-54 | 0.111 | 0.182 | -0.071 | 2.02e-13 |
| CAD | PRScs.10FCVal | PRScs.10FCVal | 0.181 | 0.181 | 0.000 | 1.00e+00 | 0.182 | 0.182 | 0.000 | 1.00e+00 |
| CAD | LDpred2.MultiPRS | PRScs.10FCVal | 0.192 | 0.181 | 0.010 | 1.34e-01 | 0.186 | 0.182 | 0.004 | 2.37e-01 |
| CAD | pT+clump.MultiPRS | PRScs.10FCVal | 0.167 | 0.181 | -0.014 | 4.05e-02 | 0.166 | 0.182 | -0.016 | 6.35e-03 |
| CAD | LDpred1.Inf | PRScs.10FCVal | 0.148 | 0.181 | -0.034 | 7.54e-28 | 0.147 | 0.182 | -0.035 | 1.93e-08 |
| CAD | LDpred1.MultiPRS | PRScs.10FCVal | 0.173 | 0.181 | -0.008 | 2.27e-01 | 0.174 | 0.182 | -0.008 | 9.95e-02 |
| CAD | pT+clump.10FCVal | PRScs.10FCVal | 0.135 | 0.181 | -0.047 | 7.26e-36 | 0.134 | 0.182 | -0.048 | 3.75e-10 |
| CAD | lassosum.PseudoVal | PRScs.10FCVal | 0.162 | 0.181 | -0.019 | 1.52e-09 | 0.160 | 0.182 | -0.022 | 5.08e-04 |
| CAD | PRScs.PseudoVal | PRScs.10FCVal | 0.181 | 0.181 | 0.000 | 8.06e-01 | 0.182 | 0.182 | 0.000 | 9.32e-01 |
| CAD | DBSLMM.PseudoVal | PRScs.10FCVal | 0.171 | 0.181 | -0.011 | 5.71e-05 | 0.164 | 0.182 | -0.018 | 1.26e-03 |
| CAD | SBLUP.Inf | PRScs.10FCVal | 0.162 | 0.181 | -0.019 | 1.82e-16 | 0.157 | 0.182 | -0.025 | 1.91e-07 |
| CAD | LDpred2.Inf | PRScs.10FCVal | 0.162 | 0.181 | -0.020 | 3.52e-16 | 0.157 | 0.182 | -0.025 | 5.18e-07 |
| CAD | lassosum.10FCVal | PRScs.10FCVal | 0.174 | 0.181 | -0.008 | 3.95e-04 | 0.171 | 0.182 | -0.011 | 1.35e-02 |
| CAD | LDpred2.10FCVal | PRScs.10FCVal | 0.188 | 0.181 | 0.006 | 2.02e-05 | 0.184 | 0.182 | 0.002 | 5.10e-01 |
| CAD | lassosum.MultiPRS | PRScs.10FCVal | 0.187 | 0.181 | 0.006 | 3.95e-01 | 0.186 | 0.182 | 0.004 | 2.86e-01 |
| CAD | LDpred1.10FCVal | PRScs.10FCVal | 0.172 | 0.181 | -0.009 | 2.25e-04 | 0.172 | 0.182 | -0.010 | 5.38e-02 |
| CAD | All.MultiPRS | PRScs.10FCVal | 0.191 | 0.181 | 0.010 | 1.43e-01 | 0.190 | 0.182 | 0.008 | 2.77e-02 |
| CAD | SBayesR.PseudoVal | PRScs.10FCVal | 0.159 | 0.181 | -0.022 | 2.00e-10 | 0.160 | 0.182 | -0.022 | 1.29e-03 |
| CAD | PRScs.MultiPRS | PRScs.10FCVal | 0.186 | 0.181 | 0.005 | 5.12e-01 | 0.186 | 0.182 | 0.004 | 6.57e-02 |
| CAD | All.MultiPRS | PRScs.PseudoVal | 0.191 | 0.181 | 0.010 | 1.28e-01 | 0.190 | 0.182 | 0.008 | 2.50e-02 |
| CAD | pT+clump.MultiPRS | PRScs.PseudoVal | 0.167 | 0.181 | -0.014 | 4.66e-02 | 0.166 | 0.182 | -0.015 | 6.98e-03 |
| CAD | LDpred2.PseudoVal | PRScs.PseudoVal | 0.107 | 0.181 | -0.074 | 5.44e-54 | 0.111 | 0.182 | -0.071 | 2.35e-13 |
| CAD | SBayesR.PseudoVal | PRScs.PseudoVal | 0.159 | 0.181 | -0.022 | 4.15e-10 | 0.160 | 0.182 | -0.022 | 1.52e-03 |
| CAD | LDpred2.MultiPRS | PRScs.PseudoVal | 0.192 | 0.181 | 0.011 | 1.20e-01 | 0.186 | 0.182 | 0.004 | 2.07e-01 |
| CAD | SBLUP.Inf | PRScs.PseudoVal | 0.162 | 0.181 | -0.019 | 1.44e-15 | 0.157 | 0.182 | -0.025 | 2.83e-07 |
| CAD | LDpred1.Inf | PRScs.PseudoVal | 0.148 | 0.181 | -0.033 | 1.80e-27 | 0.147 | 0.182 | -0.034 | 1.92e-08 |
| CAD | PRScs.10FCVal | PRScs.PseudoVal | 0.181 | 0.181 | 0.000 | 8.06e-01 | 0.182 | 0.182 | 0.000 | 9.32e-01 |
| CAD | LDpred2.Inf | PRScs.PseudoVal | 0.162 | 0.181 | -0.019 | 2.46e-15 | 0.157 | 0.182 | -0.024 | 7.55e-07 |
| CAD | PRScs.PseudoVal | PRScs.PseudoVal | 0.181 | 0.181 | 0.000 | 1.00e+00 | 0.182 | 0.182 | 0.000 | 1.00e+00 |
| CAD | LDpred2.10FCVal | PRScs.PseudoVal | 0.188 | 0.181 | 0.007 | 5.17e-06 | 0.184 | 0.182 | 0.002 | 4.55e-01 |
| CAD | pT+clump.10FCVal | PRScs.PseudoVal | 0.135 | 0.181 | -0.046 | 3.89e-35 | 0.134 | 0.182 | -0.048 | 4.87e-10 |
| CAD | PRScs.MultiPRS | PRScs.PseudoVal | 0.186 | 0.181 | 0.005 | 4.76e-01 | 0.186 | 0.182 | 0.004 | 6.60e-02 |
| CAD | lassosum.PseudoVal | PRScs.PseudoVal | 0.162 | 0.181 | -0.019 | 3.52e-09 | 0.160 | 0.182 | -0.022 | 6.59e-04 |
| CAD | lassosum.10FCVal | PRScs.PseudoVal | 0.174 | 0.181 | -0.007 | 8.71e-04 | 0.171 | 0.182 | -0.010 | 1.80e-02 |
| CAD | LDpred1.MultiPRS | PRScs.PseudoVal | 0.173 | 0.181 | -0.008 | 2.49e-01 | 0.174 | 0.182 | -0.008 | 1.06e-01 |
| CAD | lassosum.MultiPRS | PRScs.PseudoVal | 0.187 | 0.181 | 0.006 | 3.65e-01 | 0.186 | 0.182 | 0.004 | 2.61e-01 |
| CAD | LDpred1.10FCVal | PRScs.PseudoVal | 0.172 | 0.181 | -0.009 | 3.27e-04 | 0.172 | 0.182 | -0.009 | 5.67e-02 |
| CAD | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.171 | 0.181 | -0.011 | 8.13e-05 | 0.164 | 0.182 | -0.017 | 1.35e-03 |
| CAD | All.MultiPRS | pT+clump.MultiPRS | 0.191 | 0.167 | 0.024 | 5.79e-04 | 0.190 | 0.166 | 0.024 | 4.72e-06 |
| CAD | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.159 | 0.167 | -0.008 | 2.62e-01 | 0.160 | 0.166 | -0.007 | 3.42e-01 |
| CAD | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.173 | 0.167 | 0.006 | 4.00e-01 | 0.174 | 0.166 | 0.008 | 2.06e-01 |
| CAD | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.167 | 0.167 | 0.000 | 1.00e+00 | 0.166 | 0.166 | 0.000 | 1.00e+00 |
| CAD | lassosum.PseudoVal | pT+clump.MultiPRS | 0.162 | 0.167 | -0.005 | 4.64e-01 | 0.160 | 0.166 | -0.006 | 3.08e-01 |
| CAD | PRScs.10FCVal | pT+clump.MultiPRS | 0.181 | 0.167 | 0.014 | 4.05e-02 | 0.182 | 0.166 | 0.016 | 6.35e-03 |
| CAD | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.171 | 0.167 | 0.003 | 6.39e-01 | 0.164 | 0.166 | -0.002 | 7.54e-01 |
| CAD | SBLUP.Inf | pT+clump.MultiPRS | 0.162 | 0.167 | -0.005 | 4.54e-01 | 0.157 | 0.166 | -0.009 | 2.10e-01 |
| CAD | LDpred2.Inf | pT+clump.MultiPRS | 0.162 | 0.167 | -0.005 | 4.31e-01 | 0.157 | 0.166 | -0.009 | 2.36e-01 |
| CAD | pT+clump.10FCVal | pT+clump.MultiPRS | 0.135 | 0.167 | -0.033 | 2.49e-06 | 0.134 | 0.166 | -0.032 | 2.00e-07 |
| CAD | LDpred2.10FCVal | pT+clump.MultiPRS | 0.188 | 0.167 | 0.021 | 2.88e-03 | 0.184 | 0.166 | 0.018 | 1.04e-03 |
| CAD | PRScs.PseudoVal | pT+clump.MultiPRS | 0.181 | 0.167 | 0.014 | 4.66e-02 | 0.182 | 0.166 | 0.015 | 6.98e-03 |
| CAD | LDpred1.10FCVal | pT+clump.MultiPRS | 0.172 | 0.167 | 0.005 | 4.88e-01 | 0.172 | 0.166 | 0.006 | 3.47e-01 |
| CAD | LDpred1.Inf | pT+clump.MultiPRS | 0.148 | 0.167 | -0.020 | 4.66e-03 | 0.147 | 0.166 | -0.019 | 1.87e-02 |
| CAD | PRScs.MultiPRS | pT+clump.MultiPRS | 0.186 | 0.167 | 0.019 | 6.89e-03 | 0.186 | 0.166 | 0.020 | 1.85e-04 |
| CAD | lassosum.10FCVal | pT+clump.MultiPRS | 0.174 | 0.167 | 0.007 | 3.42e-01 | 0.171 | 0.166 | 0.005 | 4.12e-01 |
| CAD | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.107 | 0.167 | -0.060 | 5.92e-18 | 0.111 | 0.166 | -0.055 | 1.63e-08 |
| CAD | lassosum.MultiPRS | pT+clump.MultiPRS | 0.187 | 0.167 | 0.020 | 3.66e-03 | 0.186 | 0.166 | 0.019 | 1.03e-04 |
| CAD | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.192 | 0.167 | 0.024 | 3.88e-04 | 0.186 | 0.166 | 0.020 | 2.15e-04 |
| CAD | LDpred2.10FCVal | pT+clump.10FCVal | 0.188 | 0.135 | 0.053 | 2.35e-49 | 0.184 | 0.134 | 0.050 | 9.99e-12 |
| CAD | pT+clump.MultiPRS | pT+clump.10FCVal | 0.167 | 0.135 | 0.033 | 2.49e-06 | 0.166 | 0.134 | 0.032 | 2.00e-07 |
| CAD | LDpred2.Inf | pT+clump.10FCVal | 0.162 | 0.135 | 0.027 | 2.87e-16 | 0.157 | 0.134 | 0.023 | 5.01e-04 |
| CAD | SBayesR.PseudoVal | pT+clump.10FCVal | 0.159 | 0.135 | 0.025 | 3.38e-06 | 0.160 | 0.134 | 0.026 | 1.85e-02 |
| CAD | PRScs.MultiPRS | pT+clump.10FCVal | 0.186 | 0.135 | 0.051 | 1.35e-13 | 0.186 | 0.134 | 0.052 | 3.08e-12 |
| CAD | SBLUP.Inf | pT+clump.10FCVal | 0.162 | 0.135 | 0.028 | 3.67e-17 | 0.157 | 0.134 | 0.023 | 4.60e-04 |
| CAD | lassosum.MultiPRS | pT+clump.10FCVal | 0.187 | 0.135 | 0.053 | 3.25e-14 | 0.186 | 0.134 | 0.052 | 6.58e-12 |
| CAD | PRScs.10FCVal | pT+clump.10FCVal | 0.181 | 0.135 | 0.047 | 7.26e-36 | 0.182 | 0.134 | 0.048 | 3.75e-10 |
| CAD | LDpred2.MultiPRS | pT+clump.10FCVal | 0.192 | 0.135 | 0.057 | 1.59e-16 | 0.186 | 0.134 | 0.052 | 4.26e-11 |
| CAD | lassosum.10FCVal | pT+clump.10FCVal | 0.174 | 0.135 | 0.039 | 1.13e-30 | 0.171 | 0.134 | 0.037 | 5.16e-08 |
| CAD | LDpred2.PseudoVal | pT+clump.10FCVal | 0.107 | 0.135 | -0.028 | 5.25e-07 | 0.111 | 0.134 | -0.023 | 3.98e-02 |
| CAD | LDpred1.MultiPRS | pT+clump.10FCVal | 0.173 | 0.135 | 0.039 | 2.70e-08 | 0.174 | 0.134 | 0.040 | 5.74e-06 |
| CAD | pT+clump.10FCVal | pT+clump.10FCVal | 0.135 | 0.135 | 0.000 | 1.00e+00 | 0.134 | 0.134 | 0.000 | 1.00e+00 |
| CAD | All.MultiPRS | pT+clump.10FCVal | 0.191 | 0.135 | 0.056 | 2.96e-17 | 0.190 | 0.134 | 0.056 | 7.05e-13 |
| CAD | LDpred1.Inf | pT+clump.10FCVal | 0.148 | 0.135 | 0.013 | 9.34e-04 | 0.147 | 0.134 | 0.013 | 1.00e-01 |
| CAD | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.171 | 0.135 | 0.036 | 1.19e-14 | 0.164 | 0.134 | 0.030 | 1.35e-03 |
| CAD | LDpred1.10FCVal | pT+clump.10FCVal | 0.172 | 0.135 | 0.038 | 9.68e-17 | 0.172 | 0.134 | 0.038 | 3.33e-05 |
| CAD | lassosum.PseudoVal | pT+clump.10FCVal | 0.162 | 0.135 | 0.028 | 3.12e-08 | 0.160 | 0.134 | 0.026 | 1.05e-02 |
| CAD | PRScs.PseudoVal | pT+clump.10FCVal | 0.181 | 0.135 | 0.046 | 3.89e-35 | 0.182 | 0.134 | 0.048 | 4.87e-10 |
| CAD | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.167 | 0.159 | 0.008 | 2.62e-01 | 0.166 | 0.160 | 0.007 | 3.42e-01 |
| CAD | LDpred1.10FCVal | SBayesR.PseudoVal | 0.172 | 0.159 | 0.013 | 1.80e-05 | 0.172 | 0.160 | 0.013 | 3.25e-02 |
| CAD | PRScs.10FCVal | SBayesR.PseudoVal | 0.181 | 0.159 | 0.022 | 2.00e-10 | 0.182 | 0.160 | 0.022 | 1.29e-03 |
| CAD | LDpred2.10FCVal | SBayesR.PseudoVal | 0.188 | 0.159 | 0.028 | 7.04e-17 | 0.184 | 0.160 | 0.024 | 4.16e-04 |
| CAD | lassosum.10FCVal | SBayesR.PseudoVal | 0.174 | 0.159 | 0.014 | 4.68e-04 | 0.171 | 0.160 | 0.012 | 1.57e-01 |
| CAD | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.107 | 0.159 | -0.053 | 1.47e-25 | 0.111 | 0.160 | -0.049 | 1.54e-06 |
| CAD | lassosum.MultiPRS | SBayesR.PseudoVal | 0.187 | 0.159 | 0.028 | 5.61e-05 | 0.186 | 0.160 | 0.026 | 2.64e-05 |
| CAD | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.192 | 0.159 | 0.032 | 2.97e-06 | 0.186 | 0.160 | 0.026 | 2.31e-05 |
| CAD | All.MultiPRS | SBayesR.PseudoVal | 0.191 | 0.159 | 0.032 | 1.82e-06 | 0.190 | 0.160 | 0.030 | 4.73e-07 |
| CAD | PRScs.PseudoVal | SBayesR.PseudoVal | 0.181 | 0.159 | 0.022 | 4.15e-10 | 0.182 | 0.160 | 0.022 | 1.52e-03 |
| CAD | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.173 | 0.159 | 0.014 | 4.90e-02 | 0.174 | 0.160 | 0.014 | 1.98e-02 |
| CAD | pT+clump.10FCVal | SBayesR.PseudoVal | 0.135 | 0.159 | -0.025 | 3.38e-06 | 0.134 | 0.160 | -0.026 | 1.85e-02 |
| CAD | lassosum.PseudoVal | SBayesR.PseudoVal | 0.162 | 0.159 | 0.003 | 3.22e-01 | 0.160 | 0.160 | 0.000 | 9.68e-01 |
| CAD | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.159 | 0.159 | 0.000 | 1.00e+00 | 0.160 | 0.160 | 0.000 | 1.00e+00 |
| CAD | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.171 | 0.159 | 0.011 | 2.87e-05 | 0.164 | 0.160 | 0.005 | 3.77e-01 |
| CAD | SBLUP.Inf | SBayesR.PseudoVal | 0.162 | 0.159 | 0.003 | 5.91e-01 | 0.157 | 0.160 | -0.003 | 7.92e-01 |
| CAD | LDpred2.Inf | SBayesR.PseudoVal | 0.162 | 0.159 | 0.002 | 6.33e-01 | 0.157 | 0.160 | -0.002 | 8.20e-01 |
| CAD | LDpred1.Inf | SBayesR.PseudoVal | 0.148 | 0.159 | -0.012 | 1.57e-02 | 0.147 | 0.160 | -0.012 | 2.03e-01 |
| CAD | PRScs.MultiPRS | SBayesR.PseudoVal | 0.186 | 0.159 | 0.026 | 1.28e-04 | 0.186 | 0.160 | 0.026 | 6.63e-05 |
| CAD | LDpred2.10FCVal | SBLUP.Inf | 0.188 | 0.162 | 0.026 | 2.86e-31 | 0.184 | 0.157 | 0.027 | 1.60e-09 |
| CAD | PRScs.PseudoVal | SBLUP.Inf | 0.181 | 0.162 | 0.019 | 1.44e-15 | 0.182 | 0.157 | 0.025 | 2.83e-07 |
| CAD | LDpred1.10FCVal | SBLUP.Inf | 0.172 | 0.162 | 0.010 | 5.47e-03 | 0.172 | 0.157 | 0.015 | 3.80e-02 |
| CAD | LDpred2.Inf | SBLUP.Inf | 0.162 | 0.162 | 0.000 | 5.79e-01 | 0.157 | 0.157 | 0.000 | 7.37e-01 |
| CAD | SBayesR.PseudoVal | SBLUP.Inf | 0.159 | 0.162 | -0.003 | 5.91e-01 | 0.160 | 0.157 | 0.003 | 7.92e-01 |
| CAD | lassosum.10FCVal | SBLUP.Inf | 0.174 | 0.162 | 0.012 | 3.21e-07 | 0.171 | 0.157 | 0.014 | 1.92e-03 |
| CAD | pT+clump.MultiPRS | SBLUP.Inf | 0.167 | 0.162 | 0.005 | 4.54e-01 | 0.166 | 0.157 | 0.009 | 2.10e-01 |
| CAD | lassosum.MultiPRS | SBLUP.Inf | 0.187 | 0.162 | 0.025 | 2.69e-04 | 0.186 | 0.157 | 0.028 | 3.19e-07 |
| CAD | PRScs.10FCVal | SBLUP.Inf | 0.181 | 0.162 | 0.019 | 1.82e-16 | 0.182 | 0.157 | 0.025 | 1.91e-07 |
| CAD | PRScs.MultiPRS | SBLUP.Inf | 0.186 | 0.162 | 0.024 | 5.64e-04 | 0.186 | 0.157 | 0.029 | 3.18e-09 |
| CAD | LDpred1.MultiPRS | SBLUP.Inf | 0.173 | 0.162 | 0.011 | 1.10e-01 | 0.174 | 0.157 | 0.017 | 9.91e-03 |
| CAD | LDpred2.PseudoVal | SBLUP.Inf | 0.107 | 0.162 | -0.055 | 1.36e-25 | 0.111 | 0.157 | -0.046 | 1.53e-05 |
| CAD | LDpred1.Inf | SBLUP.Inf | 0.148 | 0.162 | -0.014 | 7.47e-10 | 0.147 | 0.157 | -0.010 | 3.51e-02 |
| CAD | LDpred2.MultiPRS | SBLUP.Inf | 0.192 | 0.162 | 0.030 | 1.72e-05 | 0.186 | 0.157 | 0.029 | 7.35e-08 |
| CAD | All.MultiPRS | SBLUP.Inf | 0.191 | 0.162 | 0.029 | 1.17e-05 | 0.190 | 0.157 | 0.033 | 1.03e-08 |
| CAD | SBLUP.Inf | SBLUP.Inf | 0.162 | 0.162 | 0.000 | 1.00e+00 | 0.157 | 0.157 | 0.000 | 1.00e+00 |
| CAD | DBSLMM.PseudoVal | SBLUP.Inf | 0.171 | 0.162 | 0.008 | 2.86e-02 | 0.164 | 0.157 | 0.007 | 3.48e-01 |
| CAD | pT+clump.10FCVal | SBLUP.Inf | 0.135 | 0.162 | -0.028 | 3.67e-17 | 0.134 | 0.157 | -0.023 | 4.60e-04 |
| CAD | lassosum.PseudoVal | SBLUP.Inf | 0.162 | 0.162 | 0.000 | 9.82e-01 | 0.160 | 0.157 | 0.003 | 7.61e-01 |
| IBD | LDpred2.PseudoVal | All.MultiPRS | 0.100 | 0.146 | -0.046 | 6.78e-11 | 0.103 | 0.151 | -0.048 | 3.06e-10 |
| IBD | pT+clump.10FCVal | All.MultiPRS | 0.112 | 0.146 | -0.034 | 1.23e-06 | 0.112 | 0.151 | -0.038 | 1.18e-08 |
| IBD | LDpred2.MultiPRS | All.MultiPRS | 0.143 | 0.146 | -0.003 | 4.68e-03 | 0.148 | 0.151 | -0.003 | 1.30e-01 |
| IBD | PRScs.PseudoVal | All.MultiPRS | 0.130 | 0.146 | -0.016 | 2.37e-02 | 0.128 | 0.151 | -0.023 | 8.64e-07 |
| IBD | LDpred1.Inf | All.MultiPRS | 0.100 | 0.146 | -0.045 | 9.52e-11 | 0.104 | 0.151 | -0.046 | 2.48e-09 |
| IBD | PRScs.MultiPRS | All.MultiPRS | 0.135 | 0.146 | -0.010 | 1.64e-08 | 0.137 | 0.151 | -0.013 | 2.70e-04 |
| IBD | LDpred2.Inf | All.MultiPRS | 0.096 | 0.146 | -0.050 | 1.80e-12 | 0.099 | 0.151 | -0.052 | 8.38e-11 |
| IBD | lassosum.10FCVal | All.MultiPRS | 0.134 | 0.146 | -0.011 | 1.08e-01 | 0.136 | 0.151 | -0.015 | 3.09e-04 |
| IBD | LDpred2.10FCVal | All.MultiPRS | 0.143 | 0.146 | -0.003 | 7.13e-01 | 0.145 | 0.151 | -0.005 | 2.18e-02 |
| IBD | lassosum.MultiPRS | All.MultiPRS | 0.142 | 0.146 | -0.003 | 5.78e-03 | 0.146 | 0.151 | -0.005 | 2.40e-02 |
| IBD | LDpred1.10FCVal | All.MultiPRS | 0.130 | 0.146 | -0.016 | 2.46e-02 | 0.134 | 0.151 | -0.016 | 5.90e-04 |
| IBD | All.MultiPRS | All.MultiPRS | 0.146 | 0.146 | 0.000 | 1.00e+00 | 0.151 | 0.151 | 0.000 | 1.00e+00 |
| IBD | SBayesR.PseudoVal | All.MultiPRS | 0.122 | 0.146 | -0.023 | 8.33e-04 | 0.130 | 0.151 | -0.021 | 1.68e-04 |
| IBD | LDpred1.MultiPRS | All.MultiPRS | 0.131 | 0.146 | -0.014 | 3.85e-10 | 0.137 | 0.151 | -0.013 | 2.45e-03 |
| IBD | pT+clump.MultiPRS | All.MultiPRS | 0.128 | 0.146 | -0.018 | 1.20e-02 | 0.137 | 0.151 | -0.014 | 4.39e-03 |
| IBD | lassosum.PseudoVal | All.MultiPRS | 0.134 | 0.146 | -0.012 | 9.17e-02 | 0.137 | 0.151 | -0.014 | 6.95e-04 |
| IBD | PRScs.10FCVal | All.MultiPRS | 0.129 | 0.146 | -0.017 | 1.65e-02 | 0.133 | 0.151 | -0.018 | 1.04e-04 |
| IBD | DBSLMM.PseudoVal | All.MultiPRS | 0.131 | 0.146 | -0.015 | 3.45e-02 | 0.135 | 0.151 | -0.016 | 4.88e-04 |
| IBD | SBLUP.Inf | All.MultiPRS | 0.098 | 0.146 | -0.048 | 8.60e-12 | 0.102 | 0.151 | -0.049 | 5.67e-10 |
| IBD | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.122 | 0.131 | -0.009 | 5.95e-03 | 0.130 | 0.135 | -0.005 | 4.09e-01 |
| IBD | LDpred2.Inf | DBSLMM.PseudoVal | 0.096 | 0.131 | -0.035 | 1.11e-15 | 0.099 | 0.135 | -0.036 | 2.78e-05 |
| IBD | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.112 | 0.131 | -0.019 | 3.83e-07 | 0.112 | 0.135 | -0.023 | 2.20e-03 |
| IBD | PRScs.10FCVal | DBSLMM.PseudoVal | 0.129 | 0.131 | -0.002 | 4.92e-01 | 0.133 | 0.135 | -0.002 | 6.91e-01 |
| IBD | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.130 | 0.131 | -0.001 | 7.21e-01 | 0.128 | 0.135 | -0.007 | 2.01e-01 |
| IBD | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.128 | 0.131 | -0.003 | 6.96e-01 | 0.137 | 0.135 | 0.002 | 7.37e-01 |
| IBD | LDpred1.Inf | DBSLMM.PseudoVal | 0.100 | 0.131 | -0.031 | 6.18e-13 | 0.104 | 0.135 | -0.031 | 2.85e-04 |
| IBD | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.142 | 0.131 | 0.012 | 9.45e-02 | 0.146 | 0.135 | 0.010 | 3.19e-02 |
| IBD | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.143 | 0.131 | 0.012 | 1.05e-06 | 0.145 | 0.135 | 0.010 | 4.05e-02 |
| IBD | lassosum.10FCVal | DBSLMM.PseudoVal | 0.134 | 0.131 | 0.004 | 2.77e-01 | 0.136 | 0.135 | 0.000 | 9.50e-01 |
| IBD | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.130 | 0.131 | -0.001 | 7.63e-01 | 0.134 | 0.135 | -0.001 | 8.85e-01 |
| IBD | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.131 | 0.131 | 0.001 | 9.31e-01 | 0.137 | 0.135 | 0.002 | 7.29e-01 |
| IBD | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.135 | 0.131 | 0.005 | 5.18e-01 | 0.137 | 0.135 | 0.002 | 5.89e-01 |
| IBD | All.MultiPRS | DBSLMM.PseudoVal | 0.146 | 0.131 | 0.015 | 3.45e-02 | 0.151 | 0.135 | 0.016 | 4.88e-04 |
| IBD | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.100 | 0.131 | -0.031 | 5.01e-14 | 0.103 | 0.135 | -0.032 | 8.99e-05 |
| IBD | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.131 | 0.131 | 0.000 | 1.00e+00 | 0.135 | 0.135 | 0.000 | 1.00e+00 |
| IBD | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.143 | 0.131 | 0.012 | 8.02e-02 | 0.148 | 0.135 | 0.013 | 5.37e-03 |
| IBD | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.134 | 0.131 | 0.003 | 3.11e-01 | 0.137 | 0.135 | 0.001 | 8.16e-01 |
| IBD | SBLUP.Inf | DBSLMM.PseudoVal | 0.098 | 0.131 | -0.033 | 2.73e-14 | 0.102 | 0.135 | -0.034 | 1.06e-04 |
| IBD | SBayesR.PseudoVal | lassosum.MultiPRS | 0.122 | 0.142 | -0.020 | 3.76e-03 | 0.130 | 0.146 | -0.016 | 6.98e-03 |
| IBD | LDpred2.10FCVal | lassosum.MultiPRS | 0.143 | 0.142 | 0.001 | 9.39e-01 | 0.145 | 0.146 | 0.000 | 9.51e-01 |
| IBD | LDpred1.10FCVal | lassosum.MultiPRS | 0.130 | 0.142 | -0.013 | 7.10e-02 | 0.134 | 0.146 | -0.011 | 2.48e-02 |
| IBD | LDpred2.Inf | lassosum.MultiPRS | 0.096 | 0.142 | -0.046 | 3.78e-11 | 0.099 | 0.146 | -0.047 | 5.28e-09 |
| IBD | PRScs.10FCVal | lassosum.MultiPRS | 0.129 | 0.142 | -0.014 | 5.04e-02 | 0.133 | 0.146 | -0.013 | 7.49e-03 |
| IBD | PRScs.MultiPRS | lassosum.MultiPRS | 0.135 | 0.142 | -0.007 | 2.30e-04 | 0.137 | 0.146 | -0.008 | 3.75e-02 |
| IBD | pT+clump.MultiPRS | lassosum.MultiPRS | 0.128 | 0.142 | -0.014 | 3.84e-02 | 0.137 | 0.146 | -0.008 | 9.83e-02 |
| IBD | lassosum.MultiPRS | lassosum.MultiPRS | 0.142 | 0.142 | 0.000 | 1.00e+00 | 0.146 | 0.146 | 0.000 | 1.00e+00 |
| IBD | lassosum.PseudoVal | lassosum.MultiPRS | 0.134 | 0.142 | -0.009 | 2.14e-01 | 0.137 | 0.146 | -0.009 | 8.44e-03 |
| IBD | LDpred2.MultiPRS | lassosum.MultiPRS | 0.143 | 0.142 | 0.001 | 7.26e-01 | 0.148 | 0.146 | 0.002 | 4.40e-01 |
| IBD | LDpred1.MultiPRS | lassosum.MultiPRS | 0.131 | 0.142 | -0.011 | 4.87e-06 | 0.137 | 0.146 | -0.008 | 7.81e-02 |
| IBD | LDpred2.PseudoVal | lassosum.MultiPRS | 0.100 | 0.142 | -0.043 | 1.15e-09 | 0.103 | 0.146 | -0.043 | 2.14e-08 |
| IBD | LDpred1.Inf | lassosum.MultiPRS | 0.100 | 0.142 | -0.042 | 1.59e-09 | 0.104 | 0.146 | -0.041 | 1.42e-07 |
| IBD | pT+clump.10FCVal | lassosum.MultiPRS | 0.112 | 0.142 | -0.031 | 1.06e-05 | 0.112 | 0.146 | -0.033 | 2.99e-06 |
| IBD | lassosum.10FCVal | lassosum.MultiPRS | 0.134 | 0.142 | -0.008 | 2.45e-01 | 0.136 | 0.146 | -0.010 | 2.62e-03 |
| IBD | SBLUP.Inf | lassosum.MultiPRS | 0.098 | 0.142 | -0.045 | 1.65e-10 | 0.102 | 0.146 | -0.044 | 2.46e-08 |
| IBD | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.131 | 0.142 | -0.012 | 9.45e-02 | 0.135 | 0.146 | -0.010 | 3.19e-02 |
| IBD | All.MultiPRS | lassosum.MultiPRS | 0.146 | 0.142 | 0.003 | 5.78e-03 | 0.151 | 0.146 | 0.005 | 2.40e-02 |
| IBD | PRScs.PseudoVal | lassosum.MultiPRS | 0.130 | 0.142 | -0.013 | 6.88e-02 | 0.128 | 0.146 | -0.018 | 2.05e-04 |
| IBD | LDpred1.Inf | lassosum.PseudoVal | 0.100 | 0.134 | -0.034 | 8.55e-12 | 0.104 | 0.137 | -0.032 | 1.04e-03 |
| IBD | PRScs.10FCVal | lassosum.PseudoVal | 0.129 | 0.134 | -0.005 | 1.37e-01 | 0.133 | 0.137 | -0.004 | 5.86e-01 |
| IBD | LDpred2.MultiPRS | lassosum.PseudoVal | 0.143 | 0.134 | 0.009 | 1.87e-01 | 0.148 | 0.137 | 0.011 | 5.80e-03 |
| IBD | lassosum.PseudoVal | lassosum.PseudoVal | 0.134 | 0.134 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| IBD | PRScs.PseudoVal | lassosum.PseudoVal | 0.130 | 0.134 | -0.004 | 2.35e-01 | 0.128 | 0.137 | -0.009 | 2.00e-01 |
| IBD | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.131 | 0.134 | -0.003 | 3.11e-01 | 0.135 | 0.137 | -0.001 | 8.16e-01 |
| IBD | SBLUP.Inf | lassosum.PseudoVal | 0.098 | 0.134 | -0.036 | 9.64e-13 | 0.102 | 0.137 | -0.035 | 5.22e-04 |
| IBD | LDpred2.Inf | lassosum.PseudoVal | 0.096 | 0.134 | -0.038 | 8.86e-14 | 0.099 | 0.137 | -0.038 | 1.91e-04 |
| IBD | pT+clump.10FCVal | lassosum.PseudoVal | 0.112 | 0.134 | -0.022 | 3.58e-11 | 0.112 | 0.137 | -0.024 | 2.77e-04 |
| IBD | LDpred2.10FCVal | lassosum.PseudoVal | 0.143 | 0.134 | 0.009 | 6.85e-07 | 0.145 | 0.137 | 0.009 | 1.86e-02 |
| IBD | lassosum.MultiPRS | lassosum.PseudoVal | 0.142 | 0.134 | 0.009 | 2.14e-01 | 0.146 | 0.137 | 0.009 | 8.44e-03 |
| IBD | LDpred1.10FCVal | lassosum.PseudoVal | 0.130 | 0.134 | -0.004 | 2.60e-01 | 0.134 | 0.137 | -0.002 | 7.44e-01 |
| IBD | All.MultiPRS | lassosum.PseudoVal | 0.146 | 0.134 | 0.012 | 9.17e-02 | 0.151 | 0.137 | 0.014 | 6.95e-04 |
| IBD | SBayesR.PseudoVal | lassosum.PseudoVal | 0.122 | 0.134 | -0.012 | 5.37e-05 | 0.130 | 0.137 | -0.007 | 2.48e-01 |
| IBD | lassosum.10FCVal | lassosum.PseudoVal | 0.134 | 0.134 | 0.001 | 7.74e-01 | 0.136 | 0.137 | -0.001 | 7.92e-01 |
| IBD | pT+clump.MultiPRS | lassosum.PseudoVal | 0.128 | 0.134 | -0.006 | 4.10e-01 | 0.137 | 0.137 | 0.001 | 9.24e-01 |
| IBD | LDpred2.PseudoVal | lassosum.PseudoVal | 0.100 | 0.134 | -0.034 | 2.12e-12 | 0.103 | 0.137 | -0.034 | 4.99e-04 |
| IBD | LDpred1.MultiPRS | lassosum.PseudoVal | 0.131 | 0.134 | -0.002 | 7.28e-01 | 0.137 | 0.137 | 0.001 | 9.26e-01 |
| IBD | PRScs.MultiPRS | lassosum.PseudoVal | 0.135 | 0.134 | 0.001 | 8.31e-01 | 0.137 | 0.137 | 0.001 | 8.65e-01 |
| IBD | lassosum.10FCVal | lassosum.10FCVal | 0.134 | 0.134 | 0.000 | 1.00e+00 | 0.136 | 0.136 | 0.000 | 1.00e+00 |
| IBD | pT+clump.MultiPRS | lassosum.10FCVal | 0.128 | 0.134 | -0.006 | 3.66e-01 | 0.137 | 0.136 | 0.002 | 7.90e-01 |
| IBD | LDpred2.Inf | lassosum.10FCVal | 0.096 | 0.134 | -0.038 | 1.28e-19 | 0.099 | 0.136 | -0.037 | 1.46e-05 |
| IBD | SBayesR.PseudoVal | lassosum.10FCVal | 0.122 | 0.134 | -0.012 | 3.13e-04 | 0.130 | 0.136 | -0.006 | 4.07e-01 |
| IBD | PRScs.MultiPRS | lassosum.10FCVal | 0.135 | 0.134 | 0.001 | 8.93e-01 | 0.137 | 0.136 | 0.002 | 7.13e-01 |
| IBD | SBLUP.Inf | lassosum.10FCVal | 0.098 | 0.134 | -0.037 | 2.49e-18 | 0.102 | 0.136 | -0.034 | 5.12e-05 |
| IBD | lassosum.MultiPRS | lassosum.10FCVal | 0.142 | 0.134 | 0.008 | 2.45e-01 | 0.146 | 0.136 | 0.010 | 2.62e-03 |
| IBD | PRScs.10FCVal | lassosum.10FCVal | 0.129 | 0.134 | -0.006 | 4.49e-02 | 0.133 | 0.136 | -0.003 | 6.32e-01 |
| IBD | LDpred2.MultiPRS | lassosum.10FCVal | 0.143 | 0.134 | 0.009 | 2.15e-01 | 0.148 | 0.136 | 0.012 | 3.63e-03 |
| IBD | PRScs.PseudoVal | lassosum.10FCVal | 0.130 | 0.134 | -0.005 | 9.96e-02 | 0.128 | 0.136 | -0.008 | 1.69e-01 |
| IBD | LDpred2.PseudoVal | lassosum.10FCVal | 0.100 | 0.134 | -0.035 | 1.07e-17 | 0.103 | 0.136 | -0.033 | 5.44e-05 |
| IBD | LDpred1.MultiPRS | lassosum.10FCVal | 0.131 | 0.134 | -0.003 | 6.70e-01 | 0.137 | 0.136 | 0.002 | 7.73e-01 |
| IBD | pT+clump.10FCVal | lassosum.10FCVal | 0.112 | 0.134 | -0.023 | 2.18e-08 | 0.112 | 0.136 | -0.023 | 3.88e-03 |
| IBD | All.MultiPRS | lassosum.10FCVal | 0.146 | 0.134 | 0.011 | 1.08e-01 | 0.151 | 0.136 | 0.015 | 3.09e-04 |
| IBD | LDpred1.Inf | lassosum.10FCVal | 0.100 | 0.134 | -0.034 | 2.23e-16 | 0.104 | 0.136 | -0.031 | 1.82e-04 |
| IBD | DBSLMM.PseudoVal | lassosum.10FCVal | 0.131 | 0.134 | -0.004 | 2.77e-01 | 0.135 | 0.136 | 0.000 | 9.50e-01 |
| IBD | LDpred1.10FCVal | lassosum.10FCVal | 0.130 | 0.134 | -0.004 | 1.23e-01 | 0.134 | 0.136 | -0.001 | 8.26e-01 |
| IBD | lassosum.PseudoVal | lassosum.10FCVal | 0.134 | 0.134 | -0.001 | 7.74e-01 | 0.137 | 0.136 | 0.001 | 7.92e-01 |
| IBD | LDpred2.10FCVal | lassosum.10FCVal | 0.143 | 0.134 | 0.009 | 5.93e-05 | 0.145 | 0.136 | 0.010 | 2.26e-02 |
| IBD | LDpred2.10FCVal | LDpred1.MultiPRS | 0.143 | 0.131 | 0.012 | 9.54e-02 | 0.145 | 0.137 | 0.008 | 1.09e-01 |
| IBD | PRScs.PseudoVal | LDpred1.MultiPRS | 0.130 | 0.131 | -0.002 | 8.16e-01 | 0.128 | 0.137 | -0.009 | 1.37e-02 |
| IBD | LDpred2.Inf | LDpred1.MultiPRS | 0.096 | 0.131 | -0.035 | 5.13e-07 | 0.099 | 0.137 | -0.038 | 3.73e-11 |
| IBD | pT+clump.10FCVal | LDpred1.MultiPRS | 0.112 | 0.131 | -0.020 | 4.81e-03 | 0.112 | 0.137 | -0.025 | 3.53e-03 |
| IBD | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.122 | 0.131 | -0.009 | 1.91e-01 | 0.130 | 0.137 | -0.007 | 2.90e-01 |
| IBD | All.MultiPRS | LDpred1.MultiPRS | 0.146 | 0.131 | 0.014 | 3.85e-10 | 0.151 | 0.137 | 0.013 | 2.45e-03 |
| IBD | LDpred1.10FCVal | LDpred1.MultiPRS | 0.130 | 0.131 | -0.002 | 8.28e-01 | 0.134 | 0.137 | -0.003 | 2.83e-02 |
| IBD | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.131 | 0.131 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| IBD | PRScs.10FCVal | LDpred1.MultiPRS | 0.129 | 0.131 | -0.003 | 7.13e-01 | 0.133 | 0.137 | -0.004 | 2.59e-01 |
| IBD | lassosum.PseudoVal | LDpred1.MultiPRS | 0.134 | 0.131 | 0.002 | 7.28e-01 | 0.137 | 0.137 | -0.001 | 9.26e-01 |
| IBD | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.128 | 0.131 | -0.003 | 6.32e-01 | 0.137 | 0.137 | 0.000 | 9.93e-01 |
| IBD | LDpred1.Inf | LDpred1.MultiPRS | 0.100 | 0.131 | -0.031 | 8.98e-06 | 0.104 | 0.137 | -0.033 | 1.21e-11 |
| IBD | lassosum.MultiPRS | LDpred1.MultiPRS | 0.142 | 0.131 | 0.011 | 4.87e-06 | 0.146 | 0.137 | 0.008 | 7.81e-02 |
| IBD | PRScs.MultiPRS | LDpred1.MultiPRS | 0.135 | 0.131 | 0.004 | 4.74e-02 | 0.137 | 0.137 | 0.000 | 9.29e-01 |
| IBD | lassosum.10FCVal | LDpred1.MultiPRS | 0.134 | 0.131 | 0.003 | 6.70e-01 | 0.136 | 0.137 | -0.002 | 7.73e-01 |
| IBD | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.100 | 0.131 | -0.032 | 6.87e-06 | 0.103 | 0.137 | -0.034 | 1.36e-10 |
| IBD | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.131 | 0.131 | -0.001 | 9.31e-01 | 0.135 | 0.137 | -0.002 | 7.29e-01 |
| IBD | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.143 | 0.131 | 0.012 | 2.01e-07 | 0.148 | 0.137 | 0.011 | 1.36e-02 |
| IBD | SBLUP.Inf | LDpred1.MultiPRS | 0.098 | 0.131 | -0.034 | 1.59e-06 | 0.102 | 0.137 | -0.036 | 5.31e-10 |
| IBD | All.MultiPRS | LDpred1.Inf | 0.146 | 0.100 | 0.045 | 9.52e-11 | 0.151 | 0.104 | 0.046 | 2.48e-09 |
| IBD | LDpred2.PseudoVal | LDpred1.Inf | 0.100 | 0.100 | 0.000 | 7.79e-01 | 0.103 | 0.104 | -0.001 | 6.52e-01 |
| IBD | SBayesR.PseudoVal | LDpred1.Inf | 0.122 | 0.100 | 0.022 | 5.19e-06 | 0.130 | 0.104 | 0.026 | 8.42e-03 |
| IBD | LDpred2.MultiPRS | LDpred1.Inf | 0.143 | 0.100 | 0.043 | 1.00e-09 | 0.148 | 0.104 | 0.044 | 3.44e-08 |
| IBD | PRScs.PseudoVal | LDpred1.Inf | 0.130 | 0.100 | 0.030 | 3.08e-26 | 0.128 | 0.104 | 0.023 | 2.67e-05 |
| IBD | LDpred1.Inf | LDpred1.Inf | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.104 | 0.104 | 0.000 | 1.00e+00 |
| IBD | pT+clump.10FCVal | LDpred1.Inf | 0.112 | 0.100 | 0.011 | 3.66e-02 | 0.112 | 0.104 | 0.008 | 4.73e-01 |
| IBD | PRScs.MultiPRS | LDpred1.Inf | 0.135 | 0.100 | 0.035 | 5.71e-07 | 0.137 | 0.104 | 0.033 | 1.28e-06 |
| IBD | lassosum.PseudoVal | LDpred1.Inf | 0.134 | 0.100 | 0.034 | 8.55e-12 | 0.137 | 0.104 | 0.032 | 1.04e-03 |
| IBD | pT+clump.MultiPRS | LDpred1.Inf | 0.128 | 0.100 | 0.028 | 6.84e-05 | 0.137 | 0.104 | 0.033 | 9.33e-05 |
| IBD | LDpred1.MultiPRS | LDpred1.Inf | 0.131 | 0.100 | 0.031 | 8.98e-06 | 0.137 | 0.104 | 0.033 | 1.21e-11 |
| IBD | PRScs.10FCVal | LDpred1.Inf | 0.129 | 0.100 | 0.029 | 1.33e-23 | 0.133 | 0.104 | 0.029 | 4.76e-07 |
| IBD | LDpred2.Inf | LDpred1.Inf | 0.096 | 0.100 | -0.004 | 9.05e-03 | 0.099 | 0.104 | -0.005 | 8.81e-02 |
| IBD | SBLUP.Inf | LDpred1.Inf | 0.098 | 0.100 | -0.003 | 1.14e-01 | 0.102 | 0.104 | -0.003 | 3.85e-01 |
| IBD | DBSLMM.PseudoVal | LDpred1.Inf | 0.131 | 0.100 | 0.031 | 6.18e-13 | 0.135 | 0.104 | 0.031 | 2.85e-04 |
| IBD | lassosum.MultiPRS | LDpred1.Inf | 0.142 | 0.100 | 0.042 | 1.59e-09 | 0.146 | 0.104 | 0.041 | 1.42e-07 |
| IBD | LDpred1.10FCVal | LDpred1.Inf | 0.130 | 0.100 | 0.030 | 3.19e-46 | 0.134 | 0.104 | 0.030 | 6.93e-13 |
| IBD | lassosum.10FCVal | LDpred1.Inf | 0.134 | 0.100 | 0.034 | 2.23e-16 | 0.136 | 0.104 | 0.031 | 1.82e-04 |
| IBD | LDpred2.10FCVal | LDpred1.Inf | 0.143 | 0.100 | 0.043 | 5.37e-23 | 0.145 | 0.104 | 0.041 | 2.09e-06 |
| IBD | LDpred1.Inf | LDpred1.10FCVal | 0.100 | 0.130 | -0.030 | 3.19e-46 | 0.104 | 0.134 | -0.030 | 6.93e-13 |
| IBD | PRScs.MultiPRS | LDpred1.10FCVal | 0.135 | 0.130 | 0.005 | 4.37e-01 | 0.137 | 0.134 | 0.003 | 4.20e-01 |
| IBD | LDpred1.10FCVal | LDpred1.10FCVal | 0.130 | 0.130 | 0.000 | 1.00e+00 | 0.134 | 0.134 | 0.000 | 1.00e+00 |
| IBD | All.MultiPRS | LDpred1.10FCVal | 0.146 | 0.130 | 0.016 | 2.46e-02 | 0.151 | 0.134 | 0.016 | 5.90e-04 |
| IBD | SBayesR.PseudoVal | LDpred1.10FCVal | 0.122 | 0.130 | -0.008 | 3.13e-02 | 0.130 | 0.134 | -0.004 | 5.44e-01 |
| IBD | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.131 | 0.130 | 0.001 | 7.63e-01 | 0.135 | 0.134 | 0.001 | 8.85e-01 |
| IBD | pT+clump.MultiPRS | LDpred1.10FCVal | 0.128 | 0.130 | -0.002 | 7.94e-01 | 0.137 | 0.134 | 0.003 | 6.46e-01 |
| IBD | lassosum.MultiPRS | LDpred1.10FCVal | 0.142 | 0.130 | 0.013 | 7.10e-02 | 0.146 | 0.134 | 0.011 | 2.48e-02 |
| IBD | PRScs.10FCVal | LDpred1.10FCVal | 0.129 | 0.130 | -0.001 | 5.65e-01 | 0.133 | 0.134 | -0.001 | 7.01e-01 |
| IBD | lassosum.10FCVal | LDpred1.10FCVal | 0.134 | 0.130 | 0.004 | 1.23e-01 | 0.136 | 0.134 | 0.001 | 8.26e-01 |
| IBD | SBLUP.Inf | LDpred1.10FCVal | 0.098 | 0.130 | -0.032 | 1.52e-34 | 0.102 | 0.134 | -0.033 | 3.45e-10 |
| IBD | LDpred2.PseudoVal | LDpred1.10FCVal | 0.100 | 0.130 | -0.030 | 7.70e-36 | 0.103 | 0.134 | -0.031 | 7.10e-11 |
| IBD | pT+clump.10FCVal | LDpred1.10FCVal | 0.112 | 0.130 | -0.018 | 4.74e-05 | 0.112 | 0.134 | -0.022 | 1.27e-02 |
| IBD | LDpred2.MultiPRS | LDpred1.10FCVal | 0.143 | 0.130 | 0.013 | 5.97e-02 | 0.148 | 0.134 | 0.014 | 3.70e-03 |
| IBD | PRScs.PseudoVal | LDpred1.10FCVal | 0.130 | 0.130 | 0.000 | 9.50e-01 | 0.128 | 0.134 | -0.006 | 7.48e-02 |
| IBD | LDpred2.10FCVal | LDpred1.10FCVal | 0.143 | 0.130 | 0.013 | 2.26e-06 | 0.145 | 0.134 | 0.011 | 4.34e-02 |
| IBD | LDpred1.MultiPRS | LDpred1.10FCVal | 0.131 | 0.130 | 0.002 | 8.28e-01 | 0.137 | 0.134 | 0.003 | 2.83e-02 |
| IBD | LDpred2.Inf | LDpred1.10FCVal | 0.096 | 0.130 | -0.034 | 1.35e-37 | 0.099 | 0.134 | -0.035 | 1.75e-11 |
| IBD | lassosum.PseudoVal | LDpred1.10FCVal | 0.134 | 0.130 | 0.004 | 2.60e-01 | 0.137 | 0.134 | 0.002 | 7.44e-01 |
| IBD | LDpred1.Inf | LDpred2.MultiPRS | 0.100 | 0.143 | -0.043 | 1.00e-09 | 0.104 | 0.148 | -0.044 | 3.44e-08 |
| IBD | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.131 | 0.143 | -0.012 | 8.02e-02 | 0.135 | 0.148 | -0.013 | 5.37e-03 |
| IBD | SBLUP.Inf | LDpred2.MultiPRS | 0.098 | 0.143 | -0.045 | 1.03e-10 | 0.102 | 0.148 | -0.046 | 7.79e-09 |
| IBD | LDpred2.Inf | LDpred2.MultiPRS | 0.096 | 0.143 | -0.047 | 2.32e-11 | 0.099 | 0.148 | -0.049 | 9.28e-10 |
| IBD | pT+clump.10FCVal | LDpred2.MultiPRS | 0.112 | 0.143 | -0.031 | 7.28e-06 | 0.112 | 0.148 | -0.036 | 2.91e-07 |
| IBD | LDpred2.10FCVal | LDpred2.MultiPRS | 0.143 | 0.143 | 0.000 | 9.97e-01 | 0.145 | 0.148 | -0.003 | 5.07e-02 |
| IBD | PRScs.PseudoVal | LDpred2.MultiPRS | 0.130 | 0.143 | -0.013 | 5.78e-02 | 0.128 | 0.148 | -0.020 | 1.22e-05 |
| IBD | LDpred1.10FCVal | LDpred2.MultiPRS | 0.130 | 0.143 | -0.013 | 5.97e-02 | 0.134 | 0.148 | -0.014 | 3.70e-03 |
| IBD | All.MultiPRS | LDpred2.MultiPRS | 0.146 | 0.143 | 0.003 | 4.68e-03 | 0.151 | 0.148 | 0.003 | 1.30e-01 |
| IBD | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.122 | 0.143 | -0.021 | 2.93e-03 | 0.130 | 0.148 | -0.018 | 6.92e-04 |
| IBD | lassosum.10FCVal | LDpred2.MultiPRS | 0.134 | 0.143 | -0.009 | 2.15e-01 | 0.136 | 0.148 | -0.012 | 3.63e-03 |
| IBD | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.128 | 0.143 | -0.015 | 3.18e-02 | 0.137 | 0.148 | -0.011 | 3.66e-02 |
| IBD | lassosum.MultiPRS | LDpred2.MultiPRS | 0.142 | 0.143 | -0.001 | 7.26e-01 | 0.146 | 0.148 | -0.002 | 4.40e-01 |
| IBD | PRScs.10FCVal | LDpred2.MultiPRS | 0.129 | 0.143 | -0.014 | 4.20e-02 | 0.133 | 0.148 | -0.015 | 7.61e-04 |
| IBD | PRScs.MultiPRS | LDpred2.MultiPRS | 0.135 | 0.143 | -0.008 | 1.43e-05 | 0.137 | 0.148 | -0.010 | 2.96e-03 |
| IBD | lassosum.PseudoVal | LDpred2.MultiPRS | 0.134 | 0.143 | -0.009 | 1.87e-01 | 0.137 | 0.148 | -0.011 | 5.80e-03 |
| IBD | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.100 | 0.143 | -0.043 | 7.24e-10 | 0.103 | 0.148 | -0.045 | 2.56e-09 |
| IBD | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.131 | 0.143 | -0.012 | 2.01e-07 | 0.137 | 0.148 | -0.011 | 1.36e-02 |
| IBD | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.148 | 0.148 | 0.000 | 1.00e+00 |
| IBD | LDpred1.Inf | LDpred2.10FCVal | 0.100 | 0.143 | -0.043 | 5.37e-23 | 0.104 | 0.145 | -0.041 | 2.09e-06 |
| IBD | PRScs.MultiPRS | LDpred2.10FCVal | 0.135 | 0.143 | -0.008 | 2.69e-01 | 0.137 | 0.145 | -0.008 | 6.25e-02 |
| IBD | LDpred2.MultiPRS | LDpred2.10FCVal | 0.143 | 0.143 | 0.000 | 9.97e-01 | 0.148 | 0.145 | 0.003 | 5.07e-02 |
| IBD | PRScs.PseudoVal | LDpred2.10FCVal | 0.130 | 0.143 | -0.013 | 8.11e-07 | 0.128 | 0.145 | -0.018 | 1.05e-03 |
| IBD | LDpred2.10FCVal | LDpred2.10FCVal | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.145 | 0.145 | 0.000 | 1.00e+00 |
| IBD | lassosum.MultiPRS | LDpred2.10FCVal | 0.142 | 0.143 | -0.001 | 9.39e-01 | 0.146 | 0.145 | 0.000 | 9.51e-01 |
| IBD | LDpred1.10FCVal | LDpred2.10FCVal | 0.130 | 0.143 | -0.013 | 2.26e-06 | 0.134 | 0.145 | -0.011 | 4.34e-02 |
| IBD | All.MultiPRS | LDpred2.10FCVal | 0.146 | 0.143 | 0.003 | 7.13e-01 | 0.151 | 0.145 | 0.005 | 2.18e-02 |
| IBD | SBayesR.PseudoVal | LDpred2.10FCVal | 0.122 | 0.143 | -0.021 | 8.56e-15 | 0.130 | 0.145 | -0.015 | 4.05e-03 |
| IBD | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.131 | 0.143 | -0.012 | 1.05e-06 | 0.135 | 0.145 | -0.010 | 4.05e-02 |
| IBD | pT+clump.MultiPRS | LDpred2.10FCVal | 0.128 | 0.143 | -0.015 | 3.22e-02 | 0.137 | 0.145 | -0.008 | 1.27e-01 |
| IBD | lassosum.PseudoVal | LDpred2.10FCVal | 0.134 | 0.143 | -0.009 | 6.85e-07 | 0.137 | 0.145 | -0.009 | 1.86e-02 |
| IBD | PRScs.10FCVal | LDpred2.10FCVal | 0.129 | 0.143 | -0.014 | 7.53e-08 | 0.133 | 0.145 | -0.012 | 1.75e-02 |
| IBD | lassosum.10FCVal | LDpred2.10FCVal | 0.134 | 0.143 | -0.009 | 5.93e-05 | 0.136 | 0.145 | -0.010 | 2.26e-02 |
| IBD | SBLUP.Inf | LDpred2.10FCVal | 0.098 | 0.143 | -0.045 | 2.29e-24 | 0.102 | 0.145 | -0.044 | 7.02e-07 |
| IBD | LDpred2.PseudoVal | LDpred2.10FCVal | 0.100 | 0.143 | -0.043 | 8.00e-25 | 0.103 | 0.145 | -0.042 | 3.76e-07 |
| IBD | pT+clump.10FCVal | LDpred2.10FCVal | 0.112 | 0.143 | -0.031 | 1.07e-20 | 0.112 | 0.145 | -0.033 | 7.17e-07 |
| IBD | LDpred2.Inf | LDpred2.10FCVal | 0.096 | 0.143 | -0.047 | 3.12e-26 | 0.099 | 0.145 | -0.046 | 1.31e-07 |
| IBD | LDpred1.MultiPRS | LDpred2.10FCVal | 0.131 | 0.143 | -0.012 | 9.54e-02 | 0.137 | 0.145 | -0.008 | 1.09e-01 |
| IBD | lassosum.10FCVal | LDpred2.PseudoVal | 0.134 | 0.100 | 0.035 | 1.07e-17 | 0.136 | 0.103 | 0.033 | 5.44e-05 |
| IBD | SBLUP.Inf | LDpred2.PseudoVal | 0.098 | 0.100 | -0.002 | 1.13e-02 | 0.102 | 0.103 | -0.001 | 4.08e-01 |
| IBD | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.122 | 0.100 | 0.023 | 3.30e-06 | 0.130 | 0.103 | 0.027 | 5.52e-03 |
| IBD | All.MultiPRS | LDpred2.PseudoVal | 0.146 | 0.100 | 0.046 | 6.78e-11 | 0.151 | 0.103 | 0.048 | 3.06e-10 |
| IBD | PRScs.PseudoVal | LDpred2.PseudoVal | 0.130 | 0.100 | 0.030 | 3.68e-33 | 0.128 | 0.103 | 0.025 | 6.09e-07 |
| IBD | lassosum.MultiPRS | LDpred2.PseudoVal | 0.142 | 0.100 | 0.043 | 1.15e-09 | 0.146 | 0.103 | 0.043 | 2.14e-08 |
| IBD | PRScs.MultiPRS | LDpred2.PseudoVal | 0.135 | 0.100 | 0.036 | 4.27e-07 | 0.137 | 0.103 | 0.035 | 8.23e-08 |
| IBD | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.143 | 0.100 | 0.043 | 7.24e-10 | 0.148 | 0.103 | 0.045 | 2.56e-09 |
| IBD | PRScs.10FCVal | LDpred2.PseudoVal | 0.129 | 0.100 | 0.029 | 5.76e-30 | 0.133 | 0.103 | 0.030 | 2.87e-09 |
| IBD | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.103 | 0.103 | 0.000 | 1.00e+00 |
| IBD | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.128 | 0.100 | 0.028 | 5.25e-05 | 0.137 | 0.103 | 0.034 | 3.40e-05 |
| IBD | pT+clump.10FCVal | LDpred2.PseudoVal | 0.112 | 0.100 | 0.012 | 2.91e-02 | 0.112 | 0.103 | 0.009 | 3.94e-01 |
| IBD | lassosum.PseudoVal | LDpred2.PseudoVal | 0.134 | 0.100 | 0.034 | 2.12e-12 | 0.137 | 0.103 | 0.034 | 4.99e-04 |
| IBD | LDpred1.Inf | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 7.79e-01 | 0.104 | 0.103 | 0.001 | 6.52e-01 |
| IBD | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.131 | 0.100 | 0.032 | 6.87e-06 | 0.137 | 0.103 | 0.034 | 1.36e-10 |
| IBD | LDpred1.10FCVal | LDpred2.PseudoVal | 0.130 | 0.100 | 0.030 | 7.70e-36 | 0.134 | 0.103 | 0.031 | 7.10e-11 |
| IBD | LDpred2.Inf | LDpred2.PseudoVal | 0.096 | 0.100 | -0.004 | 4.59e-18 | 0.099 | 0.103 | -0.004 | 2.49e-06 |
| IBD | LDpred2.10FCVal | LDpred2.PseudoVal | 0.143 | 0.100 | 0.043 | 8.00e-25 | 0.145 | 0.103 | 0.042 | 3.76e-07 |
| IBD | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.131 | 0.100 | 0.031 | 5.01e-14 | 0.135 | 0.103 | 0.032 | 8.99e-05 |
| IBD | pT+clump.MultiPRS | LDpred2.Inf | 0.128 | 0.096 | 0.032 | 4.85e-06 | 0.137 | 0.099 | 0.038 | 1.02e-05 |
| IBD | lassosum.MultiPRS | LDpred2.Inf | 0.142 | 0.096 | 0.046 | 3.78e-11 | 0.146 | 0.099 | 0.047 | 5.28e-09 |
| IBD | SBayesR.PseudoVal | LDpred2.Inf | 0.122 | 0.096 | 0.026 | 2.07e-07 | 0.130 | 0.099 | 0.031 | 2.23e-03 |
| IBD | All.MultiPRS | LDpred2.Inf | 0.146 | 0.096 | 0.050 | 1.80e-12 | 0.151 | 0.099 | 0.052 | 8.38e-11 |
| IBD | SBLUP.Inf | LDpred2.Inf | 0.098 | 0.096 | 0.002 | 9.96e-03 | 0.102 | 0.099 | 0.003 | 3.32e-02 |
| IBD | LDpred2.PseudoVal | LDpred2.Inf | 0.100 | 0.096 | 0.004 | 4.59e-18 | 0.103 | 0.099 | 0.004 | 2.49e-06 |
| IBD | PRScs.MultiPRS | LDpred2.Inf | 0.135 | 0.096 | 0.039 | 2.40e-08 | 0.137 | 0.099 | 0.039 | 2.49e-08 |
| IBD | LDpred2.MultiPRS | LDpred2.Inf | 0.143 | 0.096 | 0.047 | 2.32e-11 | 0.148 | 0.099 | 0.049 | 9.28e-10 |
| IBD | PRScs.10FCVal | LDpred2.Inf | 0.129 | 0.096 | 0.033 | 1.97e-32 | 0.133 | 0.099 | 0.034 | 4.51e-10 |
| IBD | LDpred1.Inf | LDpred2.Inf | 0.100 | 0.096 | 0.004 | 9.05e-03 | 0.104 | 0.099 | 0.005 | 8.81e-02 |
| IBD | LDpred1.MultiPRS | LDpred2.Inf | 0.131 | 0.096 | 0.035 | 5.13e-07 | 0.137 | 0.099 | 0.038 | 3.73e-11 |
| IBD | pT+clump.10FCVal | LDpred2.Inf | 0.112 | 0.096 | 0.016 | 5.82e-03 | 0.112 | 0.099 | 0.013 | 2.39e-01 |
| IBD | lassosum.PseudoVal | LDpred2.Inf | 0.134 | 0.096 | 0.038 | 8.86e-14 | 0.137 | 0.099 | 0.038 | 1.91e-04 |
| IBD | PRScs.PseudoVal | LDpred2.Inf | 0.130 | 0.096 | 0.034 | 8.96e-36 | 0.128 | 0.099 | 0.029 | 7.38e-08 |
| IBD | DBSLMM.PseudoVal | LDpred2.Inf | 0.131 | 0.096 | 0.035 | 1.11e-15 | 0.135 | 0.099 | 0.036 | 2.78e-05 |
| IBD | LDpred1.10FCVal | LDpred2.Inf | 0.130 | 0.096 | 0.034 | 1.35e-37 | 0.134 | 0.099 | 0.035 | 1.75e-11 |
| IBD | LDpred2.Inf | LDpred2.Inf | 0.096 | 0.096 | 0.000 | 1.00e+00 | 0.099 | 0.099 | 0.000 | 1.00e+00 |
| IBD | lassosum.10FCVal | LDpred2.Inf | 0.134 | 0.096 | 0.038 | 1.28e-19 | 0.136 | 0.099 | 0.037 | 1.46e-05 |
| IBD | LDpred2.10FCVal | LDpred2.Inf | 0.143 | 0.096 | 0.047 | 3.12e-26 | 0.145 | 0.099 | 0.046 | 1.31e-07 |
| IBD | PRScs.10FCVal | PRScs.MultiPRS | 0.129 | 0.135 | -0.007 | 3.53e-01 | 0.133 | 0.137 | -0.005 | 1.19e-01 |
| IBD | LDpred1.10FCVal | PRScs.MultiPRS | 0.130 | 0.135 | -0.005 | 4.37e-01 | 0.134 | 0.137 | -0.003 | 4.20e-01 |
| IBD | lassosum.MultiPRS | PRScs.MultiPRS | 0.142 | 0.135 | 0.007 | 2.30e-04 | 0.146 | 0.137 | 0.008 | 3.75e-02 |
| IBD | LDpred2.MultiPRS | PRScs.MultiPRS | 0.143 | 0.135 | 0.008 | 1.43e-05 | 0.148 | 0.137 | 0.010 | 2.96e-03 |
| IBD | All.MultiPRS | PRScs.MultiPRS | 0.146 | 0.135 | 0.010 | 1.64e-08 | 0.151 | 0.137 | 0.013 | 2.70e-04 |
| IBD | PRScs.PseudoVal | PRScs.MultiPRS | 0.130 | 0.135 | -0.006 | 4.28e-01 | 0.128 | 0.137 | -0.010 | 9.56e-04 |
| IBD | LDpred1.MultiPRS | PRScs.MultiPRS | 0.131 | 0.135 | -0.004 | 4.74e-02 | 0.137 | 0.137 | 0.000 | 9.29e-01 |
| IBD | pT+clump.MultiPRS | PRScs.MultiPRS | 0.128 | 0.135 | -0.007 | 2.98e-01 | 0.137 | 0.137 | 0.000 | 9.43e-01 |
| IBD | lassosum.PseudoVal | PRScs.MultiPRS | 0.134 | 0.135 | -0.001 | 8.31e-01 | 0.137 | 0.137 | -0.001 | 8.65e-01 |
| IBD | SBayesR.PseudoVal | PRScs.MultiPRS | 0.122 | 0.135 | -0.013 | 6.18e-02 | 0.130 | 0.137 | -0.008 | 2.44e-01 |
| IBD | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.131 | 0.135 | -0.005 | 5.18e-01 | 0.135 | 0.137 | -0.002 | 5.89e-01 |
| IBD | SBLUP.Inf | PRScs.MultiPRS | 0.098 | 0.135 | -0.038 | 8.40e-08 | 0.102 | 0.137 | -0.036 | 1.73e-07 |
| IBD | LDpred2.Inf | PRScs.MultiPRS | 0.096 | 0.135 | -0.039 | 2.40e-08 | 0.099 | 0.137 | -0.039 | 2.49e-08 |
| IBD | pT+clump.10FCVal | PRScs.MultiPRS | 0.112 | 0.135 | -0.024 | 7.24e-04 | 0.112 | 0.137 | -0.025 | 1.37e-03 |
| IBD | LDpred2.10FCVal | PRScs.MultiPRS | 0.143 | 0.135 | 0.008 | 2.69e-01 | 0.145 | 0.137 | 0.008 | 6.25e-02 |
| IBD | lassosum.10FCVal | PRScs.MultiPRS | 0.134 | 0.135 | -0.001 | 8.93e-01 | 0.136 | 0.137 | -0.002 | 7.13e-01 |
| IBD | LDpred2.PseudoVal | PRScs.MultiPRS | 0.100 | 0.135 | -0.036 | 4.27e-07 | 0.103 | 0.137 | -0.035 | 8.23e-08 |
| IBD | LDpred1.Inf | PRScs.MultiPRS | 0.100 | 0.135 | -0.035 | 5.71e-07 | 0.104 | 0.137 | -0.033 | 1.28e-06 |
| IBD | PRScs.MultiPRS | PRScs.MultiPRS | 0.135 | 0.135 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| IBD | LDpred2.PseudoVal | PRScs.10FCVal | 0.100 | 0.129 | -0.029 | 5.76e-30 | 0.103 | 0.133 | -0.030 | 2.87e-09 |
| IBD | PRScs.10FCVal | PRScs.10FCVal | 0.129 | 0.129 | 0.000 | 1.00e+00 | 0.133 | 0.133 | 0.000 | 1.00e+00 |
| IBD | LDpred2.MultiPRS | PRScs.10FCVal | 0.143 | 0.129 | 0.014 | 4.20e-02 | 0.148 | 0.133 | 0.015 | 7.61e-04 |
| IBD | pT+clump.MultiPRS | PRScs.10FCVal | 0.128 | 0.129 | -0.001 | 9.13e-01 | 0.137 | 0.133 | 0.004 | 5.02e-01 |
| IBD | LDpred1.Inf | PRScs.10FCVal | 0.100 | 0.129 | -0.029 | 1.33e-23 | 0.104 | 0.133 | -0.029 | 4.76e-07 |
| IBD | LDpred1.MultiPRS | PRScs.10FCVal | 0.131 | 0.129 | 0.003 | 7.13e-01 | 0.137 | 0.133 | 0.004 | 2.59e-01 |
| IBD | pT+clump.10FCVal | PRScs.10FCVal | 0.112 | 0.129 | -0.017 | 1.25e-04 | 0.112 | 0.133 | -0.021 | 2.03e-02 |
| IBD | lassosum.PseudoVal | PRScs.10FCVal | 0.134 | 0.129 | 0.005 | 1.37e-01 | 0.137 | 0.133 | 0.004 | 5.86e-01 |
| IBD | PRScs.PseudoVal | PRScs.10FCVal | 0.130 | 0.129 | 0.001 | 5.16e-01 | 0.128 | 0.133 | -0.005 | 8.03e-02 |
| IBD | DBSLMM.PseudoVal | PRScs.10FCVal | 0.131 | 0.129 | 0.002 | 4.92e-01 | 0.135 | 0.133 | 0.002 | 6.91e-01 |
| IBD | SBLUP.Inf | PRScs.10FCVal | 0.098 | 0.129 | -0.031 | 7.19e-30 | 0.102 | 0.133 | -0.031 | 6.60e-09 |
| IBD | LDpred2.Inf | PRScs.10FCVal | 0.096 | 0.129 | -0.033 | 1.97e-32 | 0.099 | 0.133 | -0.034 | 4.51e-10 |
| IBD | lassosum.10FCVal | PRScs.10FCVal | 0.134 | 0.129 | 0.006 | 4.49e-02 | 0.136 | 0.133 | 0.003 | 6.32e-01 |
| IBD | LDpred2.10FCVal | PRScs.10FCVal | 0.143 | 0.129 | 0.014 | 7.53e-08 | 0.145 | 0.133 | 0.012 | 1.75e-02 |
| IBD | lassosum.MultiPRS | PRScs.10FCVal | 0.142 | 0.129 | 0.014 | 5.04e-02 | 0.146 | 0.133 | 0.013 | 7.49e-03 |
| IBD | LDpred1.10FCVal | PRScs.10FCVal | 0.130 | 0.129 | 0.001 | 5.65e-01 | 0.134 | 0.133 | 0.001 | 7.01e-01 |
| IBD | All.MultiPRS | PRScs.10FCVal | 0.146 | 0.129 | 0.017 | 1.65e-02 | 0.151 | 0.133 | 0.018 | 1.04e-04 |
| IBD | SBayesR.PseudoVal | PRScs.10FCVal | 0.122 | 0.129 | -0.007 | 7.72e-02 | 0.130 | 0.133 | -0.003 | 6.98e-01 |
| IBD | PRScs.MultiPRS | PRScs.10FCVal | 0.135 | 0.129 | 0.007 | 3.53e-01 | 0.137 | 0.133 | 0.005 | 1.19e-01 |
| IBD | All.MultiPRS | PRScs.PseudoVal | 0.146 | 0.130 | 0.016 | 2.37e-02 | 0.151 | 0.128 | 0.023 | 8.64e-07 |
| IBD | pT+clump.MultiPRS | PRScs.PseudoVal | 0.128 | 0.130 | -0.002 | 8.07e-01 | 0.137 | 0.128 | 0.009 | 1.44e-01 |
| IBD | LDpred2.PseudoVal | PRScs.PseudoVal | 0.100 | 0.130 | -0.030 | 3.68e-33 | 0.103 | 0.128 | -0.025 | 6.09e-07 |
| IBD | SBayesR.PseudoVal | PRScs.PseudoVal | 0.122 | 0.130 | -0.008 | 4.60e-02 | 0.130 | 0.128 | 0.002 | 7.75e-01 |
| IBD | LDpred2.MultiPRS | PRScs.PseudoVal | 0.143 | 0.130 | 0.013 | 5.78e-02 | 0.148 | 0.128 | 0.020 | 1.22e-05 |
| IBD | SBLUP.Inf | PRScs.PseudoVal | 0.098 | 0.130 | -0.032 | 3.04e-33 | 0.102 | 0.128 | -0.026 | 6.93e-07 |
| IBD | LDpred1.Inf | PRScs.PseudoVal | 0.100 | 0.130 | -0.030 | 3.08e-26 | 0.104 | 0.128 | -0.023 | 2.67e-05 |
| IBD | PRScs.10FCVal | PRScs.PseudoVal | 0.129 | 0.130 | -0.001 | 5.16e-01 | 0.133 | 0.128 | 0.005 | 8.03e-02 |
| IBD | LDpred2.Inf | PRScs.PseudoVal | 0.096 | 0.130 | -0.034 | 8.96e-36 | 0.099 | 0.128 | -0.029 | 7.38e-08 |
| IBD | PRScs.PseudoVal | PRScs.PseudoVal | 0.130 | 0.130 | 0.000 | 1.00e+00 | 0.128 | 0.128 | 0.000 | 1.00e+00 |
| IBD | LDpred2.10FCVal | PRScs.PseudoVal | 0.143 | 0.130 | 0.013 | 8.11e-07 | 0.145 | 0.128 | 0.018 | 1.05e-03 |
| IBD | pT+clump.10FCVal | PRScs.PseudoVal | 0.112 | 0.130 | -0.018 | 6.32e-05 | 0.112 | 0.128 | -0.016 | 8.35e-02 |
| IBD | PRScs.MultiPRS | PRScs.PseudoVal | 0.135 | 0.130 | 0.006 | 4.28e-01 | 0.137 | 0.128 | 0.010 | 9.56e-04 |
| IBD | lassosum.PseudoVal | PRScs.PseudoVal | 0.134 | 0.130 | 0.004 | 2.35e-01 | 0.137 | 0.128 | 0.009 | 2.00e-01 |
| IBD | lassosum.10FCVal | PRScs.PseudoVal | 0.134 | 0.130 | 0.005 | 9.96e-02 | 0.136 | 0.128 | 0.008 | 1.69e-01 |
| IBD | LDpred1.MultiPRS | PRScs.PseudoVal | 0.131 | 0.130 | 0.002 | 8.16e-01 | 0.137 | 0.128 | 0.009 | 1.37e-02 |
| IBD | lassosum.MultiPRS | PRScs.PseudoVal | 0.142 | 0.130 | 0.013 | 6.88e-02 | 0.146 | 0.128 | 0.018 | 2.05e-04 |
| IBD | LDpred1.10FCVal | PRScs.PseudoVal | 0.130 | 0.130 | 0.000 | 9.50e-01 | 0.134 | 0.128 | 0.006 | 7.48e-02 |
| IBD | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.131 | 0.130 | 0.001 | 7.21e-01 | 0.135 | 0.128 | 0.007 | 2.01e-01 |
| IBD | All.MultiPRS | pT+clump.MultiPRS | 0.146 | 0.128 | 0.018 | 1.20e-02 | 0.151 | 0.137 | 0.014 | 4.39e-03 |
| IBD | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.122 | 0.128 | -0.006 | 4.08e-01 | 0.130 | 0.137 | -0.007 | 3.00e-01 |
| IBD | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.131 | 0.128 | 0.003 | 6.32e-01 | 0.137 | 0.137 | 0.000 | 9.93e-01 |
| IBD | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.128 | 0.128 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| IBD | lassosum.PseudoVal | pT+clump.MultiPRS | 0.134 | 0.128 | 0.006 | 4.10e-01 | 0.137 | 0.137 | -0.001 | 9.24e-01 |
| IBD | PRScs.10FCVal | pT+clump.MultiPRS | 0.129 | 0.128 | 0.001 | 9.13e-01 | 0.133 | 0.137 | -0.004 | 5.02e-01 |
| IBD | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.131 | 0.128 | 0.003 | 6.96e-01 | 0.135 | 0.137 | -0.002 | 7.37e-01 |
| IBD | SBLUP.Inf | pT+clump.MultiPRS | 0.098 | 0.128 | -0.030 | 1.38e-05 | 0.102 | 0.137 | -0.036 | 3.59e-05 |
| IBD | LDpred2.Inf | pT+clump.MultiPRS | 0.096 | 0.128 | -0.032 | 4.85e-06 | 0.099 | 0.137 | -0.038 | 1.02e-05 |
| IBD | pT+clump.10FCVal | pT+clump.MultiPRS | 0.112 | 0.128 | -0.016 | 1.93e-02 | 0.112 | 0.137 | -0.025 | 9.88e-07 |
| IBD | LDpred2.10FCVal | pT+clump.MultiPRS | 0.143 | 0.128 | 0.015 | 3.22e-02 | 0.145 | 0.137 | 0.008 | 1.27e-01 |
| IBD | PRScs.PseudoVal | pT+clump.MultiPRS | 0.130 | 0.128 | 0.002 | 8.07e-01 | 0.128 | 0.137 | -0.009 | 1.44e-01 |
| IBD | LDpred1.10FCVal | pT+clump.MultiPRS | 0.130 | 0.128 | 0.002 | 7.94e-01 | 0.134 | 0.137 | -0.003 | 6.46e-01 |
| IBD | LDpred1.Inf | pT+clump.MultiPRS | 0.100 | 0.128 | -0.028 | 6.84e-05 | 0.104 | 0.137 | -0.033 | 9.33e-05 |
| IBD | PRScs.MultiPRS | pT+clump.MultiPRS | 0.135 | 0.128 | 0.007 | 2.98e-01 | 0.137 | 0.137 | 0.000 | 9.43e-01 |
| IBD | lassosum.10FCVal | pT+clump.MultiPRS | 0.134 | 0.128 | 0.006 | 3.66e-01 | 0.136 | 0.137 | -0.002 | 7.90e-01 |
| IBD | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.100 | 0.128 | -0.028 | 5.25e-05 | 0.103 | 0.137 | -0.034 | 3.40e-05 |
| IBD | lassosum.MultiPRS | pT+clump.MultiPRS | 0.142 | 0.128 | 0.014 | 3.84e-02 | 0.146 | 0.137 | 0.008 | 9.83e-02 |
| IBD | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.143 | 0.128 | 0.015 | 3.18e-02 | 0.148 | 0.137 | 0.011 | 3.66e-02 |
| IBD | LDpred2.10FCVal | pT+clump.10FCVal | 0.143 | 0.112 | 0.031 | 1.07e-20 | 0.145 | 0.112 | 0.033 | 7.17e-07 |
| IBD | pT+clump.MultiPRS | pT+clump.10FCVal | 0.128 | 0.112 | 0.016 | 1.93e-02 | 0.137 | 0.112 | 0.025 | 9.88e-07 |
| IBD | LDpred2.Inf | pT+clump.10FCVal | 0.096 | 0.112 | -0.016 | 5.82e-03 | 0.099 | 0.112 | -0.013 | 2.39e-01 |
| IBD | SBayesR.PseudoVal | pT+clump.10FCVal | 0.122 | 0.112 | 0.011 | 4.15e-03 | 0.130 | 0.112 | 0.018 | 1.55e-02 |
| IBD | PRScs.MultiPRS | pT+clump.10FCVal | 0.135 | 0.112 | 0.024 | 7.24e-04 | 0.137 | 0.112 | 0.025 | 1.37e-03 |
| IBD | SBLUP.Inf | pT+clump.10FCVal | 0.098 | 0.112 | -0.014 | 1.37e-02 | 0.102 | 0.112 | -0.011 | 3.47e-01 |
| IBD | lassosum.MultiPRS | pT+clump.10FCVal | 0.142 | 0.112 | 0.031 | 1.06e-05 | 0.146 | 0.112 | 0.033 | 2.99e-06 |
| IBD | PRScs.10FCVal | pT+clump.10FCVal | 0.129 | 0.112 | 0.017 | 1.25e-04 | 0.133 | 0.112 | 0.021 | 2.03e-02 |
| IBD | LDpred2.MultiPRS | pT+clump.10FCVal | 0.143 | 0.112 | 0.031 | 7.28e-06 | 0.148 | 0.112 | 0.036 | 2.91e-07 |
| IBD | lassosum.10FCVal | pT+clump.10FCVal | 0.134 | 0.112 | 0.023 | 2.18e-08 | 0.136 | 0.112 | 0.023 | 3.88e-03 |
| IBD | LDpred2.PseudoVal | pT+clump.10FCVal | 0.100 | 0.112 | -0.012 | 2.91e-02 | 0.103 | 0.112 | -0.009 | 3.94e-01 |
| IBD | LDpred1.MultiPRS | pT+clump.10FCVal | 0.131 | 0.112 | 0.020 | 4.81e-03 | 0.137 | 0.112 | 0.025 | 3.53e-03 |
| IBD | pT+clump.10FCVal | pT+clump.10FCVal | 0.112 | 0.112 | 0.000 | 1.00e+00 | 0.112 | 0.112 | 0.000 | 1.00e+00 |
| IBD | All.MultiPRS | pT+clump.10FCVal | 0.146 | 0.112 | 0.034 | 1.23e-06 | 0.151 | 0.112 | 0.038 | 1.18e-08 |
| IBD | LDpred1.Inf | pT+clump.10FCVal | 0.100 | 0.112 | -0.011 | 3.66e-02 | 0.104 | 0.112 | -0.008 | 4.73e-01 |
| IBD | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.131 | 0.112 | 0.019 | 3.83e-07 | 0.135 | 0.112 | 0.023 | 2.20e-03 |
| IBD | LDpred1.10FCVal | pT+clump.10FCVal | 0.130 | 0.112 | 0.018 | 4.74e-05 | 0.134 | 0.112 | 0.022 | 1.27e-02 |
| IBD | lassosum.PseudoVal | pT+clump.10FCVal | 0.134 | 0.112 | 0.022 | 3.58e-11 | 0.137 | 0.112 | 0.024 | 2.77e-04 |
| IBD | PRScs.PseudoVal | pT+clump.10FCVal | 0.130 | 0.112 | 0.018 | 6.32e-05 | 0.128 | 0.112 | 0.016 | 8.35e-02 |
| IBD | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.128 | 0.122 | 0.006 | 4.08e-01 | 0.137 | 0.130 | 0.007 | 3.00e-01 |
| IBD | LDpred1.10FCVal | SBayesR.PseudoVal | 0.130 | 0.122 | 0.008 | 3.13e-02 | 0.134 | 0.130 | 0.004 | 5.44e-01 |
| IBD | PRScs.10FCVal | SBayesR.PseudoVal | 0.129 | 0.122 | 0.007 | 7.72e-02 | 0.133 | 0.130 | 0.003 | 6.98e-01 |
| IBD | LDpred2.10FCVal | SBayesR.PseudoVal | 0.143 | 0.122 | 0.021 | 8.56e-15 | 0.145 | 0.130 | 0.015 | 4.05e-03 |
| IBD | lassosum.10FCVal | SBayesR.PseudoVal | 0.134 | 0.122 | 0.012 | 3.13e-04 | 0.136 | 0.130 | 0.006 | 4.07e-01 |
| IBD | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.100 | 0.122 | -0.023 | 3.30e-06 | 0.103 | 0.130 | -0.027 | 5.52e-03 |
| IBD | lassosum.MultiPRS | SBayesR.PseudoVal | 0.142 | 0.122 | 0.020 | 3.76e-03 | 0.146 | 0.130 | 0.016 | 6.98e-03 |
| IBD | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.143 | 0.122 | 0.021 | 2.93e-03 | 0.148 | 0.130 | 0.018 | 6.92e-04 |
| IBD | All.MultiPRS | SBayesR.PseudoVal | 0.146 | 0.122 | 0.023 | 8.33e-04 | 0.151 | 0.130 | 0.021 | 1.68e-04 |
| IBD | PRScs.PseudoVal | SBayesR.PseudoVal | 0.130 | 0.122 | 0.008 | 4.60e-02 | 0.128 | 0.130 | -0.002 | 7.75e-01 |
| IBD | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.131 | 0.122 | 0.009 | 1.91e-01 | 0.137 | 0.130 | 0.007 | 2.90e-01 |
| IBD | pT+clump.10FCVal | SBayesR.PseudoVal | 0.112 | 0.122 | -0.011 | 4.15e-03 | 0.112 | 0.130 | -0.018 | 1.55e-02 |
| IBD | lassosum.PseudoVal | SBayesR.PseudoVal | 0.134 | 0.122 | 0.012 | 5.37e-05 | 0.137 | 0.130 | 0.007 | 2.48e-01 |
| IBD | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.122 | 0.122 | 0.000 | 1.00e+00 | 0.130 | 0.130 | 0.000 | 1.00e+00 |
| IBD | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.131 | 0.122 | 0.009 | 5.95e-03 | 0.135 | 0.130 | 0.005 | 4.09e-01 |
| IBD | SBLUP.Inf | SBayesR.PseudoVal | 0.098 | 0.122 | -0.025 | 1.19e-06 | 0.102 | 0.130 | -0.028 | 5.27e-03 |
| IBD | LDpred2.Inf | SBayesR.PseudoVal | 0.096 | 0.122 | -0.026 | 2.07e-07 | 0.099 | 0.130 | -0.031 | 2.23e-03 |
| IBD | LDpred1.Inf | SBayesR.PseudoVal | 0.100 | 0.122 | -0.022 | 5.19e-06 | 0.104 | 0.130 | -0.026 | 8.42e-03 |
| IBD | PRScs.MultiPRS | SBayesR.PseudoVal | 0.135 | 0.122 | 0.013 | 6.18e-02 | 0.137 | 0.130 | 0.008 | 2.44e-01 |
| IBD | LDpred2.10FCVal | SBLUP.Inf | 0.143 | 0.098 | 0.045 | 2.29e-24 | 0.145 | 0.102 | 0.044 | 7.02e-07 |
| IBD | PRScs.PseudoVal | SBLUP.Inf | 0.130 | 0.098 | 0.032 | 3.04e-33 | 0.128 | 0.102 | 0.026 | 6.93e-07 |
| IBD | LDpred1.10FCVal | SBLUP.Inf | 0.130 | 0.098 | 0.032 | 1.52e-34 | 0.134 | 0.102 | 0.033 | 3.45e-10 |
| IBD | LDpred2.Inf | SBLUP.Inf | 0.096 | 0.098 | -0.002 | 9.96e-03 | 0.099 | 0.102 | -0.003 | 3.32e-02 |
| IBD | SBayesR.PseudoVal | SBLUP.Inf | 0.122 | 0.098 | 0.025 | 1.19e-06 | 0.130 | 0.102 | 0.028 | 5.27e-03 |
| IBD | lassosum.10FCVal | SBLUP.Inf | 0.134 | 0.098 | 0.037 | 2.49e-18 | 0.136 | 0.102 | 0.034 | 5.12e-05 |
| IBD | pT+clump.MultiPRS | SBLUP.Inf | 0.128 | 0.098 | 0.030 | 1.38e-05 | 0.137 | 0.102 | 0.036 | 3.59e-05 |
| IBD | lassosum.MultiPRS | SBLUP.Inf | 0.142 | 0.098 | 0.045 | 1.65e-10 | 0.146 | 0.102 | 0.044 | 2.46e-08 |
| IBD | PRScs.10FCVal | SBLUP.Inf | 0.129 | 0.098 | 0.031 | 7.19e-30 | 0.133 | 0.102 | 0.031 | 6.60e-09 |
| IBD | PRScs.MultiPRS | SBLUP.Inf | 0.135 | 0.098 | 0.038 | 8.40e-08 | 0.137 | 0.102 | 0.036 | 1.73e-07 |
| IBD | LDpred1.MultiPRS | SBLUP.Inf | 0.131 | 0.098 | 0.034 | 1.59e-06 | 0.137 | 0.102 | 0.036 | 5.31e-10 |
| IBD | LDpred2.PseudoVal | SBLUP.Inf | 0.100 | 0.098 | 0.002 | 1.13e-02 | 0.103 | 0.102 | 0.001 | 4.08e-01 |
| IBD | LDpred1.Inf | SBLUP.Inf | 0.100 | 0.098 | 0.003 | 1.14e-01 | 0.104 | 0.102 | 0.003 | 3.85e-01 |
| IBD | LDpred2.MultiPRS | SBLUP.Inf | 0.143 | 0.098 | 0.045 | 1.03e-10 | 0.148 | 0.102 | 0.046 | 7.79e-09 |
| IBD | All.MultiPRS | SBLUP.Inf | 0.146 | 0.098 | 0.048 | 8.60e-12 | 0.151 | 0.102 | 0.049 | 5.67e-10 |
| IBD | SBLUP.Inf | SBLUP.Inf | 0.098 | 0.098 | 0.000 | 1.00e+00 | 0.102 | 0.102 | 0.000 | 1.00e+00 |
| IBD | DBSLMM.PseudoVal | SBLUP.Inf | 0.131 | 0.098 | 0.033 | 2.73e-14 | 0.135 | 0.102 | 0.034 | 1.06e-04 |
| IBD | pT+clump.10FCVal | SBLUP.Inf | 0.112 | 0.098 | 0.014 | 1.37e-02 | 0.112 | 0.102 | 0.011 | 3.47e-01 |
| IBD | lassosum.PseudoVal | SBLUP.Inf | 0.134 | 0.098 | 0.036 | 9.64e-13 | 0.137 | 0.102 | 0.035 | 5.22e-04 |
| MultiScler | LDpred2.PseudoVal | All.MultiPRS | 0.101 | 0.133 | -0.032 | 4.28e-06 | 0.088 | 0.121 | -0.033 | 4.34e-05 |
| MultiScler | pT+clump.10FCVal | All.MultiPRS | 0.090 | 0.133 | -0.043 | 8.56e-10 | 0.092 | 0.121 | -0.029 | 4.14e-04 |
| MultiScler | LDpred2.MultiPRS | All.MultiPRS | 0.130 | 0.133 | -0.003 | 8.80e-03 | 0.117 | 0.121 | -0.004 | 1.26e-01 |
| MultiScler | PRScs.PseudoVal | All.MultiPRS | 0.103 | 0.133 | -0.030 | 1.73e-05 | 0.101 | 0.121 | -0.020 | 2.48e-03 |
| MultiScler | LDpred1.Inf | All.MultiPRS | 0.089 | 0.133 | -0.045 | 2.09e-10 | 0.081 | 0.121 | -0.040 | 7.74e-07 |
| MultiScler | PRScs.MultiPRS | All.MultiPRS | 0.118 | 0.133 | -0.015 | 7.83e-08 | 0.108 | 0.121 | -0.013 | 1.71e-02 |
| MultiScler | LDpred2.Inf | All.MultiPRS | 0.092 | 0.133 | -0.041 | 3.97e-09 | 0.078 | 0.121 | -0.043 | 1.43e-06 |
| MultiScler | lassosum.10FCVal | All.MultiPRS | 0.115 | 0.133 | -0.018 | 1.01e-02 | 0.102 | 0.121 | -0.019 | 2.67e-03 |
| MultiScler | LDpred2.10FCVal | All.MultiPRS | 0.112 | 0.133 | -0.021 | 2.19e-03 | 0.101 | 0.121 | -0.020 | 2.34e-04 |
| MultiScler | lassosum.MultiPRS | All.MultiPRS | 0.126 | 0.133 | -0.007 | 2.93e-04 | 0.124 | 0.121 | 0.003 | 4.05e-01 |
| MultiScler | LDpred1.10FCVal | All.MultiPRS | 0.103 | 0.133 | -0.030 | 1.40e-05 | 0.095 | 0.121 | -0.026 | 6.90e-05 |
| MultiScler | All.MultiPRS | All.MultiPRS | 0.133 | 0.133 | 0.000 | 1.00e+00 | 0.121 | 0.121 | 0.000 | 1.00e+00 |
| MultiScler | SBayesR.PseudoVal | All.MultiPRS | 0.100 | 0.133 | -0.034 | 1.49e-06 | 0.084 | 0.121 | -0.037 | 1.99e-06 |
| MultiScler | LDpred1.MultiPRS | All.MultiPRS | 0.103 | 0.133 | -0.030 | 1.34e-19 | 0.096 | 0.121 | -0.025 | 1.32e-04 |
| MultiScler | pT+clump.MultiPRS | All.MultiPRS | 0.114 | 0.133 | -0.019 | 6.93e-03 | 0.116 | 0.121 | -0.005 | 3.17e-01 |
| MultiScler | lassosum.PseudoVal | All.MultiPRS | 0.100 | 0.133 | -0.033 | 2.34e-06 | 0.106 | 0.121 | -0.015 | 2.27e-02 |
| MultiScler | PRScs.10FCVal | All.MultiPRS | 0.118 | 0.133 | -0.016 | 2.63e-02 | 0.104 | 0.121 | -0.017 | 6.52e-03 |
| MultiScler | DBSLMM.PseudoVal | All.MultiPRS | 0.090 | 0.133 | -0.043 | 7.21e-10 | 0.085 | 0.121 | -0.036 | 1.06e-05 |
| MultiScler | SBLUP.Inf | All.MultiPRS | 0.093 | 0.133 | -0.040 | 8.51e-09 | 0.079 | 0.121 | -0.042 | 1.61e-06 |
| MultiScler | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.100 | 0.090 | 0.010 | 1.22e-01 | 0.084 | 0.085 | -0.001 | 9.40e-01 |
| MultiScler | LDpred2.Inf | DBSLMM.PseudoVal | 0.092 | 0.090 | 0.002 | 7.64e-01 | 0.078 | 0.085 | -0.006 | 6.18e-01 |
| MultiScler | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.090 | 0.090 | 0.000 | 9.47e-01 | 0.092 | 0.085 | 0.007 | 3.21e-01 |
| MultiScler | PRScs.10FCVal | DBSLMM.PseudoVal | 0.118 | 0.090 | 0.028 | 3.09e-08 | 0.104 | 0.085 | 0.019 | 5.58e-02 |
| MultiScler | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.103 | 0.090 | 0.013 | 2.44e-09 | 0.101 | 0.085 | 0.017 | 1.79e-04 |
| MultiScler | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.114 | 0.090 | 0.024 | 5.00e-04 | 0.116 | 0.085 | 0.031 | 3.13e-04 |
| MultiScler | LDpred1.Inf | DBSLMM.PseudoVal | 0.089 | 0.090 | -0.001 | 7.87e-01 | 0.081 | 0.085 | -0.004 | 7.01e-01 |
| MultiScler | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.126 | 0.090 | 0.036 | 2.94e-07 | 0.124 | 0.085 | 0.040 | 2.57e-07 |
| MultiScler | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.112 | 0.090 | 0.022 | 1.69e-13 | 0.101 | 0.085 | 0.016 | 6.56e-03 |
| MultiScler | lassosum.10FCVal | DBSLMM.PseudoVal | 0.115 | 0.090 | 0.025 | 3.61e-07 | 0.102 | 0.085 | 0.017 | 7.84e-02 |
| MultiScler | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.103 | 0.090 | 0.013 | 3.38e-04 | 0.095 | 0.085 | 0.010 | 1.53e-01 |
| MultiScler | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.103 | 0.090 | 0.013 | 6.98e-02 | 0.096 | 0.085 | 0.011 | 1.24e-01 |
| MultiScler | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.118 | 0.090 | 0.028 | 5.80e-05 | 0.108 | 0.085 | 0.023 | 4.38e-04 |
| MultiScler | All.MultiPRS | DBSLMM.PseudoVal | 0.133 | 0.090 | 0.043 | 7.21e-10 | 0.121 | 0.085 | 0.036 | 1.06e-05 |
| MultiScler | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.101 | 0.090 | 0.011 | 6.67e-02 | 0.088 | 0.085 | 0.003 | 7.77e-01 |
| MultiScler | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.090 | 0.090 | 0.000 | 1.00e+00 | 0.085 | 0.085 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.130 | 0.090 | 0.040 | 1.40e-08 | 0.117 | 0.085 | 0.032 | 1.71e-04 |
| MultiScler | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.100 | 0.090 | 0.010 | 1.26e-05 | 0.106 | 0.085 | 0.021 | 1.02e-05 |
| MultiScler | SBLUP.Inf | DBSLMM.PseudoVal | 0.093 | 0.090 | 0.003 | 6.58e-01 | 0.079 | 0.085 | -0.006 | 6.47e-01 |
| MultiScler | SBayesR.PseudoVal | lassosum.MultiPRS | 0.100 | 0.126 | -0.026 | 1.64e-04 | 0.084 | 0.124 | -0.041 | 7.10e-06 |
| MultiScler | LDpred2.10FCVal | lassosum.MultiPRS | 0.112 | 0.126 | -0.014 | 4.28e-02 | 0.101 | 0.124 | -0.023 | 2.36e-04 |
| MultiScler | LDpred1.10FCVal | lassosum.MultiPRS | 0.103 | 0.126 | -0.023 | 9.32e-04 | 0.095 | 0.124 | -0.029 | 3.53e-07 |
| MultiScler | LDpred2.Inf | lassosum.MultiPRS | 0.092 | 0.126 | -0.034 | 1.26e-06 | 0.078 | 0.124 | -0.046 | 3.23e-08 |
| MultiScler | PRScs.10FCVal | lassosum.MultiPRS | 0.118 | 0.126 | -0.008 | 2.36e-01 | 0.104 | 0.124 | -0.021 | 8.05e-05 |
| MultiScler | PRScs.MultiPRS | lassosum.MultiPRS | 0.118 | 0.126 | -0.008 | 4.13e-05 | 0.108 | 0.124 | -0.016 | 1.70e-05 |
| MultiScler | pT+clump.MultiPRS | lassosum.MultiPRS | 0.114 | 0.126 | -0.012 | 9.69e-02 | 0.116 | 0.124 | -0.009 | 1.30e-01 |
| MultiScler | lassosum.MultiPRS | lassosum.MultiPRS | 0.126 | 0.126 | 0.000 | 1.00e+00 | 0.124 | 0.124 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.PseudoVal | lassosum.MultiPRS | 0.100 | 0.126 | -0.026 | 2.26e-04 | 0.106 | 0.124 | -0.019 | 3.38e-04 |
| MultiScler | LDpred2.MultiPRS | lassosum.MultiPRS | 0.130 | 0.126 | 0.004 | 1.20e-01 | 0.117 | 0.124 | -0.007 | 1.25e-01 |
| MultiScler | LDpred1.MultiPRS | lassosum.MultiPRS | 0.103 | 0.126 | -0.023 | 1.99e-14 | 0.096 | 0.124 | -0.028 | 1.65e-06 |
| MultiScler | LDpred2.PseudoVal | lassosum.MultiPRS | 0.101 | 0.126 | -0.025 | 3.72e-04 | 0.088 | 0.124 | -0.036 | 9.10e-07 |
| MultiScler | LDpred1.Inf | lassosum.MultiPRS | 0.089 | 0.126 | -0.037 | 1.03e-07 | 0.081 | 0.124 | -0.044 | 6.13e-09 |
| MultiScler | pT+clump.10FCVal | lassosum.MultiPRS | 0.090 | 0.126 | -0.036 | 3.50e-07 | 0.092 | 0.124 | -0.033 | 3.14e-04 |
| MultiScler | lassosum.10FCVal | lassosum.MultiPRS | 0.115 | 0.126 | -0.011 | 1.24e-01 | 0.102 | 0.124 | -0.022 | 1.99e-06 |
| MultiScler | SBLUP.Inf | lassosum.MultiPRS | 0.093 | 0.126 | -0.033 | 2.37e-06 | 0.079 | 0.124 | -0.045 | 3.28e-08 |
| MultiScler | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.090 | 0.126 | -0.036 | 2.94e-07 | 0.085 | 0.124 | -0.040 | 2.57e-07 |
| MultiScler | All.MultiPRS | lassosum.MultiPRS | 0.133 | 0.126 | 0.007 | 2.93e-04 | 0.121 | 0.124 | -0.003 | 4.05e-01 |
| MultiScler | PRScs.PseudoVal | lassosum.MultiPRS | 0.103 | 0.126 | -0.023 | 1.11e-03 | 0.101 | 0.124 | -0.023 | 9.43e-06 |
| MultiScler | LDpred1.Inf | lassosum.PseudoVal | 0.089 | 0.100 | -0.012 | 1.63e-02 | 0.081 | 0.106 | -0.025 | 9.08e-03 |
| MultiScler | PRScs.10FCVal | lassosum.PseudoVal | 0.118 | 0.100 | 0.017 | 3.41e-05 | 0.104 | 0.106 | -0.002 | 8.12e-01 |
| MultiScler | LDpred2.MultiPRS | lassosum.PseudoVal | 0.130 | 0.100 | 0.030 | 2.34e-05 | 0.117 | 0.106 | 0.011 | 1.10e-01 |
| MultiScler | lassosum.PseudoVal | lassosum.PseudoVal | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.106 | 0.106 | 0.000 | 1.00e+00 |
| MultiScler | PRScs.PseudoVal | lassosum.PseudoVal | 0.103 | 0.100 | 0.003 | 9.12e-02 | 0.101 | 0.106 | -0.004 | 1.99e-01 |
| MultiScler | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.090 | 0.100 | -0.010 | 1.26e-05 | 0.085 | 0.106 | -0.021 | 1.02e-05 |
| MultiScler | SBLUP.Inf | lassosum.PseudoVal | 0.093 | 0.100 | -0.007 | 2.03e-01 | 0.079 | 0.106 | -0.027 | 2.03e-02 |
| MultiScler | LDpred2.Inf | lassosum.PseudoVal | 0.092 | 0.100 | -0.008 | 1.57e-01 | 0.078 | 0.106 | -0.027 | 1.88e-02 |
| MultiScler | pT+clump.10FCVal | lassosum.PseudoVal | 0.090 | 0.100 | -0.010 | 1.75e-02 | 0.092 | 0.106 | -0.014 | 9.87e-02 |
| MultiScler | LDpred2.10FCVal | lassosum.PseudoVal | 0.112 | 0.100 | 0.012 | 5.34e-06 | 0.101 | 0.106 | -0.005 | 3.48e-01 |
| MultiScler | lassosum.MultiPRS | lassosum.PseudoVal | 0.126 | 0.100 | 0.026 | 2.26e-04 | 0.124 | 0.106 | 0.019 | 3.38e-04 |
| MultiScler | LDpred1.10FCVal | lassosum.PseudoVal | 0.103 | 0.100 | 0.003 | 3.97e-01 | 0.095 | 0.106 | -0.011 | 7.21e-02 |
| MultiScler | All.MultiPRS | lassosum.PseudoVal | 0.133 | 0.100 | 0.033 | 2.34e-06 | 0.121 | 0.106 | 0.015 | 2.27e-02 |
| MultiScler | SBayesR.PseudoVal | lassosum.PseudoVal | 0.100 | 0.100 | -0.001 | 9.27e-01 | 0.084 | 0.106 | -0.022 | 5.37e-02 |
| MultiScler | lassosum.10FCVal | lassosum.PseudoVal | 0.115 | 0.100 | 0.015 | 1.19e-04 | 0.102 | 0.106 | -0.004 | 6.45e-01 |
| MultiScler | pT+clump.MultiPRS | lassosum.PseudoVal | 0.114 | 0.100 | 0.014 | 4.24e-02 | 0.116 | 0.106 | 0.010 | 2.05e-01 |
| MultiScler | LDpred2.PseudoVal | lassosum.PseudoVal | 0.101 | 0.100 | 0.001 | 8.77e-01 | 0.088 | 0.106 | -0.018 | 1.01e-01 |
| MultiScler | LDpred1.MultiPRS | lassosum.PseudoVal | 0.103 | 0.100 | 0.003 | 7.10e-01 | 0.096 | 0.106 | -0.010 | 1.26e-01 |
| MultiScler | PRScs.MultiPRS | lassosum.PseudoVal | 0.118 | 0.100 | 0.018 | 9.90e-03 | 0.108 | 0.106 | 0.002 | 6.47e-01 |
| MultiScler | lassosum.10FCVal | lassosum.10FCVal | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.102 | 0.102 | 0.000 | 1.00e+00 |
| MultiScler | pT+clump.MultiPRS | lassosum.10FCVal | 0.114 | 0.115 | -0.001 | 9.05e-01 | 0.116 | 0.102 | 0.014 | 6.52e-02 |
| MultiScler | LDpred2.Inf | lassosum.10FCVal | 0.092 | 0.115 | -0.023 | 5.11e-11 | 0.078 | 0.102 | -0.024 | 8.32e-04 |
| MultiScler | SBayesR.PseudoVal | lassosum.10FCVal | 0.100 | 0.115 | -0.016 | 1.35e-03 | 0.084 | 0.102 | -0.018 | 5.88e-02 |
| MultiScler | PRScs.MultiPRS | lassosum.10FCVal | 0.118 | 0.115 | 0.003 | 6.66e-01 | 0.108 | 0.102 | 0.006 | 2.50e-01 |
| MultiScler | SBLUP.Inf | lassosum.10FCVal | 0.093 | 0.115 | -0.022 | 1.71e-10 | 0.079 | 0.102 | -0.023 | 9.70e-04 |
| MultiScler | lassosum.MultiPRS | lassosum.10FCVal | 0.126 | 0.115 | 0.011 | 1.24e-01 | 0.124 | 0.102 | 0.022 | 1.99e-06 |
| MultiScler | PRScs.10FCVal | lassosum.10FCVal | 0.118 | 0.115 | 0.002 | 2.18e-01 | 0.104 | 0.102 | 0.002 | 6.91e-01 |
| MultiScler | LDpred2.MultiPRS | lassosum.10FCVal | 0.130 | 0.115 | 0.015 | 3.75e-02 | 0.117 | 0.102 | 0.015 | 2.62e-02 |
| MultiScler | PRScs.PseudoVal | lassosum.10FCVal | 0.103 | 0.115 | -0.012 | 1.19e-03 | 0.101 | 0.102 | -0.001 | 9.09e-01 |
| MultiScler | LDpred2.PseudoVal | lassosum.10FCVal | 0.101 | 0.115 | -0.014 | 5.11e-06 | 0.088 | 0.102 | -0.014 | 2.54e-02 |
| MultiScler | LDpred1.MultiPRS | lassosum.10FCVal | 0.103 | 0.115 | -0.012 | 7.70e-02 | 0.096 | 0.102 | -0.006 | 3.93e-01 |
| MultiScler | pT+clump.10FCVal | lassosum.10FCVal | 0.090 | 0.115 | -0.025 | 5.56e-06 | 0.092 | 0.102 | -0.011 | 3.44e-01 |
| MultiScler | All.MultiPRS | lassosum.10FCVal | 0.133 | 0.115 | 0.018 | 1.01e-02 | 0.121 | 0.102 | 0.019 | 2.67e-03 |
| MultiScler | LDpred1.Inf | lassosum.10FCVal | 0.089 | 0.115 | -0.027 | 1.75e-13 | 0.081 | 0.102 | -0.021 | 2.99e-03 |
| MultiScler | DBSLMM.PseudoVal | lassosum.10FCVal | 0.090 | 0.115 | -0.025 | 3.61e-07 | 0.085 | 0.102 | -0.017 | 7.84e-02 |
| MultiScler | LDpred1.10FCVal | lassosum.10FCVal | 0.103 | 0.115 | -0.012 | 3.88e-04 | 0.095 | 0.102 | -0.007 | 2.99e-01 |
| MultiScler | lassosum.PseudoVal | lassosum.10FCVal | 0.100 | 0.115 | -0.015 | 1.19e-04 | 0.106 | 0.102 | 0.004 | 6.45e-01 |
| MultiScler | LDpred2.10FCVal | lassosum.10FCVal | 0.112 | 0.115 | -0.003 | 4.52e-01 | 0.101 | 0.102 | -0.001 | 9.02e-01 |
| MultiScler | LDpred2.10FCVal | LDpred1.MultiPRS | 0.112 | 0.103 | 0.009 | 1.97e-01 | 0.101 | 0.096 | 0.005 | 4.71e-01 |
| MultiScler | PRScs.PseudoVal | LDpred1.MultiPRS | 0.103 | 0.103 | 0.000 | 9.63e-01 | 0.101 | 0.096 | 0.005 | 3.37e-01 |
| MultiScler | LDpred2.Inf | LDpred1.MultiPRS | 0.092 | 0.103 | -0.011 | 1.23e-01 | 0.078 | 0.096 | -0.018 | 5.93e-02 |
| MultiScler | pT+clump.10FCVal | LDpred1.MultiPRS | 0.090 | 0.103 | -0.013 | 7.51e-02 | 0.092 | 0.096 | -0.005 | 6.48e-01 |
| MultiScler | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.100 | 0.103 | -0.003 | 6.55e-01 | 0.084 | 0.096 | -0.012 | 2.59e-01 |
| MultiScler | All.MultiPRS | LDpred1.MultiPRS | 0.133 | 0.103 | 0.030 | 1.34e-19 | 0.121 | 0.096 | 0.025 | 1.32e-04 |
| MultiScler | LDpred1.10FCVal | LDpred1.MultiPRS | 0.103 | 0.103 | 0.000 | 9.94e-01 | 0.095 | 0.096 | -0.001 | 4.87e-01 |
| MultiScler | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.103 | 0.103 | 0.000 | 1.00e+00 | 0.096 | 0.096 | 0.000 | 1.00e+00 |
| MultiScler | PRScs.10FCVal | LDpred1.MultiPRS | 0.118 | 0.103 | 0.015 | 3.41e-02 | 0.104 | 0.096 | 0.008 | 2.57e-01 |
| MultiScler | lassosum.PseudoVal | LDpred1.MultiPRS | 0.100 | 0.103 | -0.003 | 7.10e-01 | 0.106 | 0.096 | 0.010 | 1.26e-01 |
| MultiScler | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.114 | 0.103 | 0.012 | 9.81e-02 | 0.116 | 0.096 | 0.019 | 1.80e-02 |
| MultiScler | LDpred1.Inf | LDpred1.MultiPRS | 0.089 | 0.103 | -0.014 | 4.37e-02 | 0.081 | 0.096 | -0.016 | 3.80e-03 |
| MultiScler | lassosum.MultiPRS | LDpred1.MultiPRS | 0.126 | 0.103 | 0.023 | 1.99e-14 | 0.124 | 0.096 | 0.028 | 1.65e-06 |
| MultiScler | PRScs.MultiPRS | LDpred1.MultiPRS | 0.118 | 0.103 | 0.015 | 9.87e-10 | 0.108 | 0.096 | 0.012 | 1.43e-02 |
| MultiScler | lassosum.10FCVal | LDpred1.MultiPRS | 0.115 | 0.103 | 0.012 | 7.70e-02 | 0.102 | 0.096 | 0.006 | 3.93e-01 |
| MultiScler | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.101 | 0.103 | -0.002 | 7.99e-01 | 0.088 | 0.096 | -0.008 | 3.44e-01 |
| MultiScler | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.090 | 0.103 | -0.013 | 6.98e-02 | 0.085 | 0.096 | -0.011 | 1.24e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.130 | 0.103 | 0.027 | 6.54e-16 | 0.117 | 0.096 | 0.021 | 1.14e-03 |
| MultiScler | SBLUP.Inf | LDpred1.MultiPRS | 0.093 | 0.103 | -0.010 | 1.57e-01 | 0.079 | 0.096 | -0.017 | 6.52e-02 |
| MultiScler | All.MultiPRS | LDpred1.Inf | 0.133 | 0.089 | 0.045 | 2.09e-10 | 0.121 | 0.081 | 0.040 | 7.74e-07 |
| MultiScler | LDpred2.PseudoVal | LDpred1.Inf | 0.101 | 0.089 | 0.012 | 1.53e-04 | 0.088 | 0.081 | 0.008 | 2.53e-01 |
| MultiScler | SBayesR.PseudoVal | LDpred1.Inf | 0.100 | 0.089 | 0.011 | 3.06e-02 | 0.084 | 0.081 | 0.003 | 7.59e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred1.Inf | 0.130 | 0.089 | 0.041 | 4.49e-09 | 0.117 | 0.081 | 0.036 | 5.01e-06 |
| MultiScler | PRScs.PseudoVal | LDpred1.Inf | 0.103 | 0.089 | 0.015 | 8.65e-04 | 0.101 | 0.081 | 0.021 | 1.73e-02 |
| MultiScler | LDpred1.Inf | LDpred1.Inf | 0.089 | 0.089 | 0.000 | 1.00e+00 | 0.081 | 0.081 | 0.000 | 1.00e+00 |
| MultiScler | pT+clump.10FCVal | LDpred1.Inf | 0.090 | 0.089 | 0.002 | 7.76e-01 | 0.092 | 0.081 | 0.011 | 3.58e-01 |
| MultiScler | PRScs.MultiPRS | LDpred1.Inf | 0.118 | 0.089 | 0.030 | 2.45e-05 | 0.108 | 0.081 | 0.027 | 1.37e-04 |
| MultiScler | lassosum.PseudoVal | LDpred1.Inf | 0.100 | 0.089 | 0.012 | 1.63e-02 | 0.106 | 0.081 | 0.025 | 9.08e-03 |
| MultiScler | pT+clump.MultiPRS | LDpred1.Inf | 0.114 | 0.089 | 0.026 | 2.28e-04 | 0.116 | 0.081 | 0.035 | 1.55e-04 |
| MultiScler | LDpred1.MultiPRS | LDpred1.Inf | 0.103 | 0.089 | 0.014 | 4.37e-02 | 0.096 | 0.081 | 0.016 | 3.80e-03 |
| MultiScler | PRScs.10FCVal | LDpred1.Inf | 0.118 | 0.089 | 0.029 | 3.77e-19 | 0.104 | 0.081 | 0.023 | 3.46e-04 |
| MultiScler | LDpred2.Inf | LDpred1.Inf | 0.092 | 0.089 | 0.003 | 3.45e-01 | 0.078 | 0.081 | -0.002 | 7.47e-01 |
| MultiScler | SBLUP.Inf | LDpred1.Inf | 0.093 | 0.089 | 0.004 | 2.29e-01 | 0.079 | 0.081 | -0.002 | 8.05e-01 |
| MultiScler | DBSLMM.PseudoVal | LDpred1.Inf | 0.090 | 0.089 | 0.001 | 7.87e-01 | 0.085 | 0.081 | 0.004 | 7.01e-01 |
| MultiScler | lassosum.MultiPRS | LDpred1.Inf | 0.126 | 0.089 | 0.037 | 1.03e-07 | 0.124 | 0.081 | 0.044 | 6.13e-09 |
| MultiScler | LDpred1.10FCVal | LDpred1.Inf | 0.103 | 0.089 | 0.014 | 1.11e-07 | 0.095 | 0.081 | 0.014 | 7.42e-03 |
| MultiScler | lassosum.10FCVal | LDpred1.Inf | 0.115 | 0.089 | 0.027 | 1.75e-13 | 0.102 | 0.081 | 0.021 | 2.99e-03 |
| MultiScler | LDpred2.10FCVal | LDpred1.Inf | 0.112 | 0.089 | 0.023 | 3.29e-06 | 0.101 | 0.081 | 0.020 | 3.93e-02 |
| MultiScler | LDpred1.Inf | LDpred1.10FCVal | 0.089 | 0.103 | -0.014 | 1.11e-07 | 0.081 | 0.095 | -0.014 | 7.42e-03 |
| MultiScler | PRScs.MultiPRS | LDpred1.10FCVal | 0.118 | 0.103 | 0.015 | 2.73e-02 | 0.108 | 0.095 | 0.013 | 3.72e-03 |
| MultiScler | LDpred1.10FCVal | LDpred1.10FCVal | 0.103 | 0.103 | 0.000 | 1.00e+00 | 0.095 | 0.095 | 0.000 | 1.00e+00 |
| MultiScler | All.MultiPRS | LDpred1.10FCVal | 0.133 | 0.103 | 0.030 | 1.40e-05 | 0.121 | 0.095 | 0.026 | 6.90e-05 |
| MultiScler | SBayesR.PseudoVal | LDpred1.10FCVal | 0.100 | 0.103 | -0.003 | 5.77e-01 | 0.084 | 0.095 | -0.011 | 3.17e-01 |
| MultiScler | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.090 | 0.103 | -0.013 | 3.38e-04 | 0.085 | 0.095 | -0.010 | 1.53e-01 |
| MultiScler | pT+clump.MultiPRS | LDpred1.10FCVal | 0.114 | 0.103 | 0.012 | 9.58e-02 | 0.116 | 0.095 | 0.021 | 1.16e-02 |
| MultiScler | lassosum.MultiPRS | LDpred1.10FCVal | 0.126 | 0.103 | 0.023 | 9.32e-04 | 0.124 | 0.095 | 0.029 | 3.53e-07 |
| MultiScler | PRScs.10FCVal | LDpred1.10FCVal | 0.118 | 0.103 | 0.015 | 1.07e-05 | 0.104 | 0.095 | 0.009 | 1.83e-01 |
| MultiScler | lassosum.10FCVal | LDpred1.10FCVal | 0.115 | 0.103 | 0.012 | 3.88e-04 | 0.102 | 0.095 | 0.007 | 2.99e-01 |
| MultiScler | SBLUP.Inf | LDpred1.10FCVal | 0.093 | 0.103 | -0.010 | 3.65e-02 | 0.079 | 0.095 | -0.016 | 9.09e-02 |
| MultiScler | LDpred2.PseudoVal | LDpred1.10FCVal | 0.101 | 0.103 | -0.002 | 6.85e-01 | 0.088 | 0.095 | -0.007 | 4.29e-01 |
| MultiScler | pT+clump.10FCVal | LDpred1.10FCVal | 0.090 | 0.103 | -0.012 | 1.02e-02 | 0.092 | 0.095 | -0.003 | 7.38e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred1.10FCVal | 0.130 | 0.103 | 0.027 | 1.16e-04 | 0.117 | 0.095 | 0.022 | 6.29e-04 |
| MultiScler | PRScs.PseudoVal | LDpred1.10FCVal | 0.103 | 0.103 | 0.000 | 8.82e-01 | 0.101 | 0.095 | 0.006 | 1.97e-01 |
| MultiScler | LDpred2.10FCVal | LDpred1.10FCVal | 0.112 | 0.103 | 0.009 | 7.45e-03 | 0.101 | 0.095 | 0.006 | 3.58e-01 |
| MultiScler | LDpred1.MultiPRS | LDpred1.10FCVal | 0.103 | 0.103 | 0.000 | 9.94e-01 | 0.096 | 0.095 | 0.001 | 4.87e-01 |
| MultiScler | LDpred2.Inf | LDpred1.10FCVal | 0.092 | 0.103 | -0.011 | 2.39e-02 | 0.078 | 0.095 | -0.017 | 8.30e-02 |
| MultiScler | lassosum.PseudoVal | LDpred1.10FCVal | 0.100 | 0.103 | -0.003 | 3.97e-01 | 0.106 | 0.095 | 0.011 | 7.21e-02 |
| MultiScler | LDpred1.Inf | LDpred2.MultiPRS | 0.089 | 0.130 | -0.041 | 4.49e-09 | 0.081 | 0.117 | -0.036 | 5.01e-06 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.090 | 0.130 | -0.040 | 1.40e-08 | 0.085 | 0.117 | -0.032 | 1.71e-04 |
| MultiScler | SBLUP.Inf | LDpred2.MultiPRS | 0.093 | 0.130 | -0.037 | 1.38e-07 | 0.079 | 0.117 | -0.038 | 7.07e-06 |
| MultiScler | LDpred2.Inf | LDpred2.MultiPRS | 0.092 | 0.130 | -0.038 | 6.85e-08 | 0.078 | 0.117 | -0.039 | 6.03e-06 |
| MultiScler | pT+clump.10FCVal | LDpred2.MultiPRS | 0.090 | 0.130 | -0.040 | 1.67e-08 | 0.092 | 0.117 | -0.025 | 6.11e-03 |
| MultiScler | LDpred2.10FCVal | LDpred2.MultiPRS | 0.112 | 0.130 | -0.018 | 1.02e-02 | 0.101 | 0.117 | -0.016 | 1.06e-03 |
| MultiScler | PRScs.PseudoVal | LDpred2.MultiPRS | 0.103 | 0.130 | -0.027 | 1.41e-04 | 0.101 | 0.117 | -0.016 | 1.94e-02 |
| MultiScler | LDpred1.10FCVal | LDpred2.MultiPRS | 0.103 | 0.130 | -0.027 | 1.16e-04 | 0.095 | 0.117 | -0.022 | 6.29e-04 |
| MultiScler | All.MultiPRS | LDpred2.MultiPRS | 0.133 | 0.130 | 0.003 | 8.80e-03 | 0.121 | 0.117 | 0.004 | 1.26e-01 |
| MultiScler | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.100 | 0.130 | -0.030 | 1.59e-05 | 0.084 | 0.117 | -0.033 | 9.98e-05 |
| MultiScler | lassosum.10FCVal | LDpred2.MultiPRS | 0.115 | 0.130 | -0.015 | 3.75e-02 | 0.102 | 0.117 | -0.015 | 2.62e-02 |
| MultiScler | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.114 | 0.130 | -0.015 | 2.76e-02 | 0.116 | 0.117 | -0.001 | 8.38e-01 |
| MultiScler | lassosum.MultiPRS | LDpred2.MultiPRS | 0.126 | 0.130 | -0.004 | 1.20e-01 | 0.124 | 0.117 | 0.007 | 1.25e-01 |
| MultiScler | PRScs.10FCVal | LDpred2.MultiPRS | 0.118 | 0.130 | -0.012 | 8.38e-02 | 0.104 | 0.117 | -0.013 | 4.68e-02 |
| MultiScler | PRScs.MultiPRS | LDpred2.MultiPRS | 0.118 | 0.130 | -0.012 | 1.14e-04 | 0.108 | 0.117 | -0.009 | 1.24e-01 |
| MultiScler | lassosum.PseudoVal | LDpred2.MultiPRS | 0.100 | 0.130 | -0.030 | 2.34e-05 | 0.106 | 0.117 | -0.011 | 1.10e-01 |
| MultiScler | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.101 | 0.130 | -0.029 | 4.02e-05 | 0.088 | 0.117 | -0.029 | 1.63e-04 |
| MultiScler | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.103 | 0.130 | -0.027 | 6.54e-16 | 0.096 | 0.117 | -0.021 | 1.14e-03 |
| MultiScler | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.130 | 0.130 | 0.000 | 1.00e+00 | 0.117 | 0.117 | 0.000 | 1.00e+00 |
| MultiScler | LDpred1.Inf | LDpred2.10FCVal | 0.089 | 0.112 | -0.023 | 3.29e-06 | 0.081 | 0.101 | -0.020 | 3.93e-02 |
| MultiScler | PRScs.MultiPRS | LDpred2.10FCVal | 0.118 | 0.112 | 0.006 | 3.60e-01 | 0.108 | 0.101 | 0.007 | 2.87e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred2.10FCVal | 0.130 | 0.112 | 0.018 | 1.02e-02 | 0.117 | 0.101 | 0.016 | 1.06e-03 |
| MultiScler | PRScs.PseudoVal | LDpred2.10FCVal | 0.103 | 0.112 | -0.009 | 1.62e-03 | 0.101 | 0.101 | 0.000 | 9.66e-01 |
| MultiScler | LDpred2.10FCVal | LDpred2.10FCVal | 0.112 | 0.112 | 0.000 | 1.00e+00 | 0.101 | 0.101 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.MultiPRS | LDpred2.10FCVal | 0.126 | 0.112 | 0.014 | 4.28e-02 | 0.124 | 0.101 | 0.023 | 2.36e-04 |
| MultiScler | LDpred1.10FCVal | LDpred2.10FCVal | 0.103 | 0.112 | -0.009 | 7.45e-03 | 0.095 | 0.101 | -0.006 | 3.58e-01 |
| MultiScler | All.MultiPRS | LDpred2.10FCVal | 0.133 | 0.112 | 0.021 | 2.19e-03 | 0.121 | 0.101 | 0.020 | 2.34e-04 |
| MultiScler | SBayesR.PseudoVal | LDpred2.10FCVal | 0.100 | 0.112 | -0.012 | 1.79e-02 | 0.084 | 0.101 | -0.017 | 9.64e-02 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.090 | 0.112 | -0.022 | 1.69e-13 | 0.085 | 0.101 | -0.016 | 6.56e-03 |
| MultiScler | pT+clump.MultiPRS | LDpred2.10FCVal | 0.114 | 0.112 | 0.003 | 7.16e-01 | 0.116 | 0.101 | 0.015 | 7.21e-02 |
| MultiScler | lassosum.PseudoVal | LDpred2.10FCVal | 0.100 | 0.112 | -0.012 | 5.34e-06 | 0.106 | 0.101 | 0.005 | 3.48e-01 |
| MultiScler | PRScs.10FCVal | LDpred2.10FCVal | 0.118 | 0.112 | 0.006 | 2.13e-01 | 0.104 | 0.101 | 0.003 | 7.70e-01 |
| MultiScler | lassosum.10FCVal | LDpred2.10FCVal | 0.115 | 0.112 | 0.003 | 4.52e-01 | 0.102 | 0.101 | 0.001 | 9.02e-01 |
| MultiScler | SBLUP.Inf | LDpred2.10FCVal | 0.093 | 0.112 | -0.019 | 1.17e-03 | 0.079 | 0.101 | -0.022 | 5.85e-02 |
| MultiScler | LDpred2.PseudoVal | LDpred2.10FCVal | 0.101 | 0.112 | -0.011 | 4.67e-02 | 0.088 | 0.101 | -0.013 | 2.36e-01 |
| MultiScler | pT+clump.10FCVal | LDpred2.10FCVal | 0.090 | 0.112 | -0.022 | 3.00e-08 | 0.092 | 0.101 | -0.009 | 2.45e-01 |
| MultiScler | LDpred2.Inf | LDpred2.10FCVal | 0.092 | 0.112 | -0.020 | 7.23e-04 | 0.078 | 0.101 | -0.023 | 5.37e-02 |
| MultiScler | LDpred1.MultiPRS | LDpred2.10FCVal | 0.103 | 0.112 | -0.009 | 1.97e-01 | 0.096 | 0.101 | -0.005 | 4.71e-01 |
| MultiScler | lassosum.10FCVal | LDpred2.PseudoVal | 0.115 | 0.101 | 0.014 | 5.11e-06 | 0.102 | 0.088 | 0.014 | 2.54e-02 |
| MultiScler | SBLUP.Inf | LDpred2.PseudoVal | 0.093 | 0.101 | -0.008 | 4.96e-30 | 0.079 | 0.088 | -0.009 | 1.32e-10 |
| MultiScler | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.100 | 0.101 | -0.001 | 7.85e-01 | 0.084 | 0.088 | -0.004 | 6.59e-01 |
| MultiScler | All.MultiPRS | LDpred2.PseudoVal | 0.133 | 0.101 | 0.032 | 4.28e-06 | 0.121 | 0.088 | 0.033 | 4.34e-05 |
| MultiScler | PRScs.PseudoVal | LDpred2.PseudoVal | 0.103 | 0.101 | 0.002 | 6.63e-01 | 0.101 | 0.088 | 0.013 | 1.77e-01 |
| MultiScler | lassosum.MultiPRS | LDpred2.PseudoVal | 0.126 | 0.101 | 0.025 | 3.72e-04 | 0.124 | 0.088 | 0.036 | 9.10e-07 |
| MultiScler | PRScs.MultiPRS | LDpred2.PseudoVal | 0.118 | 0.101 | 0.017 | 1.41e-02 | 0.108 | 0.088 | 0.020 | 7.88e-03 |
| MultiScler | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.130 | 0.101 | 0.029 | 4.02e-05 | 0.117 | 0.088 | 0.029 | 1.63e-04 |
| MultiScler | PRScs.10FCVal | LDpred2.PseudoVal | 0.118 | 0.101 | 0.017 | 6.94e-12 | 0.104 | 0.088 | 0.016 | 1.52e-03 |
| MultiScler | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.101 | 0.101 | 0.000 | 1.00e+00 | 0.088 | 0.088 | 0.000 | 1.00e+00 |
| MultiScler | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.114 | 0.101 | 0.013 | 5.58e-02 | 0.116 | 0.088 | 0.027 | 2.40e-03 |
| MultiScler | pT+clump.10FCVal | LDpred2.PseudoVal | 0.090 | 0.101 | -0.011 | 8.27e-02 | 0.092 | 0.088 | 0.003 | 7.84e-01 |
| MultiScler | lassosum.PseudoVal | LDpred2.PseudoVal | 0.100 | 0.101 | -0.001 | 8.77e-01 | 0.106 | 0.088 | 0.018 | 1.01e-01 |
| MultiScler | LDpred1.Inf | LDpred2.PseudoVal | 0.089 | 0.101 | -0.012 | 1.53e-04 | 0.081 | 0.088 | -0.008 | 2.53e-01 |
| MultiScler | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.103 | 0.101 | 0.002 | 7.99e-01 | 0.096 | 0.088 | 0.008 | 3.44e-01 |
| MultiScler | LDpred1.10FCVal | LDpred2.PseudoVal | 0.103 | 0.101 | 0.002 | 6.85e-01 | 0.095 | 0.088 | 0.007 | 4.29e-01 |
| MultiScler | LDpred2.Inf | LDpred2.PseudoVal | 0.092 | 0.101 | -0.009 | 4.19e-33 | 0.078 | 0.088 | -0.010 | 9.82e-11 |
| MultiScler | LDpred2.10FCVal | LDpred2.PseudoVal | 0.112 | 0.101 | 0.011 | 4.67e-02 | 0.101 | 0.088 | 0.013 | 2.36e-01 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.090 | 0.101 | -0.011 | 6.67e-02 | 0.085 | 0.088 | -0.003 | 7.77e-01 |
| MultiScler | pT+clump.MultiPRS | LDpred2.Inf | 0.114 | 0.092 | 0.022 | 1.36e-03 | 0.116 | 0.078 | 0.037 | 9.97e-05 |
| MultiScler | lassosum.MultiPRS | LDpred2.Inf | 0.126 | 0.092 | 0.034 | 1.26e-06 | 0.124 | 0.078 | 0.046 | 3.23e-08 |
| MultiScler | SBayesR.PseudoVal | LDpred2.Inf | 0.100 | 0.092 | 0.008 | 1.17e-01 | 0.084 | 0.078 | 0.005 | 5.81e-01 |
| MultiScler | All.MultiPRS | LDpred2.Inf | 0.133 | 0.092 | 0.041 | 3.97e-09 | 0.121 | 0.078 | 0.043 | 1.43e-06 |
| MultiScler | SBLUP.Inf | LDpred2.Inf | 0.093 | 0.092 | 0.001 | 3.83e-13 | 0.079 | 0.078 | 0.001 | 2.41e-02 |
| MultiScler | LDpred2.PseudoVal | LDpred2.Inf | 0.101 | 0.092 | 0.009 | 4.19e-33 | 0.088 | 0.078 | 0.010 | 9.82e-11 |
| MultiScler | PRScs.MultiPRS | LDpred2.Inf | 0.118 | 0.092 | 0.026 | 1.83e-04 | 0.108 | 0.078 | 0.030 | 5.67e-04 |
| MultiScler | LDpred2.MultiPRS | LDpred2.Inf | 0.130 | 0.092 | 0.038 | 6.85e-08 | 0.117 | 0.078 | 0.039 | 6.03e-06 |
| MultiScler | PRScs.10FCVal | LDpred2.Inf | 0.118 | 0.092 | 0.026 | 9.94e-19 | 0.104 | 0.078 | 0.025 | 1.63e-05 |
| MultiScler | LDpred1.Inf | LDpred2.Inf | 0.089 | 0.092 | -0.003 | 3.45e-01 | 0.081 | 0.078 | 0.002 | 7.47e-01 |
| MultiScler | LDpred1.MultiPRS | LDpred2.Inf | 0.103 | 0.092 | 0.011 | 1.23e-01 | 0.096 | 0.078 | 0.018 | 5.93e-02 |
| MultiScler | pT+clump.10FCVal | LDpred2.Inf | 0.090 | 0.092 | -0.002 | 7.92e-01 | 0.092 | 0.078 | 0.013 | 3.10e-01 |
| MultiScler | lassosum.PseudoVal | LDpred2.Inf | 0.100 | 0.092 | 0.008 | 1.57e-01 | 0.106 | 0.078 | 0.027 | 1.88e-02 |
| MultiScler | PRScs.PseudoVal | LDpred2.Inf | 0.103 | 0.092 | 0.011 | 3.72e-02 | 0.101 | 0.078 | 0.023 | 3.27e-02 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.Inf | 0.090 | 0.092 | -0.002 | 7.64e-01 | 0.085 | 0.078 | 0.006 | 6.18e-01 |
| MultiScler | LDpred1.10FCVal | LDpred2.Inf | 0.103 | 0.092 | 0.011 | 2.39e-02 | 0.095 | 0.078 | 0.017 | 8.30e-02 |
| MultiScler | LDpred2.Inf | LDpred2.Inf | 0.092 | 0.092 | 0.000 | 1.00e+00 | 0.078 | 0.078 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.10FCVal | LDpred2.Inf | 0.115 | 0.092 | 0.023 | 5.11e-11 | 0.102 | 0.078 | 0.024 | 8.32e-04 |
| MultiScler | LDpred2.10FCVal | LDpred2.Inf | 0.112 | 0.092 | 0.020 | 7.23e-04 | 0.101 | 0.078 | 0.023 | 5.37e-02 |
| MultiScler | PRScs.10FCVal | PRScs.MultiPRS | 0.118 | 0.118 | -0.001 | 9.35e-01 | 0.104 | 0.108 | -0.004 | 3.15e-01 |
| MultiScler | LDpred1.10FCVal | PRScs.MultiPRS | 0.103 | 0.118 | -0.015 | 2.73e-02 | 0.095 | 0.108 | -0.013 | 3.72e-03 |
| MultiScler | lassosum.MultiPRS | PRScs.MultiPRS | 0.126 | 0.118 | 0.008 | 4.13e-05 | 0.124 | 0.108 | 0.016 | 1.70e-05 |
| MultiScler | LDpred2.MultiPRS | PRScs.MultiPRS | 0.130 | 0.118 | 0.012 | 1.14e-04 | 0.117 | 0.108 | 0.009 | 1.24e-01 |
| MultiScler | All.MultiPRS | PRScs.MultiPRS | 0.133 | 0.118 | 0.015 | 7.83e-08 | 0.121 | 0.108 | 0.013 | 1.71e-02 |
| MultiScler | PRScs.PseudoVal | PRScs.MultiPRS | 0.103 | 0.118 | -0.015 | 3.11e-02 | 0.101 | 0.108 | -0.007 | 5.56e-02 |
| MultiScler | LDpred1.MultiPRS | PRScs.MultiPRS | 0.103 | 0.118 | -0.015 | 9.87e-10 | 0.096 | 0.108 | -0.012 | 1.43e-02 |
| MultiScler | pT+clump.MultiPRS | PRScs.MultiPRS | 0.114 | 0.118 | -0.004 | 5.82e-01 | 0.116 | 0.108 | 0.008 | 2.68e-01 |
| MultiScler | lassosum.PseudoVal | PRScs.MultiPRS | 0.100 | 0.118 | -0.018 | 9.90e-03 | 0.106 | 0.108 | -0.002 | 6.47e-01 |
| MultiScler | SBayesR.PseudoVal | PRScs.MultiPRS | 0.100 | 0.118 | -0.019 | 7.82e-03 | 0.084 | 0.108 | -0.024 | 2.42e-02 |
| MultiScler | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.090 | 0.118 | -0.028 | 5.80e-05 | 0.085 | 0.108 | -0.023 | 4.38e-04 |
| MultiScler | SBLUP.Inf | PRScs.MultiPRS | 0.093 | 0.118 | -0.025 | 3.00e-04 | 0.079 | 0.108 | -0.029 | 6.11e-04 |
| MultiScler | LDpred2.Inf | PRScs.MultiPRS | 0.092 | 0.118 | -0.026 | 1.83e-04 | 0.078 | 0.108 | -0.030 | 5.67e-04 |
| MultiScler | pT+clump.10FCVal | PRScs.MultiPRS | 0.090 | 0.118 | -0.028 | 6.70e-05 | 0.092 | 0.108 | -0.016 | 8.26e-02 |
| MultiScler | LDpred2.10FCVal | PRScs.MultiPRS | 0.112 | 0.118 | -0.006 | 3.60e-01 | 0.101 | 0.108 | -0.007 | 2.87e-01 |
| MultiScler | lassosum.10FCVal | PRScs.MultiPRS | 0.115 | 0.118 | -0.003 | 6.66e-01 | 0.102 | 0.108 | -0.006 | 2.50e-01 |
| MultiScler | LDpred2.PseudoVal | PRScs.MultiPRS | 0.101 | 0.118 | -0.017 | 1.41e-02 | 0.088 | 0.108 | -0.020 | 7.88e-03 |
| MultiScler | LDpred1.Inf | PRScs.MultiPRS | 0.089 | 0.118 | -0.030 | 2.45e-05 | 0.081 | 0.108 | -0.027 | 1.37e-04 |
| MultiScler | PRScs.MultiPRS | PRScs.MultiPRS | 0.118 | 0.118 | 0.000 | 1.00e+00 | 0.108 | 0.108 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.PseudoVal | PRScs.10FCVal | 0.101 | 0.118 | -0.017 | 6.94e-12 | 0.088 | 0.104 | -0.016 | 1.52e-03 |
| MultiScler | PRScs.10FCVal | PRScs.10FCVal | 0.118 | 0.118 | 0.000 | 1.00e+00 | 0.104 | 0.104 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.MultiPRS | PRScs.10FCVal | 0.130 | 0.118 | 0.012 | 8.38e-02 | 0.117 | 0.104 | 0.013 | 4.68e-02 |
| MultiScler | pT+clump.MultiPRS | PRScs.10FCVal | 0.114 | 0.118 | -0.003 | 6.38e-01 | 0.116 | 0.104 | 0.012 | 1.09e-01 |
| MultiScler | LDpred1.Inf | PRScs.10FCVal | 0.089 | 0.118 | -0.029 | 3.77e-19 | 0.081 | 0.104 | -0.023 | 3.46e-04 |
| MultiScler | LDpred1.MultiPRS | PRScs.10FCVal | 0.103 | 0.118 | -0.015 | 3.41e-02 | 0.096 | 0.104 | -0.008 | 2.57e-01 |
| MultiScler | pT+clump.10FCVal | PRScs.10FCVal | 0.090 | 0.118 | -0.027 | 8.85e-07 | 0.092 | 0.104 | -0.012 | 2.80e-01 |
| MultiScler | lassosum.PseudoVal | PRScs.10FCVal | 0.100 | 0.118 | -0.017 | 3.41e-05 | 0.106 | 0.104 | 0.002 | 8.12e-01 |
| MultiScler | PRScs.PseudoVal | PRScs.10FCVal | 0.103 | 0.118 | -0.015 | 9.11e-05 | 0.101 | 0.104 | -0.002 | 7.39e-01 |
| MultiScler | DBSLMM.PseudoVal | PRScs.10FCVal | 0.090 | 0.118 | -0.028 | 3.09e-08 | 0.085 | 0.104 | -0.019 | 5.58e-02 |
| MultiScler | SBLUP.Inf | PRScs.10FCVal | 0.093 | 0.118 | -0.025 | 4.27e-18 | 0.079 | 0.104 | -0.025 | 1.81e-05 |
| MultiScler | LDpred2.Inf | PRScs.10FCVal | 0.092 | 0.118 | -0.026 | 9.94e-19 | 0.078 | 0.104 | -0.025 | 1.63e-05 |
| MultiScler | lassosum.10FCVal | PRScs.10FCVal | 0.115 | 0.118 | -0.002 | 2.18e-01 | 0.102 | 0.104 | -0.002 | 6.91e-01 |
| MultiScler | LDpred2.10FCVal | PRScs.10FCVal | 0.112 | 0.118 | -0.006 | 2.13e-01 | 0.101 | 0.104 | -0.003 | 7.70e-01 |
| MultiScler | lassosum.MultiPRS | PRScs.10FCVal | 0.126 | 0.118 | 0.008 | 2.36e-01 | 0.124 | 0.104 | 0.021 | 8.05e-05 |
| MultiScler | LDpred1.10FCVal | PRScs.10FCVal | 0.103 | 0.118 | -0.015 | 1.07e-05 | 0.095 | 0.104 | -0.009 | 1.83e-01 |
| MultiScler | All.MultiPRS | PRScs.10FCVal | 0.133 | 0.118 | 0.016 | 2.63e-02 | 0.121 | 0.104 | 0.017 | 6.52e-03 |
| MultiScler | SBayesR.PseudoVal | PRScs.10FCVal | 0.100 | 0.118 | -0.018 | 3.42e-04 | 0.084 | 0.104 | -0.020 | 4.69e-02 |
| MultiScler | PRScs.MultiPRS | PRScs.10FCVal | 0.118 | 0.118 | 0.001 | 9.35e-01 | 0.108 | 0.104 | 0.004 | 3.15e-01 |
| MultiScler | All.MultiPRS | PRScs.PseudoVal | 0.133 | 0.103 | 0.030 | 1.73e-05 | 0.121 | 0.101 | 0.020 | 2.48e-03 |
| MultiScler | pT+clump.MultiPRS | PRScs.PseudoVal | 0.114 | 0.103 | 0.011 | 1.07e-01 | 0.116 | 0.101 | 0.014 | 6.14e-02 |
| MultiScler | LDpred2.PseudoVal | PRScs.PseudoVal | 0.101 | 0.103 | -0.002 | 6.63e-01 | 0.088 | 0.101 | -0.013 | 1.77e-01 |
| MultiScler | SBayesR.PseudoVal | PRScs.PseudoVal | 0.100 | 0.103 | -0.003 | 5.50e-01 | 0.084 | 0.101 | -0.017 | 1.33e-01 |
| MultiScler | LDpred2.MultiPRS | PRScs.PseudoVal | 0.130 | 0.103 | 0.027 | 1.41e-04 | 0.117 | 0.101 | 0.016 | 1.94e-02 |
| MultiScler | SBLUP.Inf | PRScs.PseudoVal | 0.093 | 0.103 | -0.010 | 5.29e-02 | 0.079 | 0.101 | -0.022 | 3.54e-02 |
| MultiScler | LDpred1.Inf | PRScs.PseudoVal | 0.089 | 0.103 | -0.015 | 8.65e-04 | 0.081 | 0.101 | -0.021 | 1.73e-02 |
| MultiScler | PRScs.10FCVal | PRScs.PseudoVal | 0.118 | 0.103 | 0.015 | 9.11e-05 | 0.104 | 0.101 | 0.002 | 7.39e-01 |
| MultiScler | LDpred2.Inf | PRScs.PseudoVal | 0.092 | 0.103 | -0.011 | 3.72e-02 | 0.078 | 0.101 | -0.023 | 3.27e-02 |
| MultiScler | PRScs.PseudoVal | PRScs.PseudoVal | 0.103 | 0.103 | 0.000 | 1.00e+00 | 0.101 | 0.101 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.10FCVal | PRScs.PseudoVal | 0.112 | 0.103 | 0.009 | 1.62e-03 | 0.101 | 0.101 | 0.000 | 9.66e-01 |
| MultiScler | pT+clump.10FCVal | PRScs.PseudoVal | 0.090 | 0.103 | -0.013 | 2.29e-03 | 0.092 | 0.101 | -0.010 | 2.60e-01 |
| MultiScler | PRScs.MultiPRS | PRScs.PseudoVal | 0.118 | 0.103 | 0.015 | 3.11e-02 | 0.108 | 0.101 | 0.007 | 5.56e-02 |
| MultiScler | lassosum.PseudoVal | PRScs.PseudoVal | 0.100 | 0.103 | -0.003 | 9.12e-02 | 0.106 | 0.101 | 0.004 | 1.99e-01 |
| MultiScler | lassosum.10FCVal | PRScs.PseudoVal | 0.115 | 0.103 | 0.012 | 1.19e-03 | 0.102 | 0.101 | 0.001 | 9.09e-01 |
| MultiScler | LDpred1.MultiPRS | PRScs.PseudoVal | 0.103 | 0.103 | 0.000 | 9.63e-01 | 0.096 | 0.101 | -0.005 | 3.37e-01 |
| MultiScler | lassosum.MultiPRS | PRScs.PseudoVal | 0.126 | 0.103 | 0.023 | 1.11e-03 | 0.124 | 0.101 | 0.023 | 9.43e-06 |
| MultiScler | LDpred1.10FCVal | PRScs.PseudoVal | 0.103 | 0.103 | 0.000 | 8.82e-01 | 0.095 | 0.101 | -0.006 | 1.97e-01 |
| MultiScler | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.090 | 0.103 | -0.013 | 2.44e-09 | 0.085 | 0.101 | -0.017 | 1.79e-04 |
| MultiScler | All.MultiPRS | pT+clump.MultiPRS | 0.133 | 0.114 | 0.019 | 6.93e-03 | 0.121 | 0.116 | 0.005 | 3.17e-01 |
| MultiScler | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.100 | 0.114 | -0.015 | 3.58e-02 | 0.084 | 0.116 | -0.032 | 1.82e-04 |
| MultiScler | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.103 | 0.114 | -0.012 | 9.81e-02 | 0.096 | 0.116 | -0.019 | 1.80e-02 |
| MultiScler | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.116 | 0.116 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.PseudoVal | pT+clump.MultiPRS | 0.100 | 0.114 | -0.014 | 4.24e-02 | 0.106 | 0.116 | -0.010 | 2.05e-01 |
| MultiScler | PRScs.10FCVal | pT+clump.MultiPRS | 0.118 | 0.114 | 0.003 | 6.38e-01 | 0.104 | 0.116 | -0.012 | 1.09e-01 |
| MultiScler | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.090 | 0.114 | -0.024 | 5.00e-04 | 0.085 | 0.116 | -0.031 | 3.13e-04 |
| MultiScler | SBLUP.Inf | pT+clump.MultiPRS | 0.093 | 0.114 | -0.022 | 2.09e-03 | 0.079 | 0.116 | -0.037 | 1.11e-04 |
| MultiScler | LDpred2.Inf | pT+clump.MultiPRS | 0.092 | 0.114 | -0.022 | 1.36e-03 | 0.078 | 0.116 | -0.037 | 9.97e-05 |
| MultiScler | pT+clump.10FCVal | pT+clump.MultiPRS | 0.090 | 0.114 | -0.024 | 5.61e-04 | 0.092 | 0.116 | -0.024 | 3.30e-04 |
| MultiScler | LDpred2.10FCVal | pT+clump.MultiPRS | 0.112 | 0.114 | -0.003 | 7.16e-01 | 0.101 | 0.116 | -0.015 | 7.21e-02 |
| MultiScler | PRScs.PseudoVal | pT+clump.MultiPRS | 0.103 | 0.114 | -0.011 | 1.07e-01 | 0.101 | 0.116 | -0.014 | 6.14e-02 |
| MultiScler | LDpred1.10FCVal | pT+clump.MultiPRS | 0.103 | 0.114 | -0.012 | 9.58e-02 | 0.095 | 0.116 | -0.021 | 1.16e-02 |
| MultiScler | LDpred1.Inf | pT+clump.MultiPRS | 0.089 | 0.114 | -0.026 | 2.28e-04 | 0.081 | 0.116 | -0.035 | 1.55e-04 |
| MultiScler | PRScs.MultiPRS | pT+clump.MultiPRS | 0.118 | 0.114 | 0.004 | 5.82e-01 | 0.108 | 0.116 | -0.008 | 2.68e-01 |
| MultiScler | lassosum.10FCVal | pT+clump.MultiPRS | 0.115 | 0.114 | 0.001 | 9.05e-01 | 0.102 | 0.116 | -0.014 | 6.52e-02 |
| MultiScler | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.101 | 0.114 | -0.013 | 5.58e-02 | 0.088 | 0.116 | -0.027 | 2.40e-03 |
| MultiScler | lassosum.MultiPRS | pT+clump.MultiPRS | 0.126 | 0.114 | 0.012 | 9.69e-02 | 0.124 | 0.116 | 0.009 | 1.30e-01 |
| MultiScler | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.130 | 0.114 | 0.015 | 2.76e-02 | 0.117 | 0.116 | 0.001 | 8.38e-01 |
| MultiScler | LDpred2.10FCVal | pT+clump.10FCVal | 0.112 | 0.090 | 0.022 | 3.00e-08 | 0.101 | 0.092 | 0.009 | 2.45e-01 |
| MultiScler | pT+clump.MultiPRS | pT+clump.10FCVal | 0.114 | 0.090 | 0.024 | 5.61e-04 | 0.116 | 0.092 | 0.024 | 3.30e-04 |
| MultiScler | LDpred2.Inf | pT+clump.10FCVal | 0.092 | 0.090 | 0.002 | 7.92e-01 | 0.078 | 0.092 | -0.013 | 3.10e-01 |
| MultiScler | SBayesR.PseudoVal | pT+clump.10FCVal | 0.100 | 0.090 | 0.009 | 6.97e-02 | 0.084 | 0.092 | -0.008 | 4.68e-01 |
| MultiScler | PRScs.MultiPRS | pT+clump.10FCVal | 0.118 | 0.090 | 0.028 | 6.70e-05 | 0.108 | 0.092 | 0.016 | 8.26e-02 |
| MultiScler | SBLUP.Inf | pT+clump.10FCVal | 0.093 | 0.090 | 0.003 | 6.86e-01 | 0.079 | 0.092 | -0.013 | 3.28e-01 |
| MultiScler | lassosum.MultiPRS | pT+clump.10FCVal | 0.126 | 0.090 | 0.036 | 3.50e-07 | 0.124 | 0.092 | 0.033 | 3.14e-04 |
| MultiScler | PRScs.10FCVal | pT+clump.10FCVal | 0.118 | 0.090 | 0.027 | 8.85e-07 | 0.104 | 0.092 | 0.012 | 2.80e-01 |
| MultiScler | LDpred2.MultiPRS | pT+clump.10FCVal | 0.130 | 0.090 | 0.040 | 1.67e-08 | 0.117 | 0.092 | 0.025 | 6.11e-03 |
| MultiScler | lassosum.10FCVal | pT+clump.10FCVal | 0.115 | 0.090 | 0.025 | 5.56e-06 | 0.102 | 0.092 | 0.011 | 3.44e-01 |
| MultiScler | LDpred2.PseudoVal | pT+clump.10FCVal | 0.101 | 0.090 | 0.011 | 8.27e-02 | 0.088 | 0.092 | -0.003 | 7.84e-01 |
| MultiScler | LDpred1.MultiPRS | pT+clump.10FCVal | 0.103 | 0.090 | 0.013 | 7.51e-02 | 0.096 | 0.092 | 0.005 | 6.48e-01 |
| MultiScler | pT+clump.10FCVal | pT+clump.10FCVal | 0.090 | 0.090 | 0.000 | 1.00e+00 | 0.092 | 0.092 | 0.000 | 1.00e+00 |
| MultiScler | All.MultiPRS | pT+clump.10FCVal | 0.133 | 0.090 | 0.043 | 8.56e-10 | 0.121 | 0.092 | 0.029 | 4.14e-04 |
| MultiScler | LDpred1.Inf | pT+clump.10FCVal | 0.089 | 0.090 | -0.002 | 7.76e-01 | 0.081 | 0.092 | -0.011 | 3.58e-01 |
| MultiScler | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.090 | 0.090 | 0.000 | 9.47e-01 | 0.085 | 0.092 | -0.007 | 3.21e-01 |
| MultiScler | LDpred1.10FCVal | pT+clump.10FCVal | 0.103 | 0.090 | 0.012 | 1.02e-02 | 0.095 | 0.092 | 0.003 | 7.38e-01 |
| MultiScler | lassosum.PseudoVal | pT+clump.10FCVal | 0.100 | 0.090 | 0.010 | 1.75e-02 | 0.106 | 0.092 | 0.014 | 9.87e-02 |
| MultiScler | PRScs.PseudoVal | pT+clump.10FCVal | 0.103 | 0.090 | 0.013 | 2.29e-03 | 0.101 | 0.092 | 0.010 | 2.60e-01 |
| MultiScler | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.114 | 0.100 | 0.015 | 3.58e-02 | 0.116 | 0.084 | 0.032 | 1.82e-04 |
| MultiScler | LDpred1.10FCVal | SBayesR.PseudoVal | 0.103 | 0.100 | 0.003 | 5.77e-01 | 0.095 | 0.084 | 0.011 | 3.17e-01 |
| MultiScler | PRScs.10FCVal | SBayesR.PseudoVal | 0.118 | 0.100 | 0.018 | 3.42e-04 | 0.104 | 0.084 | 0.020 | 4.69e-02 |
| MultiScler | LDpred2.10FCVal | SBayesR.PseudoVal | 0.112 | 0.100 | 0.012 | 1.79e-02 | 0.101 | 0.084 | 0.017 | 9.64e-02 |
| MultiScler | lassosum.10FCVal | SBayesR.PseudoVal | 0.115 | 0.100 | 0.016 | 1.35e-03 | 0.102 | 0.084 | 0.018 | 5.88e-02 |
| MultiScler | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.101 | 0.100 | 0.001 | 7.85e-01 | 0.088 | 0.084 | 0.004 | 6.59e-01 |
| MultiScler | lassosum.MultiPRS | SBayesR.PseudoVal | 0.126 | 0.100 | 0.026 | 1.64e-04 | 0.124 | 0.084 | 0.041 | 7.10e-06 |
| MultiScler | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.130 | 0.100 | 0.030 | 1.59e-05 | 0.117 | 0.084 | 0.033 | 9.98e-05 |
| MultiScler | All.MultiPRS | SBayesR.PseudoVal | 0.133 | 0.100 | 0.034 | 1.49e-06 | 0.121 | 0.084 | 0.037 | 1.99e-06 |
| MultiScler | PRScs.PseudoVal | SBayesR.PseudoVal | 0.103 | 0.100 | 0.003 | 5.50e-01 | 0.101 | 0.084 | 0.017 | 1.33e-01 |
| MultiScler | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.103 | 0.100 | 0.003 | 6.55e-01 | 0.096 | 0.084 | 0.012 | 2.59e-01 |
| MultiScler | pT+clump.10FCVal | SBayesR.PseudoVal | 0.090 | 0.100 | -0.009 | 6.97e-02 | 0.092 | 0.084 | 0.008 | 4.68e-01 |
| MultiScler | lassosum.PseudoVal | SBayesR.PseudoVal | 0.100 | 0.100 | 0.001 | 9.27e-01 | 0.106 | 0.084 | 0.022 | 5.37e-02 |
| MultiScler | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.084 | 0.084 | 0.000 | 1.00e+00 |
| MultiScler | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.090 | 0.100 | -0.010 | 1.22e-01 | 0.085 | 0.084 | 0.001 | 9.40e-01 |
| MultiScler | SBLUP.Inf | SBayesR.PseudoVal | 0.093 | 0.100 | -0.007 | 1.67e-01 | 0.079 | 0.084 | -0.005 | 6.20e-01 |
| MultiScler | LDpred2.Inf | SBayesR.PseudoVal | 0.092 | 0.100 | -0.008 | 1.17e-01 | 0.078 | 0.084 | -0.005 | 5.81e-01 |
| MultiScler | LDpred1.Inf | SBayesR.PseudoVal | 0.089 | 0.100 | -0.011 | 3.06e-02 | 0.081 | 0.084 | -0.003 | 7.59e-01 |
| MultiScler | PRScs.MultiPRS | SBayesR.PseudoVal | 0.118 | 0.100 | 0.019 | 7.82e-03 | 0.108 | 0.084 | 0.024 | 2.42e-02 |
| MultiScler | LDpred2.10FCVal | SBLUP.Inf | 0.112 | 0.093 | 0.019 | 1.17e-03 | 0.101 | 0.079 | 0.022 | 5.85e-02 |
| MultiScler | PRScs.PseudoVal | SBLUP.Inf | 0.103 | 0.093 | 0.010 | 5.29e-02 | 0.101 | 0.079 | 0.022 | 3.54e-02 |
| MultiScler | LDpred1.10FCVal | SBLUP.Inf | 0.103 | 0.093 | 0.010 | 3.65e-02 | 0.095 | 0.079 | 0.016 | 9.09e-02 |
| MultiScler | LDpred2.Inf | SBLUP.Inf | 0.092 | 0.093 | -0.001 | 3.83e-13 | 0.078 | 0.079 | -0.001 | 2.41e-02 |
| MultiScler | SBayesR.PseudoVal | SBLUP.Inf | 0.100 | 0.093 | 0.007 | 1.67e-01 | 0.084 | 0.079 | 0.005 | 6.20e-01 |
| MultiScler | lassosum.10FCVal | SBLUP.Inf | 0.115 | 0.093 | 0.022 | 1.71e-10 | 0.102 | 0.079 | 0.023 | 9.70e-04 |
| MultiScler | pT+clump.MultiPRS | SBLUP.Inf | 0.114 | 0.093 | 0.022 | 2.09e-03 | 0.116 | 0.079 | 0.037 | 1.11e-04 |
| MultiScler | lassosum.MultiPRS | SBLUP.Inf | 0.126 | 0.093 | 0.033 | 2.37e-06 | 0.124 | 0.079 | 0.045 | 3.28e-08 |
| MultiScler | PRScs.10FCVal | SBLUP.Inf | 0.118 | 0.093 | 0.025 | 4.27e-18 | 0.104 | 0.079 | 0.025 | 1.81e-05 |
| MultiScler | PRScs.MultiPRS | SBLUP.Inf | 0.118 | 0.093 | 0.025 | 3.00e-04 | 0.108 | 0.079 | 0.029 | 6.11e-04 |
| MultiScler | LDpred1.MultiPRS | SBLUP.Inf | 0.103 | 0.093 | 0.010 | 1.57e-01 | 0.096 | 0.079 | 0.017 | 6.52e-02 |
| MultiScler | LDpred2.PseudoVal | SBLUP.Inf | 0.101 | 0.093 | 0.008 | 4.96e-30 | 0.088 | 0.079 | 0.009 | 1.32e-10 |
| MultiScler | LDpred1.Inf | SBLUP.Inf | 0.089 | 0.093 | -0.004 | 2.29e-01 | 0.081 | 0.079 | 0.002 | 8.05e-01 |
| MultiScler | LDpred2.MultiPRS | SBLUP.Inf | 0.130 | 0.093 | 0.037 | 1.38e-07 | 0.117 | 0.079 | 0.038 | 7.07e-06 |
| MultiScler | All.MultiPRS | SBLUP.Inf | 0.133 | 0.093 | 0.040 | 8.51e-09 | 0.121 | 0.079 | 0.042 | 1.61e-06 |
| MultiScler | SBLUP.Inf | SBLUP.Inf | 0.093 | 0.093 | 0.000 | 1.00e+00 | 0.079 | 0.079 | 0.000 | 1.00e+00 |
| MultiScler | DBSLMM.PseudoVal | SBLUP.Inf | 0.090 | 0.093 | -0.003 | 6.58e-01 | 0.085 | 0.079 | 0.006 | 6.47e-01 |
| MultiScler | pT+clump.10FCVal | SBLUP.Inf | 0.090 | 0.093 | -0.003 | 6.86e-01 | 0.092 | 0.079 | 0.013 | 3.28e-01 |
| MultiScler | lassosum.PseudoVal | SBLUP.Inf | 0.100 | 0.093 | 0.007 | 2.03e-01 | 0.106 | 0.079 | 0.027 | 2.03e-02 |
| RheuArth | LDpred2.PseudoVal | All.MultiPRS | 0.091 | 0.185 | -0.094 | 1.39e-41 | 0.077 | 0.161 | -0.085 | 3.01e-19 |
| RheuArth | pT+clump.10FCVal | All.MultiPRS | 0.111 | 0.185 | -0.074 | 4.10e-26 | 0.103 | 0.161 | -0.058 | 4.53e-11 |
| RheuArth | LDpred2.MultiPRS | All.MultiPRS | 0.151 | 0.185 | -0.034 | 1.71e-32 | 0.127 | 0.161 | -0.034 | 3.84e-09 |
| RheuArth | PRScs.PseudoVal | All.MultiPRS | 0.162 | 0.185 | -0.022 | 1.13e-03 | 0.129 | 0.161 | -0.032 | 1.52e-12 |
| RheuArth | LDpred1.Inf | All.MultiPRS | 0.136 | 0.185 | -0.048 | 2.35e-12 | 0.111 | 0.161 | -0.050 | 8.32e-14 |
| RheuArth | PRScs.MultiPRS | All.MultiPRS | 0.166 | 0.185 | -0.019 | 1.02e-21 | 0.133 | 0.161 | -0.028 | 7.09e-12 |
| RheuArth | LDpred2.Inf | All.MultiPRS | 0.085 | 0.185 | -0.100 | 7.60e-47 | 0.072 | 0.161 | -0.089 | 7.28e-20 |
| RheuArth | lassosum.10FCVal | All.MultiPRS | 0.162 | 0.185 | -0.023 | 7.57e-04 | 0.130 | 0.161 | -0.032 | 2.31e-12 |
| RheuArth | LDpred2.10FCVal | All.MultiPRS | 0.151 | 0.185 | -0.034 | 1.08e-06 | 0.128 | 0.161 | -0.033 | 4.06e-08 |
| RheuArth | lassosum.MultiPRS | All.MultiPRS | 0.183 | 0.185 | -0.002 | 7.54e-01 | 0.150 | 0.161 | -0.011 | 9.70e-04 |
| RheuArth | LDpred1.10FCVal | All.MultiPRS | 0.136 | 0.185 | -0.048 | 2.27e-12 | 0.112 | 0.161 | -0.050 | 1.88e-13 |
| RheuArth | All.MultiPRS | All.MultiPRS | 0.185 | 0.185 | 0.000 | 1.00e+00 | 0.161 | 0.161 | 0.000 | 1.00e+00 |
| RheuArth | SBayesR.PseudoVal | All.MultiPRS | 0.124 | 0.185 | -0.061 | 2.02e-18 | 0.119 | 0.161 | -0.042 | 2.05e-07 |
| RheuArth | LDpred1.MultiPRS | All.MultiPRS | 0.145 | 0.185 | -0.039 | 4.71e-39 | 0.119 | 0.161 | -0.042 | 4.49e-12 |
| RheuArth | pT+clump.MultiPRS | All.MultiPRS | 0.131 | 0.185 | -0.054 | 1.01e-14 | 0.124 | 0.161 | -0.037 | 2.25e-07 |
| RheuArth | lassosum.PseudoVal | All.MultiPRS | 0.114 | 0.185 | -0.070 | 3.66e-24 | 0.089 | 0.161 | -0.072 | 3.27e-17 |
| RheuArth | PRScs.10FCVal | All.MultiPRS | 0.161 | 0.185 | -0.024 | 6.00e-04 | 0.128 | 0.161 | -0.034 | 9.61e-13 |
| RheuArth | DBSLMM.PseudoVal | All.MultiPRS | 0.142 | 0.185 | -0.043 | 7.64e-10 | 0.111 | 0.161 | -0.051 | 3.42e-18 |
| RheuArth | SBLUP.Inf | All.MultiPRS | 0.112 | 0.185 | -0.073 | 7.80e-26 | 0.097 | 0.161 | -0.065 | 8.79e-15 |
| RheuArth | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.124 | 0.142 | -0.018 | 7.22e-04 | 0.119 | 0.111 | 0.008 | 4.44e-01 |
| RheuArth | LDpred2.Inf | DBSLMM.PseudoVal | 0.085 | 0.142 | -0.057 | 2.08e-22 | 0.072 | 0.111 | -0.039 | 1.03e-03 |
| RheuArth | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.111 | 0.142 | -0.031 | 6.56e-09 | 0.103 | 0.111 | -0.007 | 4.95e-01 |
| RheuArth | PRScs.10FCVal | DBSLMM.PseudoVal | 0.161 | 0.142 | 0.019 | 6.05e-19 | 0.128 | 0.111 | 0.017 | 8.66e-05 |
| RheuArth | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.162 | 0.142 | 0.020 | 2.04e-21 | 0.129 | 0.111 | 0.018 | 2.45e-05 |
| RheuArth | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.131 | 0.142 | -0.011 | 1.13e-01 | 0.124 | 0.111 | 0.013 | 2.00e-01 |
| RheuArth | LDpred1.Inf | DBSLMM.PseudoVal | 0.136 | 0.142 | -0.006 | 1.56e-01 | 0.111 | 0.111 | 0.000 | 9.75e-01 |
| RheuArth | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.183 | 0.142 | 0.040 | 5.92e-09 | 0.150 | 0.111 | 0.039 | 4.38e-11 |
| RheuArth | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.151 | 0.142 | 0.009 | 1.78e-02 | 0.128 | 0.111 | 0.018 | 2.01e-02 |
| RheuArth | lassosum.10FCVal | DBSLMM.PseudoVal | 0.162 | 0.142 | 0.019 | 1.48e-12 | 0.130 | 0.111 | 0.019 | 7.03e-04 |
| RheuArth | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.136 | 0.142 | -0.006 | 1.60e-01 | 0.112 | 0.111 | 0.001 | 9.21e-01 |
| RheuArth | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.145 | 0.142 | 0.003 | 6.47e-01 | 0.119 | 0.111 | 0.008 | 2.79e-01 |
| RheuArth | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.166 | 0.142 | 0.023 | 7.53e-04 | 0.133 | 0.111 | 0.022 | 6.67e-06 |
| RheuArth | All.MultiPRS | DBSLMM.PseudoVal | 0.185 | 0.142 | 0.043 | 7.64e-10 | 0.161 | 0.111 | 0.051 | 3.42e-18 |
| RheuArth | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.091 | 0.142 | -0.051 | 5.15e-19 | 0.077 | 0.111 | -0.034 | 2.97e-03 |
| RheuArth | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.142 | 0.142 | 0.000 | 1.00e+00 | 0.111 | 0.111 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.151 | 0.142 | 0.009 | 2.16e-01 | 0.127 | 0.111 | 0.017 | 3.37e-02 |
| RheuArth | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.114 | 0.142 | -0.028 | 3.51e-08 | 0.089 | 0.111 | -0.021 | 3.52e-02 |
| RheuArth | SBLUP.Inf | DBSLMM.PseudoVal | 0.112 | 0.142 | -0.030 | 8.95e-09 | 0.097 | 0.111 | -0.014 | 1.80e-01 |
| RheuArth | SBayesR.PseudoVal | lassosum.MultiPRS | 0.124 | 0.183 | -0.058 | 4.17e-17 | 0.119 | 0.150 | -0.031 | 4.64e-04 |
| RheuArth | LDpred2.10FCVal | lassosum.MultiPRS | 0.151 | 0.183 | -0.031 | 5.78e-06 | 0.128 | 0.150 | -0.022 | 2.24e-03 |
| RheuArth | LDpred1.10FCVal | lassosum.MultiPRS | 0.136 | 0.183 | -0.046 | 2.97e-11 | 0.112 | 0.150 | -0.039 | 1.72e-08 |
| RheuArth | LDpred2.Inf | lassosum.MultiPRS | 0.085 | 0.183 | -0.098 | 2.27e-44 | 0.072 | 0.150 | -0.078 | 4.72e-15 |
| RheuArth | PRScs.10FCVal | lassosum.MultiPRS | 0.161 | 0.183 | -0.022 | 1.94e-03 | 0.128 | 0.150 | -0.022 | 4.05e-07 |
| RheuArth | PRScs.MultiPRS | lassosum.MultiPRS | 0.166 | 0.183 | -0.017 | 1.37e-02 | 0.133 | 0.150 | -0.017 | 1.07e-05 |
| RheuArth | pT+clump.MultiPRS | lassosum.MultiPRS | 0.131 | 0.183 | -0.051 | 1.21e-13 | 0.124 | 0.150 | -0.026 | 1.26e-03 |
| RheuArth | lassosum.MultiPRS | lassosum.MultiPRS | 0.183 | 0.183 | 0.000 | 1.00e+00 | 0.150 | 0.150 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.PseudoVal | lassosum.MultiPRS | 0.114 | 0.183 | -0.068 | 9.32e-23 | 0.089 | 0.150 | -0.061 | 2.33e-13 |
| RheuArth | LDpred2.MultiPRS | lassosum.MultiPRS | 0.151 | 0.183 | -0.032 | 4.42e-06 | 0.127 | 0.150 | -0.023 | 1.09e-03 |
| RheuArth | LDpred1.MultiPRS | lassosum.MultiPRS | 0.145 | 0.183 | -0.037 | 7.83e-08 | 0.119 | 0.150 | -0.031 | 7.91e-07 |
| RheuArth | LDpred2.PseudoVal | lassosum.MultiPRS | 0.091 | 0.183 | -0.091 | 2.73e-39 | 0.077 | 0.150 | -0.074 | 3.15e-14 |
| RheuArth | LDpred1.Inf | lassosum.MultiPRS | 0.136 | 0.183 | -0.046 | 3.03e-11 | 0.111 | 0.150 | -0.039 | 9.27e-09 |
| RheuArth | pT+clump.10FCVal | lassosum.MultiPRS | 0.111 | 0.183 | -0.071 | 7.40e-25 | 0.103 | 0.150 | -0.047 | 1.39e-06 |
| RheuArth | lassosum.10FCVal | lassosum.MultiPRS | 0.162 | 0.183 | -0.021 | 2.35e-03 | 0.130 | 0.150 | -0.020 | 1.55e-08 |
| RheuArth | SBLUP.Inf | lassosum.MultiPRS | 0.112 | 0.183 | -0.071 | 3.93e-24 | 0.097 | 0.150 | -0.054 | 2.26e-10 |
| RheuArth | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.142 | 0.183 | -0.040 | 5.92e-09 | 0.111 | 0.150 | -0.039 | 4.38e-11 |
| RheuArth | All.MultiPRS | lassosum.MultiPRS | 0.185 | 0.183 | 0.002 | 7.54e-01 | 0.161 | 0.150 | 0.011 | 9.70e-04 |
| RheuArth | PRScs.PseudoVal | lassosum.MultiPRS | 0.162 | 0.183 | -0.020 | 3.47e-03 | 0.129 | 0.150 | -0.021 | 7.80e-07 |
| RheuArth | LDpred1.Inf | lassosum.PseudoVal | 0.136 | 0.114 | 0.022 | 2.15e-08 | 0.111 | 0.089 | 0.022 | 5.69e-03 |
| RheuArth | PRScs.10FCVal | lassosum.PseudoVal | 0.161 | 0.114 | 0.047 | 2.29e-23 | 0.128 | 0.089 | 0.038 | 4.46e-05 |
| RheuArth | LDpred2.MultiPRS | lassosum.PseudoVal | 0.151 | 0.114 | 0.036 | 1.66e-07 | 0.127 | 0.089 | 0.038 | 7.30e-05 |
| RheuArth | lassosum.PseudoVal | lassosum.PseudoVal | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.089 | 0.089 | 0.000 | 1.00e+00 |
| RheuArth | PRScs.PseudoVal | lassosum.PseudoVal | 0.162 | 0.114 | 0.048 | 4.55e-25 | 0.129 | 0.089 | 0.040 | 1.94e-05 |
| RheuArth | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.142 | 0.114 | 0.028 | 3.51e-08 | 0.111 | 0.089 | 0.021 | 3.52e-02 |
| RheuArth | SBLUP.Inf | lassosum.PseudoVal | 0.112 | 0.114 | -0.002 | 5.39e-01 | 0.097 | 0.089 | 0.007 | 3.37e-01 |
| RheuArth | LDpred2.Inf | lassosum.PseudoVal | 0.085 | 0.114 | -0.029 | 1.23e-11 | 0.072 | 0.089 | -0.017 | 4.76e-02 |
| RheuArth | pT+clump.10FCVal | lassosum.PseudoVal | 0.111 | 0.114 | -0.003 | 6.14e-01 | 0.103 | 0.089 | 0.014 | 2.56e-01 |
| RheuArth | LDpred2.10FCVal | lassosum.PseudoVal | 0.151 | 0.114 | 0.037 | 8.81e-14 | 0.128 | 0.089 | 0.039 | 9.95e-05 |
| RheuArth | lassosum.MultiPRS | lassosum.PseudoVal | 0.183 | 0.114 | 0.068 | 9.32e-23 | 0.150 | 0.089 | 0.061 | 2.33e-13 |
| RheuArth | LDpred1.10FCVal | lassosum.PseudoVal | 0.136 | 0.114 | 0.022 | 1.67e-08 | 0.112 | 0.089 | 0.022 | 4.24e-03 |
| RheuArth | All.MultiPRS | lassosum.PseudoVal | 0.185 | 0.114 | 0.070 | 3.66e-24 | 0.161 | 0.089 | 0.072 | 3.27e-17 |
| RheuArth | SBayesR.PseudoVal | lassosum.PseudoVal | 0.124 | 0.114 | 0.010 | 4.28e-02 | 0.119 | 0.089 | 0.030 | 2.34e-03 |
| RheuArth | lassosum.10FCVal | lassosum.PseudoVal | 0.162 | 0.114 | 0.047 | 4.34e-27 | 0.130 | 0.089 | 0.041 | 4.65e-06 |
| RheuArth | pT+clump.MultiPRS | lassosum.PseudoVal | 0.131 | 0.114 | 0.017 | 1.61e-02 | 0.124 | 0.089 | 0.035 | 4.18e-04 |
| RheuArth | LDpred2.PseudoVal | lassosum.PseudoVal | 0.091 | 0.114 | -0.023 | 3.18e-08 | 0.077 | 0.089 | -0.013 | 1.32e-01 |
| RheuArth | LDpred1.MultiPRS | lassosum.PseudoVal | 0.145 | 0.114 | 0.031 | 8.34e-06 | 0.119 | 0.089 | 0.030 | 2.27e-04 |
| RheuArth | PRScs.MultiPRS | lassosum.PseudoVal | 0.166 | 0.114 | 0.051 | 1.80e-13 | 0.133 | 0.089 | 0.044 | 3.08e-08 |
| RheuArth | lassosum.10FCVal | lassosum.10FCVal | 0.162 | 0.162 | 0.000 | 1.00e+00 | 0.130 | 0.130 | 0.000 | 1.00e+00 |
| RheuArth | pT+clump.MultiPRS | lassosum.10FCVal | 0.131 | 0.162 | -0.030 | 1.25e-05 | 0.124 | 0.130 | -0.006 | 5.22e-01 |
| RheuArth | LDpred2.Inf | lassosum.10FCVal | 0.085 | 0.162 | -0.076 | 2.73e-45 | 0.072 | 0.130 | -0.058 | 1.31e-07 |
| RheuArth | SBayesR.PseudoVal | lassosum.10FCVal | 0.124 | 0.162 | -0.037 | 1.17e-13 | 0.119 | 0.130 | -0.011 | 2.92e-01 |
| RheuArth | PRScs.MultiPRS | lassosum.10FCVal | 0.166 | 0.162 | 0.004 | 5.61e-01 | 0.133 | 0.130 | 0.003 | 3.74e-01 |
| RheuArth | SBLUP.Inf | lassosum.10FCVal | 0.112 | 0.162 | -0.050 | 2.47e-26 | 0.097 | 0.130 | -0.033 | 4.26e-04 |
| RheuArth | lassosum.MultiPRS | lassosum.10FCVal | 0.183 | 0.162 | 0.021 | 2.35e-03 | 0.150 | 0.130 | 0.020 | 1.55e-08 |
| RheuArth | PRScs.10FCVal | lassosum.10FCVal | 0.161 | 0.162 | 0.000 | 8.14e-01 | 0.128 | 0.130 | -0.002 | 5.84e-01 |
| RheuArth | LDpred2.MultiPRS | lassosum.10FCVal | 0.151 | 0.162 | -0.011 | 1.20e-01 | 0.127 | 0.130 | -0.002 | 7.25e-01 |
| RheuArth | PRScs.PseudoVal | lassosum.10FCVal | 0.162 | 0.162 | 0.001 | 6.61e-01 | 0.129 | 0.130 | -0.001 | 8.17e-01 |
| RheuArth | LDpred2.PseudoVal | lassosum.10FCVal | 0.091 | 0.162 | -0.070 | 9.12e-41 | 0.077 | 0.130 | -0.053 | 5.63e-07 |
| RheuArth | LDpred1.MultiPRS | lassosum.10FCVal | 0.145 | 0.162 | -0.016 | 1.94e-02 | 0.119 | 0.130 | -0.011 | 1.02e-01 |
| RheuArth | pT+clump.10FCVal | lassosum.10FCVal | 0.111 | 0.162 | -0.050 | 3.22e-22 | 0.103 | 0.130 | -0.027 | 1.18e-02 |
| RheuArth | All.MultiPRS | lassosum.10FCVal | 0.185 | 0.162 | 0.023 | 7.57e-04 | 0.161 | 0.130 | 0.032 | 2.31e-12 |
| RheuArth | LDpred1.Inf | lassosum.10FCVal | 0.136 | 0.162 | -0.025 | 1.26e-12 | 0.111 | 0.130 | -0.019 | 9.06e-03 |
| RheuArth | DBSLMM.PseudoVal | lassosum.10FCVal | 0.142 | 0.162 | -0.019 | 1.48e-12 | 0.111 | 0.130 | -0.019 | 7.03e-04 |
| RheuArth | LDpred1.10FCVal | lassosum.10FCVal | 0.136 | 0.162 | -0.025 | 3.16e-12 | 0.112 | 0.130 | -0.018 | 1.29e-02 |
| RheuArth | lassosum.PseudoVal | lassosum.10FCVal | 0.114 | 0.162 | -0.047 | 4.34e-27 | 0.089 | 0.130 | -0.041 | 4.65e-06 |
| RheuArth | LDpred2.10FCVal | lassosum.10FCVal | 0.151 | 0.162 | -0.010 | 2.81e-03 | 0.128 | 0.130 | -0.002 | 8.33e-01 |
| RheuArth | LDpred2.10FCVal | LDpred1.MultiPRS | 0.151 | 0.145 | 0.006 | 4.04e-01 | 0.128 | 0.119 | 0.009 | 2.39e-01 |
| RheuArth | PRScs.PseudoVal | LDpred1.MultiPRS | 0.162 | 0.145 | 0.017 | 1.43e-02 | 0.129 | 0.119 | 0.010 | 9.98e-02 |
| RheuArth | LDpred2.Inf | LDpred1.MultiPRS | 0.085 | 0.145 | -0.060 | 4.34e-18 | 0.072 | 0.119 | -0.047 | 9.38e-09 |
| RheuArth | pT+clump.10FCVal | LDpred1.MultiPRS | 0.111 | 0.145 | -0.034 | 1.01e-06 | 0.103 | 0.119 | -0.016 | 1.69e-01 |
| RheuArth | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.124 | 0.145 | -0.021 | 2.31e-03 | 0.119 | 0.119 | 0.000 | 9.95e-01 |
| RheuArth | All.MultiPRS | LDpred1.MultiPRS | 0.185 | 0.145 | 0.039 | 4.71e-39 | 0.161 | 0.119 | 0.042 | 4.49e-12 |
| RheuArth | LDpred1.10FCVal | LDpred1.MultiPRS | 0.136 | 0.145 | -0.009 | 1.92e-01 | 0.112 | 0.119 | -0.007 | 1.54e-02 |
| RheuArth | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.145 | 0.145 | 0.000 | 1.00e+00 | 0.119 | 0.119 | 0.000 | 1.00e+00 |
| RheuArth | PRScs.10FCVal | LDpred1.MultiPRS | 0.161 | 0.145 | 0.016 | 2.30e-02 | 0.128 | 0.119 | 0.009 | 1.62e-01 |
| RheuArth | lassosum.PseudoVal | LDpred1.MultiPRS | 0.114 | 0.145 | -0.031 | 8.34e-06 | 0.089 | 0.119 | -0.030 | 2.27e-04 |
| RheuArth | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.131 | 0.145 | -0.014 | 4.16e-02 | 0.124 | 0.119 | 0.005 | 5.93e-01 |
| RheuArth | LDpred1.Inf | LDpred1.MultiPRS | 0.136 | 0.145 | -0.009 | 1.93e-01 | 0.111 | 0.119 | -0.008 | 1.11e-02 |
| RheuArth | lassosum.MultiPRS | LDpred1.MultiPRS | 0.183 | 0.145 | 0.037 | 7.83e-08 | 0.150 | 0.119 | 0.031 | 7.91e-07 |
| RheuArth | PRScs.MultiPRS | LDpred1.MultiPRS | 0.166 | 0.145 | 0.020 | 4.04e-15 | 0.133 | 0.119 | 0.014 | 6.35e-03 |
| RheuArth | lassosum.10FCVal | LDpred1.MultiPRS | 0.162 | 0.145 | 0.016 | 1.94e-02 | 0.130 | 0.119 | 0.011 | 1.02e-01 |
| RheuArth | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.091 | 0.145 | -0.054 | 5.95e-15 | 0.077 | 0.119 | -0.042 | 7.86e-08 |
| RheuArth | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.142 | 0.145 | -0.003 | 6.47e-01 | 0.111 | 0.119 | -0.008 | 2.79e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.151 | 0.145 | 0.005 | 1.40e-01 | 0.127 | 0.119 | 0.008 | 2.68e-01 |
| RheuArth | SBLUP.Inf | LDpred1.MultiPRS | 0.112 | 0.145 | -0.033 | 1.54e-06 | 0.097 | 0.119 | -0.022 | 5.58e-04 |
| RheuArth | All.MultiPRS | LDpred1.Inf | 0.185 | 0.136 | 0.048 | 2.35e-12 | 0.161 | 0.111 | 0.050 | 8.32e-14 |
| RheuArth | LDpred2.PseudoVal | LDpred1.Inf | 0.091 | 0.136 | -0.045 | 1.23e-38 | 0.077 | 0.111 | -0.034 | 7.15e-07 |
| RheuArth | SBayesR.PseudoVal | LDpred1.Inf | 0.124 | 0.136 | -0.012 | 5.16e-03 | 0.119 | 0.111 | 0.008 | 3.59e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred1.Inf | 0.151 | 0.136 | 0.014 | 3.74e-02 | 0.127 | 0.111 | 0.016 | 3.76e-02 |
| RheuArth | PRScs.PseudoVal | LDpred1.Inf | 0.162 | 0.136 | 0.026 | 1.28e-14 | 0.129 | 0.111 | 0.018 | 8.33e-03 |
| RheuArth | LDpred1.Inf | LDpred1.Inf | 0.136 | 0.136 | 0.000 | 1.00e+00 | 0.111 | 0.111 | 0.000 | 1.00e+00 |
| RheuArth | pT+clump.10FCVal | LDpred1.Inf | 0.111 | 0.136 | -0.025 | 1.17e-05 | 0.103 | 0.111 | -0.008 | 5.06e-01 |
| RheuArth | PRScs.MultiPRS | LDpred1.Inf | 0.166 | 0.136 | 0.029 | 2.41e-05 | 0.133 | 0.111 | 0.022 | 1.22e-04 |
| RheuArth | lassosum.PseudoVal | LDpred1.Inf | 0.114 | 0.136 | -0.022 | 2.15e-08 | 0.089 | 0.111 | -0.022 | 5.69e-03 |
| RheuArth | pT+clump.MultiPRS | LDpred1.Inf | 0.131 | 0.136 | -0.005 | 4.56e-01 | 0.124 | 0.111 | 0.013 | 1.63e-01 |
| RheuArth | LDpred1.MultiPRS | LDpred1.Inf | 0.145 | 0.136 | 0.009 | 1.93e-01 | 0.119 | 0.111 | 0.008 | 1.11e-02 |
| RheuArth | PRScs.10FCVal | LDpred1.Inf | 0.161 | 0.136 | 0.025 | 1.17e-12 | 0.128 | 0.111 | 0.017 | 1.74e-02 |
| RheuArth | LDpred2.Inf | LDpred1.Inf | 0.085 | 0.136 | -0.051 | 6.20e-46 | 0.072 | 0.111 | -0.039 | 6.42e-08 |
| RheuArth | SBLUP.Inf | LDpred1.Inf | 0.112 | 0.136 | -0.024 | 9.79e-20 | 0.097 | 0.111 | -0.014 | 6.78e-03 |
| RheuArth | DBSLMM.PseudoVal | LDpred1.Inf | 0.142 | 0.136 | 0.006 | 1.56e-01 | 0.111 | 0.111 | 0.000 | 9.75e-01 |
| RheuArth | lassosum.MultiPRS | LDpred1.Inf | 0.183 | 0.136 | 0.046 | 3.03e-11 | 0.150 | 0.111 | 0.039 | 9.27e-09 |
| RheuArth | LDpred1.10FCVal | LDpred1.Inf | 0.136 | 0.136 | 0.000 | 9.77e-01 | 0.112 | 0.111 | 0.001 | 7.53e-01 |
| RheuArth | lassosum.10FCVal | LDpred1.Inf | 0.162 | 0.136 | 0.025 | 1.26e-12 | 0.130 | 0.111 | 0.019 | 9.06e-03 |
| RheuArth | LDpred2.10FCVal | LDpred1.Inf | 0.151 | 0.136 | 0.015 | 2.94e-04 | 0.128 | 0.111 | 0.017 | 3.88e-02 |
| RheuArth | LDpred1.Inf | LDpred1.10FCVal | 0.136 | 0.136 | 0.000 | 9.77e-01 | 0.111 | 0.112 | -0.001 | 7.53e-01 |
| RheuArth | PRScs.MultiPRS | LDpred1.10FCVal | 0.166 | 0.136 | 0.029 | 2.37e-05 | 0.133 | 0.112 | 0.022 | 2.28e-04 |
| RheuArth | LDpred1.10FCVal | LDpred1.10FCVal | 0.136 | 0.136 | 0.000 | 1.00e+00 | 0.112 | 0.112 | 0.000 | 1.00e+00 |
| RheuArth | All.MultiPRS | LDpred1.10FCVal | 0.185 | 0.136 | 0.048 | 2.27e-12 | 0.161 | 0.112 | 0.050 | 1.88e-13 |
| RheuArth | SBayesR.PseudoVal | LDpred1.10FCVal | 0.124 | 0.136 | -0.012 | 5.12e-03 | 0.119 | 0.112 | 0.007 | 3.91e-01 |
| RheuArth | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.142 | 0.136 | 0.006 | 1.60e-01 | 0.111 | 0.112 | -0.001 | 9.21e-01 |
| RheuArth | pT+clump.MultiPRS | LDpred1.10FCVal | 0.131 | 0.136 | -0.005 | 4.58e-01 | 0.124 | 0.112 | 0.012 | 1.81e-01 |
| RheuArth | lassosum.MultiPRS | LDpred1.10FCVal | 0.183 | 0.136 | 0.046 | 2.97e-11 | 0.150 | 0.112 | 0.039 | 1.72e-08 |
| RheuArth | PRScs.10FCVal | LDpred1.10FCVal | 0.161 | 0.136 | 0.025 | 2.93e-12 | 0.128 | 0.112 | 0.016 | 2.36e-02 |
| RheuArth | lassosum.10FCVal | LDpred1.10FCVal | 0.162 | 0.136 | 0.025 | 3.16e-12 | 0.130 | 0.112 | 0.018 | 1.29e-02 |
| RheuArth | SBLUP.Inf | LDpred1.10FCVal | 0.112 | 0.136 | -0.024 | 6.80e-20 | 0.097 | 0.112 | -0.015 | 4.75e-03 |
| RheuArth | LDpred2.PseudoVal | LDpred1.10FCVal | 0.091 | 0.136 | -0.045 | 2.89e-39 | 0.077 | 0.112 | -0.035 | 4.04e-07 |
| RheuArth | pT+clump.10FCVal | LDpred1.10FCVal | 0.111 | 0.136 | -0.025 | 1.25e-05 | 0.103 | 0.112 | -0.008 | 4.74e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred1.10FCVal | 0.151 | 0.136 | 0.014 | 3.71e-02 | 0.127 | 0.112 | 0.016 | 4.62e-02 |
| RheuArth | PRScs.PseudoVal | LDpred1.10FCVal | 0.162 | 0.136 | 0.026 | 4.09e-14 | 0.129 | 0.112 | 0.017 | 1.20e-02 |
| RheuArth | LDpred2.10FCVal | LDpred1.10FCVal | 0.151 | 0.136 | 0.015 | 3.09e-04 | 0.128 | 0.112 | 0.017 | 4.70e-02 |
| RheuArth | LDpred1.MultiPRS | LDpred1.10FCVal | 0.145 | 0.136 | 0.009 | 1.92e-01 | 0.119 | 0.112 | 0.007 | 1.54e-02 |
| RheuArth | LDpred2.Inf | LDpred1.10FCVal | 0.085 | 0.136 | -0.051 | 1.35e-46 | 0.072 | 0.112 | -0.040 | 3.55e-08 |
| RheuArth | lassosum.PseudoVal | LDpred1.10FCVal | 0.114 | 0.136 | -0.022 | 1.67e-08 | 0.089 | 0.112 | -0.022 | 4.24e-03 |
| RheuArth | LDpred1.Inf | LDpred2.MultiPRS | 0.136 | 0.151 | -0.014 | 3.74e-02 | 0.111 | 0.127 | -0.016 | 3.76e-02 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.142 | 0.151 | -0.009 | 2.16e-01 | 0.111 | 0.127 | -0.017 | 3.37e-02 |
| RheuArth | SBLUP.Inf | LDpred2.MultiPRS | 0.112 | 0.151 | -0.039 | 2.51e-08 | 0.097 | 0.127 | -0.031 | 3.36e-04 |
| RheuArth | LDpred2.Inf | LDpred2.MultiPRS | 0.085 | 0.151 | -0.066 | 4.35e-21 | 0.072 | 0.127 | -0.055 | 8.96e-10 |
| RheuArth | pT+clump.10FCVal | LDpred2.MultiPRS | 0.111 | 0.151 | -0.040 | 1.42e-08 | 0.103 | 0.127 | -0.024 | 4.90e-02 |
| RheuArth | LDpred2.10FCVal | LDpred2.MultiPRS | 0.151 | 0.151 | 0.000 | 9.59e-01 | 0.128 | 0.127 | 0.001 | 5.19e-01 |
| RheuArth | PRScs.PseudoVal | LDpred2.MultiPRS | 0.162 | 0.151 | 0.012 | 9.50e-02 | 0.129 | 0.127 | 0.002 | 8.07e-01 |
| RheuArth | LDpred1.10FCVal | LDpred2.MultiPRS | 0.136 | 0.151 | -0.014 | 3.71e-02 | 0.112 | 0.127 | -0.016 | 4.62e-02 |
| RheuArth | All.MultiPRS | LDpred2.MultiPRS | 0.185 | 0.151 | 0.034 | 1.71e-32 | 0.161 | 0.127 | 0.034 | 3.84e-09 |
| RheuArth | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.124 | 0.151 | -0.027 | 1.31e-04 | 0.119 | 0.127 | -0.008 | 4.55e-01 |
| RheuArth | lassosum.10FCVal | LDpred2.MultiPRS | 0.162 | 0.151 | 0.011 | 1.20e-01 | 0.130 | 0.127 | 0.002 | 7.25e-01 |
| RheuArth | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.131 | 0.151 | -0.020 | 4.78e-03 | 0.124 | 0.127 | -0.003 | 7.59e-01 |
| RheuArth | lassosum.MultiPRS | LDpred2.MultiPRS | 0.183 | 0.151 | 0.032 | 4.42e-06 | 0.150 | 0.127 | 0.023 | 1.09e-03 |
| RheuArth | PRScs.10FCVal | LDpred2.MultiPRS | 0.161 | 0.151 | 0.010 | 1.36e-01 | 0.128 | 0.127 | 0.000 | 9.45e-01 |
| RheuArth | PRScs.MultiPRS | LDpred2.MultiPRS | 0.166 | 0.151 | 0.015 | 2.59e-06 | 0.133 | 0.127 | 0.006 | 3.67e-01 |
| RheuArth | lassosum.PseudoVal | LDpred2.MultiPRS | 0.114 | 0.151 | -0.036 | 1.66e-07 | 0.089 | 0.127 | -0.038 | 7.30e-05 |
| RheuArth | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.091 | 0.151 | -0.060 | 1.11e-17 | 0.077 | 0.127 | -0.051 | 3.13e-09 |
| RheuArth | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.145 | 0.151 | -0.005 | 1.40e-01 | 0.119 | 0.127 | -0.008 | 2.68e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.151 | 0.151 | 0.000 | 1.00e+00 | 0.127 | 0.127 | 0.000 | 1.00e+00 |
| RheuArth | LDpred1.Inf | LDpred2.10FCVal | 0.136 | 0.151 | -0.015 | 2.94e-04 | 0.111 | 0.128 | -0.017 | 3.88e-02 |
| RheuArth | PRScs.MultiPRS | LDpred2.10FCVal | 0.166 | 0.151 | 0.014 | 3.70e-02 | 0.133 | 0.128 | 0.005 | 4.62e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred2.10FCVal | 0.151 | 0.151 | 0.000 | 9.59e-01 | 0.127 | 0.128 | -0.001 | 5.19e-01 |
| RheuArth | PRScs.PseudoVal | LDpred2.10FCVal | 0.162 | 0.151 | 0.011 | 5.79e-04 | 0.129 | 0.128 | 0.001 | 9.20e-01 |
| RheuArth | LDpred2.10FCVal | LDpred2.10FCVal | 0.151 | 0.151 | 0.000 | 1.00e+00 | 0.128 | 0.128 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.MultiPRS | LDpred2.10FCVal | 0.183 | 0.151 | 0.031 | 5.78e-06 | 0.150 | 0.128 | 0.022 | 2.24e-03 |
| RheuArth | LDpred1.10FCVal | LDpred2.10FCVal | 0.136 | 0.151 | -0.015 | 3.09e-04 | 0.112 | 0.128 | -0.017 | 4.70e-02 |
| RheuArth | All.MultiPRS | LDpred2.10FCVal | 0.185 | 0.151 | 0.034 | 1.08e-06 | 0.161 | 0.128 | 0.033 | 4.06e-08 |
| RheuArth | SBayesR.PseudoVal | LDpred2.10FCVal | 0.124 | 0.151 | -0.027 | 2.27e-06 | 0.119 | 0.128 | -0.009 | 4.23e-01 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.142 | 0.151 | -0.009 | 1.78e-02 | 0.111 | 0.128 | -0.018 | 2.01e-02 |
| RheuArth | pT+clump.MultiPRS | LDpred2.10FCVal | 0.131 | 0.151 | -0.020 | 4.19e-03 | 0.124 | 0.128 | -0.004 | 6.98e-01 |
| RheuArth | lassosum.PseudoVal | LDpred2.10FCVal | 0.114 | 0.151 | -0.037 | 8.81e-14 | 0.089 | 0.128 | -0.039 | 9.95e-05 |
| RheuArth | PRScs.10FCVal | LDpred2.10FCVal | 0.161 | 0.151 | 0.010 | 2.16e-03 | 0.128 | 0.128 | -0.001 | 9.37e-01 |
| RheuArth | lassosum.10FCVal | LDpred2.10FCVal | 0.162 | 0.151 | 0.010 | 2.81e-03 | 0.130 | 0.128 | 0.002 | 8.33e-01 |
| RheuArth | SBLUP.Inf | LDpred2.10FCVal | 0.112 | 0.151 | -0.039 | 3.17e-17 | 0.097 | 0.128 | -0.032 | 7.23e-04 |
| RheuArth | LDpred2.PseudoVal | LDpred2.10FCVal | 0.091 | 0.151 | -0.060 | 6.14e-38 | 0.077 | 0.128 | -0.052 | 4.18e-08 |
| RheuArth | pT+clump.10FCVal | LDpred2.10FCVal | 0.111 | 0.151 | -0.040 | 3.66e-11 | 0.103 | 0.128 | -0.025 | 4.03e-02 |
| RheuArth | LDpred2.Inf | LDpred2.10FCVal | 0.085 | 0.151 | -0.066 | 6.68e-42 | 0.072 | 0.128 | -0.056 | 1.22e-08 |
| RheuArth | LDpred1.MultiPRS | LDpred2.10FCVal | 0.145 | 0.151 | -0.006 | 4.04e-01 | 0.119 | 0.128 | -0.009 | 2.39e-01 |
| RheuArth | lassosum.10FCVal | LDpred2.PseudoVal | 0.162 | 0.091 | 0.070 | 9.12e-41 | 0.130 | 0.077 | 0.053 | 5.63e-07 |
| RheuArth | SBLUP.Inf | LDpred2.PseudoVal | 0.112 | 0.091 | 0.021 | 1.47e-46 | 0.097 | 0.077 | 0.020 | 5.03e-12 |
| RheuArth | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.124 | 0.091 | 0.033 | 2.30e-14 | 0.119 | 0.077 | 0.042 | 1.03e-06 |
| RheuArth | All.MultiPRS | LDpred2.PseudoVal | 0.185 | 0.091 | 0.094 | 1.39e-41 | 0.161 | 0.077 | 0.085 | 3.01e-19 |
| RheuArth | PRScs.PseudoVal | LDpred2.PseudoVal | 0.162 | 0.091 | 0.071 | 1.45e-41 | 0.129 | 0.077 | 0.052 | 7.39e-07 |
| RheuArth | lassosum.MultiPRS | LDpred2.PseudoVal | 0.183 | 0.091 | 0.091 | 2.73e-39 | 0.150 | 0.077 | 0.074 | 3.15e-14 |
| RheuArth | PRScs.MultiPRS | LDpred2.PseudoVal | 0.166 | 0.091 | 0.074 | 7.81e-27 | 0.133 | 0.077 | 0.057 | 1.96e-09 |
| RheuArth | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.151 | 0.091 | 0.060 | 1.11e-17 | 0.127 | 0.077 | 0.051 | 3.13e-09 |
| RheuArth | PRScs.10FCVal | LDpred2.PseudoVal | 0.161 | 0.091 | 0.070 | 2.78e-39 | 0.128 | 0.077 | 0.051 | 1.81e-06 |
| RheuArth | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.091 | 0.091 | 0.000 | 1.00e+00 | 0.077 | 0.077 | 0.000 | 1.00e+00 |
| RheuArth | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.131 | 0.091 | 0.040 | 1.21e-08 | 0.124 | 0.077 | 0.047 | 6.01e-06 |
| RheuArth | pT+clump.10FCVal | LDpred2.PseudoVal | 0.111 | 0.091 | 0.020 | 2.26e-03 | 0.103 | 0.077 | 0.027 | 4.23e-02 |
| RheuArth | lassosum.PseudoVal | LDpred2.PseudoVal | 0.114 | 0.091 | 0.023 | 3.18e-08 | 0.089 | 0.077 | 0.013 | 1.32e-01 |
| RheuArth | LDpred1.Inf | LDpred2.PseudoVal | 0.136 | 0.091 | 0.045 | 1.23e-38 | 0.111 | 0.077 | 0.034 | 7.15e-07 |
| RheuArth | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.145 | 0.091 | 0.054 | 5.95e-15 | 0.119 | 0.077 | 0.042 | 7.86e-08 |
| RheuArth | LDpred1.10FCVal | LDpred2.PseudoVal | 0.136 | 0.091 | 0.045 | 2.89e-39 | 0.112 | 0.077 | 0.035 | 4.04e-07 |
| RheuArth | LDpred2.Inf | LDpred2.PseudoVal | 0.085 | 0.091 | -0.006 | 4.76e-70 | 0.072 | 0.077 | -0.004 | 1.35e-10 |
| RheuArth | LDpred2.10FCVal | LDpred2.PseudoVal | 0.151 | 0.091 | 0.060 | 6.14e-38 | 0.128 | 0.077 | 0.052 | 4.18e-08 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.142 | 0.091 | 0.051 | 5.15e-19 | 0.111 | 0.077 | 0.034 | 2.97e-03 |
| RheuArth | pT+clump.MultiPRS | LDpred2.Inf | 0.131 | 0.085 | 0.046 | 5.56e-11 | 0.124 | 0.072 | 0.052 | 9.22e-07 |
| RheuArth | lassosum.MultiPRS | LDpred2.Inf | 0.183 | 0.085 | 0.098 | 2.27e-44 | 0.150 | 0.072 | 0.078 | 4.72e-15 |
| RheuArth | SBayesR.PseudoVal | LDpred2.Inf | 0.124 | 0.085 | 0.039 | 1.46e-19 | 0.119 | 0.072 | 0.047 | 5.94e-08 |
| RheuArth | All.MultiPRS | LDpred2.Inf | 0.185 | 0.085 | 0.100 | 7.60e-47 | 0.161 | 0.072 | 0.089 | 7.28e-20 |
| RheuArth | SBLUP.Inf | LDpred2.Inf | 0.112 | 0.085 | 0.027 | 1.54e-68 | 0.097 | 0.072 | 0.024 | 1.44e-15 |
| RheuArth | LDpred2.PseudoVal | LDpred2.Inf | 0.091 | 0.085 | 0.006 | 4.76e-70 | 0.077 | 0.072 | 0.004 | 1.35e-10 |
| RheuArth | PRScs.MultiPRS | LDpred2.Inf | 0.166 | 0.085 | 0.080 | 4.66e-31 | 0.133 | 0.072 | 0.061 | 4.06e-10 |
| RheuArth | LDpred2.MultiPRS | LDpred2.Inf | 0.151 | 0.085 | 0.066 | 4.35e-21 | 0.127 | 0.072 | 0.055 | 8.96e-10 |
| RheuArth | PRScs.10FCVal | LDpred2.Inf | 0.161 | 0.085 | 0.076 | 1.05e-43 | 0.128 | 0.072 | 0.056 | 4.44e-07 |
| RheuArth | LDpred1.Inf | LDpred2.Inf | 0.136 | 0.085 | 0.051 | 6.20e-46 | 0.111 | 0.072 | 0.039 | 6.42e-08 |
| RheuArth | LDpred1.MultiPRS | LDpred2.Inf | 0.145 | 0.085 | 0.060 | 4.34e-18 | 0.119 | 0.072 | 0.047 | 9.38e-09 |
| RheuArth | pT+clump.10FCVal | LDpred2.Inf | 0.111 | 0.085 | 0.026 | 7.88e-05 | 0.103 | 0.072 | 0.031 | 1.83e-02 |
| RheuArth | lassosum.PseudoVal | LDpred2.Inf | 0.114 | 0.085 | 0.029 | 1.23e-11 | 0.089 | 0.072 | 0.017 | 4.76e-02 |
| RheuArth | PRScs.PseudoVal | LDpred2.Inf | 0.162 | 0.085 | 0.077 | 4.19e-46 | 0.129 | 0.072 | 0.057 | 1.74e-07 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.Inf | 0.142 | 0.085 | 0.057 | 2.08e-22 | 0.111 | 0.072 | 0.039 | 1.03e-03 |
| RheuArth | LDpred1.10FCVal | LDpred2.Inf | 0.136 | 0.085 | 0.051 | 1.35e-46 | 0.112 | 0.072 | 0.040 | 3.55e-08 |
| RheuArth | LDpred2.Inf | LDpred2.Inf | 0.085 | 0.085 | 0.000 | 1.00e+00 | 0.072 | 0.072 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.10FCVal | LDpred2.Inf | 0.162 | 0.085 | 0.076 | 2.73e-45 | 0.130 | 0.072 | 0.058 | 1.31e-07 |
| RheuArth | LDpred2.10FCVal | LDpred2.Inf | 0.151 | 0.085 | 0.066 | 6.68e-42 | 0.128 | 0.072 | 0.056 | 1.22e-08 |
| RheuArth | PRScs.10FCVal | PRScs.MultiPRS | 0.161 | 0.166 | -0.004 | 5.20e-01 | 0.128 | 0.133 | -0.005 | 2.00e-02 |
| RheuArth | LDpred1.10FCVal | PRScs.MultiPRS | 0.136 | 0.166 | -0.029 | 2.37e-05 | 0.112 | 0.133 | -0.022 | 2.28e-04 |
| RheuArth | lassosum.MultiPRS | PRScs.MultiPRS | 0.183 | 0.166 | 0.017 | 1.37e-02 | 0.150 | 0.133 | 0.017 | 1.07e-05 |
| RheuArth | LDpred2.MultiPRS | PRScs.MultiPRS | 0.151 | 0.166 | -0.015 | 2.59e-06 | 0.127 | 0.133 | -0.006 | 3.67e-01 |
| RheuArth | All.MultiPRS | PRScs.MultiPRS | 0.185 | 0.166 | 0.019 | 1.02e-21 | 0.161 | 0.133 | 0.028 | 7.09e-12 |
| RheuArth | PRScs.PseudoVal | PRScs.MultiPRS | 0.162 | 0.166 | -0.003 | 6.41e-01 | 0.129 | 0.133 | -0.004 | 4.66e-02 |
| RheuArth | LDpred1.MultiPRS | PRScs.MultiPRS | 0.145 | 0.166 | -0.020 | 4.04e-15 | 0.119 | 0.133 | -0.014 | 6.35e-03 |
| RheuArth | pT+clump.MultiPRS | PRScs.MultiPRS | 0.131 | 0.166 | -0.034 | 7.69e-07 | 0.124 | 0.133 | -0.009 | 3.36e-01 |
| RheuArth | lassosum.PseudoVal | PRScs.MultiPRS | 0.114 | 0.166 | -0.051 | 1.80e-13 | 0.089 | 0.133 | -0.044 | 3.08e-08 |
| RheuArth | SBayesR.PseudoVal | PRScs.MultiPRS | 0.124 | 0.166 | -0.041 | 2.42e-09 | 0.119 | 0.133 | -0.014 | 1.57e-01 |
| RheuArth | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.142 | 0.166 | -0.023 | 7.53e-04 | 0.111 | 0.133 | -0.022 | 6.67e-06 |
| RheuArth | SBLUP.Inf | PRScs.MultiPRS | 0.112 | 0.166 | -0.054 | 1.12e-14 | 0.097 | 0.133 | -0.037 | 6.73e-06 |
| RheuArth | LDpred2.Inf | PRScs.MultiPRS | 0.085 | 0.166 | -0.080 | 4.66e-31 | 0.072 | 0.133 | -0.061 | 4.06e-10 |
| RheuArth | pT+clump.10FCVal | PRScs.MultiPRS | 0.111 | 0.166 | -0.054 | 6.47e-15 | 0.103 | 0.133 | -0.030 | 6.80e-03 |
| RheuArth | LDpred2.10FCVal | PRScs.MultiPRS | 0.151 | 0.166 | -0.014 | 3.70e-02 | 0.128 | 0.133 | -0.005 | 4.62e-01 |
| RheuArth | lassosum.10FCVal | PRScs.MultiPRS | 0.162 | 0.166 | -0.004 | 5.61e-01 | 0.130 | 0.133 | -0.003 | 3.74e-01 |
| RheuArth | LDpred2.PseudoVal | PRScs.MultiPRS | 0.091 | 0.166 | -0.074 | 7.81e-27 | 0.077 | 0.133 | -0.057 | 1.96e-09 |
| RheuArth | LDpred1.Inf | PRScs.MultiPRS | 0.136 | 0.166 | -0.029 | 2.41e-05 | 0.111 | 0.133 | -0.022 | 1.22e-04 |
| RheuArth | PRScs.MultiPRS | PRScs.MultiPRS | 0.166 | 0.166 | 0.000 | 1.00e+00 | 0.133 | 0.133 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.PseudoVal | PRScs.10FCVal | 0.091 | 0.161 | -0.070 | 2.78e-39 | 0.077 | 0.128 | -0.051 | 1.81e-06 |
| RheuArth | PRScs.10FCVal | PRScs.10FCVal | 0.161 | 0.161 | 0.000 | 1.00e+00 | 0.128 | 0.128 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.MultiPRS | PRScs.10FCVal | 0.151 | 0.161 | -0.010 | 1.36e-01 | 0.127 | 0.128 | 0.000 | 9.45e-01 |
| RheuArth | pT+clump.MultiPRS | PRScs.10FCVal | 0.131 | 0.161 | -0.030 | 1.69e-05 | 0.124 | 0.128 | -0.004 | 7.08e-01 |
| RheuArth | LDpred1.Inf | PRScs.10FCVal | 0.136 | 0.161 | -0.025 | 1.17e-12 | 0.111 | 0.128 | -0.017 | 1.74e-02 |
| RheuArth | LDpred1.MultiPRS | PRScs.10FCVal | 0.145 | 0.161 | -0.016 | 2.30e-02 | 0.119 | 0.128 | -0.009 | 1.62e-01 |
| RheuArth | pT+clump.10FCVal | PRScs.10FCVal | 0.111 | 0.161 | -0.050 | 1.59e-20 | 0.103 | 0.128 | -0.024 | 2.54e-02 |
| RheuArth | lassosum.PseudoVal | PRScs.10FCVal | 0.114 | 0.161 | -0.047 | 2.29e-23 | 0.089 | 0.128 | -0.038 | 4.46e-05 |
| RheuArth | PRScs.PseudoVal | PRScs.10FCVal | 0.162 | 0.161 | 0.001 | 9.33e-02 | 0.129 | 0.128 | 0.001 | 4.34e-01 |
| RheuArth | DBSLMM.PseudoVal | PRScs.10FCVal | 0.142 | 0.161 | -0.019 | 6.05e-19 | 0.111 | 0.128 | -0.017 | 8.66e-05 |
| RheuArth | SBLUP.Inf | PRScs.10FCVal | 0.112 | 0.161 | -0.049 | 7.77e-25 | 0.097 | 0.128 | -0.031 | 1.14e-03 |
| RheuArth | LDpred2.Inf | PRScs.10FCVal | 0.085 | 0.161 | -0.076 | 1.05e-43 | 0.072 | 0.128 | -0.056 | 4.44e-07 |
| RheuArth | lassosum.10FCVal | PRScs.10FCVal | 0.162 | 0.161 | 0.000 | 8.14e-01 | 0.130 | 0.128 | 0.002 | 5.84e-01 |
| RheuArth | LDpred2.10FCVal | PRScs.10FCVal | 0.151 | 0.161 | -0.010 | 2.16e-03 | 0.128 | 0.128 | 0.001 | 9.37e-01 |
| RheuArth | lassosum.MultiPRS | PRScs.10FCVal | 0.183 | 0.161 | 0.022 | 1.94e-03 | 0.150 | 0.128 | 0.022 | 4.05e-07 |
| RheuArth | LDpred1.10FCVal | PRScs.10FCVal | 0.136 | 0.161 | -0.025 | 2.93e-12 | 0.112 | 0.128 | -0.016 | 2.36e-02 |
| RheuArth | All.MultiPRS | PRScs.10FCVal | 0.185 | 0.161 | 0.024 | 6.00e-04 | 0.161 | 0.128 | 0.034 | 9.61e-13 |
| RheuArth | SBayesR.PseudoVal | PRScs.10FCVal | 0.124 | 0.161 | -0.037 | 2.74e-12 | 0.119 | 0.128 | -0.009 | 4.16e-01 |
| RheuArth | PRScs.MultiPRS | PRScs.10FCVal | 0.166 | 0.161 | 0.004 | 5.20e-01 | 0.133 | 0.128 | 0.005 | 2.00e-02 |
| RheuArth | All.MultiPRS | PRScs.PseudoVal | 0.185 | 0.162 | 0.022 | 1.13e-03 | 0.161 | 0.129 | 0.032 | 1.52e-12 |
| RheuArth | pT+clump.MultiPRS | PRScs.PseudoVal | 0.131 | 0.162 | -0.031 | 7.57e-06 | 0.124 | 0.129 | -0.005 | 6.19e-01 |
| RheuArth | LDpred2.PseudoVal | PRScs.PseudoVal | 0.091 | 0.162 | -0.071 | 1.45e-41 | 0.077 | 0.129 | -0.052 | 7.39e-07 |
| RheuArth | SBayesR.PseudoVal | PRScs.PseudoVal | 0.124 | 0.162 | -0.038 | 3.26e-13 | 0.119 | 0.129 | -0.010 | 3.50e-01 |
| RheuArth | LDpred2.MultiPRS | PRScs.PseudoVal | 0.151 | 0.162 | -0.012 | 9.50e-02 | 0.127 | 0.129 | -0.002 | 8.07e-01 |
| RheuArth | SBLUP.Inf | PRScs.PseudoVal | 0.112 | 0.162 | -0.050 | 9.11e-27 | 0.097 | 0.129 | -0.032 | 5.86e-04 |
| RheuArth | LDpred1.Inf | PRScs.PseudoVal | 0.136 | 0.162 | -0.026 | 1.28e-14 | 0.111 | 0.129 | -0.018 | 8.33e-03 |
| RheuArth | PRScs.10FCVal | PRScs.PseudoVal | 0.161 | 0.162 | -0.001 | 9.33e-02 | 0.128 | 0.129 | -0.001 | 4.34e-01 |
| RheuArth | LDpred2.Inf | PRScs.PseudoVal | 0.085 | 0.162 | -0.077 | 4.19e-46 | 0.072 | 0.129 | -0.057 | 1.74e-07 |
| RheuArth | PRScs.PseudoVal | PRScs.PseudoVal | 0.162 | 0.162 | 0.000 | 1.00e+00 | 0.129 | 0.129 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.10FCVal | PRScs.PseudoVal | 0.151 | 0.162 | -0.011 | 5.79e-04 | 0.128 | 0.129 | -0.001 | 9.20e-01 |
| RheuArth | pT+clump.10FCVal | PRScs.PseudoVal | 0.111 | 0.162 | -0.051 | 1.73e-21 | 0.103 | 0.129 | -0.026 | 1.89e-02 |
| RheuArth | PRScs.MultiPRS | PRScs.PseudoVal | 0.166 | 0.162 | 0.003 | 6.41e-01 | 0.133 | 0.129 | 0.004 | 4.66e-02 |
| RheuArth | lassosum.PseudoVal | PRScs.PseudoVal | 0.114 | 0.162 | -0.048 | 4.55e-25 | 0.089 | 0.129 | -0.040 | 1.94e-05 |
| RheuArth | lassosum.10FCVal | PRScs.PseudoVal | 0.162 | 0.162 | -0.001 | 6.61e-01 | 0.130 | 0.129 | 0.001 | 8.17e-01 |
| RheuArth | LDpred1.MultiPRS | PRScs.PseudoVal | 0.145 | 0.162 | -0.017 | 1.43e-02 | 0.119 | 0.129 | -0.010 | 9.98e-02 |
| RheuArth | lassosum.MultiPRS | PRScs.PseudoVal | 0.183 | 0.162 | 0.020 | 3.47e-03 | 0.150 | 0.129 | 0.021 | 7.80e-07 |
| RheuArth | LDpred1.10FCVal | PRScs.PseudoVal | 0.136 | 0.162 | -0.026 | 4.09e-14 | 0.112 | 0.129 | -0.017 | 1.20e-02 |
| RheuArth | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.142 | 0.162 | -0.020 | 2.04e-21 | 0.111 | 0.129 | -0.018 | 2.45e-05 |
| RheuArth | All.MultiPRS | pT+clump.MultiPRS | 0.185 | 0.131 | 0.054 | 1.01e-14 | 0.161 | 0.124 | 0.037 | 2.25e-07 |
| RheuArth | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.124 | 0.131 | -0.007 | 3.21e-01 | 0.119 | 0.124 | -0.005 | 4.74e-01 |
| RheuArth | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.145 | 0.131 | 0.014 | 4.16e-02 | 0.119 | 0.124 | -0.005 | 5.93e-01 |
| RheuArth | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.131 | 0.131 | 0.000 | 1.00e+00 | 0.124 | 0.124 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.PseudoVal | pT+clump.MultiPRS | 0.114 | 0.131 | -0.017 | 1.61e-02 | 0.089 | 0.124 | -0.035 | 4.18e-04 |
| RheuArth | PRScs.10FCVal | pT+clump.MultiPRS | 0.161 | 0.131 | 0.030 | 1.69e-05 | 0.128 | 0.124 | 0.004 | 7.08e-01 |
| RheuArth | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.142 | 0.131 | 0.011 | 1.13e-01 | 0.111 | 0.124 | -0.013 | 2.00e-01 |
| RheuArth | SBLUP.Inf | pT+clump.MultiPRS | 0.112 | 0.131 | -0.019 | 6.28e-03 | 0.097 | 0.124 | -0.027 | 3.57e-03 |
| RheuArth | LDpred2.Inf | pT+clump.MultiPRS | 0.085 | 0.131 | -0.046 | 5.56e-11 | 0.072 | 0.124 | -0.052 | 9.22e-07 |
| RheuArth | pT+clump.10FCVal | pT+clump.MultiPRS | 0.111 | 0.131 | -0.020 | 4.36e-03 | 0.103 | 0.124 | -0.021 | 2.56e-04 |
| RheuArth | LDpred2.10FCVal | pT+clump.MultiPRS | 0.151 | 0.131 | 0.020 | 4.19e-03 | 0.128 | 0.124 | 0.004 | 6.98e-01 |
| RheuArth | PRScs.PseudoVal | pT+clump.MultiPRS | 0.162 | 0.131 | 0.031 | 7.57e-06 | 0.129 | 0.124 | 0.005 | 6.19e-01 |
| RheuArth | LDpred1.10FCVal | pT+clump.MultiPRS | 0.136 | 0.131 | 0.005 | 4.58e-01 | 0.112 | 0.124 | -0.012 | 1.81e-01 |
| RheuArth | LDpred1.Inf | pT+clump.MultiPRS | 0.136 | 0.131 | 0.005 | 4.56e-01 | 0.111 | 0.124 | -0.013 | 1.63e-01 |
| RheuArth | PRScs.MultiPRS | pT+clump.MultiPRS | 0.166 | 0.131 | 0.034 | 7.69e-07 | 0.133 | 0.124 | 0.009 | 3.36e-01 |
| RheuArth | lassosum.10FCVal | pT+clump.MultiPRS | 0.162 | 0.131 | 0.030 | 1.25e-05 | 0.130 | 0.124 | 0.006 | 5.22e-01 |
| RheuArth | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.091 | 0.131 | -0.040 | 1.21e-08 | 0.077 | 0.124 | -0.047 | 6.01e-06 |
| RheuArth | lassosum.MultiPRS | pT+clump.MultiPRS | 0.183 | 0.131 | 0.051 | 1.21e-13 | 0.150 | 0.124 | 0.026 | 1.26e-03 |
| RheuArth | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.151 | 0.131 | 0.020 | 4.78e-03 | 0.127 | 0.124 | 0.003 | 7.59e-01 |
| RheuArth | LDpred2.10FCVal | pT+clump.10FCVal | 0.151 | 0.111 | 0.040 | 3.66e-11 | 0.128 | 0.103 | 0.025 | 4.03e-02 |
| RheuArth | pT+clump.MultiPRS | pT+clump.10FCVal | 0.131 | 0.111 | 0.020 | 4.36e-03 | 0.124 | 0.103 | 0.021 | 2.56e-04 |
| RheuArth | LDpred2.Inf | pT+clump.10FCVal | 0.085 | 0.111 | -0.026 | 7.88e-05 | 0.072 | 0.103 | -0.031 | 1.83e-02 |
| RheuArth | SBayesR.PseudoVal | pT+clump.10FCVal | 0.124 | 0.111 | 0.013 | 4.47e-03 | 0.119 | 0.103 | 0.016 | 8.06e-02 |
| RheuArth | PRScs.MultiPRS | pT+clump.10FCVal | 0.166 | 0.111 | 0.054 | 6.47e-15 | 0.133 | 0.103 | 0.030 | 6.80e-03 |
| RheuArth | SBLUP.Inf | pT+clump.10FCVal | 0.112 | 0.111 | 0.001 | 8.97e-01 | 0.097 | 0.103 | -0.007 | 5.84e-01 |
| RheuArth | lassosum.MultiPRS | pT+clump.10FCVal | 0.183 | 0.111 | 0.071 | 7.40e-25 | 0.150 | 0.103 | 0.047 | 1.39e-06 |
| RheuArth | PRScs.10FCVal | pT+clump.10FCVal | 0.161 | 0.111 | 0.050 | 1.59e-20 | 0.128 | 0.103 | 0.024 | 2.54e-02 |
| RheuArth | LDpred2.MultiPRS | pT+clump.10FCVal | 0.151 | 0.111 | 0.040 | 1.42e-08 | 0.127 | 0.103 | 0.024 | 4.90e-02 |
| RheuArth | lassosum.10FCVal | pT+clump.10FCVal | 0.162 | 0.111 | 0.050 | 3.22e-22 | 0.130 | 0.103 | 0.027 | 1.18e-02 |
| RheuArth | LDpred2.PseudoVal | pT+clump.10FCVal | 0.091 | 0.111 | -0.020 | 2.26e-03 | 0.077 | 0.103 | -0.027 | 4.23e-02 |
| RheuArth | LDpred1.MultiPRS | pT+clump.10FCVal | 0.145 | 0.111 | 0.034 | 1.01e-06 | 0.119 | 0.103 | 0.016 | 1.69e-01 |
| RheuArth | pT+clump.10FCVal | pT+clump.10FCVal | 0.111 | 0.111 | 0.000 | 1.00e+00 | 0.103 | 0.103 | 0.000 | 1.00e+00 |
| RheuArth | All.MultiPRS | pT+clump.10FCVal | 0.185 | 0.111 | 0.074 | 4.10e-26 | 0.161 | 0.103 | 0.058 | 4.53e-11 |
| RheuArth | LDpred1.Inf | pT+clump.10FCVal | 0.136 | 0.111 | 0.025 | 1.17e-05 | 0.111 | 0.103 | 0.008 | 5.06e-01 |
| RheuArth | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.142 | 0.111 | 0.031 | 6.56e-09 | 0.111 | 0.103 | 0.007 | 4.95e-01 |
| RheuArth | LDpred1.10FCVal | pT+clump.10FCVal | 0.136 | 0.111 | 0.025 | 1.25e-05 | 0.112 | 0.103 | 0.008 | 4.74e-01 |
| RheuArth | lassosum.PseudoVal | pT+clump.10FCVal | 0.114 | 0.111 | 0.003 | 6.14e-01 | 0.089 | 0.103 | -0.014 | 2.56e-01 |
| RheuArth | PRScs.PseudoVal | pT+clump.10FCVal | 0.162 | 0.111 | 0.051 | 1.73e-21 | 0.129 | 0.103 | 0.026 | 1.89e-02 |
| RheuArth | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.131 | 0.124 | 0.007 | 3.21e-01 | 0.124 | 0.119 | 0.005 | 4.74e-01 |
| RheuArth | LDpred1.10FCVal | SBayesR.PseudoVal | 0.136 | 0.124 | 0.012 | 5.12e-03 | 0.112 | 0.119 | -0.007 | 3.91e-01 |
| RheuArth | PRScs.10FCVal | SBayesR.PseudoVal | 0.161 | 0.124 | 0.037 | 2.74e-12 | 0.128 | 0.119 | 0.009 | 4.16e-01 |
| RheuArth | LDpred2.10FCVal | SBayesR.PseudoVal | 0.151 | 0.124 | 0.027 | 2.27e-06 | 0.128 | 0.119 | 0.009 | 4.23e-01 |
| RheuArth | lassosum.10FCVal | SBayesR.PseudoVal | 0.162 | 0.124 | 0.037 | 1.17e-13 | 0.130 | 0.119 | 0.011 | 2.92e-01 |
| RheuArth | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.091 | 0.124 | -0.033 | 2.30e-14 | 0.077 | 0.119 | -0.042 | 1.03e-06 |
| RheuArth | lassosum.MultiPRS | SBayesR.PseudoVal | 0.183 | 0.124 | 0.058 | 4.17e-17 | 0.150 | 0.119 | 0.031 | 4.64e-04 |
| RheuArth | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.151 | 0.124 | 0.027 | 1.31e-04 | 0.127 | 0.119 | 0.008 | 4.55e-01 |
| RheuArth | All.MultiPRS | SBayesR.PseudoVal | 0.185 | 0.124 | 0.061 | 2.02e-18 | 0.161 | 0.119 | 0.042 | 2.05e-07 |
| RheuArth | PRScs.PseudoVal | SBayesR.PseudoVal | 0.162 | 0.124 | 0.038 | 3.26e-13 | 0.129 | 0.119 | 0.010 | 3.50e-01 |
| RheuArth | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.145 | 0.124 | 0.021 | 2.31e-03 | 0.119 | 0.119 | 0.000 | 9.95e-01 |
| RheuArth | pT+clump.10FCVal | SBayesR.PseudoVal | 0.111 | 0.124 | -0.013 | 4.47e-03 | 0.103 | 0.119 | -0.016 | 8.06e-02 |
| RheuArth | lassosum.PseudoVal | SBayesR.PseudoVal | 0.114 | 0.124 | -0.010 | 4.28e-02 | 0.089 | 0.119 | -0.030 | 2.34e-03 |
| RheuArth | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.124 | 0.124 | 0.000 | 1.00e+00 | 0.119 | 0.119 | 0.000 | 1.00e+00 |
| RheuArth | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.142 | 0.124 | 0.018 | 7.22e-04 | 0.111 | 0.119 | -0.008 | 4.44e-01 |
| RheuArth | SBLUP.Inf | SBayesR.PseudoVal | 0.112 | 0.124 | -0.012 | 1.36e-03 | 0.097 | 0.119 | -0.022 | 3.49e-03 |
| RheuArth | LDpred2.Inf | SBayesR.PseudoVal | 0.085 | 0.124 | -0.039 | 1.46e-19 | 0.072 | 0.119 | -0.047 | 5.94e-08 |
| RheuArth | LDpred1.Inf | SBayesR.PseudoVal | 0.136 | 0.124 | 0.012 | 5.16e-03 | 0.111 | 0.119 | -0.008 | 3.59e-01 |
| RheuArth | PRScs.MultiPRS | SBayesR.PseudoVal | 0.166 | 0.124 | 0.041 | 2.42e-09 | 0.133 | 0.119 | 0.014 | 1.57e-01 |
| RheuArth | LDpred2.10FCVal | SBLUP.Inf | 0.151 | 0.112 | 0.039 | 3.17e-17 | 0.128 | 0.097 | 0.032 | 7.23e-04 |
| RheuArth | PRScs.PseudoVal | SBLUP.Inf | 0.162 | 0.112 | 0.050 | 9.11e-27 | 0.129 | 0.097 | 0.032 | 5.86e-04 |
| RheuArth | LDpred1.10FCVal | SBLUP.Inf | 0.136 | 0.112 | 0.024 | 6.80e-20 | 0.112 | 0.097 | 0.015 | 4.75e-03 |
| RheuArth | LDpred2.Inf | SBLUP.Inf | 0.085 | 0.112 | -0.027 | 1.54e-68 | 0.072 | 0.097 | -0.024 | 1.44e-15 |
| RheuArth | SBayesR.PseudoVal | SBLUP.Inf | 0.124 | 0.112 | 0.012 | 1.36e-03 | 0.119 | 0.097 | 0.022 | 3.49e-03 |
| RheuArth | lassosum.10FCVal | SBLUP.Inf | 0.162 | 0.112 | 0.050 | 2.47e-26 | 0.130 | 0.097 | 0.033 | 4.26e-04 |
| RheuArth | pT+clump.MultiPRS | SBLUP.Inf | 0.131 | 0.112 | 0.019 | 6.28e-03 | 0.124 | 0.097 | 0.027 | 3.57e-03 |
| RheuArth | lassosum.MultiPRS | SBLUP.Inf | 0.183 | 0.112 | 0.071 | 3.93e-24 | 0.150 | 0.097 | 0.054 | 2.26e-10 |
| RheuArth | PRScs.10FCVal | SBLUP.Inf | 0.161 | 0.112 | 0.049 | 7.77e-25 | 0.128 | 0.097 | 0.031 | 1.14e-03 |
| RheuArth | PRScs.MultiPRS | SBLUP.Inf | 0.166 | 0.112 | 0.054 | 1.12e-14 | 0.133 | 0.097 | 0.037 | 6.73e-06 |
| RheuArth | LDpred1.MultiPRS | SBLUP.Inf | 0.145 | 0.112 | 0.033 | 1.54e-06 | 0.119 | 0.097 | 0.022 | 5.58e-04 |
| RheuArth | LDpred2.PseudoVal | SBLUP.Inf | 0.091 | 0.112 | -0.021 | 1.47e-46 | 0.077 | 0.097 | -0.020 | 5.03e-12 |
| RheuArth | LDpred1.Inf | SBLUP.Inf | 0.136 | 0.112 | 0.024 | 9.79e-20 | 0.111 | 0.097 | 0.014 | 6.78e-03 |
| RheuArth | LDpred2.MultiPRS | SBLUP.Inf | 0.151 | 0.112 | 0.039 | 2.51e-08 | 0.127 | 0.097 | 0.031 | 3.36e-04 |
| RheuArth | All.MultiPRS | SBLUP.Inf | 0.185 | 0.112 | 0.073 | 7.80e-26 | 0.161 | 0.097 | 0.065 | 8.79e-15 |
| RheuArth | SBLUP.Inf | SBLUP.Inf | 0.112 | 0.112 | 0.000 | 1.00e+00 | 0.097 | 0.097 | 0.000 | 1.00e+00 |
| RheuArth | DBSLMM.PseudoVal | SBLUP.Inf | 0.142 | 0.112 | 0.030 | 8.95e-09 | 0.111 | 0.097 | 0.014 | 1.80e-01 |
| RheuArth | pT+clump.10FCVal | SBLUP.Inf | 0.111 | 0.112 | -0.001 | 8.97e-01 | 0.103 | 0.097 | 0.007 | 5.84e-01 |
| RheuArth | lassosum.PseudoVal | SBLUP.Inf | 0.114 | 0.112 | 0.002 | 5.39e-01 | 0.089 | 0.097 | -0.007 | 3.37e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | All.MultiPRS | 0.139 | 0.186 | -0.047 | 7.98e-12 | 0.133 | 0.181 | -0.048 | 1.76e-10 |
| Prostate_Cancer | pT+clump.10FCVal | All.MultiPRS | 0.152 | 0.186 | -0.034 | 8.40e-07 | 0.166 | 0.181 | -0.015 | 2.88e-02 |
| Prostate_Cancer | LDpred2.MultiPRS | All.MultiPRS | 0.183 | 0.186 | -0.003 | 6.19e-01 | 0.175 | 0.181 | -0.006 | 9.08e-02 |
| Prostate_Cancer | PRScs.PseudoVal | All.MultiPRS | 0.183 | 0.186 | -0.004 | 5.80e-01 | 0.172 | 0.181 | -0.009 | 1.53e-02 |
| Prostate_Cancer | LDpred1.Inf | All.MultiPRS | 0.143 | 0.186 | -0.043 | 3.80e-10 | 0.133 | 0.181 | -0.048 | 5.21e-11 |
| Prostate_Cancer | PRScs.MultiPRS | All.MultiPRS | 0.184 | 0.186 | -0.003 | 7.14e-01 | 0.173 | 0.181 | -0.008 | 2.74e-02 |
| Prostate_Cancer | LDpred2.Inf | All.MultiPRS | 0.135 | 0.186 | -0.051 | 1.64e-13 | 0.129 | 0.181 | -0.052 | 2.66e-11 |
| Prostate_Cancer | lassosum.10FCVal | All.MultiPRS | 0.180 | 0.186 | -0.006 | 3.57e-01 | 0.177 | 0.181 | -0.004 | 3.70e-01 |
| Prostate_Cancer | LDpred2.10FCVal | All.MultiPRS | 0.183 | 0.186 | -0.003 | 6.39e-01 | 0.173 | 0.181 | -0.008 | 3.74e-02 |
| Prostate_Cancer | lassosum.MultiPRS | All.MultiPRS | 0.187 | 0.186 | 0.001 | 9.28e-01 | 0.181 | 0.181 | 0.000 | 9.65e-01 |
| Prostate_Cancer | LDpred1.10FCVal | All.MultiPRS | 0.165 | 0.186 | -0.021 | 2.24e-03 | 0.152 | 0.181 | -0.029 | 2.30e-07 |
| Prostate_Cancer | All.MultiPRS | All.MultiPRS | 0.186 | 0.186 | 0.000 | 1.00e+00 | 0.181 | 0.181 | 0.000 | 1.00e+00 |
| Prostate_Cancer | SBayesR.PseudoVal | All.MultiPRS | 0.176 | 0.186 | -0.010 | 1.48e-01 | 0.171 | 0.181 | -0.010 | 2.71e-02 |
| Prostate_Cancer | LDpred1.MultiPRS | All.MultiPRS | 0.164 | 0.186 | -0.022 | 1.53e-03 | 0.149 | 0.181 | -0.032 | 2.66e-08 |
| Prostate_Cancer | pT+clump.MultiPRS | All.MultiPRS | 0.168 | 0.186 | -0.018 | 8.51e-03 | 0.175 | 0.181 | -0.007 | 2.34e-01 |
| Prostate_Cancer | lassosum.PseudoVal | All.MultiPRS | 0.168 | 0.186 | -0.018 | 8.99e-03 | 0.164 | 0.181 | -0.017 | 1.72e-03 |
| Prostate_Cancer | PRScs.10FCVal | All.MultiPRS | 0.181 | 0.186 | -0.006 | 4.20e-01 | 0.176 | 0.181 | -0.005 | 2.50e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | All.MultiPRS | 0.176 | 0.186 | -0.010 | 1.49e-01 | 0.166 | 0.181 | -0.015 | 1.07e-03 |
| Prostate_Cancer | SBLUP.Inf | All.MultiPRS | 0.137 | 0.186 | -0.050 | 8.19e-13 | 0.128 | 0.181 | -0.053 | 1.01e-11 |
| Prostate_Cancer | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.176 | 0.176 | 0.000 | 9.91e-01 | 0.171 | 0.166 | 0.005 | 3.65e-01 |
| Prostate_Cancer | LDpred2.Inf | DBSLMM.PseudoVal | 0.135 | 0.176 | -0.041 | 3.72e-34 | 0.129 | 0.166 | -0.037 | 4.57e-08 |
| Prostate_Cancer | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.152 | 0.176 | -0.024 | 7.26e-11 | 0.166 | 0.166 | 0.001 | 9.45e-01 |
| Prostate_Cancer | PRScs.10FCVal | DBSLMM.PseudoVal | 0.181 | 0.176 | 0.004 | 1.62e-02 | 0.176 | 0.166 | 0.011 | 3.19e-03 |
| Prostate_Cancer | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.183 | 0.176 | 0.006 | 5.43e-04 | 0.172 | 0.166 | 0.006 | 8.06e-02 |
| Prostate_Cancer | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.168 | 0.176 | -0.008 | 2.33e-01 | 0.175 | 0.166 | 0.009 | 1.02e-01 |
| Prostate_Cancer | LDpred1.Inf | DBSLMM.PseudoVal | 0.143 | 0.176 | -0.033 | 3.58e-26 | 0.133 | 0.166 | -0.033 | 1.87e-07 |
| Prostate_Cancer | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.187 | 0.176 | 0.011 | 1.26e-01 | 0.181 | 0.166 | 0.016 | 1.88e-04 |
| Prostate_Cancer | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.183 | 0.176 | 0.007 | 1.39e-03 | 0.173 | 0.166 | 0.008 | 7.03e-02 |
| Prostate_Cancer | lassosum.10FCVal | DBSLMM.PseudoVal | 0.180 | 0.176 | 0.004 | 1.64e-01 | 0.177 | 0.166 | 0.012 | 2.45e-02 |
| Prostate_Cancer | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.165 | 0.176 | -0.011 | 7.93e-06 | 0.152 | 0.166 | -0.014 | 4.95e-03 |
| Prostate_Cancer | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.164 | 0.176 | -0.012 | 8.52e-02 | 0.149 | 0.166 | -0.016 | 8.68e-04 |
| Prostate_Cancer | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.184 | 0.176 | 0.007 | 2.84e-01 | 0.173 | 0.166 | 0.007 | 2.83e-02 |
| Prostate_Cancer | All.MultiPRS | DBSLMM.PseudoVal | 0.186 | 0.176 | 0.010 | 1.49e-01 | 0.181 | 0.166 | 0.015 | 1.07e-03 |
| Prostate_Cancer | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.139 | 0.176 | -0.037 | 1.19e-30 | 0.133 | 0.166 | -0.033 | 3.58e-07 |
| Prostate_Cancer | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.176 | 0.176 | 0.000 | 1.00e+00 | 0.166 | 0.166 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.183 | 0.176 | 0.007 | 3.46e-01 | 0.175 | 0.166 | 0.009 | 2.19e-02 |
| Prostate_Cancer | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.168 | 0.176 | -0.008 | 1.21e-02 | 0.164 | 0.166 | -0.002 | 7.63e-01 |
| Prostate_Cancer | SBLUP.Inf | DBSLMM.PseudoVal | 0.137 | 0.176 | -0.040 | 6.96e-33 | 0.128 | 0.166 | -0.038 | 1.26e-08 |
| Prostate_Cancer | SBayesR.PseudoVal | lassosum.MultiPRS | 0.176 | 0.187 | -0.011 | 1.26e-01 | 0.171 | 0.181 | -0.010 | 5.96e-02 |
| Prostate_Cancer | LDpred2.10FCVal | lassosum.MultiPRS | 0.183 | 0.187 | -0.004 | 5.77e-01 | 0.173 | 0.181 | -0.008 | 1.60e-02 |
| Prostate_Cancer | LDpred1.10FCVal | lassosum.MultiPRS | 0.165 | 0.187 | -0.022 | 1.73e-03 | 0.152 | 0.181 | -0.029 | 1.16e-08 |
| Prostate_Cancer | LDpred2.Inf | lassosum.MultiPRS | 0.135 | 0.187 | -0.052 | 9.67e-14 | 0.129 | 0.181 | -0.052 | 2.18e-12 |
| Prostate_Cancer | PRScs.10FCVal | lassosum.MultiPRS | 0.181 | 0.187 | -0.006 | 3.72e-01 | 0.176 | 0.181 | -0.005 | 1.58e-01 |
| Prostate_Cancer | PRScs.MultiPRS | lassosum.MultiPRS | 0.184 | 0.187 | -0.003 | 4.26e-02 | 0.173 | 0.181 | -0.008 | 6.54e-03 |
| Prostate_Cancer | pT+clump.MultiPRS | lassosum.MultiPRS | 0.168 | 0.187 | -0.019 | 6.46e-03 | 0.175 | 0.181 | -0.007 | 1.86e-01 |
| Prostate_Cancer | lassosum.MultiPRS | lassosum.MultiPRS | 0.187 | 0.187 | 0.000 | 1.00e+00 | 0.181 | 0.181 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.PseudoVal | lassosum.MultiPRS | 0.168 | 0.187 | -0.019 | 7.06e-03 | 0.164 | 0.181 | -0.018 | 1.66e-04 |
| Prostate_Cancer | LDpred2.MultiPRS | lassosum.MultiPRS | 0.183 | 0.187 | -0.004 | 5.56e-01 | 0.175 | 0.181 | -0.007 | 4.75e-02 |
| Prostate_Cancer | LDpred1.MultiPRS | lassosum.MultiPRS | 0.164 | 0.187 | -0.023 | 7.43e-18 | 0.149 | 0.181 | -0.032 | 8.09e-10 |
| Prostate_Cancer | LDpred2.PseudoVal | lassosum.MultiPRS | 0.139 | 0.187 | -0.048 | 4.82e-12 | 0.133 | 0.181 | -0.048 | 1.32e-11 |
| Prostate_Cancer | LDpred1.Inf | lassosum.MultiPRS | 0.143 | 0.187 | -0.044 | 2.42e-10 | 0.133 | 0.181 | -0.048 | 2.52e-12 |
| Prostate_Cancer | pT+clump.10FCVal | lassosum.MultiPRS | 0.152 | 0.187 | -0.035 | 5.96e-07 | 0.166 | 0.181 | -0.015 | 2.26e-02 |
| Prostate_Cancer | lassosum.10FCVal | lassosum.MultiPRS | 0.180 | 0.187 | -0.007 | 3.14e-01 | 0.177 | 0.181 | -0.004 | 9.85e-02 |
| Prostate_Cancer | SBLUP.Inf | lassosum.MultiPRS | 0.137 | 0.187 | -0.050 | 4.67e-13 | 0.128 | 0.181 | -0.053 | 6.18e-13 |
| Prostate_Cancer | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.176 | 0.187 | -0.011 | 1.26e-01 | 0.166 | 0.181 | -0.016 | 1.88e-04 |
| Prostate_Cancer | All.MultiPRS | lassosum.MultiPRS | 0.186 | 0.187 | -0.001 | 9.28e-01 | 0.181 | 0.181 | 0.000 | 9.65e-01 |
| Prostate_Cancer | PRScs.PseudoVal | lassosum.MultiPRS | 0.183 | 0.187 | -0.004 | 5.21e-01 | 0.172 | 0.181 | -0.009 | 4.79e-03 |
| Prostate_Cancer | LDpred1.Inf | lassosum.PseudoVal | 0.143 | 0.168 | -0.025 | 5.16e-07 | 0.133 | 0.164 | -0.031 | 2.09e-03 |
| Prostate_Cancer | PRScs.10FCVal | lassosum.PseudoVal | 0.181 | 0.168 | 0.013 | 1.55e-06 | 0.176 | 0.164 | 0.013 | 1.27e-02 |
| Prostate_Cancer | LDpred2.MultiPRS | lassosum.PseudoVal | 0.183 | 0.168 | 0.015 | 3.46e-02 | 0.175 | 0.164 | 0.011 | 3.85e-02 |
| Prostate_Cancer | lassosum.PseudoVal | lassosum.PseudoVal | 0.168 | 0.168 | 0.000 | 1.00e+00 | 0.164 | 0.164 | 0.000 | 1.00e+00 |
| Prostate_Cancer | PRScs.PseudoVal | lassosum.PseudoVal | 0.183 | 0.168 | 0.014 | 1.14e-07 | 0.172 | 0.164 | 0.008 | 1.26e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.176 | 0.168 | 0.008 | 1.21e-02 | 0.166 | 0.164 | 0.002 | 7.63e-01 |
| Prostate_Cancer | SBLUP.Inf | lassosum.PseudoVal | 0.137 | 0.168 | -0.031 | 4.95e-09 | 0.128 | 0.164 | -0.036 | 8.48e-04 |
| Prostate_Cancer | LDpred2.Inf | lassosum.PseudoVal | 0.135 | 0.168 | -0.033 | 8.14e-10 | 0.129 | 0.164 | -0.035 | 1.11e-03 |
| Prostate_Cancer | pT+clump.10FCVal | lassosum.PseudoVal | 0.152 | 0.168 | -0.016 | 9.05e-08 | 0.166 | 0.164 | 0.002 | 6.78e-01 |
| Prostate_Cancer | LDpred2.10FCVal | lassosum.PseudoVal | 0.183 | 0.168 | 0.015 | 3.31e-09 | 0.173 | 0.164 | 0.010 | 5.28e-02 |
| Prostate_Cancer | lassosum.MultiPRS | lassosum.PseudoVal | 0.187 | 0.168 | 0.019 | 7.06e-03 | 0.181 | 0.164 | 0.018 | 1.66e-04 |
| Prostate_Cancer | LDpred1.10FCVal | lassosum.PseudoVal | 0.165 | 0.168 | -0.003 | 4.37e-01 | 0.152 | 0.164 | -0.012 | 1.26e-01 |
| Prostate_Cancer | All.MultiPRS | lassosum.PseudoVal | 0.186 | 0.168 | 0.018 | 8.99e-03 | 0.181 | 0.164 | 0.017 | 1.72e-03 |
| Prostate_Cancer | SBayesR.PseudoVal | lassosum.PseudoVal | 0.176 | 0.168 | 0.008 | 3.42e-04 | 0.171 | 0.164 | 0.007 | 9.86e-02 |
| Prostate_Cancer | lassosum.10FCVal | lassosum.PseudoVal | 0.180 | 0.168 | 0.012 | 3.97e-07 | 0.177 | 0.164 | 0.014 | 3.45e-03 |
| Prostate_Cancer | pT+clump.MultiPRS | lassosum.PseudoVal | 0.168 | 0.168 | 0.000 | 9.88e-01 | 0.175 | 0.164 | 0.011 | 9.03e-02 |
| Prostate_Cancer | LDpred2.PseudoVal | lassosum.PseudoVal | 0.139 | 0.168 | -0.029 | 2.15e-08 | 0.133 | 0.164 | -0.031 | 2.83e-03 |
| Prostate_Cancer | LDpred1.MultiPRS | lassosum.PseudoVal | 0.164 | 0.168 | -0.004 | 5.82e-01 | 0.149 | 0.164 | -0.014 | 6.69e-02 |
| Prostate_Cancer | PRScs.MultiPRS | lassosum.PseudoVal | 0.184 | 0.168 | 0.016 | 2.52e-02 | 0.173 | 0.164 | 0.009 | 7.34e-02 |
| Prostate_Cancer | lassosum.10FCVal | lassosum.10FCVal | 0.180 | 0.180 | 0.000 | 1.00e+00 | 0.177 | 0.177 | 0.000 | 1.00e+00 |
| Prostate_Cancer | pT+clump.MultiPRS | lassosum.10FCVal | 0.168 | 0.180 | -0.012 | 8.64e-02 | 0.175 | 0.177 | -0.003 | 6.21e-01 |
| Prostate_Cancer | LDpred2.Inf | lassosum.10FCVal | 0.135 | 0.180 | -0.045 | 1.07e-25 | 0.129 | 0.177 | -0.048 | 8.90e-09 |
| Prostate_Cancer | SBayesR.PseudoVal | lassosum.10FCVal | 0.176 | 0.180 | -0.004 | 1.96e-01 | 0.171 | 0.177 | -0.006 | 2.58e-01 |
| Prostate_Cancer | PRScs.MultiPRS | lassosum.10FCVal | 0.184 | 0.180 | 0.004 | 5.82e-01 | 0.173 | 0.177 | -0.004 | 2.73e-01 |
| Prostate_Cancer | SBLUP.Inf | lassosum.10FCVal | 0.137 | 0.180 | -0.043 | 3.00e-24 | 0.128 | 0.177 | -0.049 | 4.50e-09 |
| Prostate_Cancer | lassosum.MultiPRS | lassosum.10FCVal | 0.187 | 0.180 | 0.007 | 3.14e-01 | 0.181 | 0.177 | 0.004 | 9.85e-02 |
| Prostate_Cancer | PRScs.10FCVal | lassosum.10FCVal | 0.181 | 0.180 | 0.001 | 6.91e-01 | 0.176 | 0.177 | -0.001 | 8.38e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | lassosum.10FCVal | 0.183 | 0.180 | 0.003 | 6.74e-01 | 0.175 | 0.177 | -0.003 | 4.95e-01 |
| Prostate_Cancer | PRScs.PseudoVal | lassosum.10FCVal | 0.183 | 0.180 | 0.003 | 2.05e-01 | 0.172 | 0.177 | -0.005 | 1.70e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | lassosum.10FCVal | 0.139 | 0.180 | -0.041 | 2.85e-23 | 0.133 | 0.177 | -0.044 | 4.07e-08 |
| Prostate_Cancer | LDpred1.MultiPRS | lassosum.10FCVal | 0.164 | 0.180 | -0.016 | 2.52e-02 | 0.149 | 0.177 | -0.028 | 3.52e-06 |
| Prostate_Cancer | pT+clump.10FCVal | lassosum.10FCVal | 0.152 | 0.180 | -0.028 | 9.78e-17 | 0.166 | 0.177 | -0.011 | 8.84e-02 |
| Prostate_Cancer | All.MultiPRS | lassosum.10FCVal | 0.186 | 0.180 | 0.006 | 3.57e-01 | 0.181 | 0.177 | 0.004 | 3.70e-01 |
| Prostate_Cancer | LDpred1.Inf | lassosum.10FCVal | 0.143 | 0.180 | -0.037 | 9.22e-21 | 0.133 | 0.177 | -0.044 | 1.48e-08 |
| Prostate_Cancer | DBSLMM.PseudoVal | lassosum.10FCVal | 0.176 | 0.180 | -0.004 | 1.64e-01 | 0.166 | 0.177 | -0.012 | 2.45e-02 |
| Prostate_Cancer | LDpred1.10FCVal | lassosum.10FCVal | 0.165 | 0.180 | -0.015 | 1.00e-06 | 0.152 | 0.177 | -0.025 | 2.16e-05 |
| Prostate_Cancer | lassosum.PseudoVal | lassosum.10FCVal | 0.168 | 0.180 | -0.012 | 3.97e-07 | 0.164 | 0.177 | -0.014 | 3.45e-03 |
| Prostate_Cancer | LDpred2.10FCVal | lassosum.10FCVal | 0.183 | 0.180 | 0.003 | 8.51e-02 | 0.173 | 0.177 | -0.004 | 2.60e-01 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred1.MultiPRS | 0.183 | 0.164 | 0.019 | 7.19e-03 | 0.173 | 0.149 | 0.024 | 2.11e-06 |
| Prostate_Cancer | PRScs.PseudoVal | LDpred1.MultiPRS | 0.183 | 0.164 | 0.018 | 9.17e-03 | 0.172 | 0.149 | 0.022 | 9.32e-07 |
| Prostate_Cancer | LDpred2.Inf | LDpred1.MultiPRS | 0.135 | 0.164 | -0.029 | 2.83e-05 | 0.129 | 0.149 | -0.020 | 3.15e-04 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred1.MultiPRS | 0.152 | 0.164 | -0.012 | 8.13e-02 | 0.166 | 0.149 | 0.017 | 5.06e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.176 | 0.164 | 0.012 | 8.65e-02 | 0.171 | 0.149 | 0.022 | 4.60e-03 |
| Prostate_Cancer | All.MultiPRS | LDpred1.MultiPRS | 0.186 | 0.164 | 0.022 | 1.53e-03 | 0.181 | 0.149 | 0.032 | 2.66e-08 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred1.MultiPRS | 0.165 | 0.164 | 0.001 | 9.09e-01 | 0.152 | 0.149 | 0.003 | 1.50e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.164 | 0.164 | 0.000 | 1.00e+00 | 0.149 | 0.149 | 0.000 | 1.00e+00 |
| Prostate_Cancer | PRScs.10FCVal | LDpred1.MultiPRS | 0.181 | 0.164 | 0.016 | 1.86e-02 | 0.176 | 0.149 | 0.027 | 1.05e-08 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred1.MultiPRS | 0.168 | 0.164 | 0.004 | 5.82e-01 | 0.164 | 0.149 | 0.014 | 6.69e-02 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.168 | 0.164 | 0.004 | 5.91e-01 | 0.175 | 0.149 | 0.025 | 1.19e-04 |
| Prostate_Cancer | LDpred1.Inf | LDpred1.MultiPRS | 0.143 | 0.164 | -0.021 | 2.09e-03 | 0.133 | 0.149 | -0.016 | 1.67e-04 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred1.MultiPRS | 0.187 | 0.164 | 0.023 | 7.43e-18 | 0.181 | 0.149 | 0.032 | 8.09e-10 |
| Prostate_Cancer | PRScs.MultiPRS | LDpred1.MultiPRS | 0.184 | 0.164 | 0.019 | 1.82e-17 | 0.173 | 0.149 | 0.023 | 1.75e-07 |
| Prostate_Cancer | lassosum.10FCVal | LDpred1.MultiPRS | 0.180 | 0.164 | 0.016 | 2.52e-02 | 0.177 | 0.149 | 0.028 | 3.52e-06 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.139 | 0.164 | -0.025 | 2.59e-04 | 0.133 | 0.149 | -0.016 | 2.07e-03 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.176 | 0.164 | 0.012 | 8.52e-02 | 0.166 | 0.149 | 0.016 | 8.68e-04 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.183 | 0.164 | 0.018 | 7.53e-03 | 0.175 | 0.149 | 0.025 | 4.97e-08 |
| Prostate_Cancer | SBLUP.Inf | LDpred1.MultiPRS | 0.137 | 0.164 | -0.028 | 7.05e-05 | 0.128 | 0.149 | -0.021 | 1.20e-04 |
| Prostate_Cancer | All.MultiPRS | LDpred1.Inf | 0.186 | 0.143 | 0.043 | 3.80e-10 | 0.181 | 0.133 | 0.048 | 5.21e-11 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred1.Inf | 0.139 | 0.143 | -0.004 | 7.93e-03 | 0.133 | 0.133 | 0.000 | 9.64e-01 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred1.Inf | 0.176 | 0.143 | 0.033 | 7.12e-12 | 0.171 | 0.133 | 0.038 | 7.33e-05 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred1.Inf | 0.183 | 0.143 | 0.040 | 8.54e-09 | 0.175 | 0.133 | 0.042 | 2.45e-10 |
| Prostate_Cancer | PRScs.PseudoVal | LDpred1.Inf | 0.183 | 0.143 | 0.039 | 1.16e-32 | 0.172 | 0.133 | 0.039 | 2.91e-09 |
| Prostate_Cancer | LDpred1.Inf | LDpred1.Inf | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.133 | 0.133 | 0.000 | 1.00e+00 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred1.Inf | 0.152 | 0.143 | 0.009 | 6.60e-02 | 0.166 | 0.133 | 0.033 | 8.73e-04 |
| Prostate_Cancer | PRScs.MultiPRS | LDpred1.Inf | 0.184 | 0.143 | 0.041 | 4.16e-09 | 0.173 | 0.133 | 0.040 | 1.23e-09 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred1.Inf | 0.168 | 0.143 | 0.025 | 5.16e-07 | 0.164 | 0.133 | 0.031 | 2.09e-03 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred1.Inf | 0.168 | 0.143 | 0.025 | 2.88e-04 | 0.175 | 0.133 | 0.042 | 7.25e-08 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred1.Inf | 0.164 | 0.143 | 0.021 | 2.09e-03 | 0.149 | 0.133 | 0.016 | 1.67e-04 |
| Prostate_Cancer | PRScs.10FCVal | LDpred1.Inf | 0.181 | 0.143 | 0.038 | 2.16e-28 | 0.176 | 0.133 | 0.043 | 1.26e-10 |
| Prostate_Cancer | LDpred2.Inf | LDpred1.Inf | 0.135 | 0.143 | -0.008 | 1.28e-06 | 0.129 | 0.133 | -0.004 | 2.10e-01 |
| Prostate_Cancer | SBLUP.Inf | LDpred1.Inf | 0.137 | 0.143 | -0.006 | 5.10e-05 | 0.128 | 0.133 | -0.005 | 1.15e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred1.Inf | 0.176 | 0.143 | 0.033 | 3.58e-26 | 0.166 | 0.133 | 0.033 | 1.87e-07 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred1.Inf | 0.187 | 0.143 | 0.044 | 2.42e-10 | 0.181 | 0.133 | 0.048 | 2.52e-12 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred1.Inf | 0.165 | 0.143 | 0.022 | 4.80e-22 | 0.152 | 0.133 | 0.019 | 4.30e-05 |
| Prostate_Cancer | lassosum.10FCVal | LDpred1.Inf | 0.180 | 0.143 | 0.037 | 9.22e-21 | 0.177 | 0.133 | 0.044 | 1.48e-08 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred1.Inf | 0.183 | 0.143 | 0.040 | 5.13e-29 | 0.173 | 0.133 | 0.040 | 1.43e-08 |
| Prostate_Cancer | LDpred1.Inf | LDpred1.10FCVal | 0.143 | 0.165 | -0.022 | 4.80e-22 | 0.133 | 0.152 | -0.019 | 4.30e-05 |
| Prostate_Cancer | PRScs.MultiPRS | LDpred1.10FCVal | 0.184 | 0.165 | 0.019 | 7.42e-03 | 0.173 | 0.152 | 0.021 | 1.77e-06 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred1.10FCVal | 0.165 | 0.165 | 0.000 | 1.00e+00 | 0.152 | 0.152 | 0.000 | 1.00e+00 |
| Prostate_Cancer | All.MultiPRS | LDpred1.10FCVal | 0.186 | 0.165 | 0.021 | 2.24e-03 | 0.181 | 0.152 | 0.029 | 2.30e-07 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred1.10FCVal | 0.176 | 0.165 | 0.011 | 3.98e-03 | 0.171 | 0.152 | 0.019 | 1.14e-02 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.176 | 0.165 | 0.011 | 7.93e-06 | 0.166 | 0.152 | 0.014 | 4.95e-03 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred1.10FCVal | 0.168 | 0.165 | 0.003 | 6.72e-01 | 0.175 | 0.152 | 0.023 | 5.77e-04 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred1.10FCVal | 0.187 | 0.165 | 0.022 | 1.73e-03 | 0.181 | 0.152 | 0.029 | 1.16e-08 |
| Prostate_Cancer | PRScs.10FCVal | LDpred1.10FCVal | 0.181 | 0.165 | 0.016 | 4.90e-11 | 0.176 | 0.152 | 0.024 | 1.06e-07 |
| Prostate_Cancer | lassosum.10FCVal | LDpred1.10FCVal | 0.180 | 0.165 | 0.015 | 1.00e-06 | 0.177 | 0.152 | 0.025 | 2.16e-05 |
| Prostate_Cancer | SBLUP.Inf | LDpred1.10FCVal | 0.137 | 0.165 | -0.028 | 1.14e-22 | 0.128 | 0.152 | -0.024 | 4.07e-05 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred1.10FCVal | 0.139 | 0.165 | -0.026 | 2.28e-20 | 0.133 | 0.152 | -0.019 | 7.06e-04 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred1.10FCVal | 0.152 | 0.165 | -0.013 | 2.97e-03 | 0.166 | 0.152 | 0.014 | 9.75e-02 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred1.10FCVal | 0.183 | 0.165 | 0.018 | 1.06e-02 | 0.175 | 0.152 | 0.023 | 6.83e-07 |
| Prostate_Cancer | PRScs.PseudoVal | LDpred1.10FCVal | 0.183 | 0.165 | 0.017 | 4.15e-14 | 0.172 | 0.152 | 0.020 | 8.30e-06 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred1.10FCVal | 0.183 | 0.165 | 0.018 | 1.00e-12 | 0.173 | 0.152 | 0.021 | 1.59e-05 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred1.10FCVal | 0.164 | 0.165 | -0.001 | 9.09e-01 | 0.149 | 0.152 | -0.003 | 1.50e-01 |
| Prostate_Cancer | LDpred2.Inf | LDpred1.10FCVal | 0.135 | 0.165 | -0.030 | 8.30e-24 | 0.129 | 0.152 | -0.023 | 1.11e-04 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred1.10FCVal | 0.168 | 0.165 | 0.003 | 4.37e-01 | 0.164 | 0.152 | 0.012 | 1.26e-01 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.MultiPRS | 0.143 | 0.183 | -0.040 | 8.54e-09 | 0.133 | 0.175 | -0.042 | 2.45e-10 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.176 | 0.183 | -0.007 | 3.46e-01 | 0.166 | 0.175 | -0.009 | 2.19e-02 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.MultiPRS | 0.137 | 0.183 | -0.046 | 2.81e-11 | 0.128 | 0.175 | -0.047 | 7.53e-11 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.MultiPRS | 0.135 | 0.183 | -0.048 | 6.33e-12 | 0.129 | 0.175 | -0.046 | 8.86e-11 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.MultiPRS | 0.152 | 0.183 | -0.031 | 1.01e-05 | 0.166 | 0.175 | -0.009 | 2.16e-01 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.MultiPRS | 0.183 | 0.183 | 0.000 | 9.76e-01 | 0.173 | 0.175 | -0.001 | 9.07e-02 |
| Prostate_Cancer | PRScs.PseudoVal | LDpred2.MultiPRS | 0.183 | 0.183 | 0.000 | 9.56e-01 | 0.172 | 0.175 | -0.003 | 2.51e-01 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.MultiPRS | 0.165 | 0.183 | -0.018 | 1.06e-02 | 0.152 | 0.175 | -0.023 | 6.83e-07 |
| Prostate_Cancer | All.MultiPRS | LDpred2.MultiPRS | 0.186 | 0.183 | 0.003 | 6.19e-01 | 0.181 | 0.175 | 0.006 | 9.08e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.176 | 0.183 | -0.007 | 3.43e-01 | 0.171 | 0.175 | -0.004 | 4.72e-01 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.MultiPRS | 0.180 | 0.183 | -0.003 | 6.74e-01 | 0.177 | 0.175 | 0.003 | 4.95e-01 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.168 | 0.183 | -0.015 | 3.25e-02 | 0.175 | 0.175 | 0.000 | 9.80e-01 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.MultiPRS | 0.187 | 0.183 | 0.004 | 5.56e-01 | 0.181 | 0.175 | 0.007 | 4.75e-02 |
| Prostate_Cancer | PRScs.10FCVal | LDpred2.MultiPRS | 0.181 | 0.183 | -0.002 | 7.58e-01 | 0.176 | 0.175 | 0.002 | 4.86e-01 |
| Prostate_Cancer | PRScs.MultiPRS | LDpred2.MultiPRS | 0.184 | 0.183 | 0.001 | 8.95e-01 | 0.173 | 0.175 | -0.002 | 4.40e-01 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.MultiPRS | 0.168 | 0.183 | -0.015 | 3.46e-02 | 0.164 | 0.175 | -0.011 | 3.85e-02 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.139 | 0.183 | -0.044 | 2.33e-10 | 0.133 | 0.175 | -0.042 | 4.58e-10 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.164 | 0.183 | -0.018 | 7.53e-03 | 0.149 | 0.175 | -0.025 | 4.97e-08 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.183 | 0.183 | 0.000 | 1.00e+00 | 0.175 | 0.175 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.10FCVal | 0.143 | 0.183 | -0.040 | 5.13e-29 | 0.133 | 0.173 | -0.040 | 1.43e-08 |
| Prostate_Cancer | PRScs.MultiPRS | LDpred2.10FCVal | 0.184 | 0.183 | 0.001 | 9.19e-01 | 0.173 | 0.173 | 0.000 | 8.89e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.10FCVal | 0.183 | 0.183 | 0.000 | 9.76e-01 | 0.175 | 0.173 | 0.001 | 9.07e-02 |
| Prostate_Cancer | PRScs.PseudoVal | LDpred2.10FCVal | 0.183 | 0.183 | -0.001 | 6.50e-01 | 0.172 | 0.173 | -0.001 | 5.88e-01 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.10FCVal | 0.183 | 0.183 | 0.000 | 1.00e+00 | 0.173 | 0.173 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.10FCVal | 0.187 | 0.183 | 0.004 | 5.77e-01 | 0.181 | 0.173 | 0.008 | 1.60e-02 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.10FCVal | 0.165 | 0.183 | -0.018 | 1.00e-12 | 0.152 | 0.173 | -0.021 | 1.59e-05 |
| Prostate_Cancer | All.MultiPRS | LDpred2.10FCVal | 0.186 | 0.183 | 0.003 | 6.39e-01 | 0.181 | 0.173 | 0.008 | 3.74e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.10FCVal | 0.176 | 0.183 | -0.007 | 7.85e-03 | 0.171 | 0.173 | -0.002 | 6.45e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.176 | 0.183 | -0.007 | 1.39e-03 | 0.166 | 0.173 | -0.008 | 7.03e-02 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.10FCVal | 0.168 | 0.183 | -0.015 | 3.02e-02 | 0.175 | 0.173 | 0.001 | 8.03e-01 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.10FCVal | 0.168 | 0.183 | -0.015 | 3.31e-09 | 0.164 | 0.173 | -0.010 | 5.28e-02 |
| Prostate_Cancer | PRScs.10FCVal | LDpred2.10FCVal | 0.181 | 0.183 | -0.002 | 6.78e-02 | 0.176 | 0.173 | 0.003 | 1.97e-01 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.10FCVal | 0.180 | 0.183 | -0.003 | 8.51e-02 | 0.177 | 0.173 | 0.004 | 2.60e-01 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.10FCVal | 0.137 | 0.183 | -0.046 | 1.04e-32 | 0.128 | 0.173 | -0.045 | 5.06e-09 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.10FCVal | 0.139 | 0.183 | -0.044 | 4.76e-33 | 0.133 | 0.173 | -0.040 | 2.79e-08 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.10FCVal | 0.152 | 0.183 | -0.031 | 9.77e-19 | 0.166 | 0.173 | -0.007 | 2.94e-01 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.10FCVal | 0.135 | 0.183 | -0.048 | 1.63e-35 | 0.129 | 0.173 | -0.044 | 6.30e-09 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.10FCVal | 0.164 | 0.183 | -0.019 | 7.19e-03 | 0.149 | 0.173 | -0.024 | 2.11e-06 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.PseudoVal | 0.180 | 0.139 | 0.041 | 2.85e-23 | 0.177 | 0.133 | 0.044 | 4.07e-08 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.PseudoVal | 0.137 | 0.139 | -0.002 | 1.05e-02 | 0.128 | 0.133 | -0.005 | 6.91e-03 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.176 | 0.139 | 0.037 | 5.49e-14 | 0.171 | 0.133 | 0.038 | 9.49e-05 |
| Prostate_Cancer | All.MultiPRS | LDpred2.PseudoVal | 0.186 | 0.139 | 0.047 | 7.98e-12 | 0.181 | 0.133 | 0.048 | 1.76e-10 |
| Prostate_Cancer | PRScs.PseudoVal | LDpred2.PseudoVal | 0.183 | 0.139 | 0.044 | 1.20e-35 | 0.172 | 0.133 | 0.039 | 1.26e-08 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.PseudoVal | 0.187 | 0.139 | 0.048 | 4.82e-12 | 0.181 | 0.133 | 0.048 | 1.32e-11 |
| Prostate_Cancer | PRScs.MultiPRS | LDpred2.PseudoVal | 0.184 | 0.139 | 0.045 | 1.08e-10 | 0.173 | 0.133 | 0.040 | 6.15e-09 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.183 | 0.139 | 0.044 | 2.33e-10 | 0.175 | 0.133 | 0.042 | 4.58e-10 |
| Prostate_Cancer | PRScs.10FCVal | LDpred2.PseudoVal | 0.181 | 0.139 | 0.042 | 1.82e-31 | 0.176 | 0.133 | 0.044 | 6.22e-10 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.139 | 0.139 | 0.000 | 1.00e+00 | 0.133 | 0.133 | 0.000 | 1.00e+00 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.168 | 0.139 | 0.029 | 2.64e-05 | 0.175 | 0.133 | 0.042 | 1.48e-07 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.PseudoVal | 0.152 | 0.139 | 0.013 | 1.03e-02 | 0.166 | 0.133 | 0.033 | 1.11e-03 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.PseudoVal | 0.168 | 0.139 | 0.029 | 2.15e-08 | 0.164 | 0.133 | 0.031 | 2.83e-03 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.PseudoVal | 0.143 | 0.139 | 0.004 | 7.93e-03 | 0.133 | 0.133 | 0.000 | 9.64e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.164 | 0.139 | 0.025 | 2.59e-04 | 0.149 | 0.133 | 0.016 | 2.07e-03 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.PseudoVal | 0.165 | 0.139 | 0.026 | 2.28e-20 | 0.152 | 0.133 | 0.019 | 7.06e-04 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.PseudoVal | 0.135 | 0.139 | -0.004 | 2.32e-33 | 0.129 | 0.133 | -0.004 | 1.43e-09 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.PseudoVal | 0.183 | 0.139 | 0.044 | 4.76e-33 | 0.173 | 0.133 | 0.040 | 2.79e-08 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.176 | 0.139 | 0.037 | 1.19e-30 | 0.166 | 0.133 | 0.033 | 3.58e-07 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.Inf | 0.168 | 0.135 | 0.033 | 2.17e-06 | 0.175 | 0.129 | 0.045 | 2.43e-08 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.Inf | 0.187 | 0.135 | 0.052 | 9.67e-14 | 0.181 | 0.129 | 0.052 | 2.18e-12 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.Inf | 0.176 | 0.135 | 0.041 | 8.05e-16 | 0.171 | 0.129 | 0.042 | 2.97e-05 |
| Prostate_Cancer | All.MultiPRS | LDpred2.Inf | 0.186 | 0.135 | 0.051 | 1.64e-13 | 0.181 | 0.129 | 0.052 | 2.66e-11 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.Inf | 0.137 | 0.135 | 0.001 | 5.50e-02 | 0.128 | 0.129 | -0.001 | 5.42e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.Inf | 0.139 | 0.135 | 0.004 | 2.32e-33 | 0.133 | 0.129 | 0.004 | 1.43e-09 |
| Prostate_Cancer | PRScs.MultiPRS | LDpred2.Inf | 0.184 | 0.135 | 0.049 | 2.76e-12 | 0.173 | 0.129 | 0.044 | 1.17e-09 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.Inf | 0.183 | 0.135 | 0.048 | 6.33e-12 | 0.175 | 0.129 | 0.046 | 8.86e-11 |
| Prostate_Cancer | PRScs.10FCVal | LDpred2.Inf | 0.181 | 0.135 | 0.045 | 6.93e-34 | 0.176 | 0.129 | 0.047 | 1.46e-10 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.Inf | 0.143 | 0.135 | 0.008 | 1.28e-06 | 0.133 | 0.129 | 0.004 | 2.10e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.Inf | 0.164 | 0.135 | 0.029 | 2.83e-05 | 0.149 | 0.129 | 0.020 | 3.15e-04 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.Inf | 0.152 | 0.135 | 0.017 | 1.31e-03 | 0.166 | 0.129 | 0.037 | 3.82e-04 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.Inf | 0.168 | 0.135 | 0.033 | 8.14e-10 | 0.164 | 0.129 | 0.035 | 1.11e-03 |
| Prostate_Cancer | PRScs.PseudoVal | LDpred2.Inf | 0.183 | 0.135 | 0.047 | 4.26e-38 | 0.172 | 0.129 | 0.043 | 2.77e-09 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.Inf | 0.176 | 0.135 | 0.041 | 3.72e-34 | 0.166 | 0.129 | 0.037 | 4.57e-08 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.Inf | 0.165 | 0.135 | 0.030 | 8.30e-24 | 0.152 | 0.129 | 0.023 | 1.11e-04 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.Inf | 0.135 | 0.135 | 0.000 | 1.00e+00 | 0.129 | 0.129 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.Inf | 0.180 | 0.135 | 0.045 | 1.07e-25 | 0.177 | 0.129 | 0.048 | 8.90e-09 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.Inf | 0.183 | 0.135 | 0.048 | 1.63e-35 | 0.173 | 0.129 | 0.044 | 6.30e-09 |
| Prostate_Cancer | PRScs.10FCVal | PRScs.MultiPRS | 0.181 | 0.184 | -0.003 | 6.61e-01 | 0.176 | 0.173 | 0.004 | 3.09e-02 |
| Prostate_Cancer | LDpred1.10FCVal | PRScs.MultiPRS | 0.165 | 0.184 | -0.019 | 7.42e-03 | 0.152 | 0.173 | -0.021 | 1.77e-06 |
| Prostate_Cancer | lassosum.MultiPRS | PRScs.MultiPRS | 0.187 | 0.184 | 0.003 | 4.26e-02 | 0.181 | 0.173 | 0.008 | 6.54e-03 |
| Prostate_Cancer | LDpred2.MultiPRS | PRScs.MultiPRS | 0.183 | 0.184 | -0.001 | 8.95e-01 | 0.175 | 0.173 | 0.002 | 4.40e-01 |
| Prostate_Cancer | All.MultiPRS | PRScs.MultiPRS | 0.186 | 0.184 | 0.003 | 7.14e-01 | 0.181 | 0.173 | 0.008 | 2.74e-02 |
| Prostate_Cancer | PRScs.PseudoVal | PRScs.MultiPRS | 0.183 | 0.184 | -0.001 | 8.52e-01 | 0.172 | 0.173 | -0.001 | 5.01e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | PRScs.MultiPRS | 0.164 | 0.184 | -0.019 | 1.82e-17 | 0.149 | 0.173 | -0.023 | 1.75e-07 |
| Prostate_Cancer | pT+clump.MultiPRS | PRScs.MultiPRS | 0.168 | 0.184 | -0.016 | 2.34e-02 | 0.175 | 0.173 | 0.002 | 7.50e-01 |
| Prostate_Cancer | lassosum.PseudoVal | PRScs.MultiPRS | 0.168 | 0.184 | -0.016 | 2.52e-02 | 0.164 | 0.173 | -0.009 | 7.34e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | PRScs.MultiPRS | 0.176 | 0.184 | -0.007 | 2.82e-01 | 0.171 | 0.173 | -0.002 | 7.10e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.176 | 0.184 | -0.007 | 2.84e-01 | 0.166 | 0.173 | -0.007 | 2.83e-02 |
| Prostate_Cancer | SBLUP.Inf | PRScs.MultiPRS | 0.137 | 0.184 | -0.047 | 1.21e-11 | 0.128 | 0.173 | -0.045 | 3.50e-10 |
| Prostate_Cancer | LDpred2.Inf | PRScs.MultiPRS | 0.135 | 0.184 | -0.049 | 2.76e-12 | 0.129 | 0.173 | -0.044 | 1.17e-09 |
| Prostate_Cancer | pT+clump.10FCVal | PRScs.MultiPRS | 0.152 | 0.184 | -0.032 | 5.80e-06 | 0.166 | 0.173 | -0.007 | 3.38e-01 |
| Prostate_Cancer | LDpred2.10FCVal | PRScs.MultiPRS | 0.183 | 0.184 | -0.001 | 9.19e-01 | 0.173 | 0.173 | 0.000 | 8.89e-01 |
| Prostate_Cancer | lassosum.10FCVal | PRScs.MultiPRS | 0.180 | 0.184 | -0.004 | 5.82e-01 | 0.177 | 0.173 | 0.004 | 2.73e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | PRScs.MultiPRS | 0.139 | 0.184 | -0.045 | 1.08e-10 | 0.133 | 0.173 | -0.040 | 6.15e-09 |
| Prostate_Cancer | LDpred1.Inf | PRScs.MultiPRS | 0.143 | 0.184 | -0.041 | 4.16e-09 | 0.133 | 0.173 | -0.040 | 1.23e-09 |
| Prostate_Cancer | PRScs.MultiPRS | PRScs.MultiPRS | 0.184 | 0.184 | 0.000 | 1.00e+00 | 0.173 | 0.173 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.PseudoVal | PRScs.10FCVal | 0.139 | 0.181 | -0.042 | 1.82e-31 | 0.133 | 0.176 | -0.044 | 6.22e-10 |
| Prostate_Cancer | PRScs.10FCVal | PRScs.10FCVal | 0.181 | 0.181 | 0.000 | 1.00e+00 | 0.176 | 0.176 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.MultiPRS | PRScs.10FCVal | 0.183 | 0.181 | 0.002 | 7.58e-01 | 0.175 | 0.176 | -0.002 | 4.86e-01 |
| Prostate_Cancer | pT+clump.MultiPRS | PRScs.10FCVal | 0.168 | 0.181 | -0.013 | 6.77e-02 | 0.175 | 0.176 | -0.002 | 7.36e-01 |
| Prostate_Cancer | LDpred1.Inf | PRScs.10FCVal | 0.143 | 0.181 | -0.038 | 2.16e-28 | 0.133 | 0.176 | -0.043 | 1.26e-10 |
| Prostate_Cancer | LDpred1.MultiPRS | PRScs.10FCVal | 0.164 | 0.181 | -0.016 | 1.86e-02 | 0.149 | 0.176 | -0.027 | 1.05e-08 |
| Prostate_Cancer | pT+clump.10FCVal | PRScs.10FCVal | 0.152 | 0.181 | -0.028 | 1.19e-15 | 0.166 | 0.176 | -0.010 | 1.38e-01 |
| Prostate_Cancer | lassosum.PseudoVal | PRScs.10FCVal | 0.168 | 0.181 | -0.013 | 1.55e-06 | 0.164 | 0.176 | -0.013 | 1.27e-02 |
| Prostate_Cancer | PRScs.PseudoVal | PRScs.10FCVal | 0.183 | 0.181 | 0.002 | 8.72e-02 | 0.172 | 0.176 | -0.005 | 2.11e-02 |
| Prostate_Cancer | DBSLMM.PseudoVal | PRScs.10FCVal | 0.176 | 0.181 | -0.004 | 1.62e-02 | 0.166 | 0.176 | -0.011 | 3.19e-03 |
| Prostate_Cancer | SBLUP.Inf | PRScs.10FCVal | 0.137 | 0.181 | -0.044 | 6.06e-32 | 0.128 | 0.176 | -0.048 | 6.50e-11 |
| Prostate_Cancer | LDpred2.Inf | PRScs.10FCVal | 0.135 | 0.181 | -0.045 | 6.93e-34 | 0.129 | 0.176 | -0.047 | 1.46e-10 |
| Prostate_Cancer | lassosum.10FCVal | PRScs.10FCVal | 0.180 | 0.181 | -0.001 | 6.91e-01 | 0.177 | 0.176 | 0.001 | 8.38e-01 |
| Prostate_Cancer | LDpred2.10FCVal | PRScs.10FCVal | 0.183 | 0.181 | 0.002 | 6.78e-02 | 0.173 | 0.176 | -0.003 | 1.97e-01 |
| Prostate_Cancer | lassosum.MultiPRS | PRScs.10FCVal | 0.187 | 0.181 | 0.006 | 3.72e-01 | 0.181 | 0.176 | 0.005 | 1.58e-01 |
| Prostate_Cancer | LDpred1.10FCVal | PRScs.10FCVal | 0.165 | 0.181 | -0.016 | 4.90e-11 | 0.152 | 0.176 | -0.024 | 1.06e-07 |
| Prostate_Cancer | All.MultiPRS | PRScs.10FCVal | 0.186 | 0.181 | 0.006 | 4.20e-01 | 0.181 | 0.176 | 0.005 | 2.50e-01 |
| Prostate_Cancer | SBayesR.PseudoVal | PRScs.10FCVal | 0.176 | 0.181 | -0.004 | 9.28e-02 | 0.171 | 0.176 | -0.006 | 2.80e-01 |
| Prostate_Cancer | PRScs.MultiPRS | PRScs.10FCVal | 0.184 | 0.181 | 0.003 | 6.61e-01 | 0.173 | 0.176 | -0.004 | 3.09e-02 |
| Prostate_Cancer | All.MultiPRS | PRScs.PseudoVal | 0.186 | 0.183 | 0.004 | 5.80e-01 | 0.181 | 0.172 | 0.009 | 1.53e-02 |
| Prostate_Cancer | pT+clump.MultiPRS | PRScs.PseudoVal | 0.168 | 0.183 | -0.014 | 3.75e-02 | 0.175 | 0.172 | 0.003 | 6.19e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | PRScs.PseudoVal | 0.139 | 0.183 | -0.044 | 1.20e-35 | 0.133 | 0.172 | -0.039 | 1.26e-08 |
| Prostate_Cancer | SBayesR.PseudoVal | PRScs.PseudoVal | 0.176 | 0.183 | -0.006 | 2.28e-02 | 0.171 | 0.172 | -0.001 | 8.70e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | PRScs.PseudoVal | 0.183 | 0.183 | 0.000 | 9.56e-01 | 0.175 | 0.172 | 0.003 | 2.51e-01 |
| Prostate_Cancer | SBLUP.Inf | PRScs.PseudoVal | 0.137 | 0.183 | -0.046 | 4.43e-36 | 0.128 | 0.172 | -0.044 | 1.18e-09 |
| Prostate_Cancer | LDpred1.Inf | PRScs.PseudoVal | 0.143 | 0.183 | -0.039 | 1.16e-32 | 0.133 | 0.172 | -0.039 | 2.91e-09 |
| Prostate_Cancer | PRScs.10FCVal | PRScs.PseudoVal | 0.181 | 0.183 | -0.002 | 8.72e-02 | 0.176 | 0.172 | 0.005 | 2.11e-02 |
| Prostate_Cancer | LDpred2.Inf | PRScs.PseudoVal | 0.135 | 0.183 | -0.047 | 4.26e-38 | 0.129 | 0.172 | -0.043 | 2.77e-09 |
| Prostate_Cancer | PRScs.PseudoVal | PRScs.PseudoVal | 0.183 | 0.183 | 0.000 | 1.00e+00 | 0.172 | 0.172 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.10FCVal | PRScs.PseudoVal | 0.183 | 0.183 | 0.001 | 6.50e-01 | 0.173 | 0.172 | 0.001 | 5.88e-01 |
| Prostate_Cancer | pT+clump.10FCVal | PRScs.PseudoVal | 0.152 | 0.183 | -0.030 | 5.41e-17 | 0.166 | 0.172 | -0.006 | 4.21e-01 |
| Prostate_Cancer | PRScs.MultiPRS | PRScs.PseudoVal | 0.184 | 0.183 | 0.001 | 8.52e-01 | 0.173 | 0.172 | 0.001 | 5.01e-01 |
| Prostate_Cancer | lassosum.PseudoVal | PRScs.PseudoVal | 0.168 | 0.183 | -0.014 | 1.14e-07 | 0.164 | 0.172 | -0.008 | 1.26e-01 |
| Prostate_Cancer | lassosum.10FCVal | PRScs.PseudoVal | 0.180 | 0.183 | -0.003 | 2.05e-01 | 0.177 | 0.172 | 0.005 | 1.70e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | PRScs.PseudoVal | 0.164 | 0.183 | -0.018 | 9.17e-03 | 0.149 | 0.172 | -0.022 | 9.32e-07 |
| Prostate_Cancer | lassosum.MultiPRS | PRScs.PseudoVal | 0.187 | 0.183 | 0.004 | 5.21e-01 | 0.181 | 0.172 | 0.009 | 4.79e-03 |
| Prostate_Cancer | LDpred1.10FCVal | PRScs.PseudoVal | 0.165 | 0.183 | -0.017 | 4.15e-14 | 0.152 | 0.172 | -0.020 | 8.30e-06 |
| Prostate_Cancer | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.176 | 0.183 | -0.006 | 5.43e-04 | 0.166 | 0.172 | -0.006 | 8.06e-02 |
| Prostate_Cancer | All.MultiPRS | pT+clump.MultiPRS | 0.186 | 0.168 | 0.018 | 8.51e-03 | 0.181 | 0.175 | 0.007 | 2.34e-01 |
| Prostate_Cancer | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.176 | 0.168 | 0.008 | 2.36e-01 | 0.171 | 0.175 | -0.004 | 5.68e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.164 | 0.168 | -0.004 | 5.91e-01 | 0.149 | 0.175 | -0.025 | 1.19e-04 |
| Prostate_Cancer | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.168 | 0.168 | 0.000 | 1.00e+00 | 0.175 | 0.175 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.PseudoVal | pT+clump.MultiPRS | 0.168 | 0.168 | 0.000 | 9.88e-01 | 0.164 | 0.175 | -0.011 | 9.03e-02 |
| Prostate_Cancer | PRScs.10FCVal | pT+clump.MultiPRS | 0.181 | 0.168 | 0.013 | 6.77e-02 | 0.176 | 0.175 | 0.002 | 7.36e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.176 | 0.168 | 0.008 | 2.33e-01 | 0.166 | 0.175 | -0.009 | 1.02e-01 |
| Prostate_Cancer | SBLUP.Inf | pT+clump.MultiPRS | 0.137 | 0.168 | -0.031 | 6.02e-06 | 0.128 | 0.175 | -0.046 | 1.05e-08 |
| Prostate_Cancer | LDpred2.Inf | pT+clump.MultiPRS | 0.135 | 0.168 | -0.033 | 2.17e-06 | 0.129 | 0.175 | -0.045 | 2.43e-08 |
| Prostate_Cancer | pT+clump.10FCVal | pT+clump.MultiPRS | 0.152 | 0.168 | -0.016 | 2.19e-02 | 0.166 | 0.175 | -0.008 | 5.70e-02 |
| Prostate_Cancer | LDpred2.10FCVal | pT+clump.MultiPRS | 0.183 | 0.168 | 0.015 | 3.02e-02 | 0.173 | 0.175 | -0.001 | 8.03e-01 |
| Prostate_Cancer | PRScs.PseudoVal | pT+clump.MultiPRS | 0.183 | 0.168 | 0.014 | 3.75e-02 | 0.172 | 0.175 | -0.003 | 6.19e-01 |
| Prostate_Cancer | LDpred1.10FCVal | pT+clump.MultiPRS | 0.165 | 0.168 | -0.003 | 6.72e-01 | 0.152 | 0.175 | -0.023 | 5.77e-04 |
| Prostate_Cancer | LDpred1.Inf | pT+clump.MultiPRS | 0.143 | 0.168 | -0.025 | 2.88e-04 | 0.133 | 0.175 | -0.042 | 7.25e-08 |
| Prostate_Cancer | PRScs.MultiPRS | pT+clump.MultiPRS | 0.184 | 0.168 | 0.016 | 2.34e-02 | 0.173 | 0.175 | -0.002 | 7.50e-01 |
| Prostate_Cancer | lassosum.10FCVal | pT+clump.MultiPRS | 0.180 | 0.168 | 0.012 | 8.64e-02 | 0.177 | 0.175 | 0.003 | 6.21e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.139 | 0.168 | -0.029 | 2.64e-05 | 0.133 | 0.175 | -0.042 | 1.48e-07 |
| Prostate_Cancer | lassosum.MultiPRS | pT+clump.MultiPRS | 0.187 | 0.168 | 0.019 | 6.46e-03 | 0.181 | 0.175 | 0.007 | 1.86e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.183 | 0.168 | 0.015 | 3.25e-02 | 0.175 | 0.175 | 0.000 | 9.80e-01 |
| Prostate_Cancer | LDpred2.10FCVal | pT+clump.10FCVal | 0.183 | 0.152 | 0.031 | 9.77e-19 | 0.173 | 0.166 | 0.007 | 2.94e-01 |
| Prostate_Cancer | pT+clump.MultiPRS | pT+clump.10FCVal | 0.168 | 0.152 | 0.016 | 2.19e-02 | 0.175 | 0.166 | 0.008 | 5.70e-02 |
| Prostate_Cancer | LDpred2.Inf | pT+clump.10FCVal | 0.135 | 0.152 | -0.017 | 1.31e-03 | 0.129 | 0.166 | -0.037 | 3.82e-04 |
| Prostate_Cancer | SBayesR.PseudoVal | pT+clump.10FCVal | 0.176 | 0.152 | 0.024 | 1.05e-14 | 0.171 | 0.166 | 0.005 | 4.40e-01 |
| Prostate_Cancer | PRScs.MultiPRS | pT+clump.10FCVal | 0.184 | 0.152 | 0.032 | 5.80e-06 | 0.173 | 0.166 | 0.007 | 3.38e-01 |
| Prostate_Cancer | SBLUP.Inf | pT+clump.10FCVal | 0.137 | 0.152 | -0.015 | 3.57e-03 | 0.128 | 0.166 | -0.038 | 3.00e-04 |
| Prostate_Cancer | lassosum.MultiPRS | pT+clump.10FCVal | 0.187 | 0.152 | 0.035 | 5.96e-07 | 0.181 | 0.166 | 0.015 | 2.26e-02 |
| Prostate_Cancer | PRScs.10FCVal | pT+clump.10FCVal | 0.181 | 0.152 | 0.028 | 1.19e-15 | 0.176 | 0.166 | 0.010 | 1.38e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | pT+clump.10FCVal | 0.183 | 0.152 | 0.031 | 1.01e-05 | 0.175 | 0.166 | 0.009 | 2.16e-01 |
| Prostate_Cancer | lassosum.10FCVal | pT+clump.10FCVal | 0.180 | 0.152 | 0.028 | 9.78e-17 | 0.177 | 0.166 | 0.011 | 8.84e-02 |
| Prostate_Cancer | LDpred2.PseudoVal | pT+clump.10FCVal | 0.139 | 0.152 | -0.013 | 1.03e-02 | 0.133 | 0.166 | -0.033 | 1.11e-03 |
| Prostate_Cancer | LDpred1.MultiPRS | pT+clump.10FCVal | 0.164 | 0.152 | 0.012 | 8.13e-02 | 0.149 | 0.166 | -0.017 | 5.06e-02 |
| Prostate_Cancer | pT+clump.10FCVal | pT+clump.10FCVal | 0.152 | 0.152 | 0.000 | 1.00e+00 | 0.166 | 0.166 | 0.000 | 1.00e+00 |
| Prostate_Cancer | All.MultiPRS | pT+clump.10FCVal | 0.186 | 0.152 | 0.034 | 8.40e-07 | 0.181 | 0.166 | 0.015 | 2.88e-02 |
| Prostate_Cancer | LDpred1.Inf | pT+clump.10FCVal | 0.143 | 0.152 | -0.009 | 6.60e-02 | 0.133 | 0.166 | -0.033 | 8.73e-04 |
| Prostate_Cancer | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.176 | 0.152 | 0.024 | 7.26e-11 | 0.166 | 0.166 | -0.001 | 9.45e-01 |
| Prostate_Cancer | LDpred1.10FCVal | pT+clump.10FCVal | 0.165 | 0.152 | 0.013 | 2.97e-03 | 0.152 | 0.166 | -0.014 | 9.75e-02 |
| Prostate_Cancer | lassosum.PseudoVal | pT+clump.10FCVal | 0.168 | 0.152 | 0.016 | 9.05e-08 | 0.164 | 0.166 | -0.002 | 6.78e-01 |
| Prostate_Cancer | PRScs.PseudoVal | pT+clump.10FCVal | 0.183 | 0.152 | 0.030 | 5.41e-17 | 0.172 | 0.166 | 0.006 | 4.21e-01 |
| Prostate_Cancer | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.168 | 0.176 | -0.008 | 2.36e-01 | 0.175 | 0.171 | 0.004 | 5.68e-01 |
| Prostate_Cancer | LDpred1.10FCVal | SBayesR.PseudoVal | 0.165 | 0.176 | -0.011 | 3.98e-03 | 0.152 | 0.171 | -0.019 | 1.14e-02 |
| Prostate_Cancer | PRScs.10FCVal | SBayesR.PseudoVal | 0.181 | 0.176 | 0.004 | 9.28e-02 | 0.176 | 0.171 | 0.006 | 2.80e-01 |
| Prostate_Cancer | LDpred2.10FCVal | SBayesR.PseudoVal | 0.183 | 0.176 | 0.007 | 7.85e-03 | 0.173 | 0.171 | 0.002 | 6.45e-01 |
| Prostate_Cancer | lassosum.10FCVal | SBayesR.PseudoVal | 0.180 | 0.176 | 0.004 | 1.96e-01 | 0.177 | 0.171 | 0.006 | 2.58e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.139 | 0.176 | -0.037 | 5.49e-14 | 0.133 | 0.171 | -0.038 | 9.49e-05 |
| Prostate_Cancer | lassosum.MultiPRS | SBayesR.PseudoVal | 0.187 | 0.176 | 0.011 | 1.26e-01 | 0.181 | 0.171 | 0.010 | 5.96e-02 |
| Prostate_Cancer | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.183 | 0.176 | 0.007 | 3.43e-01 | 0.175 | 0.171 | 0.004 | 4.72e-01 |
| Prostate_Cancer | All.MultiPRS | SBayesR.PseudoVal | 0.186 | 0.176 | 0.010 | 1.48e-01 | 0.181 | 0.171 | 0.010 | 2.71e-02 |
| Prostate_Cancer | PRScs.PseudoVal | SBayesR.PseudoVal | 0.183 | 0.176 | 0.006 | 2.28e-02 | 0.172 | 0.171 | 0.001 | 8.70e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.164 | 0.176 | -0.012 | 8.65e-02 | 0.149 | 0.171 | -0.022 | 4.60e-03 |
| Prostate_Cancer | pT+clump.10FCVal | SBayesR.PseudoVal | 0.152 | 0.176 | -0.024 | 1.05e-14 | 0.166 | 0.171 | -0.005 | 4.40e-01 |
| Prostate_Cancer | lassosum.PseudoVal | SBayesR.PseudoVal | 0.168 | 0.176 | -0.008 | 3.42e-04 | 0.164 | 0.171 | -0.007 | 9.86e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.176 | 0.176 | 0.000 | 1.00e+00 | 0.171 | 0.171 | 0.000 | 1.00e+00 |
| Prostate_Cancer | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.176 | 0.176 | 0.000 | 9.91e-01 | 0.166 | 0.171 | -0.005 | 3.65e-01 |
| Prostate_Cancer | SBLUP.Inf | SBayesR.PseudoVal | 0.137 | 0.176 | -0.040 | 1.15e-14 | 0.128 | 0.171 | -0.043 | 2.19e-05 |
| Prostate_Cancer | LDpred2.Inf | SBayesR.PseudoVal | 0.135 | 0.176 | -0.041 | 8.05e-16 | 0.129 | 0.171 | -0.042 | 2.97e-05 |
| Prostate_Cancer | LDpred1.Inf | SBayesR.PseudoVal | 0.143 | 0.176 | -0.033 | 7.12e-12 | 0.133 | 0.171 | -0.038 | 7.33e-05 |
| Prostate_Cancer | PRScs.MultiPRS | SBayesR.PseudoVal | 0.184 | 0.176 | 0.007 | 2.82e-01 | 0.173 | 0.171 | 0.002 | 7.10e-01 |
| Prostate_Cancer | LDpred2.10FCVal | SBLUP.Inf | 0.183 | 0.137 | 0.046 | 1.04e-32 | 0.173 | 0.128 | 0.045 | 5.06e-09 |
| Prostate_Cancer | PRScs.PseudoVal | SBLUP.Inf | 0.183 | 0.137 | 0.046 | 4.43e-36 | 0.172 | 0.128 | 0.044 | 1.18e-09 |
| Prostate_Cancer | LDpred1.10FCVal | SBLUP.Inf | 0.165 | 0.137 | 0.028 | 1.14e-22 | 0.152 | 0.128 | 0.024 | 4.07e-05 |
| Prostate_Cancer | LDpred2.Inf | SBLUP.Inf | 0.135 | 0.137 | -0.001 | 5.50e-02 | 0.129 | 0.128 | 0.001 | 5.42e-01 |
| Prostate_Cancer | SBayesR.PseudoVal | SBLUP.Inf | 0.176 | 0.137 | 0.040 | 1.15e-14 | 0.171 | 0.128 | 0.043 | 2.19e-05 |
| Prostate_Cancer | lassosum.10FCVal | SBLUP.Inf | 0.180 | 0.137 | 0.043 | 3.00e-24 | 0.177 | 0.128 | 0.049 | 4.50e-09 |
| Prostate_Cancer | pT+clump.MultiPRS | SBLUP.Inf | 0.168 | 0.137 | 0.031 | 6.02e-06 | 0.175 | 0.128 | 0.046 | 1.05e-08 |
| Prostate_Cancer | lassosum.MultiPRS | SBLUP.Inf | 0.187 | 0.137 | 0.050 | 4.67e-13 | 0.181 | 0.128 | 0.053 | 6.18e-13 |
| Prostate_Cancer | PRScs.10FCVal | SBLUP.Inf | 0.181 | 0.137 | 0.044 | 6.06e-32 | 0.176 | 0.128 | 0.048 | 6.50e-11 |
| Prostate_Cancer | PRScs.MultiPRS | SBLUP.Inf | 0.184 | 0.137 | 0.047 | 1.21e-11 | 0.173 | 0.128 | 0.045 | 3.50e-10 |
| Prostate_Cancer | LDpred1.MultiPRS | SBLUP.Inf | 0.164 | 0.137 | 0.028 | 7.05e-05 | 0.149 | 0.128 | 0.021 | 1.20e-04 |
| Prostate_Cancer | LDpred2.PseudoVal | SBLUP.Inf | 0.139 | 0.137 | 0.002 | 1.05e-02 | 0.133 | 0.128 | 0.005 | 6.91e-03 |
| Prostate_Cancer | LDpred1.Inf | SBLUP.Inf | 0.143 | 0.137 | 0.006 | 5.10e-05 | 0.133 | 0.128 | 0.005 | 1.15e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | SBLUP.Inf | 0.183 | 0.137 | 0.046 | 2.81e-11 | 0.175 | 0.128 | 0.047 | 7.53e-11 |
| Prostate_Cancer | All.MultiPRS | SBLUP.Inf | 0.186 | 0.137 | 0.050 | 8.19e-13 | 0.181 | 0.128 | 0.053 | 1.01e-11 |
| Prostate_Cancer | SBLUP.Inf | SBLUP.Inf | 0.137 | 0.137 | 0.000 | 1.00e+00 | 0.128 | 0.128 | 0.000 | 1.00e+00 |
| Prostate_Cancer | DBSLMM.PseudoVal | SBLUP.Inf | 0.176 | 0.137 | 0.040 | 6.96e-33 | 0.166 | 0.128 | 0.038 | 1.26e-08 |
| Prostate_Cancer | pT+clump.10FCVal | SBLUP.Inf | 0.152 | 0.137 | 0.015 | 3.57e-03 | 0.166 | 0.128 | 0.038 | 3.00e-04 |
| Prostate_Cancer | lassosum.PseudoVal | SBLUP.Inf | 0.168 | 0.137 | 0.031 | 4.95e-09 | 0.164 | 0.128 | 0.036 | 8.48e-04 |
| Breast_Cancer | LDpred2.PseudoVal | All.MultiPRS | 0.169 | 0.228 | -0.059 | 5.12e-19 | 0.161 | 0.215 | -0.054 | 3.92e-14 |
| Breast_Cancer | pT+clump.10FCVal | All.MultiPRS | 0.179 | 0.228 | -0.049 | 1.19e-13 | 0.167 | 0.215 | -0.048 | 4.23e-13 |
| Breast_Cancer | LDpred2.MultiPRS | All.MultiPRS | 0.224 | 0.228 | -0.003 | 6.28e-01 | 0.211 | 0.215 | -0.004 | 5.16e-02 |
| Breast_Cancer | PRScs.PseudoVal | All.MultiPRS | 0.220 | 0.228 | -0.007 | 2.70e-01 | 0.208 | 0.215 | -0.007 | 1.82e-02 |
| Breast_Cancer | LDpred1.Inf | All.MultiPRS | 0.172 | 0.228 | -0.056 | 2.56e-17 | 0.162 | 0.215 | -0.053 | 3.87e-14 |
| Breast_Cancer | PRScs.MultiPRS | All.MultiPRS | 0.222 | 0.228 | -0.006 | 4.18e-01 | 0.210 | 0.215 | -0.005 | 1.02e-01 |
| Breast_Cancer | LDpred2.Inf | All.MultiPRS | 0.168 | 0.228 | -0.060 | 1.35e-19 | 0.160 | 0.215 | -0.055 | 3.36e-14 |
| Breast_Cancer | lassosum.10FCVal | All.MultiPRS | 0.219 | 0.228 | -0.008 | 1.94e-01 | 0.199 | 0.215 | -0.016 | 4.48e-06 |
| Breast_Cancer | LDpred2.10FCVal | All.MultiPRS | 0.223 | 0.228 | -0.004 | 5.31e-01 | 0.211 | 0.215 | -0.004 | 1.28e-01 |
| Breast_Cancer | lassosum.MultiPRS | All.MultiPRS | 0.225 | 0.228 | -0.002 | 7.28e-01 | 0.212 | 0.215 | -0.003 | 1.60e-01 |
| Breast_Cancer | LDpred1.10FCVal | All.MultiPRS | 0.201 | 0.228 | -0.027 | 4.03e-05 | 0.186 | 0.215 | -0.029 | 1.35e-08 |
| Breast_Cancer | All.MultiPRS | All.MultiPRS | 0.228 | 0.228 | 0.000 | 1.00e+00 | 0.215 | 0.215 | 0.000 | 1.00e+00 |
| Breast_Cancer | SBayesR.PseudoVal | All.MultiPRS | 0.221 | 0.228 | -0.007 | 3.04e-01 | 0.205 | 0.215 | -0.010 | 2.21e-03 |
| Breast_Cancer | LDpred1.MultiPRS | All.MultiPRS | 0.211 | 0.228 | -0.016 | 1.08e-15 | 0.195 | 0.215 | -0.021 | 7.66e-07 |
| Breast_Cancer | pT+clump.MultiPRS | All.MultiPRS | 0.200 | 0.228 | -0.027 | 5.53e-05 | 0.188 | 0.215 | -0.027 | 6.94e-08 |
| Breast_Cancer | lassosum.PseudoVal | All.MultiPRS | 0.217 | 0.228 | -0.010 | 1.07e-01 | 0.199 | 0.215 | -0.016 | 1.85e-05 |
| Breast_Cancer | PRScs.10FCVal | All.MultiPRS | 0.220 | 0.228 | -0.008 | 2.27e-01 | 0.207 | 0.215 | -0.008 | 1.16e-02 |
| Breast_Cancer | DBSLMM.PseudoVal | All.MultiPRS | 0.217 | 0.228 | -0.010 | 1.21e-01 | 0.211 | 0.215 | -0.004 | 2.21e-01 |
| Breast_Cancer | SBLUP.Inf | All.MultiPRS | 0.168 | 0.228 | -0.060 | 1.18e-19 | 0.158 | 0.215 | -0.057 | 6.47e-15 |
| Breast_Cancer | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.221 | 0.217 | 0.003 | 1.87e-01 | 0.205 | 0.211 | -0.006 | 2.63e-01 |
| Breast_Cancer | LDpred2.Inf | DBSLMM.PseudoVal | 0.168 | 0.217 | -0.050 | 4.35e-63 | 0.160 | 0.211 | -0.051 | 2.59e-17 |
| Breast_Cancer | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.179 | 0.217 | -0.039 | 1.13e-26 | 0.167 | 0.211 | -0.044 | 1.63e-09 |
| Breast_Cancer | PRScs.10FCVal | DBSLMM.PseudoVal | 0.220 | 0.217 | 0.002 | 2.02e-01 | 0.207 | 0.211 | -0.004 | 2.99e-01 |
| Breast_Cancer | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.220 | 0.217 | 0.003 | 8.35e-02 | 0.208 | 0.211 | -0.003 | 3.66e-01 |
| Breast_Cancer | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.200 | 0.217 | -0.017 | 1.10e-02 | 0.188 | 0.211 | -0.023 | 3.20e-06 |
| Breast_Cancer | LDpred1.Inf | DBSLMM.PseudoVal | 0.172 | 0.217 | -0.046 | 7.43e-56 | 0.162 | 0.211 | -0.049 | 4.73e-17 |
| Breast_Cancer | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.225 | 0.217 | 0.008 | 2.57e-01 | 0.212 | 0.211 | 0.001 | 7.62e-01 |
| Breast_Cancer | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.223 | 0.217 | 0.006 | 2.79e-03 | 0.211 | 0.211 | 0.000 | 9.43e-01 |
| Breast_Cancer | lassosum.10FCVal | DBSLMM.PseudoVal | 0.219 | 0.217 | 0.002 | 5.12e-01 | 0.199 | 0.211 | -0.012 | 2.03e-02 |
| Breast_Cancer | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.201 | 0.217 | -0.017 | 6.72e-14 | 0.186 | 0.211 | -0.025 | 5.79e-08 |
| Breast_Cancer | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.211 | 0.217 | -0.006 | 3.30e-01 | 0.195 | 0.211 | -0.016 | 4.98e-04 |
| Breast_Cancer | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.222 | 0.217 | 0.005 | 5.06e-01 | 0.210 | 0.211 | 0.000 | 9.39e-01 |
| Breast_Cancer | All.MultiPRS | DBSLMM.PseudoVal | 0.228 | 0.217 | 0.010 | 1.21e-01 | 0.215 | 0.211 | 0.004 | 2.21e-01 |
| Breast_Cancer | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.169 | 0.217 | -0.049 | 7.14e-62 | 0.161 | 0.211 | -0.050 | 2.83e-17 |
| Breast_Cancer | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.217 | 0.217 | 0.000 | 1.00e+00 | 0.211 | 0.211 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.224 | 0.217 | 0.007 | 3.19e-01 | 0.211 | 0.211 | 0.000 | 9.77e-01 |
| Breast_Cancer | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.217 | 0.217 | 0.000 | 8.85e-01 | 0.199 | 0.211 | -0.012 | 4.09e-02 |
| Breast_Cancer | SBLUP.Inf | DBSLMM.PseudoVal | 0.168 | 0.217 | -0.050 | 7.26e-65 | 0.158 | 0.211 | -0.052 | 6.94e-19 |
| Breast_Cancer | SBayesR.PseudoVal | lassosum.MultiPRS | 0.221 | 0.225 | -0.004 | 5.27e-01 | 0.205 | 0.212 | -0.007 | 6.17e-02 |
| Breast_Cancer | LDpred2.10FCVal | lassosum.MultiPRS | 0.223 | 0.225 | -0.002 | 8.02e-01 | 0.211 | 0.212 | -0.001 | 7.83e-01 |
| Breast_Cancer | LDpred1.10FCVal | lassosum.MultiPRS | 0.201 | 0.225 | -0.024 | 3.39e-04 | 0.186 | 0.212 | -0.026 | 1.03e-06 |
| Breast_Cancer | LDpred2.Inf | lassosum.MultiPRS | 0.168 | 0.225 | -0.057 | 4.78e-17 | 0.160 | 0.212 | -0.052 | 1.88e-12 |
| Breast_Cancer | PRScs.10FCVal | lassosum.MultiPRS | 0.220 | 0.225 | -0.005 | 4.20e-01 | 0.207 | 0.212 | -0.005 | 1.14e-01 |
| Breast_Cancer | PRScs.MultiPRS | lassosum.MultiPRS | 0.222 | 0.225 | -0.003 | 6.40e-01 | 0.210 | 0.212 | -0.001 | 6.00e-01 |
| Breast_Cancer | pT+clump.MultiPRS | lassosum.MultiPRS | 0.200 | 0.225 | -0.025 | 2.44e-04 | 0.188 | 0.212 | -0.024 | 7.15e-07 |
| Breast_Cancer | lassosum.MultiPRS | lassosum.MultiPRS | 0.225 | 0.225 | 0.000 | 1.00e+00 | 0.212 | 0.212 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.PseudoVal | lassosum.MultiPRS | 0.217 | 0.225 | -0.008 | 2.32e-01 | 0.199 | 0.212 | -0.013 | 4.60e-05 |
| Breast_Cancer | LDpred2.MultiPRS | lassosum.MultiPRS | 0.224 | 0.225 | -0.001 | 5.62e-01 | 0.211 | 0.212 | -0.001 | 6.85e-01 |
| Breast_Cancer | LDpred1.MultiPRS | lassosum.MultiPRS | 0.211 | 0.225 | -0.014 | 3.87e-02 | 0.195 | 0.212 | -0.017 | 1.49e-04 |
| Breast_Cancer | LDpred2.PseudoVal | lassosum.MultiPRS | 0.169 | 0.225 | -0.056 | 1.58e-16 | 0.161 | 0.212 | -0.051 | 2.04e-12 |
| Breast_Cancer | LDpred1.Inf | lassosum.MultiPRS | 0.172 | 0.225 | -0.053 | 5.98e-15 | 0.162 | 0.212 | -0.050 | 2.69e-12 |
| Breast_Cancer | pT+clump.10FCVal | lassosum.MultiPRS | 0.179 | 0.225 | -0.047 | 9.73e-12 | 0.167 | 0.212 | -0.045 | 3.14e-12 |
| Breast_Cancer | lassosum.10FCVal | lassosum.MultiPRS | 0.219 | 0.225 | -0.006 | 3.71e-01 | 0.199 | 0.212 | -0.013 | 4.75e-06 |
| Breast_Cancer | SBLUP.Inf | lassosum.MultiPRS | 0.168 | 0.225 | -0.057 | 4.39e-17 | 0.158 | 0.212 | -0.054 | 2.59e-13 |
| Breast_Cancer | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.217 | 0.225 | -0.008 | 2.57e-01 | 0.211 | 0.212 | -0.001 | 7.62e-01 |
| Breast_Cancer | All.MultiPRS | lassosum.MultiPRS | 0.228 | 0.225 | 0.002 | 7.28e-01 | 0.215 | 0.212 | 0.003 | 1.60e-01 |
| Breast_Cancer | PRScs.PseudoVal | lassosum.MultiPRS | 0.220 | 0.225 | -0.005 | 4.81e-01 | 0.208 | 0.212 | -0.004 | 1.76e-01 |
| Breast_Cancer | LDpred1.Inf | lassosum.PseudoVal | 0.172 | 0.217 | -0.045 | 8.22e-25 | 0.162 | 0.199 | -0.037 | 2.34e-05 |
| Breast_Cancer | PRScs.10FCVal | lassosum.PseudoVal | 0.220 | 0.217 | 0.003 | 2.11e-01 | 0.207 | 0.199 | 0.008 | 6.16e-02 |
| Breast_Cancer | LDpred2.MultiPRS | lassosum.PseudoVal | 0.224 | 0.217 | 0.007 | 2.90e-01 | 0.211 | 0.199 | 0.011 | 4.64e-03 |
| Breast_Cancer | lassosum.PseudoVal | lassosum.PseudoVal | 0.217 | 0.217 | 0.000 | 1.00e+00 | 0.199 | 0.199 | 0.000 | 1.00e+00 |
| Breast_Cancer | PRScs.PseudoVal | lassosum.PseudoVal | 0.220 | 0.217 | 0.003 | 1.32e-01 | 0.208 | 0.199 | 0.008 | 5.69e-02 |
| Breast_Cancer | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.217 | 0.217 | 0.000 | 8.85e-01 | 0.211 | 0.199 | 0.012 | 4.09e-02 |
| Breast_Cancer | SBLUP.Inf | lassosum.PseudoVal | 0.168 | 0.217 | -0.049 | 1.43e-27 | 0.158 | 0.199 | -0.041 | 7.57e-06 |
| Breast_Cancer | LDpred2.Inf | lassosum.PseudoVal | 0.168 | 0.217 | -0.049 | 1.60e-27 | 0.160 | 0.199 | -0.039 | 1.85e-05 |
| Breast_Cancer | pT+clump.10FCVal | lassosum.PseudoVal | 0.179 | 0.217 | -0.038 | 5.41e-37 | 0.167 | 0.199 | -0.032 | 7.03e-08 |
| Breast_Cancer | LDpred2.10FCVal | lassosum.PseudoVal | 0.223 | 0.217 | 0.006 | 2.21e-03 | 0.211 | 0.199 | 0.012 | 4.64e-03 |
| Breast_Cancer | lassosum.MultiPRS | lassosum.PseudoVal | 0.225 | 0.217 | 0.008 | 2.32e-01 | 0.212 | 0.199 | 0.013 | 4.60e-05 |
| Breast_Cancer | LDpred1.10FCVal | lassosum.PseudoVal | 0.201 | 0.217 | -0.016 | 1.79e-06 | 0.186 | 0.199 | -0.013 | 5.42e-02 |
| Breast_Cancer | All.MultiPRS | lassosum.PseudoVal | 0.228 | 0.217 | 0.010 | 1.07e-01 | 0.215 | 0.199 | 0.016 | 1.85e-05 |
| Breast_Cancer | SBayesR.PseudoVal | lassosum.PseudoVal | 0.221 | 0.217 | 0.004 | 3.57e-02 | 0.205 | 0.199 | 0.006 | 1.10e-01 |
| Breast_Cancer | lassosum.10FCVal | lassosum.PseudoVal | 0.219 | 0.217 | 0.002 | 1.14e-01 | 0.199 | 0.199 | 0.000 | 9.41e-01 |
| Breast_Cancer | pT+clump.MultiPRS | lassosum.PseudoVal | 0.200 | 0.217 | -0.017 | 1.31e-02 | 0.188 | 0.199 | -0.012 | 3.21e-02 |
| Breast_Cancer | LDpred2.PseudoVal | lassosum.PseudoVal | 0.169 | 0.217 | -0.048 | 4.77e-27 | 0.161 | 0.199 | -0.039 | 2.02e-05 |
| Breast_Cancer | LDpred1.MultiPRS | lassosum.PseudoVal | 0.211 | 0.217 | -0.006 | 3.64e-01 | 0.195 | 0.199 | -0.005 | 4.05e-01 |
| Breast_Cancer | PRScs.MultiPRS | lassosum.PseudoVal | 0.222 | 0.217 | 0.005 | 4.69e-01 | 0.210 | 0.199 | 0.011 | 3.74e-03 |
| Breast_Cancer | lassosum.10FCVal | lassosum.10FCVal | 0.219 | 0.219 | 0.000 | 1.00e+00 | 0.199 | 0.199 | 0.000 | 1.00e+00 |
| Breast_Cancer | pT+clump.MultiPRS | lassosum.10FCVal | 0.200 | 0.219 | -0.019 | 5.40e-03 | 0.188 | 0.199 | -0.011 | 2.53e-02 |
| Breast_Cancer | LDpred2.Inf | lassosum.10FCVal | 0.168 | 0.219 | -0.051 | 1.14e-40 | 0.160 | 0.199 | -0.039 | 4.70e-07 |
| Breast_Cancer | SBayesR.PseudoVal | lassosum.10FCVal | 0.221 | 0.219 | 0.002 | 4.32e-01 | 0.205 | 0.199 | 0.006 | 1.88e-01 |
| Breast_Cancer | PRScs.MultiPRS | lassosum.10FCVal | 0.222 | 0.219 | 0.003 | 6.71e-01 | 0.210 | 0.199 | 0.011 | 9.27e-04 |
| Breast_Cancer | SBLUP.Inf | lassosum.10FCVal | 0.168 | 0.219 | -0.051 | 5.81e-41 | 0.158 | 0.199 | -0.041 | 1.27e-07 |
| Breast_Cancer | lassosum.MultiPRS | lassosum.10FCVal | 0.225 | 0.219 | 0.006 | 3.71e-01 | 0.212 | 0.199 | 0.013 | 4.75e-06 |
| Breast_Cancer | PRScs.10FCVal | lassosum.10FCVal | 0.220 | 0.219 | 0.001 | 7.42e-01 | 0.207 | 0.199 | 0.008 | 2.54e-02 |
| Breast_Cancer | LDpred2.MultiPRS | lassosum.10FCVal | 0.224 | 0.219 | 0.005 | 4.48e-01 | 0.211 | 0.199 | 0.012 | 1.56e-03 |
| Breast_Cancer | PRScs.PseudoVal | lassosum.10FCVal | 0.220 | 0.219 | 0.001 | 4.82e-01 | 0.208 | 0.199 | 0.009 | 1.94e-02 |
| Breast_Cancer | LDpred2.PseudoVal | lassosum.10FCVal | 0.169 | 0.219 | -0.050 | 4.09e-40 | 0.161 | 0.199 | -0.038 | 5.08e-07 |
| Breast_Cancer | LDpred1.MultiPRS | lassosum.10FCVal | 0.211 | 0.219 | -0.008 | 2.21e-01 | 0.195 | 0.199 | -0.004 | 3.53e-01 |
| Breast_Cancer | pT+clump.10FCVal | lassosum.10FCVal | 0.179 | 0.219 | -0.040 | 3.58e-34 | 0.167 | 0.199 | -0.032 | 1.31e-06 |
| Breast_Cancer | All.MultiPRS | lassosum.10FCVal | 0.228 | 0.219 | 0.008 | 1.94e-01 | 0.215 | 0.199 | 0.016 | 4.48e-06 |
| Breast_Cancer | LDpred1.Inf | lassosum.10FCVal | 0.172 | 0.219 | -0.047 | 6.72e-37 | 0.162 | 0.199 | -0.037 | 6.56e-07 |
| Breast_Cancer | DBSLMM.PseudoVal | lassosum.10FCVal | 0.217 | 0.219 | -0.002 | 5.12e-01 | 0.211 | 0.199 | 0.012 | 2.03e-02 |
| Breast_Cancer | LDpred1.10FCVal | lassosum.10FCVal | 0.201 | 0.219 | -0.018 | 5.81e-11 | 0.186 | 0.199 | -0.013 | 2.09e-02 |
| Breast_Cancer | lassosum.PseudoVal | lassosum.10FCVal | 0.217 | 0.219 | -0.002 | 1.14e-01 | 0.199 | 0.199 | 0.000 | 9.41e-01 |
| Breast_Cancer | LDpred2.10FCVal | lassosum.10FCVal | 0.223 | 0.219 | 0.004 | 7.62e-03 | 0.211 | 0.199 | 0.012 | 2.39e-04 |
| Breast_Cancer | LDpred2.10FCVal | LDpred1.MultiPRS | 0.223 | 0.211 | 0.012 | 5.72e-02 | 0.211 | 0.195 | 0.017 | 1.03e-05 |
| Breast_Cancer | PRScs.PseudoVal | LDpred1.MultiPRS | 0.220 | 0.211 | 0.009 | 1.54e-01 | 0.208 | 0.195 | 0.013 | 5.03e-04 |
| Breast_Cancer | LDpred2.Inf | LDpred1.MultiPRS | 0.168 | 0.211 | -0.043 | 4.60e-11 | 0.160 | 0.195 | -0.034 | 4.42e-08 |
| Breast_Cancer | pT+clump.10FCVal | LDpred1.MultiPRS | 0.179 | 0.211 | -0.032 | 9.31e-07 | 0.167 | 0.195 | -0.027 | 5.69e-04 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.221 | 0.211 | 0.010 | 1.36e-01 | 0.205 | 0.195 | 0.010 | 7.84e-02 |
| Breast_Cancer | All.MultiPRS | LDpred1.MultiPRS | 0.228 | 0.211 | 0.016 | 1.08e-15 | 0.215 | 0.195 | 0.021 | 7.66e-07 |
| Breast_Cancer | LDpred1.10FCVal | LDpred1.MultiPRS | 0.201 | 0.211 | -0.010 | 1.11e-01 | 0.186 | 0.195 | -0.009 | 2.74e-03 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.211 | 0.211 | 0.000 | 1.00e+00 | 0.195 | 0.195 | 0.000 | 1.00e+00 |
| Breast_Cancer | PRScs.10FCVal | LDpred1.MultiPRS | 0.220 | 0.211 | 0.009 | 1.88e-01 | 0.207 | 0.195 | 0.013 | 1.30e-03 |
| Breast_Cancer | lassosum.PseudoVal | LDpred1.MultiPRS | 0.217 | 0.211 | 0.006 | 3.64e-01 | 0.199 | 0.195 | 0.005 | 4.05e-01 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.200 | 0.211 | -0.011 | 1.06e-01 | 0.188 | 0.195 | -0.007 | 2.46e-01 |
| Breast_Cancer | LDpred1.Inf | LDpred1.MultiPRS | 0.172 | 0.211 | -0.039 | 2.07e-09 | 0.162 | 0.195 | -0.033 | 1.30e-08 |
| Breast_Cancer | lassosum.MultiPRS | LDpred1.MultiPRS | 0.225 | 0.211 | 0.014 | 3.87e-02 | 0.212 | 0.195 | 0.017 | 1.49e-04 |
| Breast_Cancer | PRScs.MultiPRS | LDpred1.MultiPRS | 0.222 | 0.211 | 0.011 | 1.12e-01 | 0.210 | 0.195 | 0.016 | 2.31e-05 |
| Breast_Cancer | lassosum.10FCVal | LDpred1.MultiPRS | 0.219 | 0.211 | 0.008 | 2.21e-01 | 0.199 | 0.195 | 0.004 | 3.53e-01 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.169 | 0.211 | -0.042 | 1.22e-10 | 0.161 | 0.195 | -0.034 | 5.76e-08 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.217 | 0.211 | 0.006 | 3.30e-01 | 0.211 | 0.195 | 0.016 | 4.98e-04 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.224 | 0.211 | 0.013 | 5.36e-02 | 0.211 | 0.195 | 0.016 | 1.43e-04 |
| Breast_Cancer | SBLUP.Inf | LDpred1.MultiPRS | 0.168 | 0.211 | -0.043 | 4.15e-11 | 0.158 | 0.195 | -0.036 | 6.15e-09 |
| Breast_Cancer | All.MultiPRS | LDpred1.Inf | 0.228 | 0.172 | 0.056 | 2.56e-17 | 0.215 | 0.162 | 0.053 | 3.87e-14 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred1.Inf | 0.169 | 0.172 | -0.003 | 1.11e-02 | 0.161 | 0.162 | -0.001 | 6.44e-01 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred1.Inf | 0.221 | 0.172 | 0.049 | 1.33e-28 | 0.205 | 0.162 | 0.043 | 1.16e-06 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred1.Inf | 0.224 | 0.172 | 0.052 | 1.84e-14 | 0.211 | 0.162 | 0.049 | 3.19e-12 |
| Breast_Cancer | PRScs.PseudoVal | LDpred1.Inf | 0.220 | 0.172 | 0.049 | 5.39e-62 | 0.208 | 0.162 | 0.046 | 4.13e-15 |
| Breast_Cancer | LDpred1.Inf | LDpred1.Inf | 0.172 | 0.172 | 0.000 | 1.00e+00 | 0.162 | 0.162 | 0.000 | 1.00e+00 |
| Breast_Cancer | pT+clump.10FCVal | LDpred1.Inf | 0.179 | 0.172 | 0.007 | 1.61e-01 | 0.167 | 0.162 | 0.005 | 5.92e-01 |
| Breast_Cancer | PRScs.MultiPRS | LDpred1.Inf | 0.222 | 0.172 | 0.050 | 2.33e-13 | 0.210 | 0.162 | 0.049 | 1.62e-14 |
| Breast_Cancer | lassosum.PseudoVal | LDpred1.Inf | 0.217 | 0.172 | 0.045 | 8.22e-25 | 0.199 | 0.162 | 0.037 | 2.34e-05 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred1.Inf | 0.200 | 0.172 | 0.028 | 3.78e-05 | 0.188 | 0.162 | 0.026 | 5.36e-04 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred1.Inf | 0.211 | 0.172 | 0.039 | 2.07e-09 | 0.195 | 0.162 | 0.033 | 1.30e-08 |
| Breast_Cancer | PRScs.10FCVal | LDpred1.Inf | 0.220 | 0.172 | 0.048 | 1.15e-54 | 0.207 | 0.162 | 0.045 | 1.32e-13 |
| Breast_Cancer | LDpred2.Inf | LDpred1.Inf | 0.168 | 0.172 | -0.004 | 1.01e-03 | 0.160 | 0.162 | -0.002 | 4.89e-01 |
| Breast_Cancer | SBLUP.Inf | LDpred1.Inf | 0.168 | 0.172 | -0.004 | 3.48e-04 | 0.158 | 0.162 | -0.004 | 1.23e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred1.Inf | 0.217 | 0.172 | 0.046 | 7.43e-56 | 0.211 | 0.162 | 0.049 | 4.73e-17 |
| Breast_Cancer | lassosum.MultiPRS | LDpred1.Inf | 0.225 | 0.172 | 0.053 | 5.98e-15 | 0.212 | 0.162 | 0.050 | 2.69e-12 |
| Breast_Cancer | LDpred1.10FCVal | LDpred1.Inf | 0.201 | 0.172 | 0.029 | 4.41e-59 | 0.186 | 0.162 | 0.024 | 1.58e-11 |
| Breast_Cancer | lassosum.10FCVal | LDpred1.Inf | 0.219 | 0.172 | 0.047 | 6.72e-37 | 0.199 | 0.162 | 0.037 | 6.56e-07 |
| Breast_Cancer | LDpred2.10FCVal | LDpred1.Inf | 0.223 | 0.172 | 0.052 | 6.15e-61 | 0.211 | 0.162 | 0.049 | 4.56e-15 |
| Breast_Cancer | LDpred1.Inf | LDpred1.10FCVal | 0.172 | 0.201 | -0.029 | 4.41e-59 | 0.162 | 0.186 | -0.024 | 1.58e-11 |
| Breast_Cancer | PRScs.MultiPRS | LDpred1.10FCVal | 0.222 | 0.201 | 0.021 | 1.89e-03 | 0.210 | 0.186 | 0.025 | 4.14e-08 |
| Breast_Cancer | LDpred1.10FCVal | LDpred1.10FCVal | 0.201 | 0.201 | 0.000 | 1.00e+00 | 0.186 | 0.186 | 0.000 | 1.00e+00 |
| Breast_Cancer | All.MultiPRS | LDpred1.10FCVal | 0.228 | 0.201 | 0.027 | 4.03e-05 | 0.215 | 0.186 | 0.029 | 1.35e-08 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred1.10FCVal | 0.221 | 0.201 | 0.020 | 7.87e-09 | 0.205 | 0.186 | 0.019 | 6.82e-03 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.217 | 0.201 | 0.017 | 6.72e-14 | 0.211 | 0.186 | 0.025 | 5.79e-08 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred1.10FCVal | 0.200 | 0.201 | -0.001 | 9.21e-01 | 0.188 | 0.186 | 0.002 | 7.90e-01 |
| Breast_Cancer | lassosum.MultiPRS | LDpred1.10FCVal | 0.225 | 0.201 | 0.024 | 3.39e-04 | 0.212 | 0.186 | 0.026 | 1.03e-06 |
| Breast_Cancer | PRScs.10FCVal | LDpred1.10FCVal | 0.220 | 0.201 | 0.019 | 3.74e-18 | 0.207 | 0.186 | 0.021 | 1.49e-06 |
| Breast_Cancer | lassosum.10FCVal | LDpred1.10FCVal | 0.219 | 0.201 | 0.018 | 5.81e-11 | 0.199 | 0.186 | 0.013 | 2.09e-02 |
| Breast_Cancer | SBLUP.Inf | LDpred1.10FCVal | 0.168 | 0.201 | -0.033 | 1.39e-57 | 0.158 | 0.186 | -0.028 | 3.98e-11 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred1.10FCVal | 0.169 | 0.201 | -0.032 | 1.66e-52 | 0.161 | 0.186 | -0.025 | 2.63e-09 |
| Breast_Cancer | pT+clump.10FCVal | LDpred1.10FCVal | 0.179 | 0.201 | -0.022 | 2.79e-07 | 0.167 | 0.186 | -0.019 | 2.75e-02 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred1.10FCVal | 0.224 | 0.201 | 0.023 | 5.72e-04 | 0.211 | 0.186 | 0.025 | 1.18e-06 |
| Breast_Cancer | PRScs.PseudoVal | LDpred1.10FCVal | 0.220 | 0.201 | 0.020 | 7.40e-22 | 0.208 | 0.186 | 0.022 | 1.50e-07 |
| Breast_Cancer | LDpred2.10FCVal | LDpred1.10FCVal | 0.223 | 0.201 | 0.023 | 1.00e-25 | 0.211 | 0.186 | 0.025 | 9.83e-09 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred1.10FCVal | 0.211 | 0.201 | 0.010 | 1.11e-01 | 0.195 | 0.186 | 0.009 | 2.74e-03 |
| Breast_Cancer | LDpred2.Inf | LDpred1.10FCVal | 0.168 | 0.201 | -0.033 | 1.52e-54 | 0.160 | 0.186 | -0.026 | 1.66e-09 |
| Breast_Cancer | lassosum.PseudoVal | LDpred1.10FCVal | 0.217 | 0.201 | 0.016 | 1.79e-06 | 0.199 | 0.186 | 0.013 | 5.42e-02 |
| Breast_Cancer | LDpred1.Inf | LDpred2.MultiPRS | 0.172 | 0.224 | -0.052 | 1.84e-14 | 0.162 | 0.211 | -0.049 | 3.19e-12 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.217 | 0.224 | -0.007 | 3.19e-01 | 0.211 | 0.211 | 0.000 | 9.77e-01 |
| Breast_Cancer | SBLUP.Inf | LDpred2.MultiPRS | 0.168 | 0.224 | -0.057 | 1.44e-16 | 0.158 | 0.211 | -0.052 | 5.15e-13 |
| Breast_Cancer | LDpred2.Inf | LDpred2.MultiPRS | 0.168 | 0.224 | -0.056 | 1.59e-16 | 0.160 | 0.211 | -0.051 | 1.28e-12 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.MultiPRS | 0.179 | 0.224 | -0.046 | 2.54e-11 | 0.167 | 0.211 | -0.044 | 2.71e-10 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.MultiPRS | 0.223 | 0.224 | -0.001 | 9.09e-01 | 0.211 | 0.211 | 0.000 | 8.35e-01 |
| Breast_Cancer | PRScs.PseudoVal | LDpred2.MultiPRS | 0.220 | 0.224 | -0.004 | 5.70e-01 | 0.208 | 0.211 | -0.003 | 3.58e-01 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.MultiPRS | 0.201 | 0.224 | -0.023 | 5.72e-04 | 0.186 | 0.211 | -0.025 | 1.18e-06 |
| Breast_Cancer | All.MultiPRS | LDpred2.MultiPRS | 0.228 | 0.224 | 0.003 | 6.28e-01 | 0.215 | 0.211 | 0.004 | 5.16e-02 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.221 | 0.224 | -0.003 | 6.20e-01 | 0.205 | 0.211 | -0.006 | 1.61e-01 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.MultiPRS | 0.219 | 0.224 | -0.005 | 4.48e-01 | 0.199 | 0.211 | -0.012 | 1.56e-03 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.200 | 0.224 | -0.024 | 4.03e-04 | 0.188 | 0.211 | -0.023 | 2.14e-05 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.MultiPRS | 0.225 | 0.224 | 0.001 | 5.62e-01 | 0.212 | 0.211 | 0.001 | 6.85e-01 |
| Breast_Cancer | PRScs.10FCVal | LDpred2.MultiPRS | 0.220 | 0.224 | -0.005 | 5.03e-01 | 0.207 | 0.211 | -0.003 | 2.72e-01 |
| Breast_Cancer | PRScs.MultiPRS | LDpred2.MultiPRS | 0.222 | 0.224 | -0.002 | 7.41e-01 | 0.210 | 0.211 | 0.000 | 9.62e-01 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.MultiPRS | 0.217 | 0.224 | -0.007 | 2.90e-01 | 0.199 | 0.211 | -0.011 | 4.64e-03 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.169 | 0.224 | -0.056 | 5.16e-16 | 0.161 | 0.211 | -0.050 | 1.46e-12 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.211 | 0.224 | -0.013 | 5.36e-02 | 0.195 | 0.211 | -0.016 | 1.43e-04 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.224 | 0.224 | 0.000 | 1.00e+00 | 0.211 | 0.211 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred1.Inf | LDpred2.10FCVal | 0.172 | 0.223 | -0.052 | 6.15e-61 | 0.162 | 0.211 | -0.049 | 4.56e-15 |
| Breast_Cancer | PRScs.MultiPRS | LDpred2.10FCVal | 0.222 | 0.223 | -0.001 | 8.29e-01 | 0.210 | 0.211 | -0.001 | 8.37e-01 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.10FCVal | 0.224 | 0.223 | 0.001 | 9.09e-01 | 0.211 | 0.211 | 0.000 | 8.35e-01 |
| Breast_Cancer | PRScs.PseudoVal | LDpred2.10FCVal | 0.220 | 0.223 | -0.003 | 2.71e-02 | 0.208 | 0.211 | -0.003 | 2.28e-01 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.10FCVal | 0.223 | 0.223 | 0.000 | 1.00e+00 | 0.211 | 0.211 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.10FCVal | 0.225 | 0.223 | 0.002 | 8.02e-01 | 0.212 | 0.211 | 0.001 | 7.83e-01 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.10FCVal | 0.201 | 0.223 | -0.023 | 1.00e-25 | 0.186 | 0.211 | -0.025 | 9.83e-09 |
| Breast_Cancer | All.MultiPRS | LDpred2.10FCVal | 0.228 | 0.223 | 0.004 | 5.31e-01 | 0.215 | 0.211 | 0.004 | 1.28e-01 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.10FCVal | 0.221 | 0.223 | -0.003 | 2.44e-01 | 0.205 | 0.211 | -0.006 | 1.76e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.217 | 0.223 | -0.006 | 2.79e-03 | 0.211 | 0.211 | 0.000 | 9.43e-01 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.10FCVal | 0.200 | 0.223 | -0.023 | 6.08e-04 | 0.188 | 0.211 | -0.023 | 6.41e-06 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.10FCVal | 0.217 | 0.223 | -0.006 | 2.21e-03 | 0.199 | 0.211 | -0.012 | 4.64e-03 |
| Breast_Cancer | PRScs.10FCVal | LDpred2.10FCVal | 0.220 | 0.223 | -0.004 | 8.00e-03 | 0.207 | 0.211 | -0.004 | 1.69e-01 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.10FCVal | 0.219 | 0.223 | -0.004 | 7.62e-03 | 0.199 | 0.211 | -0.012 | 2.39e-04 |
| Breast_Cancer | SBLUP.Inf | LDpred2.10FCVal | 0.168 | 0.223 | -0.056 | 2.84e-66 | 0.158 | 0.211 | -0.053 | 7.01e-16 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.10FCVal | 0.169 | 0.223 | -0.055 | 5.75e-68 | 0.161 | 0.211 | -0.050 | 1.61e-15 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.10FCVal | 0.179 | 0.223 | -0.045 | 9.93e-37 | 0.167 | 0.211 | -0.044 | 4.14e-10 |
| Breast_Cancer | LDpred2.Inf | LDpred2.10FCVal | 0.168 | 0.223 | -0.056 | 7.21e-69 | 0.160 | 0.211 | -0.051 | 1.57e-15 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.10FCVal | 0.211 | 0.223 | -0.012 | 5.72e-02 | 0.195 | 0.211 | -0.017 | 1.03e-05 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.PseudoVal | 0.219 | 0.169 | 0.050 | 4.09e-40 | 0.199 | 0.161 | 0.038 | 5.08e-07 |
| Breast_Cancer | SBLUP.Inf | LDpred2.PseudoVal | 0.168 | 0.169 | -0.001 | 1.60e-01 | 0.158 | 0.161 | -0.002 | 1.04e-01 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.221 | 0.169 | 0.052 | 1.73e-31 | 0.205 | 0.161 | 0.044 | 7.91e-07 |
| Breast_Cancer | All.MultiPRS | LDpred2.PseudoVal | 0.228 | 0.169 | 0.059 | 5.12e-19 | 0.215 | 0.161 | 0.054 | 3.92e-14 |
| Breast_Cancer | PRScs.PseudoVal | LDpred2.PseudoVal | 0.220 | 0.169 | 0.052 | 2.09e-66 | 0.208 | 0.161 | 0.047 | 5.51e-15 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.PseudoVal | 0.225 | 0.169 | 0.056 | 1.58e-16 | 0.212 | 0.161 | 0.051 | 2.04e-12 |
| Breast_Cancer | PRScs.MultiPRS | LDpred2.PseudoVal | 0.222 | 0.169 | 0.053 | 8.17e-15 | 0.210 | 0.161 | 0.050 | 2.45e-14 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.224 | 0.169 | 0.056 | 5.16e-16 | 0.211 | 0.161 | 0.050 | 1.46e-12 |
| Breast_Cancer | PRScs.10FCVal | LDpred2.PseudoVal | 0.220 | 0.169 | 0.051 | 2.76e-59 | 0.207 | 0.161 | 0.046 | 1.15e-13 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.169 | 0.169 | 0.000 | 1.00e+00 | 0.161 | 0.161 | 0.000 | 1.00e+00 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.200 | 0.169 | 0.031 | 5.11e-06 | 0.188 | 0.161 | 0.027 | 5.11e-04 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.PseudoVal | 0.179 | 0.169 | 0.010 | 4.74e-02 | 0.167 | 0.161 | 0.006 | 5.26e-01 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.PseudoVal | 0.217 | 0.169 | 0.048 | 4.77e-27 | 0.199 | 0.161 | 0.039 | 2.02e-05 |
| Breast_Cancer | LDpred1.Inf | LDpred2.PseudoVal | 0.172 | 0.169 | 0.003 | 1.11e-02 | 0.162 | 0.161 | 0.001 | 6.44e-01 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.211 | 0.169 | 0.042 | 1.22e-10 | 0.195 | 0.161 | 0.034 | 5.76e-08 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.PseudoVal | 0.201 | 0.169 | 0.032 | 1.66e-52 | 0.186 | 0.161 | 0.025 | 2.63e-09 |
| Breast_Cancer | LDpred2.Inf | LDpred2.PseudoVal | 0.168 | 0.169 | -0.001 | 2.98e-05 | 0.160 | 0.161 | -0.001 | 2.09e-01 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.PseudoVal | 0.223 | 0.169 | 0.055 | 5.75e-68 | 0.211 | 0.161 | 0.050 | 1.61e-15 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.217 | 0.169 | 0.049 | 7.14e-62 | 0.211 | 0.161 | 0.050 | 2.83e-17 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.Inf | 0.200 | 0.168 | 0.032 | 2.58e-06 | 0.188 | 0.160 | 0.028 | 4.44e-04 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.Inf | 0.225 | 0.168 | 0.057 | 4.78e-17 | 0.212 | 0.160 | 0.052 | 1.88e-12 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.Inf | 0.221 | 0.168 | 0.053 | 5.70e-32 | 0.205 | 0.160 | 0.045 | 7.43e-07 |
| Breast_Cancer | All.MultiPRS | LDpred2.Inf | 0.228 | 0.168 | 0.060 | 1.35e-19 | 0.215 | 0.160 | 0.055 | 3.36e-14 |
| Breast_Cancer | SBLUP.Inf | LDpred2.Inf | 0.168 | 0.168 | 0.000 | 9.24e-01 | 0.158 | 0.160 | -0.002 | 2.14e-01 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.Inf | 0.169 | 0.168 | 0.001 | 2.98e-05 | 0.161 | 0.160 | 0.001 | 2.09e-01 |
| Breast_Cancer | PRScs.MultiPRS | LDpred2.Inf | 0.222 | 0.168 | 0.054 | 2.65e-15 | 0.210 | 0.160 | 0.050 | 3.38e-14 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.Inf | 0.224 | 0.168 | 0.056 | 1.59e-16 | 0.211 | 0.160 | 0.051 | 1.28e-12 |
| Breast_Cancer | PRScs.10FCVal | LDpred2.Inf | 0.220 | 0.168 | 0.052 | 3.12e-59 | 0.207 | 0.160 | 0.047 | 1.76e-13 |
| Breast_Cancer | LDpred1.Inf | LDpred2.Inf | 0.172 | 0.168 | 0.004 | 1.01e-03 | 0.162 | 0.160 | 0.002 | 4.89e-01 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.Inf | 0.211 | 0.168 | 0.043 | 4.60e-11 | 0.195 | 0.160 | 0.034 | 4.42e-08 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.Inf | 0.179 | 0.168 | 0.011 | 3.12e-02 | 0.167 | 0.160 | 0.007 | 4.94e-01 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.Inf | 0.217 | 0.168 | 0.049 | 1.60e-27 | 0.199 | 0.160 | 0.039 | 1.85e-05 |
| Breast_Cancer | PRScs.PseudoVal | LDpred2.Inf | 0.220 | 0.168 | 0.053 | 2.64e-66 | 0.208 | 0.160 | 0.048 | 8.61e-15 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.Inf | 0.217 | 0.168 | 0.050 | 4.35e-63 | 0.211 | 0.160 | 0.051 | 2.59e-17 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.Inf | 0.201 | 0.168 | 0.033 | 1.52e-54 | 0.186 | 0.160 | 0.026 | 1.66e-09 |
| Breast_Cancer | LDpred2.Inf | LDpred2.Inf | 0.168 | 0.168 | 0.000 | 1.00e+00 | 0.160 | 0.160 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.Inf | 0.219 | 0.168 | 0.051 | 1.14e-40 | 0.199 | 0.160 | 0.039 | 4.70e-07 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.Inf | 0.223 | 0.168 | 0.056 | 7.21e-69 | 0.211 | 0.160 | 0.051 | 1.57e-15 |
| Breast_Cancer | PRScs.10FCVal | PRScs.MultiPRS | 0.220 | 0.222 | -0.002 | 7.35e-01 | 0.207 | 0.210 | -0.003 | 2.17e-02 |
| Breast_Cancer | LDpred1.10FCVal | PRScs.MultiPRS | 0.201 | 0.222 | -0.021 | 1.89e-03 | 0.186 | 0.210 | -0.025 | 4.14e-08 |
| Breast_Cancer | lassosum.MultiPRS | PRScs.MultiPRS | 0.225 | 0.222 | 0.003 | 6.40e-01 | 0.212 | 0.210 | 0.001 | 6.00e-01 |
| Breast_Cancer | LDpred2.MultiPRS | PRScs.MultiPRS | 0.224 | 0.222 | 0.002 | 7.41e-01 | 0.211 | 0.210 | 0.000 | 9.62e-01 |
| Breast_Cancer | All.MultiPRS | PRScs.MultiPRS | 0.228 | 0.222 | 0.006 | 4.18e-01 | 0.215 | 0.210 | 0.005 | 1.02e-01 |
| Breast_Cancer | PRScs.PseudoVal | PRScs.MultiPRS | 0.220 | 0.222 | -0.002 | 8.13e-01 | 0.208 | 0.210 | -0.003 | 3.31e-02 |
| Breast_Cancer | LDpred1.MultiPRS | PRScs.MultiPRS | 0.211 | 0.222 | -0.011 | 1.12e-01 | 0.195 | 0.210 | -0.016 | 2.31e-05 |
| Breast_Cancer | pT+clump.MultiPRS | PRScs.MultiPRS | 0.200 | 0.222 | -0.022 | 1.34e-03 | 0.188 | 0.210 | -0.023 | 1.31e-06 |
| Breast_Cancer | lassosum.PseudoVal | PRScs.MultiPRS | 0.217 | 0.222 | -0.005 | 4.69e-01 | 0.199 | 0.210 | -0.011 | 3.74e-03 |
| Breast_Cancer | SBayesR.PseudoVal | PRScs.MultiPRS | 0.221 | 0.222 | -0.001 | 8.69e-01 | 0.205 | 0.210 | -0.006 | 1.68e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.217 | 0.222 | -0.005 | 5.06e-01 | 0.211 | 0.210 | 0.000 | 9.39e-01 |
| Breast_Cancer | SBLUP.Inf | PRScs.MultiPRS | 0.168 | 0.222 | -0.054 | 2.49e-15 | 0.158 | 0.210 | -0.052 | 3.63e-15 |
| Breast_Cancer | LDpred2.Inf | PRScs.MultiPRS | 0.168 | 0.222 | -0.054 | 2.65e-15 | 0.160 | 0.210 | -0.050 | 3.38e-14 |
| Breast_Cancer | pT+clump.10FCVal | PRScs.MultiPRS | 0.179 | 0.222 | -0.043 | 2.60e-10 | 0.167 | 0.210 | -0.043 | 6.33e-11 |
| Breast_Cancer | LDpred2.10FCVal | PRScs.MultiPRS | 0.223 | 0.222 | 0.001 | 8.29e-01 | 0.211 | 0.210 | 0.001 | 8.37e-01 |
| Breast_Cancer | lassosum.10FCVal | PRScs.MultiPRS | 0.219 | 0.222 | -0.003 | 6.71e-01 | 0.199 | 0.210 | -0.011 | 9.27e-04 |
| Breast_Cancer | LDpred2.PseudoVal | PRScs.MultiPRS | 0.169 | 0.222 | -0.053 | 8.17e-15 | 0.161 | 0.210 | -0.050 | 2.45e-14 |
| Breast_Cancer | LDpred1.Inf | PRScs.MultiPRS | 0.172 | 0.222 | -0.050 | 2.33e-13 | 0.162 | 0.210 | -0.049 | 1.62e-14 |
| Breast_Cancer | PRScs.MultiPRS | PRScs.MultiPRS | 0.222 | 0.222 | 0.000 | 1.00e+00 | 0.210 | 0.210 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.PseudoVal | PRScs.10FCVal | 0.169 | 0.220 | -0.051 | 2.76e-59 | 0.161 | 0.207 | -0.046 | 1.15e-13 |
| Breast_Cancer | PRScs.10FCVal | PRScs.10FCVal | 0.220 | 0.220 | 0.000 | 1.00e+00 | 0.207 | 0.207 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.MultiPRS | PRScs.10FCVal | 0.224 | 0.220 | 0.005 | 5.03e-01 | 0.211 | 0.207 | 0.003 | 2.72e-01 |
| Breast_Cancer | pT+clump.MultiPRS | PRScs.10FCVal | 0.200 | 0.220 | -0.020 | 4.11e-03 | 0.188 | 0.207 | -0.020 | 7.77e-05 |
| Breast_Cancer | LDpred1.Inf | PRScs.10FCVal | 0.172 | 0.220 | -0.048 | 1.15e-54 | 0.162 | 0.207 | -0.045 | 1.32e-13 |
| Breast_Cancer | LDpred1.MultiPRS | PRScs.10FCVal | 0.211 | 0.220 | -0.009 | 1.88e-01 | 0.195 | 0.207 | -0.013 | 1.30e-03 |
| Breast_Cancer | pT+clump.10FCVal | PRScs.10FCVal | 0.179 | 0.220 | -0.041 | 8.48e-33 | 0.167 | 0.207 | -0.040 | 4.65e-09 |
| Breast_Cancer | lassosum.PseudoVal | PRScs.10FCVal | 0.217 | 0.220 | -0.003 | 2.11e-01 | 0.199 | 0.207 | -0.008 | 6.16e-02 |
| Breast_Cancer | PRScs.PseudoVal | PRScs.10FCVal | 0.220 | 0.220 | 0.001 | 4.96e-01 | 0.208 | 0.207 | 0.001 | 7.91e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | PRScs.10FCVal | 0.217 | 0.220 | -0.002 | 2.02e-01 | 0.211 | 0.207 | 0.004 | 2.99e-01 |
| Breast_Cancer | SBLUP.Inf | PRScs.10FCVal | 0.168 | 0.220 | -0.052 | 1.05e-59 | 0.158 | 0.207 | -0.049 | 1.77e-14 |
| Breast_Cancer | LDpred2.Inf | PRScs.10FCVal | 0.168 | 0.220 | -0.052 | 3.12e-59 | 0.160 | 0.207 | -0.047 | 1.76e-13 |
| Breast_Cancer | lassosum.10FCVal | PRScs.10FCVal | 0.219 | 0.220 | -0.001 | 7.42e-01 | 0.199 | 0.207 | -0.008 | 2.54e-02 |
| Breast_Cancer | LDpred2.10FCVal | PRScs.10FCVal | 0.223 | 0.220 | 0.004 | 8.00e-03 | 0.211 | 0.207 | 0.004 | 1.69e-01 |
| Breast_Cancer | lassosum.MultiPRS | PRScs.10FCVal | 0.225 | 0.220 | 0.005 | 4.20e-01 | 0.212 | 0.207 | 0.005 | 1.14e-01 |
| Breast_Cancer | LDpred1.10FCVal | PRScs.10FCVal | 0.201 | 0.220 | -0.019 | 3.74e-18 | 0.186 | 0.207 | -0.021 | 1.49e-06 |
| Breast_Cancer | All.MultiPRS | PRScs.10FCVal | 0.228 | 0.220 | 0.008 | 2.27e-01 | 0.215 | 0.207 | 0.008 | 1.16e-02 |
| Breast_Cancer | SBayesR.PseudoVal | PRScs.10FCVal | 0.221 | 0.220 | 0.001 | 5.86e-01 | 0.205 | 0.207 | -0.002 | 6.20e-01 |
| Breast_Cancer | PRScs.MultiPRS | PRScs.10FCVal | 0.222 | 0.220 | 0.002 | 7.35e-01 | 0.210 | 0.207 | 0.003 | 2.17e-02 |
| Breast_Cancer | All.MultiPRS | PRScs.PseudoVal | 0.228 | 0.220 | 0.007 | 2.70e-01 | 0.215 | 0.208 | 0.007 | 1.82e-02 |
| Breast_Cancer | pT+clump.MultiPRS | PRScs.PseudoVal | 0.200 | 0.220 | -0.020 | 2.96e-03 | 0.188 | 0.208 | -0.020 | 4.61e-05 |
| Breast_Cancer | LDpred2.PseudoVal | PRScs.PseudoVal | 0.169 | 0.220 | -0.052 | 2.09e-66 | 0.161 | 0.208 | -0.047 | 5.51e-15 |
| Breast_Cancer | SBayesR.PseudoVal | PRScs.PseudoVal | 0.221 | 0.220 | 0.000 | 8.32e-01 | 0.205 | 0.208 | -0.003 | 5.56e-01 |
| Breast_Cancer | LDpred2.MultiPRS | PRScs.PseudoVal | 0.224 | 0.220 | 0.004 | 5.70e-01 | 0.211 | 0.208 | 0.003 | 3.58e-01 |
| Breast_Cancer | SBLUP.Inf | PRScs.PseudoVal | 0.168 | 0.220 | -0.053 | 5.03e-67 | 0.158 | 0.208 | -0.049 | 6.60e-16 |
| Breast_Cancer | LDpred1.Inf | PRScs.PseudoVal | 0.172 | 0.220 | -0.049 | 5.39e-62 | 0.162 | 0.208 | -0.046 | 4.13e-15 |
| Breast_Cancer | PRScs.10FCVal | PRScs.PseudoVal | 0.220 | 0.220 | -0.001 | 4.96e-01 | 0.207 | 0.208 | -0.001 | 7.91e-01 |
| Breast_Cancer | LDpred2.Inf | PRScs.PseudoVal | 0.168 | 0.220 | -0.053 | 2.64e-66 | 0.160 | 0.208 | -0.048 | 8.61e-15 |
| Breast_Cancer | PRScs.PseudoVal | PRScs.PseudoVal | 0.220 | 0.220 | 0.000 | 1.00e+00 | 0.208 | 0.208 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.10FCVal | PRScs.PseudoVal | 0.223 | 0.220 | 0.003 | 2.71e-02 | 0.211 | 0.208 | 0.003 | 2.28e-01 |
| Breast_Cancer | pT+clump.10FCVal | PRScs.PseudoVal | 0.179 | 0.220 | -0.042 | 6.22e-33 | 0.167 | 0.208 | -0.041 | 4.21e-09 |
| Breast_Cancer | PRScs.MultiPRS | PRScs.PseudoVal | 0.222 | 0.220 | 0.002 | 8.13e-01 | 0.210 | 0.208 | 0.003 | 3.31e-02 |
| Breast_Cancer | lassosum.PseudoVal | PRScs.PseudoVal | 0.217 | 0.220 | -0.003 | 1.32e-01 | 0.199 | 0.208 | -0.008 | 5.69e-02 |
| Breast_Cancer | lassosum.10FCVal | PRScs.PseudoVal | 0.219 | 0.220 | -0.001 | 4.82e-01 | 0.199 | 0.208 | -0.009 | 1.94e-02 |
| Breast_Cancer | LDpred1.MultiPRS | PRScs.PseudoVal | 0.211 | 0.220 | -0.009 | 1.54e-01 | 0.195 | 0.208 | -0.013 | 5.03e-04 |
| Breast_Cancer | lassosum.MultiPRS | PRScs.PseudoVal | 0.225 | 0.220 | 0.005 | 4.81e-01 | 0.212 | 0.208 | 0.004 | 1.76e-01 |
| Breast_Cancer | LDpred1.10FCVal | PRScs.PseudoVal | 0.201 | 0.220 | -0.020 | 7.40e-22 | 0.186 | 0.208 | -0.022 | 1.50e-07 |
| Breast_Cancer | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.217 | 0.220 | -0.003 | 8.35e-02 | 0.211 | 0.208 | 0.003 | 3.66e-01 |
| Breast_Cancer | All.MultiPRS | pT+clump.MultiPRS | 0.228 | 0.200 | 0.027 | 5.53e-05 | 0.215 | 0.188 | 0.027 | 6.94e-08 |
| Breast_Cancer | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.221 | 0.200 | 0.021 | 2.34e-03 | 0.205 | 0.188 | 0.017 | 1.95e-03 |
| Breast_Cancer | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.211 | 0.200 | 0.011 | 1.06e-01 | 0.195 | 0.188 | 0.007 | 2.46e-01 |
| Breast_Cancer | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.200 | 0.200 | 0.000 | 1.00e+00 | 0.188 | 0.188 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.PseudoVal | pT+clump.MultiPRS | 0.217 | 0.200 | 0.017 | 1.31e-02 | 0.199 | 0.188 | 0.012 | 3.21e-02 |
| Breast_Cancer | PRScs.10FCVal | pT+clump.MultiPRS | 0.220 | 0.200 | 0.020 | 4.11e-03 | 0.207 | 0.188 | 0.020 | 7.77e-05 |
| Breast_Cancer | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.217 | 0.200 | 0.017 | 1.10e-02 | 0.211 | 0.188 | 0.023 | 3.20e-06 |
| Breast_Cancer | SBLUP.Inf | pT+clump.MultiPRS | 0.168 | 0.200 | -0.032 | 2.47e-06 | 0.158 | 0.188 | -0.029 | 1.42e-04 |
| Breast_Cancer | LDpred2.Inf | pT+clump.MultiPRS | 0.168 | 0.200 | -0.032 | 2.58e-06 | 0.160 | 0.188 | -0.028 | 4.44e-04 |
| Breast_Cancer | pT+clump.10FCVal | pT+clump.MultiPRS | 0.179 | 0.200 | -0.021 | 1.89e-03 | 0.167 | 0.188 | -0.021 | 1.49e-05 |
| Breast_Cancer | LDpred2.10FCVal | pT+clump.MultiPRS | 0.223 | 0.200 | 0.023 | 6.08e-04 | 0.211 | 0.188 | 0.023 | 6.41e-06 |
| Breast_Cancer | PRScs.PseudoVal | pT+clump.MultiPRS | 0.220 | 0.200 | 0.020 | 2.96e-03 | 0.208 | 0.188 | 0.020 | 4.61e-05 |
| Breast_Cancer | LDpred1.10FCVal | pT+clump.MultiPRS | 0.201 | 0.200 | 0.001 | 9.21e-01 | 0.186 | 0.188 | -0.002 | 7.90e-01 |
| Breast_Cancer | LDpred1.Inf | pT+clump.MultiPRS | 0.172 | 0.200 | -0.028 | 3.78e-05 | 0.162 | 0.188 | -0.026 | 5.36e-04 |
| Breast_Cancer | PRScs.MultiPRS | pT+clump.MultiPRS | 0.222 | 0.200 | 0.022 | 1.34e-03 | 0.210 | 0.188 | 0.023 | 1.31e-06 |
| Breast_Cancer | lassosum.10FCVal | pT+clump.MultiPRS | 0.219 | 0.200 | 0.019 | 5.40e-03 | 0.199 | 0.188 | 0.011 | 2.53e-02 |
| Breast_Cancer | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.169 | 0.200 | -0.031 | 5.11e-06 | 0.161 | 0.188 | -0.027 | 5.11e-04 |
| Breast_Cancer | lassosum.MultiPRS | pT+clump.MultiPRS | 0.225 | 0.200 | 0.025 | 2.44e-04 | 0.212 | 0.188 | 0.024 | 7.15e-07 |
| Breast_Cancer | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.224 | 0.200 | 0.024 | 4.03e-04 | 0.211 | 0.188 | 0.023 | 2.14e-05 |
| Breast_Cancer | LDpred2.10FCVal | pT+clump.10FCVal | 0.223 | 0.179 | 0.045 | 9.93e-37 | 0.211 | 0.167 | 0.044 | 4.14e-10 |
| Breast_Cancer | pT+clump.MultiPRS | pT+clump.10FCVal | 0.200 | 0.179 | 0.021 | 1.89e-03 | 0.188 | 0.167 | 0.021 | 1.49e-05 |
| Breast_Cancer | LDpred2.Inf | pT+clump.10FCVal | 0.168 | 0.179 | -0.011 | 3.12e-02 | 0.160 | 0.167 | -0.007 | 4.94e-01 |
| Breast_Cancer | SBayesR.PseudoVal | pT+clump.10FCVal | 0.221 | 0.179 | 0.042 | 6.15e-41 | 0.205 | 0.167 | 0.038 | 1.01e-09 |
| Breast_Cancer | PRScs.MultiPRS | pT+clump.10FCVal | 0.222 | 0.179 | 0.043 | 2.60e-10 | 0.210 | 0.167 | 0.043 | 6.33e-11 |
| Breast_Cancer | SBLUP.Inf | pT+clump.10FCVal | 0.168 | 0.179 | -0.011 | 2.98e-02 | 0.158 | 0.167 | -0.009 | 3.86e-01 |
| Breast_Cancer | lassosum.MultiPRS | pT+clump.10FCVal | 0.225 | 0.179 | 0.047 | 9.73e-12 | 0.212 | 0.167 | 0.045 | 3.14e-12 |
| Breast_Cancer | PRScs.10FCVal | pT+clump.10FCVal | 0.220 | 0.179 | 0.041 | 8.48e-33 | 0.207 | 0.167 | 0.040 | 4.65e-09 |
| Breast_Cancer | LDpred2.MultiPRS | pT+clump.10FCVal | 0.224 | 0.179 | 0.046 | 2.54e-11 | 0.211 | 0.167 | 0.044 | 2.71e-10 |
| Breast_Cancer | lassosum.10FCVal | pT+clump.10FCVal | 0.219 | 0.179 | 0.040 | 3.58e-34 | 0.199 | 0.167 | 0.032 | 1.31e-06 |
| Breast_Cancer | LDpred2.PseudoVal | pT+clump.10FCVal | 0.169 | 0.179 | -0.010 | 4.74e-02 | 0.161 | 0.167 | -0.006 | 5.26e-01 |
| Breast_Cancer | LDpred1.MultiPRS | pT+clump.10FCVal | 0.211 | 0.179 | 0.032 | 9.31e-07 | 0.195 | 0.167 | 0.027 | 5.69e-04 |
| Breast_Cancer | pT+clump.10FCVal | pT+clump.10FCVal | 0.179 | 0.179 | 0.000 | 1.00e+00 | 0.167 | 0.167 | 0.000 | 1.00e+00 |
| Breast_Cancer | All.MultiPRS | pT+clump.10FCVal | 0.228 | 0.179 | 0.049 | 1.19e-13 | 0.215 | 0.167 | 0.048 | 4.23e-13 |
| Breast_Cancer | LDpred1.Inf | pT+clump.10FCVal | 0.172 | 0.179 | -0.007 | 1.61e-01 | 0.162 | 0.167 | -0.005 | 5.92e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.217 | 0.179 | 0.039 | 1.13e-26 | 0.211 | 0.167 | 0.044 | 1.63e-09 |
| Breast_Cancer | LDpred1.10FCVal | pT+clump.10FCVal | 0.201 | 0.179 | 0.022 | 2.79e-07 | 0.186 | 0.167 | 0.019 | 2.75e-02 |
| Breast_Cancer | lassosum.PseudoVal | pT+clump.10FCVal | 0.217 | 0.179 | 0.038 | 5.41e-37 | 0.199 | 0.167 | 0.032 | 7.03e-08 |
| Breast_Cancer | PRScs.PseudoVal | pT+clump.10FCVal | 0.220 | 0.179 | 0.042 | 6.22e-33 | 0.208 | 0.167 | 0.041 | 4.21e-09 |
| Breast_Cancer | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.200 | 0.221 | -0.021 | 2.34e-03 | 0.188 | 0.205 | -0.017 | 1.95e-03 |
| Breast_Cancer | LDpred1.10FCVal | SBayesR.PseudoVal | 0.201 | 0.221 | -0.020 | 7.87e-09 | 0.186 | 0.205 | -0.019 | 6.82e-03 |
| Breast_Cancer | PRScs.10FCVal | SBayesR.PseudoVal | 0.220 | 0.221 | -0.001 | 5.86e-01 | 0.207 | 0.205 | 0.002 | 6.20e-01 |
| Breast_Cancer | LDpred2.10FCVal | SBayesR.PseudoVal | 0.223 | 0.221 | 0.003 | 2.44e-01 | 0.211 | 0.205 | 0.006 | 1.76e-01 |
| Breast_Cancer | lassosum.10FCVal | SBayesR.PseudoVal | 0.219 | 0.221 | -0.002 | 4.32e-01 | 0.199 | 0.205 | -0.006 | 1.88e-01 |
| Breast_Cancer | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.169 | 0.221 | -0.052 | 1.73e-31 | 0.161 | 0.205 | -0.044 | 7.91e-07 |
| Breast_Cancer | lassosum.MultiPRS | SBayesR.PseudoVal | 0.225 | 0.221 | 0.004 | 5.27e-01 | 0.212 | 0.205 | 0.007 | 6.17e-02 |
| Breast_Cancer | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.224 | 0.221 | 0.003 | 6.20e-01 | 0.211 | 0.205 | 0.006 | 1.61e-01 |
| Breast_Cancer | All.MultiPRS | SBayesR.PseudoVal | 0.228 | 0.221 | 0.007 | 3.04e-01 | 0.215 | 0.205 | 0.010 | 2.21e-03 |
| Breast_Cancer | PRScs.PseudoVal | SBayesR.PseudoVal | 0.220 | 0.221 | 0.000 | 8.32e-01 | 0.208 | 0.205 | 0.003 | 5.56e-01 |
| Breast_Cancer | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.211 | 0.221 | -0.010 | 1.36e-01 | 0.195 | 0.205 | -0.010 | 7.84e-02 |
| Breast_Cancer | pT+clump.10FCVal | SBayesR.PseudoVal | 0.179 | 0.221 | -0.042 | 6.15e-41 | 0.167 | 0.205 | -0.038 | 1.01e-09 |
| Breast_Cancer | lassosum.PseudoVal | SBayesR.PseudoVal | 0.217 | 0.221 | -0.004 | 3.57e-02 | 0.199 | 0.205 | -0.006 | 1.10e-01 |
| Breast_Cancer | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.221 | 0.221 | 0.000 | 1.00e+00 | 0.205 | 0.205 | 0.000 | 1.00e+00 |
| Breast_Cancer | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.217 | 0.221 | -0.003 | 1.87e-01 | 0.211 | 0.205 | 0.006 | 2.63e-01 |
| Breast_Cancer | SBLUP.Inf | SBayesR.PseudoVal | 0.168 | 0.221 | -0.053 | 8.23e-32 | 0.158 | 0.205 | -0.047 | 2.98e-07 |
| Breast_Cancer | LDpred2.Inf | SBayesR.PseudoVal | 0.168 | 0.221 | -0.053 | 5.70e-32 | 0.160 | 0.205 | -0.045 | 7.43e-07 |
| Breast_Cancer | LDpred1.Inf | SBayesR.PseudoVal | 0.172 | 0.221 | -0.049 | 1.33e-28 | 0.162 | 0.205 | -0.043 | 1.16e-06 |
| Breast_Cancer | PRScs.MultiPRS | SBayesR.PseudoVal | 0.222 | 0.221 | 0.001 | 8.69e-01 | 0.210 | 0.205 | 0.006 | 1.68e-01 |
| Breast_Cancer | LDpred2.10FCVal | SBLUP.Inf | 0.223 | 0.168 | 0.056 | 2.84e-66 | 0.211 | 0.158 | 0.053 | 7.01e-16 |
| Breast_Cancer | PRScs.PseudoVal | SBLUP.Inf | 0.220 | 0.168 | 0.053 | 5.03e-67 | 0.208 | 0.158 | 0.049 | 6.60e-16 |
| Breast_Cancer | LDpred1.10FCVal | SBLUP.Inf | 0.201 | 0.168 | 0.033 | 1.39e-57 | 0.186 | 0.158 | 0.028 | 3.98e-11 |
| Breast_Cancer | LDpred2.Inf | SBLUP.Inf | 0.168 | 0.168 | 0.000 | 9.24e-01 | 0.160 | 0.158 | 0.002 | 2.14e-01 |
| Breast_Cancer | SBayesR.PseudoVal | SBLUP.Inf | 0.221 | 0.168 | 0.053 | 8.23e-32 | 0.205 | 0.158 | 0.047 | 2.98e-07 |
| Breast_Cancer | lassosum.10FCVal | SBLUP.Inf | 0.219 | 0.168 | 0.051 | 5.81e-41 | 0.199 | 0.158 | 0.041 | 1.27e-07 |
| Breast_Cancer | pT+clump.MultiPRS | SBLUP.Inf | 0.200 | 0.168 | 0.032 | 2.47e-06 | 0.188 | 0.158 | 0.029 | 1.42e-04 |
| Breast_Cancer | lassosum.MultiPRS | SBLUP.Inf | 0.225 | 0.168 | 0.057 | 4.39e-17 | 0.212 | 0.158 | 0.054 | 2.59e-13 |
| Breast_Cancer | PRScs.10FCVal | SBLUP.Inf | 0.220 | 0.168 | 0.052 | 1.05e-59 | 0.207 | 0.158 | 0.049 | 1.77e-14 |
| Breast_Cancer | PRScs.MultiPRS | SBLUP.Inf | 0.222 | 0.168 | 0.054 | 2.49e-15 | 0.210 | 0.158 | 0.052 | 3.63e-15 |
| Breast_Cancer | LDpred1.MultiPRS | SBLUP.Inf | 0.211 | 0.168 | 0.043 | 4.15e-11 | 0.195 | 0.158 | 0.036 | 6.15e-09 |
| Breast_Cancer | LDpred2.PseudoVal | SBLUP.Inf | 0.169 | 0.168 | 0.001 | 1.60e-01 | 0.161 | 0.158 | 0.002 | 1.04e-01 |
| Breast_Cancer | LDpred1.Inf | SBLUP.Inf | 0.172 | 0.168 | 0.004 | 3.48e-04 | 0.162 | 0.158 | 0.004 | 1.23e-01 |
| Breast_Cancer | LDpred2.MultiPRS | SBLUP.Inf | 0.224 | 0.168 | 0.057 | 1.44e-16 | 0.211 | 0.158 | 0.052 | 5.15e-13 |
| Breast_Cancer | All.MultiPRS | SBLUP.Inf | 0.228 | 0.168 | 0.060 | 1.18e-19 | 0.215 | 0.158 | 0.057 | 6.47e-15 |
| Breast_Cancer | SBLUP.Inf | SBLUP.Inf | 0.168 | 0.168 | 0.000 | 1.00e+00 | 0.158 | 0.158 | 0.000 | 1.00e+00 |
| Breast_Cancer | DBSLMM.PseudoVal | SBLUP.Inf | 0.217 | 0.168 | 0.050 | 7.26e-65 | 0.211 | 0.158 | 0.052 | 6.94e-19 |
| Breast_Cancer | pT+clump.10FCVal | SBLUP.Inf | 0.179 | 0.168 | 0.011 | 2.98e-02 | 0.167 | 0.158 | 0.009 | 3.86e-01 |
| Breast_Cancer | lassosum.PseudoVal | SBLUP.Inf | 0.217 | 0.168 | 0.049 | 1.43e-27 | 0.199 | 0.158 | 0.041 | 7.57e-06 |
Show cross-validation differences between methods
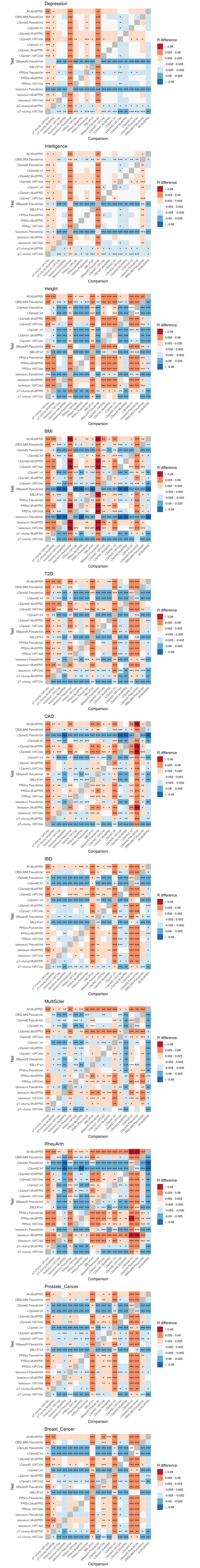
Predictive utility estimated using 10-fold cross-validation
Show test-validation differences between methods
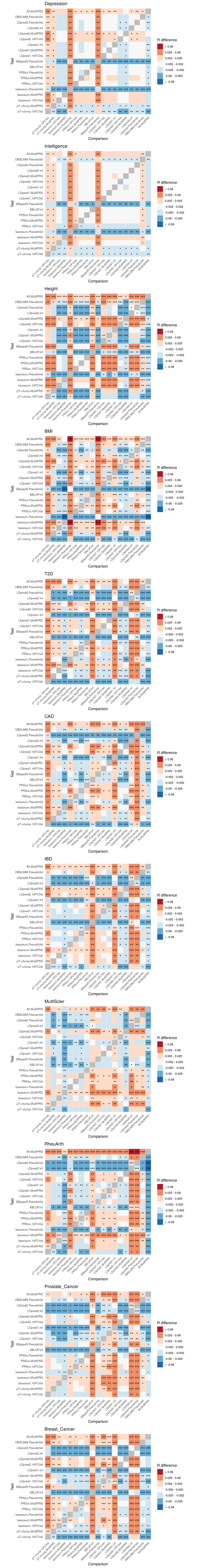
Predictive utility estimated using test set validation
4.1.2 TEDS
Show pT+clump method results
| Phenotype | Method | Model | CrossVal R (SE) | IndepVal R (SE) |
|---|---|---|---|---|
| Height21 | pT+clump (sparse) | 10FCVal | 0.387 (0.014) | 0.353 (0.028) |
| Height21 | pT+clump (sparse) | Multi-PRS | 0.4 (0.014) | 0.388 (0.028) |
| Height21 | pT+clump (non-nested) | 10FCVal | 0.326 (0.014) | 0.305 (0.029) |
| Height21 | pT+clump (non-nested) | Multi-PRS | 0.398 (0.014) | 0.386 (0.028) |
| Height21 | pT+clump (dense) | 10FCVal | 0.396 (0.014) | 0.366 (0.028) |
| Height21 | pT+clump (dense) | Multi-PRS | 0.405 (0.014) | 0.391 (0.028) |
| BMI21 | pT+clump (sparse) | 10FCVal | 0.31 (0.015) | 0.301 (0.03) |
| BMI21 | pT+clump (sparse) | Multi-PRS | 0.317 (0.015) | 0.295 (0.03) |
| BMI21 | pT+clump (non-nested) | 10FCVal | 0.214 (0.015) | 0.179 (0.03) |
| BMI21 | pT+clump (non-nested) | Multi-PRS | 0.313 (0.015) | 0.291 (0.03) |
| BMI21 | pT+clump (dense) | 10FCVal | 0.31 (0.015) | 0.302 (0.03) |
| BMI21 | pT+clump (dense) | Multi-PRS | 0.317 (0.015) | 0.295 (0.03) |
| GCSE | pT+clump (sparse) | 10FCVal | 0.373 (0.012) | 0.311 (0.025) |
| GCSE | pT+clump (sparse) | Multi-PRS | 0.381 (0.012) | 0.328 (0.025) |
| GCSE | pT+clump (non-nested) | 10FCVal | 0.256 (0.013) | 0.232 (0.025) |
| GCSE | pT+clump (non-nested) | Multi-PRS | 0.381 (0.012) | 0.328 (0.025) |
| GCSE | pT+clump (dense) | 10FCVal | 0.373 (0.012) | 0.315 (0.025) |
| GCSE | pT+clump (dense) | Multi-PRS | 0.38 (0.012) | 0.331 (0.025) |
| ADHD | pT+clump (sparse) | 10FCVal | 0.094 (0.013) | 0.15 (0.025) |
| ADHD | pT+clump (sparse) | Multi-PRS | 0.095 (0.013) | 0.162 (0.025) |
| ADHD | pT+clump (non-nested) | 10FCVal | 0.064 (0.013) | 0.105 (0.025) |
| ADHD | pT+clump (non-nested) | Multi-PRS | 0.09 (0.013) | 0.155 (0.025) |
| ADHD | pT+clump (dense) | 10FCVal | 0.095 (0.013) | 0.148 (0.025) |
| ADHD | pT+clump (dense) | Multi-PRS | 0.108 (0.018) | 0.155 (0.025) |
Show cross-validation results
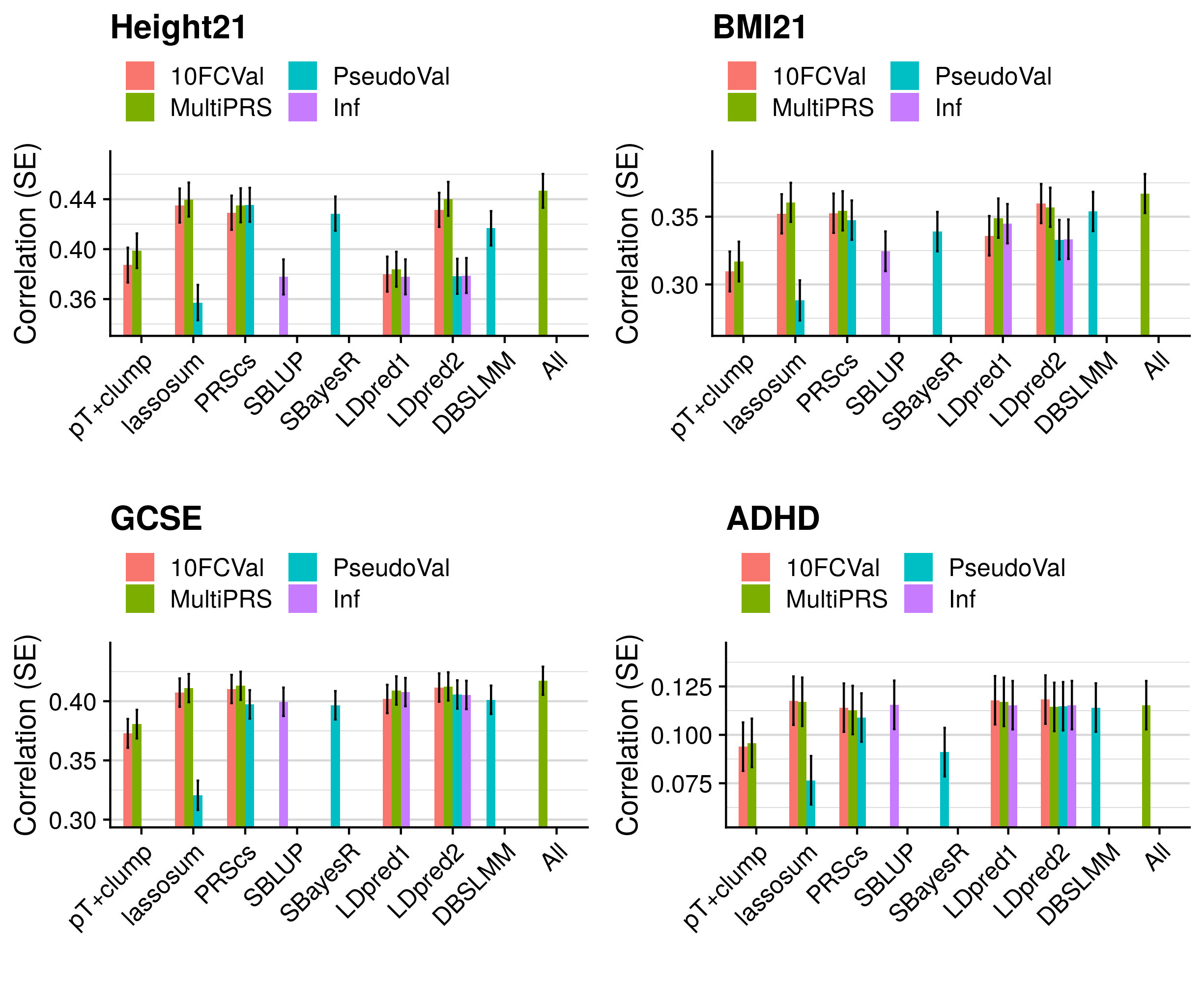
Predictive utility estimated using 10-fold cross-validation
Show test-validation results
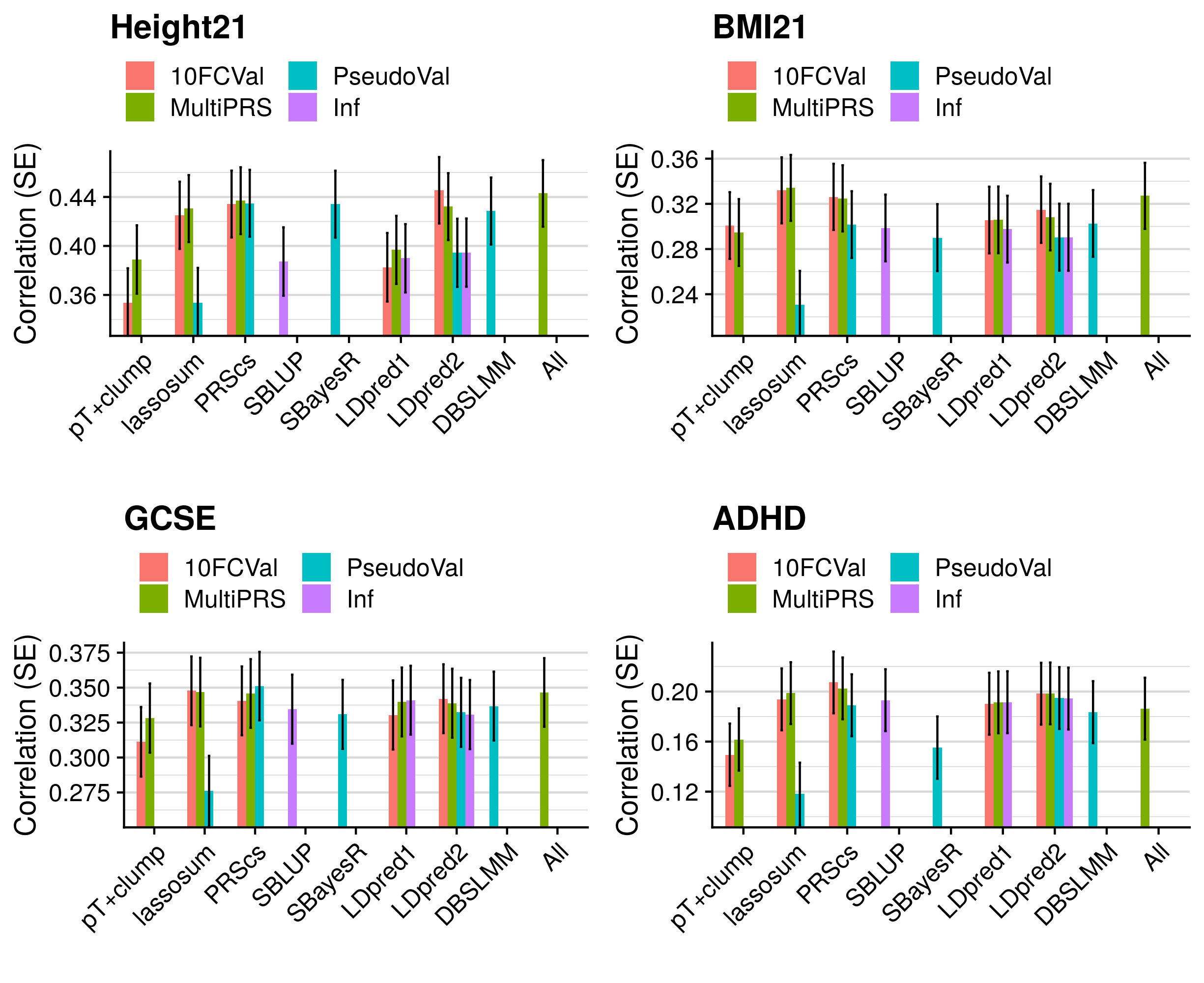
Predictive utility estimated using test set validation
Show TEDS results table
| Phenotype | Method | Model | CrossVal_R | CrossVal_R_SE | CrossVal_pval | IndepVal_R | IndepVal_R_SE | IndepVal_pval |
|---|---|---|---|---|---|---|---|---|
| Height21 | All | MultiPRS | 0.447 | 0.014 | 4.31e-213 | 0.443 | 0.027 | 1.26e-53 |
| Height21 | DBSLMM | PseudoVal | 0.417 | 0.014 | 6.59e-183 | 0.428 | 0.027 | 6.01e-50 |
| Height21 | lassosum | MultiPRS | 0.440 | 0.014 | 7.96e-206 | 0.431 | 0.027 | 1.88e-50 |
| Height21 | lassosum | 10FCVal | 0.435 | 0.014 | 6.67e-201 | 0.425 | 0.027 | 4.36e-49 |
| Height21 | lassosum | PseudoVal | 0.357 | 0.014 | 1.64e-131 | 0.354 | 0.028 | 1.63e-33 |
| Height21 | LDpred1 | MultiPRS | 0.384 | 0.014 | 2.94e-153 | 0.397 | 0.028 | 1.88e-42 |
| Height21 | LDpred1 | Inf | 0.378 | 0.014 | 4.25e-148 | 0.390 | 0.028 | 6.17e-41 |
| Height21 | LDpred1 | 10FCVal | 0.380 | 0.014 | 6.34e-150 | 0.383 | 0.028 | 2.33e-39 |
| Height21 | LDpred2 | MultiPRS | 0.440 | 0.014 | 2.09e-206 | 0.432 | 0.027 | 7.46e-51 |
| Height21 | LDpred2 | 10FCVal | 0.431 | 0.014 | 2.19e-197 | 0.445 | 0.027 | 2.71e-54 |
| Height21 | LDpred2 | PseudoVal | 0.378 | 0.014 | 1.83e-148 | 0.394 | 0.028 | 6.43e-42 |
| Height21 | LDpred2 | Inf | 0.379 | 0.014 | 5.33e-149 | 0.395 | 0.028 | 5.92e-42 |
| Height21 | PRScs | MultiPRS | 0.435 | 0.014 | 4.30e-201 | 0.437 | 0.027 | 4.40e-52 |
| Height21 | PRScs | 10FCVal | 0.429 | 0.014 | 3.85e-195 | 0.434 | 0.027 | 2.21e-51 |
| Height21 | PRScs | PseudoVal | 0.436 | 0.014 | 1.70e-201 | 0.435 | 0.027 | 1.53e-51 |
| Height21 | pT+clump | MultiPRS | 0.399 | 0.014 | 3.07e-166 | 0.389 | 0.028 | 1.01e-40 |
| Height21 | pT+clump | 10FCVal | 0.387 | 0.014 | 4.03e-156 | 0.353 | 0.028 | 1.90e-33 |
| Height21 | SBayesR | PseudoVal | 0.428 | 0.014 | 2.12e-194 | 0.434 | 0.027 | 2.34e-51 |
| Height21 | SBLUP | Inf | 0.378 | 0.014 | 4.90e-148 | 0.387 | 0.028 | 2.32e-40 |
| BMI21 | All | MultiPRS | 0.367 | 0.014 | 2.08e-133 | 0.327 | 0.029 | 1.88e-27 |
| BMI21 | DBSLMM | PseudoVal | 0.354 | 0.014 | 1.97e-123 | 0.303 | 0.030 | 1.47e-23 |
| BMI21 | lassosum | MultiPRS | 0.361 | 0.014 | 1.89e-128 | 0.334 | 0.029 | 1.21e-28 |
| BMI21 | lassosum | 10FCVal | 0.352 | 0.014 | 3.69e-122 | 0.332 | 0.029 | 2.79e-28 |
| BMI21 | lassosum | PseudoVal | 0.288 | 0.015 | 1.08e-80 | 0.231 | 0.030 | 4.64e-14 |
| BMI21 | LDpred1 | MultiPRS | 0.349 | 0.015 | 7.25e-120 | 0.306 | 0.029 | 4.85e-24 |
| BMI21 | LDpred1 | Inf | 0.345 | 0.015 | 6.27e-117 | 0.298 | 0.030 | 8.50e-23 |
| BMI21 | LDpred1 | 10FCVal | 0.336 | 0.015 | 1.04e-110 | 0.306 | 0.029 | 5.09e-24 |
| BMI21 | LDpred2 | MultiPRS | 0.357 | 0.014 | 1.02e-125 | 0.308 | 0.029 | 2.00e-24 |
| BMI21 | LDpred2 | 10FCVal | 0.360 | 0.014 | 9.14e-128 | 0.315 | 0.029 | 1.84e-25 |
| BMI21 | LDpred2 | PseudoVal | 0.333 | 0.015 | 1.09e-108 | 0.290 | 0.030 | 9.52e-22 |
| BMI21 | LDpred2 | Inf | 0.333 | 0.015 | 6.02e-109 | 0.290 | 0.030 | 9.80e-22 |
| BMI21 | PRScs | MultiPRS | 0.354 | 0.014 | 8.88e-124 | 0.325 | 0.029 | 4.41e-27 |
| BMI21 | PRScs | 10FCVal | 0.353 | 0.014 | 1.69e-122 | 0.326 | 0.029 | 2.69e-27 |
| BMI21 | PRScs | PseudoVal | 0.347 | 0.015 | 8.41e-119 | 0.302 | 0.030 | 2.10e-23 |
| BMI21 | pT+clump | MultiPRS | 0.317 | 0.015 | 4.72e-98 | 0.295 | 0.030 | 2.36e-22 |
| BMI21 | pT+clump | 10FCVal | 0.310 | 0.015 | 1.99e-93 | 0.301 | 0.030 | 2.84e-23 |
| BMI21 | SBayesR | PseudoVal | 0.339 | 0.015 | 8.16e-113 | 0.290 | 0.030 | 1.09e-21 |
| BMI21 | SBLUP | Inf | 0.324 | 0.015 | 5.56e-103 | 0.299 | 0.030 | 6.00e-23 |
| GCSE | All | MultiPRS | 0.417 | 0.012 | 1.14e-244 | 0.347 | 0.025 | 1.87e-42 |
| GCSE | DBSLMM | PseudoVal | 0.401 | 0.012 | 1.23e-224 | 0.337 | 0.025 | 4.74e-40 |
| GCSE | lassosum | MultiPRS | 0.411 | 0.012 | 7.98e-237 | 0.347 | 0.025 | 1.61e-42 |
| GCSE | lassosum | 10FCVal | 0.407 | 0.012 | 4.55e-232 | 0.348 | 0.025 | 9.42e-43 |
| GCSE | lassosum | PseudoVal | 0.321 | 0.012 | 1.50e-139 | 0.276 | 0.025 | 6.05e-27 |
| GCSE | LDpred1 | MultiPRS | 0.409 | 0.012 | 2.61e-234 | 0.340 | 0.025 | 9.04e-41 |
| GCSE | LDpred1 | Inf | 0.408 | 0.012 | 1.33e-232 | 0.341 | 0.025 | 4.43e-41 |
| GCSE | LDpred1 | 10FCVal | 0.402 | 0.012 | 1.72e-225 | 0.330 | 0.025 | 1.55e-38 |
| GCSE | LDpred2 | MultiPRS | 0.412 | 0.012 | 1.31e-238 | 0.339 | 0.025 | 1.49e-40 |
| GCSE | LDpred2 | 10FCVal | 0.412 | 0.012 | 2.30e-237 | 0.342 | 0.025 | 2.45e-41 |
| GCSE | LDpred2 | PseudoVal | 0.406 | 0.012 | 4.26e-230 | 0.332 | 0.025 | 5.58e-39 |
| GCSE | LDpred2 | Inf | 0.405 | 0.012 | 1.23e-229 | 0.331 | 0.025 | 1.34e-38 |
| GCSE | PRScs | MultiPRS | 0.413 | 0.012 | 3.03e-239 | 0.346 | 0.025 | 2.86e-42 |
| GCSE | PRScs | 10FCVal | 0.410 | 0.012 | 7.43e-236 | 0.341 | 0.025 | 5.82e-41 |
| GCSE | PRScs | PseudoVal | 0.397 | 0.012 | 5.80e-220 | 0.351 | 0.025 | 1.34e-43 |
| GCSE | pT+clump | MultiPRS | 0.381 | 0.012 | 1.40e-200 | 0.328 | 0.025 | 5.13e-38 |
| GCSE | pT+clump | 10FCVal | 0.373 | 0.012 | 6.29e-192 | 0.311 | 0.025 | 3.63e-34 |
| GCSE | SBayesR | PseudoVal | 0.397 | 0.012 | 4.85e-219 | 0.331 | 0.025 | 1.23e-38 |
| GCSE | SBLUP | Inf | 0.400 | 0.012 | 1.37e-222 | 0.335 | 0.025 | 1.61e-39 |
| ADHD | All | MultiPRS | 0.115 | 0.013 | 4.12e-20 | 0.186 | 0.025 | 8.96e-14 |
| ADHD | DBSLMM | PseudoVal | 0.114 | 0.013 | 1.00e-19 | 0.184 | 0.025 | 2.11e-13 |
| ADHD | lassosum | MultiPRS | 0.117 | 0.013 | 1.09e-20 | 0.199 | 0.025 | 1.69e-15 |
| ADHD | lassosum | PseudoVal | 0.077 | 0.013 | 1.17e-09 | 0.118 | 0.025 | 2.58e-06 |
| ADHD | lassosum | 10FCVal | 0.118 | 0.013 | 7.06e-21 | 0.194 | 0.025 | 8.52e-15 |
| ADHD | LDpred1 | MultiPRS | 0.117 | 0.013 | 1.13e-20 | 0.191 | 0.025 | 1.83e-14 |
| ADHD | LDpred1 | Inf | 0.115 | 0.013 | 4.11e-20 | 0.191 | 0.025 | 1.77e-14 |
| ADHD | LDpred1 | 10FCVal | 0.118 | 0.013 | 6.01e-21 | 0.190 | 0.025 | 2.60e-14 |
| ADHD | LDpred2 | MultiPRS | 0.114 | 0.013 | 7.89e-20 | 0.199 | 0.025 | 1.81e-15 |
| ADHD | LDpred2 | 10FCVal | 0.118 | 0.013 | 4.58e-21 | 0.198 | 0.025 | 1.97e-15 |
| ADHD | LDpred2 | PseudoVal | 0.115 | 0.013 | 6.15e-20 | 0.195 | 0.025 | 6.11e-15 |
| ADHD | LDpred2 | Inf | 0.115 | 0.013 | 3.98e-20 | 0.194 | 0.025 | 6.97e-15 |
| ADHD | PRScs | MultiPRS | 0.113 | 0.013 | 2.60e-19 | 0.203 | 0.025 | 4.72e-16 |
| ADHD | PRScs | 10FCVal | 0.114 | 0.013 | 1.06e-19 | 0.207 | 0.025 | 9.33e-17 |
| ADHD | PRScs | PseudoVal | 0.109 | 0.013 | 4.15e-18 | 0.189 | 0.025 | 3.84e-14 |
| ADHD | pT+clump | MultiPRS | 0.096 | 0.013 | 2.43e-14 | 0.162 | 0.025 | 1.09e-10 |
| ADHD | pT+clump | 10FCVal | 0.094 | 0.013 | 7.96e-14 | 0.150 | 0.025 | 2.46e-09 |
| ADHD | SBayesR | PseudoVal | 0.091 | 0.013 | 4.32e-13 | 0.155 | 0.025 | 5.89e-10 |
| ADHD | SBLUP | Inf | 0.116 | 0.013 | 3.56e-20 | 0.193 | 0.025 | 1.06e-14 |
Show cross-validation results compared to pT+clump
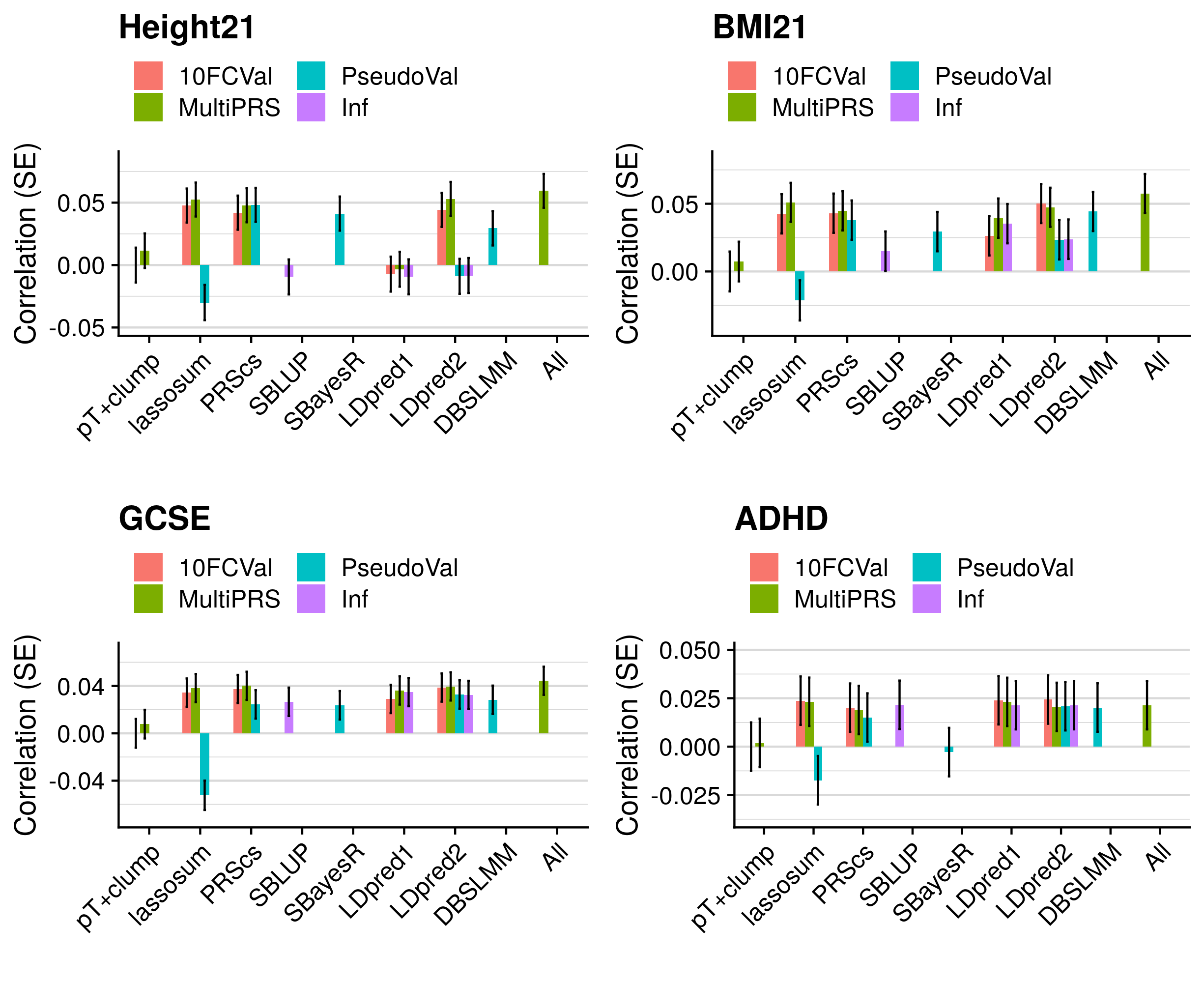
Predictive utility estimated using 10-fold cross-validation
Show test-validation results compared to pT+clump
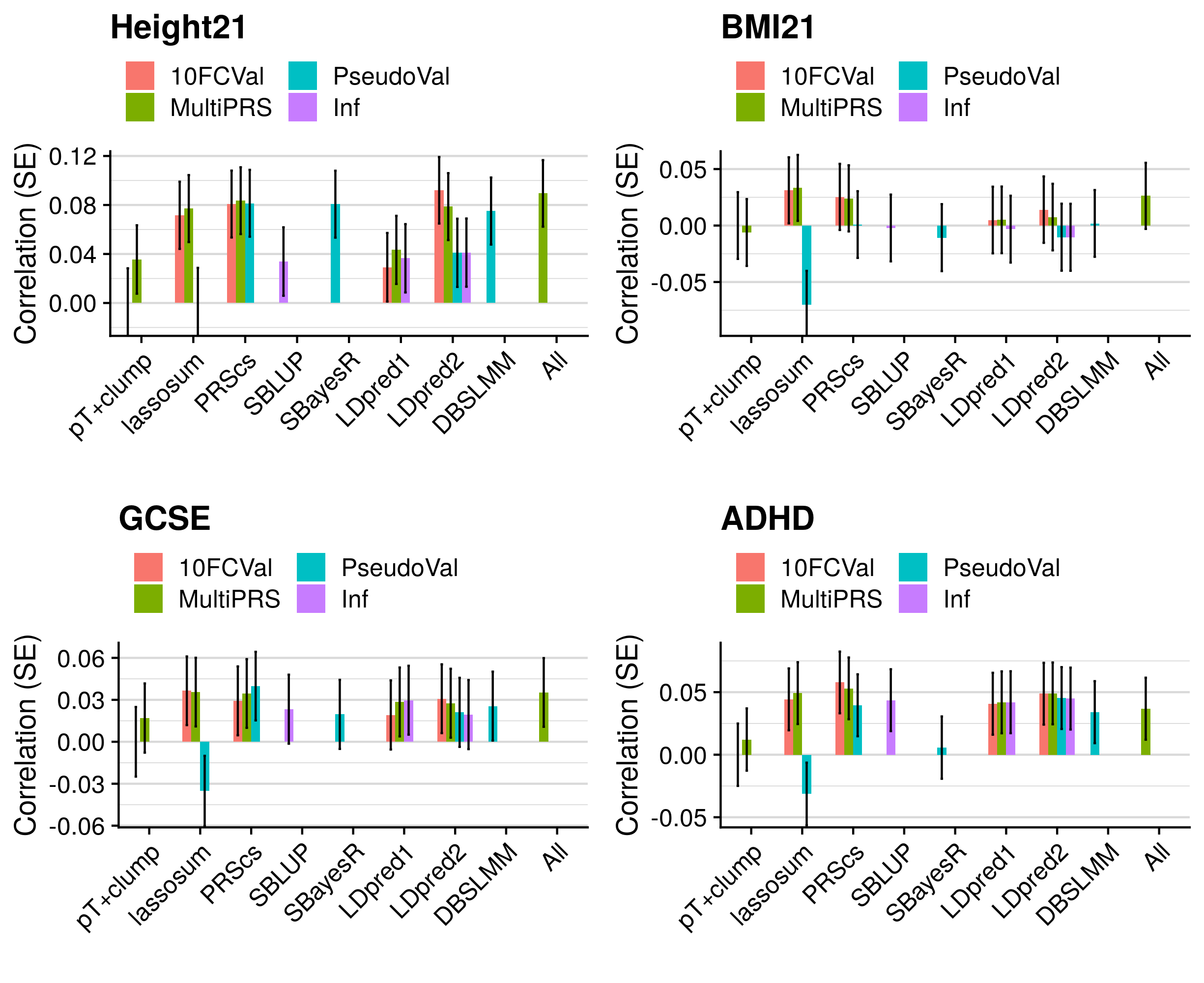
Predictive utility estimated using test set validation
Show difference results table
| Phenotype | Model 1 Test | Model 2 Test | Model 1 CrossVal R | Model 2 CrossVal R | CrossVal R Diff | CrossVal R Diff P | Model 1 IndepVal R | Model 2 IndepVal R | IndepVal R Diff | IndepVal R Diff P |
|---|---|---|---|---|---|---|---|---|---|---|
| Height21 | LDpred2.PseudoVal | All.MultiPRS | 0.378 | 0.447 | -0.068 | 2.06e-04 | 0.394 | 0.443 | -0.049 | 9.86e-04 |
| Height21 | pT+clump.10FCVal | All.MultiPRS | 0.387 | 0.447 | -0.059 | 1.38e-03 | 0.353 | 0.443 | -0.089 | 1.34e-09 |
| Height21 | LDpred2.MultiPRS | All.MultiPRS | 0.440 | 0.447 | -0.006 | 7.22e-01 | 0.432 | 0.443 | -0.011 | 1.53e-01 |
| Height21 | PRScs.PseudoVal | All.MultiPRS | 0.436 | 0.447 | -0.011 | 5.35e-01 | 0.435 | 0.443 | -0.008 | 2.60e-01 |
| Height21 | LDpred1.Inf | All.MultiPRS | 0.378 | 0.447 | -0.069 | 1.85e-04 | 0.390 | 0.443 | -0.053 | 2.98e-04 |
| Height21 | PRScs.MultiPRS | All.MultiPRS | 0.435 | 0.447 | -0.012 | 1.17e-03 | 0.437 | 0.443 | -0.006 | 3.71e-01 |
| Height21 | LDpred2.Inf | All.MultiPRS | 0.379 | 0.447 | -0.068 | 2.36e-04 | 0.395 | 0.443 | -0.048 | 9.47e-04 |
| Height21 | lassosum.PseudoVal | All.MultiPRS | 0.357 | 0.447 | -0.089 | 1.52e-06 | 0.354 | 0.443 | -0.089 | 9.71e-08 |
| Height21 | LDpred2.10FCVal | All.MultiPRS | 0.431 | 0.447 | -0.015 | 3.99e-01 | 0.445 | 0.443 | 0.003 | 7.50e-01 |
| Height21 | lassosum.MultiPRS | All.MultiPRS | 0.440 | 0.447 | -0.007 | 6.99e-01 | 0.431 | 0.443 | -0.012 | 3.78e-02 |
| Height21 | LDpred1.10FCVal | All.MultiPRS | 0.380 | 0.447 | -0.067 | 2.88e-04 | 0.383 | 0.443 | -0.060 | 7.89e-05 |
| Height21 | All.MultiPRS | All.MultiPRS | 0.447 | 0.447 | 0.000 | 1.00e+00 | 0.443 | 0.443 | 0.000 | 1.00e+00 |
| Height21 | SBayesR.PseudoVal | All.MultiPRS | 0.428 | 0.447 | -0.018 | 3.14e-01 | 0.434 | 0.443 | -0.009 | 3.23e-01 |
| Height21 | LDpred1.MultiPRS | All.MultiPRS | 0.384 | 0.447 | -0.063 | 6.29e-04 | 0.397 | 0.443 | -0.046 | 1.03e-03 |
| Height21 | pT+clump.MultiPRS | All.MultiPRS | 0.399 | 0.447 | -0.048 | 8.50e-03 | 0.389 | 0.443 | -0.054 | 7.39e-06 |
| Height21 | lassosum.10FCVal | All.MultiPRS | 0.435 | 0.447 | -0.012 | 5.14e-01 | 0.425 | 0.443 | -0.018 | 1.65e-02 |
| Height21 | PRScs.10FCVal | All.MultiPRS | 0.429 | 0.447 | -0.017 | 3.34e-01 | 0.434 | 0.443 | -0.009 | 2.44e-01 |
| Height21 | DBSLMM.PseudoVal | All.MultiPRS | 0.417 | 0.447 | -0.030 | 9.92e-02 | 0.428 | 0.443 | -0.014 | 1.51e-01 |
| Height21 | SBLUP.Inf | All.MultiPRS | 0.378 | 0.447 | -0.069 | 1.82e-04 | 0.387 | 0.443 | -0.056 | 2.84e-04 |
| Height21 | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.428 | 0.417 | 0.012 | 1.08e-01 | 0.434 | 0.428 | 0.006 | 6.83e-01 |
| Height21 | LDpred2.Inf | DBSLMM.PseudoVal | 0.379 | 0.417 | -0.038 | 1.96e-09 | 0.395 | 0.428 | -0.034 | 8.97e-03 |
| Height21 | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.387 | 0.417 | -0.029 | 1.69e-03 | 0.353 | 0.428 | -0.075 | 5.64e-05 |
| Height21 | PRScs.10FCVal | DBSLMM.PseudoVal | 0.429 | 0.417 | 0.013 | 2.65e-02 | 0.434 | 0.428 | 0.006 | 5.88e-01 |
| Height21 | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.436 | 0.417 | 0.019 | 1.39e-04 | 0.435 | 0.428 | 0.006 | 5.00e-01 |
| Height21 | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.399 | 0.417 | -0.018 | 3.30e-01 | 0.389 | 0.428 | -0.040 | 3.94e-03 |
| Height21 | LDpred1.Inf | DBSLMM.PseudoVal | 0.378 | 0.417 | -0.039 | 4.36e-10 | 0.390 | 0.428 | -0.039 | 1.86e-03 |
| Height21 | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.440 | 0.417 | 0.023 | 2.06e-01 | 0.431 | 0.428 | 0.002 | 8.45e-01 |
| Height21 | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.431 | 0.417 | 0.015 | 6.35e-03 | 0.445 | 0.428 | 0.017 | 1.08e-01 |
| Height21 | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.357 | 0.417 | -0.059 | 6.20e-08 | 0.354 | 0.428 | -0.075 | 3.14e-04 |
| Height21 | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.380 | 0.417 | -0.037 | 8.68e-08 | 0.383 | 0.428 | -0.046 | 1.34e-03 |
| Height21 | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.384 | 0.417 | -0.033 | 7.50e-02 | 0.397 | 0.428 | -0.032 | 1.13e-02 |
| Height21 | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.435 | 0.417 | 0.018 | 3.13e-01 | 0.437 | 0.428 | 0.008 | 3.43e-01 |
| Height21 | All.MultiPRS | DBSLMM.PseudoVal | 0.447 | 0.417 | 0.030 | 9.92e-02 | 0.443 | 0.428 | 0.014 | 1.51e-01 |
| Height21 | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.378 | 0.417 | -0.038 | 1.30e-09 | 0.394 | 0.428 | -0.034 | 8.94e-03 |
| Height21 | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.417 | 0.417 | 0.000 | 1.00e+00 | 0.428 | 0.428 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.440 | 0.417 | 0.024 | 1.97e-01 | 0.432 | 0.428 | 0.004 | 7.74e-01 |
| Height21 | lassosum.10FCVal | DBSLMM.PseudoVal | 0.435 | 0.417 | 0.018 | 5.36e-03 | 0.425 | 0.428 | -0.003 | 7.82e-01 |
| Height21 | SBLUP.Inf | DBSLMM.PseudoVal | 0.378 | 0.417 | -0.039 | 3.12e-10 | 0.387 | 0.428 | -0.041 | 1.27e-03 |
| Height21 | SBayesR.PseudoVal | lassosum.MultiPRS | 0.428 | 0.440 | -0.011 | 5.35e-01 | 0.434 | 0.431 | 0.004 | 7.46e-01 |
| Height21 | LDpred2.10FCVal | lassosum.MultiPRS | 0.431 | 0.440 | -0.008 | 6.49e-01 | 0.445 | 0.431 | 0.015 | 1.25e-01 |
| Height21 | LDpred1.10FCVal | lassosum.MultiPRS | 0.380 | 0.440 | -0.060 | 1.23e-03 | 0.383 | 0.431 | -0.048 | 2.96e-03 |
| Height21 | LDpred2.Inf | lassosum.MultiPRS | 0.379 | 0.440 | -0.061 | 1.07e-03 | 0.395 | 0.431 | -0.036 | 1.88e-02 |
| Height21 | PRScs.10FCVal | lassosum.MultiPRS | 0.429 | 0.440 | -0.011 | 5.61e-01 | 0.434 | 0.431 | 0.004 | 6.32e-01 |
| Height21 | PRScs.MultiPRS | lassosum.MultiPRS | 0.435 | 0.440 | -0.005 | 7.98e-01 | 0.437 | 0.431 | 0.006 | 3.51e-01 |
| Height21 | pT+clump.MultiPRS | lassosum.MultiPRS | 0.399 | 0.440 | -0.041 | 2.69e-02 | 0.389 | 0.431 | -0.042 | 1.31e-03 |
| Height21 | lassosum.MultiPRS | lassosum.MultiPRS | 0.440 | 0.440 | 0.000 | 1.00e+00 | 0.431 | 0.431 | 0.000 | 1.00e+00 |
| Height21 | lassosum.10FCVal | lassosum.MultiPRS | 0.435 | 0.440 | -0.005 | 7.90e-01 | 0.425 | 0.431 | -0.005 | 2.83e-01 |
| Height21 | LDpred2.MultiPRS | lassosum.MultiPRS | 0.440 | 0.440 | 0.001 | 9.75e-01 | 0.432 | 0.431 | 0.002 | 8.71e-01 |
| Height21 | LDpred1.MultiPRS | lassosum.MultiPRS | 0.384 | 0.440 | -0.056 | 2.53e-03 | 0.397 | 0.431 | -0.034 | 2.33e-02 |
| Height21 | LDpred2.PseudoVal | lassosum.MultiPRS | 0.378 | 0.440 | -0.061 | 9.49e-04 | 0.394 | 0.431 | -0.036 | 1.92e-02 |
| Height21 | LDpred1.Inf | lassosum.MultiPRS | 0.378 | 0.440 | -0.062 | 8.53e-04 | 0.390 | 0.431 | -0.041 | 7.00e-03 |
| Height21 | pT+clump.10FCVal | lassosum.MultiPRS | 0.387 | 0.440 | -0.052 | 4.46e-03 | 0.353 | 0.431 | -0.077 | 2.15e-06 |
| Height21 | lassosum.PseudoVal | lassosum.MultiPRS | 0.357 | 0.440 | -0.083 | 9.39e-06 | 0.354 | 0.431 | -0.077 | 1.98e-06 |
| Height21 | SBLUP.Inf | lassosum.MultiPRS | 0.378 | 0.440 | -0.062 | 8.41e-04 | 0.387 | 0.431 | -0.043 | 5.74e-03 |
| Height21 | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.417 | 0.440 | -0.023 | 2.06e-01 | 0.428 | 0.431 | -0.002 | 8.45e-01 |
| Height21 | All.MultiPRS | lassosum.MultiPRS | 0.447 | 0.440 | 0.007 | 6.99e-01 | 0.443 | 0.431 | 0.012 | 3.78e-02 |
| Height21 | PRScs.PseudoVal | lassosum.MultiPRS | 0.436 | 0.440 | -0.004 | 8.16e-01 | 0.435 | 0.431 | 0.004 | 5.60e-01 |
| Height21 | LDpred1.Inf | lassosum.10FCVal | 0.378 | 0.435 | -0.057 | 2.80e-11 | 0.390 | 0.425 | -0.035 | 3.47e-02 |
| Height21 | PRScs.10FCVal | lassosum.10FCVal | 0.429 | 0.435 | -0.006 | 1.40e-01 | 0.434 | 0.425 | 0.009 | 2.25e-01 |
| Height21 | LDpred2.MultiPRS | lassosum.10FCVal | 0.440 | 0.435 | 0.005 | 7.67e-01 | 0.432 | 0.425 | 0.007 | 4.63e-01 |
| Height21 | lassosum.10FCVal | lassosum.10FCVal | 0.435 | 0.435 | 0.000 | 1.00e+00 | 0.425 | 0.425 | 0.000 | 1.00e+00 |
| Height21 | PRScs.PseudoVal | lassosum.10FCVal | 0.436 | 0.435 | 0.001 | 8.87e-01 | 0.435 | 0.425 | 0.010 | 2.06e-01 |
| Height21 | DBSLMM.PseudoVal | lassosum.10FCVal | 0.417 | 0.435 | -0.018 | 5.36e-03 | 0.428 | 0.425 | 0.003 | 7.82e-01 |
| Height21 | SBLUP.Inf | lassosum.10FCVal | 0.378 | 0.435 | -0.057 | 1.04e-10 | 0.387 | 0.425 | -0.038 | 2.97e-02 |
| Height21 | LDpred2.Inf | lassosum.10FCVal | 0.379 | 0.435 | -0.056 | 2.98e-11 | 0.395 | 0.425 | -0.030 | 6.61e-02 |
| Height21 | pT+clump.10FCVal | lassosum.10FCVal | 0.387 | 0.435 | -0.048 | 7.29e-09 | 0.353 | 0.425 | -0.072 | 8.44e-06 |
| Height21 | LDpred2.10FCVal | lassosum.10FCVal | 0.431 | 0.435 | -0.003 | 5.20e-01 | 0.445 | 0.425 | 0.020 | 5.12e-02 |
| Height21 | lassosum.MultiPRS | lassosum.10FCVal | 0.440 | 0.435 | 0.005 | 7.90e-01 | 0.431 | 0.425 | 0.005 | 2.83e-01 |
| Height21 | LDpred1.10FCVal | lassosum.10FCVal | 0.380 | 0.435 | -0.055 | 3.22e-10 | 0.383 | 0.425 | -0.042 | 1.60e-02 |
| Height21 | All.MultiPRS | lassosum.10FCVal | 0.447 | 0.435 | 0.012 | 5.14e-01 | 0.443 | 0.425 | 0.018 | 1.65e-02 |
| Height21 | SBayesR.PseudoVal | lassosum.10FCVal | 0.428 | 0.435 | -0.006 | 2.47e-01 | 0.434 | 0.425 | 0.009 | 4.12e-01 |
| Height21 | lassosum.PseudoVal | lassosum.10FCVal | 0.357 | 0.435 | -0.078 | 3.45e-24 | 0.354 | 0.425 | -0.071 | 1.41e-06 |
| Height21 | pT+clump.MultiPRS | lassosum.10FCVal | 0.399 | 0.435 | -0.036 | 4.78e-02 | 0.389 | 0.425 | -0.036 | 6.07e-03 |
| Height21 | LDpred2.PseudoVal | lassosum.10FCVal | 0.378 | 0.435 | -0.057 | 3.18e-11 | 0.394 | 0.425 | -0.031 | 6.83e-02 |
| Height21 | LDpred1.MultiPRS | lassosum.10FCVal | 0.384 | 0.435 | -0.051 | 5.47e-03 | 0.397 | 0.425 | -0.028 | 8.65e-02 |
| Height21 | PRScs.MultiPRS | lassosum.10FCVal | 0.435 | 0.435 | 0.000 | 9.92e-01 | 0.437 | 0.425 | 0.012 | 1.15e-01 |
| Height21 | lassosum.PseudoVal | lassosum.PseudoVal | 0.357 | 0.357 | 0.000 | 1.00e+00 | 0.354 | 0.354 | 0.000 | 1.00e+00 |
| Height21 | pT+clump.MultiPRS | lassosum.PseudoVal | 0.399 | 0.357 | 0.041 | 2.80e-02 | 0.389 | 0.354 | 0.035 | 7.68e-02 |
| Height21 | LDpred2.Inf | lassosum.PseudoVal | 0.379 | 0.357 | 0.022 | 1.00e-01 | 0.395 | 0.354 | 0.041 | 1.07e-01 |
| Height21 | SBayesR.PseudoVal | lassosum.PseudoVal | 0.428 | 0.357 | 0.071 | 1.23e-20 | 0.434 | 0.354 | 0.080 | 1.23e-07 |
| Height21 | PRScs.MultiPRS | lassosum.PseudoVal | 0.435 | 0.357 | 0.078 | 3.06e-05 | 0.437 | 0.354 | 0.083 | 4.73e-07 |
| Height21 | SBLUP.Inf | lassosum.PseudoVal | 0.378 | 0.357 | 0.020 | 1.32e-01 | 0.387 | 0.354 | 0.033 | 2.01e-01 |
| Height21 | lassosum.MultiPRS | lassosum.PseudoVal | 0.440 | 0.357 | 0.083 | 9.39e-06 | 0.431 | 0.354 | 0.077 | 1.98e-06 |
| Height21 | PRScs.10FCVal | lassosum.PseudoVal | 0.429 | 0.357 | 0.072 | 2.39e-20 | 0.434 | 0.354 | 0.080 | 1.34e-07 |
| Height21 | LDpred2.MultiPRS | lassosum.PseudoVal | 0.440 | 0.357 | 0.083 | 8.31e-06 | 0.432 | 0.354 | 0.078 | 9.14e-06 |
| Height21 | PRScs.PseudoVal | lassosum.PseudoVal | 0.436 | 0.357 | 0.078 | 4.38e-19 | 0.435 | 0.354 | 0.081 | 1.75e-06 |
| Height21 | LDpred2.PseudoVal | lassosum.PseudoVal | 0.378 | 0.357 | 0.021 | 1.13e-01 | 0.394 | 0.354 | 0.041 | 1.11e-01 |
| Height21 | LDpred1.MultiPRS | lassosum.PseudoVal | 0.384 | 0.357 | 0.027 | 1.58e-01 | 0.397 | 0.354 | 0.043 | 8.58e-02 |
| Height21 | pT+clump.10FCVal | lassosum.PseudoVal | 0.387 | 0.357 | 0.030 | 4.48e-03 | 0.353 | 0.354 | 0.000 | 9.86e-01 |
| Height21 | All.MultiPRS | lassosum.PseudoVal | 0.447 | 0.357 | 0.089 | 1.52e-06 | 0.443 | 0.354 | 0.089 | 9.71e-08 |
| Height21 | LDpred1.Inf | lassosum.PseudoVal | 0.378 | 0.357 | 0.021 | 1.22e-01 | 0.390 | 0.354 | 0.036 | 1.56e-01 |
| Height21 | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.417 | 0.357 | 0.059 | 6.20e-08 | 0.428 | 0.354 | 0.075 | 3.14e-04 |
| Height21 | LDpred1.10FCVal | lassosum.PseudoVal | 0.380 | 0.357 | 0.023 | 8.22e-02 | 0.383 | 0.354 | 0.029 | 2.57e-01 |
| Height21 | lassosum.10FCVal | lassosum.PseudoVal | 0.435 | 0.357 | 0.078 | 3.45e-24 | 0.425 | 0.354 | 0.071 | 1.41e-06 |
| Height21 | LDpred2.10FCVal | lassosum.PseudoVal | 0.431 | 0.357 | 0.074 | 6.73e-13 | 0.445 | 0.354 | 0.092 | 4.02e-06 |
| Height21 | LDpred2.10FCVal | LDpred1.MultiPRS | 0.431 | 0.384 | 0.048 | 9.75e-03 | 0.445 | 0.397 | 0.049 | 1.38e-05 |
| Height21 | PRScs.PseudoVal | LDpred1.MultiPRS | 0.436 | 0.384 | 0.052 | 4.89e-03 | 0.435 | 0.397 | 0.038 | 3.68e-03 |
| Height21 | LDpred2.Inf | LDpred1.MultiPRS | 0.379 | 0.384 | -0.005 | 7.89e-01 | 0.395 | 0.397 | -0.002 | 7.95e-01 |
| Height21 | pT+clump.10FCVal | LDpred1.MultiPRS | 0.387 | 0.384 | 0.003 | 8.59e-01 | 0.353 | 0.397 | -0.043 | 6.22e-02 |
| Height21 | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.428 | 0.384 | 0.045 | 1.57e-02 | 0.434 | 0.397 | 0.037 | 5.29e-02 |
| Height21 | All.MultiPRS | LDpred1.MultiPRS | 0.447 | 0.384 | 0.063 | 6.29e-04 | 0.443 | 0.397 | 0.046 | 1.03e-03 |
| Height21 | LDpred1.10FCVal | LDpred1.MultiPRS | 0.380 | 0.384 | -0.004 | 8.34e-01 | 0.383 | 0.397 | -0.014 | 1.69e-02 |
| Height21 | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.384 | 0.384 | 0.000 | 1.00e+00 | 0.397 | 0.397 | 0.000 | 1.00e+00 |
| Height21 | PRScs.10FCVal | LDpred1.MultiPRS | 0.429 | 0.384 | 0.045 | 1.38e-02 | 0.434 | 0.397 | 0.037 | 1.33e-02 |
| Height21 | lassosum.10FCVal | LDpred1.MultiPRS | 0.435 | 0.384 | 0.051 | 5.47e-03 | 0.425 | 0.397 | 0.028 | 8.65e-02 |
| Height21 | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.399 | 0.384 | 0.015 | 4.29e-01 | 0.389 | 0.397 | -0.008 | 6.69e-01 |
| Height21 | LDpred1.Inf | LDpred1.MultiPRS | 0.378 | 0.384 | -0.006 | 7.45e-01 | 0.390 | 0.397 | -0.007 | 2.65e-01 |
| Height21 | lassosum.MultiPRS | LDpred1.MultiPRS | 0.440 | 0.384 | 0.056 | 2.53e-03 | 0.431 | 0.397 | 0.034 | 2.33e-02 |
| Height21 | PRScs.MultiPRS | LDpred1.MultiPRS | 0.435 | 0.384 | 0.051 | 5.49e-03 | 0.437 | 0.397 | 0.040 | 1.83e-03 |
| Height21 | lassosum.PseudoVal | LDpred1.MultiPRS | 0.357 | 0.384 | -0.027 | 1.58e-01 | 0.354 | 0.397 | -0.043 | 8.58e-02 |
| Height21 | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.378 | 0.384 | -0.006 | 7.63e-01 | 0.394 | 0.397 | -0.002 | 7.80e-01 |
| Height21 | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.417 | 0.384 | 0.033 | 7.50e-02 | 0.428 | 0.397 | 0.032 | 1.13e-02 |
| Height21 | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.440 | 0.384 | 0.056 | 2.61e-03 | 0.432 | 0.397 | 0.035 | 1.90e-02 |
| Height21 | SBLUP.Inf | LDpred1.MultiPRS | 0.378 | 0.384 | -0.006 | 7.42e-01 | 0.387 | 0.397 | -0.009 | 2.18e-01 |
| Height21 | All.MultiPRS | LDpred1.Inf | 0.447 | 0.378 | 0.069 | 1.85e-04 | 0.443 | 0.390 | 0.053 | 2.98e-04 |
| Height21 | LDpred2.PseudoVal | LDpred1.Inf | 0.378 | 0.378 | 0.000 | 9.06e-01 | 0.394 | 0.390 | 0.004 | 5.36e-01 |
| Height21 | SBayesR.PseudoVal | LDpred1.Inf | 0.428 | 0.378 | 0.051 | 7.00e-07 | 0.434 | 0.390 | 0.044 | 2.50e-02 |
| Height21 | LDpred2.MultiPRS | LDpred1.Inf | 0.440 | 0.378 | 0.062 | 7.58e-04 | 0.432 | 0.390 | 0.042 | 6.15e-03 |
| Height21 | PRScs.PseudoVal | LDpred1.Inf | 0.436 | 0.378 | 0.058 | 2.65e-17 | 0.435 | 0.390 | 0.045 | 6.45e-04 |
| Height21 | LDpred1.Inf | LDpred1.Inf | 0.378 | 0.378 | 0.000 | 1.00e+00 | 0.390 | 0.390 | 0.000 | 1.00e+00 |
| Height21 | pT+clump.10FCVal | LDpred1.Inf | 0.387 | 0.378 | 0.009 | 4.14e-01 | 0.353 | 0.390 | -0.036 | 1.15e-01 |
| Height21 | PRScs.MultiPRS | LDpred1.Inf | 0.435 | 0.378 | 0.057 | 1.97e-03 | 0.437 | 0.390 | 0.047 | 3.01e-04 |
| Height21 | lassosum.10FCVal | LDpred1.Inf | 0.435 | 0.378 | 0.057 | 2.80e-11 | 0.425 | 0.390 | 0.035 | 3.47e-02 |
| Height21 | pT+clump.MultiPRS | LDpred1.Inf | 0.399 | 0.378 | 0.021 | 2.63e-01 | 0.389 | 0.390 | -0.001 | 9.56e-01 |
| Height21 | LDpred1.MultiPRS | LDpred1.Inf | 0.384 | 0.378 | 0.006 | 7.45e-01 | 0.397 | 0.390 | 0.007 | 2.65e-01 |
| Height21 | PRScs.10FCVal | LDpred1.Inf | 0.429 | 0.378 | 0.051 | 1.03e-10 | 0.434 | 0.390 | 0.044 | 3.69e-03 |
| Height21 | LDpred2.Inf | LDpred1.Inf | 0.379 | 0.378 | 0.001 | 7.67e-01 | 0.395 | 0.390 | 0.005 | 5.15e-01 |
| Height21 | SBLUP.Inf | LDpred1.Inf | 0.378 | 0.378 | 0.000 | 9.82e-01 | 0.387 | 0.390 | -0.003 | 6.93e-01 |
| Height21 | DBSLMM.PseudoVal | LDpred1.Inf | 0.417 | 0.378 | 0.039 | 4.36e-10 | 0.428 | 0.390 | 0.039 | 1.86e-03 |
| Height21 | lassosum.MultiPRS | LDpred1.Inf | 0.440 | 0.378 | 0.062 | 8.53e-04 | 0.431 | 0.390 | 0.041 | 7.00e-03 |
| Height21 | LDpred1.10FCVal | LDpred1.Inf | 0.380 | 0.378 | 0.002 | 6.74e-01 | 0.383 | 0.390 | -0.007 | 4.93e-01 |
| Height21 | lassosum.PseudoVal | LDpred1.Inf | 0.357 | 0.378 | -0.021 | 1.22e-01 | 0.354 | 0.390 | -0.036 | 1.56e-01 |
| Height21 | LDpred2.10FCVal | LDpred1.Inf | 0.431 | 0.378 | 0.054 | 4.13e-19 | 0.445 | 0.390 | 0.056 | 9.40e-07 |
| Height21 | LDpred1.Inf | LDpred1.10FCVal | 0.378 | 0.380 | -0.002 | 6.74e-01 | 0.390 | 0.383 | 0.007 | 4.93e-01 |
| Height21 | PRScs.MultiPRS | LDpred1.10FCVal | 0.435 | 0.380 | 0.055 | 2.85e-03 | 0.437 | 0.383 | 0.054 | 1.84e-04 |
| Height21 | LDpred1.10FCVal | LDpred1.10FCVal | 0.380 | 0.380 | 0.000 | 1.00e+00 | 0.383 | 0.383 | 0.000 | 1.00e+00 |
| Height21 | All.MultiPRS | LDpred1.10FCVal | 0.447 | 0.380 | 0.067 | 2.88e-04 | 0.443 | 0.383 | 0.060 | 7.89e-05 |
| Height21 | SBayesR.PseudoVal | LDpred1.10FCVal | 0.428 | 0.380 | 0.048 | 1.66e-06 | 0.434 | 0.383 | 0.051 | 1.04e-02 |
| Height21 | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.417 | 0.380 | 0.037 | 8.68e-08 | 0.428 | 0.383 | 0.046 | 1.34e-03 |
| Height21 | pT+clump.MultiPRS | LDpred1.10FCVal | 0.399 | 0.380 | 0.019 | 3.16e-01 | 0.389 | 0.383 | 0.006 | 7.48e-01 |
| Height21 | lassosum.MultiPRS | LDpred1.10FCVal | 0.440 | 0.380 | 0.060 | 1.23e-03 | 0.431 | 0.383 | 0.048 | 2.96e-03 |
| Height21 | PRScs.10FCVal | LDpred1.10FCVal | 0.429 | 0.380 | 0.049 | 1.78e-09 | 0.434 | 0.383 | 0.052 | 1.73e-03 |
| Height21 | lassosum.PseudoVal | LDpred1.10FCVal | 0.357 | 0.380 | -0.023 | 8.22e-02 | 0.354 | 0.383 | -0.029 | 2.57e-01 |
| Height21 | SBLUP.Inf | LDpred1.10FCVal | 0.378 | 0.380 | -0.002 | 6.70e-01 | 0.387 | 0.383 | 0.005 | 6.67e-01 |
| Height21 | LDpred2.PseudoVal | LDpred1.10FCVal | 0.378 | 0.380 | -0.002 | 7.64e-01 | 0.394 | 0.383 | 0.012 | 3.19e-01 |
| Height21 | pT+clump.10FCVal | LDpred1.10FCVal | 0.387 | 0.380 | 0.007 | 5.35e-01 | 0.353 | 0.383 | -0.029 | 2.26e-01 |
| Height21 | LDpred2.MultiPRS | LDpred1.10FCVal | 0.440 | 0.380 | 0.060 | 1.20e-03 | 0.432 | 0.383 | 0.049 | 2.21e-03 |
| Height21 | PRScs.PseudoVal | LDpred1.10FCVal | 0.436 | 0.380 | 0.056 | 1.75e-14 | 0.435 | 0.383 | 0.052 | 4.02e-04 |
| Height21 | LDpred2.10FCVal | LDpred1.10FCVal | 0.431 | 0.380 | 0.051 | 7.60e-15 | 0.445 | 0.383 | 0.063 | 1.48e-06 |
| Height21 | LDpred1.MultiPRS | LDpred1.10FCVal | 0.384 | 0.380 | 0.004 | 8.34e-01 | 0.397 | 0.383 | 0.014 | 1.69e-02 |
| Height21 | LDpred2.Inf | LDpred1.10FCVal | 0.379 | 0.380 | -0.001 | 8.49e-01 | 0.395 | 0.383 | 0.012 | 3.15e-01 |
| Height21 | lassosum.10FCVal | LDpred1.10FCVal | 0.435 | 0.380 | 0.055 | 3.22e-10 | 0.425 | 0.383 | 0.042 | 1.60e-02 |
| Height21 | LDpred1.Inf | LDpred2.MultiPRS | 0.378 | 0.440 | -0.062 | 7.58e-04 | 0.390 | 0.432 | -0.042 | 6.15e-03 |
| Height21 | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.417 | 0.440 | -0.024 | 1.97e-01 | 0.428 | 0.432 | -0.004 | 7.74e-01 |
| Height21 | SBLUP.Inf | LDpred2.MultiPRS | 0.378 | 0.440 | -0.063 | 7.43e-04 | 0.387 | 0.432 | -0.045 | 4.87e-03 |
| Height21 | LDpred2.Inf | LDpred2.MultiPRS | 0.379 | 0.440 | -0.061 | 9.20e-04 | 0.395 | 0.432 | -0.038 | 1.11e-02 |
| Height21 | pT+clump.10FCVal | LDpred2.MultiPRS | 0.387 | 0.440 | -0.053 | 3.99e-03 | 0.353 | 0.432 | -0.079 | 1.04e-05 |
| Height21 | LDpred2.10FCVal | LDpred2.MultiPRS | 0.431 | 0.440 | -0.009 | 6.26e-01 | 0.445 | 0.432 | 0.013 | 6.92e-02 |
| Height21 | PRScs.PseudoVal | LDpred2.MultiPRS | 0.436 | 0.440 | -0.005 | 7.92e-01 | 0.435 | 0.432 | 0.003 | 7.59e-01 |
| Height21 | LDpred1.10FCVal | LDpred2.MultiPRS | 0.380 | 0.440 | -0.060 | 1.20e-03 | 0.383 | 0.432 | -0.049 | 2.21e-03 |
| Height21 | All.MultiPRS | LDpred2.MultiPRS | 0.447 | 0.440 | 0.006 | 7.22e-01 | 0.443 | 0.432 | 0.011 | 1.53e-01 |
| Height21 | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.428 | 0.440 | -0.012 | 5.15e-01 | 0.434 | 0.432 | 0.002 | 8.66e-01 |
| Height21 | lassosum.PseudoVal | LDpred2.MultiPRS | 0.357 | 0.440 | -0.083 | 8.31e-06 | 0.354 | 0.432 | -0.078 | 9.14e-06 |
| Height21 | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.399 | 0.440 | -0.042 | 2.41e-02 | 0.389 | 0.432 | -0.043 | 3.36e-03 |
| Height21 | lassosum.MultiPRS | LDpred2.MultiPRS | 0.440 | 0.440 | -0.001 | 9.75e-01 | 0.431 | 0.432 | -0.002 | 8.71e-01 |
| Height21 | PRScs.10FCVal | LDpred2.MultiPRS | 0.429 | 0.440 | -0.011 | 5.42e-01 | 0.434 | 0.432 | 0.002 | 8.22e-01 |
| Height21 | PRScs.MultiPRS | LDpred2.MultiPRS | 0.435 | 0.440 | -0.005 | 7.75e-01 | 0.437 | 0.432 | 0.005 | 5.78e-01 |
| Height21 | lassosum.10FCVal | LDpred2.MultiPRS | 0.435 | 0.440 | -0.005 | 7.67e-01 | 0.425 | 0.432 | -0.007 | 4.63e-01 |
| Height21 | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.378 | 0.440 | -0.062 | 8.14e-04 | 0.394 | 0.432 | -0.038 | 1.14e-02 |
| Height21 | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.384 | 0.440 | -0.056 | 2.61e-03 | 0.397 | 0.432 | -0.035 | 1.90e-02 |
| Height21 | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.440 | 0.440 | 0.000 | 1.00e+00 | 0.432 | 0.432 | 0.000 | 1.00e+00 |
| Height21 | LDpred1.Inf | LDpred2.10FCVal | 0.378 | 0.431 | -0.054 | 4.13e-19 | 0.390 | 0.445 | -0.056 | 9.40e-07 |
| Height21 | PRScs.MultiPRS | LDpred2.10FCVal | 0.435 | 0.431 | 0.004 | 8.40e-01 | 0.437 | 0.445 | -0.008 | 2.96e-01 |
| Height21 | LDpred2.MultiPRS | LDpred2.10FCVal | 0.440 | 0.431 | 0.009 | 6.26e-01 | 0.432 | 0.445 | -0.013 | 6.92e-02 |
| Height21 | PRScs.PseudoVal | LDpred2.10FCVal | 0.436 | 0.431 | 0.004 | 3.49e-01 | 0.435 | 0.445 | -0.011 | 2.04e-01 |
| Height21 | LDpred2.10FCVal | LDpred2.10FCVal | 0.431 | 0.431 | 0.000 | 1.00e+00 | 0.445 | 0.445 | 0.000 | 1.00e+00 |
| Height21 | lassosum.MultiPRS | LDpred2.10FCVal | 0.440 | 0.431 | 0.008 | 6.49e-01 | 0.431 | 0.445 | -0.015 | 1.25e-01 |
| Height21 | LDpred1.10FCVal | LDpred2.10FCVal | 0.380 | 0.431 | -0.051 | 7.60e-15 | 0.383 | 0.445 | -0.063 | 1.48e-06 |
| Height21 | All.MultiPRS | LDpred2.10FCVal | 0.447 | 0.431 | 0.015 | 3.99e-01 | 0.443 | 0.445 | -0.003 | 7.50e-01 |
| Height21 | SBayesR.PseudoVal | LDpred2.10FCVal | 0.428 | 0.431 | -0.003 | 6.71e-01 | 0.434 | 0.445 | -0.011 | 4.04e-01 |
| Height21 | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.417 | 0.431 | -0.015 | 6.35e-03 | 0.428 | 0.445 | -0.017 | 1.08e-01 |
| Height21 | pT+clump.MultiPRS | LDpred2.10FCVal | 0.399 | 0.431 | -0.033 | 7.43e-02 | 0.389 | 0.445 | -0.057 | 1.04e-04 |
| Height21 | lassosum.10FCVal | LDpred2.10FCVal | 0.435 | 0.431 | 0.003 | 5.20e-01 | 0.425 | 0.445 | -0.020 | 5.12e-02 |
| Height21 | PRScs.10FCVal | LDpred2.10FCVal | 0.429 | 0.431 | -0.002 | 6.60e-01 | 0.434 | 0.445 | -0.011 | 2.54e-01 |
| Height21 | lassosum.PseudoVal | LDpred2.10FCVal | 0.357 | 0.431 | -0.074 | 6.73e-13 | 0.354 | 0.445 | -0.092 | 4.02e-06 |
| Height21 | SBLUP.Inf | LDpred2.10FCVal | 0.378 | 0.431 | -0.054 | 4.70e-18 | 0.387 | 0.445 | -0.058 | 6.59e-07 |
| Height21 | LDpred2.PseudoVal | LDpred2.10FCVal | 0.378 | 0.431 | -0.053 | 6.53e-26 | 0.394 | 0.445 | -0.051 | 2.50e-07 |
| Height21 | pT+clump.10FCVal | LDpred2.10FCVal | 0.387 | 0.431 | -0.044 | 3.65e-06 | 0.353 | 0.445 | -0.092 | 1.17e-06 |
| Height21 | LDpred2.Inf | LDpred2.10FCVal | 0.379 | 0.431 | -0.053 | 2.14e-25 | 0.395 | 0.445 | -0.051 | 2.47e-07 |
| Height21 | LDpred1.MultiPRS | LDpred2.10FCVal | 0.384 | 0.431 | -0.048 | 9.75e-03 | 0.397 | 0.445 | -0.049 | 1.38e-05 |
| Height21 | lassosum.PseudoVal | LDpred2.PseudoVal | 0.357 | 0.378 | -0.021 | 1.13e-01 | 0.354 | 0.394 | -0.041 | 1.11e-01 |
| Height21 | SBLUP.Inf | LDpred2.PseudoVal | 0.378 | 0.378 | -0.001 | 8.87e-01 | 0.387 | 0.394 | -0.007 | 3.14e-01 |
| Height21 | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.428 | 0.378 | 0.050 | 7.66e-07 | 0.434 | 0.394 | 0.040 | 4.63e-02 |
| Height21 | All.MultiPRS | LDpred2.PseudoVal | 0.447 | 0.378 | 0.068 | 2.06e-04 | 0.443 | 0.394 | 0.049 | 9.86e-04 |
| Height21 | PRScs.PseudoVal | LDpred2.PseudoVal | 0.436 | 0.378 | 0.057 | 6.26e-17 | 0.435 | 0.394 | 0.040 | 2.91e-03 |
| Height21 | lassosum.MultiPRS | LDpred2.PseudoVal | 0.440 | 0.378 | 0.061 | 9.49e-04 | 0.431 | 0.394 | 0.036 | 1.92e-02 |
| Height21 | PRScs.MultiPRS | LDpred2.PseudoVal | 0.435 | 0.378 | 0.057 | 2.14e-03 | 0.437 | 0.394 | 0.043 | 1.55e-03 |
| Height21 | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.440 | 0.378 | 0.062 | 8.14e-04 | 0.432 | 0.394 | 0.038 | 1.14e-02 |
| Height21 | PRScs.10FCVal | LDpred2.PseudoVal | 0.429 | 0.378 | 0.051 | 1.04e-10 | 0.434 | 0.394 | 0.040 | 1.04e-02 |
| Height21 | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.378 | 0.378 | 0.000 | 1.00e+00 | 0.394 | 0.394 | 0.000 | 1.00e+00 |
| Height21 | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.399 | 0.378 | 0.020 | 2.73e-01 | 0.389 | 0.394 | -0.005 | 7.70e-01 |
| Height21 | pT+clump.10FCVal | LDpred2.PseudoVal | 0.387 | 0.378 | 0.009 | 4.38e-01 | 0.353 | 0.394 | -0.041 | 8.21e-02 |
| Height21 | lassosum.10FCVal | LDpred2.PseudoVal | 0.435 | 0.378 | 0.057 | 3.18e-11 | 0.425 | 0.394 | 0.031 | 6.83e-02 |
| Height21 | LDpred1.Inf | LDpred2.PseudoVal | 0.378 | 0.378 | 0.000 | 9.06e-01 | 0.390 | 0.394 | -0.004 | 5.36e-01 |
| Height21 | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.384 | 0.378 | 0.006 | 7.63e-01 | 0.397 | 0.394 | 0.002 | 7.80e-01 |
| Height21 | LDpred1.10FCVal | LDpred2.PseudoVal | 0.380 | 0.378 | 0.002 | 7.64e-01 | 0.383 | 0.394 | -0.012 | 3.19e-01 |
| Height21 | LDpred2.Inf | LDpred2.PseudoVal | 0.379 | 0.378 | 0.001 | 1.63e-01 | 0.395 | 0.394 | 0.000 | 8.52e-01 |
| Height21 | LDpred2.10FCVal | LDpred2.PseudoVal | 0.431 | 0.378 | 0.053 | 6.53e-26 | 0.445 | 0.394 | 0.051 | 2.50e-07 |
| Height21 | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.417 | 0.378 | 0.038 | 1.30e-09 | 0.428 | 0.394 | 0.034 | 8.94e-03 |
| Height21 | pT+clump.MultiPRS | LDpred2.Inf | 0.399 | 0.379 | 0.020 | 2.88e-01 | 0.389 | 0.395 | -0.006 | 7.61e-01 |
| Height21 | lassosum.MultiPRS | LDpred2.Inf | 0.440 | 0.379 | 0.061 | 1.07e-03 | 0.431 | 0.395 | 0.036 | 1.88e-02 |
| Height21 | SBayesR.PseudoVal | LDpred2.Inf | 0.428 | 0.379 | 0.050 | 8.46e-07 | 0.434 | 0.395 | 0.040 | 4.52e-02 |
| Height21 | All.MultiPRS | LDpred2.Inf | 0.447 | 0.379 | 0.068 | 2.36e-04 | 0.443 | 0.395 | 0.048 | 9.47e-04 |
| Height21 | SBLUP.Inf | LDpred2.Inf | 0.378 | 0.379 | -0.001 | 7.48e-01 | 0.387 | 0.395 | -0.007 | 3.05e-01 |
| Height21 | LDpred2.PseudoVal | LDpred2.Inf | 0.378 | 0.379 | -0.001 | 1.63e-01 | 0.394 | 0.395 | 0.000 | 8.52e-01 |
| Height21 | PRScs.MultiPRS | LDpred2.Inf | 0.435 | 0.379 | 0.056 | 2.40e-03 | 0.437 | 0.395 | 0.042 | 1.35e-03 |
| Height21 | LDpred2.MultiPRS | LDpred2.Inf | 0.440 | 0.379 | 0.061 | 9.20e-04 | 0.432 | 0.395 | 0.038 | 1.11e-02 |
| Height21 | PRScs.10FCVal | LDpred2.Inf | 0.429 | 0.379 | 0.050 | 8.68e-11 | 0.434 | 0.395 | 0.040 | 9.54e-03 |
| Height21 | LDpred1.Inf | LDpred2.Inf | 0.378 | 0.379 | -0.001 | 7.67e-01 | 0.390 | 0.395 | -0.005 | 5.15e-01 |
| Height21 | LDpred1.MultiPRS | LDpred2.Inf | 0.384 | 0.379 | 0.005 | 7.89e-01 | 0.397 | 0.395 | 0.002 | 7.95e-01 |
| Height21 | pT+clump.10FCVal | LDpred2.Inf | 0.387 | 0.379 | 0.008 | 4.67e-01 | 0.353 | 0.395 | -0.041 | 7.81e-02 |
| Height21 | lassosum.10FCVal | LDpred2.Inf | 0.435 | 0.379 | 0.056 | 2.98e-11 | 0.425 | 0.395 | 0.030 | 6.61e-02 |
| Height21 | PRScs.PseudoVal | LDpred2.Inf | 0.436 | 0.379 | 0.057 | 3.36e-17 | 0.435 | 0.395 | 0.040 | 2.52e-03 |
| Height21 | DBSLMM.PseudoVal | LDpred2.Inf | 0.417 | 0.379 | 0.038 | 1.96e-09 | 0.428 | 0.395 | 0.034 | 8.97e-03 |
| Height21 | LDpred1.10FCVal | LDpred2.Inf | 0.380 | 0.379 | 0.001 | 8.49e-01 | 0.383 | 0.395 | -0.012 | 3.15e-01 |
| Height21 | LDpred2.Inf | LDpred2.Inf | 0.379 | 0.379 | 0.000 | 1.00e+00 | 0.395 | 0.395 | 0.000 | 1.00e+00 |
| Height21 | lassosum.PseudoVal | LDpred2.Inf | 0.357 | 0.379 | -0.022 | 1.00e-01 | 0.354 | 0.395 | -0.041 | 1.07e-01 |
| Height21 | LDpred2.10FCVal | LDpred2.Inf | 0.431 | 0.379 | 0.053 | 2.14e-25 | 0.445 | 0.395 | 0.051 | 2.47e-07 |
| Height21 | PRScs.10FCVal | PRScs.MultiPRS | 0.429 | 0.435 | -0.006 | 7.45e-01 | 0.434 | 0.437 | -0.003 | 5.47e-01 |
| Height21 | LDpred1.10FCVal | PRScs.MultiPRS | 0.380 | 0.435 | -0.055 | 2.85e-03 | 0.383 | 0.437 | -0.054 | 1.84e-04 |
| Height21 | lassosum.MultiPRS | PRScs.MultiPRS | 0.440 | 0.435 | 0.005 | 7.98e-01 | 0.431 | 0.437 | -0.006 | 3.51e-01 |
| Height21 | LDpred2.MultiPRS | PRScs.MultiPRS | 0.440 | 0.435 | 0.005 | 7.75e-01 | 0.432 | 0.437 | -0.005 | 5.78e-01 |
| Height21 | All.MultiPRS | PRScs.MultiPRS | 0.447 | 0.435 | 0.012 | 1.17e-03 | 0.443 | 0.437 | 0.006 | 3.71e-01 |
| Height21 | PRScs.PseudoVal | PRScs.MultiPRS | 0.436 | 0.435 | 0.000 | 9.83e-01 | 0.435 | 0.437 | -0.002 | 1.88e-01 |
| Height21 | LDpred1.MultiPRS | PRScs.MultiPRS | 0.384 | 0.435 | -0.051 | 5.49e-03 | 0.397 | 0.437 | -0.040 | 1.83e-03 |
| Height21 | pT+clump.MultiPRS | PRScs.MultiPRS | 0.399 | 0.435 | -0.036 | 4.75e-02 | 0.389 | 0.437 | -0.048 | 2.06e-04 |
| Height21 | lassosum.10FCVal | PRScs.MultiPRS | 0.435 | 0.435 | 0.000 | 9.92e-01 | 0.425 | 0.437 | -0.012 | 1.15e-01 |
| Height21 | SBayesR.PseudoVal | PRScs.MultiPRS | 0.428 | 0.435 | -0.007 | 7.14e-01 | 0.434 | 0.437 | -0.003 | 7.93e-01 |
| Height21 | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.417 | 0.435 | -0.018 | 3.13e-01 | 0.428 | 0.437 | -0.008 | 3.43e-01 |
| Height21 | SBLUP.Inf | PRScs.MultiPRS | 0.378 | 0.435 | -0.057 | 1.94e-03 | 0.387 | 0.437 | -0.050 | 3.14e-04 |
| Height21 | LDpred2.Inf | PRScs.MultiPRS | 0.379 | 0.435 | -0.056 | 2.40e-03 | 0.395 | 0.437 | -0.042 | 1.35e-03 |
| Height21 | pT+clump.10FCVal | PRScs.MultiPRS | 0.387 | 0.435 | -0.048 | 1.02e-02 | 0.353 | 0.437 | -0.084 | 9.07e-07 |
| Height21 | LDpred2.10FCVal | PRScs.MultiPRS | 0.431 | 0.435 | -0.004 | 8.40e-01 | 0.445 | 0.437 | 0.008 | 2.96e-01 |
| Height21 | lassosum.PseudoVal | PRScs.MultiPRS | 0.357 | 0.435 | -0.078 | 3.06e-05 | 0.354 | 0.437 | -0.083 | 4.73e-07 |
| Height21 | LDpred2.PseudoVal | PRScs.MultiPRS | 0.378 | 0.435 | -0.057 | 2.14e-03 | 0.394 | 0.437 | -0.043 | 1.55e-03 |
| Height21 | LDpred1.Inf | PRScs.MultiPRS | 0.378 | 0.435 | -0.057 | 1.97e-03 | 0.390 | 0.437 | -0.047 | 3.01e-04 |
| Height21 | PRScs.MultiPRS | PRScs.MultiPRS | 0.435 | 0.435 | 0.000 | 1.00e+00 | 0.437 | 0.437 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.PseudoVal | PRScs.10FCVal | 0.378 | 0.429 | -0.051 | 1.04e-10 | 0.394 | 0.434 | -0.040 | 1.04e-02 |
| Height21 | PRScs.10FCVal | PRScs.10FCVal | 0.429 | 0.429 | 0.000 | 1.00e+00 | 0.434 | 0.434 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.MultiPRS | PRScs.10FCVal | 0.440 | 0.429 | 0.011 | 5.42e-01 | 0.432 | 0.434 | -0.002 | 8.22e-01 |
| Height21 | pT+clump.MultiPRS | PRScs.10FCVal | 0.399 | 0.429 | -0.030 | 9.68e-02 | 0.389 | 0.434 | -0.045 | 6.26e-04 |
| Height21 | LDpred1.Inf | PRScs.10FCVal | 0.378 | 0.429 | -0.051 | 1.03e-10 | 0.390 | 0.434 | -0.044 | 3.69e-03 |
| Height21 | LDpred1.MultiPRS | PRScs.10FCVal | 0.384 | 0.429 | -0.045 | 1.38e-02 | 0.397 | 0.434 | -0.037 | 1.33e-02 |
| Height21 | pT+clump.10FCVal | PRScs.10FCVal | 0.387 | 0.429 | -0.042 | 4.48e-07 | 0.353 | 0.434 | -0.081 | 9.12e-07 |
| Height21 | lassosum.10FCVal | PRScs.10FCVal | 0.435 | 0.429 | 0.006 | 1.40e-01 | 0.425 | 0.434 | -0.009 | 2.25e-01 |
| Height21 | PRScs.PseudoVal | PRScs.10FCVal | 0.436 | 0.429 | 0.006 | 2.45e-02 | 0.435 | 0.434 | 0.001 | 9.09e-01 |
| Height21 | DBSLMM.PseudoVal | PRScs.10FCVal | 0.417 | 0.429 | -0.013 | 2.65e-02 | 0.428 | 0.434 | -0.006 | 5.88e-01 |
| Height21 | SBLUP.Inf | PRScs.10FCVal | 0.378 | 0.429 | -0.051 | 4.23e-10 | 0.387 | 0.434 | -0.047 | 3.56e-03 |
| Height21 | LDpred2.Inf | PRScs.10FCVal | 0.379 | 0.429 | -0.050 | 8.68e-11 | 0.395 | 0.434 | -0.040 | 9.54e-03 |
| Height21 | lassosum.PseudoVal | PRScs.10FCVal | 0.357 | 0.429 | -0.072 | 2.39e-20 | 0.354 | 0.434 | -0.080 | 1.34e-07 |
| Height21 | LDpred2.10FCVal | PRScs.10FCVal | 0.431 | 0.429 | 0.002 | 6.60e-01 | 0.445 | 0.434 | 0.011 | 2.54e-01 |
| Height21 | lassosum.MultiPRS | PRScs.10FCVal | 0.440 | 0.429 | 0.011 | 5.61e-01 | 0.431 | 0.434 | -0.004 | 6.32e-01 |
| Height21 | LDpred1.10FCVal | PRScs.10FCVal | 0.380 | 0.429 | -0.049 | 1.78e-09 | 0.383 | 0.434 | -0.052 | 1.73e-03 |
| Height21 | All.MultiPRS | PRScs.10FCVal | 0.447 | 0.429 | 0.017 | 3.34e-01 | 0.443 | 0.434 | 0.009 | 2.44e-01 |
| Height21 | SBayesR.PseudoVal | PRScs.10FCVal | 0.428 | 0.429 | -0.001 | 8.87e-01 | 0.434 | 0.434 | 0.000 | 9.92e-01 |
| Height21 | PRScs.MultiPRS | PRScs.10FCVal | 0.435 | 0.429 | 0.006 | 7.45e-01 | 0.437 | 0.434 | 0.003 | 5.47e-01 |
| Height21 | All.MultiPRS | PRScs.PseudoVal | 0.447 | 0.436 | 0.011 | 5.35e-01 | 0.443 | 0.435 | 0.008 | 2.60e-01 |
| Height21 | pT+clump.MultiPRS | PRScs.PseudoVal | 0.399 | 0.436 | -0.037 | 4.43e-02 | 0.389 | 0.435 | -0.046 | 4.73e-04 |
| Height21 | LDpred2.PseudoVal | PRScs.PseudoVal | 0.378 | 0.436 | -0.057 | 6.26e-17 | 0.394 | 0.435 | -0.040 | 2.91e-03 |
| Height21 | SBayesR.PseudoVal | PRScs.PseudoVal | 0.428 | 0.436 | -0.007 | 2.22e-01 | 0.434 | 0.435 | -0.001 | 9.48e-01 |
| Height21 | LDpred2.MultiPRS | PRScs.PseudoVal | 0.440 | 0.436 | 0.005 | 7.92e-01 | 0.432 | 0.435 | -0.003 | 7.59e-01 |
| Height21 | SBLUP.Inf | PRScs.PseudoVal | 0.378 | 0.436 | -0.058 | 5.77e-16 | 0.387 | 0.435 | -0.048 | 6.60e-04 |
| Height21 | LDpred1.Inf | PRScs.PseudoVal | 0.378 | 0.436 | -0.058 | 2.65e-17 | 0.390 | 0.435 | -0.045 | 6.45e-04 |
| Height21 | PRScs.10FCVal | PRScs.PseudoVal | 0.429 | 0.436 | -0.006 | 2.45e-02 | 0.434 | 0.435 | -0.001 | 9.09e-01 |
| Height21 | LDpred2.Inf | PRScs.PseudoVal | 0.379 | 0.436 | -0.057 | 3.36e-17 | 0.395 | 0.435 | -0.040 | 2.52e-03 |
| Height21 | PRScs.PseudoVal | PRScs.PseudoVal | 0.436 | 0.436 | 0.000 | 1.00e+00 | 0.435 | 0.435 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.10FCVal | PRScs.PseudoVal | 0.431 | 0.436 | -0.004 | 3.49e-01 | 0.445 | 0.435 | 0.011 | 2.04e-01 |
| Height21 | pT+clump.10FCVal | PRScs.PseudoVal | 0.387 | 0.436 | -0.048 | 1.95e-08 | 0.353 | 0.435 | -0.081 | 1.95e-06 |
| Height21 | PRScs.MultiPRS | PRScs.PseudoVal | 0.435 | 0.436 | 0.000 | 9.83e-01 | 0.437 | 0.435 | 0.002 | 1.88e-01 |
| Height21 | lassosum.10FCVal | PRScs.PseudoVal | 0.435 | 0.436 | -0.001 | 8.87e-01 | 0.425 | 0.435 | -0.010 | 2.06e-01 |
| Height21 | lassosum.PseudoVal | PRScs.PseudoVal | 0.357 | 0.436 | -0.078 | 4.38e-19 | 0.354 | 0.435 | -0.081 | 1.75e-06 |
| Height21 | LDpred1.MultiPRS | PRScs.PseudoVal | 0.384 | 0.436 | -0.052 | 4.89e-03 | 0.397 | 0.435 | -0.038 | 3.68e-03 |
| Height21 | lassosum.MultiPRS | PRScs.PseudoVal | 0.440 | 0.436 | 0.004 | 8.16e-01 | 0.431 | 0.435 | -0.004 | 5.60e-01 |
| Height21 | LDpred1.10FCVal | PRScs.PseudoVal | 0.380 | 0.436 | -0.056 | 1.75e-14 | 0.383 | 0.435 | -0.052 | 4.02e-04 |
| Height21 | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.417 | 0.436 | -0.019 | 1.39e-04 | 0.428 | 0.435 | -0.006 | 5.00e-01 |
| Height21 | All.MultiPRS | pT+clump.MultiPRS | 0.447 | 0.399 | 0.048 | 8.50e-03 | 0.443 | 0.389 | 0.054 | 7.39e-06 |
| Height21 | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.428 | 0.399 | 0.030 | 1.07e-01 | 0.434 | 0.389 | 0.045 | 1.73e-03 |
| Height21 | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.384 | 0.399 | -0.015 | 4.29e-01 | 0.397 | 0.389 | 0.008 | 6.69e-01 |
| Height21 | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.399 | 0.399 | 0.000 | 1.00e+00 | 0.389 | 0.389 | 0.000 | 1.00e+00 |
| Height21 | lassosum.10FCVal | pT+clump.MultiPRS | 0.435 | 0.399 | 0.036 | 4.78e-02 | 0.425 | 0.389 | 0.036 | 6.07e-03 |
| Height21 | PRScs.10FCVal | pT+clump.MultiPRS | 0.429 | 0.399 | 0.030 | 9.68e-02 | 0.434 | 0.389 | 0.045 | 6.26e-04 |
| Height21 | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.417 | 0.399 | 0.018 | 3.30e-01 | 0.428 | 0.389 | 0.040 | 3.94e-03 |
| Height21 | SBLUP.Inf | pT+clump.MultiPRS | 0.378 | 0.399 | -0.021 | 2.62e-01 | 0.387 | 0.389 | -0.002 | 9.30e-01 |
| Height21 | LDpred2.Inf | pT+clump.MultiPRS | 0.379 | 0.399 | -0.020 | 2.88e-01 | 0.395 | 0.389 | 0.006 | 7.61e-01 |
| Height21 | pT+clump.10FCVal | pT+clump.MultiPRS | 0.387 | 0.399 | -0.011 | 5.39e-01 | 0.353 | 0.389 | -0.036 | 4.81e-05 |
| Height21 | LDpred2.10FCVal | pT+clump.MultiPRS | 0.431 | 0.399 | 0.033 | 7.43e-02 | 0.445 | 0.389 | 0.057 | 1.04e-04 |
| Height21 | PRScs.PseudoVal | pT+clump.MultiPRS | 0.436 | 0.399 | 0.037 | 4.43e-02 | 0.435 | 0.389 | 0.046 | 4.73e-04 |
| Height21 | LDpred1.10FCVal | pT+clump.MultiPRS | 0.380 | 0.399 | -0.019 | 3.16e-01 | 0.383 | 0.389 | -0.006 | 7.48e-01 |
| Height21 | LDpred1.Inf | pT+clump.MultiPRS | 0.378 | 0.399 | -0.021 | 2.63e-01 | 0.390 | 0.389 | 0.001 | 9.56e-01 |
| Height21 | PRScs.MultiPRS | pT+clump.MultiPRS | 0.435 | 0.399 | 0.036 | 4.75e-02 | 0.437 | 0.389 | 0.048 | 2.06e-04 |
| Height21 | lassosum.PseudoVal | pT+clump.MultiPRS | 0.357 | 0.399 | -0.041 | 2.80e-02 | 0.354 | 0.389 | -0.035 | 7.68e-02 |
| Height21 | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.378 | 0.399 | -0.020 | 2.73e-01 | 0.394 | 0.389 | 0.005 | 7.70e-01 |
| Height21 | lassosum.MultiPRS | pT+clump.MultiPRS | 0.440 | 0.399 | 0.041 | 2.69e-02 | 0.431 | 0.389 | 0.042 | 1.31e-03 |
| Height21 | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.440 | 0.399 | 0.042 | 2.41e-02 | 0.432 | 0.389 | 0.043 | 3.36e-03 |
| Height21 | LDpred2.10FCVal | pT+clump.10FCVal | 0.431 | 0.387 | 0.044 | 3.65e-06 | 0.445 | 0.353 | 0.092 | 1.17e-06 |
| Height21 | pT+clump.MultiPRS | pT+clump.10FCVal | 0.399 | 0.387 | 0.011 | 5.39e-01 | 0.389 | 0.353 | 0.036 | 4.81e-05 |
| Height21 | LDpred2.Inf | pT+clump.10FCVal | 0.379 | 0.387 | -0.008 | 4.67e-01 | 0.395 | 0.353 | 0.041 | 7.81e-02 |
| Height21 | SBayesR.PseudoVal | pT+clump.10FCVal | 0.428 | 0.387 | 0.041 | 7.18e-07 | 0.434 | 0.353 | 0.081 | 6.75e-07 |
| Height21 | PRScs.MultiPRS | pT+clump.10FCVal | 0.435 | 0.387 | 0.048 | 1.02e-02 | 0.437 | 0.353 | 0.084 | 9.07e-07 |
| Height21 | SBLUP.Inf | pT+clump.10FCVal | 0.378 | 0.387 | -0.010 | 4.21e-01 | 0.387 | 0.353 | 0.034 | 1.61e-01 |
| Height21 | lassosum.MultiPRS | pT+clump.10FCVal | 0.440 | 0.387 | 0.052 | 4.46e-03 | 0.431 | 0.353 | 0.077 | 2.15e-06 |
| Height21 | PRScs.10FCVal | pT+clump.10FCVal | 0.429 | 0.387 | 0.042 | 4.48e-07 | 0.434 | 0.353 | 0.081 | 9.12e-07 |
| Height21 | LDpred2.MultiPRS | pT+clump.10FCVal | 0.440 | 0.387 | 0.053 | 3.99e-03 | 0.432 | 0.353 | 0.079 | 1.04e-05 |
| Height21 | lassosum.PseudoVal | pT+clump.10FCVal | 0.357 | 0.387 | -0.030 | 4.48e-03 | 0.354 | 0.353 | 0.000 | 9.86e-01 |
| Height21 | LDpred2.PseudoVal | pT+clump.10FCVal | 0.378 | 0.387 | -0.009 | 4.38e-01 | 0.394 | 0.353 | 0.041 | 8.21e-02 |
| Height21 | LDpred1.MultiPRS | pT+clump.10FCVal | 0.384 | 0.387 | -0.003 | 8.59e-01 | 0.397 | 0.353 | 0.043 | 6.22e-02 |
| Height21 | pT+clump.10FCVal | pT+clump.10FCVal | 0.387 | 0.387 | 0.000 | 1.00e+00 | 0.353 | 0.353 | 0.000 | 1.00e+00 |
| Height21 | All.MultiPRS | pT+clump.10FCVal | 0.447 | 0.387 | 0.059 | 1.38e-03 | 0.443 | 0.353 | 0.089 | 1.34e-09 |
| Height21 | LDpred1.Inf | pT+clump.10FCVal | 0.378 | 0.387 | -0.009 | 4.14e-01 | 0.390 | 0.353 | 0.036 | 1.15e-01 |
| Height21 | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.417 | 0.387 | 0.029 | 1.69e-03 | 0.428 | 0.353 | 0.075 | 5.64e-05 |
| Height21 | LDpred1.10FCVal | pT+clump.10FCVal | 0.380 | 0.387 | -0.007 | 5.35e-01 | 0.383 | 0.353 | 0.029 | 2.26e-01 |
| Height21 | lassosum.10FCVal | pT+clump.10FCVal | 0.435 | 0.387 | 0.048 | 7.29e-09 | 0.425 | 0.353 | 0.072 | 8.44e-06 |
| Height21 | PRScs.PseudoVal | pT+clump.10FCVal | 0.436 | 0.387 | 0.048 | 1.95e-08 | 0.435 | 0.353 | 0.081 | 1.95e-06 |
| Height21 | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.399 | 0.428 | -0.030 | 1.07e-01 | 0.389 | 0.434 | -0.045 | 1.73e-03 |
| Height21 | LDpred1.10FCVal | SBayesR.PseudoVal | 0.380 | 0.428 | -0.048 | 1.66e-06 | 0.383 | 0.434 | -0.051 | 1.04e-02 |
| Height21 | PRScs.10FCVal | SBayesR.PseudoVal | 0.429 | 0.428 | 0.001 | 8.87e-01 | 0.434 | 0.434 | 0.000 | 9.92e-01 |
| Height21 | LDpred2.10FCVal | SBayesR.PseudoVal | 0.431 | 0.428 | 0.003 | 6.71e-01 | 0.445 | 0.434 | 0.011 | 4.04e-01 |
| Height21 | lassosum.PseudoVal | SBayesR.PseudoVal | 0.357 | 0.428 | -0.071 | 1.23e-20 | 0.354 | 0.434 | -0.080 | 1.23e-07 |
| Height21 | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.378 | 0.428 | -0.050 | 7.66e-07 | 0.394 | 0.434 | -0.040 | 4.63e-02 |
| Height21 | lassosum.MultiPRS | SBayesR.PseudoVal | 0.440 | 0.428 | 0.011 | 5.35e-01 | 0.431 | 0.434 | -0.004 | 7.46e-01 |
| Height21 | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.440 | 0.428 | 0.012 | 5.15e-01 | 0.432 | 0.434 | -0.002 | 8.66e-01 |
| Height21 | All.MultiPRS | SBayesR.PseudoVal | 0.447 | 0.428 | 0.018 | 3.14e-01 | 0.443 | 0.434 | 0.009 | 3.23e-01 |
| Height21 | PRScs.PseudoVal | SBayesR.PseudoVal | 0.436 | 0.428 | 0.007 | 2.22e-01 | 0.435 | 0.434 | 0.001 | 9.48e-01 |
| Height21 | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.384 | 0.428 | -0.045 | 1.57e-02 | 0.397 | 0.434 | -0.037 | 5.29e-02 |
| Height21 | pT+clump.10FCVal | SBayesR.PseudoVal | 0.387 | 0.428 | -0.041 | 7.18e-07 | 0.353 | 0.434 | -0.081 | 6.75e-07 |
| Height21 | lassosum.10FCVal | SBayesR.PseudoVal | 0.435 | 0.428 | 0.006 | 2.47e-01 | 0.425 | 0.434 | -0.009 | 4.12e-01 |
| Height21 | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.428 | 0.428 | 0.000 | 1.00e+00 | 0.434 | 0.434 | 0.000 | 1.00e+00 |
| Height21 | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.417 | 0.428 | -0.012 | 1.08e-01 | 0.428 | 0.434 | -0.006 | 6.83e-01 |
| Height21 | SBLUP.Inf | SBayesR.PseudoVal | 0.378 | 0.428 | -0.051 | 1.14e-06 | 0.387 | 0.434 | -0.047 | 2.12e-02 |
| Height21 | LDpred2.Inf | SBayesR.PseudoVal | 0.379 | 0.428 | -0.050 | 8.46e-07 | 0.395 | 0.434 | -0.040 | 4.52e-02 |
| Height21 | LDpred1.Inf | SBayesR.PseudoVal | 0.378 | 0.428 | -0.051 | 7.00e-07 | 0.390 | 0.434 | -0.044 | 2.50e-02 |
| Height21 | PRScs.MultiPRS | SBayesR.PseudoVal | 0.435 | 0.428 | 0.007 | 7.14e-01 | 0.437 | 0.434 | 0.003 | 7.93e-01 |
| Height21 | LDpred2.10FCVal | SBLUP.Inf | 0.431 | 0.378 | 0.054 | 4.70e-18 | 0.445 | 0.387 | 0.058 | 6.59e-07 |
| Height21 | PRScs.PseudoVal | SBLUP.Inf | 0.436 | 0.378 | 0.058 | 5.77e-16 | 0.435 | 0.387 | 0.048 | 6.60e-04 |
| Height21 | LDpred1.10FCVal | SBLUP.Inf | 0.380 | 0.378 | 0.002 | 6.70e-01 | 0.383 | 0.387 | -0.005 | 6.67e-01 |
| Height21 | LDpred2.Inf | SBLUP.Inf | 0.379 | 0.378 | 0.001 | 7.48e-01 | 0.395 | 0.387 | 0.007 | 3.05e-01 |
| Height21 | SBayesR.PseudoVal | SBLUP.Inf | 0.428 | 0.378 | 0.051 | 1.14e-06 | 0.434 | 0.387 | 0.047 | 2.12e-02 |
| Height21 | lassosum.PseudoVal | SBLUP.Inf | 0.357 | 0.378 | -0.020 | 1.32e-01 | 0.354 | 0.387 | -0.033 | 2.01e-01 |
| Height21 | pT+clump.MultiPRS | SBLUP.Inf | 0.399 | 0.378 | 0.021 | 2.62e-01 | 0.389 | 0.387 | 0.002 | 9.30e-01 |
| Height21 | lassosum.MultiPRS | SBLUP.Inf | 0.440 | 0.378 | 0.062 | 8.41e-04 | 0.431 | 0.387 | 0.043 | 5.74e-03 |
| Height21 | PRScs.10FCVal | SBLUP.Inf | 0.429 | 0.378 | 0.051 | 4.23e-10 | 0.434 | 0.387 | 0.047 | 3.56e-03 |
| Height21 | PRScs.MultiPRS | SBLUP.Inf | 0.435 | 0.378 | 0.057 | 1.94e-03 | 0.437 | 0.387 | 0.050 | 3.14e-04 |
| Height21 | LDpred1.MultiPRS | SBLUP.Inf | 0.384 | 0.378 | 0.006 | 7.42e-01 | 0.397 | 0.387 | 0.009 | 2.18e-01 |
| Height21 | LDpred2.PseudoVal | SBLUP.Inf | 0.378 | 0.378 | 0.001 | 8.87e-01 | 0.394 | 0.387 | 0.007 | 3.14e-01 |
| Height21 | LDpred1.Inf | SBLUP.Inf | 0.378 | 0.378 | 0.000 | 9.82e-01 | 0.390 | 0.387 | 0.003 | 6.93e-01 |
| Height21 | LDpred2.MultiPRS | SBLUP.Inf | 0.440 | 0.378 | 0.063 | 7.43e-04 | 0.432 | 0.387 | 0.045 | 4.87e-03 |
| Height21 | All.MultiPRS | SBLUP.Inf | 0.447 | 0.378 | 0.069 | 1.82e-04 | 0.443 | 0.387 | 0.056 | 2.84e-04 |
| Height21 | SBLUP.Inf | SBLUP.Inf | 0.378 | 0.378 | 0.000 | 1.00e+00 | 0.387 | 0.387 | 0.000 | 1.00e+00 |
| Height21 | DBSLMM.PseudoVal | SBLUP.Inf | 0.417 | 0.378 | 0.039 | 3.12e-10 | 0.428 | 0.387 | 0.041 | 1.27e-03 |
| Height21 | pT+clump.10FCVal | SBLUP.Inf | 0.387 | 0.378 | 0.010 | 4.21e-01 | 0.353 | 0.387 | -0.034 | 1.61e-01 |
| Height21 | lassosum.10FCVal | SBLUP.Inf | 0.435 | 0.378 | 0.057 | 1.04e-10 | 0.425 | 0.387 | 0.038 | 2.97e-02 |
| BMI21 | LDpred2.PseudoVal | All.MultiPRS | 0.333 | 0.367 | -0.034 | 8.71e-02 | 0.290 | 0.327 | -0.037 | 5.91e-03 |
| BMI21 | pT+clump.10FCVal | All.MultiPRS | 0.310 | 0.367 | -0.058 | 4.03e-03 | 0.301 | 0.327 | -0.026 | 1.31e-01 |
| BMI21 | LDpred2.MultiPRS | All.MultiPRS | 0.357 | 0.367 | -0.010 | 6.06e-01 | 0.308 | 0.327 | -0.019 | 2.28e-02 |
| BMI21 | PRScs.PseudoVal | All.MultiPRS | 0.347 | 0.367 | -0.020 | 3.19e-01 | 0.302 | 0.327 | -0.025 | 1.47e-02 |
| BMI21 | LDpred1.Inf | All.MultiPRS | 0.345 | 0.367 | -0.022 | 2.62e-01 | 0.298 | 0.327 | -0.029 | 3.19e-02 |
| BMI21 | PRScs.MultiPRS | All.MultiPRS | 0.354 | 0.367 | -0.013 | 3.62e-03 | 0.325 | 0.327 | -0.002 | 8.02e-01 |
| BMI21 | LDpred2.Inf | All.MultiPRS | 0.333 | 0.367 | -0.034 | 9.06e-02 | 0.290 | 0.327 | -0.037 | 5.37e-03 |
| BMI21 | lassosum.PseudoVal | All.MultiPRS | 0.288 | 0.367 | -0.079 | 7.24e-05 | 0.231 | 0.327 | -0.097 | 1.81e-06 |
| BMI21 | LDpred2.10FCVal | All.MultiPRS | 0.360 | 0.367 | -0.007 | 7.06e-01 | 0.315 | 0.327 | -0.012 | 1.82e-01 |
| BMI21 | lassosum.MultiPRS | All.MultiPRS | 0.361 | 0.367 | -0.007 | 7.41e-01 | 0.334 | 0.327 | 0.007 | 3.74e-01 |
| BMI21 | LDpred1.10FCVal | All.MultiPRS | 0.336 | 0.367 | -0.031 | 1.18e-01 | 0.306 | 0.327 | -0.021 | 1.32e-01 |
| BMI21 | All.MultiPRS | All.MultiPRS | 0.367 | 0.367 | 0.000 | 1.00e+00 | 0.327 | 0.327 | 0.000 | 1.00e+00 |
| BMI21 | SBayesR.PseudoVal | All.MultiPRS | 0.339 | 0.367 | -0.028 | 1.53e-01 | 0.290 | 0.327 | -0.037 | 9.43e-03 |
| BMI21 | LDpred1.MultiPRS | All.MultiPRS | 0.349 | 0.367 | -0.018 | 1.08e-03 | 0.306 | 0.327 | -0.021 | 7.29e-02 |
| BMI21 | pT+clump.MultiPRS | All.MultiPRS | 0.317 | 0.367 | -0.050 | 1.18e-02 | 0.295 | 0.327 | -0.032 | 4.17e-02 |
| BMI21 | lassosum.10FCVal | All.MultiPRS | 0.352 | 0.367 | -0.015 | 4.45e-01 | 0.332 | 0.327 | 0.005 | 6.52e-01 |
| BMI21 | PRScs.10FCVal | All.MultiPRS | 0.353 | 0.367 | -0.015 | 4.61e-01 | 0.326 | 0.327 | -0.001 | 9.27e-01 |
| BMI21 | DBSLMM.PseudoVal | All.MultiPRS | 0.354 | 0.367 | -0.013 | 5.01e-01 | 0.303 | 0.327 | -0.024 | 2.60e-02 |
| BMI21 | SBLUP.Inf | All.MultiPRS | 0.324 | 0.367 | -0.043 | 3.29e-02 | 0.299 | 0.327 | -0.028 | 5.84e-02 |
| BMI21 | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.339 | 0.354 | -0.015 | 4.11e-02 | 0.290 | 0.303 | -0.013 | 4.21e-01 |
| BMI21 | LDpred2.Inf | DBSLMM.PseudoVal | 0.333 | 0.354 | -0.020 | 3.21e-03 | 0.290 | 0.303 | -0.012 | 3.92e-01 |
| BMI21 | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.310 | 0.354 | -0.044 | 1.47e-07 | 0.301 | 0.303 | -0.002 | 9.11e-01 |
| BMI21 | PRScs.10FCVal | DBSLMM.PseudoVal | 0.353 | 0.354 | -0.001 | 7.81e-01 | 0.326 | 0.303 | 0.023 | 1.24e-02 |
| BMI21 | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.347 | 0.354 | -0.006 | 2.26e-01 | 0.302 | 0.303 | -0.001 | 9.29e-01 |
| BMI21 | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.317 | 0.354 | -0.037 | 6.51e-02 | 0.295 | 0.303 | -0.008 | 5.93e-01 |
| BMI21 | LDpred1.Inf | DBSLMM.PseudoVal | 0.345 | 0.354 | -0.009 | 1.60e-01 | 0.298 | 0.303 | -0.005 | 7.11e-01 |
| BMI21 | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.361 | 0.354 | 0.007 | 7.34e-01 | 0.334 | 0.303 | 0.031 | 6.77e-03 |
| BMI21 | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.360 | 0.354 | 0.006 | 3.34e-01 | 0.315 | 0.303 | 0.012 | 3.36e-01 |
| BMI21 | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.288 | 0.354 | -0.066 | 2.85e-11 | 0.231 | 0.303 | -0.072 | 6.61e-04 |
| BMI21 | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.336 | 0.354 | -0.018 | 1.45e-02 | 0.306 | 0.303 | 0.003 | 8.44e-01 |
| BMI21 | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.349 | 0.354 | -0.005 | 8.06e-01 | 0.306 | 0.303 | 0.003 | 8.11e-01 |
| BMI21 | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.354 | 0.354 | 0.000 | 9.81e-01 | 0.325 | 0.303 | 0.022 | 1.07e-02 |
| BMI21 | All.MultiPRS | DBSLMM.PseudoVal | 0.367 | 0.354 | 0.013 | 5.01e-01 | 0.327 | 0.303 | 0.024 | 2.60e-02 |
| BMI21 | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.333 | 0.354 | -0.021 | 2.91e-03 | 0.290 | 0.303 | -0.012 | 4.00e-01 |
| BMI21 | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.354 | 0.354 | 0.000 | 1.00e+00 | 0.303 | 0.303 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.357 | 0.354 | 0.003 | 8.76e-01 | 0.308 | 0.303 | 0.006 | 6.70e-01 |
| BMI21 | lassosum.10FCVal | DBSLMM.PseudoVal | 0.352 | 0.354 | -0.002 | 7.61e-01 | 0.332 | 0.303 | 0.029 | 1.45e-02 |
| BMI21 | SBLUP.Inf | DBSLMM.PseudoVal | 0.324 | 0.354 | -0.029 | 4.67e-05 | 0.299 | 0.303 | -0.004 | 7.86e-01 |
| BMI21 | SBayesR.PseudoVal | lassosum.MultiPRS | 0.339 | 0.361 | -0.022 | 2.82e-01 | 0.290 | 0.334 | -0.044 | 2.88e-03 |
| BMI21 | LDpred2.10FCVal | lassosum.MultiPRS | 0.360 | 0.361 | -0.001 | 9.64e-01 | 0.315 | 0.334 | -0.019 | 1.03e-01 |
| BMI21 | LDpred1.10FCVal | lassosum.MultiPRS | 0.336 | 0.361 | -0.025 | 2.19e-01 | 0.306 | 0.334 | -0.028 | 7.60e-02 |
| BMI21 | LDpred2.Inf | lassosum.MultiPRS | 0.333 | 0.361 | -0.027 | 1.75e-01 | 0.290 | 0.334 | -0.044 | 5.08e-03 |
| BMI21 | PRScs.10FCVal | lassosum.MultiPRS | 0.353 | 0.361 | -0.008 | 6.87e-01 | 0.326 | 0.334 | -0.008 | 4.38e-01 |
| BMI21 | PRScs.MultiPRS | lassosum.MultiPRS | 0.354 | 0.361 | -0.006 | 7.50e-01 | 0.325 | 0.334 | -0.009 | 2.88e-01 |
| BMI21 | pT+clump.MultiPRS | lassosum.MultiPRS | 0.317 | 0.361 | -0.044 | 2.92e-02 | 0.295 | 0.334 | -0.039 | 1.69e-02 |
| BMI21 | lassosum.MultiPRS | lassosum.MultiPRS | 0.361 | 0.361 | 0.000 | 1.00e+00 | 0.334 | 0.334 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.10FCVal | lassosum.MultiPRS | 0.352 | 0.361 | -0.008 | 6.69e-01 | 0.332 | 0.334 | -0.002 | 7.62e-01 |
| BMI21 | LDpred2.MultiPRS | lassosum.MultiPRS | 0.357 | 0.361 | -0.004 | 5.42e-01 | 0.308 | 0.334 | -0.026 | 3.80e-02 |
| BMI21 | LDpred1.MultiPRS | lassosum.MultiPRS | 0.349 | 0.361 | -0.012 | 5.57e-01 | 0.306 | 0.334 | -0.028 | 4.80e-02 |
| BMI21 | LDpred2.PseudoVal | lassosum.MultiPRS | 0.333 | 0.361 | -0.028 | 1.69e-01 | 0.290 | 0.334 | -0.044 | 5.66e-03 |
| BMI21 | LDpred1.Inf | lassosum.MultiPRS | 0.345 | 0.361 | -0.016 | 4.31e-01 | 0.298 | 0.334 | -0.037 | 1.87e-02 |
| BMI21 | pT+clump.10FCVal | lassosum.MultiPRS | 0.310 | 0.361 | -0.051 | 1.16e-02 | 0.301 | 0.334 | -0.033 | 6.72e-02 |
| BMI21 | lassosum.PseudoVal | lassosum.MultiPRS | 0.288 | 0.361 | -0.072 | 3.70e-04 | 0.231 | 0.334 | -0.104 | 4.58e-08 |
| BMI21 | SBLUP.Inf | lassosum.MultiPRS | 0.324 | 0.361 | -0.036 | 7.19e-02 | 0.299 | 0.334 | -0.035 | 2.83e-02 |
| BMI21 | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.354 | 0.361 | -0.007 | 7.34e-01 | 0.303 | 0.334 | -0.031 | 6.77e-03 |
| BMI21 | All.MultiPRS | lassosum.MultiPRS | 0.367 | 0.361 | 0.007 | 7.41e-01 | 0.327 | 0.334 | -0.007 | 3.74e-01 |
| BMI21 | PRScs.PseudoVal | lassosum.MultiPRS | 0.347 | 0.361 | -0.013 | 5.11e-01 | 0.302 | 0.334 | -0.032 | 1.34e-03 |
| BMI21 | LDpred1.Inf | lassosum.10FCVal | 0.345 | 0.352 | -0.007 | 3.03e-01 | 0.298 | 0.332 | -0.034 | 2.41e-02 |
| BMI21 | PRScs.10FCVal | lassosum.10FCVal | 0.353 | 0.352 | 0.000 | 9.26e-01 | 0.326 | 0.332 | -0.006 | 5.68e-01 |
| BMI21 | LDpred2.MultiPRS | lassosum.10FCVal | 0.357 | 0.352 | 0.005 | 8.08e-01 | 0.308 | 0.332 | -0.024 | 6.71e-02 |
| BMI21 | lassosum.10FCVal | lassosum.10FCVal | 0.352 | 0.352 | 0.000 | 1.00e+00 | 0.332 | 0.332 | 0.000 | 1.00e+00 |
| BMI21 | PRScs.PseudoVal | lassosum.10FCVal | 0.347 | 0.352 | -0.005 | 3.71e-01 | 0.302 | 0.332 | -0.030 | 6.84e-03 |
| BMI21 | DBSLMM.PseudoVal | lassosum.10FCVal | 0.354 | 0.352 | 0.002 | 7.61e-01 | 0.303 | 0.332 | -0.029 | 1.45e-02 |
| BMI21 | SBLUP.Inf | lassosum.10FCVal | 0.324 | 0.352 | -0.028 | 5.02e-04 | 0.299 | 0.332 | -0.033 | 3.58e-02 |
| BMI21 | LDpred2.Inf | lassosum.10FCVal | 0.333 | 0.352 | -0.019 | 1.35e-02 | 0.290 | 0.332 | -0.042 | 8.22e-03 |
| BMI21 | pT+clump.10FCVal | lassosum.10FCVal | 0.310 | 0.352 | -0.043 | 7.13e-07 | 0.301 | 0.332 | -0.031 | 6.73e-02 |
| BMI21 | LDpred2.10FCVal | lassosum.10FCVal | 0.360 | 0.352 | 0.008 | 1.94e-01 | 0.315 | 0.332 | -0.017 | 1.63e-01 |
| BMI21 | lassosum.MultiPRS | lassosum.10FCVal | 0.361 | 0.352 | 0.008 | 6.69e-01 | 0.334 | 0.332 | 0.002 | 7.62e-01 |
| BMI21 | LDpred1.10FCVal | lassosum.10FCVal | 0.336 | 0.352 | -0.016 | 3.88e-02 | 0.306 | 0.332 | -0.026 | 9.96e-02 |
| BMI21 | All.MultiPRS | lassosum.10FCVal | 0.367 | 0.352 | 0.015 | 4.45e-01 | 0.327 | 0.332 | -0.005 | 6.52e-01 |
| BMI21 | SBayesR.PseudoVal | lassosum.10FCVal | 0.339 | 0.352 | -0.013 | 8.95e-02 | 0.290 | 0.332 | -0.042 | 8.78e-03 |
| BMI21 | lassosum.PseudoVal | lassosum.10FCVal | 0.288 | 0.352 | -0.064 | 1.20e-11 | 0.231 | 0.332 | -0.101 | 3.46e-07 |
| BMI21 | pT+clump.MultiPRS | lassosum.10FCVal | 0.317 | 0.352 | -0.035 | 7.82e-02 | 0.295 | 0.332 | -0.037 | 1.70e-02 |
| BMI21 | LDpred2.PseudoVal | lassosum.10FCVal | 0.333 | 0.352 | -0.019 | 1.25e-02 | 0.290 | 0.332 | -0.041 | 8.97e-03 |
| BMI21 | LDpred1.MultiPRS | lassosum.10FCVal | 0.349 | 0.352 | -0.003 | 8.73e-01 | 0.306 | 0.332 | -0.026 | 6.36e-02 |
| BMI21 | PRScs.MultiPRS | lassosum.10FCVal | 0.354 | 0.352 | 0.002 | 9.11e-01 | 0.325 | 0.332 | -0.007 | 4.37e-01 |
| BMI21 | lassosum.PseudoVal | lassosum.PseudoVal | 0.288 | 0.288 | 0.000 | 1.00e+00 | 0.231 | 0.231 | 0.000 | 1.00e+00 |
| BMI21 | pT+clump.MultiPRS | lassosum.PseudoVal | 0.317 | 0.288 | 0.029 | 1.59e-01 | 0.295 | 0.231 | 0.064 | 2.82e-03 |
| BMI21 | LDpred2.Inf | lassosum.PseudoVal | 0.333 | 0.288 | 0.045 | 2.80e-04 | 0.290 | 0.231 | 0.060 | 2.26e-02 |
| BMI21 | SBayesR.PseudoVal | lassosum.PseudoVal | 0.339 | 0.288 | 0.051 | 8.27e-14 | 0.290 | 0.231 | 0.060 | 2.43e-05 |
| BMI21 | PRScs.MultiPRS | lassosum.PseudoVal | 0.354 | 0.288 | 0.066 | 9.55e-04 | 0.325 | 0.231 | 0.094 | 6.03e-07 |
| BMI21 | SBLUP.Inf | lassosum.PseudoVal | 0.324 | 0.288 | 0.036 | 4.71e-03 | 0.299 | 0.231 | 0.068 | 1.07e-02 |
| BMI21 | lassosum.MultiPRS | lassosum.PseudoVal | 0.361 | 0.288 | 0.072 | 3.70e-04 | 0.334 | 0.231 | 0.104 | 4.58e-08 |
| BMI21 | PRScs.10FCVal | lassosum.PseudoVal | 0.353 | 0.288 | 0.064 | 2.54e-09 | 0.326 | 0.231 | 0.096 | 2.09e-05 |
| BMI21 | LDpred2.MultiPRS | lassosum.PseudoVal | 0.357 | 0.288 | 0.069 | 7.14e-04 | 0.308 | 0.231 | 0.078 | 3.54e-04 |
| BMI21 | PRScs.PseudoVal | lassosum.PseudoVal | 0.347 | 0.288 | 0.059 | 5.06e-15 | 0.302 | 0.231 | 0.071 | 4.61e-06 |
| BMI21 | LDpred2.PseudoVal | lassosum.PseudoVal | 0.333 | 0.288 | 0.045 | 3.51e-04 | 0.290 | 0.231 | 0.060 | 2.39e-02 |
| BMI21 | LDpred1.MultiPRS | lassosum.PseudoVal | 0.349 | 0.288 | 0.061 | 2.49e-03 | 0.306 | 0.231 | 0.075 | 2.83e-03 |
| BMI21 | pT+clump.10FCVal | lassosum.PseudoVal | 0.310 | 0.288 | 0.021 | 9.90e-02 | 0.301 | 0.231 | 0.070 | 7.45e-03 |
| BMI21 | All.MultiPRS | lassosum.PseudoVal | 0.367 | 0.288 | 0.079 | 7.24e-05 | 0.327 | 0.231 | 0.097 | 1.81e-06 |
| BMI21 | LDpred1.Inf | lassosum.PseudoVal | 0.345 | 0.288 | 0.057 | 3.52e-06 | 0.298 | 0.231 | 0.067 | 1.01e-02 |
| BMI21 | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.354 | 0.288 | 0.066 | 2.85e-11 | 0.303 | 0.231 | 0.072 | 6.61e-04 |
| BMI21 | LDpred1.10FCVal | lassosum.PseudoVal | 0.336 | 0.288 | 0.048 | 2.04e-04 | 0.306 | 0.231 | 0.075 | 4.87e-03 |
| BMI21 | lassosum.10FCVal | lassosum.PseudoVal | 0.352 | 0.288 | 0.064 | 1.20e-11 | 0.332 | 0.231 | 0.101 | 3.46e-07 |
| BMI21 | LDpred2.10FCVal | lassosum.PseudoVal | 0.360 | 0.288 | 0.071 | 3.43e-12 | 0.315 | 0.231 | 0.084 | 8.12e-05 |
| BMI21 | LDpred2.10FCVal | LDpred1.MultiPRS | 0.360 | 0.349 | 0.011 | 5.89e-01 | 0.315 | 0.306 | 0.009 | 4.45e-01 |
| BMI21 | PRScs.PseudoVal | LDpred1.MultiPRS | 0.347 | 0.349 | -0.001 | 9.41e-01 | 0.302 | 0.306 | -0.004 | 7.70e-01 |
| BMI21 | LDpred2.Inf | LDpred1.MultiPRS | 0.333 | 0.349 | -0.016 | 4.38e-01 | 0.290 | 0.306 | -0.015 | 1.15e-01 |
| BMI21 | pT+clump.10FCVal | LDpred1.MultiPRS | 0.310 | 0.349 | -0.039 | 5.12e-02 | 0.301 | 0.306 | -0.005 | 7.60e-01 |
| BMI21 | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.339 | 0.349 | -0.010 | 6.16e-01 | 0.290 | 0.306 | -0.016 | 4.24e-01 |
| BMI21 | All.MultiPRS | LDpred1.MultiPRS | 0.367 | 0.349 | 0.018 | 1.08e-03 | 0.327 | 0.306 | 0.021 | 7.29e-02 |
| BMI21 | LDpred1.10FCVal | LDpred1.MultiPRS | 0.336 | 0.349 | -0.013 | 5.18e-01 | 0.306 | 0.306 | 0.000 | 9.87e-01 |
| BMI21 | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.349 | 0.349 | 0.000 | 1.00e+00 | 0.306 | 0.306 | 0.000 | 1.00e+00 |
| BMI21 | PRScs.10FCVal | LDpred1.MultiPRS | 0.353 | 0.349 | 0.004 | 8.55e-01 | 0.326 | 0.306 | 0.020 | 2.37e-02 |
| BMI21 | lassosum.10FCVal | LDpred1.MultiPRS | 0.352 | 0.349 | 0.003 | 8.73e-01 | 0.332 | 0.306 | 0.026 | 6.36e-02 |
| BMI21 | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.317 | 0.349 | -0.032 | 1.11e-01 | 0.295 | 0.306 | -0.011 | 5.08e-01 |
| BMI21 | LDpred1.Inf | LDpred1.MultiPRS | 0.345 | 0.349 | -0.004 | 8.37e-01 | 0.298 | 0.306 | -0.008 | 2.15e-01 |
| BMI21 | lassosum.MultiPRS | LDpred1.MultiPRS | 0.361 | 0.349 | 0.012 | 5.57e-01 | 0.334 | 0.306 | 0.028 | 4.80e-02 |
| BMI21 | PRScs.MultiPRS | LDpred1.MultiPRS | 0.354 | 0.349 | 0.005 | 2.81e-01 | 0.325 | 0.306 | 0.019 | 7.04e-02 |
| BMI21 | lassosum.PseudoVal | LDpred1.MultiPRS | 0.288 | 0.349 | -0.061 | 2.49e-03 | 0.231 | 0.306 | -0.075 | 2.83e-03 |
| BMI21 | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.333 | 0.349 | -0.016 | 4.27e-01 | 0.290 | 0.306 | -0.015 | 1.26e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.354 | 0.349 | 0.005 | 8.06e-01 | 0.303 | 0.306 | -0.003 | 8.11e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.357 | 0.349 | 0.008 | 6.86e-01 | 0.308 | 0.306 | 0.002 | 8.44e-01 |
| BMI21 | SBLUP.Inf | LDpred1.MultiPRS | 0.324 | 0.349 | -0.024 | 2.25e-01 | 0.299 | 0.306 | -0.007 | 5.15e-01 |
| BMI21 | All.MultiPRS | LDpred1.Inf | 0.367 | 0.345 | 0.022 | 2.62e-01 | 0.327 | 0.298 | 0.029 | 3.19e-02 |
| BMI21 | LDpred2.PseudoVal | LDpred1.Inf | 0.333 | 0.345 | -0.012 | 8.81e-03 | 0.290 | 0.298 | -0.007 | 4.35e-01 |
| BMI21 | SBayesR.PseudoVal | LDpred1.Inf | 0.339 | 0.345 | -0.006 | 5.43e-01 | 0.290 | 0.298 | -0.008 | 7.13e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred1.Inf | 0.357 | 0.345 | 0.012 | 5.44e-01 | 0.308 | 0.298 | 0.011 | 4.23e-01 |
| BMI21 | PRScs.PseudoVal | LDpred1.Inf | 0.347 | 0.345 | 0.003 | 7.00e-01 | 0.302 | 0.298 | 0.004 | 7.88e-01 |
| BMI21 | LDpred1.Inf | LDpred1.Inf | 0.345 | 0.345 | 0.000 | 1.00e+00 | 0.298 | 0.298 | 0.000 | 1.00e+00 |
| BMI21 | pT+clump.10FCVal | LDpred1.Inf | 0.310 | 0.345 | -0.035 | 5.93e-06 | 0.301 | 0.298 | 0.003 | 8.46e-01 |
| BMI21 | PRScs.MultiPRS | LDpred1.Inf | 0.354 | 0.345 | 0.009 | 6.34e-01 | 0.325 | 0.298 | 0.027 | 1.52e-02 |
| BMI21 | lassosum.10FCVal | LDpred1.Inf | 0.352 | 0.345 | 0.007 | 3.03e-01 | 0.332 | 0.298 | 0.034 | 2.41e-02 |
| BMI21 | pT+clump.MultiPRS | LDpred1.Inf | 0.317 | 0.345 | -0.028 | 1.65e-01 | 0.295 | 0.298 | -0.003 | 8.62e-01 |
| BMI21 | LDpred1.MultiPRS | LDpred1.Inf | 0.349 | 0.345 | 0.004 | 8.37e-01 | 0.306 | 0.298 | 0.008 | 2.15e-01 |
| BMI21 | PRScs.10FCVal | LDpred1.Inf | 0.353 | 0.345 | 0.008 | 6.80e-02 | 0.326 | 0.298 | 0.029 | 2.21e-03 |
| BMI21 | LDpred2.Inf | LDpred1.Inf | 0.333 | 0.345 | -0.011 | 8.70e-03 | 0.290 | 0.298 | -0.007 | 4.11e-01 |
| BMI21 | SBLUP.Inf | LDpred1.Inf | 0.324 | 0.345 | -0.020 | 7.45e-05 | 0.299 | 0.298 | 0.001 | 9.26e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred1.Inf | 0.354 | 0.345 | 0.009 | 1.60e-01 | 0.303 | 0.298 | 0.005 | 7.11e-01 |
| BMI21 | lassosum.MultiPRS | LDpred1.Inf | 0.361 | 0.345 | 0.016 | 4.31e-01 | 0.334 | 0.298 | 0.037 | 1.87e-02 |
| BMI21 | LDpred1.10FCVal | LDpred1.Inf | 0.336 | 0.345 | -0.009 | 6.73e-02 | 0.306 | 0.298 | 0.008 | 4.18e-01 |
| BMI21 | lassosum.PseudoVal | LDpred1.Inf | 0.288 | 0.345 | -0.057 | 3.52e-06 | 0.231 | 0.298 | -0.067 | 1.01e-02 |
| BMI21 | LDpred2.10FCVal | LDpred1.Inf | 0.360 | 0.345 | 0.015 | 1.10e-02 | 0.315 | 0.298 | 0.017 | 1.70e-01 |
| BMI21 | LDpred1.Inf | LDpred1.10FCVal | 0.345 | 0.336 | 0.009 | 6.73e-02 | 0.298 | 0.306 | -0.008 | 4.18e-01 |
| BMI21 | PRScs.MultiPRS | LDpred1.10FCVal | 0.354 | 0.336 | 0.018 | 3.59e-01 | 0.325 | 0.306 | 0.019 | 1.43e-01 |
| BMI21 | LDpred1.10FCVal | LDpred1.10FCVal | 0.336 | 0.336 | 0.000 | 1.00e+00 | 0.306 | 0.306 | 0.000 | 1.00e+00 |
| BMI21 | All.MultiPRS | LDpred1.10FCVal | 0.367 | 0.336 | 0.031 | 1.18e-01 | 0.327 | 0.306 | 0.021 | 1.32e-01 |
| BMI21 | SBayesR.PseudoVal | LDpred1.10FCVal | 0.339 | 0.336 | 0.003 | 7.68e-01 | 0.290 | 0.306 | -0.016 | 4.69e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.354 | 0.336 | 0.018 | 1.45e-02 | 0.303 | 0.306 | -0.003 | 8.44e-01 |
| BMI21 | pT+clump.MultiPRS | LDpred1.10FCVal | 0.317 | 0.336 | -0.019 | 3.45e-01 | 0.295 | 0.306 | -0.011 | 5.50e-01 |
| BMI21 | lassosum.MultiPRS | LDpred1.10FCVal | 0.361 | 0.336 | 0.025 | 2.19e-01 | 0.334 | 0.306 | 0.028 | 7.60e-02 |
| BMI21 | PRScs.10FCVal | LDpred1.10FCVal | 0.353 | 0.336 | 0.017 | 2.52e-03 | 0.326 | 0.306 | 0.020 | 7.77e-02 |
| BMI21 | lassosum.PseudoVal | LDpred1.10FCVal | 0.288 | 0.336 | -0.048 | 2.04e-04 | 0.231 | 0.306 | -0.075 | 4.87e-03 |
| BMI21 | SBLUP.Inf | LDpred1.10FCVal | 0.324 | 0.336 | -0.011 | 5.10e-02 | 0.299 | 0.306 | -0.007 | 5.66e-01 |
| BMI21 | LDpred2.PseudoVal | LDpred1.10FCVal | 0.333 | 0.336 | -0.003 | 6.12e-01 | 0.290 | 0.306 | -0.015 | 2.01e-01 |
| BMI21 | pT+clump.10FCVal | LDpred1.10FCVal | 0.310 | 0.336 | -0.026 | 2.61e-03 | 0.301 | 0.306 | -0.005 | 7.81e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred1.10FCVal | 0.357 | 0.336 | 0.021 | 2.94e-01 | 0.308 | 0.306 | 0.003 | 8.60e-01 |
| BMI21 | PRScs.PseudoVal | LDpred1.10FCVal | 0.347 | 0.336 | 0.012 | 1.41e-01 | 0.302 | 0.306 | -0.004 | 8.06e-01 |
| BMI21 | LDpred2.10FCVal | LDpred1.10FCVal | 0.360 | 0.336 | 0.024 | 5.15e-04 | 0.315 | 0.306 | 0.009 | 5.20e-01 |
| BMI21 | LDpred1.MultiPRS | LDpred1.10FCVal | 0.349 | 0.336 | 0.013 | 5.18e-01 | 0.306 | 0.306 | 0.000 | 9.87e-01 |
| BMI21 | LDpred2.Inf | LDpred1.10FCVal | 0.333 | 0.336 | -0.003 | 6.52e-01 | 0.290 | 0.306 | -0.015 | 1.91e-01 |
| BMI21 | lassosum.10FCVal | LDpred1.10FCVal | 0.352 | 0.336 | 0.016 | 3.88e-02 | 0.332 | 0.306 | 0.026 | 9.96e-02 |
| BMI21 | LDpred1.Inf | LDpred2.MultiPRS | 0.345 | 0.357 | -0.012 | 5.44e-01 | 0.298 | 0.308 | -0.011 | 4.23e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.354 | 0.357 | -0.003 | 8.76e-01 | 0.303 | 0.308 | -0.006 | 6.70e-01 |
| BMI21 | SBLUP.Inf | LDpred2.MultiPRS | 0.324 | 0.357 | -0.032 | 1.05e-01 | 0.299 | 0.308 | -0.010 | 5.15e-01 |
| BMI21 | LDpred2.Inf | LDpred2.MultiPRS | 0.333 | 0.357 | -0.024 | 2.39e-01 | 0.290 | 0.308 | -0.018 | 7.16e-02 |
| BMI21 | pT+clump.10FCVal | LDpred2.MultiPRS | 0.310 | 0.357 | -0.047 | 1.90e-02 | 0.301 | 0.308 | -0.008 | 6.80e-01 |
| BMI21 | LDpred2.10FCVal | LDpred2.MultiPRS | 0.360 | 0.357 | 0.003 | 8.90e-01 | 0.315 | 0.308 | 0.007 | 9.99e-02 |
| BMI21 | PRScs.PseudoVal | LDpred2.MultiPRS | 0.347 | 0.357 | -0.009 | 6.35e-01 | 0.302 | 0.308 | -0.007 | 5.78e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.MultiPRS | 0.336 | 0.357 | -0.021 | 2.94e-01 | 0.306 | 0.308 | -0.003 | 8.60e-01 |
| BMI21 | All.MultiPRS | LDpred2.MultiPRS | 0.367 | 0.357 | 0.010 | 6.06e-01 | 0.327 | 0.308 | 0.019 | 2.28e-02 |
| BMI21 | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.339 | 0.357 | -0.018 | 3.72e-01 | 0.290 | 0.308 | -0.018 | 2.73e-01 |
| BMI21 | lassosum.PseudoVal | LDpred2.MultiPRS | 0.288 | 0.357 | -0.069 | 7.14e-04 | 0.231 | 0.308 | -0.078 | 3.54e-04 |
| BMI21 | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.317 | 0.357 | -0.040 | 4.60e-02 | 0.295 | 0.308 | -0.014 | 4.22e-01 |
| BMI21 | lassosum.MultiPRS | LDpred2.MultiPRS | 0.361 | 0.357 | 0.004 | 5.42e-01 | 0.334 | 0.308 | 0.026 | 3.80e-02 |
| BMI21 | PRScs.10FCVal | LDpred2.MultiPRS | 0.353 | 0.357 | -0.004 | 8.26e-01 | 0.326 | 0.308 | 0.018 | 1.11e-01 |
| BMI21 | PRScs.MultiPRS | LDpred2.MultiPRS | 0.354 | 0.357 | -0.003 | 8.94e-01 | 0.325 | 0.308 | 0.017 | 1.18e-01 |
| BMI21 | lassosum.10FCVal | LDpred2.MultiPRS | 0.352 | 0.357 | -0.005 | 8.08e-01 | 0.332 | 0.308 | 0.024 | 6.71e-02 |
| BMI21 | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.333 | 0.357 | -0.024 | 2.32e-01 | 0.290 | 0.308 | -0.018 | 7.38e-02 |
| BMI21 | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.349 | 0.357 | -0.008 | 6.86e-01 | 0.306 | 0.308 | -0.002 | 8.44e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.357 | 0.357 | 0.000 | 1.00e+00 | 0.308 | 0.308 | 0.000 | 1.00e+00 |
| BMI21 | LDpred1.Inf | LDpred2.10FCVal | 0.345 | 0.360 | -0.015 | 1.10e-02 | 0.298 | 0.315 | -0.017 | 1.70e-01 |
| BMI21 | PRScs.MultiPRS | LDpred2.10FCVal | 0.354 | 0.360 | -0.005 | 7.86e-01 | 0.325 | 0.315 | 0.010 | 3.09e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred2.10FCVal | 0.357 | 0.360 | -0.003 | 8.90e-01 | 0.308 | 0.315 | -0.007 | 9.99e-02 |
| BMI21 | PRScs.PseudoVal | LDpred2.10FCVal | 0.347 | 0.360 | -0.012 | 2.38e-02 | 0.302 | 0.315 | -0.013 | 2.43e-01 |
| BMI21 | LDpred2.10FCVal | LDpred2.10FCVal | 0.360 | 0.360 | 0.000 | 1.00e+00 | 0.315 | 0.315 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.MultiPRS | LDpred2.10FCVal | 0.361 | 0.360 | 0.001 | 9.64e-01 | 0.334 | 0.315 | 0.019 | 1.03e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.10FCVal | 0.336 | 0.360 | -0.024 | 5.15e-04 | 0.306 | 0.315 | -0.009 | 5.20e-01 |
| BMI21 | All.MultiPRS | LDpred2.10FCVal | 0.367 | 0.360 | 0.007 | 7.06e-01 | 0.327 | 0.315 | 0.012 | 1.82e-01 |
| BMI21 | SBayesR.PseudoVal | LDpred2.10FCVal | 0.339 | 0.360 | -0.021 | 6.89e-03 | 0.290 | 0.315 | -0.025 | 1.24e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.354 | 0.360 | -0.006 | 3.34e-01 | 0.303 | 0.315 | -0.012 | 3.36e-01 |
| BMI21 | pT+clump.MultiPRS | LDpred2.10FCVal | 0.317 | 0.360 | -0.043 | 3.21e-02 | 0.295 | 0.315 | -0.020 | 2.28e-01 |
| BMI21 | lassosum.10FCVal | LDpred2.10FCVal | 0.352 | 0.360 | -0.008 | 1.94e-01 | 0.332 | 0.315 | 0.017 | 1.63e-01 |
| BMI21 | PRScs.10FCVal | LDpred2.10FCVal | 0.353 | 0.360 | -0.007 | 1.53e-01 | 0.326 | 0.315 | 0.011 | 2.81e-01 |
| BMI21 | lassosum.PseudoVal | LDpred2.10FCVal | 0.288 | 0.360 | -0.071 | 3.43e-12 | 0.231 | 0.315 | -0.084 | 8.12e-05 |
| BMI21 | SBLUP.Inf | LDpred2.10FCVal | 0.324 | 0.360 | -0.035 | 2.96e-07 | 0.299 | 0.315 | -0.016 | 2.55e-01 |
| BMI21 | LDpred2.PseudoVal | LDpred2.10FCVal | 0.333 | 0.360 | -0.027 | 4.85e-10 | 0.290 | 0.315 | -0.024 | 7.75e-03 |
| BMI21 | pT+clump.10FCVal | LDpred2.10FCVal | 0.310 | 0.360 | -0.050 | 1.48e-08 | 0.301 | 0.315 | -0.014 | 4.34e-01 |
| BMI21 | LDpred2.Inf | LDpred2.10FCVal | 0.333 | 0.360 | -0.026 | 7.94e-10 | 0.290 | 0.315 | -0.024 | 7.19e-03 |
| BMI21 | LDpred1.MultiPRS | LDpred2.10FCVal | 0.349 | 0.360 | -0.011 | 5.89e-01 | 0.306 | 0.315 | -0.009 | 4.45e-01 |
| BMI21 | lassosum.PseudoVal | LDpred2.PseudoVal | 0.288 | 0.333 | -0.045 | 3.51e-04 | 0.231 | 0.290 | -0.060 | 2.39e-02 |
| BMI21 | SBLUP.Inf | LDpred2.PseudoVal | 0.324 | 0.333 | -0.009 | 1.34e-01 | 0.299 | 0.290 | 0.008 | 4.95e-01 |
| BMI21 | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.339 | 0.333 | 0.006 | 5.46e-01 | 0.290 | 0.290 | 0.000 | 9.84e-01 |
| BMI21 | All.MultiPRS | LDpred2.PseudoVal | 0.367 | 0.333 | 0.034 | 8.71e-02 | 0.327 | 0.290 | 0.037 | 5.91e-03 |
| BMI21 | PRScs.PseudoVal | LDpred2.PseudoVal | 0.347 | 0.333 | 0.014 | 4.81e-02 | 0.302 | 0.290 | 0.011 | 4.77e-01 |
| BMI21 | lassosum.MultiPRS | LDpred2.PseudoVal | 0.361 | 0.333 | 0.028 | 1.69e-01 | 0.334 | 0.290 | 0.044 | 5.66e-03 |
| BMI21 | PRScs.MultiPRS | LDpred2.PseudoVal | 0.354 | 0.333 | 0.021 | 2.86e-01 | 0.325 | 0.290 | 0.034 | 5.59e-03 |
| BMI21 | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.357 | 0.333 | 0.024 | 2.32e-01 | 0.308 | 0.290 | 0.018 | 7.38e-02 |
| BMI21 | PRScs.10FCVal | LDpred2.PseudoVal | 0.353 | 0.333 | 0.020 | 1.38e-04 | 0.326 | 0.290 | 0.036 | 9.62e-04 |
| BMI21 | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.333 | 0.333 | 0.000 | 1.00e+00 | 0.290 | 0.290 | 0.000 | 1.00e+00 |
| BMI21 | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.317 | 0.333 | -0.016 | 4.23e-01 | 0.295 | 0.290 | 0.004 | 8.23e-01 |
| BMI21 | pT+clump.10FCVal | LDpred2.PseudoVal | 0.310 | 0.333 | -0.023 | 7.25e-03 | 0.301 | 0.290 | 0.010 | 5.63e-01 |
| BMI21 | lassosum.10FCVal | LDpred2.PseudoVal | 0.352 | 0.333 | 0.019 | 1.25e-02 | 0.332 | 0.290 | 0.041 | 8.97e-03 |
| BMI21 | LDpred1.Inf | LDpred2.PseudoVal | 0.345 | 0.333 | 0.012 | 8.81e-03 | 0.298 | 0.290 | 0.007 | 4.35e-01 |
| BMI21 | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.349 | 0.333 | 0.016 | 4.27e-01 | 0.306 | 0.290 | 0.015 | 1.26e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.PseudoVal | 0.336 | 0.333 | 0.003 | 6.12e-01 | 0.306 | 0.290 | 0.015 | 2.01e-01 |
| BMI21 | LDpred2.Inf | LDpred2.PseudoVal | 0.333 | 0.333 | 0.000 | 4.29e-01 | 0.290 | 0.290 | 0.000 | 9.31e-01 |
| BMI21 | LDpred2.10FCVal | LDpred2.PseudoVal | 0.360 | 0.333 | 0.027 | 4.85e-10 | 0.315 | 0.290 | 0.024 | 7.75e-03 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.354 | 0.333 | 0.021 | 2.91e-03 | 0.303 | 0.290 | 0.012 | 4.00e-01 |
| BMI21 | pT+clump.MultiPRS | LDpred2.Inf | 0.317 | 0.333 | -0.016 | 4.13e-01 | 0.295 | 0.290 | 0.004 | 8.17e-01 |
| BMI21 | lassosum.MultiPRS | LDpred2.Inf | 0.361 | 0.333 | 0.027 | 1.75e-01 | 0.334 | 0.290 | 0.044 | 5.08e-03 |
| BMI21 | SBayesR.PseudoVal | LDpred2.Inf | 0.339 | 0.333 | 0.006 | 5.67e-01 | 0.290 | 0.290 | 0.000 | 9.87e-01 |
| BMI21 | All.MultiPRS | LDpred2.Inf | 0.367 | 0.333 | 0.034 | 9.06e-02 | 0.327 | 0.290 | 0.037 | 5.37e-03 |
| BMI21 | SBLUP.Inf | LDpred2.Inf | 0.324 | 0.333 | -0.009 | 1.13e-01 | 0.299 | 0.290 | 0.008 | 4.86e-01 |
| BMI21 | LDpred2.PseudoVal | LDpred2.Inf | 0.333 | 0.333 | 0.000 | 4.29e-01 | 0.290 | 0.290 | 0.000 | 9.31e-01 |
| BMI21 | PRScs.MultiPRS | LDpred2.Inf | 0.354 | 0.333 | 0.021 | 2.95e-01 | 0.325 | 0.290 | 0.034 | 4.38e-03 |
| BMI21 | LDpred2.MultiPRS | LDpred2.Inf | 0.357 | 0.333 | 0.024 | 2.39e-01 | 0.308 | 0.290 | 0.018 | 7.16e-02 |
| BMI21 | PRScs.10FCVal | LDpred2.Inf | 0.353 | 0.333 | 0.019 | 1.31e-04 | 0.326 | 0.290 | 0.036 | 6.81e-04 |
| BMI21 | LDpred1.Inf | LDpred2.Inf | 0.345 | 0.333 | 0.011 | 8.70e-03 | 0.298 | 0.290 | 0.007 | 4.11e-01 |
| BMI21 | LDpred1.MultiPRS | LDpred2.Inf | 0.349 | 0.333 | 0.016 | 4.38e-01 | 0.306 | 0.290 | 0.015 | 1.15e-01 |
| BMI21 | pT+clump.10FCVal | LDpred2.Inf | 0.310 | 0.333 | -0.024 | 5.78e-03 | 0.301 | 0.290 | 0.010 | 5.55e-01 |
| BMI21 | lassosum.10FCVal | LDpred2.Inf | 0.352 | 0.333 | 0.019 | 1.35e-02 | 0.332 | 0.290 | 0.042 | 8.22e-03 |
| BMI21 | PRScs.PseudoVal | LDpred2.Inf | 0.347 | 0.333 | 0.014 | 4.88e-02 | 0.302 | 0.290 | 0.011 | 4.61e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.Inf | 0.354 | 0.333 | 0.020 | 3.21e-03 | 0.303 | 0.290 | 0.012 | 3.92e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.Inf | 0.336 | 0.333 | 0.003 | 6.52e-01 | 0.306 | 0.290 | 0.015 | 1.91e-01 |
| BMI21 | LDpred2.Inf | LDpred2.Inf | 0.333 | 0.333 | 0.000 | 1.00e+00 | 0.290 | 0.290 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.PseudoVal | LDpred2.Inf | 0.288 | 0.333 | -0.045 | 2.80e-04 | 0.231 | 0.290 | -0.060 | 2.26e-02 |
| BMI21 | LDpred2.10FCVal | LDpred2.Inf | 0.360 | 0.333 | 0.026 | 7.94e-10 | 0.315 | 0.290 | 0.024 | 7.19e-03 |
| BMI21 | PRScs.10FCVal | PRScs.MultiPRS | 0.353 | 0.354 | -0.002 | 9.30e-01 | 0.326 | 0.325 | 0.001 | 7.80e-01 |
| BMI21 | LDpred1.10FCVal | PRScs.MultiPRS | 0.336 | 0.354 | -0.018 | 3.59e-01 | 0.306 | 0.325 | -0.019 | 1.43e-01 |
| BMI21 | lassosum.MultiPRS | PRScs.MultiPRS | 0.361 | 0.354 | 0.006 | 7.50e-01 | 0.334 | 0.325 | 0.009 | 2.88e-01 |
| BMI21 | LDpred2.MultiPRS | PRScs.MultiPRS | 0.357 | 0.354 | 0.003 | 8.94e-01 | 0.308 | 0.325 | -0.017 | 1.18e-01 |
| BMI21 | All.MultiPRS | PRScs.MultiPRS | 0.367 | 0.354 | 0.013 | 3.62e-03 | 0.327 | 0.325 | 0.002 | 8.02e-01 |
| BMI21 | PRScs.PseudoVal | PRScs.MultiPRS | 0.347 | 0.354 | -0.007 | 7.29e-01 | 0.302 | 0.325 | -0.023 | 2.32e-04 |
| BMI21 | LDpred1.MultiPRS | PRScs.MultiPRS | 0.349 | 0.354 | -0.005 | 2.81e-01 | 0.306 | 0.325 | -0.019 | 7.04e-02 |
| BMI21 | pT+clump.MultiPRS | PRScs.MultiPRS | 0.317 | 0.354 | -0.037 | 6.11e-02 | 0.295 | 0.325 | -0.030 | 3.00e-02 |
| BMI21 | lassosum.10FCVal | PRScs.MultiPRS | 0.352 | 0.354 | -0.002 | 9.11e-01 | 0.332 | 0.325 | 0.007 | 4.37e-01 |
| BMI21 | SBayesR.PseudoVal | PRScs.MultiPRS | 0.339 | 0.354 | -0.015 | 4.39e-01 | 0.290 | 0.325 | -0.035 | 1.15e-02 |
| BMI21 | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.354 | 0.354 | 0.000 | 9.81e-01 | 0.303 | 0.325 | -0.022 | 1.07e-02 |
| BMI21 | SBLUP.Inf | PRScs.MultiPRS | 0.324 | 0.354 | -0.030 | 1.37e-01 | 0.299 | 0.325 | -0.026 | 4.47e-02 |
| BMI21 | LDpred2.Inf | PRScs.MultiPRS | 0.333 | 0.354 | -0.021 | 2.95e-01 | 0.290 | 0.325 | -0.034 | 4.38e-03 |
| BMI21 | pT+clump.10FCVal | PRScs.MultiPRS | 0.310 | 0.354 | -0.045 | 2.60e-02 | 0.301 | 0.325 | -0.024 | 1.30e-01 |
| BMI21 | LDpred2.10FCVal | PRScs.MultiPRS | 0.360 | 0.354 | 0.005 | 7.86e-01 | 0.315 | 0.325 | -0.010 | 3.09e-01 |
| BMI21 | lassosum.PseudoVal | PRScs.MultiPRS | 0.288 | 0.354 | -0.066 | 9.55e-04 | 0.231 | 0.325 | -0.094 | 6.03e-07 |
| BMI21 | LDpred2.PseudoVal | PRScs.MultiPRS | 0.333 | 0.354 | -0.021 | 2.86e-01 | 0.290 | 0.325 | -0.034 | 5.59e-03 |
| BMI21 | LDpred1.Inf | PRScs.MultiPRS | 0.345 | 0.354 | -0.009 | 6.34e-01 | 0.298 | 0.325 | -0.027 | 1.52e-02 |
| BMI21 | PRScs.MultiPRS | PRScs.MultiPRS | 0.354 | 0.354 | 0.000 | 1.00e+00 | 0.325 | 0.325 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.PseudoVal | PRScs.10FCVal | 0.333 | 0.353 | -0.020 | 1.38e-04 | 0.290 | 0.326 | -0.036 | 9.62e-04 |
| BMI21 | PRScs.10FCVal | PRScs.10FCVal | 0.353 | 0.353 | 0.000 | 1.00e+00 | 0.326 | 0.326 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.MultiPRS | PRScs.10FCVal | 0.357 | 0.353 | 0.004 | 8.26e-01 | 0.308 | 0.326 | -0.018 | 1.11e-01 |
| BMI21 | pT+clump.MultiPRS | PRScs.10FCVal | 0.317 | 0.353 | -0.036 | 7.46e-02 | 0.295 | 0.326 | -0.032 | 3.45e-02 |
| BMI21 | LDpred1.Inf | PRScs.10FCVal | 0.345 | 0.353 | -0.008 | 6.80e-02 | 0.298 | 0.326 | -0.029 | 2.21e-03 |
| BMI21 | LDpred1.MultiPRS | PRScs.10FCVal | 0.349 | 0.353 | -0.004 | 8.55e-01 | 0.306 | 0.326 | -0.020 | 2.37e-02 |
| BMI21 | pT+clump.10FCVal | PRScs.10FCVal | 0.310 | 0.353 | -0.043 | 1.39e-08 | 0.301 | 0.326 | -0.025 | 9.91e-02 |
| BMI21 | lassosum.10FCVal | PRScs.10FCVal | 0.352 | 0.353 | 0.000 | 9.26e-01 | 0.332 | 0.326 | 0.006 | 5.68e-01 |
| BMI21 | PRScs.PseudoVal | PRScs.10FCVal | 0.347 | 0.353 | -0.005 | 2.81e-01 | 0.302 | 0.326 | -0.024 | 1.49e-02 |
| BMI21 | DBSLMM.PseudoVal | PRScs.10FCVal | 0.354 | 0.353 | 0.001 | 7.81e-01 | 0.303 | 0.326 | -0.023 | 1.24e-02 |
| BMI21 | SBLUP.Inf | PRScs.10FCVal | 0.324 | 0.353 | -0.028 | 2.97e-07 | 0.299 | 0.326 | -0.028 | 1.85e-02 |
| BMI21 | LDpred2.Inf | PRScs.10FCVal | 0.333 | 0.353 | -0.019 | 1.31e-04 | 0.290 | 0.326 | -0.036 | 6.81e-04 |
| BMI21 | lassosum.PseudoVal | PRScs.10FCVal | 0.288 | 0.353 | -0.064 | 2.54e-09 | 0.231 | 0.326 | -0.096 | 2.09e-05 |
| BMI21 | LDpred2.10FCVal | PRScs.10FCVal | 0.360 | 0.353 | 0.007 | 1.53e-01 | 0.315 | 0.326 | -0.011 | 2.81e-01 |
| BMI21 | lassosum.MultiPRS | PRScs.10FCVal | 0.361 | 0.353 | 0.008 | 6.87e-01 | 0.334 | 0.326 | 0.008 | 4.38e-01 |
| BMI21 | LDpred1.10FCVal | PRScs.10FCVal | 0.336 | 0.353 | -0.017 | 2.52e-03 | 0.306 | 0.326 | -0.020 | 7.77e-02 |
| BMI21 | All.MultiPRS | PRScs.10FCVal | 0.367 | 0.353 | 0.015 | 4.61e-01 | 0.327 | 0.326 | 0.001 | 9.27e-01 |
| BMI21 | SBayesR.PseudoVal | PRScs.10FCVal | 0.339 | 0.353 | -0.014 | 9.89e-02 | 0.290 | 0.326 | -0.036 | 3.26e-02 |
| BMI21 | PRScs.MultiPRS | PRScs.10FCVal | 0.354 | 0.353 | 0.002 | 9.30e-01 | 0.325 | 0.326 | -0.001 | 7.80e-01 |
| BMI21 | All.MultiPRS | PRScs.PseudoVal | 0.367 | 0.347 | 0.020 | 3.19e-01 | 0.327 | 0.302 | 0.025 | 1.47e-02 |
| BMI21 | pT+clump.MultiPRS | PRScs.PseudoVal | 0.317 | 0.347 | -0.031 | 1.28e-01 | 0.295 | 0.302 | -0.007 | 6.28e-01 |
| BMI21 | LDpred2.PseudoVal | PRScs.PseudoVal | 0.333 | 0.347 | -0.014 | 4.81e-02 | 0.290 | 0.302 | -0.011 | 4.77e-01 |
| BMI21 | SBayesR.PseudoVal | PRScs.PseudoVal | 0.339 | 0.347 | -0.008 | 1.32e-01 | 0.290 | 0.302 | -0.012 | 2.96e-01 |
| BMI21 | LDpred2.MultiPRS | PRScs.PseudoVal | 0.357 | 0.347 | 0.009 | 6.35e-01 | 0.308 | 0.302 | 0.007 | 5.78e-01 |
| BMI21 | SBLUP.Inf | PRScs.PseudoVal | 0.324 | 0.347 | -0.023 | 3.36e-03 | 0.299 | 0.302 | -0.003 | 8.55e-01 |
| BMI21 | LDpred1.Inf | PRScs.PseudoVal | 0.345 | 0.347 | -0.003 | 7.00e-01 | 0.298 | 0.302 | -0.004 | 7.88e-01 |
| BMI21 | PRScs.10FCVal | PRScs.PseudoVal | 0.353 | 0.347 | 0.005 | 2.81e-01 | 0.326 | 0.302 | 0.024 | 1.49e-02 |
| BMI21 | LDpred2.Inf | PRScs.PseudoVal | 0.333 | 0.347 | -0.014 | 4.88e-02 | 0.290 | 0.302 | -0.011 | 4.61e-01 |
| BMI21 | PRScs.PseudoVal | PRScs.PseudoVal | 0.347 | 0.347 | 0.000 | 1.00e+00 | 0.302 | 0.302 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.10FCVal | PRScs.PseudoVal | 0.360 | 0.347 | 0.012 | 2.38e-02 | 0.315 | 0.302 | 0.013 | 2.43e-01 |
| BMI21 | pT+clump.10FCVal | PRScs.PseudoVal | 0.310 | 0.347 | -0.038 | 1.92e-05 | 0.301 | 0.302 | -0.001 | 9.62e-01 |
| BMI21 | PRScs.MultiPRS | PRScs.PseudoVal | 0.354 | 0.347 | 0.007 | 7.29e-01 | 0.325 | 0.302 | 0.023 | 2.32e-04 |
| BMI21 | lassosum.10FCVal | PRScs.PseudoVal | 0.352 | 0.347 | 0.005 | 3.71e-01 | 0.332 | 0.302 | 0.030 | 6.84e-03 |
| BMI21 | lassosum.PseudoVal | PRScs.PseudoVal | 0.288 | 0.347 | -0.059 | 5.06e-15 | 0.231 | 0.302 | -0.071 | 4.61e-06 |
| BMI21 | LDpred1.MultiPRS | PRScs.PseudoVal | 0.349 | 0.347 | 0.001 | 9.41e-01 | 0.306 | 0.302 | 0.004 | 7.70e-01 |
| BMI21 | lassosum.MultiPRS | PRScs.PseudoVal | 0.361 | 0.347 | 0.013 | 5.11e-01 | 0.334 | 0.302 | 0.032 | 1.34e-03 |
| BMI21 | LDpred1.10FCVal | PRScs.PseudoVal | 0.336 | 0.347 | -0.012 | 1.41e-01 | 0.306 | 0.302 | 0.004 | 8.06e-01 |
| BMI21 | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.354 | 0.347 | 0.006 | 2.26e-01 | 0.303 | 0.302 | 0.001 | 9.29e-01 |
| BMI21 | All.MultiPRS | pT+clump.MultiPRS | 0.367 | 0.317 | 0.050 | 1.18e-02 | 0.327 | 0.295 | 0.032 | 4.17e-02 |
| BMI21 | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.339 | 0.317 | 0.022 | 2.73e-01 | 0.290 | 0.295 | -0.005 | 8.01e-01 |
| BMI21 | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.349 | 0.317 | 0.032 | 1.11e-01 | 0.306 | 0.295 | 0.011 | 5.08e-01 |
| BMI21 | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.317 | 0.317 | 0.000 | 1.00e+00 | 0.295 | 0.295 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.10FCVal | pT+clump.MultiPRS | 0.352 | 0.317 | 0.035 | 7.82e-02 | 0.332 | 0.295 | 0.037 | 1.70e-02 |
| BMI21 | PRScs.10FCVal | pT+clump.MultiPRS | 0.353 | 0.317 | 0.036 | 7.46e-02 | 0.326 | 0.295 | 0.032 | 3.45e-02 |
| BMI21 | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.354 | 0.317 | 0.037 | 6.51e-02 | 0.303 | 0.295 | 0.008 | 5.93e-01 |
| BMI21 | SBLUP.Inf | pT+clump.MultiPRS | 0.324 | 0.317 | 0.008 | 7.08e-01 | 0.299 | 0.295 | 0.004 | 8.29e-01 |
| BMI21 | LDpred2.Inf | pT+clump.MultiPRS | 0.333 | 0.317 | 0.016 | 4.13e-01 | 0.290 | 0.295 | -0.004 | 8.17e-01 |
| BMI21 | pT+clump.10FCVal | pT+clump.MultiPRS | 0.310 | 0.317 | -0.007 | 7.17e-01 | 0.301 | 0.295 | 0.006 | 4.30e-01 |
| BMI21 | LDpred2.10FCVal | pT+clump.MultiPRS | 0.360 | 0.317 | 0.043 | 3.21e-02 | 0.315 | 0.295 | 0.020 | 2.28e-01 |
| BMI21 | PRScs.PseudoVal | pT+clump.MultiPRS | 0.347 | 0.317 | 0.031 | 1.28e-01 | 0.302 | 0.295 | 0.007 | 6.28e-01 |
| BMI21 | LDpred1.10FCVal | pT+clump.MultiPRS | 0.336 | 0.317 | 0.019 | 3.45e-01 | 0.306 | 0.295 | 0.011 | 5.50e-01 |
| BMI21 | LDpred1.Inf | pT+clump.MultiPRS | 0.345 | 0.317 | 0.028 | 1.65e-01 | 0.298 | 0.295 | 0.003 | 8.62e-01 |
| BMI21 | PRScs.MultiPRS | pT+clump.MultiPRS | 0.354 | 0.317 | 0.037 | 6.11e-02 | 0.325 | 0.295 | 0.030 | 3.00e-02 |
| BMI21 | lassosum.PseudoVal | pT+clump.MultiPRS | 0.288 | 0.317 | -0.029 | 1.59e-01 | 0.231 | 0.295 | -0.064 | 2.82e-03 |
| BMI21 | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.333 | 0.317 | 0.016 | 4.23e-01 | 0.290 | 0.295 | -0.004 | 8.23e-01 |
| BMI21 | lassosum.MultiPRS | pT+clump.MultiPRS | 0.361 | 0.317 | 0.044 | 2.92e-02 | 0.334 | 0.295 | 0.039 | 1.69e-02 |
| BMI21 | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.357 | 0.317 | 0.040 | 4.60e-02 | 0.308 | 0.295 | 0.014 | 4.22e-01 |
| BMI21 | LDpred2.10FCVal | pT+clump.10FCVal | 0.360 | 0.310 | 0.050 | 1.48e-08 | 0.315 | 0.301 | 0.014 | 4.34e-01 |
| BMI21 | pT+clump.MultiPRS | pT+clump.10FCVal | 0.317 | 0.310 | 0.007 | 7.17e-01 | 0.295 | 0.301 | -0.006 | 4.30e-01 |
| BMI21 | LDpred2.Inf | pT+clump.10FCVal | 0.333 | 0.310 | 0.024 | 5.78e-03 | 0.290 | 0.301 | -0.010 | 5.55e-01 |
| BMI21 | SBayesR.PseudoVal | pT+clump.10FCVal | 0.339 | 0.310 | 0.029 | 6.51e-03 | 0.290 | 0.301 | -0.011 | 6.24e-01 |
| BMI21 | PRScs.MultiPRS | pT+clump.10FCVal | 0.354 | 0.310 | 0.045 | 2.60e-02 | 0.325 | 0.301 | 0.024 | 1.30e-01 |
| BMI21 | SBLUP.Inf | pT+clump.10FCVal | 0.324 | 0.310 | 0.015 | 8.52e-02 | 0.299 | 0.301 | -0.002 | 9.03e-01 |
| BMI21 | lassosum.MultiPRS | pT+clump.10FCVal | 0.361 | 0.310 | 0.051 | 1.16e-02 | 0.334 | 0.301 | 0.033 | 6.72e-02 |
| BMI21 | PRScs.10FCVal | pT+clump.10FCVal | 0.353 | 0.310 | 0.043 | 1.39e-08 | 0.326 | 0.301 | 0.025 | 9.91e-02 |
| BMI21 | LDpred2.MultiPRS | pT+clump.10FCVal | 0.357 | 0.310 | 0.047 | 1.90e-02 | 0.308 | 0.301 | 0.008 | 6.80e-01 |
| BMI21 | lassosum.PseudoVal | pT+clump.10FCVal | 0.288 | 0.310 | -0.021 | 9.90e-02 | 0.231 | 0.301 | -0.070 | 7.45e-03 |
| BMI21 | LDpred2.PseudoVal | pT+clump.10FCVal | 0.333 | 0.310 | 0.023 | 7.25e-03 | 0.290 | 0.301 | -0.010 | 5.63e-01 |
| BMI21 | LDpred1.MultiPRS | pT+clump.10FCVal | 0.349 | 0.310 | 0.039 | 5.12e-02 | 0.306 | 0.301 | 0.005 | 7.60e-01 |
| BMI21 | pT+clump.10FCVal | pT+clump.10FCVal | 0.310 | 0.310 | 0.000 | 1.00e+00 | 0.301 | 0.301 | 0.000 | 1.00e+00 |
| BMI21 | All.MultiPRS | pT+clump.10FCVal | 0.367 | 0.310 | 0.058 | 4.03e-03 | 0.327 | 0.301 | 0.026 | 1.31e-01 |
| BMI21 | LDpred1.Inf | pT+clump.10FCVal | 0.345 | 0.310 | 0.035 | 5.93e-06 | 0.298 | 0.301 | -0.003 | 8.46e-01 |
| BMI21 | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.354 | 0.310 | 0.044 | 1.47e-07 | 0.303 | 0.301 | 0.002 | 9.11e-01 |
| BMI21 | LDpred1.10FCVal | pT+clump.10FCVal | 0.336 | 0.310 | 0.026 | 2.61e-03 | 0.306 | 0.301 | 0.005 | 7.81e-01 |
| BMI21 | lassosum.10FCVal | pT+clump.10FCVal | 0.352 | 0.310 | 0.043 | 7.13e-07 | 0.332 | 0.301 | 0.031 | 6.73e-02 |
| BMI21 | PRScs.PseudoVal | pT+clump.10FCVal | 0.347 | 0.310 | 0.038 | 1.92e-05 | 0.302 | 0.301 | 0.001 | 9.62e-01 |
| BMI21 | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.317 | 0.339 | -0.022 | 2.73e-01 | 0.295 | 0.290 | 0.005 | 8.01e-01 |
| BMI21 | LDpred1.10FCVal | SBayesR.PseudoVal | 0.336 | 0.339 | -0.003 | 7.68e-01 | 0.306 | 0.290 | 0.016 | 4.69e-01 |
| BMI21 | PRScs.10FCVal | SBayesR.PseudoVal | 0.353 | 0.339 | 0.014 | 9.89e-02 | 0.326 | 0.290 | 0.036 | 3.26e-02 |
| BMI21 | LDpred2.10FCVal | SBayesR.PseudoVal | 0.360 | 0.339 | 0.021 | 6.89e-03 | 0.315 | 0.290 | 0.025 | 1.24e-01 |
| BMI21 | lassosum.PseudoVal | SBayesR.PseudoVal | 0.288 | 0.339 | -0.051 | 8.27e-14 | 0.231 | 0.290 | -0.060 | 2.43e-05 |
| BMI21 | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.333 | 0.339 | -0.006 | 5.46e-01 | 0.290 | 0.290 | 0.000 | 9.84e-01 |
| BMI21 | lassosum.MultiPRS | SBayesR.PseudoVal | 0.361 | 0.339 | 0.022 | 2.82e-01 | 0.334 | 0.290 | 0.044 | 2.88e-03 |
| BMI21 | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.357 | 0.339 | 0.018 | 3.72e-01 | 0.308 | 0.290 | 0.018 | 2.73e-01 |
| BMI21 | All.MultiPRS | SBayesR.PseudoVal | 0.367 | 0.339 | 0.028 | 1.53e-01 | 0.327 | 0.290 | 0.037 | 9.43e-03 |
| BMI21 | PRScs.PseudoVal | SBayesR.PseudoVal | 0.347 | 0.339 | 0.008 | 1.32e-01 | 0.302 | 0.290 | 0.012 | 2.96e-01 |
| BMI21 | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.349 | 0.339 | 0.010 | 6.16e-01 | 0.306 | 0.290 | 0.016 | 4.24e-01 |
| BMI21 | pT+clump.10FCVal | SBayesR.PseudoVal | 0.310 | 0.339 | -0.029 | 6.51e-03 | 0.301 | 0.290 | 0.011 | 6.24e-01 |
| BMI21 | lassosum.10FCVal | SBayesR.PseudoVal | 0.352 | 0.339 | 0.013 | 8.95e-02 | 0.332 | 0.290 | 0.042 | 8.78e-03 |
| BMI21 | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.339 | 0.339 | 0.000 | 1.00e+00 | 0.290 | 0.290 | 0.000 | 1.00e+00 |
| BMI21 | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.354 | 0.339 | 0.015 | 4.11e-02 | 0.303 | 0.290 | 0.013 | 4.21e-01 |
| BMI21 | SBLUP.Inf | SBayesR.PseudoVal | 0.324 | 0.339 | -0.015 | 1.58e-01 | 0.299 | 0.290 | 0.009 | 6.89e-01 |
| BMI21 | LDpred2.Inf | SBayesR.PseudoVal | 0.333 | 0.339 | -0.006 | 5.67e-01 | 0.290 | 0.290 | 0.000 | 9.87e-01 |
| BMI21 | LDpred1.Inf | SBayesR.PseudoVal | 0.345 | 0.339 | 0.006 | 5.43e-01 | 0.298 | 0.290 | 0.008 | 7.13e-01 |
| BMI21 | PRScs.MultiPRS | SBayesR.PseudoVal | 0.354 | 0.339 | 0.015 | 4.39e-01 | 0.325 | 0.290 | 0.035 | 1.15e-02 |
| BMI21 | LDpred2.10FCVal | SBLUP.Inf | 0.360 | 0.324 | 0.035 | 2.96e-07 | 0.315 | 0.299 | 0.016 | 2.55e-01 |
| BMI21 | PRScs.PseudoVal | SBLUP.Inf | 0.347 | 0.324 | 0.023 | 3.36e-03 | 0.302 | 0.299 | 0.003 | 8.55e-01 |
| BMI21 | LDpred1.10FCVal | SBLUP.Inf | 0.336 | 0.324 | 0.011 | 5.10e-02 | 0.306 | 0.299 | 0.007 | 5.66e-01 |
| BMI21 | LDpred2.Inf | SBLUP.Inf | 0.333 | 0.324 | 0.009 | 1.13e-01 | 0.290 | 0.299 | -0.008 | 4.86e-01 |
| BMI21 | SBayesR.PseudoVal | SBLUP.Inf | 0.339 | 0.324 | 0.015 | 1.58e-01 | 0.290 | 0.299 | -0.009 | 6.89e-01 |
| BMI21 | lassosum.PseudoVal | SBLUP.Inf | 0.288 | 0.324 | -0.036 | 4.71e-03 | 0.231 | 0.299 | -0.068 | 1.07e-02 |
| BMI21 | pT+clump.MultiPRS | SBLUP.Inf | 0.317 | 0.324 | -0.008 | 7.08e-01 | 0.295 | 0.299 | -0.004 | 8.29e-01 |
| BMI21 | lassosum.MultiPRS | SBLUP.Inf | 0.361 | 0.324 | 0.036 | 7.19e-02 | 0.334 | 0.299 | 0.035 | 2.83e-02 |
| BMI21 | PRScs.10FCVal | SBLUP.Inf | 0.353 | 0.324 | 0.028 | 2.97e-07 | 0.326 | 0.299 | 0.028 | 1.85e-02 |
| BMI21 | PRScs.MultiPRS | SBLUP.Inf | 0.354 | 0.324 | 0.030 | 1.37e-01 | 0.325 | 0.299 | 0.026 | 4.47e-02 |
| BMI21 | LDpred1.MultiPRS | SBLUP.Inf | 0.349 | 0.324 | 0.024 | 2.25e-01 | 0.306 | 0.299 | 0.007 | 5.15e-01 |
| BMI21 | LDpred2.PseudoVal | SBLUP.Inf | 0.333 | 0.324 | 0.009 | 1.34e-01 | 0.290 | 0.299 | -0.008 | 4.95e-01 |
| BMI21 | LDpred1.Inf | SBLUP.Inf | 0.345 | 0.324 | 0.020 | 7.45e-05 | 0.298 | 0.299 | -0.001 | 9.26e-01 |
| BMI21 | LDpred2.MultiPRS | SBLUP.Inf | 0.357 | 0.324 | 0.032 | 1.05e-01 | 0.308 | 0.299 | 0.010 | 5.15e-01 |
| BMI21 | All.MultiPRS | SBLUP.Inf | 0.367 | 0.324 | 0.043 | 3.29e-02 | 0.327 | 0.299 | 0.028 | 5.84e-02 |
| BMI21 | SBLUP.Inf | SBLUP.Inf | 0.324 | 0.324 | 0.000 | 1.00e+00 | 0.299 | 0.299 | 0.000 | 1.00e+00 |
| BMI21 | DBSLMM.PseudoVal | SBLUP.Inf | 0.354 | 0.324 | 0.029 | 4.67e-05 | 0.303 | 0.299 | 0.004 | 7.86e-01 |
| BMI21 | pT+clump.10FCVal | SBLUP.Inf | 0.310 | 0.324 | -0.015 | 8.52e-02 | 0.301 | 0.299 | 0.002 | 9.03e-01 |
| BMI21 | lassosum.10FCVal | SBLUP.Inf | 0.352 | 0.324 | 0.028 | 5.02e-04 | 0.332 | 0.299 | 0.033 | 3.58e-02 |
| GCSE | LDpred2.PseudoVal | All.MultiPRS | 0.406 | 0.417 | -0.012 | 4.66e-01 | 0.332 | 0.347 | -0.014 | 2.51e-02 |
| GCSE | pT+clump.10FCVal | All.MultiPRS | 0.373 | 0.417 | -0.044 | 5.86e-03 | 0.311 | 0.347 | -0.035 | 3.75e-03 |
| GCSE | LDpred2.MultiPRS | All.MultiPRS | 0.412 | 0.417 | -0.005 | 1.61e-02 | 0.339 | 0.347 | -0.008 | 6.48e-02 |
| GCSE | PRScs.PseudoVal | All.MultiPRS | 0.397 | 0.417 | -0.020 | 2.11e-01 | 0.351 | 0.347 | 0.005 | 5.38e-01 |
| GCSE | LDpred1.Inf | All.MultiPRS | 0.408 | 0.417 | -0.010 | 5.46e-01 | 0.341 | 0.347 | -0.006 | 4.01e-01 |
| GCSE | PRScs.MultiPRS | All.MultiPRS | 0.413 | 0.417 | -0.004 | 7.89e-01 | 0.346 | 0.347 | -0.001 | 8.67e-01 |
| GCSE | LDpred2.Inf | All.MultiPRS | 0.405 | 0.417 | -0.012 | 4.52e-01 | 0.331 | 0.347 | -0.016 | 1.59e-02 |
| GCSE | lassosum.PseudoVal | All.MultiPRS | 0.321 | 0.417 | -0.097 | 4.07e-09 | 0.276 | 0.347 | -0.071 | 3.89e-05 |
| GCSE | LDpred2.10FCVal | All.MultiPRS | 0.412 | 0.417 | -0.006 | 7.17e-01 | 0.342 | 0.347 | -0.004 | 3.38e-01 |
| GCSE | lassosum.MultiPRS | All.MultiPRS | 0.411 | 0.417 | -0.006 | 6.98e-01 | 0.347 | 0.347 | 0.000 | 9.59e-01 |
| GCSE | LDpred1.10FCVal | All.MultiPRS | 0.402 | 0.417 | -0.015 | 3.35e-01 | 0.330 | 0.347 | -0.016 | 3.81e-02 |
| GCSE | All.MultiPRS | All.MultiPRS | 0.417 | 0.417 | 0.000 | 1.00e+00 | 0.347 | 0.347 | 0.000 | 1.00e+00 |
| GCSE | SBayesR.PseudoVal | All.MultiPRS | 0.397 | 0.417 | -0.021 | 1.96e-01 | 0.331 | 0.347 | -0.016 | 9.45e-02 |
| GCSE | LDpred1.MultiPRS | All.MultiPRS | 0.409 | 0.417 | -0.008 | 6.07e-01 | 0.340 | 0.347 | -0.007 | 2.45e-01 |
| GCSE | pT+clump.MultiPRS | All.MultiPRS | 0.381 | 0.417 | -0.037 | 2.16e-02 | 0.328 | 0.347 | -0.018 | 7.70e-02 |
| GCSE | lassosum.10FCVal | All.MultiPRS | 0.407 | 0.417 | -0.010 | 5.29e-01 | 0.348 | 0.347 | 0.001 | 8.42e-01 |
| GCSE | PRScs.10FCVal | All.MultiPRS | 0.410 | 0.417 | -0.007 | 6.61e-01 | 0.341 | 0.347 | -0.006 | 2.92e-01 |
| GCSE | DBSLMM.PseudoVal | All.MultiPRS | 0.401 | 0.417 | -0.016 | 3.14e-01 | 0.337 | 0.347 | -0.010 | 1.66e-01 |
| GCSE | SBLUP.Inf | All.MultiPRS | 0.400 | 0.417 | -0.018 | 2.66e-01 | 0.335 | 0.347 | -0.012 | 1.47e-01 |
| GCSE | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.397 | 0.401 | -0.005 | 3.44e-01 | 0.331 | 0.337 | -0.006 | 5.72e-01 |
| GCSE | LDpred2.Inf | DBSLMM.PseudoVal | 0.405 | 0.401 | 0.004 | 3.90e-01 | 0.331 | 0.337 | -0.006 | 5.44e-01 |
| GCSE | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.373 | 0.401 | -0.028 | 2.61e-05 | 0.311 | 0.337 | -0.026 | 7.14e-02 |
| GCSE | PRScs.10FCVal | DBSLMM.PseudoVal | 0.410 | 0.401 | 0.009 | 1.71e-02 | 0.341 | 0.337 | 0.004 | 6.46e-01 |
| GCSE | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.397 | 0.401 | -0.004 | 3.38e-01 | 0.351 | 0.337 | 0.014 | 9.88e-02 |
| GCSE | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.381 | 0.401 | -0.021 | 2.13e-01 | 0.328 | 0.337 | -0.009 | 4.68e-01 |
| GCSE | LDpred1.Inf | DBSLMM.PseudoVal | 0.408 | 0.401 | 0.006 | 1.45e-01 | 0.341 | 0.337 | 0.004 | 6.60e-01 |
| GCSE | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.411 | 0.401 | 0.010 | 5.35e-01 | 0.347 | 0.337 | 0.010 | 2.52e-01 |
| GCSE | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.412 | 0.401 | 0.010 | 1.61e-02 | 0.342 | 0.337 | 0.005 | 5.58e-01 |
| GCSE | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.321 | 0.401 | -0.081 | 3.95e-22 | 0.276 | 0.337 | -0.061 | 6.21e-04 |
| GCSE | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.402 | 0.401 | 0.001 | 8.86e-01 | 0.330 | 0.337 | -0.006 | 5.42e-01 |
| GCSE | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.409 | 0.401 | 0.008 | 6.22e-01 | 0.340 | 0.337 | 0.003 | 7.50e-01 |
| GCSE | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.413 | 0.401 | 0.012 | 4.59e-01 | 0.346 | 0.337 | 0.009 | 1.97e-01 |
| GCSE | All.MultiPRS | DBSLMM.PseudoVal | 0.417 | 0.401 | 0.016 | 3.14e-01 | 0.347 | 0.337 | 0.010 | 1.66e-01 |
| GCSE | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.406 | 0.401 | 0.004 | 3.40e-01 | 0.332 | 0.337 | -0.004 | 6.50e-01 |
| GCSE | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.401 | 0.401 | 0.000 | 1.00e+00 | 0.337 | 0.337 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.412 | 0.401 | 0.011 | 4.79e-01 | 0.339 | 0.337 | 0.002 | 8.05e-01 |
| GCSE | lassosum.10FCVal | DBSLMM.PseudoVal | 0.407 | 0.401 | 0.006 | 1.61e-01 | 0.348 | 0.337 | 0.011 | 2.43e-01 |
| GCSE | SBLUP.Inf | DBSLMM.PseudoVal | 0.400 | 0.401 | -0.002 | 7.40e-01 | 0.335 | 0.337 | -0.002 | 8.41e-01 |
| GCSE | SBayesR.PseudoVal | lassosum.MultiPRS | 0.397 | 0.411 | -0.015 | 3.63e-01 | 0.331 | 0.347 | -0.016 | 1.26e-01 |
| GCSE | LDpred2.10FCVal | lassosum.MultiPRS | 0.412 | 0.411 | 0.000 | 9.78e-01 | 0.342 | 0.347 | -0.005 | 4.81e-01 |
| GCSE | LDpred1.10FCVal | lassosum.MultiPRS | 0.402 | 0.411 | -0.009 | 5.66e-01 | 0.330 | 0.347 | -0.016 | 8.28e-02 |
| GCSE | LDpred2.Inf | lassosum.MultiPRS | 0.405 | 0.411 | -0.006 | 7.16e-01 | 0.331 | 0.347 | -0.016 | 7.29e-02 |
| GCSE | PRScs.10FCVal | lassosum.MultiPRS | 0.410 | 0.411 | -0.001 | 9.61e-01 | 0.341 | 0.347 | -0.006 | 3.58e-01 |
| GCSE | PRScs.MultiPRS | lassosum.MultiPRS | 0.413 | 0.411 | 0.002 | 4.70e-01 | 0.346 | 0.347 | -0.001 | 8.58e-01 |
| GCSE | pT+clump.MultiPRS | lassosum.MultiPRS | 0.381 | 0.411 | -0.030 | 6.07e-02 | 0.328 | 0.347 | -0.019 | 1.13e-01 |
| GCSE | lassosum.MultiPRS | lassosum.MultiPRS | 0.411 | 0.411 | 0.000 | 1.00e+00 | 0.347 | 0.347 | 0.000 | 1.00e+00 |
| GCSE | lassosum.10FCVal | lassosum.MultiPRS | 0.407 | 0.411 | -0.004 | 8.11e-01 | 0.348 | 0.347 | 0.001 | 7.97e-01 |
| GCSE | LDpred2.MultiPRS | lassosum.MultiPRS | 0.412 | 0.411 | 0.001 | 9.29e-01 | 0.339 | 0.347 | -0.008 | 2.75e-01 |
| GCSE | LDpred1.MultiPRS | lassosum.MultiPRS | 0.409 | 0.411 | -0.002 | 6.04e-01 | 0.340 | 0.347 | -0.007 | 3.80e-01 |
| GCSE | LDpred2.PseudoVal | lassosum.MultiPRS | 0.406 | 0.411 | -0.005 | 7.34e-01 | 0.332 | 0.347 | -0.014 | 9.52e-02 |
| GCSE | LDpred1.Inf | lassosum.MultiPRS | 0.408 | 0.411 | -0.003 | 8.32e-01 | 0.341 | 0.347 | -0.006 | 4.86e-01 |
| GCSE | pT+clump.10FCVal | lassosum.MultiPRS | 0.373 | 0.411 | -0.038 | 1.81e-02 | 0.311 | 0.347 | -0.036 | 7.92e-03 |
| GCSE | lassosum.PseudoVal | lassosum.MultiPRS | 0.321 | 0.411 | -0.091 | 3.52e-08 | 0.276 | 0.347 | -0.071 | 2.48e-05 |
| GCSE | SBLUP.Inf | lassosum.MultiPRS | 0.400 | 0.411 | -0.012 | 4.70e-01 | 0.335 | 0.347 | -0.012 | 2.25e-01 |
| GCSE | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.401 | 0.411 | -0.010 | 5.35e-01 | 0.337 | 0.347 | -0.010 | 2.52e-01 |
| GCSE | All.MultiPRS | lassosum.MultiPRS | 0.417 | 0.411 | 0.006 | 6.98e-01 | 0.347 | 0.347 | 0.000 | 9.59e-01 |
| GCSE | PRScs.PseudoVal | lassosum.MultiPRS | 0.397 | 0.411 | -0.014 | 3.89e-01 | 0.351 | 0.347 | 0.004 | 5.77e-01 |
| GCSE | LDpred1.Inf | lassosum.10FCVal | 0.408 | 0.407 | 0.000 | 9.14e-01 | 0.341 | 0.348 | -0.007 | 4.22e-01 |
| GCSE | PRScs.10FCVal | lassosum.10FCVal | 0.410 | 0.407 | 0.003 | 4.22e-01 | 0.341 | 0.348 | -0.007 | 3.70e-01 |
| GCSE | LDpred2.MultiPRS | lassosum.10FCVal | 0.412 | 0.407 | 0.005 | 7.42e-01 | 0.339 | 0.348 | -0.009 | 2.12e-01 |
| GCSE | lassosum.10FCVal | lassosum.10FCVal | 0.407 | 0.407 | 0.000 | 1.00e+00 | 0.348 | 0.348 | 0.000 | 1.00e+00 |
| GCSE | PRScs.PseudoVal | lassosum.10FCVal | 0.397 | 0.407 | -0.010 | 2.12e-03 | 0.351 | 0.348 | 0.003 | 6.16e-01 |
| GCSE | DBSLMM.PseudoVal | lassosum.10FCVal | 0.401 | 0.407 | -0.006 | 1.61e-01 | 0.337 | 0.348 | -0.011 | 2.43e-01 |
| GCSE | SBLUP.Inf | lassosum.10FCVal | 0.400 | 0.407 | -0.008 | 1.14e-01 | 0.335 | 0.348 | -0.013 | 1.98e-01 |
| GCSE | LDpred2.Inf | lassosum.10FCVal | 0.405 | 0.407 | -0.002 | 6.46e-01 | 0.331 | 0.348 | -0.017 | 5.47e-02 |
| GCSE | pT+clump.10FCVal | lassosum.10FCVal | 0.373 | 0.407 | -0.034 | 3.87e-07 | 0.311 | 0.348 | -0.036 | 9.69e-03 |
| GCSE | LDpred2.10FCVal | lassosum.10FCVal | 0.412 | 0.407 | 0.004 | 1.62e-01 | 0.342 | 0.348 | -0.006 | 3.59e-01 |
| GCSE | lassosum.MultiPRS | lassosum.10FCVal | 0.411 | 0.407 | 0.004 | 8.11e-01 | 0.347 | 0.348 | -0.001 | 7.97e-01 |
| GCSE | LDpred1.10FCVal | lassosum.10FCVal | 0.402 | 0.407 | -0.005 | 2.45e-01 | 0.330 | 0.348 | -0.017 | 7.38e-02 |
| GCSE | All.MultiPRS | lassosum.10FCVal | 0.417 | 0.407 | 0.010 | 5.29e-01 | 0.347 | 0.348 | -0.001 | 8.42e-01 |
| GCSE | SBayesR.PseudoVal | lassosum.10FCVal | 0.397 | 0.407 | -0.011 | 1.91e-02 | 0.331 | 0.348 | -0.017 | 8.58e-02 |
| GCSE | lassosum.PseudoVal | lassosum.10FCVal | 0.321 | 0.407 | -0.087 | 1.70e-32 | 0.276 | 0.348 | -0.072 | 3.58e-06 |
| GCSE | pT+clump.MultiPRS | lassosum.10FCVal | 0.381 | 0.407 | -0.027 | 1.06e-01 | 0.328 | 0.348 | -0.019 | 1.07e-01 |
| GCSE | LDpred2.PseudoVal | lassosum.10FCVal | 0.406 | 0.407 | -0.002 | 6.94e-01 | 0.332 | 0.348 | -0.015 | 6.39e-02 |
| GCSE | LDpred1.MultiPRS | lassosum.10FCVal | 0.409 | 0.407 | 0.002 | 9.10e-01 | 0.340 | 0.348 | -0.008 | 3.29e-01 |
| GCSE | PRScs.MultiPRS | lassosum.10FCVal | 0.413 | 0.407 | 0.006 | 7.18e-01 | 0.346 | 0.348 | -0.002 | 7.59e-01 |
| GCSE | lassosum.PseudoVal | lassosum.PseudoVal | 0.321 | 0.321 | 0.000 | 1.00e+00 | 0.276 | 0.276 | 0.000 | 1.00e+00 |
| GCSE | pT+clump.MultiPRS | lassosum.PseudoVal | 0.381 | 0.321 | 0.060 | 3.70e-04 | 0.328 | 0.276 | 0.052 | 5.51e-03 |
| GCSE | LDpred2.Inf | lassosum.PseudoVal | 0.405 | 0.321 | 0.085 | 1.54e-18 | 0.331 | 0.276 | 0.055 | 6.84e-03 |
| GCSE | SBayesR.PseudoVal | lassosum.PseudoVal | 0.397 | 0.321 | 0.076 | 2.14e-37 | 0.331 | 0.276 | 0.055 | 1.49e-05 |
| GCSE | PRScs.MultiPRS | lassosum.PseudoVal | 0.413 | 0.321 | 0.092 | 1.67e-08 | 0.346 | 0.276 | 0.070 | 6.81e-05 |
| GCSE | SBLUP.Inf | lassosum.PseudoVal | 0.400 | 0.321 | 0.079 | 3.28e-15 | 0.335 | 0.276 | 0.059 | 5.25e-03 |
| GCSE | lassosum.MultiPRS | lassosum.PseudoVal | 0.411 | 0.321 | 0.091 | 3.52e-08 | 0.347 | 0.276 | 0.071 | 2.48e-05 |
| GCSE | PRScs.10FCVal | lassosum.PseudoVal | 0.410 | 0.321 | 0.090 | 9.90e-22 | 0.341 | 0.276 | 0.064 | 1.14e-03 |
| GCSE | LDpred2.MultiPRS | lassosum.PseudoVal | 0.412 | 0.321 | 0.092 | 2.37e-08 | 0.339 | 0.276 | 0.063 | 2.25e-04 |
| GCSE | PRScs.PseudoVal | lassosum.PseudoVal | 0.397 | 0.321 | 0.077 | 7.69e-31 | 0.351 | 0.276 | 0.075 | 4.42e-08 |
| GCSE | LDpred2.PseudoVal | lassosum.PseudoVal | 0.406 | 0.321 | 0.085 | 2.71e-19 | 0.332 | 0.276 | 0.056 | 4.53e-03 |
| GCSE | LDpred1.MultiPRS | lassosum.PseudoVal | 0.409 | 0.321 | 0.089 | 7.46e-08 | 0.340 | 0.276 | 0.064 | 1.37e-03 |
| GCSE | pT+clump.10FCVal | lassosum.PseudoVal | 0.373 | 0.321 | 0.052 | 1.02e-06 | 0.311 | 0.276 | 0.035 | 1.22e-01 |
| GCSE | All.MultiPRS | lassosum.PseudoVal | 0.417 | 0.321 | 0.097 | 4.07e-09 | 0.347 | 0.276 | 0.071 | 3.89e-05 |
| GCSE | LDpred1.Inf | lassosum.PseudoVal | 0.408 | 0.321 | 0.087 | 2.61e-20 | 0.341 | 0.276 | 0.065 | 1.22e-03 |
| GCSE | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.401 | 0.321 | 0.081 | 3.95e-22 | 0.337 | 0.276 | 0.061 | 6.21e-04 |
| GCSE | LDpred1.10FCVal | lassosum.PseudoVal | 0.402 | 0.321 | 0.081 | 1.52e-17 | 0.330 | 0.276 | 0.054 | 6.77e-03 |
| GCSE | lassosum.10FCVal | lassosum.PseudoVal | 0.407 | 0.321 | 0.087 | 1.70e-32 | 0.348 | 0.276 | 0.072 | 3.58e-06 |
| GCSE | LDpred2.10FCVal | lassosum.PseudoVal | 0.412 | 0.321 | 0.091 | 5.61e-28 | 0.342 | 0.276 | 0.066 | 1.45e-04 |
| GCSE | LDpred2.10FCVal | LDpred1.MultiPRS | 0.412 | 0.409 | 0.002 | 8.78e-01 | 0.342 | 0.340 | 0.002 | 7.42e-01 |
| GCSE | PRScs.PseudoVal | LDpred1.MultiPRS | 0.397 | 0.409 | -0.012 | 4.63e-01 | 0.351 | 0.340 | 0.011 | 2.69e-01 |
| GCSE | LDpred2.Inf | LDpred1.MultiPRS | 0.405 | 0.409 | -0.004 | 8.13e-01 | 0.331 | 0.340 | -0.009 | 1.47e-01 |
| GCSE | pT+clump.10FCVal | LDpred1.MultiPRS | 0.373 | 0.409 | -0.036 | 2.56e-02 | 0.311 | 0.340 | -0.028 | 3.00e-02 |
| GCSE | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.397 | 0.409 | -0.013 | 4.34e-01 | 0.331 | 0.340 | -0.009 | 4.97e-01 |
| GCSE | All.MultiPRS | LDpred1.MultiPRS | 0.417 | 0.409 | 0.008 | 6.07e-01 | 0.347 | 0.340 | 0.007 | 2.45e-01 |
| GCSE | LDpred1.10FCVal | LDpred1.MultiPRS | 0.402 | 0.409 | -0.007 | 6.55e-01 | 0.330 | 0.340 | -0.009 | 7.63e-02 |
| GCSE | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.409 | 0.409 | 0.000 | 1.00e+00 | 0.340 | 0.340 | 0.000 | 1.00e+00 |
| GCSE | PRScs.10FCVal | LDpred1.MultiPRS | 0.410 | 0.409 | 0.001 | 9.38e-01 | 0.341 | 0.340 | 0.001 | 8.96e-01 |
| GCSE | lassosum.10FCVal | LDpred1.MultiPRS | 0.407 | 0.409 | -0.002 | 9.10e-01 | 0.348 | 0.340 | 0.008 | 3.29e-01 |
| GCSE | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.381 | 0.409 | -0.028 | 8.01e-02 | 0.328 | 0.340 | -0.012 | 3.54e-01 |
| GCSE | LDpred1.Inf | LDpred1.MultiPRS | 0.408 | 0.409 | -0.001 | 9.32e-01 | 0.341 | 0.340 | 0.001 | 6.81e-01 |
| GCSE | lassosum.MultiPRS | LDpred1.MultiPRS | 0.411 | 0.409 | 0.002 | 6.04e-01 | 0.347 | 0.340 | 0.007 | 3.80e-01 |
| GCSE | PRScs.MultiPRS | LDpred1.MultiPRS | 0.413 | 0.409 | 0.004 | 1.89e-01 | 0.346 | 0.340 | 0.006 | 3.37e-01 |
| GCSE | lassosum.PseudoVal | LDpred1.MultiPRS | 0.321 | 0.409 | -0.089 | 7.46e-08 | 0.276 | 0.340 | -0.064 | 1.37e-03 |
| GCSE | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.406 | 0.409 | -0.003 | 8.31e-01 | 0.332 | 0.340 | -0.007 | 2.15e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.401 | 0.409 | -0.008 | 6.22e-01 | 0.337 | 0.340 | -0.003 | 7.50e-01 |
| GCSE | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.412 | 0.409 | 0.003 | 8.30e-01 | 0.339 | 0.340 | -0.001 | 9.05e-01 |
| GCSE | SBLUP.Inf | LDpred1.MultiPRS | 0.400 | 0.409 | -0.010 | 5.52e-01 | 0.335 | 0.340 | -0.005 | 4.80e-01 |
| GCSE | All.MultiPRS | LDpred1.Inf | 0.417 | 0.408 | 0.010 | 5.46e-01 | 0.347 | 0.341 | 0.006 | 4.01e-01 |
| GCSE | LDpred2.PseudoVal | LDpred1.Inf | 0.406 | 0.408 | -0.002 | 4.65e-01 | 0.332 | 0.341 | -0.009 | 1.41e-01 |
| GCSE | SBayesR.PseudoVal | LDpred1.Inf | 0.397 | 0.408 | -0.011 | 7.41e-02 | 0.331 | 0.341 | -0.010 | 4.47e-01 |
| GCSE | LDpred2.MultiPRS | LDpred1.Inf | 0.412 | 0.408 | 0.005 | 7.63e-01 | 0.339 | 0.341 | -0.002 | 7.79e-01 |
| GCSE | PRScs.PseudoVal | LDpred1.Inf | 0.397 | 0.408 | -0.010 | 3.09e-02 | 0.351 | 0.341 | 0.010 | 3.37e-01 |
| GCSE | LDpred1.Inf | LDpred1.Inf | 0.408 | 0.408 | 0.000 | 1.00e+00 | 0.341 | 0.341 | 0.000 | 1.00e+00 |
| GCSE | pT+clump.10FCVal | LDpred1.Inf | 0.373 | 0.408 | -0.035 | 1.52e-08 | 0.311 | 0.341 | -0.030 | 2.37e-02 |
| GCSE | PRScs.MultiPRS | LDpred1.Inf | 0.413 | 0.408 | 0.005 | 7.39e-01 | 0.346 | 0.341 | 0.005 | 4.73e-01 |
| GCSE | lassosum.10FCVal | LDpred1.Inf | 0.407 | 0.408 | 0.000 | 9.14e-01 | 0.348 | 0.341 | 0.007 | 4.22e-01 |
| GCSE | pT+clump.MultiPRS | LDpred1.Inf | 0.381 | 0.408 | -0.027 | 9.98e-02 | 0.328 | 0.341 | -0.013 | 3.07e-01 |
| GCSE | LDpred1.MultiPRS | LDpred1.Inf | 0.409 | 0.408 | 0.001 | 9.32e-01 | 0.340 | 0.341 | -0.001 | 6.81e-01 |
| GCSE | PRScs.10FCVal | LDpred1.Inf | 0.410 | 0.408 | 0.003 | 3.87e-01 | 0.341 | 0.341 | 0.000 | 9.39e-01 |
| GCSE | LDpred2.Inf | LDpred1.Inf | 0.405 | 0.408 | -0.002 | 4.09e-01 | 0.331 | 0.341 | -0.010 | 9.52e-02 |
| GCSE | SBLUP.Inf | LDpred1.Inf | 0.400 | 0.408 | -0.008 | 2.27e-02 | 0.335 | 0.341 | -0.006 | 4.01e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred1.Inf | 0.401 | 0.408 | -0.006 | 1.45e-01 | 0.337 | 0.341 | -0.004 | 6.60e-01 |
| GCSE | lassosum.MultiPRS | LDpred1.Inf | 0.411 | 0.408 | 0.003 | 8.32e-01 | 0.347 | 0.341 | 0.006 | 4.86e-01 |
| GCSE | LDpred1.10FCVal | LDpred1.Inf | 0.402 | 0.408 | -0.006 | 9.78e-02 | 0.330 | 0.341 | -0.011 | 1.54e-01 |
| GCSE | lassosum.PseudoVal | LDpred1.Inf | 0.321 | 0.408 | -0.087 | 2.61e-20 | 0.276 | 0.341 | -0.065 | 1.22e-03 |
| GCSE | LDpred2.10FCVal | LDpred1.Inf | 0.412 | 0.408 | 0.004 | 2.49e-01 | 0.342 | 0.341 | 0.001 | 8.84e-01 |
| GCSE | LDpred1.Inf | LDpred1.10FCVal | 0.408 | 0.402 | 0.006 | 9.78e-02 | 0.341 | 0.330 | 0.011 | 1.54e-01 |
| GCSE | PRScs.MultiPRS | LDpred1.10FCVal | 0.413 | 0.402 | 0.011 | 4.87e-01 | 0.346 | 0.330 | 0.015 | 5.88e-02 |
| GCSE | LDpred1.10FCVal | LDpred1.10FCVal | 0.402 | 0.402 | 0.000 | 1.00e+00 | 0.330 | 0.330 | 0.000 | 1.00e+00 |
| GCSE | All.MultiPRS | LDpred1.10FCVal | 0.417 | 0.402 | 0.015 | 3.35e-01 | 0.347 | 0.330 | 0.016 | 3.81e-02 |
| GCSE | SBayesR.PseudoVal | LDpred1.10FCVal | 0.397 | 0.402 | -0.005 | 4.08e-01 | 0.331 | 0.330 | 0.000 | 9.75e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.401 | 0.402 | -0.001 | 8.86e-01 | 0.337 | 0.330 | 0.006 | 5.42e-01 |
| GCSE | pT+clump.MultiPRS | LDpred1.10FCVal | 0.381 | 0.402 | -0.021 | 1.97e-01 | 0.328 | 0.330 | -0.002 | 8.69e-01 |
| GCSE | lassosum.MultiPRS | LDpred1.10FCVal | 0.411 | 0.402 | 0.009 | 5.66e-01 | 0.347 | 0.330 | 0.016 | 8.28e-02 |
| GCSE | PRScs.10FCVal | LDpred1.10FCVal | 0.410 | 0.402 | 0.008 | 2.62e-02 | 0.341 | 0.330 | 0.010 | 2.04e-01 |
| GCSE | lassosum.PseudoVal | LDpred1.10FCVal | 0.321 | 0.402 | -0.081 | 1.52e-17 | 0.276 | 0.330 | -0.054 | 6.77e-03 |
| GCSE | SBLUP.Inf | LDpred1.10FCVal | 0.400 | 0.402 | -0.002 | 5.73e-01 | 0.335 | 0.330 | 0.004 | 6.50e-01 |
| GCSE | LDpred2.PseudoVal | LDpred1.10FCVal | 0.406 | 0.402 | 0.004 | 3.47e-01 | 0.332 | 0.330 | 0.002 | 8.25e-01 |
| GCSE | pT+clump.10FCVal | LDpred1.10FCVal | 0.373 | 0.402 | -0.029 | 1.56e-05 | 0.311 | 0.330 | -0.019 | 1.77e-01 |
| GCSE | LDpred2.MultiPRS | LDpred1.10FCVal | 0.412 | 0.402 | 0.011 | 5.06e-01 | 0.339 | 0.330 | 0.008 | 3.59e-01 |
| GCSE | PRScs.PseudoVal | LDpred1.10FCVal | 0.397 | 0.402 | -0.005 | 3.81e-01 | 0.351 | 0.330 | 0.021 | 6.81e-02 |
| GCSE | LDpred2.10FCVal | LDpred1.10FCVal | 0.412 | 0.402 | 0.010 | 2.10e-02 | 0.342 | 0.330 | 0.012 | 1.88e-01 |
| GCSE | LDpred1.MultiPRS | LDpred1.10FCVal | 0.409 | 0.402 | 0.007 | 6.55e-01 | 0.340 | 0.330 | 0.009 | 7.63e-02 |
| GCSE | LDpred2.Inf | LDpred1.10FCVal | 0.405 | 0.402 | 0.003 | 4.05e-01 | 0.331 | 0.330 | 0.000 | 9.74e-01 |
| GCSE | lassosum.10FCVal | LDpred1.10FCVal | 0.407 | 0.402 | 0.005 | 2.45e-01 | 0.348 | 0.330 | 0.017 | 7.38e-02 |
| GCSE | LDpred1.Inf | LDpred2.MultiPRS | 0.408 | 0.412 | -0.005 | 7.63e-01 | 0.341 | 0.339 | 0.002 | 7.79e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.401 | 0.412 | -0.011 | 4.79e-01 | 0.337 | 0.339 | -0.002 | 8.05e-01 |
| GCSE | SBLUP.Inf | LDpred2.MultiPRS | 0.400 | 0.412 | -0.013 | 4.17e-01 | 0.335 | 0.339 | -0.004 | 6.48e-01 |
| GCSE | LDpred2.Inf | LDpred2.MultiPRS | 0.405 | 0.412 | -0.007 | 6.51e-01 | 0.331 | 0.339 | -0.008 | 1.10e-01 |
| GCSE | pT+clump.10FCVal | LDpred2.MultiPRS | 0.373 | 0.412 | -0.040 | 1.41e-02 | 0.311 | 0.339 | -0.028 | 4.78e-02 |
| GCSE | LDpred2.10FCVal | LDpred2.MultiPRS | 0.412 | 0.412 | -0.001 | 9.50e-01 | 0.342 | 0.339 | 0.003 | 2.48e-01 |
| GCSE | PRScs.PseudoVal | LDpred2.MultiPRS | 0.397 | 0.412 | -0.015 | 3.42e-01 | 0.351 | 0.339 | 0.012 | 1.31e-01 |
| GCSE | LDpred1.10FCVal | LDpred2.MultiPRS | 0.402 | 0.412 | -0.011 | 5.06e-01 | 0.330 | 0.339 | -0.008 | 3.59e-01 |
| GCSE | All.MultiPRS | LDpred2.MultiPRS | 0.417 | 0.412 | 0.005 | 1.61e-02 | 0.347 | 0.339 | 0.008 | 6.48e-02 |
| GCSE | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.397 | 0.412 | -0.016 | 3.20e-01 | 0.331 | 0.339 | -0.008 | 4.36e-01 |
| GCSE | lassosum.PseudoVal | LDpred2.MultiPRS | 0.321 | 0.412 | -0.092 | 2.37e-08 | 0.276 | 0.339 | -0.063 | 2.25e-04 |
| GCSE | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.381 | 0.412 | -0.032 | 4.60e-02 | 0.328 | 0.339 | -0.011 | 3.77e-01 |
| GCSE | lassosum.MultiPRS | LDpred2.MultiPRS | 0.411 | 0.412 | -0.001 | 9.29e-01 | 0.347 | 0.339 | 0.008 | 2.75e-01 |
| GCSE | PRScs.10FCVal | LDpred2.MultiPRS | 0.410 | 0.412 | -0.002 | 8.90e-01 | 0.341 | 0.339 | 0.002 | 8.28e-01 |
| GCSE | PRScs.MultiPRS | LDpred2.MultiPRS | 0.413 | 0.412 | 0.001 | 9.75e-01 | 0.346 | 0.339 | 0.007 | 2.80e-01 |
| GCSE | lassosum.10FCVal | LDpred2.MultiPRS | 0.407 | 0.412 | -0.005 | 7.42e-01 | 0.348 | 0.339 | 0.009 | 2.12e-01 |
| GCSE | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.406 | 0.412 | -0.007 | 6.68e-01 | 0.332 | 0.339 | -0.007 | 1.83e-01 |
| GCSE | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.409 | 0.412 | -0.003 | 8.30e-01 | 0.340 | 0.339 | 0.001 | 9.05e-01 |
| GCSE | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.412 | 0.412 | 0.000 | 1.00e+00 | 0.339 | 0.339 | 0.000 | 1.00e+00 |
| GCSE | LDpred1.Inf | LDpred2.10FCVal | 0.408 | 0.412 | -0.004 | 2.49e-01 | 0.341 | 0.342 | -0.001 | 8.84e-01 |
| GCSE | PRScs.MultiPRS | LDpred2.10FCVal | 0.413 | 0.412 | 0.001 | 9.25e-01 | 0.346 | 0.342 | 0.004 | 5.55e-01 |
| GCSE | LDpred2.MultiPRS | LDpred2.10FCVal | 0.412 | 0.412 | 0.001 | 9.50e-01 | 0.339 | 0.342 | -0.003 | 2.48e-01 |
| GCSE | PRScs.PseudoVal | LDpred2.10FCVal | 0.397 | 0.412 | -0.014 | 2.22e-04 | 0.351 | 0.342 | 0.009 | 2.62e-01 |
| GCSE | LDpred2.10FCVal | LDpred2.10FCVal | 0.412 | 0.412 | 0.000 | 1.00e+00 | 0.342 | 0.342 | 0.000 | 1.00e+00 |
| GCSE | lassosum.MultiPRS | LDpred2.10FCVal | 0.411 | 0.412 | 0.000 | 9.78e-01 | 0.347 | 0.342 | 0.005 | 4.81e-01 |
| GCSE | LDpred1.10FCVal | LDpred2.10FCVal | 0.402 | 0.412 | -0.010 | 2.10e-02 | 0.330 | 0.342 | -0.012 | 1.88e-01 |
| GCSE | All.MultiPRS | LDpred2.10FCVal | 0.417 | 0.412 | 0.006 | 7.17e-01 | 0.347 | 0.342 | 0.004 | 3.38e-01 |
| GCSE | SBayesR.PseudoVal | LDpred2.10FCVal | 0.397 | 0.412 | -0.015 | 2.80e-03 | 0.331 | 0.342 | -0.011 | 2.89e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.401 | 0.412 | -0.010 | 1.61e-02 | 0.337 | 0.342 | -0.005 | 5.58e-01 |
| GCSE | pT+clump.MultiPRS | LDpred2.10FCVal | 0.381 | 0.412 | -0.031 | 6.06e-02 | 0.328 | 0.342 | -0.014 | 2.48e-01 |
| GCSE | lassosum.10FCVal | LDpred2.10FCVal | 0.407 | 0.412 | -0.004 | 1.62e-01 | 0.348 | 0.342 | 0.006 | 3.59e-01 |
| GCSE | PRScs.10FCVal | LDpred2.10FCVal | 0.410 | 0.412 | -0.001 | 7.35e-01 | 0.341 | 0.342 | -0.002 | 8.39e-01 |
| GCSE | lassosum.PseudoVal | LDpred2.10FCVal | 0.321 | 0.412 | -0.091 | 5.61e-28 | 0.276 | 0.342 | -0.066 | 1.45e-04 |
| GCSE | SBLUP.Inf | LDpred2.10FCVal | 0.400 | 0.412 | -0.012 | 4.75e-03 | 0.335 | 0.342 | -0.007 | 4.10e-01 |
| GCSE | LDpred2.PseudoVal | LDpred2.10FCVal | 0.406 | 0.412 | -0.006 | 7.62e-03 | 0.332 | 0.342 | -0.010 | 3.31e-02 |
| GCSE | pT+clump.10FCVal | LDpred2.10FCVal | 0.373 | 0.412 | -0.039 | 3.35e-09 | 0.311 | 0.342 | -0.031 | 2.38e-02 |
| GCSE | LDpred2.Inf | LDpred2.10FCVal | 0.405 | 0.412 | -0.006 | 7.61e-03 | 0.331 | 0.342 | -0.011 | 2.17e-02 |
| GCSE | LDpred1.MultiPRS | LDpred2.10FCVal | 0.409 | 0.412 | -0.002 | 8.78e-01 | 0.340 | 0.342 | -0.002 | 7.42e-01 |
| GCSE | lassosum.PseudoVal | LDpred2.PseudoVal | 0.321 | 0.406 | -0.085 | 2.71e-19 | 0.276 | 0.332 | -0.056 | 4.53e-03 |
| GCSE | SBLUP.Inf | LDpred2.PseudoVal | 0.400 | 0.406 | -0.006 | 1.01e-01 | 0.335 | 0.332 | 0.002 | 7.78e-01 |
| GCSE | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.397 | 0.406 | -0.009 | 1.48e-01 | 0.331 | 0.332 | -0.001 | 9.13e-01 |
| GCSE | All.MultiPRS | LDpred2.PseudoVal | 0.417 | 0.406 | 0.012 | 4.66e-01 | 0.347 | 0.332 | 0.014 | 2.51e-02 |
| GCSE | PRScs.PseudoVal | LDpred2.PseudoVal | 0.397 | 0.406 | -0.008 | 9.07e-02 | 0.351 | 0.332 | 0.019 | 7.27e-02 |
| GCSE | lassosum.MultiPRS | LDpred2.PseudoVal | 0.411 | 0.406 | 0.005 | 7.34e-01 | 0.347 | 0.332 | 0.014 | 9.52e-02 |
| GCSE | PRScs.MultiPRS | LDpred2.PseudoVal | 0.413 | 0.406 | 0.007 | 6.45e-01 | 0.346 | 0.332 | 0.013 | 6.28e-02 |
| GCSE | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.412 | 0.406 | 0.007 | 6.68e-01 | 0.339 | 0.332 | 0.007 | 1.83e-01 |
| GCSE | PRScs.10FCVal | LDpred2.PseudoVal | 0.410 | 0.406 | 0.005 | 1.77e-01 | 0.341 | 0.332 | 0.008 | 2.59e-01 |
| GCSE | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.406 | 0.406 | 0.000 | 1.00e+00 | 0.332 | 0.332 | 0.000 | 1.00e+00 |
| GCSE | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.381 | 0.406 | -0.025 | 1.29e-01 | 0.328 | 0.332 | -0.004 | 7.53e-01 |
| GCSE | pT+clump.10FCVal | LDpred2.PseudoVal | 0.373 | 0.406 | -0.033 | 5.80e-07 | 0.311 | 0.332 | -0.021 | 1.27e-01 |
| GCSE | lassosum.10FCVal | LDpred2.PseudoVal | 0.407 | 0.406 | 0.002 | 6.94e-01 | 0.348 | 0.332 | 0.015 | 6.39e-02 |
| GCSE | LDpred1.Inf | LDpred2.PseudoVal | 0.408 | 0.406 | 0.002 | 4.65e-01 | 0.341 | 0.332 | 0.009 | 1.41e-01 |
| GCSE | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.409 | 0.406 | 0.003 | 8.31e-01 | 0.340 | 0.332 | 0.007 | 2.15e-01 |
| GCSE | LDpred1.10FCVal | LDpred2.PseudoVal | 0.402 | 0.406 | -0.004 | 3.47e-01 | 0.330 | 0.332 | -0.002 | 8.25e-01 |
| GCSE | LDpred2.Inf | LDpred2.PseudoVal | 0.405 | 0.406 | 0.000 | 4.25e-01 | 0.331 | 0.332 | -0.002 | 1.18e-01 |
| GCSE | LDpred2.10FCVal | LDpred2.PseudoVal | 0.412 | 0.406 | 0.006 | 7.62e-03 | 0.342 | 0.332 | 0.010 | 3.31e-02 |
| GCSE | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.401 | 0.406 | -0.004 | 3.40e-01 | 0.337 | 0.332 | 0.004 | 6.50e-01 |
| GCSE | pT+clump.MultiPRS | LDpred2.Inf | 0.381 | 0.405 | -0.025 | 1.34e-01 | 0.328 | 0.331 | -0.002 | 8.51e-01 |
| GCSE | lassosum.MultiPRS | LDpred2.Inf | 0.411 | 0.405 | 0.006 | 7.16e-01 | 0.347 | 0.331 | 0.016 | 7.29e-02 |
| GCSE | SBayesR.PseudoVal | LDpred2.Inf | 0.397 | 0.405 | -0.009 | 1.78e-01 | 0.331 | 0.331 | 0.000 | 9.91e-01 |
| GCSE | All.MultiPRS | LDpred2.Inf | 0.417 | 0.405 | 0.012 | 4.52e-01 | 0.347 | 0.331 | 0.016 | 1.59e-02 |
| GCSE | SBLUP.Inf | LDpred2.Inf | 0.400 | 0.405 | -0.006 | 1.30e-01 | 0.335 | 0.331 | 0.004 | 6.36e-01 |
| GCSE | LDpred2.PseudoVal | LDpred2.Inf | 0.406 | 0.405 | 0.000 | 4.25e-01 | 0.332 | 0.331 | 0.002 | 1.18e-01 |
| GCSE | PRScs.MultiPRS | LDpred2.Inf | 0.413 | 0.405 | 0.008 | 6.28e-01 | 0.346 | 0.331 | 0.015 | 4.37e-02 |
| GCSE | LDpred2.MultiPRS | LDpred2.Inf | 0.412 | 0.405 | 0.007 | 6.51e-01 | 0.339 | 0.331 | 0.008 | 1.10e-01 |
| GCSE | PRScs.10FCVal | LDpred2.Inf | 0.410 | 0.405 | 0.005 | 1.46e-01 | 0.341 | 0.331 | 0.010 | 1.78e-01 |
| GCSE | LDpred1.Inf | LDpred2.Inf | 0.408 | 0.405 | 0.002 | 4.09e-01 | 0.341 | 0.331 | 0.010 | 9.52e-02 |
| GCSE | LDpred1.MultiPRS | LDpred2.Inf | 0.409 | 0.405 | 0.004 | 8.13e-01 | 0.340 | 0.331 | 0.009 | 1.47e-01 |
| GCSE | pT+clump.10FCVal | LDpred2.Inf | 0.373 | 0.405 | -0.032 | 9.19e-07 | 0.311 | 0.331 | -0.019 | 1.62e-01 |
| GCSE | lassosum.10FCVal | LDpred2.Inf | 0.407 | 0.405 | 0.002 | 6.46e-01 | 0.348 | 0.331 | 0.017 | 5.47e-02 |
| GCSE | PRScs.PseudoVal | LDpred2.Inf | 0.397 | 0.405 | -0.008 | 1.21e-01 | 0.351 | 0.331 | 0.020 | 6.27e-02 |
| GCSE | DBSLMM.PseudoVal | LDpred2.Inf | 0.401 | 0.405 | -0.004 | 3.90e-01 | 0.337 | 0.331 | 0.006 | 5.44e-01 |
| GCSE | LDpred1.10FCVal | LDpred2.Inf | 0.402 | 0.405 | -0.003 | 4.05e-01 | 0.330 | 0.331 | 0.000 | 9.74e-01 |
| GCSE | LDpred2.Inf | LDpred2.Inf | 0.405 | 0.405 | 0.000 | 1.00e+00 | 0.331 | 0.331 | 0.000 | 1.00e+00 |
| GCSE | lassosum.PseudoVal | LDpred2.Inf | 0.321 | 0.405 | -0.085 | 1.54e-18 | 0.276 | 0.331 | -0.055 | 6.84e-03 |
| GCSE | LDpred2.10FCVal | LDpred2.Inf | 0.412 | 0.405 | 0.006 | 7.61e-03 | 0.342 | 0.331 | 0.011 | 2.17e-02 |
| GCSE | PRScs.10FCVal | PRScs.MultiPRS | 0.410 | 0.413 | -0.003 | 8.65e-01 | 0.341 | 0.346 | -0.005 | 9.20e-02 |
| GCSE | LDpred1.10FCVal | PRScs.MultiPRS | 0.402 | 0.413 | -0.011 | 4.87e-01 | 0.330 | 0.346 | -0.015 | 5.88e-02 |
| GCSE | lassosum.MultiPRS | PRScs.MultiPRS | 0.411 | 0.413 | -0.002 | 4.70e-01 | 0.347 | 0.346 | 0.001 | 8.58e-01 |
| GCSE | LDpred2.MultiPRS | PRScs.MultiPRS | 0.412 | 0.413 | -0.001 | 9.75e-01 | 0.339 | 0.346 | -0.007 | 2.80e-01 |
| GCSE | All.MultiPRS | PRScs.MultiPRS | 0.417 | 0.413 | 0.004 | 7.89e-01 | 0.347 | 0.346 | 0.001 | 8.67e-01 |
| GCSE | PRScs.PseudoVal | PRScs.MultiPRS | 0.397 | 0.413 | -0.016 | 3.26e-01 | 0.351 | 0.346 | 0.005 | 4.54e-01 |
| GCSE | LDpred1.MultiPRS | PRScs.MultiPRS | 0.409 | 0.413 | -0.004 | 1.89e-01 | 0.340 | 0.346 | -0.006 | 3.37e-01 |
| GCSE | pT+clump.MultiPRS | PRScs.MultiPRS | 0.381 | 0.413 | -0.032 | 4.58e-02 | 0.328 | 0.346 | -0.018 | 1.21e-01 |
| GCSE | lassosum.10FCVal | PRScs.MultiPRS | 0.407 | 0.413 | -0.006 | 7.18e-01 | 0.348 | 0.346 | 0.002 | 7.59e-01 |
| GCSE | SBayesR.PseudoVal | PRScs.MultiPRS | 0.397 | 0.413 | -0.016 | 3.02e-01 | 0.331 | 0.346 | -0.015 | 1.59e-01 |
| GCSE | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.401 | 0.413 | -0.012 | 4.59e-01 | 0.337 | 0.346 | -0.009 | 1.97e-01 |
| GCSE | SBLUP.Inf | PRScs.MultiPRS | 0.400 | 0.413 | -0.013 | 3.99e-01 | 0.335 | 0.346 | -0.011 | 1.98e-01 |
| GCSE | LDpred2.Inf | PRScs.MultiPRS | 0.405 | 0.413 | -0.008 | 6.28e-01 | 0.331 | 0.346 | -0.015 | 4.37e-02 |
| GCSE | pT+clump.10FCVal | PRScs.MultiPRS | 0.373 | 0.413 | -0.040 | 1.30e-02 | 0.311 | 0.346 | -0.035 | 8.04e-03 |
| GCSE | LDpred2.10FCVal | PRScs.MultiPRS | 0.412 | 0.413 | -0.001 | 9.25e-01 | 0.342 | 0.346 | -0.004 | 5.55e-01 |
| GCSE | lassosum.PseudoVal | PRScs.MultiPRS | 0.321 | 0.413 | -0.092 | 1.67e-08 | 0.276 | 0.346 | -0.070 | 6.81e-05 |
| GCSE | LDpred2.PseudoVal | PRScs.MultiPRS | 0.406 | 0.413 | -0.007 | 6.45e-01 | 0.332 | 0.346 | -0.013 | 6.28e-02 |
| GCSE | LDpred1.Inf | PRScs.MultiPRS | 0.408 | 0.413 | -0.005 | 7.39e-01 | 0.341 | 0.346 | -0.005 | 4.73e-01 |
| GCSE | PRScs.MultiPRS | PRScs.MultiPRS | 0.413 | 0.413 | 0.000 | 1.00e+00 | 0.346 | 0.346 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.PseudoVal | PRScs.10FCVal | 0.406 | 0.410 | -0.005 | 1.77e-01 | 0.332 | 0.341 | -0.008 | 2.59e-01 |
| GCSE | PRScs.10FCVal | PRScs.10FCVal | 0.410 | 0.410 | 0.000 | 1.00e+00 | 0.341 | 0.341 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.MultiPRS | PRScs.10FCVal | 0.412 | 0.410 | 0.002 | 8.90e-01 | 0.339 | 0.341 | -0.002 | 8.28e-01 |
| GCSE | pT+clump.MultiPRS | PRScs.10FCVal | 0.381 | 0.410 | -0.030 | 7.06e-02 | 0.328 | 0.341 | -0.012 | 3.08e-01 |
| GCSE | LDpred1.Inf | PRScs.10FCVal | 0.408 | 0.410 | -0.003 | 3.87e-01 | 0.341 | 0.341 | 0.000 | 9.39e-01 |
| GCSE | LDpred1.MultiPRS | PRScs.10FCVal | 0.409 | 0.410 | -0.001 | 9.38e-01 | 0.340 | 0.341 | -0.001 | 8.96e-01 |
| GCSE | pT+clump.10FCVal | PRScs.10FCVal | 0.373 | 0.410 | -0.037 | 7.12e-10 | 0.311 | 0.341 | -0.029 | 2.29e-02 |
| GCSE | lassosum.10FCVal | PRScs.10FCVal | 0.407 | 0.410 | -0.003 | 4.22e-01 | 0.348 | 0.341 | 0.007 | 3.70e-01 |
| GCSE | PRScs.PseudoVal | PRScs.10FCVal | 0.397 | 0.410 | -0.013 | 3.15e-03 | 0.351 | 0.341 | 0.011 | 2.69e-01 |
| GCSE | DBSLMM.PseudoVal | PRScs.10FCVal | 0.401 | 0.410 | -0.009 | 1.71e-02 | 0.337 | 0.341 | -0.004 | 6.46e-01 |
| GCSE | SBLUP.Inf | PRScs.10FCVal | 0.400 | 0.410 | -0.011 | 5.14e-03 | 0.335 | 0.341 | -0.006 | 4.76e-01 |
| GCSE | LDpred2.Inf | PRScs.10FCVal | 0.405 | 0.410 | -0.005 | 1.46e-01 | 0.331 | 0.341 | -0.010 | 1.78e-01 |
| GCSE | lassosum.PseudoVal | PRScs.10FCVal | 0.321 | 0.410 | -0.090 | 9.90e-22 | 0.276 | 0.341 | -0.064 | 1.14e-03 |
| GCSE | LDpred2.10FCVal | PRScs.10FCVal | 0.412 | 0.410 | 0.001 | 7.35e-01 | 0.342 | 0.341 | 0.002 | 8.39e-01 |
| GCSE | lassosum.MultiPRS | PRScs.10FCVal | 0.411 | 0.410 | 0.001 | 9.61e-01 | 0.347 | 0.341 | 0.006 | 3.58e-01 |
| GCSE | LDpred1.10FCVal | PRScs.10FCVal | 0.402 | 0.410 | -0.008 | 2.62e-02 | 0.330 | 0.341 | -0.010 | 2.04e-01 |
| GCSE | All.MultiPRS | PRScs.10FCVal | 0.417 | 0.410 | 0.007 | 6.61e-01 | 0.347 | 0.341 | 0.006 | 2.92e-01 |
| GCSE | SBayesR.PseudoVal | PRScs.10FCVal | 0.397 | 0.410 | -0.014 | 2.46e-02 | 0.331 | 0.341 | -0.010 | 4.56e-01 |
| GCSE | PRScs.MultiPRS | PRScs.10FCVal | 0.413 | 0.410 | 0.003 | 8.65e-01 | 0.346 | 0.341 | 0.005 | 9.20e-02 |
| GCSE | All.MultiPRS | PRScs.PseudoVal | 0.417 | 0.397 | 0.020 | 2.11e-01 | 0.347 | 0.351 | -0.005 | 5.38e-01 |
| GCSE | pT+clump.MultiPRS | PRScs.PseudoVal | 0.381 | 0.397 | -0.017 | 3.12e-01 | 0.328 | 0.351 | -0.023 | 6.07e-02 |
| GCSE | LDpred2.PseudoVal | PRScs.PseudoVal | 0.406 | 0.397 | 0.008 | 9.07e-02 | 0.332 | 0.351 | -0.019 | 7.27e-02 |
| GCSE | SBayesR.PseudoVal | PRScs.PseudoVal | 0.397 | 0.397 | -0.001 | 8.33e-01 | 0.331 | 0.351 | -0.020 | 8.45e-03 |
| GCSE | LDpred2.MultiPRS | PRScs.PseudoVal | 0.412 | 0.397 | 0.015 | 3.42e-01 | 0.339 | 0.351 | -0.012 | 1.31e-01 |
| GCSE | SBLUP.Inf | PRScs.PseudoVal | 0.400 | 0.397 | 0.002 | 6.98e-01 | 0.335 | 0.351 | -0.017 | 1.73e-01 |
| GCSE | LDpred1.Inf | PRScs.PseudoVal | 0.408 | 0.397 | 0.010 | 3.09e-02 | 0.341 | 0.351 | -0.010 | 3.37e-01 |
| GCSE | PRScs.10FCVal | PRScs.PseudoVal | 0.410 | 0.397 | 0.013 | 3.15e-03 | 0.341 | 0.351 | -0.011 | 2.69e-01 |
| GCSE | LDpred2.Inf | PRScs.PseudoVal | 0.405 | 0.397 | 0.008 | 1.21e-01 | 0.331 | 0.351 | -0.020 | 6.27e-02 |
| GCSE | PRScs.PseudoVal | PRScs.PseudoVal | 0.397 | 0.397 | 0.000 | 1.00e+00 | 0.351 | 0.351 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.10FCVal | PRScs.PseudoVal | 0.412 | 0.397 | 0.014 | 2.22e-04 | 0.342 | 0.351 | -0.009 | 2.62e-01 |
| GCSE | pT+clump.10FCVal | PRScs.PseudoVal | 0.373 | 0.397 | -0.024 | 6.09e-04 | 0.311 | 0.351 | -0.040 | 8.58e-03 |
| GCSE | PRScs.MultiPRS | PRScs.PseudoVal | 0.413 | 0.397 | 0.016 | 3.26e-01 | 0.346 | 0.351 | -0.005 | 4.54e-01 |
| GCSE | lassosum.10FCVal | PRScs.PseudoVal | 0.407 | 0.397 | 0.010 | 2.12e-03 | 0.348 | 0.351 | -0.003 | 6.16e-01 |
| GCSE | lassosum.PseudoVal | PRScs.PseudoVal | 0.321 | 0.397 | -0.077 | 7.69e-31 | 0.276 | 0.351 | -0.075 | 4.42e-08 |
| GCSE | LDpred1.MultiPRS | PRScs.PseudoVal | 0.409 | 0.397 | 0.012 | 4.63e-01 | 0.340 | 0.351 | -0.011 | 2.69e-01 |
| GCSE | lassosum.MultiPRS | PRScs.PseudoVal | 0.411 | 0.397 | 0.014 | 3.89e-01 | 0.347 | 0.351 | -0.004 | 5.77e-01 |
| GCSE | LDpred1.10FCVal | PRScs.PseudoVal | 0.402 | 0.397 | 0.005 | 3.81e-01 | 0.330 | 0.351 | -0.021 | 6.81e-02 |
| GCSE | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.401 | 0.397 | 0.004 | 3.38e-01 | 0.337 | 0.351 | -0.014 | 9.88e-02 |
| GCSE | All.MultiPRS | pT+clump.MultiPRS | 0.417 | 0.381 | 0.037 | 2.16e-02 | 0.347 | 0.328 | 0.018 | 7.70e-02 |
| GCSE | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.397 | 0.381 | 0.016 | 3.35e-01 | 0.331 | 0.328 | 0.003 | 8.47e-01 |
| GCSE | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.409 | 0.381 | 0.028 | 8.01e-02 | 0.340 | 0.328 | 0.012 | 3.54e-01 |
| GCSE | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.381 | 0.381 | 0.000 | 1.00e+00 | 0.328 | 0.328 | 0.000 | 1.00e+00 |
| GCSE | lassosum.10FCVal | pT+clump.MultiPRS | 0.407 | 0.381 | 0.027 | 1.06e-01 | 0.348 | 0.328 | 0.019 | 1.07e-01 |
| GCSE | PRScs.10FCVal | pT+clump.MultiPRS | 0.410 | 0.381 | 0.030 | 7.06e-02 | 0.341 | 0.328 | 0.012 | 3.08e-01 |
| GCSE | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.401 | 0.381 | 0.021 | 2.13e-01 | 0.337 | 0.328 | 0.009 | 4.68e-01 |
| GCSE | SBLUP.Inf | pT+clump.MultiPRS | 0.400 | 0.381 | 0.019 | 2.51e-01 | 0.335 | 0.328 | 0.006 | 6.38e-01 |
| GCSE | LDpred2.Inf | pT+clump.MultiPRS | 0.405 | 0.381 | 0.025 | 1.34e-01 | 0.331 | 0.328 | 0.002 | 8.51e-01 |
| GCSE | pT+clump.10FCVal | pT+clump.MultiPRS | 0.373 | 0.381 | -0.008 | 6.40e-01 | 0.311 | 0.328 | -0.017 | 4.87e-03 |
| GCSE | LDpred2.10FCVal | pT+clump.MultiPRS | 0.412 | 0.381 | 0.031 | 6.06e-02 | 0.342 | 0.328 | 0.014 | 2.48e-01 |
| GCSE | PRScs.PseudoVal | pT+clump.MultiPRS | 0.397 | 0.381 | 0.017 | 3.12e-01 | 0.351 | 0.328 | 0.023 | 6.07e-02 |
| GCSE | LDpred1.10FCVal | pT+clump.MultiPRS | 0.402 | 0.381 | 0.021 | 1.97e-01 | 0.330 | 0.328 | 0.002 | 8.69e-01 |
| GCSE | LDpred1.Inf | pT+clump.MultiPRS | 0.408 | 0.381 | 0.027 | 9.98e-02 | 0.341 | 0.328 | 0.013 | 3.07e-01 |
| GCSE | PRScs.MultiPRS | pT+clump.MultiPRS | 0.413 | 0.381 | 0.032 | 4.58e-02 | 0.346 | 0.328 | 0.018 | 1.21e-01 |
| GCSE | lassosum.PseudoVal | pT+clump.MultiPRS | 0.321 | 0.381 | -0.060 | 3.70e-04 | 0.276 | 0.328 | -0.052 | 5.51e-03 |
| GCSE | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.406 | 0.381 | 0.025 | 1.29e-01 | 0.332 | 0.328 | 0.004 | 7.53e-01 |
| GCSE | lassosum.MultiPRS | pT+clump.MultiPRS | 0.411 | 0.381 | 0.030 | 6.07e-02 | 0.347 | 0.328 | 0.019 | 1.13e-01 |
| GCSE | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.412 | 0.381 | 0.032 | 4.60e-02 | 0.339 | 0.328 | 0.011 | 3.77e-01 |
| GCSE | LDpred2.10FCVal | pT+clump.10FCVal | 0.412 | 0.373 | 0.039 | 3.35e-09 | 0.342 | 0.311 | 0.031 | 2.38e-02 |
| GCSE | pT+clump.MultiPRS | pT+clump.10FCVal | 0.381 | 0.373 | 0.008 | 6.40e-01 | 0.328 | 0.311 | 0.017 | 4.87e-03 |
| GCSE | LDpred2.Inf | pT+clump.10FCVal | 0.405 | 0.373 | 0.032 | 9.19e-07 | 0.331 | 0.311 | 0.019 | 1.62e-01 |
| GCSE | SBayesR.PseudoVal | pT+clump.10FCVal | 0.397 | 0.373 | 0.024 | 3.62e-03 | 0.331 | 0.311 | 0.020 | 2.50e-01 |
| GCSE | PRScs.MultiPRS | pT+clump.10FCVal | 0.413 | 0.373 | 0.040 | 1.30e-02 | 0.346 | 0.311 | 0.035 | 8.04e-03 |
| GCSE | SBLUP.Inf | pT+clump.10FCVal | 0.400 | 0.373 | 0.027 | 6.62e-05 | 0.335 | 0.311 | 0.023 | 9.47e-02 |
| GCSE | lassosum.MultiPRS | pT+clump.10FCVal | 0.411 | 0.373 | 0.038 | 1.81e-02 | 0.347 | 0.311 | 0.036 | 7.92e-03 |
| GCSE | PRScs.10FCVal | pT+clump.10FCVal | 0.410 | 0.373 | 0.037 | 7.12e-10 | 0.341 | 0.311 | 0.029 | 2.29e-02 |
| GCSE | LDpred2.MultiPRS | pT+clump.10FCVal | 0.412 | 0.373 | 0.040 | 1.41e-02 | 0.339 | 0.311 | 0.028 | 4.78e-02 |
| GCSE | lassosum.PseudoVal | pT+clump.10FCVal | 0.321 | 0.373 | -0.052 | 1.02e-06 | 0.276 | 0.311 | -0.035 | 1.22e-01 |
| GCSE | LDpred2.PseudoVal | pT+clump.10FCVal | 0.406 | 0.373 | 0.033 | 5.80e-07 | 0.332 | 0.311 | 0.021 | 1.27e-01 |
| GCSE | LDpred1.MultiPRS | pT+clump.10FCVal | 0.409 | 0.373 | 0.036 | 2.56e-02 | 0.340 | 0.311 | 0.028 | 3.00e-02 |
| GCSE | pT+clump.10FCVal | pT+clump.10FCVal | 0.373 | 0.373 | 0.000 | 1.00e+00 | 0.311 | 0.311 | 0.000 | 1.00e+00 |
| GCSE | All.MultiPRS | pT+clump.10FCVal | 0.417 | 0.373 | 0.044 | 5.86e-03 | 0.347 | 0.311 | 0.035 | 3.75e-03 |
| GCSE | LDpred1.Inf | pT+clump.10FCVal | 0.408 | 0.373 | 0.035 | 1.52e-08 | 0.341 | 0.311 | 0.030 | 2.37e-02 |
| GCSE | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.401 | 0.373 | 0.028 | 2.61e-05 | 0.337 | 0.311 | 0.026 | 7.14e-02 |
| GCSE | LDpred1.10FCVal | pT+clump.10FCVal | 0.402 | 0.373 | 0.029 | 1.56e-05 | 0.330 | 0.311 | 0.019 | 1.77e-01 |
| GCSE | lassosum.10FCVal | pT+clump.10FCVal | 0.407 | 0.373 | 0.034 | 3.87e-07 | 0.348 | 0.311 | 0.036 | 9.69e-03 |
| GCSE | PRScs.PseudoVal | pT+clump.10FCVal | 0.397 | 0.373 | 0.024 | 6.09e-04 | 0.351 | 0.311 | 0.040 | 8.58e-03 |
| GCSE | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.381 | 0.397 | -0.016 | 3.35e-01 | 0.328 | 0.331 | -0.003 | 8.47e-01 |
| GCSE | LDpred1.10FCVal | SBayesR.PseudoVal | 0.402 | 0.397 | 0.005 | 4.08e-01 | 0.330 | 0.331 | 0.000 | 9.75e-01 |
| GCSE | PRScs.10FCVal | SBayesR.PseudoVal | 0.410 | 0.397 | 0.014 | 2.46e-02 | 0.341 | 0.331 | 0.010 | 4.56e-01 |
| GCSE | LDpred2.10FCVal | SBayesR.PseudoVal | 0.412 | 0.397 | 0.015 | 2.80e-03 | 0.342 | 0.331 | 0.011 | 2.89e-01 |
| GCSE | lassosum.PseudoVal | SBayesR.PseudoVal | 0.321 | 0.397 | -0.076 | 2.14e-37 | 0.276 | 0.331 | -0.055 | 1.49e-05 |
| GCSE | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.406 | 0.397 | 0.009 | 1.48e-01 | 0.332 | 0.331 | 0.001 | 9.13e-01 |
| GCSE | lassosum.MultiPRS | SBayesR.PseudoVal | 0.411 | 0.397 | 0.015 | 3.63e-01 | 0.347 | 0.331 | 0.016 | 1.26e-01 |
| GCSE | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.412 | 0.397 | 0.016 | 3.20e-01 | 0.339 | 0.331 | 0.008 | 4.36e-01 |
| GCSE | All.MultiPRS | SBayesR.PseudoVal | 0.417 | 0.397 | 0.021 | 1.96e-01 | 0.347 | 0.331 | 0.016 | 9.45e-02 |
| GCSE | PRScs.PseudoVal | SBayesR.PseudoVal | 0.397 | 0.397 | 0.001 | 8.33e-01 | 0.351 | 0.331 | 0.020 | 8.45e-03 |
| GCSE | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.409 | 0.397 | 0.013 | 4.34e-01 | 0.340 | 0.331 | 0.009 | 4.97e-01 |
| GCSE | pT+clump.10FCVal | SBayesR.PseudoVal | 0.373 | 0.397 | -0.024 | 3.62e-03 | 0.311 | 0.331 | -0.020 | 2.50e-01 |
| GCSE | lassosum.10FCVal | SBayesR.PseudoVal | 0.407 | 0.397 | 0.011 | 1.91e-02 | 0.348 | 0.331 | 0.017 | 8.58e-02 |
| GCSE | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.397 | 0.397 | 0.000 | 1.00e+00 | 0.331 | 0.331 | 0.000 | 1.00e+00 |
| GCSE | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.401 | 0.397 | 0.005 | 3.44e-01 | 0.337 | 0.331 | 0.006 | 5.72e-01 |
| GCSE | SBLUP.Inf | SBayesR.PseudoVal | 0.400 | 0.397 | 0.003 | 6.68e-01 | 0.335 | 0.331 | 0.004 | 7.99e-01 |
| GCSE | LDpred2.Inf | SBayesR.PseudoVal | 0.405 | 0.397 | 0.009 | 1.78e-01 | 0.331 | 0.331 | 0.000 | 9.91e-01 |
| GCSE | LDpred1.Inf | SBayesR.PseudoVal | 0.408 | 0.397 | 0.011 | 7.41e-02 | 0.341 | 0.331 | 0.010 | 4.47e-01 |
| GCSE | PRScs.MultiPRS | SBayesR.PseudoVal | 0.413 | 0.397 | 0.016 | 3.02e-01 | 0.346 | 0.331 | 0.015 | 1.59e-01 |
| GCSE | LDpred2.10FCVal | SBLUP.Inf | 0.412 | 0.400 | 0.012 | 4.75e-03 | 0.342 | 0.335 | 0.007 | 4.10e-01 |
| GCSE | PRScs.PseudoVal | SBLUP.Inf | 0.397 | 0.400 | -0.002 | 6.98e-01 | 0.351 | 0.335 | 0.017 | 1.73e-01 |
| GCSE | LDpred1.10FCVal | SBLUP.Inf | 0.402 | 0.400 | 0.002 | 5.73e-01 | 0.330 | 0.335 | -0.004 | 6.50e-01 |
| GCSE | LDpred2.Inf | SBLUP.Inf | 0.405 | 0.400 | 0.006 | 1.30e-01 | 0.331 | 0.335 | -0.004 | 6.36e-01 |
| GCSE | SBayesR.PseudoVal | SBLUP.Inf | 0.397 | 0.400 | -0.003 | 6.68e-01 | 0.331 | 0.335 | -0.004 | 7.99e-01 |
| GCSE | lassosum.PseudoVal | SBLUP.Inf | 0.321 | 0.400 | -0.079 | 3.28e-15 | 0.276 | 0.335 | -0.059 | 5.25e-03 |
| GCSE | pT+clump.MultiPRS | SBLUP.Inf | 0.381 | 0.400 | -0.019 | 2.51e-01 | 0.328 | 0.335 | -0.006 | 6.38e-01 |
| GCSE | lassosum.MultiPRS | SBLUP.Inf | 0.411 | 0.400 | 0.012 | 4.70e-01 | 0.347 | 0.335 | 0.012 | 2.25e-01 |
| GCSE | PRScs.10FCVal | SBLUP.Inf | 0.410 | 0.400 | 0.011 | 5.14e-03 | 0.341 | 0.335 | 0.006 | 4.76e-01 |
| GCSE | PRScs.MultiPRS | SBLUP.Inf | 0.413 | 0.400 | 0.013 | 3.99e-01 | 0.346 | 0.335 | 0.011 | 1.98e-01 |
| GCSE | LDpred1.MultiPRS | SBLUP.Inf | 0.409 | 0.400 | 0.010 | 5.52e-01 | 0.340 | 0.335 | 0.005 | 4.80e-01 |
| GCSE | LDpred2.PseudoVal | SBLUP.Inf | 0.406 | 0.400 | 0.006 | 1.01e-01 | 0.332 | 0.335 | -0.002 | 7.78e-01 |
| GCSE | LDpred1.Inf | SBLUP.Inf | 0.408 | 0.400 | 0.008 | 2.27e-02 | 0.341 | 0.335 | 0.006 | 4.01e-01 |
| GCSE | LDpred2.MultiPRS | SBLUP.Inf | 0.412 | 0.400 | 0.013 | 4.17e-01 | 0.339 | 0.335 | 0.004 | 6.48e-01 |
| GCSE | All.MultiPRS | SBLUP.Inf | 0.417 | 0.400 | 0.018 | 2.66e-01 | 0.347 | 0.335 | 0.012 | 1.47e-01 |
| GCSE | SBLUP.Inf | SBLUP.Inf | 0.400 | 0.400 | 0.000 | 1.00e+00 | 0.335 | 0.335 | 0.000 | 1.00e+00 |
| GCSE | DBSLMM.PseudoVal | SBLUP.Inf | 0.401 | 0.400 | 0.002 | 7.40e-01 | 0.337 | 0.335 | 0.002 | 8.41e-01 |
| GCSE | pT+clump.10FCVal | SBLUP.Inf | 0.373 | 0.400 | -0.027 | 6.62e-05 | 0.311 | 0.335 | -0.023 | 9.47e-02 |
| GCSE | lassosum.10FCVal | SBLUP.Inf | 0.407 | 0.400 | 0.008 | 1.14e-01 | 0.348 | 0.335 | 0.013 | 1.98e-01 |
| ADHD | LDpred2.PseudoVal | All.MultiPRS | 0.115 | 0.115 | -0.001 | 9.76e-01 | 0.195 | 0.186 | 0.008 | 5.00e-01 |
| ADHD | pT+clump.10FCVal | All.MultiPRS | 0.094 | 0.115 | -0.021 | 2.27e-01 | 0.150 | 0.186 | -0.037 | 5.75e-02 |
| ADHD | LDpred2.MultiPRS | All.MultiPRS | 0.114 | 0.115 | -0.001 | 9.60e-01 | 0.199 | 0.186 | 0.012 | 2.85e-01 |
| ADHD | PRScs.PseudoVal | All.MultiPRS | 0.109 | 0.115 | -0.006 | 7.18e-01 | 0.189 | 0.186 | 0.003 | 8.36e-01 |
| ADHD | LDpred1.Inf | All.MultiPRS | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.191 | 0.186 | 0.005 | 6.83e-01 |
| ADHD | PRScs.MultiPRS | All.MultiPRS | 0.113 | 0.115 | -0.002 | 8.86e-01 | 0.203 | 0.186 | 0.016 | 1.94e-01 |
| ADHD | LDpred2.Inf | All.MultiPRS | 0.115 | 0.115 | 0.000 | 9.98e-01 | 0.194 | 0.186 | 0.008 | 5.12e-01 |
| ADHD | lassosum.10FCVal | All.MultiPRS | 0.118 | 0.115 | 0.002 | 8.94e-01 | 0.194 | 0.186 | 0.007 | 5.38e-01 |
| ADHD | LDpred2.10FCVal | All.MultiPRS | 0.118 | 0.115 | 0.003 | 8.69e-01 | 0.198 | 0.186 | 0.012 | 2.97e-01 |
| ADHD | lassosum.MultiPRS | All.MultiPRS | 0.117 | 0.115 | 0.002 | 9.19e-01 | 0.199 | 0.186 | 0.012 | 1.92e-01 |
| ADHD | LDpred1.10FCVal | All.MultiPRS | 0.118 | 0.115 | 0.003 | 8.85e-01 | 0.190 | 0.186 | 0.004 | 7.38e-01 |
| ADHD | All.MultiPRS | All.MultiPRS | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.186 | 0.186 | 0.000 | 1.00e+00 |
| ADHD | SBayesR.PseudoVal | All.MultiPRS | 0.091 | 0.115 | -0.024 | 1.69e-01 | 0.155 | 0.186 | -0.031 | 1.12e-01 |
| ADHD | LDpred1.MultiPRS | All.MultiPRS | 0.117 | 0.115 | 0.002 | 9.21e-01 | 0.191 | 0.186 | 0.005 | 6.68e-01 |
| ADHD | pT+clump.MultiPRS | All.MultiPRS | 0.096 | 0.115 | -0.019 | 2.64e-01 | 0.162 | 0.186 | -0.025 | 1.61e-01 |
| ADHD | lassosum.PseudoVal | All.MultiPRS | 0.077 | 0.115 | -0.039 | 2.75e-02 | 0.118 | 0.186 | -0.068 | 2.13e-03 |
| ADHD | PRScs.10FCVal | All.MultiPRS | 0.114 | 0.115 | -0.001 | 9.43e-01 | 0.207 | 0.186 | 0.021 | 1.15e-01 |
| ADHD | DBSLMM.PseudoVal | All.MultiPRS | 0.114 | 0.115 | -0.001 | 9.46e-01 | 0.184 | 0.186 | -0.003 | 8.33e-01 |
| ADHD | SBLUP.Inf | All.MultiPRS | 0.116 | 0.115 | 0.000 | 9.91e-01 | 0.193 | 0.186 | 0.007 | 5.81e-01 |
| ADHD | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.091 | 0.114 | -0.023 | 2.17e-02 | 0.155 | 0.184 | -0.028 | 1.54e-01 |
| ADHD | LDpred2.Inf | DBSLMM.PseudoVal | 0.115 | 0.114 | 0.001 | 8.29e-01 | 0.194 | 0.184 | 0.011 | 3.41e-01 |
| ADHD | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.094 | 0.114 | -0.020 | 3.55e-02 | 0.150 | 0.184 | -0.034 | 6.94e-02 |
| ADHD | PRScs.10FCVal | DBSLMM.PseudoVal | 0.114 | 0.114 | 0.000 | 9.91e-01 | 0.207 | 0.184 | 0.024 | 4.81e-02 |
| ADHD | PRScs.PseudoVal | DBSLMM.PseudoVal | 0.109 | 0.114 | -0.005 | 3.84e-01 | 0.189 | 0.184 | 0.006 | 6.32e-01 |
| ADHD | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.096 | 0.114 | -0.018 | 3.01e-01 | 0.162 | 0.184 | -0.022 | 1.60e-01 |
| ADHD | LDpred1.Inf | DBSLMM.PseudoVal | 0.115 | 0.114 | 0.001 | 8.35e-01 | 0.191 | 0.184 | 0.008 | 4.95e-01 |
| ADHD | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.117 | 0.114 | 0.003 | 8.68e-01 | 0.199 | 0.184 | 0.015 | 1.42e-01 |
| ADHD | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.118 | 0.114 | 0.004 | 4.45e-01 | 0.198 | 0.184 | 0.015 | 1.65e-01 |
| ADHD | lassosum.10FCVal | DBSLMM.PseudoVal | 0.118 | 0.114 | 0.004 | 6.04e-01 | 0.194 | 0.184 | 0.010 | 4.43e-01 |
| ADHD | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.118 | 0.114 | 0.004 | 5.03e-01 | 0.190 | 0.184 | 0.007 | 5.46e-01 |
| ADHD | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.117 | 0.114 | 0.003 | 8.69e-01 | 0.191 | 0.184 | 0.008 | 4.78e-01 |
| ADHD | PRScs.MultiPRS | DBSLMM.PseudoVal | 0.113 | 0.114 | -0.001 | 9.42e-01 | 0.203 | 0.184 | 0.019 | 1.04e-01 |
| ADHD | All.MultiPRS | DBSLMM.PseudoVal | 0.115 | 0.114 | 0.001 | 9.46e-01 | 0.186 | 0.184 | 0.003 | 8.33e-01 |
| ADHD | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.115 | 0.114 | 0.001 | 9.08e-01 | 0.195 | 0.184 | 0.011 | 3.20e-01 |
| ADHD | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.184 | 0.184 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.114 | 0.114 | 0.000 | 9.85e-01 | 0.199 | 0.184 | 0.015 | 1.55e-01 |
| ADHD | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.077 | 0.114 | -0.038 | 7.96e-04 | 0.118 | 0.184 | -0.065 | 2.85e-03 |
| ADHD | SBLUP.Inf | DBSLMM.PseudoVal | 0.116 | 0.114 | 0.001 | 8.13e-01 | 0.193 | 0.184 | 0.010 | 4.07e-01 |
| ADHD | SBayesR.PseudoVal | lassosum.MultiPRS | 0.091 | 0.117 | -0.026 | 1.45e-01 | 0.155 | 0.199 | -0.044 | 1.99e-02 |
| ADHD | LDpred2.10FCVal | lassosum.MultiPRS | 0.118 | 0.117 | 0.001 | 9.49e-01 | 0.198 | 0.199 | 0.000 | 9.51e-01 |
| ADHD | LDpred1.10FCVal | lassosum.MultiPRS | 0.118 | 0.117 | 0.001 | 9.65e-01 | 0.190 | 0.199 | -0.008 | 2.73e-01 |
| ADHD | LDpred2.Inf | lassosum.MultiPRS | 0.115 | 0.117 | -0.002 | 9.23e-01 | 0.194 | 0.199 | -0.004 | 6.33e-01 |
| ADHD | PRScs.10FCVal | lassosum.MultiPRS | 0.114 | 0.117 | -0.003 | 8.65e-01 | 0.207 | 0.199 | 0.009 | 3.60e-01 |
| ADHD | PRScs.MultiPRS | lassosum.MultiPRS | 0.113 | 0.117 | -0.004 | 2.87e-01 | 0.203 | 0.199 | 0.004 | 6.24e-01 |
| ADHD | pT+clump.MultiPRS | lassosum.MultiPRS | 0.096 | 0.117 | -0.021 | 2.29e-01 | 0.162 | 0.199 | -0.037 | 1.07e-02 |
| ADHD | lassosum.MultiPRS | lassosum.MultiPRS | 0.117 | 0.117 | 0.000 | 1.00e+00 | 0.199 | 0.199 | 0.000 | 1.00e+00 |
| ADHD | lassosum.PseudoVal | lassosum.MultiPRS | 0.077 | 0.117 | -0.041 | 2.23e-02 | 0.118 | 0.199 | -0.081 | 1.23e-04 |
| ADHD | LDpred2.MultiPRS | lassosum.MultiPRS | 0.114 | 0.117 | -0.003 | 4.95e-01 | 0.199 | 0.199 | 0.000 | 9.78e-01 |
| ADHD | LDpred1.MultiPRS | lassosum.MultiPRS | 0.117 | 0.117 | 0.000 | 9.91e-01 | 0.191 | 0.199 | -0.007 | 3.44e-01 |
| ADHD | LDpred2.PseudoVal | lassosum.MultiPRS | 0.115 | 0.117 | -0.002 | 8.97e-01 | 0.195 | 0.199 | -0.004 | 6.79e-01 |
| ADHD | LDpred1.Inf | lassosum.MultiPRS | 0.115 | 0.117 | -0.002 | 9.22e-01 | 0.191 | 0.199 | -0.007 | 4.37e-01 |
| ADHD | pT+clump.10FCVal | lassosum.MultiPRS | 0.094 | 0.117 | -0.023 | 1.95e-01 | 0.150 | 0.199 | -0.049 | 3.02e-03 |
| ADHD | lassosum.10FCVal | lassosum.MultiPRS | 0.118 | 0.117 | 0.001 | 9.74e-01 | 0.194 | 0.199 | -0.005 | 4.25e-01 |
| ADHD | SBLUP.Inf | lassosum.MultiPRS | 0.116 | 0.117 | -0.002 | 9.30e-01 | 0.193 | 0.199 | -0.006 | 5.12e-01 |
| ADHD | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.114 | 0.117 | -0.003 | 8.68e-01 | 0.184 | 0.199 | -0.015 | 1.42e-01 |
| ADHD | All.MultiPRS | lassosum.MultiPRS | 0.115 | 0.117 | -0.002 | 9.19e-01 | 0.186 | 0.199 | -0.012 | 1.92e-01 |
| ADHD | PRScs.PseudoVal | lassosum.MultiPRS | 0.109 | 0.117 | -0.008 | 6.49e-01 | 0.189 | 0.199 | -0.010 | 2.76e-01 |
| ADHD | LDpred1.Inf | lassosum.PseudoVal | 0.115 | 0.077 | 0.039 | 1.84e-03 | 0.191 | 0.118 | 0.073 | 2.67e-03 |
| ADHD | PRScs.10FCVal | lassosum.PseudoVal | 0.114 | 0.077 | 0.038 | 3.37e-04 | 0.207 | 0.118 | 0.089 | 1.61e-05 |
| ADHD | LDpred2.MultiPRS | lassosum.PseudoVal | 0.114 | 0.077 | 0.038 | 3.25e-02 | 0.199 | 0.118 | 0.080 | 1.93e-04 |
| ADHD | lassosum.PseudoVal | lassosum.PseudoVal | 0.077 | 0.077 | 0.000 | 1.00e+00 | 0.118 | 0.118 | 0.000 | 1.00e+00 |
| ADHD | PRScs.PseudoVal | lassosum.PseudoVal | 0.109 | 0.077 | 0.032 | 2.04e-03 | 0.189 | 0.118 | 0.071 | 5.26e-04 |
| ADHD | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.114 | 0.077 | 0.038 | 7.96e-04 | 0.184 | 0.118 | 0.065 | 2.85e-03 |
| ADHD | SBLUP.Inf | lassosum.PseudoVal | 0.116 | 0.077 | 0.039 | 1.86e-03 | 0.193 | 0.118 | 0.075 | 2.13e-03 |
| ADHD | LDpred2.Inf | lassosum.PseudoVal | 0.115 | 0.077 | 0.039 | 1.92e-03 | 0.194 | 0.118 | 0.076 | 1.88e-03 |
| ADHD | pT+clump.10FCVal | lassosum.PseudoVal | 0.094 | 0.077 | 0.017 | 1.94e-01 | 0.150 | 0.118 | 0.031 | 2.28e-01 |
| ADHD | LDpred2.10FCVal | lassosum.PseudoVal | 0.118 | 0.077 | 0.042 | 1.44e-04 | 0.198 | 0.118 | 0.080 | 1.91e-04 |
| ADHD | lassosum.MultiPRS | lassosum.PseudoVal | 0.117 | 0.077 | 0.041 | 2.23e-02 | 0.199 | 0.118 | 0.081 | 1.23e-04 |
| ADHD | LDpred1.10FCVal | lassosum.PseudoVal | 0.118 | 0.077 | 0.041 | 1.14e-04 | 0.190 | 0.118 | 0.072 | 6.39e-04 |
| ADHD | All.MultiPRS | lassosum.PseudoVal | 0.115 | 0.077 | 0.039 | 2.75e-02 | 0.186 | 0.118 | 0.068 | 2.13e-03 |
| ADHD | SBayesR.PseudoVal | lassosum.PseudoVal | 0.091 | 0.077 | 0.015 | 2.23e-01 | 0.155 | 0.118 | 0.037 | 1.22e-01 |
| ADHD | lassosum.10FCVal | lassosum.PseudoVal | 0.118 | 0.077 | 0.041 | 1.96e-04 | 0.194 | 0.118 | 0.076 | 5.33e-04 |
| ADHD | pT+clump.MultiPRS | lassosum.PseudoVal | 0.096 | 0.077 | 0.019 | 2.74e-01 | 0.162 | 0.118 | 0.044 | 6.43e-02 |
| ADHD | LDpred2.PseudoVal | lassosum.PseudoVal | 0.115 | 0.077 | 0.038 | 2.13e-03 | 0.195 | 0.118 | 0.077 | 1.69e-03 |
| ADHD | LDpred1.MultiPRS | lassosum.PseudoVal | 0.117 | 0.077 | 0.040 | 2.22e-02 | 0.191 | 0.118 | 0.073 | 7.86e-04 |
| ADHD | PRScs.MultiPRS | lassosum.PseudoVal | 0.113 | 0.077 | 0.036 | 4.09e-02 | 0.203 | 0.118 | 0.084 | 8.81e-05 |
| ADHD | lassosum.10FCVal | lassosum.10FCVal | 0.118 | 0.118 | 0.000 | 1.00e+00 | 0.194 | 0.194 | 0.000 | 1.00e+00 |
| ADHD | pT+clump.MultiPRS | lassosum.10FCVal | 0.096 | 0.118 | -0.022 | 2.17e-01 | 0.162 | 0.194 | -0.032 | 4.00e-02 |
| ADHD | LDpred2.Inf | lassosum.10FCVal | 0.115 | 0.118 | -0.002 | 6.30e-01 | 0.194 | 0.194 | 0.001 | 9.48e-01 |
| ADHD | SBayesR.PseudoVal | lassosum.10FCVal | 0.091 | 0.118 | -0.027 | 5.38e-03 | 0.155 | 0.194 | -0.039 | 4.15e-02 |
| ADHD | PRScs.MultiPRS | lassosum.10FCVal | 0.113 | 0.118 | -0.005 | 7.86e-01 | 0.203 | 0.194 | 0.009 | 2.65e-01 |
| ADHD | SBLUP.Inf | lassosum.10FCVal | 0.116 | 0.118 | -0.002 | 6.31e-01 | 0.193 | 0.194 | -0.001 | 9.39e-01 |
| ADHD | lassosum.MultiPRS | lassosum.10FCVal | 0.117 | 0.118 | -0.001 | 9.74e-01 | 0.199 | 0.194 | 0.005 | 4.25e-01 |
| ADHD | PRScs.10FCVal | lassosum.10FCVal | 0.114 | 0.118 | -0.004 | 4.53e-01 | 0.207 | 0.194 | 0.014 | 1.61e-01 |
| ADHD | LDpred2.MultiPRS | lassosum.10FCVal | 0.114 | 0.118 | -0.003 | 8.57e-01 | 0.199 | 0.194 | 0.005 | 5.46e-01 |
| ADHD | PRScs.PseudoVal | lassosum.10FCVal | 0.109 | 0.118 | -0.009 | 6.51e-02 | 0.189 | 0.194 | -0.005 | 6.05e-01 |
| ADHD | LDpred2.PseudoVal | lassosum.10FCVal | 0.115 | 0.118 | -0.003 | 5.68e-01 | 0.195 | 0.194 | 0.001 | 9.18e-01 |
| ADHD | LDpred1.MultiPRS | lassosum.10FCVal | 0.117 | 0.118 | -0.001 | 9.72e-01 | 0.191 | 0.194 | -0.002 | 7.65e-01 |
| ADHD | pT+clump.10FCVal | lassosum.10FCVal | 0.094 | 0.118 | -0.024 | 2.94e-03 | 0.150 | 0.194 | -0.044 | 5.36e-03 |
| ADHD | All.MultiPRS | lassosum.10FCVal | 0.115 | 0.118 | -0.002 | 8.94e-01 | 0.186 | 0.194 | -0.007 | 5.38e-01 |
| ADHD | LDpred1.Inf | lassosum.10FCVal | 0.115 | 0.118 | -0.002 | 6.32e-01 | 0.191 | 0.194 | -0.002 | 8.15e-01 |
| ADHD | DBSLMM.PseudoVal | lassosum.10FCVal | 0.114 | 0.118 | -0.004 | 6.04e-01 | 0.184 | 0.194 | -0.010 | 4.43e-01 |
| ADHD | LDpred1.10FCVal | lassosum.10FCVal | 0.118 | 0.118 | 0.000 | 9.56e-01 | 0.190 | 0.194 | -0.003 | 6.56e-01 |
| ADHD | lassosum.PseudoVal | lassosum.10FCVal | 0.077 | 0.118 | -0.041 | 1.96e-04 | 0.118 | 0.194 | -0.076 | 5.33e-04 |
| ADHD | LDpred2.10FCVal | lassosum.10FCVal | 0.118 | 0.118 | 0.001 | 8.85e-01 | 0.198 | 0.194 | 0.005 | 5.65e-01 |
| ADHD | LDpred2.10FCVal | LDpred1.MultiPRS | 0.118 | 0.117 | 0.001 | 9.46e-01 | 0.198 | 0.191 | 0.007 | 9.43e-02 |
| ADHD | PRScs.PseudoVal | LDpred1.MultiPRS | 0.109 | 0.117 | -0.008 | 6.49e-01 | 0.189 | 0.191 | -0.002 | 7.43e-01 |
| ADHD | LDpred2.Inf | LDpred1.MultiPRS | 0.115 | 0.117 | -0.002 | 9.25e-01 | 0.194 | 0.191 | 0.003 | 5.87e-01 |
| ADHD | pT+clump.10FCVal | LDpred1.MultiPRS | 0.094 | 0.117 | -0.023 | 1.95e-01 | 0.150 | 0.191 | -0.042 | 1.01e-02 |
| ADHD | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.091 | 0.117 | -0.026 | 1.44e-01 | 0.155 | 0.191 | -0.036 | 5.87e-02 |
| ADHD | All.MultiPRS | LDpred1.MultiPRS | 0.115 | 0.117 | -0.002 | 9.21e-01 | 0.186 | 0.191 | -0.005 | 6.68e-01 |
| ADHD | LDpred1.10FCVal | LDpred1.MultiPRS | 0.118 | 0.117 | 0.001 | 9.62e-01 | 0.190 | 0.191 | -0.001 | 3.69e-01 |
| ADHD | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.117 | 0.117 | 0.000 | 1.00e+00 | 0.191 | 0.191 | 0.000 | 1.00e+00 |
| ADHD | PRScs.10FCVal | LDpred1.MultiPRS | 0.114 | 0.117 | -0.003 | 8.66e-01 | 0.207 | 0.191 | 0.016 | 2.94e-02 |
| ADHD | lassosum.PseudoVal | LDpred1.MultiPRS | 0.077 | 0.117 | -0.040 | 2.22e-02 | 0.118 | 0.191 | -0.073 | 7.86e-04 |
| ADHD | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.096 | 0.117 | -0.021 | 2.30e-01 | 0.162 | 0.191 | -0.030 | 5.96e-02 |
| ADHD | LDpred1.Inf | LDpred1.MultiPRS | 0.115 | 0.117 | -0.002 | 9.23e-01 | 0.191 | 0.191 | 0.000 | 9.83e-01 |
| ADHD | lassosum.MultiPRS | LDpred1.MultiPRS | 0.117 | 0.117 | 0.000 | 9.91e-01 | 0.199 | 0.191 | 0.007 | 3.44e-01 |
| ADHD | PRScs.MultiPRS | LDpred1.MultiPRS | 0.113 | 0.117 | -0.004 | 1.86e-01 | 0.203 | 0.191 | 0.011 | 7.52e-02 |
| ADHD | lassosum.10FCVal | LDpred1.MultiPRS | 0.118 | 0.117 | 0.001 | 9.72e-01 | 0.194 | 0.191 | 0.002 | 7.65e-01 |
| ADHD | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.115 | 0.117 | -0.002 | 8.99e-01 | 0.195 | 0.191 | 0.003 | 5.48e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.114 | 0.117 | -0.003 | 8.69e-01 | 0.184 | 0.191 | -0.008 | 4.78e-01 |
| ADHD | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.114 | 0.117 | -0.003 | 2.06e-01 | 0.199 | 0.191 | 0.007 | 7.07e-02 |
| ADHD | SBLUP.Inf | LDpred1.MultiPRS | 0.116 | 0.117 | -0.002 | 9.32e-01 | 0.193 | 0.191 | 0.002 | 7.46e-01 |
| ADHD | All.MultiPRS | LDpred1.Inf | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.186 | 0.191 | -0.005 | 6.83e-01 |
| ADHD | LDpred2.PseudoVal | LDpred1.Inf | 0.115 | 0.115 | -0.001 | 7.82e-01 | 0.195 | 0.191 | 0.003 | 4.09e-01 |
| ADHD | SBayesR.PseudoVal | LDpred1.Inf | 0.091 | 0.115 | -0.024 | 1.53e-02 | 0.155 | 0.191 | -0.036 | 7.16e-02 |
| ADHD | LDpred2.MultiPRS | LDpred1.Inf | 0.114 | 0.115 | -0.001 | 9.61e-01 | 0.199 | 0.191 | 0.007 | 2.13e-01 |
| ADHD | PRScs.PseudoVal | LDpred1.Inf | 0.109 | 0.115 | -0.006 | 1.63e-01 | 0.189 | 0.191 | -0.002 | 7.88e-01 |
| ADHD | LDpred1.Inf | LDpred1.Inf | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.191 | 0.191 | 0.000 | 1.00e+00 |
| ADHD | pT+clump.10FCVal | LDpred1.Inf | 0.094 | 0.115 | -0.021 | 8.52e-03 | 0.150 | 0.191 | -0.042 | 9.67e-03 |
| ADHD | PRScs.MultiPRS | LDpred1.Inf | 0.113 | 0.115 | -0.003 | 8.88e-01 | 0.203 | 0.191 | 0.011 | 1.59e-01 |
| ADHD | lassosum.PseudoVal | LDpred1.Inf | 0.077 | 0.115 | -0.039 | 1.84e-03 | 0.118 | 0.191 | -0.073 | 2.67e-03 |
| ADHD | pT+clump.MultiPRS | LDpred1.Inf | 0.096 | 0.115 | -0.019 | 2.70e-01 | 0.162 | 0.191 | -0.030 | 6.84e-02 |
| ADHD | LDpred1.MultiPRS | LDpred1.Inf | 0.117 | 0.115 | 0.002 | 9.23e-01 | 0.191 | 0.191 | 0.000 | 9.83e-01 |
| ADHD | PRScs.10FCVal | LDpred1.Inf | 0.114 | 0.115 | -0.001 | 7.83e-01 | 0.207 | 0.191 | 0.016 | 8.31e-02 |
| ADHD | LDpred2.Inf | LDpred1.Inf | 0.115 | 0.115 | 0.000 | 9.83e-01 | 0.194 | 0.191 | 0.003 | 4.62e-01 |
| ADHD | SBLUP.Inf | LDpred1.Inf | 0.116 | 0.115 | 0.000 | 9.24e-01 | 0.193 | 0.191 | 0.002 | 6.92e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred1.Inf | 0.114 | 0.115 | -0.001 | 8.35e-01 | 0.184 | 0.191 | -0.008 | 4.95e-01 |
| ADHD | lassosum.MultiPRS | LDpred1.Inf | 0.117 | 0.115 | 0.002 | 9.22e-01 | 0.199 | 0.191 | 0.007 | 4.37e-01 |
| ADHD | LDpred1.10FCVal | LDpred1.Inf | 0.118 | 0.115 | 0.003 | 3.81e-01 | 0.190 | 0.191 | -0.001 | 8.32e-01 |
| ADHD | lassosum.10FCVal | LDpred1.Inf | 0.118 | 0.115 | 0.002 | 6.32e-01 | 0.194 | 0.191 | 0.002 | 8.15e-01 |
| ADHD | LDpred2.10FCVal | LDpred1.Inf | 0.118 | 0.115 | 0.003 | 3.30e-01 | 0.198 | 0.191 | 0.007 | 2.54e-01 |
| ADHD | LDpred1.Inf | LDpred1.10FCVal | 0.115 | 0.118 | -0.003 | 3.81e-01 | 0.191 | 0.190 | 0.001 | 8.32e-01 |
| ADHD | PRScs.MultiPRS | LDpred1.10FCVal | 0.113 | 0.118 | -0.005 | 7.77e-01 | 0.203 | 0.190 | 0.012 | 5.23e-02 |
| ADHD | LDpred1.10FCVal | LDpred1.10FCVal | 0.118 | 0.118 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| ADHD | All.MultiPRS | LDpred1.10FCVal | 0.115 | 0.118 | -0.003 | 8.85e-01 | 0.186 | 0.190 | -0.004 | 7.38e-01 |
| ADHD | SBayesR.PseudoVal | LDpred1.10FCVal | 0.091 | 0.118 | -0.027 | 4.67e-03 | 0.155 | 0.190 | -0.035 | 6.47e-02 |
| ADHD | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.114 | 0.118 | -0.004 | 5.03e-01 | 0.184 | 0.190 | -0.007 | 5.46e-01 |
| ADHD | pT+clump.MultiPRS | LDpred1.10FCVal | 0.096 | 0.118 | -0.022 | 2.12e-01 | 0.162 | 0.190 | -0.029 | 7.00e-02 |
| ADHD | lassosum.MultiPRS | LDpred1.10FCVal | 0.117 | 0.118 | -0.001 | 9.65e-01 | 0.199 | 0.190 | 0.008 | 2.73e-01 |
| ADHD | PRScs.10FCVal | LDpred1.10FCVal | 0.114 | 0.118 | -0.004 | 2.94e-01 | 0.207 | 0.190 | 0.017 | 1.79e-02 |
| ADHD | lassosum.10FCVal | LDpred1.10FCVal | 0.118 | 0.118 | 0.000 | 9.56e-01 | 0.194 | 0.190 | 0.003 | 6.56e-01 |
| ADHD | SBLUP.Inf | LDpred1.10FCVal | 0.116 | 0.118 | -0.002 | 4.70e-01 | 0.193 | 0.190 | 0.003 | 6.49e-01 |
| ADHD | LDpred2.PseudoVal | LDpred1.10FCVal | 0.115 | 0.118 | -0.003 | 3.59e-01 | 0.195 | 0.190 | 0.005 | 4.94e-01 |
| ADHD | pT+clump.10FCVal | LDpred1.10FCVal | 0.094 | 0.118 | -0.024 | 4.01e-03 | 0.150 | 0.190 | -0.041 | 1.34e-02 |
| ADHD | LDpred2.MultiPRS | LDpred1.10FCVal | 0.114 | 0.118 | -0.003 | 8.47e-01 | 0.199 | 0.190 | 0.008 | 5.13e-02 |
| ADHD | PRScs.PseudoVal | LDpred1.10FCVal | 0.109 | 0.118 | -0.009 | 1.30e-02 | 0.189 | 0.190 | -0.001 | 8.60e-01 |
| ADHD | LDpred2.10FCVal | LDpred1.10FCVal | 0.118 | 0.118 | 0.000 | 8.73e-01 | 0.198 | 0.190 | 0.008 | 6.43e-02 |
| ADHD | LDpred1.MultiPRS | LDpred1.10FCVal | 0.117 | 0.118 | -0.001 | 9.62e-01 | 0.191 | 0.190 | 0.001 | 3.69e-01 |
| ADHD | LDpred2.Inf | LDpred1.10FCVal | 0.115 | 0.118 | -0.003 | 4.50e-01 | 0.194 | 0.190 | 0.004 | 5.24e-01 |
| ADHD | lassosum.PseudoVal | LDpred1.10FCVal | 0.077 | 0.118 | -0.041 | 1.14e-04 | 0.118 | 0.190 | -0.072 | 6.39e-04 |
| ADHD | LDpred1.Inf | LDpred2.MultiPRS | 0.115 | 0.114 | 0.001 | 9.61e-01 | 0.191 | 0.199 | -0.007 | 2.13e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.114 | 0.114 | 0.000 | 9.85e-01 | 0.184 | 0.199 | -0.015 | 1.55e-01 |
| ADHD | SBLUP.Inf | LDpred2.MultiPRS | 0.116 | 0.114 | 0.001 | 9.52e-01 | 0.193 | 0.199 | -0.005 | 2.77e-01 |
| ADHD | LDpred2.Inf | LDpred2.MultiPRS | 0.115 | 0.114 | 0.001 | 9.59e-01 | 0.194 | 0.199 | -0.004 | 3.37e-01 |
| ADHD | pT+clump.10FCVal | LDpred2.MultiPRS | 0.094 | 0.114 | -0.021 | 2.50e-01 | 0.150 | 0.199 | -0.049 | 2.99e-03 |
| ADHD | LDpred2.10FCVal | LDpred2.MultiPRS | 0.118 | 0.114 | 0.004 | 8.32e-01 | 0.198 | 0.199 | 0.000 | 8.11e-01 |
| ADHD | PRScs.PseudoVal | LDpred2.MultiPRS | 0.109 | 0.114 | -0.005 | 7.58e-01 | 0.189 | 0.199 | -0.009 | 1.72e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.MultiPRS | 0.118 | 0.114 | 0.003 | 8.47e-01 | 0.190 | 0.199 | -0.008 | 5.13e-02 |
| ADHD | All.MultiPRS | LDpred2.MultiPRS | 0.115 | 0.114 | 0.001 | 9.60e-01 | 0.186 | 0.199 | -0.012 | 2.85e-01 |
| ADHD | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.091 | 0.114 | -0.023 | 1.89e-01 | 0.155 | 0.199 | -0.043 | 1.75e-02 |
| ADHD | lassosum.10FCVal | LDpred2.MultiPRS | 0.118 | 0.114 | 0.003 | 8.57e-01 | 0.194 | 0.199 | -0.005 | 5.46e-01 |
| ADHD | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.096 | 0.114 | -0.019 | 2.92e-01 | 0.162 | 0.199 | -0.037 | 1.98e-02 |
| ADHD | lassosum.MultiPRS | LDpred2.MultiPRS | 0.117 | 0.114 | 0.003 | 4.95e-01 | 0.199 | 0.199 | 0.000 | 9.78e-01 |
| ADHD | PRScs.10FCVal | LDpred2.MultiPRS | 0.114 | 0.114 | 0.000 | 9.82e-01 | 0.207 | 0.199 | 0.009 | 2.30e-01 |
| ADHD | PRScs.MultiPRS | LDpred2.MultiPRS | 0.113 | 0.114 | -0.002 | 6.20e-01 | 0.203 | 0.199 | 0.004 | 5.34e-01 |
| ADHD | lassosum.PseudoVal | LDpred2.MultiPRS | 0.077 | 0.114 | -0.038 | 3.25e-02 | 0.118 | 0.199 | -0.080 | 1.93e-04 |
| ADHD | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.115 | 0.114 | 0.000 | 9.85e-01 | 0.195 | 0.199 | -0.004 | 4.14e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.117 | 0.114 | 0.003 | 2.06e-01 | 0.191 | 0.199 | -0.007 | 7.07e-02 |
| ADHD | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.199 | 0.199 | 0.000 | 1.00e+00 |
| ADHD | LDpred1.Inf | LDpred2.10FCVal | 0.115 | 0.118 | -0.003 | 3.30e-01 | 0.191 | 0.198 | -0.007 | 2.54e-01 |
| ADHD | PRScs.MultiPRS | LDpred2.10FCVal | 0.113 | 0.118 | -0.005 | 7.62e-01 | 0.203 | 0.198 | 0.004 | 5.11e-01 |
| ADHD | LDpred2.MultiPRS | LDpred2.10FCVal | 0.114 | 0.118 | -0.004 | 8.32e-01 | 0.199 | 0.198 | 0.000 | 8.11e-01 |
| ADHD | PRScs.PseudoVal | LDpred2.10FCVal | 0.109 | 0.118 | -0.009 | 1.19e-02 | 0.189 | 0.198 | -0.009 | 1.86e-01 |
| ADHD | LDpred2.10FCVal | LDpred2.10FCVal | 0.118 | 0.118 | 0.000 | 1.00e+00 | 0.198 | 0.198 | 0.000 | 1.00e+00 |
| ADHD | lassosum.MultiPRS | LDpred2.10FCVal | 0.117 | 0.118 | -0.001 | 9.49e-01 | 0.199 | 0.198 | 0.000 | 9.51e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.10FCVal | 0.118 | 0.118 | 0.000 | 8.73e-01 | 0.190 | 0.198 | -0.008 | 6.43e-02 |
| ADHD | All.MultiPRS | LDpred2.10FCVal | 0.115 | 0.118 | -0.003 | 8.69e-01 | 0.186 | 0.198 | -0.012 | 2.97e-01 |
| ADHD | SBayesR.PseudoVal | LDpred2.10FCVal | 0.091 | 0.118 | -0.027 | 2.98e-03 | 0.155 | 0.198 | -0.043 | 1.82e-02 |
| ADHD | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.114 | 0.118 | -0.004 | 4.45e-01 | 0.184 | 0.198 | -0.015 | 1.65e-01 |
| ADHD | pT+clump.MultiPRS | LDpred2.10FCVal | 0.096 | 0.118 | -0.022 | 2.05e-01 | 0.162 | 0.198 | -0.037 | 2.06e-02 |
| ADHD | lassosum.PseudoVal | LDpred2.10FCVal | 0.077 | 0.118 | -0.042 | 1.44e-04 | 0.118 | 0.198 | -0.080 | 1.91e-04 |
| ADHD | PRScs.10FCVal | LDpred2.10FCVal | 0.114 | 0.118 | -0.004 | 2.58e-01 | 0.207 | 0.198 | 0.009 | 2.22e-01 |
| ADHD | lassosum.10FCVal | LDpred2.10FCVal | 0.118 | 0.118 | -0.001 | 8.85e-01 | 0.194 | 0.198 | -0.005 | 5.65e-01 |
| ADHD | SBLUP.Inf | LDpred2.10FCVal | 0.116 | 0.118 | -0.003 | 3.24e-01 | 0.193 | 0.198 | -0.005 | 3.26e-01 |
| ADHD | LDpred2.PseudoVal | LDpred2.10FCVal | 0.115 | 0.118 | -0.003 | 1.74e-01 | 0.195 | 0.198 | -0.003 | 4.89e-01 |
| ADHD | pT+clump.10FCVal | LDpred2.10FCVal | 0.094 | 0.118 | -0.024 | 3.71e-03 | 0.150 | 0.198 | -0.049 | 3.13e-03 |
| ADHD | LDpred2.Inf | LDpred2.10FCVal | 0.115 | 0.118 | -0.003 | 2.27e-01 | 0.194 | 0.198 | -0.004 | 4.05e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.10FCVal | 0.117 | 0.118 | -0.001 | 9.46e-01 | 0.191 | 0.198 | -0.007 | 9.43e-02 |
| ADHD | lassosum.10FCVal | LDpred2.PseudoVal | 0.118 | 0.115 | 0.003 | 5.68e-01 | 0.194 | 0.195 | -0.001 | 9.18e-01 |
| ADHD | SBLUP.Inf | LDpred2.PseudoVal | 0.116 | 0.115 | 0.001 | 7.02e-01 | 0.193 | 0.195 | -0.002 | 6.52e-01 |
| ADHD | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.091 | 0.115 | -0.024 | 1.41e-02 | 0.155 | 0.195 | -0.040 | 3.96e-02 |
| ADHD | All.MultiPRS | LDpred2.PseudoVal | 0.115 | 0.115 | 0.001 | 9.76e-01 | 0.186 | 0.195 | -0.008 | 5.00e-01 |
| ADHD | PRScs.PseudoVal | LDpred2.PseudoVal | 0.109 | 0.115 | -0.006 | 2.03e-01 | 0.189 | 0.195 | -0.006 | 5.23e-01 |
| ADHD | lassosum.MultiPRS | LDpred2.PseudoVal | 0.117 | 0.115 | 0.002 | 8.97e-01 | 0.199 | 0.195 | 0.004 | 6.79e-01 |
| ADHD | PRScs.MultiPRS | LDpred2.PseudoVal | 0.113 | 0.115 | -0.002 | 9.13e-01 | 0.203 | 0.195 | 0.008 | 3.50e-01 |
| ADHD | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.114 | 0.115 | 0.000 | 9.85e-01 | 0.199 | 0.195 | 0.004 | 4.14e-01 |
| ADHD | PRScs.10FCVal | LDpred2.PseudoVal | 0.114 | 0.115 | -0.001 | 8.74e-01 | 0.207 | 0.195 | 0.012 | 1.69e-01 |
| ADHD | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.195 | 0.195 | 0.000 | 1.00e+00 |
| ADHD | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.096 | 0.115 | -0.019 | 2.83e-01 | 0.162 | 0.195 | -0.033 | 4.79e-02 |
| ADHD | pT+clump.10FCVal | LDpred2.PseudoVal | 0.094 | 0.115 | -0.021 | 1.31e-02 | 0.150 | 0.195 | -0.045 | 6.74e-03 |
| ADHD | lassosum.PseudoVal | LDpred2.PseudoVal | 0.077 | 0.115 | -0.038 | 2.13e-03 | 0.118 | 0.195 | -0.077 | 1.69e-03 |
| ADHD | LDpred1.Inf | LDpred2.PseudoVal | 0.115 | 0.115 | 0.001 | 7.82e-01 | 0.191 | 0.195 | -0.003 | 4.09e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.117 | 0.115 | 0.002 | 8.99e-01 | 0.191 | 0.195 | -0.003 | 5.48e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.PseudoVal | 0.118 | 0.115 | 0.003 | 3.59e-01 | 0.190 | 0.195 | -0.005 | 4.94e-01 |
| ADHD | LDpred2.Inf | LDpred2.PseudoVal | 0.115 | 0.115 | 0.001 | 5.56e-01 | 0.194 | 0.195 | 0.000 | 8.30e-01 |
| ADHD | LDpred2.10FCVal | LDpred2.PseudoVal | 0.118 | 0.115 | 0.003 | 1.74e-01 | 0.198 | 0.195 | 0.003 | 4.89e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.114 | 0.115 | -0.001 | 9.08e-01 | 0.184 | 0.195 | -0.011 | 3.20e-01 |
| ADHD | pT+clump.MultiPRS | LDpred2.Inf | 0.096 | 0.115 | -0.020 | 2.69e-01 | 0.162 | 0.194 | -0.033 | 4.47e-02 |
| ADHD | lassosum.MultiPRS | LDpred2.Inf | 0.117 | 0.115 | 0.002 | 9.23e-01 | 0.199 | 0.194 | 0.004 | 6.33e-01 |
| ADHD | SBayesR.PseudoVal | LDpred2.Inf | 0.091 | 0.115 | -0.024 | 1.22e-02 | 0.155 | 0.194 | -0.039 | 4.21e-02 |
| ADHD | All.MultiPRS | LDpred2.Inf | 0.115 | 0.115 | 0.000 | 9.98e-01 | 0.186 | 0.194 | -0.008 | 5.12e-01 |
| ADHD | SBLUP.Inf | LDpred2.Inf | 0.116 | 0.115 | 0.000 | 9.15e-01 | 0.193 | 0.194 | -0.001 | 6.44e-01 |
| ADHD | LDpred2.PseudoVal | LDpred2.Inf | 0.115 | 0.115 | -0.001 | 5.56e-01 | 0.195 | 0.194 | 0.000 | 8.30e-01 |
| ADHD | PRScs.MultiPRS | LDpred2.Inf | 0.113 | 0.115 | -0.003 | 8.87e-01 | 0.203 | 0.194 | 0.008 | 2.97e-01 |
| ADHD | LDpred2.MultiPRS | LDpred2.Inf | 0.114 | 0.115 | -0.001 | 9.59e-01 | 0.199 | 0.194 | 0.004 | 3.37e-01 |
| ADHD | PRScs.10FCVal | LDpred2.Inf | 0.114 | 0.115 | -0.001 | 7.77e-01 | 0.207 | 0.194 | 0.013 | 1.60e-01 |
| ADHD | LDpred1.Inf | LDpred2.Inf | 0.115 | 0.115 | 0.000 | 9.83e-01 | 0.191 | 0.194 | -0.003 | 4.62e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.Inf | 0.117 | 0.115 | 0.002 | 9.25e-01 | 0.191 | 0.194 | -0.003 | 5.87e-01 |
| ADHD | pT+clump.10FCVal | LDpred2.Inf | 0.094 | 0.115 | -0.021 | 8.33e-03 | 0.150 | 0.194 | -0.045 | 5.40e-03 |
| ADHD | lassosum.PseudoVal | LDpred2.Inf | 0.077 | 0.115 | -0.039 | 1.92e-03 | 0.118 | 0.194 | -0.076 | 1.88e-03 |
| ADHD | PRScs.PseudoVal | LDpred2.Inf | 0.109 | 0.115 | -0.006 | 1.62e-01 | 0.189 | 0.194 | -0.005 | 5.52e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred2.Inf | 0.114 | 0.115 | -0.001 | 8.29e-01 | 0.184 | 0.194 | -0.011 | 3.41e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.Inf | 0.118 | 0.115 | 0.003 | 4.50e-01 | 0.190 | 0.194 | -0.004 | 5.24e-01 |
| ADHD | LDpred2.Inf | LDpred2.Inf | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.194 | 0.194 | 0.000 | 1.00e+00 |
| ADHD | lassosum.10FCVal | LDpred2.Inf | 0.118 | 0.115 | 0.002 | 6.30e-01 | 0.194 | 0.194 | -0.001 | 9.48e-01 |
| ADHD | LDpred2.10FCVal | LDpred2.Inf | 0.118 | 0.115 | 0.003 | 2.27e-01 | 0.198 | 0.194 | 0.004 | 4.05e-01 |
| ADHD | PRScs.10FCVal | PRScs.MultiPRS | 0.114 | 0.113 | 0.001 | 9.45e-01 | 0.207 | 0.203 | 0.005 | 2.25e-01 |
| ADHD | LDpred1.10FCVal | PRScs.MultiPRS | 0.118 | 0.113 | 0.005 | 7.77e-01 | 0.190 | 0.203 | -0.012 | 5.23e-02 |
| ADHD | lassosum.MultiPRS | PRScs.MultiPRS | 0.117 | 0.113 | 0.004 | 2.87e-01 | 0.199 | 0.203 | -0.004 | 6.24e-01 |
| ADHD | LDpred2.MultiPRS | PRScs.MultiPRS | 0.114 | 0.113 | 0.002 | 6.20e-01 | 0.199 | 0.203 | -0.004 | 5.34e-01 |
| ADHD | All.MultiPRS | PRScs.MultiPRS | 0.115 | 0.113 | 0.002 | 8.86e-01 | 0.186 | 0.203 | -0.016 | 1.94e-01 |
| ADHD | PRScs.PseudoVal | PRScs.MultiPRS | 0.109 | 0.113 | -0.004 | 8.28e-01 | 0.189 | 0.203 | -0.014 | 8.48e-02 |
| ADHD | LDpred1.MultiPRS | PRScs.MultiPRS | 0.117 | 0.113 | 0.004 | 1.86e-01 | 0.191 | 0.203 | -0.011 | 7.52e-02 |
| ADHD | pT+clump.MultiPRS | PRScs.MultiPRS | 0.096 | 0.113 | -0.017 | 3.36e-01 | 0.162 | 0.203 | -0.041 | 6.55e-03 |
| ADHD | lassosum.PseudoVal | PRScs.MultiPRS | 0.077 | 0.113 | -0.036 | 4.09e-02 | 0.118 | 0.203 | -0.084 | 8.81e-05 |
| ADHD | SBayesR.PseudoVal | PRScs.MultiPRS | 0.091 | 0.113 | -0.022 | 2.22e-01 | 0.155 | 0.203 | -0.047 | 1.19e-02 |
| ADHD | DBSLMM.PseudoVal | PRScs.MultiPRS | 0.114 | 0.113 | 0.001 | 9.42e-01 | 0.184 | 0.203 | -0.019 | 1.04e-01 |
| ADHD | SBLUP.Inf | PRScs.MultiPRS | 0.116 | 0.113 | 0.003 | 8.80e-01 | 0.193 | 0.203 | -0.009 | 1.83e-01 |
| ADHD | LDpred2.Inf | PRScs.MultiPRS | 0.115 | 0.113 | 0.003 | 8.87e-01 | 0.194 | 0.203 | -0.008 | 2.97e-01 |
| ADHD | pT+clump.10FCVal | PRScs.MultiPRS | 0.094 | 0.113 | -0.019 | 2.90e-01 | 0.150 | 0.203 | -0.053 | 6.43e-04 |
| ADHD | LDpred2.10FCVal | PRScs.MultiPRS | 0.118 | 0.113 | 0.005 | 7.62e-01 | 0.198 | 0.203 | -0.004 | 5.11e-01 |
| ADHD | lassosum.10FCVal | PRScs.MultiPRS | 0.118 | 0.113 | 0.005 | 7.86e-01 | 0.194 | 0.203 | -0.009 | 2.65e-01 |
| ADHD | LDpred2.PseudoVal | PRScs.MultiPRS | 0.115 | 0.113 | 0.002 | 9.13e-01 | 0.195 | 0.203 | -0.008 | 3.50e-01 |
| ADHD | LDpred1.Inf | PRScs.MultiPRS | 0.115 | 0.113 | 0.003 | 8.88e-01 | 0.191 | 0.203 | -0.011 | 1.59e-01 |
| ADHD | PRScs.MultiPRS | PRScs.MultiPRS | 0.113 | 0.113 | 0.000 | 1.00e+00 | 0.203 | 0.203 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.PseudoVal | PRScs.10FCVal | 0.115 | 0.114 | 0.001 | 8.74e-01 | 0.195 | 0.207 | -0.012 | 1.69e-01 |
| ADHD | PRScs.10FCVal | PRScs.10FCVal | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.207 | 0.207 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.MultiPRS | PRScs.10FCVal | 0.114 | 0.114 | 0.000 | 9.82e-01 | 0.199 | 0.207 | -0.009 | 2.30e-01 |
| ADHD | pT+clump.MultiPRS | PRScs.10FCVal | 0.096 | 0.114 | -0.018 | 3.02e-01 | 0.162 | 0.207 | -0.046 | 5.72e-03 |
| ADHD | LDpred1.Inf | PRScs.10FCVal | 0.115 | 0.114 | 0.001 | 7.83e-01 | 0.191 | 0.207 | -0.016 | 8.31e-02 |
| ADHD | LDpred1.MultiPRS | PRScs.10FCVal | 0.117 | 0.114 | 0.003 | 8.66e-01 | 0.191 | 0.207 | -0.016 | 2.94e-02 |
| ADHD | pT+clump.10FCVal | PRScs.10FCVal | 0.094 | 0.114 | -0.020 | 2.32e-02 | 0.150 | 0.207 | -0.058 | 8.98e-04 |
| ADHD | lassosum.PseudoVal | PRScs.10FCVal | 0.077 | 0.114 | -0.038 | 3.37e-04 | 0.118 | 0.207 | -0.089 | 1.61e-05 |
| ADHD | PRScs.PseudoVal | PRScs.10FCVal | 0.109 | 0.114 | -0.005 | 2.31e-01 | 0.189 | 0.207 | -0.018 | 2.78e-02 |
| ADHD | DBSLMM.PseudoVal | PRScs.10FCVal | 0.114 | 0.114 | 0.000 | 9.91e-01 | 0.184 | 0.207 | -0.024 | 4.81e-02 |
| ADHD | SBLUP.Inf | PRScs.10FCVal | 0.116 | 0.114 | 0.001 | 7.52e-01 | 0.193 | 0.207 | -0.014 | 1.14e-01 |
| ADHD | LDpred2.Inf | PRScs.10FCVal | 0.115 | 0.114 | 0.001 | 7.77e-01 | 0.194 | 0.207 | -0.013 | 1.60e-01 |
| ADHD | lassosum.10FCVal | PRScs.10FCVal | 0.118 | 0.114 | 0.004 | 4.53e-01 | 0.194 | 0.207 | -0.014 | 1.61e-01 |
| ADHD | LDpred2.10FCVal | PRScs.10FCVal | 0.118 | 0.114 | 0.004 | 2.58e-01 | 0.198 | 0.207 | -0.009 | 2.22e-01 |
| ADHD | lassosum.MultiPRS | PRScs.10FCVal | 0.117 | 0.114 | 0.003 | 8.65e-01 | 0.199 | 0.207 | -0.009 | 3.60e-01 |
| ADHD | LDpred1.10FCVal | PRScs.10FCVal | 0.118 | 0.114 | 0.004 | 2.94e-01 | 0.190 | 0.207 | -0.017 | 1.79e-02 |
| ADHD | All.MultiPRS | PRScs.10FCVal | 0.115 | 0.114 | 0.001 | 9.43e-01 | 0.186 | 0.207 | -0.021 | 1.15e-01 |
| ADHD | SBayesR.PseudoVal | PRScs.10FCVal | 0.091 | 0.114 | -0.023 | 1.49e-02 | 0.155 | 0.207 | -0.052 | 5.50e-03 |
| ADHD | PRScs.MultiPRS | PRScs.10FCVal | 0.113 | 0.114 | -0.001 | 9.45e-01 | 0.203 | 0.207 | -0.005 | 2.25e-01 |
| ADHD | All.MultiPRS | PRScs.PseudoVal | 0.115 | 0.109 | 0.006 | 7.18e-01 | 0.186 | 0.189 | -0.003 | 8.36e-01 |
| ADHD | pT+clump.MultiPRS | PRScs.PseudoVal | 0.096 | 0.109 | -0.013 | 4.58e-01 | 0.162 | 0.189 | -0.027 | 9.13e-02 |
| ADHD | LDpred2.PseudoVal | PRScs.PseudoVal | 0.115 | 0.109 | 0.006 | 2.03e-01 | 0.195 | 0.189 | 0.006 | 5.23e-01 |
| ADHD | SBayesR.PseudoVal | PRScs.PseudoVal | 0.091 | 0.109 | -0.018 | 5.71e-02 | 0.155 | 0.189 | -0.034 | 7.14e-02 |
| ADHD | LDpred2.MultiPRS | PRScs.PseudoVal | 0.114 | 0.109 | 0.005 | 7.58e-01 | 0.199 | 0.189 | 0.009 | 1.72e-01 |
| ADHD | SBLUP.Inf | PRScs.PseudoVal | 0.116 | 0.109 | 0.007 | 1.50e-01 | 0.193 | 0.189 | 0.004 | 6.45e-01 |
| ADHD | LDpred1.Inf | PRScs.PseudoVal | 0.115 | 0.109 | 0.006 | 1.63e-01 | 0.191 | 0.189 | 0.002 | 7.88e-01 |
| ADHD | PRScs.10FCVal | PRScs.PseudoVal | 0.114 | 0.109 | 0.005 | 2.31e-01 | 0.207 | 0.189 | 0.018 | 2.78e-02 |
| ADHD | LDpred2.Inf | PRScs.PseudoVal | 0.115 | 0.109 | 0.006 | 1.62e-01 | 0.194 | 0.189 | 0.005 | 5.52e-01 |
| ADHD | PRScs.PseudoVal | PRScs.PseudoVal | 0.109 | 0.109 | 0.000 | 1.00e+00 | 0.189 | 0.189 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.10FCVal | PRScs.PseudoVal | 0.118 | 0.109 | 0.009 | 1.19e-02 | 0.198 | 0.189 | 0.009 | 1.86e-01 |
| ADHD | pT+clump.10FCVal | PRScs.PseudoVal | 0.094 | 0.109 | -0.015 | 8.48e-02 | 0.150 | 0.189 | -0.040 | 2.15e-02 |
| ADHD | PRScs.MultiPRS | PRScs.PseudoVal | 0.113 | 0.109 | 0.004 | 8.28e-01 | 0.203 | 0.189 | 0.014 | 8.48e-02 |
| ADHD | lassosum.PseudoVal | PRScs.PseudoVal | 0.077 | 0.109 | -0.032 | 2.04e-03 | 0.118 | 0.189 | -0.071 | 5.26e-04 |
| ADHD | lassosum.10FCVal | PRScs.PseudoVal | 0.118 | 0.109 | 0.009 | 6.51e-02 | 0.194 | 0.189 | 0.005 | 6.05e-01 |
| ADHD | LDpred1.MultiPRS | PRScs.PseudoVal | 0.117 | 0.109 | 0.008 | 6.49e-01 | 0.191 | 0.189 | 0.002 | 7.43e-01 |
| ADHD | lassosum.MultiPRS | PRScs.PseudoVal | 0.117 | 0.109 | 0.008 | 6.49e-01 | 0.199 | 0.189 | 0.010 | 2.76e-01 |
| ADHD | LDpred1.10FCVal | PRScs.PseudoVal | 0.118 | 0.109 | 0.009 | 1.30e-02 | 0.190 | 0.189 | 0.001 | 8.60e-01 |
| ADHD | DBSLMM.PseudoVal | PRScs.PseudoVal | 0.114 | 0.109 | 0.005 | 3.84e-01 | 0.184 | 0.189 | -0.006 | 6.32e-01 |
| ADHD | All.MultiPRS | pT+clump.MultiPRS | 0.115 | 0.096 | 0.019 | 2.64e-01 | 0.186 | 0.162 | 0.025 | 1.61e-01 |
| ADHD | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.091 | 0.096 | -0.005 | 7.87e-01 | 0.155 | 0.162 | -0.006 | 7.69e-01 |
| ADHD | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.117 | 0.096 | 0.021 | 2.30e-01 | 0.191 | 0.162 | 0.030 | 5.96e-02 |
| ADHD | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.096 | 0.096 | 0.000 | 1.00e+00 | 0.162 | 0.162 | 0.000 | 1.00e+00 |
| ADHD | lassosum.PseudoVal | pT+clump.MultiPRS | 0.077 | 0.096 | -0.019 | 2.74e-01 | 0.118 | 0.162 | -0.044 | 6.43e-02 |
| ADHD | PRScs.10FCVal | pT+clump.MultiPRS | 0.114 | 0.096 | 0.018 | 3.02e-01 | 0.207 | 0.162 | 0.046 | 5.72e-03 |
| ADHD | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.114 | 0.096 | 0.018 | 3.01e-01 | 0.184 | 0.162 | 0.022 | 1.60e-01 |
| ADHD | SBLUP.Inf | pT+clump.MultiPRS | 0.116 | 0.096 | 0.020 | 2.65e-01 | 0.193 | 0.162 | 0.031 | 4.41e-02 |
| ADHD | LDpred2.Inf | pT+clump.MultiPRS | 0.115 | 0.096 | 0.020 | 2.69e-01 | 0.194 | 0.162 | 0.033 | 4.47e-02 |
| ADHD | pT+clump.10FCVal | pT+clump.MultiPRS | 0.094 | 0.096 | -0.002 | 9.14e-01 | 0.150 | 0.162 | -0.012 | 9.24e-02 |
| ADHD | LDpred2.10FCVal | pT+clump.MultiPRS | 0.118 | 0.096 | 0.022 | 2.05e-01 | 0.198 | 0.162 | 0.037 | 2.06e-02 |
| ADHD | PRScs.PseudoVal | pT+clump.MultiPRS | 0.109 | 0.096 | 0.013 | 4.58e-01 | 0.189 | 0.162 | 0.027 | 9.13e-02 |
| ADHD | LDpred1.10FCVal | pT+clump.MultiPRS | 0.118 | 0.096 | 0.022 | 2.12e-01 | 0.190 | 0.162 | 0.029 | 7.00e-02 |
| ADHD | LDpred1.Inf | pT+clump.MultiPRS | 0.115 | 0.096 | 0.019 | 2.70e-01 | 0.191 | 0.162 | 0.030 | 6.84e-02 |
| ADHD | PRScs.MultiPRS | pT+clump.MultiPRS | 0.113 | 0.096 | 0.017 | 3.36e-01 | 0.203 | 0.162 | 0.041 | 6.55e-03 |
| ADHD | lassosum.10FCVal | pT+clump.MultiPRS | 0.118 | 0.096 | 0.022 | 2.17e-01 | 0.194 | 0.162 | 0.032 | 4.00e-02 |
| ADHD | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.115 | 0.096 | 0.019 | 2.83e-01 | 0.195 | 0.162 | 0.033 | 4.79e-02 |
| ADHD | lassosum.MultiPRS | pT+clump.MultiPRS | 0.117 | 0.096 | 0.021 | 2.29e-01 | 0.199 | 0.162 | 0.037 | 1.07e-02 |
| ADHD | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.114 | 0.096 | 0.019 | 2.92e-01 | 0.199 | 0.162 | 0.037 | 1.98e-02 |
| ADHD | LDpred2.10FCVal | pT+clump.10FCVal | 0.118 | 0.094 | 0.024 | 3.71e-03 | 0.198 | 0.150 | 0.049 | 3.13e-03 |
| ADHD | pT+clump.MultiPRS | pT+clump.10FCVal | 0.096 | 0.094 | 0.002 | 9.14e-01 | 0.162 | 0.150 | 0.012 | 9.24e-02 |
| ADHD | LDpred2.Inf | pT+clump.10FCVal | 0.115 | 0.094 | 0.021 | 8.33e-03 | 0.194 | 0.150 | 0.045 | 5.40e-03 |
| ADHD | SBayesR.PseudoVal | pT+clump.10FCVal | 0.091 | 0.094 | -0.003 | 8.12e-01 | 0.155 | 0.150 | 0.006 | 8.06e-01 |
| ADHD | PRScs.MultiPRS | pT+clump.10FCVal | 0.113 | 0.094 | 0.019 | 2.90e-01 | 0.203 | 0.150 | 0.053 | 6.43e-04 |
| ADHD | SBLUP.Inf | pT+clump.10FCVal | 0.116 | 0.094 | 0.022 | 5.10e-03 | 0.193 | 0.150 | 0.044 | 4.41e-03 |
| ADHD | lassosum.MultiPRS | pT+clump.10FCVal | 0.117 | 0.094 | 0.023 | 1.95e-01 | 0.199 | 0.150 | 0.049 | 3.02e-03 |
| ADHD | PRScs.10FCVal | pT+clump.10FCVal | 0.114 | 0.094 | 0.020 | 2.32e-02 | 0.207 | 0.150 | 0.058 | 8.98e-04 |
| ADHD | LDpred2.MultiPRS | pT+clump.10FCVal | 0.114 | 0.094 | 0.021 | 2.50e-01 | 0.199 | 0.150 | 0.049 | 2.99e-03 |
| ADHD | lassosum.10FCVal | pT+clump.10FCVal | 0.118 | 0.094 | 0.024 | 2.94e-03 | 0.194 | 0.150 | 0.044 | 5.36e-03 |
| ADHD | LDpred2.PseudoVal | pT+clump.10FCVal | 0.115 | 0.094 | 0.021 | 1.31e-02 | 0.195 | 0.150 | 0.045 | 6.74e-03 |
| ADHD | LDpred1.MultiPRS | pT+clump.10FCVal | 0.117 | 0.094 | 0.023 | 1.95e-01 | 0.191 | 0.150 | 0.042 | 1.01e-02 |
| ADHD | pT+clump.10FCVal | pT+clump.10FCVal | 0.094 | 0.094 | 0.000 | 1.00e+00 | 0.150 | 0.150 | 0.000 | 1.00e+00 |
| ADHD | All.MultiPRS | pT+clump.10FCVal | 0.115 | 0.094 | 0.021 | 2.27e-01 | 0.186 | 0.150 | 0.037 | 5.75e-02 |
| ADHD | LDpred1.Inf | pT+clump.10FCVal | 0.115 | 0.094 | 0.021 | 8.52e-03 | 0.191 | 0.150 | 0.042 | 9.67e-03 |
| ADHD | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.114 | 0.094 | 0.020 | 3.55e-02 | 0.184 | 0.150 | 0.034 | 6.94e-02 |
| ADHD | LDpred1.10FCVal | pT+clump.10FCVal | 0.118 | 0.094 | 0.024 | 4.01e-03 | 0.190 | 0.150 | 0.041 | 1.34e-02 |
| ADHD | lassosum.PseudoVal | pT+clump.10FCVal | 0.077 | 0.094 | -0.017 | 1.94e-01 | 0.118 | 0.150 | -0.031 | 2.28e-01 |
| ADHD | PRScs.PseudoVal | pT+clump.10FCVal | 0.109 | 0.094 | 0.015 | 8.48e-02 | 0.189 | 0.150 | 0.040 | 2.15e-02 |
| ADHD | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.096 | 0.091 | 0.005 | 7.87e-01 | 0.162 | 0.155 | 0.006 | 7.69e-01 |
| ADHD | LDpred1.10FCVal | SBayesR.PseudoVal | 0.118 | 0.091 | 0.027 | 4.67e-03 | 0.190 | 0.155 | 0.035 | 6.47e-02 |
| ADHD | PRScs.10FCVal | SBayesR.PseudoVal | 0.114 | 0.091 | 0.023 | 1.49e-02 | 0.207 | 0.155 | 0.052 | 5.50e-03 |
| ADHD | LDpred2.10FCVal | SBayesR.PseudoVal | 0.118 | 0.091 | 0.027 | 2.98e-03 | 0.198 | 0.155 | 0.043 | 1.82e-02 |
| ADHD | lassosum.10FCVal | SBayesR.PseudoVal | 0.118 | 0.091 | 0.027 | 5.38e-03 | 0.194 | 0.155 | 0.039 | 4.15e-02 |
| ADHD | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.115 | 0.091 | 0.024 | 1.41e-02 | 0.195 | 0.155 | 0.040 | 3.96e-02 |
| ADHD | lassosum.MultiPRS | SBayesR.PseudoVal | 0.117 | 0.091 | 0.026 | 1.45e-01 | 0.199 | 0.155 | 0.044 | 1.99e-02 |
| ADHD | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.114 | 0.091 | 0.023 | 1.89e-01 | 0.199 | 0.155 | 0.043 | 1.75e-02 |
| ADHD | All.MultiPRS | SBayesR.PseudoVal | 0.115 | 0.091 | 0.024 | 1.69e-01 | 0.186 | 0.155 | 0.031 | 1.12e-01 |
| ADHD | PRScs.PseudoVal | SBayesR.PseudoVal | 0.109 | 0.091 | 0.018 | 5.71e-02 | 0.189 | 0.155 | 0.034 | 7.14e-02 |
| ADHD | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.117 | 0.091 | 0.026 | 1.44e-01 | 0.191 | 0.155 | 0.036 | 5.87e-02 |
| ADHD | pT+clump.10FCVal | SBayesR.PseudoVal | 0.094 | 0.091 | 0.003 | 8.12e-01 | 0.150 | 0.155 | -0.006 | 8.06e-01 |
| ADHD | lassosum.PseudoVal | SBayesR.PseudoVal | 0.077 | 0.091 | -0.015 | 2.23e-01 | 0.118 | 0.155 | -0.037 | 1.22e-01 |
| ADHD | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.091 | 0.091 | 0.000 | 1.00e+00 | 0.155 | 0.155 | 0.000 | 1.00e+00 |
| ADHD | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.114 | 0.091 | 0.023 | 2.17e-02 | 0.184 | 0.155 | 0.028 | 1.54e-01 |
| ADHD | SBLUP.Inf | SBayesR.PseudoVal | 0.116 | 0.091 | 0.024 | 1.18e-02 | 0.193 | 0.155 | 0.038 | 4.90e-02 |
| ADHD | LDpred2.Inf | SBayesR.PseudoVal | 0.115 | 0.091 | 0.024 | 1.22e-02 | 0.194 | 0.155 | 0.039 | 4.21e-02 |
| ADHD | LDpred1.Inf | SBayesR.PseudoVal | 0.115 | 0.091 | 0.024 | 1.53e-02 | 0.191 | 0.155 | 0.036 | 7.16e-02 |
| ADHD | PRScs.MultiPRS | SBayesR.PseudoVal | 0.113 | 0.091 | 0.022 | 2.22e-01 | 0.203 | 0.155 | 0.047 | 1.19e-02 |
| ADHD | LDpred2.10FCVal | SBLUP.Inf | 0.118 | 0.116 | 0.003 | 3.24e-01 | 0.198 | 0.193 | 0.005 | 3.26e-01 |
| ADHD | PRScs.PseudoVal | SBLUP.Inf | 0.109 | 0.116 | -0.007 | 1.50e-01 | 0.189 | 0.193 | -0.004 | 6.45e-01 |
| ADHD | LDpred1.10FCVal | SBLUP.Inf | 0.118 | 0.116 | 0.002 | 4.70e-01 | 0.190 | 0.193 | -0.003 | 6.49e-01 |
| ADHD | LDpred2.Inf | SBLUP.Inf | 0.115 | 0.116 | 0.000 | 9.15e-01 | 0.194 | 0.193 | 0.001 | 6.44e-01 |
| ADHD | SBayesR.PseudoVal | SBLUP.Inf | 0.091 | 0.116 | -0.024 | 1.18e-02 | 0.155 | 0.193 | -0.038 | 4.90e-02 |
| ADHD | lassosum.10FCVal | SBLUP.Inf | 0.118 | 0.116 | 0.002 | 6.31e-01 | 0.194 | 0.193 | 0.001 | 9.39e-01 |
| ADHD | pT+clump.MultiPRS | SBLUP.Inf | 0.096 | 0.116 | -0.020 | 2.65e-01 | 0.162 | 0.193 | -0.031 | 4.41e-02 |
| ADHD | lassosum.MultiPRS | SBLUP.Inf | 0.117 | 0.116 | 0.002 | 9.30e-01 | 0.199 | 0.193 | 0.006 | 5.12e-01 |
| ADHD | PRScs.10FCVal | SBLUP.Inf | 0.114 | 0.116 | -0.001 | 7.52e-01 | 0.207 | 0.193 | 0.014 | 1.14e-01 |
| ADHD | PRScs.MultiPRS | SBLUP.Inf | 0.113 | 0.116 | -0.003 | 8.80e-01 | 0.203 | 0.193 | 0.009 | 1.83e-01 |
| ADHD | LDpred1.MultiPRS | SBLUP.Inf | 0.117 | 0.116 | 0.002 | 9.32e-01 | 0.191 | 0.193 | -0.002 | 7.46e-01 |
| ADHD | LDpred2.PseudoVal | SBLUP.Inf | 0.115 | 0.116 | -0.001 | 7.02e-01 | 0.195 | 0.193 | 0.002 | 6.52e-01 |
| ADHD | LDpred1.Inf | SBLUP.Inf | 0.115 | 0.116 | 0.000 | 9.24e-01 | 0.191 | 0.193 | -0.002 | 6.92e-01 |
| ADHD | LDpred2.MultiPRS | SBLUP.Inf | 0.114 | 0.116 | -0.001 | 9.52e-01 | 0.199 | 0.193 | 0.005 | 2.77e-01 |
| ADHD | All.MultiPRS | SBLUP.Inf | 0.115 | 0.116 | 0.000 | 9.91e-01 | 0.186 | 0.193 | -0.007 | 5.81e-01 |
| ADHD | SBLUP.Inf | SBLUP.Inf | 0.116 | 0.116 | 0.000 | 1.00e+00 | 0.193 | 0.193 | 0.000 | 1.00e+00 |
| ADHD | DBSLMM.PseudoVal | SBLUP.Inf | 0.114 | 0.116 | -0.001 | 8.13e-01 | 0.184 | 0.193 | -0.010 | 4.07e-01 |
| ADHD | pT+clump.10FCVal | SBLUP.Inf | 0.094 | 0.116 | -0.022 | 5.10e-03 | 0.150 | 0.193 | -0.044 | 4.41e-03 |
| ADHD | lassosum.PseudoVal | SBLUP.Inf | 0.077 | 0.116 | -0.039 | 1.86e-03 | 0.118 | 0.193 | -0.075 | 2.13e-03 |
Show cross-validation differences between methods
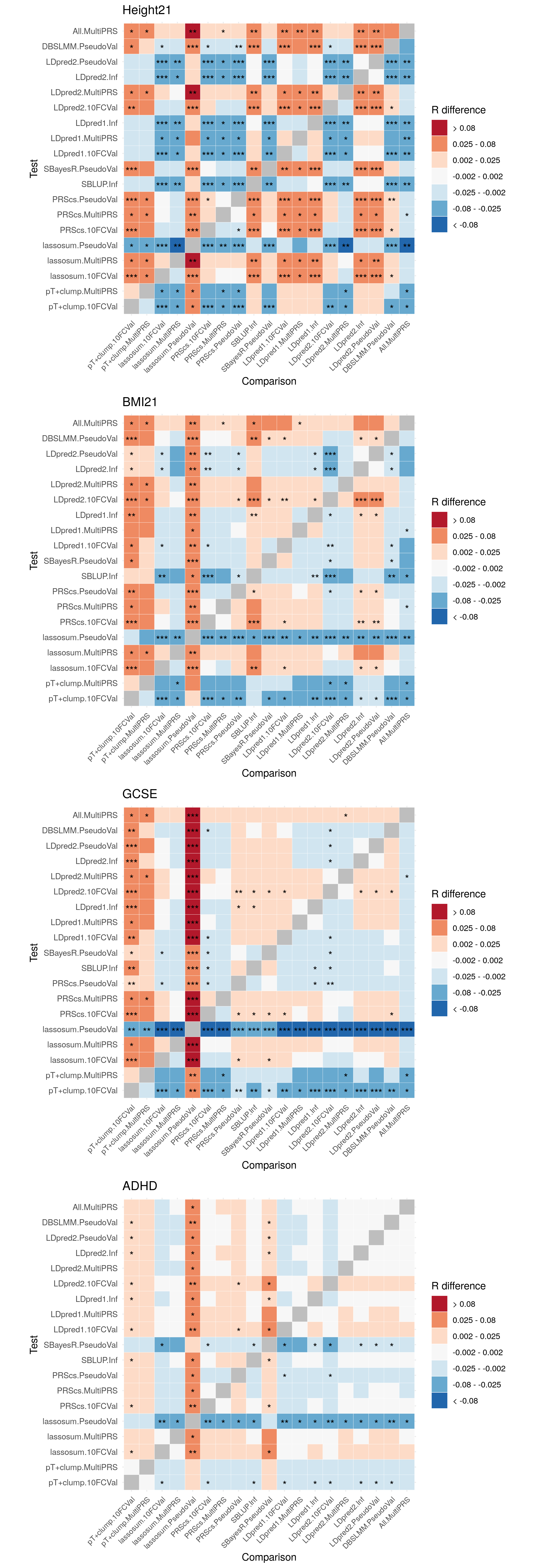
Predictive utility estimated using 10-fold cross-validation
Show test-validation differences between methods
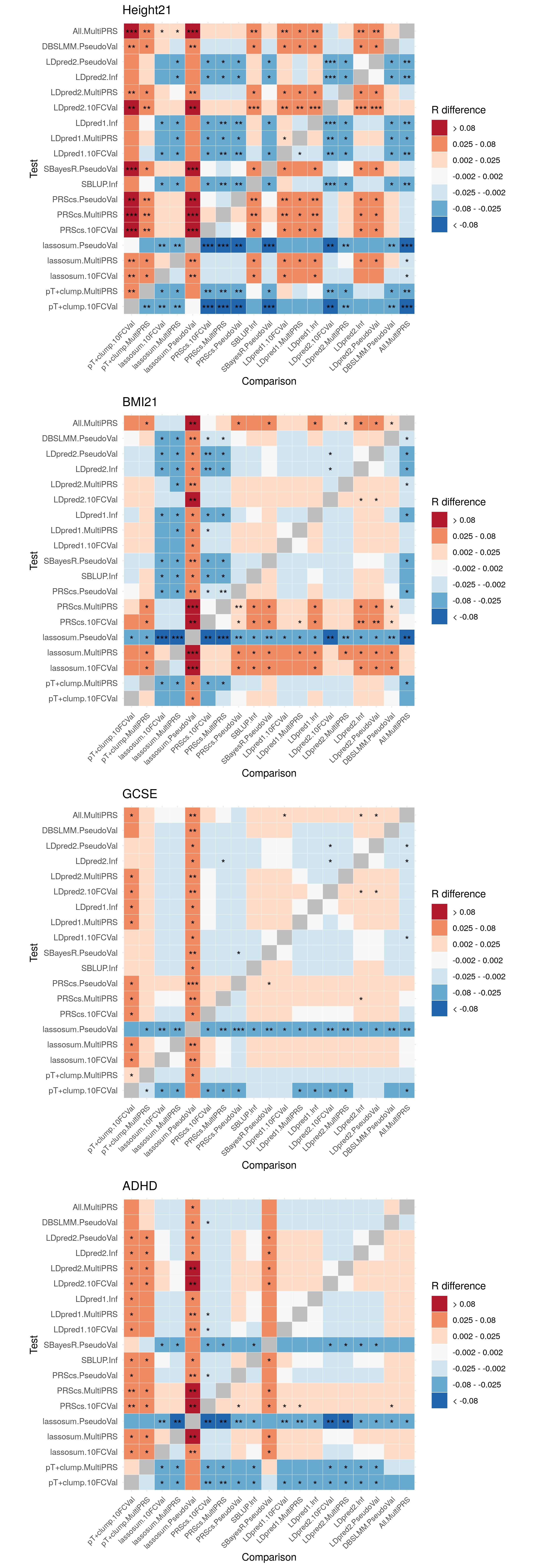
Predictive utility estimated using test set validation
4.2 Using UK Biobank as reference
4.2.1 UKB
Show cross-validation results
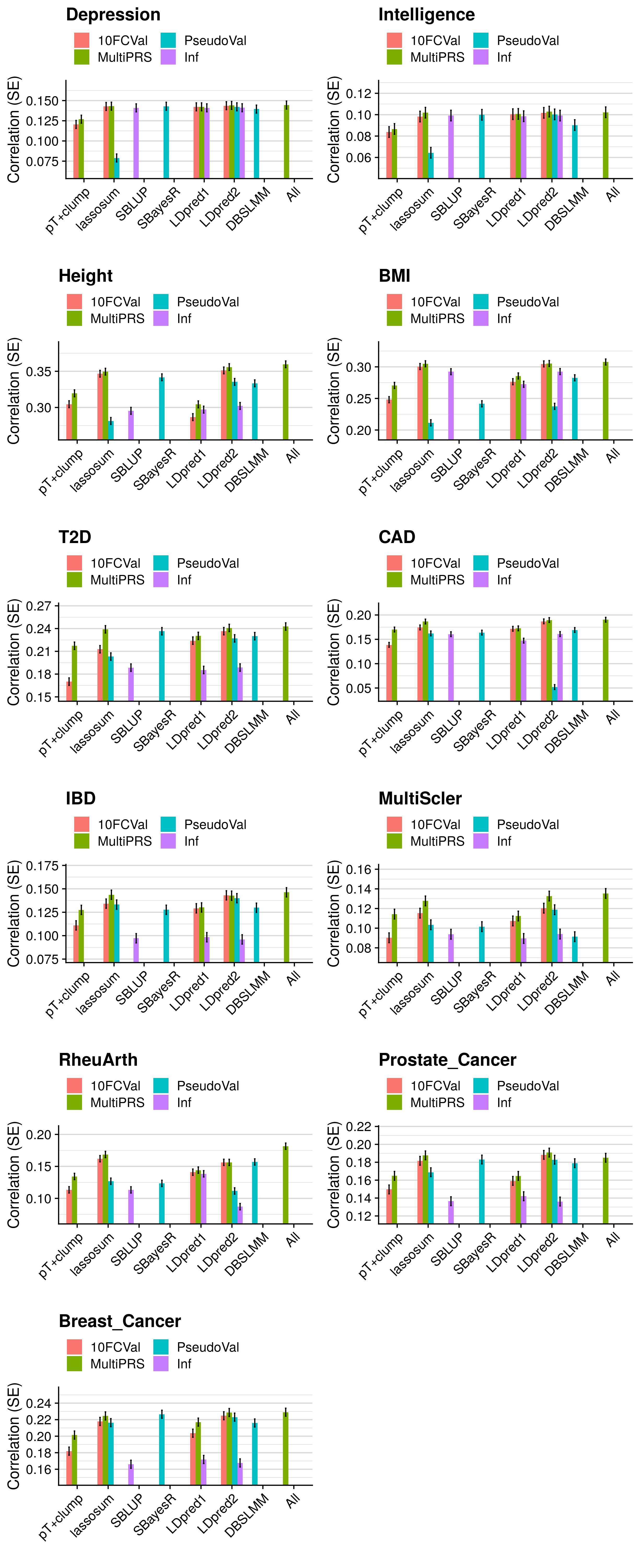
Predictive utility estimated using 10-fold cross-validation
Show test-validation results
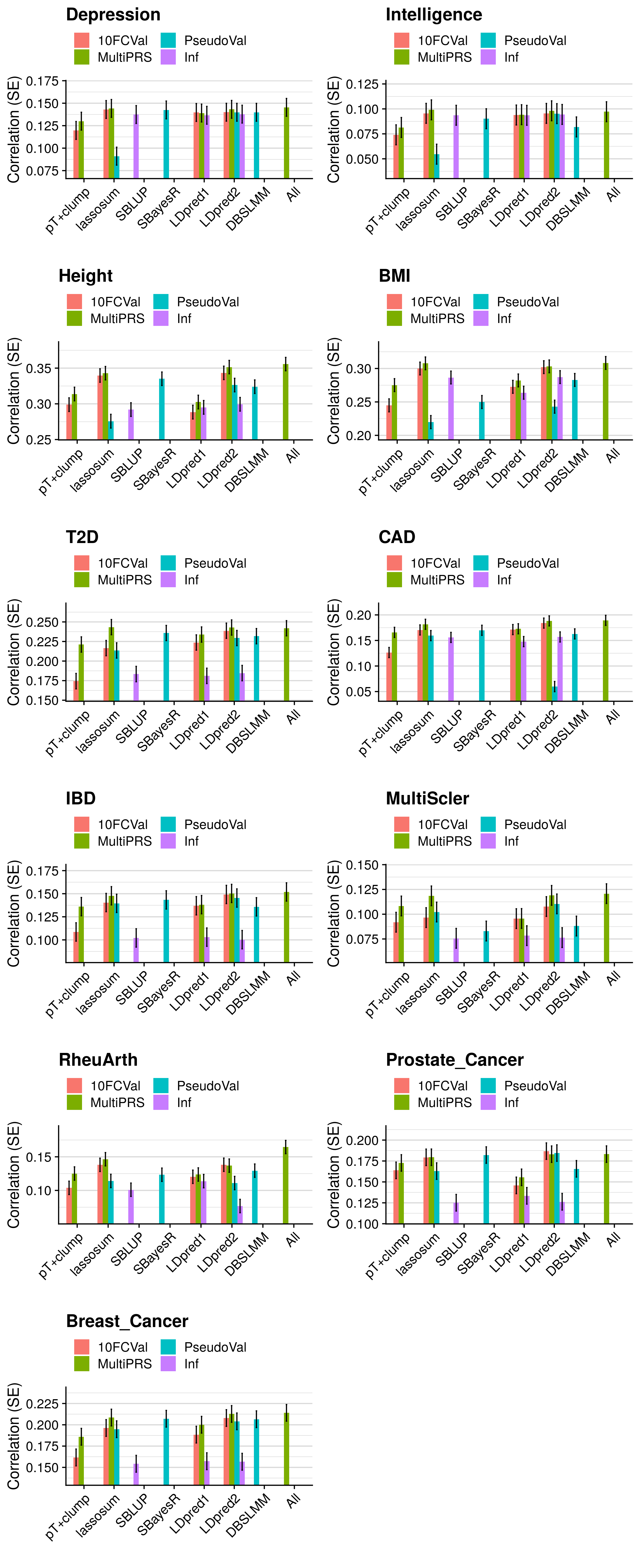
Predictive utility estimated using test set validation
Show UKBB results table
| Phenotype | Method | Model | CrossVal_R | CrossVal_R_SE | CrossVal_OR | CrossVal_LowCI | CrossVal_HighCI | Cross_LiabR2 | Cross_AUC | CrossVal_pval | IndepVal_R | IndepVal_R_SE | IndepVal_OR | IndepVal_LowCI | IndepVal_HighCI | Indep_LiabR2 | Indep_AUC | IndepVal_pval |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Depression | All | MultiPRS | 0.144 | 0.005 | 1.343 | 1.316 | 1.370 | 0.025 | 0.582 | 2.11e-185 | 0.145 | 0.010 | 1.346 | 1.292 | 1.401 | 0.025 | 0.582 | 2.29e-48 |
| Depression | DBSLMM | PseudoVal | 0.139 | 0.005 | 1.329 | 1.302 | 1.356 | 0.023 | 0.579 | 7.64e-173 | 0.140 | 0.010 | 1.330 | 1.277 | 1.385 | 0.024 | 0.579 | 7.03e-45 |
| Depression | lassosum | MultiPRS | 0.143 | 0.005 | 1.339 | 1.312 | 1.366 | 0.025 | 0.581 | 6.56e-182 | 0.144 | 0.010 | 1.342 | 1.289 | 1.398 | 0.025 | 0.581 | 1.32e-47 |
| Depression | lassosum | PseudoVal | 0.079 | 0.005 | 1.172 | 1.149 | 1.196 | 0.007 | 0.545 | 2.78e-56 | 0.091 | 0.010 | 1.202 | 1.155 | 1.250 | 0.010 | 0.551 | 6.95e-20 |
| Depression | lassosum | 10FCVal | 0.143 | 0.005 | 1.338 | 1.311 | 1.365 | 0.025 | 0.581 | 5.29e-181 | 0.143 | 0.010 | 1.339 | 1.286 | 1.394 | 0.025 | 0.580 | 7.56e-47 |
| Depression | LDpred1 | MultiPRS | 0.142 | 0.005 | 1.337 | 1.310 | 1.364 | 0.024 | 0.581 | 8.03e-180 | 0.139 | 0.010 | 1.328 | 1.275 | 1.382 | 0.023 | 0.578 | 2.40e-44 |
| Depression | LDpred1 | Inf | 0.141 | 0.005 | 1.333 | 1.306 | 1.360 | 0.024 | 0.580 | 9.03e-177 | 0.137 | 0.010 | 1.321 | 1.269 | 1.375 | 0.022 | 0.577 | 7.76e-43 |
| Depression | LDpred1 | 10FCVal | 0.142 | 0.005 | 1.336 | 1.310 | 1.364 | 0.024 | 0.581 | 7.71e-180 | 0.140 | 0.010 | 1.329 | 1.277 | 1.384 | 0.023 | 0.578 | 9.24e-45 |
| Depression | LDpred2 | MultiPRS | 0.144 | 0.005 | 1.341 | 1.315 | 1.369 | 0.025 | 0.582 | 2.76e-184 | 0.143 | 0.010 | 1.339 | 1.286 | 1.394 | 0.025 | 0.581 | 6.03e-47 |
| Depression | LDpred2 | 10FCVal | 0.144 | 0.005 | 1.340 | 1.314 | 1.368 | 0.025 | 0.581 | 2.49e-183 | 0.140 | 0.010 | 1.330 | 1.278 | 1.385 | 0.024 | 0.579 | 6.24e-45 |
| Depression | LDpred2 | PseudoVal | 0.142 | 0.005 | 1.337 | 1.310 | 1.364 | 0.024 | 0.581 | 6.41e-180 | 0.140 | 0.010 | 1.330 | 1.277 | 1.385 | 0.024 | 0.579 | 6.69e-45 |
| Depression | LDpred2 | Inf | 0.141 | 0.005 | 1.333 | 1.307 | 1.360 | 0.024 | 0.580 | 5.06e-177 | 0.138 | 0.010 | 1.324 | 1.272 | 1.379 | 0.023 | 0.578 | 1.29e-43 |
| Depression | pT+clump | MultiPRS | 0.127 | 0.005 | 1.294 | 1.268 | 1.320 | 0.019 | 0.571 | 2.65e-143 | 0.130 | 0.010 | 1.303 | 1.251 | 1.356 | 0.020 | 0.573 | 5.91e-39 |
| Depression | pT+clump | 10FCVal | 0.120 | 0.005 | 1.277 | 1.251 | 1.303 | 0.017 | 0.568 | 3.52e-129 | 0.120 | 0.010 | 1.275 | 1.225 | 1.327 | 0.017 | 0.566 | 2.98e-33 |
| Depression | SBayesR | PseudoVal | 0.143 | 0.005 | 1.339 | 1.312 | 1.366 | 0.025 | 0.581 | 1.10e-181 | 0.143 | 0.010 | 1.337 | 1.284 | 1.392 | 0.024 | 0.581 | 1.57e-46 |
| Depression | SBLUP | Inf | 0.141 | 0.005 | 1.333 | 1.306 | 1.360 | 0.024 | 0.580 | 1.02e-176 | 0.137 | 0.010 | 1.323 | 1.271 | 1.377 | 0.023 | 0.578 | 2.36e-43 |
| Intelligence | All | MultiPRS | 0.102 | 0.005 | NA | NA | NA | NA | NA | 2.03e-93 | 0.097 | 0.010 | NA | NA | NA | NA | NA | 2.05e-22 |
| Intelligence | DBSLMM | PseudoVal | 0.090 | 0.005 | NA | NA | NA | NA | NA | 3.39e-73 | 0.082 | 0.010 | NA | NA | NA | NA | NA | 2.24e-16 |
| Intelligence | lassosum | MultiPRS | 0.102 | 0.005 | NA | NA | NA | NA | NA | 9.52e-93 | 0.099 | 0.010 | NA | NA | NA | NA | NA | 3.12e-23 |
| Intelligence | lassosum | PseudoVal | 0.064 | 0.005 | NA | NA | NA | NA | NA | 5.12e-38 | 0.055 | 0.010 | NA | NA | NA | NA | NA | 4.54e-08 |
| Intelligence | lassosum | 10FCVal | 0.098 | 0.005 | NA | NA | NA | NA | NA | 2.01e-86 | 0.096 | 0.010 | NA | NA | NA | NA | NA | 1.00e-21 |
| Intelligence | LDpred1 | MultiPRS | 0.101 | 0.005 | NA | NA | NA | NA | NA | 1.22e-90 | 0.094 | 0.010 | NA | NA | NA | NA | NA | 3.67e-21 |
| Intelligence | LDpred1 | Inf | 0.099 | 0.005 | NA | NA | NA | NA | NA | 5.96e-87 | 0.094 | 0.010 | NA | NA | NA | NA | NA | 6.76e-21 |
| Intelligence | LDpred1 | 10FCVal | 0.100 | 0.005 | NA | NA | NA | NA | NA | 2.90e-90 | 0.094 | 0.010 | NA | NA | NA | NA | NA | 5.05e-21 |
| Intelligence | LDpred2 | MultiPRS | 0.103 | 0.005 | NA | NA | NA | NA | NA | 1.48e-94 | 0.098 | 0.010 | NA | NA | NA | NA | NA | 8.08e-23 |
| Intelligence | LDpred2 | 10FCVal | 0.102 | 0.005 | NA | NA | NA | NA | NA | 1.71e-92 | 0.095 | 0.010 | NA | NA | NA | NA | NA | 1.06e-21 |
| Intelligence | LDpred2 | PseudoVal | 0.100 | 0.005 | NA | NA | NA | NA | NA | 6.34e-90 | 0.095 | 0.010 | NA | NA | NA | NA | NA | 1.33e-21 |
| Intelligence | LDpred2 | Inf | 0.099 | 0.005 | NA | NA | NA | NA | NA | 6.74e-88 | 0.094 | 0.010 | NA | NA | NA | NA | NA | 2.95e-21 |
| Intelligence | pT+clump | MultiPRS | 0.087 | 0.005 | NA | NA | NA | NA | NA | 1.74e-67 | 0.081 | 0.010 | NA | NA | NA | NA | NA | 3.56e-16 |
| Intelligence | pT+clump | 10FCVal | 0.084 | 0.005 | NA | NA | NA | NA | NA | 3.02e-63 | 0.074 | 0.010 | NA | NA | NA | NA | NA | 1.23e-13 |
| Intelligence | SBayesR | PseudoVal | 0.100 | 0.005 | NA | NA | NA | NA | NA | 3.08e-89 | 0.090 | 0.010 | NA | NA | NA | NA | NA | 1.67e-19 |
| Intelligence | SBLUP | Inf | 0.099 | 0.005 | NA | NA | NA | NA | NA | 5.34e-88 | 0.094 | 0.010 | NA | NA | NA | NA | NA | 6.50e-21 |
| Height | All | MultiPRS | 0.360 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.356 | 0.009 | NA | NA | NA | NA | NA | 5.44e-296 |
| Height | DBSLMM | PseudoVal | 0.333 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.324 | 0.009 | NA | NA | NA | NA | NA | 4.24e-243 |
| Height | lassosum | MultiPRS | 0.349 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.343 | 0.009 | NA | NA | NA | NA | NA | 5.60e-274 |
| Height | lassosum | 10FCVal | 0.347 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.340 | 0.009 | NA | NA | NA | NA | NA | 1.76e-268 |
| Height | lassosum | PseudoVal | 0.281 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.276 | 0.010 | NA | NA | NA | NA | NA | 6.42e-174 |
| Height | LDpred1 | MultiPRS | 0.304 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.303 | 0.010 | NA | NA | NA | NA | NA | 1.07e-210 |
| Height | LDpred1 | Inf | 0.297 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.295 | 0.010 | NA | NA | NA | NA | NA | 4.55e-200 |
| Height | LDpred1 | 10FCVal | 0.287 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.288 | 0.010 | NA | NA | NA | NA | NA | 7.90e-191 |
| Height | LDpred2 | MultiPRS | 0.356 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.351 | 0.009 | NA | NA | NA | NA | NA | 1.32e-288 |
| Height | LDpred2 | 10FCVal | 0.351 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.343 | 0.009 | NA | NA | NA | NA | NA | 1.02e-274 |
| Height | LDpred2 | PseudoVal | 0.335 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.326 | 0.009 | NA | NA | NA | NA | NA | 9.62e-247 |
| Height | LDpred2 | Inf | 0.302 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.299 | 0.010 | NA | NA | NA | NA | NA | 3.51e-206 |
| Height | pT+clump | MultiPRS | 0.320 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.314 | 0.009 | NA | NA | NA | NA | NA | 2.69e-227 |
| Height | pT+clump | 10FCVal | 0.305 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.299 | 0.010 | NA | NA | NA | NA | NA | 2.15e-205 |
| Height | SBayesR | PseudoVal | 0.342 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.335 | 0.009 | NA | NA | NA | NA | NA | 5.79e-261 |
| Height | SBLUP | Inf | 0.295 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.292 | 0.010 | NA | NA | NA | NA | NA | 1.00e-195 |
| BMI | All | MultiPRS | 0.308 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.308 | 0.010 | NA | NA | NA | NA | NA | 3.71e-219 |
| BMI | DBSLMM | PseudoVal | 0.283 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.283 | 0.010 | NA | NA | NA | NA | NA | 2.24e-183 |
| BMI | lassosum | MultiPRS | 0.305 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.308 | 0.010 | NA | NA | NA | NA | NA | 3.16e-218 |
| BMI | lassosum | PseudoVal | 0.211 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.220 | 0.010 | NA | NA | NA | NA | NA | 1.02e-109 |
| BMI | lassosum | 10FCVal | 0.300 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.300 | 0.010 | NA | NA | NA | NA | NA | 4.96e-207 |
| BMI | LDpred1 | MultiPRS | 0.286 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.282 | 0.010 | NA | NA | NA | NA | NA | 2.36e-182 |
| BMI | LDpred1 | Inf | 0.272 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.264 | 0.010 | NA | NA | NA | NA | NA | 8.31e-159 |
| BMI | LDpred1 | 10FCVal | 0.277 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.273 | 0.010 | NA | NA | NA | NA | NA | 7.02e-170 |
| BMI | LDpred2 | MultiPRS | 0.305 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.303 | 0.010 | NA | NA | NA | NA | NA | 1.18e-211 |
| BMI | LDpred2 | 10FCVal | 0.305 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.302 | 0.010 | NA | NA | NA | NA | NA | 5.91e-210 |
| BMI | LDpred2 | PseudoVal | 0.237 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.243 | 0.010 | NA | NA | NA | NA | NA | 3.13e-134 |
| BMI | LDpred2 | Inf | 0.293 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.287 | 0.010 | NA | NA | NA | NA | NA | 4.32e-189 |
| BMI | pT+clump | MultiPRS | 0.271 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.275 | 0.010 | NA | NA | NA | NA | NA | 3.53e-173 |
| BMI | pT+clump | 10FCVal | 0.248 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.245 | 0.010 | NA | NA | NA | NA | NA | 1.90e-136 |
| BMI | SBayesR | PseudoVal | 0.241 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.250 | 0.010 | NA | NA | NA | NA | NA | 3.61e-142 |
| BMI | SBLUP | Inf | 0.292 | 0.005 | NA | NA | NA | NA | NA | 0.00e+00 | 0.287 | 0.010 | NA | NA | NA | NA | NA | 3.03e-188 |
| T2D | All | MultiPRS | 0.243 | 0.005 | 1.706 | 1.668 | 1.744 | 0.062 | 0.650 | 0.00e+00 | 0.242 | 0.010 | 1.695 | 1.622 | 1.772 | 0.061 | 0.648 | 3.82e-133 |
| T2D | DBSLMM | PseudoVal | 0.230 | 0.005 | 1.659 | 1.622 | 1.696 | 0.056 | 0.643 | 0.00e+00 | 0.232 | 0.010 | 1.659 | 1.587 | 1.734 | 0.055 | 0.642 | 4.45e-122 |
| T2D | lassosum | MultiPRS | 0.239 | 0.005 | 1.693 | 1.656 | 1.731 | 0.060 | 0.648 | 0.00e+00 | 0.243 | 0.010 | 1.702 | 1.629 | 1.780 | 0.061 | 0.649 | 9.58e-135 |
| T2D | lassosum | PseudoVal | 0.203 | 0.005 | 1.559 | 1.525 | 1.593 | 0.043 | 0.625 | 0.00e+00 | 0.214 | 0.010 | 1.590 | 1.522 | 1.661 | 0.047 | 0.630 | 1.73e-103 |
| T2D | lassosum | 10FCVal | 0.213 | 0.005 | 1.595 | 1.560 | 1.631 | 0.047 | 0.631 | 0.00e+00 | 0.217 | 0.010 | 1.604 | 1.535 | 1.676 | 0.048 | 0.633 | 1.82e-106 |
| T2D | LDpred1 | MultiPRS | 0.230 | 0.005 | 1.659 | 1.622 | 1.696 | 0.056 | 0.643 | 0.00e+00 | 0.234 | 0.010 | 1.666 | 1.594 | 1.742 | 0.057 | 0.643 | 2.56e-124 |
| T2D | LDpred1 | Inf | 0.185 | 0.005 | 1.502 | 1.469 | 1.535 | 0.036 | 0.616 | 2.97e-306 | 0.181 | 0.010 | 1.482 | 1.419 | 1.547 | 0.033 | 0.612 | 1.86e-74 |
| T2D | LDpred1 | 10FCVal | 0.224 | 0.005 | 1.636 | 1.600 | 1.673 | 0.053 | 0.639 | 0.00e+00 | 0.224 | 0.010 | 1.629 | 1.559 | 1.702 | 0.052 | 0.637 | 1.20e-113 |
| T2D | LDpred2 | MultiPRS | 0.241 | 0.005 | 1.700 | 1.662 | 1.738 | 0.061 | 0.649 | 0.00e+00 | 0.243 | 0.010 | 1.701 | 1.627 | 1.778 | 0.061 | 0.648 | 3.34e-134 |
| T2D | LDpred2 | 10FCVal | 0.237 | 0.005 | 1.682 | 1.645 | 1.720 | 0.059 | 0.646 | 0.00e+00 | 0.239 | 0.010 | 1.684 | 1.611 | 1.760 | 0.059 | 0.646 | 1.27e-129 |
| T2D | LDpred2 | PseudoVal | 0.227 | 0.005 | 1.647 | 1.611 | 1.684 | 0.054 | 0.640 | 0.00e+00 | 0.229 | 0.010 | 1.650 | 1.578 | 1.724 | 0.054 | 0.640 | 1.41e-119 |
| T2D | LDpred2 | Inf | 0.189 | 0.005 | 1.512 | 1.479 | 1.546 | 0.037 | 0.618 | 5.36e-317 | 0.185 | 0.010 | 1.493 | 1.430 | 1.559 | 0.035 | 0.614 | 2.06e-77 |
| T2D | pT+clump | MultiPRS | 0.217 | 0.005 | 1.613 | 1.578 | 1.649 | 0.050 | 0.635 | 0.00e+00 | 0.221 | 0.010 | 1.621 | 1.551 | 1.694 | 0.050 | 0.636 | 4.64e-111 |
| T2D | pT+clump | 10FCVal | 0.170 | 0.005 | 1.451 | 1.420 | 1.483 | 0.030 | 0.606 | 2.62e-257 | 0.174 | 0.010 | 1.460 | 1.398 | 1.525 | 0.031 | 0.608 | 4.30e-69 |
| T2D | SBayesR | PseudoVal | 0.236 | 0.005 | 1.683 | 1.646 | 1.721 | 0.059 | 0.647 | 0.00e+00 | 0.236 | 0.010 | 1.673 | 1.601 | 1.749 | 0.057 | 0.645 | 2.02e-126 |
| T2D | SBLUP | Inf | 0.189 | 0.005 | 1.512 | 1.479 | 1.545 | 0.037 | 0.617 | 1.26e-316 | 0.183 | 0.010 | 1.489 | 1.426 | 1.555 | 0.034 | 0.613 | 2.23e-76 |
| CAD | All | MultiPRS | 0.190 | 0.005 | 1.482 | 1.452 | 1.513 | 0.027 | 0.609 | 1.19e-322 | 0.189 | 0.010 | 1.480 | 1.420 | 1.542 | 0.027 | 0.607 | 2.06e-81 |
| CAD | DBSLMM | PseudoVal | 0.169 | 0.005 | 1.416 | 1.387 | 1.445 | 0.021 | 0.597 | 1.43e-254 | 0.163 | 0.010 | 1.396 | 1.341 | 1.454 | 0.020 | 0.592 | 2.21e-60 |
| CAD | lassosum | MultiPRS | 0.187 | 0.005 | 1.470 | 1.440 | 1.501 | 0.026 | 0.606 | 4.28e-310 | 0.182 | 0.010 | 1.455 | 1.396 | 1.516 | 0.025 | 0.602 | 6.27e-75 |
| CAD | lassosum | PseudoVal | 0.162 | 0.005 | 1.394 | 1.366 | 1.423 | 0.020 | 0.592 | 1.10e-233 | 0.160 | 0.010 | 1.386 | 1.331 | 1.444 | 0.019 | 0.589 | 5.44e-58 |
| CAD | lassosum | 10FCVal | 0.174 | 0.005 | 1.432 | 1.403 | 1.461 | 0.023 | 0.599 | 1.23e-270 | 0.171 | 0.010 | 1.420 | 1.363 | 1.479 | 0.022 | 0.595 | 3.71e-66 |
| CAD | LDpred1 | MultiPRS | 0.173 | 0.005 | 1.426 | 1.397 | 1.456 | 0.022 | 0.598 | 8.75e-265 | 0.173 | 0.010 | 1.427 | 1.370 | 1.487 | 0.022 | 0.597 | 6.01e-68 |
| CAD | LDpred1 | Inf | 0.147 | 0.005 | 1.351 | 1.324 | 1.379 | 0.016 | 0.585 | 7.18e-193 | 0.148 | 0.010 | 1.353 | 1.299 | 1.408 | 0.016 | 0.584 | 5.94e-50 |
| CAD | LDpred1 | 10FCVal | 0.172 | 0.005 | 1.423 | 1.394 | 1.453 | 0.022 | 0.598 | 4.09e-262 | 0.171 | 0.010 | 1.422 | 1.365 | 1.481 | 0.022 | 0.596 | 1.02e-66 |
| CAD | LDpred2 | MultiPRS | 0.190 | 0.005 | 1.480 | 1.450 | 1.511 | 0.027 | 0.608 | 1.32e-320 | 0.188 | 0.010 | 1.476 | 1.417 | 1.538 | 0.027 | 0.606 | 1.92e-80 |
| CAD | LDpred2 | 10FCVal | 0.187 | 0.005 | 1.471 | 1.441 | 1.502 | 0.026 | 0.607 | 4.04e-311 | 0.184 | 0.010 | 1.462 | 1.403 | 1.524 | 0.025 | 0.604 | 7.28e-77 |
| CAD | LDpred2 | PseudoVal | 0.052 | 0.005 | 1.110 | 1.088 | 1.132 | 0.002 | 0.528 | 2.57e-25 | 0.060 | 0.010 | 1.127 | 1.084 | 1.173 | 0.003 | 0.531 | 2.24e-09 |
| CAD | LDpred2 | Inf | 0.161 | 0.005 | 1.390 | 1.362 | 1.419 | 0.019 | 0.592 | 8.05e-230 | 0.157 | 0.010 | 1.379 | 1.324 | 1.436 | 0.018 | 0.590 | 4.12e-56 |
| CAD | pT+clump | MultiPRS | 0.170 | 0.005 | 1.419 | 1.390 | 1.448 | 0.022 | 0.597 | 2.05e-257 | 0.166 | 0.010 | 1.406 | 1.350 | 1.464 | 0.020 | 0.593 | 1.35e-62 |
| CAD | pT+clump | 10FCVal | 0.139 | 0.005 | 1.326 | 1.300 | 1.353 | 0.014 | 0.578 | 6.44e-171 | 0.126 | 0.010 | 1.293 | 1.242 | 1.346 | 0.012 | 0.570 | 6.73e-37 |
| CAD | SBayesR | PseudoVal | 0.164 | 0.005 | 1.399 | 1.371 | 1.428 | 0.020 | 0.593 | 1.28e-238 | 0.170 | 0.010 | 1.418 | 1.361 | 1.477 | 0.022 | 0.595 | 1.14e-65 |
| CAD | SBLUP | Inf | 0.161 | 0.005 | 1.390 | 1.362 | 1.418 | 0.019 | 0.592 | 2.92e-229 | 0.156 | 0.010 | 1.376 | 1.321 | 1.433 | 0.018 | 0.589 | 1.75e-55 |
| IBD | All | MultiPRS | 0.146 | 0.005 | 1.560 | 1.513 | 1.610 | 0.049 | 0.644 | 3.68e-190 | 0.152 | 0.010 | 1.586 | 1.489 | 1.690 | 0.054 | 0.653 | 1.10e-52 |
| IBD | DBSLMM | PseudoVal | 0.130 | 0.005 | 1.500 | 1.453 | 1.548 | 0.039 | 0.631 | 7.76e-150 | 0.136 | 0.010 | 1.534 | 1.439 | 1.635 | 0.043 | 0.636 | 2.29e-42 |
| IBD | lassosum | MultiPRS | 0.144 | 0.005 | 1.550 | 1.502 | 1.599 | 0.048 | 0.641 | 1.99e-183 | 0.148 | 0.010 | 1.573 | 1.477 | 1.676 | 0.051 | 0.650 | 6.17e-50 |
| IBD | lassosum | PseudoVal | 0.133 | 0.005 | 1.515 | 1.468 | 1.563 | 0.041 | 0.633 | 1.07e-157 | 0.140 | 0.010 | 1.542 | 1.447 | 1.644 | 0.045 | 0.643 | 1.14e-44 |
| IBD | lassosum | 10FCVal | 0.134 | 0.005 | 1.517 | 1.470 | 1.566 | 0.042 | 0.632 | 4.55e-160 | 0.140 | 0.010 | 1.546 | 1.451 | 1.648 | 0.046 | 0.644 | 3.17e-45 |
| IBD | LDpred1 | MultiPRS | 0.130 | 0.005 | 1.516 | 1.468 | 1.566 | 0.039 | 0.629 | 1.53e-150 | 0.138 | 0.010 | 1.555 | 1.457 | 1.658 | 0.044 | 0.639 | 8.27e-44 |
| IBD | LDpred1 | Inf | 0.098 | 0.005 | 1.400 | 1.353 | 1.448 | 0.022 | 0.599 | 1.74e-86 | 0.103 | 0.010 | 1.423 | 1.329 | 1.523 | 0.024 | 0.603 | 5.46e-25 |
| IBD | LDpred1 | 10FCVal | 0.129 | 0.005 | 1.502 | 1.455 | 1.551 | 0.038 | 0.628 | 1.94e-148 | 0.137 | 0.010 | 1.537 | 1.442 | 1.639 | 0.044 | 0.635 | 4.42e-43 |
| IBD | LDpred2 | MultiPRS | 0.143 | 0.005 | 1.550 | 1.502 | 1.599 | 0.047 | 0.642 | 6.23e-181 | 0.150 | 0.010 | 1.581 | 1.484 | 1.684 | 0.053 | 0.651 | 1.35e-51 |
| IBD | LDpred2 | 10FCVal | 0.143 | 0.005 | 1.549 | 1.501 | 1.598 | 0.047 | 0.642 | 8.36e-182 | 0.149 | 0.010 | 1.576 | 1.479 | 1.678 | 0.052 | 0.651 | 7.24e-51 |
| IBD | LDpred2 | PseudoVal | 0.140 | 0.005 | 1.539 | 1.491 | 1.588 | 0.045 | 0.640 | 3.57e-174 | 0.145 | 0.010 | 1.561 | 1.465 | 1.663 | 0.049 | 0.649 | 2.45e-48 |
| IBD | LDpred2 | Inf | 0.096 | 0.005 | 1.390 | 1.343 | 1.438 | 0.021 | 0.597 | 1.45e-82 | 0.100 | 0.010 | 1.414 | 1.320 | 1.515 | 0.023 | 0.601 | 9.32e-24 |
| IBD | pT+clump | MultiPRS | 0.127 | 0.005 | 1.503 | 1.455 | 1.552 | 0.037 | 0.630 | 1.42e-144 | 0.136 | 0.010 | 1.544 | 1.447 | 1.647 | 0.043 | 0.634 | 1.70e-42 |
| IBD | pT+clump | 10FCVal | 0.111 | 0.005 | 1.445 | 1.398 | 1.494 | 0.028 | 0.616 | 7.90e-110 | 0.109 | 0.010 | 1.438 | 1.346 | 1.537 | 0.027 | 0.606 | 1.29e-27 |
| IBD | SBayesR | PseudoVal | 0.128 | 0.005 | 1.487 | 1.441 | 1.535 | 0.038 | 0.629 | 7.55e-145 | 0.143 | 0.010 | 1.547 | 1.453 | 1.647 | 0.048 | 0.647 | 4.88e-47 |
| IBD | SBLUP | Inf | 0.097 | 0.005 | 1.394 | 1.347 | 1.442 | 0.022 | 0.598 | 1.32e-84 | 0.102 | 0.010 | 1.421 | 1.327 | 1.521 | 0.024 | 0.601 | 1.35e-24 |
| MultiScler | All | MultiPRS | 0.135 | 0.005 | 1.557 | 1.501 | 1.616 | 0.081 | 0.722 | 7.93e-163 | 0.121 | 0.010 | 1.506 | 1.396 | 1.624 | 0.060 | 0.699 | 8.44e-34 |
| MultiScler | DBSLMM | PseudoVal | 0.091 | 0.005 | 1.426 | 1.370 | 1.485 | 0.037 | 0.665 | 7.49e-75 | 0.088 | 0.010 | 1.408 | 1.300 | 1.525 | 0.032 | 0.637 | 1.12e-18 |
| MultiScler | lassosum | MultiPRS | 0.128 | 0.005 | 1.542 | 1.483 | 1.603 | 0.072 | 0.712 | 4.17e-145 | 0.119 | 0.010 | 1.509 | 1.398 | 1.628 | 0.057 | 0.690 | 1.29e-32 |
| MultiScler | lassosum | PseudoVal | 0.103 | 0.005 | 1.453 | 1.397 | 1.510 | 0.047 | 0.692 | 1.28e-95 | 0.102 | 0.010 | 1.442 | 1.335 | 1.558 | 0.043 | 0.674 | 1.23e-24 |
| MultiScler | lassosum | 10FCVal | 0.115 | 0.005 | 1.520 | 1.460 | 1.582 | 0.059 | 0.679 | 2.36e-118 | 0.097 | 0.010 | 1.451 | 1.339 | 1.572 | 0.038 | 0.667 | 3.71e-22 |
| MultiScler | LDpred1 | MultiPRS | 0.112 | 0.005 | 1.533 | 1.471 | 1.596 | 0.056 | 0.685 | 1.73e-112 | 0.096 | 0.010 | 1.455 | 1.342 | 1.577 | 0.037 | 0.664 | 8.91e-22 |
| MultiScler | LDpred1 | Inf | 0.090 | 0.005 | 1.484 | 1.417 | 1.553 | 0.035 | 0.647 | 5.95e-72 | 0.078 | 0.010 | 1.435 | 1.308 | 1.573 | 0.025 | 0.627 | 4.26e-15 |
| MultiScler | LDpred1 | 10FCVal | 0.107 | 0.005 | 1.495 | 1.436 | 1.557 | 0.051 | 0.681 | 7.79e-103 | 0.096 | 0.010 | 1.434 | 1.325 | 1.552 | 0.037 | 0.661 | 1.02e-21 |
| MultiScler | LDpred2 | MultiPRS | 0.133 | 0.005 | 1.549 | 1.492 | 1.607 | 0.078 | 0.721 | 1.36e-156 | 0.119 | 0.010 | 1.497 | 1.389 | 1.614 | 0.058 | 0.697 | 7.03e-33 |
| MultiScler | LDpred2 | 10FCVal | 0.120 | 0.005 | 1.501 | 1.446 | 1.558 | 0.064 | 0.713 | 6.85e-129 | 0.108 | 0.010 | 1.460 | 1.353 | 1.576 | 0.047 | 0.686 | 3.31e-27 |
| MultiScler | LDpred2 | PseudoVal | 0.119 | 0.005 | 1.518 | 1.461 | 1.577 | 0.062 | 0.714 | 1.46e-125 | 0.110 | 0.010 | 1.475 | 1.366 | 1.593 | 0.050 | 0.692 | 1.64e-28 |
| MultiScler | LDpred2 | Inf | 0.094 | 0.005 | 1.501 | 1.435 | 1.571 | 0.039 | 0.646 | 3.59e-79 | 0.076 | 0.010 | 1.426 | 1.300 | 1.565 | 0.024 | 0.632 | 2.04e-14 |
| MultiScler | pT+clump | MultiPRS | 0.114 | 0.005 | 1.509 | 1.450 | 1.570 | 0.058 | 0.695 | 1.83e-116 | 0.108 | 0.010 | 1.500 | 1.386 | 1.624 | 0.048 | 0.678 | 1.73e-27 |
| MultiScler | pT+clump | 10FCVal | 0.090 | 0.005 | 1.444 | 1.384 | 1.506 | 0.036 | 0.665 | 4.63e-73 | 0.092 | 0.010 | 1.459 | 1.342 | 1.587 | 0.034 | 0.630 | 3.25e-20 |
| MultiScler | SBayesR | PseudoVal | 0.102 | 0.005 | 1.510 | 1.447 | 1.575 | 0.046 | 0.675 | 3.72e-92 | 0.083 | 0.010 | 1.426 | 1.307 | 1.555 | 0.028 | 0.640 | 9.20e-17 |
| MultiScler | SBLUP | Inf | 0.094 | 0.005 | 1.501 | 1.435 | 1.570 | 0.039 | 0.646 | 7.41e-79 | 0.076 | 0.010 | 1.422 | 1.296 | 1.561 | 0.023 | 0.630 | 3.58e-14 |
| RheuArth | All | MultiPRS | 0.182 | 0.005 | 1.643 | 1.596 | 1.692 | 0.063 | 0.669 | 1.61e-293 | 0.165 | 0.010 | 1.577 | 1.487 | 1.672 | 0.052 | 0.649 | 1.18e-61 |
| RheuArth | DBSLMM | PseudoVal | 0.157 | 0.005 | 1.542 | 1.498 | 1.588 | 0.047 | 0.652 | 8.55e-219 | 0.129 | 0.010 | 1.454 | 1.370 | 1.542 | 0.032 | 0.613 | 1.33e-38 |
| RheuArth | lassosum | MultiPRS | 0.169 | 0.005 | 1.592 | 1.546 | 1.640 | 0.054 | 0.661 | 7.29e-253 | 0.146 | 0.010 | 1.516 | 1.428 | 1.609 | 0.041 | 0.637 | 4.89e-49 |
| RheuArth | lassosum | PseudoVal | 0.127 | 0.005 | 1.491 | 1.444 | 1.539 | 0.030 | 0.619 | 4.78e-143 | 0.114 | 0.010 | 1.431 | 1.341 | 1.526 | 0.025 | 0.614 | 2.54e-30 |
| RheuArth | lassosum | 10FCVal | 0.162 | 0.005 | 1.564 | 1.519 | 1.610 | 0.050 | 0.655 | 2.75e-233 | 0.138 | 0.010 | 1.481 | 1.396 | 1.571 | 0.036 | 0.630 | 9.72e-44 |
| RheuArth | LDpred1 | MultiPRS | 0.144 | 0.005 | 1.534 | 1.488 | 1.582 | 0.039 | 0.642 | 1.14e-184 | 0.124 | 0.010 | 1.455 | 1.368 | 1.548 | 0.029 | 0.618 | 2.20e-35 |
| RheuArth | LDpred1 | Inf | 0.138 | 0.005 | 1.519 | 1.472 | 1.566 | 0.036 | 0.636 | 3.18e-170 | 0.114 | 0.010 | 1.423 | 1.337 | 1.516 | 0.024 | 0.611 | 3.41e-30 |
| RheuArth | LDpred1 | 10FCVal | 0.141 | 0.005 | 1.518 | 1.472 | 1.565 | 0.038 | 0.639 | 1.68e-176 | 0.120 | 0.010 | 1.443 | 1.356 | 1.535 | 0.027 | 0.611 | 1.51e-33 |
| RheuArth | LDpred2 | MultiPRS | 0.156 | 0.005 | 1.582 | 1.534 | 1.630 | 0.046 | 0.649 | 7.20e-217 | 0.137 | 0.010 | 1.507 | 1.417 | 1.602 | 0.036 | 0.630 | 5.19e-43 |
| RheuArth | LDpred2 | 10FCVal | 0.156 | 0.005 | 1.580 | 1.533 | 1.629 | 0.046 | 0.650 | 3.81e-217 | 0.138 | 0.010 | 1.509 | 1.420 | 1.604 | 0.036 | 0.631 | 7.81e-44 |
| RheuArth | LDpred2 | PseudoVal | 0.112 | 0.005 | 1.429 | 1.384 | 1.476 | 0.023 | 0.620 | 5.47e-111 | 0.111 | 0.010 | 1.431 | 1.341 | 1.527 | 0.023 | 0.610 | 9.20e-29 |
| RheuArth | LDpred2 | Inf | 0.087 | 0.005 | 1.360 | 1.313 | 1.408 | 0.014 | 0.589 | 4.32e-68 | 0.077 | 0.010 | 1.314 | 1.225 | 1.409 | 0.011 | 0.580 | 1.50e-14 |
| RheuArth | pT+clump | MultiPRS | 0.134 | 0.005 | 1.509 | 1.462 | 1.557 | 0.034 | 0.635 | 3.26e-160 | 0.125 | 0.010 | 1.470 | 1.381 | 1.565 | 0.030 | 0.623 | 3.50e-36 |
| RheuArth | pT+clump | 10FCVal | 0.113 | 0.005 | 1.437 | 1.391 | 1.483 | 0.024 | 0.617 | 1.64e-114 | 0.104 | 0.010 | 1.396 | 1.309 | 1.489 | 0.020 | 0.610 | 2.25e-25 |
| RheuArth | SBayesR | PseudoVal | 0.123 | 0.005 | 1.486 | 1.439 | 1.535 | 0.029 | 0.624 | 1.51e-135 | 0.123 | 0.010 | 1.484 | 1.391 | 1.583 | 0.029 | 0.617 | 3.39e-35 |
| RheuArth | SBLUP | Inf | 0.113 | 0.005 | 1.453 | 1.405 | 1.501 | 0.024 | 0.615 | 2.59e-114 | 0.101 | 0.010 | 1.400 | 1.310 | 1.497 | 0.019 | 0.601 | 4.25e-24 |
| Prostate_Cancer | All | MultiPRS | 0.185 | 0.005 | 1.641 | 1.594 | 1.690 | 0.170 | 0.700 | 7.81e-305 | 0.183 | 0.010 | 1.637 | 1.543 | 1.738 | 0.171 | 0.703 | 3.27e-76 |
| Prostate_Cancer | DBSLMM | PseudoVal | 0.179 | 0.005 | 1.645 | 1.596 | 1.696 | 0.159 | 0.691 | 5.07e-285 | 0.166 | 0.010 | 1.592 | 1.498 | 1.691 | 0.141 | 0.693 | 1.90e-62 |
| Prostate_Cancer | lassosum | MultiPRS | 0.188 | 0.005 | 1.686 | 1.635 | 1.738 | 0.174 | 0.700 | 6.60e-314 | 0.179 | 0.010 | 1.642 | 1.547 | 1.744 | 0.165 | 0.704 | 3.84e-73 |
| Prostate_Cancer | lassosum | 10FCVal | 0.182 | 0.005 | 1.665 | 1.615 | 1.716 | 0.164 | 0.694 | 1.18e-293 | 0.179 | 0.010 | 1.639 | 1.544 | 1.740 | 0.164 | 0.703 | 4.29e-73 |
| Prostate_Cancer | lassosum | PseudoVal | 0.169 | 0.005 | 1.633 | 1.584 | 1.684 | 0.142 | 0.683 | 1.32e-253 | 0.163 | 0.010 | 1.601 | 1.506 | 1.703 | 0.136 | 0.686 | 1.88e-60 |
| Prostate_Cancer | LDpred1 | MultiPRS | 0.165 | 0.005 | 1.615 | 1.565 | 1.666 | 0.135 | 0.680 | 2.27e-241 | 0.155 | 0.010 | 1.584 | 1.488 | 1.686 | 0.124 | 0.683 | 3.85e-55 |
| Prostate_Cancer | LDpred1 | Inf | 0.142 | 0.005 | 1.552 | 1.503 | 1.602 | 0.101 | 0.658 | 2.43e-179 | 0.133 | 0.010 | 1.528 | 1.432 | 1.630 | 0.092 | 0.656 | 6.33e-41 |
| Prostate_Cancer | LDpred1 | 10FCVal | 0.159 | 0.005 | 1.590 | 1.542 | 1.640 | 0.126 | 0.675 | 3.36e-225 | 0.146 | 0.010 | 1.549 | 1.455 | 1.649 | 0.110 | 0.670 | 1.17e-48 |
| Prostate_Cancer | LDpred2 | MultiPRS | 0.191 | 0.005 | 1.669 | 1.620 | 1.720 | 0.180 | 0.704 | 0.00e+00 | 0.183 | 0.010 | 1.640 | 1.546 | 1.740 | 0.171 | 0.707 | 3.76e-76 |
| Prostate_Cancer | LDpred2 | 10FCVal | 0.188 | 0.005 | 1.667 | 1.618 | 1.718 | 0.175 | 0.703 | 8.17e-316 | 0.187 | 0.010 | 1.656 | 1.561 | 1.756 | 0.178 | 0.712 | 3.46e-79 |
| Prostate_Cancer | LDpred2 | PseudoVal | 0.183 | 0.005 | 1.656 | 1.606 | 1.707 | 0.166 | 0.698 | 1.24e-297 | 0.184 | 0.010 | 1.655 | 1.560 | 1.757 | 0.174 | 0.709 | 2.83e-77 |
| Prostate_Cancer | LDpred2 | Inf | 0.136 | 0.005 | 1.536 | 1.487 | 1.586 | 0.093 | 0.651 | 1.30e-164 | 0.126 | 0.010 | 1.504 | 1.408 | 1.606 | 0.083 | 0.654 | 7.89e-37 |
| Prostate_Cancer | pT+clump | MultiPRS | 0.165 | 0.005 | 1.625 | 1.574 | 1.676 | 0.135 | 0.681 | 1.10e-241 | 0.173 | 0.010 | 1.654 | 1.554 | 1.761 | 0.153 | 0.692 | 8.75e-68 |
| Prostate_Cancer | pT+clump | 10FCVal | 0.150 | 0.005 | 1.580 | 1.531 | 1.632 | 0.112 | 0.666 | 6.18e-199 | 0.164 | 0.010 | 1.632 | 1.530 | 1.740 | 0.138 | 0.671 | 4.43e-61 |
| Prostate_Cancer | SBayesR | PseudoVal | 0.183 | 0.005 | 1.646 | 1.598 | 1.697 | 0.166 | 0.700 | 2.48e-298 | 0.182 | 0.010 | 1.629 | 1.536 | 1.728 | 0.169 | 0.705 | 2.69e-75 |
| Prostate_Cancer | SBLUP | Inf | 0.137 | 0.005 | 1.538 | 1.489 | 1.588 | 0.093 | 0.651 | 1.27e-165 | 0.125 | 0.010 | 1.500 | 1.404 | 1.602 | 0.081 | 0.652 | 3.52e-36 |
| Breast_Cancer | All | MultiPRS | 0.229 | 0.005 | 1.722 | 1.681 | 1.765 | 0.107 | 0.667 | 0.00e+00 | 0.214 | 0.010 | 1.652 | 1.574 | 1.733 | 0.090 | 0.654 | 5.88e-104 |
| Breast_Cancer | DBSLMM | PseudoVal | 0.216 | 0.005 | 1.675 | 1.635 | 1.717 | 0.095 | 0.659 | 0.00e+00 | 0.207 | 0.010 | 1.629 | 1.552 | 1.710 | 0.083 | 0.647 | 8.47e-97 |
| Breast_Cancer | lassosum | MultiPRS | 0.225 | 0.005 | 1.707 | 1.666 | 1.749 | 0.102 | 0.664 | 0.00e+00 | 0.209 | 0.010 | 1.633 | 1.556 | 1.714 | 0.085 | 0.651 | 1.18e-98 |
| Breast_Cancer | lassosum | PseudoVal | 0.216 | 0.005 | 1.679 | 1.638 | 1.721 | 0.095 | 0.657 | 0.00e+00 | 0.195 | 0.010 | 1.585 | 1.511 | 1.663 | 0.074 | 0.642 | 4.05e-86 |
| Breast_Cancer | lassosum | 10FCVal | 0.218 | 0.005 | 1.683 | 1.642 | 1.725 | 0.096 | 0.659 | 0.00e+00 | 0.196 | 0.010 | 1.591 | 1.517 | 1.670 | 0.075 | 0.643 | 2.16e-87 |
| Breast_Cancer | LDpred1 | MultiPRS | 0.217 | 0.005 | 1.678 | 1.638 | 1.720 | 0.096 | 0.660 | 0.00e+00 | 0.200 | 0.010 | 1.606 | 1.530 | 1.685 | 0.078 | 0.644 | 8.04e-91 |
| Breast_Cancer | LDpred1 | Inf | 0.172 | 0.005 | 1.523 | 1.486 | 1.562 | 0.060 | 0.627 | 1.28e-262 | 0.157 | 0.010 | 1.464 | 1.394 | 1.537 | 0.048 | 0.612 | 2.27e-56 |
| Breast_Cancer | LDpred1 | 10FCVal | 0.204 | 0.005 | 1.630 | 1.590 | 1.670 | 0.084 | 0.650 | 0.00e+00 | 0.188 | 0.010 | 1.567 | 1.493 | 1.645 | 0.069 | 0.635 | 1.28e-80 |
| Breast_Cancer | LDpred2 | MultiPRS | 0.229 | 0.005 | 1.723 | 1.681 | 1.766 | 0.106 | 0.667 | 0.00e+00 | 0.213 | 0.010 | 1.648 | 1.570 | 1.729 | 0.089 | 0.654 | 1.02e-102 |
| Breast_Cancer | LDpred2 | 10FCVal | 0.225 | 0.005 | 1.706 | 1.665 | 1.749 | 0.103 | 0.664 | 0.00e+00 | 0.208 | 0.010 | 1.629 | 1.553 | 1.709 | 0.085 | 0.650 | 4.03e-98 |
| Breast_Cancer | LDpred2 | PseudoVal | 0.223 | 0.005 | 1.701 | 1.660 | 1.744 | 0.101 | 0.662 | 0.00e+00 | 0.204 | 0.010 | 1.615 | 1.539 | 1.694 | 0.081 | 0.647 | 1.83e-94 |
| Breast_Cancer | LDpred2 | Inf | 0.168 | 0.005 | 1.509 | 1.473 | 1.547 | 0.057 | 0.625 | 7.16e-250 | 0.157 | 0.010 | 1.462 | 1.393 | 1.535 | 0.048 | 0.614 | 6.11e-56 |
| Breast_Cancer | pT+clump | MultiPRS | 0.201 | 0.005 | 1.627 | 1.588 | 1.668 | 0.082 | 0.648 | 0.00e+00 | 0.186 | 0.010 | 1.559 | 1.485 | 1.636 | 0.068 | 0.634 | 1.72e-78 |
| Breast_Cancer | pT+clump | 10FCVal | 0.182 | 0.005 | 1.561 | 1.523 | 1.600 | 0.067 | 0.633 | 1.38e-294 | 0.162 | 0.010 | 1.480 | 1.410 | 1.554 | 0.051 | 0.620 | 1.58e-59 |
| Breast_Cancer | SBayesR | PseudoVal | 0.227 | 0.005 | 1.712 | 1.671 | 1.755 | 0.104 | 0.664 | 0.00e+00 | 0.207 | 0.010 | 1.628 | 1.551 | 1.708 | 0.084 | 0.649 | 2.54e-97 |
| Breast_Cancer | SBLUP | Inf | 0.166 | 0.005 | 1.504 | 1.468 | 1.542 | 0.056 | 0.623 | 4.63e-245 | 0.154 | 0.010 | 1.455 | 1.386 | 1.527 | 0.046 | 0.611 | 2.73e-54 |
Show cross-validation results compared to pT+clump
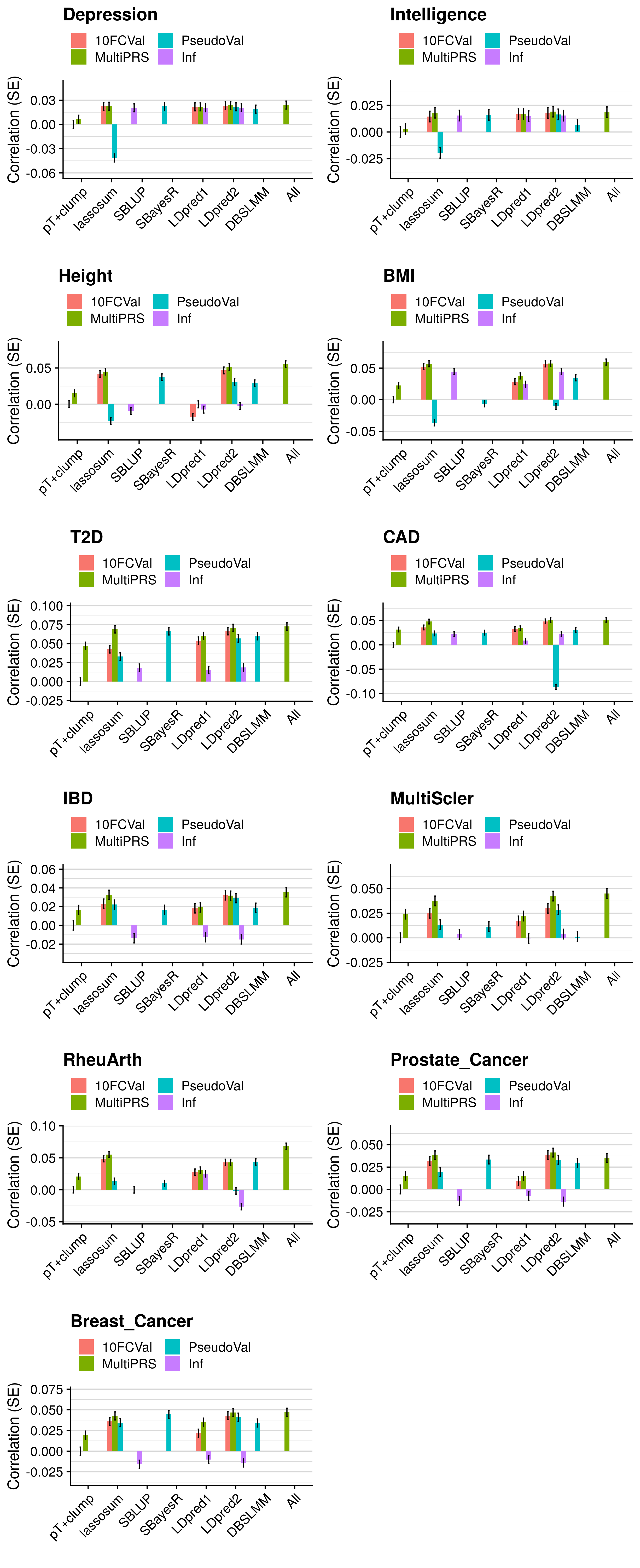
Predictive utility estimated using 10-fold cross-validation
Show test-validation results compared to pT+clump
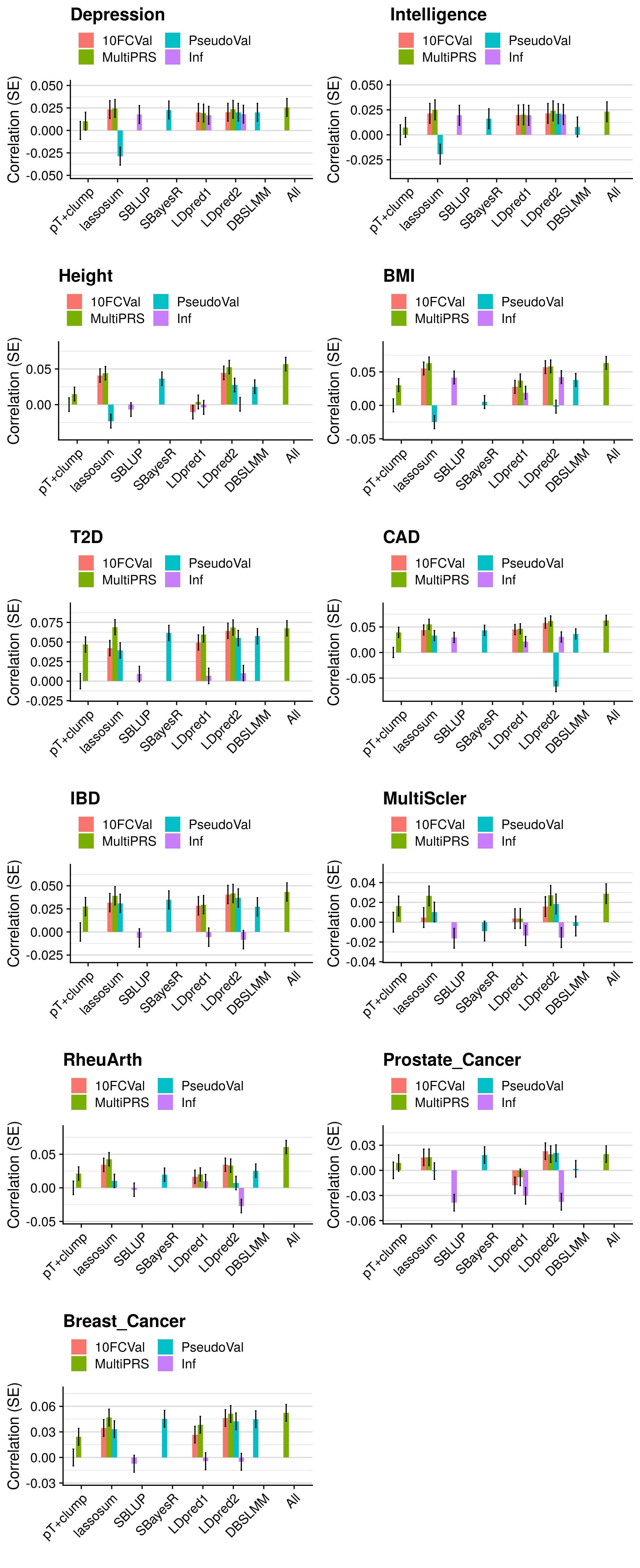
Predictive utility estimated using test set validation
Show difference results table
| Phenotype | Model 1 Test | Model 2 Test | Model 1 CrossVal R | Model 2 CrossVal R | CrossVal R Diff | CrossVal R Diff P | Model 1 IndepVal R | Model 2 IndepVal R | IndepVal R Diff | IndepVal R Diff P |
|---|---|---|---|---|---|---|---|---|---|---|
| Depression | lassosum.PseudoVal | All.MultiPRS | 0.079 | 0.144 | -0.066 | 7.03e-21 | 0.091 | 0.145 | -0.054 | 8.93e-09 |
| Depression | All.MultiPRS | All.MultiPRS | 0.144 | 0.144 | 0.000 | 1.00e+00 | 0.145 | 0.145 | 0.000 | 1.00e+00 |
| Depression | DBSLMM.PseudoVal | All.MultiPRS | 0.139 | 0.144 | -0.005 | 4.75e-01 | 0.140 | 0.145 | -0.005 | 3.89e-02 |
| Depression | LDpred1.Inf | All.MultiPRS | 0.141 | 0.144 | -0.003 | 6.25e-01 | 0.137 | 0.145 | -0.009 | 5.42e-04 |
| Depression | LDpred2.Inf | All.MultiPRS | 0.141 | 0.144 | -0.003 | 6.35e-01 | 0.138 | 0.145 | -0.008 | 1.49e-03 |
| Depression | lassosum.MultiPRS | All.MultiPRS | 0.143 | 0.144 | -0.001 | 9.44e-02 | 0.144 | 0.145 | -0.001 | 4.65e-01 |
| Depression | LDpred1.10FCVal | All.MultiPRS | 0.142 | 0.144 | -0.002 | 7.54e-01 | 0.140 | 0.145 | -0.006 | 4.36e-03 |
| Depression | pT+clump.MultiPRS | All.MultiPRS | 0.127 | 0.144 | -0.018 | 1.17e-02 | 0.130 | 0.145 | -0.015 | 1.43e-03 |
| Depression | LDpred2.MultiPRS | All.MultiPRS | 0.144 | 0.144 | 0.000 | 4.20e-01 | 0.143 | 0.145 | -0.002 | 4.33e-02 |
| Depression | pT+clump.10FCVal | All.MultiPRS | 0.120 | 0.144 | -0.024 | 6.04e-04 | 0.120 | 0.145 | -0.026 | 1.64e-05 |
| Depression | lassosum.10FCVal | All.MultiPRS | 0.143 | 0.144 | -0.002 | 8.04e-01 | 0.143 | 0.145 | -0.002 | 2.47e-01 |
| Depression | LDpred2.10FCVal | All.MultiPRS | 0.144 | 0.144 | -0.001 | 9.07e-01 | 0.140 | 0.145 | -0.005 | 1.69e-03 |
| Depression | SBayesR.PseudoVal | All.MultiPRS | 0.143 | 0.144 | -0.001 | 8.34e-01 | 0.143 | 0.145 | -0.003 | 1.19e-01 |
| Depression | LDpred1.MultiPRS | All.MultiPRS | 0.142 | 0.144 | -0.002 | 7.53e-01 | 0.139 | 0.145 | -0.006 | 3.04e-03 |
| Depression | LDpred2.PseudoVal | All.MultiPRS | 0.142 | 0.144 | -0.002 | 7.57e-01 | 0.140 | 0.145 | -0.005 | 4.81e-03 |
| Depression | SBLUP.Inf | All.MultiPRS | 0.141 | 0.144 | -0.003 | 6.23e-01 | 0.137 | 0.145 | -0.008 | 7.74e-04 |
| Depression | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.144 | 0.139 | 0.005 | 5.14e-01 | 0.143 | 0.140 | 0.003 | 2.27e-01 |
| Depression | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.120 | 0.139 | -0.019 | 1.66e-09 | 0.120 | 0.140 | -0.020 | 1.47e-03 |
| Depression | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.079 | 0.139 | -0.061 | 2.19e-36 | 0.091 | 0.140 | -0.049 | 3.04e-07 |
| Depression | All.MultiPRS | DBSLMM.PseudoVal | 0.144 | 0.139 | 0.005 | 4.75e-01 | 0.145 | 0.140 | 0.005 | 3.89e-02 |
| Depression | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.142 | 0.139 | 0.003 | 6.88e-01 | 0.139 | 0.140 | -0.001 | 7.61e-01 |
| Depression | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.142 | 0.139 | 0.003 | 3.89e-02 | 0.140 | 0.140 | 0.000 | 9.90e-01 |
| Depression | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.139 | 0.139 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| Depression | LDpred1.Inf | DBSLMM.PseudoVal | 0.141 | 0.139 | 0.002 | 2.82e-01 | 0.137 | 0.140 | -0.003 | 2.62e-01 |
| Depression | LDpred2.Inf | DBSLMM.PseudoVal | 0.141 | 0.139 | 0.002 | 2.41e-01 | 0.138 | 0.140 | -0.002 | 4.79e-01 |
| Depression | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.143 | 0.139 | 0.004 | 6.04e-01 | 0.144 | 0.140 | 0.004 | 1.98e-01 |
| Depression | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.142 | 0.139 | 0.003 | 4.34e-02 | 0.140 | 0.140 | 0.000 | 9.46e-01 |
| Depression | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.127 | 0.139 | -0.013 | 7.35e-02 | 0.130 | 0.140 | -0.010 | 7.09e-02 |
| Depression | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.143 | 0.139 | 0.004 | 2.39e-02 | 0.143 | 0.140 | 0.003 | 4.10e-01 |
| Depression | lassosum.10FCVal | DBSLMM.PseudoVal | 0.143 | 0.139 | 0.003 | 5.81e-02 | 0.143 | 0.140 | 0.003 | 3.77e-01 |
| Depression | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.144 | 0.139 | 0.004 | 2.50e-03 | 0.140 | 0.140 | 0.000 | 9.76e-01 |
| Depression | SBLUP.Inf | DBSLMM.PseudoVal | 0.141 | 0.139 | 0.002 | 2.51e-01 | 0.137 | 0.140 | -0.002 | 3.67e-01 |
| Depression | All.MultiPRS | lassosum.MultiPRS | 0.144 | 0.143 | 0.001 | 9.44e-02 | 0.145 | 0.144 | 0.001 | 4.65e-01 |
| Depression | LDpred1.MultiPRS | lassosum.MultiPRS | 0.142 | 0.143 | -0.001 | 9.06e-01 | 0.139 | 0.144 | -0.005 | 8.31e-02 |
| Depression | LDpred1.Inf | lassosum.MultiPRS | 0.141 | 0.143 | -0.002 | 7.70e-01 | 0.137 | 0.144 | -0.008 | 2.26e-02 |
| Depression | LDpred2.Inf | lassosum.MultiPRS | 0.141 | 0.143 | -0.002 | 7.81e-01 | 0.138 | 0.144 | -0.006 | 5.48e-02 |
| Depression | lassosum.MultiPRS | lassosum.MultiPRS | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.144 | 0.144 | 0.000 | 1.00e+00 |
| Depression | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.139 | 0.143 | -0.004 | 6.04e-01 | 0.140 | 0.144 | -0.004 | 1.98e-01 |
| Depression | lassosum.PseudoVal | lassosum.MultiPRS | 0.079 | 0.143 | -0.064 | 4.79e-20 | 0.091 | 0.144 | -0.053 | 2.11e-08 |
| Depression | LDpred2.MultiPRS | lassosum.MultiPRS | 0.144 | 0.143 | 0.001 | 4.58e-01 | 0.143 | 0.144 | -0.001 | 6.86e-01 |
| Depression | pT+clump.10FCVal | lassosum.MultiPRS | 0.120 | 0.143 | -0.023 | 1.22e-03 | 0.120 | 0.144 | -0.024 | 9.42e-05 |
| Depression | lassosum.10FCVal | lassosum.MultiPRS | 0.143 | 0.143 | 0.000 | 9.59e-01 | 0.143 | 0.144 | -0.001 | 3.04e-01 |
| Depression | LDpred2.10FCVal | lassosum.MultiPRS | 0.144 | 0.143 | 0.001 | 9.36e-01 | 0.140 | 0.144 | -0.004 | 1.40e-01 |
| Depression | SBayesR.PseudoVal | lassosum.MultiPRS | 0.143 | 0.143 | 0.000 | 9.90e-01 | 0.143 | 0.144 | -0.002 | 5.50e-01 |
| Depression | LDpred2.PseudoVal | lassosum.MultiPRS | 0.142 | 0.143 | -0.001 | 9.10e-01 | 0.140 | 0.144 | -0.004 | 1.48e-01 |
| Depression | SBLUP.Inf | lassosum.MultiPRS | 0.141 | 0.143 | -0.002 | 7.68e-01 | 0.137 | 0.144 | -0.007 | 3.29e-02 |
| Depression | pT+clump.MultiPRS | lassosum.MultiPRS | 0.127 | 0.143 | -0.016 | 2.02e-02 | 0.130 | 0.144 | -0.014 | 6.06e-03 |
| Depression | LDpred1.10FCVal | lassosum.MultiPRS | 0.142 | 0.143 | -0.001 | 9.07e-01 | 0.140 | 0.144 | -0.005 | 1.15e-01 |
| Depression | All.MultiPRS | lassosum.PseudoVal | 0.144 | 0.079 | 0.066 | 7.03e-21 | 0.145 | 0.091 | 0.054 | 8.93e-09 |
| Depression | LDpred1.MultiPRS | lassosum.PseudoVal | 0.142 | 0.079 | 0.063 | 1.28e-19 | 0.139 | 0.091 | 0.048 | 5.29e-06 |
| Depression | LDpred2.PseudoVal | lassosum.PseudoVal | 0.142 | 0.079 | 0.063 | 9.31e-32 | 0.140 | 0.091 | 0.049 | 4.87e-06 |
| Depression | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.139 | 0.079 | 0.061 | 2.19e-36 | 0.140 | 0.091 | 0.049 | 3.04e-07 |
| Depression | LDpred1.Inf | lassosum.PseudoVal | 0.141 | 0.079 | 0.062 | 4.87e-30 | 0.137 | 0.091 | 0.045 | 2.48e-05 |
| Depression | LDpred2.Inf | lassosum.PseudoVal | 0.141 | 0.079 | 0.062 | 2.32e-29 | 0.138 | 0.091 | 0.047 | 1.93e-05 |
| Depression | lassosum.MultiPRS | lassosum.PseudoVal | 0.143 | 0.079 | 0.064 | 4.79e-20 | 0.144 | 0.091 | 0.053 | 2.11e-08 |
| Depression | LDpred1.10FCVal | lassosum.PseudoVal | 0.142 | 0.079 | 0.063 | 3.14e-33 | 0.140 | 0.091 | 0.049 | 3.13e-06 |
| Depression | lassosum.PseudoVal | lassosum.PseudoVal | 0.079 | 0.079 | 0.000 | 1.00e+00 | 0.091 | 0.091 | 0.000 | 1.00e+00 |
| Depression | LDpred2.MultiPRS | lassosum.PseudoVal | 0.144 | 0.079 | 0.065 | 1.28e-20 | 0.143 | 0.091 | 0.052 | 4.31e-08 |
| Depression | pT+clump.10FCVal | lassosum.PseudoVal | 0.120 | 0.079 | 0.042 | 2.09e-12 | 0.120 | 0.091 | 0.029 | 1.52e-02 |
| Depression | lassosum.10FCVal | lassosum.PseudoVal | 0.143 | 0.079 | 0.064 | 3.03e-34 | 0.143 | 0.091 | 0.052 | 5.18e-07 |
| Depression | LDpred2.10FCVal | lassosum.PseudoVal | 0.144 | 0.079 | 0.065 | 1.06e-34 | 0.140 | 0.091 | 0.049 | 2.64e-06 |
| Depression | SBayesR.PseudoVal | lassosum.PseudoVal | 0.143 | 0.079 | 0.064 | 5.91e-40 | 0.143 | 0.091 | 0.051 | 7.65e-08 |
| Depression | SBLUP.Inf | lassosum.PseudoVal | 0.141 | 0.079 | 0.062 | 8.45e-30 | 0.137 | 0.091 | 0.046 | 1.92e-05 |
| Depression | pT+clump.MultiPRS | lassosum.PseudoVal | 0.127 | 0.079 | 0.048 | 7.44e-12 | 0.130 | 0.091 | 0.039 | 1.21e-04 |
| Depression | All.MultiPRS | lassosum.10FCVal | 0.144 | 0.143 | 0.002 | 8.04e-01 | 0.145 | 0.143 | 0.002 | 2.47e-01 |
| Depression | LDpred1.MultiPRS | lassosum.10FCVal | 0.142 | 0.143 | 0.000 | 9.46e-01 | 0.139 | 0.143 | -0.004 | 1.68e-01 |
| Depression | LDpred1.Inf | lassosum.10FCVal | 0.141 | 0.143 | -0.002 | 2.86e-01 | 0.137 | 0.143 | -0.006 | 4.44e-02 |
| Depression | LDpred2.Inf | lassosum.10FCVal | 0.141 | 0.143 | -0.002 | 2.90e-01 | 0.138 | 0.143 | -0.005 | 8.94e-02 |
| Depression | lassosum.MultiPRS | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 9.59e-01 | 0.144 | 0.143 | 0.001 | 3.04e-01 |
| Depression | DBSLMM.PseudoVal | lassosum.10FCVal | 0.139 | 0.143 | -0.003 | 5.81e-02 | 0.140 | 0.143 | -0.003 | 3.77e-01 |
| Depression | lassosum.PseudoVal | lassosum.10FCVal | 0.079 | 0.143 | -0.064 | 3.03e-34 | 0.091 | 0.143 | -0.052 | 5.18e-07 |
| Depression | LDpred2.MultiPRS | lassosum.10FCVal | 0.144 | 0.143 | 0.001 | 8.53e-01 | 0.143 | 0.143 | 0.000 | 9.58e-01 |
| Depression | pT+clump.10FCVal | lassosum.10FCVal | 0.120 | 0.143 | -0.022 | 6.57e-14 | 0.120 | 0.143 | -0.023 | 9.26e-05 |
| Depression | lassosum.10FCVal | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.143 | 0.143 | 0.000 | 1.00e+00 |
| Depression | LDpred2.10FCVal | lassosum.10FCVal | 0.144 | 0.143 | 0.001 | 5.15e-01 | 0.140 | 0.143 | -0.003 | 2.89e-01 |
| Depression | SBayesR.PseudoVal | lassosum.10FCVal | 0.143 | 0.143 | 0.000 | 8.61e-01 | 0.143 | 0.143 | 0.000 | 8.73e-01 |
| Depression | LDpred2.PseudoVal | lassosum.10FCVal | 0.142 | 0.143 | 0.000 | 7.51e-01 | 0.140 | 0.143 | -0.003 | 2.61e-01 |
| Depression | SBLUP.Inf | lassosum.10FCVal | 0.141 | 0.143 | -0.002 | 2.43e-01 | 0.137 | 0.143 | -0.006 | 6.04e-02 |
| Depression | pT+clump.MultiPRS | lassosum.10FCVal | 0.127 | 0.143 | -0.016 | 2.40e-02 | 0.130 | 0.143 | -0.013 | 1.29e-02 |
| Depression | LDpred1.10FCVal | lassosum.10FCVal | 0.142 | 0.143 | 0.000 | 7.35e-01 | 0.140 | 0.143 | -0.003 | 2.33e-01 |
| Depression | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.139 | 0.142 | -0.003 | 6.88e-01 | 0.140 | 0.139 | 0.001 | 7.61e-01 |
| Depression | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.144 | 0.142 | 0.002 | 8.01e-01 | 0.143 | 0.139 | 0.004 | 4.38e-02 |
| Depression | pT+clump.10FCVal | LDpred1.MultiPRS | 0.120 | 0.142 | -0.022 | 1.76e-03 | 0.120 | 0.139 | -0.019 | 1.36e-03 |
| Depression | lassosum.10FCVal | LDpred1.MultiPRS | 0.143 | 0.142 | 0.000 | 9.46e-01 | 0.143 | 0.139 | 0.004 | 1.68e-01 |
| Depression | lassosum.PseudoVal | LDpred1.MultiPRS | 0.079 | 0.142 | -0.063 | 1.28e-19 | 0.091 | 0.139 | -0.048 | 5.29e-06 |
| Depression | All.MultiPRS | LDpred1.MultiPRS | 0.144 | 0.142 | 0.002 | 7.53e-01 | 0.145 | 0.139 | 0.006 | 3.04e-03 |
| Depression | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.142 | 0.142 | 0.000 | 1.00e+00 | 0.139 | 0.139 | 0.000 | 1.00e+00 |
| Depression | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.142 | 0.142 | 0.000 | 9.96e-01 | 0.140 | 0.139 | 0.001 | 5.40e-01 |
| Depression | SBLUP.Inf | LDpred1.MultiPRS | 0.141 | 0.142 | -0.001 | 8.59e-01 | 0.137 | 0.139 | -0.002 | 2.66e-01 |
| Depression | LDpred1.Inf | LDpred1.MultiPRS | 0.141 | 0.142 | -0.001 | 8.61e-01 | 0.137 | 0.139 | -0.002 | 5.76e-02 |
| Depression | LDpred2.Inf | LDpred1.MultiPRS | 0.141 | 0.142 | -0.001 | 8.73e-01 | 0.138 | 0.139 | -0.001 | 4.58e-01 |
| Depression | lassosum.MultiPRS | LDpred1.MultiPRS | 0.143 | 0.142 | 0.001 | 9.06e-01 | 0.144 | 0.139 | 0.005 | 8.31e-02 |
| Depression | LDpred1.10FCVal | LDpred1.MultiPRS | 0.142 | 0.142 | 0.000 | 9.99e-01 | 0.140 | 0.139 | 0.001 | 4.73e-01 |
| Depression | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.127 | 0.142 | -0.015 | 2.79e-02 | 0.130 | 0.139 | -0.009 | 9.53e-02 |
| Depression | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.143 | 0.142 | 0.001 | 9.16e-01 | 0.143 | 0.139 | 0.003 | 1.55e-01 |
| Depression | LDpred2.10FCVal | LDpred1.MultiPRS | 0.144 | 0.142 | 0.001 | 8.42e-01 | 0.140 | 0.139 | 0.001 | 5.51e-01 |
| Depression | All.MultiPRS | LDpred1.Inf | 0.144 | 0.141 | 0.003 | 6.25e-01 | 0.145 | 0.137 | 0.009 | 5.42e-04 |
| Depression | LDpred1.Inf | LDpred1.Inf | 0.141 | 0.141 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| Depression | LDpred2.Inf | LDpred1.Inf | 0.141 | 0.141 | 0.000 | 8.86e-01 | 0.138 | 0.137 | 0.001 | 3.68e-01 |
| Depression | DBSLMM.PseudoVal | LDpred1.Inf | 0.139 | 0.141 | -0.002 | 2.82e-01 | 0.140 | 0.137 | 0.003 | 2.62e-01 |
| Depression | lassosum.PseudoVal | LDpred1.Inf | 0.079 | 0.141 | -0.062 | 4.87e-30 | 0.091 | 0.137 | -0.045 | 2.48e-05 |
| Depression | LDpred2.MultiPRS | LDpred1.Inf | 0.144 | 0.141 | 0.003 | 6.70e-01 | 0.143 | 0.137 | 0.007 | 5.34e-03 |
| Depression | pT+clump.10FCVal | LDpred1.Inf | 0.120 | 0.141 | -0.021 | 9.10e-12 | 0.120 | 0.137 | -0.017 | 5.54e-03 |
| Depression | lassosum.10FCVal | LDpred1.Inf | 0.143 | 0.141 | 0.002 | 2.86e-01 | 0.143 | 0.137 | 0.006 | 4.44e-02 |
| Depression | LDpred2.10FCVal | LDpred1.Inf | 0.144 | 0.141 | 0.003 | 1.27e-03 | 0.140 | 0.137 | 0.003 | 3.73e-02 |
| Depression | SBayesR.PseudoVal | LDpred1.Inf | 0.143 | 0.141 | 0.002 | 1.78e-01 | 0.143 | 0.137 | 0.006 | 3.98e-02 |
| Depression | LDpred1.MultiPRS | LDpred1.Inf | 0.142 | 0.141 | 0.001 | 8.61e-01 | 0.139 | 0.137 | 0.002 | 5.76e-02 |
| Depression | LDpred2.PseudoVal | LDpred1.Inf | 0.142 | 0.141 | 0.001 | 9.68e-02 | 0.140 | 0.137 | 0.003 | 2.78e-02 |
| Depression | SBLUP.Inf | LDpred1.Inf | 0.141 | 0.141 | 0.000 | 9.73e-01 | 0.137 | 0.137 | 0.001 | 5.14e-01 |
| Depression | pT+clump.MultiPRS | LDpred1.Inf | 0.127 | 0.141 | -0.014 | 4.37e-02 | 0.130 | 0.137 | -0.007 | 2.44e-01 |
| Depression | lassosum.MultiPRS | LDpred1.Inf | 0.143 | 0.141 | 0.002 | 7.70e-01 | 0.144 | 0.137 | 0.008 | 2.26e-02 |
| Depression | LDpred1.10FCVal | LDpred1.Inf | 0.142 | 0.141 | 0.001 | 1.07e-01 | 0.140 | 0.137 | 0.003 | 4.00e-02 |
| Depression | LDpred1.Inf | LDpred1.10FCVal | 0.141 | 0.142 | -0.001 | 1.07e-01 | 0.137 | 0.140 | -0.003 | 4.00e-02 |
| Depression | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.139 | 0.142 | -0.003 | 4.34e-02 | 0.140 | 0.140 | 0.000 | 9.46e-01 |
| Depression | lassosum.PseudoVal | LDpred1.10FCVal | 0.079 | 0.142 | -0.063 | 3.14e-33 | 0.091 | 0.140 | -0.049 | 3.13e-06 |
| Depression | LDpred2.MultiPRS | LDpred1.10FCVal | 0.144 | 0.142 | 0.002 | 8.02e-01 | 0.143 | 0.140 | 0.003 | 7.03e-02 |
| Depression | pT+clump.10FCVal | LDpred1.10FCVal | 0.120 | 0.142 | -0.022 | 3.04e-13 | 0.120 | 0.140 | -0.020 | 8.98e-04 |
| Depression | lassosum.10FCVal | LDpred1.10FCVal | 0.143 | 0.142 | 0.000 | 7.35e-01 | 0.143 | 0.140 | 0.003 | 2.33e-01 |
| Depression | LDpred2.10FCVal | LDpred1.10FCVal | 0.144 | 0.142 | 0.001 | 6.98e-02 | 0.140 | 0.140 | 0.000 | 8.60e-01 |
| Depression | All.MultiPRS | LDpred1.10FCVal | 0.144 | 0.142 | 0.002 | 7.54e-01 | 0.145 | 0.140 | 0.006 | 4.36e-03 |
| Depression | LDpred1.MultiPRS | LDpred1.10FCVal | 0.142 | 0.142 | 0.000 | 9.99e-01 | 0.139 | 0.140 | -0.001 | 4.73e-01 |
| Depression | LDpred2.PseudoVal | LDpred1.10FCVal | 0.142 | 0.142 | 0.000 | 9.64e-01 | 0.140 | 0.140 | 0.000 | 8.75e-01 |
| Depression | SBLUP.Inf | LDpred1.10FCVal | 0.141 | 0.142 | -0.001 | 1.08e-01 | 0.137 | 0.140 | -0.002 | 1.43e-01 |
| Depression | pT+clump.MultiPRS | LDpred1.10FCVal | 0.127 | 0.142 | -0.015 | 2.84e-02 | 0.130 | 0.140 | -0.010 | 6.95e-02 |
| Depression | LDpred2.Inf | LDpred1.10FCVal | 0.141 | 0.142 | -0.001 | 1.74e-01 | 0.138 | 0.140 | -0.002 | 2.66e-01 |
| Depression | lassosum.MultiPRS | LDpred1.10FCVal | 0.143 | 0.142 | 0.001 | 9.07e-01 | 0.144 | 0.140 | 0.005 | 1.15e-01 |
| Depression | LDpred1.10FCVal | LDpred1.10FCVal | 0.142 | 0.142 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| Depression | SBayesR.PseudoVal | LDpred1.10FCVal | 0.143 | 0.142 | 0.001 | 5.10e-01 | 0.143 | 0.140 | 0.003 | 2.09e-01 |
| Depression | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.144 | 0.144 | 0.000 | 1.00e+00 | 0.143 | 0.143 | 0.000 | 1.00e+00 |
| Depression | lassosum.PseudoVal | LDpred2.MultiPRS | 0.079 | 0.144 | -0.065 | 1.28e-20 | 0.091 | 0.143 | -0.052 | 4.31e-08 |
| Depression | All.MultiPRS | LDpred2.MultiPRS | 0.144 | 0.144 | 0.000 | 4.20e-01 | 0.145 | 0.143 | 0.002 | 4.33e-02 |
| Depression | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.142 | 0.144 | -0.002 | 8.01e-01 | 0.139 | 0.143 | -0.004 | 4.38e-02 |
| Depression | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.139 | 0.144 | -0.005 | 5.14e-01 | 0.140 | 0.143 | -0.003 | 2.27e-01 |
| Depression | LDpred1.Inf | LDpred2.MultiPRS | 0.141 | 0.144 | -0.003 | 6.70e-01 | 0.137 | 0.143 | -0.007 | 5.34e-03 |
| Depression | LDpred2.Inf | LDpred2.MultiPRS | 0.141 | 0.144 | -0.003 | 6.81e-01 | 0.138 | 0.143 | -0.005 | 1.20e-02 |
| Depression | lassosum.MultiPRS | LDpred2.MultiPRS | 0.143 | 0.144 | -0.001 | 4.58e-01 | 0.144 | 0.143 | 0.001 | 6.86e-01 |
| Depression | LDpred1.10FCVal | LDpred2.MultiPRS | 0.142 | 0.144 | -0.002 | 8.02e-01 | 0.140 | 0.143 | -0.003 | 7.03e-02 |
| Depression | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.127 | 0.144 | -0.017 | 1.40e-02 | 0.130 | 0.143 | -0.013 | 1.36e-02 |
| Depression | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.143 | 0.144 | -0.001 | 8.84e-01 | 0.143 | 0.143 | -0.001 | 7.11e-01 |
| Depression | pT+clump.10FCVal | LDpred2.MultiPRS | 0.120 | 0.144 | -0.024 | 7.60e-04 | 0.120 | 0.143 | -0.023 | 2.53e-04 |
| Depression | lassosum.10FCVal | LDpred2.MultiPRS | 0.143 | 0.144 | -0.001 | 8.53e-01 | 0.143 | 0.143 | 0.000 | 9.58e-01 |
| Depression | LDpred2.10FCVal | LDpred2.MultiPRS | 0.144 | 0.144 | 0.000 | 9.57e-01 | 0.140 | 0.143 | -0.003 | 1.08e-02 |
| Depression | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.142 | 0.144 | -0.002 | 8.05e-01 | 0.140 | 0.143 | -0.003 | 5.72e-02 |
| Depression | SBLUP.Inf | LDpred2.MultiPRS | 0.141 | 0.144 | -0.003 | 6.68e-01 | 0.137 | 0.143 | -0.006 | 9.31e-03 |
| Depression | LDpred2.MultiPRS | LDpred2.10FCVal | 0.144 | 0.144 | 0.000 | 9.57e-01 | 0.143 | 0.140 | 0.003 | 1.08e-02 |
| Depression | pT+clump.10FCVal | LDpred2.10FCVal | 0.120 | 0.144 | -0.023 | 5.02e-14 | 0.120 | 0.140 | -0.020 | 1.11e-03 |
| Depression | lassosum.PseudoVal | LDpred2.10FCVal | 0.079 | 0.144 | -0.065 | 1.06e-34 | 0.091 | 0.140 | -0.049 | 2.64e-06 |
| Depression | All.MultiPRS | LDpred2.10FCVal | 0.144 | 0.144 | 0.001 | 9.07e-01 | 0.145 | 0.140 | 0.005 | 1.69e-03 |
| Depression | LDpred1.MultiPRS | LDpred2.10FCVal | 0.142 | 0.144 | -0.001 | 8.42e-01 | 0.139 | 0.140 | -0.001 | 5.51e-01 |
| Depression | LDpred2.PseudoVal | LDpred2.10FCVal | 0.142 | 0.144 | -0.001 | 1.76e-03 | 0.140 | 0.140 | 0.000 | 9.56e-01 |
| Depression | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.139 | 0.144 | -0.004 | 2.50e-03 | 0.140 | 0.140 | 0.000 | 9.76e-01 |
| Depression | LDpred1.Inf | LDpred2.10FCVal | 0.141 | 0.144 | -0.003 | 1.27e-03 | 0.137 | 0.140 | -0.003 | 3.73e-02 |
| Depression | LDpred2.Inf | LDpred2.10FCVal | 0.141 | 0.144 | -0.002 | 6.49e-06 | 0.138 | 0.140 | -0.002 | 5.88e-02 |
| Depression | lassosum.MultiPRS | LDpred2.10FCVal | 0.143 | 0.144 | -0.001 | 9.36e-01 | 0.144 | 0.140 | 0.004 | 1.40e-01 |
| Depression | LDpred1.10FCVal | LDpred2.10FCVal | 0.142 | 0.144 | -0.001 | 6.98e-02 | 0.140 | 0.140 | 0.000 | 8.60e-01 |
| Depression | pT+clump.MultiPRS | LDpred2.10FCVal | 0.127 | 0.144 | -0.017 | 1.69e-02 | 0.130 | 0.140 | -0.010 | 7.15e-02 |
| Depression | SBayesR.PseudoVal | LDpred2.10FCVal | 0.143 | 0.144 | -0.001 | 5.79e-01 | 0.143 | 0.140 | 0.003 | 2.78e-01 |
| Depression | lassosum.10FCVal | LDpred2.10FCVal | 0.143 | 0.144 | -0.001 | 5.15e-01 | 0.143 | 0.140 | 0.003 | 2.89e-01 |
| Depression | LDpred2.10FCVal | LDpred2.10FCVal | 0.144 | 0.144 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| Depression | SBLUP.Inf | LDpred2.10FCVal | 0.141 | 0.144 | -0.003 | 5.14e-05 | 0.137 | 0.140 | -0.003 | 5.09e-02 |
| Depression | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.144 | 0.142 | 0.002 | 8.05e-01 | 0.143 | 0.140 | 0.003 | 5.72e-02 |
| Depression | lassosum.PseudoVal | LDpred2.PseudoVal | 0.079 | 0.142 | -0.063 | 9.31e-32 | 0.091 | 0.140 | -0.049 | 4.87e-06 |
| Depression | All.MultiPRS | LDpred2.PseudoVal | 0.144 | 0.142 | 0.002 | 7.57e-01 | 0.145 | 0.140 | 0.005 | 4.81e-03 |
| Depression | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.142 | 0.142 | 0.000 | 9.96e-01 | 0.139 | 0.140 | -0.001 | 5.40e-01 |
| Depression | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.139 | 0.142 | -0.003 | 3.89e-02 | 0.140 | 0.140 | 0.000 | 9.90e-01 |
| Depression | LDpred1.Inf | LDpred2.PseudoVal | 0.141 | 0.142 | -0.001 | 9.68e-02 | 0.137 | 0.140 | -0.003 | 2.78e-02 |
| Depression | LDpred2.Inf | LDpred2.PseudoVal | 0.141 | 0.142 | -0.001 | 5.10e-05 | 0.138 | 0.140 | -0.002 | 3.18e-04 |
| Depression | lassosum.MultiPRS | LDpred2.PseudoVal | 0.143 | 0.142 | 0.001 | 9.10e-01 | 0.144 | 0.140 | 0.004 | 1.48e-01 |
| Depression | LDpred1.10FCVal | LDpred2.PseudoVal | 0.142 | 0.142 | 0.000 | 9.64e-01 | 0.140 | 0.140 | 0.000 | 8.75e-01 |
| Depression | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.127 | 0.142 | -0.015 | 2.81e-02 | 0.130 | 0.140 | -0.010 | 6.71e-02 |
| Depression | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.143 | 0.142 | 0.001 | 5.70e-01 | 0.143 | 0.140 | 0.003 | 2.92e-01 |
| Depression | pT+clump.10FCVal | LDpred2.PseudoVal | 0.120 | 0.142 | -0.022 | 1.48e-13 | 0.120 | 0.140 | -0.020 | 6.85e-04 |
| Depression | lassosum.10FCVal | LDpred2.PseudoVal | 0.143 | 0.142 | 0.000 | 7.51e-01 | 0.143 | 0.140 | 0.003 | 2.61e-01 |
| Depression | LDpred2.10FCVal | LDpred2.PseudoVal | 0.144 | 0.142 | 0.001 | 1.76e-03 | 0.140 | 0.140 | 0.000 | 9.56e-01 |
| Depression | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.142 | 0.142 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| Depression | SBLUP.Inf | LDpred2.PseudoVal | 0.141 | 0.142 | -0.001 | 1.64e-02 | 0.137 | 0.140 | -0.003 | 1.92e-02 |
| Depression | DBSLMM.PseudoVal | LDpred2.Inf | 0.139 | 0.141 | -0.002 | 2.41e-01 | 0.140 | 0.138 | 0.002 | 4.79e-01 |
| Depression | LDpred2.MultiPRS | LDpred2.Inf | 0.144 | 0.141 | 0.003 | 6.81e-01 | 0.143 | 0.138 | 0.005 | 1.20e-02 |
| Depression | pT+clump.10FCVal | LDpred2.Inf | 0.120 | 0.141 | -0.021 | 3.74e-12 | 0.120 | 0.138 | -0.018 | 2.51e-03 |
| Depression | lassosum.10FCVal | LDpred2.Inf | 0.143 | 0.141 | 0.002 | 2.90e-01 | 0.143 | 0.138 | 0.005 | 8.94e-02 |
| Depression | lassosum.PseudoVal | LDpred2.Inf | 0.079 | 0.141 | -0.062 | 2.32e-29 | 0.091 | 0.138 | -0.047 | 1.93e-05 |
| Depression | All.MultiPRS | LDpred2.Inf | 0.144 | 0.141 | 0.003 | 6.35e-01 | 0.145 | 0.138 | 0.008 | 1.49e-03 |
| Depression | LDpred1.MultiPRS | LDpred2.Inf | 0.142 | 0.141 | 0.001 | 8.73e-01 | 0.139 | 0.138 | 0.001 | 4.58e-01 |
| Depression | LDpred2.PseudoVal | LDpred2.Inf | 0.142 | 0.141 | 0.001 | 5.10e-05 | 0.140 | 0.138 | 0.002 | 3.18e-04 |
| Depression | SBLUP.Inf | LDpred2.Inf | 0.141 | 0.141 | 0.000 | 7.79e-01 | 0.137 | 0.138 | 0.000 | 6.24e-01 |
| Depression | LDpred1.Inf | LDpred2.Inf | 0.141 | 0.141 | 0.000 | 8.86e-01 | 0.137 | 0.138 | -0.001 | 3.68e-01 |
| Depression | LDpred2.Inf | LDpred2.Inf | 0.141 | 0.141 | 0.000 | 1.00e+00 | 0.138 | 0.138 | 0.000 | 1.00e+00 |
| Depression | lassosum.MultiPRS | LDpred2.Inf | 0.143 | 0.141 | 0.002 | 7.81e-01 | 0.144 | 0.138 | 0.006 | 5.48e-02 |
| Depression | LDpred1.10FCVal | LDpred2.Inf | 0.142 | 0.141 | 0.001 | 1.74e-01 | 0.140 | 0.138 | 0.002 | 2.66e-01 |
| Depression | pT+clump.MultiPRS | LDpred2.Inf | 0.127 | 0.141 | -0.014 | 4.22e-02 | 0.130 | 0.138 | -0.008 | 1.63e-01 |
| Depression | SBayesR.PseudoVal | LDpred2.Inf | 0.143 | 0.141 | 0.002 | 2.00e-01 | 0.143 | 0.138 | 0.005 | 1.07e-01 |
| Depression | LDpred2.10FCVal | LDpred2.Inf | 0.144 | 0.141 | 0.002 | 6.49e-06 | 0.140 | 0.138 | 0.002 | 5.88e-02 |
| Depression | All.MultiPRS | pT+clump.MultiPRS | 0.144 | 0.127 | 0.018 | 1.17e-02 | 0.145 | 0.130 | 0.015 | 1.43e-03 |
| Depression | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.142 | 0.127 | 0.015 | 2.79e-02 | 0.139 | 0.130 | 0.009 | 9.53e-02 |
| Depression | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.142 | 0.127 | 0.015 | 2.81e-02 | 0.140 | 0.130 | 0.010 | 6.71e-02 |
| Depression | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.139 | 0.127 | 0.013 | 7.35e-02 | 0.140 | 0.130 | 0.010 | 7.09e-02 |
| Depression | LDpred1.Inf | pT+clump.MultiPRS | 0.141 | 0.127 | 0.014 | 4.37e-02 | 0.137 | 0.130 | 0.007 | 2.44e-01 |
| Depression | LDpred2.Inf | pT+clump.MultiPRS | 0.141 | 0.127 | 0.014 | 4.22e-02 | 0.138 | 0.130 | 0.008 | 1.63e-01 |
| Depression | lassosum.MultiPRS | pT+clump.MultiPRS | 0.143 | 0.127 | 0.016 | 2.02e-02 | 0.144 | 0.130 | 0.014 | 6.06e-03 |
| Depression | LDpred1.10FCVal | pT+clump.MultiPRS | 0.142 | 0.127 | 0.015 | 2.84e-02 | 0.140 | 0.130 | 0.010 | 6.95e-02 |
| Depression | lassosum.PseudoVal | pT+clump.MultiPRS | 0.079 | 0.127 | -0.048 | 7.44e-12 | 0.091 | 0.130 | -0.039 | 1.21e-04 |
| Depression | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.144 | 0.127 | 0.017 | 1.40e-02 | 0.143 | 0.130 | 0.013 | 1.36e-02 |
| Depression | pT+clump.10FCVal | pT+clump.MultiPRS | 0.120 | 0.127 | -0.006 | 3.56e-01 | 0.120 | 0.130 | -0.010 | 5.91e-04 |
| Depression | lassosum.10FCVal | pT+clump.MultiPRS | 0.143 | 0.127 | 0.016 | 2.40e-02 | 0.143 | 0.130 | 0.013 | 1.29e-02 |
| Depression | LDpred2.10FCVal | pT+clump.MultiPRS | 0.144 | 0.127 | 0.017 | 1.69e-02 | 0.140 | 0.130 | 0.010 | 7.15e-02 |
| Depression | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.143 | 0.127 | 0.016 | 2.18e-02 | 0.143 | 0.130 | 0.013 | 1.43e-02 |
| Depression | SBLUP.Inf | pT+clump.MultiPRS | 0.141 | 0.127 | 0.014 | 4.40e-02 | 0.137 | 0.130 | 0.007 | 1.82e-01 |
| Depression | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.127 | 0.127 | 0.000 | 1.00e+00 | 0.130 | 0.130 | 0.000 | 1.00e+00 |
| Depression | lassosum.PseudoVal | pT+clump.10FCVal | 0.079 | 0.120 | -0.042 | 2.09e-12 | 0.091 | 0.120 | -0.029 | 1.52e-02 |
| Depression | All.MultiPRS | pT+clump.10FCVal | 0.144 | 0.120 | 0.024 | 6.04e-04 | 0.145 | 0.120 | 0.026 | 1.64e-05 |
| Depression | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.139 | 0.120 | 0.019 | 1.66e-09 | 0.140 | 0.120 | 0.020 | 1.47e-03 |
| Depression | LDpred1.Inf | pT+clump.10FCVal | 0.141 | 0.120 | 0.021 | 9.10e-12 | 0.137 | 0.120 | 0.017 | 5.54e-03 |
| Depression | LDpred2.Inf | pT+clump.10FCVal | 0.141 | 0.120 | 0.021 | 3.74e-12 | 0.138 | 0.120 | 0.018 | 2.51e-03 |
| Depression | lassosum.MultiPRS | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 1.22e-03 | 0.144 | 0.120 | 0.024 | 9.42e-05 |
| Depression | LDpred1.10FCVal | pT+clump.10FCVal | 0.142 | 0.120 | 0.022 | 3.04e-13 | 0.140 | 0.120 | 0.020 | 8.98e-04 |
| Depression | pT+clump.MultiPRS | pT+clump.10FCVal | 0.127 | 0.120 | 0.006 | 3.56e-01 | 0.130 | 0.120 | 0.010 | 5.91e-04 |
| Depression | LDpred2.MultiPRS | pT+clump.10FCVal | 0.144 | 0.120 | 0.024 | 7.60e-04 | 0.143 | 0.120 | 0.023 | 2.53e-04 |
| Depression | pT+clump.10FCVal | pT+clump.10FCVal | 0.120 | 0.120 | 0.000 | 1.00e+00 | 0.120 | 0.120 | 0.000 | 1.00e+00 |
| Depression | lassosum.10FCVal | pT+clump.10FCVal | 0.143 | 0.120 | 0.022 | 6.57e-14 | 0.143 | 0.120 | 0.023 | 9.26e-05 |
| Depression | LDpred2.10FCVal | pT+clump.10FCVal | 0.144 | 0.120 | 0.023 | 5.02e-14 | 0.140 | 0.120 | 0.020 | 1.11e-03 |
| Depression | SBayesR.PseudoVal | pT+clump.10FCVal | 0.143 | 0.120 | 0.023 | 5.56e-13 | 0.143 | 0.120 | 0.023 | 2.86e-04 |
| Depression | LDpred1.MultiPRS | pT+clump.10FCVal | 0.142 | 0.120 | 0.022 | 1.76e-03 | 0.139 | 0.120 | 0.019 | 1.36e-03 |
| Depression | LDpred2.PseudoVal | pT+clump.10FCVal | 0.142 | 0.120 | 0.022 | 1.48e-13 | 0.140 | 0.120 | 0.020 | 6.85e-04 |
| Depression | SBLUP.Inf | pT+clump.10FCVal | 0.141 | 0.120 | 0.021 | 4.02e-12 | 0.137 | 0.120 | 0.018 | 3.05e-03 |
| Depression | LDpred1.Inf | SBayesR.PseudoVal | 0.141 | 0.143 | -0.002 | 1.78e-01 | 0.137 | 0.143 | -0.006 | 3.98e-02 |
| Depression | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.139 | 0.143 | -0.004 | 2.39e-02 | 0.140 | 0.143 | -0.003 | 4.10e-01 |
| Depression | lassosum.PseudoVal | SBayesR.PseudoVal | 0.079 | 0.143 | -0.064 | 5.91e-40 | 0.091 | 0.143 | -0.051 | 7.65e-08 |
| Depression | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.144 | 0.143 | 0.001 | 8.84e-01 | 0.143 | 0.143 | 0.001 | 7.11e-01 |
| Depression | pT+clump.10FCVal | SBayesR.PseudoVal | 0.120 | 0.143 | -0.023 | 5.56e-13 | 0.120 | 0.143 | -0.023 | 2.86e-04 |
| Depression | lassosum.10FCVal | SBayesR.PseudoVal | 0.143 | 0.143 | 0.000 | 8.61e-01 | 0.143 | 0.143 | 0.000 | 8.73e-01 |
| Depression | LDpred2.10FCVal | SBayesR.PseudoVal | 0.144 | 0.143 | 0.001 | 5.79e-01 | 0.140 | 0.143 | -0.003 | 2.78e-01 |
| Depression | All.MultiPRS | SBayesR.PseudoVal | 0.144 | 0.143 | 0.001 | 8.34e-01 | 0.145 | 0.143 | 0.003 | 1.19e-01 |
| Depression | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.142 | 0.143 | -0.001 | 9.16e-01 | 0.139 | 0.143 | -0.003 | 1.55e-01 |
| Depression | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.142 | 0.143 | -0.001 | 5.70e-01 | 0.140 | 0.143 | -0.003 | 2.92e-01 |
| Depression | SBLUP.Inf | SBayesR.PseudoVal | 0.141 | 0.143 | -0.002 | 1.78e-01 | 0.137 | 0.143 | -0.005 | 8.29e-02 |
| Depression | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.127 | 0.143 | -0.016 | 2.18e-02 | 0.130 | 0.143 | -0.013 | 1.43e-02 |
| Depression | LDpred2.Inf | SBayesR.PseudoVal | 0.141 | 0.143 | -0.002 | 2.00e-01 | 0.138 | 0.143 | -0.005 | 1.07e-01 |
| Depression | lassosum.MultiPRS | SBayesR.PseudoVal | 0.143 | 0.143 | 0.000 | 9.90e-01 | 0.144 | 0.143 | 0.002 | 5.50e-01 |
| Depression | LDpred1.10FCVal | SBayesR.PseudoVal | 0.142 | 0.143 | -0.001 | 5.10e-01 | 0.140 | 0.143 | -0.003 | 2.09e-01 |
| Depression | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.143 | 0.143 | 0.000 | 1.00e+00 |
| Depression | All.MultiPRS | SBLUP.Inf | 0.144 | 0.141 | 0.003 | 6.23e-01 | 0.145 | 0.137 | 0.008 | 7.74e-04 |
| Depression | LDpred1.Inf | SBLUP.Inf | 0.141 | 0.141 | 0.000 | 9.73e-01 | 0.137 | 0.137 | -0.001 | 5.14e-01 |
| Depression | LDpred2.Inf | SBLUP.Inf | 0.141 | 0.141 | 0.000 | 7.79e-01 | 0.138 | 0.137 | 0.000 | 6.24e-01 |
| Depression | DBSLMM.PseudoVal | SBLUP.Inf | 0.139 | 0.141 | -0.002 | 2.51e-01 | 0.140 | 0.137 | 0.002 | 3.67e-01 |
| Depression | lassosum.PseudoVal | SBLUP.Inf | 0.079 | 0.141 | -0.062 | 8.45e-30 | 0.091 | 0.137 | -0.046 | 1.92e-05 |
| Depression | LDpred2.MultiPRS | SBLUP.Inf | 0.144 | 0.141 | 0.003 | 6.68e-01 | 0.143 | 0.137 | 0.006 | 9.31e-03 |
| Depression | pT+clump.10FCVal | SBLUP.Inf | 0.120 | 0.141 | -0.021 | 4.02e-12 | 0.120 | 0.137 | -0.018 | 3.05e-03 |
| Depression | lassosum.10FCVal | SBLUP.Inf | 0.143 | 0.141 | 0.002 | 2.43e-01 | 0.143 | 0.137 | 0.006 | 6.04e-02 |
| Depression | LDpred2.10FCVal | SBLUP.Inf | 0.144 | 0.141 | 0.003 | 5.14e-05 | 0.140 | 0.137 | 0.003 | 5.09e-02 |
| Depression | SBayesR.PseudoVal | SBLUP.Inf | 0.143 | 0.141 | 0.002 | 1.78e-01 | 0.143 | 0.137 | 0.005 | 8.29e-02 |
| Depression | LDpred1.MultiPRS | SBLUP.Inf | 0.142 | 0.141 | 0.001 | 8.59e-01 | 0.139 | 0.137 | 0.002 | 2.66e-01 |
| Depression | LDpred2.PseudoVal | SBLUP.Inf | 0.142 | 0.141 | 0.001 | 1.64e-02 | 0.140 | 0.137 | 0.003 | 1.92e-02 |
| Depression | SBLUP.Inf | SBLUP.Inf | 0.141 | 0.141 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| Depression | pT+clump.MultiPRS | SBLUP.Inf | 0.127 | 0.141 | -0.014 | 4.40e-02 | 0.130 | 0.137 | -0.007 | 1.82e-01 |
| Depression | lassosum.MultiPRS | SBLUP.Inf | 0.143 | 0.141 | 0.002 | 7.68e-01 | 0.144 | 0.137 | 0.007 | 3.29e-02 |
| Depression | LDpred1.10FCVal | SBLUP.Inf | 0.142 | 0.141 | 0.001 | 1.08e-01 | 0.140 | 0.137 | 0.002 | 1.43e-01 |
| Intelligence | lassosum.PseudoVal | All.MultiPRS | 0.064 | 0.102 | -0.038 | 8.95e-08 | 0.055 | 0.097 | -0.043 | 4.76e-07 |
| Intelligence | All.MultiPRS | All.MultiPRS | 0.102 | 0.102 | 0.000 | 1.00e+00 | 0.097 | 0.097 | 0.000 | 1.00e+00 |
| Intelligence | DBSLMM.PseudoVal | All.MultiPRS | 0.090 | 0.102 | -0.012 | 8.98e-02 | 0.082 | 0.097 | -0.015 | 1.55e-03 |
| Intelligence | LDpred1.Inf | All.MultiPRS | 0.099 | 0.102 | -0.004 | 6.04e-01 | 0.094 | 0.097 | -0.004 | 2.74e-01 |
| Intelligence | LDpred2.Inf | All.MultiPRS | 0.099 | 0.102 | -0.003 | 6.59e-01 | 0.094 | 0.097 | -0.003 | 3.81e-01 |
| Intelligence | lassosum.MultiPRS | All.MultiPRS | 0.102 | 0.102 | 0.000 | 6.94e-01 | 0.099 | 0.097 | 0.002 | 3.09e-01 |
| Intelligence | LDpred1.10FCVal | All.MultiPRS | 0.100 | 0.102 | -0.002 | 8.02e-01 | 0.094 | 0.097 | -0.003 | 2.38e-01 |
| Intelligence | pT+clump.MultiPRS | All.MultiPRS | 0.087 | 0.102 | -0.016 | 2.63e-02 | 0.081 | 0.097 | -0.016 | 4.05e-03 |
| Intelligence | LDpred2.MultiPRS | All.MultiPRS | 0.103 | 0.102 | 0.001 | 9.29e-01 | 0.098 | 0.097 | 0.001 | 7.16e-01 |
| Intelligence | pT+clump.10FCVal | All.MultiPRS | 0.084 | 0.102 | -0.018 | 8.92e-03 | 0.074 | 0.097 | -0.023 | 2.53e-04 |
| Intelligence | lassosum.10FCVal | All.MultiPRS | 0.098 | 0.102 | -0.004 | 5.74e-01 | 0.096 | 0.097 | -0.002 | 6.41e-01 |
| Intelligence | LDpred2.10FCVal | All.MultiPRS | 0.102 | 0.102 | -0.001 | 9.42e-01 | 0.095 | 0.097 | -0.002 | 4.83e-01 |
| Intelligence | SBayesR.PseudoVal | All.MultiPRS | 0.100 | 0.102 | -0.002 | 7.39e-01 | 0.090 | 0.097 | -0.007 | 2.56e-02 |
| Intelligence | LDpred1.MultiPRS | All.MultiPRS | 0.101 | 0.102 | -0.002 | 8.23e-01 | 0.094 | 0.097 | -0.003 | 2.60e-01 |
| Intelligence | LDpred2.PseudoVal | All.MultiPRS | 0.100 | 0.102 | -0.002 | 7.81e-01 | 0.095 | 0.097 | -0.002 | 4.89e-01 |
| Intelligence | SBLUP.Inf | All.MultiPRS | 0.099 | 0.102 | -0.003 | 6.65e-01 | 0.094 | 0.097 | -0.004 | 2.57e-01 |
| Intelligence | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.103 | 0.090 | 0.013 | 7.38e-02 | 0.098 | 0.082 | 0.016 | 1.97e-03 |
| Intelligence | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.084 | 0.090 | -0.007 | 9.44e-02 | 0.074 | 0.082 | -0.008 | 3.05e-01 |
| Intelligence | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.064 | 0.090 | -0.026 | 1.66e-08 | 0.055 | 0.082 | -0.027 | 3.10e-03 |
| Intelligence | All.MultiPRS | DBSLMM.PseudoVal | 0.102 | 0.090 | 0.012 | 8.98e-02 | 0.097 | 0.082 | 0.015 | 1.55e-03 |
| Intelligence | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.101 | 0.090 | 0.010 | 1.21e-01 | 0.094 | 0.082 | 0.012 | 1.43e-02 |
| Intelligence | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.100 | 0.090 | 0.010 | 4.57e-05 | 0.095 | 0.082 | 0.013 | 6.29e-03 |
| Intelligence | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.090 | 0.090 | 0.000 | 1.00e+00 | 0.082 | 0.082 | 0.000 | 1.00e+00 |
| Intelligence | LDpred1.Inf | DBSLMM.PseudoVal | 0.099 | 0.090 | 0.008 | 1.63e-03 | 0.094 | 0.082 | 0.012 | 2.61e-02 |
| Intelligence | LDpred2.Inf | DBSLMM.PseudoVal | 0.099 | 0.090 | 0.009 | 4.77e-04 | 0.094 | 0.082 | 0.012 | 1.29e-02 |
| Intelligence | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.102 | 0.090 | 0.012 | 1.00e-01 | 0.099 | 0.082 | 0.017 | 5.67e-04 |
| Intelligence | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.100 | 0.090 | 0.010 | 5.33e-05 | 0.094 | 0.082 | 0.012 | 1.79e-02 |
| Intelligence | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.087 | 0.090 | -0.004 | 6.01e-01 | 0.081 | 0.082 | -0.001 | 9.37e-01 |
| Intelligence | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.100 | 0.090 | 0.010 | 3.90e-04 | 0.090 | 0.082 | 0.008 | 1.32e-01 |
| Intelligence | lassosum.10FCVal | DBSLMM.PseudoVal | 0.098 | 0.090 | 0.008 | 5.50e-03 | 0.096 | 0.082 | 0.014 | 1.72e-02 |
| Intelligence | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.102 | 0.090 | 0.011 | 7.33e-07 | 0.095 | 0.082 | 0.014 | 3.44e-03 |
| Intelligence | SBLUP.Inf | DBSLMM.PseudoVal | 0.099 | 0.090 | 0.009 | 4.11e-04 | 0.094 | 0.082 | 0.012 | 1.98e-02 |
| Intelligence | All.MultiPRS | lassosum.MultiPRS | 0.102 | 0.102 | 0.000 | 6.94e-01 | 0.097 | 0.099 | -0.002 | 3.09e-01 |
| Intelligence | LDpred1.MultiPRS | lassosum.MultiPRS | 0.101 | 0.102 | -0.001 | 8.65e-01 | 0.094 | 0.099 | -0.005 | 1.18e-01 |
| Intelligence | LDpred1.Inf | lassosum.MultiPRS | 0.099 | 0.102 | -0.003 | 6.41e-01 | 0.094 | 0.099 | -0.005 | 1.31e-01 |
| Intelligence | LDpred2.Inf | lassosum.MultiPRS | 0.099 | 0.102 | -0.003 | 6.97e-01 | 0.094 | 0.099 | -0.005 | 1.63e-01 |
| Intelligence | lassosum.MultiPRS | lassosum.MultiPRS | 0.102 | 0.102 | 0.000 | 1.00e+00 | 0.099 | 0.099 | 0.000 | 1.00e+00 |
| Intelligence | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.090 | 0.102 | -0.012 | 1.00e-01 | 0.082 | 0.099 | -0.017 | 5.67e-04 |
| Intelligence | lassosum.PseudoVal | lassosum.MultiPRS | 0.064 | 0.102 | -0.037 | 1.18e-07 | 0.055 | 0.099 | -0.044 | 1.21e-07 |
| Intelligence | LDpred2.MultiPRS | lassosum.MultiPRS | 0.103 | 0.102 | 0.001 | 8.86e-01 | 0.098 | 0.099 | -0.001 | 7.87e-01 |
| Intelligence | pT+clump.10FCVal | lassosum.MultiPRS | 0.084 | 0.102 | -0.018 | 1.04e-02 | 0.074 | 0.099 | -0.025 | 1.36e-04 |
| Intelligence | lassosum.10FCVal | lassosum.MultiPRS | 0.098 | 0.102 | -0.004 | 6.11e-01 | 0.096 | 0.099 | -0.003 | 2.32e-01 |
| Intelligence | LDpred2.10FCVal | lassosum.MultiPRS | 0.102 | 0.102 | 0.000 | 9.84e-01 | 0.095 | 0.099 | -0.004 | 1.67e-01 |
| Intelligence | SBayesR.PseudoVal | lassosum.MultiPRS | 0.100 | 0.102 | -0.002 | 7.79e-01 | 0.090 | 0.099 | -0.009 | 1.54e-02 |
| Intelligence | LDpred2.PseudoVal | lassosum.MultiPRS | 0.100 | 0.102 | -0.002 | 8.22e-01 | 0.095 | 0.099 | -0.004 | 2.03e-01 |
| Intelligence | SBLUP.Inf | lassosum.MultiPRS | 0.099 | 0.102 | -0.003 | 7.03e-01 | 0.094 | 0.099 | -0.005 | 1.00e-01 |
| Intelligence | pT+clump.MultiPRS | lassosum.MultiPRS | 0.087 | 0.102 | -0.015 | 3.00e-02 | 0.081 | 0.099 | -0.018 | 2.04e-03 |
| Intelligence | LDpred1.10FCVal | lassosum.MultiPRS | 0.100 | 0.102 | -0.001 | 8.43e-01 | 0.094 | 0.099 | -0.005 | 1.02e-01 |
| Intelligence | All.MultiPRS | lassosum.PseudoVal | 0.102 | 0.064 | 0.038 | 8.95e-08 | 0.097 | 0.055 | 0.043 | 4.76e-07 |
| Intelligence | LDpred1.MultiPRS | lassosum.PseudoVal | 0.101 | 0.064 | 0.036 | 9.47e-08 | 0.094 | 0.055 | 0.040 | 2.54e-06 |
| Intelligence | LDpred2.PseudoVal | lassosum.PseudoVal | 0.100 | 0.064 | 0.036 | 3.13e-14 | 0.095 | 0.055 | 0.041 | 1.64e-05 |
| Intelligence | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.090 | 0.064 | 0.026 | 1.66e-08 | 0.082 | 0.055 | 0.027 | 3.10e-03 |
| Intelligence | LDpred1.Inf | lassosum.PseudoVal | 0.099 | 0.064 | 0.034 | 6.88e-12 | 0.094 | 0.055 | 0.039 | 8.86e-05 |
| Intelligence | LDpred2.Inf | lassosum.PseudoVal | 0.099 | 0.064 | 0.035 | 2.34e-12 | 0.094 | 0.055 | 0.040 | 5.65e-05 |
| Intelligence | lassosum.MultiPRS | lassosum.PseudoVal | 0.102 | 0.064 | 0.037 | 1.18e-07 | 0.099 | 0.055 | 0.044 | 1.21e-07 |
| Intelligence | LDpred1.10FCVal | lassosum.PseudoVal | 0.100 | 0.064 | 0.036 | 9.81e-17 | 0.094 | 0.055 | 0.039 | 5.66e-06 |
| Intelligence | lassosum.PseudoVal | lassosum.PseudoVal | 0.064 | 0.064 | 0.000 | 1.00e+00 | 0.055 | 0.055 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.MultiPRS | lassosum.PseudoVal | 0.103 | 0.064 | 0.038 | 4.26e-08 | 0.098 | 0.055 | 0.043 | 6.57e-07 |
| Intelligence | pT+clump.10FCVal | lassosum.PseudoVal | 0.084 | 0.064 | 0.019 | 2.57e-04 | 0.074 | 0.055 | 0.019 | 6.68e-02 |
| Intelligence | lassosum.10FCVal | lassosum.PseudoVal | 0.098 | 0.064 | 0.034 | 5.72e-13 | 0.096 | 0.055 | 0.041 | 1.31e-05 |
| Intelligence | LDpred2.10FCVal | lassosum.PseudoVal | 0.102 | 0.064 | 0.037 | 1.25e-19 | 0.095 | 0.055 | 0.041 | 6.72e-07 |
| Intelligence | SBayesR.PseudoVal | lassosum.PseudoVal | 0.100 | 0.064 | 0.036 | 1.09e-18 | 0.090 | 0.055 | 0.036 | 9.83e-06 |
| Intelligence | SBLUP.Inf | lassosum.PseudoVal | 0.099 | 0.064 | 0.035 | 2.11e-12 | 0.094 | 0.055 | 0.039 | 7.96e-05 |
| Intelligence | pT+clump.MultiPRS | lassosum.PseudoVal | 0.087 | 0.064 | 0.022 | 1.60e-03 | 0.081 | 0.055 | 0.027 | 2.40e-03 |
| Intelligence | All.MultiPRS | lassosum.10FCVal | 0.102 | 0.098 | 0.004 | 5.74e-01 | 0.097 | 0.096 | 0.002 | 6.41e-01 |
| Intelligence | LDpred1.MultiPRS | lassosum.10FCVal | 0.101 | 0.098 | 0.002 | 7.18e-01 | 0.094 | 0.096 | -0.001 | 7.06e-01 |
| Intelligence | LDpred1.Inf | lassosum.10FCVal | 0.099 | 0.098 | 0.000 | 8.64e-01 | 0.094 | 0.096 | -0.002 | 5.74e-01 |
| Intelligence | LDpred2.Inf | lassosum.10FCVal | 0.099 | 0.098 | 0.001 | 6.01e-01 | 0.094 | 0.096 | -0.001 | 7.27e-01 |
| Intelligence | lassosum.MultiPRS | lassosum.10FCVal | 0.102 | 0.098 | 0.004 | 6.11e-01 | 0.099 | 0.096 | 0.003 | 2.32e-01 |
| Intelligence | DBSLMM.PseudoVal | lassosum.10FCVal | 0.090 | 0.098 | -0.008 | 5.50e-03 | 0.082 | 0.096 | -0.014 | 1.72e-02 |
| Intelligence | lassosum.PseudoVal | lassosum.10FCVal | 0.064 | 0.098 | -0.034 | 5.72e-13 | 0.055 | 0.096 | -0.041 | 1.31e-05 |
| Intelligence | LDpred2.MultiPRS | lassosum.10FCVal | 0.103 | 0.098 | 0.005 | 5.13e-01 | 0.098 | 0.096 | 0.003 | 5.75e-01 |
| Intelligence | pT+clump.10FCVal | lassosum.10FCVal | 0.084 | 0.098 | -0.014 | 3.17e-07 | 0.074 | 0.096 | -0.022 | 1.27e-04 |
| Intelligence | lassosum.10FCVal | lassosum.10FCVal | 0.098 | 0.098 | 0.000 | 1.00e+00 | 0.096 | 0.096 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.10FCVal | lassosum.10FCVal | 0.102 | 0.098 | 0.003 | 5.35e-02 | 0.095 | 0.096 | 0.000 | 9.86e-01 |
| Intelligence | SBayesR.PseudoVal | lassosum.10FCVal | 0.100 | 0.098 | 0.002 | 4.10e-01 | 0.090 | 0.096 | -0.005 | 1.67e-01 |
| Intelligence | LDpred2.PseudoVal | lassosum.10FCVal | 0.100 | 0.098 | 0.002 | 1.69e-01 | 0.095 | 0.096 | 0.000 | 9.19e-01 |
| Intelligence | SBLUP.Inf | lassosum.10FCVal | 0.099 | 0.098 | 0.001 | 5.75e-01 | 0.094 | 0.096 | -0.002 | 5.42e-01 |
| Intelligence | pT+clump.MultiPRS | lassosum.10FCVal | 0.087 | 0.098 | -0.012 | 9.69e-02 | 0.081 | 0.096 | -0.014 | 9.48e-03 |
| Intelligence | LDpred1.10FCVal | lassosum.10FCVal | 0.100 | 0.098 | 0.002 | 1.95e-01 | 0.094 | 0.096 | -0.002 | 6.22e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.090 | 0.101 | -0.010 | 1.21e-01 | 0.082 | 0.094 | -0.012 | 1.43e-02 |
| Intelligence | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.103 | 0.101 | 0.002 | 7.55e-01 | 0.098 | 0.094 | 0.004 | 2.57e-01 |
| Intelligence | pT+clump.10FCVal | LDpred1.MultiPRS | 0.084 | 0.101 | -0.017 | 1.21e-02 | 0.074 | 0.094 | -0.020 | 1.88e-03 |
| Intelligence | lassosum.10FCVal | LDpred1.MultiPRS | 0.098 | 0.101 | -0.002 | 7.18e-01 | 0.096 | 0.094 | 0.001 | 7.06e-01 |
| Intelligence | lassosum.PseudoVal | LDpred1.MultiPRS | 0.064 | 0.101 | -0.036 | 9.47e-08 | 0.055 | 0.094 | -0.040 | 2.54e-06 |
| Intelligence | All.MultiPRS | LDpred1.MultiPRS | 0.102 | 0.101 | 0.002 | 8.23e-01 | 0.097 | 0.094 | 0.003 | 2.60e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.101 | 0.101 | 0.000 | 1.00e+00 | 0.094 | 0.094 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.100 | 0.101 | 0.000 | 9.51e-01 | 0.095 | 0.094 | 0.001 | 6.24e-01 |
| Intelligence | SBLUP.Inf | LDpred1.MultiPRS | 0.099 | 0.101 | -0.002 | 8.21e-01 | 0.094 | 0.094 | -0.001 | 8.14e-01 |
| Intelligence | LDpred1.Inf | LDpred1.MultiPRS | 0.099 | 0.101 | -0.002 | 7.51e-01 | 0.094 | 0.094 | -0.001 | 7.67e-01 |
| Intelligence | LDpred2.Inf | LDpred1.MultiPRS | 0.099 | 0.101 | -0.002 | 8.15e-01 | 0.094 | 0.094 | 0.000 | 9.28e-01 |
| Intelligence | lassosum.MultiPRS | LDpred1.MultiPRS | 0.102 | 0.101 | 0.001 | 8.65e-01 | 0.099 | 0.094 | 0.005 | 1.18e-01 |
| Intelligence | LDpred1.10FCVal | LDpred1.MultiPRS | 0.100 | 0.101 | 0.000 | 9.74e-01 | 0.094 | 0.094 | 0.000 | 7.75e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.087 | 0.101 | -0.014 | 4.55e-02 | 0.081 | 0.094 | -0.013 | 2.59e-02 |
| Intelligence | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.100 | 0.101 | -0.001 | 9.05e-01 | 0.090 | 0.094 | -0.004 | 1.40e-01 |
| Intelligence | LDpred2.10FCVal | LDpred1.MultiPRS | 0.102 | 0.101 | 0.001 | 8.76e-01 | 0.095 | 0.094 | 0.001 | 5.07e-01 |
| Intelligence | All.MultiPRS | LDpred1.Inf | 0.102 | 0.099 | 0.004 | 6.04e-01 | 0.097 | 0.094 | 0.004 | 2.74e-01 |
| Intelligence | LDpred1.Inf | LDpred1.Inf | 0.099 | 0.099 | 0.000 | 1.00e+00 | 0.094 | 0.094 | 0.000 | 1.00e+00 |
| Intelligence | LDpred2.Inf | LDpred1.Inf | 0.099 | 0.099 | 0.001 | 5.23e-01 | 0.094 | 0.094 | 0.001 | 6.05e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred1.Inf | 0.090 | 0.099 | -0.008 | 1.63e-03 | 0.082 | 0.094 | -0.012 | 2.61e-02 |
| Intelligence | lassosum.PseudoVal | LDpred1.Inf | 0.064 | 0.099 | -0.034 | 6.88e-12 | 0.055 | 0.094 | -0.039 | 8.86e-05 |
| Intelligence | LDpred2.MultiPRS | LDpred1.Inf | 0.103 | 0.099 | 0.004 | 5.41e-01 | 0.098 | 0.094 | 0.005 | 2.53e-01 |
| Intelligence | pT+clump.10FCVal | LDpred1.Inf | 0.084 | 0.099 | -0.015 | 3.24e-06 | 0.074 | 0.094 | -0.020 | 1.85e-03 |
| Intelligence | lassosum.10FCVal | LDpred1.Inf | 0.098 | 0.099 | 0.000 | 8.64e-01 | 0.096 | 0.094 | 0.002 | 5.74e-01 |
| Intelligence | LDpred2.10FCVal | LDpred1.Inf | 0.102 | 0.099 | 0.003 | 3.31e-02 | 0.095 | 0.094 | 0.002 | 5.09e-01 |
| Intelligence | SBayesR.PseudoVal | LDpred1.Inf | 0.100 | 0.099 | 0.001 | 4.85e-01 | 0.090 | 0.094 | -0.003 | 3.58e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred1.Inf | 0.101 | 0.099 | 0.002 | 7.51e-01 | 0.094 | 0.094 | 0.001 | 7.67e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred1.Inf | 0.100 | 0.099 | 0.002 | 6.60e-02 | 0.095 | 0.094 | 0.002 | 3.53e-01 |
| Intelligence | SBLUP.Inf | LDpred1.Inf | 0.099 | 0.099 | 0.001 | 4.75e-01 | 0.094 | 0.094 | 0.000 | 9.81e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred1.Inf | 0.087 | 0.099 | -0.012 | 8.87e-02 | 0.081 | 0.094 | -0.012 | 4.90e-02 |
| Intelligence | lassosum.MultiPRS | LDpred1.Inf | 0.102 | 0.099 | 0.003 | 6.41e-01 | 0.099 | 0.094 | 0.005 | 1.31e-01 |
| Intelligence | LDpred1.10FCVal | LDpred1.Inf | 0.100 | 0.099 | 0.002 | 7.36e-02 | 0.094 | 0.094 | 0.000 | 8.85e-01 |
| Intelligence | LDpred1.Inf | LDpred1.10FCVal | 0.099 | 0.100 | -0.002 | 7.36e-02 | 0.094 | 0.094 | 0.000 | 8.85e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.090 | 0.100 | -0.010 | 5.33e-05 | 0.082 | 0.094 | -0.012 | 1.79e-02 |
| Intelligence | lassosum.PseudoVal | LDpred1.10FCVal | 0.064 | 0.100 | -0.036 | 9.81e-17 | 0.055 | 0.094 | -0.039 | 5.66e-06 |
| Intelligence | LDpred2.MultiPRS | LDpred1.10FCVal | 0.103 | 0.100 | 0.002 | 7.33e-01 | 0.098 | 0.094 | 0.004 | 2.41e-01 |
| Intelligence | pT+clump.10FCVal | LDpred1.10FCVal | 0.084 | 0.100 | -0.017 | 2.79e-07 | 0.074 | 0.094 | -0.020 | 1.99e-03 |
| Intelligence | lassosum.10FCVal | LDpred1.10FCVal | 0.098 | 0.100 | -0.002 | 1.95e-01 | 0.096 | 0.094 | 0.002 | 6.22e-01 |
| Intelligence | LDpred2.10FCVal | LDpred1.10FCVal | 0.102 | 0.100 | 0.001 | 2.06e-01 | 0.095 | 0.094 | 0.002 | 4.10e-01 |
| Intelligence | All.MultiPRS | LDpred1.10FCVal | 0.102 | 0.100 | 0.002 | 8.02e-01 | 0.097 | 0.094 | 0.003 | 2.38e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred1.10FCVal | 0.101 | 0.100 | 0.000 | 9.74e-01 | 0.094 | 0.094 | 0.000 | 7.75e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred1.10FCVal | 0.100 | 0.100 | 0.000 | 8.57e-01 | 0.095 | 0.094 | 0.001 | 5.15e-01 |
| Intelligence | SBLUP.Inf | LDpred1.10FCVal | 0.099 | 0.100 | -0.001 | 3.15e-01 | 0.094 | 0.094 | 0.000 | 9.18e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred1.10FCVal | 0.087 | 0.100 | -0.014 | 4.88e-02 | 0.081 | 0.094 | -0.012 | 3.06e-02 |
| Intelligence | LDpred2.Inf | LDpred1.10FCVal | 0.099 | 0.100 | -0.001 | 2.97e-01 | 0.094 | 0.094 | 0.001 | 8.27e-01 |
| Intelligence | lassosum.MultiPRS | LDpred1.10FCVal | 0.102 | 0.100 | 0.001 | 8.43e-01 | 0.099 | 0.094 | 0.005 | 1.02e-01 |
| Intelligence | LDpred1.10FCVal | LDpred1.10FCVal | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.094 | 0.094 | 0.000 | 1.00e+00 |
| Intelligence | SBayesR.PseudoVal | LDpred1.10FCVal | 0.100 | 0.100 | -0.001 | 6.76e-01 | 0.090 | 0.094 | -0.004 | 1.79e-01 |
| Intelligence | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.103 | 0.103 | 0.000 | 1.00e+00 | 0.098 | 0.098 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.PseudoVal | LDpred2.MultiPRS | 0.064 | 0.103 | -0.038 | 4.26e-08 | 0.055 | 0.098 | -0.043 | 6.57e-07 |
| Intelligence | All.MultiPRS | LDpred2.MultiPRS | 0.102 | 0.103 | -0.001 | 9.29e-01 | 0.097 | 0.098 | -0.001 | 7.16e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.101 | 0.103 | -0.002 | 7.55e-01 | 0.094 | 0.098 | -0.004 | 2.57e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.090 | 0.103 | -0.013 | 7.38e-02 | 0.082 | 0.098 | -0.016 | 1.97e-03 |
| Intelligence | LDpred1.Inf | LDpred2.MultiPRS | 0.099 | 0.103 | -0.004 | 5.41e-01 | 0.094 | 0.098 | -0.005 | 2.53e-01 |
| Intelligence | LDpred2.Inf | LDpred2.MultiPRS | 0.099 | 0.103 | -0.004 | 5.94e-01 | 0.094 | 0.098 | -0.004 | 3.09e-01 |
| Intelligence | lassosum.MultiPRS | LDpred2.MultiPRS | 0.102 | 0.103 | -0.001 | 8.86e-01 | 0.099 | 0.098 | 0.001 | 7.87e-01 |
| Intelligence | LDpred1.10FCVal | LDpred2.MultiPRS | 0.100 | 0.103 | -0.002 | 7.33e-01 | 0.094 | 0.098 | -0.004 | 2.41e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.087 | 0.103 | -0.016 | 2.18e-02 | 0.081 | 0.098 | -0.017 | 8.35e-03 |
| Intelligence | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.100 | 0.103 | -0.003 | 6.72e-01 | 0.090 | 0.098 | -0.008 | 4.77e-02 |
| Intelligence | pT+clump.10FCVal | LDpred2.MultiPRS | 0.084 | 0.103 | -0.019 | 6.51e-03 | 0.074 | 0.098 | -0.024 | 7.49e-04 |
| Intelligence | lassosum.10FCVal | LDpred2.MultiPRS | 0.098 | 0.103 | -0.005 | 5.13e-01 | 0.096 | 0.098 | -0.003 | 5.75e-01 |
| Intelligence | LDpred2.10FCVal | LDpred2.MultiPRS | 0.102 | 0.103 | -0.001 | 8.70e-01 | 0.095 | 0.098 | -0.003 | 3.91e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.100 | 0.103 | -0.003 | 7.12e-01 | 0.095 | 0.098 | -0.003 | 3.96e-01 |
| Intelligence | SBLUP.Inf | LDpred2.MultiPRS | 0.099 | 0.103 | -0.004 | 6.00e-01 | 0.094 | 0.098 | -0.004 | 2.20e-01 |
| Intelligence | LDpred2.MultiPRS | LDpred2.10FCVal | 0.103 | 0.102 | 0.001 | 8.70e-01 | 0.098 | 0.095 | 0.003 | 3.91e-01 |
| Intelligence | pT+clump.10FCVal | LDpred2.10FCVal | 0.084 | 0.102 | -0.018 | 1.35e-07 | 0.074 | 0.095 | -0.021 | 1.47e-03 |
| Intelligence | lassosum.PseudoVal | LDpred2.10FCVal | 0.064 | 0.102 | -0.037 | 1.25e-19 | 0.055 | 0.095 | -0.041 | 6.72e-07 |
| Intelligence | All.MultiPRS | LDpred2.10FCVal | 0.102 | 0.102 | 0.001 | 9.42e-01 | 0.097 | 0.095 | 0.002 | 4.83e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.10FCVal | 0.101 | 0.102 | -0.001 | 8.76e-01 | 0.094 | 0.095 | -0.001 | 5.07e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred2.10FCVal | 0.100 | 0.102 | -0.001 | 1.26e-01 | 0.095 | 0.095 | 0.000 | 9.03e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.090 | 0.102 | -0.011 | 7.33e-07 | 0.082 | 0.095 | -0.014 | 3.44e-03 |
| Intelligence | LDpred1.Inf | LDpred2.10FCVal | 0.099 | 0.102 | -0.003 | 3.31e-02 | 0.094 | 0.095 | -0.002 | 5.09e-01 |
| Intelligence | LDpred2.Inf | LDpred2.10FCVal | 0.099 | 0.102 | -0.003 | 3.35e-02 | 0.094 | 0.095 | -0.001 | 6.65e-01 |
| Intelligence | lassosum.MultiPRS | LDpred2.10FCVal | 0.102 | 0.102 | 0.000 | 9.84e-01 | 0.099 | 0.095 | 0.004 | 1.67e-01 |
| Intelligence | LDpred1.10FCVal | LDpred2.10FCVal | 0.100 | 0.102 | -0.001 | 2.06e-01 | 0.094 | 0.095 | -0.002 | 4.10e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred2.10FCVal | 0.087 | 0.102 | -0.015 | 3.15e-02 | 0.081 | 0.095 | -0.014 | 1.67e-02 |
| Intelligence | SBayesR.PseudoVal | LDpred2.10FCVal | 0.100 | 0.102 | -0.002 | 1.81e-01 | 0.090 | 0.095 | -0.005 | 5.17e-02 |
| Intelligence | lassosum.10FCVal | LDpred2.10FCVal | 0.098 | 0.102 | -0.003 | 5.35e-02 | 0.096 | 0.095 | 0.000 | 9.86e-01 |
| Intelligence | LDpred2.10FCVal | LDpred2.10FCVal | 0.102 | 0.102 | 0.000 | 1.00e+00 | 0.095 | 0.095 | 0.000 | 1.00e+00 |
| Intelligence | SBLUP.Inf | LDpred2.10FCVal | 0.099 | 0.102 | -0.003 | 4.24e-02 | 0.094 | 0.095 | -0.002 | 4.51e-01 |
| Intelligence | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.103 | 0.100 | 0.003 | 7.12e-01 | 0.098 | 0.095 | 0.003 | 3.96e-01 |
| Intelligence | lassosum.PseudoVal | LDpred2.PseudoVal | 0.064 | 0.100 | -0.036 | 3.13e-14 | 0.055 | 0.095 | -0.041 | 1.64e-05 |
| Intelligence | All.MultiPRS | LDpred2.PseudoVal | 0.102 | 0.100 | 0.002 | 7.81e-01 | 0.097 | 0.095 | 0.002 | 4.89e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.101 | 0.100 | 0.000 | 9.51e-01 | 0.094 | 0.095 | -0.001 | 6.24e-01 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.090 | 0.100 | -0.010 | 4.57e-05 | 0.082 | 0.095 | -0.013 | 6.29e-03 |
| Intelligence | LDpred1.Inf | LDpred2.PseudoVal | 0.099 | 0.100 | -0.002 | 6.60e-02 | 0.094 | 0.095 | -0.002 | 3.53e-01 |
| Intelligence | LDpred2.Inf | LDpred2.PseudoVal | 0.099 | 0.100 | -0.001 | 1.81e-03 | 0.094 | 0.095 | -0.001 | 2.60e-01 |
| Intelligence | lassosum.MultiPRS | LDpred2.PseudoVal | 0.102 | 0.100 | 0.002 | 8.22e-01 | 0.099 | 0.095 | 0.004 | 2.03e-01 |
| Intelligence | LDpred1.10FCVal | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 8.57e-01 | 0.094 | 0.095 | -0.001 | 5.15e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.087 | 0.100 | -0.014 | 5.18e-02 | 0.081 | 0.095 | -0.014 | 1.83e-02 |
| Intelligence | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 8.12e-01 | 0.090 | 0.095 | -0.005 | 1.19e-01 |
| Intelligence | pT+clump.10FCVal | LDpred2.PseudoVal | 0.084 | 0.100 | -0.016 | 1.11e-07 | 0.074 | 0.095 | -0.021 | 5.47e-04 |
| Intelligence | lassosum.10FCVal | LDpred2.PseudoVal | 0.098 | 0.100 | -0.002 | 1.69e-01 | 0.096 | 0.095 | 0.000 | 9.19e-01 |
| Intelligence | LDpred2.10FCVal | LDpred2.PseudoVal | 0.102 | 0.100 | 0.001 | 1.26e-01 | 0.095 | 0.095 | 0.000 | 9.03e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.095 | 0.095 | 0.000 | 1.00e+00 |
| Intelligence | SBLUP.Inf | LDpred2.PseudoVal | 0.099 | 0.100 | -0.001 | 2.27e-02 | 0.094 | 0.095 | -0.002 | 8.13e-02 |
| Intelligence | DBSLMM.PseudoVal | LDpred2.Inf | 0.090 | 0.099 | -0.009 | 4.77e-04 | 0.082 | 0.094 | -0.012 | 1.29e-02 |
| Intelligence | LDpred2.MultiPRS | LDpred2.Inf | 0.103 | 0.099 | 0.004 | 5.94e-01 | 0.098 | 0.094 | 0.004 | 3.09e-01 |
| Intelligence | pT+clump.10FCVal | LDpred2.Inf | 0.084 | 0.099 | -0.015 | 1.14e-06 | 0.074 | 0.094 | -0.020 | 1.05e-03 |
| Intelligence | lassosum.10FCVal | LDpred2.Inf | 0.098 | 0.099 | -0.001 | 6.01e-01 | 0.096 | 0.094 | 0.001 | 7.27e-01 |
| Intelligence | lassosum.PseudoVal | LDpred2.Inf | 0.064 | 0.099 | -0.035 | 2.34e-12 | 0.055 | 0.094 | -0.040 | 5.65e-05 |
| Intelligence | All.MultiPRS | LDpred2.Inf | 0.102 | 0.099 | 0.003 | 6.59e-01 | 0.097 | 0.094 | 0.003 | 3.81e-01 |
| Intelligence | LDpred1.MultiPRS | LDpred2.Inf | 0.101 | 0.099 | 0.002 | 8.15e-01 | 0.094 | 0.094 | 0.000 | 9.28e-01 |
| Intelligence | LDpred2.PseudoVal | LDpred2.Inf | 0.100 | 0.099 | 0.001 | 1.81e-03 | 0.095 | 0.094 | 0.001 | 2.60e-01 |
| Intelligence | SBLUP.Inf | LDpred2.Inf | 0.099 | 0.099 | 0.000 | 8.40e-01 | 0.094 | 0.094 | -0.001 | 1.42e-01 |
| Intelligence | LDpred1.Inf | LDpred2.Inf | 0.099 | 0.099 | -0.001 | 5.23e-01 | 0.094 | 0.094 | -0.001 | 6.05e-01 |
| Intelligence | LDpred2.Inf | LDpred2.Inf | 0.099 | 0.099 | 0.000 | 1.00e+00 | 0.094 | 0.094 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.MultiPRS | LDpred2.Inf | 0.102 | 0.099 | 0.003 | 6.97e-01 | 0.099 | 0.094 | 0.005 | 1.63e-01 |
| Intelligence | LDpred1.10FCVal | LDpred2.Inf | 0.100 | 0.099 | 0.001 | 2.97e-01 | 0.094 | 0.094 | -0.001 | 8.27e-01 |
| Intelligence | pT+clump.MultiPRS | LDpred2.Inf | 0.087 | 0.099 | -0.013 | 7.50e-02 | 0.081 | 0.094 | -0.013 | 3.39e-02 |
| Intelligence | SBayesR.PseudoVal | LDpred2.Inf | 0.100 | 0.099 | 0.001 | 6.83e-01 | 0.090 | 0.094 | -0.004 | 2.52e-01 |
| Intelligence | LDpred2.10FCVal | LDpred2.Inf | 0.102 | 0.099 | 0.003 | 3.35e-02 | 0.095 | 0.094 | 0.001 | 6.65e-01 |
| Intelligence | All.MultiPRS | pT+clump.MultiPRS | 0.102 | 0.087 | 0.016 | 2.63e-02 | 0.097 | 0.081 | 0.016 | 4.05e-03 |
| Intelligence | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.101 | 0.087 | 0.014 | 4.55e-02 | 0.094 | 0.081 | 0.013 | 2.59e-02 |
| Intelligence | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.100 | 0.087 | 0.014 | 5.18e-02 | 0.095 | 0.081 | 0.014 | 1.83e-02 |
| Intelligence | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.090 | 0.087 | 0.004 | 6.01e-01 | 0.082 | 0.081 | 0.001 | 9.37e-01 |
| Intelligence | LDpred1.Inf | pT+clump.MultiPRS | 0.099 | 0.087 | 0.012 | 8.87e-02 | 0.094 | 0.081 | 0.012 | 4.90e-02 |
| Intelligence | LDpred2.Inf | pT+clump.MultiPRS | 0.099 | 0.087 | 0.013 | 7.50e-02 | 0.094 | 0.081 | 0.013 | 3.39e-02 |
| Intelligence | lassosum.MultiPRS | pT+clump.MultiPRS | 0.102 | 0.087 | 0.015 | 3.00e-02 | 0.099 | 0.081 | 0.018 | 2.04e-03 |
| Intelligence | LDpred1.10FCVal | pT+clump.MultiPRS | 0.100 | 0.087 | 0.014 | 4.88e-02 | 0.094 | 0.081 | 0.012 | 3.06e-02 |
| Intelligence | lassosum.PseudoVal | pT+clump.MultiPRS | 0.064 | 0.087 | -0.022 | 1.60e-03 | 0.055 | 0.081 | -0.027 | 2.40e-03 |
| Intelligence | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.103 | 0.087 | 0.016 | 2.18e-02 | 0.098 | 0.081 | 0.017 | 8.35e-03 |
| Intelligence | pT+clump.10FCVal | pT+clump.MultiPRS | 0.084 | 0.087 | -0.003 | 6.87e-01 | 0.074 | 0.081 | -0.007 | 1.15e-02 |
| Intelligence | lassosum.10FCVal | pT+clump.MultiPRS | 0.098 | 0.087 | 0.012 | 9.69e-02 | 0.096 | 0.081 | 0.014 | 9.48e-03 |
| Intelligence | LDpred2.10FCVal | pT+clump.MultiPRS | 0.102 | 0.087 | 0.015 | 3.15e-02 | 0.095 | 0.081 | 0.014 | 1.67e-02 |
| Intelligence | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.100 | 0.087 | 0.013 | 5.90e-02 | 0.090 | 0.081 | 0.009 | 1.39e-01 |
| Intelligence | SBLUP.Inf | pT+clump.MultiPRS | 0.099 | 0.087 | 0.013 | 7.37e-02 | 0.094 | 0.081 | 0.012 | 4.41e-02 |
| Intelligence | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.087 | 0.087 | 0.000 | 1.00e+00 | 0.081 | 0.081 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.PseudoVal | pT+clump.10FCVal | 0.064 | 0.084 | -0.019 | 2.57e-04 | 0.055 | 0.074 | -0.019 | 6.68e-02 |
| Intelligence | All.MultiPRS | pT+clump.10FCVal | 0.102 | 0.084 | 0.018 | 8.92e-03 | 0.097 | 0.074 | 0.023 | 2.53e-04 |
| Intelligence | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.090 | 0.084 | 0.007 | 9.44e-02 | 0.082 | 0.074 | 0.008 | 3.05e-01 |
| Intelligence | LDpred1.Inf | pT+clump.10FCVal | 0.099 | 0.084 | 0.015 | 3.24e-06 | 0.094 | 0.074 | 0.020 | 1.85e-03 |
| Intelligence | LDpred2.Inf | pT+clump.10FCVal | 0.099 | 0.084 | 0.015 | 1.14e-06 | 0.094 | 0.074 | 0.020 | 1.05e-03 |
| Intelligence | lassosum.MultiPRS | pT+clump.10FCVal | 0.102 | 0.084 | 0.018 | 1.04e-02 | 0.099 | 0.074 | 0.025 | 1.36e-04 |
| Intelligence | LDpred1.10FCVal | pT+clump.10FCVal | 0.100 | 0.084 | 0.017 | 2.79e-07 | 0.094 | 0.074 | 0.020 | 1.99e-03 |
| Intelligence | pT+clump.MultiPRS | pT+clump.10FCVal | 0.087 | 0.084 | 0.003 | 6.87e-01 | 0.081 | 0.074 | 0.007 | 1.15e-02 |
| Intelligence | LDpred2.MultiPRS | pT+clump.10FCVal | 0.103 | 0.084 | 0.019 | 6.51e-03 | 0.098 | 0.074 | 0.024 | 7.49e-04 |
| Intelligence | pT+clump.10FCVal | pT+clump.10FCVal | 0.084 | 0.084 | 0.000 | 1.00e+00 | 0.074 | 0.074 | 0.000 | 1.00e+00 |
| Intelligence | lassosum.10FCVal | pT+clump.10FCVal | 0.098 | 0.084 | 0.014 | 3.17e-07 | 0.096 | 0.074 | 0.022 | 1.27e-04 |
| Intelligence | LDpred2.10FCVal | pT+clump.10FCVal | 0.102 | 0.084 | 0.018 | 1.35e-07 | 0.095 | 0.074 | 0.021 | 1.47e-03 |
| Intelligence | SBayesR.PseudoVal | pT+clump.10FCVal | 0.100 | 0.084 | 0.016 | 2.95e-06 | 0.090 | 0.074 | 0.016 | 1.84e-02 |
| Intelligence | LDpred1.MultiPRS | pT+clump.10FCVal | 0.101 | 0.084 | 0.017 | 1.21e-02 | 0.094 | 0.074 | 0.020 | 1.88e-03 |
| Intelligence | LDpred2.PseudoVal | pT+clump.10FCVal | 0.100 | 0.084 | 0.016 | 1.11e-07 | 0.095 | 0.074 | 0.021 | 5.47e-04 |
| Intelligence | SBLUP.Inf | pT+clump.10FCVal | 0.099 | 0.084 | 0.015 | 7.46e-07 | 0.094 | 0.074 | 0.020 | 1.45e-03 |
| Intelligence | LDpred1.Inf | SBayesR.PseudoVal | 0.099 | 0.100 | -0.001 | 4.85e-01 | 0.094 | 0.090 | 0.003 | 3.58e-01 |
| Intelligence | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.090 | 0.100 | -0.010 | 3.90e-04 | 0.082 | 0.090 | -0.008 | 1.32e-01 |
| Intelligence | lassosum.PseudoVal | SBayesR.PseudoVal | 0.064 | 0.100 | -0.036 | 1.09e-18 | 0.055 | 0.090 | -0.036 | 9.83e-06 |
| Intelligence | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.103 | 0.100 | 0.003 | 6.72e-01 | 0.098 | 0.090 | 0.008 | 4.77e-02 |
| Intelligence | pT+clump.10FCVal | SBayesR.PseudoVal | 0.084 | 0.100 | -0.016 | 2.95e-06 | 0.074 | 0.090 | -0.016 | 1.84e-02 |
| Intelligence | lassosum.10FCVal | SBayesR.PseudoVal | 0.098 | 0.100 | -0.002 | 4.10e-01 | 0.096 | 0.090 | 0.005 | 1.67e-01 |
| Intelligence | LDpred2.10FCVal | SBayesR.PseudoVal | 0.102 | 0.100 | 0.002 | 1.81e-01 | 0.095 | 0.090 | 0.005 | 5.17e-02 |
| Intelligence | All.MultiPRS | SBayesR.PseudoVal | 0.102 | 0.100 | 0.002 | 7.39e-01 | 0.097 | 0.090 | 0.007 | 2.56e-02 |
| Intelligence | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.101 | 0.100 | 0.001 | 9.05e-01 | 0.094 | 0.090 | 0.004 | 1.40e-01 |
| Intelligence | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.100 | 0.100 | 0.000 | 8.12e-01 | 0.095 | 0.090 | 0.005 | 1.19e-01 |
| Intelligence | SBLUP.Inf | SBayesR.PseudoVal | 0.099 | 0.100 | -0.001 | 7.09e-01 | 0.094 | 0.090 | 0.003 | 3.59e-01 |
| Intelligence | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.087 | 0.100 | -0.013 | 5.90e-02 | 0.081 | 0.090 | -0.009 | 1.39e-01 |
| Intelligence | LDpred2.Inf | SBayesR.PseudoVal | 0.099 | 0.100 | -0.001 | 6.83e-01 | 0.094 | 0.090 | 0.004 | 2.52e-01 |
| Intelligence | lassosum.MultiPRS | SBayesR.PseudoVal | 0.102 | 0.100 | 0.002 | 7.79e-01 | 0.099 | 0.090 | 0.009 | 1.54e-02 |
| Intelligence | LDpred1.10FCVal | SBayesR.PseudoVal | 0.100 | 0.100 | 0.001 | 6.76e-01 | 0.094 | 0.090 | 0.004 | 1.79e-01 |
| Intelligence | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.100 | 0.100 | 0.000 | 1.00e+00 | 0.090 | 0.090 | 0.000 | 1.00e+00 |
| Intelligence | All.MultiPRS | SBLUP.Inf | 0.102 | 0.099 | 0.003 | 6.65e-01 | 0.097 | 0.094 | 0.004 | 2.57e-01 |
| Intelligence | LDpred1.Inf | SBLUP.Inf | 0.099 | 0.099 | -0.001 | 4.75e-01 | 0.094 | 0.094 | 0.000 | 9.81e-01 |
| Intelligence | LDpred2.Inf | SBLUP.Inf | 0.099 | 0.099 | 0.000 | 8.40e-01 | 0.094 | 0.094 | 0.001 | 1.42e-01 |
| Intelligence | DBSLMM.PseudoVal | SBLUP.Inf | 0.090 | 0.099 | -0.009 | 4.11e-04 | 0.082 | 0.094 | -0.012 | 1.98e-02 |
| Intelligence | lassosum.PseudoVal | SBLUP.Inf | 0.064 | 0.099 | -0.035 | 2.11e-12 | 0.055 | 0.094 | -0.039 | 7.96e-05 |
| Intelligence | LDpred2.MultiPRS | SBLUP.Inf | 0.103 | 0.099 | 0.004 | 6.00e-01 | 0.098 | 0.094 | 0.004 | 2.20e-01 |
| Intelligence | pT+clump.10FCVal | SBLUP.Inf | 0.084 | 0.099 | -0.015 | 7.46e-07 | 0.074 | 0.094 | -0.020 | 1.45e-03 |
| Intelligence | lassosum.10FCVal | SBLUP.Inf | 0.098 | 0.099 | -0.001 | 5.75e-01 | 0.096 | 0.094 | 0.002 | 5.42e-01 |
| Intelligence | LDpred2.10FCVal | SBLUP.Inf | 0.102 | 0.099 | 0.003 | 4.24e-02 | 0.095 | 0.094 | 0.002 | 4.51e-01 |
| Intelligence | SBayesR.PseudoVal | SBLUP.Inf | 0.100 | 0.099 | 0.001 | 7.09e-01 | 0.090 | 0.094 | -0.003 | 3.59e-01 |
| Intelligence | LDpred1.MultiPRS | SBLUP.Inf | 0.101 | 0.099 | 0.002 | 8.21e-01 | 0.094 | 0.094 | 0.001 | 8.14e-01 |
| Intelligence | LDpred2.PseudoVal | SBLUP.Inf | 0.100 | 0.099 | 0.001 | 2.27e-02 | 0.095 | 0.094 | 0.002 | 8.13e-02 |
| Intelligence | SBLUP.Inf | SBLUP.Inf | 0.099 | 0.099 | 0.000 | 1.00e+00 | 0.094 | 0.094 | 0.000 | 1.00e+00 |
| Intelligence | pT+clump.MultiPRS | SBLUP.Inf | 0.087 | 0.099 | -0.013 | 7.37e-02 | 0.081 | 0.094 | -0.012 | 4.41e-02 |
| Intelligence | lassosum.MultiPRS | SBLUP.Inf | 0.102 | 0.099 | 0.003 | 7.03e-01 | 0.099 | 0.094 | 0.005 | 1.00e-01 |
| Intelligence | LDpred1.10FCVal | SBLUP.Inf | 0.100 | 0.099 | 0.001 | 3.15e-01 | 0.094 | 0.094 | 0.000 | 9.18e-01 |
| Height | lassosum.10FCVal | All.MultiPRS | 0.347 | 0.360 | -0.013 | 4.14e-02 | 0.340 | 0.356 | -0.016 | 8.37e-10 |
| Height | All.MultiPRS | All.MultiPRS | 0.360 | 0.360 | 0.000 | 1.00e+00 | 0.356 | 0.356 | 0.000 | 1.00e+00 |
| Height | DBSLMM.PseudoVal | All.MultiPRS | 0.333 | 0.360 | -0.026 | 4.12e-05 | 0.324 | 0.356 | -0.032 | 1.48e-17 |
| Height | LDpred1.Inf | All.MultiPRS | 0.297 | 0.360 | -0.063 | 3.97e-22 | 0.295 | 0.356 | -0.061 | 1.51e-27 |
| Height | LDpred2.Inf | All.MultiPRS | 0.302 | 0.360 | -0.058 | 5.40e-19 | 0.299 | 0.356 | -0.056 | 1.07e-25 |
| Height | lassosum.MultiPRS | All.MultiPRS | 0.349 | 0.360 | -0.010 | 1.12e-01 | 0.343 | 0.356 | -0.013 | 1.05e-07 |
| Height | LDpred1.10FCVal | All.MultiPRS | 0.287 | 0.360 | -0.073 | 2.27e-29 | 0.288 | 0.356 | -0.067 | 7.25e-29 |
| Height | pT+clump.MultiPRS | All.MultiPRS | 0.320 | 0.360 | -0.040 | 4.95e-10 | 0.314 | 0.356 | -0.042 | 2.27e-20 |
| Height | LDpred2.MultiPRS | All.MultiPRS | 0.356 | 0.360 | -0.004 | 2.18e-07 | 0.351 | 0.356 | -0.004 | 4.46e-03 |
| Height | pT+clump.10FCVal | All.MultiPRS | 0.305 | 0.360 | -0.055 | 1.31e-17 | 0.299 | 0.356 | -0.057 | 2.13e-26 |
| Height | lassosum.PseudoVal | All.MultiPRS | 0.281 | 0.360 | -0.078 | 1.68e-33 | 0.276 | 0.356 | -0.080 | 6.46e-38 |
| Height | LDpred2.10FCVal | All.MultiPRS | 0.351 | 0.360 | -0.008 | 1.84e-01 | 0.343 | 0.356 | -0.012 | 2.80e-08 |
| Height | SBayesR.PseudoVal | All.MultiPRS | 0.342 | 0.360 | -0.018 | 4.95e-03 | 0.335 | 0.356 | -0.021 | 3.39e-11 |
| Height | LDpred1.MultiPRS | All.MultiPRS | 0.304 | 0.360 | -0.055 | 1.47e-17 | 0.303 | 0.356 | -0.053 | 1.45e-24 |
| Height | LDpred2.PseudoVal | All.MultiPRS | 0.335 | 0.360 | -0.024 | 1.43e-04 | 0.326 | 0.356 | -0.029 | 2.29e-16 |
| Height | SBLUP.Inf | All.MultiPRS | 0.295 | 0.360 | -0.064 | 3.07e-23 | 0.292 | 0.356 | -0.064 | 6.29e-30 |
| Height | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.356 | 0.333 | 0.022 | 4.71e-04 | 0.351 | 0.324 | 0.027 | 2.22e-13 |
| Height | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.305 | 0.333 | -0.029 | 1.05e-19 | 0.299 | 0.324 | -0.025 | 8.71e-05 |
| Height | lassosum.10FCVal | DBSLMM.PseudoVal | 0.347 | 0.333 | 0.013 | 2.21e-11 | 0.340 | 0.324 | 0.016 | 9.25e-05 |
| Height | All.MultiPRS | DBSLMM.PseudoVal | 0.360 | 0.333 | 0.026 | 4.12e-05 | 0.356 | 0.324 | 0.032 | 1.48e-17 |
| Height | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.304 | 0.333 | -0.029 | 8.32e-06 | 0.303 | 0.324 | -0.021 | 1.06e-08 |
| Height | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.335 | 0.333 | 0.002 | 2.31e-01 | 0.326 | 0.324 | 0.002 | 4.78e-01 |
| Height | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.333 | 0.333 | 0.000 | 1.00e+00 | 0.324 | 0.324 | 0.000 | 1.00e+00 |
| Height | LDpred1.Inf | DBSLMM.PseudoVal | 0.297 | 0.333 | -0.036 | 9.92e-84 | 0.295 | 0.324 | -0.029 | 1.36e-14 |
| Height | LDpred2.Inf | DBSLMM.PseudoVal | 0.302 | 0.333 | -0.031 | 5.87e-65 | 0.299 | 0.324 | -0.025 | 1.98e-11 |
| Height | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.349 | 0.333 | 0.016 | 1.22e-02 | 0.343 | 0.324 | 0.019 | 8.74e-08 |
| Height | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.287 | 0.333 | -0.047 | 5.70e-105 | 0.288 | 0.324 | -0.036 | 1.75e-16 |
| Height | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.320 | 0.333 | -0.014 | 3.32e-02 | 0.314 | 0.324 | -0.010 | 3.26e-02 |
| Height | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.342 | 0.333 | 0.008 | 4.55e-09 | 0.335 | 0.324 | 0.011 | 9.59e-05 |
| Height | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.281 | 0.333 | -0.052 | 1.35e-42 | 0.276 | 0.324 | -0.048 | 2.73e-10 |
| Height | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.351 | 0.333 | 0.018 | 3.89e-24 | 0.343 | 0.324 | 0.019 | 4.79e-08 |
| Height | SBLUP.Inf | DBSLMM.PseudoVal | 0.295 | 0.333 | -0.038 | 1.63e-95 | 0.292 | 0.324 | -0.032 | 2.32e-18 |
| Height | All.MultiPRS | lassosum.MultiPRS | 0.360 | 0.349 | 0.010 | 1.12e-01 | 0.356 | 0.343 | 0.013 | 1.05e-07 |
| Height | LDpred1.MultiPRS | lassosum.MultiPRS | 0.304 | 0.349 | -0.045 | 2.29e-71 | 0.303 | 0.343 | -0.040 | 2.80e-15 |
| Height | LDpred1.Inf | lassosum.MultiPRS | 0.297 | 0.349 | -0.052 | 7.65e-16 | 0.295 | 0.343 | -0.048 | 1.05e-18 |
| Height | LDpred2.Inf | lassosum.MultiPRS | 0.302 | 0.349 | -0.047 | 3.02e-13 | 0.299 | 0.343 | -0.044 | 2.93e-16 |
| Height | lassosum.MultiPRS | lassosum.MultiPRS | 0.349 | 0.349 | 0.000 | 1.00e+00 | 0.343 | 0.343 | 0.000 | 1.00e+00 |
| Height | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.333 | 0.349 | -0.016 | 1.22e-02 | 0.324 | 0.343 | -0.019 | 8.74e-08 |
| Height | lassosum.10FCVal | lassosum.MultiPRS | 0.347 | 0.349 | -0.003 | 6.56e-01 | 0.340 | 0.343 | -0.003 | 1.84e-02 |
| Height | LDpred2.MultiPRS | lassosum.MultiPRS | 0.356 | 0.349 | 0.006 | 3.23e-01 | 0.351 | 0.343 | 0.009 | 1.96e-03 |
| Height | pT+clump.10FCVal | lassosum.MultiPRS | 0.305 | 0.349 | -0.045 | 3.42e-12 | 0.299 | 0.343 | -0.044 | 5.98e-16 |
| Height | lassosum.PseudoVal | lassosum.MultiPRS | 0.281 | 0.349 | -0.068 | 8.05e-26 | 0.276 | 0.343 | -0.067 | 6.10e-30 |
| Height | LDpred2.10FCVal | lassosum.MultiPRS | 0.351 | 0.349 | 0.002 | 7.89e-01 | 0.343 | 0.343 | 0.000 | 8.89e-01 |
| Height | SBayesR.PseudoVal | lassosum.MultiPRS | 0.342 | 0.349 | -0.008 | 2.25e-01 | 0.335 | 0.343 | -0.008 | 2.32e-02 |
| Height | LDpred2.PseudoVal | lassosum.MultiPRS | 0.335 | 0.349 | -0.014 | 2.73e-02 | 0.326 | 0.343 | -0.017 | 5.78e-05 |
| Height | SBLUP.Inf | lassosum.MultiPRS | 0.295 | 0.349 | -0.054 | 8.80e-17 | 0.292 | 0.343 | -0.051 | 6.23e-21 |
| Height | pT+clump.MultiPRS | lassosum.MultiPRS | 0.320 | 0.349 | -0.030 | 3.54e-06 | 0.314 | 0.343 | -0.029 | 3.64e-11 |
| Height | LDpred1.10FCVal | lassosum.MultiPRS | 0.287 | 0.349 | -0.063 | 5.99e-22 | 0.288 | 0.343 | -0.055 | 7.32e-21 |
| Height | All.MultiPRS | lassosum.10FCVal | 0.360 | 0.347 | 0.013 | 4.14e-02 | 0.356 | 0.340 | 0.016 | 8.37e-10 |
| Height | LDpred1.MultiPRS | lassosum.10FCVal | 0.304 | 0.347 | -0.042 | 6.77e-11 | 0.303 | 0.340 | -0.037 | 4.73e-12 |
| Height | LDpred2.PseudoVal | lassosum.10FCVal | 0.335 | 0.347 | -0.011 | 7.05e-08 | 0.326 | 0.340 | -0.013 | 1.72e-03 |
| Height | DBSLMM.PseudoVal | lassosum.10FCVal | 0.333 | 0.347 | -0.013 | 2.21e-11 | 0.324 | 0.340 | -0.016 | 9.25e-05 |
| Height | LDpred1.Inf | lassosum.10FCVal | 0.297 | 0.347 | -0.050 | 2.04e-69 | 0.295 | 0.340 | -0.045 | 5.72e-15 |
| Height | LDpred2.Inf | lassosum.10FCVal | 0.302 | 0.347 | -0.045 | 1.11e-57 | 0.299 | 0.340 | -0.040 | 8.17e-13 |
| Height | lassosum.MultiPRS | lassosum.10FCVal | 0.349 | 0.347 | 0.003 | 6.56e-01 | 0.343 | 0.340 | 0.003 | 1.84e-02 |
| Height | LDpred1.10FCVal | lassosum.10FCVal | 0.287 | 0.347 | -0.060 | 2.53e-89 | 0.288 | 0.340 | -0.051 | 3.56e-17 |
| Height | lassosum.10FCVal | lassosum.10FCVal | 0.347 | 0.347 | 0.000 | 1.00e+00 | 0.340 | 0.340 | 0.000 | 1.00e+00 |
| Height | LDpred2.MultiPRS | lassosum.10FCVal | 0.356 | 0.347 | 0.009 | 1.51e-01 | 0.351 | 0.340 | 0.012 | 3.24e-05 |
| Height | pT+clump.10FCVal | lassosum.10FCVal | 0.305 | 0.347 | -0.042 | 1.75e-51 | 0.299 | 0.340 | -0.041 | 4.20e-13 |
| Height | lassosum.PseudoVal | lassosum.10FCVal | 0.281 | 0.347 | -0.065 | 8.97e-111 | 0.276 | 0.340 | -0.064 | 9.46e-28 |
| Height | LDpred2.10FCVal | lassosum.10FCVal | 0.351 | 0.347 | 0.005 | 3.60e-03 | 0.343 | 0.340 | 0.004 | 2.42e-01 |
| Height | SBayesR.PseudoVal | lassosum.10FCVal | 0.342 | 0.347 | -0.005 | 6.81e-03 | 0.335 | 0.340 | -0.005 | 2.16e-01 |
| Height | SBLUP.Inf | lassosum.10FCVal | 0.295 | 0.347 | -0.051 | 1.25e-73 | 0.292 | 0.340 | -0.048 | 7.40e-17 |
| Height | pT+clump.MultiPRS | lassosum.10FCVal | 0.320 | 0.347 | -0.027 | 2.82e-05 | 0.314 | 0.340 | -0.026 | 6.71e-09 |
| Height | All.MultiPRS | lassosum.PseudoVal | 0.360 | 0.281 | 0.078 | 1.68e-33 | 0.356 | 0.276 | 0.080 | 6.46e-38 |
| Height | LDpred1.MultiPRS | lassosum.PseudoVal | 0.304 | 0.281 | 0.023 | 4.91e-04 | 0.303 | 0.276 | 0.027 | 3.06e-03 |
| Height | LDpred1.Inf | lassosum.PseudoVal | 0.297 | 0.281 | 0.016 | 7.29e-04 | 0.295 | 0.276 | 0.019 | 3.86e-02 |
| Height | LDpred2.Inf | lassosum.PseudoVal | 0.302 | 0.281 | 0.021 | 6.88e-06 | 0.299 | 0.276 | 0.024 | 1.08e-02 |
| Height | lassosum.MultiPRS | lassosum.PseudoVal | 0.349 | 0.281 | 0.068 | 8.05e-26 | 0.343 | 0.276 | 0.067 | 6.10e-30 |
| Height | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.333 | 0.281 | 0.052 | 1.35e-42 | 0.324 | 0.276 | 0.048 | 2.73e-10 |
| Height | lassosum.10FCVal | lassosum.PseudoVal | 0.347 | 0.281 | 0.065 | 8.97e-111 | 0.340 | 0.276 | 0.064 | 9.46e-28 |
| Height | LDpred2.MultiPRS | lassosum.PseudoVal | 0.356 | 0.281 | 0.074 | 2.28e-30 | 0.351 | 0.276 | 0.076 | 5.30e-29 |
| Height | pT+clump.10FCVal | lassosum.PseudoVal | 0.305 | 0.281 | 0.023 | 7.20e-11 | 0.299 | 0.276 | 0.023 | 1.16e-03 |
| Height | lassosum.PseudoVal | lassosum.PseudoVal | 0.281 | 0.281 | 0.000 | 1.00e+00 | 0.276 | 0.276 | 0.000 | 1.00e+00 |
| Height | LDpred2.10FCVal | lassosum.PseudoVal | 0.351 | 0.281 | 0.070 | 3.04e-77 | 0.343 | 0.276 | 0.068 | 3.38e-19 |
| Height | SBayesR.PseudoVal | lassosum.PseudoVal | 0.342 | 0.281 | 0.060 | 1.86e-52 | 0.335 | 0.276 | 0.059 | 7.44e-14 |
| Height | LDpred2.PseudoVal | lassosum.PseudoVal | 0.335 | 0.281 | 0.054 | 3.02e-37 | 0.326 | 0.276 | 0.051 | 2.86e-09 |
| Height | SBLUP.Inf | lassosum.PseudoVal | 0.295 | 0.281 | 0.014 | 2.77e-03 | 0.292 | 0.276 | 0.016 | 8.48e-02 |
| Height | pT+clump.MultiPRS | lassosum.PseudoVal | 0.320 | 0.281 | 0.038 | 6.27e-09 | 0.314 | 0.276 | 0.038 | 6.25e-08 |
| Height | LDpred1.10FCVal | lassosum.PseudoVal | 0.287 | 0.281 | 0.005 | 2.61e-01 | 0.288 | 0.276 | 0.013 | 1.84e-01 |
| Height | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.333 | 0.304 | 0.029 | 8.32e-06 | 0.324 | 0.303 | 0.021 | 1.06e-08 |
| Height | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.356 | 0.304 | 0.052 | 2.19e-15 | 0.351 | 0.303 | 0.049 | 6.12e-24 |
| Height | pT+clump.10FCVal | LDpred1.MultiPRS | 0.305 | 0.304 | 0.000 | 9.73e-01 | 0.299 | 0.303 | -0.004 | 6.31e-01 |
| Height | lassosum.PseudoVal | LDpred1.MultiPRS | 0.281 | 0.304 | -0.023 | 4.91e-04 | 0.276 | 0.303 | -0.027 | 3.06e-03 |
| Height | lassosum.10FCVal | LDpred1.MultiPRS | 0.347 | 0.304 | 0.042 | 6.77e-11 | 0.340 | 0.303 | 0.037 | 4.73e-12 |
| Height | All.MultiPRS | LDpred1.MultiPRS | 0.360 | 0.304 | 0.055 | 1.47e-17 | 0.356 | 0.303 | 0.053 | 1.45e-24 |
| Height | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.304 | 0.304 | 0.000 | 1.00e+00 | 0.303 | 0.303 | 0.000 | 1.00e+00 |
| Height | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.335 | 0.304 | 0.031 | 1.98e-06 | 0.326 | 0.303 | 0.024 | 5.93e-14 |
| Height | SBLUP.Inf | LDpred1.MultiPRS | 0.295 | 0.304 | -0.009 | 1.77e-01 | 0.292 | 0.303 | -0.011 | 3.62e-05 |
| Height | LDpred1.Inf | LDpred1.MultiPRS | 0.297 | 0.304 | -0.007 | 2.73e-01 | 0.295 | 0.303 | -0.007 | 5.76e-04 |
| Height | LDpred2.Inf | LDpred1.MultiPRS | 0.302 | 0.304 | -0.002 | 7.41e-01 | 0.299 | 0.303 | -0.003 | 2.20e-01 |
| Height | lassosum.MultiPRS | LDpred1.MultiPRS | 0.349 | 0.304 | 0.045 | 2.29e-71 | 0.343 | 0.303 | 0.040 | 2.80e-15 |
| Height | LDpred1.10FCVal | LDpred1.MultiPRS | 0.287 | 0.304 | -0.018 | 7.78e-03 | 0.288 | 0.303 | -0.014 | 1.50e-05 |
| Height | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.320 | 0.304 | 0.015 | 1.96e-02 | 0.314 | 0.303 | 0.011 | 6.22e-02 |
| Height | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.342 | 0.304 | 0.037 | 9.02e-09 | 0.335 | 0.303 | 0.033 | 2.05e-21 |
| Height | LDpred2.10FCVal | LDpred1.MultiPRS | 0.351 | 0.304 | 0.047 | 4.72e-13 | 0.343 | 0.303 | 0.041 | 1.70e-21 |
| Height | All.MultiPRS | LDpred1.Inf | 0.360 | 0.297 | 0.063 | 3.97e-22 | 0.356 | 0.295 | 0.061 | 1.51e-27 |
| Height | LDpred1.Inf | LDpred1.Inf | 0.297 | 0.297 | 0.000 | 1.00e+00 | 0.295 | 0.295 | 0.000 | 1.00e+00 |
| Height | LDpred2.Inf | LDpred1.Inf | 0.302 | 0.297 | 0.005 | 1.76e-10 | 0.299 | 0.295 | 0.004 | 5.96e-03 |
| Height | DBSLMM.PseudoVal | LDpred1.Inf | 0.333 | 0.297 | 0.036 | 9.92e-84 | 0.324 | 0.295 | 0.029 | 1.36e-14 |
| Height | lassosum.10FCVal | LDpred1.Inf | 0.347 | 0.297 | 0.050 | 2.04e-69 | 0.340 | 0.295 | 0.045 | 5.72e-15 |
| Height | LDpred2.MultiPRS | LDpred1.Inf | 0.356 | 0.297 | 0.059 | 1.24e-19 | 0.351 | 0.295 | 0.056 | 9.59e-27 |
| Height | pT+clump.10FCVal | LDpred1.Inf | 0.305 | 0.297 | 0.007 | 5.75e-02 | 0.299 | 0.295 | 0.004 | 6.35e-01 |
| Height | lassosum.PseudoVal | LDpred1.Inf | 0.281 | 0.297 | -0.016 | 7.29e-04 | 0.276 | 0.295 | -0.019 | 3.86e-02 |
| Height | LDpred2.10FCVal | LDpred1.Inf | 0.351 | 0.297 | 0.054 | 1.34e-120 | 0.343 | 0.295 | 0.048 | 3.39e-25 |
| Height | SBayesR.PseudoVal | LDpred1.Inf | 0.342 | 0.297 | 0.045 | 4.07e-136 | 0.335 | 0.295 | 0.040 | 2.27e-28 |
| Height | LDpred1.MultiPRS | LDpred1.Inf | 0.304 | 0.297 | 0.007 | 2.73e-01 | 0.303 | 0.295 | 0.007 | 5.76e-04 |
| Height | LDpred2.PseudoVal | LDpred1.Inf | 0.335 | 0.297 | 0.038 | 2.99e-119 | 0.326 | 0.295 | 0.031 | 3.20e-21 |
| Height | SBLUP.Inf | LDpred1.Inf | 0.295 | 0.297 | -0.002 | 3.31e-02 | 0.292 | 0.295 | -0.003 | 4.65e-02 |
| Height | pT+clump.MultiPRS | LDpred1.Inf | 0.320 | 0.297 | 0.023 | 6.01e-04 | 0.314 | 0.295 | 0.019 | 2.57e-03 |
| Height | lassosum.MultiPRS | LDpred1.Inf | 0.349 | 0.297 | 0.052 | 7.65e-16 | 0.343 | 0.295 | 0.048 | 1.05e-18 |
| Height | LDpred1.10FCVal | LDpred1.Inf | 0.287 | 0.297 | -0.010 | 5.90e-16 | 0.288 | 0.295 | -0.007 | 1.02e-02 |
| Height | LDpred1.Inf | LDpred1.10FCVal | 0.297 | 0.287 | 0.010 | 5.90e-16 | 0.295 | 0.288 | 0.007 | 1.02e-02 |
| Height | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.333 | 0.287 | 0.047 | 5.70e-105 | 0.324 | 0.288 | 0.036 | 1.75e-16 |
| Height | lassosum.10FCVal | LDpred1.10FCVal | 0.347 | 0.287 | 0.060 | 2.53e-89 | 0.340 | 0.288 | 0.051 | 3.56e-17 |
| Height | LDpred2.MultiPRS | LDpred1.10FCVal | 0.356 | 0.287 | 0.069 | 1.86e-26 | 0.351 | 0.288 | 0.063 | 1.43e-28 |
| Height | pT+clump.10FCVal | LDpred1.10FCVal | 0.305 | 0.287 | 0.018 | 1.21e-05 | 0.299 | 0.288 | 0.010 | 2.05e-01 |
| Height | lassosum.PseudoVal | LDpred1.10FCVal | 0.281 | 0.287 | -0.005 | 2.61e-01 | 0.276 | 0.288 | -0.013 | 1.84e-01 |
| Height | LDpred2.10FCVal | LDpred1.10FCVal | 0.351 | 0.287 | 0.065 | 1.97e-143 | 0.343 | 0.288 | 0.055 | 5.93e-27 |
| Height | All.MultiPRS | LDpred1.10FCVal | 0.360 | 0.287 | 0.073 | 2.27e-29 | 0.356 | 0.288 | 0.067 | 7.25e-29 |
| Height | LDpred1.MultiPRS | LDpred1.10FCVal | 0.304 | 0.287 | 0.018 | 7.78e-03 | 0.303 | 0.288 | 0.014 | 1.50e-05 |
| Height | LDpred2.PseudoVal | LDpred1.10FCVal | 0.335 | 0.287 | 0.049 | 1.15e-139 | 0.326 | 0.288 | 0.038 | 2.46e-22 |
| Height | SBLUP.Inf | LDpred1.10FCVal | 0.295 | 0.287 | 0.009 | 1.86e-11 | 0.292 | 0.288 | 0.004 | 1.74e-01 |
| Height | pT+clump.MultiPRS | LDpred1.10FCVal | 0.320 | 0.287 | 0.033 | 5.53e-07 | 0.314 | 0.288 | 0.025 | 1.23e-04 |
| Height | LDpred2.Inf | LDpred1.10FCVal | 0.302 | 0.287 | 0.015 | 1.44e-30 | 0.299 | 0.288 | 0.011 | 5.01e-05 |
| Height | lassosum.MultiPRS | LDpred1.10FCVal | 0.349 | 0.287 | 0.063 | 5.99e-22 | 0.343 | 0.288 | 0.055 | 7.32e-21 |
| Height | LDpred1.10FCVal | LDpred1.10FCVal | 0.287 | 0.287 | 0.000 | 1.00e+00 | 0.288 | 0.288 | 0.000 | 1.00e+00 |
| Height | SBayesR.PseudoVal | LDpred1.10FCVal | 0.342 | 0.287 | 0.055 | 2.65e-154 | 0.335 | 0.288 | 0.047 | 2.46e-28 |
| Height | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.356 | 0.356 | 0.000 | 1.00e+00 | 0.351 | 0.351 | 0.000 | 1.00e+00 |
| Height | lassosum.10FCVal | LDpred2.MultiPRS | 0.347 | 0.356 | -0.009 | 1.51e-01 | 0.340 | 0.351 | -0.012 | 3.24e-05 |
| Height | All.MultiPRS | LDpred2.MultiPRS | 0.360 | 0.356 | 0.004 | 2.18e-07 | 0.356 | 0.351 | 0.004 | 4.46e-03 |
| Height | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.304 | 0.356 | -0.052 | 2.19e-15 | 0.303 | 0.351 | -0.049 | 6.12e-24 |
| Height | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.333 | 0.356 | -0.022 | 4.71e-04 | 0.324 | 0.351 | -0.027 | 2.22e-13 |
| Height | LDpred1.Inf | LDpred2.MultiPRS | 0.297 | 0.356 | -0.059 | 1.24e-19 | 0.295 | 0.351 | -0.056 | 9.59e-27 |
| Height | LDpred2.Inf | LDpred2.MultiPRS | 0.302 | 0.356 | -0.054 | 1.05e-16 | 0.299 | 0.351 | -0.052 | 1.14e-24 |
| Height | lassosum.MultiPRS | LDpred2.MultiPRS | 0.349 | 0.356 | -0.006 | 3.23e-01 | 0.343 | 0.351 | -0.009 | 1.96e-03 |
| Height | LDpred1.10FCVal | LDpred2.MultiPRS | 0.287 | 0.356 | -0.069 | 1.86e-26 | 0.288 | 0.351 | -0.063 | 1.43e-28 |
| Height | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.320 | 0.356 | -0.036 | 1.94e-08 | 0.314 | 0.351 | -0.038 | 1.20e-15 |
| Height | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.342 | 0.356 | -0.014 | 2.74e-02 | 0.335 | 0.351 | -0.016 | 7.51e-10 |
| Height | pT+clump.10FCVal | LDpred2.MultiPRS | 0.305 | 0.356 | -0.051 | 2.05e-15 | 0.299 | 0.351 | -0.053 | 2.82e-19 |
| Height | lassosum.PseudoVal | LDpred2.MultiPRS | 0.281 | 0.356 | -0.074 | 2.28e-30 | 0.276 | 0.351 | -0.076 | 5.30e-29 |
| Height | LDpred2.10FCVal | LDpred2.MultiPRS | 0.351 | 0.356 | -0.005 | 4.69e-01 | 0.343 | 0.351 | -0.008 | 4.00e-08 |
| Height | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.335 | 0.356 | -0.020 | 1.39e-03 | 0.326 | 0.351 | -0.025 | 1.76e-16 |
| Height | SBLUP.Inf | LDpred2.MultiPRS | 0.295 | 0.356 | -0.060 | 1.10e-20 | 0.292 | 0.351 | -0.059 | 1.79e-29 |
| Height | LDpred2.MultiPRS | LDpred2.10FCVal | 0.356 | 0.351 | 0.005 | 4.69e-01 | 0.351 | 0.343 | 0.008 | 4.00e-08 |
| Height | pT+clump.10FCVal | LDpred2.10FCVal | 0.305 | 0.351 | -0.047 | 1.14e-51 | 0.299 | 0.343 | -0.045 | 1.32e-12 |
| Height | lassosum.10FCVal | LDpred2.10FCVal | 0.347 | 0.351 | -0.005 | 3.60e-03 | 0.340 | 0.343 | -0.004 | 2.42e-01 |
| Height | All.MultiPRS | LDpred2.10FCVal | 0.360 | 0.351 | 0.008 | 1.84e-01 | 0.356 | 0.343 | 0.012 | 2.80e-08 |
| Height | LDpred1.MultiPRS | LDpred2.10FCVal | 0.304 | 0.351 | -0.047 | 4.72e-13 | 0.303 | 0.343 | -0.041 | 1.70e-21 |
| Height | LDpred2.PseudoVal | LDpred2.10FCVal | 0.335 | 0.351 | -0.016 | 4.79e-51 | 0.326 | 0.343 | -0.017 | 6.38e-16 |
| Height | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.333 | 0.351 | -0.018 | 3.89e-24 | 0.324 | 0.343 | -0.019 | 4.79e-08 |
| Height | LDpred1.Inf | LDpred2.10FCVal | 0.297 | 0.351 | -0.054 | 1.34e-120 | 0.295 | 0.343 | -0.048 | 3.39e-25 |
| Height | LDpred2.Inf | LDpred2.10FCVal | 0.302 | 0.351 | -0.049 | 6.45e-109 | 0.299 | 0.343 | -0.044 | 3.82e-23 |
| Height | lassosum.MultiPRS | LDpred2.10FCVal | 0.349 | 0.351 | -0.002 | 7.89e-01 | 0.343 | 0.343 | 0.000 | 8.89e-01 |
| Height | LDpred1.10FCVal | LDpred2.10FCVal | 0.287 | 0.351 | -0.065 | 1.97e-143 | 0.288 | 0.343 | -0.055 | 5.93e-27 |
| Height | pT+clump.MultiPRS | LDpred2.10FCVal | 0.320 | 0.351 | -0.032 | 9.45e-07 | 0.314 | 0.343 | -0.030 | 1.69e-09 |
| Height | SBayesR.PseudoVal | LDpred2.10FCVal | 0.342 | 0.351 | -0.009 | 1.27e-18 | 0.335 | 0.343 | -0.008 | 1.26e-04 |
| Height | lassosum.PseudoVal | LDpred2.10FCVal | 0.281 | 0.351 | -0.070 | 3.04e-77 | 0.276 | 0.343 | -0.068 | 3.38e-19 |
| Height | LDpred2.10FCVal | LDpred2.10FCVal | 0.351 | 0.351 | 0.000 | 1.00e+00 | 0.343 | 0.343 | 0.000 | 1.00e+00 |
| Height | SBLUP.Inf | LDpred2.10FCVal | 0.295 | 0.351 | -0.056 | 1.43e-129 | 0.292 | 0.343 | -0.051 | 1.05e-28 |
| Height | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.356 | 0.335 | 0.020 | 1.39e-03 | 0.351 | 0.326 | 0.025 | 1.76e-16 |
| Height | lassosum.10FCVal | LDpred2.PseudoVal | 0.347 | 0.335 | 0.011 | 7.05e-08 | 0.340 | 0.326 | 0.013 | 1.72e-03 |
| Height | All.MultiPRS | LDpred2.PseudoVal | 0.360 | 0.335 | 0.024 | 1.43e-04 | 0.356 | 0.326 | 0.029 | 2.29e-16 |
| Height | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.304 | 0.335 | -0.031 | 1.98e-06 | 0.303 | 0.326 | -0.024 | 5.93e-14 |
| Height | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.333 | 0.335 | -0.002 | 2.31e-01 | 0.324 | 0.326 | -0.002 | 4.78e-01 |
| Height | LDpred1.Inf | LDpred2.PseudoVal | 0.297 | 0.335 | -0.038 | 2.99e-119 | 0.295 | 0.326 | -0.031 | 3.20e-21 |
| Height | LDpred2.Inf | LDpred2.PseudoVal | 0.302 | 0.335 | -0.033 | 6.08e-109 | 0.299 | 0.326 | -0.027 | 3.36e-19 |
| Height | lassosum.MultiPRS | LDpred2.PseudoVal | 0.349 | 0.335 | 0.014 | 2.73e-02 | 0.343 | 0.326 | 0.017 | 5.78e-05 |
| Height | LDpred1.10FCVal | LDpred2.PseudoVal | 0.287 | 0.335 | -0.049 | 1.15e-139 | 0.288 | 0.326 | -0.038 | 2.46e-22 |
| Height | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.320 | 0.335 | -0.016 | 1.52e-02 | 0.314 | 0.326 | -0.013 | 2.36e-02 |
| Height | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.342 | 0.335 | 0.006 | 1.74e-10 | 0.335 | 0.326 | 0.009 | 1.32e-05 |
| Height | pT+clump.10FCVal | LDpred2.PseudoVal | 0.305 | 0.335 | -0.031 | 4.62e-18 | 0.299 | 0.326 | -0.027 | 1.40e-04 |
| Height | lassosum.PseudoVal | LDpred2.PseudoVal | 0.281 | 0.335 | -0.054 | 3.02e-37 | 0.276 | 0.326 | -0.051 | 2.86e-09 |
| Height | LDpred2.10FCVal | LDpred2.PseudoVal | 0.351 | 0.335 | 0.016 | 4.79e-51 | 0.343 | 0.326 | 0.017 | 6.38e-16 |
| Height | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.335 | 0.335 | 0.000 | 1.00e+00 | 0.326 | 0.326 | 0.000 | 1.00e+00 |
| Height | SBLUP.Inf | LDpred2.PseudoVal | 0.295 | 0.335 | -0.040 | 5.39e-142 | 0.292 | 0.326 | -0.034 | 1.72e-27 |
| Height | DBSLMM.PseudoVal | LDpred2.Inf | 0.333 | 0.302 | 0.031 | 5.87e-65 | 0.324 | 0.299 | 0.025 | 1.98e-11 |
| Height | LDpred2.MultiPRS | LDpred2.Inf | 0.356 | 0.302 | 0.054 | 1.05e-16 | 0.351 | 0.299 | 0.052 | 1.14e-24 |
| Height | pT+clump.10FCVal | LDpred2.Inf | 0.305 | 0.302 | 0.002 | 5.44e-01 | 0.299 | 0.299 | -0.001 | 9.45e-01 |
| Height | lassosum.PseudoVal | LDpred2.Inf | 0.281 | 0.302 | -0.021 | 6.88e-06 | 0.276 | 0.299 | -0.024 | 1.08e-02 |
| Height | lassosum.10FCVal | LDpred2.Inf | 0.347 | 0.302 | 0.045 | 1.11e-57 | 0.340 | 0.299 | 0.040 | 8.17e-13 |
| Height | All.MultiPRS | LDpred2.Inf | 0.360 | 0.302 | 0.058 | 5.40e-19 | 0.356 | 0.299 | 0.056 | 1.07e-25 |
| Height | LDpred1.MultiPRS | LDpred2.Inf | 0.304 | 0.302 | 0.002 | 7.41e-01 | 0.303 | 0.299 | 0.003 | 2.20e-01 |
| Height | LDpred2.PseudoVal | LDpred2.Inf | 0.335 | 0.302 | 0.033 | 6.08e-109 | 0.326 | 0.299 | 0.027 | 3.36e-19 |
| Height | SBLUP.Inf | LDpred2.Inf | 0.295 | 0.302 | -0.007 | 9.77e-28 | 0.292 | 0.299 | -0.007 | 2.22e-09 |
| Height | LDpred1.Inf | LDpred2.Inf | 0.297 | 0.302 | -0.005 | 1.76e-10 | 0.295 | 0.299 | -0.004 | 5.96e-03 |
| Height | LDpred2.Inf | LDpred2.Inf | 0.302 | 0.302 | 0.000 | 1.00e+00 | 0.299 | 0.299 | 0.000 | 1.00e+00 |
| Height | lassosum.MultiPRS | LDpred2.Inf | 0.349 | 0.302 | 0.047 | 3.02e-13 | 0.343 | 0.299 | 0.044 | 2.93e-16 |
| Height | LDpred1.10FCVal | LDpred2.Inf | 0.287 | 0.302 | -0.015 | 1.44e-30 | 0.288 | 0.299 | -0.011 | 5.01e-05 |
| Height | pT+clump.MultiPRS | LDpred2.Inf | 0.320 | 0.302 | 0.017 | 7.72e-03 | 0.314 | 0.299 | 0.014 | 2.19e-02 |
| Height | SBayesR.PseudoVal | LDpred2.Inf | 0.342 | 0.302 | 0.040 | 3.63e-112 | 0.335 | 0.299 | 0.036 | 6.09e-24 |
| Height | LDpred2.10FCVal | LDpred2.Inf | 0.351 | 0.302 | 0.049 | 6.45e-109 | 0.343 | 0.299 | 0.044 | 3.82e-23 |
| Height | All.MultiPRS | pT+clump.MultiPRS | 0.360 | 0.320 | 0.040 | 4.95e-10 | 0.356 | 0.314 | 0.042 | 2.27e-20 |
| Height | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.304 | 0.320 | -0.015 | 1.96e-02 | 0.303 | 0.314 | -0.011 | 6.22e-02 |
| Height | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.335 | 0.320 | 0.016 | 1.52e-02 | 0.326 | 0.314 | 0.013 | 2.36e-02 |
| Height | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.333 | 0.320 | 0.014 | 3.32e-02 | 0.324 | 0.314 | 0.010 | 3.26e-02 |
| Height | LDpred1.Inf | pT+clump.MultiPRS | 0.297 | 0.320 | -0.023 | 6.01e-04 | 0.295 | 0.314 | -0.019 | 2.57e-03 |
| Height | LDpred2.Inf | pT+clump.MultiPRS | 0.302 | 0.320 | -0.017 | 7.72e-03 | 0.299 | 0.314 | -0.014 | 2.19e-02 |
| Height | lassosum.MultiPRS | pT+clump.MultiPRS | 0.349 | 0.320 | 0.030 | 3.54e-06 | 0.343 | 0.314 | 0.029 | 3.64e-11 |
| Height | LDpred1.10FCVal | pT+clump.MultiPRS | 0.287 | 0.320 | -0.033 | 5.53e-07 | 0.288 | 0.314 | -0.025 | 1.23e-04 |
| Height | lassosum.10FCVal | pT+clump.MultiPRS | 0.347 | 0.320 | 0.027 | 2.82e-05 | 0.340 | 0.314 | 0.026 | 6.71e-09 |
| Height | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.356 | 0.320 | 0.036 | 1.94e-08 | 0.351 | 0.314 | 0.038 | 1.20e-15 |
| Height | pT+clump.10FCVal | pT+clump.MultiPRS | 0.305 | 0.320 | -0.015 | 2.17e-02 | 0.299 | 0.314 | -0.015 | 4.59e-07 |
| Height | lassosum.PseudoVal | pT+clump.MultiPRS | 0.281 | 0.320 | -0.038 | 6.27e-09 | 0.276 | 0.314 | -0.038 | 6.25e-08 |
| Height | LDpred2.10FCVal | pT+clump.MultiPRS | 0.351 | 0.320 | 0.032 | 9.45e-07 | 0.343 | 0.314 | 0.030 | 1.69e-09 |
| Height | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.342 | 0.320 | 0.022 | 6.23e-04 | 0.335 | 0.314 | 0.021 | 7.74e-06 |
| Height | SBLUP.Inf | pT+clump.MultiPRS | 0.295 | 0.320 | -0.024 | 2.27e-04 | 0.292 | 0.314 | -0.022 | 4.55e-04 |
| Height | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.320 | 0.320 | 0.000 | 1.00e+00 | 0.314 | 0.314 | 0.000 | 1.00e+00 |
| Height | lassosum.10FCVal | pT+clump.10FCVal | 0.347 | 0.305 | 0.042 | 1.75e-51 | 0.340 | 0.299 | 0.041 | 4.20e-13 |
| Height | All.MultiPRS | pT+clump.10FCVal | 0.360 | 0.305 | 0.055 | 1.31e-17 | 0.356 | 0.299 | 0.057 | 2.13e-26 |
| Height | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.333 | 0.305 | 0.029 | 1.05e-19 | 0.324 | 0.299 | 0.025 | 8.71e-05 |
| Height | LDpred1.Inf | pT+clump.10FCVal | 0.297 | 0.305 | -0.007 | 5.75e-02 | 0.295 | 0.299 | -0.004 | 6.35e-01 |
| Height | LDpred2.Inf | pT+clump.10FCVal | 0.302 | 0.305 | -0.002 | 5.44e-01 | 0.299 | 0.299 | 0.001 | 9.45e-01 |
| Height | lassosum.MultiPRS | pT+clump.10FCVal | 0.349 | 0.305 | 0.045 | 3.42e-12 | 0.343 | 0.299 | 0.044 | 5.98e-16 |
| Height | LDpred1.10FCVal | pT+clump.10FCVal | 0.287 | 0.305 | -0.018 | 1.21e-05 | 0.288 | 0.299 | -0.010 | 2.05e-01 |
| Height | pT+clump.MultiPRS | pT+clump.10FCVal | 0.320 | 0.305 | 0.015 | 2.17e-02 | 0.314 | 0.299 | 0.015 | 4.59e-07 |
| Height | LDpred2.MultiPRS | pT+clump.10FCVal | 0.356 | 0.305 | 0.051 | 2.05e-15 | 0.351 | 0.299 | 0.053 | 2.82e-19 |
| Height | pT+clump.10FCVal | pT+clump.10FCVal | 0.305 | 0.305 | 0.000 | 1.00e+00 | 0.299 | 0.299 | 0.000 | 1.00e+00 |
| Height | lassosum.PseudoVal | pT+clump.10FCVal | 0.281 | 0.305 | -0.023 | 7.20e-11 | 0.276 | 0.299 | -0.023 | 1.16e-03 |
| Height | LDpred2.10FCVal | pT+clump.10FCVal | 0.351 | 0.305 | 0.047 | 1.14e-51 | 0.343 | 0.299 | 0.045 | 1.32e-12 |
| Height | SBayesR.PseudoVal | pT+clump.10FCVal | 0.342 | 0.305 | 0.037 | 1.69e-31 | 0.335 | 0.299 | 0.036 | 1.46e-08 |
| Height | LDpred1.MultiPRS | pT+clump.10FCVal | 0.304 | 0.305 | 0.000 | 9.73e-01 | 0.303 | 0.299 | 0.004 | 6.31e-01 |
| Height | LDpred2.PseudoVal | pT+clump.10FCVal | 0.335 | 0.305 | 0.031 | 4.62e-18 | 0.326 | 0.299 | 0.027 | 1.40e-04 |
| Height | SBLUP.Inf | pT+clump.10FCVal | 0.295 | 0.305 | -0.009 | 2.11e-02 | 0.292 | 0.299 | -0.007 | 3.89e-01 |
| Height | LDpred1.Inf | SBayesR.PseudoVal | 0.297 | 0.342 | -0.045 | 4.07e-136 | 0.295 | 0.335 | -0.040 | 2.27e-28 |
| Height | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.333 | 0.342 | -0.008 | 4.55e-09 | 0.324 | 0.335 | -0.011 | 9.59e-05 |
| Height | lassosum.10FCVal | SBayesR.PseudoVal | 0.347 | 0.342 | 0.005 | 6.81e-03 | 0.340 | 0.335 | 0.005 | 2.16e-01 |
| Height | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.356 | 0.342 | 0.014 | 2.74e-02 | 0.351 | 0.335 | 0.016 | 7.51e-10 |
| Height | pT+clump.10FCVal | SBayesR.PseudoVal | 0.305 | 0.342 | -0.037 | 1.69e-31 | 0.299 | 0.335 | -0.036 | 1.46e-08 |
| Height | lassosum.PseudoVal | SBayesR.PseudoVal | 0.281 | 0.342 | -0.060 | 1.86e-52 | 0.276 | 0.335 | -0.059 | 7.44e-14 |
| Height | LDpred2.10FCVal | SBayesR.PseudoVal | 0.351 | 0.342 | 0.009 | 1.27e-18 | 0.343 | 0.335 | 0.008 | 1.26e-04 |
| Height | All.MultiPRS | SBayesR.PseudoVal | 0.360 | 0.342 | 0.018 | 4.95e-03 | 0.356 | 0.335 | 0.021 | 3.39e-11 |
| Height | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.304 | 0.342 | -0.037 | 9.02e-09 | 0.303 | 0.335 | -0.033 | 2.05e-21 |
| Height | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.335 | 0.342 | -0.006 | 1.74e-10 | 0.326 | 0.335 | -0.009 | 1.32e-05 |
| Height | SBLUP.Inf | SBayesR.PseudoVal | 0.295 | 0.342 | -0.046 | 1.17e-145 | 0.292 | 0.335 | -0.043 | 1.21e-32 |
| Height | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.320 | 0.342 | -0.022 | 6.23e-04 | 0.314 | 0.335 | -0.021 | 7.74e-06 |
| Height | LDpred2.Inf | SBayesR.PseudoVal | 0.302 | 0.342 | -0.040 | 3.63e-112 | 0.299 | 0.335 | -0.036 | 6.09e-24 |
| Height | lassosum.MultiPRS | SBayesR.PseudoVal | 0.349 | 0.342 | 0.008 | 2.25e-01 | 0.343 | 0.335 | 0.008 | 2.32e-02 |
| Height | LDpred1.10FCVal | SBayesR.PseudoVal | 0.287 | 0.342 | -0.055 | 2.65e-154 | 0.288 | 0.335 | -0.047 | 2.46e-28 |
| Height | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.342 | 0.342 | 0.000 | 1.00e+00 | 0.335 | 0.335 | 0.000 | 1.00e+00 |
| Height | All.MultiPRS | SBLUP.Inf | 0.360 | 0.295 | 0.064 | 3.07e-23 | 0.356 | 0.292 | 0.064 | 6.29e-30 |
| Height | LDpred1.Inf | SBLUP.Inf | 0.297 | 0.295 | 0.002 | 3.31e-02 | 0.295 | 0.292 | 0.003 | 4.65e-02 |
| Height | LDpred2.Inf | SBLUP.Inf | 0.302 | 0.295 | 0.007 | 9.77e-28 | 0.299 | 0.292 | 0.007 | 2.22e-09 |
| Height | DBSLMM.PseudoVal | SBLUP.Inf | 0.333 | 0.295 | 0.038 | 1.63e-95 | 0.324 | 0.292 | 0.032 | 2.32e-18 |
| Height | lassosum.10FCVal | SBLUP.Inf | 0.347 | 0.295 | 0.051 | 1.25e-73 | 0.340 | 0.292 | 0.048 | 7.40e-17 |
| Height | LDpred2.MultiPRS | SBLUP.Inf | 0.356 | 0.295 | 0.060 | 1.10e-20 | 0.351 | 0.292 | 0.059 | 1.79e-29 |
| Height | pT+clump.10FCVal | SBLUP.Inf | 0.305 | 0.295 | 0.009 | 2.11e-02 | 0.299 | 0.292 | 0.007 | 3.89e-01 |
| Height | lassosum.PseudoVal | SBLUP.Inf | 0.281 | 0.295 | -0.014 | 2.77e-03 | 0.276 | 0.292 | -0.016 | 8.48e-02 |
| Height | LDpred2.10FCVal | SBLUP.Inf | 0.351 | 0.295 | 0.056 | 1.43e-129 | 0.343 | 0.292 | 0.051 | 1.05e-28 |
| Height | SBayesR.PseudoVal | SBLUP.Inf | 0.342 | 0.295 | 0.046 | 1.17e-145 | 0.335 | 0.292 | 0.043 | 1.21e-32 |
| Height | LDpred1.MultiPRS | SBLUP.Inf | 0.304 | 0.295 | 0.009 | 1.77e-01 | 0.303 | 0.292 | 0.011 | 3.62e-05 |
| Height | LDpred2.PseudoVal | SBLUP.Inf | 0.335 | 0.295 | 0.040 | 5.39e-142 | 0.326 | 0.292 | 0.034 | 1.72e-27 |
| Height | SBLUP.Inf | SBLUP.Inf | 0.295 | 0.295 | 0.000 | 1.00e+00 | 0.292 | 0.292 | 0.000 | 1.00e+00 |
| Height | pT+clump.MultiPRS | SBLUP.Inf | 0.320 | 0.295 | 0.024 | 2.27e-04 | 0.314 | 0.292 | 0.022 | 4.55e-04 |
| Height | lassosum.MultiPRS | SBLUP.Inf | 0.349 | 0.295 | 0.054 | 8.80e-17 | 0.343 | 0.292 | 0.051 | 6.23e-21 |
| Height | LDpred1.10FCVal | SBLUP.Inf | 0.287 | 0.295 | -0.009 | 1.86e-11 | 0.288 | 0.292 | -0.004 | 1.74e-01 |
| BMI | lassosum.PseudoVal | All.MultiPRS | 0.211 | 0.308 | -0.096 | 1.02e-46 | 0.220 | 0.308 | -0.088 | 1.27e-30 |
| BMI | All.MultiPRS | All.MultiPRS | 0.308 | 0.308 | 0.000 | 1.00e+00 | 0.308 | 0.308 | 0.000 | 1.00e+00 |
| BMI | DBSLMM.PseudoVal | All.MultiPRS | 0.283 | 0.308 | -0.025 | 1.48e-04 | 0.283 | 0.308 | -0.025 | 1.61e-10 |
| BMI | LDpred1.Inf | All.MultiPRS | 0.272 | 0.308 | -0.035 | 9.24e-08 | 0.264 | 0.308 | -0.045 | 3.12e-21 |
| BMI | LDpred2.Inf | All.MultiPRS | 0.293 | 0.308 | -0.015 | 2.14e-02 | 0.287 | 0.308 | -0.021 | 1.94e-11 |
| BMI | lassosum.MultiPRS | All.MultiPRS | 0.305 | 0.308 | -0.003 | 6.71e-01 | 0.308 | 0.308 | -0.001 | 6.87e-01 |
| BMI | LDpred1.10FCVal | All.MultiPRS | 0.277 | 0.308 | -0.031 | 2.30e-06 | 0.273 | 0.308 | -0.036 | 8.40e-16 |
| BMI | pT+clump.MultiPRS | All.MultiPRS | 0.271 | 0.308 | -0.037 | 1.76e-08 | 0.275 | 0.308 | -0.033 | 2.01e-12 |
| BMI | LDpred2.MultiPRS | All.MultiPRS | 0.305 | 0.308 | -0.002 | 4.49e-04 | 0.303 | 0.308 | -0.005 | 1.92e-04 |
| BMI | pT+clump.10FCVal | All.MultiPRS | 0.248 | 0.308 | -0.060 | 2.54e-19 | 0.245 | 0.308 | -0.063 | 1.79e-26 |
| BMI | lassosum.10FCVal | All.MultiPRS | 0.300 | 0.308 | -0.007 | 2.73e-01 | 0.300 | 0.308 | -0.008 | 3.12e-04 |
| BMI | LDpred2.10FCVal | All.MultiPRS | 0.305 | 0.308 | -0.003 | 6.46e-01 | 0.302 | 0.308 | -0.006 | 3.17e-04 |
| BMI | SBayesR.PseudoVal | All.MultiPRS | 0.241 | 0.308 | -0.066 | 2.45e-23 | 0.250 | 0.308 | -0.058 | 6.71e-20 |
| BMI | LDpred1.MultiPRS | All.MultiPRS | 0.286 | 0.308 | -0.022 | 8.15e-04 | 0.282 | 0.308 | -0.026 | 6.09e-12 |
| BMI | LDpred2.PseudoVal | All.MultiPRS | 0.237 | 0.308 | -0.070 | 5.88e-26 | 0.243 | 0.308 | -0.065 | 3.77e-23 |
| BMI | SBLUP.Inf | All.MultiPRS | 0.292 | 0.308 | -0.015 | 1.97e-02 | 0.287 | 0.308 | -0.022 | 7.97e-12 |
| BMI | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.305 | 0.283 | 0.023 | 6.13e-04 | 0.303 | 0.283 | 0.020 | 5.43e-07 |
| BMI | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.248 | 0.283 | -0.035 | 1.05e-21 | 0.245 | 0.283 | -0.038 | 2.53e-07 |
| BMI | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.211 | 0.283 | -0.071 | 3.32e-107 | 0.220 | 0.283 | -0.063 | 5.43e-22 |
| BMI | All.MultiPRS | DBSLMM.PseudoVal | 0.308 | 0.283 | 0.025 | 1.48e-04 | 0.308 | 0.283 | 0.025 | 1.61e-10 |
| BMI | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.286 | 0.283 | 0.003 | 6.59e-01 | 0.282 | 0.283 | -0.001 | 8.64e-01 |
| BMI | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.237 | 0.283 | -0.045 | 4.54e-56 | 0.243 | 0.283 | -0.040 | 5.39e-12 |
| BMI | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.283 | 0.283 | 0.000 | 1.00e+00 | 0.283 | 0.283 | 0.000 | 1.00e+00 |
| BMI | LDpred1.Inf | DBSLMM.PseudoVal | 0.272 | 0.283 | -0.010 | 1.97e-04 | 0.264 | 0.283 | -0.019 | 7.19e-04 |
| BMI | LDpred2.Inf | DBSLMM.PseudoVal | 0.293 | 0.283 | 0.010 | 8.13e-05 | 0.287 | 0.283 | 0.004 | 4.05e-01 |
| BMI | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.305 | 0.283 | 0.022 | 7.65e-04 | 0.308 | 0.283 | 0.025 | 7.44e-09 |
| BMI | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.277 | 0.283 | -0.006 | 3.64e-03 | 0.273 | 0.283 | -0.010 | 1.70e-02 |
| BMI | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.271 | 0.283 | -0.012 | 6.85e-02 | 0.275 | 0.283 | -0.008 | 1.60e-01 |
| BMI | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.241 | 0.283 | -0.041 | 2.84e-48 | 0.250 | 0.283 | -0.033 | 6.28e-09 |
| BMI | lassosum.10FCVal | DBSLMM.PseudoVal | 0.300 | 0.283 | 0.018 | 1.91e-14 | 0.300 | 0.283 | 0.017 | 2.77e-04 |
| BMI | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.305 | 0.283 | 0.022 | 1.39e-29 | 0.302 | 0.283 | 0.019 | 1.21e-06 |
| BMI | SBLUP.Inf | DBSLMM.PseudoVal | 0.292 | 0.283 | 0.010 | 1.07e-04 | 0.287 | 0.283 | 0.004 | 4.76e-01 |
| BMI | All.MultiPRS | lassosum.MultiPRS | 0.308 | 0.305 | 0.003 | 6.71e-01 | 0.308 | 0.308 | 0.001 | 6.87e-01 |
| BMI | LDpred1.MultiPRS | lassosum.MultiPRS | 0.286 | 0.305 | -0.019 | 3.49e-03 | 0.282 | 0.308 | -0.026 | 6.42e-10 |
| BMI | LDpred1.Inf | lassosum.MultiPRS | 0.272 | 0.305 | -0.032 | 9.58e-07 | 0.264 | 0.308 | -0.044 | 4.32e-19 |
| BMI | LDpred2.Inf | lassosum.MultiPRS | 0.293 | 0.305 | -0.012 | 6.14e-02 | 0.287 | 0.308 | -0.021 | 5.70e-09 |
| BMI | lassosum.MultiPRS | lassosum.MultiPRS | 0.305 | 0.305 | 0.000 | 1.00e+00 | 0.308 | 0.308 | 0.000 | 1.00e+00 |
| BMI | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.283 | 0.305 | -0.022 | 7.65e-04 | 0.283 | 0.308 | -0.025 | 7.44e-09 |
| BMI | lassosum.PseudoVal | lassosum.MultiPRS | 0.211 | 0.305 | -0.093 | 2.47e-44 | 0.220 | 0.308 | -0.088 | 1.11e-30 |
| BMI | LDpred2.MultiPRS | lassosum.MultiPRS | 0.305 | 0.305 | 0.000 | 9.55e-01 | 0.303 | 0.308 | -0.004 | 7.62e-02 |
| BMI | pT+clump.10FCVal | lassosum.MultiPRS | 0.248 | 0.305 | -0.057 | 1.41e-17 | 0.245 | 0.308 | -0.063 | 2.38e-27 |
| BMI | lassosum.10FCVal | lassosum.MultiPRS | 0.300 | 0.305 | -0.004 | 5.04e-01 | 0.300 | 0.308 | -0.008 | 8.54e-06 |
| BMI | LDpred2.10FCVal | lassosum.MultiPRS | 0.305 | 0.305 | 0.000 | 9.74e-01 | 0.302 | 0.308 | -0.006 | 2.64e-02 |
| BMI | SBayesR.PseudoVal | lassosum.MultiPRS | 0.241 | 0.305 | -0.063 | 1.90e-21 | 0.250 | 0.308 | -0.058 | 3.34e-18 |
| BMI | LDpred2.PseudoVal | lassosum.MultiPRS | 0.237 | 0.305 | -0.067 | 5.08e-24 | 0.243 | 0.308 | -0.065 | 3.22e-21 |
| BMI | SBLUP.Inf | lassosum.MultiPRS | 0.292 | 0.305 | -0.013 | 5.72e-02 | 0.287 | 0.308 | -0.021 | 7.98e-10 |
| BMI | pT+clump.MultiPRS | lassosum.MultiPRS | 0.271 | 0.305 | -0.034 | 2.07e-07 | 0.275 | 0.308 | -0.033 | 1.45e-12 |
| BMI | LDpred1.10FCVal | lassosum.MultiPRS | 0.277 | 0.305 | -0.028 | 1.79e-05 | 0.273 | 0.308 | -0.035 | 7.43e-14 |
| BMI | All.MultiPRS | lassosum.PseudoVal | 0.308 | 0.211 | 0.096 | 1.02e-46 | 0.308 | 0.220 | 0.088 | 1.27e-30 |
| BMI | LDpred1.MultiPRS | lassosum.PseudoVal | 0.286 | 0.211 | 0.074 | 3.30e-28 | 0.282 | 0.220 | 0.062 | 1.50e-17 |
| BMI | LDpred2.PseudoVal | lassosum.PseudoVal | 0.237 | 0.211 | 0.026 | 3.92e-43 | 0.243 | 0.220 | 0.023 | 1.57e-09 |
| BMI | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.283 | 0.211 | 0.071 | 3.32e-107 | 0.283 | 0.220 | 0.063 | 5.43e-22 |
| BMI | LDpred1.Inf | lassosum.PseudoVal | 0.272 | 0.211 | 0.061 | 1.50e-43 | 0.264 | 0.220 | 0.044 | 1.01e-06 |
| BMI | LDpred2.Inf | lassosum.PseudoVal | 0.293 | 0.211 | 0.081 | 2.19e-67 | 0.287 | 0.220 | 0.067 | 1.33e-12 |
| BMI | lassosum.MultiPRS | lassosum.PseudoVal | 0.305 | 0.211 | 0.093 | 2.47e-44 | 0.308 | 0.220 | 0.088 | 1.11e-30 |
| BMI | LDpred1.10FCVal | lassosum.PseudoVal | 0.277 | 0.211 | 0.065 | 1.19e-103 | 0.273 | 0.220 | 0.053 | 9.37e-18 |
| BMI | lassosum.PseudoVal | lassosum.PseudoVal | 0.211 | 0.211 | 0.000 | 1.00e+00 | 0.220 | 0.220 | 0.000 | 1.00e+00 |
| BMI | LDpred2.MultiPRS | lassosum.PseudoVal | 0.305 | 0.211 | 0.094 | 1.87e-44 | 0.303 | 0.220 | 0.083 | 2.87e-27 |
| BMI | pT+clump.10FCVal | lassosum.PseudoVal | 0.248 | 0.211 | 0.037 | 7.00e-14 | 0.245 | 0.220 | 0.025 | 1.19e-02 |
| BMI | lassosum.10FCVal | lassosum.PseudoVal | 0.300 | 0.211 | 0.089 | 2.14e-102 | 0.300 | 0.220 | 0.080 | 1.55e-21 |
| BMI | LDpred2.10FCVal | lassosum.PseudoVal | 0.305 | 0.211 | 0.093 | 9.34e-130 | 0.302 | 0.220 | 0.082 | 7.58e-26 |
| BMI | SBayesR.PseudoVal | lassosum.PseudoVal | 0.241 | 0.211 | 0.030 | 2.87e-35 | 0.250 | 0.220 | 0.030 | 8.21e-10 |
| BMI | SBLUP.Inf | lassosum.PseudoVal | 0.292 | 0.211 | 0.081 | 5.69e-67 | 0.287 | 0.220 | 0.067 | 2.09e-12 |
| BMI | pT+clump.MultiPRS | lassosum.PseudoVal | 0.271 | 0.211 | 0.059 | 3.17e-18 | 0.275 | 0.220 | 0.055 | 2.02e-12 |
| BMI | All.MultiPRS | lassosum.10FCVal | 0.308 | 0.300 | 0.007 | 2.73e-01 | 0.308 | 0.300 | 0.008 | 3.12e-04 |
| BMI | LDpred1.MultiPRS | lassosum.10FCVal | 0.286 | 0.300 | -0.015 | 2.45e-02 | 0.282 | 0.300 | -0.018 | 1.45e-05 |
| BMI | LDpred1.Inf | lassosum.10FCVal | 0.272 | 0.300 | -0.028 | 5.61e-41 | 0.264 | 0.300 | -0.036 | 1.72e-17 |
| BMI | LDpred2.Inf | lassosum.10FCVal | 0.293 | 0.300 | -0.008 | 3.61e-09 | 0.287 | 0.300 | -0.013 | 1.83e-06 |
| BMI | lassosum.MultiPRS | lassosum.10FCVal | 0.305 | 0.300 | 0.004 | 5.04e-01 | 0.308 | 0.300 | 0.008 | 8.54e-06 |
| BMI | DBSLMM.PseudoVal | lassosum.10FCVal | 0.283 | 0.300 | -0.018 | 1.91e-14 | 0.283 | 0.300 | -0.017 | 2.77e-04 |
| BMI | lassosum.PseudoVal | lassosum.10FCVal | 0.211 | 0.300 | -0.089 | 2.14e-102 | 0.220 | 0.300 | -0.080 | 1.55e-21 |
| BMI | LDpred2.MultiPRS | lassosum.10FCVal | 0.305 | 0.300 | 0.005 | 4.67e-01 | 0.303 | 0.300 | 0.003 | 2.52e-01 |
| BMI | pT+clump.10FCVal | lassosum.10FCVal | 0.248 | 0.300 | -0.052 | 1.71e-69 | 0.245 | 0.300 | -0.055 | 2.11e-20 |
| BMI | lassosum.10FCVal | lassosum.10FCVal | 0.300 | 0.300 | 0.000 | 1.00e+00 | 0.300 | 0.300 | 0.000 | 1.00e+00 |
| BMI | LDpred2.10FCVal | lassosum.10FCVal | 0.305 | 0.300 | 0.004 | 1.09e-03 | 0.302 | 0.300 | 0.002 | 4.29e-01 |
| BMI | SBayesR.PseudoVal | lassosum.10FCVal | 0.241 | 0.300 | -0.059 | 1.17e-57 | 0.250 | 0.300 | -0.050 | 1.81e-11 |
| BMI | LDpred2.PseudoVal | lassosum.10FCVal | 0.237 | 0.300 | -0.063 | 5.24e-63 | 0.243 | 0.300 | -0.057 | 8.68e-14 |
| BMI | SBLUP.Inf | lassosum.10FCVal | 0.292 | 0.300 | -0.008 | 5.76e-10 | 0.287 | 0.300 | -0.013 | 2.68e-07 |
| BMI | pT+clump.MultiPRS | lassosum.10FCVal | 0.271 | 0.300 | -0.030 | 6.31e-06 | 0.275 | 0.300 | -0.025 | 9.80e-07 |
| BMI | LDpred1.10FCVal | lassosum.10FCVal | 0.277 | 0.300 | -0.024 | 1.72e-24 | 0.273 | 0.300 | -0.027 | 8.08e-09 |
| BMI | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.283 | 0.286 | -0.003 | 6.59e-01 | 0.283 | 0.282 | 0.001 | 8.64e-01 |
| BMI | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.305 | 0.286 | 0.020 | 2.88e-03 | 0.303 | 0.282 | 0.021 | 1.26e-08 |
| BMI | pT+clump.10FCVal | LDpred1.MultiPRS | 0.248 | 0.286 | -0.038 | 1.97e-08 | 0.245 | 0.282 | -0.037 | 9.20e-08 |
| BMI | lassosum.10FCVal | LDpred1.MultiPRS | 0.300 | 0.286 | 0.015 | 2.45e-02 | 0.300 | 0.282 | 0.018 | 1.45e-05 |
| BMI | lassosum.PseudoVal | LDpred1.MultiPRS | 0.211 | 0.286 | -0.074 | 3.30e-28 | 0.220 | 0.282 | -0.062 | 1.50e-17 |
| BMI | All.MultiPRS | LDpred1.MultiPRS | 0.308 | 0.286 | 0.022 | 8.15e-04 | 0.308 | 0.282 | 0.026 | 6.09e-12 |
| BMI | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.286 | 0.286 | 0.000 | 1.00e+00 | 0.282 | 0.282 | 0.000 | 1.00e+00 |
| BMI | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.237 | 0.286 | -0.048 | 7.07e-13 | 0.243 | 0.282 | -0.039 | 1.01e-09 |
| BMI | SBLUP.Inf | LDpred1.MultiPRS | 0.292 | 0.286 | 0.007 | 3.09e-01 | 0.287 | 0.282 | 0.004 | 3.15e-01 |
| BMI | LDpred1.Inf | LDpred1.MultiPRS | 0.272 | 0.286 | -0.013 | 4.78e-02 | 0.264 | 0.282 | -0.018 | 8.04e-11 |
| BMI | LDpred2.Inf | LDpred1.MultiPRS | 0.293 | 0.286 | 0.007 | 2.94e-01 | 0.287 | 0.282 | 0.005 | 2.53e-01 |
| BMI | lassosum.MultiPRS | LDpred1.MultiPRS | 0.305 | 0.286 | 0.019 | 3.49e-03 | 0.308 | 0.282 | 0.026 | 6.42e-10 |
| BMI | LDpred1.10FCVal | LDpred1.MultiPRS | 0.277 | 0.286 | -0.009 | 1.71e-01 | 0.273 | 0.282 | -0.010 | 4.92e-05 |
| BMI | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.271 | 0.286 | -0.015 | 2.34e-02 | 0.275 | 0.282 | -0.007 | 2.09e-01 |
| BMI | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.241 | 0.286 | -0.044 | 4.57e-11 | 0.250 | 0.282 | -0.032 | 8.25e-08 |
| BMI | LDpred2.10FCVal | LDpred1.MultiPRS | 0.305 | 0.286 | 0.019 | 3.89e-03 | 0.302 | 0.282 | 0.020 | 4.91e-08 |
| BMI | All.MultiPRS | LDpred1.Inf | 0.308 | 0.272 | 0.035 | 9.24e-08 | 0.308 | 0.264 | 0.045 | 3.12e-21 |
| BMI | LDpred1.Inf | LDpred1.Inf | 0.272 | 0.272 | 0.000 | 1.00e+00 | 0.264 | 0.264 | 0.000 | 1.00e+00 |
| BMI | LDpred2.Inf | LDpred1.Inf | 0.293 | 0.272 | 0.020 | 2.76e-26 | 0.287 | 0.264 | 0.023 | 1.46e-09 |
| BMI | DBSLMM.PseudoVal | LDpred1.Inf | 0.283 | 0.272 | 0.010 | 1.97e-04 | 0.283 | 0.264 | 0.019 | 7.19e-04 |
| BMI | lassosum.PseudoVal | LDpred1.Inf | 0.211 | 0.272 | -0.061 | 1.50e-43 | 0.220 | 0.264 | -0.044 | 1.01e-06 |
| BMI | LDpred2.MultiPRS | LDpred1.Inf | 0.305 | 0.272 | 0.033 | 6.60e-07 | 0.303 | 0.264 | 0.039 | 3.08e-18 |
| BMI | pT+clump.10FCVal | LDpred1.Inf | 0.248 | 0.272 | -0.024 | 4.18e-12 | 0.245 | 0.264 | -0.019 | 8.04e-03 |
| BMI | lassosum.10FCVal | LDpred1.Inf | 0.300 | 0.272 | 0.028 | 5.61e-41 | 0.300 | 0.264 | 0.036 | 1.72e-17 |
| BMI | LDpred2.10FCVal | LDpred1.Inf | 0.305 | 0.272 | 0.032 | 3.18e-53 | 0.302 | 0.264 | 0.038 | 5.11e-19 |
| BMI | SBayesR.PseudoVal | LDpred1.Inf | 0.241 | 0.272 | -0.031 | 1.84e-15 | 0.250 | 0.264 | -0.014 | 7.83e-02 |
| BMI | LDpred1.MultiPRS | LDpred1.Inf | 0.286 | 0.272 | 0.013 | 4.78e-02 | 0.282 | 0.264 | 0.018 | 8.04e-11 |
| BMI | LDpred2.PseudoVal | LDpred1.Inf | 0.237 | 0.272 | -0.035 | 1.03e-17 | 0.243 | 0.264 | -0.021 | 1.22e-02 |
| BMI | SBLUP.Inf | LDpred1.Inf | 0.292 | 0.272 | 0.020 | 2.88e-26 | 0.287 | 0.264 | 0.023 | 2.97e-09 |
| BMI | pT+clump.MultiPRS | LDpred1.Inf | 0.271 | 0.272 | -0.002 | 7.75e-01 | 0.275 | 0.264 | 0.011 | 7.03e-02 |
| BMI | lassosum.MultiPRS | LDpred1.Inf | 0.305 | 0.272 | 0.032 | 9.58e-07 | 0.308 | 0.264 | 0.044 | 4.32e-19 |
| BMI | LDpred1.10FCVal | LDpred1.Inf | 0.277 | 0.272 | 0.004 | 3.37e-02 | 0.273 | 0.264 | 0.009 | 2.44e-02 |
| BMI | LDpred1.Inf | LDpred1.10FCVal | 0.272 | 0.277 | -0.004 | 3.37e-02 | 0.264 | 0.273 | -0.009 | 2.44e-02 |
| BMI | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.283 | 0.277 | 0.006 | 3.64e-03 | 0.283 | 0.273 | 0.010 | 1.70e-02 |
| BMI | lassosum.PseudoVal | LDpred1.10FCVal | 0.211 | 0.277 | -0.065 | 1.19e-103 | 0.220 | 0.273 | -0.053 | 9.37e-18 |
| BMI | LDpred2.MultiPRS | LDpred1.10FCVal | 0.305 | 0.277 | 0.029 | 1.32e-05 | 0.303 | 0.273 | 0.031 | 4.66e-13 |
| BMI | pT+clump.10FCVal | LDpred1.10FCVal | 0.248 | 0.277 | -0.028 | 1.51e-14 | 0.245 | 0.273 | -0.028 | 2.23e-04 |
| BMI | lassosum.10FCVal | LDpred1.10FCVal | 0.300 | 0.277 | 0.024 | 1.72e-24 | 0.300 | 0.273 | 0.027 | 8.08e-09 |
| BMI | LDpred2.10FCVal | LDpred1.10FCVal | 0.305 | 0.277 | 0.028 | 2.51e-44 | 0.302 | 0.273 | 0.029 | 6.42e-13 |
| BMI | All.MultiPRS | LDpred1.10FCVal | 0.308 | 0.277 | 0.031 | 2.30e-06 | 0.308 | 0.273 | 0.036 | 8.40e-16 |
| BMI | LDpred1.MultiPRS | LDpred1.10FCVal | 0.286 | 0.277 | 0.009 | 1.71e-01 | 0.282 | 0.273 | 0.010 | 4.92e-05 |
| BMI | LDpred2.PseudoVal | LDpred1.10FCVal | 0.237 | 0.277 | -0.039 | 1.36e-43 | 0.243 | 0.273 | -0.030 | 2.53e-07 |
| BMI | SBLUP.Inf | LDpred1.10FCVal | 0.292 | 0.277 | 0.016 | 2.08e-09 | 0.287 | 0.273 | 0.014 | 9.42e-03 |
| BMI | pT+clump.MultiPRS | LDpred1.10FCVal | 0.271 | 0.277 | -0.006 | 3.71e-01 | 0.275 | 0.273 | 0.003 | 6.63e-01 |
| BMI | LDpred2.Inf | LDpred1.10FCVal | 0.293 | 0.277 | 0.016 | 1.24e-09 | 0.287 | 0.273 | 0.015 | 6.63e-03 |
| BMI | lassosum.MultiPRS | LDpred1.10FCVal | 0.305 | 0.277 | 0.028 | 1.79e-05 | 0.308 | 0.273 | 0.035 | 7.43e-14 |
| BMI | LDpred1.10FCVal | LDpred1.10FCVal | 0.277 | 0.277 | 0.000 | 1.00e+00 | 0.273 | 0.273 | 0.000 | 1.00e+00 |
| BMI | SBayesR.PseudoVal | LDpred1.10FCVal | 0.241 | 0.277 | -0.035 | 3.92e-35 | 0.250 | 0.273 | -0.023 | 7.29e-05 |
| BMI | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.305 | 0.305 | 0.000 | 1.00e+00 | 0.303 | 0.303 | 0.000 | 1.00e+00 |
| BMI | lassosum.PseudoVal | LDpred2.MultiPRS | 0.211 | 0.305 | -0.094 | 1.87e-44 | 0.220 | 0.303 | -0.083 | 2.87e-27 |
| BMI | All.MultiPRS | LDpred2.MultiPRS | 0.308 | 0.305 | 0.002 | 4.49e-04 | 0.308 | 0.303 | 0.005 | 1.92e-04 |
| BMI | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.286 | 0.305 | -0.020 | 2.88e-03 | 0.282 | 0.303 | -0.021 | 1.26e-08 |
| BMI | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.283 | 0.305 | -0.023 | 6.13e-04 | 0.283 | 0.303 | -0.020 | 5.43e-07 |
| BMI | LDpred1.Inf | LDpred2.MultiPRS | 0.272 | 0.305 | -0.033 | 6.60e-07 | 0.264 | 0.303 | -0.039 | 3.08e-18 |
| BMI | LDpred2.Inf | LDpred2.MultiPRS | 0.293 | 0.305 | -0.013 | 5.32e-02 | 0.287 | 0.303 | -0.016 | 1.80e-08 |
| BMI | lassosum.MultiPRS | LDpred2.MultiPRS | 0.305 | 0.305 | 0.000 | 9.55e-01 | 0.308 | 0.303 | 0.004 | 7.62e-02 |
| BMI | LDpred1.10FCVal | LDpred2.MultiPRS | 0.277 | 0.305 | -0.029 | 1.32e-05 | 0.273 | 0.303 | -0.031 | 4.66e-13 |
| BMI | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.271 | 0.305 | -0.035 | 1.40e-07 | 0.275 | 0.303 | -0.028 | 6.58e-09 |
| BMI | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.241 | 0.305 | -0.064 | 9.18e-22 | 0.250 | 0.303 | -0.053 | 6.15e-17 |
| BMI | pT+clump.10FCVal | LDpred2.MultiPRS | 0.248 | 0.305 | -0.057 | 6.83e-18 | 0.245 | 0.303 | -0.058 | 1.19e-21 |
| BMI | lassosum.10FCVal | LDpred2.MultiPRS | 0.300 | 0.305 | -0.005 | 4.67e-01 | 0.300 | 0.303 | -0.003 | 2.52e-01 |
| BMI | LDpred2.10FCVal | LDpred2.MultiPRS | 0.305 | 0.305 | -0.001 | 9.28e-01 | 0.302 | 0.303 | -0.001 | 2.68e-01 |
| BMI | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.237 | 0.305 | -0.068 | 2.67e-24 | 0.243 | 0.303 | -0.060 | 1.37e-20 |
| BMI | SBLUP.Inf | LDpred2.MultiPRS | 0.292 | 0.305 | -0.013 | 4.95e-02 | 0.287 | 0.303 | -0.017 | 2.75e-08 |
| BMI | LDpred2.MultiPRS | LDpred2.10FCVal | 0.305 | 0.305 | 0.001 | 9.28e-01 | 0.303 | 0.302 | 0.001 | 2.68e-01 |
| BMI | pT+clump.10FCVal | LDpred2.10FCVal | 0.248 | 0.305 | -0.057 | 5.53e-78 | 0.245 | 0.302 | -0.057 | 5.09e-21 |
| BMI | lassosum.PseudoVal | LDpred2.10FCVal | 0.211 | 0.305 | -0.093 | 9.34e-130 | 0.220 | 0.302 | -0.082 | 7.58e-26 |
| BMI | All.MultiPRS | LDpred2.10FCVal | 0.308 | 0.305 | 0.003 | 6.46e-01 | 0.308 | 0.302 | 0.006 | 3.17e-04 |
| BMI | LDpred1.MultiPRS | LDpred2.10FCVal | 0.286 | 0.305 | -0.019 | 3.89e-03 | 0.282 | 0.302 | -0.020 | 4.91e-08 |
| BMI | LDpred2.PseudoVal | LDpred2.10FCVal | 0.237 | 0.305 | -0.067 | 1.17e-91 | 0.243 | 0.302 | -0.059 | 2.06e-18 |
| BMI | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.283 | 0.305 | -0.022 | 1.39e-29 | 0.283 | 0.302 | -0.019 | 1.21e-06 |
| BMI | LDpred1.Inf | LDpred2.10FCVal | 0.272 | 0.305 | -0.032 | 3.18e-53 | 0.264 | 0.302 | -0.038 | 5.11e-19 |
| BMI | LDpred2.Inf | LDpred2.10FCVal | 0.293 | 0.305 | -0.012 | 9.74e-23 | 0.287 | 0.302 | -0.015 | 3.20e-09 |
| BMI | lassosum.MultiPRS | LDpred2.10FCVal | 0.305 | 0.305 | 0.000 | 9.74e-01 | 0.308 | 0.302 | 0.006 | 2.64e-02 |
| BMI | LDpred1.10FCVal | LDpred2.10FCVal | 0.277 | 0.305 | -0.028 | 2.51e-44 | 0.273 | 0.302 | -0.029 | 6.42e-13 |
| BMI | pT+clump.MultiPRS | LDpred2.10FCVal | 0.271 | 0.305 | -0.034 | 2.56e-07 | 0.275 | 0.302 | -0.027 | 3.13e-08 |
| BMI | SBayesR.PseudoVal | LDpred2.10FCVal | 0.241 | 0.305 | -0.063 | 9.47e-83 | 0.250 | 0.302 | -0.052 | 4.22e-15 |
| BMI | lassosum.10FCVal | LDpred2.10FCVal | 0.300 | 0.305 | -0.004 | 1.09e-03 | 0.300 | 0.302 | -0.002 | 4.29e-01 |
| BMI | LDpred2.10FCVal | LDpred2.10FCVal | 0.305 | 0.305 | 0.000 | 1.00e+00 | 0.302 | 0.302 | 0.000 | 1.00e+00 |
| BMI | SBLUP.Inf | LDpred2.10FCVal | 0.292 | 0.305 | -0.012 | 1.37e-20 | 0.287 | 0.302 | -0.016 | 8.32e-09 |
| BMI | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.305 | 0.237 | 0.068 | 2.67e-24 | 0.303 | 0.243 | 0.060 | 1.37e-20 |
| BMI | lassosum.PseudoVal | LDpred2.PseudoVal | 0.211 | 0.237 | -0.026 | 3.92e-43 | 0.220 | 0.243 | -0.023 | 1.57e-09 |
| BMI | All.MultiPRS | LDpred2.PseudoVal | 0.308 | 0.237 | 0.070 | 5.88e-26 | 0.308 | 0.243 | 0.065 | 3.77e-23 |
| BMI | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.286 | 0.237 | 0.048 | 7.07e-13 | 0.282 | 0.243 | 0.039 | 1.01e-09 |
| BMI | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.283 | 0.237 | 0.045 | 4.54e-56 | 0.283 | 0.243 | 0.040 | 5.39e-12 |
| BMI | LDpred1.Inf | LDpred2.PseudoVal | 0.272 | 0.237 | 0.035 | 1.03e-17 | 0.264 | 0.243 | 0.021 | 1.22e-02 |
| BMI | LDpred2.Inf | LDpred2.PseudoVal | 0.293 | 0.237 | 0.055 | 8.25e-40 | 0.287 | 0.243 | 0.044 | 1.90e-07 |
| BMI | lassosum.MultiPRS | LDpred2.PseudoVal | 0.305 | 0.237 | 0.067 | 5.08e-24 | 0.308 | 0.243 | 0.065 | 3.22e-21 |
| BMI | LDpred1.10FCVal | LDpred2.PseudoVal | 0.277 | 0.237 | 0.039 | 1.36e-43 | 0.273 | 0.243 | 0.030 | 2.53e-07 |
| BMI | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.271 | 0.237 | 0.033 | 9.45e-07 | 0.275 | 0.243 | 0.032 | 9.74e-06 |
| BMI | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.241 | 0.237 | 0.004 | 6.68e-03 | 0.250 | 0.243 | 0.007 | 2.04e-02 |
| BMI | pT+clump.10FCVal | LDpred2.PseudoVal | 0.248 | 0.237 | 0.011 | 2.04e-02 | 0.245 | 0.243 | 0.002 | 8.32e-01 |
| BMI | lassosum.10FCVal | LDpred2.PseudoVal | 0.300 | 0.237 | 0.063 | 5.24e-63 | 0.300 | 0.243 | 0.057 | 8.68e-14 |
| BMI | LDpred2.10FCVal | LDpred2.PseudoVal | 0.305 | 0.237 | 0.067 | 1.17e-91 | 0.302 | 0.243 | 0.059 | 2.06e-18 |
| BMI | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.237 | 0.237 | 0.000 | 1.00e+00 | 0.243 | 0.243 | 0.000 | 1.00e+00 |
| BMI | SBLUP.Inf | LDpred2.PseudoVal | 0.292 | 0.237 | 0.055 | 3.13e-39 | 0.287 | 0.243 | 0.044 | 3.08e-07 |
| BMI | DBSLMM.PseudoVal | LDpred2.Inf | 0.283 | 0.293 | -0.010 | 8.13e-05 | 0.283 | 0.287 | -0.004 | 4.05e-01 |
| BMI | LDpred2.MultiPRS | LDpred2.Inf | 0.305 | 0.293 | 0.013 | 5.32e-02 | 0.303 | 0.287 | 0.016 | 1.80e-08 |
| BMI | pT+clump.10FCVal | LDpred2.Inf | 0.248 | 0.293 | -0.045 | 5.82e-46 | 0.245 | 0.287 | -0.042 | 1.29e-11 |
| BMI | lassosum.10FCVal | LDpred2.Inf | 0.300 | 0.293 | 0.008 | 3.61e-09 | 0.300 | 0.287 | 0.013 | 1.83e-06 |
| BMI | lassosum.PseudoVal | LDpred2.Inf | 0.211 | 0.293 | -0.081 | 2.19e-67 | 0.220 | 0.287 | -0.067 | 1.33e-12 |
| BMI | All.MultiPRS | LDpred2.Inf | 0.308 | 0.293 | 0.015 | 2.14e-02 | 0.308 | 0.287 | 0.021 | 1.94e-11 |
| BMI | LDpred1.MultiPRS | LDpred2.Inf | 0.286 | 0.293 | -0.007 | 2.94e-01 | 0.282 | 0.287 | -0.005 | 2.53e-01 |
| BMI | LDpred2.PseudoVal | LDpred2.Inf | 0.237 | 0.293 | -0.055 | 8.25e-40 | 0.243 | 0.287 | -0.044 | 1.90e-07 |
| BMI | SBLUP.Inf | LDpred2.Inf | 0.292 | 0.293 | 0.000 | 6.16e-01 | 0.287 | 0.287 | -0.001 | 4.62e-01 |
| BMI | LDpred1.Inf | LDpred2.Inf | 0.272 | 0.293 | -0.020 | 2.76e-26 | 0.264 | 0.287 | -0.023 | 1.46e-09 |
| BMI | LDpred2.Inf | LDpred2.Inf | 0.293 | 0.293 | 0.000 | 1.00e+00 | 0.287 | 0.287 | 0.000 | 1.00e+00 |
| BMI | lassosum.MultiPRS | LDpred2.Inf | 0.305 | 0.293 | 0.012 | 6.14e-02 | 0.308 | 0.287 | 0.021 | 5.70e-09 |
| BMI | LDpred1.10FCVal | LDpred2.Inf | 0.277 | 0.293 | -0.016 | 1.24e-09 | 0.273 | 0.287 | -0.015 | 6.63e-03 |
| BMI | pT+clump.MultiPRS | LDpred2.Inf | 0.271 | 0.293 | -0.022 | 9.03e-04 | 0.275 | 0.287 | -0.012 | 3.12e-02 |
| BMI | SBayesR.PseudoVal | LDpred2.Inf | 0.241 | 0.293 | -0.051 | 2.55e-36 | 0.250 | 0.287 | -0.037 | 5.73e-06 |
| BMI | LDpred2.10FCVal | LDpred2.Inf | 0.305 | 0.293 | 0.012 | 9.74e-23 | 0.302 | 0.287 | 0.015 | 3.20e-09 |
| BMI | All.MultiPRS | pT+clump.MultiPRS | 0.308 | 0.271 | 0.037 | 1.76e-08 | 0.308 | 0.275 | 0.033 | 2.01e-12 |
| BMI | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.286 | 0.271 | 0.015 | 2.34e-02 | 0.282 | 0.275 | 0.007 | 2.09e-01 |
| BMI | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.237 | 0.271 | -0.033 | 9.45e-07 | 0.243 | 0.275 | -0.032 | 9.74e-06 |
| BMI | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.283 | 0.271 | 0.012 | 6.85e-02 | 0.283 | 0.275 | 0.008 | 1.60e-01 |
| BMI | LDpred1.Inf | pT+clump.MultiPRS | 0.272 | 0.271 | 0.002 | 7.75e-01 | 0.264 | 0.275 | -0.011 | 7.03e-02 |
| BMI | LDpred2.Inf | pT+clump.MultiPRS | 0.293 | 0.271 | 0.022 | 9.03e-04 | 0.287 | 0.275 | 0.012 | 3.12e-02 |
| BMI | lassosum.MultiPRS | pT+clump.MultiPRS | 0.305 | 0.271 | 0.034 | 2.07e-07 | 0.308 | 0.275 | 0.033 | 1.45e-12 |
| BMI | LDpred1.10FCVal | pT+clump.MultiPRS | 0.277 | 0.271 | 0.006 | 3.71e-01 | 0.273 | 0.275 | -0.003 | 6.63e-01 |
| BMI | lassosum.PseudoVal | pT+clump.MultiPRS | 0.211 | 0.271 | -0.059 | 3.17e-18 | 0.220 | 0.275 | -0.055 | 2.02e-12 |
| BMI | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.305 | 0.271 | 0.035 | 1.40e-07 | 0.303 | 0.275 | 0.028 | 6.58e-09 |
| BMI | pT+clump.10FCVal | pT+clump.MultiPRS | 0.248 | 0.271 | -0.023 | 7.86e-04 | 0.245 | 0.275 | -0.030 | 2.91e-14 |
| BMI | lassosum.10FCVal | pT+clump.MultiPRS | 0.300 | 0.271 | 0.030 | 6.31e-06 | 0.300 | 0.275 | 0.025 | 9.80e-07 |
| BMI | LDpred2.10FCVal | pT+clump.MultiPRS | 0.305 | 0.271 | 0.034 | 2.56e-07 | 0.302 | 0.275 | 0.027 | 3.13e-08 |
| BMI | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.241 | 0.271 | -0.029 | 1.61e-05 | 0.250 | 0.275 | -0.025 | 4.01e-04 |
| BMI | SBLUP.Inf | pT+clump.MultiPRS | 0.292 | 0.271 | 0.022 | 1.01e-03 | 0.287 | 0.275 | 0.011 | 3.64e-02 |
| BMI | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.271 | 0.271 | 0.000 | 1.00e+00 | 0.275 | 0.275 | 0.000 | 1.00e+00 |
| BMI | lassosum.PseudoVal | pT+clump.10FCVal | 0.211 | 0.248 | -0.037 | 7.00e-14 | 0.220 | 0.245 | -0.025 | 1.19e-02 |
| BMI | All.MultiPRS | pT+clump.10FCVal | 0.308 | 0.248 | 0.060 | 2.54e-19 | 0.308 | 0.245 | 0.063 | 1.79e-26 |
| BMI | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.283 | 0.248 | 0.035 | 1.05e-21 | 0.283 | 0.245 | 0.038 | 2.53e-07 |
| BMI | LDpred1.Inf | pT+clump.10FCVal | 0.272 | 0.248 | 0.024 | 4.18e-12 | 0.264 | 0.245 | 0.019 | 8.04e-03 |
| BMI | LDpred2.Inf | pT+clump.10FCVal | 0.293 | 0.248 | 0.045 | 5.82e-46 | 0.287 | 0.245 | 0.042 | 1.29e-11 |
| BMI | lassosum.MultiPRS | pT+clump.10FCVal | 0.305 | 0.248 | 0.057 | 1.41e-17 | 0.308 | 0.245 | 0.063 | 2.38e-27 |
| BMI | LDpred1.10FCVal | pT+clump.10FCVal | 0.277 | 0.248 | 0.028 | 1.51e-14 | 0.273 | 0.245 | 0.028 | 2.23e-04 |
| BMI | pT+clump.MultiPRS | pT+clump.10FCVal | 0.271 | 0.248 | 0.023 | 7.86e-04 | 0.275 | 0.245 | 0.030 | 2.91e-14 |
| BMI | LDpred2.MultiPRS | pT+clump.10FCVal | 0.305 | 0.248 | 0.057 | 6.83e-18 | 0.303 | 0.245 | 0.058 | 1.19e-21 |
| BMI | pT+clump.10FCVal | pT+clump.10FCVal | 0.248 | 0.248 | 0.000 | 1.00e+00 | 0.245 | 0.245 | 0.000 | 1.00e+00 |
| BMI | lassosum.10FCVal | pT+clump.10FCVal | 0.300 | 0.248 | 0.052 | 1.71e-69 | 0.300 | 0.245 | 0.055 | 2.11e-20 |
| BMI | LDpred2.10FCVal | pT+clump.10FCVal | 0.305 | 0.248 | 0.057 | 5.53e-78 | 0.302 | 0.245 | 0.057 | 5.09e-21 |
| BMI | SBayesR.PseudoVal | pT+clump.10FCVal | 0.241 | 0.248 | -0.007 | 1.41e-01 | 0.250 | 0.245 | 0.005 | 5.80e-01 |
| BMI | LDpred1.MultiPRS | pT+clump.10FCVal | 0.286 | 0.248 | 0.038 | 1.97e-08 | 0.282 | 0.245 | 0.037 | 9.20e-08 |
| BMI | LDpred2.PseudoVal | pT+clump.10FCVal | 0.237 | 0.248 | -0.011 | 2.04e-02 | 0.243 | 0.245 | -0.002 | 8.32e-01 |
| BMI | SBLUP.Inf | pT+clump.10FCVal | 0.292 | 0.248 | 0.044 | 2.35e-47 | 0.287 | 0.245 | 0.042 | 1.02e-11 |
| BMI | LDpred1.Inf | SBayesR.PseudoVal | 0.272 | 0.241 | 0.031 | 1.84e-15 | 0.264 | 0.250 | 0.014 | 7.83e-02 |
| BMI | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.283 | 0.241 | 0.041 | 2.84e-48 | 0.283 | 0.250 | 0.033 | 6.28e-09 |
| BMI | lassosum.PseudoVal | SBayesR.PseudoVal | 0.211 | 0.241 | -0.030 | 2.87e-35 | 0.220 | 0.250 | -0.030 | 8.21e-10 |
| BMI | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.305 | 0.241 | 0.064 | 9.18e-22 | 0.303 | 0.250 | 0.053 | 6.15e-17 |
| BMI | pT+clump.10FCVal | SBayesR.PseudoVal | 0.248 | 0.241 | 0.007 | 1.41e-01 | 0.245 | 0.250 | -0.005 | 5.80e-01 |
| BMI | lassosum.10FCVal | SBayesR.PseudoVal | 0.300 | 0.241 | 0.059 | 1.17e-57 | 0.300 | 0.250 | 0.050 | 1.81e-11 |
| BMI | LDpred2.10FCVal | SBayesR.PseudoVal | 0.305 | 0.241 | 0.063 | 9.47e-83 | 0.302 | 0.250 | 0.052 | 4.22e-15 |
| BMI | All.MultiPRS | SBayesR.PseudoVal | 0.308 | 0.241 | 0.066 | 2.45e-23 | 0.308 | 0.250 | 0.058 | 6.71e-20 |
| BMI | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.286 | 0.241 | 0.044 | 4.57e-11 | 0.282 | 0.250 | 0.032 | 8.25e-08 |
| BMI | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.237 | 0.241 | -0.004 | 6.68e-03 | 0.243 | 0.250 | -0.007 | 2.04e-02 |
| BMI | SBLUP.Inf | SBayesR.PseudoVal | 0.292 | 0.241 | 0.051 | 6.07e-36 | 0.287 | 0.250 | 0.037 | 8.33e-06 |
| BMI | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.271 | 0.241 | 0.029 | 1.61e-05 | 0.275 | 0.250 | 0.025 | 4.01e-04 |
| BMI | LDpred2.Inf | SBayesR.PseudoVal | 0.293 | 0.241 | 0.051 | 2.55e-36 | 0.287 | 0.250 | 0.037 | 5.73e-06 |
| BMI | lassosum.MultiPRS | SBayesR.PseudoVal | 0.305 | 0.241 | 0.063 | 1.90e-21 | 0.308 | 0.250 | 0.058 | 3.34e-18 |
| BMI | LDpred1.10FCVal | SBayesR.PseudoVal | 0.277 | 0.241 | 0.035 | 3.92e-35 | 0.273 | 0.250 | 0.023 | 7.29e-05 |
| BMI | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.241 | 0.241 | 0.000 | 1.00e+00 | 0.250 | 0.250 | 0.000 | 1.00e+00 |
| BMI | All.MultiPRS | SBLUP.Inf | 0.308 | 0.292 | 0.015 | 1.97e-02 | 0.308 | 0.287 | 0.022 | 7.97e-12 |
| BMI | LDpred1.Inf | SBLUP.Inf | 0.272 | 0.292 | -0.020 | 2.88e-26 | 0.264 | 0.287 | -0.023 | 2.97e-09 |
| BMI | LDpred2.Inf | SBLUP.Inf | 0.293 | 0.292 | 0.000 | 6.16e-01 | 0.287 | 0.287 | 0.001 | 4.62e-01 |
| BMI | DBSLMM.PseudoVal | SBLUP.Inf | 0.283 | 0.292 | -0.010 | 1.07e-04 | 0.283 | 0.287 | -0.004 | 4.76e-01 |
| BMI | lassosum.PseudoVal | SBLUP.Inf | 0.211 | 0.292 | -0.081 | 5.69e-67 | 0.220 | 0.287 | -0.067 | 2.09e-12 |
| BMI | LDpred2.MultiPRS | SBLUP.Inf | 0.305 | 0.292 | 0.013 | 4.95e-02 | 0.303 | 0.287 | 0.017 | 2.75e-08 |
| BMI | pT+clump.10FCVal | SBLUP.Inf | 0.248 | 0.292 | -0.044 | 2.35e-47 | 0.245 | 0.287 | -0.042 | 1.02e-11 |
| BMI | lassosum.10FCVal | SBLUP.Inf | 0.300 | 0.292 | 0.008 | 5.76e-10 | 0.300 | 0.287 | 0.013 | 2.68e-07 |
| BMI | LDpred2.10FCVal | SBLUP.Inf | 0.305 | 0.292 | 0.012 | 1.37e-20 | 0.302 | 0.287 | 0.016 | 8.32e-09 |
| BMI | SBayesR.PseudoVal | SBLUP.Inf | 0.241 | 0.292 | -0.051 | 6.07e-36 | 0.250 | 0.287 | -0.037 | 8.33e-06 |
| BMI | LDpred1.MultiPRS | SBLUP.Inf | 0.286 | 0.292 | -0.007 | 3.09e-01 | 0.282 | 0.287 | -0.004 | 3.15e-01 |
| BMI | LDpred2.PseudoVal | SBLUP.Inf | 0.237 | 0.292 | -0.055 | 3.13e-39 | 0.243 | 0.287 | -0.044 | 3.08e-07 |
| BMI | SBLUP.Inf | SBLUP.Inf | 0.292 | 0.292 | 0.000 | 1.00e+00 | 0.287 | 0.287 | 0.000 | 1.00e+00 |
| BMI | pT+clump.MultiPRS | SBLUP.Inf | 0.271 | 0.292 | -0.022 | 1.01e-03 | 0.275 | 0.287 | -0.011 | 3.64e-02 |
| BMI | lassosum.MultiPRS | SBLUP.Inf | 0.305 | 0.292 | 0.013 | 5.72e-02 | 0.308 | 0.287 | 0.021 | 7.98e-10 |
| BMI | LDpred1.10FCVal | SBLUP.Inf | 0.277 | 0.292 | -0.016 | 2.08e-09 | 0.273 | 0.287 | -0.014 | 9.42e-03 |
| T2D | lassosum.PseudoVal | All.MultiPRS | 0.203 | 0.243 | -0.040 | 5.82e-09 | 0.214 | 0.242 | -0.028 | 1.87e-06 |
| T2D | All.MultiPRS | All.MultiPRS | 0.243 | 0.243 | 0.000 | 1.00e+00 | 0.242 | 0.242 | 0.000 | 1.00e+00 |
| T2D | DBSLMM.PseudoVal | All.MultiPRS | 0.230 | 0.243 | -0.013 | 6.00e-02 | 0.232 | 0.242 | -0.010 | 7.36e-03 |
| T2D | LDpred1.Inf | All.MultiPRS | 0.185 | 0.243 | -0.057 | 4.33e-17 | 0.181 | 0.242 | -0.061 | 8.14e-19 |
| T2D | LDpred2.Inf | All.MultiPRS | 0.189 | 0.243 | -0.054 | 2.13e-15 | 0.185 | 0.242 | -0.057 | 1.11e-17 |
| T2D | lassosum.MultiPRS | All.MultiPRS | 0.239 | 0.243 | -0.004 | 5.83e-01 | 0.243 | 0.242 | 0.001 | 5.94e-01 |
| T2D | LDpred1.10FCVal | All.MultiPRS | 0.224 | 0.243 | -0.019 | 5.98e-03 | 0.224 | 0.242 | -0.018 | 3.29e-05 |
| T2D | pT+clump.MultiPRS | All.MultiPRS | 0.217 | 0.243 | -0.025 | 1.82e-04 | 0.221 | 0.242 | -0.021 | 1.67e-05 |
| T2D | LDpred2.MultiPRS | All.MultiPRS | 0.241 | 0.243 | -0.002 | 7.81e-01 | 0.243 | 0.242 | 0.001 | 7.13e-01 |
| T2D | pT+clump.10FCVal | All.MultiPRS | 0.170 | 0.243 | -0.073 | 2.70e-26 | 0.174 | 0.242 | -0.068 | 2.06e-18 |
| T2D | lassosum.10FCVal | All.MultiPRS | 0.213 | 0.243 | -0.030 | 1.04e-05 | 0.217 | 0.242 | -0.025 | 1.69e-06 |
| T2D | LDpred2.10FCVal | All.MultiPRS | 0.237 | 0.243 | -0.006 | 3.65e-01 | 0.239 | 0.242 | -0.003 | 3.19e-01 |
| T2D | SBayesR.PseudoVal | All.MultiPRS | 0.236 | 0.243 | -0.006 | 3.57e-01 | 0.236 | 0.242 | -0.006 | 5.79e-02 |
| T2D | LDpred1.MultiPRS | All.MultiPRS | 0.230 | 0.243 | -0.012 | 6.90e-02 | 0.234 | 0.242 | -0.008 | 3.31e-02 |
| T2D | LDpred2.PseudoVal | All.MultiPRS | 0.227 | 0.243 | -0.016 | 2.11e-02 | 0.229 | 0.242 | -0.012 | 2.79e-03 |
| T2D | SBLUP.Inf | All.MultiPRS | 0.189 | 0.243 | -0.054 | 1.82e-15 | 0.183 | 0.242 | -0.059 | 2.92e-18 |
| T2D | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.241 | 0.230 | 0.011 | 1.07e-01 | 0.243 | 0.232 | 0.011 | 2.33e-04 |
| T2D | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.170 | 0.230 | -0.060 | 6.44e-45 | 0.174 | 0.232 | -0.057 | 7.63e-12 |
| T2D | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.203 | 0.230 | -0.027 | 3.47e-14 | 0.214 | 0.232 | -0.018 | 9.11e-03 |
| T2D | All.MultiPRS | DBSLMM.PseudoVal | 0.243 | 0.230 | 0.013 | 6.00e-02 | 0.242 | 0.232 | 0.010 | 7.36e-03 |
| T2D | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.230 | 0.230 | 0.000 | 9.57e-01 | 0.234 | 0.232 | 0.002 | 5.96e-01 |
| T2D | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.227 | 0.230 | -0.003 | 2.77e-01 | 0.229 | 0.232 | -0.002 | 6.56e-01 |
| T2D | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.230 | 0.230 | 0.000 | 1.00e+00 | 0.232 | 0.232 | 0.000 | 1.00e+00 |
| T2D | LDpred1.Inf | DBSLMM.PseudoVal | 0.185 | 0.230 | -0.045 | 2.65e-52 | 0.181 | 0.232 | -0.051 | 1.44e-18 |
| T2D | LDpred2.Inf | DBSLMM.PseudoVal | 0.189 | 0.230 | -0.041 | 5.45e-47 | 0.185 | 0.232 | -0.047 | 9.65e-17 |
| T2D | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.239 | 0.230 | 0.009 | 1.83e-01 | 0.243 | 0.232 | 0.012 | 3.99e-04 |
| T2D | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.224 | 0.230 | -0.006 | 2.48e-03 | 0.224 | 0.232 | -0.008 | 3.52e-02 |
| T2D | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.217 | 0.230 | -0.013 | 6.40e-02 | 0.221 | 0.232 | -0.011 | 2.98e-02 |
| T2D | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.236 | 0.230 | 0.007 | 1.10e-03 | 0.236 | 0.232 | 0.004 | 3.01e-01 |
| T2D | lassosum.10FCVal | DBSLMM.PseudoVal | 0.213 | 0.230 | -0.017 | 4.22e-09 | 0.217 | 0.232 | -0.015 | 8.97e-03 |
| T2D | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.237 | 0.230 | 0.007 | 3.02e-04 | 0.239 | 0.232 | 0.007 | 5.31e-02 |
| T2D | SBLUP.Inf | DBSLMM.PseudoVal | 0.189 | 0.230 | -0.041 | 3.08e-47 | 0.183 | 0.232 | -0.048 | 1.53e-17 |
| T2D | All.MultiPRS | lassosum.MultiPRS | 0.243 | 0.239 | 0.004 | 5.83e-01 | 0.242 | 0.243 | -0.001 | 5.94e-01 |
| T2D | LDpred1.MultiPRS | lassosum.MultiPRS | 0.230 | 0.239 | -0.009 | 2.03e-01 | 0.234 | 0.243 | -0.009 | 2.50e-03 |
| T2D | LDpred1.Inf | lassosum.MultiPRS | 0.185 | 0.239 | -0.054 | 4.10e-15 | 0.181 | 0.243 | -0.062 | 9.28e-22 |
| T2D | LDpred2.Inf | lassosum.MultiPRS | 0.189 | 0.239 | -0.050 | 1.49e-13 | 0.185 | 0.243 | -0.059 | 1.37e-19 |
| T2D | lassosum.MultiPRS | lassosum.MultiPRS | 0.239 | 0.239 | 0.000 | 1.00e+00 | 0.243 | 0.243 | 0.000 | 1.00e+00 |
| T2D | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.230 | 0.239 | -0.009 | 1.83e-01 | 0.232 | 0.243 | -0.012 | 3.99e-04 |
| T2D | lassosum.PseudoVal | lassosum.MultiPRS | 0.203 | 0.239 | -0.036 | 1.54e-07 | 0.214 | 0.243 | -0.030 | 1.82e-08 |
| T2D | LDpred2.MultiPRS | lassosum.MultiPRS | 0.241 | 0.239 | 0.002 | 7.88e-01 | 0.243 | 0.243 | 0.000 | 8.38e-01 |
| T2D | pT+clump.10FCVal | lassosum.MultiPRS | 0.170 | 0.239 | -0.069 | 1.07e-23 | 0.174 | 0.243 | -0.069 | 4.63e-20 |
| T2D | lassosum.10FCVal | lassosum.MultiPRS | 0.213 | 0.239 | -0.026 | 1.18e-04 | 0.217 | 0.243 | -0.027 | 3.08e-09 |
| T2D | LDpred2.10FCVal | lassosum.MultiPRS | 0.237 | 0.239 | -0.002 | 7.21e-01 | 0.239 | 0.243 | -0.005 | 8.85e-02 |
| T2D | SBayesR.PseudoVal | lassosum.MultiPRS | 0.236 | 0.239 | -0.003 | 7.10e-01 | 0.236 | 0.243 | -0.008 | 8.48e-03 |
| T2D | LDpred2.PseudoVal | lassosum.MultiPRS | 0.227 | 0.239 | -0.012 | 7.91e-02 | 0.229 | 0.243 | -0.014 | 5.03e-04 |
| T2D | SBLUP.Inf | lassosum.MultiPRS | 0.189 | 0.239 | -0.051 | 1.33e-13 | 0.183 | 0.243 | -0.060 | 1.96e-20 |
| T2D | pT+clump.MultiPRS | lassosum.MultiPRS | 0.217 | 0.239 | -0.022 | 1.40e-03 | 0.221 | 0.243 | -0.022 | 7.21e-07 |
| T2D | LDpred1.10FCVal | lassosum.MultiPRS | 0.224 | 0.239 | -0.015 | 2.80e-02 | 0.224 | 0.243 | -0.020 | 2.11e-07 |
| T2D | All.MultiPRS | lassosum.PseudoVal | 0.243 | 0.203 | 0.040 | 5.82e-09 | 0.242 | 0.214 | 0.028 | 1.87e-06 |
| T2D | LDpred1.MultiPRS | lassosum.PseudoVal | 0.230 | 0.203 | 0.027 | 6.44e-05 | 0.234 | 0.214 | 0.020 | 3.44e-04 |
| T2D | LDpred2.PseudoVal | lassosum.PseudoVal | 0.227 | 0.203 | 0.024 | 1.02e-27 | 0.229 | 0.214 | 0.016 | 3.14e-04 |
| T2D | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.230 | 0.203 | 0.027 | 3.47e-14 | 0.232 | 0.214 | 0.018 | 9.11e-03 |
| T2D | LDpred1.Inf | lassosum.PseudoVal | 0.185 | 0.203 | -0.018 | 5.81e-04 | 0.181 | 0.214 | -0.032 | 1.35e-03 |
| T2D | LDpred2.Inf | lassosum.PseudoVal | 0.189 | 0.203 | -0.014 | 5.33e-03 | 0.185 | 0.214 | -0.029 | 4.83e-03 |
| T2D | lassosum.MultiPRS | lassosum.PseudoVal | 0.239 | 0.203 | 0.036 | 1.54e-07 | 0.243 | 0.214 | 0.030 | 1.82e-08 |
| T2D | LDpred1.10FCVal | lassosum.PseudoVal | 0.224 | 0.203 | 0.021 | 8.45e-09 | 0.224 | 0.214 | 0.010 | 1.58e-01 |
| T2D | lassosum.PseudoVal | lassosum.PseudoVal | 0.203 | 0.203 | 0.000 | 1.00e+00 | 0.214 | 0.214 | 0.000 | 1.00e+00 |
| T2D | LDpred2.MultiPRS | lassosum.PseudoVal | 0.241 | 0.203 | 0.038 | 2.49e-08 | 0.243 | 0.214 | 0.029 | 2.60e-08 |
| T2D | pT+clump.10FCVal | lassosum.PseudoVal | 0.170 | 0.203 | -0.033 | 3.11e-26 | 0.174 | 0.214 | -0.039 | 3.26e-10 |
| T2D | lassosum.10FCVal | lassosum.PseudoVal | 0.213 | 0.203 | 0.010 | 1.01e-03 | 0.217 | 0.214 | 0.003 | 6.04e-01 |
| T2D | LDpred2.10FCVal | lassosum.PseudoVal | 0.237 | 0.203 | 0.033 | 3.61e-35 | 0.239 | 0.214 | 0.025 | 2.79e-06 |
| T2D | SBayesR.PseudoVal | lassosum.PseudoVal | 0.236 | 0.203 | 0.033 | 3.89e-37 | 0.236 | 0.214 | 0.022 | 2.01e-05 |
| T2D | SBLUP.Inf | lassosum.PseudoVal | 0.189 | 0.203 | -0.015 | 4.99e-03 | 0.183 | 0.214 | -0.030 | 3.28e-03 |
| T2D | pT+clump.MultiPRS | lassosum.PseudoVal | 0.217 | 0.203 | 0.014 | 3.80e-02 | 0.221 | 0.214 | 0.008 | 2.06e-01 |
| T2D | All.MultiPRS | lassosum.10FCVal | 0.243 | 0.213 | 0.030 | 1.04e-05 | 0.242 | 0.217 | 0.025 | 1.69e-06 |
| T2D | LDpred1.MultiPRS | lassosum.10FCVal | 0.230 | 0.213 | 0.018 | 9.79e-03 | 0.234 | 0.217 | 0.017 | 2.43e-05 |
| T2D | LDpred1.Inf | lassosum.10FCVal | 0.185 | 0.213 | -0.027 | 2.14e-15 | 0.181 | 0.217 | -0.036 | 1.48e-07 |
| T2D | LDpred2.Inf | lassosum.10FCVal | 0.189 | 0.213 | -0.024 | 2.83e-12 | 0.185 | 0.217 | -0.032 | 2.44e-06 |
| T2D | lassosum.MultiPRS | lassosum.10FCVal | 0.239 | 0.213 | 0.026 | 1.18e-04 | 0.243 | 0.217 | 0.027 | 3.08e-09 |
| T2D | DBSLMM.PseudoVal | lassosum.10FCVal | 0.230 | 0.213 | 0.017 | 4.22e-09 | 0.232 | 0.217 | 0.015 | 8.97e-03 |
| T2D | lassosum.PseudoVal | lassosum.10FCVal | 0.203 | 0.213 | -0.010 | 1.01e-03 | 0.214 | 0.217 | -0.003 | 6.04e-01 |
| T2D | LDpred2.MultiPRS | lassosum.10FCVal | 0.241 | 0.213 | 0.028 | 3.22e-05 | 0.243 | 0.217 | 0.026 | 2.69e-09 |
| T2D | pT+clump.10FCVal | lassosum.10FCVal | 0.170 | 0.213 | -0.043 | 5.95e-23 | 0.174 | 0.217 | -0.042 | 1.01e-06 |
| T2D | lassosum.10FCVal | lassosum.10FCVal | 0.213 | 0.213 | 0.000 | 1.00e+00 | 0.217 | 0.217 | 0.000 | 1.00e+00 |
| T2D | LDpred2.10FCVal | lassosum.10FCVal | 0.237 | 0.213 | 0.024 | 6.42e-38 | 0.239 | 0.217 | 0.022 | 1.74e-09 |
| T2D | SBayesR.PseudoVal | lassosum.10FCVal | 0.236 | 0.213 | 0.024 | 5.06e-26 | 0.236 | 0.217 | 0.019 | 1.67e-05 |
| T2D | LDpred2.PseudoVal | lassosum.10FCVal | 0.227 | 0.213 | 0.014 | 8.89e-11 | 0.229 | 0.217 | 0.013 | 3.29e-03 |
| T2D | SBLUP.Inf | lassosum.10FCVal | 0.189 | 0.213 | -0.024 | 1.78e-12 | 0.183 | 0.217 | -0.033 | 8.70e-07 |
| T2D | pT+clump.MultiPRS | lassosum.10FCVal | 0.217 | 0.213 | 0.005 | 5.00e-01 | 0.221 | 0.217 | 0.005 | 4.70e-01 |
| T2D | LDpred1.10FCVal | lassosum.10FCVal | 0.224 | 0.213 | 0.011 | 6.76e-07 | 0.224 | 0.217 | 0.007 | 1.14e-01 |
| T2D | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.230 | 0.230 | 0.000 | 9.57e-01 | 0.232 | 0.234 | -0.002 | 5.96e-01 |
| T2D | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.241 | 0.230 | 0.010 | 1.18e-01 | 0.243 | 0.234 | 0.009 | 7.05e-04 |
| T2D | pT+clump.10FCVal | LDpred1.MultiPRS | 0.170 | 0.230 | -0.060 | 1.83e-18 | 0.174 | 0.234 | -0.060 | 3.51e-13 |
| T2D | lassosum.10FCVal | LDpred1.MultiPRS | 0.213 | 0.230 | -0.018 | 9.79e-03 | 0.217 | 0.234 | -0.017 | 2.43e-05 |
| T2D | lassosum.PseudoVal | LDpred1.MultiPRS | 0.203 | 0.230 | -0.027 | 6.44e-05 | 0.214 | 0.234 | -0.020 | 3.44e-04 |
| T2D | All.MultiPRS | LDpred1.MultiPRS | 0.243 | 0.230 | 0.012 | 6.90e-02 | 0.242 | 0.234 | 0.008 | 3.31e-02 |
| T2D | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.230 | 0.230 | 0.000 | 1.00e+00 | 0.234 | 0.234 | 0.000 | 1.00e+00 |
| T2D | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.227 | 0.230 | -0.003 | 6.29e-01 | 0.229 | 0.234 | -0.004 | 2.73e-01 |
| T2D | SBLUP.Inf | LDpred1.MultiPRS | 0.189 | 0.230 | -0.042 | 1.04e-09 | 0.183 | 0.234 | -0.050 | 4.23e-17 |
| T2D | LDpred1.Inf | LDpred1.MultiPRS | 0.185 | 0.230 | -0.045 | 5.72e-11 | 0.181 | 0.234 | -0.053 | 3.39e-20 |
| T2D | LDpred2.Inf | LDpred1.MultiPRS | 0.189 | 0.230 | -0.042 | 1.15e-09 | 0.185 | 0.234 | -0.049 | 2.63e-16 |
| T2D | lassosum.MultiPRS | LDpred1.MultiPRS | 0.239 | 0.230 | 0.009 | 2.03e-01 | 0.243 | 0.234 | 0.009 | 2.50e-03 |
| T2D | LDpred1.10FCVal | LDpred1.MultiPRS | 0.224 | 0.230 | -0.006 | 3.57e-01 | 0.224 | 0.234 | -0.010 | 1.12e-06 |
| T2D | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.217 | 0.230 | -0.013 | 5.50e-02 | 0.221 | 0.234 | -0.013 | 1.21e-02 |
| T2D | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.236 | 0.230 | 0.006 | 3.67e-01 | 0.236 | 0.234 | 0.002 | 5.14e-01 |
| T2D | LDpred2.10FCVal | LDpred1.MultiPRS | 0.237 | 0.230 | 0.006 | 3.58e-01 | 0.239 | 0.234 | 0.005 | 1.90e-02 |
| T2D | All.MultiPRS | LDpred1.Inf | 0.243 | 0.185 | 0.057 | 4.33e-17 | 0.242 | 0.181 | 0.061 | 8.14e-19 |
| T2D | LDpred1.Inf | LDpred1.Inf | 0.185 | 0.185 | 0.000 | 1.00e+00 | 0.181 | 0.181 | 0.000 | 1.00e+00 |
| T2D | LDpred2.Inf | LDpred1.Inf | 0.189 | 0.185 | 0.003 | 3.04e-03 | 0.185 | 0.181 | 0.004 | 8.52e-02 |
| T2D | DBSLMM.PseudoVal | LDpred1.Inf | 0.230 | 0.185 | 0.045 | 2.65e-52 | 0.232 | 0.181 | 0.051 | 1.44e-18 |
| T2D | lassosum.PseudoVal | LDpred1.Inf | 0.203 | 0.185 | 0.018 | 5.81e-04 | 0.214 | 0.181 | 0.032 | 1.35e-03 |
| T2D | LDpred2.MultiPRS | LDpred1.Inf | 0.241 | 0.185 | 0.055 | 3.81e-16 | 0.243 | 0.181 | 0.062 | 1.89e-22 |
| T2D | pT+clump.10FCVal | LDpred1.Inf | 0.170 | 0.185 | -0.015 | 6.96e-03 | 0.174 | 0.181 | -0.007 | 5.51e-01 |
| T2D | lassosum.10FCVal | LDpred1.Inf | 0.213 | 0.185 | 0.027 | 2.14e-15 | 0.217 | 0.181 | 0.036 | 1.48e-07 |
| T2D | LDpred2.10FCVal | LDpred1.Inf | 0.237 | 0.185 | 0.051 | 5.22e-64 | 0.239 | 0.181 | 0.058 | 2.30e-22 |
| T2D | SBayesR.PseudoVal | LDpred1.Inf | 0.236 | 0.185 | 0.051 | 2.02e-51 | 0.236 | 0.181 | 0.055 | 1.24e-16 |
| T2D | LDpred1.MultiPRS | LDpred1.Inf | 0.230 | 0.185 | 0.045 | 5.72e-11 | 0.234 | 0.181 | 0.053 | 3.39e-20 |
| T2D | LDpred2.PseudoVal | LDpred1.Inf | 0.227 | 0.185 | 0.042 | 3.17e-25 | 0.229 | 0.181 | 0.048 | 8.23e-10 |
| T2D | SBLUP.Inf | LDpred1.Inf | 0.189 | 0.185 | 0.003 | 2.68e-03 | 0.183 | 0.181 | 0.002 | 2.39e-01 |
| T2D | pT+clump.MultiPRS | LDpred1.Inf | 0.217 | 0.185 | 0.032 | 3.23e-06 | 0.221 | 0.181 | 0.040 | 7.86e-07 |
| T2D | lassosum.MultiPRS | LDpred1.Inf | 0.239 | 0.185 | 0.054 | 4.10e-15 | 0.243 | 0.181 | 0.062 | 9.28e-22 |
| T2D | LDpred1.10FCVal | LDpred1.Inf | 0.224 | 0.185 | 0.039 | 4.57e-90 | 0.224 | 0.181 | 0.043 | 5.83e-30 |
| T2D | LDpred1.Inf | LDpred1.10FCVal | 0.185 | 0.224 | -0.039 | 4.57e-90 | 0.181 | 0.224 | -0.043 | 5.83e-30 |
| T2D | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.230 | 0.224 | 0.006 | 2.48e-03 | 0.232 | 0.224 | 0.008 | 3.52e-02 |
| T2D | lassosum.PseudoVal | LDpred1.10FCVal | 0.203 | 0.224 | -0.021 | 8.45e-09 | 0.214 | 0.224 | -0.010 | 1.58e-01 |
| T2D | LDpred2.MultiPRS | LDpred1.10FCVal | 0.241 | 0.224 | 0.017 | 1.30e-02 | 0.243 | 0.224 | 0.019 | 2.14e-08 |
| T2D | pT+clump.10FCVal | LDpred1.10FCVal | 0.170 | 0.224 | -0.054 | 1.56e-31 | 0.174 | 0.224 | -0.049 | 7.56e-08 |
| T2D | lassosum.10FCVal | LDpred1.10FCVal | 0.213 | 0.224 | -0.011 | 6.76e-07 | 0.217 | 0.224 | -0.007 | 1.14e-01 |
| T2D | LDpred2.10FCVal | LDpred1.10FCVal | 0.237 | 0.224 | 0.013 | 3.78e-18 | 0.239 | 0.224 | 0.015 | 1.01e-07 |
| T2D | All.MultiPRS | LDpred1.10FCVal | 0.243 | 0.224 | 0.019 | 5.98e-03 | 0.242 | 0.224 | 0.018 | 3.29e-05 |
| T2D | LDpred1.MultiPRS | LDpred1.10FCVal | 0.230 | 0.224 | 0.006 | 3.57e-01 | 0.234 | 0.224 | 0.010 | 1.12e-06 |
| T2D | LDpred2.PseudoVal | LDpred1.10FCVal | 0.227 | 0.224 | 0.003 | 2.53e-01 | 0.229 | 0.224 | 0.006 | 2.67e-01 |
| T2D | SBLUP.Inf | LDpred1.10FCVal | 0.189 | 0.224 | -0.036 | 1.08e-60 | 0.183 | 0.224 | -0.040 | 8.61e-22 |
| T2D | pT+clump.MultiPRS | LDpred1.10FCVal | 0.217 | 0.224 | -0.007 | 3.22e-01 | 0.221 | 0.224 | -0.003 | 6.62e-01 |
| T2D | LDpred2.Inf | LDpred1.10FCVal | 0.189 | 0.224 | -0.035 | 1.00e-59 | 0.185 | 0.224 | -0.039 | 2.16e-20 |
| T2D | lassosum.MultiPRS | LDpred1.10FCVal | 0.239 | 0.224 | 0.015 | 2.80e-02 | 0.243 | 0.224 | 0.020 | 2.11e-07 |
| T2D | LDpred1.10FCVal | LDpred1.10FCVal | 0.224 | 0.224 | 0.000 | 1.00e+00 | 0.224 | 0.224 | 0.000 | 1.00e+00 |
| T2D | SBayesR.PseudoVal | LDpred1.10FCVal | 0.236 | 0.224 | 0.012 | 1.42e-10 | 0.236 | 0.224 | 0.012 | 1.39e-03 |
| T2D | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.241 | 0.241 | 0.000 | 1.00e+00 | 0.243 | 0.243 | 0.000 | 1.00e+00 |
| T2D | lassosum.PseudoVal | LDpred2.MultiPRS | 0.203 | 0.241 | -0.038 | 2.49e-08 | 0.214 | 0.243 | -0.029 | 2.60e-08 |
| T2D | All.MultiPRS | LDpred2.MultiPRS | 0.243 | 0.241 | 0.002 | 7.81e-01 | 0.242 | 0.243 | -0.001 | 7.13e-01 |
| T2D | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.230 | 0.241 | -0.010 | 1.18e-01 | 0.234 | 0.243 | -0.009 | 7.05e-04 |
| T2D | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.230 | 0.241 | -0.011 | 1.07e-01 | 0.232 | 0.243 | -0.011 | 2.33e-04 |
| T2D | LDpred1.Inf | LDpred2.MultiPRS | 0.185 | 0.241 | -0.055 | 3.81e-16 | 0.181 | 0.243 | -0.062 | 1.89e-22 |
| T2D | LDpred2.Inf | LDpred2.MultiPRS | 0.189 | 0.241 | -0.052 | 1.51e-14 | 0.185 | 0.243 | -0.058 | 1.41e-20 |
| T2D | lassosum.MultiPRS | LDpred2.MultiPRS | 0.239 | 0.241 | -0.002 | 7.88e-01 | 0.243 | 0.243 | 0.000 | 8.38e-01 |
| T2D | LDpred1.10FCVal | LDpred2.MultiPRS | 0.224 | 0.241 | -0.017 | 1.30e-02 | 0.224 | 0.243 | -0.019 | 2.14e-08 |
| T2D | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.217 | 0.241 | -0.024 | 5.53e-04 | 0.221 | 0.243 | -0.022 | 8.74e-07 |
| T2D | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.236 | 0.241 | -0.004 | 5.17e-01 | 0.236 | 0.243 | -0.007 | 8.31e-04 |
| T2D | pT+clump.10FCVal | LDpred2.MultiPRS | 0.170 | 0.241 | -0.071 | 3.09e-25 | 0.174 | 0.243 | -0.069 | 1.09e-20 |
| T2D | lassosum.10FCVal | LDpred2.MultiPRS | 0.213 | 0.241 | -0.028 | 3.22e-05 | 0.217 | 0.243 | -0.026 | 2.69e-09 |
| T2D | LDpred2.10FCVal | LDpred2.MultiPRS | 0.237 | 0.241 | -0.004 | 5.27e-01 | 0.239 | 0.243 | -0.004 | 1.19e-02 |
| T2D | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.227 | 0.241 | -0.014 | 4.08e-02 | 0.229 | 0.243 | -0.013 | 1.49e-05 |
| T2D | SBLUP.Inf | LDpred2.MultiPRS | 0.189 | 0.241 | -0.052 | 1.35e-14 | 0.183 | 0.243 | -0.059 | 4.66e-21 |
| T2D | LDpred2.MultiPRS | LDpred2.10FCVal | 0.241 | 0.237 | 0.004 | 5.27e-01 | 0.243 | 0.239 | 0.004 | 1.19e-02 |
| T2D | pT+clump.10FCVal | LDpred2.10FCVal | 0.170 | 0.237 | -0.066 | 5.34e-65 | 0.174 | 0.239 | -0.064 | 1.30e-16 |
| T2D | lassosum.PseudoVal | LDpred2.10FCVal | 0.203 | 0.237 | -0.033 | 3.61e-35 | 0.214 | 0.239 | -0.025 | 2.79e-06 |
| T2D | All.MultiPRS | LDpred2.10FCVal | 0.243 | 0.237 | 0.006 | 3.65e-01 | 0.242 | 0.239 | 0.003 | 3.19e-01 |
| T2D | LDpred1.MultiPRS | LDpred2.10FCVal | 0.230 | 0.237 | -0.006 | 3.58e-01 | 0.234 | 0.239 | -0.005 | 1.90e-02 |
| T2D | LDpred2.PseudoVal | LDpred2.10FCVal | 0.227 | 0.237 | -0.010 | 3.84e-10 | 0.229 | 0.239 | -0.009 | 2.01e-03 |
| T2D | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.230 | 0.237 | -0.007 | 3.02e-04 | 0.232 | 0.239 | -0.007 | 5.31e-02 |
| T2D | LDpred1.Inf | LDpred2.10FCVal | 0.185 | 0.237 | -0.051 | 5.22e-64 | 0.181 | 0.239 | -0.058 | 2.30e-22 |
| T2D | LDpred2.Inf | LDpred2.10FCVal | 0.189 | 0.237 | -0.048 | 7.18e-58 | 0.185 | 0.239 | -0.054 | 2.15e-20 |
| T2D | lassosum.MultiPRS | LDpred2.10FCVal | 0.239 | 0.237 | 0.002 | 7.21e-01 | 0.243 | 0.239 | 0.005 | 8.85e-02 |
| T2D | LDpred1.10FCVal | LDpred2.10FCVal | 0.224 | 0.237 | -0.013 | 3.78e-18 | 0.224 | 0.239 | -0.015 | 1.01e-07 |
| T2D | pT+clump.MultiPRS | LDpred2.10FCVal | 0.217 | 0.237 | -0.019 | 4.68e-03 | 0.221 | 0.239 | -0.018 | 2.73e-04 |
| T2D | SBayesR.PseudoVal | LDpred2.10FCVal | 0.236 | 0.237 | 0.000 | 9.30e-01 | 0.236 | 0.239 | -0.003 | 2.01e-01 |
| T2D | lassosum.10FCVal | LDpred2.10FCVal | 0.213 | 0.237 | -0.024 | 6.42e-38 | 0.217 | 0.239 | -0.022 | 1.74e-09 |
| T2D | LDpred2.10FCVal | LDpred2.10FCVal | 0.237 | 0.237 | 0.000 | 1.00e+00 | 0.239 | 0.239 | 0.000 | 1.00e+00 |
| T2D | SBLUP.Inf | LDpred2.10FCVal | 0.189 | 0.237 | -0.048 | 2.29e-57 | 0.183 | 0.239 | -0.055 | 5.49e-21 |
| T2D | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.241 | 0.227 | 0.014 | 4.08e-02 | 0.243 | 0.229 | 0.013 | 1.49e-05 |
| T2D | lassosum.PseudoVal | LDpred2.PseudoVal | 0.203 | 0.227 | -0.024 | 1.02e-27 | 0.214 | 0.229 | -0.016 | 3.14e-04 |
| T2D | All.MultiPRS | LDpred2.PseudoVal | 0.243 | 0.227 | 0.016 | 2.11e-02 | 0.242 | 0.229 | 0.012 | 2.79e-03 |
| T2D | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.230 | 0.227 | 0.003 | 6.29e-01 | 0.234 | 0.229 | 0.004 | 2.73e-01 |
| T2D | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.230 | 0.227 | 0.003 | 2.77e-01 | 0.232 | 0.229 | 0.002 | 6.56e-01 |
| T2D | LDpred1.Inf | LDpred2.PseudoVal | 0.185 | 0.227 | -0.042 | 3.17e-25 | 0.181 | 0.229 | -0.048 | 8.23e-10 |
| T2D | LDpred2.Inf | LDpred2.PseudoVal | 0.189 | 0.227 | -0.038 | 8.77e-22 | 0.185 | 0.229 | -0.045 | 1.20e-08 |
| T2D | lassosum.MultiPRS | LDpred2.PseudoVal | 0.239 | 0.227 | 0.012 | 7.91e-02 | 0.243 | 0.229 | 0.014 | 5.03e-04 |
| T2D | LDpred1.10FCVal | LDpred2.PseudoVal | 0.224 | 0.227 | -0.003 | 2.53e-01 | 0.224 | 0.229 | -0.006 | 2.67e-01 |
| T2D | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.217 | 0.227 | -0.010 | 1.54e-01 | 0.221 | 0.229 | -0.008 | 1.16e-01 |
| T2D | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.236 | 0.227 | 0.009 | 1.42e-11 | 0.236 | 0.229 | 0.006 | 2.19e-02 |
| T2D | pT+clump.10FCVal | LDpred2.PseudoVal | 0.170 | 0.227 | -0.057 | 5.05e-67 | 0.174 | 0.229 | -0.055 | 5.90e-17 |
| T2D | lassosum.10FCVal | LDpred2.PseudoVal | 0.213 | 0.227 | -0.014 | 8.89e-11 | 0.217 | 0.229 | -0.013 | 3.29e-03 |
| T2D | LDpred2.10FCVal | LDpred2.PseudoVal | 0.237 | 0.227 | 0.010 | 3.84e-10 | 0.239 | 0.229 | 0.009 | 2.01e-03 |
| T2D | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.227 | 0.227 | 0.000 | 1.00e+00 | 0.229 | 0.229 | 0.000 | 1.00e+00 |
| T2D | SBLUP.Inf | LDpred2.PseudoVal | 0.189 | 0.227 | -0.039 | 9.37e-22 | 0.183 | 0.229 | -0.046 | 5.36e-09 |
| T2D | DBSLMM.PseudoVal | LDpred2.Inf | 0.230 | 0.189 | 0.041 | 5.45e-47 | 0.232 | 0.185 | 0.047 | 9.65e-17 |
| T2D | LDpred2.MultiPRS | LDpred2.Inf | 0.241 | 0.189 | 0.052 | 1.51e-14 | 0.243 | 0.185 | 0.058 | 1.41e-20 |
| T2D | pT+clump.10FCVal | LDpred2.Inf | 0.170 | 0.189 | -0.019 | 1.20e-03 | 0.174 | 0.185 | -0.010 | 3.63e-01 |
| T2D | lassosum.10FCVal | LDpred2.Inf | 0.213 | 0.189 | 0.024 | 2.83e-12 | 0.217 | 0.185 | 0.032 | 2.44e-06 |
| T2D | lassosum.PseudoVal | LDpred2.Inf | 0.203 | 0.189 | 0.014 | 5.33e-03 | 0.214 | 0.185 | 0.029 | 4.83e-03 |
| T2D | All.MultiPRS | LDpred2.Inf | 0.243 | 0.189 | 0.054 | 2.13e-15 | 0.242 | 0.185 | 0.057 | 1.11e-17 |
| T2D | LDpred1.MultiPRS | LDpred2.Inf | 0.230 | 0.189 | 0.042 | 1.15e-09 | 0.234 | 0.185 | 0.049 | 2.63e-16 |
| T2D | LDpred2.PseudoVal | LDpred2.Inf | 0.227 | 0.189 | 0.038 | 8.77e-22 | 0.229 | 0.185 | 0.045 | 1.20e-08 |
| T2D | SBLUP.Inf | LDpred2.Inf | 0.189 | 0.189 | 0.000 | 7.93e-01 | 0.183 | 0.185 | -0.001 | 1.28e-01 |
| T2D | LDpred1.Inf | LDpred2.Inf | 0.185 | 0.189 | -0.003 | 3.04e-03 | 0.181 | 0.185 | -0.004 | 8.52e-02 |
| T2D | LDpred2.Inf | LDpred2.Inf | 0.189 | 0.189 | 0.000 | 1.00e+00 | 0.185 | 0.185 | 0.000 | 1.00e+00 |
| T2D | lassosum.MultiPRS | LDpred2.Inf | 0.239 | 0.189 | 0.050 | 1.49e-13 | 0.243 | 0.185 | 0.059 | 1.37e-19 |
| T2D | LDpred1.10FCVal | LDpred2.Inf | 0.224 | 0.189 | 0.035 | 1.00e-59 | 0.224 | 0.185 | 0.039 | 2.16e-20 |
| T2D | pT+clump.MultiPRS | LDpred2.Inf | 0.217 | 0.189 | 0.029 | 2.79e-05 | 0.221 | 0.185 | 0.037 | 7.10e-06 |
| T2D | SBayesR.PseudoVal | LDpred2.Inf | 0.236 | 0.189 | 0.048 | 2.26e-44 | 0.236 | 0.185 | 0.051 | 1.92e-14 |
| T2D | LDpred2.10FCVal | LDpred2.Inf | 0.237 | 0.189 | 0.048 | 7.18e-58 | 0.239 | 0.185 | 0.054 | 2.15e-20 |
| T2D | All.MultiPRS | pT+clump.MultiPRS | 0.243 | 0.217 | 0.025 | 1.82e-04 | 0.242 | 0.221 | 0.021 | 1.67e-05 |
| T2D | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.230 | 0.217 | 0.013 | 5.50e-02 | 0.234 | 0.221 | 0.013 | 1.21e-02 |
| T2D | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.227 | 0.217 | 0.010 | 1.54e-01 | 0.229 | 0.221 | 0.008 | 1.16e-01 |
| T2D | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.230 | 0.217 | 0.013 | 6.40e-02 | 0.232 | 0.221 | 0.011 | 2.98e-02 |
| T2D | LDpred1.Inf | pT+clump.MultiPRS | 0.185 | 0.217 | -0.032 | 3.23e-06 | 0.181 | 0.221 | -0.040 | 7.86e-07 |
| T2D | LDpred2.Inf | pT+clump.MultiPRS | 0.189 | 0.217 | -0.029 | 2.79e-05 | 0.185 | 0.221 | -0.037 | 7.10e-06 |
| T2D | lassosum.MultiPRS | pT+clump.MultiPRS | 0.239 | 0.217 | 0.022 | 1.40e-03 | 0.243 | 0.221 | 0.022 | 7.21e-07 |
| T2D | LDpred1.10FCVal | pT+clump.MultiPRS | 0.224 | 0.217 | 0.007 | 3.22e-01 | 0.224 | 0.221 | 0.003 | 6.62e-01 |
| T2D | lassosum.PseudoVal | pT+clump.MultiPRS | 0.203 | 0.217 | -0.014 | 3.80e-02 | 0.214 | 0.221 | -0.008 | 2.06e-01 |
| T2D | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.241 | 0.217 | 0.024 | 5.53e-04 | 0.243 | 0.221 | 0.022 | 8.74e-07 |
| T2D | pT+clump.10FCVal | pT+clump.MultiPRS | 0.170 | 0.217 | -0.047 | 8.48e-12 | 0.174 | 0.221 | -0.047 | 4.13e-13 |
| T2D | lassosum.10FCVal | pT+clump.MultiPRS | 0.213 | 0.217 | -0.005 | 5.00e-01 | 0.217 | 0.221 | -0.005 | 4.70e-01 |
| T2D | LDpred2.10FCVal | pT+clump.MultiPRS | 0.237 | 0.217 | 0.019 | 4.68e-03 | 0.239 | 0.221 | 0.018 | 2.73e-04 |
| T2D | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.236 | 0.217 | 0.019 | 4.96e-03 | 0.236 | 0.221 | 0.015 | 1.84e-03 |
| T2D | SBLUP.Inf | pT+clump.MultiPRS | 0.189 | 0.217 | -0.029 | 2.60e-05 | 0.183 | 0.221 | -0.038 | 3.43e-06 |
| T2D | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.217 | 0.217 | 0.000 | 1.00e+00 | 0.221 | 0.221 | 0.000 | 1.00e+00 |
| T2D | lassosum.PseudoVal | pT+clump.10FCVal | 0.203 | 0.170 | 0.033 | 3.11e-26 | 0.214 | 0.174 | 0.039 | 3.26e-10 |
| T2D | All.MultiPRS | pT+clump.10FCVal | 0.243 | 0.170 | 0.073 | 2.70e-26 | 0.242 | 0.174 | 0.068 | 2.06e-18 |
| T2D | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.230 | 0.170 | 0.060 | 6.44e-45 | 0.232 | 0.174 | 0.057 | 7.63e-12 |
| T2D | LDpred1.Inf | pT+clump.10FCVal | 0.185 | 0.170 | 0.015 | 6.96e-03 | 0.181 | 0.174 | 0.007 | 5.51e-01 |
| T2D | LDpred2.Inf | pT+clump.10FCVal | 0.189 | 0.170 | 0.019 | 1.20e-03 | 0.185 | 0.174 | 0.010 | 3.63e-01 |
| T2D | lassosum.MultiPRS | pT+clump.10FCVal | 0.239 | 0.170 | 0.069 | 1.07e-23 | 0.243 | 0.174 | 0.069 | 4.63e-20 |
| T2D | LDpred1.10FCVal | pT+clump.10FCVal | 0.224 | 0.170 | 0.054 | 1.56e-31 | 0.224 | 0.174 | 0.049 | 7.56e-08 |
| T2D | pT+clump.MultiPRS | pT+clump.10FCVal | 0.217 | 0.170 | 0.047 | 8.48e-12 | 0.221 | 0.174 | 0.047 | 4.13e-13 |
| T2D | LDpred2.MultiPRS | pT+clump.10FCVal | 0.241 | 0.170 | 0.071 | 3.09e-25 | 0.243 | 0.174 | 0.069 | 1.09e-20 |
| T2D | pT+clump.10FCVal | pT+clump.10FCVal | 0.170 | 0.170 | 0.000 | 1.00e+00 | 0.174 | 0.174 | 0.000 | 1.00e+00 |
| T2D | lassosum.10FCVal | pT+clump.10FCVal | 0.213 | 0.170 | 0.043 | 5.95e-23 | 0.217 | 0.174 | 0.042 | 1.01e-06 |
| T2D | LDpred2.10FCVal | pT+clump.10FCVal | 0.237 | 0.170 | 0.066 | 5.34e-65 | 0.239 | 0.174 | 0.064 | 1.30e-16 |
| T2D | SBayesR.PseudoVal | pT+clump.10FCVal | 0.236 | 0.170 | 0.066 | 2.78e-74 | 0.236 | 0.174 | 0.061 | 2.70e-17 |
| T2D | LDpred1.MultiPRS | pT+clump.10FCVal | 0.230 | 0.170 | 0.060 | 1.83e-18 | 0.234 | 0.174 | 0.060 | 3.51e-13 |
| T2D | LDpred2.PseudoVal | pT+clump.10FCVal | 0.227 | 0.170 | 0.057 | 5.05e-67 | 0.229 | 0.174 | 0.055 | 5.90e-17 |
| T2D | SBLUP.Inf | pT+clump.10FCVal | 0.189 | 0.170 | 0.018 | 1.27e-03 | 0.183 | 0.174 | 0.009 | 4.24e-01 |
| T2D | LDpred1.Inf | SBayesR.PseudoVal | 0.185 | 0.236 | -0.051 | 2.02e-51 | 0.181 | 0.236 | -0.055 | 1.24e-16 |
| T2D | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.230 | 0.236 | -0.007 | 1.10e-03 | 0.232 | 0.236 | -0.004 | 3.01e-01 |
| T2D | lassosum.PseudoVal | SBayesR.PseudoVal | 0.203 | 0.236 | -0.033 | 3.89e-37 | 0.214 | 0.236 | -0.022 | 2.01e-05 |
| T2D | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.241 | 0.236 | 0.004 | 5.17e-01 | 0.243 | 0.236 | 0.007 | 8.31e-04 |
| T2D | pT+clump.10FCVal | SBayesR.PseudoVal | 0.170 | 0.236 | -0.066 | 2.78e-74 | 0.174 | 0.236 | -0.061 | 2.70e-17 |
| T2D | lassosum.10FCVal | SBayesR.PseudoVal | 0.213 | 0.236 | -0.024 | 5.06e-26 | 0.217 | 0.236 | -0.019 | 1.67e-05 |
| T2D | LDpred2.10FCVal | SBayesR.PseudoVal | 0.237 | 0.236 | 0.000 | 9.30e-01 | 0.239 | 0.236 | 0.003 | 2.01e-01 |
| T2D | All.MultiPRS | SBayesR.PseudoVal | 0.243 | 0.236 | 0.006 | 3.57e-01 | 0.242 | 0.236 | 0.006 | 5.79e-02 |
| T2D | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.230 | 0.236 | -0.006 | 3.67e-01 | 0.234 | 0.236 | -0.002 | 5.14e-01 |
| T2D | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.227 | 0.236 | -0.009 | 1.42e-11 | 0.229 | 0.236 | -0.006 | 2.19e-02 |
| T2D | SBLUP.Inf | SBayesR.PseudoVal | 0.189 | 0.236 | -0.048 | 2.02e-44 | 0.183 | 0.236 | -0.052 | 5.16e-15 |
| T2D | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.217 | 0.236 | -0.019 | 4.96e-03 | 0.221 | 0.236 | -0.015 | 1.84e-03 |
| T2D | LDpred2.Inf | SBayesR.PseudoVal | 0.189 | 0.236 | -0.048 | 2.26e-44 | 0.185 | 0.236 | -0.051 | 1.92e-14 |
| T2D | lassosum.MultiPRS | SBayesR.PseudoVal | 0.239 | 0.236 | 0.003 | 7.10e-01 | 0.243 | 0.236 | 0.008 | 8.48e-03 |
| T2D | LDpred1.10FCVal | SBayesR.PseudoVal | 0.224 | 0.236 | -0.012 | 1.42e-10 | 0.224 | 0.236 | -0.012 | 1.39e-03 |
| T2D | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.236 | 0.236 | 0.000 | 1.00e+00 | 0.236 | 0.236 | 0.000 | 1.00e+00 |
| T2D | All.MultiPRS | SBLUP.Inf | 0.243 | 0.189 | 0.054 | 1.82e-15 | 0.242 | 0.183 | 0.059 | 2.92e-18 |
| T2D | LDpred1.Inf | SBLUP.Inf | 0.185 | 0.189 | -0.003 | 2.68e-03 | 0.181 | 0.183 | -0.002 | 2.39e-01 |
| T2D | LDpred2.Inf | SBLUP.Inf | 0.189 | 0.189 | 0.000 | 7.93e-01 | 0.185 | 0.183 | 0.001 | 1.28e-01 |
| T2D | DBSLMM.PseudoVal | SBLUP.Inf | 0.230 | 0.189 | 0.041 | 3.08e-47 | 0.232 | 0.183 | 0.048 | 1.53e-17 |
| T2D | lassosum.PseudoVal | SBLUP.Inf | 0.203 | 0.189 | 0.015 | 4.99e-03 | 0.214 | 0.183 | 0.030 | 3.28e-03 |
| T2D | LDpred2.MultiPRS | SBLUP.Inf | 0.241 | 0.189 | 0.052 | 1.35e-14 | 0.243 | 0.183 | 0.059 | 4.66e-21 |
| T2D | pT+clump.10FCVal | SBLUP.Inf | 0.170 | 0.189 | -0.018 | 1.27e-03 | 0.174 | 0.183 | -0.009 | 4.24e-01 |
| T2D | lassosum.10FCVal | SBLUP.Inf | 0.213 | 0.189 | 0.024 | 1.78e-12 | 0.217 | 0.183 | 0.033 | 8.70e-07 |
| T2D | LDpred2.10FCVal | SBLUP.Inf | 0.237 | 0.189 | 0.048 | 2.29e-57 | 0.239 | 0.183 | 0.055 | 5.49e-21 |
| T2D | SBayesR.PseudoVal | SBLUP.Inf | 0.236 | 0.189 | 0.048 | 2.02e-44 | 0.236 | 0.183 | 0.052 | 5.16e-15 |
| T2D | LDpred1.MultiPRS | SBLUP.Inf | 0.230 | 0.189 | 0.042 | 1.04e-09 | 0.234 | 0.183 | 0.050 | 4.23e-17 |
| T2D | LDpred2.PseudoVal | SBLUP.Inf | 0.227 | 0.189 | 0.039 | 9.37e-22 | 0.229 | 0.183 | 0.046 | 5.36e-09 |
| T2D | SBLUP.Inf | SBLUP.Inf | 0.189 | 0.189 | 0.000 | 1.00e+00 | 0.183 | 0.183 | 0.000 | 1.00e+00 |
| T2D | pT+clump.MultiPRS | SBLUP.Inf | 0.217 | 0.189 | 0.029 | 2.60e-05 | 0.221 | 0.183 | 0.038 | 3.43e-06 |
| T2D | lassosum.MultiPRS | SBLUP.Inf | 0.239 | 0.189 | 0.051 | 1.33e-13 | 0.243 | 0.183 | 0.060 | 1.96e-20 |
| T2D | LDpred1.10FCVal | SBLUP.Inf | 0.224 | 0.189 | 0.036 | 1.08e-60 | 0.224 | 0.183 | 0.040 | 8.61e-22 |
| CAD | lassosum.PseudoVal | All.MultiPRS | 0.162 | 0.190 | -0.028 | 1.84e-05 | 0.160 | 0.189 | -0.030 | 3.91e-08 |
| CAD | All.MultiPRS | All.MultiPRS | 0.190 | 0.190 | 0.000 | 1.00e+00 | 0.189 | 0.189 | 0.000 | 1.00e+00 |
| CAD | DBSLMM.PseudoVal | All.MultiPRS | 0.169 | 0.190 | -0.021 | 1.31e-03 | 0.163 | 0.189 | -0.027 | 5.79e-08 |
| CAD | LDpred1.Inf | All.MultiPRS | 0.147 | 0.190 | -0.043 | 1.02e-10 | 0.148 | 0.189 | -0.042 | 6.41e-10 |
| CAD | LDpred2.Inf | All.MultiPRS | 0.161 | 0.190 | -0.029 | 7.72e-06 | 0.157 | 0.189 | -0.033 | 8.81e-09 |
| CAD | lassosum.MultiPRS | All.MultiPRS | 0.187 | 0.190 | -0.004 | 5.90e-01 | 0.182 | 0.189 | -0.008 | 7.51e-04 |
| CAD | LDpred1.10FCVal | All.MultiPRS | 0.172 | 0.190 | -0.019 | 4.53e-03 | 0.171 | 0.189 | -0.018 | 5.44e-05 |
| CAD | pT+clump.MultiPRS | All.MultiPRS | 0.170 | 0.190 | -0.020 | 3.60e-03 | 0.166 | 0.189 | -0.024 | 4.50e-07 |
| CAD | LDpred2.MultiPRS | All.MultiPRS | 0.190 | 0.190 | -0.001 | 9.30e-01 | 0.188 | 0.189 | -0.001 | 5.05e-01 |
| CAD | pT+clump.10FCVal | All.MultiPRS | 0.139 | 0.190 | -0.052 | 8.34e-15 | 0.126 | 0.189 | -0.063 | 7.79e-18 |
| CAD | lassosum.10FCVal | All.MultiPRS | 0.174 | 0.190 | -0.016 | 1.54e-02 | 0.171 | 0.189 | -0.019 | 8.48e-06 |
| CAD | LDpred2.10FCVal | All.MultiPRS | 0.187 | 0.190 | -0.003 | 6.02e-01 | 0.184 | 0.189 | -0.005 | 2.23e-02 |
| CAD | SBayesR.PseudoVal | All.MultiPRS | 0.164 | 0.190 | -0.026 | 5.85e-05 | 0.170 | 0.189 | -0.019 | 2.94e-04 |
| CAD | LDpred1.MultiPRS | All.MultiPRS | 0.173 | 0.190 | -0.018 | 1.02e-02 | 0.173 | 0.189 | -0.017 | 1.44e-04 |
| CAD | LDpred2.PseudoVal | All.MultiPRS | 0.052 | 0.190 | -0.138 | 1.89e-90 | 0.060 | 0.189 | -0.130 | 9.45e-28 |
| CAD | SBLUP.Inf | All.MultiPRS | 0.161 | 0.190 | -0.030 | 6.73e-06 | 0.156 | 0.189 | -0.033 | 3.43e-09 |
| CAD | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.190 | 0.169 | 0.020 | 3.03e-03 | 0.188 | 0.163 | 0.025 | 8.10e-07 |
| CAD | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.139 | 0.169 | -0.031 | 2.73e-11 | 0.126 | 0.163 | -0.036 | 6.89e-05 |
| CAD | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.162 | 0.169 | -0.007 | 3.10e-02 | 0.160 | 0.163 | -0.003 | 6.13e-01 |
| CAD | All.MultiPRS | DBSLMM.PseudoVal | 0.190 | 0.169 | 0.021 | 1.31e-03 | 0.189 | 0.163 | 0.027 | 5.79e-08 |
| CAD | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.173 | 0.169 | 0.003 | 6.28e-01 | 0.173 | 0.163 | 0.010 | 7.89e-02 |
| CAD | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.052 | 0.169 | -0.117 | 1.05e-81 | 0.060 | 0.163 | -0.103 | 6.58e-17 |
| CAD | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.169 | 0.169 | 0.000 | 1.00e+00 | 0.163 | 0.163 | 0.000 | 1.00e+00 |
| CAD | LDpred1.Inf | DBSLMM.PseudoVal | 0.147 | 0.169 | -0.022 | 2.50e-07 | 0.148 | 0.163 | -0.015 | 7.54e-02 |
| CAD | LDpred2.Inf | DBSLMM.PseudoVal | 0.161 | 0.169 | -0.008 | 3.55e-02 | 0.157 | 0.163 | -0.006 | 4.59e-01 |
| CAD | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.187 | 0.169 | 0.017 | 1.20e-02 | 0.182 | 0.163 | 0.019 | 8.49e-04 |
| CAD | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.172 | 0.169 | 0.002 | 3.80e-01 | 0.171 | 0.163 | 0.008 | 1.33e-01 |
| CAD | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.170 | 0.169 | 0.001 | 8.92e-01 | 0.166 | 0.163 | 0.003 | 6.15e-01 |
| CAD | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.164 | 0.169 | -0.005 | 3.49e-02 | 0.170 | 0.163 | 0.007 | 1.66e-01 |
| CAD | lassosum.10FCVal | DBSLMM.PseudoVal | 0.174 | 0.169 | 0.005 | 1.51e-01 | 0.171 | 0.163 | 0.008 | 2.91e-01 |
| CAD | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.187 | 0.169 | 0.018 | 1.17e-11 | 0.184 | 0.163 | 0.021 | 5.78e-05 |
| CAD | SBLUP.Inf | DBSLMM.PseudoVal | 0.161 | 0.169 | -0.009 | 2.97e-02 | 0.156 | 0.163 | -0.007 | 3.88e-01 |
| CAD | All.MultiPRS | lassosum.MultiPRS | 0.190 | 0.187 | 0.004 | 5.90e-01 | 0.189 | 0.182 | 0.008 | 7.51e-04 |
| CAD | LDpred1.MultiPRS | lassosum.MultiPRS | 0.173 | 0.187 | -0.014 | 4.12e-02 | 0.173 | 0.182 | -0.009 | 5.62e-02 |
| CAD | LDpred1.Inf | lassosum.MultiPRS | 0.147 | 0.187 | -0.039 | 1.45e-08 | 0.148 | 0.182 | -0.034 | 5.03e-07 |
| CAD | LDpred2.Inf | lassosum.MultiPRS | 0.161 | 0.187 | -0.026 | 1.91e-04 | 0.157 | 0.182 | -0.025 | 1.16e-05 |
| CAD | lassosum.MultiPRS | lassosum.MultiPRS | 0.187 | 0.187 | 0.000 | 1.00e+00 | 0.182 | 0.182 | 0.000 | 1.00e+00 |
| CAD | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.169 | 0.187 | -0.017 | 1.20e-02 | 0.163 | 0.182 | -0.019 | 8.49e-04 |
| CAD | lassosum.PseudoVal | lassosum.MultiPRS | 0.162 | 0.187 | -0.024 | 4.12e-04 | 0.160 | 0.182 | -0.022 | 1.35e-05 |
| CAD | LDpred2.MultiPRS | lassosum.MultiPRS | 0.190 | 0.187 | 0.003 | 6.53e-01 | 0.188 | 0.182 | 0.007 | 1.93e-02 |
| CAD | pT+clump.10FCVal | lassosum.MultiPRS | 0.139 | 0.187 | -0.048 | 5.13e-12 | 0.126 | 0.182 | -0.055 | 1.76e-13 |
| CAD | lassosum.10FCVal | lassosum.MultiPRS | 0.174 | 0.187 | -0.012 | 7.79e-02 | 0.171 | 0.182 | -0.011 | 1.16e-03 |
| CAD | LDpred2.10FCVal | lassosum.MultiPRS | 0.187 | 0.187 | 0.000 | 9.65e-01 | 0.184 | 0.182 | 0.002 | 4.28e-01 |
| CAD | SBayesR.PseudoVal | lassosum.MultiPRS | 0.164 | 0.187 | -0.023 | 1.02e-03 | 0.170 | 0.182 | -0.012 | 4.36e-02 |
| CAD | LDpred2.PseudoVal | lassosum.MultiPRS | 0.052 | 0.187 | -0.135 | 2.19e-82 | 0.060 | 0.182 | -0.122 | 4.64e-23 |
| CAD | SBLUP.Inf | lassosum.MultiPRS | 0.161 | 0.187 | -0.026 | 1.70e-04 | 0.156 | 0.182 | -0.026 | 4.54e-06 |
| CAD | pT+clump.MultiPRS | lassosum.MultiPRS | 0.170 | 0.187 | -0.016 | 1.74e-02 | 0.166 | 0.182 | -0.016 | 1.48e-03 |
| CAD | LDpred1.10FCVal | lassosum.MultiPRS | 0.172 | 0.187 | -0.015 | 3.09e-02 | 0.171 | 0.182 | -0.010 | 2.96e-02 |
| CAD | All.MultiPRS | lassosum.PseudoVal | 0.190 | 0.162 | 0.028 | 1.84e-05 | 0.189 | 0.160 | 0.030 | 3.91e-08 |
| CAD | LDpred1.MultiPRS | lassosum.PseudoVal | 0.173 | 0.162 | 0.010 | 1.33e-01 | 0.173 | 0.160 | 0.013 | 1.96e-02 |
| CAD | LDpred2.PseudoVal | lassosum.PseudoVal | 0.052 | 0.162 | -0.110 | 2.80e-70 | 0.060 | 0.160 | -0.100 | 1.07e-15 |
| CAD | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.169 | 0.162 | 0.007 | 3.10e-02 | 0.163 | 0.160 | 0.003 | 6.13e-01 |
| CAD | LDpred1.Inf | lassosum.PseudoVal | 0.147 | 0.162 | -0.015 | 1.76e-03 | 0.148 | 0.160 | -0.012 | 2.16e-01 |
| CAD | LDpred2.Inf | lassosum.PseudoVal | 0.161 | 0.162 | -0.001 | 7.73e-01 | 0.157 | 0.160 | -0.003 | 7.76e-01 |
| CAD | lassosum.MultiPRS | lassosum.PseudoVal | 0.187 | 0.162 | 0.024 | 4.12e-04 | 0.182 | 0.160 | 0.022 | 1.35e-05 |
| CAD | LDpred1.10FCVal | lassosum.PseudoVal | 0.172 | 0.162 | 0.010 | 5.63e-04 | 0.171 | 0.160 | 0.012 | 3.17e-02 |
| CAD | lassosum.PseudoVal | lassosum.PseudoVal | 0.162 | 0.162 | 0.000 | 1.00e+00 | 0.160 | 0.160 | 0.000 | 1.00e+00 |
| CAD | LDpred2.MultiPRS | lassosum.PseudoVal | 0.190 | 0.162 | 0.028 | 6.60e-05 | 0.188 | 0.160 | 0.029 | 3.29e-08 |
| CAD | pT+clump.10FCVal | lassosum.PseudoVal | 0.139 | 0.162 | -0.023 | 5.06e-12 | 0.126 | 0.160 | -0.033 | 1.17e-06 |
| CAD | lassosum.10FCVal | lassosum.PseudoVal | 0.174 | 0.162 | 0.012 | 1.75e-04 | 0.171 | 0.160 | 0.011 | 9.49e-02 |
| CAD | LDpred2.10FCVal | lassosum.PseudoVal | 0.187 | 0.162 | 0.025 | 1.34e-17 | 0.184 | 0.160 | 0.024 | 2.54e-05 |
| CAD | SBayesR.PseudoVal | lassosum.PseudoVal | 0.164 | 0.162 | 0.002 | 4.93e-01 | 0.170 | 0.160 | 0.010 | 3.58e-02 |
| CAD | SBLUP.Inf | lassosum.PseudoVal | 0.161 | 0.162 | -0.002 | 7.38e-01 | 0.156 | 0.160 | -0.004 | 7.01e-01 |
| CAD | pT+clump.MultiPRS | lassosum.PseudoVal | 0.170 | 0.162 | 0.008 | 2.49e-01 | 0.166 | 0.160 | 0.006 | 3.14e-01 |
| CAD | All.MultiPRS | lassosum.10FCVal | 0.190 | 0.174 | 0.016 | 1.54e-02 | 0.189 | 0.171 | 0.019 | 8.48e-06 |
| CAD | LDpred1.MultiPRS | lassosum.10FCVal | 0.173 | 0.174 | -0.002 | 7.85e-01 | 0.173 | 0.171 | 0.002 | 6.81e-01 |
| CAD | LDpred1.Inf | lassosum.10FCVal | 0.147 | 0.174 | -0.027 | 1.52e-18 | 0.148 | 0.171 | -0.023 | 2.10e-04 |
| CAD | LDpred2.Inf | lassosum.10FCVal | 0.161 | 0.174 | -0.014 | 9.99e-09 | 0.157 | 0.171 | -0.014 | 3.91e-03 |
| CAD | lassosum.MultiPRS | lassosum.10FCVal | 0.187 | 0.174 | 0.012 | 7.79e-02 | 0.182 | 0.171 | 0.011 | 1.16e-03 |
| CAD | DBSLMM.PseudoVal | lassosum.10FCVal | 0.169 | 0.174 | -0.005 | 1.51e-01 | 0.163 | 0.171 | -0.008 | 2.91e-01 |
| CAD | lassosum.PseudoVal | lassosum.10FCVal | 0.162 | 0.174 | -0.012 | 1.75e-04 | 0.160 | 0.171 | -0.011 | 9.49e-02 |
| CAD | LDpred2.MultiPRS | lassosum.10FCVal | 0.190 | 0.174 | 0.015 | 2.68e-02 | 0.188 | 0.171 | 0.018 | 8.01e-06 |
| CAD | pT+clump.10FCVal | lassosum.10FCVal | 0.139 | 0.174 | -0.036 | 5.49e-22 | 0.126 | 0.171 | -0.044 | 2.29e-09 |
| CAD | lassosum.10FCVal | lassosum.10FCVal | 0.174 | 0.174 | 0.000 | 1.00e+00 | 0.171 | 0.171 | 0.000 | 1.00e+00 |
| CAD | LDpred2.10FCVal | lassosum.10FCVal | 0.187 | 0.174 | 0.012 | 2.99e-12 | 0.184 | 0.171 | 0.013 | 1.68e-04 |
| CAD | SBayesR.PseudoVal | lassosum.10FCVal | 0.164 | 0.174 | -0.011 | 4.67e-03 | 0.170 | 0.171 | -0.001 | 9.32e-01 |
| CAD | LDpred2.PseudoVal | lassosum.10FCVal | 0.052 | 0.174 | -0.122 | 6.34e-90 | 0.060 | 0.171 | -0.111 | 8.09e-20 |
| CAD | SBLUP.Inf | lassosum.10FCVal | 0.161 | 0.174 | -0.014 | 4.45e-09 | 0.156 | 0.171 | -0.015 | 1.92e-03 |
| CAD | pT+clump.MultiPRS | lassosum.10FCVal | 0.170 | 0.174 | -0.004 | 5.35e-01 | 0.166 | 0.171 | -0.005 | 4.49e-01 |
| CAD | LDpred1.10FCVal | lassosum.10FCVal | 0.172 | 0.174 | -0.003 | 3.55e-01 | 0.171 | 0.171 | 0.001 | 9.02e-01 |
| CAD | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.169 | 0.173 | -0.003 | 6.28e-01 | 0.163 | 0.173 | -0.010 | 7.89e-02 |
| CAD | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.190 | 0.173 | 0.017 | 1.30e-02 | 0.188 | 0.173 | 0.015 | 1.52e-04 |
| CAD | pT+clump.10FCVal | LDpred1.MultiPRS | 0.139 | 0.173 | -0.034 | 1.06e-06 | 0.126 | 0.173 | -0.046 | 9.11e-09 |
| CAD | lassosum.10FCVal | LDpred1.MultiPRS | 0.174 | 0.173 | 0.002 | 7.85e-01 | 0.171 | 0.173 | -0.002 | 6.81e-01 |
| CAD | lassosum.PseudoVal | LDpred1.MultiPRS | 0.162 | 0.173 | -0.010 | 1.33e-01 | 0.160 | 0.173 | -0.013 | 1.96e-02 |
| CAD | All.MultiPRS | LDpred1.MultiPRS | 0.190 | 0.173 | 0.018 | 1.02e-02 | 0.189 | 0.173 | 0.017 | 1.44e-04 |
| CAD | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.173 | 0.173 | 0.000 | 1.00e+00 | 0.173 | 0.173 | 0.000 | 1.00e+00 |
| CAD | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.052 | 0.173 | -0.121 | 9.77e-67 | 0.060 | 0.173 | -0.113 | 3.87e-20 |
| CAD | SBLUP.Inf | LDpred1.MultiPRS | 0.161 | 0.173 | -0.012 | 8.40e-02 | 0.156 | 0.173 | -0.017 | 1.17e-02 |
| CAD | LDpred1.Inf | LDpred1.MultiPRS | 0.147 | 0.173 | -0.025 | 2.69e-04 | 0.148 | 0.173 | -0.025 | 5.30e-06 |
| CAD | LDpred2.Inf | LDpred1.MultiPRS | 0.161 | 0.173 | -0.012 | 8.91e-02 | 0.157 | 0.173 | -0.016 | 1.87e-02 |
| CAD | lassosum.MultiPRS | LDpred1.MultiPRS | 0.187 | 0.173 | 0.014 | 4.12e-02 | 0.182 | 0.173 | 0.009 | 5.62e-02 |
| CAD | LDpred1.10FCVal | LDpred1.MultiPRS | 0.172 | 0.173 | -0.001 | 9.00e-01 | 0.171 | 0.173 | -0.002 | 9.11e-02 |
| CAD | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.170 | 0.173 | -0.002 | 7.29e-01 | 0.166 | 0.173 | -0.007 | 2.48e-01 |
| CAD | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.164 | 0.173 | -0.009 | 2.08e-01 | 0.170 | 0.173 | -0.003 | 5.54e-01 |
| CAD | LDpred2.10FCVal | LDpred1.MultiPRS | 0.187 | 0.173 | 0.014 | 3.69e-02 | 0.184 | 0.173 | 0.011 | 6.92e-03 |
| CAD | All.MultiPRS | LDpred1.Inf | 0.190 | 0.147 | 0.043 | 1.02e-10 | 0.189 | 0.148 | 0.042 | 6.41e-10 |
| CAD | LDpred1.Inf | LDpred1.Inf | 0.147 | 0.147 | 0.000 | 1.00e+00 | 0.148 | 0.148 | 0.000 | 1.00e+00 |
| CAD | LDpred2.Inf | LDpred1.Inf | 0.161 | 0.147 | 0.013 | 4.92e-09 | 0.157 | 0.148 | 0.009 | 4.81e-02 |
| CAD | DBSLMM.PseudoVal | LDpred1.Inf | 0.169 | 0.147 | 0.022 | 2.50e-07 | 0.163 | 0.148 | 0.015 | 7.54e-02 |
| CAD | lassosum.PseudoVal | LDpred1.Inf | 0.162 | 0.147 | 0.015 | 1.76e-03 | 0.160 | 0.148 | 0.012 | 2.16e-01 |
| CAD | LDpred2.MultiPRS | LDpred1.Inf | 0.190 | 0.147 | 0.042 | 9.71e-10 | 0.188 | 0.148 | 0.040 | 4.12e-10 |
| CAD | pT+clump.10FCVal | LDpred1.Inf | 0.139 | 0.147 | -0.009 | 6.58e-02 | 0.126 | 0.148 | -0.021 | 2.25e-02 |
| CAD | lassosum.10FCVal | LDpred1.Inf | 0.174 | 0.147 | 0.027 | 1.52e-18 | 0.171 | 0.148 | 0.023 | 2.10e-04 |
| CAD | LDpred2.10FCVal | LDpred1.Inf | 0.187 | 0.147 | 0.040 | 1.62e-40 | 0.184 | 0.148 | 0.036 | 8.85e-10 |
| CAD | SBayesR.PseudoVal | LDpred1.Inf | 0.164 | 0.147 | 0.017 | 2.38e-04 | 0.170 | 0.148 | 0.022 | 1.36e-02 |
| CAD | LDpred1.MultiPRS | LDpred1.Inf | 0.173 | 0.147 | 0.025 | 2.69e-04 | 0.173 | 0.148 | 0.025 | 5.30e-06 |
| CAD | LDpred2.PseudoVal | LDpred1.Inf | 0.052 | 0.147 | -0.095 | 1.55e-52 | 0.060 | 0.148 | -0.088 | 1.73e-12 |
| CAD | SBLUP.Inf | LDpred1.Inf | 0.161 | 0.147 | 0.013 | 4.83e-09 | 0.156 | 0.148 | 0.008 | 7.19e-02 |
| CAD | pT+clump.MultiPRS | LDpred1.Inf | 0.170 | 0.147 | 0.023 | 1.01e-03 | 0.166 | 0.148 | 0.018 | 2.49e-02 |
| CAD | lassosum.MultiPRS | LDpred1.Inf | 0.187 | 0.147 | 0.039 | 1.45e-08 | 0.182 | 0.148 | 0.034 | 5.03e-07 |
| CAD | LDpred1.10FCVal | LDpred1.Inf | 0.172 | 0.147 | 0.024 | 3.18e-15 | 0.171 | 0.148 | 0.023 | 1.49e-04 |
| CAD | LDpred1.Inf | LDpred1.10FCVal | 0.147 | 0.172 | -0.024 | 3.18e-15 | 0.148 | 0.171 | -0.023 | 1.49e-04 |
| CAD | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.169 | 0.172 | -0.002 | 3.80e-01 | 0.163 | 0.171 | -0.008 | 1.33e-01 |
| CAD | lassosum.PseudoVal | LDpred1.10FCVal | 0.162 | 0.172 | -0.010 | 5.63e-04 | 0.160 | 0.171 | -0.012 | 3.17e-02 |
| CAD | LDpred2.MultiPRS | LDpred1.10FCVal | 0.190 | 0.172 | 0.018 | 8.95e-03 | 0.188 | 0.171 | 0.017 | 5.89e-05 |
| CAD | pT+clump.10FCVal | LDpred1.10FCVal | 0.139 | 0.172 | -0.033 | 4.70e-16 | 0.126 | 0.171 | -0.045 | 3.09e-08 |
| CAD | lassosum.10FCVal | LDpred1.10FCVal | 0.174 | 0.172 | 0.003 | 3.55e-01 | 0.171 | 0.171 | -0.001 | 9.02e-01 |
| CAD | LDpred2.10FCVal | LDpred1.10FCVal | 0.187 | 0.172 | 0.015 | 4.60e-12 | 0.184 | 0.171 | 0.013 | 3.67e-03 |
| CAD | All.MultiPRS | LDpred1.10FCVal | 0.190 | 0.172 | 0.019 | 4.53e-03 | 0.189 | 0.171 | 0.018 | 5.44e-05 |
| CAD | LDpred1.MultiPRS | LDpred1.10FCVal | 0.173 | 0.172 | 0.001 | 9.00e-01 | 0.173 | 0.171 | 0.002 | 9.11e-02 |
| CAD | LDpred2.PseudoVal | LDpred1.10FCVal | 0.052 | 0.172 | -0.120 | 6.93e-83 | 0.060 | 0.171 | -0.112 | 2.11e-19 |
| CAD | SBLUP.Inf | LDpred1.10FCVal | 0.161 | 0.172 | -0.011 | 2.07e-03 | 0.156 | 0.171 | -0.015 | 3.39e-02 |
| CAD | pT+clump.MultiPRS | LDpred1.10FCVal | 0.170 | 0.172 | -0.002 | 8.24e-01 | 0.166 | 0.171 | -0.005 | 3.74e-01 |
| CAD | LDpred2.Inf | LDpred1.10FCVal | 0.161 | 0.172 | -0.011 | 2.84e-03 | 0.157 | 0.171 | -0.014 | 4.90e-02 |
| CAD | lassosum.MultiPRS | LDpred1.10FCVal | 0.187 | 0.172 | 0.015 | 3.09e-02 | 0.182 | 0.171 | 0.010 | 2.96e-02 |
| CAD | LDpred1.10FCVal | LDpred1.10FCVal | 0.172 | 0.172 | 0.000 | 1.00e+00 | 0.171 | 0.171 | 0.000 | 1.00e+00 |
| CAD | SBayesR.PseudoVal | LDpred1.10FCVal | 0.164 | 0.172 | -0.008 | 1.06e-03 | 0.170 | 0.171 | -0.001 | 7.73e-01 |
| CAD | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.190 | 0.190 | 0.000 | 1.00e+00 | 0.188 | 0.188 | 0.000 | 1.00e+00 |
| CAD | lassosum.PseudoVal | LDpred2.MultiPRS | 0.162 | 0.190 | -0.028 | 6.60e-05 | 0.160 | 0.188 | -0.029 | 3.29e-08 |
| CAD | All.MultiPRS | LDpred2.MultiPRS | 0.190 | 0.190 | 0.001 | 9.30e-01 | 0.189 | 0.188 | 0.001 | 5.05e-01 |
| CAD | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.173 | 0.190 | -0.017 | 1.30e-02 | 0.173 | 0.188 | -0.015 | 1.52e-04 |
| CAD | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.169 | 0.190 | -0.020 | 3.03e-03 | 0.163 | 0.188 | -0.025 | 8.10e-07 |
| CAD | LDpred1.Inf | LDpred2.MultiPRS | 0.147 | 0.190 | -0.042 | 9.71e-10 | 0.148 | 0.188 | -0.040 | 4.12e-10 |
| CAD | LDpred2.Inf | LDpred2.MultiPRS | 0.161 | 0.190 | -0.029 | 3.03e-05 | 0.157 | 0.188 | -0.031 | 2.95e-09 |
| CAD | lassosum.MultiPRS | LDpred2.MultiPRS | 0.187 | 0.190 | -0.003 | 6.53e-01 | 0.182 | 0.188 | -0.007 | 1.93e-02 |
| CAD | LDpred1.10FCVal | LDpred2.MultiPRS | 0.172 | 0.190 | -0.018 | 8.95e-03 | 0.171 | 0.188 | -0.017 | 5.89e-05 |
| CAD | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.170 | 0.190 | -0.020 | 4.61e-03 | 0.166 | 0.188 | -0.022 | 7.91e-06 |
| CAD | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.164 | 0.190 | -0.026 | 1.78e-04 | 0.170 | 0.188 | -0.018 | 5.37e-04 |
| CAD | pT+clump.10FCVal | LDpred2.MultiPRS | 0.139 | 0.190 | -0.051 | 1.65e-13 | 0.126 | 0.188 | -0.062 | 6.16e-18 |
| CAD | lassosum.10FCVal | LDpred2.MultiPRS | 0.174 | 0.190 | -0.015 | 2.68e-02 | 0.171 | 0.188 | -0.018 | 8.01e-06 |
| CAD | LDpred2.10FCVal | LDpred2.MultiPRS | 0.187 | 0.190 | -0.003 | 6.85e-01 | 0.184 | 0.188 | -0.004 | 2.13e-03 |
| CAD | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.052 | 0.190 | -0.138 | 7.34e-87 | 0.060 | 0.188 | -0.129 | 1.75e-27 |
| CAD | SBLUP.Inf | LDpred2.MultiPRS | 0.161 | 0.190 | -0.029 | 2.66e-05 | 0.156 | 0.188 | -0.032 | 9.35e-10 |
| CAD | LDpred2.MultiPRS | LDpred2.10FCVal | 0.190 | 0.187 | 0.003 | 6.85e-01 | 0.188 | 0.184 | 0.004 | 2.13e-03 |
| CAD | pT+clump.10FCVal | LDpred2.10FCVal | 0.139 | 0.187 | -0.048 | 2.20e-38 | 0.126 | 0.184 | -0.058 | 7.14e-15 |
| CAD | lassosum.PseudoVal | LDpred2.10FCVal | 0.162 | 0.187 | -0.025 | 1.34e-17 | 0.160 | 0.184 | -0.024 | 2.54e-05 |
| CAD | All.MultiPRS | LDpred2.10FCVal | 0.190 | 0.187 | 0.003 | 6.02e-01 | 0.189 | 0.184 | 0.005 | 2.23e-02 |
| CAD | LDpred1.MultiPRS | LDpred2.10FCVal | 0.173 | 0.187 | -0.014 | 3.69e-02 | 0.173 | 0.184 | -0.011 | 6.92e-03 |
| CAD | LDpred2.PseudoVal | LDpred2.10FCVal | 0.052 | 0.187 | -0.135 | 2.92e-117 | 0.060 | 0.184 | -0.124 | 3.62e-26 |
| CAD | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.169 | 0.187 | -0.018 | 1.17e-11 | 0.163 | 0.184 | -0.021 | 5.78e-05 |
| CAD | LDpred1.Inf | LDpred2.10FCVal | 0.147 | 0.187 | -0.040 | 1.62e-40 | 0.148 | 0.184 | -0.036 | 8.85e-10 |
| CAD | LDpred2.Inf | LDpred2.10FCVal | 0.161 | 0.187 | -0.026 | 2.29e-31 | 0.157 | 0.184 | -0.027 | 1.93e-09 |
| CAD | lassosum.MultiPRS | LDpred2.10FCVal | 0.187 | 0.187 | 0.000 | 9.65e-01 | 0.182 | 0.184 | -0.002 | 4.28e-01 |
| CAD | LDpred1.10FCVal | LDpred2.10FCVal | 0.172 | 0.187 | -0.015 | 4.60e-12 | 0.171 | 0.184 | -0.013 | 3.67e-03 |
| CAD | pT+clump.MultiPRS | LDpred2.10FCVal | 0.170 | 0.187 | -0.017 | 1.50e-02 | 0.166 | 0.184 | -0.018 | 4.99e-04 |
| CAD | SBayesR.PseudoVal | LDpred2.10FCVal | 0.164 | 0.187 | -0.023 | 8.35e-16 | 0.170 | 0.184 | -0.014 | 1.47e-02 |
| CAD | lassosum.10FCVal | LDpred2.10FCVal | 0.174 | 0.187 | -0.012 | 2.99e-12 | 0.171 | 0.184 | -0.013 | 1.68e-04 |
| CAD | LDpred2.10FCVal | LDpred2.10FCVal | 0.187 | 0.187 | 0.000 | 1.00e+00 | 0.184 | 0.184 | 0.000 | 1.00e+00 |
| CAD | SBLUP.Inf | LDpred2.10FCVal | 0.161 | 0.187 | -0.026 | 4.48e-32 | 0.156 | 0.184 | -0.028 | 4.55e-10 |
| CAD | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.190 | 0.052 | 0.138 | 7.34e-87 | 0.188 | 0.060 | 0.129 | 1.75e-27 |
| CAD | lassosum.PseudoVal | LDpred2.PseudoVal | 0.162 | 0.052 | 0.110 | 2.80e-70 | 0.160 | 0.060 | 0.100 | 1.07e-15 |
| CAD | All.MultiPRS | LDpred2.PseudoVal | 0.190 | 0.052 | 0.138 | 1.89e-90 | 0.189 | 0.060 | 0.130 | 9.45e-28 |
| CAD | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.173 | 0.052 | 0.121 | 9.77e-67 | 0.173 | 0.060 | 0.113 | 3.87e-20 |
| CAD | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.169 | 0.052 | 0.117 | 1.05e-81 | 0.163 | 0.060 | 0.103 | 6.58e-17 |
| CAD | LDpred1.Inf | LDpred2.PseudoVal | 0.147 | 0.052 | 0.095 | 1.55e-52 | 0.148 | 0.060 | 0.088 | 1.73e-12 |
| CAD | LDpred2.Inf | LDpred2.PseudoVal | 0.161 | 0.052 | 0.109 | 8.61e-70 | 0.157 | 0.060 | 0.097 | 3.76e-15 |
| CAD | lassosum.MultiPRS | LDpred2.PseudoVal | 0.187 | 0.052 | 0.135 | 2.19e-82 | 0.182 | 0.060 | 0.122 | 4.64e-23 |
| CAD | LDpred1.10FCVal | LDpred2.PseudoVal | 0.172 | 0.052 | 0.120 | 6.93e-83 | 0.171 | 0.060 | 0.112 | 2.11e-19 |
| CAD | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.170 | 0.052 | 0.118 | 3.02e-64 | 0.166 | 0.060 | 0.106 | 1.72e-19 |
| CAD | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.164 | 0.052 | 0.112 | 8.19e-77 | 0.170 | 0.060 | 0.110 | 7.78e-20 |
| CAD | pT+clump.10FCVal | LDpred2.PseudoVal | 0.139 | 0.052 | 0.087 | 3.89e-49 | 0.126 | 0.060 | 0.067 | 1.49e-08 |
| CAD | lassosum.10FCVal | LDpred2.PseudoVal | 0.174 | 0.052 | 0.122 | 6.34e-90 | 0.171 | 0.060 | 0.111 | 8.09e-20 |
| CAD | LDpred2.10FCVal | LDpred2.PseudoVal | 0.187 | 0.052 | 0.135 | 2.92e-117 | 0.184 | 0.060 | 0.124 | 3.62e-26 |
| CAD | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.052 | 0.052 | 0.000 | 1.00e+00 | 0.060 | 0.060 | 0.000 | 1.00e+00 |
| CAD | SBLUP.Inf | LDpred2.PseudoVal | 0.161 | 0.052 | 0.109 | 2.79e-68 | 0.156 | 0.060 | 0.096 | 1.28e-14 |
| CAD | DBSLMM.PseudoVal | LDpred2.Inf | 0.169 | 0.161 | 0.008 | 3.55e-02 | 0.163 | 0.157 | 0.006 | 4.59e-01 |
| CAD | LDpred2.MultiPRS | LDpred2.Inf | 0.190 | 0.161 | 0.029 | 3.03e-05 | 0.188 | 0.157 | 0.031 | 2.95e-09 |
| CAD | pT+clump.10FCVal | LDpred2.Inf | 0.139 | 0.161 | -0.022 | 9.96e-07 | 0.126 | 0.157 | -0.030 | 7.51e-04 |
| CAD | lassosum.10FCVal | LDpred2.Inf | 0.174 | 0.161 | 0.014 | 9.99e-09 | 0.171 | 0.157 | 0.014 | 3.91e-03 |
| CAD | lassosum.PseudoVal | LDpred2.Inf | 0.162 | 0.161 | 0.001 | 7.73e-01 | 0.160 | 0.157 | 0.003 | 7.76e-01 |
| CAD | All.MultiPRS | LDpred2.Inf | 0.190 | 0.161 | 0.029 | 7.72e-06 | 0.189 | 0.157 | 0.033 | 8.81e-09 |
| CAD | LDpred1.MultiPRS | LDpred2.Inf | 0.173 | 0.161 | 0.012 | 8.91e-02 | 0.173 | 0.157 | 0.016 | 1.87e-02 |
| CAD | LDpred2.PseudoVal | LDpred2.Inf | 0.052 | 0.161 | -0.109 | 8.61e-70 | 0.060 | 0.157 | -0.097 | 3.76e-15 |
| CAD | SBLUP.Inf | LDpred2.Inf | 0.161 | 0.161 | 0.000 | 6.43e-01 | 0.156 | 0.157 | -0.001 | 2.91e-01 |
| CAD | LDpred1.Inf | LDpred2.Inf | 0.147 | 0.161 | -0.013 | 4.92e-09 | 0.148 | 0.157 | -0.009 | 4.81e-02 |
| CAD | LDpred2.Inf | LDpred2.Inf | 0.161 | 0.161 | 0.000 | 1.00e+00 | 0.157 | 0.157 | 0.000 | 1.00e+00 |
| CAD | lassosum.MultiPRS | LDpred2.Inf | 0.187 | 0.161 | 0.026 | 1.91e-04 | 0.182 | 0.157 | 0.025 | 1.16e-05 |
| CAD | LDpred1.10FCVal | LDpred2.Inf | 0.172 | 0.161 | 0.011 | 2.84e-03 | 0.171 | 0.157 | 0.014 | 4.90e-02 |
| CAD | pT+clump.MultiPRS | LDpred2.Inf | 0.170 | 0.161 | 0.009 | 1.77e-01 | 0.166 | 0.157 | 0.009 | 2.30e-01 |
| CAD | SBayesR.PseudoVal | LDpred2.Inf | 0.164 | 0.161 | 0.003 | 5.07e-01 | 0.170 | 0.157 | 0.013 | 1.56e-01 |
| CAD | LDpred2.10FCVal | LDpred2.Inf | 0.187 | 0.161 | 0.026 | 2.29e-31 | 0.184 | 0.157 | 0.027 | 1.93e-09 |
| CAD | All.MultiPRS | pT+clump.MultiPRS | 0.190 | 0.170 | 0.020 | 3.60e-03 | 0.189 | 0.166 | 0.024 | 4.50e-07 |
| CAD | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.173 | 0.170 | 0.002 | 7.29e-01 | 0.173 | 0.166 | 0.007 | 2.48e-01 |
| CAD | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.052 | 0.170 | -0.118 | 3.02e-64 | 0.060 | 0.166 | -0.106 | 1.72e-19 |
| CAD | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.169 | 0.170 | -0.001 | 8.92e-01 | 0.163 | 0.166 | -0.003 | 6.15e-01 |
| CAD | LDpred1.Inf | pT+clump.MultiPRS | 0.147 | 0.170 | -0.023 | 1.01e-03 | 0.148 | 0.166 | -0.018 | 2.49e-02 |
| CAD | LDpred2.Inf | pT+clump.MultiPRS | 0.161 | 0.170 | -0.009 | 1.77e-01 | 0.157 | 0.166 | -0.009 | 2.30e-01 |
| CAD | lassosum.MultiPRS | pT+clump.MultiPRS | 0.187 | 0.170 | 0.016 | 1.74e-02 | 0.182 | 0.166 | 0.016 | 1.48e-03 |
| CAD | LDpred1.10FCVal | pT+clump.MultiPRS | 0.172 | 0.170 | 0.002 | 8.24e-01 | 0.171 | 0.166 | 0.005 | 3.74e-01 |
| CAD | lassosum.PseudoVal | pT+clump.MultiPRS | 0.162 | 0.170 | -0.008 | 2.49e-01 | 0.160 | 0.166 | -0.006 | 3.14e-01 |
| CAD | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.190 | 0.170 | 0.020 | 4.61e-03 | 0.188 | 0.166 | 0.022 | 7.91e-06 |
| CAD | pT+clump.10FCVal | pT+clump.MultiPRS | 0.139 | 0.170 | -0.031 | 6.11e-06 | 0.126 | 0.166 | -0.039 | 9.07e-11 |
| CAD | lassosum.10FCVal | pT+clump.MultiPRS | 0.174 | 0.170 | 0.004 | 5.35e-01 | 0.171 | 0.166 | 0.005 | 4.49e-01 |
| CAD | LDpred2.10FCVal | pT+clump.MultiPRS | 0.187 | 0.170 | 0.017 | 1.50e-02 | 0.184 | 0.166 | 0.018 | 4.99e-04 |
| CAD | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.164 | 0.170 | -0.006 | 3.63e-01 | 0.170 | 0.166 | 0.004 | 5.30e-01 |
| CAD | SBLUP.Inf | pT+clump.MultiPRS | 0.161 | 0.170 | -0.010 | 1.68e-01 | 0.156 | 0.166 | -0.010 | 1.81e-01 |
| CAD | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.170 | 0.170 | 0.000 | 1.00e+00 | 0.166 | 0.166 | 0.000 | 1.00e+00 |
| CAD | lassosum.PseudoVal | pT+clump.10FCVal | 0.162 | 0.139 | 0.023 | 5.06e-12 | 0.160 | 0.126 | 0.033 | 1.17e-06 |
| CAD | All.MultiPRS | pT+clump.10FCVal | 0.190 | 0.139 | 0.052 | 8.34e-15 | 0.189 | 0.126 | 0.063 | 7.79e-18 |
| CAD | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.169 | 0.139 | 0.031 | 2.73e-11 | 0.163 | 0.126 | 0.036 | 6.89e-05 |
| CAD | LDpred1.Inf | pT+clump.10FCVal | 0.147 | 0.139 | 0.009 | 6.58e-02 | 0.148 | 0.126 | 0.021 | 2.25e-02 |
| CAD | LDpred2.Inf | pT+clump.10FCVal | 0.161 | 0.139 | 0.022 | 9.96e-07 | 0.157 | 0.126 | 0.030 | 7.51e-04 |
| CAD | lassosum.MultiPRS | pT+clump.10FCVal | 0.187 | 0.139 | 0.048 | 5.13e-12 | 0.182 | 0.126 | 0.055 | 1.76e-13 |
| CAD | LDpred1.10FCVal | pT+clump.10FCVal | 0.172 | 0.139 | 0.033 | 4.70e-16 | 0.171 | 0.126 | 0.045 | 3.09e-08 |
| CAD | pT+clump.MultiPRS | pT+clump.10FCVal | 0.170 | 0.139 | 0.031 | 6.11e-06 | 0.166 | 0.126 | 0.039 | 9.07e-11 |
| CAD | LDpred2.MultiPRS | pT+clump.10FCVal | 0.190 | 0.139 | 0.051 | 1.65e-13 | 0.188 | 0.126 | 0.062 | 6.16e-18 |
| CAD | pT+clump.10FCVal | pT+clump.10FCVal | 0.139 | 0.139 | 0.000 | 1.00e+00 | 0.126 | 0.126 | 0.000 | 1.00e+00 |
| CAD | lassosum.10FCVal | pT+clump.10FCVal | 0.174 | 0.139 | 0.036 | 5.49e-22 | 0.171 | 0.126 | 0.044 | 2.29e-09 |
| CAD | LDpred2.10FCVal | pT+clump.10FCVal | 0.187 | 0.139 | 0.048 | 2.20e-38 | 0.184 | 0.126 | 0.058 | 7.14e-15 |
| CAD | SBayesR.PseudoVal | pT+clump.10FCVal | 0.164 | 0.139 | 0.025 | 6.34e-09 | 0.170 | 0.126 | 0.044 | 5.28e-07 |
| CAD | LDpred1.MultiPRS | pT+clump.10FCVal | 0.173 | 0.139 | 0.034 | 1.06e-06 | 0.173 | 0.126 | 0.046 | 9.11e-09 |
| CAD | LDpred2.PseudoVal | pT+clump.10FCVal | 0.052 | 0.139 | -0.087 | 3.89e-49 | 0.060 | 0.126 | -0.067 | 1.49e-08 |
| CAD | SBLUP.Inf | pT+clump.10FCVal | 0.161 | 0.139 | 0.022 | 1.02e-06 | 0.156 | 0.126 | 0.030 | 9.66e-04 |
| CAD | LDpred1.Inf | SBayesR.PseudoVal | 0.147 | 0.164 | -0.017 | 2.38e-04 | 0.148 | 0.170 | -0.022 | 1.36e-02 |
| CAD | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.169 | 0.164 | 0.005 | 3.49e-02 | 0.163 | 0.170 | -0.007 | 1.66e-01 |
| CAD | lassosum.PseudoVal | SBayesR.PseudoVal | 0.162 | 0.164 | -0.002 | 4.93e-01 | 0.160 | 0.170 | -0.010 | 3.58e-02 |
| CAD | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.190 | 0.164 | 0.026 | 1.78e-04 | 0.188 | 0.170 | 0.018 | 5.37e-04 |
| CAD | pT+clump.10FCVal | SBayesR.PseudoVal | 0.139 | 0.164 | -0.025 | 6.34e-09 | 0.126 | 0.170 | -0.044 | 5.28e-07 |
| CAD | lassosum.10FCVal | SBayesR.PseudoVal | 0.174 | 0.164 | 0.011 | 4.67e-03 | 0.171 | 0.170 | 0.001 | 9.32e-01 |
| CAD | LDpred2.10FCVal | SBayesR.PseudoVal | 0.187 | 0.164 | 0.023 | 8.35e-16 | 0.184 | 0.170 | 0.014 | 1.47e-02 |
| CAD | All.MultiPRS | SBayesR.PseudoVal | 0.190 | 0.164 | 0.026 | 5.85e-05 | 0.189 | 0.170 | 0.019 | 2.94e-04 |
| CAD | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.173 | 0.164 | 0.009 | 2.08e-01 | 0.173 | 0.170 | 0.003 | 5.54e-01 |
| CAD | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.052 | 0.164 | -0.112 | 8.19e-77 | 0.060 | 0.170 | -0.110 | 7.78e-20 |
| CAD | SBLUP.Inf | SBayesR.PseudoVal | 0.161 | 0.164 | -0.003 | 4.77e-01 | 0.156 | 0.170 | -0.014 | 1.26e-01 |
| CAD | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.170 | 0.164 | 0.006 | 3.63e-01 | 0.166 | 0.170 | -0.004 | 5.30e-01 |
| CAD | LDpred2.Inf | SBayesR.PseudoVal | 0.161 | 0.164 | -0.003 | 5.07e-01 | 0.157 | 0.170 | -0.013 | 1.56e-01 |
| CAD | lassosum.MultiPRS | SBayesR.PseudoVal | 0.187 | 0.164 | 0.023 | 1.02e-03 | 0.182 | 0.170 | 0.012 | 4.36e-02 |
| CAD | LDpred1.10FCVal | SBayesR.PseudoVal | 0.172 | 0.164 | 0.008 | 1.06e-03 | 0.171 | 0.170 | 0.001 | 7.73e-01 |
| CAD | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.164 | 0.164 | 0.000 | 1.00e+00 | 0.170 | 0.170 | 0.000 | 1.00e+00 |
| CAD | All.MultiPRS | SBLUP.Inf | 0.190 | 0.161 | 0.030 | 6.73e-06 | 0.189 | 0.156 | 0.033 | 3.43e-09 |
| CAD | LDpred1.Inf | SBLUP.Inf | 0.147 | 0.161 | -0.013 | 4.83e-09 | 0.148 | 0.156 | -0.008 | 7.19e-02 |
| CAD | LDpred2.Inf | SBLUP.Inf | 0.161 | 0.161 | 0.000 | 6.43e-01 | 0.157 | 0.156 | 0.001 | 2.91e-01 |
| CAD | DBSLMM.PseudoVal | SBLUP.Inf | 0.169 | 0.161 | 0.009 | 2.97e-02 | 0.163 | 0.156 | 0.007 | 3.88e-01 |
| CAD | lassosum.PseudoVal | SBLUP.Inf | 0.162 | 0.161 | 0.002 | 7.38e-01 | 0.160 | 0.156 | 0.004 | 7.01e-01 |
| CAD | LDpred2.MultiPRS | SBLUP.Inf | 0.190 | 0.161 | 0.029 | 2.66e-05 | 0.188 | 0.156 | 0.032 | 9.35e-10 |
| CAD | pT+clump.10FCVal | SBLUP.Inf | 0.139 | 0.161 | -0.022 | 1.02e-06 | 0.126 | 0.156 | -0.030 | 9.66e-04 |
| CAD | lassosum.10FCVal | SBLUP.Inf | 0.174 | 0.161 | 0.014 | 4.45e-09 | 0.171 | 0.156 | 0.015 | 1.92e-03 |
| CAD | LDpred2.10FCVal | SBLUP.Inf | 0.187 | 0.161 | 0.026 | 4.48e-32 | 0.184 | 0.156 | 0.028 | 4.55e-10 |
| CAD | SBayesR.PseudoVal | SBLUP.Inf | 0.164 | 0.161 | 0.003 | 4.77e-01 | 0.170 | 0.156 | 0.014 | 1.26e-01 |
| CAD | LDpred1.MultiPRS | SBLUP.Inf | 0.173 | 0.161 | 0.012 | 8.40e-02 | 0.173 | 0.156 | 0.017 | 1.17e-02 |
| CAD | LDpred2.PseudoVal | SBLUP.Inf | 0.052 | 0.161 | -0.109 | 2.79e-68 | 0.060 | 0.156 | -0.096 | 1.28e-14 |
| CAD | SBLUP.Inf | SBLUP.Inf | 0.161 | 0.161 | 0.000 | 1.00e+00 | 0.156 | 0.156 | 0.000 | 1.00e+00 |
| CAD | pT+clump.MultiPRS | SBLUP.Inf | 0.170 | 0.161 | 0.010 | 1.68e-01 | 0.166 | 0.156 | 0.010 | 1.81e-01 |
| CAD | lassosum.MultiPRS | SBLUP.Inf | 0.187 | 0.161 | 0.026 | 1.70e-04 | 0.182 | 0.156 | 0.026 | 4.54e-06 |
| CAD | LDpred1.10FCVal | SBLUP.Inf | 0.172 | 0.161 | 0.011 | 2.07e-03 | 0.171 | 0.156 | 0.015 | 3.39e-02 |
| IBD | lassosum.PseudoVal | All.MultiPRS | 0.133 | 0.146 | -0.013 | 6.05e-02 | 0.140 | 0.152 | -0.012 | 4.36e-03 |
| IBD | All.MultiPRS | All.MultiPRS | 0.146 | 0.146 | 0.000 | 1.00e+00 | 0.152 | 0.152 | 0.000 | 1.00e+00 |
| IBD | DBSLMM.PseudoVal | All.MultiPRS | 0.130 | 0.146 | -0.016 | 1.87e-02 | 0.136 | 0.152 | -0.016 | 6.91e-04 |
| IBD | LDpred1.Inf | All.MultiPRS | 0.098 | 0.146 | -0.048 | 8.57e-12 | 0.103 | 0.152 | -0.049 | 4.15e-10 |
| IBD | LDpred2.Inf | All.MultiPRS | 0.096 | 0.146 | -0.050 | 8.73e-13 | 0.100 | 0.152 | -0.052 | 8.29e-11 |
| IBD | lassosum.MultiPRS | All.MultiPRS | 0.144 | 0.146 | -0.003 | 2.09e-02 | 0.148 | 0.152 | -0.004 | 7.23e-02 |
| IBD | LDpred1.10FCVal | All.MultiPRS | 0.129 | 0.146 | -0.017 | 1.47e-02 | 0.137 | 0.152 | -0.015 | 2.38e-03 |
| IBD | pT+clump.MultiPRS | All.MultiPRS | 0.127 | 0.146 | -0.019 | 7.14e-03 | 0.136 | 0.152 | -0.016 | 9.79e-04 |
| IBD | LDpred2.MultiPRS | All.MultiPRS | 0.143 | 0.146 | -0.004 | 2.30e-04 | 0.150 | 0.152 | -0.002 | 4.02e-01 |
| IBD | pT+clump.10FCVal | All.MultiPRS | 0.111 | 0.146 | -0.035 | 4.48e-07 | 0.109 | 0.152 | -0.043 | 1.80e-10 |
| IBD | lassosum.10FCVal | All.MultiPRS | 0.134 | 0.146 | -0.012 | 8.34e-02 | 0.140 | 0.152 | -0.011 | 6.29e-03 |
| IBD | LDpred2.10FCVal | All.MultiPRS | 0.143 | 0.146 | -0.003 | 6.41e-01 | 0.149 | 0.152 | -0.003 | 2.76e-01 |
| IBD | SBayesR.PseudoVal | All.MultiPRS | 0.128 | 0.146 | -0.019 | 7.54e-03 | 0.143 | 0.152 | -0.009 | 6.34e-02 |
| IBD | LDpred1.MultiPRS | All.MultiPRS | 0.130 | 0.146 | -0.016 | 3.11e-12 | 0.138 | 0.152 | -0.014 | 2.74e-03 |
| IBD | LDpred2.PseudoVal | All.MultiPRS | 0.140 | 0.146 | -0.006 | 3.67e-01 | 0.145 | 0.152 | -0.007 | 2.11e-02 |
| IBD | SBLUP.Inf | All.MultiPRS | 0.097 | 0.146 | -0.049 | 2.97e-12 | 0.102 | 0.152 | -0.050 | 2.75e-10 |
| IBD | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.143 | 0.130 | 0.013 | 6.62e-02 | 0.150 | 0.136 | 0.014 | 2.22e-03 |
| IBD | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.111 | 0.130 | -0.019 | 2.68e-07 | 0.109 | 0.136 | -0.027 | 1.73e-04 |
| IBD | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.133 | 0.130 | 0.003 | 2.54e-01 | 0.140 | 0.136 | 0.004 | 5.21e-01 |
| IBD | All.MultiPRS | DBSLMM.PseudoVal | 0.146 | 0.130 | 0.016 | 1.87e-02 | 0.152 | 0.136 | 0.016 | 6.91e-04 |
| IBD | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.130 | 0.130 | 0.000 | 9.65e-01 | 0.138 | 0.136 | 0.002 | 6.86e-01 |
| IBD | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.140 | 0.130 | 0.010 | 2.82e-04 | 0.145 | 0.136 | 0.010 | 8.69e-02 |
| IBD | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.130 | 0.130 | 0.000 | 1.00e+00 | 0.136 | 0.136 | 0.000 | 1.00e+00 |
| IBD | LDpred1.Inf | DBSLMM.PseudoVal | 0.098 | 0.130 | -0.031 | 2.52e-13 | 0.103 | 0.136 | -0.033 | 1.32e-04 |
| IBD | LDpred2.Inf | DBSLMM.PseudoVal | 0.096 | 0.130 | -0.034 | 1.38e-14 | 0.100 | 0.136 | -0.036 | 4.87e-05 |
| IBD | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.144 | 0.130 | 0.014 | 4.77e-02 | 0.148 | 0.136 | 0.012 | 2.08e-02 |
| IBD | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.129 | 0.130 | -0.001 | 8.40e-01 | 0.137 | 0.136 | 0.001 | 8.45e-01 |
| IBD | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.127 | 0.130 | -0.002 | 7.41e-01 | 0.136 | 0.136 | 0.000 | 9.70e-01 |
| IBD | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.128 | 0.130 | -0.002 | 4.59e-01 | 0.143 | 0.136 | 0.008 | 2.05e-01 |
| IBD | lassosum.10FCVal | DBSLMM.PseudoVal | 0.134 | 0.130 | 0.004 | 1.85e-01 | 0.140 | 0.136 | 0.005 | 4.80e-01 |
| IBD | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.143 | 0.130 | 0.013 | 3.34e-07 | 0.149 | 0.136 | 0.013 | 9.22e-03 |
| IBD | SBLUP.Inf | DBSLMM.PseudoVal | 0.097 | 0.130 | -0.033 | 1.41e-13 | 0.102 | 0.136 | -0.034 | 1.24e-04 |
| IBD | All.MultiPRS | lassosum.MultiPRS | 0.146 | 0.144 | 0.003 | 2.09e-02 | 0.152 | 0.148 | 0.004 | 7.23e-02 |
| IBD | LDpred1.MultiPRS | lassosum.MultiPRS | 0.130 | 0.144 | -0.014 | 3.12e-08 | 0.138 | 0.148 | -0.010 | 4.54e-02 |
| IBD | LDpred1.Inf | lassosum.MultiPRS | 0.098 | 0.144 | -0.045 | 1.03e-10 | 0.103 | 0.148 | -0.045 | 1.04e-08 |
| IBD | LDpred2.Inf | lassosum.MultiPRS | 0.096 | 0.144 | -0.048 | 1.18e-11 | 0.100 | 0.148 | -0.048 | 2.35e-09 |
| IBD | lassosum.MultiPRS | lassosum.MultiPRS | 0.144 | 0.144 | 0.000 | 1.00e+00 | 0.148 | 0.148 | 0.000 | 1.00e+00 |
| IBD | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.130 | 0.144 | -0.014 | 4.77e-02 | 0.136 | 0.148 | -0.012 | 2.08e-02 |
| IBD | lassosum.PseudoVal | lassosum.MultiPRS | 0.133 | 0.144 | -0.010 | 1.33e-01 | 0.140 | 0.148 | -0.008 | 2.65e-02 |
| IBD | LDpred2.MultiPRS | lassosum.MultiPRS | 0.143 | 0.144 | -0.001 | 5.57e-01 | 0.150 | 0.148 | 0.002 | 4.55e-01 |
| IBD | pT+clump.10FCVal | lassosum.MultiPRS | 0.111 | 0.144 | -0.033 | 2.95e-06 | 0.109 | 0.148 | -0.039 | 5.45e-08 |
| IBD | lassosum.10FCVal | lassosum.MultiPRS | 0.134 | 0.144 | -0.009 | 1.75e-01 | 0.140 | 0.148 | -0.007 | 2.94e-02 |
| IBD | LDpred2.10FCVal | lassosum.MultiPRS | 0.143 | 0.144 | -0.001 | 9.27e-01 | 0.149 | 0.148 | 0.001 | 6.78e-01 |
| IBD | SBayesR.PseudoVal | lassosum.MultiPRS | 0.128 | 0.144 | -0.016 | 2.15e-02 | 0.143 | 0.148 | -0.004 | 3.78e-01 |
| IBD | LDpred2.PseudoVal | lassosum.MultiPRS | 0.140 | 0.144 | -0.004 | 5.98e-01 | 0.145 | 0.148 | -0.002 | 5.49e-01 |
| IBD | SBLUP.Inf | lassosum.MultiPRS | 0.097 | 0.144 | -0.046 | 3.76e-11 | 0.102 | 0.148 | -0.046 | 6.21e-09 |
| IBD | pT+clump.MultiPRS | lassosum.MultiPRS | 0.127 | 0.144 | -0.016 | 2.06e-02 | 0.136 | 0.148 | -0.012 | 2.19e-02 |
| IBD | LDpred1.10FCVal | lassosum.MultiPRS | 0.129 | 0.144 | -0.014 | 3.86e-02 | 0.137 | 0.148 | -0.011 | 3.26e-02 |
| IBD | All.MultiPRS | lassosum.PseudoVal | 0.146 | 0.133 | 0.013 | 6.05e-02 | 0.152 | 0.140 | 0.012 | 4.36e-03 |
| IBD | LDpred1.MultiPRS | lassosum.PseudoVal | 0.130 | 0.133 | -0.003 | 6.61e-01 | 0.138 | 0.140 | -0.001 | 8.25e-01 |
| IBD | LDpred2.PseudoVal | lassosum.PseudoVal | 0.140 | 0.133 | 0.007 | 2.66e-04 | 0.145 | 0.140 | 0.006 | 1.22e-01 |
| IBD | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.130 | 0.133 | -0.003 | 2.54e-01 | 0.136 | 0.140 | -0.004 | 5.21e-01 |
| IBD | LDpred1.Inf | lassosum.PseudoVal | 0.098 | 0.133 | -0.035 | 1.92e-12 | 0.103 | 0.140 | -0.037 | 1.94e-04 |
| IBD | LDpred2.Inf | lassosum.PseudoVal | 0.096 | 0.133 | -0.037 | 3.13e-13 | 0.100 | 0.140 | -0.039 | 9.86e-05 |
| IBD | lassosum.MultiPRS | lassosum.PseudoVal | 0.144 | 0.133 | 0.010 | 1.33e-01 | 0.148 | 0.140 | 0.008 | 2.65e-02 |
| IBD | LDpred1.10FCVal | lassosum.PseudoVal | 0.129 | 0.133 | -0.004 | 2.45e-01 | 0.137 | 0.140 | -0.003 | 7.02e-01 |
| IBD | lassosum.PseudoVal | lassosum.PseudoVal | 0.133 | 0.133 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| IBD | LDpred2.MultiPRS | lassosum.PseudoVal | 0.143 | 0.133 | 0.010 | 1.74e-01 | 0.150 | 0.140 | 0.011 | 8.21e-03 |
| IBD | pT+clump.10FCVal | lassosum.PseudoVal | 0.111 | 0.133 | -0.022 | 2.09e-11 | 0.109 | 0.140 | -0.031 | 2.25e-06 |
| IBD | lassosum.10FCVal | lassosum.PseudoVal | 0.134 | 0.133 | 0.001 | 5.89e-01 | 0.140 | 0.140 | 0.001 | 8.09e-01 |
| IBD | LDpred2.10FCVal | lassosum.PseudoVal | 0.143 | 0.133 | 0.010 | 6.59e-06 | 0.149 | 0.140 | 0.010 | 2.60e-02 |
| IBD | SBayesR.PseudoVal | lassosum.PseudoVal | 0.128 | 0.133 | -0.006 | 3.33e-02 | 0.143 | 0.140 | 0.004 | 4.66e-01 |
| IBD | SBLUP.Inf | lassosum.PseudoVal | 0.097 | 0.133 | -0.036 | 1.77e-12 | 0.102 | 0.140 | -0.037 | 2.07e-04 |
| IBD | pT+clump.MultiPRS | lassosum.PseudoVal | 0.127 | 0.133 | -0.006 | 4.17e-01 | 0.136 | 0.140 | -0.004 | 5.45e-01 |
| IBD | All.MultiPRS | lassosum.10FCVal | 0.146 | 0.134 | 0.012 | 8.34e-02 | 0.152 | 0.140 | 0.011 | 6.29e-03 |
| IBD | LDpred1.MultiPRS | lassosum.10FCVal | 0.130 | 0.134 | -0.004 | 5.61e-01 | 0.138 | 0.140 | -0.002 | 6.72e-01 |
| IBD | LDpred1.Inf | lassosum.10FCVal | 0.098 | 0.134 | -0.036 | 2.06e-17 | 0.103 | 0.140 | -0.037 | 8.33e-06 |
| IBD | LDpred2.Inf | lassosum.10FCVal | 0.096 | 0.134 | -0.038 | 5.63e-19 | 0.100 | 0.140 | -0.040 | 2.32e-06 |
| IBD | lassosum.MultiPRS | lassosum.10FCVal | 0.144 | 0.134 | 0.009 | 1.75e-01 | 0.148 | 0.140 | 0.007 | 2.94e-02 |
| IBD | DBSLMM.PseudoVal | lassosum.10FCVal | 0.130 | 0.134 | -0.004 | 1.85e-01 | 0.136 | 0.140 | -0.005 | 4.80e-01 |
| IBD | lassosum.PseudoVal | lassosum.10FCVal | 0.133 | 0.134 | -0.001 | 5.89e-01 | 0.140 | 0.140 | -0.001 | 8.09e-01 |
| IBD | LDpred2.MultiPRS | lassosum.10FCVal | 0.143 | 0.134 | 0.008 | 2.24e-01 | 0.150 | 0.140 | 0.010 | 1.80e-02 |
| IBD | pT+clump.10FCVal | lassosum.10FCVal | 0.111 | 0.134 | -0.023 | 6.20e-09 | 0.109 | 0.140 | -0.032 | 5.42e-05 |
| IBD | lassosum.10FCVal | lassosum.10FCVal | 0.134 | 0.134 | 0.000 | 1.00e+00 | 0.140 | 0.140 | 0.000 | 1.00e+00 |
| IBD | LDpred2.10FCVal | lassosum.10FCVal | 0.143 | 0.134 | 0.009 | 2.90e-06 | 0.149 | 0.140 | 0.009 | 2.12e-02 |
| IBD | SBayesR.PseudoVal | lassosum.10FCVal | 0.128 | 0.134 | -0.007 | 2.11e-02 | 0.143 | 0.140 | 0.003 | 6.16e-01 |
| IBD | LDpred2.PseudoVal | lassosum.10FCVal | 0.140 | 0.134 | 0.006 | 9.74e-03 | 0.145 | 0.140 | 0.005 | 2.78e-01 |
| IBD | SBLUP.Inf | lassosum.10FCVal | 0.097 | 0.134 | -0.037 | 4.93e-18 | 0.102 | 0.140 | -0.038 | 6.02e-06 |
| IBD | pT+clump.MultiPRS | lassosum.10FCVal | 0.127 | 0.134 | -0.007 | 3.39e-01 | 0.136 | 0.140 | -0.004 | 4.32e-01 |
| IBD | LDpred1.10FCVal | lassosum.10FCVal | 0.129 | 0.134 | -0.005 | 8.23e-02 | 0.137 | 0.140 | -0.003 | 5.42e-01 |
| IBD | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.130 | 0.130 | 0.000 | 9.65e-01 | 0.136 | 0.138 | -0.002 | 6.86e-01 |
| IBD | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.143 | 0.130 | 0.013 | 8.80e-09 | 0.150 | 0.138 | 0.012 | 5.10e-03 |
| IBD | pT+clump.10FCVal | LDpred1.MultiPRS | 0.111 | 0.130 | -0.019 | 6.27e-03 | 0.109 | 0.138 | -0.030 | 3.59e-04 |
| IBD | lassosum.10FCVal | LDpred1.MultiPRS | 0.134 | 0.130 | 0.004 | 5.61e-01 | 0.140 | 0.138 | 0.002 | 6.72e-01 |
| IBD | lassosum.PseudoVal | LDpred1.MultiPRS | 0.133 | 0.130 | 0.003 | 6.61e-01 | 0.140 | 0.138 | 0.001 | 8.25e-01 |
| IBD | All.MultiPRS | LDpred1.MultiPRS | 0.146 | 0.130 | 0.016 | 3.11e-12 | 0.152 | 0.138 | 0.014 | 2.74e-03 |
| IBD | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.130 | 0.130 | 0.000 | 1.00e+00 | 0.138 | 0.138 | 0.000 | 1.00e+00 |
| IBD | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.140 | 0.130 | 0.010 | 1.57e-01 | 0.145 | 0.138 | 0.007 | 2.01e-01 |
| IBD | SBLUP.Inf | LDpred1.MultiPRS | 0.097 | 0.130 | -0.033 | 2.98e-06 | 0.102 | 0.138 | -0.036 | 1.27e-09 |
| IBD | LDpred1.Inf | LDpred1.MultiPRS | 0.098 | 0.130 | -0.032 | 6.20e-06 | 0.103 | 0.138 | -0.035 | 5.33e-12 |
| IBD | LDpred2.Inf | LDpred1.MultiPRS | 0.096 | 0.130 | -0.034 | 1.29e-06 | 0.100 | 0.138 | -0.038 | 1.94e-10 |
| IBD | lassosum.MultiPRS | LDpred1.MultiPRS | 0.144 | 0.130 | 0.014 | 3.12e-08 | 0.148 | 0.138 | 0.010 | 4.54e-02 |
| IBD | LDpred1.10FCVal | LDpred1.MultiPRS | 0.129 | 0.130 | -0.001 | 8.96e-01 | 0.137 | 0.138 | -0.001 | 4.56e-01 |
| IBD | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.127 | 0.130 | -0.003 | 7.08e-01 | 0.136 | 0.138 | -0.002 | 7.07e-01 |
| IBD | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.128 | 0.130 | -0.002 | 7.21e-01 | 0.143 | 0.138 | 0.005 | 3.38e-01 |
| IBD | LDpred2.10FCVal | LDpred1.MultiPRS | 0.143 | 0.130 | 0.013 | 6.46e-02 | 0.149 | 0.138 | 0.011 | 4.92e-03 |
| IBD | All.MultiPRS | LDpred1.Inf | 0.146 | 0.098 | 0.048 | 8.57e-12 | 0.152 | 0.103 | 0.049 | 4.15e-10 |
| IBD | LDpred1.Inf | LDpred1.Inf | 0.098 | 0.098 | 0.000 | 1.00e+00 | 0.103 | 0.103 | 0.000 | 1.00e+00 |
| IBD | LDpred2.Inf | LDpred1.Inf | 0.096 | 0.098 | -0.002 | 1.40e-01 | 0.100 | 0.103 | -0.003 | 3.79e-01 |
| IBD | DBSLMM.PseudoVal | LDpred1.Inf | 0.130 | 0.098 | 0.031 | 2.52e-13 | 0.136 | 0.103 | 0.033 | 1.32e-04 |
| IBD | lassosum.PseudoVal | LDpred1.Inf | 0.133 | 0.098 | 0.035 | 1.92e-12 | 0.140 | 0.103 | 0.037 | 1.94e-04 |
| IBD | LDpred2.MultiPRS | LDpred1.Inf | 0.143 | 0.098 | 0.044 | 2.75e-10 | 0.150 | 0.103 | 0.047 | 2.87e-09 |
| IBD | pT+clump.10FCVal | LDpred1.Inf | 0.111 | 0.098 | 0.013 | 1.99e-02 | 0.109 | 0.103 | 0.006 | 6.03e-01 |
| IBD | lassosum.10FCVal | LDpred1.Inf | 0.134 | 0.098 | 0.036 | 2.06e-17 | 0.140 | 0.103 | 0.037 | 8.33e-06 |
| IBD | LDpred2.10FCVal | LDpred1.Inf | 0.143 | 0.098 | 0.045 | 7.19e-33 | 0.149 | 0.103 | 0.046 | 6.70e-10 |
| IBD | SBayesR.PseudoVal | LDpred1.Inf | 0.128 | 0.098 | 0.029 | 6.35e-12 | 0.143 | 0.103 | 0.040 | 2.28e-06 |
| IBD | LDpred1.MultiPRS | LDpred1.Inf | 0.130 | 0.098 | 0.032 | 6.20e-06 | 0.138 | 0.103 | 0.035 | 5.33e-12 |
| IBD | LDpred2.PseudoVal | LDpred1.Inf | 0.140 | 0.098 | 0.042 | 9.50e-20 | 0.145 | 0.103 | 0.042 | 3.57e-06 |
| IBD | SBLUP.Inf | LDpred1.Inf | 0.097 | 0.098 | -0.001 | 4.92e-01 | 0.102 | 0.103 | -0.001 | 7.84e-01 |
| IBD | pT+clump.MultiPRS | LDpred1.Inf | 0.127 | 0.098 | 0.029 | 3.14e-05 | 0.136 | 0.103 | 0.033 | 5.55e-05 |
| IBD | lassosum.MultiPRS | LDpred1.Inf | 0.144 | 0.098 | 0.045 | 1.03e-10 | 0.148 | 0.103 | 0.045 | 1.04e-08 |
| IBD | LDpred1.10FCVal | LDpred1.Inf | 0.129 | 0.098 | 0.031 | 9.32e-46 | 0.137 | 0.103 | 0.034 | 3.86e-15 |
| IBD | LDpred1.Inf | LDpred1.10FCVal | 0.098 | 0.129 | -0.031 | 9.32e-46 | 0.103 | 0.137 | -0.034 | 3.86e-15 |
| IBD | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.130 | 0.129 | 0.001 | 8.40e-01 | 0.136 | 0.137 | -0.001 | 8.45e-01 |
| IBD | lassosum.PseudoVal | LDpred1.10FCVal | 0.133 | 0.129 | 0.004 | 2.45e-01 | 0.140 | 0.137 | 0.003 | 7.02e-01 |
| IBD | LDpred2.MultiPRS | LDpred1.10FCVal | 0.143 | 0.129 | 0.013 | 5.42e-02 | 0.150 | 0.137 | 0.013 | 4.77e-03 |
| IBD | pT+clump.10FCVal | LDpred1.10FCVal | 0.111 | 0.129 | -0.018 | 3.40e-05 | 0.109 | 0.137 | -0.028 | 1.04e-03 |
| IBD | lassosum.10FCVal | LDpred1.10FCVal | 0.134 | 0.129 | 0.005 | 8.23e-02 | 0.140 | 0.137 | 0.003 | 5.42e-01 |
| IBD | LDpred2.10FCVal | LDpred1.10FCVal | 0.143 | 0.129 | 0.014 | 9.42e-11 | 0.149 | 0.137 | 0.012 | 3.99e-03 |
| IBD | All.MultiPRS | LDpred1.10FCVal | 0.146 | 0.129 | 0.017 | 1.47e-02 | 0.152 | 0.137 | 0.015 | 2.38e-03 |
| IBD | LDpred1.MultiPRS | LDpred1.10FCVal | 0.130 | 0.129 | 0.001 | 8.96e-01 | 0.138 | 0.137 | 0.001 | 4.56e-01 |
| IBD | LDpred2.PseudoVal | LDpred1.10FCVal | 0.140 | 0.129 | 0.011 | 4.25e-04 | 0.145 | 0.137 | 0.008 | 1.69e-01 |
| IBD | SBLUP.Inf | LDpred1.10FCVal | 0.097 | 0.129 | -0.032 | 2.01e-32 | 0.102 | 0.137 | -0.035 | 6.36e-11 |
| IBD | pT+clump.MultiPRS | LDpred1.10FCVal | 0.127 | 0.129 | -0.002 | 8.08e-01 | 0.136 | 0.137 | -0.001 | 8.72e-01 |
| IBD | LDpred2.Inf | LDpred1.10FCVal | 0.096 | 0.129 | -0.033 | 1.12e-34 | 0.100 | 0.137 | -0.037 | 6.92e-12 |
| IBD | lassosum.MultiPRS | LDpred1.10FCVal | 0.144 | 0.129 | 0.014 | 3.86e-02 | 0.148 | 0.137 | 0.011 | 3.26e-02 |
| IBD | LDpred1.10FCVal | LDpred1.10FCVal | 0.129 | 0.129 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| IBD | SBayesR.PseudoVal | LDpred1.10FCVal | 0.128 | 0.129 | -0.002 | 5.75e-01 | 0.143 | 0.137 | 0.006 | 2.63e-01 |
| IBD | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.150 | 0.150 | 0.000 | 1.00e+00 |
| IBD | lassosum.PseudoVal | LDpred2.MultiPRS | 0.133 | 0.143 | -0.010 | 1.74e-01 | 0.140 | 0.150 | -0.011 | 8.21e-03 |
| IBD | All.MultiPRS | LDpred2.MultiPRS | 0.146 | 0.143 | 0.004 | 2.30e-04 | 0.152 | 0.150 | 0.002 | 4.02e-01 |
| IBD | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.130 | 0.143 | -0.013 | 8.80e-09 | 0.138 | 0.150 | -0.012 | 5.10e-03 |
| IBD | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.130 | 0.143 | -0.013 | 6.62e-02 | 0.136 | 0.150 | -0.014 | 2.22e-03 |
| IBD | LDpred1.Inf | LDpred2.MultiPRS | 0.098 | 0.143 | -0.044 | 2.75e-10 | 0.103 | 0.150 | -0.047 | 2.87e-09 |
| IBD | LDpred2.Inf | LDpred2.MultiPRS | 0.096 | 0.143 | -0.047 | 3.28e-11 | 0.100 | 0.150 | -0.050 | 4.79e-10 |
| IBD | lassosum.MultiPRS | LDpred2.MultiPRS | 0.144 | 0.143 | 0.001 | 5.57e-01 | 0.148 | 0.150 | -0.002 | 4.55e-01 |
| IBD | LDpred1.10FCVal | LDpred2.MultiPRS | 0.129 | 0.143 | -0.013 | 5.42e-02 | 0.137 | 0.150 | -0.013 | 4.77e-03 |
| IBD | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.127 | 0.143 | -0.015 | 2.96e-02 | 0.136 | 0.150 | -0.014 | 6.14e-03 |
| IBD | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.128 | 0.143 | -0.015 | 3.12e-02 | 0.143 | 0.150 | -0.007 | 9.69e-02 |
| IBD | pT+clump.10FCVal | LDpred2.MultiPRS | 0.111 | 0.143 | -0.032 | 5.89e-06 | 0.109 | 0.150 | -0.042 | 1.01e-09 |
| IBD | lassosum.10FCVal | LDpred2.MultiPRS | 0.134 | 0.143 | -0.008 | 2.24e-01 | 0.140 | 0.150 | -0.010 | 1.80e-02 |
| IBD | LDpred2.10FCVal | LDpred2.MultiPRS | 0.143 | 0.143 | 0.000 | 9.61e-01 | 0.149 | 0.150 | -0.001 | 4.47e-01 |
| IBD | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.140 | 0.143 | -0.003 | 6.99e-01 | 0.145 | 0.150 | -0.005 | 1.09e-02 |
| IBD | SBLUP.Inf | LDpred2.MultiPRS | 0.097 | 0.143 | -0.045 | 1.02e-10 | 0.102 | 0.150 | -0.048 | 2.15e-09 |
| IBD | LDpred2.MultiPRS | LDpred2.10FCVal | 0.143 | 0.143 | 0.000 | 9.61e-01 | 0.150 | 0.149 | 0.001 | 4.47e-01 |
| IBD | pT+clump.10FCVal | LDpred2.10FCVal | 0.111 | 0.143 | -0.032 | 5.93e-18 | 0.109 | 0.149 | -0.041 | 2.40e-08 |
| IBD | lassosum.PseudoVal | LDpred2.10FCVal | 0.133 | 0.143 | -0.010 | 6.59e-06 | 0.140 | 0.149 | -0.010 | 2.60e-02 |
| IBD | All.MultiPRS | LDpred2.10FCVal | 0.146 | 0.143 | 0.003 | 6.41e-01 | 0.152 | 0.149 | 0.003 | 2.76e-01 |
| IBD | LDpred1.MultiPRS | LDpred2.10FCVal | 0.130 | 0.143 | -0.013 | 6.46e-02 | 0.138 | 0.149 | -0.011 | 4.92e-03 |
| IBD | LDpred2.PseudoVal | LDpred2.10FCVal | 0.140 | 0.143 | -0.003 | 2.01e-02 | 0.145 | 0.149 | -0.004 | 1.35e-01 |
| IBD | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.130 | 0.143 | -0.013 | 3.34e-07 | 0.136 | 0.149 | -0.013 | 9.22e-03 |
| IBD | LDpred1.Inf | LDpred2.10FCVal | 0.098 | 0.143 | -0.045 | 7.19e-33 | 0.103 | 0.149 | -0.046 | 6.70e-10 |
| IBD | LDpred2.Inf | LDpred2.10FCVal | 0.096 | 0.143 | -0.047 | 6.30e-36 | 0.100 | 0.149 | -0.049 | 6.16e-11 |
| IBD | lassosum.MultiPRS | LDpred2.10FCVal | 0.144 | 0.143 | 0.001 | 9.27e-01 | 0.148 | 0.149 | -0.001 | 6.78e-01 |
| IBD | LDpred1.10FCVal | LDpred2.10FCVal | 0.129 | 0.143 | -0.014 | 9.42e-11 | 0.137 | 0.149 | -0.012 | 3.99e-03 |
| IBD | pT+clump.MultiPRS | LDpred2.10FCVal | 0.127 | 0.143 | -0.016 | 2.64e-02 | 0.136 | 0.149 | -0.013 | 1.25e-02 |
| IBD | SBayesR.PseudoVal | LDpred2.10FCVal | 0.128 | 0.143 | -0.015 | 3.45e-11 | 0.143 | 0.149 | -0.006 | 2.05e-01 |
| IBD | lassosum.10FCVal | LDpred2.10FCVal | 0.134 | 0.143 | -0.009 | 2.90e-06 | 0.140 | 0.149 | -0.009 | 2.12e-02 |
| IBD | LDpred2.10FCVal | LDpred2.10FCVal | 0.143 | 0.143 | 0.000 | 1.00e+00 | 0.149 | 0.149 | 0.000 | 1.00e+00 |
| IBD | SBLUP.Inf | LDpred2.10FCVal | 0.097 | 0.143 | -0.046 | 4.63e-34 | 0.102 | 0.149 | -0.047 | 3.33e-10 |
| IBD | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.143 | 0.140 | 0.003 | 6.99e-01 | 0.150 | 0.145 | 0.005 | 1.09e-02 |
| IBD | lassosum.PseudoVal | LDpred2.PseudoVal | 0.133 | 0.140 | -0.007 | 2.66e-04 | 0.140 | 0.145 | -0.006 | 1.22e-01 |
| IBD | All.MultiPRS | LDpred2.PseudoVal | 0.146 | 0.140 | 0.006 | 3.67e-01 | 0.152 | 0.145 | 0.007 | 2.11e-02 |
| IBD | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.130 | 0.140 | -0.010 | 1.57e-01 | 0.138 | 0.145 | -0.007 | 2.01e-01 |
| IBD | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.130 | 0.140 | -0.010 | 2.82e-04 | 0.136 | 0.145 | -0.010 | 8.69e-02 |
| IBD | LDpred1.Inf | LDpred2.PseudoVal | 0.098 | 0.140 | -0.042 | 9.50e-20 | 0.103 | 0.145 | -0.042 | 3.57e-06 |
| IBD | LDpred2.Inf | LDpred2.PseudoVal | 0.096 | 0.140 | -0.044 | 3.83e-21 | 0.100 | 0.145 | -0.045 | 1.12e-06 |
| IBD | lassosum.MultiPRS | LDpred2.PseudoVal | 0.144 | 0.140 | 0.004 | 5.98e-01 | 0.148 | 0.145 | 0.002 | 5.49e-01 |
| IBD | LDpred1.10FCVal | LDpred2.PseudoVal | 0.129 | 0.140 | -0.011 | 4.25e-04 | 0.137 | 0.145 | -0.008 | 1.69e-01 |
| IBD | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.127 | 0.140 | -0.012 | 7.40e-02 | 0.136 | 0.145 | -0.009 | 1.08e-01 |
| IBD | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.128 | 0.140 | -0.012 | 1.01e-07 | 0.143 | 0.145 | -0.002 | 6.60e-01 |
| IBD | pT+clump.10FCVal | LDpred2.PseudoVal | 0.111 | 0.140 | -0.029 | 2.26e-18 | 0.109 | 0.145 | -0.037 | 2.09e-08 |
| IBD | lassosum.10FCVal | LDpred2.PseudoVal | 0.134 | 0.140 | -0.006 | 9.74e-03 | 0.140 | 0.145 | -0.005 | 2.78e-01 |
| IBD | LDpred2.10FCVal | LDpred2.PseudoVal | 0.143 | 0.140 | 0.003 | 2.01e-02 | 0.149 | 0.145 | 0.004 | 1.35e-01 |
| IBD | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.140 | 0.140 | 0.000 | 1.00e+00 | 0.145 | 0.145 | 0.000 | 1.00e+00 |
| IBD | SBLUP.Inf | LDpred2.PseudoVal | 0.097 | 0.140 | -0.043 | 5.17e-20 | 0.102 | 0.145 | -0.043 | 3.15e-06 |
| IBD | DBSLMM.PseudoVal | LDpred2.Inf | 0.130 | 0.096 | 0.034 | 1.38e-14 | 0.136 | 0.100 | 0.036 | 4.87e-05 |
| IBD | LDpred2.MultiPRS | LDpred2.Inf | 0.143 | 0.096 | 0.047 | 3.28e-11 | 0.150 | 0.100 | 0.050 | 4.79e-10 |
| IBD | pT+clump.10FCVal | LDpred2.Inf | 0.111 | 0.096 | 0.015 | 8.14e-03 | 0.109 | 0.100 | 0.008 | 4.56e-01 |
| IBD | lassosum.10FCVal | LDpred2.Inf | 0.134 | 0.096 | 0.038 | 5.63e-19 | 0.140 | 0.100 | 0.040 | 2.32e-06 |
| IBD | lassosum.PseudoVal | LDpred2.Inf | 0.133 | 0.096 | 0.037 | 3.13e-13 | 0.140 | 0.100 | 0.039 | 9.86e-05 |
| IBD | All.MultiPRS | LDpred2.Inf | 0.146 | 0.096 | 0.050 | 8.73e-13 | 0.152 | 0.100 | 0.052 | 8.29e-11 |
| IBD | LDpred1.MultiPRS | LDpred2.Inf | 0.130 | 0.096 | 0.034 | 1.29e-06 | 0.138 | 0.100 | 0.038 | 1.94e-10 |
| IBD | LDpred2.PseudoVal | LDpred2.Inf | 0.140 | 0.096 | 0.044 | 3.83e-21 | 0.145 | 0.100 | 0.045 | 1.12e-06 |
| IBD | SBLUP.Inf | LDpred2.Inf | 0.097 | 0.096 | 0.001 | 1.23e-02 | 0.102 | 0.100 | 0.002 | 5.05e-02 |
| IBD | LDpred1.Inf | LDpred2.Inf | 0.098 | 0.096 | 0.002 | 1.40e-01 | 0.103 | 0.100 | 0.003 | 3.79e-01 |
| IBD | LDpred2.Inf | LDpred2.Inf | 0.096 | 0.096 | 0.000 | 1.00e+00 | 0.100 | 0.100 | 0.000 | 1.00e+00 |
| IBD | lassosum.MultiPRS | LDpred2.Inf | 0.144 | 0.096 | 0.048 | 1.18e-11 | 0.148 | 0.100 | 0.048 | 2.35e-09 |
| IBD | LDpred1.10FCVal | LDpred2.Inf | 0.129 | 0.096 | 0.033 | 1.12e-34 | 0.137 | 0.100 | 0.037 | 6.92e-12 |
| IBD | pT+clump.MultiPRS | LDpred2.Inf | 0.127 | 0.096 | 0.031 | 7.12e-06 | 0.136 | 0.100 | 0.036 | 2.73e-05 |
| IBD | SBayesR.PseudoVal | LDpred2.Inf | 0.128 | 0.096 | 0.032 | 1.95e-12 | 0.143 | 0.100 | 0.043 | 1.71e-06 |
| IBD | LDpred2.10FCVal | LDpred2.Inf | 0.143 | 0.096 | 0.047 | 6.30e-36 | 0.149 | 0.100 | 0.049 | 6.16e-11 |
| IBD | All.MultiPRS | pT+clump.MultiPRS | 0.146 | 0.127 | 0.019 | 7.14e-03 | 0.152 | 0.136 | 0.016 | 9.79e-04 |
| IBD | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.130 | 0.127 | 0.003 | 7.08e-01 | 0.138 | 0.136 | 0.002 | 7.07e-01 |
| IBD | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.140 | 0.127 | 0.012 | 7.40e-02 | 0.145 | 0.136 | 0.009 | 1.08e-01 |
| IBD | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.130 | 0.127 | 0.002 | 7.41e-01 | 0.136 | 0.136 | 0.000 | 9.70e-01 |
| IBD | LDpred1.Inf | pT+clump.MultiPRS | 0.098 | 0.127 | -0.029 | 3.14e-05 | 0.103 | 0.136 | -0.033 | 5.55e-05 |
| IBD | LDpred2.Inf | pT+clump.MultiPRS | 0.096 | 0.127 | -0.031 | 7.12e-06 | 0.100 | 0.136 | -0.036 | 2.73e-05 |
| IBD | lassosum.MultiPRS | pT+clump.MultiPRS | 0.144 | 0.127 | 0.016 | 2.06e-02 | 0.148 | 0.136 | 0.012 | 2.19e-02 |
| IBD | LDpred1.10FCVal | pT+clump.MultiPRS | 0.129 | 0.127 | 0.002 | 8.08e-01 | 0.137 | 0.136 | 0.001 | 8.72e-01 |
| IBD | lassosum.PseudoVal | pT+clump.MultiPRS | 0.133 | 0.127 | 0.006 | 4.17e-01 | 0.140 | 0.136 | 0.004 | 5.45e-01 |
| IBD | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.143 | 0.127 | 0.015 | 2.96e-02 | 0.150 | 0.136 | 0.014 | 6.14e-03 |
| IBD | pT+clump.10FCVal | pT+clump.MultiPRS | 0.111 | 0.127 | -0.017 | 1.84e-02 | 0.109 | 0.136 | -0.027 | 1.36e-07 |
| IBD | lassosum.10FCVal | pT+clump.MultiPRS | 0.134 | 0.127 | 0.007 | 3.39e-01 | 0.140 | 0.136 | 0.004 | 4.32e-01 |
| IBD | LDpred2.10FCVal | pT+clump.MultiPRS | 0.143 | 0.127 | 0.016 | 2.64e-02 | 0.149 | 0.136 | 0.013 | 1.25e-02 |
| IBD | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.128 | 0.127 | 0.000 | 9.86e-01 | 0.143 | 0.136 | 0.007 | 2.52e-01 |
| IBD | SBLUP.Inf | pT+clump.MultiPRS | 0.097 | 0.127 | -0.030 | 1.57e-05 | 0.102 | 0.136 | -0.034 | 6.41e-05 |
| IBD | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.127 | 0.127 | 0.000 | 1.00e+00 | 0.136 | 0.136 | 0.000 | 1.00e+00 |
| IBD | lassosum.PseudoVal | pT+clump.10FCVal | 0.133 | 0.111 | 0.022 | 2.09e-11 | 0.140 | 0.109 | 0.031 | 2.25e-06 |
| IBD | All.MultiPRS | pT+clump.10FCVal | 0.146 | 0.111 | 0.035 | 4.48e-07 | 0.152 | 0.109 | 0.043 | 1.80e-10 |
| IBD | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.130 | 0.111 | 0.019 | 2.68e-07 | 0.136 | 0.109 | 0.027 | 1.73e-04 |
| IBD | LDpred1.Inf | pT+clump.10FCVal | 0.098 | 0.111 | -0.013 | 1.99e-02 | 0.103 | 0.109 | -0.006 | 6.03e-01 |
| IBD | LDpred2.Inf | pT+clump.10FCVal | 0.096 | 0.111 | -0.015 | 8.14e-03 | 0.100 | 0.109 | -0.008 | 4.56e-01 |
| IBD | lassosum.MultiPRS | pT+clump.10FCVal | 0.144 | 0.111 | 0.033 | 2.95e-06 | 0.148 | 0.109 | 0.039 | 5.45e-08 |
| IBD | LDpred1.10FCVal | pT+clump.10FCVal | 0.129 | 0.111 | 0.018 | 3.40e-05 | 0.137 | 0.109 | 0.028 | 1.04e-03 |
| IBD | pT+clump.MultiPRS | pT+clump.10FCVal | 0.127 | 0.111 | 0.017 | 1.84e-02 | 0.136 | 0.109 | 0.027 | 1.36e-07 |
| IBD | LDpred2.MultiPRS | pT+clump.10FCVal | 0.143 | 0.111 | 0.032 | 5.89e-06 | 0.150 | 0.109 | 0.042 | 1.01e-09 |
| IBD | pT+clump.10FCVal | pT+clump.10FCVal | 0.111 | 0.111 | 0.000 | 1.00e+00 | 0.109 | 0.109 | 0.000 | 1.00e+00 |
| IBD | lassosum.10FCVal | pT+clump.10FCVal | 0.134 | 0.111 | 0.023 | 6.20e-09 | 0.140 | 0.109 | 0.032 | 5.42e-05 |
| IBD | LDpred2.10FCVal | pT+clump.10FCVal | 0.143 | 0.111 | 0.032 | 5.93e-18 | 0.149 | 0.109 | 0.041 | 2.40e-08 |
| IBD | SBayesR.PseudoVal | pT+clump.10FCVal | 0.128 | 0.111 | 0.017 | 8.70e-06 | 0.143 | 0.109 | 0.035 | 2.07e-06 |
| IBD | LDpred1.MultiPRS | pT+clump.10FCVal | 0.130 | 0.111 | 0.019 | 6.27e-03 | 0.138 | 0.109 | 0.030 | 3.59e-04 |
| IBD | LDpred2.PseudoVal | pT+clump.10FCVal | 0.140 | 0.111 | 0.029 | 2.26e-18 | 0.145 | 0.109 | 0.037 | 2.09e-08 |
| IBD | SBLUP.Inf | pT+clump.10FCVal | 0.097 | 0.111 | -0.014 | 1.54e-02 | 0.102 | 0.109 | -0.006 | 5.65e-01 |
| IBD | LDpred1.Inf | SBayesR.PseudoVal | 0.098 | 0.128 | -0.029 | 6.35e-12 | 0.103 | 0.143 | -0.040 | 2.28e-06 |
| IBD | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.130 | 0.128 | 0.002 | 4.59e-01 | 0.136 | 0.143 | -0.008 | 2.05e-01 |
| IBD | lassosum.PseudoVal | SBayesR.PseudoVal | 0.133 | 0.128 | 0.006 | 3.33e-02 | 0.140 | 0.143 | -0.004 | 4.66e-01 |
| IBD | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.143 | 0.128 | 0.015 | 3.12e-02 | 0.150 | 0.143 | 0.007 | 9.69e-02 |
| IBD | pT+clump.10FCVal | SBayesR.PseudoVal | 0.111 | 0.128 | -0.017 | 8.70e-06 | 0.109 | 0.143 | -0.035 | 2.07e-06 |
| IBD | lassosum.10FCVal | SBayesR.PseudoVal | 0.134 | 0.128 | 0.007 | 2.11e-02 | 0.140 | 0.143 | -0.003 | 6.16e-01 |
| IBD | LDpred2.10FCVal | SBayesR.PseudoVal | 0.143 | 0.128 | 0.015 | 3.45e-11 | 0.149 | 0.143 | 0.006 | 2.05e-01 |
| IBD | All.MultiPRS | SBayesR.PseudoVal | 0.146 | 0.128 | 0.019 | 7.54e-03 | 0.152 | 0.143 | 0.009 | 6.34e-02 |
| IBD | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.130 | 0.128 | 0.002 | 7.21e-01 | 0.138 | 0.143 | -0.005 | 3.38e-01 |
| IBD | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.140 | 0.128 | 0.012 | 1.01e-07 | 0.145 | 0.143 | 0.002 | 6.60e-01 |
| IBD | SBLUP.Inf | SBayesR.PseudoVal | 0.097 | 0.128 | -0.030 | 1.54e-11 | 0.102 | 0.143 | -0.041 | 4.96e-06 |
| IBD | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.127 | 0.128 | 0.000 | 9.86e-01 | 0.136 | 0.143 | -0.007 | 2.52e-01 |
| IBD | LDpred2.Inf | SBayesR.PseudoVal | 0.096 | 0.128 | -0.032 | 1.95e-12 | 0.100 | 0.143 | -0.043 | 1.71e-06 |
| IBD | lassosum.MultiPRS | SBayesR.PseudoVal | 0.144 | 0.128 | 0.016 | 2.15e-02 | 0.148 | 0.143 | 0.004 | 3.78e-01 |
| IBD | LDpred1.10FCVal | SBayesR.PseudoVal | 0.129 | 0.128 | 0.002 | 5.75e-01 | 0.137 | 0.143 | -0.006 | 2.63e-01 |
| IBD | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.128 | 0.128 | 0.000 | 1.00e+00 | 0.143 | 0.143 | 0.000 | 1.00e+00 |
| IBD | All.MultiPRS | SBLUP.Inf | 0.146 | 0.097 | 0.049 | 2.97e-12 | 0.152 | 0.102 | 0.050 | 2.75e-10 |
| IBD | LDpred1.Inf | SBLUP.Inf | 0.098 | 0.097 | 0.001 | 4.92e-01 | 0.103 | 0.102 | 0.001 | 7.84e-01 |
| IBD | LDpred2.Inf | SBLUP.Inf | 0.096 | 0.097 | -0.001 | 1.23e-02 | 0.100 | 0.102 | -0.002 | 5.05e-02 |
| IBD | DBSLMM.PseudoVal | SBLUP.Inf | 0.130 | 0.097 | 0.033 | 1.41e-13 | 0.136 | 0.102 | 0.034 | 1.24e-04 |
| IBD | lassosum.PseudoVal | SBLUP.Inf | 0.133 | 0.097 | 0.036 | 1.77e-12 | 0.140 | 0.102 | 0.037 | 2.07e-04 |
| IBD | LDpred2.MultiPRS | SBLUP.Inf | 0.143 | 0.097 | 0.045 | 1.02e-10 | 0.150 | 0.102 | 0.048 | 2.15e-09 |
| IBD | pT+clump.10FCVal | SBLUP.Inf | 0.111 | 0.097 | 0.014 | 1.54e-02 | 0.109 | 0.102 | 0.006 | 5.65e-01 |
| IBD | lassosum.10FCVal | SBLUP.Inf | 0.134 | 0.097 | 0.037 | 4.93e-18 | 0.140 | 0.102 | 0.038 | 6.02e-06 |
| IBD | LDpred2.10FCVal | SBLUP.Inf | 0.143 | 0.097 | 0.046 | 4.63e-34 | 0.149 | 0.102 | 0.047 | 3.33e-10 |
| IBD | SBayesR.PseudoVal | SBLUP.Inf | 0.128 | 0.097 | 0.030 | 1.54e-11 | 0.143 | 0.102 | 0.041 | 4.96e-06 |
| IBD | LDpred1.MultiPRS | SBLUP.Inf | 0.130 | 0.097 | 0.033 | 2.98e-06 | 0.138 | 0.102 | 0.036 | 1.27e-09 |
| IBD | LDpred2.PseudoVal | SBLUP.Inf | 0.140 | 0.097 | 0.043 | 5.17e-20 | 0.145 | 0.102 | 0.043 | 3.15e-06 |
| IBD | SBLUP.Inf | SBLUP.Inf | 0.097 | 0.097 | 0.000 | 1.00e+00 | 0.102 | 0.102 | 0.000 | 1.00e+00 |
| IBD | pT+clump.MultiPRS | SBLUP.Inf | 0.127 | 0.097 | 0.030 | 1.57e-05 | 0.136 | 0.102 | 0.034 | 6.41e-05 |
| IBD | lassosum.MultiPRS | SBLUP.Inf | 0.144 | 0.097 | 0.046 | 3.76e-11 | 0.148 | 0.102 | 0.046 | 6.21e-09 |
| IBD | LDpred1.10FCVal | SBLUP.Inf | 0.129 | 0.097 | 0.032 | 2.01e-32 | 0.137 | 0.102 | 0.035 | 6.36e-11 |
| MultiScler | lassosum.PseudoVal | All.MultiPRS | 0.103 | 0.135 | -0.032 | 5.21e-06 | 0.102 | 0.121 | -0.019 | 5.54e-03 |
| MultiScler | All.MultiPRS | All.MultiPRS | 0.135 | 0.135 | 0.000 | 1.00e+00 | 0.121 | 0.121 | 0.000 | 1.00e+00 |
| MultiScler | DBSLMM.PseudoVal | All.MultiPRS | 0.091 | 0.135 | -0.044 | 3.53e-10 | 0.088 | 0.121 | -0.033 | 6.81e-05 |
| MultiScler | LDpred1.Inf | All.MultiPRS | 0.090 | 0.135 | -0.046 | 6.81e-11 | 0.078 | 0.121 | -0.042 | 4.17e-07 |
| MultiScler | LDpred2.Inf | All.MultiPRS | 0.094 | 0.135 | -0.041 | 3.76e-09 | 0.076 | 0.121 | -0.044 | 5.44e-07 |
| MultiScler | lassosum.MultiPRS | All.MultiPRS | 0.128 | 0.135 | -0.008 | 1.56e-04 | 0.119 | 0.121 | -0.002 | 5.77e-01 |
| MultiScler | LDpred1.10FCVal | All.MultiPRS | 0.107 | 0.135 | -0.028 | 6.59e-05 | 0.096 | 0.121 | -0.025 | 1.08e-04 |
| MultiScler | pT+clump.MultiPRS | All.MultiPRS | 0.114 | 0.135 | -0.021 | 2.61e-03 | 0.108 | 0.121 | -0.012 | 2.32e-02 |
| MultiScler | LDpred2.MultiPRS | All.MultiPRS | 0.133 | 0.135 | -0.003 | 1.33e-03 | 0.119 | 0.121 | -0.002 | 2.87e-01 |
| MultiScler | pT+clump.10FCVal | All.MultiPRS | 0.090 | 0.135 | -0.045 | 1.18e-10 | 0.092 | 0.121 | -0.029 | 4.11e-04 |
| MultiScler | lassosum.10FCVal | All.MultiPRS | 0.115 | 0.135 | -0.020 | 4.13e-03 | 0.097 | 0.121 | -0.024 | 1.42e-04 |
| MultiScler | LDpred2.10FCVal | All.MultiPRS | 0.120 | 0.135 | -0.015 | 3.18e-02 | 0.108 | 0.121 | -0.013 | 3.84e-03 |
| MultiScler | SBayesR.PseudoVal | All.MultiPRS | 0.102 | 0.135 | -0.034 | 1.27e-06 | 0.083 | 0.121 | -0.038 | 5.56e-07 |
| MultiScler | LDpred1.MultiPRS | All.MultiPRS | 0.112 | 0.135 | -0.023 | 1.42e-13 | 0.096 | 0.121 | -0.025 | 4.26e-05 |
| MultiScler | LDpred2.PseudoVal | All.MultiPRS | 0.119 | 0.135 | -0.017 | 1.74e-02 | 0.110 | 0.121 | -0.010 | 1.28e-02 |
| MultiScler | SBLUP.Inf | All.MultiPRS | 0.094 | 0.135 | -0.042 | 3.21e-09 | 0.076 | 0.121 | -0.045 | 3.67e-07 |
| MultiScler | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.133 | 0.091 | 0.041 | 3.65e-09 | 0.119 | 0.088 | 0.031 | 7.58e-05 |
| MultiScler | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.090 | 0.091 | -0.001 | 7.36e-01 | 0.092 | 0.088 | 0.004 | 5.67e-01 |
| MultiScler | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.103 | 0.091 | 0.012 | 2.28e-07 | 0.102 | 0.088 | 0.014 | 3.17e-03 |
| MultiScler | All.MultiPRS | DBSLMM.PseudoVal | 0.135 | 0.091 | 0.044 | 3.53e-10 | 0.121 | 0.088 | 0.033 | 6.81e-05 |
| MultiScler | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.112 | 0.091 | 0.021 | 2.85e-03 | 0.096 | 0.088 | 0.008 | 3.09e-01 |
| MultiScler | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.119 | 0.091 | 0.027 | 2.92e-11 | 0.110 | 0.088 | 0.022 | 7.23e-03 |
| MultiScler | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.091 | 0.091 | 0.000 | 1.00e+00 | 0.088 | 0.088 | 0.000 | 1.00e+00 |
| MultiScler | LDpred1.Inf | DBSLMM.PseudoVal | 0.090 | 0.091 | -0.002 | 7.30e-01 | 0.078 | 0.088 | -0.010 | 3.60e-01 |
| MultiScler | LDpred2.Inf | DBSLMM.PseudoVal | 0.094 | 0.091 | 0.003 | 6.74e-01 | 0.076 | 0.088 | -0.012 | 3.57e-01 |
| MultiScler | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.128 | 0.091 | 0.036 | 2.09e-07 | 0.119 | 0.088 | 0.030 | 6.93e-05 |
| MultiScler | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.107 | 0.091 | 0.016 | 1.47e-05 | 0.096 | 0.088 | 0.007 | 3.11e-01 |
| MultiScler | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.114 | 0.091 | 0.023 | 1.01e-03 | 0.108 | 0.088 | 0.020 | 1.81e-02 |
| MultiScler | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.102 | 0.091 | 0.010 | 1.02e-01 | 0.083 | 0.088 | -0.005 | 6.88e-01 |
| MultiScler | lassosum.10FCVal | DBSLMM.PseudoVal | 0.115 | 0.091 | 0.024 | 7.66e-07 | 0.097 | 0.088 | 0.009 | 3.77e-01 |
| MultiScler | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.120 | 0.091 | 0.029 | 5.00e-27 | 0.108 | 0.088 | 0.020 | 3.35e-04 |
| MultiScler | SBLUP.Inf | DBSLMM.PseudoVal | 0.094 | 0.091 | 0.002 | 6.96e-01 | 0.076 | 0.088 | -0.012 | 3.28e-01 |
| MultiScler | All.MultiPRS | lassosum.MultiPRS | 0.135 | 0.128 | 0.008 | 1.56e-04 | 0.121 | 0.119 | 0.002 | 5.77e-01 |
| MultiScler | LDpred1.MultiPRS | lassosum.MultiPRS | 0.112 | 0.128 | -0.015 | 1.86e-07 | 0.096 | 0.119 | -0.023 | 1.10e-04 |
| MultiScler | LDpred1.Inf | lassosum.MultiPRS | 0.090 | 0.128 | -0.038 | 5.29e-08 | 0.078 | 0.119 | -0.040 | 1.39e-07 |
| MultiScler | LDpred2.Inf | lassosum.MultiPRS | 0.094 | 0.128 | -0.034 | 1.56e-06 | 0.076 | 0.119 | -0.042 | 2.91e-07 |
| MultiScler | lassosum.MultiPRS | lassosum.MultiPRS | 0.128 | 0.128 | 0.000 | 1.00e+00 | 0.119 | 0.119 | 0.000 | 1.00e+00 |
| MultiScler | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.091 | 0.128 | -0.036 | 2.09e-07 | 0.088 | 0.119 | -0.030 | 6.93e-05 |
| MultiScler | lassosum.PseudoVal | lassosum.MultiPRS | 0.103 | 0.128 | -0.024 | 5.13e-04 | 0.102 | 0.119 | -0.016 | 1.39e-03 |
| MultiScler | LDpred2.MultiPRS | lassosum.MultiPRS | 0.133 | 0.128 | 0.005 | 1.98e-02 | 0.119 | 0.119 | 0.001 | 9.05e-01 |
| MultiScler | pT+clump.10FCVal | lassosum.MultiPRS | 0.090 | 0.128 | -0.038 | 8.61e-08 | 0.092 | 0.119 | -0.027 | 3.25e-03 |
| MultiScler | lassosum.10FCVal | lassosum.MultiPRS | 0.115 | 0.128 | -0.012 | 7.49e-02 | 0.097 | 0.119 | -0.022 | 1.66e-06 |
| MultiScler | LDpred2.10FCVal | lassosum.MultiPRS | 0.120 | 0.128 | -0.007 | 2.89e-01 | 0.108 | 0.119 | -0.011 | 5.45e-02 |
| MultiScler | SBayesR.PseudoVal | lassosum.MultiPRS | 0.102 | 0.128 | -0.026 | 1.75e-04 | 0.083 | 0.119 | -0.036 | 2.18e-05 |
| MultiScler | LDpred2.PseudoVal | lassosum.MultiPRS | 0.119 | 0.128 | -0.009 | 1.97e-01 | 0.110 | 0.119 | -0.008 | 1.48e-01 |
| MultiScler | SBLUP.Inf | lassosum.MultiPRS | 0.094 | 0.128 | -0.034 | 1.36e-06 | 0.076 | 0.119 | -0.043 | 1.85e-07 |
| MultiScler | pT+clump.MultiPRS | lassosum.MultiPRS | 0.114 | 0.128 | -0.013 | 5.50e-02 | 0.108 | 0.119 | -0.010 | 7.50e-02 |
| MultiScler | LDpred1.10FCVal | lassosum.MultiPRS | 0.107 | 0.128 | -0.020 | 3.65e-03 | 0.096 | 0.119 | -0.023 | 5.29e-05 |
| MultiScler | All.MultiPRS | lassosum.PseudoVal | 0.135 | 0.103 | 0.032 | 5.21e-06 | 0.121 | 0.102 | 0.019 | 5.54e-03 |
| MultiScler | LDpred1.MultiPRS | lassosum.PseudoVal | 0.112 | 0.103 | 0.009 | 2.06e-01 | 0.096 | 0.102 | -0.007 | 3.04e-01 |
| MultiScler | LDpred2.PseudoVal | lassosum.PseudoVal | 0.119 | 0.103 | 0.015 | 1.16e-06 | 0.110 | 0.102 | 0.008 | 1.84e-01 |
| MultiScler | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.091 | 0.103 | -0.012 | 2.28e-07 | 0.088 | 0.102 | -0.014 | 3.17e-03 |
| MultiScler | LDpred1.Inf | lassosum.PseudoVal | 0.090 | 0.103 | -0.014 | 3.81e-03 | 0.078 | 0.102 | -0.024 | 1.32e-02 |
| MultiScler | LDpred2.Inf | lassosum.PseudoVal | 0.094 | 0.103 | -0.009 | 9.86e-02 | 0.076 | 0.102 | -0.026 | 2.48e-02 |
| MultiScler | lassosum.MultiPRS | lassosum.PseudoVal | 0.128 | 0.103 | 0.024 | 5.13e-04 | 0.119 | 0.102 | 0.016 | 1.39e-03 |
| MultiScler | LDpred1.10FCVal | lassosum.PseudoVal | 0.107 | 0.103 | 0.004 | 2.18e-01 | 0.096 | 0.102 | -0.007 | 2.89e-01 |
| MultiScler | lassosum.PseudoVal | lassosum.PseudoVal | 0.103 | 0.103 | 0.000 | 1.00e+00 | 0.102 | 0.102 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.MultiPRS | lassosum.PseudoVal | 0.133 | 0.103 | 0.029 | 2.89e-05 | 0.119 | 0.102 | 0.017 | 9.00e-03 |
| MultiScler | pT+clump.10FCVal | lassosum.PseudoVal | 0.090 | 0.103 | -0.013 | 1.21e-03 | 0.092 | 0.102 | -0.010 | 2.21e-01 |
| MultiScler | lassosum.10FCVal | lassosum.PseudoVal | 0.115 | 0.103 | 0.012 | 2.06e-03 | 0.097 | 0.102 | -0.006 | 4.57e-01 |
| MultiScler | LDpred2.10FCVal | lassosum.PseudoVal | 0.120 | 0.103 | 0.017 | 1.14e-13 | 0.108 | 0.102 | 0.006 | 2.21e-01 |
| MultiScler | SBayesR.PseudoVal | lassosum.PseudoVal | 0.102 | 0.103 | -0.002 | 7.30e-01 | 0.083 | 0.102 | -0.019 | 8.51e-02 |
| MultiScler | SBLUP.Inf | lassosum.PseudoVal | 0.094 | 0.103 | -0.010 | 9.21e-02 | 0.076 | 0.102 | -0.027 | 2.11e-02 |
| MultiScler | pT+clump.MultiPRS | lassosum.PseudoVal | 0.114 | 0.103 | 0.011 | 1.20e-01 | 0.108 | 0.102 | 0.006 | 4.28e-01 |
| MultiScler | All.MultiPRS | lassosum.10FCVal | 0.135 | 0.115 | 0.020 | 4.13e-03 | 0.121 | 0.097 | 0.024 | 1.42e-04 |
| MultiScler | LDpred1.MultiPRS | lassosum.10FCVal | 0.112 | 0.115 | -0.003 | 6.77e-01 | 0.096 | 0.097 | -0.001 | 9.00e-01 |
| MultiScler | LDpred1.Inf | lassosum.10FCVal | 0.090 | 0.115 | -0.026 | 1.60e-12 | 0.078 | 0.097 | -0.018 | 1.38e-02 |
| MultiScler | LDpred2.Inf | lassosum.10FCVal | 0.094 | 0.115 | -0.021 | 1.49e-09 | 0.076 | 0.097 | -0.020 | 4.82e-03 |
| MultiScler | lassosum.MultiPRS | lassosum.10FCVal | 0.128 | 0.115 | 0.012 | 7.49e-02 | 0.119 | 0.097 | 0.022 | 1.66e-06 |
| MultiScler | DBSLMM.PseudoVal | lassosum.10FCVal | 0.091 | 0.115 | -0.024 | 7.66e-07 | 0.088 | 0.097 | -0.009 | 3.77e-01 |
| MultiScler | lassosum.PseudoVal | lassosum.10FCVal | 0.103 | 0.115 | -0.012 | 2.06e-03 | 0.102 | 0.097 | 0.006 | 4.57e-01 |
| MultiScler | LDpred2.MultiPRS | lassosum.10FCVal | 0.133 | 0.115 | 0.017 | 1.27e-02 | 0.119 | 0.097 | 0.022 | 7.35e-04 |
| MultiScler | pT+clump.10FCVal | lassosum.10FCVal | 0.090 | 0.115 | -0.025 | 3.52e-06 | 0.092 | 0.097 | -0.005 | 6.70e-01 |
| MultiScler | lassosum.10FCVal | lassosum.10FCVal | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.097 | 0.097 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.10FCVal | lassosum.10FCVal | 0.120 | 0.115 | 0.005 | 2.28e-01 | 0.108 | 0.097 | 0.011 | 1.76e-01 |
| MultiScler | SBayesR.PseudoVal | lassosum.10FCVal | 0.102 | 0.115 | -0.014 | 1.99e-03 | 0.083 | 0.097 | -0.014 | 1.26e-01 |
| MultiScler | LDpred2.PseudoVal | lassosum.10FCVal | 0.119 | 0.115 | 0.003 | 3.86e-01 | 0.110 | 0.097 | 0.014 | 8.02e-02 |
| MultiScler | SBLUP.Inf | lassosum.10FCVal | 0.094 | 0.115 | -0.021 | 1.23e-09 | 0.076 | 0.097 | -0.021 | 3.62e-03 |
| MultiScler | pT+clump.MultiPRS | lassosum.10FCVal | 0.114 | 0.115 | -0.001 | 8.94e-01 | 0.108 | 0.097 | 0.012 | 9.64e-02 |
| MultiScler | LDpred1.10FCVal | lassosum.10FCVal | 0.107 | 0.115 | -0.008 | 1.87e-02 | 0.096 | 0.097 | -0.001 | 8.78e-01 |
| MultiScler | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.091 | 0.112 | -0.021 | 2.85e-03 | 0.088 | 0.096 | -0.008 | 3.09e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.133 | 0.112 | 0.020 | 9.21e-11 | 0.119 | 0.096 | 0.023 | 1.53e-04 |
| MultiScler | pT+clump.10FCVal | LDpred1.MultiPRS | 0.090 | 0.112 | -0.022 | 1.66e-03 | 0.092 | 0.096 | -0.004 | 6.97e-01 |
| MultiScler | lassosum.10FCVal | LDpred1.MultiPRS | 0.115 | 0.112 | 0.003 | 6.77e-01 | 0.097 | 0.096 | 0.001 | 9.00e-01 |
| MultiScler | lassosum.PseudoVal | LDpred1.MultiPRS | 0.103 | 0.112 | -0.009 | 2.06e-01 | 0.102 | 0.096 | 0.007 | 3.04e-01 |
| MultiScler | All.MultiPRS | LDpred1.MultiPRS | 0.135 | 0.112 | 0.023 | 1.42e-13 | 0.121 | 0.096 | 0.025 | 4.26e-05 |
| MultiScler | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.112 | 0.112 | 0.000 | 1.00e+00 | 0.096 | 0.096 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.119 | 0.112 | 0.006 | 3.60e-01 | 0.110 | 0.096 | 0.015 | 4.73e-02 |
| MultiScler | SBLUP.Inf | LDpred1.MultiPRS | 0.094 | 0.112 | -0.019 | 8.26e-03 | 0.076 | 0.096 | -0.020 | 3.61e-02 |
| MultiScler | LDpred1.Inf | LDpred1.MultiPRS | 0.090 | 0.112 | -0.023 | 1.18e-03 | 0.078 | 0.096 | -0.017 | 3.87e-03 |
| MultiScler | LDpred2.Inf | LDpred1.MultiPRS | 0.094 | 0.112 | -0.018 | 8.94e-03 | 0.076 | 0.096 | -0.019 | 4.31e-02 |
| MultiScler | lassosum.MultiPRS | LDpred1.MultiPRS | 0.128 | 0.112 | 0.015 | 1.86e-07 | 0.119 | 0.096 | 0.023 | 1.10e-04 |
| MultiScler | LDpred1.10FCVal | LDpred1.MultiPRS | 0.107 | 0.112 | -0.005 | 4.78e-01 | 0.096 | 0.096 | 0.000 | 9.63e-01 |
| MultiScler | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.114 | 0.112 | 0.002 | 7.76e-01 | 0.108 | 0.096 | 0.013 | 1.14e-01 |
| MultiScler | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.102 | 0.112 | -0.011 | 1.23e-01 | 0.083 | 0.096 | -0.013 | 2.15e-01 |
| MultiScler | LDpred2.10FCVal | LDpred1.MultiPRS | 0.120 | 0.112 | 0.008 | 2.55e-01 | 0.108 | 0.096 | 0.012 | 6.07e-02 |
| MultiScler | All.MultiPRS | LDpred1.Inf | 0.135 | 0.090 | 0.046 | 6.81e-11 | 0.121 | 0.078 | 0.042 | 4.17e-07 |
| MultiScler | LDpred1.Inf | LDpred1.Inf | 0.090 | 0.090 | 0.000 | 1.00e+00 | 0.078 | 0.078 | 0.000 | 1.00e+00 |
| MultiScler | LDpred2.Inf | LDpred1.Inf | 0.094 | 0.090 | 0.004 | 2.06e-01 | 0.076 | 0.078 | -0.002 | 7.82e-01 |
| MultiScler | DBSLMM.PseudoVal | LDpred1.Inf | 0.091 | 0.090 | 0.002 | 7.30e-01 | 0.088 | 0.078 | 0.010 | 3.60e-01 |
| MultiScler | lassosum.PseudoVal | LDpred1.Inf | 0.103 | 0.090 | 0.014 | 3.81e-03 | 0.102 | 0.078 | 0.024 | 1.32e-02 |
| MultiScler | LDpred2.MultiPRS | LDpred1.Inf | 0.133 | 0.090 | 0.043 | 7.69e-10 | 0.119 | 0.078 | 0.041 | 9.66e-07 |
| MultiScler | pT+clump.10FCVal | LDpred1.Inf | 0.090 | 0.090 | 0.001 | 9.04e-01 | 0.092 | 0.078 | 0.014 | 2.52e-01 |
| MultiScler | lassosum.10FCVal | LDpred1.Inf | 0.115 | 0.090 | 0.026 | 1.60e-12 | 0.097 | 0.078 | 0.018 | 1.38e-02 |
| MultiScler | LDpred2.10FCVal | LDpred1.Inf | 0.120 | 0.090 | 0.031 | 3.46e-10 | 0.108 | 0.078 | 0.029 | 2.69e-03 |
| MultiScler | SBayesR.PseudoVal | LDpred1.Inf | 0.102 | 0.090 | 0.012 | 1.20e-02 | 0.083 | 0.078 | 0.005 | 6.27e-01 |
| MultiScler | LDpred1.MultiPRS | LDpred1.Inf | 0.112 | 0.090 | 0.023 | 1.18e-03 | 0.096 | 0.078 | 0.017 | 3.87e-03 |
| MultiScler | LDpred2.PseudoVal | LDpred1.Inf | 0.119 | 0.090 | 0.029 | 4.03e-09 | 0.110 | 0.078 | 0.032 | 1.18e-03 |
| MultiScler | SBLUP.Inf | LDpred1.Inf | 0.094 | 0.090 | 0.004 | 2.27e-01 | 0.076 | 0.078 | -0.003 | 7.06e-01 |
| MultiScler | pT+clump.MultiPRS | LDpred1.Inf | 0.114 | 0.090 | 0.025 | 3.91e-04 | 0.108 | 0.078 | 0.030 | 1.29e-03 |
| MultiScler | lassosum.MultiPRS | LDpred1.Inf | 0.128 | 0.090 | 0.038 | 5.29e-08 | 0.119 | 0.078 | 0.040 | 1.39e-07 |
| MultiScler | LDpred1.10FCVal | LDpred1.Inf | 0.107 | 0.090 | 0.018 | 2.12e-13 | 0.096 | 0.078 | 0.017 | 4.69e-04 |
| MultiScler | LDpred1.Inf | LDpred1.10FCVal | 0.090 | 0.107 | -0.018 | 2.12e-13 | 0.078 | 0.096 | -0.017 | 4.69e-04 |
| MultiScler | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.091 | 0.107 | -0.016 | 1.47e-05 | 0.088 | 0.096 | -0.007 | 3.11e-01 |
| MultiScler | lassosum.PseudoVal | LDpred1.10FCVal | 0.103 | 0.107 | -0.004 | 2.18e-01 | 0.102 | 0.096 | 0.007 | 2.89e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred1.10FCVal | 0.133 | 0.107 | 0.025 | 2.98e-04 | 0.119 | 0.096 | 0.023 | 1.82e-04 |
| MultiScler | pT+clump.10FCVal | LDpred1.10FCVal | 0.090 | 0.107 | -0.017 | 3.98e-04 | 0.092 | 0.096 | -0.004 | 7.13e-01 |
| MultiScler | lassosum.10FCVal | LDpred1.10FCVal | 0.115 | 0.107 | 0.008 | 1.87e-02 | 0.097 | 0.096 | 0.001 | 8.78e-01 |
| MultiScler | LDpred2.10FCVal | LDpred1.10FCVal | 0.120 | 0.107 | 0.013 | 1.08e-04 | 0.108 | 0.096 | 0.012 | 6.61e-02 |
| MultiScler | All.MultiPRS | LDpred1.10FCVal | 0.135 | 0.107 | 0.028 | 6.59e-05 | 0.121 | 0.096 | 0.025 | 1.08e-04 |
| MultiScler | LDpred1.MultiPRS | LDpred1.10FCVal | 0.112 | 0.107 | 0.005 | 4.78e-01 | 0.096 | 0.096 | 0.000 | 9.63e-01 |
| MultiScler | LDpred2.PseudoVal | LDpred1.10FCVal | 0.119 | 0.107 | 0.011 | 3.90e-03 | 0.110 | 0.096 | 0.015 | 5.69e-02 |
| MultiScler | SBLUP.Inf | LDpred1.10FCVal | 0.094 | 0.107 | -0.014 | 2.86e-03 | 0.076 | 0.096 | -0.020 | 3.01e-02 |
| MultiScler | pT+clump.MultiPRS | LDpred1.10FCVal | 0.114 | 0.107 | 0.007 | 3.18e-01 | 0.108 | 0.096 | 0.013 | 1.13e-01 |
| MultiScler | LDpred2.Inf | LDpred1.10FCVal | 0.094 | 0.107 | -0.013 | 3.24e-03 | 0.076 | 0.096 | -0.019 | 3.63e-02 |
| MultiScler | lassosum.MultiPRS | LDpred1.10FCVal | 0.128 | 0.107 | 0.020 | 3.65e-03 | 0.119 | 0.096 | 0.023 | 5.29e-05 |
| MultiScler | LDpred1.10FCVal | LDpred1.10FCVal | 0.107 | 0.107 | 0.000 | 1.00e+00 | 0.096 | 0.096 | 0.000 | 1.00e+00 |
| MultiScler | SBayesR.PseudoVal | LDpred1.10FCVal | 0.102 | 0.107 | -0.006 | 2.61e-01 | 0.083 | 0.096 | -0.013 | 2.26e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.133 | 0.133 | 0.000 | 1.00e+00 | 0.119 | 0.119 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.PseudoVal | LDpred2.MultiPRS | 0.103 | 0.133 | -0.029 | 2.89e-05 | 0.102 | 0.119 | -0.017 | 9.00e-03 |
| MultiScler | All.MultiPRS | LDpred2.MultiPRS | 0.135 | 0.133 | 0.003 | 1.33e-03 | 0.121 | 0.119 | 0.002 | 2.87e-01 |
| MultiScler | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.112 | 0.133 | -0.020 | 9.21e-11 | 0.096 | 0.119 | -0.023 | 1.53e-04 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.091 | 0.133 | -0.041 | 3.65e-09 | 0.088 | 0.119 | -0.031 | 7.58e-05 |
| MultiScler | LDpred1.Inf | LDpred2.MultiPRS | 0.090 | 0.133 | -0.043 | 7.69e-10 | 0.078 | 0.119 | -0.041 | 9.66e-07 |
| MultiScler | LDpred2.Inf | LDpred2.MultiPRS | 0.094 | 0.133 | -0.039 | 3.35e-08 | 0.076 | 0.119 | -0.043 | 1.12e-06 |
| MultiScler | lassosum.MultiPRS | LDpred2.MultiPRS | 0.128 | 0.133 | -0.005 | 1.98e-02 | 0.119 | 0.119 | -0.001 | 9.05e-01 |
| MultiScler | LDpred1.10FCVal | LDpred2.MultiPRS | 0.107 | 0.133 | -0.025 | 2.98e-04 | 0.096 | 0.119 | -0.023 | 1.82e-04 |
| MultiScler | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.114 | 0.133 | -0.018 | 8.43e-03 | 0.108 | 0.119 | -0.011 | 6.66e-02 |
| MultiScler | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.102 | 0.133 | -0.031 | 7.85e-06 | 0.083 | 0.119 | -0.036 | 9.10e-06 |
| MultiScler | pT+clump.10FCVal | LDpred2.MultiPRS | 0.090 | 0.133 | -0.042 | 1.33e-09 | 0.092 | 0.119 | -0.027 | 8.30e-04 |
| MultiScler | lassosum.10FCVal | LDpred2.MultiPRS | 0.115 | 0.133 | -0.017 | 1.27e-02 | 0.097 | 0.119 | -0.022 | 7.35e-04 |
| MultiScler | LDpred2.10FCVal | LDpred2.MultiPRS | 0.120 | 0.133 | -0.012 | 7.64e-02 | 0.108 | 0.119 | -0.011 | 5.46e-03 |
| MultiScler | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.119 | 0.133 | -0.014 | 4.53e-02 | 0.110 | 0.119 | -0.009 | 3.50e-02 |
| MultiScler | SBLUP.Inf | LDpred2.MultiPRS | 0.094 | 0.133 | -0.039 | 2.88e-08 | 0.076 | 0.119 | -0.043 | 7.51e-07 |
| MultiScler | LDpred2.MultiPRS | LDpred2.10FCVal | 0.133 | 0.120 | 0.012 | 7.64e-02 | 0.119 | 0.108 | 0.011 | 5.46e-03 |
| MultiScler | pT+clump.10FCVal | LDpred2.10FCVal | 0.090 | 0.120 | -0.030 | 2.31e-19 | 0.092 | 0.108 | -0.016 | 2.38e-02 |
| MultiScler | lassosum.PseudoVal | LDpred2.10FCVal | 0.103 | 0.120 | -0.017 | 1.14e-13 | 0.102 | 0.108 | -0.006 | 2.21e-01 |
| MultiScler | All.MultiPRS | LDpred2.10FCVal | 0.135 | 0.120 | 0.015 | 3.18e-02 | 0.121 | 0.108 | 0.013 | 3.84e-03 |
| MultiScler | LDpred1.MultiPRS | LDpred2.10FCVal | 0.112 | 0.120 | -0.008 | 2.55e-01 | 0.096 | 0.108 | -0.012 | 6.07e-02 |
| MultiScler | LDpred2.PseudoVal | LDpred2.10FCVal | 0.119 | 0.120 | -0.002 | 4.87e-01 | 0.110 | 0.108 | 0.003 | 5.47e-01 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.091 | 0.120 | -0.029 | 5.00e-27 | 0.088 | 0.108 | -0.020 | 3.35e-04 |
| MultiScler | LDpred1.Inf | LDpred2.10FCVal | 0.090 | 0.120 | -0.031 | 3.46e-10 | 0.078 | 0.108 | -0.029 | 2.69e-03 |
| MultiScler | LDpred2.Inf | LDpred2.10FCVal | 0.094 | 0.120 | -0.026 | 3.16e-06 | 0.076 | 0.108 | -0.031 | 5.76e-03 |
| MultiScler | lassosum.MultiPRS | LDpred2.10FCVal | 0.128 | 0.120 | 0.007 | 2.89e-01 | 0.119 | 0.108 | 0.011 | 5.45e-02 |
| MultiScler | LDpred1.10FCVal | LDpred2.10FCVal | 0.107 | 0.120 | -0.013 | 1.08e-04 | 0.096 | 0.108 | -0.012 | 6.61e-02 |
| MultiScler | pT+clump.MultiPRS | LDpred2.10FCVal | 0.114 | 0.120 | -0.006 | 3.90e-01 | 0.108 | 0.108 | 0.001 | 9.32e-01 |
| MultiScler | SBayesR.PseudoVal | LDpred2.10FCVal | 0.102 | 0.120 | -0.019 | 2.03e-04 | 0.083 | 0.108 | -0.025 | 1.49e-02 |
| MultiScler | lassosum.10FCVal | LDpred2.10FCVal | 0.115 | 0.120 | -0.005 | 2.28e-01 | 0.097 | 0.108 | -0.011 | 1.76e-01 |
| MultiScler | LDpred2.10FCVal | LDpred2.10FCVal | 0.120 | 0.120 | 0.000 | 1.00e+00 | 0.108 | 0.108 | 0.000 | 1.00e+00 |
| MultiScler | SBLUP.Inf | LDpred2.10FCVal | 0.094 | 0.120 | -0.027 | 2.71e-06 | 0.076 | 0.108 | -0.032 | 4.75e-03 |
| MultiScler | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.133 | 0.119 | 0.014 | 4.53e-02 | 0.119 | 0.110 | 0.009 | 3.50e-02 |
| MultiScler | lassosum.PseudoVal | LDpred2.PseudoVal | 0.103 | 0.119 | -0.015 | 1.16e-06 | 0.102 | 0.110 | -0.008 | 1.84e-01 |
| MultiScler | All.MultiPRS | LDpred2.PseudoVal | 0.135 | 0.119 | 0.017 | 1.74e-02 | 0.121 | 0.110 | 0.010 | 1.28e-02 |
| MultiScler | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.112 | 0.119 | -0.006 | 3.60e-01 | 0.096 | 0.110 | -0.015 | 4.73e-02 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.091 | 0.119 | -0.027 | 2.92e-11 | 0.088 | 0.110 | -0.022 | 7.23e-03 |
| MultiScler | LDpred1.Inf | LDpred2.PseudoVal | 0.090 | 0.119 | -0.029 | 4.03e-09 | 0.078 | 0.110 | -0.032 | 1.18e-03 |
| MultiScler | LDpred2.Inf | LDpred2.PseudoVal | 0.094 | 0.119 | -0.025 | 3.44e-06 | 0.076 | 0.110 | -0.034 | 1.43e-03 |
| MultiScler | lassosum.MultiPRS | LDpred2.PseudoVal | 0.128 | 0.119 | 0.009 | 1.97e-01 | 0.119 | 0.110 | 0.008 | 1.48e-01 |
| MultiScler | LDpred1.10FCVal | LDpred2.PseudoVal | 0.107 | 0.119 | -0.011 | 3.90e-03 | 0.096 | 0.110 | -0.015 | 5.69e-02 |
| MultiScler | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.114 | 0.119 | -0.004 | 5.27e-01 | 0.108 | 0.110 | -0.002 | 7.55e-01 |
| MultiScler | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.102 | 0.119 | -0.017 | 1.85e-05 | 0.083 | 0.110 | -0.027 | 6.94e-04 |
| MultiScler | pT+clump.10FCVal | LDpred2.PseudoVal | 0.090 | 0.119 | -0.029 | 2.28e-12 | 0.092 | 0.110 | -0.019 | 2.96e-02 |
| MultiScler | lassosum.10FCVal | LDpred2.PseudoVal | 0.115 | 0.119 | -0.003 | 3.86e-01 | 0.097 | 0.110 | -0.014 | 8.02e-02 |
| MultiScler | LDpred2.10FCVal | LDpred2.PseudoVal | 0.120 | 0.119 | 0.002 | 4.87e-01 | 0.108 | 0.110 | -0.003 | 5.47e-01 |
| MultiScler | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.119 | 0.119 | 0.000 | 1.00e+00 | 0.110 | 0.110 | 0.000 | 1.00e+00 |
| MultiScler | SBLUP.Inf | LDpred2.PseudoVal | 0.094 | 0.119 | -0.025 | 2.96e-06 | 0.076 | 0.110 | -0.035 | 1.14e-03 |
| MultiScler | DBSLMM.PseudoVal | LDpred2.Inf | 0.091 | 0.094 | -0.003 | 6.74e-01 | 0.088 | 0.076 | 0.012 | 3.57e-01 |
| MultiScler | LDpred2.MultiPRS | LDpred2.Inf | 0.133 | 0.094 | 0.039 | 3.35e-08 | 0.119 | 0.076 | 0.043 | 1.12e-06 |
| MultiScler | pT+clump.10FCVal | LDpred2.Inf | 0.090 | 0.094 | -0.004 | 5.53e-01 | 0.092 | 0.076 | 0.016 | 2.28e-01 |
| MultiScler | lassosum.10FCVal | LDpred2.Inf | 0.115 | 0.094 | 0.021 | 1.49e-09 | 0.097 | 0.076 | 0.020 | 4.82e-03 |
| MultiScler | lassosum.PseudoVal | LDpred2.Inf | 0.103 | 0.094 | 0.009 | 9.86e-02 | 0.102 | 0.076 | 0.026 | 2.48e-02 |
| MultiScler | All.MultiPRS | LDpred2.Inf | 0.135 | 0.094 | 0.041 | 3.76e-09 | 0.121 | 0.076 | 0.044 | 5.44e-07 |
| MultiScler | LDpred1.MultiPRS | LDpred2.Inf | 0.112 | 0.094 | 0.018 | 8.94e-03 | 0.096 | 0.076 | 0.019 | 4.31e-02 |
| MultiScler | LDpred2.PseudoVal | LDpred2.Inf | 0.119 | 0.094 | 0.025 | 3.44e-06 | 0.110 | 0.076 | 0.034 | 1.43e-03 |
| MultiScler | SBLUP.Inf | LDpred2.Inf | 0.094 | 0.094 | 0.000 | 1.95e-01 | 0.076 | 0.076 | -0.001 | 1.48e-02 |
| MultiScler | LDpred1.Inf | LDpred2.Inf | 0.090 | 0.094 | -0.004 | 2.06e-01 | 0.078 | 0.076 | 0.002 | 7.82e-01 |
| MultiScler | LDpred2.Inf | LDpred2.Inf | 0.094 | 0.094 | 0.000 | 1.00e+00 | 0.076 | 0.076 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.MultiPRS | LDpred2.Inf | 0.128 | 0.094 | 0.034 | 1.56e-06 | 0.119 | 0.076 | 0.042 | 2.91e-07 |
| MultiScler | LDpred1.10FCVal | LDpred2.Inf | 0.107 | 0.094 | 0.013 | 3.24e-03 | 0.096 | 0.076 | 0.019 | 3.63e-02 |
| MultiScler | pT+clump.MultiPRS | LDpred2.Inf | 0.114 | 0.094 | 0.020 | 3.68e-03 | 0.108 | 0.076 | 0.032 | 6.82e-04 |
| MultiScler | SBayesR.PseudoVal | LDpred2.Inf | 0.102 | 0.094 | 0.008 | 8.38e-02 | 0.083 | 0.076 | 0.007 | 4.48e-01 |
| MultiScler | LDpred2.10FCVal | LDpred2.Inf | 0.120 | 0.094 | 0.026 | 3.16e-06 | 0.108 | 0.076 | 0.031 | 5.76e-03 |
| MultiScler | All.MultiPRS | pT+clump.MultiPRS | 0.135 | 0.114 | 0.021 | 2.61e-03 | 0.121 | 0.108 | 0.012 | 2.32e-02 |
| MultiScler | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.112 | 0.114 | -0.002 | 7.76e-01 | 0.096 | 0.108 | -0.013 | 1.14e-01 |
| MultiScler | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.119 | 0.114 | 0.004 | 5.27e-01 | 0.110 | 0.108 | 0.002 | 7.55e-01 |
| MultiScler | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.091 | 0.114 | -0.023 | 1.01e-03 | 0.088 | 0.108 | -0.020 | 1.81e-02 |
| MultiScler | LDpred1.Inf | pT+clump.MultiPRS | 0.090 | 0.114 | -0.025 | 3.91e-04 | 0.078 | 0.108 | -0.030 | 1.29e-03 |
| MultiScler | LDpred2.Inf | pT+clump.MultiPRS | 0.094 | 0.114 | -0.020 | 3.68e-03 | 0.076 | 0.108 | -0.032 | 6.82e-04 |
| MultiScler | lassosum.MultiPRS | pT+clump.MultiPRS | 0.128 | 0.114 | 0.013 | 5.50e-02 | 0.119 | 0.108 | 0.010 | 7.50e-02 |
| MultiScler | LDpred1.10FCVal | pT+clump.MultiPRS | 0.107 | 0.114 | -0.007 | 3.18e-01 | 0.096 | 0.108 | -0.013 | 1.13e-01 |
| MultiScler | lassosum.PseudoVal | pT+clump.MultiPRS | 0.103 | 0.114 | -0.011 | 1.20e-01 | 0.102 | 0.108 | -0.006 | 4.28e-01 |
| MultiScler | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.133 | 0.114 | 0.018 | 8.43e-03 | 0.119 | 0.108 | 0.011 | 6.66e-02 |
| MultiScler | pT+clump.10FCVal | pT+clump.MultiPRS | 0.090 | 0.114 | -0.024 | 5.58e-04 | 0.092 | 0.108 | -0.016 | 1.45e-02 |
| MultiScler | lassosum.10FCVal | pT+clump.MultiPRS | 0.115 | 0.114 | 0.001 | 8.94e-01 | 0.097 | 0.108 | -0.012 | 9.64e-02 |
| MultiScler | LDpred2.10FCVal | pT+clump.MultiPRS | 0.120 | 0.114 | 0.006 | 3.90e-01 | 0.108 | 0.108 | -0.001 | 9.32e-01 |
| MultiScler | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.102 | 0.114 | -0.013 | 6.84e-02 | 0.083 | 0.108 | -0.025 | 1.42e-03 |
| MultiScler | SBLUP.Inf | pT+clump.MultiPRS | 0.094 | 0.114 | -0.021 | 3.37e-03 | 0.076 | 0.108 | -0.033 | 5.23e-04 |
| MultiScler | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.108 | 0.108 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.PseudoVal | pT+clump.10FCVal | 0.103 | 0.090 | 0.013 | 1.21e-03 | 0.102 | 0.092 | 0.010 | 2.21e-01 |
| MultiScler | All.MultiPRS | pT+clump.10FCVal | 0.135 | 0.090 | 0.045 | 1.18e-10 | 0.121 | 0.092 | 0.029 | 4.11e-04 |
| MultiScler | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.091 | 0.090 | 0.001 | 7.36e-01 | 0.088 | 0.092 | -0.004 | 5.67e-01 |
| MultiScler | LDpred1.Inf | pT+clump.10FCVal | 0.090 | 0.090 | -0.001 | 9.04e-01 | 0.078 | 0.092 | -0.014 | 2.52e-01 |
| MultiScler | LDpred2.Inf | pT+clump.10FCVal | 0.094 | 0.090 | 0.004 | 5.53e-01 | 0.076 | 0.092 | -0.016 | 2.28e-01 |
| MultiScler | lassosum.MultiPRS | pT+clump.10FCVal | 0.128 | 0.090 | 0.038 | 8.61e-08 | 0.119 | 0.092 | 0.027 | 3.25e-03 |
| MultiScler | LDpred1.10FCVal | pT+clump.10FCVal | 0.107 | 0.090 | 0.017 | 3.98e-04 | 0.096 | 0.092 | 0.004 | 7.13e-01 |
| MultiScler | pT+clump.MultiPRS | pT+clump.10FCVal | 0.114 | 0.090 | 0.024 | 5.58e-04 | 0.108 | 0.092 | 0.016 | 1.45e-02 |
| MultiScler | LDpred2.MultiPRS | pT+clump.10FCVal | 0.133 | 0.090 | 0.042 | 1.33e-09 | 0.119 | 0.092 | 0.027 | 8.30e-04 |
| MultiScler | pT+clump.10FCVal | pT+clump.10FCVal | 0.090 | 0.090 | 0.000 | 1.00e+00 | 0.092 | 0.092 | 0.000 | 1.00e+00 |
| MultiScler | lassosum.10FCVal | pT+clump.10FCVal | 0.115 | 0.090 | 0.025 | 3.52e-06 | 0.097 | 0.092 | 0.005 | 6.70e-01 |
| MultiScler | LDpred2.10FCVal | pT+clump.10FCVal | 0.120 | 0.090 | 0.030 | 2.31e-19 | 0.108 | 0.092 | 0.016 | 2.38e-02 |
| MultiScler | SBayesR.PseudoVal | pT+clump.10FCVal | 0.102 | 0.090 | 0.011 | 3.31e-02 | 0.083 | 0.092 | -0.009 | 4.15e-01 |
| MultiScler | LDpred1.MultiPRS | pT+clump.10FCVal | 0.112 | 0.090 | 0.022 | 1.66e-03 | 0.096 | 0.092 | 0.004 | 6.97e-01 |
| MultiScler | LDpred2.PseudoVal | pT+clump.10FCVal | 0.119 | 0.090 | 0.029 | 2.28e-12 | 0.110 | 0.092 | 0.019 | 2.96e-02 |
| MultiScler | SBLUP.Inf | pT+clump.10FCVal | 0.094 | 0.090 | 0.004 | 5.73e-01 | 0.076 | 0.092 | -0.016 | 2.07e-01 |
| MultiScler | LDpred1.Inf | SBayesR.PseudoVal | 0.090 | 0.102 | -0.012 | 1.20e-02 | 0.078 | 0.083 | -0.005 | 6.27e-01 |
| MultiScler | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.091 | 0.102 | -0.010 | 1.02e-01 | 0.088 | 0.083 | 0.005 | 6.88e-01 |
| MultiScler | lassosum.PseudoVal | SBayesR.PseudoVal | 0.103 | 0.102 | 0.002 | 7.30e-01 | 0.102 | 0.083 | 0.019 | 8.51e-02 |
| MultiScler | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.133 | 0.102 | 0.031 | 7.85e-06 | 0.119 | 0.083 | 0.036 | 9.10e-06 |
| MultiScler | pT+clump.10FCVal | SBayesR.PseudoVal | 0.090 | 0.102 | -0.011 | 3.31e-02 | 0.092 | 0.083 | 0.009 | 4.15e-01 |
| MultiScler | lassosum.10FCVal | SBayesR.PseudoVal | 0.115 | 0.102 | 0.014 | 1.99e-03 | 0.097 | 0.083 | 0.014 | 1.26e-01 |
| MultiScler | LDpred2.10FCVal | SBayesR.PseudoVal | 0.120 | 0.102 | 0.019 | 2.03e-04 | 0.108 | 0.083 | 0.025 | 1.49e-02 |
| MultiScler | All.MultiPRS | SBayesR.PseudoVal | 0.135 | 0.102 | 0.034 | 1.27e-06 | 0.121 | 0.083 | 0.038 | 5.56e-07 |
| MultiScler | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.112 | 0.102 | 0.011 | 1.23e-01 | 0.096 | 0.083 | 0.013 | 2.15e-01 |
| MultiScler | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.119 | 0.102 | 0.017 | 1.85e-05 | 0.110 | 0.083 | 0.027 | 6.94e-04 |
| MultiScler | SBLUP.Inf | SBayesR.PseudoVal | 0.094 | 0.102 | -0.008 | 7.68e-02 | 0.076 | 0.083 | -0.007 | 4.01e-01 |
| MultiScler | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.114 | 0.102 | 0.013 | 6.84e-02 | 0.108 | 0.083 | 0.025 | 1.42e-03 |
| MultiScler | LDpred2.Inf | SBayesR.PseudoVal | 0.094 | 0.102 | -0.008 | 8.38e-02 | 0.076 | 0.083 | -0.007 | 4.48e-01 |
| MultiScler | lassosum.MultiPRS | SBayesR.PseudoVal | 0.128 | 0.102 | 0.026 | 1.75e-04 | 0.119 | 0.083 | 0.036 | 2.18e-05 |
| MultiScler | LDpred1.10FCVal | SBayesR.PseudoVal | 0.107 | 0.102 | 0.006 | 2.61e-01 | 0.096 | 0.083 | 0.013 | 2.26e-01 |
| MultiScler | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.102 | 0.102 | 0.000 | 1.00e+00 | 0.083 | 0.083 | 0.000 | 1.00e+00 |
| MultiScler | All.MultiPRS | SBLUP.Inf | 0.135 | 0.094 | 0.042 | 3.21e-09 | 0.121 | 0.076 | 0.045 | 3.67e-07 |
| MultiScler | LDpred1.Inf | SBLUP.Inf | 0.090 | 0.094 | -0.004 | 2.27e-01 | 0.078 | 0.076 | 0.003 | 7.06e-01 |
| MultiScler | LDpred2.Inf | SBLUP.Inf | 0.094 | 0.094 | 0.000 | 1.95e-01 | 0.076 | 0.076 | 0.001 | 1.48e-02 |
| MultiScler | DBSLMM.PseudoVal | SBLUP.Inf | 0.091 | 0.094 | -0.002 | 6.96e-01 | 0.088 | 0.076 | 0.012 | 3.28e-01 |
| MultiScler | lassosum.PseudoVal | SBLUP.Inf | 0.103 | 0.094 | 0.010 | 9.21e-02 | 0.102 | 0.076 | 0.027 | 2.11e-02 |
| MultiScler | LDpred2.MultiPRS | SBLUP.Inf | 0.133 | 0.094 | 0.039 | 2.88e-08 | 0.119 | 0.076 | 0.043 | 7.51e-07 |
| MultiScler | pT+clump.10FCVal | SBLUP.Inf | 0.090 | 0.094 | -0.004 | 5.73e-01 | 0.092 | 0.076 | 0.016 | 2.07e-01 |
| MultiScler | lassosum.10FCVal | SBLUP.Inf | 0.115 | 0.094 | 0.021 | 1.23e-09 | 0.097 | 0.076 | 0.021 | 3.62e-03 |
| MultiScler | LDpred2.10FCVal | SBLUP.Inf | 0.120 | 0.094 | 0.027 | 2.71e-06 | 0.108 | 0.076 | 0.032 | 4.75e-03 |
| MultiScler | SBayesR.PseudoVal | SBLUP.Inf | 0.102 | 0.094 | 0.008 | 7.68e-02 | 0.083 | 0.076 | 0.007 | 4.01e-01 |
| MultiScler | LDpred1.MultiPRS | SBLUP.Inf | 0.112 | 0.094 | 0.019 | 8.26e-03 | 0.096 | 0.076 | 0.020 | 3.61e-02 |
| MultiScler | LDpred2.PseudoVal | SBLUP.Inf | 0.119 | 0.094 | 0.025 | 2.96e-06 | 0.110 | 0.076 | 0.035 | 1.14e-03 |
| MultiScler | SBLUP.Inf | SBLUP.Inf | 0.094 | 0.094 | 0.000 | 1.00e+00 | 0.076 | 0.076 | 0.000 | 1.00e+00 |
| MultiScler | pT+clump.MultiPRS | SBLUP.Inf | 0.114 | 0.094 | 0.021 | 3.37e-03 | 0.108 | 0.076 | 0.033 | 5.23e-04 |
| MultiScler | lassosum.MultiPRS | SBLUP.Inf | 0.128 | 0.094 | 0.034 | 1.36e-06 | 0.119 | 0.076 | 0.043 | 1.85e-07 |
| MultiScler | LDpred1.10FCVal | SBLUP.Inf | 0.107 | 0.094 | 0.014 | 2.86e-03 | 0.096 | 0.076 | 0.020 | 3.01e-02 |
| RheuArth | lassosum.PseudoVal | All.MultiPRS | 0.127 | 0.182 | -0.055 | 2.97e-15 | 0.114 | 0.165 | -0.051 | 3.90e-11 |
| RheuArth | All.MultiPRS | All.MultiPRS | 0.182 | 0.182 | 0.000 | 1.00e+00 | 0.165 | 0.165 | 0.000 | 1.00e+00 |
| RheuArth | DBSLMM.PseudoVal | All.MultiPRS | 0.157 | 0.182 | -0.025 | 3.58e-04 | 0.129 | 0.165 | -0.035 | 1.64e-11 |
| RheuArth | LDpred1.Inf | All.MultiPRS | 0.138 | 0.182 | -0.043 | 4.24e-10 | 0.114 | 0.165 | -0.051 | 1.50e-14 |
| RheuArth | LDpred2.Inf | All.MultiPRS | 0.087 | 0.182 | -0.095 | 3.39e-42 | 0.077 | 0.165 | -0.088 | 1.47e-19 |
| RheuArth | lassosum.MultiPRS | All.MultiPRS | 0.169 | 0.182 | -0.013 | 8.61e-16 | 0.146 | 0.165 | -0.018 | 3.08e-08 |
| RheuArth | LDpred1.10FCVal | All.MultiPRS | 0.141 | 0.182 | -0.041 | 4.15e-09 | 0.120 | 0.165 | -0.044 | 4.11e-12 |
| RheuArth | pT+clump.MultiPRS | All.MultiPRS | 0.134 | 0.182 | -0.047 | 8.71e-12 | 0.125 | 0.165 | -0.039 | 9.79e-09 |
| RheuArth | LDpred2.MultiPRS | All.MultiPRS | 0.156 | 0.182 | -0.025 | 2.30e-24 | 0.137 | 0.165 | -0.028 | 3.10e-08 |
| RheuArth | pT+clump.10FCVal | All.MultiPRS | 0.113 | 0.182 | -0.068 | 1.16e-22 | 0.104 | 0.165 | -0.061 | 9.53e-13 |
| RheuArth | lassosum.10FCVal | All.MultiPRS | 0.162 | 0.182 | -0.020 | 4.64e-03 | 0.138 | 0.165 | -0.027 | 4.02e-10 |
| RheuArth | LDpred2.10FCVal | All.MultiPRS | 0.156 | 0.182 | -0.025 | 2.55e-04 | 0.138 | 0.165 | -0.026 | 1.23e-07 |
| RheuArth | SBayesR.PseudoVal | All.MultiPRS | 0.123 | 0.182 | -0.058 | 4.89e-17 | 0.123 | 0.165 | -0.041 | 4.27e-07 |
| RheuArth | LDpred1.MultiPRS | All.MultiPRS | 0.144 | 0.182 | -0.037 | 2.89e-39 | 0.124 | 0.165 | -0.041 | 2.63e-12 |
| RheuArth | LDpred2.PseudoVal | All.MultiPRS | 0.112 | 0.182 | -0.070 | 7.53e-24 | 0.111 | 0.165 | -0.054 | 2.22e-11 |
| RheuArth | SBLUP.Inf | All.MultiPRS | 0.113 | 0.182 | -0.068 | 6.39e-23 | 0.101 | 0.165 | -0.064 | 1.90e-14 |
| RheuArth | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.156 | 0.157 | -0.001 | 9.21e-01 | 0.137 | 0.129 | 0.007 | 3.06e-01 |
| RheuArth | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.113 | 0.157 | -0.044 | 3.16e-18 | 0.104 | 0.129 | -0.026 | 1.20e-02 |
| RheuArth | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.127 | 0.157 | -0.030 | 1.82e-10 | 0.114 | 0.129 | -0.015 | 1.03e-01 |
| RheuArth | All.MultiPRS | DBSLMM.PseudoVal | 0.182 | 0.157 | 0.025 | 3.58e-04 | 0.165 | 0.129 | 0.035 | 1.64e-11 |
| RheuArth | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.144 | 0.157 | -0.013 | 6.59e-02 | 0.124 | 0.129 | -0.006 | 4.07e-01 |
| RheuArth | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.112 | 0.157 | -0.045 | 3.45e-26 | 0.111 | 0.129 | -0.018 | 3.10e-02 |
| RheuArth | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.157 | 0.157 | 0.000 | 1.00e+00 | 0.129 | 0.129 | 0.000 | 1.00e+00 |
| RheuArth | LDpred1.Inf | DBSLMM.PseudoVal | 0.138 | 0.157 | -0.019 | 2.73e-06 | 0.114 | 0.129 | -0.016 | 5.22e-02 |
| RheuArth | LDpred2.Inf | DBSLMM.PseudoVal | 0.087 | 0.157 | -0.070 | 3.82e-32 | 0.077 | 0.129 | -0.053 | 1.05e-05 |
| RheuArth | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.169 | 0.157 | 0.012 | 9.03e-02 | 0.146 | 0.129 | 0.017 | 5.10e-04 |
| RheuArth | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.141 | 0.157 | -0.016 | 4.81e-06 | 0.120 | 0.129 | -0.009 | 2.01e-01 |
| RheuArth | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.134 | 0.157 | -0.023 | 1.12e-03 | 0.125 | 0.129 | -0.004 | 6.58e-01 |
| RheuArth | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.123 | 0.157 | -0.033 | 2.24e-09 | 0.123 | 0.129 | -0.006 | 5.92e-01 |
| RheuArth | lassosum.10FCVal | DBSLMM.PseudoVal | 0.162 | 0.157 | 0.005 | 3.37e-02 | 0.138 | 0.129 | 0.009 | 7.72e-02 |
| RheuArth | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.156 | 0.157 | -0.001 | 8.73e-01 | 0.138 | 0.129 | 0.009 | 2.38e-01 |
| RheuArth | SBLUP.Inf | DBSLMM.PseudoVal | 0.113 | 0.157 | -0.044 | 1.48e-16 | 0.101 | 0.129 | -0.028 | 7.76e-03 |
| RheuArth | All.MultiPRS | lassosum.MultiPRS | 0.182 | 0.169 | 0.013 | 8.61e-16 | 0.165 | 0.146 | 0.018 | 3.08e-08 |
| RheuArth | LDpred1.MultiPRS | lassosum.MultiPRS | 0.144 | 0.169 | -0.024 | 4.20e-21 | 0.124 | 0.146 | -0.023 | 1.59e-05 |
| RheuArth | LDpred1.Inf | lassosum.MultiPRS | 0.138 | 0.169 | -0.030 | 1.26e-05 | 0.114 | 0.146 | -0.033 | 6.97e-08 |
| RheuArth | LDpred2.Inf | lassosum.MultiPRS | 0.087 | 0.169 | -0.082 | 8.05e-32 | 0.077 | 0.146 | -0.070 | 1.65e-12 |
| RheuArth | lassosum.MultiPRS | lassosum.MultiPRS | 0.169 | 0.169 | 0.000 | 1.00e+00 | 0.146 | 0.146 | 0.000 | 1.00e+00 |
| RheuArth | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.157 | 0.169 | -0.012 | 9.03e-02 | 0.129 | 0.146 | -0.017 | 5.10e-04 |
| RheuArth | lassosum.PseudoVal | lassosum.MultiPRS | 0.127 | 0.169 | -0.042 | 1.77e-09 | 0.114 | 0.146 | -0.032 | 3.72e-06 |
| RheuArth | LDpred2.MultiPRS | lassosum.MultiPRS | 0.156 | 0.169 | -0.012 | 5.55e-05 | 0.137 | 0.146 | -0.010 | 1.21e-01 |
| RheuArth | pT+clump.10FCVal | lassosum.MultiPRS | 0.113 | 0.169 | -0.055 | 2.08e-15 | 0.104 | 0.146 | -0.043 | 1.52e-05 |
| RheuArth | lassosum.10FCVal | lassosum.MultiPRS | 0.162 | 0.169 | -0.007 | 3.38e-01 | 0.138 | 0.146 | -0.008 | 3.16e-04 |
| RheuArth | LDpred2.10FCVal | lassosum.MultiPRS | 0.156 | 0.169 | -0.012 | 7.48e-02 | 0.138 | 0.146 | -0.008 | 1.83e-01 |
| RheuArth | SBayesR.PseudoVal | lassosum.MultiPRS | 0.123 | 0.169 | -0.045 | 7.20e-11 | 0.123 | 0.146 | -0.023 | 1.07e-02 |
| RheuArth | LDpred2.PseudoVal | lassosum.MultiPRS | 0.112 | 0.169 | -0.057 | 2.48e-16 | 0.111 | 0.146 | -0.035 | 8.35e-05 |
| RheuArth | SBLUP.Inf | lassosum.MultiPRS | 0.113 | 0.169 | -0.055 | 1.44e-15 | 0.101 | 0.146 | -0.045 | 3.70e-08 |
| RheuArth | pT+clump.MultiPRS | lassosum.MultiPRS | 0.134 | 0.169 | -0.034 | 7.44e-07 | 0.125 | 0.146 | -0.021 | 8.60e-03 |
| RheuArth | LDpred1.10FCVal | lassosum.MultiPRS | 0.141 | 0.169 | -0.028 | 6.31e-05 | 0.120 | 0.146 | -0.026 | 7.25e-06 |
| RheuArth | All.MultiPRS | lassosum.PseudoVal | 0.182 | 0.127 | 0.055 | 2.97e-15 | 0.165 | 0.114 | 0.051 | 3.90e-11 |
| RheuArth | LDpred1.MultiPRS | lassosum.PseudoVal | 0.144 | 0.127 | 0.017 | 1.28e-02 | 0.124 | 0.114 | 0.010 | 1.88e-01 |
| RheuArth | LDpred2.PseudoVal | lassosum.PseudoVal | 0.112 | 0.127 | -0.015 | 6.26e-03 | 0.111 | 0.114 | -0.003 | 7.80e-01 |
| RheuArth | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.157 | 0.127 | 0.030 | 1.82e-10 | 0.129 | 0.114 | 0.015 | 1.03e-01 |
| RheuArth | LDpred1.Inf | lassosum.PseudoVal | 0.138 | 0.127 | 0.012 | 1.17e-03 | 0.114 | 0.114 | 0.000 | 9.71e-01 |
| RheuArth | LDpred2.Inf | lassosum.PseudoVal | 0.087 | 0.127 | -0.040 | 1.98e-22 | 0.077 | 0.114 | -0.037 | 6.67e-06 |
| RheuArth | lassosum.MultiPRS | lassosum.PseudoVal | 0.169 | 0.127 | 0.042 | 1.77e-09 | 0.146 | 0.114 | 0.032 | 3.72e-06 |
| RheuArth | LDpred1.10FCVal | lassosum.PseudoVal | 0.141 | 0.127 | 0.014 | 2.77e-04 | 0.120 | 0.114 | 0.006 | 4.27e-01 |
| RheuArth | lassosum.PseudoVal | lassosum.PseudoVal | 0.127 | 0.127 | 0.000 | 1.00e+00 | 0.114 | 0.114 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.MultiPRS | lassosum.PseudoVal | 0.156 | 0.127 | 0.029 | 2.44e-05 | 0.137 | 0.114 | 0.023 | 9.48e-03 |
| RheuArth | pT+clump.10FCVal | lassosum.PseudoVal | 0.113 | 0.127 | -0.013 | 2.35e-02 | 0.104 | 0.114 | -0.010 | 3.87e-01 |
| RheuArth | lassosum.10FCVal | lassosum.PseudoVal | 0.162 | 0.127 | 0.035 | 3.86e-19 | 0.138 | 0.114 | 0.024 | 2.29e-03 |
| RheuArth | LDpred2.10FCVal | lassosum.PseudoVal | 0.156 | 0.127 | 0.029 | 6.70e-12 | 0.138 | 0.114 | 0.024 | 5.19e-03 |
| RheuArth | SBayesR.PseudoVal | lassosum.PseudoVal | 0.123 | 0.127 | -0.003 | 4.19e-01 | 0.123 | 0.114 | 0.009 | 2.72e-01 |
| RheuArth | SBLUP.Inf | lassosum.PseudoVal | 0.113 | 0.127 | -0.014 | 1.17e-04 | 0.101 | 0.114 | -0.013 | 6.46e-02 |
| RheuArth | pT+clump.MultiPRS | lassosum.PseudoVal | 0.134 | 0.127 | 0.007 | 2.87e-01 | 0.125 | 0.114 | 0.011 | 2.31e-01 |
| RheuArth | All.MultiPRS | lassosum.10FCVal | 0.182 | 0.162 | 0.020 | 4.64e-03 | 0.165 | 0.138 | 0.027 | 4.02e-10 |
| RheuArth | LDpred1.MultiPRS | lassosum.10FCVal | 0.144 | 0.162 | -0.018 | 1.00e-02 | 0.124 | 0.138 | -0.014 | 2.36e-02 |
| RheuArth | LDpred1.Inf | lassosum.10FCVal | 0.138 | 0.162 | -0.024 | 2.75e-11 | 0.114 | 0.138 | -0.024 | 7.85e-04 |
| RheuArth | LDpred2.Inf | lassosum.10FCVal | 0.087 | 0.162 | -0.075 | 6.19e-44 | 0.077 | 0.138 | -0.061 | 1.91e-08 |
| RheuArth | lassosum.MultiPRS | lassosum.10FCVal | 0.169 | 0.162 | 0.007 | 3.38e-01 | 0.146 | 0.138 | 0.008 | 3.16e-04 |
| RheuArth | DBSLMM.PseudoVal | lassosum.10FCVal | 0.157 | 0.162 | -0.005 | 3.37e-02 | 0.129 | 0.138 | -0.009 | 7.72e-02 |
| RheuArth | lassosum.PseudoVal | lassosum.10FCVal | 0.127 | 0.162 | -0.035 | 3.86e-19 | 0.114 | 0.138 | -0.024 | 2.29e-03 |
| RheuArth | LDpred2.MultiPRS | lassosum.10FCVal | 0.156 | 0.162 | -0.006 | 4.05e-01 | 0.137 | 0.138 | -0.001 | 8.50e-01 |
| RheuArth | pT+clump.10FCVal | lassosum.10FCVal | 0.113 | 0.162 | -0.049 | 8.27e-22 | 0.104 | 0.138 | -0.034 | 8.28e-04 |
| RheuArth | lassosum.10FCVal | lassosum.10FCVal | 0.162 | 0.162 | 0.000 | 1.00e+00 | 0.138 | 0.138 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.10FCVal | lassosum.10FCVal | 0.156 | 0.162 | -0.006 | 7.38e-02 | 0.138 | 0.138 | 0.000 | 9.81e-01 |
| RheuArth | SBayesR.PseudoVal | lassosum.10FCVal | 0.123 | 0.162 | -0.039 | 2.27e-14 | 0.123 | 0.138 | -0.015 | 1.50e-01 |
| RheuArth | LDpred2.PseudoVal | lassosum.10FCVal | 0.112 | 0.162 | -0.050 | 1.66e-30 | 0.111 | 0.138 | -0.027 | 2.10e-03 |
| RheuArth | SBLUP.Inf | lassosum.10FCVal | 0.113 | 0.162 | -0.049 | 3.80e-25 | 0.101 | 0.138 | -0.037 | 1.01e-04 |
| RheuArth | pT+clump.MultiPRS | lassosum.10FCVal | 0.134 | 0.162 | -0.028 | 6.46e-05 | 0.125 | 0.138 | -0.013 | 1.37e-01 |
| RheuArth | LDpred1.10FCVal | lassosum.10FCVal | 0.141 | 0.162 | -0.021 | 1.66e-10 | 0.120 | 0.138 | -0.018 | 7.91e-03 |
| RheuArth | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.157 | 0.144 | 0.013 | 6.59e-02 | 0.129 | 0.124 | 0.006 | 4.07e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.156 | 0.144 | 0.012 | 7.85e-04 | 0.137 | 0.124 | 0.013 | 7.00e-02 |
| RheuArth | pT+clump.10FCVal | LDpred1.MultiPRS | 0.113 | 0.144 | -0.031 | 1.01e-05 | 0.104 | 0.124 | -0.020 | 7.48e-02 |
| RheuArth | lassosum.10FCVal | LDpred1.MultiPRS | 0.162 | 0.144 | 0.018 | 1.00e-02 | 0.138 | 0.124 | 0.014 | 2.36e-02 |
| RheuArth | lassosum.PseudoVal | LDpred1.MultiPRS | 0.127 | 0.144 | -0.017 | 1.28e-02 | 0.114 | 0.124 | -0.010 | 1.88e-01 |
| RheuArth | All.MultiPRS | LDpred1.MultiPRS | 0.182 | 0.144 | 0.037 | 2.89e-39 | 0.165 | 0.124 | 0.041 | 2.63e-12 |
| RheuArth | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.144 | 0.144 | 0.000 | 1.00e+00 | 0.124 | 0.124 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.112 | 0.144 | -0.033 | 3.04e-06 | 0.111 | 0.124 | -0.013 | 2.04e-01 |
| RheuArth | SBLUP.Inf | LDpred1.MultiPRS | 0.113 | 0.144 | -0.031 | 8.80e-06 | 0.101 | 0.124 | -0.023 | 5.88e-04 |
| RheuArth | LDpred1.Inf | LDpred1.MultiPRS | 0.138 | 0.144 | -0.006 | 4.06e-01 | 0.114 | 0.124 | -0.010 | 8.81e-04 |
| RheuArth | LDpred2.Inf | LDpred1.MultiPRS | 0.087 | 0.144 | -0.057 | 2.38e-16 | 0.077 | 0.124 | -0.047 | 1.39e-08 |
| RheuArth | lassosum.MultiPRS | LDpred1.MultiPRS | 0.169 | 0.144 | 0.024 | 4.20e-21 | 0.146 | 0.124 | 0.023 | 1.59e-05 |
| RheuArth | LDpred1.10FCVal | LDpred1.MultiPRS | 0.141 | 0.144 | -0.003 | 6.42e-01 | 0.120 | 0.124 | -0.003 | 1.98e-01 |
| RheuArth | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.134 | 0.144 | -0.010 | 1.55e-01 | 0.125 | 0.124 | 0.001 | 8.76e-01 |
| RheuArth | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.123 | 0.144 | -0.021 | 2.84e-03 | 0.123 | 0.124 | 0.000 | 9.68e-01 |
| RheuArth | LDpred2.10FCVal | LDpred1.MultiPRS | 0.156 | 0.144 | 0.012 | 7.94e-02 | 0.138 | 0.124 | 0.015 | 4.39e-02 |
| RheuArth | All.MultiPRS | LDpred1.Inf | 0.182 | 0.138 | 0.043 | 4.24e-10 | 0.165 | 0.114 | 0.051 | 1.50e-14 |
| RheuArth | LDpred1.Inf | LDpred1.Inf | 0.138 | 0.138 | 0.000 | 1.00e+00 | 0.114 | 0.114 | 0.000 | 1.00e+00 |
| RheuArth | LDpred2.Inf | LDpred1.Inf | 0.087 | 0.138 | -0.051 | 3.18e-46 | 0.077 | 0.114 | -0.037 | 3.05e-07 |
| RheuArth | DBSLMM.PseudoVal | LDpred1.Inf | 0.157 | 0.138 | 0.019 | 2.73e-06 | 0.129 | 0.114 | 0.016 | 5.22e-02 |
| RheuArth | lassosum.PseudoVal | LDpred1.Inf | 0.127 | 0.138 | -0.012 | 1.17e-03 | 0.114 | 0.114 | 0.000 | 9.71e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred1.Inf | 0.156 | 0.138 | 0.018 | 1.03e-02 | 0.137 | 0.114 | 0.023 | 3.02e-03 |
| RheuArth | pT+clump.10FCVal | LDpred1.Inf | 0.113 | 0.138 | -0.025 | 1.24e-05 | 0.104 | 0.114 | -0.010 | 3.87e-01 |
| RheuArth | lassosum.10FCVal | LDpred1.Inf | 0.162 | 0.138 | 0.024 | 2.75e-11 | 0.138 | 0.114 | 0.024 | 7.85e-04 |
| RheuArth | LDpred2.10FCVal | LDpred1.Inf | 0.156 | 0.138 | 0.018 | 1.69e-06 | 0.138 | 0.114 | 0.024 | 1.45e-03 |
| RheuArth | SBayesR.PseudoVal | LDpred1.Inf | 0.123 | 0.138 | -0.015 | 9.09e-05 | 0.123 | 0.114 | 0.010 | 2.18e-01 |
| RheuArth | LDpred1.MultiPRS | LDpred1.Inf | 0.144 | 0.138 | 0.006 | 4.06e-01 | 0.124 | 0.114 | 0.010 | 8.81e-04 |
| RheuArth | LDpred2.PseudoVal | LDpred1.Inf | 0.112 | 0.138 | -0.027 | 3.67e-07 | 0.111 | 0.114 | -0.003 | 7.88e-01 |
| RheuArth | SBLUP.Inf | LDpred1.Inf | 0.113 | 0.138 | -0.025 | 7.00e-21 | 0.101 | 0.114 | -0.013 | 1.74e-02 |
| RheuArth | pT+clump.MultiPRS | LDpred1.Inf | 0.134 | 0.138 | -0.004 | 5.52e-01 | 0.125 | 0.114 | 0.011 | 2.31e-01 |
| RheuArth | lassosum.MultiPRS | LDpred1.Inf | 0.169 | 0.138 | 0.030 | 1.26e-05 | 0.146 | 0.114 | 0.033 | 6.97e-08 |
| RheuArth | LDpred1.10FCVal | LDpred1.Inf | 0.141 | 0.138 | 0.003 | 2.47e-01 | 0.120 | 0.114 | 0.006 | 1.43e-01 |
| RheuArth | LDpred1.Inf | LDpred1.10FCVal | 0.138 | 0.141 | -0.003 | 2.47e-01 | 0.114 | 0.120 | -0.006 | 1.43e-01 |
| RheuArth | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.157 | 0.141 | 0.016 | 4.81e-06 | 0.129 | 0.120 | 0.009 | 2.01e-01 |
| RheuArth | lassosum.PseudoVal | LDpred1.10FCVal | 0.127 | 0.141 | -0.014 | 2.77e-04 | 0.114 | 0.120 | -0.006 | 4.27e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred1.10FCVal | 0.156 | 0.141 | 0.015 | 2.77e-02 | 0.137 | 0.120 | 0.017 | 2.87e-02 |
| RheuArth | pT+clump.10FCVal | LDpred1.10FCVal | 0.113 | 0.141 | -0.028 | 7.20e-07 | 0.104 | 0.120 | -0.016 | 1.44e-01 |
| RheuArth | lassosum.10FCVal | LDpred1.10FCVal | 0.162 | 0.141 | 0.021 | 1.66e-10 | 0.138 | 0.120 | 0.018 | 7.91e-03 |
| RheuArth | LDpred2.10FCVal | LDpred1.10FCVal | 0.156 | 0.141 | 0.015 | 3.77e-05 | 0.138 | 0.120 | 0.018 | 1.75e-02 |
| RheuArth | All.MultiPRS | LDpred1.10FCVal | 0.182 | 0.141 | 0.041 | 4.15e-09 | 0.165 | 0.120 | 0.044 | 4.11e-12 |
| RheuArth | LDpred1.MultiPRS | LDpred1.10FCVal | 0.144 | 0.141 | 0.003 | 6.42e-01 | 0.124 | 0.120 | 0.003 | 1.98e-01 |
| RheuArth | LDpred2.PseudoVal | LDpred1.10FCVal | 0.112 | 0.141 | -0.029 | 4.42e-09 | 0.111 | 0.120 | -0.009 | 3.55e-01 |
| RheuArth | SBLUP.Inf | LDpred1.10FCVal | 0.113 | 0.141 | -0.028 | 4.12e-14 | 0.101 | 0.120 | -0.019 | 8.96e-03 |
| RheuArth | pT+clump.MultiPRS | LDpred1.10FCVal | 0.134 | 0.141 | -0.007 | 3.37e-01 | 0.125 | 0.120 | 0.005 | 6.14e-01 |
| RheuArth | LDpred2.Inf | LDpred1.10FCVal | 0.087 | 0.141 | -0.054 | 1.75e-34 | 0.077 | 0.120 | -0.044 | 8.72e-07 |
| RheuArth | lassosum.MultiPRS | LDpred1.10FCVal | 0.169 | 0.141 | 0.028 | 6.31e-05 | 0.146 | 0.120 | 0.026 | 7.25e-06 |
| RheuArth | LDpred1.10FCVal | LDpred1.10FCVal | 0.141 | 0.141 | 0.000 | 1.00e+00 | 0.120 | 0.120 | 0.000 | 1.00e+00 |
| RheuArth | SBayesR.PseudoVal | LDpred1.10FCVal | 0.123 | 0.141 | -0.018 | 9.34e-05 | 0.123 | 0.120 | 0.003 | 7.36e-01 |
| RheuArth | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.156 | 0.156 | 0.000 | 1.00e+00 | 0.137 | 0.137 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.PseudoVal | LDpred2.MultiPRS | 0.127 | 0.156 | -0.029 | 2.44e-05 | 0.114 | 0.137 | -0.023 | 9.48e-03 |
| RheuArth | All.MultiPRS | LDpred2.MultiPRS | 0.182 | 0.156 | 0.025 | 2.30e-24 | 0.165 | 0.137 | 0.028 | 3.10e-08 |
| RheuArth | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.144 | 0.156 | -0.012 | 7.85e-04 | 0.124 | 0.137 | -0.013 | 7.00e-02 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.157 | 0.156 | 0.001 | 9.21e-01 | 0.129 | 0.137 | -0.007 | 3.06e-01 |
| RheuArth | LDpred1.Inf | LDpred2.MultiPRS | 0.138 | 0.156 | -0.018 | 1.03e-02 | 0.114 | 0.137 | -0.023 | 3.02e-03 |
| RheuArth | LDpred2.Inf | LDpred2.MultiPRS | 0.087 | 0.156 | -0.069 | 3.87e-23 | 0.077 | 0.137 | -0.060 | 1.19e-10 |
| RheuArth | lassosum.MultiPRS | LDpred2.MultiPRS | 0.169 | 0.156 | 0.012 | 5.55e-05 | 0.146 | 0.137 | 0.010 | 1.21e-01 |
| RheuArth | LDpred1.10FCVal | LDpred2.MultiPRS | 0.141 | 0.156 | -0.015 | 2.77e-02 | 0.120 | 0.137 | -0.017 | 2.87e-02 |
| RheuArth | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.134 | 0.156 | -0.022 | 1.62e-03 | 0.125 | 0.137 | -0.012 | 2.26e-01 |
| RheuArth | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.123 | 0.156 | -0.033 | 2.48e-06 | 0.123 | 0.137 | -0.014 | 1.90e-01 |
| RheuArth | pT+clump.10FCVal | LDpred2.MultiPRS | 0.113 | 0.156 | -0.043 | 8.15e-10 | 0.104 | 0.137 | -0.033 | 2.97e-03 |
| RheuArth | lassosum.10FCVal | LDpred2.MultiPRS | 0.162 | 0.156 | 0.006 | 4.05e-01 | 0.138 | 0.137 | 0.001 | 8.50e-01 |
| RheuArth | LDpred2.10FCVal | LDpred2.MultiPRS | 0.156 | 0.156 | 0.000 | 9.89e-01 | 0.138 | 0.137 | 0.001 | 1.82e-01 |
| RheuArth | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.112 | 0.156 | -0.045 | 1.54e-10 | 0.111 | 0.137 | -0.026 | 1.31e-04 |
| RheuArth | SBLUP.Inf | LDpred2.MultiPRS | 0.113 | 0.156 | -0.043 | 7.02e-10 | 0.101 | 0.137 | -0.036 | 4.40e-05 |
| RheuArth | LDpred2.MultiPRS | LDpred2.10FCVal | 0.156 | 0.156 | 0.000 | 9.89e-01 | 0.137 | 0.138 | -0.001 | 1.82e-01 |
| RheuArth | pT+clump.10FCVal | LDpred2.10FCVal | 0.113 | 0.156 | -0.043 | 1.02e-14 | 0.104 | 0.138 | -0.034 | 2.01e-03 |
| RheuArth | lassosum.PseudoVal | LDpred2.10FCVal | 0.127 | 0.156 | -0.029 | 6.70e-12 | 0.114 | 0.138 | -0.024 | 5.19e-03 |
| RheuArth | All.MultiPRS | LDpred2.10FCVal | 0.182 | 0.156 | 0.025 | 2.55e-04 | 0.165 | 0.138 | 0.026 | 1.23e-07 |
| RheuArth | LDpred1.MultiPRS | LDpred2.10FCVal | 0.144 | 0.156 | -0.012 | 7.94e-02 | 0.124 | 0.138 | -0.015 | 4.39e-02 |
| RheuArth | LDpred2.PseudoVal | LDpred2.10FCVal | 0.112 | 0.156 | -0.045 | 6.36e-44 | 0.111 | 0.138 | -0.027 | 2.45e-05 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.157 | 0.156 | 0.001 | 8.73e-01 | 0.129 | 0.138 | -0.009 | 2.38e-01 |
| RheuArth | LDpred1.Inf | LDpred2.10FCVal | 0.138 | 0.156 | -0.018 | 1.69e-06 | 0.114 | 0.138 | -0.024 | 1.45e-03 |
| RheuArth | LDpred2.Inf | LDpred2.10FCVal | 0.087 | 0.156 | -0.069 | 4.33e-54 | 0.077 | 0.138 | -0.061 | 1.27e-11 |
| RheuArth | lassosum.MultiPRS | LDpred2.10FCVal | 0.169 | 0.156 | 0.012 | 7.48e-02 | 0.146 | 0.138 | 0.008 | 1.83e-01 |
| RheuArth | LDpred1.10FCVal | LDpred2.10FCVal | 0.141 | 0.156 | -0.015 | 3.77e-05 | 0.120 | 0.138 | -0.018 | 1.75e-02 |
| RheuArth | pT+clump.MultiPRS | LDpred2.10FCVal | 0.134 | 0.156 | -0.022 | 1.55e-03 | 0.125 | 0.138 | -0.013 | 1.73e-01 |
| RheuArth | SBayesR.PseudoVal | LDpred2.10FCVal | 0.123 | 0.156 | -0.033 | 4.30e-11 | 0.123 | 0.138 | -0.015 | 1.39e-01 |
| RheuArth | lassosum.10FCVal | LDpred2.10FCVal | 0.162 | 0.156 | 0.006 | 7.38e-02 | 0.138 | 0.138 | 0.000 | 9.81e-01 |
| RheuArth | LDpred2.10FCVal | LDpred2.10FCVal | 0.156 | 0.156 | 0.000 | 1.00e+00 | 0.138 | 0.138 | 0.000 | 1.00e+00 |
| RheuArth | SBLUP.Inf | LDpred2.10FCVal | 0.113 | 0.156 | -0.043 | 1.49e-24 | 0.101 | 0.138 | -0.037 | 1.29e-05 |
| RheuArth | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.156 | 0.112 | 0.045 | 1.54e-10 | 0.137 | 0.111 | 0.026 | 1.31e-04 |
| RheuArth | lassosum.PseudoVal | LDpred2.PseudoVal | 0.127 | 0.112 | 0.015 | 6.26e-03 | 0.114 | 0.111 | 0.003 | 7.80e-01 |
| RheuArth | All.MultiPRS | LDpred2.PseudoVal | 0.182 | 0.112 | 0.070 | 7.53e-24 | 0.165 | 0.111 | 0.054 | 2.22e-11 |
| RheuArth | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.144 | 0.112 | 0.033 | 3.04e-06 | 0.124 | 0.111 | 0.013 | 2.04e-01 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.157 | 0.112 | 0.045 | 3.45e-26 | 0.129 | 0.111 | 0.018 | 3.10e-02 |
| RheuArth | LDpred1.Inf | LDpred2.PseudoVal | 0.138 | 0.112 | 0.027 | 3.67e-07 | 0.114 | 0.111 | 0.003 | 7.88e-01 |
| RheuArth | LDpred2.Inf | LDpred2.PseudoVal | 0.087 | 0.112 | -0.025 | 6.96e-05 | 0.077 | 0.111 | -0.034 | 6.09e-03 |
| RheuArth | lassosum.MultiPRS | LDpred2.PseudoVal | 0.169 | 0.112 | 0.057 | 2.48e-16 | 0.146 | 0.111 | 0.035 | 8.35e-05 |
| RheuArth | LDpred1.10FCVal | LDpred2.PseudoVal | 0.141 | 0.112 | 0.029 | 4.42e-09 | 0.120 | 0.111 | 0.009 | 3.55e-01 |
| RheuArth | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.134 | 0.112 | 0.023 | 1.22e-03 | 0.125 | 0.111 | 0.014 | 2.07e-01 |
| RheuArth | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.123 | 0.112 | 0.012 | 5.44e-02 | 0.123 | 0.111 | 0.012 | 3.17e-01 |
| RheuArth | pT+clump.10FCVal | LDpred2.PseudoVal | 0.113 | 0.112 | 0.002 | 7.52e-01 | 0.104 | 0.111 | -0.007 | 5.26e-01 |
| RheuArth | lassosum.10FCVal | LDpred2.PseudoVal | 0.162 | 0.112 | 0.050 | 1.66e-30 | 0.138 | 0.111 | 0.027 | 2.10e-03 |
| RheuArth | LDpred2.10FCVal | LDpred2.PseudoVal | 0.156 | 0.112 | 0.045 | 6.36e-44 | 0.138 | 0.111 | 0.027 | 2.45e-05 |
| RheuArth | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.112 | 0.112 | 0.000 | 1.00e+00 | 0.111 | 0.111 | 0.000 | 1.00e+00 |
| RheuArth | SBLUP.Inf | LDpred2.PseudoVal | 0.113 | 0.112 | 0.002 | 7.77e-01 | 0.101 | 0.111 | -0.010 | 4.06e-01 |
| RheuArth | DBSLMM.PseudoVal | LDpred2.Inf | 0.157 | 0.087 | 0.070 | 3.82e-32 | 0.129 | 0.077 | 0.053 | 1.05e-05 |
| RheuArth | LDpred2.MultiPRS | LDpred2.Inf | 0.156 | 0.087 | 0.069 | 3.87e-23 | 0.137 | 0.077 | 0.060 | 1.19e-10 |
| RheuArth | pT+clump.10FCVal | LDpred2.Inf | 0.113 | 0.087 | 0.026 | 6.56e-05 | 0.104 | 0.077 | 0.027 | 4.01e-02 |
| RheuArth | lassosum.10FCVal | LDpred2.Inf | 0.162 | 0.087 | 0.075 | 6.19e-44 | 0.138 | 0.077 | 0.061 | 1.91e-08 |
| RheuArth | lassosum.PseudoVal | LDpred2.Inf | 0.127 | 0.087 | 0.040 | 1.98e-22 | 0.114 | 0.077 | 0.037 | 6.67e-06 |
| RheuArth | All.MultiPRS | LDpred2.Inf | 0.182 | 0.087 | 0.095 | 3.39e-42 | 0.165 | 0.077 | 0.088 | 1.47e-19 |
| RheuArth | LDpred1.MultiPRS | LDpred2.Inf | 0.144 | 0.087 | 0.057 | 2.38e-16 | 0.124 | 0.077 | 0.047 | 1.39e-08 |
| RheuArth | LDpred2.PseudoVal | LDpred2.Inf | 0.112 | 0.087 | 0.025 | 6.96e-05 | 0.111 | 0.077 | 0.034 | 6.09e-03 |
| RheuArth | SBLUP.Inf | LDpred2.Inf | 0.113 | 0.087 | 0.026 | 6.79e-71 | 0.101 | 0.077 | 0.024 | 1.81e-16 |
| RheuArth | LDpred1.Inf | LDpred2.Inf | 0.138 | 0.087 | 0.051 | 3.18e-46 | 0.114 | 0.077 | 0.037 | 3.05e-07 |
| RheuArth | LDpred2.Inf | LDpred2.Inf | 0.087 | 0.087 | 0.000 | 1.00e+00 | 0.077 | 0.077 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.MultiPRS | LDpred2.Inf | 0.169 | 0.087 | 0.082 | 8.05e-32 | 0.146 | 0.077 | 0.070 | 1.65e-12 |
| RheuArth | LDpred1.10FCVal | LDpred2.Inf | 0.141 | 0.087 | 0.054 | 1.75e-34 | 0.120 | 0.077 | 0.044 | 8.72e-07 |
| RheuArth | pT+clump.MultiPRS | LDpred2.Inf | 0.134 | 0.087 | 0.047 | 1.67e-11 | 0.125 | 0.077 | 0.048 | 4.84e-06 |
| RheuArth | SBayesR.PseudoVal | LDpred2.Inf | 0.123 | 0.087 | 0.036 | 5.54e-28 | 0.123 | 0.077 | 0.047 | 3.47e-12 |
| RheuArth | LDpred2.10FCVal | LDpred2.Inf | 0.156 | 0.087 | 0.069 | 4.33e-54 | 0.138 | 0.077 | 0.061 | 1.27e-11 |
| RheuArth | All.MultiPRS | pT+clump.MultiPRS | 0.182 | 0.134 | 0.047 | 8.71e-12 | 0.165 | 0.125 | 0.039 | 9.79e-09 |
| RheuArth | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.144 | 0.134 | 0.010 | 1.55e-01 | 0.124 | 0.125 | -0.001 | 8.76e-01 |
| RheuArth | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.112 | 0.134 | -0.023 | 1.22e-03 | 0.111 | 0.125 | -0.014 | 2.07e-01 |
| RheuArth | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.157 | 0.134 | 0.023 | 1.12e-03 | 0.129 | 0.125 | 0.004 | 6.58e-01 |
| RheuArth | LDpred1.Inf | pT+clump.MultiPRS | 0.138 | 0.134 | 0.004 | 5.52e-01 | 0.114 | 0.125 | -0.011 | 2.31e-01 |
| RheuArth | LDpred2.Inf | pT+clump.MultiPRS | 0.087 | 0.134 | -0.047 | 1.67e-11 | 0.077 | 0.125 | -0.048 | 4.84e-06 |
| RheuArth | lassosum.MultiPRS | pT+clump.MultiPRS | 0.169 | 0.134 | 0.034 | 7.44e-07 | 0.146 | 0.125 | 0.021 | 8.60e-03 |
| RheuArth | LDpred1.10FCVal | pT+clump.MultiPRS | 0.141 | 0.134 | 0.007 | 3.37e-01 | 0.120 | 0.125 | -0.005 | 6.14e-01 |
| RheuArth | lassosum.PseudoVal | pT+clump.MultiPRS | 0.127 | 0.134 | -0.007 | 2.87e-01 | 0.114 | 0.125 | -0.011 | 2.31e-01 |
| RheuArth | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.156 | 0.134 | 0.022 | 1.62e-03 | 0.137 | 0.125 | 0.012 | 2.26e-01 |
| RheuArth | pT+clump.10FCVal | pT+clump.MultiPRS | 0.113 | 0.134 | -0.021 | 2.78e-03 | 0.104 | 0.125 | -0.021 | 1.64e-04 |
| RheuArth | lassosum.10FCVal | pT+clump.MultiPRS | 0.162 | 0.134 | 0.028 | 6.46e-05 | 0.138 | 0.125 | 0.013 | 1.37e-01 |
| RheuArth | LDpred2.10FCVal | pT+clump.MultiPRS | 0.156 | 0.134 | 0.022 | 1.55e-03 | 0.138 | 0.125 | 0.013 | 1.73e-01 |
| RheuArth | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.123 | 0.134 | -0.011 | 1.21e-01 | 0.123 | 0.125 | -0.002 | 8.15e-01 |
| RheuArth | SBLUP.Inf | pT+clump.MultiPRS | 0.113 | 0.134 | -0.021 | 2.68e-03 | 0.101 | 0.125 | -0.024 | 1.10e-02 |
| RheuArth | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.134 | 0.134 | 0.000 | 1.00e+00 | 0.125 | 0.125 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.PseudoVal | pT+clump.10FCVal | 0.127 | 0.113 | 0.013 | 2.35e-02 | 0.114 | 0.104 | 0.010 | 3.87e-01 |
| RheuArth | All.MultiPRS | pT+clump.10FCVal | 0.182 | 0.113 | 0.068 | 1.16e-22 | 0.165 | 0.104 | 0.061 | 9.53e-13 |
| RheuArth | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.157 | 0.113 | 0.044 | 3.16e-18 | 0.129 | 0.104 | 0.026 | 1.20e-02 |
| RheuArth | LDpred1.Inf | pT+clump.10FCVal | 0.138 | 0.113 | 0.025 | 1.24e-05 | 0.114 | 0.104 | 0.010 | 3.87e-01 |
| RheuArth | LDpred2.Inf | pT+clump.10FCVal | 0.087 | 0.113 | -0.026 | 6.56e-05 | 0.077 | 0.104 | -0.027 | 4.01e-02 |
| RheuArth | lassosum.MultiPRS | pT+clump.10FCVal | 0.169 | 0.113 | 0.055 | 2.08e-15 | 0.146 | 0.104 | 0.043 | 1.52e-05 |
| RheuArth | LDpred1.10FCVal | pT+clump.10FCVal | 0.141 | 0.113 | 0.028 | 7.20e-07 | 0.120 | 0.104 | 0.016 | 1.44e-01 |
| RheuArth | pT+clump.MultiPRS | pT+clump.10FCVal | 0.134 | 0.113 | 0.021 | 2.78e-03 | 0.125 | 0.104 | 0.021 | 1.64e-04 |
| RheuArth | LDpred2.MultiPRS | pT+clump.10FCVal | 0.156 | 0.113 | 0.043 | 8.15e-10 | 0.137 | 0.104 | 0.033 | 2.97e-03 |
| RheuArth | pT+clump.10FCVal | pT+clump.10FCVal | 0.113 | 0.113 | 0.000 | 1.00e+00 | 0.104 | 0.104 | 0.000 | 1.00e+00 |
| RheuArth | lassosum.10FCVal | pT+clump.10FCVal | 0.162 | 0.113 | 0.049 | 8.27e-22 | 0.138 | 0.104 | 0.034 | 8.28e-04 |
| RheuArth | LDpred2.10FCVal | pT+clump.10FCVal | 0.156 | 0.113 | 0.043 | 1.02e-14 | 0.138 | 0.104 | 0.034 | 2.01e-03 |
| RheuArth | SBayesR.PseudoVal | pT+clump.10FCVal | 0.123 | 0.113 | 0.010 | 5.15e-02 | 0.123 | 0.104 | 0.020 | 5.55e-02 |
| RheuArth | LDpred1.MultiPRS | pT+clump.10FCVal | 0.144 | 0.113 | 0.031 | 1.01e-05 | 0.124 | 0.104 | 0.020 | 7.48e-02 |
| RheuArth | LDpred2.PseudoVal | pT+clump.10FCVal | 0.112 | 0.113 | -0.002 | 7.52e-01 | 0.111 | 0.104 | 0.007 | 5.26e-01 |
| RheuArth | SBLUP.Inf | pT+clump.10FCVal | 0.113 | 0.113 | 0.000 | 9.87e-01 | 0.101 | 0.104 | -0.003 | 8.18e-01 |
| RheuArth | LDpred1.Inf | SBayesR.PseudoVal | 0.138 | 0.123 | 0.015 | 9.09e-05 | 0.114 | 0.123 | -0.010 | 2.18e-01 |
| RheuArth | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.157 | 0.123 | 0.033 | 2.24e-09 | 0.129 | 0.123 | 0.006 | 5.92e-01 |
| RheuArth | lassosum.PseudoVal | SBayesR.PseudoVal | 0.127 | 0.123 | 0.003 | 4.19e-01 | 0.114 | 0.123 | -0.009 | 2.72e-01 |
| RheuArth | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.156 | 0.123 | 0.033 | 2.48e-06 | 0.137 | 0.123 | 0.014 | 1.90e-01 |
| RheuArth | pT+clump.10FCVal | SBayesR.PseudoVal | 0.113 | 0.123 | -0.010 | 5.15e-02 | 0.104 | 0.123 | -0.020 | 5.55e-02 |
| RheuArth | lassosum.10FCVal | SBayesR.PseudoVal | 0.162 | 0.123 | 0.039 | 2.27e-14 | 0.138 | 0.123 | 0.015 | 1.50e-01 |
| RheuArth | LDpred2.10FCVal | SBayesR.PseudoVal | 0.156 | 0.123 | 0.033 | 4.30e-11 | 0.138 | 0.123 | 0.015 | 1.39e-01 |
| RheuArth | All.MultiPRS | SBayesR.PseudoVal | 0.182 | 0.123 | 0.058 | 4.89e-17 | 0.165 | 0.123 | 0.041 | 4.27e-07 |
| RheuArth | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.144 | 0.123 | 0.021 | 2.84e-03 | 0.124 | 0.123 | 0.000 | 9.68e-01 |
| RheuArth | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.112 | 0.123 | -0.012 | 5.44e-02 | 0.111 | 0.123 | -0.012 | 3.17e-01 |
| RheuArth | SBLUP.Inf | SBayesR.PseudoVal | 0.113 | 0.123 | -0.010 | 3.02e-04 | 0.101 | 0.123 | -0.022 | 8.97e-05 |
| RheuArth | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.134 | 0.123 | 0.011 | 1.21e-01 | 0.125 | 0.123 | 0.002 | 8.15e-01 |
| RheuArth | LDpred2.Inf | SBayesR.PseudoVal | 0.087 | 0.123 | -0.036 | 5.54e-28 | 0.077 | 0.123 | -0.047 | 3.47e-12 |
| RheuArth | lassosum.MultiPRS | SBayesR.PseudoVal | 0.169 | 0.123 | 0.045 | 7.20e-11 | 0.146 | 0.123 | 0.023 | 1.07e-02 |
| RheuArth | LDpred1.10FCVal | SBayesR.PseudoVal | 0.141 | 0.123 | 0.018 | 9.34e-05 | 0.120 | 0.123 | -0.003 | 7.36e-01 |
| RheuArth | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.123 | 0.123 | 0.000 | 1.00e+00 | 0.123 | 0.123 | 0.000 | 1.00e+00 |
| RheuArth | All.MultiPRS | SBLUP.Inf | 0.182 | 0.113 | 0.068 | 6.39e-23 | 0.165 | 0.101 | 0.064 | 1.90e-14 |
| RheuArth | LDpred1.Inf | SBLUP.Inf | 0.138 | 0.113 | 0.025 | 7.00e-21 | 0.114 | 0.101 | 0.013 | 1.74e-02 |
| RheuArth | LDpred2.Inf | SBLUP.Inf | 0.087 | 0.113 | -0.026 | 6.79e-71 | 0.077 | 0.101 | -0.024 | 1.81e-16 |
| RheuArth | DBSLMM.PseudoVal | SBLUP.Inf | 0.157 | 0.113 | 0.044 | 1.48e-16 | 0.129 | 0.101 | 0.028 | 7.76e-03 |
| RheuArth | lassosum.PseudoVal | SBLUP.Inf | 0.127 | 0.113 | 0.014 | 1.17e-04 | 0.114 | 0.101 | 0.013 | 6.46e-02 |
| RheuArth | LDpred2.MultiPRS | SBLUP.Inf | 0.156 | 0.113 | 0.043 | 7.02e-10 | 0.137 | 0.101 | 0.036 | 4.40e-05 |
| RheuArth | pT+clump.10FCVal | SBLUP.Inf | 0.113 | 0.113 | 0.000 | 9.87e-01 | 0.104 | 0.101 | 0.003 | 8.18e-01 |
| RheuArth | lassosum.10FCVal | SBLUP.Inf | 0.162 | 0.113 | 0.049 | 3.80e-25 | 0.138 | 0.101 | 0.037 | 1.01e-04 |
| RheuArth | LDpred2.10FCVal | SBLUP.Inf | 0.156 | 0.113 | 0.043 | 1.49e-24 | 0.138 | 0.101 | 0.037 | 1.29e-05 |
| RheuArth | SBayesR.PseudoVal | SBLUP.Inf | 0.123 | 0.113 | 0.010 | 3.02e-04 | 0.123 | 0.101 | 0.022 | 8.97e-05 |
| RheuArth | LDpred1.MultiPRS | SBLUP.Inf | 0.144 | 0.113 | 0.031 | 8.80e-06 | 0.124 | 0.101 | 0.023 | 5.88e-04 |
| RheuArth | LDpred2.PseudoVal | SBLUP.Inf | 0.112 | 0.113 | -0.002 | 7.77e-01 | 0.111 | 0.101 | 0.010 | 4.06e-01 |
| RheuArth | SBLUP.Inf | SBLUP.Inf | 0.113 | 0.113 | 0.000 | 1.00e+00 | 0.101 | 0.101 | 0.000 | 1.00e+00 |
| RheuArth | pT+clump.MultiPRS | SBLUP.Inf | 0.134 | 0.113 | 0.021 | 2.68e-03 | 0.125 | 0.101 | 0.024 | 1.10e-02 |
| RheuArth | lassosum.MultiPRS | SBLUP.Inf | 0.169 | 0.113 | 0.055 | 1.44e-15 | 0.146 | 0.101 | 0.045 | 3.70e-08 |
| RheuArth | LDpred1.10FCVal | SBLUP.Inf | 0.141 | 0.113 | 0.028 | 4.12e-14 | 0.120 | 0.101 | 0.019 | 8.96e-03 |
| Prostate_Cancer | lassosum.10FCVal | All.MultiPRS | 0.182 | 0.185 | -0.003 | 6.23e-01 | 0.179 | 0.183 | -0.004 | 4.18e-01 |
| Prostate_Cancer | All.MultiPRS | All.MultiPRS | 0.185 | 0.185 | 0.000 | 1.00e+00 | 0.183 | 0.183 | 0.000 | 1.00e+00 |
| Prostate_Cancer | DBSLMM.PseudoVal | All.MultiPRS | 0.179 | 0.185 | -0.006 | 3.80e-01 | 0.166 | 0.183 | -0.018 | 3.09e-04 |
| Prostate_Cancer | LDpred1.Inf | All.MultiPRS | 0.142 | 0.185 | -0.043 | 5.36e-10 | 0.133 | 0.183 | -0.050 | 1.00e-10 |
| Prostate_Cancer | LDpred2.Inf | All.MultiPRS | 0.136 | 0.185 | -0.049 | 1.63e-12 | 0.126 | 0.183 | -0.057 | 1.65e-12 |
| Prostate_Cancer | lassosum.MultiPRS | All.MultiPRS | 0.188 | 0.185 | 0.003 | 6.95e-01 | 0.179 | 0.183 | -0.004 | 3.20e-01 |
| Prostate_Cancer | LDpred1.10FCVal | All.MultiPRS | 0.159 | 0.185 | -0.026 | 1.85e-04 | 0.146 | 0.183 | -0.037 | 8.28e-09 |
| Prostate_Cancer | pT+clump.MultiPRS | All.MultiPRS | 0.165 | 0.185 | -0.020 | 3.63e-03 | 0.173 | 0.183 | -0.011 | 8.22e-02 |
| Prostate_Cancer | LDpred2.MultiPRS | All.MultiPRS | 0.191 | 0.185 | 0.006 | 4.00e-01 | 0.183 | 0.183 | 0.000 | 9.78e-01 |
| Prostate_Cancer | pT+clump.10FCVal | All.MultiPRS | 0.150 | 0.185 | -0.035 | 3.09e-07 | 0.164 | 0.183 | -0.019 | 7.43e-03 |
| Prostate_Cancer | lassosum.PseudoVal | All.MultiPRS | 0.169 | 0.185 | -0.016 | 1.97e-02 | 0.163 | 0.183 | -0.020 | 4.26e-04 |
| Prostate_Cancer | LDpred2.10FCVal | All.MultiPRS | 0.188 | 0.185 | 0.003 | 6.35e-01 | 0.187 | 0.183 | 0.004 | 3.31e-01 |
| Prostate_Cancer | SBayesR.PseudoVal | All.MultiPRS | 0.183 | 0.185 | -0.002 | 7.76e-01 | 0.182 | 0.183 | -0.001 | 7.93e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | All.MultiPRS | 0.165 | 0.185 | -0.020 | 3.40e-03 | 0.155 | 0.183 | -0.028 | 4.05e-06 |
| Prostate_Cancer | LDpred2.PseudoVal | All.MultiPRS | 0.183 | 0.185 | -0.002 | 7.52e-01 | 0.184 | 0.183 | 0.001 | 7.67e-01 |
| Prostate_Cancer | SBLUP.Inf | All.MultiPRS | 0.137 | 0.185 | -0.048 | 2.53e-12 | 0.125 | 0.183 | -0.058 | 6.20e-13 |
| Prostate_Cancer | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.191 | 0.179 | 0.012 | 8.48e-02 | 0.183 | 0.166 | 0.017 | 5.57e-06 |
| Prostate_Cancer | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.150 | 0.179 | -0.029 | 4.34e-16 | 0.164 | 0.166 | -0.002 | 7.95e-01 |
| Prostate_Cancer | lassosum.10FCVal | DBSLMM.PseudoVal | 0.182 | 0.179 | 0.003 | 3.03e-01 | 0.179 | 0.166 | 0.014 | 7.46e-03 |
| Prostate_Cancer | All.MultiPRS | DBSLMM.PseudoVal | 0.185 | 0.179 | 0.006 | 3.80e-01 | 0.183 | 0.166 | 0.018 | 3.09e-04 |
| Prostate_Cancer | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.165 | 0.179 | -0.014 | 4.10e-02 | 0.155 | 0.166 | -0.010 | 3.69e-02 |
| Prostate_Cancer | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.183 | 0.179 | 0.004 | 1.66e-01 | 0.184 | 0.166 | 0.019 | 5.60e-04 |
| Prostate_Cancer | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.179 | 0.179 | 0.000 | 1.00e+00 | 0.166 | 0.166 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred1.Inf | DBSLMM.PseudoVal | 0.142 | 0.179 | -0.037 | 2.01e-32 | 0.133 | 0.166 | -0.032 | 2.29e-07 |
| Prostate_Cancer | LDpred2.Inf | DBSLMM.PseudoVal | 0.136 | 0.179 | -0.043 | 9.44e-40 | 0.126 | 0.166 | -0.039 | 1.15e-09 |
| Prostate_Cancer | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.188 | 0.179 | 0.009 | 2.05e-01 | 0.179 | 0.166 | 0.014 | 6.01e-04 |
| Prostate_Cancer | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.159 | 0.179 | -0.020 | 4.63e-15 | 0.146 | 0.166 | -0.020 | 7.65e-05 |
| Prostate_Cancer | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.165 | 0.179 | -0.014 | 4.19e-02 | 0.173 | 0.166 | 0.007 | 1.67e-01 |
| Prostate_Cancer | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.183 | 0.179 | 0.004 | 8.71e-02 | 0.182 | 0.166 | 0.016 | 5.23e-04 |
| Prostate_Cancer | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.169 | 0.179 | -0.010 | 1.71e-03 | 0.163 | 0.166 | -0.003 | 6.70e-01 |
| Prostate_Cancer | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.188 | 0.179 | 0.009 | 2.15e-05 | 0.187 | 0.166 | 0.021 | 7.74e-07 |
| Prostate_Cancer | SBLUP.Inf | DBSLMM.PseudoVal | 0.137 | 0.179 | -0.042 | 2.36e-39 | 0.125 | 0.166 | -0.041 | 3.33e-10 |
| Prostate_Cancer | All.MultiPRS | lassosum.MultiPRS | 0.185 | 0.188 | -0.003 | 6.95e-01 | 0.183 | 0.179 | 0.004 | 3.20e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | lassosum.MultiPRS | 0.165 | 0.188 | -0.023 | 1.15e-17 | 0.155 | 0.179 | -0.024 | 6.81e-06 |
| Prostate_Cancer | LDpred1.Inf | lassosum.MultiPRS | 0.142 | 0.188 | -0.046 | 4.39e-11 | 0.133 | 0.179 | -0.046 | 5.20e-11 |
| Prostate_Cancer | LDpred2.Inf | lassosum.MultiPRS | 0.136 | 0.188 | -0.052 | 9.45e-14 | 0.126 | 0.179 | -0.053 | 5.97e-13 |
| Prostate_Cancer | lassosum.MultiPRS | lassosum.MultiPRS | 0.188 | 0.188 | 0.000 | 1.00e+00 | 0.179 | 0.179 | 0.000 | 1.00e+00 |
| Prostate_Cancer | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.179 | 0.188 | -0.009 | 2.05e-01 | 0.166 | 0.179 | -0.014 | 6.01e-04 |
| Prostate_Cancer | lassosum.10FCVal | lassosum.MultiPRS | 0.182 | 0.188 | -0.006 | 3.78e-01 | 0.179 | 0.179 | 0.000 | 9.82e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | lassosum.MultiPRS | 0.191 | 0.188 | 0.003 | 6.54e-01 | 0.183 | 0.179 | 0.004 | 2.72e-01 |
| Prostate_Cancer | pT+clump.10FCVal | lassosum.MultiPRS | 0.150 | 0.188 | -0.038 | 4.17e-08 | 0.164 | 0.179 | -0.016 | 1.81e-02 |
| Prostate_Cancer | lassosum.PseudoVal | lassosum.MultiPRS | 0.169 | 0.188 | -0.019 | 6.55e-03 | 0.163 | 0.179 | -0.017 | 4.39e-04 |
| Prostate_Cancer | LDpred2.10FCVal | lassosum.MultiPRS | 0.188 | 0.188 | 0.001 | 9.35e-01 | 0.187 | 0.179 | 0.007 | 3.18e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | lassosum.MultiPRS | 0.183 | 0.188 | -0.005 | 4.99e-01 | 0.182 | 0.179 | 0.003 | 5.53e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | lassosum.MultiPRS | 0.183 | 0.188 | -0.005 | 4.80e-01 | 0.184 | 0.179 | 0.005 | 2.74e-01 |
| Prostate_Cancer | SBLUP.Inf | lassosum.MultiPRS | 0.137 | 0.188 | -0.051 | 1.47e-13 | 0.125 | 0.179 | -0.054 | 1.74e-13 |
| Prostate_Cancer | pT+clump.MultiPRS | lassosum.MultiPRS | 0.165 | 0.188 | -0.023 | 9.48e-04 | 0.173 | 0.179 | -0.007 | 1.61e-01 |
| Prostate_Cancer | LDpred1.10FCVal | lassosum.MultiPRS | 0.159 | 0.188 | -0.029 | 3.68e-05 | 0.146 | 0.179 | -0.034 | 3.37e-09 |
| Prostate_Cancer | All.MultiPRS | lassosum.10FCVal | 0.185 | 0.182 | 0.003 | 6.23e-01 | 0.183 | 0.179 | 0.004 | 4.18e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | lassosum.10FCVal | 0.165 | 0.182 | -0.017 | 1.53e-02 | 0.155 | 0.179 | -0.024 | 1.33e-04 |
| Prostate_Cancer | LDpred2.PseudoVal | lassosum.10FCVal | 0.183 | 0.182 | 0.001 | 5.82e-01 | 0.184 | 0.179 | 0.005 | 2.39e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | lassosum.10FCVal | 0.179 | 0.182 | -0.003 | 3.03e-01 | 0.166 | 0.179 | -0.014 | 7.46e-03 |
| Prostate_Cancer | LDpred1.Inf | lassosum.10FCVal | 0.142 | 0.182 | -0.040 | 5.13e-23 | 0.133 | 0.179 | -0.046 | 6.92e-09 |
| Prostate_Cancer | LDpred2.Inf | lassosum.10FCVal | 0.136 | 0.182 | -0.046 | 6.55e-27 | 0.126 | 0.179 | -0.053 | 2.07e-10 |
| Prostate_Cancer | lassosum.MultiPRS | lassosum.10FCVal | 0.188 | 0.182 | 0.006 | 3.78e-01 | 0.179 | 0.179 | 0.000 | 9.82e-01 |
| Prostate_Cancer | LDpred1.10FCVal | lassosum.10FCVal | 0.159 | 0.182 | -0.022 | 5.59e-11 | 0.146 | 0.179 | -0.034 | 6.58e-07 |
| Prostate_Cancer | lassosum.10FCVal | lassosum.10FCVal | 0.182 | 0.182 | 0.000 | 1.00e+00 | 0.179 | 0.179 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.MultiPRS | lassosum.10FCVal | 0.191 | 0.182 | 0.009 | 1.82e-01 | 0.183 | 0.179 | 0.004 | 3.37e-01 |
| Prostate_Cancer | pT+clump.10FCVal | lassosum.10FCVal | 0.150 | 0.182 | -0.032 | 1.08e-21 | 0.164 | 0.179 | -0.016 | 1.90e-02 |
| Prostate_Cancer | lassosum.PseudoVal | lassosum.10FCVal | 0.169 | 0.182 | -0.013 | 3.05e-07 | 0.163 | 0.179 | -0.016 | 1.02e-03 |
| Prostate_Cancer | LDpred2.10FCVal | lassosum.10FCVal | 0.188 | 0.182 | 0.007 | 1.23e-04 | 0.187 | 0.179 | 0.007 | 2.94e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | lassosum.10FCVal | 0.183 | 0.182 | 0.001 | 5.40e-01 | 0.182 | 0.179 | 0.003 | 5.67e-01 |
| Prostate_Cancer | SBLUP.Inf | lassosum.10FCVal | 0.137 | 0.182 | -0.045 | 1.81e-26 | 0.125 | 0.179 | -0.054 | 8.47e-11 |
| Prostate_Cancer | pT+clump.MultiPRS | lassosum.10FCVal | 0.165 | 0.182 | -0.017 | 1.56e-02 | 0.173 | 0.179 | -0.007 | 1.99e-01 |
| Prostate_Cancer | All.MultiPRS | lassosum.PseudoVal | 0.185 | 0.169 | 0.016 | 1.97e-02 | 0.183 | 0.163 | 0.020 | 4.26e-04 |
| Prostate_Cancer | LDpred1.MultiPRS | lassosum.PseudoVal | 0.165 | 0.169 | -0.004 | 5.55e-01 | 0.155 | 0.163 | -0.007 | 3.52e-01 |
| Prostate_Cancer | LDpred1.Inf | lassosum.PseudoVal | 0.142 | 0.169 | -0.027 | 1.00e-07 | 0.133 | 0.163 | -0.030 | 3.17e-03 |
| Prostate_Cancer | LDpred2.Inf | lassosum.PseudoVal | 0.136 | 0.169 | -0.033 | 7.79e-10 | 0.126 | 0.163 | -0.037 | 5.21e-04 |
| Prostate_Cancer | lassosum.MultiPRS | lassosum.PseudoVal | 0.188 | 0.169 | 0.019 | 6.55e-03 | 0.179 | 0.163 | 0.017 | 4.39e-04 |
| Prostate_Cancer | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.179 | 0.169 | 0.010 | 1.71e-03 | 0.166 | 0.163 | 0.003 | 6.70e-01 |
| Prostate_Cancer | lassosum.10FCVal | lassosum.PseudoVal | 0.182 | 0.169 | 0.013 | 3.05e-07 | 0.179 | 0.163 | 0.016 | 1.02e-03 |
| Prostate_Cancer | LDpred2.MultiPRS | lassosum.PseudoVal | 0.191 | 0.169 | 0.022 | 1.48e-03 | 0.183 | 0.163 | 0.020 | 7.60e-05 |
| Prostate_Cancer | pT+clump.10FCVal | lassosum.PseudoVal | 0.150 | 0.169 | -0.019 | 2.55e-11 | 0.164 | 0.163 | 0.001 | 8.80e-01 |
| Prostate_Cancer | lassosum.PseudoVal | lassosum.PseudoVal | 0.169 | 0.169 | 0.000 | 1.00e+00 | 0.163 | 0.163 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.10FCVal | lassosum.PseudoVal | 0.188 | 0.169 | 0.019 | 3.05e-14 | 0.187 | 0.163 | 0.024 | 1.97e-06 |
| Prostate_Cancer | SBayesR.PseudoVal | lassosum.PseudoVal | 0.183 | 0.169 | 0.014 | 3.43e-08 | 0.182 | 0.163 | 0.019 | 1.62e-04 |
| Prostate_Cancer | LDpred2.PseudoVal | lassosum.PseudoVal | 0.183 | 0.169 | 0.014 | 9.26e-09 | 0.184 | 0.163 | 0.022 | 5.41e-06 |
| Prostate_Cancer | SBLUP.Inf | lassosum.PseudoVal | 0.137 | 0.169 | -0.032 | 1.52e-09 | 0.125 | 0.163 | -0.038 | 3.71e-04 |
| Prostate_Cancer | pT+clump.MultiPRS | lassosum.PseudoVal | 0.165 | 0.169 | -0.004 | 5.65e-01 | 0.173 | 0.163 | 0.010 | 1.10e-01 |
| Prostate_Cancer | LDpred1.10FCVal | lassosum.PseudoVal | 0.159 | 0.169 | -0.010 | 2.66e-02 | 0.146 | 0.163 | -0.017 | 4.85e-02 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.179 | 0.165 | 0.014 | 4.10e-02 | 0.166 | 0.155 | 0.010 | 3.69e-02 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.191 | 0.165 | 0.026 | 1.70e-04 | 0.183 | 0.155 | 0.028 | 3.06e-07 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred1.MultiPRS | 0.150 | 0.165 | -0.015 | 3.00e-02 | 0.164 | 0.155 | 0.008 | 3.30e-01 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred1.MultiPRS | 0.169 | 0.165 | 0.004 | 5.55e-01 | 0.163 | 0.155 | 0.007 | 3.52e-01 |
| Prostate_Cancer | lassosum.10FCVal | LDpred1.MultiPRS | 0.182 | 0.165 | 0.017 | 1.53e-02 | 0.179 | 0.155 | 0.024 | 1.33e-04 |
| Prostate_Cancer | All.MultiPRS | LDpred1.MultiPRS | 0.185 | 0.165 | 0.020 | 3.40e-03 | 0.183 | 0.155 | 0.028 | 4.05e-06 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.165 | 0.165 | 0.000 | 1.00e+00 | 0.155 | 0.155 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.183 | 0.165 | 0.018 | 9.30e-03 | 0.184 | 0.155 | 0.029 | 3.17e-05 |
| Prostate_Cancer | SBLUP.Inf | LDpred1.MultiPRS | 0.137 | 0.165 | -0.028 | 4.87e-05 | 0.125 | 0.155 | -0.030 | 6.56e-09 |
| Prostate_Cancer | LDpred1.Inf | LDpred1.MultiPRS | 0.142 | 0.165 | -0.023 | 1.09e-03 | 0.133 | 0.155 | -0.022 | 2.17e-07 |
| Prostate_Cancer | LDpred2.Inf | LDpred1.MultiPRS | 0.136 | 0.165 | -0.029 | 3.76e-05 | 0.126 | 0.155 | -0.029 | 2.32e-08 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred1.MultiPRS | 0.188 | 0.165 | 0.023 | 1.15e-17 | 0.179 | 0.155 | 0.024 | 6.81e-06 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred1.MultiPRS | 0.159 | 0.165 | -0.006 | 4.20e-01 | 0.146 | 0.155 | -0.010 | 1.07e-03 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.165 | 0.165 | 0.000 | 9.88e-01 | 0.173 | 0.155 | 0.017 | 7.77e-03 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.183 | 0.165 | 0.018 | 8.42e-03 | 0.182 | 0.155 | 0.027 | 2.07e-05 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred1.MultiPRS | 0.188 | 0.165 | 0.024 | 7.06e-04 | 0.187 | 0.155 | 0.031 | 2.27e-08 |
| Prostate_Cancer | All.MultiPRS | LDpred1.Inf | 0.185 | 0.142 | 0.043 | 5.36e-10 | 0.183 | 0.133 | 0.050 | 1.00e-10 |
| Prostate_Cancer | LDpred1.Inf | LDpred1.Inf | 0.142 | 0.142 | 0.000 | 1.00e+00 | 0.133 | 0.133 | 0.000 | 1.00e+00 |
| Prostate_Cancer | LDpred2.Inf | LDpred1.Inf | 0.136 | 0.142 | -0.006 | 5.84e-05 | 0.126 | 0.133 | -0.007 | 1.47e-02 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred1.Inf | 0.179 | 0.142 | 0.037 | 2.01e-32 | 0.166 | 0.133 | 0.032 | 2.29e-07 |
| Prostate_Cancer | lassosum.10FCVal | LDpred1.Inf | 0.182 | 0.142 | 0.040 | 5.13e-23 | 0.179 | 0.133 | 0.046 | 6.92e-09 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred1.Inf | 0.191 | 0.142 | 0.049 | 1.61e-12 | 0.183 | 0.133 | 0.050 | 1.06e-11 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred1.Inf | 0.150 | 0.142 | 0.008 | 1.31e-01 | 0.164 | 0.133 | 0.030 | 2.23e-03 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred1.Inf | 0.169 | 0.142 | 0.027 | 1.00e-07 | 0.163 | 0.133 | 0.030 | 3.17e-03 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred1.Inf | 0.188 | 0.142 | 0.046 | 1.46e-33 | 0.187 | 0.133 | 0.053 | 1.48e-12 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred1.Inf | 0.183 | 0.142 | 0.041 | 1.16e-23 | 0.182 | 0.133 | 0.049 | 2.28e-09 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred1.Inf | 0.165 | 0.142 | 0.023 | 1.09e-03 | 0.155 | 0.133 | 0.022 | 2.17e-07 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred1.Inf | 0.183 | 0.142 | 0.041 | 9.73e-20 | 0.184 | 0.133 | 0.051 | 7.76e-09 |
| Prostate_Cancer | SBLUP.Inf | LDpred1.Inf | 0.137 | 0.142 | -0.006 | 1.61e-04 | 0.125 | 0.133 | -0.008 | 4.82e-03 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred1.Inf | 0.165 | 0.142 | 0.023 | 1.01e-03 | 0.173 | 0.133 | 0.039 | 4.34e-07 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred1.Inf | 0.188 | 0.142 | 0.046 | 4.39e-11 | 0.179 | 0.133 | 0.046 | 5.20e-11 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred1.Inf | 0.159 | 0.142 | 0.017 | 3.49e-44 | 0.146 | 0.133 | 0.012 | 5.26e-07 |
| Prostate_Cancer | LDpred1.Inf | LDpred1.10FCVal | 0.142 | 0.159 | -0.017 | 3.49e-44 | 0.133 | 0.146 | -0.012 | 5.26e-07 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.179 | 0.159 | 0.020 | 4.63e-15 | 0.166 | 0.146 | 0.020 | 7.65e-05 |
| Prostate_Cancer | lassosum.10FCVal | LDpred1.10FCVal | 0.182 | 0.159 | 0.022 | 5.59e-11 | 0.179 | 0.146 | 0.034 | 6.58e-07 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred1.10FCVal | 0.191 | 0.159 | 0.032 | 4.43e-06 | 0.183 | 0.146 | 0.037 | 2.93e-10 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred1.10FCVal | 0.150 | 0.159 | -0.010 | 3.71e-02 | 0.164 | 0.146 | 0.018 | 4.74e-02 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred1.10FCVal | 0.169 | 0.159 | 0.010 | 2.66e-02 | 0.163 | 0.146 | 0.017 | 4.85e-02 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred1.10FCVal | 0.188 | 0.159 | 0.029 | 3.83e-20 | 0.187 | 0.146 | 0.041 | 3.40e-11 |
| Prostate_Cancer | All.MultiPRS | LDpred1.10FCVal | 0.185 | 0.159 | 0.026 | 1.85e-04 | 0.183 | 0.146 | 0.037 | 8.28e-09 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred1.10FCVal | 0.165 | 0.159 | 0.006 | 4.20e-01 | 0.155 | 0.146 | 0.010 | 1.07e-03 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred1.10FCVal | 0.183 | 0.159 | 0.024 | 1.29e-09 | 0.184 | 0.146 | 0.039 | 4.23e-07 |
| Prostate_Cancer | SBLUP.Inf | LDpred1.10FCVal | 0.137 | 0.159 | -0.023 | 2.49e-30 | 0.125 | 0.146 | -0.021 | 1.26e-07 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred1.10FCVal | 0.165 | 0.159 | 0.006 | 4.11e-01 | 0.173 | 0.146 | 0.027 | 8.35e-05 |
| Prostate_Cancer | LDpred2.Inf | LDpred1.10FCVal | 0.136 | 0.159 | -0.023 | 8.45e-31 | 0.126 | 0.146 | -0.020 | 6.58e-07 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred1.10FCVal | 0.188 | 0.159 | 0.029 | 3.68e-05 | 0.179 | 0.146 | 0.034 | 3.37e-09 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred1.10FCVal | 0.159 | 0.159 | 0.000 | 1.00e+00 | 0.146 | 0.146 | 0.000 | 1.00e+00 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred1.10FCVal | 0.183 | 0.159 | 0.024 | 4.40e-12 | 0.182 | 0.146 | 0.036 | 1.02e-07 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.191 | 0.191 | 0.000 | 1.00e+00 | 0.183 | 0.183 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.MultiPRS | 0.182 | 0.191 | -0.009 | 1.82e-01 | 0.179 | 0.183 | -0.004 | 3.37e-01 |
| Prostate_Cancer | All.MultiPRS | LDpred2.MultiPRS | 0.185 | 0.191 | -0.006 | 4.00e-01 | 0.183 | 0.183 | 0.000 | 9.78e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.165 | 0.191 | -0.026 | 1.70e-04 | 0.155 | 0.183 | -0.028 | 3.06e-07 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.179 | 0.191 | -0.012 | 8.48e-02 | 0.166 | 0.183 | -0.017 | 5.57e-06 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.MultiPRS | 0.142 | 0.191 | -0.049 | 1.61e-12 | 0.133 | 0.183 | -0.050 | 1.06e-11 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.MultiPRS | 0.136 | 0.191 | -0.055 | 2.39e-15 | 0.126 | 0.183 | -0.057 | 9.19e-14 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.MultiPRS | 0.188 | 0.191 | -0.003 | 6.54e-01 | 0.179 | 0.183 | -0.004 | 2.72e-01 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.MultiPRS | 0.159 | 0.191 | -0.032 | 4.43e-06 | 0.146 | 0.183 | -0.037 | 2.93e-10 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.165 | 0.191 | -0.026 | 1.73e-04 | 0.173 | 0.183 | -0.010 | 4.15e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.183 | 0.191 | -0.008 | 2.60e-01 | 0.182 | 0.183 | -0.001 | 7.39e-01 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.MultiPRS | 0.150 | 0.191 | -0.041 | 2.51e-09 | 0.164 | 0.183 | -0.019 | 2.67e-03 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.MultiPRS | 0.169 | 0.191 | -0.022 | 1.48e-03 | 0.163 | 0.183 | -0.020 | 7.60e-05 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.MultiPRS | 0.188 | 0.191 | -0.003 | 7.13e-01 | 0.187 | 0.183 | 0.004 | 6.03e-02 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.183 | 0.191 | -0.008 | 2.48e-01 | 0.184 | 0.183 | 0.001 | 6.57e-01 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.MultiPRS | 0.137 | 0.191 | -0.054 | 3.83e-15 | 0.125 | 0.183 | -0.058 | 5.47e-14 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.10FCVal | 0.191 | 0.188 | 0.003 | 7.13e-01 | 0.183 | 0.187 | -0.004 | 6.03e-02 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.10FCVal | 0.150 | 0.188 | -0.039 | 1.72e-34 | 0.164 | 0.187 | -0.023 | 2.18e-04 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.10FCVal | 0.182 | 0.188 | -0.007 | 1.23e-04 | 0.179 | 0.187 | -0.007 | 2.94e-02 |
| Prostate_Cancer | All.MultiPRS | LDpred2.10FCVal | 0.185 | 0.188 | -0.003 | 6.35e-01 | 0.183 | 0.187 | -0.004 | 3.31e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.10FCVal | 0.165 | 0.188 | -0.024 | 7.06e-04 | 0.155 | 0.187 | -0.031 | 2.27e-08 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.10FCVal | 0.183 | 0.188 | -0.005 | 8.27e-06 | 0.184 | 0.187 | -0.002 | 3.47e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.179 | 0.188 | -0.009 | 2.15e-05 | 0.166 | 0.187 | -0.021 | 7.74e-07 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.10FCVal | 0.142 | 0.188 | -0.046 | 1.46e-33 | 0.133 | 0.187 | -0.053 | 1.48e-12 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.10FCVal | 0.136 | 0.188 | -0.052 | 1.83e-38 | 0.126 | 0.187 | -0.060 | 2.06e-14 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.10FCVal | 0.188 | 0.188 | -0.001 | 9.35e-01 | 0.179 | 0.187 | -0.007 | 3.18e-02 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.10FCVal | 0.159 | 0.188 | -0.029 | 3.83e-20 | 0.146 | 0.187 | -0.041 | 3.40e-11 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.10FCVal | 0.165 | 0.188 | -0.023 | 7.22e-04 | 0.173 | 0.187 | -0.014 | 5.51e-03 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.10FCVal | 0.183 | 0.188 | -0.005 | 3.05e-04 | 0.182 | 0.187 | -0.005 | 1.21e-01 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.10FCVal | 0.169 | 0.188 | -0.019 | 3.05e-14 | 0.163 | 0.187 | -0.024 | 1.97e-06 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.10FCVal | 0.188 | 0.188 | 0.000 | 1.00e+00 | 0.187 | 0.187 | 0.000 | 1.00e+00 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.10FCVal | 0.137 | 0.188 | -0.052 | 4.26e-37 | 0.125 | 0.187 | -0.062 | 1.36e-14 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.191 | 0.183 | 0.008 | 2.48e-01 | 0.183 | 0.184 | -0.001 | 6.57e-01 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.PseudoVal | 0.182 | 0.183 | -0.001 | 5.82e-01 | 0.179 | 0.184 | -0.005 | 2.39e-01 |
| Prostate_Cancer | All.MultiPRS | LDpred2.PseudoVal | 0.185 | 0.183 | 0.002 | 7.52e-01 | 0.183 | 0.184 | -0.001 | 7.67e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.165 | 0.183 | -0.018 | 9.30e-03 | 0.155 | 0.184 | -0.029 | 3.17e-05 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.179 | 0.183 | -0.004 | 1.66e-01 | 0.166 | 0.184 | -0.019 | 5.60e-04 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.PseudoVal | 0.142 | 0.183 | -0.041 | 9.73e-20 | 0.133 | 0.184 | -0.051 | 7.76e-09 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.PseudoVal | 0.136 | 0.183 | -0.047 | 2.60e-23 | 0.126 | 0.184 | -0.058 | 3.14e-10 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.PseudoVal | 0.188 | 0.183 | 0.005 | 4.80e-01 | 0.179 | 0.184 | -0.005 | 2.74e-01 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.PseudoVal | 0.159 | 0.183 | -0.024 | 1.29e-09 | 0.146 | 0.184 | -0.039 | 4.23e-07 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.165 | 0.183 | -0.018 | 9.62e-03 | 0.173 | 0.184 | -0.012 | 4.06e-02 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.183 | 0.183 | 0.000 | 8.88e-01 | 0.182 | 0.184 | -0.002 | 4.47e-01 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.PseudoVal | 0.150 | 0.183 | -0.033 | 1.79e-28 | 0.164 | 0.184 | -0.021 | 4.81e-04 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.PseudoVal | 0.169 | 0.183 | -0.014 | 9.26e-09 | 0.163 | 0.184 | -0.022 | 5.41e-06 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.PseudoVal | 0.188 | 0.183 | 0.005 | 8.27e-06 | 0.187 | 0.184 | 0.002 | 3.47e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.183 | 0.183 | 0.000 | 1.00e+00 | 0.184 | 0.184 | 0.000 | 1.00e+00 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.PseudoVal | 0.137 | 0.183 | -0.046 | 1.39e-22 | 0.125 | 0.184 | -0.059 | 1.99e-10 |
| Prostate_Cancer | DBSLMM.PseudoVal | LDpred2.Inf | 0.179 | 0.136 | 0.043 | 9.44e-40 | 0.166 | 0.126 | 0.039 | 1.15e-09 |
| Prostate_Cancer | LDpred2.MultiPRS | LDpred2.Inf | 0.191 | 0.136 | 0.055 | 2.39e-15 | 0.183 | 0.126 | 0.057 | 9.19e-14 |
| Prostate_Cancer | pT+clump.10FCVal | LDpred2.Inf | 0.150 | 0.136 | 0.014 | 1.00e-02 | 0.164 | 0.126 | 0.038 | 3.21e-04 |
| Prostate_Cancer | lassosum.PseudoVal | LDpred2.Inf | 0.169 | 0.136 | 0.033 | 7.79e-10 | 0.163 | 0.126 | 0.037 | 5.21e-04 |
| Prostate_Cancer | lassosum.10FCVal | LDpred2.Inf | 0.182 | 0.136 | 0.046 | 6.55e-27 | 0.179 | 0.126 | 0.053 | 2.07e-10 |
| Prostate_Cancer | All.MultiPRS | LDpred2.Inf | 0.185 | 0.136 | 0.049 | 1.63e-12 | 0.183 | 0.126 | 0.057 | 1.65e-12 |
| Prostate_Cancer | LDpred1.MultiPRS | LDpred2.Inf | 0.165 | 0.136 | 0.029 | 3.76e-05 | 0.155 | 0.126 | 0.029 | 2.32e-08 |
| Prostate_Cancer | LDpred2.PseudoVal | LDpred2.Inf | 0.183 | 0.136 | 0.047 | 2.60e-23 | 0.184 | 0.126 | 0.058 | 3.14e-10 |
| Prostate_Cancer | SBLUP.Inf | LDpred2.Inf | 0.137 | 0.136 | 0.000 | 4.13e-01 | 0.125 | 0.126 | -0.001 | 2.51e-01 |
| Prostate_Cancer | LDpred1.Inf | LDpred2.Inf | 0.142 | 0.136 | 0.006 | 5.84e-05 | 0.133 | 0.126 | 0.007 | 1.47e-02 |
| Prostate_Cancer | LDpred2.Inf | LDpred2.Inf | 0.136 | 0.136 | 0.000 | 1.00e+00 | 0.126 | 0.126 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.MultiPRS | LDpred2.Inf | 0.188 | 0.136 | 0.052 | 9.45e-14 | 0.179 | 0.126 | 0.053 | 5.97e-13 |
| Prostate_Cancer | LDpred1.10FCVal | LDpred2.Inf | 0.159 | 0.136 | 0.023 | 8.45e-31 | 0.146 | 0.126 | 0.020 | 6.58e-07 |
| Prostate_Cancer | pT+clump.MultiPRS | LDpred2.Inf | 0.165 | 0.136 | 0.029 | 3.34e-05 | 0.173 | 0.126 | 0.046 | 1.29e-08 |
| Prostate_Cancer | SBayesR.PseudoVal | LDpred2.Inf | 0.183 | 0.136 | 0.047 | 1.75e-27 | 0.182 | 0.126 | 0.056 | 7.32e-11 |
| Prostate_Cancer | LDpred2.10FCVal | LDpred2.Inf | 0.188 | 0.136 | 0.052 | 1.83e-38 | 0.187 | 0.126 | 0.060 | 2.06e-14 |
| Prostate_Cancer | All.MultiPRS | pT+clump.MultiPRS | 0.185 | 0.165 | 0.020 | 3.63e-03 | 0.183 | 0.173 | 0.011 | 8.22e-02 |
| Prostate_Cancer | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.165 | 0.165 | 0.000 | 9.88e-01 | 0.155 | 0.173 | -0.017 | 7.77e-03 |
| Prostate_Cancer | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.183 | 0.165 | 0.018 | 9.62e-03 | 0.184 | 0.173 | 0.012 | 4.06e-02 |
| Prostate_Cancer | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.179 | 0.165 | 0.014 | 4.19e-02 | 0.166 | 0.173 | -0.007 | 1.67e-01 |
| Prostate_Cancer | LDpred1.Inf | pT+clump.MultiPRS | 0.142 | 0.165 | -0.023 | 1.01e-03 | 0.133 | 0.173 | -0.039 | 4.34e-07 |
| Prostate_Cancer | LDpred2.Inf | pT+clump.MultiPRS | 0.136 | 0.165 | -0.029 | 3.34e-05 | 0.126 | 0.173 | -0.046 | 1.29e-08 |
| Prostate_Cancer | lassosum.MultiPRS | pT+clump.MultiPRS | 0.188 | 0.165 | 0.023 | 9.48e-04 | 0.179 | 0.173 | 0.007 | 1.61e-01 |
| Prostate_Cancer | LDpred1.10FCVal | pT+clump.MultiPRS | 0.159 | 0.165 | -0.006 | 4.11e-01 | 0.146 | 0.173 | -0.027 | 8.35e-05 |
| Prostate_Cancer | lassosum.10FCVal | pT+clump.MultiPRS | 0.182 | 0.165 | 0.017 | 1.56e-02 | 0.179 | 0.173 | 0.007 | 1.99e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.191 | 0.165 | 0.026 | 1.73e-04 | 0.183 | 0.173 | 0.010 | 4.15e-02 |
| Prostate_Cancer | pT+clump.10FCVal | pT+clump.MultiPRS | 0.150 | 0.165 | -0.015 | 2.85e-02 | 0.164 | 0.173 | -0.009 | 4.81e-02 |
| Prostate_Cancer | lassosum.PseudoVal | pT+clump.MultiPRS | 0.169 | 0.165 | 0.004 | 5.65e-01 | 0.163 | 0.173 | -0.010 | 1.10e-01 |
| Prostate_Cancer | LDpred2.10FCVal | pT+clump.MultiPRS | 0.188 | 0.165 | 0.023 | 7.22e-04 | 0.187 | 0.173 | 0.014 | 5.51e-03 |
| Prostate_Cancer | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.183 | 0.165 | 0.018 | 8.71e-03 | 0.182 | 0.173 | 0.009 | 8.27e-02 |
| Prostate_Cancer | SBLUP.Inf | pT+clump.MultiPRS | 0.137 | 0.165 | -0.028 | 4.33e-05 | 0.125 | 0.173 | -0.048 | 5.93e-09 |
| Prostate_Cancer | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.165 | 0.165 | 0.000 | 1.00e+00 | 0.173 | 0.173 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.10FCVal | pT+clump.10FCVal | 0.182 | 0.150 | 0.032 | 1.08e-21 | 0.179 | 0.164 | 0.016 | 1.90e-02 |
| Prostate_Cancer | All.MultiPRS | pT+clump.10FCVal | 0.185 | 0.150 | 0.035 | 3.09e-07 | 0.183 | 0.164 | 0.019 | 7.43e-03 |
| Prostate_Cancer | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.179 | 0.150 | 0.029 | 4.34e-16 | 0.166 | 0.164 | 0.002 | 7.95e-01 |
| Prostate_Cancer | LDpred1.Inf | pT+clump.10FCVal | 0.142 | 0.150 | -0.008 | 1.31e-01 | 0.133 | 0.164 | -0.030 | 2.23e-03 |
| Prostate_Cancer | LDpred2.Inf | pT+clump.10FCVal | 0.136 | 0.150 | -0.014 | 1.00e-02 | 0.126 | 0.164 | -0.038 | 3.21e-04 |
| Prostate_Cancer | lassosum.MultiPRS | pT+clump.10FCVal | 0.188 | 0.150 | 0.038 | 4.17e-08 | 0.179 | 0.164 | 0.016 | 1.81e-02 |
| Prostate_Cancer | LDpred1.10FCVal | pT+clump.10FCVal | 0.159 | 0.150 | 0.010 | 3.71e-02 | 0.146 | 0.164 | -0.018 | 4.74e-02 |
| Prostate_Cancer | pT+clump.MultiPRS | pT+clump.10FCVal | 0.165 | 0.150 | 0.015 | 2.85e-02 | 0.173 | 0.164 | 0.009 | 4.81e-02 |
| Prostate_Cancer | LDpred2.MultiPRS | pT+clump.10FCVal | 0.191 | 0.150 | 0.041 | 2.51e-09 | 0.183 | 0.164 | 0.019 | 2.67e-03 |
| Prostate_Cancer | pT+clump.10FCVal | pT+clump.10FCVal | 0.150 | 0.150 | 0.000 | 1.00e+00 | 0.164 | 0.164 | 0.000 | 1.00e+00 |
| Prostate_Cancer | lassosum.PseudoVal | pT+clump.10FCVal | 0.169 | 0.150 | 0.019 | 2.55e-11 | 0.163 | 0.164 | -0.001 | 8.80e-01 |
| Prostate_Cancer | LDpred2.10FCVal | pT+clump.10FCVal | 0.188 | 0.150 | 0.039 | 1.72e-34 | 0.187 | 0.164 | 0.023 | 2.18e-04 |
| Prostate_Cancer | SBayesR.PseudoVal | pT+clump.10FCVal | 0.183 | 0.150 | 0.033 | 3.60e-27 | 0.182 | 0.164 | 0.018 | 3.11e-03 |
| Prostate_Cancer | LDpred1.MultiPRS | pT+clump.10FCVal | 0.165 | 0.150 | 0.015 | 3.00e-02 | 0.155 | 0.164 | -0.008 | 3.30e-01 |
| Prostate_Cancer | LDpred2.PseudoVal | pT+clump.10FCVal | 0.183 | 0.150 | 0.033 | 1.79e-28 | 0.184 | 0.164 | 0.021 | 4.81e-04 |
| Prostate_Cancer | SBLUP.Inf | pT+clump.10FCVal | 0.137 | 0.150 | -0.013 | 1.31e-02 | 0.125 | 0.164 | -0.039 | 2.29e-04 |
| Prostate_Cancer | LDpred1.Inf | SBayesR.PseudoVal | 0.142 | 0.183 | -0.041 | 1.16e-23 | 0.133 | 0.182 | -0.049 | 2.28e-09 |
| Prostate_Cancer | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.179 | 0.183 | -0.004 | 8.71e-02 | 0.166 | 0.182 | -0.016 | 5.23e-04 |
| Prostate_Cancer | lassosum.10FCVal | SBayesR.PseudoVal | 0.182 | 0.183 | -0.001 | 5.40e-01 | 0.179 | 0.182 | -0.003 | 5.67e-01 |
| Prostate_Cancer | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.191 | 0.183 | 0.008 | 2.60e-01 | 0.183 | 0.182 | 0.001 | 7.39e-01 |
| Prostate_Cancer | pT+clump.10FCVal | SBayesR.PseudoVal | 0.150 | 0.183 | -0.033 | 3.60e-27 | 0.164 | 0.182 | -0.018 | 3.11e-03 |
| Prostate_Cancer | lassosum.PseudoVal | SBayesR.PseudoVal | 0.169 | 0.183 | -0.014 | 3.43e-08 | 0.163 | 0.182 | -0.019 | 1.62e-04 |
| Prostate_Cancer | LDpred2.10FCVal | SBayesR.PseudoVal | 0.188 | 0.183 | 0.005 | 3.05e-04 | 0.187 | 0.182 | 0.005 | 1.21e-01 |
| Prostate_Cancer | All.MultiPRS | SBayesR.PseudoVal | 0.185 | 0.183 | 0.002 | 7.76e-01 | 0.183 | 0.182 | 0.001 | 7.93e-01 |
| Prostate_Cancer | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.165 | 0.183 | -0.018 | 8.42e-03 | 0.155 | 0.182 | -0.027 | 2.07e-05 |
| Prostate_Cancer | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.183 | 0.183 | 0.000 | 8.88e-01 | 0.184 | 0.182 | 0.002 | 4.47e-01 |
| Prostate_Cancer | SBLUP.Inf | SBayesR.PseudoVal | 0.137 | 0.183 | -0.047 | 1.29e-26 | 0.125 | 0.182 | -0.057 | 4.45e-11 |
| Prostate_Cancer | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.165 | 0.183 | -0.018 | 8.71e-03 | 0.173 | 0.182 | -0.009 | 8.27e-02 |
| Prostate_Cancer | LDpred2.Inf | SBayesR.PseudoVal | 0.136 | 0.183 | -0.047 | 1.75e-27 | 0.126 | 0.182 | -0.056 | 7.32e-11 |
| Prostate_Cancer | lassosum.MultiPRS | SBayesR.PseudoVal | 0.188 | 0.183 | 0.005 | 4.99e-01 | 0.179 | 0.182 | -0.003 | 5.53e-01 |
| Prostate_Cancer | LDpred1.10FCVal | SBayesR.PseudoVal | 0.159 | 0.183 | -0.024 | 4.40e-12 | 0.146 | 0.182 | -0.036 | 1.02e-07 |
| Prostate_Cancer | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.183 | 0.183 | 0.000 | 1.00e+00 | 0.182 | 0.182 | 0.000 | 1.00e+00 |
| Prostate_Cancer | All.MultiPRS | SBLUP.Inf | 0.185 | 0.137 | 0.048 | 2.53e-12 | 0.183 | 0.125 | 0.058 | 6.20e-13 |
| Prostate_Cancer | LDpred1.Inf | SBLUP.Inf | 0.142 | 0.137 | 0.006 | 1.61e-04 | 0.133 | 0.125 | 0.008 | 4.82e-03 |
| Prostate_Cancer | LDpred2.Inf | SBLUP.Inf | 0.136 | 0.137 | 0.000 | 4.13e-01 | 0.126 | 0.125 | 0.001 | 2.51e-01 |
| Prostate_Cancer | DBSLMM.PseudoVal | SBLUP.Inf | 0.179 | 0.137 | 0.042 | 2.36e-39 | 0.166 | 0.125 | 0.041 | 3.33e-10 |
| Prostate_Cancer | lassosum.10FCVal | SBLUP.Inf | 0.182 | 0.137 | 0.045 | 1.81e-26 | 0.179 | 0.125 | 0.054 | 8.47e-11 |
| Prostate_Cancer | LDpred2.MultiPRS | SBLUP.Inf | 0.191 | 0.137 | 0.054 | 3.83e-15 | 0.183 | 0.125 | 0.058 | 5.47e-14 |
| Prostate_Cancer | pT+clump.10FCVal | SBLUP.Inf | 0.150 | 0.137 | 0.013 | 1.31e-02 | 0.164 | 0.125 | 0.039 | 2.29e-04 |
| Prostate_Cancer | lassosum.PseudoVal | SBLUP.Inf | 0.169 | 0.137 | 0.032 | 1.52e-09 | 0.163 | 0.125 | 0.038 | 3.71e-04 |
| Prostate_Cancer | LDpred2.10FCVal | SBLUP.Inf | 0.188 | 0.137 | 0.052 | 4.26e-37 | 0.187 | 0.125 | 0.062 | 1.36e-14 |
| Prostate_Cancer | SBayesR.PseudoVal | SBLUP.Inf | 0.183 | 0.137 | 0.047 | 1.29e-26 | 0.182 | 0.125 | 0.057 | 4.45e-11 |
| Prostate_Cancer | LDpred1.MultiPRS | SBLUP.Inf | 0.165 | 0.137 | 0.028 | 4.87e-05 | 0.155 | 0.125 | 0.030 | 6.56e-09 |
| Prostate_Cancer | LDpred2.PseudoVal | SBLUP.Inf | 0.183 | 0.137 | 0.046 | 1.39e-22 | 0.184 | 0.125 | 0.059 | 1.99e-10 |
| Prostate_Cancer | SBLUP.Inf | SBLUP.Inf | 0.137 | 0.137 | 0.000 | 1.00e+00 | 0.125 | 0.125 | 0.000 | 1.00e+00 |
| Prostate_Cancer | pT+clump.MultiPRS | SBLUP.Inf | 0.165 | 0.137 | 0.028 | 4.33e-05 | 0.173 | 0.125 | 0.048 | 5.93e-09 |
| Prostate_Cancer | lassosum.MultiPRS | SBLUP.Inf | 0.188 | 0.137 | 0.051 | 1.47e-13 | 0.179 | 0.125 | 0.054 | 1.74e-13 |
| Prostate_Cancer | LDpred1.10FCVal | SBLUP.Inf | 0.159 | 0.137 | 0.023 | 2.49e-30 | 0.146 | 0.125 | 0.021 | 1.26e-07 |
| Breast_Cancer | lassosum.PseudoVal | All.MultiPRS | 0.216 | 0.229 | -0.013 | 6.37e-02 | 0.195 | 0.214 | -0.019 | 6.57e-08 |
| Breast_Cancer | All.MultiPRS | All.MultiPRS | 0.229 | 0.229 | 0.000 | 1.00e+00 | 0.214 | 0.214 | 0.000 | 1.00e+00 |
| Breast_Cancer | DBSLMM.PseudoVal | All.MultiPRS | 0.216 | 0.229 | -0.013 | 5.63e-02 | 0.207 | 0.214 | -0.007 | 2.81e-02 |
| Breast_Cancer | LDpred1.Inf | All.MultiPRS | 0.172 | 0.229 | -0.057 | 9.02e-17 | 0.157 | 0.214 | -0.057 | 2.35e-16 |
| Breast_Cancer | LDpred2.Inf | All.MultiPRS | 0.168 | 0.229 | -0.061 | 4.81e-19 | 0.157 | 0.214 | -0.057 | 6.95e-16 |
| Breast_Cancer | lassosum.MultiPRS | All.MultiPRS | 0.225 | 0.229 | -0.004 | 5.09e-01 | 0.209 | 0.214 | -0.006 | 9.42e-03 |
| Breast_Cancer | LDpred1.10FCVal | All.MultiPRS | 0.204 | 0.229 | -0.025 | 2.05e-04 | 0.188 | 0.214 | -0.026 | 4.46e-08 |
| Breast_Cancer | pT+clump.MultiPRS | All.MultiPRS | 0.201 | 0.229 | -0.028 | 5.40e-05 | 0.186 | 0.214 | -0.028 | 1.22e-09 |
| Breast_Cancer | LDpred2.MultiPRS | All.MultiPRS | 0.229 | 0.229 | 0.000 | 9.57e-01 | 0.213 | 0.214 | -0.001 | 9.90e-02 |
| Breast_Cancer | pT+clump.10FCVal | All.MultiPRS | 0.182 | 0.229 | -0.047 | 6.64e-12 | 0.162 | 0.214 | -0.052 | 3.39e-16 |
| Breast_Cancer | lassosum.10FCVal | All.MultiPRS | 0.218 | 0.229 | -0.011 | 1.01e-01 | 0.196 | 0.214 | -0.018 | 8.76e-08 |
| Breast_Cancer | LDpred2.10FCVal | All.MultiPRS | 0.225 | 0.229 | -0.004 | 5.30e-01 | 0.208 | 0.214 | -0.006 | 2.03e-03 |
| Breast_Cancer | SBayesR.PseudoVal | All.MultiPRS | 0.227 | 0.229 | -0.002 | 7.16e-01 | 0.207 | 0.214 | -0.007 | 5.39e-04 |
| Breast_Cancer | LDpred1.MultiPRS | All.MultiPRS | 0.217 | 0.229 | -0.012 | 7.84e-02 | 0.200 | 0.214 | -0.014 | 5.10e-05 |
| Breast_Cancer | LDpred2.PseudoVal | All.MultiPRS | 0.223 | 0.229 | -0.006 | 3.81e-01 | 0.204 | 0.214 | -0.010 | 6.35e-05 |
| Breast_Cancer | SBLUP.Inf | All.MultiPRS | 0.166 | 0.229 | -0.063 | 5.68e-20 | 0.154 | 0.214 | -0.060 | 8.92e-17 |
| Breast_Cancer | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.229 | 0.216 | 0.013 | 6.24e-02 | 0.213 | 0.207 | 0.006 | 8.69e-02 |
| Breast_Cancer | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.182 | 0.216 | -0.034 | 5.79e-22 | 0.162 | 0.207 | -0.045 | 2.95e-10 |
| Breast_Cancer | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.216 | 0.216 | 0.000 | 8.90e-01 | 0.195 | 0.207 | -0.012 | 3.72e-02 |
| Breast_Cancer | All.MultiPRS | DBSLMM.PseudoVal | 0.229 | 0.216 | 0.013 | 5.63e-02 | 0.214 | 0.207 | 0.007 | 2.81e-02 |
| Breast_Cancer | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.217 | 0.216 | 0.001 | 8.71e-01 | 0.200 | 0.207 | -0.006 | 1.28e-01 |
| Breast_Cancer | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.223 | 0.216 | 0.007 | 2.95e-03 | 0.204 | 0.207 | -0.003 | 6.01e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.216 | 0.216 | 0.000 | 1.00e+00 | 0.207 | 0.207 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred1.Inf | DBSLMM.PseudoVal | 0.172 | 0.216 | -0.044 | 3.62e-51 | 0.157 | 0.207 | -0.049 | 3.55e-17 |
| Breast_Cancer | LDpred2.Inf | DBSLMM.PseudoVal | 0.168 | 0.216 | -0.048 | 8.74e-60 | 0.157 | 0.207 | -0.050 | 4.40e-17 |
| Breast_Cancer | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.225 | 0.216 | 0.009 | 2.09e-01 | 0.209 | 0.207 | 0.002 | 6.09e-01 |
| Breast_Cancer | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.204 | 0.216 | -0.012 | 2.33e-09 | 0.188 | 0.207 | -0.018 | 1.38e-05 |
| Breast_Cancer | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.201 | 0.216 | -0.015 | 3.22e-02 | 0.186 | 0.207 | -0.021 | 1.76e-05 |
| Breast_Cancer | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.227 | 0.216 | 0.011 | 2.54e-08 | 0.207 | 0.207 | 0.001 | 8.85e-01 |
| Breast_Cancer | lassosum.10FCVal | DBSLMM.PseudoVal | 0.218 | 0.216 | 0.002 | 4.57e-01 | 0.196 | 0.207 | -0.010 | 4.27e-02 |
| Breast_Cancer | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.225 | 0.216 | 0.009 | 6.35e-06 | 0.208 | 0.207 | 0.001 | 7.19e-01 |
| Breast_Cancer | SBLUP.Inf | DBSLMM.PseudoVal | 0.166 | 0.216 | -0.050 | 9.50e-64 | 0.154 | 0.207 | -0.052 | 1.16e-18 |
| Breast_Cancer | All.MultiPRS | lassosum.MultiPRS | 0.229 | 0.225 | 0.004 | 5.09e-01 | 0.214 | 0.209 | 0.006 | 9.42e-03 |
| Breast_Cancer | LDpred1.MultiPRS | lassosum.MultiPRS | 0.217 | 0.225 | -0.008 | 2.70e-01 | 0.200 | 0.209 | -0.008 | 2.45e-02 |
| Breast_Cancer | LDpred1.Inf | lassosum.MultiPRS | 0.172 | 0.225 | -0.053 | 1.74e-14 | 0.157 | 0.209 | -0.051 | 5.50e-13 |
| Breast_Cancer | LDpred2.Inf | lassosum.MultiPRS | 0.168 | 0.225 | -0.057 | 1.28e-16 | 0.157 | 0.209 | -0.052 | 1.01e-12 |
| Breast_Cancer | lassosum.MultiPRS | lassosum.MultiPRS | 0.225 | 0.225 | 0.000 | 1.00e+00 | 0.209 | 0.209 | 0.000 | 1.00e+00 |
| Breast_Cancer | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.216 | 0.225 | -0.009 | 2.09e-01 | 0.207 | 0.209 | -0.002 | 6.09e-01 |
| Breast_Cancer | lassosum.PseudoVal | lassosum.MultiPRS | 0.216 | 0.225 | -0.008 | 2.30e-01 | 0.195 | 0.209 | -0.014 | 2.07e-06 |
| Breast_Cancer | LDpred2.MultiPRS | lassosum.MultiPRS | 0.229 | 0.225 | 0.004 | 5.44e-01 | 0.213 | 0.209 | 0.004 | 8.58e-02 |
| Breast_Cancer | pT+clump.10FCVal | lassosum.MultiPRS | 0.182 | 0.225 | -0.043 | 4.67e-10 | 0.162 | 0.209 | -0.047 | 2.73e-13 |
| Breast_Cancer | lassosum.10FCVal | lassosum.MultiPRS | 0.218 | 0.225 | -0.007 | 3.25e-01 | 0.196 | 0.209 | -0.012 | 9.44e-07 |
| Breast_Cancer | LDpred2.10FCVal | lassosum.MultiPRS | 0.225 | 0.225 | 0.000 | 9.77e-01 | 0.208 | 0.209 | -0.001 | 8.42e-01 |
| Breast_Cancer | SBayesR.PseudoVal | lassosum.MultiPRS | 0.227 | 0.225 | 0.002 | 7.69e-01 | 0.207 | 0.209 | -0.001 | 6.53e-01 |
| Breast_Cancer | LDpred2.PseudoVal | lassosum.MultiPRS | 0.223 | 0.225 | -0.002 | 8.25e-01 | 0.204 | 0.209 | -0.004 | 1.87e-01 |
| Breast_Cancer | SBLUP.Inf | lassosum.MultiPRS | 0.166 | 0.225 | -0.059 | 1.81e-17 | 0.154 | 0.209 | -0.054 | 1.05e-13 |
| Breast_Cancer | pT+clump.MultiPRS | lassosum.MultiPRS | 0.201 | 0.225 | -0.023 | 6.96e-04 | 0.186 | 0.209 | -0.023 | 1.70e-06 |
| Breast_Cancer | LDpred1.10FCVal | lassosum.MultiPRS | 0.204 | 0.225 | -0.021 | 2.20e-03 | 0.188 | 0.209 | -0.020 | 4.60e-05 |
| Breast_Cancer | All.MultiPRS | lassosum.PseudoVal | 0.229 | 0.216 | 0.013 | 6.37e-02 | 0.214 | 0.195 | 0.019 | 6.57e-08 |
| Breast_Cancer | LDpred1.MultiPRS | lassosum.PseudoVal | 0.217 | 0.216 | 0.001 | 9.18e-01 | 0.200 | 0.195 | 0.005 | 2.89e-01 |
| Breast_Cancer | LDpred2.PseudoVal | lassosum.PseudoVal | 0.223 | 0.216 | 0.007 | 8.20e-04 | 0.204 | 0.195 | 0.009 | 2.08e-02 |
| Breast_Cancer | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.216 | 0.216 | 0.000 | 8.90e-01 | 0.207 | 0.195 | 0.012 | 3.72e-02 |
| Breast_Cancer | LDpred1.Inf | lassosum.PseudoVal | 0.172 | 0.216 | -0.045 | 9.46e-24 | 0.157 | 0.195 | -0.038 | 2.40e-05 |
| Breast_Cancer | LDpred2.Inf | lassosum.PseudoVal | 0.168 | 0.216 | -0.049 | 1.46e-26 | 0.157 | 0.195 | -0.038 | 3.28e-05 |
| Breast_Cancer | lassosum.MultiPRS | lassosum.PseudoVal | 0.225 | 0.216 | 0.008 | 2.30e-01 | 0.209 | 0.195 | 0.014 | 2.07e-06 |
| Breast_Cancer | LDpred1.10FCVal | lassosum.PseudoVal | 0.204 | 0.216 | -0.013 | 1.46e-04 | 0.188 | 0.195 | -0.006 | 3.47e-01 |
| Breast_Cancer | lassosum.PseudoVal | lassosum.PseudoVal | 0.216 | 0.216 | 0.000 | 1.00e+00 | 0.195 | 0.195 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.MultiPRS | lassosum.PseudoVal | 0.229 | 0.216 | 0.012 | 7.07e-02 | 0.213 | 0.195 | 0.018 | 8.28e-07 |
| Breast_Cancer | pT+clump.10FCVal | lassosum.PseudoVal | 0.182 | 0.216 | -0.034 | 9.09e-31 | 0.162 | 0.195 | -0.033 | 2.39e-08 |
| Breast_Cancer | lassosum.10FCVal | lassosum.PseudoVal | 0.218 | 0.216 | 0.001 | 2.48e-01 | 0.196 | 0.195 | 0.001 | 5.79e-01 |
| Breast_Cancer | LDpred2.10FCVal | lassosum.PseudoVal | 0.225 | 0.216 | 0.008 | 4.36e-05 | 0.208 | 0.195 | 0.013 | 1.40e-03 |
| Breast_Cancer | SBayesR.PseudoVal | lassosum.PseudoVal | 0.227 | 0.216 | 0.010 | 2.77e-06 | 0.207 | 0.195 | 0.012 | 4.46e-03 |
| Breast_Cancer | SBLUP.Inf | lassosum.PseudoVal | 0.166 | 0.216 | -0.050 | 4.71e-28 | 0.154 | 0.195 | -0.041 | 1.08e-05 |
| Breast_Cancer | pT+clump.MultiPRS | lassosum.PseudoVal | 0.201 | 0.216 | -0.015 | 2.77e-02 | 0.186 | 0.195 | -0.009 | 9.55e-02 |
| Breast_Cancer | All.MultiPRS | lassosum.10FCVal | 0.229 | 0.218 | 0.011 | 1.01e-01 | 0.214 | 0.196 | 0.018 | 8.76e-08 |
| Breast_Cancer | LDpred1.MultiPRS | lassosum.10FCVal | 0.217 | 0.218 | -0.001 | 9.01e-01 | 0.200 | 0.196 | 0.004 | 3.68e-01 |
| Breast_Cancer | LDpred1.Inf | lassosum.10FCVal | 0.172 | 0.218 | -0.046 | 4.13e-34 | 0.157 | 0.196 | -0.039 | 2.63e-07 |
| Breast_Cancer | LDpred2.Inf | lassosum.10FCVal | 0.168 | 0.218 | -0.050 | 3.27e-38 | 0.157 | 0.196 | -0.040 | 3.89e-07 |
| Breast_Cancer | lassosum.MultiPRS | lassosum.10FCVal | 0.225 | 0.218 | 0.007 | 3.25e-01 | 0.209 | 0.196 | 0.012 | 9.44e-07 |
| Breast_Cancer | DBSLMM.PseudoVal | lassosum.10FCVal | 0.216 | 0.218 | -0.002 | 4.57e-01 | 0.207 | 0.196 | 0.010 | 4.27e-02 |
| Breast_Cancer | lassosum.PseudoVal | lassosum.10FCVal | 0.216 | 0.218 | -0.001 | 2.48e-01 | 0.195 | 0.196 | -0.001 | 5.79e-01 |
| Breast_Cancer | LDpred2.MultiPRS | lassosum.10FCVal | 0.229 | 0.218 | 0.011 | 1.12e-01 | 0.213 | 0.196 | 0.016 | 1.13e-06 |
| Breast_Cancer | pT+clump.10FCVal | lassosum.10FCVal | 0.182 | 0.218 | -0.036 | 5.79e-28 | 0.162 | 0.196 | -0.035 | 1.38e-07 |
| Breast_Cancer | lassosum.10FCVal | lassosum.10FCVal | 0.218 | 0.218 | 0.000 | 1.00e+00 | 0.196 | 0.196 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.10FCVal | lassosum.10FCVal | 0.225 | 0.218 | 0.007 | 1.43e-05 | 0.208 | 0.196 | 0.012 | 2.81e-04 |
| Breast_Cancer | SBayesR.PseudoVal | lassosum.10FCVal | 0.227 | 0.218 | 0.009 | 9.02e-06 | 0.207 | 0.196 | 0.011 | 6.08e-03 |
| Breast_Cancer | LDpred2.PseudoVal | lassosum.10FCVal | 0.223 | 0.218 | 0.005 | 2.96e-03 | 0.204 | 0.196 | 0.008 | 2.84e-02 |
| Breast_Cancer | SBLUP.Inf | lassosum.10FCVal | 0.166 | 0.218 | -0.052 | 2.51e-40 | 0.154 | 0.196 | -0.042 | 8.06e-08 |
| Breast_Cancer | pT+clump.MultiPRS | lassosum.10FCVal | 0.201 | 0.218 | -0.016 | 1.56e-02 | 0.186 | 0.196 | -0.010 | 4.10e-02 |
| Breast_Cancer | LDpred1.10FCVal | lassosum.10FCVal | 0.204 | 0.218 | -0.014 | 1.77e-07 | 0.188 | 0.196 | -0.008 | 1.56e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.216 | 0.217 | -0.001 | 8.71e-01 | 0.207 | 0.200 | 0.006 | 1.28e-01 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.229 | 0.217 | 0.012 | 8.85e-02 | 0.213 | 0.200 | 0.013 | 3.20e-04 |
| Breast_Cancer | pT+clump.10FCVal | LDpred1.MultiPRS | 0.182 | 0.217 | -0.035 | 1.07e-07 | 0.162 | 0.200 | -0.038 | 2.10e-07 |
| Breast_Cancer | lassosum.10FCVal | LDpred1.MultiPRS | 0.218 | 0.217 | 0.001 | 9.01e-01 | 0.196 | 0.200 | -0.004 | 3.68e-01 |
| Breast_Cancer | lassosum.PseudoVal | LDpred1.MultiPRS | 0.216 | 0.217 | -0.001 | 9.18e-01 | 0.195 | 0.200 | -0.005 | 2.89e-01 |
| Breast_Cancer | All.MultiPRS | LDpred1.MultiPRS | 0.229 | 0.217 | 0.012 | 7.84e-02 | 0.214 | 0.200 | 0.014 | 5.10e-05 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.217 | 0.217 | 0.000 | 1.00e+00 | 0.200 | 0.200 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.223 | 0.217 | 0.006 | 3.55e-01 | 0.204 | 0.200 | 0.004 | 3.73e-01 |
| Breast_Cancer | SBLUP.Inf | LDpred1.MultiPRS | 0.166 | 0.217 | -0.051 | 9.79e-15 | 0.154 | 0.200 | -0.046 | 5.07e-12 |
| Breast_Cancer | LDpred1.Inf | LDpred1.MultiPRS | 0.172 | 0.217 | -0.045 | 6.24e-12 | 0.157 | 0.200 | -0.043 | 4.89e-12 |
| Breast_Cancer | LDpred2.Inf | LDpred1.MultiPRS | 0.168 | 0.217 | -0.049 | 6.52e-14 | 0.157 | 0.200 | -0.043 | 4.70e-11 |
| Breast_Cancer | lassosum.MultiPRS | LDpred1.MultiPRS | 0.225 | 0.217 | 0.008 | 2.70e-01 | 0.209 | 0.200 | 0.008 | 2.45e-02 |
| Breast_Cancer | LDpred1.10FCVal | LDpred1.MultiPRS | 0.204 | 0.217 | -0.013 | 3.86e-02 | 0.188 | 0.200 | -0.012 | 3.80e-04 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.201 | 0.217 | -0.016 | 2.16e-02 | 0.186 | 0.200 | -0.014 | 8.49e-03 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.227 | 0.217 | 0.010 | 1.43e-01 | 0.207 | 0.200 | 0.007 | 7.69e-02 |
| Breast_Cancer | LDpred2.10FCVal | LDpred1.MultiPRS | 0.225 | 0.217 | 0.008 | 2.34e-01 | 0.208 | 0.200 | 0.008 | 2.33e-02 |
| Breast_Cancer | All.MultiPRS | LDpred1.Inf | 0.229 | 0.172 | 0.057 | 9.02e-17 | 0.214 | 0.157 | 0.057 | 2.35e-16 |
| Breast_Cancer | LDpred1.Inf | LDpred1.Inf | 0.172 | 0.172 | 0.000 | 1.00e+00 | 0.157 | 0.157 | 0.000 | 1.00e+00 |
| Breast_Cancer | LDpred2.Inf | LDpred1.Inf | 0.168 | 0.172 | -0.004 | 2.55e-04 | 0.157 | 0.157 | -0.001 | 7.90e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred1.Inf | 0.216 | 0.172 | 0.044 | 3.62e-51 | 0.207 | 0.157 | 0.049 | 3.55e-17 |
| Breast_Cancer | lassosum.PseudoVal | LDpred1.Inf | 0.216 | 0.172 | 0.045 | 9.46e-24 | 0.195 | 0.157 | 0.038 | 2.40e-05 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred1.Inf | 0.229 | 0.172 | 0.057 | 1.00e-16 | 0.213 | 0.157 | 0.056 | 1.66e-15 |
| Breast_Cancer | pT+clump.10FCVal | LDpred1.Inf | 0.182 | 0.172 | 0.010 | 3.93e-02 | 0.162 | 0.157 | 0.004 | 6.51e-01 |
| Breast_Cancer | lassosum.10FCVal | LDpred1.Inf | 0.218 | 0.172 | 0.046 | 4.13e-34 | 0.196 | 0.157 | 0.039 | 2.63e-07 |
| Breast_Cancer | LDpred2.10FCVal | LDpred1.Inf | 0.225 | 0.172 | 0.053 | 3.32e-61 | 0.208 | 0.157 | 0.051 | 3.16e-15 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred1.Inf | 0.227 | 0.172 | 0.055 | 4.25e-59 | 0.207 | 0.157 | 0.050 | 1.96e-13 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred1.Inf | 0.217 | 0.172 | 0.045 | 6.24e-12 | 0.200 | 0.157 | 0.043 | 4.89e-12 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred1.Inf | 0.223 | 0.172 | 0.051 | 1.64e-43 | 0.204 | 0.157 | 0.047 | 3.48e-10 |
| Breast_Cancer | SBLUP.Inf | LDpred1.Inf | 0.166 | 0.172 | -0.006 | 2.88e-07 | 0.154 | 0.157 | -0.003 | 1.88e-01 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred1.Inf | 0.201 | 0.172 | 0.030 | 1.78e-05 | 0.186 | 0.157 | 0.029 | 1.15e-04 |
| Breast_Cancer | lassosum.MultiPRS | LDpred1.Inf | 0.225 | 0.172 | 0.053 | 1.74e-14 | 0.209 | 0.157 | 0.051 | 5.50e-13 |
| Breast_Cancer | LDpred1.10FCVal | LDpred1.Inf | 0.204 | 0.172 | 0.032 | 1.32e-85 | 0.188 | 0.157 | 0.031 | 4.63e-22 |
| Breast_Cancer | LDpred1.Inf | LDpred1.10FCVal | 0.172 | 0.204 | -0.032 | 1.32e-85 | 0.157 | 0.188 | -0.031 | 4.63e-22 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.216 | 0.204 | 0.012 | 2.33e-09 | 0.207 | 0.188 | 0.018 | 1.38e-05 |
| Breast_Cancer | lassosum.PseudoVal | LDpred1.10FCVal | 0.216 | 0.204 | 0.013 | 1.46e-04 | 0.195 | 0.188 | 0.006 | 3.47e-01 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred1.10FCVal | 0.229 | 0.204 | 0.025 | 2.34e-04 | 0.213 | 0.188 | 0.024 | 3.11e-07 |
| Breast_Cancer | pT+clump.10FCVal | LDpred1.10FCVal | 0.182 | 0.204 | -0.022 | 2.43e-07 | 0.162 | 0.188 | -0.027 | 1.46e-03 |
| Breast_Cancer | lassosum.10FCVal | LDpred1.10FCVal | 0.218 | 0.204 | 0.014 | 1.77e-07 | 0.196 | 0.188 | 0.008 | 1.56e-01 |
| Breast_Cancer | LDpred2.10FCVal | LDpred1.10FCVal | 0.225 | 0.204 | 0.021 | 1.60e-23 | 0.208 | 0.188 | 0.019 | 5.02e-06 |
| Breast_Cancer | All.MultiPRS | LDpred1.10FCVal | 0.229 | 0.204 | 0.025 | 2.05e-04 | 0.214 | 0.188 | 0.026 | 4.46e-08 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred1.10FCVal | 0.217 | 0.204 | 0.013 | 3.86e-02 | 0.200 | 0.188 | 0.012 | 3.80e-04 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred1.10FCVal | 0.223 | 0.204 | 0.019 | 6.94e-13 | 0.204 | 0.188 | 0.016 | 4.54e-03 |
| Breast_Cancer | SBLUP.Inf | LDpred1.10FCVal | 0.166 | 0.204 | -0.038 | 4.96e-86 | 0.154 | 0.188 | -0.034 | 5.88e-19 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred1.10FCVal | 0.201 | 0.204 | -0.002 | 7.42e-01 | 0.186 | 0.188 | -0.003 | 6.72e-01 |
| Breast_Cancer | LDpred2.Inf | LDpred1.10FCVal | 0.168 | 0.204 | -0.036 | 1.16e-79 | 0.157 | 0.188 | -0.032 | 8.91e-17 |
| Breast_Cancer | lassosum.MultiPRS | LDpred1.10FCVal | 0.225 | 0.204 | 0.021 | 2.20e-03 | 0.209 | 0.188 | 0.020 | 4.60e-05 |
| Breast_Cancer | LDpred1.10FCVal | LDpred1.10FCVal | 0.204 | 0.204 | 0.000 | 1.00e+00 | 0.188 | 0.188 | 0.000 | 1.00e+00 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred1.10FCVal | 0.227 | 0.204 | 0.023 | 2.36e-22 | 0.207 | 0.188 | 0.019 | 9.70e-05 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.229 | 0.229 | 0.000 | 1.00e+00 | 0.213 | 0.213 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.MultiPRS | 0.216 | 0.229 | -0.012 | 7.07e-02 | 0.195 | 0.213 | -0.018 | 8.28e-07 |
| Breast_Cancer | All.MultiPRS | LDpred2.MultiPRS | 0.229 | 0.229 | 0.000 | 9.57e-01 | 0.214 | 0.213 | 0.001 | 9.90e-02 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.217 | 0.229 | -0.012 | 8.85e-02 | 0.200 | 0.213 | -0.013 | 3.20e-04 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.216 | 0.229 | -0.013 | 6.24e-02 | 0.207 | 0.213 | -0.006 | 8.69e-02 |
| Breast_Cancer | LDpred1.Inf | LDpred2.MultiPRS | 0.172 | 0.229 | -0.057 | 1.00e-16 | 0.157 | 0.213 | -0.056 | 1.66e-15 |
| Breast_Cancer | LDpred2.Inf | LDpred2.MultiPRS | 0.168 | 0.229 | -0.061 | 5.20e-19 | 0.157 | 0.213 | -0.056 | 2.85e-15 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.MultiPRS | 0.225 | 0.229 | -0.004 | 5.44e-01 | 0.209 | 0.213 | -0.004 | 8.58e-02 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.MultiPRS | 0.204 | 0.229 | -0.025 | 2.34e-04 | 0.188 | 0.213 | -0.024 | 3.11e-07 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.201 | 0.229 | -0.027 | 6.06e-05 | 0.186 | 0.213 | -0.027 | 2.36e-08 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.227 | 0.229 | -0.002 | 7.55e-01 | 0.207 | 0.213 | -0.006 | 1.46e-02 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.MultiPRS | 0.182 | 0.229 | -0.047 | 8.40e-12 | 0.162 | 0.213 | -0.051 | 4.12e-15 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.MultiPRS | 0.218 | 0.229 | -0.011 | 1.12e-01 | 0.196 | 0.213 | -0.016 | 1.13e-06 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.MultiPRS | 0.225 | 0.229 | -0.004 | 5.64e-01 | 0.208 | 0.213 | -0.005 | 6.77e-03 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.223 | 0.229 | -0.006 | 4.09e-01 | 0.204 | 0.213 | -0.009 | 2.14e-04 |
| Breast_Cancer | SBLUP.Inf | LDpred2.MultiPRS | 0.166 | 0.229 | -0.063 | 6.09e-20 | 0.154 | 0.213 | -0.059 | 5.14e-16 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.10FCVal | 0.229 | 0.225 | 0.004 | 5.64e-01 | 0.213 | 0.208 | 0.005 | 6.77e-03 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.10FCVal | 0.182 | 0.225 | -0.043 | 7.44e-36 | 0.162 | 0.208 | -0.046 | 1.88e-11 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.10FCVal | 0.216 | 0.225 | -0.008 | 4.36e-05 | 0.195 | 0.208 | -0.013 | 1.40e-03 |
| Breast_Cancer | All.MultiPRS | LDpred2.10FCVal | 0.229 | 0.225 | 0.004 | 5.30e-01 | 0.214 | 0.208 | 0.006 | 2.03e-03 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.10FCVal | 0.217 | 0.225 | -0.008 | 2.34e-01 | 0.200 | 0.208 | -0.008 | 2.33e-02 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.10FCVal | 0.223 | 0.225 | -0.002 | 4.61e-02 | 0.204 | 0.208 | -0.004 | 2.47e-02 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.216 | 0.225 | -0.009 | 6.35e-06 | 0.207 | 0.208 | -0.001 | 7.19e-01 |
| Breast_Cancer | LDpred1.Inf | LDpred2.10FCVal | 0.172 | 0.225 | -0.053 | 3.32e-61 | 0.157 | 0.208 | -0.051 | 3.16e-15 |
| Breast_Cancer | LDpred2.Inf | LDpred2.10FCVal | 0.168 | 0.225 | -0.057 | 3.47e-70 | 0.157 | 0.208 | -0.051 | 2.35e-15 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.10FCVal | 0.225 | 0.225 | 0.000 | 9.77e-01 | 0.209 | 0.208 | 0.001 | 8.42e-01 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.10FCVal | 0.204 | 0.225 | -0.021 | 1.60e-23 | 0.188 | 0.208 | -0.019 | 5.02e-06 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.10FCVal | 0.201 | 0.225 | -0.023 | 6.03e-04 | 0.186 | 0.208 | -0.022 | 1.57e-05 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.10FCVal | 0.227 | 0.225 | 0.002 | 1.36e-01 | 0.207 | 0.208 | -0.001 | 7.32e-01 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.10FCVal | 0.218 | 0.225 | -0.007 | 1.43e-05 | 0.196 | 0.208 | -0.012 | 2.81e-04 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.10FCVal | 0.225 | 0.225 | 0.000 | 1.00e+00 | 0.208 | 0.208 | 0.000 | 1.00e+00 |
| Breast_Cancer | SBLUP.Inf | LDpred2.10FCVal | 0.166 | 0.225 | -0.059 | 1.74e-71 | 0.154 | 0.208 | -0.054 | 4.14e-16 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.229 | 0.223 | 0.006 | 4.09e-01 | 0.213 | 0.204 | 0.009 | 2.14e-04 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.PseudoVal | 0.216 | 0.223 | -0.007 | 8.20e-04 | 0.195 | 0.204 | -0.009 | 2.08e-02 |
| Breast_Cancer | All.MultiPRS | LDpred2.PseudoVal | 0.229 | 0.223 | 0.006 | 3.81e-01 | 0.214 | 0.204 | 0.010 | 6.35e-05 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.217 | 0.223 | -0.006 | 3.55e-01 | 0.200 | 0.204 | -0.004 | 3.73e-01 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.216 | 0.223 | -0.007 | 2.95e-03 | 0.207 | 0.204 | 0.003 | 6.01e-01 |
| Breast_Cancer | LDpred1.Inf | LDpred2.PseudoVal | 0.172 | 0.223 | -0.051 | 1.64e-43 | 0.157 | 0.204 | -0.047 | 3.48e-10 |
| Breast_Cancer | LDpred2.Inf | LDpred2.PseudoVal | 0.168 | 0.223 | -0.055 | 8.73e-50 | 0.157 | 0.204 | -0.047 | 3.08e-10 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.PseudoVal | 0.225 | 0.223 | 0.002 | 8.25e-01 | 0.209 | 0.204 | 0.004 | 1.87e-01 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.PseudoVal | 0.204 | 0.223 | -0.019 | 6.94e-13 | 0.188 | 0.204 | -0.016 | 4.54e-03 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.201 | 0.223 | -0.022 | 1.46e-03 | 0.186 | 0.204 | -0.018 | 9.64e-04 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.227 | 0.223 | 0.003 | 1.38e-02 | 0.207 | 0.204 | 0.003 | 2.90e-01 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.PseudoVal | 0.182 | 0.223 | -0.041 | 3.83e-35 | 0.162 | 0.204 | -0.042 | 2.09e-10 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.PseudoVal | 0.218 | 0.223 | -0.005 | 2.96e-03 | 0.196 | 0.204 | -0.008 | 2.84e-02 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.PseudoVal | 0.225 | 0.223 | 0.002 | 4.61e-02 | 0.208 | 0.204 | 0.004 | 2.47e-02 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.223 | 0.223 | 0.000 | 1.00e+00 | 0.204 | 0.204 | 0.000 | 1.00e+00 |
| Breast_Cancer | SBLUP.Inf | LDpred2.PseudoVal | 0.166 | 0.223 | -0.057 | 2.59e-51 | 0.154 | 0.204 | -0.050 | 6.69e-11 |
| Breast_Cancer | DBSLMM.PseudoVal | LDpred2.Inf | 0.216 | 0.168 | 0.048 | 8.74e-60 | 0.207 | 0.157 | 0.050 | 4.40e-17 |
| Breast_Cancer | LDpred2.MultiPRS | LDpred2.Inf | 0.229 | 0.168 | 0.061 | 5.20e-19 | 0.213 | 0.157 | 0.056 | 2.85e-15 |
| Breast_Cancer | pT+clump.10FCVal | LDpred2.Inf | 0.182 | 0.168 | 0.014 | 4.86e-03 | 0.162 | 0.157 | 0.005 | 6.22e-01 |
| Breast_Cancer | lassosum.10FCVal | LDpred2.Inf | 0.218 | 0.168 | 0.050 | 3.27e-38 | 0.196 | 0.157 | 0.040 | 3.89e-07 |
| Breast_Cancer | lassosum.PseudoVal | LDpred2.Inf | 0.216 | 0.168 | 0.049 | 1.46e-26 | 0.195 | 0.157 | 0.038 | 3.28e-05 |
| Breast_Cancer | All.MultiPRS | LDpred2.Inf | 0.229 | 0.168 | 0.061 | 4.81e-19 | 0.214 | 0.157 | 0.057 | 6.95e-16 |
| Breast_Cancer | LDpred1.MultiPRS | LDpred2.Inf | 0.217 | 0.168 | 0.049 | 6.52e-14 | 0.200 | 0.157 | 0.043 | 4.70e-11 |
| Breast_Cancer | LDpred2.PseudoVal | LDpred2.Inf | 0.223 | 0.168 | 0.055 | 8.73e-50 | 0.204 | 0.157 | 0.047 | 3.08e-10 |
| Breast_Cancer | SBLUP.Inf | LDpred2.Inf | 0.166 | 0.168 | -0.002 | 2.58e-03 | 0.154 | 0.157 | -0.002 | 2.79e-02 |
| Breast_Cancer | LDpred1.Inf | LDpred2.Inf | 0.172 | 0.168 | 0.004 | 2.55e-04 | 0.157 | 0.157 | 0.001 | 7.90e-01 |
| Breast_Cancer | LDpred2.Inf | LDpred2.Inf | 0.168 | 0.168 | 0.000 | 1.00e+00 | 0.157 | 0.157 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.MultiPRS | LDpred2.Inf | 0.225 | 0.168 | 0.057 | 1.28e-16 | 0.209 | 0.157 | 0.052 | 1.01e-12 |
| Breast_Cancer | LDpred1.10FCVal | LDpred2.Inf | 0.204 | 0.168 | 0.036 | 1.16e-79 | 0.188 | 0.157 | 0.032 | 8.91e-17 |
| Breast_Cancer | pT+clump.MultiPRS | LDpred2.Inf | 0.201 | 0.168 | 0.034 | 9.84e-07 | 0.186 | 0.157 | 0.029 | 1.91e-04 |
| Breast_Cancer | SBayesR.PseudoVal | LDpred2.Inf | 0.227 | 0.168 | 0.059 | 1.50e-65 | 0.207 | 0.157 | 0.050 | 4.23e-13 |
| Breast_Cancer | LDpred2.10FCVal | LDpred2.Inf | 0.225 | 0.168 | 0.057 | 3.47e-70 | 0.208 | 0.157 | 0.051 | 2.35e-15 |
| Breast_Cancer | All.MultiPRS | pT+clump.MultiPRS | 0.229 | 0.201 | 0.028 | 5.40e-05 | 0.214 | 0.186 | 0.028 | 1.22e-09 |
| Breast_Cancer | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.217 | 0.201 | 0.016 | 2.16e-02 | 0.200 | 0.186 | 0.014 | 8.49e-03 |
| Breast_Cancer | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.223 | 0.201 | 0.022 | 1.46e-03 | 0.204 | 0.186 | 0.018 | 9.64e-04 |
| Breast_Cancer | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.216 | 0.201 | 0.015 | 3.22e-02 | 0.207 | 0.186 | 0.021 | 1.76e-05 |
| Breast_Cancer | LDpred1.Inf | pT+clump.MultiPRS | 0.172 | 0.201 | -0.030 | 1.78e-05 | 0.157 | 0.186 | -0.029 | 1.15e-04 |
| Breast_Cancer | LDpred2.Inf | pT+clump.MultiPRS | 0.168 | 0.201 | -0.034 | 9.84e-07 | 0.157 | 0.186 | -0.029 | 1.91e-04 |
| Breast_Cancer | lassosum.MultiPRS | pT+clump.MultiPRS | 0.225 | 0.201 | 0.023 | 6.96e-04 | 0.209 | 0.186 | 0.023 | 1.70e-06 |
| Breast_Cancer | LDpred1.10FCVal | pT+clump.MultiPRS | 0.204 | 0.201 | 0.002 | 7.42e-01 | 0.188 | 0.186 | 0.003 | 6.72e-01 |
| Breast_Cancer | lassosum.PseudoVal | pT+clump.MultiPRS | 0.216 | 0.201 | 0.015 | 2.77e-02 | 0.195 | 0.186 | 0.009 | 9.55e-02 |
| Breast_Cancer | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.229 | 0.201 | 0.027 | 6.06e-05 | 0.213 | 0.186 | 0.027 | 2.36e-08 |
| Breast_Cancer | pT+clump.10FCVal | pT+clump.MultiPRS | 0.182 | 0.201 | -0.019 | 4.57e-03 | 0.162 | 0.186 | -0.024 | 1.14e-07 |
| Breast_Cancer | lassosum.10FCVal | pT+clump.MultiPRS | 0.218 | 0.201 | 0.016 | 1.56e-02 | 0.196 | 0.186 | 0.010 | 4.10e-02 |
| Breast_Cancer | LDpred2.10FCVal | pT+clump.MultiPRS | 0.225 | 0.201 | 0.023 | 6.03e-04 | 0.208 | 0.186 | 0.022 | 1.57e-05 |
| Breast_Cancer | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.227 | 0.201 | 0.025 | 2.17e-04 | 0.207 | 0.186 | 0.021 | 2.74e-05 |
| Breast_Cancer | SBLUP.Inf | pT+clump.MultiPRS | 0.166 | 0.201 | -0.035 | 2.92e-07 | 0.154 | 0.186 | -0.032 | 4.40e-05 |
| Breast_Cancer | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.201 | 0.201 | 0.000 | 1.00e+00 | 0.186 | 0.186 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.PseudoVal | pT+clump.10FCVal | 0.216 | 0.182 | 0.034 | 9.09e-31 | 0.195 | 0.162 | 0.033 | 2.39e-08 |
| Breast_Cancer | All.MultiPRS | pT+clump.10FCVal | 0.229 | 0.182 | 0.047 | 6.64e-12 | 0.214 | 0.162 | 0.052 | 3.39e-16 |
| Breast_Cancer | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.216 | 0.182 | 0.034 | 5.79e-22 | 0.207 | 0.162 | 0.045 | 2.95e-10 |
| Breast_Cancer | LDpred1.Inf | pT+clump.10FCVal | 0.172 | 0.182 | -0.010 | 3.93e-02 | 0.157 | 0.162 | -0.004 | 6.51e-01 |
| Breast_Cancer | LDpred2.Inf | pT+clump.10FCVal | 0.168 | 0.182 | -0.014 | 4.86e-03 | 0.157 | 0.162 | -0.005 | 6.22e-01 |
| Breast_Cancer | lassosum.MultiPRS | pT+clump.10FCVal | 0.225 | 0.182 | 0.043 | 4.67e-10 | 0.209 | 0.162 | 0.047 | 2.73e-13 |
| Breast_Cancer | LDpred1.10FCVal | pT+clump.10FCVal | 0.204 | 0.182 | 0.022 | 2.43e-07 | 0.188 | 0.162 | 0.027 | 1.46e-03 |
| Breast_Cancer | pT+clump.MultiPRS | pT+clump.10FCVal | 0.201 | 0.182 | 0.019 | 4.57e-03 | 0.186 | 0.162 | 0.024 | 1.14e-07 |
| Breast_Cancer | LDpred2.MultiPRS | pT+clump.10FCVal | 0.229 | 0.182 | 0.047 | 8.40e-12 | 0.213 | 0.162 | 0.051 | 4.12e-15 |
| Breast_Cancer | pT+clump.10FCVal | pT+clump.10FCVal | 0.182 | 0.182 | 0.000 | 1.00e+00 | 0.162 | 0.162 | 0.000 | 1.00e+00 |
| Breast_Cancer | lassosum.10FCVal | pT+clump.10FCVal | 0.218 | 0.182 | 0.036 | 5.79e-28 | 0.196 | 0.162 | 0.035 | 1.38e-07 |
| Breast_Cancer | LDpred2.10FCVal | pT+clump.10FCVal | 0.225 | 0.182 | 0.043 | 7.44e-36 | 0.208 | 0.162 | 0.046 | 1.88e-11 |
| Breast_Cancer | SBayesR.PseudoVal | pT+clump.10FCVal | 0.227 | 0.182 | 0.045 | 1.54e-40 | 0.207 | 0.162 | 0.045 | 1.15e-11 |
| Breast_Cancer | LDpred1.MultiPRS | pT+clump.10FCVal | 0.217 | 0.182 | 0.035 | 1.07e-07 | 0.200 | 0.162 | 0.038 | 2.10e-07 |
| Breast_Cancer | LDpred2.PseudoVal | pT+clump.10FCVal | 0.223 | 0.182 | 0.041 | 3.83e-35 | 0.204 | 0.162 | 0.042 | 2.09e-10 |
| Breast_Cancer | SBLUP.Inf | pT+clump.10FCVal | 0.166 | 0.182 | -0.016 | 1.60e-03 | 0.154 | 0.162 | -0.007 | 4.64e-01 |
| Breast_Cancer | LDpred1.Inf | SBayesR.PseudoVal | 0.172 | 0.227 | -0.055 | 4.25e-59 | 0.157 | 0.207 | -0.050 | 1.96e-13 |
| Breast_Cancer | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.216 | 0.227 | -0.011 | 2.54e-08 | 0.207 | 0.207 | -0.001 | 8.85e-01 |
| Breast_Cancer | lassosum.PseudoVal | SBayesR.PseudoVal | 0.216 | 0.227 | -0.010 | 2.77e-06 | 0.195 | 0.207 | -0.012 | 4.46e-03 |
| Breast_Cancer | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.229 | 0.227 | 0.002 | 7.55e-01 | 0.213 | 0.207 | 0.006 | 1.46e-02 |
| Breast_Cancer | pT+clump.10FCVal | SBayesR.PseudoVal | 0.182 | 0.227 | -0.045 | 1.54e-40 | 0.162 | 0.207 | -0.045 | 1.15e-11 |
| Breast_Cancer | lassosum.10FCVal | SBayesR.PseudoVal | 0.218 | 0.227 | -0.009 | 9.02e-06 | 0.196 | 0.207 | -0.011 | 6.08e-03 |
| Breast_Cancer | LDpred2.10FCVal | SBayesR.PseudoVal | 0.225 | 0.227 | -0.002 | 1.36e-01 | 0.208 | 0.207 | 0.001 | 7.32e-01 |
| Breast_Cancer | All.MultiPRS | SBayesR.PseudoVal | 0.229 | 0.227 | 0.002 | 7.16e-01 | 0.214 | 0.207 | 0.007 | 5.39e-04 |
| Breast_Cancer | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.217 | 0.227 | -0.010 | 1.43e-01 | 0.200 | 0.207 | -0.007 | 7.69e-02 |
| Breast_Cancer | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.223 | 0.227 | -0.003 | 1.38e-02 | 0.204 | 0.207 | -0.003 | 2.90e-01 |
| Breast_Cancer | SBLUP.Inf | SBayesR.PseudoVal | 0.166 | 0.227 | -0.061 | 1.17e-67 | 0.154 | 0.207 | -0.053 | 5.60e-14 |
| Breast_Cancer | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.201 | 0.227 | -0.025 | 2.17e-04 | 0.186 | 0.207 | -0.021 | 2.74e-05 |
| Breast_Cancer | LDpred2.Inf | SBayesR.PseudoVal | 0.168 | 0.227 | -0.059 | 1.50e-65 | 0.157 | 0.207 | -0.050 | 4.23e-13 |
| Breast_Cancer | lassosum.MultiPRS | SBayesR.PseudoVal | 0.225 | 0.227 | -0.002 | 7.69e-01 | 0.209 | 0.207 | 0.001 | 6.53e-01 |
| Breast_Cancer | LDpred1.10FCVal | SBayesR.PseudoVal | 0.204 | 0.227 | -0.023 | 2.36e-22 | 0.188 | 0.207 | -0.019 | 9.70e-05 |
| Breast_Cancer | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.227 | 0.227 | 0.000 | 1.00e+00 | 0.207 | 0.207 | 0.000 | 1.00e+00 |
| Breast_Cancer | All.MultiPRS | SBLUP.Inf | 0.229 | 0.166 | 0.063 | 5.68e-20 | 0.214 | 0.154 | 0.060 | 8.92e-17 |
| Breast_Cancer | LDpred1.Inf | SBLUP.Inf | 0.172 | 0.166 | 0.006 | 2.88e-07 | 0.157 | 0.154 | 0.003 | 1.88e-01 |
| Breast_Cancer | LDpred2.Inf | SBLUP.Inf | 0.168 | 0.166 | 0.002 | 2.58e-03 | 0.157 | 0.154 | 0.002 | 2.79e-02 |
| Breast_Cancer | DBSLMM.PseudoVal | SBLUP.Inf | 0.216 | 0.166 | 0.050 | 9.50e-64 | 0.207 | 0.154 | 0.052 | 1.16e-18 |
| Breast_Cancer | lassosum.PseudoVal | SBLUP.Inf | 0.216 | 0.166 | 0.050 | 4.71e-28 | 0.195 | 0.154 | 0.041 | 1.08e-05 |
| Breast_Cancer | LDpred2.MultiPRS | SBLUP.Inf | 0.229 | 0.166 | 0.063 | 6.09e-20 | 0.213 | 0.154 | 0.059 | 5.14e-16 |
| Breast_Cancer | pT+clump.10FCVal | SBLUP.Inf | 0.182 | 0.166 | 0.016 | 1.60e-03 | 0.162 | 0.154 | 0.007 | 4.64e-01 |
| Breast_Cancer | lassosum.10FCVal | SBLUP.Inf | 0.218 | 0.166 | 0.052 | 2.51e-40 | 0.196 | 0.154 | 0.042 | 8.06e-08 |
| Breast_Cancer | LDpred2.10FCVal | SBLUP.Inf | 0.225 | 0.166 | 0.059 | 1.74e-71 | 0.208 | 0.154 | 0.054 | 4.14e-16 |
| Breast_Cancer | SBayesR.PseudoVal | SBLUP.Inf | 0.227 | 0.166 | 0.061 | 1.17e-67 | 0.207 | 0.154 | 0.053 | 5.60e-14 |
| Breast_Cancer | LDpred1.MultiPRS | SBLUP.Inf | 0.217 | 0.166 | 0.051 | 9.79e-15 | 0.200 | 0.154 | 0.046 | 5.07e-12 |
| Breast_Cancer | LDpred2.PseudoVal | SBLUP.Inf | 0.223 | 0.166 | 0.057 | 2.59e-51 | 0.204 | 0.154 | 0.050 | 6.69e-11 |
| Breast_Cancer | SBLUP.Inf | SBLUP.Inf | 0.166 | 0.166 | 0.000 | 1.00e+00 | 0.154 | 0.154 | 0.000 | 1.00e+00 |
| Breast_Cancer | pT+clump.MultiPRS | SBLUP.Inf | 0.201 | 0.166 | 0.035 | 2.92e-07 | 0.186 | 0.154 | 0.032 | 4.40e-05 |
| Breast_Cancer | lassosum.MultiPRS | SBLUP.Inf | 0.225 | 0.166 | 0.059 | 1.81e-17 | 0.209 | 0.154 | 0.054 | 1.05e-13 |
| Breast_Cancer | LDpred1.10FCVal | SBLUP.Inf | 0.204 | 0.166 | 0.038 | 4.96e-86 | 0.188 | 0.154 | 0.034 | 5.88e-19 |
Show cross-validation differences between methods
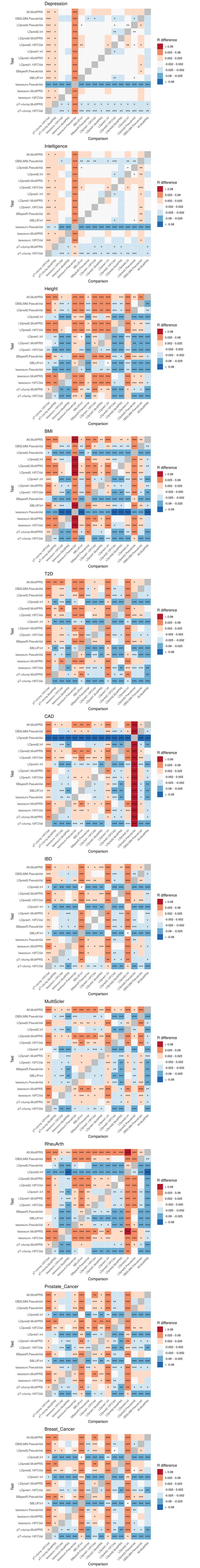
Predictive utility estimated using 10-fold cross-validation
Show test-validation differences between methods
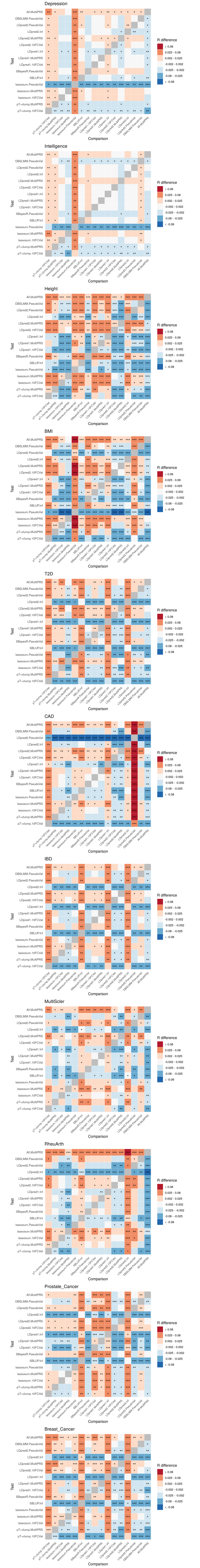
Predictive utility estimated using test set validation
4.2.2 TEDS
Show cross-validation results
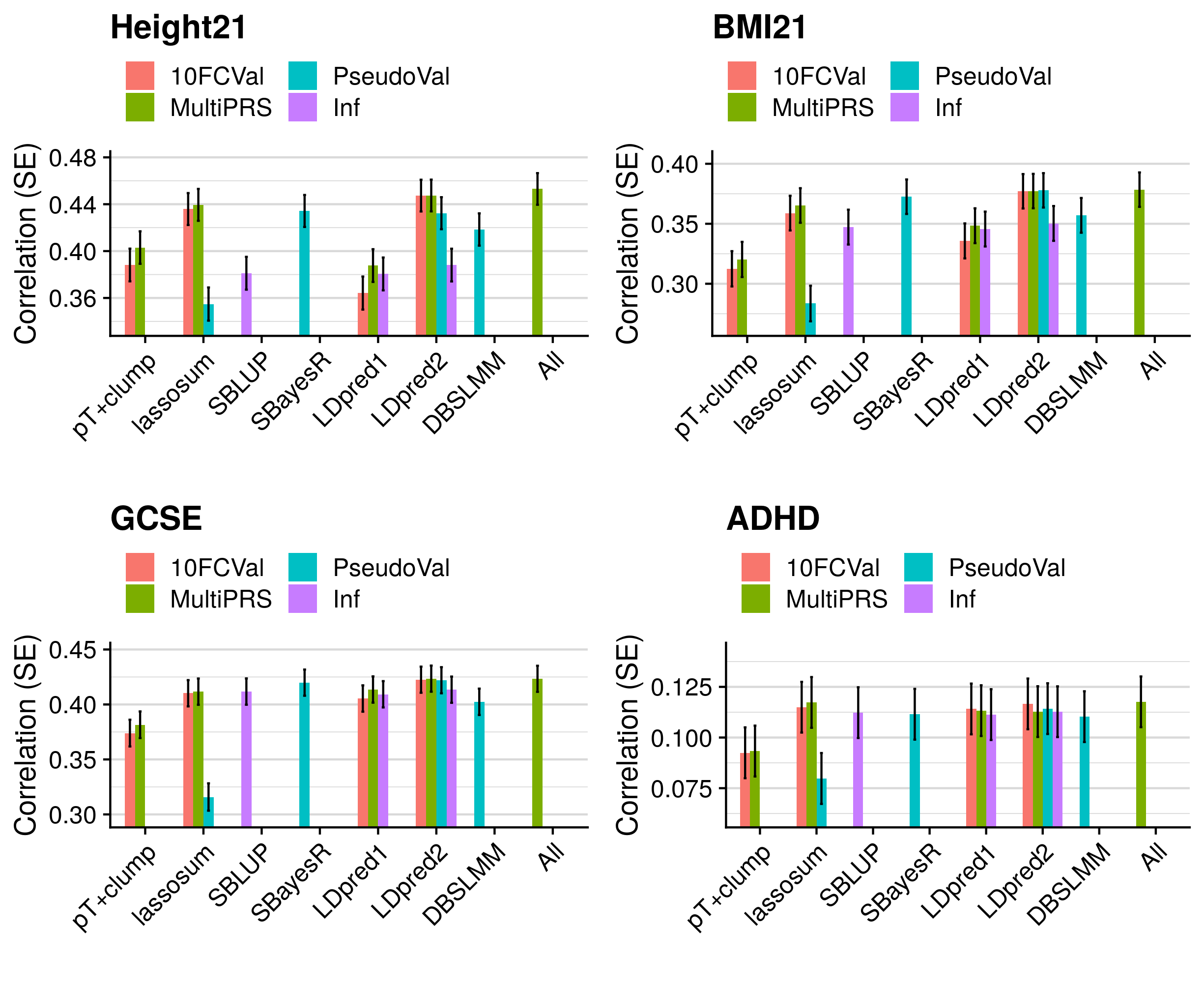
Predictive utility estimated using 10-fold cross-validation
Show test-validation results
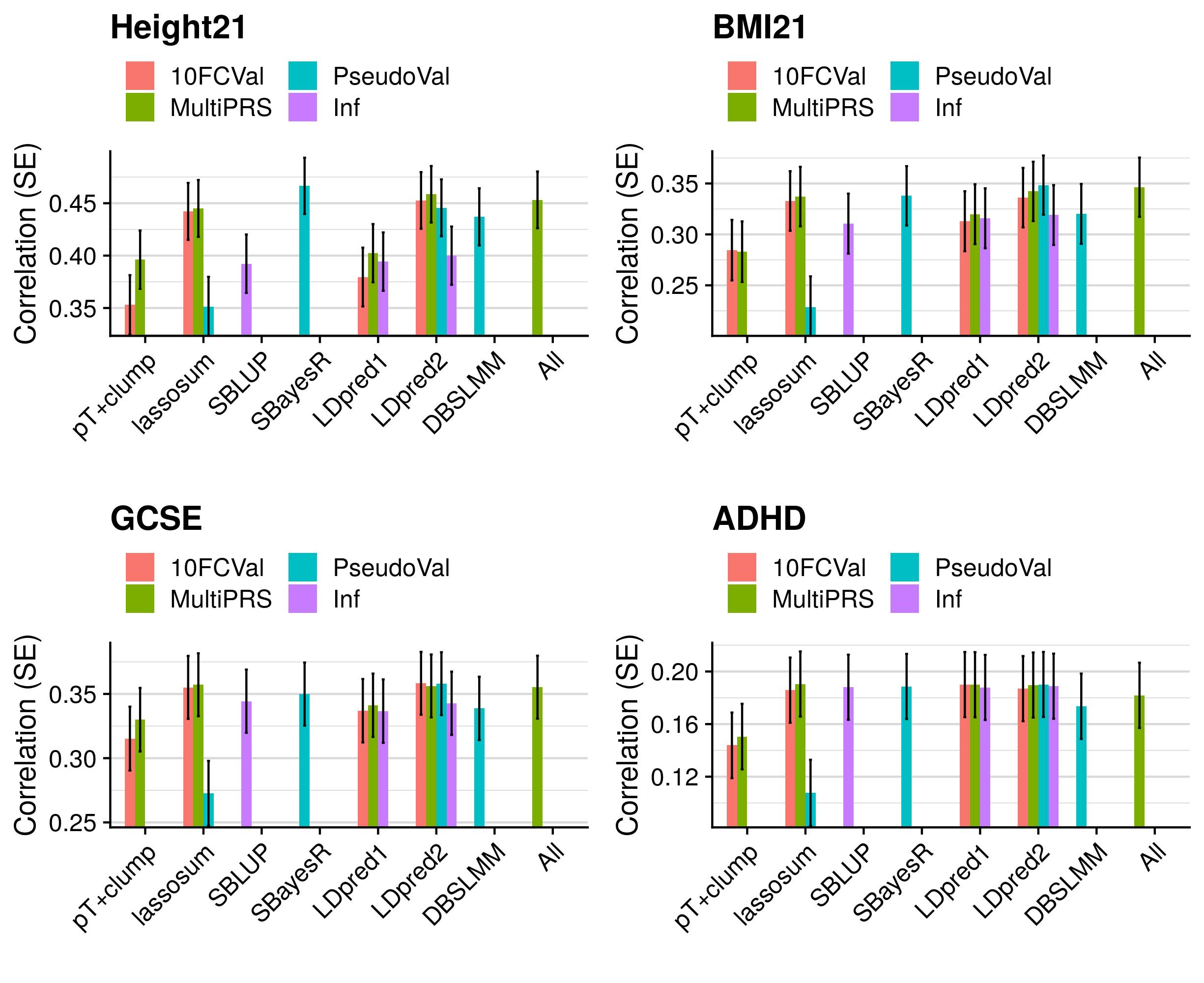
Predictive utility estimated using test set validation
Show TEDS results table
| Phenotype | Method | Model | CrossVal_R | CrossVal_R_SE | CrossVal_pval | IndepVal_R | IndepVal_R_SE | IndepVal_pval |
|---|---|---|---|---|---|---|---|---|
| Height21 | All | MultiPRS | 0.453 | 0.013 | 6.83e-220 | 0.453 | 0.027 | 2.22e-56 |
| Height21 | DBSLMM | PseudoVal | 0.418 | 0.014 | 1.37e-184 | 0.437 | 0.027 | 4.29e-52 |
| Height21 | lassosum | MultiPRS | 0.439 | 0.014 | 1.52e-205 | 0.445 | 0.027 | 3.44e-54 |
| Height21 | lassosum | 10FCVal | 0.436 | 0.014 | 7.09e-202 | 0.442 | 0.027 | 1.89e-53 |
| Height21 | lassosum | PseudoVal | 0.355 | 0.014 | 1.25e-129 | 0.351 | 0.028 | 4.66e-33 |
| Height21 | LDpred1 | MultiPRS | 0.388 | 0.014 | 1.75e-156 | 0.402 | 0.028 | 1.03e-43 |
| Height21 | LDpred1 | Inf | 0.381 | 0.014 | 2.24e-150 | 0.394 | 0.028 | 6.70e-42 |
| Height21 | LDpred1 | 10FCVal | 0.364 | 0.014 | 5.44e-137 | 0.380 | 0.028 | 1.04e-38 |
| Height21 | LDpred2 | MultiPRS | 0.447 | 0.014 | 6.63e-214 | 0.459 | 0.027 | 7.83e-58 |
| Height21 | LDpred2 | 10FCVal | 0.447 | 0.014 | 8.32e-214 | 0.453 | 0.027 | 3.19e-56 |
| Height21 | LDpred2 | PseudoVal | 0.432 | 0.014 | 2.85e-198 | 0.446 | 0.027 | 2.43e-54 |
| Height21 | LDpred2 | Inf | 0.388 | 0.014 | 7.57e-157 | 0.400 | 0.028 | 3.63e-43 |
| Height21 | pT+clump | MultiPRS | 0.403 | 0.014 | 3.92e-170 | 0.396 | 0.028 | 2.65e-42 |
| Height21 | pT+clump | 10FCVal | 0.388 | 0.014 | 6.69e-157 | 0.353 | 0.028 | 2.30e-33 |
| Height21 | SBayesR | PseudoVal | 0.434 | 0.014 | 3.34e-200 | 0.467 | 0.027 | 4.48e-60 |
| Height21 | SBLUP | Inf | 0.381 | 0.014 | 7.17e-151 | 0.392 | 0.028 | 1.87e-41 |
| BMI21 | All | MultiPRS | 0.378 | 0.014 | 2.63e-142 | 0.346 | 0.029 | 8.61e-31 |
| BMI21 | DBSLMM | PseudoVal | 0.357 | 0.014 | 9.10e-126 | 0.320 | 0.029 | 2.64e-26 |
| BMI21 | lassosum | MultiPRS | 0.365 | 0.014 | 5.66e-132 | 0.337 | 0.029 | 3.64e-29 |
| BMI21 | lassosum | 10FCVal | 0.359 | 0.014 | 3.67e-127 | 0.333 | 0.029 | 2.02e-28 |
| BMI21 | lassosum | PseudoVal | 0.284 | 0.015 | 4.71e-78 | 0.229 | 0.030 | 7.65e-14 |
| BMI21 | LDpred1 | MultiPRS | 0.348 | 0.015 | 1.92e-119 | 0.320 | 0.029 | 2.92e-26 |
| BMI21 | LDpred1 | Inf | 0.346 | 0.015 | 1.84e-117 | 0.316 | 0.029 | 1.31e-25 |
| BMI21 | LDpred1 | 10FCVal | 0.336 | 0.015 | 1.51e-110 | 0.313 | 0.029 | 3.78e-25 |
| BMI21 | LDpred2 | MultiPRS | 0.377 | 0.014 | 2.33e-141 | 0.342 | 0.029 | 4.72e-30 |
| BMI21 | LDpred2 | 10FCVal | 0.377 | 0.014 | 3.20e-141 | 0.336 | 0.029 | 5.51e-29 |
| BMI21 | LDpred2 | PseudoVal | 0.378 | 0.014 | 7.56e-142 | 0.348 | 0.029 | 3.72e-31 |
| BMI21 | LDpred2 | Inf | 0.350 | 0.014 | 7.86e-121 | 0.319 | 0.029 | 3.98e-26 |
| BMI21 | pT+clump | MultiPRS | 0.320 | 0.015 | 3.18e-100 | 0.283 | 0.030 | 1.11e-20 |
| BMI21 | pT+clump | 10FCVal | 0.312 | 0.015 | 3.06e-95 | 0.285 | 0.030 | 6.78e-21 |
| BMI21 | SBayesR | PseudoVal | 0.373 | 0.014 | 1.25e-137 | 0.338 | 0.029 | 2.75e-29 |
| BMI21 | SBLUP | Inf | 0.347 | 0.015 | 1.29e-118 | 0.311 | 0.029 | 8.83e-25 |
| GCSE | All | MultiPRS | 0.423 | 0.012 | 1.55e-252 | 0.355 | 0.024 | 1.13e-44 |
| GCSE | DBSLMM | PseudoVal | 0.402 | 0.012 | 4.19e-226 | 0.339 | 0.025 | 1.59e-40 |
| GCSE | lassosum | MultiPRS | 0.412 | 0.012 | 1.52e-237 | 0.357 | 0.024 | 3.63e-45 |
| GCSE | lassosum | 10FCVal | 0.410 | 0.012 | 1.01e-235 | 0.355 | 0.024 | 1.23e-44 |
| GCSE | lassosum | PseudoVal | 0.316 | 0.012 | 2.47e-135 | 0.273 | 0.025 | 2.65e-26 |
| GCSE | LDpred1 | MultiPRS | 0.414 | 0.012 | 5.57e-240 | 0.341 | 0.025 | 4.01e-41 |
| GCSE | LDpred1 | Inf | 0.409 | 0.012 | 1.48e-234 | 0.337 | 0.025 | 5.22e-40 |
| GCSE | LDpred1 | 10FCVal | 0.405 | 0.012 | 9.03e-230 | 0.337 | 0.025 | 4.32e-40 |
| GCSE | LDpred2 | MultiPRS | 0.423 | 0.012 | 1.03e-252 | 0.356 | 0.024 | 6.26e-45 |
| GCSE | LDpred2 | 10FCVal | 0.423 | 0.012 | 1.89e-251 | 0.358 | 0.024 | 1.82e-45 |
| GCSE | LDpred2 | PseudoVal | 0.422 | 0.012 | 7.71e-251 | 0.358 | 0.024 | 2.17e-45 |
| GCSE | LDpred2 | Inf | 0.413 | 0.012 | 8.70e-240 | 0.343 | 0.025 | 1.66e-41 |
| GCSE | pT+clump | MultiPRS | 0.382 | 0.012 | 1.35e-201 | 0.330 | 0.025 | 2.04e-38 |
| GCSE | pT+clump | 10FCVal | 0.374 | 0.012 | 4.00e-193 | 0.315 | 0.025 | 4.83e-35 |
| GCSE | SBayesR | PseudoVal | 0.420 | 0.012 | 5.56e-248 | 0.350 | 0.025 | 2.67e-43 |
| GCSE | SBLUP | Inf | 0.412 | 0.012 | 1.25e-237 | 0.344 | 0.025 | 6.57e-42 |
| ADHD | All | MultiPRS | 0.118 | 0.013 | 7.28e-21 | 0.182 | 0.025 | 3.45e-13 |
| ADHD | DBSLMM | PseudoVal | 0.110 | 0.013 | 1.60e-18 | 0.174 | 0.025 | 3.99e-12 |
| ADHD | lassosum | MultiPRS | 0.117 | 0.013 | 9.18e-21 | 0.190 | 0.025 | 2.42e-14 |
| ADHD | lassosum | PseudoVal | 0.080 | 0.013 | 2.23e-10 | 0.108 | 0.025 | 1.79e-05 |
| ADHD | lassosum | 10FCVal | 0.115 | 0.013 | 5.35e-20 | 0.186 | 0.025 | 1.03e-13 |
| ADHD | LDpred1 | MultiPRS | 0.113 | 0.013 | 1.87e-19 | 0.190 | 0.025 | 2.86e-14 |
| ADHD | LDpred1 | Inf | 0.111 | 0.013 | 7.76e-19 | 0.188 | 0.025 | 5.50e-14 |
| ADHD | LDpred1 | 10FCVal | 0.114 | 0.013 | 1.02e-19 | 0.190 | 0.025 | 2.77e-14 |
| ADHD | LDpred2 | MultiPRS | 0.113 | 0.013 | 2.67e-19 | 0.190 | 0.025 | 3.07e-14 |
| ADHD | LDpred2 | 10FCVal | 0.117 | 0.013 | 1.58e-20 | 0.187 | 0.025 | 7.27e-14 |
| ADHD | LDpred2 | PseudoVal | 0.114 | 0.013 | 9.02e-20 | 0.190 | 0.025 | 2.71e-14 |
| ADHD | LDpred2 | Inf | 0.113 | 0.013 | 2.76e-19 | 0.189 | 0.025 | 4.07e-14 |
| ADHD | pT+clump | MultiPRS | 0.093 | 0.013 | 1.13e-13 | 0.151 | 0.025 | 1.91e-09 |
| ADHD | pT+clump | 10FCVal | 0.092 | 0.013 | 1.89e-13 | 0.144 | 0.025 | 9.71e-09 |
| ADHD | SBayesR | PseudoVal | 0.111 | 0.013 | 6.80e-19 | 0.189 | 0.025 | 4.37e-14 |
| ADHD | SBLUP | Inf | 0.112 | 0.013 | 3.85e-19 | 0.188 | 0.025 | 5.20e-14 |
Show cross-validation results compared to pT+clump
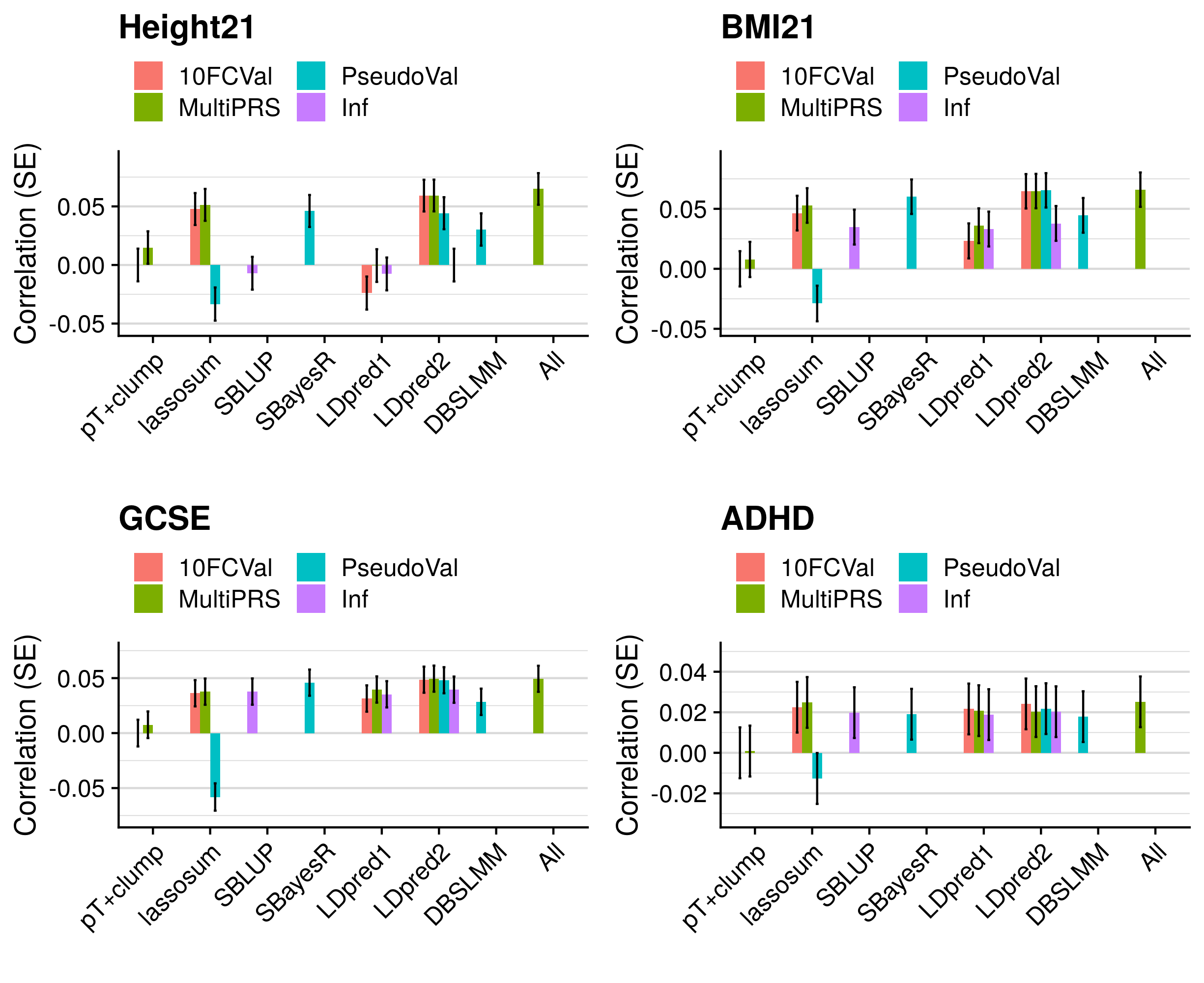
Predictive utility estimated using 10-fold cross-validation
Show test-validation results compared to pT+clump
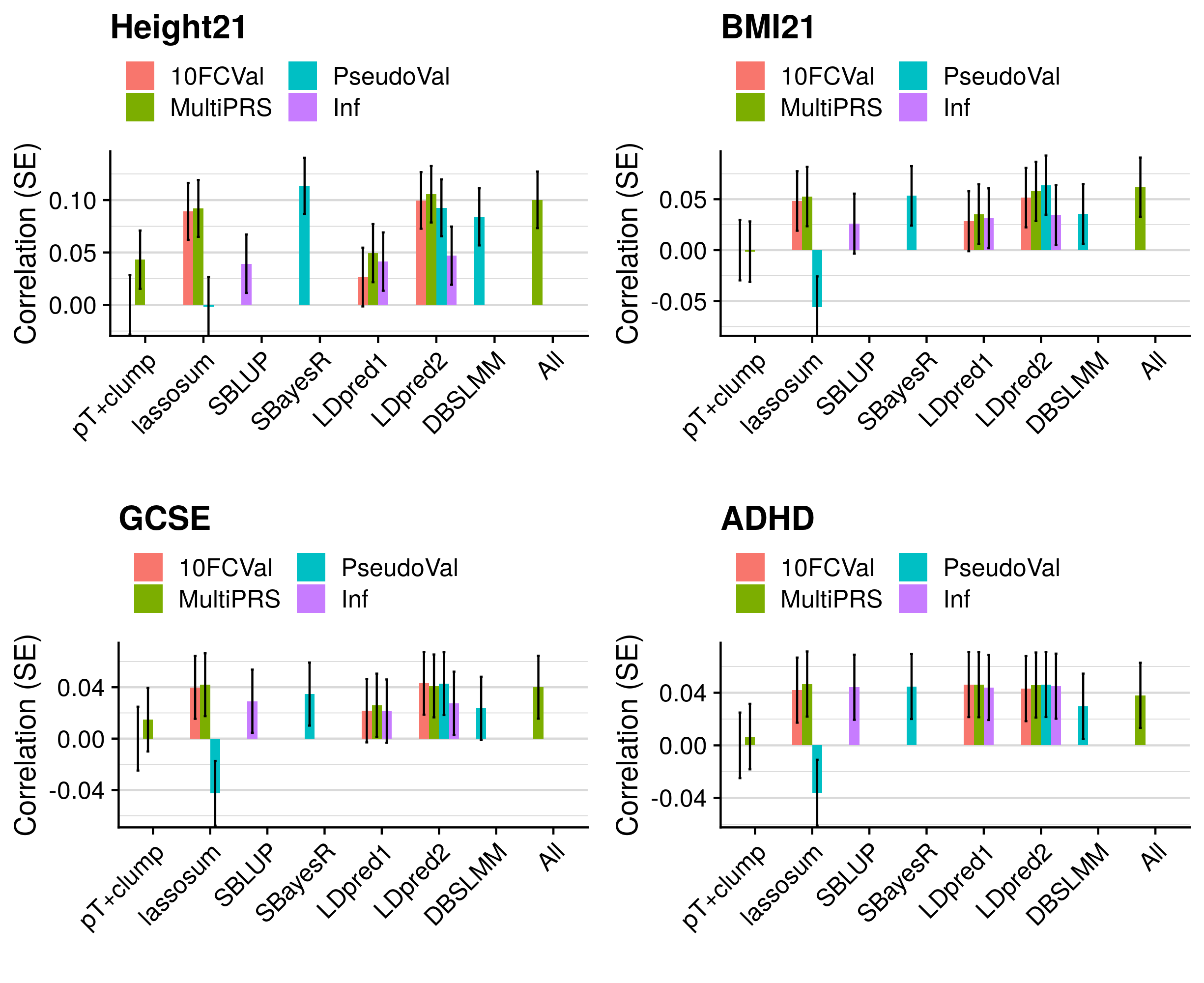
Predictive utility estimated using test set validation
Show difference results table
| Phenotype | Model 1 Test | Model 2 Test | Model 1 CrossVal R | Model 2 CrossVal R | CrossVal R Diff | CrossVal R Diff P | Model 1 IndepVal R | Model 2 IndepVal R | IndepVal R Diff | IndepVal R Diff P |
|---|---|---|---|---|---|---|---|---|---|---|
| Height21 | lassosum.10FCVal | All.MultiPRS | 0.436 | 0.453 | -0.017 | 3.39e-01 | 0.442 | 0.453 | -0.011 | 1.50e-01 |
| Height21 | All.MultiPRS | All.MultiPRS | 0.453 | 0.453 | 0.000 | 1.00e+00 | 0.453 | 0.453 | 0.000 | 1.00e+00 |
| Height21 | DBSLMM.PseudoVal | All.MultiPRS | 0.418 | 0.453 | -0.035 | 5.46e-02 | 0.437 | 0.453 | -0.016 | 1.22e-01 |
| Height21 | LDpred1.Inf | All.MultiPRS | 0.381 | 0.453 | -0.073 | 7.71e-05 | 0.394 | 0.453 | -0.059 | 8.51e-05 |
| Height21 | LDpred2.Inf | All.MultiPRS | 0.388 | 0.453 | -0.065 | 3.71e-04 | 0.400 | 0.453 | -0.053 | 2.35e-04 |
| Height21 | lassosum.MultiPRS | All.MultiPRS | 0.439 | 0.453 | -0.014 | 9.80e-05 | 0.445 | 0.453 | -0.008 | 2.33e-01 |
| Height21 | LDpred1.10FCVal | All.MultiPRS | 0.364 | 0.453 | -0.089 | 1.49e-06 | 0.380 | 0.453 | -0.074 | 3.88e-06 |
| Height21 | pT+clump.MultiPRS | All.MultiPRS | 0.403 | 0.453 | -0.050 | 6.57e-03 | 0.396 | 0.453 | -0.057 | 1.00e-06 |
| Height21 | LDpred2.MultiPRS | All.MultiPRS | 0.447 | 0.453 | -0.006 | 7.53e-01 | 0.459 | 0.453 | 0.005 | 2.31e-01 |
| Height21 | pT+clump.10FCVal | All.MultiPRS | 0.388 | 0.453 | -0.065 | 3.99e-04 | 0.353 | 0.453 | -0.100 | 1.16e-11 |
| Height21 | lassosum.PseudoVal | All.MultiPRS | 0.355 | 0.453 | -0.098 | 9.99e-08 | 0.351 | 0.453 | -0.102 | 7.91e-09 |
| Height21 | LDpred2.10FCVal | All.MultiPRS | 0.447 | 0.453 | -0.006 | 7.50e-01 | 0.453 | 0.453 | -0.001 | 9.12e-01 |
| Height21 | SBayesR.PseudoVal | All.MultiPRS | 0.434 | 0.453 | -0.019 | 2.94e-01 | 0.467 | 0.453 | 0.013 | 7.74e-02 |
| Height21 | LDpred1.MultiPRS | All.MultiPRS | 0.388 | 0.453 | -0.065 | 3.65e-04 | 0.402 | 0.453 | -0.051 | 2.08e-04 |
| Height21 | LDpred2.PseudoVal | All.MultiPRS | 0.432 | 0.453 | -0.021 | 2.49e-01 | 0.446 | 0.453 | -0.008 | 3.84e-01 |
| Height21 | SBLUP.Inf | All.MultiPRS | 0.381 | 0.453 | -0.072 | 8.46e-05 | 0.392 | 0.453 | -0.061 | 4.66e-05 |
| Height21 | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.447 | 0.418 | 0.029 | 1.05e-01 | 0.459 | 0.437 | 0.022 | 4.70e-02 |
| Height21 | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.388 | 0.418 | -0.030 | 1.21e-03 | 0.353 | 0.437 | -0.084 | 5.41e-06 |
| Height21 | lassosum.10FCVal | DBSLMM.PseudoVal | 0.436 | 0.418 | 0.017 | 5.51e-03 | 0.442 | 0.437 | 0.005 | 6.64e-01 |
| Height21 | All.MultiPRS | DBSLMM.PseudoVal | 0.453 | 0.418 | 0.035 | 5.46e-02 | 0.453 | 0.437 | 0.016 | 1.22e-01 |
| Height21 | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.388 | 0.418 | -0.031 | 9.84e-02 | 0.402 | 0.437 | -0.035 | 4.38e-03 |
| Height21 | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.432 | 0.418 | 0.014 | 6.57e-03 | 0.446 | 0.437 | 0.009 | 4.07e-01 |
| Height21 | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.418 | 0.418 | 0.000 | 1.00e+00 | 0.437 | 0.437 | 0.000 | 1.00e+00 |
| Height21 | LDpred1.Inf | DBSLMM.PseudoVal | 0.381 | 0.418 | -0.038 | 4.65e-10 | 0.394 | 0.437 | -0.043 | 5.00e-04 |
| Height21 | LDpred2.Inf | DBSLMM.PseudoVal | 0.388 | 0.418 | -0.030 | 2.22e-07 | 0.400 | 0.437 | -0.037 | 2.24e-03 |
| Height21 | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.439 | 0.418 | 0.021 | 2.46e-01 | 0.445 | 0.437 | 0.008 | 4.31e-01 |
| Height21 | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.364 | 0.418 | -0.054 | 4.24e-15 | 0.380 | 0.437 | -0.057 | 3.78e-05 |
| Height21 | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.403 | 0.418 | -0.015 | 4.03e-01 | 0.396 | 0.437 | -0.041 | 1.99e-03 |
| Height21 | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.434 | 0.418 | 0.016 | 6.40e-04 | 0.467 | 0.437 | 0.030 | 1.23e-03 |
| Height21 | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.355 | 0.418 | -0.064 | 7.07e-09 | 0.351 | 0.437 | -0.086 | 4.07e-05 |
| Height21 | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.447 | 0.418 | 0.029 | 1.75e-07 | 0.453 | 0.437 | 0.016 | 1.56e-01 |
| Height21 | SBLUP.Inf | DBSLMM.PseudoVal | 0.381 | 0.418 | -0.037 | 2.01e-10 | 0.392 | 0.437 | -0.045 | 2.25e-04 |
| Height21 | All.MultiPRS | lassosum.MultiPRS | 0.453 | 0.439 | 0.014 | 9.80e-05 | 0.453 | 0.445 | 0.008 | 2.33e-01 |
| Height21 | LDpred1.MultiPRS | lassosum.MultiPRS | 0.388 | 0.439 | -0.052 | 5.01e-03 | 0.402 | 0.445 | -0.043 | 2.81e-03 |
| Height21 | LDpred1.Inf | lassosum.MultiPRS | 0.381 | 0.439 | -0.059 | 1.43e-03 | 0.394 | 0.445 | -0.051 | 7.45e-04 |
| Height21 | LDpred2.Inf | lassosum.MultiPRS | 0.388 | 0.439 | -0.051 | 5.20e-03 | 0.400 | 0.445 | -0.045 | 2.37e-03 |
| Height21 | lassosum.MultiPRS | lassosum.MultiPRS | 0.439 | 0.439 | 0.000 | 1.00e+00 | 0.445 | 0.445 | 0.000 | 1.00e+00 |
| Height21 | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.418 | 0.439 | -0.021 | 2.46e-01 | 0.437 | 0.445 | -0.008 | 4.31e-01 |
| Height21 | lassosum.10FCVal | lassosum.MultiPRS | 0.436 | 0.439 | -0.004 | 8.43e-01 | 0.442 | 0.445 | -0.003 | 4.62e-01 |
| Height21 | LDpred2.MultiPRS | lassosum.MultiPRS | 0.447 | 0.439 | 0.008 | 6.55e-01 | 0.459 | 0.445 | 0.014 | 1.37e-01 |
| Height21 | pT+clump.10FCVal | lassosum.MultiPRS | 0.388 | 0.439 | -0.051 | 5.34e-03 | 0.353 | 0.445 | -0.092 | 1.61e-08 |
| Height21 | lassosum.PseudoVal | lassosum.MultiPRS | 0.355 | 0.439 | -0.085 | 5.03e-06 | 0.351 | 0.445 | -0.094 | 2.85e-09 |
| Height21 | LDpred2.10FCVal | lassosum.MultiPRS | 0.447 | 0.439 | 0.008 | 6.60e-01 | 0.453 | 0.445 | 0.008 | 3.94e-01 |
| Height21 | SBayesR.PseudoVal | lassosum.MultiPRS | 0.434 | 0.439 | -0.005 | 7.72e-01 | 0.467 | 0.445 | 0.022 | 2.36e-02 |
| Height21 | LDpred2.PseudoVal | lassosum.MultiPRS | 0.432 | 0.439 | -0.007 | 6.93e-01 | 0.446 | 0.445 | 0.001 | 9.59e-01 |
| Height21 | SBLUP.Inf | lassosum.MultiPRS | 0.381 | 0.439 | -0.058 | 1.55e-03 | 0.392 | 0.445 | -0.053 | 5.17e-04 |
| Height21 | pT+clump.MultiPRS | lassosum.MultiPRS | 0.403 | 0.439 | -0.036 | 4.94e-02 | 0.396 | 0.445 | -0.049 | 1.27e-04 |
| Height21 | LDpred1.10FCVal | lassosum.MultiPRS | 0.364 | 0.439 | -0.075 | 5.16e-05 | 0.380 | 0.445 | -0.066 | 5.42e-05 |
| Height21 | All.MultiPRS | lassosum.10FCVal | 0.453 | 0.436 | 0.017 | 3.39e-01 | 0.453 | 0.442 | 0.011 | 1.50e-01 |
| Height21 | LDpred1.MultiPRS | lassosum.10FCVal | 0.388 | 0.436 | -0.048 | 8.94e-03 | 0.402 | 0.442 | -0.040 | 1.35e-02 |
| Height21 | LDpred2.PseudoVal | lassosum.10FCVal | 0.432 | 0.436 | -0.004 | 5.72e-01 | 0.446 | 0.442 | 0.003 | 7.87e-01 |
| Height21 | DBSLMM.PseudoVal | lassosum.10FCVal | 0.418 | 0.436 | -0.017 | 5.51e-03 | 0.437 | 0.442 | -0.005 | 6.64e-01 |
| Height21 | LDpred1.Inf | lassosum.10FCVal | 0.381 | 0.436 | -0.055 | 1.70e-10 | 0.394 | 0.442 | -0.048 | 4.92e-03 |
| Height21 | LDpred2.Inf | lassosum.10FCVal | 0.388 | 0.436 | -0.048 | 1.61e-08 | 0.400 | 0.442 | -0.042 | 1.21e-02 |
| Height21 | lassosum.MultiPRS | lassosum.10FCVal | 0.439 | 0.436 | 0.004 | 8.43e-01 | 0.445 | 0.442 | 0.003 | 4.62e-01 |
| Height21 | LDpred1.10FCVal | lassosum.10FCVal | 0.364 | 0.436 | -0.072 | 7.42e-15 | 0.380 | 0.442 | -0.063 | 5.25e-04 |
| Height21 | lassosum.10FCVal | lassosum.10FCVal | 0.436 | 0.436 | 0.000 | 1.00e+00 | 0.442 | 0.442 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.MultiPRS | lassosum.10FCVal | 0.447 | 0.436 | 0.012 | 5.16e-01 | 0.459 | 0.442 | 0.016 | 9.35e-02 |
| Height21 | pT+clump.10FCVal | lassosum.10FCVal | 0.388 | 0.436 | -0.048 | 1.06e-08 | 0.353 | 0.442 | -0.089 | 3.47e-08 |
| Height21 | lassosum.PseudoVal | lassosum.10FCVal | 0.355 | 0.436 | -0.081 | 1.11e-23 | 0.351 | 0.442 | -0.091 | 5.83e-09 |
| Height21 | LDpred2.10FCVal | lassosum.10FCVal | 0.447 | 0.436 | 0.011 | 1.35e-02 | 0.453 | 0.442 | 0.010 | 2.63e-01 |
| Height21 | SBayesR.PseudoVal | lassosum.10FCVal | 0.434 | 0.436 | -0.002 | 7.63e-01 | 0.467 | 0.442 | 0.024 | 2.28e-02 |
| Height21 | SBLUP.Inf | lassosum.10FCVal | 0.381 | 0.436 | -0.055 | 2.42e-10 | 0.392 | 0.442 | -0.050 | 3.74e-03 |
| Height21 | pT+clump.MultiPRS | lassosum.10FCVal | 0.403 | 0.436 | -0.033 | 7.29e-02 | 0.396 | 0.442 | -0.046 | 4.49e-04 |
| Height21 | All.MultiPRS | lassosum.PseudoVal | 0.453 | 0.355 | 0.098 | 9.99e-08 | 0.453 | 0.351 | 0.102 | 7.91e-09 |
| Height21 | LDpred1.MultiPRS | lassosum.PseudoVal | 0.388 | 0.355 | 0.033 | 8.38e-02 | 0.402 | 0.351 | 0.051 | 4.25e-02 |
| Height21 | LDpred1.Inf | lassosum.PseudoVal | 0.381 | 0.355 | 0.026 | 5.60e-02 | 0.394 | 0.351 | 0.043 | 9.92e-02 |
| Height21 | LDpred2.Inf | lassosum.PseudoVal | 0.388 | 0.355 | 0.033 | 1.23e-02 | 0.400 | 0.351 | 0.049 | 5.98e-02 |
| Height21 | lassosum.MultiPRS | lassosum.PseudoVal | 0.439 | 0.355 | 0.085 | 5.03e-06 | 0.445 | 0.351 | 0.094 | 2.85e-09 |
| Height21 | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.418 | 0.355 | 0.064 | 7.07e-09 | 0.437 | 0.351 | 0.086 | 4.07e-05 |
| Height21 | lassosum.10FCVal | lassosum.PseudoVal | 0.436 | 0.355 | 0.081 | 1.11e-23 | 0.442 | 0.351 | 0.091 | 5.83e-09 |
| Height21 | LDpred2.MultiPRS | lassosum.PseudoVal | 0.447 | 0.355 | 0.093 | 5.19e-07 | 0.459 | 0.351 | 0.107 | 1.61e-07 |
| Height21 | pT+clump.10FCVal | lassosum.PseudoVal | 0.388 | 0.355 | 0.033 | 1.74e-03 | 0.353 | 0.351 | 0.002 | 9.38e-01 |
| Height21 | lassosum.PseudoVal | lassosum.PseudoVal | 0.355 | 0.355 | 0.000 | 1.00e+00 | 0.351 | 0.351 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.10FCVal | lassosum.PseudoVal | 0.447 | 0.355 | 0.093 | 1.24e-18 | 0.453 | 0.351 | 0.101 | 7.30e-07 |
| Height21 | SBayesR.PseudoVal | lassosum.PseudoVal | 0.434 | 0.355 | 0.079 | 8.89e-13 | 0.467 | 0.351 | 0.115 | 6.48e-08 |
| Height21 | LDpred2.PseudoVal | lassosum.PseudoVal | 0.432 | 0.355 | 0.078 | 8.78e-11 | 0.446 | 0.351 | 0.094 | 5.16e-05 |
| Height21 | SBLUP.Inf | lassosum.PseudoVal | 0.381 | 0.355 | 0.026 | 5.17e-02 | 0.392 | 0.351 | 0.041 | 1.20e-01 |
| Height21 | pT+clump.MultiPRS | lassosum.PseudoVal | 0.403 | 0.355 | 0.048 | 1.08e-02 | 0.396 | 0.351 | 0.045 | 2.81e-02 |
| Height21 | LDpred1.10FCVal | lassosum.PseudoVal | 0.364 | 0.355 | 0.009 | 4.97e-01 | 0.380 | 0.351 | 0.028 | 2.88e-01 |
| Height21 | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.418 | 0.388 | 0.031 | 9.84e-02 | 0.437 | 0.402 | 0.035 | 4.38e-03 |
| Height21 | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.447 | 0.388 | 0.060 | 9.89e-04 | 0.459 | 0.402 | 0.056 | 1.20e-05 |
| Height21 | pT+clump.10FCVal | LDpred1.MultiPRS | 0.388 | 0.388 | 0.000 | 9.80e-01 | 0.353 | 0.402 | -0.049 | 2.96e-02 |
| Height21 | lassosum.PseudoVal | LDpred1.MultiPRS | 0.355 | 0.388 | -0.033 | 8.38e-02 | 0.351 | 0.402 | -0.051 | 4.25e-02 |
| Height21 | lassosum.10FCVal | LDpred1.MultiPRS | 0.436 | 0.388 | 0.048 | 8.94e-03 | 0.442 | 0.402 | 0.040 | 1.35e-02 |
| Height21 | All.MultiPRS | LDpred1.MultiPRS | 0.453 | 0.388 | 0.065 | 3.65e-04 | 0.453 | 0.402 | 0.051 | 2.08e-04 |
| Height21 | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.388 | 0.388 | 0.000 | 1.00e+00 | 0.402 | 0.402 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.432 | 0.388 | 0.045 | 1.56e-02 | 0.446 | 0.402 | 0.043 | 9.52e-06 |
| Height21 | SBLUP.Inf | LDpred1.MultiPRS | 0.381 | 0.388 | -0.007 | 7.28e-01 | 0.392 | 0.402 | -0.010 | 1.81e-01 |
| Height21 | LDpred1.Inf | LDpred1.MultiPRS | 0.381 | 0.388 | -0.007 | 7.05e-01 | 0.394 | 0.402 | -0.008 | 2.10e-01 |
| Height21 | LDpred2.Inf | LDpred1.MultiPRS | 0.388 | 0.388 | 0.000 | 9.82e-01 | 0.400 | 0.402 | -0.002 | 7.52e-01 |
| Height21 | lassosum.MultiPRS | LDpred1.MultiPRS | 0.439 | 0.388 | 0.052 | 5.01e-03 | 0.445 | 0.402 | 0.043 | 2.81e-03 |
| Height21 | LDpred1.10FCVal | LDpred1.MultiPRS | 0.364 | 0.388 | -0.023 | 2.18e-01 | 0.380 | 0.402 | -0.023 | 1.18e-02 |
| Height21 | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.403 | 0.388 | 0.015 | 4.13e-01 | 0.396 | 0.402 | -0.006 | 7.18e-01 |
| Height21 | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.434 | 0.388 | 0.047 | 1.17e-02 | 0.467 | 0.402 | 0.064 | 3.18e-09 |
| Height21 | LDpred2.10FCVal | LDpred1.MultiPRS | 0.447 | 0.388 | 0.060 | 1.14e-03 | 0.453 | 0.402 | 0.050 | 1.33e-04 |
| Height21 | All.MultiPRS | LDpred1.Inf | 0.453 | 0.381 | 0.073 | 7.71e-05 | 0.453 | 0.394 | 0.059 | 8.51e-05 |
| Height21 | LDpred1.Inf | LDpred1.Inf | 0.381 | 0.381 | 0.000 | 1.00e+00 | 0.394 | 0.394 | 0.000 | 1.00e+00 |
| Height21 | LDpred2.Inf | LDpred1.Inf | 0.388 | 0.381 | 0.008 | 2.82e-03 | 0.400 | 0.394 | 0.006 | 2.74e-01 |
| Height21 | DBSLMM.PseudoVal | LDpred1.Inf | 0.418 | 0.381 | 0.038 | 4.65e-10 | 0.437 | 0.394 | 0.043 | 5.00e-04 |
| Height21 | lassosum.10FCVal | LDpred1.Inf | 0.436 | 0.381 | 0.055 | 1.70e-10 | 0.442 | 0.394 | 0.048 | 4.92e-03 |
| Height21 | LDpred2.MultiPRS | LDpred1.Inf | 0.447 | 0.381 | 0.067 | 2.43e-04 | 0.459 | 0.394 | 0.064 | 3.44e-06 |
| Height21 | pT+clump.10FCVal | LDpred1.Inf | 0.388 | 0.381 | 0.008 | 5.09e-01 | 0.353 | 0.394 | -0.041 | 7.59e-02 |
| Height21 | lassosum.PseudoVal | LDpred1.Inf | 0.355 | 0.381 | -0.026 | 5.60e-02 | 0.351 | 0.394 | -0.043 | 9.92e-02 |
| Height21 | LDpred2.10FCVal | LDpred1.Inf | 0.447 | 0.381 | 0.067 | 1.08e-20 | 0.453 | 0.394 | 0.058 | 4.17e-05 |
| Height21 | SBayesR.PseudoVal | LDpred1.Inf | 0.434 | 0.381 | 0.054 | 2.43e-20 | 0.467 | 0.394 | 0.072 | 3.56e-10 |
| Height21 | LDpred1.MultiPRS | LDpred1.Inf | 0.388 | 0.381 | 0.007 | 7.05e-01 | 0.402 | 0.394 | 0.008 | 2.10e-01 |
| Height21 | LDpred2.PseudoVal | LDpred1.Inf | 0.432 | 0.381 | 0.052 | 2.30e-23 | 0.446 | 0.394 | 0.051 | 7.51e-07 |
| Height21 | SBLUP.Inf | LDpred1.Inf | 0.381 | 0.381 | 0.001 | 8.25e-01 | 0.392 | 0.394 | -0.002 | 6.94e-01 |
| Height21 | pT+clump.MultiPRS | LDpred1.Inf | 0.403 | 0.381 | 0.023 | 2.29e-01 | 0.396 | 0.394 | 0.002 | 9.18e-01 |
| Height21 | lassosum.MultiPRS | LDpred1.Inf | 0.439 | 0.381 | 0.059 | 1.43e-03 | 0.445 | 0.394 | 0.051 | 7.45e-04 |
| Height21 | LDpred1.10FCVal | LDpred1.Inf | 0.364 | 0.381 | -0.016 | 1.21e-04 | 0.380 | 0.394 | -0.015 | 7.66e-02 |
| Height21 | LDpred1.Inf | LDpred1.10FCVal | 0.381 | 0.364 | 0.016 | 1.21e-04 | 0.394 | 0.380 | 0.015 | 7.66e-02 |
| Height21 | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.418 | 0.364 | 0.054 | 4.24e-15 | 0.437 | 0.380 | 0.057 | 3.78e-05 |
| Height21 | lassosum.10FCVal | LDpred1.10FCVal | 0.436 | 0.364 | 0.072 | 7.42e-15 | 0.442 | 0.380 | 0.063 | 5.25e-04 |
| Height21 | LDpred2.MultiPRS | LDpred1.10FCVal | 0.447 | 0.364 | 0.083 | 5.74e-06 | 0.459 | 0.380 | 0.079 | 9.10e-08 |
| Height21 | pT+clump.10FCVal | LDpred1.10FCVal | 0.388 | 0.364 | 0.024 | 4.79e-02 | 0.353 | 0.380 | -0.027 | 2.73e-01 |
| Height21 | lassosum.PseudoVal | LDpred1.10FCVal | 0.355 | 0.364 | -0.009 | 4.97e-01 | 0.351 | 0.380 | -0.028 | 2.88e-01 |
| Height21 | LDpred2.10FCVal | LDpred1.10FCVal | 0.447 | 0.364 | 0.083 | 2.11e-26 | 0.453 | 0.380 | 0.073 | 1.50e-06 |
| Height21 | All.MultiPRS | LDpred1.10FCVal | 0.453 | 0.364 | 0.089 | 1.49e-06 | 0.453 | 0.380 | 0.074 | 3.88e-06 |
| Height21 | LDpred1.MultiPRS | LDpred1.10FCVal | 0.388 | 0.364 | 0.023 | 2.18e-01 | 0.402 | 0.380 | 0.023 | 1.18e-02 |
| Height21 | LDpred2.PseudoVal | LDpred1.10FCVal | 0.432 | 0.364 | 0.068 | 3.24e-29 | 0.446 | 0.380 | 0.066 | 1.70e-08 |
| Height21 | SBLUP.Inf | LDpred1.10FCVal | 0.381 | 0.364 | 0.017 | 7.68e-05 | 0.392 | 0.380 | 0.013 | 1.34e-01 |
| Height21 | pT+clump.MultiPRS | LDpred1.10FCVal | 0.403 | 0.364 | 0.039 | 3.89e-02 | 0.396 | 0.380 | 0.017 | 3.84e-01 |
| Height21 | LDpred2.Inf | LDpred1.10FCVal | 0.388 | 0.364 | 0.024 | 4.73e-08 | 0.400 | 0.380 | 0.020 | 1.76e-02 |
| Height21 | lassosum.MultiPRS | LDpred1.10FCVal | 0.439 | 0.364 | 0.075 | 5.16e-05 | 0.445 | 0.380 | 0.066 | 5.42e-05 |
| Height21 | LDpred1.10FCVal | LDpred1.10FCVal | 0.364 | 0.364 | 0.000 | 1.00e+00 | 0.380 | 0.380 | 0.000 | 1.00e+00 |
| Height21 | SBayesR.PseudoVal | LDpred1.10FCVal | 0.434 | 0.364 | 0.070 | 4.03e-26 | 0.467 | 0.380 | 0.087 | 1.67e-11 |
| Height21 | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.447 | 0.447 | 0.000 | 1.00e+00 | 0.459 | 0.459 | 0.000 | 1.00e+00 |
| Height21 | lassosum.10FCVal | LDpred2.MultiPRS | 0.436 | 0.447 | -0.012 | 5.16e-01 | 0.442 | 0.459 | -0.016 | 9.35e-02 |
| Height21 | All.MultiPRS | LDpred2.MultiPRS | 0.453 | 0.447 | 0.006 | 7.53e-01 | 0.453 | 0.459 | -0.005 | 2.31e-01 |
| Height21 | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.388 | 0.447 | -0.060 | 9.89e-04 | 0.402 | 0.459 | -0.056 | 1.20e-05 |
| Height21 | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.418 | 0.447 | -0.029 | 1.05e-01 | 0.437 | 0.459 | -0.022 | 4.70e-02 |
| Height21 | LDpred1.Inf | LDpred2.MultiPRS | 0.381 | 0.447 | -0.067 | 2.43e-04 | 0.394 | 0.459 | -0.064 | 3.44e-06 |
| Height21 | LDpred2.Inf | LDpred2.MultiPRS | 0.388 | 0.447 | -0.059 | 1.11e-03 | 0.400 | 0.459 | -0.059 | 7.28e-06 |
| Height21 | lassosum.MultiPRS | LDpred2.MultiPRS | 0.439 | 0.447 | -0.008 | 6.55e-01 | 0.445 | 0.459 | -0.014 | 1.37e-01 |
| Height21 | LDpred1.10FCVal | LDpred2.MultiPRS | 0.364 | 0.447 | -0.083 | 5.74e-06 | 0.380 | 0.459 | -0.079 | 9.10e-08 |
| Height21 | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.403 | 0.447 | -0.044 | 1.46e-02 | 0.396 | 0.459 | -0.062 | 6.60e-06 |
| Height21 | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.434 | 0.447 | -0.013 | 4.59e-01 | 0.467 | 0.459 | 0.008 | 1.92e-01 |
| Height21 | pT+clump.10FCVal | LDpred2.MultiPRS | 0.388 | 0.447 | -0.059 | 1.10e-03 | 0.353 | 0.459 | -0.106 | 2.98e-09 |
| Height21 | lassosum.PseudoVal | LDpred2.MultiPRS | 0.355 | 0.447 | -0.093 | 5.19e-07 | 0.351 | 0.459 | -0.107 | 1.61e-07 |
| Height21 | LDpred2.10FCVal | LDpred2.MultiPRS | 0.447 | 0.447 | 0.000 | 9.96e-01 | 0.453 | 0.459 | -0.006 | 5.27e-02 |
| Height21 | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.432 | 0.447 | -0.015 | 3.97e-01 | 0.446 | 0.459 | -0.013 | 3.94e-02 |
| Height21 | SBLUP.Inf | LDpred2.MultiPRS | 0.381 | 0.447 | -0.066 | 2.71e-04 | 0.392 | 0.459 | -0.066 | 1.26e-06 |
| Height21 | LDpred2.MultiPRS | LDpred2.10FCVal | 0.447 | 0.447 | 0.000 | 9.96e-01 | 0.459 | 0.453 | 0.006 | 5.27e-02 |
| Height21 | pT+clump.10FCVal | LDpred2.10FCVal | 0.388 | 0.447 | -0.059 | 7.02e-11 | 0.353 | 0.453 | -0.100 | 1.99e-08 |
| Height21 | lassosum.10FCVal | LDpred2.10FCVal | 0.436 | 0.447 | -0.011 | 1.35e-02 | 0.442 | 0.453 | -0.010 | 2.63e-01 |
| Height21 | All.MultiPRS | LDpred2.10FCVal | 0.453 | 0.447 | 0.006 | 7.50e-01 | 0.453 | 0.453 | 0.001 | 9.12e-01 |
| Height21 | LDpred1.MultiPRS | LDpred2.10FCVal | 0.388 | 0.447 | -0.060 | 1.14e-03 | 0.402 | 0.453 | -0.050 | 1.33e-04 |
| Height21 | LDpred2.PseudoVal | LDpred2.10FCVal | 0.432 | 0.447 | -0.015 | 2.82e-06 | 0.446 | 0.453 | -0.007 | 2.71e-01 |
| Height21 | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.418 | 0.447 | -0.029 | 1.75e-07 | 0.437 | 0.453 | -0.016 | 1.56e-01 |
| Height21 | LDpred1.Inf | LDpred2.10FCVal | 0.381 | 0.447 | -0.067 | 1.08e-20 | 0.394 | 0.453 | -0.058 | 4.17e-05 |
| Height21 | LDpred2.Inf | LDpred2.10FCVal | 0.388 | 0.447 | -0.059 | 2.54e-18 | 0.400 | 0.453 | -0.053 | 9.95e-05 |
| Height21 | lassosum.MultiPRS | LDpred2.10FCVal | 0.439 | 0.447 | -0.008 | 6.60e-01 | 0.445 | 0.453 | -0.008 | 3.94e-01 |
| Height21 | LDpred1.10FCVal | LDpred2.10FCVal | 0.364 | 0.447 | -0.083 | 2.11e-26 | 0.380 | 0.453 | -0.073 | 1.50e-06 |
| Height21 | pT+clump.MultiPRS | LDpred2.10FCVal | 0.403 | 0.447 | -0.044 | 1.53e-02 | 0.396 | 0.453 | -0.057 | 3.82e-05 |
| Height21 | SBayesR.PseudoVal | LDpred2.10FCVal | 0.434 | 0.447 | -0.013 | 9.07e-05 | 0.467 | 0.453 | 0.014 | 2.82e-02 |
| Height21 | lassosum.PseudoVal | LDpred2.10FCVal | 0.355 | 0.447 | -0.093 | 1.24e-18 | 0.351 | 0.453 | -0.101 | 7.30e-07 |
| Height21 | LDpred2.10FCVal | LDpred2.10FCVal | 0.447 | 0.447 | 0.000 | 1.00e+00 | 0.453 | 0.453 | 0.000 | 1.00e+00 |
| Height21 | SBLUP.Inf | LDpred2.10FCVal | 0.381 | 0.447 | -0.066 | 1.08e-20 | 0.392 | 0.453 | -0.060 | 1.82e-05 |
| Height21 | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.447 | 0.432 | 0.015 | 3.97e-01 | 0.459 | 0.446 | 0.013 | 3.94e-02 |
| Height21 | lassosum.10FCVal | LDpred2.PseudoVal | 0.436 | 0.432 | 0.004 | 5.72e-01 | 0.442 | 0.446 | -0.003 | 7.87e-01 |
| Height21 | All.MultiPRS | LDpred2.PseudoVal | 0.453 | 0.432 | 0.021 | 2.49e-01 | 0.453 | 0.446 | 0.008 | 3.84e-01 |
| Height21 | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.388 | 0.432 | -0.045 | 1.56e-02 | 0.402 | 0.446 | -0.043 | 9.52e-06 |
| Height21 | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.418 | 0.432 | -0.014 | 6.57e-03 | 0.437 | 0.446 | -0.009 | 4.07e-01 |
| Height21 | LDpred1.Inf | LDpred2.PseudoVal | 0.381 | 0.432 | -0.052 | 2.30e-23 | 0.394 | 0.446 | -0.051 | 7.51e-07 |
| Height21 | LDpred2.Inf | LDpred2.PseudoVal | 0.388 | 0.432 | -0.044 | 4.41e-21 | 0.400 | 0.446 | -0.046 | 8.90e-07 |
| Height21 | lassosum.MultiPRS | LDpred2.PseudoVal | 0.439 | 0.432 | 0.007 | 6.93e-01 | 0.445 | 0.446 | -0.001 | 9.59e-01 |
| Height21 | LDpred1.10FCVal | LDpred2.PseudoVal | 0.364 | 0.432 | -0.068 | 3.24e-29 | 0.380 | 0.446 | -0.066 | 1.70e-08 |
| Height21 | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.403 | 0.432 | -0.029 | 1.11e-01 | 0.396 | 0.446 | -0.050 | 1.33e-03 |
| Height21 | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.434 | 0.432 | 0.002 | 5.62e-01 | 0.467 | 0.446 | 0.021 | 6.99e-04 |
| Height21 | pT+clump.10FCVal | LDpred2.PseudoVal | 0.388 | 0.432 | -0.044 | 2.10e-05 | 0.353 | 0.446 | -0.093 | 7.36e-06 |
| Height21 | lassosum.PseudoVal | LDpred2.PseudoVal | 0.355 | 0.432 | -0.078 | 8.78e-11 | 0.351 | 0.446 | -0.094 | 5.16e-05 |
| Height21 | LDpred2.10FCVal | LDpred2.PseudoVal | 0.447 | 0.432 | 0.015 | 2.82e-06 | 0.453 | 0.446 | 0.007 | 2.71e-01 |
| Height21 | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.432 | 0.432 | 0.000 | 1.00e+00 | 0.446 | 0.446 | 0.000 | 1.00e+00 |
| Height21 | SBLUP.Inf | LDpred2.PseudoVal | 0.381 | 0.432 | -0.051 | 6.61e-25 | 0.392 | 0.446 | -0.053 | 5.62e-08 |
| Height21 | DBSLMM.PseudoVal | LDpred2.Inf | 0.418 | 0.388 | 0.030 | 2.22e-07 | 0.437 | 0.400 | 0.037 | 2.24e-03 |
| Height21 | LDpred2.MultiPRS | LDpred2.Inf | 0.447 | 0.388 | 0.059 | 1.11e-03 | 0.459 | 0.400 | 0.059 | 7.28e-06 |
| Height21 | pT+clump.10FCVal | LDpred2.Inf | 0.388 | 0.388 | 0.000 | 9.96e-01 | 0.353 | 0.400 | -0.047 | 4.48e-02 |
| Height21 | lassosum.PseudoVal | LDpred2.Inf | 0.355 | 0.388 | -0.033 | 1.23e-02 | 0.351 | 0.400 | -0.049 | 5.98e-02 |
| Height21 | lassosum.10FCVal | LDpred2.Inf | 0.436 | 0.388 | 0.048 | 1.61e-08 | 0.442 | 0.400 | 0.042 | 1.21e-02 |
| Height21 | All.MultiPRS | LDpred2.Inf | 0.453 | 0.388 | 0.065 | 3.71e-04 | 0.453 | 0.400 | 0.053 | 2.35e-04 |
| Height21 | LDpred1.MultiPRS | LDpred2.Inf | 0.388 | 0.388 | 0.000 | 9.82e-01 | 0.402 | 0.400 | 0.002 | 7.52e-01 |
| Height21 | LDpred2.PseudoVal | LDpred2.Inf | 0.432 | 0.388 | 0.044 | 4.41e-21 | 0.446 | 0.400 | 0.046 | 8.90e-07 |
| Height21 | SBLUP.Inf | LDpred2.Inf | 0.381 | 0.388 | -0.007 | 5.17e-04 | 0.392 | 0.400 | -0.008 | 5.59e-02 |
| Height21 | LDpred1.Inf | LDpred2.Inf | 0.381 | 0.388 | -0.008 | 2.82e-03 | 0.394 | 0.400 | -0.006 | 2.74e-01 |
| Height21 | LDpred2.Inf | LDpred2.Inf | 0.388 | 0.388 | 0.000 | 1.00e+00 | 0.400 | 0.400 | 0.000 | 1.00e+00 |
| Height21 | lassosum.MultiPRS | LDpred2.Inf | 0.439 | 0.388 | 0.051 | 5.20e-03 | 0.445 | 0.400 | 0.045 | 2.37e-03 |
| Height21 | LDpred1.10FCVal | LDpred2.Inf | 0.364 | 0.388 | -0.024 | 4.73e-08 | 0.380 | 0.400 | -0.020 | 1.76e-02 |
| Height21 | pT+clump.MultiPRS | LDpred2.Inf | 0.403 | 0.388 | 0.015 | 4.23e-01 | 0.396 | 0.400 | -0.004 | 8.30e-01 |
| Height21 | SBayesR.PseudoVal | LDpred2.Inf | 0.434 | 0.388 | 0.046 | 3.61e-16 | 0.467 | 0.400 | 0.067 | 2.22e-09 |
| Height21 | LDpred2.10FCVal | LDpred2.Inf | 0.447 | 0.388 | 0.059 | 2.54e-18 | 0.453 | 0.400 | 0.053 | 9.95e-05 |
| Height21 | All.MultiPRS | pT+clump.MultiPRS | 0.453 | 0.403 | 0.050 | 6.57e-03 | 0.453 | 0.396 | 0.057 | 1.00e-06 |
| Height21 | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.388 | 0.403 | -0.015 | 4.13e-01 | 0.402 | 0.396 | 0.006 | 7.18e-01 |
| Height21 | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.432 | 0.403 | 0.029 | 1.11e-01 | 0.446 | 0.396 | 0.050 | 1.33e-03 |
| Height21 | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.418 | 0.403 | 0.015 | 4.03e-01 | 0.437 | 0.396 | 0.041 | 1.99e-03 |
| Height21 | LDpred1.Inf | pT+clump.MultiPRS | 0.381 | 0.403 | -0.023 | 2.29e-01 | 0.394 | 0.396 | -0.002 | 9.18e-01 |
| Height21 | LDpred2.Inf | pT+clump.MultiPRS | 0.388 | 0.403 | -0.015 | 4.23e-01 | 0.400 | 0.396 | 0.004 | 8.30e-01 |
| Height21 | lassosum.MultiPRS | pT+clump.MultiPRS | 0.439 | 0.403 | 0.036 | 4.94e-02 | 0.445 | 0.396 | 0.049 | 1.27e-04 |
| Height21 | LDpred1.10FCVal | pT+clump.MultiPRS | 0.364 | 0.403 | -0.039 | 3.89e-02 | 0.380 | 0.396 | -0.017 | 3.84e-01 |
| Height21 | lassosum.10FCVal | pT+clump.MultiPRS | 0.436 | 0.403 | 0.033 | 7.29e-02 | 0.442 | 0.396 | 0.046 | 4.49e-04 |
| Height21 | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.447 | 0.403 | 0.044 | 1.46e-02 | 0.459 | 0.396 | 0.062 | 6.60e-06 |
| Height21 | pT+clump.10FCVal | pT+clump.MultiPRS | 0.388 | 0.403 | -0.015 | 4.26e-01 | 0.353 | 0.396 | -0.043 | 3.12e-06 |
| Height21 | lassosum.PseudoVal | pT+clump.MultiPRS | 0.355 | 0.403 | -0.048 | 1.08e-02 | 0.351 | 0.396 | -0.045 | 2.81e-02 |
| Height21 | LDpred2.10FCVal | pT+clump.MultiPRS | 0.447 | 0.403 | 0.044 | 1.53e-02 | 0.453 | 0.396 | 0.057 | 3.82e-05 |
| Height21 | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.434 | 0.403 | 0.031 | 8.92e-02 | 0.467 | 0.396 | 0.070 | 1.78e-07 |
| Height21 | SBLUP.Inf | pT+clump.MultiPRS | 0.381 | 0.403 | -0.022 | 2.41e-01 | 0.392 | 0.396 | -0.004 | 8.28e-01 |
| Height21 | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.403 | 0.403 | 0.000 | 1.00e+00 | 0.396 | 0.396 | 0.000 | 1.00e+00 |
| Height21 | lassosum.10FCVal | pT+clump.10FCVal | 0.436 | 0.388 | 0.048 | 1.06e-08 | 0.442 | 0.353 | 0.089 | 3.47e-08 |
| Height21 | All.MultiPRS | pT+clump.10FCVal | 0.453 | 0.388 | 0.065 | 3.99e-04 | 0.453 | 0.353 | 0.100 | 1.16e-11 |
| Height21 | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.418 | 0.388 | 0.030 | 1.21e-03 | 0.437 | 0.353 | 0.084 | 5.41e-06 |
| Height21 | LDpred1.Inf | pT+clump.10FCVal | 0.381 | 0.388 | -0.008 | 5.09e-01 | 0.394 | 0.353 | 0.041 | 7.59e-02 |
| Height21 | LDpred2.Inf | pT+clump.10FCVal | 0.388 | 0.388 | 0.000 | 9.96e-01 | 0.400 | 0.353 | 0.047 | 4.48e-02 |
| Height21 | lassosum.MultiPRS | pT+clump.10FCVal | 0.439 | 0.388 | 0.051 | 5.34e-03 | 0.445 | 0.353 | 0.092 | 1.61e-08 |
| Height21 | LDpred1.10FCVal | pT+clump.10FCVal | 0.364 | 0.388 | -0.024 | 4.79e-02 | 0.380 | 0.353 | 0.027 | 2.73e-01 |
| Height21 | pT+clump.MultiPRS | pT+clump.10FCVal | 0.403 | 0.388 | 0.015 | 4.26e-01 | 0.396 | 0.353 | 0.043 | 3.12e-06 |
| Height21 | LDpred2.MultiPRS | pT+clump.10FCVal | 0.447 | 0.388 | 0.059 | 1.10e-03 | 0.459 | 0.353 | 0.106 | 2.98e-09 |
| Height21 | pT+clump.10FCVal | pT+clump.10FCVal | 0.388 | 0.388 | 0.000 | 1.00e+00 | 0.353 | 0.353 | 0.000 | 1.00e+00 |
| Height21 | lassosum.PseudoVal | pT+clump.10FCVal | 0.355 | 0.388 | -0.033 | 1.74e-03 | 0.351 | 0.353 | -0.002 | 9.38e-01 |
| Height21 | LDpred2.10FCVal | pT+clump.10FCVal | 0.447 | 0.388 | 0.059 | 7.02e-11 | 0.453 | 0.353 | 0.100 | 1.99e-08 |
| Height21 | SBayesR.PseudoVal | pT+clump.10FCVal | 0.434 | 0.388 | 0.046 | 6.52e-07 | 0.467 | 0.353 | 0.114 | 6.16e-10 |
| Height21 | LDpred1.MultiPRS | pT+clump.10FCVal | 0.388 | 0.388 | 0.000 | 9.80e-01 | 0.402 | 0.353 | 0.049 | 2.96e-02 |
| Height21 | LDpred2.PseudoVal | pT+clump.10FCVal | 0.432 | 0.388 | 0.044 | 2.10e-05 | 0.446 | 0.353 | 0.093 | 7.36e-06 |
| Height21 | SBLUP.Inf | pT+clump.10FCVal | 0.381 | 0.388 | -0.007 | 5.45e-01 | 0.392 | 0.353 | 0.039 | 9.29e-02 |
| Height21 | LDpred1.Inf | SBayesR.PseudoVal | 0.381 | 0.434 | -0.054 | 2.43e-20 | 0.394 | 0.467 | -0.072 | 3.56e-10 |
| Height21 | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.418 | 0.434 | -0.016 | 6.40e-04 | 0.437 | 0.467 | -0.030 | 1.23e-03 |
| Height21 | lassosum.10FCVal | SBayesR.PseudoVal | 0.436 | 0.434 | 0.002 | 7.63e-01 | 0.442 | 0.467 | -0.024 | 2.28e-02 |
| Height21 | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.447 | 0.434 | 0.013 | 4.59e-01 | 0.459 | 0.467 | -0.008 | 1.92e-01 |
| Height21 | pT+clump.10FCVal | SBayesR.PseudoVal | 0.388 | 0.434 | -0.046 | 6.52e-07 | 0.353 | 0.467 | -0.114 | 6.16e-10 |
| Height21 | lassosum.PseudoVal | SBayesR.PseudoVal | 0.355 | 0.434 | -0.079 | 8.89e-13 | 0.351 | 0.467 | -0.115 | 6.48e-08 |
| Height21 | LDpred2.10FCVal | SBayesR.PseudoVal | 0.447 | 0.434 | 0.013 | 9.07e-05 | 0.453 | 0.467 | -0.014 | 2.82e-02 |
| Height21 | All.MultiPRS | SBayesR.PseudoVal | 0.453 | 0.434 | 0.019 | 2.94e-01 | 0.453 | 0.467 | -0.013 | 7.74e-02 |
| Height21 | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.388 | 0.434 | -0.047 | 1.17e-02 | 0.402 | 0.467 | -0.064 | 3.18e-09 |
| Height21 | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.432 | 0.434 | -0.002 | 5.62e-01 | 0.446 | 0.467 | -0.021 | 6.99e-04 |
| Height21 | SBLUP.Inf | SBayesR.PseudoVal | 0.381 | 0.434 | -0.053 | 5.25e-20 | 0.392 | 0.467 | -0.074 | 1.15e-10 |
| Height21 | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.403 | 0.434 | -0.031 | 8.92e-02 | 0.396 | 0.467 | -0.070 | 1.78e-07 |
| Height21 | LDpred2.Inf | SBayesR.PseudoVal | 0.388 | 0.434 | -0.046 | 3.61e-16 | 0.400 | 0.467 | -0.067 | 2.22e-09 |
| Height21 | lassosum.MultiPRS | SBayesR.PseudoVal | 0.439 | 0.434 | 0.005 | 7.72e-01 | 0.445 | 0.467 | -0.022 | 2.36e-02 |
| Height21 | LDpred1.10FCVal | SBayesR.PseudoVal | 0.364 | 0.434 | -0.070 | 4.03e-26 | 0.380 | 0.467 | -0.087 | 1.67e-11 |
| Height21 | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.434 | 0.434 | 0.000 | 1.00e+00 | 0.467 | 0.467 | 0.000 | 1.00e+00 |
| Height21 | All.MultiPRS | SBLUP.Inf | 0.453 | 0.381 | 0.072 | 8.46e-05 | 0.453 | 0.392 | 0.061 | 4.66e-05 |
| Height21 | LDpred1.Inf | SBLUP.Inf | 0.381 | 0.381 | -0.001 | 8.25e-01 | 0.394 | 0.392 | 0.002 | 6.94e-01 |
| Height21 | LDpred2.Inf | SBLUP.Inf | 0.388 | 0.381 | 0.007 | 5.17e-04 | 0.400 | 0.392 | 0.008 | 5.59e-02 |
| Height21 | DBSLMM.PseudoVal | SBLUP.Inf | 0.418 | 0.381 | 0.037 | 2.01e-10 | 0.437 | 0.392 | 0.045 | 2.25e-04 |
| Height21 | lassosum.10FCVal | SBLUP.Inf | 0.436 | 0.381 | 0.055 | 2.42e-10 | 0.442 | 0.392 | 0.050 | 3.74e-03 |
| Height21 | LDpred2.MultiPRS | SBLUP.Inf | 0.447 | 0.381 | 0.066 | 2.71e-04 | 0.459 | 0.392 | 0.066 | 1.26e-06 |
| Height21 | pT+clump.10FCVal | SBLUP.Inf | 0.388 | 0.381 | 0.007 | 5.45e-01 | 0.353 | 0.392 | -0.039 | 9.29e-02 |
| Height21 | lassosum.PseudoVal | SBLUP.Inf | 0.355 | 0.381 | -0.026 | 5.17e-02 | 0.351 | 0.392 | -0.041 | 1.20e-01 |
| Height21 | LDpred2.10FCVal | SBLUP.Inf | 0.447 | 0.381 | 0.066 | 1.08e-20 | 0.453 | 0.392 | 0.060 | 1.82e-05 |
| Height21 | SBayesR.PseudoVal | SBLUP.Inf | 0.434 | 0.381 | 0.053 | 5.25e-20 | 0.467 | 0.392 | 0.074 | 1.15e-10 |
| Height21 | LDpred1.MultiPRS | SBLUP.Inf | 0.388 | 0.381 | 0.007 | 7.28e-01 | 0.402 | 0.392 | 0.010 | 1.81e-01 |
| Height21 | LDpred2.PseudoVal | SBLUP.Inf | 0.432 | 0.381 | 0.051 | 6.61e-25 | 0.446 | 0.392 | 0.053 | 5.62e-08 |
| Height21 | SBLUP.Inf | SBLUP.Inf | 0.381 | 0.381 | 0.000 | 1.00e+00 | 0.392 | 0.392 | 0.000 | 1.00e+00 |
| Height21 | pT+clump.MultiPRS | SBLUP.Inf | 0.403 | 0.381 | 0.022 | 2.41e-01 | 0.396 | 0.392 | 0.004 | 8.28e-01 |
| Height21 | lassosum.MultiPRS | SBLUP.Inf | 0.439 | 0.381 | 0.058 | 1.55e-03 | 0.445 | 0.392 | 0.053 | 5.17e-04 |
| Height21 | LDpred1.10FCVal | SBLUP.Inf | 0.364 | 0.381 | -0.017 | 7.68e-05 | 0.380 | 0.392 | -0.013 | 1.34e-01 |
| BMI21 | lassosum.10FCVal | All.MultiPRS | 0.359 | 0.378 | -0.020 | 3.17e-01 | 0.333 | 0.346 | -0.014 | 1.87e-01 |
| BMI21 | All.MultiPRS | All.MultiPRS | 0.378 | 0.378 | 0.000 | 1.00e+00 | 0.346 | 0.346 | 0.000 | 1.00e+00 |
| BMI21 | DBSLMM.PseudoVal | All.MultiPRS | 0.357 | 0.378 | -0.021 | 2.76e-01 | 0.320 | 0.346 | -0.026 | 1.84e-02 |
| BMI21 | LDpred1.Inf | All.MultiPRS | 0.346 | 0.378 | -0.033 | 9.76e-02 | 0.316 | 0.346 | -0.031 | 1.83e-02 |
| BMI21 | LDpred2.Inf | All.MultiPRS | 0.350 | 0.378 | -0.028 | 1.55e-01 | 0.319 | 0.346 | -0.027 | 2.38e-02 |
| BMI21 | lassosum.MultiPRS | All.MultiPRS | 0.365 | 0.378 | -0.013 | 4.96e-01 | 0.337 | 0.346 | -0.009 | 2.59e-01 |
| BMI21 | LDpred1.10FCVal | All.MultiPRS | 0.336 | 0.378 | -0.043 | 3.11e-02 | 0.313 | 0.346 | -0.033 | 1.86e-02 |
| BMI21 | pT+clump.MultiPRS | All.MultiPRS | 0.320 | 0.378 | -0.058 | 3.22e-03 | 0.283 | 0.346 | -0.063 | 6.74e-05 |
| BMI21 | LDpred2.MultiPRS | All.MultiPRS | 0.377 | 0.378 | -0.001 | 5.77e-01 | 0.342 | 0.346 | -0.004 | 3.42e-01 |
| BMI21 | pT+clump.10FCVal | All.MultiPRS | 0.312 | 0.378 | -0.066 | 9.60e-04 | 0.285 | 0.346 | -0.062 | 4.35e-04 |
| BMI21 | lassosum.PseudoVal | All.MultiPRS | 0.284 | 0.378 | -0.095 | 1.83e-06 | 0.229 | 0.346 | -0.118 | 1.15e-08 |
| BMI21 | LDpred2.10FCVal | All.MultiPRS | 0.377 | 0.378 | -0.001 | 9.44e-01 | 0.336 | 0.346 | -0.010 | 2.98e-02 |
| BMI21 | SBayesR.PseudoVal | All.MultiPRS | 0.373 | 0.378 | -0.006 | 7.63e-01 | 0.338 | 0.346 | -0.009 | 2.37e-01 |
| BMI21 | LDpred1.MultiPRS | All.MultiPRS | 0.348 | 0.378 | -0.030 | 1.71e-07 | 0.320 | 0.346 | -0.027 | 2.42e-02 |
| BMI21 | LDpred2.PseudoVal | All.MultiPRS | 0.378 | 0.378 | -0.001 | 9.77e-01 | 0.348 | 0.346 | 0.002 | 7.10e-01 |
| BMI21 | SBLUP.Inf | All.MultiPRS | 0.347 | 0.378 | -0.031 | 1.15e-01 | 0.311 | 0.346 | -0.036 | 4.62e-03 |
| BMI21 | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.377 | 0.357 | 0.020 | 3.03e-01 | 0.342 | 0.320 | 0.022 | 3.38e-02 |
| BMI21 | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.312 | 0.357 | -0.045 | 3.27e-08 | 0.285 | 0.320 | -0.036 | 2.72e-02 |
| BMI21 | lassosum.10FCVal | DBSLMM.PseudoVal | 0.359 | 0.357 | 0.002 | 7.34e-01 | 0.333 | 0.320 | 0.013 | 2.61e-01 |
| BMI21 | All.MultiPRS | DBSLMM.PseudoVal | 0.378 | 0.357 | 0.021 | 2.76e-01 | 0.346 | 0.320 | 0.026 | 1.84e-02 |
| BMI21 | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.348 | 0.357 | -0.009 | 6.64e-01 | 0.320 | 0.320 | 0.000 | 9.82e-01 |
| BMI21 | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.378 | 0.357 | 0.021 | 4.87e-05 | 0.348 | 0.320 | 0.028 | 6.77e-03 |
| BMI21 | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.357 | 0.357 | 0.000 | 1.00e+00 | 0.320 | 0.320 | 0.000 | 1.00e+00 |
| BMI21 | LDpred1.Inf | DBSLMM.PseudoVal | 0.346 | 0.357 | -0.011 | 5.84e-02 | 0.316 | 0.320 | -0.004 | 7.22e-01 |
| BMI21 | LDpred2.Inf | DBSLMM.PseudoVal | 0.350 | 0.357 | -0.007 | 2.63e-01 | 0.319 | 0.320 | -0.001 | 9.28e-01 |
| BMI21 | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.365 | 0.357 | 0.008 | 6.74e-01 | 0.337 | 0.320 | 0.017 | 1.42e-01 |
| BMI21 | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.336 | 0.357 | -0.021 | 2.44e-03 | 0.313 | 0.320 | -0.007 | 6.07e-01 |
| BMI21 | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.320 | 0.357 | -0.037 | 6.66e-02 | 0.283 | 0.320 | -0.037 | 9.66e-03 |
| BMI21 | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.373 | 0.357 | 0.016 | 3.39e-04 | 0.338 | 0.320 | 0.018 | 4.58e-02 |
| BMI21 | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.284 | 0.357 | -0.073 | 4.86e-14 | 0.229 | 0.320 | -0.092 | 1.15e-05 |
| BMI21 | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.377 | 0.357 | 0.020 | 1.44e-04 | 0.336 | 0.320 | 0.016 | 1.42e-01 |
| BMI21 | SBLUP.Inf | DBSLMM.PseudoVal | 0.347 | 0.357 | -0.010 | 1.03e-01 | 0.311 | 0.320 | -0.010 | 4.23e-01 |
| BMI21 | All.MultiPRS | lassosum.MultiPRS | 0.378 | 0.365 | 0.013 | 4.96e-01 | 0.346 | 0.337 | 0.009 | 2.59e-01 |
| BMI21 | LDpred1.MultiPRS | lassosum.MultiPRS | 0.348 | 0.365 | -0.017 | 3.88e-01 | 0.320 | 0.337 | -0.017 | 2.02e-01 |
| BMI21 | LDpred1.Inf | lassosum.MultiPRS | 0.346 | 0.365 | -0.020 | 3.16e-01 | 0.316 | 0.337 | -0.021 | 1.39e-01 |
| BMI21 | LDpred2.Inf | lassosum.MultiPRS | 0.350 | 0.365 | -0.015 | 4.45e-01 | 0.319 | 0.337 | -0.018 | 2.08e-01 |
| BMI21 | lassosum.MultiPRS | lassosum.MultiPRS | 0.365 | 0.365 | 0.000 | 1.00e+00 | 0.337 | 0.337 | 0.000 | 1.00e+00 |
| BMI21 | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.357 | 0.365 | -0.008 | 6.74e-01 | 0.320 | 0.337 | -0.017 | 1.42e-01 |
| BMI21 | lassosum.10FCVal | lassosum.MultiPRS | 0.359 | 0.365 | -0.006 | 7.46e-01 | 0.333 | 0.337 | -0.004 | 5.09e-01 |
| BMI21 | LDpred2.MultiPRS | lassosum.MultiPRS | 0.377 | 0.365 | 0.012 | 5.35e-01 | 0.342 | 0.337 | 0.005 | 6.11e-01 |
| BMI21 | pT+clump.10FCVal | lassosum.MultiPRS | 0.312 | 0.365 | -0.053 | 7.53e-03 | 0.285 | 0.337 | -0.053 | 2.36e-03 |
| BMI21 | lassosum.PseudoVal | lassosum.MultiPRS | 0.284 | 0.365 | -0.082 | 4.66e-05 | 0.229 | 0.337 | -0.109 | 9.77e-08 |
| BMI21 | LDpred2.10FCVal | lassosum.MultiPRS | 0.377 | 0.365 | 0.012 | 5.43e-01 | 0.336 | 0.337 | -0.001 | 9.17e-01 |
| BMI21 | SBayesR.PseudoVal | lassosum.MultiPRS | 0.373 | 0.365 | 0.007 | 7.08e-01 | 0.338 | 0.337 | 0.001 | 9.43e-01 |
| BMI21 | LDpred2.PseudoVal | lassosum.MultiPRS | 0.378 | 0.365 | 0.013 | 5.16e-01 | 0.348 | 0.337 | 0.011 | 2.69e-01 |
| BMI21 | SBLUP.Inf | lassosum.MultiPRS | 0.347 | 0.365 | -0.018 | 3.57e-01 | 0.311 | 0.337 | -0.027 | 6.04e-02 |
| BMI21 | pT+clump.MultiPRS | lassosum.MultiPRS | 0.320 | 0.365 | -0.045 | 2.41e-02 | 0.283 | 0.337 | -0.054 | 6.29e-04 |
| BMI21 | LDpred1.10FCVal | lassosum.MultiPRS | 0.336 | 0.365 | -0.030 | 1.34e-01 | 0.313 | 0.337 | -0.024 | 1.20e-01 |
| BMI21 | All.MultiPRS | lassosum.10FCVal | 0.378 | 0.359 | 0.020 | 3.17e-01 | 0.346 | 0.333 | 0.014 | 1.87e-01 |
| BMI21 | LDpred1.MultiPRS | lassosum.10FCVal | 0.348 | 0.359 | -0.011 | 5.96e-01 | 0.320 | 0.333 | -0.013 | 2.94e-01 |
| BMI21 | LDpred2.PseudoVal | lassosum.10FCVal | 0.378 | 0.359 | 0.019 | 3.52e-05 | 0.348 | 0.333 | 0.016 | 9.89e-02 |
| BMI21 | DBSLMM.PseudoVal | lassosum.10FCVal | 0.357 | 0.359 | -0.002 | 7.34e-01 | 0.320 | 0.333 | -0.013 | 2.61e-01 |
| BMI21 | LDpred1.Inf | lassosum.10FCVal | 0.346 | 0.359 | -0.013 | 3.83e-02 | 0.316 | 0.333 | -0.017 | 1.93e-01 |
| BMI21 | LDpred2.Inf | lassosum.10FCVal | 0.350 | 0.359 | -0.009 | 1.76e-01 | 0.319 | 0.333 | -0.014 | 2.89e-01 |
| BMI21 | lassosum.MultiPRS | lassosum.10FCVal | 0.365 | 0.359 | 0.006 | 7.46e-01 | 0.337 | 0.333 | 0.004 | 5.09e-01 |
| BMI21 | LDpred1.10FCVal | lassosum.10FCVal | 0.336 | 0.359 | -0.023 | 1.25e-03 | 0.313 | 0.333 | -0.020 | 1.70e-01 |
| BMI21 | lassosum.10FCVal | lassosum.10FCVal | 0.359 | 0.359 | 0.000 | 1.00e+00 | 0.333 | 0.333 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.MultiPRS | lassosum.10FCVal | 0.377 | 0.359 | 0.018 | 3.48e-01 | 0.342 | 0.333 | 0.009 | 3.35e-01 |
| BMI21 | pT+clump.10FCVal | lassosum.10FCVal | 0.312 | 0.359 | -0.046 | 3.12e-09 | 0.285 | 0.333 | -0.048 | 2.17e-03 |
| BMI21 | lassosum.PseudoVal | lassosum.10FCVal | 0.284 | 0.359 | -0.075 | 4.60e-14 | 0.229 | 0.333 | -0.104 | 1.12e-06 |
| BMI21 | LDpred2.10FCVal | lassosum.10FCVal | 0.377 | 0.359 | 0.018 | 1.57e-04 | 0.336 | 0.333 | 0.003 | 7.43e-01 |
| BMI21 | SBayesR.PseudoVal | lassosum.10FCVal | 0.373 | 0.359 | 0.014 | 3.06e-03 | 0.338 | 0.333 | 0.005 | 6.01e-01 |
| BMI21 | SBLUP.Inf | lassosum.10FCVal | 0.347 | 0.359 | -0.012 | 6.55e-02 | 0.311 | 0.333 | -0.022 | 7.99e-02 |
| BMI21 | pT+clump.MultiPRS | lassosum.10FCVal | 0.320 | 0.359 | -0.039 | 5.38e-02 | 0.283 | 0.333 | -0.050 | 7.63e-04 |
| BMI21 | All.MultiPRS | lassosum.PseudoVal | 0.378 | 0.284 | 0.095 | 1.83e-06 | 0.346 | 0.229 | 0.118 | 1.15e-08 |
| BMI21 | LDpred1.MultiPRS | lassosum.PseudoVal | 0.348 | 0.284 | 0.065 | 1.26e-03 | 0.320 | 0.229 | 0.091 | 2.85e-04 |
| BMI21 | LDpred1.Inf | lassosum.PseudoVal | 0.346 | 0.284 | 0.062 | 4.58e-07 | 0.316 | 0.229 | 0.087 | 7.00e-04 |
| BMI21 | LDpred2.Inf | lassosum.PseudoVal | 0.350 | 0.284 | 0.067 | 8.23e-08 | 0.319 | 0.229 | 0.090 | 5.64e-04 |
| BMI21 | lassosum.MultiPRS | lassosum.PseudoVal | 0.365 | 0.284 | 0.082 | 4.66e-05 | 0.337 | 0.229 | 0.109 | 9.77e-08 |
| BMI21 | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.357 | 0.284 | 0.073 | 4.86e-14 | 0.320 | 0.229 | 0.092 | 1.15e-05 |
| BMI21 | lassosum.10FCVal | lassosum.PseudoVal | 0.359 | 0.284 | 0.075 | 4.60e-14 | 0.333 | 0.229 | 0.104 | 1.12e-06 |
| BMI21 | LDpred2.MultiPRS | lassosum.PseudoVal | 0.377 | 0.284 | 0.094 | 2.46e-06 | 0.342 | 0.229 | 0.114 | 8.46e-08 |
| BMI21 | pT+clump.10FCVal | lassosum.PseudoVal | 0.312 | 0.284 | 0.029 | 2.47e-02 | 0.285 | 0.229 | 0.056 | 3.34e-02 |
| BMI21 | lassosum.PseudoVal | lassosum.PseudoVal | 0.284 | 0.284 | 0.000 | 1.00e+00 | 0.229 | 0.229 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.10FCVal | lassosum.PseudoVal | 0.377 | 0.284 | 0.094 | 1.57e-20 | 0.336 | 0.229 | 0.108 | 2.33e-07 |
| BMI21 | SBayesR.PseudoVal | lassosum.PseudoVal | 0.373 | 0.284 | 0.089 | 3.73e-19 | 0.338 | 0.229 | 0.109 | 1.41e-07 |
| BMI21 | LDpred2.PseudoVal | lassosum.PseudoVal | 0.378 | 0.284 | 0.094 | 9.27e-18 | 0.348 | 0.229 | 0.120 | 1.60e-07 |
| BMI21 | SBLUP.Inf | lassosum.PseudoVal | 0.347 | 0.284 | 0.064 | 3.01e-07 | 0.311 | 0.229 | 0.082 | 1.51e-03 |
| BMI21 | pT+clump.MultiPRS | lassosum.PseudoVal | 0.320 | 0.284 | 0.037 | 7.23e-02 | 0.283 | 0.229 | 0.054 | 1.19e-02 |
| BMI21 | LDpred1.10FCVal | lassosum.PseudoVal | 0.336 | 0.284 | 0.052 | 4.73e-05 | 0.313 | 0.229 | 0.084 | 1.66e-03 |
| BMI21 | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.357 | 0.348 | 0.009 | 6.64e-01 | 0.320 | 0.320 | 0.000 | 9.82e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.377 | 0.348 | 0.029 | 6.37e-08 | 0.342 | 0.320 | 0.022 | 3.75e-02 |
| BMI21 | pT+clump.10FCVal | LDpred1.MultiPRS | 0.312 | 0.348 | -0.036 | 7.54e-02 | 0.285 | 0.320 | -0.035 | 2.55e-02 |
| BMI21 | lassosum.PseudoVal | LDpred1.MultiPRS | 0.284 | 0.348 | -0.065 | 1.26e-03 | 0.229 | 0.320 | -0.091 | 2.85e-04 |
| BMI21 | lassosum.10FCVal | LDpred1.MultiPRS | 0.359 | 0.348 | 0.011 | 5.96e-01 | 0.333 | 0.320 | 0.013 | 2.94e-01 |
| BMI21 | All.MultiPRS | LDpred1.MultiPRS | 0.378 | 0.348 | 0.030 | 1.71e-07 | 0.346 | 0.320 | 0.027 | 2.42e-02 |
| BMI21 | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.348 | 0.348 | 0.000 | 1.00e+00 | 0.320 | 0.320 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.378 | 0.348 | 0.029 | 1.36e-01 | 0.348 | 0.320 | 0.029 | 2.62e-03 |
| BMI21 | SBLUP.Inf | LDpred1.MultiPRS | 0.347 | 0.348 | -0.001 | 9.54e-01 | 0.311 | 0.320 | -0.009 | 2.02e-01 |
| BMI21 | LDpred1.Inf | LDpred1.MultiPRS | 0.346 | 0.348 | -0.003 | 8.90e-01 | 0.316 | 0.320 | -0.004 | 4.89e-01 |
| BMI21 | LDpred2.Inf | LDpred1.MultiPRS | 0.350 | 0.348 | 0.002 | 9.24e-01 | 0.319 | 0.320 | -0.001 | 9.01e-01 |
| BMI21 | lassosum.MultiPRS | LDpred1.MultiPRS | 0.365 | 0.348 | 0.017 | 3.88e-01 | 0.337 | 0.320 | 0.017 | 2.02e-01 |
| BMI21 | LDpred1.10FCVal | LDpred1.MultiPRS | 0.336 | 0.348 | -0.013 | 5.29e-01 | 0.313 | 0.320 | -0.007 | 3.90e-01 |
| BMI21 | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.320 | 0.348 | -0.028 | 1.60e-01 | 0.283 | 0.320 | -0.037 | 2.24e-02 |
| BMI21 | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.373 | 0.348 | 0.024 | 2.22e-01 | 0.338 | 0.320 | 0.018 | 7.63e-02 |
| BMI21 | LDpred2.10FCVal | LDpred1.MultiPRS | 0.377 | 0.348 | 0.029 | 1.47e-01 | 0.336 | 0.320 | 0.016 | 1.62e-01 |
| BMI21 | All.MultiPRS | LDpred1.Inf | 0.378 | 0.346 | 0.033 | 9.76e-02 | 0.346 | 0.316 | 0.031 | 1.83e-02 |
| BMI21 | LDpred1.Inf | LDpred1.Inf | 0.346 | 0.346 | 0.000 | 1.00e+00 | 0.316 | 0.316 | 0.000 | 1.00e+00 |
| BMI21 | LDpred2.Inf | LDpred1.Inf | 0.350 | 0.346 | 0.005 | 8.99e-02 | 0.319 | 0.316 | 0.003 | 5.77e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred1.Inf | 0.357 | 0.346 | 0.011 | 5.84e-02 | 0.320 | 0.316 | 0.004 | 7.22e-01 |
| BMI21 | lassosum.10FCVal | LDpred1.Inf | 0.359 | 0.346 | 0.013 | 3.83e-02 | 0.333 | 0.316 | 0.017 | 1.93e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred1.Inf | 0.377 | 0.346 | 0.032 | 1.11e-01 | 0.342 | 0.316 | 0.026 | 2.44e-02 |
| BMI21 | pT+clump.10FCVal | LDpred1.Inf | 0.312 | 0.346 | -0.033 | 1.48e-05 | 0.285 | 0.316 | -0.031 | 4.54e-02 |
| BMI21 | lassosum.PseudoVal | LDpred1.Inf | 0.284 | 0.346 | -0.062 | 4.58e-07 | 0.229 | 0.316 | -0.087 | 7.00e-04 |
| BMI21 | LDpred2.10FCVal | LDpred1.Inf | 0.377 | 0.346 | 0.031 | 2.22e-07 | 0.336 | 0.316 | 0.020 | 1.07e-01 |
| BMI21 | SBayesR.PseudoVal | LDpred1.Inf | 0.373 | 0.346 | 0.027 | 1.97e-07 | 0.338 | 0.316 | 0.022 | 3.80e-02 |
| BMI21 | LDpred1.MultiPRS | LDpred1.Inf | 0.348 | 0.346 | 0.003 | 8.90e-01 | 0.320 | 0.316 | 0.004 | 4.89e-01 |
| BMI21 | LDpred2.PseudoVal | LDpred1.Inf | 0.378 | 0.346 | 0.032 | 1.85e-10 | 0.348 | 0.316 | 0.033 | 1.68e-03 |
| BMI21 | SBLUP.Inf | LDpred1.Inf | 0.347 | 0.346 | 0.002 | 6.10e-01 | 0.311 | 0.316 | -0.005 | 4.37e-01 |
| BMI21 | pT+clump.MultiPRS | LDpred1.Inf | 0.320 | 0.346 | -0.025 | 2.08e-01 | 0.283 | 0.316 | -0.033 | 4.27e-02 |
| BMI21 | lassosum.MultiPRS | LDpred1.Inf | 0.365 | 0.346 | 0.020 | 3.16e-01 | 0.337 | 0.316 | 0.021 | 1.39e-01 |
| BMI21 | LDpred1.10FCVal | LDpred1.Inf | 0.336 | 0.346 | -0.010 | 2.76e-02 | 0.313 | 0.316 | -0.003 | 7.55e-01 |
| BMI21 | LDpred1.Inf | LDpred1.10FCVal | 0.346 | 0.336 | 0.010 | 2.76e-02 | 0.316 | 0.313 | 0.003 | 7.55e-01 |
| BMI21 | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.357 | 0.336 | 0.021 | 2.44e-03 | 0.320 | 0.313 | 0.007 | 6.07e-01 |
| BMI21 | lassosum.10FCVal | LDpred1.10FCVal | 0.359 | 0.336 | 0.023 | 1.25e-03 | 0.333 | 0.313 | 0.020 | 1.70e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred1.10FCVal | 0.377 | 0.336 | 0.042 | 3.64e-02 | 0.342 | 0.313 | 0.029 | 2.66e-02 |
| BMI21 | pT+clump.10FCVal | LDpred1.10FCVal | 0.312 | 0.336 | -0.023 | 5.67e-03 | 0.285 | 0.313 | -0.028 | 9.40e-02 |
| BMI21 | lassosum.PseudoVal | LDpred1.10FCVal | 0.284 | 0.336 | -0.052 | 4.73e-05 | 0.229 | 0.313 | -0.084 | 1.66e-03 |
| BMI21 | LDpred2.10FCVal | LDpred1.10FCVal | 0.377 | 0.336 | 0.041 | 2.70e-09 | 0.336 | 0.313 | 0.023 | 9.86e-02 |
| BMI21 | All.MultiPRS | LDpred1.10FCVal | 0.378 | 0.336 | 0.043 | 3.11e-02 | 0.346 | 0.313 | 0.033 | 1.86e-02 |
| BMI21 | LDpred1.MultiPRS | LDpred1.10FCVal | 0.348 | 0.336 | 0.013 | 5.29e-01 | 0.320 | 0.313 | 0.007 | 3.90e-01 |
| BMI21 | LDpred2.PseudoVal | LDpred1.10FCVal | 0.378 | 0.336 | 0.042 | 2.54e-12 | 0.348 | 0.313 | 0.035 | 3.12e-03 |
| BMI21 | SBLUP.Inf | LDpred1.10FCVal | 0.347 | 0.336 | 0.011 | 8.16e-03 | 0.311 | 0.313 | -0.002 | 7.98e-01 |
| BMI21 | pT+clump.MultiPRS | LDpred1.10FCVal | 0.320 | 0.336 | -0.015 | 4.44e-01 | 0.283 | 0.313 | -0.030 | 9.06e-02 |
| BMI21 | LDpred2.Inf | LDpred1.10FCVal | 0.350 | 0.336 | 0.015 | 9.52e-04 | 0.319 | 0.313 | 0.006 | 5.03e-01 |
| BMI21 | lassosum.MultiPRS | LDpred1.10FCVal | 0.365 | 0.336 | 0.030 | 1.34e-01 | 0.337 | 0.313 | 0.024 | 1.20e-01 |
| BMI21 | LDpred1.10FCVal | LDpred1.10FCVal | 0.336 | 0.336 | 0.000 | 1.00e+00 | 0.313 | 0.313 | 0.000 | 1.00e+00 |
| BMI21 | SBayesR.PseudoVal | LDpred1.10FCVal | 0.373 | 0.336 | 0.037 | 3.72e-09 | 0.338 | 0.313 | 0.025 | 4.83e-02 |
| BMI21 | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.377 | 0.377 | 0.000 | 1.00e+00 | 0.342 | 0.342 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.10FCVal | LDpred2.MultiPRS | 0.359 | 0.377 | -0.018 | 3.48e-01 | 0.333 | 0.342 | -0.009 | 3.35e-01 |
| BMI21 | All.MultiPRS | LDpred2.MultiPRS | 0.378 | 0.377 | 0.001 | 5.77e-01 | 0.346 | 0.342 | 0.004 | 3.42e-01 |
| BMI21 | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.348 | 0.377 | -0.029 | 6.37e-08 | 0.320 | 0.342 | -0.022 | 3.75e-02 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.357 | 0.377 | -0.020 | 3.03e-01 | 0.320 | 0.342 | -0.022 | 3.38e-02 |
| BMI21 | LDpred1.Inf | LDpred2.MultiPRS | 0.346 | 0.377 | -0.032 | 1.11e-01 | 0.316 | 0.342 | -0.026 | 2.44e-02 |
| BMI21 | LDpred2.Inf | LDpred2.MultiPRS | 0.350 | 0.377 | -0.027 | 1.73e-01 | 0.319 | 0.342 | -0.023 | 2.83e-02 |
| BMI21 | lassosum.MultiPRS | LDpred2.MultiPRS | 0.365 | 0.377 | -0.012 | 5.35e-01 | 0.337 | 0.342 | -0.005 | 6.11e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.MultiPRS | 0.336 | 0.377 | -0.042 | 3.64e-02 | 0.313 | 0.342 | -0.029 | 2.66e-02 |
| BMI21 | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.320 | 0.377 | -0.057 | 3.87e-03 | 0.283 | 0.342 | -0.059 | 1.06e-04 |
| BMI21 | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.373 | 0.377 | -0.005 | 8.09e-01 | 0.338 | 0.342 | -0.004 | 4.91e-01 |
| BMI21 | pT+clump.10FCVal | LDpred2.MultiPRS | 0.312 | 0.377 | -0.065 | 1.19e-03 | 0.285 | 0.342 | -0.058 | 6.51e-04 |
| BMI21 | lassosum.PseudoVal | LDpred2.MultiPRS | 0.284 | 0.377 | -0.094 | 2.46e-06 | 0.229 | 0.342 | -0.114 | 8.46e-08 |
| BMI21 | LDpred2.10FCVal | LDpred2.MultiPRS | 0.377 | 0.377 | 0.000 | 9.93e-01 | 0.336 | 0.342 | -0.006 | 1.84e-02 |
| BMI21 | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.378 | 0.377 | 0.001 | 9.75e-01 | 0.348 | 0.342 | 0.006 | 5.86e-02 |
| BMI21 | SBLUP.Inf | LDpred2.MultiPRS | 0.347 | 0.377 | -0.030 | 1.30e-01 | 0.311 | 0.342 | -0.032 | 5.34e-03 |
| BMI21 | LDpred2.MultiPRS | LDpred2.10FCVal | 0.377 | 0.377 | 0.000 | 9.93e-01 | 0.342 | 0.336 | 0.006 | 1.84e-02 |
| BMI21 | pT+clump.10FCVal | LDpred2.10FCVal | 0.312 | 0.377 | -0.065 | 9.82e-14 | 0.285 | 0.336 | -0.052 | 2.90e-03 |
| BMI21 | lassosum.10FCVal | LDpred2.10FCVal | 0.359 | 0.377 | -0.018 | 1.57e-04 | 0.333 | 0.336 | -0.003 | 7.43e-01 |
| BMI21 | All.MultiPRS | LDpred2.10FCVal | 0.378 | 0.377 | 0.001 | 9.44e-01 | 0.346 | 0.336 | 0.010 | 2.98e-02 |
| BMI21 | LDpred1.MultiPRS | LDpred2.10FCVal | 0.348 | 0.377 | -0.029 | 1.47e-01 | 0.320 | 0.336 | -0.016 | 1.62e-01 |
| BMI21 | LDpred2.PseudoVal | LDpred2.10FCVal | 0.378 | 0.377 | 0.001 | 7.03e-01 | 0.348 | 0.336 | 0.012 | 4.57e-03 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.357 | 0.377 | -0.020 | 1.44e-04 | 0.320 | 0.336 | -0.016 | 1.42e-01 |
| BMI21 | LDpred1.Inf | LDpred2.10FCVal | 0.346 | 0.377 | -0.031 | 2.22e-07 | 0.316 | 0.336 | -0.020 | 1.07e-01 |
| BMI21 | LDpred2.Inf | LDpred2.10FCVal | 0.350 | 0.377 | -0.027 | 1.37e-06 | 0.319 | 0.336 | -0.017 | 1.39e-01 |
| BMI21 | lassosum.MultiPRS | LDpred2.10FCVal | 0.365 | 0.377 | -0.012 | 5.43e-01 | 0.337 | 0.336 | 0.001 | 9.17e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.10FCVal | 0.336 | 0.377 | -0.041 | 2.70e-09 | 0.313 | 0.336 | -0.023 | 9.86e-02 |
| BMI21 | pT+clump.MultiPRS | LDpred2.10FCVal | 0.320 | 0.377 | -0.057 | 4.30e-03 | 0.283 | 0.336 | -0.053 | 5.66e-04 |
| BMI21 | SBayesR.PseudoVal | LDpred2.10FCVal | 0.373 | 0.377 | -0.005 | 1.59e-01 | 0.338 | 0.336 | 0.002 | 7.88e-01 |
| BMI21 | lassosum.PseudoVal | LDpred2.10FCVal | 0.284 | 0.377 | -0.094 | 1.57e-20 | 0.229 | 0.336 | -0.108 | 2.33e-07 |
| BMI21 | LDpred2.10FCVal | LDpred2.10FCVal | 0.377 | 0.377 | 0.000 | 1.00e+00 | 0.336 | 0.336 | 0.000 | 1.00e+00 |
| BMI21 | SBLUP.Inf | LDpred2.10FCVal | 0.347 | 0.377 | -0.030 | 9.07e-07 | 0.311 | 0.336 | -0.026 | 3.72e-02 |
| BMI21 | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.377 | 0.378 | -0.001 | 9.75e-01 | 0.342 | 0.348 | -0.006 | 5.86e-02 |
| BMI21 | lassosum.10FCVal | LDpred2.PseudoVal | 0.359 | 0.378 | -0.019 | 3.52e-05 | 0.333 | 0.348 | -0.016 | 9.89e-02 |
| BMI21 | All.MultiPRS | LDpred2.PseudoVal | 0.378 | 0.378 | 0.001 | 9.77e-01 | 0.346 | 0.348 | -0.002 | 7.10e-01 |
| BMI21 | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.348 | 0.378 | -0.029 | 1.36e-01 | 0.320 | 0.348 | -0.029 | 2.62e-03 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.357 | 0.378 | -0.021 | 4.87e-05 | 0.320 | 0.348 | -0.028 | 6.77e-03 |
| BMI21 | LDpred1.Inf | LDpred2.PseudoVal | 0.346 | 0.378 | -0.032 | 1.85e-10 | 0.316 | 0.348 | -0.033 | 1.68e-03 |
| BMI21 | LDpred2.Inf | LDpred2.PseudoVal | 0.350 | 0.378 | -0.028 | 2.23e-10 | 0.319 | 0.348 | -0.029 | 9.00e-04 |
| BMI21 | lassosum.MultiPRS | LDpred2.PseudoVal | 0.365 | 0.378 | -0.013 | 5.16e-01 | 0.337 | 0.348 | -0.011 | 2.69e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.PseudoVal | 0.336 | 0.378 | -0.042 | 2.54e-12 | 0.313 | 0.348 | -0.035 | 3.12e-03 |
| BMI21 | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.320 | 0.378 | -0.058 | 3.77e-03 | 0.283 | 0.348 | -0.065 | 2.59e-05 |
| BMI21 | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.373 | 0.378 | -0.005 | 9.83e-02 | 0.338 | 0.348 | -0.011 | 1.10e-01 |
| BMI21 | pT+clump.10FCVal | LDpred2.PseudoVal | 0.312 | 0.378 | -0.065 | 1.40e-15 | 0.285 | 0.348 | -0.064 | 9.00e-05 |
| BMI21 | lassosum.PseudoVal | LDpred2.PseudoVal | 0.284 | 0.378 | -0.094 | 9.27e-18 | 0.229 | 0.348 | -0.120 | 1.60e-07 |
| BMI21 | LDpred2.10FCVal | LDpred2.PseudoVal | 0.377 | 0.378 | -0.001 | 7.03e-01 | 0.336 | 0.348 | -0.012 | 4.57e-03 |
| BMI21 | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.378 | 0.378 | 0.000 | 1.00e+00 | 0.348 | 0.348 | 0.000 | 1.00e+00 |
| BMI21 | SBLUP.Inf | LDpred2.PseudoVal | 0.347 | 0.378 | -0.031 | 6.87e-10 | 0.311 | 0.348 | -0.038 | 1.29e-04 |
| BMI21 | DBSLMM.PseudoVal | LDpred2.Inf | 0.357 | 0.350 | 0.007 | 2.63e-01 | 0.320 | 0.319 | 0.001 | 9.28e-01 |
| BMI21 | LDpred2.MultiPRS | LDpred2.Inf | 0.377 | 0.350 | 0.027 | 1.73e-01 | 0.342 | 0.319 | 0.023 | 2.83e-02 |
| BMI21 | pT+clump.10FCVal | LDpred2.Inf | 0.312 | 0.350 | -0.038 | 1.76e-06 | 0.285 | 0.319 | -0.034 | 3.17e-02 |
| BMI21 | lassosum.PseudoVal | LDpred2.Inf | 0.284 | 0.350 | -0.067 | 8.23e-08 | 0.229 | 0.319 | -0.090 | 5.64e-04 |
| BMI21 | lassosum.10FCVal | LDpred2.Inf | 0.359 | 0.350 | 0.009 | 1.76e-01 | 0.333 | 0.319 | 0.014 | 2.89e-01 |
| BMI21 | All.MultiPRS | LDpred2.Inf | 0.378 | 0.350 | 0.028 | 1.55e-01 | 0.346 | 0.319 | 0.027 | 2.38e-02 |
| BMI21 | LDpred1.MultiPRS | LDpred2.Inf | 0.348 | 0.350 | -0.002 | 9.24e-01 | 0.320 | 0.319 | 0.001 | 9.01e-01 |
| BMI21 | LDpred2.PseudoVal | LDpred2.Inf | 0.378 | 0.350 | 0.028 | 2.23e-10 | 0.348 | 0.319 | 0.029 | 9.00e-04 |
| BMI21 | SBLUP.Inf | LDpred2.Inf | 0.347 | 0.350 | -0.003 | 2.58e-01 | 0.311 | 0.319 | -0.008 | 1.29e-01 |
| BMI21 | LDpred1.Inf | LDpred2.Inf | 0.346 | 0.350 | -0.005 | 8.99e-02 | 0.316 | 0.319 | -0.003 | 5.77e-01 |
| BMI21 | LDpred2.Inf | LDpred2.Inf | 0.350 | 0.350 | 0.000 | 1.00e+00 | 0.319 | 0.319 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.MultiPRS | LDpred2.Inf | 0.365 | 0.350 | 0.015 | 4.45e-01 | 0.337 | 0.319 | 0.018 | 2.08e-01 |
| BMI21 | LDpred1.10FCVal | LDpred2.Inf | 0.336 | 0.350 | -0.015 | 9.52e-04 | 0.313 | 0.319 | -0.006 | 5.03e-01 |
| BMI21 | pT+clump.MultiPRS | LDpred2.Inf | 0.320 | 0.350 | -0.030 | 1.35e-01 | 0.283 | 0.319 | -0.036 | 3.27e-02 |
| BMI21 | SBayesR.PseudoVal | LDpred2.Inf | 0.373 | 0.350 | 0.022 | 1.30e-05 | 0.338 | 0.319 | 0.019 | 7.64e-02 |
| BMI21 | LDpred2.10FCVal | LDpred2.Inf | 0.377 | 0.350 | 0.027 | 1.37e-06 | 0.336 | 0.319 | 0.017 | 1.39e-01 |
| BMI21 | All.MultiPRS | pT+clump.MultiPRS | 0.378 | 0.320 | 0.058 | 3.22e-03 | 0.346 | 0.283 | 0.063 | 6.74e-05 |
| BMI21 | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.348 | 0.320 | 0.028 | 1.60e-01 | 0.320 | 0.283 | 0.037 | 2.24e-02 |
| BMI21 | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.378 | 0.320 | 0.058 | 3.77e-03 | 0.348 | 0.283 | 0.065 | 2.59e-05 |
| BMI21 | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.357 | 0.320 | 0.037 | 6.66e-02 | 0.320 | 0.283 | 0.037 | 9.66e-03 |
| BMI21 | LDpred1.Inf | pT+clump.MultiPRS | 0.346 | 0.320 | 0.025 | 2.08e-01 | 0.316 | 0.283 | 0.033 | 4.27e-02 |
| BMI21 | LDpred2.Inf | pT+clump.MultiPRS | 0.350 | 0.320 | 0.030 | 1.35e-01 | 0.319 | 0.283 | 0.036 | 3.27e-02 |
| BMI21 | lassosum.MultiPRS | pT+clump.MultiPRS | 0.365 | 0.320 | 0.045 | 2.41e-02 | 0.337 | 0.283 | 0.054 | 6.29e-04 |
| BMI21 | LDpred1.10FCVal | pT+clump.MultiPRS | 0.336 | 0.320 | 0.015 | 4.44e-01 | 0.313 | 0.283 | 0.030 | 9.06e-02 |
| BMI21 | lassosum.10FCVal | pT+clump.MultiPRS | 0.359 | 0.320 | 0.039 | 5.38e-02 | 0.333 | 0.283 | 0.050 | 7.63e-04 |
| BMI21 | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.377 | 0.320 | 0.057 | 3.87e-03 | 0.342 | 0.283 | 0.059 | 1.06e-04 |
| BMI21 | pT+clump.10FCVal | pT+clump.MultiPRS | 0.312 | 0.320 | -0.008 | 7.00e-01 | 0.285 | 0.283 | 0.002 | 8.48e-01 |
| BMI21 | lassosum.PseudoVal | pT+clump.MultiPRS | 0.284 | 0.320 | -0.037 | 7.23e-02 | 0.229 | 0.283 | -0.054 | 1.19e-02 |
| BMI21 | LDpred2.10FCVal | pT+clump.MultiPRS | 0.377 | 0.320 | 0.057 | 4.30e-03 | 0.336 | 0.283 | 0.053 | 5.66e-04 |
| BMI21 | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.373 | 0.320 | 0.052 | 8.71e-03 | 0.338 | 0.283 | 0.055 | 3.75e-05 |
| BMI21 | SBLUP.Inf | pT+clump.MultiPRS | 0.347 | 0.320 | 0.027 | 1.80e-01 | 0.311 | 0.283 | 0.028 | 9.34e-02 |
| BMI21 | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.320 | 0.320 | 0.000 | 1.00e+00 | 0.283 | 0.283 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.10FCVal | pT+clump.10FCVal | 0.359 | 0.312 | 0.046 | 3.12e-09 | 0.333 | 0.285 | 0.048 | 2.17e-03 |
| BMI21 | All.MultiPRS | pT+clump.10FCVal | 0.378 | 0.312 | 0.066 | 9.60e-04 | 0.346 | 0.285 | 0.062 | 4.35e-04 |
| BMI21 | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.357 | 0.312 | 0.045 | 3.27e-08 | 0.320 | 0.285 | 0.036 | 2.72e-02 |
| BMI21 | LDpred1.Inf | pT+clump.10FCVal | 0.346 | 0.312 | 0.033 | 1.48e-05 | 0.316 | 0.285 | 0.031 | 4.54e-02 |
| BMI21 | LDpred2.Inf | pT+clump.10FCVal | 0.350 | 0.312 | 0.038 | 1.76e-06 | 0.319 | 0.285 | 0.034 | 3.17e-02 |
| BMI21 | lassosum.MultiPRS | pT+clump.10FCVal | 0.365 | 0.312 | 0.053 | 7.53e-03 | 0.337 | 0.285 | 0.053 | 2.36e-03 |
| BMI21 | LDpred1.10FCVal | pT+clump.10FCVal | 0.336 | 0.312 | 0.023 | 5.67e-03 | 0.313 | 0.285 | 0.028 | 9.40e-02 |
| BMI21 | pT+clump.MultiPRS | pT+clump.10FCVal | 0.320 | 0.312 | 0.008 | 7.00e-01 | 0.283 | 0.285 | -0.002 | 8.48e-01 |
| BMI21 | LDpred2.MultiPRS | pT+clump.10FCVal | 0.377 | 0.312 | 0.065 | 1.19e-03 | 0.342 | 0.285 | 0.058 | 6.51e-04 |
| BMI21 | pT+clump.10FCVal | pT+clump.10FCVal | 0.312 | 0.312 | 0.000 | 1.00e+00 | 0.285 | 0.285 | 0.000 | 1.00e+00 |
| BMI21 | lassosum.PseudoVal | pT+clump.10FCVal | 0.284 | 0.312 | -0.029 | 2.47e-02 | 0.229 | 0.285 | -0.056 | 3.34e-02 |
| BMI21 | LDpred2.10FCVal | pT+clump.10FCVal | 0.377 | 0.312 | 0.065 | 9.82e-14 | 0.336 | 0.285 | 0.052 | 2.90e-03 |
| BMI21 | SBayesR.PseudoVal | pT+clump.10FCVal | 0.373 | 0.312 | 0.060 | 8.61e-16 | 0.338 | 0.285 | 0.053 | 3.99e-04 |
| BMI21 | LDpred1.MultiPRS | pT+clump.10FCVal | 0.348 | 0.312 | 0.036 | 7.54e-02 | 0.320 | 0.285 | 0.035 | 2.55e-02 |
| BMI21 | LDpred2.PseudoVal | pT+clump.10FCVal | 0.378 | 0.312 | 0.065 | 1.40e-15 | 0.348 | 0.285 | 0.064 | 9.00e-05 |
| BMI21 | SBLUP.Inf | pT+clump.10FCVal | 0.347 | 0.312 | 0.035 | 5.52e-06 | 0.311 | 0.285 | 0.026 | 9.79e-02 |
| BMI21 | LDpred1.Inf | SBayesR.PseudoVal | 0.346 | 0.373 | -0.027 | 1.97e-07 | 0.316 | 0.338 | -0.022 | 3.80e-02 |
| BMI21 | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.357 | 0.373 | -0.016 | 3.39e-04 | 0.320 | 0.338 | -0.018 | 4.58e-02 |
| BMI21 | lassosum.10FCVal | SBayesR.PseudoVal | 0.359 | 0.373 | -0.014 | 3.06e-03 | 0.333 | 0.338 | -0.005 | 6.01e-01 |
| BMI21 | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.377 | 0.373 | 0.005 | 8.09e-01 | 0.342 | 0.338 | 0.004 | 4.91e-01 |
| BMI21 | pT+clump.10FCVal | SBayesR.PseudoVal | 0.312 | 0.373 | -0.060 | 8.61e-16 | 0.285 | 0.338 | -0.053 | 3.99e-04 |
| BMI21 | lassosum.PseudoVal | SBayesR.PseudoVal | 0.284 | 0.373 | -0.089 | 3.73e-19 | 0.229 | 0.338 | -0.109 | 1.41e-07 |
| BMI21 | LDpred2.10FCVal | SBayesR.PseudoVal | 0.377 | 0.373 | 0.005 | 1.59e-01 | 0.336 | 0.338 | -0.002 | 7.88e-01 |
| BMI21 | All.MultiPRS | SBayesR.PseudoVal | 0.378 | 0.373 | 0.006 | 7.63e-01 | 0.346 | 0.338 | 0.009 | 2.37e-01 |
| BMI21 | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.348 | 0.373 | -0.024 | 2.22e-01 | 0.320 | 0.338 | -0.018 | 7.63e-02 |
| BMI21 | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.378 | 0.373 | 0.005 | 9.83e-02 | 0.348 | 0.338 | 0.011 | 1.10e-01 |
| BMI21 | SBLUP.Inf | SBayesR.PseudoVal | 0.347 | 0.373 | -0.025 | 1.30e-06 | 0.311 | 0.338 | -0.027 | 1.03e-02 |
| BMI21 | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.320 | 0.373 | -0.052 | 8.71e-03 | 0.283 | 0.338 | -0.055 | 3.75e-05 |
| BMI21 | LDpred2.Inf | SBayesR.PseudoVal | 0.350 | 0.373 | -0.022 | 1.30e-05 | 0.319 | 0.338 | -0.019 | 7.64e-02 |
| BMI21 | lassosum.MultiPRS | SBayesR.PseudoVal | 0.365 | 0.373 | -0.007 | 7.08e-01 | 0.337 | 0.338 | -0.001 | 9.43e-01 |
| BMI21 | LDpred1.10FCVal | SBayesR.PseudoVal | 0.336 | 0.373 | -0.037 | 3.72e-09 | 0.313 | 0.338 | -0.025 | 4.83e-02 |
| BMI21 | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.373 | 0.373 | 0.000 | 1.00e+00 | 0.338 | 0.338 | 0.000 | 1.00e+00 |
| BMI21 | All.MultiPRS | SBLUP.Inf | 0.378 | 0.347 | 0.031 | 1.15e-01 | 0.346 | 0.311 | 0.036 | 4.62e-03 |
| BMI21 | LDpred1.Inf | SBLUP.Inf | 0.346 | 0.347 | -0.002 | 6.10e-01 | 0.316 | 0.311 | 0.005 | 4.37e-01 |
| BMI21 | LDpred2.Inf | SBLUP.Inf | 0.350 | 0.347 | 0.003 | 2.58e-01 | 0.319 | 0.311 | 0.008 | 1.29e-01 |
| BMI21 | DBSLMM.PseudoVal | SBLUP.Inf | 0.357 | 0.347 | 0.010 | 1.03e-01 | 0.320 | 0.311 | 0.010 | 4.23e-01 |
| BMI21 | lassosum.10FCVal | SBLUP.Inf | 0.359 | 0.347 | 0.012 | 6.55e-02 | 0.333 | 0.311 | 0.022 | 7.99e-02 |
| BMI21 | LDpred2.MultiPRS | SBLUP.Inf | 0.377 | 0.347 | 0.030 | 1.30e-01 | 0.342 | 0.311 | 0.032 | 5.34e-03 |
| BMI21 | pT+clump.10FCVal | SBLUP.Inf | 0.312 | 0.347 | -0.035 | 5.52e-06 | 0.285 | 0.311 | -0.026 | 9.79e-02 |
| BMI21 | lassosum.PseudoVal | SBLUP.Inf | 0.284 | 0.347 | -0.064 | 3.01e-07 | 0.229 | 0.311 | -0.082 | 1.51e-03 |
| BMI21 | LDpred2.10FCVal | SBLUP.Inf | 0.377 | 0.347 | 0.030 | 9.07e-07 | 0.336 | 0.311 | 0.026 | 3.72e-02 |
| BMI21 | SBayesR.PseudoVal | SBLUP.Inf | 0.373 | 0.347 | 0.025 | 1.30e-06 | 0.338 | 0.311 | 0.027 | 1.03e-02 |
| BMI21 | LDpred1.MultiPRS | SBLUP.Inf | 0.348 | 0.347 | 0.001 | 9.54e-01 | 0.320 | 0.311 | 0.009 | 2.02e-01 |
| BMI21 | LDpred2.PseudoVal | SBLUP.Inf | 0.378 | 0.347 | 0.031 | 6.87e-10 | 0.348 | 0.311 | 0.038 | 1.29e-04 |
| BMI21 | SBLUP.Inf | SBLUP.Inf | 0.347 | 0.347 | 0.000 | 1.00e+00 | 0.311 | 0.311 | 0.000 | 1.00e+00 |
| BMI21 | pT+clump.MultiPRS | SBLUP.Inf | 0.320 | 0.347 | -0.027 | 1.80e-01 | 0.283 | 0.311 | -0.028 | 9.34e-02 |
| BMI21 | lassosum.MultiPRS | SBLUP.Inf | 0.365 | 0.347 | 0.018 | 3.57e-01 | 0.337 | 0.311 | 0.027 | 6.04e-02 |
| BMI21 | LDpred1.10FCVal | SBLUP.Inf | 0.336 | 0.347 | -0.011 | 8.16e-03 | 0.313 | 0.311 | 0.002 | 7.98e-01 |
| GCSE | lassosum.10FCVal | All.MultiPRS | 0.410 | 0.423 | -0.013 | 4.07e-01 | 0.355 | 0.355 | 0.000 | 9.84e-01 |
| GCSE | All.MultiPRS | All.MultiPRS | 0.423 | 0.423 | 0.000 | 1.00e+00 | 0.355 | 0.355 | 0.000 | 1.00e+00 |
| GCSE | DBSLMM.PseudoVal | All.MultiPRS | 0.402 | 0.423 | -0.021 | 1.86e-01 | 0.339 | 0.355 | -0.017 | 2.25e-02 |
| GCSE | LDpred1.Inf | All.MultiPRS | 0.409 | 0.423 | -0.014 | 3.75e-01 | 0.337 | 0.355 | -0.019 | 5.70e-03 |
| GCSE | LDpred2.Inf | All.MultiPRS | 0.413 | 0.423 | -0.010 | 5.31e-01 | 0.343 | 0.355 | -0.013 | 2.80e-02 |
| GCSE | lassosum.MultiPRS | All.MultiPRS | 0.412 | 0.423 | -0.012 | 4.95e-05 | 0.357 | 0.355 | 0.002 | 7.55e-01 |
| GCSE | LDpred1.10FCVal | All.MultiPRS | 0.405 | 0.423 | -0.018 | 2.58e-01 | 0.337 | 0.355 | -0.018 | 2.38e-02 |
| GCSE | pT+clump.MultiPRS | All.MultiPRS | 0.382 | 0.423 | -0.042 | 9.82e-03 | 0.330 | 0.355 | -0.025 | 1.78e-02 |
| GCSE | LDpred2.MultiPRS | All.MultiPRS | 0.423 | 0.423 | 0.000 | 8.75e-01 | 0.356 | 0.355 | 0.001 | 5.67e-01 |
| GCSE | pT+clump.10FCVal | All.MultiPRS | 0.374 | 0.423 | -0.049 | 2.08e-03 | 0.315 | 0.355 | -0.040 | 1.27e-03 |
| GCSE | lassosum.PseudoVal | All.MultiPRS | 0.316 | 0.423 | -0.108 | 5.11e-11 | 0.273 | 0.355 | -0.083 | 2.96e-06 |
| GCSE | LDpred2.10FCVal | All.MultiPRS | 0.423 | 0.423 | -0.001 | 9.58e-01 | 0.358 | 0.355 | 0.003 | 3.42e-01 |
| GCSE | SBayesR.PseudoVal | All.MultiPRS | 0.420 | 0.423 | -0.004 | 8.24e-01 | 0.350 | 0.355 | -0.005 | 2.23e-01 |
| GCSE | LDpred1.MultiPRS | All.MultiPRS | 0.414 | 0.423 | -0.010 | 5.39e-01 | 0.341 | 0.355 | -0.014 | 1.39e-02 |
| GCSE | LDpred2.PseudoVal | All.MultiPRS | 0.422 | 0.423 | -0.001 | 9.34e-01 | 0.358 | 0.355 | 0.003 | 3.53e-01 |
| GCSE | SBLUP.Inf | All.MultiPRS | 0.412 | 0.423 | -0.012 | 4.63e-01 | 0.344 | 0.355 | -0.011 | 7.74e-02 |
| GCSE | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.423 | 0.402 | 0.021 | 1.84e-01 | 0.356 | 0.339 | 0.018 | 1.77e-02 |
| GCSE | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.374 | 0.402 | -0.028 | 2.01e-05 | 0.315 | 0.339 | -0.024 | 8.81e-02 |
| GCSE | lassosum.10FCVal | DBSLMM.PseudoVal | 0.410 | 0.402 | 0.008 | 7.96e-02 | 0.355 | 0.339 | 0.016 | 8.16e-02 |
| GCSE | All.MultiPRS | DBSLMM.PseudoVal | 0.423 | 0.402 | 0.021 | 1.86e-01 | 0.355 | 0.339 | 0.017 | 2.25e-02 |
| GCSE | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.414 | 0.402 | 0.011 | 4.92e-01 | 0.341 | 0.339 | 0.002 | 7.91e-01 |
| GCSE | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.422 | 0.402 | 0.020 | 3.13e-07 | 0.358 | 0.339 | 0.019 | 1.67e-02 |
| GCSE | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.402 | 0.402 | 0.000 | 1.00e+00 | 0.339 | 0.339 | 0.000 | 1.00e+00 |
| GCSE | LDpred1.Inf | DBSLMM.PseudoVal | 0.409 | 0.402 | 0.007 | 1.11e-01 | 0.337 | 0.339 | -0.002 | 8.19e-01 |
| GCSE | LDpred2.Inf | DBSLMM.PseudoVal | 0.413 | 0.402 | 0.011 | 9.10e-03 | 0.343 | 0.339 | 0.004 | 6.60e-01 |
| GCSE | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.412 | 0.402 | 0.009 | 5.61e-01 | 0.357 | 0.339 | 0.018 | 3.52e-02 |
| GCSE | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.405 | 0.402 | 0.003 | 5.35e-01 | 0.337 | 0.339 | -0.002 | 8.64e-01 |
| GCSE | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.382 | 0.402 | -0.021 | 2.04e-01 | 0.330 | 0.339 | -0.009 | 4.34e-01 |
| GCSE | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.420 | 0.402 | 0.017 | 1.24e-06 | 0.350 | 0.339 | 0.011 | 1.37e-01 |
| GCSE | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.316 | 0.402 | -0.087 | 2.65e-25 | 0.273 | 0.339 | -0.066 | 1.64e-04 |
| GCSE | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.423 | 0.402 | 0.020 | 2.79e-07 | 0.358 | 0.339 | 0.020 | 1.83e-02 |
| GCSE | SBLUP.Inf | DBSLMM.PseudoVal | 0.412 | 0.402 | 0.009 | 2.45e-02 | 0.344 | 0.339 | 0.006 | 5.25e-01 |
| GCSE | All.MultiPRS | lassosum.MultiPRS | 0.423 | 0.412 | 0.012 | 4.95e-05 | 0.355 | 0.357 | -0.002 | 7.55e-01 |
| GCSE | LDpred1.MultiPRS | lassosum.MultiPRS | 0.414 | 0.412 | 0.002 | 9.04e-01 | 0.341 | 0.357 | -0.016 | 5.37e-02 |
| GCSE | LDpred1.Inf | lassosum.MultiPRS | 0.409 | 0.412 | -0.002 | 8.81e-01 | 0.337 | 0.357 | -0.021 | 1.33e-02 |
| GCSE | LDpred2.Inf | lassosum.MultiPRS | 0.413 | 0.412 | 0.002 | 9.11e-01 | 0.343 | 0.357 | -0.014 | 7.59e-02 |
| GCSE | lassosum.MultiPRS | lassosum.MultiPRS | 0.412 | 0.412 | 0.000 | 1.00e+00 | 0.357 | 0.357 | 0.000 | 1.00e+00 |
| GCSE | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.402 | 0.412 | -0.009 | 5.61e-01 | 0.339 | 0.357 | -0.018 | 3.52e-02 |
| GCSE | lassosum.10FCVal | lassosum.MultiPRS | 0.410 | 0.412 | -0.001 | 9.27e-01 | 0.355 | 0.357 | -0.002 | 3.85e-01 |
| GCSE | LDpred2.MultiPRS | lassosum.MultiPRS | 0.423 | 0.412 | 0.012 | 4.53e-05 | 0.356 | 0.357 | -0.001 | 8.81e-01 |
| GCSE | pT+clump.10FCVal | lassosum.MultiPRS | 0.374 | 0.412 | -0.038 | 1.93e-02 | 0.315 | 0.357 | -0.042 | 1.21e-03 |
| GCSE | lassosum.PseudoVal | lassosum.MultiPRS | 0.316 | 0.412 | -0.096 | 5.77e-09 | 0.273 | 0.357 | -0.085 | 9.13e-07 |
| GCSE | LDpred2.10FCVal | lassosum.MultiPRS | 0.423 | 0.412 | 0.011 | 4.92e-01 | 0.358 | 0.357 | 0.001 | 8.41e-01 |
| GCSE | SBayesR.PseudoVal | lassosum.MultiPRS | 0.420 | 0.412 | 0.008 | 6.05e-01 | 0.350 | 0.357 | -0.007 | 2.72e-01 |
| GCSE | LDpred2.PseudoVal | lassosum.MultiPRS | 0.422 | 0.412 | 0.010 | 5.12e-01 | 0.358 | 0.357 | 0.001 | 8.92e-01 |
| GCSE | SBLUP.Inf | lassosum.MultiPRS | 0.412 | 0.412 | 0.000 | 9.97e-01 | 0.344 | 0.357 | -0.013 | 1.08e-01 |
| GCSE | pT+clump.MultiPRS | lassosum.MultiPRS | 0.382 | 0.412 | -0.030 | 6.39e-02 | 0.330 | 0.357 | -0.027 | 1.72e-02 |
| GCSE | LDpred1.10FCVal | lassosum.MultiPRS | 0.405 | 0.412 | -0.006 | 6.95e-01 | 0.337 | 0.357 | -0.020 | 3.66e-02 |
| GCSE | All.MultiPRS | lassosum.10FCVal | 0.423 | 0.410 | 0.013 | 4.07e-01 | 0.355 | 0.355 | 0.000 | 9.84e-01 |
| GCSE | LDpred1.MultiPRS | lassosum.10FCVal | 0.414 | 0.410 | 0.003 | 8.35e-01 | 0.341 | 0.355 | -0.014 | 1.17e-01 |
| GCSE | LDpred2.PseudoVal | lassosum.10FCVal | 0.422 | 0.410 | 0.012 | 2.61e-04 | 0.358 | 0.355 | 0.003 | 6.71e-01 |
| GCSE | DBSLMM.PseudoVal | lassosum.10FCVal | 0.402 | 0.410 | -0.008 | 7.96e-02 | 0.339 | 0.355 | -0.016 | 8.16e-02 |
| GCSE | LDpred1.Inf | lassosum.10FCVal | 0.409 | 0.410 | -0.001 | 8.28e-01 | 0.337 | 0.355 | -0.019 | 3.95e-02 |
| GCSE | LDpred2.Inf | lassosum.10FCVal | 0.413 | 0.410 | 0.003 | 4.34e-01 | 0.343 | 0.355 | -0.012 | 1.54e-01 |
| GCSE | lassosum.MultiPRS | lassosum.10FCVal | 0.412 | 0.410 | 0.001 | 9.27e-01 | 0.357 | 0.355 | 0.002 | 3.85e-01 |
| GCSE | LDpred1.10FCVal | lassosum.10FCVal | 0.405 | 0.410 | -0.005 | 3.08e-01 | 0.337 | 0.355 | -0.018 | 7.39e-02 |
| GCSE | lassosum.10FCVal | lassosum.10FCVal | 0.410 | 0.410 | 0.000 | 1.00e+00 | 0.355 | 0.355 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.MultiPRS | lassosum.10FCVal | 0.423 | 0.410 | 0.013 | 4.02e-01 | 0.356 | 0.355 | 0.001 | 8.68e-01 |
| GCSE | pT+clump.10FCVal | lassosum.10FCVal | 0.374 | 0.410 | -0.036 | 1.98e-09 | 0.315 | 0.355 | -0.040 | 1.58e-03 |
| GCSE | lassosum.PseudoVal | lassosum.10FCVal | 0.316 | 0.410 | -0.094 | 7.29e-28 | 0.273 | 0.355 | -0.082 | 7.10e-06 |
| GCSE | LDpred2.10FCVal | lassosum.10FCVal | 0.423 | 0.410 | 0.012 | 6.62e-05 | 0.358 | 0.355 | 0.003 | 6.23e-01 |
| GCSE | SBayesR.PseudoVal | lassosum.10FCVal | 0.420 | 0.410 | 0.010 | 5.89e-03 | 0.350 | 0.355 | -0.005 | 4.88e-01 |
| GCSE | SBLUP.Inf | lassosum.10FCVal | 0.412 | 0.410 | 0.002 | 7.09e-01 | 0.344 | 0.355 | -0.011 | 2.09e-01 |
| GCSE | pT+clump.MultiPRS | lassosum.10FCVal | 0.382 | 0.410 | -0.029 | 8.07e-02 | 0.330 | 0.355 | -0.025 | 2.75e-02 |
| GCSE | All.MultiPRS | lassosum.PseudoVal | 0.423 | 0.316 | 0.108 | 5.11e-11 | 0.355 | 0.273 | 0.083 | 2.96e-06 |
| GCSE | LDpred1.MultiPRS | lassosum.PseudoVal | 0.414 | 0.316 | 0.098 | 5.43e-09 | 0.341 | 0.273 | 0.069 | 5.88e-04 |
| GCSE | LDpred1.Inf | lassosum.PseudoVal | 0.409 | 0.316 | 0.093 | 8.82e-23 | 0.337 | 0.273 | 0.064 | 1.37e-03 |
| GCSE | LDpred2.Inf | lassosum.PseudoVal | 0.413 | 0.316 | 0.098 | 8.07e-24 | 0.343 | 0.273 | 0.070 | 5.69e-04 |
| GCSE | lassosum.MultiPRS | lassosum.PseudoVal | 0.412 | 0.316 | 0.096 | 5.77e-09 | 0.357 | 0.273 | 0.085 | 9.13e-07 |
| GCSE | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.402 | 0.316 | 0.087 | 2.65e-25 | 0.339 | 0.273 | 0.066 | 1.64e-04 |
| GCSE | lassosum.10FCVal | lassosum.PseudoVal | 0.410 | 0.316 | 0.094 | 7.29e-28 | 0.355 | 0.273 | 0.082 | 7.10e-06 |
| GCSE | LDpred2.MultiPRS | lassosum.PseudoVal | 0.423 | 0.316 | 0.108 | 4.73e-11 | 0.356 | 0.273 | 0.084 | 1.96e-06 |
| GCSE | pT+clump.10FCVal | lassosum.PseudoVal | 0.374 | 0.316 | 0.058 | 6.52e-08 | 0.315 | 0.273 | 0.043 | 6.49e-02 |
| GCSE | lassosum.PseudoVal | lassosum.PseudoVal | 0.316 | 0.316 | 0.000 | 1.00e+00 | 0.273 | 0.273 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.10FCVal | lassosum.PseudoVal | 0.423 | 0.316 | 0.107 | 8.33e-36 | 0.358 | 0.273 | 0.086 | 1.60e-06 |
| GCSE | SBayesR.PseudoVal | lassosum.PseudoVal | 0.420 | 0.316 | 0.104 | 3.50e-36 | 0.350 | 0.273 | 0.077 | 1.01e-05 |
| GCSE | LDpred2.PseudoVal | lassosum.PseudoVal | 0.422 | 0.316 | 0.106 | 5.29e-33 | 0.358 | 0.273 | 0.085 | 3.87e-06 |
| GCSE | SBLUP.Inf | lassosum.PseudoVal | 0.412 | 0.316 | 0.096 | 1.06e-23 | 0.344 | 0.273 | 0.072 | 3.51e-04 |
| GCSE | pT+clump.MultiPRS | lassosum.PseudoVal | 0.382 | 0.316 | 0.066 | 9.99e-05 | 0.330 | 0.273 | 0.057 | 2.92e-03 |
| GCSE | LDpred1.10FCVal | lassosum.PseudoVal | 0.405 | 0.316 | 0.090 | 9.91e-20 | 0.337 | 0.273 | 0.064 | 1.90e-03 |
| GCSE | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.402 | 0.414 | -0.011 | 4.92e-01 | 0.339 | 0.341 | -0.002 | 7.91e-01 |
| GCSE | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.423 | 0.414 | 0.010 | 5.33e-01 | 0.356 | 0.341 | 0.015 | 2.09e-02 |
| GCSE | pT+clump.10FCVal | LDpred1.MultiPRS | 0.374 | 0.414 | -0.040 | 1.59e-02 | 0.315 | 0.341 | -0.026 | 4.58e-02 |
| GCSE | lassosum.PseudoVal | LDpred1.MultiPRS | 0.316 | 0.414 | -0.098 | 5.43e-09 | 0.273 | 0.341 | -0.069 | 5.88e-04 |
| GCSE | lassosum.10FCVal | LDpred1.MultiPRS | 0.410 | 0.414 | -0.003 | 8.35e-01 | 0.355 | 0.341 | 0.014 | 1.17e-01 |
| GCSE | All.MultiPRS | LDpred1.MultiPRS | 0.423 | 0.414 | 0.010 | 5.39e-01 | 0.355 | 0.341 | 0.014 | 1.39e-02 |
| GCSE | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.414 | 0.414 | 0.000 | 1.00e+00 | 0.341 | 0.341 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.422 | 0.414 | 0.008 | 6.00e-01 | 0.358 | 0.341 | 0.017 | 1.19e-02 |
| GCSE | SBLUP.Inf | LDpred1.MultiPRS | 0.412 | 0.414 | -0.002 | 9.08e-01 | 0.344 | 0.341 | 0.003 | 5.03e-01 |
| GCSE | LDpred1.Inf | LDpred1.MultiPRS | 0.409 | 0.414 | -0.004 | 7.90e-01 | 0.337 | 0.341 | -0.005 | 3.13e-01 |
| GCSE | LDpred2.Inf | LDpred1.MultiPRS | 0.413 | 0.414 | 0.000 | 9.92e-01 | 0.343 | 0.341 | 0.002 | 7.49e-01 |
| GCSE | lassosum.MultiPRS | LDpred1.MultiPRS | 0.412 | 0.414 | -0.002 | 9.04e-01 | 0.357 | 0.341 | 0.016 | 5.37e-02 |
| GCSE | LDpred1.10FCVal | LDpred1.MultiPRS | 0.405 | 0.414 | -0.008 | 6.15e-01 | 0.337 | 0.341 | -0.004 | 4.62e-01 |
| GCSE | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.382 | 0.414 | -0.032 | 4.81e-02 | 0.330 | 0.341 | -0.011 | 3.57e-01 |
| GCSE | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.420 | 0.414 | 0.006 | 6.98e-01 | 0.350 | 0.341 | 0.009 | 2.36e-01 |
| GCSE | LDpred2.10FCVal | LDpred1.MultiPRS | 0.423 | 0.414 | 0.009 | 5.80e-01 | 0.358 | 0.341 | 0.017 | 1.27e-02 |
| GCSE | All.MultiPRS | LDpred1.Inf | 0.423 | 0.409 | 0.014 | 3.75e-01 | 0.355 | 0.337 | 0.019 | 5.70e-03 |
| GCSE | LDpred1.Inf | LDpred1.Inf | 0.409 | 0.409 | 0.000 | 1.00e+00 | 0.337 | 0.337 | 0.000 | 1.00e+00 |
| GCSE | LDpred2.Inf | LDpred1.Inf | 0.413 | 0.409 | 0.004 | 3.42e-02 | 0.343 | 0.337 | 0.006 | 1.39e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred1.Inf | 0.402 | 0.409 | -0.007 | 1.11e-01 | 0.339 | 0.337 | 0.002 | 8.19e-01 |
| GCSE | lassosum.10FCVal | LDpred1.Inf | 0.410 | 0.409 | 0.001 | 8.28e-01 | 0.355 | 0.337 | 0.019 | 3.95e-02 |
| GCSE | LDpred2.MultiPRS | LDpred1.Inf | 0.423 | 0.409 | 0.014 | 3.70e-01 | 0.356 | 0.337 | 0.020 | 4.06e-03 |
| GCSE | pT+clump.10FCVal | LDpred1.Inf | 0.374 | 0.409 | -0.035 | 5.56e-09 | 0.315 | 0.337 | -0.021 | 9.35e-02 |
| GCSE | lassosum.PseudoVal | LDpred1.Inf | 0.316 | 0.409 | -0.093 | 8.82e-23 | 0.273 | 0.337 | -0.064 | 1.37e-03 |
| GCSE | LDpred2.10FCVal | LDpred1.Inf | 0.423 | 0.409 | 0.013 | 8.40e-05 | 0.358 | 0.337 | 0.022 | 2.52e-03 |
| GCSE | SBayesR.PseudoVal | LDpred1.Inf | 0.420 | 0.409 | 0.011 | 4.02e-03 | 0.350 | 0.337 | 0.013 | 8.36e-02 |
| GCSE | LDpred1.MultiPRS | LDpred1.Inf | 0.414 | 0.409 | 0.004 | 7.90e-01 | 0.341 | 0.337 | 0.005 | 3.13e-01 |
| GCSE | LDpred2.PseudoVal | LDpred1.Inf | 0.422 | 0.409 | 0.013 | 1.40e-04 | 0.358 | 0.337 | 0.021 | 2.60e-03 |
| GCSE | SBLUP.Inf | LDpred1.Inf | 0.412 | 0.409 | 0.002 | 1.91e-01 | 0.344 | 0.337 | 0.008 | 5.09e-02 |
| GCSE | pT+clump.MultiPRS | LDpred1.Inf | 0.382 | 0.409 | -0.028 | 9.07e-02 | 0.330 | 0.337 | -0.007 | 5.79e-01 |
| GCSE | lassosum.MultiPRS | LDpred1.Inf | 0.412 | 0.409 | 0.002 | 8.81e-01 | 0.357 | 0.337 | 0.021 | 1.33e-02 |
| GCSE | LDpred1.10FCVal | LDpred1.Inf | 0.405 | 0.409 | -0.004 | 2.03e-01 | 0.337 | 0.337 | 0.000 | 9.59e-01 |
| GCSE | LDpred1.Inf | LDpred1.10FCVal | 0.409 | 0.405 | 0.004 | 2.03e-01 | 0.337 | 0.337 | 0.000 | 9.59e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.402 | 0.405 | -0.003 | 5.35e-01 | 0.339 | 0.337 | 0.002 | 8.64e-01 |
| GCSE | lassosum.10FCVal | LDpred1.10FCVal | 0.410 | 0.405 | 0.005 | 3.08e-01 | 0.355 | 0.337 | 0.018 | 7.39e-02 |
| GCSE | LDpred2.MultiPRS | LDpred1.10FCVal | 0.423 | 0.405 | 0.018 | 2.55e-01 | 0.356 | 0.337 | 0.019 | 1.71e-02 |
| GCSE | pT+clump.10FCVal | LDpred1.10FCVal | 0.374 | 0.405 | -0.031 | 1.51e-06 | 0.315 | 0.337 | -0.022 | 1.15e-01 |
| GCSE | lassosum.PseudoVal | LDpred1.10FCVal | 0.316 | 0.405 | -0.090 | 9.91e-20 | 0.273 | 0.337 | -0.064 | 1.90e-03 |
| GCSE | LDpred2.10FCVal | LDpred1.10FCVal | 0.423 | 0.405 | 0.017 | 1.64e-05 | 0.358 | 0.337 | 0.021 | 1.28e-02 |
| GCSE | All.MultiPRS | LDpred1.10FCVal | 0.423 | 0.405 | 0.018 | 2.58e-01 | 0.355 | 0.337 | 0.018 | 2.38e-02 |
| GCSE | LDpred1.MultiPRS | LDpred1.10FCVal | 0.414 | 0.405 | 0.008 | 6.15e-01 | 0.341 | 0.337 | 0.004 | 4.62e-01 |
| GCSE | LDpred2.PseudoVal | LDpred1.10FCVal | 0.422 | 0.405 | 0.017 | 2.07e-05 | 0.358 | 0.337 | 0.021 | 1.21e-02 |
| GCSE | SBLUP.Inf | LDpred1.10FCVal | 0.412 | 0.405 | 0.006 | 1.92e-02 | 0.344 | 0.337 | 0.007 | 2.02e-01 |
| GCSE | pT+clump.MultiPRS | LDpred1.10FCVal | 0.382 | 0.405 | -0.024 | 1.45e-01 | 0.330 | 0.337 | -0.007 | 5.96e-01 |
| GCSE | LDpred2.Inf | LDpred1.10FCVal | 0.413 | 0.405 | 0.008 | 4.69e-03 | 0.343 | 0.337 | 0.006 | 3.35e-01 |
| GCSE | lassosum.MultiPRS | LDpred1.10FCVal | 0.412 | 0.405 | 0.006 | 6.95e-01 | 0.357 | 0.337 | 0.020 | 3.66e-02 |
| GCSE | LDpred1.10FCVal | LDpred1.10FCVal | 0.405 | 0.405 | 0.000 | 1.00e+00 | 0.337 | 0.337 | 0.000 | 1.00e+00 |
| GCSE | SBayesR.PseudoVal | LDpred1.10FCVal | 0.420 | 0.405 | 0.014 | 7.00e-04 | 0.350 | 0.337 | 0.013 | 1.53e-01 |
| GCSE | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.423 | 0.423 | 0.000 | 1.00e+00 | 0.356 | 0.356 | 0.000 | 1.00e+00 |
| GCSE | lassosum.10FCVal | LDpred2.MultiPRS | 0.410 | 0.423 | -0.013 | 4.02e-01 | 0.355 | 0.356 | -0.001 | 8.68e-01 |
| GCSE | All.MultiPRS | LDpred2.MultiPRS | 0.423 | 0.423 | 0.000 | 8.75e-01 | 0.355 | 0.356 | -0.001 | 5.67e-01 |
| GCSE | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.414 | 0.423 | -0.010 | 5.33e-01 | 0.341 | 0.356 | -0.015 | 2.09e-02 |
| GCSE | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.402 | 0.423 | -0.021 | 1.84e-01 | 0.339 | 0.356 | -0.018 | 1.77e-02 |
| GCSE | LDpred1.Inf | LDpred2.MultiPRS | 0.409 | 0.423 | -0.014 | 3.70e-01 | 0.337 | 0.356 | -0.020 | 4.06e-03 |
| GCSE | LDpred2.Inf | LDpred2.MultiPRS | 0.413 | 0.423 | -0.010 | 5.26e-01 | 0.343 | 0.356 | -0.014 | 1.58e-02 |
| GCSE | lassosum.MultiPRS | LDpred2.MultiPRS | 0.412 | 0.423 | -0.012 | 4.53e-05 | 0.357 | 0.356 | 0.001 | 8.81e-01 |
| GCSE | LDpred1.10FCVal | LDpred2.MultiPRS | 0.405 | 0.423 | -0.018 | 2.55e-01 | 0.337 | 0.356 | -0.019 | 1.71e-02 |
| GCSE | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.382 | 0.423 | -0.042 | 9.62e-03 | 0.330 | 0.356 | -0.026 | 1.86e-02 |
| GCSE | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.420 | 0.423 | -0.004 | 8.17e-01 | 0.350 | 0.356 | -0.006 | 1.85e-01 |
| GCSE | pT+clump.10FCVal | LDpred2.MultiPRS | 0.374 | 0.423 | -0.050 | 2.02e-03 | 0.315 | 0.356 | -0.041 | 1.51e-03 |
| GCSE | lassosum.PseudoVal | LDpred2.MultiPRS | 0.316 | 0.423 | -0.108 | 4.73e-11 | 0.273 | 0.356 | -0.084 | 1.96e-06 |
| GCSE | LDpred2.10FCVal | LDpred2.MultiPRS | 0.423 | 0.423 | -0.001 | 9.51e-01 | 0.358 | 0.356 | 0.002 | 4.70e-01 |
| GCSE | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.422 | 0.423 | -0.001 | 9.27e-01 | 0.358 | 0.356 | 0.002 | 4.97e-01 |
| GCSE | SBLUP.Inf | LDpred2.MultiPRS | 0.412 | 0.423 | -0.012 | 4.58e-01 | 0.344 | 0.356 | -0.012 | 5.65e-02 |
| GCSE | LDpred2.MultiPRS | LDpred2.10FCVal | 0.423 | 0.423 | 0.001 | 9.51e-01 | 0.356 | 0.358 | -0.002 | 4.70e-01 |
| GCSE | pT+clump.10FCVal | LDpred2.10FCVal | 0.374 | 0.423 | -0.049 | 3.83e-15 | 0.315 | 0.358 | -0.043 | 8.13e-04 |
| GCSE | lassosum.10FCVal | LDpred2.10FCVal | 0.410 | 0.423 | -0.012 | 6.62e-05 | 0.355 | 0.358 | -0.003 | 6.23e-01 |
| GCSE | All.MultiPRS | LDpred2.10FCVal | 0.423 | 0.423 | 0.001 | 9.58e-01 | 0.355 | 0.358 | -0.003 | 3.42e-01 |
| GCSE | LDpred1.MultiPRS | LDpred2.10FCVal | 0.414 | 0.423 | -0.009 | 5.80e-01 | 0.341 | 0.358 | -0.017 | 1.27e-02 |
| GCSE | LDpred2.PseudoVal | LDpred2.10FCVal | 0.422 | 0.423 | 0.000 | 6.87e-01 | 0.358 | 0.358 | 0.000 | 9.06e-01 |
| GCSE | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.402 | 0.423 | -0.020 | 2.79e-07 | 0.339 | 0.358 | -0.020 | 1.83e-02 |
| GCSE | LDpred1.Inf | LDpred2.10FCVal | 0.409 | 0.423 | -0.013 | 8.40e-05 | 0.337 | 0.358 | -0.022 | 2.52e-03 |
| GCSE | LDpred2.Inf | LDpred2.10FCVal | 0.413 | 0.423 | -0.009 | 2.10e-03 | 0.343 | 0.358 | -0.016 | 1.29e-02 |
| GCSE | lassosum.MultiPRS | LDpred2.10FCVal | 0.412 | 0.423 | -0.011 | 4.92e-01 | 0.357 | 0.358 | -0.001 | 8.41e-01 |
| GCSE | LDpred1.10FCVal | LDpred2.10FCVal | 0.405 | 0.423 | -0.017 | 1.64e-05 | 0.337 | 0.358 | -0.021 | 1.28e-02 |
| GCSE | pT+clump.MultiPRS | LDpred2.10FCVal | 0.382 | 0.423 | -0.041 | 1.22e-02 | 0.330 | 0.358 | -0.028 | 1.23e-02 |
| GCSE | SBayesR.PseudoVal | LDpred2.10FCVal | 0.420 | 0.423 | -0.003 | 2.39e-01 | 0.350 | 0.358 | -0.008 | 8.44e-02 |
| GCSE | lassosum.PseudoVal | LDpred2.10FCVal | 0.316 | 0.423 | -0.107 | 8.33e-36 | 0.273 | 0.358 | -0.086 | 1.60e-06 |
| GCSE | LDpred2.10FCVal | LDpred2.10FCVal | 0.423 | 0.423 | 0.000 | 1.00e+00 | 0.358 | 0.358 | 0.000 | 1.00e+00 |
| GCSE | SBLUP.Inf | LDpred2.10FCVal | 0.412 | 0.423 | -0.011 | 6.98e-04 | 0.344 | 0.358 | -0.014 | 3.85e-02 |
| GCSE | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.423 | 0.422 | 0.001 | 9.27e-01 | 0.356 | 0.358 | -0.002 | 4.97e-01 |
| GCSE | lassosum.10FCVal | LDpred2.PseudoVal | 0.410 | 0.422 | -0.012 | 2.61e-04 | 0.355 | 0.358 | -0.003 | 6.71e-01 |
| GCSE | All.MultiPRS | LDpred2.PseudoVal | 0.423 | 0.422 | 0.001 | 9.34e-01 | 0.355 | 0.358 | -0.003 | 3.53e-01 |
| GCSE | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.414 | 0.422 | -0.008 | 6.00e-01 | 0.341 | 0.358 | -0.017 | 1.19e-02 |
| GCSE | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.402 | 0.422 | -0.020 | 3.13e-07 | 0.339 | 0.358 | -0.019 | 1.67e-02 |
| GCSE | LDpred1.Inf | LDpred2.PseudoVal | 0.409 | 0.422 | -0.013 | 1.40e-04 | 0.337 | 0.358 | -0.021 | 2.60e-03 |
| GCSE | LDpred2.Inf | LDpred2.PseudoVal | 0.413 | 0.422 | -0.009 | 2.22e-03 | 0.343 | 0.358 | -0.015 | 1.02e-02 |
| GCSE | lassosum.MultiPRS | LDpred2.PseudoVal | 0.412 | 0.422 | -0.010 | 5.12e-01 | 0.357 | 0.358 | -0.001 | 8.92e-01 |
| GCSE | LDpred1.10FCVal | LDpred2.PseudoVal | 0.405 | 0.422 | -0.017 | 2.07e-05 | 0.337 | 0.358 | -0.021 | 1.21e-02 |
| GCSE | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.382 | 0.422 | -0.041 | 1.31e-02 | 0.330 | 0.358 | -0.028 | 1.47e-02 |
| GCSE | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.420 | 0.422 | -0.002 | 3.15e-01 | 0.350 | 0.358 | -0.008 | 7.45e-02 |
| GCSE | pT+clump.10FCVal | LDpred2.PseudoVal | 0.374 | 0.422 | -0.048 | 1.31e-14 | 0.315 | 0.358 | -0.043 | 9.82e-04 |
| GCSE | lassosum.PseudoVal | LDpred2.PseudoVal | 0.316 | 0.422 | -0.106 | 5.29e-33 | 0.273 | 0.358 | -0.085 | 3.87e-06 |
| GCSE | LDpred2.10FCVal | LDpred2.PseudoVal | 0.423 | 0.422 | 0.000 | 6.87e-01 | 0.358 | 0.358 | 0.000 | 9.06e-01 |
| GCSE | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.422 | 0.422 | 0.000 | 1.00e+00 | 0.358 | 0.358 | 0.000 | 1.00e+00 |
| GCSE | SBLUP.Inf | LDpred2.PseudoVal | 0.412 | 0.422 | -0.010 | 9.20e-04 | 0.344 | 0.358 | -0.014 | 3.67e-02 |
| GCSE | DBSLMM.PseudoVal | LDpred2.Inf | 0.402 | 0.413 | -0.011 | 9.10e-03 | 0.339 | 0.343 | -0.004 | 6.60e-01 |
| GCSE | LDpred2.MultiPRS | LDpred2.Inf | 0.423 | 0.413 | 0.010 | 5.26e-01 | 0.356 | 0.343 | 0.014 | 1.58e-02 |
| GCSE | pT+clump.10FCVal | LDpred2.Inf | 0.374 | 0.413 | -0.039 | 1.47e-10 | 0.315 | 0.343 | -0.028 | 3.09e-02 |
| GCSE | lassosum.PseudoVal | LDpred2.Inf | 0.316 | 0.413 | -0.098 | 8.07e-24 | 0.273 | 0.343 | -0.070 | 5.69e-04 |
| GCSE | lassosum.10FCVal | LDpred2.Inf | 0.410 | 0.413 | -0.003 | 4.34e-01 | 0.355 | 0.343 | 0.012 | 1.54e-01 |
| GCSE | All.MultiPRS | LDpred2.Inf | 0.423 | 0.413 | 0.010 | 5.31e-01 | 0.355 | 0.343 | 0.013 | 2.80e-02 |
| GCSE | LDpred1.MultiPRS | LDpred2.Inf | 0.414 | 0.413 | 0.000 | 9.92e-01 | 0.341 | 0.343 | -0.002 | 7.49e-01 |
| GCSE | LDpred2.PseudoVal | LDpred2.Inf | 0.422 | 0.413 | 0.009 | 2.22e-03 | 0.358 | 0.343 | 0.015 | 1.02e-02 |
| GCSE | SBLUP.Inf | LDpred2.Inf | 0.412 | 0.413 | -0.002 | 2.15e-01 | 0.344 | 0.343 | 0.002 | 5.61e-01 |
| GCSE | LDpred1.Inf | LDpred2.Inf | 0.409 | 0.413 | -0.004 | 3.42e-02 | 0.337 | 0.343 | -0.006 | 1.39e-01 |
| GCSE | LDpred2.Inf | LDpred2.Inf | 0.413 | 0.413 | 0.000 | 1.00e+00 | 0.343 | 0.343 | 0.000 | 1.00e+00 |
| GCSE | lassosum.MultiPRS | LDpred2.Inf | 0.412 | 0.413 | -0.002 | 9.11e-01 | 0.357 | 0.343 | 0.014 | 7.59e-02 |
| GCSE | LDpred1.10FCVal | LDpred2.Inf | 0.405 | 0.413 | -0.008 | 4.69e-03 | 0.337 | 0.343 | -0.006 | 3.35e-01 |
| GCSE | pT+clump.MultiPRS | LDpred2.Inf | 0.382 | 0.413 | -0.032 | 5.13e-02 | 0.330 | 0.343 | -0.013 | 2.93e-01 |
| GCSE | SBayesR.PseudoVal | LDpred2.Inf | 0.420 | 0.413 | 0.006 | 7.92e-02 | 0.350 | 0.343 | 0.007 | 3.43e-01 |
| GCSE | LDpred2.10FCVal | LDpred2.Inf | 0.423 | 0.413 | 0.009 | 2.10e-03 | 0.358 | 0.343 | 0.016 | 1.29e-02 |
| GCSE | All.MultiPRS | pT+clump.MultiPRS | 0.423 | 0.382 | 0.042 | 9.82e-03 | 0.355 | 0.330 | 0.025 | 1.78e-02 |
| GCSE | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.414 | 0.382 | 0.032 | 4.81e-02 | 0.341 | 0.330 | 0.011 | 3.57e-01 |
| GCSE | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.422 | 0.382 | 0.041 | 1.31e-02 | 0.358 | 0.330 | 0.028 | 1.47e-02 |
| GCSE | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.402 | 0.382 | 0.021 | 2.04e-01 | 0.339 | 0.330 | 0.009 | 4.34e-01 |
| GCSE | LDpred1.Inf | pT+clump.MultiPRS | 0.409 | 0.382 | 0.028 | 9.07e-02 | 0.337 | 0.330 | 0.007 | 5.79e-01 |
| GCSE | LDpred2.Inf | pT+clump.MultiPRS | 0.413 | 0.382 | 0.032 | 5.13e-02 | 0.343 | 0.330 | 0.013 | 2.93e-01 |
| GCSE | lassosum.MultiPRS | pT+clump.MultiPRS | 0.412 | 0.382 | 0.030 | 6.39e-02 | 0.357 | 0.330 | 0.027 | 1.72e-02 |
| GCSE | LDpred1.10FCVal | pT+clump.MultiPRS | 0.405 | 0.382 | 0.024 | 1.45e-01 | 0.337 | 0.330 | 0.007 | 5.96e-01 |
| GCSE | lassosum.10FCVal | pT+clump.MultiPRS | 0.410 | 0.382 | 0.029 | 8.07e-02 | 0.355 | 0.330 | 0.025 | 2.75e-02 |
| GCSE | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.423 | 0.382 | 0.042 | 9.62e-03 | 0.356 | 0.330 | 0.026 | 1.86e-02 |
| GCSE | pT+clump.10FCVal | pT+clump.MultiPRS | 0.374 | 0.382 | -0.008 | 6.48e-01 | 0.315 | 0.330 | -0.015 | 1.26e-02 |
| GCSE | lassosum.PseudoVal | pT+clump.MultiPRS | 0.316 | 0.382 | -0.066 | 9.99e-05 | 0.273 | 0.330 | -0.057 | 2.92e-03 |
| GCSE | LDpred2.10FCVal | pT+clump.MultiPRS | 0.423 | 0.382 | 0.041 | 1.22e-02 | 0.358 | 0.330 | 0.028 | 1.23e-02 |
| GCSE | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.420 | 0.382 | 0.038 | 1.92e-02 | 0.350 | 0.330 | 0.020 | 7.53e-02 |
| GCSE | SBLUP.Inf | pT+clump.MultiPRS | 0.412 | 0.382 | 0.030 | 6.54e-02 | 0.344 | 0.330 | 0.014 | 2.19e-01 |
| GCSE | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.382 | 0.382 | 0.000 | 1.00e+00 | 0.330 | 0.330 | 0.000 | 1.00e+00 |
| GCSE | lassosum.10FCVal | pT+clump.10FCVal | 0.410 | 0.374 | 0.036 | 1.98e-09 | 0.355 | 0.315 | 0.040 | 1.58e-03 |
| GCSE | All.MultiPRS | pT+clump.10FCVal | 0.423 | 0.374 | 0.049 | 2.08e-03 | 0.355 | 0.315 | 0.040 | 1.27e-03 |
| GCSE | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.402 | 0.374 | 0.028 | 2.01e-05 | 0.339 | 0.315 | 0.024 | 8.81e-02 |
| GCSE | LDpred1.Inf | pT+clump.10FCVal | 0.409 | 0.374 | 0.035 | 5.56e-09 | 0.337 | 0.315 | 0.021 | 9.35e-02 |
| GCSE | LDpred2.Inf | pT+clump.10FCVal | 0.413 | 0.374 | 0.039 | 1.47e-10 | 0.343 | 0.315 | 0.028 | 3.09e-02 |
| GCSE | lassosum.MultiPRS | pT+clump.10FCVal | 0.412 | 0.374 | 0.038 | 1.93e-02 | 0.357 | 0.315 | 0.042 | 1.21e-03 |
| GCSE | LDpred1.10FCVal | pT+clump.10FCVal | 0.405 | 0.374 | 0.031 | 1.51e-06 | 0.337 | 0.315 | 0.022 | 1.15e-01 |
| GCSE | pT+clump.MultiPRS | pT+clump.10FCVal | 0.382 | 0.374 | 0.008 | 6.48e-01 | 0.330 | 0.315 | 0.015 | 1.26e-02 |
| GCSE | LDpred2.MultiPRS | pT+clump.10FCVal | 0.423 | 0.374 | 0.050 | 2.02e-03 | 0.356 | 0.315 | 0.041 | 1.51e-03 |
| GCSE | pT+clump.10FCVal | pT+clump.10FCVal | 0.374 | 0.374 | 0.000 | 1.00e+00 | 0.315 | 0.315 | 0.000 | 1.00e+00 |
| GCSE | lassosum.PseudoVal | pT+clump.10FCVal | 0.316 | 0.374 | -0.058 | 6.52e-08 | 0.273 | 0.315 | -0.043 | 6.49e-02 |
| GCSE | LDpred2.10FCVal | pT+clump.10FCVal | 0.423 | 0.374 | 0.049 | 3.83e-15 | 0.358 | 0.315 | 0.043 | 8.13e-04 |
| GCSE | SBayesR.PseudoVal | pT+clump.10FCVal | 0.420 | 0.374 | 0.046 | 3.47e-13 | 0.350 | 0.315 | 0.035 | 8.78e-03 |
| GCSE | LDpred1.MultiPRS | pT+clump.10FCVal | 0.414 | 0.374 | 0.040 | 1.59e-02 | 0.341 | 0.315 | 0.026 | 4.58e-02 |
| GCSE | LDpred2.PseudoVal | pT+clump.10FCVal | 0.422 | 0.374 | 0.048 | 1.31e-14 | 0.358 | 0.315 | 0.043 | 9.82e-04 |
| GCSE | SBLUP.Inf | pT+clump.10FCVal | 0.412 | 0.374 | 0.038 | 2.31e-10 | 0.344 | 0.315 | 0.029 | 1.90e-02 |
| GCSE | LDpred1.Inf | SBayesR.PseudoVal | 0.409 | 0.420 | -0.011 | 4.02e-03 | 0.337 | 0.350 | -0.013 | 8.36e-02 |
| GCSE | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.402 | 0.420 | -0.017 | 1.24e-06 | 0.339 | 0.350 | -0.011 | 1.37e-01 |
| GCSE | lassosum.10FCVal | SBayesR.PseudoVal | 0.410 | 0.420 | -0.010 | 5.89e-03 | 0.355 | 0.350 | 0.005 | 4.88e-01 |
| GCSE | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.423 | 0.420 | 0.004 | 8.17e-01 | 0.356 | 0.350 | 0.006 | 1.85e-01 |
| GCSE | pT+clump.10FCVal | SBayesR.PseudoVal | 0.374 | 0.420 | -0.046 | 3.47e-13 | 0.315 | 0.350 | -0.035 | 8.78e-03 |
| GCSE | lassosum.PseudoVal | SBayesR.PseudoVal | 0.316 | 0.420 | -0.104 | 3.50e-36 | 0.273 | 0.350 | -0.077 | 1.01e-05 |
| GCSE | LDpred2.10FCVal | SBayesR.PseudoVal | 0.423 | 0.420 | 0.003 | 2.39e-01 | 0.358 | 0.350 | 0.008 | 8.44e-02 |
| GCSE | All.MultiPRS | SBayesR.PseudoVal | 0.423 | 0.420 | 0.004 | 8.24e-01 | 0.355 | 0.350 | 0.005 | 2.23e-01 |
| GCSE | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.414 | 0.420 | -0.006 | 6.98e-01 | 0.341 | 0.350 | -0.009 | 2.36e-01 |
| GCSE | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.422 | 0.420 | 0.002 | 3.15e-01 | 0.358 | 0.350 | 0.008 | 7.45e-02 |
| GCSE | SBLUP.Inf | SBayesR.PseudoVal | 0.412 | 0.420 | -0.008 | 2.46e-02 | 0.344 | 0.350 | -0.006 | 4.58e-01 |
| GCSE | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.382 | 0.420 | -0.038 | 1.92e-02 | 0.330 | 0.350 | -0.020 | 7.53e-02 |
| GCSE | LDpred2.Inf | SBayesR.PseudoVal | 0.413 | 0.420 | -0.006 | 7.92e-02 | 0.343 | 0.350 | -0.007 | 3.43e-01 |
| GCSE | lassosum.MultiPRS | SBayesR.PseudoVal | 0.412 | 0.420 | -0.008 | 6.05e-01 | 0.357 | 0.350 | 0.007 | 2.72e-01 |
| GCSE | LDpred1.10FCVal | SBayesR.PseudoVal | 0.405 | 0.420 | -0.014 | 7.00e-04 | 0.337 | 0.350 | -0.013 | 1.53e-01 |
| GCSE | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.420 | 0.420 | 0.000 | 1.00e+00 | 0.350 | 0.350 | 0.000 | 1.00e+00 |
| GCSE | All.MultiPRS | SBLUP.Inf | 0.423 | 0.412 | 0.012 | 4.63e-01 | 0.355 | 0.344 | 0.011 | 7.74e-02 |
| GCSE | LDpred1.Inf | SBLUP.Inf | 0.409 | 0.412 | -0.002 | 1.91e-01 | 0.337 | 0.344 | -0.008 | 5.09e-02 |
| GCSE | LDpred2.Inf | SBLUP.Inf | 0.413 | 0.412 | 0.002 | 2.15e-01 | 0.343 | 0.344 | -0.002 | 5.61e-01 |
| GCSE | DBSLMM.PseudoVal | SBLUP.Inf | 0.402 | 0.412 | -0.009 | 2.45e-02 | 0.339 | 0.344 | -0.006 | 5.25e-01 |
| GCSE | lassosum.10FCVal | SBLUP.Inf | 0.410 | 0.412 | -0.002 | 7.09e-01 | 0.355 | 0.344 | 0.011 | 2.09e-01 |
| GCSE | LDpred2.MultiPRS | SBLUP.Inf | 0.423 | 0.412 | 0.012 | 4.58e-01 | 0.356 | 0.344 | 0.012 | 5.65e-02 |
| GCSE | pT+clump.10FCVal | SBLUP.Inf | 0.374 | 0.412 | -0.038 | 2.31e-10 | 0.315 | 0.344 | -0.029 | 1.90e-02 |
| GCSE | lassosum.PseudoVal | SBLUP.Inf | 0.316 | 0.412 | -0.096 | 1.06e-23 | 0.273 | 0.344 | -0.072 | 3.51e-04 |
| GCSE | LDpred2.10FCVal | SBLUP.Inf | 0.423 | 0.412 | 0.011 | 6.98e-04 | 0.358 | 0.344 | 0.014 | 3.85e-02 |
| GCSE | SBayesR.PseudoVal | SBLUP.Inf | 0.420 | 0.412 | 0.008 | 2.46e-02 | 0.350 | 0.344 | 0.006 | 4.58e-01 |
| GCSE | LDpred1.MultiPRS | SBLUP.Inf | 0.414 | 0.412 | 0.002 | 9.08e-01 | 0.341 | 0.344 | -0.003 | 5.03e-01 |
| GCSE | LDpred2.PseudoVal | SBLUP.Inf | 0.422 | 0.412 | 0.010 | 9.20e-04 | 0.358 | 0.344 | 0.014 | 3.67e-02 |
| GCSE | SBLUP.Inf | SBLUP.Inf | 0.412 | 0.412 | 0.000 | 1.00e+00 | 0.344 | 0.344 | 0.000 | 1.00e+00 |
| GCSE | pT+clump.MultiPRS | SBLUP.Inf | 0.382 | 0.412 | -0.030 | 6.54e-02 | 0.330 | 0.344 | -0.014 | 2.19e-01 |
| GCSE | lassosum.MultiPRS | SBLUP.Inf | 0.412 | 0.412 | 0.000 | 9.97e-01 | 0.357 | 0.344 | 0.013 | 1.08e-01 |
| GCSE | LDpred1.10FCVal | SBLUP.Inf | 0.405 | 0.412 | -0.006 | 1.92e-02 | 0.337 | 0.344 | -0.007 | 2.02e-01 |
| ADHD | lassosum.PseudoVal | All.MultiPRS | 0.080 | 0.118 | -0.038 | 3.12e-02 | 0.108 | 0.182 | -0.074 | 4.42e-04 |
| ADHD | All.MultiPRS | All.MultiPRS | 0.118 | 0.118 | 0.000 | 1.00e+00 | 0.182 | 0.182 | 0.000 | 1.00e+00 |
| ADHD | DBSLMM.PseudoVal | All.MultiPRS | 0.110 | 0.118 | -0.007 | 6.77e-01 | 0.174 | 0.182 | -0.008 | 5.24e-01 |
| ADHD | LDpred1.Inf | All.MultiPRS | 0.111 | 0.118 | -0.006 | 7.21e-01 | 0.188 | 0.182 | 0.006 | 6.21e-01 |
| ADHD | LDpred2.Inf | All.MultiPRS | 0.113 | 0.118 | -0.005 | 7.82e-01 | 0.189 | 0.182 | 0.007 | 5.52e-01 |
| ADHD | lassosum.MultiPRS | All.MultiPRS | 0.117 | 0.118 | 0.000 | 9.86e-01 | 0.190 | 0.182 | 0.009 | 2.91e-01 |
| ADHD | LDpred1.10FCVal | All.MultiPRS | 0.114 | 0.118 | -0.004 | 8.41e-01 | 0.190 | 0.182 | 0.008 | 4.74e-01 |
| ADHD | pT+clump.MultiPRS | All.MultiPRS | 0.093 | 0.118 | -0.024 | 1.65e-01 | 0.151 | 0.182 | -0.031 | 5.79e-02 |
| ADHD | LDpred2.MultiPRS | All.MultiPRS | 0.113 | 0.118 | -0.005 | 7.82e-01 | 0.190 | 0.182 | 0.008 | 4.48e-01 |
| ADHD | pT+clump.10FCVal | All.MultiPRS | 0.092 | 0.118 | -0.025 | 1.53e-01 | 0.144 | 0.182 | -0.038 | 4.18e-02 |
| ADHD | lassosum.10FCVal | All.MultiPRS | 0.115 | 0.118 | -0.003 | 8.80e-01 | 0.186 | 0.182 | 0.004 | 7.23e-01 |
| ADHD | LDpred2.10FCVal | All.MultiPRS | 0.117 | 0.118 | -0.001 | 9.54e-01 | 0.187 | 0.182 | 0.005 | 6.28e-01 |
| ADHD | SBayesR.PseudoVal | All.MultiPRS | 0.111 | 0.118 | -0.006 | 7.28e-01 | 0.189 | 0.182 | 0.007 | 5.57e-01 |
| ADHD | LDpred1.MultiPRS | All.MultiPRS | 0.113 | 0.118 | -0.004 | 8.06e-01 | 0.190 | 0.182 | 0.008 | 4.63e-01 |
| ADHD | LDpred2.PseudoVal | All.MultiPRS | 0.114 | 0.118 | -0.003 | 8.49e-01 | 0.190 | 0.182 | 0.008 | 4.55e-01 |
| ADHD | SBLUP.Inf | All.MultiPRS | 0.112 | 0.118 | -0.005 | 7.62e-01 | 0.188 | 0.182 | 0.006 | 5.97e-01 |
| ADHD | LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.113 | 0.110 | 0.002 | 8.88e-01 | 0.190 | 0.174 | 0.016 | 1.43e-01 |
| ADHD | pT+clump.10FCVal | DBSLMM.PseudoVal | 0.092 | 0.110 | -0.018 | 6.69e-02 | 0.144 | 0.174 | -0.030 | 1.22e-01 |
| ADHD | lassosum.PseudoVal | DBSLMM.PseudoVal | 0.080 | 0.110 | -0.030 | 6.19e-03 | 0.108 | 0.174 | -0.066 | 2.97e-03 |
| ADHD | All.MultiPRS | DBSLMM.PseudoVal | 0.118 | 0.110 | 0.007 | 6.77e-01 | 0.182 | 0.174 | 0.008 | 5.24e-01 |
| ADHD | LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.113 | 0.110 | 0.003 | 8.65e-01 | 0.190 | 0.174 | 0.016 | 1.70e-01 |
| ADHD | LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.114 | 0.110 | 0.004 | 4.90e-01 | 0.190 | 0.174 | 0.017 | 1.48e-01 |
| ADHD | DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.110 | 0.110 | 0.000 | 1.00e+00 | 0.174 | 0.174 | 0.000 | 1.00e+00 |
| ADHD | LDpred1.Inf | DBSLMM.PseudoVal | 0.111 | 0.110 | 0.001 | 8.66e-01 | 0.188 | 0.174 | 0.014 | 2.43e-01 |
| ADHD | LDpred2.Inf | DBSLMM.PseudoVal | 0.113 | 0.110 | 0.002 | 6.80e-01 | 0.189 | 0.174 | 0.015 | 1.96e-01 |
| ADHD | lassosum.MultiPRS | DBSLMM.PseudoVal | 0.117 | 0.110 | 0.007 | 6.92e-01 | 0.190 | 0.174 | 0.017 | 1.36e-01 |
| ADHD | LDpred1.10FCVal | DBSLMM.PseudoVal | 0.114 | 0.110 | 0.004 | 5.12e-01 | 0.190 | 0.174 | 0.016 | 1.63e-01 |
| ADHD | pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.093 | 0.110 | -0.017 | 3.37e-01 | 0.151 | 0.174 | -0.023 | 1.58e-01 |
| ADHD | SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.111 | 0.110 | 0.001 | 8.40e-01 | 0.189 | 0.174 | 0.015 | 2.03e-01 |
| ADHD | lassosum.10FCVal | DBSLMM.PseudoVal | 0.115 | 0.110 | 0.005 | 5.02e-01 | 0.186 | 0.174 | 0.012 | 3.69e-01 |
| ADHD | LDpred2.10FCVal | DBSLMM.PseudoVal | 0.117 | 0.110 | 0.006 | 2.74e-01 | 0.187 | 0.174 | 0.013 | 2.41e-01 |
| ADHD | SBLUP.Inf | DBSLMM.PseudoVal | 0.112 | 0.110 | 0.002 | 7.41e-01 | 0.188 | 0.174 | 0.014 | 2.26e-01 |
| ADHD | All.MultiPRS | lassosum.MultiPRS | 0.118 | 0.117 | 0.000 | 9.86e-01 | 0.182 | 0.190 | -0.009 | 2.91e-01 |
| ADHD | LDpred1.MultiPRS | lassosum.MultiPRS | 0.113 | 0.117 | -0.004 | 8.18e-01 | 0.190 | 0.190 | -0.001 | 9.52e-01 |
| ADHD | LDpred1.Inf | lassosum.MultiPRS | 0.111 | 0.117 | -0.006 | 7.37e-01 | 0.188 | 0.190 | -0.003 | 7.91e-01 |
| ADHD | LDpred2.Inf | lassosum.MultiPRS | 0.113 | 0.117 | -0.005 | 7.98e-01 | 0.189 | 0.190 | -0.002 | 8.60e-01 |
| ADHD | lassosum.MultiPRS | lassosum.MultiPRS | 0.117 | 0.117 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| ADHD | DBSLMM.PseudoVal | lassosum.MultiPRS | 0.110 | 0.117 | -0.007 | 6.92e-01 | 0.174 | 0.190 | -0.017 | 1.36e-01 |
| ADHD | lassosum.PseudoVal | lassosum.MultiPRS | 0.080 | 0.117 | -0.038 | 3.46e-02 | 0.108 | 0.190 | -0.083 | 3.77e-05 |
| ADHD | LDpred2.MultiPRS | lassosum.MultiPRS | 0.113 | 0.117 | -0.005 | 2.20e-01 | 0.190 | 0.190 | -0.001 | 9.14e-01 |
| ADHD | pT+clump.10FCVal | lassosum.MultiPRS | 0.092 | 0.117 | -0.025 | 1.63e-01 | 0.144 | 0.190 | -0.047 | 4.28e-03 |
| ADHD | lassosum.10FCVal | lassosum.MultiPRS | 0.115 | 0.117 | -0.002 | 8.95e-01 | 0.186 | 0.190 | -0.005 | 4.70e-01 |
| ADHD | LDpred2.10FCVal | lassosum.MultiPRS | 0.117 | 0.117 | -0.001 | 9.68e-01 | 0.187 | 0.190 | -0.004 | 6.24e-01 |
| ADHD | SBayesR.PseudoVal | lassosum.MultiPRS | 0.111 | 0.117 | -0.006 | 7.44e-01 | 0.189 | 0.190 | -0.002 | 8.22e-01 |
| ADHD | LDpred2.PseudoVal | lassosum.MultiPRS | 0.114 | 0.117 | -0.003 | 8.64e-01 | 0.190 | 0.190 | 0.000 | 9.66e-01 |
| ADHD | SBLUP.Inf | lassosum.MultiPRS | 0.112 | 0.117 | -0.005 | 7.78e-01 | 0.188 | 0.190 | -0.002 | 7.87e-01 |
| ADHD | pT+clump.MultiPRS | lassosum.MultiPRS | 0.093 | 0.117 | -0.024 | 1.74e-01 | 0.151 | 0.190 | -0.040 | 3.71e-03 |
| ADHD | LDpred1.10FCVal | lassosum.MultiPRS | 0.114 | 0.117 | -0.003 | 8.57e-01 | 0.190 | 0.190 | 0.000 | 9.62e-01 |
| ADHD | All.MultiPRS | lassosum.PseudoVal | 0.118 | 0.080 | 0.038 | 3.12e-02 | 0.182 | 0.108 | 0.074 | 4.42e-04 |
| ADHD | LDpred1.MultiPRS | lassosum.PseudoVal | 0.113 | 0.080 | 0.033 | 5.66e-02 | 0.190 | 0.108 | 0.082 | 3.66e-04 |
| ADHD | LDpred2.PseudoVal | lassosum.PseudoVal | 0.114 | 0.080 | 0.034 | 4.45e-03 | 0.190 | 0.108 | 0.082 | 5.76e-04 |
| ADHD | DBSLMM.PseudoVal | lassosum.PseudoVal | 0.110 | 0.080 | 0.030 | 6.19e-03 | 0.174 | 0.108 | 0.066 | 2.97e-03 |
| ADHD | LDpred1.Inf | lassosum.PseudoVal | 0.111 | 0.080 | 0.032 | 1.19e-02 | 0.188 | 0.108 | 0.080 | 1.27e-03 |
| ADHD | LDpred2.Inf | lassosum.PseudoVal | 0.113 | 0.080 | 0.033 | 8.85e-03 | 0.189 | 0.108 | 0.081 | 1.12e-03 |
| ADHD | lassosum.MultiPRS | lassosum.PseudoVal | 0.117 | 0.080 | 0.038 | 3.46e-02 | 0.190 | 0.108 | 0.083 | 3.77e-05 |
| ADHD | LDpred1.10FCVal | lassosum.PseudoVal | 0.114 | 0.080 | 0.034 | 4.13e-03 | 0.190 | 0.108 | 0.082 | 5.27e-04 |
| ADHD | lassosum.PseudoVal | lassosum.PseudoVal | 0.080 | 0.080 | 0.000 | 1.00e+00 | 0.108 | 0.108 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.MultiPRS | lassosum.PseudoVal | 0.113 | 0.080 | 0.033 | 6.24e-02 | 0.190 | 0.108 | 0.082 | 1.21e-04 |
| ADHD | pT+clump.10FCVal | lassosum.PseudoVal | 0.092 | 0.080 | 0.013 | 3.45e-01 | 0.144 | 0.108 | 0.036 | 1.77e-01 |
| ADHD | lassosum.10FCVal | lassosum.PseudoVal | 0.115 | 0.080 | 0.035 | 3.37e-03 | 0.186 | 0.108 | 0.078 | 9.92e-04 |
| ADHD | LDpred2.10FCVal | lassosum.PseudoVal | 0.117 | 0.080 | 0.037 | 6.55e-04 | 0.187 | 0.108 | 0.079 | 1.96e-04 |
| ADHD | SBayesR.PseudoVal | lassosum.PseudoVal | 0.111 | 0.080 | 0.032 | 1.60e-03 | 0.189 | 0.108 | 0.081 | 5.62e-05 |
| ADHD | SBLUP.Inf | lassosum.PseudoVal | 0.112 | 0.080 | 0.032 | 9.88e-03 | 0.188 | 0.108 | 0.080 | 1.22e-03 |
| ADHD | pT+clump.MultiPRS | lassosum.PseudoVal | 0.093 | 0.080 | 0.014 | 4.44e-01 | 0.151 | 0.108 | 0.043 | 7.00e-02 |
| ADHD | All.MultiPRS | lassosum.10FCVal | 0.118 | 0.115 | 0.003 | 8.80e-01 | 0.182 | 0.186 | -0.004 | 7.23e-01 |
| ADHD | LDpred1.MultiPRS | lassosum.10FCVal | 0.113 | 0.115 | -0.002 | 9.23e-01 | 0.190 | 0.186 | 0.004 | 6.35e-01 |
| ADHD | LDpred1.Inf | lassosum.10FCVal | 0.111 | 0.115 | -0.004 | 4.06e-01 | 0.188 | 0.186 | 0.002 | 8.15e-01 |
| ADHD | LDpred2.Inf | lassosum.10FCVal | 0.113 | 0.115 | -0.002 | 5.91e-01 | 0.189 | 0.186 | 0.003 | 7.12e-01 |
| ADHD | lassosum.MultiPRS | lassosum.10FCVal | 0.117 | 0.115 | 0.002 | 8.95e-01 | 0.190 | 0.186 | 0.005 | 4.70e-01 |
| ADHD | DBSLMM.PseudoVal | lassosum.10FCVal | 0.110 | 0.115 | -0.005 | 5.02e-01 | 0.174 | 0.186 | -0.012 | 3.69e-01 |
| ADHD | lassosum.PseudoVal | lassosum.10FCVal | 0.080 | 0.115 | -0.035 | 3.37e-03 | 0.108 | 0.186 | -0.078 | 9.92e-04 |
| ADHD | LDpred2.MultiPRS | lassosum.10FCVal | 0.113 | 0.115 | -0.002 | 9.02e-01 | 0.190 | 0.186 | 0.004 | 6.09e-01 |
| ADHD | pT+clump.10FCVal | lassosum.10FCVal | 0.092 | 0.115 | -0.022 | 2.72e-03 | 0.144 | 0.186 | -0.042 | 4.85e-03 |
| ADHD | lassosum.10FCVal | lassosum.10FCVal | 0.115 | 0.115 | 0.000 | 1.00e+00 | 0.186 | 0.186 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.10FCVal | lassosum.10FCVal | 0.117 | 0.115 | 0.002 | 6.72e-01 | 0.187 | 0.186 | 0.001 | 8.81e-01 |
| ADHD | SBayesR.PseudoVal | lassosum.10FCVal | 0.111 | 0.115 | -0.003 | 4.91e-01 | 0.189 | 0.186 | 0.003 | 7.80e-01 |
| ADHD | LDpred2.PseudoVal | lassosum.10FCVal | 0.114 | 0.115 | -0.001 | 8.53e-01 | 0.190 | 0.186 | 0.004 | 5.63e-01 |
| ADHD | SBLUP.Inf | lassosum.10FCVal | 0.112 | 0.115 | -0.003 | 4.75e-01 | 0.188 | 0.186 | 0.002 | 7.63e-01 |
| ADHD | pT+clump.MultiPRS | lassosum.10FCVal | 0.093 | 0.115 | -0.022 | 2.20e-01 | 0.151 | 0.186 | -0.035 | 1.52e-02 |
| ADHD | LDpred1.10FCVal | lassosum.10FCVal | 0.114 | 0.115 | -0.001 | 8.32e-01 | 0.190 | 0.186 | 0.004 | 6.08e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.110 | 0.113 | -0.003 | 8.65e-01 | 0.174 | 0.190 | -0.016 | 1.70e-01 |
| ADHD | LDpred2.MultiPRS | LDpred1.MultiPRS | 0.113 | 0.113 | 0.000 | 9.78e-01 | 0.190 | 0.190 | 0.000 | 9.65e-01 |
| ADHD | pT+clump.10FCVal | LDpred1.MultiPRS | 0.092 | 0.113 | -0.021 | 2.36e-01 | 0.144 | 0.190 | -0.046 | 4.68e-03 |
| ADHD | lassosum.10FCVal | LDpred1.MultiPRS | 0.115 | 0.113 | 0.002 | 9.23e-01 | 0.186 | 0.190 | -0.004 | 6.35e-01 |
| ADHD | lassosum.PseudoVal | LDpred1.MultiPRS | 0.080 | 0.113 | -0.033 | 5.66e-02 | 0.108 | 0.190 | -0.082 | 3.66e-04 |
| ADHD | All.MultiPRS | LDpred1.MultiPRS | 0.118 | 0.113 | 0.004 | 8.06e-01 | 0.182 | 0.190 | -0.008 | 4.63e-01 |
| ADHD | LDpred1.MultiPRS | LDpred1.MultiPRS | 0.113 | 0.113 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.PseudoVal | LDpred1.MultiPRS | 0.114 | 0.113 | 0.001 | 9.55e-01 | 0.190 | 0.190 | 0.000 | 9.72e-01 |
| ADHD | SBLUP.Inf | LDpred1.MultiPRS | 0.112 | 0.113 | -0.001 | 9.55e-01 | 0.188 | 0.190 | -0.002 | 7.11e-01 |
| ADHD | LDpred1.Inf | LDpred1.MultiPRS | 0.111 | 0.113 | -0.002 | 9.11e-01 | 0.188 | 0.190 | -0.002 | 6.32e-01 |
| ADHD | LDpred2.Inf | LDpred1.MultiPRS | 0.113 | 0.113 | -0.001 | 9.76e-01 | 0.189 | 0.190 | -0.001 | 8.29e-01 |
| ADHD | lassosum.MultiPRS | LDpred1.MultiPRS | 0.117 | 0.113 | 0.004 | 8.18e-01 | 0.190 | 0.190 | 0.001 | 9.52e-01 |
| ADHD | LDpred1.10FCVal | LDpred1.MultiPRS | 0.114 | 0.113 | 0.001 | 9.62e-01 | 0.190 | 0.190 | 0.000 | 9.71e-01 |
| ADHD | pT+clump.MultiPRS | LDpred1.MultiPRS | 0.093 | 0.113 | -0.020 | 2.61e-01 | 0.151 | 0.190 | -0.039 | 1.11e-02 |
| ADHD | SBayesR.PseudoVal | LDpred1.MultiPRS | 0.111 | 0.113 | -0.002 | 9.19e-01 | 0.189 | 0.190 | -0.001 | 8.57e-01 |
| ADHD | LDpred2.10FCVal | LDpred1.MultiPRS | 0.117 | 0.113 | 0.003 | 8.49e-01 | 0.187 | 0.190 | -0.003 | 5.99e-01 |
| ADHD | All.MultiPRS | LDpred1.Inf | 0.118 | 0.111 | 0.006 | 7.21e-01 | 0.182 | 0.188 | -0.006 | 6.21e-01 |
| ADHD | LDpred1.Inf | LDpred1.Inf | 0.111 | 0.111 | 0.000 | 1.00e+00 | 0.188 | 0.188 | 0.000 | 1.00e+00 |
| ADHD | LDpred2.Inf | LDpred1.Inf | 0.113 | 0.111 | 0.001 | 3.99e-01 | 0.189 | 0.188 | 0.001 | 7.78e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred1.Inf | 0.110 | 0.111 | -0.001 | 8.66e-01 | 0.174 | 0.188 | -0.014 | 2.43e-01 |
| ADHD | lassosum.PseudoVal | LDpred1.Inf | 0.080 | 0.111 | -0.032 | 1.19e-02 | 0.108 | 0.188 | -0.080 | 1.27e-03 |
| ADHD | LDpred2.MultiPRS | LDpred1.Inf | 0.113 | 0.111 | 0.001 | 9.34e-01 | 0.190 | 0.188 | 0.002 | 7.64e-01 |
| ADHD | pT+clump.10FCVal | LDpred1.Inf | 0.092 | 0.111 | -0.019 | 2.00e-02 | 0.144 | 0.188 | -0.044 | 6.02e-03 |
| ADHD | lassosum.10FCVal | LDpred1.Inf | 0.115 | 0.111 | 0.004 | 4.06e-01 | 0.186 | 0.188 | -0.002 | 8.15e-01 |
| ADHD | LDpred2.10FCVal | LDpred1.Inf | 0.117 | 0.111 | 0.005 | 1.08e-01 | 0.187 | 0.188 | -0.001 | 8.93e-01 |
| ADHD | SBayesR.PseudoVal | LDpred1.Inf | 0.111 | 0.111 | 0.000 | 9.67e-01 | 0.189 | 0.188 | 0.001 | 9.33e-01 |
| ADHD | LDpred1.MultiPRS | LDpred1.Inf | 0.113 | 0.111 | 0.002 | 9.11e-01 | 0.190 | 0.188 | 0.002 | 6.32e-01 |
| ADHD | LDpred2.PseudoVal | LDpred1.Inf | 0.114 | 0.111 | 0.003 | 1.14e-01 | 0.190 | 0.188 | 0.002 | 5.51e-01 |
| ADHD | SBLUP.Inf | LDpred1.Inf | 0.112 | 0.111 | 0.001 | 5.89e-01 | 0.188 | 0.188 | 0.000 | 9.61e-01 |
| ADHD | pT+clump.MultiPRS | LDpred1.Inf | 0.093 | 0.111 | -0.018 | 3.08e-01 | 0.151 | 0.188 | -0.037 | 1.86e-02 |
| ADHD | lassosum.MultiPRS | LDpred1.Inf | 0.117 | 0.111 | 0.006 | 7.37e-01 | 0.190 | 0.188 | 0.003 | 7.91e-01 |
| ADHD | LDpred1.10FCVal | LDpred1.Inf | 0.114 | 0.111 | 0.003 | 5.98e-02 | 0.190 | 0.188 | 0.002 | 4.54e-01 |
| ADHD | LDpred1.Inf | LDpred1.10FCVal | 0.111 | 0.114 | -0.003 | 5.98e-02 | 0.188 | 0.190 | -0.002 | 4.54e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred1.10FCVal | 0.110 | 0.114 | -0.004 | 5.12e-01 | 0.174 | 0.190 | -0.016 | 1.63e-01 |
| ADHD | lassosum.PseudoVal | LDpred1.10FCVal | 0.080 | 0.114 | -0.034 | 4.13e-03 | 0.108 | 0.190 | -0.082 | 5.27e-04 |
| ADHD | LDpred2.MultiPRS | LDpred1.10FCVal | 0.113 | 0.114 | -0.001 | 9.41e-01 | 0.190 | 0.190 | 0.000 | 9.47e-01 |
| ADHD | pT+clump.10FCVal | LDpred1.10FCVal | 0.092 | 0.114 | -0.022 | 7.45e-03 | 0.144 | 0.190 | -0.046 | 4.07e-03 |
| ADHD | lassosum.10FCVal | LDpred1.10FCVal | 0.115 | 0.114 | 0.001 | 8.32e-01 | 0.186 | 0.190 | -0.004 | 6.08e-01 |
| ADHD | LDpred2.10FCVal | LDpred1.10FCVal | 0.117 | 0.114 | 0.003 | 3.47e-01 | 0.187 | 0.190 | -0.003 | 5.78e-01 |
| ADHD | All.MultiPRS | LDpred1.10FCVal | 0.118 | 0.114 | 0.004 | 8.41e-01 | 0.182 | 0.190 | -0.008 | 4.74e-01 |
| ADHD | LDpred1.MultiPRS | LDpred1.10FCVal | 0.113 | 0.114 | -0.001 | 9.62e-01 | 0.190 | 0.190 | 0.000 | 9.71e-01 |
| ADHD | LDpred2.PseudoVal | LDpred1.10FCVal | 0.114 | 0.114 | 0.000 | 9.26e-01 | 0.190 | 0.190 | 0.000 | 9.86e-01 |
| ADHD | SBLUP.Inf | LDpred1.10FCVal | 0.112 | 0.114 | -0.002 | 3.55e-01 | 0.188 | 0.190 | -0.002 | 6.08e-01 |
| ADHD | pT+clump.MultiPRS | LDpred1.10FCVal | 0.093 | 0.114 | -0.021 | 2.39e-01 | 0.151 | 0.190 | -0.040 | 1.08e-02 |
| ADHD | LDpred2.Inf | LDpred1.10FCVal | 0.113 | 0.114 | -0.001 | 4.91e-01 | 0.189 | 0.190 | -0.001 | 7.55e-01 |
| ADHD | lassosum.MultiPRS | LDpred1.10FCVal | 0.117 | 0.114 | 0.003 | 8.57e-01 | 0.190 | 0.190 | 0.000 | 9.62e-01 |
| ADHD | LDpred1.10FCVal | LDpred1.10FCVal | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| ADHD | SBayesR.PseudoVal | LDpred1.10FCVal | 0.111 | 0.114 | -0.003 | 4.93e-01 | 0.189 | 0.190 | -0.001 | 8.48e-01 |
| ADHD | LDpred2.MultiPRS | LDpred2.MultiPRS | 0.113 | 0.113 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| ADHD | lassosum.PseudoVal | LDpred2.MultiPRS | 0.080 | 0.113 | -0.033 | 6.24e-02 | 0.108 | 0.190 | -0.082 | 1.21e-04 |
| ADHD | All.MultiPRS | LDpred2.MultiPRS | 0.118 | 0.113 | 0.005 | 7.82e-01 | 0.182 | 0.190 | -0.008 | 4.48e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.MultiPRS | 0.113 | 0.113 | 0.000 | 9.78e-01 | 0.190 | 0.190 | 0.000 | 9.65e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.110 | 0.113 | -0.002 | 8.88e-01 | 0.174 | 0.190 | -0.016 | 1.43e-01 |
| ADHD | LDpred1.Inf | LDpred2.MultiPRS | 0.111 | 0.113 | -0.001 | 9.34e-01 | 0.188 | 0.190 | -0.002 | 7.64e-01 |
| ADHD | LDpred2.Inf | LDpred2.MultiPRS | 0.113 | 0.113 | 0.000 | 9.98e-01 | 0.189 | 0.190 | -0.001 | 8.62e-01 |
| ADHD | lassosum.MultiPRS | LDpred2.MultiPRS | 0.117 | 0.113 | 0.005 | 2.20e-01 | 0.190 | 0.190 | 0.001 | 9.14e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.MultiPRS | 0.114 | 0.113 | 0.001 | 9.41e-01 | 0.190 | 0.190 | 0.000 | 9.47e-01 |
| ADHD | pT+clump.MultiPRS | LDpred2.MultiPRS | 0.093 | 0.113 | -0.019 | 2.71e-01 | 0.151 | 0.190 | -0.039 | 8.00e-03 |
| ADHD | SBayesR.PseudoVal | LDpred2.MultiPRS | 0.111 | 0.113 | -0.001 | 9.42e-01 | 0.189 | 0.190 | -0.001 | 8.55e-01 |
| ADHD | pT+clump.10FCVal | LDpred2.MultiPRS | 0.092 | 0.113 | -0.020 | 2.54e-01 | 0.144 | 0.190 | -0.046 | 4.49e-03 |
| ADHD | lassosum.10FCVal | LDpred2.MultiPRS | 0.115 | 0.113 | 0.002 | 9.02e-01 | 0.186 | 0.190 | -0.004 | 6.09e-01 |
| ADHD | LDpred2.10FCVal | LDpred2.MultiPRS | 0.117 | 0.113 | 0.004 | 8.30e-01 | 0.187 | 0.190 | -0.003 | 4.19e-02 |
| ADHD | LDpred2.PseudoVal | LDpred2.MultiPRS | 0.114 | 0.113 | 0.001 | 9.34e-01 | 0.190 | 0.190 | 0.000 | 9.15e-01 |
| ADHD | SBLUP.Inf | LDpred2.MultiPRS | 0.112 | 0.113 | -0.001 | 9.77e-01 | 0.188 | 0.190 | -0.002 | 7.53e-01 |
| ADHD | LDpred2.MultiPRS | LDpred2.10FCVal | 0.113 | 0.117 | -0.004 | 8.30e-01 | 0.190 | 0.187 | 0.003 | 4.19e-02 |
| ADHD | pT+clump.10FCVal | LDpred2.10FCVal | 0.092 | 0.117 | -0.024 | 3.07e-03 | 0.144 | 0.187 | -0.043 | 7.38e-03 |
| ADHD | lassosum.PseudoVal | LDpred2.10FCVal | 0.080 | 0.117 | -0.037 | 6.55e-04 | 0.108 | 0.187 | -0.079 | 1.96e-04 |
| ADHD | All.MultiPRS | LDpred2.10FCVal | 0.118 | 0.117 | 0.001 | 9.54e-01 | 0.182 | 0.187 | -0.005 | 6.28e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.10FCVal | 0.113 | 0.117 | -0.003 | 8.49e-01 | 0.190 | 0.187 | 0.003 | 5.99e-01 |
| ADHD | LDpred2.PseudoVal | LDpred2.10FCVal | 0.114 | 0.117 | -0.002 | 2.79e-01 | 0.190 | 0.187 | 0.003 | 4.67e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred2.10FCVal | 0.110 | 0.117 | -0.006 | 2.74e-01 | 0.174 | 0.187 | -0.013 | 2.41e-01 |
| ADHD | LDpred1.Inf | LDpred2.10FCVal | 0.111 | 0.117 | -0.005 | 1.08e-01 | 0.188 | 0.187 | 0.001 | 8.93e-01 |
| ADHD | LDpred2.Inf | LDpred2.10FCVal | 0.113 | 0.117 | -0.004 | 1.77e-01 | 0.189 | 0.187 | 0.002 | 7.46e-01 |
| ADHD | lassosum.MultiPRS | LDpred2.10FCVal | 0.117 | 0.117 | 0.001 | 9.68e-01 | 0.190 | 0.187 | 0.004 | 6.24e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.10FCVal | 0.114 | 0.117 | -0.003 | 3.47e-01 | 0.190 | 0.187 | 0.003 | 5.78e-01 |
| ADHD | pT+clump.MultiPRS | LDpred2.10FCVal | 0.093 | 0.117 | -0.023 | 1.87e-01 | 0.151 | 0.187 | -0.036 | 1.36e-02 |
| ADHD | SBayesR.PseudoVal | LDpred2.10FCVal | 0.111 | 0.117 | -0.005 | 1.09e-01 | 0.189 | 0.187 | 0.002 | 7.99e-01 |
| ADHD | lassosum.10FCVal | LDpred2.10FCVal | 0.115 | 0.117 | -0.002 | 6.72e-01 | 0.186 | 0.187 | -0.001 | 8.81e-01 |
| ADHD | LDpred2.10FCVal | LDpred2.10FCVal | 0.117 | 0.117 | 0.000 | 1.00e+00 | 0.187 | 0.187 | 0.000 | 1.00e+00 |
| ADHD | SBLUP.Inf | LDpred2.10FCVal | 0.112 | 0.117 | -0.004 | 1.38e-01 | 0.188 | 0.187 | 0.001 | 8.53e-01 |
| ADHD | LDpred2.MultiPRS | LDpred2.PseudoVal | 0.113 | 0.114 | -0.001 | 9.34e-01 | 0.190 | 0.190 | 0.000 | 9.15e-01 |
| ADHD | lassosum.PseudoVal | LDpred2.PseudoVal | 0.080 | 0.114 | -0.034 | 4.45e-03 | 0.108 | 0.190 | -0.082 | 5.76e-04 |
| ADHD | All.MultiPRS | LDpred2.PseudoVal | 0.118 | 0.114 | 0.003 | 8.49e-01 | 0.182 | 0.190 | -0.008 | 4.55e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.PseudoVal | 0.113 | 0.114 | -0.001 | 9.55e-01 | 0.190 | 0.190 | 0.000 | 9.72e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.110 | 0.114 | -0.004 | 4.90e-01 | 0.174 | 0.190 | -0.017 | 1.48e-01 |
| ADHD | LDpred1.Inf | LDpred2.PseudoVal | 0.111 | 0.114 | -0.003 | 1.14e-01 | 0.188 | 0.190 | -0.002 | 5.51e-01 |
| ADHD | LDpred2.Inf | LDpred2.PseudoVal | 0.113 | 0.114 | -0.002 | 4.74e-02 | 0.189 | 0.190 | -0.001 | 3.99e-01 |
| ADHD | lassosum.MultiPRS | LDpred2.PseudoVal | 0.117 | 0.114 | 0.003 | 8.64e-01 | 0.190 | 0.190 | 0.000 | 9.66e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.PseudoVal | 0.114 | 0.114 | 0.000 | 9.26e-01 | 0.190 | 0.190 | 0.000 | 9.86e-01 |
| ADHD | pT+clump.MultiPRS | LDpred2.PseudoVal | 0.093 | 0.114 | -0.021 | 2.36e-01 | 0.151 | 0.190 | -0.040 | 9.04e-03 |
| ADHD | SBayesR.PseudoVal | LDpred2.PseudoVal | 0.111 | 0.114 | -0.003 | 4.92e-01 | 0.189 | 0.190 | -0.002 | 8.48e-01 |
| ADHD | pT+clump.10FCVal | LDpred2.PseudoVal | 0.092 | 0.114 | -0.022 | 6.31e-03 | 0.144 | 0.190 | -0.046 | 3.18e-03 |
| ADHD | lassosum.10FCVal | LDpred2.PseudoVal | 0.115 | 0.114 | 0.001 | 8.53e-01 | 0.186 | 0.190 | -0.004 | 5.63e-01 |
| ADHD | LDpred2.10FCVal | LDpred2.PseudoVal | 0.117 | 0.114 | 0.002 | 2.79e-01 | 0.187 | 0.190 | -0.003 | 4.67e-01 |
| ADHD | LDpred2.PseudoVal | LDpred2.PseudoVal | 0.114 | 0.114 | 0.000 | 1.00e+00 | 0.190 | 0.190 | 0.000 | 1.00e+00 |
| ADHD | SBLUP.Inf | LDpred2.PseudoVal | 0.112 | 0.114 | -0.002 | 8.47e-02 | 0.188 | 0.190 | -0.002 | 3.58e-01 |
| ADHD | DBSLMM.PseudoVal | LDpred2.Inf | 0.110 | 0.113 | -0.002 | 6.80e-01 | 0.174 | 0.189 | -0.015 | 1.96e-01 |
| ADHD | LDpred2.MultiPRS | LDpred2.Inf | 0.113 | 0.113 | 0.000 | 9.98e-01 | 0.190 | 0.189 | 0.001 | 8.62e-01 |
| ADHD | pT+clump.10FCVal | LDpred2.Inf | 0.092 | 0.113 | -0.020 | 1.19e-02 | 0.144 | 0.189 | -0.045 | 4.49e-03 |
| ADHD | lassosum.10FCVal | LDpred2.Inf | 0.115 | 0.113 | 0.002 | 5.91e-01 | 0.186 | 0.189 | -0.003 | 7.12e-01 |
| ADHD | lassosum.PseudoVal | LDpred2.Inf | 0.080 | 0.113 | -0.033 | 8.85e-03 | 0.108 | 0.189 | -0.081 | 1.12e-03 |
| ADHD | All.MultiPRS | LDpred2.Inf | 0.118 | 0.113 | 0.005 | 7.82e-01 | 0.182 | 0.189 | -0.007 | 5.52e-01 |
| ADHD | LDpred1.MultiPRS | LDpred2.Inf | 0.113 | 0.113 | 0.001 | 9.76e-01 | 0.190 | 0.189 | 0.001 | 8.29e-01 |
| ADHD | LDpred2.PseudoVal | LDpred2.Inf | 0.114 | 0.113 | 0.002 | 4.74e-02 | 0.190 | 0.189 | 0.001 | 3.99e-01 |
| ADHD | SBLUP.Inf | LDpred2.Inf | 0.112 | 0.113 | 0.000 | 6.55e-01 | 0.188 | 0.189 | -0.001 | 6.98e-01 |
| ADHD | LDpred1.Inf | LDpred2.Inf | 0.111 | 0.113 | -0.001 | 3.99e-01 | 0.188 | 0.189 | -0.001 | 7.78e-01 |
| ADHD | LDpred2.Inf | LDpred2.Inf | 0.113 | 0.113 | 0.000 | 1.00e+00 | 0.189 | 0.189 | 0.000 | 1.00e+00 |
| ADHD | lassosum.MultiPRS | LDpred2.Inf | 0.117 | 0.113 | 0.005 | 7.98e-01 | 0.190 | 0.189 | 0.002 | 8.60e-01 |
| ADHD | LDpred1.10FCVal | LDpred2.Inf | 0.114 | 0.113 | 0.001 | 4.91e-01 | 0.190 | 0.189 | 0.001 | 7.55e-01 |
| ADHD | pT+clump.MultiPRS | LDpred2.Inf | 0.093 | 0.113 | -0.019 | 2.72e-01 | 0.151 | 0.189 | -0.038 | 1.42e-02 |
| ADHD | SBayesR.PseudoVal | LDpred2.Inf | 0.111 | 0.113 | -0.001 | 7.83e-01 | 0.189 | 0.189 | 0.000 | 9.80e-01 |
| ADHD | LDpred2.10FCVal | LDpred2.Inf | 0.117 | 0.113 | 0.004 | 1.77e-01 | 0.187 | 0.189 | -0.002 | 7.46e-01 |
| ADHD | All.MultiPRS | pT+clump.MultiPRS | 0.118 | 0.093 | 0.024 | 1.65e-01 | 0.182 | 0.151 | 0.031 | 5.79e-02 |
| ADHD | LDpred1.MultiPRS | pT+clump.MultiPRS | 0.113 | 0.093 | 0.020 | 2.61e-01 | 0.190 | 0.151 | 0.039 | 1.11e-02 |
| ADHD | LDpred2.PseudoVal | pT+clump.MultiPRS | 0.114 | 0.093 | 0.021 | 2.36e-01 | 0.190 | 0.151 | 0.040 | 9.04e-03 |
| ADHD | DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.110 | 0.093 | 0.017 | 3.37e-01 | 0.174 | 0.151 | 0.023 | 1.58e-01 |
| ADHD | LDpred1.Inf | pT+clump.MultiPRS | 0.111 | 0.093 | 0.018 | 3.08e-01 | 0.188 | 0.151 | 0.037 | 1.86e-02 |
| ADHD | LDpred2.Inf | pT+clump.MultiPRS | 0.113 | 0.093 | 0.019 | 2.72e-01 | 0.189 | 0.151 | 0.038 | 1.42e-02 |
| ADHD | lassosum.MultiPRS | pT+clump.MultiPRS | 0.117 | 0.093 | 0.024 | 1.74e-01 | 0.190 | 0.151 | 0.040 | 3.71e-03 |
| ADHD | LDpred1.10FCVal | pT+clump.MultiPRS | 0.114 | 0.093 | 0.021 | 2.39e-01 | 0.190 | 0.151 | 0.040 | 1.08e-02 |
| ADHD | lassosum.PseudoVal | pT+clump.MultiPRS | 0.080 | 0.093 | -0.014 | 4.44e-01 | 0.108 | 0.151 | -0.043 | 7.00e-02 |
| ADHD | LDpred2.MultiPRS | pT+clump.MultiPRS | 0.113 | 0.093 | 0.019 | 2.71e-01 | 0.190 | 0.151 | 0.039 | 8.00e-03 |
| ADHD | pT+clump.10FCVal | pT+clump.MultiPRS | 0.092 | 0.093 | -0.001 | 9.62e-01 | 0.144 | 0.151 | -0.007 | 3.29e-01 |
| ADHD | lassosum.10FCVal | pT+clump.MultiPRS | 0.115 | 0.093 | 0.022 | 2.20e-01 | 0.186 | 0.151 | 0.035 | 1.52e-02 |
| ADHD | LDpred2.10FCVal | pT+clump.MultiPRS | 0.117 | 0.093 | 0.023 | 1.87e-01 | 0.187 | 0.151 | 0.036 | 1.36e-02 |
| ADHD | SBayesR.PseudoVal | pT+clump.MultiPRS | 0.111 | 0.093 | 0.018 | 3.03e-01 | 0.189 | 0.151 | 0.038 | 1.58e-02 |
| ADHD | SBLUP.Inf | pT+clump.MultiPRS | 0.112 | 0.093 | 0.019 | 2.83e-01 | 0.188 | 0.151 | 0.038 | 1.28e-02 |
| ADHD | pT+clump.MultiPRS | pT+clump.MultiPRS | 0.093 | 0.093 | 0.000 | 1.00e+00 | 0.151 | 0.151 | 0.000 | 1.00e+00 |
| ADHD | lassosum.PseudoVal | pT+clump.10FCVal | 0.080 | 0.092 | -0.013 | 3.45e-01 | 0.108 | 0.144 | -0.036 | 1.77e-01 |
| ADHD | All.MultiPRS | pT+clump.10FCVal | 0.118 | 0.092 | 0.025 | 1.53e-01 | 0.182 | 0.144 | 0.038 | 4.18e-02 |
| ADHD | DBSLMM.PseudoVal | pT+clump.10FCVal | 0.110 | 0.092 | 0.018 | 6.69e-02 | 0.174 | 0.144 | 0.030 | 1.22e-01 |
| ADHD | LDpred1.Inf | pT+clump.10FCVal | 0.111 | 0.092 | 0.019 | 2.00e-02 | 0.188 | 0.144 | 0.044 | 6.02e-03 |
| ADHD | LDpred2.Inf | pT+clump.10FCVal | 0.113 | 0.092 | 0.020 | 1.19e-02 | 0.189 | 0.144 | 0.045 | 4.49e-03 |
| ADHD | lassosum.MultiPRS | pT+clump.10FCVal | 0.117 | 0.092 | 0.025 | 1.63e-01 | 0.190 | 0.144 | 0.047 | 4.28e-03 |
| ADHD | LDpred1.10FCVal | pT+clump.10FCVal | 0.114 | 0.092 | 0.022 | 7.45e-03 | 0.190 | 0.144 | 0.046 | 4.07e-03 |
| ADHD | pT+clump.MultiPRS | pT+clump.10FCVal | 0.093 | 0.092 | 0.001 | 9.62e-01 | 0.151 | 0.144 | 0.007 | 3.29e-01 |
| ADHD | LDpred2.MultiPRS | pT+clump.10FCVal | 0.113 | 0.092 | 0.020 | 2.54e-01 | 0.190 | 0.144 | 0.046 | 4.49e-03 |
| ADHD | pT+clump.10FCVal | pT+clump.10FCVal | 0.092 | 0.092 | 0.000 | 1.00e+00 | 0.144 | 0.144 | 0.000 | 1.00e+00 |
| ADHD | lassosum.10FCVal | pT+clump.10FCVal | 0.115 | 0.092 | 0.022 | 2.72e-03 | 0.186 | 0.144 | 0.042 | 4.85e-03 |
| ADHD | LDpred2.10FCVal | pT+clump.10FCVal | 0.117 | 0.092 | 0.024 | 3.07e-03 | 0.187 | 0.144 | 0.043 | 7.38e-03 |
| ADHD | SBayesR.PseudoVal | pT+clump.10FCVal | 0.111 | 0.092 | 0.019 | 3.36e-02 | 0.189 | 0.144 | 0.045 | 1.09e-02 |
| ADHD | LDpred1.MultiPRS | pT+clump.10FCVal | 0.113 | 0.092 | 0.021 | 2.36e-01 | 0.190 | 0.144 | 0.046 | 4.68e-03 |
| ADHD | LDpred2.PseudoVal | pT+clump.10FCVal | 0.114 | 0.092 | 0.022 | 6.31e-03 | 0.190 | 0.144 | 0.046 | 3.18e-03 |
| ADHD | SBLUP.Inf | pT+clump.10FCVal | 0.112 | 0.092 | 0.020 | 1.02e-02 | 0.188 | 0.144 | 0.044 | 3.63e-03 |
| ADHD | LDpred1.Inf | SBayesR.PseudoVal | 0.111 | 0.111 | 0.000 | 9.67e-01 | 0.188 | 0.189 | -0.001 | 9.33e-01 |
| ADHD | DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.110 | 0.111 | -0.001 | 8.40e-01 | 0.174 | 0.189 | -0.015 | 2.03e-01 |
| ADHD | lassosum.PseudoVal | SBayesR.PseudoVal | 0.080 | 0.111 | -0.032 | 1.60e-03 | 0.108 | 0.189 | -0.081 | 5.62e-05 |
| ADHD | LDpred2.MultiPRS | SBayesR.PseudoVal | 0.113 | 0.111 | 0.001 | 9.42e-01 | 0.190 | 0.189 | 0.001 | 8.55e-01 |
| ADHD | pT+clump.10FCVal | SBayesR.PseudoVal | 0.092 | 0.111 | -0.019 | 3.36e-02 | 0.144 | 0.189 | -0.045 | 1.09e-02 |
| ADHD | lassosum.10FCVal | SBayesR.PseudoVal | 0.115 | 0.111 | 0.003 | 4.91e-01 | 0.186 | 0.189 | -0.003 | 7.80e-01 |
| ADHD | LDpred2.10FCVal | SBayesR.PseudoVal | 0.117 | 0.111 | 0.005 | 1.09e-01 | 0.187 | 0.189 | -0.002 | 7.99e-01 |
| ADHD | All.MultiPRS | SBayesR.PseudoVal | 0.118 | 0.111 | 0.006 | 7.28e-01 | 0.182 | 0.189 | -0.007 | 5.57e-01 |
| ADHD | LDpred1.MultiPRS | SBayesR.PseudoVal | 0.113 | 0.111 | 0.002 | 9.19e-01 | 0.190 | 0.189 | 0.001 | 8.57e-01 |
| ADHD | LDpred2.PseudoVal | SBayesR.PseudoVal | 0.114 | 0.111 | 0.003 | 4.92e-01 | 0.190 | 0.189 | 0.002 | 8.48e-01 |
| ADHD | SBLUP.Inf | SBayesR.PseudoVal | 0.112 | 0.111 | 0.001 | 8.64e-01 | 0.188 | 0.189 | -0.001 | 9.50e-01 |
| ADHD | pT+clump.MultiPRS | SBayesR.PseudoVal | 0.093 | 0.111 | -0.018 | 3.03e-01 | 0.151 | 0.189 | -0.038 | 1.58e-02 |
| ADHD | LDpred2.Inf | SBayesR.PseudoVal | 0.113 | 0.111 | 0.001 | 7.83e-01 | 0.189 | 0.189 | 0.000 | 9.80e-01 |
| ADHD | lassosum.MultiPRS | SBayesR.PseudoVal | 0.117 | 0.111 | 0.006 | 7.44e-01 | 0.190 | 0.189 | 0.002 | 8.22e-01 |
| ADHD | LDpred1.10FCVal | SBayesR.PseudoVal | 0.114 | 0.111 | 0.003 | 4.93e-01 | 0.190 | 0.189 | 0.001 | 8.48e-01 |
| ADHD | SBayesR.PseudoVal | SBayesR.PseudoVal | 0.111 | 0.111 | 0.000 | 1.00e+00 | 0.189 | 0.189 | 0.000 | 1.00e+00 |
| ADHD | All.MultiPRS | SBLUP.Inf | 0.118 | 0.112 | 0.005 | 7.62e-01 | 0.182 | 0.188 | -0.006 | 5.97e-01 |
| ADHD | LDpred1.Inf | SBLUP.Inf | 0.111 | 0.112 | -0.001 | 5.89e-01 | 0.188 | 0.188 | 0.000 | 9.61e-01 |
| ADHD | LDpred2.Inf | SBLUP.Inf | 0.113 | 0.112 | 0.000 | 6.55e-01 | 0.189 | 0.188 | 0.001 | 6.98e-01 |
| ADHD | DBSLMM.PseudoVal | SBLUP.Inf | 0.110 | 0.112 | -0.002 | 7.41e-01 | 0.174 | 0.188 | -0.014 | 2.26e-01 |
| ADHD | lassosum.PseudoVal | SBLUP.Inf | 0.080 | 0.112 | -0.032 | 9.88e-03 | 0.108 | 0.188 | -0.080 | 1.22e-03 |
| ADHD | LDpred2.MultiPRS | SBLUP.Inf | 0.113 | 0.112 | 0.001 | 9.77e-01 | 0.190 | 0.188 | 0.002 | 7.53e-01 |
| ADHD | pT+clump.10FCVal | SBLUP.Inf | 0.092 | 0.112 | -0.020 | 1.02e-02 | 0.144 | 0.188 | -0.044 | 3.63e-03 |
| ADHD | lassosum.10FCVal | SBLUP.Inf | 0.115 | 0.112 | 0.003 | 4.75e-01 | 0.186 | 0.188 | -0.002 | 7.63e-01 |
| ADHD | LDpred2.10FCVal | SBLUP.Inf | 0.117 | 0.112 | 0.004 | 1.38e-01 | 0.187 | 0.188 | -0.001 | 8.53e-01 |
| ADHD | SBayesR.PseudoVal | SBLUP.Inf | 0.111 | 0.112 | -0.001 | 8.64e-01 | 0.189 | 0.188 | 0.001 | 9.50e-01 |
| ADHD | LDpred1.MultiPRS | SBLUP.Inf | 0.113 | 0.112 | 0.001 | 9.55e-01 | 0.190 | 0.188 | 0.002 | 7.11e-01 |
| ADHD | LDpred2.PseudoVal | SBLUP.Inf | 0.114 | 0.112 | 0.002 | 8.47e-02 | 0.190 | 0.188 | 0.002 | 3.58e-01 |
| ADHD | SBLUP.Inf | SBLUP.Inf | 0.112 | 0.112 | 0.000 | 1.00e+00 | 0.188 | 0.188 | 0.000 | 1.00e+00 |
| ADHD | pT+clump.MultiPRS | SBLUP.Inf | 0.093 | 0.112 | -0.019 | 2.83e-01 | 0.151 | 0.188 | -0.038 | 1.28e-02 |
| ADHD | lassosum.MultiPRS | SBLUP.Inf | 0.117 | 0.112 | 0.005 | 7.78e-01 | 0.190 | 0.188 | 0.002 | 7.87e-01 |
| ADHD | LDpred1.10FCVal | SBLUP.Inf | 0.114 | 0.112 | 0.002 | 3.55e-01 | 0.190 | 0.188 | 0.002 | 6.08e-01 |
Show cross-validation differences between methods
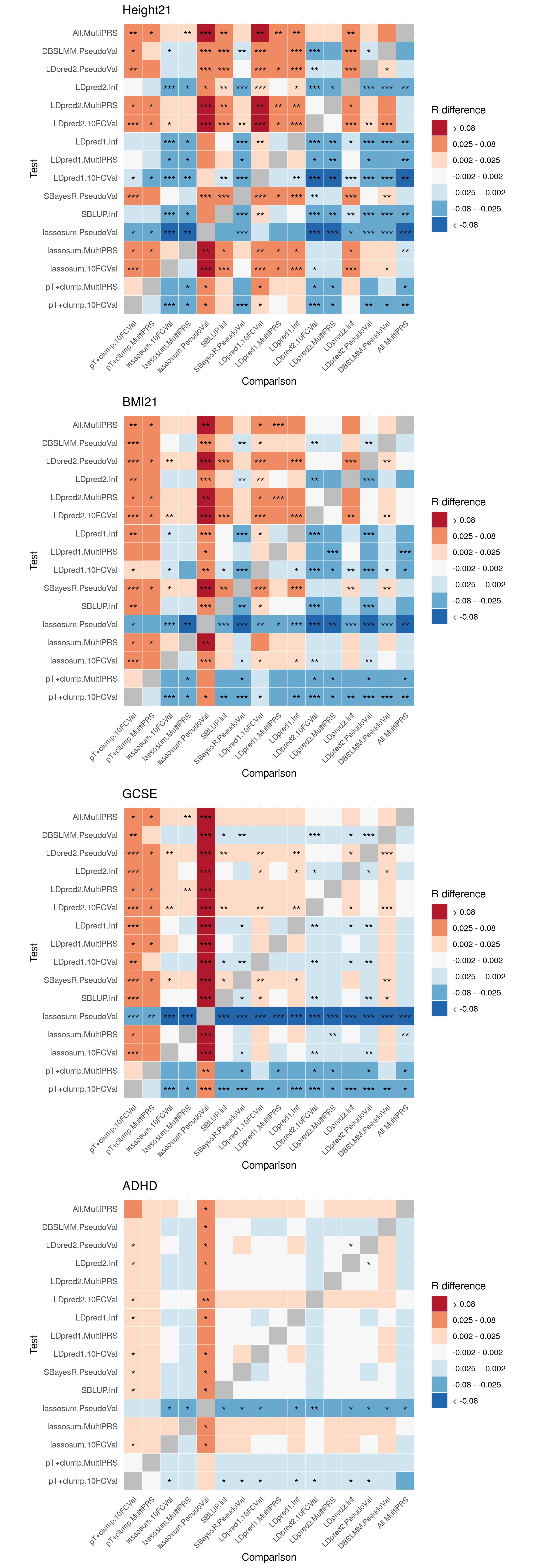
Predictive utility estimated using 10-fold cross-validation
Show test-validation differences between methods
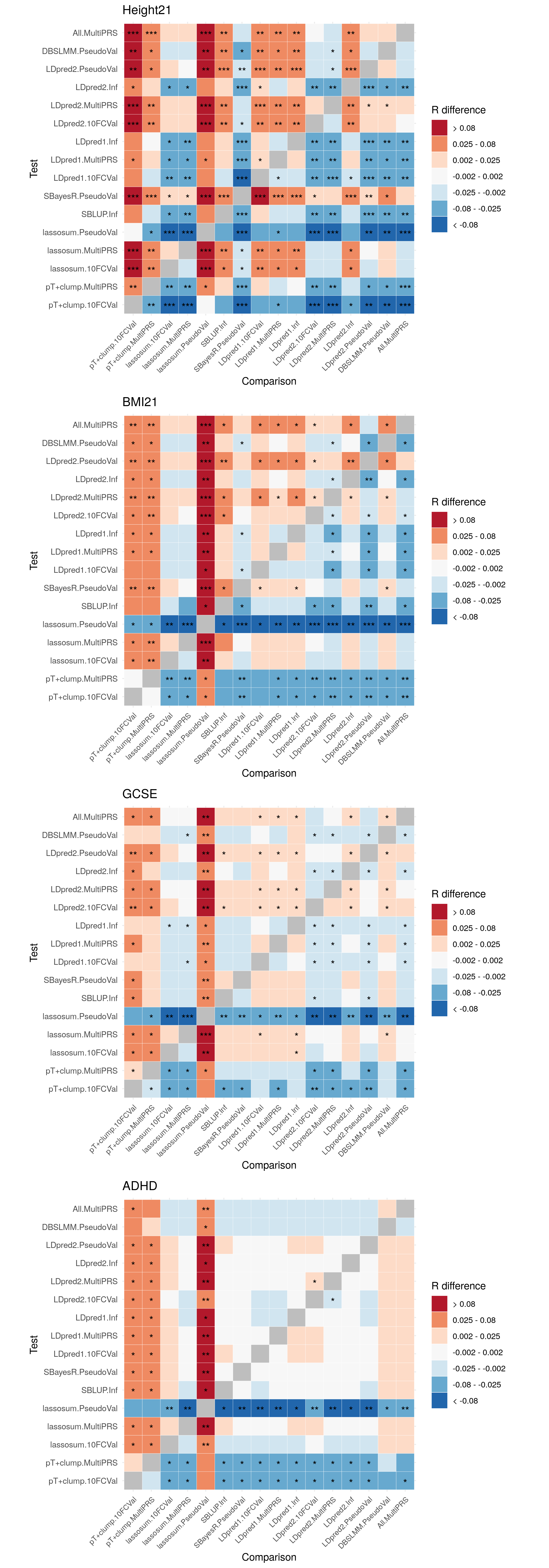
Predictive utility estimated using test set validation
4.3 Further analysis of GCTB
Show SBayesR approaches used
| GWAS | Reference | Process | h2 (SE) | perVariant | Complete | Converge | Used |
|---|---|---|---|---|---|---|---|
| DEPR06 | 1KG | Original | 0.035 (0.002) | TRUE | TRUE | TRUE | TRUE |
| DEPR06 | 1KG | P<0.4 | 0.092 (0.003) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | 1KG | v2.03 | 0.035 (0.002) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | 1KG | v2.03 robust | 0.043 (0.003) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | UKB | Original | 0.057 (0.002) | TRUE | TRUE | TRUE | TRUE |
| DEPR06 | UKB | P<0.4 | 0.138 (0.003) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | UKB | v2.03 | 0.057 (0.002) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | UKB | v2.03 robust | 0.057 (0.002) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | GCTB | Original | 0.058 (0.002) | TRUE | TRUE | TRUE | TRUE |
| DEPR06 | GCTB | P<0.4 | 0.14 (0.003) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | GCTB | v2.03 | 0.059 (0.002) | TRUE | TRUE | TRUE | FALSE |
| DEPR06 | GCTB | v2.03 robust | 0.058 (0.002) | TRUE | TRUE | TRUE | FALSE |
| COLL01 | 1KG | Original | 0.113 (0.007) | FALSE | TRUE | TRUE | TRUE |
| COLL01 | 1KG | P<0.4 | 0.343 (0.01) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | 1KG | v2.03 | 0.115 (0.007) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | 1KG | v2.03 robust | 0.112 (0.031) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | UKB | Original | 0.162 (0.007) | FALSE | TRUE | TRUE | TRUE |
| COLL01 | UKB | P<0.4 | 0.504 (0.013) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | UKB | v2.03 | 0.165 (0.008) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | UKB | v2.03 robust | 0.141 (0.006) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | GCTB | Original | 0.168 (0.007) | FALSE | TRUE | TRUE | TRUE |
| COLL01 | GCTB | P<0.4 | 0.531 (0.014) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | GCTB | v2.03 | 0.171 (0.007) | FALSE | TRUE | TRUE | FALSE |
| COLL01 | GCTB | v2.03 robust | 0.146 (0.007) | FALSE | TRUE | TRUE | FALSE |
| BODY04 | 1KG | Original | 0.248 (0.059) | TRUE | TRUE | TRUE | TRUE |
| BODY04 | 1KG | P<0.4 | 0.192 (0.016) | TRUE | TRUE | TRUE | FALSE |
| BODY04 | 1KG | v2.03 | 0.255 (0.064) | TRUE | TRUE | TRUE | FALSE |
| BODY04 | 1KG | v2.03 robust | 0.048 (0.003) | TRUE | TRUE | TRUE | FALSE |
| BODY04 | UKB | Original | 1.397 (0.68) | TRUE | TRUE | FALSE | FALSE |
| BODY04 | UKB | P<0.4 | 1.048 (0.42) | TRUE | TRUE | TRUE | TRUE |
| BODY04 | UKB | v2.03 | 1.399 (0.68) | TRUE | TRUE | FALSE | FALSE |
| BODY04 | UKB | v2.03 robust | 0.061 (0.002) | TRUE | TRUE | TRUE | FALSE |
| BODY04 | GCTB | Original | 0.92 (0.43) | TRUE | TRUE | TRUE | TRUE |
| BODY04 | GCTB | P<0.4 | 0.701 (0.265) | TRUE | TRUE | TRUE | FALSE |
| BODY04 | GCTB | v2.03 | 0.957 (0.461) | TRUE | TRUE | TRUE | FALSE |
| BODY04 | GCTB | v2.03 robust | 0.059 (0.002) | TRUE | TRUE | TRUE | FALSE |
| HEIG03 | 1KG | Original | 0.718 (0.109) | TRUE | TRUE | TRUE | TRUE |
| HEIG03 | 1KG | P<0.4 | 0.56 (0.057) | TRUE | TRUE | TRUE | FALSE |
| HEIG03 | 1KG | v2.03 | 0.719 (0.11) | TRUE | TRUE | TRUE | FALSE |
| HEIG03 | 1KG | v2.03 robust | 0.251 (0.005) | TRUE | TRUE | TRUE | FALSE |
| HEIG03 | UKB | Original | 3.876 (1.436) | TRUE | TRUE | FALSE | FALSE |
| HEIG03 | UKB | P<0.4 | 3.163 (1.174) | TRUE | TRUE | FALSE | FALSE |
| HEIG03 | UKB | v2.03 | 3.877 (1.437) | TRUE | TRUE | FALSE | FALSE |
| HEIG03 | UKB | v2.03 robust | 0.433 (0.004) | TRUE | TRUE | TRUE | TRUE |
| HEIG03 | GCTB | Original | 2.806 (1.142) | TRUE | FALSE | FALSE | FALSE |
| HEIG03 | GCTB | P<0.4 | 2.424 (0.887) | TRUE | FALSE | FALSE | FALSE |
| HEIG03 | GCTB | v2.03 | 2.958 (1.157) | TRUE | TRUE | FALSE | FALSE |
| HEIG03 | GCTB | v2.03 robust | 0.44 (0.004) | TRUE | TRUE | TRUE | TRUE |
| DIAB05 | 1KG | Original | 0.065 (0.005) | TRUE | TRUE | TRUE | TRUE |
| DIAB05 | 1KG | P<0.4 | 0.225 (0.007) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | 1KG | v2.03 | 0.066 (0.006) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | 1KG | v2.03 robust | 0.059 (0.006) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | UKB | Original | 0.093 (0.005) | TRUE | TRUE | TRUE | TRUE |
| DIAB05 | UKB | P<0.4 | 0.34 (0.009) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | UKB | v2.03 | 0.094 (0.005) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | UKB | v2.03 robust | 0.084 (0.005) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | GCTB | Original | 0.092 (0.005) | TRUE | TRUE | TRUE | TRUE |
| DIAB05 | GCTB | P<0.4 | 0.344 (0.009) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | GCTB | v2.03 | 0.093 (0.005) | TRUE | TRUE | TRUE | FALSE |
| DIAB05 | GCTB | v2.03 robust | 0.085 (0.005) | TRUE | TRUE | TRUE | FALSE |
| COAD01 | 1KG | Original | 0.033 (0.003) | FALSE | TRUE | TRUE | TRUE |
| COAD01 | 1KG | P<0.4 | 0.156 (0.005) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | 1KG | v2.03 | 0.036 (0.003) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | 1KG | v2.03 robust | 0.035 (0.004) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | UKB | Original | 0.077 (0.019) | FALSE | TRUE | TRUE | TRUE |
| COAD01 | UKB | P<0.4 | 0.247 (0.02) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | UKB | v2.03 | 0.078 (0.019) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | UKB | v2.03 robust | 0.043 (0.003) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | GCTB | Original | 0.077 (0.021) | FALSE | TRUE | TRUE | TRUE |
| COAD01 | GCTB | P<0.4 | 0.232 (0.007) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | GCTB | v2.03 | 0.072 (0.016) | FALSE | TRUE | TRUE | FALSE |
| COAD01 | GCTB | v2.03 robust | 0.042 (0.003) | FALSE | TRUE | TRUE | FALSE |
| CROH01 | 1KG | Original | 0.774 (0.246) | FALSE | TRUE | TRUE | TRUE |
| CROH01 | 1KG | P<0.4 | 1.468 (0.047) | FALSE | TRUE | FALSE | FALSE |
| CROH01 | 1KG | v2.03 | 0.905 (0.301) | FALSE | TRUE | TRUE | FALSE |
| CROH01 | 1KG | v2.03 robust | 0.389 (0.042) | FALSE | TRUE | TRUE | FALSE |
| CROH01 | UKB | Original | 0.64 (0.141) | FALSE | FALSE | FALSE | FALSE |
| CROH01 | UKB | P<0.4 | 2.073 (0.058) | FALSE | TRUE | FALSE | FALSE |
| CROH01 | UKB | v2.03 | 0.589 (0.056) | FALSE | TRUE | TRUE | TRUE |
| CROH01 | UKB | v2.03 robust | 0.476 (0.031) | FALSE | TRUE | TRUE | FALSE |
| CROH01 | GCTB | Original | 0.835 (0.12) | FALSE | FALSE | FALSE | FALSE |
| CROH01 | GCTB | P<0.4 | 2.12 (0.057) | FALSE | TRUE | FALSE | FALSE |
| CROH01 | GCTB | v2.03 | 0.933 (0.164) | FALSE | TRUE | TRUE | TRUE |
| CROH01 | GCTB | v2.03 robust | 0.485 (0.027) | FALSE | TRUE | TRUE | FALSE |
| SCLE03 | 1KG | Original | 0.689 (0.077) | FALSE | TRUE | TRUE | TRUE |
| SCLE03 | 1KG | P<0.4 | 1.345 (0.051) | FALSE | TRUE | FALSE | FALSE |
| SCLE03 | 1KG | v2.03 | 0.713 (0.085) | FALSE | TRUE | TRUE | FALSE |
| SCLE03 | 1KG | v2.03 robust | 0.239 (0.024) | FALSE | TRUE | TRUE | FALSE |
| SCLE03 | UKB | Original | 0.741 (0.094) | FALSE | FALSE | FALSE | FALSE |
| SCLE03 | UKB | P<0.4 | 1.784 (0.049) | FALSE | FALSE | FALSE | FALSE |
| SCLE03 | UKB | v2.03 | 0.742 (0.092) | FALSE | TRUE | TRUE | TRUE |
| SCLE03 | UKB | v2.03 robust | 0.303 (0.021) | FALSE | TRUE | TRUE | FALSE |
| SCLE03 | GCTB | Original | 0.767 (0.126) | FALSE | FALSE | FALSE | FALSE |
| SCLE03 | GCTB | P<0.4 | 1.845 (0.051) | FALSE | FALSE | FALSE | FALSE |
| SCLE03 | GCTB | v2.03 | 0.697 (0.08) | FALSE | TRUE | TRUE | TRUE |
| SCLE03 | GCTB | v2.03 robust | 0.289 (0.022) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | 1KG | Original | 0.244 (0.011) | FALSE | TRUE | TRUE | TRUE |
| RHEU02 | 1KG | P<0.4 | 0.536 (0.018) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | 1KG | v2.03 | 0.244 (0.011) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | 1KG | v2.03 robust | 0.226 (0.015) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | UKB | Original | 0.358 (0.021) | FALSE | TRUE | TRUE | TRUE |
| RHEU02 | UKB | P<0.4 | 0.778 (0.022) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | UKB | v2.03 | 0.365 (0.033) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | UKB | v2.03 robust | 0.352 (0.016) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | GCTB | Original | 0.358 (0.012) | FALSE | TRUE | TRUE | TRUE |
| RHEU02 | GCTB | P<0.4 | 0.787 (0.022) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | GCTB | v2.03 | 0.36 (0.012) | FALSE | TRUE | TRUE | FALSE |
| RHEU02 | GCTB | v2.03 robust | 0.366 (0.016) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | 1KG | Original | 0.063 (0.002) | FALSE | TRUE | TRUE | TRUE |
| EDUC03 | 1KG | P<0.4 | 0.093 (0.002) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | 1KG | v2.03 | 0.063 (0.002) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | 1KG | v2.03 robust | 0.063 (0.002) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | UKB | Original | 0.106 (0.001) | FALSE | TRUE | TRUE | TRUE |
| EDUC03 | UKB | P<0.4 | 0.143 (0.002) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | UKB | v2.03 | 0.106 (0.001) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | UKB | v2.03 robust | 0.106 (0.001) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | GCTB | Original | 0.108 (0.001) | FALSE | TRUE | TRUE | TRUE |
| EDUC03 | GCTB | P<0.4 | 0.143 (0.002) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | GCTB | v2.03 | 0.108 (0.001) | FALSE | TRUE | TRUE | FALSE |
| EDUC03 | GCTB | v2.03 robust | 0.109 (0.001) | FALSE | TRUE | TRUE | FALSE |
| ADHD04 | 1KG | Original | 0.254 (0.013) | FALSE | TRUE | TRUE | TRUE |
| ADHD04 | 1KG | P<0.4 | 0.902 (0.027) | FALSE | TRUE | TRUE | FALSE |
| ADHD04 | 1KG | v2.03 | 0.26 (0.012) | FALSE | TRUE | TRUE | FALSE |
| ADHD04 | 1KG | v2.03 robust | 0.313 (0.042) | FALSE | TRUE | TRUE | FALSE |
| ADHD04 | UKB | Original | 0.378 (0.016) | FALSE | TRUE | TRUE | TRUE |
| ADHD04 | UKB | P<0.4 | 1.274 (0.033) | FALSE | TRUE | FALSE | FALSE |
| ADHD04 | UKB | v2.03 | 0.385 (0.015) | FALSE | TRUE | TRUE | FALSE |
| ADHD04 | UKB | v2.03 robust | 0.384 (0.015) | FALSE | TRUE | TRUE | FALSE |
| ADHD04 | GCTB | Original | 0.39 (0.016) | FALSE | TRUE | TRUE | TRUE |
| ADHD04 | GCTB | P<0.4 | 1.295 (0.034) | FALSE | TRUE | FALSE | FALSE |
| ADHD04 | GCTB | v2.03 | 0.395 (0.016) | FALSE | TRUE | TRUE | FALSE |
| ADHD04 | GCTB | v2.03 robust | 0.39 (0.016) | FALSE | TRUE | TRUE | FALSE |
| BODY11 | 1KG | Original | 0.123 (0.002) | TRUE | TRUE | TRUE | TRUE |
| BODY11 | 1KG | P<0.4 | 0.158 (0.002) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | 1KG | v2.03 | 0.124 (0.002) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | 1KG | v2.03 robust | 0.123 (0.003) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | UKB | Original | 0.213 (0.003) | TRUE | TRUE | TRUE | TRUE |
| BODY11 | UKB | P<0.4 | 0.251 (0.002) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | UKB | v2.03 | 0.211 (0.002) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | UKB | v2.03 robust | 0.211 (0.002) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | GCTB | Original | 0.217 (0.003) | TRUE | TRUE | TRUE | TRUE |
| BODY11 | GCTB | P<0.4 | 0.253 (0.002) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | GCTB | v2.03 | 0.217 (0.003) | TRUE | TRUE | TRUE | FALSE |
| BODY11 | GCTB | v2.03 robust | 0.216 (0.002) | TRUE | TRUE | TRUE | FALSE |
| PRCA01 | 1KG | Original | 0.124 (0.004) | FALSE | TRUE | TRUE | TRUE |
| PRCA01 | 1KG | P<0.4 | 0.244 (0.005) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | 1KG | v2.03 | 0.126 (0.004) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | 1KG | v2.03 robust | 0.124 (0.005) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | UKB | Original | 0.241 (0.021) | FALSE | TRUE | TRUE | TRUE |
| PRCA01 | UKB | P<0.4 | 0.351 (0.007) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | UKB | v2.03 | 0.242 (0.022) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | UKB | v2.03 robust | 0.161 (0.004) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | GCTB | Original | 0.195 (0.02) | FALSE | TRUE | TRUE | TRUE |
| PRCA01 | GCTB | P<0.4 | 0.357 (0.007) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | GCTB | v2.03 | 0.197 (0.021) | FALSE | TRUE | TRUE | FALSE |
| PRCA01 | GCTB | v2.03 robust | 0.163 (0.005) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | 1KG | Original | 0.107 (0.003) | FALSE | TRUE | TRUE | TRUE |
| BRCA01 | 1KG | P<0.4 | 0.209 (0.004) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | 1KG | v2.03 | 0.107 (0.003) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | 1KG | v2.03 robust | 0.106 (0.004) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | UKB | Original | 0.158 (0.003) | FALSE | TRUE | TRUE | TRUE |
| BRCA01 | UKB | P<0.4 | 0.304 (0.005) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | UKB | v2.03 | 0.158 (0.003) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | UKB | v2.03 robust | 0.158 (0.004) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | GCTB | Original | 0.16 (0.003) | FALSE | TRUE | TRUE | TRUE |
| BRCA01 | GCTB | P<0.4 | 0.308 (0.005) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | GCTB | v2.03 | 0.16 (0.003) | FALSE | TRUE | TRUE | FALSE |
| BRCA01 | GCTB | v2.03 robust | 0.16 (0.004) | FALSE | TRUE | TRUE | FALSE |
Compare SBayesR results
# Create a table indicating whether per variant sample size is available
Files<-c('UKBB','TEDS')
Samples<-c('UKBB','TEDS')
source('/users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Target_scoring.config')
for(file in 1:length(Files)){
if(Samples[file] == 'UKBB'){
pheno<-c('Depression','Intelligence','BMI','Height','T2D','CAD','IBD','MultiScler','RheuArth','Breast_Cancer','Prostate_Cancer')
gwas<-c('DEPR06','COLL01','BODY04','HEIG03','DIAB05','COAD01','CROH01','SCLE03','RHEU02','BRCA01','PRCA01')
} else {
pheno<-c('Height21', 'BMI21', 'GCSE', 'ADHD')
gwas<-c('HEIG03', 'BODY11', 'EDUC03', 'ADHD04')
}
res<-NULL
for(i in 1:length(pheno)){
res_eval_1KG_i<-NULL
res_eval_1KG_P4_i<-NULL
res_eval_1KG_GCTB_203_i<-NULL
res_eval_1KG_GCTB_203_robust_i<-NULL
res_eval_UKBB_i<-NULL
res_eval_UKBB_P4_i<-NULL
res_eval_UKBB_GCTB_203_i<-NULL
res_eval_UKBB_GCTB_203_robust_i<-NULL
res_eval_GCTB_i<-NULL
res_eval_GCTB_P4_i<-NULL
res_eval_GCTB_GCTB_203_i<-NULL
res_eval_GCTB_GCTB_203_robust_i<-NULL
# 1KG ref
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_1KG_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_1KG_i<-res_eval_1KG_i[2,]
res_eval_1KG_i$Model<-"v2.02 (Default)"
res_eval_1KG_i$Ref<-'1KG'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_P4.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_1KG_P4_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_P4.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_1KG_P4_i<-res_eval_1KG_P4_i[2,]
res_eval_1KG_P4_i$Model<-"v2.02 (P<0.4)"
res_eval_1KG_P4_i$Ref<-'1KG'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTB_203.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_1KG_GCTB_203_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTB_203.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_1KG_GCTB_203_i<-res_eval_1KG_GCTB_203_i[2,]
res_eval_1KG_GCTB_203_i$Model<-"v2.03 (Default)"
res_eval_1KG_GCTB_203_i$Ref<-'1KG'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTB_203_robust.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_1KG_GCTB_203_robust_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTB_203_robust.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_1KG_GCTB_203_robust_i<-res_eval_1KG_GCTB_203_robust_i[2,]
res_eval_1KG_GCTB_203_robust_i$Model<-"v2.03 (Robust)"
res_eval_1KG_GCTB_203_robust_i$Ref<-'1KG'
}
# UKB ref
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_UKBB_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_UKBB_i<-res_eval_UKBB_i[2,]
res_eval_UKBB_i$Model<-"v2.02 (Default)"
res_eval_UKBB_i$Ref<-'UKB'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'_P4.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_UKBB_P4_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'_P4.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_UKBB_P4_i<-res_eval_UKBB_P4_i[2,]
res_eval_UKBB_P4_i$Model<-"v2.02 (P<0.4)"
res_eval_UKBB_P4_i$Ref<-'UKB'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'_GCTB_203.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_UKBB_GCTB_203_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'_GCTB_203.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_UKBB_GCTB_203_i<-res_eval_UKBB_GCTB_203_i[2,]
res_eval_UKBB_GCTB_203_i$Model<-"v2.03 (Default)"
res_eval_UKBB_GCTB_203_i$Ref<-'UKB'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'_GCTB_203_robust.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_UKBB_GCTB_203_robust_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.UKBB_ref.',gwas[i],'_GCTB_203_robust.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_UKBB_GCTB_203_robust_i<-res_eval_UKBB_GCTB_203_robust_i[2,]
res_eval_UKBB_GCTB_203_robust_i$Model<-"v2.03 (Robust)"
res_eval_UKBB_GCTB_203_robust_i$Ref<-'UKB'
}
# GCTB ref
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_GCTB_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_GCTB_i<-res_eval_GCTB_i[2,]
res_eval_GCTB_i$Model<-"v2.02 (Default)"
res_eval_GCTB_i$Ref<-'GCTB'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref_P4.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_GCTB_P4_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref_P4.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_GCTB_P4_i<-res_eval_GCTB_P4_i[2,]
res_eval_GCTB_P4_i$Model<-"v2.02 (P<0.4)"
res_eval_GCTB_P4_i$Ref<-'GCTB'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref_GCTB_203.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_GCTB_GCTB_203_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref_GCTB_203.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_GCTB_GCTB_203_i<-res_eval_GCTB_GCTB_203_i[2,]
res_eval_GCTB_GCTB_203_i$Model<-"v2.03 (Default)"
res_eval_GCTB_GCTB_203_i$Ref<-'GCTB'
}
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR.pred_eval.txt'))){
res_eval_GCTB_GCTB_203_robust_i<-read.table(paste0('/users/k1806347/brc_scratch/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/',pheno[i],'/Association_withPRSs/',Samples[file],'.w_hm3.',gwas[i],'_GCTBref_GCTB_203_robust.EUR-PRSs_SBayesR.pred_eval.txt'), header=T, stringsAsFactors=F)
res_eval_GCTB_GCTB_203_robust_i<-res_eval_GCTB_GCTB_203_robust_i[2,]
res_eval_GCTB_GCTB_203_robust_i$Model<-"v2.03 (Robust)"
res_eval_GCTB_GCTB_203_robust_i$Ref<-'GCTB'
}
res_i<-do.call(rbind, list(res_eval_1KG_i,res_eval_1KG_P4_i, res_eval_1KG_GCTB_203_i,res_eval_1KG_GCTB_203_robust_i,res_eval_UKBB_i,res_eval_UKBB_P4_i,res_eval_UKBB_GCTB_203_i,res_eval_UKBB_GCTB_203_robust_i,res_eval_GCTB_i,res_eval_GCTB_P4_i,res_eval_GCTB_GCTB_203_i,res_eval_GCTB_GCTB_203_robust_i))
res_i$Phenotype<-pheno[i]
res_i$Cross_LiabR2<-NULL
res_i$Indep_LiabR2<-NULL
res_i$CrossVal_pval<-NULL
res_i$IndepVal_pval<-NULL
res_i$Cross_AUC<-NULL
res_i$Indep_AUC<-NULL
res_i$CrossVal_OR<-NULL
res_i$IndepVal_OR<-NULL
res_i$CrossVal_HighCI<-NULL
res_i$IndepVal_HighCI<-NULL
res_i$CrossVal_LowCI<-NULL
res_i$IndepVal_LowCI<-NULL
res_i$CrossVal_Ncas<-NULL
res_i$IndepVal_Ncas<-NULL
res_i$CrossVal_Ncon<-NULL
res_i$IndepVal_Ncon<-NULL
res<-rbind(res, res_i)
}
write.csv(res, paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_SBayesR_',Samples[file],'.csv'), row.names=F)
res$Ref<-factor(res$Ref, levels=c('1KG','UKB','GCTB'))
res$Model<-factor(res$Model, levels=c("v2.02 (Default)","v2.02 (P<0.4)","v2.03 (Default)","v2.03 (Robust)"))
plots_CrossVal<-list()
plots_IndepVal<-list()
for(i in 1:length(pheno)){
plots_CrossVal[[pheno[i]]] <- ggplot( res[res$Phenotype==pheno[i],], aes(x=Ref, y=CrossVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=CrossVal_R-CrossVal_R_SE, ymax=CrossVal_R+CrossVal_R_SE), width=.4, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res$CrossVal_R[res$Phenotype==pheno[i]])-0.05, max(res$CrossVal_R[res$Phenotype==pheno[i]])+0.05), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
plots_IndepVal[[pheno[i]]] <- ggplot( res[res$Phenotype==pheno[i],], aes(x=Ref, y=IndepVal_R, fill=Model)) +
geom_bar(stat="identity", position=position_dodge(preserve = "single"), width = 0.7) +
geom_errorbar(aes(ymin=IndepVal_R-IndepVal_R_SE, ymax=IndepVal_R+IndepVal_R_SE), width=.4, position=position_dodge(width = 0.7, preserve = "single")) +
labs(y="Correlation (SE)", x='', title=pheno[i]) +
coord_cartesian(ylim=c(min(res$IndepVal_R[res$Phenotype==pheno[i]])-0.05, max(res$IndepVal_R[res$Phenotype==pheno[i]])+0.05), clip="on") +
theme_half_open() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
background_grid(major = 'y', minor = 'y')
}
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_SBayesR_CrossVal_',Samples[file],'.png'), units='px', res=300, width=2250, height=800*ceiling(length(pheno)/2))
print(plot_grid(plotlist=plots_CrossVal, ncol = 2))
dev.off()
png(paste0('/scratch/users/k1806347/Analyses/PRS_comparison/',Samples[file],'_outcomes_for_prediction/PRS_SBayesR_IndepVal_',Samples[file],'.png'), units='px', res=300, width=2250, height=800*ceiling(length(pheno)/2))
print(plot_grid(plotlist=plots_IndepVal, ncol = 2))
dev.off()
}
# This sensitivity analysis shows the new robust implementation of SBayesR is more reliable, though can perform worse when using the smaller 1KG Phase 3 reference. I think we should compare methods using the robust implmentation of SBayesR.Show UKBB cross-validation results
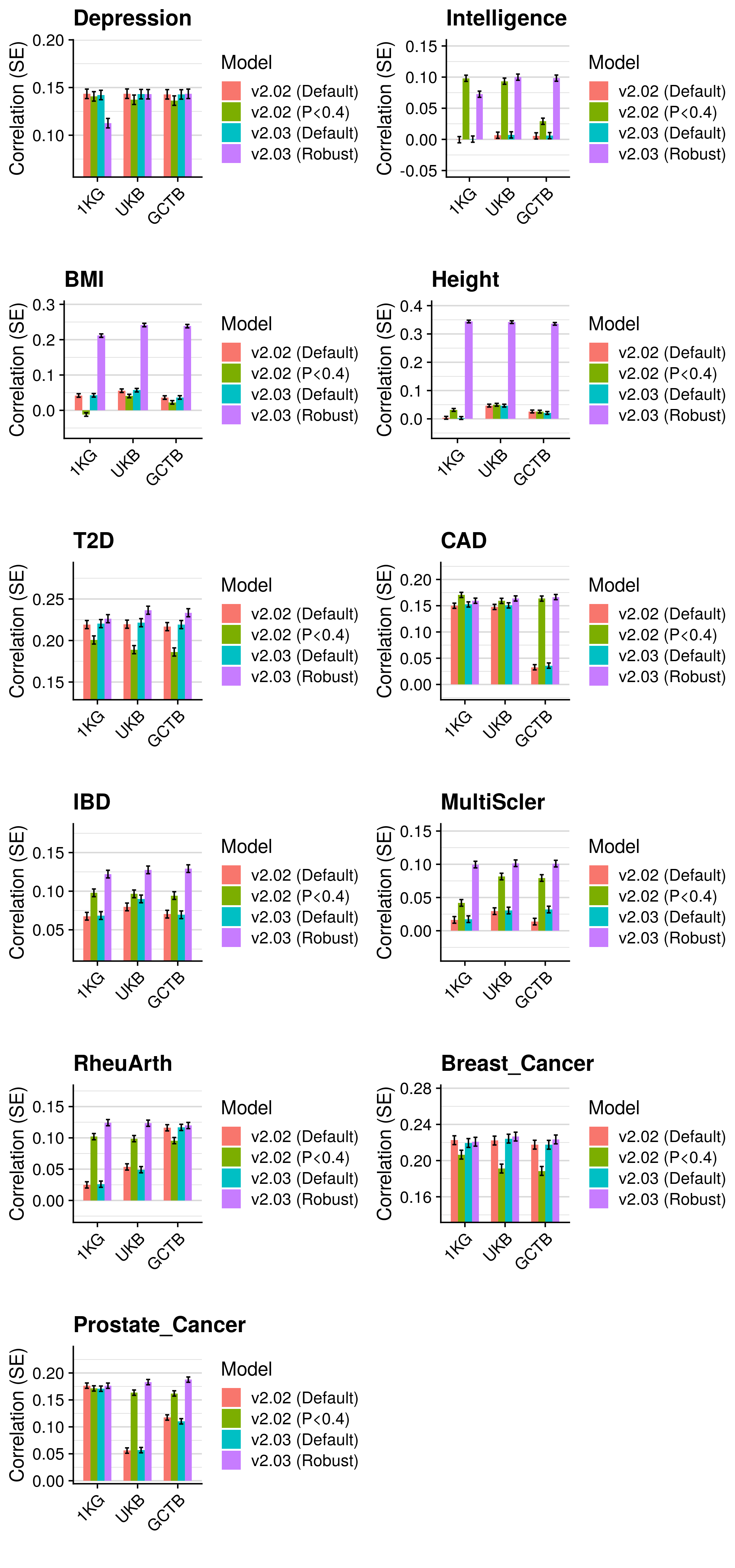
Predictive utility estimated using 10-fold cross-validation
Show UKBB test-validation results
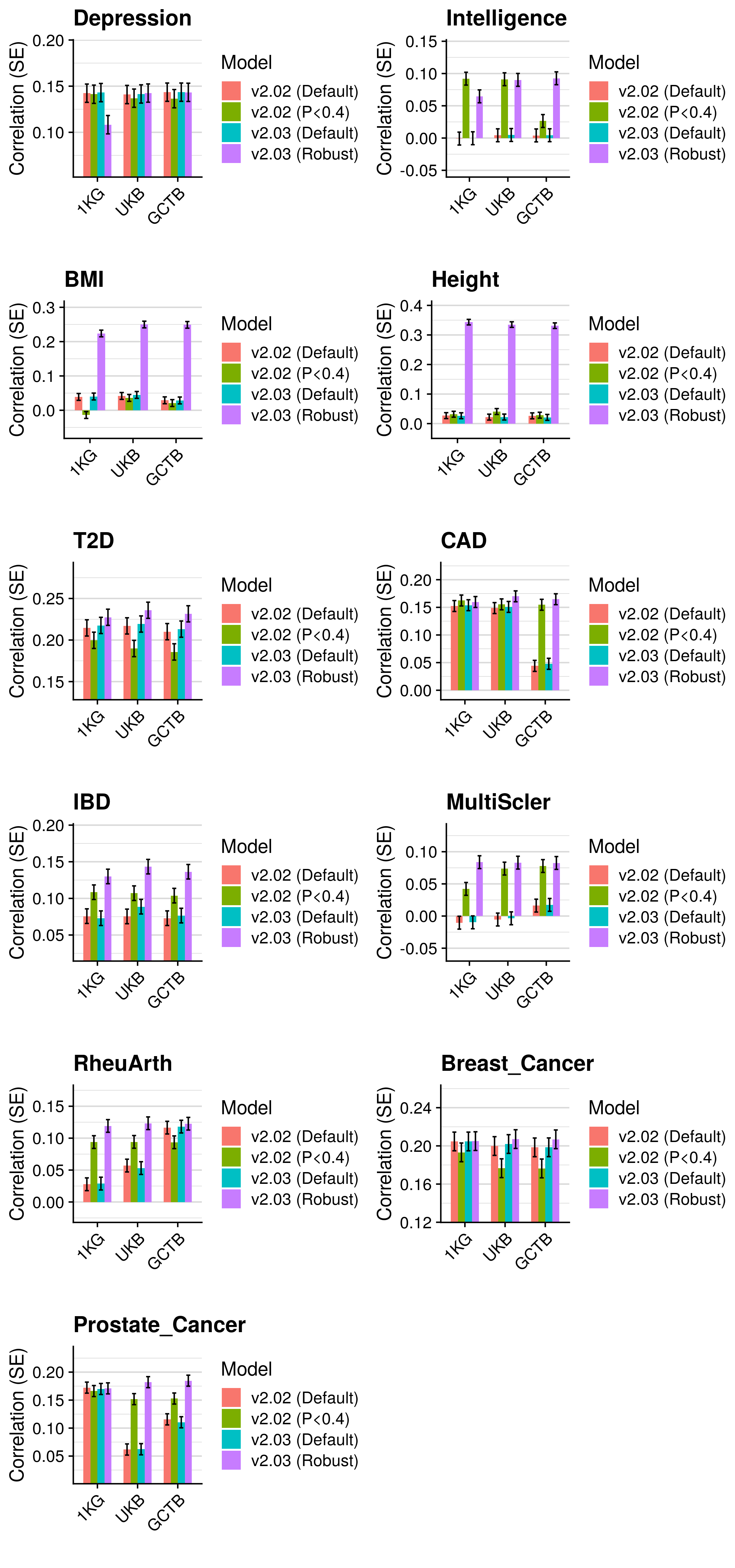
Predictive utility estimated using test set validation
Show TEDS cross-validation results
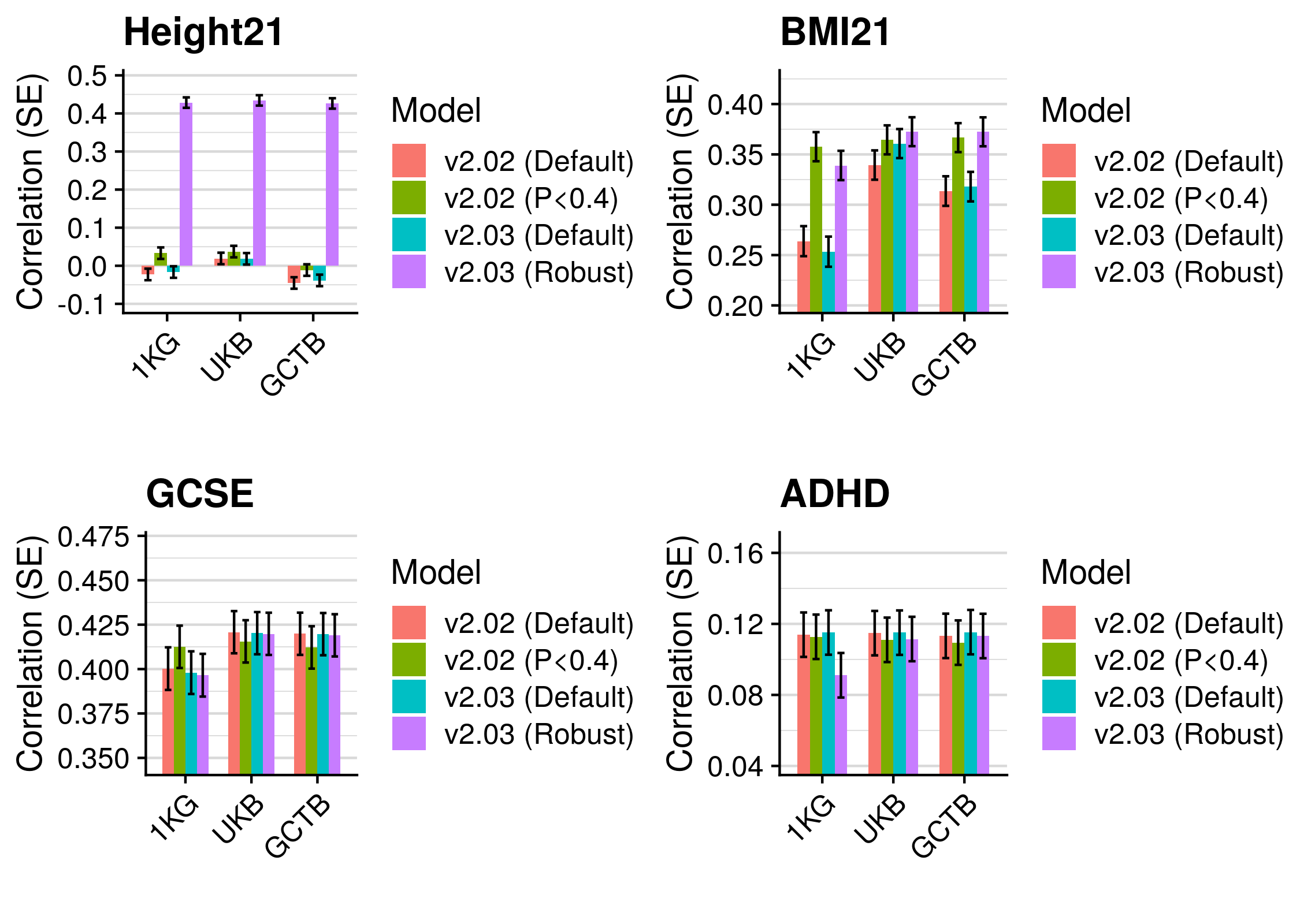
Predictive utility estimated using 10-fold cross-validation
Show TEDS test-validation results
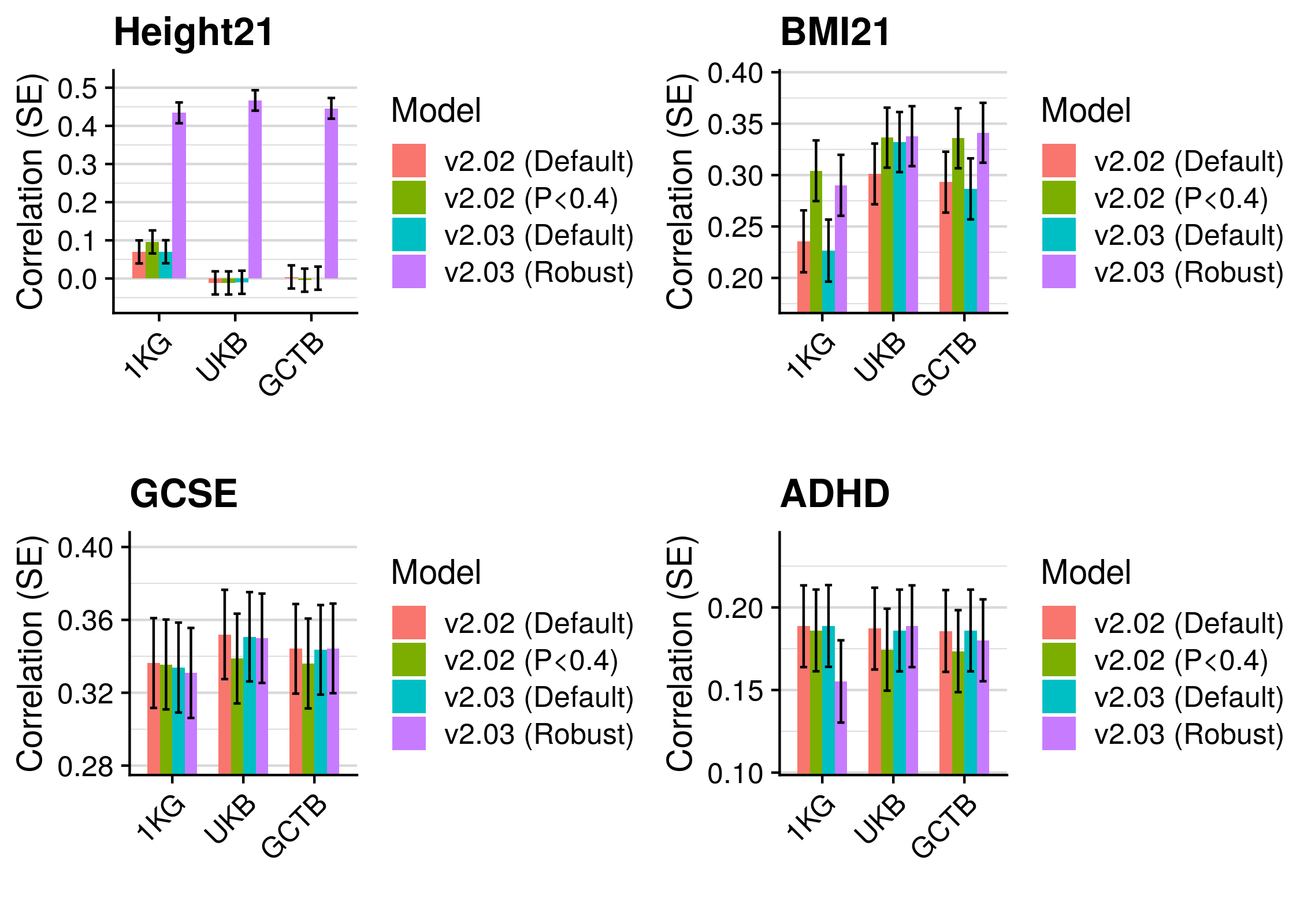
Predictive utility estimated using test set validation
4.4 Average performance
Show cross-validation results
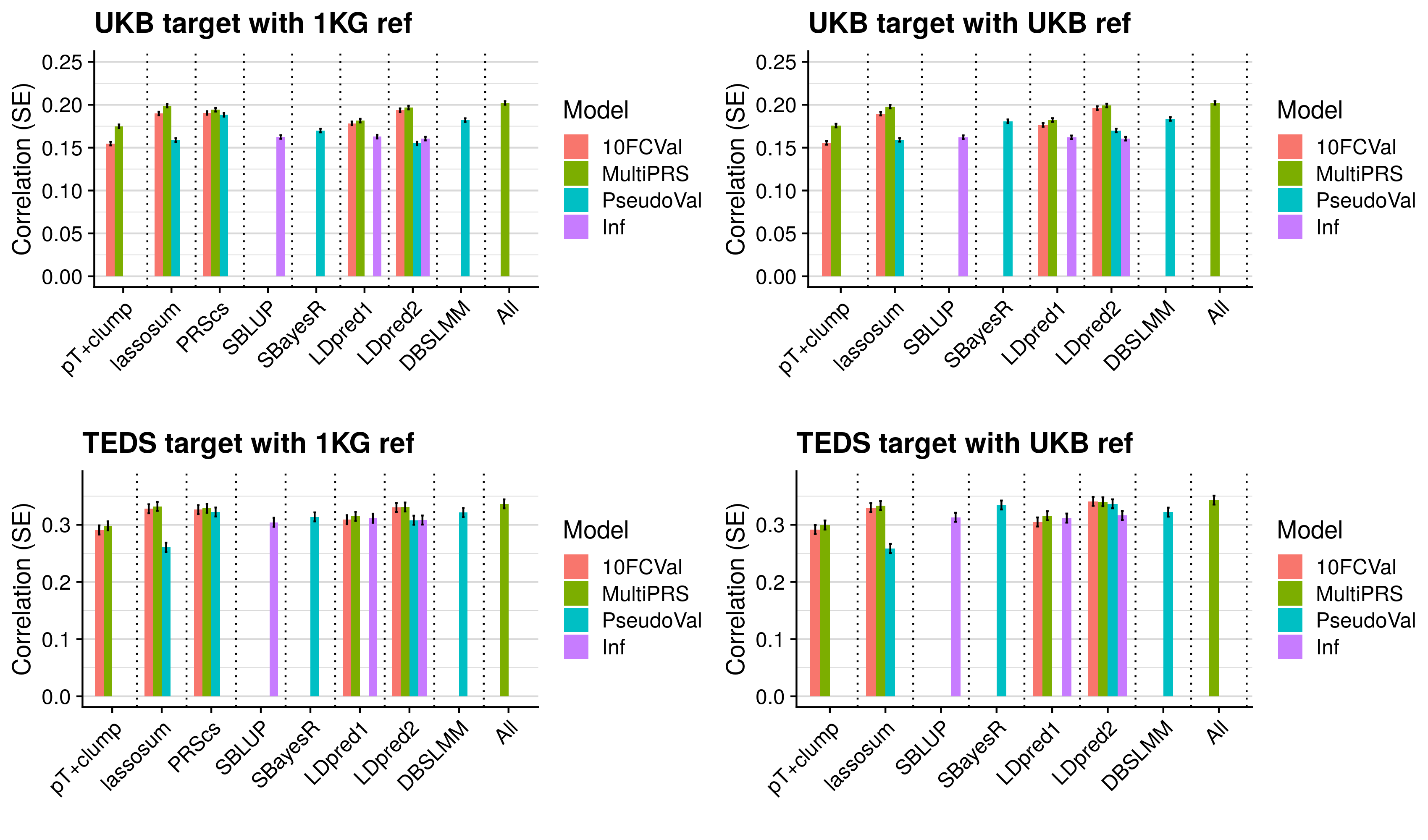
Predictive utility estimated using 10-fold cross-validation
Show test-validation results
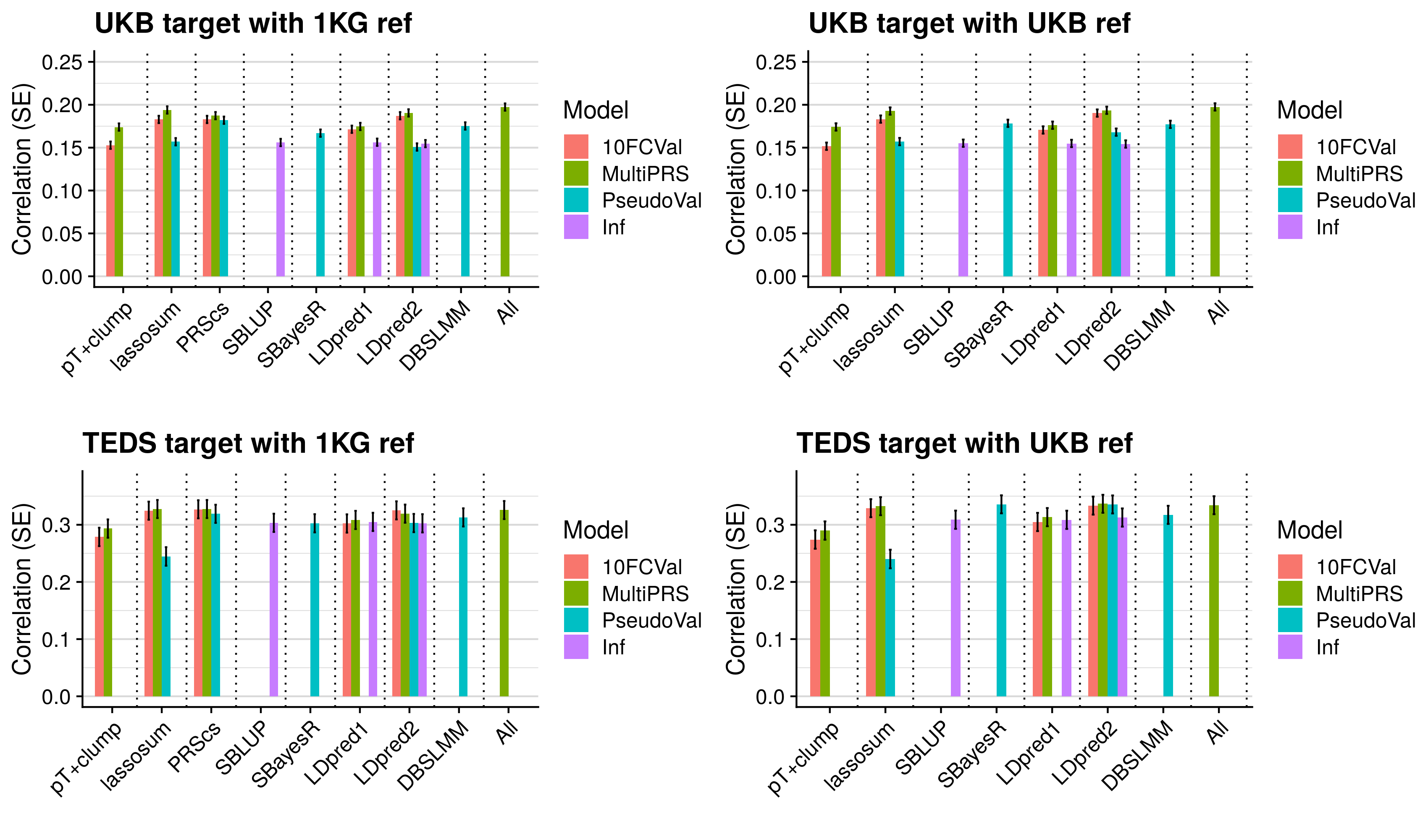
Predictive utility estimated using test set validation
Show table: UKBB (1KG ref)
| Method | Model | CrossVal R (SE) | IndepVal R (SE) |
|---|---|---|---|
| pT+clump | 10FCVal | 0.155 (0.002) | 0.153 (0.004) |
| pT+clump | MultiPRS | 0.175 (0.002) | 0.174 (0.004) |
| lassosum | 10FCVal | 0.19 (0.002) | 0.183 (0.004) |
| lassosum | MultiPRS | 0.199 (0.002) | 0.194 (0.004) |
| lassosum | PseudoVal | 0.159 (0.002) | 0.157 (0.004) |
| PRScs | 10FCVal | 0.19 (0.002) | 0.183 (0.004) |
| PRScs | MultiPRS | 0.194 (0.002) | 0.187 (0.004) |
| PRScs | PseudoVal | 0.188 (0.002) | 0.182 (0.004) |
| SBLUP | Inf | 0.162 (0.002) | 0.156 (0.004) |
| SBayesR | PseudoVal | 0.17 (0.002) | 0.167 (0.004) |
| LDpred1 | 10FCVal | 0.178 (0.002) | 0.171 (0.004) |
| LDpred1 | MultiPRS | 0.181 (0.002) | 0.175 (0.004) |
| LDpred1 | Inf | 0.163 (0.002) | 0.156 (0.004) |
| LDpred2 | 10FCVal | 0.194 (0.002) | 0.187 (0.004) |
| LDpred2 | MultiPRS | 0.197 (0.002) | 0.191 (0.004) |
| LDpred2 | PseudoVal | 0.155 (0.002) | 0.151 (0.004) |
| LDpred2 | Inf | 0.161 (0.002) | 0.155 (0.004) |
| DBSLMM | PseudoVal | 0.182 (0.002) | 0.175 (0.004) |
| All | MultiPRS | 0.202 (0.002) | 0.197 (0.004) |
Show table: UKBB (UKBB ref)
| Method | Model | CrossVal R (SE) | IndepVal R (SE) |
|---|---|---|---|
| pT+clump | 10FCVal | 0.156 (0.002) | 0.152 (0.004) |
| pT+clump | MultiPRS | 0.176 (0.002) | 0.174 (0.004) |
| lassosum | 10FCVal | 0.19 (0.002) | 0.183 (0.004) |
| lassosum | MultiPRS | 0.198 (0.002) | 0.193 (0.004) |
| lassosum | PseudoVal | 0.159 (0.002) | 0.157 (0.004) |
| SBLUP | Inf | 0.162 (0.002) | 0.155 (0.004) |
| SBayesR | PseudoVal | 0.181 (0.002) | 0.178 (0.004) |
| LDpred1 | 10FCVal | 0.177 (0.002) | 0.171 (0.004) |
| LDpred1 | MultiPRS | 0.182 (0.002) | 0.176 (0.004) |
| LDpred1 | Inf | 0.162 (0.002) | 0.155 (0.004) |
| LDpred2 | 10FCVal | 0.196 (0.002) | 0.19 (0.004) |
| LDpred2 | MultiPRS | 0.199 (0.002) | 0.194 (0.004) |
| LDpred2 | PseudoVal | 0.17 (0.002) | 0.168 (0.004) |
| LDpred2 | Inf | 0.16 (0.002) | 0.154 (0.004) |
| DBSLMM | PseudoVal | 0.183 (0.002) | 0.177 (0.004) |
| All | MultiPRS | 0.202 (0.002) | 0.197 (0.004) |
Show table: TEDS (1KG ref)
| Method | Model | CrossVal R (SE) | IndepVal R (SE) |
|---|---|---|---|
| pT+clump | 10FCVal | 0.291 (0.008) | 0.279 (0.016) |
| pT+clump | MultiPRS | 0.298 (0.008) | 0.293 (0.016) |
| lassosum | 10FCVal | 0.328 (0.008) | 0.325 (0.016) |
| lassosum | MultiPRS | 0.332 (0.008) | 0.328 (0.016) |
| lassosum | PseudoVal | 0.261 (0.008) | 0.245 (0.016) |
| PRScs | 10FCVal | 0.327 (0.008) | 0.327 (0.016) |
| PRScs | MultiPRS | 0.329 (0.008) | 0.328 (0.016) |
| PRScs | PseudoVal | 0.322 (0.008) | 0.319 (0.016) |
| SBLUP | Inf | 0.304 (0.008) | 0.303 (0.016) |
| SBayesR | PseudoVal | 0.314 (0.008) | 0.303 (0.016) |
| LDpred1 | 10FCVal | 0.309 (0.008) | 0.302 (0.016) |
| LDpred1 | MultiPRS | 0.315 (0.008) | 0.308 (0.016) |
| LDpred1 | Inf | 0.311 (0.008) | 0.305 (0.016) |
| LDpred2 | 10FCVal | 0.33 (0.008) | 0.325 (0.016) |
| LDpred2 | MultiPRS | 0.331 (0.008) | 0.319 (0.016) |
| LDpred2 | PseudoVal | 0.308 (0.008) | 0.303 (0.016) |
| LDpred2 | Inf | 0.308 (0.008) | 0.303 (0.016) |
| DBSLMM | PseudoVal | 0.321 (0.008) | 0.313 (0.016) |
| All | MultiPRS | 0.337 (0.008) | 0.326 (0.016) |
Show table: TEDS (UKBB ref)
| Method | Model | CrossVal R (SE) | IndepVal R (SE) |
|---|---|---|---|
| pT+clump | 10FCVal | 0.292 (0.008) | 0.274 (0.016) |
| pT+clump | MultiPRS | 0.3 (0.008) | 0.29 (0.016) |
| lassosum | 10FCVal | 0.33 (0.008) | 0.329 (0.016) |
| lassosum | MultiPRS | 0.333 (0.008) | 0.332 (0.016) |
| lassosum | PseudoVal | 0.258 (0.008) | 0.24 (0.016) |
| SBLUP | Inf | 0.313 (0.008) | 0.309 (0.016) |
| SBayesR | PseudoVal | 0.335 (0.008) | 0.336 (0.016) |
| LDpred1 | 10FCVal | 0.305 (0.008) | 0.305 (0.016) |
| LDpred1 | MultiPRS | 0.316 (0.008) | 0.313 (0.016) |
| LDpred1 | Inf | 0.312 (0.008) | 0.309 (0.016) |
| LDpred2 | 10FCVal | 0.341 (0.008) | 0.334 (0.016) |
| LDpred2 | MultiPRS | 0.34 (0.008) | 0.337 (0.016) |
| LDpred2 | PseudoVal | 0.337 (0.008) | 0.336 (0.016) |
| LDpred2 | Inf | 0.316 (0.008) | 0.313 (0.016) |
| DBSLMM | PseudoVal | 0.322 (0.008) | 0.317 (0.016) |
| All | MultiPRS | 0.343 (0.008) | 0.334 (0.016) |
4.4.1 Average performance relative to pT+clump
Show cross-validation results
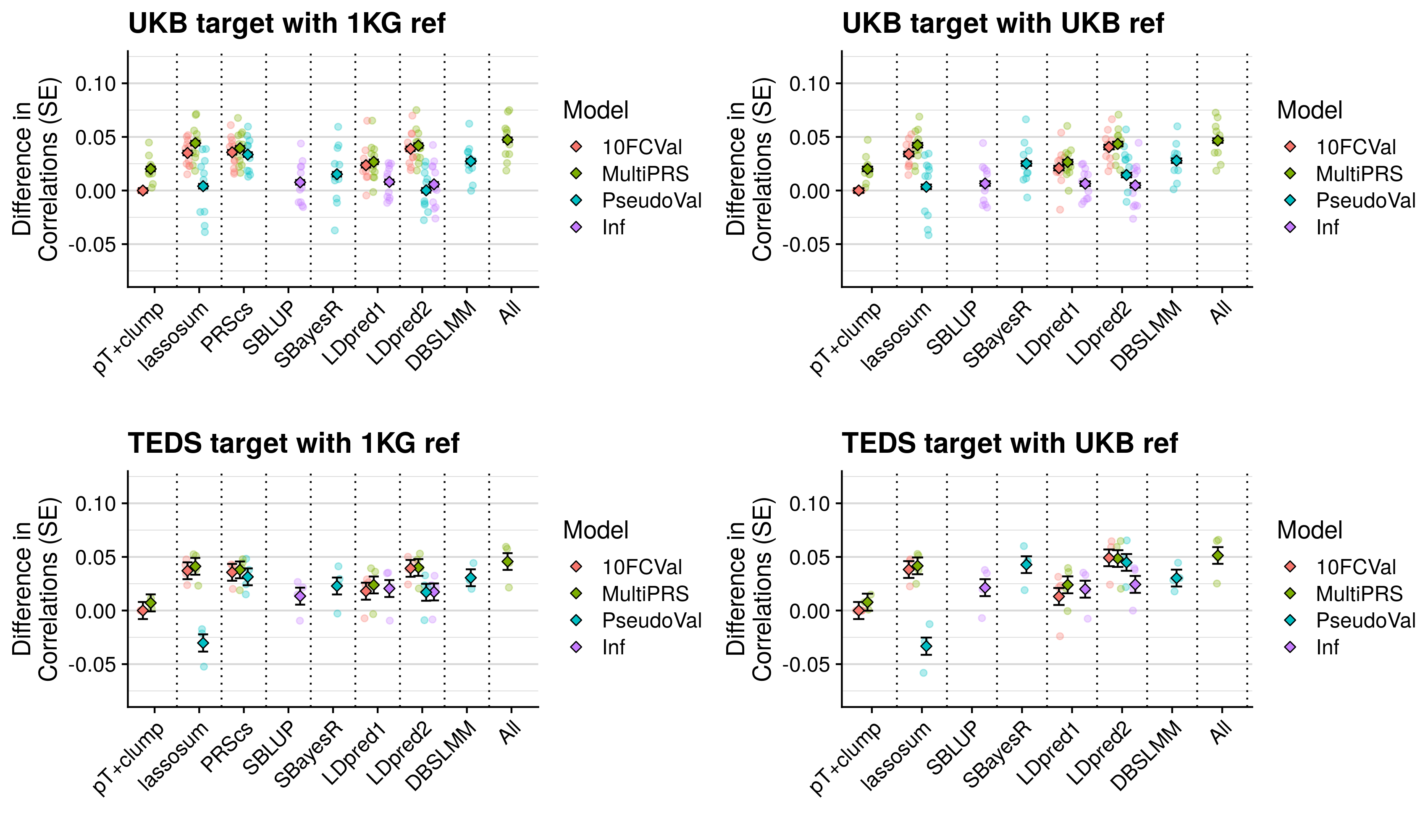
Predictive utility estimated using 10-fold cross-validation
Show test-validation results
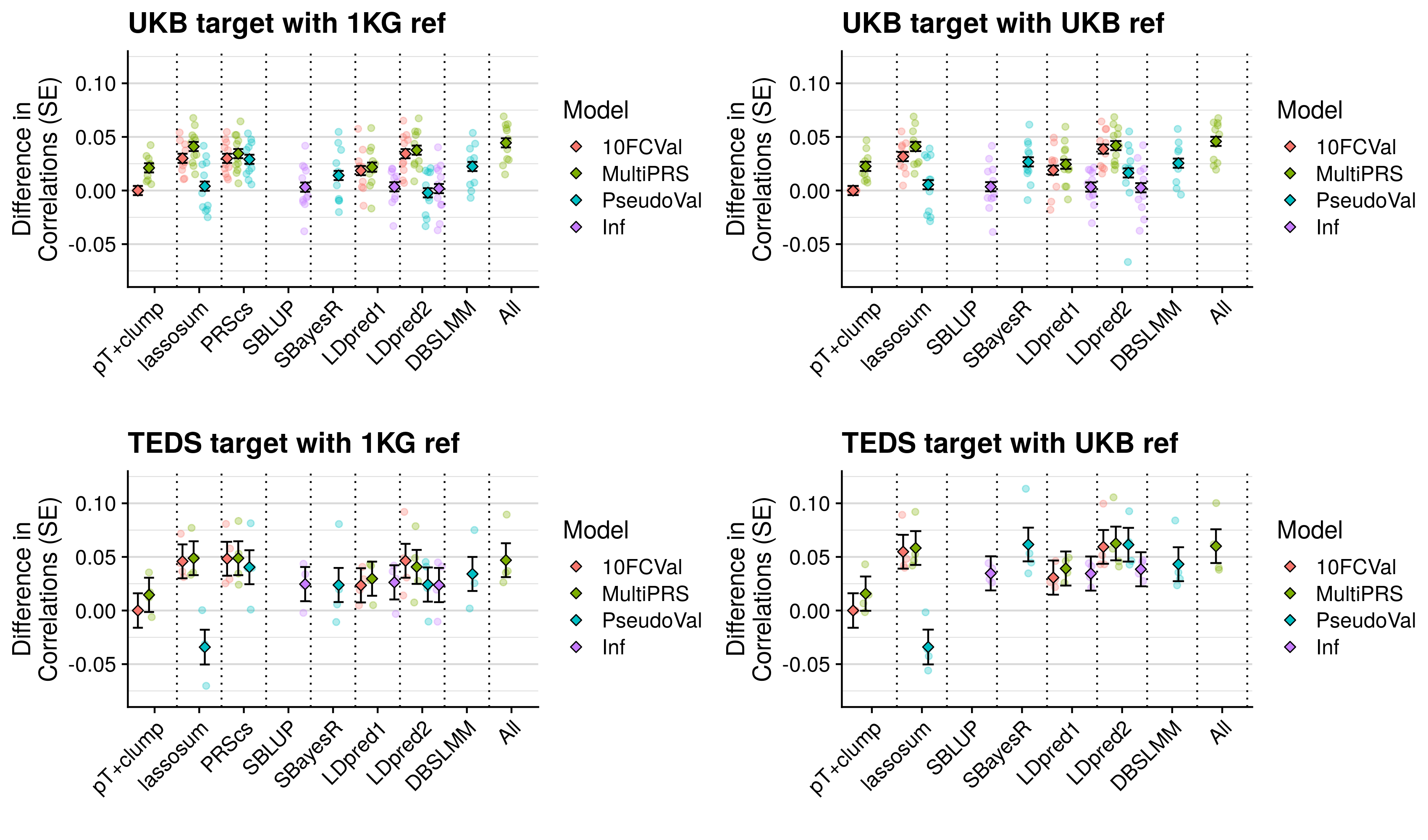
Predictive utility estimated using test set validation
Show by polygenicity cross-validation results
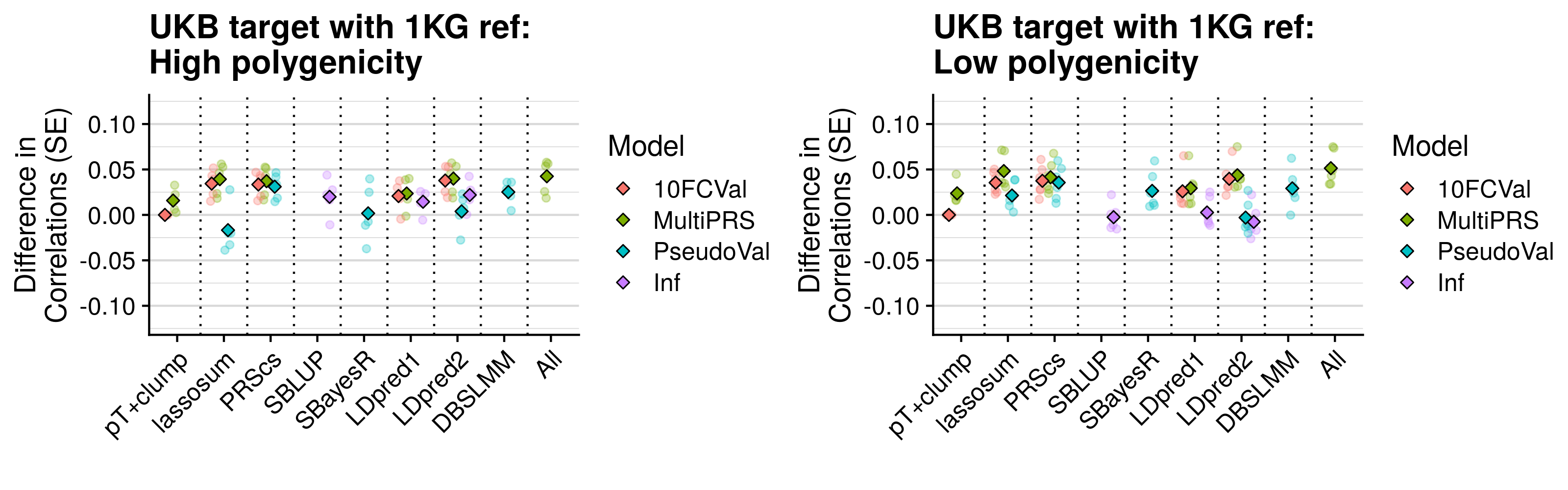
Predictive utility estimated using 10-fold cross-validation
Show by polygenicity test-validation results
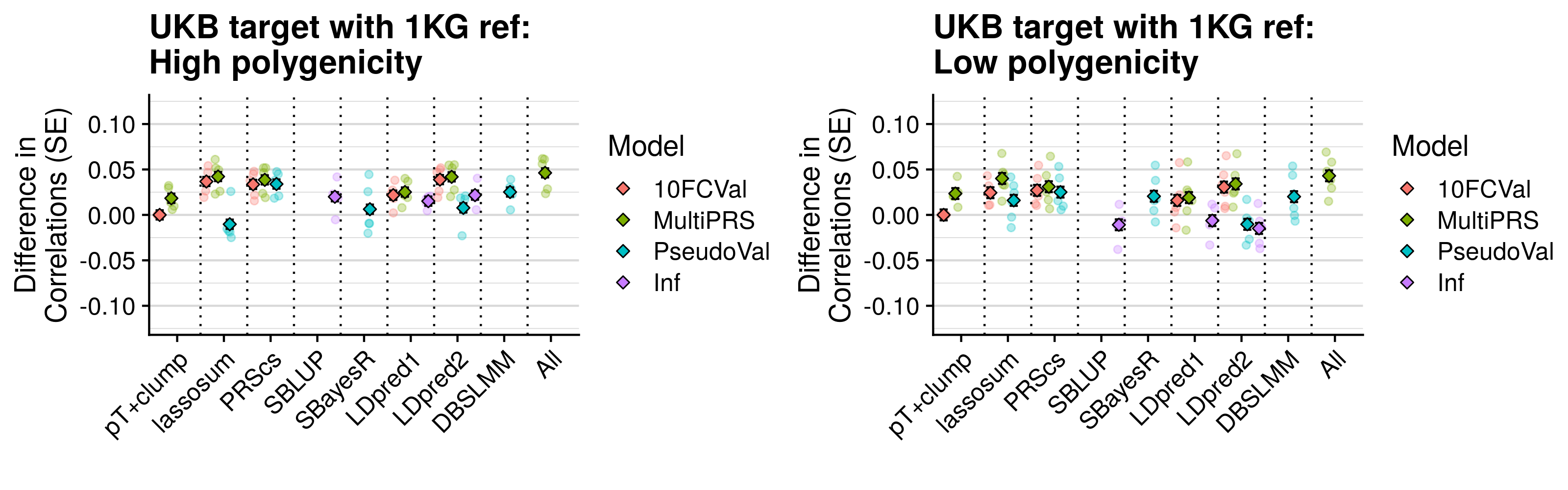
Predictive utility estimated using test set validation
Show cross-validation results with PCs regressed
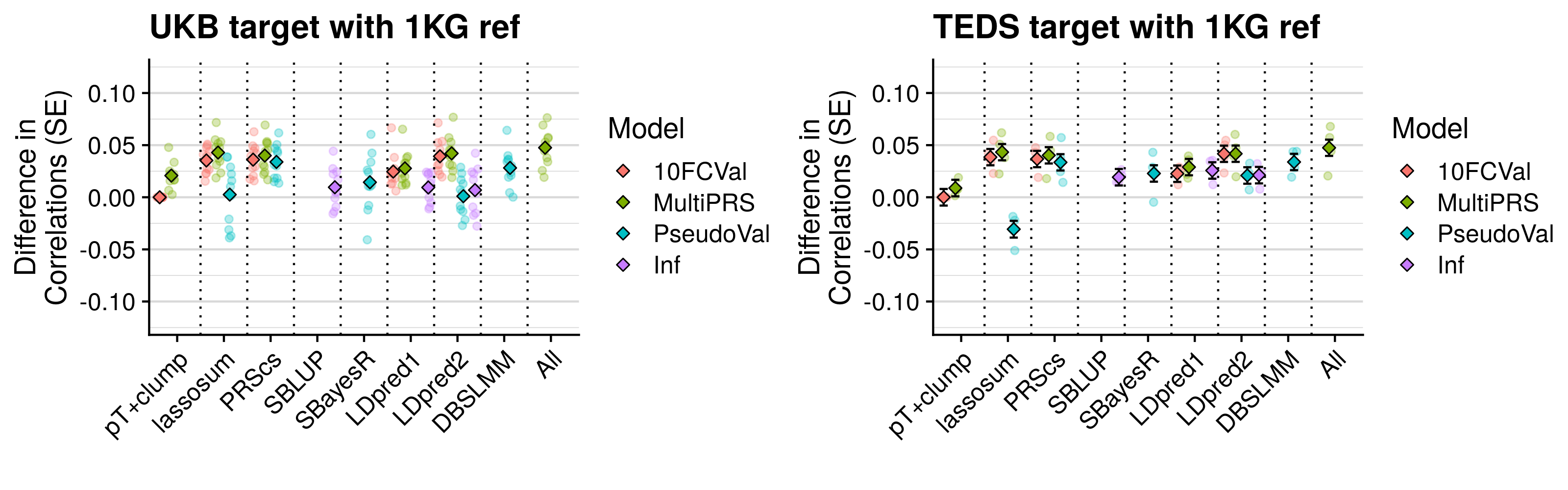
Predictive utility estimated using 10-fold cross-validation
Show test-validation results with PCs regressed
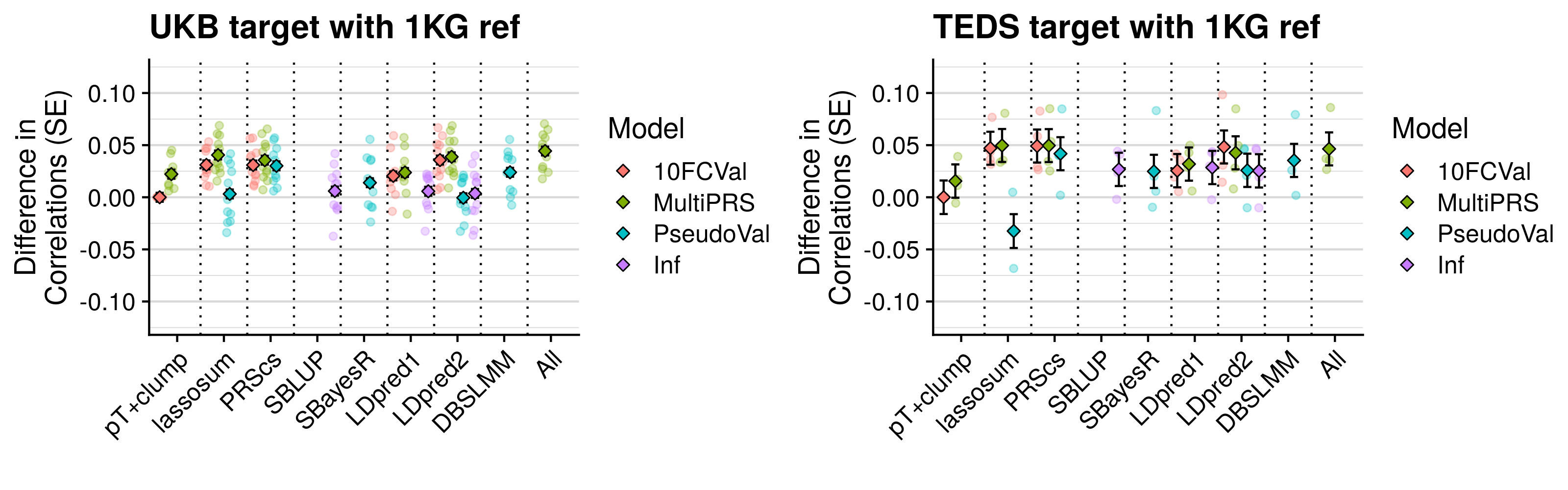
Predictive utility estimated using test set validation
4.4.2 Average performance difference across all methods
Show cross-validation results
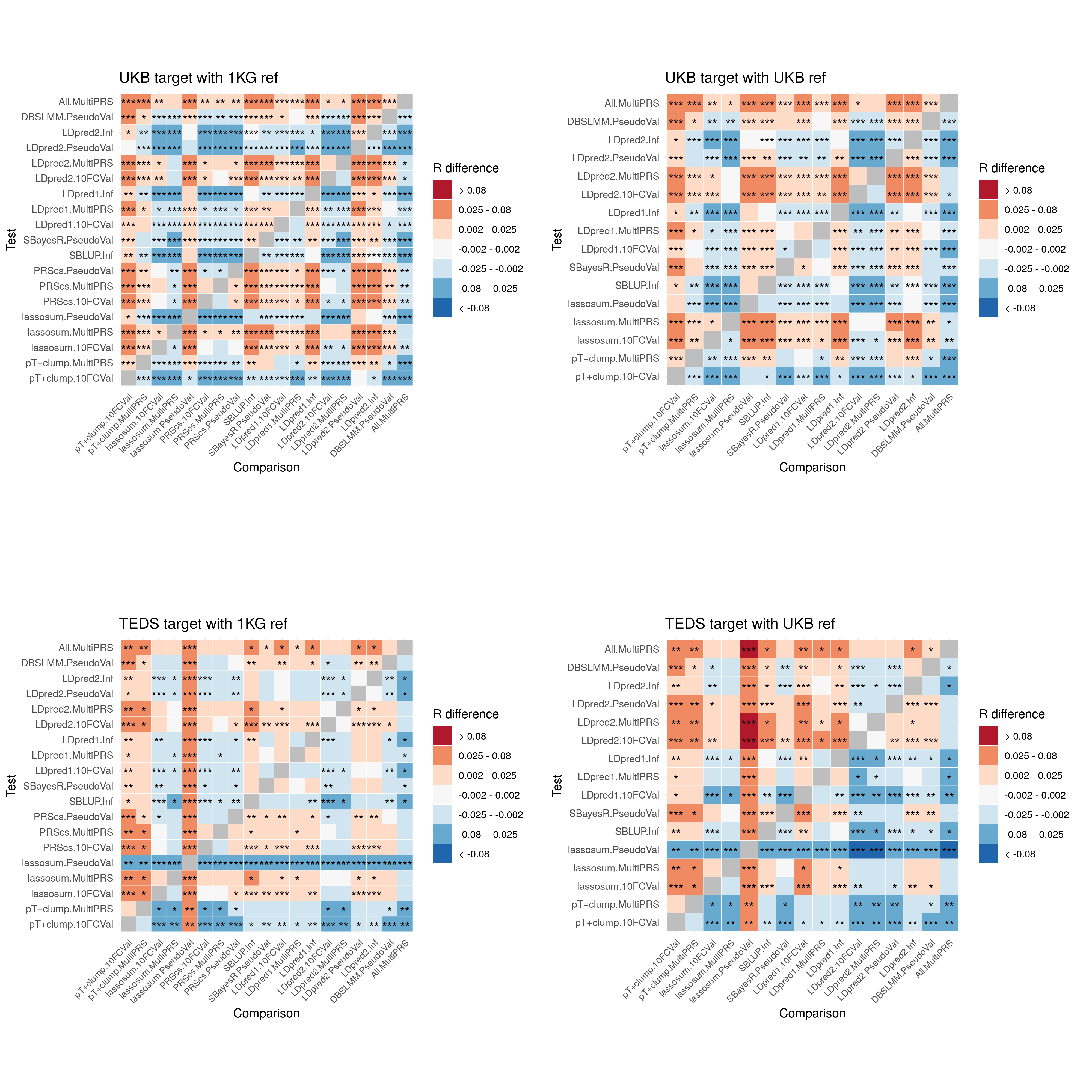
Predictive utility estimated using 10-fold cross-validation
Show test-validation results

Predictive utility estimated using test set validation
Show table: UKBB (1KG ref)
| Reference | Test | CrossVal Diff | CrossVal Diff (%) | CrossVal Diff Z | CrossVal Diff P | IndepVal Diff | IndepVal Diff (%) | IndepVal Diff Z | IndepVal Diff P |
|---|---|---|---|---|---|---|---|---|---|
| pT+clump.10FCVal | pT+clump.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| pT+clump.MultiPRS | pT+clump.10FCVal | -0.020 | -13.020 | -6.724 | 1.77e-11 | -0.021 | -13.831 | -9.574 | 1.03e-21 |
| lassosum.10FCVal | pT+clump.10FCVal | -0.035 | -22.615 | -19.871 | 7.24e-88 | -0.030 | -19.649 | -8.484 | 2.18e-17 |
| lassosum.MultiPRS | pT+clump.10FCVal | -0.044 | -28.560 | -14.796 | 1.55e-49 | -0.041 | -26.861 | -12.592 | 2.33e-36 |
| lassosum.PseudoVal | pT+clump.10FCVal | -0.004 | -2.616 | -2.119 | 3.41e-02 | -0.004 | -2.652 | -1.057 | 2.91e-01 |
| PRScs.10FCVal | pT+clump.10FCVal | -0.036 | -23.037 | -19.236 | 1.84e-82 | -0.030 | -19.638 | -8.054 | 7.98e-16 |
| PRScs.MultiPRS | pT+clump.10FCVal | -0.039 | -25.496 | -13.216 | 7.11e-40 | -0.034 | -22.541 | -9.904 | 3.99e-23 |
| PRScs.PseudoVal | pT+clump.10FCVal | -0.034 | -21.666 | -18.786 | 9.88e-79 | -0.029 | -19.050 | -8.098 | 5.59e-16 |
| SBLUP.Inf | pT+clump.10FCVal | -0.008 | -4.963 | -3.517 | 4.36e-04 | -0.003 | -2.059 | -0.720 | 4.72e-01 |
| SBayesR.PseudoVal | pT+clump.10FCVal | -0.015 | -9.829 | -8.242 | 1.70e-16 | -0.014 | -9.186 | -3.781 | 1.56e-04 |
| LDpred1.10FCVal | pT+clump.10FCVal | -0.024 | -15.258 | -12.312 | 7.80e-35 | -0.019 | -12.165 | -4.830 | 1.36e-06 |
| LDpred1.MultiPRS | pT+clump.10FCVal | -0.027 | -17.272 | -8.939 | 3.92e-19 | -0.022 | -14.272 | -5.782 | 7.40e-09 |
| LDpred1.Inf | pT+clump.10FCVal | -0.008 | -5.186 | -3.784 | 1.54e-04 | -0.003 | -2.262 | -0.812 | 4.17e-01 |
| LDpred2.10FCVal | pT+clump.10FCVal | -0.039 | -25.109 | -23.022 | 2.79e-117 | -0.034 | -22.480 | -10.097 | 5.71e-24 |
| LDpred2.MultiPRS | pT+clump.10FCVal | -0.042 | -27.112 | -14.034 | 9.72e-45 | -0.038 | -24.631 | -10.750 | 5.90e-27 |
| LDpred2.PseudoVal | pT+clump.10FCVal | 0.000 | -0.101 | -0.069 | 9.45e-01 | 0.002 | 1.382 | 0.462 | 6.44e-01 |
| LDpred2.Inf | pT+clump.10FCVal | -0.006 | -3.751 | -2.610 | 9.04e-03 | -0.002 | -1.185 | -0.407 | 6.84e-01 |
| DBSLMM.PseudoVal | pT+clump.10FCVal | -0.027 | -17.677 | -15.912 | 5.21e-57 | -0.022 | -14.656 | -6.480 | 9.17e-11 |
| All.MultiPRS | pT+clump.10FCVal | -0.047 | -30.588 | -15.977 | 1.83e-57 | -0.044 | -29.098 | -14.192 | 1.02e-45 |
| pT+clump.10FCVal | pT+clump.MultiPRS | 0.020 | 11.520 | 6.724 | 1.77e-11 | 0.021 | 12.151 | 9.574 | 1.03e-21 |
| pT+clump.MultiPRS | pT+clump.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.10FCVal | pT+clump.MultiPRS | -0.015 | -8.490 | -4.978 | 6.41e-07 | -0.009 | -5.111 | -3.228 | 1.25e-03 |
| lassosum.MultiPRS | pT+clump.MultiPRS | -0.024 | -13.750 | -8.081 | 6.40e-16 | -0.020 | -11.446 | -8.216 | 2.10e-16 |
| lassosum.PseudoVal | pT+clump.MultiPRS | 0.016 | 9.205 | 5.371 | 7.84e-08 | 0.017 | 9.821 | 5.233 | 1.66e-07 |
| PRScs.10FCVal | pT+clump.MultiPRS | -0.016 | -8.863 | -5.197 | 2.02e-07 | -0.009 | -5.101 | -3.098 | 1.95e-03 |
| PRScs.MultiPRS | pT+clump.MultiPRS | -0.019 | -11.039 | -6.477 | 9.38e-11 | -0.013 | -7.652 | -5.048 | 4.47e-07 |
| PRScs.PseudoVal | pT+clump.MultiPRS | -0.013 | -7.651 | -4.484 | 7.33e-06 | -0.008 | -4.584 | -2.758 | 5.82e-03 |
| SBLUP.Inf | pT+clump.MultiPRS | 0.012 | 7.128 | 4.164 | 3.13e-05 | 0.018 | 10.342 | 5.236 | 1.64e-07 |
| SBayesR.PseudoVal | pT+clump.MultiPRS | 0.005 | 2.823 | 1.650 | 9.90e-02 | 0.007 | 4.081 | 2.302 | 2.13e-02 |
| LDpred1.10FCVal | pT+clump.MultiPRS | -0.003 | -1.981 | -1.159 | 2.46e-01 | 0.003 | 1.464 | 0.860 | 3.90e-01 |
| LDpred1.MultiPRS | pT+clump.MultiPRS | -0.007 | -3.762 | -2.201 | 2.77e-02 | -0.001 | -0.387 | -0.230 | 8.18e-01 |
| LDpred1.Inf | pT+clump.MultiPRS | 0.012 | 6.931 | 4.049 | 5.13e-05 | 0.018 | 10.163 | 5.153 | 2.56e-07 |
| LDpred2.10FCVal | pT+clump.MultiPRS | -0.019 | -10.697 | -6.274 | 3.52e-10 | -0.013 | -7.598 | -4.577 | 4.73e-06 |
| LDpred2.MultiPRS | pT+clump.MultiPRS | -0.022 | -12.469 | -7.324 | 2.41e-13 | -0.017 | -9.487 | -5.975 | 2.30e-09 |
| LDpred2.PseudoVal | pT+clump.MultiPRS | 0.020 | 11.430 | 6.666 | 2.63e-11 | 0.023 | 13.365 | 6.493 | 8.42e-11 |
| LDpred2.Inf | pT+clump.MultiPRS | 0.014 | 8.201 | 4.789 | 1.68e-06 | 0.019 | 11.110 | 5.447 | 5.13e-08 |
| DBSLMM.PseudoVal | pT+clump.MultiPRS | -0.007 | -4.120 | -2.413 | 1.58e-02 | -0.001 | -0.724 | -0.425 | 6.71e-01 |
| All.MultiPRS | pT+clump.MultiPRS | -0.027 | -15.545 | -9.134 | 6.58e-20 | -0.023 | -13.411 | -10.002 | 1.50e-23 |
| pT+clump.10FCVal | lassosum.10FCVal | 0.035 | 18.444 | 19.871 | 7.24e-88 | 0.030 | 16.422 | 8.484 | 2.18e-17 |
| pT+clump.MultiPRS | lassosum.10FCVal | 0.015 | 7.825 | 4.978 | 6.41e-07 | 0.009 | 4.862 | 3.228 | 1.25e-03 |
| lassosum.10FCVal | lassosum.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.MultiPRS | lassosum.10FCVal | -0.009 | -4.849 | -3.092 | 1.99e-03 | -0.011 | -6.027 | -7.878 | 3.34e-15 |
| lassosum.PseudoVal | lassosum.10FCVal | 0.031 | 16.310 | 21.066 | 1.62e-98 | 0.026 | 14.206 | 8.831 | 1.04e-18 |
| PRScs.10FCVal | lassosum.10FCVal | -0.001 | -0.344 | -0.747 | 4.55e-01 | 0.000 | 0.009 | 0.010 | 9.92e-01 |
| PRScs.MultiPRS | lassosum.10FCVal | -0.004 | -2.350 | -1.501 | 1.33e-01 | -0.004 | -2.417 | -2.520 | 1.17e-02 |
| PRScs.PseudoVal | lassosum.10FCVal | 0.001 | 0.774 | 1.445 | 1.49e-01 | 0.001 | 0.501 | 0.450 | 6.53e-01 |
| SBLUP.Inf | lassosum.10FCVal | 0.027 | 14.396 | 18.505 | 1.88e-76 | 0.027 | 14.702 | 9.096 | 9.39e-20 |
| SBayesR.PseudoVal | lassosum.10FCVal | 0.020 | 10.428 | 11.949 | 6.54e-33 | 0.016 | 8.745 | 4.807 | 1.53e-06 |
| LDpred1.10FCVal | lassosum.10FCVal | 0.011 | 6.000 | 9.228 | 2.75e-20 | 0.011 | 6.255 | 4.633 | 3.61e-06 |
| LDpred1.MultiPRS | lassosum.10FCVal | 0.008 | 4.357 | 2.782 | 5.41e-03 | 0.008 | 4.494 | 3.488 | 4.86e-04 |
| LDpred1.Inf | lassosum.10FCVal | 0.027 | 14.214 | 18.843 | 3.35e-79 | 0.027 | 14.532 | 9.266 | 1.93e-20 |
| LDpred2.10FCVal | lassosum.10FCVal | -0.004 | -2.035 | -3.526 | 4.21e-04 | -0.004 | -2.366 | -1.980 | 4.77e-02 |
| LDpred2.MultiPRS | lassosum.10FCVal | -0.007 | -3.668 | -2.341 | 1.93e-02 | -0.008 | -4.163 | -3.655 | 2.57e-04 |
| LDpred2.PseudoVal | lassosum.10FCVal | 0.035 | 18.361 | 21.630 | 9.31e-104 | 0.032 | 17.578 | 9.951 | 2.49e-23 |
| LDpred2.Inf | lassosum.10FCVal | 0.029 | 15.385 | 18.957 | 3.88e-80 | 0.028 | 15.432 | 9.146 | 5.91e-20 |
| DBSLMM.PseudoVal | lassosum.10FCVal | 0.008 | 4.027 | 5.679 | 1.35e-08 | 0.008 | 4.174 | 2.823 | 4.76e-03 |
| All.MultiPRS | lassosum.10FCVal | -0.012 | -6.503 | -4.192 | 2.76e-05 | -0.014 | -7.897 | -7.869 | 3.57e-15 |
| pT+clump.10FCVal | lassosum.MultiPRS | 0.044 | 22.215 | 14.796 | 1.55e-49 | 0.041 | 21.173 | 12.592 | 2.33e-36 |
| pT+clump.MultiPRS | lassosum.MultiPRS | 0.024 | 12.088 | 8.081 | 6.40e-16 | 0.020 | 10.270 | 8.216 | 2.10e-16 |
| lassosum.10FCVal | lassosum.MultiPRS | 0.009 | 4.625 | 3.092 | 1.99e-03 | 0.011 | 5.684 | 7.878 | 3.34e-15 |
| lassosum.MultiPRS | lassosum.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.PseudoVal | lassosum.MultiPRS | 0.040 | 20.181 | 13.452 | 3.00e-41 | 0.037 | 19.083 | 14.155 | 1.74e-45 |
| PRScs.10FCVal | lassosum.MultiPRS | 0.009 | 4.296 | 2.872 | 4.08e-03 | 0.011 | 5.693 | 6.405 | 1.50e-10 |
| PRScs.MultiPRS | lassosum.MultiPRS | 0.005 | 2.383 | 2.364 | 1.81e-02 | 0.007 | 3.405 | 4.748 | 2.05e-06 |
| PRScs.PseudoVal | lassosum.MultiPRS | 0.011 | 5.362 | 3.583 | 3.40e-04 | 0.012 | 6.157 | 6.830 | 8.51e-12 |
| SBLUP.Inf | lassosum.MultiPRS | 0.037 | 18.355 | 12.218 | 2.49e-34 | 0.038 | 19.551 | 13.030 | 8.30e-39 |
| SBayesR.PseudoVal | lassosum.MultiPRS | 0.029 | 14.570 | 9.733 | 2.19e-22 | 0.027 | 13.932 | 9.165 | 4.94e-20 |
| LDpred1.10FCVal | lassosum.MultiPRS | 0.021 | 10.347 | 6.901 | 5.15e-12 | 0.022 | 11.584 | 10.273 | 9.28e-25 |
| LDpred1.MultiPRS | lassosum.MultiPRS | 0.017 | 8.781 | 7.264 | 3.76e-13 | 0.019 | 9.923 | 9.198 | 3.64e-20 |
| LDpred1.Inf | lassosum.MultiPRS | 0.036 | 18.181 | 12.100 | 1.06e-33 | 0.038 | 19.390 | 13.424 | 4.40e-41 |
| LDpred2.10FCVal | lassosum.MultiPRS | 0.005 | 2.684 | 1.797 | 7.24e-02 | 0.007 | 3.453 | 3.632 | 2.81e-04 |
| LDpred2.MultiPRS | lassosum.MultiPRS | 0.002 | 1.126 | 1.039 | 2.99e-01 | 0.003 | 1.758 | 2.019 | 4.34e-02 |
| LDpred2.PseudoVal | lassosum.MultiPRS | 0.044 | 22.137 | 14.727 | 4.31e-49 | 0.043 | 22.263 | 13.892 | 7.06e-44 |
| LDpred2.Inf | lassosum.MultiPRS | 0.038 | 19.298 | 12.843 | 9.36e-38 | 0.039 | 20.240 | 12.884 | 5.53e-38 |
| DBSLMM.PseudoVal | lassosum.MultiPRS | 0.017 | 8.466 | 5.653 | 1.58e-08 | 0.019 | 9.621 | 8.448 | 2.96e-17 |
| All.MultiPRS | lassosum.MultiPRS | -0.003 | -1.577 | -1.392 | 1.64e-01 | -0.003 | -1.763 | -2.904 | 3.68e-03 |
| pT+clump.10FCVal | lassosum.PseudoVal | 0.004 | 2.549 | 2.119 | 3.41e-02 | 0.004 | 2.583 | 1.057 | 2.91e-01 |
| pT+clump.MultiPRS | lassosum.PseudoVal | -0.016 | -10.139 | -5.371 | 7.84e-08 | -0.017 | -10.891 | -5.233 | 1.66e-07 |
| lassosum.10FCVal | lassosum.PseudoVal | -0.031 | -19.489 | -21.066 | 1.62e-98 | -0.026 | -16.559 | -8.831 | 1.04e-18 |
| lassosum.MultiPRS | lassosum.PseudoVal | -0.040 | -25.283 | -13.452 | 3.00e-41 | -0.037 | -23.584 | -14.155 | 1.74e-45 |
| lassosum.PseudoVal | lassosum.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| PRScs.10FCVal | lassosum.PseudoVal | -0.032 | -19.901 | -19.392 | 9.04e-84 | -0.026 | -16.548 | -7.976 | 1.51e-15 |
| PRScs.MultiPRS | lassosum.PseudoVal | -0.035 | -22.297 | -11.869 | 1.71e-32 | -0.030 | -19.376 | -10.910 | 1.03e-27 |
| PRScs.PseudoVal | lassosum.PseudoVal | -0.029 | -18.565 | -20.610 | 2.25e-94 | -0.025 | -15.974 | -8.784 | 1.58e-18 |
| SBLUP.Inf | lassosum.PseudoVal | -0.004 | -2.288 | -1.715 | 8.63e-02 | 0.001 | 0.578 | 0.214 | 8.30e-01 |
| SBayesR.PseudoVal | lassosum.PseudoVal | -0.011 | -7.029 | -6.858 | 6.99e-12 | -0.010 | -6.366 | -3.061 | 2.21e-03 |
| LDpred1.10FCVal | lassosum.PseudoVal | -0.020 | -12.320 | -12.368 | 3.90e-35 | -0.015 | -9.267 | -4.616 | 3.91e-06 |
| LDpred1.MultiPRS | lassosum.PseudoVal | -0.023 | -14.283 | -7.599 | 2.98e-14 | -0.018 | -11.320 | -5.600 | 2.15e-08 |
| LDpred1.Inf | lassosum.PseudoVal | -0.004 | -2.505 | -1.949 | 5.13e-02 | 0.001 | 0.379 | 0.146 | 8.84e-01 |
| LDpred2.10FCVal | lassosum.PseudoVal | -0.035 | -21.920 | -24.441 | 6.26e-132 | -0.030 | -19.316 | -10.639 | 1.96e-26 |
| LDpred2.MultiPRS | lassosum.PseudoVal | -0.038 | -23.872 | -12.691 | 6.67e-37 | -0.034 | -21.411 | -11.505 | 1.24e-30 |
| LDpred2.PseudoVal | lassosum.PseudoVal | 0.004 | 2.450 | 1.918 | 5.52e-02 | 0.006 | 3.930 | 1.520 | 1.29e-01 |
| LDpred2.Inf | lassosum.PseudoVal | -0.002 | -1.106 | -0.817 | 4.14e-01 | 0.002 | 1.429 | 0.521 | 6.02e-01 |
| DBSLMM.PseudoVal | lassosum.PseudoVal | -0.023 | -14.677 | -14.968 | 1.19e-50 | -0.018 | -11.694 | -5.877 | 4.17e-09 |
| All.MultiPRS | lassosum.PseudoVal | -0.043 | -27.259 | -14.635 | 1.68e-48 | -0.040 | -25.763 | -14.291 | 2.50e-46 |
| pT+clump.10FCVal | PRScs.10FCVal | 0.036 | 18.724 | 19.236 | 1.84e-82 | 0.030 | 16.415 | 8.054 | 7.98e-16 |
| pT+clump.MultiPRS | PRScs.10FCVal | 0.016 | 8.142 | 5.197 | 2.02e-07 | 0.009 | 4.854 | 3.098 | 1.95e-03 |
| lassosum.10FCVal | PRScs.10FCVal | 0.001 | 0.343 | 0.747 | 4.55e-01 | 0.000 | -0.009 | -0.010 | 9.92e-01 |
| lassosum.MultiPRS | PRScs.10FCVal | -0.009 | -4.489 | -2.872 | 4.08e-03 | -0.011 | -6.037 | -6.405 | 1.50e-10 |
| lassosum.PseudoVal | PRScs.10FCVal | 0.032 | 16.598 | 19.392 | 9.04e-84 | 0.026 | 14.198 | 7.976 | 1.51e-15 |
| PRScs.10FCVal | PRScs.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| PRScs.MultiPRS | PRScs.10FCVal | -0.004 | -1.999 | -1.281 | 2.00e-01 | -0.004 | -2.427 | -4.123 | 3.75e-05 |
| PRScs.PseudoVal | PRScs.10FCVal | 0.002 | 1.114 | 2.797 | 5.16e-03 | 0.001 | 0.492 | 0.593 | 5.54e-01 |
| SBLUP.Inf | PRScs.10FCVal | 0.028 | 14.690 | 22.036 | 1.30e-107 | 0.027 | 14.694 | 10.594 | 3.19e-26 |
| SBayesR.PseudoVal | PRScs.10FCVal | 0.020 | 10.735 | 12.128 | 7.48e-34 | 0.016 | 8.736 | 4.712 | 2.45e-06 |
| LDpred1.10FCVal | PRScs.10FCVal | 0.012 | 6.322 | 10.950 | 6.62e-28 | 0.011 | 6.247 | 5.219 | 1.80e-07 |
| LDpred1.MultiPRS | PRScs.10FCVal | 0.009 | 4.686 | 3.002 | 2.68e-03 | 0.008 | 4.485 | 3.899 | 9.66e-05 |
| LDpred1.Inf | PRScs.10FCVal | 0.028 | 14.508 | 22.021 | 1.81e-107 | 0.027 | 14.524 | 10.615 | 2.54e-26 |
| LDpred2.10FCVal | PRScs.10FCVal | -0.003 | -1.684 | -2.912 | 3.59e-03 | -0.004 | -2.375 | -1.982 | 4.75e-02 |
| LDpred2.MultiPRS | PRScs.10FCVal | -0.006 | -3.312 | -2.121 | 3.39e-02 | -0.008 | -4.173 | -3.855 | 1.16e-04 |
| LDpred2.PseudoVal | PRScs.10FCVal | 0.035 | 18.641 | 25.153 | 1.30e-139 | 0.032 | 17.570 | 11.380 | 5.23e-30 |
| LDpred2.Inf | PRScs.10FCVal | 0.030 | 15.675 | 22.427 | 2.15e-111 | 0.028 | 15.425 | 10.597 | 3.08e-26 |
| DBSLMM.PseudoVal | PRScs.10FCVal | 0.008 | 4.357 | 7.086 | 1.38e-12 | 0.008 | 4.165 | 3.247 | 1.17e-03 |
| All.MultiPRS | PRScs.10FCVal | -0.012 | -6.137 | -3.972 | 7.14e-05 | -0.014 | -7.907 | -7.912 | 2.54e-15 |
| pT+clump.10FCVal | PRScs.MultiPRS | 0.039 | 20.316 | 13.216 | 7.11e-40 | 0.034 | 18.395 | 9.904 | 3.99e-23 |
| pT+clump.MultiPRS | PRScs.MultiPRS | 0.019 | 9.942 | 6.477 | 9.38e-11 | 0.013 | 7.108 | 5.048 | 4.47e-07 |
| lassosum.10FCVal | PRScs.MultiPRS | 0.004 | 2.296 | 1.501 | 1.33e-01 | 0.004 | 2.360 | 2.520 | 1.17e-02 |
| lassosum.MultiPRS | PRScs.MultiPRS | -0.005 | -2.441 | -2.364 | 1.81e-02 | -0.007 | -3.525 | -4.748 | 2.05e-06 |
| lassosum.PseudoVal | PRScs.MultiPRS | 0.035 | 18.232 | 11.869 | 1.71e-32 | 0.030 | 16.231 | 10.910 | 1.03e-27 |
| PRScs.10FCVal | PRScs.MultiPRS | 0.004 | 1.960 | 1.281 | 2.00e-01 | 0.004 | 2.369 | 4.123 | 3.75e-05 |
| PRScs.MultiPRS | PRScs.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| PRScs.PseudoVal | PRScs.MultiPRS | 0.006 | 3.052 | 1.996 | 4.59e-02 | 0.005 | 2.850 | 4.988 | 6.09e-07 |
| SBLUP.Inf | PRScs.MultiPRS | 0.032 | 16.361 | 10.660 | 1.56e-26 | 0.031 | 16.715 | 11.450 | 2.36e-30 |
| SBayesR.PseudoVal | PRScs.MultiPRS | 0.024 | 12.484 | 8.155 | 3.50e-16 | 0.020 | 10.899 | 6.351 | 2.13e-10 |
| LDpred1.10FCVal | PRScs.MultiPRS | 0.016 | 8.158 | 5.325 | 1.01e-07 | 0.016 | 8.468 | 8.527 | 1.50e-17 |
| LDpred1.MultiPRS | PRScs.MultiPRS | 0.013 | 6.553 | 6.298 | 3.02e-10 | 0.013 | 6.748 | 7.074 | 1.51e-12 |
| LDpred1.Inf | PRScs.MultiPRS | 0.031 | 16.184 | 10.547 | 5.26e-26 | 0.031 | 16.549 | 12.276 | 1.22e-34 |
| LDpred2.10FCVal | PRScs.MultiPRS | 0.001 | 0.308 | 0.202 | 8.40e-01 | 0.000 | 0.050 | 0.052 | 9.59e-01 |
| LDpred2.MultiPRS | PRScs.MultiPRS | -0.003 | -1.288 | -1.149 | 2.51e-01 | -0.003 | -1.705 | -1.864 | 6.23e-02 |
| LDpred2.PseudoVal | PRScs.MultiPRS | 0.039 | 20.236 | 13.167 | 1.35e-39 | 0.037 | 19.523 | 12.337 | 5.72e-35 |
| LDpred2.Inf | PRScs.MultiPRS | 0.034 | 17.327 | 11.288 | 1.50e-29 | 0.033 | 17.428 | 11.333 | 8.98e-30 |
| DBSLMM.PseudoVal | PRScs.MultiPRS | 0.012 | 6.231 | 4.071 | 4.68e-05 | 0.012 | 6.435 | 6.210 | 5.29e-10 |
| All.MultiPRS | PRScs.MultiPRS | -0.008 | -4.057 | -3.926 | 8.65e-05 | -0.010 | -5.350 | -6.565 | 5.20e-11 |
| pT+clump.10FCVal | PRScs.PseudoVal | 0.034 | 17.808 | 18.786 | 9.88e-79 | 0.029 | 16.001 | 8.098 | 5.59e-16 |
| pT+clump.MultiPRS | PRScs.PseudoVal | 0.013 | 7.107 | 4.484 | 7.33e-06 | 0.008 | 4.383 | 2.758 | 5.82e-03 |
| lassosum.10FCVal | PRScs.PseudoVal | -0.001 | -0.780 | -1.445 | 1.49e-01 | -0.001 | -0.504 | -0.450 | 6.53e-01 |
| lassosum.MultiPRS | PRScs.PseudoVal | -0.011 | -5.666 | -3.583 | 3.40e-04 | -0.012 | -6.561 | -6.830 | 8.51e-12 |
| lassosum.PseudoVal | PRScs.PseudoVal | 0.029 | 15.658 | 20.610 | 2.25e-94 | 0.025 | 13.774 | 8.784 | 1.58e-18 |
| PRScs.10FCVal | PRScs.PseudoVal | -0.002 | -1.127 | -2.797 | 5.16e-03 | -0.001 | -0.495 | -0.593 | 5.54e-01 |
| PRScs.MultiPRS | PRScs.PseudoVal | -0.006 | -3.148 | -1.996 | 4.59e-02 | -0.005 | -2.933 | -4.988 | 6.09e-07 |
| PRScs.PseudoVal | PRScs.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBLUP.Inf | PRScs.PseudoVal | 0.026 | 13.728 | 17.804 | 6.54e-71 | 0.026 | 14.272 | 8.945 | 3.73e-19 |
| SBayesR.PseudoVal | PRScs.PseudoVal | 0.018 | 9.729 | 10.739 | 6.70e-27 | 0.015 | 8.285 | 4.385 | 1.16e-05 |
| LDpred1.10FCVal | PRScs.PseudoVal | 0.010 | 5.267 | 10.132 | 3.98e-24 | 0.011 | 5.783 | 5.375 | 7.67e-08 |
| LDpred1.MultiPRS | PRScs.PseudoVal | 0.007 | 3.612 | 2.287 | 2.22e-02 | 0.007 | 4.013 | 3.774 | 1.61e-04 |
| LDpred1.Inf | PRScs.PseudoVal | 0.026 | 13.545 | 19.530 | 6.08e-85 | 0.026 | 14.101 | 9.822 | 9.03e-23 |
| LDpred2.10FCVal | PRScs.PseudoVal | -0.005 | -2.830 | -5.564 | 2.64e-08 | -0.005 | -2.882 | -2.731 | 6.31e-03 |
| LDpred2.MultiPRS | PRScs.PseudoVal | -0.008 | -4.476 | -2.833 | 4.61e-03 | -0.009 | -4.688 | -4.250 | 2.14e-05 |
| LDpred2.PseudoVal | PRScs.PseudoVal | 0.033 | 17.725 | 22.100 | 3.14e-108 | 0.031 | 17.163 | 10.328 | 5.29e-25 |
| LDpred2.Inf | PRScs.PseudoVal | 0.028 | 14.725 | 18.304 | 7.65e-75 | 0.027 | 15.006 | 9.004 | 2.18e-19 |
| DBSLMM.PseudoVal | PRScs.PseudoVal | 0.006 | 3.279 | 6.561 | 5.36e-11 | 0.007 | 3.691 | 3.546 | 3.92e-04 |
| All.MultiPRS | PRScs.PseudoVal | -0.014 | -7.333 | -4.690 | 2.73e-06 | -0.015 | -8.440 | -8.253 | 1.55e-16 |
| pT+clump.10FCVal | SBLUP.Inf | 0.008 | 4.729 | 3.517 | 4.36e-04 | 0.003 | 2.017 | 0.720 | 4.72e-01 |
| pT+clump.MultiPRS | SBLUP.Inf | -0.012 | -7.675 | -4.164 | 3.13e-05 | -0.018 | -11.535 | -5.236 | 1.64e-07 |
| lassosum.10FCVal | SBLUP.Inf | -0.027 | -16.817 | -18.505 | 1.88e-76 | -0.027 | -17.236 | -9.096 | 9.39e-20 |
| lassosum.MultiPRS | SBLUP.Inf | -0.037 | -22.481 | -12.218 | 2.49e-34 | -0.038 | -24.302 | -13.030 | 8.30e-39 |
| lassosum.PseudoVal | SBLUP.Inf | 0.004 | 2.237 | 1.715 | 8.63e-02 | -0.001 | -0.581 | -0.214 | 8.30e-01 |
| PRScs.10FCVal | SBLUP.Inf | -0.028 | -17.219 | -22.036 | 1.30e-107 | -0.027 | -17.225 | -10.594 | 3.19e-26 |
| PRScs.MultiPRS | SBLUP.Inf | -0.032 | -19.562 | -10.660 | 1.56e-26 | -0.031 | -20.070 | -11.450 | 2.36e-30 |
| PRScs.PseudoVal | SBLUP.Inf | -0.026 | -15.913 | -17.804 | 6.54e-71 | -0.026 | -16.648 | -8.945 | 3.73e-19 |
| SBLUP.Inf | SBLUP.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBayesR.PseudoVal | SBLUP.Inf | -0.008 | -4.635 | -3.881 | 1.04e-04 | -0.011 | -6.984 | -2.802 | 5.07e-03 |
| LDpred1.10FCVal | SBLUP.Inf | -0.016 | -9.808 | -12.976 | 1.68e-38 | -0.015 | -9.902 | -6.292 | 3.13e-10 |
| LDpred1.MultiPRS | SBLUP.Inf | -0.019 | -11.727 | -6.385 | 1.71e-10 | -0.019 | -11.967 | -7.307 | 2.73e-13 |
| LDpred1.Inf | SBLUP.Inf | 0.000 | -0.212 | -0.397 | 6.91e-01 | 0.000 | -0.199 | -0.179 | 8.58e-01 |
| LDpred2.10FCVal | SBLUP.Inf | -0.031 | -19.193 | -19.484 | 1.50e-84 | -0.031 | -20.010 | -9.751 | 1.83e-22 |
| LDpred2.MultiPRS | SBLUP.Inf | -0.034 | -21.102 | -11.480 | 1.66e-30 | -0.035 | -22.117 | -11.881 | 1.48e-32 |
| LDpred2.PseudoVal | SBLUP.Inf | 0.008 | 4.632 | 9.685 | 3.51e-22 | 0.005 | 3.372 | 3.358 | 7.86e-04 |
| LDpred2.Inf | SBLUP.Inf | 0.002 | 1.155 | 6.110 | 9.97e-10 | 0.001 | 0.857 | 2.173 | 2.98e-02 |
| DBSLMM.PseudoVal | SBLUP.Inf | -0.020 | -12.112 | -11.279 | 1.66e-29 | -0.019 | -12.343 | -5.510 | 3.59e-08 |
| All.MultiPRS | SBLUP.Inf | -0.040 | -24.413 | -13.405 | 5.68e-41 | -0.041 | -26.494 | -14.031 | 1.01e-44 |
| pT+clump.10FCVal | SBayesR.PseudoVal | 0.015 | 8.949 | 8.242 | 1.70e-16 | 0.014 | 8.413 | 3.781 | 1.56e-04 |
| pT+clump.MultiPRS | SBayesR.PseudoVal | -0.005 | -2.905 | -1.650 | 9.90e-02 | -0.007 | -4.254 | -2.302 | 2.13e-02 |
| lassosum.10FCVal | SBayesR.PseudoVal | -0.020 | -11.642 | -11.949 | 6.54e-33 | -0.016 | -9.583 | -4.807 | 1.53e-06 |
| lassosum.MultiPRS | SBayesR.PseudoVal | -0.029 | -17.055 | -9.733 | 2.19e-22 | -0.027 | -16.187 | -9.165 | 4.94e-20 |
| lassosum.PseudoVal | SBayesR.PseudoVal | 0.011 | 6.568 | 6.858 | 6.99e-12 | 0.010 | 5.985 | 3.061 | 2.21e-03 |
| PRScs.10FCVal | SBayesR.PseudoVal | -0.020 | -12.026 | -12.128 | 7.48e-34 | -0.016 | -9.573 | -4.712 | 2.45e-06 |
| PRScs.MultiPRS | SBayesR.PseudoVal | -0.024 | -14.265 | -8.155 | 3.50e-16 | -0.020 | -12.232 | -6.351 | 2.13e-10 |
| PRScs.PseudoVal | SBayesR.PseudoVal | -0.018 | -10.778 | -10.739 | 6.70e-27 | -0.015 | -9.034 | -4.385 | 1.16e-05 |
| SBLUP.Inf | SBayesR.PseudoVal | 0.008 | 4.430 | 3.881 | 1.04e-04 | 0.011 | 6.528 | 2.802 | 5.07e-03 |
| SBayesR.PseudoVal | SBayesR.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.10FCVal | SBayesR.PseudoVal | -0.008 | -4.943 | -5.008 | 5.51e-07 | -0.005 | -2.728 | -1.353 | 1.76e-01 |
| LDpred1.MultiPRS | SBayesR.PseudoVal | -0.012 | -6.777 | -3.868 | 1.10e-04 | -0.008 | -4.658 | -2.316 | 2.06e-02 |
| LDpred1.Inf | SBayesR.PseudoVal | 0.007 | 4.227 | 3.665 | 2.47e-04 | 0.011 | 6.341 | 2.695 | 7.04e-03 |
| LDpred2.10FCVal | SBayesR.PseudoVal | -0.024 | -13.913 | -14.632 | 1.76e-48 | -0.020 | -12.175 | -6.250 | 4.11e-10 |
| LDpred2.MultiPRS | SBayesR.PseudoVal | -0.027 | -15.737 | -8.980 | 2.70e-19 | -0.024 | -14.145 | -7.804 | 5.98e-15 |
| LDpred2.PseudoVal | SBayesR.PseudoVal | 0.015 | 8.857 | 7.975 | 1.52e-15 | 0.016 | 9.679 | 4.277 | 1.90e-05 |
| LDpred2.Inf | SBayesR.PseudoVal | 0.009 | 5.534 | 4.781 | 1.74e-06 | 0.012 | 7.328 | 3.105 | 1.91e-03 |
| DBSLMM.PseudoVal | SBayesR.PseudoVal | -0.012 | -7.145 | -7.132 | 9.89e-13 | -0.008 | -5.009 | -2.431 | 1.50e-02 |
| All.MultiPRS | SBayesR.PseudoVal | -0.032 | -18.901 | -10.897 | 1.19e-27 | -0.030 | -18.236 | -11.200 | 4.06e-29 |
| pT+clump.10FCVal | LDpred1.10FCVal | 0.024 | 13.238 | 12.312 | 7.80e-35 | 0.019 | 10.845 | 4.830 | 1.36e-06 |
| pT+clump.MultiPRS | LDpred1.10FCVal | 0.003 | 1.942 | 1.159 | 2.46e-01 | -0.003 | -1.486 | -0.860 | 3.90e-01 |
| lassosum.10FCVal | LDpred1.10FCVal | -0.011 | -6.383 | -9.228 | 2.75e-20 | -0.011 | -6.673 | -4.633 | 3.61e-06 |
| lassosum.MultiPRS | LDpred1.10FCVal | -0.021 | -11.541 | -6.901 | 5.15e-12 | -0.022 | -13.102 | -10.273 | 9.28e-25 |
| lassosum.PseudoVal | LDpred1.10FCVal | 0.020 | 10.969 | 12.368 | 3.90e-35 | 0.015 | 8.481 | 4.616 | 3.91e-06 |
| PRScs.10FCVal | LDpred1.10FCVal | -0.012 | -6.749 | -10.950 | 6.62e-28 | -0.011 | -6.663 | -5.219 | 1.80e-07 |
| PRScs.MultiPRS | LDpred1.10FCVal | -0.016 | -8.883 | -5.325 | 1.01e-07 | -0.016 | -9.251 | -8.527 | 1.50e-17 |
| PRScs.PseudoVal | LDpred1.10FCVal | -0.010 | -5.560 | -10.132 | 3.98e-24 | -0.011 | -6.138 | -5.375 | 7.67e-08 |
| SBLUP.Inf | LDpred1.10FCVal | 0.016 | 8.932 | 12.976 | 1.68e-38 | 0.015 | 9.010 | 6.292 | 3.13e-10 |
| SBayesR.PseudoVal | LDpred1.10FCVal | 0.008 | 4.710 | 5.008 | 5.51e-07 | 0.005 | 2.656 | 1.353 | 1.76e-01 |
| LDpred1.10FCVal | LDpred1.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.MultiPRS | LDpred1.10FCVal | -0.003 | -1.747 | -1.047 | 2.95e-01 | -0.003 | -1.879 | -4.086 | 4.39e-05 |
| LDpred1.Inf | LDpred1.10FCVal | 0.016 | 8.738 | 18.234 | 2.79e-74 | 0.015 | 8.829 | 8.840 | 9.58e-19 |
| LDpred2.10FCVal | LDpred1.10FCVal | -0.015 | -8.547 | -13.324 | 1.67e-40 | -0.016 | -9.197 | -6.889 | 5.60e-12 |
| LDpred2.MultiPRS | LDpred1.10FCVal | -0.018 | -10.285 | -6.158 | 7.36e-10 | -0.019 | -11.114 | -8.540 | 1.34e-17 |
| LDpred2.PseudoVal | LDpred1.10FCVal | 0.023 | 13.150 | 18.108 | 2.78e-73 | 0.021 | 12.078 | 7.998 | 1.27e-15 |
| LDpred2.Inf | LDpred1.10FCVal | 0.018 | 9.984 | 13.602 | 3.90e-42 | 0.017 | 9.789 | 6.405 | 1.50e-10 |
| DBSLMM.PseudoVal | LDpred1.10FCVal | -0.004 | -2.098 | -2.989 | 2.80e-03 | -0.004 | -2.221 | -1.517 | 1.29e-01 |
| All.MultiPRS | LDpred1.10FCVal | -0.024 | -13.301 | -8.041 | 8.93e-16 | -0.026 | -15.096 | -11.747 | 7.30e-32 |
| pT+clump.10FCVal | LDpred1.MultiPRS | 0.027 | 14.728 | 8.939 | 3.92e-19 | 0.022 | 12.490 | 5.782 | 7.40e-09 |
| pT+clump.MultiPRS | LDpred1.MultiPRS | 0.007 | 3.626 | 2.201 | 2.77e-02 | 0.001 | 0.386 | 0.230 | 8.18e-01 |
| lassosum.10FCVal | LDpred1.MultiPRS | -0.008 | -4.556 | -2.782 | 5.41e-03 | -0.008 | -4.706 | -3.488 | 4.86e-04 |
| lassosum.MultiPRS | LDpred1.MultiPRS | -0.017 | -9.626 | -7.264 | 3.76e-13 | -0.019 | -11.016 | -9.198 | 3.64e-20 |
| lassosum.PseudoVal | LDpred1.MultiPRS | 0.023 | 12.498 | 7.599 | 2.98e-14 | 0.018 | 10.169 | 5.600 | 2.15e-08 |
| PRScs.10FCVal | LDpred1.MultiPRS | -0.009 | -4.916 | -3.002 | 2.68e-03 | -0.008 | -4.696 | -3.899 | 9.66e-05 |
| PRScs.MultiPRS | LDpred1.MultiPRS | -0.013 | -7.013 | -6.298 | 3.02e-10 | -0.013 | -7.236 | -7.074 | 1.51e-12 |
| PRScs.PseudoVal | LDpred1.MultiPRS | -0.007 | -3.747 | -2.287 | 2.22e-02 | -0.007 | -4.181 | -3.774 | 1.61e-04 |
| SBLUP.Inf | LDpred1.MultiPRS | 0.019 | 10.496 | 6.385 | 1.71e-10 | 0.019 | 10.688 | 7.307 | 2.73e-13 |
| SBayesR.PseudoVal | LDpred1.MultiPRS | 0.012 | 6.347 | 3.868 | 1.10e-04 | 0.008 | 4.451 | 2.316 | 2.06e-02 |
| LDpred1.10FCVal | LDpred1.MultiPRS | 0.003 | 1.717 | 1.047 | 2.95e-01 | 0.003 | 1.844 | 4.086 | 4.39e-05 |
| LDpred1.MultiPRS | LDpred1.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.Inf | LDpred1.MultiPRS | 0.019 | 10.306 | 6.270 | 3.62e-10 | 0.018 | 10.510 | 10.161 | 2.95e-24 |
| LDpred2.10FCVal | LDpred1.MultiPRS | -0.012 | -6.683 | -4.086 | 4.38e-05 | -0.013 | -7.183 | -5.730 | 1.00e-08 |
| LDpred2.MultiPRS | LDpred1.MultiPRS | -0.015 | -8.391 | -6.405 | 1.51e-10 | -0.016 | -9.065 | -7.435 | 1.04e-13 |
| LDpred2.PseudoVal | LDpred1.MultiPRS | 0.027 | 14.642 | 8.895 | 5.82e-19 | 0.024 | 13.699 | 8.793 | 1.46e-18 |
| LDpred2.Inf | LDpred1.MultiPRS | 0.021 | 11.530 | 7.012 | 2.34e-12 | 0.020 | 11.453 | 7.384 | 1.53e-13 |
| DBSLMM.PseudoVal | LDpred1.MultiPRS | -0.001 | -0.345 | -0.210 | 8.33e-01 | -0.001 | -0.336 | -0.238 | 8.12e-01 |
| All.MultiPRS | LDpred1.MultiPRS | -0.021 | -11.355 | -10.071 | 7.44e-24 | -0.023 | -12.974 | -10.918 | 9.41e-28 |
| pT+clump.10FCVal | LDpred1.Inf | 0.008 | 4.931 | 3.784 | 1.54e-04 | 0.003 | 2.212 | 0.812 | 4.17e-01 |
| pT+clump.MultiPRS | LDpred1.Inf | -0.012 | -7.447 | -4.049 | 5.13e-05 | -0.018 | -11.313 | -5.153 | 2.56e-07 |
| lassosum.10FCVal | LDpred1.Inf | -0.027 | -16.569 | -18.843 | 3.35e-79 | -0.027 | -17.002 | -9.266 | 1.93e-20 |
| lassosum.MultiPRS | LDpred1.Inf | -0.036 | -22.221 | -12.100 | 1.06e-33 | -0.038 | -24.054 | -13.424 | 4.40e-41 |
| lassosum.PseudoVal | LDpred1.Inf | 0.004 | 2.444 | 1.949 | 5.13e-02 | -0.001 | -0.381 | -0.146 | 8.84e-01 |
| PRScs.10FCVal | LDpred1.Inf | -0.028 | -16.970 | -22.021 | 1.81e-107 | -0.027 | -16.992 | -10.615 | 2.54e-26 |
| PRScs.MultiPRS | LDpred1.Inf | -0.031 | -19.309 | -10.547 | 5.26e-26 | -0.031 | -19.830 | -12.276 | 1.22e-34 |
| PRScs.PseudoVal | LDpred1.Inf | -0.026 | -15.667 | -19.530 | 6.08e-85 | -0.026 | -16.416 | -9.822 | 9.03e-23 |
| SBLUP.Inf | LDpred1.Inf | 0.000 | 0.212 | 0.397 | 6.91e-01 | 0.000 | 0.199 | 0.179 | 8.58e-01 |
| SBayesR.PseudoVal | LDpred1.Inf | -0.007 | -4.414 | -3.665 | 2.47e-04 | -0.011 | -6.771 | -2.695 | 7.04e-03 |
| LDpred1.10FCVal | LDpred1.Inf | -0.016 | -9.575 | -18.234 | 2.79e-74 | -0.015 | -9.683 | -8.840 | 9.58e-19 |
| LDpred1.MultiPRS | LDpred1.Inf | -0.019 | -11.490 | -6.270 | 3.62e-10 | -0.018 | -11.744 | -10.161 | 2.95e-24 |
| LDpred1.Inf | LDpred1.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.10FCVal | LDpred1.Inf | -0.031 | -18.941 | -20.398 | 1.76e-92 | -0.031 | -19.771 | -10.217 | 1.66e-24 |
| LDpred2.MultiPRS | LDpred1.Inf | -0.034 | -20.845 | -11.368 | 6.03e-30 | -0.034 | -21.873 | -11.953 | 6.25e-33 |
| LDpred2.PseudoVal | LDpred1.Inf | 0.008 | 4.834 | 7.086 | 1.38e-12 | 0.006 | 3.564 | 2.502 | 1.24e-02 |
| LDpred2.Inf | LDpred1.Inf | 0.002 | 1.365 | 2.338 | 1.94e-02 | 0.002 | 1.054 | 0.866 | 3.87e-01 |
| DBSLMM.PseudoVal | LDpred1.Inf | -0.019 | -11.874 | -12.152 | 5.57e-34 | -0.019 | -12.119 | -5.940 | 2.85e-09 |
| All.MultiPRS | LDpred1.Inf | -0.039 | -24.149 | -13.283 | 2.91e-40 | -0.041 | -26.242 | -14.524 | 8.58e-48 |
| pT+clump.10FCVal | LDpred2.10FCVal | 0.039 | 20.070 | 23.022 | 2.79e-117 | 0.034 | 18.354 | 10.097 | 5.71e-24 |
| pT+clump.MultiPRS | LDpred2.10FCVal | 0.019 | 9.663 | 6.274 | 3.52e-10 | 0.013 | 7.061 | 4.577 | 4.73e-06 |
| lassosum.10FCVal | LDpred2.10FCVal | 0.004 | 1.994 | 3.526 | 4.21e-04 | 0.004 | 2.311 | 1.980 | 4.77e-02 |
| lassosum.MultiPRS | LDpred2.10FCVal | -0.005 | -2.758 | -1.797 | 7.24e-02 | -0.007 | -3.576 | -3.632 | 2.81e-04 |
| lassosum.PseudoVal | LDpred2.10FCVal | 0.035 | 17.979 | 24.441 | 6.26e-132 | 0.030 | 16.189 | 10.639 | 1.96e-26 |
| PRScs.10FCVal | LDpred2.10FCVal | 0.003 | 1.657 | 2.912 | 3.59e-03 | 0.004 | 2.320 | 1.982 | 4.75e-02 |
| PRScs.MultiPRS | LDpred2.10FCVal | -0.001 | -0.309 | -0.202 | 8.40e-01 | 0.000 | -0.050 | -0.052 | 9.59e-01 |
| PRScs.PseudoVal | LDpred2.10FCVal | 0.005 | 2.752 | 5.564 | 2.64e-08 | 0.005 | 2.801 | 2.731 | 6.31e-03 |
| SBLUP.Inf | LDpred2.10FCVal | 0.031 | 16.103 | 19.484 | 1.50e-84 | 0.031 | 16.673 | 9.751 | 1.83e-22 |
| SBayesR.PseudoVal | LDpred2.10FCVal | 0.024 | 12.214 | 14.632 | 1.76e-48 | 0.020 | 10.854 | 6.250 | 4.11e-10 |
| LDpred1.10FCVal | LDpred2.10FCVal | 0.015 | 7.874 | 13.324 | 1.67e-40 | 0.016 | 8.422 | 6.889 | 5.60e-12 |
| LDpred1.MultiPRS | LDpred2.10FCVal | 0.012 | 6.265 | 4.086 | 4.38e-05 | 0.013 | 6.701 | 5.730 | 1.00e-08 |
| LDpred1.Inf | LDpred2.10FCVal | 0.031 | 15.925 | 20.398 | 1.76e-92 | 0.031 | 16.507 | 10.217 | 1.66e-24 |
| LDpred2.10FCVal | LDpred2.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.MultiPRS | LDpred2.10FCVal | -0.003 | -1.601 | -1.044 | 2.97e-01 | -0.003 | -1.756 | -3.628 | 2.86e-04 |
| LDpred2.PseudoVal | LDpred2.10FCVal | 0.039 | 19.989 | 23.852 | 9.63e-126 | 0.036 | 19.483 | 11.222 | 3.18e-29 |
| LDpred2.Inf | LDpred2.10FCVal | 0.033 | 17.072 | 20.616 | 1.98e-94 | 0.033 | 17.387 | 10.142 | 3.60e-24 |
| DBSLMM.PseudoVal | LDpred2.10FCVal | 0.012 | 5.941 | 10.442 | 1.59e-25 | 0.012 | 6.388 | 5.413 | 6.21e-08 |
| All.MultiPRS | LDpred2.10FCVal | -0.008 | -4.379 | -2.887 | 3.89e-03 | -0.010 | -5.403 | -6.547 | 5.88e-11 |
| pT+clump.10FCVal | LDpred2.MultiPRS | 0.042 | 21.329 | 14.034 | 9.72e-45 | 0.038 | 19.763 | 10.750 | 5.90e-27 |
| pT+clump.MultiPRS | LDpred2.MultiPRS | 0.022 | 11.087 | 7.324 | 2.41e-13 | 0.017 | 8.665 | 5.975 | 2.30e-09 |
| lassosum.10FCVal | LDpred2.MultiPRS | 0.007 | 3.538 | 2.341 | 1.93e-02 | 0.008 | 3.997 | 3.655 | 2.57e-04 |
| lassosum.MultiPRS | LDpred2.MultiPRS | -0.002 | -1.139 | -1.039 | 2.99e-01 | -0.003 | -1.789 | -2.019 | 4.34e-02 |
| lassosum.PseudoVal | LDpred2.MultiPRS | 0.038 | 19.272 | 12.691 | 6.67e-37 | 0.034 | 17.635 | 11.505 | 1.24e-30 |
| PRScs.10FCVal | LDpred2.MultiPRS | 0.006 | 3.206 | 2.121 | 3.39e-02 | 0.008 | 4.006 | 3.855 | 1.16e-04 |
| PRScs.MultiPRS | LDpred2.MultiPRS | 0.003 | 1.271 | 1.149 | 2.51e-01 | 0.003 | 1.676 | 1.864 | 6.23e-02 |
| PRScs.PseudoVal | LDpred2.MultiPRS | 0.008 | 4.285 | 2.833 | 4.61e-03 | 0.009 | 4.478 | 4.250 | 2.14e-05 |
| SBLUP.Inf | LDpred2.MultiPRS | 0.034 | 17.425 | 11.480 | 1.66e-30 | 0.035 | 18.111 | 11.881 | 1.48e-32 |
| SBayesR.PseudoVal | LDpred2.MultiPRS | 0.027 | 13.597 | 8.980 | 2.70e-19 | 0.024 | 12.392 | 7.804 | 5.98e-15 |
| LDpred1.10FCVal | LDpred2.MultiPRS | 0.018 | 9.326 | 6.158 | 7.36e-10 | 0.019 | 10.002 | 8.540 | 1.34e-17 |
| LDpred1.MultiPRS | LDpred2.MultiPRS | 0.015 | 7.742 | 6.405 | 1.51e-10 | 0.016 | 8.311 | 7.435 | 1.04e-13 |
| LDpred1.Inf | LDpred2.MultiPRS | 0.034 | 17.249 | 11.368 | 6.03e-30 | 0.034 | 17.948 | 11.953 | 6.25e-33 |
| LDpred2.10FCVal | LDpred2.MultiPRS | 0.003 | 1.576 | 1.044 | 2.97e-01 | 0.003 | 1.725 | 3.628 | 2.86e-04 |
| LDpred2.MultiPRS | LDpred2.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.PseudoVal | LDpred2.MultiPRS | 0.042 | 21.250 | 13.985 | 1.94e-44 | 0.040 | 20.872 | 13.446 | 3.27e-41 |
| LDpred2.Inf | LDpred2.MultiPRS | 0.036 | 18.379 | 12.106 | 9.83e-34 | 0.036 | 18.812 | 12.328 | 6.43e-35 |
| DBSLMM.PseudoVal | LDpred2.MultiPRS | 0.015 | 7.423 | 4.904 | 9.37e-07 | 0.015 | 8.004 | 6.402 | 1.54e-10 |
| All.MultiPRS | LDpred2.MultiPRS | -0.005 | -2.734 | -2.581 | 9.85e-03 | -0.007 | -3.584 | -5.795 | 6.85e-09 |
| pT+clump.10FCVal | LDpred2.PseudoVal | 0.000 | 0.101 | 0.069 | 9.45e-01 | -0.002 | -1.402 | -0.462 | 6.44e-01 |
| pT+clump.MultiPRS | LDpred2.PseudoVal | -0.020 | -12.905 | -6.666 | 2.63e-11 | -0.023 | -15.427 | -6.493 | 8.42e-11 |
| lassosum.10FCVal | LDpred2.PseudoVal | -0.035 | -22.491 | -21.630 | 9.31e-104 | -0.032 | -21.327 | -9.951 | 2.49e-23 |
| lassosum.MultiPRS | LDpred2.PseudoVal | -0.044 | -28.430 | -14.727 | 4.31e-49 | -0.043 | -28.639 | -13.892 | 7.06e-44 |
| lassosum.PseudoVal | LDpred2.PseudoVal | -0.004 | -2.512 | -1.918 | 5.52e-02 | -0.006 | -4.091 | -1.520 | 1.29e-01 |
| PRScs.10FCVal | LDpred2.PseudoVal | -0.035 | -22.913 | -25.153 | 1.30e-139 | -0.032 | -21.315 | -11.380 | 5.23e-30 |
| PRScs.MultiPRS | LDpred2.PseudoVal | -0.039 | -25.369 | -13.167 | 1.35e-39 | -0.037 | -24.259 | -12.337 | 5.72e-35 |
| PRScs.PseudoVal | LDpred2.PseudoVal | -0.033 | -21.543 | -22.100 | 3.14e-108 | -0.031 | -20.718 | -10.328 | 5.29e-25 |
| SBLUP.Inf | LDpred2.PseudoVal | -0.008 | -4.857 | -9.685 | 3.51e-22 | -0.005 | -3.489 | -3.358 | 7.86e-04 |
| SBayesR.PseudoVal | LDpred2.PseudoVal | -0.015 | -9.718 | -7.975 | 1.52e-15 | -0.016 | -10.717 | -4.277 | 1.90e-05 |
| LDpred1.10FCVal | LDpred2.PseudoVal | -0.023 | -15.142 | -18.108 | 2.78e-73 | -0.021 | -13.737 | -7.998 | 1.27e-15 |
| LDpred1.MultiPRS | LDpred2.PseudoVal | -0.027 | -17.153 | -8.895 | 5.82e-19 | -0.024 | -15.874 | -8.793 | 1.46e-18 |
| LDpred1.Inf | LDpred2.PseudoVal | -0.008 | -5.080 | -7.086 | 1.38e-12 | -0.006 | -3.696 | -2.502 | 1.24e-02 |
| LDpred2.10FCVal | LDpred2.PseudoVal | -0.039 | -24.983 | -23.852 | 9.63e-126 | -0.036 | -24.197 | -11.222 | 3.18e-29 |
| LDpred2.MultiPRS | LDpred2.PseudoVal | -0.042 | -26.984 | -13.985 | 1.94e-44 | -0.040 | -26.378 | -13.446 | 3.27e-41 |
| LDpred2.PseudoVal | LDpred2.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.Inf | LDpred2.PseudoVal | -0.006 | -3.646 | -8.244 | 1.67e-16 | -0.004 | -2.603 | -2.831 | 4.64e-03 |
| DBSLMM.PseudoVal | LDpred2.PseudoVal | -0.027 | -17.558 | -15.306 | 6.92e-53 | -0.025 | -16.263 | -6.881 | 5.93e-12 |
| All.MultiPRS | LDpred2.PseudoVal | -0.047 | -30.456 | -15.914 | 5.03e-57 | -0.047 | -30.907 | -14.972 | 1.11e-50 |
| pT+clump.10FCVal | LDpred2.Inf | 0.006 | 3.615 | 2.610 | 9.04e-03 | 0.002 | 1.171 | 0.407 | 6.84e-01 |
| pT+clump.MultiPRS | LDpred2.Inf | -0.014 | -8.933 | -4.789 | 1.68e-06 | -0.019 | -12.499 | -5.447 | 5.13e-08 |
| lassosum.10FCVal | LDpred2.Inf | -0.029 | -18.182 | -18.957 | 3.88e-80 | -0.028 | -18.249 | -9.146 | 5.91e-20 |
| lassosum.MultiPRS | LDpred2.Inf | -0.038 | -23.912 | -12.843 | 9.36e-38 | -0.039 | -25.375 | -12.884 | 5.53e-38 |
| lassosum.PseudoVal | LDpred2.Inf | 0.002 | 1.094 | 0.817 | 4.14e-01 | -0.002 | -1.450 | -0.521 | 6.02e-01 |
| PRScs.10FCVal | LDpred2.Inf | -0.030 | -18.589 | -22.427 | 2.15e-111 | -0.028 | -18.238 | -10.597 | 3.08e-26 |
| PRScs.MultiPRS | LDpred2.Inf | -0.034 | -20.959 | -11.288 | 1.50e-29 | -0.033 | -21.107 | -11.333 | 8.98e-30 |
| PRScs.PseudoVal | LDpred2.Inf | -0.028 | -17.267 | -18.304 | 7.65e-75 | -0.027 | -17.656 | -9.004 | 2.18e-19 |
| SBLUP.Inf | LDpred2.Inf | -0.002 | -1.169 | -6.110 | 9.97e-10 | -0.001 | -0.864 | -2.173 | 2.98e-02 |
| SBayesR.PseudoVal | LDpred2.Inf | -0.009 | -5.858 | -4.781 | 1.74e-06 | -0.012 | -7.908 | -3.105 | 1.91e-03 |
| LDpred1.10FCVal | LDpred2.Inf | -0.018 | -11.091 | -13.602 | 3.90e-42 | -0.017 | -10.852 | -6.405 | 1.50e-10 |
| LDpred1.MultiPRS | LDpred2.Inf | -0.021 | -13.032 | -7.012 | 2.34e-12 | -0.020 | -12.934 | -7.384 | 1.53e-13 |
| LDpred1.Inf | LDpred2.Inf | -0.002 | -1.383 | -2.338 | 1.94e-02 | -0.002 | -1.065 | -0.866 | 3.87e-01 |
| LDpred2.10FCVal | LDpred2.Inf | -0.033 | -20.586 | -20.616 | 1.98e-94 | -0.033 | -21.046 | -10.142 | 3.60e-24 |
| LDpred2.MultiPRS | LDpred2.Inf | -0.036 | -22.517 | -12.106 | 9.83e-34 | -0.036 | -23.172 | -12.328 | 6.43e-35 |
| LDpred2.PseudoVal | LDpred2.Inf | 0.006 | 3.518 | 8.244 | 1.67e-16 | 0.004 | 2.537 | 2.831 | 4.64e-03 |
| LDpred2.Inf | LDpred2.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| DBSLMM.PseudoVal | LDpred2.Inf | -0.022 | -13.422 | -11.964 | 5.48e-33 | -0.021 | -13.313 | -5.706 | 1.16e-08 |
| All.MultiPRS | LDpred2.Inf | -0.042 | -25.867 | -14.038 | 9.15e-45 | -0.043 | -27.586 | -13.973 | 2.28e-44 |
| pT+clump.10FCVal | DBSLMM.PseudoVal | 0.027 | 15.021 | 15.912 | 5.21e-57 | 0.022 | 12.782 | 6.480 | 9.17e-11 |
| pT+clump.MultiPRS | DBSLMM.PseudoVal | 0.007 | 3.957 | 2.413 | 1.58e-02 | 0.001 | 0.719 | 0.425 | 6.71e-01 |
| lassosum.10FCVal | DBSLMM.PseudoVal | -0.008 | -4.197 | -5.679 | 1.35e-08 | -0.008 | -4.355 | -2.823 | 4.76e-03 |
| lassosum.MultiPRS | DBSLMM.PseudoVal | -0.017 | -9.249 | -5.653 | 1.58e-08 | -0.019 | -10.645 | -8.448 | 2.96e-17 |
| lassosum.PseudoVal | DBSLMM.PseudoVal | 0.023 | 12.798 | 14.968 | 1.19e-50 | 0.018 | 10.470 | 5.877 | 4.17e-09 |
| PRScs.10FCVal | DBSLMM.PseudoVal | -0.008 | -4.555 | -7.086 | 1.38e-12 | -0.008 | -4.346 | -3.247 | 1.17e-03 |
| PRScs.MultiPRS | DBSLMM.PseudoVal | -0.012 | -6.645 | -4.071 | 4.68e-05 | -0.012 | -6.878 | -6.210 | 5.29e-10 |
| PRScs.PseudoVal | DBSLMM.PseudoVal | -0.006 | -3.390 | -6.561 | 5.36e-11 | -0.007 | -3.832 | -3.546 | 3.92e-04 |
| SBLUP.Inf | DBSLMM.PseudoVal | 0.020 | 10.803 | 11.279 | 1.66e-29 | 0.019 | 10.987 | 5.510 | 3.59e-08 |
| SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.012 | 6.669 | 7.132 | 9.89e-13 | 0.008 | 4.770 | 2.431 | 1.50e-02 |
| LDpred1.10FCVal | DBSLMM.PseudoVal | 0.004 | 2.055 | 2.989 | 2.80e-03 | 0.004 | 2.172 | 1.517 | 1.29e-01 |
| LDpred1.MultiPRS | DBSLMM.PseudoVal | 0.001 | 0.344 | 0.210 | 8.33e-01 | 0.001 | 0.334 | 0.238 | 8.12e-01 |
| LDpred1.Inf | DBSLMM.PseudoVal | 0.019 | 10.614 | 12.152 | 5.57e-34 | 0.019 | 10.809 | 5.940 | 2.85e-09 |
| LDpred2.10FCVal | DBSLMM.PseudoVal | -0.012 | -6.316 | -10.442 | 1.59e-25 | -0.012 | -6.824 | -5.413 | 6.21e-08 |
| LDpred2.MultiPRS | DBSLMM.PseudoVal | -0.015 | -8.019 | -4.904 | 9.37e-07 | -0.015 | -8.700 | -6.402 | 1.54e-10 |
| LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.027 | 14.935 | 15.306 | 6.92e-53 | 0.025 | 13.988 | 6.881 | 5.93e-12 |
| LDpred2.Inf | DBSLMM.PseudoVal | 0.022 | 11.834 | 11.964 | 5.48e-33 | 0.021 | 11.749 | 5.706 | 1.16e-08 |
| DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| All.MultiPRS | DBSLMM.PseudoVal | -0.020 | -10.972 | -6.779 | 1.21e-11 | -0.022 | -12.596 | -10.100 | 5.55e-24 |
| pT+clump.10FCVal | All.MultiPRS | 0.047 | 23.423 | 15.977 | 1.83e-57 | 0.044 | 22.539 | 14.192 | 1.02e-45 |
| pT+clump.MultiPRS | All.MultiPRS | 0.027 | 13.453 | 9.134 | 6.58e-20 | 0.023 | 11.825 | 10.002 | 1.50e-23 |
| lassosum.10FCVal | All.MultiPRS | 0.012 | 6.106 | 4.192 | 2.76e-05 | 0.014 | 7.319 | 7.869 | 3.57e-15 |
| lassosum.MultiPRS | All.MultiPRS | 0.003 | 1.553 | 1.392 | 1.64e-01 | 0.003 | 1.733 | 2.904 | 3.68e-03 |
| lassosum.PseudoVal | All.MultiPRS | 0.043 | 21.420 | 14.635 | 1.68e-48 | 0.040 | 20.485 | 14.291 | 2.50e-46 |
| PRScs.10FCVal | All.MultiPRS | 0.012 | 5.782 | 3.972 | 7.14e-05 | 0.014 | 7.327 | 7.912 | 2.54e-15 |
| PRScs.MultiPRS | All.MultiPRS | 0.008 | 3.899 | 3.926 | 8.65e-05 | 0.010 | 5.079 | 6.565 | 5.20e-11 |
| PRScs.PseudoVal | All.MultiPRS | 0.014 | 6.832 | 4.690 | 2.73e-06 | 0.015 | 7.783 | 8.253 | 1.55e-16 |
| SBLUP.Inf | All.MultiPRS | 0.040 | 19.623 | 13.405 | 5.68e-41 | 0.041 | 20.945 | 14.031 | 1.01e-44 |
| SBayesR.PseudoVal | All.MultiPRS | 0.032 | 15.897 | 10.897 | 1.19e-27 | 0.030 | 15.424 | 11.200 | 4.06e-29 |
| LDpred1.10FCVal | All.MultiPRS | 0.024 | 11.739 | 8.041 | 8.93e-16 | 0.026 | 13.116 | 11.747 | 7.30e-32 |
| LDpred1.MultiPRS | All.MultiPRS | 0.021 | 10.197 | 10.071 | 7.44e-24 | 0.023 | 11.484 | 10.918 | 9.41e-28 |
| LDpred1.Inf | All.MultiPRS | 0.039 | 19.452 | 13.283 | 2.91e-40 | 0.041 | 20.787 | 14.524 | 8.58e-48 |
| LDpred2.10FCVal | All.MultiPRS | 0.008 | 4.195 | 2.887 | 3.89e-03 | 0.010 | 5.126 | 6.547 | 5.88e-11 |
| LDpred2.MultiPRS | All.MultiPRS | 0.005 | 2.662 | 2.581 | 9.85e-03 | 0.007 | 3.460 | 5.795 | 6.85e-09 |
| LDpred2.PseudoVal | All.MultiPRS | 0.047 | 23.346 | 15.914 | 5.03e-57 | 0.047 | 23.610 | 14.972 | 1.11e-50 |
| LDpred2.Inf | All.MultiPRS | 0.042 | 20.551 | 14.038 | 9.15e-45 | 0.043 | 21.622 | 13.973 | 2.28e-44 |
| DBSLMM.PseudoVal | All.MultiPRS | 0.020 | 9.887 | 6.779 | 1.21e-11 | 0.022 | 11.187 | 10.100 | 5.55e-24 |
| All.MultiPRS | All.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
Show table: UKBB (UKBB ref)
| Reference | Test | CrossVal Diff | CrossVal Diff (%) | CrossVal Diff Z | CrossVal Diff P | IndepVal Diff | IndepVal Diff (%) | IndepVal Diff Z | IndepVal Diff P |
|---|---|---|---|---|---|---|---|---|---|
| pT+clump.10FCVal | pT+clump.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| pT+clump.10FCVal | pT+clump.MultiPRS | -0.020 | -12.956 | -6.725 | 1.75e-11 | -0.022 | -14.832 | -10.134 | 3.90e-24 |
| pT+clump.10FCVal | lassosum.10FCVal | -0.034 | -21.879 | -19.538 | 5.27e-85 | -0.032 | -20.866 | -9.057 | 1.33e-19 |
| pT+clump.10FCVal | lassosum.MultiPRS | -0.042 | -27.196 | -14.153 | 1.80e-45 | -0.041 | -27.070 | -12.564 | 3.34e-36 |
| pT+clump.10FCVal | lassosum.PseudoVal | -0.003 | -2.238 | -1.900 | 5.74e-02 | -0.005 | -3.594 | -1.484 | 1.38e-01 |
| pT+clump.10FCVal | SBLUP.Inf | -0.007 | -4.216 | -2.965 | 3.03e-03 | -0.004 | -2.340 | -0.801 | 4.23e-01 |
| pT+clump.10FCVal | SBayesR.PseudoVal | -0.025 | -16.178 | -14.037 | 9.21e-45 | -0.027 | -17.640 | -7.447 | 9.55e-14 |
| pT+clump.10FCVal | LDpred1.10FCVal | -0.021 | -13.452 | -10.971 | 5.29e-28 | -0.019 | -12.510 | -4.957 | 7.16e-07 |
| pT+clump.10FCVal | LDpred1.MultiPRS | -0.027 | -17.087 | -8.919 | 4.70e-19 | -0.025 | -16.170 | -6.671 | 2.55e-11 |
| pT+clump.10FCVal | LDpred1.Inf | -0.006 | -4.109 | -2.989 | 2.80e-03 | -0.003 | -2.136 | -0.756 | 4.50e-01 |
| pT+clump.10FCVal | LDpred2.10FCVal | -0.041 | -26.039 | -24.945 | 2.43e-137 | -0.039 | -25.505 | -11.861 | 1.88e-32 |
| pT+clump.10FCVal | LDpred2.MultiPRS | -0.043 | -27.918 | -14.549 | 5.92e-48 | -0.042 | -27.643 | -12.766 | 2.54e-37 |
| pT+clump.10FCVal | LDpred2.PseudoVal | -0.014 | -9.284 | -8.202 | 2.36e-16 | -0.016 | -10.839 | -4.632 | 3.62e-06 |
| pT+clump.10FCVal | LDpred2.Inf | -0.005 | -3.122 | -2.159 | 3.08e-02 | -0.003 | -1.716 | -0.578 | 5.63e-01 |
| pT+clump.10FCVal | DBSLMM.PseudoVal | -0.028 | -17.895 | -16.591 | 8.16e-62 | -0.026 | -16.834 | -7.580 | 3.47e-14 |
| pT+clump.10FCVal | All.MultiPRS | -0.047 | -29.944 | -15.651 | 3.29e-55 | -0.046 | -30.233 | -14.824 | 1.02e-49 |
| pT+clump.MultiPRS | pT+clump.10FCVal | 0.020 | 11.470 | 6.725 | 1.75e-11 | 0.022 | 12.916 | 10.134 | 3.90e-24 |
| pT+clump.MultiPRS | pT+clump.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| pT+clump.MultiPRS | lassosum.10FCVal | -0.014 | -7.899 | -4.655 | 3.24e-06 | -0.009 | -5.255 | -3.399 | 6.77e-04 |
| pT+clump.MultiPRS | lassosum.MultiPRS | -0.022 | -12.606 | -7.439 | 1.01e-13 | -0.019 | -10.657 | -7.752 | 9.08e-15 |
| pT+clump.MultiPRS | lassosum.PseudoVal | 0.017 | 9.489 | 5.563 | 2.66e-08 | 0.017 | 9.786 | 5.321 | 1.03e-07 |
| pT+clump.MultiPRS | SBLUP.Inf | 0.014 | 7.737 | 4.543 | 5.55e-06 | 0.019 | 10.879 | 5.522 | 3.36e-08 |
| pT+clump.MultiPRS | SBayesR.PseudoVal | -0.005 | -2.852 | -1.676 | 9.38e-02 | -0.004 | -2.445 | -1.536 | 1.25e-01 |
| pT+clump.MultiPRS | LDpred1.10FCVal | -0.001 | -0.439 | -0.258 | 7.96e-01 | 0.004 | 2.022 | 1.182 | 2.37e-01 |
| pT+clump.MultiPRS | LDpred1.MultiPRS | -0.006 | -3.657 | -2.153 | 3.13e-02 | -0.002 | -1.165 | -0.708 | 4.79e-01 |
| pT+clump.MultiPRS | LDpred1.Inf | 0.014 | 7.833 | 4.599 | 4.24e-06 | 0.019 | 11.056 | 5.612 | 2.00e-08 |
| pT+clump.MultiPRS | LDpred2.10FCVal | -0.020 | -11.582 | -6.828 | 8.63e-12 | -0.016 | -9.295 | -6.073 | 1.26e-09 |
| pT+clump.MultiPRS | LDpred2.MultiPRS | -0.023 | -13.246 | -7.814 | 5.53e-15 | -0.019 | -11.156 | -7.578 | 3.52e-14 |
| pT+clump.MultiPRS | LDpred2.PseudoVal | 0.006 | 3.251 | 1.908 | 5.64e-02 | 0.006 | 3.477 | 1.905 | 5.68e-02 |
| pT+clump.MultiPRS | LDpred2.Inf | 0.015 | 8.706 | 5.110 | 3.23e-07 | 0.020 | 11.422 | 5.644 | 1.66e-08 |
| pT+clump.MultiPRS | DBSLMM.PseudoVal | -0.008 | -4.372 | -2.573 | 1.01e-02 | -0.003 | -1.743 | -1.053 | 2.92e-01 |
| pT+clump.MultiPRS | All.MultiPRS | -0.026 | -15.040 | -8.879 | 6.75e-19 | -0.023 | -13.412 | -10.185 | 2.33e-24 |
| lassosum.10FCVal | pT+clump.10FCVal | 0.034 | 17.951 | 19.538 | 5.27e-85 | 0.032 | 17.264 | 9.057 | 1.33e-19 |
| lassosum.10FCVal | pT+clump.MultiPRS | 0.014 | 7.321 | 4.655 | 3.24e-06 | 0.009 | 4.993 | 3.399 | 6.77e-04 |
| lassosum.10FCVal | lassosum.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.10FCVal | lassosum.MultiPRS | -0.008 | -4.363 | -2.779 | 5.45e-03 | -0.009 | -5.132 | -7.183 | 6.83e-13 |
| lassosum.10FCVal | lassosum.PseudoVal | 0.031 | 16.115 | 20.975 | 1.11e-97 | 0.026 | 14.290 | 9.002 | 2.22e-19 |
| lassosum.10FCVal | SBLUP.Inf | 0.027 | 14.492 | 18.554 | 7.57e-77 | 0.028 | 15.328 | 9.456 | 3.19e-21 |
| lassosum.10FCVal | SBayesR.PseudoVal | 0.009 | 4.678 | 6.222 | 4.91e-10 | 0.005 | 2.670 | 1.706 | 8.80e-02 |
| lassosum.10FCVal | LDpred1.10FCVal | 0.013 | 6.914 | 10.722 | 8.03e-27 | 0.013 | 6.914 | 5.172 | 2.32e-07 |
| lassosum.10FCVal | LDpred1.MultiPRS | 0.007 | 3.932 | 2.517 | 1.18e-02 | 0.007 | 3.886 | 3.058 | 2.22e-03 |
| lassosum.10FCVal | LDpred1.Inf | 0.028 | 14.580 | 19.256 | 1.27e-82 | 0.028 | 15.496 | 9.850 | 6.83e-23 |
| lassosum.10FCVal | LDpred2.10FCVal | -0.006 | -3.413 | -6.343 | 2.25e-10 | -0.007 | -3.838 | -3.466 | 5.28e-04 |
| lassosum.10FCVal | LDpred2.MultiPRS | -0.009 | -4.955 | -3.161 | 1.57e-03 | -0.010 | -5.607 | -5.133 | 2.86e-07 |
| lassosum.10FCVal | LDpred2.PseudoVal | 0.020 | 10.334 | 14.051 | 7.57e-45 | 0.015 | 8.296 | 5.456 | 4.86e-08 |
| lassosum.10FCVal | LDpred2.Inf | 0.029 | 15.390 | 19.026 | 1.04e-80 | 0.029 | 15.845 | 9.437 | 3.83e-21 |
| lassosum.10FCVal | DBSLMM.PseudoVal | 0.006 | 3.269 | 4.710 | 2.48e-06 | 0.006 | 3.336 | 2.316 | 2.05e-02 |
| lassosum.10FCVal | All.MultiPRS | -0.013 | -6.618 | -4.238 | 2.26e-05 | -0.014 | -7.750 | -7.769 | 7.93e-15 |
| lassosum.MultiPRS | pT+clump.10FCVal | 0.042 | 21.381 | 14.153 | 1.80e-45 | 0.041 | 21.303 | 12.564 | 3.34e-36 |
| lassosum.MultiPRS | pT+clump.MultiPRS | 0.022 | 11.195 | 7.439 | 1.01e-13 | 0.019 | 9.631 | 7.752 | 9.08e-15 |
| lassosum.MultiPRS | lassosum.10FCVal | 0.008 | 4.180 | 2.779 | 5.45e-03 | 0.009 | 4.882 | 7.183 | 6.83e-13 |
| lassosum.MultiPRS | lassosum.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.MultiPRS | lassosum.PseudoVal | 0.039 | 19.622 | 13.005 | 1.15e-38 | 0.036 | 18.474 | 13.953 | 3.02e-44 |
| lassosum.MultiPRS | SBLUP.Inf | 0.036 | 18.066 | 11.967 | 5.30e-33 | 0.038 | 19.462 | 12.949 | 2.39e-38 |
| lassosum.MultiPRS | SBayesR.PseudoVal | 0.017 | 8.663 | 5.756 | 8.60e-09 | 0.014 | 7.421 | 5.651 | 1.60e-08 |
| lassosum.MultiPRS | LDpred1.10FCVal | 0.021 | 10.805 | 7.169 | 7.54e-13 | 0.022 | 11.458 | 10.252 | 1.16e-24 |
| lassosum.MultiPRS | LDpred1.MultiPRS | 0.016 | 7.947 | 7.480 | 7.43e-14 | 0.017 | 8.578 | 8.221 | 2.02e-16 |
| lassosum.MultiPRS | LDpred1.Inf | 0.036 | 18.151 | 12.022 | 2.71e-33 | 0.038 | 19.622 | 13.682 | 1.29e-42 |
| lassosum.MultiPRS | LDpred2.10FCVal | 0.002 | 0.910 | 0.606 | 5.45e-01 | 0.002 | 1.231 | 1.391 | 1.64e-01 |
| lassosum.MultiPRS | LDpred2.MultiPRS | -0.001 | -0.568 | -0.526 | 5.99e-01 | -0.001 | -0.451 | -0.550 | 5.82e-01 |
| lassosum.MultiPRS | LDpred2.PseudoVal | 0.028 | 14.082 | 9.345 | 9.22e-21 | 0.025 | 12.773 | 9.486 | 2.40e-21 |
| lassosum.MultiPRS | LDpred2.Inf | 0.037 | 18.927 | 12.534 | 4.86e-36 | 0.038 | 19.953 | 12.703 | 5.70e-37 |
| lassosum.MultiPRS | DBSLMM.PseudoVal | 0.014 | 7.312 | 4.858 | 1.19e-06 | 0.016 | 8.055 | 7.186 | 6.66e-13 |
| lassosum.MultiPRS | All.MultiPRS | -0.004 | -2.161 | -2.255 | 2.42e-02 | -0.005 | -2.489 | -4.060 | 4.90e-05 |
| lassosum.PseudoVal | pT+clump.10FCVal | 0.003 | 2.189 | 1.900 | 5.74e-02 | 0.005 | 3.470 | 1.484 | 1.38e-01 |
| lassosum.PseudoVal | pT+clump.MultiPRS | -0.017 | -10.484 | -5.563 | 2.66e-08 | -0.017 | -10.848 | -5.321 | 1.03e-07 |
| lassosum.PseudoVal | lassosum.10FCVal | -0.031 | -19.211 | -20.975 | 1.11e-97 | -0.026 | -16.673 | -9.002 | 2.22e-19 |
| lassosum.PseudoVal | lassosum.MultiPRS | -0.039 | -24.412 | -13.005 | 1.15e-38 | -0.036 | -22.661 | -13.953 | 3.02e-44 |
| lassosum.PseudoVal | lassosum.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.PseudoVal | SBLUP.Inf | -0.003 | -1.935 | -1.461 | 1.44e-01 | 0.002 | 1.211 | 0.452 | 6.51e-01 |
| lassosum.PseudoVal | SBayesR.PseudoVal | -0.022 | -13.634 | -13.869 | 9.73e-44 | -0.021 | -13.558 | -6.825 | 8.82e-12 |
| lassosum.PseudoVal | LDpred1.10FCVal | -0.017 | -10.969 | -10.752 | 5.78e-27 | -0.014 | -8.606 | -4.178 | 2.94e-05 |
| lassosum.PseudoVal | LDpred1.MultiPRS | -0.023 | -14.524 | -7.759 | 8.56e-15 | -0.019 | -12.139 | -6.167 | 6.97e-10 |
| lassosum.PseudoVal | LDpred1.Inf | -0.003 | -1.830 | -1.437 | 1.51e-01 | 0.002 | 1.407 | 0.547 | 5.84e-01 |
| lassosum.PseudoVal | LDpred2.10FCVal | -0.037 | -23.280 | -26.410 | 1.05e-153 | -0.033 | -21.151 | -11.864 | 1.83e-32 |
| lassosum.PseudoVal | LDpred2.MultiPRS | -0.040 | -25.118 | -13.393 | 6.63e-41 | -0.036 | -23.214 | -12.904 | 4.26e-38 |
| lassosum.PseudoVal | LDpred2.PseudoVal | -0.011 | -6.892 | -6.800 | 1.05e-11 | -0.011 | -6.993 | -3.415 | 6.37e-04 |
| lassosum.PseudoVal | LDpred2.Inf | -0.001 | -0.865 | -0.642 | 5.21e-01 | 0.003 | 1.813 | 0.665 | 5.06e-01 |
| lassosum.PseudoVal | DBSLMM.PseudoVal | -0.024 | -15.314 | -15.924 | 4.32e-57 | -0.020 | -12.780 | -6.561 | 5.35e-11 |
| lassosum.PseudoVal | All.MultiPRS | -0.043 | -27.100 | -14.502 | 1.18e-47 | -0.040 | -25.714 | -14.423 | 3.71e-47 |
| SBLUP.Inf | pT+clump.10FCVal | 0.007 | 4.046 | 2.965 | 3.03e-03 | 0.004 | 2.286 | 0.801 | 4.23e-01 |
| SBLUP.Inf | pT+clump.MultiPRS | -0.014 | -8.386 | -4.543 | 5.55e-06 | -0.019 | -12.207 | -5.522 | 3.36e-08 |
| SBLUP.Inf | lassosum.10FCVal | -0.027 | -16.948 | -18.554 | 7.57e-77 | -0.028 | -18.103 | -9.456 | 3.19e-21 |
| SBLUP.Inf | lassosum.MultiPRS | -0.036 | -22.050 | -11.967 | 5.30e-33 | -0.038 | -24.165 | -12.949 | 2.39e-38 |
| SBLUP.Inf | lassosum.PseudoVal | 0.003 | 1.898 | 1.461 | 1.44e-01 | -0.002 | -1.226 | -0.452 | 6.51e-01 |
| SBLUP.Inf | SBLUP.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBLUP.Inf | SBayesR.PseudoVal | -0.019 | -11.477 | -12.012 | 3.06e-33 | -0.023 | -14.951 | -7.473 | 7.83e-14 |
| SBLUP.Inf | LDpred1.10FCVal | -0.014 | -8.862 | -11.626 | 3.05e-31 | -0.015 | -9.938 | -6.223 | 4.89e-10 |
| SBLUP.Inf | LDpred1.MultiPRS | -0.020 | -12.350 | -6.734 | 1.65e-11 | -0.021 | -13.514 | -8.056 | 7.91e-16 |
| SBLUP.Inf | LDpred1.Inf | 0.000 | 0.103 | 0.196 | 8.45e-01 | 0.000 | 0.198 | 0.179 | 8.58e-01 |
| SBLUP.Inf | LDpred2.10FCVal | -0.034 | -20.940 | -22.638 | 1.82e-113 | -0.035 | -22.636 | -11.689 | 1.45e-31 |
| SBLUP.Inf | LDpred2.MultiPRS | -0.037 | -22.743 | -12.352 | 4.74e-35 | -0.038 | -24.725 | -12.961 | 2.03e-38 |
| SBLUP.Inf | LDpred2.PseudoVal | -0.008 | -4.863 | -4.109 | 3.97e-05 | -0.013 | -8.305 | -3.357 | 7.88e-04 |
| SBLUP.Inf | LDpred2.Inf | 0.002 | 1.050 | 6.837 | 8.06e-12 | 0.001 | 0.610 | 1.898 | 5.77e-02 |
| SBLUP.Inf | DBSLMM.PseudoVal | -0.021 | -13.125 | -12.177 | 4.12e-34 | -0.022 | -14.163 | -6.265 | 3.72e-10 |
| SBLUP.Inf | All.MultiPRS | -0.040 | -24.687 | -13.461 | 2.65e-41 | -0.042 | -27.256 | -14.302 | 2.12e-46 |
| SBayesR.PseudoVal | pT+clump.10FCVal | 0.025 | 13.925 | 14.037 | 9.21e-45 | 0.027 | 14.995 | 7.447 | 9.55e-14 |
| SBayesR.PseudoVal | pT+clump.MultiPRS | 0.005 | 2.773 | 1.676 | 9.38e-02 | 0.004 | 2.387 | 1.536 | 1.25e-01 |
| SBayesR.PseudoVal | lassosum.10FCVal | -0.009 | -4.908 | -6.222 | 4.91e-10 | -0.005 | -2.743 | -1.706 | 8.80e-02 |
| SBayesR.PseudoVal | lassosum.MultiPRS | -0.017 | -9.484 | -5.756 | 8.60e-09 | -0.014 | -8.016 | -5.651 | 1.60e-08 |
| SBayesR.PseudoVal | lassosum.PseudoVal | 0.022 | 11.998 | 13.869 | 9.73e-44 | 0.021 | 11.939 | 6.825 | 8.82e-12 |
| SBayesR.PseudoVal | SBLUP.Inf | 0.019 | 10.296 | 12.012 | 3.06e-33 | 0.023 | 13.006 | 7.473 | 7.83e-14 |
| SBayesR.PseudoVal | SBayesR.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBayesR.PseudoVal | LDpred1.10FCVal | 0.004 | 2.346 | 3.065 | 2.17e-03 | 0.008 | 4.361 | 2.801 | 5.09e-03 |
| SBayesR.PseudoVal | LDpred1.MultiPRS | -0.001 | -0.783 | -0.477 | 6.33e-01 | 0.002 | 1.249 | 0.843 | 3.99e-01 |
| SBayesR.PseudoVal | LDpred1.Inf | 0.019 | 10.388 | 11.672 | 1.78e-31 | 0.024 | 13.179 | 7.298 | 2.93e-13 |
| SBayesR.PseudoVal | LDpred2.10FCVal | -0.015 | -8.488 | -11.790 | 4.41e-32 | -0.012 | -6.686 | -4.556 | 5.21e-06 |
| SBayesR.PseudoVal | LDpred2.MultiPRS | -0.018 | -10.106 | -6.141 | 8.22e-10 | -0.015 | -8.503 | -6.224 | 4.85e-10 |
| SBayesR.PseudoVal | LDpred2.PseudoVal | 0.011 | 5.933 | 7.668 | 1.75e-14 | 0.010 | 5.781 | 3.674 | 2.39e-04 |
| SBayesR.PseudoVal | LDpred2.Inf | 0.020 | 11.237 | 12.864 | 7.18e-38 | 0.024 | 13.536 | 7.631 | 2.33e-14 |
| SBayesR.PseudoVal | DBSLMM.PseudoVal | -0.003 | -1.478 | -1.714 | 8.65e-02 | 0.001 | 0.685 | 0.389 | 6.97e-01 |
| SBayesR.PseudoVal | All.MultiPRS | -0.021 | -11.850 | -7.228 | 4.90e-13 | -0.019 | -10.705 | -8.325 | 8.40e-17 |
| LDpred1.10FCVal | pT+clump.10FCVal | 0.021 | 11.857 | 10.971 | 5.29e-28 | 0.019 | 11.119 | 4.957 | 7.16e-07 |
| LDpred1.10FCVal | pT+clump.MultiPRS | 0.001 | 0.437 | 0.258 | 7.96e-01 | -0.004 | -2.064 | -1.182 | 2.37e-01 |
| LDpred1.10FCVal | lassosum.10FCVal | -0.013 | -7.428 | -10.722 | 8.03e-27 | -0.013 | -7.427 | -5.172 | 2.32e-07 |
| LDpred1.10FCVal | lassosum.MultiPRS | -0.021 | -12.114 | -7.169 | 7.54e-13 | -0.022 | -12.941 | -10.252 | 1.16e-24 |
| LDpred1.10FCVal | lassosum.PseudoVal | 0.017 | 9.885 | 10.752 | 5.78e-27 | 0.014 | 7.924 | 4.178 | 2.94e-05 |
| LDpred1.10FCVal | SBLUP.Inf | 0.014 | 8.141 | 11.626 | 3.05e-31 | 0.015 | 9.040 | 6.223 | 4.89e-10 |
| LDpred1.10FCVal | SBayesR.PseudoVal | -0.004 | -2.402 | -3.065 | 2.17e-03 | -0.008 | -4.559 | -2.801 | 5.09e-03 |
| LDpred1.10FCVal | LDpred1.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.10FCVal | LDpred1.MultiPRS | -0.006 | -3.204 | -1.906 | 5.67e-02 | -0.006 | -3.253 | -5.635 | 1.75e-08 |
| LDpred1.10FCVal | LDpred1.Inf | 0.015 | 8.236 | 16.927 | 2.84e-64 | 0.016 | 9.220 | 9.127 | 7.05e-20 |
| LDpred1.10FCVal | LDpred2.10FCVal | -0.020 | -11.094 | -18.082 | 4.46e-73 | -0.020 | -11.550 | -9.115 | 7.89e-20 |
| LDpred1.10FCVal | LDpred2.MultiPRS | -0.023 | -12.751 | -7.556 | 4.15e-14 | -0.023 | -13.450 | -10.224 | 1.55e-24 |
| LDpred1.10FCVal | LDpred2.PseudoVal | 0.006 | 3.674 | 4.265 | 2.00e-05 | 0.003 | 1.485 | 0.834 | 4.04e-01 |
| LDpred1.10FCVal | LDpred2.Inf | 0.016 | 9.105 | 12.397 | 2.71e-35 | 0.016 | 9.594 | 6.299 | 3.00e-10 |
| LDpred1.10FCVal | DBSLMM.PseudoVal | -0.007 | -3.916 | -5.736 | 9.70e-09 | -0.007 | -3.843 | -2.708 | 6.76e-03 |
| LDpred1.10FCVal | All.MultiPRS | -0.026 | -14.537 | -8.647 | 5.29e-18 | -0.027 | -15.752 | -11.997 | 3.69e-33 |
| LDpred1.MultiPRS | pT+clump.10FCVal | 0.027 | 14.593 | 8.919 | 4.70e-19 | 0.025 | 13.919 | 6.671 | 2.55e-11 |
| LDpred1.MultiPRS | pT+clump.MultiPRS | 0.006 | 3.528 | 2.153 | 3.13e-02 | 0.002 | 1.152 | 0.708 | 4.79e-01 |
| LDpred1.MultiPRS | lassosum.10FCVal | -0.007 | -4.093 | -2.517 | 1.18e-02 | -0.007 | -4.043 | -3.058 | 2.22e-03 |
| LDpred1.MultiPRS | lassosum.MultiPRS | -0.016 | -8.634 | -7.480 | 7.43e-14 | -0.017 | -9.383 | -8.221 | 2.02e-16 |
| LDpred1.MultiPRS | lassosum.PseudoVal | 0.023 | 12.682 | 7.759 | 8.56e-15 | 0.019 | 10.825 | 6.167 | 6.97e-10 |
| LDpred1.MultiPRS | SBLUP.Inf | 0.020 | 10.992 | 6.734 | 1.65e-11 | 0.021 | 11.905 | 8.056 | 7.91e-16 |
| LDpred1.MultiPRS | SBayesR.PseudoVal | 0.001 | 0.777 | 0.477 | 6.33e-01 | -0.002 | -1.265 | -0.843 | 3.99e-01 |
| LDpred1.MultiPRS | LDpred1.10FCVal | 0.006 | 3.104 | 1.906 | 5.67e-02 | 0.006 | 3.151 | 5.635 | 1.75e-08 |
| LDpred1.MultiPRS | LDpred1.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.MultiPRS | LDpred1.Inf | 0.020 | 11.084 | 6.789 | 1.13e-11 | 0.021 | 12.080 | 11.268 | 1.89e-29 |
| LDpred1.MultiPRS | LDpred2.10FCVal | -0.014 | -7.646 | -4.709 | 2.49e-06 | -0.014 | -8.036 | -7.068 | 1.57e-12 |
| LDpred1.MultiPRS | LDpred2.MultiPRS | -0.017 | -9.251 | -7.125 | 1.04e-12 | -0.017 | -9.876 | -8.372 | 5.66e-17 |
| LDpred1.MultiPRS | LDpred2.PseudoVal | 0.012 | 6.664 | 4.088 | 4.35e-05 | 0.008 | 4.589 | 2.785 | 5.35e-03 |
| LDpred1.MultiPRS | LDpred2.Inf | 0.022 | 11.927 | 7.305 | 2.78e-13 | 0.022 | 12.442 | 8.018 | 1.07e-15 |
| LDpred1.MultiPRS | DBSLMM.PseudoVal | -0.001 | -0.690 | -0.424 | 6.72e-01 | -0.001 | -0.572 | -0.420 | 6.74e-01 |
| LDpred1.MultiPRS | All.MultiPRS | -0.020 | -10.981 | -8.540 | 1.34e-17 | -0.021 | -12.105 | -10.450 | 1.46e-25 |
| LDpred1.Inf | pT+clump.10FCVal | 0.006 | 3.947 | 2.989 | 2.80e-03 | 0.003 | 2.092 | 0.756 | 4.50e-01 |
| LDpred1.Inf | pT+clump.MultiPRS | -0.014 | -8.498 | -4.599 | 4.24e-06 | -0.019 | -12.430 | -5.612 | 2.00e-08 |
| LDpred1.Inf | lassosum.10FCVal | -0.028 | -17.069 | -19.256 | 1.27e-82 | -0.028 | -18.338 | -9.850 | 6.83e-23 |
| LDpred1.Inf | lassosum.MultiPRS | -0.036 | -22.176 | -12.022 | 2.71e-33 | -0.038 | -24.412 | -13.682 | 1.29e-42 |
| LDpred1.Inf | lassosum.PseudoVal | 0.003 | 1.797 | 1.437 | 1.51e-01 | -0.002 | -1.427 | -0.547 | 5.84e-01 |
| LDpred1.Inf | SBLUP.Inf | 0.000 | -0.103 | -0.196 | 8.45e-01 | 0.000 | -0.199 | -0.179 | 8.58e-01 |
| LDpred1.Inf | SBayesR.PseudoVal | -0.019 | -11.592 | -11.672 | 1.78e-31 | -0.024 | -15.179 | -7.298 | 2.93e-13 |
| LDpred1.Inf | LDpred1.10FCVal | -0.015 | -8.975 | -16.927 | 2.84e-64 | -0.016 | -10.157 | -9.127 | 7.05e-20 |
| LDpred1.Inf | LDpred1.MultiPRS | -0.020 | -12.466 | -6.789 | 1.13e-11 | -0.021 | -13.740 | -11.268 | 1.89e-29 |
| LDpred1.Inf | LDpred1.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.Inf | LDpred2.10FCVal | -0.034 | -21.065 | -23.701 | 3.52e-124 | -0.035 | -22.880 | -12.284 | 1.11e-34 |
| LDpred1.Inf | LDpred2.MultiPRS | -0.037 | -22.870 | -12.410 | 2.32e-35 | -0.039 | -24.973 | -13.301 | 2.28e-40 |
| LDpred1.Inf | LDpred2.PseudoVal | -0.008 | -4.971 | -4.384 | 1.16e-05 | -0.013 | -8.521 | -3.591 | 3.29e-04 |
| LDpred1.Inf | LDpred2.Inf | 0.002 | 0.948 | 1.653 | 9.83e-02 | 0.001 | 0.412 | 0.342 | 7.32e-01 |
| LDpred1.Inf | DBSLMM.PseudoVal | -0.021 | -13.242 | -13.584 | 4.96e-42 | -0.022 | -14.390 | -7.034 | 2.01e-12 |
| LDpred1.Inf | All.MultiPRS | -0.040 | -24.816 | -13.511 | 1.34e-41 | -0.043 | -27.509 | -14.911 | 2.82e-50 |
| LDpred2.10FCVal | pT+clump.10FCVal | 0.041 | 20.660 | 24.945 | 2.43e-137 | 0.039 | 20.322 | 11.861 | 1.88e-32 |
| LDpred2.10FCVal | pT+clump.MultiPRS | 0.020 | 10.380 | 6.828 | 8.63e-12 | 0.016 | 8.504 | 6.073 | 1.26e-09 |
| LDpred2.10FCVal | lassosum.10FCVal | 0.006 | 3.301 | 6.343 | 2.25e-10 | 0.007 | 3.696 | 3.466 | 5.28e-04 |
| LDpred2.10FCVal | lassosum.MultiPRS | -0.002 | -0.918 | -0.606 | 5.45e-01 | -0.002 | -1.247 | -1.391 | 1.64e-01 |
| LDpred2.10FCVal | lassosum.PseudoVal | 0.037 | 18.884 | 26.410 | 1.05e-153 | 0.033 | 17.458 | 11.864 | 1.83e-32 |
| LDpred2.10FCVal | SBLUP.Inf | 0.034 | 17.314 | 22.638 | 1.82e-113 | 0.035 | 18.458 | 11.689 | 1.45e-31 |
| LDpred2.10FCVal | SBayesR.PseudoVal | 0.015 | 7.824 | 11.790 | 4.41e-32 | 0.012 | 6.267 | 4.556 | 5.21e-06 |
| LDpred2.10FCVal | LDpred1.10FCVal | 0.020 | 9.986 | 18.082 | 4.46e-73 | 0.020 | 10.354 | 9.115 | 7.89e-20 |
| LDpred2.10FCVal | LDpred1.MultiPRS | 0.014 | 7.103 | 4.709 | 2.49e-06 | 0.014 | 7.438 | 7.068 | 1.57e-12 |
| LDpred2.10FCVal | LDpred1.Inf | 0.034 | 17.400 | 23.701 | 3.52e-124 | 0.035 | 18.620 | 12.284 | 1.11e-34 |
| LDpred2.10FCVal | LDpred2.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.10FCVal | LDpred2.MultiPRS | -0.003 | -1.491 | -0.985 | 3.25e-01 | -0.003 | -1.703 | -3.887 | 1.02e-04 |
| LDpred2.10FCVal | LDpred2.PseudoVal | 0.026 | 13.293 | 25.675 | 2.21e-145 | 0.022 | 11.686 | 10.937 | 7.68e-28 |
| LDpred2.10FCVal | LDpred2.Inf | 0.036 | 18.182 | 23.582 | 5.94e-123 | 0.036 | 18.955 | 11.900 | 1.19e-32 |
| LDpred2.10FCVal | DBSLMM.PseudoVal | 0.013 | 6.462 | 11.787 | 4.56e-32 | 0.013 | 6.909 | 6.090 | 1.13e-09 |
| LDpred2.10FCVal | All.MultiPRS | -0.006 | -3.099 | -2.056 | 3.98e-02 | -0.007 | -3.767 | -5.191 | 2.09e-07 |
| LDpred2.MultiPRS | pT+clump.10FCVal | 0.043 | 21.825 | 14.549 | 5.92e-48 | 0.042 | 21.656 | 12.766 | 2.54e-37 |
| LDpred2.MultiPRS | pT+clump.MultiPRS | 0.023 | 11.697 | 7.814 | 5.53e-15 | 0.019 | 10.037 | 7.578 | 3.52e-14 |
| LDpred2.MultiPRS | lassosum.10FCVal | 0.009 | 4.721 | 3.161 | 1.57e-03 | 0.010 | 5.309 | 5.133 | 2.86e-07 |
| LDpred2.MultiPRS | lassosum.MultiPRS | 0.001 | 0.565 | 0.526 | 5.99e-01 | 0.001 | 0.449 | 0.550 | 5.82e-01 |
| LDpred2.MultiPRS | lassosum.PseudoVal | 0.040 | 20.076 | 13.393 | 6.63e-41 | 0.036 | 18.841 | 12.904 | 4.26e-38 |
| LDpred2.MultiPRS | SBLUP.Inf | 0.037 | 18.529 | 12.352 | 4.74e-35 | 0.038 | 19.824 | 12.961 | 2.03e-38 |
| LDpred2.MultiPRS | SBayesR.PseudoVal | 0.018 | 9.178 | 6.141 | 8.22e-10 | 0.015 | 7.837 | 6.224 | 4.85e-10 |
| LDpred2.MultiPRS | LDpred1.10FCVal | 0.023 | 11.309 | 7.556 | 4.15e-14 | 0.023 | 11.856 | 10.224 | 1.55e-24 |
| LDpred2.MultiPRS | LDpred1.MultiPRS | 0.017 | 8.467 | 7.125 | 1.04e-12 | 0.017 | 8.988 | 8.372 | 5.66e-17 |
| LDpred2.MultiPRS | LDpred1.Inf | 0.037 | 18.613 | 12.410 | 2.32e-35 | 0.039 | 19.983 | 13.301 | 2.28e-40 |
| LDpred2.MultiPRS | LDpred2.10FCVal | 0.003 | 1.469 | 0.985 | 3.25e-01 | 0.003 | 1.675 | 3.887 | 1.02e-04 |
| LDpred2.MultiPRS | LDpred2.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.MultiPRS | LDpred2.PseudoVal | 0.029 | 14.567 | 9.733 | 2.19e-22 | 0.025 | 13.165 | 12.270 | 1.32e-34 |
| LDpred2.MultiPRS | LDpred2.Inf | 0.039 | 19.385 | 12.919 | 3.50e-38 | 0.039 | 20.312 | 13.172 | 1.27e-39 |
| LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.016 | 7.836 | 5.239 | 1.61e-07 | 0.016 | 8.468 | 7.201 | 6.00e-13 |
| LDpred2.MultiPRS | All.MultiPRS | -0.003 | -1.584 | -1.847 | 6.47e-02 | -0.004 | -2.029 | -3.895 | 9.83e-05 |
| LDpred2.PseudoVal | pT+clump.10FCVal | 0.014 | 8.496 | 8.202 | 2.36e-16 | 0.016 | 9.779 | 4.632 | 3.62e-06 |
| LDpred2.PseudoVal | pT+clump.MultiPRS | -0.006 | -3.360 | -1.908 | 5.64e-02 | -0.006 | -3.602 | -1.905 | 5.68e-02 |
| LDpred2.PseudoVal | lassosum.10FCVal | -0.020 | -11.525 | -14.051 | 7.57e-45 | -0.015 | -9.047 | -5.456 | 4.86e-08 |
| LDpred2.PseudoVal | lassosum.MultiPRS | -0.028 | -16.390 | -9.345 | 9.22e-21 | -0.025 | -14.644 | -9.486 | 2.40e-21 |
| LDpred2.PseudoVal | lassosum.PseudoVal | 0.011 | 6.448 | 6.800 | 1.05e-11 | 0.011 | 6.536 | 3.415 | 6.37e-04 |
| LDpred2.PseudoVal | SBLUP.Inf | 0.008 | 4.637 | 4.109 | 3.97e-05 | 0.013 | 7.668 | 3.357 | 7.88e-04 |
| LDpred2.PseudoVal | SBayesR.PseudoVal | -0.011 | -6.307 | -7.668 | 1.75e-14 | -0.010 | -6.136 | -3.674 | 2.39e-04 |
| LDpred2.PseudoVal | LDpred1.10FCVal | -0.006 | -3.814 | -4.265 | 2.00e-05 | -0.003 | -1.508 | -0.834 | 4.04e-01 |
| LDpred2.PseudoVal | LDpred1.MultiPRS | -0.012 | -7.140 | -4.088 | 4.35e-05 | -0.008 | -4.810 | -2.785 | 5.35e-03 |
| LDpred2.PseudoVal | LDpred1.Inf | 0.008 | 4.736 | 4.384 | 1.16e-05 | 0.013 | 7.852 | 3.591 | 3.29e-04 |
| LDpred2.PseudoVal | LDpred2.10FCVal | -0.026 | -15.331 | -25.675 | 2.21e-145 | -0.022 | -13.232 | -10.937 | 7.68e-28 |
| LDpred2.PseudoVal | LDpred2.MultiPRS | -0.029 | -17.051 | -9.733 | 2.19e-22 | -0.025 | -15.161 | -12.270 | 1.32e-34 |
| LDpred2.PseudoVal | LDpred2.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.PseudoVal | LDpred2.Inf | 0.010 | 5.639 | 4.981 | 6.33e-07 | 0.014 | 8.231 | 3.590 | 3.31e-04 |
| LDpred2.PseudoVal | DBSLMM.PseudoVal | -0.013 | -7.879 | -9.195 | 3.74e-20 | -0.009 | -5.409 | -3.114 | 1.84e-03 |
| LDpred2.PseudoVal | All.MultiPRS | -0.032 | -18.905 | -10.803 | 3.34e-27 | -0.029 | -17.497 | -12.906 | 4.15e-38 |
| LDpred2.Inf | pT+clump.10FCVal | 0.005 | 3.028 | 2.159 | 3.08e-02 | 0.003 | 1.687 | 0.578 | 5.63e-01 |
| LDpred2.Inf | pT+clump.MultiPRS | -0.015 | -9.536 | -5.110 | 3.23e-07 | -0.020 | -12.895 | -5.644 | 1.66e-08 |
| LDpred2.Inf | lassosum.10FCVal | -0.029 | -18.189 | -19.026 | 1.04e-80 | -0.029 | -18.828 | -9.437 | 3.83e-21 |
| LDpred2.Inf | lassosum.MultiPRS | -0.037 | -23.345 | -12.534 | 4.86e-36 | -0.038 | -24.926 | -12.703 | 5.70e-37 |
| LDpred2.Inf | lassosum.PseudoVal | 0.001 | 0.857 | 0.642 | 5.21e-01 | -0.003 | -1.847 | -0.665 | 5.06e-01 |
| LDpred2.Inf | SBLUP.Inf | -0.002 | -1.061 | -6.837 | 8.06e-12 | -0.001 | -0.613 | -1.898 | 5.77e-02 |
| LDpred2.Inf | SBayesR.PseudoVal | -0.020 | -12.660 | -12.864 | 7.18e-38 | -0.024 | -15.656 | -7.631 | 2.33e-14 |
| LDpred2.Inf | LDpred1.10FCVal | -0.016 | -10.017 | -12.397 | 2.71e-35 | -0.016 | -10.612 | -6.299 | 3.00e-10 |
| LDpred2.Inf | LDpred1.MultiPRS | -0.022 | -13.542 | -7.305 | 2.78e-13 | -0.022 | -14.211 | -8.018 | 1.07e-15 |
| LDpred2.Inf | LDpred1.Inf | -0.002 | -0.957 | -1.653 | 9.83e-02 | -0.001 | -0.414 | -0.342 | 7.32e-01 |
| LDpred2.Inf | LDpred2.10FCVal | -0.036 | -22.223 | -23.582 | 5.94e-123 | -0.036 | -23.388 | -11.900 | 1.19e-32 |
| LDpred2.Inf | LDpred2.MultiPRS | -0.039 | -24.046 | -12.919 | 3.50e-38 | -0.039 | -25.490 | -13.172 | 1.27e-39 |
| LDpred2.Inf | LDpred2.PseudoVal | -0.010 | -5.976 | -4.981 | 6.33e-07 | -0.014 | -8.969 | -3.590 | 3.31e-04 |
| LDpred2.Inf | LDpred2.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.Inf | DBSLMM.PseudoVal | -0.023 | -14.325 | -12.770 | 2.41e-37 | -0.023 | -14.863 | -6.344 | 2.24e-10 |
| LDpred2.Inf | All.MultiPRS | -0.042 | -26.010 | -14.031 | 1.01e-44 | -0.043 | -28.036 | -14.145 | 1.99e-45 |
| DBSLMM.PseudoVal | pT+clump.10FCVal | 0.028 | 15.179 | 16.591 | 8.16e-62 | 0.026 | 14.409 | 7.580 | 3.47e-14 |
| DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.008 | 4.189 | 2.573 | 1.01e-02 | 0.003 | 1.714 | 1.053 | 2.92e-01 |
| DBSLMM.PseudoVal | lassosum.10FCVal | -0.006 | -3.379 | -4.710 | 2.48e-06 | -0.006 | -3.451 | -2.316 | 2.05e-02 |
| DBSLMM.PseudoVal | lassosum.MultiPRS | -0.014 | -7.889 | -4.858 | 1.19e-06 | -0.016 | -8.761 | -7.186 | 6.66e-13 |
| DBSLMM.PseudoVal | lassosum.PseudoVal | 0.024 | 13.280 | 15.924 | 4.32e-57 | 0.020 | 11.332 | 6.561 | 5.35e-11 |
| DBSLMM.PseudoVal | SBLUP.Inf | 0.021 | 11.602 | 12.177 | 4.12e-34 | 0.022 | 12.406 | 6.265 | 3.72e-10 |
| DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.003 | 1.457 | 1.714 | 8.65e-02 | -0.001 | -0.690 | -0.389 | 6.97e-01 |
| DBSLMM.PseudoVal | LDpred1.10FCVal | 0.007 | 3.768 | 5.736 | 9.70e-09 | 0.007 | 3.701 | 2.708 | 6.76e-03 |
| DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.001 | 0.685 | 0.424 | 6.72e-01 | 0.001 | 0.568 | 0.420 | 6.74e-01 |
| DBSLMM.PseudoVal | LDpred1.Inf | 0.021 | 11.694 | 13.584 | 4.96e-42 | 0.022 | 12.580 | 7.034 | 2.01e-12 |
| DBSLMM.PseudoVal | LDpred2.10FCVal | -0.013 | -6.908 | -11.787 | 4.56e-32 | -0.013 | -7.422 | -6.090 | 1.13e-09 |
| DBSLMM.PseudoVal | LDpred2.MultiPRS | -0.016 | -8.502 | -5.239 | 1.61e-07 | -0.016 | -9.251 | -7.201 | 6.00e-13 |
| DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.013 | 7.303 | 9.195 | 3.74e-20 | 0.009 | 5.131 | 3.114 | 1.84e-03 |
| DBSLMM.PseudoVal | LDpred2.Inf | 0.023 | 12.530 | 12.770 | 2.41e-37 | 0.023 | 12.940 | 6.344 | 2.24e-10 |
| DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| DBSLMM.PseudoVal | All.MultiPRS | -0.019 | -10.221 | -6.325 | 2.53e-10 | -0.020 | -11.468 | -9.376 | 6.83e-21 |
| All.MultiPRS | pT+clump.10FCVal | 0.047 | 23.044 | 15.651 | 3.29e-55 | 0.046 | 23.215 | 14.824 | 1.02e-49 |
| All.MultiPRS | pT+clump.MultiPRS | 0.026 | 13.073 | 8.879 | 6.75e-19 | 0.023 | 11.826 | 10.185 | 2.33e-24 |
| All.MultiPRS | lassosum.10FCVal | 0.013 | 6.207 | 4.238 | 2.26e-05 | 0.014 | 7.192 | 7.769 | 7.93e-15 |
| All.MultiPRS | lassosum.MultiPRS | 0.004 | 2.115 | 2.255 | 2.42e-02 | 0.005 | 2.429 | 4.060 | 4.90e-05 |
| All.MultiPRS | lassosum.PseudoVal | 0.043 | 21.322 | 14.502 | 1.18e-47 | 0.040 | 20.455 | 14.423 | 3.71e-47 |
| All.MultiPRS | SBLUP.Inf | 0.040 | 19.799 | 13.461 | 2.65e-41 | 0.042 | 21.418 | 14.302 | 2.12e-46 |
| All.MultiPRS | SBayesR.PseudoVal | 0.021 | 10.594 | 7.228 | 4.90e-13 | 0.019 | 9.670 | 8.325 | 8.40e-17 |
| All.MultiPRS | LDpred1.10FCVal | 0.026 | 12.692 | 8.647 | 5.29e-18 | 0.027 | 13.609 | 11.997 | 3.69e-33 |
| All.MultiPRS | LDpred1.MultiPRS | 0.020 | 9.894 | 8.540 | 1.34e-17 | 0.021 | 10.798 | 10.450 | 1.46e-25 |
| All.MultiPRS | LDpred1.Inf | 0.040 | 19.882 | 13.511 | 1.34e-41 | 0.043 | 21.574 | 14.911 | 2.82e-50 |
| All.MultiPRS | LDpred2.10FCVal | 0.006 | 3.005 | 2.056 | 3.98e-02 | 0.007 | 3.630 | 5.191 | 2.09e-07 |
| All.MultiPRS | LDpred2.MultiPRS | 0.003 | 1.559 | 1.847 | 6.47e-02 | 0.004 | 1.989 | 3.895 | 9.83e-05 |
| All.MultiPRS | LDpred2.PseudoVal | 0.032 | 15.899 | 10.803 | 3.34e-27 | 0.029 | 14.892 | 12.906 | 4.15e-38 |
| All.MultiPRS | LDpred2.Inf | 0.042 | 20.641 | 14.031 | 1.01e-44 | 0.043 | 21.897 | 14.145 | 1.99e-45 |
| All.MultiPRS | DBSLMM.PseudoVal | 0.019 | 9.273 | 6.325 | 2.53e-10 | 0.020 | 10.288 | 9.376 | 6.83e-21 |
| All.MultiPRS | All.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
Show table: TEDS (1KG ref)
| Reference | Test | CrossVal Diff | CrossVal Diff (%) | CrossVal Diff Z | CrossVal Diff P | IndepVal Diff | IndepVal Diff (%) | IndepVal Diff Z | IndepVal Diff P |
|---|---|---|---|---|---|---|---|---|---|
| pT+clump.10FCVal | pT+clump.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| pT+clump.10FCVal | pT+clump.MultiPRS | -0.007 | -2.450 | -0.651 | 5.15e-01 | -0.015 | -5.242 | -3.302 | 9.60e-04 |
| pT+clump.10FCVal | lassosum.10FCVal | -0.037 | -12.753 | -7.873 | 3.47e-15 | -0.046 | -16.458 | -4.868 | 1.13e-06 |
| pT+clump.10FCVal | lassosum.MultiPRS | -0.041 | -14.173 | -3.796 | 1.47e-04 | -0.049 | -17.504 | -5.066 | 4.05e-07 |
| pT+clump.10FCVal | lassosum.PseudoVal | 0.030 | 10.405 | 4.221 | 2.43e-05 | 0.034 | 12.244 | 2.364 | 1.81e-02 |
| pT+clump.10FCVal | PRScs.10FCVal | -0.036 | -12.253 | -7.738 | 1.01e-14 | -0.048 | -17.323 | -5.209 | 1.90e-07 |
| pT+clump.10FCVal | PRScs.MultiPRS | -0.038 | -13.036 | -3.491 | 4.82e-04 | -0.049 | -17.502 | -5.334 | 9.63e-08 |
| pT+clump.10FCVal | PRScs.PseudoVal | -0.031 | -10.801 | -6.316 | 2.69e-10 | -0.040 | -14.492 | -4.017 | 5.91e-05 |
| pT+clump.10FCVal | SBLUP.Inf | -0.013 | -4.610 | -2.582 | 9.82e-03 | -0.025 | -8.842 | -2.328 | 1.99e-02 |
| pT+clump.10FCVal | SBayesR.PseudoVal | -0.023 | -7.865 | -3.861 | 1.13e-04 | -0.024 | -8.541 | -2.008 | 4.46e-02 |
| pT+clump.10FCVal | LDpred1.10FCVal | -0.018 | -6.199 | -3.412 | 6.45e-04 | -0.024 | -8.432 | -2.182 | 2.91e-02 |
| pT+clump.10FCVal | LDpred1.MultiPRS | -0.024 | -8.198 | -2.189 | 2.86e-02 | -0.030 | -10.647 | -2.887 | 3.88e-03 |
| pT+clump.10FCVal | LDpred1.Inf | -0.021 | -7.055 | -4.094 | 4.24e-05 | -0.026 | -9.419 | -2.562 | 1.04e-02 |
| pT+clump.10FCVal | LDpred2.10FCVal | -0.039 | -13.519 | -7.938 | 2.05e-15 | -0.046 | -16.649 | -4.654 | 3.25e-06 |
| pT+clump.10FCVal | LDpred2.MultiPRS | -0.040 | -13.803 | -3.700 | 2.15e-04 | -0.041 | -14.599 | -4.107 | 4.01e-05 |
| pT+clump.10FCVal | LDpred2.PseudoVal | -0.017 | -5.854 | -3.241 | 1.19e-03 | -0.024 | -8.698 | -2.268 | 2.33e-02 |
| pT+clump.10FCVal | LDpred2.Inf | -0.017 | -5.959 | -3.342 | 8.31e-04 | -0.024 | -8.525 | -2.251 | 2.44e-02 |
| pT+clump.10FCVal | DBSLMM.PseudoVal | -0.031 | -10.509 | -6.001 | 1.96e-09 | -0.034 | -12.239 | -3.349 | 8.12e-04 |
| pT+clump.10FCVal | All.MultiPRS | -0.046 | -15.710 | -4.227 | 2.36e-05 | -0.047 | -16.845 | -4.893 | 9.92e-07 |
| pT+clump.MultiPRS | pT+clump.10FCVal | 0.007 | 2.392 | 0.651 | 5.15e-01 | 0.015 | 4.981 | 3.302 | 9.60e-04 |
| pT+clump.MultiPRS | pT+clump.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| pT+clump.MultiPRS | lassosum.10FCVal | -0.030 | -10.056 | -2.770 | 5.61e-03 | -0.031 | -10.657 | -3.681 | 2.32e-04 |
| pT+clump.MultiPRS | lassosum.MultiPRS | -0.034 | -11.443 | -3.152 | 1.62e-03 | -0.034 | -11.651 | -4.084 | 4.42e-05 |
| pT+clump.MultiPRS | lassosum.PseudoVal | 0.037 | 12.548 | 3.394 | 6.88e-04 | 0.049 | 16.615 | 3.880 | 1.04e-04 |
| pT+clump.MultiPRS | PRScs.10FCVal | -0.029 | -9.568 | -2.635 | 8.41e-03 | -0.034 | -11.480 | -3.949 | 7.85e-05 |
| pT+clump.MultiPRS | PRScs.MultiPRS | -0.031 | -10.333 | -2.855 | 4.30e-03 | -0.034 | -11.649 | -4.283 | 1.84e-05 |
| pT+clump.MultiPRS | PRScs.PseudoVal | -0.024 | -8.151 | -2.241 | 2.50e-02 | -0.026 | -8.789 | -3.061 | 2.21e-03 |
| pT+clump.MultiPRS | SBLUP.Inf | -0.006 | -2.108 | -0.576 | 5.65e-01 | -0.010 | -3.420 | -1.014 | 3.10e-01 |
| pT+clump.MultiPRS | SBayesR.PseudoVal | -0.016 | -5.285 | -1.450 | 1.47e-01 | -0.009 | -3.134 | -0.889 | 3.74e-01 |
| pT+clump.MultiPRS | LDpred1.10FCVal | -0.011 | -3.659 | -1.000 | 3.17e-01 | -0.009 | -3.031 | -0.888 | 3.75e-01 |
| pT+clump.MultiPRS | LDpred1.MultiPRS | -0.017 | -5.610 | -1.540 | 1.24e-01 | -0.015 | -5.136 | -1.598 | 1.10e-01 |
| pT+clump.MultiPRS | LDpred1.Inf | -0.013 | -4.495 | -1.231 | 2.18e-01 | -0.012 | -3.969 | -1.221 | 2.22e-01 |
| pT+clump.MultiPRS | LDpred2.10FCVal | -0.032 | -10.804 | -2.976 | 2.92e-03 | -0.032 | -10.839 | -3.594 | 3.26e-04 |
| pT+clump.MultiPRS | LDpred2.MultiPRS | -0.033 | -11.081 | -3.067 | 2.17e-03 | -0.026 | -8.891 | -2.926 | 3.43e-03 |
| pT+clump.MultiPRS | LDpred2.PseudoVal | -0.010 | -3.322 | -0.909 | 3.63e-01 | -0.010 | -3.284 | -0.967 | 3.34e-01 |
| pT+clump.MultiPRS | LDpred2.Inf | -0.010 | -3.425 | -0.937 | 3.49e-01 | -0.009 | -3.119 | -0.929 | 3.53e-01 |
| pT+clump.MultiPRS | DBSLMM.PseudoVal | -0.023 | -7.866 | -2.160 | 3.08e-02 | -0.020 | -6.648 | -2.322 | 2.02e-02 |
| pT+clump.MultiPRS | All.MultiPRS | -0.039 | -12.943 | -3.610 | 3.07e-04 | -0.032 | -11.025 | -3.817 | 1.35e-04 |
| lassosum.10FCVal | pT+clump.10FCVal | 0.037 | 11.311 | 7.873 | 3.47e-15 | 0.046 | 14.132 | 4.868 | 1.13e-06 |
| lassosum.10FCVal | pT+clump.MultiPRS | 0.030 | 9.137 | 2.770 | 5.61e-03 | 0.031 | 9.631 | 3.681 | 2.32e-04 |
| lassosum.10FCVal | lassosum.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.10FCVal | lassosum.MultiPRS | -0.004 | -1.260 | -0.386 | 7.00e-01 | -0.003 | -0.899 | -0.879 | 3.79e-01 |
| lassosum.10FCVal | lassosum.PseudoVal | 0.067 | 20.539 | 12.555 | 3.73e-36 | 0.080 | 24.645 | 7.323 | 2.43e-13 |
| lassosum.10FCVal | PRScs.10FCVal | 0.001 | 0.444 | 0.554 | 5.80e-01 | -0.002 | -0.743 | -0.450 | 6.53e-01 |
| lassosum.10FCVal | PRScs.MultiPRS | -0.001 | -0.251 | -0.077 | 9.39e-01 | -0.003 | -0.896 | -0.629 | 5.29e-01 |
| lassosum.10FCVal | PRScs.PseudoVal | 0.006 | 1.731 | 2.195 | 2.82e-02 | 0.005 | 1.688 | 1.043 | 2.97e-01 |
| lassosum.10FCVal | SBLUP.Inf | 0.024 | 7.222 | 6.026 | 1.68e-09 | 0.021 | 6.540 | 2.709 | 6.75e-03 |
| lassosum.10FCVal | SBayesR.PseudoVal | 0.014 | 4.335 | 3.387 | 7.07e-04 | 0.022 | 6.798 | 2.585 | 9.75e-03 |
| lassosum.10FCVal | LDpred1.10FCVal | 0.019 | 5.812 | 5.036 | 4.76e-07 | 0.022 | 6.892 | 2.901 | 3.72e-03 |
| lassosum.10FCVal | LDpred1.MultiPRS | 0.013 | 4.040 | 1.234 | 2.17e-01 | 0.016 | 4.990 | 2.301 | 2.14e-02 |
| lassosum.10FCVal | LDpred1.Inf | 0.017 | 5.053 | 4.512 | 6.43e-06 | 0.020 | 6.044 | 2.618 | 8.84e-03 |
| lassosum.10FCVal | LDpred2.10FCVal | -0.002 | -0.680 | -0.819 | 4.13e-01 | -0.001 | -0.164 | -0.097 | 9.23e-01 |
| lassosum.10FCVal | LDpred2.MultiPRS | -0.003 | -0.931 | -0.285 | 7.76e-01 | 0.005 | 1.596 | 0.915 | 3.60e-01 |
| lassosum.10FCVal | LDpred2.PseudoVal | 0.020 | 6.119 | 5.293 | 1.20e-07 | 0.022 | 6.663 | 2.824 | 4.74e-03 |
| lassosum.10FCVal | LDpred2.Inf | 0.020 | 6.026 | 5.260 | 1.44e-07 | 0.022 | 6.812 | 2.910 | 3.61e-03 |
| lassosum.10FCVal | DBSLMM.PseudoVal | 0.007 | 1.990 | 1.867 | 6.19e-02 | 0.012 | 3.623 | 1.665 | 9.59e-02 |
| lassosum.10FCVal | All.MultiPRS | -0.009 | -2.623 | -0.809 | 4.19e-01 | -0.001 | -0.333 | -0.194 | 8.46e-01 |
| lassosum.MultiPRS | pT+clump.10FCVal | 0.041 | 12.414 | 3.796 | 1.47e-04 | 0.049 | 14.897 | 5.066 | 4.05e-07 |
| lassosum.MultiPRS | pT+clump.MultiPRS | 0.034 | 10.268 | 3.152 | 1.62e-03 | 0.034 | 10.435 | 4.084 | 4.42e-05 |
| lassosum.MultiPRS | lassosum.10FCVal | 0.004 | 1.244 | 0.386 | 7.00e-01 | 0.003 | 0.891 | 0.879 | 3.79e-01 |
| lassosum.MultiPRS | lassosum.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.MultiPRS | lassosum.PseudoVal | 0.071 | 21.528 | 6.547 | 5.86e-11 | 0.083 | 25.316 | 7.541 | 4.67e-14 |
| lassosum.MultiPRS | PRScs.10FCVal | 0.006 | 1.682 | 0.521 | 6.02e-01 | 0.001 | 0.154 | 0.098 | 9.22e-01 |
| lassosum.MultiPRS | PRScs.MultiPRS | 0.003 | 0.996 | 0.448 | 6.54e-01 | 0.000 | 0.002 | 0.002 | 9.99e-01 |
| lassosum.MultiPRS | PRScs.PseudoVal | 0.010 | 2.954 | 0.913 | 3.61e-01 | 0.008 | 2.564 | 1.633 | 1.02e-01 |
| lassosum.MultiPRS | SBLUP.Inf | 0.028 | 8.376 | 2.568 | 1.02e-02 | 0.024 | 7.372 | 3.184 | 1.45e-03 |
| lassosum.MultiPRS | SBayesR.PseudoVal | 0.018 | 5.525 | 1.705 | 8.82e-02 | 0.025 | 7.628 | 2.974 | 2.94e-03 |
| lassosum.MultiPRS | LDpred1.10FCVal | 0.023 | 6.984 | 2.147 | 3.18e-02 | 0.025 | 7.721 | 3.401 | 6.72e-04 |
| lassosum.MultiPRS | LDpred1.MultiPRS | 0.017 | 5.234 | 2.297 | 2.16e-02 | 0.019 | 5.836 | 2.824 | 4.75e-03 |
| lassosum.MultiPRS | LDpred1.Inf | 0.021 | 6.234 | 1.917 | 5.53e-02 | 0.023 | 6.881 | 3.115 | 1.84e-03 |
| lassosum.MultiPRS | LDpred2.10FCVal | 0.002 | 0.573 | 0.177 | 8.59e-01 | 0.002 | 0.728 | 0.445 | 6.56e-01 |
| lassosum.MultiPRS | LDpred2.MultiPRS | 0.001 | 0.325 | 0.155 | 8.77e-01 | 0.008 | 2.472 | 1.459 | 1.45e-01 |
| lassosum.MultiPRS | LDpred2.PseudoVal | 0.024 | 7.287 | 2.237 | 2.53e-02 | 0.025 | 7.495 | 3.315 | 9.16e-04 |
| lassosum.MultiPRS | LDpred2.Inf | 0.024 | 7.195 | 2.209 | 2.72e-02 | 0.025 | 7.642 | 3.409 | 6.53e-04 |
| lassosum.MultiPRS | DBSLMM.PseudoVal | 0.011 | 3.210 | 0.994 | 3.20e-01 | 0.015 | 4.481 | 2.392 | 1.68e-02 |
| lassosum.MultiPRS | All.MultiPRS | -0.004 | -1.346 | -0.421 | 6.73e-01 | 0.002 | 0.561 | 0.425 | 6.71e-01 |
| lassosum.PseudoVal | pT+clump.10FCVal | -0.030 | -11.614 | -4.221 | 2.43e-05 | -0.034 | -13.952 | -2.364 | 1.81e-02 |
| lassosum.PseudoVal | pT+clump.MultiPRS | -0.037 | -14.349 | -3.394 | 6.88e-04 | -0.049 | -19.925 | -3.880 | 1.04e-04 |
| lassosum.PseudoVal | lassosum.10FCVal | -0.067 | -25.848 | -12.555 | 3.73e-36 | -0.080 | -32.706 | -7.323 | 2.43e-13 |
| lassosum.PseudoVal | lassosum.MultiPRS | -0.071 | -27.433 | -6.547 | 5.86e-11 | -0.083 | -33.898 | -7.541 | 4.67e-14 |
| lassosum.PseudoVal | lassosum.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.PseudoVal | PRScs.10FCVal | -0.066 | -25.289 | -11.309 | 1.19e-29 | -0.082 | -33.692 | -6.927 | 4.29e-12 |
| lassosum.PseudoVal | PRScs.MultiPRS | -0.068 | -26.164 | -6.272 | 3.58e-10 | -0.083 | -33.895 | -7.374 | 1.65e-13 |
| lassosum.PseudoVal | PRScs.PseudoVal | -0.062 | -23.669 | -12.255 | 1.57e-34 | -0.075 | -30.466 | -7.446 | 9.64e-14 |
| lassosum.PseudoVal | SBLUP.Inf | -0.044 | -16.760 | -5.991 | 2.09e-09 | -0.059 | -24.027 | -4.016 | 5.93e-05 |
| lassosum.PseudoVal | SBayesR.PseudoVal | -0.053 | -20.392 | -10.739 | 6.69e-27 | -0.058 | -23.684 | -5.743 | 9.32e-09 |
| lassosum.PseudoVal | LDpred1.10FCVal | -0.048 | -18.533 | -7.027 | 2.11e-12 | -0.058 | -23.560 | -4.144 | 3.41e-05 |
| lassosum.PseudoVal | LDpred1.MultiPRS | -0.054 | -20.764 | -4.956 | 7.21e-07 | -0.064 | -26.084 | -4.659 | 3.17e-06 |
| lassosum.PseudoVal | LDpred1.Inf | -0.051 | -19.489 | -7.198 | 6.12e-13 | -0.060 | -24.685 | -4.217 | 2.47e-05 |
| lassosum.PseudoVal | LDpred2.10FCVal | -0.070 | -26.703 | -11.687 | 1.48e-31 | -0.081 | -32.924 | -6.717 | 1.85e-11 |
| lassosum.PseudoVal | LDpred2.MultiPRS | -0.070 | -27.019 | -6.446 | 1.15e-10 | -0.075 | -30.588 | -6.375 | 1.83e-10 |
| lassosum.PseudoVal | LDpred2.PseudoVal | -0.047 | -18.147 | -6.658 | 2.78e-11 | -0.058 | -23.863 | -4.069 | 4.73e-05 |
| lassosum.PseudoVal | LDpred2.Inf | -0.048 | -18.265 | -6.693 | 2.19e-11 | -0.058 | -23.666 | -4.032 | 5.54e-05 |
| lassosum.PseudoVal | DBSLMM.PseudoVal | -0.061 | -23.343 | -10.096 | 5.76e-24 | -0.068 | -27.898 | -5.597 | 2.17e-08 |
| lassosum.PseudoVal | All.MultiPRS | -0.076 | -29.148 | -7.016 | 2.28e-12 | -0.081 | -33.147 | -7.041 | 1.91e-12 |
| PRScs.10FCVal | pT+clump.10FCVal | 0.036 | 10.915 | 7.738 | 1.01e-14 | 0.048 | 14.766 | 5.209 | 1.90e-07 |
| PRScs.10FCVal | pT+clump.MultiPRS | 0.029 | 8.732 | 2.635 | 8.41e-03 | 0.034 | 10.297 | 3.949 | 7.85e-05 |
| PRScs.10FCVal | lassosum.10FCVal | -0.001 | -0.446 | -0.554 | 5.80e-01 | 0.002 | 0.738 | 0.450 | 6.53e-01 |
| PRScs.10FCVal | lassosum.MultiPRS | -0.006 | -1.711 | -0.521 | 6.02e-01 | -0.001 | -0.154 | -0.098 | 9.22e-01 |
| PRScs.10FCVal | lassosum.PseudoVal | 0.066 | 20.185 | 11.309 | 1.19e-29 | 0.082 | 25.201 | 6.927 | 4.29e-12 |
| PRScs.10FCVal | PRScs.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| PRScs.10FCVal | PRScs.MultiPRS | -0.002 | -0.698 | -0.213 | 8.31e-01 | 0.000 | -0.152 | -0.205 | 8.37e-01 |
| PRScs.10FCVal | PRScs.PseudoVal | 0.004 | 1.293 | 1.697 | 8.98e-02 | 0.008 | 2.413 | 1.529 | 1.26e-01 |
| PRScs.10FCVal | SBLUP.Inf | 0.022 | 6.808 | 6.677 | 2.44e-11 | 0.024 | 7.229 | 3.506 | 4.55e-04 |
| PRScs.10FCVal | SBayesR.PseudoVal | 0.013 | 3.909 | 2.864 | 4.18e-03 | 0.024 | 7.486 | 2.707 | 6.80e-03 |
| PRScs.10FCVal | LDpred1.10FCVal | 0.018 | 5.393 | 5.523 | 3.33e-08 | 0.025 | 7.579 | 3.801 | 1.44e-04 |
| PRScs.10FCVal | LDpred1.MultiPRS | 0.012 | 3.612 | 1.098 | 2.72e-01 | 0.019 | 5.691 | 3.286 | 1.02e-03 |
| PRScs.10FCVal | LDpred1.Inf | 0.015 | 4.630 | 5.044 | 4.57e-07 | 0.022 | 6.737 | 3.661 | 2.51e-04 |
| PRScs.10FCVal | LDpred2.10FCVal | -0.004 | -1.128 | -1.428 | 1.53e-01 | 0.002 | 0.575 | 0.358 | 7.21e-01 |
| PRScs.10FCVal | LDpred2.MultiPRS | -0.005 | -1.381 | -0.421 | 6.74e-01 | 0.008 | 2.322 | 1.430 | 1.53e-01 |
| PRScs.10FCVal | LDpred2.PseudoVal | 0.019 | 5.700 | 5.895 | 3.75e-09 | 0.024 | 7.352 | 3.762 | 1.69e-04 |
| PRScs.10FCVal | LDpred2.Inf | 0.018 | 5.607 | 5.857 | 4.71e-09 | 0.025 | 7.499 | 3.881 | 1.04e-04 |
| PRScs.10FCVal | DBSLMM.PseudoVal | 0.005 | 1.553 | 1.698 | 8.95e-02 | 0.014 | 4.334 | 2.360 | 1.83e-02 |
| PRScs.10FCVal | All.MultiPRS | -0.010 | -3.080 | -0.944 | 3.45e-01 | 0.001 | 0.408 | 0.237 | 8.13e-01 |
| PRScs.MultiPRS | pT+clump.10FCVal | 0.038 | 11.533 | 3.491 | 4.82e-04 | 0.049 | 14.895 | 5.334 | 9.63e-08 |
| PRScs.MultiPRS | pT+clump.MultiPRS | 0.031 | 9.365 | 2.855 | 4.30e-03 | 0.034 | 10.433 | 4.283 | 1.84e-05 |
| PRScs.MultiPRS | lassosum.10FCVal | 0.001 | 0.251 | 0.077 | 9.39e-01 | 0.003 | 0.888 | 0.629 | 5.29e-01 |
| PRScs.MultiPRS | lassosum.MultiPRS | -0.003 | -1.006 | -0.448 | 6.54e-01 | 0.000 | -0.002 | -0.002 | 9.99e-01 |
| PRScs.MultiPRS | lassosum.PseudoVal | 0.068 | 20.738 | 6.272 | 3.58e-10 | 0.083 | 25.315 | 7.374 | 1.65e-13 |
| PRScs.MultiPRS | PRScs.10FCVal | 0.002 | 0.693 | 0.213 | 8.31e-01 | 0.000 | 0.152 | 0.205 | 8.37e-01 |
| PRScs.MultiPRS | PRScs.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| PRScs.MultiPRS | PRScs.PseudoVal | 0.007 | 1.978 | 0.607 | 5.44e-01 | 0.008 | 2.561 | 2.229 | 2.58e-02 |
| PRScs.MultiPRS | SBLUP.Inf | 0.025 | 7.454 | 2.267 | 2.34e-02 | 0.024 | 7.370 | 3.772 | 1.62e-04 |
| PRScs.MultiPRS | SBayesR.PseudoVal | 0.015 | 4.575 | 1.404 | 1.60e-01 | 0.025 | 7.626 | 3.017 | 2.56e-03 |
| PRScs.MultiPRS | LDpred1.10FCVal | 0.020 | 6.049 | 1.843 | 6.54e-02 | 0.025 | 7.719 | 3.981 | 6.86e-05 |
| PRScs.MultiPRS | LDpred1.MultiPRS | 0.014 | 4.280 | 2.714 | 6.64e-03 | 0.019 | 5.834 | 3.528 | 4.19e-04 |
| PRScs.MultiPRS | LDpred1.Inf | 0.017 | 5.291 | 1.615 | 1.06e-01 | 0.023 | 6.879 | 3.888 | 1.01e-04 |
| PRScs.MultiPRS | LDpred2.10FCVal | -0.001 | -0.427 | -0.131 | 8.95e-01 | 0.002 | 0.726 | 0.515 | 6.06e-01 |
| PRScs.MultiPRS | LDpred2.MultiPRS | -0.002 | -0.678 | -0.251 | 8.02e-01 | 0.008 | 2.470 | 1.675 | 9.38e-02 |
| PRScs.MultiPRS | LDpred2.PseudoVal | 0.021 | 6.354 | 1.937 | 5.27e-02 | 0.025 | 7.492 | 3.966 | 7.30e-05 |
| PRScs.MultiPRS | LDpred2.Inf | 0.021 | 6.261 | 1.908 | 5.64e-02 | 0.025 | 7.640 | 4.121 | 3.78e-05 |
| PRScs.MultiPRS | DBSLMM.PseudoVal | 0.007 | 2.236 | 0.686 | 4.93e-01 | 0.015 | 4.479 | 2.682 | 7.33e-03 |
| PRScs.MultiPRS | All.MultiPRS | -0.008 | -2.365 | -1.066 | 2.86e-01 | 0.002 | 0.559 | 0.364 | 7.16e-01 |
| PRScs.PseudoVal | pT+clump.10FCVal | 0.031 | 9.748 | 6.316 | 2.69e-10 | 0.040 | 12.658 | 4.017 | 5.91e-05 |
| PRScs.PseudoVal | pT+clump.MultiPRS | 0.024 | 7.536 | 2.241 | 2.50e-02 | 0.026 | 8.079 | 3.061 | 2.21e-03 |
| PRScs.PseudoVal | lassosum.10FCVal | -0.006 | -1.762 | -2.195 | 2.82e-02 | -0.005 | -1.717 | -1.043 | 2.97e-01 |
| PRScs.PseudoVal | lassosum.MultiPRS | -0.010 | -3.044 | -0.913 | 3.61e-01 | -0.008 | -2.631 | -1.633 | 1.02e-01 |
| PRScs.PseudoVal | lassosum.PseudoVal | 0.062 | 19.139 | 12.255 | 1.57e-34 | 0.075 | 23.352 | 7.446 | 9.64e-14 |
| PRScs.PseudoVal | PRScs.10FCVal | -0.004 | -1.310 | -1.697 | 8.98e-02 | -0.008 | -2.473 | -1.529 | 1.26e-01 |
| PRScs.PseudoVal | PRScs.MultiPRS | -0.007 | -2.018 | -0.607 | 5.44e-01 | -0.008 | -2.629 | -2.229 | 2.58e-02 |
| PRScs.PseudoVal | PRScs.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| PRScs.PseudoVal | SBLUP.Inf | 0.018 | 5.587 | 4.776 | 1.79e-06 | 0.016 | 4.935 | 2.038 | 4.16e-02 |
| PRScs.PseudoVal | SBayesR.PseudoVal | 0.009 | 2.650 | 2.275 | 2.29e-02 | 0.017 | 5.198 | 2.215 | 2.67e-02 |
| PRScs.PseudoVal | LDpred1.10FCVal | 0.013 | 4.153 | 3.715 | 2.03e-04 | 0.017 | 5.293 | 2.255 | 2.41e-02 |
| PRScs.PseudoVal | LDpred1.MultiPRS | 0.008 | 2.349 | 0.704 | 4.81e-01 | 0.011 | 3.358 | 1.593 | 1.11e-01 |
| PRScs.PseudoVal | LDpred1.Inf | 0.011 | 3.380 | 3.175 | 1.50e-03 | 0.014 | 4.431 | 1.976 | 4.81e-02 |
| PRScs.PseudoVal | LDpred2.10FCVal | -0.008 | -2.453 | -3.054 | 2.26e-03 | -0.006 | -1.884 | -1.151 | 2.50e-01 |
| PRScs.PseudoVal | LDpred2.MultiPRS | -0.009 | -2.709 | -0.813 | 4.16e-01 | 0.000 | -0.094 | -0.055 | 9.56e-01 |
| PRScs.PseudoVal | LDpred2.PseudoVal | 0.014 | 4.465 | 4.064 | 4.82e-05 | 0.016 | 5.061 | 2.209 | 2.72e-02 |
| PRScs.PseudoVal | LDpred2.Inf | 0.014 | 4.370 | 3.996 | 6.44e-05 | 0.017 | 5.212 | 2.286 | 2.23e-02 |
| PRScs.PseudoVal | DBSLMM.PseudoVal | 0.001 | 0.264 | 0.280 | 7.79e-01 | 0.006 | 1.968 | 1.018 | 3.09e-01 |
| PRScs.PseudoVal | All.MultiPRS | -0.014 | -4.431 | -1.341 | 1.80e-01 | -0.007 | -2.055 | -1.121 | 2.62e-01 |
| SBLUP.Inf | pT+clump.10FCVal | 0.013 | 4.407 | 2.582 | 9.82e-03 | 0.025 | 8.123 | 2.328 | 1.99e-02 |
| SBLUP.Inf | pT+clump.MultiPRS | 0.006 | 2.065 | 0.576 | 5.65e-01 | 0.010 | 3.307 | 1.014 | 3.10e-01 |
| SBLUP.Inf | lassosum.10FCVal | -0.024 | -7.784 | -6.026 | 1.68e-09 | -0.021 | -6.997 | -2.709 | 6.75e-03 |
| SBLUP.Inf | lassosum.MultiPRS | -0.028 | -9.142 | -2.568 | 1.02e-02 | -0.024 | -7.959 | -3.184 | 1.45e-03 |
| SBLUP.Inf | lassosum.PseudoVal | 0.044 | 14.354 | 5.991 | 2.09e-09 | 0.059 | 19.372 | 4.016 | 5.93e-05 |
| SBLUP.Inf | PRScs.10FCVal | -0.022 | -7.305 | -6.677 | 2.44e-11 | -0.024 | -7.793 | -3.506 | 4.55e-04 |
| SBLUP.Inf | PRScs.MultiPRS | -0.025 | -8.055 | -2.267 | 2.34e-02 | -0.024 | -7.956 | -3.772 | 1.62e-04 |
| SBLUP.Inf | PRScs.PseudoVal | -0.018 | -5.918 | -4.776 | 1.79e-06 | -0.016 | -5.191 | -2.038 | 4.16e-02 |
| SBLUP.Inf | SBLUP.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBLUP.Inf | SBayesR.PseudoVal | -0.009 | -3.111 | -1.704 | 8.83e-02 | 0.001 | 0.276 | 0.075 | 9.41e-01 |
| SBLUP.Inf | LDpred1.10FCVal | -0.005 | -1.519 | -1.650 | 9.89e-02 | 0.001 | 0.376 | 0.198 | 8.43e-01 |
| SBLUP.Inf | LDpred1.MultiPRS | -0.010 | -3.429 | -0.961 | 3.37e-01 | -0.005 | -1.659 | -1.057 | 2.91e-01 |
| SBLUP.Inf | LDpred1.Inf | -0.007 | -2.337 | -3.294 | 9.87e-04 | -0.002 | -0.531 | -0.357 | 7.21e-01 |
| SBLUP.Inf | LDpred2.10FCVal | -0.026 | -8.516 | -8.507 | 1.79e-17 | -0.022 | -7.173 | -3.547 | 3.90e-04 |
| SBLUP.Inf | LDpred2.MultiPRS | -0.027 | -8.787 | -2.474 | 1.34e-02 | -0.016 | -5.290 | -2.324 | 2.01e-02 |
| SBLUP.Inf | LDpred2.PseudoVal | -0.004 | -1.189 | -1.567 | 1.17e-01 | 0.000 | 0.132 | 0.084 | 9.33e-01 |
| SBLUP.Inf | LDpred2.Inf | -0.004 | -1.289 | -1.747 | 8.06e-02 | 0.001 | 0.291 | 0.189 | 8.50e-01 |
| SBLUP.Inf | DBSLMM.PseudoVal | -0.017 | -5.639 | -4.706 | 2.52e-06 | -0.009 | -3.121 | -1.261 | 2.07e-01 |
| SBLUP.Inf | All.MultiPRS | -0.032 | -10.611 | -3.001 | 2.69e-03 | -0.022 | -7.353 | -2.940 | 3.29e-03 |
| SBayesR.PseudoVal | pT+clump.10FCVal | 0.023 | 7.292 | 3.861 | 1.13e-04 | 0.024 | 7.869 | 2.008 | 4.46e-02 |
| SBayesR.PseudoVal | pT+clump.MultiPRS | 0.016 | 5.020 | 1.450 | 1.47e-01 | 0.009 | 3.039 | 0.889 | 3.74e-01 |
| SBayesR.PseudoVal | lassosum.10FCVal | -0.014 | -4.531 | -3.387 | 7.07e-04 | -0.022 | -7.294 | -2.585 | 9.75e-03 |
| SBayesR.PseudoVal | lassosum.MultiPRS | -0.018 | -5.848 | -1.705 | 8.82e-02 | -0.025 | -8.258 | -2.974 | 2.94e-03 |
| SBayesR.PseudoVal | lassosum.PseudoVal | 0.053 | 16.938 | 10.739 | 6.69e-27 | 0.058 | 19.149 | 5.743 | 9.32e-09 |
| SBayesR.PseudoVal | PRScs.10FCVal | -0.013 | -4.067 | -2.864 | 4.18e-03 | -0.024 | -8.092 | -2.707 | 6.80e-03 |
| SBayesR.PseudoVal | PRScs.MultiPRS | -0.015 | -4.794 | -1.404 | 1.60e-01 | -0.025 | -8.256 | -3.017 | 2.56e-03 |
| SBayesR.PseudoVal | PRScs.PseudoVal | -0.009 | -2.722 | -2.275 | 2.29e-02 | -0.017 | -5.483 | -2.215 | 2.67e-02 |
| SBayesR.PseudoVal | SBLUP.Inf | 0.009 | 3.018 | 1.704 | 8.83e-02 | -0.001 | -0.277 | -0.075 | 9.41e-01 |
| SBayesR.PseudoVal | SBayesR.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBayesR.PseudoVal | LDpred1.10FCVal | 0.005 | 1.544 | 0.893 | 3.72e-01 | 0.000 | 0.100 | 0.027 | 9.78e-01 |
| SBayesR.PseudoVal | LDpred1.MultiPRS | -0.001 | -0.308 | -0.090 | 9.28e-01 | -0.006 | -1.940 | -0.553 | 5.80e-01 |
| SBayesR.PseudoVal | LDpred1.Inf | 0.002 | 0.751 | 0.438 | 6.61e-01 | -0.002 | -0.809 | -0.223 | 8.24e-01 |
| SBayesR.PseudoVal | LDpred2.10FCVal | -0.016 | -5.242 | -3.788 | 1.52e-04 | -0.023 | -7.470 | -2.561 | 1.04e-02 |
| SBayesR.PseudoVal | LDpred2.MultiPRS | -0.017 | -5.505 | -1.603 | 1.09e-01 | -0.017 | -5.582 | -1.951 | 5.11e-02 |
| SBayesR.PseudoVal | LDpred2.PseudoVal | 0.006 | 1.865 | 1.089 | 2.76e-01 | 0.000 | -0.145 | -0.040 | 9.68e-01 |
| SBayesR.PseudoVal | LDpred2.Inf | 0.006 | 1.767 | 1.032 | 3.02e-01 | 0.000 | 0.015 | 0.004 | 9.97e-01 |
| SBayesR.PseudoVal | DBSLMM.PseudoVal | -0.008 | -2.451 | -1.721 | 8.53e-02 | -0.010 | -3.407 | -1.140 | 2.54e-01 |
| SBayesR.PseudoVal | All.MultiPRS | -0.023 | -7.273 | -2.140 | 3.24e-02 | -0.023 | -7.651 | -2.849 | 4.39e-03 |
| LDpred1.10FCVal | pT+clump.10FCVal | 0.018 | 5.837 | 3.412 | 6.45e-04 | 0.024 | 7.776 | 2.182 | 2.91e-02 |
| LDpred1.10FCVal | pT+clump.MultiPRS | 0.011 | 3.530 | 1.000 | 3.17e-01 | 0.009 | 2.942 | 0.888 | 3.75e-01 |
| LDpred1.10FCVal | lassosum.10FCVal | -0.019 | -6.171 | -5.036 | 4.76e-07 | -0.022 | -7.402 | -2.901 | 3.72e-03 |
| LDpred1.10FCVal | lassosum.MultiPRS | -0.023 | -7.509 | -2.147 | 3.18e-02 | -0.025 | -8.367 | -3.401 | 6.72e-04 |
| LDpred1.10FCVal | lassosum.PseudoVal | 0.048 | 15.635 | 7.027 | 2.11e-12 | 0.058 | 19.068 | 4.144 | 3.41e-05 |
| LDpred1.10FCVal | PRScs.10FCVal | -0.018 | -5.700 | -5.523 | 3.33e-08 | -0.025 | -8.200 | -3.801 | 1.44e-04 |
| LDpred1.10FCVal | PRScs.MultiPRS | -0.020 | -6.438 | -1.843 | 6.54e-02 | -0.025 | -8.364 | -3.981 | 6.86e-05 |
| LDpred1.10FCVal | PRScs.PseudoVal | -0.013 | -4.333 | -3.715 | 2.03e-04 | -0.017 | -5.589 | -2.255 | 2.41e-02 |
| LDpred1.10FCVal | SBLUP.Inf | 0.005 | 1.496 | 1.650 | 9.89e-02 | -0.001 | -0.378 | -0.198 | 8.43e-01 |
| LDpred1.10FCVal | SBayesR.PseudoVal | -0.005 | -1.569 | -0.893 | 3.72e-01 | 0.000 | -0.100 | -0.027 | 9.78e-01 |
| LDpred1.10FCVal | LDpred1.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.10FCVal | LDpred1.MultiPRS | -0.006 | -1.882 | -0.536 | 5.92e-01 | -0.006 | -2.043 | -1.903 | 5.71e-02 |
| LDpred1.10FCVal | LDpred1.Inf | -0.002 | -0.806 | -1.012 | 3.11e-01 | -0.003 | -0.911 | -0.546 | 5.85e-01 |
| LDpred1.10FCVal | LDpred2.10FCVal | -0.021 | -6.893 | -7.013 | 2.33e-12 | -0.023 | -7.578 | -3.691 | 2.23e-04 |
| LDpred1.10FCVal | LDpred2.MultiPRS | -0.022 | -7.160 | -2.047 | 4.07e-02 | -0.017 | -5.688 | -2.509 | 1.21e-02 |
| LDpred1.10FCVal | LDpred2.PseudoVal | 0.001 | 0.325 | 0.354 | 7.23e-01 | -0.001 | -0.245 | -0.128 | 8.98e-01 |
| LDpred1.10FCVal | LDpred2.Inf | 0.001 | 0.226 | 0.247 | 8.05e-01 | 0.000 | -0.086 | -0.045 | 9.64e-01 |
| LDpred1.10FCVal | DBSLMM.PseudoVal | -0.013 | -4.058 | -3.406 | 6.58e-04 | -0.011 | -3.511 | -1.389 | 1.65e-01 |
| LDpred1.10FCVal | All.MultiPRS | -0.028 | -8.956 | -2.576 | 1.00e-02 | -0.023 | -7.759 | -3.205 | 1.35e-03 |
| LDpred1.MultiPRS | pT+clump.10FCVal | 0.024 | 7.577 | 2.189 | 2.86e-02 | 0.030 | 9.622 | 2.887 | 3.88e-03 |
| LDpred1.MultiPRS | pT+clump.MultiPRS | 0.017 | 5.312 | 1.540 | 1.24e-01 | 0.015 | 4.885 | 1.598 | 1.10e-01 |
| LDpred1.MultiPRS | lassosum.10FCVal | -0.013 | -4.210 | -1.234 | 2.17e-01 | -0.016 | -5.252 | -2.301 | 2.14e-02 |
| LDpred1.MultiPRS | lassosum.MultiPRS | -0.017 | -5.523 | -2.297 | 2.16e-02 | -0.019 | -6.197 | -2.824 | 4.75e-03 |
| LDpred1.MultiPRS | lassosum.PseudoVal | 0.054 | 17.194 | 4.956 | 7.21e-07 | 0.064 | 20.688 | 4.659 | 3.17e-06 |
| LDpred1.MultiPRS | PRScs.10FCVal | -0.012 | -3.748 | -1.098 | 2.72e-01 | -0.019 | -6.034 | -3.286 | 1.02e-03 |
| LDpred1.MultiPRS | PRScs.MultiPRS | -0.014 | -4.472 | -2.714 | 6.64e-03 | -0.019 | -6.195 | -3.528 | 4.19e-04 |
| LDpred1.MultiPRS | PRScs.PseudoVal | -0.008 | -2.406 | -0.704 | 4.81e-01 | -0.011 | -3.475 | -1.593 | 1.11e-01 |
| LDpred1.MultiPRS | SBLUP.Inf | 0.010 | 3.316 | 0.961 | 3.37e-01 | 0.005 | 1.632 | 1.057 | 2.91e-01 |
| LDpred1.MultiPRS | SBayesR.PseudoVal | 0.001 | 0.307 | 0.090 | 9.28e-01 | 0.006 | 1.903 | 0.553 | 5.80e-01 |
| LDpred1.MultiPRS | LDpred1.10FCVal | 0.006 | 1.847 | 0.536 | 5.92e-01 | 0.006 | 2.002 | 1.903 | 5.71e-02 |
| LDpred1.MultiPRS | LDpred1.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.MultiPRS | LDpred1.Inf | 0.003 | 1.056 | 0.307 | 7.59e-01 | 0.003 | 1.110 | 1.125 | 2.61e-01 |
| LDpred1.MultiPRS | LDpred2.10FCVal | -0.015 | -4.918 | -1.441 | 1.50e-01 | -0.017 | -5.424 | -3.206 | 1.35e-03 |
| LDpred1.MultiPRS | LDpred2.MultiPRS | -0.016 | -5.180 | -1.833 | 6.68e-02 | -0.011 | -3.572 | -1.825 | 6.80e-02 |
| LDpred1.MultiPRS | LDpred2.PseudoVal | 0.007 | 2.167 | 0.629 | 5.29e-01 | 0.005 | 1.762 | 1.194 | 2.32e-01 |
| LDpred1.MultiPRS | LDpred2.Inf | 0.007 | 2.069 | 0.601 | 5.48e-01 | 0.006 | 1.918 | 1.308 | 1.91e-01 |
| LDpred1.MultiPRS | DBSLMM.PseudoVal | -0.007 | -2.136 | -0.625 | 5.32e-01 | -0.004 | -1.439 | -0.647 | 5.18e-01 |
| LDpred1.MultiPRS | All.MultiPRS | -0.022 | -6.943 | -2.461 | 1.38e-02 | -0.017 | -5.602 | -2.653 | 7.98e-03 |
| LDpred1.Inf | pT+clump.10FCVal | 0.021 | 6.590 | 4.094 | 4.24e-05 | 0.026 | 8.608 | 2.562 | 1.04e-02 |
| LDpred1.Inf | pT+clump.MultiPRS | 0.013 | 4.301 | 1.231 | 2.18e-01 | 0.012 | 3.817 | 1.221 | 2.22e-01 |
| LDpred1.Inf | lassosum.10FCVal | -0.017 | -5.322 | -4.512 | 6.43e-06 | -0.020 | -6.433 | -2.618 | 8.84e-03 |
| LDpred1.Inf | lassosum.MultiPRS | -0.021 | -6.649 | -1.917 | 5.53e-02 | -0.023 | -7.389 | -3.115 | 1.84e-03 |
| LDpred1.Inf | lassosum.PseudoVal | 0.051 | 16.310 | 7.198 | 6.12e-13 | 0.060 | 19.798 | 4.217 | 2.47e-05 |
| LDpred1.Inf | PRScs.10FCVal | -0.015 | -4.855 | -5.044 | 4.57e-07 | -0.022 | -7.224 | -3.661 | 2.51e-04 |
| LDpred1.Inf | PRScs.MultiPRS | -0.017 | -5.587 | -1.615 | 1.06e-01 | -0.023 | -7.387 | -3.888 | 1.01e-04 |
| LDpred1.Inf | PRScs.PseudoVal | -0.011 | -3.499 | -3.175 | 1.50e-03 | -0.014 | -4.636 | -1.976 | 4.81e-02 |
| LDpred1.Inf | SBLUP.Inf | 0.007 | 2.284 | 3.294 | 9.87e-04 | 0.002 | 0.528 | 0.357 | 7.21e-01 |
| LDpred1.Inf | SBayesR.PseudoVal | -0.002 | -0.756 | -0.438 | 6.61e-01 | 0.002 | 0.803 | 0.223 | 8.24e-01 |
| LDpred1.Inf | LDpred1.10FCVal | 0.002 | 0.800 | 1.012 | 3.11e-01 | 0.003 | 0.902 | 0.546 | 5.85e-01 |
| LDpred1.Inf | LDpred1.MultiPRS | -0.003 | -1.067 | -0.307 | 7.59e-01 | -0.003 | -1.122 | -1.125 | 2.61e-01 |
| LDpred1.Inf | LDpred1.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.Inf | LDpred2.10FCVal | -0.019 | -6.038 | -6.879 | 6.02e-12 | -0.020 | -6.607 | -3.603 | 3.14e-04 |
| LDpred1.Inf | LDpred2.MultiPRS | -0.020 | -6.303 | -1.821 | 6.86e-02 | -0.014 | -4.734 | -2.254 | 2.42e-02 |
| LDpred1.Inf | LDpred2.PseudoVal | 0.003 | 1.123 | 1.779 | 7.53e-02 | 0.002 | 0.659 | 0.505 | 6.13e-01 |
| LDpred1.Inf | LDpred2.Inf | 0.003 | 1.024 | 1.642 | 1.01e-01 | 0.002 | 0.817 | 0.634 | 5.26e-01 |
| LDpred1.Inf | DBSLMM.PseudoVal | -0.010 | -3.226 | -2.950 | 3.18e-03 | -0.008 | -2.577 | -1.113 | 2.66e-01 |
| LDpred1.Inf | All.MultiPRS | -0.025 | -8.084 | -2.348 | 1.89e-02 | -0.021 | -6.787 | -2.908 | 3.64e-03 |
| LDpred2.10FCVal | pT+clump.10FCVal | 0.039 | 11.909 | 7.938 | 2.05e-15 | 0.046 | 14.273 | 4.654 | 3.25e-06 |
| LDpred2.10FCVal | pT+clump.MultiPRS | 0.032 | 9.751 | 2.976 | 2.92e-03 | 0.032 | 9.779 | 3.594 | 3.26e-04 |
| LDpred2.10FCVal | lassosum.10FCVal | 0.002 | 0.675 | 0.819 | 4.13e-01 | 0.001 | 0.164 | 0.097 | 9.23e-01 |
| LDpred2.10FCVal | lassosum.MultiPRS | -0.002 | -0.576 | -0.177 | 8.59e-01 | -0.002 | -0.733 | -0.445 | 6.56e-01 |
| LDpred2.10FCVal | lassosum.PseudoVal | 0.070 | 21.075 | 11.687 | 1.48e-31 | 0.081 | 24.769 | 6.717 | 1.85e-11 |
| LDpred2.10FCVal | PRScs.10FCVal | 0.004 | 1.116 | 1.428 | 1.53e-01 | -0.002 | -0.578 | -0.358 | 7.21e-01 |
| LDpred2.10FCVal | PRScs.MultiPRS | 0.001 | 0.425 | 0.131 | 8.95e-01 | -0.002 | -0.731 | -0.515 | 6.06e-01 |
| LDpred2.10FCVal | PRScs.PseudoVal | 0.008 | 2.395 | 3.054 | 2.26e-03 | 0.006 | 1.849 | 1.151 | 2.50e-01 |
| LDpred2.10FCVal | SBLUP.Inf | 0.026 | 7.848 | 8.507 | 1.79e-17 | 0.022 | 6.693 | 3.547 | 3.90e-04 |
| LDpred2.10FCVal | SBayesR.PseudoVal | 0.016 | 4.981 | 3.788 | 1.52e-04 | 0.023 | 6.951 | 2.561 | 1.04e-02 |
| LDpred2.10FCVal | LDpred1.10FCVal | 0.021 | 6.448 | 7.013 | 2.33e-12 | 0.023 | 7.044 | 3.691 | 2.23e-04 |
| LDpred2.10FCVal | LDpred1.MultiPRS | 0.015 | 4.688 | 1.441 | 1.50e-01 | 0.017 | 5.145 | 3.206 | 1.35e-03 |
| LDpred2.10FCVal | LDpred1.Inf | 0.019 | 5.694 | 6.879 | 6.02e-12 | 0.020 | 6.198 | 3.603 | 3.14e-04 |
| LDpred2.10FCVal | LDpred2.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.10FCVal | LDpred2.MultiPRS | -0.001 | -0.250 | -0.077 | 9.39e-01 | 0.006 | 1.757 | 2.344 | 1.91e-02 |
| LDpred2.10FCVal | LDpred2.PseudoVal | 0.022 | 6.753 | 10.504 | 8.27e-26 | 0.022 | 6.816 | 5.125 | 2.97e-07 |
| LDpred2.10FCVal | LDpred2.Inf | 0.022 | 6.660 | 10.381 | 3.04e-25 | 0.023 | 6.965 | 5.248 | 1.53e-07 |
| LDpred2.10FCVal | DBSLMM.PseudoVal | 0.009 | 2.652 | 2.776 | 5.51e-03 | 0.012 | 3.781 | 1.916 | 5.53e-02 |
| LDpred2.10FCVal | All.MultiPRS | -0.006 | -1.930 | -0.599 | 5.49e-01 | -0.001 | -0.168 | -0.108 | 9.14e-01 |
| LDpred2.MultiPRS | pT+clump.10FCVal | 0.040 | 12.129 | 3.700 | 2.15e-04 | 0.041 | 12.739 | 4.107 | 4.01e-05 |
| LDpred2.MultiPRS | pT+clump.MultiPRS | 0.033 | 9.975 | 3.067 | 2.17e-03 | 0.026 | 8.165 | 2.926 | 3.43e-03 |
| LDpred2.MultiPRS | lassosum.10FCVal | 0.003 | 0.922 | 0.285 | 7.76e-01 | -0.005 | -1.622 | -0.915 | 3.60e-01 |
| LDpred2.MultiPRS | lassosum.MultiPRS | -0.001 | -0.326 | -0.155 | 8.77e-01 | -0.008 | -2.535 | -1.459 | 1.45e-01 |
| LDpred2.MultiPRS | lassosum.PseudoVal | 0.070 | 21.272 | 6.446 | 1.15e-10 | 0.075 | 23.423 | 6.375 | 1.83e-10 |
| LDpred2.MultiPRS | PRScs.10FCVal | 0.005 | 1.362 | 0.421 | 6.74e-01 | -0.008 | -2.377 | -1.430 | 1.53e-01 |
| LDpred2.MultiPRS | PRScs.MultiPRS | 0.002 | 0.673 | 0.251 | 8.02e-01 | -0.008 | -2.533 | -1.675 | 9.38e-02 |
| LDpred2.MultiPRS | PRScs.PseudoVal | 0.009 | 2.638 | 0.813 | 4.16e-01 | 0.000 | 0.094 | 0.055 | 9.56e-01 |
| LDpred2.MultiPRS | SBLUP.Inf | 0.027 | 8.077 | 2.474 | 1.34e-02 | 0.016 | 5.024 | 2.324 | 2.01e-02 |
| LDpred2.MultiPRS | SBayesR.PseudoVal | 0.017 | 5.217 | 1.603 | 1.09e-01 | 0.017 | 5.287 | 1.951 | 5.11e-02 |
| LDpred2.MultiPRS | LDpred1.10FCVal | 0.022 | 6.681 | 2.047 | 4.07e-02 | 0.017 | 5.382 | 2.509 | 1.21e-02 |
| LDpred2.MultiPRS | LDpred1.MultiPRS | 0.016 | 4.925 | 1.833 | 6.68e-02 | 0.011 | 3.449 | 1.825 | 6.80e-02 |
| LDpred2.MultiPRS | LDpred1.Inf | 0.020 | 5.929 | 1.821 | 6.86e-02 | 0.014 | 4.520 | 2.254 | 2.42e-02 |
| LDpred2.MultiPRS | LDpred2.10FCVal | 0.001 | 0.249 | 0.077 | 9.39e-01 | -0.006 | -1.789 | -2.344 | 1.91e-02 |
| LDpred2.MultiPRS | LDpred2.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.MultiPRS | LDpred2.PseudoVal | 0.023 | 6.985 | 2.142 | 3.22e-02 | 0.016 | 5.150 | 3.078 | 2.09e-03 |
| LDpred2.MultiPRS | LDpred2.Inf | 0.023 | 6.892 | 2.113 | 3.46e-02 | 0.017 | 5.301 | 3.188 | 1.43e-03 |
| LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.010 | 2.894 | 0.893 | 3.72e-01 | 0.007 | 2.060 | 0.987 | 3.24e-01 |
| LDpred2.MultiPRS | All.MultiPRS | -0.006 | -1.676 | -0.630 | 5.29e-01 | -0.006 | -1.960 | -1.308 | 1.91e-01 |
| LDpred2.PseudoVal | pT+clump.10FCVal | 0.017 | 5.530 | 3.241 | 1.19e-03 | 0.024 | 8.002 | 2.268 | 2.33e-02 |
| LDpred2.PseudoVal | pT+clump.MultiPRS | 0.010 | 3.215 | 0.909 | 3.63e-01 | 0.010 | 3.179 | 0.967 | 3.34e-01 |
| LDpred2.PseudoVal | lassosum.10FCVal | -0.020 | -6.518 | -5.293 | 1.20e-07 | -0.022 | -7.139 | -2.824 | 4.74e-03 |
| LDpred2.PseudoVal | lassosum.MultiPRS | -0.024 | -7.860 | -2.237 | 2.53e-02 | -0.025 | -8.102 | -3.315 | 9.16e-04 |
| LDpred2.PseudoVal | lassosum.PseudoVal | 0.047 | 15.360 | 6.658 | 2.78e-11 | 0.058 | 19.266 | 4.069 | 4.73e-05 |
| LDpred2.PseudoVal | PRScs.10FCVal | -0.019 | -6.045 | -5.895 | 3.75e-09 | -0.024 | -7.935 | -3.762 | 1.69e-04 |
| LDpred2.PseudoVal | PRScs.MultiPRS | -0.021 | -6.785 | -1.937 | 5.27e-02 | -0.025 | -8.099 | -3.966 | 7.30e-05 |
| LDpred2.PseudoVal | PRScs.PseudoVal | -0.014 | -4.674 | -4.064 | 4.82e-05 | -0.016 | -5.330 | -2.209 | 2.72e-02 |
| LDpred2.PseudoVal | SBLUP.Inf | 0.004 | 1.175 | 1.567 | 1.17e-01 | 0.000 | -0.132 | -0.084 | 9.33e-01 |
| LDpred2.PseudoVal | SBayesR.PseudoVal | -0.006 | -1.900 | -1.089 | 2.76e-01 | 0.000 | 0.145 | 0.040 | 9.68e-01 |
| LDpred2.PseudoVal | LDpred1.10FCVal | -0.001 | -0.326 | -0.354 | 7.23e-01 | 0.001 | 0.245 | 0.128 | 8.98e-01 |
| LDpred2.PseudoVal | LDpred1.MultiPRS | -0.007 | -2.215 | -0.629 | 5.29e-01 | -0.005 | -1.793 | -1.194 | 2.32e-01 |
| LDpred2.PseudoVal | LDpred1.Inf | -0.003 | -1.135 | -1.779 | 7.53e-02 | -0.002 | -0.664 | -0.505 | 6.13e-01 |
| LDpred2.PseudoVal | LDpred2.10FCVal | -0.022 | -7.242 | -10.504 | 8.27e-26 | -0.022 | -7.315 | -5.125 | 2.97e-07 |
| LDpred2.PseudoVal | LDpred2.MultiPRS | -0.023 | -7.509 | -2.142 | 3.22e-02 | -0.016 | -5.429 | -3.078 | 2.09e-03 |
| LDpred2.PseudoVal | LDpred2.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.PseudoVal | LDpred2.Inf | 0.000 | -0.100 | -0.811 | 4.17e-01 | 0.000 | 0.159 | 0.637 | 5.24e-01 |
| LDpred2.PseudoVal | DBSLMM.PseudoVal | -0.014 | -4.398 | -3.830 | 1.28e-04 | -0.010 | -3.257 | -1.358 | 1.74e-01 |
| LDpred2.PseudoVal | All.MultiPRS | -0.029 | -9.311 | -2.668 | 7.63e-03 | -0.023 | -7.495 | -3.236 | 1.21e-03 |
| LDpred2.Inf | pT+clump.10FCVal | 0.017 | 5.624 | 3.342 | 8.31e-04 | 0.024 | 7.855 | 2.251 | 2.44e-02 |
| LDpred2.Inf | pT+clump.MultiPRS | 0.010 | 3.311 | 0.937 | 3.49e-01 | 0.009 | 3.025 | 0.929 | 3.53e-01 |
| LDpred2.Inf | lassosum.10FCVal | -0.020 | -6.412 | -5.260 | 1.44e-07 | -0.022 | -7.310 | -2.910 | 3.61e-03 |
| LDpred2.Inf | lassosum.MultiPRS | -0.024 | -7.752 | -2.209 | 2.72e-02 | -0.025 | -8.274 | -3.409 | 6.53e-04 |
| LDpred2.Inf | lassosum.PseudoVal | 0.048 | 15.444 | 6.693 | 2.19e-11 | 0.058 | 19.137 | 4.032 | 5.54e-05 |
| LDpred2.Inf | PRScs.10FCVal | -0.018 | -5.940 | -5.857 | 4.71e-09 | -0.025 | -8.108 | -3.881 | 1.04e-04 |
| LDpred2.Inf | PRScs.MultiPRS | -0.021 | -6.679 | -1.908 | 5.64e-02 | -0.025 | -8.272 | -4.121 | 3.78e-05 |
| LDpred2.Inf | PRScs.PseudoVal | -0.014 | -4.570 | -3.996 | 6.44e-05 | -0.017 | -5.498 | -2.286 | 2.23e-02 |
| LDpred2.Inf | SBLUP.Inf | 0.004 | 1.273 | 1.747 | 8.06e-02 | -0.001 | -0.292 | -0.189 | 8.50e-01 |
| LDpred2.Inf | SBayesR.PseudoVal | -0.006 | -1.799 | -1.032 | 3.02e-01 | 0.000 | -0.015 | -0.004 | 9.97e-01 |
| LDpred2.Inf | LDpred1.10FCVal | -0.001 | -0.227 | -0.247 | 8.05e-01 | 0.000 | 0.086 | 0.045 | 9.64e-01 |
| LDpred2.Inf | LDpred1.MultiPRS | -0.007 | -2.113 | -0.601 | 5.48e-01 | -0.006 | -1.955 | -1.308 | 1.91e-01 |
| LDpred2.Inf | LDpred1.Inf | -0.003 | -1.035 | -1.642 | 1.01e-01 | -0.002 | -0.824 | -0.634 | 5.26e-01 |
| LDpred2.Inf | LDpred2.10FCVal | -0.022 | -7.135 | -10.381 | 3.04e-25 | -0.023 | -7.486 | -5.248 | 1.53e-07 |
| LDpred2.Inf | LDpred2.MultiPRS | -0.023 | -7.402 | -2.113 | 3.46e-02 | -0.017 | -5.597 | -3.188 | 1.43e-03 |
| LDpred2.Inf | LDpred2.PseudoVal | 0.000 | 0.099 | 0.811 | 4.17e-01 | 0.000 | -0.159 | -0.637 | 5.24e-01 |
| LDpred2.Inf | LDpred2.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.Inf | DBSLMM.PseudoVal | -0.013 | -4.294 | -3.730 | 1.92e-04 | -0.010 | -3.422 | -1.424 | 1.55e-01 |
| LDpred2.Inf | All.MultiPRS | -0.028 | -9.203 | -2.640 | 8.30e-03 | -0.023 | -7.667 | -3.328 | 8.76e-04 |
| DBSLMM.PseudoVal | pT+clump.10FCVal | 0.031 | 9.510 | 6.001 | 1.96e-09 | 0.034 | 10.904 | 3.349 | 8.12e-04 |
| DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.023 | 7.292 | 2.160 | 3.08e-02 | 0.020 | 6.234 | 2.322 | 2.02e-02 |
| DBSLMM.PseudoVal | lassosum.10FCVal | -0.007 | -2.031 | -1.867 | 6.19e-02 | -0.012 | -3.759 | -1.665 | 9.59e-02 |
| DBSLMM.PseudoVal | lassosum.MultiPRS | -0.011 | -3.316 | -0.994 | 3.20e-01 | -0.015 | -4.691 | -2.392 | 1.68e-02 |
| DBSLMM.PseudoVal | lassosum.PseudoVal | 0.061 | 18.925 | 10.096 | 5.76e-24 | 0.068 | 21.813 | 5.597 | 2.17e-08 |
| DBSLMM.PseudoVal | PRScs.10FCVal | -0.005 | -1.578 | -1.698 | 8.95e-02 | -0.014 | -4.530 | -2.360 | 1.83e-02 |
| DBSLMM.PseudoVal | PRScs.MultiPRS | -0.007 | -2.287 | -0.686 | 4.93e-01 | -0.015 | -4.689 | -2.682 | 7.33e-03 |
| DBSLMM.PseudoVal | PRScs.PseudoVal | -0.001 | -0.264 | -0.280 | 7.79e-01 | -0.006 | -2.008 | -1.018 | 3.09e-01 |
| DBSLMM.PseudoVal | SBLUP.Inf | 0.017 | 5.338 | 4.706 | 2.52e-06 | 0.009 | 3.027 | 1.261 | 2.07e-01 |
| DBSLMM.PseudoVal | SBayesR.PseudoVal | 0.008 | 2.392 | 1.721 | 8.53e-02 | 0.010 | 3.295 | 1.140 | 2.54e-01 |
| DBSLMM.PseudoVal | LDpred1.10FCVal | 0.013 | 3.900 | 3.406 | 6.58e-04 | 0.011 | 3.392 | 1.389 | 1.65e-01 |
| DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.007 | 2.091 | 0.625 | 5.32e-01 | 0.004 | 1.418 | 0.647 | 5.18e-01 |
| DBSLMM.PseudoVal | LDpred1.Inf | 0.010 | 3.125 | 2.950 | 3.18e-03 | 0.008 | 2.512 | 1.113 | 2.66e-01 |
| DBSLMM.PseudoVal | LDpred2.10FCVal | -0.009 | -2.724 | -2.776 | 5.51e-03 | -0.012 | -3.929 | -1.916 | 5.53e-02 |
| DBSLMM.PseudoVal | LDpred2.MultiPRS | -0.010 | -2.981 | -0.893 | 3.72e-01 | -0.007 | -2.103 | -0.987 | 3.24e-01 |
| DBSLMM.PseudoVal | LDpred2.PseudoVal | 0.014 | 4.213 | 3.830 | 1.28e-04 | 0.010 | 3.155 | 1.358 | 1.74e-01 |
| DBSLMM.PseudoVal | LDpred2.Inf | 0.013 | 4.117 | 3.730 | 1.92e-04 | 0.010 | 3.309 | 1.424 | 1.55e-01 |
| DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| DBSLMM.PseudoVal | All.MultiPRS | -0.015 | -4.707 | -1.418 | 1.56e-01 | -0.013 | -4.104 | -2.065 | 3.89e-02 |
| All.MultiPRS | pT+clump.10FCVal | 0.046 | 13.577 | 4.227 | 2.36e-05 | 0.047 | 14.417 | 4.893 | 9.92e-07 |
| All.MultiPRS | pT+clump.MultiPRS | 0.039 | 11.459 | 3.610 | 3.07e-04 | 0.032 | 9.930 | 3.817 | 1.35e-04 |
| All.MultiPRS | lassosum.10FCVal | 0.009 | 2.556 | 0.809 | 4.19e-01 | 0.001 | 0.332 | 0.194 | 8.46e-01 |
| All.MultiPRS | lassosum.MultiPRS | 0.004 | 1.328 | 0.421 | 6.73e-01 | -0.002 | -0.564 | -0.425 | 6.71e-01 |
| All.MultiPRS | lassosum.PseudoVal | 0.076 | 22.570 | 7.016 | 2.28e-12 | 0.081 | 24.895 | 7.041 | 1.91e-12 |
| All.MultiPRS | PRScs.10FCVal | 0.010 | 2.988 | 0.944 | 3.45e-01 | -0.001 | -0.409 | -0.237 | 8.13e-01 |
| All.MultiPRS | PRScs.MultiPRS | 0.008 | 2.311 | 1.066 | 2.86e-01 | -0.002 | -0.562 | -0.364 | 7.16e-01 |
| All.MultiPRS | PRScs.PseudoVal | 0.014 | 4.243 | 1.341 | 1.80e-01 | 0.007 | 2.014 | 1.121 | 2.62e-01 |
| All.MultiPRS | SBLUP.Inf | 0.032 | 9.593 | 3.001 | 2.69e-03 | 0.022 | 6.850 | 2.940 | 3.29e-03 |
| All.MultiPRS | SBayesR.PseudoVal | 0.023 | 6.780 | 2.140 | 3.24e-02 | 0.023 | 7.107 | 2.849 | 4.39e-03 |
| All.MultiPRS | LDpred1.10FCVal | 0.028 | 8.220 | 2.576 | 1.00e-02 | 0.023 | 7.200 | 3.205 | 1.35e-03 |
| All.MultiPRS | LDpred1.MultiPRS | 0.022 | 6.492 | 2.461 | 1.38e-02 | 0.017 | 5.305 | 2.653 | 7.98e-03 |
| All.MultiPRS | LDpred1.Inf | 0.025 | 7.480 | 2.348 | 1.89e-02 | 0.021 | 6.355 | 2.908 | 3.64e-03 |
| All.MultiPRS | LDpred2.10FCVal | 0.006 | 1.893 | 0.599 | 5.49e-01 | 0.001 | 0.168 | 0.108 | 9.14e-01 |
| All.MultiPRS | LDpred2.MultiPRS | 0.006 | 1.649 | 0.630 | 5.29e-01 | 0.006 | 1.922 | 1.308 | 1.91e-01 |
| All.MultiPRS | LDpred2.PseudoVal | 0.029 | 8.518 | 2.668 | 7.63e-03 | 0.023 | 6.973 | 3.236 | 1.21e-03 |
| All.MultiPRS | LDpred2.Inf | 0.028 | 8.427 | 2.640 | 8.30e-03 | 0.023 | 7.121 | 3.328 | 8.76e-04 |
| All.MultiPRS | DBSLMM.PseudoVal | 0.015 | 4.495 | 1.418 | 1.56e-01 | 0.013 | 3.942 | 2.065 | 3.89e-02 |
| All.MultiPRS | All.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
Show table: TEDS (UKBB ref)
| Reference | Test | CrossVal Diff | CrossVal Diff (%) | CrossVal Diff Z | CrossVal Diff P | IndepVal Diff | IndepVal Diff (%) | IndepVal Diff Z | IndepVal Diff P |
|---|---|---|---|---|---|---|---|---|---|
| pT+clump.10FCVal | pT+clump.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| pT+clump.10FCVal | pT+clump.MultiPRS | -0.008 | -2.666 | -0.710 | 4.78e-01 | -0.016 | -5.743 | -3.548 | 3.88e-04 |
| pT+clump.10FCVal | lassosum.10FCVal | -0.038 | -13.102 | -8.644 | 5.41e-18 | -0.055 | -20.007 | -6.195 | 5.81e-10 |
| pT+clump.10FCVal | lassosum.MultiPRS | -0.042 | -14.280 | -3.869 | 1.09e-04 | -0.058 | -21.268 | -6.214 | 5.15e-10 |
| pT+clump.10FCVal | lassosum.PseudoVal | 0.033 | 11.401 | 4.629 | 3.67e-06 | 0.034 | 12.416 | 2.330 | 1.98e-02 |
| pT+clump.10FCVal | SBLUP.Inf | -0.021 | -7.308 | -4.336 | 1.45e-05 | -0.035 | -12.645 | -3.484 | 4.94e-04 |
| pT+clump.10FCVal | SBayesR.PseudoVal | -0.043 | -14.660 | -8.970 | 2.97e-19 | -0.062 | -22.463 | -6.435 | 1.23e-10 |
| pT+clump.10FCVal | LDpred1.10FCVal | -0.013 | -4.489 | -2.502 | 1.23e-02 | -0.031 | -11.204 | -2.906 | 3.67e-03 |
| pT+clump.10FCVal | LDpred1.MultiPRS | -0.024 | -8.213 | -2.196 | 2.81e-02 | -0.039 | -14.294 | -3.884 | 1.03e-04 |
| pT+clump.10FCVal | LDpred1.Inf | -0.020 | -6.826 | -4.004 | 6.23e-05 | -0.034 | -12.583 | -3.422 | 6.23e-04 |
| pT+clump.10FCVal | LDpred2.10FCVal | -0.049 | -16.839 | -10.291 | 7.78e-25 | -0.059 | -21.652 | -6.229 | 4.70e-10 |
| pT+clump.10FCVal | LDpred2.MultiPRS | -0.048 | -16.618 | -4.506 | 6.59e-06 | -0.063 | -22.811 | -6.589 | 4.43e-11 |
| pT+clump.10FCVal | LDpred2.PseudoVal | -0.045 | -15.379 | -9.205 | 3.41e-20 | -0.061 | -22.391 | -6.294 | 3.09e-10 |
| pT+clump.10FCVal | LDpred2.Inf | -0.024 | -8.356 | -4.851 | 1.23e-06 | -0.038 | -14.036 | -3.798 | 1.46e-04 |
| pT+clump.10FCVal | DBSLMM.PseudoVal | -0.030 | -10.380 | -5.998 | 2.00e-09 | -0.043 | -15.757 | -4.281 | 1.86e-05 |
| pT+clump.10FCVal | All.MultiPRS | -0.051 | -17.602 | -4.779 | 1.76e-06 | -0.060 | -21.898 | -6.290 | 3.17e-10 |
| pT+clump.MultiPRS | pT+clump.10FCVal | 0.008 | 2.597 | 0.710 | 4.78e-01 | 0.016 | 5.431 | 3.548 | 3.88e-04 |
| pT+clump.MultiPRS | pT+clump.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| pT+clump.MultiPRS | lassosum.10FCVal | -0.030 | -10.165 | -2.815 | 4.88e-03 | -0.039 | -13.488 | -4.844 | 1.27e-06 |
| pT+clump.MultiPRS | lassosum.MultiPRS | -0.034 | -11.312 | -3.135 | 1.72e-03 | -0.043 | -14.681 | -5.276 | 1.32e-07 |
| pT+clump.MultiPRS | lassosum.PseudoVal | 0.041 | 13.702 | 3.720 | 1.99e-04 | 0.050 | 17.173 | 3.911 | 9.18e-05 |
| pT+clump.MultiPRS | SBLUP.Inf | -0.014 | -4.521 | -1.245 | 2.13e-01 | -0.019 | -6.527 | -2.092 | 3.65e-02 |
| pT+clump.MultiPRS | SBayesR.PseudoVal | -0.035 | -11.683 | -3.241 | 1.19e-03 | -0.046 | -15.811 | -5.683 | 1.32e-08 |
| pT+clump.MultiPRS | LDpred1.10FCVal | -0.005 | -1.775 | -0.488 | 6.26e-01 | -0.015 | -5.164 | -1.540 | 1.24e-01 |
| pT+clump.MultiPRS | LDpred1.MultiPRS | -0.016 | -5.403 | -1.491 | 1.36e-01 | -0.023 | -8.086 | -2.576 | 1.00e-02 |
| pT+clump.MultiPRS | LDpred1.Inf | -0.012 | -4.052 | -1.116 | 2.65e-01 | -0.019 | -6.468 | -2.042 | 4.11e-02 |
| pT+clump.MultiPRS | LDpred2.10FCVal | -0.041 | -13.805 | -3.835 | 1.26e-04 | -0.044 | -15.044 | -5.280 | 1.29e-07 |
| pT+clump.MultiPRS | LDpred2.MultiPRS | -0.041 | -13.590 | -3.796 | 1.47e-04 | -0.047 | -16.140 | -5.681 | 1.34e-08 |
| pT+clump.MultiPRS | LDpred2.PseudoVal | -0.037 | -12.383 | -3.437 | 5.88e-04 | -0.046 | -15.743 | -5.312 | 1.08e-07 |
| pT+clump.MultiPRS | LDpred2.Inf | -0.017 | -5.542 | -1.528 | 1.26e-01 | -0.023 | -7.843 | -2.446 | 1.44e-02 |
| pT+clump.MultiPRS | DBSLMM.PseudoVal | -0.023 | -7.513 | -2.075 | 3.80e-02 | -0.027 | -9.470 | -3.315 | 9.16e-04 |
| pT+clump.MultiPRS | All.MultiPRS | -0.044 | -14.548 | -4.064 | 4.83e-05 | -0.044 | -15.277 | -5.340 | 9.28e-08 |
| lassosum.10FCVal | pT+clump.10FCVal | 0.038 | 11.585 | 8.644 | 5.41e-18 | 0.055 | 16.671 | 6.195 | 5.81e-10 |
| lassosum.10FCVal | pT+clump.MultiPRS | 0.030 | 9.227 | 2.815 | 4.88e-03 | 0.039 | 11.885 | 4.844 | 1.27e-06 |
| lassosum.10FCVal | lassosum.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.10FCVal | lassosum.MultiPRS | -0.003 | -1.041 | -0.322 | 7.48e-01 | -0.003 | -1.051 | -1.171 | 2.42e-01 |
| lassosum.10FCVal | lassosum.PseudoVal | 0.071 | 21.665 | 12.167 | 4.68e-34 | 0.089 | 27.017 | 7.384 | 1.54e-13 |
| lassosum.10FCVal | SBLUP.Inf | 0.017 | 5.123 | 4.902 | 9.51e-07 | 0.020 | 6.134 | 2.918 | 3.52e-03 |
| lassosum.10FCVal | SBayesR.PseudoVal | -0.005 | -1.377 | -1.641 | 1.01e-01 | -0.007 | -2.047 | -1.199 | 2.31e-01 |
| lassosum.10FCVal | LDpred1.10FCVal | 0.025 | 7.616 | 6.616 | 3.68e-11 | 0.024 | 7.335 | 3.149 | 1.64e-03 |
| lassosum.10FCVal | LDpred1.MultiPRS | 0.014 | 4.323 | 1.326 | 1.85e-01 | 0.016 | 4.761 | 2.275 | 2.29e-02 |
| lassosum.10FCVal | LDpred1.Inf | 0.018 | 5.549 | 5.136 | 2.81e-07 | 0.020 | 6.186 | 2.840 | 4.50e-03 |
| lassosum.10FCVal | LDpred2.10FCVal | -0.011 | -3.304 | -4.461 | 8.15e-06 | -0.005 | -1.371 | -0.906 | 3.65e-01 |
| lassosum.10FCVal | LDpred2.MultiPRS | -0.010 | -3.108 | -0.966 | 3.34e-01 | -0.008 | -2.337 | -1.525 | 1.27e-01 |
| lassosum.10FCVal | LDpred2.PseudoVal | -0.007 | -2.013 | -2.482 | 1.31e-02 | -0.007 | -1.987 | -1.208 | 2.27e-01 |
| lassosum.10FCVal | LDpred2.Inf | 0.014 | 4.197 | 3.989 | 6.63e-05 | 0.016 | 4.975 | 2.336 | 1.95e-02 |
| lassosum.10FCVal | DBSLMM.PseudoVal | 0.008 | 2.407 | 2.289 | 2.21e-02 | 0.012 | 3.541 | 1.675 | 9.39e-02 |
| lassosum.10FCVal | All.MultiPRS | -0.013 | -3.978 | -1.239 | 2.15e-01 | -0.005 | -1.576 | -0.954 | 3.40e-01 |
| lassosum.MultiPRS | pT+clump.10FCVal | 0.042 | 12.495 | 3.869 | 1.09e-04 | 0.058 | 17.538 | 6.214 | 5.15e-10 |
| lassosum.MultiPRS | pT+clump.MultiPRS | 0.034 | 10.162 | 3.135 | 1.72e-03 | 0.043 | 12.802 | 5.276 | 1.32e-07 |
| lassosum.MultiPRS | lassosum.10FCVal | 0.003 | 1.030 | 0.322 | 7.48e-01 | 0.003 | 1.040 | 1.171 | 2.42e-01 |
| lassosum.MultiPRS | lassosum.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.MultiPRS | lassosum.PseudoVal | 0.075 | 22.472 | 6.889 | 5.62e-12 | 0.092 | 27.776 | 8.332 | 7.97e-17 |
| lassosum.MultiPRS | SBLUP.Inf | 0.020 | 6.101 | 1.897 | 5.78e-02 | 0.024 | 7.110 | 3.408 | 6.55e-04 |
| lassosum.MultiPRS | SBayesR.PseudoVal | -0.001 | -0.333 | -0.104 | 9.17e-01 | -0.003 | -0.986 | -0.642 | 5.21e-01 |
| lassosum.MultiPRS | LDpred1.10FCVal | 0.029 | 8.567 | 2.654 | 7.94e-03 | 0.028 | 8.299 | 3.655 | 2.57e-04 |
| lassosum.MultiPRS | LDpred1.MultiPRS | 0.018 | 5.308 | 1.656 | 9.77e-02 | 0.019 | 5.751 | 2.843 | 4.47e-03 |
| lassosum.MultiPRS | LDpred1.Inf | 0.022 | 6.522 | 2.027 | 4.27e-02 | 0.024 | 7.162 | 3.345 | 8.24e-04 |
| lassosum.MultiPRS | LDpred2.10FCVal | -0.007 | -2.240 | -0.703 | 4.82e-01 | -0.001 | -0.317 | -0.221 | 8.25e-01 |
| lassosum.MultiPRS | LDpred2.MultiPRS | -0.007 | -2.046 | -0.938 | 3.48e-01 | -0.004 | -1.272 | -0.881 | 3.78e-01 |
| lassosum.MultiPRS | LDpred2.PseudoVal | -0.003 | -0.962 | -0.301 | 7.63e-01 | -0.003 | -0.926 | -0.572 | 5.67e-01 |
| lassosum.MultiPRS | LDpred2.Inf | 0.017 | 5.184 | 1.613 | 1.07e-01 | 0.020 | 5.963 | 2.836 | 4.57e-03 |
| lassosum.MultiPRS | DBSLMM.PseudoVal | 0.011 | 3.413 | 1.068 | 2.86e-01 | 0.015 | 4.544 | 2.405 | 1.62e-02 |
| lassosum.MultiPRS | All.MultiPRS | -0.010 | -2.907 | -1.319 | 1.87e-01 | -0.002 | -0.520 | -0.393 | 6.95e-01 |
| lassosum.PseudoVal | pT+clump.10FCVal | -0.033 | -12.868 | -4.629 | 3.67e-06 | -0.034 | -14.176 | -2.330 | 1.98e-02 |
| lassosum.PseudoVal | pT+clump.MultiPRS | -0.041 | -15.877 | -3.720 | 1.99e-04 | -0.050 | -20.733 | -3.911 | 9.18e-05 |
| lassosum.PseudoVal | lassosum.10FCVal | -0.071 | -27.657 | -12.167 | 4.68e-34 | -0.089 | -37.018 | -7.384 | 1.54e-13 |
| lassosum.PseudoVal | lassosum.MultiPRS | -0.075 | -28.985 | -6.889 | 5.62e-12 | -0.092 | -38.458 | -8.332 | 7.97e-17 |
| lassosum.PseudoVal | lassosum.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| lassosum.PseudoVal | SBLUP.Inf | -0.055 | -21.117 | -7.627 | 2.40e-14 | -0.069 | -28.613 | -4.756 | 1.97e-06 |
| lassosum.PseudoVal | SBayesR.PseudoVal | -0.076 | -29.415 | -12.975 | 1.70e-38 | -0.096 | -39.823 | -8.059 | 7.71e-16 |
| lassosum.PseudoVal | LDpred1.10FCVal | -0.046 | -17.935 | -6.431 | 1.27e-10 | -0.065 | -26.968 | -4.450 | 8.60e-06 |
| lassosum.PseudoVal | LDpred1.MultiPRS | -0.057 | -22.138 | -5.223 | 1.76e-07 | -0.073 | -30.496 | -5.271 | 1.36e-07 |
| lassosum.PseudoVal | LDpred1.Inf | -0.053 | -20.573 | -7.474 | 7.76e-14 | -0.069 | -28.542 | -4.764 | 1.90e-06 |
| lassosum.PseudoVal | LDpred2.10FCVal | -0.082 | -31.874 | -13.830 | 1.67e-43 | -0.093 | -38.897 | -7.780 | 7.28e-15 |
| lassosum.PseudoVal | LDpred2.MultiPRS | -0.082 | -31.625 | -7.559 | 4.05e-14 | -0.097 | -40.220 | -8.026 | 1.01e-15 |
| lassosum.PseudoVal | LDpred2.PseudoVal | -0.078 | -30.226 | -11.926 | 8.62e-33 | -0.095 | -39.741 | -7.224 | 5.05e-13 |
| lassosum.PseudoVal | LDpred2.Inf | -0.058 | -22.299 | -8.062 | 7.49e-16 | -0.073 | -30.202 | -5.005 | 5.59e-07 |
| lassosum.PseudoVal | DBSLMM.PseudoVal | -0.064 | -24.584 | -10.594 | 3.19e-26 | -0.077 | -32.166 | -6.351 | 2.14e-10 |
| lassosum.PseudoVal | All.MultiPRS | -0.085 | -32.735 | -7.842 | 4.44e-15 | -0.094 | -39.178 | -8.120 | 4.66e-16 |
| SBLUP.Inf | pT+clump.10FCVal | 0.021 | 6.810 | 4.336 | 1.45e-05 | 0.035 | 11.226 | 3.484 | 4.94e-04 |
| SBLUP.Inf | pT+clump.MultiPRS | 0.014 | 4.326 | 1.245 | 2.13e-01 | 0.019 | 6.127 | 2.092 | 3.65e-02 |
| SBLUP.Inf | lassosum.10FCVal | -0.017 | -5.400 | -4.902 | 9.51e-07 | -0.020 | -6.535 | -2.918 | 3.52e-03 |
| SBLUP.Inf | lassosum.MultiPRS | -0.020 | -6.497 | -1.897 | 5.78e-02 | -0.024 | -7.655 | -3.408 | 6.55e-04 |
| SBLUP.Inf | lassosum.PseudoVal | 0.055 | 17.435 | 7.627 | 2.40e-14 | 0.069 | 22.248 | 4.756 | 1.97e-06 |
| SBLUP.Inf | SBLUP.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBLUP.Inf | SBayesR.PseudoVal | -0.021 | -6.852 | -7.504 | 6.21e-14 | -0.027 | -8.716 | -4.695 | 2.66e-06 |
| SBLUP.Inf | LDpred1.10FCVal | 0.008 | 2.627 | 4.100 | 4.14e-05 | 0.004 | 1.279 | 0.953 | 3.41e-01 |
| SBLUP.Inf | LDpred1.MultiPRS | -0.003 | -0.844 | -0.244 | 8.08e-01 | -0.005 | -1.463 | -1.227 | 2.20e-01 |
| SBLUP.Inf | LDpred1.Inf | 0.001 | 0.449 | 0.988 | 3.23e-01 | 0.000 | 0.055 | 0.058 | 9.54e-01 |
| SBLUP.Inf | LDpred2.10FCVal | -0.028 | -8.882 | -9.487 | 2.37e-21 | -0.025 | -7.996 | -4.184 | 2.86e-05 |
| SBLUP.Inf | LDpred2.MultiPRS | -0.027 | -8.676 | -2.537 | 1.12e-02 | -0.028 | -9.024 | -5.003 | 5.65e-07 |
| SBLUP.Inf | LDpred2.PseudoVal | -0.024 | -7.521 | -10.698 | 1.03e-26 | -0.027 | -8.652 | -6.058 | 1.37e-09 |
| SBLUP.Inf | LDpred2.Inf | -0.003 | -0.976 | -2.797 | 5.17e-03 | -0.004 | -1.235 | -1.728 | 8.40e-02 |
| SBLUP.Inf | DBSLMM.PseudoVal | -0.009 | -2.862 | -2.729 | 6.35e-03 | -0.009 | -2.763 | -1.278 | 2.01e-01 |
| SBLUP.Inf | All.MultiPRS | -0.030 | -9.593 | -2.810 | 4.95e-03 | -0.025 | -8.214 | -3.734 | 1.89e-04 |
| SBayesR.PseudoVal | pT+clump.10FCVal | 0.043 | 12.786 | 8.970 | 2.97e-19 | 0.062 | 18.343 | 6.435 | 1.23e-10 |
| SBayesR.PseudoVal | pT+clump.MultiPRS | 0.035 | 10.461 | 3.241 | 1.19e-03 | 0.046 | 13.653 | 5.683 | 1.32e-08 |
| SBayesR.PseudoVal | lassosum.10FCVal | 0.005 | 1.359 | 1.641 | 1.01e-01 | 0.007 | 2.006 | 1.199 | 2.31e-01 |
| SBayesR.PseudoVal | lassosum.MultiPRS | 0.001 | 0.332 | 0.104 | 9.17e-01 | 0.003 | 0.976 | 0.642 | 5.21e-01 |
| SBayesR.PseudoVal | lassosum.PseudoVal | 0.076 | 22.729 | 12.975 | 1.70e-38 | 0.096 | 28.481 | 8.059 | 7.71e-16 |
| SBayesR.PseudoVal | SBLUP.Inf | 0.021 | 6.412 | 7.504 | 6.21e-14 | 0.027 | 8.017 | 4.695 | 2.66e-06 |
| SBayesR.PseudoVal | SBayesR.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| SBayesR.PseudoVal | LDpred1.10FCVal | 0.030 | 8.871 | 9.484 | 2.45e-21 | 0.031 | 9.194 | 4.888 | 1.02e-06 |
| SBayesR.PseudoVal | LDpred1.MultiPRS | 0.019 | 5.623 | 1.752 | 7.98e-02 | 0.022 | 6.671 | 4.195 | 2.73e-05 |
| SBayesR.PseudoVal | LDpred1.Inf | 0.023 | 6.833 | 8.076 | 6.71e-16 | 0.027 | 8.068 | 4.726 | 2.29e-06 |
| SBayesR.PseudoVal | LDpred2.10FCVal | -0.006 | -1.900 | -3.549 | 3.87e-04 | 0.002 | 0.662 | 0.618 | 5.36e-01 |
| SBayesR.PseudoVal | LDpred2.MultiPRS | -0.006 | -1.707 | -0.539 | 5.90e-01 | -0.001 | -0.284 | -0.273 | 7.85e-01 |
| SBayesR.PseudoVal | LDpred2.PseudoVal | -0.002 | -0.627 | -1.096 | 2.73e-01 | 0.000 | 0.059 | 0.052 | 9.59e-01 |
| SBayesR.PseudoVal | LDpred2.Inf | 0.018 | 5.499 | 6.534 | 6.42e-11 | 0.023 | 6.881 | 4.064 | 4.82e-05 |
| SBayesR.PseudoVal | DBSLMM.PseudoVal | 0.012 | 3.733 | 4.487 | 7.23e-06 | 0.018 | 5.476 | 3.267 | 1.09e-03 |
| SBayesR.PseudoVal | All.MultiPRS | -0.009 | -2.565 | -0.811 | 4.17e-01 | 0.002 | 0.461 | 0.331 | 7.40e-01 |
| LDpred1.10FCVal | pT+clump.10FCVal | 0.013 | 4.296 | 2.502 | 1.23e-02 | 0.031 | 10.075 | 2.906 | 3.67e-03 |
| LDpred1.10FCVal | pT+clump.MultiPRS | 0.005 | 1.744 | 0.488 | 6.26e-01 | 0.015 | 4.911 | 1.540 | 1.24e-01 |
| LDpred1.10FCVal | lassosum.10FCVal | -0.025 | -8.244 | -6.616 | 3.68e-11 | -0.024 | -7.916 | -3.149 | 1.64e-03 |
| LDpred1.10FCVal | lassosum.MultiPRS | -0.029 | -9.370 | -2.654 | 7.94e-03 | -0.028 | -9.050 | -3.655 | 2.57e-04 |
| LDpred1.10FCVal | lassosum.PseudoVal | 0.046 | 15.207 | 6.431 | 1.27e-10 | 0.065 | 21.240 | 4.450 | 8.60e-06 |
| LDpred1.10FCVal | SBLUP.Inf | -0.008 | -2.698 | -4.100 | 4.14e-05 | -0.004 | -1.296 | -0.953 | 3.41e-01 |
| LDpred1.10FCVal | SBayesR.PseudoVal | -0.030 | -9.734 | -9.484 | 2.45e-21 | -0.031 | -10.124 | -4.888 | 1.02e-06 |
| LDpred1.10FCVal | LDpred1.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.10FCVal | LDpred1.MultiPRS | -0.011 | -3.564 | -1.000 | 3.17e-01 | -0.008 | -2.778 | -2.153 | 3.13e-02 |
| LDpred1.10FCVal | LDpred1.Inf | -0.007 | -2.237 | -3.373 | 7.43e-04 | -0.004 | -1.240 | -0.914 | 3.61e-01 |
| LDpred1.10FCVal | LDpred2.10FCVal | -0.036 | -11.820 | -11.011 | 3.38e-28 | -0.029 | -9.395 | -4.340 | 1.42e-05 |
| LDpred1.10FCVal | LDpred2.MultiPRS | -0.035 | -11.608 | -3.297 | 9.76e-04 | -0.032 | -10.437 | -5.075 | 3.88e-07 |
| LDpred1.10FCVal | LDpred2.PseudoVal | -0.032 | -10.422 | -11.647 | 2.36e-31 | -0.031 | -10.060 | -5.608 | 2.05e-08 |
| LDpred1.10FCVal | LDpred2.Inf | -0.011 | -3.701 | -5.495 | 3.92e-08 | -0.008 | -2.547 | -1.859 | 6.31e-02 |
| LDpred1.10FCVal | DBSLMM.PseudoVal | -0.017 | -5.638 | -4.702 | 2.57e-06 | -0.012 | -4.094 | -1.669 | 9.51e-02 |
| LDpred1.10FCVal | All.MultiPRS | -0.038 | -12.549 | -3.571 | 3.56e-04 | -0.029 | -9.616 | -3.957 | 7.57e-05 |
| LDpred1.MultiPRS | pT+clump.10FCVal | 0.024 | 7.590 | 2.196 | 2.81e-02 | 0.039 | 12.506 | 3.884 | 1.03e-04 |
| LDpred1.MultiPRS | pT+clump.MultiPRS | 0.016 | 5.126 | 1.491 | 1.36e-01 | 0.023 | 7.481 | 2.576 | 1.00e-02 |
| LDpred1.MultiPRS | lassosum.10FCVal | -0.014 | -4.518 | -1.326 | 1.85e-01 | -0.016 | -4.999 | -2.275 | 2.29e-02 |
| LDpred1.MultiPRS | lassosum.MultiPRS | -0.018 | -5.606 | -1.656 | 9.77e-02 | -0.019 | -6.102 | -2.843 | 4.47e-03 |
| LDpred1.MultiPRS | lassosum.PseudoVal | 0.057 | 18.126 | 5.223 | 1.76e-07 | 0.073 | 23.369 | 5.271 | 1.36e-07 |
| LDpred1.MultiPRS | SBLUP.Inf | 0.003 | 0.837 | 0.244 | 8.08e-01 | 0.005 | 1.442 | 1.227 | 2.20e-01 |
| LDpred1.MultiPRS | SBayesR.PseudoVal | -0.019 | -5.958 | -1.752 | 7.98e-02 | -0.022 | -7.148 | -4.195 | 2.73e-05 |
| LDpred1.MultiPRS | LDpred1.10FCVal | 0.011 | 3.442 | 1.000 | 3.17e-01 | 0.008 | 2.703 | 2.153 | 3.13e-02 |
| LDpred1.MultiPRS | LDpred1.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.MultiPRS | LDpred1.Inf | 0.004 | 1.282 | 0.373 | 7.09e-01 | 0.005 | 1.497 | 1.491 | 1.36e-01 |
| LDpred1.MultiPRS | LDpred2.10FCVal | -0.025 | -7.971 | -2.349 | 1.88e-02 | -0.020 | -6.438 | -3.573 | 3.53e-04 |
| LDpred1.MultiPRS | LDpred2.MultiPRS | -0.025 | -7.767 | -2.765 | 5.69e-03 | -0.023 | -7.452 | -4.366 | 1.26e-05 |
| LDpred1.MultiPRS | LDpred2.PseudoVal | -0.021 | -6.622 | -1.947 | 5.15e-02 | -0.022 | -7.085 | -4.814 | 1.48e-06 |
| LDpred1.MultiPRS | LDpred2.Inf | 0.000 | -0.131 | -0.038 | 9.69e-01 | 0.001 | 0.225 | 0.194 | 8.46e-01 |
| LDpred1.MultiPRS | DBSLMM.PseudoVal | -0.006 | -2.002 | -0.585 | 5.58e-01 | -0.004 | -1.280 | -0.595 | 5.52e-01 |
| LDpred1.MultiPRS | All.MultiPRS | -0.027 | -8.676 | -3.060 | 2.21e-03 | -0.021 | -6.653 | -3.305 | 9.49e-04 |
| LDpred1.Inf | pT+clump.10FCVal | 0.020 | 6.390 | 4.004 | 6.23e-05 | 0.034 | 11.177 | 3.422 | 6.23e-04 |
| LDpred1.Inf | pT+clump.MultiPRS | 0.012 | 3.894 | 1.116 | 2.65e-01 | 0.019 | 6.075 | 2.042 | 4.11e-02 |
| LDpred1.Inf | lassosum.10FCVal | -0.018 | -5.875 | -5.136 | 2.81e-07 | -0.020 | -6.594 | -2.840 | 4.50e-03 |
| LDpred1.Inf | lassosum.MultiPRS | -0.022 | -6.977 | -2.027 | 4.27e-02 | -0.024 | -7.714 | -3.345 | 8.24e-04 |
| LDpred1.Inf | lassosum.PseudoVal | 0.053 | 17.063 | 7.474 | 7.76e-14 | 0.069 | 22.205 | 4.764 | 1.90e-06 |
| LDpred1.Inf | SBLUP.Inf | -0.001 | -0.451 | -0.988 | 3.23e-01 | 0.000 | -0.055 | -0.058 | 9.54e-01 |
| LDpred1.Inf | SBayesR.PseudoVal | -0.023 | -7.334 | -8.076 | 6.71e-16 | -0.027 | -8.776 | -4.726 | 2.29e-06 |
| LDpred1.Inf | LDpred1.10FCVal | 0.007 | 2.188 | 3.373 | 7.43e-04 | 0.004 | 1.225 | 0.914 | 3.61e-01 |
| LDpred1.Inf | LDpred1.MultiPRS | -0.004 | -1.299 | -0.373 | 7.09e-01 | -0.005 | -1.519 | -1.491 | 1.36e-01 |
| LDpred1.Inf | LDpred1.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred1.Inf | LDpred2.10FCVal | -0.029 | -9.373 | -9.716 | 2.58e-22 | -0.025 | -8.055 | -4.048 | 5.16e-05 |
| LDpred1.Inf | LDpred2.MultiPRS | -0.029 | -9.166 | -2.667 | 7.65e-03 | -0.028 | -9.085 | -4.812 | 1.50e-06 |
| LDpred1.Inf | LDpred2.PseudoVal | -0.025 | -8.006 | -10.591 | 3.30e-26 | -0.027 | -8.712 | -5.591 | 2.26e-08 |
| LDpred1.Inf | LDpred2.Inf | -0.004 | -1.432 | -3.330 | 8.70e-04 | -0.004 | -1.291 | -1.441 | 1.50e-01 |
| LDpred1.Inf | DBSLMM.PseudoVal | -0.010 | -3.326 | -3.104 | 1.91e-03 | -0.009 | -2.819 | -1.270 | 2.04e-01 |
| LDpred1.Inf | All.MultiPRS | -0.031 | -10.087 | -2.938 | 3.31e-03 | -0.026 | -8.274 | -3.654 | 2.58e-04 |
| LDpred2.10FCVal | pT+clump.10FCVal | 0.049 | 14.412 | 10.291 | 7.78e-25 | 0.059 | 17.798 | 6.229 | 4.70e-10 |
| LDpred2.10FCVal | pT+clump.MultiPRS | 0.041 | 12.130 | 3.835 | 1.26e-04 | 0.044 | 13.077 | 5.280 | 1.29e-07 |
| LDpred2.10FCVal | lassosum.10FCVal | 0.011 | 3.198 | 4.461 | 8.15e-06 | 0.005 | 1.352 | 0.906 | 3.65e-01 |
| LDpred2.10FCVal | lassosum.MultiPRS | 0.007 | 2.191 | 0.703 | 4.82e-01 | 0.001 | 0.316 | 0.221 | 8.25e-01 |
| LDpred2.10FCVal | lassosum.PseudoVal | 0.082 | 24.170 | 13.830 | 1.67e-43 | 0.093 | 28.004 | 7.780 | 7.28e-15 |
| LDpred2.10FCVal | SBLUP.Inf | 0.028 | 8.158 | 9.487 | 2.37e-21 | 0.025 | 7.404 | 4.184 | 2.86e-05 |
| LDpred2.10FCVal | SBayesR.PseudoVal | 0.006 | 1.865 | 3.549 | 3.87e-04 | -0.002 | -0.667 | -0.618 | 5.36e-01 |
| LDpred2.10FCVal | LDpred1.10FCVal | 0.036 | 10.570 | 11.011 | 3.38e-28 | 0.029 | 8.588 | 4.340 | 1.42e-05 |
| LDpred2.10FCVal | LDpred1.MultiPRS | 0.025 | 7.383 | 2.349 | 1.88e-02 | 0.020 | 6.049 | 3.573 | 3.53e-04 |
| LDpred2.10FCVal | LDpred1.Inf | 0.029 | 8.570 | 9.716 | 2.58e-22 | 0.025 | 7.455 | 4.048 | 5.16e-05 |
| LDpred2.10FCVal | LDpred2.10FCVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.10FCVal | LDpred2.MultiPRS | 0.001 | 0.189 | 0.061 | 9.51e-01 | -0.003 | -0.953 | -2.138 | 3.25e-02 |
| LDpred2.10FCVal | LDpred2.PseudoVal | 0.004 | 1.250 | 3.302 | 9.59e-04 | -0.002 | -0.608 | -0.772 | 4.40e-01 |
| LDpred2.10FCVal | LDpred2.Inf | 0.025 | 7.261 | 8.982 | 2.66e-19 | 0.021 | 6.260 | 3.712 | 2.06e-04 |
| LDpred2.10FCVal | DBSLMM.PseudoVal | 0.019 | 5.529 | 6.164 | 7.07e-10 | 0.016 | 4.846 | 2.598 | 9.38e-03 |
| LDpred2.10FCVal | All.MultiPRS | -0.002 | -0.652 | -0.211 | 8.33e-01 | -0.001 | -0.202 | -0.180 | 8.57e-01 |
| LDpred2.MultiPRS | pT+clump.10FCVal | 0.048 | 14.250 | 4.506 | 6.59e-06 | 0.063 | 18.574 | 6.589 | 4.43e-11 |
| LDpred2.MultiPRS | pT+clump.MultiPRS | 0.041 | 11.964 | 3.796 | 1.47e-04 | 0.047 | 13.897 | 5.681 | 1.34e-08 |
| LDpred2.MultiPRS | lassosum.10FCVal | 0.010 | 3.015 | 0.966 | 3.34e-01 | 0.008 | 2.283 | 1.525 | 1.27e-01 |
| LDpred2.MultiPRS | lassosum.MultiPRS | 0.007 | 2.005 | 0.938 | 3.48e-01 | 0.004 | 1.256 | 0.881 | 3.78e-01 |
| LDpred2.MultiPRS | lassosum.PseudoVal | 0.082 | 24.026 | 7.559 | 4.05e-14 | 0.097 | 28.683 | 8.026 | 1.01e-15 |
| LDpred2.MultiPRS | SBLUP.Inf | 0.027 | 7.983 | 2.537 | 1.12e-02 | 0.028 | 8.277 | 5.003 | 5.65e-07 |
| LDpred2.MultiPRS | SBayesR.PseudoVal | 0.006 | 1.679 | 0.539 | 5.90e-01 | 0.001 | 0.283 | 0.273 | 7.85e-01 |
| LDpred2.MultiPRS | LDpred1.10FCVal | 0.035 | 10.401 | 3.297 | 9.76e-04 | 0.032 | 9.451 | 5.075 | 3.88e-07 |
| LDpred2.MultiPRS | LDpred1.MultiPRS | 0.025 | 7.207 | 2.765 | 5.69e-03 | 0.023 | 6.935 | 4.366 | 1.26e-05 |
| LDpred2.MultiPRS | LDpred1.Inf | 0.029 | 8.397 | 2.667 | 7.65e-03 | 0.028 | 8.328 | 4.812 | 1.50e-06 |
| LDpred2.MultiPRS | LDpred2.10FCVal | -0.001 | -0.190 | -0.061 | 9.51e-01 | 0.003 | 0.944 | 2.138 | 3.25e-02 |
| LDpred2.MultiPRS | LDpred2.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.MultiPRS | LDpred2.PseudoVal | 0.004 | 1.063 | 0.341 | 7.33e-01 | 0.001 | 0.342 | 0.480 | 6.31e-01 |
| LDpred2.MultiPRS | LDpred2.Inf | 0.024 | 7.085 | 2.253 | 2.42e-02 | 0.024 | 7.145 | 4.591 | 4.41e-06 |
| LDpred2.MultiPRS | DBSLMM.PseudoVal | 0.018 | 5.350 | 1.714 | 8.66e-02 | 0.019 | 5.744 | 3.268 | 1.08e-03 |
| LDpred2.MultiPRS | All.MultiPRS | -0.003 | -0.843 | -0.443 | 6.58e-01 | 0.003 | 0.743 | 0.738 | 4.61e-01 |
| LDpred2.PseudoVal | pT+clump.10FCVal | 0.045 | 13.329 | 9.205 | 3.41e-20 | 0.061 | 18.295 | 6.294 | 3.09e-10 |
| LDpred2.PseudoVal | pT+clump.MultiPRS | 0.037 | 11.018 | 3.437 | 5.88e-04 | 0.046 | 13.602 | 5.312 | 1.08e-07 |
| LDpred2.PseudoVal | lassosum.10FCVal | 0.007 | 1.973 | 2.482 | 1.31e-02 | 0.007 | 1.948 | 1.208 | 2.27e-01 |
| LDpred2.PseudoVal | lassosum.MultiPRS | 0.003 | 0.953 | 0.301 | 7.63e-01 | 0.003 | 0.918 | 0.572 | 5.67e-01 |
| LDpred2.PseudoVal | lassosum.PseudoVal | 0.078 | 23.210 | 11.926 | 8.62e-33 | 0.095 | 28.439 | 7.224 | 5.05e-13 |
| LDpred2.PseudoVal | SBLUP.Inf | 0.024 | 6.995 | 10.698 | 1.03e-26 | 0.027 | 7.963 | 6.058 | 1.37e-09 |
| LDpred2.PseudoVal | SBayesR.PseudoVal | 0.002 | 0.623 | 1.096 | 2.73e-01 | 0.000 | -0.059 | -0.052 | 9.59e-01 |
| LDpred2.PseudoVal | LDpred1.10FCVal | 0.032 | 9.438 | 11.647 | 2.36e-31 | 0.031 | 9.140 | 5.608 | 2.05e-08 |
| LDpred2.PseudoVal | LDpred1.MultiPRS | 0.021 | 6.210 | 1.947 | 5.15e-02 | 0.022 | 6.616 | 4.814 | 1.48e-06 |
| LDpred2.PseudoVal | LDpred1.Inf | 0.025 | 7.413 | 10.591 | 3.30e-26 | 0.027 | 8.014 | 5.591 | 2.26e-08 |
| LDpred2.PseudoVal | LDpred2.10FCVal | -0.004 | -1.266 | -3.302 | 9.59e-04 | 0.002 | 0.604 | 0.772 | 4.40e-01 |
| LDpred2.PseudoVal | LDpred2.MultiPRS | -0.004 | -1.074 | -0.341 | 7.33e-01 | -0.001 | -0.343 | -0.480 | 6.31e-01 |
| LDpred2.PseudoVal | LDpred2.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.PseudoVal | LDpred2.Inf | 0.020 | 6.087 | 10.372 | 3.32e-25 | 0.023 | 6.826 | 5.722 | 1.05e-08 |
| LDpred2.PseudoVal | DBSLMM.PseudoVal | 0.015 | 4.333 | 4.919 | 8.69e-07 | 0.018 | 5.420 | 3.014 | 2.58e-03 |
| LDpred2.PseudoVal | All.MultiPRS | -0.006 | -1.927 | -0.613 | 5.40e-01 | 0.001 | 0.403 | 0.312 | 7.55e-01 |
| LDpred2.Inf | pT+clump.10FCVal | 0.024 | 7.711 | 4.851 | 1.23e-06 | 0.038 | 12.309 | 3.798 | 1.46e-04 |
| LDpred2.Inf | pT+clump.MultiPRS | 0.017 | 5.251 | 1.528 | 1.26e-01 | 0.023 | 7.272 | 2.446 | 1.44e-02 |
| LDpred2.Inf | lassosum.10FCVal | -0.014 | -4.381 | -3.989 | 6.63e-05 | -0.016 | -5.235 | -2.336 | 1.95e-02 |
| LDpred2.Inf | lassosum.MultiPRS | -0.017 | -5.467 | -1.613 | 1.07e-01 | -0.020 | -6.341 | -2.836 | 4.57e-03 |
| LDpred2.Inf | lassosum.PseudoVal | 0.058 | 18.233 | 8.062 | 7.49e-16 | 0.073 | 23.196 | 5.005 | 5.59e-07 |
| LDpred2.Inf | SBLUP.Inf | 0.003 | 0.967 | 2.797 | 5.17e-03 | 0.004 | 1.220 | 1.728 | 8.40e-02 |
| LDpred2.Inf | SBayesR.PseudoVal | -0.018 | -5.819 | -6.534 | 6.42e-11 | -0.023 | -7.389 | -4.064 | 4.82e-05 |
| LDpred2.Inf | LDpred1.10FCVal | 0.011 | 3.569 | 5.495 | 3.92e-08 | 0.008 | 2.484 | 1.859 | 6.31e-02 |
| LDpred2.Inf | LDpred1.MultiPRS | 0.000 | 0.131 | 0.038 | 9.69e-01 | -0.001 | -0.226 | -0.194 | 8.46e-01 |
| LDpred2.Inf | LDpred1.Inf | 0.004 | 1.412 | 3.330 | 8.70e-04 | 0.004 | 1.275 | 1.441 | 1.50e-01 |
| LDpred2.Inf | LDpred2.10FCVal | -0.025 | -7.830 | -8.982 | 2.66e-19 | -0.021 | -6.678 | -3.712 | 2.06e-04 |
| LDpred2.Inf | LDpred2.MultiPRS | -0.024 | -7.625 | -2.253 | 2.42e-02 | -0.024 | -7.694 | -4.591 | 4.41e-06 |
| LDpred2.Inf | LDpred2.PseudoVal | -0.020 | -6.482 | -10.372 | 3.32e-25 | -0.023 | -7.326 | -5.722 | 1.05e-08 |
| LDpred2.Inf | LDpred2.Inf | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| LDpred2.Inf | DBSLMM.PseudoVal | -0.006 | -1.868 | -1.799 | 7.21e-02 | -0.005 | -1.509 | -0.701 | 4.83e-01 |
| LDpred2.Inf | All.MultiPRS | -0.027 | -8.533 | -2.527 | 1.15e-02 | -0.022 | -6.894 | -3.277 | 1.05e-03 |
| DBSLMM.PseudoVal | pT+clump.10FCVal | 0.030 | 9.404 | 5.998 | 2.00e-09 | 0.043 | 13.612 | 4.281 | 1.86e-05 |
| DBSLMM.PseudoVal | pT+clump.MultiPRS | 0.023 | 6.988 | 2.075 | 3.80e-02 | 0.027 | 8.650 | 3.315 | 9.16e-04 |
| DBSLMM.PseudoVal | lassosum.10FCVal | -0.008 | -2.467 | -2.289 | 2.21e-02 | -0.012 | -3.671 | -1.675 | 9.39e-02 |
| DBSLMM.PseudoVal | lassosum.MultiPRS | -0.011 | -3.533 | -1.068 | 2.86e-01 | -0.015 | -4.761 | -2.405 | 1.62e-02 |
| DBSLMM.PseudoVal | lassosum.PseudoVal | 0.064 | 19.733 | 10.594 | 3.19e-26 | 0.077 | 24.338 | 6.351 | 2.14e-10 |
| DBSLMM.PseudoVal | SBLUP.Inf | 0.009 | 2.783 | 2.729 | 6.35e-03 | 0.009 | 2.688 | 1.278 | 2.01e-01 |
| DBSLMM.PseudoVal | SBayesR.PseudoVal | -0.012 | -3.878 | -4.487 | 7.23e-06 | -0.018 | -5.793 | -3.267 | 1.09e-03 |
| DBSLMM.PseudoVal | LDpred1.10FCVal | 0.017 | 5.337 | 4.702 | 2.57e-06 | 0.012 | 3.933 | 1.669 | 9.51e-02 |
| DBSLMM.PseudoVal | LDpred1.MultiPRS | 0.006 | 1.963 | 0.585 | 5.58e-01 | 0.004 | 1.264 | 0.595 | 5.52e-01 |
| DBSLMM.PseudoVal | LDpred1.Inf | 0.010 | 3.219 | 3.104 | 1.91e-03 | 0.009 | 2.742 | 1.270 | 2.04e-01 |
| DBSLMM.PseudoVal | LDpred2.10FCVal | -0.019 | -5.852 | -6.164 | 7.07e-10 | -0.016 | -5.092 | -2.598 | 9.38e-03 |
| DBSLMM.PseudoVal | LDpred2.MultiPRS | -0.018 | -5.652 | -1.714 | 8.66e-02 | -0.019 | -6.094 | -3.268 | 1.08e-03 |
| DBSLMM.PseudoVal | LDpred2.PseudoVal | -0.015 | -4.529 | -4.919 | 8.69e-07 | -0.018 | -5.731 | -3.014 | 2.58e-03 |
| DBSLMM.PseudoVal | LDpred2.Inf | 0.006 | 1.834 | 1.799 | 7.21e-02 | 0.005 | 1.486 | 0.701 | 4.83e-01 |
| DBSLMM.PseudoVal | DBSLMM.PseudoVal | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |
| DBSLMM.PseudoVal | All.MultiPRS | -0.021 | -6.543 | -1.985 | 4.72e-02 | -0.017 | -5.305 | -2.673 | 7.51e-03 |
| All.MultiPRS | pT+clump.10FCVal | 0.051 | 14.967 | 4.779 | 1.76e-06 | 0.060 | 17.964 | 6.290 | 3.17e-10 |
| All.MultiPRS | pT+clump.MultiPRS | 0.044 | 12.700 | 4.064 | 4.83e-05 | 0.044 | 13.252 | 5.340 | 9.28e-08 |
| All.MultiPRS | lassosum.10FCVal | 0.013 | 3.826 | 1.239 | 2.15e-01 | 0.005 | 1.551 | 0.954 | 3.40e-01 |
| All.MultiPRS | lassosum.MultiPRS | 0.010 | 2.825 | 1.319 | 1.87e-01 | 0.002 | 0.517 | 0.393 | 6.95e-01 |
| All.MultiPRS | lassosum.PseudoVal | 0.085 | 24.662 | 7.842 | 4.44e-15 | 0.094 | 28.149 | 8.120 | 4.66e-16 |
| All.MultiPRS | SBLUP.Inf | 0.030 | 8.753 | 2.810 | 4.95e-03 | 0.025 | 7.591 | 3.734 | 1.89e-04 |
| All.MultiPRS | SBayesR.PseudoVal | 0.009 | 2.501 | 0.811 | 4.17e-01 | -0.002 | -0.464 | -0.331 | 7.40e-01 |
| All.MultiPRS | LDpred1.10FCVal | 0.038 | 11.150 | 3.571 | 3.56e-04 | 0.029 | 8.773 | 3.957 | 7.57e-05 |
| All.MultiPRS | LDpred1.MultiPRS | 0.027 | 7.983 | 3.060 | 2.21e-03 | 0.021 | 6.238 | 3.305 | 9.49e-04 |
| All.MultiPRS | LDpred1.Inf | 0.031 | 9.163 | 2.938 | 3.31e-03 | 0.026 | 7.642 | 3.654 | 2.58e-04 |
| All.MultiPRS | LDpred2.10FCVal | 0.002 | 0.648 | 0.211 | 8.33e-01 | 0.001 | 0.202 | 0.180 | 8.57e-01 |
| All.MultiPRS | LDpred2.MultiPRS | 0.003 | 0.836 | 0.443 | 6.58e-01 | -0.003 | -0.749 | -0.738 | 4.61e-01 |
| All.MultiPRS | LDpred2.PseudoVal | 0.006 | 1.890 | 0.613 | 5.40e-01 | -0.001 | -0.405 | -0.312 | 7.55e-01 |
| All.MultiPRS | LDpred2.Inf | 0.027 | 7.862 | 2.527 | 1.15e-02 | 0.022 | 6.449 | 3.277 | 1.05e-03 |
| All.MultiPRS | DBSLMM.PseudoVal | 0.021 | 6.141 | 1.985 | 4.72e-02 | 0.017 | 5.038 | 2.673 | 7.51e-03 |
| All.MultiPRS | All.MultiPRS | 0.000 | 0.000 | 0.000 | 1.00e+00 | 0.000 | 0.000 | 0.000 | 1.00e+00 |