Evaluate local rG weighted polygenic scores
1 Introduction
Here we will test whether weighting PGS by local heritability and genetic correlations with the target phenotype improves prediction. We will use three target outcomes: Depression, BMI, and Intelligence. We will collect GWAS for traits geneticaly correlated with the target outcomes, derive polygenic scores using the leading PRS method (LDAK MegaPRS), and then evaluate their predictive utility before and after weighting by local genetic correlation (estimated using HESS or LAVA).
A preprint of this work can be found here.
2 Methods
2.1 GWAS sumstats
2.1.1 Depression
Target trait GWAS: Wray et al. excl UKB
Secondary GWAS: Schizophreina (PGC2), Bipolar disorder (PGC2), Autism, Smoking, ADHD, Educationtional attainment, Anxiety, age at menarche
2.1.2 BMI
Target trait GWAS: Locke et al.
Secondary GWAS: Obesity, Childhood obesity, Waist circumference, coronary artery disease, age at menarche, Educationtional attainment, type 2 diabetes, fasting insulin.
2.1.3 Intelligence
Target trait GWAS: Intelligence
Secondary GWAS: Educational attainment, high IQ, Childhood IQ, ADHD, Depression, Schizophrenia, Autism, Longevity
Create table listing GWAS
# I need the latest csv of the database to create the table
gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
pheno<-read.csv('/users/k1806347/brc_scratch/Data/GWAS_sumstats/QC_sumstats_list_031218.csv', stringsAsFactors=F)
pheno<-pheno[pheno$Code %in% gwas,]
pheno<-pheno[c('Code','sample_size_discovery','Ncases','Ncontrols','PMID')]
pheno<-pheno[match(gwas, pheno$Code),]
pheno$Code<-gwas
pheno$Phenotype<-c('Major Depression','Schizophrenia','Bipolar Disorder','Autism','Smoking (Ever/Never Regular)','Attention Deficit Hyperactivity Disorder','College Completion','Type-2 Diabetes','Anxiety (Factor Score)','Age at Menarche','Obesity Class 1','Waist Circumference','Coronary Arety Disease','Fasting Insulin (Age- & Sex-adjusted)','Childhood Intelligence','Intelligence','BMI')
pheno$PMID[pheno$Code == 'BIPO02']<-'31043756'
pheno$PMID[pheno$Code == 'AUTI07']<-'30804558'
pheno$PMID[pheno$Code == 'ADHD05']<-'30478444'
pheno$PMID[pheno$Code == 'INTE03']<-'29942086'
names(pheno)<-c('Code','N','Ncase','Ncon','PMID','Phenotype')
pheno$N<-gsub(',','',pheno$N)
pheno$Ncase<-gsub(',','',pheno$Ncase)
pheno$Ncon<-gsub(',','',pheno$Ncon)
pheno$Ncase[pheno$Code == 'BIPO02']<-20352
pheno$Ncon[pheno$Code == 'BIPO02']<-31358
pheno$Ncase[pheno$Code == 'AUTI07']<-18381
pheno$Ncon[pheno$Code == 'AUTI07']<-27969
pheno$N<-as.numeric(pheno$N)
pheno$Ncase<-as.numeric(pheno$Ncase)
pheno$Ncon<-as.numeric(pheno$Ncon)
pheno$N[is.na(pheno$N)]<-pheno$Ncase[is.na(pheno$N)]+pheno$Ncon[is.na(pheno$N)]
pheno<-pheno[,c('Code','Phenotype','N','Ncase','Ncon','PMID')]
write.csv(pheno, '/users/k1806347/brc_scratch/Analyses/local_rg_pgs/gwas_lis.csv', row.names=F, quote=F)index
Show table of GWAS sumstats
| Code | Phenotype | N | Ncase | Ncon | PMID |
|---|---|---|---|---|---|
| DEPR06 | Major Depression | 431394 | 116404 | 314990 | 29700475 |
| SCHI02 | Schizophrenia | 77096 | 33640 | 43456 | 25056061 |
| BIPO02 | Bipolar Disorder | 51710 | 20352 | 31358 | 31043756 |
| AUTI07 | Autism | 46350 | 18381 | 27969 | 30804558 |
| SMOK04 | Smoking (Ever/Never Regular) | 74035 | 41969 | 32066 | 20418890 |
| ADHD05 | Attention Deficit Hyperactivity Disorder | 53293 | 19099 | 34194 | 30478444 |
| COLL01 | College Completion | 95427 | NA | NA | 23722424 |
| DIAB05 | Type-2 Diabetes | 159208 | 26676 | 132532 | 28566273 |
| ANXI02 | Anxiety (Factor Score) | 17310 | NA | NA | 26754954 |
| MENA01F | Age at Menarche | 132989 | NA | NA | 25231870 |
| OBES01 | Obesity Class 1 | 98697 | 32858 | 65839 | 23563607 |
| WAIS01 | Waist Circumference | 231927 | NA | NA | 25673412 |
| COAD01 | Coronary Arety Disease | 184305 | 60801 | 123504 | 26343387 |
| GLYC05 | Fasting Insulin (Age- & Sex-adjusted) | 38238 | NA | NA | 20081858 |
| INTE01 | Childhood Intelligence | 12441 | NA | NA | 23358156 |
| INTE03 | Intelligence | 269867 | NA | NA | 29942086 |
| BODY04 | BMI | 322154 | NA | NA | 25673413 |
2.1.4 GWAS Quality Control
Show code
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
# Create directory
mkdir -p /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats
# Create file listing GWAS that haven't been processed.
> /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/todo.txt
for gwas in $(echo DEPR06 SCHI02 BIPO02 AUTI07 SMOK04 ADHD05 COLL01 DIAB05 ANXI02 MENA01F OBES01 WAIS01 COAD01 GLYC05 INTE01 INTE03 BODY04);do
if [ ! -f /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/${gwas}.cleaned.gz ]; then
echo $gwas >> /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/todo.txt
fi
done
# Create shell script to run using sbatch
cat > /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 5G
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(sed "${SLURM_ARRAY_TASK_ID}q;d" /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/todo.txt)
echo ${gwas}
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/sumstat_cleaner/sumstat_cleaner.R \
--sumstats ${gwas_rep_cleaned}/${gwas}.gz \
--ref_plink_chr ${Geno_1KG_dir}/1KGPhase3.w_hm3.chr \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--output /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/${gwas}.cleaned
EOF
sbatch --array 1-$(wc -l /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/todo.txt | cut -d' ' -f1)%5 /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/sbatch.sh
# GLYC05 contains MAF so FREQ doesn't correspond to particular allele. As a result we are loosing many SNPs. Remove FREQ column and re-QC.
cp ${gwas_rep_cleaned}/GLYC05.gz /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/GLYC05_tmp.gz
zcat /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/GLYC05_tmp.gz | cut -f 1-3,5-8 | gzip > /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/GLYC05_tmp2.gz
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/sumstat_cleaner/sumstat_cleaner.R \
--sumstats /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/GLYC05_tmp2.gz \
--ref_plink_chr ${Geno_1KG_dir}/1KGPhase3.w_hm3.chr \
--ref_freq_chr ${Geno_1KG_dir}/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr \
--output /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/GLYC05.cleaned
rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/GLYC05_tmp.gz
rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/GLYC05_tmp2.gz2.2 Estimate genome-wide and local genetic correlation
Show code
# Create phenotype data.frame
# Leave number of cases and control as NA as when set as bainry LAVA seems to take a lot longer.
# We can convert the heritability estimatest to the liability model later if we want to.
gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
pheno<-data.frame(phenotype=gwas,
cases=NA,
controls=NA,
prevalence=NA,
filename=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/', gwas,'.cleaned.gz'))
dir.create('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/')
write.table(pheno, '/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/input.info.txt', col.names=T, row.names=F, quote=F)##########
# Prepare sample overlap file
##########
# Munge all sumstats
for gwas in $(echo DEPR06 SCHI02 BIPO02 AUTI07 SMOK04 ADHD05 COLL01 DIAB05 ANXI02 MENA01F OBES01 WAIS01 COAD01 GLYC05 INTE01 INTE03 BODY04); do
sbatch -p brc,shared ~/brc_scratch/Software/munge_sumstats.sh \
--sumstats /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/${gwas}.cleaned.gz \
--merge-alleles /users/k1806347/brc_scratch/Data/ldsc/w_hm3.snplist \
--out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/${gwas}.cleaned
done
# Run bivariate LDSC for all traits
mkdir /users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/sample_overlap
for gwas1 in $(echo DEPR06 SCHI02 BIPO02 AUTI07 SMOK04 ADHD05 COLL01 DIAB05 ANXI02 MENA01F OBES01 WAIS01 COAD01 GLYC05 INTE01 INTE03 BODY04); do
for gwas2 in $(echo DEPR06 SCHI02 BIPO02 AUTI07 SMOK04 ADHD05 COLL01 DIAB05 ANXI02 MENA01F OBES01 WAIS01 COAD01 GLYC05 INTE01 INTE03 BODY04); do
sbatch -p brc,shared ~/brc_scratch/Software/ldsc.sh \
--rg /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/${gwas1}.cleaned.sumstats.gz,/users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/${gwas2}.cleaned.sumstats.gz \
--ref-ld-chr /users/k1806347/brc_scratch/Data/ldsc/eur_w_ld_chr/ \
--w-ld-chr /users/k1806347/brc_scratch/Data/ldsc/eur_w_ld_chr/ \
--out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/sample_overlap/${gwas1}_${gwas2}_rg
done
done
# Collate results
cd /users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/sample_overlap/
FILES=($(ls *_rg.log))
N=$(echo ${#FILES[@]})
for I in ${FILES[@]}; do
PHEN=$(echo $I | sed 's/_rg\.log//')
# subset log files to relevant output
tail -n 5 $I | head -n 2 > $PHEN.rg
# add to single data set
if [[ $I == ${FILES[0]} ]]; then
cat $PHEN.rg > all.rg
else
cat $PHEN.rg | sed '1d' >> all.rg
fi
done
scor = read.table("/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/sample_overlap/all.rg",header=T) # read in
scor = scor[,c("p1","p2","gcov_int")] # retain key headers
scor$p1 = gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/','',gsub(".cleaned.sumstats.gz","",scor$p1))
scor$p2 = gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/','',gsub(".cleaned.sumstats.gz","",scor$p2))
phen = unique(scor$p1)
n = length(phen)
mat = matrix(NA,n,n) # create matrix
rownames(mat) = colnames(mat) = phen # set col/rownames
for (i in phen) {
for (j in phen) {
mat[i,j] = subset(scor, p1==i & p2==j)$gcov_int
}
}
if (!all(t(mat)==mat)) { mat[lower.tri(mat)] = t(mat)[lower.tri(mat)] } # sometimes there might be small differences in gcov_int depending on which phenotype was analysed as the outcome / predictor
mat = round(cov2cor(mat),5) # standardise
write.table(mat, "/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/sample_overlap/sample.overlap.txt", quote=F) # save#####
# Create R script to run LAVA across genome
#####
cat > /users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/lava.R << 'EOF'
# command line arguments, specifying input/output file names and phenotype subset
arg = commandArgs(T); ref.prefix = arg[1]; loc.file = arg[2]; info.file = arg[3]; sample.overlap.file = arg[4]; phenos = unlist(strsplit(arg[5],",")); out.fname = arg[6]
### Load package
library(LAVA)
### Read in data
loci = read.loci(loc.file); n.loc = nrow(loci)
input = process.input(info.file, sample.overlap.file, ref.prefix, phenos)
print(paste("Starting LAVA analysis for",n.loc,"loci"))
progress = ceiling(quantile(1:n.loc, seq(.05,1,.05))) # (if you want to print the progress)
### Analyse
u=b=list()
for (i in 1:n.loc) {
if (i %in% progress) print(paste("..",names(progress[which(progress==i)]))) # (printing progress)
locus = process.locus(loci[i,], input) # process locus
# It is possible that the locus cannot be defined for various reasons (e.g. too few SNPs), so the !is.null(locus) check is necessary before calling the analysis functions.
if (!is.null(locus)) {
# extract some general locus info for the output
loc.info = data.frame(locus = locus$id, chr = locus$chr, start = locus$start, stop = locus$stop, n.snps = locus$n.snps, n.pcs = locus$K)
# run the univariate and bivariate tests
loc.out = run.univ.bivar(locus)
u[[i]] = cbind(loc.info, loc.out$univ)
if(!is.null(loc.out$bivar)) b[[i]] = cbind(loc.info, loc.out$bivar)
}
}
# save the output
write.table(do.call(rbind,u), paste0(out.fname,".univ.lava"), row.names=F,quote=F,col.names=T)
write.table(do.call(rbind,b), paste0(out.fname,".bivar.lava"), row.names=F,quote=F,col.names=T)
print(paste0("Done! Analysis output written to ",out.fname,".*.lava"))
EOF
########
# Run LAVA for each target GWAS
########
for target_gwas in $(echo DEPR06 BODY04 INTE03); do
for secondary_gwas in $(echo DEPR06 SCHI02 BIPO02 AUTI07 SMOK04 ADHD05 COLL01 DIAB05 ANXI02 MENA01F OBES01 WAIS01 COAD01 GLYC05 INTE01 INTE03 BODY04); do
if [ ${target_gwas} != ${secondary_gwas} ]; then
mkdir -p /users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/${target_gwas}
sbatch -p brc,shared --mem 10G /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/lava.R \
/users/k1806347/brc_scratch/Data/1KG/Phase3/1KGPhase3.w_hm3.GW \
/users/k1806347/brc_scratch/Data/LAVA/blocks_s2500_m25_f1_w200.GRCh37_hg19.locfile \
/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/input.info.txt \
/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/sample_overlap/sample.overlap.txt \
${target_gwas},${secondary_gwas} \
/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/${target_gwas}/${target_gwas}.${secondary_gwas}
fi
done
done
# Notes: If the target phenotype GWAS does not have a large sample, then heritability within a locus may not be significant, and therefore rho cannot be accuratly estimated. This means there may also be an advantage to using unweighted secondary PGS as they may contain information from shared loci that cannot be identified using LAVA.
# I should create an info file with the correct number of cases and controls
# This takes approximately 2.5 hours per pair of traits2.3 Characterise the genome-wide and local genetic correlation results
Show code
library(data.table)
library(cowplot)
library(ggplot2)
target_gwas<-c('DEPR06','BODY04','INTE03')
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
ncase<-c(116404,33640,20352,18381,41969,19099,NA,26676,NA,NA,32858,NA,60801,NA,NA,NA,NA)
ncon<-c(314990,43456,31358,27969,32066,34194,NA,132532,NA,NA,65839,NA,123504,NA,NA,NA,NA)
prev<-c(0.15,0.01,0.015,0.012,0.5,0.05,NA,0.05,NA,NA,0.13,NA,0.03,NA,NA,NA,NA)
des<-NULL
all_res<-NULL
for(target_gwas_i in target_gwas){
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
univ_res<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/',target_gwas_i,'/',target_gwas_i,'.',secondary_gwas_i,'.univ.lava'))
bivar_res<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/',target_gwas_i,'/',target_gwas_i,'.',secondary_gwas_i,'.bivar.lava'))
# Convert heritability to liability scale
h2l_R2 <- function(k, r2, p) {
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
h2l_R2 = C*r2 / (1 + C*theta*r2)
return(h2l_R2)
}
univ_res$h2.liab<-NA
for(trait in c(target_gwas_i, secondary_gwas_i)){
if(!is.na(ncase[secondary_gwas == trait])){
univ_res$h2.liab[univ_res$phen == trait]<-h2l_R2(k=prev[secondary_gwas == trait], r2=univ_res$h2.obs[univ_res$phen == trait], p=ncase[secondary_gwas == trait]/(ncase[secondary_gwas == trait]+ncon[secondary_gwas == trait]))
} else {
univ_res$h2.liab[univ_res$phen == trait]<-univ_res$h2.obs[univ_res$phen == trait]
}
}
# Calculate SE for h2
univ_res$z<-abs(qnorm(univ_res$p))
univ_res$se<-univ_res$h2.liab/univ_res$z
bivar_res<-bivar_res[!is.na(bivar_res$p),]
bivar_res$Direction<-'+'
bivar_res$Direction[bivar_res$rho < 0]<-'-'
bivar_res$p.fdr<-p.adjust(bivar_res$p, method='fdr')
univ_res_wide<-merge(univ_res[univ_res$phen == target_gwas_i,c('locus','chr','start','stop','h2.obs','se','z','p')],univ_res[univ_res$phen == secondary_gwas_i,c('locus','h2.obs','se','z','p')], by='locus')
tmp<-merge(univ_res_wide, bivar_res[,c('locus','rho','p','phen1','phen2')], by='locus')
all_res<-rbind(all_res, tmp)
des<-rbind(des, data.frame(target=target_gwas_i,
secondary=secondary_gwas_i,
n_loci=nrow(bivar_res),
n_snps=sum(bivar_res$n.snps),
n_sig_loci=nrow(bivar_res[bivar_res$p < 0.05,]),
n_sig_snps=sum(bivar_res[bivar_res$p < 0.05,]$n.snps),
n_sig_loci_pos=nrow(bivar_res[bivar_res$p < 0.05 & bivar_res$Direction == '+',]),
n_sig_loci_neg=nrow(bivar_res[bivar_res$p < 0.05 & bivar_res$Direction == '-',]),
n_sig_fdr_loci=nrow(bivar_res[bivar_res$p.fdr < 0.05,]),
n_sig_fdr_snps=sum(bivar_res[bivar_res$p.fdr < 0.05,]$n.snps),
n_sig_fdr_loci_pos=nrow(bivar_res[bivar_res$p.fdr < 0.05 & bivar_res$Direction == '+',]),
n_sig_fdr_loci_neg=nrow(bivar_res[bivar_res$p.fdr < 0.05 & bivar_res$Direction == '-',])))
}
}
write.table(des, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/lava_descript.csv'), row.names = F, quote=F)
write.table(all_res, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/all_res.csv'), row.names = F, quote=F)2.4 Generate polygenic scores
Show code
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(echo DEPR06 SCHI02 BIPO02 AUTI07 SMOK04 ADHD05 COLL01 DIAB05 ANXI02 MENA01F OBES01 WAIS01 COAD01 GLYC05 INTE01 INTE03 BODY04)
# Create directory
mkdir -p /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK
# Create file listing GWAS that haven't been processed.
> /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/todo.txt
for gwas_i in $(echo DEPR06 SCHI02 BIPO02 AUTI07 SMOK04 ADHD05 COLL01 DIAB05 ANXI02 MENA01F OBES01 WAIS01 COAD01 GLYC05 INTE01 INTE03 BODY04);do
if [ ! -f /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/${gwas_i}/1KGPhase3.w_hm3.${gwas_i}.EUR.scale ]; then
echo ${gwas_i} >> /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/todo.txt
fi
done
# Create shell script to run using sbatch
cat > /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/sbatch.sh << 'EOF'
#!/bin/sh
#SBATCH -p shared,brc
#SBATCH --mem 20G
#SBATCH -n 5
#SBATCH --nodes 1
#SBATCH -J LDAK
. /users/k1806347/brc_scratch/Software/MyGit/GenoPred/config_used/Pipeline_prep.config
gwas=$(awk -v var="$SLURM_ARRAY_TASK_ID" 'NR == var {print $1}' /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/todo.txt)
echo ${gwas}
/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/ldak_mega_prs/ldak_mega_prs.R \
--ref_plink ${Geno_1KG_dir}/1KGPhase3.w_hm3.GW \
--ref_keep ${Geno_1KG_dir}/keep_files/EUR_samples.keep \
--sumstats /users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/${gwas}.cleaned.gz \
--plink1 ${plink1_9} \
--plink2 ${plink2} \
--ldak /users/k1806347/brc_scratch/Software/ldak5.1.linux \
--ldak_map /users/k1806347/brc_scratch/Data/LDAK/genetic_map_b37 \
--ldak_tag /users/k1806347/brc_scratch/Data/LDAK/bld/ \
--ldak_highld /users/k1806347/brc_scratch/Data/LDAK/highld.txt \
--memory 5000 \
--n_cores 5 \
--output /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/${gwas}/1KGPhase3.w_hm3.${gwas} \
--ref_pop_scale ${Geno_1KG_dir}/super_pop_keep.list
EOF
sbatch --array 1-$(wc -l /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/todo.txt | cut -d' ' -f1)%6 /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/sbatch.sh
2.5 Reweight polygenic scores
Show code
library(data.table)
# Read in reference bim for RSID coordinates
ref_bim<-fread('/users/k1806347/brc_scratch/Data/1KG/Phase3/1KGPhase3.w_hm3.GW.bim')
names(ref_bim)<-c('CHR','SNP','POS','BP','A1','A2')
# Read in the rho estimates
target_gwas<-c('DEPR06','BODY04','INTE03')
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
ncase<-c(116404,33640,20352,18381,41969,19099,NA,26676,NA,NA,32858,NA,60801,NA,NA,NA,NA)
ncon<-c(314990,43456,31358,27969,32066,34194,NA,132532,NA,NA,65839,NA,123504,NA,NA,NA,NA)
prev<-c(0.15,0.01,0.015,0.012,0.5,0.05,NA,0.05,NA,NA,0.13,NA,0.03,NA,NA,NA,NA)
univ_res_all<-list()
for(target_gwas_i in target_gwas){
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
univ_res<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/',target_gwas_i,'/',target_gwas_i,'.',secondary_gwas_i,'.univ.lava'))
# Remove loci with non-significant heritability
univ_res<-univ_res[univ_res$p < 0.05,]
# Convert heritability to liability scale
h2l_R2 <- function(k, r2, p) {
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
h2l_R2 = C*r2 / (1 + C*theta*r2)
return(h2l_R2)
}
univ_res$h2.liab<-NA
for(trait in c(target_gwas_i, secondary_gwas_i)){
if(!is.na(ncase[secondary_gwas == trait])){
univ_res$h2.liab[univ_res$phen == trait]<-h2l_R2(k=prev[secondary_gwas == trait], r2=univ_res$h2.obs[univ_res$phen == trait], p=ncase[secondary_gwas == trait]/(ncase[secondary_gwas == trait]+ncon[secondary_gwas == trait]))
} else {
univ_res$h2.liab[univ_res$phen == trait]<-univ_res$h2.obs[univ_res$phen == trait]
}
}
univ_res_all[[paste0(target_gwas_i,'_',secondary_gwas_i)]]<-univ_res
}
}
bivar_res_all<-NULL
for(target_gwas_i in target_gwas){
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
bivar_res<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/',target_gwas_i,'/',target_gwas_i,'.',secondary_gwas_i,'.bivar.lava'))
bivar_res<-bivar_res[!is.na(bivar_res$p),]
bivar_res$p.fdr<-p.adjust(bivar_res$p, method='fdr')
bivar_res_all<-rbind(bivar_res_all, bivar_res)
}
}
# Read in the score files and reweight by rho
for(target_gwas_i in target_gwas){
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
# Write out the pseudoval model for each GWAS
pseudo<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudoval.txt'))
pseudo$V1[pseudo$V2 == max(pseudo$V2)][1]
score<-fread(paste0('zcat /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".score.gz | cut -d' ' -f 1,2,",as.numeric(gsub('Score_','',pseudo$V1[pseudo$V2 == max(pseudo$V2)][1]))+2))
names(score)<-c('SNP','A1','SCORE')
fwrite(score[,c('SNP','A1','SCORE'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.score"), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.score.gz"))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.score.gz"))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.score"))
# Now reweight the score for by rho and h2
score_bim<-merge(score, ref_bim[,c('SNP','CHR','BP'),with=F], by='SNP')
univ_res<-univ_res_all[[paste0(target_gwas_i,'_', secondary_gwas_i)]]
bivar_res<-bivar_res_all[bivar_res_all$phen1 == target_gwas_i & bivar_res_all$phen2 == secondary_gwas_i,]
score_bim_reweight<-NULL
for(locus in unique(bivar_res$locus)){
tmp<-score_bim[score_bim$CHR == bivar_res$chr[bivar_res$locus == locus] & score_bim$BP >= bivar_res$start[bivar_res$locus == locus] & score_bim$BP <= bivar_res$stop[bivar_res$locus == locus],]
tmp$SCORE_rho<-tmp$SCORE*bivar_res$rho[bivar_res$locus == locus]
tmp$SCORE_rho_h2<-(tmp$SCORE*bivar_res$rho[bivar_res$locus == locus])/(univ_res$h2.liab[univ_res$locus == locus & univ_res$phen == secondary_gwas_i] / univ_res$h2.liab[univ_res$locus == locus & univ_res$phen == target_gwas_i])
# Create score only considering nominally significant rho
if(bivar_res$p[bivar_res$locus == locus] < 0.05){
tmp$SCORE_sig<-tmp$SCORE
} else {
tmp$SCORE_sig<-0
}
if(bivar_res$p[bivar_res$locus == locus] < 0.05){
tmp$SCORE_rho_sig<-tmp$SCORE_rho
} else {
tmp$SCORE_rho_sig<-0
}
if(bivar_res$p[bivar_res$locus == locus] < 0.05){
tmp$SCORE_rho_sig_h2<-tmp$SCORE_rho_h2
} else {
tmp$SCORE_rho_sig_h2<-0
}
# Create score only considering fdr significant rho
if(bivar_res$p.fdr[bivar_res$locus == locus] < 0.05){
tmp$SCORE_fdr<-tmp$SCORE
} else {
tmp$SCORE_fdr<-0
}
if(bivar_res$p.fdr[bivar_res$locus == locus] < 0.05){
tmp$SCORE_rho_fdr<-tmp$SCORE_rho
} else {
tmp$SCORE_rho_fdr<-0
}
if(bivar_res$p.fdr[bivar_res$locus == locus] < 0.05){
tmp$SCORE_rho_fdr_h2<-tmp$SCORE_rho_h2
} else {
tmp$SCORE_rho_fdr_h2<-0
}
tmp<-tmp[,c('SNP','A1','SCORE','SCORE_rho','SCORE_rho_h2','SCORE_sig','SCORE_rho_sig','SCORE_rho_sig_h2','SCORE_fdr','SCORE_rho_fdr','SCORE_rho_fdr_h2'), with=F]
score_bim_reweight<-rbind(score_bim_reweight, tmp)
}
# Write out sig h2 restricted but unweighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_unweighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_unweighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_unweighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_unweighted_",target_gwas_i,'.score'))
# Write out sig rho restricted but unweighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_sig'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_unweighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_unweighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_unweighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_unweighted_",target_gwas_i,'.score'))
# Write out fdr sig rho restricted but unweighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_fdr'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_unweighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_unweighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_unweighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_unweighted_",target_gwas_i,'.score'))
# Write out sig h2 restricted and rho weighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_rho'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_",target_gwas_i,'.score'))
# Write out sig rho restricted and rho weighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_rho_sig'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_",target_gwas_i,'.score'))
# Write out fdr sig rho restricted and rho weighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_rho_fdr'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_",target_gwas_i,'.score'))
# Write out sig h2 restricted and rho and h2 weighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_rho_h2'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_",target_gwas_i,'.score'))
# Write out sig rho restricted and rho and h2 weighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_rho_sig_h2'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_",target_gwas_i,'.score'))
# Write out fdr rho restricted and rho and h2 weighted
fwrite(score_bim_reweight[,c('SNP','A1','SCORE_rho_fdr_h2'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_",target_gwas_i,'.score'))
# Combine reweighted PGS with full PGS
score_full_reweight<-score
for(type in names(score_bim_reweight)[grepl('SCORE',names(score_bim_reweight)) & grepl('rho',names(score_bim_reweight))]){
tmp<-merge(score, score_bim_reweight[,c('SNP',type), with=F], by='SNP', all=T)
tmp[[type]][which(tmp[[type]] == 0 | is.na(tmp[[type]]))]<-tmp[['SCORE']][which(tmp[[type]] == 0 | is.na(tmp[[type]]))]
tmp<-tmp[match(score_full_reweight$SNP, tmp$SNP),]
score_full_reweight[[type]]<-tmp[[type]]
}
# Write out sig h2 restricted and rho weighted with unadjusted SNPs
fwrite(score_full_reweight[,c('SNP','A1','SCORE_rho'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score'))
# Write out sig rho restricted and rho weighted with unadjusted SNPs
fwrite(score_full_reweight[,c('SNP','A1','SCORE_rho_sig'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score'))
# Write out fdr sig rho restricted and rho weighted with unadjusted SNPs
fwrite(score_full_reweight[,c('SNP','A1','SCORE_rho_fdr'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_weighted_with_unadjusted_",target_gwas_i,'.score'))
# Write out sig h2 restricted and rho and h2 weighted with unadjusted SNPs
fwrite(score_full_reweight[,c('SNP','A1','SCORE_rho_h2'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score'))
# Write out sig rho restricted and rho and h2 weighted with unadjusted SNPs
fwrite(score_full_reweight[,c('SNP','A1','SCORE_rho_sig_h2'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score'))
# Write out fdr rho restricted and rho and h2 weighted with unadjusted SNPs
fwrite(score_full_reweight[,c('SNP','A1','SCORE_rho_fdr_h2'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_rho_h2_weighted_with_unadjusted_",target_gwas_i,'.score'))
# Make score files for SNPs that aren't reweighted
score_unadjust<-score
for(type in names(score_bim_reweight)[grepl('SCORE',names(score_bim_reweight)) & !(grepl('rho',names(score_bim_reweight)))]){
tmp<-score$SCORE
tmp[score$SNP %in% score_bim_reweight$SNP[which(score_bim_reweight[[type]] != 0)]]<-0
score_unadjust[[paste0(type,'_remainder')]]<-tmp
}
# Write out unadjusted for SNPs not in sig h2 loci
fwrite(score_unadjust[,c('SNP','A1','SCORE_remainder'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_remainder_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_remainder_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_remainder_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.h2_restricted_remainder_",target_gwas_i,'.score'))
# Write out unadjusted for SNPs not in sig rho loci
fwrite(score_unadjust[,c('SNP','A1','SCORE_sig_remainder'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_remainder_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_remainder_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_remainder_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_restricted_remainder_",target_gwas_i,'.score'))
# Write out unadjusted for SNPs not in fdr sig rho loci
fwrite(score_unadjust[,c('SNP','A1','SCORE_fdr_remainder'),with=F], paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_remainder_",target_gwas_i,'.score'), quote=F, sep=' ', na='NA')
if(file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_remainder_",target_gwas_i,'.score.gz'))){
system(paste0('rm /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_remainder_",target_gwas_i,'.score.gz'))
}
system(paste0('gzip /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,".pseudo.rho_fdr_restricted_remainder_",target_gwas_i,'.score'))
}
}
# Process new score files with external_scorer script to create scale files
# Read in the rho estimates
target_gwas<-c('DEPR06','BODY04','INTE03')
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
for(target_gwas_i in target_gwas){
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted','rho_restricted_rho_weighted_with_unadjusted','rho_fdr_restricted_rho_weighted_with_unadjusted','h2_restricted_rho_weighted_with_unadjusted','rho_restricted_rho_h2_weighted_with_unadjusted','rho_fdr_restricted_rho_h2_weighted_with_unadjusted','h2_restricted_rho_h2_weighted_with_unadjusted','rho_restricted_remainder','rho_fdr_restricted_remainder','h2_restricted_remainder')){
dir.create(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/processed/',secondary_gwas_i,'.pseudo.',type,'_',target_gwas_i), recursive=T)
system(paste0('sbatch -p brc,shared --mem 10G /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/external_score_processor/external_score_processor_plink2.R --ref_plink_chr /users/k1806347/brc_scratch/Data/1KG/Phase3/1KGPhase3.w_hm3.chr --score /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo.',type,'_',target_gwas_i,'.score.gz --plink2 /users/k1806347/brc_scratch/Software/plink2 --output /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/processed/',secondary_gwas_i,'.pseudo.',type,'_',target_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo.',type,'_',target_gwas_i,' --ref_pop_scale /users/k1806347/brc_scratch/Data/1KG/Phase3/super_pop_keep.list'))
}
}
}
for(secondary_gwas_i in secondary_gwas){
dir.create(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/processed/',secondary_gwas_i,'.pseudo'), recursive=T)
system(paste0('/users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/external_score_processor/external_score_processor_plink2.R --ref_plink_chr /users/k1806347/brc_scratch/Data/1KG/Phase3/1KGPhase3.w_hm3.chr --score /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo.score.gz --plink2 /users/k1806347/brc_scratch/Software/plink2 --output /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/processed/',secondary_gwas_i,'.pseudo/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo --ref_pop_scale /users/k1806347/brc_scratch/Data/1KG/Phase3/super_pop_keep.list'))
}2.6 Perform polygenic scoring
Show code
# Read in the rho estimates
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
for(i in 1:length(target_gwas)){
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted','rho_restricted_rho_weighted_with_unadjusted','rho_fdr_restricted_rho_weighted_with_unadjusted','h2_restricted_rho_weighted_with_unadjusted','rho_restricted_rho_h2_weighted_with_unadjusted','rho_fdr_restricted_rho_h2_weighted_with_unadjusted','h2_restricted_rho_h2_weighted_with_unadjusted','rho_restricted_remainder','rho_fdr_restricted_remainder','h2_restricted_remainder')){
if(!file.exists(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs.profiles'))){
system(paste0('sbatch -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer_plink2.R --target_plink_chr /users/k1806347/brc_scratch/Data/UKBB/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr --target_keep /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --ref_score /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo.',type,'_',target_gwas_i,'.score.gz --ref_scale /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/processed/',secondary_gwas_i,'.pseudo.',type,'_',target_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo.',type,'_',target_gwas_i,'.EUR.scale --ref_freq_chr /users/k1806347/brc_scratch/Data/1KG/Phase3/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr --plink2 /users/k1806347/brc_scratch/Software/plink2_alpha/plink2 --output /users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs'))
}
}
}
}
for(secondary_gwas_i in secondary_gwas){
system(paste0('sbatch -p brc,shared --mem 20G /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Scaled_polygenic_scorer/Scaled_polygenic_scorer_plink2.R --target_plink_chr /users/k1806347/brc_scratch/Data/UKBB/Genotype/Harmonised/UKBB.w_hm3.QCd.AllSNP.chr --target_keep /users/k1806347/brc_scratch/Data/UKBB/Projected_PCs/Ancestry_idenitfier/UKBB.w_hm3.AllAncestry.EUR.keep --ref_score /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo.score.gz --ref_scale /users/k1806347/brc_scratch/Analyses/local_rg_pgs/score_files/LDAK/',secondary_gwas_i,'/processed/',secondary_gwas_i,'.pseudo/1KGPhase3.w_hm3.',secondary_gwas_i,'.pseudo.EUR.scale --ref_freq_chr /users/k1806347/brc_scratch/Data/1KG/Phase3/freq_files/EUR/1KGPhase3.w_hm3.EUR.chr --plink2 /users/k1806347/brc_scratch/Software/plink2_alpha/plink2 --output /users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/prs'))
}2.7 Evaluate polygenic scores
Show code
############
# First test whether inclusion of secondary PRS improve prediction over target phenotype gwas (i.e. replicate Krapohl et al.)
############
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
# use for loop to read all values and indexes
for(i in 1:length(target_gwas)){
pred_file<-NULL
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
target_pop_prev_i<-target_pop_prev[i]
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',target_gwas_i,'/prs.profiles'),
group='target')
pred_file<-rbind(pred_file, tmp)
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/prs.profiles'),
group='secondary')
pred_file<-rbind(pred_file, tmp)
}
dir.create(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i), recursive=T)
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_1.txt'), col.names=T, row.names=F, quote=F)
system(paste0('sbatch --mem 10G -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2_nested.R --pheno /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_1 --assoc T --outcome_pop_prev ',target_pop_prev_i,' --predictors /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_1.txt'))
}
############
# Compare prs restricted by h2 and rho significance, and unweighted and weighted by rho, and rho and h2
############
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
# use for loop to read all values and indexes
for(i in 1:length(target_gwas)){
pred_file<-NULL
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
target_pop_prev_i<-target_pop_prev[i]
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted')){
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
}
}
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_2.txt'), col.names=T, row.names=F, quote=F)
system(paste0('sbatch --mem 10G -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2_nested.R --pheno /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_2 --assoc T --outcome_pop_prev ',target_pop_prev_i,' --predictors /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_2.txt'))
}
############
# Test whether adding weighted PRS improves prediction over the standard approach (i.e. Krapohl et al.)
############
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
# use for loop to read all values and indexes
for(i in 1:length(target_gwas)){
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted')){
pred_file<-NULL
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
target_pop_prev_i<-target_pop_prev[i]
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',target_gwas_i,'/prs.profiles'),
group='baseline')
pred_file<-rbind(pred_file, tmp)
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/prs.profiles'),
group='baseline')
pred_file<-rbind(pred_file, tmp)
# Add in weighted prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
}
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_3_',type,'.txt'), col.names=T, row.names=F, quote=F)
system(paste0('sbatch --mem 10G -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2_nested.R --pheno /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_3_',type,' --assoc T --outcome_pop_prev ',target_pop_prev_i,' --predictors /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_3_',type,'.txt'))
}
}
############
# Compare unadjusted PRS to reweighted PRS with unadjusted SNPs
############
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
# use for loop to read all values and indexes
for(i in 1:length(target_gwas)){
for(type in c('rho_restricted_rho_weighted_with_unadjusted','rho_fdr_restricted_rho_weighted_with_unadjusted','h2_restricted_rho_weighted_with_unadjusted','rho_restricted_rho_h2_weighted_with_unadjusted','rho_fdr_restricted_rho_h2_weighted_with_unadjusted','h2_restricted_rho_h2_weighted_with_unadjusted')){
pred_file<-NULL
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
target_pop_prev_i<-target_pop_prev[i]
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/prs.profiles'),
group='standard')
pred_file<-rbind(pred_file, tmp)
# Add in weighted prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
}
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_4_',type,'.txt'), col.names=T, row.names=F, quote=F)
system(paste0('sbatch --mem 10G -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2_nested.R --pheno /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_4_',type,' --assoc T --outcome_pop_prev ',target_pop_prev_i,' --predictors /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_4_',type,'.txt'))
}
}
############
# Compare unadjusted PRS with target PRS to reweighted PRS with unadjusted SNPs and target PRS
############
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
# use for loop to read all values and indexes
for(i in 1:length(target_gwas)){
for(type in c('rho_restricted_rho_weighted_with_unadjusted','rho_fdr_restricted_rho_weighted_with_unadjusted','h2_restricted_rho_weighted_with_unadjusted','rho_restricted_rho_h2_weighted_with_unadjusted','rho_fdr_restricted_rho_h2_weighted_with_unadjusted','h2_restricted_rho_h2_weighted_with_unadjusted')){
pred_file<-NULL
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
target_pop_prev_i<-target_pop_prev[i]
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',target_gwas_i,'/prs.profiles'),
group='baseline')
pred_file<-rbind(pred_file, tmp)
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/prs.profiles'),
group='baseline')
pred_file<-rbind(pred_file, tmp)
# Add in weighted prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
}
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_5_',type,'.txt'), col.names=T, row.names=F, quote=F)
system(paste0('sbatch --mem 10G -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2_nested.R --pheno /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_5_',type,' --assoc T --outcome_pop_prev ',target_pop_prev_i,' --predictors /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_5_',type,'.txt'))
}
}
############
# Compare secondary PGS weighted and the remainder (seperately) to unadjusted secondary PGS
############
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
# use for loop to read all values and indexes
for(i in 1:length(target_gwas)){
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted')){
pred_file<-NULL
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
target_pop_prev_i<-target_pop_prev[i]
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/prs.profiles'),
group='standard')
pred_file<-rbind(pred_file, tmp)
# Add in weighted prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
# Add in remainder of weighted prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',gsub('restricted_.*','restricted_remainder',type),'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
}
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_6_',type,'.txt'), col.names=T, row.names=F, quote=F)
system(paste0('sbatch --mem 10G -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2_nested.R --pheno /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_6_',type,' --assoc T --outcome_pop_prev ',target_pop_prev_i,' --predictors /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_6_',type,'.txt'))
}
}
############
# Compare secondary PGS weighted and the remainder (seperately) +target PGS to unadjusted secondary PGS + target PGS
############
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
# use for loop to read all values and indexes
for(i in 1:length(target_gwas)){
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted')){
pred_file<-NULL
target_gwas_i<-target_gwas[i]
target_pheno_i<-target_pheno[i]
target_pop_prev_i<-target_pop_prev[i]
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',target_gwas_i,'/prs.profiles'),
group='baseline')
pred_file<-rbind(pred_file, tmp)
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',target_gwas_i,'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
for(secondary_gwas_i in secondary_gwas[secondary_gwas != target_gwas_i]){
# Add in prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/prs.profiles'),
group='baseline')
pred_file<-rbind(pred_file, tmp)
# Add in weighted prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',type,'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
# Add in remainder of weighted prs for target phenotype
tmp<-data.frame(predictors=paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/',secondary_gwas_i,'/',target_gwas_i,'/',gsub('restricted_.*','restricted_remainder',type),'/prs.profiles'),
group=type)
pred_file<-rbind(pred_file, tmp)
}
write.table(pred_file, paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_7_',type,'.txt'), col.names=T, row.names=F, quote=F)
system(paste0('sbatch --mem 10G -p brc,shared /users/k1806347/brc_scratch/Software/Rscript.sh /users/k1806347/brc_scratch/Software/MyGit/GenoPred/Scripts/Model_builder/Model_builder_V2_nested.R --pheno /users/k1806347/brc_scratch/Data/UKBB/Phenotype/PRS_comp_subset/UKBB.',target_pheno_i,'.txt --out /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_7_',type,' --assoc T --outcome_pop_prev ',target_pop_prev_i,' --predictors /users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_7_',type,'.txt'))
}
}3 Results
3.1 Heritability/Genetic correlation results
Show code
target_gwas<-c('DEPR06','BODY04','INTE03')
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
ncase<-c(116404,33640,20352,18381,41969,19099,NA,26676,NA,NA,32858,NA,60801,NA,NA,NA,NA)
ncon<-c(314990,43456,31358,27969,32066,34194,NA,132532,NA,NA,65839,NA,123504,NA,NA,NA,NA)
prev<-c(0.15,0.01,0.015,0.012,0.5,0.05,NA,0.05,NA,NA,0.13,NA,0.03,NA,NA,NA,NA)
# Read in LDSC results
scor = read.table("/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/sample_overlap/all.rg",header=T)
###
# Tabulate SNP-heritability results
###
hsq<-scor[,c("p1","p2","h2_obs","h2_obs_se")]
hsq<-hsq[hsq$p1 == hsq$p2,]
hsq$p2<-NULL
hsq$p1<-gsub('.cleaned.*','',gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/','',hsq$p1))
# Calculate p-value
hsq$p<-pnorm(-abs(hsq$h2_obs/hsq$h2_obs_se))
# Convert heritability to liability scale
h2l_R2 <- function(k, r2, p) {
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
h2l_R2 = C*r2 / (1 + C*theta*r2)
return(h2l_R2)
}
se_h2l_R2 <- function(k,h2,se, p) {
x= qnorm(1-k)
z= dnorm(x)
i=z/k
C= k*(1-k)*k*(1-k)/(z^2*p*(1-p))
theta= i*((p-k)/(1-k))*(i*((p-k)/(1-k))-x)
se_h2l_R2 = C*(1-h2*theta)*se
return(se_h2l_R2)
}
hsq<-hsq[match(secondary_gwas, hsq$p1),]
hsq$h2_liab<-hsq$h2_obs
hsq$h2_liab_se<-hsq$h2_obs_se
for(i in which(!is.na(prev))){
hsq$h2_liab[i]<-h2l_R2(k=prev[i], r2 = hsq$h2_obs[i], p = (ncase/(ncase+ncon))[i])
hsq$h2_liab_se[i]<-se_h2l_R2(k=prev[i], h2 = hsq$h2_obs[i], se=hsq$h2_obs_se[i], p = (ncase/(ncase+ncon))[i])
}
hsq$label<-paste0(round(hsq$h2_liab,3), "\n(", round(hsq$h2_liab_se, 3), ")")
###
# Tabulate rG estimates
###
rg<-scor[,c("p1","p2","rg","se")]
rg$p1<-gsub('.cleaned.*','',gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/','',rg$p1))
rg$p2<-gsub('.cleaned.*','',gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/GWAS_sumstats/','',rg$p2))
rg$label<-paste0(round(rg$rg,3), "\n(", round(rg$se, 3), ")")
# Calculate p
rg$p<-2*pnorm(-abs(rg$rg/rg$se))
###
# Tabulate local rG estimates
###
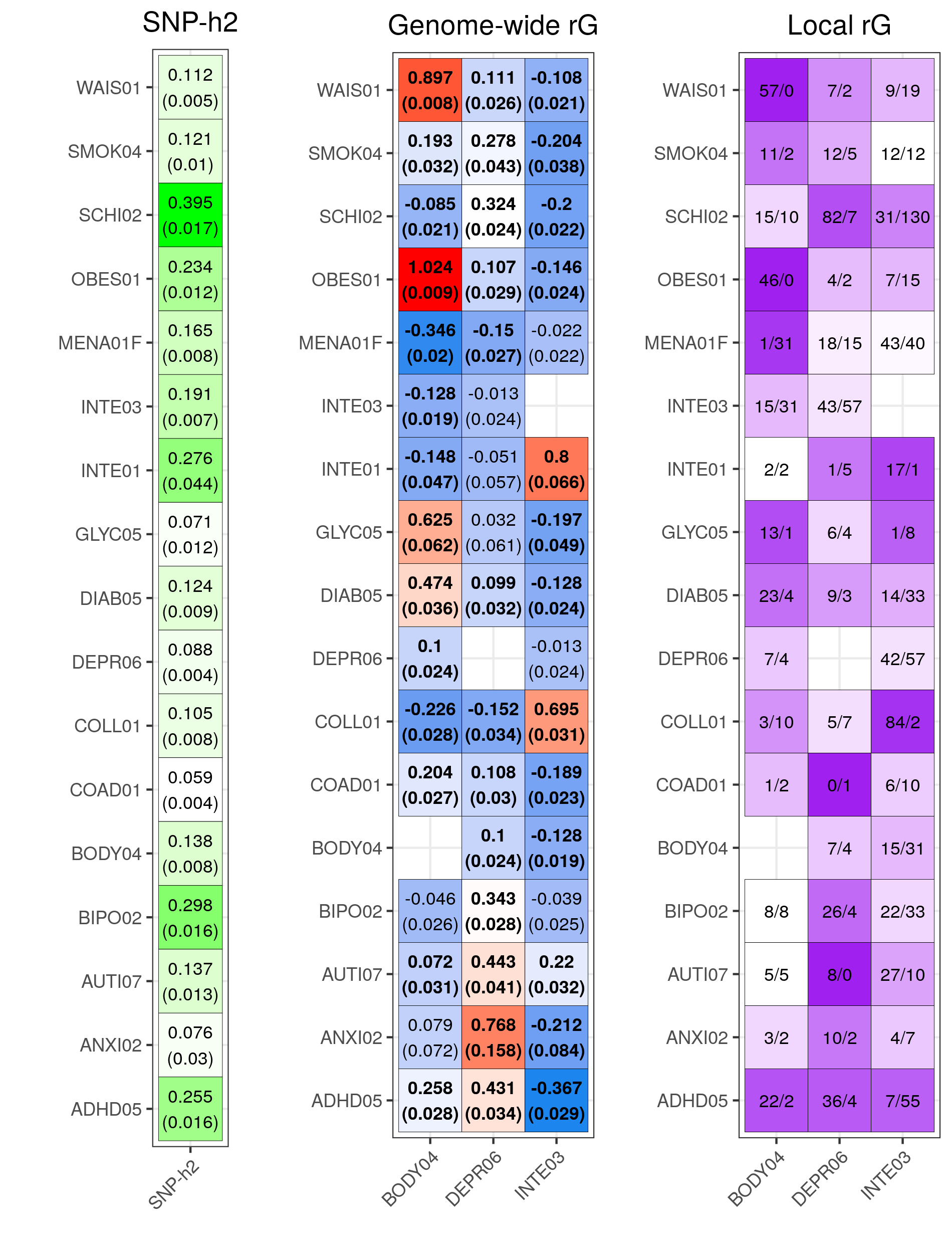
lava_res<-fread('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/lava_descript.csv')
lava_res$label<-paste0(lava_res$n_sig_fdr_loci_pos, '/', lava_res$n_sig_fdr_loci_neg)
lava_res$prop_conc<-lava_res$n_sig_fdr_loci_pos/lava_res$n_sig_fdr_loci
lava_res$prop_conc[lava_res$prop_conc < 0.5]<-1-lava_res$prop_conc[lava_res$prop_conc < 0.5]
###
# Create plot
###
library(ggplot2)
library(cowplot)
hsq_plot<-ggplot(data = hsq, aes(x = 'SNP-h2', y = p1)) +
theme_bw() +
geom_tile(aes(fill = h2_liab), colour = 'black') +
scale_fill_gradientn(colours=c("white","green"), na.value = 'grey',name = "hsq") +
theme(axis.text.x = element_text(angle = 45, hjust = 1),plot.title = element_text(hjust = 0.5), legend.position = "none") +
geom_text(aes(label=label), color="black", size=3) +
labs(x ="", y = "", title='SNP-h2') +
coord_fixed()
rg_tmp<-rg[rg$p1 %in% target_gwas,]
rg_tmp<-rg_tmp[rg_tmp$p1 != rg_tmp$p2,]
rg_plot<-ggplot(data = rg_tmp, aes(x = p1, y = p2)) +
theme_bw() +
geom_tile(aes(fill = rg), colour = 'black') +
scale_fill_gradientn(colours=c("dodgerblue2","white","red"), na.value = 'grey',name = "rG") +
theme(axis.text.x = element_text(angle = 45, hjust = 1),plot.title = element_text(hjust = 0.5), legend.position = "none") +
geom_text(aes(label=label), color="black", size=3) +
geom_text(data = rg_tmp[rg_tmp$p<0.05,], aes(x = p1, y = p2, label=label), color="black", size=3, fontface = "bold") +
labs(x ="", y = "", title='Genome-wide rG') +
coord_fixed()
local_rg_plot<-ggplot(data = lava_res, aes(x = target, y = secondary)) +
theme_bw() +
geom_tile(aes(fill = prop_conc), colour = 'black') +
scale_fill_gradientn(colours=c("white","purple"), na.value = 'grey',name = "Direction\nconcordance") +
theme(axis.text.x = element_text(angle = 45, hjust = 1),plot.title = element_text(hjust = 0.5), legend.position = "none") +
geom_text(aes(label=label), color="black", size=3) +
labs(x ="", y = "", title='Local rG') +
coord_fixed()
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/hsq_rg_local.png'), units='px', res=300, width=1900, height=2500)
plot_grid(plotlist=list(hsq_plot, rg_plot, local_rg_plot), rel_widths=c(0.9,1.2,1.2), nrow=1)
dev.off()Show SNP-h2, rG and local rG figure
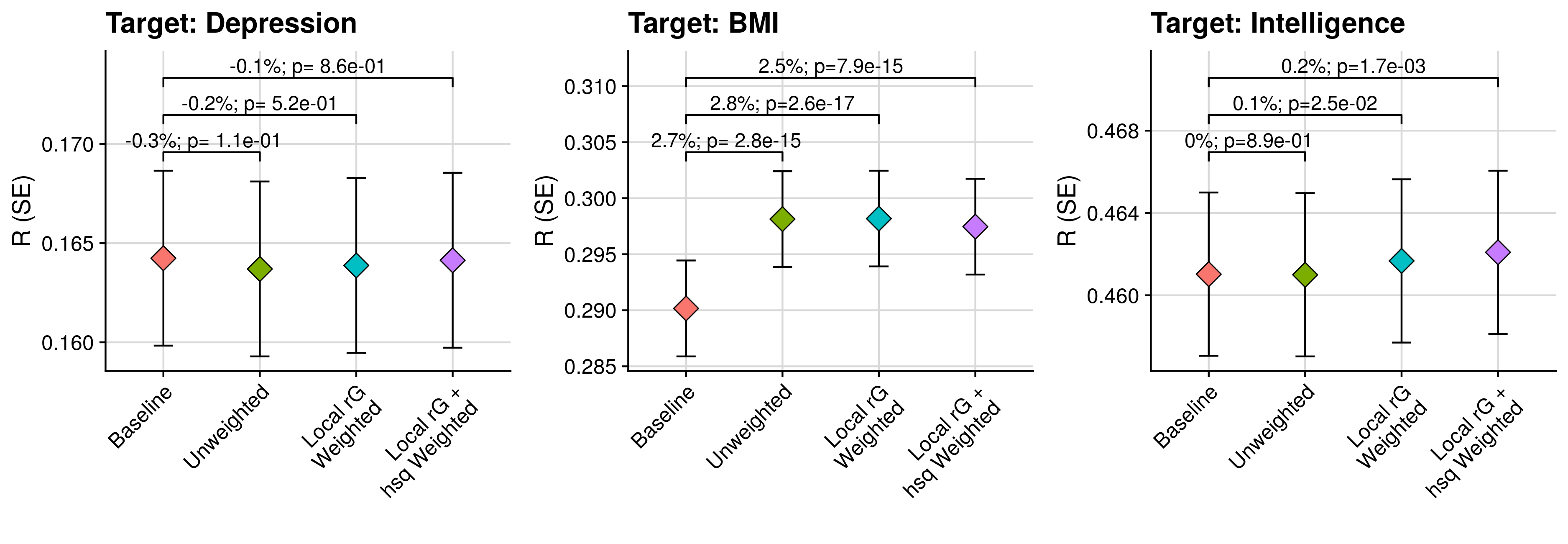
3.2 Comparison of models
Show code
library(data.table)
library(ggplot2)
library(cowplot)
target_gwas<-c('DEPR06','BODY04','INTE03')
target_pheno<-c('Depression','BMI','Intelligence')
target_pop_prev<-c(0.15,NA,NA)
secondary_gwas<-c('DEPR06','SCHI02','BIPO02','AUTI07','SMOK04','ADHD05','COLL01','DIAB05','ANXI02','MENA01F','OBES01','WAIS01','COAD01','GLYC05','INTE01','INTE03','BODY04')
#########
# Plot results testing gain from seconadary PGS over target PGS alone
#########
test<-1
eval_plot_list<-list()
for(i in 1:length(target_gwas)){
target_pheno_i<-target_pheno[i]
tmp_eval<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.pred_eval.txt'))
tmp_comp<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.pred_comp.txt'))
tmp_eval$Model<-c('Target\nPGS','Secondary\nPGS','All')
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
tmp_comp$R_diff_pval<-format(tmp_comp$R_diff_pval, scientific = TRUE, digits = 2)
tmp_comp$R_diff_pval[tmp_comp$R_diff_pval != "0.0e+00"]<-paste0('=',tmp_comp$R_diff_pval[tmp_comp$R_diff_pval != "0.0e+00"])
tmp_comp$R_diff_pval[tmp_comp$R_diff_pval == "0.0e+00"]<-'<1e-300'
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[2]), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[6]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_list[[paste0(i,'_',test)]]<-eval_plot
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'.png'), units='px', res=300, width=4600, height=1500)
print(plot_grid(plotlist=eval_plot_list, nrow=1))
dev.off()
#########
# Compare results from stratified/weighted secondary PGS
#########
test<-2
eval_plot_list<-list()
assoc_plot_brief_list<-list()
assoc_plot_brief_unres_list<-list()
lava_plot_list<-list()
for(i in 1:length(target_gwas)){
target_pheno_i<-target_pheno[i]
target_gwas_i<-target_gwas[i]
tmp_assoc<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.assoc.txt'))
tmp_assoc$Filter<-gsub('.restricted.*','',gsub('Group_','',tmp_assoc$Predictor))
tmp_assoc$Filter[tmp_assoc$Filter == 'rho']<-'rho-p < 0.05'
tmp_assoc$Filter[tmp_assoc$Filter == 'rho.fdr']<-'rho-fdr.p < 0.05'
tmp_assoc$Filter[tmp_assoc$Filter == 'h2']<-'hsq-p < 0.05'
tmp_assoc$Filter[tmp_assoc$Filter == 'All_group']<-'All'
tmp_assoc$Filter<-factor(tmp_assoc$Filter, levels=c('hsq-p < 0.05','rho-p < 0.05','rho-fdr.p < 0.05','All'))
tmp_assoc$Weighting<-gsub('.PredFile.*','',gsub('.*.restricted.','',tmp_assoc$Predictor))
tmp_assoc$Weighting[tmp_assoc$Weighting == 'unweighted']<-'Unweighted'
tmp_assoc$Weighting[tmp_assoc$Weighting == 'rho.weighted']<-'rho*BETA2'
tmp_assoc$Weighting[tmp_assoc$Weighting == 'rho.h2.weighted']<-"rho*BETA2/(hsq2/hsq1)"
tmp_assoc$Weighting<-factor(tmp_assoc$Weighting, levels=c('Unweighted','rho*BETA2','rho*BETA2/(hsq2/hsq1)','All'))
tmp_assoc$Test<-paste0(tmp_assoc$Filter,': ',tmp_assoc$Weighting)
tmp_assoc$Test<-factor(tmp_assoc$Test, levels=c("hsq-p < 0.05: Unweighted","rho-p < 0.05: Unweighted","rho-fdr.p < 0.05: Unweighted","hsq-p < 0.05: rho*BETA2","rho-p < 0.05: rho*BETA2","rho-fdr.p < 0.05: rho*BETA2","hsq-p < 0.05: rho*BETA2/(hsq2/hsq1)","rho-p < 0.05: rho*BETA2/(hsq2/hsq1)","rho-fdr.p < 0.05: rho*BETA2/(hsq2/hsq1)"))
tmp_pred<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.txt'))
gsub('/','',gsub(paste0(target_gwas_i,'.*'),'',gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/','',tmp_pred$predictors)))
tmp_assoc$GWAS<-gsub('/','',gsub(paste0(target_gwas_i,'.*'),'',gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/','',tmp_pred$predictors)))
tmp_assoc$GWAS<-factor(tmp_assoc$GWAS, levels=unique(tmp_assoc$GWAS))
assoc_plot<-ggplot(tmp_assoc, aes(x=GWAS, y=abs(BETA), fill=Weighting)) +
geom_bar(stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=abs(BETA)-SE, ymax=abs(BETA)+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="Absolute-R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)) +
facet_grid(rows = vars(Filter))
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.png'), units='px', res=300, width=4500, height=2000)
print(assoc_plot)
dev.off()
assoc_plot_brief<-ggplot(tmp_assoc[tmp_assoc$Filter == 'rho-p < 0.05',], aes(x=GWAS, y=abs(BETA), fill=Weighting)) +
geom_bar(stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=abs(BETA)-SE, ymax=abs(BETA)+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="Absolute-R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
assoc_plot_brief_list[[i]]<-assoc_plot_brief
# Compare with unrestricted PGS associations
tmp_assoc_unres<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',1,'.assoc.txt'))
tmp_pred_unres<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',1,'.txt'))
tmp_assoc_unres$GWAS<-gsub('/prs.profiles','',gsub('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/prs/','',tmp_pred_unres$predictors))
tmp_assoc_unres$GWAS<-factor(tmp_assoc_unres$GWAS, levels=unique(tmp_assoc_unres$GWAS))
tmp_assoc_unres$Filter<-'Standard'
tmp_assoc_unres$Weighting<-'Standard'
tmp_assoc_unres$Test<-'Standard'
tmp_assoc_unres<-tmp_assoc_unres[tmp_assoc_unres$GWAS != target_gwas_i,]
tmp_assoc_both<-rbind(tmp_assoc_unres,tmp_assoc)
assoc_plot_brief_unres<-ggplot(tmp_assoc_both[tmp_assoc_both$Filter == 'rho-p < 0.05' | tmp_assoc_both$Filter == 'Standard',], aes(x=GWAS, y=abs(BETA), fill=Weighting)) +
geom_bar(stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=abs(BETA)-SE, ymax=abs(BETA)+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="Absolute-R (SE)", x='', fill='Type') +
theme_half_open() +
background_grid() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
assoc_plot_brief_unres_list[[i]]<-assoc_plot_brief_unres
lava_res<-fread('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/lava/results/lava_descript.csv')
lava_res<-lava_res[lava_res$target == target_gwas_i,]
tmp_assoc_com<-merge(tmp_assoc[tmp_assoc$Test == 'rho-p < 0.05: Unweighted',],tmp_assoc[tmp_assoc$Test == 'rho-p < 0.05: rho*BETA2',], by='GWAS')
tmp_assoc_com$R_diff<-abs(tmp_assoc_com$BETA.y)-abs(tmp_assoc_com$BETA.x)
tmp_assoc_com<-tmp_assoc_com[,c('GWAS','R_diff'),with=F]
lava_res<-merge(tmp_assoc_com, lava_res, by.x='GWAS', by.y='secondary')
lava_res$prop_conc<-lava_res$n_sig_loci_pos/lava_res$n_sig_loci
lava_res$prop_conc[lava_res$prop_conc < 0.5]<-1-lava_res$prop_conc[lava_res$prop_conc < 0.5]
lava_res$non_uniform<-1-lava_res$prop_conc
lava_plot<-ggplot(lava_res, aes(x=non_uniform, y=R_diff)) +
geom_point(aes(size=lava_res$n_sig_loci)) +
scale_size_continuous(range = c(1, 3)) +
labs(title=paste0('Target: ',target_pheno_i), y="R improvement\nwhen weighting", x='Proportion Inconsistent', size='N loci') +
theme_half_open() +
background_grid()
lava_plot_list[[i]]<-lava_plot
tmp_eval<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.pred_eval.txt'))
tmp_comp<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.pred_comp.txt'))
tmp_eval$Filter<-gsub('.restricted.*','',tmp_eval$Model)
tmp_eval$Filter[tmp_eval$Filter == 'rho']<-'rho-p < 0.05'
tmp_eval$Filter[tmp_eval$Filter == 'rho.fdr']<-'rho-fdr.p < 0.05'
tmp_eval$Filter[tmp_eval$Filter == 'h2']<-'hsq-p < 0.05'
tmp_eval$Filter[tmp_eval$Filter == 'All_group']<-'All'
tmp_eval$Filter<-factor(tmp_eval$Filter, levels=c('hsq-p < 0.05','rho-p < 0.05','rho-fdr.p < 0.05','All'))
tmp_eval$Weighting<-gsub('_group','',gsub('.*.restricted.','',tmp_eval$Model))
tmp_eval$Weighting[tmp_eval$Weighting == 'unweighted']<-'Unweighted'
tmp_eval$Weighting[tmp_eval$Weighting == 'rho.weighted']<-'rho*BETA2'
tmp_eval$Weighting[tmp_eval$Weighting == 'rho.h2.weighted']<-"rho*BETA2/(hsq2/hsq1)"
tmp_eval$Weighting<-factor(tmp_eval$Weighting, levels=c('Unweighted','rho*BETA2','rho*BETA2/(hsq2/hsq1)','All'))
eval_plot<-ggplot(tmp_eval, aes(x=Filter, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Weighting), stat="identity", position=position_dodge()) +
geom_errorbar(data=tmp_eval, aes(ymin=R-SE, ymax=R+SE, group=Weighting), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid()
eval_plot_list[[paste0(i,'_',test)]]<-eval_plot
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'.png'), units='px', res=300, width=2500, height=2500)
print(plot_grid(plotlist=eval_plot_list, ncol=1))
dev.off()
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'_assoc.png'), units='px', res=300, width=2200, height=2500)
print(plot_grid(plotlist=assoc_plot_brief_list, ncol=1))
dev.off()
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'_assoc_with_unres.png'), units='px', res=300, width=2200, height=2500)
print(plot_grid(plotlist=assoc_plot_brief_unres_list, ncol=1))
dev.off()
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'_lava.png'), units='px', res=300, width=1500, height=2000)
print(plot_grid(plotlist=lava_plot_list, ncol=1))
dev.off()
#########
# Plot results testing gain from seconadary PGS over target PGS alone
#########
test<-3
eval_plot_all_list<-list()
tmp_eval_all<-list()
tmp_comp_all<-list()
for(i in 1:length(target_gwas)){
eval_plot_list<-list()
tmp_eval_i<-list()
tmp_comp_i<-list()
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted')){
target_pheno_i<-target_pheno[i]
tmp_eval<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_eval.txt'))
tmp_comp<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_comp.txt'))
tmp_comp$R_diff_pval<-format(tmp_comp$R_diff_pval, scientific = TRUE, digits = 2)
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0]<-paste0('=',tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0])
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) == 0]<-'<1e-300'
Filter<-gsub('_restricted.*','', type)
if(Filter == 'rho'){Filter<-'rho-p < 0.05'}
if(Filter == 'rho.fdr'){Filter<-'rho-fdr.p < 0.05'}
if(Filter == 'h2'){Filter<-'hsq-p < 0.05'}
Weighting<-gsub('_group','',gsub('.*restricted_','', type))
if(Weighting == 'unweighted'){Weighting<-'Unweighted'}
if(Weighting == 'rho_weighted'){Weighting<-'local rg\nweighted'}
if(Weighting == 'rho_h2_weighted'){Weighting<-'local rg +\nhsq weighted'}
tmp_eval$Model<-c('Target PGS +\nSecondary PGS',paste0(Filter,'\n',Weighting),'All')
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
if(grepl('rho_restricted',type)){
tmp_eval$Model<-gsub('rho-p < 0.05\n','',tmp_eval$Model)
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_all_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
tmp_eval$Phenotype<-target_pheno_i
tmp_eval$Type<-type
tmp_eval_i[[paste0(i,'_',test,'_',type)]]<-tmp_eval
tmp_comp$Phenotype<-target_pheno_i
tmp_comp$Type<-type
tmp_comp_i[[paste0(i,'_',test,'_',type)]]<-tmp_comp
}
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.png'), units='px', res=300, width=5000, height=3000)
print(plot_grid(plotlist=eval_plot_list, ncol=3))
dev.off()
tmp_eval_all[[i]]<-do.call(rbind,tmp_eval_i)
tmp_eval_all[[i]]$Ncase<-NULL
tmp_eval_all[[i]]$Ncont<-NULL
tmp_eval_all[[i]]$R2o<-NULL
tmp_eval_all[[i]]$R2l<-NULL
tmp_comp_all[[i]]<-do.call(rbind,tmp_comp_i)
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'.png'), units='px', res=300, width=4750, height=3000)
print(plot_grid(plotlist=eval_plot_all_list, ncol=3))
dev.off()
###
# Combine h2_resricted results and make simplified plot
###
tmp_eval_all_tab<-do.call(rbind, tmp_eval_all)
tmp_comp_all_tab<-do.call(rbind, tmp_comp_all)
# Remove secondary PGS only rows
tmp_eval_all_tab<-tmp_eval_all_tab[tmp_eval_all_tab$Model == 'Target PGS +\nSecondary PGS' | tmp_eval_all_tab$Model == 'All' ,]
tmp_comp_all_tab<-tmp_comp_all_tab[(tmp_comp_all_tab$Model_1 == 'All' & tmp_comp_all_tab$Model_2 == 'baseline'),]
# Rename 'All' Models
tmp_eval_all_tab$Model<-as.character(tmp_eval_all_tab$Model)
tmp_eval_all_tab$Model[tmp_eval_all_tab$Model == 'All']<-paste0(tmp_eval_all_tab$Model[tmp_eval_all_tab$Model == 'All'],'_',tmp_eval_all_tab$Type[tmp_eval_all_tab$Model == 'All'])
# Remove duplicated Target PGS + Secondary PGS results
tmp_eval_all_tab<-tmp_eval_all_tab[!duplicated(paste0(tmp_eval_all_tab$Model,tmp_eval_all_tab$Phenotype)),]
# Rename models
tmp_eval_all_tab$Model<-rep(c('Baseline','Unweighted','Local rG\nWeighted','Local rG +\nhsq Weighted'),3)
tmp_eval_all_tab$Model<-factor(tmp_eval_all_tab$Model, levels=c('Baseline','Unweighted','Local rG\nWeighted','Local rG +\nhsq Weighted'))
eval_plot_brief_all_list<-NULL
for(i in 1:length(target_gwas)){
target_pheno_i<-target_pheno[i]
tmp_eval_all_tab_i<-tmp_eval_all_tab[tmp_eval_all_tab$Phenotype == target_pheno_i,]
tmp_comp_all_tab_i<-tmp_comp_all_tab[tmp_comp_all_tab$Phenotype == target_pheno_i,]
eval_plot<-ggplot(tmp_eval_all_tab_i, aes(x=Model, y=R)) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
geom_point(aes(fill=Model), stat="identity", position=position_dodge(1), size=5, shape=23, colour='black') +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none", axis.text.x = element_text(angle = 45, hjust=1))
ylim_range<-max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)-min(tmp_eval_all_tab_i$R-tmp_eval_all_tab_i$SE)
paths_1<-data.frame(x=c(1,1,2,2),
y=c(max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(1*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(1*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(1*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(1*(ylim_range/5))))
paths_3<-data.frame(x=c(1,1,4,4),
y=c(max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(2*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(2*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(2*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(2*(ylim_range/5))))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(ylim_range/15),label=paste0(round((tmp_comp_all_tab_i$R_diff/tmp_comp_all_tab_i$Model_2_R)[1] * 100,1),'%; p', tmp_comp_all_tab_i$R_diff_pval[1],' '), size=4) +
annotate("text",x=2,y=max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(ylim_range/15)+(1*(ylim_range/5)),label=paste0(round((tmp_comp_all_tab_i$R_diff/tmp_comp_all_tab_i$Model_2_R)[2] * 100,1),'%; p', tmp_comp_all_tab_i$R_diff_pval[2]), size=4) +
annotate("text",x=2.5,y=max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(ylim_range/15)+(2*(ylim_range/5)),label=paste0(round((tmp_comp_all_tab_i$R_diff/tmp_comp_all_tab_i$Model_2_R)[3] * 100,1),'%; p', tmp_comp_all_tab_i$R_diff_pval[3]), size=4)
eval_plot_brief_all_list[[i]]<-eval_plot
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'_brief.png'), units='px', res=300, width=3750, height=1300)
print(plot_grid(plotlist=eval_plot_brief_all_list, ncol=3))
dev.off()
#########
# Plot results testing gain from reweighted with unadjusted seconadary PGS
#########
for(test in 4:5){
eval_plot_all_list<-list()
for(i in 1:length(target_gwas)){
eval_plot_list<-list()
for(type in c('rho_restricted_rho_weighted_with_unadjusted','rho_fdr_restricted_rho_weighted_with_unadjusted','h2_restricted_rho_weighted_with_unadjusted','rho_restricted_rho_h2_weighted_with_unadjusted','rho_fdr_restricted_rho_h2_weighted_with_unadjusted','h2_restricted_rho_h2_weighted_with_unadjusted')){
target_pheno_i<-target_pheno[i]
tmp_eval<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_eval.txt'))
tmp_comp<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_comp.txt'))
tmp_comp$R_diff_pval<-format(tmp_comp$R_diff_pval, scientific = TRUE, digits = 2)
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0]<-paste0('=',tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0])
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) == 0]<-'<1e-300'
Filter<-gsub('_restricted.*','', type)
if(Filter == 'rho'){Filter<-'rho-p < 0.05'}
if(Filter == 'rho.fdr'){Filter<-'rho-fdr.p < 0.05'}
if(Filter == 'h2'){Filter<-'hsq-p < 0.05'}
Weighting<-gsub('_with_unadjusted','',gsub('.*restricted_','', type))
if(Weighting == 'unweighted'){Weighting<-'Unweighted'}
if(Weighting == 'rho_weighted'){Weighting<-'local rg\nweighted'}
if(Weighting == 'rho_h2_weighted'){Weighting<-'local rg +\nhsq weighted'}
tmp_eval$Model<-c('Standard',paste0(Filter,'\n',Weighting),'All')
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
if(grepl('rho_restricted',type)){
tmp_eval$Model<-gsub('rho-p < 0.05\n','',tmp_eval$Model)
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_all_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
}
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.png'), units='px', res=300, width=5000, height=3000)
print(plot_grid(plotlist=eval_plot_list, ncol=3))
dev.off()
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'.png'), units='px', res=300, width=4750, height=3000)
print(plot_grid(plotlist=eval_plot_all_list, ncol=3))
dev.off()
}
#########
# Plot results testing gain from seconadary PGS over target PGS alone
#########
test<-6
eval_plot_all_list<-list()
tmp_eval_all<-list()
tmp_comp_all<-list()
for(i in 1:length(target_gwas)){
eval_plot_list<-list()
tmp_eval_i<-list()
tmp_comp_i<-list()
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted')){
target_pheno_i<-target_pheno[i]
tmp_eval<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_eval.txt'))
tmp_comp<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_comp.txt'))
tmp_comp$R_diff_pval<-format(tmp_comp$R_diff_pval, scientific = TRUE, digits = 2)
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0]<-paste0('=',tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0])
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) == 0]<-'<1e-300'
Filter<-gsub('_restricted.*','', type)
if(Filter == 'rho'){Filter<-'rho-p < 0.05'}
if(Filter == 'rho.fdr'){Filter<-'rho-fdr.p < 0.05'}
if(Filter == 'h2'){Filter<-'hsq-p < 0.05'}
Weighting<-gsub('_group','',gsub('.*restricted_','', type))
if(Weighting == 'unweighted'){Weighting<-'Unweighted'}
if(Weighting == 'rho_weighted'){Weighting<-'local rg\nweighted'}
if(Weighting == 'rho_h2_weighted'){Weighting<-'local rg +\nhsq weighted'}
tmp_eval$Model<-c('Standard',paste0(Filter,'\n',Weighting),'All')
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
if(grepl('rho_restricted',type)){
tmp_eval$Model<-gsub('rho-p < 0.05\n','',tmp_eval$Model)
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_all_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
tmp_eval$Phenotype<-target_pheno_i
tmp_eval$Type<-type
tmp_eval_i[[paste0(i,'_',test,'_',type)]]<-tmp_eval
tmp_comp$Phenotype<-target_pheno_i
tmp_comp$Type<-type
tmp_comp_i[[paste0(i,'_',test,'_',type)]]<-tmp_comp
}
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.png'), units='px', res=300, width=5000, height=3000)
print(plot_grid(plotlist=eval_plot_list, ncol=3))
dev.off()
tmp_eval_all[[i]]<-do.call(rbind,tmp_eval_i)
tmp_eval_all[[i]]$Ncase<-NULL
tmp_eval_all[[i]]$Ncont<-NULL
tmp_eval_all[[i]]$R2o<-NULL
tmp_eval_all[[i]]$R2l<-NULL
tmp_comp_all[[i]]<-do.call(rbind,tmp_comp_i)
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'.png'), units='px', res=300, width=4750, height=3000)
print(plot_grid(plotlist=eval_plot_all_list, ncol=3))
dev.off()
# Make brief version of the plot
tmp_eval_all_tab<-do.call(rbind, tmp_eval_all)
tmp_comp_all_tab<-do.call(rbind, tmp_comp_all)
# Remove secondary PGS only rows
tmp_eval_all_tab<-tmp_eval_all_tab[tmp_eval_all_tab$Model == 'Standard' | tmp_eval_all_tab$Model == 'All' ,]
tmp_comp_all_tab<-tmp_comp_all_tab[(tmp_comp_all_tab$Model_1 == 'All' & tmp_comp_all_tab$Model_2 == 'standard'),]
# Rename 'All' Models
tmp_eval_all_tab$Model<-as.character(tmp_eval_all_tab$Model)
tmp_eval_all_tab$Model[tmp_eval_all_tab$Model == 'All']<-paste0(tmp_eval_all_tab$Model[tmp_eval_all_tab$Model == 'All'],'_',tmp_eval_all_tab$Type[tmp_eval_all_tab$Model == 'All'])
# Remove duplicated Target PGS + Secondary PGS results
tmp_eval_all_tab<-tmp_eval_all_tab[!duplicated(paste0(tmp_eval_all_tab$Model,tmp_eval_all_tab$Phenotype)),]
# Rename models
tmp_eval_all_tab$Model<-rep(c('Standard','Unweighted','Local rG\nWeighted','Local rG +\nhsq Weighted'),3)
tmp_eval_all_tab$Model<-factor(tmp_eval_all_tab$Model, levels=c('Standard','Unweighted','Local rG\nWeighted','Local rG +\nhsq Weighted'))
eval_plot_brief_all_list<-NULL
for(i in 1:length(target_gwas)){
target_pheno_i<-target_pheno[i]
tmp_eval_all_tab_i<-tmp_eval_all_tab[tmp_eval_all_tab$Phenotype == target_pheno_i,]
tmp_comp_all_tab_i<-tmp_comp_all_tab[tmp_comp_all_tab$Phenotype == target_pheno_i,]
eval_plot<-ggplot(tmp_eval_all_tab_i, aes(x=Model, y=R)) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
geom_point(aes(fill=Model), stat="identity", position=position_dodge(1), size=5, shape=23, colour='black') +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none", axis.text.x = element_text(angle = 45, hjust=1))
ylim_range<-max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)-min(tmp_eval_all_tab_i$R-tmp_eval_all_tab_i$SE)
paths_1<-data.frame(x=c(1,1,2,2),
y=c(max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(1*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(1*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(1*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(1*(ylim_range/5))))
paths_3<-data.frame(x=c(1,1,4,4),
y=c(max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(2*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(2*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(2*(ylim_range/5)),
max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/20)+(2*(ylim_range/5))))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(ylim_range/15),label=paste0(round((tmp_comp_all_tab_i$R_diff/tmp_comp_all_tab_i$Model_2_R)[1] * 100,1),'%; p', tmp_comp_all_tab_i$R_diff_pval[1],' '), size=4) +
annotate("text",x=2,y=max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(ylim_range/15)+(1*(ylim_range/5)),label=paste0(round((tmp_comp_all_tab_i$R_diff/tmp_comp_all_tab_i$Model_2_R)[2] * 100,1),'%; p', tmp_comp_all_tab_i$R_diff_pval[2]), size=4) +
annotate("text",x=2.5,y=max(tmp_eval_all_tab_i$R+tmp_eval_all_tab_i$SE)+(ylim_range/10)+(ylim_range/15)+(2*(ylim_range/5)),label=paste0(round((tmp_comp_all_tab_i$R_diff/tmp_comp_all_tab_i$Model_2_R)[3] * 100,1),'%; p', tmp_comp_all_tab_i$R_diff_pval[3]), size=4)
eval_plot_brief_all_list[[i]]<-eval_plot
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'_brief.png'), units='px', res=300, width=3750, height=1300)
print(plot_grid(plotlist=eval_plot_brief_all_list, ncol=3))
dev.off()
#########
# Plot results testing gain from seconadary PGS over target PGS alone
#########
test<-7
eval_plot_all_list<-list()
for(i in 1:length(target_gwas)){
eval_plot_list<-list()
for(type in c('rho_restricted_unweighted','rho_fdr_restricted_unweighted','h2_restricted_unweighted','rho_restricted_rho_weighted','rho_fdr_restricted_rho_weighted','h2_restricted_rho_weighted','rho_restricted_rho_h2_weighted','rho_fdr_restricted_rho_h2_weighted','h2_restricted_rho_h2_weighted')){
target_pheno_i<-target_pheno[i]
tmp_eval<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_eval.txt'))
tmp_comp<-fread(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'_',type,'.pred_comp.txt'))
tmp_comp$R_diff_pval<-format(tmp_comp$R_diff_pval, scientific = TRUE, digits = 2)
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0]<-paste0('=',tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) != 0])
tmp_comp$R_diff_pval[as.numeric(tmp_comp$R_diff_pval) == 0]<-'<1e-300'
Filter<-gsub('_restricted.*','', type)
if(Filter == 'rho'){Filter<-'rho-p < 0.05'}
if(Filter == 'rho.fdr'){Filter<-'rho-fdr.p < 0.05'}
if(Filter == 'h2'){Filter<-'hsq-p < 0.05'}
Weighting<-gsub('_group','',gsub('.*restricted_','', type))
if(Weighting == 'unweighted'){Weighting<-'Unweighted'}
if(Weighting == 'rho_weighted'){Weighting<-'local rg\nweighted'}
if(Weighting == 'rho_h2_weighted'){Weighting<-'local rg +\nhsq weighted'}
tmp_eval$Model<-c('Target PGS +\nSecondary PGS',paste0(Filter,'\n',Weighting),'All')
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
if(grepl('rho_restricted',type)){
tmp_eval$Model<-gsub('rho-p < 0.05\n','',tmp_eval$Model)
tmp_eval$Model<-factor(tmp_eval$Model, levels=tmp_eval$Model)
eval_plot<-ggplot(tmp_eval, aes(x=Model, y=R)) +
geom_bar(data=tmp_eval, aes(fill=Model), stat="identity", position=position_dodge()) +
geom_errorbar(aes(ymin=R-SE, ymax=R+SE), width=.2, position=position_dodge(.9)) +
labs(title=paste0('Target: ',target_pheno_i), y="R (SE)", x='') +
theme_half_open() +
background_grid() +
theme(legend.position = "none")
if(min(tmp_eval$R-tmp_eval$SE) < 0){
ylim_range<-max(tmp_eval$R+tmp_eval$SE)-min(tmp_eval$R-tmp_eval$SE)
} else {
ylim_range<-max(tmp_eval$R+tmp_eval$SE)
}
paths_1<-data.frame(x=c(1,1,1.95,1.95),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_2<-data.frame(x=c(2.05,2.05,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)))
paths_3<-data.frame(x=c(1,1,3,3),
y=c(max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5),
max(tmp_eval$R+tmp_eval$SE)+(ylim_range/20)+(ylim_range/5)))
eval_plot<-eval_plot +
geom_path(data=paths_1, aes(x=x,y=y), colour='black') +
geom_path(data=paths_2, aes(x=x,y=y), colour='black') +
geom_path(data=paths_3, aes(x=x,y=y), colour='black')
eval_plot<-eval_plot +
annotate("text",x=1.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[4] * 100,1),'%; p', tmp_comp$R_diff_pval[4],' '), size=4) +
annotate("text",x=2.5,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[8] * 100,1),'%; p', tmp_comp$R_diff_pval[8]), size=4) +
annotate("text",x=2,y=max(tmp_eval$R+tmp_eval$SE)+(ylim_range/10)+(ylim_range/5)+(ylim_range/10),label=paste0(round((tmp_comp$R_diff/tmp_comp$Model_2_R)[7] * 100,1),'%; p', tmp_comp$R_diff_pval[7]), size=4)
eval_plot_all_list[[paste0(i,'_',test,'_',type)]]<-eval_plot
}
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/',target_pheno_i,'/test_',test,'.png'), units='px', res=300, width=5000, height=3000)
print(plot_grid(plotlist=eval_plot_list, ncol=3))
dev.off()
}
png(paste0('/users/k1806347/brc_scratch/Analyses/local_rg_pgs/assoc/test_',test,'.png'), units='px', res=300, width=4750, height=3000)
print(plot_grid(plotlist=eval_plot_all_list, ncol=3))
dev.off()Show gain provided by secondary PGS over target PGS
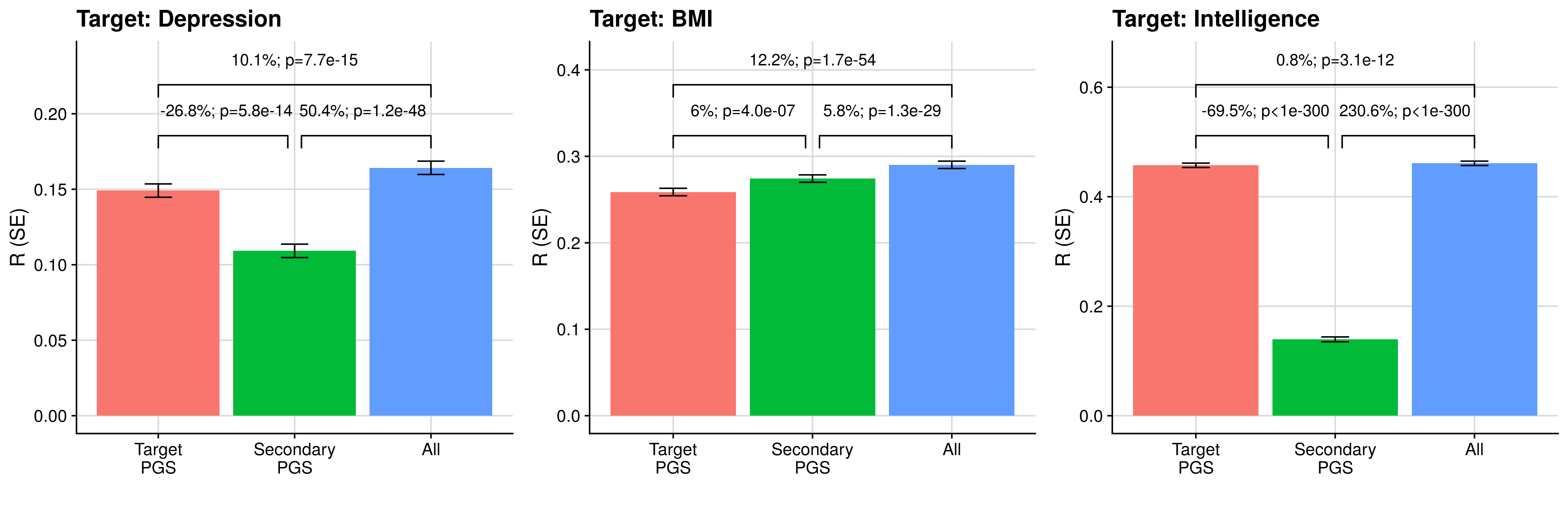
Show association between phenotype and each reweighted PGS
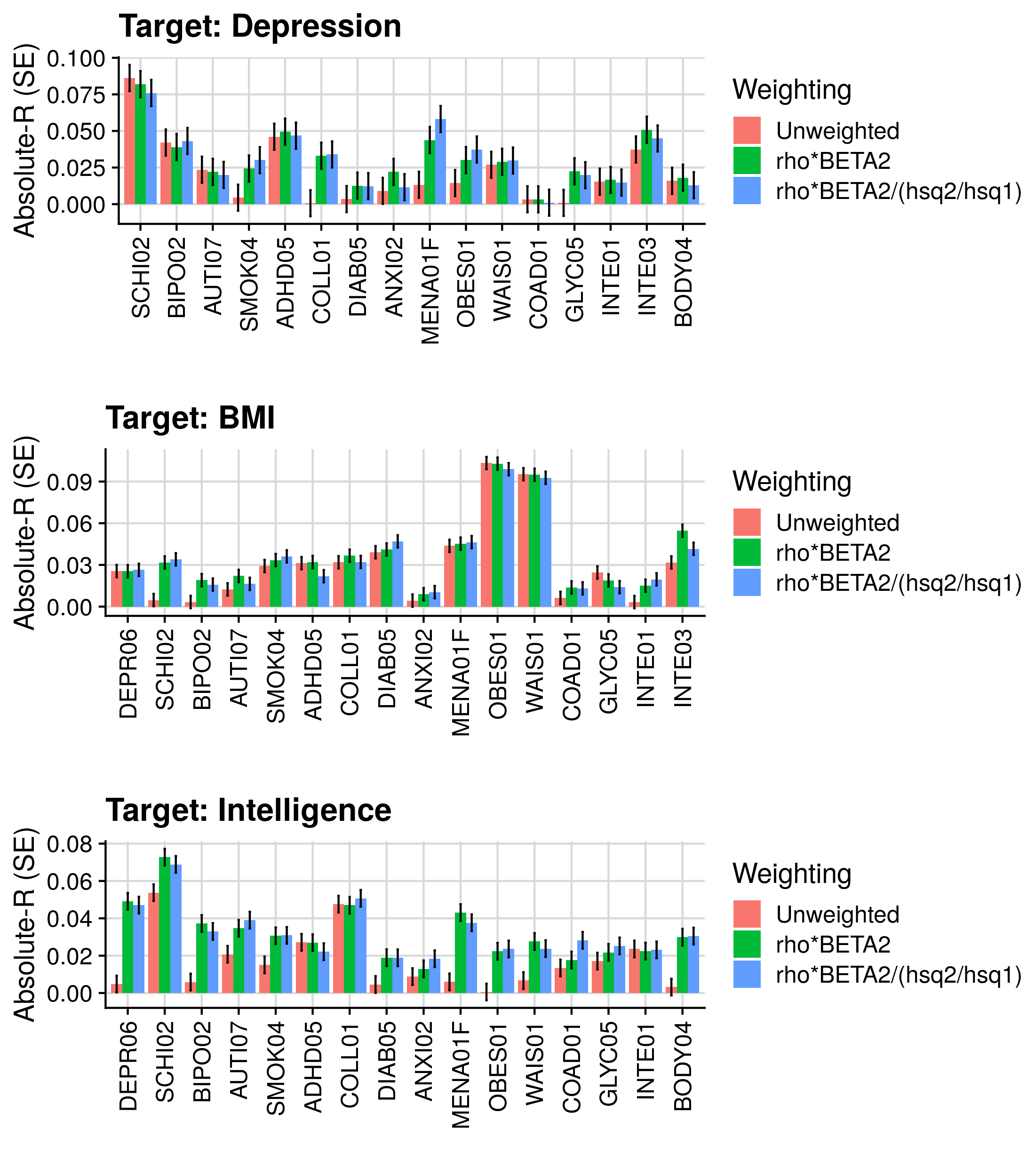
Show relationship between gain in secondary PGS prediction and inconsistency of local rG directions
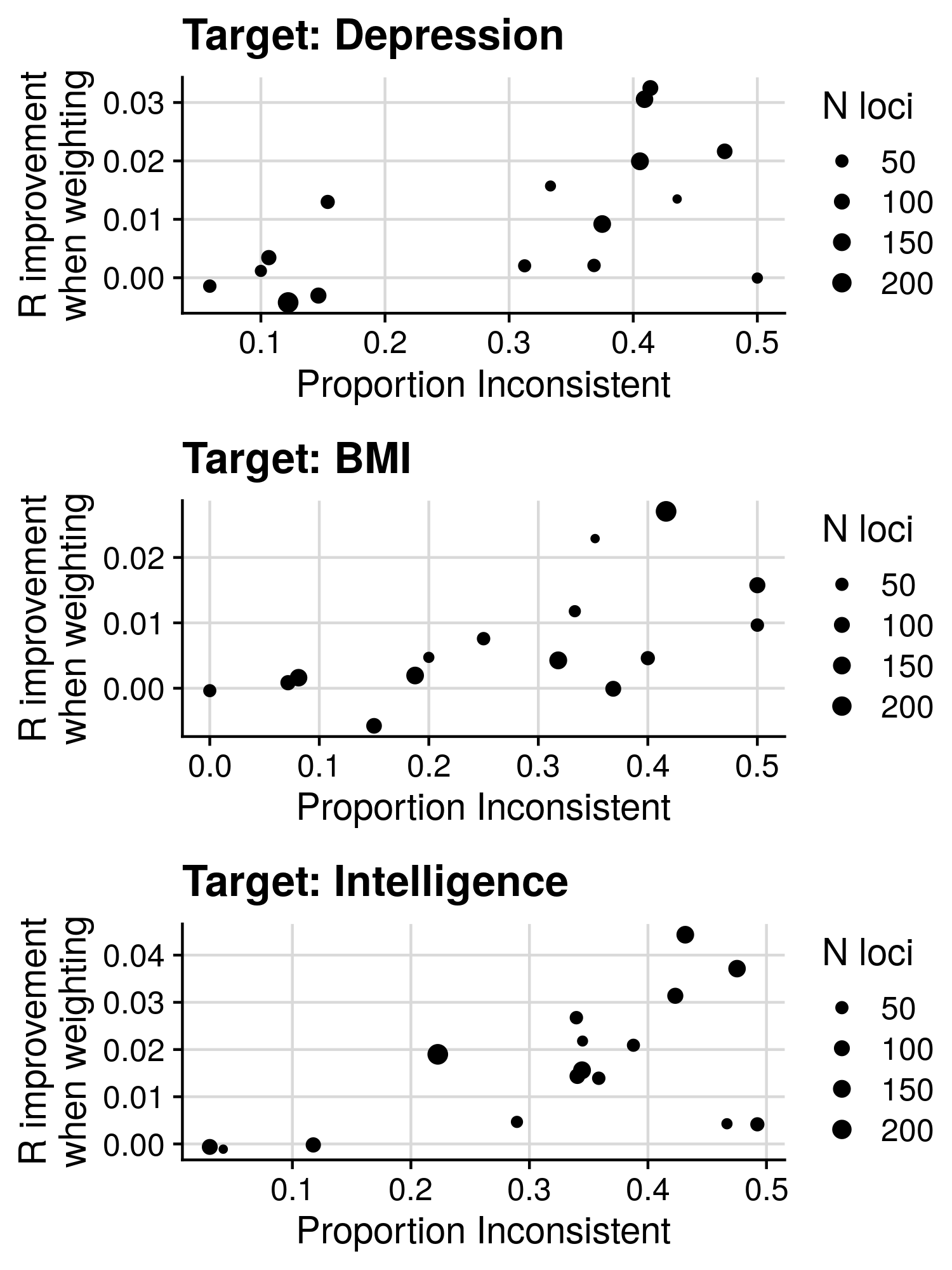
Show gain from local hsq/rG weighted secondary PGS over standard secondary PGS
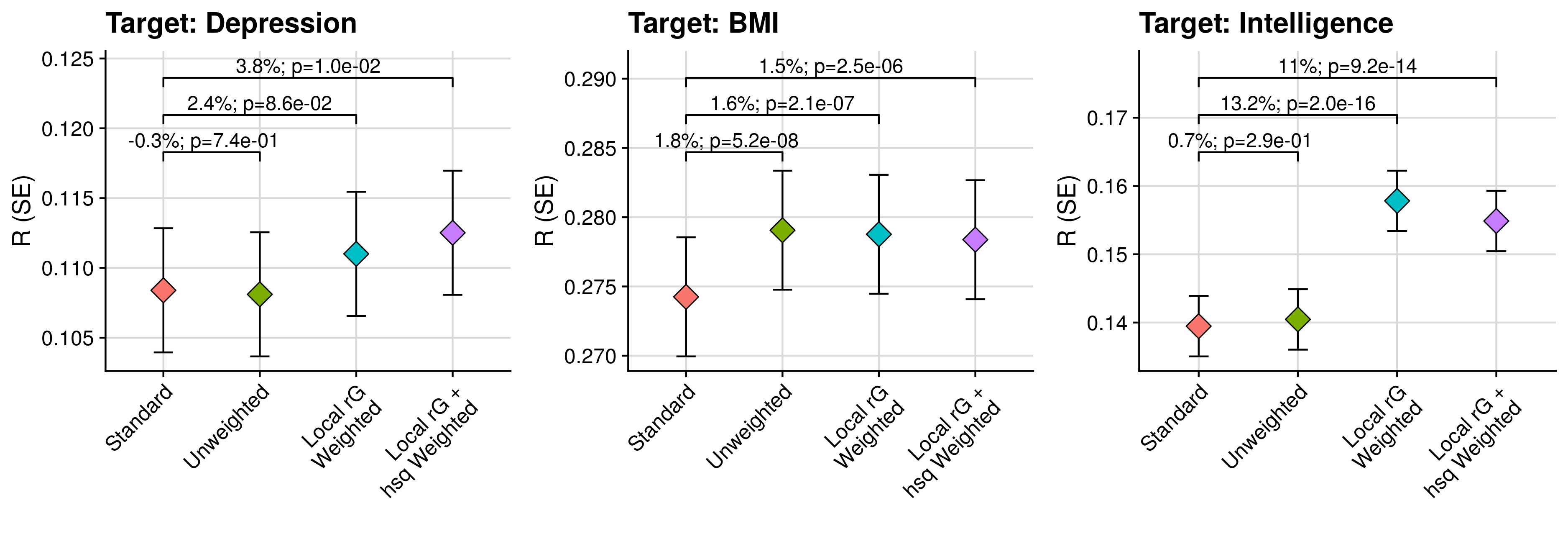
Show gain from local hsq/rG weighted PGS over target PGS and standard secondary PGS