{
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# StackingCVRegressor"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"An ensemble-learning meta-regressor for stacking regression"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"> from mlxtend.regressor import StackingCVRegressor"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Overview"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Stacking is an ensemble learning technique to combine multiple regression models via a meta-regressor. The `StackingCVRegressor` extends the standard stacking algorithm (implemented as [`StackingRegressor`](StackingRegressor.md)) using out-of-fold predictions to prepare the input data for the level-2 classifier.\n",
"\n",
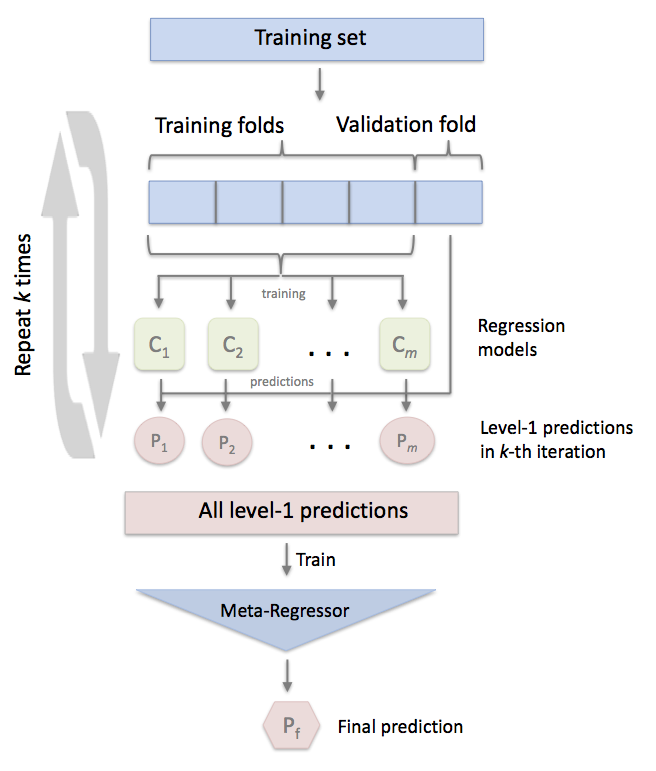
"In the standard stacking procedure, the first-level regressors are fit to the same training set that is used prepare the inputs for the second-level regressor, which may lead to overfitting. The `StackingCVRegressor`, however, uses the concept of out-of-fold predictions: the dataset is split into k folds, and in k successive rounds, k-1 folds are used to fit the first level regressor. In each round, the first-level regressors are then applied to the remaining 1 subset that was not used for model fitting in each iteration. The resulting predictions are then stacked and provided -- as input data -- to the second-level regressor. After the training of the `StackingCVRegressor`, the first-level regressors are fit to the entire dataset for optimal predicitons."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
""
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### References\n"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"- Breiman, Leo. \"[Stacked regressions.](http://link.springer.com/article/10.1023/A:1018046112532#page-1)\" Machine learning 24.1 (1996): 49-64.\n",
"- Analogous implementation: [`StackingCVClassifier`](../classifier/StackingCVClassifier.md)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Example 1: Boston Housing Data Predictions"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"In this example we evaluate some basic prediction models on the boston housing dataset and see how the $R^2$ and MSE scores are affected by combining the models with `StackingCVRegressor`. The code output below demonstrates that the stacked model performs the best on this dataset -- slightly better than the best single regression model."
]
},
{
"cell_type": "code",
"execution_count": 1,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"5-fold cross validation scores:\n",
"\n",
"R^2 Score: 0.45 (+/- 0.29) [SVM]\n",
"R^2 Score: 0.43 (+/- 0.14) [Lasso]\n",
"R^2 Score: 0.52 (+/- 0.28) [Random Forest]\n",
"R^2 Score: 0.58 (+/- 0.24) [StackingClassifier]\n"
]
}
],
"source": [
"from mlxtend.regressor import StackingCVRegressor\n",
"from sklearn.datasets import load_boston\n",
"from sklearn.svm import SVR\n",
"from sklearn.linear_model import Lasso\n",
"from sklearn.ensemble import RandomForestRegressor\n",
"from sklearn.model_selection import cross_val_score\n",
"import numpy as np\n",
"\n",
"RANDOM_SEED = 42\n",
"\n",
"X, y = load_boston(return_X_y=True)\n",
"\n",
"svr = SVR(kernel='linear')\n",
"lasso = Lasso()\n",
"rf = RandomForestRegressor(n_estimators=5, \n",
" random_state=RANDOM_SEED)\n",
"\n",
"# The StackingCVRegressor uses scikit-learn's check_cv\n",
"# internally, which doesn't support a random seed. Thus\n",
"# NumPy's random seed need to be specified explicitely for\n",
"# deterministic behavior\n",
"np.random.seed(RANDOM_SEED)\n",
"stack = StackingCVRegressor(regressors=(svr, lasso, rf),\n",
" meta_regressor=lasso)\n",
"\n",
"print('5-fold cross validation scores:\\n')\n",
"\n",
"for clf, label in zip([svr, lasso, rf, stack], ['SVM', 'Lasso', \n",
" 'Random Forest', \n",
" 'StackingClassifier']):\n",
" scores = cross_val_score(clf, X, y, cv=5)\n",
" print(\"R^2 Score: %0.2f (+/- %0.2f) [%s]\" % (\n",
" scores.mean(), scores.std(), label))"
]
},
{
"cell_type": "code",
"execution_count": 2,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"5-fold cross validation scores:\n",
"\n",
"Neg. MSE Score: -33.69 (+/- 22.36) [SVM]\n",
"Neg. MSE Score: -35.53 (+/- 16.99) [Lasso]\n",
"Neg. MSE Score: -27.32 (+/- 16.62) [Random Forest]\n",
"Neg. MSE Score: -25.64 (+/- 18.11) [StackingClassifier]\n"
]
}
],
"source": [
"# The StackingCVRegressor uses scikit-learn's check_cv\n",
"# internally, which doesn't support a random seed. Thus\n",
"# NumPy's random seed need to be specified explicitely for\n",
"# deterministic behavior\n",
"np.random.seed(RANDOM_SEED)\n",
"stack = StackingCVRegressor(regressors=(svr, lasso, rf),\n",
" meta_regressor=lasso)\n",
"\n",
"print('5-fold cross validation scores:\\n')\n",
"\n",
"for clf, label in zip([svr, lasso, rf, stack], ['SVM', 'Lasso', \n",
" 'Random Forest', \n",
" 'StackingClassifier']):\n",
" scores = cross_val_score(clf, X, y, cv=5, scoring='neg_mean_squared_error')\n",
" print(\"Neg. MSE Score: %0.2f (+/- %0.2f) [%s]\" % (\n",
" scores.mean(), scores.std(), label))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## Example 2: GridSearchCV with Stacking"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"In this second example we demonstrate how `StackingCVRegressor` works in combination with `GridSearchCV`. The stack still allows tuning hyper parameters of the base and meta models!\n",
"\n",
"To set up a parameter grid for scikit-learn's `GridSearch`, we simply provide the estimator's names in the parameter grid -- in the special case of the meta-regressor, we append the `'meta-'` prefix.\n"
]
},
{
"cell_type": "code",
"execution_count": 3,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Best: 0.673590 using {'lasso__alpha': 0.4, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.3}\n"
]
}
],
"source": [
"from mlxtend.regressor import StackingCVRegressor\n",
"from sklearn.datasets import load_boston\n",
"from sklearn.linear_model import Lasso\n",
"from sklearn.linear_model import Ridge\n",
"from sklearn.ensemble import RandomForestRegressor\n",
"from sklearn.model_selection import GridSearchCV\n",
"\n",
"X, y = load_boston(return_X_y=True)\n",
"\n",
"ridge = Ridge()\n",
"lasso = Lasso()\n",
"rf = RandomForestRegressor(random_state=RANDOM_SEED)\n",
"\n",
"# The StackingCVRegressor uses scikit-learn's check_cv\n",
"# internally, which doesn't support a random seed. Thus\n",
"# NumPy's random seed need to be specified explicitely for\n",
"# deterministic behavior\n",
"np.random.seed(RANDOM_SEED)\n",
"stack = StackingCVRegressor(regressors=(lasso, ridge),\n",
" meta_regressor=rf, \n",
" use_features_in_secondary=True)\n",
"\n",
"params = {'lasso__alpha': [0.1, 1.0, 10.0],\n",
" 'ridge__alpha': [0.1, 1.0, 10.0]}\n",
"\n",
"grid = GridSearchCV(\n",
" estimator=stack, \n",
" param_grid={\n",
" 'lasso__alpha': [x/5.0 for x in range(1, 10)],\n",
" 'ridge__alpha': [x/20.0 for x in range(1, 10)],\n",
" 'meta-randomforestregressor__n_estimators': [10, 100]\n",
" }, \n",
" cv=5,\n",
" refit=True\n",
")\n",
"\n",
"grid.fit(X, y)\n",
"\n",
"print(\"Best: %f using %s\" % (grid.best_score_, grid.best_params_))"
]
},
{
"cell_type": "code",
"execution_count": 4,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"0.622 +/- 0.10 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.05}\n",
"0.649 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.1}\n",
"0.650 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.15}\n",
"0.667 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.2}\n",
"0.629 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.25}\n",
"0.663 +/- 0.08 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.3}\n",
"0.633 +/- 0.08 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.35}\n",
"0.637 +/- 0.08 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.4}\n",
"0.649 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.45}\n",
"0.653 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 100, 'ridge__alpha': 0.05}\n",
"0.648 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 100, 'ridge__alpha': 0.1}\n",
"0.645 +/- 0.09 {'lasso__alpha': 0.2, 'meta-randomforestregressor__n_estimators': 100, 'ridge__alpha': 0.15}\n",
"...\n",
"Best parameters: {'lasso__alpha': 0.4, 'meta-randomforestregressor__n_estimators': 10, 'ridge__alpha': 0.3}\n",
"Accuracy: 0.67\n"
]
}
],
"source": [
"cv_keys = ('mean_test_score', 'std_test_score', 'params')\n",
"\n",
"for r, _ in enumerate(grid.cv_results_['mean_test_score']):\n",
" print(\"%0.3f +/- %0.2f %r\"\n",
" % (grid.cv_results_[cv_keys[0]][r],\n",
" grid.cv_results_[cv_keys[1]][r] / 2.0,\n",
" grid.cv_results_[cv_keys[2]][r]))\n",
" if r > 10:\n",
" break\n",
"print('...')\n",
"\n",
"print('Best parameters: %s' % grid.best_params_)\n",
"print('Accuracy: %.2f' % grid.best_score_)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"**Note**\n",
"\n",
"The `StackingCVRegressor` also enables grid search over the `regressors` argument. However, due to the current implementation of `GridSearchCV` in scikit-learn, it is not possible to search over both, differenct classifiers and classifier parameters at the same time. For instance, while the following parameter dictionary works\n",
"\n",
" params = {'randomforestregressor__n_estimators': [1, 100],\n",
" 'regressors': [(regr1, regr1, regr1), (regr2, regr3)]}\n",
" \n",
"it will use the instance settings of `regr1`, `regr2`, and `regr3` and not overwrite it with the `'n_estimators'` settings from `'randomforestregressor__n_estimators': [1, 100]`."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"## API"
]
},
{
"cell_type": "code",
"execution_count": 5,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"## StackingCVRegressor\n",
"\n",
"*StackingCVRegressor(regressors, meta_regressor, cv=5, shuffle=True, use_features_in_secondary=False)*\n",
"\n",
"A 'Stacking Cross-Validation' regressor for scikit-learn estimators.\n",
"\n",
"New in mlxtend v0.7.0\n",
"\n",
"**Notes**\n",
"\n",
"The StackingCVRegressor uses scikit-learn's check_cv\n",
"internally, which doesn't support a random seed. Thus\n",
"NumPy's random seed need to be specified explicitely for\n",
"deterministic behavior, for instance, by setting\n",
"np.random.seed(RANDOM_SEED)\n",
"prior to fitting the StackingCVRegressor\n",
"\n",
"**Parameters**\n",
"\n",
"- `regressors` : array-like, shape = [n_regressors]\n",
"\n",
" A list of classifiers.\n",
" Invoking the `fit` method on the `StackingCVRegressor` will fit clones\n",
" of these original regressors that will\n",
" be stored in the class attribute `self.regr_`.\n",
"\n",
"- `meta_regressor` : object\n",
"\n",
" The meta-classifier to be fitted on the ensemble of\n",
" classifiers\n",
"\n",
"- `cv` : int, cross-validation generator or iterable, optional (default: 5)\n",
"\n",
" Determines the cross-validation splitting strategy.\n",
" Possible inputs for cv are:\n",
" - None, to use the default 5-fold cross validation,\n",
" - integer, to specify the number of folds in a `KFold`,\n",
" - An object to be used as a cross-validation generator.\n",
" - An iterable yielding train, test splits.\n",
" For integer/None inputs, it will use `KFold` cross-validation\n",
"\n",
"- `use_features_in_secondary` : bool (default: False)\n",
"\n",
" If True, the meta-classifier will be trained both on\n",
" the predictions of the original regressors and the\n",
" original dataset.\n",
" If False, the meta-regressor will be trained only on\n",
" the predictions of the original regressors.\n",
"\n",
"- `shuffle` : bool (default: True)\n",
"\n",
" If True, and the `cv` argument is integer, the training data will\n",
" be shuffled at fitting stage prior to cross-validation. If the `cv`\n",
" argument is a specific cross validation technique, this argument is\n",
" omitted.\n",
"\n",
"### Methods\n",
"\n",
"
\n",
"\n",
"*fit(X, y, groups=None)*\n",
"\n",
"Fit ensemble regressors and the meta-regressor.\n",
"\n",
"**Parameters**\n",
"\n",
"- `X` : numpy array, shape = [n_samples, n_features]\n",
"\n",
" Training vectors, where n_samples is the number of samples and\n",
" n_features is the number of features.\n",
"\n",
"\n",
"- `y` : numpy array, shape = [n_samples]\n",
"\n",
" Target values.\n",
"\n",
"\n",
"- `groups` : numpy array/None, shape = [n_samples]\n",
"\n",
" The group that each sample belongs to. This is used by specific\n",
" folding strategies such as GroupKFold()\n",
"\n",
"**Returns**\n",
"\n",
"- `self` : object\n",
"\n",
"\n",
"
\n",
"\n",
"*fit_transform(X, y=None, **fit_params)*\n",
"\n",
"Fit to data, then transform it.\n",
"\n",
"Fits transformer to X and y with optional parameters fit_params\n",
"and returns a transformed version of X.\n",
"\n",
"**Parameters**\n",
"\n",
"- `X` : numpy array of shape [n_samples, n_features]\n",
"\n",
" Training set.\n",
"\n",
"\n",
"- `y` : numpy array of shape [n_samples]\n",
"\n",
" Target values.\n",
"\n",
"**Returns**\n",
"\n",
"- `X_new` : numpy array of shape [n_samples, n_features_new]\n",
"\n",
" Transformed array.\n",
"\n",
"
\n",
"\n",
"*get_params(deep=True)*\n",
"\n",
"Get parameters for this estimator.\n",
"\n",
"**Parameters**\n",
"\n",
"- `deep` : boolean, optional\n",
"\n",
" If True, will return the parameters for this estimator and\n",
" contained subobjects that are estimators.\n",
"\n",
"**Returns**\n",
"\n",
"- `params` : mapping of string to any\n",
"\n",
" Parameter names mapped to their values.\n",
"\n",
"
\n",
"\n",
"*predict(X)*\n",
"\n",
"None\n",
"\n",
"
\n",
"\n",
"*score(X, y, sample_weight=None)*\n",
"\n",
"Returns the coefficient of determination R^2 of the prediction.\n",
"\n",
"The coefficient R^2 is defined as (1 - u/v), where u is the regression\n",
"sum of squares ((y_true - y_pred) ** 2).sum() and v is the residual\n",
"sum of squares ((y_true - y_true.mean()) ** 2).sum().\n",
"\n",
"Best possible score is 1.0 and it can be negative (because the\n",
"\n",
"model can be arbitrarily worse). A constant model that always\n",
"predicts the expected value of y, disregarding the input features,\n",
"would get a R^2 score of 0.0.\n",
"\n",
"**Parameters**\n",
"\n",
"- `X` : array-like, shape = (n_samples, n_features)\n",
"\n",
" Test samples.\n",
"\n",
"\n",
"- `y` : array-like, shape = (n_samples) or (n_samples, n_outputs)\n",
"\n",
" True values for X.\n",
"\n",
"\n",
"- `sample_weight` : array-like, shape = [n_samples], optional\n",
"\n",
" Sample weights.\n",
"\n",
"**Returns**\n",
"\n",
"- `score` : float\n",
"\n",
" R^2 of self.predict(X) wrt. y.\n",
"\n",
"
\n",
"\n",
"*set_params(**params)*\n",
"\n",
"Set the parameters of this estimator.\n",
"\n",
"The method works on simple estimators as well as on nested objects\n",
"(such as pipelines). The latter have parameters of the form\n",
"``__`` so that it's possible to update each\n",
"component of a nested object.\n",
"\n",
"**Returns**\n",
"\n",
"self\n",
"\n",
"\n"
]
}
],
"source": [
"with open('../../api_modules/mlxtend.regressor/StackingCVRegressor.md', 'r') as f:\n",
" print(f.read())"
]
}
],
"metadata": {
"anaconda-cloud": {},
"kernelspec": {
"display_name": "Python 3",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.6.1"
}
},
"nbformat": 4,
"nbformat_minor": 1
}