[](https://cran.r-project.org/package=iCellR)
[](https://cran.r-project.org/package=iCellR)
[](https://www.gnu.org/licenses/old-licenses/gpl-2.0.en.html)
# Single (i) Cell R package (iCellR)
iCellR is an interactive R package designed to facilitate the analysis and visualization of high-throughput single-cell sequencing data. It supports a variety of single-cell technologies, including `scRNA-Seq`, `scVDJ-Seq`, `scATAC-Seq`, `CITE-Seq`, and `Spatial Transcriptomics` (ST).
Maintainer: [Alireza Khodadadi-Jamayran](https://scholar.google.com/scholar?q=author:%22Khodadadi-Jamayran%20A%22)
### News (April 2021)
Use the latest version of iCellR (v1.6.4) for scATAC-seq and Spatial Transcriptomics (ST) analyses. Leverage the [i.score](https://github.com/rezakj/iCellR/wiki/i.score) function for `scoring cells based on gene signatures` using methods such as `Tirosh, Mean, Sum, GSVA, ssgsea, Zscore, and Plage`.
### News (July 2020)
Explore iCellR version 1.5.5, now featuring tools for cell cycle analysis `(phases G0, G1S, G2M, M, G1M, and S)`. See example [phase](https://genome.med.nyu.edu/results/external/iCellR/example1/All_cellcycle.png), New Pseudotime Abstract KNetL (PAK map) functionality added – visualize pseudotime progression [(PAK map)](https://genome.med.nyu.edu/results/external/iCellR/example1/pseudotime.KNetL.png). Perform gene-gene correlation analysis using updated visualization tools. [correlations](https://genome.med.nyu.edu/results/external/iCellR/example1/gene-gene.correlation.png).
### News (May 2020)
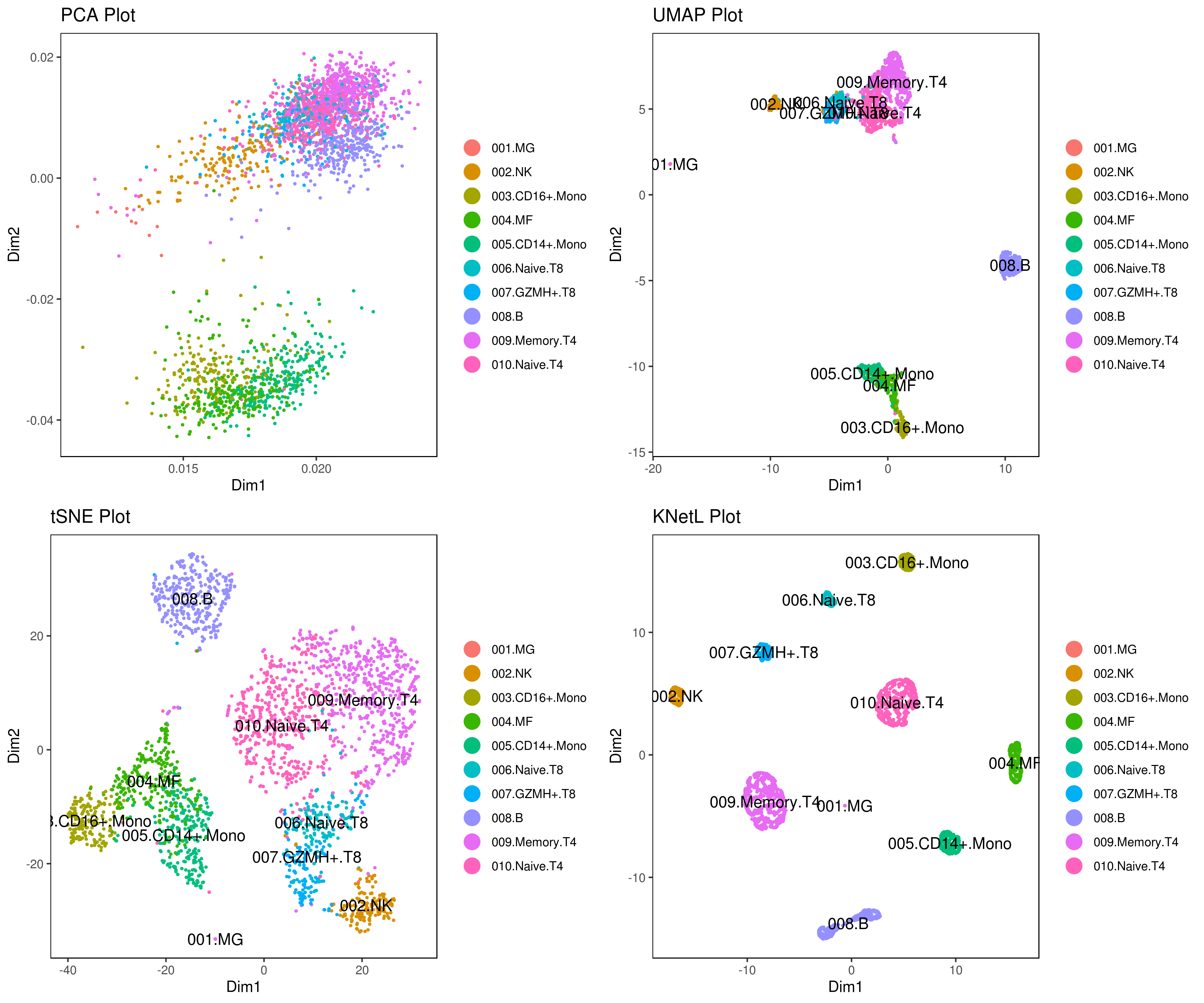
Explore the `KNetL map`, an advanced adjustable and dynamic dimensionality reduction method [KNetL map](https://genome.med.nyu.edu/results/external/iCellR/example1/Allclusts.Annotated.png)  KNetL (pronounced “nettle”) offers enhanced zooming capabilities [KNetL](https://www.biorxiv.org/content/10.1101/2020.05.05.078550v1.full) to show significantly more detail compared to tSNE and UMAP.
### News (April 2020)
Introducing `imputation and coverage correction (CC)` methods for improved gene-gene correlation analysis. ([CC](https://genome.med.nyu.edu/results/external/iCellR/example1/gene-gene.correlation.png)). Perform `batch alignment using iCellR's CPCA` and CCCA tools (CCCA and [CPCA](https://genome.med.nyu.edu/results/external/iCellR/example2/AllCondsClusts.png)) [methods](https://www.biorxiv.org/content/10.1101/2020.03.31.019109v1.full). Expanded databases for cell type prediction now include ImmGen and MCA.
### News (Sep. 2018)
`scSeqR` has been renamed to `iCellR`, and scSeqR has been discontinued. Please use iCellR moving forward, as scSeqR is no longer supported. `UMAP` is added to iCellR. Interactive `cell gating` has been added, allowing users to select cells directly within HTML plots using Plotly.
### Tutorials and manual
- Link to `manual` [Manual](https://cran.r-project.org/web/packages/iCellR/iCellR.pdf) and Comprehensive R Archive Network [(CRAN)](https://cran.r-project.org/web/packages/iCellR/index.html).
- Gor `getting started` and `tutorials` go to our [Wiki page](https://github.com/rezakj/iCellR/wiki).
- Link to a video tutorial for CITE-Seq and scRNA-Seq analysis: [Video](https://vimeo.com/337822487)
- All you need to know about KNetL map: [Video](https://youtu.be/tkoPTVciQm0)
- If you are using `FlowJo` or `SeqGeq`, they offer plugins for iCellR and other single-cell analysis tools. You can find the list of all plugins here: https://www.flowjo.com/exchange/#/ . Specifically, the iCellR plugin can be found here: https://www.flowjo.com/exchange/#/plugin/profile?id=34. Additionally, a SeqGeq Differential Expression (DE) tutorial is available to guide you through the process: [SeqGeq DE tutorial](https://www.youtube.com/watch?v=gXFmWRpdwow)
For `citing iCellR` use this [PMID: 34353854](https://cancerdiscovery.aacrjournals.org/content/early/2021/07/28/2159-8290.CD-21-0369)
iCellR publications: [PMID: 35660135](https://pubmed.ncbi.nlm.nih.gov/35660135/) (scRNA-seq/KNetL) [PMID: 35180378](https://pubmed.ncbi.nlm.nih.gov/35180378/) (CITE-seq/KNetL), [PMID: 34911733](https://pubmed.ncbi.nlm.nih.gov/34911733/) (i.score and cell ranking), [PMID: 34963055](https://www.cell.com/cell/fulltext/S0092-8674(21)01427-6?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0092867421014276%3Fshowall%3Dtrue) (scRNA-seq), [PMID 31744829](https://www.ncbi.nlm.nih.gov/pubmed/31744829) (scRNA-seq), [PMID: 31934613](https://www.ncbi.nlm.nih.gov/pubmed/31934613) (bulk RNA-seq from TCGA), [PMID: 32550269](https://pubmed.ncbi.nlm.nih.gov/32550269/) (scVDJ-seq), [PMID: 34135081](https://jasn.asnjournals.org/content/32/8/1987), [PMID: 33593073](https://www.ahajournals.org/doi/10.1161/CIRCRESAHA.120.317914), [PMID: 34634466](https://pubmed.ncbi.nlm.nih.gov/34634466/), [PMID: 35302059](https://pubmed.ncbi.nlm.nih.gov/35302059/), [PMID: 34353854](https://cancerdiscovery.aacrjournals.org/content/early/2021/07/28/2159-8290.CD-21-0369)
Single (i) Cell R package (iCellR)
KNetL (pronounced “nettle”) offers enhanced zooming capabilities [KNetL](https://www.biorxiv.org/content/10.1101/2020.05.05.078550v1.full) to show significantly more detail compared to tSNE and UMAP.
### News (April 2020)
Introducing `imputation and coverage correction (CC)` methods for improved gene-gene correlation analysis. ([CC](https://genome.med.nyu.edu/results/external/iCellR/example1/gene-gene.correlation.png)). Perform `batch alignment using iCellR's CPCA` and CCCA tools (CCCA and [CPCA](https://genome.med.nyu.edu/results/external/iCellR/example2/AllCondsClusts.png)) [methods](https://www.biorxiv.org/content/10.1101/2020.03.31.019109v1.full). Expanded databases for cell type prediction now include ImmGen and MCA.
### News (Sep. 2018)
`scSeqR` has been renamed to `iCellR`, and scSeqR has been discontinued. Please use iCellR moving forward, as scSeqR is no longer supported. `UMAP` is added to iCellR. Interactive `cell gating` has been added, allowing users to select cells directly within HTML plots using Plotly.
### Tutorials and manual
- Link to `manual` [Manual](https://cran.r-project.org/web/packages/iCellR/iCellR.pdf) and Comprehensive R Archive Network [(CRAN)](https://cran.r-project.org/web/packages/iCellR/index.html).
- Gor `getting started` and `tutorials` go to our [Wiki page](https://github.com/rezakj/iCellR/wiki).
- Link to a video tutorial for CITE-Seq and scRNA-Seq analysis: [Video](https://vimeo.com/337822487)
- All you need to know about KNetL map: [Video](https://youtu.be/tkoPTVciQm0)
- If you are using `FlowJo` or `SeqGeq`, they offer plugins for iCellR and other single-cell analysis tools. You can find the list of all plugins here: https://www.flowjo.com/exchange/#/ . Specifically, the iCellR plugin can be found here: https://www.flowjo.com/exchange/#/plugin/profile?id=34. Additionally, a SeqGeq Differential Expression (DE) tutorial is available to guide you through the process: [SeqGeq DE tutorial](https://www.youtube.com/watch?v=gXFmWRpdwow)
For `citing iCellR` use this [PMID: 34353854](https://cancerdiscovery.aacrjournals.org/content/early/2021/07/28/2159-8290.CD-21-0369)
iCellR publications: [PMID: 35660135](https://pubmed.ncbi.nlm.nih.gov/35660135/) (scRNA-seq/KNetL) [PMID: 35180378](https://pubmed.ncbi.nlm.nih.gov/35180378/) (CITE-seq/KNetL), [PMID: 34911733](https://pubmed.ncbi.nlm.nih.gov/34911733/) (i.score and cell ranking), [PMID: 34963055](https://www.cell.com/cell/fulltext/S0092-8674(21)01427-6?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0092867421014276%3Fshowall%3Dtrue) (scRNA-seq), [PMID 31744829](https://www.ncbi.nlm.nih.gov/pubmed/31744829) (scRNA-seq), [PMID: 31934613](https://www.ncbi.nlm.nih.gov/pubmed/31934613) (bulk RNA-seq from TCGA), [PMID: 32550269](https://pubmed.ncbi.nlm.nih.gov/32550269/) (scVDJ-seq), [PMID: 34135081](https://jasn.asnjournals.org/content/32/8/1987), [PMID: 33593073](https://www.ahajournals.org/doi/10.1161/CIRCRESAHA.120.317914), [PMID: 34634466](https://pubmed.ncbi.nlm.nih.gov/34634466/), [PMID: 35302059](https://pubmed.ncbi.nlm.nih.gov/35302059/), [PMID: 34353854](https://cancerdiscovery.aacrjournals.org/content/early/2021/07/28/2159-8290.CD-21-0369)
Single (i) Cell R package (iCellR)






***
## For `getting started` and `tutorials` go to our [Wiki page](https://github.com/rezakj/iCellR/wiki).













