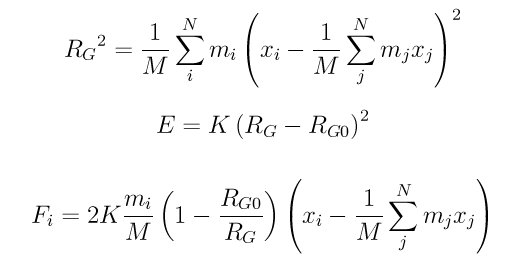
Syntax:
fix ID group-ID spring/rg K RG0
if RG0 = NULL, use the current RG as the target value
Examples:
fix 1 protein spring/rg 5.0 10.0 fix 2 micelle spring/rg 5.0 NULL
Description:
Apply a harmonic restraining force to atoms in the group to affect their central moment about the center of mass (radius of gyration). This fix is useful to encourage a protein or polymer to fold/unfold and also when sampling along the radius of gyration as a reaction coordinate (i.e. for protein folding).
The radius of gyration is defined as RG in the first formula. The energy of the constraint and associated force on each atom is given by the second and third formulas, when the group is at a different RG than the target value RG0.
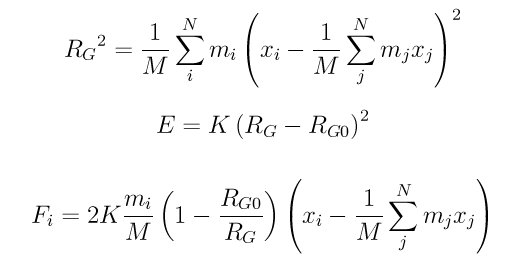
The (xi - center-of-mass) term is computed taking into account periodic boundary conditions, m_i is the mass of the atom, and M is the mass of the entire group. Note that K is thus a force constant for the aggregate force on the group of atoms, not a per-atom force.
If RG0 is specified as NULL, then the RG of the group is computed at the time the fix is specified, and that value is used as the target.
Restart, fix_modify, output, run start/stop, minimize info:
No information about this fix is written to binary restart files. None of the fix_modify options are relevant to this fix. No global or per-atom quantities are stored by this fix for access by various output commands. No parameter of this fix can be used with the start/stop keywords of the run command. This fix is not invoked during energy minimization.
Restrictions: none
Related commands:
fix spring, fix spring/self fix drag, fix smd
Default: none