 \n",
"\n",
"
\n",
"\n",
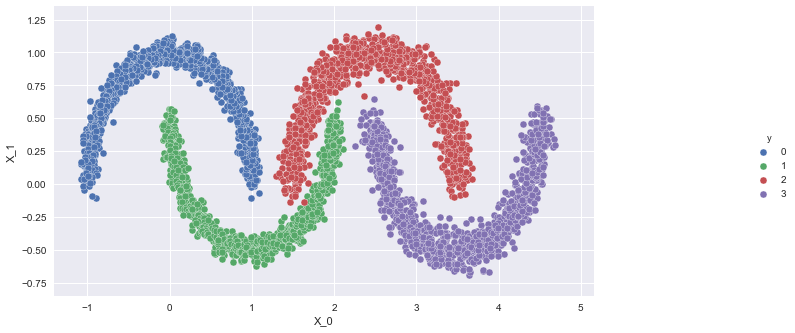
"| X_0 | \n", "\t\t\tX_1 | \n", "\t\t\ty | \n", "\t\t
| X[:,0] | \n", "\t\t\tX[:,1] | \n", "\t\t\ty | \n", "\t\t
use the pd.DataFrame() command and sns.pairplot(x_vars, y_vars, hue, data)
" ] }, { "cell_type": "code", "execution_count": null, "metadata": { "tags": [ "s1", "l1", "ans" ] }, "outputs": [], "source": [ "moon_df = pd.DataFrame({'X_0':X[:,0], 'X_1':X[:,1], 'y':y})\r\n", "g = sns.pairplot(x_vars=\"X_0\", y_vars=\"X_1\", hue=\"y\", data = moon_df)\r\n", "g.fig.set_size_inches(14, 6)\r\n", "sns.despine()" ] }, { "cell_type": "code", "execution_count": null, "metadata": { "tags": [ "s1", "hid", "l1" ] }, "outputs": [], "source": [ "ref_tmp_var = False\n", "\n", "\n", "try:\n", " ref_assert_var = False\n", " moon_df_ = pd.DataFrame({'X_0':X[:,0], 'X_1':X[:,1], 'y':y})\n", " \n", " import numpy as np\r\n", " \r\n", " if np.all(moon_df['X_0'] == moon_df_['X_0']) and np.all(moon_df['X_1'] == moon_df_['X_1']) and np.all(moon_df['y'] == moon_df_['y']):\r\n", " ref_assert_var = True\r\n", " out = g\r\n", " else:\r\n", " ref_assert_var = False\n", " \n", "except Exception:\n", " print('Please follow the instructions given and use the same variables provided in the instructions.')\n", "else:\n", " if ref_assert_var:\n", " ref_tmp_var = True\n", " else:\n", " print('Please follow the instructions given and use the same variables provided in the instructions.')\n", "\n", "\n", "assert ref_tmp_var" ] }, { "cell_type": "markdown", "metadata": { "tags": [ "l2", "content", "s2" ] }, "source": [ "\n", "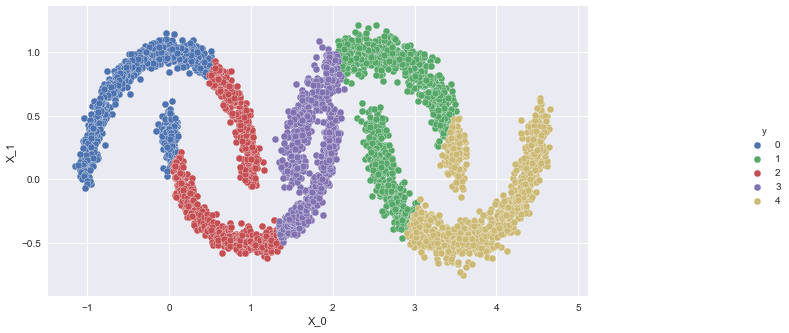 \n",
"\n",
"
\n",
"\n",
"Use the code from GMMs.
" ] }, { "cell_type": "code", "execution_count": null, "metadata": { "tags": [ "l2", "s2", "ans" ] }, "outputs": [], "source": [ "mixture_model = GaussianMixture(n_components = 4, covariance_type = 'spherical')\r\n", "mixture_model.fit(X)\r\n", "moon_df['gmm_clus'] = mixture_model.predict(X)\r\n", "# Plot the clusters\r\n", "g = sns.pairplot(x_vars=\"X_0\", y_vars=\"X_1\", hue=\"gmm_clus\", data = moon_df)\r\n", "g.fig.set_size_inches(14, 6)\r\n", "sns.despine()" ] }, { "cell_type": "code", "execution_count": null, "metadata": { "tags": [ "l2", "hid", "s2" ] }, "outputs": [], "source": [ "ref_tmp_var = False\n", "\n", "\n", "try:\n", " ref_assert_var = False\n", " mixture_model_ = GaussianMixture(n_components = 4, covariance_type = 'spherical')\r\n", " mixture_model_.fit(X)\r\n", " y_hat = mixture_model_.predict(X)\n", " \n", " import numpy as np\r\n", " \r\n", " if len(y_hat) == len(moon_df['gmm_clus']):\r\n", " ref_assert_var = True\r\n", " out = g\r\n", " else:\r\n", " ref_assert_var = False\n", " \n", "except Exception:\n", " print('Please follow the instructions given and use the same variables provided in the instructions.')\n", "else:\n", " if ref_assert_var:\n", " ref_tmp_var = True\n", " else:\n", " print('Please follow the instructions given and use the same variables provided in the instructions.')\n", "\n", "\n", "assert ref_tmp_var" ] } ], "metadata": { "executed_sections": [], "rf_version": 1 }, "nbformat": 4, "nbformat_minor": 2 }