---
title: Introduction to HPCC Cluster and Linux
linkTitle: "Linux & HPCC Cluster"
weight: 3
type: docs
---
## HPCC Cluster Overview
The HPCC Cluster (formerly called biocluster) is a shared research computing system available at UCR. The HPCC website is available [here](http://hpcc.ucr.edu/index.html).
### What Is a Computer Cluster?
* A computer cluster is an assembly of CPU units, so called computer nodes that work together to perform many computations in parallel. To achieve this, an internal network (e.g. Infiniband interconnect) connects the nodes to a larger unit, while one or more head nodes controls the load and traffic across the entire system.
* Usually, users log into one of the head nodes via `ssh` to submit their computing requests to a queuing system provided by resource management and scheduling software, such as Slurm, SGE, or TORQUE/MAUI. The queuing system distributes the processes to the computer nodes in a controlled fashion.
* Because the head node controls the entire system, users should never run computing jobs on the head node directly!
* For code testing purposes, one can log into one of the nodes via `srun` (see below) and run jobs interactively.
### Hardware Infrastructure
### Computer nodes
- Over 12,000 CPU cores
- 130 Intel, AMD and GPU nodes
- 32-128 CPU cores per node
- 256-4,096 GB of RAM per node
- 64 GPUs including NVIDIA K80, P100, A100 and H100
### Interconnect
- HDR IB @200Gbs
### Storage
- Parallel GPFS storage system with 5.0 PB usable space
- File system scales to over 50 PB
- Backup of same architecture and similar amount
### User traffic
- Computing tasks need to be submitted via `sbatch` or `srun`
- HPCC Cluster headnode only for login, not for computing tasks!
- Monitor cluster activity: `squeue` or `jobMonitor` (`qstatMonitor`)
### Manuals
- [HPCC Cluster Manual](https://hpcc.ucr.edu/manuals/hpc_cluster/)
- [Linux Manual](https://hpcc.ucr.edu/manuals/linux_basics/)
## Linux Basics
### Log into HPCC Cluster via SSH Terminal
Terminal-based login is the most feature-rich method for accessing remote Linux systems. Web-based alternatives via [JupyterHub](https://hpcc.ucr.edu/manuals/access/login/#b-web-based-access) and
[RStudio Server](https://hpcc.ucr.edu/manuals/access/login/#b-web-based-access) are also possible. To access the HPCC
cluster with the standard `ssh` protocol, users want to follow steps 1-3. Only
step 1 is required after setting up SSH Key-based access.
__1.__ Type the following `ssh` login command from a terminal application, where `` needs to be replaced by the actual account name of a user. The `<>` characters indicate a placeholder and need to be removed. Next, press enter to execute the `ssh` command.
```bash
ssh -X @cluster.hpcc.ucr.edu
```
After this a user is logged in to one of the headnodes, which are `skylark` or `bluejay`.
The `-X` argument enables X11 support, which is required for opening GUI applications on remote systems.
__2.__ Type your password and hit enter. Note, when typing the password the cursor will not move and nothing is printed to the screen. If SSH Key access is enabled, both the password and Duo steps will be skipped automatically during the log in process.
__3.__ Follow the [Duo multifactor authenication](https://hpcc.ucr.edu/manuals/access/login/#passwordduo) instructions printed to the screen. As external users do not have access to UCR's Duo system, they can only log in via the alternative SSH Key method. How to enable SSH Keys is described [here](https://hpcc.ucr.edu/manuals/access/login/#ssh-keys). Note, Duo will be bypassed if SSH Key based login is enabled. This can be more conveniet than Duo when accessing the cluster frequently.
If the login is performed via a GUI application, which is an option in MobaXterm,
then one can provide the same login information given under the above `ssh`
commad in the corresponding fields of the login window as follows:
+ Host name: `cluster.hpcc.ucr.edu`
+ User name: ...
+ Password: ...
__Importantly__, after the first login into a new account (or a password reset),
users need to change their password with the `passwd` command and then follow
the on-screen instructions. This requires to enter the current password once
and the new password twice. New passwords need to be at least 8 characters
long and meet at least 3 of the following requirments: lowercase character,
uppercase character, number, and punctuation character.
__What to do if password/Duo is not working?__ If this happens then most often the login is blocked because a password was typed too many times incorrectly, or
not changed after the first login (see above). To correct this, please request a password reset by emailing [support@hpcc.ucr.edu](mailto:support@hpcc.ucr.edu).
Remember, password/Duo based access is only possible if a user's UCR NetID matches the corresponding HPCC username. If this is not the case then
UCR users can request to change their HPCC user account name to their NetID or use the [SSH key](https://hpcc.ucr.edu/manuals/access/login/#ssh-keys) based access method.
#### Terminal Options
Various ssh terminal applications are available for all major operating systems. Examples include:
+ macOS: built-in macOS [Terminal](https://support.apple.com/guide/terminal/welcome/mac) or [iTerm2](https://iterm2.com/)
+ Windows: [MobaXterm](http://mobaxterm.mobatek.net/) is a very feature rich terminal option for Windows users. [Putty](http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html) is an alternative, but outdated and not recommended anymore. [Here](https://mobaxterm.mobatek.net/demo.html) are annimated usage introductions for MobaXterm.
+ Additional useful manuals for MobaXterm are here: [SSH-Key Generation (HPCC Manual)](https://hpcc.ucr.edu/manuals/hpc_cluster/sshkeys/sshkeys_winos/#create-ssh-keys-mobaxterm), [SSH Key Generation (Main MobaXterm Manual)](https://mobaxterm.mobatek.net/documentation.html#6_3) and [Connect to Remote Systems (UNL Manual)](https://hcc.unl.edu/docs/connecting/mobaxterm/).
+ __Note__, when using MobaXterm, it is important to check that the usage of a persistent home directory has been enabled. For this, go to `Settings` -> `General` -> `Persistent home directory`. If the latter field shows ``, then specify a suitable location for your home directory.
+ To find out where MobaXterm stores its files (incl. SSH Keys), type in the MobaXterm terminal `open ~`. This will open a user's home directory in the Windows file browser.
+ Linux: a wide range of Terminal applications is available for Linux. Usually, the default terminal available on a Linux distribution will be sufficient.
+ ChromeOS: after enabling Linux apps on Chromebooks one can use the default terminal that is similar to those on Linux systems.
+ Additional login information can be found on the corresponding HPCC manuals:
+ Login page: [here](https://hpcc.ucr.edu/manuals/hpc_cluster/login/)
+ SSH Keys: [here](https://hpcc.ucr.edu/manuals/access/login/#ssh-keys)
+ Duo Multifactor Authenication: [here](https://hpcc.ucr.edu/manuals/access/login/#passwordduo)
+ UCR Duo Manual: [here](https://its.ucr.edu/sites/g/files/rcwecm321/files/2018-06/Multi-Factor%20Authentication%20Handout.pdf)
#### Remote Graphics Support
X11 support is included in the terminal applications of most OSs. This includes MobaXterm on Windows, Linux and ChromeOS terminals. On macOS systems, users need to run XQuartz
in the background to enable X11 graphics display support. XQuartz can be downloaded from [here](https://www.xquartz.org/) (also see this video [here](https://www.youtube.com/watch?v=uS4zTqfwSSQ)).
Note, XQuartz is optional if remote graphics support is not needed.
### Important Linux Commands
The following provides a short overview of important shell commands. Much more detailed information can be found on [HPCC's Linux tutorials](https://hpcc.ucr.edu/manuals/linux_basics/).
Finding help
```sh
man
```
List content of directories
```sh
ls
ls -l or ll # includes details about files and directories
ls -al # includes hidden files starting with a dot in their name
ll -d # lists permissions of specfic directory or file
```
Print current working directory
```sh
pwd
pwd -P # returns physical location in case one followed symbolic link
```
Search in files and directories
```sh
grep
```
Word count
```sh
wc
```
Create directory
```sh
mkdir
```
Change directory
```sh
cd # changes pwd to specified path
cd # changes pwd to root of home directory
cd - # switches to previous directory
```
Change directory
```sh
cd
```
Delete files and directories
```sh
rm
```
Move and rename files
```sh
mv
```
Copy files from internet to `pwd`
```sh
wget
```
Viewing files
```sh
less
```
### File Exchange
__GUI applications__
+ Windows: [WinSCP](http://winscp.net/eng/index.php) or [MobaXterm](https://mobaxterm.mobatek.net/features.html)
+ Mac OS X: [CyberDuck](http://cyberduck.en.softonic.com/mac)
+ Win/OS X/Linux: [FileZilla](https://filezilla-project.org/)

![]() Fig 1: FileZilla settings with an SSH key. For generating SSH keys see here.
Fig 1: FileZilla settings with an SSH key. For generating SSH keys see here.
__SCP: via command-line__ ([Manual](https://linux.die.net/man/1/scp))
Advantages of this method include: batch up/downloads and ease of automation.
```sh
scp file user@remotehost:/home/user/ # From local to remote
scp user@remotehost:/home/user/file . # From remote to local
```
__RSYNC: via command-line__ ([Manual](https://linux.die.net/man/1/rsync))
Advantages of this method include: same as SCP plus differential update options and viewing of directory content.
Print (view) content of remote directory
```sh
rsync user@remotehost:~/somedirectory/*
```
Download directory or file(s)
```sh
rsync -avzhe ssh user@remotehost:~/somedirectory .
# -a: recursive archive mode (thus -r not required), also preserves permissions, time stamps, etc
# -v: verbose
# -z: compress data during transfer
# -h: print messages in human-readable format
# -e: specifies transfer protocol; using ssh here provides encryption during transfer
# --delete: files that were deleted on source will be deleted also in backup-destination
# -n: for testing use this dry-run option, but drop '-e ssh' in this case
```
Upload directory or file(s)
```sh
rsync -avzhe ssh somedirectory user@hostname:~/
```
### Check Integrity of Files
To check the integrity of files (_e.g._ after downloading or copying them),
one can use their hash (checksum) values created by `md5sum`. These hash
values are specific to a file and very small in size. If a hash value for a
data file is identical with the hash value of the downloaded copy, then the
downloaded copy is usually identical with the source file. The following routine
assumes that a file named `myfile1.txt` was downloaded along with its
checksum (here `*.md5` created for testing). Next, the checksum values are compared.
```sh
md5sum myfile1.txt # generates checksum
md5sum myfile1.txt > myfile1.md5 # saves checksum to file
```
```
4c1ac93e1be5f77451fa909653b2404c myfile1.txt
```
```sh
md5sum -c myfile1.md5 # checks checksum value
```
```
myfile1.txt: OK
```
### Compare Differences Among Directories
Differences in files and content of two directories can be identified with
the `diff` command. The following also shows how to exclude certain files
in this comparison, here a file called `.git`.
```sh
diff -r --exclude=".git" dir1/ dir2/
```
### STD IN/OUT/ERR, Redirect & Wildcards
Wildcard `*` to specify many files
```sh
file.*
```
Redirect `ls` output to file
```sh
ls > file
```
Specify file as input to command
```sh
command < myfile
```
Append output of command to file
```sh
command >> myfile
```
Pipe `STDOUT` of one command to another command
```sh
command1 | command2
```
Turn off progress info
```sh
command > /dev/null
```
Pipe output of `grep` to `wc`
```sh
grep pattern file | wc
```
Print `STDERR` to file
```sh
grep pattern nonexistingfile 2 > mystderr
```
### Homework Assignment (HW2)
See HW2 page [here](https://girke.bioinformatics.ucr.edu/GEN242/assignments/homework/hw02/hw02/).
### Permissions and ownership
List directories and files
```sh
ls -al
ls -ld # lists only specified dir/file
```
The previous command shows something like this for each file/dir: `drwxrwxrwx`. The
meaning of this syntax is as follows:
* `d`: directory
* `rwx`: read, write and execute permissions, respectively
* first triplet: user permissions (u)
* second triplet: group permissions (g)
* third triplet: world permissions (o)
Example for assigning write and execute permissions to user, group and world
```sh
chmod ugo+rx my_file
```
* `+` causes the permissions selected to be added
* `-` causes them to be removed
* `=` causes them to be the only permissions that the file has.
When performing the same operation on many files with subdirectories then one can
use `-R` for recursive behavior.
```sh
chmod -R ugo+rx my_dir
```
Since directories have to be executable the capital `X` option can be useful which
applies only to directories but not to files. The following will assign `drwxr-xr-x` to directories
and `-rw-r--r--` to files and hidden files.
```sh
chmod -R ugo-x,u+rwX,go+rX,go-w ./* ./.[!.]*
```
Syntax for changing user & group ownership
```sh
chown :
```
### Symbolic Links
Symbolic links are short nicknames to files and directories that save typing of their full paths.
```sh
ln -s original_filename new_nickname
```
## Software and module system
* Over 2,000 software tools are currently installed on the HPCC Cluster
* Custom installs in user accounts via various mechanisms, e.g. environment management systems such as [conda](https://conda.io/projects/conda/en/latest/index.html)
* Most common research databases used in bioinformatics are available
* Support of most common programming languages used in research computing
* A module system is used to facilitate the management of software tools. This includes any number of versions of each software.
* New software install requests can be sent to support@hpcc.ucr.edu.
* To use software manged under the module system, users need to learn using some basic commands. The most common commands are listed below.
Print available modules
```sh
module avail
```
Print available modules starting with letter 'R'
```sh
module avail R
```
Load default module R
```sh
module load R
```
Unload specific module R
```sh
module unload R/4.2.0
```
Load specific R version
```sh
module unload R/4.1.2
```
List loaded modules
```sh
module list
```
## Installs and package management with Conda:
See [here](https://hpcc.ucr.edu/manuals/hpc_cluster/package_manage/).
## Big data storage
Each user account on HPCC Cluster comes only with 20GB of disk space. Much more disk space is
available in a dedicated `bigdata` directory. How much space depends on the subscription
of each user group. The path of `bigdata` and `bigdata-shared` is as follows:
* `/bigdata/labname/username`
* `/bigdata/labname/shared`
All lab members share the same bigdata pool. The course number `gen242` is used as `labname`
for user accounts adminstered under GEN242 (here /bigdata/gen242/).
The disk usage of `home` and `bigdata` can be monitored on the [HPCC Cluster Dashboard](https://dashboard.hpcc.ucr.edu/).
## Queuing system: `Slurm`
The HPCC cluster uses `Slurm` as queuing and load balancing system. To control user traffic, any
type of compute intensive jobs need to be submitted via `sbatch` or `srun` (see below) to the computer
nodes. Much more detailed information on this topic can be found on these sites:
+ [UCR HPCC Manual](http://hpcc.ucr.edu/manuals_linux-cluster_jobs.html)
+ [Slurm Documentation](https://slurm.schedmd.com/documentation.html)
+ [Torque/Slurm Comparison](http://www.nersc.gov/users/computational-systems/cori/running-jobs/for-edison-users/torque-moab-vs-slurm-comparisons/)
+ [Switching from Torque to Slurm](https://sites.google.com/a/case.edu/hpc-upgraded-cluster/slurm-cluster-commands)
+ [Slurm Quick Start Tutorial](http://www.ceci-hpc.be/slurm_tutorial.html)

![]() Fig 2: Overview of Slurm on HPCC cluster.
### Job submission with `sbatch`
Print information about queues/partitions available on a cluster.
```sh
sinfo
```
Compute jobs are submitted with `sbatch` via a submission script (here `script_name.sh`).
```sh
sbatch script_name.sh
```
The following sample submission script (`script_name.sh`) executes an R script named `my_script.R`.
```sh
#!/bin/bash -l
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --mem-per-cpu=1G
#SBATCH --time=1-00:15:00 # 1 day and 15 minutes
#SBATCH --mail-user=useremail@address.com
#SBATCH --mail-type=ALL
#SBATCH --job-name="some_test"
#SBATCH --partition="gen242" # Choose alternative partitions from: intel, batch, highmem, gpu, short
#SBATCH --account="gen242" # Same as above
Rscript my_script.R
```
`STDOUT` and `STDERROR` of jobs will be written to files named
`slurm-.out` or to a custom file specified under `#SBATCH --output` in
the submission script.
### Interactive sessions with `srun`
This option logs a user in to a computer node of a specified partition (queue), while Slurm monitors and controls the resource request.
```sh
srun --pty bash -l
```
Interactive session with specific resource requests. Additional information about partitions is [here](https://hpcc.ucr.edu/manuals/hpc_cluster/jobs/).
```sh
srun --x11 --partition=gen242 --account=gen242 --mem=2gb --cpus-per-task 4 --ntasks 1 --time 1:00:00 --pty bash -l
```
The argument `--mem` limits the amount of RAM, `--cpus` the number of CPU
cores, `--time` the time how long a session will be active. Under
`--parition` one can choose among different queues and node architectures.
Current options under `--partition` for most users of the HPCC cluster are: `intel`, `batch`, `highmem`, `gpu`,
and `short`. The latter has a time limit of 2 hours. Note, `--x11` will only work when logged in with X11 support.
This requires the `-X` argument when logging in via `ssh` (see above). On macOS systems X11 support is provided
by [XQuartz](https://www.xquartz.org/) which needs to be installed and running on a system prior to loging in to
a remote system. If X11 support is not available or broken then one can still connect via `srun` by dropping the
`--x11` argument form the `srun` command.
To run your most frequently used `srun` command quickly, one can place the following line
in a user's `~/.bashrc` file. After the next login or sourcing the `.bashrc` file, one can
execute the command with its alias, here: `srun_gen242`. In this case `echo` is used to print the command to the screen rather
than executing it right away. This way one can copy and paste it, make changes as needed and
then execute it.
```sh
alias srun_gen242='echo "srun --x11 --partition=gen242 --account=gen242 --mem=20gb --cpus-per-task 8 --ntasks 1 --time 20:00:00 --pty bash -l"'
```
### Monitoring jobs with `squeue`
List all jobs in queue
```sh
squeue
```
List jobs of a specific user
```sh
squeue -u
```
Print more detailed information about a job
```sh
scontrol show job
scontrol show jobid -dd
```
Custom command to summarize and visualize cluster activity
```sh
jobMonitor
```
### Deleting and altering jobs
Delete a single job
```sh
scancel -i
```
Delete all jobs of a user
```sh
scancel -u
```
Delete all jobs of a certain name
```sh
scancel --name
```
Altering jobs with `scontrol update`. The below example changes the walltime (``) of a specific job (``).
```sh
scontrol update jobid= TimeLimit=
```
### Resource limits
Resourse limits for users can be viewed as follows.
```sh
sacctmgr show account $GROUP format=Account,User,Partition,GrpCPUs,GrpMem,GrpNodes --ass | grep $USER
```
Similarly, one can view the limits of the group a user belongs to.
```sh
sacctmgr show account $GROUP format=Account,User,Partition,GrpCPUs,GrpMem,GrpNodes,GrpTRES%30 --ass | head -3
```
## Text/code editors
The following list includes examples of several widely used code editors.
* __Vi/Vim/Neovim__: Non-graphical (terminal-based) editor. Vi is guaranteed to be available on any system. Vim and Nvim (Neovim) are the improved versions of vi.
* __Emacs__: Non-graphical or window-based editor. You still need to know keystroke commands to use it. Installed on all Linux distributions and on most other Unix systems.
* __Pico__: Simple terminal-based editor available on most versions of Unix. Uses keystroke commands, but they are listed in logical fashion at bottom of screen.
* __Nano__: A simple terminal-based editor which is default on modern Debian systems.
* __Atom__: Modern text editor developed by GitHub project.
### Why does it matter?
To work efficiently on remote systems like a computer cluster, it is essential
to learn how to work in a pure command-line interface. GUI environments like
RStudio and similar coding environments are not suitable for this. In addition,
there is a lot of value of knowing how to work in an environment that is not
restricted to a specific programming language. Therefore, this class embraces
RStudio where it is useful, but for working on remote systems like HPCC Cluster, it
uses Nvim and Tmux. Both are useful for many programming languages.
Combinded with the `nvim-r` plugin they also provide a powerful command-line working
environment for R. The following provides a brief introduction to this environment.
### Vim overview
The following opens a file (here `myfile`) with nvim (or vim)
```sh
nvim myfile.txt # for neovim (or 'vim myfile.txt' for vim)
```
Once you are in Nvim, there are three main modes: normal, insert and command mode. The most important commands for switching between the three modes are:
* `i`: The `i` key brings you from the normal mode to the insert mode. The latter is used for typing.
* `Esc`: The `Esc` key brings you from the insert mode back to the normal mode.
* `:`: The `:` key starts the command mode at the bottom of the screen.
Use the arrow keys to move your cursor in the text. Using `Fn Up/Down key` allows to page through
the text quicker. In the following command overview, all commands starting with `:` need to be typed in the command mode.
All other commands are typed in the normal mode after pushing the `Esc` key.
Important modifier keys to control vim/nvim
* `:w`: save changes to file. If you are in editing mode you have to hit `Esc` first.
* `:q`: quit file that has not been changed
* `:wq`: save and quit file
* `:!q`: quit file without saving any changes
### Useful resources for learning vim/nvim
* [Interactive Vim Tutorial](http://www.openvim.com)
* [Official Vim Documentation](http://vimdoc.sourceforge.net/)
* [HPCC Vim/Nvim Overview](https://hpcc.ucr.edu/manuals/hpc_cluster/terminalide/#vimnvim-overview)
* [HPCC Linux Manual (old)](http://hpcc.ucr.edu/manuals_linux-basics_vim.html)
## Nvim-R-Tmux essentials
Terminal-based Working Environment for R: [Nvim-R-Tmux](https://gist.github.com/tgirke/7a7c197b443243937f68c422e5471899).
Fig 2: Overview of Slurm on HPCC cluster.
### Job submission with `sbatch`
Print information about queues/partitions available on a cluster.
```sh
sinfo
```
Compute jobs are submitted with `sbatch` via a submission script (here `script_name.sh`).
```sh
sbatch script_name.sh
```
The following sample submission script (`script_name.sh`) executes an R script named `my_script.R`.
```sh
#!/bin/bash -l
#SBATCH --nodes=1
#SBATCH --ntasks=1
#SBATCH --cpus-per-task=1
#SBATCH --mem-per-cpu=1G
#SBATCH --time=1-00:15:00 # 1 day and 15 minutes
#SBATCH --mail-user=useremail@address.com
#SBATCH --mail-type=ALL
#SBATCH --job-name="some_test"
#SBATCH --partition="gen242" # Choose alternative partitions from: intel, batch, highmem, gpu, short
#SBATCH --account="gen242" # Same as above
Rscript my_script.R
```
`STDOUT` and `STDERROR` of jobs will be written to files named
`slurm-.out` or to a custom file specified under `#SBATCH --output` in
the submission script.
### Interactive sessions with `srun`
This option logs a user in to a computer node of a specified partition (queue), while Slurm monitors and controls the resource request.
```sh
srun --pty bash -l
```
Interactive session with specific resource requests. Additional information about partitions is [here](https://hpcc.ucr.edu/manuals/hpc_cluster/jobs/).
```sh
srun --x11 --partition=gen242 --account=gen242 --mem=2gb --cpus-per-task 4 --ntasks 1 --time 1:00:00 --pty bash -l
```
The argument `--mem` limits the amount of RAM, `--cpus` the number of CPU
cores, `--time` the time how long a session will be active. Under
`--parition` one can choose among different queues and node architectures.
Current options under `--partition` for most users of the HPCC cluster are: `intel`, `batch`, `highmem`, `gpu`,
and `short`. The latter has a time limit of 2 hours. Note, `--x11` will only work when logged in with X11 support.
This requires the `-X` argument when logging in via `ssh` (see above). On macOS systems X11 support is provided
by [XQuartz](https://www.xquartz.org/) which needs to be installed and running on a system prior to loging in to
a remote system. If X11 support is not available or broken then one can still connect via `srun` by dropping the
`--x11` argument form the `srun` command.
To run your most frequently used `srun` command quickly, one can place the following line
in a user's `~/.bashrc` file. After the next login or sourcing the `.bashrc` file, one can
execute the command with its alias, here: `srun_gen242`. In this case `echo` is used to print the command to the screen rather
than executing it right away. This way one can copy and paste it, make changes as needed and
then execute it.
```sh
alias srun_gen242='echo "srun --x11 --partition=gen242 --account=gen242 --mem=20gb --cpus-per-task 8 --ntasks 1 --time 20:00:00 --pty bash -l"'
```
### Monitoring jobs with `squeue`
List all jobs in queue
```sh
squeue
```
List jobs of a specific user
```sh
squeue -u
```
Print more detailed information about a job
```sh
scontrol show job
scontrol show jobid -dd
```
Custom command to summarize and visualize cluster activity
```sh
jobMonitor
```
### Deleting and altering jobs
Delete a single job
```sh
scancel -i
```
Delete all jobs of a user
```sh
scancel -u
```
Delete all jobs of a certain name
```sh
scancel --name
```
Altering jobs with `scontrol update`. The below example changes the walltime (``) of a specific job (``).
```sh
scontrol update jobid= TimeLimit=
```
### Resource limits
Resourse limits for users can be viewed as follows.
```sh
sacctmgr show account $GROUP format=Account,User,Partition,GrpCPUs,GrpMem,GrpNodes --ass | grep $USER
```
Similarly, one can view the limits of the group a user belongs to.
```sh
sacctmgr show account $GROUP format=Account,User,Partition,GrpCPUs,GrpMem,GrpNodes,GrpTRES%30 --ass | head -3
```
## Text/code editors
The following list includes examples of several widely used code editors.
* __Vi/Vim/Neovim__: Non-graphical (terminal-based) editor. Vi is guaranteed to be available on any system. Vim and Nvim (Neovim) are the improved versions of vi.
* __Emacs__: Non-graphical or window-based editor. You still need to know keystroke commands to use it. Installed on all Linux distributions and on most other Unix systems.
* __Pico__: Simple terminal-based editor available on most versions of Unix. Uses keystroke commands, but they are listed in logical fashion at bottom of screen.
* __Nano__: A simple terminal-based editor which is default on modern Debian systems.
* __Atom__: Modern text editor developed by GitHub project.
### Why does it matter?
To work efficiently on remote systems like a computer cluster, it is essential
to learn how to work in a pure command-line interface. GUI environments like
RStudio and similar coding environments are not suitable for this. In addition,
there is a lot of value of knowing how to work in an environment that is not
restricted to a specific programming language. Therefore, this class embraces
RStudio where it is useful, but for working on remote systems like HPCC Cluster, it
uses Nvim and Tmux. Both are useful for many programming languages.
Combinded with the `nvim-r` plugin they also provide a powerful command-line working
environment for R. The following provides a brief introduction to this environment.
### Vim overview
The following opens a file (here `myfile`) with nvim (or vim)
```sh
nvim myfile.txt # for neovim (or 'vim myfile.txt' for vim)
```
Once you are in Nvim, there are three main modes: normal, insert and command mode. The most important commands for switching between the three modes are:
* `i`: The `i` key brings you from the normal mode to the insert mode. The latter is used for typing.
* `Esc`: The `Esc` key brings you from the insert mode back to the normal mode.
* `:`: The `:` key starts the command mode at the bottom of the screen.
Use the arrow keys to move your cursor in the text. Using `Fn Up/Down key` allows to page through
the text quicker. In the following command overview, all commands starting with `:` need to be typed in the command mode.
All other commands are typed in the normal mode after pushing the `Esc` key.
Important modifier keys to control vim/nvim
* `:w`: save changes to file. If you are in editing mode you have to hit `Esc` first.
* `:q`: quit file that has not been changed
* `:wq`: save and quit file
* `:!q`: quit file without saving any changes
### Useful resources for learning vim/nvim
* [Interactive Vim Tutorial](http://www.openvim.com)
* [Official Vim Documentation](http://vimdoc.sourceforge.net/)
* [HPCC Vim/Nvim Overview](https://hpcc.ucr.edu/manuals/hpc_cluster/terminalide/#vimnvim-overview)
* [HPCC Linux Manual (old)](http://hpcc.ucr.edu/manuals_linux-basics_vim.html)
## Nvim-R-Tmux essentials
Terminal-based Working Environment for R: [Nvim-R-Tmux](https://gist.github.com/tgirke/7a7c197b443243937f68c422e5471899).
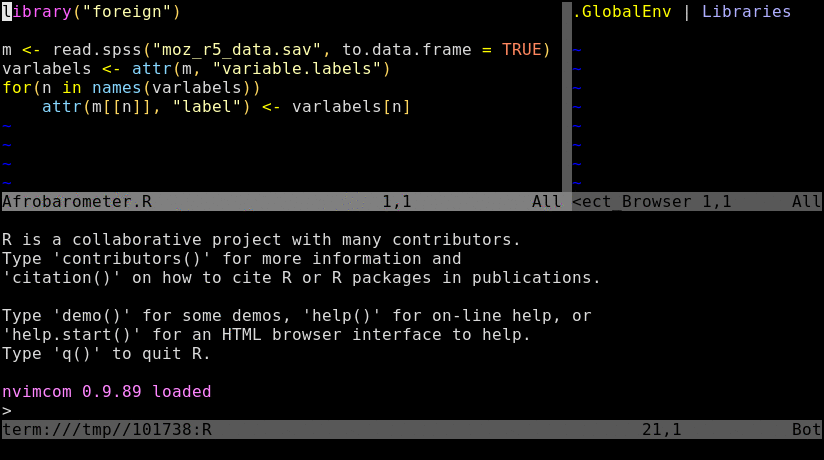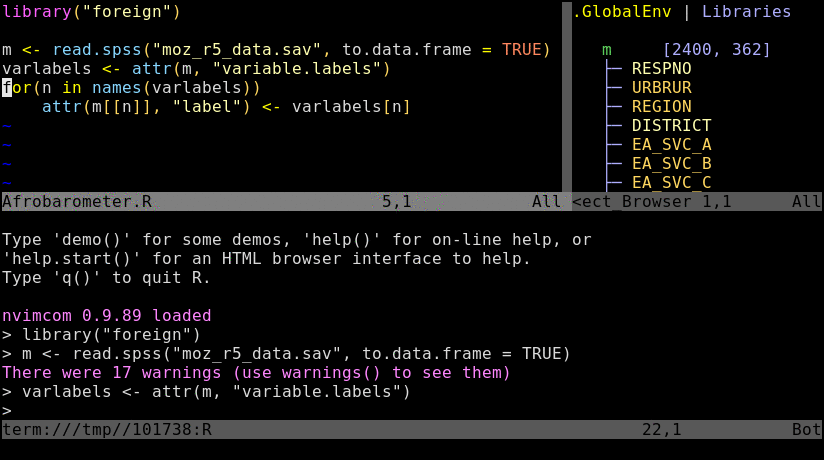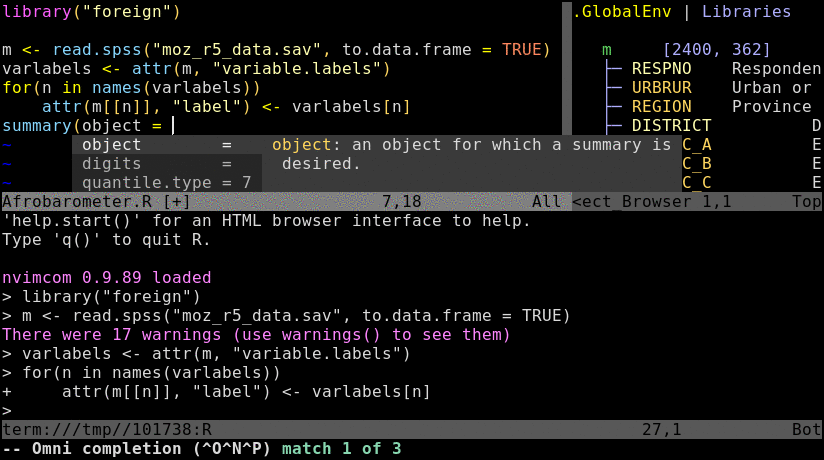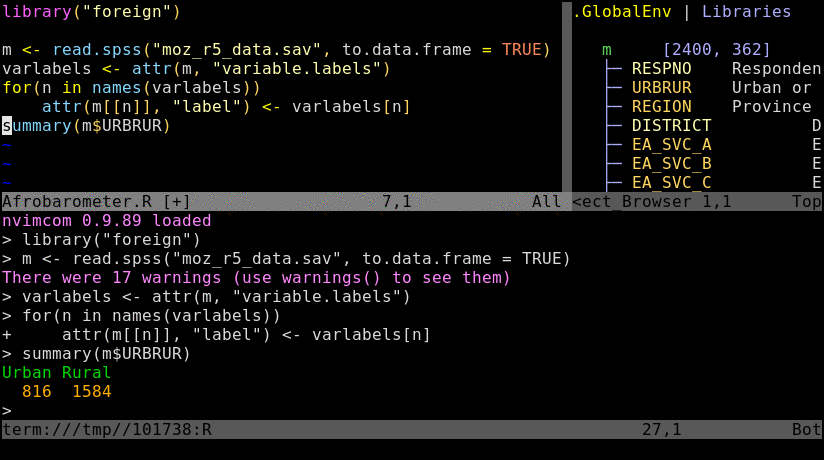 Nvim-R-Tmux IDE for R
### Basics
Tmux is a terminal multiplexer that allows to split terminal windows and to detach/reattach to
existing terminal sessions. Combinded with the `nvim-r` plugin it provides a powerful command-line working
environment for R where users can send code from a script to the R console or command-line.
Both tmux and the `nvim-r` plugin need to be installed on a system. On HPCC Cluster both are configured
in each user account. If this is not the case then follow the quick configuration instructions given in the following subsection.
### Quick configuration in user accounts of UCR's HPCC
Skip these steps if Nvim-R-Tmux is already configured in your account. Or follow the [detailed
instructions](https://github.com/tgirke/Nvim-R_Tmux) to install Nvim-R-Tmux from scratch on your own system.
1. Log in to your user account on HPCC and execute `Install_Nvim-R_Tmux` (old version: `install_nvimRtmux`). Alternatively, follow these step-by-step [install commands](https://github.com/tgirke/Nvim-R_Tmux).
2. To enable the nvim-R-tmux environment, log out and in again.
3. Follow usage instructions of next section.
### Basic usage of Nvim-R-Tmux
The official and much more detailed user manual for `Nvim-R` is available [here](https://github.com/jalvesaq/Nvim-R/blob/master/doc/Nvim-R.txt).
The following gives a short introduction into the basic usage of Nvim-R-Tmux. The short overview is on this [slide](https://docs.google.com/presentation/d/1AYVON1pS-vBe5NLp89L2HMBZZjr6L-Z20zN6ZG9r7L0/edit#slide=id.p):
__1. Start tmux session__
Note, running Nvim from within a tmux session is optional. One can skip this step if tmux functionality is not required. However, when working on remote systems, the option of re-attaching to terminal sessions is often indispensable. When using `tmux` it is important to remember on which head node it was started (on HPCC: `skylark` or `bluejay`), since it can only be restarted from the head node where it is running.
```sh
tmux # starts a new tmux session
tmux a # attaches to an existing session
```
__2. Open nvim-connected R session__
Open a `*.R` or `*.Rmd` file with `nvim` and intialize a connected R session with `\rf`. This command can be remapped to other key combinations, e.g. uncommenting lines 10-12 in `.config/nvim/init.vim` will remap it to the `F2` key. Note, the resulting split window among Nvim and R behaves like a split viewport in `nvim` or `vim` meaning the usage of `Ctrl-w w` followed by `i` and `Esc` is important for navigation.
```sh
nvim myscript.R # or *.Rmd file
```
__3. Send R code from nvim to the R pane__
Single lines of code can be sent from nvim to the R console by pressing the space bar. To send
several lines at once, one can select them in nvim's visual mode and then hit the space bar.
Please note, the default command for sending code lines in the nvim-r-plugin is `\l`. This key
binding has been remapped in the provided `.config/nvim/init.vim` file to the space bar. Most other key bindings (shortcuts) still start with the `\` as LocalLeader, _e.g._ `\rh` opens the help for a function/object where the curser is located in nvim. More details on this are given below.
### Important keybindings for nvim
The main advantages of Neovim compared to Vim are its better performance and its built-in terminal emulator facilitating the communication among Neovim and interactive programming environments such as R. Since the Vim and Neovim environments are managed independently, one can run them in parallel on the same system without interfering with each other. The usage of Neovim is almost identical to Vim.
In the following keybinding syntax, keys connected with a dash `-` need to be pressed at the same time, _e.g._ `Ctrl-w` or `Ctrl-a`. Any key after that (space separated) needs to be pressed after releasing the combined keys. So `Ctrl-w w` means: press `Ctrl` and `w` simultaneously, then release both keys, and then press `w`. Usually, these key combinations need to be pressed in a short sequence to work.
__Nvim commands__
* `\rf`: opens vim-connected R session. If you do this the first time in your user account, you might be asked to create an `R` directory under `~/`. If so approve this action by pressing `y`.
* `spacebar`: sends code from vim to R; here remapped in `init.vim` from default `\l`
* `\ce`: send code chunk where cursor is and show commands in console.
* `\ch`: send all code chunks above cursor including current one. For many more options, see section 4 in main Nvim-R manual [here](https://raw.githubusercontent.com/jalvesaq/Nvim-R/master/doc/Nvim-R.txt).
* `:split` or `:vsplit`: splits viewport (similar to pane split in tmux)
* `gz`: maximizes size of viewport in normal mode (similar to Tmux's `Ctrl-a z` zoom utility)
* `Ctrl-w w`: jumps cursor to R viewport and back; toggle between insert (`i`) and command (`Esc`) mode is required for navigation and controlling the environment.
* `Ctrl-w r`: swaps viewports
* `Ctrl-w =`: resizes splits to equal size
* `:vsplit term://bash` or `:terminal`: opens terminal in split mode or in a separate window, respectively.
* `:resize <+5 or -5>`: resizes height by specified value
* `:vertical resize <+5 or -5>`: resizes width by specified value
* `Ctrl-w H` or `Ctrl-w K`: toggles between horizontal/vertical splits
* `Ctrl-spacebar`: omni completion for R objects/functions when nvim is in insert mode. Note, this has been remapped in `init.vim` from difficult to type default `Ctrl-x Ctrl-o`.
* `:set mouse=n`: enables mouse support and `: set mouse-=n` disables it
* `:set mouse=a`: enables mouse support and `: set mouse-=a` disables it. The `a` option applies it to all vim modes, which sometimes results in unexpected behavior. On some systems the `n` option may be more suitable. To enable mouse support by default, include `set mouse=n` in Nvim’s config file located in a user’s home under `~/.config/nvim/init.vim`.
* `:h nvim-R`: opens nvim-R's user manual; navigation works the same as for any Vim/Nvim help document
* `/` or `?`: searches in text forward or backward, respectively.
* `:%s/search_pattern/replace_pattern/cg`: replacement syntax. The `c` flag at the end will ask for confirmation of each replacement. Drop it if this is not required.
* `:%s/\s\+$//`: removes trailing spaces at end of lines
* `yy`, `dd` and `p`: deletes, copies and pastes lines where the cursor is. See vim's visual mode [here](https://opensource.com/article/19/2/getting-started-vim-visual-mode) for more options.
* `:set spell`: turns on spell checking, and `:set nospell` turns it off. Misspelled words are highlighted. To get spelling suggestions, place the cursor on a misspelled word and then press `z=`. Suggestions can be accepted by the given numbers and then pressing `Enter`. To jump to the next or previous misspelled word, type use `]s` or `[s`, respectively. New (custom) words can be added to the spelling dictionary with `zg`.
* `:Rhelp fct_name`: opens help for a function from nvim's command mode with text completion support
* `Ctrl-s and Ctrl-x`: freezes/unfreezes vim (some systems)
* `:set tabstop=20`: quick and simple way to view tab delimited tables
An extended listing of important nvim commands is [here](https://hpcc.ucr.edu/manuals/hpc_cluster/terminalide/#vimnvim-overview).
### File browser built into vim: `NERDtree`
NERDtree provides file browser functionality for Vim. To enable it, the NERDtree plugin needs to be installed. It is included in the account configuration
with `Install_Nvim-R_Tmux` mentioned [above](https://bit.ly/43Ci6MF). To use NERDtree, open
a file with vim/nvim and then type in normal mode `zz`.
The basic NERDtree usage is explained [here](https://github.com/tgirke/Nvim-R_Tmux#33-basic-nerdtree-usage).
### Important keybindings for tmux
__Pane-level commands__
* `Ctrl-a %`: splits pane vertically
* `Ctrl-a "`: splits pane horizontally
* `Ctrl-a o`: jumps cursor to next pane
* `Ctrl-a Ctrl-o`: swaps panes
* `Ctrl-a `: rotates pane arrangement
* `Ctrl-a Alt `: resizes to left or right
* `Ctrl-a Esc `: resizes to left or right
* `Ctrl-a : set -g mouse on`: turns on mouse support
__Window-level comands__
* `Ctrl-a n`: switches to next tmux window
* `Ctrl-a Ctrl-a`: switches to previous tmux window
* `Ctrl-a c`: creates a new tmux window
* `Ctrl-a 1`: switches to specific tmux window selected by number
__Session-level comands__
* `Ctrl-a d`: detaches from current session
* `Ctrl-a s`: switch between available tmux sesssions
* `$ tmux new -s `: starts new session with a specific name
* `$ tmux ls`: lists available tmux session(s)
* `$ tmux attach -t `: attaches to specific tmux session
* `$ tmux attach`: reattaches to session
* `$ tmux kill-session -t `: kills a specific tmux session
* `Ctrl-a : kill-session`: kills a session from tmux command mode that can be initiated with `Ctrl-a :`
Mouse support in tmux can be enabled with the following command.
* `Ctrl-a : set -g mouse on`
To turn mouse support on by default, include on a separate line of `~/.tmux.conf` this command: `set -g mouse on`
## Nvim IDEs for other languages
For other languages, such as Bash, Python and Ruby, one can use the
[vimcmdline](https://github.com/jalvesaq/vimcmdline) plugin for nvim (or vim). To
install it, one needs to copy from the `vimcmdline` resository the directories
`ftplugin`, `plugin` and `syntax` and their files to `~/.config/nvim/`. For
user accounts of UCR’s HPCC, the above install script `install_nvimRtmux` includes the
install of `vimcmdline` (since 09-Jun-18).
The usage of `vimcmdline` is very similar to `nvim-R`. To start a connected terminal session, one
opens with nvim a code file with the extension of a given language (_e.g._ `*.sh` for Bash or `*.py` for Python),
while the corresponding interactive interpreter session is initiated
by pressing the key sequence `\s` (corresponds to `\rf` under `nvim-R`). Subsequently, code lines can be sent
with the space bar. More details are available [here](https://github.com/jalvesaq/vimcmdline).
Nvim-R-Tmux IDE for R
### Basics
Tmux is a terminal multiplexer that allows to split terminal windows and to detach/reattach to
existing terminal sessions. Combinded with the `nvim-r` plugin it provides a powerful command-line working
environment for R where users can send code from a script to the R console or command-line.
Both tmux and the `nvim-r` plugin need to be installed on a system. On HPCC Cluster both are configured
in each user account. If this is not the case then follow the quick configuration instructions given in the following subsection.
### Quick configuration in user accounts of UCR's HPCC
Skip these steps if Nvim-R-Tmux is already configured in your account. Or follow the [detailed
instructions](https://github.com/tgirke/Nvim-R_Tmux) to install Nvim-R-Tmux from scratch on your own system.
1. Log in to your user account on HPCC and execute `Install_Nvim-R_Tmux` (old version: `install_nvimRtmux`). Alternatively, follow these step-by-step [install commands](https://github.com/tgirke/Nvim-R_Tmux).
2. To enable the nvim-R-tmux environment, log out and in again.
3. Follow usage instructions of next section.
### Basic usage of Nvim-R-Tmux
The official and much more detailed user manual for `Nvim-R` is available [here](https://github.com/jalvesaq/Nvim-R/blob/master/doc/Nvim-R.txt).
The following gives a short introduction into the basic usage of Nvim-R-Tmux. The short overview is on this [slide](https://docs.google.com/presentation/d/1AYVON1pS-vBe5NLp89L2HMBZZjr6L-Z20zN6ZG9r7L0/edit#slide=id.p):
__1. Start tmux session__
Note, running Nvim from within a tmux session is optional. One can skip this step if tmux functionality is not required. However, when working on remote systems, the option of re-attaching to terminal sessions is often indispensable. When using `tmux` it is important to remember on which head node it was started (on HPCC: `skylark` or `bluejay`), since it can only be restarted from the head node where it is running.
```sh
tmux # starts a new tmux session
tmux a # attaches to an existing session
```
__2. Open nvim-connected R session__
Open a `*.R` or `*.Rmd` file with `nvim` and intialize a connected R session with `\rf`. This command can be remapped to other key combinations, e.g. uncommenting lines 10-12 in `.config/nvim/init.vim` will remap it to the `F2` key. Note, the resulting split window among Nvim and R behaves like a split viewport in `nvim` or `vim` meaning the usage of `Ctrl-w w` followed by `i` and `Esc` is important for navigation.
```sh
nvim myscript.R # or *.Rmd file
```
__3. Send R code from nvim to the R pane__
Single lines of code can be sent from nvim to the R console by pressing the space bar. To send
several lines at once, one can select them in nvim's visual mode and then hit the space bar.
Please note, the default command for sending code lines in the nvim-r-plugin is `\l`. This key
binding has been remapped in the provided `.config/nvim/init.vim` file to the space bar. Most other key bindings (shortcuts) still start with the `\` as LocalLeader, _e.g._ `\rh` opens the help for a function/object where the curser is located in nvim. More details on this are given below.
### Important keybindings for nvim
The main advantages of Neovim compared to Vim are its better performance and its built-in terminal emulator facilitating the communication among Neovim and interactive programming environments such as R. Since the Vim and Neovim environments are managed independently, one can run them in parallel on the same system without interfering with each other. The usage of Neovim is almost identical to Vim.
In the following keybinding syntax, keys connected with a dash `-` need to be pressed at the same time, _e.g._ `Ctrl-w` or `Ctrl-a`. Any key after that (space separated) needs to be pressed after releasing the combined keys. So `Ctrl-w w` means: press `Ctrl` and `w` simultaneously, then release both keys, and then press `w`. Usually, these key combinations need to be pressed in a short sequence to work.
__Nvim commands__
* `\rf`: opens vim-connected R session. If you do this the first time in your user account, you might be asked to create an `R` directory under `~/`. If so approve this action by pressing `y`.
* `spacebar`: sends code from vim to R; here remapped in `init.vim` from default `\l`
* `\ce`: send code chunk where cursor is and show commands in console.
* `\ch`: send all code chunks above cursor including current one. For many more options, see section 4 in main Nvim-R manual [here](https://raw.githubusercontent.com/jalvesaq/Nvim-R/master/doc/Nvim-R.txt).
* `:split` or `:vsplit`: splits viewport (similar to pane split in tmux)
* `gz`: maximizes size of viewport in normal mode (similar to Tmux's `Ctrl-a z` zoom utility)
* `Ctrl-w w`: jumps cursor to R viewport and back; toggle between insert (`i`) and command (`Esc`) mode is required for navigation and controlling the environment.
* `Ctrl-w r`: swaps viewports
* `Ctrl-w =`: resizes splits to equal size
* `:vsplit term://bash` or `:terminal`: opens terminal in split mode or in a separate window, respectively.
* `:resize <+5 or -5>`: resizes height by specified value
* `:vertical resize <+5 or -5>`: resizes width by specified value
* `Ctrl-w H` or `Ctrl-w K`: toggles between horizontal/vertical splits
* `Ctrl-spacebar`: omni completion for R objects/functions when nvim is in insert mode. Note, this has been remapped in `init.vim` from difficult to type default `Ctrl-x Ctrl-o`.
* `:set mouse=n`: enables mouse support and `: set mouse-=n` disables it
* `:set mouse=a`: enables mouse support and `: set mouse-=a` disables it. The `a` option applies it to all vim modes, which sometimes results in unexpected behavior. On some systems the `n` option may be more suitable. To enable mouse support by default, include `set mouse=n` in Nvim’s config file located in a user’s home under `~/.config/nvim/init.vim`.
* `:h nvim-R`: opens nvim-R's user manual; navigation works the same as for any Vim/Nvim help document
* `/` or `?`: searches in text forward or backward, respectively.
* `:%s/search_pattern/replace_pattern/cg`: replacement syntax. The `c` flag at the end will ask for confirmation of each replacement. Drop it if this is not required.
* `:%s/\s\+$//`: removes trailing spaces at end of lines
* `yy`, `dd` and `p`: deletes, copies and pastes lines where the cursor is. See vim's visual mode [here](https://opensource.com/article/19/2/getting-started-vim-visual-mode) for more options.
* `:set spell`: turns on spell checking, and `:set nospell` turns it off. Misspelled words are highlighted. To get spelling suggestions, place the cursor on a misspelled word and then press `z=`. Suggestions can be accepted by the given numbers and then pressing `Enter`. To jump to the next or previous misspelled word, type use `]s` or `[s`, respectively. New (custom) words can be added to the spelling dictionary with `zg`.
* `:Rhelp fct_name`: opens help for a function from nvim's command mode with text completion support
* `Ctrl-s and Ctrl-x`: freezes/unfreezes vim (some systems)
* `:set tabstop=20`: quick and simple way to view tab delimited tables
An extended listing of important nvim commands is [here](https://hpcc.ucr.edu/manuals/hpc_cluster/terminalide/#vimnvim-overview).
### File browser built into vim: `NERDtree`
NERDtree provides file browser functionality for Vim. To enable it, the NERDtree plugin needs to be installed. It is included in the account configuration
with `Install_Nvim-R_Tmux` mentioned [above](https://bit.ly/43Ci6MF). To use NERDtree, open
a file with vim/nvim and then type in normal mode `zz`.
The basic NERDtree usage is explained [here](https://github.com/tgirke/Nvim-R_Tmux#33-basic-nerdtree-usage).
### Important keybindings for tmux
__Pane-level commands__
* `Ctrl-a %`: splits pane vertically
* `Ctrl-a "`: splits pane horizontally
* `Ctrl-a o`: jumps cursor to next pane
* `Ctrl-a Ctrl-o`: swaps panes
* `Ctrl-a `: rotates pane arrangement
* `Ctrl-a Alt `: resizes to left or right
* `Ctrl-a Esc `: resizes to left or right
* `Ctrl-a : set -g mouse on`: turns on mouse support
__Window-level comands__
* `Ctrl-a n`: switches to next tmux window
* `Ctrl-a Ctrl-a`: switches to previous tmux window
* `Ctrl-a c`: creates a new tmux window
* `Ctrl-a 1`: switches to specific tmux window selected by number
__Session-level comands__
* `Ctrl-a d`: detaches from current session
* `Ctrl-a s`: switch between available tmux sesssions
* `$ tmux new -s `: starts new session with a specific name
* `$ tmux ls`: lists available tmux session(s)
* `$ tmux attach -t `: attaches to specific tmux session
* `$ tmux attach`: reattaches to session
* `$ tmux kill-session -t `: kills a specific tmux session
* `Ctrl-a : kill-session`: kills a session from tmux command mode that can be initiated with `Ctrl-a :`
Mouse support in tmux can be enabled with the following command.
* `Ctrl-a : set -g mouse on`
To turn mouse support on by default, include on a separate line of `~/.tmux.conf` this command: `set -g mouse on`
## Nvim IDEs for other languages
For other languages, such as Bash, Python and Ruby, one can use the
[vimcmdline](https://github.com/jalvesaq/vimcmdline) plugin for nvim (or vim). To
install it, one needs to copy from the `vimcmdline` resository the directories
`ftplugin`, `plugin` and `syntax` and their files to `~/.config/nvim/`. For
user accounts of UCR’s HPCC, the above install script `install_nvimRtmux` includes the
install of `vimcmdline` (since 09-Jun-18).
The usage of `vimcmdline` is very similar to `nvim-R`. To start a connected terminal session, one
opens with nvim a code file with the extension of a given language (_e.g._ `*.sh` for Bash or `*.py` for Python),
while the corresponding interactive interpreter session is initiated
by pressing the key sequence `\s` (corresponds to `\rf` under `nvim-R`). Subsequently, code lines can be sent
with the space bar. More details are available [here](https://github.com/jalvesaq/vimcmdline).


