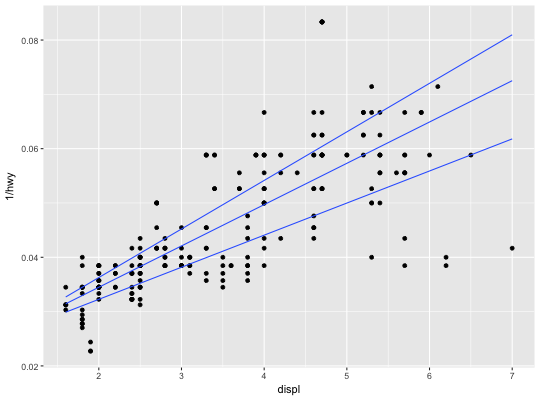
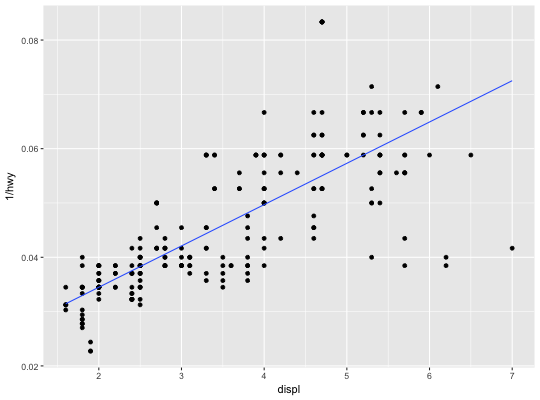
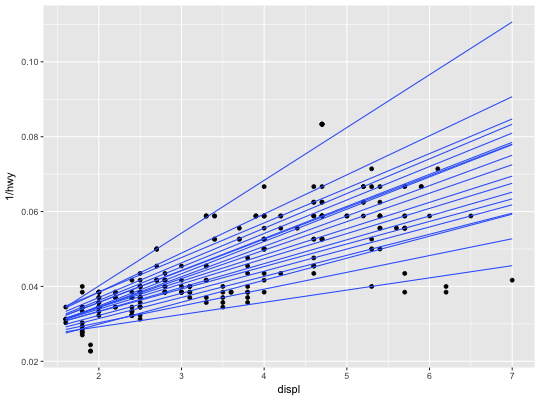
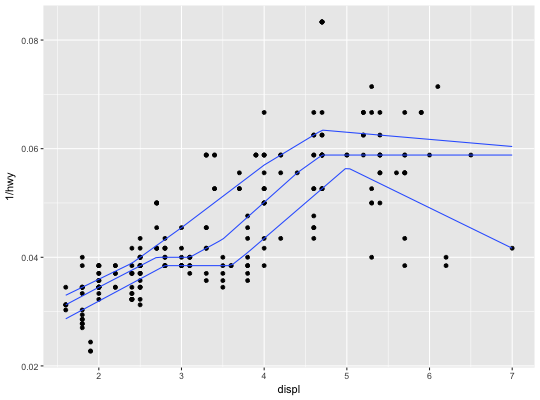
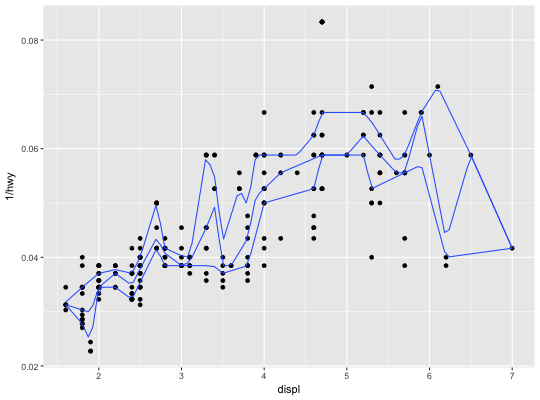
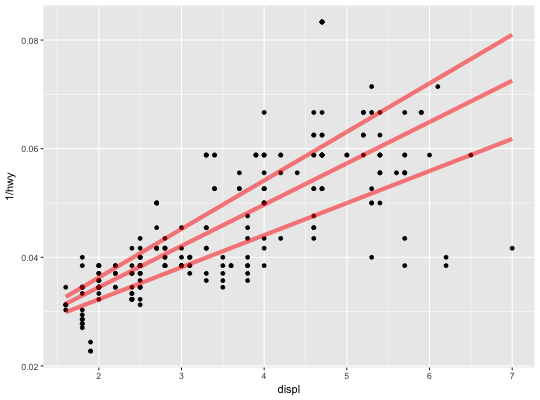
Quantile regression
This fits a quantile regression to the data and draws the fitted quantiles
with lines. This is as a continuous analogue to geom_boxplot.
geom_quantile(mapping = NULL, data = NULL, stat = "quantile", position = "identity", ..., lineend = "butt", linejoin = "round", linemitre = 1, na.rm = FALSE, show.legend = NA, inherit.aes = TRUE) stat_quantile(mapping = NULL, data = NULL, geom = "quantile", position = "identity", ..., quantiles = c(0.25, 0.5, 0.75), formula = NULL, method = "rq", method.args = list(), na.rm = FALSE, show.legend = NA, inherit.aes = TRUE)
Arguments
| mapping | Set of aesthetic mappings created by |
|---|---|
| data | The data to be displayed in this layer. There are three options: If A A |
| position | Position adjustment, either as a string, or the result of a call to a position adjustment function. |
| ... | other arguments passed on to |
| lineend | Line end style (round, butt, square) |
| linejoin | Line join style (round, mitre, bevel) |
| linemitre | Line mitre limit (number greater than 1) |
| na.rm | If |
| show.legend | logical. Should this layer be included in the legends?
|
| inherit.aes | If |
| geom, stat | Use to override the default connection between
|
| quantiles | conditional quantiles of y to calculate and display |
| formula | formula relating y variables to x variables |
| method | Quantile regression method to use. Currently only supports
|
| method.args | List of additional arguments passed on to the modelling
function defined by |
Aesthetics
geom_quantile understands the following aesthetics (required aesthetics are in bold):
xyalphacolourgrouplinetypesizeweight
Computed variables
- quantile
quantile of distribution
Examples
#>#> #>#> #> #>#> #>#> #> #>#>#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'm + geom_quantile(quantiles = 0.5)#>#> Warning: partial match of 'coef' to 'coefficients'q10 <- seq(0.05, 0.95, by = 0.05) m + geom_quantile(quantiles = q10)#>#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'# You can also use rqss to fit smooth quantiles m + geom_quantile(method = "rqss")#>#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'factor' to 'factors'# Note that rqss doesn't pick a smoothing constant automatically, so # you'll need to tweak lambda yourself m + geom_quantile(method = "rqss", lambda = 0.1)#>#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'factor' to 'factors'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'factor' to 'factors'# Set aesthetics to fixed value m + geom_quantile(colour = "red", size = 2, alpha = 0.5)#>#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'#> Warning: partial match of 'coef' to 'coefficients'