---
title: "tidymodels"
subtitle: "Biostat 203B"
author: "Dr. Hua Zhou @ UCLA"
date: today
format:
html:
theme: cosmo
embed-resources: true
number-sections: true
toc: true
toc-depth: 4
toc-location: left
code-fold: false
engine: knitr
knitr:
opts_chunk:
fig.align: 'center'
# fig.width: 6
# fig.height: 4
message: FALSE
cache: false
---
# Overview
- A typical data science project:
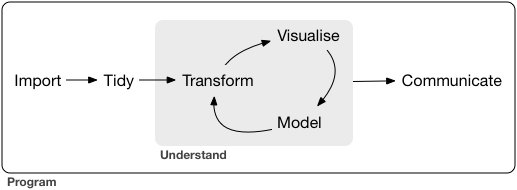
- [tidymodels](https://www.tidymodels.org/) is an ecosystem for:
1. Feature engineering: coding qualitative predictors, transformation of predictors (e.g., log), extracting key features from raw variables (e.g., getting the day of the week out of a date variable), interaction terms, ... ([recipes](https://recipes.tidymodels.org/reference/index.html) package);
2. Build and fit a model ([parsnip](https://parsnip.tidymodels.org/index.html) package);
3. Evaluate model using resampling (such as cross-validation) ([tune](https://tune.tidymodels.org/) and [dial](https://dials.tidymodels.org/) packages);
4. Tuning model parameters.

- tidymodels is the R analog of [sklearn.pipeline](https://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html) in Python and [MLJ.jl](https://alan-turing-institute.github.io/MLJ.jl/dev/) in Julia.
# Heart data example
We illustrate a binary classification example using a dataset from the Cleveland Clinic Foundation for Heart Disease.
## Logistic regression (with enet regularization) workflow
[qmd](https://raw.githubusercontent.com/ucla-biostat-203b/2024winter/master/slides/18-tidymodels/workflow_logit_heart.qmd), [html](https://ucla-biostat-203b.github.io/2024winter/slides/18-tidymodels/workflow_logit_heart.html)
## Random forest workflow
[qmd](https://raw.githubusercontent.com/ucla-biostat-203b/2024winter/master/slides/18-tidymodels/workflow_rf_heart.qmd), [html](https://ucla-biostat-203b.github.io/2024winter/slides/18-tidymodels/workflow_rf_heart.html)
## Boosting (XGBoost) workflow
[qmd](https://raw.githubusercontent.com/ucla-biostat-203b/2024winter/master/slides/18-tidymodels/workflow_xgboost_heart.qmd), [html](https://ucla-biostat-203b.github.io/2024winter/slides/18-tidymodels/workflow_xgboost_heart.html)
## SVM (with radial basis kernel) workflow
[qmd](https://raw.githubusercontent.com/ucla-biostat-203b/2024winter/master/slides/18-tidymodels/workflow_svmrbf_heart.qmd), [html](https://ucla-biostat-203b.github.io/2024winter/slides/18-tidymodels/workflow_svmrbf_heart.html)
## Multi-layer perceptron (MLP) workflow
[qmd](https://raw.githubusercontent.com/ucla-biostat-203b/2024winter/master/slides/18-tidymodels/workflow_mlp_heart.qmd), [html](https://ucla-biostat-203b.github.io/2024winter/slides/18-tidymodels/workflow_mlp_heart.html)
## Ensemble (model stacking) workflow
> We differentiate **homogenous ensemble** (e.g., bagging, boosting) from **heterogeneous ensemble** (e.g., stacking). The former uses the same type of model (e.g., random forest) to build multiple models and then combine them. The latter uses different types of models (e.g., random forest, SVM, and neural network) to build multiple models and then combine them.
[qmd](https://raw.githubusercontent.com/ucla-biostat-203b/2024winter/master/slides/18-tidymodels/workflow_stack_heart.qmd), [html](https://ucla-biostat-203b.github.io/2024winter/slides/18-tidymodels/workflow_stack_heart.html)


