---
title: Plotting in Julia
subtitle: Biostat/Biomath M257
author: Dr. Hua Zhou @ UCLA
date: today
format:
html:
theme: cosmo
embed-resources: true
number-sections: true
toc: true
toc-depth: 4
toc-location: left
code-fold: false
jupyter:
jupytext:
formats: 'ipynb,qmd'
text_representation:
extension: .qmd
format_name: quarto
format_version: '1.0'
jupytext_version: 1.14.5
kernelspec:
display_name: Julia 1.8.5
language: julia
name: julia-1.8
---
System information (for reproducibility):
```{julia}
versioninfo()
```
Load packages:
```{julia}
using Pkg
Pkg.activate(pwd())
Pkg.instantiate()
Pkg.status()
```
```{julia}
using Gadfly, Plots, PlotlyJS, PyPlot, Random, RCall
```
# Plotting in Julia
**Warning**: The time to first plot can be frustratingly long in Julia because of the JIT compilation.
The five most popular options (currently) for plotting in Julia are
- [Plots.jl](https://github.com/JuliaPlots/Plots.jl)
- Defines an unified interface for plotting
- Maps arguments to different plotting "backends"
- PyPlot, GR, PlotlyJS, and many more
- For a complete list of backends:
- Mapping of attributes to backends:
- First runs can be slowish. I found the GR backend fastest and most stable.
- [Makie.jl](https://github.com/JuliaPlots/Makie.jl)
- A pure Julia solution. Very extensible. Check [GeneticsMakie.jl](https://github.com/mmkim1210/GeneticsMakie.jl).
- R users can just use [RCall.jl](https://github.com/JuliaInterop/RCall.jl) package to plot using ggplot2 in R.
- [Gadfly.jl](https://github.com/GiovineItalia/Gadfly.jl)
- Julia equivalent of `ggplot2` in R
- [PyPlot.jl](https://github.com/JuliaPy/PyPlot.jl)
- Wrapper for Python's matplotlib
## Gadfly.jl
To demonstrate Gadfly, we will go through an example and compare it to ggplot2.
```{julia}
using RCall
R"""
library(ggplot2)
library(dplyr)
df <- ToothGrowth %>%
group_by(supp, dose) %>%
summarise(se = sd(len) / n(), len = mean(len), n = n())
ggplot(df, aes(x = dose, y = len, group = supp, color = supp)) +
geom_line() +
geom_point() +
geom_errorbar(aes(ymin = len - se, ymax = len + se), width = 0.1, alpha = 0.5,
position = position_dodge(0.005)) +
scale_color_manual(values = c(VC = "skyblue", OJ = "orange")) +
labs(x = "Dose", y = "Tooth Length")
"""
```
```{julia}
@rget df # retrieve dataframe from R to Julia workspace
using Gadfly
df[!, :ymin] = df[!, :len] - df[!, :se]
df[!, :ymax] = df[!, :len] + df[!, :se]
Gadfly.plot(df,
x = :dose,
y = :len,
color = :supp,
Geom.point,
Guide.xlabel("Dose"),
Guide.ylabel("Tooth Length"),
Guide.xticks(ticks = [0.5, 1.0, 1.5, 2.0]),
Geom.line,
Geom.errorbar,
ymin = :ymin,
ymax = :ymax,
Scale.color_discrete_manual("orange", "skyblue"))
```
Both offer more customized options
```{julia}
R"""
ggplot(df, aes(x = dose, y = len, group = supp, color = supp)) +
geom_line() +
geom_point() +
geom_errorbar(aes(ymin = len - se, ymax = len + se), width = 0.1, alpha = 0.5,
position = position_dodge(0.005)) +
theme(legend.position = c(0.8,0.1),
legend.key = element_blank(),
axis.text.x = element_text(angle = 0, size = 11),
axis.ticks = element_blank(),
panel.grid.major = element_blank(),
legend.text=element_text(size = 11),
panel.border = element_blank(),
panel.grid.minor = element_blank(),
panel.background = element_blank(),
axis.line = element_line(color = 'black',size = 0.3),
plot.title = element_text(hjust = 0.5)) +
scale_color_manual(values = c(VC = "skyblue", OJ = "orange")) +
labs(x = "Dose", y = "Tooth Length")
"""
```
```{julia}
Gadfly.plot(df, x = :dose, y = :len, color = :supp, Geom.point,
Guide.xlabel("Dose"), Guide.ylabel("Tooth Length"),
Guide.xticks(ticks = [0.5, 1.0, 1.5, 2.0]),
Theme(panel_fill = nothing, highlight_width = 0mm, point_size = 0.5mm,
key_position = :inside,
grid_line_width = 0mm, panel_stroke = colorant"black"),
Geom.line, Geom.errorbar, ymin = :ymin, ymax = :ymax,
Scale.color_discrete_manual("orange", "skyblue"))
```
## Plots.jl
We demonstrate Plots.jl below:
```{julia}
# Pkg.add("Plots")
using Plots, Random
Random.seed!(123) # set seed
x = cumsum(randn(50, 2), dims=1);
```
```{julia}
# Pkg.add("PyPlot")
pyplot() # set the backend to PyPlot
Plots.plot(x, title="Random walk", xlab="time")
```
```{julia}
gr() # change backend to GR
Plots.plot(x, title="Random walk", xlab="time")
```
```
gr()
@gif for i in 1:20
Plots.plot(x -> sin(x) / (.2i), 0, i, xlim=(0, 20), ylim=(-.75, .75))
scatter!(x -> cos(x) * .01 * i, 0, i, m=1)
end;
```
produces following animation.
 ```{julia}
# Pkg.add("PlotlyJS")
plotlyjs() # change backend to PlotlyJS
Plots.plot(x, title="Random walk", xlab="time")
```
```{julia}
# Pkg.add("PlotlyJS")
plotlyjs() # change backend to PlotlyJS
Plots.plot(x, title="Random walk", xlab="time")
```
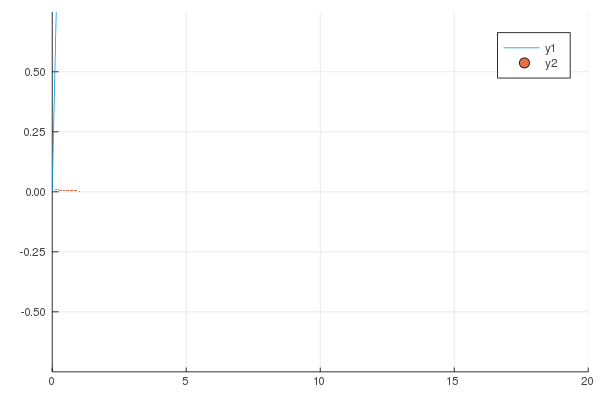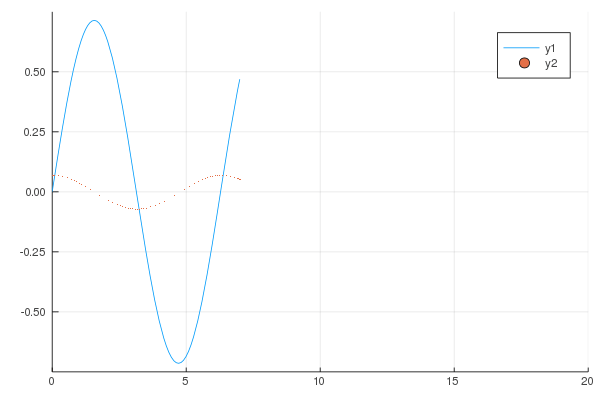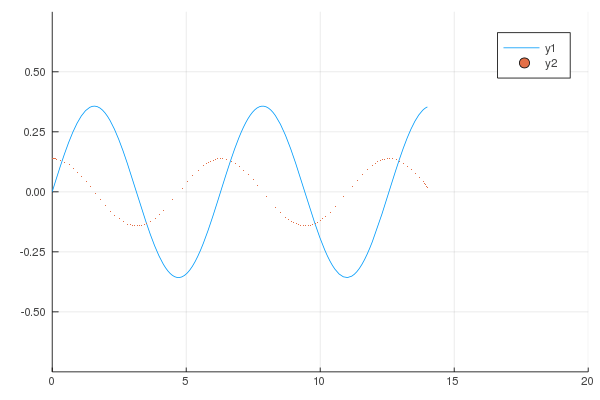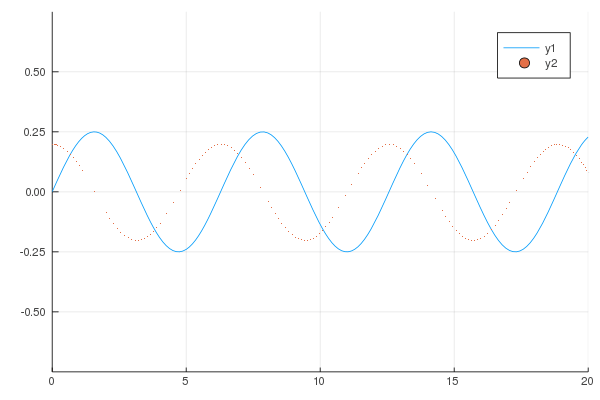 ```{julia}
# Pkg.add("PlotlyJS")
plotlyjs() # change backend to PlotlyJS
Plots.plot(x, title="Random walk", xlab="time")
```
```{julia}
# Pkg.add("PlotlyJS")
plotlyjs() # change backend to PlotlyJS
Plots.plot(x, title="Random walk", xlab="time")
```