---
execute:
echo: true
message: false
warning: false
fig-format: "svg"
format:
revealjs:
theme: lecture_styles.scss
highlight-style: a11y-dark
controls: true
controls-tutorial: true
reference-location: margin
slide-number: true
code-link: true
chalkboard: true
incremental: false
smaller: true
preview-links: true
code-line-numbers: true
history: false
progress: true
link-external-icon: true
code-annotations: hover
pointer:
color: "#b18eb1"
revealjs-plugins:
- pointer
---
```{r}
#| echo: false
#| cache: false
require(downlit)
require(xml2)
require(tidyverse)
knitr::opts_chunk$set(comment = ">")
```
## {#title-slide data-menu-title="Writing Functions" background="#1e4655" background-image="../../images/csss-logo.png" background-position="center top 5%" background-size="50%"}
[Writing Functions]{.custom-title}
[CS&SS 508 • Lecture 8]{.custom-subtitle}
[{{< var lectures.eight >}}]{.custom-subtitle2}
[Victoria Sass]{.custom-subtitle3}
# Roadmap {.section-title background-color="#99a486"}
------------------------------------------------------------------------
::: columns
::: {.column width="50%"}
### Last time, we learned:
- Types of Data
- Strings
- Pattern Matching & Regular Expressions
:::
::: {.column width="50%"}
::: fragment
### Today, we will cover:
- Function Basics
- Types of Functions
- Vector Functions
- Dataframe Functions
- Plot Functions
- Function Style Guide
:::
:::
:::
# Function Basics {.section-title background-color="#99a486"}
## Why Functions?
R (as well as mathematics in general) is full of functions!
. . .
We use functions to:
- Compute summary statistics (`mean()`, `sd()`, `min()`)
- Fit models to data (`lm(Fertility ~ Agriculture, data = swiss)`)
- Read in data (`read_csv()`)
- Create visualizations (`ggplot()`)
- And a lot more!!
## Examples of Existing Functions
::: incremental
- `mean()`:
- Input: a vector
- Output: a single number
- `dplyr::filter()`:
- Input: a data frame, logical conditions
- Output: a data frame with rows removed using those conditions
- `readr::read_csv()`:
- Input: a file path, optionally variable names or types
- Output: a data frame containing info read in from file
:::
. . .
Each function requires **inputs**, and returns **outputs**
## Why Write Your Own Functions?
::: {.incremental}
* Functions allow you to automate common tasks in a more powerful and general way than copy-and-pasting
* As requirements change, you only need to update code in one place, instead of many.
* You eliminate the chance of making incidental mistakes compared to when you copy and paste (i.e. updating a variable name in one place, but not in another).
* It makes it easier to reuse work from project-to-project, increasing your productivity over time.
* If well named, your function can make your overall code easier to understand.
:::
## Plan your Function before Writing
Before you can write effective code, you need to know *exactly* what you want:
::: incremental
- **Goal:** Do I want a single value? vector? one observation per person? per year?
- **Current State:** What do I currently have? data frame, vector? long or wide format?
- **Translate:** How can I take what I have and turn it into my goal?
- Sketch out the steps!
- Break it down into little operations
:::
. . .
**As we become more advanced coders, this concept is key!!**
**Remember:** *When you're stuck, try searching your problem on Google!!*
## Simple, Motivating Example
:::: {.columns}
::: {.column width="62%"}
```{r}
#| echo: false
set.seed(5000)
```
```{r}
#| eval: false
set.seed(5000) # <1>
df <- tibble(
a = rnorm(5),
b = rnorm(5),
c = rnorm(5),
d = rnorm(5)
)
df
df |> mutate(
a = (a - min(a, na.rm = TRUE)) /
(max(a, na.rm = TRUE) - min(a, na.rm = TRUE)),
b = (b - min(b, na.rm = TRUE)) /
(max(b, na.rm = TRUE) - min(a, na.rm = TRUE)),
c = (c - min(c, na.rm = TRUE)) /
(max(c, na.rm = TRUE) - min(c, na.rm = TRUE)),
d = (d - min(d, na.rm = TRUE)) /
(max(d, na.rm = TRUE) - min(d, na.rm = TRUE))
)
```
1. `set.seed()` allows you to reproduce the same numbers from any random number-generating process (i.e. `rnorm()`)
::: {.incremental .fragment fragment-index=3}
* What do you think this code does?
* Are there any typos?
* Could we write this more efficiently as a function?
:::
:::
::: {.column width="38%"}
::: {.fragment fragment-index=1}
```{r}
#| echo: false
set.seed(5000)
df <- tibble(
a = rnorm(5),
b = rnorm(5),
c = rnorm(5),
d = rnorm(5)
)
df
```
:::
\
::: {.fragment fragment-index=2}
```{r}
#| echo: false
df |> mutate(
a = (a - min(a, na.rm = TRUE)) /
(max(a, na.rm = TRUE) - min(a, na.rm = TRUE)),
b = (b - min(b, na.rm = TRUE)) /
(max(b, na.rm = TRUE) - min(a, na.rm = TRUE)),
c = (c - min(c, na.rm = TRUE)) /
(max(c, na.rm = TRUE) - min(c, na.rm = TRUE)),
d = (d - min(d, na.rm = TRUE)) /
(max(d, na.rm = TRUE) - min(d, na.rm = TRUE))
)
```
:::
:::
::::
## Writing a Function
To write a function you need to first analyse your repeated code to figure what parts are constant and what parts vary.
. . .
Let's look at the contents of the mutate from the last slide again.
. . .
```{r}
#| eval: false
(a - min(a, na.rm = TRUE)) / (max(a, na.rm = TRUE) - min(a, na.rm = TRUE))
(b - min(b, na.rm = TRUE)) / (max(b, na.rm = TRUE) - min(b, na.rm = TRUE))
(c - min(c, na.rm = TRUE)) / (max(c, na.rm = TRUE) - min(c, na.rm = TRUE))
(d - min(d, na.rm = TRUE)) / (max(d, na.rm = TRUE) - min(d, na.rm = TRUE))
```
. . .
There's quite a bit of repetition here and only a few elements that change.
. . .
We can see how concise our code can be if we replace the varying part with 🟪:
```{r}
#| eval: false
(🟪 - min(🟪, na.rm = TRUE)) / (max(🟪, na.rm = TRUE) - min(🟪, na.rm = TRUE))
```
## Anatomy of a Function
To turn our code into a function we need three things:
::: incremental
- **Name**: What you call the function so you can use it later. The more explanatory this is the easier your code will be to understand.
- **Argument(s)** (aka input(s), parameter(s)): What the user passes to the function that affects how it works. This is what [varies]{.custom-red} across calls.
- **Body**: The code that’s [repeated]{.custom-red} across all the calls.
:::
. . .
**Function Template**
```{r}
#| eval: false
NAME <- function(ARGUMENT1, ARGUMENT2 = DEFAULT){ # <1>
BODY
}
```
1. In this example, `ARGUMENT1` and `ARGUMENT2` values won't exist outside of the function. `ARGUMENT2` is an optional argument as it's been given a default value to use if the user does not specify one.
. . .
For our current example, this would be:
```{r}
rescale01 <- function(x) { # <2>
(x - min(x, na.rm = TRUE)) / (max(x, na.rm = TRUE) - min(x, na.rm = TRUE))
}
```
2. You can name the placeholder value(s) whatever you want but `x` is the conventional name for a numeric vector so we'll use `x` here.
## Testing Your Function
::: {.fragment}
[It's good practice to test a few simple inputs to make sure your function works as expected.]{style="font-size: 85%;"}
:::
::: {.fragment}
```{r}
rescale01(c(-10, 0, 10))
rescale01(c(1, 2, 3, NA, 5))
```
:::
::: {.fragment}
[Now we can rewrite our original code in a much simpler way!^[We'll see how we can simplify this even further next week!]]{style="font-size: 85%;"}
:::
::: {.fragment}
```{r}
df |> mutate(a = rescale01(a),
b = rescale01(b),
c = rescale01(c),
d = rescale01(d))
```
:::
## Improving Your Function
Writing a function is often an iterative process: you'll write the core of the function and then notice the ways it can be made more efficient or that it needs to include additional syntax to handle a specific use-case.
:::: {.columns}
::: {.column width="42%"}
::: {.fragment}
For instance, you might observe that our function does some unnecessary computational repetition by evaluating `min()` twice and `max()` once when both can be computed once with `range()`.
:::
::: {.fragment}
```{r}
#| code-line-numbers: false
rescale01 <- function(x) {
rng <- range(x, na.rm = TRUE)
(x - rng[1]) / (rng[2] - rng[1])
}
```
:::
:::
::: {.column width="58%"}
::: {.fragment}
Or you might find out through trial and error that our function doesn't handle infinite values well.
```{r}
#| code-line-numbers: false
x <- c(1:10, Inf)
rescale01(x)
```
:::
::: {.fragment}
Updating it to exclude infinite values makes it more general as it accounts for more use cases.
```{r}
#| code-line-numbers: false
rescale01 <- function(x) {
rng <- range(x, na.rm = TRUE, finite = TRUE)
(x - rng[1]) / (rng[2] - rng[1])
}
```
:::
:::
::::
# Vector Functions {.section-title background-color="#99a486"}
## What are Vector Functions?
The function we just created is a vector function!
. . .
Vector functions are simply functions that take one or more vectors as input and return a vector as output.
. . .
There are two types of vector functions: mutate functions and summary functions.
::: {.fragment}
#### Mutate Functions
* Return an output the same length as the input
* Therefore, these functions work well within `mutate()` and `filter()`
:::
::: {.fragment}
#### Summary Functions
* Return a single value
* Therefore well suited for use in `summarize()`
:::
## Examples of Mutate Functions
. . .
```{r}
z_score <- function(x) {
(x - mean(x, na.rm = TRUE)) / sd(x, na.rm = TRUE) # <1>
}
ages <- c(25, 82, 73, 44, 5)
z_score(ages)
```
1. Rescales a vector to have a mean of zero and a standard deviation of one.
. . .
```{r}
clamp <- function(x, min, max) {
case_when( # <2>
x < min ~ min, # <2>
x > max ~ max, # <2>
.default = x # <2>
)
}
clamp(1:10, min = 3, max = 7)
```
2. Ensures all values of a vector lie in between a minimum or a maximum.
. . .
```{r}
first_upper <- function(x) {
str_sub(x, 1, 1) <- str_to_upper(str_sub(x, 1, 1)) # <3>
x # <3>
}
first_upper("hi there, how's your day going?")
```
3. Make the first character of a sentence upper case.
## Examples of Summarize Functions
. . .
```{r}
cv <- function(x, na.rm = FALSE) {
sd(x, na.rm = na.rm) / mean(x, na.rm = na.rm) # <1>
}
cv(runif(100, min = 0, max = 50))
```
1. Calculation for the coefficient of variation, which divides the standard deviation by the mean.
. . .
```{r}
n_missing <- function(x) {
sum(is.na(x)) # <2>
}
var <- sample(c(seq(1, 20, 1), NA, NA), size = 100, replace = TRUE) # <3>
n_missing(var)
```
2. Calculates the number of missing values ([Source](https://twitter.com/gbganalyst/status/1571619641390252033)).
3. Creating a random sample of 100 values with a mix of integers from 1 to 100 and `NA` values.
. . .
```{r}
mape <- function(actual, predicted) {
sum(abs((actual - predicted) / actual)) / length(actual) # <4>
}
model1 <- lm(dist ~ speed, data = cars)
mape(cars$dist, model1$fitted.values) # <5>
```
4. Calculates the mean absolute percentage error which measures the average magnitude of error produced by a model, or how far off predictions are on average.
5. This tells us that the average absolute percentage difference between the predicted values and the actual values is ~ 38%.
# Data Frame Functions {.section-title background-color="#99a486"}
## What are Data Frame Functions?
Vector functions are useful for pulling out code that’s repeated within a dplyr verb.
. . .
But if you are building a long pipeline that is used repeatedly you'll want to write a dataframe function.
. . .
Data frame functions work like dplyr verbs: they take a data frame as the first argument, some extra arguments that say what to do with it, and return a data frame or a vector.
#### Example
```{r}
#| error: true
#| output-location: fragment
grouped_mean <- function(df, group_var, mean_var) {
df |>
group_by(group_var) |> # <1>
summarize(mean(mean_var)) # <1>
}
diamonds |> grouped_mean(cut, carat)
```
1. The goal of this function is to compute the mean of `mean_var` grouped by `group_var.`
::: {.fragment}
Uh oh, what happened?
:::
## Tidy Evaluation
Tidy evaluation is what allows us to refer to the names of variables inside a data frame without any special treatment.
. . .
This is the reason we don't have to use the `$` operator and can just call the variables directly and `tidyverse` functions know what we're referring to.
. . .
::: {.panel-tabset}
### Base `R`
```{r}
#| eval: false
diamonds[diamonds$cut == "Ideal" & diamonds$price < 1000, ]
```
### `tidyverse`
```{r}
#| eval: false
diamonds |> filter(cut == "Ideal" & price < 1000)
```
:::
. . .
Most of the time tidy evaluation does exactly what we want it to do.
. . .
The downside of tidy evaluation comes when we want to wrap up repeated tidyverse code into a function.
. . .
Here we need some way to tell the functions within *our function* not to treat our argument names as the name of the variables, but instead *look inside them* for the variable we actually want to use.
## Embracing
The tidy evaluation solution to this issue is called embracing, which means wrapping variable names in two sets of curly braces (i.e. `var` becomes `{{{ var }}}`).
. . .
Embracing a variable tells `dplyr` to use the value stored inside the argument, not the argument as the literal variable name.
. . .
```{r}
#| output-location: fragment
grouped_mean <- function(df, group_var, mean_var) {
df |>
group_by({{ group_var }}) |>
summarize(mean({{ mean_var }}))
}
diamonds |> grouped_mean(cut, carat)
```
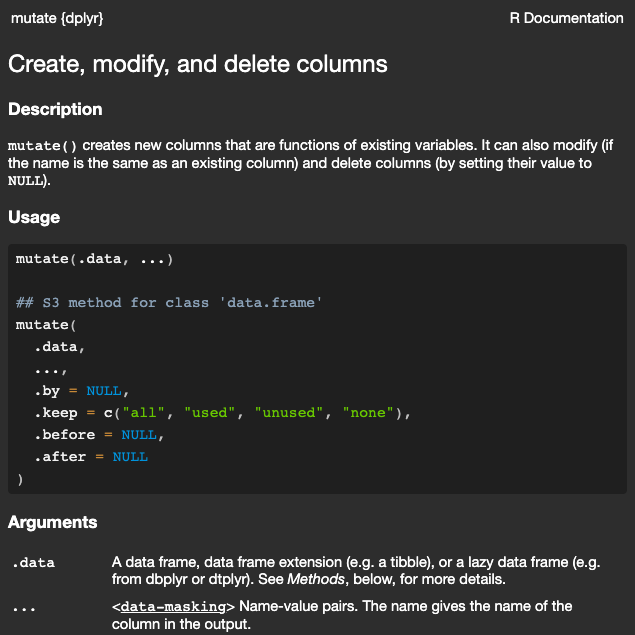
## When to Embrace? [{{< fa scroll >}}]{style="color:#99a486"} {.scrollable}
Look up the documentation of the function!
. . .
The two most common sub-types of tidy evaluation are **data-masking**^[Used in functions like `arrange()`, `filter()`, and `summarize()` that compute with variables.] and **tidy-selection**^[Used in functions like `select()`, `relocate()`, and `rename()` that select variables.].
{fig-align="center"}
## Data Frame Function Examples
```{r}
#| output-location: fragment
summary6 <- function(data, var) {
data |> summarize( # <1>
min = min({{ var }}, na.rm = TRUE), # <1>
mean = mean({{ var }}, na.rm = TRUE), # <1>
median = median({{ var }}, na.rm = TRUE), # <1>
max = max({{ var }}, na.rm = TRUE), # <1>
n = n(), # <1>
n_miss = sum(is.na({{ var }})), # <1>
.groups = "drop" # <2>
)
}
diamonds |> summary6(carat)
```
1. The goal of this function is to compute six common summary statistics for a specified variable of a dataset.
2. Whenever you wrap `summarize()` in a helper function it’s good practice to set `.groups = "drop"` to both avoid the message and leave the data in an ungrouped state.
## Data Frame Function Examples
```{r}
#| output-location: fragment
count_prop <- function(df, var, sort = FALSE) {
df |> # <1>
count({{ var }}, sort = sort) |> # <1>
mutate(prop = n / sum(n)) # <1>
}
diamonds |> count_prop(clarity)
```
1. This function is a variation of `count()` which also calculates the proportion ([Source](https://twitter.com/Diabb6/status/1571635146658402309)).
::: {.aside}
[For more functions like this one, check out the `tabyl()` function and additional formatting options in the `janitor` package.]{.fragment}
:::
# Plot Functions {.section-title background-color="#99a486"}
## What are Plot Functions?
:::: {.columns}
::: {.column width="60%"}
What if you have a lot of similar plots to create? You can use a function to eliminate redundency.
::: {.fragment}
The same technique can be used if you want to write a function that returns a plot since `aes()` is a data-masking function.
:::
::: {.fragment}
Simply use embracing within the `aes()` call to `ggplot()`!
:::
::: {.fragment}
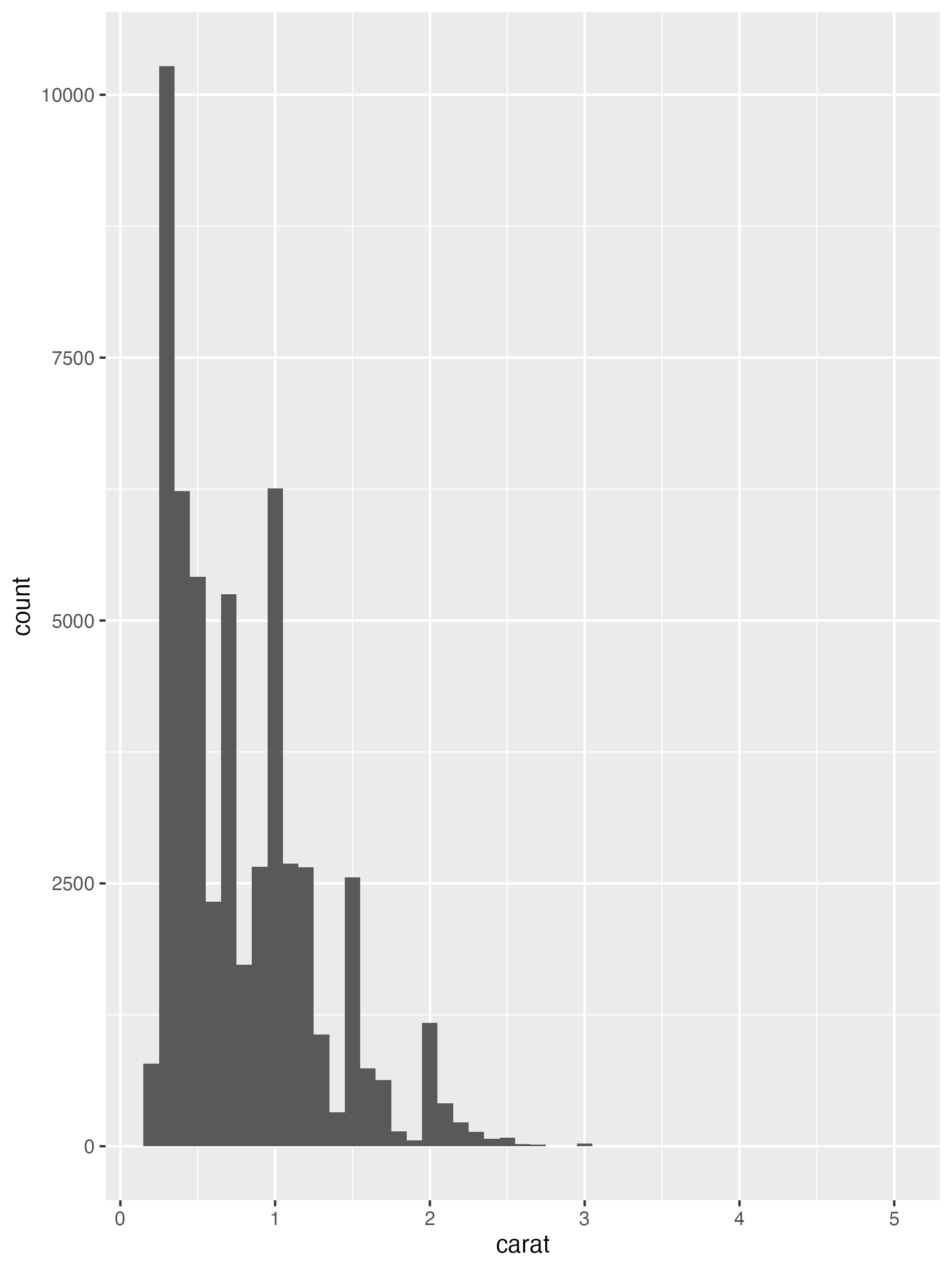
```{r}
#| output: false
histogram <- function(df, var, binwidth = NULL) {
df |>
ggplot(aes(x = {{ var }})) + # <1>
geom_histogram(binwidth = binwidth) # <1>
}
diamonds |> histogram(carat, 0.1) # <2>
```
1. This is a useful function for quickly getting histograms of a specified binwidth from a dataset.
2. Note that `histogram()` returns a ggplot2 plot, meaning you can still add on additional components if you want. Just remember to switch from `|>` to `+`.
:::
:::
::: {.column width="40%"}
```{r}
#| echo: false
ggsave("histogram.png", width = 6, height = 8, units = "in")
```
::: {.fragment}
{.absolute right=5, top=25}
:::
:::
::::
## Data Manipulation & Plotting
You might want to create a function that has a bit of data manipulation **_and_** returns a plot.
. . .
```{r}
#| output-location: fragment
#| fig-align: center
sorted_bars <- function(df, var) { # <1>
df |>
mutate({{ var }} := fct_rev(fct_infreq({{ var }}))) |> # <2>
ggplot(aes(y = {{ var }})) +
geom_bar()
}
diamonds |> sorted_bars(clarity)
```
1. This function creates a vertical bar chart where you automatically sort the bars in frequency order using `fct_infreq()`.
2. `:=` (commonly referred to as the “walrus operator”) is used here because we are generating the variable name based on user-supplied data. `R`’s syntax doesn’t allow anything to the left of `=` except for a single, literal name. To work around this problem, we use the special operator `:=` which tidy evaluation treats in exactly the same way as `=`.
## Functions that Label
:::: {.columns}
::: {.column width="60%"}
What if we want to add labels using our function?
::: {.fragment fragment-index=1}
For that we need to use the low-level package `rlang` that’s used by just about every other package in the tidyverse because it implements tidy evaluation (as well as many other useful tools).
:::
:::
::: {.column width="40%"}
:::
::::
. . .
Let's take our histogram example from before:
```{r}
#| output: false
#| echo: true
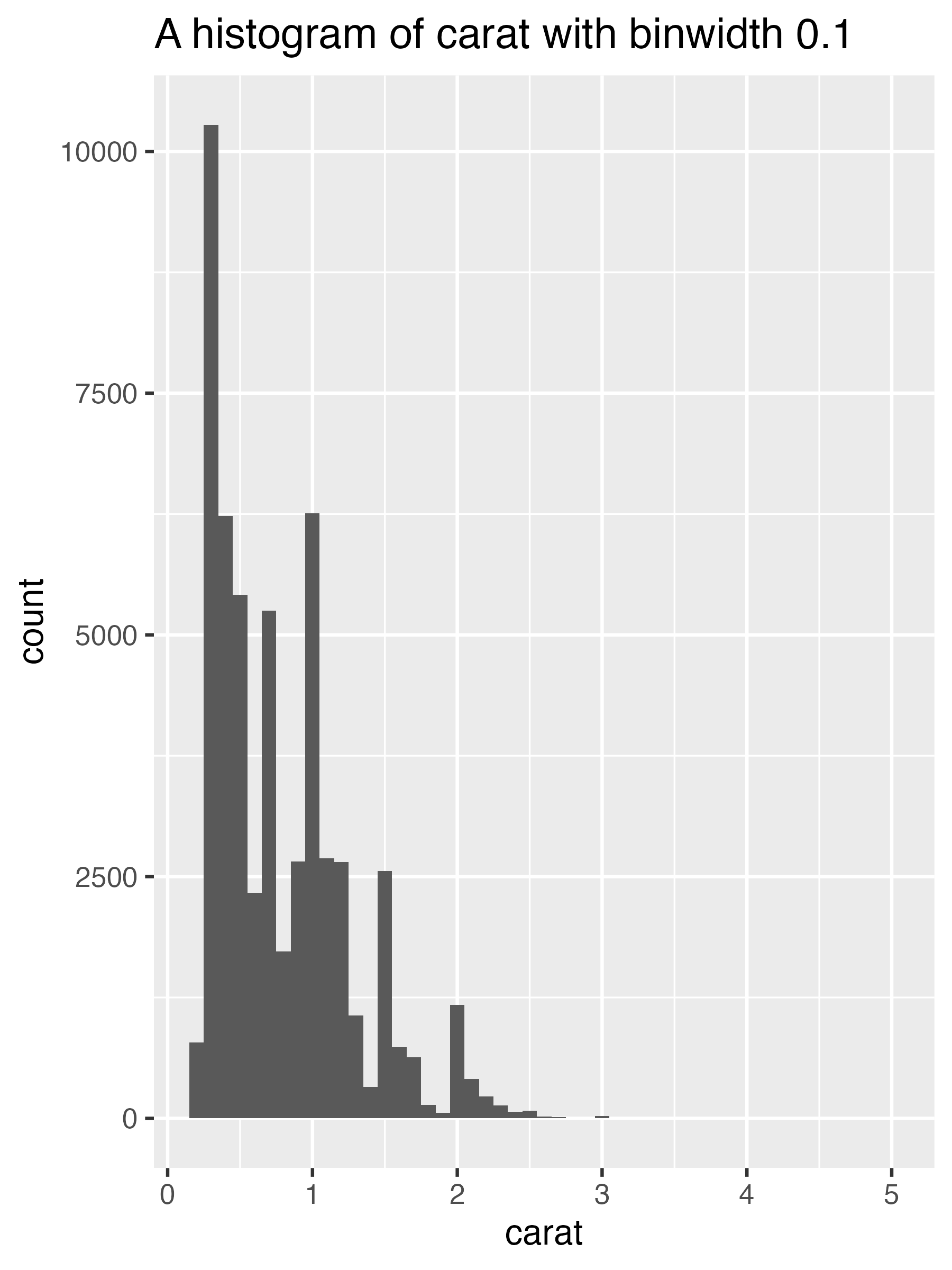
histogram <- function(df, var, binwidth) {
label <- rlang::englue("A histogram of {{ var }} with binwidth { binwidth }") # <1>
df |>
ggplot(aes(x = {{ var }})) +
geom_histogram(binwidth = binwidth) +
labs(title = label)
}
diamonds |> histogram(carat, 0.1)
```
1. `rlang::englue()` works similarly to `str_glue()`, so any value wrapped in
`{ }` will be inserted into the string. But it also understands `{{{ }}}`, which automatically inserts the appropriate variable name.
. . .
```{r}
#| echo: false
#| output: false
histogram <- function(df, var, binwidth) {
label <- rlang::englue("A histogram of {{var}} with binwidth { binwidth }")
df |>
ggplot(aes(x = {{ var }})) +
geom_histogram(binwidth = binwidth) +
labs(title = label)
}
diamonds |> histogram(carat, 0.1) + theme_gray(base_size = 16)
ggsave("histogram_label.png", width = 6, height = 8, units = "in")
```
{.absolute right=50 top=0 width="325" height="400"}
# Function Style Guide {.section-title background-color="#99a486"}
## Best Practices
::: {.incremental}
* Make function names descriptive; again longer is better due to RStudio's auto-complete feature.
* Generally, function names should be verbs, and arguments should be nouns.
- Some exceptions: computation of a well-known noun (i.e. `mean()`), accessing a property of an object (i.e. `coef()`)
* `function()` should always be followed by squiggly brackets (`{}`), and the contents should be indented by an additional two spaces^[This makes it easier to see the hierarchy in your code by skimming the left-hand margin.].
* You should put extra spaces inside of `{{ }}`. This makes it very obvious that something unusual is happening.
:::
::: aside
You can read the official tidyverse style guide for functions [here](https://style.tidyverse.org/functions).
:::
# Lab {.section-title background-color="#99a486"}
## Writing Functions
1. Practice turning the following code snippets into functions. Think about what each function does. What would you call it? How many arguments does it need?
. . .
```{r}
#| eval: false
mean(is.na(x))
mean(is.na(y))
mean(is.na(z))
x / sum(x, na.rm = TRUE)
y / sum(y, na.rm = TRUE)
z / sum(z, na.rm = TRUE)
round(x / sum(x, na.rm = TRUE) * 100, 1)
round(y / sum(y, na.rm = TRUE) * 100, 1)
round(z / sum(z, na.rm = TRUE) * 100, 1)
```
. . .
2. Bonus: Write a function that takes a name as an input (i.e. a character string) and returns a greeting based on the current time of day. **Hint**: use a time argument that defaults to `lubridate::now()`. That will make it easier to test your function.
## Answers
1. Practice turning the following code snippets into functions. Think about what each function does. What would you call it? How many arguments does it need?
```{r}
#| eval: false
mean(is.na(x))
mean(is.na(y))
mean(is.na(z))
```
. . .
```{r}
#| output-location: fragment
prop_na <- function(x){
mean(is.na(x))
}
set.seed(50) # <1>
values <- sample(c(seq(1, 10, 1), NA), 5, replace = TRUE)
values
```
1. `set.seed()` is a function that can be used to create reproducible results when writing code that involves creating variables that take on random values.
```{r}
#| output-location: fragment
prop_na(values)
```
This code calculates the proportion of `NA` values in a vector. I would call it `prop_na()` which would take a single argument, `x`, and return a single numeric value, between 0 and 1.
## Answers
1. Practice turning the following code snippets into functions. Think about what each function does. What would you call it? How many arguments does it need?
```{r}
#| eval: false
x / sum(x, na.rm = TRUE)
y / sum(y, na.rm = TRUE)
z / sum(z, na.rm = TRUE)
```
. . .
\
```{r}
#| output-location: fragment
sums_to_one <- function(x, na.rm = FALSE) {
x / sum(x, na.rm = na.rm)
}
sums_to_one(values)
```
```{r}
#| output-location: fragment
sums_to_one(values, na.rm = TRUE)
```
This code standardizes a vector so that it sums to one. It takes a numeric vector and an optional specification for removing `NA`s. While the original code had `na.rm = TRUE`, it's best to set the default to `FALSE` which will alert the user if `NA`s are present by returning `NA`.
## Answers
1. Practice turning the following code snippets into functions. Think about what each function does. What would you call it? How many arguments does it need?
```{r}
#| eval: false
round(x / sum(x, na.rm = TRUE) * 100, 1)
round(y / sum(y, na.rm = TRUE) * 100, 1)
round(z / sum(z, na.rm = TRUE) * 100, 1)
```
\
. . .
```{r}
#| output-location: fragment
pct_vec <- function(x, na.rm = FALSE){
round(x / sum(x, na.rm = na.rm) * 100, 1)
}
pct_vec(values, na.rm = TRUE)
```
This code takes a numeric vector and finds what each value represents as a percentage of the sum of the entire vector and rounds it to the first decimal place. There is also an optional `na.rm` argument set to `FALSE` by default.
## Answers
2. Bonus: Write a function that takes a name as an input (i.e. a character string) and returns a greeting based on the current time of day. **Hint 1**: use a time argument that defaults to `lubridate::now()`. That will make it easier to test your function. **Hint 2**: Use `rlang::englue` to combine your greetings with the name input.
. . .
```{r}
#| output-location: fragment
greet <- function(name, time = now()){ # <1>
hr <- hour(time)
greeting <- case_when(hr < 12 & hr >= 5 ~ rlang::englue("Good morning {name}."), # <2>
hr < 17 & hr >= 12 ~ rlang::englue("Good afternoon {name}."), # <2>
hr >= 17 ~ rlang::englue("Good evening {name}."), # <2>
.default = rlang::englue("Why are you awake rn, {name}???")) # <2>
return(greeting) # <3>
}
greet("Vic") # <4>
```
1. By default this function will take the current time to determine the specific greeting.
2. Using `englue()` allows you to include user-specified values with `{ }`.
3. `return()` or `print()` or simply calling the variable `greeting` is necessary for the function to work as expected.
4. The last time this lecture (and therefore this code) was rendered was at `r lubridate::now()`
```{r}
#| output-location: fragment
greet("Vic", time = ymd_h("2025-11-25 2am"))
```
# Homework{.section-title background-color="#1e4655"}
## {data-menu-title="Homework 8" background-iframe="https://vsass.github.io/CSSS508/Homework/HW8/homework8.html" background-interactive=TRUE}