Chapter 6: WRF Data Assimilation
Table of Contents
- Introduction
- Installing WRFDA
- Installing WRFNL and WRFPLUS
- Running Observation Preprocessor (OBSPROC)
- Running WRFDA
- Radiance Data Assimilations in WRFDA
- WRFDA Diagnostics
- Updating WRF boundary conditions
- Running gen_be
- Additional WRFDA Exercises
- Hybrid Data Assimilation
- Description of Namelist Variables
Introduction
Data assimilation is the technique by which observations are combined with a NWP product (the first guess or background forecast) and their respective error statistics to provide an improved estimate (the analysis) of the atmospheric (or oceanic, Jovian, whatever) state. Variational (Var) data assimilation achieves this through the iterative minimization of a prescribed cost (or penalty) function. Differences between the analysis and observations/first guess are penalized (damped) according to their perceived error. The difference between three-dimensional (3D-Var) and four-dimensional (4D-Var) data assimilation is the use of a numerical forecast model in the latter.
The MMM Division of NCAR supports a unified (global/regional, multi-model, 3/4D-Var) model-space data assimilation system (WRFDA) for use by NCAR staff and collaborators, and is also freely available to the general community, together with further documentation, test results, plans etc., from the WRFDA web-page http://www.mmm.ucar.edu/wrf/users/wrfda/Docs/user_guide_V3.2/users_guide_chap6.htm.
Various components of the WRFDA system are shown in blue in the sketch below, together with their relationship with rest of the WRF system.
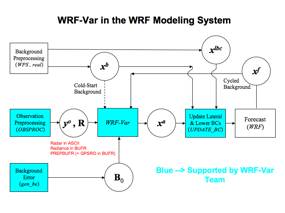
xb: first guess either from previous WRF forecast or from WPS/REAL output.
xlbc: lateral boundary from WPS/REAL output.
xa: analysis from WRFDA data assimilation system.
xf: WRF forecast output.
yo: observations processed by OBSPROC. (note: PREPBUFR input, Radar and Radiance data don’t go through OBSPROC)
B0: background error statistics from generic BE data (CV3) or gen_be.
R: observational and representative error statistics.
In this chapter, you will learn how to run the various components of WRFDA system. For the training purpose, you are supplied with a test case including the following input data: a) observation file (in the format prior to OBSPROC), b) WRF NetCDF background file (WPS/REAL output used as a first guess of the analysis), and c) Background error statistics (estimate of errors in the background file). You can download the test dataset from http://www.mmm.ucar.edu/wrf/users/wrfda/download/testdata.html. In your own work, you have to create all these input files yourselves. See the section Running Observation Preprocessor for creating your observation files. See section Running gen_be for generating your background error statistics file if you want to use cv_options=5.
Before using your own data, we suggest that you start by running through the WRFDA related programs at least once using the supplied test case. This serves two purposes: First, you can learn how to run the programs with data we have tested ourselves, and second you can test whether your computer is adequate to run the entire modeling system. After you have done the tutorial, you can try running other, more computationally intensive, case studies and experimenting with some of the many namelist variables.
WARNING: It is impossible to test every code upgrade with every permutation of computer, compiler, number of processors, case, namelist option, etc. The “namelist” options that are supported are indicated in the “WRFDA/var/README.namelist” and these are the default options.
Running with your own domain. Hopefully, our test cases will have prepared you for the variety of ways in which you may wish to run WRFDA. Please inform us about your experiences.
As a professional courtesy, we request that you include the following reference in any publications that makes use of any component of the community WRFDA system:
Barker, D.M., W. Huang, Y.R. Guo, and Q.N. Xiao., 2004: A Three-Dimensional (3DVAR) Data Assimilation System For Use With MM5: Implementation and Initial Results. Mon. Wea. Rev., 132, 897-914.
Huang, X.Y., Q. Xiao, D.M. Barker, X. Zhang, J. Michalakes, W. Huang, T. Henderson, J. Bray, Y. Chen, Z. Ma, J. Dudhia, Y. Guo, X. Zhang, D.J. Won, H.C. Lin, and Y.H. Kuo, 2009: Four-Dimensional Variational Data Assimilation for WRF: Formulation and Preliminary Results. Mon. Wea. Rev., 137, 299–314.
Running WRFDA requires a Fortran 90 compiler. We have currently tested the WRFDA on the following platforms: IBM (XLF), SGI Altix (INTEL), PC/Linux (PGI, INTEL, GFORTRAN), and Apple (G95/PGI). Please let us know if this does not meet your requirements, and we will attempt to add other machines to our list of supported architectures as resources allow. Although we are interested to hear of your experiences on modifying compile options, we do not yet recommend making changes to the configure file used to compile WRFDA.
Installing WRFDA
a. Obtaining WRFDA Source Code
Users can download the WRFDA source code from http://www.mmm.ucar.edu/wrf/users/wrfda/download/get_source.html.
After the tar file is unzipped (gunzip WRFDAV3.2.tar.gz) and untarred (untar WRFDAV3.2.tar), the directory WRFDA should be created; this directory contains the WRFDA source, external libraries, and fixed files. The following is a list of the system components and the content for each directory:
|
Directory Name |
Content |
|
var/da |
WRFDA source code |
|
var/run |
Fixed input files required by WRFDA, such as background error covariances, and radiance related files CRTM coefficients, radiance_info and VARBC.in. |
|
var/external
|
Library needed by WRFDA, include crtm, bufr, lapack, blas |
|
var/obsproc |
Obsproc source code , namelist, and observation error file. |
|
var/gen_be
|
Source code of generate background error |
|
var/build |
Build all .exe files.
|
b. Compile WRFDA and Libraries
Start with V3.1.1, to compile the WRFDA code, it is necessary to have installed the NetCDF library. The NetCDF library is the only mandatory library to install WRFDA, if only conventional observational data from LITTLE_R format file is to be used.
Only if you intend to use observational data with PREPBUFR format, an environment variables is needed to be set like (using the C-shell),
> setenv BUFR 1
In addition to BUFR library, if you intend to assimilate satellite radiance data with CRTM (V2.0.2),
> setenv CRTM 1
The CRTM will be compiled with WRFDA together. You don’t need to install the CRTM separately any more since CRTM V2.0.2. However, if you intend to use RTTOV (8.7) to assimilate radiance data, which still have to be installed separately. RTTOV (8.7) can be downloaded from http://www.metoffice.gov.uk/science/creating/working_together/nwpsaf_public.html. The additional necessary environment variables needed are set (again using the C-shell), by commands looking something like
> setenv RTTOV /usr/local/rttov87
(Note: make a linkage of $RTTOV/librttov.a to
$RTTOV/src/librttov8.7.a)
Note: Make sure the required libraries were all compiled using the same compiler that will be used to build WRFDA, since the libraries produced by one compiler may not be compatible with code compiled with another.
Assuming all required libraries are available and the WRFDA source code is ready, start to install the WRFDA as following step:
To configure WRFDA, enter the WRFDA directory and type
> ./configure wrfda
A list of configuration options for your computer should appear. Each option combines a compiler type and a parallelism option; since the configuration script doesn’t check which compilers are actually available, be sure to only select among the options for compilers that are available on your system. The parallelism option allows for a single-processor (serial) compilation, shared-memory parallel (smpar) compilation, distributed-memory parallel (dmpar) compilation and distributed-memory with shared-memory parallel (sm+dm) compilation. For example, on a Macintosh computer, the above steps look like:
> ./configure wrfda
checking for perl5... no
checking for perl... found /usr/bin/perl (perl)
Will use NETCDF in dir: /users/noname/work/external/g95/netcdf-3.6.1
PHDF5 not set in environment. Will configure WRF for use without.
$JASPERLIB or $JASPERINC not found in environment, configuring to build without grib2 I/O...
------------------------------------------------------------------------
Please select from among the following supported platforms.
1. Darwin (MACOS) PGI compiler with pgcc (serial)
2. Darwin (MACOS) PGI compiler with pgcc (smpar)
3. Darwin (MACOS) PGI compiler with pgcc (dmpar)
4. Darwin (MACOS) PGI compiler with pgcc (dm+sm)
5. Darwin (MACOS) intel compiler with icc (serial)
6. Darwin (MACOS) intel compiler with icc (smpar)
7. Darwin (MACOS) intel compiler with icc (dmpar)
8. Darwin (MACOS) intel compiler with icc (dm+sm)
9. Darwin (MACOS) intel compiler with cc (serial)
10. Darwin (MACOS) intel compiler with cc (smpar)
11. Darwin (MACOS) intel compiler with cc (dmpar)
12. Darwin (MACOS) intel compiler with cc (dm+sm)
13. Darwin (MACOS) g95 with gcc (serial)
14. Darwin (MACOS) g95 with gcc (dmpar)
15. Darwin (MACOS) xlf (serial)
16. Darwin (MACOS) xlf (dmpar)
Enter selection [1-10] : 13
------------------------------------------------------------------------
Compile for nesting? (0=no nesting, 1=basic, 2=preset moves, 3=vortex following) [default 0]:
Configuration successful. To build the model type compile .
……
After running the configuration script and choosing a compilation option, a configure.wrf file will be created. Because of the variety of ways that a computer can be configured, if the WRFDA build ultimately fails, there is a chance that minor modifications to the configure.wrf file may be needed.
Note: WRF compiles with –r4 option while WRFDA compiles with –r8. For this reason, WRF and WRFDA cannot reside and be compiled under the same directory.
Hint: It is helpful to start with something simple, such as the serial build. If it is successful, move on to build dmpar code. Remember to type ‘clean –a’ between each build.
To compile the code, type
> ./compile all_wrfvar >&! compile.out
Successful compilation of ‘all_wrfvar” will produce 32 executables in the var/build directory which are linked in var/da directory, as well as obsproc.exe in var/obsproc/src directory. You can list these executables by issuing the command (from WRFDA directory)
> ls -l var/build/*exe var/obsproc/src/obsproc.exe
-rwxr-xr-x 1 noname users 641048 Mar 23 09:28 var/build/da_advance_time.exe
-rwxr-xr-x 1 noname users 954016 Mar 23 09:29 var/build/da_bias_airmass.exe
-rwxr-xr-x 1 noname users 721140 Mar 23 09:29 var/build/da_bias_scan.exe
-rwxr-xr-x 1 noname users 686652 Mar 23 09:29 var/build/da_bias_sele.exe
-rwxr-xr-x 1 noname users 700772 Mar 23 09:29 var/build/da_bias_verif.exe
-rwxr-xr-x 1 noname users 895300 Mar 23 09:29 var/build/da_rad_diags.exe
-rwxr-xr-x 1 noname users 742660 Mar 23 09:29 var/build/da_tune_obs_desroziers.exe
-rwxr-xr-x 1 noname users 942948 Mar 23 09:29 var/build/da_tune_obs_hollingsworth1.exe
-rwxr-xr-x 1 noname users 913904 Mar 23 09:29 var/build/da_tune_obs_hollingsworth2.exe
-rwxr-xr-x 1 noname users 943000 Mar 23 09:28 var/build/da_update_bc.exe
-rwxr-xr-x 1 noname users 1125892 Mar 23 09:29 var/build/da_verif_anal.exe
-rwxr-xr-x 1 noname users 705200 Mar 23 09:29 var/build/da_verif_obs.exe
-rwxr-xr-x 1 noname users 46602708 Mar 23 09:28 var/build/da_wrfvar.exe
-rwxr-xr-x 1 noname users 1938628 Mar 23 09:29 var/build/gen_be_cov2d.exe
-rwxr-xr-x 1 noname users 1938628 Mar 23 09:29 var/build/gen_be_cov3d.exe
-rwxr-xr-x 1 noname users 1930436 Mar 23 09:29 var/build/gen_be_diags.exe
-rwxr-xr-x 1 noname users 1942724 Mar 23 09:29 var/build/gen_be_diags_read.exe
-rwxr-xr-x 1 noname users 1941268 Mar 23 09:29 var/build/gen_be_ensmean.exe
-rwxr-xr-x 1 noname users 1955192 Mar 23 09:29 var/build/gen_be_ensrf.exe
-rwxr-xr-x 1 noname users 1979588 Mar 23 09:28 var/build/gen_be_ep1.exe
-rwxr-xr-x 1 noname users 1961948 Mar 23 09:28 var/build/gen_be_ep2.exe
-rwxr-xr-x 1 noname users 1945360 Mar 23 09:29 var/build/gen_be_etkf.exe
-rwxr-xr-x 1 noname users 1990936 Mar 23 09:28 var/build/gen_be_stage0_wrf.exe
-rwxr-xr-x 1 noname users 1955012 Mar 23 09:28 var/build/gen_be_stage1.exe
-rwxr-xr-x 1 noname users 1967296 Mar 23 09:28 var/build/gen_be_stage1_1dvar.exe
-rwxr-xr-x 1 noname users 1950916 Mar 23 09:28 var/build/gen_be_stage2.exe
-rwxr-xr-x 1 noname users 2160796 Mar 23 09:29 var/build/gen_be_stage2_1dvar.exe
-rwxr-xr-x 1 noname users 1942724 Mar 23 09:29 var/build/gen_be_stage2a.exe
-rwxr-xr-x 1 noname users 1950916 Mar 23 09:29 var/build/gen_be_stage3.exe
-rwxr-xr-x 1 noname users 1938628 Mar 23 09:29 var/build/gen_be_stage4_global.exe
-rwxr-xr-x 1 noname users 1938732 Mar 23 09:29 var/build/gen_be_stage4_regional.exe
-rwxr-xr-x 1 noname users 1094740 Mar 23 09:29 var/build/gen_be_vertloc.exe
-rwxr-xr-x 1 noname users 1752352 Mar 23 09:29 var/obsproc/src/obsproc.exe
da_wrfvar.exe is the main executable for running WRFDA. Make sure it is created after the compilation. Sometimes (unfortunately) it is possible that other utilities get successfully compiled, while the main da_wrfvar.exe fails; please check the compilation log file carefully to figure out the problem.
The basic gen_be utility for regional model consists of gen_be_stage0_wrf.exe, gen_be_stage1.exe, gen_be_stage2.exe, gen_be_stage2a.exe, gen_be_stage3.exe, gen_be_stage4_regional.exe, and gen_be_diags.exe.
da_updated_bc.exe is used for updating WRF boundary condition after a new WRFDA analysis is generated.
da_advance_time.exe is a very handy and useful tool for date/time manipulation. Type “da_advance_time.exe” to see its usage instruction.
In addition to the executables for running WRFDA and gen_be, obsproc.exe (the executable for preparing conventional data for WRFDA) compilation is also included in “./compile all_wrfvar”.
Go to /external/bufr and /external/crtm to check if the libbufr.a and libcrtm.a were generated if you use BUFR and CRTM library.
c. Clean Compilation
To remove all object files and executables, type:
clean
To remove all build files, including configure.wrfda, type:
clean -a
The clean command is recommended if compilation fails or configuration file is changed.
Installing WRFNL and WRFPLUS (For 4D-Var only)
If you intend to run WRF 4D-Var, it is necessary to have installed the WRFNL (WRF nonlinear model) and WRFPLUS (WRF adjoint and tangent linear model). WRFNL is a modified version of WRF V3.2 and can only be used for 4D-Var purposes. WRFPLUS contains the adjoint and tangent linear models based on a simplified WRF model, which only includes some simple physical processes such as vertical diffusion and large-scale condensation.
To install WRFNL:
- Get the WRF zipped tar file from:
http://www.mmm.ucar.edu/wrf/users/download/get_source.html
- Unzip and untar the file, name the directory WRFNL
> cd WRFNL
> gzip -cd WRFV3.TAR.gz | tar -xf - ; mv WRFV3 WRFNL
- Get the WRFNL patch zipped tar file from:
http://www.mmm.ucar.edu/wrf/users/wrfda/download/wrfnl.html
- unzip and untar the WRFNL patch file
> gzip -cd WRFNL3.2_PATCH.tar.gz | tar -xf -
> ./configure
serial means single processor
dmpar means Distributed Memory Parallel (MPI)
smpar is not supported for 4D-Var
Please select 0 for the second option for no nesting
- Compile the WRFNL
> ./compile em_real
> ls -ls main/*.exe
If you built
the real-data case, you should see wrf.exe
To install WRFPLUS:
- Get the WRFPLUS zipped tar file from:
http://www.mmm.ucar.edu/wrf/users/wrfda/download/wrfplus.html
- Unzip and untar the file to WRFPLUS
> gzip -cd WRFPLUS3.2.tar.gz | tar -xf -
> cd WRFPLUS
> ./configure wrfplus
serial means single processor
dmpar means Distributed Memory Parallel (MPI)
Note: wrfplus was tested on following platforms:
IBM AIX: xlfrte 11.1.0.5
Linux : pgf90 6.2-5 64-bit target on x86-64 Linux (environmental variable PGHPF_ZMEM=yes is needed)
Mac OS (Intel) : g95 0.91!
- Compile WRFPLUS
> ./compile wrf
> ls -ls main/*.exe
You should see wrfplus.exe
Running Observation Preprocessor (OBSPROC)
The OBSPROC program reads observations in LITTLE_R format (a legendary ASCII format, in use since MM5 era). Please refer to the documentation at http://www.mmm.ucar.edu/mm5/mm5v3/data/how_to_get_rawdata.html for LITTLE_R format description. For your applications, you will have to prepare your own observation files. Please see http://www.mmm.ucar.edu/mm5/mm5v3/data/free_data.html for the sources of some freely available observations and the program for converting the observations to LITTLE_R format. Because the raw observation data files could be in any of formats, such as ASCII, BUFR, PREPBUFR, MADIS, HDF, etc. Further more, for each of formats, there may be the different versions. To make WRFDA system as general as possible, the LITTLE_R format ASCII file was adopted as an intermediate observation data format for WRFDA system. Some extensions were made in the LITTLE_R format for WRFDA applications. More complete description of LITTLE_R format and conventional observation data sources for WRFDA could be found from the web page: 2010 Winter Tutorial by clicking “Observation Pre-processing”. The conversion of the user-specific-source data to the LITTLE_R format observation data file is the users’ task.
The purposes of OBSPROC are:
· Remove observations outside the time range and domain (horizontal and top).
· Re-order and merge duplicate (in time and location) data reports.
· Retrieve pressure or height based on observed information using the hydrostatic assumption.
· Check vertical consistency and super adiabatic for multi-level observations.
· Assign observational errors based on a pre-specified error file.
· Write out the observation file to be used by WRFDA in ASCII or BUFR format.
The OBSPROC program—obsproc.exe should be found under the directory WRFDA/var/obsproc/src if “compile all_wrfvar” was completed successfully.
a. Prepare observational data for 3D-Var
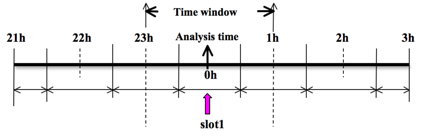
To prepare the observation file, for example, at the analysis time 0h for 3D-Var, all the observations between ±1h (or ±1.5h) will be processed, as illustrated in following figure, which means that the observations between 23h and 1h are treated as the observations at 0h.
Before running obsproc.exe, create the required namelist file namelist.obsproc (see WRFDA/var/obsproc/README.namelist, or the section Description of Namelist Variables for details).
For your reference, an example file named “namelist_obsproc.3dvar.wrfvar-tut” has already been created in the var/obsproc directory. Thus, proceed as follows.
> cp namelist.obsproc.3dvar.wrfvar-tut namelist.obsproc
Next, edit the namelist file namelist.obsproc by changing the following variables to accommodate your experiments.
&record1
obs_gts_filename='obs.2008020512'
&record2
time_window_min = '2008-02-05_11:00:00',: The earliest time edge as ccyy-mm-dd_hh:mn:ss
time_analysis = '2008-02-05_12:00:00', : The analysis time as ccyy-mm-dd_hh:mn:ss
time_window_max = '2008-02-05_13:00:00',: The latest time edge as ccyy-mm-dd_hh:mn:ss
&record6,7,8
Edit all the domain setting according with your own experiment. You may pay special attention on NESTIX and NESTJX, which is described in the section Description of Namelist Variables for details).
&record9
use_for = '3DVAR', ; used for 3D-Var, default
To run OBSPROC, type
> obsproc.exe >&! obsproc.out
Once obsproc.exe has completed successfully, you will see an observation data file, obs_gts_2008-02-05_12:00:00.3DVAR, in the obsproc directory. This is the input observation file to WRFDA.
obs_gts_2008-02-05_12:00:00.3DVAR is an ASCII file that contains a header section (listed below) followed by observations. The meanings and format of observations in the file are described in the last six lines of the header section.
TOTAL = 9066, MISS. =-888888.,
SYNOP = 757, METAR = 2416, SHIP = 145, BUOY = 250, BOGUS = 0, TEMP = 86,
AMDAR = 19, AIREP = 205, TAMDAR= 0, PILOT = 85, SATEM = 106, SATOB = 2556,
GPSPW = 187, GPSZD = 0, GPSRF = 3, GPSEP = 0, SSMT1 = 0, SSMT2 = 0,
TOVS = 0, QSCAT = 2190, PROFL = 61, AIRSR = 0, OTHER = 0,
PHIC = 40.00, XLONC = -95.00, TRUE1 = 30.00, TRUE2 = 60.00, XIM11 = 1.00, XJM11 = 1.00,
base_temp= 290.00, base_lapse= 50.00, PTOP = 1000., base_pres=100000., base_tropo_pres= 20000., base_strat_temp= 215.,
IXC = 60, JXC = 90, IPROJ = 1, IDD = 1, MAXNES= 1,
NESTIX= 60,
NESTJX= 90,
NUMC = 1,
DIS = 60.00,
NESTI = 1,
NESTJ = 1,
INFO = PLATFORM, DATE, NAME, LEVELS, LATITUDE, LONGITUDE, ELEVATION, ID.
SRFC = SLP, PW (DATA,QC,ERROR).
EACH = PRES, SPEED, DIR, HEIGHT, TEMP, DEW PT, HUMID (DATA,QC,ERROR)*LEVELS.
INFO_FMT = (A12,1X,A19,1X,A40,1X,I6,3(F12.3,11X),6X,A40)
SRFC_FMT = (F12.3,I4,F7.2,F12.3,I4,F7.3)
EACH_FMT = (3(F12.3,I4,F7.2),11X,3(F12.3,I4,F7.2),11X,3(F12.3,I4,F7.2))
#------------------------------------------------------------------------------#
…… observations ………
Before running WRFDA, you may like to learn more about various types of data that will be passed to WRFDA for this case, for example, their geographical distribution, etc. This file is in ASCII format and so you can easily view it. To have a graphical view about the content of this file, there is a “MAP_plot” utility to look at the data distribution for each type of observations. To use this utility, proceed as follows.
> cd MAP_plot
> make
We have prepared some configure.user.ibm/linux/mac/… files for some platforms, when “make” is typed, the Makefile will use one of them to determine the compiler and compiler option. Please modify the Makefile and configure.user.xxx to accommodate the complier on your platform. Successful compilation will produce Map.exe. Note: The successful compilation of Map.exe requires pre-installed NCARG Graphics libraries under $(NCARG_ROOT)/lib.
Modify the script Map.csh to set the time window and full path of input observation file (obs_gts_2008-02-05_12:00:00.3DVAR). You will need to set the following strings in this script as follows:
Map_plot = /users/noname/WRFDA/var/obsproc/MAP_plot
TIME_WINDOW_MIN = ‘2008020511’
TIME_ANALYSIS = ‘2008020512’
TIME_WINDOW_MAX
= ‘2008020513’
OBSDATA
= ../obs_gts_2008-02-05_12:00:00.3DVAR
Next, type
> Map.csh
When the job has completed, you will have a gmeta file gmeta.{analysis_time} corresponding to analysis_time=2008020512. This contains plots of data distribution for each type of observations contained in the OBS data file: obs_gts_2008-02-05_12:00:00.3DVAR. To view this, type
> idt gmeta.2008020512
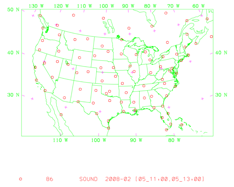
It will display (panel by panel) geographical distribution of various types of data. Following is the geographic distribution of “sonde” observations for this case.
There is an alternative way to plot the observation by using ncl script: WRFDA/var/graphics/ncl/plot_ob_ascii_loc.ncl. However, with this way, you need to provide the first guess file to the ncl script, and have ncl installed in your system.
b. Prepare observational data for 4D-Var
To prepare the observation file, for example, at the analysis time 0h for 4D-Var, all observations from 0h to 6h will be processed and grouped in 7 sub-windows from slot1 to slot7, as illustrated in following figure. NOTE: The “Analysis time” in the figure below is not the actual analysis time (0h), it just indicates the time_analysis setting in the namelist file, and is set to three hours later than the actual analysis time. The actual analysis time is still 0h.
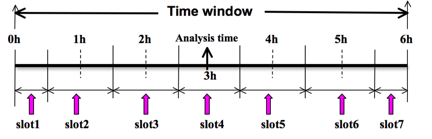
An example file named “namelist_obsproc.4dvar.wrfvar-tut” has already been created in the var/obsproc directory. Thus, proceed as follows:
> cp namelist.obsproc.4dvar.wrfvar-tut namelist.obsproc
In the namelist file, you need to change the following variables to accommodate your experiments. In this test case, the actual analysis time is 2008-02-05_12:00:00, but in namelist, the time_analysis should be set to 3 hours later. The different value of time_analysis will make the different number of time slots before and after time_analysis. For example, if you set time_analysis = 2008-02-05_16:00:00, and set the num_slots_past = 4 and time_slots_ahead=2. The final results will be same as before.
&record1
obs_gts_filename='obs.2008020512'
&record2
time_window_min = '2008-02-05_12:00:00',: The earliest time edge as ccyy-mm-dd_hh:mn:ss
time_analysis = '2008-02-05_15:00:00', : The analysis time as ccyy-mm-dd_hh:mn:ss
time_window_max = '2008-02-05_18:00:00',: The latest time edge as ccyy-mm-dd_hh:mn:ss
&record6,7,8
Edit all the domain setting according with your own experiment. You may pay special attention on NESTIX and NESTJX, which is described in the section Description of Namelist Variables for details).
&record9
use_for = '4DVAR', ; used for 3D-Var, default
; num_slots_past and num_slots_ahead are used ONLY for FGAT and 4DVAR:
num_slots_past = 3, ; the number of time slots before time_analysis
num_slots_ahead = 3, ; the number of time slots after time_analysis
To run OBSPROC, type
> obsproc.exe >&! obsproc.out
Once obsproc.exe has completed successfully, you will see 7 observation data files:
obs_gts_2008-02-05_12:00:00.4DVAR
obs_gts_2008-02-05_13:00:00.4DVAR
obs_gts_2008-02-05_14:00:00.4DVAR
obs_gts_2008-02-05_15:00:00.4DVAR
obs_gts_2008-02-05_16:00:00.4DVAR
obs_gts_2008-02-05_17:00:00.4DVAR
obs_gts_2008-02-05_18:00:00.4DVAR
They are the input observation files to WRF 4D-Var. You can also use “MAP_Plot” to view the geographic distribution of different observations at different time slots.
Running WRFDA
a. Download Test Data
The WRFDA system requires three input files to run:
a) A WRF first guess and boundary input files output from either WPS/real (cold-start)
or WRF forecast (warm-start)
b) Observations (in ASCII format, PREBUFR or BUFR for radiance)
c) A background error statistics file (containing background error covariance)
The following table summarizes the above info:
|
Input Data |
Format |
Created By |
|
First Guess
|
NETCDF |
WRF Preprocessing System (WPS) and real.exe or WRF |
|
Observations |
ASCII (PREPBUFR also possible) |
Observation Preprocessor (OBSPROC) |
|
Background Error Statistics |
Binary |
/Default CV3 |
In the test case, you will store data in a directory defined by the environment variable $DAT_DIR. This directory can be at any location and it should have read access. Type
> setenv DAT_DIR your_choice_of_dat_dir
Here, "your_choice_of_dat_dir" is the directory where the WRFDA input data is stored. Create this directory if it does not exist, and type
> cd $DAT_DIR
Download the test data for a “Tutorial” case valid at 12 UTC 5th February 2008 from http://www.mmm.ucar.edu/wrf/users/wrfda/download/testdata.html
Once you have downloaded “WRFDAV3.2-testdata.tar.gz” file to $DAT_DIR, extract it by typing
> gunzip WRFDAV3.2-testdata.tar.gz
>
tar -xvf WRFDAV3.2-testdata.tar
Now you should find the following three sub-directories/files under “$DAT_DIR”
ob/2008020512/ob.2008020512.gz
# Observation data in “little_r” format
rc/2008020512/wrfinput_d01
# First guess file
rc/2008020512/wrfbdy_d01
# lateral boundary file
be/be.dat
# Background error file
......
You should first go through the section “Running Observation Preprocessor (OBSPROC)” and have a WRF-3D-Var-ready observation file (obs_gts_2008-02-05_12:00:00.3DVAR) generated in your OBSPROC working directory. You could then copy or move obs_gts_2008-02-05_12:00:00.3DVAR to be in $DAT_DIR/ob/2008020512/ob.ascii.
If you want to try 4D-Var, please go through the section “Running Observation Preprocessor (OBSPROC)” and have the WRF-4D-Var-ready observation files (obs_gts_2008-02-05_12:00:00.4DVAR,……). You could copy or move the observation files to $DAT_DIR/ob using following commands:
> mv obs_gts_2008-02-05_12:00:00.4DVAR $DAT_DIR/ob/2008020512/ob.ascii+
> mv obs_gts_2008-02-05_13:00:00.4DVAR $DAT_DIR/ob/2008020513/ob.ascii
> mv obs_gts_2008-02-05_14:00:00.4DVAR $DAT_DIR/ob/2008020514/ob.ascii
> mv obs_gts_2008-02-05_15:00:00.4DVAR $DAT_DIR/ob/2008020515/ob.ascii
> mv obs_gts_2008-02-05_16:00:00.4DVAR $DAT_DIR/ob/2008020516/ob.ascii
> mv obs_gts_2008-02-05_17:00:00.4DVAR $DAT_DIR/ob/2008020517/ob.ascii
> mv obs_gts_2008-02-05_18:00:00.4DVAR $DAT_DIR/ob/2008020518/ob.ascii-
At this point you have three of the input files (first guess, observation and background error statistics files in directory $DAT_DIR) required to run WRFDA, and have successfully downloaded and compiled the WRFDA code. If this is correct, you are ready to learn how to run WRFDA.
b. Run the Case—3D-Var
The data for this case is valid at 12 UTC 5th February 2008. The first guess comes from the NCEP FNL (Final) Operational Global Analysis data, passed through the WRF-WPS and real programs.
To run WRF 3D-Var, first create and cd to a working directory, for example, WRFDA/var/test/tutorial, and then follow the steps below:
> cd WRFDA/var/test/tutorial
> ln -sf WRFDA/run/LANDUSE.TBL ./LANDUSE.TBL
> ln -sf $DAT_DIR/rc/2008020512/wrfinput_d01 ./fg (link first guess file as fg)
> ln -sf WRFDA/var/obsproc/obs_gts_2008-02-05_12:00:00.3DVAR ./ob.ascii (link OBSPROC processed observation file as ob.ascii)
> ln -sf $DAT_DIR/be/be.dat ./be.dat (link background error statistics as be.dat)
> ln -sf WRFDA/var/da/da_wrfvar.exe ./da_wrfvar.exe (link executable)
We will begin by editing the file, namelist.input, which is a very basic namelist.input for running the tutorial test case is shown below and provided as WRFDA/var/test/tutorial/namelist.input. Only the time and domain settings need to be specified in this case, if we are using the default settings provided in WRFDA/Registry/Registry.wrfvar)
&wrfvar1
print_detail_grad=false,
/
&wrfvar2
/
&wrfvar3
/
&wrfvar4
/
&wrfvar5
/
&wrfvar6
/
&wrfvar7
/
&wrfvar8
/
&wrfvar9
/
&wrfvar10
/
&wrfvar11
/
&wrfvar12
/
&wrfvar13
/
&wrfvar14
/
&wrfvar15
/
&wrfvar16
/
&wrfvar17
/
&wrfvar18
analysis_date="2008-02-05_12:00:00.0000",
/
&wrfvar19
/
&wrfvar20
/
&wrfvar21
time_window_min="2008-02-05_11:00:00.0000",
/
&wrfvar22
time_window_max="2008-02-05_13:00:00.0000",
/
&wrfvar23
/
&time_control
start_year=2008,
start_month=02,
start_day=05,
start_hour=12,
end_year=2008,
end_month=02,
end_day=05,
end_hour=12,
/
&dfi_control
/
&domains
e_we=90,
e_sn=60,
e_vert=41,
dx=60000,
dy=60000,
/
&physics
mp_physics=3,
ra_lw_physics=1,
ra_sw_physics=1,
radt=60,
sf_sfclay_physics=1,
sf_surface_physics=1,
bl_pbl_physics=1,
cu_physics=1,
cudt=5,
num_soil_layers=5, (IMPORTANT: it’s essential to make sure the setting here is consistent with the number in your first guess file)
mp_zero_out=2,
co2tf=0,
/
&fdda
/
&dynamics
/
&bdy_control
/
&grib2
/
&namelist_quilt
/
> da_wrfvar.exe >&! wrfda.log
The file wrfda.log (or rsl.out.0000 if run in distributed-memory mode) contains important WRFDA runtime log information. Always check the log after a WRFDA run:
*** VARIATIONAL ANALYSIS ***
DYNAMICS OPTION: Eulerian Mass Coordinate
WRF NUMBER OF TILES = 1
Set up observations (ob)
Using ASCII format observation input
scan obs ascii
end scan obs ascii
Observation summary
ob time 1
sound 85 global, 85 local
synop 531 global, 525 local
pilot 84 global, 84 local
satem 78 global, 78 local
geoamv 736 global, 719 local
polaramv 0 global, 0 local
airep 132 global, 131 local
gpspw 183 global, 183 local
gpsrf 0 global, 0 local
metar 1043 global, 1037 local
ships 86 global, 82 local
ssmi_rv 0 global, 0 local
ssmi_tb 0 global, 0 local
ssmt1 0 global, 0 local
ssmt2 0 global, 0 local
qscat 0 global, 0 local
profiler 61 global, 61 local
buoy 216 global, 216 local
bogus 0 global, 0 local
pseudo 0 global, 0 local
radar 0 global, 0 local
radiance 0 global, 0 local
airs retrieval 0 global, 0 local
sonde_sfc 85 global, 85 local
mtgirs 0 global, 0 local
tamdar 0 global, 0 local
Set up background errors for regional application
WRF-Var dry control variables are:psi, chi_u, t_u and psfc
Humidity control variable is q/qsg
Using the averaged regression coefficients for unbalanced part
Vertical truncation for psi = 15( 99.00%)
Vertical truncation for chi_u = 20( 99.00%)
Vertical truncation for t_u = 29( 99.00%)
Vertical truncation for rh = 22( 99.00%)
Calculate innovation vector(iv)
Minimize cost function using CG method
For this run cost function diagnostics will not be written
Starting outer iteration : 1
Starting cost function: 2.28356084D+04, Gradient= 2.23656955D+02
For this outer iteration gradient target is: 2.23656955D+00
----------------------------------------------------------
Iter Gradient Step
1 1.82455068D+02 7.47025772D-02
2 1.64971618D+02 8.05531077D-02
3 1.13694365D+02 7.22382618D-02
4 7.87359568D+01 7.51905761D-02
5 5.71607218D+01 7.94572516D-02
6 4.18746777D+01 8.30731280D-02
7 2.95722963D+01 6.13223951D-02
8 2.34205172D+01 9.05920463D-02
9 1.63772518D+01 6.48090044D-02
10 1.09735524D+01 7.71148550D-02
11 8.22748934D+00 8.81041046D-02
12 5.65846963D+00 7.89528133D-02
13 4.15664769D+00 7.45589721D-02
14 3.16925808D+00 8.35300020D-02
----------------------------------------------------------
Inner iteration stopped after 15 iterations
Final: 15 iter, J= 1.76436785D+04, g= 2.06098421D+00
----------------------------------------------------------
Diagnostics
Final cost function J = 17643.68
Total number of obs. = 26726
Final value of J = 17643.67853
Final value of Jo = 15284.64894
Final value of Jb = 2359.02958
Final value of Jc = 0.00000
Final value of Je = 0.00000
Final value of Jp = 0.00000
Final J / total num_obs = 0.66017
Jb factor used(1) = 1.00000
Jb factor used(2) = 1.00000
Jb factor used(3) = 1.00000
Jb factor used(4) = 1.00000
Jb factor used(5) = 1.00000
Jb factor used = 1.00000
Je factor used = 1.00000
VarBC factor used = 1.00000
*** WRF-Var completed successfully ***
A file called namelist.output (which contains the complete namelist settings) will be generated after a successful da_wrfvar.exe run. The settings appearing in namelist.output, but not specified in your namelist.input, are the default values from WRFDA/Registry/Registry.wrfvar.
After successful completion of job, wrfvar_output (the WRFDA analysis file, i.e. the new initial condition for WRF) should appear in the working directory along with a number of diagnostic files. Various text diagnostics output files will be explained in the next section (WRFDA Diagnostics).
In order to understand the role of various important WRFDA options, try re-running WRFDA by changing different namelist options. Such as making WRFDA convergence criteria more stringent. This is achieved by reducing the value of the convergence criteria “EPS” to e.g. 0.0001 by adding "EPS=0.0001" in the namelist.input record &wrfvar6. See section (WRFDA additional exercises) for more namelist options
c. Run the Case—4D-Var
To run WRF 4D-Var, first create and cd to a working directory, for example, WRFDA/var/test/4dvar; next assuming that we are using the C-shell, set the working directories for the three WRF 4D-Var components WRFDA, WRFNL and WRFPLUS thusly
> setenv WRFDA_DIR /ptmp/$user/WRFDA
> setenv WRFNL_DIR /ptmp/$user/WRFNL
> setenv WRFPLUS_DIR /ptmp/$user/WRFPLUS
Assume the analysis date is 2008020512 and the test data directories are:
> setenv DATA_DIR /ptmp/$user/DATA
> ls –lr $DATA_DIR
ob/2008020512
ob/2008020513
ob/2008020514
ob/2008020515
ob/2008020516
ob/2008020517
ob/2008020518
rc/2008020512
be
Note: Currently, WRF 4D-Var can only run with the observation data processed by OBSPROC, and cannot work with PREPBUFR format data; Although WRF-4DVar is able to assimilate satellite radiance BUFR data, but this capability is still under testing.
Assume the working directory is:
> setenv WORK_DIR $WRFDA_DIR/var/test/4dvar
Then follow the steps below:
1) Link the executables.
> cd $WORK_DIR
> ln -fs $WRFDA_DIR/var/da/da_wrfvar.exe .
> cd $WORK_DIR/nl
> ln -fs $WRFNL_DIR/main/wrf.exe .
> cd $WORK_DIR/ad
> ln -fs $WRFPLUS_DIR/main/wrfplus.exe .
> cd $WORK_DIR/tl
> ln -fs $WRFPLUS_DIR/main/wrfplus.exe .
2) Link the observational data, first guess and BE. (Currently, only LITTLE_R formatted observational data is supported in 4D-Var, PREPBUFR observational data is not supported)
> cd $WORK_DIR
> ln -fs $DATA_DIR/ob/2008020512/ob.ascii+ ob01.ascii
> ln -fs $DATA_DIR/ob/2008020513/ob.ascii ob02.ascii
> ln -fs $DATA_DIR/ob/2008020514/ob.ascii ob03.ascii
> ln -fs $DATA_DIR/ob/2008020515/ob.ascii ob04.ascii
> ln -fs $DATA_DIR/ob/2008020516/ob.ascii ob05.ascii
> ln -fs $DATA_DIR/ob/2008020517/ob.ascii ob06.ascii
> ln -fs $DATA_DIR/ob/2008020518/ob.ascii- ob07.ascii
> ln -fs $DATA_DIR/rc/2008020512/wrfinput_d01 .
> ln -fs $DATA_DIR/rc/2008020512/wrfbdy_d01 .
> ln -fs wrfinput_d01 fg
> ln -fs wrfinput_d01 fg01
> ln -fs $DATA_DIR/be/be.dat .
3) Establish the miscellaneous links.
> cd $WORK_DIR
> ln -fs nl/nl_d01_2008-02-05_13:00:00 fg02
> ln -fs nl/nl_d01_2008-02-05_14:00:00 fg03
> ln -fs nl/nl_d01_2008-02-05_15:00:00 fg04
> ln -fs nl/nl_d01_2008-02-05_16:00:00 fg05
> ln -fs nl/nl_d01_2008-02-05_17:00:00 fg06
> ln -fs nl/nl_d01_2008-02-05_18:00:00 fg07
> ln -fs ad/ad_d01_2008-02-05_12:00:00 gr01
> ln -fs tl/tl_d01_2008-02-05_13:00:00 tl02
> ln -fs tl/tl_d01_2008-02-05_14:00:00 tl03
> ln -fs tl/tl_d01_2008-02-05_15:00:00 tl04
> ln -fs tl/tl_d01_2008-02-05_16:00:00 tl05
> ln -fs tl/tl_d01_2008-02-05_17:00:00 tl06
> ln -fs tl/tl_d01_2008-02-05_18:00:00 tl07
> cd $WORK_DIR/ad
> ln -fs ../af01 auxinput3_d01_2008-02-05_12:00:00
> ln -fs ../af02 auxinput3_d01_2008-02-05_13:00:00
> ln -fs ../af03 auxinput3_d01_2008-02-05_14:00:00
> ln -fs ../af04 auxinput3_d01_2008-02-05_15:00:00
> ln -fs ../af05 auxinput3_d01_2008-02-05_16:00:00
> ln -fs ../af06 auxinput3_d01_2008-02-05_17:00:00
> ln -fs ../af07 auxinput3_d01_2008-02-05_18:00:00
4) Run in single processor mode (serial compilation required for WRFDA, WRFNL and WRFPLUS)
Edit $WORK_DIR/namelist.input to match your experiment settings.
> cp $WORK_DIR/nl/namelist.input.serial $WORK_DIR/nl/namelist.input
Edit $WORK_DIR/nl/namelist.input to match your experiment settings.
> cp $WORK_DIR/ad/namelist.input.serial $WORK_DIR/ad/namelist.input
> cp $WORK_DIR/tl/namelist.input.serial $WORK_DIR/tl/namelist.input
Edit $WORK_DIR/ad/namelist.input and $WORK_DIR/tl/namelist.input to match your experiment settings, but only change following variables:
&time_control
run_hours=06,
start_year=2008,
start_month=02,
start_day=05,
start_hour=12,
end_year=2008,
end_month=02,
end_day=05,
end_hour=18,
......
&domains
time_step=360, # NOTE:MUST BE THE SAME WITH WHICH IN $WORK_DIR/nl/namelist.input
e_we=90,
e_sn=60,
e_vert=41,
dx=60000,
dy=60000,
......
> cd $WORK_DIR
> setenv NUM_PROCS 1
> ./da_wrfvar.exe >&! wrfda.log
5) Run with multiple processors with MPMD mode. (dmpar compilation required for WRFDA, WRFNL and WRFPLUS)
Edit $WORK_DIR/namelist.input to match your experiment settings.
> cp $WORK_DIR/nl/namelist.input.parallel $WORK_DIR/nl/namelist.input
Edit $WORK_DIR/nl/namelist.input to match your experiment settings.
> cp $WORK_DIR/ad/namelist.input.parallel $WORK_DIR/ad/namelist.input
> cp $WORK_DIR/tl/namelist.input.parallel $WORK_DIR/tl/namelist.input
Edit $WORK_DIR/ad/namelist.input and $WORK_DIR/tl/namelist.input to match your experiment settings.
Currently, parallel WRF 4D-Var is a MPMD (Multiple Program Multiple Data) application. Because there are so many parallel configurations across the platforms, it is very difficult to define a generic way to run the WRF 4D-Var parallel. As an example, to launch the three WRF 4D-Var executables as a concurrent parallel job on a 16 processor cluster, use:
> mpirun –np 4 da_wrfvar.exe: -np 8 ad/wrfplus.exe: -np 4 nl/wrf.exe
In the above example, 4 processors are assigned to run WRFDA, 4 processors are assigned to run WRFNL and 8 processors for WRFPLUS due to high computational cost in adjoint code.
The file wrfda.log (or rsl.out.0000 if running in parallel mode) contains important WRF-4DVar runtime log information. Always check the log after a WRF-4DVar run.
Radiance Data Assimilations in WRFDA
This section gives a brief description for various aspects related to radiance assimilation in WRFDA. Each aspect is described mainly from the viewpoint of usage rather than more technical and scientific details, which will appear in separated technical report and scientific paper. Namelist parameters controlling different aspects of radiance assimilation will be detailed in the following sections. It should be noted that this section does not cover general aspects of the WRFDA assimilation. These can be found in other sections of chapter 6 of this users guide or other WRFDA documentation.
a. Running WRFDA with radiances
In addition to the basic input files (LANDUSE.TBL, fg, ob.ascii, be.dat) mentioned in “Running WRFDA” section, the following extra files are required for radiances: radiance data in NCEP BUFR format, radiance_info files, VARBC.in, RTM (CRTM or RTTOV) coefficient files.
Edit namelist.input (Pay special attention to &wrfvar4, &wrfvar14, &wrfvar21, and &wrfvar22 for radiance-related options. A very basic namelist.input for running the radiance test case is provided as WRFDA/var/test/radiance/namelist.input)
> ln -sf ${DAT_DIR}/gdas1.t00z.1bamua.tm00.bufr_d ./amsua.bufr
> ln -sf ${DAT_DIR}/gdas1.t00z.1bamub.tm00.bufr_d ./amsub.bufr
> ln -sf WRFDA/var/run/radiance_info ./radiance_info # (radiance_info is a directory)
> ln -sf WRFDA/var/run/VARBC.in ./VARBC.in
(CRTM only) > ln -sf WRFDA/var/run/crtm_coeffs ./crtm_coeffs #(crtm_coeffs is a directory)
(RTTOV only) > ln -sf rttov87/rtcoef_rttov7/* . # (a list of rtcoef* files)
See the following sections for more details on each aspect.
b. Radiance Data Ingest
Currently, the ingest interface for NCEP BUFR radiance data is implemented in WRFDA. The radiance data are available through NCEP’s public ftp server ftp://ftp.ncep.noaa.gov/pub/data/nccf/com/gfs/prod/gdas.${yyyymmddhh} in near real-time (with 6-hour delay) and can meet requirements both for research purposes and some real-time applications.
So far, WRFDA can read data from the NOAA ATOVS instruments (HIRS, AMSU-A, AMSU-B and MHS), the EOS Aqua instruments (AIRS, AMSU-A) and DMSP instruments (SSMIS). Note that NCEP radiance BUFR files are separated by instrument names (i.e., each file for one type instrument) and each file contains global radiance (generally converted to brightness temperature) within 6-hour assimilation window from multi-platforms. For running WRFDA, users need to rename NCEP corresponding BUFR files (table 1) to hirs3.bufr (including HIRS data from NOAA-15/16/17), hirs4.bufr (including HIRS data from NOAA-18, METOP-2), amsua.bufr (including AMSU-A data from NOAA-15/16/18, METOP-2), amsub.bufr (including AMSU-B data from NOAA-15/16/17), mhs.bufr (including MHS data from NOAA-18 and METOP-2), airs.bufr (including AIRS and AMSU-A data from EOS-AQUA) and ssmis.bufr (SSMIS data from DMSP-16, AFWA provided) for WRFDA filename convention. Note that airs.bufr file contains not only AIRS data but also AMSU-A, which is collocated with AIRS pixels (1 AMSU-A pixels collocated with 9 AIRS pixels). Users must place these files in the working directory where WRFDA executable is located. It should also be mentioned that WRFDA reads these BUFR radiance files directly without use if any separate pre-processing program is used. All processing of radiance data, such as quality control, thinning and bias correction and so on, is carried out inside WRFDA. This is different from conventional observation assimilation, which requires a pre-processing package (OBSPROC) to generate WRFDA readable ASCII files. For reading the radiance BUFR files, WRFDA must be compiled with the NCEP BUFR library (see http://www.nco.ncep.noaa.gov/sib/decoders/BUFRLIB/).
Table 1: NCEP and WRFDA radiance BUFR file naming convention
|
NCEP BUFR file names |
WRFDA naming convention |
|
gdas1.t00z.1bamua.tm00.bufr_d |
amsua.bufr |
|
gdas1.t00z.1bamub.tm00.bufr_d |
amsub.bufr |
|
gdas1.t00z.1bhrs3.tm00.bufr_d |
hirs3.bufr |
|
gdas1.t00z.1bhrs4.tm00.bufr_d |
hirs4.bufr |
|
gdas1.t00z.1bmhs.tm00.bufr_d |
mhs.bufr |
|
gdas1.t00z.airsev.tm00.bufr_d |
airs.bufr |
Namelist parameters are used to control the reading of corresponding BUFR files into WRFDA. For instance, USE_AMSUAOBS, USE_AMSUBOBS, USE_HIRS3OBS, USE_HIRS4OBS, USE_MHSOBS, USE_AIRSOBS, USE_EOS_AMSUAOBS and USE_SSMISOBS control whether or not the respective file is read. These are logical parameters that are assigned to FALSE by default; therefore they must be set to true to read the respective observation file. Also note that these parameters only control whether the data is read, not whether the data included in the files is to be assimilated. This is controlled by other namelist parameters explained in the next section.
NCEP BUFR files downloaded from NCEP’s public ftp server ftp://ftp.ncep.noaa.gov/pub/data/nccf/com/gfs/prod/gdas.${yyyymmddhh} are Fortran-blocked on big-endian machine and can be directly used on big-endian machines (for example, IBM). For most Linux clusters with Intel platforms, users need to first unblock the BUFR files, and then reblock them. The utility for blocking/unblocking is available from http://www.nco.ncep.noaa.gov/sib/decoders/BUFRLIB/toc/cwordsh
c. Radiative Transfer Model
The core component for direct radiance assimilation is to incorporate a radiative transfer model (RTM, should be accurate enough yet fast) into the WRFDA system as one part of observation operators. Two widely used RTMs in NWP community, RTTOV8* (developed by EUMETSAT in Europe), and CRTM (developed by the Joint Center for Satellite Data Assimilation (JCSDA) in US), are already implemented in WRFDA system with a flexible and consistent user interface. Selecting which RTM to be used is controlled by a simple namelist parameter RTM_OPTION (1 for RTTOV, the default, and 2 for CRTM). WRFDA is designed to be able to compile with only one of two RTM libraries or without RTM libraries (for those not interested in radiance assimilation) by the definition of environment variables “CRTM” and “RTTOV” (see Installing WRFDA section).
Both RTMs can calculate radiances for almost all available instruments aboard various satellite platforms in orbit. An important feature of WRFDA design is that all data structures related to radiance assimilation are dynamically allocated during running time according to simple namelist setup. The instruments to be assimilated are controlled at run time by four integer namelist parameters: RTMINIT_NSENSOR (the total number of sensors to be assimilated), RTMINIT_PLATFORM (the platforms IDs array to be assimilated with dimension RTMINIT_NSENSOR, e.g., 1 for NOAA, 9 for EOS, 10 for METOP and 2 for DMSP), RTMINIT_SATID (satellite IDs array) and RTMINIT_SENSOR (sensor IDs array, e.g., 0 for HIRS, 3 for AMSU-A, 4 for AMSU-B, 15 for MHS, 10 for SSMIS, 11 for AIRS). For instance, the configuration for assimilating 12 sensors from 7 satellites (what WRFDA can assimilated currently) will be
RTMINIT_NSENSOR = 12 # 5 AMSUA; 3 AMSUB; 2 MHS; 1 AIRS; 1 SSMIS
RTMINIT_PLATFORM = 1,1,1,9,10, 1,1,1, 1,10, 9, 2
RTMINIT_SATID = 15,16,18,2,2, 15,16,17, 18,2, 2, 16
RTMINIT_SENSOR = 3,3,3,3,3, 4,4,4, 15,15, 11, 10
The instrument triplets (platform, satellite and sensor ID) in the namelist can be ranked in any order. More detail about the convention of instrument triplet can be found at the tables 2 and 3 in RTTOV8/9 Users Guide (http://www.metoffice.gov.uk/research/interproj/nwpsaf/rtm/rttov8_ug.pdf Or http://www.metoffice.gov.uk/research/interproj/nwpsaf/rtm/rttov9_files/users_guide_91_v1.6.pdf)
CRTM uses a different instrument naming method. A convert routine inside WRFDA is already created to make CRTM use the same instrument triplet as RTTOV such that the user interface remains the same for RTTOV and CRTM.
When running WRFDA with radiance assimilation switched on (RTTOV or CRTM), a set of RTM coefficient files need to be loaded. For RTTOV option, RTTOV coefficient files are to be directly copied or linked under the working directory; for CRTM option, CRTM coefficient files are to be copied or linked to a sub-directory “crtm_coeffs” under the working directory. Only coefficients listed in namelist are needed. Potentially WRFDA can assimilate all sensors as long as the corresponding coefficient files are provided with RTTOV and CRTM. In addition, necessary developments on corresponding data interface, quality control and bias correction are also important to make radiance data assimilated properly. However, a modular design of radiance relevant routines already facilitates much to add more instruments in WRFDA.
RTTOV packages are not distributed with WRFDA due to license and support issues. Users are encouraged to contact the corresponding team for obtaining RTMs. See following links for more information.
http://www.metoffice.gov.uk/research/interproj/nwpsaf/rtm/index.html .
CRTM pakages are now distributed with WRFDA, which locate in the WRFDA/var/external/crtm. Users can still find it on the following link:
ftp://ftp.emc.ncep.noaa.gov/jcsda/CRTM.
d. Channel Selection
Channel selection in WRFDA is controlled by radiance ‘info’ files located in the sub-directory ‘radiance_info’ under the working directory. These files are separated by satellites and sensors, e.g., noaa-15-amsua.info, noaa-16-amsub.info, dmsp-16-ssmis.info and so on. An example for 5 channels from noaa-15-amsub.info is shown below. The fourth column is used by WRFDA to control if assimilating corresponding channel. Channels with the value “-1” indicates that the channel is “not assimilated” (channels 1, 2 and 4 in this case), with the value “1” means “assimilated” (channels 3 and 5). The sixth column is used by WRFDA to set the observation error for each channel. Other columns are not used by WRFDA. It should be mentioned that these error values might not necessarily be optimal for your applications; It is user’s responsibility to obtain the optimal error statistics for your own applications.
sensor channel IR/MW use idum varch polarisation(0:vertical;1:horizontal)
415 1 1 -1 0 0.5500000000E+01 0.0000000000E+00
415 2 1 -1 0 0.3750000000E+01 0.0000000000E+00
415 3 1 1 0 0.3500000000E+01 0.0000000000E+00
415 4 1 -1 0 0.3200000000E+01 0.0000000000E+00
415 5 1 1 0 0.2500000000E+01 0.0000000000E+00
e. Bias Correction
Satellite radiance is generally considered biased with respect to a reference (e.g., background or analysis field in NWP assimilation) due to system error of observation itself, reference field and RTM. Bias correction is a necessary step prior to assimilating radiance data. In WRFDA, there are two ways of performing bias correction. One is based on Harris and Kelly (2001) method and is carried out using a set of coefficient files pre-calculated with an off-line statistics package, which will apply to a training dataset for a month-long period. The other is Variational Bias Correction (VarBC). Only VarBC is introduced here and recommended for users because of its relative simplicity in usage.
f. Variational Bias Correction
Getting started with VarBC
To use VarBC, set namelist option USE_VARBC to TRUE and have a VARBC.in file in the working directory. VARBC.in is a VarBC setup file in ASCII format. A template is provided with the WRFDA package (WRFDA/var/run/VARBC.in).
Input and Output files
All VarBC input is passed through one single ASCII file called VARBC.in file. Once WRFDA has run with the VarBC option switched on, it will produce a VARBC.out file which looks very much like the VARBC.in file you provided. This output file will then be used as input file for the next assimilation cycle.
Coldstart
Coldstarting means starting the VarBC from scratch i.e. when you do not know the values of the bias parameters.
The Coldstart is a routine in WRFDA. The bias predictor statistics (mean and standard deviation) are computed automatically and will be used to normalize the bias parameters. All coldstarted bias parameters are set to zero, except the first bias parameter (= simple offset), which is set to the mode (=peak) of the distribution of the (uncorrected) innovations for the given channel.
A threshold of number of observations can be set through a namelist option VARBC_NOBSMIN (default = 10), under which it is considered that not enough observations are present to keep the Coldstart values (i.e. bias predictor statistics and bias parameter values) for the next cycle. In this case, the next cycle will do another Coldstart.
Background Constraint for the bias parameters
The background constraint controls the inertia you want to impose on the predictors (i.e. the smoothing in the predictor time series). It corresponds to an extra term in the WRFDA cost function.
It is defined through an integer number in the VARBC.in file. This number is related to a number of observations: the bigger the number, the more inertia constraint. If these numbers are set to zero, the predictors can evolve without any constraint.
Scaling factor
The VarBC uses a specific preconditioning, which can be scaled through a namelist option VARBC_FACTOR (default = 1.0).
Offline bias correction
The analysis of the VarBC parameters can be performed "offline", i.e. independently from the main WRFDA analysis. No extra code is needed, just set the following MAX_VERT_VAR* namelist variables to be 0, which will disable the standard control variable and only keep the VarBC control variable.
MAX_VERT_VAR1=0.0
MAX_VERT_VAR2=0.0
MAX_VERT_VAR3=0.0
MAX_VERT_VAR4=0.0
MAX_VERT_VAR5=0.0
Freeze VarBC
In certain circumstances, you might want to keep the VarBC bias parameters constant in time (="frozen"). In this case, the bias correction is read and applied to the innovations, but it is not updated during the minimization. This can easily be achieved by setting the namelist options:
USE_VARBC=false
FREEZE_VARBC=true
Passive observations
Some observations are useful for preprocessing (e.g. Quality Control, Cloud detection) but you might not want to assimilate them. If you still need to estimate their bias correction, these observations need to go through the VarBC code in the minimization. For this purpose, the VarBC uses a separate threshold on the QC values, called "qc_varbc_bad". This threshold is currently set to the same value as "qc_bad", but can easily be changed to any ad hoc value.
g. Other namelist variables to control radiance assimilation
RAD_MONITORING (30)
Integer array of dimension RTMINIT_NSENSER, where 0 for assimilating mode, 1 for monitoring mode (only calculate innovation).
THINNING
Logical, TRUE will perform thinning on radiance data.
THINNING_MESH (30)
Real array with dimension RTMINIT_NSENSOR, values indicate thinning mesh (in KM) for different sensors.
QC_RAD
Logical, control if perform quality control, always set to TRUE.
WRITE_IV_RAD_ASCII
Logical, control if output Observation minus Background files which are in ASCII format and separated by sensors and processors.
WRITE_OA_RAD_ASCII
Logical, control if output Observation minus Analysis files (including also O minus B) which are ASCII format and separated by sensors and processors.
USE_ERROR_FACTOR_RAD
Logical, controls use of a radiance error tuning factor file “radiance_error.factor”, which is created with empirical values or generated using variational tunning method (Desroziers and Ivanov, 2001)
ONLY_SEA_RAD
Logical, controls whether only assimilating radiance over water.
TIME_WINDOW_MIN
String, e.g., "2007-08-15_03:00:00.0000", start time of assimilation time window
TIME_WINDOW_MAX
String, e.g., "2007-08-15_09:00:00.0000", end time of assimilation time window
CRTM_ATMOSPHERE
Integer, used by CRTM to choose climatology reference profile used above model top (up to 0.01hPa).
0: Invalid (default, use U.S. Standard Atmosphere)
1: Tropical
2: Midlatitude summer
3: Midlatitude winter
4: Subarctic summer
5: Subarctic winter
6: U.S. Standard Atmosphere
USE_ANTCORR (30)
Logical array with dimension RTMINIT_NSENSER, control if performing Antenna Correction in CRTM.
AIRS_WARMEST_FOV
Logical, controls whether using the observation brightness temperature for AIRS Window channel #914 as criterium for GSI thinning.
USE_CRTM_KMATRIX
Logical, controls whether using CRTM K matrix rather than calling CRTM TL and AD routines for gradient calculation.
h. Diagnostics and Monitoring
(1) Monitoring capability within WRFDA.
Run WRFDA with the rad_monitoring namelist parameter in record wrfvar14 in namelist.input.
0 means assimilating mode, innovations (O minus B) are calculated and data are used in minimization.
1 means monitoring mode: innovations are calculated for diagnostics and monitoring. Data are not used in minimization.
Number of rad_monitoring should correspond to number of rtminit_nsensor. If rad_monitoring is not set, then default value of 0 will be used for all sensors.
(2) Outputing radiance diagnostics from WRFDA
Run WRFDA with the following namelist variables in record wrfvar14 in namelist.input.
write_iv_rad_ascii=.true.
to write out (observation-background) and other diagnostics information in plain-text files with prefix inv followed by instrument name and processor id. For example, 01_inv_noaa-17-amsub.0000 (01 is outerloop index, 0000 is processor index)
write_oa_rad_ascii=.true.
to write out (observation-background), (observation-analysis) and other diagnostics information in plain-text files with prefix oma followed by instrument name and processor id. For example, 01_oma_noaa-18-mhs.0001
Each processor writes out information of one instrument in one file in the WRFDA working directory.
(3) Radiance diagnostics data processing
A Fortran90 program is used to collect the 01_inv* or 01_oma* files and write out in netCDF format (one instrument in one file with prefix diags followed by instrument name, analysis date, and suffix .nc)) for easier data viewing, handling and plotting with netCDF utilities and NCL scripts.
(4) Radiance diagnostics plotting
NCL scripts (WRFDA/var/graphics/ncl/plot_rad_diags.ncl and WRFDA/var/graphics/ncl/advance_cymdh.ncl) are used for plotting. The NCL script can be run from a shell script, or run stand-alone with interactive ncl command (need to edit the NCL script and set the plot options. Also the path of advance_cymdh.ncl, a date advancing script loaded in the main NCL plotting script, may need to be modified).
Step (3) and (4) can be done by running a single ksh script (WRFDA/var/scripts/da_rad_diags.ksh) with proper settings. In addition to the settings of directories and what instruments to plot, there are some useful plotting options, explained below.
|
export OUT_TYPE=ncgm |
ncgm or pdf pdf will be much slower than ncgm and generate huge output if plots are not split. But pdf has higher resolution than ncgm. |
|
export PLOT_STATS_ONLY=false |
true or false true: only statistics of OMB/OMA vs channels and OMB/OMA vs dates will be plotted. false: data coverage, scatter plots (before and after bias correction), histograms (before and after bias correction), and statistics will be plotted. |
|
export PLOT_OPT=sea_only |
all, sea_only, land_only |
|
export PLOT_QCED=false
|
true or false true: plot only quality-controlled data false: plot all data |
|
export PLOT_HISTO=false |
true or false: switch for histogram plots |
|
export PLOT_SCATT=true |
true or false: switch for scatter plots |
|
export PLOT_EMISS=false |
true or false: switch for emissivity plots |
|
export PLOT_SPLIT=false |
true or false true: one frame in each file false: all frames in one file |
|
export PLOT_CLOUDY=false
|
true or false true: plot cloudy data. Cloudy data to be plotted are defined by PLOT_CLOUDY_OPT (si or clwp), CLWP_VALUE, SI_VALUE settings. |
|
export PLOT_CLOUDY_OPT=si |
si or clwp clwp: cloud liquid water path from model si: scatter index from obs, for amsua, amsub and mhs only |
|
export CLWP_VALUE=0.2 |
only plot points with clwp >= clwp_value (when clwp_value > 0) clwp > clwp_value (when clwp_value = 0) |
|
export SI_VALUE=3.0 |
|
(5) evolution of VarBC parameters
NCL scripts (WRFDA/var/graphics/ncl/plot_rad_varbc_param.ncl and WRFDA/var/graphics/ncl/advance_cymdh.ncl) are used for plotting evolutions of VarBC parameters.
WRFDA Diagnostics
WRFDA produces a number of diagnostic files that contain useful information on how the data assimilation has performed. This section will introduce you to some of these files, and what to look for.
Having run WRFDA, it is important to check a number of output files to see if the assimilation appears sensible. The WRFDA package, which includes lots of useful scripts may be downloaded from http://www.mmm.ucar.edu/wrf/users/wrfda/download/tools.html
The content of some useful diagnostic files are as follows:
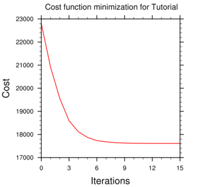
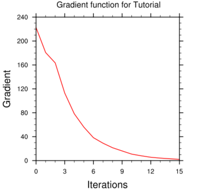
cost_fn and grad_fn: These files hold (in ASCII format) WRFDA cost and gradient function values, respectively, for the first and last iterations. However, if you run with PRINT_DETAIL_GRAD=true, these values will be listed for each iteration; this can be helpful for visualization purposes. The NCL script WRFDA/var/graphcs/ncl/plot_cost_grad_fn.ncl may be used to plot the content of cost_fn and grad_fn, if these files are generated with PRINT_DETAIL_GRAD=true.