General Homework Guidelines
GitHub prerequisites
I assume you can pull from and push to GitHub from RStudio.
Homework workflow
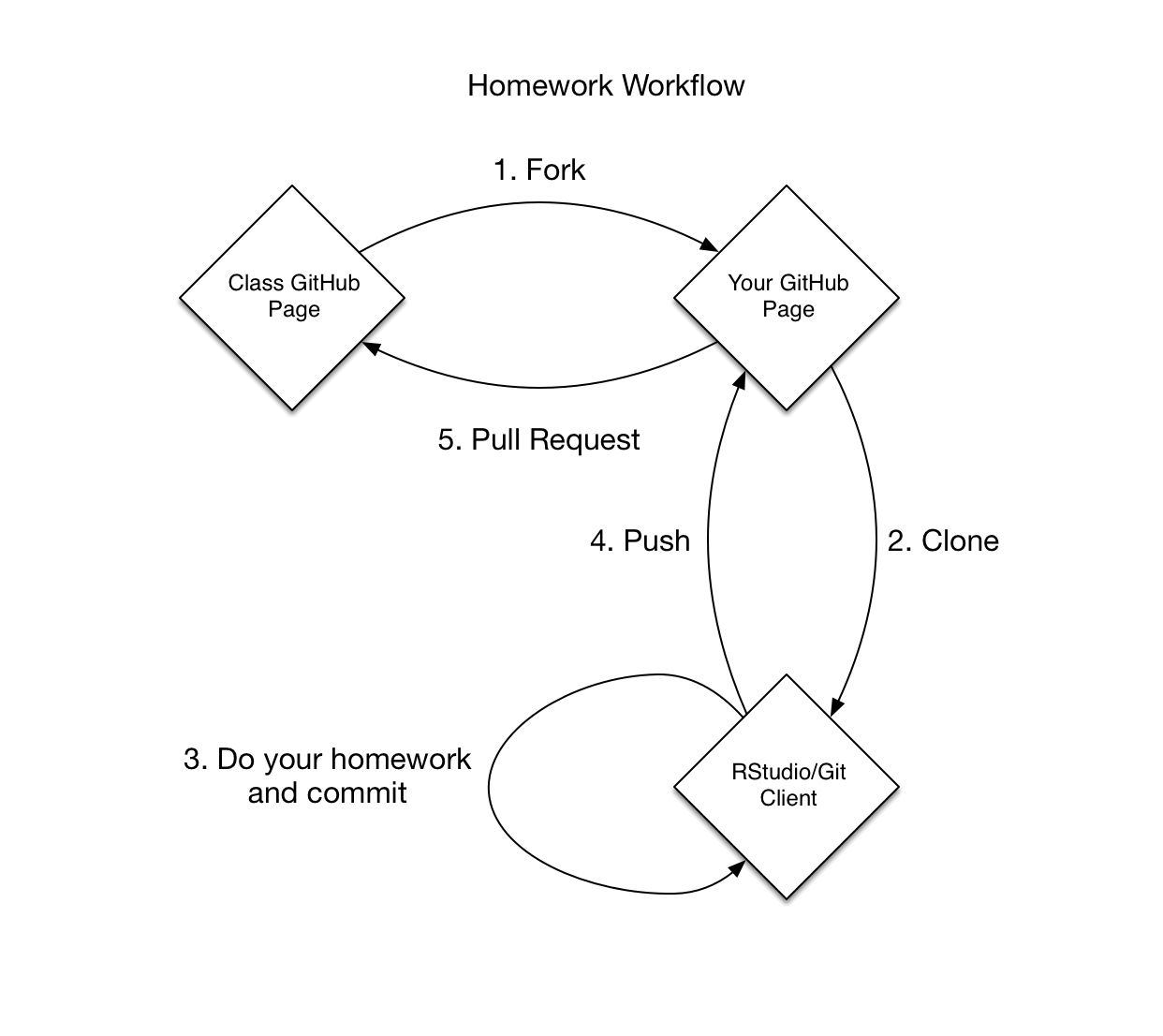
Homework assignments will be stored in separate Git repositories under the uc-cfss organization on GitHub. To complete a homework assignment, you need to:
- Fork the repository
- Clone the repository to your computer
- Modify the files and commit changes to complete your solution.
- Push/sync the changes up to GitHub.
- Create a pull request on the original repository to turn in the assignment. Make sure to include your name in the pull request.
Which files to commit
- Always commit the main source document, e.g., the R script or R Markdown or Markdown document. Commit early, commit often!
- For R Markdown source, also commit the intermediate Markdown (
.md) file and any accompaying files, such as figures.- Some purists would say intermediate and downstream products do NOT belong in the repo. After all, you can always recreate them from source, right? But here in reality, it turns out to be incredibly handy to have this in the repo.
- Commit the end product file. For homework submissions this is generally the Markdown file (
.md) because your output format isgithub_documentas well as all the graphs generated from the code chunks. For other projects, this might be an HTML (.html) or PDF (.pdf) file.- See above comment re: version control purists vs. pragmatists.
- You may not want to commit the Markdown and/or HTML until the work is fairly advanced, maybe even until submission. Once these enter the repo, you really should recompile them each time you commit changes to the R Markdown source, so that the Git history reflects the way these files should evolve as an ensemble.
- Never ever edit the intermediate/output documents “by hand”. Only edit the source and then regenerate the downstream products from that.
Make your work shine!
Here are some minor tweaks that can make a big difference in how awesome your product is.
Make it easy for people to access your work
Reduce the friction for graders to get the hard-working source code (the .R or .Rmd file) and the front-facing report (.md or .html).
- Create a
README.mdin the homework’s main directory to serve as the landing page for your submission. Whenever anyone visits this repo, this will be automatically rendered nicely! In particular, hyperlinks will work. - With this
README.mdfile, create annotated links to the documents graders will need to access. Such as:- Your main R Markdown document
- The Markdown product that comes from knitting your main R Markdown document
- Remember GitHub will render this into pseudo-HTML automagically
- Remember the figures in
_files/need to be available in the repo in order to appear here
Linking to HTML files in the repo
Simply visiting an HTML file in a GitHub repo just shows ugly HTML source. You need to do a little extra work to see this rendered as a proper webpage.
- Navigate to the HTML file on GitHub. Get the URL of the page, which should look something like this:
https://github.com/uc-cfss/uc-cfss.github.io/blob/master/hw00_homework_guidelines.html. Copy that URL! - Create a link to that in the usual Markdown way BUT prepend
http://htmlpreview.github.io/?to the URL. So the URL in your link should look something like this:http://htmlpreview.github.io/?https://github.com/uc-cfss/uc-cfss.github.io/blob/master/hw00_homework_guidelines.html. You can learn more about this preview facility here. - This sort of link would be fabulous to include in
README.md.
Make it easy for others to run your code
- In exactly one, very early R chunk, load any necessary packages, so your dependencies are obvious.
- In exactly one, very early R chunk, import anything coming from an external file. This will make it easy for someone to see which data files are required, edit to reflect their locals paths if necessary, etc. There are situations where you might not keep data in the repo itself.
- In exactly one, very last R chunk, report your session information. This prints version information about R, the operating system, and loaded packages so the reader knows the state of your machine when you rendered the R Markdown document. An R chunk with
devtools::session_info()will produce something that looks like this:
## Session info -------------------------------------------------------------## setting value
## version R version 3.4.3 (2017-11-30)
## system x86_64, darwin15.6.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## tz America/Chicago
## date 2018-04-05## Packages -----------------------------------------------------------------## package * version date source
## backports 1.1.2 2017-12-13 CRAN (R 3.4.3)
## base * 3.4.3 2017-12-07 local
## compiler 3.4.3 2017-12-07 local
## datasets * 3.4.3 2017-12-07 local
## devtools 1.13.5 2018-02-18 CRAN (R 3.4.3)
## digest 0.6.15 2018-01-28 CRAN (R 3.4.3)
## evaluate 0.10.1 2017-06-24 CRAN (R 3.4.1)
## graphics * 3.4.3 2017-12-07 local
## grDevices * 3.4.3 2017-12-07 local
## htmltools 0.3.6 2017-04-28 CRAN (R 3.4.0)
## knitr 1.20 2018-02-20 CRAN (R 3.4.3)
## magrittr 1.5 2014-11-22 CRAN (R 3.4.0)
## memoise 1.1.0 2017-04-21 CRAN (R 3.4.0)
## methods * 3.4.3 2017-12-07 local
## Rcpp 0.12.16 2018-03-13 CRAN (R 3.4.4)
## rmarkdown 1.9 2018-03-01 CRAN (R 3.4.3)
## rprojroot 1.3-2 2018-01-03 CRAN (R 3.4.3)
## stats * 3.4.3 2017-12-07 local
## stringi 1.1.7 2018-03-12 CRAN (R 3.4.3)
## stringr 1.3.0 2018-02-19 CRAN (R 3.4.3)
## tools 3.4.3 2017-12-07 local
## utils * 3.4.3 2017-12-07 local
## withr 2.1.2 2018-03-15 CRAN (R 3.4.4)
## yaml 2.1.18 2018-03-08 CRAN (R 3.4.4)- Pretend you are someone else. Clone a fresh copy of your own repo from GitHub, fire up a new RStudio session and try to knit your R Markdown file. Does it “just work”? It should!
Make pretty tables
Instead of just printing an object with R, you could format the info in an attractive table. Some leads:
- The
kable()function fromknitr. - Also look into the packages
xtable,pander.
Not sure what #rstats package to use to make your lovely Reproducible Table? I made a flowchart for you! #knitr pic.twitter.com/5qC5EBADN4
— Andrew MacDonald (@polesasunder) May 7, 2014
Acknowledgments
- This page is derived in part from “UBC STAT 545A and 547M”, licensed under the CC BY-NC 3.0 Creative Commons License.
This work is licensed under the CC BY-NC 4.0 Creative Commons License.