Basic example
Here is the magick of ggplot2: the ability to map a variable to marker features. Here, the marker color depends on its value in the field called Species in the input data frame.
Note that the legend is built automatically.
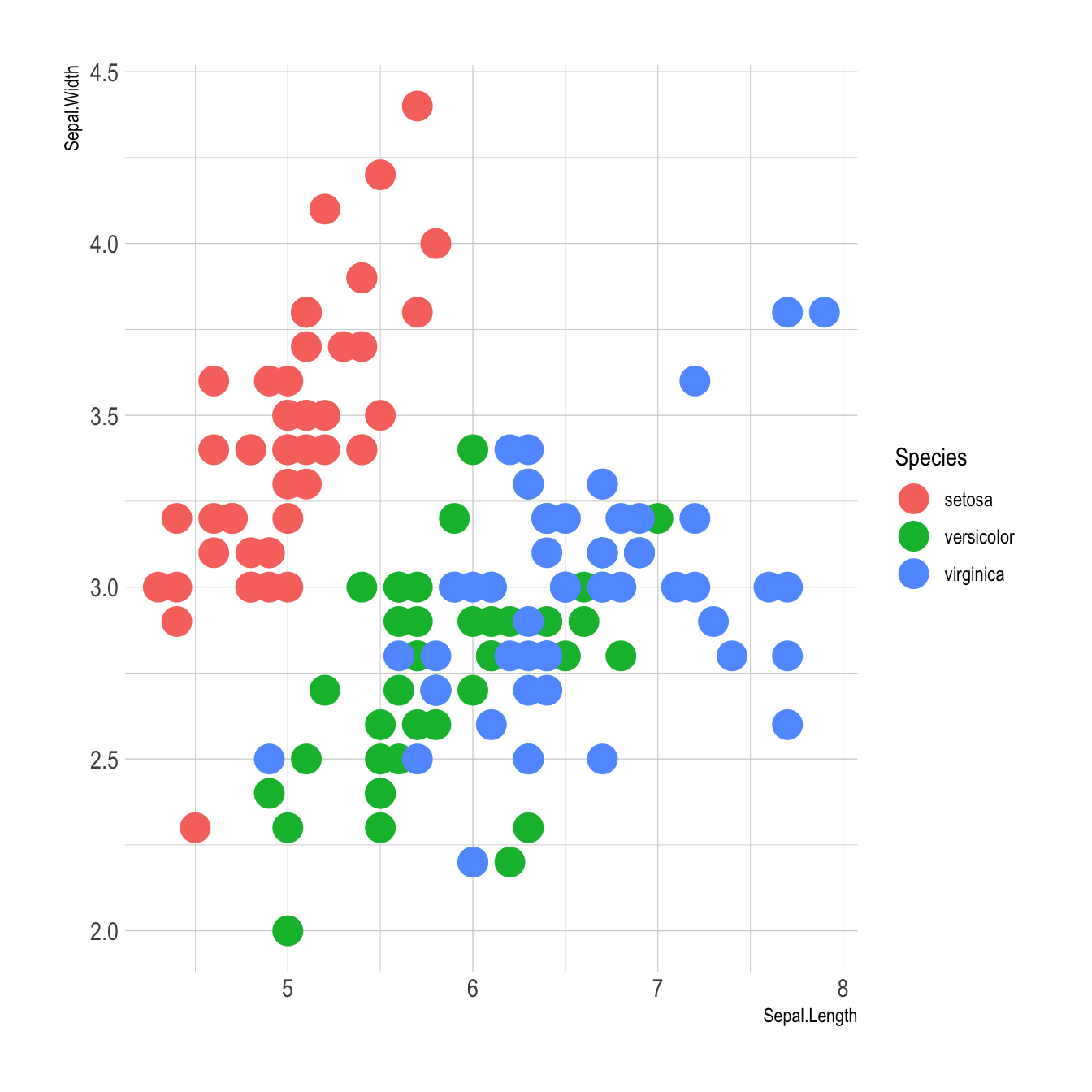
# load ggplot2
library(ggplot2)
library(hrbrthemes)
# mtcars dataset is natively available in R
# head(mtcars)
# A basic scatterplot with color depending on Species
ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, color=Species)) +
geom_point(size=6) +
theme_ipsum()Works with any aesthetics
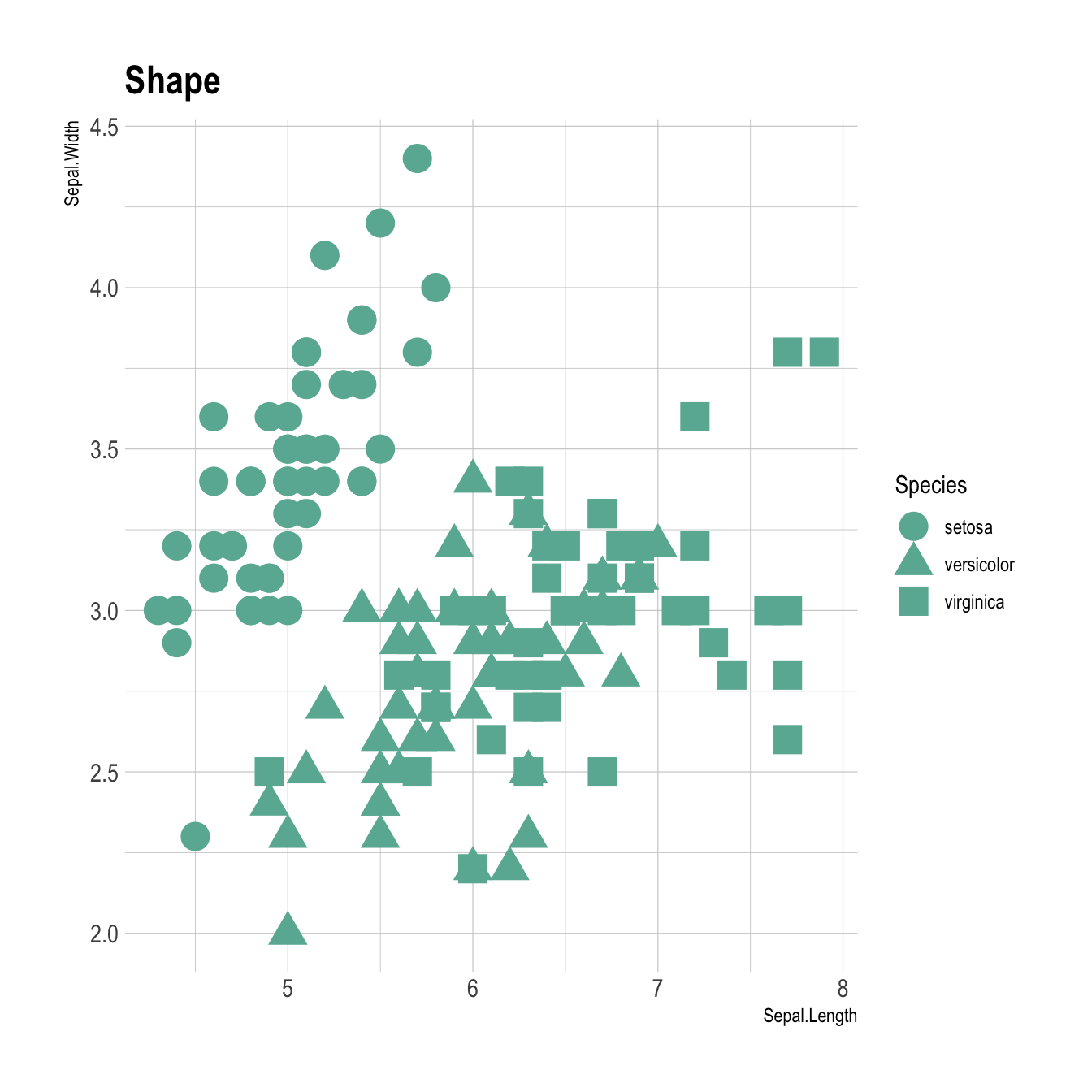
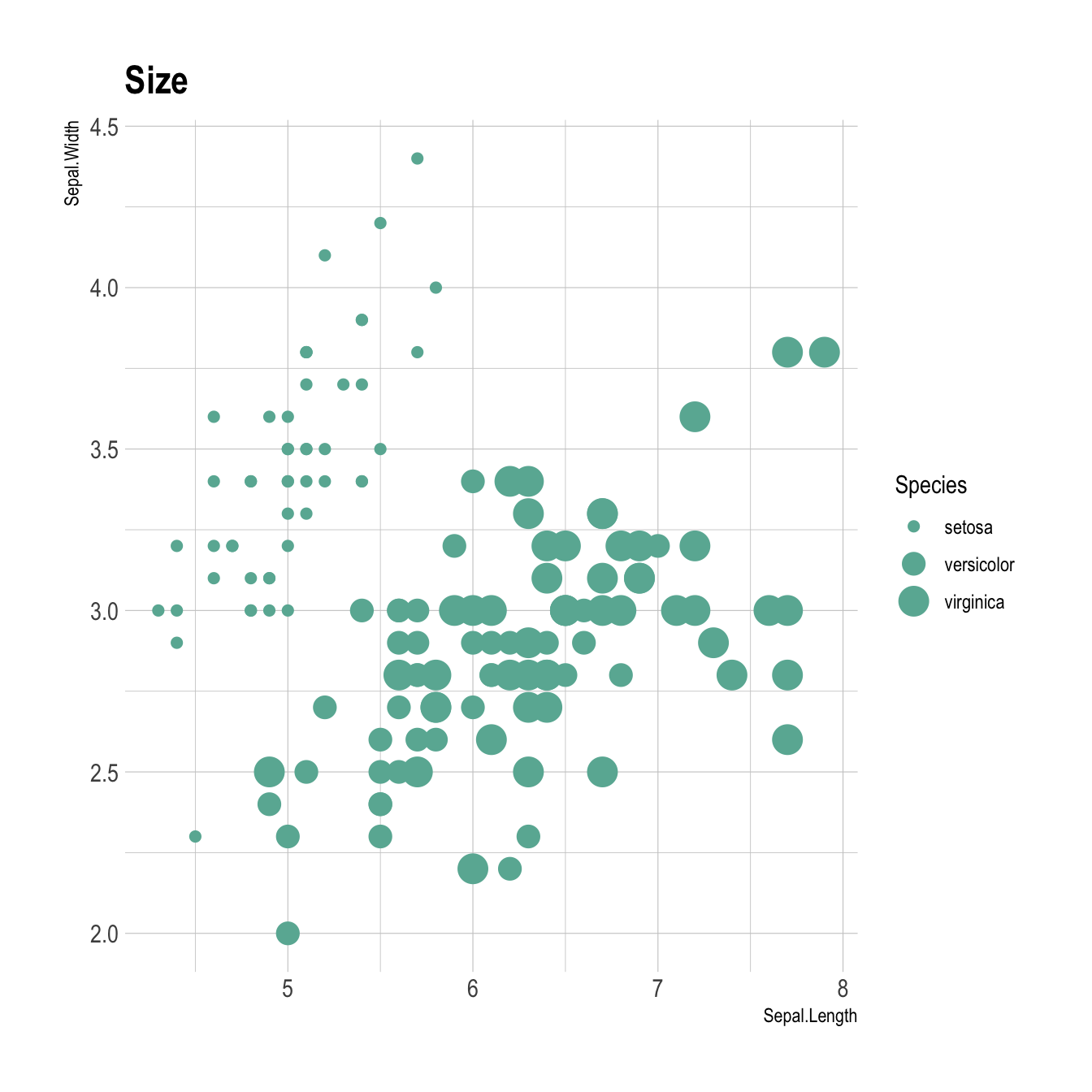
You can map variables to any marker features. For instance, specie is represente below using transparency (left), shape (middle) and size (right).
# load ggplot2
library(ggplot2)
library(hrbrthemes)
# Transparency
ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, alpha=Species)) +
geom_point(size=6, color="#69b3a2") +
theme_ipsum()
# Shape
ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, shape=Species)) +
geom_point(size=6) +
theme_ipsum()
# Size
ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, shape=Species)) +
geom_point(size=6) +
theme_ipsum()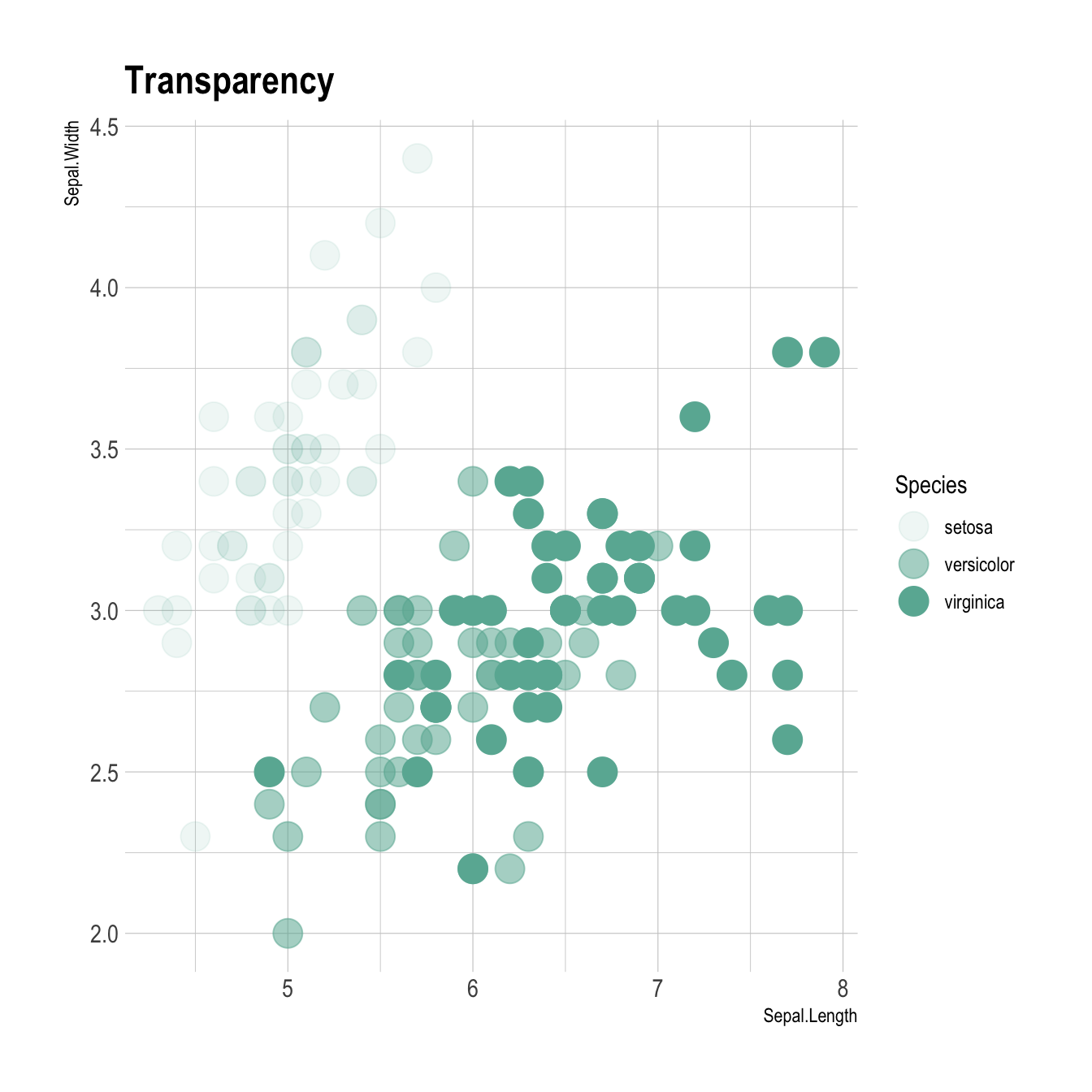
Mapping to several features
Last but not least, note that you can map one or several variables to one or several features. Here, shape, transparency, size and color all depends on the marker Species value.
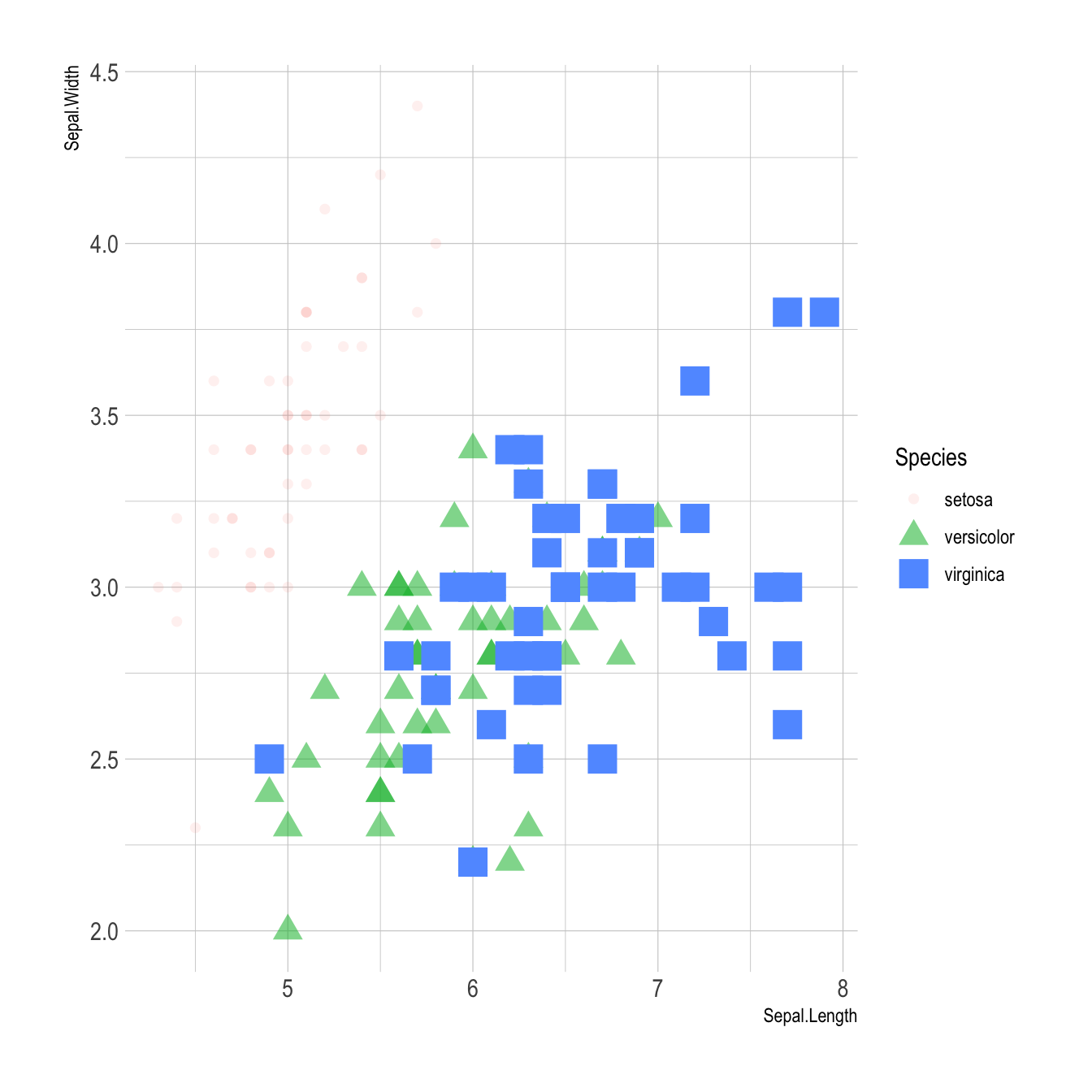
# load ggplot2
library(ggplot2)
library(hrbrthemes)
# A basic scatterplot with color depending on Species
ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, shape=Species, alpha=Species, size=Species, color=Species)) +
geom_point() +
theme_ipsum()




