Descriptive analysis in R with the package metan
Tiago Olivoto
2023-03-06
Source:vignettes/vignettes_descriptive.Rmd
vignettes_descriptive.RmdGetting started
metan provides a simple and intuitive pipe-friendly
framework for performing descriptive statistics. A set
of functions can be used to compute the most used descriptive
statistics quickly. In this tutorial, we will use the data example
data_ge2 as motivation examples. See the section Rendering engine to know how HTML tables were
generated.
Let’s start with a very simple (but widely used) example: to compute
the mean of a numeric variable (say, PH) from a data set (data_ge2).
Using the R base functions stats::mean(), the solution
would look like to:
mean(data_ge2$PH)
# [1] 2.484813Consider that now we want to compute the mean value of PH for each
level of the factor GEN. In other words, to compute the mean value of PH
for each genotype. The solution using stats::aggregate() is
then:
aggr <- aggregate(PH ~ GEN, data = data_ge2, FUN = mean)
print_table(aggr)Statistics by levels of a factor
Using the metan’s function mean_by() the
amount of code needed is dramatically reduced. To compute the grand mean
for all numeric variables of data_ge, we simple use:
ov_mean <-
mean_by(data_ge2)
print_table(ov_mean)To compute the mean values for each level of the factor GEN, we then
need to add the grouping variable GEN in the argument
....
ov_mean2 <-
mean_by(data_ge2, GEN)
print_table(ov_mean2)The following _by() functions are available for
computing the main descriptive statistics by levels of a factor.
Useful functions
Other useful functions are also implemented. All of them works
naturally with %>%, handle grouped data with
dplyr::group_by() and multiple variables (all numeric
variables from .data by default).
-
av_dev()computes the average absolute deviation. -
ci_mean_t()computes t-inverval (95%) for the mean. -
ci_mean_z()computes z-inverval (95%) for the mean. -
cv()computes the coefficient of variation. -
freq_table()Computes frequency fable. -
hm_mean(),gm_mean()computes the harmonic and geometric means, respectively. The harmonic mean is the reciprocal of the arithmetic mean of the reciprocals. The geometric mean is the nth root of n products. -
kurt()computes the kurtosis like used in SAS and SPSS. -
range_data()Computes the range of the values. -
sd_amo(),sd_pop()Computes sample and populational standard deviation, respectively. -
sem()computes the standard error of the mean. -
skew()computes the skewness like used in SAS and SPSS. -
sum_dev()computes the sum of the absolute deviations. -
sum_sq_dev()computes the sum of the squared deviations. -
var_amo(),var_pop()computes sample and populational variance. -
valid_n()Return the valid (not NA) length of a data.
Let’s show some examples. Note that select helpers can be used to select variables based on their names.
# Confidence t-interval 0.95 for the mean # Variables with name width greater than 3 characters # Grouped by levels of ENV data_ge2 %>% group_by(ENV) %>% ci_mean_t(width_greater_than(3)) %>% print_table()The wrapper function desc_stat()
To compute all statistics at once we can use
desc_stat(). This is a wrapper function around the above
ones and may be used to compute measures of central tendency, position,
and dispersion. By default (stats = "main"), seven
statistics (coefficient of variation, maximum, mean, median, minimum,
sample standard deviation, standard error and confidence interval of the
mean) are computed. Other allowed values are "all" to show
all the statistics, "robust" to show robust statistics,
"quantile" to show quantile statistics, or chose one (or
more) statistics using a comma-separated vector with the statistic
names, e.g., stats = c("mean, cv"). We can also use
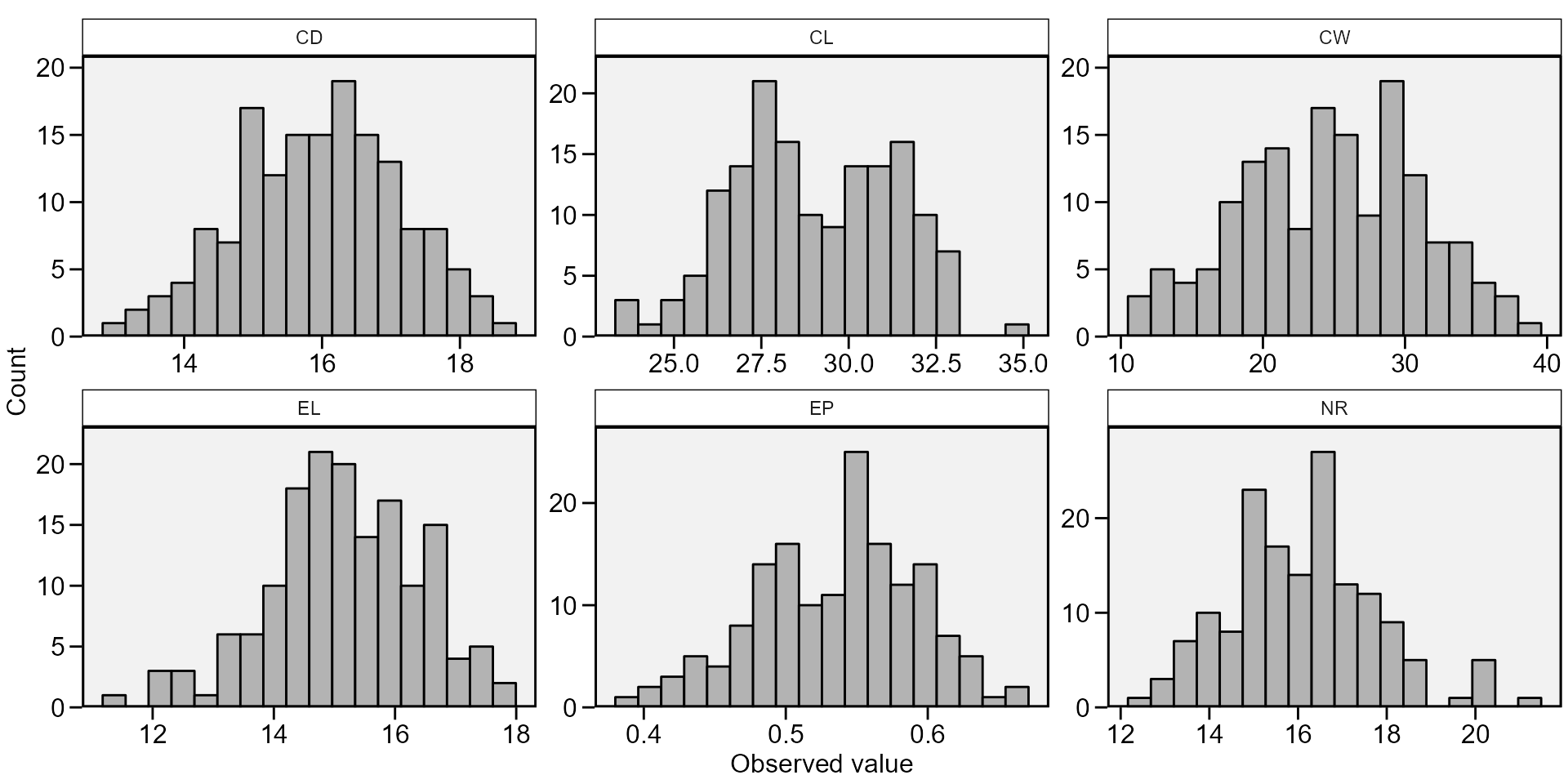
hist = TRUE to create a histogram for each variable. Here,
select helpers can also be used in the argument ....
All statistics for all numeric variables
all <- desc_stat(data_ge2, stats = "all")
print_table(all)Statistics by levels of factors
To compute the statistics for each level of a factor, use the
argument by. In addition, it is possible to select the
statistics to compute using the argument stats, that is a
single statistic name, e.g., "mean", or a a comma-separated
vector of names with " at the beginning and end of vector
only. Note that the statistic names ARE NOTE case
sensitive, i.e., both "mean", "Mean", or
"MEAN" are recognized. Comma or spaces can be used to
separate the statistics’ names.
- All options bellow will work:
stats = c("mean, se, cv, max, min")stats = c("mean se cv max min")stats = c("MEAN, Se, CV max Min")
stats_c <-
desc_stat(data_ge2,
contains("C"),
stats = ("mean, se, cv, max, min"),
by = ENV)
print_table(stats_c)We may convert the results above into a wider format by
using the function desc_wider()
desc_wider(stats_c, mean) %>%
print_table()To compute the descriptive statistics by more than one grouping
variable, we need to pass a grouped data to the argument
.data with the function group_by(). Let’s
compute the mean, the standard error of the mean and the sample size for
the variables EP and EL for all combinations
of the factors ENV and GEN.
stats_grp <-
data_ge2 %>%
group_by(ENV, GEN) %>%
desc_stat(EP, EL,
stats = c("mean, se, n"))
print_table(stats_grp)Rendering engine
This vignette was built with pkgdown. All tables were produced
with the package DT using the
following function.
library(DT) # Used to make the tables
# Function to make HTML tables
print_table <- function(table, rownames = FALSE, digits = 3, ...){
df <- datatable(table, rownames = rownames, extensions = 'Buttons',
options = list(scrollX = TRUE,
dom = '<<t>Bp>',
buttons = c('copy', 'excel', 'pdf', 'print')), ...)
num_cols <- c(as.numeric(which(sapply(table, class) == "numeric")))
if(length(num_cols) > 0){
formatSignif(df, columns = num_cols, digits = digits)
} else{
df
}
}