Cross-validation for estimation of all AMMI-family models
cv_ammif provides a complete cross-validation of replicate-based data
using AMMI-family models. By default, the first validation is carried out
considering the AMMIF (all possible axis used). Considering this model, the
original dataset is split up into two datasets: training set and validation
set. The 'training' set has all combinations (genotype x environment) with
N-1 replications. The 'validation' set has the remaining replication. The
splitting of the dataset into modeling and validation sets depends on the
design informed. For Completely Randomized Block Design (default), and
alpha-lattice design (declaring block arguments), complete replicates
are selected within environments. The remained replicate serves as validation
data. If design = 'RCD' is informed, completely randomly samples are
made for each genotype-by-environment combination (Olivoto et al. 2019). The
estimated values for each member of the AMMI-family model are compared with
the 'validation' data. The Root Mean Square Prediction Difference (RMSPD) is
computed. At the end of boots, a list is returned.
IMPORTANT: If the data set is unbalanced (i.e., any genotype missing in any environment) the function will return an error. An error is also observed if any combination of genotype-environment has a different number of replications than observed in the trial.
Arguments
- .data
The dataset containing the columns related to Environments, Genotypes, replication/block and response variable(s).
- env
The name of the column that contains the levels of the environments.
- gen
The name of the column that contains the levels of the genotypes.
- rep
The name of the column that contains the levels of the replications/blocks. AT LEAST THREE REPLICATES ARE REQUIRED TO PERFORM THE CROSS-VALIDATION.
- resp
The response variable.
- nboot
The number of resamples to be used in the cross-validation. Defaults to 200.
- block
Defaults to
NULL. In this case, a randomized complete block design is considered. If block is informed, then a resolvable alpha-lattice design (Patterson and Williams, 1976) is employed. All effects, except the error, are assumed to be fixed.- design
The experimental design used in each environment. Defaults to
RCBD(Randomized complete Block Design). For Completely Randomized Designs informdesign = 'CRD'.- verbose
A logical argument to define if a progress bar is shown. Default is
TRUE.
Value
An object of class cv_ammif with the following items:
RMSPD: A vector with nboot-estimates of the Root Mean Squared Prediction Difference between predicted and validating data.
RMSPDmean: The mean of RMSPDmean estimates.
Estimated: A data frame that contain the values (predicted, observed, validation) of the last loop.
Modeling: The dataset used as modeling data in the last loop
Testing: The dataset used as testing data in the last loop.
References
Patterson, H.D., and E.R. Williams. 1976. A new class of resolvable incomplete block designs. Biometrika 63:83-92.
Author
Tiago Olivoto tiagoolivoto@gmail.com
Examples
# \donttest{
library(metan)
model <- cv_ammif(data_ge2,
env = ENV,
gen = GEN,
rep = REP,
resp = EH,
nboot = 5)
#> 1 of 5 sets using AMMIF |== | 5% 00:00:00
2 of 5 sets using AMMIF |==== | 10% 00:00:01
3 of 5 sets using AMMIF |====== | 15% 00:00:01
4 of 5 sets using AMMIF |======== | 20% 00:00:02
5 of 5 sets using AMMIF |========== | 25% 00:00:02
1 of 5 sets using AMMI2 |============ | 30% 00:00:03
2 of 5 sets using AMMI2 |============== | 35% 00:00:04
3 of 5 sets using AMMI2 |================ | 40% 00:00:04
4 of 5 sets using AMMI2 |================== | 45% 00:00:05
5 of 5 sets using AMMI2 |==================== | 50% 00:00:05
1 of 5 sets using AMMI1 |====================== | 55% 00:00:06
2 of 5 sets using AMMI1 |======================== | 60% 00:00:06
3 of 5 sets using AMMI1 |========================== | 65% 00:00:07
4 of 5 sets using AMMI1 |============================ | 70% 00:00:08
5 of 5 sets using AMMI1 |============================== | 75% 00:00:08
1 of 5 sets using AMMI0 |================================ | 80% 00:00:09
2 of 5 sets using AMMI0 |================================== | 85% 00:00:09
3 of 5 sets using AMMI0 |==================================== | 90% 00:00:10
4 of 5 sets using AMMI0 |====================================== | 95% 00:00:10
5 of 5 sets using AMMI0 |========================================| 100% 00:00:11
plot(model)
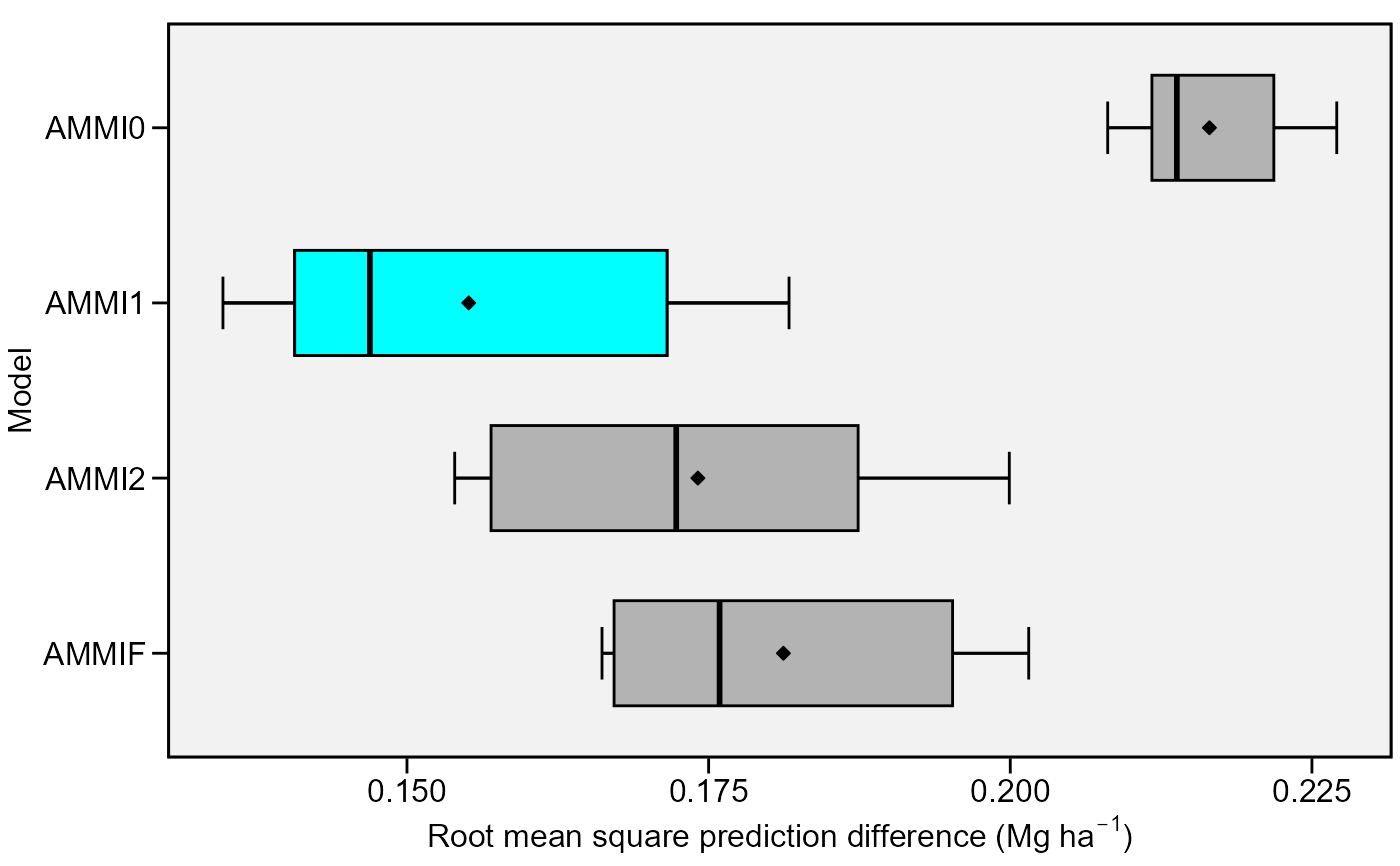 # }
# }
