Produces genotype plus genotype-by-environment model based on a multi-environment trial dataset containing at least the columns for genotypes, environments and one response variable or a two-way table.
Usage
gge(
.data,
env,
gen,
resp,
centering = "environment",
scaling = "none",
svp = "environment",
by = NULL,
...
)Arguments
- .data
The dataset containing the columns related to Environments, Genotypes and the response variable(s).
- env
The name of the column that contains the levels of the environments.
- gen
The name of the column that contains the levels of the genotypes.
- resp
The response variable(s). To analyze multiple variables in a single procedure a vector of variables may be used. For example
resp = c(var1, var2, var3). Select helpers are also supported.- centering
The centering method. Must be one of the
'none | 0', for no centering;'global | 1', for global centered (E+G+GE);'environment | 2'(default), for environment-centered (G+GE); or'double | 3', for double centered (GE). A biplot cannot be produced with models produced without centering.- scaling
The scaling method. Must be one of the
'none | 0'(default), for no scaling; or'sd | 1', where each value is divided by the standard deviation of its corresponding environment (column). This will put all environments roughly he same rang of values.- svp
The method for singular value partitioning. Must be one of the
'genotype | 1', (The singular value is entirely partitioned into the genotype eigenvectors, also called row metric preserving);'environment | 2', default, (The singular value is entirely partitioned into the environment eigenvectors, also called column metric preserving); or'symmetrical | 3'(The singular value is symmetrically partitioned into the genotype and the environment eigenvectors This SVP is most often used in AMMI analysis and other biplot analysis, but it is not ideal for visualizing either the relationship among genotypes or that among the environments).- by
One variable (factor) to compute the function by. It is a shortcut to
dplyr::group_by().This is especially useful, for example, when the researcher want to produce GGE biplots for each level of a categorical variable. In this case, an object of class gge_grouped is returned.- ...
Arguments passed to the function
impute_missing_val()for imputation of missing values in case of unbalanced data.
Value
The function returns a list of class gge containing the following objects
coordgen The coordinates for genotypes for all components.
coordenv The coordinates for environments for all components.
eigenvalues The vector of eigenvalues.
totalvar The overall variance.
labelgen The name of the genotypes.
labelenv The names of the environments.
labelaxes The axes labels.
ge_mat The data used to produce the model (scaled and centered).
centering The centering method.
scaling The scaling method.
svp The singular value partitioning method.
d The factor used to generate in which the ranges of genotypes and environments are comparable when singular value partitioning is set to 'genotype' or 'environment'.
grand_mean The grand mean of the trial.
mean_gen A vector with the means of the genotypes.
mean_env A vector with the means of the environments.
scale_var The scaling vector when the scaling method is
'sd'.
References
Yan, W., and M.S. Kang. 2003. GGE biplot analysis: a graphical tool for breeders, geneticists, and agronomists. CRC Press.
Author
Tiago Olivoto tiagoolivoto@gmail.com
Examples
# \donttest{
library(metan)
mod <- gge(data_ge, ENV, GEN, GY)
plot(mod)
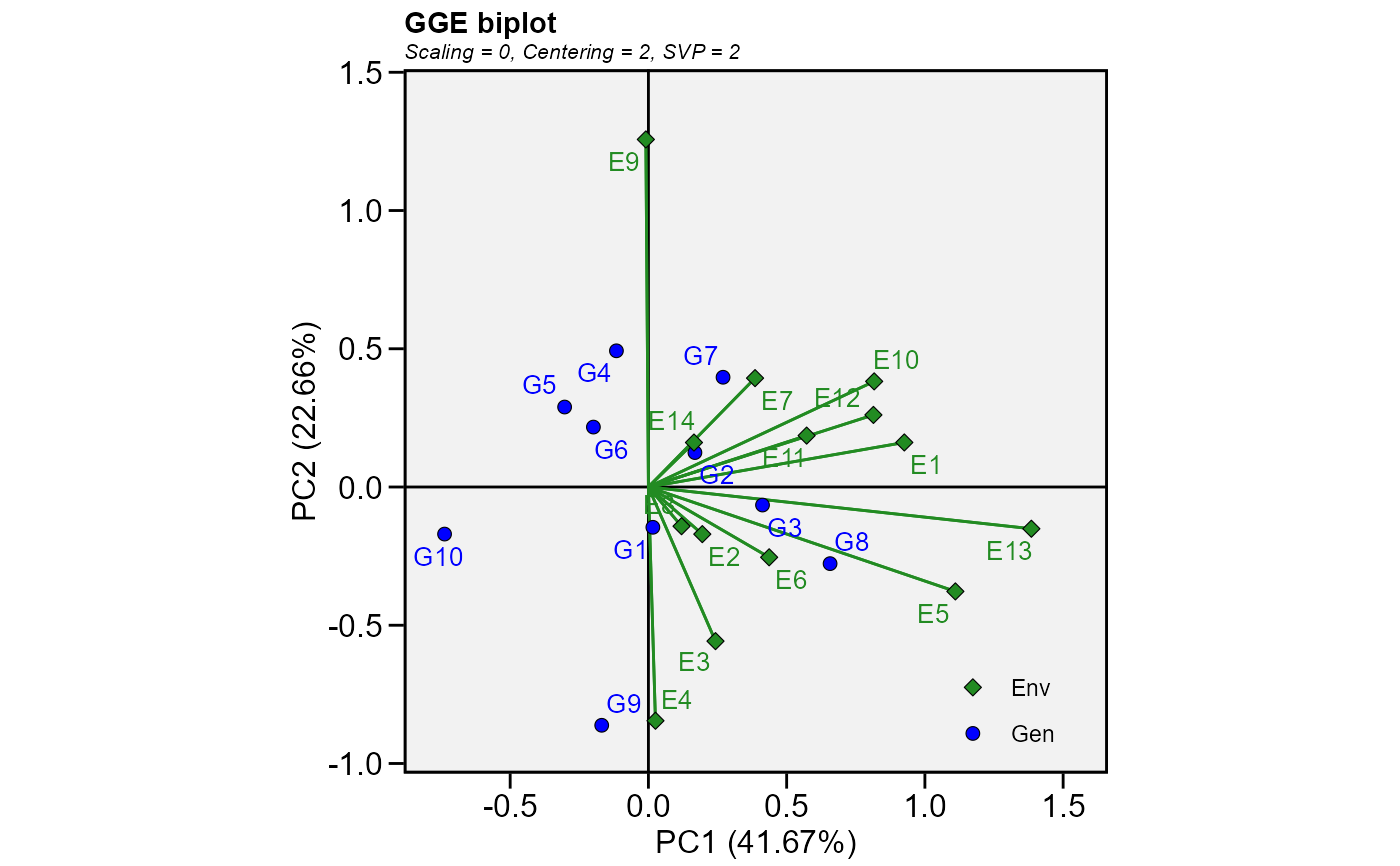 # GGE model for all numeric variables
mod2 <- gge(data_ge2, ENV, GEN, resp = everything())
plot(mod2, var = "ED")
# GGE model for all numeric variables
mod2 <- gge(data_ge2, ENV, GEN, resp = everything())
plot(mod2, var = "ED")
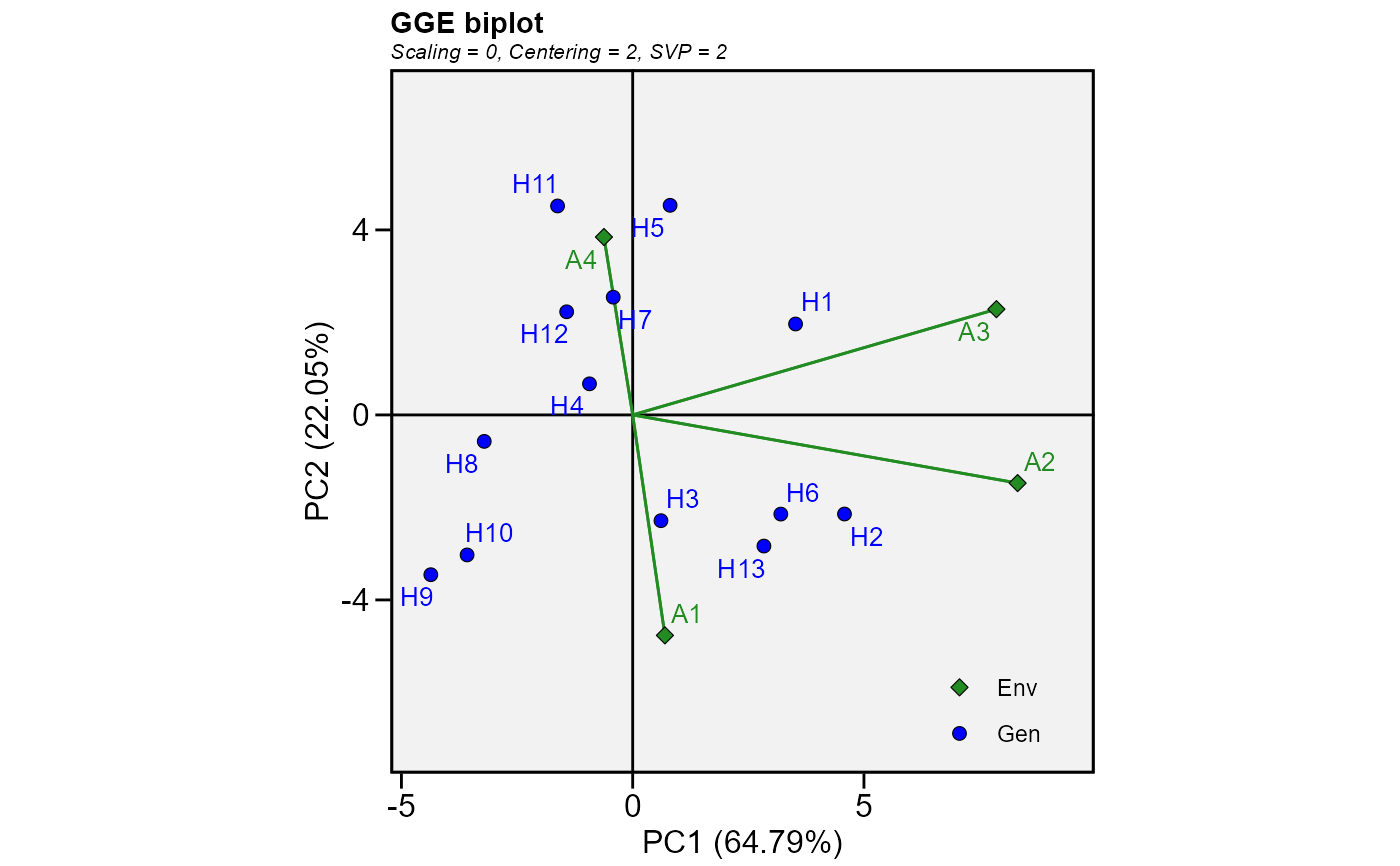 # If we have a two-way table with the mean values for
# genotypes and environments
table <- make_mat(data_ge, GEN, ENV, GY) %>% round(2)
table
#> E1 E10 E11 E12 E13 E14 E2 E3 E4 E5 E6 E7 E8 E9
#> G1 2.37 2.31 1.36 1.34 3.00 1.53 3.04 4.08 3.49 4.17 2.81 1.90 2.27 2.78
#> G10 1.97 1.54 0.90 1.02 1.83 1.86 3.15 4.11 4.27 3.37 2.48 2.24 2.70 3.15
#> G2 2.90 2.30 1.49 1.99 3.03 1.43 3.23 4.57 3.72 3.83 2.54 1.99 2.05 3.36
#> G3 2.89 2.34 1.57 1.76 3.47 2.06 3.61 4.13 4.13 4.13 2.98 2.16 2.85 3.29
#> G4 2.59 2.17 1.37 1.53 2.64 1.86 3.19 3.85 3.30 3.78 2.70 1.98 2.30 3.72
#> G5 2.19 2.14 1.33 1.69 2.57 1.78 3.14 3.74 3.38 3.47 2.43 1.66 2.71 3.30
#> G6 2.30 2.21 1.50 1.39 2.91 1.80 3.29 3.43 3.40 3.57 2.34 1.76 2.54 3.04
#> G7 2.77 2.44 1.36 1.95 3.18 1.94 2.61 4.10 3.02 4.05 2.67 2.55 2.58 3.14
#> G8 2.90 2.57 1.68 2.00 3.52 1.99 3.44 4.11 4.14 4.81 2.91 2.26 2.88 2.83
#> G9 2.33 1.74 1.13 1.41 2.95 1.57 3.09 4.51 3.90 3.93 2.77 1.39 2.49 1.94
make_long(table) %>%
gge(ENV, GEN, Y) %>%
plot()
# If we have a two-way table with the mean values for
# genotypes and environments
table <- make_mat(data_ge, GEN, ENV, GY) %>% round(2)
table
#> E1 E10 E11 E12 E13 E14 E2 E3 E4 E5 E6 E7 E8 E9
#> G1 2.37 2.31 1.36 1.34 3.00 1.53 3.04 4.08 3.49 4.17 2.81 1.90 2.27 2.78
#> G10 1.97 1.54 0.90 1.02 1.83 1.86 3.15 4.11 4.27 3.37 2.48 2.24 2.70 3.15
#> G2 2.90 2.30 1.49 1.99 3.03 1.43 3.23 4.57 3.72 3.83 2.54 1.99 2.05 3.36
#> G3 2.89 2.34 1.57 1.76 3.47 2.06 3.61 4.13 4.13 4.13 2.98 2.16 2.85 3.29
#> G4 2.59 2.17 1.37 1.53 2.64 1.86 3.19 3.85 3.30 3.78 2.70 1.98 2.30 3.72
#> G5 2.19 2.14 1.33 1.69 2.57 1.78 3.14 3.74 3.38 3.47 2.43 1.66 2.71 3.30
#> G6 2.30 2.21 1.50 1.39 2.91 1.80 3.29 3.43 3.40 3.57 2.34 1.76 2.54 3.04
#> G7 2.77 2.44 1.36 1.95 3.18 1.94 2.61 4.10 3.02 4.05 2.67 2.55 2.58 3.14
#> G8 2.90 2.57 1.68 2.00 3.52 1.99 3.44 4.11 4.14 4.81 2.91 2.26 2.88 2.83
#> G9 2.33 1.74 1.13 1.41 2.95 1.57 3.09 4.51 3.90 3.93 2.77 1.39 2.49 1.94
make_long(table) %>%
gge(ENV, GEN, Y) %>%
plot()
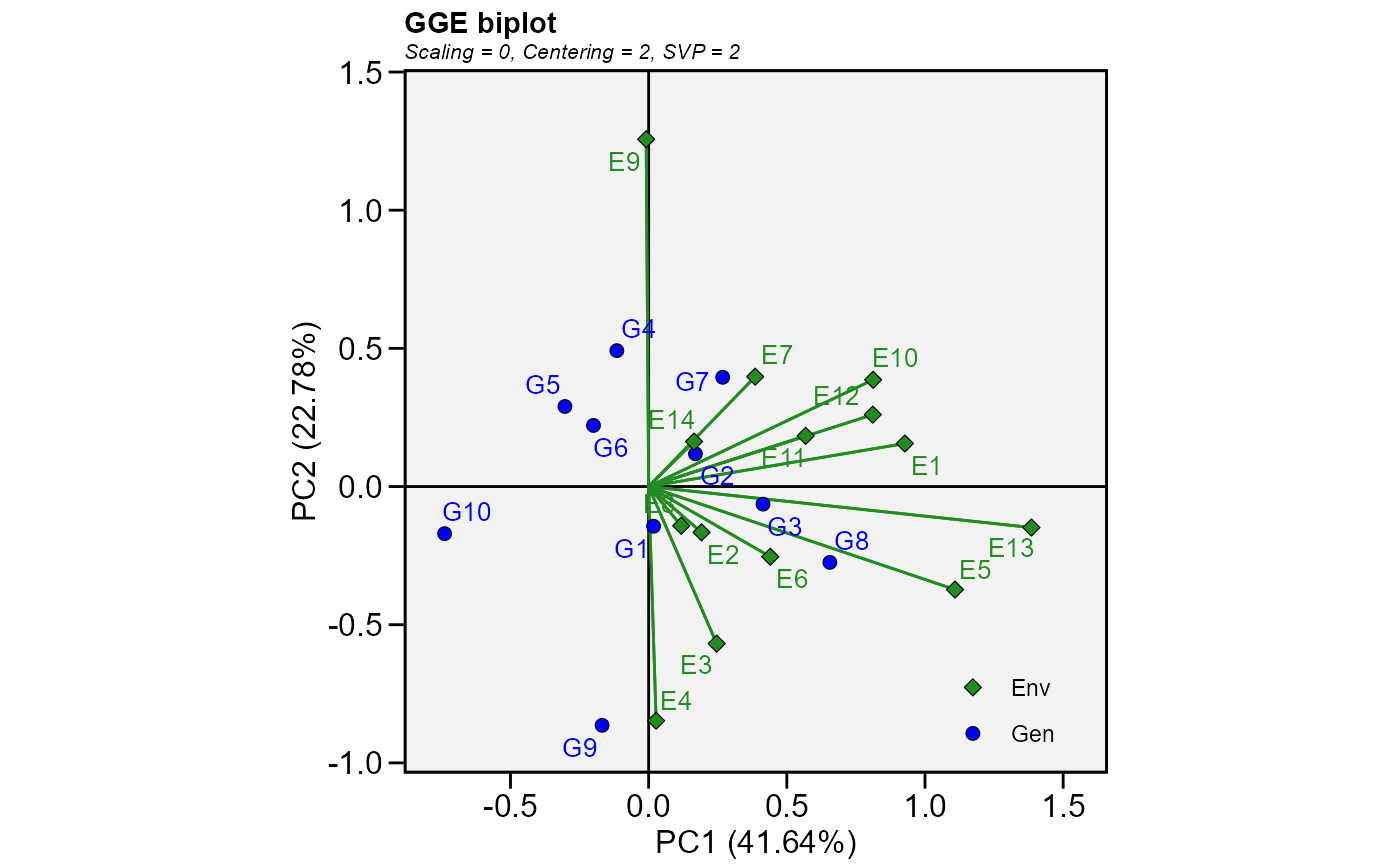 # }
# }
