Performs a Mantel test between two correlation/distance matrices. The function calculates the correlation between two matrices, the Z-score that is is the sum of the products of the corresponding elements of the matrices and a two-tailed p-value (null hypothesis: \[r = 0\]).
Arguments
- mat1, mat2
A correlation matrix or an object of class
dist.- nboot
The number of permutations to be used. Defaults to
1000.- plot
if
plot = TRUE, plots the density estimate of the permutation distribution along with the observed Z-score as a vertical line.
Value
mantel_rThe correlation between the two matrices.z_scoreThe Z-score.p-valueThe quantile of the observed Z-score. in the permutation distribution.
Author
Tiago Olivoto tiagoolivoto@gmail.com
Examples
# \donttest{
library(metan)
# Test if the correlation of traits (data_ge2 dataset)
# changes between A1 and A2 levels of factor ENV
A1 <- corr_coef(data_ge2 %>% subset(ENV == "A1"))[["cor"]]
A2 <- corr_coef(data_ge2 %>% subset(ENV == "A2"))[["cor"]]
mantel_test(A1, A2, plot = TRUE)
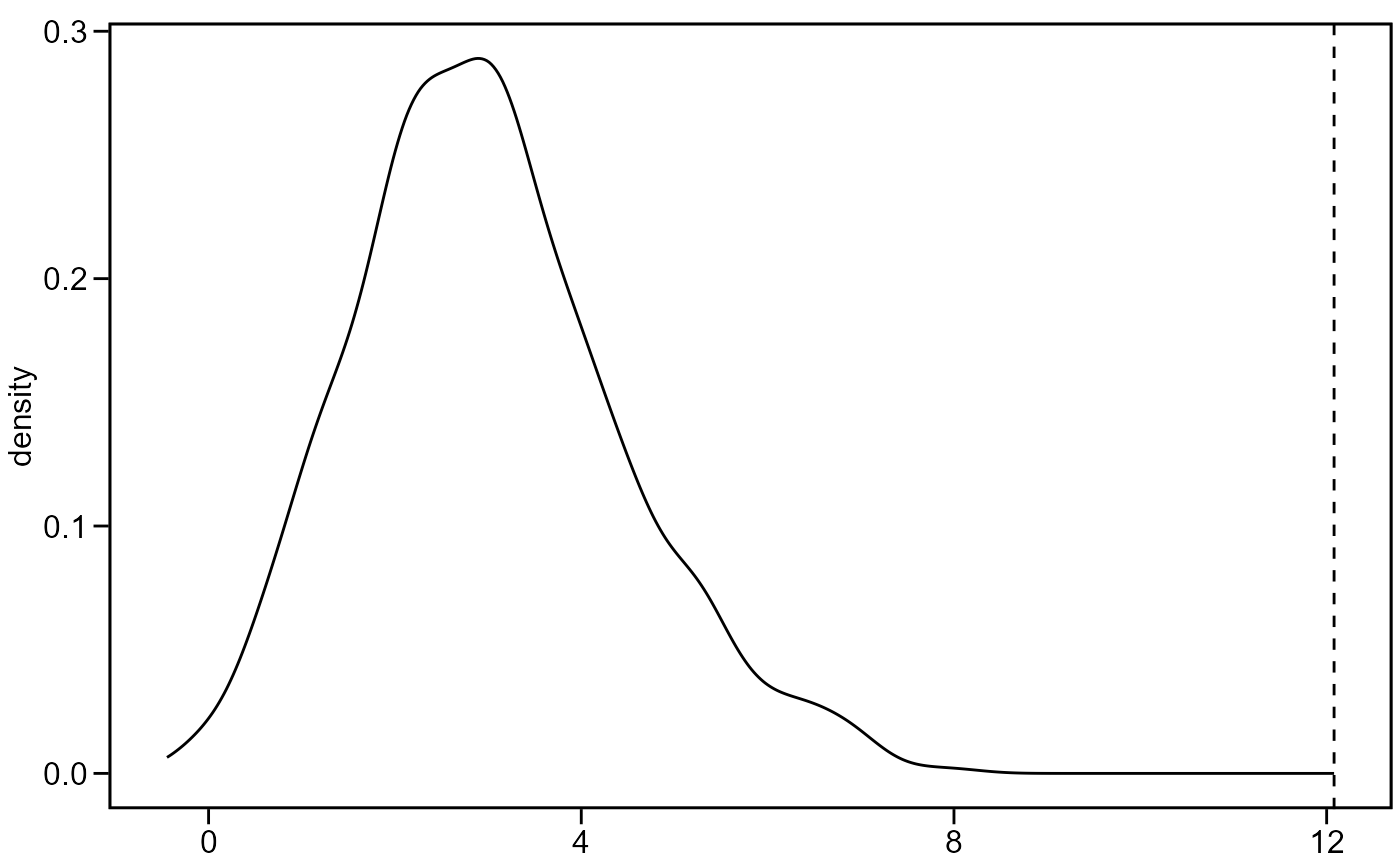 #> $mantel_r
#> [1] 0.7154291
#>
#> $z_score
#> [1] 12.07974
#>
#> $p_value
#> [1] 0.000999001
#>
# }
#> $mantel_r
#> [1] 0.7154291
#>
#> $z_score
#> [1] 12.07974
#>
#> $p_value
#> [1] 0.000999001
#>
# }
