Note
Go to the end to download the full example code or to run this example in your browser via JupyterLite or Binder.
Imputing missing values before building an estimator#
Missing values can be replaced by the mean, the median or the most frequent
value using the basic SimpleImputer.
In this example we will investigate different imputation techniques:
imputation by the constant value 0
imputation by the mean value of each feature
k nearest neighbor imputation
iterative imputation
In all the cases, for each feature, we add a new feature indicating the missingness.
We will use two datasets: Diabetes dataset which consists of 10 feature variables collected from diabetes patients with an aim to predict disease progression and California housing dataset for which the target is the median house value for California districts.
As neither of these datasets have missing values, we will remove some
values to create new versions with artificially missing data. The performance
of
RandomForestRegressor on the full original dataset
is then compared the performance on the altered datasets with the artificially
missing values imputed using different techniques.
# Authors: The scikit-learn developers
# SPDX-License-Identifier: BSD-3-Clause
Download the data and make missing values sets#
First we download the two datasets. Diabetes dataset is shipped with scikit-learn. It has 442 entries, each with 10 features. California housing dataset is much larger with 20640 entries and 8 features. It needs to be downloaded. We will only use the first 300 entries for the sake of speeding up the calculations but feel free to use the whole dataset.
import numpy as np
from sklearn.datasets import fetch_california_housing, load_diabetes
X_diabetes, y_diabetes = load_diabetes(return_X_y=True)
X_california, y_california = fetch_california_housing(return_X_y=True)
X_diabetes = X_diabetes[:300]
y_diabetes = y_diabetes[:300]
X_california = X_california[:300]
y_california = y_california[:300]
def add_missing_values(X_full, y_full, rng):
n_samples, n_features = X_full.shape
# Add missing values in 75% of the lines
missing_rate = 0.75
n_missing_samples = int(n_samples * missing_rate)
missing_samples = np.zeros(n_samples, dtype=bool)
missing_samples[:n_missing_samples] = True
rng.shuffle(missing_samples)
missing_features = rng.randint(0, n_features, n_missing_samples)
X_missing = X_full.copy()
X_missing[missing_samples, missing_features] = np.nan
y_missing = y_full.copy()
return X_missing, y_missing
rng = np.random.RandomState(42)
X_miss_diabetes, y_miss_diabetes = add_missing_values(X_diabetes, y_diabetes, rng)
X_miss_california, y_miss_california = add_missing_values(
X_california, y_california, rng
)
Impute the missing data and score#
Now we will write a function which will score the results on the differently
imputed data, including the case of no imputation for full data.
We will use RandomForestRegressor for the target
regression.
from sklearn.ensemble import RandomForestRegressor
# To use the experimental IterativeImputer, we need to explicitly ask for it:
from sklearn.experimental import enable_iterative_imputer # noqa: F401
from sklearn.impute import IterativeImputer, KNNImputer, SimpleImputer
from sklearn.model_selection import cross_val_score
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import RobustScaler
N_SPLITS = 4
def get_score(X, y, imputer=None):
regressor = RandomForestRegressor(random_state=0)
if imputer is not None:
estimator = make_pipeline(imputer, regressor)
else:
estimator = regressor
scores = cross_val_score(
estimator, X, y, scoring="neg_mean_squared_error", cv=N_SPLITS
)
return scores.mean(), scores.std()
x_labels = []
mses_diabetes = np.zeros(5)
stds_diabetes = np.zeros(5)
mses_california = np.zeros(5)
stds_california = np.zeros(5)
Estimate the score#
First, we want to estimate the score on the original data:
mses_diabetes[0], stds_diabetes[0] = get_score(X_diabetes, y_diabetes)
mses_california[0], stds_california[0] = get_score(X_california, y_california)
x_labels.append("Full Data")
Replace missing values by 0#
Now we will estimate the score on the data where the missing values are replaced by 0:
imputer = SimpleImputer(strategy="constant", fill_value=0, add_indicator=True)
mses_diabetes[1], stds_diabetes[1] = get_score(
X_miss_diabetes, y_miss_diabetes, imputer
)
mses_california[1], stds_california[1] = get_score(
X_miss_california, y_miss_california, imputer
)
x_labels.append("Zero Imputation")
Impute missing values with mean#
imputer = SimpleImputer(strategy="mean", add_indicator=True)
mses_diabetes[2], stds_diabetes[2] = get_score(
X_miss_diabetes, y_miss_diabetes, imputer
)
mses_california[2], stds_california[2] = get_score(
X_miss_california, y_miss_california, imputer
)
x_labels.append("Mean Imputation")
kNN-imputation of the missing values#
KNNImputer imputes missing values using the weighted
or unweighted mean of the desired number of nearest neighbors. If your features
have vastly different scales (as in the California housing dataset),
consider re-scaling them to potentially improve performance.
imputer = KNNImputer(add_indicator=True)
mses_diabetes[3], stds_diabetes[3] = get_score(
X_miss_diabetes, y_miss_diabetes, imputer
)
mses_california[3], stds_california[3] = get_score(
X_miss_california, y_miss_california, make_pipeline(RobustScaler(), imputer)
)
x_labels.append("KNN Imputation")
Iterative imputation of the missing values#
Another option is the IterativeImputer. This uses
round-robin regression, modeling each feature with missing values as a
function of other features, in turn. We use the class’s default choice
of the regressor model (BayesianRidge)
to predict missing feature values. The performance of the predictor
may be negatively affected by vastly different scales of the features,
so we re-scale the features in the California housing dataset.
imputer = IterativeImputer(add_indicator=True)
mses_diabetes[4], stds_diabetes[4] = get_score(
X_miss_diabetes, y_miss_diabetes, imputer
)
mses_california[4], stds_california[4] = get_score(
X_miss_california, y_miss_california, make_pipeline(RobustScaler(), imputer)
)
x_labels.append("Iterative Imputation")
mses_diabetes = mses_diabetes * -1
mses_california = mses_california * -1
Plot the results#
Finally we are going to visualize the score:
import matplotlib.pyplot as plt
n_bars = len(mses_diabetes)
xval = np.arange(n_bars)
colors = ["r", "g", "b", "orange", "black"]
# plot diabetes results
plt.figure(figsize=(12, 6))
ax1 = plt.subplot(121)
for j in xval:
ax1.barh(
j,
mses_diabetes[j],
xerr=stds_diabetes[j],
color=colors[j],
alpha=0.6,
align="center",
)
ax1.set_title("Imputation Techniques with Diabetes Data")
ax1.set_xlim(left=np.min(mses_diabetes) * 0.9, right=np.max(mses_diabetes) * 1.1)
ax1.set_yticks(xval)
ax1.set_xlabel("MSE")
ax1.invert_yaxis()
ax1.set_yticklabels(x_labels)
# plot california dataset results
ax2 = plt.subplot(122)
for j in xval:
ax2.barh(
j,
mses_california[j],
xerr=stds_california[j],
color=colors[j],
alpha=0.6,
align="center",
)
ax2.set_title("Imputation Techniques with California Data")
ax2.set_yticks(xval)
ax2.set_xlabel("MSE")
ax2.invert_yaxis()
ax2.set_yticklabels([""] * n_bars)
plt.show()
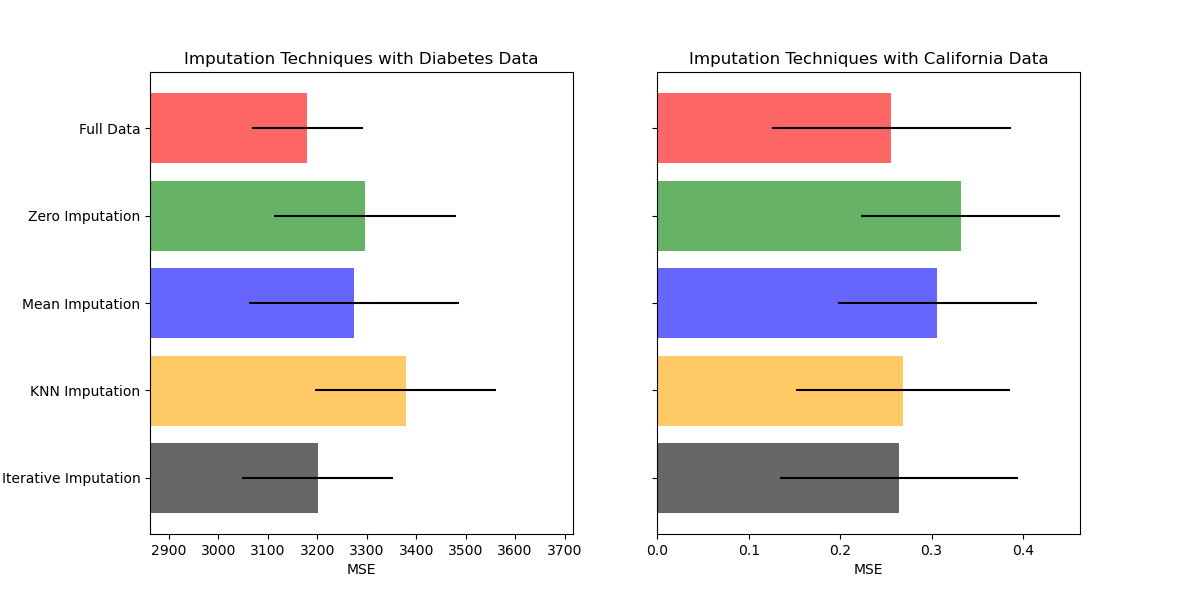
You can also try different techniques. For instance, the median is a more robust estimator for data with high magnitude variables which could dominate results (otherwise known as a ‘long tail’).
Total running time of the script: (0 minutes 8.145 seconds)
Related examples
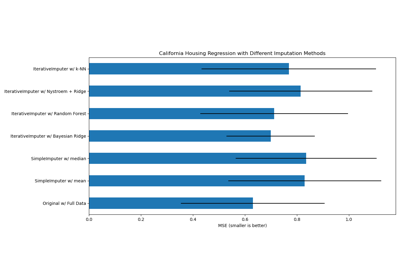
Imputing missing values with variants of IterativeImputer
Concentration Prior Type Analysis of Variation Bayesian Gaussian Mixture