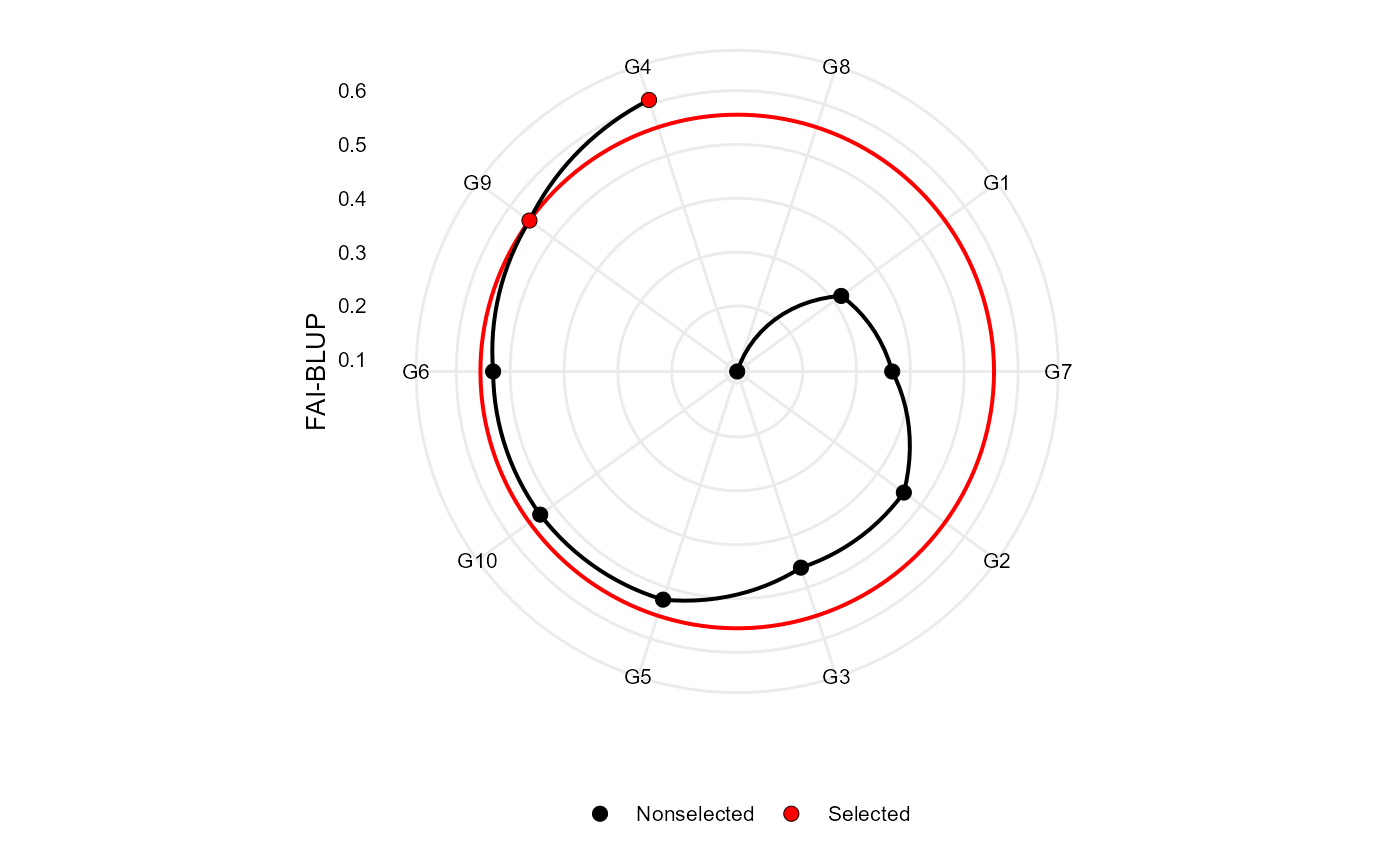
Plot the multitrait index based on factor analysis and ideotype-design proposed by Rocha et al. (2018).
Usage
# S3 method for fai_blup
plot(
x,
ideotype = 1,
SI = 15,
radar = TRUE,
arrange.label = FALSE,
size.point = 2.5,
size.line = 0.7,
size.text = 10,
col.sel = "red",
col.nonsel = "black",
...
)Arguments
- x
An object of class
waasb- ideotype
The ideotype to be plotted. Default is 1.
- SI
An integer (0-100). The selection intensity in percentage of the total number of genotypes.
- radar
Logical argument. If true (default) a radar plot is generated after using
coord_polar().- arrange.label
Logical argument. If
TRUE, the labels are arranged to avoid text overlapping. This becomes useful when the number of genotypes is large, say, more than 30.- size.point
The size of the point in graphic. Defaults to 2.5.
- size.line
The size of the line in graphic. Defaults to 0.7.
- size.text
The size for the text in the plot. Defaults to 10.
- col.sel
The colour for selected genotypes. Defaults to
"red".- col.nonsel
The colour for nonselected genotypes. Defaults to
"black".- ...
Other arguments to be passed from ggplot2::theme().
References
Rocha, J.R.A.S.C.R, J.C. Machado, and P.C.S. Carneiro. 2018. Multitrait index based on factor analysis and ideotype-design: proposal and application on elephant grass breeding for bioenergy. GCB Bioenergy 10:52-60. doi:10.1111/gcbb.12443
Author
Tiago Olivoto tiagoolivoto@gmail.com
Examples
# \donttest{
library(metan)
mod <- waasb(data_ge,
env = ENV,
gen = GEN,
rep = REP,
resp = c(GY, HM))
#> Evaluating trait GY |====================== | 50% 00:00:02
Evaluating trait HM |============================================| 100% 00:00:05
#> Method: REML/BLUP
#> Random effects: GEN, GEN:ENV
#> Fixed effects: ENV, REP(ENV)
#> Denominador DF: Satterthwaite's method
#> ---------------------------------------------------------------------------
#> P-values for Likelihood Ratio Test of the analyzed traits
#> ---------------------------------------------------------------------------
#> model GY HM
#> COMPLETE NA NA
#> GEN 1.11e-05 5.07e-03
#> GEN:ENV 2.15e-11 2.27e-15
#> ---------------------------------------------------------------------------
#> All variables with significant (p < 0.05) genotype-vs-environment interaction
FAI <- fai_blup(mod,
DI = c('max, max'),
UI = c('min, min'))
#>
#> -----------------------------------------------------------------------------------
#> Principal Component Analysis
#> -----------------------------------------------------------------------------------
#> eigen.values cumulative.var
#> PC1 1.1 55.23
#> PC2 0.9 100.00
#>
#> -----------------------------------------------------------------------------------
#> Factor Analysis
#> -----------------------------------------------------------------------------------
#> FA1 comunalits
#> GY -0.74 0.55
#> HM 0.74 0.55
#>
#> -----------------------------------------------------------------------------------
#> Comunalit Mean: 0.5523038
#> Selection differential
#> -----------------------------------------------------------------------------------
#> VAR Factor Xo Xs SD SDperc sense goal
#> 1 GY 1 2.674242 2.594199 -0.08004274 -2.9931005 increase 0
#> 2 HM 1 48.088286 48.005568 -0.08271774 -0.1720122 increase 0
#>
#> -----------------------------------------------------------------------------------
#> Selected genotypes
#> G4 G9
#> -----------------------------------------------------------------------------------
plot(FAI)
 # }
# }
