Chapter 3 Exporting tree with data
3.1 Introduction
The treeio package supports parsing
various phylogenetic tree file formats including software outputs that contain
evolutionary evidences. Some of the formats are just log file
(e.g. PAML
and r8s outputs), while some of the others are
non-standard formats (e.g. BEAST
and MrBayes outputs that introduce square
bracket, which was reserved to store comment in standard Nexus format, to store
inferences). With treeio, we are
now able to parse these files to extract phylogenetic tree and map associated
data on the tree structure. Exporting tree structure is easy, users can use
as.phyo method defined treeio to
convert treedata object to phylo object then using write.tree or
write.nexus implemented
in ape package
(Paradis, Claude, and Strimmer 2004) to export the tree structure as Newick text or Nexus file.
This is quite useful for converting non-standard formats to standard format and
for extracting tree from software outputs, such as log file.
However, exporting tree with associated data is still challenging. These associated data can be parsed from analysis programs or obtained from external sources (e.g. phenotypic data, experimental data and clinical data). The major obstacle here is that there is no standard format that designed for storing tree with data. NeXML (Vos et al. 2012) maybe the most flexible format, however it is currently not widely supported. Most of the analysis programs in this field rely extensively on Newick string and Nexus format. In my opinion, although BEAST Nexus format8 may not be the best solution, it is currently a good approach for storing heterogeneous associated data. The beauty of the format is that all the annotate elements are stored within square bracket, which is reserved for comments. So that the file can be parsed as standard Nexus by ignoring annotate elements and existing programs should be able to read them.
3.2 Exporting Tree Data to BEAST Nexus Format
3.2.1 Exporting/converting software output
The treeio package provides
write.beast to export treedata object as BEAST Nexus file (Bouckaert et al. 2014).
With treeio, it is easy to convert
software output to BEAST format if the output can be parsed
by treeio (see Chapter 1). For example, we can
convert NHX file to BEAST file and use NHX tags to color the tree using
FigTree9 (Figure 3.1A) or convert CODEML output and use
dN/dS, dN or dS to color the tree in FigTree (Figure 3.1B).
Here is an example of converting NHX file to BEAST format:
nhxfile <- system.file("extdata/NHX", "phyldog.nhx", package="treeio")
nhx <- read.nhx(nhxfile)
# write.beast(nhx, file = "phyldog.tree")
write.beast(nhx)#NEXUS
[R-package treeio, Wed May 13 18:07:19 2020]
BEGIN TAXA;
DIMENSIONS NTAX = 16;
TAXLABELS
Prayidae_D27SS7@2825365
Kephyes_ovata@2606431
Chuniphyes_multidentata@1277217
Apolemia_sp_@1353964
Bargmannia_amoena@263997
Bargmannia_elongata@946788
Physonect_sp_@2066767
Stephalia_dilata@2960089
Frillagalma_vityazi@1155031
Resomia_ornicephala@3111757
Lychnagalma_utricularia@2253871
Nanomia_bijuga@717864
Cordagalma_sp_@1525873
Rhizophysa_filiformis@3073669
Hydra_magnipapillata@52244
Ectopleura_larynx@3556167
;
END;
BEGIN TREES;
TRANSLATE
1 Prayidae_D27SS7@2825365,
2 Kephyes_ovata@2606431,
3 Chuniphyes_multidentata@1277217,
4 Apolemia_sp_@1353964,
5 Bargmannia_amoena@263997,
6 Bargmannia_elongata@946788,
7 Physonect_sp_@2066767,
8 Stephalia_dilata@2960089,
9 Frillagalma_vityazi@1155031,
10 Resomia_ornicephala@3111757,
11 Lychnagalma_utricularia@2253871,
12 Nanomia_bijuga@717864,
13 Cordagalma_sp_@1525873,
14 Rhizophysa_filiformis@3073669,
15 Hydra_magnipapillata@52244,
16 Ectopleura_larynx@3556167
;
TREE * UNTITLED = [&R] (((1[&Ev=S,S=58,ND=0]:0.0682841,(2[&Ev=S,S=69,ND=1]:0.0193941,3[&Ev=S,S=70,ND=2]:0.0121378)[&Ev=S,S=60,ND=3]:0.0217782)[&Ev=S,S=36,ND=4]:0.0607598,((4[&Ev=S,S=31,ND=9]:0.11832,(((5[&Ev=S,S=37,ND=10]:0.0144549,6[&Ev=S,S=38,ND=11]:0.0149723)[&Ev=S,S=33,ND=12]:0.0925388,7[&Ev=S,S=61,ND=13]:0.077429)[&Ev=S,S=24,ND=14]:0.0274637,(8[&Ev=S,S=52,ND=15]:0.0761163,((9[&Ev=S,S=53,ND=16]:0.0906068,10[&Ev=S,S=54,ND=17]:1e-06)[&Ev=S,S=45,ND=18]:1e-06,((11[&Ev=S,S=65,ND=19]:0.120851,12[&Ev=S,S=71,ND=20]:0.133939)[&Ev=S,S=56,ND=21]:1e-06,13[&Ev=S,S=64,ND=22]:0.0693814)[&Ev=S,S=46,ND=23]:1e-06)[&Ev=S,S=40,ND=24]:0.0333823)[&Ev=S,S=35,ND=25]:1e-06)[&Ev=D,S=24,ND=26]:0.0431861)[&Ev=S,S=19,ND=27]:1e-06,14[&Ev=S,S=26,ND=28]:0.22283)[&Ev=S,S=17,ND=29]:0.0292362)[&Ev=D,S=17,ND=8]:0.185603,(15[&Ev=S,S=16,ND=5]:0.0621782,16[&Ev=S,S=15,ND=6]:0.332505)[&Ev=S,S=12,ND=7]:0.185603)[&Ev=S,S=9,ND=30];
END;Another example of converting CodeML output to BEAST format:
mlcfile <- system.file("extdata/PAML_Codeml", "mlc", package="treeio")
ml <- read.codeml_mlc(mlcfile)
# write.beast(ml, file = "codeml.tree")
write.beast(ml)#NEXUS
[R-package treeio, Wed May 13 18:07:19 2020]
BEGIN TAXA;
DIMENSIONS NTAX = 15;
TAXLABELS
A
B
C
D
E
F
G
H
I
J
K
L
M
N
O
;
END;
BEGIN TREES;
TRANSLATE
1 A,
2 B,
3 C,
4 D,
5 E,
6 F,
7 G,
8 H,
9 I,
10 J,
11 K,
12 L,
13 M,
14 N,
15 O
;
TREE * UNTITLED = [&U] (11[&t=0.082,N=1514.9,S=633.1,dN_vs_dS=0.0224,dN=0.002,dS=0.0878,N_x_dN=3,S_x_dS=55.6]:0.081785,14[&t=0.062,N=1514.9,S=633.1,dN_vs_dS=0.0095,dN=7e-04,dS=0.0689,N_x_dN=1,S_x_dS=43.6]:0.062341,(4[&t=0.082,N=1514.9,S=633.1,dN_vs_dS=0.0385,dN=0.0033,dS=0.0849,N_x_dN=5,S_x_dS=53.8]:0.082021,(12[&t=0.006,N=1514.9,S=633.1,dN_vs_dS=1e-04,dN=0,dS=0.0062,N_x_dN=0,S_x_dS=3.9]:0.005508,(10[&t=0.014,N=1514.9,S=633.1,dN_vs_dS=0.0457,dN=7e-04,dS=0.0143,N_x_dN=1,S_x_dS=9]:0.013996,(7[&t=0.046,N=1514.9,S=633.1,dN_vs_dS=0.1621,dN=0.006,dS=0.0373,N_x_dN=9.2,S_x_dS=23.6]:0.045746,((3[&t=0.028,N=1514.9,S=633.1,dN_vs_dS=0.0461,dN=0.0013,dS=0.0282,N_x_dN=2,S_x_dS=17.9]:0.02773,(5[&t=0.031,N=1514.9,S=633.1,dN_vs_dS=0.0641,dN=0.002,dS=0.0305,N_x_dN=3,S_x_dS=19.3]:0.031104,15[&t=0.048,N=1514.9,S=633.1,dN_vs_dS=0.0538,dN=0.0026,dS=0.0485,N_x_dN=4,S_x_dS=30.7]:0.048389)23[&t=0.008,N=1514.9,S=633.1,dN_vs_dS=1e-04,dN=0,dS=0.0094,N_x_dN=0,S_x_dS=6]:0.008328)22[&t=0.016,N=1514.9,S=633.1,dN_vs_dS=0.0395,dN=7e-04,dS=0.0165,N_x_dN=1,S_x_dS=10.4]:0.015959,(8[&t=0.021,N=1514.9,S=633.1,dN_vs_dS=0.1028,dN=0.002,dS=0.0191,N_x_dN=3,S_x_dS=12.1]:0.021007,(9[&t=0.015,N=1514.9,S=633.1,dN_vs_dS=1e-04,dN=0,dS=0.0167,N_x_dN=0,S_x_dS=10.6]:0.014739,(2[&t=0.032,N=1514.9,S=633.1,dN_vs_dS=1e-04,dN=0,dS=0.0358,N_x_dN=0,S_x_dS=22.7]:0.031643,(1[&t=0.01,N=1514.9,S=633.1,dN_vs_dS=0.0646,dN=7e-04,dS=0.0101,N_x_dN=1,S_x_dS=6.4]:0.01034,(6[&t=0.007,N=1514.9,S=633.1,dN_vs_dS=0.298,dN=0.0013,dS=0.0044,N_x_dN=2,S_x_dS=2.8]:0.006649,13[&t=0.009,N=1514.9,S=633.1,dN_vs_dS=0.0738,dN=7e-04,dS=0.0088,N_x_dN=1,S_x_dS=5.6]:0.009195)28[&t=0.028,N=1514.9,S=633.1,dN_vs_dS=0.0453,dN=0.0013,dS=0.0289,N_x_dN=2,S_x_dS=18.3]:0.028303)27[&t=0.008,N=1514.9,S=633.1,dN_vs_dS=0.0863,dN=7e-04,dS=0.0076,N_x_dN=1,S_x_dS=4.8]:0.008072)26[&t=0.003,N=1514.9,S=633.1,dN_vs_dS=1.5591,dN=0.0013,dS=8e-04,N_x_dN=2,S_x_dS=0.5]:0.0035)25[&t=0.02,N=1514.9,S=633.1,dN_vs_dS=1e-04,dN=0,dS=0.023,N_x_dN=0,S_x_dS=14.6]:0.020359)24[&t=0.001,N=1514.9,S=633.1,dN_vs_dS=1e-04,dN=0,dS=6e-04,N_x_dN=0,S_x_dS=0.4]:0.000555)21[&t=0.024,N=1514.9,S=633.1,dN_vs_dS=0.0549,dN=0.0013,dS=0.0237,N_x_dN=2,S_x_dS=15]:0.023675)20[&t=0.046,N=1514.9,S=633.1,dN_vs_dS=0.0419,dN=0.002,dS=0.047,N_x_dN=3,S_x_dS=29.8]:0.045745)19[&t=0.015,N=1514.9,S=633.1,dN_vs_dS=1e-04,dN=0,dS=0.0166,N_x_dN=0,S_x_dS=10.5]:0.014684)18[&t=0.059,N=1514.9,S=633.1,dN_vs_dS=0.0964,dN=0.0053,dS=0.0545,N_x_dN=8,S_x_dS=34.5]:0.059308)17[&t=0.232,N=1514.9,S=633.1,dN_vs_dS=0.0129,dN=0.0033,dS=0.2541,N_x_dN=5,S_x_dS=160.9]:0.231628)16;
END;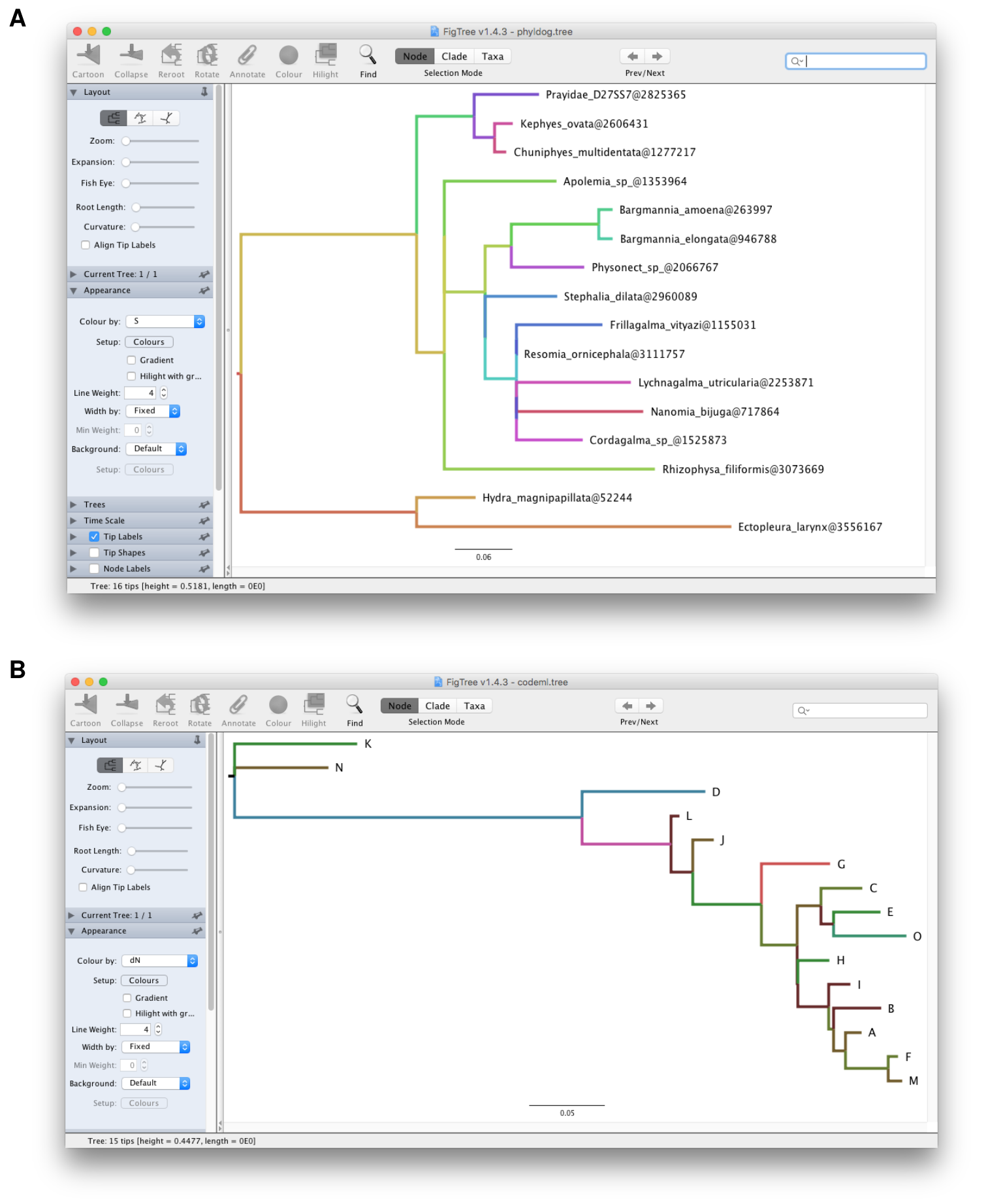
Figure 3.1: Visualizing BEAST file in FigTree. Directly visualizing NHX file (A) and CodeML output (B) in FigTree is not supported. treeio can convert these files to BEAST compatible NEXUS format which can be directly opened in FigTree and visualized annotated data.
3.2.2 Combining tree with external data
Using the utilities provided
by tidytree and treeio, it is easy to link
external data onto the corresponding phylogeny. The write.beast function enable users
to export the tree with external data to a single tree file.
phylo <- as.phylo(nhx)
## print the newick text
write.tree(phylo)[1] "(((Prayidae_D27SS7@2825365:0.0682841,(Kephyes_ovata@2606431:0.0193941,Chuniphyes_multidentata@1277217:0.0121378):0.0217782):0.0607598,((Apolemia_sp_@1353964:0.11832,(((Bargmannia_amoena@263997:0.0144549,Bargmannia_elongata@946788:0.0149723):0.0925388,Physonect_sp_@2066767:0.077429):0.0274637,(Stephalia_dilata@2960089:0.0761163,((Frillagalma_vityazi@1155031:0.0906068,Resomia_ornicephala@3111757:1e-06):1e-06,((Lychnagalma_utricularia@2253871:0.120851,Nanomia_bijuga@717864:0.133939):1e-06,Cordagalma_sp_@1525873:0.0693814):1e-06):0.0333823):1e-06):0.0431861):1e-06,Rhizophysa_filiformis@3073669:0.22283):0.0292362):0.185603,(Hydra_magnipapillata@52244:0.0621782,Ectopleura_larynx@3556167:0.332505):0.185603);"N <- Nnode2(phylo)
fake_data <- tibble(node = 1:N, fake_trait = rnorm(N), another_trait = runif(N))
fake_tree <- full_join(phylo, fake_data, by = "node")
write.beast(fake_tree)#NEXUS
[R-package treeio, Wed May 13 18:07:21 2020]
BEGIN TAXA;
DIMENSIONS NTAX = 16;
TAXLABELS
Prayidae_D27SS7@2825365
Kephyes_ovata@2606431
Chuniphyes_multidentata@1277217
Apolemia_sp_@1353964
Bargmannia_amoena@263997
Bargmannia_elongata@946788
Physonect_sp_@2066767
Stephalia_dilata@2960089
Frillagalma_vityazi@1155031
Resomia_ornicephala@3111757
Lychnagalma_utricularia@2253871
Nanomia_bijuga@717864
Cordagalma_sp_@1525873
Rhizophysa_filiformis@3073669
Hydra_magnipapillata@52244
Ectopleura_larynx@3556167
;
END;
BEGIN TREES;
TRANSLATE
1 Prayidae_D27SS7@2825365,
2 Kephyes_ovata@2606431,
3 Chuniphyes_multidentata@1277217,
4 Apolemia_sp_@1353964,
5 Bargmannia_amoena@263997,
6 Bargmannia_elongata@946788,
7 Physonect_sp_@2066767,
8 Stephalia_dilata@2960089,
9 Frillagalma_vityazi@1155031,
10 Resomia_ornicephala@3111757,
11 Lychnagalma_utricularia@2253871,
12 Nanomia_bijuga@717864,
13 Cordagalma_sp_@1525873,
14 Rhizophysa_filiformis@3073669,
15 Hydra_magnipapillata@52244,
16 Ectopleura_larynx@3556167
;
TREE * UNTITLED = [&R] (((1[&fake_trait=-0.118140158479532,another_trait=0.358319966588169]:0.0682841,(2[&fake_trait=-1.02308336346315,another_trait=0.178163790609688]:0.0193941,3[&fake_trait=0.103599672229184,another_trait=0.93473564600572]:0.0121378)[&fake_trait=-0.59581303574252,another_trait=0.922105083474889]:0.0217782)[&fake_trait=-0.300012313113108,another_trait=0.119739749003202]:0.0607598,(14[&fake_trait=0.589053316158453,another_trait=0.565819646697491]:0.22283,(4[&fake_trait=-0.765460180119788,another_trait=0.888429019600153]:0.11832,((7[&fake_trait=-0.306908277375037,another_trait=0.650574740953743]:0.077429,(5[&fake_trait=-0.357587376956114,another_trait=0.547185462666675]:0.0144549,6[&fake_trait=0.582576884787801,another_trait=0.00538489152677357]:0.0149723)[&fake_trait=1.68690137848482,another_trait=0.634304485516623]:0.0925388)[&fake_trait=1.00444824814337,another_trait=0.420116872293875]:0.0274637,(8[&fake_trait=1.10912385375853,another_trait=0.583255837904289]:0.0761163,((9[&fake_trait=-0.707380205642954,another_trait=0.808900895994157]:0.0906068,10[&fake_trait=0.838100693609065,another_trait=0.0142157829832286]:1e-06)[&fake_trait=-0.509864481867074,another_trait=0.939013464609161]:1e-06,(13[&fake_trait=1.56583127248603,another_trait=0.0454649559687823]:0.0693814,(11[&fake_trait=1.45882763141046,another_trait=0.516774294432253]:0.120851,12[&fake_trait=2.43471798636817,another_trait=0.00467536062933505]:0.133939)[&fake_trait=-1.07656365665571,another_trait=0.518969761673361]:1e-06)[&fake_trait=-1.48426105861817,another_trait=0.044428545050323]:1e-06)[&fake_trait=0.409198668366587,another_trait=0.538205695804209]:0.0333823)[&fake_trait=-0.692527146833782,another_trait=0.58515489147976]:1e-06)[&fake_trait=-1.41216094329287,another_trait=0.935482121771201]:0.0431861)[&fake_trait=1.65980403458888,another_trait=0.125082567567006]:1e-06)[&fake_trait=0.476325081878056,another_trait=0.262554442277178]:0.0292362)[&fake_trait=1.15832764983724,another_trait=0.646949481917545]:0.185603,(15[&fake_trait=0.704447323258862,another_trait=0.663062628591433]:0.0621782,16[&fake_trait=-0.659190065346089,another_trait=0.505218442762271]:0.332505)[&fake_trait=-1.04282102019802,another_trait=0.641324875177816]:0.185603)[&fake_trait=-0.479609047683268,another_trait=0.519890103489161];
END;After merging, the fake_trait and another_trait stored in fake_data will be linked to the tree, phylo, and store in the treedata object, the fake_tree. The write.beast() function export the tree with associated data to a single BEAST format file. The associated data can be used to visualized the tree using ggtree (Figure 5.7) or FigTree (Figure 3.1).
3.2.3 Merging tree data from different sources
Not only Newick tree text can be combined with associated data, but also tree data obtained from software output can be combined with external data, as well as different tree objects can be merged together (for details, see Chapter 2).
## combine tree object with data
tree_with_data <- full_join(nhx, fake_data, by = "node")
tree_with_data## 'treedata' S4 object that stored information of
## '/home/ygc/R/library/treeio/extdata/NHX/phyldog.nhx'.
##
## ...@ phylo:
## Phylogenetic tree with 16 tips and 15 internal nodes.
##
## Tip labels:
## Prayidae_D27SS7@2825365, Kephyes_ovata@2606431, Chuniphyes_multidentata@1277217, Apolemia_sp_@1353964, Bargmannia_amoena@263997, Bargmannia_elongata@946788, ...
##
## Rooted; includes branch lengths.
##
## with the following features available:
## 'Ev', 'S', 'ND', 'fake_trait', 'another_trait'.## merge two tree object
tree2 <- merge_tree(nhx, fake_tree)
tree2## 'treedata' S4 object that stored information of
## '/home/ygc/R/library/treeio/extdata/NHX/phyldog.nhx'.
##
## ...@ phylo:
## Phylogenetic tree with 16 tips and 15 internal nodes.
##
## Tip labels:
## Prayidae_D27SS7@2825365, Kephyes_ovata@2606431, Chuniphyes_multidentata@1277217, Apolemia_sp_@1353964, Bargmannia_amoena@263997, Bargmannia_elongata@946788, ...
##
## Rooted; includes branch lengths.
##
## with the following features available:
## 'Ev', 'S', 'ND', 'fake_trait', 'another_trait'.identical(tree_with_data, tree2)## [1] TRUEAfter merging data from different sources, the tree with the associated data can be exported into a single file.
write.beast(tree2)#NEXUS
[R-package treeio, Wed May 13 18:07:22 2020]
BEGIN TAXA;
DIMENSIONS NTAX = 16;
TAXLABELS
Prayidae_D27SS7@2825365
Kephyes_ovata@2606431
Chuniphyes_multidentata@1277217
Apolemia_sp_@1353964
Bargmannia_amoena@263997
Bargmannia_elongata@946788
Physonect_sp_@2066767
Stephalia_dilata@2960089
Frillagalma_vityazi@1155031
Resomia_ornicephala@3111757
Lychnagalma_utricularia@2253871
Nanomia_bijuga@717864
Cordagalma_sp_@1525873
Rhizophysa_filiformis@3073669
Hydra_magnipapillata@52244
Ectopleura_larynx@3556167
;
END;
BEGIN TREES;
TRANSLATE
1 Prayidae_D27SS7@2825365,
2 Kephyes_ovata@2606431,
3 Chuniphyes_multidentata@1277217,
4 Apolemia_sp_@1353964,
5 Bargmannia_amoena@263997,
6 Bargmannia_elongata@946788,
7 Physonect_sp_@2066767,
8 Stephalia_dilata@2960089,
9 Frillagalma_vityazi@1155031,
10 Resomia_ornicephala@3111757,
11 Lychnagalma_utricularia@2253871,
12 Nanomia_bijuga@717864,
13 Cordagalma_sp_@1525873,
14 Rhizophysa_filiformis@3073669,
15 Hydra_magnipapillata@52244,
16 Ectopleura_larynx@3556167
;
TREE * UNTITLED = [&R] (((1[&Ev=S,S=58,ND=0,fake_trait=-0.118140158479532,another_trait=0.358319966588169]:0.0682841,(2[&Ev=S,S=69,ND=1,fake_trait=-1.02308336346315,another_trait=0.178163790609688]:0.0193941,3[&Ev=S,S=70,ND=2,fake_trait=0.103599672229184,another_trait=0.93473564600572]:0.0121378)[&Ev=S,S=60,ND=3,fake_trait=-0.59581303574252,another_trait=0.922105083474889]:0.0217782)[&Ev=S,S=36,ND=4,fake_trait=-0.300012313113108,another_trait=0.119739749003202]:0.0607598,((4[&Ev=S,S=31,ND=9,fake_trait=-0.765460180119788,another_trait=0.888429019600153]:0.11832,(((5[&Ev=S,S=37,ND=10,fake_trait=-0.357587376956114,another_trait=0.547185462666675]:0.0144549,6[&Ev=S,S=38,ND=11,fake_trait=0.582576884787801,another_trait=0.00538489152677357]:0.0149723)[&Ev=S,S=33,ND=12,fake_trait=1.68690137848482,another_trait=0.634304485516623]:0.0925388,7[&Ev=S,S=61,ND=13,fake_trait=-0.306908277375037,another_trait=0.650574740953743]:0.077429)[&Ev=S,S=24,ND=14,fake_trait=1.00444824814337,another_trait=0.420116872293875]:0.0274637,(8[&Ev=S,S=52,ND=15,fake_trait=1.10912385375853,another_trait=0.583255837904289]:0.0761163,((9[&Ev=S,S=53,ND=16,fake_trait=-0.707380205642954,another_trait=0.808900895994157]:0.0906068,10[&Ev=S,S=54,ND=17,fake_trait=0.838100693609065,another_trait=0.0142157829832286]:1e-06)[&Ev=S,S=45,ND=18,fake_trait=-0.509864481867074,another_trait=0.939013464609161]:1e-06,((11[&Ev=S,S=65,ND=19,fake_trait=1.45882763141046,another_trait=0.516774294432253]:0.120851,12[&Ev=S,S=71,ND=20,fake_trait=2.43471798636817,another_trait=0.00467536062933505]:0.133939)[&Ev=S,S=56,ND=21,fake_trait=-1.07656365665571,another_trait=0.518969761673361]:1e-06,13[&Ev=S,S=64,ND=22,fake_trait=1.56583127248603,another_trait=0.0454649559687823]:0.0693814)[&Ev=S,S=46,ND=23,fake_trait=-1.48426105861817,another_trait=0.044428545050323]:1e-06)[&Ev=S,S=40,ND=24,fake_trait=0.409198668366587,another_trait=0.538205695804209]:0.0333823)[&Ev=S,S=35,ND=25,fake_trait=-0.692527146833782,another_trait=0.58515489147976]:1e-06)[&Ev=D,S=24,ND=26,fake_trait=-1.41216094329287,another_trait=0.935482121771201]:0.0431861)[&Ev=S,S=19,ND=27,fake_trait=1.65980403458888,another_trait=0.125082567567006]:1e-06,14[&Ev=S,S=26,ND=28,fake_trait=0.589053316158453,another_trait=0.565819646697491]:0.22283)[&Ev=S,S=17,ND=29,fake_trait=0.476325081878056,another_trait=0.262554442277178]:0.0292362)[&Ev=D,S=17,ND=8,fake_trait=1.15832764983724,another_trait=0.646949481917545]:0.185603,(15[&Ev=S,S=16,ND=5,fake_trait=0.704447323258862,another_trait=0.663062628591433]:0.0621782,16[&Ev=S,S=15,ND=6,fake_trait=-0.659190065346089,another_trait=0.505218442762271]:0.332505)[&Ev=S,S=12,ND=7,fake_trait=-1.04282102019802,another_trait=0.641324875177816]:0.185603)[&Ev=S,S=9,ND=30,fake_trait=-0.479609047683268,another_trait=0.519890103489161];
END;The output BEAST Nexus file can be imported into R using the read.beast
function and all the associated data can be used to annotate the tree
using ggtree (Yu et al. 2017).
outfile <- tempfile(fileext = ".tree")
write.beast(tree2, file = outfile)
read.beast(outfile)## 'treedata' S4 object that stored information of
## '/tmp/RtmpvGnYhp/file55a55620d472e.tree'.
##
## ...@ phylo:
## Phylogenetic tree with 16 tips and 15 internal nodes.
##
## Tip labels:
## Prayidae_D27SS7@2825365, Kephyes_ovata@2606431, Chuniphyes_multidentata@1277217, Apolemia_sp_@1353964, Bargmannia_amoena@263997, Bargmannia_elongata@946788, ...
##
## Rooted; includes branch lengths.
##
## with the following features available:
## 'another_trait', 'Ev', 'fake_trait', 'ND', 'S'.3.3 Exporting Tree Data to jtree Format
The treeio package provides
write.beast to export treedata to BEAST Nexus file. This is quite useful
to convert file format, combine tree with data and merging tree data from
different sources as we demonstrated in
session 3.2.
The treeio package also supplies
read.beast function to parse output file of write.beast. Although
with treeio, the R community has the ability to
manipulate BEAST Nexus format and process tree data, there is still lacking
library/package for parsing BEAST file in other programming language.
JSON (JavaScript Object Notation) is a lightweight data-interchange format and widely supported in almost all modern programming languages. To make it easy to import tree with data in other programming languages, treeio supports exporting tree with data in jtree format, which is JSON-based and can be easy to parse using any languages that supports JSON.
write.jtree(tree2){
"tree": "(((Prayidae_D27SS7@2825365:0.0682841{1},(Kephyes_ovata@2606431:0.0193941{2},Chuniphyes_multidentata@1277217:0.0121378{3}):0.0217782{20}):0.0607598{19},((Apolemia_sp_@1353964:0.11832{4},(((Bargmannia_amoena@263997:0.0144549{5},Bargmannia_elongata@946788:0.0149723{6}):0.0925388{25},Physonect_sp_@2066767:0.077429{7}):0.0274637{24},(Stephalia_dilata@2960089:0.0761163{8},((Frillagalma_vityazi@1155031:0.0906068{9},Resomia_ornicephala@3111757:1{10}e-06):1{28}e-06,((Lychnagalma_utricularia@2253871:0.120851{11},Nanomia_bijuga@717864:0.133939{12}):1{30}e-06,Cordagalma_sp_@1525873:0.0693814{13}):1{29}e-06):0.0333823{27}):1{26}e-06):0.0431861{23}):1{22}e-06,Rhizophysa_filiformis@3073669:0.22283{14}):0.0292362{21}):0.185603{18},(Hydra_magnipapillata@52244:0.0621782{15},Ectopleura_larynx@3556167:0.332505{16}):0.185603{31}){17};",
"data":[
{
"edge_num": 1,
"Ev": "S",
"S": "58",
"ND": 0,
"fake_trait": -0.1181,
"another_trait": 0.3583
},
{
"edge_num": 2,
"Ev": "S",
"S": "69",
"ND": 1,
"fake_trait": -1.0231,
"another_trait": 0.1782
},
{
"edge_num": 3,
"Ev": "S",
"S": "70",
"ND": 2,
"fake_trait": 0.1036,
"another_trait": 0.9347
},
{
"edge_num": 4,
"Ev": "S",
"S": "31",
"ND": 9,
"fake_trait": -0.7655,
"another_trait": 0.8884
},
{
"edge_num": 5,
"Ev": "S",
"S": "37",
"ND": 10,
"fake_trait": -0.3576,
"another_trait": 0.5472
},
{
"edge_num": 6,
"Ev": "S",
"S": "38",
"ND": 11,
"fake_trait": 0.5826,
"another_trait": 0.0054
},
{
"edge_num": 7,
"Ev": "S",
"S": "61",
"ND": 13,
"fake_trait": -0.3069,
"another_trait": 0.6506
},
{
"edge_num": 8,
"Ev": "S",
"S": "52",
"ND": 15,
"fake_trait": 1.1091,
"another_trait": 0.5833
},
{
"edge_num": 9,
"Ev": "S",
"S": "53",
"ND": 16,
"fake_trait": -0.7074,
"another_trait": 0.8089
},
{
"edge_num": 10,
"Ev": "S",
"S": "54",
"ND": 17,
"fake_trait": 0.8381,
"another_trait": 0.0142
},
{
"edge_num": 11,
"Ev": "S",
"S": "65",
"ND": 19,
"fake_trait": 1.4588,
"another_trait": 0.5168
},
{
"edge_num": 12,
"Ev": "S",
"S": "71",
"ND": 20,
"fake_trait": 2.4347,
"another_trait": 0.0047
},
{
"edge_num": 13,
"Ev": "S",
"S": "64",
"ND": 22,
"fake_trait": 1.5658,
"another_trait": 0.0455
},
{
"edge_num": 14,
"Ev": "S",
"S": "26",
"ND": 28,
"fake_trait": 0.5891,
"another_trait": 0.5658
},
{
"edge_num": 15,
"Ev": "S",
"S": "16",
"ND": 5,
"fake_trait": 0.7044,
"another_trait": 0.6631
},
{
"edge_num": 16,
"Ev": "S",
"S": "15",
"ND": 6,
"fake_trait": -0.6592,
"another_trait": 0.5052
},
{
"edge_num": 17,
"Ev": "S",
"S": "9",
"ND": 30,
"fake_trait": -0.4796,
"another_trait": 0.5199
},
{
"edge_num": 18,
"Ev": "D",
"S": "17",
"ND": 8,
"fake_trait": 1.1583,
"another_trait": 0.6469
},
{
"edge_num": 19,
"Ev": "S",
"S": "36",
"ND": 4,
"fake_trait": -0.3,
"another_trait": 0.1197
},
{
"edge_num": 20,
"Ev": "S",
"S": "60",
"ND": 3,
"fake_trait": -0.5958,
"another_trait": 0.9221
},
{
"edge_num": 21,
"Ev": "S",
"S": "17",
"ND": 29,
"fake_trait": 0.4763,
"another_trait": 0.2626
},
{
"edge_num": 22,
"Ev": "S",
"S": "19",
"ND": 27,
"fake_trait": 1.6598,
"another_trait": 0.1251
},
{
"edge_num": 23,
"Ev": "D",
"S": "24",
"ND": 26,
"fake_trait": -1.4122,
"another_trait": 0.9355
},
{
"edge_num": 24,
"Ev": "S",
"S": "24",
"ND": 14,
"fake_trait": 1.0044,
"another_trait": 0.4201
},
{
"edge_num": 25,
"Ev": "S",
"S": "33",
"ND": 12,
"fake_trait": 1.6869,
"another_trait": 0.6343
},
{
"edge_num": 26,
"Ev": "S",
"S": "35",
"ND": 25,
"fake_trait": -0.6925,
"another_trait": 0.5852
},
{
"edge_num": 27,
"Ev": "S",
"S": "40",
"ND": 24,
"fake_trait": 0.4092,
"another_trait": 0.5382
},
{
"edge_num": 28,
"Ev": "S",
"S": "45",
"ND": 18,
"fake_trait": -0.5099,
"another_trait": 0.939
},
{
"edge_num": 29,
"Ev": "S",
"S": "46",
"ND": 23,
"fake_trait": -1.4843,
"another_trait": 0.0444
},
{
"edge_num": 30,
"Ev": "S",
"S": "56",
"ND": 21,
"fake_trait": -1.0766,
"another_trait": 0.519
},
{
"edge_num": 31,
"Ev": "S",
"S": "12",
"ND": 7,
"fake_trait": -1.0428,
"another_trait": 0.6413
}
],
"metadata": {"info": "R-package treeio", "data": "Wed May 13 18:07:22 2020"}
}The jtree format is based on JSON and can be parsed using JSON parser.
jtree_file <- tempfile(fileext = '.jtree')
write.jtree(tree2, file = jtree_file)
jsonlite::fromJSON(jtree_file)$tree
[1] "(((Prayidae_D27SS7@2825365:0.0682841{1},(Kephyes_ovata@2606431:0.0193941{2},Chuniphyes_multidentata@1277217:0.0121378{3}):0.0217782{20}):0.0607598{19},((Apolemia_sp_@1353964:0.11832{4},(((Bargmannia_amoena@263997:0.0144549{5},Bargmannia_elongata@946788:0.0149723{6}):0.0925388{25},Physonect_sp_@2066767:0.077429{7}):0.0274637{24},(Stephalia_dilata@2960089:0.0761163{8},((Frillagalma_vityazi@1155031:0.0906068{9},Resomia_ornicephala@3111757:1{10}e-06):1{28}e-06,((Lychnagalma_utricularia@2253871:0.120851{11},Nanomia_bijuga@717864:0.133939{12}):1{30}e-06,Cordagalma_sp_@1525873:0.0693814{13}):1{29}e-06):0.0333823{27}):1{26}e-06):0.0431861{23}):1{22}e-06,Rhizophysa_filiformis@3073669:0.22283{14}):0.0292362{21}):0.185603{18},(Hydra_magnipapillata@52244:0.0621782{15},Ectopleura_larynx@3556167:0.332505{16}):0.185603{31}){17};"
$data
edge_num Ev S ND fake_trait another_trait
1 1 S 58 0 -0.1181 0.3583
2 2 S 69 1 -1.0231 0.1782
3 3 S 70 2 0.1036 0.9347
4 4 S 31 9 -0.7655 0.8884
5 5 S 37 10 -0.3576 0.5472
6 6 S 38 11 0.5826 0.0054
7 7 S 61 13 -0.3069 0.6506
8 8 S 52 15 1.1091 0.5833
9 9 S 53 16 -0.7074 0.8089
10 10 S 54 17 0.8381 0.0142
11 11 S 65 19 1.4588 0.5168
12 12 S 71 20 2.4347 0.0047
13 13 S 64 22 1.5658 0.0455
14 14 S 26 28 0.5891 0.5658
15 15 S 16 5 0.7044 0.6631
16 16 S 15 6 -0.6592 0.5052
17 17 S 9 30 -0.4796 0.5199
18 18 D 17 8 1.1583 0.6469
19 19 S 36 4 -0.3000 0.1197
20 20 S 60 3 -0.5958 0.9221
21 21 S 17 29 0.4763 0.2626
22 22 S 19 27 1.6598 0.1251
23 23 D 24 26 -1.4122 0.9355
24 24 S 24 14 1.0044 0.4201
25 25 S 33 12 1.6869 0.6343
26 26 S 35 25 -0.6925 0.5852
27 27 S 40 24 0.4092 0.5382
28 28 S 45 18 -0.5099 0.9390
29 29 S 46 23 -1.4843 0.0444
30 30 S 56 21 -1.0766 0.5190
31 31 S 12 7 -1.0428 0.6413
$metadata
$metadata$info
[1] "R-package treeio"
$metadata$data
[1] "Wed May 13 18:07:22 2020"The jtree file can be directly imported as a treedata object using
read.jtree provided also
in treeio package (see also session 1.3).
read.jtree(jtree_file)## 'treedata' S4 object that stored information of
## '/tmp/RtmpvGnYhp/file55a554496727b.jtree'.
##
## ...@ phylo:
## Phylogenetic tree with 16 tips and 15 internal nodes.
##
## Tip labels:
## Prayidae_D27SS7@2825365, Kephyes_ovata@2606431, Chuniphyes_multidentata@1277217, Apolemia_sp_@1353964, Bargmannia_amoena@263997, Bargmannia_elongata@946788, ...
##
## Rooted; includes branch lengths.
##
## with the following features available:
## 'Ev', 'S', 'ND', 'fake_trait', 'another_trait'.3.4 Summary
Phylogenetic tree associated data is often stored in a separate file and need expertise to map the data to the tree structure. Lacking standardization to store and represent phylogeny and associated data makes it difficult for researchers to access and integrate the phylogenetic data into their studies. The treeio package provides functions to import phylogeny with associated data from a number of sources, including analysis finding from commonly used software and external data such as experimental, clinical or meta data. These tree + data can be exported into a single file as a BEAST or jtree formats, and the output file can be parsed back to R by treeio and the data is easy to access. The input and output utilities supplied by treeio package lay the foundation for phylogenetic data integration for downstream comparative study and visualization.
References
Bouckaert, Remco, Joseph Heled, Denise Kühnert, Tim Vaughan, Chieh-Hsi Wu, Dong Xie, Marc A. Suchard, Andrew Rambaut, and Alexei J. Drummond. 2014. “BEAST 2: A Software Platform for Bayesian Evolutionary Analysis.” PLoS Comput Biol 10 (4): e1003537. https://doi.org/10.1371/journal.pcbi.1003537.
Paradis, Emmanuel, Julien Claude, and Korbinian Strimmer. 2004. “APE: Analyses of Phylogenetics and Evolution in R Language.” Bioinformatics 20 (2): 289–90. https://doi.org/10.1093/bioinformatics/btg412.
Vos, Rutger A., James P. Balhoff, Jason A. Caravas, Mark T. Holder, Hilmar Lapp, Wayne P. Maddison, Peter E. Midford, et al. 2012. “NeXML: Rich, Extensible, and Verifiable Representation of Comparative Data and Metadata.” Systematic Biology 61 (4): 675–89. https://doi.org/10.1093/sysbio/sys025.
Yu, Guangchuang, David K. Smith, Huachen Zhu, Yi Guan, and Tommy Tsan-Yuk Lam. 2017. “Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data.” Methods in Ecology and Evolution 8 (1): 28–36. https://doi.org/10.1111/2041-210X.12628.