Note
Go to the end to download the full example code.
Plotting sensor layouts of MEG systems#
Show sensor layouts of different MEG systems.
# Author: Eric Larson <larson.eric.d@gmail.com>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
from pathlib import Path
import mne
from mne.datasets import sample, spm_face, testing
from mne.io import (
read_raw_artemis123,
read_raw_bti,
read_raw_ctf,
read_raw_fif,
read_raw_kit,
)
from mne.viz import plot_alignment, set_3d_title
print(__doc__)
root_path = Path(mne.__file__).parent.absolute()
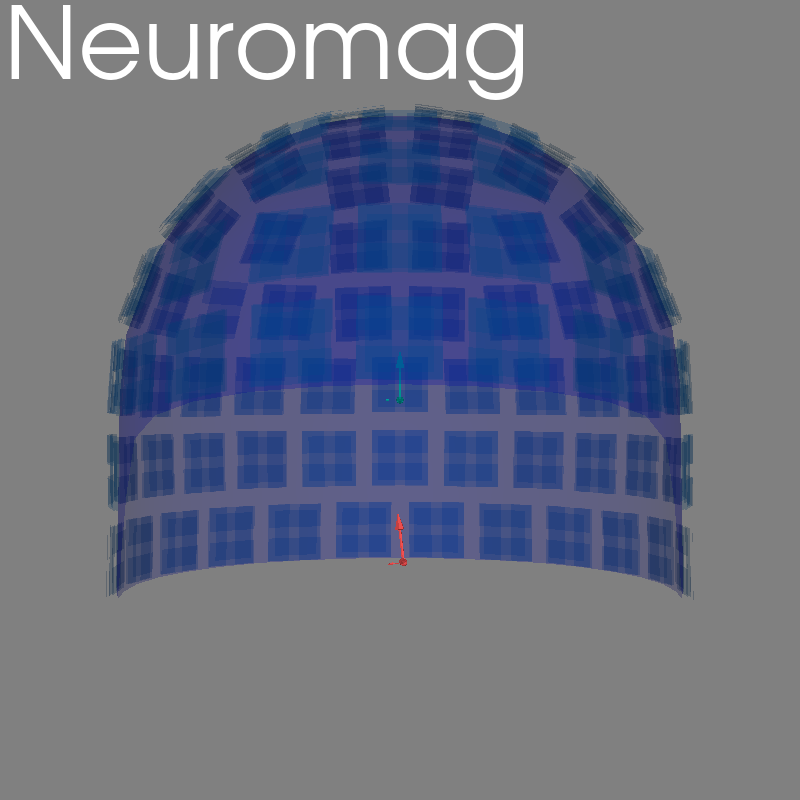
Neuromag#
kwargs = dict(eeg=False, coord_frame="meg", show_axes=True, verbose=True)
raw = read_raw_fif(sample.data_path() / "MEG" / "sample" / "sample_audvis_raw.fif")
fig = plot_alignment(raw.info, meg=("helmet", "sensors"), **kwargs)
set_3d_title(figure=fig, title="Neuromag")
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Getting helmet for system 306m
Channel types:: grad: 203, mag: 102
CTF#
raw = read_raw_ctf(
spm_face.data_path() / "MEG" / "spm" / "SPM_CTF_MEG_example_faces1_3D.ds"
)
fig = plot_alignment(raw.info, meg=("helmet", "sensors", "ref"), **kwargs)
set_3d_title(figure=fig, title="CTF 275")

ds directory : /home/circleci/mne_data/MNE-spm-face/MEG/spm/SPM_CTF_MEG_example_faces1_3D.ds
res4 data read.
hc data read.
Separate EEG position data file not present.
Quaternion matching (desired vs. transformed):
-0.90 72.01 0.00 mm <-> -0.90 72.01 0.00 mm (orig : -43.09 61.46 -252.17 mm) diff = 0.000 mm
0.90 -72.01 0.00 mm <-> 0.90 -72.01 0.00 mm (orig : 53.49 -45.24 -258.02 mm) diff = 0.000 mm
98.30 0.00 0.00 mm <-> 98.30 -0.00 0.00 mm (orig : 78.60 72.16 -241.87 mm) diff = 0.000 mm
Coordinate transformations established.
Polhemus data for 3 HPI coils added
Device coordinate locations for 3 HPI coils added
Measurement info composed.
Finding samples for /home/circleci/mne_data/MNE-spm-face/MEG/spm/SPM_CTF_MEG_example_faces1_3D.ds/SPM_CTF_MEG_example_faces1_3D.meg4:
System clock channel is available, checking which samples are valid.
1 x 324474 = 324474 samples from 340 chs
Current compensation grade : 3
Getting helmet for system CTF_275
Channel types:: ref_meg: 29, mag: 274
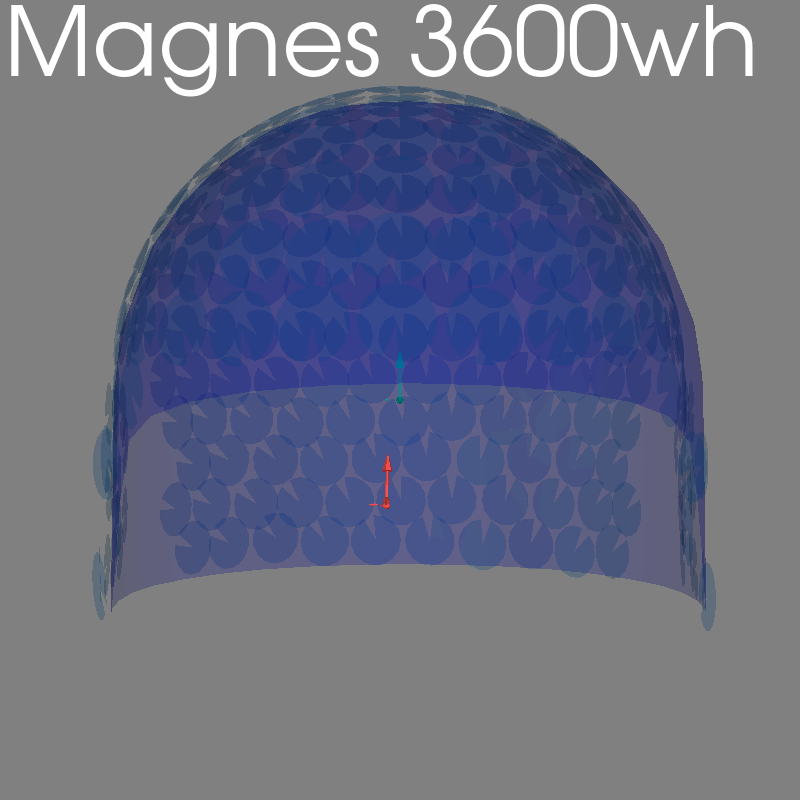
BTi#
bti_path = root_path / "io" / "bti" / "tests" / "data"
raw = read_raw_bti(
bti_path / "test_pdf_linux",
bti_path / "test_config_linux",
bti_path / "test_hs_linux",
)
fig = plot_alignment(raw.info, meg=("helmet", "sensors", "ref"), **kwargs)
set_3d_title(figure=fig, title="Magnes 3600wh")
Reading 4D PDF file /home/circleci/project/mne/io/bti/tests/data/test_pdf_linux...
Creating Neuromag info structure ...
... Setting channel info structure.
... putting coil transforms in Neuromag coordinates
... Reading digitization points from /home/circleci/project/mne/io/bti/tests/data/test_hs_linux
Currently direct inclusion of 4D weight tables is not supported. For critical use cases please take into account the MNE command "mne_create_comp_data" to include weights as printed out by the 4D "print_table" routine.
Getting helmet for system Magnes_3600wh
Channel types:: mag: 248, ref_meg: 23
KIT#
kit_path = root_path / "io" / "kit" / "tests" / "data"
raw = read_raw_kit(kit_path / "test.sqd")
fig = plot_alignment(raw.info, meg=("helmet", "sensors"), **kwargs)
set_3d_title(figure=fig, title="KIT")

Extracting SQD Parameters from /home/circleci/project/mne/io/kit/tests/data/test.sqd...
Creating Raw.info structure...
Setting channel info structure...
Creating Info structure...
Ready.
Getting helmet for system KIT (derived from 157 MEG channel locations)
Channel types:: mag: 157
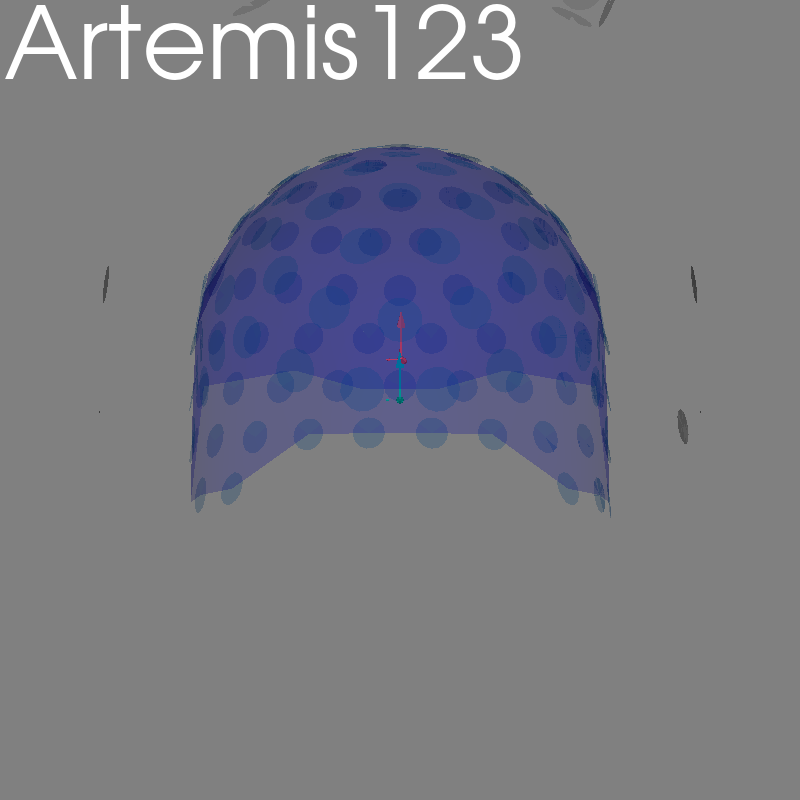
Artemis123#
raw = read_raw_artemis123(
testing.data_path()
/ "ARTEMIS123"
/ "Artemis_Data_2017-04-14-10h-38m-59s_Phantom_1k_HPI_1s.bin"
)
fig = plot_alignment(raw.info, meg=("helmet", "sensors", "ref"), **kwargs)
set_3d_title(figure=fig, title="Artemis123")
Reading header...
Loading mne loc file /home/circleci/project/mne/io/artemis123/resources/Artemis123_mneLoc.csv
Using 4 HPI coils: 140 150 160 170 Hz
Line interference frequencies: Hz
Using time window: 250.0 ms
Fitting 3 HPI coil locations at up to 1 time points (0.3 s duration)
0%| | cHPI amplitudes : 0/1 [00:00<?, ?it/s]
100%|██████████| cHPI amplitudes : 1/1 [00:00<00:00, 35.85it/s]
100%|██████████| cHPI amplitudes : 1/1 [00:00<00:00, 35.46it/s]
Computing 1863 HPI location guesses (1 cm grid in a 8.2 cm sphere)
HPIFIT: 3 coils digitized in order 1 2 3
HPI consistency of isotrak and hpifit is OK.
0%| | cHPI locations : 0/1 [00:00<?, ?it/s]
100%|██████████| cHPI locations : 1/1 [00:00<00:00, 3.30it/s]
100%|██████████| cHPI locations : 1/1 [00:00<00:00, 3.29it/s]
HPI coil 1 - location goodness of fit (0.997)
HPI coil 2 - location goodness of fit (0.999)
HPI coil 3 - location goodness of fit (0.999)
Assuming Cardinal HPIs
Getting helmet for system ARTEMIS123 (derived from 123 MEG channel locations)
Channel types:: mag: 119, ref_meg: 11
Total running time of the script: (0 minutes 22.737 seconds)