mne.io.read_raw_ctf#
- mne.io.read_raw_ctf(directory, system_clock='truncate', preload=False, clean_names=False, verbose=None) RawCTF[source]#
Raw object from CTF directory.
- Parameters:
- directorypath-like
Path to the CTF data (ending in
'.ds').- system_clock
str How to treat the system clock. Use “truncate” (default) to truncate the data file when the system clock drops to zero, and use “ignore” to ignore the system clock (e.g., if head positions are measured multiple times during a recording).
- preloadbool or
str(defaultFalse) Preload data into memory for data manipulation and faster indexing. If True, the data will be preloaded into memory (fast, requires large amount of memory). If preload is a string, preload is the file name of a memory-mapped file which is used to store the data on the hard drive (slower, requires less memory).
- clean_namesbool, optional
If True main channel names and compensation channel names will be cleaned from CTF suffixes. The default is False.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- Returns:
- rawinstance of
RawCTF The raw data.
- rawinstance of
Notes
New in v0.11.
To read in the Polhemus digitization data (for example, from a .pos file), include the file in the CTF directory. The points will then automatically be read into the
mne.io.Rawinstance viamne.io.read_raw_ctf.
Examples using mne.io.read_raw_ctf#
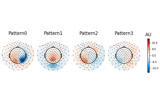
Compute spatial filters with Spatio-Spectral Decomposition (SSD)

Annotate movement artifacts and reestimate dev_head_t
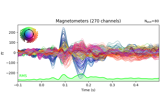
Working with CTF data: the Brainstorm auditory dataset