mne.Dipole#
- class mne.Dipole(times, pos, amplitude, ori, gof, name=None, conf=None, khi2=None, nfree=None, *, verbose=None)[source]#
Dipole class for sequential dipole fits.
Note
This class should usually not be instantiated directly via
mne.Dipole(...). Instead, use one of the functions listed in the See Also section below.Used to store positions, orientations, amplitudes, times, goodness of fit of dipoles, typically obtained with Neuromag/xfit, mne_dipole_fit or certain inverse solvers. Note that dipole position vectors are given in the head coordinate frame.
- Parameters:
- times
array, shape (n_dipoles,) The time instants at which each dipole was fitted (s).
- pos
array, shape (n_dipoles, 3) The dipoles positions (m) in head coordinates.
- amplitude
array, shape (n_dipoles,) The amplitude of the dipoles (Am).
- ori
array, shape (n_dipoles, 3) The dipole orientations (normalized to unit length).
- gof
array, shape (n_dipoles,) The goodness of fit.
- name
str|None Name of the dipole.
- conf
dict Confidence limits in dipole orientation for “vol” in m^3 (volume), “depth” in m (along the depth axis), “long” in m (longitudinal axis), “trans” in m (transverse axis), “qlong” in Am, and “qtrans” in Am (currents). The current confidence limit in the depth direction is assumed to be zero (although it can be non-zero when a BEM is used).
New in v0.15.
- khi2
array, shape (n_dipoles,) The χ^2 values for the fits.
New in v0.15.
- nfree
array, shape (n_dipoles,) The number of free parameters for each fit.
New in v0.15.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- times
- Attributes:
amplitudeThe amplitude of the dipoles (Am).
confConfidence limits in dipole orientation.
gofThe goodness of fit.
khi2The χ^2 values for the fits.
nameName of the dipole.
nfreeThe number of free parameters for each fit.
oriThe dipole orientations (normalized to unit length).
posThe dipoles positions (m) in head coordinates.
timesTime vector in seconds.
Methods
__getitem__(item)Get a time slice.
__len__()Return the number of dipoles.
copy()Copy the Dipoles object.
crop([tmin, tmax, include_tmax, verbose])Crop data to a given time interval.
plot_amplitudes([color, show])Plot the dipole amplitudes as a function of time.
plot_locations(trans, subject[, ...])Plot dipole locations.
save(fname[, overwrite, verbose])Save dipole in a
.dipor.bdipfile.time_as_index(times[, use_rounding])Convert time to indices.
to_mni(subject, trans[, subjects_dir, verbose])Convert dipole location from head to MNI coordinates.
to_mri(subject, trans[, subjects_dir, verbose])Convert dipole location from head to MRI surface RAS coordinates.
to_volume_labels(trans[, subject, aseg, ...])Find an ROI in atlas for the dipole positions.
See also
Notes
This class is for sequential dipole fits, where the position changes as a function of time. For fixed dipole fits, where the position is fixed as a function of time, use
mne.DipoleFixed.- __getitem__(item)[source]#
Get a time slice.
- Parameters:
- itemarray_like or
slice The slice of time points to use.
- itemarray_like or
- Returns:
- dipinstance of
Dipole The sliced dipole.
- dipinstance of
- __len__()[source]#
Return the number of dipoles.
- Returns:
- len
int The number of dipoles.
- len
Examples
This can be used as:
>>> len(dipoles) 10
- property amplitude#
The amplitude of the dipoles (Am).
- property conf#
Confidence limits in dipole orientation.
- crop(tmin=None, tmax=None, include_tmax=True, verbose=None)[source]#
Crop data to a given time interval.
- Parameters:
- tmin
float|None Start time of selection in seconds.
- tmax
float|None End time of selection in seconds.
- include_tmaxbool
If True (default), include tmax. If False, exclude tmax (similar to how Python indexing typically works).
New in v0.19.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- tmin
- Returns:
- selfinstance of
Dipole The cropped instance.
- selfinstance of
Examples using
crop:
- property gof#
The goodness of fit.
- property khi2#
The χ^2 values for the fits.
- property name#
Name of the dipole.
- property nfree#
The number of free parameters for each fit.
- property ori#
The dipole orientations (normalized to unit length).
- plot_amplitudes(color='k', show=True)[source]#
Plot the dipole amplitudes as a function of time.
- Parameters:
- Returns:
- fig
matplotlib.figure.Figure The figure object containing the plot.
- fig
- plot_locations(trans, subject, subjects_dir=None, mode='orthoview', coord_frame='mri', idx='gof', show_all=True, ax=None, block=False, show=True, scale=None, color=None, *, highlight_color='r', fig=None, title=None, head_source='seghead', surf='pial', width=None, verbose=None)[source]#
Plot dipole locations.
If mode is set to ‘arrow’ or ‘sphere’, only the location of the first time point of each dipole is shown else use the show_all parameter.
- Parameters:
- trans
dict|None The mri to head trans. Can be None with mode set to ‘3d’.
- subject
str|None The FreeSurfer subject name (will be used to set the FreeSurfer environment variable
SUBJECT). Can beNonewith mode set to'3d'.- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- mode
str Can be:
'arrow'or'sphere'Plot in 3D mode using PyVista with the given glyph type.
'orthoview'Plot in matplotlib
Axes3Dusing matplotlib with MRI slices shown on the sides of a cube, with the dipole(s) shown as arrows extending outward from a dot (i.e., the arrows pivot on the tail).'outlines'Plot in matplotlib
Axesusing a quiver of arrows for the dipoles in three axes (axial, coronal, and sagittal views), with the arrow pivoting in the middle of the arrow.
Changed in version 1.1: Added support for
'outlines'.- coord_frame
str Coordinate frame to use: ‘head’ or ‘mri’. Can also be ‘mri_rotated’ when mode equals
'outlines'. Defaults to ‘mri’.New in v0.14.0.
Changed in version 1.1: Added support for
'mri_rotated'.- idx
int| ‘gof’ | ‘amplitude’ Index of the initially plotted dipole. Can also be ‘gof’ to plot the dipole with highest goodness of fit value or ‘amplitude’ to plot the dipole with the highest amplitude. The dipoles can also be browsed through using up/down arrow keys or mouse scroll. Defaults to ‘gof’. Only used if mode equals ‘orthoview’.
New in v0.14.0.
- show_allbool
Whether to always plot all the dipoles. If
True(default), the active dipole is plotted as a red dot and its location determines the shown MRI slices. The non-active dipoles are plotted as small blue dots. IfFalse, only the active dipole is plotted. Only used ifmode='orthoview'.New in v0.14.0.
- axinstance of matplotlib
Axes3D|listof matplotlibAxes|None Axes to plot into. If None (default), axes will be created. If mode equals
'orthoview', must be a singleAxes3D. If mode equals'outlines', must be a list of threeAxes.New in v0.14.0.
- blockbool
Whether to halt program execution until the figure is closed. Defaults to False. Only used if mode equals ‘orthoview’.
New in v0.14.0.
- showbool
Show figure if True. Defaults to True. Only used if mode equals ‘orthoview’.
- scale
float The scale (size in meters) of the dipoles if
modeis not'orthoview'. The default is 0.03 when mode is'outlines'and 0.005 otherwise.- color
tuple The color of the dipoles. The default (None) will use
'y'if mode is'orthoview'andshow_allis True, else ‘r’. Can also be a list of colors to use when mode is'outlines'.Changed in version 0.19.0: Color is now passed in orthoview mode.
- highlight_colorcolor
The highlight color. Only used in orthoview mode with
show_all=True.New in v0.19.0.
- figinstance of
Figure3D|None 3D figure in which to plot the alignment. If
None, creates a new 600x600 pixel figure with black background. Only used when mode is'arrow'or'sphere'.New in v0.19.0.
- title
str|None The title of the figure if
mode='orthoview'(ignored for all other modes). IfNone, dipole number and its properties (amplitude, orientation etc.) will be shown. Defaults toNone.New in v0.21.0.
- head_source
str|listofstr Head source(s) to use. See the
sourceoption ofmne.get_head_surf()for more information. Only used when mode equals'outlines'.New in v1.1.
- surf
str|None Brain surface to show outlines for, can be
'white','pial', orNone. Only used when mode is'outlines'.New in v1.1.
- width
float|None Width of the matplotlib quiver arrow, see
matplotlib.axes.Axes.quiver(). If None (default), when mode is'outlines'0.015 will be used, and when mode is'orthoview'the matplotlib default is used.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- trans
- Returns:
- figinstance of
Figure3Dormatplotlib.figure.Figure The PyVista figure or matplotlib Figure.
- figinstance of
Notes
New in v0.9.0.
Examples using
plot_locations: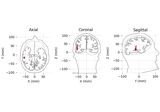
Source localization with equivalent current dipole (ECD) fit
Source localization with equivalent current dipole (ECD) fit
- property pos#
The dipoles positions (m) in head coordinates.
- save(fname, overwrite=False, *, verbose=None)[source]#
Save dipole in a
.dipor.bdipfile.The
.[b]dipformat is formne.Dipoleobjects, that is, fixed-position dipole fits. For these fits, the amplitude, orientation, and position vary as a function of time.- Parameters:
- fnamepath-like
The name of the
.dipor.bdipfile.- overwritebool
If True (default False), overwrite the destination file if it exists.
New in v0.20.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
See also
Notes
Changed in version 0.20: Support for writing bdip (Xfit binary) files.
- property times#
Time vector in seconds.
- to_mni(subject, trans, subjects_dir=None, verbose=None)[source]#
Convert dipole location from head to MNI coordinates.
- Parameters:
- subject
str The FreeSurfer subject name.
- trans
str|dict| instance ofTransform If str, the path to the head<->MRI transform
*-trans.fiffile produced during coregistration. Can also be'fsaverage'to use the built-in fsaverage transformation.- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- subject
- Returns:
- pos_mni
array, shape (n_pos, 3) The MNI coordinates (in mm) of pos.
- pos_mni
Examples using
to_mni:
Source localization with equivalent current dipole (ECD) fit
Source localization with equivalent current dipole (ECD) fit
- to_mri(subject, trans, subjects_dir=None, verbose=None)[source]#
Convert dipole location from head to MRI surface RAS coordinates.
- Parameters:
- subject
str The FreeSurfer subject name.
- trans
str|dict| instance ofTransform If str, the path to the head<->MRI transform
*-trans.fiffile produced during coregistration. Can also be'fsaverage'to use the built-in fsaverage transformation.- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- subject
- Returns:
- pos_mri
array, shape (n_pos, 3) The Freesurfer surface RAS coordinates (in mm) of pos.
- pos_mri
Examples using
to_mri:
Source localization with equivalent current dipole (ECD) fit
Source localization with equivalent current dipole (ECD) fit
- to_volume_labels(trans, subject='fsaverage', aseg='aparc+aseg', subjects_dir=None, verbose=None)[source]#
Find an ROI in atlas for the dipole positions.
- Parameters:
- transpath-like |
dict| instance ofTransform|"fsaverage"|None If str, the path to the head<->MRI transform
*-trans.fiffile produced during coregistration. Can also be'fsaverage'to use the built-in fsaverage transformation. If trans is None, an identity matrix is assumed.Changed in version 0.19: Support for ‘fsaverage’ argument.
- subject
str The FreeSurfer subject name.
- aseg
str The anatomical segmentation file. Default
autousesaparc+asegif available andwmparcif not. This may be any anatomical segmentation file in the mri subdirectory of the Freesurfer subject directory.Changed in version 1.8: Added support for the new default
'auto'.- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- transpath-like |
- Returns:
- labels
list List of anatomical region names from anatomical segmentation atlas.
- labels
Notes
New in v0.24.
Examples using
to_volume_labels:
Source localization with equivalent current dipole (ECD) fit
Source localization with equivalent current dipole (ECD) fit
Examples using mne.Dipole#

Compute sparse inverse solution with mixed norm: MxNE and irMxNE

Computing source timecourses with an XFit-like multi-dipole model

Source localization with equivalent current dipole (ECD) fit
The role of dipole orientations in distributed source localization