Note
Go to the end to download the full example code.
Annotating continuous data#
This tutorial describes adding annotations to a Raw object,
and how annotations are used in later stages of data processing.
As usual we’ll start by importing the modules we need, loading some
example data, and (since we won’t actually analyze the
raw data in this tutorial) cropping the Raw object to just 60
seconds before loading it into RAM to save memory:
# Authors: The MNE-Python contributors.
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import os
from datetime import timedelta
import mne
sample_data_folder = mne.datasets.sample.data_path()
sample_data_raw_file = os.path.join(
sample_data_folder, "MEG", "sample", "sample_audvis_raw.fif"
)
raw = mne.io.read_raw_fif(sample_data_raw_file, verbose=False)
raw.crop(tmax=60).load_data()
Reading 0 ... 36037 = 0.000 ... 60.000 secs...
Annotations in MNE-Python are a way of storing short strings of
information about temporal spans of a Raw object. Below the
surface, Annotations are list-like objects,
where each element comprises three pieces of information: an onset time
(in seconds), a duration (also in seconds), and a description (a text
string). Additionally, the Annotations object itself also keeps
track of orig_time, which is a POSIX timestamp denoting a real-world
time relative to which the annotation onsets should be interpreted.
Creating annotations programmatically#
If you know in advance what spans of the Raw object you want
to annotate, Annotations can be created programmatically, and
you can even pass lists or arrays to the Annotations
constructor to annotate multiple spans at once:
my_annot = mne.Annotations(
onset=[3, 5, 7], # in seconds
duration=[1, 0.5, 0.25], # in seconds, too
description=["AAA", "BBB", "CCC"],
)
print(my_annot)
<Annotations | 3 segments: AAA (1), BBB (1), CCC (1)>
Notice that orig_time is None, because we haven’t specified it. In
those cases, when you add the annotations to a Raw object,
it is assumed that the orig_time matches the time of the first sample of
the recording, so orig_time will be set to match the recording
measurement date (raw.info['meas_date']).
raw.set_annotations(my_annot)
print(raw.annotations)
# convert meas_date (a tuple of seconds, microseconds) into a float:
meas_date = raw.info["meas_date"]
orig_time = raw.annotations.orig_time
print(meas_date == orig_time)
<Annotations | 3 segments: AAA (1), BBB (1), CCC (1)>
True
Since the example data comes from a Neuromag system that starts counting
sample numbers before the recording begins, adding my_annot to the
Raw object also involved another automatic change: an offset
equalling the time of the first recorded sample (raw.first_samp /
raw.info['sfreq']) was added to the onset values of each annotation
(see Time, sample number, and sample index for more info on raw.first_samp):
time_of_first_sample = raw.first_samp / raw.info["sfreq"]
print(my_annot.onset + time_of_first_sample)
print(raw.annotations.onset)
[45.95597083 47.95597083 49.95597083]
[45.95597083 47.95597083 49.95597083]
If you know that your annotation onsets are relative to some other time, you
can set orig_time before you call set_annotations(),
and the onset times will get adjusted based on the time difference between
your specified orig_time and raw.info['meas_date'], but without the
additional adjustment for raw.first_samp. orig_time can be specified
in various ways (see the documentation of Annotations for the
options); here we’ll use an ISO 8601 formatted string, and set it to be 50
seconds later than raw.info['meas_date'].
time_format = "%Y-%m-%d %H:%M:%S.%f"
new_orig_time = (meas_date + timedelta(seconds=50)).strftime(time_format)
print(new_orig_time)
later_annot = mne.Annotations(
onset=[3, 5, 7],
duration=[1, 0.5, 0.25],
description=["DDD", "EEE", "FFF"],
orig_time=new_orig_time,
)
raw2 = raw.copy().set_annotations(later_annot)
print(later_annot.onset)
print(raw2.annotations.onset)
2002-12-03 19:02:00.720100
[3. 5. 7.]
[53. 55. 57.]
Note
If your annotations fall outside the range of data times in the
Raw object, the annotations outside the data range will
not be added to raw.annotations, and a warning will be issued.
Now that your annotations have been added to a Raw object,
you can see them when you visualize the Raw object:
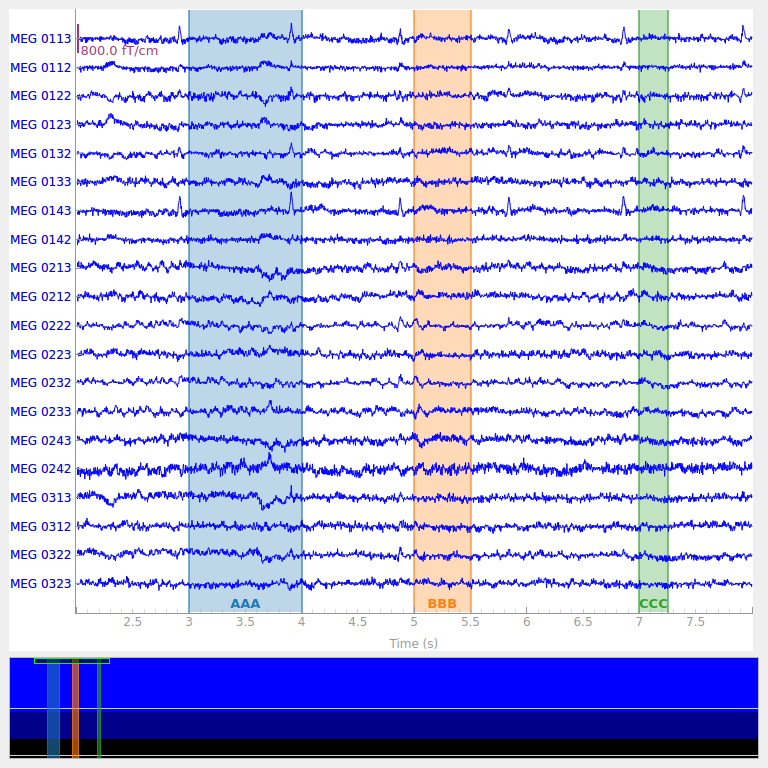
The three annotations appear as differently colored rectangles because they
have different description values (which are printed along the top
edge of the plot area). Notice also that colored spans appear in the small
scroll bar at the bottom of the plot window, making it easy to quickly view
where in a Raw object the annotations are so you can easily
browse through the data to find and examine them.
Annotating Raw objects interactively#
Annotations can also be added to a Raw object interactively
by clicking-and-dragging the mouse in the plot window. To do this, you must
first enter “annotation mode” by pressing a while the plot window is
focused; this will bring up the annotation controls:
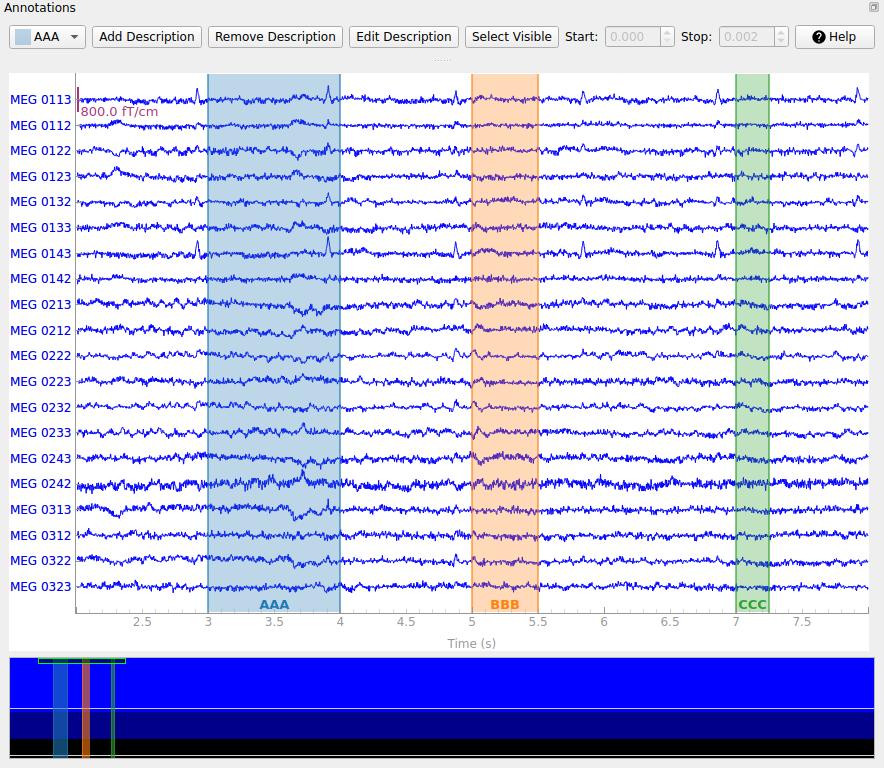
The drop-down-menu on the left determines which existing label will be created by the next click-and-drag operation in the main plot window. New annotation descriptions can be added by clicking the Add description button; the new description will be added to the list of descriptions and automatically selected. The following functions relate to which description is currently selected in the drop-down-menu: With Remove description you can remove description including the annotations. With Edit description you can edit the description of either only one annotation (the one currently selected) or all annotations of a description. With Set Visible you can show or hide descriptions.
During interactive annotation it is also possible to adjust the start and end times of existing annotations, by clicking-and-dragging on the left or right edges of the highlighting rectangle corresponding to that annotation. When an annotation is selected (the background of the label at the bottom changes to darker) the values for start and stop are visible in two spinboxes and can also be edited there.
Warning
Calling set_annotations() replaces any annotations
currently stored in the Raw object, so be careful when
working with annotations that were created interactively (you could lose
a lot of work if you accidentally overwrite your interactive
annotations). A good safeguard is to run
interactive_annot = raw.annotations after you finish an interactive
annotation session, so that the annotations are stored in a separate
variable outside the Raw object.
How annotations affect preprocessing and analysis#
You may have noticed that the description for new labels in the annotation
controls window defaults to BAD_. The reason for this is that annotation
is often used to mark bad temporal spans of data (such as movement artifacts
or environmental interference that cannot be removed in other ways such as
projection or filtering). Several
MNE-Python operations
are “annotation aware” and will avoid using data that is annotated with a
description that begins with “bad” or “BAD”; such operations typically have a
boolean reject_by_annotation parameter. Examples of such operations are
independent components analysis (mne.preprocessing.ICA), functions
for finding heartbeat and blink artifacts
(find_ecg_events(),
find_eog_events()), and creation of epoched data
from continuous data (mne.Epochs). See Rejecting bad data spans and breaks
for details.
Operations on Annotations objects#
Annotations objects can be combined by simply adding them with
the + operator, as long as they share the same orig_time:
new_annot = mne.Annotations(onset=3.75, duration=0.75, description="AAA")
raw.set_annotations(my_annot + new_annot)
raw.plot(start=2, duration=6)
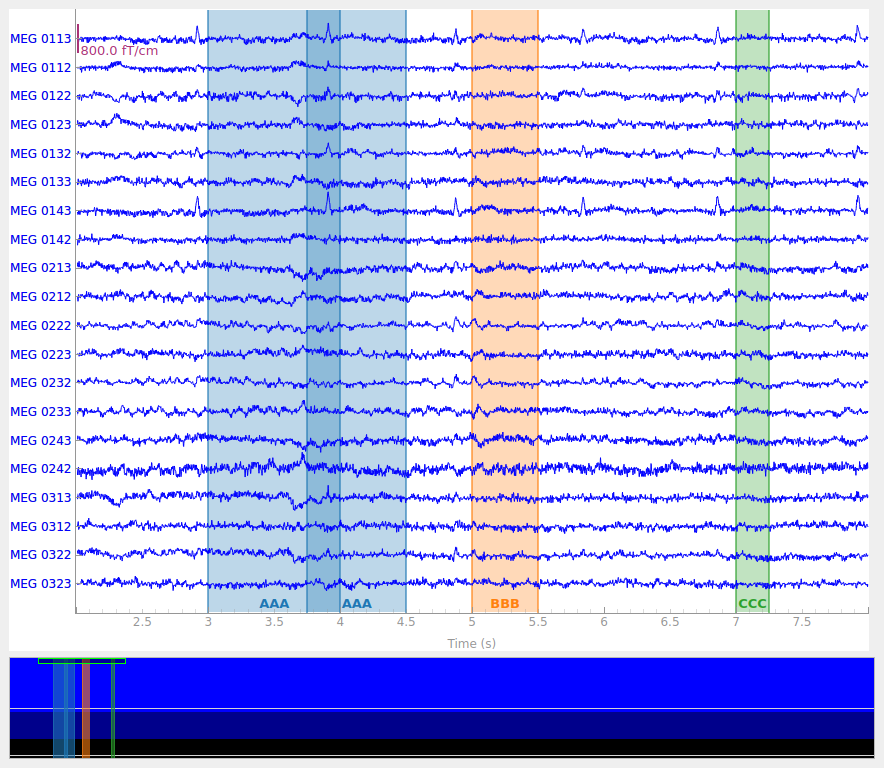
Notice that it is possible to create overlapping annotations, even when they share the same description. This is not possible when annotating interactively; click-and-dragging to create a new annotation that overlaps with an existing annotation with the same description will cause the old and new annotations to be merged.
Individual annotations can be accessed by indexing an
Annotations object, and subsets of the annotations can be
achieved by either slicing or indexing with a list, tuple, or array of
indices:
print(raw.annotations[0]) # just the first annotation
print(raw.annotations[:2]) # the first two annotations
print(raw.annotations[(3, 2)]) # the fourth and third annotations
OrderedDict({'onset': np.float64(45.95597082905339), 'duration': np.float64(1.0), 'description': np.str_('AAA'), 'orig_time': datetime.datetime(2002, 12, 3, 19, 1, 10, 720100, tzinfo=datetime.timezone.utc), 'extras': {}})
<Annotations | 2 segments: AAA (2)>
<Annotations | 2 segments: BBB (1), CCC (1)>
You can also iterate over the annotations within an Annotations
object:
'AAA' goes from 45.95597082905339 to 46.95597082905339
'AAA' goes from 46.70597082905339 to 47.45597082905339
'BBB' goes from 47.95597082905339 to 48.45597082905339
'CCC' goes from 49.95597082905339 to 50.20597082905339
Note that iterating, indexing and slicing Annotations all
return a copy, so changes to an indexed, sliced, or iterated element will not
modify the original Annotations object.
# later_annot WILL be changed, because we're modifying the first element of
# later_annot.onset directly:
later_annot.onset[0] = 99
# later_annot WILL NOT be changed, because later_annot[0] returns a copy
# before the 'onset' field is changed:
later_annot[0]["onset"] = 77
print(later_annot[0]["onset"])
99.0
Reading and writing Annotations to/from a file#
Annotations objects have a save() method
which can write .fif, .csv, and .txt formats (the
format to write is inferred from the file extension in the filename you
provide). Be aware that the format of the onset information that is written
to the file depends on the file extension. While .csv files store the
onset as timestamps, .txt files write floats (in seconds). There is a
corresponding read_annotations() function to load them from disk:
raw.annotations.save("saved-annotations.csv", overwrite=True)
annot_from_file = mne.read_annotations("saved-annotations.csv")
print(annot_from_file)
<Annotations | 4 segments: AAA (2), BBB (1), CCC (1)>
Total running time of the script: (0 minutes 4.677 seconds)