Note
Go to the end to download the full example code.
Generate simulated evoked data#
Use simulate_sparse_stc() to simulate evoked data.
# Author: Daniel Strohmeier <daniel.strohmeier@tu-ilmenau.de>
# Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import matplotlib.pyplot as plt
import numpy as np
import mne
from mne.datasets import sample
from mne.simulation import simulate_evoked, simulate_sparse_stc
from mne.time_frequency import fit_iir_model_raw
from mne.viz import plot_sparse_source_estimates
print(__doc__)
Load real data as templates
data_path = sample.data_path()
meg_path = data_path / "MEG" / "sample"
raw = mne.io.read_raw_fif(meg_path / "sample_audvis_raw.fif")
proj = mne.read_proj(meg_path / "sample_audvis_ecg-proj.fif")
raw.add_proj(proj)
raw.info["bads"] = ["MEG 2443", "EEG 053"] # mark bad channels
fwd_fname = meg_path / "sample_audvis-meg-eeg-oct-6-fwd.fif"
ave_fname = meg_path / "sample_audvis-no-filter-ave.fif"
cov_fname = meg_path / "sample_audvis-cov.fif"
fwd = mne.read_forward_solution(fwd_fname)
fwd = mne.pick_types_forward(fwd, meg=True, eeg=True, exclude=raw.info["bads"])
cov = mne.read_cov(cov_fname)
info = mne.io.read_info(ave_fname)
label_names = ["Aud-lh", "Aud-rh"]
labels = [mne.read_label(meg_path / "labels" / f"{ln}.label") for ln in label_names]
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Read a total of 6 projection items:
ECG-planar-999--0.200-0.400-PCA-01 (1 x 203) idle
ECG-planar-999--0.200-0.400-PCA-02 (1 x 203) idle
ECG-axial-999--0.200-0.400-PCA-01 (1 x 102) idle
ECG-axial-999--0.200-0.400-PCA-02 (1 x 102) idle
ECG-eeg-999--0.200-0.400-PCA-01 (1 x 59) idle
ECG-eeg-999--0.200-0.400-PCA-02 (1 x 59) idle
6 projection items deactivated
Reading forward solution from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-eeg-oct-6-fwd.fif...
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Desired named matrix (kind = 3523 (FIFF_MNE_FORWARD_SOLUTION_GRAD)) not available
Read MEG forward solution (7498 sources, 306 channels, free orientations)
Desired named matrix (kind = 3523 (FIFF_MNE_FORWARD_SOLUTION_GRAD)) not available
Read EEG forward solution (7498 sources, 60 channels, free orientations)
Forward solutions combined: MEG, EEG
Source spaces transformed to the forward solution coordinate frame
364 out of 366 channels remain after picking
366 x 366 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
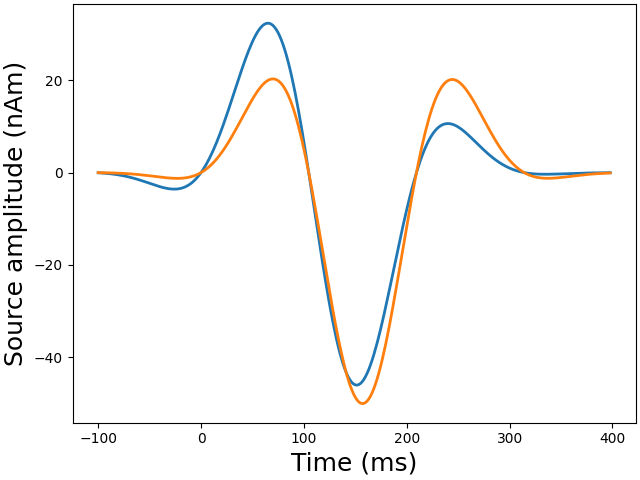
Generate source time courses from 2 dipoles and the corresponding evoked data
times = np.arange(300, dtype=np.float64) / raw.info["sfreq"] - 0.1
rng = np.random.RandomState(42)
def data_fun(times):
"""Generate random source time courses."""
return (
50e-9
* np.sin(30.0 * times)
* np.exp(-((times - 0.15 + 0.05 * rng.randn(1)) ** 2) / 0.01)
)
stc = simulate_sparse_stc(
fwd["src"],
n_dipoles=2,
times=times,
random_state=42,
labels=labels,
data_fun=data_fun,
)
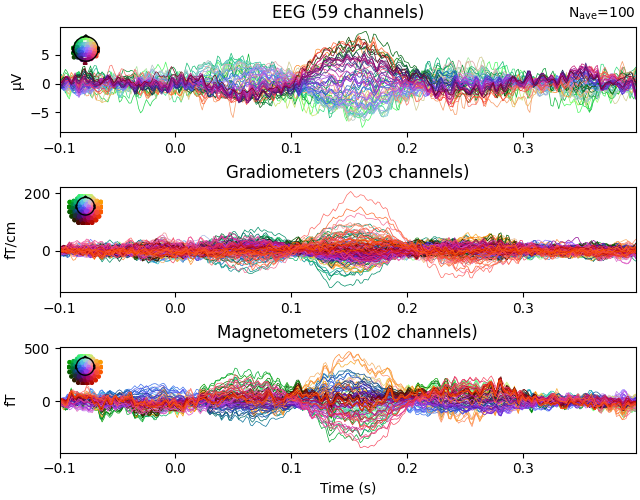
Generate noisy evoked data
picks = mne.pick_types(raw.info, meg=True, exclude="bads")
iir_filter = fit_iir_model_raw(raw, order=5, picks=picks, tmin=60, tmax=180)[1]
nave = 100 # simulate average of 100 epochs
evoked = simulate_evoked(
fwd, stc, info, cov, nave=nave, use_cps=True, iir_filter=iir_filter
)
Average patch normals will be employed in the rotation to the local surface coordinates....
Converting to surface-based source orientations...
[done]
Projecting source estimate to sensor space...
[done]
4 projection items deactivated
Created an SSP operator (subspace dimension = 4)
4 projection items activated
SSP projectors applied...
Plot
plot_sparse_source_estimates(
fwd["src"], stc, bgcolor=(1, 1, 1), opacity=0.5, high_resolution=True
)
plt.figure()
plt.psd(evoked.data[0])
evoked.plot(time_unit="s")
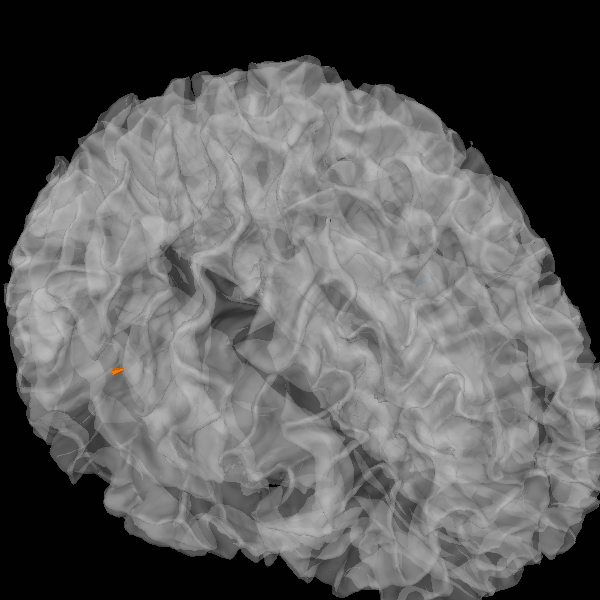
Total number of active sources: [ 89235 233092]
Total running time of the script: (0 minutes 8.458 seconds)