Note
Go to the end to download the full example code.
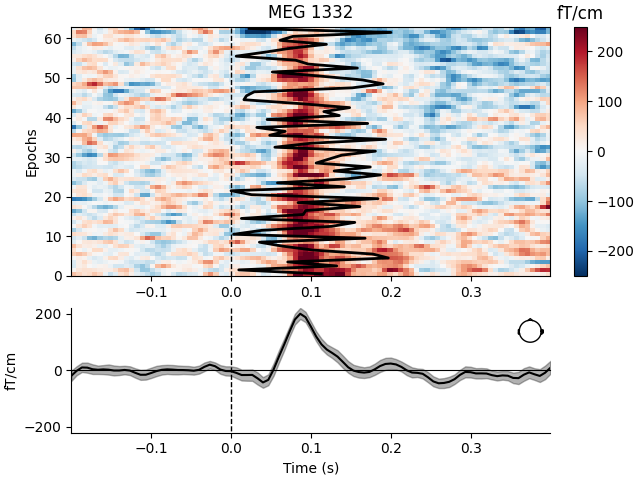
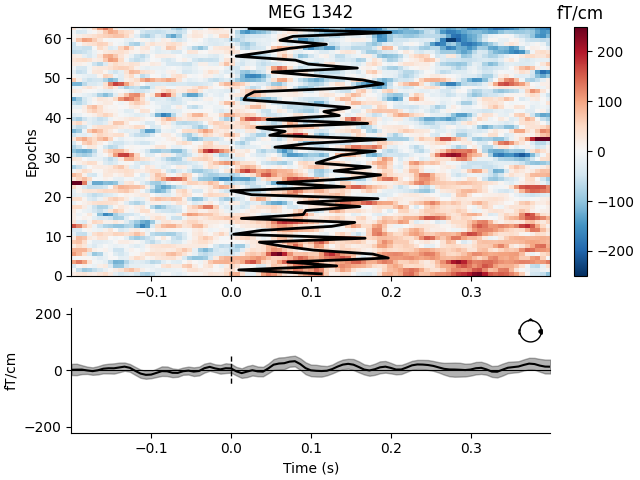
Visualize channel over epochs as an image#
This will produce what is sometimes called an event related potential / field (ERP/ERF) image.
Two images are produced, one with a good channel and one with a channel that does not show any evoked field.
It is also demonstrated how to reorder the epochs using a 1D spectral embedding as described in [1].
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import matplotlib.pyplot as plt
import numpy as np
import mne
from mne import io
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
Set parameters
meg_path = data_path / "MEG" / "sample"
raw_fname = meg_path / "sample_audvis_filt-0-40_raw.fif"
event_fname = meg_path / "sample_audvis_filt-0-40_raw-eve.fif"
event_id, tmin, tmax = 1, -0.2, 0.4
# Setup for reading the raw data
raw = io.read_raw_fif(raw_fname)
events = mne.read_events(event_fname)
# Set up pick list: EEG + MEG - bad channels (modify to your needs)
raw.info["bads"] = ["MEG 2443", "EEG 053"]
# Create epochs, here for gradiometers + EOG only for simplicity
epochs = mne.Epochs(
raw,
events,
event_id,
tmin,
tmax,
proj=True,
picks=("grad", "eog"),
baseline=(None, 0),
preload=True,
reject=dict(grad=4000e-13, eog=150e-6),
)
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 72 events and 91 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
9 bad epochs dropped
Show event-related fields images
# and order with spectral reordering
# If you don't have scikit-learn installed set order_func to None
from sklearn.manifold import spectral_embedding # noqa
from sklearn.metrics.pairwise import rbf_kernel # noqa
def order_func(times, data):
this_data = data[:, (times > 0.0) & (times < 0.350)]
this_data /= np.sqrt(np.sum(this_data**2, axis=1))[:, np.newaxis]
return np.argsort(
spectral_embedding(
rbf_kernel(this_data, gamma=1.0), n_components=1, random_state=0
).ravel()
)
good_pick = 97 # channel with a clear evoked response
bad_pick = 98 # channel with no evoked response
# We'll also plot a sample time onset for each trial
plt_times = np.linspace(0, 0.2, len(epochs))
plt.close("all")
mne.viz.plot_epochs_image(
epochs,
[good_pick, bad_pick],
sigma=0.5,
order=order_func,
vmin=-250,
vmax=250,
overlay_times=plt_times,
show=True,
)
Not setting metadata
63 matching events found
No baseline correction applied
0 projection items activated
Not setting metadata
63 matching events found
No baseline correction applied
0 projection items activated
References#
Total running time of the script: (0 minutes 2.651 seconds)