pCreode
Input saved to /scratch/irc/personal/robrechtc/tmp//RtmpcLES5h/input:
expression.csv
params.json
calculating densities for datapoints: 0 -> 91
****Always check density overlay for radius fit****
Number of data points in downsample = 88
Performing 10 independent runs, may take some time
Number of data points in downsample = 88
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
Traceback (most recent call last):
File "/code/run.py", line 46, in
num_runs = params["num_runs"]
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 662, in pCreode
aligned_ind = consensus_alignment( down, hi_pl_ind.copy(), data, density, noise)
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 374, in consensus_alignment
new_nodes[jj] = gp[jj]
IndexError: index 4 is out of bounds for axis 0 with size 4
Input saved to /scratch/irc/personal/robrechtc/tmp//RtmpcQ9nbg/input:
expression.csv
params.json
calculating densities for datapoints: 0 -> 93
****Always check density overlay for radius fit****
Number of data points in downsample = 92
Performing 10 independent runs, may take some time
Number of data points in downsample = 92
Constructing density kNN
finding endstates
Number of endstates found -> 8
hierarchical placing
consensus aligning
Traceback (most recent call last):
File "/code/run.py", line 46, in
num_runs = params["num_runs"]
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 662, in pCreode
aligned_ind = consensus_alignment( down, hi_pl_ind.copy(), data, density, noise)
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 374, in consensus_alignment
new_nodes[jj] = gp[jj]
IndexError: index 2 is out of bounds for axis 0 with size 2
Input saved to /scratch/irc/personal/robrechtc/tmp//RtmpL5AFog/input:
expression.csv
params.json
calculating densities for datapoints: 0 -> 237
****Always check density overlay for radius fit****
Number of data points in downsample = 238
Performing 10 independent runs, may take some time
Number of data points in downsample = 238
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
Traceback (most recent call last):
File "/code/run.py", line 46, in
num_runs = params["num_runs"]
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 662, in pCreode
aligned_ind = consensus_alignment( down, hi_pl_ind.copy(), data, density, noise)
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 351, in consensus_alignment
rand_ind = np.random.choice( range( num_cells), num_cells, replace=False)
File "mtrand.pyx", line 1126, in mtrand.RandomState.choice
ValueError: a must be non-empty
Input saved to /scratch/irc/personal/robrechtc/tmp//RtmpwVprIg/input:
expression.csv
params.json
calculating densities for datapoints: 0 -> 354
****Always check density overlay for radius fit****
Number of data points in downsample = 343
Performing 10 independent runs, may take some time
Number of data points in downsample = 343
Constructing density kNN
finding endstates
Number of endstates found -> 6
hierarchical placing
consensus aligning
Traceback (most recent call last):
File "/code/run.py", line 46, in
num_runs = params["num_runs"]
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 662, in pCreode
aligned_ind = consensus_alignment( down, hi_pl_ind.copy(), data, density, noise)
File "/usr/local/lib/python2.7/site-packages/pcreode/functions.py", line 374, in consensus_alignment
new_nodes[jj] = gp[jj]
IndexError: index 12 is out of bounds for axis 0 with size 12
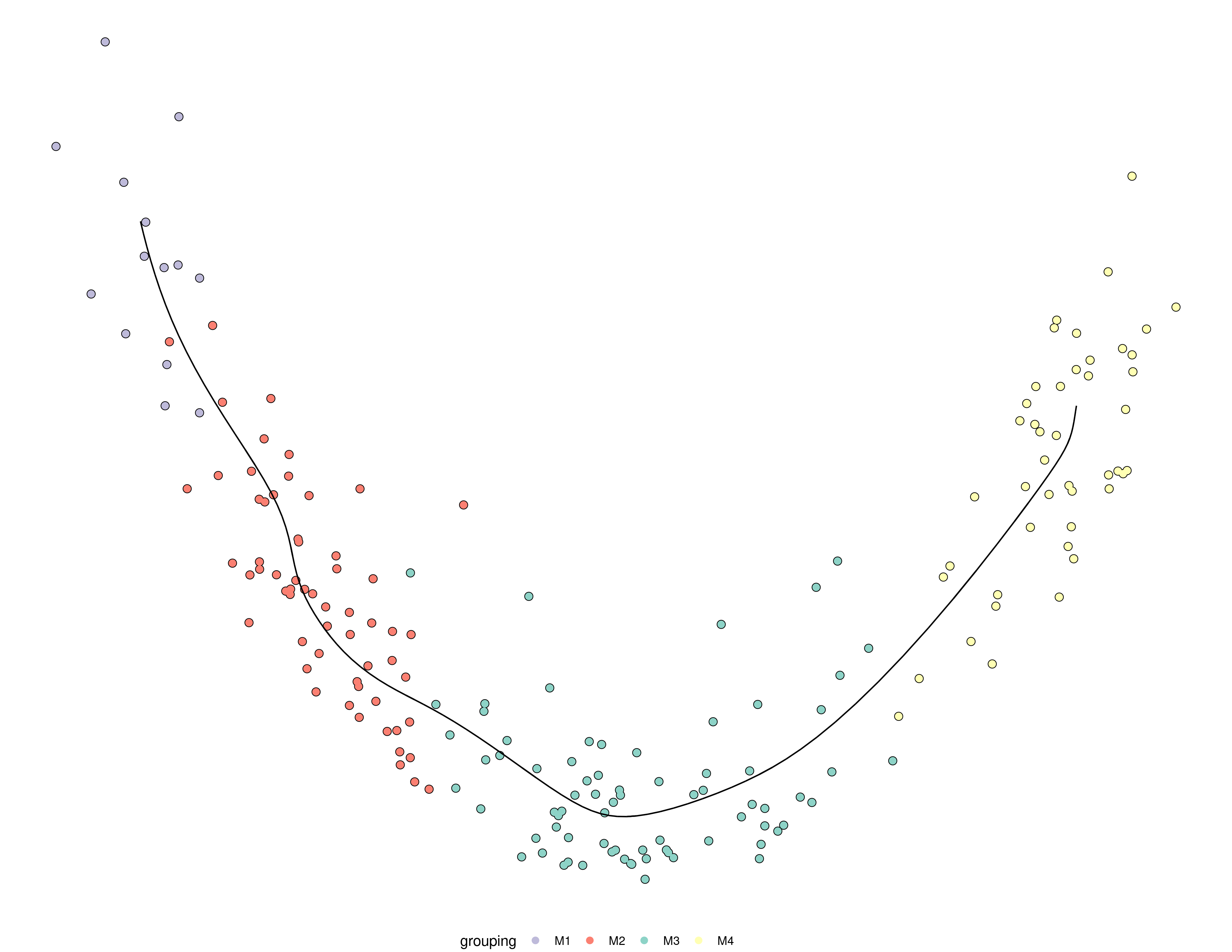

Input saved to /scratch/irc/personal/robrechtc/tmp//RtmpcY8OgV/input:
expression.csv
params.json
calculating densities for datapoints: 0 -> 193
****Always check density overlay for radius fit****
Number of data points in downsample = 135
Performing 10 independent runs, may take some time
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 1
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 2
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 3
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 4
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 5
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 6
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 7
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 8
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 9
Number of data points in downsample = 135
Constructing density kNN
finding endstates
Number of endstates found -> 4
hierarchical placing
consensus aligning
saving files for run_num 10
scoring graph 1
scoring graph 2
scoring graph 3
scoring graph 4
scoring graph 5
scoring graph 6
scoring graph 7
scoring graph 8
scoring graph 9
Most representative graph IDs from first to worst [0 1 2 3 4 5 6 7 8 9]output saved in /scratch/irc/personal/robrechtc/tmp//RtmpcY8OgV/output:
dimred_milestones.csv
dimred.csv
milestone_network.csv
timings.json
