- Cover
- Acknowledgments
- 1 Preface
- 2 (R)e-Introduction to statistics
- 2.1 Histograms, boxplots, and density curves
- 2.2 Pirate-plots
- 2.3 Models, hypotheses, and permutations for the two sample mean situation
- 2.4 Permutation testing for the two sample mean situation
- 2.5 Hypothesis testing (general)
- 2.6 Connecting randomization (nonparametric) and parametric tests
- 2.7 Second example of permutation tests
- 2.8 Reproducibility Crisis: Moving beyond p < 0.05, publication bias, and multiple testing issues
- 2.9 Confidence intervals and bootstrapping
- 2.10 Bootstrap confidence intervals for difference in GPAs
- 2.11 Chapter summary
- 2.12 Summary of important R code
- 2.13 Practice problems
- 3 One-Way ANOVA
- 3.1 Situation
- 3.2 Linear model for One-Way ANOVA (cell-means and reference-coding)
- 3.3 One-Way ANOVA Sums of Squares, Mean Squares, and F-test
- 3.4 ANOVA model diagnostics including QQ-plots
- 3.5 Guinea pig tooth growth One-Way ANOVA example
- 3.6 Multiple (pair-wise) comparisons using Tukey’s HSD and the compact letter display
- 3.7 Pair-wise comparisons for the Overtake data
- 3.8 Chapter summary
- 3.9 Summary of important R code
- 3.10 Practice problems
- 4 Two-Way ANOVA
- 4.1 Situation
- 4.2 Designing a two-way experiment and visualizing results
- 4.3 Two-Way ANOVA models and hypothesis tests
- 4.4 Guinea pig tooth growth analysis with Two-Way ANOVA
- 4.5 Observational study example: The Psychology of Debt
- 4.6 Pushing Two-Way ANOVA to the limit: Un-replicated designs and Estimability
- 4.7 Chapter summary
- 4.8 Summary of important R code
- 4.9 Practice problems
- 5 Chi-square tests
- 5.1 Situation, contingency tables, and tableplots
- 5.2 Homogeneity test hypotheses
- 5.3 Independence test hypotheses
- 5.4 Models for R by C tables
- 5.5 Permutation tests for the \(X^2\) statistic
- 5.6 Chi-square distribution for the \(X^2\) statistic
- 5.7 Examining residuals for the source of differences
- 5.8 General protocol for \(X^2\) tests
- 5.9 Political party and voting results: Complete analysis
- 5.10 Is cheating and lying related in students?
- 5.11 Analyzing a stratified random sample of California schools
- 5.12 Chapter summary
- 5.13 Summary of important R commands
- 5.14 Practice problems
- 6 Correlation and Simple Linear Regression
- 6.1 Relationships between two quantitative variables
- 6.2 Estimating the correlation coefficient
- 6.3 Relationships between variables by groups
- 6.4 Inference for the correlation coefficient
- 6.5 Are tree diameters related to tree heights?
- 6.6 Describing relationships with a regression model
- 6.7 Least Squares Estimation
- 6.8 Measuring the strength of regressions: R2
- 6.9 Outliers: leverage and influence
- 6.10 Residual diagnostics – setting the stage for inference
- 6.11 Old Faithful discharge and waiting times
- 6.12 Chapter summary
- 6.13 Summary of important R code
- 6.14 Practice problems
- 7 Simple linear regression inference
- 7.1 Model
- 7.2 Confidence interval and hypothesis tests for the slope and intercept
- 7.3 Bozeman temperature trend
- 7.4 Randomization-based inferences for the slope coefficient
- 7.5 Transformations part I: Linearizing relationships
- 7.6 Transformations part II: Impacts on SLR interpretations: log(y), log(x), & both log(y) & log(x)
- 7.7 Confidence interval for the mean and prediction intervals for a new observation
- 7.8 Chapter summary
- 7.9 Summary of important R code
- 7.10 Practice problems
- 8 Multiple linear regression
- 8.1 Going from SLR to MLR
- 8.2 Validity conditions in MLR
- 8.3 Interpretation of MLR terms
- 8.4 Comparing multiple regression models
- 8.5 General recommendations for MLR interpretations and VIFs
- 8.6 MLR inference: Parameter inferences using the t-distribution
- 8.7 Overall F-test in multiple linear regression
- 8.8 Case study: First year college GPA and SATs
- 8.9 Different intercepts for different groups: MLR with indicator variables
- 8.10 Additive MLR with more than two groups: Headache example
- 8.11 Different slopes and different intercepts
- 8.12 F-tests for MLR models with quantitative and categorical variables and interactions
- 8.13 AICs for model selection
- 8.14 Case study: Forced expiratory volume model selection using AICs
- 8.15 Chapter summary
- 8.16 Summary of important R code
- 8.17 Practice problems
- 9 Case studies
- 9.1 Overview of material covered
- 9.2 The impact of simulated chronic nitrogen deposition on the biomass and N2-fixation activity of two boreal feather moss–cyanobacteria associations
- 9.3 Ants learn to rely on more informative attributes during decision-making
- 9.4 Multi-variate models are essential for understanding vertebrate diversification in deep time
- 9.5 What do didgeridoos really do about sleepiness?
- 9.6 General summary
- References
3.6 Multiple (pair-wise) comparisons using Tukey’s HSD and the compact letter display
With evidence that the true means are likely not all equal, many researchers want to explore which groups show evidence of differing from one another. This provides information on the source of the overall difference that was detected and detailed information on which groups differed from one another. Because this is a shot-gun/unfocused sort of approach, some people think it is an over-used procedure. Others feel that it is an important method of addressing detailed questions about group comparisons in a valid and safe way. For example, we might want to know if OJ is different from VC at the 0.5 mg/day dosage level and these methods will allow us to get an answer to this sort of question. It also will test for differences between the OJ.0.5 and VC.2 groups and every other pair of levels that you can construct (15 total!). This method actually takes us back to the methods in Chapter 2 where we compared the means of two groups except that we need to deal with potentially many pair-wise comparisons, making an adjustment to account for that inflation in Type I errors that occurs due to many tests being performed at the same time. A commonly used method to make all the pair-wise comparisons that includes a correction for doing this is called Tukey’s Honest Significant Difference (Tukey’s HSD) method64. The name suggests that not using it could lead to a dishonest answer and that it will give you an honest result. It is more that if you don’t do some sort of correction for all the tests you are performing, you might find some spurious65 results. There are other methods that could be used to do a similar correction and also provide “honest” inferences; we are just going to learn one of them. Tukey’s method employs a different correction from the Bonferroni method discussed in Chapter 2 but also controls the family-wise error rate across all the pairs being compared.
In pair-wise comparisons between all the pairs of means in a One-Way ANOVA, the number of
tests is based on the number of pairs. We can calculate the number of tests using
\(J\) choose 2, \(\begin{pmatrix}J\\2\end{pmatrix}\), to get the number of unique pairs of
size 2 that we can make out of \(J\) individual treatment levels. We don’t need to
explore the combinatorics formula for this, as the choose function in R can give us the
answers:
## [1] 3## [1] 6## [1] 10## [1] 15## [1] 21So if you have three groups (the smallest number where we have to worry about more than one pair), there are three unique pairs to compare. For six groups, like in the guinea pig study, we have to consider 15 tests to compare all the unique pairs of groups and with seven groups, there are 21 tests. Once there are more than two groups to compare, it seems like we should be worried about inflated family-wise error rates. Fortunately, the Tukey’s HSD method controls the family-wise error rate at your specified level (say 0.05) across any number of pair-wise comparisons. This means that the overall rate of at least one Type I error across all the tests is controlled at the specified significance level, often 5%. To do this, each test must use a slightly more conservative cut-off than if just one test is performed and the procedure helps us figure out how much more conservative we need to be.
Tukey’s HSD starts with focusing on the difference between the groups with the largest and smallest means (\(\bar{y}_{max}-\bar{y}_{min}\)). If \((\bar{y}_{max}-\bar{y}_{min}) \le \text{Margin of Error}\) for the difference in the means, then all other pairwise differences, say \(\vert \bar{y}_j - \bar{y}_{j'}\vert\), for two groups \(j\) and \(j'\), will be less than or equal to that margin of error. This also means that any confidence intervals for any difference in the means will contain 0. Tukey’s HSD selects a critical value so that (\(\bar{y}_{max}-\bar{y}_{min}\)) will be less than the margin of error in 95% of data sets drawn from populations with a common mean. This implies that in 95% of data sets in which all the population means are the same, all confidence intervals for differences in pairs of means will contain 0. Tukey’s HSD provides confidence intervals for the difference in true means between groups \(j\) and \(j'\), \(\mu_j-\mu_{j'}\), for all pairs where \(j \ne j'\), using
\[(\bar{y}_j - \bar{y}_{j'}) \mp \frac{q^*}{\sqrt{2}}\sqrt{\text{MS}_E\left(\frac{1}{n_j}+ \frac{1}{n_{j'}}\right)}\]
where
\(\frac{q^*}{\sqrt{2}}\sqrt{\text{MS}_E\left(\frac{1}{n_j}+\frac{1}{n_{j'}}\right)}\)
is the margin of error for the intervals. The distribution used
to find the multiplier, \(q^*\), for the confidence intervals is available in the
qtukey function and generally provides a slightly larger multiplier than the
regular \(t^*\) from our two-sample \(t\)-based confidence interval discussed in
Chapter 2. The formula otherwise is very similar to the one used in Chapter 2 with the SE for the difference in the means based on a measure of residual variance (here \(MS_E\)) times \(\left(\frac{1}{n_j}+\frac{1}{n_{j'}}\right)\) which weights the results based on the relative sample sizes in the groups.
We will use the confint, cld, and plot functions
applied to output from the glht function (all from the multcomp package;
Hothorn, Bretz, and Westfall (2008), (Hothorn, Bretz, and Westfall 2019)) to get the required comparisons from our
ANOVA model.
Unfortunately, its code format is a little complicated – but there are
just two places to modify the code: include the model name and after mcp
(stands for multiple comparison procedure) in the linfct option, you need to include the
explanatory variable name as VARIABLENAME="Tukey". The last part is to get the
Tukey HSD multiple comparisons run on our explanatory variable66. Once we obtain the
intervals using the confint function or using plot applied to the stored results, we can use them to test \(H_0: \mu_j = \mu_{j'} \text{ vs } H_A: \mu_j \ne \mu_{j'}\)
by assessing whether 0 is in the confidence interval for each pair. If 0 is in the
interval, then there is weak evidence against the null hypothesis for that pair, so we not detect a difference in that pair and not conclude that there is a difference. If 0 is not in the
interval, then we have strong evidence against \(H_0\) for that pair, detect a difference, and conclude that there is a difference in that pair at the specified family-wise significance
level. You will see a switch to using the
word “detection” to describe null hypotheses that we find strong evidence against as it
can help to compactly write up these complicated results. The following code provides the numerical
and graphical67 results of applying Tukey’s HSD
to the linear model for the Guinea Pig data:
(ref:fig3-19) Graphical display of pair-wise comparisons from Tukey’s HSD for the Guinea Pig data. Any confidence intervals that do not contain 0 provide strong evidence against the null hypothesis of no difference in the true means for that pair of groups.
##
## Simultaneous Confidence Intervals
##
## Multiple Comparisons of Means: Tukey Contrasts
##
##
## Fit: lm(formula = len ~ Treat, data = ToothGrowth)
##
## Quantile = 2.955
## 95% family-wise confidence level
##
##
## Linear Hypotheses:
## Estimate lwr upr
## VC.0.5 - OJ.0.5 == 0 -5.2500 -10.0490 -0.4510
## OJ.1 - OJ.0.5 == 0 9.4700 4.6710 14.2690
## VC.1 - OJ.0.5 == 0 3.5400 -1.2590 8.3390
## OJ.2 - OJ.0.5 == 0 12.8300 8.0310 17.6290
## VC.2 - OJ.0.5 == 0 12.9100 8.1110 17.7090
## OJ.1 - VC.0.5 == 0 14.7200 9.9210 19.5190
## VC.1 - VC.0.5 == 0 8.7900 3.9910 13.5890
## OJ.2 - VC.0.5 == 0 18.0800 13.2810 22.8790
## VC.2 - VC.0.5 == 0 18.1600 13.3610 22.9590
## VC.1 - OJ.1 == 0 -5.9300 -10.7290 -1.1310
## OJ.2 - OJ.1 == 0 3.3600 -1.4390 8.1590
## VC.2 - OJ.1 == 0 3.4400 -1.3590 8.2390
## OJ.2 - VC.1 == 0 9.2900 4.4910 14.0890
## VC.2 - VC.1 == 0 9.3700 4.5710 14.1690
## VC.2 - OJ.2 == 0 0.0800 -4.7190 4.8790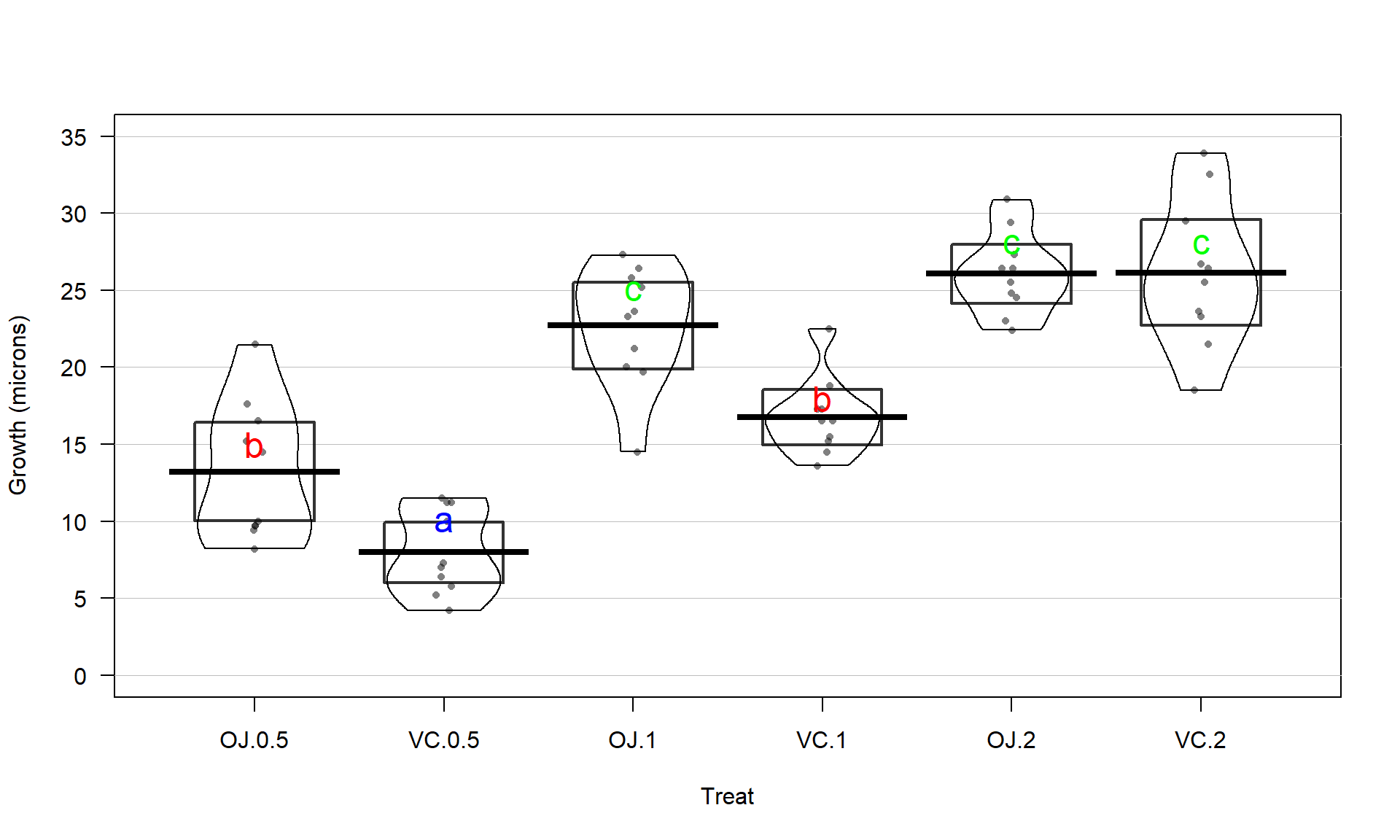
Figure 2.45: (ref:fig3-19)
Figure 2.45 contains confidence intervals for the difference in
the means for all 15 pairs of groups. For example, the first row in the plot contains
the confidence interval for comparing VC.0.5 and OJ.0.5 (VC.0.5 minus OJ.0.5). In the numerical output, you can find that this 95%
family-wise confidence interval goes from -10.05 to -0.45 microns (lwr and
upr in the numerical output provide the CI endpoints). This interval does not
contain 0 since its upper end point is -0.45 microns and so we can now say that
there is strong evidence against the null hypothesis of no difference in this pair and that we detect that OJ and VC have different true mean growth rates at the 0.5 mg
dosage level. We can go further and say that we are 95% confident that the difference
in the true mean tooth growth between VC.0.5 and OJ.0.5 (VC.0.5-OJ.0.5) is between
-10.05 and -0.45 microns, after adjusting for comparing all the pairs of groups. The center of this CI is -5.25 which is \(\hat{\tau}_2\) and the estimate difference between VC.0.5 and the baseline category of OJ.0.5. That means we can get an un-adjusted 95% confidence interval from the confint function to compare to this adjusted CI. The interval that does not account for all the comparisons goes from -8.51 to -1.99 microns (second row out confint output), showing the increased width needed in Tukey’s interval to control the family-wise error rate when many pairs are being compared. With 14 other intervals, we obviously can’t give them all this much attention…
## 2.5 % 97.5 %
## (Intercept) 10.9276907 15.532309
## TreatVC.0.5 -8.5059571 -1.994043
## TreatOJ.1 6.2140429 12.725957
## TreatVC.1 0.2840429 6.795957
## TreatOJ.2 9.5740429 16.085957
## TreatVC.2 9.6540429 16.165957If you put all these pair-wise tests together, you can generate an overall interpretation of Tukey’s HSD results that discusses sets of groups that are not detectably different from one another and those groups that were distinguished from other sets of groups. To do this, start with listing out the groups that are not detectably different (CIs contain 0), which, here, only occurs for four of the pairs. The CIs that contain 0 are for the pairs VC.1 and OJ.0.5, OJ.2 and OJ.1, VC.2 and OJ.1, and, finally, VC.2 and OJ.2. So VC.2, OJ.1, and OJ.2 are all not detectably different from each other and VC.1 and OJ.0.5 are also not detectably different. If you look carefully, VC.0.5 is detected as different from every other group. So there are basically three sets of groups that can be grouped together as “similar”: VC.2, OJ.1, and OJ.2; VC.1 and OJ.0.5; and VC.0.5. Sometimes groups overlap with some levels not being detectably different from other levels that belong to different groups and the story is not as clear as it is in this case. An example of this sort of overlap is seen in the next section.
There is a method that many researchers use to more efficiently generate and
report these sorts of results that is called a compact letter display
(CLD, Piepho (2004))68. The cld function can be applied to the results from
glht to generate the CLD that we can use to provide a “simple” summary of
the sets of groups. In this discussion, we define a set as a union of different
groups that can contain one or more members and the member of these groups are
the different treatment levels.
## OJ.0.5 VC.0.5 OJ.1 VC.1 OJ.2 VC.2
## "b" "a" "c" "b" "c" "c"Groups with the same letter are not detectably different (are in the same set) and groups that are detectably different get different letters (are in different sets). Groups can have more than one letter to reflect “overlap” between the sets of groups and sometimes a set of groups contains only a single treatment level (VC.0.5 is a set of size 1). Note that if the groups have the same letter, this does not mean they are the same, just that there is insufficient evidence to declare a difference for that pair. If we consider the previous output for the CLD, the “a” set contains VC.0.5, the “b” set contains OJ.0.5 and VC.1, and the “c” set contains OJ.1, OJ.2, and VC.2. These are exactly the groups of treatment levels that we obtained by going through all fifteen pairwise results.
One benefit of this work is that the CLD letters can be added to a plot (such as the pirate-plot) to
help fully report the results and understand the sorts of differences Tukey’s
HSD detected. The code with text involves placing text on the figure. In the text function, the x and y axis locations are specified (x-axis goes from 1 to 6 for the 6 categories) as well as the text to add (the CLD here). Some trial and error for locations may be needed to get the letters to be easily seen in a given pirate-plot.
Figure 2.46 enhances the discussion by showing that the
“a” group with VC.0.5 had the lowest average tooth
growth, the “b” group had intermediate tooth growth
for treatments OJ.0.5 and VC.1, and the highest growth rates came from
OJ.1, OJ.2, and VC.2. Even though VC.2 had the highest average growth rate,
we are not able to prove that its true mean is any higher
than the other groups labeled with “c”. Hopefully the
ease of getting to the story of the Tukey’s HSD results from a plot like this
explains why it is common to report results using these methods instead of
reporting 15 confidence intervals for all the pair-wise differences, either in a table or the plot.
(ref:fig3-20) Pirate-plot of odontoblast growth by group with Tukey’s HSD compact letter display. Note some extra pirate-plot options are used to enhance focus on the CLD results.
pirateplot(len~Treat, data=ToothGrowth, ylab="Growth (microns)", inf.method="ci", theme=2,inf.f.o = 0,point.o = .5) #Options theme=2,inf.f.o = 0,point.o = .5 added to focus on CLD
text(x=2,y=10,"a",col="blue",cex=1.5) #CLD added
text(x=c(1,4),y=c(15,18),"b",col="red",cex=1.5)
text(x=c(3,5,6),y=c(25,28,28),"c",col="green",cex=1.5)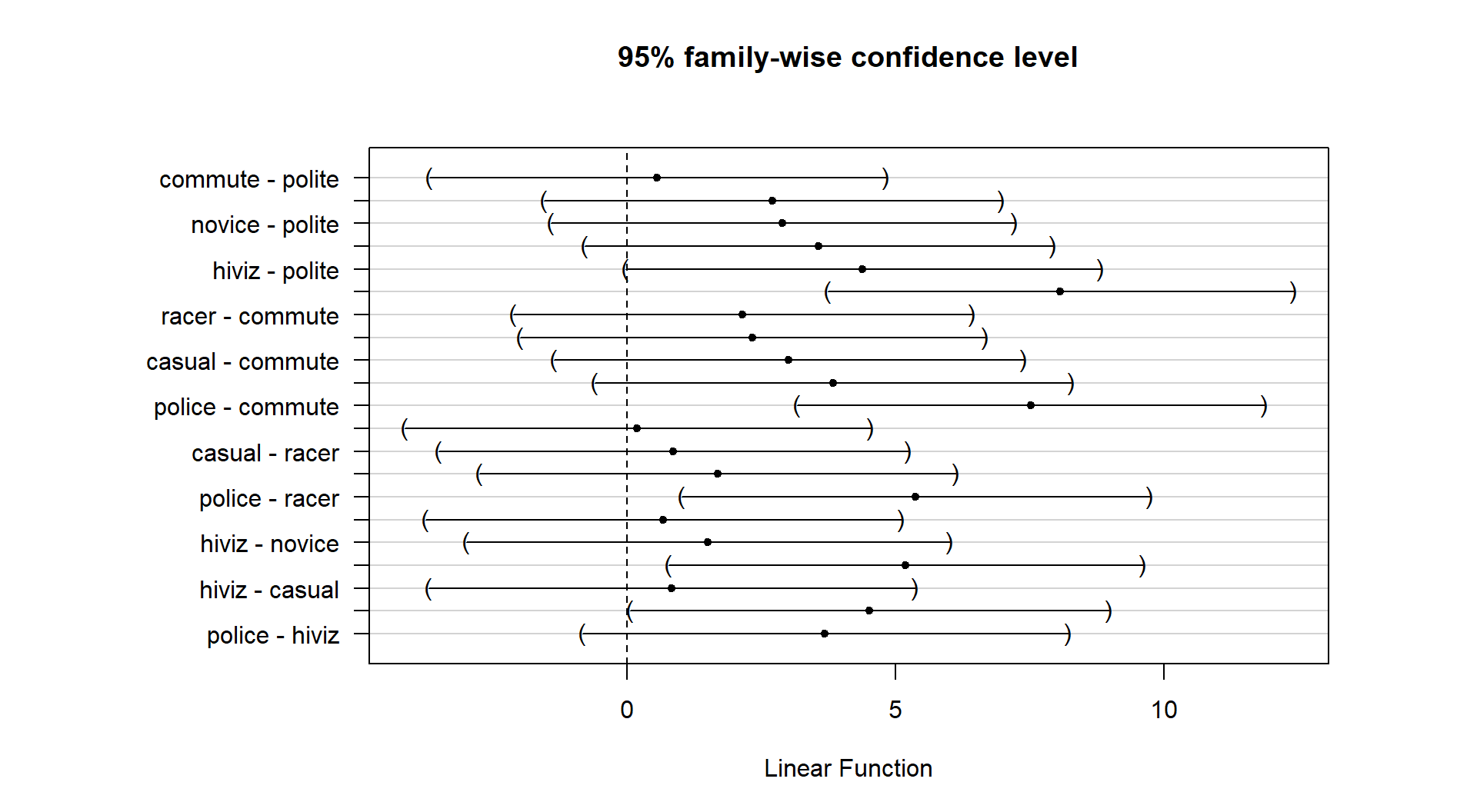
Figure 2.46: (ref:fig3-20)
There are just a couple of other details to mention on this set of methods. First,
note that we interpret the set of confidence intervals simultaneously: We are
95% confident that ALL the intervals contain the respective differences in
the true means (this is a family-wise interpretation). These intervals are
adjusted from our regular two-sample \(t\) intervals that came from lm from Chapter 2
to allow this stronger interpretation. Specifically,
they are wider. Second, if sample sizes are unequal in the groups, Tukey’s HSD
is conservative and provides a family-wise error rate that is lower than the
nominal (or specified) level. In other words, it fails less often than expected
and the intervals provided are a little wider than needed, containing all the
pairwise differences at higher than the nominal confidence level of (typically)
95%. Third, this is a parametric approach and violations of normality and
constant variance will push the method in the other direction, potentially
making the technique dangerously liberal. Nonparametric approaches to this
problem are also possible, but will not be considered here.
Tukey’s HSD results can also be displayed as p-values for each pair-wise test result. This is a little less common but can allow you to directly assess the strength of evidence for a particular pair instead of using the detected/not result that the family-wise CIs provide. But the family-wise CIs are useful for exploring the size of the differences in the pairs and we need to simplify things to detect/not in these situations because there are so many tests. But if you want to see the Tukey HSD p-values, you can use the summary function on the object that is storing the results:
##
## Simultaneous Tests for General Linear Hypotheses
##
## Multiple Comparisons of Means: Tukey Contrasts
##
##
## Fit: lm(formula = len ~ Treat, data = ToothGrowth)
##
## Linear Hypotheses:
## Estimate Std. Error t value Pr(>|t|)
## VC.0.5 - OJ.0.5 == 0 -5.250 1.624 -3.233 0.02424
## OJ.1 - OJ.0.5 == 0 9.470 1.624 5.831 < 0.001
## VC.1 - OJ.0.5 == 0 3.540 1.624 2.180 0.26411
## OJ.2 - OJ.0.5 == 0 12.830 1.624 7.900 < 0.001
## VC.2 - OJ.0.5 == 0 12.910 1.624 7.949 < 0.001
## OJ.1 - VC.0.5 == 0 14.720 1.624 9.064 < 0.001
## VC.1 - VC.0.5 == 0 8.790 1.624 5.413 < 0.001
## OJ.2 - VC.0.5 == 0 18.080 1.624 11.133 < 0.001
## VC.2 - VC.0.5 == 0 18.160 1.624 11.182 < 0.001
## VC.1 - OJ.1 == 0 -5.930 1.624 -3.651 0.00739
## OJ.2 - OJ.1 == 0 3.360 1.624 2.069 0.31868
## VC.2 - OJ.1 == 0 3.440 1.624 2.118 0.29372
## OJ.2 - VC.1 == 0 9.290 1.624 5.720 < 0.001
## VC.2 - VC.1 == 0 9.370 1.624 5.770 < 0.001
## VC.2 - OJ.2 == 0 0.080 1.624 0.049 1.00000
## (Adjusted p values reported -- single-step method)These reinforce the strong evidence for many of the pairs and less strong evidence for four pairs that were not detected to be different. So these p-values provide another method to employ to report the Tukey’s HSD results – you would only need to report and explore the confidence intervals or the p-values, not both.
Tukey’s HSD does not require you to find a small p-value from your overall \(F\)-test to employ the methods but if you apply it to situations with p-values larger than your a priori significance level, you are unlikely to find any pairs that are detected as being different. Some statisticians suggest that you shouldn’t employ follow-up tests such as Tukey’s HSD when there is not sufficient evidence to reject the overall null hypothesis and would be able to reasonably criticize the following results.
References
Hothorn, Torsten, Frank Bretz, and Peter Westfall. 2008. “Simultaneous Inference in General Parametric Models.” Biometrical Journal 50 (3): 346–63.
Hothorn, Torsten, Frank Bretz, and Peter Westfall. 2019. Multcomp: Simultaneous Inference in General Parametric Models. https://CRAN.R-project.org/package=multcomp.
Piepho, Hans-Peter. 2004. “An Algorithm for a Letter-Based Representation of All-Pairwise Comparisons.” Journal of Computational and Graphical Statistics 13 (2): 456–66.
When this procedure is used with unequal group sizes it is also sometimes called Tukey-Kramer’s method.↩
We often use “spurious” to describe falsely rejected null hypotheses, but they are also called false detections.↩
In more complex models, this code can be used to create pair-wise comparisons on one of many explanatory variables.↩
The plot of results usually contains all the labels of groups but if the labels are long or there many groups, sometimes the row labels are hard to see even with re-sizing the plot to make it taller in RStudio. The numerical output is useful as a guide to help you read the plot in those situations.↩
Note that this method is implemented slightly differently than explained here in some software packages so if you see this in a journal article, read the discussion carefully.↩