- Cover
- Acknowledgments
- 1 Preface
- 2 (R)e-Introduction to statistics
- 2.1 Histograms, boxplots, and density curves
- 2.2 Pirate-plots
- 2.3 Models, hypotheses, and permutations for the two sample mean situation
- 2.4 Permutation testing for the two sample mean situation
- 2.5 Hypothesis testing (general)
- 2.6 Connecting randomization (nonparametric) and parametric tests
- 2.7 Second example of permutation tests
- 2.8 Reproducibility Crisis: Moving beyond p < 0.05, publication bias, and multiple testing issues
- 2.9 Confidence intervals and bootstrapping
- 2.10 Bootstrap confidence intervals for difference in GPAs
- 2.11 Chapter summary
- 2.12 Summary of important R code
- 2.13 Practice problems
- 3 One-Way ANOVA
- 3.1 Situation
- 3.2 Linear model for One-Way ANOVA (cell-means and reference-coding)
- 3.3 One-Way ANOVA Sums of Squares, Mean Squares, and F-test
- 3.4 ANOVA model diagnostics including QQ-plots
- 3.5 Guinea pig tooth growth One-Way ANOVA example
- 3.6 Multiple (pair-wise) comparisons using Tukey’s HSD and the compact letter display
- 3.7 Pair-wise comparisons for the Overtake data
- 3.8 Chapter summary
- 3.9 Summary of important R code
- 3.10 Practice problems
- 4 Two-Way ANOVA
- 4.1 Situation
- 4.2 Designing a two-way experiment and visualizing results
- 4.3 Two-Way ANOVA models and hypothesis tests
- 4.4 Guinea pig tooth growth analysis with Two-Way ANOVA
- 4.5 Observational study example: The Psychology of Debt
- 4.6 Pushing Two-Way ANOVA to the limit: Un-replicated designs and Estimability
- 4.7 Chapter summary
- 4.8 Summary of important R code
- 4.9 Practice problems
- 5 Chi-square tests
- 5.1 Situation, contingency tables, and tableplots
- 5.2 Homogeneity test hypotheses
- 5.3 Independence test hypotheses
- 5.4 Models for R by C tables
- 5.5 Permutation tests for the \(X^2\) statistic
- 5.6 Chi-square distribution for the \(X^2\) statistic
- 5.7 Examining residuals for the source of differences
- 5.8 General protocol for \(X^2\) tests
- 5.9 Political party and voting results: Complete analysis
- 5.10 Is cheating and lying related in students?
- 5.11 Analyzing a stratified random sample of California schools
- 5.12 Chapter summary
- 5.13 Summary of important R commands
- 5.14 Practice problems
- 6 Correlation and Simple Linear Regression
- 6.1 Relationships between two quantitative variables
- 6.2 Estimating the correlation coefficient
- 6.3 Relationships between variables by groups
- 6.4 Inference for the correlation coefficient
- 6.5 Are tree diameters related to tree heights?
- 6.6 Describing relationships with a regression model
- 6.7 Least Squares Estimation
- 6.8 Measuring the strength of regressions: R2
- 6.9 Outliers: leverage and influence
- 6.10 Residual diagnostics – setting the stage for inference
- 6.11 Old Faithful discharge and waiting times
- 6.12 Chapter summary
- 6.13 Summary of important R code
- 6.14 Practice problems
- 7 Simple linear regression inference
- 7.1 Model
- 7.2 Confidence interval and hypothesis tests for the slope and intercept
- 7.3 Bozeman temperature trend
- 7.4 Randomization-based inferences for the slope coefficient
- 7.5 Transformations part I: Linearizing relationships
- 7.6 Transformations part II: Impacts on SLR interpretations: log(y), log(x), & both log(y) & log(x)
- 7.7 Confidence interval for the mean and prediction intervals for a new observation
- 7.8 Chapter summary
- 7.9 Summary of important R code
- 7.10 Practice problems
- 8 Multiple linear regression
- 8.1 Going from SLR to MLR
- 8.2 Validity conditions in MLR
- 8.3 Interpretation of MLR terms
- 8.4 Comparing multiple regression models
- 8.5 General recommendations for MLR interpretations and VIFs
- 8.6 MLR inference: Parameter inferences using the t-distribution
- 8.7 Overall F-test in multiple linear regression
- 8.8 Case study: First year college GPA and SATs
- 8.9 Different intercepts for different groups: MLR with indicator variables
- 8.10 Additive MLR with more than two groups: Headache example
- 8.11 Different slopes and different intercepts
- 8.12 F-tests for MLR models with quantitative and categorical variables and interactions
- 8.13 AICs for model selection
- 8.14 Case study: Forced expiratory volume model selection using AICs
- 8.15 Chapter summary
- 8.16 Summary of important R code
- 8.17 Practice problems
- 9 Case studies
- 9.1 Overview of material covered
- 9.2 The impact of simulated chronic nitrogen deposition on the biomass and N2-fixation activity of two boreal feather moss–cyanobacteria associations
- 9.3 Ants learn to rely on more informative attributes during decision-making
- 9.4 Multi-variate models are essential for understanding vertebrate diversification in deep time
- 9.5 What do didgeridoos really do about sleepiness?
- 9.6 General summary
- References
6.6 Describing relationships with a regression model
When the relationship appears to
be relatively linear, it makes sense to estimate and then interpret a line to
represent the relationship between the variables. This line is called a
regression line and involves finding a line that best fits (explains
variation in) the response variable for the
given values of the explanatory variable.
For regression, it matters which
variable you choose for \(x\) and which you choose for \(y\) – for correlation
it did not matter. This regression line describes the “effect” of \(x\) on
\(y\) and also provides an equation for predicting values of \(y\) for given
values of \(x\). The Beers and BAC data provide a nice example to start
our exploration of regression models. The beer consumption is a clear
explanatory variable,
detectable in the story because (1) it was randomly assigned to subjects and
(2) basic science supports beer consumption amount being an explanatory
variable for BAC. In some situations, this will not be so clear, but look for
random assignment or scientific logic to guide your choices of variables as
explanatory or response98. Regression lines are actually provided by
default in the scatterplot function with the reg.line=T option or just
omitting reg.line=F from the previous versions of the code since it is a
default option to provide the lines.
(ref:fig6-13) Scatterplot with estimated regression line for the Beers and BAC data. Horizontal dashed lines for the predicted BAC for 4 and 5 beers consumed.
scatterplot(BAC~Beers, ylim=c(0,.2), xlim=c(0,9), data=BB,
boxplot=F, main="Scatterplot with regression line",
lwd=2, smooth=F)
abline(v=1:9, col="grey")
abline(h=c(0.05914,0.0771), col="blue", lty=2, lwd=2)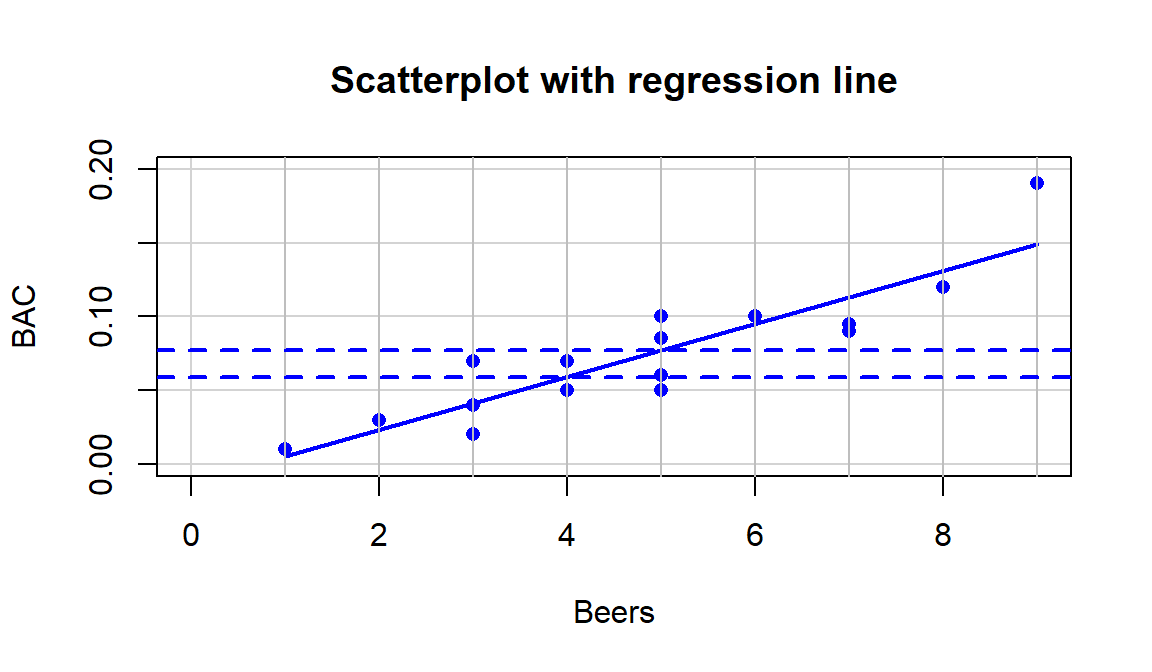
Figure 2.112: (ref:fig6-13)
The equation for a line is \(y=a+bx\), or maybe \(y=mx+b\). In the version
\(mx+b\) you learned that \(m\) is a slope coefficient that relates a
change in \(x\) to changes in \(y\) and that \(b\) is a y-intercept (the
value of \(y\) when \(x\) is 0). In Figure 2.112, two extra
horizontal lines are added to help you see the defining characteristics of the line. The
slope, whatever letter you use, is the change in \(y\) for a one-unit
increase in \(x\). Here, the slope is the change in BAC for a 1 beer
increase in Beers, such as the change from 4 to 5 beers. The
y-values (blue, dashed lines) for Beers = 4 and 5 go from 0.059 to
0.077. This means that for a 1 beer increase (+1 unit change in \(x\)), the
BAC goes up by \(0.077-0.059=0.018\) (+0.018 unit change in \(y\)).
We can also try to find the y-intercept on the graph by looking for the
BAC level for 0 Beers consumed. The y-value (BAC) ends up
being around -0.01 if you extend the regression line to Beers=0.
You might assume that the BAC should be 0 for Beers=0 but the
researchers did not observe any students at 0 Beers, so we don’t
really know what the BAC might be at this value. We have to
use our line to predict this value. This ends up providing a
prediction below 0 – an impossible value for BAC. If the
y-intercept were positive, it would suggest that the students
has a BAC over 0 even without drinking.
The numbers reported were very
accurate because we weren’t using the plot alone to generate the
values – we
were using a linear model to estimate the equation to
describe the
relationship between Beers and BAC. In statistics, we estimate
“\(m\)” and “\(b\)”. We also write the equation starting with the y-intercept
and use slightly different notation that allows us to extend to more
complicated models with more variables.
Specifically, the estimated
regression equation is \(\hat{y} = b_0 + b_1x\), where
\(\hat{y}\) is the estimated value of \(y\) for a given \(x\),
\(b_0\) is the estimated y-intercept (predicted value of \(y\) when \(x\) is 0),
\(b_1\) is the estimated slope coefficient, and
\(x\) is the explanatory variable.
One of the differences between when you learned equations in algebra classes and our situation is that the line is not a perfect description of the relationship between \(x\) and \(y\) – it is an “on average” description and will usually leave differences between the line and the observations, which we call residuals \((e = y-\hat{y})\). We worked with residuals in the ANOVA99 material. The residuals describe the vertical distance in the scatterplot between our model (regression line) and the actual observed data point. The lack of a perfect fit of the line to the observations distinguishes statistical equations from those you learned in math classes. The equations work the same, but we have to modify interpretations of the coefficients to reflect this.
We also tie this estimated model to a theoretical or population regression model:
\[y_i = \beta_0 + \beta_1x_i+\varepsilon_i\]
where:
\(y_i\) is the observed response for the \(i^{th}\) observation,
\(x_i\) is the observed value of the explanatory variable for the \(i^{th}\) observation,
\(\beta_0 + \beta_1x_i\) is the true mean function evaluated at \(x_i\),
\(\beta_0\) is the true (or population) y-intercept,
\(\beta_1\) is the true (or population) slope coefficient, and
the deviations, \(\varepsilon_i\), are assumed to be independent and normally distributed with mean 0 and standard deviation \(\sigma\) or, more compactly, \(\varepsilon_i \sim N(0,\sigma^2)\).
This presents another version of the linear model from Chapters 2, ??, and ??, now with a quantitative explanatory variable instead of categorical explanatory variable(s). This chapter focuses mostly on the estimated regression coefficients, but remember that we are doing statistics and our desire is to make inferences to a larger population. So, estimated coefficients, \(b_0\) and \(b_1\), are approximations to theoretical coefficients, \(\beta_0\) and \(\beta_1\). In other words, \(b_0\) and \(b_1\) are the statistics that try to estimate the true population parameters \(\beta_0\) and \(\beta_1\), respectively.
To get estimated regression coefficients, we use the lm function
and our standard lm(y~x, data=...) setup.
This is the same function
used to estimate our ANOVA models and much of this
will look familiar. In fact, the ties between ANOVA and regression are
deep and fundamental but not the topic of this section. For the Beers
and BAC example, the estimated regression coefficients can be
found from:
##
## Call:
## lm(formula = BAC ~ Beers, data = BB)
##
## Coefficients:
## (Intercept) Beers
## -0.01270 0.01796More often, we will extract these from the coefficient table produced
by a model summary:
##
## Call:
## lm(formula = BAC ~ Beers, data = BB)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.027118 -0.017350 0.001773 0.008623 0.041027
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -0.012701 0.012638 -1.005 0.332
## Beers 0.017964 0.002402 7.480 2.97e-06
##
## Residual standard error: 0.02044 on 14 degrees of freedom
## Multiple R-squared: 0.7998, Adjusted R-squared: 0.7855
## F-statistic: 55.94 on 1 and 14 DF, p-value: 2.969e-06From either version of the output, you can find the estimated y-intercept
in the (Intercept) part of the output and the slope coefficient in the
Beers part of the output. So \(b_0 = -0.0127\), \(b_1=0.01796\), and
the estimated regression equation is
\[\widehat{\text{BAC}}_i = -0.0127 + 0.01796\cdot\text{Beers}_i.\]
This is the equation that was plotted in Figure 2.112. In writing out the equation, it is good to replace \(x\) and \(y\) with the variable names to make the predictor and response variables clear. If you prefer to write all equations with \(\boldsymbol{x}\) and \(\boldsymbol{y}\), you need to define \(\boldsymbol{x}\) and \(\boldsymbol{y}\) or else these equations are not clearly defined.
There is a general interpretation for the slope coefficient that you will need to master. In general, we interpret the slope coefficient as:
- Slope interpretation (general): For a 1 [unit of X] increase in X, we expect, on average, a \(\boldsymbol{b_1}\) [unit of Y] change in Y.
Figure 2.113 can help you think about the different sorts of slope coefficients we might need to interpret, both providing changes in the response variable for 1 unit increases in the predictor variable.
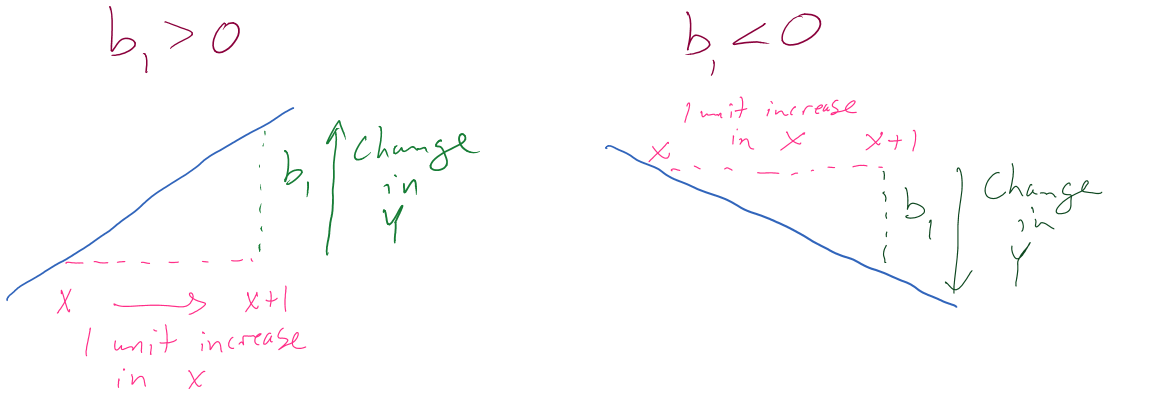
Figure 2.113: Diagram of interpretation of slope coefficients.
Applied to this problem, for each additional 1 beer consumed, we expect a 0.018 gram per dL change in the BAC on average. Using “change” in the interpretation for what happened in the response allows you to use the same template for the interpretation even with negative slopes – be careful about saying “decrease” when the slope is negative as you can create a double-negative and end up implying an increase… Note also that you need to carefully incorporate the units of \(x\) and the units of \(y\) to make the interpretation clear. For example, if the change in BAC for 1 beer increase is 0.018, then we could also modify the size of the change in \(x\) to be a 10 beer increase and then the estimated change in BAC is \(10*0.018 = 0.18\) g/dL. Both are correct as long as you are clear about the change in \(x\) you are talking about. Typically, we will just use the units used in the original variables and only change the scale of “change in \(x\)” when it provides an interpretation we are particularly interested in.
Similarly, the general interpretation for a y-intercept is:
- Y-intercept interpretation (general): For X= 0 [units of X], we expect, on average, \(\boldsymbol{b_0}\) [units of Y] in Y.
Again, applied to the BAC data set: For 0 beers for Beers consumed,
we expect, on
average, -0.012 g/dL BAC. The y-intercept interpretation is often less
interesting than the slope interpretation but can be interesting in some
situations. Here, it is predicting average BAC for Beers=0, which
is a value outside the scope of the \(x\text{'s}\) (Beers was observed
between 1 and 9). Prediction outside the scope of the predictor values is
called extrapolation. Extrapolation is dangerous at best and
misleading at worst. That said, if you are asked to
interpret the y-intercept you should still interpret it, but it is also good to
note if it is outside of the region where we had observations on the
explanatory variable. Another example is useful for practicing how to do these
interpretations.
In the Australian Athlete data, we
saw a weak negative relationship between Body Fat (% body weight that
is fat) and Hematocrit (% red blood cells in the blood). The scatterplot
in Figure 2.114 shows just the
results for the female athletes along with the regression line which has a
negative slope coefficient. The estimated regression coefficients are found
using the lm function:
aisR2 <- ais[-c(56,166), c("Ht","Hc","Bfat","Sex")]
m2 <- lm(Hc~Bfat, data=subset(aisR2,Sex==1)) #Results for Females ##
## Call:
## lm(formula = Hc ~ Bfat, data = subset(aisR2, Sex == 1))
##
## Residuals:
## Min 1Q Median 3Q Max
## -5.2399 -2.2132 -0.1061 1.8917 6.6453
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 42.01378 0.93269 45.046 <2e-16
## Bfat -0.08504 0.05067 -1.678 0.0965
##
## Residual standard error: 2.598 on 97 degrees of freedom
## Multiple R-squared: 0.02822, Adjusted R-squared: 0.0182
## F-statistic: 2.816 on 1 and 97 DF, p-value: 0.09653scatterplot(Hc~Bfat, data=subset(aisR2,Sex==1), smooth=F,
main="Scatterplot of Body Fat vs Hematocrit for Female Athletes",
ylab="Hc (% blood)", xlab="Body fat (% weight)")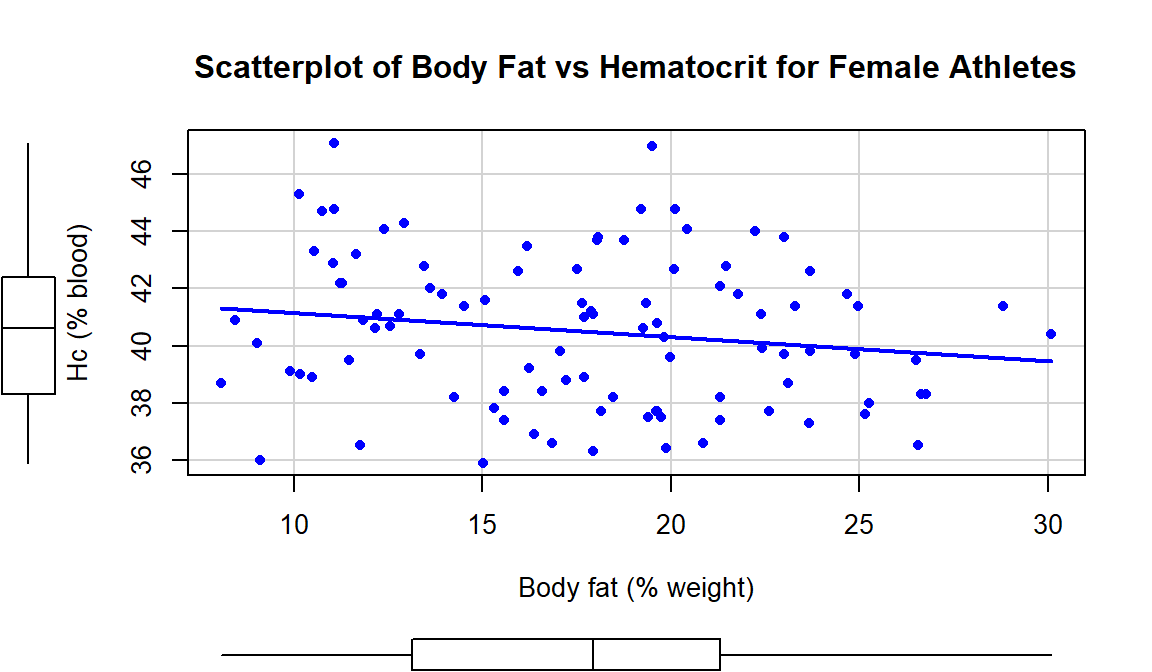
Figure 2.114: Scatterplot of Hematocrit versus Body Fat for female athletes.
Based on these results, the estimated regression equation is \(\widehat{\text{Hc}}_i = 42.014 - 0.085\cdot\text{BodyFat}_i\) with \(b_0 = 42.014\) and \(b_1 = 0.085\). The slope coefficient interpretation is: For a one percent increase in body fat, we expect, on average, a -0.085% (blood) change in Hematocrit for Australian female athletes. For the y-intercept, the interpretation is: For a 0% body fat female athlete, we expect a Hematocrit of 42.014% on average. Again, this y-intercept involves extrapolation to a region of \(x\)’s that we did not observed. None of the athletes had body fat below 5% so we don’t know what would happen to the hematocrit of an athlete that had no body fat except that it probably would not continue to follow a linear relationship.
Even with clear scientific logic, we sometimes make choices to flip the model directions to facilitate different types of analyses. In Vsevolozhskaya et al. (2014) we looked at genomic differences based on obesity groups, even though we were really interested in exploring how gene-level differences explained differences in obesity.↩
The residuals from these methods and ANOVA are the same because they all come from linear models but are completely different from the standardized residuals used in the Chi-square material in Chapter ??.↩